
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 November 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1278715
This article is part of the Research Topic Evaluation of Preharvest and Postharvest Factors on Forage Crop Quality, Physiology, and Ensiling Characteristics View all 11 articles
 Xiaochen Deng1
Xiaochen Deng1 Yushan Jia2*
Yushan Jia2* Gentu Ge1,2
Gentu Ge1,2 Zhijun Wang1,2
Zhijun Wang1,2 Mingjian Liu1,2
Mingjian Liu1,2 Jian Bao1,2
Jian Bao1,2 Muqier Zhao1,2
Muqier Zhao1,2 Qiang Si1,2
Qiang Si1,2 Yichao Liu1,2
Yichao Liu1,2 Weixuan Zhao1,2
Weixuan Zhao1,2Objective: This study aimed to analyze the fermentation quality, microbial community, and volatile metabolites of oat silage harvested at two different stages, while examining the correlation between microorganisms and volatile metabolites.
Methods: Oats were harvested at two growth stages (pre-heading [PRH] and post-heading [POH] stages), followed by 90 days of natural fermentation, with 6 replicates per treatment. Pre- and post-silage samples were randomly selected for nutrient composition, fermentation parameters, microbial population, and high-throughput sequencing analysis. Volatile metabolomics analysis was also performed on samples after 90 days of fermentation to detect differences in flavor quality after silage.
Results: The effect of growth stage on the nutrient content of oats was significant, with pre-heading oats having higher crude protein and post-heading oats having higher water soluble carbohydrates content (p < 0.05). Following a 90-day fermentation period, the pH and ammonia nitrogen/total nitrogen levels in the PRH-90 (silage from pre-heading oats after 90 days of fermentation) group demonstrated a significant decrease (p < 0.05), whereas the lactic acid content was notably higher compared to the POH-90 (silage from post-heading oats after 90 days of fermentation) group (p <0.05). Lactiplantibacillus dominated in the PRH-90 group and Enterococcus dominated in the POH-90 group, with abundances of (> 86%) and (> 87%), respectively. The differential volatile metabolites of the two treatment groups were dominated by esters and terpenoids, and the differences in flavor were mainly concentrated in sweet, green, and fruity odors. The results of Kyoto encyclopedia of genes and genomes pathway enrichment analysis demonstrated three major metabolic pathways: phenylpropanoid biosynthesis, phenylalanine metabolism, and biosynthesis of secondary metabolites. Specific microorganisms were significantly correlated with flavor indicators and flavor metabolites. Lactiplantibacillus was significantly positively correlated with flavor substances indicating sweet and fruity flavors, contributing to good flavor, while Enterococcus was significantly and positively correlated with flavor substances indicating bad flavors.
Conclusion: In summary, growth stage had significant effects on nutritional components, fermentation parameters and flavor quality of oats, with the fermentation process dominated by Lactiplantibacillus leading to good flavor, while the fermentation process dominated by Enterococcus led to the development of poor flavor.
As an important grain-feed crop for livestock, oats (Avena sativa L.) are widely cultivated worldwide and have the advantages of high nutritional value, high grass yield, and good palatability (Zhou et al., 1999). Ensiling oats, a common preservation method, can be preserve oats for a long period of time, and also improves palatability and the organoleptic quality of oats (Nilsson and Rydin, 1963).
Ensiling refers to a technique of reducing the pH value of raw materials through microbial fermentation, inhibiting the growth and reproduction of detrimental microorganisms, in order to maximize the preservation of its nutrients and extend the shelf life of the raw material (Mcdonald et al., 1991). As a crop rich in protein, fiber, and trace elements, oats can be better preserved and utilized for its nutrient content after ensiling. However, microbial activity and metabolites during ensiling may have an impact on the quality and flavor of oats (Limin et al., 2018).
In recent years, microbiomics and volatile metabolomics have emerged as important branches of modern biotechnology, finding extensive applications in the fields of food science and nutrition (Shi et al., 2022; Zhao et al., 2022). Microbiomics provides insight into the species, abundance and functions of different microorganisms through the study of microbial communities, which in turn reveals their association with the quality and flavor of fermented products (Paraskevi et al., 2020). Volatile metabolomics, on the other hand, can explore the flavor characteristics of fermented products and their relationship with microbial metabolic activities by analyzing the composition and variation of volatile compounds in food (Iijima, 2014). The investigation of microbial communities and volatile metabolites during oat ensiling is essential to understand the complex biochemical processes involved in the ensiling process. Microorganisms, particularly lactic acid bacteria, assume a crucial role in the fermentation process, producing organic acids and other metabolites that contribute to the preservation and flavor development of silage oats (Liu M. et al., 2022). In addition, the composition and abundance of volatile compounds may vary depending on silage conditions, oat fertility and microbial interactions, ultimately affecting the sensory characteristics of the final product. Previous studies have focused on oat fertility, moisture content, and exogenous additives (Gardner and Wiggans, 1961; Mustafa and Seguin, 2003; Jian et al., 2015; Gomes et al., 2019; Xu et al., 2022), with fewer studies on oat silage odor after oat silage fermentation, and no in-depth studies on the relationship between microbial activity and flavor development during the fermentation process.
The objective of this study is to investigate the alterations in quality and flavor of oats following the silage process at different stages using microbiomics and volatile metabolomics. Through the analysis of microbial communities and volatile metabolites, we aim to identify the patterns and dynamics of microorganisms and volatile compounds during oat ensiling, which can lay a scientific foundation for the improvement of oat ensiling techniques and enhancing silage quality.
The oats were harvested on August 22, 2022 (pre-heading stage) and September 6, 2022 (post-heading stage), respectively, in Ar Horqin Banner, Chifeng City, Inner Mongolia Autonomous Region, China (43°21′43″-45°24′20″N, 119°02′15″-121°01′E). A part of the harvested oat material was directly returned to the laboratory for the determination of fresh oat material, while the other part was naturally air-dried to the substance with rough 70% moisture content, and then the sample was cut into 2-3cm in length. The experiment was divided into four groups with six bags (250×360mm polythene plastic bag) per group, each bag containing 400 g without any additives. The bags vacuum sealed for storage (Type: DZ-500/2E; Hefei Hanjie Packaging Machinery Inkjet Co., Ltd., Hefei, China) and opened and sampled after 90 d of storage at room temperature for the subsequent analysis. The different treatment groups are named as follows: PRH-fm, fresh oat material at pre-heading stage; POH-fm, fresh oat material at post-heading stage; PRH-90, silage from pre-heading oats after 90 days of fermentation; POH-90, silage from post-heading oats after 90 days of fermentation.
Samples of 10 g each were collected from each treatment group at 0 days and 90 days. Subsequently, 90 mL of distilled water was added, and the mixture was homogenized for two minutes by a homogenizer (Model: HX-4, Shanghai Huxi Industrial Co., Ltd., China). After filtration of the resulting extract, the pH value was measured using an acidity meter (Model: S400-B, Mettler-Toledo, LLC, America). The remaining materials were placed in an oven at 115°C for 15 minutes and then dried at 65°C for 48 hours for weighing dry matter (DM) content. The dried materials were ground into powder and stored separately. The Kjeldahl method was employed to analyze crude protein (CP) content (Sun et al., 2021). An ANKOM fiber analyzer was utilized to quantify neutral detergent fiber (NDF) and the acid detergent fiber (ADF) (Model: A2000i; Beijing Anke Borui Technology Co., Ltd., China), and the measurement of soluble carbohydrates (WSC) was performed through anthrone-sulfuric acid colorimetry (Murphy, 1958). Determination of lactic acid (LA) and acetic acid (AA) in silage after 90 days of fermentation was accomplished by high performance liquid chromatography (Fu et al., 2022). Determination of ammonia nitrogen (NH3-N) concentration used the phenol hypochlorite method according to Broderick and Kang (1980).
Ten g of fresh and silage oat samples were collected, and 90 mL of sterile water was added. The mixture was homogenized for 2 minutes using a homogenizer (Model: HX-4, Shanghai Huxi Industrial Co., Ltd., China), and the resulting bacterial solution was obtained after filtration. Culture media (Guangzhou Huankai Microbial Science and Technology Co., Ltd., Guangzhou, China) were used to isolate and enumerate various microorganisms. The culture medium for lactic acid bacteria was De Man Rogosa Sharpe agar culture medium, while nutrient agar culture medium was used for aerobic bacteria, iron-methylene blue agar culture medium for coliform bacteria, and potato glucose agar culture medium for mold and yeast. The quantification of microbial communities was performed using the plate counting method. The number of colonies was the number of viable microorganisms in the colony forming unit (cfu)/g fresh substance (FM). The number of viable microorganisms in colony forming unit (cfu)/g of fresh matter (FM) was determined by counting the colonies.
Total microbial genomic DNA was extracted from homogenized experimental fresh oat samples and silage oat samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s instructions. The quality and concentration of DNA were determined by 1.0% agarose gel electrophoresis and a NanoDrop2000 spectrophotometer (Thermo Scientific, United States). The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 799F and 1193R (Liu et al., 2016) by T100 Thermal Cycler PCR thermocycler (BIO-RAD, USA). The PCR reaction mixture consisted of 4 μL of 5× Fast Pfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of Fast Pfu polymerase, 0.2 μL of BSA, 10 ng of template DNA, and ddH2O, resulting in a final volume of 20 µL. Following the manufacturer’s instructions, the PCR product was extracted and purified from a 2% agarose gel using the PCR Clean-Up Kit (YuHua, Shanghai, China). Subsequently, the purified product was quantified using the Qubit 4.0 system, as per the manufacturer’s protocol (Thermo Fisher Scientific, USA). Sequencing data for the 16S rRNA gene sequence of the oat samples in the two treatment groups were uploaded and stored in NCBI BioProject, and the accession number can be found under PRJNA1005624.
The oat materials were harvested, weighed, and promptly frozen in liquid nitrogen for preservation. Subsequently, they were stored at -80°C until required for analysis. To prepare the samples, the frozen oat materials were ground into a fine powder using liquid nitrogen.
For each analysis, 500 mg (1 mL) of the powdered oat sample was immediately transferred into a 20 mL headspace vial (Agilent, Palo Alto, CA, USA). The vial was supplemented with 10μL of saturated NaCl solution (50μg/mL) to prevent enzyme reactions. The vials were tightly sealed using crimp-top caps equipped with TFE-silicone headspace septa (Agilent). Prior to solid-phase microextraction (SPME) analysis, each vial was placed in an oven set at 60°C for 5 minutes. Subsequently, a 120 µm DVB/CWR/PDMS fiber (Agilent) was exposed to the headspace of the sample for 15 minutes at 60°C.
