- 1State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 2Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
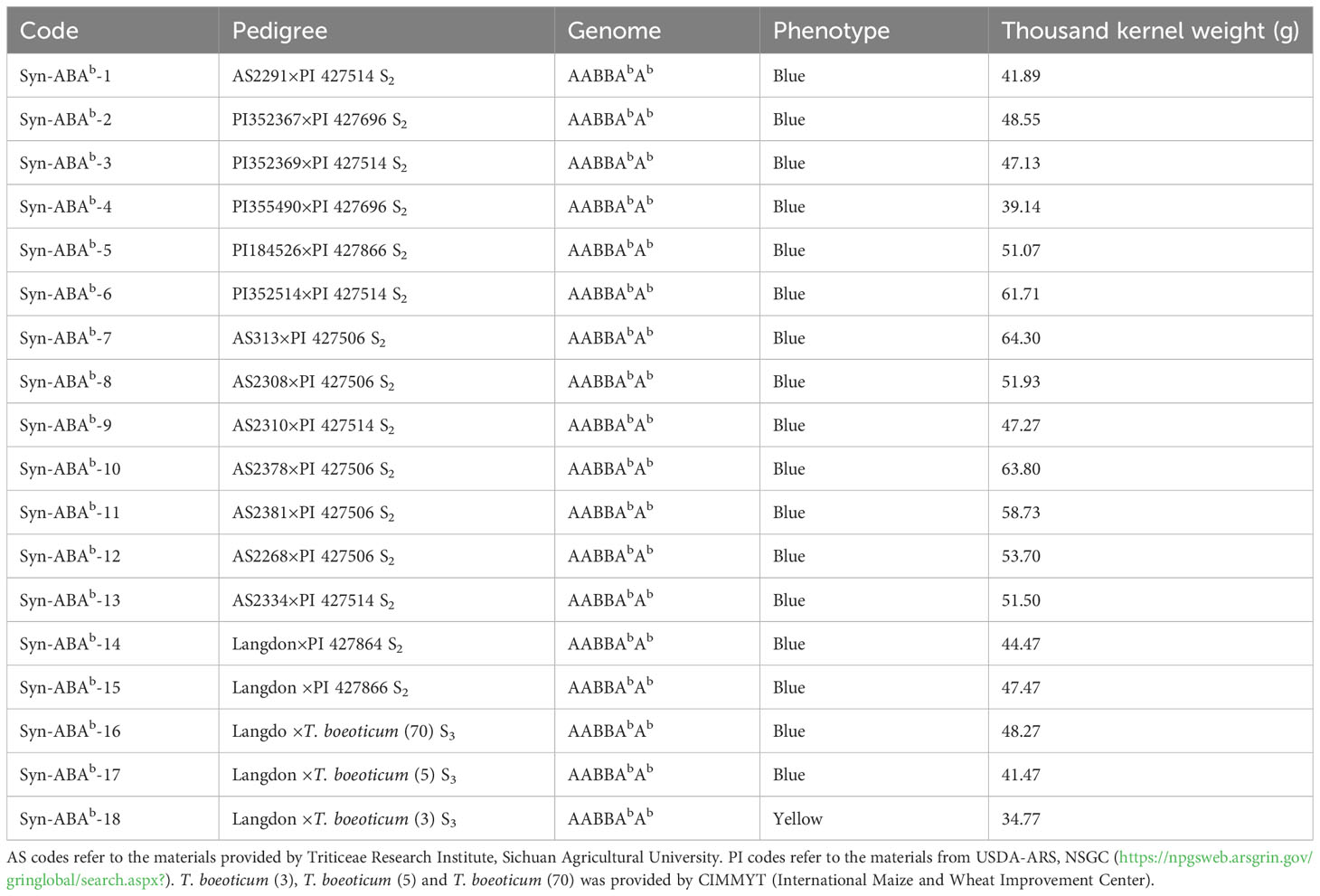
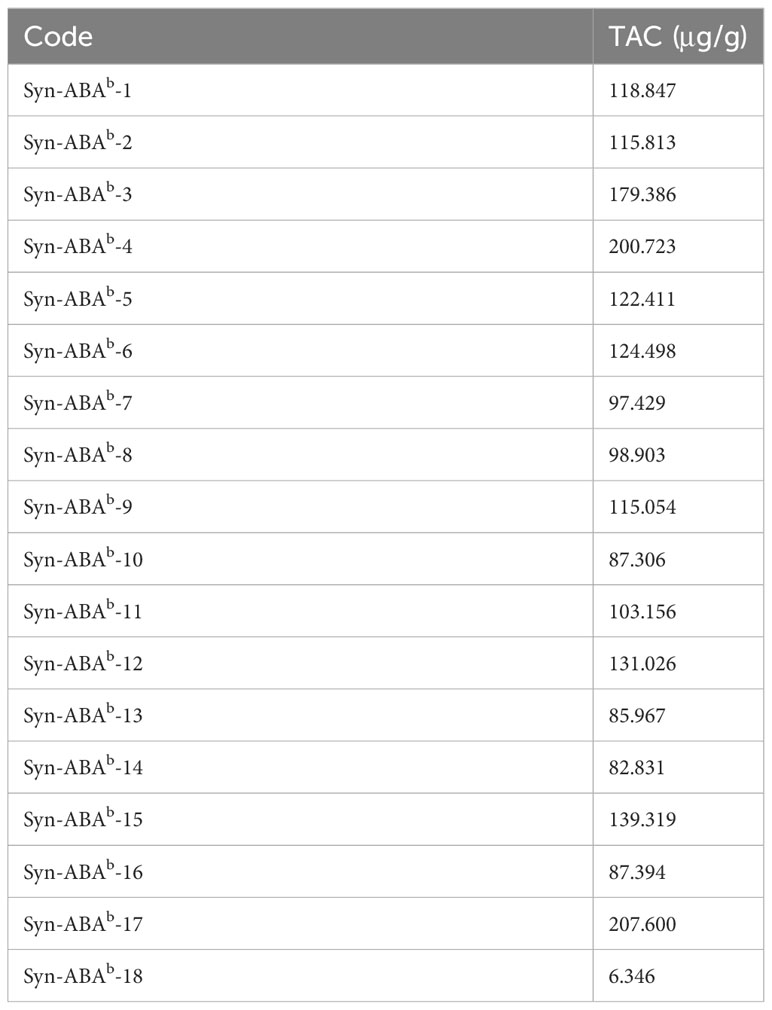
Triticum boeoticum Boiss. (AbAb, 2n = 2x = 14) is a wheat-related species with the blue aleurone trait. In this study, 18 synthetic Triticum turgidum-Triticum boeoticum amphiploids were identified, which were derived from crosses between T. boeoticum and T. turgidum. Three probes (Oligo-pTa535, Oligo-pSc119.2, and Oligo-pTa713) for multicolor fluorescence in situ hybridization (mc-FISH) were combined with genomic in situ hybridization (GISH) to identify chromosomal composition. Seven nutritional indices (anthocyanins, protein, total essential amino acids TEAA, Fe, Zn, Mn and Cu) were measured, and the nutritional components of 18 synthetic amphiploids were comprehensively ranked by principal component analysis (PCA). The results showed that all three synthetic amphiploids used for cytological identification contained 42 chromosomes, including 14 A, 14 B, and 14 Ab chromosomes. The average anthocyanin content was 82.830 μg/g to 207.606 μg/g in the whole meal of the 17 blue-grained lines (Syn-ABAb-1 to Syn-ABAb-17), which was obviously higher than that in the yellow-grained line Syn-ABAb-18 (6.346 μg/g). The crude protein content was between 154.406 and 180.517 g/kg, and the EAA content was 40.193-63.558 mg/g. The Fe, Zn, Mn and Cu levels in the 17 blue-grained lines were 60.55 to 97.41 mg/kg, 60.55-97.41 mg/kg, 35.11 to 65.20 mg/kg and 5.74 to 7.22 mg/kg, respectively, which were higher than those in the yellow-grained line. The contribution of the first three principal components reached 84%. The first principal component was mainly anthocyanins, Fe, Zn and Mn. The second principal component contained protein and amino acids, and the third component contained only Cu. The top 5 Triticum turgidum-Triticum boeoticum amphiploids were Syn-ABAb-11, Syn-ABAb-17, Syn-ABAb-5, Syn-ABAb-8 and Syn-ABAb-4. These amphidiploids exhibited the potential to serve as candidates for hybridization with common wheat, as indicated by comprehensive score rankings, toward enhancing the nutritional quality of wheat.
