
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 11 October 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1279468
This article is part of the Research TopicGene Regulatory Networks involved in the Molecular Response to Drought, Salt and Osmotic Stresses in CropsView all 7 articles
Karst lands provide a poor substrate to support plant growth, as they are low in nutrients and water content. Common buckwheat (Fagopyrum esculentum) is becoming a popular crop for its gluten-free grains and their high levels of phenolic compounds, but buckwheat yields are affected by high water requirements during grain filling. Here, we describe a wild population of drought-tolerant Fagopyrum leptopodum and its potential for enhancing drought tolerance in cultivated buckwheat. We determined that the expression of a gene encoding a Basic leucine zipper (bZIP) transcription factor, FlbZIP12, from F. leptopodum is induced by abiotic stresses, including treatment with the phytohormone abscisic acid, salt, and polyethylene glycol. In addition, we show that overexpressing FlbZIP12 in Tartary buckwheat (Fagopyrum tataricum) root hairs promoted drought tolerance by increasing the activities of the enzymes superoxide dismutase and catalase, decreasing malondialdehyde content, and upregulating the expression of stress-related genes. Notably, FlbZIP12 overexpression induced the expression of key genes involved in flavonoid biosynthesis. We also determined that FlbZIP12 interacts with protein kinases from the FlSnRK2 family in vitro and in vivo. Taken together, our results provide a theoretical basis for improving drought tolerance in buckwheat via modulating the expression of FlbZIP12 and flavonoid contents.
The Fagopyrum genus, belonging to the Polygonaceae family, is an annual or perennial, herbaceous or semi-shrub dicotyledonous plant (Zhang et al., 2017; Fan et al., 2021). Common buckwheat (F. esculentum) is widely cultivated in Asia, Europe, and the Americas, whereas Tartary buckwheat (F. tataricum) and tall buckwheat (F. cymosum, also known as F. dibotrys) are mainly cultivated in China (Li et al., 2022). In recent years, buckwheat has become a popular crop for its gluten-free grains and their high levels of phenolic compounds, thus making it suitable for people suffering from celiac disease and other gluten sensitivities (Siracusa et al., 2017).
Compared to other crops such as maize (Zea mays), rice (Oryza sativa), and wheat (Triticum aestivum), buckwheat is not widely cultivated (Aubert et al., 2020), and has low production (1700–2500 kg hm−2) (Kreft et al., 2020). The grain yields achieved by current buckwheat varieties thus cannot meet the increasing market demand. Buckwheat is widely planted in mountainous areas (Park et al., 2011), including in southwestern China with less precipitation than in other parts of the country. The high water requirement during seed maturation of buckwheat poses a severe threat to yield.
Plants have evolved sophisticated strategies to cope with drought stress. Indeed, accumulation of the phytohormone abscisic acid (ABA) triggers multiple physiological responses through ABA signaling (Soma et al., 2023), such as inhibiting seed germination and promoting stomatal closure to decrease transpiration. In addition, plants produce a variety of pigments, such as flavonoids, carotenoids, and anthocyanins (Liu et al., 2023; Mishra, 2023). These strategies are not exclusive and multiple strategies can be employed simultaneously to enhance drought tolerance.
The core components of the ABA signaling pathway comprise the ABA receptors PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTOR (PYR/PYL/RCAR), type 2C protein phosphatases (PP2Cs), and sucrose non-fermenting1 (SNF1)-related protein kinase2 (SnRK2) (Ma et al., 2009; Park et al., 2009). SnRK2s phosphorylate downstream targets that include ion channels (SLOW ANION CHANNEL-ASSOCIATED1 [SLAC1] and POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA1 [KAT1]) and an NADPH oxidase (RESPIRATORY BURST OXIDASE HOMOLOG F [RBOHF]), thereby regulating stomatal movement (Joshi-Saha et al., 2011). Several basic leucine zipper (bZIP) transcription factors, for example ABA-INSENSITIVE5 (ABI5), are phosphorylated by SnRK2s and then activate the expression of drought-related genes (Fujii et al., 2007). In rice, OsbZIP46 is a positive regulator of ABA signaling and overexpressing OsbZIP46 enhances drought tolerance (Tang et al., 2012; Chang et al., 2017). The Arabidopsis bZIP-type ABF1-4 and IDD14 transcription factor form the complex to promote their transcriptional activities, and then regulate drought-stress responses (Liu et al., 2022). The ABA signaling pathway is well known in Arabidopsis (Arabidopsis thaliana), rice, and maize, but is rather unclear in the Fagopyrum genus.
Different from the cultivated buckwheat species, F. leptopodum is mainly distributed in rocks and in dry, hot valley areas, indicating that it may be highly drought tolerant and able to withstand stressful growth conditions (Fan et al., 2021; Li et al., 2022). Currently, only the sequence of the chloroplast genome is reported for this species (Fan et al., 2021). An investigation of the mechanisms by which F. leptopodum can grow in such inhospitable conditions would pave the way for the rapid identification of useful genes with direct application to crop breeding.
In order to improve the drought tolerance of cultivated buckwheat species, we investigated the wild buckwheat population. We found that the poor habitat of a wild population of F. leptopodum and discovered that the population is decreasing in southwestern China. Through molecular and physiological analyses, we show that the transcription factor FlbZIP12 interacts with the kinases FlSnRK2.2 and FlSnRK2.6 to confer drought tolerance in F. leptopodum. These findings not only elucidate the mechanism of drought tolerance in F. leptopodum, but provide a good start to pay attention to this species.
The habitat of F. leptopodum was investigated in southwest China. Mature seeds were collected in the wild. Mature dry seeds from five populations were then sown in a greenhouse to perform morphological analysis, including inflorescence, flowers, and pollens.
Seeds from Muli in Table 1 were sown in growth chambers to analyze phenotype and perform physiological analysis. Pollen analysis was performed by scanning electron microscopy as described previously (Zhang et al., 2007). Plants in soil cultures (vermiculite: nutrient soil = 3:1) were grown in growth chambers. F. leptopodum seedlings were grown on Murashige and Skoog (MS) medium in a growth chamber at 25°C, under a 16-h light/8-h dark photoperiod, with 60% relative humidity, and 250 μmol photons m−2 s−1 light intensity. Ten-day-old seedlings were transferred to the MS supplemented with or without 25% (w/v) polyethylene glycol 6000 (PEG6000), 200 mM NaCl, or 50 μM ABA for 0.5, 1, 2, 3, 6, 12 or 24 h.
We used MEGA11 for the multiple sequence analysis and alignment as well as the phylogenetic tree analysis (Tamura et al., 2021). The Neighbor-Joining (NJ) method (1000 times) was used to construct the phylogenetic tree as previously described (Ma et al., 2019).
Total RNA was extracted from the seedlings using an RNAout Kit (TIANGEN, China). Reverse transcription was performed with 1 µg of total RNA using Hifair® II 1st Strand cDNA Synthesis SuperMix (Yeasen, Shanghai, China). QPCR was performed on a Bio-Rad real-time instrument (CFX96) with the following cycling conditions: 96°C for 2 min, 35 cycles of 96°C for 30 s and 55°C for 30 s. QPCR was performed using Hieff qPCR SYBR Green Master Mix (Yeasen, Shanghai, China). At least three repeated were performed. The relative expression levels were calculated by comparing the target genes’ cycle thresholds (CTs) with the reference gene FlH3 (the paralog of FtH3, histone H3). Data quantifcation was carried out with the CFX Manager software using the 2-ΔΔCt method (Darwish et al., 2022). The primers used for RT-qPCR are listed in Table S1.
