- 1Department of Crop Sciences, University of Illinois at Urbana-Champaign, Urbana, IL, United States
- 2School of Agriculture, Research Center for Perennial Rice Engineering and Technology in Yunnan, Yunnan University, Kunming, China
- 3Yunnan Seed Laboratory & Yunnan Key Laboratory for Rice Genetic Improvement, Food Crops Research Institute, Yunnan Academy of Agricultural Sciences (YAAS), Kunming, China
- 4T.T. Chang Genetic Resources Center, International Rice Research Institute (IRRI), Los Baños, Philippines
- 5CGIAR Genebank Initiative, Salisbury, United Kingdom
The undomesticated rice relative Oryza longistaminata is a valuable genetic resource for the improvement of the domesticated Asian rice, Oryza sativa. To facilitate the conservation, management, and use of O. longistaminata germplasm, we sought to quantify the population structure and diversity of this species across its geographic range, which includes most of sub-Saharan Africa, and to determine phylogenetic relationships to other AA-genome species of rice present in Africa, including the prevalence of interspecific hybridization between O. longistaminata and O. sativa. Though past plant breeding efforts to introgress genes from O. longistaminata have improved biotic stress resistance, ratooning ability, and yield in O. sativa, progress has been limited by substantial breeding barriers. Nevertheless, despite the strong breeding barriers observed by plant breeders who have attempted this interspecific cross, there have been multiple reports of spontaneous hybrids of O. sativa and O. longistaminata (aka “Obake”) obtained from natural populations in Africa. However, the frequency and extent of such natural introgressions and their effect on the evolution of O. longistaminata had not been previously investigated. We studied 190 O. longistaminata accessions, primarily from the International Rice Research Institute genebank collection, along with 309 O. sativa, 25 Oryza barthii, and 83 Oryza glaberrima control outgroups, and 17 control interspecific O. sativa/O. longistaminata hybrids. We analyzed the materials using 178,651 single-nucleotide polymorphisms (SNPs) and seven plastid microsatellite markers. This study identified three genetic subpopulations of O. longistaminata, which correspond geographically to Northwestern Africa, Pan-Africa, and Southern Africa. We confirmed that O. longistaminata is, perhaps counterintuitively, more closely related to the Asian species, O. sativa, than the African species O. barthii and O. glaberrima. We identified 19 recent spontaneous interspecific hybrid individuals between O. sativa and O. longistaminata in the germplasm sampled. Notably, the recent introgression between O. sativa and O. longistaminata has been bidirectional. Moreover, low levels of O. sativa alleles admixed in many predominantly O. longistaminata accessions suggest that introgression also occurred in the distant past, but only in Southern Africa.
Introduction
Global consumption of Asian domesticated rice, Oryza sativa, is projected to rise from 439 million tons to 555 million tons between 2010 and 2035 (Seck et al., 2012). To keep pace with demand, annual rice yield growth must increase by 0.2%–0.5% (Mohanty et al., 2013). The rice wild relative Oryza longistaminata is a valuable genetic resource for trait improvement of Asian domesticated rice, O. sativa (Khush et al., 1991; Ramos et al., 2016). A perennial, cross-pollinated species native to all of sub-Saharan Africa, O. longistaminata is highly diverse (Bezançon et al., 1977; Kiambi et al., 2018; Kiambi et al., 2005; Melaku et al., 2013). Of the eight AA-genome species that make up the primary germplasm pool of domesticated rice, O. longistaminata is one of the most genetically distinct from O. sativa (Zhu et al., 2014; Wambugu et al., 2015). O. longistaminata is distantly related to the two other AA-genome species of rice indigenous to Africa—a domesticated species, Oryza glaberrima, and its wild progenitor, Oryza barthii (Zhu et al., 2014; Wambugu et al., 2015). Though past breeding efforts have introgressed valuable traits from O. longistaminata to domesticated Asian rice, including bacterial blight resistance, perennial habit, and floral traits conducive to outcrossing, the development of genetic resources for this understudied wild species would assist further progress (Ronald et al., 1992; Sanni et al., 2013; Atwell et al., 2014; Gichuhi et al., 2016; Zhang et al., 2017; Prahalada et al., 2021; Zhang et al., 2023). Understanding O. longistaminata population structure and genetic diversity based on high genome coverage across the full geographic range of this species would facilitate conservation planning, germplasm management, and improvement of domesticated rice (Wambugu et al., 2013).
Because hybridization with O. sativa is complicated by substantial breeding barriers (e.g., endosperm abortion, F1 sterility, and hybrid breakdown), manipulating the crossability between these two species could accelerate introgression efforts (Chu and Oka, 1970a; Chu and Oka, 1970b; Causse and Ghesquiere, 1991). The success rate of laboratory efforts at hybridization has varied from less than 1 in 1,000 (Ramos, IRRI, Philippines, pers. comm.) to 1 in 50 (Chu and Oka, 1970a; Kaushal and Ravi, 1998). Recovering interspecific progeny has typically required embryo rescue (Causse and Ghesquiere, 1991; Khush et al., 1991; Tao and Sripichitt, 2000; Ramos et al., 2016). In contrast to the strong breeding barriers observed in the laboratory, however, Bezançon et al. (1977); Chu and Oka (1970b); Ghesquière (1986); Kanya (2010), and Kilewa (2014) reported the occurrence of spontaneous hybrids between O. sativa and O. longistaminata, especially along the edges of rice fields in Africa. Chu and Oka (1970b) named these spontaneous hybrids between O. sativa and O. longistaminata, “Obake”, a Japanese word for a spirit that is in a state of change or transformation, which is an apropos metaphor for a hybrid swarm. However, it is unclear how frequent are such spontaneous hybrids between O. sativa and O. longistaminata in Africa and if their occurrence is only recent or a long-extant phenomenon, especially given that O. sativa is not native to Africa. On an applied level, identification of natural hybrids of O. sativa and O. longistaminata, including backcross generations, could facilitate introgression efforts if plant breeders can identify individuals that have O. longistaminata genes, but which are also highly crossable with O. sativa. Given the broad geographic range of O. longistaminata, the species’ great potential as a source of traits for improving domesticated rice, and the need for a continent-wide and genome-wide population genetics analysis, the present study was conducted to 1) characterize the population structure of O. longistaminata throughout its native range in sub-Saharan Africa using 190 accessions primarily from the International Rice Research Institute (IRRI) genebank, 2) quantify genetic diversity of O. longistaminata overall and among its genetic groups, and 3) determine if previous reports of spontaneous interspecific hybrids between O. sativa and O. longistaminata in Africa can be confirmed in the IRRI germplasm collection and to conduct an Africa-wide quantitative assessment of recent and past introgressions.
Materials and methods
Plant materials
We studied 190 accessions of O. longistaminata, collected from much of the species’ native geographic range in sub-Saharan Africa; this included all of the available and viable accessions in the IRRI genebank and one accession from Yunnan University (Table S1). On average, these O. longistaminata accessions have undergone three seed-increases in a screenhouse since acquisition by the IRRI genebank. Up to two individuals from each O. longistaminata accession were sampled, totaling 365 individuals. Seeds were aseptically germinated, and O. longistaminata plants were grown to maturity in a greenhouse in Urbana, IL. Atypical phenotypes in the O. longistaminata individuals were recorded (e.g., lack of rhizomes, filled grains produced by selfing, and short stature; Vaughan, 1994). Control outgroups of O. sativa (n = 309), O. barthii (n = 25), and O. glaberrima (n = 83) were obtained as seeds from the USDA National Plant Germplasm System or Dr. Fengyi Hu’s perennial rice breeding program at Yunnan University, or their sequence data only were accessed via the 3k Rice Genomes Project or the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), with one individual per accession studied for these inbreeding species (Table S1). The control outgroup species were chosen because they comprise, along with O. longistaminata, all of the AA-genome Oryza species known to exist in Africa. Additionally, an O. sativa/O. longistaminata F1 hybrid from a controlled cross, two of its F2 progeny, and 14 recombinant inbred lines (RILs) were included in the study as controls for comparison with putative interspecific hybrids observed in the O. longistaminata accessions. The interspecific hybrid controls were not counted in the O. longistaminata sample size. The control interspecific individuals were from a cross between the Thai O. sativa ssp. indica cultivar RD-23 and a Nigerian O. longistaminata accession (Dayun and Sripichitt, 2000; Hu et al., 2003), both of which were included in the study. In total, 799 individuals were studied: 365 O. longistaminata, 309 O. sativa, 25 O. barthii, 83 O. glaberrima, and 17 known O. sativa/O. longistaminata hybrids or RILs. In a few accessions of all species, the species was mislabeled in the germplasm source, and sample sizes reflect the species identity observed in our study (Table S1). As discussed extensively, some genotypes labeled as O. longistaminata were putative interspecific hybrids with O. sativa; for simplicity, these are included in the O. longistaminata sample size (n = 365).