Introduction
Wheat holds significant importance as a primary energy source in many regions globally. Its unique processing qualities facilitate the preparation of various products, such as bread, cookies, pasta, and noodles (John and Babu, 2021). In this era of rapid economic growth, the nutritional perspective on food security is gaining increasing attention (Li et al., 2006; Tian et al., 2018). The development of biofortified colored wheats (black, purple, and blue) provides nutritional and functional health benefits to the already energy-rich wheat (Garg et al., 2022). Currently, the pursuit of high-quality wheat is equally important as that of high-yielding wheat, and biofortified colored wheat offers a novel opportunity.
Anthocyanins are plant secondary metabolites belonging to the polyphenol class of natural water-soluble pigments. They contribute to the vibrant colors observed in various parts of plants, such as the fruits, leaves, seed coat, and flowers, with hues ranging from orange−red to blue−violet (Ficco et al., 2014; Khlestkina et al., 2015; Wallace and Giusti, 2015). Anthocyanins have strong antioxidant potential, as they protect cells from free radical damage by neutralizing and scavenging free radicals (Cerletti et al., 2017). Moreover, they can disrupt the growth cycle of cancer cells. Studies have demonstrated that anthocyanins can also prevent heart disease, inflammation, and cancer and exhibit anti-aging and anti-diabetes properties (Bagchi et al., 2004; Shipp and Abdel-Aal, 2010; Lin et al., 2017).
Micronutrient deficiencies in the human diet have long been a serious concern in global public health (Bhullar and Gruissem, 2013), particularly for grain-based diets, which not only lack a variety of micronutrients but also have micronutrients with reduced bioavailability to the human body. Malnutrition resulting from micronutrient deficiencies, especially zinc and iron deficiencies, affects a significant portion of the global population, with iron deficiency-related anemia being one of the most prevalent health disorders worldwide (Cakmak, 2008; Gupta et al., 2021; Loskutov and Khlestkina, 2021). The World Health Organization (WHO) estimates that over 2 billion people worldwide experience deficiencies in Fe/Zn (https://www.who.int/health-topics/micronutrients#tab=tab_1). Biofortification, which involves increasing the micronutrient content and concentration of cereals, is an important approach to improving nutritional quality. Improving the levels of micronutrients such as iron and zinc in wheat through the use of breeding technology is considered an effective strategy for increasing the intake of essential micronutrients via food (Morgounov et al., 2007).
T. boeoticum Boiss. (Ab Ab, 2n=2x=14) is an important source of the blue aleurone trait (Zeven, 1991), with the associated candidate gene being TbMYC4A (Liu et al., 2021). T. boeoticum Boiss. has attracted the interest of consumers and the food industry due to its beneficial nutritional characteristics. It exhibits low dietary fiber content but is abundant in protein, lipids (mainly unsaturated fatty acids), fructans, carotenoids, and micronutrients (including iron and zinc) (Hidalgo and Brandolini, 2014; Lachman et al., 2017). T. boeoticum Boiss. exhibits greater tolerance to diverse environmental stressors than common wheat, rendering it a valuable genetic reservoir for fortifying stress resistance in wheat cultivars (Budak et al., 2013). Nax2 and Nax1 represent salt-tolerance genes exclusive to T. boeoticum Boiss.; incorporating them into common wheat amplifies its salt tolerance (Munns et al., 2012; Tounsi et al., 2016). To date, three powdery mildew resistance genes—Pm25, PmTb7A.1, and potentially allelic variant PmTb7A.2 have been discerned in T. boeoticum Boiss., the latter possibly being linked to Pm1 (Shi et al., 1998; Elkot et al., 2015). Successfully, resistance genes Sr22 (for leaf rust) and QYrtb.pau-5A, along with YrZ15-1370 (for stripe rust), have been introduced into common wheat (Paull et al., 1994; Chhuneja et al., 2008; Zhang et al., 2021). Artificial amphiploids play a significant role in wheat breeding programs as well as in genetic and evolutionary research (Gill et al., 1988; Megyeri et al., 2011; Ahmed et al., 2014; Badaeva et al., 2016; Li et al., 2018). To overcome the challenge of obtaining offspring from T. boeoticum Boiss and common wheat, we developed 17 new blue-grained synthetic Triticum turgidum-Triticum boeoticum amphiploids (AABBAbAb, 2n = 6x = 42). Cytological identification and nutritional characteristic analysis were conducted on these lines, which are crucial for transferring favorable genes from T. boeoticum Boiss. to common wheat, facilitating the improvement of the latter with beneficial traits.
Materials and methods
Plant materials
Seventeen new blue-grained synthetic Triticum turgidum-Triticum boeoticum amphiploids, Syn-ABAb-1 to Syn-ABAb-17 (AABBAbAb, 2n = 6x = 42), and one yellow-grained synthetic Triticum turgidum-Triticum boeoticum amphiploid, Syn-ABAb-18 (AABBAbAb, 2n = 6x = 42), were used in this study (Table 1). Eighteen amphiploids and their parents were sown at the Wenjiang Experimental Station of Sichuan Agricultural University in October 2022. Individual plants were grown 10 cm apart within rows, with 30 cm between rows of 1 m long. 182.25 kg of N-P2O5-K2O compound fertilizer with a ratio of 15:15:15 is added per acre. The thousand kernel weight of the 18 amphiploids ranged from 34.77 to 64.30g (Table 1), and the grains displayed a relatively plump shape. A suitable quantity of seeds was randomly selected for the determination of nutritional composition.
Cytological observation
Multicolor fluorescence in situ hybridization (mc-FISH) was performed using the methods described by Tang et al. (2014). Oligo-pTa535, Oligo-pSc119.2, and Oligo-pTa713 were utilized as probes to distinguish individual chromosomes of Syn-ABAb-1 to Syn-ABAb-18. All probes were labeled with either FAM (6-carboxyfluorescein) or TAMRA (6-carboxytetramethylrhodamine) by Tsingke Biotechnology Co., Ltd. Genomic in situ hybridization (GISH) was employed to identify the A and B genomes of synthetic amphiploids. The total genomic DNA of T. boeoticum G52 (A genome) was labeled with an Atto488 NT labeling kit (green) (Jena Bioscience) as a probe, and the total genomic DNA of A. speltoides (B genome) was used for blocking. The identification of Ab chromosomes was performed as described by Feng et al. (2020). Hybridization signals were visualized and captured using an Olympus BX-63 epifluorescence microscope equipped with a Photometric SenSys DP70 CCD camera (Olympus, Tokyo, Japan).
PCR amplification
Genomic DNA was extracted from fresh leaves using a plant genomic DNA kit (Tsingke Biotechnology Co., Ltd.). To identify the T. boeoticum blue aleurone layer 2 (Ba2) candidate gene (Liu et al., 2021), the TbMYC4Aa1-F and TbMYC4Aa1-R (forward, 5′- CGCTATCAGCTCCCAGTCAG -3′; reverse, 5′- CATCCTTCCACGCCAGAACT-3′) primers were used. The PCR system and amplification program for primers used in the study were as follows: 1 µl of DNA (200 ng/ml), 0.5 µl of F/R primers (10 mM), 5 µl of 2× Taq Master Mix (Tsingke Biotechnology Co., Ltd.), and double-distilled water to 10 µl. The PCR products were examined by 2% agarose electrophoresis.
Total anthocyanin content
Total anthocyanins in the seeds of the synthetic amphiploids were extracted according to the method described by Wang et al. (2016). All samples were ground in liquid nitrogen and crushed, and approximately 0.5 g of sample powder was accurately weighed for the experiment. To determine the total anthocyanin content (TAC), the absorbance was measured at 530 nm using a microplate reader (Multiskan GO, Thermo Scientific, MA, USA). A standard curve was constructed for cyanidin 3-sophoroside chloride (Sigma) by also measuring the absorbance at 530 nm. The total anthocyanin content of the synthetic amphiploids was quantified in cyanidin 3-sophoroside chloride equivalents.
Crude protein and amino acid determination
The protein content was analyzed according to the Chinese National Standards: GB 5009.5-2016 (China) by using the Kjeldahl method. Accurately weigh 0.2g of each sample for measurement. A factor of 6.25 was used to calculate the crude protein content (Salo-väänänen and Koivistoinen, 1996).
