
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 October 2023
Sec. Functional Plant Ecology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1289240
This article is part of the Research TopicDispersal Ecology of Land Plants: Striving Towards a More Universal UnderstandingView all 5 articles
Introduction: The dramatic fluctuations of climate conditions since the late Tertiary era have resulted in major species range shifts. These movements were conditioned by geographic barriers and species dispersal capacities. In land plants, gene flow occurs through the movement of male gametes (sperm cells, pollen grains), which carry nDNA, and diaspores (spores, seeds), which carry both cpDNA and nDNA, making them an ideal model to compare the imprints of past climate change on the spatial genetic structures of different genomic compartments. Based on a meta-analysis of cpDNA and nDNA sequence data in western Europe, we test the hypotheses that nDNA genetic structures are similar in bryophytes and spermatophytes due to the similar size of spores and pollen grains, whereas genetic structures derived from the analysis of cpDNA are significantly stronger in spermatophytes than in bryophytes due to the substantially larger size of seeds as compared to spores.
Methods: Sequence data at 1-4 loci were retrieved for 11 bryophyte and 17 spermatophyte species across their entire European range. Genetic structures between and within southern and northern populations were analyzed through F and N statistics and Mantel tests.
Results and discussion: Gst and Nst between southern and northern Europe derived from cpDNA were significantly higher, and the proportion of significant tests was higher in spermatophytes than in bryophytes. This suggests that in the latter, migrations across mountain ranges were sufficient to maintain a homogenous allelic structure across Europe, evidencing the minor role played by mountain ranges in bryophyte migrations. With nDNA, patterns of genetic structure did not significantly differ between bryophytes and spermatophytes, in line with the hypothesis that spores and pollen grains exhibit similar dispersal capacities due to their size similarity. Stronger levels of genetic differentiation between southern and northern Europe, and within southern Europe, in spermatophytes than in bryophytes, caused by higher long-distance dispersal capacities of spores as compared to seeds, may account for the strikingly higher levels of endemism in spermatophytes than in bryophytes in the Mediterranean biodiversity hotspot.
In land plants, assessing dispersal is a challenging issue that can be approached either directly or indirectly (Bullock et al., 2006). Direct techniques involve actual measurements of diaspore dispersal and subsequent modelling of the probability of dispersal as a function of the distance from the source (the dispersal kernel) (see Clobert et al., 2012 for review). Indirect techniques are based on inferences from spatial genetic structures (e.g. Vekemans and Hardy, 2004), which are shaped by the dispersal of male gametes (sperm cells, pollen grains) and diaspores (spores, asexual diaspores, seeds). In spermatophytes and bryophytes, chloroplast DNA (cpDNA) is mostly maternally inherited and therefore dispersed only via seeds, spores and asexual diaspores. Nuclear DNA (nDNA), in contrast, has a biparental inheritance. It is thus dispersed by both seeds and pollen grains in spermatophytes, and spores, asexual diaspores and sperm cells in bryophytes. The contribution of gametes and diaspores to gene flow can hence be disentangled by using markers of different inheritance.
In spermatophytes, variations in dispersal capacities are intimately linked to morphological traits of diaspores related to particular dispersal vectors, referred to as dispersal syndromes (Arjona et al., 2018). In anemochorous species in particular, the range, at which seeds disperse, is correlated with their ornamentation patterns but also with their size, smaller particles dispersing further than larger ones due to their lower settling velocity, and thus, longer expected airborne time (Bullock et al., 2006). Seed size ranges between 0.05 mm (Arditti and Ghani, 2000) to tens of centimetres (Morgan et al., 2017), largely exceeding the size of pollen grains, which range between 10 and about 100 µm in diameter (Hao et al., 2020). Seeds and pollen grains have different potential for dispersal (Santos et al., 2018), so that genetic variation in cpDNA is typically much more spatially structured than in nDNA (Petit et al., 2005). In bryophytes, sperm cells must swim to the archegonia through a continuous film of water, so that fertilization distances are typically very short, <30 cm in mosses up to about 20 m in complex thalloid liverworts equipped with specialized antheridial ‘splash cups’ (Pressel and Duckett, 2019). Therefore, one can hypothesize that, in bryophytes, gene flow mostly occurs via spores, which mostly measure about 10-20 µm in diameter, whose release is facilitated by the hygroscopic movements of specific structures, namely the peristome in mosses and elaters in liverworts, and which are mostly wind-dispersed. The long-distance dispersal capacities of bryophyte spores have had a substantial impact on their biogeographic history and extant distribution patterns, as evidenced by their much lower rates of endemism, larger, trans-oceanic distribution ranges (Patiño and Vanderpoorten, 2018), and lower species turn-over among communities (Patiño et al., 2014b; Mouton et al., 2023) than in spermatophytes. To our knowledge, however, no study has so far attempted at determining whether the putatively higher dispersal capacities of bryophytes as compared to spermatophytes has impacted their spatial genetic structure.
The massive species distribution range shifts that took place due to fluctuations of climate conditions since the late Tertiary era (Hewitt, 1996; Comes and Kadereit, 1998; Hewitt, 1999; Hewitt, 2000; Hewitt, 2003; Petit et al., 2003; Hewitt, 2004; Lumibao et al., 2017) offer an appealing framework to assess how plant species migrated as a response to climate change. In Europe, palaeontological and phylogeographic evidence suggest that species either persisted in scattered southern refugia, wherein populations evolved in isolation, generating high genetic differentiation among them (Hewitt, 1999; Hewitt, 2000; Hewitt, 2004; Médail and Diadema, 2009), or in micro-refugia located in the steppe zone South of the ice sheet (Bhagwat and Willis, 2008). During warmer periods, populations expanded northward from southern refugia or northern micro-refugia, generating admixed populations with high genetic diversity at mid-latitudes, and genetically depauperate populations at high latitudes resulting from long-distance dispersal events and associated founder effects (Hewitt, 1999; Petit et al., 2003).
These movements have been hampered by East-West oriented mountain ranges, limiting genetic exchange between populations located South and North of the barrier and resulting in differences in allele frequencies among populations (Petit et al., 2003). When dispersal was limited to such an extent, that dispersal rates have been lower than migration rates, sister alleles tended to be distributed closer to each other than distantly related alleles, generating phylogeographic signal (Pons and Petit, 1996) potentially leading, ultimately, to allopatric speciation.
Here, we performed a meta-analysis of the spatial genetic structures of bryophytes and spermatophytes across Europe to address the following questions and test the following hypotheses:
- To what extent have mountain ranges hampered post-glacial colonization of the continent in bryophytes and spermatophytes? We hypothesize that genetic divergence between southern and northern populations is higher in spermatophytes than in bryophytes (H1). We further hypothesize that these differences are reflected by the presence of a phylogeographical signal in spermatophytes, wherein sister alleles are more likely to occur in sympatry, whereas in bryophytes, dispersal rates would exceed mutation rates, breaking the phylogenetic structure of genetic variation across the continent (H2).
