- 1School of Breeding and Multiplication (Sanya Institute of Breeding and Multiplication), Hainan University, Sanya, China
- 2Key Laboratory of Biology and Genetic Resources of Tropical Crops, Ministry of Agriculture, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Hainan Institute for Tropical Agricultural Resources, Haikou, China
Phosphatidylethanolamine binding protein (PEBP) plays an important role in regulating flowering time and morphogenesis of plants. However, the identification and functional analysis of PEBP gene in pineapple (AcPEBP) have not been systematically studied. The pineapple genome contained 11 PEBP family members, which were subsequently classified into three subfamilies (FT-like, TFL-like and MFT-like) based on phylogenetic relationships. The arrangement of these 11 shows an unequal pattern across the six chromosomes of pineapple the pineapple genome. The anticipated outcomes of the promoter cis-acting elements indicate that the PEBP gene is subject to regulation by diverse light signals and endogenous hormones such as ethylene. The findings from transcriptome examination and quantitative real-time polymerase chain reaction (qRT-PCR) indicate that FT-like members AcFT3 and AcFT4 display a heightened expression level, specifically within the floral structures. The expression of AcFT3 and AcFT4 increases sharply and remains at a high level after 4 days of ethylene induction, while the expression of AcFT7 and AcMFT1 decreases gradually during the flowering process. Additionally, AcFT3, AcFT4 and AcFT7 show specific expression in different floral organs of pineapple. These outcomes imply that members belonging to the FT-like subfamily may have a significant impact on the process of bud differentiation and flower development. Through transcriptional activation analysis, it was determined that AcFT4 possesses transcriptional activation capability and is situated in the nucleus and peripheral cytoplasm. Overexpression of AcFT4 in Arabidopsis resulted in the promotion of early flowering by 6-7 days. The protein interaction prediction network identified potential flower regulators, including CO, AP1, LFY and SOC1, that may interact with PEBP proteins. This study explores flower development in pineapple, thereby serving as a valuable reference for future research endeavors in this domain.
Introduction
Phosphatidylethanolamine-binding proteins (PEBP) have been found to have significant involvement in flower induction, regulation of flowering time, and seed germination in plants (Zhang et al., 2020; Jin et al., 2021). PEBP has been categorized into three subfamilies, namely FLOWERING LOCUS T-like (FT-like), TERMINAL FLOWER 1 (TFL1-like) and MOTHER OF FT and TFL1 (MFT-like). Within the FT-Like subfamily, there are two members, namely Flowering Locus T (FT) and TWIN SISTER OF FT (TSF). FT serves as an integral factor in the flowering pathway and plays a crucial role in the regulation of the flowering process (Adeyemo et al., 2019). Under conditions of extended daylight, the expression of FT is induced, resulting in the formation of the FT-FD complex with FD. This complex then activates the expression of APETALA1 (AP1) and LEAFY (LFY) genes, thereby facilitating the transition to the flowering stage (Abe et al., 2005; Wigge et al., 2005). In rice, Hd3a and RFT1 play a role in activating the expression of AP1 homologs, namely OsMADS14 and OsMADS15, thereby promoting flowering (Komiya et al., 2008). Furthermore, FT homologs have been extensively identified and studied in various other plant species, such as Lilium (Yan et al., 2021), Cucumis melo L (Zhang et al., 2020; Zhang and Zhang. 2020), sugarcane (Venail et al., 2022), Perilla frutescens (Xu et al., 2022) and Dendrobium huoshanense (Song et al., 2021). There is an increasing body of evidence indicating the significant involvement of FT genes in the flowering mechanism of plants. In the case of grapes, the expression of VvFT serves as an indicator that the developmental transition from juvenile to adult stage has occurred (Carmona et al., 2007). In apples, MdFT1 and MdFT2 exhibit distinct expression patterns, yet both possess the capability to initiate early flowering in apple plants (Kotoda et al., 2010). In cassava, the Arabidopsis FT gene accelerates flower bud formation and increases lateral branching, and its overexpression also significantly increases flower yield and prolongs flower development life (Adeyemo et al., 2017). Furthermore, the translocation expression of CsFTL3 in chrysanthemum has been found to stimulate early flowering in both Arabidopsis and chrysanthemum plants (Oda et al., 2012). These findings suggest that the role of the FT gene in flower induction is conserved across various plant species.
MFT, acknowledged as the most ancient constituent of the PEBP family, governs the process of seed germination by means of the abscisic acid (ABA) and gibberellin (GA) signaling pathways in Arabidopsis (Xi et al., 2010). The TFL1-like subfamily encompasses TERMINAL FLOWER 1 (TFL1), BROTHER OF FT AND TFL1 (BFT), and CENTRORADIALIS (CEN). Both the TFL1-like and FT-like subfamilies participate in the management of flowering. Nevertheless, in comparison to FT, TFL1 exhibits inhibitory characteristics towards the flowering process (Zhu et al., 2020; Song et al., 2021). In cucumber, the antagonistic interaction between CsTFL1 and CsFT, along with the competitive binding with the CsNOT2a-CsFDP complex, hinders cucumber growth and the development of terminal flowers (Wen et al., 2019). Similarly, in rice, the TFL1-like protein RCN inhibits flower development by competitively binding 14-3-3 and FD proteins to Hd3a/RFT1 (Kaneko-Suzuki et al., 2018). Additionally, the overexpression of MiTFL1-1 and MiTFL1-2 genes in mango has been found to significantly impede the flowering in Arabidopsis (Gafni et al., 2022).
The pineapple (Ananas comosus (L.) Merr.) is a tropical fruit that thrives in tropical and subtropical regions. The timing of its flowering significantly impacts both its yield and the optimal time for harvesting. While the PEBP family of genes is recognized for its crucial role in regulating flowering time and flower development (Jin et al., 2021; Kim et al., 2022), the specific characteristics of the PEBP gene family in pineapple and the specific members involved in the flowering process remain uncertain. This study comprehensively analyzed and identified all members of the PEBP gene family in the pineapple genome, encompassing their phylogenetic relationships, chromosome localization, gene structure, conserved motifs and promoter cis-acting elements. Additionally, considering the pivotal role of the PEBP gene in flower development, the expression pattern of AcPEBP and the expression characteristics of FT-like subfamily members were further investigated during flowering. Ultimately, this study provides a theoretical basis for future investigations into the regulatory role of PEBP genes in pineapple flowering.
Materials and methods
Identification of AcPEBP genes in pineapple
Download pineapple gene sequence data from the pineapple genome database (http://pineapple.angiosperms.org/) (Xu et al., 2018). Using the Arabidopsis PEBP protein sequence as a query, the amino acid sequence of Arabidopsis PEBP is obtained from the TAIR database (http://www.arabidopsis.org/) (Garcia-Hernandez et al., 2002). The pineapple database was searched with Blastp, and candidate pineapple AcPEBP family gene sequences were identified. After removing redundant sequences, the candidate AcPEBP family genes were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/cdd) to confirm the presence of the PEBP gene domains. The ExPASy website (http://web.expasy.org/protparam/) is to estimate the molecular weight (MW), isoelectric point (PI) and grand average of hydropathicity (GRAVY) for proteins found in pineapples (Wilkins et al., 1999). The subcellular localization prediction was performed using ProtComp v.9.0 (http://linux1.softberry.com/cgi-bin/programs/proloc/protcomppl.pl) (Li et al., 2020).
Phylogenetic analyses
A multiple sequence alignment was conducted using ClustalX software for pineapple, Arabidopsis, and rice amino acid sequences. Based on 1000 bootstrap analyses performed using MEGA7.0 software with the neighbor-joining (NJ) algorithm (Kumar et al., 2016), the phylogenetic tree was constructed. The tree was visually enhanced and beautified using the chiplot website (https://www.chiplot.online/).
Conserved motifs and exon-intron structure analysis
Based on the whole-genome GFF annotation file, the analysis of exons and introns was performed using the Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn). Based on the conserved domain information obtained from NCBI (https://www.ncbi.nlm.nih.gov/), the conserved motifs of the AcPEBP gene family were determined using the MEME online tool (http://meme-suite.org/tools/meme) (Bailey et al., 2009). The results of gene structure and conserved motif analysis were visualized using the TBtools software (Chen et al., 2020).
Cis-element identification and protein interaction prediction analysis
The promoter sequence of the AcPEBP gene, spanning 2000bp upstream of the transcription start site, was submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for the speculation of cis-regulatory elements in the promoter region (Lescot et al., 2002). The protein-protein interaction (PPI) of AcPEBP was prophesied using the STRING website (https://cn.string-db.org/) (Szklarczyk et al., 2021).
Pineapple AcPEBP synteny analysis by chromosome location and duplication event
By utilizing the genome sequence and general feature format (GFF) file, the TBtools software was employed to retrieve the chromosomal positions of the pineapple PEBP family genes, enabling the creation of a gene localization information map. Obtain the genome sequences of maize, banana, rice, tomato, grape and Arabidopsis by downloading them from the Ensembl plants database (http://plants.ensembl.org/index.html). Collinearity files between each pair of species were obtained using the MCScanX software (http://chibba.pgml.uga.edu/mcscan2/) (Wang et al., 2012). The TBtools software was used to generate the collinearity analysis plot.
Expression profiling of AcPEBP genes by RNA-Seq
The expression data for the AcPEBP genes in various pineapple tissues were obtained from the pineapple transcriptome dataset. Furthermore, the transcriptome data for pineapple were publicly deposited in the National Genomics Data Center database (https://ngdc.cncb.ac.cn/gsa/ (accessed on 6 May 2022)), The assigned accession of the submission is CRA006826.The quantification of transcript abundance for each AcPEBP gene was performed using the Fragments Per Kilobase of exon per Million mapped reads (FPKM) metric. The visualization of RNA-seq data was achieved by generating heatmaps using TBtools.