Molecular markers
The materials were analyzed using 178,651 single-nucleotide polymorphisms (SNPs) and seven plastid microsatellite markers. DNA was extracted from lyophilized seedling-stage leaf tissue using the cetyltrimethylammonium bromide (CTAB) method with minor modifications for the samples sequenced in-house for this study (Fulton et al., 1995). To identify SNPs, restriction site-associated DNA sequencing (RAD-seq) libraries were prepared according to Clark et al. (2014) based on the method of Poland et al. (2012). In brief, DNA from each individual was digested with the restriction enzymes PstI-HF and MspI followed by ligation to barcoded adapters; then, the samples were pooled, and 200–500-bp fragments were selected and amplified by polymerase chain reaction (PCR). Libraries were sequenced on an Illumina Hi-Seq 2000 for 100-bp single-end reads at the Roy J. Carver Biotechnology Center at the University of Illinois. Demultiplexed reads were aligned to the Nipponbare IRGSP-1.0 reference genome using Bowtie 2.2.4 (Langmead et al., 2009). For all individuals in the study, SNPs were called with samtools 1.7 and bcftools 1.7. SNPs were filtered in vcftools 0.1.15 and TASSEL 5.2 such that at each site, only the two most common alleles of each variant were retained, the maximum proportion of heterozygous calls at a site was 0.5, and allelic read depths ranged from 7 to 400 (Bradbury et al., 2007). Individuals with >40% missing calls were dropped from further study. For each set of individuals used in a given iteration of population structure analysis, the SNPs were filtered to require a minimum minor allele frequency of 0.01 and a minimum site count equal to 66% of the number of individuals in the dataset.
Seven plastid markers were amplified from the DNA to check for consistency between maternal and nuclear genotypes, as well as to confirm the directionality of crosses that produced putative interspecific hybrids (Table S2). Some plastid markers were described by Ishii and McCouch (2000), and others were developed from data described by Yin et al. (2015) and Wambugu et al. (2015). The PCR products were size-separated by capillary electrophoresis with Genescan LIZ500 size standard on a 3730xL DNA Analyzer (Applied Biosystems, Foster City, CA, USA) at the Core DNA Sequencing Facility at the University of Illinois. Markers were scored with the software STRand 2.4.59.
Genetic data analysis
To identify genetic subpopulations and assign individuals to those groups, two complementary analyses of population structure were conducted using the ADMIXTURE model in the software STRUCTURE 2.3.4 (Falush et al., 2003) and discriminant analysis of principal components (DAPC) in the R package adegenet (Jombart et al., 2010; Jombart and Ahmed, 2011). Three replications of ADMIXTURE at each K = 1 through K = 9 were run with a burn-in of 10,000 Markov chain Monte Carlo (MCMC) repetitions followed by 50,000 default MCMC repetitions, and the Evanno method as implemented by StructureHarvester was used to identify the optimal number of clusters (Earl and vonHoldt, 2012). For each individual, the proportion of the genome that originated from each cluster, or Q value, was reported from STRUCTURE (Pritchard et al., 2000). For DAPC, principal component analysis was first conducted with the glPca function, and then the find.clusters function was used to make initial groupings with the n.start option set to 500 to ensure convergence; dapc was used to assign individuals to each cluster. The number of clusters with the minimum Bayesian information criterion (BIC) was chosen as optimal. To distinguish putative interspecific hybridization of O. longistaminata from model noise or variation in individual relatedness to a shared common ancestor, putative O. longistaminata interspecific hybrids were defined to have 4.4%–95.6% admixture with control outgroups. The thresholds were chosen because the minimum admixture detected in control O. sativa/O. longistaminata RILs was 4.5% (Table S1). To further determine the ancestry of putative hybrids, three replications of STRUCTURE were run at K = 3 with the USEPOPINFO and PFROMPOPFLAGONLY options set to 1, MIGRPRIOR set to 0, and a burn-in of 10,000 MCMC repetitions followed by 50,000 default MCMC repetitions. Population information was specified only for individuals that had ancestry totally within one group (Table S1). Neighbor-joining trees were generated in the R package ape (Popescu et al., 2012) to observe local topologies. Genetic distance was calculated in TASSEL 5.2.30, and all trees were rooted using root.phylo in the R package ape at the O. barthii individual from accession SRR3231693, which had the maximum pairwise genetic distance observed. Geographic maps were drawn in ArcGIS 10.3.1 (ESRI). A plastid haplotype network was constructed with seven segregating microsatellites for 312 O. longistaminata individuals, 15 putative interspecific hybrids, 5 O. sativa individuals, and 3 O. barthii individuals with no missing data following Clark et al. (2014).
Spatial principal component analysis (sPCA) was conducted in the R package adegenet to identify spatial patterns in genetic variation. To reduce computation time, SNPs were thinned to a minimum distance of 100 bp, and only SNPs with a minor allele frequency greater than 0.01 that had been sampled in all individuals were used, yielding 3,974 SNPs. Genotypes were pooled by accession collection site, and allele frequency was estimated per collection site before analysis; a connectivity network of the collection sites was generated using the minimum spanning method. The lagged principal scores were interpolated by the natural neighbor method in ArcGIS 10.3.1.
Estimates of FST (genetic differentiation among subpopulations), FIS (inbreeding coefficient), and D (Nei’s genetic diversity or expected heterozygosity) were calculated, adjusted for sample size following Nei and Chesser (1983), and averaged across loci using a custom R script. Pairwise estimates of genetic differentiation among genetic groups were estimated with Jost’s D (Jost, 2008) using the pairwiseJostDnumeric R function (Clark, 2016).
Results
Three O. longistaminata genetic groups and one interspecific hybrid group identified
O. longistaminata was differentiated from the control outgroups O. sativa, O. barthii, and O. glaberrima in the STRUCTURE and DAPC analyses (nind = 799, nSNPs = 178,651; Figure 1). The DAPC analysis identified genetic groups with higher resolution, and the optimal number of groups was six: three groups of O. longistaminata, two groups of O. sativa, and one group comprising the African species O. glaberrima and O. barthii (Figure 1A). In contrast, K = 2 was optimal in the STRUCTURE analysis with one cluster including all O. longistaminata individuals and the other including all of the outgroup species, O. sativa, O. barthii, and O. glaberrima (Figure 1A). Notably, 19 individuals that had greater than 4.5% admixture with the outgroup species in STRUCTURE were putative recent interspecific hybrids (Figures 1, 2; Table S1; Table 1). Many of the putative interspecific hybrids had phenotypes that were atypical for O. longistaminata (Table S3).
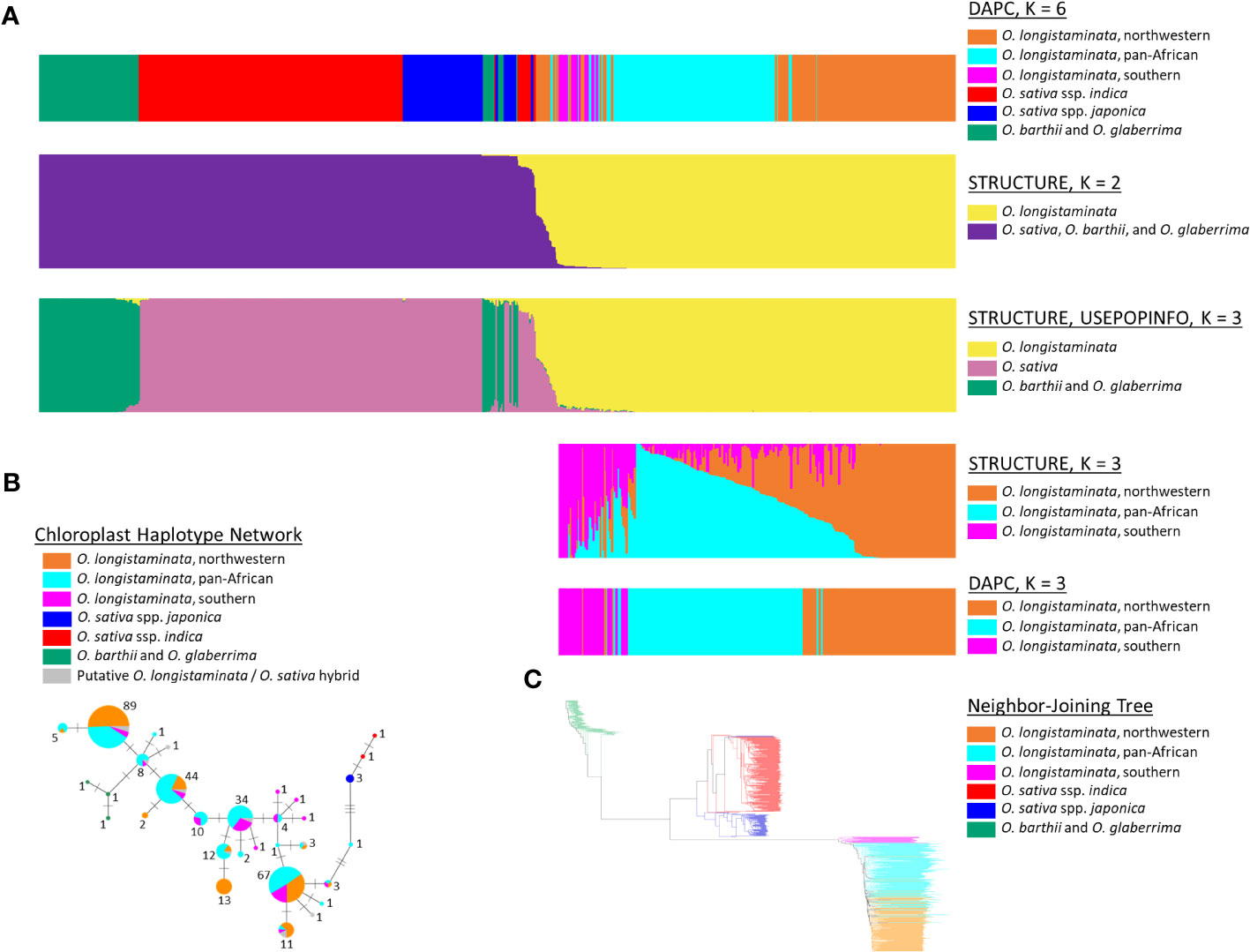
Figure 1 Population structure of Oryza longistaminata and phylogenetic relationships of O. longistaminata to AA-genome species Oryza sativa, Oryza barthii, and Oryza glaberrima. (A) Results for STRUCTURE, discriminant analysis of principal components (DAPC), and STRUCTURE with USEPOPINFO analyses of O. longistaminata (n = 346), O. sativa (n = 309), O. barthii (n = 25), O. glaberrima (n = 83), known O. sativa/O. longistaminata hybrids or recombinant inbred lines (RILs) (n = 17), and putative interspecific hybrids of O. sativa and O. longistaminata (n = 19). (B) Chloroplast haplotype network for 312 O. longistaminata individuals, 15 putative interspecific hybrids of O. longistaminata and O. sativa, 5 O. sativa individuals, and 3 O. barthii individuals. Each haplotype node is represented by a pie chart, the size of which is scaled to the number of individuals carrying the haplotype (shown adjacent) and the colors of which represent the proportion of individuals at that haplotype that carry a given nuclear genotype. (C) Neighbor-joining tree of all individuals studied except interspecific hybrids (both control and putative); individual edges are colored by DAPC-defined nuclear genotypes.