- To what extent have differences in dispersal capacities between bryophytes and spermatophytes shaped differences in their spatial genetic structure on both sides of the mountain barriers? We hypothesize that the slope of the regression between genetic similarity and geographic distance is significantly higher in spermatophytes than in bryophytes and that distance is a better predictor of genetic similarity in spermatophytes than in bryophytes due to faster recolonization by random long-distance dispersal events since the last glacial maximum in the latter (H3).
- These differences vary depending on the analysis of cpDNA vs nDNA variation. We hypothesize that H1, H2 and H3 apply using cpDNA markers, whereas the spatial genetic structure derived from the analysis of nDNA markers are similar between bryophytes and spermatophytes due to similar sizes of spores and pollen grains (H4).
This study focused on Western Europe, as circumscribed eastwards by the easternmost margin of the Alps, i.e., ~15.00 decimal degrees of longitude. The area was divided into South and North, delimited by the Pyrenees and the Alps (Figure 1). A search of plant phylogeographic and population genetic structure in Europe was performed using Scopus, employing phylogeography or phylogeographic, genetic structure or geographic structure, Europe or European, spermatophyte or bryophyte, as keywords. We focused on studies employing DNA sequence data, which allow for an assessment of the genetic divergence among alleles, and hence, to test for phylogeographical signal (H2). From an initial number of 188 datasets, 160 were removed due to limited sampling (here set at a minimum of 15 specimens at each locus), leaving a total of 17 and 11 spermatophyte and bryophyte datasets, respectively (Table S1).
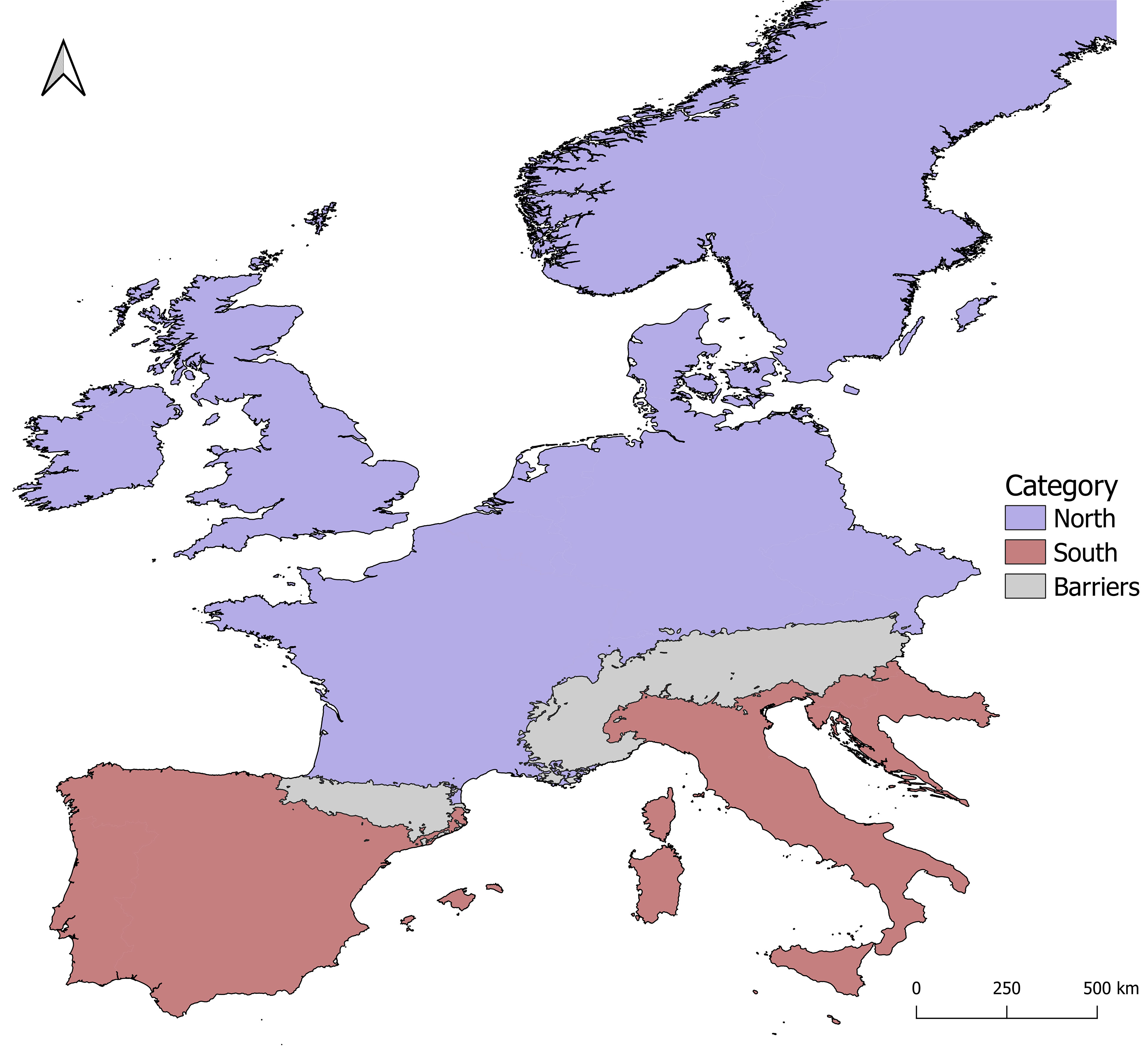
Figure 1 Study area (western Europe) and its division into northern and southern regions on both sides on the mountain ranges of the Alps and the Pyrenees.
Individual sequences were retrieved from GenBank and aligned with the ‘muscle’ algorithm (Edgar, 2004) as implemented in Seaview v5.0.4 (Gouy et al., 2010). In all analyses, cpDNA data were concatenated and considered as one locus, whereas each nuclear locus was considered individually. Regions of ambiguous alignment were removed with Gblocks (v0.91b) (Talavera and Castresana, 2007) as follows: the minimum length of a block after gap cleaning was set to 5. Positions with a gap in less than 50% of the sequences were selected in the final alignment if they were within an appropriate block. All segments including more than 8 contiguous nonconserved positions were rejected. Finally, the minimum number of sequences for a flank position was 85%. Pairwise genetic distances between haplotypes were computed using Kimura’s 2-parameters distance substitution model in the R environment using the package ape 5.6-2 (Paradis and Schliep, 2019). Indels were treated as missing data.
To test the impact of geographic barriers on genetic structure, Gst and Nst were computed between the northern and southern areas. A standard deviation was computed for multilocus datasets by a Jackknife test, wherein Gst and Nst were recalculated after successively pruning one locus from the data at a time as implemented by Spagedi (Hardy and Vekemans, 2002). Presence of a geographic partitioning of allele frequencies between the North and the South was tested by 1000 random permutations of individuals among regions. To test the hypothesis that Gst is, on average, higher in spermatophytes than in bryophytes (H1), t-tests assuming unequal variances were implemented.