Quantitative analysis of amino acids was performed using electrospray ionization ultra-performance liquid chromatography tandem mass spectrometry (UPLC−MS/MS) (Waterval et al., 2009). Weigh 0.05g of each sample for quantitative and qualitative analysis of amino acids. Methanol, acetonitrile, and formic acid were purchased from ANPEL. Hydrochloric acid was purchased from Sinoreagent. AccQ•Tag reagent was purchased from Waters.
The sample extracts were analyzed using a UPLC–Orbitrap-MS system (UPLC, Vanquish; MS, QE). The UPLC analytical conditions were as follows: column, Waters ACQUITY UPLC BEH C18 (1.7 μm, 50*2.1 mm); column temperature, 55°C; flow rate, 0.5 mL/min; injection volume, 1 μL; solvent system, water (0.1% formic acid):acetonitrile (0.1% formic acid); gradient program, 95:5 V/V at 0 min, 90:10 V/V at 5.5 min, 75:25 V/V at 7.5 min, 40:60 V/V at 8 min, 95:5 V/V at 8.5 min, 95:5 V/V at 13 min. HRMS data were recorded on a Q Exactive hybrid Q–Orbitrap mass spectrometer equipped with a heated ESI source (Thermo Fisher Scientific) utilizing the full MS acquisition methods. The ESI source parameters were set as follows: spray voltage, 3 kV; sheath gas pressure, 40 arb; aux gas pressure, 10 arb; sweep gas pressure, 0 arb; capillary temperature, 320°C; and aux gas heater temperature, 350°C.
Data were acquired on the Q Exactive instrument using Xcalibur 4.1 (Thermo Scientific) and processed using TraceFinder™4.1 Clinical (Thermo Scientific). The quantified data were output in Excel format. The data are available via ProteomeXchange with identifier PXD046077.
Amino acid analysis
The amino acid ratio coefficient method evaluates food protein content by calculating the ratio of amino acid(s) (RAA), the amino acid ratio coefficient (RC), and the amino acid ratio coefficient score (SRC) in the sample. The RAA, RC, and SRC were calculated according to the following formula from the FAO/WHO/UNU (2007).
where CV is the coefficient of variation of the RC.
Micronutrients analysis
The whole meal micronutrient concentrations (Fe, Zn, Cu, Mn) were determined using inductively coupled plasma atomic emission spectroscopy (ICP−OES, Thermo Scientific iCAP6300, USA) at wavelengths of 259.5 nm (Fe), 213.8 nm (Zn), 324.7 nm (Cu), and 259.3 nm (Mn), following the guidelines of the National Standard for Food Safety GB 5009.268-2016 (China). A 0.5 g sample was accurately weighed for the experiment.
Principal component analysis
Initially, the correlations between diverse nutritional indicators within the samples were computed. Following this, principal component analysis (PCA) was performed, and components with eigenvalues surpassing 1 were isolated as principal components. Subsequently, a comprehensive evaluation model was established to compute the composite scores for the 18 synthetic amphiploids. PCA was performed using the R package psych v2.3.6 (Revelle and Revelle, 2015). Images in the analysis process were generated using the R package ggplot2 v.2.2.1 (Wickham, 2011) for plotting. Differences were considered significant at p ≤ 0.05.
Statistical analysis
All the above nutrients were analyzed in triplicate, and all analyses were carried out in SPSS 20.0 (SPSS 20.0 for Windows; SPSS Inc., Chicago, IL, USA). Differences in nutritional indicators between blue-grained and yellow-grained amphidiploids were evaluated by independent sample t-test. Significant differences were considered at p<0.05.
Results
Chromosomal observations
Cytological identification was conducted by analysis of the chromosomes of three synthetic Triticum turgidum-Triticum boeoticum amphiploids (Syn-ABAb-14 to Syn-ABAb-16). Subsequently, mc-FISH identification was conducted to clarify their chromosomal composition. To enhance signal acquisition, we used Oligo-pTa535 (green), Oligo-pSc119.2 (red), and Oligo-pTa713 (yellow) as probes for identifying the three lines. Oligo-pSc119.2 effectively identified the chromosomes of the B genome, exhibiting signals on chromosomes 4A and 5A as well. Oligo-pTa535 exhibited distinctive signals at various locations across all A genomes, concurrently identifying chromosomes 1Ab, 2Ab, 3Ab, 4 Ab, 5 Ab, and 7 Ab within the Ab genome; however, the signal intensity was not particularly strong. In contrast, the probe Oligo-pTa713 clearly exhibited noticeable signals on chromosomes 3Ab and 6Ab within the Ab genome. These three probes successfully discriminated the 42 chromosomes of synthetic amphiploids (Figures 1A, B, D, E, G, H). The GISH results revealed that these three amphiploids contained 28 A genome and Ab genome chromosomes (green) (Figures 1C, F, I). In summary, these amphiploids contained 14 A, 14 B, and 14 Ab chromosomes.
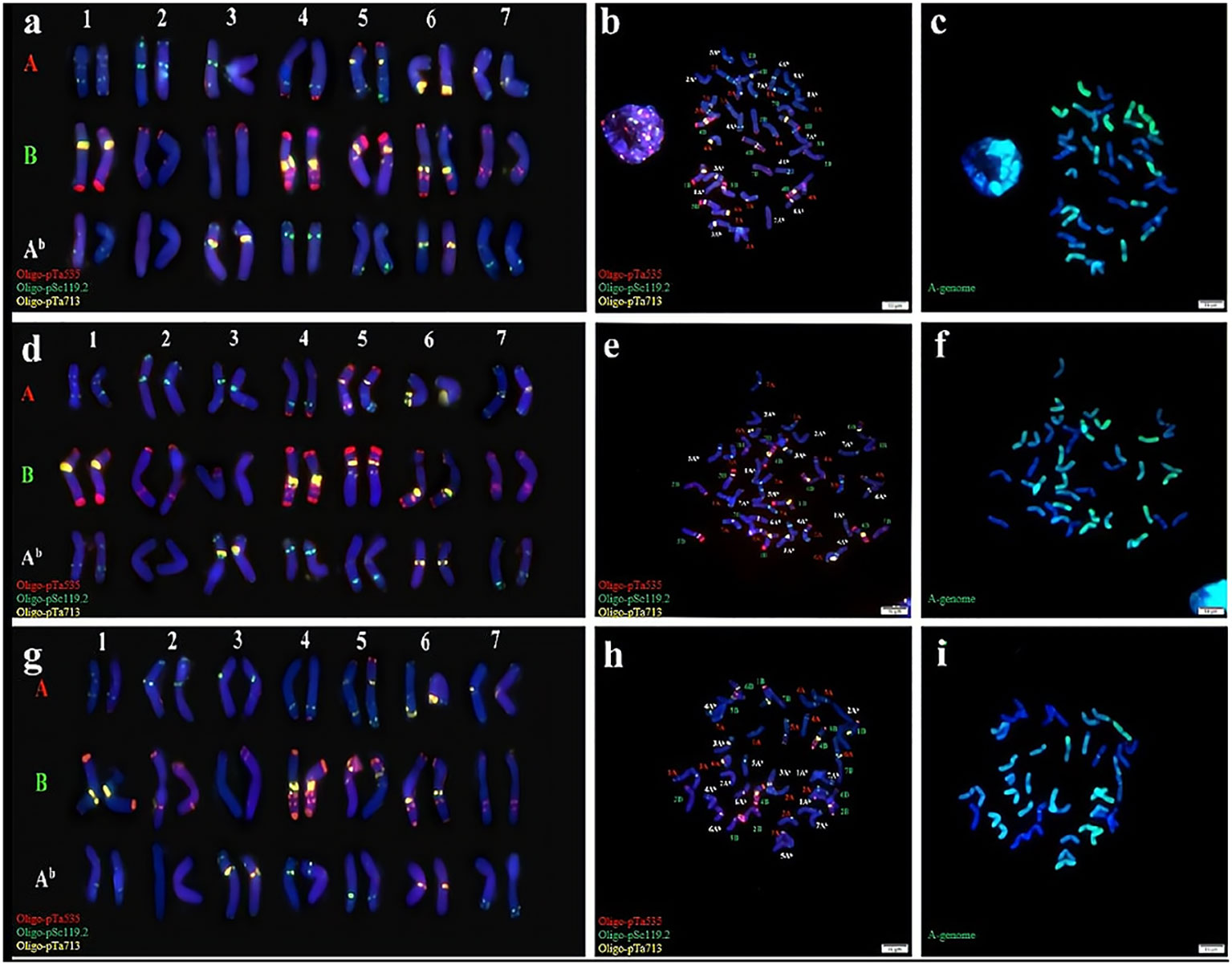
Figure 1 Cytogenetic identification of synthetic amphiploids Syn-ABAb-14, Syn-ABAb-15 and Syn-ABAb-16. FISH identification of (A, B), Syn-ABAb-14; (D, E), Syn-ABAb-15; and (G, H), Syn-ABAb-16 using the three synthetic oligonucleotide probes Oligo-pTa535 (green), Oligo-pSc119.2 (red), and Oligo-pTa713 (yellow). Panels (C, F, I) show the GISH patterns of the metaphase chromosomes of Syn-ABAb-14, Syn-ABAb-15 and Syn-ABAb-16. T. boeoticum genomic DNA (green) was used as a probe for GISH.
