
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Plant Sci. , 13 September 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1277625
This article is part of the Research Topic Genomics and Gene Editing of Orphan Plants View all 5 articles
Editorial on the Research Topic
Genomics and gene editing of orphan plants
Orphan crops are of vital importance for global food security. These crops have the capacity to provide nutritious sustenance and show strong adaptability to changing climates. The cultivation and conservation of these crops contribute to safeguarding biodiversity and supporting small-scale farmers by offering diverse income opportunities. These plants display durability to survive under challenging environmental conditions such as drought, extreme temperatures, and pest exposure. Scientists can enhance crop quality and productivity by utilizing controlled breeding methods and advanced genetic engineering techniques like the CRISPR/Cas system (Figure 1). Moreover, gene editing tools like TALENs and ZFNs have been employed to tackle the unique difficulties, such as diseases, nutrition, breeding, etc., posed by orphan crops (Venezia and Creasey Krainer, 2021). Fragrant Rice is known for its smell because of its defective badh2 allele encoding betaine aldehyde dehydrogenase (BADH2), which produces 2-acetyl-1-pyrroline (2AP). TALENs were engineered to disrupt the OsBADH2 gene, increasing the 2AP content through homozygous mutations. Therefore, targeted mutagenesis using TALENs is useful in creating important agronomic traits (Shan et al., 2015). Genomics and gene editing technologies accelerate breeding by enabling scientists to identify and select genes underlying desired traits quickly (Chen et al., 2019), while efficient transformation processes are crucial for applying Cas9 to target sequences, enhancing the accuracy and precision of gene editing techniques (Cattivelli et al., 2008; Movahedi et al., 2023). Plants with specific traits are developed by minimizing off-target effects and ensuring successful Cas9 conjugation with a guide RNA molecule, which helps Cas9 locate and bind to the corresponding DNA sequence. The deployment of this technology has dramatically accelerated breeding programs by swiftly incorporating helpful traits into plant varieties (Wolter et al., 2019). Phyllostachys edulis holds significant global importance as a monopodial bamboo species. The lack of a genetic transformation system poses challenges in elucidating the functionalities of genes that govern crucial traits and conducting molecular breeding in Moso bamboo. Researchers have achieved a 5% increase in plant regeneration and transformation efficiency using plant growth regulators and antibiotic screening. CRISPR/Cas genetic changes have been used to enhance desirable traits in Moso bamboo, such as pest resistance (Huang et al.). Researchers can improve crop productivity, plant quality, yield, and nutritional content by targeting specific genes with CRISPR/Cas (Curtin et al., 2018). This technology offers several advantages for orphan crops, including transferring valuable traits such as disease and stress resistance (Sedeek et al., 2019). Successful application of the CRISPR/Cas system has been demonstrated in sesame, where cytochrome P450 genes involved in sesamin and sesamolin biosynthesis were successfully targeted, resulting in expected genetic modifications (You et al.). Sequencing the genomes of different orphan plant varieties provides the basis for identifying essential genes responsible for desirable traits (Gasparini et al., 2021). Genomic insights into orphan plants offer opportunities to manipulate genes to enhance traits like taste, aroma, and nutritional value. For instance, in Physalis pruinosa, genomic resources and efficient transformation techniques were developed (Lemmon et al., 2018). Through CRISPR/Cas genome editing, orthologues of tomato domestication and improved genes were mutated, significantly enhancing plant architecture, flower production, and fruit size. This rapid development of targeted allelic diversity and novel breeding germplasm showcases the potential for improving plants distantly related to crops (Lemmon et al., 2018). By specifically targeting genes involved in secondary metabolite synthesis, CRISPR/Cas editing has the potential to enhance the nutritional value of orphan plants by improving flavor profiles and increasing the production of beneficial compounds. In one study, Cas9 was employed to inactivate the kauniolide synthase genes in Cichorium intybus, interrupting sesquiterpene lactone biosynthesis. These findings highlight the potential of CRISPR/Cas in enhancing the nutritional value of chicory taproots by altering the biosynthesis of sesquiterpene lactones (Cankar et al.). Moreover, genomics and gene editing technologies, like CRISPR/Cas, advance agriculture by editing plant genomes, enhancing sustainability, and reducing reliance on chemical pesticides and fertilizers (Qaim, 2020). However, it is crucial to address the regulatory and ethical considerations surrounding gene editing in orphan crops (Sprink et al., 2016). Public engagement and science communication are necessary to fully realize the potential of genomics and gene editing and promote responsible and sustainable agricultural practices. Gene knock-in using CRISPR-Cas in plants is significant, as it enables the insertion of favorite genes into specific locations within the plant genome (Zhang et al., 2017; Movahedi et al., 2022). Site-specific gene knock-in using CRISPR/Cas enables precise and targeted insertion of desired genes into specific loci within the plant genome, making it highly significant in plant biology and crop enhancement. This technique has been applied to Chlamydomonas reinhardtii to facilitate the insertion of an optimized foreign bacterial phytase gene at a specific site (Shahabadi et al.). These findings indicate that CRISPR/Cas can be used to insert a foreign gene into the nucleus, showing its effectiveness in accurately adding genes to specific locations without causing adverse effects on gene expression.