Plant materials and treatments
Pineapple (Ananas comosus cv. Comte de Paris) was sourced from the Zhanjiang Subtropical Crop Research Institute in China. Plant materials approximately 30 cm long were selected when the pineapples were around 15 months old. The plants were induced with 30 mL of 200 mg/L-1 ethephon, while an equivalent amount of water was used as a control for the untreated materials. Apical buds of pineapple were collected at different time points, including 0 h, 12 h, 1 d, 2 d, 4 d, 1 w, 2 w, 3 w, 4 w, 5 w, 6 w and 7 w after treatment. Each sample was collected with three separate biological replicates. The specimens were rapidly placed in liquid nitrogen and then kept at -80°C in a freezer for later RNA extraction.
RNA extraction and real-time quantitative PCR analysis
Total RNA was extracted from pineapple samples using a polysaccharide polyphenol plant RNA extraction kit (China Huatuanyang Biotechnology Co., LTD.), and the quality and concentration of total RNA were determined by NanoDropTM One/OneC (Thermo Fisher Scientific, USA), an ultra-fine spectrophotometer. Total RNA First Strand cDNA was synthesized by RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). qRT-PCR was performed using the SYBR Green qPCR Master MIX Kit (Thermo Fisher Scientific, USA). Employing the AcActin gene as the internal reference gene, qRT-PCR was performed to calculate the relative expression level of the AcPEBP gene through the 2-ΔΔCt method (Rao et al., 2013). Each sample was replicated three times.
Subcellular localization of PEBP proteins
To validate the subcellular localization of the AcPEBP protein in plant cells, the full-length sequence of AcFT4 was cloned into the pCAMBIA2300-GFP vector to generate the fusion construct (2300 GFP-AcFT4). The fusion construct and control vector were transformed into the Agrobacterium tumefaciens strain GV3101 (ANGYUBio, China). Subsequently, they were used for transformation into healthy tobacco leaves. After 2-3 d, the transient expression of AcFT4-GFP was observed using a confocal laser scanning microscope (Zeiss, Germany).
Transcriptional activity of PEBP proteins
The pGBKT7-AcFT4 fusion vector was constructed and transferred into the yeast strain AH109 for the analysis of AcFT4 protein’s transcriptional activation activity (AngYuBio, China). The transformed colonies were streaked on selective medium SD/−Trp and incubated at 30°C for 48-72 h. Subsequently, the colonies were further selected on SD/−Trp/−His/−Ade medium, and X-α-Gal was added to observe if the AcFT4 protein exhibited transcriptional activation activity.
Expression vector construction and plant transformation
The agrobacterium containing the recombinant plasmid was introduced into Arabidopsis flowers through the floral dip method (Zhang et al., 2006). Arabidopsis plants were cultured at room temperature (22°C) until they matured. Subsequently, transgenic positive plants were selected on MS medium supplemented with 50 mg/mL kanamycin. Healthy and green Arabidopsis plants were chosen for rapid PCR-based DNA screening to confirm positive transgenic individuals. The selected positive seedlings were further screened for T3 generation, to obtain homozygous transgenic lines, and the phenotypes of the transgenic homozygous lines were observed.
Results
Characterization and identification of PEBP proteins in pineapple
The BLASTP search was conducted in the pineapple genome database, utilizing the Arabidopsis PEBP protein as the query. Following the elimination of redundant members, a total of 11 AcPEBP genes were identified and subsequently named based on their resemblance to homologous genes in Arabidopsis. These genes encompass AcFT1-AcFT7, AcTFL1a-AcTFL1b and AcMFT1-AcMFT2. As depicted in Table 1, the AcPEBP family consists of 11 members, exhibiting varying protein lengths that range from 110 amino acids (AcMFT2) to 238 amino acids (AcFT6). The molecular weights of the proteins in this family vary from 12.34 kDa (AcMFT2) to 26.93 kDa (AcFT6). The isoelectric points of these proteins range from 6.96 (AcMFT1) to 9.61 (AcFT1). All members of this protein family exhibit a negative GRAVY value, indicating their hydrophobic nature. Subcellular localization prediction analysis revealed that, with the exception of AcMFT1, which is localized in Chloroplasts and Cytoplasm, the other AcPEBP member proteins are predicted to be located in the nucleus. Additionally, AcFT1 and AcMFT2 are also distributed in the Cytoplasm.
Phylogenetic analysis
In order to examin the evolutionary relationships among AcPEBP members, a phylogenetic tree (Figure 1) was generated using the neighbor-joining technique with MEGA software. This analysis was based on the full-length amino acid sequences obtained from pineapple, Arabidopsis, and rice. The resulting phylogenetic tree classified the 11 AcPEBP proteins, 5 AtPEBP proteins, and 19 OsPEBP proteins into three distinct subfamilies, namely FT-like, TFL-like and MFT-like, as depicted in Figure 1. Among the AcPEBP family members, the FT-like subfamily displayed the greatest abundance of members, with a total of 7 AcPEBP proteins. In contrast, both the TFL-like and MFT-like subfamilies consisted of only 2 members each.
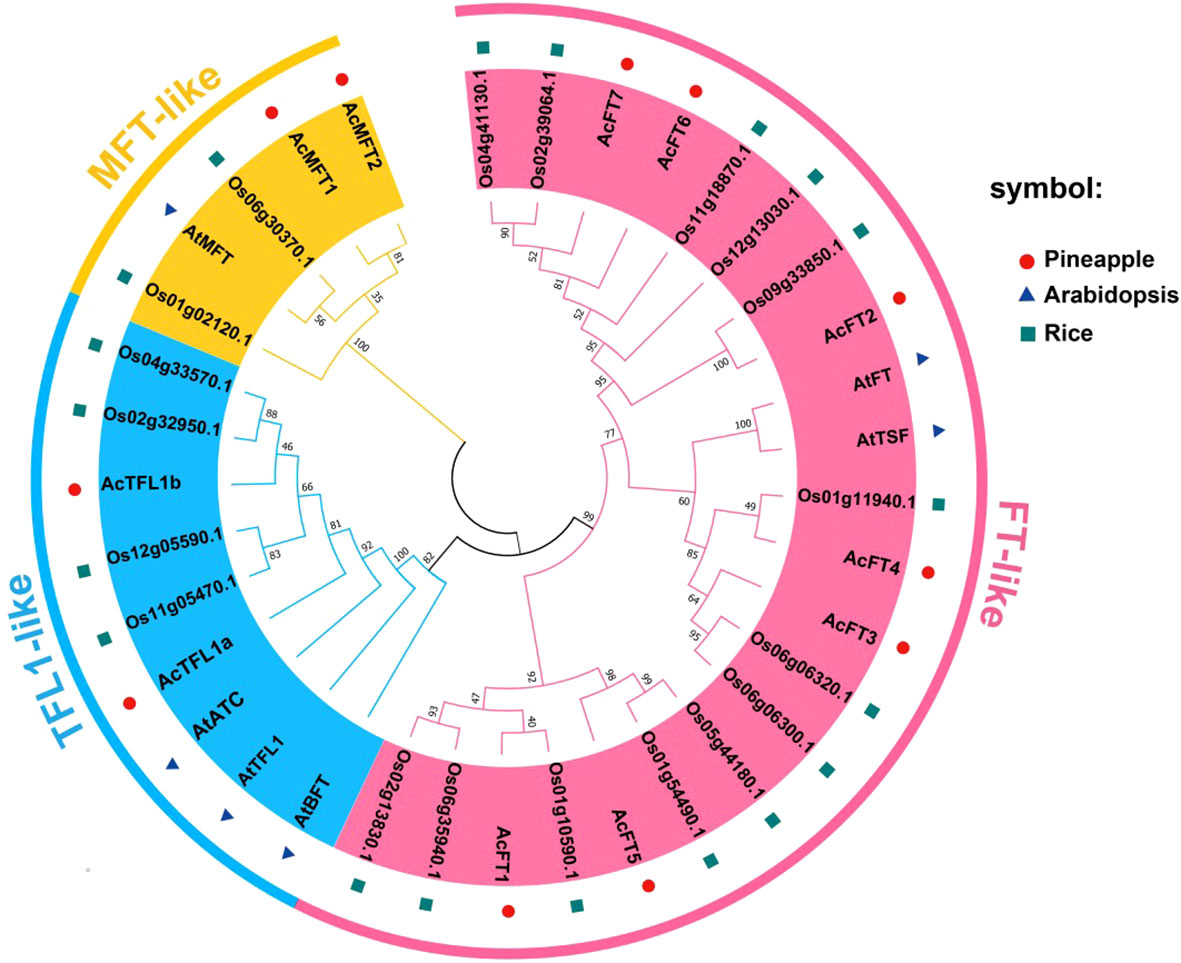
Figure 1 The phylogenetic analysis of PEBP proteins among pineapple, Arabidopsis and rice. The phylo-genetic tree was constructed using MEGA7.0 program. (The neighbor-joining (NJ) algorithm, bootstrap value = 1000). The phylogenetic connections and structural characteristics of the 11 AcPEBP genes.
Gene structures, conserved domains, and motif analysis
To acquire a more thorough comprehension of the AcPEBP gene members, we utilized the MEME website to forecast the conserved motifs present within the AcPEBP family. As illustrated in Figure 2, the number of conserved motifs in each AcPEBP protein ranges from Motif 4 to Motif 8. All members include motifs Motif 2, Motif 4, and Motif 5, while a majority of the members exhibit motifs 1 through 5 (Figure 2B). This implies that the AcPEBP protein demonstrates a notable degree of conservation among its constituents. Figure 2C shows that the quantity of introns in the AcPEBP gene family, ranging from 2 to 5. Among them, the MFT subfamily exhibits the highest number of introns in AcMFT1, reaching 5, while AcMFT2 within the same subfamily displays the lowest count, with only 2 introns. Furthermore, other subfamily members share similarities in terms of the amount of introns and exons, suggesting a potential correlation between genetic structure and specific biological functions, regulatory pathways, or evolutionary relationships (Liu et al., 2021).
