- School of Agricultural Science and Engineering, Liaocheng University, Liaocheng, China
Actin-depolymerizing factors (ADFs) are highly conserved small-molecule actin-binding proteins found throughout eukaryotic cells. In land plants, ADFs form a small gene family that displays functional redundancy despite variations among its individual members. ADF can bind to actin monomers or polymerized microfilaments and regulate dynamic changes in the cytoskeletal framework through specialized biochemical activities, such as severing, depolymerizing, and bundling. The involvement of ADFs in modulating the microfilaments’ dynamic changes has significant implications for various physiological processes, including plant growth, development, and stress response. The current body of research has greatly advanced our comprehension of the involvement of ADFs in the regulation of plant responses to both biotic and abiotic stresses, particularly with respect to the molecular regulatory mechanisms that govern ADF activity during the transmission of stress signals. Stress has the capacity to directly modify the transcription levels of ADF genes, as well as indirectly regulate their expression through transcription factors such as MYB, C-repeat binding factors, ABF, and 14-3-3 proteins. Furthermore, apart from their role in regulating actin dynamics, ADFs possess the ability to modulate the stress response by influencing downstream genes associated with pathogen resistance and abiotic stress response. This paper provides a comprehensive overview of the current advancements in plant ADF gene research and suggests that the identification of plant ADF family genes across a broader spectrum, thorough analysis of ADF gene regulation in stress resistance of plants, and manipulation of ADF genes through genome-editing techniques to enhance plant stress resistance are crucial avenues for future investigation in this field.
Introduction
Plants are sessile growth organisms that face various unfavorable biotic and abiotic stresses during their life cycle (Verma et al., 2016). Biotic and abiotic stresses refer to biological or environmental factors that are detrimental to the survival and growth of plants at all phenological stages and can cause significant damage to agricultural production (Zhu, 2016; Li et al., 2019). Plants use numerous physiological and biochemical mechanisms to mitigate the impact of adverse conditions on their growth and survival (Hasanuzzaman et al., 2013; Zhao et al., 2020).
The actin cytoskeleton is an essential component of the plant cellular skeletal system. It not only maintains the shape of the cell by providing it with a three-dimensional structure but also participates in regulating various life activities, including cell motility, growth, division, differentiation, organelle movement, endocytosis, exocytosis, and responses to physiological and environmental signals (Staiger, 2000; Pollard and Cooper, 2009; Henty-Ridilla et al., 2013; Wang and Mao, 2019). In cells, actin exists in two forms, i.e., globular actin (G-actin), which is generally present as a monomer, and filamentous actin (F-actin), which exists as polymerized filaments (also known as microfilaments). The latter is the form that primarily performs biological functions (Staiger, 2000; Pollard, 2016). During microfilament formation, three G-actin molecules initially aggregate to form a nuclei (called nucleation), following which other G-actin molecules are gradually added to the ends of the nuclei to elongate the filament (called elongation) (Pollard and Cooper, 2009). The elongation rates considerably differ between the two ends, with the faster-elongating end termed the “barbed end” and the slower-elongating end termed the “pointed end” (Li et al., 2015). Different microfilaments subsequently crosslink to form a three-dimensional network structure or align in parallel to each other to form thicker bundles of microfilaments (Uribe and Jay, 2009).
In response to physiological or environmental signals, the two cellular forms of actin constantly polymerize and depolymerize, resulting in highly dynamic changes in microfilaments, ensuring a rapid cellular response (Pollard and Cooper, 2009). The microfilaments and their dynamic changes play an important role in regulates plant stress tolerance (Li et al., 2015; Porter and Day, 2016; Wang et al., 2022). For example, when actin polymerization is blocked with the inhibitor latrunculin B, plants are more susceptible to pathogenic and nonpathogenic bacteria (Henty-Ridilla et al., 2013), and numerous studies have also revealed that actin dynamics correlated with the plant response to abiotic stress, such as cold (Pokorna et al., 2004), heat (Müller et al., 2007; Malerba et al., 2010), salt (Wang et al., 2010), and alkaline (Zhou et al., 2010). Understanding the regulation of microfilament dynamics will enrich our understanding of plant stress response.
Actin polymerization, depolymerization, crosslinking, and bundling are processes regulated by a series of actin-binding proteins (ABPs), and hundreds of ABPs have been discovered in eukaryotes (Li et al., 2015; Porter and Day, 2016). In general, ABPs interact with actin and regulate their dynamic changes, thereby participating in various physiological activities of the cell (Pollard, 2016; Augustine et al., 2021). Actin-depolymerizing factor (ADF) is a small-sized (15-22 kDa) and highly conserved ABP ubiquitously exist in eukaryotic cells (Maciver and Hussey, 2002). The first ADF was isolated from chicken embryo brain cells, and the authors found that the isolated protein is distinct from other ABPs in its isoelectric point and has the capacity to depolymerize F-actin (Bamburg et al., 1980). Subsequently, ADF genes have been cloned from various eukaryotes, including fungi, animals and plants (Maciver and Hussey, 2002; Inada, 2017). ADF can bind both G-actin and F-actin and regulate remodeling of microfilament framework via its specialized biochemical activities (Hussey et al., 2002; Andrianantoandro and Pollard, 2006). Initially, ADF was found to severs or depolymerizes F-actin into shorter fragments or G-actin monomers, which provides new sites for actin filament initiation and supplies additional actin monomers for further polymerization (Bamburg et al., 1980; Maciver et al., 1991; Carlier et al., 1997). Subsequently, the biochemical activity of ADF was found to depend on the local concentration in cells (Andrianantoandro and Pollard, 2006). Low concentrations of ADF favor severing or deploymerizing whereas high concentrations favor actin nucleation as well as accelerate Pi release from ADP-Pi subunits in filaments and dissociation of branches formed by actin-related protein2/3 complex (Blanchoin and Pollard, 1999; Blanchoin et al., 2000; Andrianantoandro and Pollard, 2006). This range of biochemical activities makes ADF an important factor for regulating dynamic changes in actin filaments, which involves in most of the cellular processes of eukaryotes (Staiger, 2000; Pollard, 2016). Hence, ADFs widely participates in numerous plant growth and development processes, including flowering (Burgos-Rivera et al., 2008), pollen development and pollen tube growth (Chen et al., 2002; Daher and Geitmann, 2012; Zheng et al., 2013), cell elongation and secondary cell wall formation (Wang et al., 2009a), and responses to various biotic and abiotic stresses (Huang et al., 2012; Tang et al., 2016; Inada, 2017; Zhang et al., 2017).
Previous studies have reviewed plant ADF family genes in terms of evolutionary classification, expression profiles, transcriptional regulation, biochemical activity, and biological function (Hussey et al., 2002; Maciver and Hussey, 2002; Inada, 2017). However, there are many papers published and significant progress has been made since then, especially those regarding the molecular mechanisms underlying its involvement in signal responses to stress conditions. Recently, the publication of new plant genome sequences has led to the systematic reporting of ADF gene families from a dozen plant species (Feng et al., 2006; Ruzicka et al., 2007; Huang et al., 2012; Khatun et al., 2016; Huang et al., 2020; Xu et al., 2021; Sun et al., 2023). Furthermore, research on plant ADF genes and their involvement in stress responses has gradually received more attention, with significant progress being made in recent years. In this paper, we reviewed the research progress of the responses and molecular regulatory mechanisms of plant ADF genes to different forms of biotic and abiotic stresses. This review aims to provide a thorough understanding of the role played by ADF genes in plant stress responses and the molecular regulatory mechanisms that underlie them, and offers suggestions for future research directions in this field.
Expression profiles and biochemical activity diversification of plant ADF genes
Varying numbers of ADF genes in different species
Although ADF genes exist in all eukaryotes, the number of these genes considerably varies among species. Single-cell eukaryotes and animal genomes contain no more than three members of the ADF family. For example, yeast (Saccharomyces cerevisiae), roundworm (Caenorhabditis elegans), and the alga Chlamydomonas reinhardtii all have only one ADF gene (Gunning et al., 2015). Three ADF members are present in the genomes of zebrafish (Danio rerio), jungle fowl (Gallus gallus), and humans (Homo sapiens) (Gunning et al., 2015). Conversely, land plants possess an expanded ADF gene family. For example, eleven ADF genes have been identified in Arabidopsis thaliana (Feng et al., 2006; Ruzicka et al., 2007), rice (Oryza sativa; Feng et al., 2006; Huang et al., 2012), and tomato (Solanum lycopersicum; Khatun et al., 2016) each; eight in cucumber (Cucumis sativus; Liu et al., 2016) and Antarctic hairgrass (Deschampsia antarctica; Byun et al., 2021) each; nine in common bean (Phaseolus vulgaris; Ortega-Ortega et al., 2020); ten in pigeon pea (Cajanus albicans; Cao et al., 2020); thirteen in maize (Zea mays; Huang et al., 2020); fourteen in poplar (Populus trichocarpa; Roy-Zokan et al., 2015); eighteen in soybean (Glycine max; Sun et al., 2023); twenty-five in wheat (Triticum aestivum; Xu et al., 2021); twenty-seven in banana (Musa acuminata; Nan et al., 2017); and thirty-seven in upland cotton (Gossypium hirsutum; Sun et al., 2021). In contrast to single-cell eukaryotes and animals, plants exhibit a multitude of distinct and functionally specialized actin filament systems, alongside a larger actin gene family (McDowell et al., 1996; Zhang et al., 2010; Gunning et al., 2015). Likewise, an increased number of genes has been observed in numerous ABP gene families, such as profilin, formin, and villin (Bao et al., 2011; Gunning et al., 2015; Duan et al., 2021; Zhou et al., 2023). The diverse members within these extensive gene families, believed to have originated from gene duplication events, are presumed to be expressed in a highly differential manner, specific to tissues, environmental conditions, and temporal factors (McDowell et al., 1996; Bao et al., 2011). This expression pattern enables plants to dynamically restructure the actin cytoskeleton in response to evolving requirements throughout their growth and development processes (Gunning et al., 2015). Regarding plant ADF genes, the expansion of the gene family may facilitate their expression in intricate biological profiles, enabling differentiation into various biological functions, as elucidated in numerous subsequent articles.