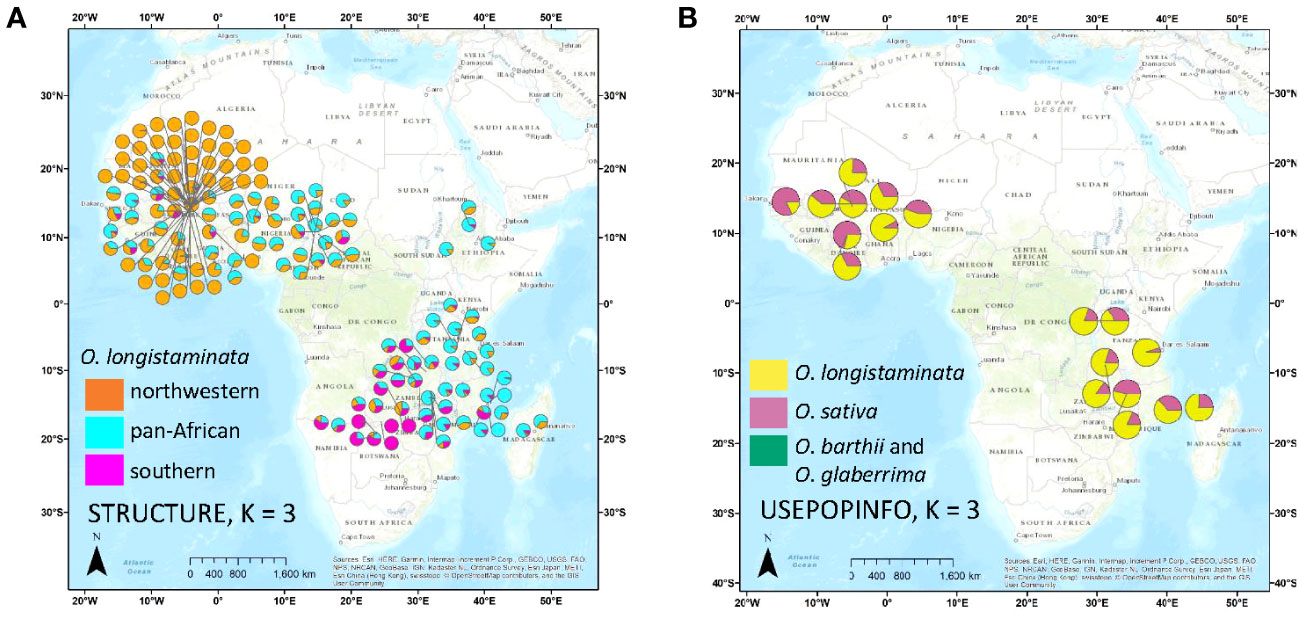
Figure 2 Maps of accession origins and STRUCTURE results for Oryza longistaminata. Ancestry of each accession is represented by a pie chart showing average Q for each genetic group for a given STRUCTURE run. (A) Ancestry at K = 3 of the O. longistaminata individuals <4.5% admixed with control outgroups, Oryza sativa, Oryza barthii, and Oryza glaberrima. (B) Ancestry as determined by STRUCTURE with USEPOPINFO of the O. longistaminata individuals >4.5% admixed with control outgroups.

Table 1 Putative Oryza sativa/Oryza longistaminata progeny with >4.5% O. sativa ancestry and known F1, F2, and RIL control interspecific hybrids derived from a controlled cross.
To investigate population structure within O. longistaminata without potential bias from outgroup species and putative interspecific hybrids, a second set of the DAPC and STRUCTURE analyses was conducted on only the O. longistaminata individuals with less than 4.5% interspecific admixture (nind = 351, nSNPs = 75,371; Figures 1A, 2A). The three O. longistaminata genetic groups observed in the prior analysis were again identified with DAPC (Figure 1A). Geographic maps showed that the three DAPC groups corresponded to Northwestern Africa, Pan-Africa, and Southern Africa (Figure 2A). Compared to the DAPC of all individuals, the DAPC of O. longistaminata with less than 4.5% interspecific admixture led to the reassignment of 25 individuals from the Pan-Africa group to the Southern Africa group (Table S1). The STRUCTURE analysis of the O. longistaminata subset identified K = 2 as optimal. Individuals without intraspecific admixture were concentrated in Northwestern Africa and Southern Africa, but most individuals were admixed between the two O. longistaminata groups. At STRUCTURE K = 3, all but five individuals in the Pan-Africa group were admixed intraspecifically with other O. longistaminata groups (Figure 1A; Table S1). At K = 3, the overall group membership of individuals did not change between DAPC and STRUCTURE (Figure 1A). The three O. longistaminata DAPC groups also formed distinct clades within the neighbor-joining tree (Figure 1C). An individual’s proportion of intraspecific admixture with each of the three O. longistaminata groups was moderately correlated with latitude (r2 = 0.37) and longitude (r2 = 0.44; Table S1).
The plastid haplotype network, which was constructed for O. longistaminata, the putative hybrids, and control O. sativa and O. barthii, consisted of 31 unique haplotypes (Figure 1B). The haplotype network was colored to indicate DAPC-defined groups based on nuclear genotypes (Figure 1B). As expected, plastid haplotypes did not overlap among O. longistaminata, O. sativa, and O. barthii (Figure 1B). The topology of the network differed from the neighbor-joining tree in that O. longistaminata fell between the two outgroups (Figures 1B, C). Among the O. longistaminata individuals that had less than 4.5% admixture with O. sativa, 25 unique haplotypes were identified. Some plastid haplotypes were more commonly found in a given O. longistaminata DAPC group, but no high-frequency haplotypes were specific to a nuclear genotype group. All of the putative recent interspecific hybrids had O. longistaminata plastid haplotypes (Figure 1B). In contrast, the control interspecific hybrids had an O. sativa plastid haplotype, as expected. The recent interspecific hybrids had nine of the 25 O. longistaminata-specific plastid haplotypes, with no particular overrepresentation of a single haplotype (Figure 1B).
Spatial principal component analysis of 3,974 SNPs at 153 geographic sites showed the overall geographic patterns of genetic structure observed with DAPC and STRUCTURE (Figure 3). Two eigenvectors were retained for analysis. The first eigenvector accounted for 11.3% of the genetic variation between sites and differentiated individuals in the northwest from all of the others (Figure 3A). The second eigenvector represented 3.1% of genetic variation between sites and differentiated individuals in the extreme south of the geographic distribution from all others (Figure 3B).