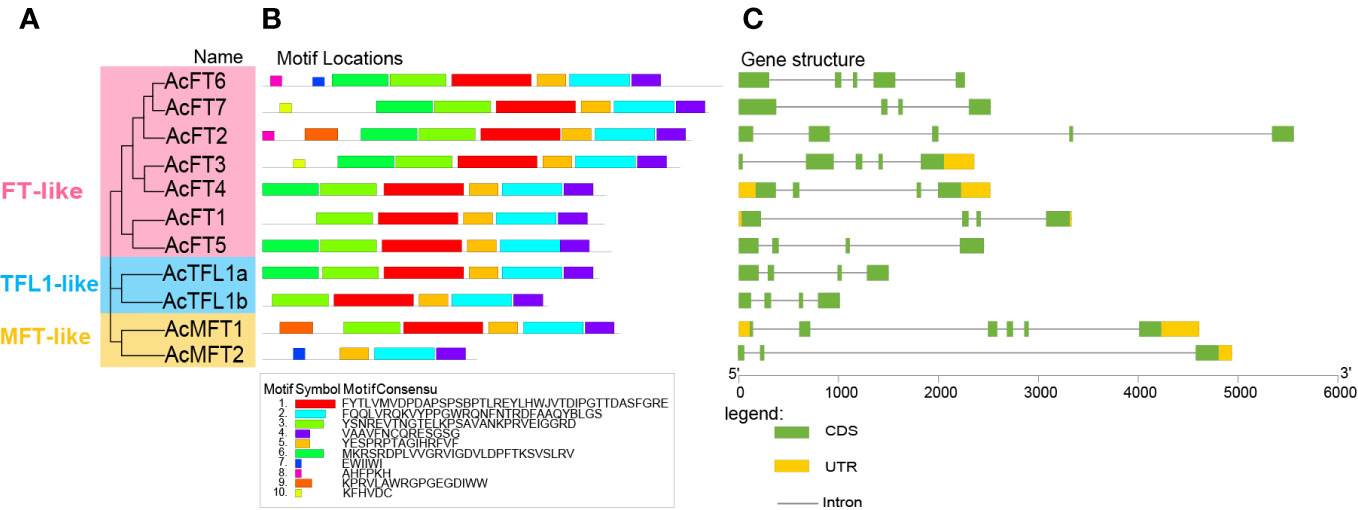
Figure 2 Phylogenetic connections and structural characteristics of the 11 AcPEBP genes. (A) Phylogenetic analysis using the Neighbor-Joining method for the PEBP family in pineapple. (B) The patterns of predicted motifs within the AcPEBP proteins. (C) Gene structures of PEBP family genes in pineapple. Yellow icons represent untranslated regions (UTR), green icons represent coding sequences (CDS), and lines connecting UTR and CDS represent introns (a noncoding segments of a gene or mRNA).
Cis-element analysis of AcPEBPs
For the purpose of comprehending the potential roles and expression regulatory mechanisms of the AcPEBP gene, cis-acting elements located within the gene’s promoter region (upstream 2 kb region) were predicted using the PlantCARE website (Figure 3). A total of 27 cis-acting elements were identified, encompassing 7 light-responsive elements, 9 plant hormone-responsive elements, 5 stress-related elements and 6 growth and development-responsive elements. The plant hormone-responsive elements, including encompassing gibberellin-responsive elements (TATC-box, P-box and GARE motif), auxin-responsive elements (TGA-element and AuxRR core), salicylic acid-responsive elements (TCA-element), abscisic acid-responsive elements (ABRE), as well as CGTCA and TGACG motifs involved in MeJA (methyl jasmonate) responses, are the most abundant. Additionally, the promoters of 10 AcPEBP gene family members contain light-responsive elements Box 4 and G-box. This observation suggests that the AcPEBP gene family may exhibit responsiveness to both hormonal and light signals.
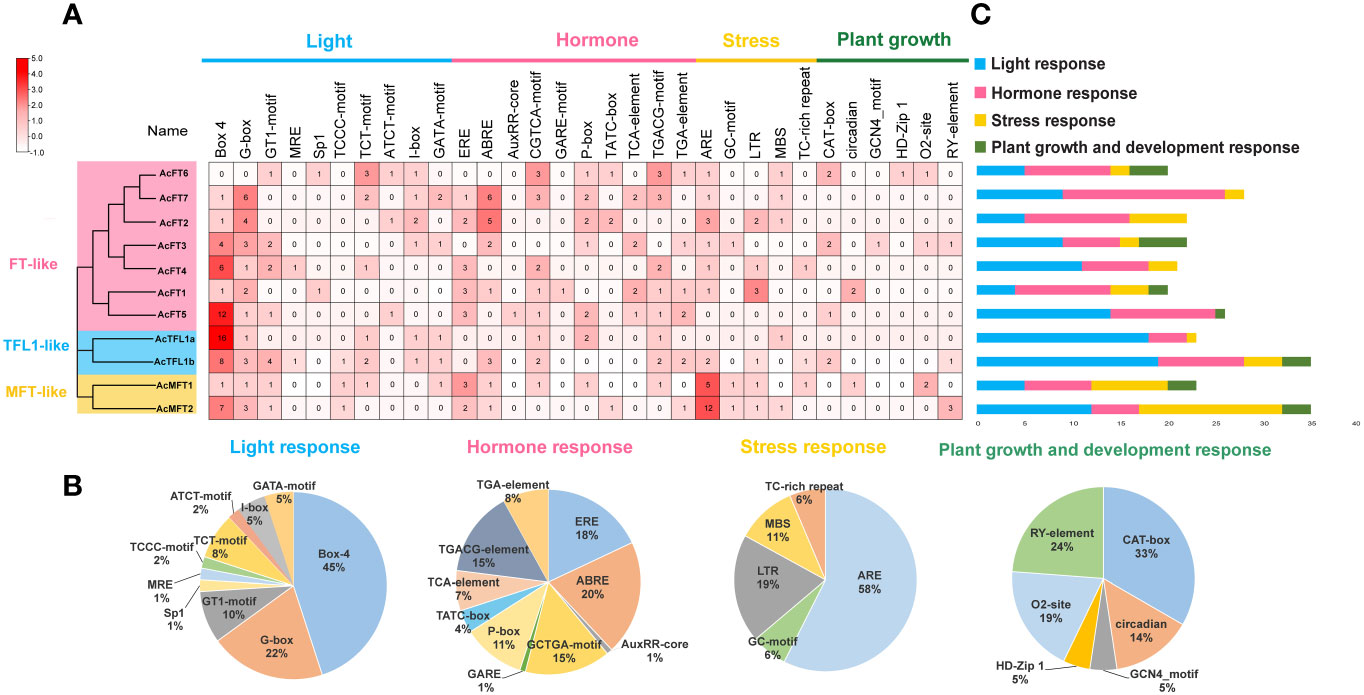
Figure 3 Analysis of cis-elements within the promoters of AcPEBP genes. (A) The cis-element analysis of PEBP promoter region. (B) The count of various types of elements in AcPEBP promoters was tallied and is represented using different colors. (C) The pie chart shows the proportion of each cis-element of the four types of response elements.
Chromosomal position and gene duplication of the AcPEBP gene
In order to comprehend the distribution characteristics and gene replication of the PEBP gene, the genomic annotation information of pineapple was employed to ascertain the chromosomal localization of the AcPEBP gene. As depicted in Figure 4, the 11 genes exhibit an uneven distribution across 6 chromosomes, constituting approximately 1/4 of the 25 chromosomes present in pineapple. Notably, chromosome 5 harbors the highest number of three AcPEBP genes; whereas chromosomes 1, 10 and 17 each contain two AcPEBP genes, there is only one AcPEBP gene on chromosome 16.
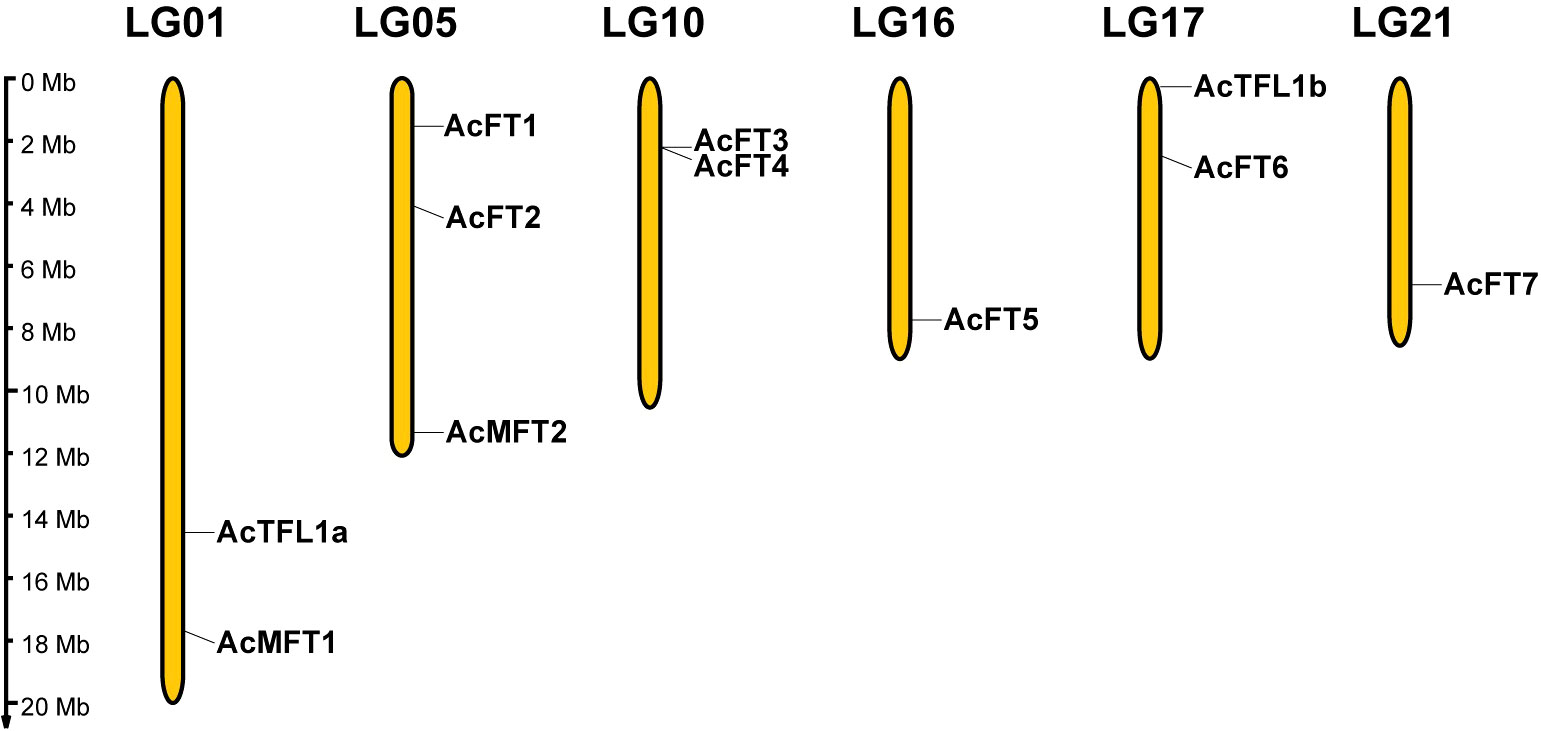
Figure 4 Distribution of AcPEBP genes on the pineapple chromosomes. The yellow vertical lines indicate the pineapple chromosome (Chr), the number is above the Chr. The left scale bar is the length of the chromosome.