The identification and quantification of volatile organic compounds (VOCs) were carried out using an Agilent Model 8890 gas chromatograph coupled with a 7000D mass spectrometer (Agilent). The GC system was equipped with a DB-5MS (5% phenyl-polydimethylsiloxane) capillary column measuring 30 m × 0.25 mm × 0.25 μm. Helium gas was utilized as the carrier gas, flowing at a linear velocity of 1.2 mL/min. The injector temperature was maintained at 250°C, while the detector temperature was set at 280°C. To achieve separation and analysis, the oven temperature was programmed as follows: initial temperature of 40°C for 3.5 minutes, followed by a ramp of 10°C/min to 100°C, then a ramp of 7°C/min to 180°C, and finally a ramp of 25°C/min to 280°C. The temperature was held at 280°C for 5 minutes.
Significant differences in the test materials were analyzed using SAS 9.2, and the 0.05 level was considered to be the least significance level between the treatment groups. Unsupervised principal component analysis (PCA) was conducted using the prcomp statistical function in R (www.r-project.org). The hierarchical cluster analysis (HCA) results for samples and metabolites were generated using the Complex Heatmap R package. Microbiota and metabolome data were performed using an online platform of Majorbio Cloud Platform (https://cloud.majorbio.com/page/tools/).
The nutritional components, fermentation products, and microbial populations of whole oats before and after silage at different stages are shown in Table 1. Distinct variations were observed in the nutritional components, fermentation products, and microbial population of oats between the pre-heading and post-heading stages. The NDF (628.27 ± 8.56), ADF (381.98 ± 17.10), and WSC (50.63 ± 1.40) of fresh oats at the post-heading stage significantly increased compared to the pre-heading stage, while CP (10.38 ± 0.41) significantly decreased (p < 0.05). Following a 90-day fermentation period, there was a significant decrease in the content of the CP, NDF, ADF, and WSC in silage oats harvested at both pre-heading and post-heading stages (p < 0.05). Silage oats at pre-heading stage had lower NDF (464.71 ± 11.83), ADF (296.53 ± 0.55), WSC (17.54 ± 0.51), but higher CP (10.80 ± 0.31) than at post-heading stage (p < 0.05). Comparing the fermentation products of the two stages, pH, AA, and NH3-N were significantly lower in pre-heading oats after ensiling compared to post-heading oats, and LA and LA/AA were found to be significantly higher in pre-heading oats compared to post-heading oats (p < 0.05). The results of microbial plate count analysis revealed no statistically significant differences in the populations of aerobic bacteria and coliform bacteria attached to fresh oats at the two stages (p < 0.05), while lactic acid bacteria and yeast attached to fresh oats at the pre-heading stage were lower than those attached to fresh oats at post-heading stage (p < 0.05). After ensiling, the populations of lactic acid bacteria and aerobic bacteria increased significantly in oats at the pre-heading stage, while the opposite was observed in oats at the post-heading stage (p < 0.05). No presence of molds was detected in any of the treatment groups.
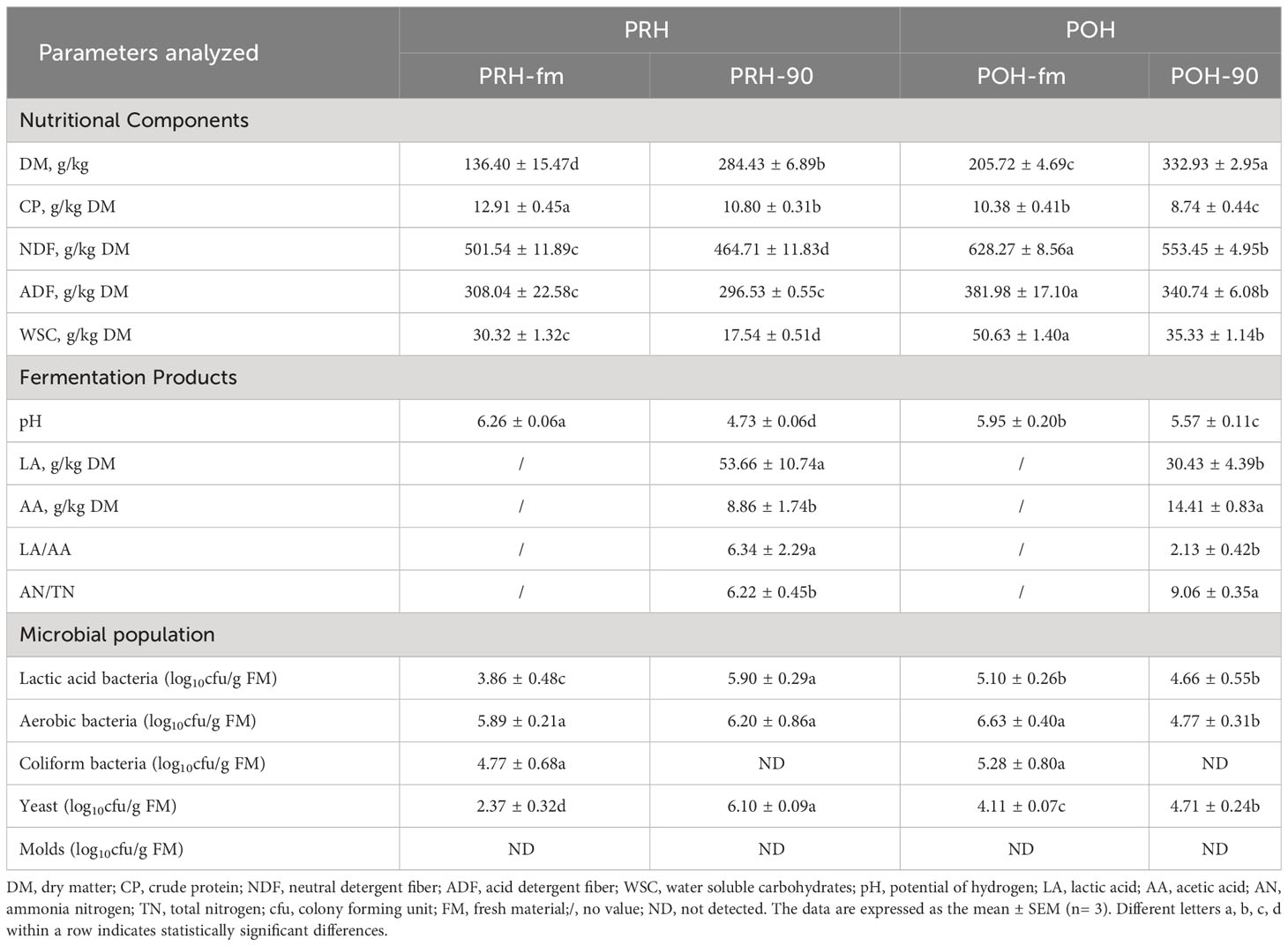
Table 1 Nutritional components, fermentation products, and microbial populations of whole oats harvested at different stages before and after ensiling.
Figure 1 presents the microbiota composition at the phylum and genus levels in both fresh oats and oat silage. At the phylum level, Proteobacteria dominated the fresh oat samples harvested at both two stages, with abundances of (> 68%) and (> 92%). Firmicutes and unclassified d Bacteria were the next most dominant phylum found in the fresh oat samples. It was noteworthy that the abundances of Bacteroidota and Actinobacteria in the fresh oat samples were (> 11%) and (> 1%) at the pre-heading stage, while they were less abundant or not detected in fresh oat samples at the post-heading stage. After 90 days of fermentation, the most abundant phylum was Firmicutes, and almost all 16S sequences belonged to the phylum of Firmicutes in both stages (PRH-90 – 99.48% vs POH-90 – 99.43%).
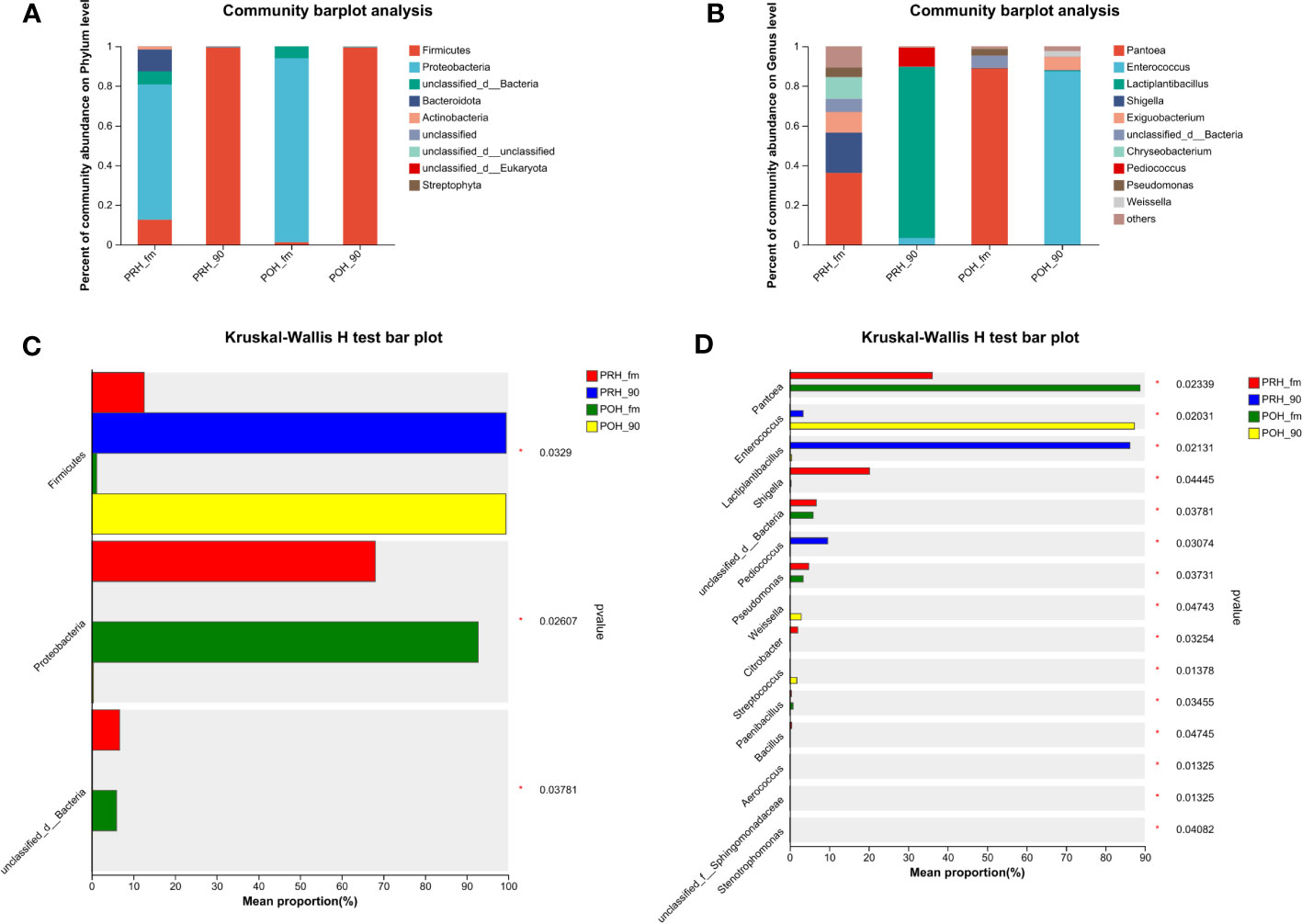
Figure 1 Relative abundance (A, B) and statistical comparison of relative abundance (C, D) of fresh and silage oat bacterial community at the phylum and genus levels. (A) Relative abundance at the phylum level. (B) Relative abundance at the genus level. (C) Statistical comparison of the relative abundance at the phylum level. (D) Statistical comparison of the relative abundance at the genus level. PRH-fm, the epiphytic microbiota of fresh oats at the pre-heading stage; PRH-90, the epiphytic microbiota of pre-heading oats after 90 days of fermentation; POH-fm, the epiphytic microbiota of fresh oats at the post-heading stage; POH-90, the epiphytic microbiota of post-heading oats after 90 days of fermentation.