Expression profiles of ADF genes in Arabidopsis
Among all land plants, the expression characteristics and biological functions of ADF genes in Arabidopsis have been the most extensively studied. Phylogenetic analysis reveals that the eleven AtADF genes can be divided into four groups (I–IV), with group II further divided into subgroups II-a and II-b (Feng et al., 2006; Ruzicka et al., 2007). Within each group, AtADF genes demonstrate comparable tissue-specific expression patterns, although notable disparities in expression characteristics exist among members across distinct groups. Group I comprises four genes: AtADF1, AtADF2, AtADF3, and AtADF4. These genes are stably expressed at high levels in all plant tissues/organs except in pollen. Overall, AtADF3 exhibits the highest expression level. Group II comprises four genes: AtADF7, AtADF8, AtADF10, and AtADF11, which are preferentially expressed in cell types demonstrating polarized growth characteristics. AtADF7 and AtADF10 are members of the subgroup II-a and are specifically expressed in mature pollen grains and pollen tubes, whereas AtADF8 and AtADF11 (subgroup II-b) are specifically expressed in root hairs and root epidermal cells that can differentiate into root hairs. Group III comprises only two genes, AtADF5 and AtADF9, which exhibit lower expression levels in vegetative tissues but are highly expressed in cells undergoing rapid growth or differentiation, including callus tissues, young leaves, and meristematic regions. Group IV only contains one gene, AtADF6, which is stably expressed in all tissues, including pollen (Ruzicka et al., 2007). In a study by Dong et al. (2001), ADF gene promoter–GUS fusions were employed for genetic transformation in Arabidopsis, and the authors found that AtADF1 and AtADF6 were expressed in the vascular tissues of all organs, while AtADF5 was only expressed in the root apical meristem. Immunocytochemical analysis further revealed that proteins encoded by group I genes are localized to the nucleus and cytoplasm simultaneously, while proteins encoded by group II genes are mainly localized to the cytoplasm of pollen tubes and the apical regions of root hairs (Ruzicka et al., 2007). These results indicate that the expression and localization of ADFs are precisely regulated, and different ADFs are required to function in distinct tissue types and subcellular locations.
Expression profiles of ADF genes in several crops
Previous studies have reported that ADF genes in other plants exhibit tissue-specific expression characteristics similar to those found in Arabidopsis. Here we take the expression patterns of ADF genes in several crops, including rice, maize, wheat, cotton, tomato, and soybean, as examples. OsADF2, OsADF4, OsADF5, and OsADF11 are persistently expressed in the roots, stems, leaves, sheaths, spikelets, and seeds of rice, while OsADF9 is specifically expressed in spikelets during the heading stage (Huang et al., 2012). ZmADF3, ZmADF4, ZmADF5, ZmADF6 and ZmADF10 showed relatively higher expression in all tissues of maize, whereas ZmADF1, ZmADF2, ZmADF7, ZmADF12, and ZmADF13 showed high expression levels in reproductive organs such as tassel, anther, and pollen (Huang et al., 2020). Of the twenty-five TaADF genes in wheat, nine of them exhibit anther-specific expression, while the others are diversely expressed in different tissues (Xu et al., 2021). In upland cotton, GhADF6 and GhADF8 are predominantly expressed in petals while GhADF7 is highly expressed in anthers (Li et al., 2010). Among the nine SlADF genes in tomato, SlADF1, SlADF3 and SlADF10 are predominately expressed in flowers and specifically in the stamen compared to other parts (Khatun et al., 2016). In soybean, our lab used genome-wide identification techniques to show that the soybean ADF gene family displays tissue-specific expression patterns very similar to those found in Arabidopsis (Sun et al., 2023). In short, GmADF genes in groups I and IV are expressed throughout the soybean plant, those of group II are specifically expressed in flowers, while the expression level of genes in group III is lower than that in groups I and IV (Sun et al., 2023).
Biochemical activity diversification of plant ADFs
The expansion and diversification of the expression patterns of ADF gene family members in land plants imply that their biochemical activities or biological functions may have been differentiated during evolution (Ren and Xiang, 2007; Tholl et al., 2011). Biochemical experiments have shown that nine AtADF members in groups I, II, and IV of Arabidopsis can sever or depolymerize F-actin, with the four members of group I being the most active (Nan et al., 2017). The two AtADF members in group III do not show severing or depolymerizing activities, but instead have the ability to promote F-actin bundling (Tholl et al., 2011; Nan et al., 2017). Three crucial amino acid alterations were confirmed to be responsible for these divergent biochemical activities. Taking AtADF9 from the group III as an example, the 3rd Leu, 4th Lys, and 18th Lys (the corresponding amino acid residue in AtADFs from other three groups are Met, Ala, Leu/Thr, respectively) are necessary for its F-actin bundling activity (Nan et al., 2017). By comparing variations in the amino acid sequences of the Arabidopsis protein and its homologs in other plants, Nan et al. (2017) suggested that this biochemical activity divergence may be widely present in angiosperms.
Function of plant ADF genes in biotic stress
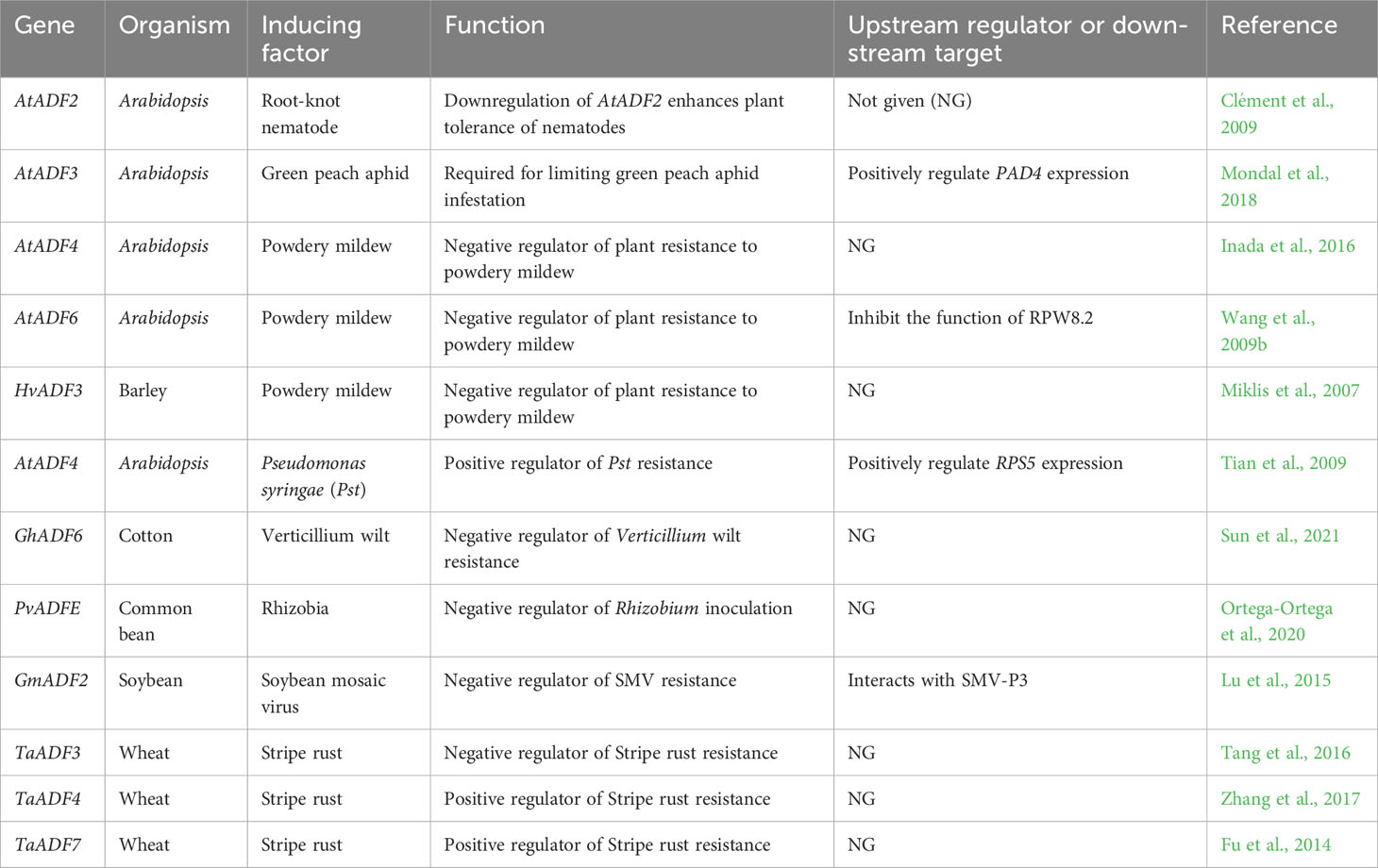
Biological stress of plants refers to the inhibition of their growth, development, and survival caused by biological factors such as pests, bacteria, fungi, viruses, etc. (Verma et al., 2016; Jiang et al., 2017). These harmful animals or microbes attack numerous agricultural crops, causing devastating effects on plant productivity and yield (Leonard et al., 2017). Increasing studies showed that plant ADF genes and actin cytoskeleton dynamics are widely involved in plant responses to biotic stress (Table 1). Understanding the biological function and the regulatory mechanism of these ADFs is essential for the development of biotic stress-tolerant crops. In this section, we will summarize the research progress of ADF gene in plant response to biotic stress.
Pest resistance
Resistance to root-knot nematode
The root-knot nematode (Meloidogyne incognita) is a highly specialized and polyphagous plant-pathogenic nematode. Its second-stage juveniles can penetrate plant root apical meristems via stylets. Thereafter, they migrate within the plant and establish parasitic relationships with vascular tissues, leading to the formation of giant cells and production of galls (Fuller et al., 2008). Moreover, the cytoskeletal system of giant cells undergoes rearrangement during its altered development (Jammes et al., 2005). The expression levels of five ADF genes (i.e., AtADF2–AtADF6) were higher in the galls of infected roots compared with those of an uninfected control. Of these genes, AtADF2 exhibited a nearly three-fold increase in expression 2–3 weeks following nematode infection, and its expression was concentrated in giant cells. Moreover, AtADF2 knockdown in Arabidopsis increased the bundling of actin filaments, resulting in delayed giant cell development and decreased nematode reproduction. Thus, these findings imply that AtADF2 positively regulates plant resistance to root-knot nematodes (Clément et al., 2009). Similarly, in cucumber five of eight CcADF genes demonstrated increased expression in nematode-induced galls, suggesting that CcADF genes may facilitate nematode feeding on cucumber roots (Liu et al., 2016).