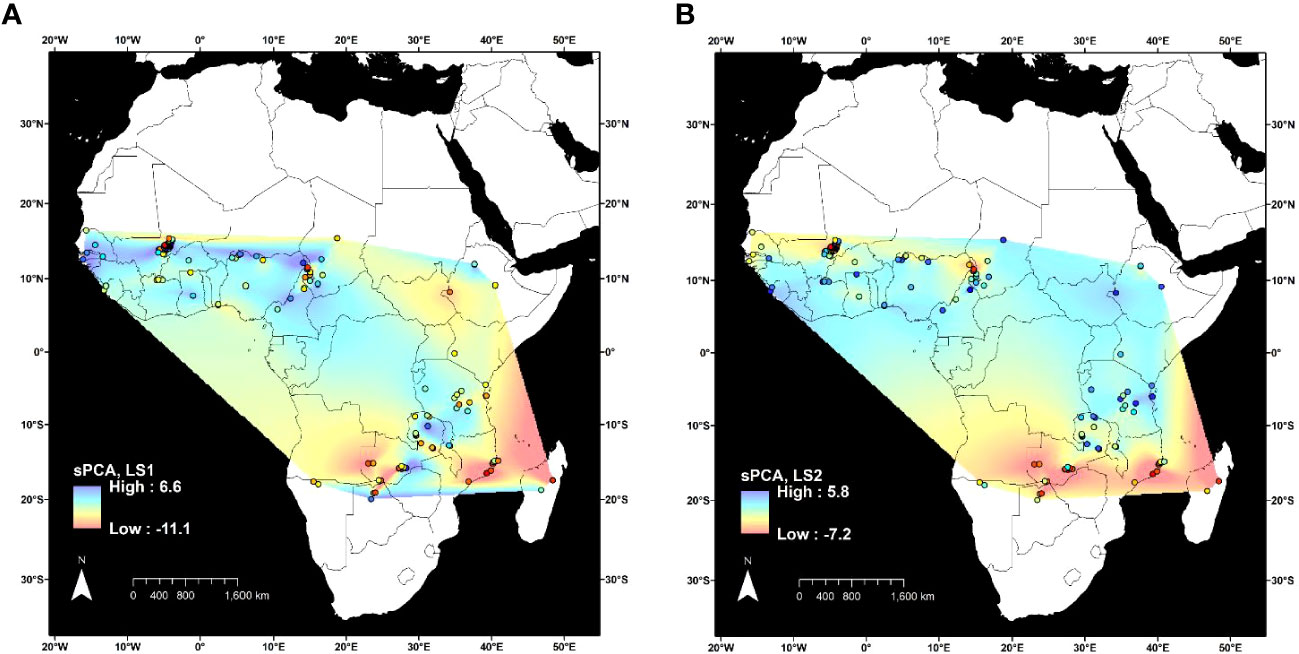
Figure 3 Spatial principal component analysis (sPCA) of 3,974 variants in 346 Oryza longistaminata individuals that were <4.5% admixed with control outgroups Oryza sativa, Oryza glaberrima, and Oryza barthii. Lagged principal component scores for two retained components were plotted for each collection site (circles outlined in black), and values were interpolated between sites by the natural neighbor method. (A) First lagged principal component score. (B) Second lagged principal component score.
Because all but 16 of the 126 individuals in the Northwestern Africa O. longistaminata group were from a 64,000-km2 region of Mali, it is possible that high geographic sampling density led to the distinction of this group (Figure 2A). To examine the effects of uneven geographic sampling, STRUCTURE and DAPC were rerun using genetic data from a set of O. longistaminata accessions filtered to a minimum geographic distance of 25 km (nind = 173, nSNPs = 74,793). STRUCTURE results still showed optimal K = 2 with individuals of singular group ancestry falling in Northwestern and Southern groups. However, DAPC indicated optimal K = 2, and the formerly distinct Northwestern group merged with the Pan-Africa group (data not shown). It is possible that denser geographic sampling of the entire species would maintain the Northwestern group and additionally reveal further substructure in other regions. For example, the sPCA suggested that there may be a distinct northeastern group despite sparse sampling from the northeast in the current dataset (Figure 3B).
Genetic diversity was similar among the three O. longistaminata DAPC groups, with the Northwestern Africa group having the lowest estimate of D (Table 2). Overall, genetic differentiation among the O. longistaminata groups, as indicated by FST and Jost’s D, was low (Tables 2, 3). The Southern Africa group was the most diverse, the most differentiated from the other O. longistaminata groups, and also the most closely related to the outgroup species (Tables 2, 3; Figure 1C). In contrast to the Southern Africa group’s high genetic diversity, the group’s inbreeding coefficient was approximately twice that observed for the other O. longistaminata groups (Table 2). The Pan-Africa subpopulation was the least differentiated from the whole. Among all pairwise comparisons of the O. longistaminata groups, the Northwestern Africa and Southern Africa groups were the most genetically differentiated (Table 3).
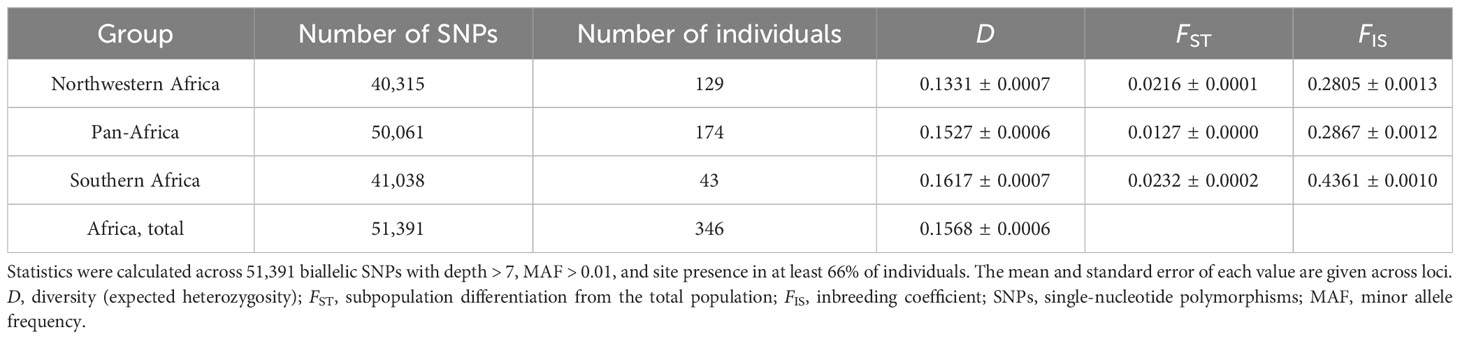
Table 2 Diversity statistics for Oryza longistaminata genetic groups identified by discriminant analysis of principal components (DAPC) based on 346 individuals that were <4.5% admixed with control outgroups.
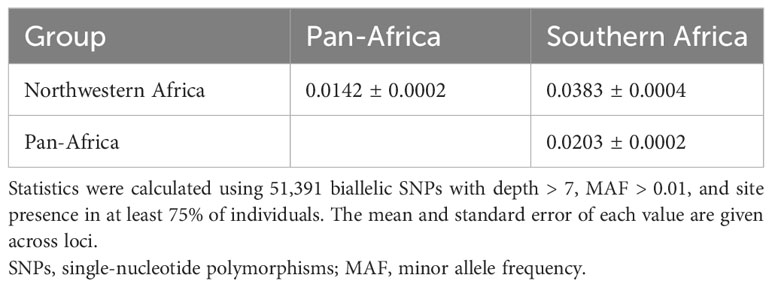
Table 3 Pairwise Jost’s D statistic showing differentiation between Oryza longistaminata genetic groups identified with discriminant analysis of principal components (DAPC) based on 346 individuals that were <4.5% admixed with control outgroups.
O. sativa/O. longistaminata hybrids and bidirectional introgression
To further investigate the ancestry of the previously identified putative recent interspecific hybrids (>4.5% interspecific admixture) and the O. longistaminata observed to have a low proportion of interspecific admixture (<4.5%), a STRUCTURE analysis was conducted with USEPOPINFO (nind = 799, nSNPs = 178,651; Figure 1A). Individuals with zero interspecific admixture were assigned to three predefined groups—O. sativa, O. glaberrima–O. barthii, and O. longistaminata—and used to assign ancestry from those groups to other individuals (Table S1). As controls, the analysis included a known O. sativa/O. longistaminata F1 hybrid from a controlled cross, two of its F2 progeny, and 14 RILs. As expected, the known interspecific hybrids from controlled crosses were correctly identified as hybrids of O. sativa and O. longistaminata (Table 1). The analysis indicated that all individuals previously observed to have more than 4.5% interspecific admixture had ancestry predominantly from O. sativa (Asian) and O. longistaminata (African) and negligible ancestry from O. barthii (African) and O. glaberrima (African; Figure 1A, Table 1). Ancestry from O. sativa was 70% or higher in two individuals (Table 1), indicating introgression of O. longistaminata genes into Asian domesticated rice; however, higher ancestry from O. longistaminata than O. sativa was observed in the other 17 interspecific individuals, indicating introgression of genes from the domesticated Asian species into the undomesticated African species. Four individuals had Q values similar to the F1 and F2 controls, and several other individuals had ancestry ratios near those expected in the BC1 and BC2 generation for introgression into both O. longistaminata and O. sativa (Table 1). The putative early-generation hybrids between O. sativa and O. longistaminata may have been from recent crossing events and were from areas of sub-Saharan Africa where O. sativa is currently cultivated (Food and Agriculture Organization of the United Nations, 2023).