Genus level compositions of the bacterial community in fresh oats and oat silage are described in Figure 1B. The most abundant genus in fresh oats was Pantoea at both stages, but there was a difference in its abundance (PRH-fm – 36.11% vs POH-fm – 88.74%). Chryseobacterium was detected in fresh oats at the pre-heading stage, while it was less abundant or not detected in fresh oats at the post-heading stage. Several other major genera detected in fresh oats were Shigella, Exiguobacterium, and Pseudomonas. Following a 90-day fermentation period, silages from raw materials harvested at different stages presented distinct microbial abundances. In pre-heading oat silage, the abundance of Lactiplantibacillus was the highest and dominant (> 86%), followed by Pediococcus (9.62%) and Enterococcus (3.37%), while in post-heading oat silage, the abundance of Enterococcus was the highest and dominant (> 87%), followed by Exiguobacterium (6.63%), Weisella (2.87%), and Lactiplantibacillus (0.46%).
As shown in Figures 1C, D, the relative abundance comparison bar chart of species was tested through the Kruskal-Wallis rank sum test. Following the ensiling process, there was a significant increase observed in the abundance of Firmicutes, whereas the abundance of Proteobacteria and unclassified d Bacteria exhibited a significant decrease compared to fresh oats (p < 0.05). Among the genera analyzed, Pantoea, Enterococcus, and Lactiplantibacillus exhibited the most significant variations in abundance at the genus level in Figure 1D. The abundance of Pantoea exhibited a significant increase following the ensiling process (p < 0.05). Enterococcus, Lactiplantibacillus, and Pediococcus significantly increased in pre-heading oat silage, and Enterococcus, Weisella, and Streptococcus significantly increased in post-heading oat silage (p < 0.05).
To identify key biomarkers in oat silage, bacterial communities in oat silage were subjected to LEfSe analysis with a linear discriminant analysis threshold of 2.0. Among the bacterial community (Figures 2A, B), a total of 29 bacterial species, including notable genera such as Bacillus, Lactiplantibacillus, and Enterococcus, were identified as significantly different between the four groups. Three genera (Bacillus, Kineococcus, and Pseudomonas) in the PRH-fm group exhibited significant differences compared to the other three groups; two genera (Pantoea and Paenibacillus) in the POH-fm group exhibited significant differences compared to the other three groups; and two genera (Lactiplantibacillus and Pediococcus) in the PRE-90 group and (Enterococcus and Weissella) in the POH-90 group exhibited significant differences compared to the other three groups.
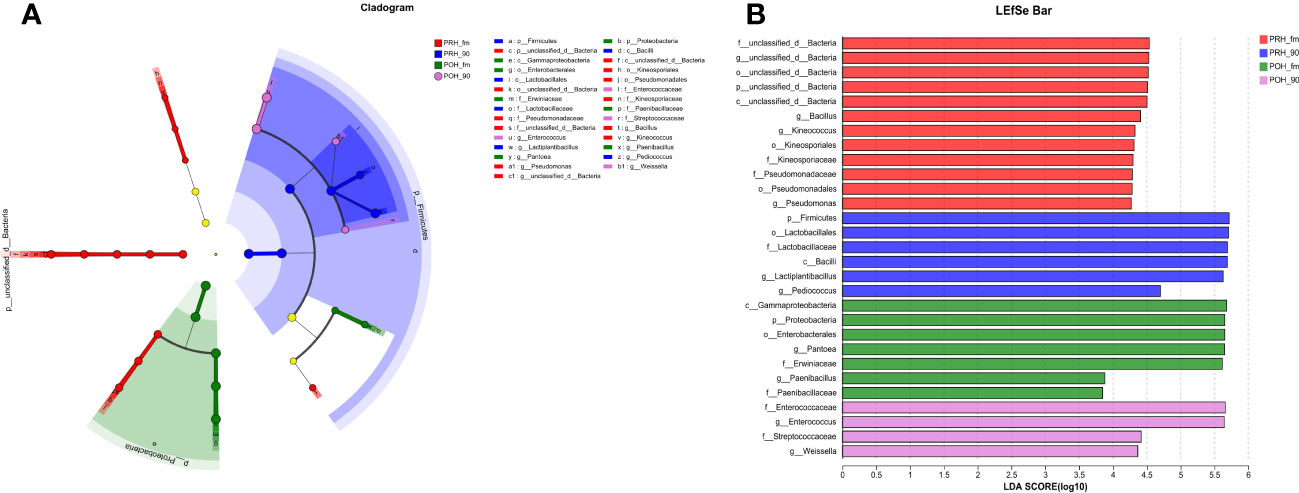
Figure 2 Evolutionary branch and LDA value distribution in fresh and silage oats harvested at different stages at the genus level OTU tables. (A) Evolutionary branch diagram of various species. The circles radiating from the inside to the outside represent classification levels ranging from phylum to genus. Different colored nodes indicate microbial taxa that are significantly enriched in the corresponding groups and that significantly affect the differences between groups. Light yellow nodes indicate microbial taxa that are not significantly different in any of the different groups or have no significant effect on the differences between groups. (B) LDA value distribution of different species (default score = 4). For LDA scores obtained by LDA analysis (linear regression analysis), the larger the LDA score, the greater the effect of species abundance on the differential effect. PRH-fm, the epiphytic microbiota of fresh oats at the pre-heading stage; PRH-90, the epiphytic microbiota of pre-heading oats after 90 days of fermentation; POH-fm, the epiphytic microbiota of fresh oats at the post-heading stage; POH-90, the epiphytic microbiota of post-heading oats after 90 days of fermentation.
The Spearman correlation heatmap which reveals the associations between fermentative metabolites and the microbial composition in oat silage is presented in Figure 3. The results show that the primary microorganisms engaged in the fermentation process were Curtobacterium, Planomicrobium, Aerococcus, Streptococcus, Exiguobacterium, Pediococcus, and Lactiplantibacillus. The CP content exhibited a significant negative correlation with Curtobacterium at p < 0.01 and Exiguobacterium at p < 0.05. The NDF content showed a positive correlation with Curtobacterium, Aerococcus, Streptococcus, and Enterococcus, while a negative correlation was observed with Lactiplantibacillus (p < 0.05). At a significance level of p < 0.001, the ADF content exhibited positive correlations with Planomicrobium, Aerococcus, and Streptococcus, while showing a negative correlation with Pediococcus. The WSC content was negatively correlated with Curtobacterium at p < 0.001 and Exiguobacterium at p < 0.01, while a positive correlation was observed with Lactiplantibacillus at p < 0.05. Aerococcus and Streptococcus showed positive associations with pH (p < 0.001), AA (p < 0.05), and AN/TN (p < 0.01), while exhibiting negative associations with LA (p < 0.05). Exiguobacterium was negatively associated with LA (p < 0.01), while a positive correlation was observed with AA at p < 0.01 and AN/TN at p < 0.05. Lactiplantibacillus exhibited a negative association with pH at p < 0.05, AA at p < 0.001, and AN/TN at p < 0.01, while demonstrating a positive association with LA and LA/AA (p < 0.001). It was noteworthy that Planomicrobium exhibited a positive correlation with pH (p < 0.05), AA (p > 0.05), and AN/TN (p > 0.05), but was negatively associated with LA and LA/AA (p > 0.05), while the opposite was true for Pediococcus.
