
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 December 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1293958
 Qiyu Xu1,2,3†
Qiyu Xu1,2,3† Tao Zhu1,2,3†
Tao Zhu1,2,3† Ruifeng Zhao4
Ruifeng Zhao4 Yang Zhao5
Yang Zhao5 Yangkai Duan1,2,3
Yangkai Duan1,2,3 Xiang Liu1,2,3
Xiang Liu1,2,3 Guodong Luan1,2,3
Guodong Luan1,2,3 Ruibo Hu1,2,3
Ruibo Hu1,2,3 Sanyuan Tang6
Sanyuan Tang6 Xinrong Ma4
Xinrong Ma4 Yan Liu5
Yan Liu5 Shengjun Li1,2,3*
Shengjun Li1,2,3* Xuefeng Lu1,2,3*
Xuefeng Lu1,2,3*Salt stress detrimentally impacts plant growth, imperiling crop yield and food quality. Ameliorating plant resilience and productivity in saline environments is critical for global food security. Here, we report the positive effect of Arthrospira (Spirulina) on plant growth and salt tolerance in Arabidopsis and sweet sorghum. Arthrospira application greatly promotes seed germination and seedling growth in both species under salt stress conditions in a dosage-dependent manner. Application of 6 mg Arthrospira per plate significantly enhances K+/Na+ equilibrium and reactive oxygen species (ROS) scavenging in Arabidopsis, reducing salt-induced toxicity. The primary root length, survival rate, chlorophyll content, photosynthesis, plant height, biomass and yield were all improved in both species. Concurrently, Arthrospira demonstrated the synthesis of compatible solutes, such as trehalose (Tre) and glucosylglycerol (GG), contributing to heightened stress tolerance when co-cultivated with Arabidopsis on plates. Transcriptome analysis revealed dramatic up-/down- regulation of genes involved in phytohormone signal transduction, chlorophyll and photosynthesis metabolism, and phenylpropanoid metabolism in Arabidopsis. Furthermore, the application of Arthrospira exerted a positive influence on the rhizosphere bacteriome structure in sweet sorghum, crucial for nutrient cycling and soil health enhancement. Our findings uncovered the underlying mechanisms of algae-plants interaction in saline soil, proposing strategies to enhance crop productivity and soil quality, thereby addressing the urgent need for sustainable agriculture practices to mitigate salinity’s repercussions amidst climate change challenges.
Salt stress is a significant constraint on agriculture (Pan et al., 2021), posing conceivable repercussions for approximately 50% of the world’s arable land by 2050 (Kaashyap et al., 2017). Sodium chloride (Na+) primarily causes ion toxicity to plants, resulting in osmotic stress, K+/Na+ imbalance, which disrupt water and nutrient uptake (Gul et al., 2019; Gadelha et al., 2021). In addition, the imposition of salt stress triggers the synthesis of reactive oxygen species (ROS), instigating potential harm to vital cellular constituents, including DNA, proteins, and lipids. This cascade of events culminates in the attenuation of plant growth and, in severe cases, mortality (Zahra et al., 2022). Salt stress also destroys the structure of chloroplast, photosystem, and electron transfer system, and restricts gas exchange by reducing stomatal opening, which in turn reduces the photosynthetic rate (Negrão et al., 2017; Gadelha et al., 2021). Plants have developed diverse mechanisms to acclimate and endure saline stress conditions, encompassing ion equilibrium, accumulation of osmoprotectants, and orchestration of hormonal regulatory pathways (Ma et al., 2019; Wei et al., 2020).
In recent years, the interaction between plants and microorganisms, such as Rhizobia and Arbuscular Mycorrhizal Fungi (AMF), has emerged as a substantiated contributor to both plant growth and stress resilience (Durán et al., 2018; de Vries et al., 2020). Microorganisms produce amino acids, proteins, metabolisms, thereby furnishing nutrients that contribute to and positively impact the growth of plants (Berendsen et al., 2012; Di Lelio et al., 2023). Furthermore, certain soil microorganisms improve soil quality and ecology by enhancing water and nutrient retention (Abinandan et al., 2019). Additionally, many microorganisms regulate plant metabolism and physiological processes by facilitating nutrient absorption and utilization of N, P, and K, and improving photosynthetic efficiency, leading to significant increase of crop yield (Colla and Rouphael, 2020). Moreover, under stress conditions, microorganisms can accumulate large amounts of osmoregulatory substances, thereby augmenting the plant’s resilience against both biotic and abiotic stressors (Renuka et al., 2018; Abinandan et al., 2019).
Microalgae serve as organic fertilizer in agriculture, reducing chemical fertilizer usage and improving product quality. These biofertilizers play a crucial role in establishing a coherent pathway toward sustainable development (Lu and Valtchev, 2023). Cyanobacteria, which are primitive oxygen-producing photosynthetic organisms, are essential members of rhizobacteria and commonly found in biological soil crust communities. They possess diverse taxonomy and biochemistry, enabling them to synthesize various bioactive molecules like sulfated polysaccharides, amino acids, phenolics, vitamins, phytohormones (auxin, cytokinin, gibberellin, abscisic acid), and compatible solutes. These molecules effectively enhance crop development while conferring resilience against abiotic stressors (Luo et al., 2022). Arthrospira, also known as Spirulina, is a salt-tolerant genus of cyanobacteria with applications spanning industries such as health food, wastewater treatment, pharmaceutics, perfumery, and medicine (FAO, 2022; Hamouda et al., 2022). Nonetheless, the understanding of Arthrospira-based biofertilizers, especially their mechanisms of action, remains limited.
Arthrospira has been proven to improve soil mineral conditions and increase crop yields as a microalgae fertilizer (Álvarez-Gómez et al., 2016; Ismaiel et al., 2022; Pei and Yu, 2022). A recent discovery unveiled a range of physiologically active chemicals in Arthrospira extract that could function as plant growth regulators, promoting seedling development and salt stress tolerance in Triticum aestivum (Hamouda et al., 2022). Arthrospira is also capable of producing intracellular compatible solutes, such as trehalose (Tre) and glucosylglycerol (GG), to adapt to higher extracellular salinity (Warr et al., 1985; Klähn and Hagemann, 2011; Luo et al., 2022). Consequently, the application of Arthrospira is expected to impact the composition of rhizosphere bacteria in high-salt environments, highlighting its potential as a sustainable biofertilizer for sustainable agriculture (Rocha et al., 2020).
Despite several reports highlighting the contribution of Arthrospira to both plant growth and stress tolerance (Mutale-joan et al., 2021), the comprehensive comprehension of the underlying mechanisms has yet to be achieved. Numerous studies focusing on Arabidopsis have elucidated signaling pathways regulating plant growth and stress responses (Zhang et al., 2020; Zhao et al., 2020). Notably, the ABA signaling pathway is pivotal in seed germination, plant development, and senescence, essential for plant survival under adverse conditions like drought and salinity (Liu et al., 2022). The conservative structures and regulatory proteins in Arabidopsis establish it as a pivotal model for comprehending the interactions between Arthrospira and crops. Sorghum (Sorghum bicolor (L.) Moench) is the fifth most important cereal crop, providing human foods, animal feeds, and bioenergy. Notably, sorghum exhibits exceptional tolerance to abiotic stresses, such as salinity and drought, making it an ideal crop in the face of changing climate conditions (Osman et al., 2022). In this study, by comprehensive analysis of cellular components, we found that Arthrospira is capable of synthesizing compatible solutes, among them GG and Tre, and confirmed their positive effect in plant salt tolerance. The transcriptomicc data indicated that Arthrospira modifies the expression of genes involved in phytohormone signaling to regulate salt tolerance. Notably, based on 16S rRNA amplicon sequencing and soil nutrient measurements, the Arthrospira application also improves soil quality by increasing the abundance of beneficial microbes, water content and soil nutrients. Taken together, this study illuminates the underlying mechanisms of Arthrospira-plant interaction under salt stress.
The Arabidopsis thaliana (Col-0) and Sorghum bicolor (M-81E) were used as experimental materials and grown in growth chambers. Arabidopsis seeds were subjected to surface sterilization by immersion in a 10% (V/V) NaClO solution for 7 minutes. Subsequently, these seeds were placed onto half-strength Murashige and Skoog (½ MS) medium (pH 5.8), supplemented with 1% (w/v) sucrose and 1% (w/v) agar. Following a 3-day stratification period at 4°C in darkness, the plates were translocated to a growth chamber set at 22°C, operating under long-day conditions (16 hours of light and 8 hours of darkness per photoperiod). The light intensity within the growth chamber was maintained at 90 μmol photons m−2 s−1. Sorghum seeds were thoroughly surface sterilized and germinated in a growth chamber, maintaining a photoperiod of 16 hours of light (28°C) followed by 8 hours of darkness (23°C). The light intensity in the chamber was set at 600 μmol photons m−2 s−1. The seedlings of both species were then transferred to either new plates or potted soil with the same growth conditions for subsequent tests after 3-5 days.
In Arabidopsis, salt stress treatment was initiated around three weeks post-germination. Each pot was irrigated with 50 mL of a 200 mM NaCl solution every two days. Arthrospira treatment commenced one week after the initiation of salt stress. For sweet sorghum, salt stress was induced at the elongation stage, and each pot was irrigated with 1 L of 200 mM NaCl solution every three days. Arthrospira treatment was implemented two weeks after the initiation of salt stress.
Cultivation of Arthrospira platensis was conducted under photoautotrophic conditions, utilizing the standard Zarrouk culture medium. The cultivation environment was illuminated with white light at an intensity ranging from 30 to 50 µmol photons m−2 s−1, while maintaining a temperature of 30°C. Arthrospira at the exponential growth stage was filtered with nylon mesh (48 μm pore size), washed with sterilized water, and resuspended in ½ MS medium or tap water in proper concentration as needed. For cell density quantification, a volume of 2 mL from the Arthrospira culture was extracted for analysis. The optical density of the cells was quantified at 730 nm (OD730) employing a spectrophotometer (Metash, China). The biomass content of Arthrospira was calculated using a calibration curve correlating the OD730 values to biomass fresh weight (FW) concentrations with the following formula: Biomass (g/L) = 7.752*OD730 – 1.216 (Supplementary Figure 1).
For co-cultivation, the 1/2 MS plates (approximately 20 mL in volume) were inoculated with sterilized water containing various concentrations (1.2, 6, 12, 24 mg FW for Arabidopsis and 12, 24, 72, 96, 120, 300 mg FW for sorghum) of Arthrospira. For soil treatment, a 1:1 volume ratio of nutrient and regular soil was mixed in a vertical plastic pot. The dimensions of the pots utilized were as follows: 7.4 cm (height) × 6.8 cm (top diameter) × 5 cm (bottom diameter) for Arabidopsis, and 31 cm (height) × 33 cm (width) × 23 cm (bottom diameter) for sorghum. Arthrospira was sprayed onto the soil surfaces at different concentrations (6, 60, 120, 300 mg FW for Arabidopsis and 6, 30 g for sorghum) once a week for three consecutive weeks.
Arthrospira’s crude protein content was assessed through the Kjeldahl method, adhering to the protocol outlined (Barbano et al., 1990). The calculation of Total Kjeldahl Nitrogen (TKN) and crude protein content was achieved by multiplication using a factor of 6.25 (López et al., 2010). The extraction of total lipids was performed utilizing a chloroform and methanol mixture (2:1, v/v), followed by GC-MS analysis, in accordance with our previously established method (Duan et al., 2011). Furthermore, elemental analysis was executed employing Inductively Coupled Plasma Mass Spectrometry (ICP-MS, Shimadzu, Japan) (Dawczynski et al., 2007). The mono- and disaccharides (i.e., glucose, fructose, sucrose, trehalose and glucosylglycerol) were determined by HPLC (High Performance Liquid Chromatography, Shimadzu, Japan) (Lee et al., 2012).
The germination rate and germination-related parameters were determined according to the following equations:
The number of germinated seeds (Gt) was recorded along with the corresponding germination days (Dt). Post-germination, the length of the embryonic shoot (S) and the length of the primary root (L) were measured. Measurements were conducted using ImageJ software (Version 1.53t).
The sorghum stem diameter was measured with a caliper using the basal internode 5 cm above the soil surface. Sorghum leaves directly attached to the fourth internode were harvested and their fresh weight was measured utilizing an electronic balance.