Resistance to aphids
Aphids (Hemiptera: Aphididae), a diverse family of ~250 different species, are pests who feed on plants and affect plant growth and productivity via removing nutrients from sieve elements, altering source–sink relationships, and spreading viral diseases (Goggin, 2007). Arabidopsis atadf3 mutants were more susceptible to green peach aphids (GPAs; Myzus persicae Sülzer) infestation compared with wild type plants. GPAs fed faster and for a longer duration on atadf3 mutants, and their populations could therefore reproduce more quickly. Introducing AtADF3 into atadf3 mutant plants rescued the resistance to GPAs, indicating that AtADF3 has a critical role in limiting GPAs infestation (Mondal et al., 2018). By monitoring aphid feeding behavior, the authors found that the AtADF3 expression hinders with the ability of GPAs to find and feed from sieve elements. PAD4 (phytoalexin-deficient 4) is an important regulatory factor in Arabidopsis defense against peach aphids, negatively regulating aphid feeding and fecundity (Louis et al., 2010). Further research confirmed that PAD4 is a critical downstream player of the AtADF3-dependent defense mechanism (Mondal et al., 2018).
Resistance to corn borer
The corn borer is a major maize pest in many regions of the world, where it severely affects its yield by feeding on organs such as leaves, stems, and male and female inflorescences (Meihls et al., 2012). A recent genome-wide association analysis revealed that ZmADF4 is significantly associated with resistance to the Mediterranean corn borer (Sesamia nonagrioides) in the stem (Samayoa et al., 2015). The biological function of ZmADF4 in maize resistance to corn borer is worth further exploration.
Fungal stress
Resistance to powdery mildew
Powdery mildew is a obligate biotrophic fungal pathogen that seriously threatening over 10,000 plant species, including crops, vegetables, trees, and ornamental plants (Hirose et al., 2005; Hückelhoven and Panstruga, 2011). ADF genes from different plants have been found to play different roles in regulating powdery mildew resistance. In Arabidopsis, the four Group I AtADF genes have been found to play a negative regulatory role regarding resistance to Golovinomyces orontii (G.orontii), with AtADF4 exhibiting the most significant effect. In atadf4 mutants and atadf1-4 quadruple knockdown plants, researchers identified an accumulation of hydrogen peroxide and cell-specific death at the sites of G.orontii infection. In addition, they also found an increase in the abundance of microfilaments, and the plants show enhanced resistance against powdery mildew (Inada et al., 2016). RPW8.2 (resistance to powdery mildew 8.2) is an atypical mildew resistance protein found in Arabidopsis (Xiao et al., 2001). Overexpression of AtADF6 (belongs to group IV) inhibited its localization to the membrane surrounding the powdery mildew fungal haustorium, which is required for inducing resistance against powdery mildew. Thus, this evidence indicates that AtADF6 may play a negative role toward powdery mildew resistance (Wang et al., 2009b). In contrast, the overexpression of AtADF5 (belongs to group III) had no effect on RPW8.2 localization, implying the existence of functional diversification among ADF members in plant response to powdery mildew (Wang et al., 2009b). It is reasonable to presume that the functional diversification between AtADF5 and AtADF6 may resulted from their difference in biochemical activity (Nan et al., 2017) or expression profile (Dong et al., 2001). Ectopic expression of HvADF3 in barley (Hordeum vulgare) epidermal cells was found to disrupt the integrity of the actin cytoskeleton in cells. This in turn enhanced fungal entry and lead to increased susceptibility to the barley pathogen Blumeria graminis f. sp. hordei (Bgh) (Miklis et al., 2007). Moreover, transient overexpression of AtADF1, AtADF5, AtADF6, AtADF7, and AtADF12 was found to significantly increase the entry rate of Bgh in barley, while the overexpression of AtADF2, AtADF3, AtADF4 and AtADF9 had no significant effect.
Resistance to stripe rust
In wheat, stripe rust caused by Puccinia striiformis f. sp. tritici is a widespread and devastating disease (Hovmøller, 2007). Different wheat ADF genes were found to exhibit varied response patterns against different physiological races of this pathogen, and may therefore play different roles in regulating stripe rust resistance. For example, the avirulent race CYR23 strongly induced the expression of TaADF4 and TaADF7 in wheat, while the virulent race CYR31 induced their expression to a lesser extent. Silencing TaADF4 and TaADF7 in wheat lines inoculated with CYR23 led to significant changes in microfilament structures, reduced accumulation of reactive oxygen species (ROS), and weakened hypersensitive reactions. Taken together, these effects indicate that TaADF4 and TaADF7 positively regulate wheat resistance against non-adapted races of stripe rust by modulating microfilament dynamics (Fu et al., 2014; Zhang et al., 2017). In contrast to the response patterns of these genes, TaADF3 showed elevated expression levels upon CYR31 induction but showed significantly decreased expression levels following CYR23 inoculation. Silencing TaADF3 enhanced wheat resistance to CYR31 while reducing both ROS accumulation and hypersensitive reactions (Tang et al., 2016).
Resistance to verticillium wilt
Verticillium wilt is mainly caused by Verticillium dahliae or Verticillium alboatrum, two soil-borne vascular fungal pathogens that severely affect cotton production (Klosterman et al., 2009). After infection by Verticillium dahliae, the expression of GhADF6, a gene homologous to AtADF6 found in upland cotton (Gossypium hirsutum), was downregulated in root epidermal cells. Silencing of GhADF6 increased the abundance of microfilaments in root epidermal cells, and the plants showed enhanced resistance to Verticillium dahlia. Thus, GhADF6 likely plays a negative regulatory role with respect to cotton verticillium wilt resistance (Sun et al., 2021). Taken together, these findings highlight the complex regulatory roles of ADF genes regarding plant defense against fungal diseases.
Bacterial stress
Participation in Innate immunity caused by bacterial MAMP
Innate immunity is the first line of host defense against microbial invasion and is evolutionally conserved in all multicellular organisms, which is activated by pattern-recognition receptors (PRRs) that recognize microbe-associated molecular patterns (MAMPs) (Deng et al., 2020). MAMPs mediated microfilament rearrangement, as featured by increased abundance and remodeling of microfilament, plays an important role in plant innate immune signal transduction (Henty-Ridilla et al., 2014). Within minutes of treatment with a bacterial MAMP, elf26 (a conserved 26-amino acid peptide from bacterial elongation factor), a dose- and time-dependent increase in actin filament abundance was detected in epidermal cells throughout the Arabidopsis hypocotyl. However, actin architecture and dynamics in an atadf4 mutant fail to respond to elf26 treatment, suggested that AtADF4 plays a key role in modulating actin dynamics by participating in innate immune signal transduction caused by bacteria in plants (Henty-Ridilla et al., 2014).
Resistance to Pst DC3000 expressing AvrPphB
Pseudomonas syringae pv tomato (Pst) is a hemibiotrophic bacterial pathogen. The Arabidopsis mutant atadf4 shows abnormal microfilament dynamics and increased susceptibility to that of Pst DC3000 expressing the incompatible effector AvrPphB, but not to strains expressing AvrRps2 or AvrB. Moreover, a transgenic experiment showed that AtADF4 is able to restore the resistance that is compromised in the atadf4 mutant, thereby indicating that AtADF4 is required for the resistance to Pst DC3000 expressing AvrPphB (Tian et al., 2009). RPS5 (resistance to Pseudomonas syringae 5) is a gene that encodes a resistance protein capable of recognizing AvrPphB and activating downstream defense signals in Arabidopsis (Chisholm et al., 2006). Subsequent studies have reported that the increased susceptibility of atadf4 mutant to Pst DC3000 expressing AvrPphB is associated with decreased RPS5 expression, suggesting that AtADF4 may regulate plant resistance to Pst DC3000 expressing AvrPphB via the coordinated regulation of microfilament dynamics and R-gene transcription (Porter et al., 2012).
Participation in interaction with rhizobia
Rhizobia is a class of gram-negative soil bacteria that includes Rhizobium, Bradyrhizobium, Sinorhizobium, and Azorhizobium. These bacteria can form symbiotic nitrogen-fixing nodules with leguminous plants and increase nitrogen fixation in arable fields by as much as 30% (Mus et al., 2016). PvADFE is one of the nine ADF genes found in common bean, primarily expressed in roots and nodules inoculated with Rhizobium tropici (Ortega-Ortega et al., 2020). In addition, PvADFE silencing increases the number and size of nodules and enhances nitrogen fixation activity. Conversely, the overexpression of this gene resulted in the opposite phenotype. In addition, the expression levels of two genes related to nodulation development and signaling, NIN and ENOD2, were significantly decreased in the roots of plants overexpressing PvADFE, thereby indicating that PvADFE plays a negative regulatory role in rhizobial infection and nodulation of common bean (Ortega-Ortega et al., 2020).
Viral stress
Viruses are molecular parasites that complete the entire life cycle by utilizing the resources of host cells. Many crucial functions of plants are affected by viruses, including nutrient absorption, nutrient translocation, photosynthesis, growth, and development (Gergerich and Dolja, 2006). In an infected plants, virus-encoded movement proteins and cellular factors allow viruses to move within infected cells (local movement) and long distances through the vascular system (systemic movement) (Garcia-Ruiz, 2018). A great deal of attention is given to understanding the fundamental mechanism of viral infections as well as factors involved in gene regulation during viral infections (Garcia-Ruiz, 2018). Microfilament has been reported to play an important role in the process of virus infection (Chen et al., 2010; Tilsner et al., 2012; Porter and Day, 2016).