The full-length coding sequence of FlbZIP12 (1329bp) was amplified from buckwheat seedlings using primeSTAR (TaKaRa) with the following cycling conditions: 30 cycles of 98°C for 10 s, 55°C for 5 s and 72°C for 10 s. The PCR product was ligated into the pCAMBIA2300 vector to obtain pCAMBIA2300-FlbZIP12-GFP. The plasmids pCAMBIA2300-FlbZIP12-GFP and pCAMBIA2300 (negative control) were transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101. Then, overnight cultures from each Agrobacterium colony were harvested and resuspended in infiltration buffer containing 0.1 mM acetosyringone, 2 mM Na3PO4, and 10 mM MES-KOH pH 5.6. The cultures were individually infiltrated into the leaves of 4-week-old Nicotiana benthamiana plants. At 36 h following infiltration, fluorescence signals were observed using a confocal laser scanning microscope (Leica DM4 B).
Induction and transformation of F. leptopodum hairy roots were carried out as described previously (Yao et al., 2020). The plasmid pCAMBIA2300-FlbZIP12-GFP was transformed into Agrobacterium strain ACC10060, with pCAMBIA2300-GFP used as the control. Briefly, seeds were sown on half-strength solid MS medium and allowed to germinate and grow for 12 days. A 0.8–1 cm section of the hypocotyl was excised and placed on solid MS medium for 24 h. Then, the explants were soaked in an Agrobacterium culture harboring the respective plasmid for 15 min and then placed on MS medium containing 100 μM acetosyringone for 3 days in the dark. Subsequently, the explants were removed from the dark conditions and placed on hairy root induction medium containing 500 mg/L cephalosporins and 50 mg/L kanamycin for 7 days. After the hairy roots had formed, the explants were transferred to fresh medium (containing 50 mg/L kanamycin) for subculture. The hairy root cultures were incubated in the dark at 25°C on a shaker at 80 rpm.
Superoxide dismutase (SOD) activity, Catalase (CAT), and malondialdehyde (MDA) content measurements of buckwheat after PEG6000 were performed according to the instructions of the reagent kit (Nanjing Jiancheng Bioengineering Institute, China).
The full-length FlbZIP12 coding sequence was cloned into pGADT7. The full-length coding sequences of FlSnRK2.2, FlSnRK2.3, and FlSnRK2.6 were individually cloned into the pGBKT7 vector. All primer sequences are listed in Table S1. Protein interaction assays were performed by co-transforming the bait and each prey plasmid into yeast (Saccharomyces cerevisiae) strain AH109 according to a previously described protocol (Zhou et al., 2017). Selected positive closes were transferred to selective medium lacking Adenine, Histidine, Leucine, and Tryptophan (Ade, His, Leu, and Trp). Then, yeast cells were incubated at 30°C for 4 days. The empty vectors were co-transformed as negative controls, and the interaction of AtCARK3 and AtRCAR12 was used as a positive control (Wang et al., 2020a).
For the BiFC assay, the full-length coding sequences of FlSnRK2.2, FlSnRK2.6, and FlbZIP12 without the stop codon were individually cloned into the pSPYCE and pSPYNE vectors, respectively. These vectors were introduced into Agrobacterium strain GV3101. Positive Agrobacterium colonies were incubated overnight in LB medium with the appropriate antibiotics and resuspended in infiltration buffer (0.1 mM acetosyringone, 2 mM Na3PO4, and 10 mM MES-KOH pH 5.6) to identical concentrations (OD600 = 0.8). The Agrobacterium cells were infiltrated into N. benthamiana leaves using a needleless syringe. After 2 days, cells with YFP fluorescence were observed and imaged with a confocal laser-scanning microscope.
We investigated the distribution of F. leptopodum in southwest China, where we discovered only five populations (Table 1). The distribution of the species ranges from 888 to 2503 m in altitude, East longitude from 100°56′0239″ to 103°35′2255″, North longitude from 26°16′5844″ to 31°30′2127″ (Table 1). We noticed morphological differences between plants grown in different habitats (Figure 1). For example, we observed deep red leaves for all plants grown in the wild, whereas they developed green leaves in a growth chamber, which is consistent with a previous survey (Fan et al., 2021). According to herbarium records, F. leptopodum was previously collected from three sites, where we found no F. leptopodum specimens, indicating habitat loss. We did however collect one sample from the buckwheat variety F. leptopodum var. grossii, exhibiting sparse flowering on racemes, which is consistent with a previous survey (Hu et al., 2016). Similar to the different appearance of leaves, we observed pink, white, or pale-pink flowers on F. leptopodum in the field, but these plants produced white flowers when grown in the growth chamber (Figure 2A).

Figure 1 Habitat of F. leptopodum in southwestern China (Muli, Sichuan). The white arrows indicate F. leptopodum.
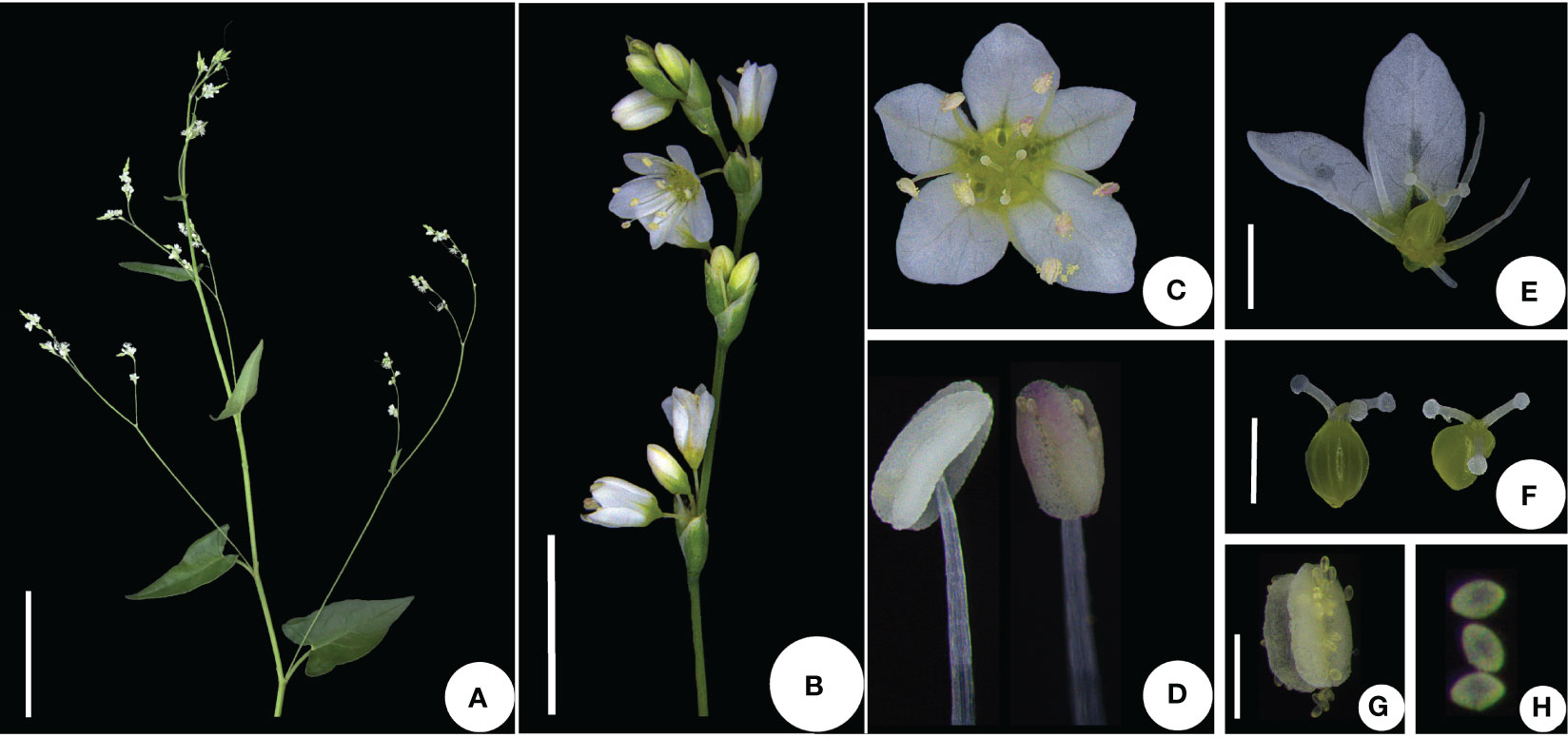
Figure 2 Flower structure of F leptopodum. (A) Compound raceme; (B) raceme; (C) mature flower; (D) stamen; (E) partial structure of a flower (showing a superior ovary); (F) pistil (showing the stigma morphology); (G) anthers; (H) pollens. Scale bars: A, 5 cm; B, 5 mm; D, 2 mm; E, 400 μm; F, 1 mm; G, 400 μm; H, 200 μm; I, 50 μm.