We sought for phylogeographic signal in the data by testing the hypothesis that Nst > Gst (H2). Nst is a measure of genetic differentiation among populations analogous to Gst but taking into account the phylogenetic relationships among alleles (Pons and Petit, 1996). An interesting property is that Nst > Gst when phylogeographical signal exists; that is, when distinct alleles sampled within populations are phylogenetically closer, on average, than alleles sampled from different populations (Pons and Petit, 1996). The hypothesis that Nst > Gst was tested by computing the distribution of the null hypothesis (no phylogenetic structure of allelic differentiation among regions) by conducting 1000 permutations of the matrix of genetic distance among alleles.
To test our third hypothesis (H3) that the slope of the regression between genetic similarity and geographic distance is significantly higher in spermatophytes than in bryophytes and that distance is a better predictor of genetic similarity in spermatophytes than in bryophytes, we regressed pairwise kinship coefficients Fij (Ritland, 1996) between individuals and pairwise geographic distances. In these conditions, populations were not continuous, and presence of a significant slope was interpreted in terms of a significant geographic structure of genetic variation resulting from either presence of a geographic barrier or actual isolation-by-distance. We determined whether log-transforming the distance significantly improved the r2 of the Mantel test by comparing the r2 obtained using linear and log-transformed distances using a paired t-test. Significance of the slope was tested by 1000 random permutations of individuals among localities (Mantel test). We further estimated the standard deviation of the slope by implementing a Jackknife test as described above.
To test the hypothesis that the spatial genetic structures of bryophytes and spermatophytes differ with cpDNA due to the larger size of seeds as compared to spores, but not with nDNA due to the similar size of spores and pollen grains (H4), all analyses were performed for cpDNA, nDNA, and both markers combined, except the Mantel tests between geographic distance and Fij derived from nDNA due to missing geographic coordinates for individuals in two spermatophyte species (Arabidopsis thaliana and Beta vulgaris), leaving only four spermatophyte species with nDNA data available for this comparison.
Gst between populations of northern and southern Europe ranged in bryophytes between 0 and 0.17 (excluding an outlier value of 0.77 in Pulvigera lyellii (Hook. & Taylor) Plášek, Sawicki & Ochyra due to the very clear segregation of alleles, see Figure S7), 0.02 and 0.29, and 0.05 and 0.25 with cpDNA markers, nDNA markers, and both combined (Table 1), respectively. In spermatophytes, the range of Gst was 0-0.51, 0.02-0.14 and 0.02-0.29 with cpDNA markers, nDNA markers, and both combined, respectively. (Table 1). On average, Gst was significantly higher in spermatophytes than in bryophytes with cpDNA (Gst = 0.06 ± 0.05 and 0.16 ± 0.17 in bryophytes and in spermatophytes, respectively, p<0.05), but not with nDNA (Gst = 0.09 ± 0.11 and 0.09 ± 0.05 in bryophytes and in spermatophytes, respectively, p=0.46) and both markers combined (Gst = 0.12 ± 0.10 and 0.11 ± 0.11 in bryophytes and in spermatophytes, respectively, p=0.43). Significant Gst values were found in two out of the 11 bryophyte species considered for cpDNA markers, nDNA markers, and both combined (Table 1). A higher proportion of significant test was obtained in spermatophytes, wherein 8, 2 and 2 Gst values derived from the analysis of cpDNA markers, nDNA markers, and both combined, respectively, were found (Table 1).
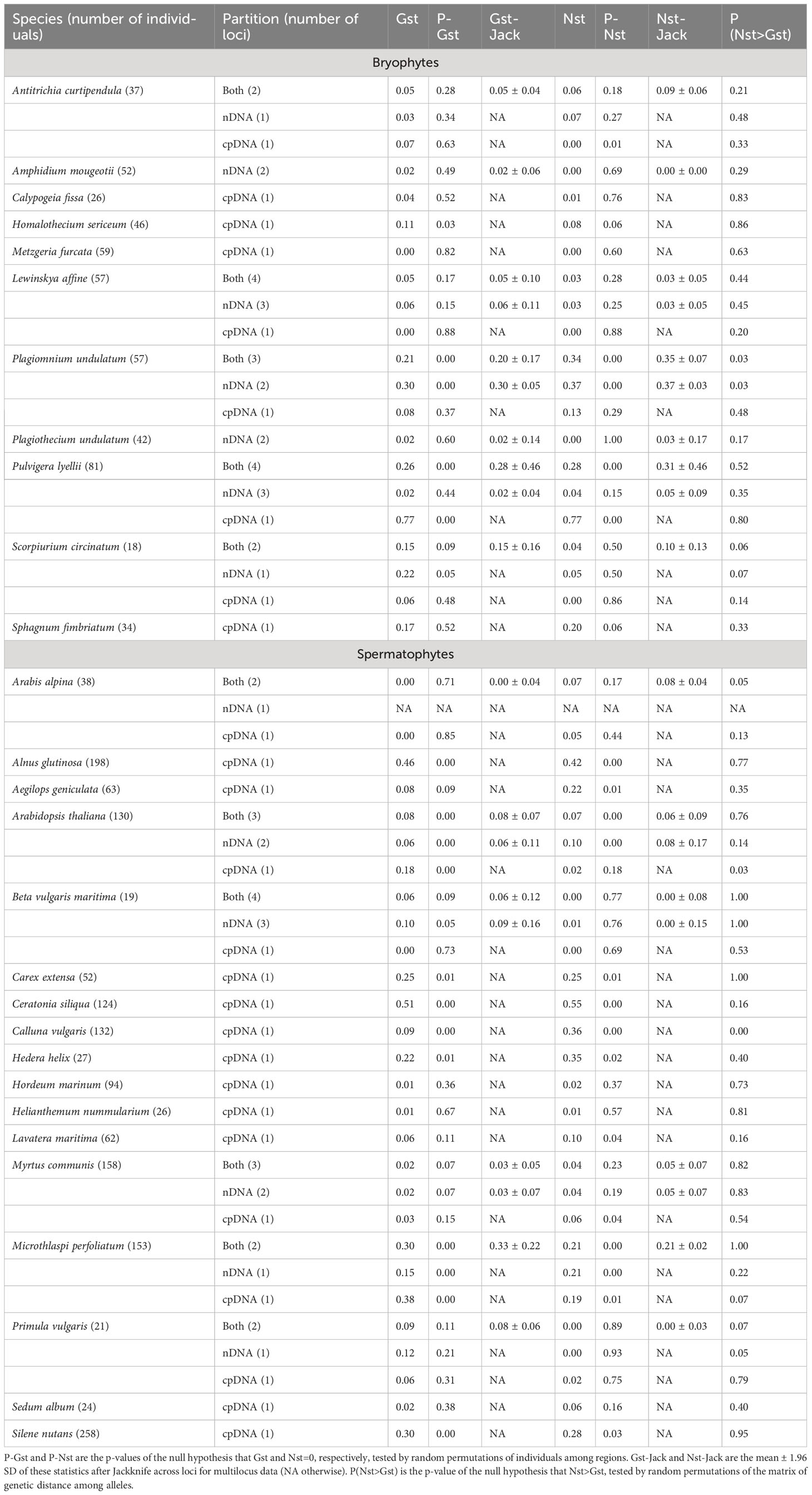
Table 1 Summary statistics of the genetic structure (Gst, Nst) of 11 bryophyte and 17 spermatophyte species between northern and southern Europe based on cpDNA, nDNA sequence data and both combined.