Soybean mosaic virus (SMV), which belongs to the Potyvirus genus, is one of the most prevalent and destructive viral pathogens in soybean cultivation regions around the world. Mosaic and necrosis symptoms are common on the leaves of soybean plants that are infected with SMV (Hill et al., 2007). The P3 protein of SMV (SMV-P3) plays a major role in its replication and movement, and also responsible for symptom development in SMV-infected plants (Hajimorad et al., 2018). SMV-P3 exhibits strong variability and complex functionality, which is consistent with the symptoms of soybean mosaic disease (Hajimorad et al., 2018). By screening soybean cDNA library, Lu et al. (2015) found that an ADF, GmADF2, interacts with SMV-P3, and this interaction is further confirmed using bimolecular fluorescence complementation assay. Further experiments showed that the interaction between GmADF2 and SMV-P3 is occurred in both the cytomembrane and cytoskeleton of plant cells, indicated the GmADF2 was trailed by SMV-P3 (Lu et al., 2015). These results suggested that GmADF2 is an important host factor for SMV-P3 and may promote its intercellular movement, thus plays a crucial role for the virus to establish infection (Lu et al., 2015).
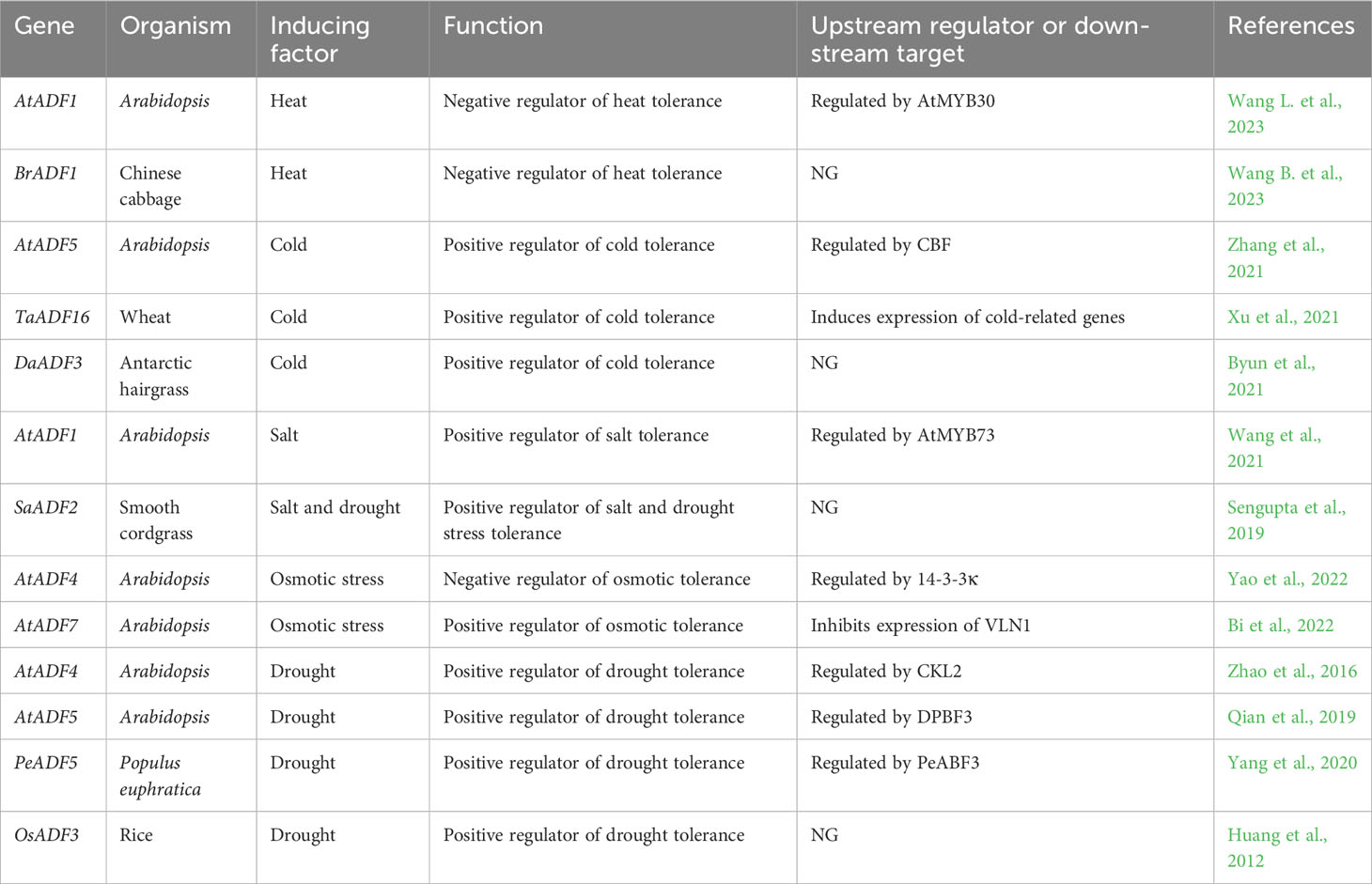
Role of ADF genes in abiotic stress resistance
In addition to biotic stresses, abiotic stresses like cold, heat, drought, salinity, flooding and nutrient deficiency are the major limiting factors for crop yields (Saijo and Loo, 2020). Abiotic stress factors can individually or collectively affect plant growth and development (Zhu, 2016). Plant ADF genes are widely involved in various abiotic stress responses (Figure 1). However, their modes of response and functions vary among plant species and tissues (Table 2). In this section, we will review the functions and molecular regulatory mechanisms of plant ADF genes in regulating abiotic stress in plants.
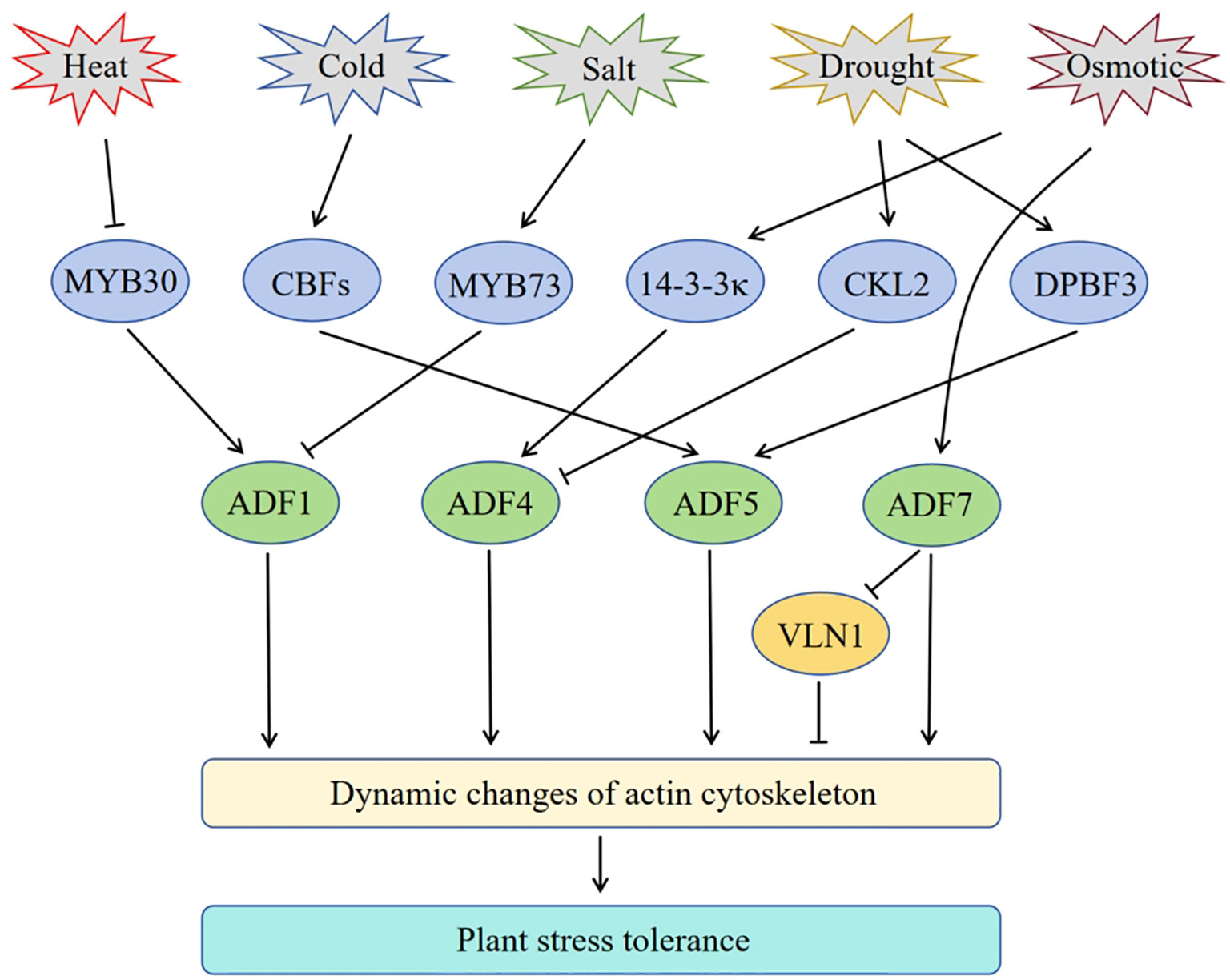
Figure 1 Schematic of the involvement of Arabidopsis ADF genes in plant response to abiotic stress. Arrows represent positive regulation, and bar ends mean inhibitory action.