We analyzed the inflorescence and flowers of F. leptopodum plants germinated from seeds collected at different sites and cultivated on nutrient-rich soil (vermiculite:soil = 3:1). These plants produced dense or sparse panicles (Figure 2B). The pistils and stamens were unequal in length in all flowers, indicating that F. leptopodum is a cross-pollinating species (Figure 2C). Flowers radiated symmetrically, with a calyx composed of five sepals and a simple, five-part perianth (Figure 2D). Each flower contained eight stamens, arranged in two whorls, with five stamens in the outer whorl and three stamens in the inner whorl (Figure 2E). Each flower also consisted of three styles terminating with a capitate stigma, and mature disc flowers were glandular (Figure 2F). The ovary was superior and consisted of three carpels, which were combined into a single-loculed ovary (Figures 2G, H).
We also examined pollen grains by scanning electron microscopy. The pollen grains were spherical or elongated, with three well-defined grooves, but lacking a sulcus (Figures 3A-F). The outer wall decoration mesh exhibited edges and corners, some mesh being elongated, and the mesh ridge showed clear peaks (Figure 3F).
Based on the habitat of F. leptopodum, we speculated that it would display strong drought tolerance compared to common buckwheat or Tartary buckwheat. Thus, exploring drought tolerance genes in this species is crucial for improving drought responses in buckwheat. When plants suffer from water scarcity, ABA accumulates to promote multiple physiological responses through the ABA signaling pathway (Leung and Giraudat, 1998; Cutler et al., 2010). ABA-related transcription factors have important roles in ABA signaling transduction (Raghavendra et al., 2010; Zhao et al., 2020). Here, we cloned a gene homologous to ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTORs (ABFs) from Arabidopsis. We named FlbZIP12, which displayed high similarity with FtbZIP12 (Figure S1A, B). We cloned the FlbZIP12 coding sequence in-frame and upstream of the green fluorescent protein (GFP) sequence and transiently expressed the resulting construct in N. benthamiana leaves. The FlbZIP12-GFP fusion protein mainly localized in the nucleus (Figure 4A).
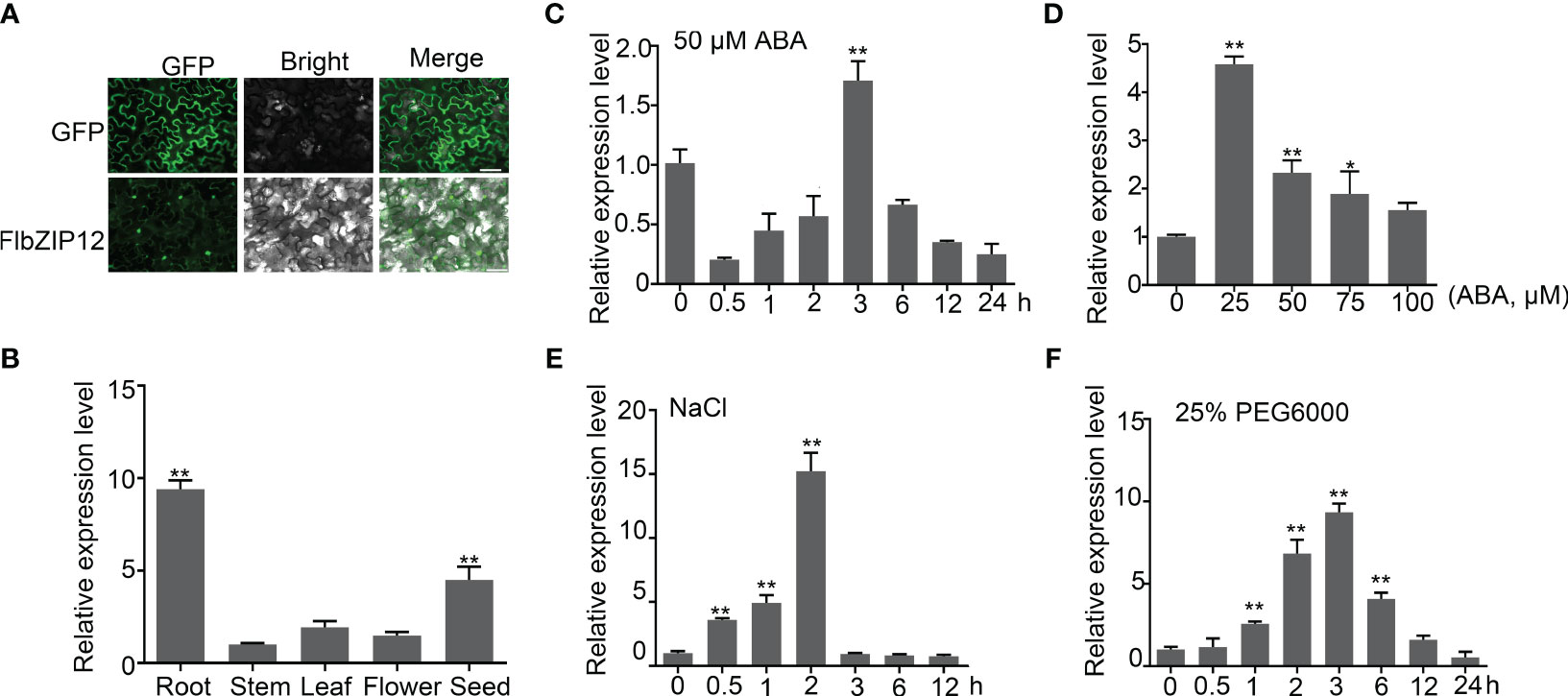
Figure 4 FlbZIP12 expression is induced by abiotic stresses. (A) Localization of FlbZIP12 in N. benthamiana epidermal cells. pCAMBIA2300-FlbZIP12-GFP or the empty vector was transiently expressed in N. benthamiana cells, and then GFP signals were observed using a Leica fluorescence microscope. At least three leaves were observed. Scale bars, 50 μm. (B) Relative FlbZIP12 expression among different tissues in F leptopodum as determined by RT-qPCR. The expression in stems was set to 1. (C) Relative FlbZIP12 expression in 10-d-old seedlings after treatment with 50 μM ABA at the indicated time. (D-F) Relative FlbZIP12 expression induced by various concentrations of ABA for 3 h (D); 200 mM NaCl (E); and 25% PEG6000 (F). FlH3 was used as an internal control. Values are means ± SD (n = 3). Significant differences are indicated (*P < 0.05; **P < 0.01; one-way ANOVA with Tukey’s post-test).