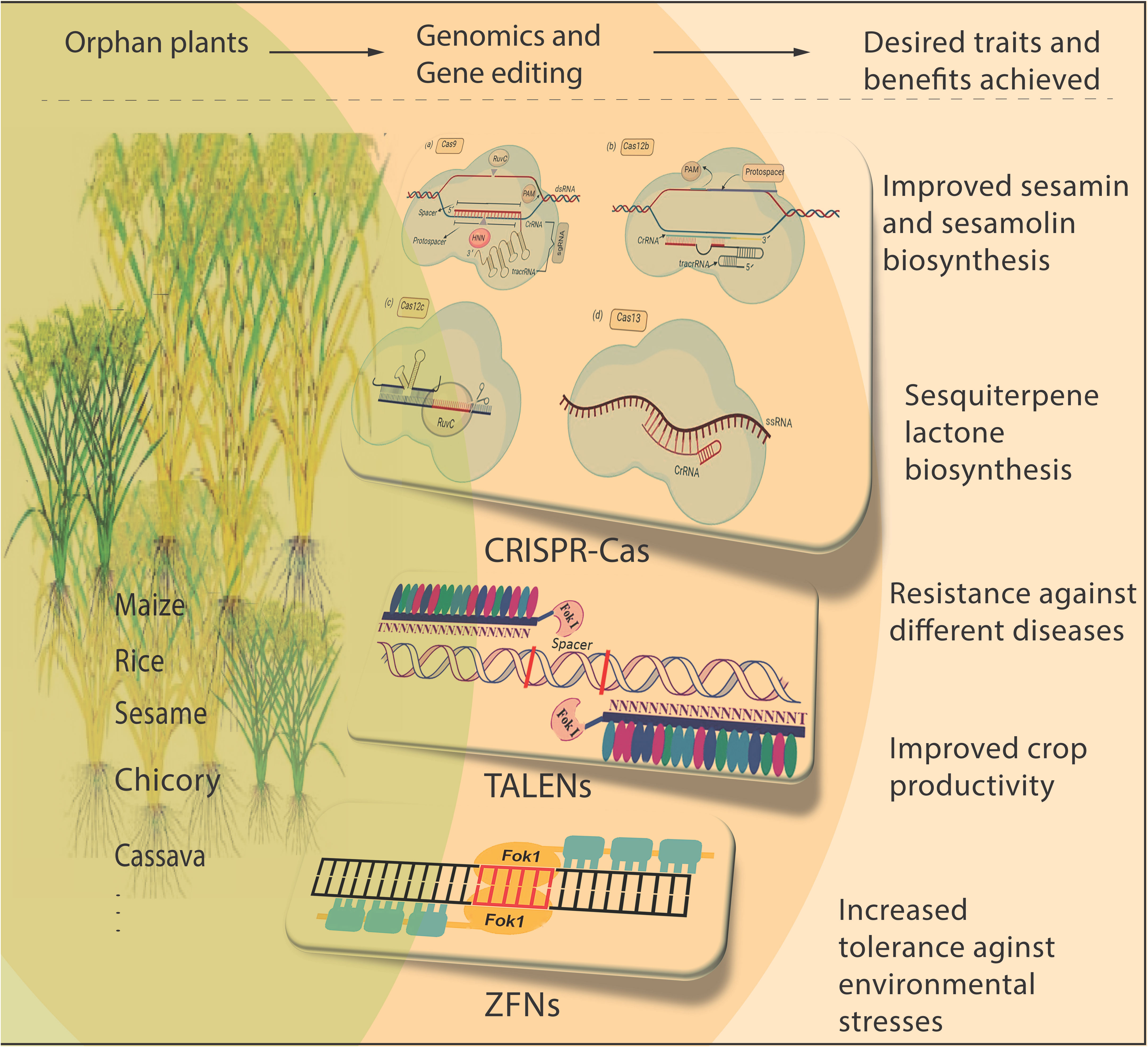
Figure 1 Pictorial diagram illustrating genomics and gene editing technologies for improving desirable traits in orphan crops. Genome editing technologies are being developed to enhance the productivity and disease resistance of orphan crops like rice, maize, cassava chicory, etc. For instance, sesame can benefit from sesamin and sesamolin biosynthesis, while chicory can benefit from sesquiterpene lactone biosynthesis. The CRISPR/Cas, TALENs, and ZFN technologies have transformed the field of plant genetics by providing precise tools to modify specific traits by altering the genome editing of plants.