To unravel the expansion mechanism of the pineapple PEBP gene family, gene collinearity analysis was performed. The results, as shown in Figure 5, revealed that 11 AcPEBP genes were involved in 4 segmental duplication events (AcFT1/AcFT5, AcFT6/AcFT7, AcFT2/AcFT7, AcFT2/AcTFL2), as well as 1 tandem duplication event (AcFT3/AcFT4) (Figure 5A). With the exception of AcFT2/AcTFL2, the majority of genes implicated in the duplication events belong to the FT-like subfamily. This observation suggests that gene duplication appears to be the primary mechanism driving the expansion of the pineapple FT-like subfamily. To enhance comprehension of the evolutionary trajectory and source of the AcPEBP gene family, collinearity maps were generated comparing pineapple with three dicotyledonous plants (Arabidopsis, grape and tomato), as well as three monocotyledonous plants (banana, maize and rice) (Figure 5B). Pineapple displays 13, 20 and 19 homologous gene pairs with banana, maize and rice, respectively, notably higher count compared to the homologous gene pairs observed between pineapple and Arabidopsis (1), tomato (1) and grape (2). This phenomenon can be attributed to the fact that pineapples pertain to the category of monocotyledonous plants, thereby establishing a more intimate evolutionary connection with other species falling under the same category, while exhibiting a comparatively distant evolutionary association with dicotyledonous species.
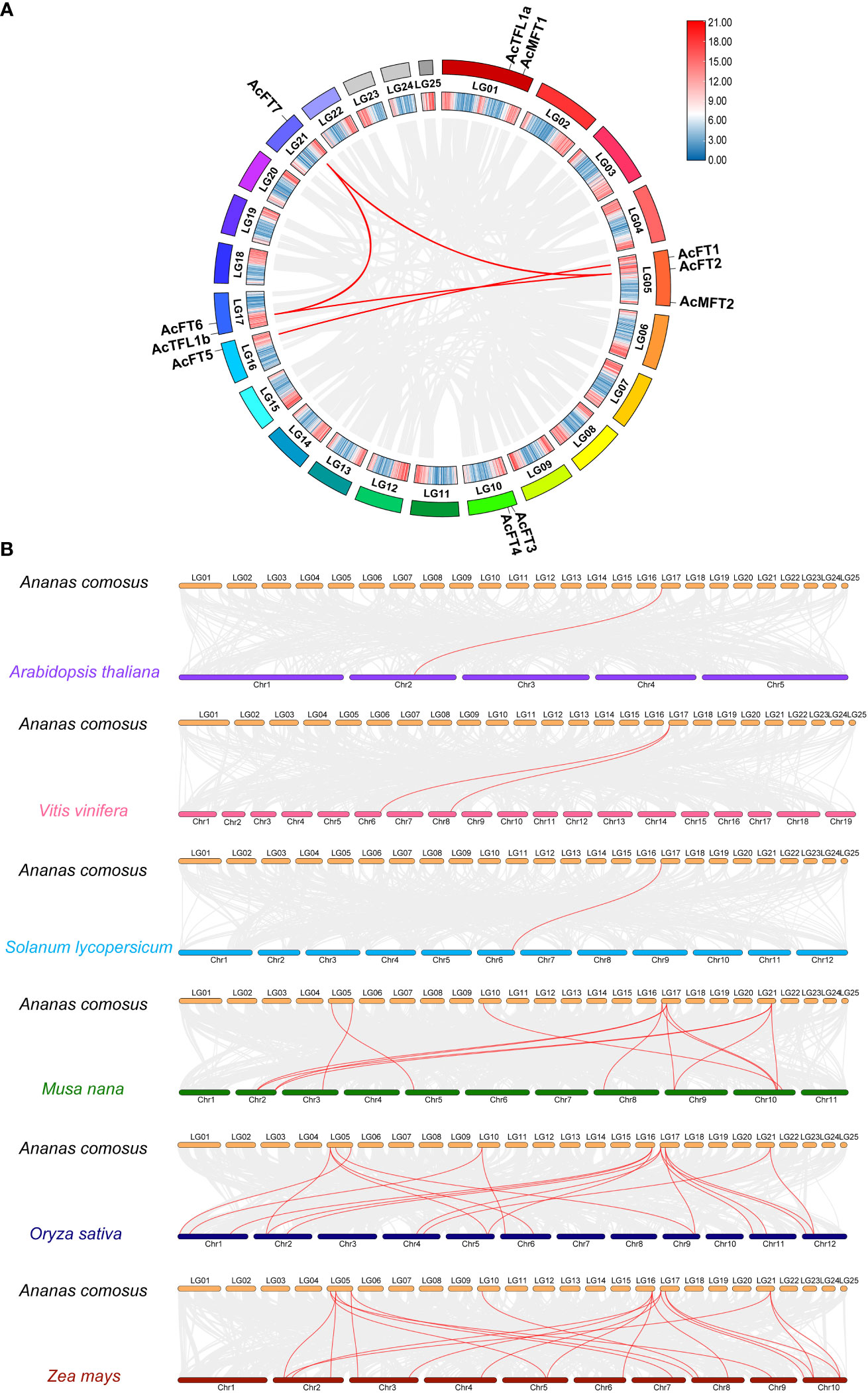
Figure 5 Duplication analysis and synlinearity analysis. (A) Duplicate analysis of AcPEBP gene in pineapple. (B) Synteny analysis of AcPEBP gene between pineapple and other plants (Arabidopsis, grape, tomato, banana, rice and corn). The red line indicates homologous gene pairs.
AcPEBP protein interaction network
The construction of the protein-protein interaction network of AcPEBP was based on the interactions of homologous proteins in Arabidopsis thaliana (Figure 6). Noteworthy findings were observed within this network. Specifically, in the FT interaction network (Figure 6A), CO, FD, and AP1 as crucial components in the Arabidopsis FT floral-dependent regulation (Komeda, 2004). Additionally, AcTFL1 may interact with VIN3, CAL, AGL8, AGL24, SOC1, and LFY (Figure 6B). The regulation of these genes likely entails specific nodes within the flowering regulatory network that synchronize the timing of flowering and orchestrate the progression of the flowering process (Azpeitia et al., 2021). DECOY and MLE2.7 appear in the interaction networks of three subfamilies (Figure 6), suggesting their potential significance in plant growth, development, and other biological processes. Consequently, conducting a thorough investigation into the pivotal factors governing the regulation of FT will yield a more comprehensive comprehension of the flowering regulatory mechanism exhibited by PEBP family constituents in pineapple.
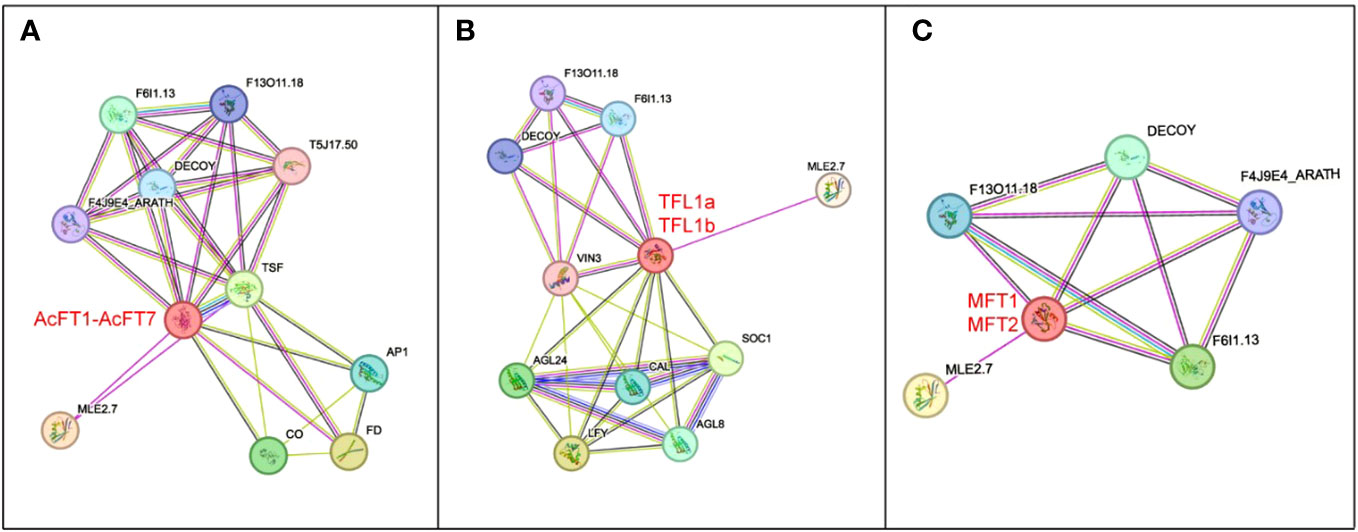
Figure 6 Depicts the predicted protein-protein interaction regulatory network of AcPEBP. (A-C) Respectively represent the network predictions for AcFTs, AcTFLs and AcMFTs, along with their interacting proteins.