We analyzed the FlbZIP12 expression profile by reverse-transcription quantitative PCR (RT-qPCR): FlbZIP12 was highly expressed in roots, at lower levels in seeds, and at very low levels in stems (Figure 4B). Further, FlbZIP12 expression was induced in seedlings treated with 20 μM ABA for 3 h, and was upregulated about 2-fold (Figure 4C, D). In the presence of 25% polyethylene glycol 6000 (PEG6000) to simulate osmotic stress, FlbZIP12 expression significantly increased at 1 h into the treatment relative to untreated control seedlings (Figure 4E). Seedlings treated with 200 mM NaCl showed a 15-fold increase in FlbZIP12 expression at 2 h into treatment, compared to untreated control seedlings (Figure 4F). These results indicate that FlbZIP12 is involved in abiotic stress.
To explore the functions of FlbZIP12 in planta, we overexpressed FlbZIP12-GFP in Tartary buckwheat hairy roots, yielding three independent lines (#2, #5, and #9) with higher FlbZIP12 expression levels than the empty vector negative control (pCAMBIA2300-GFP) (Figure 5A, B). As these materials are not well suited to perform drought tolerance assays, we tested the effects of simulated drought by treatment with 25% PEG6000.
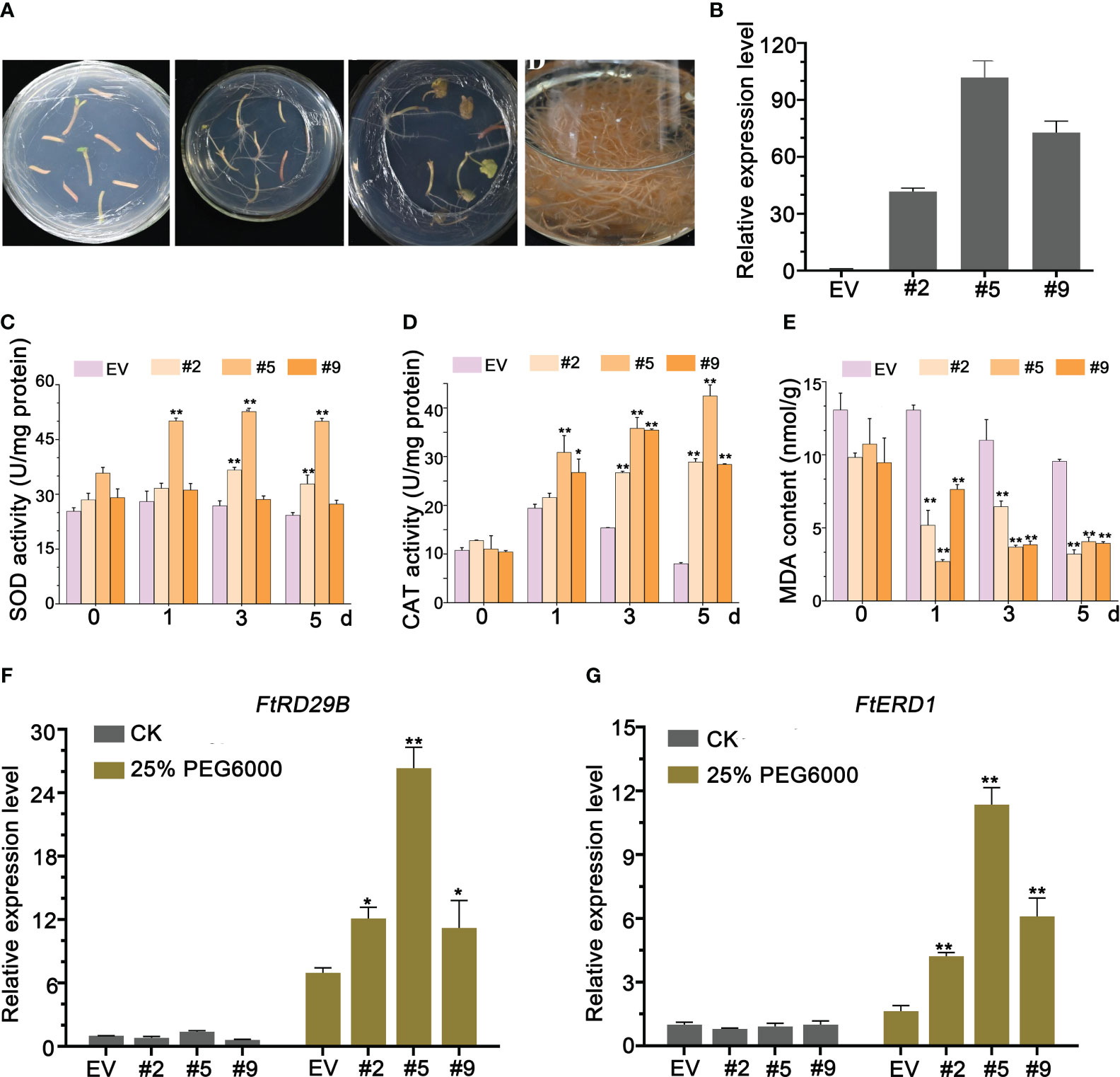
Figure 5 FlbZIP12 overexpression enhances drought tolerance. (A) Transformation of Tartary buckwheat hairy roots. (B) Relative FlbZIP12 expression in 28-d-old hairy root lines (#2, #5, and #9) and control plants (transformed with the empty vector). (C-E) Oxidative response after 25% PEG6000 treatment. (C) SOD activity; (D) CAT activity; (E) MDA content. (F, G) Relative expression levels of stress-related genes in hairy root lines treated with 25% PEG6000 for 5 d The expression level of FlbZIP12 in the empty vector control was set to 1 (B, F, G). FlH3 was used as an internal control (B, F, G). Values are means ± SD (n = 3, B–G). Significant differences between the empty vector (EV) control and FlbZIP12-overexpressing hairy root lines are shown (*P < 0.05; **P < 0.01; one-way ANOVA with Tukey’s post-test).
Reactive oxygen species (ROS) are volatile and short-lived molecules playing important roles in several physiological functions, including acclimation to drought, salinity, or osmotic stresses (Jin et al., 2022). ROS scavenging in plants is mediated by an array of enzymes including superoxide dismutase (SOD) and catalase (CAT) (Gill and Tuteja, 2010; Sewelam et al., 2016). Thus, we measured SOD and CAT activity in transgenic lines treated with PEG6000 for 1, 3, or 5 h, using untreated samples as 0-h controls. FlbZIP12 overexpression resulted in a significant increase of SOD and CAT activities compared to the control (Figure 5C, D). Furthermore, the SOD and CAT activities were strongest in OE#5 plants, which also had the highest FlbZIP12 expression level among the three lines (Figure 5C, D). We also measured the levels of malondialdehyde (MDA), the product of lipid oxidation and a marker of oxidative stress, in these hairy roots. FlbZIP12 overexpression was associated with lower oxidative damage in response to PEG6000 treatment relative to the control (Figure 5E). These data indicate that FlbZIP12 decreases ROS levels, leading to decreased MDA content in response to unsuitable environments in F. leptopodum. Similarly, oxidative stress was previously observed in response to water stress in buckwheat species (Aubert et al., 2020).