Nst in bryophytes ranged between 0 and 0.20 (excluding an outlier value of 0.77 in Pulvigera lyellii), 0 and 0.37, and 0 and 0.34 with cpDNA markers, nDNA markers, and both combined, respectively (Figure 1). In spermatophytes, the range of Nst was 0-0.55, 0-0.21 and 0-0.21 with cpDNA markers, nDNA markers, and both combined (Figure 1). On average, Nst between the North and the South were significantly higher in spermatophytes than in bryophytes with cpDNA (Nst=0.05 ± 0.08 and 0.17 ± 0.17 in bryophytes and spermatophytes, respectively, p<0.05), but not with nDNA and both markers combined (Table 1). Significant Nst values were found in 2 cpDNA, 1 nDNA and 2 combined bryophyte datasets. Again, the proportion of significant Nst values was higher in spermatophytes, with 10, 1 and 1 significant tests for cpDNA, nDNA, and both combined, respectively.
A significant phylogeographical signal (Nst>Gst) was found in 1 and 2 of the bryophyte and spermatophytes species investigated, respectively (Table 1).
Log-transforming geographic distances in Mantel tests did not improve the r2 as compared to linear distances (for bryophytes: r2 linear=0.02 ± 0.04, r2 log=0.06 ± 0.13, p=0.28 in the South; r2 linear=0.01 ± 0.01, r2 log=0.01 ± 0.01, p=0.29 in the North; for spermatophytes: r2 linear=0.09 ± 0.09, r2 log=0.11 ± 0.09, p=0.28 in the South; r2 linear=0.07 ± 0.12, r2 log=0.06 ± 0.12, p=0.46 in the North). The average (± SD) slope of the regression between Fij derived from cpDNA markers and geographic distance was about four times steeper in spermatophytes than in bryophytes in the South (b=-7.16 10-5 ± 9.35 10-5 in bryophytes, b=-2.94 10-4 ± 3.0 10-4 in spermatophytes, p<0.05), but did not significantly differ in the North (b=-6.68 10-5 ± 6.74 10-5 in bryophytes, b=-7.13 10-5 ± 1.2 10-4 in spermatophytes, p=0.49) (Figure 2; Table S2). The r2 of these regressions was higher in spermatophytes than in bryophytes, but the difference was significant only in the South (South: r2 = 0.09 ± 0.09 and r2 = 0.02 ± 0.04 in spermatophytes and in bryophytes, respectively, p<0.05; North: r2 = 0.07 ± 0.12 and r2 = 0.01 ± 0.01 in spermatophytes and in bryophytes, respectively, p=0.07) (Table S2).
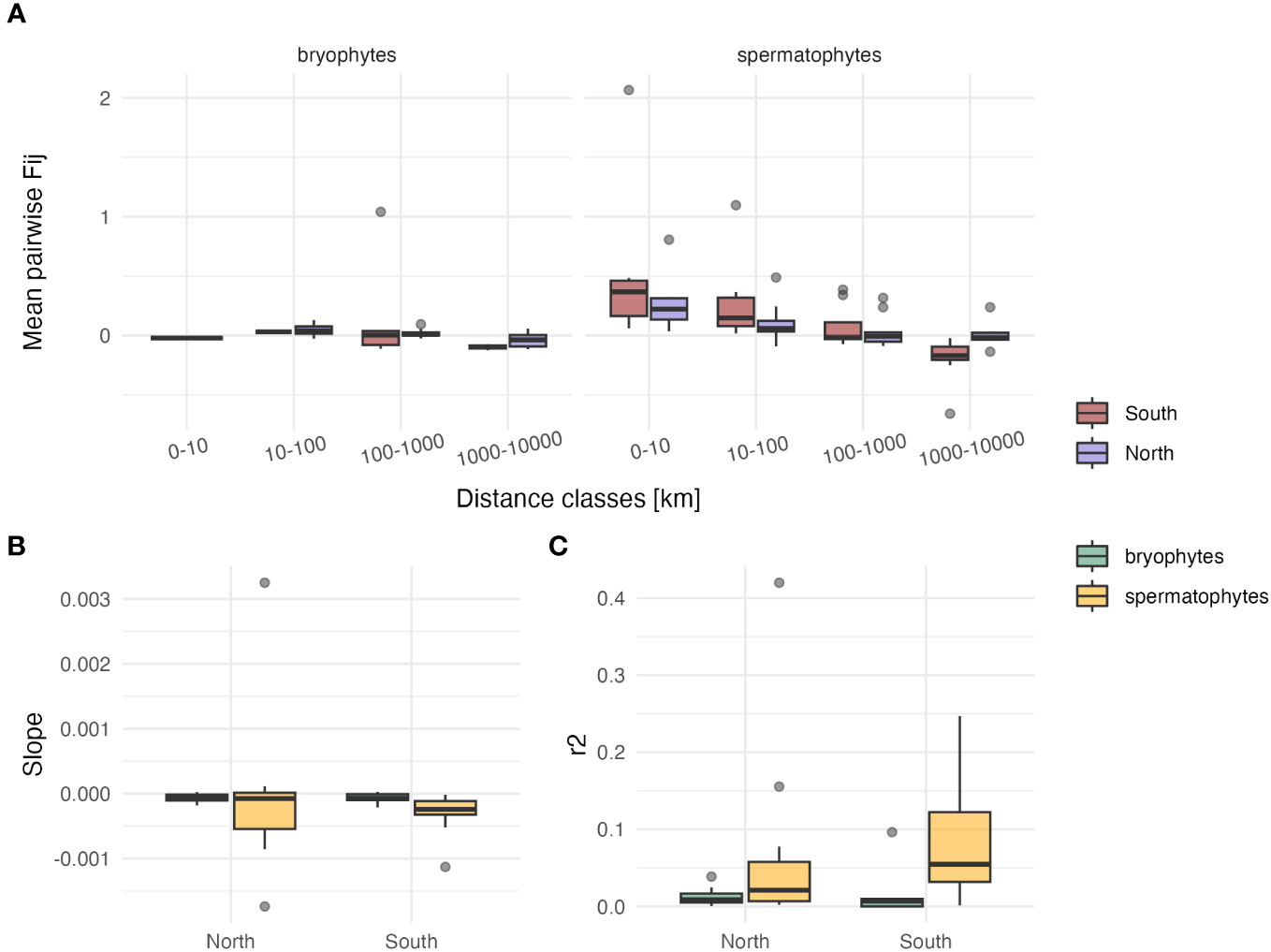
Figure 2 Comparison of isolation by distance patterns among individuals based on cpDNA sequence data between bryophytes and spermatophytes in southern (n=5 and n=12 in bryophytes and spermatophytes, respectively) and northern Europe (n=8 and n=12 in bryophytes and spermatophytes, respectively). (A) Box-plots (showing the first and third quartiles (upper and lower bounds), second quartile (center), 1.5* interquartile range (whiskers) and minima–maxima beyond the whiskers) of the mean kinship coefficient (Fij) between pairs of individuals as a function of geographic distance. (B) Box-plots of the slope of the regression between Fij and geographic distances. (C) Box-plots of the r2 of this relationship.
