
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 09 October 2023
Sec. Plant Symbiotic Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1277262
This article is part of the Research Topic Plant-Rhizobia Symbiosis and Nitrogen Fixation in Legumes View all 12 articles
Rhizobia are soil bacteria that can establish a nitrogen-fixing symbiosis with legume plants. As horizontally transmitted symbionts, the life cycle of rhizobia includes a free-living phase in the soil and a plant-associated symbiotic phase. Throughout this life cycle, rhizobia are exposed to a myriad of other microorganisms that interact with them, modulating their fitness and symbiotic performance. In this review, we describe the diversity of interactions between rhizobia and other microorganisms that can occur in the rhizosphere, during the initiation of nodulation, and within nodules. Some of these rhizobia-microbe interactions are indirect, and occur when the presence of some microbes modifies plant physiology in a way that feeds back on rhizobial fitness. We further describe how these interactions can impose significant selective pressures on rhizobia and modify their evolutionary trajectories. More extensive investigations on the eco-evolutionary dynamics of rhizobia in complex biotic environments will likely reveal fascinating new aspects of this well-studied symbiotic interaction and provide critical knowledge for future agronomical applications.
Ecological systems are complex. They involve a multitude of organisms that can interact with each other. These interactions, ranging from antagonism to mutualism, strongly influence the fitness of each individual and, consequently, the structure of the communities in which they live. Microbial communities, termed microbiomes, in particular have received much attention because of the fundamental role they play in Earth’s biogeochemical cycles and in plant and animal health. Common mechanisms governing interactions within microbiomes include competition for resources, predation, the production of antagonistic/toxic molecules, cross-feeding processes, the production of public goods, or the formation of protection structures such as biofilms (Konopka, 2009; Pierce and Dutton, 2022). These interactions are particularly prevalent and significant in dense host-associated microbial communities, such as the mammalian gut or the rhizosphere (Hassani et al., 2018; Coyte and Rakoff-Nahoum, 2019; Kern et al., 2021; Chepsergon and Moleleki, 2023). In these ecosystems, positive or negative interactions between microbiome members can allow or inhibit, respectively, the proliferation of pathogens or beneficial microbes, with important effects on host health. For example, some rhizospheric bacteria were shown to inhibit the growth of fungal pathogens and protect plants against disease (Carrión et al., 2018; Durán et al., 2018).
Notable members of the rhizosphere community are rhizobia. These bacteria are able to form mutualistic associations with legume plants, during which they fix atmospheric nitrogen to the benefit of the host, in exchange for carbon compounds from photosynthesis (Poole et al., 2018). Rhizobia are gram-negative bacteria classified in 18 different genera of Alpha- and Beta-proteobacteria including Rhizobium, Sinorhizobium, Bradyrhizobium, Mesorhizobium, Azorhizobium, Cupriavidus, and Paraburkholderia (Masson-Boivin et al., 2009; Tang and Capela, 2020). Rhizobia are horizontally-transmitted symbionts. Their biphasic life cycle is composed of a free-living saprophytic phase, where rhizobia are part of the soil and rhizosphere microbiomes, and a symbiotic phase in their host. Soil bacteria are attracted to the germinating seeds or the mature roots following the perception of chemoattractants present in plant exudates (Compton and Scharf, 2021). In the plant, rhizobia are hosted in specific root organs, called nodules. Nodule formation is initiated by the exchange of compatible signals between rhizobia and legumes (Walker et al., 2020). In most rhizobia, the expression of nod genes, responsible for the synthesis of lipochito-oligosaccharides called Nod Factors (NF), is induced by specific flavonoids exuded by host plants. NF, whose structures vary between rhizobium strains, are then specifically recognised by plant receptors. The perception of NF allows the entry of bacteria in root tissues, where they start to proliferate extracellularly. NF perception and downstream signaling also trigger a plant development program, which leads to nodule organogenesis. In most legumes of the Papilionoideae and Mimosoid clades (Sprent, 2009; De Faria et al., 2022), rhizobia are then engulfed in the cytoplasm of nodule cells, where they form structures surrounded by the plant plasma membrane called symbiosomes. Rhizobia differentiate into bacteroids that fix nitrogen, and persist for several weeks or months within nodule cells. In legumes of the Inverted Repeat-Lacking Clade and Dalbergioid clade, this differentiation is terminal, meaning that bacteroids cannot resume growth after nodule senescence (Mergaert et al., 2006; Czernic et al., 2015; Montiel et al., 2016). During nodule senescence, undifferentiated bacteria and non-terminally differentiated bacteroids present in nodules are released and can recolonise the soil and the rhizosphere. The ability of a given rhizobial strain to successfully complete the different steps of this complex life cycle will determine how fit it is in its current environment. Studying the different factors governing rhizobial fitness is critical to understand the diversity, ecology and evolution of these important plant symbionts.