Blue aleurone layer 2 candidate gene PCR testing
The objective was to ascertain the presence of the candidate gene TbMYC4A from T. boeoticum within the 17 blue-grained amphiploids. For this purpose, specific primers (TbMYC4Aa1-F and TbMYC4Aa1-R) were employed in the testing process, utilizing T. turgidum (negative control) and T. boeoticum (positive control) as controls. The results revealed that the 17 blue-grained amphiploids, akin to T. boeoticum, exhibited amplification of a 310 bp band (Figure 2). In contrast, T. turgidum did not display this band, suggesting that the 17 blue-grained amphiploids harbored the candidate gene TbMYC4A from T. boeoticum.

Figure 2 Distribution of TbMYC4A in Syn-ABAb-1 to Syn-ABAb-17. Lanes 1 and 2: T. turgidum; lanes 3 and 4: T. boeoticum; lanes 5 to 21 correspond to materials from Syn-ABAb-1 to Syn-ABAb-17, respectively.
Total anthocyanin content
The grains of Syn-ABAb-1 to Syn-ABAb-17 were noticeably blue compared to those of Syn-ABAb-18, indicating the inheritance of the blue aleurone trait from T. boeoticum (Figure 3). After measurement, the TAC of the 17 blue-grained amphiploids ranged from 82.830 μg/g to 207.606 μg/g, which was significantly higher than that of Syn-ABAb-18 (yellow-grained) (6.346 μg/g) (Table 2).
Crude protein and amino acid analysis
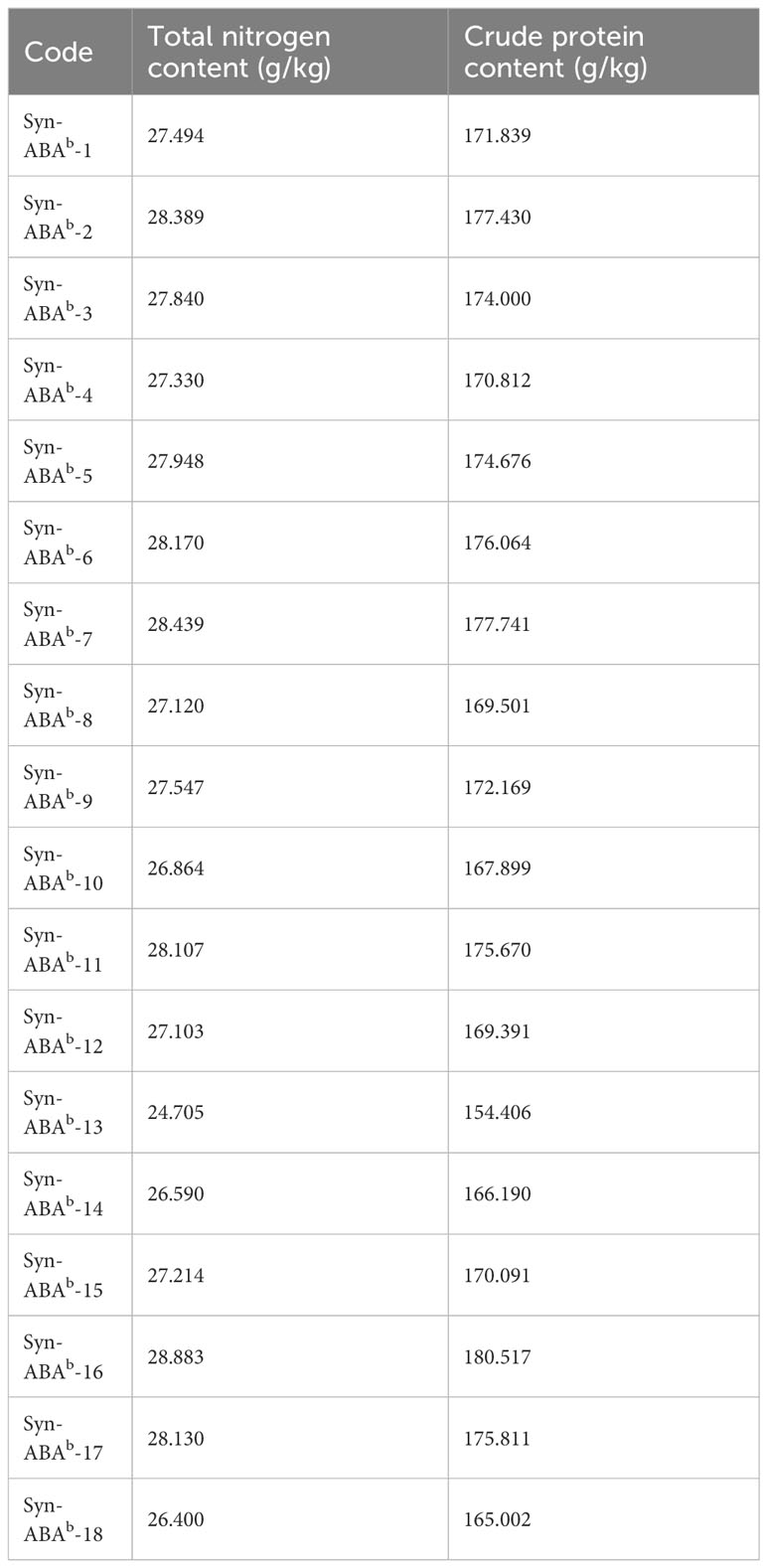
The crude protein content of the 18 amphiploids ranged from 154.406 g/kg to 180.517 g/kg (Table 3). Notably, the crude protein content of 12 of these samples surpassed 170 g/kg. Only one line exhibited a protein content lower than that of Syn-ABAb-18.
The amino acid content of the 18 amphiploids is presented in Table S1. These amphiploids exhibit similar amino acid profiles, encompassing seventeen types of amino acids, of which seven are essential amino acids (EAAs). Among these, the most abundant amino acids closely resemble those found in common wheat, specifically glutamic acid and proline. In comparison with the EAAs outlined in the reference protein pattern recommended by the FAO/WHO/UNU (2007), lysine emerged as the first limiting amino acid in the examined material, with RAA scores ranging from 0.55 to 0.88 across the 18 amphiploids. According to the theory of amino acid balance, the nutritional value of wheat increases as the SRC approaches 100. The SRC values of the 18 amphiploids ranged from 57.60 to 64.57, indicating a relatively well-balanced protein quality (Table S2).