Notably, the results from STRUCTURE with the USEPOPINFO option indicated that all of the individuals in the Southern Africa O. longistaminata group had low-level admixture with O. sativa (mean = 3.7%; range, 2.4%–5.3%) and also with O. barthii–O. glaberrima (mean = 1.3%; range, 0.8%–2.0%). Not all individuals showing interspecific admixture belonged to the Southern group. However, O. sativa ancestry of less than 4.5% was rare in the Northwestern Africa individuals (mean, 0.0%; range, 0.0%–2.5%) and Pan-Africa individuals (mean, 0.4%; range, 0.0%–3.8%). Ancestry from O. barthii–O. glaberrima was also rare in Northwestern Africa (mean, 0.0%; range, 0.0%–0.7%) and Pan-Africa (mean = 0.1%, range = 0.0%–1.5%) O. longistaminata groups.
Discussion
Our study adds support to the current literature consensus that O. longistaminata is more closely related to the Asian species O. sativa than the African species O. barthii and O. glaberrima. We report for the first time that there appear to be three main genetic subpopulations in the sampled O. longistaminata, with populations primarily structured by geographic distance. Moreover, we identified recent spontaneous interspecific hybrids of O. sativa and O. longistaminata in the IRRI genebank collection, which can be immediately useful for breeding. A further novel and notable contribution of this study was the observation of low-level admixture between O. sativa and O. longistaminata in Southern Africa only, which was likely the result of ancient hybridization upon the introduction of O. sativa to Madagascar approximately 1,000 years before the present. Additionally, we provided new nuclear and chloroplast markers for the species.
Relationship between O. longistaminata and the other AA-genome species in Africa
Our study indicated that the undomesticated African rice, O. longistaminata, shared a more recent common ancestor with Asian domesticated rice, O. sativa, than the African O. glaberrima–O. barthii group had (Figure 1C). Most previously conducted studies, including those with the highest genome coverages and sample sizes, have concluded that O. longistaminata is more closely related to O. sativa than to O. barthii and O. glaberrima (Wambugu et al., 2015; Zhu et al., 2014; Cheng et al., 2002; Ren et al., 2003; Kwon et al., 2005; Duan et al., 2007; Marathi et al., 2015); only two studies found O. longistaminata to be more closely related to O. barthii and O. glaberrima than O. sativa (Park et al., 2003; Yin et al., 2015). Our study adds support to the current consensus because it uses the highest genome coverage and the greatest number of individuals of any study to date. A more recent divergence between O. longistaminata and its Asian relative O. sativa than its African relatives, O. glaberrima and O. barthii, suggests that Africa has two distinct lineages of native AA-genome rice species, likely associated with independent migration events.
Population structure in O. longistaminata
To our knowledge, this is the first study to evaluate the O. longistaminata population structure across most of sub-Saharan Africa using densely spaced genome-wide molecular markers. Three genetic groups of O. longistaminata were identified (Figure 1A), two of which were associated with distinct geographic regions of Africa (Northwestern and Southern; Figure 2A), which will be useful information for conserving germplasm of this species and for using this wild relative to improve domesticated rice. Though differentiation among the three O. longistaminata populations was low, geographic distance appeared to be the main factor associated with genetic differentiation; sharp barriers to gene flow were not observed (Figure 3). STRUCTURE, DAPC, sPCA, and Jost’s D consistently indicated that the extremes of differentiation in O. longistaminata were between the Northwestern and Southern populations (Figures 1, 2; Table 3). Similarly, the individuals of the Pan-Africa group showed a gradient of admixture with each of the other genetic groups roughly according to their geographic proximity to each other. These observations were consistent with the species’ biology, which includes perennation with dispersal of rhizomes along rivers during floods (Kleynhans et al., 2007; Green and El-Moghraby, 2009), obligate outcrossing due to self-incompatibility (Ghesquière, 1986), dispersal of seed by birds (Gupta, 2004), and adaptation to tropical environments that were relatively stable during periods of glaciation (Maley, 1996).
The mechanisms of genetic differentiation within O. longistaminata likely included drift and perhaps isolation associated with differences in flowering time. Populations at the edges of a species’ geographic range, such as the Northwestern and Southern populations, can have low population densities that leave individuals with few neighbors with which to outcross; over time, lower effective population size accelerates genetic drift and increases inbreeding. Consistent with this scenario, the Southern Africa O. longistaminata group had substantially greater inbreeding than the other groups, though it also had unexpectedly high genetic diversity (Table 2). Given that the Northwestern and Southern populations are separated by more than 20 degrees of latitude and the equator, variation in flowering time could also lead to isolation. Variation in flowering time could be due to genetic differences in day-length sensitivity, the timing of the growing season, or a combination of both. O. longistaminata has been previously observed to be a short-day plant, though some individuals (most commonly observed near domesticated rice fields) are insensitive to photoperiod (Ghesquière, 1986). In the Urbana, IL greenhouse (40°N), most accessions flowered only during the short days of late autumn and winter, with substantial differences in flowering time among accessions; however, some flowered during the long days of summer (i.e., were apparently day-neutral). Similarly, O. longistaminata accessions collected in Ethiopia (8–12°N) were observed to flower during long days at Jinghong Rice Breeding Station in China (20°N; G. Melaku, Addis Ababa University, Ethiopia, pers. comm.).
Origins of the Southern Africa O. longistaminata group
The Southern Africa group was unique among the three O. longistaminata groups in that all individuals had low-level estimated admixture with O. sativa (Figure 1; Table S1). Two hypotheses were proposed to account for the apparent interspecific admixture that is a defining feature of the Southern Africa group: 1) this group is more similar to the ancestral O. longistaminata population than the other groups, and the apparent admixture with O. sativa is an artifact that represents alleles in common to the ancestral O. longistaminata population and its Asian AA-genome relatives, with many of the alleles subsequently lost in the more derived Pan-Africa and Northwestern Africa groups; 2) the low level of admixture with O. sativa in the Southern Africa O. longistaminata group was the result of ancient interspecific hybridizations and subsequent introgression. However, these hypotheses are not mutually exclusive: it is also possible that the Southern Africa O. longistaminata group could be the center of origin for this species and was also the first to subsequently hybridize with O. sativa.
The proximity of the Southern Africa group to the outgroup species in the neighbor-joining tree could be consistent with the first hypothesis of greater similarity to the ancestral O. longistaminata population, but it would also be expected if the admixture was actually the result of interspecific hybridization (Figure 1C). Additionally, the ordered decrease in genetic diversity of the three O. longistaminata groups from Southern Africa to Pan-Africa to Northwestern Africa could indicate that the species radiated north and west from a southern center of diversity; however, introgression of alleles from another species could explain the high diversity observed in the Southern group (Table 2). The Southern Africa group individuals not only had low levels of ancestry with O. sativa but also had low levels of ancestry with the O. barthii–O. glaberrima group (Figure 1; Table S1). It would have been unlikely that the Southern Africa group individuals hybridized with both O. sativa and O. glaberrima, given that O. glaberrima is primarily cultivated in Northwestern Africa and that O. barthii is sympatric with all of the O. longistaminata groups identified (Vaughan, 1994). Thus, the ordered increase in apparent O. barthii–O. glaberrima ancestry from the posited basal Southern group to the intermediate Pan-Africa group to the most derived Northwestern group would be consistent with an ordered decline in the populations’ relatedness to the AA-genome common ancestral population. This could suggest that apparent coancestry with both O. sativa and O. barthii–O. glaberrima is due to residual relatedness to a common ancestor. However, the observed low-level admixture with O. barthii–O. glaberrima in the Southern Africa O. longistaminata group was also similar to levels observed in the 19 recent hybrids between O. sativa and O. longistaminata (>4.5% admixture) regardless of their geographic origin (Figure 1), which indicates that this likely represents baseline relatedness of O. barthii–O. glaberrima and O. sativa to a common ancestor. Thus, the common-ancestry hypothesis would not account for the greater observed admixture with O. sativa (mean = 3.7%; range, 2.4%–5.3%) than O. barthii–O. glaberrima (mean = 1.3%; range, 0.8%–2.0%) in the Southern Africa O. longistaminata group. Instead, we would expect many O. longistaminata individuals that are introgressed with O. sativa to also show a lower level of coancestry with O. barthii–O. glaberrima, representing alleles derived from a common ancestor.