Roughly 100 mg of Arabidopsis leaves were weighed and subsequently pulverized into a fine powder within liquid nitrogen. The powdered sample was mixed with 1 ml of extraction solution and vortexed for 3-5 minutes. The resultant mixture underwent centrifugation at 8000g and 4°C for 10 minutes, leading to the collection of the supernatant. The levels of hydrogen peroxide (H2O2) and malondialdehyde (MDA), the enzyme activities of catalase (CAT), superoxide dismutase (SOD), peroxidase (POD) and ascorbate peroxidase (APX) in the leaves, were determined according to manufacturer-specified protocols using PYHA1, PYHA2, PMHA2, PMHA4, PMHA1 and PMHA3, respectively (Molfarming, China).
The absolute chlorophyll contents of leaves were measured using the spectrophotometer after 95% ethanol extracts, as described previously (Ling et al., 2011). SPAD values were also recorded with a DP-TYS-4N meter (Tuopu, China).
Concomitant chlorophyll fluorescence measurements were conducted employing a MAXI-IMAGING-PAM system (Heinz Walz GmbH, Effeltrich, Germany). To ascertain the maximal fluorescence yield (Fm’), a saturating pulse of red light (1,000 μmol photons m−2 s−1) was administered. The calculation of the electron transport rate (ETR), indicative of the actual photon flux propelling photosystem II (PSII), was determined using the subsequent equation:
Here, Fs denotes ‘steady-state’ fluorescence, Fm’ represents the maximal fluorescence achieved during a saturating light flash, f signifies the fraction of absorbed quanta utilized by photosystem II (PSII), I stands for the incident photon flux density, and αleaf accounts for leaf absorptance.
After a 30-minute period of darkness, leaves were positioned on a black cardboard surface prior to image acquisition. Imaging was carried out utilizing the NightSHADE LB985 in vivo imaging system, incorporating a CCD camera (Berthold, Germany). The LED channels were configured to wavelengths of 470, 660, and 730 nm, each set at an intensity of 40 μmol photons m−2 s−1. The measurement of delayed fluorescence extended over a 120-second duration, employing a 4×4-pixel binning. Subsequent data analysis was performed using the indigo software (Berthold, Germany).
Photosynthetic gas exchange was evaluated using the CIRAS-3 (PP Systems, Inc., USA) equipped with a 3-cm² leaf chamber. The second fully expanded leaf of each plant was selected for analysis, and parameters including intercellular carbon dioxide concentration, stomatal conductance, and photosynthetic rate were quantified. Illumination was maintained at 2,000 μmol photons m-2 s-1 of photosynthetically active radiation. The reference CO2 concentration was set around 700 μmol CO2 mol-1 of air (μg mL-1). The temperature of the chamber was held constant at 30°C, while the flow rate was maintained at 250 µmol s-1. All measurements were executed during a sunny day from 10:00 AM to 2:00 PM.
Approximately 100 mg of leaf or stem tissues were powdered using liquid nitrogen. Total sugar extraction was performed and quantified through an ion chromatograph (Thermo Fisher, USA), following a previously outlined protocol with minor adaptations (Xu Q, et al., 2018). The separation column employed was a CarboPac PA 10 column, and the mobile phase comprised a 200 mM NaOH solution, running at a flow rate of 1 mL min-1. For amperometric detection, a gold electrode served as the working electrode, and an Ag/AgCl electrode functioned as the reference electrode. The elution time spanned 40 min. Standard curves were generated using a combination of three sugars (i.e., Glc, Fru, and Suc). The relative concentrations of these distinct sugars were quantified through the peak area normalization method, executed within the Chromeleon software (Thermo Fisher, USA).
Arabidopsis leaves were pulverized using liquid nitrogen. Subsequently, 100 mg of the resulting powder was solubilized in 1 M HCl. The supernatant was collected and diluted with 1 M HCl to an appropriate concentration after filtering the residue (Fang et al., 2018). The quantification of Na+ and K+ content was executed using a flame photometer (Jingke, China).
Arabidopsis leaves were meticulously pulverized into a fine powder using liquid nitrogen. Subsequently, the hormones were extracted utilizing an isopropanol/water/hydrochloric acid solution. This solution was employed to enhance solubility in the organic solvent and to inactivate specific enzymes. The sample was then concentrated through dichloromethane and nitrogen gas blowdown, and the ABA content was measured using HPLC-MS/MS (Shimadzu, Japan; AB SCIEX, USA).
Total RNA extraction was carried out utilizing Trizol (Invitrogen), adhering to the manufacturer’s stipulations. Evaluation of RNA integrity was performed through employment of the Agilent 2100 Bioanalyzer (Agilent, USA). Subsequently, samples possessing an RNA Integrity Number (RIN) value equal to or greater than 7 were selected for subsequent analyses. Library preparation was undertaken using the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, USA). The transcriptomic sequencing and subsequent analytical procedures were conducted by OE Biotech Company Limited (Shanghai, China).
After harvesting the sorghum in pots, the non-rhizosphere soils were discarded. Sorghum roots were vigorously shaken to release tightly attached rhizosphere soils and mixed evenly to form a homogenized sample for each plant. DNA extraction from rhizosphere soil samples was executed using the E.Z.N.A.® Soil DNA Kit (Omega, USA). The V1-V9 region of the bacterial 16S rRNA gene underwent PCR amplification using 27F (5’-AGRGTTYGATYMTGGCTCAG-3’) and 1492R (5’-RGYTACCTTGTTACGACTT-3’) primers. Following amplification, SMRTbell libraries were prepared through blunt ligation (Pacific Biosciences, USA). All amplicon sequencing and subsequent analytical procedures were performed by Biozeron Biotechnology Company Limited (Shanghai, China).
Soil pH was evaluated through the utilization of a soil slurry and a pH meter (METTLER, USA). The traditional gravimetric method was employed to ascertain soil water content (SWC). The concentrations of total nitrogen (TN), Olsen-P, and Avail. K were determined utilizing a soil nutrient meter (YN-4000, China).
For the synthesis of first-strand cDNA, 1 μg of total RNA was employed in conjunction with the PrimeScript RT reagent Kit (TransGen Biotech, China). Quantification of transcript levels in selected genes was performed via qRT-PCR, using the SYBR Green PCR Master Mix kit (TransGen Biotech, China) and the QuantStudio1 real-time detection system (Thermo Fisher, USA), following the manufacturer’s guidelines. The UBQ9 gene was utilized as an internal control for normalization. Three biological replicates of each treatment underwent RNA extraction, and for each sample, three technical replicates were conducted. A comprehensive list of the primers employed is available in Supplementary Table S1.
All outcomes reported in this study were expressed as the mean ± standard deviation (SD). Each experiment was carried out in triplicate. Data analysis was performed using SPSS 26.0 (IBM Corp, Armonk, NY, USA), applying one-way ANOVA (Analysis of Variance) analysis alongside the student’s t-test or the Duncan test as appropriate. Significance levels are denoted as *P<0.05, **P<0.01, and ***P<0.001. In instances where there are no significant differences, letters are sorted alphabetically, commencing from the highest values, with matching letters signifying a lack of significant distinction.
To uncover the effect of Arthrospira on plant response under salt stress, we supplied various concentrations of Arthrospira to the medium containing 150 mM NaCl and investigated the seed germination and seedling growth of Arabidopsis. Under salt stress condition, the seed germination was significantly inhibited, while this impact was largely rescued by the application of Arthrospira in a dosage-dependent manner, with the optimal concentration of 6 mg FW per plate (Figures 1A–C; Supplementary Figures 2A–E). The germination, seedling vigor and cotyledon greening rate were all improved upon Arthrospira application (Figures 2C–E). In addition, the seedling growth, including root length, survival rate and the leaf weight, was dramatically increased after Arthrospira application (Figures 1D–F; Supplementary Figures 2F–H). Furthermore, Arthrospira reduced the Na+ accumulation and improved K+ content in Arabidopsis leaves, thus increased the K+/Na+ ratio and alleviated the ion toxicity caused by salt stress (Figure 1G; Supplementary Figures 2I, J). Importantly, Arthrospira application significantly enhanced the activity of antioxidant enzyme activity, such as SOD (superoxide dismutase), POD (peroxidase), CAT (catalase), and APX (ascorbate peroxidase), which attenuate the toxic effects of H2O2 and MDA induced by salt stress (Figures 1H–M).
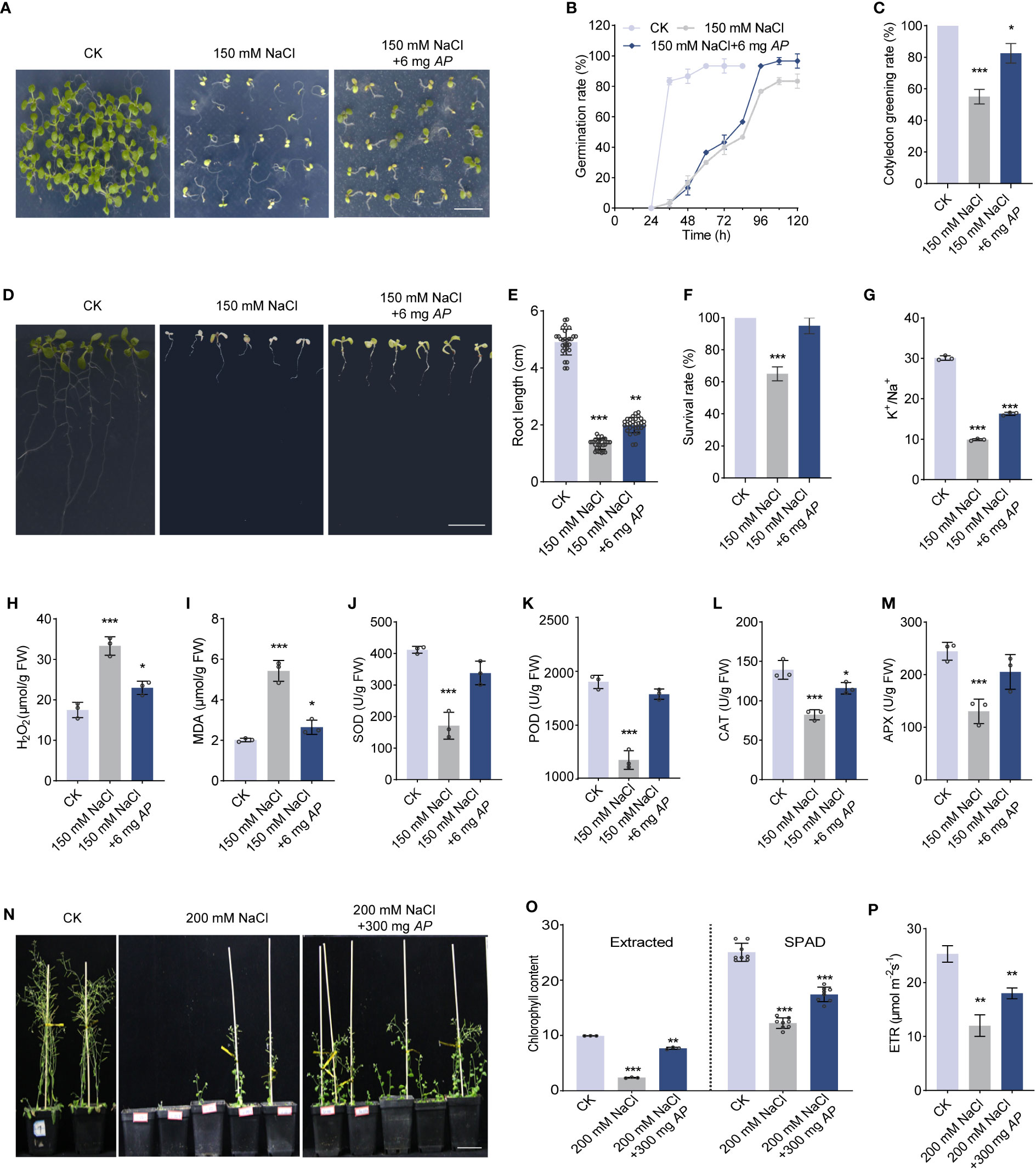
Figure 1 Arthrospira improves Arabidopsis germination and growth under salt stress. (A) Phenotype of 7-day-old Arabidopsis cultivated on ½ MS medium with or without 150 mM NaCl containing 6 mg Arthrospira. (B-C) germination rate (B) and cotyledon greening rate (C) were compared for seedlings grown in ½ MS medium. (D) Phenotypes of 4-day-old Arabidopsis seedlings initially grown on ½ MS medium, were subsequently exposed to 150 mM NaCl supplemented with 6 mg Arthrospira for another 7 days. (E-M) Comparison of root length (E), survival rate (F), K+/Na+ ratio (G), H2O2 content (H), MDA content (I), antioxidant enzyme activity for SOD (J), POD (K), CAT (L) and APX (M) of Arabidopsis grown in ½ MS medium. (N) Phenotypes of 4-day-old Arabidopsis seedlings initially grown on ½ MS medium, were subsequently exposed to soil with 200 mM NaCl for 5 weeks containing 300 mg Arthrospira. (O-P) Comparison of chlorophyll content (O) and relative photosynthetic electron transfer rate (ETR) (P) for Arabidopsis grown in soil. AP, Arthrospira platensis. Scar bars, 1 cm in A and D, 5 cm in N. *P<0.05, **P<0.01, and ***P<0.001.