In addition, FlbZIP12, as a transcription factor, directly or indirectly activates the transcription of stress-responsive genes, including RESPONSIVE TO DESICCATION29B (RD29B) and EARLY RESPONSIVE TO DEHYDRATION STRESS1 (ERD1). Thus, we performed an RT-qPCR analysis to test mRNA levels for these two genes in our hairy root transgenic lines. After PEG6000 treatment, the RD29B and ERD1 transcript levels were upregulated in the FlbZIP12 overexpressing lines compared to the negative control line (Figure 5F, G). However, there was no significant difference among hairy roots without treatment. There results suggest that FlbZIP12 regulates the expression of stress-related genes in response to drought or osmotic stress.
FlbZIP12 shares high sequence similarity to the Tartary buckwheat transcription factor FtbZIP5 (Figure S1). In a previous study, FtbZIP5 was shown to interact with FtSnRK2.6 in yeast (S. cerevisiae) (Li et al., 2020). Thus, we hypothesized that FlbZIP12 might interact with FlSnRK2s, including FlSnRK2.2, FlSnRK2.3, and FlSnRK2.6. To test this notion, we cloned the coding sequences of several FlSnRK2s. We then performed yeast two-hybrid assays using FlbZIP12 as bait: FlbZIP12 interacted with FlSnRK2.2 and FlSnRK2.6, but not with FlSnRK2.3 (Figure 6A).
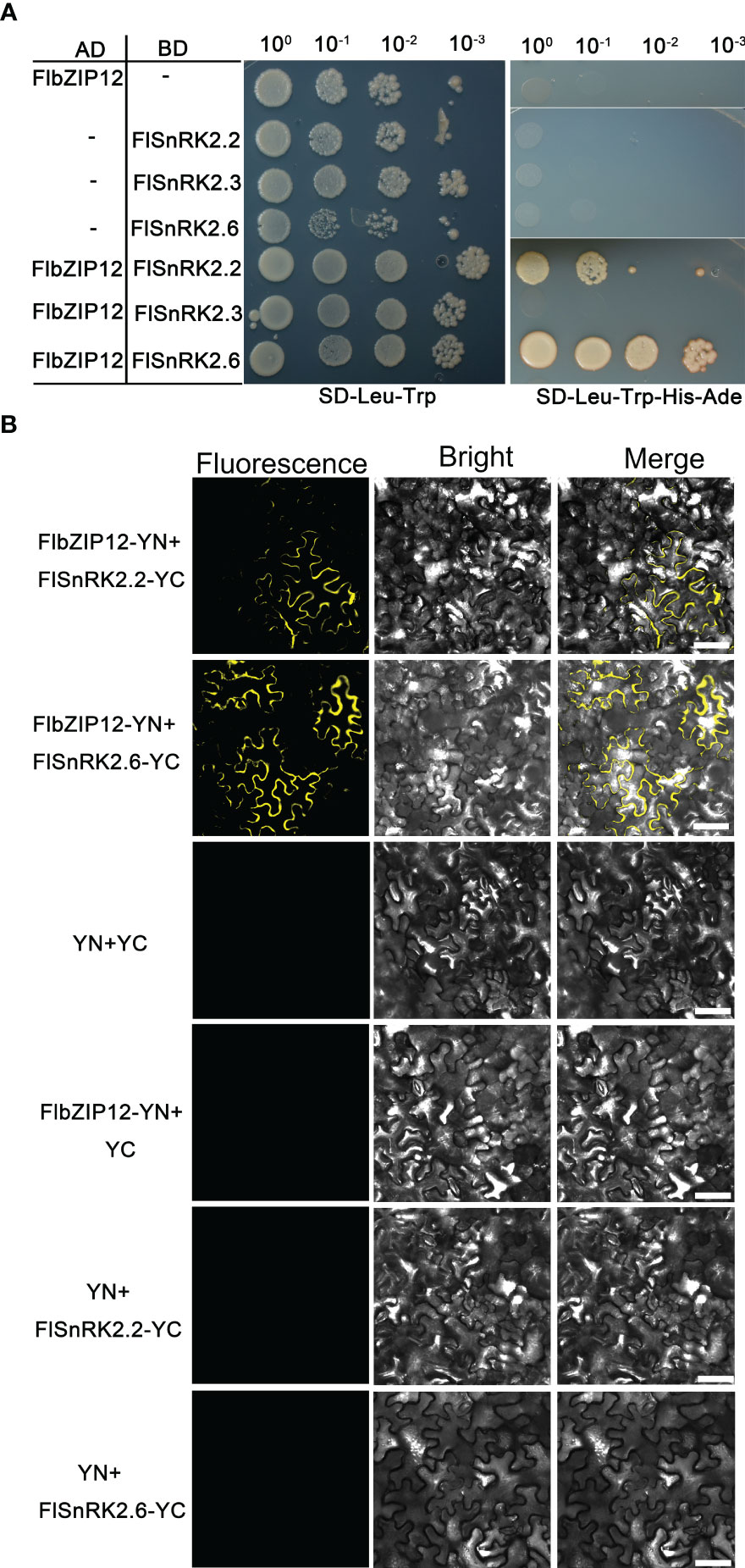
Figure 6 FlbZIP12 interacts with FlSnRK2s. (A) FlbZIP12 interacts with FlSnRK2.2 and FlSnRK2.6 in yeast. Yeast cells co-expressing a construct encoding FlbZIP12 fused to the GAL4 AD and a construct encoding FlSnRK2.2, FlSnRK2.3, or FlSnRK2.6 fused to the GAL4 BD were spotted onto synthetic defined (SD) medium lacking Leu and Trp (–Leu–Trp) to select positive transformants and on SD medium –Leu–Trp–His–Ade to test for protein–protein interaction. Growth was monitored after 4 d (B) Bimolecular fluorescence complementation assays showing the interactions of FlbZIP12 and SnRK2s in N. benthamiana leaves. N‐terminal YFP and C‐terminal YFP were fused to FlbZIP12 or FlSnRK2s, respectively, and the fusion constructs were co‐expressed in N. benthamiana leaves followed by observation on a confocal microscope. Scale bar, 30 μm. At least three leaves were observed.
To validate the interaction of FlbZIP12 with FlSnRK2.2 and FlSnRK2.6 in planta, we conducted bimolecular fluorescence complementation assays. To this end, we cloned the FlbZIP12 coding sequence in-frame and upstream of the sequence encoding the N‐terminal half of yellow fluorescent protein (YFP), yielding FlbZIP12-NYFP; similarly, we cloned the FlSnRK2.2 and FlSnRK2.6 coding sequences in-frame and upstream of the sequence encoding the C‐terminal half of YFP, resulting in FlSnRK2.2-CYFP and FlSnRK2.6-CYFP. We then transiently expressed the corresponding constructs in N. benthamiana leaf cells. We detected fluorescence signals in the cytosol, indicating that FlbZIP12 interacts with FlSnRK2.2 and FlSnRK2.6; by contrast, we detected no fluorescence for the negative control co-expressing the empty vectors NYFP and CYFP (Figure 6B). These finding indicate that the interaction of bZIP-type transcription factors with SnRK2s is conserved in F. leptopodum.