While differences in long-distance dispersal capacities between bryophytes and spermatophytes have long been discussed in terms of community assembly rules (for instance, in terms of ecological vs historical factors shaping variation of species composition (Patiño et al., 2013) or of variation of the shape of the species-area relationship due to differences in turnover rates among communities (Patiño et al., 2014b), the present results provide a first explicit basis of comparison to determine how such differences have shaped differences in their extant patterns of genetic structure. In agreement with our first hypothesis (H1), Gst and Nst between populations located in southern and northern Europe derived from cpDNA were significantly higher, and the proportion of significant tests was higher in spermatophytes than in bryophytes. In spermatophytes, the stronger North/South geographic structure is consistent with the idea that mountain ranges hampered post-glacial seed dispersal (Petit et al., 2003). In bryophytes, the fact, that a significant difference in allele frequencies between the North and the South (Gst>0) was found in only two out of the 11 investigated species, indicates that migrations across mountain ranges separating the North and the South were sufficient to maintain a homogenous allelic structure across Europe, evidencing the minor role played by mountain ranges in bryophyte migrations. These results are reminiscent of the minor role played by geographic barriers in mosses, wherein populations disjunct on both sides of an oceanic barrier exhibited the same levels of genetic differentiation as populations separated by similar distances but located on the same continent (Grundmann et al., 2007; Hutsemékers et al., 2011; Kyrkjeeide et al., 2016).
Nevertheless, and in contrast with our second hypothesis (H2), evidence for phylogeographical signal in the data was virtually absent for both bryophytes and spermatophytes. While in bryophytes, absence of a phylogeographical signal in the data was expected due to the large contribution of migrants of extra-European origin (Ledent et al., 2019), thereby breaking any phylogenetic structure of the spatial variation of alleles, we initially expected to observe sympatric diversification on both sides of the mountain barriers in spermatophytes. While such a signal was reported in European spermatophytes in specific areas, where they evolve in isolation (e.g., Epipactis helleborine (L.) Crantz in the Italian peninsula (Tranchida-Lombardo et al., 2011), Salvia officinalis L. in the Balkans (Stojanović et al., 2015), the absence of such a signal in the present results illustrates the fact that recolonization of northern areas did not take place from the diversification of a common ancestor, but instead, from several recolonization events from different refugia (Petit et al., 2003).
There was, however, a notable exception in the patterns described above, caused by the strikingly high value of the Gst of 0.77 derived from the analysis of cpDNA variation in the moss Pulvigera lyellii due to the almost complete allele segregation between northern and southern Europe (Figure S7), while Gst ranged between 0.00 and 0.05 across the tree nDNA loci sequenced for that species. By comparison, Korpelainen et al. (2005) reported an average Gst derived from the analysis of sequence data of 0.12 ± 0.11 in a review of the levels of genetic differentiation in bryophytes. While a higher structure in cpDNA is expected in spermatophytes due to different sizes, and hence, dispersal capacities between seeds and pollen grains, no such difference is expected in bryophytes, wherein sperm cells have an extremely limited dispersal range, with a maximum fertilization distance of <30cm in mosses up to about 20m in complex thalloid liverworts equipped with specialized antheridial ‘splash cups’ (Pressel and Duckett, 2019), thereby hardly contributing to gene flow at large spatial scales. We suggest that two processes may account for the striking level of cpDNA variation in P. lyellii. First, genetic drift, which is stronger in the chloroplast than in the nuclear genome (Ness et al., 2016), may not be balanced by migration. A dioicous moss species, P. lyellii indeed reproduces almost exclusively vegetatively in Europe (Zuijlen et al., 2023) via specialized asexual propagules, which have long been thought, due to their larger size and sensitivity to harsh conditions, to disperse much less effectively than spores (see Patiño and Vanderpoorten, 2018 for review). Such a hypothesis is, however, weakened by two lines of evidence. First, the idea, that vegetative diaspores are involved in short-distance dispersal, whereas spores are involved in long-distance dispersal, has been challenged (Pohjamo et al., 2006; Laenen et al., 2016). Second, other dioicous species included in the present study and also largely failing to reproduce sexually, such as Plagiomnium undulatum (Hedw.) T.J. Kop., did not display the same levels of spatial structure in cpDNA. Alternatively, the high cpDNA structure in P. lyellii could be explained in terms of selection. Recent evidence suggests that thermal adaptation may occur even in species with high gene flow (Evans et al., 2023), raising the hypothesis that, as opposed to previous assumptions (see Patiño et al., 2014a for review), bryophyte species may adapt locally, which would have considerable implications regarding predictions of their sensitivity to climate change that have assumed climate niche conservatism (Zanatta et al., 2020).