Bolstering the interspecific hybridization hypothesis is the observation that the current pattern of low-level O. sativa admixture in O. longistaminata (ubiquitous in the Southern Africa group but rare in the Northwestern Africa and Pan-Africa groups) mirrors the historical timing and location of Asian domesticated rice cultivation in Africa. Intriguingly, recent archaeological and molecular genetic data indicate that O. sativa was introduced to southeastern Africa via Madagascar and the Comoros Islands by farmers who migrated from Southeast Asia across the Indian Ocean as early as ~1,000 years before present (Mather et al., 2010; Crowther et al., 2016), which could account for a low level of ancient interspecific admixture in the current Southern Africa O. longistaminata group. However, in Northwestern Africa, O. glaberrima was domesticated from the indigenous wild O. barthii ~3,000 years ago (Linares, 2002). Though Asian domesticated rice, O. sativa, was likely introduced into West Africa by Europeans as early as the mid-16th century, extensive production of Asian rice in West Africa and its replacement of African rice cultivars primarily occurred recently, in the second half of the 20th century (Linares, 2002). Thus, opportunities for hybridization between O. sativa and O. longistaminata in West Africa were likely infrequent until modern times. However, the distribution of O. glaberrima was limited to West Africa. Thus, in southeastern Africa, the introduction of O. sativa directly from Asia ~1,000 years ago (Mather et al., 2010; Nayar, 2014; Crowther et al., 2016) would not have faced competition for cropping space from pre-existing domesticated rice. The archeological record indicates that these ancient rice-growing farmer-migrants from Asia to southeast Africa maintained the knowledge and practice of growing Asian domesticated rice in their new home, as many of their descendants in Madagascar still do today. Given that our study (Figure 2) and others (Chu and Oka, 1970b; Kanya, 2010; Kilewa, 2014) have documented with molecular genetic data recent bidirectional introgression between O. sativa and O. longistaminata, throughout the latter’s natural geographic range (i.e., not limited to Southern Africa), then it would be reasonable to expect that similar ancient sympatric populations would have also produced interspecific progenies where they existed. Moreover, the ancient hybridization hypothesis is consistent with both the high genetic diversity and high inbreeding estimates observed for the Southern Africa O. longistaminata group because introgression of alleles from another species would be expected to increase genetic diversity, and O. sativa/O. longistaminata F1 hybrids are often self-compatible (Ghesquière, 1986; Hu et al., 2003). High rates of inbreeding would not typically be expected if the population represented a center of origin and diversity. Thus, the data from our study suggest that interspecific hybridization and introgression in Northwestern Africa are predominantly recent, whereas in Southern Africa, both recent and likely ancient introgressions have occurred. If the ancient hybridization hypothesis is correct, then the Pan-Africa group may be the most genetically similar to the ancestral O. longistaminata population because it is genetically diverse yet relatively free of low-level introgressions from O. sativa.
Implications of spontaneous O. sativa/O. longistaminata hybridization and bidirectional introgression for germplasm conservation and breeding
The current study and at least three prior studies (Chu and Oka, 1970b; Kanya, 2010; Kilewa, 2014) have produced molecular evidence of recent spontaneous hybridizations between O. sativa and O. longistaminata, lending credence to studies that documented interspecific hybrids without confirmation with molecular markers (Bezançon et al., 1977; Ghesquière, 1986; Kiambi et al., 2018; Kiambi et al., 2005). Moreover, spontaneous progeny derived from backcrosses to each of the parent species have also been observed (Table 1). In contrast to findings of spontaneous interspecific hybridization, plant breeders have typically regarded the cross between O. sativa and O. longistaminata as exceptionally difficult; to rescue interspecific progeny from the abortive endosperm, 1- to 2-week-old embryos or ovules were cultured in vitro (Ramos et al., 2016; Dayun and Sripichitt, 2000). The current study identified 19 early-generation interspecific progeny (>4.5% admixed) from Côte D’Ivoire, Mali, Nigeria, Tanzania, Mozambique, Malawi, and Zambia out of 365 individuals with O. longistaminata ancestry (Table 1). This discovery rate of 5.2% was approximately double the ~2.5% maximum reported from controlled crosses thus far (Kaushal and Ravi, 1998). The present study cannot eliminate the possibility that the recent hybridization events that led to the observed interspecific progeny occurred during seed increases of O. longistaminata accessions in the IRRI screenhouse, but even if so, the rate of hybrid recovery was exceptional compared to that of controlled crosses. The greater frequency of interspecific progeny backcrossed to O. longistaminata than to O. sativa observed in this study could be explained by observational bias because interspecific individuals with greater ancestry from O. sativa would be less likely to exhibit phenotypes typical of O. longistaminata, and therefore explorers or germplasm curators may have been less likely to sample or retain them. However, it is also possible that backcrosses to O. sativa are typically less fit (e.g., exhibit hybrid breakdown) than backcrosses to O. longistaminata. Consistent with the trends observed here, Kilewa (2014) reported that in Tanzania, the spontaneous production of F1 interspecific hybrids from O. sativa fields where wild O. longistaminata grew sympatrically averaged 7.3% for O. longistaminata/O. sativa crosses and 2.6% for O. sativa/O. longistaminata crosses. Additionally, Sacks et al. (2006) observed that seed-set for crosses between an O. longistaminata/O. sativa F1 backcrossed to O. sativa varied among 11 cultivar backcross parents. Given that few controlled crosses between O. sativa and O. longistaminata have been attempted, it is possible that there are substantial differences in general and/or specific interspecific crossability within one or both parental species that could be used to advantage by plant breeders to facilitate introgression for rice improvement.
Although targeted introgression of genes from O. longistaminata into domesticated Asian rice may be desirable in breeding, the introgression of genes from domesticated Asian rice into wild populations of the African O. longistaminata may be considered undesirable genetic pollution (Maxted and Guarino, 2006). Data from our study indicate that interspecific hybridization and introgression in Northwestern Africa are predominantly recent, whereas in Southern Africa, both recent and ancient introgressions have likely occurred. If historical regional differences in the timing of Asian rice cultivation explain why ancient introgressions of O. sativa genes into wild O. longistaminata are common in populations from Southern Africa but not from West Africa, then in the future, the West Africa populations of O. longistaminata will likely accumulate a greater proportion of genes from O. sativa than currently observed.
Given that obtaining O. sativa/O. longistaminata F1 progeny from controlled crosses has required considerable effort (Causse and Ghesquiere, 1991; Khush et al., 1991; Dayun and Sripichitt, 2000; Ramos, IRRI, Philippines, pers. comm.; Kaushal and Ravi, 1998; Ramos et al., 2016), which has been a major barrier to using O. longistaminata for rice improvement, the 19 early-generation interspecific progenies identified in the current study are a valuable resource for breeding improved cultivars of domesticated Asian rice. It should also be possible to mine additional genotypes with O. sativa introgressions from O. longistaminata germplasm collections, thus greatly facilitating breeding. Armed with this knowledge, including which O. longistaminata accessions in the IRRI genebank harbor interspecific hybrids with O. sativa (Tables 1, S1), rice breeders should take full advantage of this newly discovered opportunity. Furthermore, introgression efficiency from controlled crosses can likely be improved by screening O. longistaminata and O. sativa germplasm—including close relatives of the observed interspecific hybrids—for interspecific crossability to identify individuals with relatively high general crossability to the other species. Moreover, among the natural interspecific hybrids in the IRRI genebank may be individuals with exceptional crossability to both parental species, O. sativa and O. longistaminata, and would thus be valuable bridging lines. Given the high genetic and geographic diversity of O. longistaminata, along with its well-documented biotic stress tolerance (Thottappilly and Rossel, 1993; Soriano et al., 1999; Rakotomalala, 2001; Gupta, 2004; Du, 2008; Panigrahi and Rajamani, 2008), likely abiotic stress tolerance (Liu et al., 2004; Giuliani et al., 2013; Atwell et al., 2014), and other known traits of agronomic value (Marathi et al., 2015; Gichuhi et al., 2016; Reinhold-Hurek et al., 2015; Brar, 2005; Ramos et al., 2016), we expect that further selective introgression of O. longistaminata genes into O. sativa will result in new cultivars of great value to humanity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ML: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft. LC: Supervision, Data curation, Writing – review & editing. SZ: Resources, Writing – review & editing, Investigation. FH: Resources, Writing – review & editing, Supervision. DT: Resources, Writing – review & editing. RH: Resources, Supervision, Writing – review & editing. ES: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by U.S. Borlaug Fellows in Global Food Security Graduate Research Grant, Purdue Center for Global Food Security, USAID and University of Illinois at Urbana-Champaign Campus Research Board.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1278196/full#supplementary-material
References
Atwell, B. J., Wang, H., Scafaro, A. P. (2014). Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 215–216 (February), 48–58. doi: 10.1016/j.plantsci.2013.10.007
Bezançon, G., Bozza, J. L., Koffi, G., Second, G. (1977, January). Variability of Oryza longistaminata and the sativa complex of Oryza in Africa. Ecological and evolutive aspects. In Meeting on African Rice Species pp. 25–26.