All along their life cycle, rhizobia interact with other microorganisms composing the soil, rhizosphere and nodule microbiomes, and are therefore involved in a diversity of interactions that affect their fitness either directly or indirectly through plant-mediated mechanisms. This review focuses on how the microbial community context, i.e. the ecological interactions between rhizobia and other microorganisms, contributes to determining rhizobial fitness. We first discuss the notion of fitness in the case of rhizobia and then describe how the diverse rhizobia-microorganisms (including rhizobia-rhizobia) interactions affect the fitness of rhizobia and their evolutionary dynamics.
Fitness is a central notion in evolutionary biology, as it measures how well a genotype performs in terms of survival and reproduction in a given environment. Yet, experimental measurements of fitness can be challenging. Even in seemingly simple systems (Vasi et al., 1994), bacterial fitness is dependent on several phenotypic traits (called ‘fitness components’) that will determine a genotype’s performance at the different steps of the life cycle (Orr, 2009). However, the life history traits that are measured and considered as best fitness proxies can differ between studies and experimenters. This is typically the case for rhizobia. The different measurable fitness components include (i) proliferation and survival in the soil and the rhizosphere, (ii) nodulation proficiency and competitiveness, (iii) proliferation and survival within nodules, and (iv) bacterial release from nodules during senescence. Below we highlight salient aspects of some of these different fitness measures.
Understandably, literature on rhizobial fitness has put a lot of emphasis on bacterial traits governing the association with host plants. First, the nodulation step is a major determinant of fitness for rhizobia, since it represents a strong selective bottleneck for rhizobial populations and rhizobia founding nodules will leave many more descendants than those staying in the rhizosphere (Denison and Kiers, 2011). Although nodules are usually considered to be founded by one single bacterium, mixed nodules, hosting several different rhizobial strains, can also be found in proportions that vary depending on the plant growth substrate or rhizobial density (Checcucci et al., 2016; Daubech et al., 2017; Mendoza-Suárez et al., 2020). The ability of rhizobia to form nodules can be measured during single inoculation experiments (nodulation proficiency) or in co-inoculation experiments (nodulation competitiveness). There can be significant discrepancies between these two types of assays, as the outcome of nodule occupancy following co-inoculations is unpredictable from data in single inoculation. Indeed nodulation competitiveness is a complex trait that involves a large number of bacterial genes and functions (see section 4) and that is not fully understood yet (Younginger and Friesen, 2019; Mendoza-Suárez et al., 2021). Rhizobial strains can be more competitive for nodulation if (i) they are more efficient at colonising the rhizosphere (for example by growing faster on the available nutrient sources or by producing antimicrobial compounds that inhibit the growth of sensitive competitor strains), (ii) they are faster to reach the root and induce nodulation, and/or (iii) they show an optimal compatibility with the host plant (Handelsman et al., 1984; Kiers et al., 2013; Boivin and Lepetit, 2020; Mendoza-Suárez et al., 2021). Moreover, when the co-inoculated strains have different nitrogen fixation efficiencies, the absolute and relative numbers of nodules formed by one plant can be modulated by the mechanism of auto-regulation of nodulation (AON, see section 5). Indeed, in single inoculations, strains that are fixing large amounts of nitrogen may form a relatively small number of nodules (since the nitrogen needs of the plants will be covered with few nodules), while strains that fix low amounts of nitrogen may form a large number of nodules. Yet, co-inoculating these two types of strains may modulate the number of nodules formed by each strain (Daubech et al., 2017; Westhoek et al., 2017).