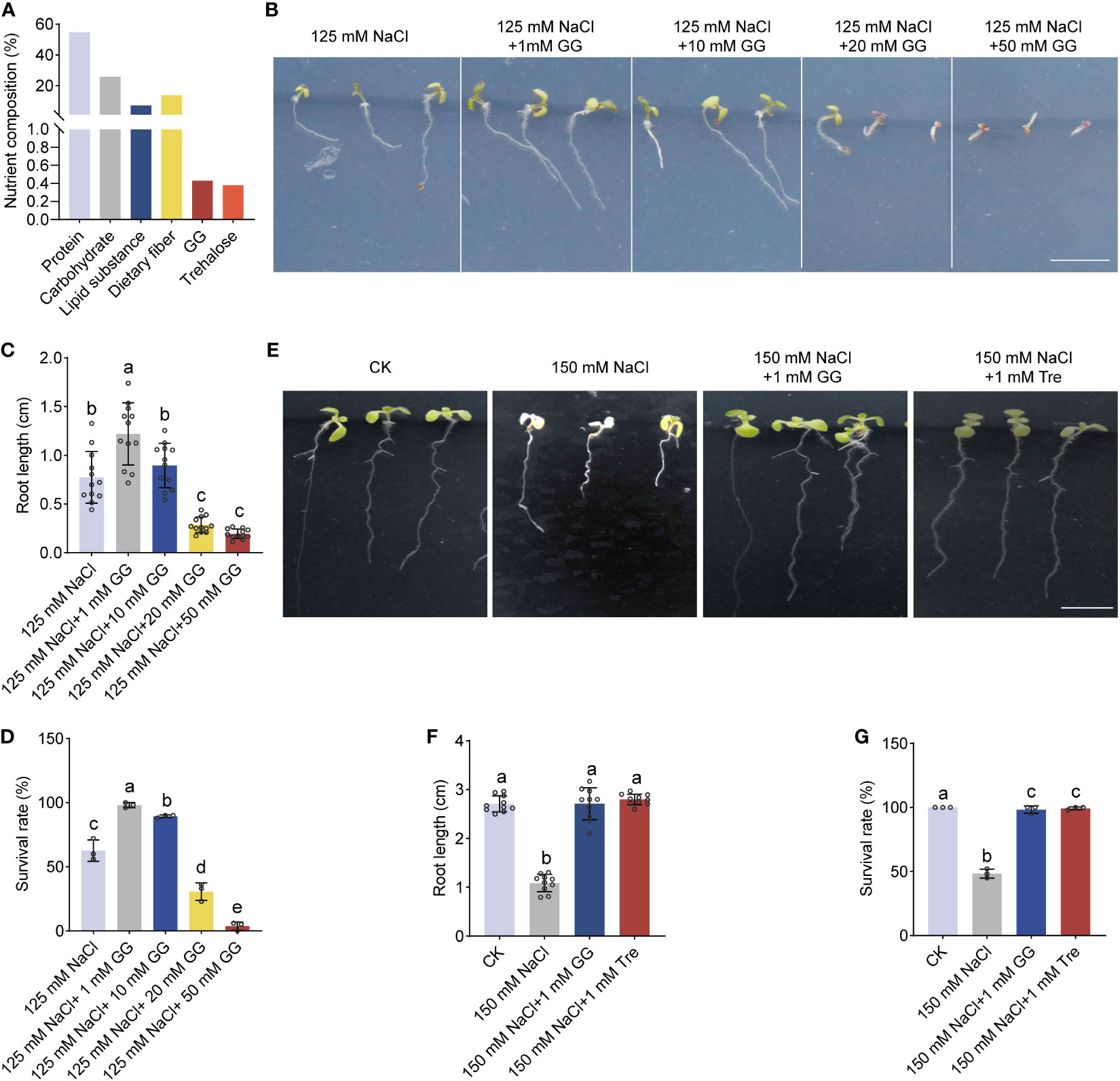
Figure 2 The effect of GG and Tre on seedling growth of Arabidopsis under salt stress. (A) Comprehensive analysis of the entire cellular components of Arthrospira. (B) Phenotypes of 4-day-old Arabidopsis seedlings initially grown on ½ MS medium, were subsequently exposed to 150 mM NaCl supplemented with various concentration of GG. (C-D) Comparison of the root length (C) and survival rate (D) for Arabidopsis grown in ½ MS medium in (B, E) P Phenotypes of 4-day-old Arabidopsis seedlings initially grown on ½ MS medium, were subsequently exposed to 150 mM NaCl supplemented with 1 mM GG or Tre. (F, G) Comparison of root length (F) and survival rate (G) for Arabidopsis grown on ½ MS medium in (E) AP, Arthrospira platensis. GG, Glucosylglycerol. Tre, Trehalose. Scar bars, 1 cm.
Subsequently, we evaluated the effect of Arthrospira on plant growth in salt-containing soil. The result showed that plant growth and photosynthetic performance, including chlorophyll content and photosynthetic electron transfer rate (ETR), were significantly improved upon Arthrospira application (Figures 1N–P; Supplementary Figures 3A–C). As soluble sugars in plants are required for salt and osmotic stress response (Saddhe et al., 2021), we examined sugar content using ion chromatography. The content of total sugar, especially sucrose, was increased when subjected to salt stress (Supplementary Figure 3D). However, Arthrospira suppressed the increment of sugar, suggesting that Arthrospira alleviates the damage of salt stress to Arabidopsis (Supplementary Figure 3D).
In order to investigate the fundamental mechanism by which Arthrospira enhances plant resilience in the face of salt-induced stress, we analyzed the whole cell components of Arthrospira grown under 200 mM NaCl conditions (Supplementary Figure 4). The result showed that glucosylglycerol (GG) and trehalose (Tre) were synthesized and accumulated, with the amount of 0.43% and 0.38% (dry weight), respectively (Figure 2A; Supplementary Table S2). GG and Tre belong to the category of compatible solutes, previously documented for their capacity to enhance both plant growth and stress resilience (Tan et al., 2016; Yang et al., 2022). According to the contents of compatible solutes, 6-mg application of Arthrospira produces approximately 2.36 mM GG and 1.76 mM Tre, respectively (Supplementary Table S3). We next treated Arabidopsis with different concentrations of GG and Tre. The result also demonstrated that 1-10 mM GG promotes plant salt tolerance, while a high level of GG inhibits this effect (Figures 2B–D). In addition, 1 mM GG and 1 mM Tre showed a significant increment of root growth and survival rate under salt stress conditions (Figures 2E–G), supporting their positive promotion in plant salt tolerance.
To better understand the plant’s response on a transcriptional level, we conducted an RNA-seq analysis using Arabidopsis seedlings grown in the conditions of normal (CK), 150 mM NaCl with (S_AP) or without (S) 6 mg FW Arthrospira (Supplementary Figure 5A). After data filtering, each sample generated around 7 Gb of data with an average genome alignment of 98.68% (Supplementary Table S4). The principal component analysis (PCA) and correlation analysis showed that all samples were separated into three groups on the PC1 x PC2 score plot with a very strong correlation (Supplementary Figures 5B, C).
Under salt-stressed conditions, 5296 differently expressed genes (DEGs) were identified compared with the normal condition (CK-S). Once applied with Arthrospira, 2989 genes were affected (S_AP-S) (Figure 3A; Supplementary Figures 6A, B). Among the DEGs, 660 and 2967 genes were specifically detected in the comparison groups of S_AP-S and CK-S, respectively (Figures 3B, C; Supplementary Table S5). Notably, 2,329 overlapped DEGs, including 1,146 up-regulated and 1,183 down-regulated ones, were detected in the two comparison groups (Figures 3B–D; Supplementary Table S5). To verify RNA-seq data, ten DEGs, including three genes involved in chlorophyll metabolism, two in photosynthesis, three in hormone signal transduction, and two in phenylpropanoid metabolism, were chosen for qRT-PCR validation. The qRT-PCR result exhibited congruence with the RNA-seq results (Figure 3E), demonstrating a high correlation coefficient of 0.97 (Figure 3F), demonstrating the reproducibility and dependability of the transcriptomic data. Importantly, the expression level of genes required for salt response, including DREB (dehydration-responsive element-binding proteins), LEA (late embryonic development abundant protein), and ERD (early responsive to dehydration), were dramatically affected by salt and Arthrospira (Figure 3G), demonstrating that Arthrospira application recovered the impairment of salt stress by fine-tuning the genome-wide transcriptional level.