Expression profiles of AcPEBP in different tissues in pineapple
To explore the expression patterns of the AcPEBP gene family across various tissues, transcriptomic data to determine the expression levels of PEBP genes in pineapple buds, flowers, fruits, leaves and roots. Our findings revealed distinct tissue specificity in the expression of AcPEBP family members (Figure 7). Notably, AcFT3 and AcFT4 exhibited significantly higher expression levels in flowers and buds compared to other tissue sites. Additionally, AcFT6 and AcMFT1 displayed the highest expression levels in fruits and leaves, respectively;AcMFT1 and AcFT7 also exhibited elevated expression levels in roots. In addition to these members, AcFT1, AcFT2, AcFT5, AcTFL1a, AcTFL1b and AcMFT2 are almost not expressed. Consequently, it can be inferred that AcFT3 and AcFT4 likely assume a significant function in the process of flowering and flower development in pineapple.
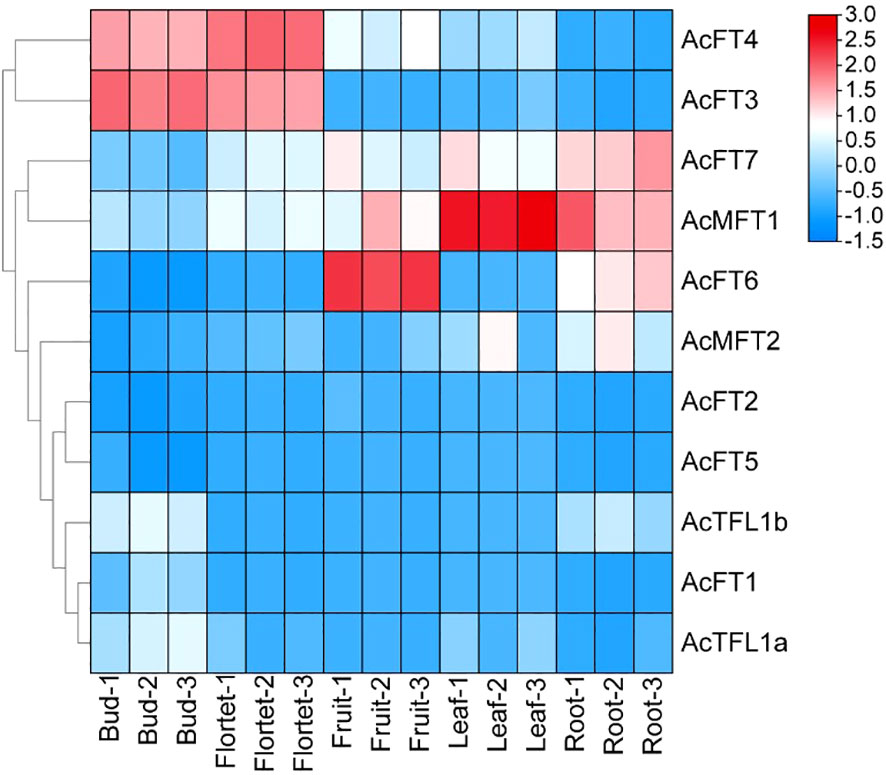
Figure 7 Expression profile of AcPEBP genes in different tissues of pineapple. The labels of the X-axis represent the different organs of the pineapple. The Y-axis refers to the deduced FPKM value normalized with Log2.
Expression patterns of AcPEBP during flower induction and the flowering process in pineapple
To enhance the credibility of AcPEBP in flower induction and flowering processes, a thorough investigation was conducted to assess the expression patterns of AcPEBP at different stages of flower development. As illustrated in Figure 8, the AcPEBP gene exhibited distinct patterns of expression. However, of greater interests to us is the observation that the members exhibiting elevated expression levels during the late stages of flower development in pineapple are predominantly concentrated within the FT-like subfamily. For example, during the course of the flowering process, the expression levels of AcFT3 and AcFT4 exhibited a gradual increment in conjunction with the development of flower buds (Figure 8A). qRT-PCR findings indicated that the expression levels of AcFT3 and AcFT4 genes in flower bud differentiation gradually increased after 4 days of ethethylene treatment and culminating in a peak at 5 to 7 weeks (Figure 8C). Furthermore, it is noteworthy that the fifth week following ethylene catalysis emerges as a pivotal phase for flower organ differentiation. Subsequently, an in-depth examination of the expression patterns of the AcPEPB gene in various flower organs of the pineapple was conducted (Figure 8B). The expression levels of AcFT7, AcFT3 and AcFT4 in different flower organs of pineapple were significantly increased.The concordance between the RNA-seq data and the qRT-PCR analysis results provides additional validation for the experimental findings. These findings suggest that the FT-like subfamily within the PEBP family may have a significant role in the pineapple flowering process.
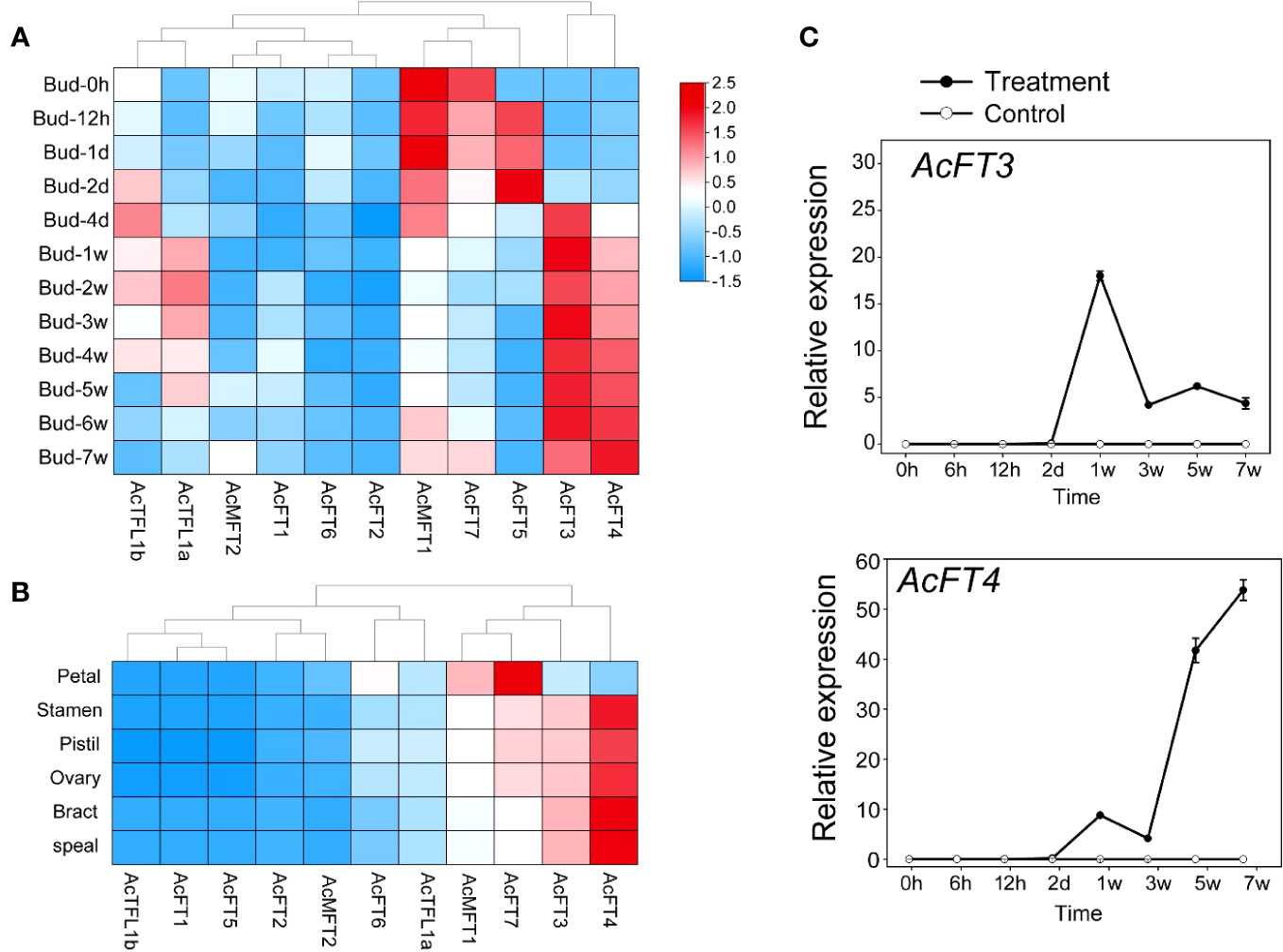
Figure 8 Expression profile of AcPEBP genes in flower induction and flowering process. (A) A heatmap illustrating the transcriptional levels of AcPEBP genes as determined by RNA-seq. (B) Expression pattern analysis of AcPEBP genes across various floral organs in pineapple. (C) Validation of AcPEBP gene expressions using qRT-PCR.
Localization within cells and transcriptional activity of AcFT4
To elucidate the potential functions of AcFT4 in the transcriptional regulatory system, a recombinant AcFT4-GFP protein was synthesized and temporarily expressed in the leaves of tobacco plant (Nicotiana benthamiana). GFP originating from the AcFT4-GFP fusion protein were observed in both the nucleus and peripheral cytoplasm (Figure 9), which is consistent with the results observed in other species (Harig et al., 2012; Liu et al., 2022). To investigate whether the AcFT4 protein has transcriptional activity, the coding sequence of AcFT4 was integrated with the GAL4 DNA-binding domain encoding sequence in the pGBKT7 vector. The resulting recombinant construct was introduced into the yeast strain AH109. The findings revealed robust growth of yeast colonies on the SD/-Trp medium (Figure 10). In addition to negative controls, blue yeast colonies transformed with pGBT7-AcFT4 were observed on SD/-Trp/-His/-Ade medium. This observation indicates that AcFT4 may indeed participate in the transcriptional regulatory system.