A stronger geographic structuring of cpDNA genetic variation in spermatophytes than in bryophytes was also found within southern Europe. In fact, in agreement with our third hypothesis (H3), the slope of the regression between kinship coefficients derived from cpDNA variation and geographic distance in southern Europe was about four times higher in spermatophytes than in bryophytes, and geographic distance was a significantly better predictor of this relationship, as evidenced by significantly higher r2 in spermatophytes than in bryophytes. In northern Europe in contrast, the differences of genetic structure between bryophytes and spermatophytes, as expressed by the slope and r2 of the regression between genetic similarity and geographic distance, was not significant. In spermatophytes, the strong levels of genetic divergence among populations point to an evolution in isolation in the different peninsulas over long periods of time (Petit et al., 2003). In bryophytes, genetic differentiation among populations within the Mediterranean is less pronounced due to the comparatively higher dispersal capacities of the group, especially for migrations above the sea (Hutsemékers et al., 2011; Kyrkjeeide et al., 2016), as further suggested by the comparatively low Fst values reported (for example, Fst=0.13 between E and W Mediterranean in Homalothecium meridionale (M. Fleisch. & Warnst.) Hedenäs (Désamoré et al., 2012), Fst=0.18 and 0.10 across the Mediterraneo-Atlantic region in Rhynchostegium riparioides (Hedw.) Cardot and Radula lindenbergiana Gottsche ex C. Hartm., respectively (Hutsemékers et al., 2011; Laenen et al., 2011)). In the North, the lack of significance of the genetic structure between bryophytes and spermatophytes suggests that a rapid post-glacial recolonization took place in both groups in the absence of major geographic barriers. Altogether, stronger levels of genetic differentiation between southern and northern Europe, and within southern Europe, in spermatophytes than in bryophytes, may account for the strikingly higher levels of endemism in the former than in the latter. This is especially true in the Mediterranean, the world’s second largest biodiversity hotspot, wherein 22% of spermatophyte species are endemic to the region (Lopez-Alvarado and Farris, 2022). In bryophytes, by comparison, there are 23 moss species endemic to the Mediterranean (Hodgetts and Lockhart, 2020) out of 1168 moss species in the area (Ros et al., 2013), resulting in an endemism rate of <2%, and none of the 403 liverwort and hornwort species occurring in the Mediterranean (Ros et al., 2007) is endemic.
In agreement with our fourth hypothesis, the spatial genetic structure of spermatophytes was significantly stronger than those of bryophytes based on the analysis of cpDNA, but not nDNA. Seeds, which carry cpDNA, are in fact several orders of magnitude larger than bryophyte spores. The latter typically measure 10-20 µm on average (Patiño and Vanderpoorten, 2018) and are much smaller than even the smallest seeds, whose size ranges between 0.05 and 6 mm (Arditti and Ghani, 2000). Consequently, the settling velocity, a key parameter of the ability of a particle to disperse in the air (Katul et al., 2005), is much lower in bryophyte spores, wherein it ranges between 0.49 and 8.5 cm/s (Zanatta et al., 2016), than in seeds, wherein, even in ‘dust’ orchid seeds or in seeds with anemochorous adaptations, such as in Asteraceae, settling velocities range between 9 and 40 cm/s (Zotz et al., 2016) and 0.25 and 1.01 m/s (Andersen, 1992). Pollen grains, in contrast, typically measure 20-40 µm, ranging between 10 and about 100 µm (Hao et al., 2020) and exhibit settling velocities of a few cm/s (Hirose and Osada, 2016), i.e., within the range reported for bryophyte spores. The similar genetic structure observed for bryophyte spores and pollen grains, which both carry nDNA, is thus consistent with their similarity in size and dispersal capacities.
The comparative analysis of the genetic structure between northern and southern Europe in bryophytes and spermatophytes presented here suggests that mountain ranges of the Alps and the Pyrenees were a significantly more severe impediment for post-glacial migrations in the latter than in the former. The higher genetic structures found in European spermatophytes as compared to bryophytes offers an appealing explanation for the strikingly lower rates of endemism that characterize bryophytes, especially in areas recognized as biodiversity hotspots such as the Mediterranean. A lower dispersal capacity of spermatophytes has also implications for their ability to migrate as a response to ongoing climate change. The results presented here must, however, be interpreted with caution, as our review revealed a quite limited number of phylogeographic studies for plant species distributed across southern and northern Europe based on DNA sequence variation. In particular, the number of phylogeographic studies for bryophytes in the Mediterranean is extremely limited. Nevertheless, our results reveal intriguing patterns regarding the relative dispersal capacities of pollen grains, spores and seeds, opening the door to a large-scale comparative analysis based on large numbers of markers that can now relatively easily be generated with NGS techniques.
Publicly available datasets were analyzed in this study. This data can be found here: https://doi.org/10.1111/j.1095-8339.2007.00775.x; https://doi.org/10.1111/ele.13254; https://doi.org/10.1111/j.1469-8137.2006.01870.x; https://doi.org/10.1111/j.1365-294X.2005.02848.x; https://doi.org/10.1111/mec.13348; https://doi.org/10.1111/j.1469-8137.2010.03328.x; https://doi.org/10.1111/j.1365-294X.2007.03615.x; https://doi.org/10.1002/ece3.3774; https://doi.org/10.1111/j.1365-294X.2009.04449.x ;https://doi.org/10.1111/jbi.13726; https://doi.org/10.1093/botlinnean/boab043; https://doi.org/10.1093/botlinnean/boab043 ;https://doi.org/10.1111/j.1365-294X.2007.03228.x; https://link.springer.com/article/10.1007/s00606-016-1299-1#Sec12; https://doi.org/10.1093/botlinnean/boy025; https://doi.org/10.1093/botlinnean/boy032; https://doi.org/10.1016/j.flora.2019.02.012; https://pbsociety.org.pl/journals/index.php/asbp/article/view/asbp.89313;https://doi.org/10.1371/journal.pone.0179961; https://doi.org/10.1016/j.ppees.2018.10.003.
TF: Writing – original draft, Writing – review & editing. AL: Writing – review & editing. FC: Writing – review & editing. AV: Writing – original draft, Writing – review & editing, Conceptualization.
The authors declare financial support was received for the research, authorship, and/or publication of this article. AV is a research director of the Belgian Funds for Scientific Research (FRS-FNRS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor MB declared a past co-authorship with the author AV.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1289240/full#supplementary-material
Supplementary Table 1 | Datasets employed in the comparative analysis of the spatial genetic structure of bryophytes and spermatophytes in western Europe: species selected, literature source, number of loci and sample size (southern-northern).
Supplementary Table 2 | Slope, p-value, and mean ± 1.96 SD of the slope after Jackknife across loci (for multilocus data, NA otherwise) of the regression between pairwise kinship coefficient Fij derived from cpDNA sequence data and geographic distances in selected bryophyte and spermatophyte species in northern and southern Europe.