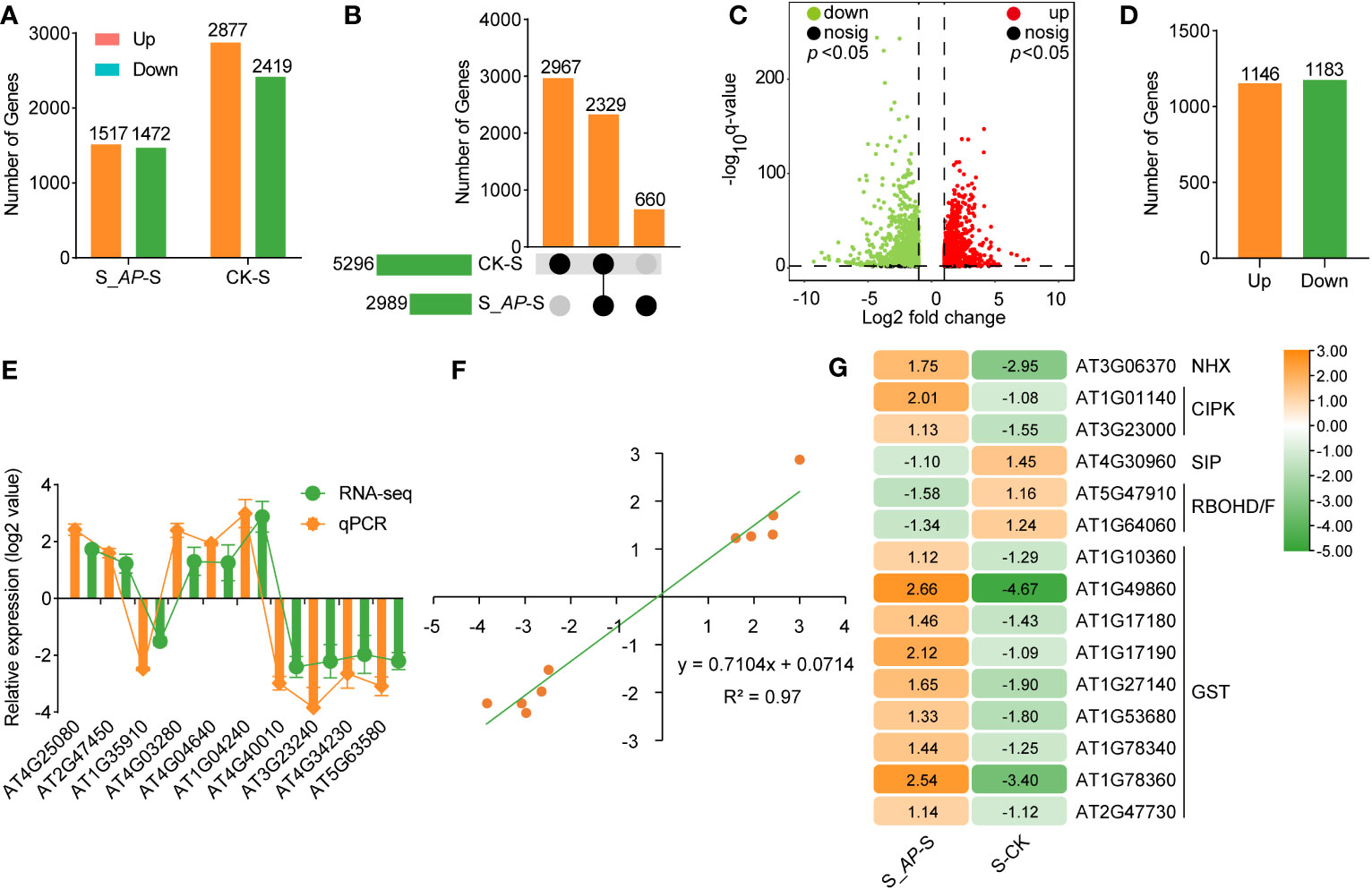
Figure 3 RNA-seq analysis of Arabidopsis seedlings cultivated in ½ MS medium with 150 mM NaCl containing 6 mg Arthrospira. (A) The count of DEGs in S_AP-S and CK-S was assessed. (B) Upset Venn diagrams of DEGs in S_AP-S and CK-S. (C) Volcano plot of log2-fold change values of shared DEGs between S_AP-S and S-CK group. (D) Summary of shared DEGs in both S_AP-S and CK-S. (E) Log2-fold change values of selected DEGs in RNA-seq and qRT-PCR. (F) Scatter plot demonstrates the consistency of RNA-seq and qRT-PCR data. X axis represents log2(S_AP/S) and log2(CK/S) of RNA-seq, Y axis represents log2(S_AP/S) and log2(CK/S) of qPCR. (G) Hierarchical cluster analysis of several classic salt-responsive genes in S_AP-S and S-CK. The Orange and green bars or dots indicate up- and down-regulated DEGs in (A, C, D, G) respectively. CK represents Arabidopsis seedlings grown on normal ½ MS medium condition; S represents Arabidopsis seedlings grown on ½ MS medium containing 150 mM NaCl, and S_AP represents Arabidopsis seedlings grown on ½ MS medium containing 150 mM NaCl and 6 mg Arthrospira. AP, Arthrospira platensis. DEGs, differentially expressed genes.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were carried out to elucidate the functional roles of genes and potential signaling pathways associated with salt stress- and Arthrospira-induced. Most DEGs were specifically enriched in plant growth regulation, photosynthesis, and phenylpropanoid metabolism (Figure 4A; Supplementary Figure 7). Importantly, genes required for responses to hormones, including auxin, ABA, and ethylene were enriched (Figure 4A; Supplementary Figure 8). ABA and ethylene are the important hormones involved in abiotic stress response, while auxin plays a critical role in plant growth (Yu et al., 2019; Waadt et al., 2022). In the RNA-seq data, the expression of genes encoding the rate-limiting enzymes in ABA biosynthesis, 9-cis-epoxy carotenoid dioxygenase (NCED), and the cytochrome P450, family 707, subfamily A (CYP707A) that involved in ABA hydroxylation were significantly up-regulated upon salt stress (Figure 4B). However, once Arthrospira was added, their expression levels were significantly reduced (Figure 4B). In line with this, endogenous ABA abundance was elevated by salt stress but decreased by Arthrospira (Figure 4C; Supplementary Figure 9). Moreover, the expression patterns of Type 2C protein phosphatases (PP2Cs) and Snf1-Related Kinases 2 (SnRK2s), pivotal constituents of the ABA signaling pathway, were influenced by the presence of Arthrospira (Figure 4B). The expression level of ABA response genes, such as RD20, RD29, RAB18 and ABI5, was reduced under Arthrospira treatment (Figure 4D). These findings indicate that Arthrospira might ameliorate the adverse consequences of elevated salt stress through modulation of the plant phytohormone pathway.
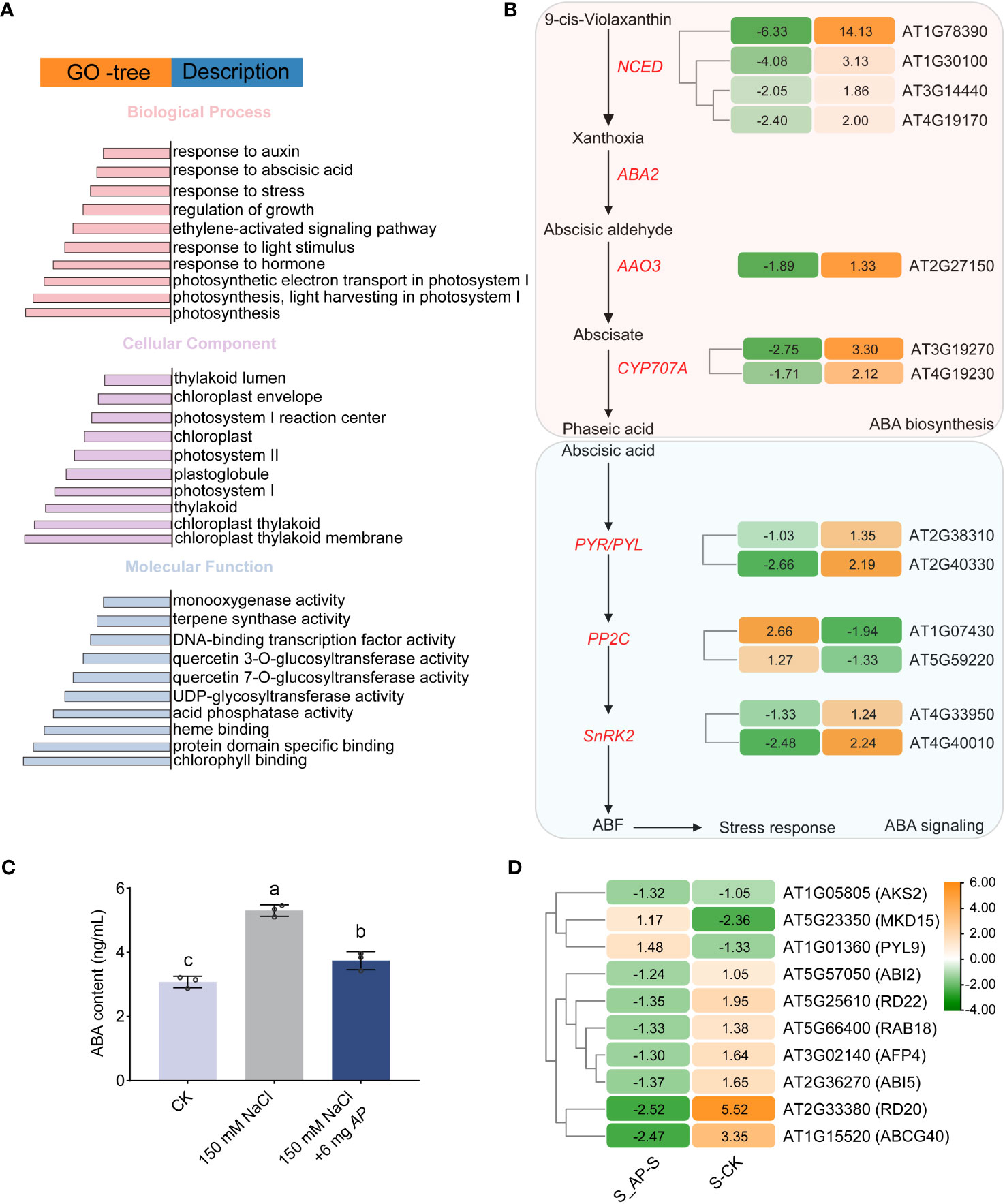
Figure 4 The effect of Arthrospira on plant ABA signal transduction pathway of Arabidopsis seedlings under salt stress. (A) GO enrichment analysis of all shared DEGs. (B) Abscisic acid (ABA) biosynthesis and signal transduction cascades in Arabidopsis seedlings. (C) ABA content in Arabidopsis seedlings cultivated in ½ MS medium with 150 mM NaCl with or without 6 mg Arthrospira. (D) Hierarchical cluster analysis of genes in downstream response of ABA signaling pathway in S_AP-S and CK-S. The Orange and green bars indicate up- and down-regulated DEGs in B and D, respectively. Each colored cell represents the log2 fold change values. CK represents Arabidopsis seedlings cultivated in ½ MS medium condition; S represents Arabidopsis seedlings cultivated in ½ MS medium with 150 mM NaCl, and S_AP represents Arabidopsis seedlings cultivated in ½ MS medium with 150 mM NaCl and 6 mg Arthrospira. AP, Arthrospira platensis. GO, gene ontology. ABA, Abscisic Acid.
Furthermore, a significant enrichment of genes involved in growth regulation was also observed (Supplementary Figure 10). Photosynthesis provides the primary source of organic compounds that are essential for plant growth and fitness. Our data demonstrated that genes required for chlorophyll biosynthesis (Chla and Chlb) were upregulated, while those involved in chlorophyll degradation were significantly downregulated after Arthrospira application, leading to increased chlorophyll levels (Supplementary Figure 11). Additionally, the genes encoding proteins related to photosystem I (PSI), photosystem II (PSII), antenna proteins, cytochrome b6f complex (PETC), and photosynthetic electron transport showed an increased abundance in Arthrospira condition (Supplementary Figure 11).
Since intracellular K+/Na+ homeostasis is crucial for salt tolerance, we first analyzed the transcriptional level of genes involved in Na+ and K+ transportation. Based on the RNA-seq data, no notable distinction was observed in the genes implicated in salt overly sensitive (SOS) and high-affinity potassium transporter (HKT). However, the expression of genes regulating Na+/H+ antiporter (NHX4) and K+ transporters (high-affinity K, HAK5) was inhibited by salt stress but induced by Arthrospira (Figure 5A). In addition, several CBL-interacting protein kinases (CIPKs) associated with the SOS pathway were also increased by the application of Arthrospira (Figure 5A), implying that the potential regulation of K+/Na+ homeostasis might involve the modulation of gene expression related to Na+ or K+ transport.
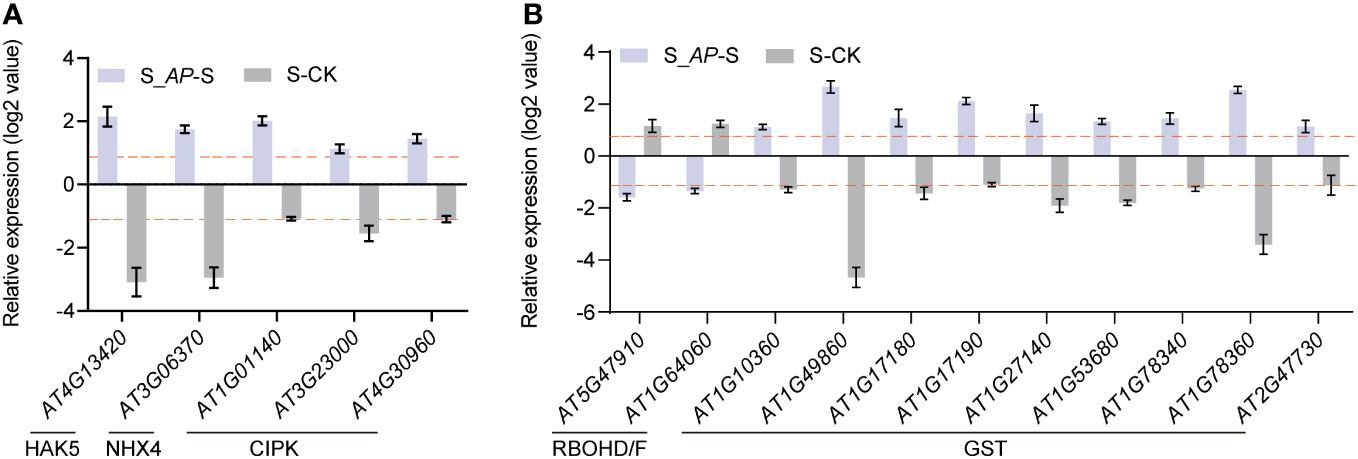
Figure 5 The expression profiles of salt transport- and ROS-related genes. (A) Expression levels of HAK5, NHX4 and CIPKs in Arabidopsis seedlings analyzed in RNA-seq. (B) Expression levels of ROS production genes (RBOHD/F) and ROS scavenging genes (GSTs) in Arabidopsis seedlings analyzed in RNA-seq. The log2-fold change values in S_AP-S and CK-S are presented. CK represents Arabidopsis cultivated in ½ MS medium condition; S represents Arabidopsis cultivated in ½ MS medium with 150 mM NaCl, and S_AP represents Arabidopsis cultivated in ½ MS medium with 150 mM NaCl and 6 mg Arthrospira.
