
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 December 2023
Sec. Plant Systematics and Evolution
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1283292
Introduction: During plant evolution, intracellular DNA transfer (IDT) occurs not only from organelles to the nucleus but also between organelles. To further comprehend these events, both organelle genomes and transcriptomes are needed.
Methods: In this study, we constructed organelle genomes and transcriptomes for two Dystaenia species and described their dynamic IDTs between their nuclear and mitochondrial genomes, or plastid and mitochondrial genomes (plastome and mitogenome).
Results and Discussion: We identified the putative functional transfers of the mitochondrial genes 5′ rpl2, rps10, rps14, rps19, and sdh3 to the nucleus in both Dystaenia species and detected two transcripts for the rpl2 and sdh3 genes. Additional transcriptomes from the Apicaceae species also provided evidence for the transfers and duplications of these mitochondrial genes, showing lineage-specific patterns. Intrageneric variations of the IDT were found between the Dystaenia organelle genomes. Recurrent plastid-to-mitochondrion DNA transfer events were only identified in the D. takeshimana mitogenome, and a pair of mitochondrial DNAs of plastid origin (MIPTs) may generate minor alternative isoforms. We only found a mitochondrion-to-plastid DNA transfer event in the D. ibukiensis plastome. This event may be linked to inverted repeat boundary shifts in its plastome. We inferred that the insertion region involved an MIPT that had already acquired a plastid sequence in its mitogenome via IDT. We propose that the MIPT acts as a homologous region pairing between the donor and recipient sequences. Our results provide insight into the evolution of organelle genomes across the family Apiaceae.
Dystaenia Kitag. is a genus of perennial herbs in the family Apiaceae (also known as Umbelliferae), which is endemic to Japan and Korea. It has two species: D. ibukiensis (Y.Yabe) Kitag. and D. takeshimana (Nakai) Kitag. (Kitagawa, 1937). This genus has attracted the attention of biologists because of its evolutionary patterns and processes in oceanic islands (Pfosser et al., 2005). This is because D. takeshimana is native to Ulleungdo, an oceanic island in Korea, and likely originated via anagenetic speciation from the Japanese species D. ibukiensis (Pfosser et al., 2005). Oceanic islands exhibit two modes of speciation: anagenesis and cladogenesis (Takayama et al., 2015). Among the oceanic islands, Ulleungdo has a higher level of endemism derived via anagenetic (88%) than cladogenetic speciation (Stuessy et al., 2006). Molecular phylogenetic studies based on nuclear internal transcribed spacer (ITS) and trnL-F regions have shown that Dystaenia is a monophyletic group, but its sister group remains unknown (Choi et al., 1998; Pfosser et al., 2005). Multilocus datasets from three genomic sequences are required to better understand the evolutionary history of Dystaenia.
Recently, next-generation sequencing platforms have generated deep coverage enabling the assembly of plastid and mitochondrial genomes. Comparative genomics of organelles helps shed new light on evolutionary events across the tree of life and provides valuable sources for phylogenetic studies. The plastid genome (plastome) of angiosperms generally has a conserved quadripartite structure with a pair of inverted repeat (IR), large single-copy (LSC), and small single-copy (SSC) regions (Ruhlman and Jansen, 2021). Angiosperm plastomes range from 120 kb to 170 kb in length and contain 79 proteins, 30 transfer RNA (tRNA), and four ribosomal RNA (rRNA) genes. In contrast, the mitochondrial genome (mitogenome) of angiosperms exhibits a dynamic structure with circular, linear, and branched molecules (Sloan, 2013). Angiosperm mitogenomes range from 222 kb to 983 kb in length and contain 41 proteins, 14 tRNA, and three rRNA genes (Mower et al., 2012). Furthermore, organellar phylogenomic analysis based on genome-scale data provides new insights into the origins of species that have undergone complex evolutionary histories with incomplete lineage sorting and hybridization (Liu et al., 2019; Park and Park, 2020; Wu et al., 2022).
The family Apiaceae comprises approximately 3,575–3,820 species in 442–466 genera with a cosmopolitan distribution (Christenhusz and Byng, 2016; Plunkett et al., 2018). This family includes many economically important medicinal species and exhibits extensive morphological diversity (Kljuykov et al., 2019; Wang et al., 2022). The family Apiaceae is classified into four subfamilies: Apioideae, Azorelloideae, Mackinlayoideae, and Saniculoideae. The Apioideae is the largest subfamily of Apiaceae, which contains approximately 84% of species and 85% of genera (Wen et al., 2021). Phylogenetic analyses based on nuclear ribosomal and plastid DNA sequences have identified 21 tribes and 20 clades (Downie et al., 2010). Plastid phylogenomic analysis has revealed a well-resolved relationship within the Apioideae subfamily (Wen et al., 2021). However, previous studies have shown incongruence between topologies based on chloroplast and nuclear sequence datasets (Zhou et al., 2009; Downie et al., 2010). Chloroplast capture resulting from hybridization or introgression has been suggested as a possible reason for phylogenetic incongruence (Rieseberg and Soltis, 1991; Stegemann et al., 2012). Whole-genome duplication and hybridization are involved in the evolutionary history of this family (Ren et al., 2018; Jia et al., 2023). Numerous nuclear single-copy genes from transcriptome data have been utilized to conduct a phylogenetic analysis of Apioideae, showing that this family has undergone a rapid evolutionary divergence and incomplete lineage sorting (Wen et al., 2020). To date, the complete plastomes of 439 species from 83 genera have been sequenced [National Center for Biotechnology Information (NCBI), accessed 26 June 2023], 421 species from the subfamily Apioideae, 16 species from the subfamily Saniculoideae, one species from the subfamily Azorelloideae, and one species from the subfamily Mackinlayoideae. The published plastomes of Apiaceae range from 141 kb to 179 kb and contain a full set of protein-coding genes. The plastomes have a conserved quadripartite structure, but the IR boundaries have shifted during Apiaceae genome evolution (Wang et al., 2021a; Wen et al., 2021; Yuan et al., 2021; Samigullin et al., 2022). The most notable feature among Apiaceae plastomes is the plastid DNA of mitochondrial origin (PLMT). Mitochondrion-to-plastid DNA transfers have been reported in five lineages of this family: Crithmum and Petroselinum (Downie and Jansen, 2014; Downie and Jansen, 2015) and Caucalis, Cuminum, and Daucus (Iorizzo et al., 2012; Spooner et al., 2017). In contrast to plastomes, there are thus far only five species from the subfamily Apioideae with sequenced mitogenomes: Apium graveolens (Cheng et al., 2021), Bupleurum chinense (Qiao et al., 2022), Coriandrum sativum (Wang et al., 2021b), Daucus carota (Iorizzo et al., 2012), and Foeniculum vulgare (Palumbo et al., 2020). These mitogenomes range from 281 kb to 435 kb in size, with circular molecules. Losses of six mitochondria-encoded genes (rpl2, rps10, rps14, rps19, sdh3, and sdh4) were observed in all five mitogenomes.
As part of our ongoing research on the evolution of the genus Dystaenia, we determined the plastid and mitochondrial genomes of two species: D. ibukiensis and D. takeshimana. A comparison of the two-organelle genomes revealed intrageneric variations in size, structure, and gene content in Dystaenia. We demonstrated the functional replacement of rpl2, rps10, rps14, rps19, and sdh3 from the mitochondria to the nucleus and also compared intracellular DNA transfer (IDT) between the plastid and mitochondrial genomes. Interestingly, we identified a mitochondrion-to-plastid DNA transfer event in the D. ibukiensis plastome and discussed the evolutionary history of PLMT. Our results provide new insights into the evolution of Apiaceae organelle genomes, including intercompartmental transfers.
Fresh leaves of D. takeshimana and D. ibukiensis were collected from Ulleungdo, Geongbuk, South Korea, and Mt. Ibuki, Shiga, Japan, respectively. Total genomic DNAs were isolated using the Exgene Plant SV Mini Kit (GenAll, Seoul, South Korea), following the manufacturer’s protocol. Total RNA was isolated, as described by Breitler et al. (2016). After gel electrophoresis and qualitative assessment, DNA and RNA samples were sequenced using the Illumina HiSeq2500 platform (Illumina, San Diego, CA, USA). Approximately 43 Gb and 20 Gb of 150-bp paired-end (PE) reads were generated from 550-bp insert libraries from the DNAs of D. takeshimana and D. ibukiensis, respectively. The DNAs from D. takeshimana and D. ibukiensis were used for long reads generated from four and two flow cells on the Oxford Nanopore Technologies (ONT) GridION platform (ONT, Oxford, UK), and 68 Gb and 12 Gb of ONT reads were produced, respectively. RNAs from D. takeshimana and D. ibukiensis were sequenced by the Illumina platform, generating 12 Gb and 8 Gb of 150-bp PE reads, respectively.
Multiple assemblies for both species were generated using Canu v2.2 (Koren et al., 2017), MaSuRCA v4.0.5 (Zimin et al., 2017), SPAdes v3.15.3 (Antipov et al., 2016), and Velvet v1.2.10 (Zerbino and Birney, 2008) based on long- and short-read data. For example, single-type platform assemblies have been created using Canu for ONT reads and Velvet for Illumina reads. The default parameters were used for Canu. We used pairwise combinations of k-mers (99–145) and expected coverage values (50, 100, 150, 200, 500, and 1,000) without the scaffolding option for Velvet assemblies. Hybrid assemblies were generated using MaSuRCA and SPAdes, combining Illumina and ONT reads. The default parameters were used for MaSuRCA. For the SPAde assemblies, independent runs were executed with multiple coverage cutoffs (10, 50, 100, 200, and 500) using the “careful” option. All de novo assemblies were performed on a 64-core Linux workstation with 2,048 GB of memory. Plastid and mitochondrial contigs were identified using a BLAST-like algorithm in Geneious Prime 2022.2 (www.geneious.com) with Liriodendron tulipifera plastome and mitogenome sequences (NC_008326 and NC_021152) as queries. The identified organellar contigs were manually aligned, and a consensus genome sequence was generated for each by tracking and end-inspecting the organellar contigs. The coverage depth of the whole plastome and mitogenome sequences was evaluated by mapping Illumina PE and ONT reads using Bowtie v2.4.2 (Langmead and Salzberg, 2012) and BWA v0.7.17 (Li, 2013). To predict all tRNA genes in the organelle genomes, ARAGORN v1.2.38 (Laslett and Canback, 2004) and tRNAscan-SE v2.0.9 (Chan et al., 2021) were used. Circular or linear organellar genomes were generated using OGDRAW v1.3.1 (Greiner et al., 2019). The newly sequenced genomes were deposited in GenBank with accession numbers OR231235-OR231238.