Determination of the micronutrients
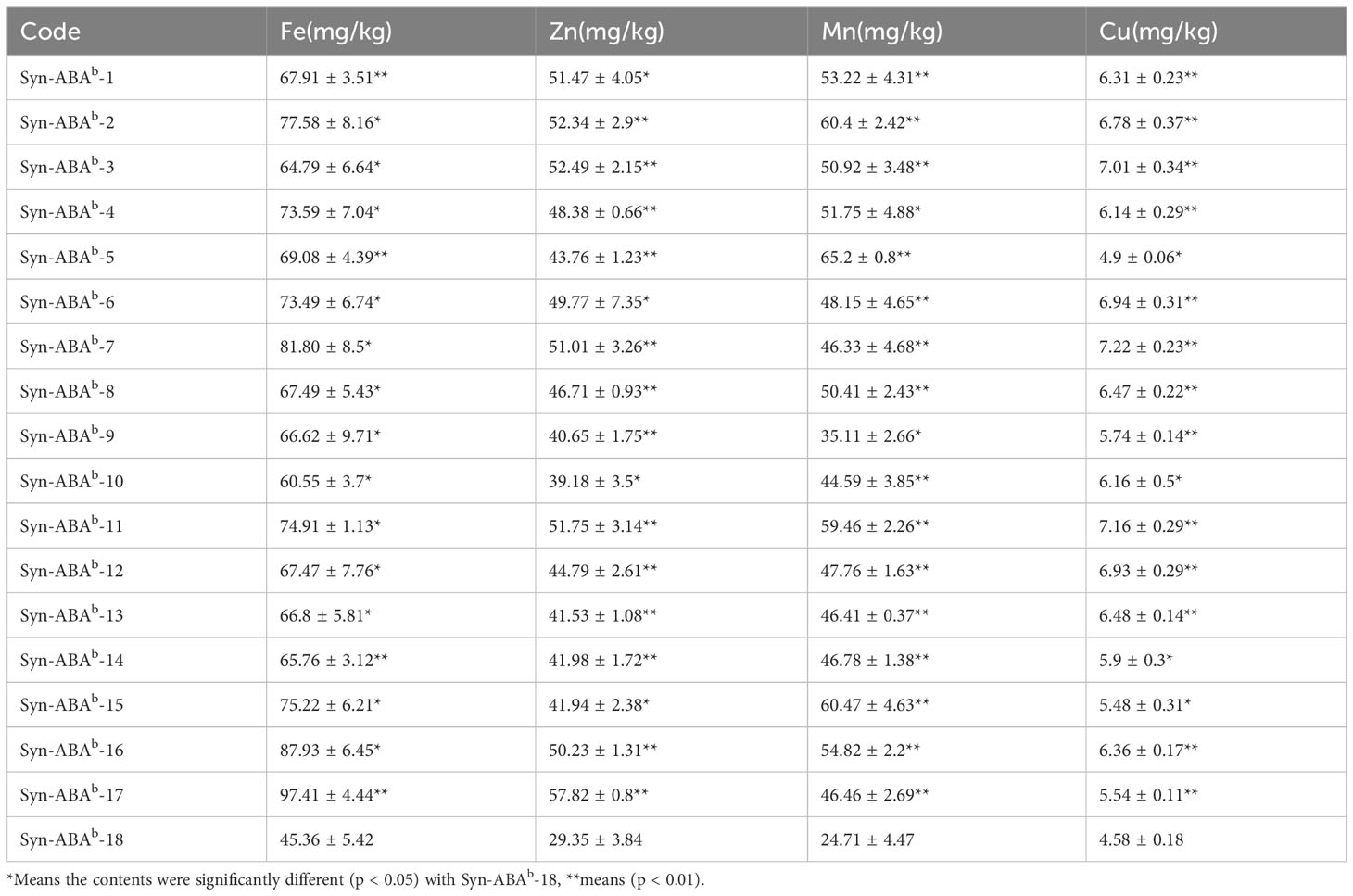
Table 4 presents the microelement content in the 18 amphiploids. Among the four elements measured, the levels in the 17 blue-grained amphiploids were higher than those in the yellow-grained amphiploid. The iron (Fe) content in the blue-grained materials ranged from 60.55 to 97.41 mg/kg, representing a 33.48-114.70% increase compared to the yellow-grained amphiploid. Regarding the zinc (Zn) content, the range was 39.18-57.82 mg/kg, reflecting an increase of 33.48-97.01%. The manganese (Mn) content, ranging from 35.11 to 65.20 mg/kg, exhibited an increase of 42.07-163.87%. Last, the copper (Cu) content ranged from 5.74 to 7.22 mg/kg, indicating an increase of 25.28-57.62%.
Principal component analysis
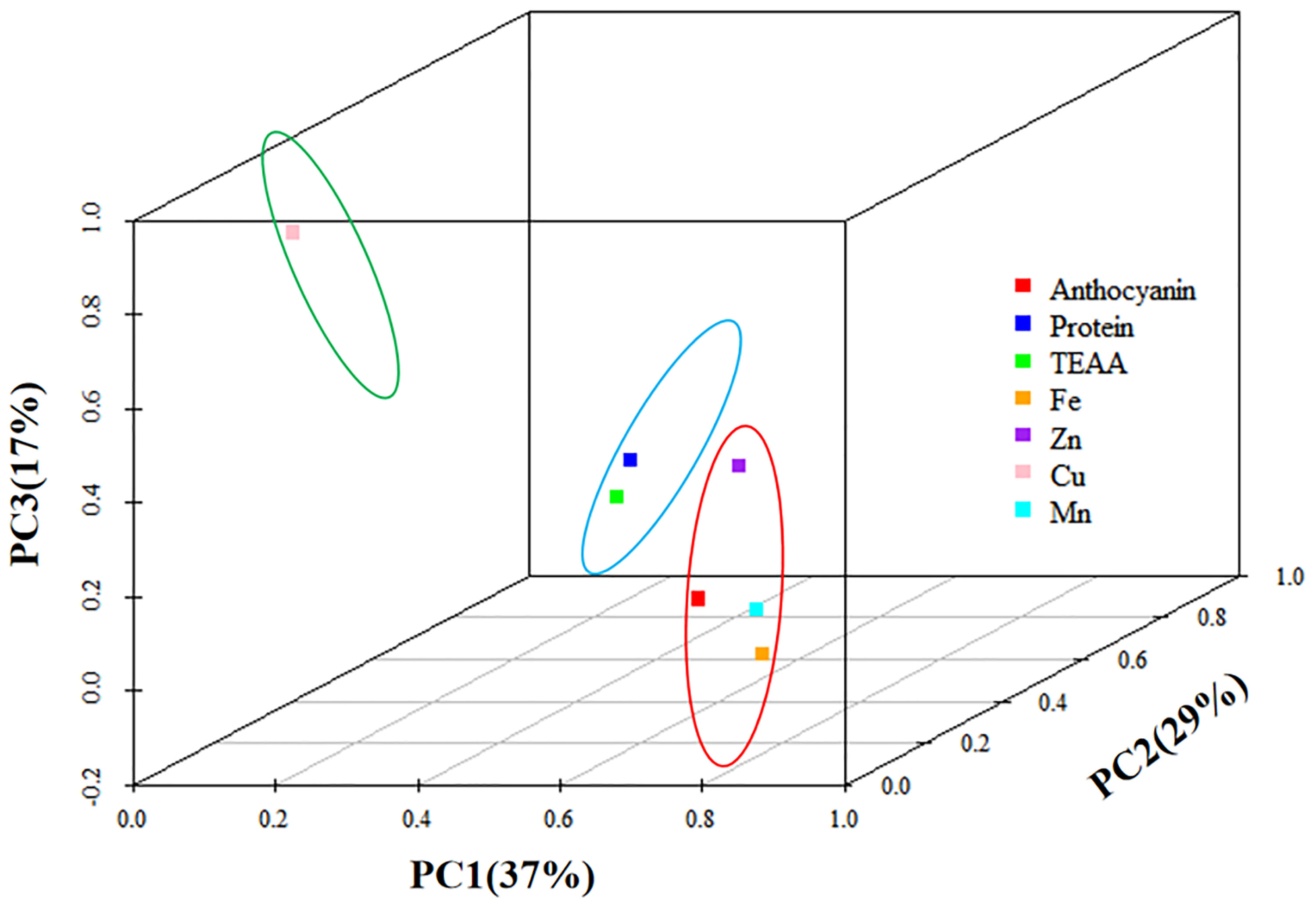
The contribution of the first three principal components reached 84% (Table S3), representing most of the index information, and could be used as a comprehensive index to evaluate the nutritional quality of the 18 materials. The maximum variance contribution method was used for rotation during factor analysis, finally obtaining the rotated eigenvalue, variance contribution rate, and cumulative variance contribution rate.