Moreover, the exogenous addition of Arthrospira notably reduced the levels of H2O2 and MDA in Arabidopsis (Figures 1H, I), which are crucial components in reactive oxygen species (ROS). The reduced ROS level may result from decreased synthesis or enhanced scavenging. Indeed, the analysis of RNA-seq data revealed that the genes responsible for ROS synthesis, namely RBOHD and RBOHF, were significantly induced, whereas their expression markedly decreased after Arthrospira application (Figure 5B). Additionally, genes encoding GST subfamily which is required for ROS elimination presented the opposite trend when treated with salt and Arthrospira (Figure 5B), indicating that Arthrospira modified both ROS synthesis and scavenging processes.
Given Arthrospira’s demonstrated amelioration of salt stress impact on Arabidopsis, we sought to ascertain its potential in enhancing stress tolerance in sweet sorghum, a globally significant economic crop. To this end, we first compared the germination rate under conditions involving NaCl and Arthrospira. Salt stress inhibited the germination speed, germination index, and seedling vigor index (Figure 6A; Supplementary Figures 12A-C). On the contrary, Arthrospira has the ability to decrease the negative effect of salt in a dosage-dependent manner (Figure 6A; Supplementary Figures 12A–C). Next, we examined the plant performance of soil-grown sweet sorghum to Arthrospira application. The result indicated that Arthrospira greatly promoted the tolerance and growth of sorghum plants, including plant height, photosynthesis, ETR, and delayed fluorescence (Figures 6B–F; Supplementary Figure 12D). Most importantly, the important agronomic traits of sweet sorghum, such as stem diameter and total sugar content, were also improved by Arthrospira (Figures 6G, H; Supplementary Figure 12E). The kernel size and weight were also increased up to 20% after the application of Arthrospira (Figures 6I, J; Supplementary Figure 12F). Overall, these findings indicated that Arthrospira promotes the germination and growth of sweet sorghum under salt stress, implying its potential application in other crops.
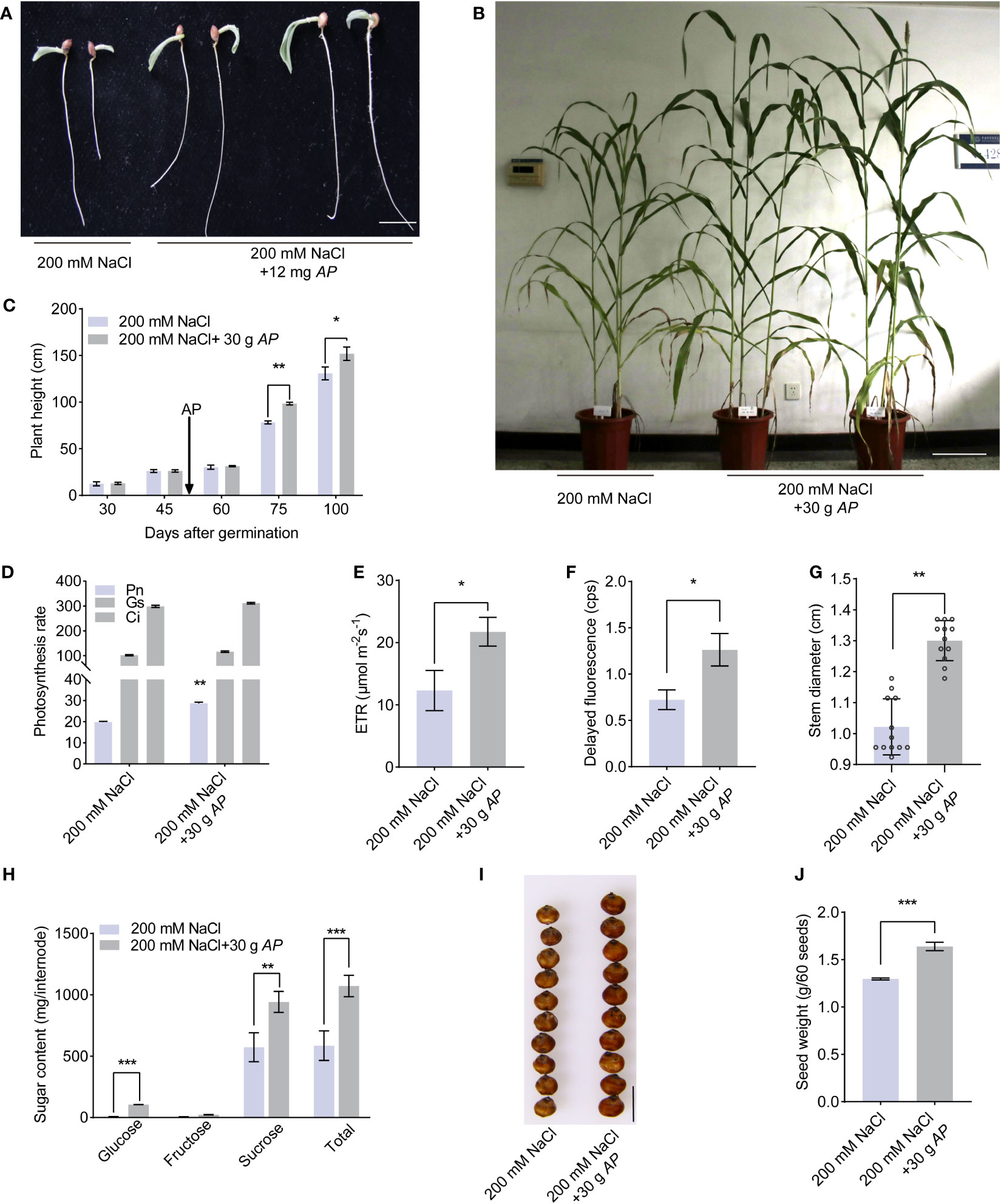
Figure 6 Arthrospira treatment improves sweet sorghum germination and growth under salt stress. (A) Phenotype of 7-day-old sorghum seedlings grow on filter paper with 200 mM NaCl containing 12 mg Arthrospira. (B) Phenotype of 100-day-old sorghum grown on soil supplemented with 200 mM NaCl containing 30 g Arthrospira. (C-G) Comparison of the plant height (C), photosynthesis rate (D), ETR (E), delayed fluorescence (F) and stem diameter (G) under slat stress with or without 30 g Arthrospira. (H) Sugar content of sorghum internode. (I) Photograph of sweet sorghum seeds under 200 mM NaCl with or without 30 g Arthrospira. (J) Seed weight of sorghum in (I) AP, Arthrospira platensis. Scar bars, 1 cm in A and H, 10 cm in B. *P<0.05, **P<0.01, and ***P<0.001.
As mentioned above, Arthrospira generates a lot of components which may improve soil quality (Figure 2A). To test this, the physical and chemical characteristics of soils grown sorghum with or without Arthrospira were examined. The pH and available phosphorus (Olsen-P) showed no significant differences (Supplementary Figures 13A, B), likely due to the soil’s inherent weak acidity. However, water content increased by 17.1% (Figure 7A), total nitrogen (TN) content increased by 11% (Figure 7B), and available potassium (Avail. K) increased by 5% (Figure 7C). To investigate whether these changes were related to the rhizosphere community, high-throughput amplicon sequencing of 16S rRNA was performed using rhizosphere soils after sorghum was harvested. A marginal enhancement in the alpha diversity of rhizosphere bacteria was observed with Arthrospira application under 200 mM NaCl stress (Supplementary Table S6). This was reflected by the Chao1 index, the richness of bacterial communities as well as the ACE index. The prevalent bacterial phyla with relative abundance surpassing 0.5% encompassed Proteobacteria, Bacteroidetes, Planctomycetes, Acidobacteria, Chloroflexi, Actinobacteria, Gemmatimonadetes, Verrucomicrobia, Firmicutes, Spirochaetes, Cyanobacteria, and Armatimonadetes. These phyla collectively constituted 92.3% and 93.4% of the entire rhizosphere bacterial community in the untreated control and Arthrospira-treated specimens, respectively. Noteworthy alterations were apparent in the relative distribution of numerous dominant phyla subsequent to the application of Arthrospira. For instance, the predominant phylum Proteobacteria reduced from 47.3% to 33.4% and the phylum Acidobacteria decreased from 12.5% to 9.7%. On the contrary, Planctomycetes increased from 7.9% to 15.4% and Bacteroidetes increased from 10.9% to 16.8% (Supplementary Figures 13C, D). Following that, an examination was conducted to assess alterations in rhizosphere bacterial communities at the genus level. The findings indicated that the top 50 soil bacterial genera could be generally separated into two major clusters (Figure 7D). The upper cluster represents those with a reduced relative abundance after Arthrospira application, including Rhodanobacter, Hypericibacter, Dongia, and Luteitalea with good repeatability among parallel samples. The lower cluster represents those with increased relative abundance after Arthrospira application, including Steroidobacteraceae, Flexilinea, Caldilinea, Niastella, Ohtaekwangia, and Bythopirellula with good repeatability among parallel samples. Compared to that of bulk soil, microbial diversity in the rhizosphere generally was reduced as these environments are with excess available carbon. Many copiotrophs, such as Bacteroidetes and Proteobacteria, are the main phyla that were thought to be enriched (Ling et al., 2022). Our results reveal that the abundance of Proteobacteria and Bacteroidetes not only significantly changed but also in a reverse direction upon Arthrospira application in saline soil after sorghum growth, indicating a remarkable perturbation on the beneficial interaction among soil microorganisms, plants, and to some extent, the soil properties. As Arthrospira belongs to the phylum Cyanobacteria, then the abundance of rhizosphere cyanobacteria at the genus level was also further compared. Arthrospira amplicons were not detected within the bacterial communities of the rhizosphere. The sorghum rhizosphere cyanobacterial genera that dominate under salt stress conditions were identified as Nodosilinea, Microcystis, Dolichospermum, and Raphidiopsis. Notably, the application of Arthrospira led to a two-fold increase in Nodosilinea (Figure 7E). Members of this genus are featured by forming nodules along the length of their filament, which are capable of biological nitrogen fixation (Li and Brand, 2007; Perkerson Iii et al., 2011). Taken together, Arthrospira application might benefit plants and soil by reshaping the structure of rhizosphere bacteriome which could help to modulate nutrient cycling.