Rcorrector v1.0.4 (Song and Florea, 2015) was used to correct sequencing errors in raw reads from D. takeshimana and D. ibukiensis RNA sequencing (RNA-seq). Two Dystaenia transcriptomes were assembled de novo using Trinity v2.13.2 (Grabherr et al., 2011) with the “trimmomatic” option. The completeness of the assemblies was examined using Benchmarking Universal Single-Copy Orthologs (BUSCO) v5.2.2 (Manni et al., 2021) with the lineage “eudicots_odb10” (2,326 orthologs; 2019-11-20). RNA-seq data were obtained from the NCBI Sequence Read Archive for Apium graveolens (SRR1023730), Bupleurum chinense (SRR8863755), Coriandrum sativum (SRR8863732), and Saposhnikovia divaricate (SRR8863754); four additional transcriptomes were assembled as described above.
Repetitive sequences in Dystaenia organelle genomes were identified using ROUSFinder.py (Wynn and Christensen, 2019). The two plastomes and two mitogenomes were aligned using the “progressive Mauve algorithm in Mauve v2.3.182 (Darling et al., 2004) in Geneious Prime. Organellar protein-coding genes were collected from five species, each with available genomes. Individual gene alignments were generated using the “Translation Align” approach with MAFFT v7.49 in Geneious Prime. A maximum likelihood (ML) tree was constructed from a concatenated alignment of the 24-gene dataset using IQ-TREE2 v2.2.03, with a best-fit model (-m TEST) and 1,000 ultra-fast bootstrap replicates (-B 1000).
To investigate PLMTs and mitochondrial DNAs of plastid origin (MIPTs), we performed a reciprocal “BLASTN” searches between the plastid genome and its mitochondrial counterpart with an e-value cutoff of 1 × 10−6, requiring at least 80% sequence identity and a minimum length of 50 bp. In addition, the CENSOR web server (Kohany et al., 2006) was used to search the mitochondrial genomes for putative nuclear transposable elements (TEs) with default parameters and “green plants” as a reference sequence source. To identify functional intracellular gene transfer (IGT) to the nucleus, candidate transcripts were verified by BLASTN searches for organellar genes against their transcriptomes. The conserved domain of the predicted open reading frame (ORF) was annotated by CD searches against the Conserved Domain Database (CDD) v3.19 (Lu et al., 2019). LOCALIZER v1.0.4 (Sperschneider et al., 2017) and TargetP v2.0 (Almagro Armenteros et al., 2019) were used to predict the presence of N-terminal presequences [chloroplast transit peptide (cTP) and mitochondrial targeting peptide (mTP)] and their potential cleavage sites. The transcript sequences were used as queries in “BLASTN” against the de novo genome sequence of two Dystaenia species. The sequenced genes were deposited in GenBank (Supplementary Table 1). Phylogenetic trees were constructed using ML methods as described above.
The plastomes of D. takeshimana (147,706 bp) and D. ibukiensis (153,487 bp) were assembled into a typical quadripartite structure, with the LSC and SSC separated by two IRs (Figure 1). Of the two species, D. takeshimana had the largest LSC (93,013 bp) and the smallest IR (18,568 bp). The average coverage of D. takeshimana and D. ibukiensis plastomes was 3,994× and 771× for Illumina and 5,450× and 420× for ONT, respectively (Table 1, Supplementary Figure 1). The D. takeshimana plastome showed syntenic conservation with the other angiosperms, whereas the D. ibukiensis plastome showed inversion and relocation (Figure 1). Mauve alignment of the two Dysteania species revealed three locally collinear blocks with two breakpoints (Supplementary Figure 2A). Both plastomes contained the same set of genes encoding 79 proteins, 30 tRNAs, and four rRNAs (Figure 1, Table 1). The average guanine-cytosine (GC) content in the D. takeshimana and D. ibukiensis plastomes was 37.5% and 37.7%, respectively, and contained six and seven small repeat pairs (<100 bp), covering 0.1% and 0.3% of the D. takeshimana and D. ibukiensis plastomes, respectively (Supplementary Table 2).
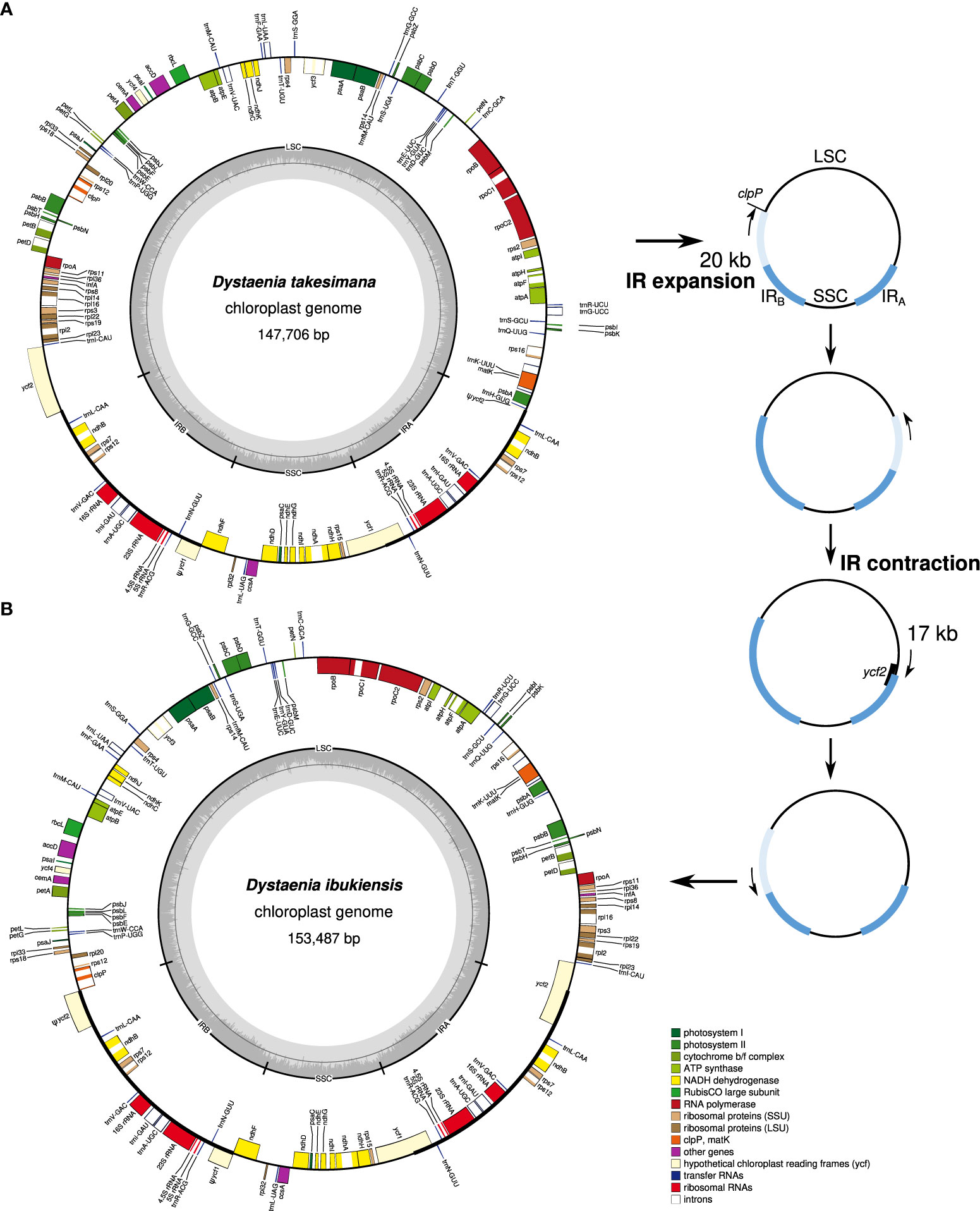
Figure 1 Plastid genome maps of Dystaenia. (A) D. takeshimana. (B) D. ibukiensis. Genes on the inside and outside of the map are transcribed in clockwise and counterclockwise directions, respectively. The hypothetical model of plastome rearrangement by IR expansion and contraction in the genus Dystaenia (right).
The D. takeshimana mitogenome (282,211 bp) was assembled into a single circular molecule (Figure 2A). In contrast, the D. ibukiensis mitogenome (281,432 bp) was assembled into two circular molecules (235,140 bp and 37,165 bp) with a linear molecule (9,127 bp) connected to the two circles (Figure 2B). The average coverage of the D. takeshimana and D. ibukiensis mitogenomes was 335× and 313× for Illumina and 475× and 186× for ONT, respectively (Table 1, Supplementary Figure 1). Comparative analysis revealed significant structural variation between the two mitogenomes (Supplementary Figure 2B), although 91% of the D. takeshimana mitogenome was homologous to the D. ibukiensis mitogenome. However, two mitogenomes shared a loss of synteny, in which eight of the 14 ancestral gene clusters were missing (Supplementary Figure 2C). Both mitogenomes contained the same set of genes encoding 35 proteins, 18 tRNAs (including five plastid-derived tRNAs), and three rRNAs (Figure 2, Table 1). Both species lack four genes, 5′ ribosomal protein subunits L2 (rpl2), S10 (rps10), S19 (rps19), and succinate dehydrogenase 3 (sdh3), in their mitogenomes. In addition, both mitogenomes had a truncated sdh4 gene at the C-terminus, with only a portion of the conserved domain. The ribosomal protein subunit L5 (rpl5) gene was truncated at the N-terminus in only D. ibukiensis mitogenome. The average GC contents from D. takeshimana and D. ibukiensis mitogenomes were 44.6% and 44.3%, respectively. Two mitogenomes contained 79 and 55 repeat pairs, covering 33.5% and 25.3% of the D. takeshimana and D. ibukiensis mitogenomes, respectively (Figure 3, Supplementary Table 3). The D. takeshimana mitogenome contained 20 large (> 1,000 bp) repeats, ranging from 1,034 bp to 5,706 bp, and the D. ibukiensis mitogenome contained 13 large repeats, ranging from 2,140 bp to 6,502 bp (Supplementary Table 3).