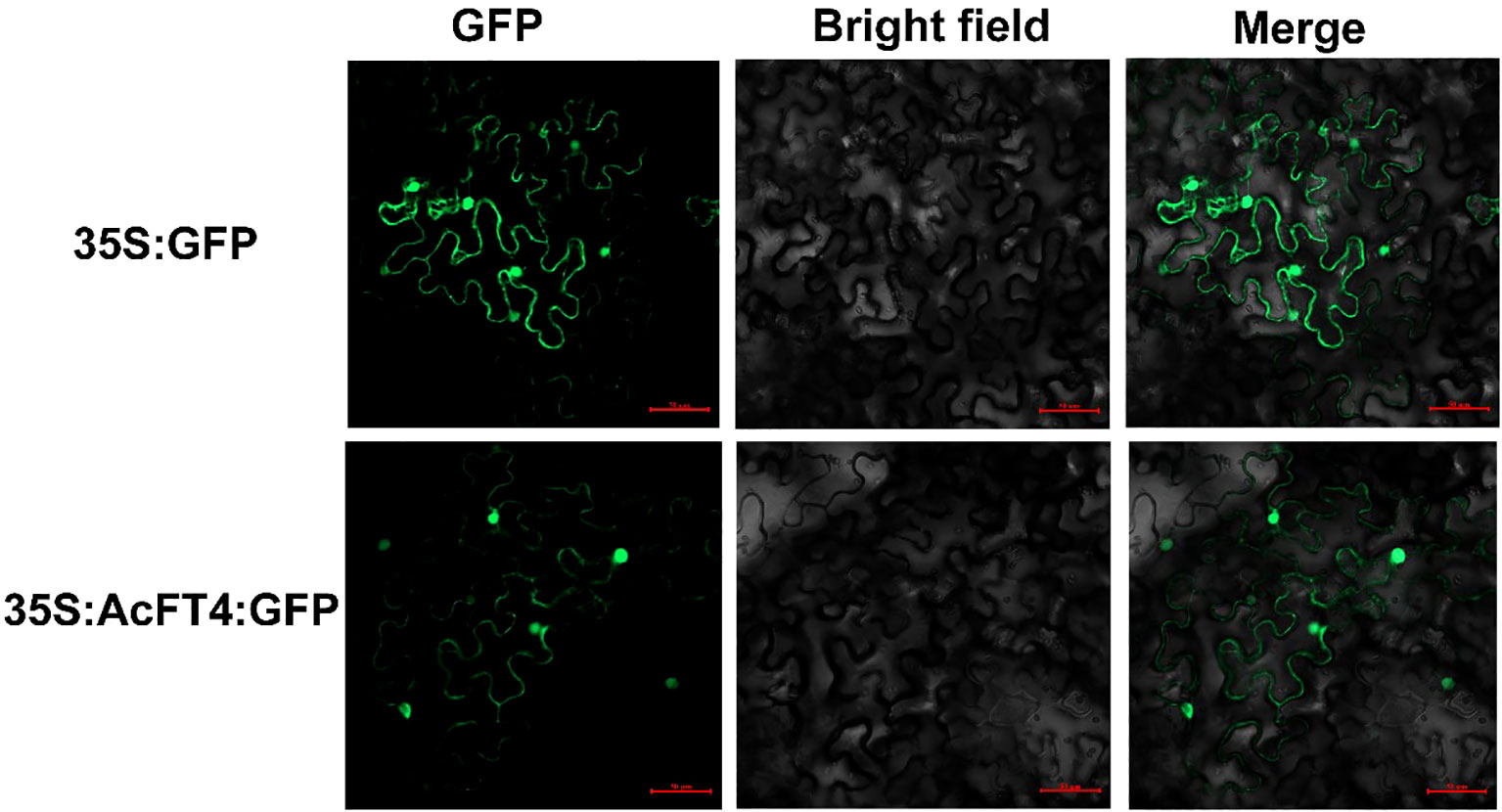
Figure 9 Subcellular localization of AcPEBP genes. Each candidate gene was individually inserted into the pCAMBIA2300-GFP vector. The empty vector was used as the control.
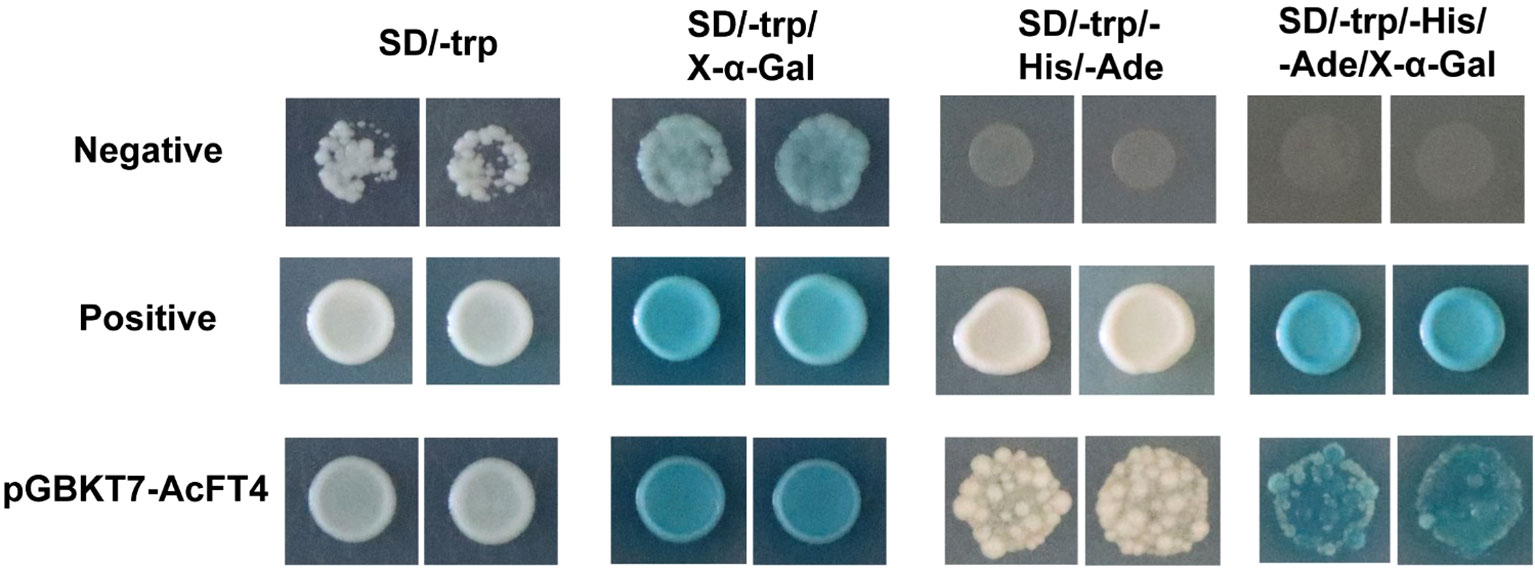
Figure 10 Assessment of the transcriptional activity of AcFT4 in yeast. The negative control involved the transformation of the empty vector pGBKT7 into yeast. The positive control consisted of the co-transformation of pGADT7-T and pGBKT7-53 into yeast.
Over-expression of AcFT4 in Arabidopsis strongly accelerated flowering
The 35S::AcFT4 construct was introduced into wild-type Arabidopsis thaliana to assess whether AcFT4 induces flower development. The empty vector with the CaMV35S promoter was transformed into Arabidopsis plants as the negative control (35S-COL). After kanamycin screening and PCR identification, three T3 transgenic plants were selected for further analysis, taking into consideration the expression level of AcFT4. Compared with the controls, all transgenic strains showed different degrees of early flowering (Figures 11A–C). Transgenic plants overexpressing AcFT4 had an earlier flowering time of 6-7 days and had 5-6 fewer rosette leaves at flowering (Figures 11B, C). These data suggest that overexpression of AcFT4 can promote flowering in transgenic Arabidopsis thaliana.
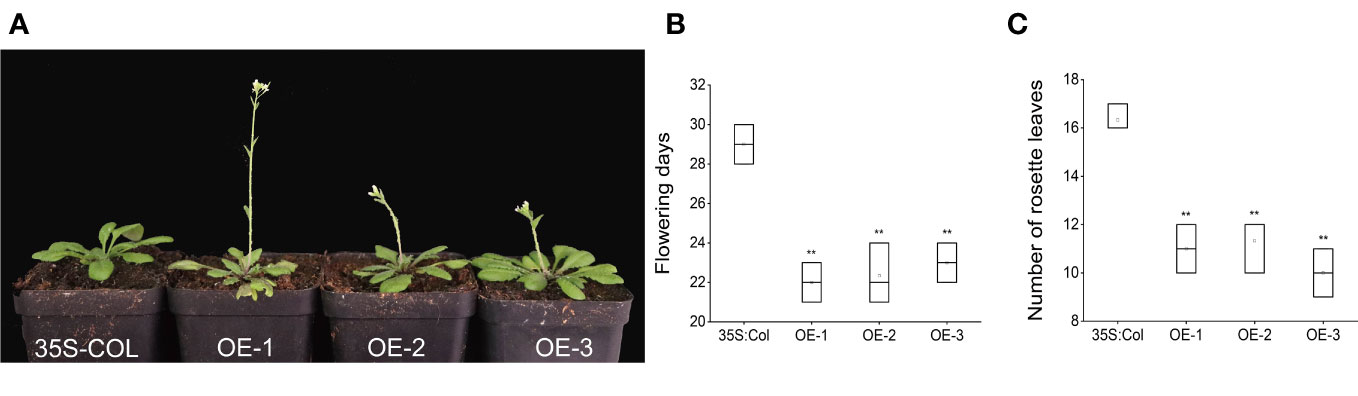
Figure 11 AcPEBP plants display an early flowering phenotype. (A) Transgenic lines. (B) Flowering days. (C) Number of rosette leaves. Values represent the mean ± SD of three independent biological replicates. The asterisk denotes a significantly different compared with controls (**P < 0.05, based on t-test).