To summarize, combining genomics and gene editing offers excellent opportunities to tackle worldwide concerns regarding food security. It also provides a means to enhance the flavor, fragrance, and nutritional content of underutilized plants. These technologies facilitate the engineering of improved crop varieties with beneficial characteristics, leading to better nutrition and overall health.
AM: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing. BP: Validation, Writing – review & editing. SK: Validation, Writing – review & editing.
Our gratitude goes out to all the authors for their exceptional contributions, as well as the reviewers and other associate editors for their valuable feedback and inputs, which were pivotal in the success of this Research Topic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cattivelli, L., Rizza, F., Badeck, F.-W., Mazzucotelli, E., Mastrangelo, A. M., Francia, E., et al. (2008). Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crops Res. 105, 1–14. doi: 10.1016/j.fcr.2007.07.004
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Curtin, S. J., Xiong, Y., Michno, J.-M., Campbell, B. W., Stec, A. O., Čermák, T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 16, 1125–1137. doi: 10.1111/pbi.12857
Gasparini, K., Moreira, J. D. R., Peres, L. E. P., Zsögön, A. (2021). De novo domestication of wild species to create crops with increased resilience and nutritional value. Curr. Opin. Plant Biol. 60, 102006. doi: 10.1016/j.pbi.2021.102006
Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., et al. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Movahedi, A., Wei, H., Kadkhodaei, S., Sun, W., Zhuge, Q., Yang, L., et al. (2023). CRISPR-mediated genome editing in poplar issued by efficient transformation. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1159615
Movahedi, A., Wei, H., Zhou, X., Fountain, J. C., Chen, Z.-H., Mu, Z., et al. (2022). Precise exogenous insertion and sequence replacements in poplar by simultaneous HDR overexpression and NHEJ suppression using CRISPR-Cas9. Hortic. Res. 9, uhac154. doi: 10.1093/hr/uhac154
Qaim, M. (2020). Role of new plant breeding technologies for food security and sustainable agricultural development. Appl. Economic Perspect. Policy 42, 129–150. doi: 10.1002/aepp.13044
Sedeek, K. E. M., Mahas, A., Mahfouz, M. (2019). Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00114
Shan, Q., Zhang, Y., Chen, K., Zhang, K., Gao, C. (2015). Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13, 791–800. doi: 10.1111/pbi.12312
Sprink, T., Eriksson, D., Schiemann, J., Hartung, F. (2016). Regulatory hurdles for genome editing: process- vs. product-based approaches in different regulatory contexts. Plant Cell Rep. 35, 1493–1506. doi: 10.1007/s00299-016-1990-2
Venezia, M., Creasey Krainer, K. M. (2021). Current advancements and limitations of gene editing in orphan crops. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.742932
Wolter, F., Schindele, P., Puchta, H. (2019). Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19, 176. doi: 10.1186/s12870-019-1775-1
Keywords: CRISPR-Cas, orphan crops, genomics, gene editing, genetic improvement
Citation: Movahedi A, Pucker B and Kadkhodaei S (2023) Editorial: Genomics and gene editing of orphan plants. Front. Plant Sci. 14:1277625. doi: 10.3389/fpls.2023.1277625
Received: 14 August 2023; Accepted: 04 September 2023;
Published: 13 September 2023.
Edited and Reviewed by:
Channakeshavaiah Chikkaputtaiah, North East Institute of Science and Technology (CSIR), IndiaCopyright © 2023 Movahedi, Pucker and Kadkhodaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ali Movahedi, YWxpX21vdmFoZWRpQG5qZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.