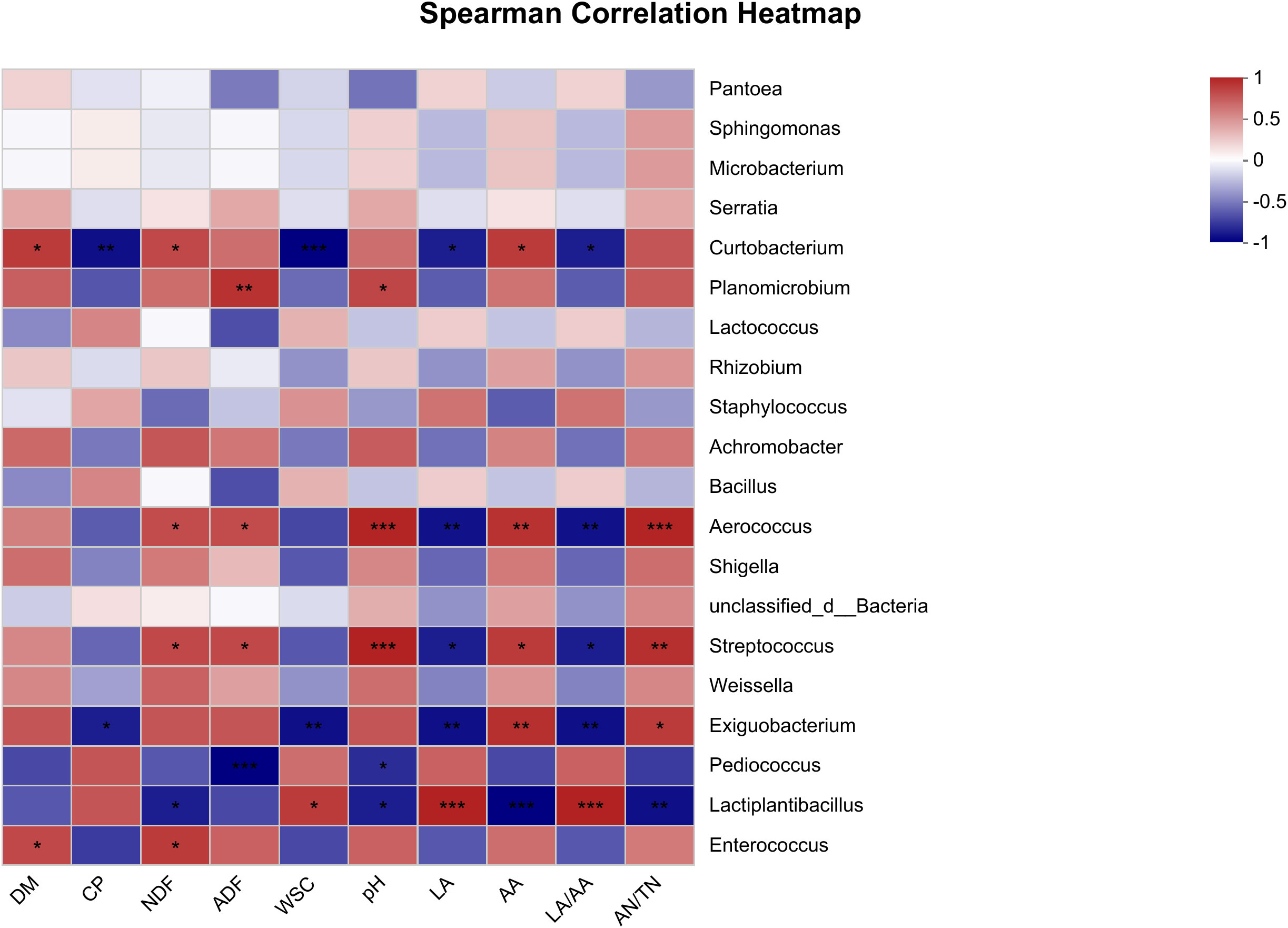
Figure 3 Spearman correlation heatmap of fermentative metabolites and bacterial community (top 20 genera) in oat silage fermented for 90 days. R-values are shown in different colors in the graph, with red indicating positive correlation (0 < r < 1) and blue indicating negative correlation (-1 < R < 0). The p-values are marked with * if 0.01 < p ≤ 0.05; **, 0.001 < p ≤ 0.01; ***, p ≤ 0.001.
In the current study, an extensive volatile metabolomics analysis was performed on oat silage samples collected at both pre-heading and post-heading stages, leading to the detection of a total of 662 volatile metabolites (Supplementary Table 1), including 30 acids, 50 alcohols, 44 aldehydes, 15 amines, 31 aromatics, 122 esters, 2 ethers, 3 halogenated hydrocarbons, 98 heterocyclic compounds, 60 hydrocarbons, 60 ketone, 7 nitrogen compounds, 14 phenols, 5 sulfur compounds, 119 terpenoids, and 2 others.
Unsupervised PCA was used to assess differences in volatile metabolites after fermentation of oats harvested at different stages. PCA effectively distinguished the PRH-90 and POH-90 samples from the QC samples, as depicted in Figure 4C. Additionally, the data points of the QC group exhibited a high concentration, suggesting a high level of repeatability in the sample collection process. Moreover, the oat silage samples were categorized into distinct regions based on the first principal component (PC1) and the second principal component (PC2), underscoring the impact of fermentation. PC1 (57.49%) and PC2 (13.78%) collectively accounted for 71.27% of the total variance (Figure 4A). Notably, the PRH-90 and POH-90 groups were significantly separated, indicating the substantial alteration of volatile metabolites in silage oats induced by fermentation.
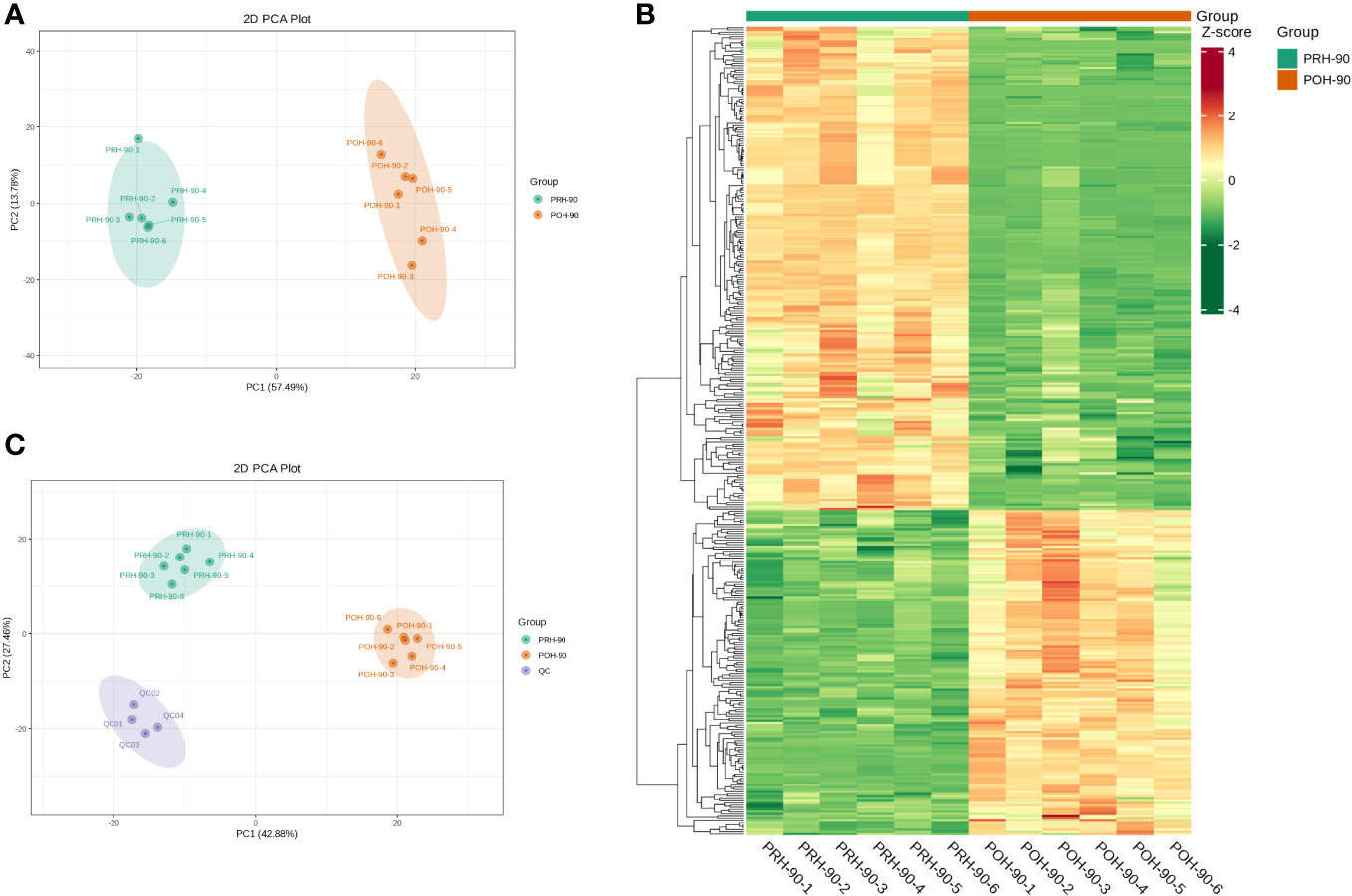
Figure 4 Multivariate statistical analysis of volatile metabolites from silage oats (PRH - 90 and POH - 90). (A) PCA scored plot of all silage oat samples. (B) Hierarchical cluster diagram of all silage oat samples. (C) PCA scored plot of all silage oat samples and the QC sample. PRH-90, volatile metabolites of pre-heading oats after 90 days of fermentation; POH-90, volatile metabolites of post-heading oats after 90 days of fermentation.
Hierarchical cluster analysis (HCA) was conducted on the accumulation patterns of metabolites across different samples, resulting in the generation of an overall clustering map of the samples (Figure 4B). The heatmap clearly illustrated the distinct profiles of PRH-90 and POH-90, emphasizing the significant influence of microorganism activities during fermentation on the metabolites of oat silage.
Figure 5A shows the radar chart of the top 10 sensory flavors selected for the highest number of annotations for the differential volatile metabolites and the annotated sensory flavor profiles obtained based on the screening criteria identified in the PRH-90 and POH-90 groups. The differential flavor metabolites were mainly enriched in sweet, green and fruity odors, with the most flavor substances indicating sweet odor reaching 56. Figure 5B shows the association network diagram of the top 10 sensory flavors with the corresponding differential flavor metabolites. Sweet odor was associated with Benzeneacetic acid, 2-methylpropyl ester, 2-Propenoic acid, 3-phenyl-, 2-Buten-1-ol, 3-methyl-, acetate, etc. The odor of green was associated with 2-Nonenal, (E)-, 2-Propenal, 3-(2-furanyl)-, Butanoic acid, 2-methyl-, hexyl ester, etc. The odor of fruity was associated with 2-Buten-1-ol, 3-methyl-, acetate, Butanoic acid, 2-methyl-, hexyl ester, Formic acid, octyl ester, etc. The above were only the top few differential volatile metabolites with the highest VIP value annotated to a sensory flavor profile.