In the first principal component, anthocyanins had higher loads, along with Fe, Zn, and Mn, indicating a high correlation (Figure 4). The second component mainly contained protein and essential amino acids, while the third component contained only Cu. Table S4 shows the rotational component matrix of the factor analysis; Figure 5 presents the rotational load diagram of seven nutrient indices from the PCA. Table S5 shows the weight score of each index factor in the 18 amphiploids. The scores of the three principal components were calculated by using the standardized data of 7 different nutrient indices, and the comprehensive nutrient scores of the 18 amphiploids were calculated by using the contribution rate of rotation variance as the coefficient. The principal components (Y1, Y2, Y3) and comprehensive score (Y) were calculated according to the formulas below:
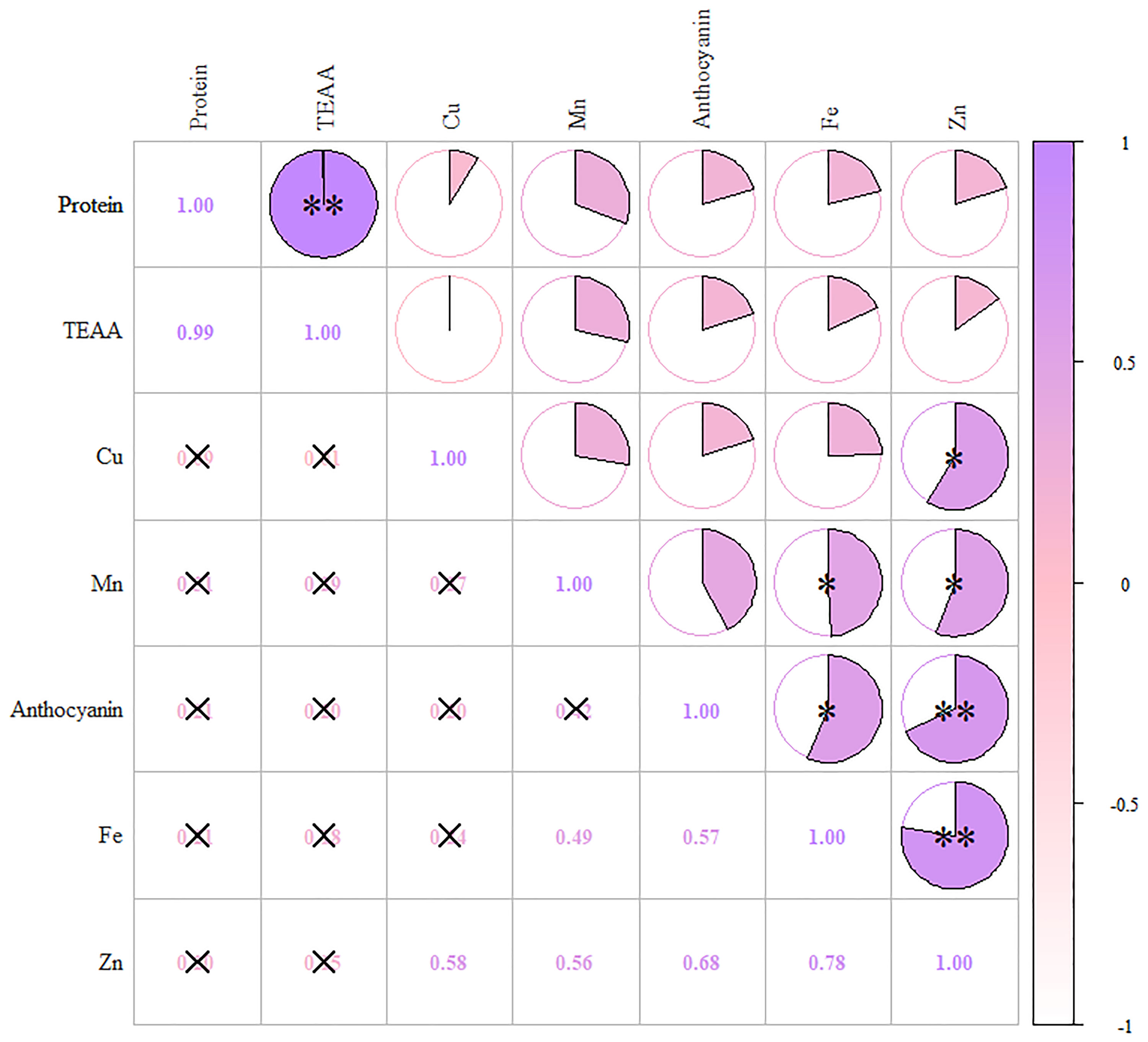
Figure 4 Correlation heatmap of anthocyanins, protein, TEAA, Fe, Zn, Mn and Cu. The color scale from white (low) to purple (high) indicates the correlation coefficient. *means significantly different at p < 0.05, **means significantly different at p < 0.01.
The comprehensive evaluation scores and rankings resulting from the factor analysis of the seven nutrient indices in the 18 amphiploids served as crucial foundations for assessing their nutritional profile. As shown in Table S6, after reviewing the comprehensive scores and rankings, the only yellow-grained amphiploid among the 18 amphiploids ranked last. Among the top five samples, Syn-ABAb-5, Syn-ABAb-8, and Syn-ABAb-11 exhibited elevated scores within the second principal component. This implies heightened levels of protein and essential amino acids, rendering them viable candidates for the cultivation of high-protein and high-amino-acid-content wheat varieties. Conversely, Syn-ABAb-4 and Syn-ABAb-17 exhibited superior scores within the first principal component. This positioning qualifies them as suitable parents for biofortification breeding, enriching wheat with anthocyanins, Fe, and Zn.
Discussion
T. boeoticum serves as a secondary gene pool for wheat; however, its integration into wheat breeding has been hindered by limited chromosomal pairing between T. boeoticum and common wheat (Feldman and Sears, 1981). Generating amphidiploids offers an effective approach to address this problem. Numerous previous studies have aimed to create amphidiploids by crossing wheat with related species from the genera Aegilops, Secale, Thinopyrum, and Triticum (Nemeth et al., 2015). These amphidiploids are sought as a source of genetic variation for enhancing wheat (Kroupin et al., 2019). In recent years, fluorescence in situ hybridization (FISH) has gained prominence as a crucial technique for cytological identification. In this study, three probes, Oligo-pTa535, Oligo-pSc119.2, and Oligo-pTa713, were used simultaneously to identify A, Ab and B genomes, and this was combined with GISH to verify the results. The probes exhibited distinct signals on the chromosomes of the three genomes. These probes can be used as cytological markers in future wheat breeding programs.
Anthocyanins have garnered growing interest among nutritionists and food scientists due to their antioxidant activity. The blue wheat variety Skorpion cultivated in the Czech Republic exhibited a total anthocyanin content of 9.26 mg/kg (Bartl et al., 2015). The anthocyanin content in the whole meal of three Canadian blue wheat species was 135.64, 168.62, and 194.36 μg/g (Francavilla and Joye, 2022). Liu et al. (2022) reported the TAC of the 4Ab (4B) disomic substitution line Z18-1244 as 67.24 μg/g. In this study, the TAC of 17 blue-grained amphidiploids was determined to be between 82.830 μg/g and 207.606 μg/g. Variations in anthocyanin content across studies are evident, likely stemming from distinctions in varieties and growth conditions, including soil, weather, and light exposure periods, all of which influence anthocyanin levels (He et al., 2010). The protein content of durum wheat is 12-16%, which is generally higher than that of common wheat (8-14%) (Marcotuli et al., 2020). The protein content of the 18 amphidiploids developed within this investigation ranged from 15.44% to 18.05%. The essential amino acid content plays a pivotal role in food quality assessment (Tian et al., 2018). The SRCs of the amphidiploids were between 57.60 and 64.57, indicating that their amino acid composition was reasonable, and the content of the first limiting amino acid, lysine (3.151-5.027 μg/g), in grains was relatively high. Approximately two billion people worldwide experience micronutrient deficiencies, notably, Fe and Zn deficiencies, and elevating the micronutrients content in grains is an effective solution for this problem (Çakmak et al., 2004). The iron (Fe) and zinc (Zn) concentrations were evaluated for 66 spring and common winter wheat genotypes derived from Central Asian breeding programs. Specifically, the iron content ranged from 25 mg/kg to 56 mg/kg (average 38 mg/kg), while the zinc content ranged between 20 mg/kg and 39 mg/kg (average 28 mg/kg) (Morgounov et al., 2007). The Fe and Zn levels in the blue-grained amphidiploids ranged from 60.55 to 97.41 mg/kg and 39.18 to 57.82 mg/kg, respectively, exceeding the average amount in Asian wheat.