We have observed high levels of flavones in plants grown in the wild. Thus, we tested whether FlbZIP12 activates the expression of flavone biosynthesis genes in hairy roots overexpressing FlbZIP12. The biosynthesis pathway consists of phenylalanine ammonia-lyase (PAL), followed by reactions catalyzed by cinnamate 4-hydroxylase (C4H), 4-coumarate coenzyme A ligase (4CL), chalcone synthase (CHS), flavone 3-hydroxylase (F3H), flavonoid 3-hydroxylase (F3′H), and flavonol synthase (FLS). All of these flavone biosynthesis genes were significantly more highly expressed in the overexpression line than in the empty vector control line (Figure 7). Indeed, FtC4H reached 60-fold higher levels in OE#5 hairy roots compared to the empty vector control line (Figure 7). These results suggest that FlbZIP12 triggers flavonoid biosynthesis in F. leptopodum.
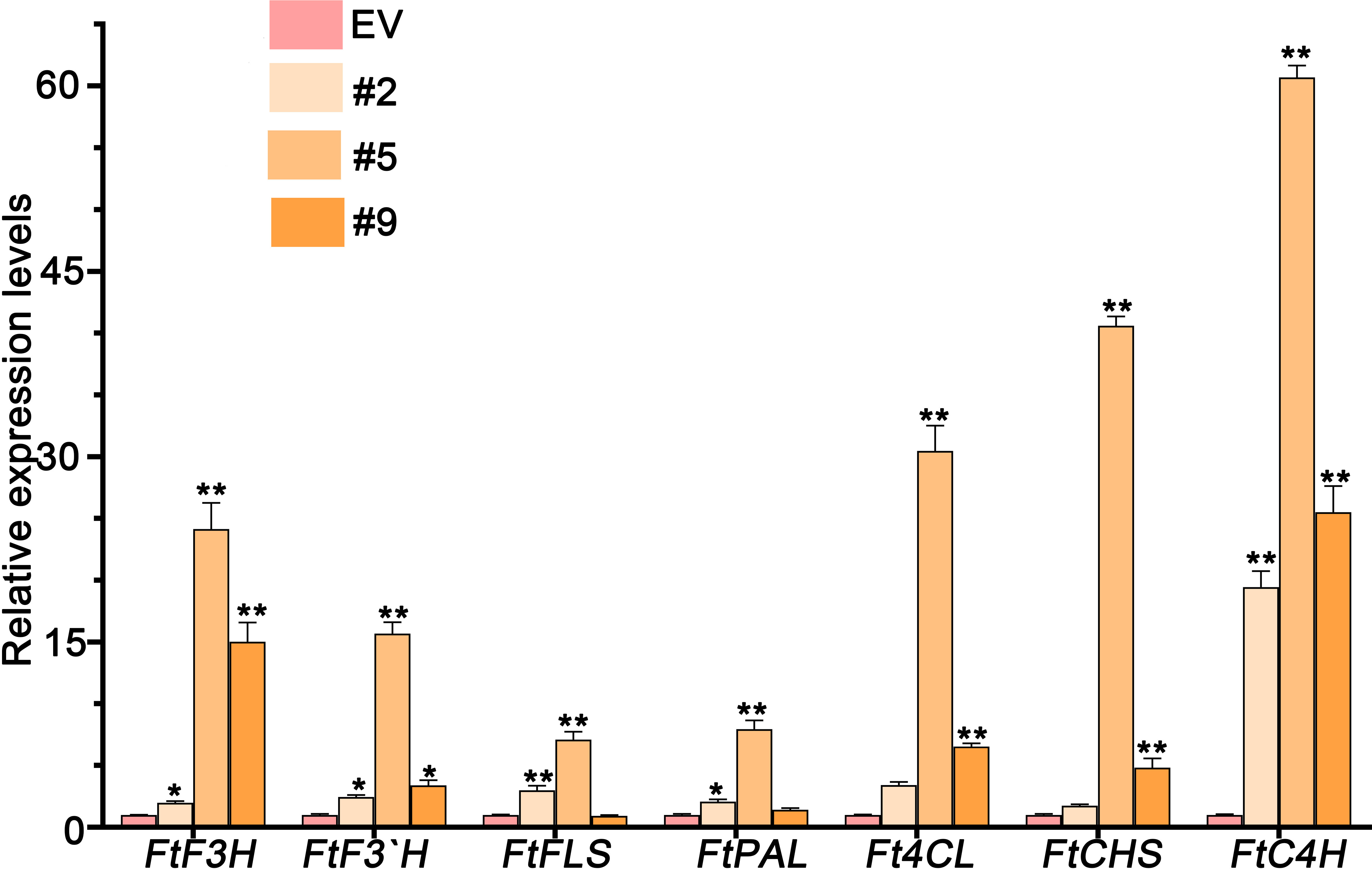
Figure 7 Relative expression levels of flavonoid biosynthesis genes (FtF3H, FtF3′H, FtFLS, FtPAL, Ft4CL, FtCHS, and FtC4H) in transgenic hairy root lines and empty vector control (EV) hairy roots. Gene expression levels in the EV roots were set to 1. FlH3 was used as an internal control. Values are means ± SD of three replicates. Significant differences between the EV control and FlbZIP12-overexpressing hairy root lines are shown (*P < 0.05; **P < 0.01; one-way ANOVA with Tukey’s post-test).
The wild species, F. leptopodum, is mainly distributed in water deficit and soil infertile (Figure 1). It is suitable for a pioneer species for Karst land or ecological remediation of mining area. For example, Zabelia tyaihyonii is a familiar shrub, growing in the karst forest (Choi et al., 2023). The mining area is polluted by metals, including arsenic, cadmium, and aluminum. Previous studies have shown that Tartary buckwheat combats Al toxicity by rapidly secreting oxalate from roots, externally chelating Al and internally forming nonphytotoxic Al complexes with citrate or oxalate (Wang et al., 2015; Zhu et al., 2015; Zhang et al., 2017). RNA-Seq data indicate that membrane transporters and transcription factors cooperatively response to Al stress (Zhang et al., 2017).
ABA signaling Here, we found that FlbZIP12 interacted with FlSnRK2s (Figure 6). SnRK2.2 and SnRK2.3 have been proposed to have a role in the long-term stomatal response to drought conditions (Virlouvet and Fromm, 2015), but are also rapidly activated by ABA and contribute to rapid ABA responses in guard cells (Zhang et al., 2020). The bZIP transcription factors can regulate the transcriptional expressions of stress-related genes by binding specifically to cis-regulatory elements in the promoters, thereby modulating plant stress resistance (Baoxiang et al., 2022). We thus propose that SnRK2.2 and SnRK2.6 activate the bZIP transcription factors, AREB (ABFs), and ABI5 to promote the expression of stress-related genes (Wang et al., 2020b). A previous report showed that FtbZIP83 in Tartary buckwheat also interacts with SnRK2.3 and SnRK2.6 in yeast cells, and overexpressing FtbZIP85 in Arabidopsis enhanced drought and salt tolerance via the ABA-dependent signaling pathway (Li et al., 2019). In grapevine (Vitis vinifera), the bZIP-type transcription factor ABF2 also mediates ROS scavenging ability and cell membrane integrity in response to osmotic stress (Liu et al., 2019). Therefore, FlbZIP12, as a positive regulator, would have the similar function of ABF2 in the ABA signaling pathway in plants.