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23 (19), 2633–2635. doi: 10.1093/bioinformatics/btm308
Brar, D. S. (2005). Broadening the gene pool of rice through introgression from wild species. International Rice Research Institute 2005, 157.
Causse, M. A., Ghesquiere, A. (1991). “Prospective use of Oryza longistaminata for rice breeding,” in Rice Genetics II: Proceedings of the Second International Rice Genetics Symposium (Manila: International Rice Research Institute), 81–89.
Cheng, C., Tsuchimoto, S., Ohtsubo, H., Ohtsubo, E. (2002). Evolutionary relationships among rice species with AA genome based on SINE insertion analysis. Genes Genet. Syst. 77 (5), 323–334. doi: 10.1266/ggs.77.323
Chu, Y., Oka, H. (1970a). The genetic basis of crossing barriers between Oryza perennis subsp. barthii and its related taxa. Evolution 24 (1), 35–144. doi: 10.1111/j.1558-5646.1970.tb01746.x
Chu, Y., Oka, H. (1970b). Introgression across isolating barriers in wild and cultivated oryza species. Evolution 24 (2), 344–355. doi: 10.1111/j.1558-5646.1970.tb01766.x
Clark, L. V. (2016) pairwiseJostDnumeric function. Available at: https://github.com/lvclark/R_genetics_conv/blob/master/pairwiseJostDnumeric.R.
Clark, L. V., Brummer, J. E., Głowacka, K., Hall, M. C., Heo, K., Peng, J., et al. (2014). A footprint of past climate change on the diversity and population structure of Miscanthus sinensis. Ann. Bot. 114 (1), 97–107. doi: 10.1093/aob/mcu084
Crowther, A., Lucas, L., Helm, R., Horton, M., Shipton, C., Wright, H. T., et al. (2016). Ancient crops provide first archaeological signature of the westward Austronesian expansion. Proc. Natl. Acad. Sci. United States America 113 (24), 6635–6640. doi: 10.1073/pnas.1522714113
Dayun, T., Sripichitt, P. (2000). Preliminary report on transfer traits of vegetative propagation from wild rice species to oryza sativa via distant hybridization and embryo rescue. Agric. Nat. Resour. 34 (1), 1–11.
Du, M. (2008). Transfer of Resistance to Yellow Stem Borer (Scirpophaga incertalus Walker) from Wild Species into Rice (Oryza sativa L.) and Molecular Characterization of Introgression. (Los Banos, Philippines: University of the Phillipines Los Banos).
Duan, S., Lu, B., Li, Z., Tong, J., Kong, J., Yao, W., et al. (2007). Phylogenetic analysis of AA-genome Oryza species (Poaceae) based on chloroplast, mitochondrial, and nuclear DNA sequences. Biochem. Genet. 45 (1–2), 113–129. doi: 10.1007/s10528-006-9062-x
Earl, D. A., vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4 (2), 359–361. doi: 10.1007/s12686-011-9548-7
Falush, D., Stephens, M., Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164 (4), 1567–1587. doi: 10.1111/j.1471-8286.2007.01758.x
Food and Agriculture Organization of the United Nations (2023). FAOSTAT statistical database (Rome: FAO).
Fulton, T. M., Chunwongse, J., Tanksley, S. D. (1995). Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13 (3), 207–209. doi: 10.1007/BF02670897
Ghesquière, A. (1986). “Evolution of Oryza longistaminata,” in Rice Genetics: Proceedings of the International Rice Genetics Symposium (Manila: International Rice Research Institute), 15–25. doi: 10.1017/CBO9781107415324.004
Gichuhi, E., Himi, E., Takahashi, H., Maekawa, M. (2016). Characterization and QTL analysis of Oryza longistaminata introgression line, pLIA-1, derived from a cross between Oryza longistaminata and Oryza sativa (Taichung 65) under non-fertilized conditions. J Rice Res. 4 (174), 2. doi: 10.4172/2375-4338.1000174
Giuliani, R., Koteyeva, N., Voznesenskaya, E., Evans, M. A., Cousins, A. B., Edwards, G. E. (2013). Coordination of leaf photosynthesis, transpiration, and structural traits in rice and wild relatives (genus Oryza). Plant Physiol. 162 (3), 1632–1651.
Green, J., El-Moghraby, A. (2009). “Swamps of the Upper White Nile,” in The Nile: origin, environments, limnology and human use. Ed. Dumont, H. (Dordrecht: Springer Netherlands), 193–204. doi: 10.1007/978-1-4020-9726-3_10
Gupta, A. (2004). WIPO-UNEP study on the role of intellectual property rights in the sharing of benefits arising from the use of biological resources and associated traditional knowledge (Vol. 769). WIPO.
Hu, F. Y., Tao, D. Y., Sacks, E., Fu, B. Y., Xu, P., Li, J., et al. (2003). Convergent evolution of perenniality in rice and sorghum. Proc. Natl. Acad. Sci. United States America 100 (7), 4050–4054. doi: 10.1073/pnas.0630531100
Ishii, T., McCouch, S. R. (2000). Microsatellites and microsynteny in the chloroplast genomes of Oryza and eight other Gramineae species. Theor. Appl. Genet. 100, 1257–1266. doi: 10.1007/s001220051432
Jombart, T., Ahmed, I. (2011). adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27 (21), 3070–3071. doi: 10.1093/bioinformatics/btr521
Jombart, T., Devillard, S., Balloux, F., Falush, D., Stephens, M., Pritchard, J., et al. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11 (1), 94. doi: 10.1186/1471-2156-11-94
Jost, L. (2008). GST and its relatives do not measure differentiation. Mol. Ecol. 17 (18), 4015–4026.
Kanya, J. I. (2010). Gene flow between cultivated rice (oryza sativa) and the wild rice (oryza longistaminata), and its potential ecological consequences in Kenya. Doctoral dissertation. (Kenya: University of Nairobi).
Kaushal, P., Ravi (1998). Crossability of wild species of Oryza with O. sativa cvs PR 106 and Pusa Basmati 1 for transfer of bacterial leaf blight resistance through interspecific hybridization. J. Agric. Sci. 130 (4), 423–430. doi: 10.1017/S0021859698005383
Khush, G., Bacalangco, E., Ogawa, T. (1991). 18. A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. Newslett. 7, 121–122.
Kiambi, D. K., Ford-Lloyd, B. V., Jackson, M. T., Guarino, L., Maxted, N., Newbury, H. J. (2018). Collection of wild rice (Oryza L.) in east and southern Africa in responsed to genetic erosion. PGR Newslett. 142 (142), 10–20.
Kiambi, D., Newbury, H., Ford-Lloyd, B., Dawson, I. (2005). Contrasting genetic diversity among Oryza longistaminata (A. Chev et Roehr) populations from different geographic origins using AFLP. Afr. J. Biotechnol. 4 (4), 308–317.
Kilewa, R. A. (2014). Distribution of wild rice species and hybridization between cultivated rice and Oryza longistaminata in Tanzania (Adelaide, Australia: University of Adelaide).
Kleynhans, M. T., James, C. S., Birkhead, A. L. (2007). Hydrologic and hydraulic modelling of the Nyl River floodplain Part 3: Applications to assess ecological impact. Water SA 33 (1), 21–25. doi: 10.4314/wsa.v33i1.47867
Kwon, S.-J., Park, K.-C., Kim, J.-H., Lee, J. K., Kim, N.-S. (2005). Rim 2/Hipa CACTA transposon display: a new genetic marker technique in Oryza species. BMC Genet. 6, 15. doi: 10.1186/1471-2156-6-15
Langmead, B., Trapnell, C., Pop, M., Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10 (3), R25. doi: 10.1186/gb-2009-10-3-r25
Linares, O. F. (2002). African rice (Oryza glaberrima): History and future potential. Proc. Natl. Acad. Sci. 99 (25), 16360–16365. doi: 10.1073/pnas.252604599
Liu, L., Lafitte, R., Guan, D. (2004). Wild Oryza species as potential sources of drought-adaptive traits. Euphytica 138 (2), 149–161. doi: 10.1023/B:EUPH.0000046801.27042.14
Maley, J. (1996). “The African rain forest - main characteristics of changes in vegetation and climate from the Upper Cretaceous to the Quaternary,” in Essays on the Ecology of the Guinea-Congo Rainforest. Eds. Alexander, R., Swaine, I., Watling, M. (Edinburgh, Scotland: Proceedings of the Royal Society of Edinburgh), 31–73.
Marathi, B., Ramos, J., Hechanova, S. L., Oane, R. H., Jena, K. K. (2015). SNP genotyping and characterization of pistil traits revealing a distinct phylogenetic relationship among the species of Oryza. Euphytica 201 (1), 131–148. doi: 10.1007/s10681-014-1213-2
Mather, K. A., Molina, J., Flowers, J. M., Rubinstein, S., Rauh, B. L., Lawton-Rauh, A., et al. (2010). Migration, isolation and hybridization in island crop populations: the case of Madagascar rice. Mol. Ecol. 19 (22), 4892–4905. doi: 10.1111/j.1365-294X.2010.04845.x
Maxted, N., Guarino, L. (2006). Genetic erosion and genetic pollution of crop wild relatives. in Genetic erosion and pollution assessment methodologies. Proceedings of PGR Forum Workshop, vol. 5, 35–45.