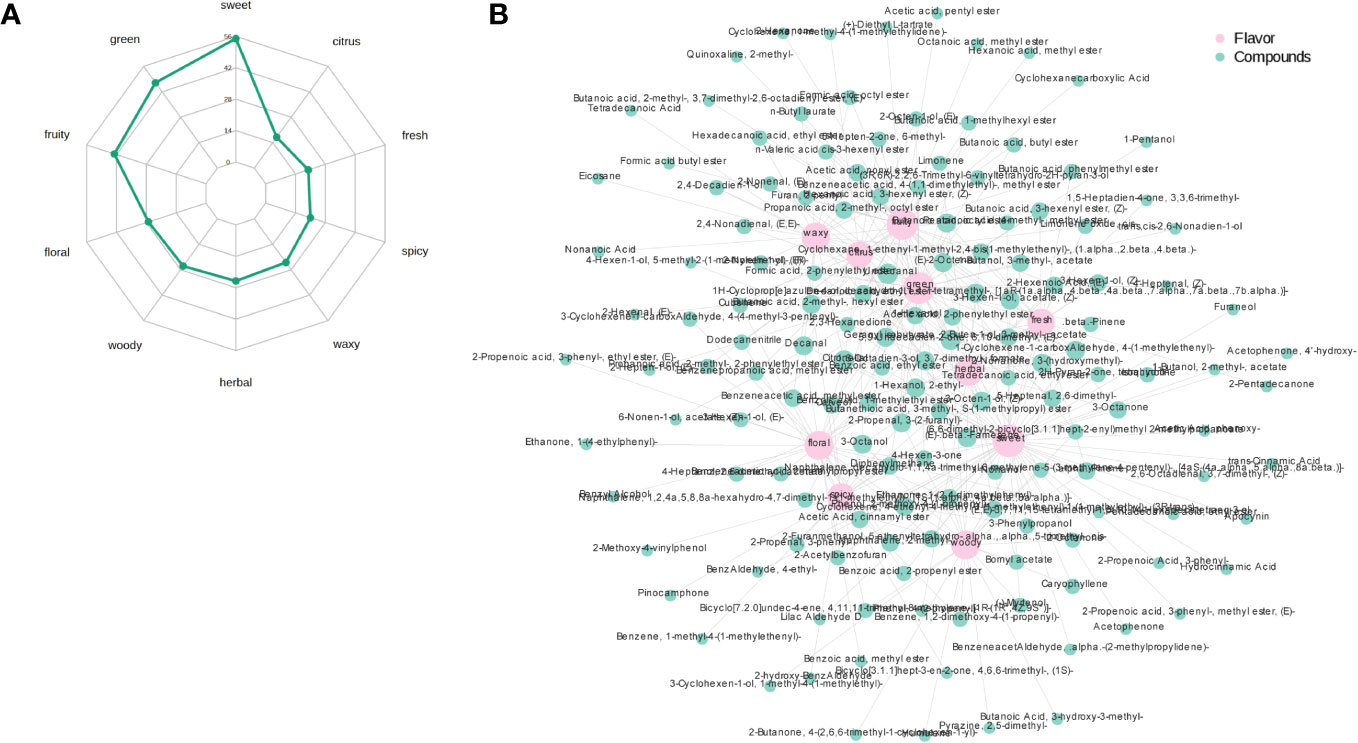
Figure 5 Radar chart of the sensory flavor profile of differential volatile metabolites and network chart of sensory flavor association with differential volatile metabolites. (A) Radar chart of the sensory flavor profile of differential volatile metabolites. The numbers corresponding to green dots are the number of differential metabolites annotated to that sensory flavor profile. (B) Network chart of sensory flavor association with differential volatile metabolites. The pink circle indicates the sensory flavor profile, the green circle indicates the differential metabolite, and the line between the two colored circles represents the differential metabolite annotated to that sensory flavor profile. When the number of annotated differential metabolites exceeds 50, the top 50 differential metabolites with the largest VIP values are displayed.
In order to elucidate the disparities in volatile metabolites between oats at different fermentation stages, orthogonal partial least squares discrimination analysis (OPLS-DA) was conducted. This analysis aimed to identify the different volatile metabolites in PRH-90 and POH-90 samples using the criteria of VIP > 1 and a P-value < 0.05. A total of 363 differential volatile metabolites were screened between the PRH-90 and POH-90 samples (Supplementary Table 2). Among them, 146 volatile metabolites were up-regulated while 217 volatile metabolites were down-regulated, and these differentially expressed metabolites were visualized in a volcano plot to depict the distribution of volatile metabolites with significant changes (Figure 6A). Figure 6B showed the grouping ring of differential metabolite classes between PRH-90 and POH-90. The classes that account for a relatively high proportion of the differential volatile metabolites were ester (19.28%), terpenoids (17.36%), heterocyclic compound (13.77%), ketone (9.64%), hydrocarbons (7.71%), alcohol (7.16%), aldehyde (7.16%), and acid (5.79%).
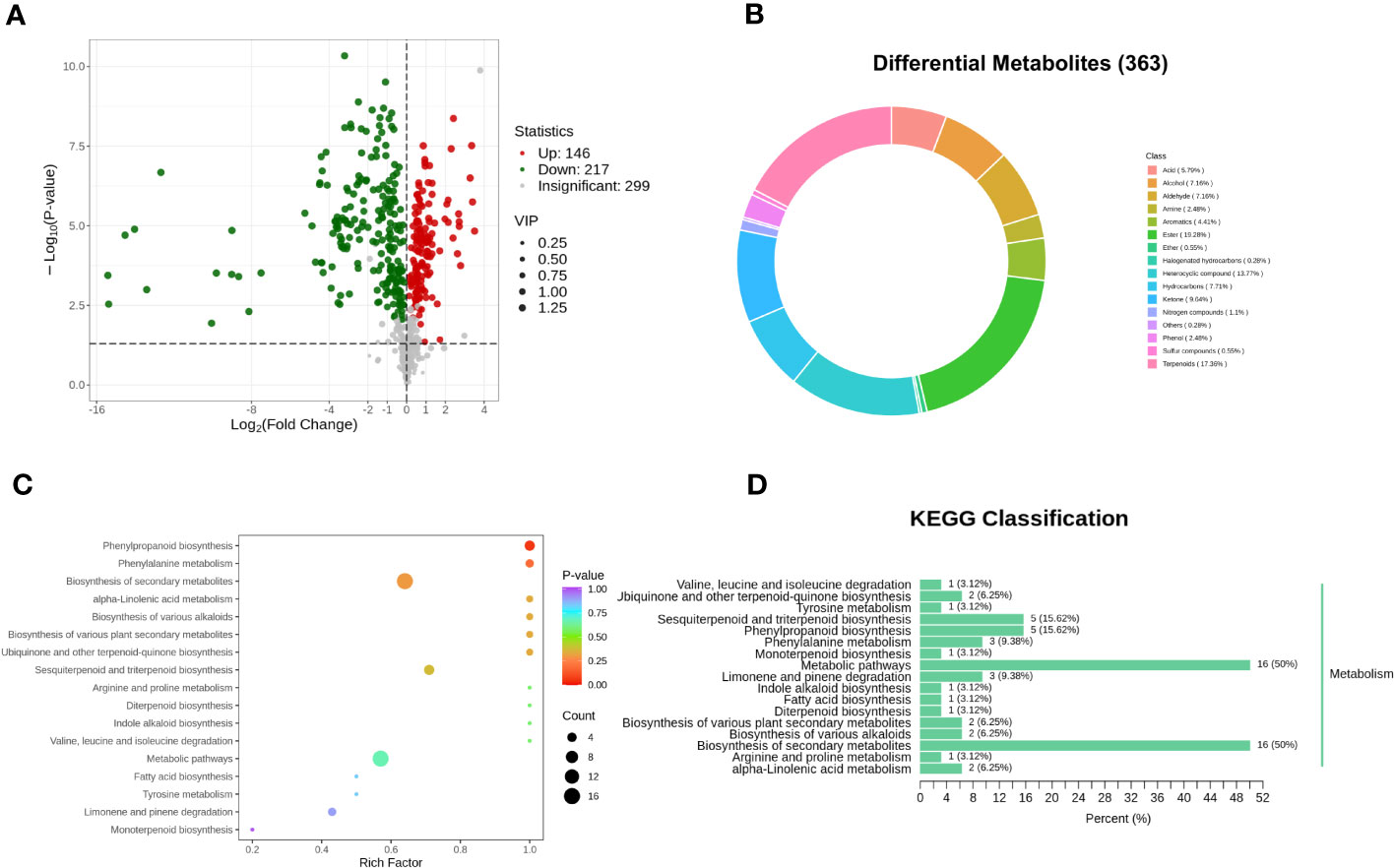
Figure 6 Differential volatile metabolites and metabolic pathways associated with differential volatile metabolites between PRH-90 and POH-90. (A) Volcano plot of differential volatile metabolites. (B) Grouping rings of differential volatile metabolites in terms of the number of classes. (C) Overview of KEGG pathway of the differential volatile metabolites. (D) Differential volatile metabolite KEGG classification plot.
Pathway enrichment analysis of 363 differential volatile metabolites was performed using the KEGG database. A total of 32 differential volatile metabolites were identified, distributed in 17 metabolic pathways. Subsequently, we performed KEGG pathway enrichment analysis (Figures 6C, D) to determine the differences in metabolic pathways between the two groups of samples. The results of KEGG pathway enrichment analysis demonstrated three major metabolic pathways: phenylpropanoid biosynthesis, phenylalanine metabolism, and biosynthesis of secondary metabolites.
To clarify the effect of microbial activity on the flavor development of oats throughout fermentation, we screened the top 50 major differential volatile metabolites (VIP > 1 and P-value < 0.05) to obtain 28 flavor metabolites (Supplementary Table 3). Subsequently, Spearman correlation analysis was conducted to examine the correlation between the top 20 microorganisms in abundance and flavor metabolites (Figure 7). The results show that a total of 11 genera were related to the first 10 sensory flavor characteristics with higher annotations. In addition, 8 genera were associated with more than 9 flavor metabolites and 3 genera were associated with more than 4 flavor metabolites of less than 9 species.
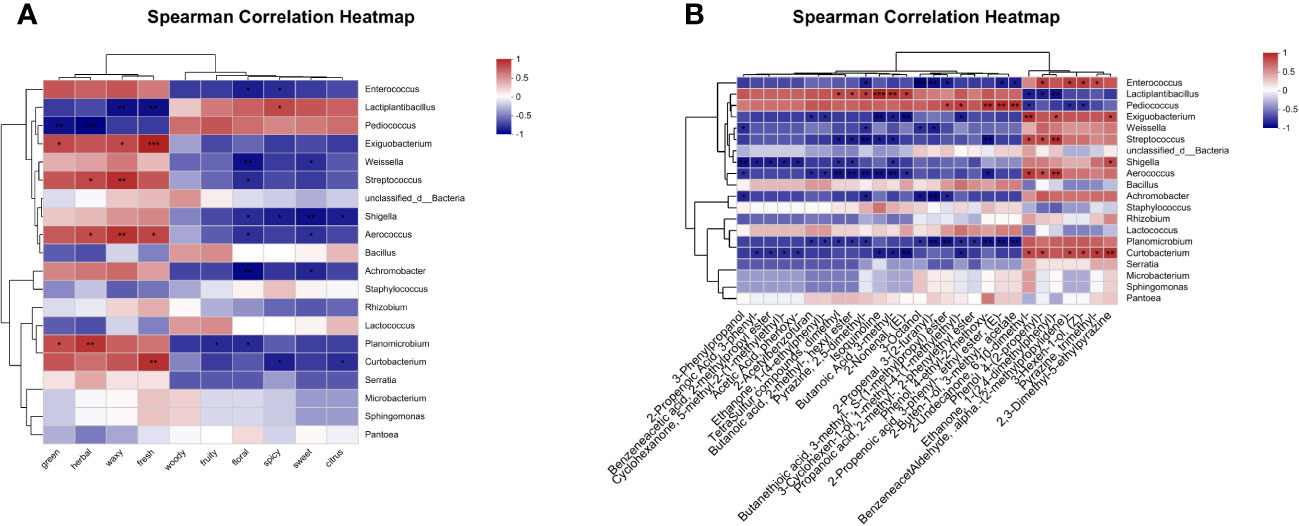
Figure 7 Correlation heatmap of bacterial community (top 20 genera) and differential metabolites after 90 days of fermentation. (A) Correlation between sweet, green, fruity, floral, woody, herbal, waxy, spicy, fresh, and citrus, as well as microorganisms. (B) Correlation between 28 flavor metabolites and microorganisms.