Temperature stress
Cold stress
Cold stress, including chilling (cold temperatures of above 0°C) and freezing stress (below 0°C), causes plant growth to slow down, stagnate, and retrogress, thereby reducing the yield (Zhang et al., 2019). Aside from membrane rigidification, ROS accumulation, protein destabilization, and metabolic disequilibrium, cold stress has also been reported to disrupt the microfilament of plant cells and interfere with all cellular processes (Fan et al., 2015; Liu et al., 2018). The expression profiles of AtADF genes in response to temperature stress had been reported to diversely among different groups. The expression levels of AtADF genes of Group I, III, and IV have been found to be significantly induced by cold or heat stress, with two group III ADF genes (i.e., AtADF5 and AtADF9) responding the most strongly (Fan et al., 2015; Fan et al., 2016). In Arabidopsis plants exposed to cold stress, the survival rate of atadf5 mutants significantly decreased relative to the wild type. Moreover, mutant plants showed disordered actin cytoskeleton in root epidermal cells, suggesting that AtADF5 plays an important role in mediating cold stress tolerance in Arabidopsis (Zhang et al., 2021). In the face of cold stress, plants depend on C-repeat binding factors (CBFs) as their key molecular switches (Liu et al., 2018). CBFs can activate the expression of AtADF5 by binding to CRT/DRE elements in its promoter, and AtADF5 can in turn regulate dynamic changes in the actin cytoskeleton to modulate the cold response of Arabidopsis plants (Zhang et al., 2021). In freezing-tolerant wheat cultivars, the ADF gene Wcor719 can be specifically induced by exposure to low temperatures, while the expression levels of this gene do not significantly change in freezing-sensitive cultivars. Moreover, its expression level is also insensitive to high temperature, salt stress, mechanical damage, and abscisic acid (ABA) (Danyluk et al., 1996; Ouellet et al., 2001). In contrast, wheat TaADF4 is induced by heat stress, but its expression levels are significantly decreased in response to low temperature or salt stress (Zhang et al., 2017). A genome-wide analysis showed that 25 TaADF genes exist in the genome of the “Chinese Spring” wheat cultivar, and that cold stress can affect the expression levels of seven TaADF genes, six of which (i.e., TaADFs 13, 16, 17, 18, 21, and 22) are upregulated (Xu et al., 2021). In Arabidopsis, the heterologous expression of TaADF16, the most highly expressed and upregulated ADF gene in response to cold stress, can enhance plant cold stress resistance by accelerating ROS scavenging and by altering osmotic regulation in cells (Xu et al., 2021). Moreover, the expression levels of seven cold stress-responsive genes were found to be significantly higher in a TaADF16-overexpressing line than in the wild type regardless of whether the transgenic Arabidopsis plants were exposed to cold conditions. This indicates that the overexpression of TaADF16 can generally induce the expression of cold-related genes (Xu et al., 2021). Antarctic hairgrass is the only monocotyledonous flowering plant in Antarctica and its genome contains eight ADF genes. Cold stress can induce the expression of five DaADF genes, with DaADF3 showing the most significant cold stress response (Byun et al., 2021). In rice, plants that overexpress DaADF3 exhibit improved cold stress resistance, as measured via survival rate, leaf chlorophyll content, and electrolyte leakage along with changes in microfilament organization in the root tips (Byun et al., 2021).
Heat stress
Heat stress is commonly defined as the increase in temperature beyond a specific threshold level for a duration that is adequate to induce irreversible harm to the growth and development of plants (Bita and Gerats, 2013). The effects of heat stress on plants and cells are numerous. For example, high temperatures alter membrane fluidity and denaturate proteins which impair enzyme function (Malerba et al., 2010). Recently, ADF genes have also been found to be involved in plant tolerance to high temperatures. For example, AtADF1 expression was repressed by high temperatures, and atadf1 mutant seedlings exhibited greater actin filament stability and faster growth than the wild type (Wang L. et al., 2023). Conversely, AtADF1 overexpression showed the opposite phenotype. Further experiments revealed that AtADF1 transcription is regulated by AtMYB30, a key transcription factor involved in responses to various forms of abiotic stress, including heat (Liao et al., 2017). This finding indicates that AtADF1 is a target gene in the AtMYB30-mediated plant response to abiotic stress (Wang B. et al., 2023). Similarly, the authors found that BrADF1 from Chinese cabbage (Brassica rapa), a gene that is highly homologous to AtADF1, regulates F-actin dynamics and plant tolerance to heat stress in a manner similar to that of AtADF1 (Wang L. et al., 2023).
Salt stress
Salt stress refers to the adverse effect of excessive soluble salts in soil on plant growth and development, which has both osmotic and ionic or ion-toxicity effects on cells (Zhu, 2016). More than one third of the world’s irrigated lands are affected by salinization, a worldwide problem that threatens the growth and yield of crops (Zhao et al., 2020). In Arabidopsis, the expression levels of AtADF1 rapidly increase in response to salt stress, and the survival rate of the atadf1 mutant decreases significantly compared with the wild type under salt stress. Moreover, mutant plants exhibit cytoskeletal changes, including increased microfilament bundles in cells, while AtADF1-overexpressing plants exhibit opposite macroscopic and microscopic phenotypes. These results indicate that AtADF1 positively regulates plant salt tolerance by promoting actin depolymerization (Wang et al., 2021). Moreover, AtMYB73 is a negative regulatory factor for salt stress in Arabidopsis (Kim et al., 2013), and further experiments have revealed that AtMYB73 negatively regulates AtADF1 expression (Wang et al., 2021). Therefore, AtADF1 is likely an important player in the AtMYB73-mediated salt stress response pathway (Wang et al., 2021). Smooth cordgrass (Spartina alterniflora) is a perennial grass halophyte that has adapted to salt and drought conditions owing to specific alleles for genes involved in stress tolerance (Baisakh et al., 2008). The SaADF2 of smooth cordgrass is homologous to the OsADF2 of rice. Although the sequence similarity between their proteins exceeds 95%, six amino acid differences (i.e. the 6th Ser, 19th Asp, 25th Leu, 118th Gln, 132nd Pro and 133rd Thr in OsADF2 were substituted by Thr, Asn, His, His, Ser and Ser in SaADF2, respectively) may responsible for the substantial differences in their three-dimensional structures (Sengupta et al., 2019). Biochemical analysis revealed that SaADF2 displays greater actin-binding affinity and can depolymerize microfilaments more efficiently than OsADF2, which enhances cellular actin dynamics in cells. In rice, SaADF2 overexpression engenders greater drought and salt tolerance compared with that in the wild type and OsADF2-overexpression lines (Sengupta et al., 2019). A detailed biochemical investigation is required to determine the specific amino acid(s) that play a critical role in ADF’s biochemical activity. This knowledge could present an opportunity to utilize genome-editing technology for performing site-specific mutations, enabling the manipulation of ADF activity in crop breeding practices for enhancing stress tolerance.
Osmotic stress
Osmotic stress, often caused by drought and high salinity, occurs when soil contains excess soluble salt that prevents water absorption of plants (Yoshida et al., 2014). Microfilament cytoskeleton had been confirmed to participates, and plays a crucial role, in responses to osmotic stress in plants (Wang et al., 2010). In Arabidopsis seedlings subjected to osmotic stress, expression level of AtADF4 considerably increased. In addition, the survival rate of atadf4 mutants was higher than that of the wild type, while the survival rate of AtADF4-overexpressing lines decreased. Thus, these results indicate that AtADF4 plays a negative regulatory role in the plant response to osmotic stress (Yao et al., 2022). Further experiments demonstrated that a phosphopeptide-binding protein, 14-3-3κ, acts as an upstream regulator of AtADF4 to regulate the Arabidopsis response to osmotic stress (Yao et al., 2022). Root hairs are important organs for plants to absorb nutrients and water. Osmotic stress can induce AtADF7 expression, which in turn is responsible for inhibiting the expression of the actin-bundling protein VILLIN1 (VLN1) in root cells, thereby reducing microfilament bundles in root cells and promoting root hair growth. These findings indicate that the AtADF7–VLN1 pathway is essential for root hair formation under osmotic stress tolerance, and plays critical role in enhancing plant osmotic stress tolerance (Bi et al., 2022).
Drought stress
Drought is an adverse environmental stress that hampers normal growth, disrupts water relations, and decreases water-use efficiency in plants. To adapt to drought stress, plants have developed intricate mechanisms, one of which involves regulating the opening and closing of stomata (Saradadevi et al., 2017). Stomata are pores found on the epidermis of aerial parts of plants and are responsible for absorbing carbon dioxide and releasing water vapor (Jiang et al., 2012). The stomatal aperture is finely tuned to prevailing environmental conditions by a pair of guard cells surrounding each pore (Schroeder et al., 2001). Stomatal movement mediated by ABA is particularly important for plant adaptations to drought conditions. Microfilament dynamics play a crucial role in regulating the opening and closing of the stomata, involving the alteration of the radial orientation of the actin filaments during open stomata changes to a longitudinal orientation characteristic of closed stomata during stomatal closure (Zhao et al., 2011).
In Arabidopsis, AtADF1 overexpression leads to disorganized microfilament bundles in guard cells, in turn resulting in abnormal stomatal closure following ABA treatment (Dong et al., 2001). Arabidopsis casein kinase 1-like protein 2 (CKL2) plays an important regulatory role in ABA- and drought-induced stomatal closure. CKL2 can inhibit the depolymerization activity of AtADF4 via phosphorylation, thereby rendering the microfilament cytoskeleton more stable in guard cells and regulating stomatal opening or closing (Zhao et al., 2016). ABA and drought stress also induce AtADF5 expression in Arabidopsis seedlings. Compared with the wild type, an atadf5 mutant showed reduced microfilament bundles in cells, delayed stomatal closure, intensified leaf dehydration, and decreased survival rates under drought conditions (Qian et al., 2019). Further biochemical experiments demonstrated that DPBF3, an ABA-responsive element-binding factor (ABF/AREB), can activate AtADF5 expression via ABA-responsive core elements in its promoter region. AtADF5 regulates stomatal movement by modulating the rearrangement of microfilament structures through its F-actin bundling activity, which improves plants’ adaptability to drought stress. Therefore, AtADF5 is an important player in the ABF/AREB-mediated pathway facilitating plant responses to drought stress (Qian et al., 2019). In Populus euphratica, PeABF3 is a transcription factor involved in ABA signaling response and its expression is induced by drought and ABA. PeABF3 can activate the expression of PeADF5, which facilitates ABA-induced stomatal movement by promoting actin cytoskeletal rearrangement and enhancing drought resistance (Yang et al., 2020). Furthermore, in rice the exogenous application of ABA or various stress conditions induces OsADF3 expression in the root tips and lateral roots. Moreover, the heterologous expression of OsADF3 in Arabidopsis enhanced its drought tolerance, as evidenced by improved germination rate, primary root length, and survival rate. In addition, several drought-tolerance responsive genes are upregulated under drought stress, suggesting that OsADF3 may exert regulatory effects upstream of these genes (Huang et al., 2012). The above studies indicate that plant ADF genes are important factors in regulating stomatal movement and play an important role in enhancing plant drought resistance. Further exploration and research on these drought resistant ADF genes and homologous ADF genes in other plants, especially crops, will provide important genetic resources for the cultivation of drought resistant crops.