Melaku, G., Haileselassie, T., Feyissa, T., Kiboi, S. (2013). Genetic diversity of the African wild rice (Oryza longistaminata Chev. et Roehr) from Ethiopia as revealed by SSR markers. Genet. Resour. Crop Evol. 60 (3), 1047–1056. doi: 10.1007/s10722-012-9900-0
Mohanty, S., Wassmann, R., Nelson, A., Moya, P., Jagadish, S. V. K. (2013). Rice and climate change: significance for food security and vulnerability. International Rice Research Institute 14, 1–14.
Nayar, N. M. (2014). “The Origin of African Rice,” in Origin and Phylogeny of Rices (Waltham, Massachusetts, USA: Elsevier), 117–168. doi: 10.1016/B978-0-12-417177-0.00005-X
Nei, M., Chesser, R. K. (1983). Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 47 (3), 253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x
Panigrahi, D., Rajamani, S. (2008). Genetic evaluation of the wild oryza species for resistance against the yellow stem borer, scirophaga incertulas wlk. J. Plant Prot. Environ. 5 (1), 26–29.
Park, K. C., Kim, N. H., Cho, Y. S., Kang, K. H., Lee, J. K., Kim, N. S. (2003). Genetic variations of AA genome Oryza species measured by MITE-AFLP. Theor. Appl. Genet. 107 (2), 203–209. doi: 10.1007/s00122-003-1252-x
Poland, J. A., Brown, P. J., Sorrells, M. E., Jannink, J. L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PloS One 7 (2). doi: 10.1371/journal.pone.0032253
Popescu, A. A., Huber, K. T., Paradis, E. (2012). Ape 3.0: New tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics 28 (11), 1536–1537. doi: 10.1093/bioinformatics/bts184
Prahalada, G. D., Marathi, B., Vinarao, R., Kim, S. R., Diocton, R., Ramos, J., et al. (2021). QTL mapping of a novel genomic region associated with high out-crossing rate derived from Oryza longistaminata and development of new CMS lines in rice, O. sativa L. Rice 14, 1–16. doi: 10.1186/s12284-021-00521-9
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 (2), 945–959.
Rakotomalala, M. (2001). “Case History on Development and Deployment of Resistant Varieties to Control RYMV in Madagascar,” in Plant Virology in Sub-Saharan Africa. Eds. Hughes, B., Odu, J. (Ibadan: International Institute of Tropical Agriculture), 423–341.
Ramos, J. M., Furuta, T., Uehara, K., Chihiro, N., Angeles-Shim, R. B., Shim, J., et al. (2016). Development of chromosome segment substitution lines (CSSLs) of Oryza longistaminata A. Chev. & R??hr in the background of the elite japonica rice cultivar, Taichung 65 and their evaluation for yield traits. Euphytica 210 (2), 151–163. doi: 10.1007/s10681-016-1685-3
Reinhold-Hurek, B., Hurek, T., Hu, L., Wang, Q., Yang, H. (2015). Transgenic plants for nitrogen fixation (Geneva, Switzerland: World Intellectual Property Association). (WO2015036593A2).
Ren, F., Lu, B.-R., Li, S., Huang, J., Zhu, Y. (2003). A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. TAG Theor. Appl. Genet. 108 (1), 113–120. doi: 10.1007/s00122-003-1414-x
Ronald, P. C., Albano, B., Tabien, R., Abenes, L., Wu, K. S., McCouch, S., et al. (1992). Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Gen. Genetics: MGG 236 (1), 113–120. doi: 10.1007/BF00279649
Sacks, E. J., Schmit, V., McNally, K. L., Sta. Cruz, M. T. (2006). Fertility in an interspecific rice population and its effect on selection for rhizome length. Field Crops Res. 95 (1), 30–38. doi: 10.1016/j.fcr.2005.01.026
Sanni, K. A., Tia, D. D., Ojo, D. K., Ogunbayo, A. S., Sikirou, M., Hamilton, N. R. S. (2013). Diversity of rice and related wild species in Africa. In Realizing Africa's rice promise (Wallingford UK: CABI), 87–94. doi: 10.1079/9781845938123.0087
Seck, P. A., Diagne, A., Mohanty, S., Wopereis, M. C. S. (2012). Crops that feed the world 7: Rice. Food Secur. 4 (1), 7–24. doi: 10.1007/s12571-012-0168-1
Soriano, I. R., Schmit, V., Brar, D. S., Prot, J.-C., Reversat, G. (1999). Resistance to rice root-knot nematode Meloidogyne graminicola identified in Oryza longistaminata and O. glaberrima. Nematology 1 (4), 395–398. doi: 10.1163/156854199508397
Tao, D., Sripichitt, P. (2000). Preliminary Report on Transfer Traits of Vegetative Propagation from Wild Rice Species to Oryza sativa via Distant Hybridization and Embryo Rescue. Kasetsart Journal: Natural Sci. 34 (1), 117–124.
Thottappilly, G., Rossel, H. (1993). Evaluation of resistance to rice yellow mottle virus in oryza species. Indian J. Virol. 9 (1), 65–73.
Vaughan, D. A. (1994). The Wild Relatives of Rice: A Genetic Resources Handbook. 1st ed (Manila: International Rice Research Institute).
Wambugu, P. W., Brozynska, M., Furtado, A., Waters, D. L., Henry, R. J. (2015). Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 5 (1), 13957. doi: 10.1038/srep13957
Wambugu, P. W., Furtado, A., Waters, D., Nyamongo, D. O., Henry, R. J. (2013). Conservation and utilization of African Oryza genetic resources. Rice (New York N.Y.) 6 (1), 29. doi: 10.1186/1939-8433-6-29
Yin, H., Akimoto, M., Kaewcheenchai, R., Sotowa, M., Ishii, T., Ishikawa, R. (2015). Inconsistent diversities between nuclear and plastid genomes of AA genome species in the genus Oryza. Genes Genet. Syst. 90 (5), 269–281. doi: 10.1266/ggs.14-00063
Zhang, S., Hu, J., Yang, C., Liu, H., Yang, F., Zhou, J., et al. (2017). Genotype by environment interactions for grain yield of perennial rice derivatives (Oryza sativa L./Oryza longistaminata) in southern China and Laos. Field Crops Res. 207, 62–70. doi: 10.1016/j.fcr.2017.03.007
Zhang, S., Huang, G., Zhang, Y., Lv, X., Wan, K., Liang, J., et al. (2023). Sustained productivity and agronomic potential of perennial rice. Nat. Sustainability 6, 28–38. doi: 10.1038/s41893-022-00997-3
Zhu, T., Xu, P. Z., Liu, J. P., Peng, S., Mo, X. C., Gao, L. Z. (2014). Phylogenetic relationships and genome divergence among the AA- genome species of the genus Oryza as revealed by 53 nuclear genes and 16 intergenic regions. Mol. Phylogenet. Evol. 70 (1), 348–361. doi: 10.1016/j.ympev.2013.10.008
Keywords: Oryza longistaminata, Oryza sativa (domesticated Asian rice), Oryza glaberrima (domesticated African rice), interspecific hybridization, crop wild relatives (CWR), AA genome, germplasm conservation, population genetics
Citation: Labroo MR, Clark LV, Zhang S, Hu F, Tao D, Hamilton RS and Sacks EJ (2023) Solving the mystery of Obake rice in Africa: population structure analyses of Oryza longistaminata reveal three genetic groups and evidence of both recent and ancient introgression with O. sativa. Front. Plant Sci. 14:1278196. doi: 10.3389/fpls.2023.1278196
Received: 15 August 2023; Accepted: 17 October 2023;
Published: 15 November 2023.
Edited by:
Ahmed Amri, International Center for Agriculture Research in the Dry Areas (ICARDA), EthiopiaReviewed by:
Stefania Grando, Consultant, Ascoli Piceno, ItalyYogesh Vikal, Punjab Agricultural University, India
Copyright © 2023 Labroo, Clark, Zhang, Hu, Tao, Hamilton and Sacks. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erik J. Sacks, ZXNhY2tzQGlsbGlub2lzLmVkdQ==
†Present addresses: Marlee R. Labroo, Bayer Crop Science, Chesterfield, MO, United States
Lindsay V. Clark, Research Scientific Computing, Seattle Children’s Research Institute, Seattle, WA
 Marlee R. Labroo
Marlee R. Labroo Lindsay V. Clark
Lindsay V. Clark Shilai Zhang
Shilai Zhang Fengyi Hu
Fengyi Hu Dayun Tao
Dayun Tao Ruaraidh Sackville Hamilton
Ruaraidh Sackville Hamilton Erik J. Sacks
Erik J. Sacks