Figure 7A illustrates the correlations between bacterial genus levels and the top 10 sensory flavor characteristics with extensive annotations. Enterococcus was positively correlated with green, herbal, waxy, and fresh odors (p > 0.05), while demonstrating a significant negative correlation with floral and spicy odors (p < 0.05). Lactiplantibacillus showed a positive correlation with spicy odor, but was negatively correlated with waxy and fresh odors (p < 0.05). Pediococcus showed a negative correlation with green (p < 0.01) and herbal (p < 0.001) odors. Exiguobacterium showed a positive correlation with green, waxy, and fresh odors (p < 0.05). Streptococcus showed a positive correlation with herbal and waxy odors, but was negatively correlated with floral odor (p < 0.05).
The correlations between bacterial genus levels and 28 flavor metabolites are shown in Figure 7B. Enterococcus showed a negative correlation with Pyrazine, 2,5-dimethyl-, 3-Octanol, 2-Propenal, 3-(2-furanyl)-, Butanethioic acid, 3-methyl-, S-(1-methylpropyl) ester, 2-Propenoic acid, 3-phenyl-, ethyl ester, (E)-, and 2-Buten-1-ol, 3-methyl-, acetate, but was positively correlated with Phenol, 4-(2-propenyl)-, Benzeneacetaldehyde,.alpha.-(2-methylpropylidene)-, 3-Hexen-1-ol, (Z)-, and Pyrazine, trimethyl- (p < 0.05). Lactiplantibacillus was positively correlated with TetraSulfur compounds, dimethyl, Butanoic acid, 2-methyl-, hexyl ester, Pyrazine, 2,5-dimethyl-, Isoquinoline, Butanoic Acid, 3-methyl-, and 2-Nonenal, (E)-, but negatively correlated with 2-Undecanone, 6,10-dimethyl-, Phenol, 4-(2-propenyl)-, and Ethanone, 1-(2,4-dimethylphenyl)- (p < 0.05). Pediococcus showed a positive correlation with Butanethioic acid, 3-methyl-, S-(1-methylpropyl) ester, 3-Cyclohexen-1-ol, 1-methyl-4-(1-methylethyl)-, Phenol, 4-ethyl-2-methoxy-, 2-Propenoic acid, 3-phenyl-, ethyl ester, (E)-, and 2-Buten-1-ol, 3-methyl-, acetate, but was negatively correlated with 2-Undecanone, 6,10-dimethyl-, Benzeneacetaldehyde,.alpha.-(2-methylpropylidene)-, and 3-Hexen-1-ol, (Z)- (p < 0.05). Exiguobacterium was negatively correlated with 2-Acetylbenzofuran, Ethanone, 1-(4-ethylphenyl)-, Isoquinoline, Butanoic acid, 3-methyl-, 2-Nonenal, (E)-, and 3-Cyclohexen-1-ol, 1-methyl-4-(1-methylethyl)-, but negatively correlated with 2-Undecanone, 6,10-dimethyl-, Ethanone, 1-(2,4-dimethylphenyl)-, and 2,3-Dimethyl-5-ethylpyrazine (p < 0.05). Streptococcus was negatively correlated with Tetrasulfur compounds, dimethyl, Butanoic acid, 2-methyl-, hexyl ester, Pyrazine, 2,5-dimethyl-, Isoquinoline, Butanoic acid, 3-methyl-, and Phenol, 4-ethyl-2-methoxy-, but positively correlated with 2-Undecanone, 6,10-dimethyl-, Phenol, 4-(2-propenyl)-, and Ethanone, 1-(2,4-dimethylphenyl)- (p < 0.05).
Nutritional components, fermentation products, and microbial population differed in oats harvested at pre-heading and post-heading stages. As the fertility of oats increased, water content decreased and dry matter accumulation increased, which led to a gradual decrease in nutritional components, as indicated by the decrease in CP, NDF, and ADF contents. The increase of WSC content in post-heading oats may be due to the gradual accumulation of starch in the seeds as the plant matures (David et al., 2010). However, the WSC content in fresh oats at both stages was lower than the value suggested by Amer et al. (2012), and may not provide sufficient fermentation substrate for lactic acid bacteria, leading to slow growth and inability of lactic acid bacteria to dominate fermentation process. The microbial community attached to the raw material plays a crucial role in determining the quality of silage (Lin et al., 1992). In this study, a low number of lactic acid bacteria were found to be attached to fresh oats, but the count of aerobic bacteria was high, and a large number of coliform bacteria were attached to fresh oats, which are adverse for silage fermentation. Epiphytic populations of yeasts and molds were within the range of values typically reported before ensiling (Pahlow et al., 2003).
After ensiling, CP, NDF, ADF and WSC contents showed a decrease. The decrease in CP content is due to the hydrolysis of true proteins into peptide nitrogen, free amino acid nitrogen, NH3-N and non-protein nitrogen such as amines through the synergistic action of plant proteases and microbial enzymes (Bommert and Whipple, 2018). The decrease in NDF, ADF and WSC content was due to the decomposition of some cellulose and hemicellulose during silage fermentation, which were consumed as substrates together with WSC by microorganisms (Dean et al., 2005). The pH value serves as a crucial indicator of silage quality, providing insights into the preservation status and the degree of decomposition caused by undesirable microorganisms (Ren et al., 2022). In the present study, the pH level of pre-heading oat silage was significantly lower than that of post-heading oat silage (4.73 vs 5.57), which may be due to the high content of unfermentable components such as NDF and ADF in oats at the post-heading stage, which in turn affects the fermentation of oats by microorganisms. Fresh oats at the post-heading stage contained more miscellaneous bacteria, which led to the inability of lactic acid bacteria to dominate in the fermentation and high pH due to poor microbial fermentation. Correspondingly, compared to oats ensiled at the pre-heading stage, oat silage at the post-heading stage contained lower LA (30.43 g/kg DM) and higher AN/TN (9.06). The ratio of AN/TN in silage indicated the extent of protein and amino acid decomposition, and lower values of this ratio indicate less protein decomposition and better silage quality (Yuan et al., 2012). The differences in LA, AA, and AN/TN among the oat silages of both two stages in this study may be related to the type and number of microorganisms attached to the oats at the time of harvesting.
Further microbial sequencing analysis confirmed the experimental results. Throughout the fermentation process of 90 days, there was a shift in the dominant bacterial phylum from Proteobacteria and Bacteroidota to Firmicutes, which is a common occurrence in silage (Siran et al., 2020; Bai et al., 2023). This shift could be attributed to the acidic and anaerobic conditions during the ensiling process, which created a more favorable environment for the proliferation of Firmicutes (Keshri et al., 2018). The bacterial community composition at the genus level was significantly different between the two treatment groups, with Lactiplantibacillus dominating the PRH-90 group and Enterococcus dominating the POH-90 group. The reason for this finding may be due to the different raw materials and the different microorganisms attached to the raw materials. The PRH-90 group exhibited successful fermentation due to the significant role of Lactiplantibacillus in inhibiting the growth of undesirable microorganisms, reducing the ammoniacal nitrogen content, and enhancing the quality of the silage (Du et al., 2022). The POH-90 group contained more undesirable microorganisms on the raw materials and the pH dropped slowly, failing to provide the acidic environment needed for a well-fermented silage. Wang et al. (2019) reported that the naturally fermented treatment group exhibited a higher abundance of Enterococcus, which resulted in increased AA and NH3-N content, while negatively impacting silage quality. Similarly, in this study, the POH-90 group had poorer silage quality due to the dominance of Enterococcus in the fermentation, resulting in lower LA content, and higher pH, AA content and NH3-N/TN compared to the PRH-90 group.
Because the main process of silage fermentation is due to the activities of microorganisms, investigating the relationship between nutritional components, fermentation products, and microorganisms is vital for comprehending their role in the fermentation process. In the present study, a notable association was identified between eight genera and nutritional components, as well as fermentation products. The highest abundance of Lactiplantibacillus was found in the PRH-90 group and was positively correlated with WSC and LA, and negatively correlated with pH, AA and AN/TN. This showed that Lactiplantibacillus could better preserve WSC, increase LA, and lower pH and AN/TN, thus achieving good nutrient preservation and improving silage quality (Dong et al., 2022). Lower pH and AN/TN are usually better, as low pH could inhibit harmful microorganisms, and AN/TN could reflect the degree of protein degradation during silage process. Well-fermented silage feed generally has low pH and AN/TN (Filya, 2003). Pediococcus was the second most abundant genera detected in the PRH-90 group and also showed a negative correlation with pH, probably due to its synergistic effect with Lactiplantibacillus. Liu L. et al. (2022) reported that the dominance of Lactiplantibacillus on day 31 of the fermentation process was accompanied by a significant rise in the abundance of Pediococcus. In the POH-90 group, Enterococcus was the most abundant genera, but there was no significant association with indicators of silage quality. Cai (1999) inoculated samples with Enterococcus as an additive and found that Enterococcus did not improve the fermentation quality of the silage. The detection of Exiguobacterium, Streptococcus, and Aerococcus in the POH-90 group samples showed a positive correlation with the level of AN/TN, while Streptococcus and Aerococcus also showed a positive correlation with pH, which may explain the higher levels of pH and AN/TN in the POH-90 group compared to the PRH-90 group.