Discussion
The PEBP family in plants ethanolamine-binding proteins that are highly conserved. These genes are crucial in regulating various processes such as the transition to flowering, seed development and dormancy, and the formation of inflorescence structure in higher plants (Yue et al., 2021; Chen F. et al., 2021; Périlleux et al., 2019). Despite the extensive research on the PEBP family in other plant species, the function of this family in pineapples remains inadequately investigated. Therefore, there is a pressing need for further investigation into the function of the AcPEBP family in pineapples. In this study, 11 PEBP genes were discovered within the pineapple genome database. Similar to Arabidopsis and rice, the PEBP gene family within the pineapple genome was also divided into three subfamilies: FT-like, TFL1-like and FT-like. This is also consistent with the classification of fruit tree crops such as grapes (Carmona et al., 2007). However, there are significant differences in the number of PEBP family members between species, potentially attributed to gene duplication or deletion events during the course of evolution. A previous study suggested that exon and intron structures were involved in the evolution of plant species (Zhang and Li, 2017). In particular, most genes in the FT/TFL1 subfamily predominantly features a gene structure comprising four exons and three introns (Figure 2C), and they show a similar pattern of conserved motif distribution. This suggests that these genes are conserved in evolution. However, in contrast to the FT/TFL1 subfamily, the AcMFT2 gene had five introns, while the AcMFT1 gene had only two introns. The presence of exon-intron acquisition and deletion within the MFT-like subgroup of pineapple, along with substantial alterations in the conserved pattern, suggests potential functional diversity among MFT-like genes. (Xi and Yu, 2010; Song et al., 2020).
At present, it is generally believed that the expression pattern of genes is typically associated with their biological functions (Dong et al., 2020). Previous studies have demonstrated that Arabidopsis AtFT gene exhibits expressed in both reproductive and vegetative organs (Kobayashi et al., 1999). RoFT gene of rose is expressed in flower bud tips and buds (Remay et al., 2009). The expression of the VvFT gene was detected during the development of grape inflorescence (Carmona et al., 2007). In apples, MdFT1 primarily assumes a significant role in apple fruit development, of whereas MdFT2 is predominantly expressed in the reproductive organs of apple, including flower buds and young fruits. The NtFT5 gene in tobacco is expressed in tobacco flowers (Wang et al., 2018). In this study, the expression pattern of the AcPEBP gene family exhibited certain tissue specificity. Specifically, AcFT7 and AcFT6 were highly expressed in the root and fruit of pineapple, respectively, and AcMFT1 exhibited high expression levels in the root and leaf tissues. These genes likely play pivotal roles within the contexts of roots, leaves and fruits. It is worth noting that AcFT3 and AcFT4 not only exhibited higher transcription levels in flower buds compared to other tissues, but also displayed elevated expression levels in flowers (Figure 7). After that, the expression of AcFT3 and AcFT4 was observed to be significantly specific across five distinct flower organs of pineapple (Figure 8B). This suggests that the AcFT genes potentially exert a significant regulatory on the process of flower bud formation and flowering in pineapple. In order to further verify the results, the expression of the FT gene in pineapple flower buds was analyzed assessed through qRT-PCR. The results showed that the expression level of AcFT4 began to increase within just 4 days after ethylene treatment and reached a peak in late flower development (Figure 8C). This indicates that members of the pineapple PEBP family might play an important role in the flowering process of pineapple.
Simultaneously, examination of promoter cis-acting elements has suggested the possible participation of light-responsive and hormone-responsive within the intricate flowering regulatory network. (Figure 6). This phenomenon has been previously observed in various plant species. The peanut FT-like gene exhibits higher elevated expression levels in short-day conditions and is involved in the regulation of flowering time under short-day conditions (Jin et al., 2019).In Dendrobium huoshanense, gibberellin acid (GA) induces strong expression of DhFT3 and DhFT10, while the expression of DhTFL1 swiftly declines following GA treatment. This may have different regulatory roles in flowering control (Song et al., 2021). The short-day plant Pharbitis nil FT homolog PnFT2 may be stimulated through the application of salicylic acid (SA) (Wada et al., 2010). In a recent groundbreaking discovery, Chen Y. et al. revealed that ERF1 functions as a regulatory factor in ethylene signaling, directly suppressing the expression of FT in Arabidopsis during a complex flowering cascade, delaying flowering (Chen Y. et al., 2021). Interestingly, treatment of pineapple plants with the ethylene-releasing agent ethephon has been demonstrated to induce early flowering of pineapples (Espinosa et al., 2017). Therefore, we posit that the flowering pathway in pineapples may differ from that of the model plant Arabidopsis.
FT-like genes in plants play a crucial role in regulating flowering (Takagi et al., 2023). This study demonstrated that the overexpression of AcFT4 in Arabidopsis resulted in early flowering (Figure 11). Transgenic lines showed flowering time 6-7 days earlier than empty vector plants with CaMV35S promoters. This finding suggests that AcFT4 acts as a flower promoting factor, consistent with other species. For instance, the ectopic expression of ZCN8 in maize accelerates flowering in transgenic Arabidopsis (Danilevskaya et al., 2008). Overexpression of MiFT1, MiFT2 and MiFT3 in mango has been found to induce early flowering phenotypes in Arabidopsis (Fan et al., 2020). In strawberries, FveFT2 is a non-photoperiodic florigen that allows short-day plants to flower, and its excessive expression greatly enhances the flowering phenomenon (Gaston et al., 2021). In addition to the function of FT itself, key components CO, FD and LFY in the known flowering regulation network of FT genes were found in the protein interaction prediction network (Figure 6) (Blümel et al., 2015). These genes collaborate within signaling pathway, collectively orchestrating the process of flowering. Nevertheless, whether the FT gene in pineapple functions through these flowering regulatory factors still requires further investigation. This will contribute to a more comprehensive understanding of the molecular mechanisms of PEBP members in the pineapple flowering signaling pathway.
Conclusions
In this study, 11 members of the PEBP family were identified from the pineapple genome annotation file. Analysis of cis-acting elements suggests that AcPEBP genes may be influenced by a complex network of hormonal and light regulation. The analysis of gene expression showed that AcFT3 and AcFT4 were specifically highly expressed in the later stages of flower development and in floral organs, suggesting that these genes may be involved in the process of pineapple flowering induction. Following this, the subcellular localization and transcriptional activation activities of AcFT4 were subsequently confirmed. AcFT4 exhibits transcriptional activity and is localized in various cellular compartments, including the nucleus and peripheral cytoplasm. The overexpression of AcFT4 in transgenic Arabidopsis thaliana resulted in an expedited flowering process. In addition, important components of floral-dependent regulation of FT, CO, LFY and AP1, were found in the protein interaction network. In conclusion, the genomic analysis of AcPEBP family provides a new basis for the study of flower development related functions of PEBP family members in pineapple.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
HZ: Writing – review & editing, Funding acquisition. XZ: Data curation, Software, Writing – original draft. YO: Conceptualization, Writing – review & editing. LZ: Data curation, Investigation, Resources, Software, Writing – review & editing. ZL: Investigation, Resources, Writing – review & editing. YW: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The project was funded by the National Natural Science Fund of China (32160687 and 32360723), the major science and technology project of Hainan Province (ZDKJ2021014), the Natural Science Foundation of Hainan Province (321RC467), and the Scientific Research Start-up Fund Project of Hainan University (KYQD-ZR-20090).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Adeyemo, O. S., Chavarriaga, P., Tohme, J., Fregene, M., Davis, S. J., Setter, T. L. (2017). Overexpression of Arabidopsis FLOWERING LOCUS T (FT) gene improves floral development in cassava (Manihot esculenta, Crantz). PloS One 12, e0181460. doi: 10.1371/journal.pone.0181460
Adeyemo, O. S., Hyde, P. T., Setter, T. L. (2019). Identification of FT family genes that respond to photoperiod, temperature and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reprod. 32, 181–191. doi: 10.1007/s00497-018-00354-5
Azpeitia, E., Tichtinsky, G., Le Masson, M., Serrano-Mislata, A., Lucas, J., Gregis, V., et al. (2021). Cauliflower fractal forms arise from perturbations of floral gene networks. Science 373, 192–197. doi: 10.1126/science.abg5999
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Blümel, M., Dally, N., Jung, C. (2015). Flowering time regulation in crops—what did we learn from Arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Carmona, M. J., Calonje, M., Martínez-Zapater, J. M. (2007). The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 63, 637–650. doi: 10.1007/s11103-006-9113-z