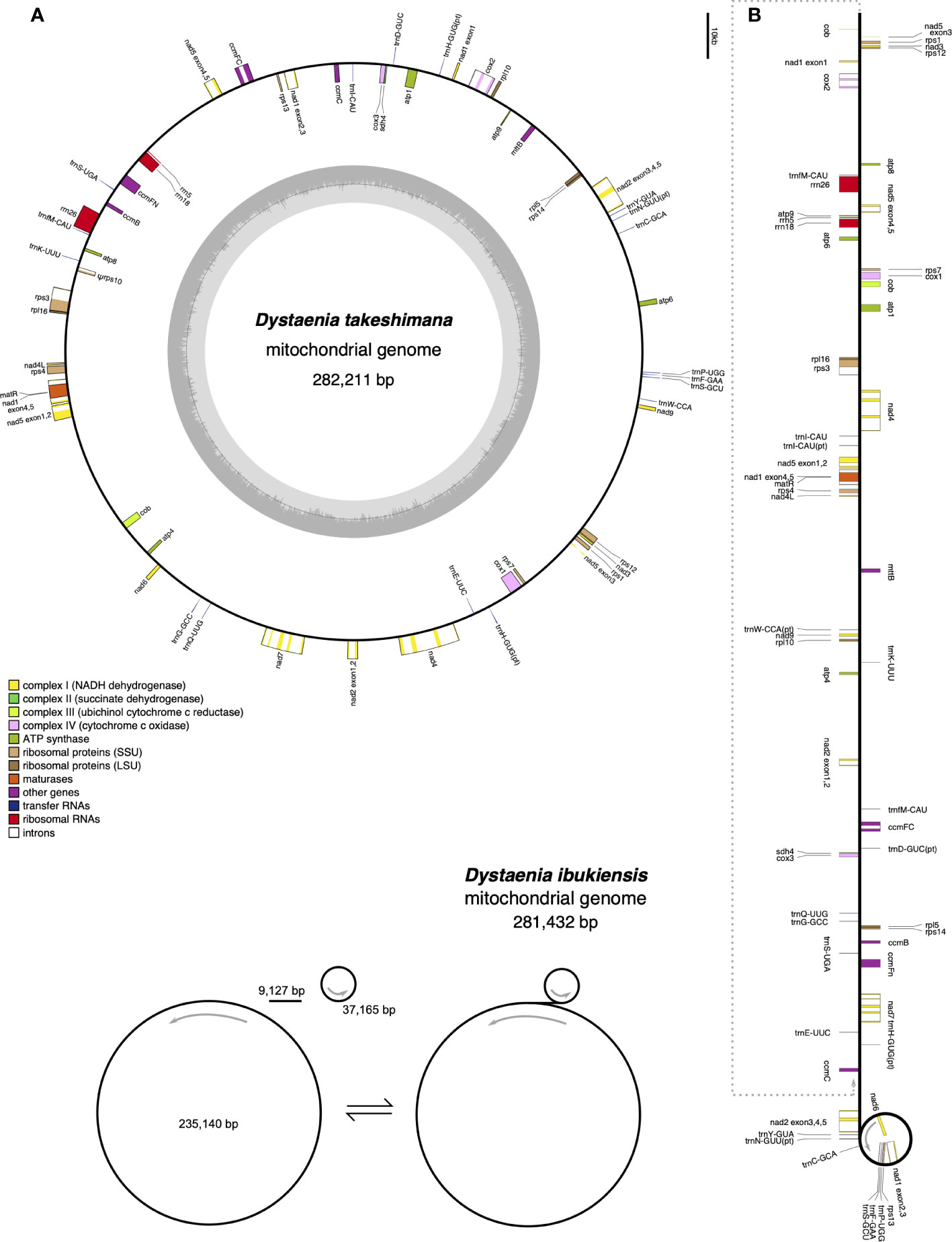
Figure 2 Mitochondrial genome maps of Dystaenia. (A) D. takeshimana. (B) D. ibukiensis. Genes on the inside and outside of the map are transcribed in clockwise and counterclockwise directions, respectively.
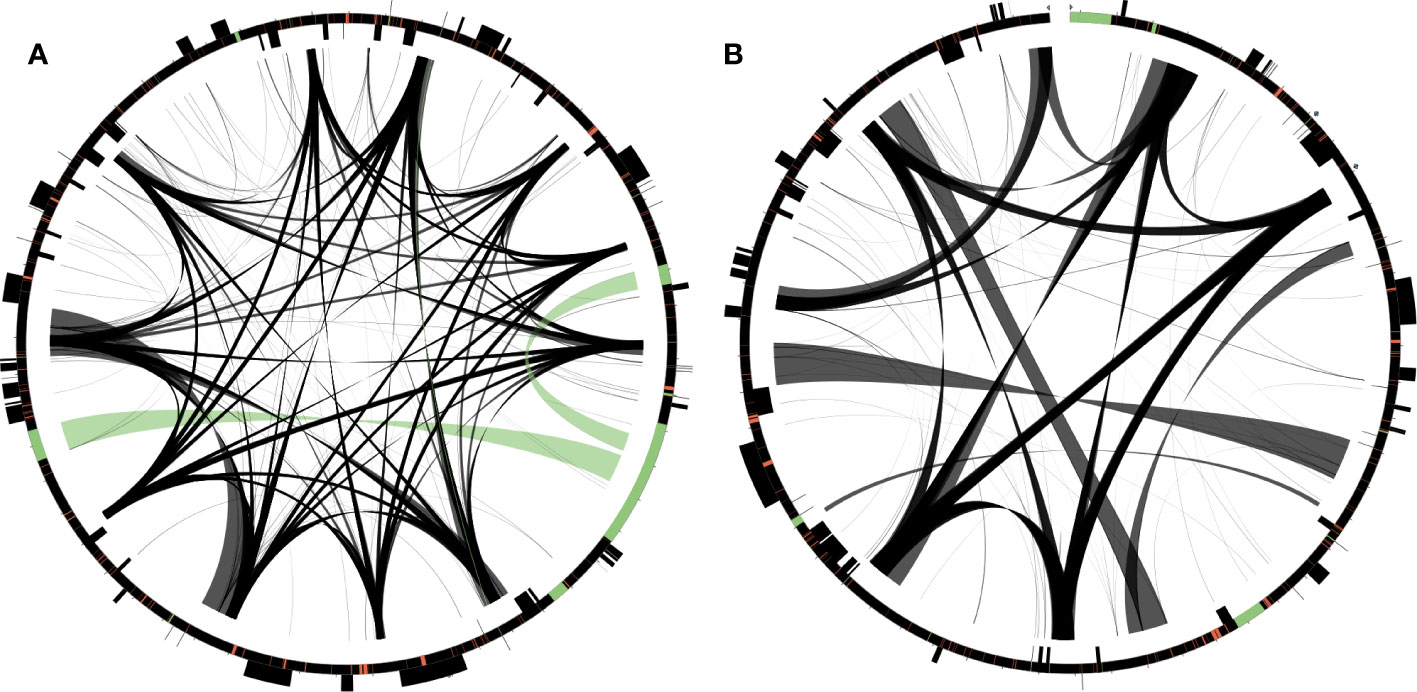
Figure 3 Distribution of repetitive DNA in two Dystaenia mitogenomes. (A) D. takeshimana. (B) D. ibukiensis. Black lines within circular maps indicate the positions of the pairs of repeats, with crossed connecting lines denoting reverse repeats. Black boxes on the inner and outer circle indicate the positions of mitochondrial genes. Green and red lines inside of the circular maps indicate mitochondrial DNAs of plastid origin and transposable elements, respectively.
Although losses of the mitochondrial-encoded 5′ rpl2, rps10, rps19, and sdh3 genes were only identified in the Dystaenia organelle genomes, we performed BLASTN searches using 41 mitochondrial and 79 plastid gene sequences against each transcriptome dataset. BUSCO assessment of the transcriptomes resulted in 83.6% (D. takeshimana) and 62.1% (D. ibukiensis) (Supplementary Figure 3). We identified multiple transcripts with high sequence identities to the four genes containing the targeting sequences and conserved domain (Table 2, Supplementary Figure 4). Moreover, we identified a nuclear transcript for mitochondrial rps14, which is present in the mitogenome (Table 2, Supplementary Figure 4). In particular, two nuclear transcripts for the mitochondrial 5′ rpl2 and sdh3 were identified in both Dystaenia transcriptomes (Table 2, Supplementary Figure 4). To test the potential gene split transfer of truncated sdh4, we performed BLASTN searches using Amborella mitochondrial sdh4 as the query. One and two transcripts were identified in the D. takeshimana and D. ibukiensis transcriptome, respectively (Supplementary Figure 4). These transcripts contained a portion of the conserved peptidylprolyl isomerase (PPIase) domain upstream of the sdh4 domain. However, TargetP failed to predict the mitochondrial target sequences. No nuclear transcript was identified for the truncated mitochondrial rpl5 gene from the D. ibukienesis mitogenome.
Examining the draft D. takeshimana de novo genome assembly identified the generic structure of the nuclear genes (Figure 4A). This analysis revealed several exons and introns in the five nuclear genes. The nuclear-encoded RPS14 is intronless, whereas the 5′ RPL2, RPS10, and RPS19 contain two exons, whereas SDH3 contains four exons (Figure 4A). Variations in the intron length of each 5′ RPL2 and SDH3 gene were identified (Figure 4A). In the case of the nuclear-encoded RPS14, we found extra copies with 70.9% and 65.3% nucleotide sequence identity on the other D. takeshimana and D. ibukiensis nuclear genome scaffolds, respectively. The D. takeshimana nuclear genome contains an intact ORF; the D. ibukiensis nuclear genome has multiple internal stop codons. We constructed a phylogenetic tree using 24 mitochondrial genes from seven available Apiaceae mitogenomes (Figure 4B). The phylogenetic distribution of gene loss showed that all analyzed species shared the loss of mitochondrial rpl2, rps19, and sdh3 genes. The loss of sdh4 occurred independently in D. carota subsp. sativus and B. chinense, and loss of rps10 occurred independently in D. carota subsp. sativus and in the two Dystaenia species/S. divaricate clade. The loss of rps14 occurred independently in A. graveolens and D. carota subsp. sativus. The truncation of sdh4 occurred in the common ancestor of A. graveolens, C. sativum, the two Dystaenia species, and S. divaricate. The truncation of rpl5 is unique to the D. ibukiensis mitogenome. Comparison of the five high-quality transcriptomes of A. graveolens, B. chinense, C. sativum, D. carota subsp. sativus, and S. divaricate (Supplementary Figure 3) provided additional evidence for multiple gene transfers from the mitochondria to the nucleus (Figure 5, Supplementary Table 4). Similar to the two Dystaenia species, S. divaricate contains two nuclear transcripts for 5′ RPL2, and A. graveolens, C. sativum, and S. divaricate contain two copies of the SDH3 transcripts (Figure 5). Two copies of RPS10 are unique to D. carota subsp. sativus. One transcript for 5′ RPL2, RPS14, and RPS19 was found in the A. graveolens, C. sativum, and D. carota subsp. sativus transcriptomes. The B. chinense transcriptome contains 5′ RPL2, RPS19, and SDH3. Similar to the two Dystaenia species, multiple transcripts of mitochondrial sdh4, which also contains a portion of the conserved peptidylprolyl isomerase (PPIase) and sdh4 domains without mitochondrial targeting sequences, were found in three transcriptomes (A. graveolens, B. chinense, and D. carota subsp. Sativus; Supplementary Figure 5). However, transcripts from C. sativum and S. divaricate transcriptomes included a 5′ extension of 113 bp and 126 bp, respectively, with an incomplete ORF at the N-terminus. The first 32 and 42 amino acids of these ORFs (when “ATA” is translated as a start codon) were predicted as an mTP by LOCALIZER (0.974 and 0.889).