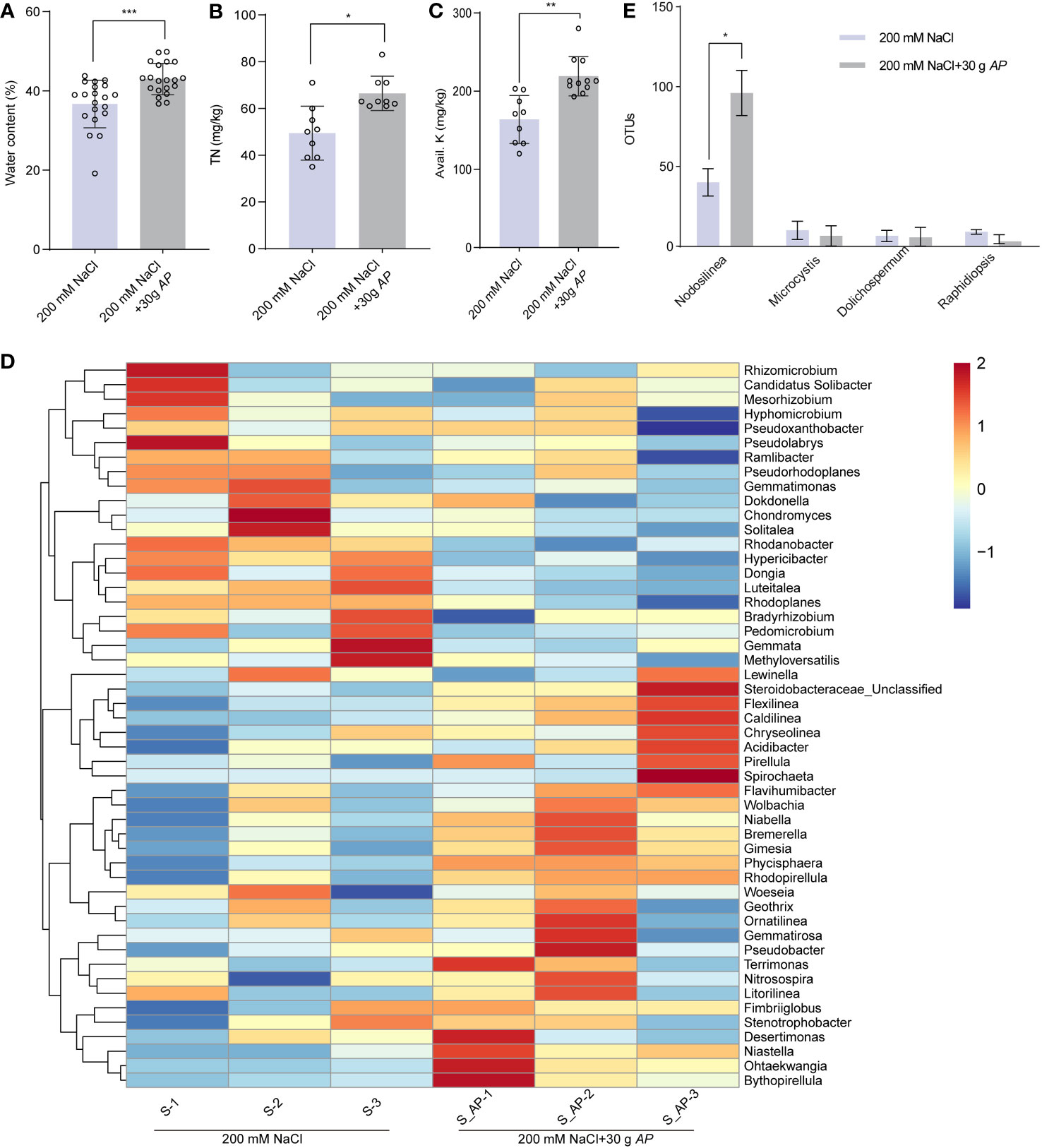
Figure 7 Arthrospira alters the soil chemical properties and structure of rhizosphere. (A-C) Water content (A), total nitrogen (TN) (B), and Avail. K (C) in soil where sorghum grown with or without 30 g Arthrospira. (D) Heat map displays the genus-level relative abundance of rhizosphere bacterial communities with and without Arthrospira treatment. (E) Histogram shows the relative abundance of rhizosphere cyanobacteria with and without Arthrospira treatment. *P<0.05, **P<0.01, and ***P<0.001.
In this study, we report the molecular mechanism and the potential application of Arthrospira in plant improvement under salinity stress. Arthrospira significantly improves the resistance to salt stress of Arabidopsis and sweet sorghum in a dosage dependent manner, whether in the medium or soil. This effect might be due to the biosynthesis of GG and Tre in Arthrospira upon salt stress (Figures 1, 6; Supplementary Figures 2, 3, 12). Additionally, transcriptomic sequencing revealed that Arthrospira modulates the gene expression related to stress adaptation and developmental regulation. Most importantly, Arthrospira application significantly modifies the rhizosphere structure, thereby improving soil quality, which could provide an approach to develop a sustainable agriculture in the world.
During salt stress conditions, preserving plant morphology and yield hinges upon a range of physiological determinants and intrinsic regulatory mechanisms, encompassing ROS scavenging, ion transportation, and hormone homeostasis (Zhu, 2016; Zhao et al., 2020). The physiological stability of plants relies to a great extent on the dynamic balance of Na+ and K+ within their cells (Jiang et al., 2012). High levels of Na+ inhibit K+ absorption, disrupt ion equilibrium, and hinder plant growth. In this investigation, the application of Arthrospira resulted in a noteworthy reduction in Na+ content while concurrently elevating K+ levels, thus maintained K+/Na+ homeostasis under salt stress (Figure 1; Supplementary Figure 2). RNA-seq analysis and endogenous ion measurements revealed that Arthrospira application significantly upregulated the expression of HAK5 (AT4G13420) and NHX4 (AT3G06370) under salt stress, as well as CIPK kinases associated with the SOS pathway (Figure 5). NHX4 encodes a Na+/H+ antiporter accountable for compartmentalizing Na+ from cytoplasm into the vacuole (Wang et al., 2016), the expression pattern indicating that application of Arthrospira could enhance the Na+ compartmentalization and promotes vacuolar sequestration. HAK5 mediates K+ absorption through the plasma membrane and maintains K+/Na+ homeostasis under salt stress (Jung et al., 2009; Nieves-Cordones et al., 2019). Besides, CIPK9 (AT1G01140) and CIPK6 (AT4G30960, SIP3) were reported to interact with SOS2 and SOS3, respectively, to modulate K+ homeostasis (Guo et al., 2001; Pandey et al., 2007). Furthermore, CIPK6 has the capacity to directly regulate the potassium channel AKT2 (Held et al., 2011), while CIPK9 interacts with HAK5 to regulate K+ uptake (Lara et al., 2020). These observations imply that Arthrospira exerts stronger regulatory effects on K+ compared to Na+, enhancing the control of K+/Na+ ratios during salt stress and mitigating ion toxicity in plants by fine-tuning genes involved in ion homeostasis, thus enhancing salt tolerance.
Secondary stress induced by salt stress, particularly oxidative stress, significantly inhibits plant development. Salt stress triggers the outbreak of ROS within plant cells, leading to detrimental oxidative conditions that damage cell membrane, cause lipid peroxidation, and generate aldehyde compounds like malondialdehyde (MDA) (Wang et al., 2008; Olsen et al., 2013). The balance between ROS production and clearance system largely regulates their action within plant cells. In comparison to the salt stress group, the addition of Arthrospira was found to decrease H2O2 and MDA level while increasing the activity of multiple antioxidant enzymes (Figure 1), indicating a potential modulation of genes involved in ROS homeostasis. This was further confirmed through RNA-seq analysis of gene expression related to ROS and antioxidant enzyme activities. Arthrospira supplementation significantly reduced the expression of ROS synthesis-related gene RBOHD/F (Figure 5). Although multiple antioxidant enzyme activities were significantly affected, only GST-like genes showed significant enrichment in the RNA-seq analysis (Figure 5). Moreover, overexpressing of poplar HAK6, the homologous gene of HAKA5 in Arabidopsis, enhanced K+/Na+ control during salt stress and promoted maintenance of ROS homeostasis (Xu et al., 2023). Therefore, we propose that exogenous Arthrospira facilitates the maintenance of ROS balance within plants during salt stress through the dual effects of controlling K+/Na+ levels and enhancing antioxidant enzyme activities.
Arthrospira hydrolysates have emerged as promising biostimulants for various plant species, enhancing growth, photosynthetic efficiency and yield (Santini et al., 2022; Shedeed et al., 2022). In this study, the presence of compatible substances like GG and Tre in the lysate was suggestive of their potential pivotal contribution in enhancing plant salt tolerance and promoting growth (Figure 2). Previous studies have shown that exogenously Tre influences downstream gene expression of ABA signaling, thereby modulating salt tolerance in tomato plants without affecting ABA synthesis (Yu et al., 2019). Our investigation further revealed that the application of Arthrospira not only altered endogenous ABA levels but also regulated downstream gene expression under salt stress conditions (Figure 4), highlighting the complex effects imparted by Arthrospira supplementation. Notably, under salt stress, Arthrospira-treated plants exhibited reduced ABA content compared to the control, selectively repressing AtNCED while promoting upregulation of CYP707A (Figure 4), indicating Arthrospira’s potential role in negatively regulating ABA metabolism, consistent with prior studies in soybean and apple (Li et al., 2015; Asaf et al., 2017). The significant transcriptional upregulation of SnRK2s, crucial in ABA signaling, along with downstream gene expression (Figure 4), suggests that Arthrospira treatment actively stimulates the ABA signaling pathway, potentially initiating stomatal regulation in response to salt stress, as observed in tomato and Arabidopsis transgenic plants (Sun et al., 2011; Yu et al., 2019). CIPK6, previously identified as a kinase regulating ion homeostasis, is also involved in the ABA pathway by mediating ABA signal transduction and influencing downstream gene expression (Chen et al., 2013). Therefore, precise regulation of ABA level and downstream signaling is of great significance for simultaneously enhancing salt tolerance and growth. It is pertinent to acknowledge that the regulation of growth may also be achieved through comprehensive control of auxin, chloroplasts, and photosynthesis (Supplementary Figures 8, 10, 11). Moreover, although the biosynthetic pathway for GG is absent in higher plants, studies have successfully transformed Arabidopsis and potato with ggpPS gene from the γ-proteobacterium Azotobacter vinelandii, encoding a combined GG-phosphate synthase/phosphatase (Klähn et al., 2009; Sievers et al., 2013). It has been observed that low concentrations of GG in Arabidopsis leaves enhance salt tolerance without impacting growth, while moderate or high levels of GG inhibited plant growth without improving salt tolerance (Klähn et al., 2009). This aligns with our findings that GG exhibits a dosage-dependent effect on plant salt stress and growth (Figure 2). Despite previous research have highlighted the significant role of Tre in processes like ABA and ROS regulation, the involvement of GG in these mechanisms has not been investigated. Our study provides additional insights into the essential functions of both compounds. While Tre and GG exhibited notable enhancements in plant salt tolerance in agar plates, the impracticality of their direct application to soil-grown plants arises from the high cost of extraction and purification from Arthrospira. Nevertheless, Arthrospira holds promise for application in farm fields, particularly those with high salinity, to enhance crop tolerance and improve soil quality. Collectively, these findings suggest that the addition of Arthrospira may achieve a better balance between improving resistance and promoting growth by finely regulating multiple pathways.
Arthrospira addition significantly improved soil properties, nutrient status (Figure 7), and root-associated bacteria, known for their beneficial role in nutrient acquisition and plant pathogen defense. Under salt stress or with the addition of chemical fertilizer (Wu et al., 2021; Yukun et al., 2021), the alpha diversity of rhizosphere bacteria in sorghum significantly decreases. However, in this study, we observed that the bacterial richness within the rhizosphere soil under 200 mM NaCl stress exhibited no noteworthy distinction when compared to the Arthrospira-treated soil under analogous circumstances (Supplementary Table S6). This suggests that Arthrospira treatment can enhance soil properties and crop yield by maintaining the richness of rhizobacteria. Although the assembly patterns of the rhizosphere bacterial community vary among plant species, they can exhibit great similarities within the same plant species even in different environments (Xu J, et al., 2018; Trivedi et al., 2020). Our taxonomic analysis revealed that the dominant phyla of sorghum rhizobacteria are broadly consistent with findings from other studies (Supplementary Figures 13C, D) (Yukun et al., 2021). Proteobacteria, the most abundant phylum, tends to be more abundant in high-salinity soil (Yukun et al., 2021). Intriguingly, we found Arthrospira treatment significantly reduces the abundance of Proteobacteria under salt-stress conditions (Supplementary Figures 13C, D). This beneficial alteration indicates Arthrospira application could lead to fundamental changes in soil microbial-driven functions. The increased relative abundances of Planctomycetes and Bacteroidetes (Ravinath et al., 2022), two phyla of representative plant growth-promoting bacteria, demonstrates the important role of Arthrospira application on sorghum performance from another perspective, beyond salt tolerance. Certain Planctomycetes, such as Pirellula, have demonstrated the capability to enhance plant growth by generating hydrogen sulfide (H2S), a factor that exerts a favorable influence on root development. Additionally, this process is conducive to ABA-mediated responses to abiotic stress, notably by instigating stomatal closure (Ravinath et al., 2022). This finding aligns with our transcriptome analysis of Arabidopsis (Figure 5A). Additionally, we did observe the enrichment of Pirellula at the genus level (Figure 7D). The bacteroidetes genus Niastella possesses phosphate solubilization trait through its alkaline phosphatase activity, and its relative abundance also obviously increased (Figure 7D). While Arthrospira is unlikely to colonize the sorghum rhizosphere, it enhances the abundance of Nodosilinea, likely to contributing to nitrogen fixation and regulating sulfur metabolism (Perkerson Iii et al., 2011; Carrión et al., 2017).