The odor of silage could reflect the quality of silage more directly, and volatile metabolomics has been widely used to detect flavor chemistry (Qualley and Dudareva, 2009). Ester and terpenoids were found to be the main volatiles of silage in this study, which is consistent with other studies (Di Cagno et al., 2017; Weiss, 2017). Analysis of differential volatiles in the PRH-90 and POH-90 groups detected a total of 363 differential volatiles, indicating that the flavors of the PRH-90 and POH-90 groups were significantly different, which is consistent with the PCA results, where the two groups were clearly separated, implying that the two groups were significantly different. Compared with the PRH-90 group, the content of many esters in the POH-90 group decreased significantly, while flavor substances indicating sweetness and fruitiness, such as Benzoic acid, ethyl ester, 1-Butanol, 3-methyl-, acetate, Pentanoic acid, 4-methyl-, methyl ester, Formic acid, octyl ester, and Benzeneacetic acid, 2-methylpropyl ester, which could make the fermentation more intense in terms of fruity and floral odors, were the main contributing components to the aroma of the silage (Shukui et al., 2007; Fernando et al., 2008; Sympoura et al., 2009; José, 2014). Terpenoids have been previously associated with citrus and pine odors, and an increase in the content of these compounds may enhance the fragrance (Beaulieu et al., 2015). The content of terpenoids with citrus and pine odors (2,6-Octadienal, 3,7-dimethyl-, (Z)-, Limonene oxide, cis-, 2,7-Octadien-4-ol, 2-methyl-6-methylene-, (S)-, etc.) in the POH-90 group also decreased. In addition, there was a decrease in flavor substances among acids, alcohols, ketones, aldehydes and heterocyclic compounds contributing components to the aroma. KEGG pathway enrichment analysis demonstrated metabolic pathways that may be associated with differential metabolite synthesis. The most significant metabolic pathways included phenylpropanoid biosynthesis, phenylalanine metabolism, and biosynthesis of secondary metabolites. The phenylpropanoid biosynthesis pathway is related to phenolic acid substances, and phenylalanine metabolism is an important metabolic pathway in the winemaking process and is connected to the generation of aromatic compounds (Fairbairn et al., 2017; Wang et al., 2022). In this experiment, the content of substances such as Phenol, 2-methoxy-4-(1-propenyl)-, trans-Cinnamic acid, and 2-Propenoic acid, 3-phenyl-, which represent sweet and floral odors, decreased in the POH-90 group. These substances are usually influenced by the phenylpropanoid biosynthesis and phenylalanine metabolism pathway. The biosynthesis of secondary metabolites pathway was related to the synthesis of terpenoids, and the large number of terpenoids detected in this experiment may have been influenced by the biosynthesis pathway of secondary metabolites (De et al., 2011). These findings could potentially elucidate the variation in odors observed between the PRH-90 and POH-90 groups following fermentation.
The formation of metabolites in silage is intricately linked to the presence and activities of microorganisms, and this experiment further analyzed the relationship between the dominant microorganisms (top 20 genera) and odors as well as the main differential flavor substances. The dominant genera (Lactiplantibacillus and Pediococcus) in the well-fermented PRH-90 group were positively correlated with odors that mostly indicated good flavor such as sweet, fruity, floral, and citrus, while the dominant genera (Enterococcus, Exiguobacterium, Weisella, and Streptococcus) in the poorly fermented POH-90 group were negatively correlated with odors that indicated poor flavor such as green, herbal, waxy and fresh, indicating that microorganisms play a pivotal role in shaping the final flavor of silage through their activities during the fermentation process. Butanoic acid, 2-methyl-, hexyl ester, 2-Nonenal, (E)- and 2-Propenoic acid, 3-phenyl-, ethyl ester, (E)- have been reported as aroma substances with a large impact on flavor (Palma-Harris et al., 2001; Young et al., 2004; Pu et al., 2021), and in this experiment they may have been influenced by the fermentation of Lactiplantibacillus and Pediococcus, and had an important effect on the formation of fruit odor in silage. 2-Propenoic acid, 3-phenyl-, ethyl ester, (E)- and 2-Buten-1-ol, 3-methyl-, acetate have been reported as aroma substances that, contribute significantly to the fermentation consequent aroma, and their synthesis may have been promoted by the fermentation of Pediococcus in this experiment, while their synthesis may have been inhibited by the fermentation of Enterococcus. Previous studies have shown that Lactiplantibacillus and Pediococcus contribute to the synthesis of aroma substances during fermentation, suppress undesirable odors and improve the flavor of fermented products, while the fermentation process dominated by Enterococcus could produce undesirable odors (Miller et al., 2018; Huipeng et al., 2020; Zhen et al., 2020). However, our results could not fully elucidate the role of microorganisms in the metabolism of flavor substances, and the metabolic mechanisms of flavor substances and the role of microorganisms in them need to be further explored and elucidated.
To investigate the changes in silage quality and flavor of oats harvested at pre-heading and post-heading stages, microbial diversity and volatile metabolites were analyzed throughout the oat fermentation process. The results indicated that PRH-90 group was dominated by Lactiplantibacillus with lower pH and AN/TN and better fermentation quality compared to POH-90 group, while POH-90 group was dominated by Enterococcus with higher pH and AN/TN and poorer fermentation quality compared to PRH-90 group. Esters and terpenoids were compounds that significantly contributed to flavor during oat fermentation. The differences in flavor between the two groups of oats after ensiling were mainly concentrated in sweet, green, and fruity odors. Changes in the relative abundance of major flavor metabolites were highly correlated with microorganisms. Lactiplantibacillus and Enterococcus, as the predominant genera in the oat fermentation process, are considered to contribute significantly to the flavor quality of silage oats. The results of this study provide insights into the relationship between microorganisms and flavor development during oat fermentation, and help to further understand the regulatory mechanisms of silage flavor formation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA data: PRJNA1005624 (including 12 SRA (sequence read archive) accession numbers: SRX21369993- SRX21370004).
XD: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Validation, Visualization, Writing – original draft. YJ: Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing. GG: Methodology, Project administration, Supervision, Writing – review & editing. ZW: Methodology, Writing – review & editing. ML: Methodology, Writing – review & editing, Conceptualization, Resources, Supervision. JB: Methodology, Writing – review & editing. MZ: Methodology, Writing – review & editing. QS: Methodology, Writing – review & editing. YL: Methodology, Writing – review & editing. WZ: Methodology, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Dairy Technology Innovation Center Creates Key Projects (2021 – National Dairy Center - 1).
The authors would like to thank the National Dairy Technology Innovation Center Creates Key Projects (2021 – National Dairy Center - 1) for the funding of this research and thank the staff of Wuhan Metware Biotechnology Co., Ltd (Wuhan, China) for their support during the metabolite data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that this study received funding from the National Dairy Technology Innovation Center Creates Key Projects (2021 – National Dairy Center - 1). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1278715/full#supplementary-material
Supplementary Table 1 | Volatile metabolites detected by GC-MS.
Supplementary Table 2 | Differential volatile metabolites detected by GC-MS.
Supplementary Table 3 | The flavor substances in the top 50 differential metabolites identified by VIP (VIP > 1.0) and P-value (P-value < 0.05).
Supplementary Table 4 | Flavor substances contained in the top 10 highest number of sensory flavor profiles annotated to differential metabolites identified by the comparison group based on screening criteria.
Amer, S., Hassanat, F., Berthiaume, R., Seguin, P., Mustafa, A. F. (2012). Effects of water soluble carbohydrate content on ensiling characteristics, chemical composition and in vitro gas production of forage millet and forage sorghum silages. Anim. Feed Sci. Technol. 177, 23–29. doi: 10.1016/j.anifeedsci.2012.07.024
Bai, B., Qiu, R., Wang, Z., Liu, Y., Bao, J., Sun, L., et al. (2023). Effects of cellulase and lactic acid bacteria on ensiling performance and bacterial community of caragana korshinskii silage. Microorganisms 11 (2), 337. doi: 10.3390/microorganisms11020337
Beaulieu, J. C., Lloyd, S. W., Preece, J. E., Moersfelder, J. W., Stein-Chisholm, R. E., Obando-Ulloa, J. M. (2015). Physicochemical properties and aroma volatile profiles in a diverse collection of California-grown pomegranate (Punica granatum L.) germplasm. Food Chem. 181, 354–364. doi: 10.1016/j.foodchem.2015.02.026
Bommert, P., Whipple, C. (2018). Grass inflorescence architecture and meristem determinacy. Semin. Cell Dev. Biol. 79, 37–47. doi: 10.1016/j.semcdb.2017.10.004
Broderick, G. A., Kang, J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media1. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Cai, Y. (1999). Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J. Dairy Sci. 82, 2466–2471. doi: 10.3168/jds.S0022-0302(99)75498-6
David, D. B. d, Nörnberg, J. L., de Azevedo, E. B., Brüning, G., Kessler, J. D., Skonieski, F. R. (2010). Nutritional value of black and white oat cultivars ensiled in two phenological stages. Rev. Bras. Zootecnia 39, 1409–1417. doi: 10.1590/S1516-35982010000700003
De, P. J. C., Vallim, M. A., Laneuville, T. V. (2011). What are and where are the bioactive terpenoids metabolites from Dictyotaceae (Phaeophyceae). Rev. Bras. Farmacognosia 21, 216–228. doi: 10.1590/S0102-695X2011005000079
Dean, D. B., Adesogan, A. T., Krueger, N., Littell, R. C. (2005). Effect of fibrolytic enzymes on the fermentation characteristics, aerobic stability, and digestibility of Bermudagrass silage. J. Dairy Sci. 88, 994–1003. doi: 10.3168/jds.S0022-0302(05)72767-3