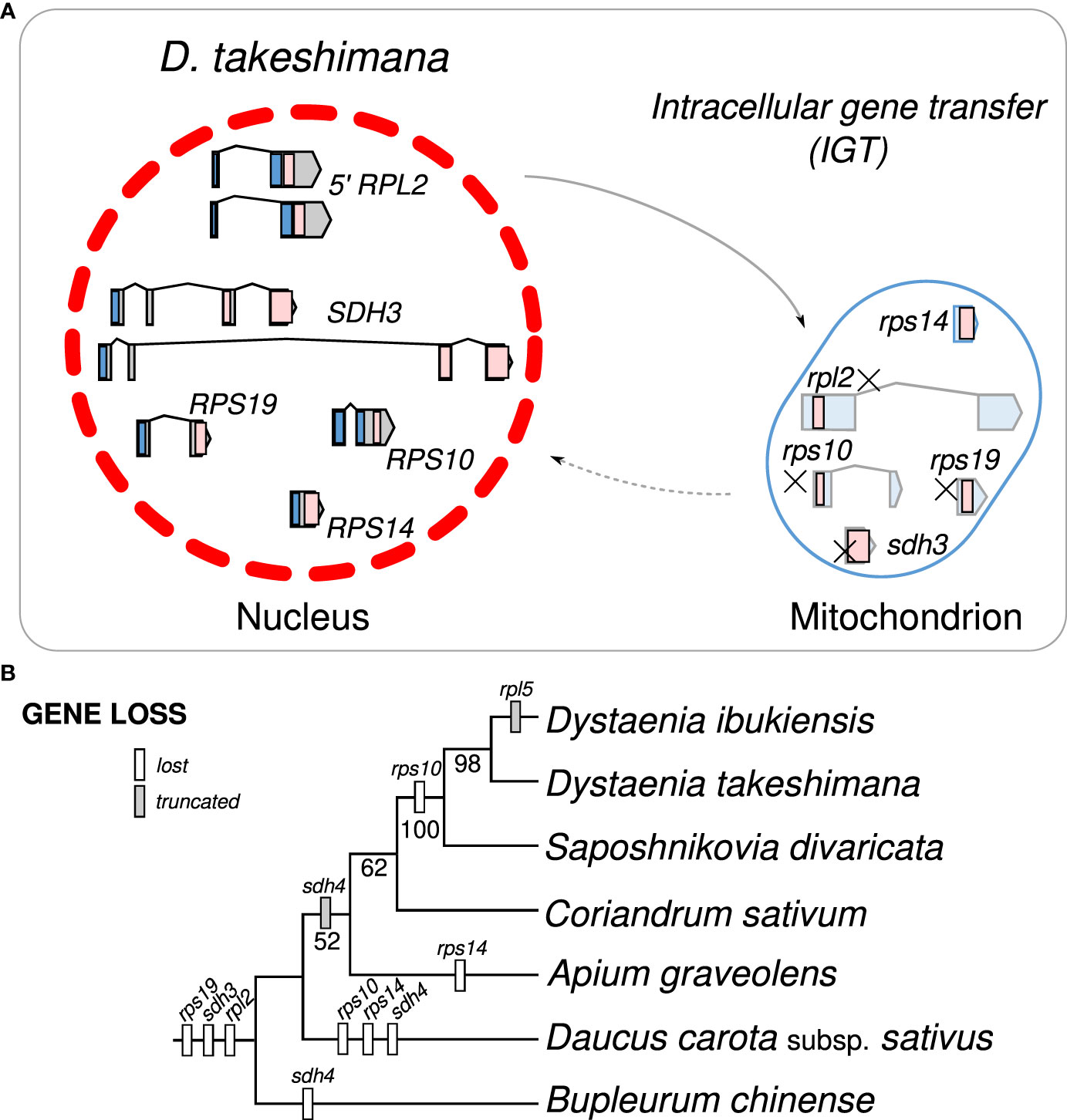
Figure 4 Intracellular gene transfer events to the nucleus in Dystaenia with related species. (A) Schematic diagram of mitochondrial gene transfers from mitochondrial genome to the D. takeshimana nuclear genome. (B) Phylogenetic distribution of gene content among seven Apiaceae species.
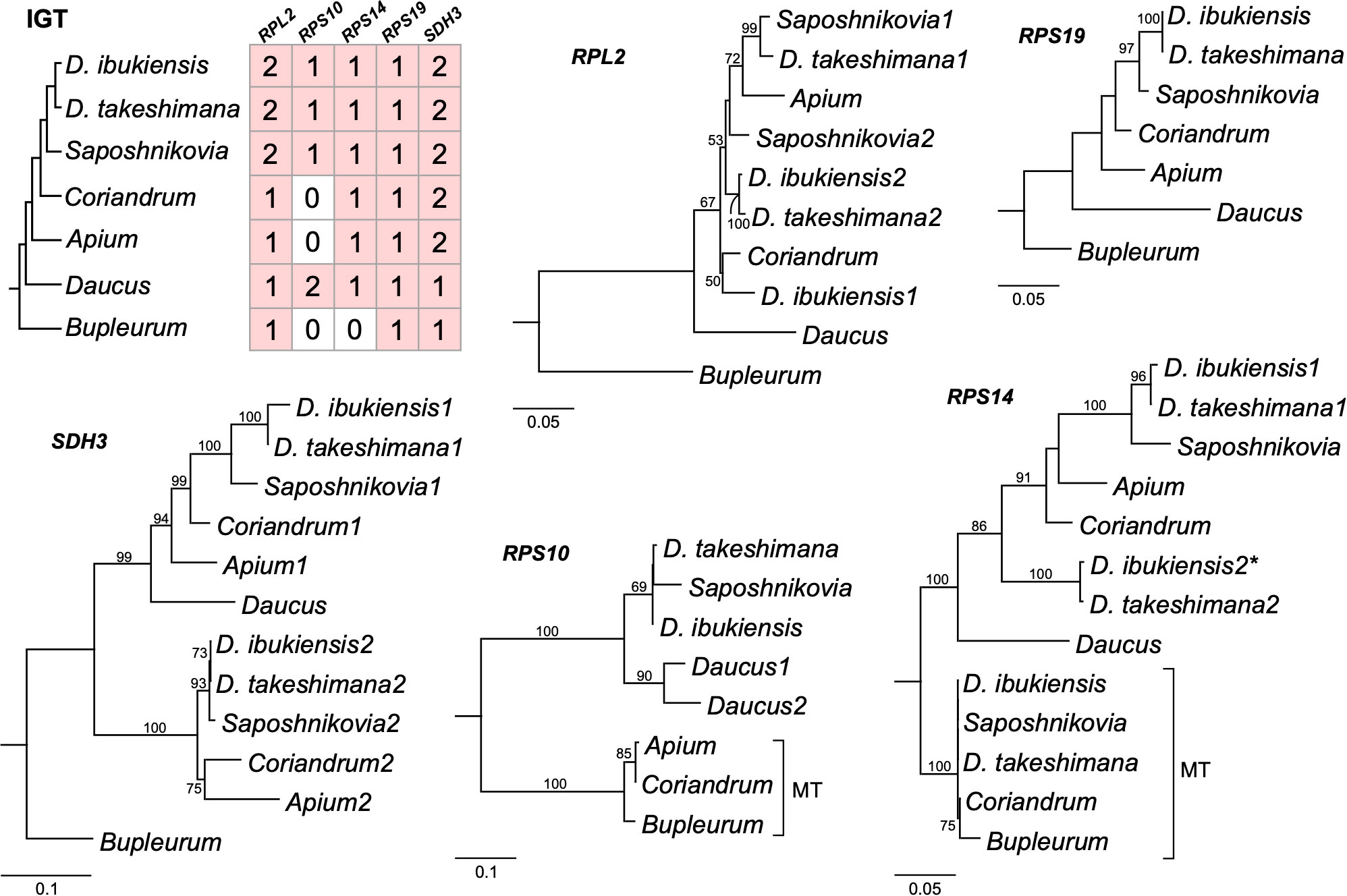
Figure 5 Phylogenetic trees for the nuclear-encoded genes from seven Apiaceae species. A species phylogeny and copy number variation in each nuclear-encoded genes (right). Maximum-likelihood trees based on each nuclear gene. The numbers “1” and “2” after each species in the phylograms represent paralogs. Bootstrap support values > 50% are shown at nodes. MT, mitochondrial gene; asterisk, pseudogene.
We further generated a phylogenetic tree of each gene (5′ RPL2, RPS10, RPS14, RPS19, and SDH3) (Figure 5). The phylograms based on the 5′ RPL2 and RPS10 matrix conflicted with the 24-gene data matrix of the phylogenetic tree. However, the phylogenetic analyses of RPS19, SDH3, and RPS14 were consistent with the 24-gene data matrix of the phylogenetic tree, although A. graveolens and C. sativum were conflicting. Although it was difficult to infer the evolutionary history of the gene duplication events for the 5′ RPL2, it was possible for the SDH3 and RPS14 based on robust relationships with high bootstrap values. For example, phylogenetic analysis of the nuclear-encoded SDH3 copies suggested that duplication events occurred in the common ancestors of Dystaenia, A. graveolens, C. sativum, D. carota subsp. sativus, and S. divaricate, and the loss of one copy occurred independently in D. carota subsp. sativus (Figures 4, 5). Phylogenetic analysis of the nuclear-encoded RPS14 copies suggested that the IGT occurred in the common ancestor of the two Dystaenia species, A. graveolens, C. sativum, D. carota subsp. sativus, and S. divaricate, and then stochastic losses occurred after the duplication events (Figures 4, 5).