Andersen, M. C. (1992). An analysis of variability in seed settling velocities of several wind-dispersed asteraceae. Am. J. Bot. 79 (10), 1087–1091. doi: 10.1002/j.15372197.1992.tb13702.x
Arditti, J., Ghani, A. K. A. (2000). Tansley Review No. 110. Numerical and physical properties of orchid seeds and their biological implications. New Phytol. 145 (3), 367–421. doi: 10.1046/j.14698137.2000.00587.x
Arjona, Y., Nogales, M., Heleno, R., Vargas, P. (2018). Long-distance dispersal syndromes matter: diaspore–trait effect on shaping plant distribution across the Canary Islands. Ecography (Copenhagen) 41 (5), 805–814. doi: 10.1111/ecog.02624
Bhagwat, S. A., Willis, K. J. (2008). Species persistence in northerly glacial refugia of Europe: a matter of chance or biogeographical traits. J. biogeography 35 (3), 464–482. doi: 10.1111/j.13652699.2007.01861.x
Bullock, J. M., Shea, K., Skarpaas, O. (2006). Measuring plant dispersal: an introduction to field methods and experimental design. Plant Ecol. 186 (2), 217–234. doi: 10.1007/s11258-006-91245
Clobert, J., Baguette, M., Benton, T. G., Bullock, J. M. (2012). Dispersal Ecology and Evolution (Oxford: Oxford University Press). doi: 10.1093/acprof:oso/9780199608898.001.0001
Comes, H. P., Kadereit, J. W. (1998). The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 3 (11), 432–438. doi: 10.1016/S1360-1385(98)01327-2
Désamoré, A., Laenen, B., Stech, M., Papp, B., Hedenäs, L., Mateo, R., et al. (2012). How do temperate bryophytes face the challenge of a changing environment? Lessons from the past and predictions for the future. Global Change Biol. 18 (9), 2915–2924. doi: 10.1111/j.1365-2486.2012.02752.x
Edgar, R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5 (1), 113–113. doi: 10.1186/1471-2105-5-113
Evans, A., de Kort, H., Brys, R., Duffy, K. J., Jersáková, J., Kull, T., et al. (2023). Historical biogeography and local adaptation explain population genetic structure in a widespread terrestrial orchid. Ann. Bot. 131 (4), 623–634. doi: 10.1093/aob/mcad010
Gouy, M., Guindon, S., Gascuel, O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27 (2), 221–224. doi: 10.1093/molbev/msp259
Grundmann, M., Ansell, E. W., Russell, S. J., Koch, M. A., Vogel, J. C. (2007). Genetic structure of the widespread and common Mediterranean bryophyte Pleurochaete squarrosa (Brid.) Lindb. (Pottiaceae) – evidence from nuclear and plastidic DNA sequence variation and allozymes. Mol. Ecol. 16 (4), 709–722. doi: 10.1111/j.1365294X.2007.03181.x
Hao, K., Tian, Z.-X., Wang, Z.-C., Huang, S.-Q. (2020). Pollen grain size associated with pollinator feeding strategy. Proc. R. Society. B Biol. Sci. 287 (1933), 20201191–20201191. doi: 10.1098/rspb.2020.1191
Hardy, O. J., Vekemans, X. (2002). spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. notes. 2 (4), 618–620. doi: 10.1046/j.1471-8286.2002.00305.x
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Society. 58 (3), 247–276. doi: 10.1111/j.1095-8312.1996.tb01434.x
Hewitt, G. M. (1999). Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 68 (1–2), 87–112. doi: 10.1006/bijl.1999.0332
Hewitt, G. (2000). The genetic legacy of the Quaternary ice ages. Nat. (London) 405 (6789), 907–913. doi: 10.1038/35016000
Hewitt, G. (2003). “‘18 - Ice ages: Species distributions, and evolution’,” in Evolution on Planet Earth. Eds. Rothschild, L. J., Lister, A. M. (London: Academic Press), 339–361. doi: 10.1016/B978-012598655-7/50045-8
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. Biol. Sci. 359 (1442), 183–195. doi: 10.1098/rstb.2003.1388
Hirose, Y., Osada, K. (2016). Terminal settling velocity and physical properties of pollen grains in still air. Aerobiologia 32 (3), 385–394. doi: 10.1007/s10453-015-9408-0
Hodgetts, N., Lockhart, N. (2020). Checklist and country status of European bryophytes – update 2020 (Gaeltacht, Ireland: Irish Wildlife Manuals, No. 84. National Parks and Wildlife Service, Department of Arts, Heritage).
Hutsemékers, V., Shaw, A. J., Szvovenyi, P., Gonzalez-Mancebo, J. M., Muñoz, J., Vanderpoorten, A. (2011). Oceanic islands are not sinks of biodiversity in spore-producing plants. Proc. Natl. Acad. Sci. - PNAS 108 (47), 18989–18994. doi: 10.1073/pnas.1109119108
Katul, G. G., Porporato, A., Nathan, R., Siqueira, M., Soons, M. B., Poggi, D., et al. (2005). Mechanistic analytical models for long-distance seed dispersal by wind. Am. Nat. 166 (3), 368–381. doi: 10.1086/432589
Korpelainen, H., Pohjamo, M., Laaka-Lindberg, S. (2005). ‘How efficiently does bryophyte dispersal lead to gene flow?’. J. Hattori Botanical Lab. 97, 195–205.
Kyrkjeeide, M. O., Hassel, K., Flatberg, K. I., Shaw, A. J., Brochmann, C., Stenøien, H. K. (2016). Long-distance dispersal and barriers shape genetic structure of peatmosses (Sphagnum) across the Northern Hemisphere. J. biogeography 43 (6), 1215–1226. doi: 10.1111/jbi.12716
Laenen, B., Désamoré, A., Devos, N., Shaw, A. J., Carine, M. A., Gonzalez-Mancebo, J. M., et al. (2011). Macaronesia: a source of hidden genetic diversity for post-glacial recolonization of western Europe in the leafy liverwort Radula lindenbergiana: Hidden diversity in Macaronesia. J. biogeography 38 (4), 631–639. doi: 10.1111/j.1365-2699.2010.02440.x
Laenen, B., Machac, A., Gradstein, S. R., Shaw, B., Patiño, J., Désamoré, A., et al. (2016). Geographical range in liverworts: does sex really matter? J. biogeography 43 (3), 627–635. doi: 10.1111/jbi.12661
Ledent, A., Désamoré, A., Laenen, B., Mardulyn, P., McDaniel, S. F., Zanatta, F., et al. (2019). No borders during the post-glacial assembly of European bryophytes. Ecol. Lett. 22 (6), 973–986. doi: 10.1111/ele.13254
Lopez-Alvarado, J., Farris, E. (2022). Ecology and evolution of plants in the mediterranean basin: perspectives and challenges’. Plants (Basel) 11 (12), 1584. doi: 10.3390/plants11121584
Lumibao, C. Y., Hoban, S. M., McLachlan, J. (2017). Ice ages leave genetic diversity “hotspots” in Europe but not in Eastern North America. Ecol. Lett. 20 (11), 1459–1468. doi: 10.1111/ele.12853
Médail, F., Diadema, K. (2009). Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. biogeography 36 (7), 1333–1345. doi: 10.1111/j.1365-2699.2008.02051.x
Morgan, E. J., Kaiser-Bunbury, C. N., Edwards, P. J., Fleischer-Dogley, F., Kettle, C. J. (2017). Tracing coco de mer’s reproductive history: Pollen and nutrient limitations reduce fecundity. Ecol. Evol. 7 (19), 7765–7776. doi: 10.1002/ece3.3312
Mouton, L., Patiño, J., Carine, M., Rumsey, F., Menezes de Sequeira, M., González-Mancebo, J. M., et al. (2023). Patterns and drivers of beta diversity across geographic scales and lineages in the Macaronesian flora. J. biogeography 50 (5), 858–869. doi: 10.1111/jbi.14580
Ness, R. W., Kraemer, S. A., Colegrave, N., Keightley, P. D. (2016). Direct estimate of the spontaneous mutation rate uncovers the effects of drift and recombination in the Chlamydomonas reinhardtii plastid genome. Mol. Biol. Evol. 33 (3), 800–808. doi: 10.1093/molbev/msv272
Paradis, E., Schliep, K. (2019). pe 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35 (3), 526–528. doi: 10.1093/bioinformatics/bty633
Patiño, J., Carine, M., Fernández-Palacios, J. M., Otto, R., Schaefer, H., Vanderpoorten, A. (2014b). The anagenetic world of spore-producing land plants. New Phytol. 201 (1), 305–311. doi: 10.1111/nph.12480
Patiño, J., Guilhaumon, F., Triantis, K. A., Gradstein, S. R., Hedenäs, L., González-Mancebo, J. M., et al. (2013). Accounting for data heterogeneity in patterns of biodiversity: an application of linear mixed effect models to the oceanic island biogeography of spore-producing plants. Ecography (Copenhagen) 36(8), 904–913. doi: 10.1111/j.1600-0587.2012.00020.x
Patiño, J., Vanderpoorten, A. (2018). ‘Bryophyte biogeography’. Critical Reviews in Plant Sciences 37, 175–209.