Summary and future prospects
As an important type of actin-binding protein, ADFs are widely involved in dynamic changes to the microfilaments of cells. Accordingly, they play a crucial role in plant growth, development, and stress response. In this study, we provide a systematic summary of the involvement of ADFs in the regulation of both biotic and abiotic stresses in plants. This includes the expression patterns of ADF genes in response to various stresses, their regulatory role with respect to plant stress responses, and the molecular mechanisms by which ADFs regulate stress tolerance. Research suggests that stress conditions not only directly regulate the transcription levels of ADF genes, but many transcription factors (including members of the MYB, ABF, and CBF TF families) are also involved in regulating the expression of ADF genes in response to different forms of stress. Furthermore, ADFs not only directly regulate the polymerization, depolymerization, and arrangement of microfilaments in cells, but also indirectly affect plant stress responses by influencing the expression of various other stress-related genes. Taken together, these results indicate that ADF participates in precise and complex mechanisms to regulate plant stress responses.
However, to date our understanding of the role of ADF genes in the regulation of plant stress responses remains insufficient. First, there is currently limited systematic information regarding the ADF gene family in plants, since data exists for fewer than 20 plant species. Previous studies have shown that ADFs exist in land plants as members of diversified gene families, and their expression patterns and biochemical activities exhibit obvious inter-group specificity. Genomic and transcriptomic studies provide a convenient way to comprehensively identify and characterize the ADF gene family in additional plant species. Moreover, systematic analysis of the plant ADF gene family will provide important information for further investigation of the biological functions of different ADF genes. Second, previous studies have found that the expression levels of many plant ADF genes change in response to stress, which suggests that ADF genes play an important role in plant stress responses. At the molecular regulation level of the ADF gene regulation of stress responses, Arabidopsis thaliana has received an overwhelming share of the research attention and has made significant progress in understanding ADF genes in this model system. However, there has been limited research on other plants, especially on agriculturally important crops, in which the specific functions and molecular regulatory mechanisms of ADF genes remain largely unclear. This hinders their application for crop improvement. Future in-depth study of stress-related ADF genes in crop species is critical for molecular breeding and genetic engineering.
Finally, studies of SaADF2 in salt- and drought-tolerant smooth cordgrass suggest that some key amino acids in ADF influence its biochemical activity, and can thereby be manipulated to exert stronger regulatory effects on specific plant stress responses. This study suggests that there may be beneficial ADF alleles in plant species with strong stress resistances that may enhance crop resilience. In the future, detailed investigation of these genes and exploration of advantageous protein variant sites may make it possible to use genome-editing techniques to modify ADF genes for stress-tolerant crop breeding.
Author contributions
YS: Funding acquisition, Writing – original draft, Writing – review and editing. MS: Writing – original draft. DW: Writing – original draft. YG: Writing – original draft. QS: Writing – original draft. PL: Writing – original draft. JY: Investigation, Writing – original draft. PC: Investigation, Writing – original draft. SG: Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by National Natural Science Foundation of China (32101788), the Industrial Promotion Project of Shandong Agricultural Science and Technology Park (2019YQ035), the Open Project of Liaocheng University Landscape Architecture Discipline (319462212), the Higher Educational Program of Shandong Province for Introduction and Cultivation of Young Innovative Talents (S202310447030; CXCY2023267), and the Doctoral Research Start-up Foundation of Liaocheng University (318052021).
Acknowledgments
We appreciate the linguistic assistance provided by ZYEdit during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andrianantoandro, E., Pollard, T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/Cofilin. Mol. Cell 24, 13–23. doi: 10.1016/j.molcel.2006.08.006
Augustine, S. M., Cherian, A. V., Seiling, K., Di Fiore, S., Raven, N., Commandeur, U., et al. (2021). Targeted mutagenesis in Nicotiana tabacum ADF gene using shockwave-mediated ribonucleoprotein delivery increases osmotic stress tolerance. Physiol. Plant 173, 993–1007. doi: 10.1111/ppl.13499
Baisakh, N., Subudhi, P. K., Varadwaj, P. (2008). Primary responses to salt stress in a halophyte, smooth cordgrass (Spartina alterniflora Loisel.). Funct. Integr. Genomics 8, 287–300. doi: 10.1007/s10142-008-0075-x
Bamburg, J. R., Harris, H. E., Weeds, A. G. (1980). Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 121, 178–182. doi: 10.1016/0014-5793(80)81292-0
Bao, Y., Hu, G., Flagel, L. E., Salmon, A., Bezanilla, M., Paterson, A. H., et al. (2011). Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proc. Natl. Acad. Sci. U.S.A. 108 (52), 21152–21157. doi: 10.1073/pnas.1115926109
Bi, S., Li, M., Liu, C., Liu, X., Cheng, J., Wang, L., et al. (2022). Actin depolymerizing factor ADF7 inhibits actin bundling protein VILLIN1 to regulate root hair formation in response to osmotic stress in Arabidopsis. PloS Genet. 18, e1010338. doi: 10.1371/journal.pgen.1010338
Bita, C. E., Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00273
Blanchoin, L., Pollard, T. D. (1999). Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546. doi: 10.1074/jbc.274.22.15538
Blanchoin, L., Pollard, T. D., Mullins, R. D. (2000). Interaction of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282. doi: 10.1016/S0960-9822(00)00749-1
Burgos-Rivera, B., Ruzicka, D. R., Deal, R. B., McKinney, E. C., King-Reid, L., Meagher, R. B. (2008). ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 68, 619–632. doi: 10.1007/s11103-008-9398-1
Byun, M. Y., Cui, L. H., Lee, A., Oh, H. G., Yoo, Y. H., Lee, J., et al. (2021). Abiotic stress-induced actin depolymerizing factor 3 from Deschampsia Antarctica enhanced cold tolerance when constitutively expressed in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.734500
Cao, H., Amin, R., Niu, L., Song, Z., Dong, B., Li, H., et al. (2020). Multidimensional analysis of actin depolymerising factor family in pigeon pea under different environmental stress revealed specific response genes in each subgroup. Funct. Plant Biol. 48, 180–194. doi: 10.1071/FP20190
Carlier, M. F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G. X., et al. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322. doi: 10.1083/jcb.136.6.1307
Chen, C., Wong, E. I., Vidali, L., Estavillo, A., Hepler, P. K., Wu, H. M., et al. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell. 14, 2175–2190. doi: 10.1105/tpc.003038
Chen, C., Zhang, Y., Zhu, L., Yuan, M. (2010). The actin cytoskeleton is involved in the regulation of the plasmodesmal size exclusion limit. Plant Signal Behav. 5 (12), 1663–1665. doi: 10.4161/psb.5.12.14018
Chisholm, S. T., Coaker, G., Day, B., Staskawicz, B. J. (2006). Host-Microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Clément, M., Ketelaar, T., Rodiuc, N., Banora, M. Y., Smertenko, A., Engler, G., et al. (2009). Actin-depolymerizing factor2-mediated actin dynamics are essential for rootknot nematode infection of Arabidopsis. Plant Cell 21, 2963–2979. doi: 10.1105/tpc.109.069104
Daher, F. B., Geitmann, A. (2012). Actin depolymerizing factors ADF7 and ADF10 play distinct roles during pollen development and pollen tube growth. Plant Signal. Behav. 7, 879–881. doi: 10.4161/psb.20436
Danyluk, J., Carpentier, E., Sarhan, F. (1996). Identification and characterization of a low temperature regulated gene encoding an actin-binding protein from wheat. FEBS Lett. 389, 324–327. doi: 10.1016/0014-5793(96)00599-6
Deng, M., Tam, J. W., Wang, L., Liang, K., Li, S., Zhang, L., et al. (2020). TRAF3IP3 negatively regulates cytosolic RNA induced anti-viral signaling by promoting TBK1 K48 ubiquitination. Nat. Commun. 11 (1), 2193. doi: 10.1038/s41467-020-16014-0
Dong, C. H., Kost, B., Xia, G., Chua, N. H. (2001). Molecular identification and characterization of the Arabidopsis AtADF1, AtADF5 and AtADF6 genes. Plant Mol. Biol. 45, 517–527. doi: 10.1023/A:1010687911374
Duan, W. J., Liu, Z. H., Bai, J. F., Yuan, S. H., Li, Y. M., Lu, F. K., et al. (2021). Comprehensive analysis of formin gene family highlights candidate genes related to pollen cytoskeleton and male fertility in wheat (Triticum aestivum L.). BMC Genomics 22 (1), 570. doi: 10.1186/s12864-021-07878-7
Fan, T. T., Ni, J. J., Dong, W. C., An, L. Z., Xiang, Y., Cao, S. Q. (2015). Effect of low temperature on profilins and ADFs transcription and actin cytoskeleton reorganization in Arabidopsis. Biol. Plant 59, 793–796. doi: 10.1007/s10535-015-0546-6
Fan, T. T., Wang, R., Xiang, Y., An, L., Cao, S. (2016). Heat stress induces actin cytoskeletal reorganization and transcript profiles of vegetative profilins and actin depolymerizing factors (ADFs) in Arabidopsis. Acta Physiol. Plant 38, 37–49. doi: 10.1007/s11738-016-2061-6