MYB-bHLH-WD40 (MBW) have been known as the important regulator in flavonoid metabolisms (Hichri et al., 2011). Apart from this, bZIP transcription factors are also involved in flavonoid biosynthesis. In Ginkgo biloba, overexpressing GbbZIP08 in Nicotiana tabacum promotes flavonoid accumulation via up-regulating the transcription levels of flavonoid-synthesis-related genes (Han et al., 2023). Here, we found that overexpressing FlbZIP12 in hairy roots also could upregulate expressions of flavonoid-synthesis-related genes.
Plants respond to drought stress by regulating ABA signal transduction and flavonoid biosynthesis (Gao et al., 2023; Zhang et al., 2023). We showed here that FlbZIP12 may interact with the kinases SnRK2.2 and SnRK2.6 to upregulate RD29B and ERD1 expression to enhance F. leptopodum drought tolerance. Moreover, FlbZIP12 may increase flavonoid contents by inducing the expression of flavone biosynthesis genes for acclimation to unsuitable environments. However, more evidence is required on the mechanisms associated with the ABA-dependent signaling pathway in response to drought stress, as well as an understanding of the influence of anthocyanin or rutin biosynthesis.
Phytohormones such as ABA and jasmonic acid play the key roles in inducing cellular signaling pathways related to stress response of plants (Kuromori et al., 2018; Heidari et al., 2020). Many signaling pathways and ion transporters are directly or indirectly induced by ABA in response to abiotic stresses (Faraji et al., 2021; Hashemipetroudi et al., 2023). Besides, bZIP gene family is among the important transcription factors that are induced by upstream factors such as ABA and will control the expression patterns of a large number of target genes.
Primers used in this study can be found in the Supplementary Materials.
AW: Investigation, Project administration, Resources, Writing – review & editing. YL: Investigation, Writing – review & editing, Formal Analysis. QL: Investigation, Writing – review & editing. XL: Writing – original draft. XZ: Writing – review & editing. JK: Writing – review & editing. ZL: Data curation, Writing – review & editing. YY: Data curation, Writing – review & editing. JW: Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Science & Technology Department of Sichuan Province (2022ZHXC0061, 2023YFN0019), Innovation Team of Triticeae Crops of Sichuan Province: Breeding and application of new Tartary buckwheat varieties (2019scyz02), the cooperation project of dazhou City - Sichuan University (2021CDDZ-15-SCU)
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1279468/full#supplementary-material
Aubert, L., Konradova, D., Barris, S., Quinet, M. (2020). Different drought resistance mechanisms between two buckwheat species Fagopyrum esculentum and Fagopyrum tataricum. Physiol. Plant 172, 577–586. doi: 10.1111/ppl.13248
Baoxiang, W., Bo, X., Yan, L., Jingfang, L., Zhiguang, S., Ming, C., et al. (2022). A Novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice (Oryza sativa L.). Plant Sci. 323, 111393. doi: 10.1016/j.plantsci.2022.111393
Chang, Y., Nguyen, B. H., Xie, Y., Xiao, B., Tang, N., Zhu, W., et al. (2017). Co-overexpression of the constitutively active form of osbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in rice. Front. Plant Sci. 8, 1102. doi: 10.3389/fpls.2017.01102
Choi, I. S., Han, E. K., Wojciechowski, M. F., Heo, T. I., Park, J. S., Yang, J. C., et al. (2023). The genetic structure and demographic history of Zabelia tyaihyonii, endemic to Korean limestone karst forests, based on genome-wide SNP markers. Ecol. Evol. 13, e10252. doi: 10.1002/ece3.10252
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Darwish, D. B. E., Ali, M., Abdelkawy, A. M., Zayed, M., Alatawy, M., Nagah, A. (2022). Constitutive overexpression of GsIMaT2 gene from wild soybean enhances rhizobia interaction and increase nodulation in soybean (Glycine max). BMC Plant Biol. 22, 431. doi: 10.1186/s12870-022-03811-6
Fan, Y., Jin, Y., Ding, M., Tang, Y., Cheng, J., Zhang, K., et al. (2021). The complete chloroplast genome sequences of eight fagopyrum species: insights into genome evolution and phylogenetic relationships. Front. Plant Sci. 12, 799904. doi: 10.3389/fpls.2021.799904
Faraji, S., Heidari, P., Amouei, H., Filiz, E., Abdullah, Poczai, P. (2021). Investigation and Computational Analysis of the Sulfotransferase (SOT) Gene Family in Potato (Solanum tuberosum): Insights into Sulfur Adjustment for Proper Development and Stimuli Responses. Plants (Basel). 10, 2597. doi: 10.3390/plants10122597
Fujii, H., Verslues, P. E., Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 19, 485–494. doi: 10.1105/tpc.106.048538
Gao, J., Zhang, Y., Xu, C., Wang, X., Wang, P., Huang, S. (2023). Abscisic acid collaborates with lignin and flavonoid to improve pre-silking drought tolerance by tuning stem elongation and ear development in maize (Zea mays L.). Plant J. 114, 437–454. doi: 10.1111/tpj.16147
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. biochem.: PPB. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Han, H., Dong, L., Zhang, W., Liao, Y., Wang, L., Wang, Q., et al. (2023). Ginkgo biloba GbbZIP08 transcription factor is involved in the regulation of flavonoid biosynthesis. J. Plant Physiol. 287, 154054. doi: 10.1016/j.jplph.2023.154054
Hashemipetroudi, S. H., Arab, M., Heidari, P., Kuhlmann, M. (2023). Genome-wide analysis of the laccase (LAC) gene family in Aeluropus littoralis: A focus on identification, evolution and expression patterns in response to abiotic stresses and ABA treatment. Front. Plant Sci. 14, 1112354. doi: 10.3389/fpls.2023.1112354
Heidari, P., Mazloomi, F., Nussbaumer, T., Barcaccia, G. (2020). Insights into the SAM Synthetase Gene Family and Its Roles in Tomato Seedlings under Abiotic Stresses and Hormone Treatments. Plants (Basel) 9, 586. doi: 10.3390/plants9050586
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Hu, Y., Zhang, Z., Wu, B., Gao, J., Li, Y. J. B. S. (2016). Genetic relationships of buckwheat species based on the sequence analysis of ITS and ndhF-rpl. Biodivers. Sci. 24, 296–303. doi: 10.17520/biods.2015259
Jin, M. K., Yang, Y. T., Zhao, C. X., Huang, X. R., Chen, H. M., Zhao, W. L., et al. (2022). ROS as a key player in quinolone antibiotic stress on Arabidopsis thaliana: From the perspective of photosystem function, oxidative stress and phyllosphere microbiome. Sci. Total Environ. 848, 157821. doi: 10.1016/j.scitotenv.2022.157821
Joshi-Saha, A., Valon, C., Leung, J. (2011). Abscisic acid signal off the STARting block. Mol. Plant 4, 562–580. doi: 10.1093/mp/ssr055