Research has previously indicated that colored wheat tends to accumulate higher levels of micronutrients than white wheat. Tian et al. (2018) pointed out that colored wheat exhibited increased levels of Zn, Fe, and Mg, by 108.54%-142.68%, 8.57%-42.86%, and 5.31%-40.63%, respectively. Guo et al. (2013) observed higher levels of Fe, Zn, Mn, Cu, Se, Mg, K, and P in black wheat. This study obtained results similar to those of previous reports. Correlation analysis and PCA showed that anthocyanins had a high correlation with Fe and Zn, with high loads in the first principal component. This suggests a close association between anthocyanins and Fe and Zn in blue-grained wheat. Since the created amphidiploids came from different parents, we also constructed a nutritional evaluation system based on 7 indices to evaluate, compare and rank these amphiploids. Among the top five amphiploids, Syn-ABAb-5, Syn-ABAb-8, and Syn-ABAb-11 had high protein and amino acid levels, while Syn-ABAb-4 and Syn-ABAb-17 had high anthocyanin and micronutrients levels. Therefore, the created amphidiploids exhibited the potential to serve as parent candidates for hybridization with common wheat, guided by comprehensive score rankings, toward enhancing the nutritional quality of wheat.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository and accession number(s) can be found below: PRIDE database (https://www.ebi.ac.uk/pride/), accession number PXD04607.
Author contributions
XL: Data curation, Investigation, Methodology, Software, Writing – original draft. XJ: Investigation, Software, Methodology, Writing – review & editing. JZ: Investigation, Software, Writing – review & editing. HY: Investigation, Software, Writing – review & editing. MS: Investigation, Software, Writing – review & editing. LW: Investigation, Software, Writing – review & editing. YM: Investigation, Software, Writing – review & editing. LC: Software, Investigation, Writing – review & editing. KZ: Investigation, Software, Writing – review & editing. MH: Software, Supervision, Writing – review & editing. BJ: Investigation, Methodology, Writing – review & editing. LH: Investigation, Supervision, Writing – review & editing. SN: Investigation, Supervision, Writing – review & editing. XJC: Investigation, Data curation, Writing – review & editing. XC: Investigation, Software, Writing – review & editing. DL: Supervision, Methodology, Writing – review & editing. LZ: Conceptualization, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Sichuan Science and Technology Program (2022ZDZX0016) and the Key Research and Development Program of Sichuan Province, China (2021YFYZ0002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1285847/full#supplementary-material
References
Ahmed, S., Bux, H., Rasheed, A., Gul Kazi, A., Rauf, A., Mahmood, T., et al. (2014). Stripe rust resistance in Triticum durum–T. monococcum and T. durum–T. urartu amphiploids. Australas. Plant Pathol. 43, 109–113. doi: 10.1007/s13313-013-0237-8
Badaeva, E. D., Ruban, A. S., Zoshchuk, S. A., Surzhikov, S. A., Knüpffer, H., Kilian, B. (2016). Molecular cytogenetic characterization of Triticum timopheevii chromosomes provides new insight on genome evolution of T. zhukovskyi. Plant Syst. Evol. 302, 943–956. doi: 10.1007/s00606-016-1309-3
Bagchi, D., Sen, C. K., Bagchi, M., Atalay, M. (2004). Anti-angiogenic, antioxidant, and anti-carcinogenic properties of a novel anthocyanin-rich berry extract formula. Biochem. (Moscow). 69, 75–80. doi: 10.1023/B:BIRY.0000016355.19999.93
Bartl, P., Albreht, A., Skrt, M., Tremlová, B., Ošt’ádalová, M., Šmejkal, K., et al. (2015). Anthocyanins in purple and blue wheat grains and in resulting bread: quantity, composition, and thermal stability. Int. J. Food Sci. Nutr. 66, 514–519. doi: 10.3109/09637486.2015.1056108
Bhullar, N. K., Gruissem, W. (2013). Nutritional enhancement of rice for human health: the contribution of biotechnology. Biotechnol. Adv. 31, 50–57. doi: 10.1016/j.biotechadv.2012.02.001
Budak, H., Kantar, M., Yucebilgili Kurtoglu, K. (2013). Drought tolerance in modern and wild wheat. Sci. World J. 4, 66–75. doi: 10.1155/2013/548246
Cakmak, I. (2008). Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil. 302, 1–17. doi: 10.1007/s11104-007-9466-3
Çakmak, İ., Torun, A., Millet, E., Feldman, M., Fahima, T., Korol, A., et al. (2004). Triticum dicoccoides: an important genetic resource for increasing zinc and iron concentration in modern cultivated wheat. Soil Sci. Plant Nutr. 50 (7), 1047–1054. doi: 10.1080/00380768.2004.10408573
Cerletti, C., De Curtis, A., Bracone, F., Digesù, C., Morganti, A. G., Iacoviello, L., et al. (2017). Dietary anthocyanins and health: data from FLORA and ATHENA EU projects. Br. J. Clin. Pharmacol. 83, 103–106. doi: 10.1111/bcp.12943
Chhuneja, P., Kaur, S., Garg, T., Ghai, M., Kaur, S., Prashar, M., et al. (2008). Mapping of adult plant stripe rust resistance genes in diploid: a genome wheat species and their transfer to bread wheat. Theor. Appl. Genet. 116, 313–324. doi: 10.1007/s00122-007-0668-0
Elkot, A. F. A., Chhuneja, P., Kaur, S., Saluja, M., Keller, B., Singh, K. (2015). Marker assisted transfer of two powdery mildew resistance genes PmTb7A.1 and PmTb7A.2 from Triticum boeoticum (Boiss.) to Triticum aestivum (L.). PloS One 10, e0128297. doi: 10.1371/journal.pone.0128297
FAO/WHO/UNU (2007). “Protein and amino acid requirements in human nutrition. Report of a joint WHO/FAO/UNU expert consultation,” in WHO technical report series No. 935(Geneva, Switzerland: WHO).