Feng, Y., Liu, Q., Xue, Q. (2006). Comparative study of rice and Arabidopsis actin-depolymerizing factors gene families. J. Plant Physiol. 163, 69–79. doi: 10.1016/j.jplph.2005.01.015
Fu, Y., Duan, X., Tang, C., Li, X., Voegele, R. T., Wang, X., et al. (2014). TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. Plant J. 78, 16–30. doi: 10.1111/tpj.12457
Fuller, V. L., Lilley, C. J., Urwin, P. E. (2008). Nematode resistance. New Phytol. 180, 27–44. doi: 10.1111/j.1469-8137.2008.02508.x
Garcia-Ruiz, H. (2018). Susceptibility genes to plant viruses. Viruses 10 (9), 484. doi: 10.3390/v10090484
Gergerich, R. C., Dolja, V. V. (2006). Introduction to plant viruses, the invisible foe. PHI. 0414, 1. doi: 10.1094/PHI-I-2006-0414-01
Goggin, F. L. (2007). Plant–aphid interactions: molecular and ecological perspectives. Curr. Opin. Plant Biol. 10, 399–408. doi: 10.1016/j.pbi.2007.06.004
Gunning, P. W., Ghoshdastider, U., Whitaker, S., Popp, D., Robinson, R. C. (2015). The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 128, 2009–2019. doi: 10.1242/jcs.165563
Hajimorad, M. R., Domier, L. L., Tolin, S. A., Whitham, S. A., Saghai Maroof, M. A. (2018). Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 19, 1563–1579. doi: 10.1111/mpp.12644
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
Henty-Ridilla, J. L., Li, J., Blanchoin, L., Staiger, C. J. (2013). Actin dynamics in the cortical array of plant cells. Curr. Opin. Plant Biol. 16, 678–687. doi: 10.1016/j.pbi.2013.10.012
Henty-Ridilla, J. L., Li, J., Day, B., Staiger, C. J. (2014). ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell. 26 (1), 340–352. doi: 10.1105/tpc.113.122499
Hill, J. H., Koval, N. C., Gaska, J. M., Grau, C. R. (2007). Identification of field tolerance to bean pod mottle and soybean mosaic viruses in soybean. Crop Sci. 47 (1), 212–218. doi: 10.2135/cropsci2006.03.0157
Hirose, S., Tanda, S., Kiss, L., Grigaliunaite, B., Havrylenko, M., Takamatsu, S. (2005). Molecular phylogeny and evolution of the maple powdery mildew (Sawadaea, Erysiphaceae) inferred from nuclear rDNA sequences. Mycol. Res. 109, 912–922. doi: 10.1017/s0953756205003527
Hovmøller, M. S. (2007). Sources of seedling and adult plant resistance to Puccinia striiformis f.sp. tritici in European wheats. Plant Breed. 126, 225–233. doi: 10.1111/j.1439-0523.2007.01369.x
Huang, J., Sun, W., Ren, J. X., Yang, R. C., Fan, J. S., Li, Y. F., et al. (2020). Genome-wide identification and characterization of actin-depolymerizing factor (ADF) family genes and expression analysis of responses to various stresses in Zea mays L. Int. J. Mol. Sci. 21, 1751–1768. doi: 10.3390/ijms21051751
Huang, Y. C., Huang, W. L., Hong, C. Y., Lur, H. S., Chang, M. C. (2012). Comprehensive analysis of differentially expressed rice actin depolymerizing factor gene family and heterologous overexpression of OsADF3 confers Arabidopsis thaliana drought tolerance. Rice 5, 33. doi: 10.1186/1939-8433-5-33
Hückelhoven, R., Panstruga, R. (2011). Cell biology of the plant–powdery mildew interaction. Curr. Opin. Plant Biol. 14, 738–746. doi: 10.1016/j.pbi.2011.08.002
Hussey, P. J., Allwood, E. G., Smertenko, A. P. (2002). Actin–binding proteins in the Arabidopsis genome database: properties of functionally distinct plant actin–depolymerizing factors/cofilins. Philos. Trans. R. Soc Lond. B. Biol. Sci. 357, 791–798. doi: 10.1098/rstb.2002.1086
Inada, N. (2017). Plant actin depolymerizing factor: actin microfilament disassembly and more. J. Plant Res. 130, 227–238. doi: 10.1007/s10265-016-0899-8
Inada, N., Higaki, T., Hasezawa, S. (2016). Nuclear function of subclass I actin-depolymerizing factor contributes to susceptibility in Arabidopsis to an adapted powdery mildew fungus. Plant Physiol. 170, 1420–1434. doi: 10.1104/pp.15.01265
Jammes, F., Lecomte, P., de Almeida-Engler, J., Bitton, F., Martin-Magniette, M. L., Renou, J. P., et al. (2005). Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 44, 447–458. doi: 10.1111/j.1365-313X.2005.02532.x
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59 (2), 86–101. doi: 10.1111/jipb.12513
Jiang, K., Sorefan, K., Deeks, M. J., Bevan, M. W., Hussey, P. J., Hetherington, A. M. (2012). The ARP2/3 complex mediates guard cell actin reorganization and stomatal movement in Arabidopsis. Plant Cell 24, 2031–2040. doi: 10.1105/tpc.112.096263
Khatun, K., Robin, A. H., Park, J. I., Kim, C. K., Lim, K. B., Kim, M. B., et al. (2016). Genome-wide identification, characterization and expression profiling of ADF family genes in Solanum lycopersicum L. Genes (Basel). 7, 79–97. doi: 10.3390/genes7100079
Kim, J. H., Nguyen, N. H., Jeong, C. Y., Nguyen, N. T., Hong, S. W., Lee, H. (2013). Loss of the R2R3 MYB, AtMYB73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 170, 1461–1465. doi: 10.1016/j.jplph.2013.05.011
Klosterman, S. J., Atallah, Z. K., Vallad, G. E., Subbarao, K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. doi: 10.1146/annurev-phyto-080508-081748
Leonard, S., Hommais, F., Nasser, W., Reverchon, S. (2017). Plant-phytopathogen interactions: bacterial responses to environmental and plant stimuli. Environ. Microbiol. 19 (5), 1689–1716. doi: 10.1111/1462-2920.13611
Li, J., Blanchoin, L., Staiger, C. J. (2015). Signaling to actin stochastic dynamics. Annu. Rev. Plant Biol. 66, 415–440. doi: 10.1146/annurev-arplant-050213-040327
Li, J., Han, G., Sun, C., Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 14, 1613131. doi: 10.1080/15592324.2019.1613131
Li, X. B., Xu, D., Wang, X. L., Huang, G. Q., Luo, J., Li, D. D., et al. (2010). Three cotton genes preferentially expressed in flower tissues encode actin-depolymerizing factors which are involved in F-actin dynamics in cells. J. Exp. Bot. 61, 41–53. doi: 10.1093/jxb/erp280
Liao, C., Zheng, Y., Guo, Y. (2017). MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 216, 163–177. doi: 10.1111/nph.14679
Liu, B., Liu, X., Liu, Y., Xue, S., Cai, Y., Yang, S., et al. (2016). The infection of cucumber (Cucumis sativus L.) roots by Meloidogyne incognita alters the expression of actin-depolymerizing factor (ADF) genes, particularly in association with giant cell formation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01393
Liu, J., Shi, Y., Yang, S. (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795. doi: 10.1111/jipb.12657
Louis, J., Leung, Q., Pegadaraju, V., Reese, J., Shah, J. (2010). PAD4-dependent antibiosis contributes to the ssi2 -conferred hyper-resistance to the green peach aphid. Mol. Plant-Microbe Interact. 23, 618–627. doi: 10.1094/MPMI-23-5-0618
Lu, L., Wu, G., Xu, X., Luan, H., Zhi, H., Cui, J., et al. (2015). Soybean actin-depolymerizing factor 2 interacts with Soybean mosaic virus-encoded P3 protein. Virus Genes 50, 333–339. doi: 10.1007/s11262-014-1150-0
Maciver, S. K., Hussey, P. J. (2002). The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 3, reviews3007. doi: 10.1186/gb-2002-3-5-reviews3007
Maciver, S. K., Zot, H. G., Pollard, T. D. (1991). Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 115, 1611–1620. doi: 10.1083/jcb.115.6.1611
Malerba, M., Crosti, P., Cerana, R. (2010). Effect of heat stress on actin cytoskeleton and endoplasmic reticulum of tobacco BY-2 cultured cells and its inhibition by Co2+. Protoplasma. 239, 23–30. doi: 10.1007/s00709-009-0078-z
McDowell, J. M., Huang, S. R., Mckinney, E. C., An, Y. Q., Meagher, R. B. (1996). Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142, 587–602. doi: 10.1073/pnas.79.2.495
Meihls, L. N., Kaur, H., Jander, G. (2012). Natural variation in maize defense against insect herbivores. Cold Spring Harb. Symp. Quant. Biol. 77, 269–283. doi: 10.1101/sqb.2012.77.014662
Miklis, M., Consonni, C., Bhat, R. A., Lipka, V., Schulze-Lefert, P., Panstruga, R. (2007). Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 144, 1132–1143. doi: 10.1104/pp.107.098897
Mondal, H. A., Louis, J., Archer, L., Patel, M., Nalam, V. J., Sarowar, S., et al. (2018). Arabidopsis ACTIN-DEPOLYMERIZING FACTOR3 is required for controlling aphid feeding from the fhloem. Plant Physiol. 176, 879–890. doi: 10.1104/pp.17.01438
Müller, J., Menzel, D., Samaj, J. (2007). Cell-type-specific disruption and recovery of the cytoskeleton in Arabidopsis thaliana epidermal root cells upon heat shock stress. Protoplasma. 230, 231–242. doi: 10.1007/s00709-006-0239-2
Mus, F., Crook, M. B., Garcia, K., Garcia Costas, A., Geddes, B. A., Kouri, E. D., et al. (2016). Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 82, 3698–3710. doi: 10.1128/AEM.01055-16
Nan, Q., Qian, D., Niu, Y., He, Y., Tong, S., Niu, Z., et al. (2017). Plant actin-depolymerizing factors possess opposing biochemical properties arising from key amino acid changes throughout evolution. Plant Cell 29, 395–408. doi: 10.1105/tpc.16.00690
Ortega-Ortega, Y., Carrasco-Castilla, J., Juárez-Verdayes, M. A., Toscano-Morales, R., Fonseca-García, C., Nava, N., et al. (2020). Actin depolymerizing factor modulates rhizobial infection and nodule organogenesis in common bean. Int. J. Mol. Sci. 21, 1970. doi: 10.3390/ijms21061970
Ouellet, F., Carpentier, É., Cope, M. J., Monroy, A. F., Sarhan, F. (2001). Regulation of a wheat actin-depolymerizing factor during cold acclimation. Plant Physiol. 125, 360–368. doi: 10.1104/pp.125.1.360
Pokorna, J., Schwarzerova, K., Zelenkova, S., Petrasek, J., Janotova, I., Capkova, V., et al. (2004). Sites of actin filament initiation and reorganization in cold treated tobacco cells. Plant Cell Environ. 27, 641–653. doi: 10.1111/j.1365-3040.2004.01186.x