Kreft, I., Zhou, M., Golob, A., Germ, M., Likar, M., Dziedzic, K., et al. (2020). Breeding buckwheat for nutritional quality. Breed. Sci. 70, 67–73. doi: 10.1270/jsbbs.19016
Kuromori, T., Seo, M., Shinozaki, K. (2018). ABA transport and plant water stress responses. Trends Plant Sci. 23, 513–522. doi: 10.1016/j.tplants.2018.04.001
Leung, J., Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Phys. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Li, Q. J., Liu, Y., Wang, A. H., Chen, Q. F., Wang, J. M., Peng, L., et al. (2022). Plastome comparison and phylogenomics of Fagopyrum (Polygonaceae): insights into sequence differences between Fagopyrum and its related taxa. BMC Plant Biol. 22, 339. doi: 10.1186/s12870-022-03715-5
Li, Q., Wu, Q., Wang, A., Lv, B., Dong, Q., Yao, Y., et al. (2019). Tartary buckwheat transcription factor FtbZIP83 improves the drought/salt tolerance of Arabidopsis via an ABA-mediated pathway. Plant Physiol. Biochem.: PPB. 144, 312–323. doi: 10.1016/j.plaphy.2019.10.003
Li, Q., Zhao, H., Wang, X., Kang, J., Lv, B., Dong, Q., et al. (2020). Tartary buckwheat transcription factor ftbZIP5, regulated by ftSnRK2.6, can improve salt/drought resistance in transgenic arabidopsis. Int. J. Mol. Sci. 21, 1123. doi: 10.3390/ijms21031123
Liu, J., Chu, J., Ma, C., Jiang, Y., Ma, Y., Xiong, J., et al. (2019). Overexpression of an ABA-dependent grapevine bZIP transcription factor, VvABF2, enhances osmotic stress in Arabidopsis. Plant Cell Rep. 38, 587–596. doi: 10.1007/s00299-019-02389-y
Liu, J., Shu, D., Tan, Z., Ma, M., Guo, N., Gao, S., et al. (2022). The Arabidopsis IDD14 transcription factor interacts with bZIP-type ABFs/AREBs and cooperatively regulates ABA-mediated drought tolerance. New Phytol. 236, 929–942. doi: 10.1111/nph.18381
Liu, S., Wang, J., Liu, Z., Yang, Y., Li, X. (2023). FtbZIP85 is involved in the accumulation of proanthocyanidin by regulating the transcription of ftDFR in tartary buckwheat. Curr. Issues Mol. Biol. 45, 3375–3390. doi: 10.3390/cimb45040221
Ma, Z., Liu, M., Sun, W., Huang, L., Wu, Q., Bu, T., et al. (2019). Genome-wide identification and expression analysis of the trihelix transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 19, 344. doi: 10.1186/s12870-019-1957-x
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mishra, D. (2023). How pink is too pink: A tussle between plant and nature. Physiol. Plant 175, e13895. doi: 10.1111/ppl.13895
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Park, N. I., Li, X., Suzuki, T., Kim, S. J., Woo, S. H., Park, C. H., et al. (2011). Differential expression of anthocyanin biosynthetic genes and anthocyanin accumulation in Tartary Buckwheat cultivars ‘Hokkai t8’ and ‘Hokkai t10’. J. Agric. Food Chem. 59, 2356–2361. doi: 10.1021/jf200020b
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Sewelam, N., Kazan, K., Schenk, P. M. (2016). Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. Sci. 7, 187. doi: 10.3389/fpls.2016.00187
Siracusa, L., Gresta, F., Sperlinga, E., Ruberto, G. (2017). Effect of sowing time and soil water content on grain yield and phenolic profile of four buckwheat (Fagopyrum esculentum Moench.) varieties in a Mediterranean environment. J. Food Compost. Anal. 62, 1–7. doi: 10.1016/j.jfca.2017.04.005
Soma, F., Takahashi, F., Kidokoro, S., Kameoka, H., Suzuki, T., Uga, Y., et al. (2023). Constitutively active B2 Raf-like kinases are required for drought-responsive gene expression upstream of ABA-activated SnRK2 kinases. Proc. Natl. Acad. Sci. U S A. 120, e2221863120. doi: 10.1073/pnas.2221863120
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, N., Zhang, H., Li, X., Xiao, J., Xiong, L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Virlouvet, L., Fromm, M. (2015). Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 205, 596–607. doi: 10.1111/nph.13080
Wang, H., Chen, R. F., Iwashita, T., Shen, R. F., Ma, J. F. (2015). Physiological characterization of aluminum tolerance and accumulation in tartary and wild buckwheat. New Phytol. 205, 273–279. doi: 10.1111/nph.13011
Wang, H.-H., Qiu, Y., Yu, Q., Zhang, Q., Li, X., Wang, J., et al. (2020a). Close arrangement of CARK3 and PMEIL affects ABA-mediated pollen sterility in Arabidopsis thaliana. Plant Cell Enviro. 43, 2699–2711. doi: 10.1111/pce.13871
Wang, W., Wang, X., Wang, Y., Zhou, G., Wang, C., Hussain, S., et al. (2020b). SlEAD1, an EAR motif-containing ABA down-regulated novel transcription repressor regulates ABA response in tomato. GM Crops Food. 11, 275–289. doi: 10.1080/21645698.2020.1790287
Yao, P., Huang, Y., Dong, Q., Wan, M., Wang, A., Chen, Y., et al. (2020). FtMYB6, a light-induced SG7 R2R3-MYB transcription factor, promotes flavonol biosynthesis in tartary buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 68, 13685–13696. doi: 10.1021/acs.jafc.0c03037
Zhang, S., He, C., Wei, L., Jian, S., Liu, N. (2023). Transcriptome and metabolome analysis reveals key genes and secondary metabolites of Casuarina equisetifolia ssp. incana in response to drought stress. BMC Plant Biol. 23, 200. doi: 10.1186/s12870-023-04206-x
Zhang, L., Li, X., Ma, B., Gao, Q., Du, H., Han, Y., et al. (2017). The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol. Plant 10, 1224–1237. doi: 10.1016/j.molp.2017.08.013
Zhang, L., Takahashi, Y., Hsu, P. K., Kollist, H., Merilo, E., Krysan, P. J., et al. (2020). FRET kinase sensor development reveals SnRK2/OST1 activation by ABA but not by MeJA and high CO(2) during stomatal closure. Elife 28, 9. doi: 10.7554/eLife.56351
Zhang, Z. B., Zhu, J., Gao, J. F., Wang, C., Li, H., Li, H., et al. (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52, 528–538. doi: 10.1111/j.1365-313X.2007.03254.x
Zhao, H., Nie, K., Zhou, H., Yan, X., Zhan, Q., Zheng, Y., et al. (2020). ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 228, 596–608. doi: 10.1111/nph.16713
Zhou, M., Sun, Z., Ding, M., Logacheva, M. D., Kreft, I., Wang, D., et al. (2017). FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytol. 216, 814–828. doi: 10.1111/nph.14692
Keywords: Fagopyrum leptopodum, drought stress, flavonoids, ABA signaling, bZIP transcription factor
Citation: Wang A, Liu Y, Li Q, Li X, Zhang X, Kong J, Liu Z, Yang Y and Wang J (2023) FlbZIP12 gene enhances drought tolerance via modulating flavonoid biosynthesis in Fagopyrum leptopodum. Front. Plant Sci. 14:1279468. doi: 10.3389/fpls.2023.1279468
Received: 22 August 2023; Accepted: 21 September 2023;
Published: 11 October 2023.
Edited by:
Antonio Ferrante, University of Milan, ItalyReviewed by:
Parviz Heidari, Shahrood University of Technology, IranCopyright © 2023 Wang, Liu, Li, Li, Zhang, Kong, Liu, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmei Wang, d2FuZ2ppYW5tZWlAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.