Overall, this study highlights Arthrospira’s beneficial impact on plant growth and soil enhancement under salt stress. Mechanisms encompass: (i) Reducing sodium accumulation, fostering potassium uptake for ion balance; (ii) Boosting plant antioxidants, mitigating ROS-induced stress; (iii) Enhancing photosynthesis efficiency via gene alteration, augmenting energy and nutrient production; (iv) Modulating plant hormones (e.g., ABA, auxin), regulating stomatal closure and stress-responsive genes; (v) Influencing rhizosphere microbes, favoring beneficial organisms for nutrient cycling and soil health (Figure 8). Arthrospira emerges as a promising biofertilizer for global saline agriculture, advancing plant growth, salt stress relief, and soil sustainability.
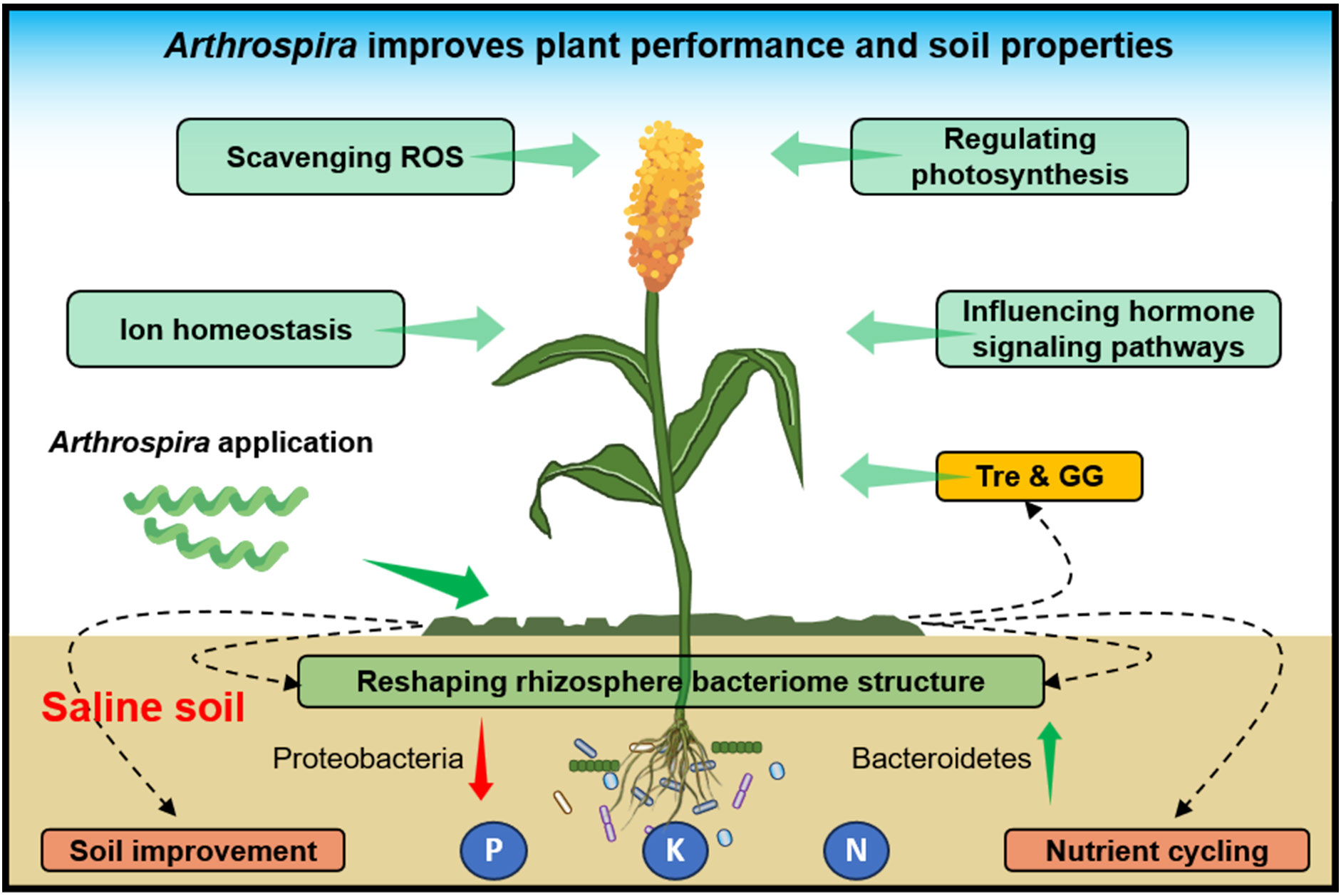
Figure 8 Schematic model of Arthrospira promotes plant growth and soil properties under high salinity environments. Shown Arthrospira’s positive impact on plant growth, soil enhancement under salt stress, achieved through mechanisms like ion balance, enhanced antioxidants, improved photosynthesis, hormone modulation, and microbial influence, positions it as a promising biofertilizer for sustainable agriculture in saline environments.
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) repository, accession number PRJNA995389 for RNA-seq and PRJNA1002101 for 16S rRNA sequencing. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
QX: Conceptualization, Data curation, Methodology, Visualization, Writing – original draft. TZ: Conceptualization, Data curation, Methodology, Validation, Writing – original draft. RZ: Data curation, Methodology, Software, Writing – original draft. YZ: Data curation, Methodology, Software, Writing – original draft. YD: Data curation, Methodology, Software, Writing – original draft. XL: Data curation, Methodology, Software, Writing – original draft. GL: Data curation, Methodology, Software, Writing – original draft. RH: Data curation, Methodology, Software, Writing – original draft. ST: Data curation, Methodology, Software, Writing – original draft. XM: Data curation, Methodology, Software, Writing – original draft. YL: Data curation, Methodology, Software, Writing – original draft. SL: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. XFL: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (Grant number 2021YFA0909700), the Innovation Fund of Shandong Energy Institute (SEI I202130), the Joint Funds of the Key Laboratory of Biofuels, Chinese Academy of Sciences (Y21804110Z), the QIBEBT International Cooperation Project (QIBEBT ICP202301) and the Shandong Taishan Scholarship (to XFL and GL).
We thank Na Sui from Shandong Normal University for providing M-81E seeds.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1293958/full#supplementary-material
Abinandan, S., Subashchandrabose, S. R., Venkateswarlu, K., Megharaj, M. (2019). Soil microalgae and cyanobacteria: the biotechnological potential in the maintenance of soil fertility and health. Crit. Rev. Biotechnol. 39, 981–998. doi: 10.1080/07388551.2019.1654972
Álvarez-Gómez, F., Korbee, N., Figueroa, F. L. (2016). Analysis of antioxidant capacity and bioactive compounds in marine macroalgal and lichenic extracts using different solvents and evaluation methods. Cienc. Marinas 42, 271–288. doi: 10.7773/cm.v42i4.2677
Asaf, S., Khan, A. L., Khan, M. A., Imran, Q. M., Yun, B., Lee, I. (2017). Osmoprotective functions conferred to soybean plants via inoculation with Sphingomonas sp. LK11 exogenous trehalose. Microbiological Res. 205, 135–145. doi: 10.1016/j.micres.2017.08.009
Barbano, D. M., Clark, J. L., Dunham, C. E., Flemin, R. J. (1990). Kjeldahl method for determination of total nitrogen content of milk: collaborative study. J. Assoc. Off. Analytical Chem. 73, 849–859. doi: 10.1093/jaoac/73.6.849
Berendsen, R. L., Pieterse, C. M., Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Carrión, O., Pratscher, J., Curson, A. R., Williams, B. T., Rostant, W. G., Murrell, J. C., et al. (2017). Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 11, 2379–2390. doi: 10.1038/ismej.2017.105
Chen, L., Wang, Q.-Q., Zhou, L., Ren, F., Li, D.-D., Li, X.-B. (2013). Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol. Biol. Rep. 40, 4759–4767. doi: 10.1007/s11033-013-2572-9
Colla, G., Rouphael, Y. (2020). Microalgae: new source of plant biostimulants. Agronomy 10, 1240. doi: 10.3390/agronomy10091240
Dawczynski, C., Schäfer, U., Leiterer, M., Jahreis, G. (2007). Nutritional and toxicological importance of macro, trace, and ultra-trace elements in algae food products. J. Agric. Food Chem. 55, 10470–10475. doi: 10.1021/jf0721500
de Vries, F. T., Griffiths, R. I., Knight, C. G., Nicolitch, O., Williams, A. (2020). Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 368, 270–274. doi: 10.1126/science.aaz5192
Di Lelio, I., Forni, G., Magoga, G., Brunetti, M., Bruno, D., Becchimanzi, A., et al. (2023). A soil fungus confers plant resistance against a phytophagous insect by disrupting the symbiotic role of its gut microbiota. Proc. Natl. Acad. Sci. 120, e2216922120. doi: 10.1073/pnas.2216922120
Duan, Y., Zhu, Z., Cai, K., Tan, X., Lu, X. (2011). De novo biosynthesis of biodiesel by Escherichia coli in optimized fed-batch cultivation. PloS One 6, e20265. doi: 10.1371/journal.pone.0020265
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983. e914. doi: 10.1016/j.cell.2018.10.020
Fang, Y. J., Tan, Y. H., Xiong, Z. L. (2018). Determination of na+, K+ Content in rice leaf (Flame photometric method). Bio-protocol 101, e1010149. doi: 10.21769/BioProtoc.1010149
FAO (2022). The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation (Rome: FAO). doi: 10.4060/cc0461en
Gadelha, C. G., Coutinho, Í. A. C., de Paiva Pinheiro, S. K., de Castro Miguel, E., de Carvalho, H. H., de Sousa Lopes, L., et al. (2021). Sodium uptake and transport regulation, and photosynthetic efficiency maintenance as the basis of differential salt tolerance in rice cultivars. Environ. Exp. Bot. 192, 104654. doi: 10.1016/j.envexpbot.2021.104654
Gul, M., Wakeel, A., Steffens, D., Lindberg, S. (2019). Potassium-induced decrease in cytosolic Na+ alleviates deleterious effects of salt stress on wheat (Triticum aestivum L.). Plant Biol. 21, 825–831. doi: 10.1111/plb.12999
Guo, Y., Halfter, U., Ishitani, M., Zhu, J. K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. doi: 10.1105/TPC.010021
Hamouda, R. A., Shehawy, M. A., Mohy El Din, S. M., Albalwe, F. M., Albalawi, H. M. R., Hussein, M. H. (2022). Protective role of Spirulina platensis liquid extract against salinity stress effects on Triticum aestivum L. Green Process. Synthesis 11, 648–658. doi: 10.1515/gps-2022-0065
Held, K., Pascaud, F., Eckert, C., Gajdanowicz, P., Hashimoto, K., Corratgé-Faillie, C., et al. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21, 1116–1130. doi: 10.1038/cr.2011.50
Ismaiel, S. A. R., Khedr, F. G., Metwally, A. G., Soror, A. F. S. (2022). Effect of biostimulants on soil characteristics, plant growth and yield of Pea (Pisum sativum L.) under field conditions. Plant Sci. Today 9, 650–657. doi: 10.14719/pst.1748
Jiang, C., Belfield, E. J., Mithani, A., Visscher, A., Ragoussis, J., Mott, R., et al. (2012). ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 31, 4359–4370. doi: 10.1038/emboj.2012.273
Jung, J. Y., Shin, R., Schachtman, D. P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21, 607–621. doi: 10.1105/tpc.108.063099
Kaashyap, M., Ford, R., Bohra, A., Kuvalekar, A., Mantri, N. (2017). Improving salt tolerance of chickpea using modern genomics tools and molecular breeding. Curr. Genomics 18, 557–567. doi: 10.2174/1389202918666170705155252
Klähn, S., Hagemann, M. (2011). Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 13, 551–562. doi: 10.1111/j.1462-2920.2010.02366.x
Klähn, S., Marquardt, D. M., Rollwitz, I., Hagemann, M. (2009). Expression of the ggpPS gene for glucosylglycerol biosynthesis from Azotobacter vinelandii improves the salt tolerance of Arabidopsis thaliana. J. Exp. Bot. 60, 1679–1689. doi: 10.1093/jxb/erp030
Lara, A., Ródenas, R., Andrés, Z., Martínez, V., Quintero, F. J., Nieves-Cordones, M., et al. (2020). Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9. J. Exp. Bot. 71, 5053–5060. doi: 10.1093/jxb/eraa212
Lee, M. C., Chen, Y. C., Peng, T. C. (2012). Two-stage culture method for optimized polysaccharide production in Spirulina platensis. J. Sci. Food Agric. 92, 1562–1569. doi: 10.1002/jsfa.4743
Li, Z., Brand, J. (2007). Leptolyngbya nodulosa sp. nov.(Oscillatoriaceae), a subtropical marine cyanobacterium that produces a unique multicellular structure. Phycologia 46, 396–401. doi: 10.2216/06-89.1
Li, C., Tan, D., Liang, D., Chang, C., Jia, D., Ma, F. (2015). Melatonin mediates the regulation of ABA metabolism, free-radical scavenging, and stomatal behaviour in two Malus species under drought stress. J. Exp. Bot. 66, 669–680. doi: 10.1093/jxb/eru476
Ling, Q., Huang, W., Jarvis, P. (2011). Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynthesis Res. 107, 209–214. doi: 10.1007/s11120-010-9606-0