Pollard, T. D. (2016). Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226. doi: 10.1101/cshperspect.a018226
Pollard, T. D., Cooper, J. A. (2009). Actin, a central player in cell shape and movement. Science 326, 1208–1212. doi: 10.1126/science.1175862
Porter, K., Day, B. (2016). From filaments to function: the role of the plant actin cytoskeleton in pathogen perception, signaling and immunity. J. Integr. Plant Biol. 58, 299–311. doi: 10.1111/jipb.12445
Porter, K., Shimono, M., Tian, M., Day, B. (2012). Arabidopsis actin-depolymerizing factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PloS Pathog. 8, e1003006. doi: 10.1371/journal.ppat.1003006
Qian, D., Zhang, Z., He, J., Zhang, P., Ou, X., Li, T., et al. (2019). Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 70, 435–446. doi: 10.1093/jxb/ery385
Ren, H., Xiang, Y. (2007). The function of actin-binding proteins in pollen tube growth. Protoplasma 230, 171–182. doi: 10.1007/s00709-006-0231-x
Roy-Zokan, E. M., Dyer, K. A., Meagher, R. B. (2015). Phylogenetic patterns of codon evolution in the ACTIN-DEPOLYMERIZING FACTOR/COFILIN (ADF/CFL) gene family. PloS One 10, e0145917. doi: 10.1371/journal.pone.0145917
Ruzicka, D. R., Kandasamy, M. K., McKinney, E. C., Burgos-Rivera, B., Meagher, R. B. (2007). The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 52, 460–472. doi: 10.1111/j.1365-313X.2007.03257.x
Saijo, Y., Loo, E. P. (2020). Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 225 (1), 87–104. doi: 10.1111/nph.15989
Samayoa, L., Malvar, R., Olukolu, B. A., Holland, J. B., Butrón, A. (2015). Genome-wide association study reveals a set of genes associated with resistance to the Mediterranean corn borer (Sesamia nonagrioides L.) in a maize diversity panel. BMC Plant Biol. 15, 35. doi: 10.1186/s12870-014-0403-3
Saradadevi, R., Palta, J. A., Siddique, K. H. (2017). ABA-mediated stomatal response in regulating water use during the development of terminal drought in wheat. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01251
Schroeder, J. I., Allen, G. J., Hugouvieux, V., Kwak, J. M., Waner, D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 627–658. doi: 10.1146/annurev.arplant.52.1.627
Sengupta, S., Mangu, V., Sanchez, L., Bedre, R., Joshi, R., Rajasekaran, K., et al. (2019). An actin depolymerizing factor from the halophyte smooth cordgrass, Spartina alterniflora (SaADF2) is superior to its rice homolog (OsADF2) in conferring drought and salt tolerance when constitutively overexpressed in rice. Plant Biotechnol. J. 17, 188–205. doi: 10.1111/pbi.12957
Staiger, C. J. (2000). Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 257–288. doi: 10.1146/annurev.arplant.51.1.257
Sun, Y. W., Wang, D. Y., Shi, M. M., Gong, Y. J., Yin, S. W., Jiao, Y. X., et al. (2023). Genome-wide identification of actin-depolymerizing factor gene family and their expression patterns under various abiotic stresses in soybean (Glycine max). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1236175
Sun, Y. D., Zhong, M. M., Li, Y. B., Zhang, R. H., Su, L., Xia, G. X., et al. (2021). GhADF6-mediated actin reorganization is associated with defence against Verticillium dahliae infection in cotton. Mol. Plant Pathol. 22, 1656–1667. doi: 10.1111/mpp.13137
Tang, C., Deng, L., Chang, D., Chen, S., Wang, X., Kang, Z. (2016). TaADF3, an actin-depolymerizing factor, negatively modulates wheat resistance against Puccinia striiformis. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01214
Tholl, S., Moreau, F., Hoffmann, C., Arumugam, K., Dieterle, M., Moes, D., et al. (2011). Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 585, 1821–1827. doi: 10.1016/j.febslet.2011.05.019
Tian, M., Chaudhry, F., Ruzicka, D. R., Meagher, R. B., Staiger, C. J., Day, B. (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 150, 815–824. doi: 10.1104/pp.109.137604
Tilsner, J., Linnik, O., Wright, K. M., Bell, K., Roberts, A. G., Lacomme, C., et al. (2012). The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol. 158 (3), 1359–1370. doi: 10.1104/pp.111.189605
Uribe, R., Jay, D. (2009). A review of actin binding proteins: new perspectives. Mol. Biol. Rep. 36, 121–125. doi: 10.1007/s11033-007-9159-2
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Wang, L., Cheng, J., Bi, S., Wang, J., Cheng, X., Liu, S., et al. (2023). Actin depolymerization factor ADF1 regulated by MYB30 plays an important role in plant thermal adaptation. Int. J. Mol. Sci. 24, 5675. doi: 10.3390/ijms24065675
Wang, J., Lian, N., Zhang, Y., Man, Y., Chen, L., Yang, H., et al. (2022). The cytoskeleton in plant immunity: dynamics, regulation, and function. Int. J. Mol. Sci. 23 (24), 15553. doi: 10.3390/ijms232415553
Wang, X., Mao, T. (2019). Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr. Opin. Plant Biol. 52, 86–96. doi: 10.1016/j.pbi.2019.08.002
Wang, L., Qiu, T., Yue, J., Guo, N., He, Y., Han, X., et al. (2021). Arabidopsis ADF1 is regulated by MYB73 and is involved in response to salt stress affecting actin filament organization. Plant Cell Physiol. 62, 1387–1395. doi: 10.1093/pcp/pcab081
Wang, H., Wang, J., Gao, P., Jiao, G., Li, Y., Wang, G., et al. (2009a). Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol. J. 7, 13–23. doi: 10.1111/j.1467-7652.2008.00367.x
Wang, W., Wen, Y., Berkey, R., Xiao, S. (2009b). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21, 2898–2913. doi: 10.1105/tpc.109.067587
Wang, C., Zhang, L., Yuan, M., Ge, Y., Liu, Y., Fan, J., et al. (2010). The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biol. 12, 70–98. doi: 10.1111/j.1438-8677.2009.00201.x
Wang, B., Zou, M., Pan, Q., Li, J. (2023). Analysis of actin array rearrangement during the plant response to bacterial stimuli. Methods Mol. Biol. 2604, 263–270. doi: 10.1007/978-1-0716-2867-6_21
Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M., et al. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. doi: 10.1126/science.291.5501.118
Xu, K., Zhao, Y., Zhao, S., Liu, H., Wang, W., Zhang, S., et al. (2021). Genome-wide identification and low temperature responsive pattern of actin depolymerizing factor (ADF) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.618984
Yang, Y., Li, H. G., Wang, J., Wang, H. L., He, F., Su, Y., et al. (2020). ABF3 enhances drought tolerance via promoting ABA-induced stomatal closure by directly regulating ADF5 in Populus euphratica. J. Exp. Bot. 71, 7270–7285. doi: 10.1093/jxb/eraa383
Yao, H., Li, X., Peng, L., Hua, X., Zhang, Q., Li, K., et al. (2022). Binding of 14-3-3κ to ADF4 is involved in the regulation of hypocotyl growth and response to osmotic stress in Arabidopsis. Plant Sci. 320, 111261. doi: 10.1016/j.plantsci.2022.111261
Yoshida, T., Mogami, J., Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Zhang, D., Du, Q., Xu, B., Zhang, Z., Li, B. (2010). The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol. Genet. Genomics 284 (2), 105–119. doi: 10.1007/s00438-010-0552-5
Zhang, B., Hua, Y., Wang, J., Huo, Y., Shimono, M., Day, B., et al. (2017). TaADF4, an actin-depolymerizing factor from wheat, is required for resistance to the stripe rust pathogen Puccinia striiformis f. sp. tritici. Plant J. 89, 1210–1224. doi: 10.1111/tpj.13459
Zhang, J., Li, X. M., Lin, H. X., Chong, K. (2019). Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 70, 753–780. doi: 10.1146/annurev-arplant-050718-100016
Zhang, P., Qian, D., Luo, C., Niu, Y., Li, T., Li, C., et al. (2021). Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.635533
Zhao, S., Jiang, Y., Zhao, Y., Huang, S., Yuan, M., Zhao, Y., et al. (2016). CASEIN KINASE1-LIKE PROTEIN2 regulates actin filament stability and stomatal closure via phosphorylation of Actin depolymerizing factor. Plant Cell 28, 1422–1439. doi: 10.1105/tpc.16.00078
Zhao, C., Zhang, H., Song, C., Zhu, J. K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zhao, Y., Zhao, S., Mao, T., Qu, X., Cao, W., Zhang, L., et al. (2011). The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 23, 2314–2330. doi: 10.1105/tpc.111.086546
Zheng, Y., Xie, Y., Jiang, Y., Qu, X., Huang, S. (2013). Arabidopsis actin depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25, 3405–3423. doi: 10.1105/tpc.113.117820
Zhou, Y., He, L., Zhou, S., Wu, Q., Zhou, X., Mao, Y., et al. (2023). Genome-wide identification and expression analysis of the VILLIN gene family in soybean. Plants-Basel. 12 (11), 2101. doi: 10.3390/plants12112101
Zhou, Y., Yang, Z., Guo, G., Guo, Y. (2010). Microfilament dynamics is required for root growth under alkaline stress in Arabidopsis. J. Integr. Plant Biol. 52, 952–958. doi: 10.1111/j.1744-7909.2010.00981.x
Keywords: actin-depolymerizing factor, microfilament, plant growth and development, biotic stress, abiotic stress, stress response, research progress
Citation: Sun Y, Shi M, Wang D, Gong Y, Sha Q, Lv P, Yang J, Chu P and Guo S (2023) Research progress on the roles of actin-depolymerizing factor in plant stress responses. Front. Plant Sci. 14:1278311. doi: 10.3389/fpls.2023.1278311
Received: 16 August 2023; Accepted: 01 November 2023;
Published: 16 November 2023.
Edited by:
Sushil Satish Chhapekar, University of Missouri, United StatesReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandNoriko Inada, Osaka Metropolitan University, Japan
Copyright © 2023 Sun, Shi, Wang, Gong, Sha, Lv, Yang, Chu and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongwang Sun, c3VueW9uZ3dhbmdAbGN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yongwang Sun
Yongwang Sun Mengmeng Shi
Mengmeng Shi Deying Wang
Deying Wang Yujie Gong
Yujie Gong Qi Sha
Qi Sha Peng Lv
Peng Lv Jing Yang
Jing Yang Shangjing Guo
Shangjing Guo