The D. takeshimana and D. ibukiensis mitogenomes contained 29,079 bp and 12,684 bp of MIPTs (Table 1, Supplementary Table 5), accounting for 10.3% and 4.5% of each mitogenome, respectively. MIPTs were widely scattered across mitogenomes (Figure 3). The 16 insertion regions of D. takeshimana contained three intact protein-coding genes (ndhB, rps7, and rps12), three rRNAs (16S), 10 tRNAs genes (three of which had two copies), several partial genes (ndhB, petG, psaB, rpoC1, and ycf2), and intergenic spacer regions (Supplementary Table 5). The 14 insertion regions in D. ibukiensis contained one intact protein-coding gene (matK), three rRNAs (23S, 4.5S, and 5S), seven tRNAs (two of which had two copies), one pseudogene, several partial genes (clpP, psbA, psbB, and rpoB), and intergenic spacer regions (Supplementary Table 5). Both mitogenomes shared multiple fragments of tRNA genes (trnD-GUC, trnH-GUG, trnI-CAU, trnN-GUU, and trnW-CCA), partial rpoB and rrn23 genes, and a large fragment (part of the trnI-GAU, intact trnA-UGC, and rrn23 genes). The rpoB fragment (215 bp) from the D. ibukiensis mitogenome was located upstream of the truncated rpl5 gene. In the D. takeshimana mitogenome, five MIPTs were associated with three repeat pairs (79 bp, 2736 bp, and 4322 bp). The small repeat pairs included trnH-GUG, and two large repeat pairs contained two partial fragments (a partial fragment of ndhB, intact rps7, and rps12; a partial fragment of ndhB and ycf2 and intact trnL-CAA) from the largest MIPT (Figure 3A). In addition to plastid-derived sequences, the D. takeshimana and D. ibukiensis mitogenomes contained 16,272 bp of (5.8%) and 15,522 bp (5.5%) of TEs, respectively (Supplementary Table 6), the majority of which were LTR retrotransposons (50% and 52%). TEs in both mitogenomes were inserted into the genic and intergenic regions (Figure 3).
The regions that Mauve did not align indicated that the sequence lacked detectable homologous regions between the two plastomes (Supplementary Figure 2A). According to the possible scenario for IR boundary shifts, the region is associated with the end of the boundary (Figures 1, 6). Nucleotide sequence alignment of the trnH-psbB intergenic spacer from the D. ibukiensis plastome with two intergenic spacers (clpP-psbB and trnH-ycf1) from the D. takeshimana plastome revealed the insertion of a 2,411-bp fragment. The GC content of the insertion region was 46.5%, whereas the remaining LSC regions had a GC content of 36.2%. Illumina reads from D. ibukiensis mapped to its plastome confirmed the insertion of trnH and psbB resulting from uniform coverage. The primers designed to amplify this region yielded PCR products of the expected size (Supplementary Figure 6).
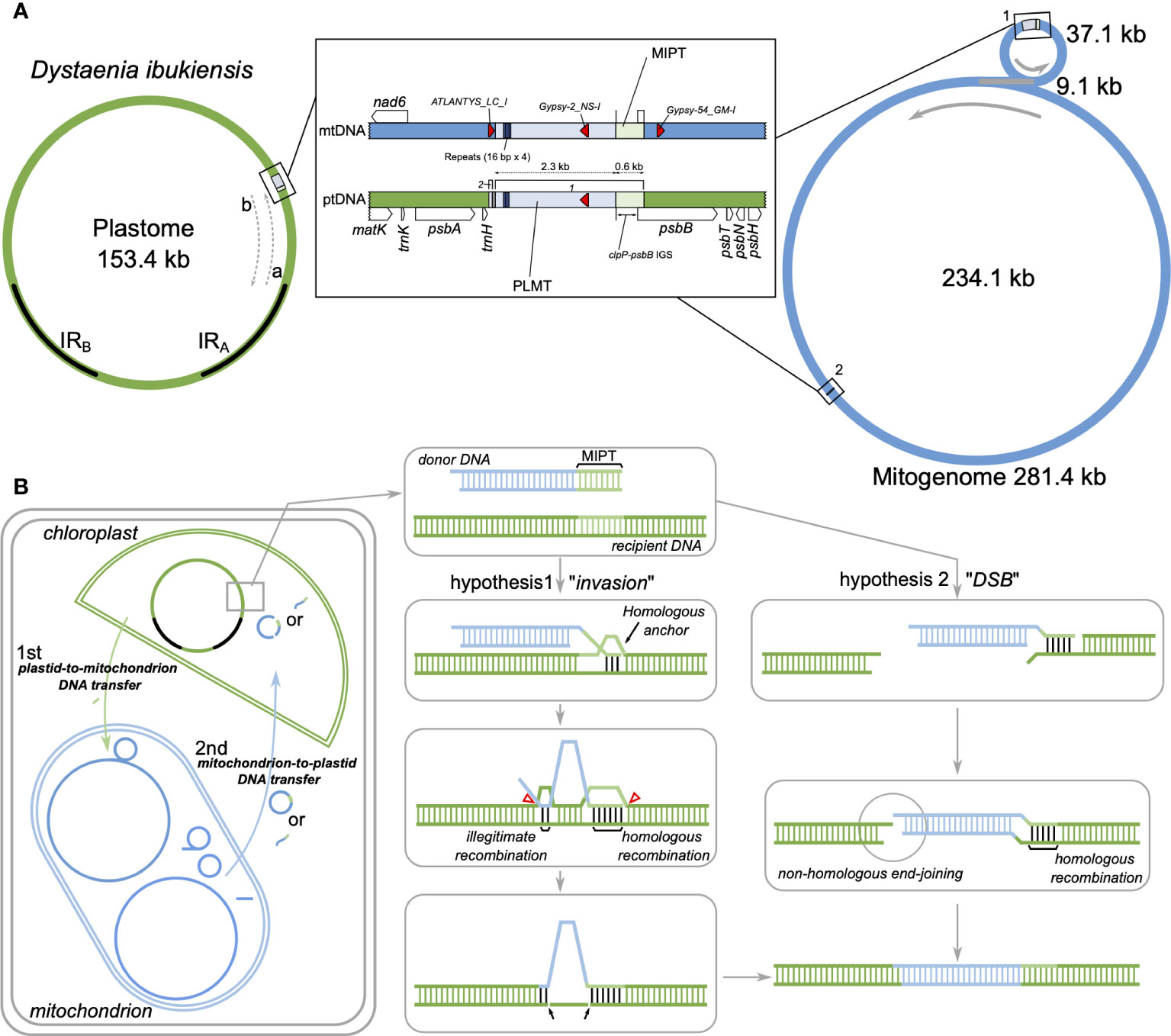
Figure 6 Intracellular DNA transfer events to the plastids in Dystaenia. (A) Schematic diagram of genomic regions surrounding the plastid DNA of mitochondrial origin (PLMT) and mitochondrial DNA of plastid origin (MIPT) from D. ibukiensis organelle genomes. Genome maps are based on the results of Figures 1, 2. The IRA and IRB indicate inverted repeats in the plastome. (B) Models of intracellular DNA transfer events in D. ibukiensis organelle genomes. The hypothetical models illustrate the potential integration of PLMT from the mitogenome into the plastome through invasion or double-stranded break (DSB) mechanism after plastid-to-mitochondrion DNA transfer.
To identify possible mitochondrion-to-plastid DNA transfer [plastid DNA of mitochondrial origin (PLMT)], the inserted region sequences of the D. ibukiensis plastome were used to query the D. ibukiensis mitogenome using “BLASTN” searches. BLAST (> 30 bp in length) returned two hits to the two non-coding regions of the mitogenome (Figure 6A, Supplementary Figure 7). A large fragment (2,294 bp) was located upstream from the nad6 gene in a small mitochondrial circle with 99.3% nucleotide identity. Compared with our MIPT analysis, a 579-bp sequence adjacent to the large fragment revealed MIPT. A small fragment (49 bp) was located upstream from the atp4 gene in a large mitochondrial circle with 100% nucleotide identity. The short sequences (10 bp, “AGAAAGGCCC”) at the end of the small fragment were identical to the sequences downstream of the trnH gene in the D. takeshimana plastome, indicating that the short sequences were D. ibukiensis plastid DNA. Three hits to TEs (ATLANTYS_LC_I, Gypsy-2_LC_I, and Gypsy-54_GM-I) were detected in the mitogenome, whereas the plastome contained only one TE (Figure 6A). PLMT had three repetitive sequences of “CCTACGTATGCCTATG”; however, the mitogenome contained four copies (Figure 6A). No PLMT hits were identified from reciprocal BLAST searches between the D. takeshimana plastid and mitochondrial genomes, indicating that the mitochondrion-to-plastid DNA transfer event was unique to the D. ibukiensis plastome.
Plastids and mitochondria originate from cyanobacterial and alpha-proteobacterial endosymbiont ancestors within plant cells, respectively (Lang et al., 1999; Keeling, 2010). The angiosperm organelle genomes are typically assembled into circular maps (Mower et al., 2012; Smith and Keeling, 2015; Ruhlman and Jansen, 2021). However, mitogenomes often exhibit subcircular, linear, and branched chromosomes (Bendich, 2007; Sloan, 2013; Smith and Keeling, 2015). The presence of recombination activities with repeats (particularly large repeats) contribute to variations in the mitogenome structural organization (Kühn and Gualberto, 2012). Complete plastid and mitochondrial genomic sequences are required to understand organelle genome evolution better. Here, we generated the complete plastid and mitochondrial genomes of two Dystaenia species and showed contrasting patterns of genomic structure within organellar genome evolution. For example, compared with other Apiaceae plastomes, the D. takeshimana plastome was highly conserved, whereas D. ibukiensis underwent relocation and inversion events of a large region (17 kb, a part of ycf2 through psbB genes) (Figure 1). We inferred that the structural changes in the D. ibukiensis plastome were likely caused by a series of IR expansions and contractions (Figure 1): first, an expansion at the IRB/LSC boundary to the clpP occurred, followed by a contraction at the IRA/LSC boundary, from the clpP to the middle of the ycf2 gene. In the plastomes of Apiaceae, the inversion of only a small fragment (490 bp, trnY-trnD-trnE) has been documented in Peucedanum (Liu et al., 2022) and Angelica (Wang et al., 2021a). It is well known that the expansion and contraction of the IR play a key role in the evolution of Apiaceae plastomes. In D. ibukiensis, IR expansion and contraction resulted in the relocation of a large gene block (Figure 1). The occurrence of large events in the D. ibukiensis plastome is a unique feature of the family Apiaceae. In particular, the D. ibukiensis plastome harbored foreign DNA transferred from its mitochondrial counterpart (Figure 6). Incorporating foreign DNA into the D. ibukiensis plastome is a valuable case study for the evolution of plastomes in Apiaceae.