Di Cagno, R., Filannino, P., Gobbetti, M. (2017). Lactic acid fermentation drives the optimal volatile flavor-aroma profile of pomegranate juice. Int. J. Food Microbiol. 248, 56–62. doi: 10.1016/j.ijfoodmicro.2017.02.014
Dong, J., Li, S., Chen, X., Sun, Z., Sun, Y., Zhen, Y., et al. (2022). Effects of Lactiplantibacillus plantarum inoculation on the quality and bacterial community of whole-crop corn silage at different harvest stages. Chem. Biol. Technol. Agric. 9, 57. doi: 10.1186/s40538-022-00326-y
Du, Z., Sun, L., Lin, Y., Chen, C., Yang, F., Cai, Y. (2022). Use of Napier grass and rice straw hay as exogenous additive improves microbial community and fermentation quality of paper mulberry silage. Anim. Feed Sci. Technol. 285, 115219. doi: 10.1016/j.anifeedsci.2022.115219
Fairbairn, S., McKinnon, A., Musarurwa, H. T., Ferreira, A. C., Bauer, F. F. (2017). The impact of single amino acids on growth and volatile aroma production by saccharomyces cerevisiae strains. Front. Microbiol. 8, 2554. doi: 10.3389/fmicb.2017.02554
Fernando, V., José, V.G., Salvador, G., Salvador, V., Paloma, M. (2008). Rational selection of non-Saccharomyces wine yeasts for mixed starters based on ester formation and enological traits. Food Microbiol. 25, 778–785. doi: 10.1016/j.fm.2008.04.015
Filya, I. (2003). The effect of Lactobacillus buchneri and Lactobacillus plantarum on the fermentation, aerobic stability, and ruminal degradability of low dry matter corn and sorghum silages. J. dairy Sci. 86, 3575–3581. doi: 10.3168/jds.S0022-0302(03)73963-0
Fu, Z., Sun, L., Hou, M., Hao, J., Lu, Q., Liu, T., et al. (2022). Effects of different harvest frequencies on microbial community and metabolomic properties of annual ryegrass silage. Front. Microbiol. 13, 971449. doi: 10.3389/fmicb.2022.971449
Gardner, F. P., Wiggans, S. C. (1961). Yield, moisture, and protein composition of spring oats cut for silage at different stages of maturity1. Agron. J. 53, 251–254. doi: 10.2134/agronj1961.00021962005300040014x
Gomes, A. L. M., Jacovaci, F. A., Bolson, D. C., Nussio, L. G., Jobim, C. C., Daniel, J. L. P. (2019). Effects of light wilting and heterolactic inoculant on the formation of volatile organic compounds, fermentative losses and aerobic stability of oat silage. Anim. Feed Sci. Technol 247, 194–198. doi: 10.1016/j.anifeedsci.2018.11.016
Huipeng, L., He, Z., Wang, X., Song, G., Chen, H., Lin, X., et al. (2020). Bacterial profiles and volatile flavor compounds in commercial Suancai with varying salt concentration from Northeastern China. Food Res. Int. 137, 109384. doi: 10.1016/j.foodres.2020.109384
Iijima, Y. (2014). Recent advances in the application of metabolomics to studies of biogenic volatile organic compounds (BVOC) produced by plant. Metabolites 4, 699–721. doi: 10.3390/metabo4030699
Jian, G., Yang, C., Liu, G.-h. (2015). Nutritional evaluation of fresh and wilted mixed silage of naked oats (Avena nuda) and alfalfa (Medicago sativa). Int. J. Agric. Biol. 17, 761–766. doi: 10.17957/IJAB/14.0003
José, M.L. (2014). Influence of the type of fiber coating and extraction time on foal dry-cured loin volatile compounds extracted by solid-phase microextraction (SPME). Meat Sci. 96, 179–186. doi: 10.1016/j.meatsci.2013.06.017
Keshri, J., Chen, Y., Pinto, R., Kroupitski, Y., Weinberg, Z. G., Sela, S. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
Limin, K., Shaver, R. D., Grant, R. J., Schmidt, R. J. (2018). Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 101, 4020–4033. doi: 10.3168/jds.2017-13909
Lin, C., Bolsen, K. K., Brent, B. E., Fung, D. Y. C. (1992). Epiphytic lactic acid bacteria succession during the pre-ensiling and ensiling periods of alfalfa and maize. J. Appl. Bacteriol 73, 375–387. doi: 10.1111/j.1365-2672.1992.tb04992.x
Liu, L., Tao, Y., Li, Y., Deng, X., Liu, G., Yao, Y., et al. (2022). Isolation and characterization of bacteria that produce quorum sensing molecules during the fermentation and deterioration of pickles. Int. J. Food Microbiol. 379, 109869. doi: 10.1016/j.ijfoodmicro.2022.109869
Liu, M., Wang, Y., Wang, Z., Bao, J., Zhao, M., Ge, G., et al. (2022). Effects of isolated LAB on chemical composition, fermentation quality and bacterial community of stipa grandis silage. Microorganisms 10, 2463. doi: 10.3390/microorganisms10122463
Liu, C., Zhao, D., Ma, W., Guo, Y., Wang, A., Wang, Q., et al. (2016). Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 100, 1421–1426. doi: 10.1007/s00253-015-7039-6
Mcdonald, P., Henderson, A. R., Heron, S. J. E. (1991). The biochemistry of silage. Exp. Agric. 28 (1), 125–125. doi: 10.1016/0377-8401(82)90026-8
Miller, P., Haveroen, M. E., Solichová, K., Merkl, R., McMullen, L. M., Míková, K., et al. (2018). Shelf life extension of liquid whole eggs by heat and bacteriocin treatment. Czech J. Food Sci. 28, 280–289. doi: 10.17221/142/2010-CJFS
Murphy, R. P. (1958). A method for the extraction of plant samples and the determination of total soluble carbohydrates. J. Sci. Food Agric. 9, 714–717. doi: 10.1002/jsfa.2740091104
Mustafa, A. F., Seguin, P. (2003). Effects of stage of maturity on ensiling characteristics and ruminal nutrient degradability of oat silage. Arch. Anim. Nutr. 57, 347–358. doi: 10.1080/00039420310001607707
Nilsson, R., Rydin, C. (1963). Fermentation as a means of preserving organic materials. Acta Chemica Scandinavica 17, 174–179. doi: 10.3891/acta.chem.scand.17s-0174
Pahlow, G., Muck, R. E., Driehuis, F., Oude Elferink, S. J. W. H., Spoelstra, S. F. (2003). Microbiology of ensiling. Agronomy 42, 33. doi: 10.2134/agronmonogr42.c2
Palma-Harris, C., McFeeters, R. F., Fleming, H. P. (2001). Solid-phase microextraction (SPME) technique for measurement of generation of fresh cucumber flavor compounds. J. Agric. Food Chem. 49, 4203–4207. doi: 10.1021/jf010182w
Paraskevi, B., Chrysanthi, M., Kambouris Manousos, E., Ioannis, G. (2020). “Chapter 12 - microbiomic prospects in fermented food and beverage technology,” in Translational and Applied Genomics (London: Academic Press), 245–277.
Pu, D., Shan, Y., Duan, W., Huang, Y., Liang, L., Yan, Y., et al. (2021). Characterization of the key aroma compounds in the fruit of litsea pungens hemsl. (LPH) by GC-MS/O, OAV, and sensory techniques. J. Food Qual. 4, 1–9. doi: 10.1155/2021/6668606
Qualley, A. V., Dudareva, N. (2009). Metabolomics of plant volatiles. in dmitry A. Belostotsky (ed.). Plant Syst. Biol. 553, 329–343. doi: 10.1007/978-1-60327-563-7_17
Ren, X., Tian, H., Zhao, K., Li, D., Xiao, Z., Yu, Y., et al. (2022). Research on pH Value Detection Method during Maize Silage Secondary Fermentation Based on Computer Vision. Agriculture 12, 1623. doi: 10.3390/agriculture12101623
Shi, N., Li, L., Wu, Y., Xiang, H., Li, C., Chen, S., et al. (2022). Exploring the roles of microorganisms and metabolites in the fermentation of sea bass (Lateolabrax japonicas) based on high-throughput sequencing and untargeted metabolomics. Food Sci. Technol. 167, 113795.
Shukui, Z., Lu, X., Ji, K., Guo, K., Li, Y., Wu, C., et al. (2007). Characterization of flavor compounds in Chinese liquor Moutai by comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry. Analytica Chimica Acta 597, 340–348. doi: 10.1016/j.aca.2007.07.007
Siran, W., J, Z., Dong, Z., Li, J., Kaka, N. A., Shao, T. (2020). Sequencing and microbiota transplantation to determine the role of microbiota on the fermentation type of oat silage. Bioresource Technol. 309, 123371. doi: 10.1016/j.biortech.2020.123371
Sun, L., Na, N., Li, X., Li, Z., Wang, C., Wu, X., et al. (2021). Impact of packing density on the bacterial community, fermentation, and in vitro digestibility of whole-crop barley silage. Agriculture 11, 672. doi: 10.3390/agriculture11070672
Sympoura, F., Cornu, A., Tournayre, P., Massouras, T., Berdagué, J. L., Martin., B. (2009). Odor compounds in cheese made from the milk of cows supplemented with extruded linseed and α-tocopherol. J. Dairy Sci. 92, 3040–3048. doi: 10.3168/jds.2008-1802
Wang, Y., Chen, X., Wang, C., He, L., Zhou, W., Yang, F., et al. (2019). The bacterial community and fermentation quality of mulberry (Morus alba) leaf silage with or without Lactobacillus casei and sucrose. Bioresour Technol. 293, 122059. doi: 10.1016/j.biortech.2019.122059
Wang, Y., Li, C., Lin, J., Sun, Y., Wei, S., Wu, L. (2022). The impact of different withering approaches on the metabolism of flavor compounds in oolong tea leaves. Foods 11, 3601. doi: 10.3390/foods11223601
Weiss, K. (2017). Volatile organic compounds in silages–Effects of management factors on their formation: A review. Slovak J. Anim. Sci. 50, 55–67.
Xu, J., Zhang, K., Lin, Y., Li, M., Wang, X., Yu, Q., et al. (2022). Effect of cellulase and lactic acid bacteria on the fermentation quality, carbohydrate conversion, and microbial community of ensiling oat with different moisture contents. Front. Microbiol. 13, 1013258. doi: 10.3389/fmicb.2022.1013258
Young, J. C., Chu, C. L., Lu, X., Zhu, H. (2004). Ester variability in apple varieties as determined by solid-phase microextraction and gas chromatography-mass spectrometry. J. Agric. Food Chem. 52, 8086–8093. doi: 10.1021/jf049364r
Yuan, X., Yu, C., Shimojo, M., Shao., T. (2012). Improvement of fermentation and nutritive quality of straw-grass silage by inclusion of wet hulless-barley distillers’ Grains in tibet. Asian-Australas J. Anim. Sci. 25, 479–485. doi: 10.5713/ajas.2011.11435
Zhao, D., Hu, J., Chen, W. (2022). Analysis of the relationship between microorganisms and flavor development in dry-cured grass carp by high-throughput sequencing, volatile flavor analysis and metabolomics. Food Chem. 368, 130889. doi: 10.1016/j.foodchem.2021.130889
Zhen, H., Chen, H., Wang, X., Lin, X., Ji, C., Li, S., et al. (2020). Effects of different temperatures on bacterial diversity and volatile flavor compounds during the fermentation of suancai, a traditional fermented vegetable food from northeastern China. Food Sci. Technol. 118, 108773.
Keywords: oats, microbial community, volatile metabolites, flavor, fermentation quality
Citation: Deng X, Jia Y, Ge G, Wang Z, Liu M, Bao J, Zhao M, Si Q, Liu Y and Zhao W (2023) Microbiomics and volatile metabolomics-based investigation of changes in quality and flavor of oat (Avena sativa L.) silage at different stages. Front. Plant Sci. 14:1278715. doi: 10.3389/fpls.2023.1278715
Received: 16 August 2023; Accepted: 23 October 2023;
Published: 06 November 2023.
Edited by:
Siran Wang, Nanjing Agricultural University, ChinaReviewed by:
Yin Wan, Nanchang University, ChinaCopyright © 2023 Deng, Jia, Ge, Wang, Liu, Bao, Zhao, Si, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yushan Jia, anlzX25tQHNpbmEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.