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Li, Y., Li, X., Li, W., Xu, J., Cao, H., et al. (2021). Ectopic expression of the Arabidopsis florigen gene FLOWERING LOCUS T in seeds enhances seed dormancy via the GA and DOG1 pathways. Plant J. 107, 909–924. doi: 10.1111/tpj.15354
Chen, Y., Zhang, L., Zhang, H., Chen, L., Yu, D. (2021). ERF1 delays flowering through direct inhibition of FLOWERING LOCUS T expression in Arabidopsis. J. Integr. Plant Biol. 63, 1712–1723. doi: 10.1111/jipb.13144
Danilevskaya, O. N., Meng, X., Hou, Z., Ananiev, E. V., Simmons, C. R. (2008). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146, 250–264. doi: 10.1104/pp.107.109538
Dong, L., Lu, Y., Liu, S. (2020). Genome-wide member identification, phylogeny and expression analysis of PEBP gene family in wheat and its progenitors. PeerJ 8, e10483. doi: 10.7717/peerj.10483
Espinosa, M. E. Á., Moreira, R. O., Lima, A. A., Ságio, S. A., Barreto, H. G., Luiz, S. L. P., et al. (2017). Early histological, hormonal, and molecular changes during pineapple (Ananas comosus (L.) Merrill) artificial flowering induction. J. Plant Physiol. 209, 11–19. doi: 10.1016/j.jplph.2016.11.009
Fan, Z. Y., He, X. H., Fan, Y., Yu, H. X., Wang, Y. H., Xie, X. J., et al. (2020). Isolation and functional characterization of three MiFTs genes from mango. Plant Physiol. Biochem. 155, 169–176. doi: 10.1016/j.plaphy.2020.07.009
Gafni, I., Rai, A. C., Halon, E., Zviran, T., Sisai, I., Samach., A., et al. (2022). Expression profiling of four mango FT/TFL1-encoding genes under different fruit load conditions, and their involvement in flowering regulation. Plants (Basel) 11, 2409. doi: 10.3390/plants11182409
Garcia-Hernandez, M., Berardini, T. Z., Chen, G., Crist, D., Doyle, A., Huala, E., et al. (2002). TAIR: a resource for integrated Arabidopsis data. Funct. Integr. Genomics 2, 239–253. doi: 10.1007/s10142-002-0077-z
Gaston, A., Potier, A., Alonso, M., Sabbadini, S., Delmas, F., Tenreira, T., et al. (2021). The FveFT2 florigen/FveTFL1 antiflorigen balance is critical for the control of seasonal flowering in strawberry while FveFT3 modulates axillary meristem fate and yield. New Phytologist 232, 372–387. doi: 10.1111/nph.17557
Harig, L., Beinecke, F. A., Oltmanns, J., Muth, J., Müller, O., Rüping, B., et al. (2012). Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 72, 908–921. doi: 10.1111/j.1365-313X.2012.05125.x
Jin, S., Nasim, Z., Susila, H., Ahn, J. H. (2021). Evolution and functional diversification of FLOWERING LOCUS T/TERMINAL FLOWER 1 family genes in plants. Semin. Cell Dev. Biol. 109, 20–30. doi: 10.1016/j.semcdb.2020.05.007
Jin, H., Tang, X., Xing, M., Zhu, H., Sui, J., Cai, C., et al. (2019). Molecular and transcriptional characterization of phosphatidyl ethanolamine-binding proteins in wild peanuts Arachis duranensis and Arachis ipaensis. BMC Plant Biol. 19, 484. doi: 10.1186/s12870-019-2113-3
Kaneko-Suzuki, M., Kurihara-Ishikawa, R., Okushita-Terakawa, C., Kojima, C., Nagano-Fujiwara, M., Ohki, I., et al. (2018). TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 59, 458–468. doi: 10.1093/pcp/pcy021
Kim, G., Rim, Y., Cho, H., Hyun, T. K. (2022). Identification and functional characterization of FLOWERING LOCUS T in Platycodon grandiflorus. Plants (Basel) 11, 325. doi: 10.3390/plants11030325
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Komeda, Y. (2004). Genetic regulation of time to flower in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55, 521–535. doi: 10.1146/annurev.arplant.55.031903.141644
Komiya, R., Ikegami, A., Tamaki, S., Yokoi, S., Shimamoto, K. (2008). Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774. doi: 10.1242/dev.008631
Kotoda, N., Hayashi, H., Suzuki, M., Igarashi, M., Hatsuyama, Y., Kidou, S., et al. (2010). Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus x domestica Borkh.). Plant Cell Physiol. 51, 561–575. doi: 10.1093/pcp/pcq021
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, H., Ran, K., Dong, Q., Zhao, Q., Shi, S. (2020). Cloning, sequencing, and expression analysis of 32 NAC transcription factors (MdNAC) in apple. PeerJ 8, e8249. doi: 10.7717/peerj.8249
Liu, J., Wang, J., Wang, M., Zhao, J., Zheng, Y., Zhang, T., et al. (2021). Genome-wide analysis of the R2R3-MYB gene family in Fragaria × ananassa and its function identification during anthocyanins biosynthesis in pink-flowered strawberry. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.702160
Liu, X., Zhao, D., Ou, C., Hao, W., Zhao, Z., Zhuang, F. (2022). Genome-wide identification and characterization profile of phosphatidy ethanolamine-binding protein family genes in carrot. Front. Genet. 13. doi: 10.3389/fgene.2022.1047890
Oda, A., Narumi, T., Li, T., Kando, T., Higuchi, Y., Sumitomo, K., et al. (2012). CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 63, 1461–1477. doi: 10.1093/jxb/err387
Périlleux, C., Bouché, F., Randoux, M., Orman-Ligeza, B. (2019). Turning meristems into fortresses. Trends Plant Sci. 24, 431–442. doi: 10.1016/j.tplants.2019.02.004
Rao, X., Huang, X., Zhou, Z., Lin, X. (2013). An improvement of the 2ˆ (-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 3, 71–85. doi: 10.1089/cmb.2012.0279
Remay, A., Lalanne, D., Thouroude, T., Le Couviour, F., Hibrand-Saint Oyant, L., Foucher, F. (2009). A survey of flowering genes reveals the role of gibberellins in floral control in rose. Theor. Appl. Genet. 119, 767–781. doi: 10.1007/s00122-009-1087-1
Song, C., Li, G., Dai, J., Deng, H. (2021). Genome-wide analysis of PEBP genes in Dendrobium huoshanense: unveiling the antagonistic functions of FT/TFL1 in flowering time. Front. Genet. 12. doi: 10.3389/fgene.2021.687689
Song, S., Wang, G., Wu, H., Fan, X., Liang, L., Zhao, H., et al. (2020). OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 103, 532–546. doi: 10.1111/tpj.14748
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi: 10.1093/nar/gkaa1074
Takagi, H., Hempton, A. K., Imaizumi, T. (2023). Photoperiodic flowering in Arabidopsis: Multilayered regulatory mechanisms of CONSTANS and the florigen FLOWERING LOCUS T. Plant Commun. 4, 100552. doi: 10.1016/j.xplc.2023.100552
Venail, J., da Silva Santos, P. H., Manechini, J. R., Alves, L. C., Scarpari, M., Falcão, T., et al. (2022). Analysis of the PEBP gene family and identification of a novel FLOWERING LOCUS T orthologue in sugarcane. J. Exp. Bot. 5, 2035–2049. doi: 10.1093/jxb/erab539
Wada, K. C., Yamada, M., Shiraya, T., Takeno, K. (2010). Salicylic acid and the flowering gene FLOWERING LOCUS T homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil. J. Plant Physiol. 167, 447–452. doi: 10.1016/j.jplph.2009.10.006
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, G., Wang, P., Gao, Y., Li, Y., Wu, L., Gao, J., et al. (2018). Isolation and functional characterization of a novel FLOWERING LOCUS T homolog (NtFT5) in Nicotiana tabacum. J. Plant Physiol. 231, 393–401. doi: 10.1016/j.jplph.2018.10.021
Wen, C., Zhao, W., Liu, W., Yang, L., Wang, Y., Liu, X., et al. (2019). CsTFL1 inhibits determinate growth and terminal flower formation through interaction with CsNOT2a in cucumber. Development 146, dev180166. doi: 10.1242/dev.180166
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xi, W., Yu, H. (2010). MOTHER OF FT AND TFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signal Behav. 5, 1315–1317. doi: 10.4161/psb.5.10.13161
Xu, H., Guo, X., Hao, Y., Lu, G., Li, D., Lu, J., et al. (2022). Genome-wide characterization of PEBP gene family in Perilla frutescens and PfFT1 promotes flowering time in Arabidopsis thaliana. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1026696
Xu, H., Yu, Q., Shi, Y., Hua, X., Tang, H., Yang, L., et al. (2018). PGD: pineapple genomics database. Hortic. Res. 5, 66. doi: 10.1038/s41438-018-0078-2
Yan, X., Cao, Q. Z., He, H. B., Wang, L. J., Jia., G. X. (2021). Functional analysis and expression patterns of members of the FLOWERING LOCUS T (FT) gene family in Lilium. Plant Physiol. Biochem. 163, 250–260. doi: 10.1016/j.plaphy.2021.03.056
Yue, L., Li, X., Fang, C., Chen, L., Yang, H., Yang, J., et al. (2021). FT5a interferes with the Dt1-AP1 feedback loop to control flowering time and shoot determinacy in soybean. J. Integr. Plant Biol. 63, 1004–1020. doi: 10.1111/jipb.13070
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, Q., Li, C. (2017). Comparisons of copy number, genomic structure, and conserved motifs for α-amylase genes from barley, rice, and wheat. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01727
Zhang, B., Li, C., Li, Y., Yu, H. (2020). Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat. Plants. 6, 1146–1157. doi: 10.1038/s41477-020-0749-5
Zhang, H., Zhang, Y. (2020). Molecularcloning and functional characterization of CmFT (FLOWERING LOCUS T) from Cucumis melo L. J. Genet. 99, 1–8. doi: 10.1007/s12041-020-1191-1
Keywords: pineapple (Ananas comosus (L.) Merr.), PEBP, genome-wide, expression profiles, flowering
Citation: Zhang X, Ouyang Y, Zhao L, Li Z, Zhang H and Wei Y (2023) Genome-wide identification of PEBP gene family in pineapple reveal its potential functions in flowering. Front. Plant Sci. 14:1277436. doi: 10.3389/fpls.2023.1277436
Received: 14 August 2023; Accepted: 11 October 2023;
Published: 26 October 2023.
Edited by:
Eleni Tani, Agricultural University of Athens, GreeceReviewed by:
Xiaomei Zhang, Chinese Academy of Agricultural Sciences, ChinaHulya Ilbi, Ege University, Türkiye
Copyright © 2023 Zhang, Ouyang, Zhao, Li, Zhang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongna Zhang, MTM2OTI0NzY5NzlAMTM5LmNvbQ==; Yongzan Wei, d3l6NDYyNkAxNjMuY29t
 Xiaohan Zhang1
Xiaohan Zhang1 Hongna Zhang
Hongna Zhang Yongzan Wei
Yongzan Wei