The D. takeshimana mitogenome mapped a circular molecule similar to those of Apiaceae mitogenomes (A. graveolens, B. chinense, C. sativum, D. carota subsp. sativus, and F. vulgare), whereas the D. ibukiensis mitogenome mapped two molecules with substoichiometric linear (Figure 2). Similar to D. ibukiensis, a recent study demonstrated that the Aragoa cleefii mitogenome from Plantaginaceae exhibits a high level of complexity, characterized by the presence of two circular maps connected by a substoichiometric linear (Mower et al., 2021). Large recombinogenic repeats are commonly found in plant mitogenomes. Although the D. takeshimana mitogenome (20) had more large repeat pairs (>1 kb) than the D. ibukiensis mitogenome (13), all D. ibukiensis repeat pairs were >2 kb. The presence of large repeats (>1 kb) in both Dystaenia mitogenomes indicates the existence of multiple major or minor alternative isoforms, as confirmed by contigs generated from hybrid assemblies of short and long reads. Therefore, additional chromosomal configuration may be present in the two Dystaenia mitochondria. Interestingly, we observed the same fragments of plastid-derived sequences in the D. takeshimana mitogenome (Figure 3), identifying a pair of repeats. The three MIPTs can be mediators of homologous recombination in the D. takeshimana mitogenome. It is likely that the fragments moved independently from the plastome and were inserted into the mitogenome or that two large fragments of plastid sequences were inserted into the mitogenome, one of which was split by genomic recombination.
Our analyses showed that gene loss and transfer to the nucleus during the evolution of Apicaeae organelles occurred only in mitogenomes. In angiosperm mitogenomes, a high frequency of 15 ribosomal proteins and two sdh gene losses have been documented (Adams et al., 2002). In the common ancestor of eudicots, two protein-coding genes, rps2 and rps11, were lost (Adams et al., 2002). The successful transfers of 10 mitochondrial genes (5′ rpl2, rpl5, rps4, rps7, rps10, rps12, rps14, rps19, sdh3, and sdh4) into the nucleus have been reported in multiple lineages (Adams et al., 2001; Park et al., 2014; Park et al., 2015; Park and Park, 2020). Comparative analyses of Dystaenia and related species indicated that three mitochondrial genes (rpl2, rps19, and sdh3) were transferred to the nucleus in the common ancestor of all analyzed Apiaceae species; one (rps10) was unique to the specific lineages, Dystaenia/Sposhnikovia clades and D. carota subsp. sativus; and rps14 was transferred to the nucleus in the common ancestor of all analyzed Apiaceae species except for Bupleurum (Figures 4, 5). The split transfer of the 3′ end of the mitochondrial rpl2 gene to the nucleus occurred in the common ancestor of core eudicots (Adams et al., 2001), in which the 5′ portion of rpl2 was present in the mitogenome. Subsequently, multiple transfers of 5′ rpl2 to the nucleus were documented in three lineages: Medicago Soybean (Adams et al., 2001) and Geranium (Park et al., 2015). We also found the 5′ RPL2 transcripts in multiple transcriptomes from Apiaceae, resulting in the complete transfer of the mitochondrial rpl2 to the nucleus during Apiaceae evolution. In particular, the two Dystaenia species and S. divaricate contained two copies of 5′ RPL2 genes, and the phylogenetic analysis indicated that its origin and evolutionary history remain unclear. In the case of RPS19, all analyzed species contained one copy, and the topology was consistent with the ML tree based on the 24 mitochondrial gene sets, although some support values at the nodes were weak.
The phylogenetic relationships of SDH3 among the analyzed Apiaceae lineages provide a good example of how mitochondrial genes are transferred to the nucleus and duplicated (Figure 5). The phylogenetic analysis of nuclear-encoded SDH3 copies suggested that a single transfer to the nucleus occurred in the common ancestor of the seven species. Duplication events occurred in the common ancestors of Dystaenia, A. graveolens, C. sativum, D. carota subsp. sativus, and S. divaricate, and, subsequently, the loss of one copy occurred independently in D. carota subsp. sativus. The nuclear-encoded RPS14 gene was found in all transcriptomes except Bupleurum, suggesting that functional replacement occurred in the common ancestors of Dystaenia, A. graveolens, C. sativum, D. carota subsp. sativus, and S. divaricate, although only the loss of mitochondrial rps14 occurred independently in A. graveolens and D. carota subsp. sativus. The coexistence of mitochondrial and nuclear rps14 homologs within Dystaenia, C. sativum, and S. divaricate indicates that successful functional replacement of the nucleus was necessary before the original mitochondrial copy was lost. In the case of Dystaenia rps14, two copies were detected in the nucleus. Phylogenetic analysis suggests that two independent transfers of mitochondrial rps14 to the nucleus occurred in the common ancestor of Dystaenia and that the copies acquired a targeting sequence. However, the second copy from both species may fail to function as a replacement because the targeting sequence from the second copy of D. takeshimana has a weak signal (Table 2), and the second copy of D. ibukiensis loses the targeting sequence and has multiple internal stop codons. Similar to Dystaenia, the Rhazya stricta genome contains two nuclear copies of rps14 and one mitochondrial copy; one of the nuclear copies is a pseudogene (Park et al., 2014). However, phylogenetic analysis supports a single transfer of mitochondrial rps14 to the Rhazya nuclear genome (Park et al., 2014).
Although multiple transcripts for mitochondrial sdh4 contained parts of the PPIase and sdh4 domains, our analyses did not find sufficient evidence for mitochondrial target peptides to shuttle the product back to the mitochondria. However, the Arabidopsis thaliana mitogenome also contains a truncated sdh4 gene at the C-terminus, previously annotated as a pseudogene. The nuclear-encoded SDH4 gene presence, which contains an mTP and a partial domain of sdh4, suggests the possibility of split gene transfer and fission of sdh4 in the mitogenome. MIPT was located upstream of the truncated rpl5 gene, suggesting that the transfer event disrupted the ORF of rpl5, resulting in a loss of functionality. However, our analysis found no evidence for IGT of the truncated mitochondrial rpl5 in the D. ibukiensis mitogenome. The evolutionary fate of mitochondrial rpl5 in D. ibukiensis and the fate of mitochondrial sdh4 among the analyzed genera were unclear from the present data. Additional deep genome sequencing will be required to address this question.
In addition to IGT to the nucleus, inter-compartmental transfers between plant organelle genomes have been documented; plastid-to-mitochondrial DNA transfers are common, but transfers in the opposite direction are rare (Mower et al., 2012). Within angiosperm plastomes, mitochondrion-to-plastid DNA transfers have been documented in five families: Anacardium in Anacardiaceae (Rabah et al., 2017); Caucalis, Crithmum, Cuminum, Daucus, and Petroselinum in Apiaceae (Iorizzo et al., 2012; Downie and Jansen, 2014; Downie and Jansen, 2015; Spooner et al., 2017), the tribe Asclepiadeae of Apocynaceae (Straub et al., 2013); Convallaria in Asparagaceae (Raman et al., 2019); and Paspalum and Pariana in Poaceae (Ma et al., 2015; Burke et al., 2016). Most fragments of mitochondrial DNA were inserted into their plastome-IR region, except for Crithmum, Petroselinum, and tribe Asclepiadeae. The foreign DNA of Crithmum and Petroselinum was found between the IRA and LSC gene trnH-GUG, and the tribe Asclepiadeae plastomes contained PLMT sequences between rps2 and rpoC2 genes in LSC. We discovered a mitochondrial insertion in the D. ibukiensis plastome trnH-psbB intergenic spacer in the LSC and inferred that the insertion sequences were associated with IR boundary shifts (expansion and contraction) (Figure 6). The complete D. ibukiensis mitogenome provided strong evidence for the occurrence of DNA transfer into the plastid counterpart, showing that the PLMT was fragmented into two in the mitogenome (Figure 6). However, the mitochondrial DNA was likely to transfer intact as plant mitogenomes have a high frequency of genomic rearrangements between recombinationally active repeats. Only D. carota and Asclepias syriaca had complete mitogenomes, which confirmed that the PLMTs comprise two or three pieces in their mitogenomes, also suggesting a single DNA transfer event (Iorizzo et al., 2012; Straub et al., 2013).
The integration of mitochondrial DNA into the D. ibukiensis plastome can be comprehensively explained by considering the hypothesis of invasion and DSB (Figure 6). We propose two hypotheses to explain how the mitochondrial DNA fragment was integrated into the D. ibukiensis plastome (Figure 6B). In both hypotheses, plastid-to-mitochondrial DNA transfer occurred first, followed by mitochondrion-to-plastid DNA transfer. In this case, the transfer portion of the mitochondrial DNA could be a circular or linear molecule containing MIPT, which acts as a homologous region pairing between the donor and recipient sequences. After mitochondrial DNA is imported into the plastid, it can be integrated into the plastome by two different processes: 1) a displacement loop (D-loop) associated with IR boundary shifts facilitates pairing with a homologous anchor at an MIPT site and strand invasion to initiate the recombination repair process; and 2) fusion between the donor and recipient DNA occurs within or at the end of the interacting microhomologous sequences (illegitimate recombination), followed by the deletion of a segment of the recipient DNA. Finally, the integration of mitochondrial sequences is completed by DNA replication, and a double-strand break (DSB) occurs before the interaction with the plastome. Next, integration occurs as a single-stranded DNA molecule is annealed at an MIPT site to a complementary single-stranded overhang present at a DSB site in the plastome. Finally, integration is completed by second-strand synthesis and ligation via non-homologous end-joining.