Patiño, J., Weigelt, P., Weigelt, P., Solymos, P., Guilhaumon, F., Kreft, H., Triantis, K., et al. (2014a). Differences in species-area relationships among the major lineages of land plants: a macroecological perspective. Global Ecol. biogeography 23 (11), 1275–1283. doi: 10.1111/geb.12230
Petit, R. J., Aguinalde, I., de Beaulieu, J. L., Bittkau, C., Brewer, S., Cheddadi, R., et al. (2003). Glacial refugia; hotspots but not melting pots of genetic diversity. Sci. (American Assoc. Advancement Science) 300 (5625), 1563–1565. doi: 10.1126/science.1083264
Petit, R. J., Duminil, J., Fineschi, S., Hampe, A., Salvini, D., Vendramin, G. G., et al. (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14 (3), 689–701. doi: 10.1111/j.1365-294X.2004.02410.x
Pohjamo, M., Laaka-Lindberg, S., Ovaskainen, O., Korpelainen, H. (2006). Dispersal potential of spores and asexual propagules in the epixylic hepatic Anastrophyllum hellerianum. Evolutionary Ecol. 20 (5), 415–430. doi: 10.1007/s10682-006-0011-2
Pons, O., Petit, R. J. (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genet. (Austin) 144 (3), 1237–1245. doi: 10.1093/genetics/144.3.1237
Pressel, S., Duckett, J. G. (2019). ‘Do motile spermatozoids limit the effectiveness of sexual reproduction in bryophytes? Not in the liverwort Marchantia polymorpha’. J. systematics evolution : JSE 57 (4), 371–381. doi: 10.1111/jse.12528
Ritland, K. (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genetical Res. 67 (2), 175–185. doi: 10.1017/S0016672300033620
Ros, R. M., Mazimpaka, V., Abou-Salama, U., Aleffi, M., Blockeel, T. L., Brugués, M., et al. (2007). Hepatics and anthocerotes of the mediterranean, an annotated checklist. Cryptogamie Bryologie 28 (4), 351–437.
Ros, R. M., Mazimpaka, V., Abou-Salama, U., Aleffi, M., Blockeel, T. L., Brugués, M., et al. (2013). Mosses of the mediterranean, an annotated checklist. Cryptogamie. Bryologie 34 (2), 99–283. doi: 10.7872/cryb.v34.iss2.2013.99
Santos dos, J., Galarda Varassin, I., Cunha Muschner, V., Ovaskainen, O. (2018). Estimating seed and pollen dispersal kernels from genetic data demonstrates a high pollen dispersal capacity for an endangered palm species. Am. J. Bot. 105 (11), 1802–1812. doi: 10.1002/ajb2.1176
Stojanović, D., Aleksić, J. M., Jančić, I., Jančić, R. (2015). A Mediterranean medicinal plant in the continental Balkans: A plastid DNA-based phylogeographic survey of Salvia officinalis (Lamiaceae) and its conservation implications. Willdenowia 45 (1), 103–118. doi: 10.3372/wi.45.45112
Talavera, G., Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biol. 56 (4), 564–577. doi: 10.1080/10635150701472164
Tranchida-Lombardo, V., Cafasso, D., Cristaudo, A., Cozzolino, S. (2011). Phylogeographic patterns, genetic affinities and morphological differentiation between Epipactis helleborine and related lineages in a Mediterranean glacial refugium. Ann. Bot. 107 (3), 427–436. doi: 10.1093/aob/mcq256
Vekemans, X., Hardy, O. J. (2004). New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. ecology. 13 (4), 921–935. doi: 10.1046/j.1365-294X.2004.02076.x
Zanatta, F., Patiño, J., Lebeau, F., Massinon, M., Hylander, K., Degreef, J., et al. (2016). Measuring spore settling velocity for an improved assessment of dispersal rates in mosses. Ann. Bot. 118 (2), 197–206. doi: 10.1093/aob/mcw092
Zanatta, F., Engler, R., Collart, F., Broennimann, O., Mateo, R. G., Papp, B., et al. (2020). Bryophytes are predicted to lag behind future climate change despite their high dispersal capacities. Nat. Commun. 11 (1), 5601–5601. doi: 10.1038/s41467-020-19410-8
Zotz, G., Weichgrebe, T., Happatz, H., Einzmann, H. (2016). Measuring the terminal velocity of tiny diaspores. Seed Sci. Res. 26 (3), 222–230. doi: 10.1017/S0960258516000155
Keywords: spatial genetic structure, geographic barriers, phylogeographical signal, GST, NST, Mediterranean, nDNA, cpDNA
Citation: Fichant T, Ledent A, Collart F and Vanderpoorten A (2023) Dispersal capacities of pollen, seeds and spores: insights from comparative analyses of spatial genetic structures in bryophytes and spermatophytes. Front. Plant Sci. 14:1289240. doi: 10.3389/fpls.2023.1289240
Received: 05 September 2023; Accepted: 17 October 2023;
Published: 30 October 2023.
Edited by:
M. Teresa Boquete, University of Santiago de Compostela, SpainReviewed by:
Jesús Muñoz, Spanish National Research Council (CSIC), SpainCopyright © 2023 Fichant, Ledent, Collart and Vanderpoorten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Vanderpoorten, YS52YW5kZXJwb29ydGVuQHVsaWVnZS5iZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.