Ling, N., Wang, T., Kuzyakov, Y. (2022). Rhizosphere bacteriome structure and functions. Nat. Commun. 13, 836. doi: 10.1038/s41467-022-28448-9
Liu, G., Liang, J., Lou, L., Tian, M., Zhang, X., Liu, L., et al. (2022). The deubiquitinases UBP12 and UBP13 integrate with the E3 ubiquitin ligase XBAT35. 2 to modulate VPS23A stability in ABA signaling. Sci. Adv. 8, eabl5765. doi: 10.1126/sciadv.abl5765
López, C. V. G., García, M., Fernández, F. G. A., Bustos, C. S., Chisti, Y., Sevilla, J. M. F. (2010). Protein measurements of microalgal and cyanobacterial biomass. Bioresource Technol. 101, 7587–7591. doi: 10.1016/j.biortech.2010.04.077
Lu, X., Valtchev, V. (2023). Green Carbon: Towards a greener world. Green Carbon 1, 1. doi: 10.1016/j.greenca.2023.06.001
Luo, Q., Duan, Y., Lu, X. (2022). Biological sources, metabolism, and production of glucosylglycerols, a group of natural glucosides of biotechnological interest. Biotechnol. Adv. 59, 107964. doi: 10.1016/j.biotechadv.2022.107964
Ma, L., Ye, J., Yang, Y., Lin, H., Yue, L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48, 697–709. e695. doi: 10.1016/j.devcel.2019.02.010
Mutale-joan, C., Rachidi, F., Mohamed, H. A., Mernissi, N. E., Aasfar, A., Barakate, M., et al. (2021). Microalgae-cyanobacteria–based biostimulant effect on salinity tolerance mechanisms, nutrient uptake, and tomato plant growth under salt stress. J. Appl. Phycology 33, 3779–3795. doi: 10.1007/s10811-021-02559-0
Negrão, S., Schmöckel, S., Tester, M. (2017). Evaluating physiological responses of plants to salinity stress. Ann. Bot. 119, 1–11. doi: 10.1093/aob/mcw191
Nieves-Cordones, M., Lara, A., Ródenas, R., Amo, J., Rivero, R. M., Martínez, V., et al. (2019). Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants. Plant Cell Environ. 42, 2357–2371. doi: 10.1111/pce.13573
Olsen, L. F., Issinger, O.-G., Guerra, B. (2013). The Yin and Yang of redox regulation. Redox Rep. 18, 245–252. doi: 10.1179/1351000213Y.0000000059
Osman, M. E. F. M., Dirar, A. I., Konozy, E. H. E. (2022). Genome-wide screening of lectin putative genes from Sorghum bicolor L., distribution in QTLs and a probable implications of lectins in abiotic stress tolerance. BMC Plant Biol. 22, 1–15. doi: 10.1186/s12870-022-03792-6
Pan, T., Liu, M., Kreslavski, V. D., Zharmukhamedov, S. K., Nie, C., Yu, M., et al. (2021). Non-stomatal limitation of photosynthesis by soil salinity. Crit. Rev. Environ. Sci. Technol. 51, 791–825. doi: 10.1080/10643389.2020.1735231
Pandey, G. K., Cheong, Y. H., Kim, B. G., Grant, J. J., Li, L., Luan, S. (2007). CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res. 17, 411–421. doi: 10.1038/cr.2007.39
Pei, H., Yu, Z. (2022). Microalgae: a revolution for salt-affected soil remediation. Trends Biotechnol. 41, 147–149. doi: 10.1016/j.tibtech.2022.08.009
Perkerson Iii, R. B., Johansen, J. R., Kovácik, L., Brand, J., Kaštovský, J., Casamatta, D. A. (2011). A unique pseudanabaenalean (cyanobacteria) genus nodosilinea gen. Nov. Based on morphological and molecular data (1). J. Phycology 47, 1397–1412. doi: 10.1111/j.1529-8817.2011.01077.x
Ravinath, R., Das, A. J., Usha, T., Ramesh, N., Middha, S. K. (2022). Targeted metagenome sequencing reveals the abundance of Planctomycetes and Bacteroidetes in the rhizosphere of pomegranate. Arch. Microbiol. 204, 481. doi: 10.1007/s00203-022-03100-8
Renuka, N., Guldhe, A., Prasanna, R., Singh, P., Bux, F. (2018). Microalgae as multi-functional options in modern agriculture: current trends, prospects and challenges. Biotechnol. Adv. 36, 1255–1273. doi: 10.1016/j.biotechadv.2018.04.004
Rocha, F., Esteban Lucas-Borja, M., Pereira, P., Muñoz-Rojas, M. (2020). Cyanobacteria as a nature-based biotechnological tool for restoring salt-affected soils. Agronomy 10, 1321. doi: 10.3390/agronomy10091321
Saddhe, A. A., Manuka, R., Penna, S. (2021). Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiologia Plantarum 171, 739–755. doi: 10.1111/ppl.13283
Santini, G., Rodolfi, L., Biondi, N., Sampietro, G., Tredici, M. R. (2022). Effects of cyanobacterial-based biostimulants on plant growth and development: a case study on basil (Ocimum basilicum L.). J. Appl. Phycology 34, 2063–2073. doi: 10.1007/s10811-022-02781-4
Shedeed, Z. A., Gheda, S., Elsanadily, S., Alharbi, K., Osman, M. E. (2022). Spirulina platensis biofertilization for enhancing growth, photosynthetic capacity and yield of Lupinus luteus. Agriculture 12, 781. doi: 10.3390/agriculture12060781
Sievers, N., Muders, K., Henneberg, M., Klähn, S., Effmert, M., Junghans, H., et al. (2013). Establishing glucosylglycerol synthesis in potato (Solanum tuberosum L. cv. Albatros) by expression of the ggpPS gene from Azotobacter vinelandii. J. Plant Sci. Mol. Breed. 2, 10.7243. doi: 10.7243/2050-2389-2-1
Sun, L., Wang, Y., Chen, P., Ren, J., Ji, K., Li, Q., et al. (2011). Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J. Exp. Bot. 62, 5659–5669. doi: 10.1093/jxb/err252
Tan, X., Luo, Q., Lu, X. (2016). Biosynthesis, biotechnological production, and applications of glucosylglycerols. Appl. Microbiol. Biotechnol. 100, 6131–6139. doi: 10.1007/s00253-016-7608-3
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi: 10.1038/s41580-022-00479-6
Wang, R., Chen, S., Zhou, X., Shen, X., Deng, L., Zhu, H., et al. (2008). Ionic homeostasis and reactive oxygen species control in leaves and xylem sap of two poplars subjected to NaCl stress. Tree Physiol. 28, 947–957. doi: 10.1093/treephys/28.6.947
Wang, C., Yang, Y., Wang, H., Ran, X., Li, B., Zhang, J., et al. (2016). Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 14, 1838–1851. doi: 10.1111/pbi.12544
Warr, S. R., Reed, R. H., Chudek, J. A., Foster, R., Stewart, W. D. (1985). Osmotic adjustment in Spirulina platensis. Planta 163, 424–429. doi: 10.1007/BF00395153
Wei, M., Zhuang, Y., Li, H., Li, P., Huo, H., Shu, D., et al. (2020). The cloning and characterization of hypersensitive to salt stress mutant, affected in quinolinate synthase, highlights the involvement of NAD in stress-induced accumulation of ABA and proline. Plant J. 102, 85–98. doi: 10.1111/tpj.14613
Wu, A.-L., Jiao, X.-Y., Wang, J.-S., Dong, E.-W., Guo, J., Wang, L.-G., et al. (2021). Sorghum rhizosphere effects reduced soil bacterial diversity by recruiting specific bacterial species under low nitrogen stress. Sci. Total Environ. 770, 144742. doi: 10.1016/j.scitotenv.2020.144742
Xu, J., Zhang, Y., Zhang, P., Trivedi, P., Riera, N., Wang, Y., et al. (2018). The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 9, 4894. doi: 10.1038/s41467-018-07343-2
Xu, Q., Chen, S., Yunjuan, R., Chen, S., Liesche, J. (2018). Regulation of sucrose transporters and phloem loading in response to environmental cues. Plant Physiol. 176, 930–945. doi: 10.1104/pp.17.01088
Xu, W., Wang, Y., Xie, J., Tan, S., Wang, H., Zhao, Y., et al. (2023). Growth-regulating factor 15-mediated gene regulatory network enhances salt tolerance in poplar. Plant Physiol. 191, 2367–2384. doi: 10.1093/plphys/kiac600
Yang, Y., Xie, J., Li, J., Zhang, J., Zhang, X., Yao, Y., et al. (2022). Trehalose alleviates salt tolerance by improving photosynthetic performance and maintaining mineral ion homeostasis in tomato plants. Frontier Plant Sci. 13, 974507. doi: 10.3389/fpls.2022.974507
Yu, W., Zhao, R., Wang, L., Zhang, S., Li, R., Sheng, J., et al. (2019). ABA signaling rather than ABA metabolism is involved in trehalose-induced drought tolerance in tomato plants. Planta 250, 643–655. doi: 10.1007/s00425-019-03195-2
Yukun, G., Jianghui, C., Genzeng, R., Shilin, W., Puyuan, Y., Congpei, Y., et al. (2021). Changes in the root-associated bacteria of sorghum are driven by the combined effects of salt and sorghum development. Environ. Microbiome 16, 1–15. doi: 10.1186/s40793-021-00383-0
Zahra, N., Al Hinai, M. S., Hafeez, M. B., Rehman, A., Wahid, A., Siddique, K. H., et al. (2022). Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol. Biochem. 178, 55–69. doi: 10.1016/j.plaphy.2022.03.003
Zhang, H., Zhao, Y., Zhu, J. K. (2020). Thriving under stress: how plants balance growth and the stress response. Dev. Cell 55, 529–543. doi: 10.1016/j.devcel.2020.10.012
Zhao, C., Zhang, H., Song, C., Zhu, J.-K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 24, 100017. doi: 10.1016/j.xinn.2020.100017
Keywords: Arabidopsis, Arthrospira biofertilizer, rhizosphere bacteria, salt stress, soil properties, sweet sorghum, transcriptome
Citation: Xu Q, Zhu T, Zhao R, Zhao Y, Duan Y, Liu X, Luan G, Hu R, Tang S, Ma X, Liu Y, Li S and Lu X (2023) Arthrospira promotes plant growth and soil properties under high salinity environments. Front. Plant Sci. 14:1293958. doi: 10.3389/fpls.2023.1293958
Received: 14 September 2023; Accepted: 17 November 2023;
Published: 05 December 2023.
Edited by:
Yongfeng Guo, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Piyada Theerakulpisut, Khon Kaen University, ThailandCopyright © 2023 Xu, Zhu, Zhao, Zhao, Duan, Liu, Luan, Hu, Tang, Ma, Liu, Li and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengjun Li, bGlfc2pAcWliZWJ0LmFjLmNu; Xuefeng Lu, bHZ4ZkBxaWJlYnQuYWMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.