The double D-loop strategy has been widely recognized as a fundamental aspect of plastid DNA replication (Heinhorst and Cannon, 1993). One potential mechanism that could contribute to IR expansion in the plastome was also associated with a DSB, which is subsequently followed by strand invasion, expansion, and recombination within the IR region (Goulding et al., 1996). Homology-facilitated illegitimate recombination between short regions has been observed in plastomes (Ogihara et al., 1988; Maréchal et al., 2009). Previous studies have shown that illegitimate recombination sites have GC-rich microhomologies of 3 bp to 10 bp (Prudhomme et al., 2002; De Vries et al., 2004). We present two hypotheses to explain how mitochondrial DNA can integrate into the plastome (Figure 6). In both hypotheses, identifying MIPT sequences adjacent to the transferred sequences in the D. ibukiensis mitogenome strongly indicated that homologous recombination plays a crucial role in facilitating the integration of its plastome. Both invasion and DSB are possible scenarios. The formation of a D-loop bubble is the predominant mechanism in plastid DNA and can facilitate homologous DNA pairing. The PLMT region in the D. ibukiensis plastome is associated with IR expansion, and DSB repair can occur via nonhomologous end-joining. Furthermore, identifying microhomologous sequences at the ends of the transferred sequences in the D. ibukiensis plastome suggested the possibility of illegitimate recombination. TE can be mobilized within a genome (Huang et al., 2012), and LTR retrotransposons can target tRNA genes. In the D. ibukiensis mitogenome, three TEs were detected around the transferred DNA, one within the transferred DNA, and two in either of the flanking regions (Figure 6), which were categorized as gypsy LTR retrotransposons. PLMT in the D. ibukiensis plastome is located upstream of the trnH-GUG. Thus, mitochondria-located PLMT may move from the mitogenome to the plastome. Two distinct mechanisms have been postulated to underlie these events: mitochondrial integration into the D. carota and A. syriaca plastomes were inferred by the likely mechanism of non-LTR retrotransposons and repair of a DSB by homologous recombination, respectively (Iorizzo et al., 2012; Straub et al., 2013). A physical connection between chloroplasts and mitochondria may facilitate the exchange of materials between these organelles. Stromules are dynamic tubular structures extending from the chloroplast surface of plant cells (Köhler et al., 1997; Hanson and Sattarzadeh, 2008). Stromules have been observed to interact with other organelles, including mitochondria, peroxisomes, and the endoplasmic reticulum (ER) (Hanson and Conklin, 2020), suggesting a potential role in inter-organelle communication and the exchange of molecules. The ER is a prevalent mediator in facilitating interactions among the other organelles (Mathur et al., 2022), suggesting that the delivered DNA may be able to be imported into the chloroplast through the ER. A previous study has shown that circular or linear foreign DNA can enter the chloroplast envelope under stressful environmental conditions (Cerutti and Jagendorf, 1995).
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) GenBank, https://www.ncbi.nlm.nih.gov/genbank/, OR231235-OR231238, OR756216-OR756231, OR764728-OR764735, OR771027-OR771028, OR771084-OR771087.
SJunP: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing - original draft. SJ-P: Conceptualization, Funding acquisition, Project administration, Writing - review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2018R1A2B6006885 to SJ-P).
The authors thank Sunmi Park for assistance with the PCR and Dr. Kutae Park for assistance sampling of D. ibukiensis. We also thank two reviewers for comments on an earlier version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1283292/full#supplementary-material
Adams, K. L., Ong, H. C., Palmer, J. D. (2001). Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 18 (12), 2289–2297. doi: 10.1093/oxfordjournals.molbev.a003775
Adams, K. L., Qiu, Y.-L., Stoutemyer, M., Palmer, J. D. (2002). Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. 99 (15), 9905–9912. doi: 10.1073/pnas.042694899
Almagro Armenteros, J. J., Salvatore, M., Emanuelsson, O., Winther, O., von Heijne, G., Elofsson, A., et al. (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2 (5), e201900429. doi: 10.26508/lsa.201900429
Antipov, D., Korobeynikov, A., McLean, J. S., Pevzner, P. A. (2016). hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32 (7), 1009–1015. doi: 10.1093/bioinformatics/btv688
Bendich, A. J. (2007). The size and form of chromosomes are constant in the nucleus, but highly variable in bacteria, mitochondria and chloroplasts. Bioessays 29 (5), 474–483. doi: 10.1002/bies.20576
Breitler, J. C., Campa, C., Georget, F., Bertrand, B., Etienne, H. (2016). A single-step method for RNA isolation from tropical crops in the field. Sci. Rep. 6(1), 38368. doi: 10.1038/srep38368
Burke, S. V., Wysocki, W. P., Zuloaga, F. O., Craine, J. M., Pires, J. C., Edger, P. P., et al. (2016). Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae). BMC Plant Biol. 16 (1), 140. doi: 10.1186/s12870-016-0823-3
Cerutti, H., Jagendorf, A. (1995). Movement of DNA across the chloroplast envelope: Implications for the transfer of promiscuous DNA. Photosynth Res. 46 (1-2), 329–337. doi: 10.1007/bf00020448
Chan, P. P., Lin, B. Y., Mak, A. J., Lowe, T. M. (2021). tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49 (16), 9077–9096. doi: 10.1093/nar/gkab688
Cheng, Q., Wang, P., Li, T., Liu, J., Zhang, Y., Wang, Y., et al. (2021). Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in celery (Apium graveolens L.). Int. J. Mol. Sci. 22 (16), 8584. doi: 10.3390/ijms22168584
Choi, H. K., Kim, Y. D., Sun, B. Y., Shin, H. C. (1998). Phylogeny of Dystaenia in subfamily Apioideae (Family Apiaceae) based on ITS sequences. Korean J. Pl. Taxon 28 (2), 139–149. doi: 10.11110/kjpt.1998.28.2.139
Christenhusz, M. J., Byng, J. W. (2016). The number of known plants species in the world and its annual increase. Phytotaxa 261 (3), 201–217. doi: 10.11646/phytotaxa.261.3.1
Darling, A. C. E., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14 (7), 1394–1403. doi: 10.1101/gr.2289704
De Vries, J., Herzfeld, T., Wackernagel, W. (2004). Transfer of plastid DNA from tobacco to the soil bacterium Acinetobacter sp. by natural transformation. Mol. Microbiol. 53 (1), 323–334. doi: 10.1111/j.1365-2958.2004.04132.x
Downie, S. R., Jansen, R. (2014). Another first for the Apiaceae: evidence for mitochondrial DNA transfer into the plastid genome. J. Fac. Pharm. İst. Univ. 44, 131–144.
Downie, S. R., Jansen, R. K. (2015). A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40 (1), 336–351, 316. doi: 10.1600/036364415X686620
Downie, S. R., Spalik, K., Katz-Downie, D. S., Reduron, J.-P. (2010). Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Div. Evol. 128 (1), 111. doi: 10.1127/1869-6155/2010/0128-0005
Goulding, S. E., Wolfe, K., Olmstead, R., Morden, C. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. MGG 252 (1), 195–206. doi: 10.1007/BF02173220
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 (7), 644–652. doi: 10.1038/nbt.1883
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47 (W1), W59–W64. doi: 10.1093/nar/gkz238
Hanson, M. R., Conklin, P. L. (2020). Stromules, functional extensions of plastids within the plant cell. Curr. Opin. Plant Biol. 58, 25–32. doi: 10.1016/j.pbi.2020.10.005
Hanson, M. R., Sattarzadeh, A. (2008). Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environ. 31 (5), 646–657. doi: 10.1111/j.1365-3040.2007.01768.x
Heinhorst, S., Cannon, G. C. (1993). DNA replication in chloroplasts. J. Cell Sci. 104, 1–9. doi: 10.1242/jcs.104.1.1
Huang, C. R. L., Burns, K. H., Boeke, J. D. (2012). Active transposition in genomes. Annu. Rev. Genet. 46 (1), 651–675. doi: 10.1146/annurev-genet-110711-155616
Iorizzo, M., Senalik, D., Szklarczyk, M., Grzebelus, D., Spooner, D., Simon, P. (2012). De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 12 (1), 61. doi: 10.1186/1471-2229-12-61
Jia, Y., Liu, M.-L., López-Pujol, J., Jia, R.-W., Kou, Y.-X., Yue, M., et al. (2023). The hybridization origin of the Chinese endemic herb genus Notopterygium (Apiaceae): Evidence from population genomics and ecological niche analysis. Mol. Phylogenet. Evol. 182, 107736. doi: 10.1016/j.ympev.2023.107736
Keeling, P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc B: Biol. 365 (1541), 729–748. doi: 10.1098/rstb.2009.0103
Kitagawa, M. (1937). Miscellaneous notes on Apiaceae (Umbelliferæ) of Japan and manchuria: II. Bot. Mag. (Tokyo) 51, 805–812. doi: 10.15281/jplantres1887.51.805
Kljuykov, E. V., Petrova, S. E., Degtjareva, G. V., Zakharova, E. A., Samigullin, T. H., Tilney, P. M. (2019). A taxonomic survey of monocotylar Apiaceae and the implications of their morphological diversity for their systematics and evolution. Bot. J. Linn. 192 (3), 449–473. doi: 10.1093/botlinnean/boz095
Kohany, O., Gentles, A. J., Hankus, L., Jurka, J. (2006). Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinf. 7 (1), 1–7. doi: 10.1186/1471-2105-7-474
Köhler, R. H., Cao, J., Zipfel, W. R., Webb, W. W., Hanson, M. R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276 (5321), 2039–2042. doi: 10.1126/science.276.5321.2039
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27 (5), 722–736. doi: 10.1101/gr.215087.116
Kühn, K., Gualberto, J. M. (2012). “Chapter nine - recombination in the stability, repair and evolution of the mitochondrial genome,” in Adv. Bot. Res. Ed. Maréchal-Drouard, L. (Cambridge, MA: Academic Press), 215–252. doi: 10.1016/B978-0-12-394279-1.00009-0
Lang, B. F., Gray, M. W., Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33 (1), 351–397. doi: 10.1146/annurev.genet.33.1.351
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Laslett, D., Canback, B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32 (1), 11–16. doi: 10.1093/nar/gkh152
Li, H. (2013). Aligning sequence reads, clone sequneces and assembly contigs with BWA-MEM. arXiv preprint arXiv. doi: 10.48550/arXiv.1303.3997
Liu, Y., Johnson, M. G., Cox, C. J., Medina, R., Devos, N., Vanderpoorten, A., et al. (2019). Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat. Commun. 10 (1), 1485. doi: 10.1038/s41467-019-09454-w
Liu, C.-K., Lei, J.-Q., Jiang, Q.-P., Zhou, S.-D., He, X.-J. (2022). The complete plastomes of seven Peucedanum plants: comparative and phylogenetic analyses for the Peucedanum genus. BMC Plant Biol. 22 (1), 101. doi: 10.1186/s12870-022-03488-x
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2019). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48 (D1), D265–D268. doi: 10.1093/nar/gkz991
Ma, P.-F., Zhang, Y.-X., Guo, Z.-H., Li, D.-Z. (2015). Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci. Rep. 5 (1), 11608. doi: 10.1038/srep11608
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Maréchal, A., Parent, J.-S., Véronneau-Lafortune, F., Joyeux, A., Lang, B. F., Brisson, N. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. 106 (34), 14693–14698. doi: 10.1073/pnas.0901710106
Mathur, J., Kroeker, O. F., Lobbezoo, M., Mathur, N. (2022). The ER is a common mediator for the behavior and interactions of other organelles. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.846970
Mower, J. P., Hanley, L., Wolff, K., Pabón-Mora, N., González, F. (2021). Complete mitogenomes of two Aragoa species and phylogeny of plantagineae (Plantaginaceae, lamiales) using mitochondrial genes and the nuclear ribosomal RNA repeat. Plants 10 (12), 2673. doi: 10.3390/plants10122673
Mower, J. P., Sloan, D. B., Alverson, A. J. (2012). "Plant mitochondrial genome diversity: the genomics revolution," in Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. Ed. JH Wendel (New York, NY, Springer), 123–144.