Feldman, M., Sears, E. R. (1981). The wild gene resources of wheat. Sci. Am. 244, 102–113. doi: 10.1038/scientificamerican0181-102
Feng, Z., Zhang, M., Liu, X., Liang, D., Liu, X., Hao, M., et al. (2020). FISH karyotype comparison between Ab-and A-genome chromosomes using oligonucleotide probes. J. Appl. Genet. 61, 313–322. doi: 10.1007/s13353-020-00555-7
Ficco, D. B., De Simone, V., Colecchia, S. A., Pecorella, I., Platani, C., Nigro, F., et al. (2014). Genetic variability in anthocyanin composition and nutritional properties of blue, purple, and red bread (Triticum aestivum L.) and durum (Triticum turgidum L. ssp. turgidum convar. durum) wheats. J. Agric. Food Chem. 62, 8686–8695. doi: 10.1021/jf5003683
Francavilla, A., Joye, I. J. (2022). Anthocyanin content of crackers and bread made with purple and blue wheat varieties. Molecules. 27, 7180. doi: 10.3390/molecules27217180
Garg, M., Kaur, S., Sharma, A., Kumari, A., Tiwari, V., Sharma, S., et al. (2022). Rising demand for healthy foods-anthocyanin biofortified colored wheat is a new research trend. Front. Nutr. 9, 878221. doi: 10.3389/fnut.2022.878221
Gill, R. S., Dhaliwal, H. S., Multani, D. S. (1988). Synthesis and evaluation of Triticum durum—T. monococcum amphiploids. Theor. Appl. Genet. 75, 912–916. doi: 10.1007/BF00258053
Guo, Z., Zhang, Z., Xu, P., Guo, Y. (2013). Analysis of nutrient composition of purple wheat. Cereal Res. Commun. 41, 293–303. doi: 10.1556/CRC.2012.0037
Gupta, P., Balyan, H., Sharma, S., Kumar, R. (2021). Biofortification and bioavailability of Zn, Fe and Se in wheat: present status and future prospects. Theor. Appl. Genet. 134, 1–35. doi: 10.1007/s00122-020-03709-7
He, F., Mu, L., Yan, G.-L., Liang, N.-N., Pan, Q.-H., Wang, J., et al. (2010). Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules. 15, 9057–9091. doi: 10.3390/molecules15129057
Hidalgo, A., Brandolini, A. (2014). Nutritional properties of einkorn wheat (Triticum monococcum L.). J. Sci. Food Agric. 94, 601–612. doi: 10.1002/jsfa.6382
John, D. A., Babu, G. R. (2021). Lessons from the aftermaths of green revolution on food system and health. Front. Sustain Food Syst. 5, 644559. doi: 10.3389/fsufs.2021.644559
Khlestkina, E., Shoeva, O. Y., Gordeeva, E. (2015). Flavonoid biosynthesis genes in wheat. Russ. J. Genet.: Appl. Res. 5, 268–278. doi: 10.1134/S2079059715030077
Kroupin, P. Y., Divashuk, M., Karlov, G. (2019). Gene resources of perennial wild cereals involved in breeding to improve wheat crop. Selskokhoziaistvennaia Biol. 54, 409–425. doi: 10.15389/agrobiology.2019.3.409eng
Lachman, J., Martinek, P., Kotíková, Z., Orsák, M., Šulc, M. (2017). Genetics and chemistry of pigments in wheat grain–A review. J. Cereal Sci. 74, 145–154. doi: 10.1016/j.jcs.2017.02.007
Li, W., Beta, T., Sun, S., Corke, H. (2006). Protein characteristics of Chinese black-grained wheat. Food Chem. 98, 463–472. doi: 10.1016/j.foodchem.2005.06.020
Li, H. Y., Liu, X. J., Zhang, M. H., Feng, Z., Liu, D. C., Ayliffe, M., et al. (2018). Development and identification of new synthetic T. turgidum–T. monococcum amphiploids. Plant Genet. Resour. 16 (6), 555–563. doi: 10.1017/S1479262118000175
Lin, B.-W., Gong, C.-C., Song, H.-F., Cui, Y.-Y. (2017). Effects of anthocyanins on the prevention and treatment of cancer. Br. J. Pharmacol. 174, 1226–1243. doi: 10.1111/bph.13627
Liu, X., Zhang, M., Jiang, X., Li, H., Jia, Z., Hao, M., et al. (2021). TbMYC4A is a candidate gene controlling the blue aleurone trait in a wheat-Triticum boeoticum substitution line. Front. Plant Sci. 12, 762265. doi: 10.3389/fpls.2021.762265
Liu, X., Zhang, M., Wang, D., Peng, T., Liu, X., Hao, M., et al. (2022). Anthocyanin composition and nutritional properties of a blue-grained wheat-Triticum boeoticum substitution line. Cereal Res. Commun. 50, 501–507. doi: 10.1007/s42976-021-00199-1
Loskutov, I. G., Khlestkina, E. K. (2021). Wheat, barley, and oat breeding for health benefit components in grain. Plants. 10 (1), 86. doi: 10.3390/plants10010086
Marcotuli, I., Colasuonno, P., Hsieh, Y. S., Fincher, G. B., Gadaleta, A. (2020). Non-starch polysaccharides in durum wheat: a review. Int. J. Mol. Sci. 21, 2933. doi: 10.3390/ijms21082933
Megyeri, M., Mikó, P., Molnár, I., Kovács, G. (2011). Development of synthetic amphiploids based of Triticum turgidum × T. monococcum crosses to improve the adaptability of cereals. Acta Agron. Hung. 59, 267–274. doi: 10.1556/AAgr.59.2011.3.11
Morgounov, A., Gómez-Becerra, H. F., Abugalieva, A., Dzhunusova, M., Yessimbekova, M., Muminjanov, H., et al. (2007). Iron and zinc grain density in common wheat grown in Central Asia. Euphytica. 155, 193–203. doi: 10.1007/s10681-006-9321-2
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Nemeth, C., Yang, C., Kasprzak, P., Hubbart, S., Scholefield, D., Mehra, S., et al. (2015). Generation of amphidiploids from hybrids of wheat and related species from the genera Aegilops, Secale, Thinopyrum, and Triticum as a source of genetic variation for wheat improvement. Genome. 58, 71–79. doi: 10.1139/gen-2015-0002
Paull, J. G., Pallotta, M. A., Langridge, P., The, T. T. (1994). RFLP markers associated with Sr22 and recombination between chromosome 7A of bread wheat and the diploid species Triticum boeoticum. Theor. Appl. Genet. 89, 1039–1045. doi: 10.1007/BF00224536
Revelle, W., Revelle, M. W. (2015). Package ‘psych’. Compr. R Arch. Netw. 337, 338. Available at: https://personality-project.org/r/psych-manual.pdf.
Salo-väänänen, P. P., Koivistoinen, P. E. (1996). Determination of protein in foods: comparison of net protein and crude protein (N × 6.25) values. Food Chem. 57, 27–31. doi: 10.1016/0308-8146(96)00157-4
Shi, A. N., Leath, S., Murphy, J. P. (1998). A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology. 88, 144–147. doi: 10.1094/PHYTO.1998.88.2.144
Shipp, J., Abdel-Aal, E. S. M. (2010). Food applications and physiological effects of anthocyanins as functional food ingredients. Open Food Sci. J. 4, 7–22. doi: 10.2174/1874256401004010007
Tang, Z., Yang, Z., Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119. 2, pTa-535, pTa71, CCS1, and pAWRC. 1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Tian, S., Chen, Z., Wei, Y. (2018). Measurement of colour-grained wheat nutrient compounds and the application of combination technology in dough. J. Cereal Sci. 83, 63–67. doi: 10.1016/j.jcs.2018.07.018
Tounsi, S., Amar, S. B., Masmoudi, H., Sentenac, H., Brini, F., Very, A.-A. (2016). Characterization of two HKT1;4 transporters from Triticum monococcum to elucidate the determinants of the wheat salt tolerance Nax1 QTL. Plant Cell Physiol. 57, 2047–2057. doi: 10.1093/pcp/pcw123
Wallace, T. C., Giusti, M. M. (2015). Anthocyanins. Adv. Nutr. 6, 620–622. doi: 10.3945/an.115.009233
Wang, H., Yang, J., Zhang, M., Fan, W., Firon, N., Pattanaik, S., et al. (2016). Altered phenylpropanoid metabolism in the maize Lc-expressed sweet potato (Ipomoea batatas) affects storage root development. Sci. Rep. 6 (1), 18645. doi: 10.1038/srep18645
Waterval, W. A. H., Scheijen, J. L. J. M., Ortmans-Ploemen, M. M. J. C., Habets-van Der Poel, C. D., Bierau, J. (2009). Quantitative UPLC-MS/MS analysis of underivatised amino acids in body fluids is a reliable tool for the diagnosis and follow-up of patients with inborn errors of metabolism. Clin. Chim. Acta 407, 36–42. doi: 10.1016/j.cca.2009.06.023
Wickham, H. (2011). ggplot2. Wiley Interdiscip. Rev. Comput. Statist. 3, 180–185. doi: 10.1002/wics.147
Zeven, A. (1991). Wheats with purple and blue grains: a review. Euphytica 56, 243–258. doi: 10.1007/BF00042371
Keywords: Triticum boeoticum, synthetic amphiploid, blue aleurone, anthocyanins, amino acid, micronutrients, PCA
Citation: Liu X, Jiang X, Zhang J, Ye H, Shen M, Wu L, Miao Y, Chen L, Zhou K, Hao M, Jiang B, Huang L, Ning S, Chen X, Chen X, Liu D and Zhang L (2023) Molecular cytogenetic identification and nutritional composition evaluation of newly synthesized Triticum turgidum-Triticum boeoticum amphiploids (AABBAbAb). Front. Plant Sci. 14:1285847. doi: 10.3389/fpls.2023.1285847
Received: 30 August 2023; Accepted: 22 November 2023;
Published: 07 December 2023.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Hakan Ozkan, Çukurova University, TürkiyePenko Spetsov, Agricultural Academy (CAB), Bulgaria
Copyright © 2023 Liu, Jiang, Zhang, Ye, Shen, Wu, Miao, Chen, Zhou, Hao, Jiang, Huang, Ning, Chen, Chen, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianquan Zhang, emhhbmdsaWFucXVhbjE5NzdAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
‡ORCID: Lianquan Zhang, orcid.org/0000-0001-9005-8705
 Xin Liu1,2†
Xin Liu1,2† Yongping Miao
Yongping Miao Ming Hao
Ming Hao Bo Jiang
Bo Jiang Lin Huang
Lin Huang Shunzong Ning
Shunzong Ning Dengcai Liu
Dengcai Liu Lianquan Zhang
Lianquan Zhang