Ogihara, Y., Terachi, T., Sasakuma, T. (1988). Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. 85 (22), 8573–8577. doi: 10.1073/pnas.85.22.8573
Palumbo, F., Vitulo, N., Vannozzi, A., Magon, G., Barcaccia, G. (2020). The Mitochondrial Genome Assembly of Fennel (Foeniculum vulgare) Reveals Two Different atp6 Gene Sequences in Cytoplasmic Male Sterile Accessions. Int. J. Mol. Sci. 21 (13), 4664. doi: 10.3390/ijms21134664
Park, S., Grewe, F., Zhu, A., Ruhlman, T. A., Sabir, J., Mower, J. P., et al. (2015). Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytol. 208 (2), 570–583. doi: 10.1111/nph.13467
Park, S., Park, S. (2020). Large-scale phylogenomics reveals ancient introgression in Asian Hepatica and new insights into the origin of the insular endemic Hepatica maxima. Sci. Rep. 10 (1), 16288. doi: 10.1038/s41598-020-73397-2
Park, S., Ruhlman, T. A., Sabir, J. S. M., Mutwakil, M. H. Z., Baeshen, M. N., Sabir, M. J., et al. (2014). Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics 15 (1), 405. doi: 10.1186/1471-2164-15-405
Pfosser, M., Jakubowsky, G., Schlüter, P. M., Fer, T., Kato, H., Stuessy, T. F., et al. (2005). Evolution of Dystaenia takeshimana (Apiaceae), endemic to Ullung Island, Korea. Plant Syst. Evol. 256 (1), 159–170. doi: 10.1007/s00606-005-0374-9
Plunkett, G. M., Pimenov, M. G., Reduron, J.-P., Kljuykov, E. V., van Wyk, B.-E., Ostroumova, T. A., et al. (2018). “Apiaceae,” in Flowering plants. Eudicots: apiales, gentianales (except rubiaceae). Eds. Kadereit, J. W., Bittrich, V. (Cham: Springer International Publishing), 9–206.
Prudhomme, M., Libante, V., Claverys, J.-P. (2002). Homologous recombination at the border: Insertion-deletions and the trapping of foreign DNA in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. 99 (4), 2100–2105. doi: 10.1073/pnas.032262999
Qiao, Y., Zhang, X., Li, Z., Song, Y., Sun, Z. (2022). Assembly and comparative analysis of the complete mitochondrial genome of Bupleurum chinense DC. BMC Genomics 23 (1), 664. doi: 10.1186/s12864-022-08892-z
Rabah, S. O., Lee, C., Hajrah, N. H., Makki, R. M., Alharby, H. F., Alhebshi, A. M., et al. (2017). Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome 10 (3). doi: 10.3835/plantgenome2017.03.0020
Raman, G., Park, S., Lee, E. M., Park, S. (2019). Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Sci. Rep. 9 (1), 5028. doi: 10.1038/s41598-019-41377-w
Ren, R., Wang, H., Guo, C., Zhang, N., Zeng, L., Chen, Y., et al. (2018). Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 11 (3), 414–428. doi: 10.1016/j.molp.2018.01.002
Rieseberg, L. H., Soltis, D. (1991). Phylogenetic consequences of cytoplasmic gene flow in plants. Zurich Switzerland: Evolutionary Trends in Plants.
Ruhlman, T. A., Jansen, R. K. (2021). “Plastid genomes of flowering plants: essential principles,” in Chloroplast biotechnology: methods and protocols. Ed. Maliga, P. (New York, NY: Springer US), 3–47.
Samigullin, T., Logacheva, M., Terentieva, E., Degtjareva, G., Pimenov, M., Valiejo-Roman, C. (2022). Plastid phylogenomic analysis of tordylieae tribe (Apiaceae, apioideae). Plants 11 (5), 709. doi: 10.3390/plants11050709
Sloan, D. B. (2013). One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 200 (4), 978–985. doi: 10.1111/nph.12395
Smith, D. R., Keeling, P. J. (2015). Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. 112 (33), 10177–10184. doi: 10.1073/pnas.1422049112
Song, L., Florea, L. (2015). Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. GigaScience 4 (1), 48. doi: 10.1186/s13742-015-0089-y
Sperschneider, J., Catanzariti, A.-M., DeBoer, K., Petre, B., Gardiner, D. M., Singh, K. B., et al. (2017). LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 7 (1), 44598. doi: 10.1038/srep44598
Spooner, D. M., Ruess, H., Iorizzo, M., Senalik, D., Simon, P. (2017). Entire plastid phylogeny of the carrot genus (Daucus, Apiaceae): Concordance with nuclear data and mitochondrial and nuclear DNA insertions to the plastid. Am. J. Bot. 104 (2), 296–312. doi: 10.3732/ajb.1600415
Stegemann, S., Keuthe, M., Greiner, S., Bock, R. (2012). Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. 109 (7), 2434–2438. doi: 10.1073/pnas.1114076109
Straub, S. C. K., Cronn, R. C., Edwards, C., Fishbein, M., Liston, A. (2013). Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol. Evol. 5 (10), 1872–1885. doi: 10.1093/gbe/evt140
Stuessy, T. F., Jakubowsky, G., Gómez, R. S., Pfosser, M., Schlüter, P. M., Fer, T., et al. (2006). Anagenetic evolution in island plants. J. Biogeogr. 33 (7), 1259–1265. doi: 10.1111/j.1365-2699.2006.01504.x
Takayama, K., López-Sepúlveda, P., Greimler, J., Crawford, D. J., Peñailillo, P., Baeza, M., et al. (2015). Genetic consequences of cladogenetic vs. anagenetic speciation in endemic plants of oceanic islands. AoB Plants 7. doi: 10.1093/aobpla/plv102
Wang, Y., Lan, Q., Zhao, X., Wang, L., Yu, W., Wang, B., et al. (2021b). The complete mitochondrial genome of Coriandrum sativum. Mitochondrial DNA B Resour. 6 (8), 2391–2392. doi: 10.1080/23802359.2021.1951131
Wang, X.-J., Luo, Q., Li, T., Meng, P.-H., Pu, Y.-T., Liu, J.-X., et al. (2022). Origin, evolution, breeding, and omics of Apiaceae: a family of vegetables and medicinal plants. Hortic. Res. 9. doi: 10.1093/hr/uhac076
Wang, M., Wang, X., Sun, J., Wang, Y., Ge, Y., Dong, W., et al. (2021a). Phylogenomic and evolutionary dynamics of inverted repeats across Angelica plastomes. BMC Plant Biol. 21 (1), 26. doi: 10.1186/s12870-020-02801-w
Wen, J., Xie, D.-F., Price, M., Ren, T., Deng, Y.-Q., Gui, L.-J., et al. (2021). Backbone phylogeny and evolution of Apioideae (Apiaceae): New insights from phylogenomic analyses of plastome data. Mol. Phylogenet. Evol. 161, 107183. doi: 10.1016/j.ympev.2021.107183
Wen, J., Yu, Y., Xie, D.-F., Peng, C., Liu, Q., Zhou, S.-D., et al. (2020). A transcriptome-based study on the phylogeny and evolution of the taxonomically controversial subfamily Apioideae (Apiaceae). Ann. Bot. 125 (6), 937–953. doi: 10.1093/aob/mcaa011
Wu, H., Yang, J.-B., Liu, J.-X., Li, D.-Z., Ma, P.-F. (2022). Organelle phylogenomics and extensive conflicting phylogenetic signals in the monocot order poales. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.824672
Wynn, E. L., Christensen, A. C. (2019). Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3: Genes Genom. Genet. 9 (2), 549–559. doi: 10.1534/g3.118.200948
Yuan, C., Sha, X., Xiong, M., Zhong, W., Wei, Y., Li, M., et al. (2021). Uncovering dynamic evolution in the plastid genome of seven Ligusticum species provides insights into species discrimination and phylogenetic implications. Sci. Rep. 11 (1), 988. doi: 10.1038/s41598-020-80225-0
Zerbino, D. R., Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18 (5), 821–829. doi: 10.1101/gr.074492.107
Zhou, J., Gong, X., Downie, S. R., Peng, H. (2009). Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol. Phylogenet. Evol. 53 (1), 56–68. doi: 10.1016/j.ympev.2009.05.029
Keywords: plastid genome, mitochondrial genome, gene duplication, intracellular transfer, inversion
Citation: Park S and Park SJ (2023) Intrageneric structural variation in organelle genomes from the genus Dystaenia (Apiaceae): genome rearrangement and mitochondrion-to-plastid DNA transfer. Front. Plant Sci. 14:1283292. doi: 10.3389/fpls.2023.1283292
Received: 30 August 2023; Accepted: 14 November 2023;
Published: 05 December 2023.
Edited by:
Manjusha Verma, National Bureau of Plant Genetic Resources (ICAR), IndiaReviewed by:
Karsten Liere, Amedes Genetics, GermanyCopyright © 2023 Park and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: SeonJoo Park, c2pwYXJrMDFAeW51LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.