Second, bacterial proliferation within nodules can be assessed by harvesting, crushing surface-sterilised nodules and plating the resulting suspensions on selective media. An important aspect of these experiments is that only viable nodule bacteria will be detected. In certain legumes, bacteroids undergo a process of 'terminal differentiation' characterised by drastic morphological and physiological changes as well as a loss of viability (Mergaert et al., 2006). As a result, the proportion of viable bacteria within nodules can vary from less than 1% in pea or Medicago to almost 100% in plants where bacteroids are not terminally differentiated such as soybean, with intermediate cases (Gresshoff and Rolfe, 1978; Mergaert et al., 2006; Marchetti et al., 2011). When terminal differentiation occurs, viable bacteria that can be recovered from nodules most likely arise from bacteria that were still located in infection threads or intercellular spaces at the time of harvest.
In addition, the proliferation and/or viability of rhizobia within nodule cells can be affected by their nitrogen fixation activity. Since the first discovery that soybean can ‘sanction’ non-fixing rhizobia (Kiers et al., 2003) and thereby promotes fitness of nitrogen-fixing ones, ample experimental data from several model species has accumulated to support the idea that there is a positive correlation between nitrogen fixation and rhizobial fitness (Kiers et al., 2006; Oono et al., 2011; Friesen, 2012; Kimbrel et al., 2013; Berrabah et al., 2015; Daubech et al., 2017; Quides et al., 2017; Burghardt et al., 2018; Westhoek et al., 2021; Batstone et al., 2022; Epstein et al., 2023). Plants can even discriminate fixing versus non-fixing strains in mixed nodules, and only target non-fixing bacteria for premature degeneration (Daubech et al., 2017; Regus et al., 2017). However, there are exceptions, where non-fixing rhizobia show higher fitness than fixing ones [(Crook et al., 2012; Porter and Simms, 2014; Gano‐Cohen et al., 2019) but see (Frederickson, 2020; Wendlandt et al., 2022) to ponder two of these examples]. Nodule size or weight are sometimes used as convenient proxies for bacterial fitness, but in certain cases, these values should be interpreted with caution. Indeed, the efficiency of nitrogen fixation (and the occurrence of plant ‘sanctions’, see below) or the accumulation of storage compounds can modify the relationship between nodule weight and the number of viable bacteria per nodule (Oono et al., 2011; Ratcliff et al., 2011). Another study showed, using a large collection of natural S. meliloti isolates, that nodule weight was positively correlated to the symbiotic fitness of the bacteria but only in one of the two plant genotypes tested (Batstone et al., 2022).
At the final stage of the symbiosis, nodules senesce and bacteria return to the soil where they can survive for months or even years before re-infecting a new host (Denison and Kiers, 2011). This extended phase of the rhizobial life cycle is crucial for the ecology and evolution of these bacteria, but remains largely understudied. In aged nodules of M. sativa, a population of saprophytic bacteria develop in the proximal zone (Timmers et al., 2000). A recent transcriptomic analysis showed that cell division and the general stress response are activated in rhizobia from senescent nodules (Sauviac et al., 2022). These results indicate that exiting nodules is an active process for rhizobia, but there are, to our knowledge, no study that analysed the determinants of rhizobial fitness during this phase. To persist in the soil, some bacteria may rely on previously accumulated carbon storage particles, such as polymers of poly-3-hydroxybutyrate (PHB) (Müller-Santos et al., 2021). Among rhizobial strains, some, but not all (Trainer and Charles, 2006; Chen et al., 2023), were shown to accumulate high amounts of PHB in nodules, which can represent up to 50% of the dry weight of the cells (Bergersen et al., 1995; Tavernier et al., 1997). This stored PHB can support several divisions of bacteria or a much longer survival of dormant cells without the need for any other carbon sources (Muller and Denison, 2018). Therefore, measuring PHB content in bacteroids could be a good proxy for estimating the ability of rhizobia to reproduce in soil, and an indicator of the ‘quality’ of the progenies released from nodules (Ratcliff et al., 2008; Ratcliff et al., 2011; Muller and Denison, 2018). Alternative polymers such as glycogen can also be produced by some rhizobia in nodules to store carbon and energy (Lodwig et al., 2005; Wang et al., 2007a). These processes, which consume significant amounts of energy resources, are thought to divert energy from nitrogen fixation (Lodwig and Poole, 2003) and thus induce plant sanctions (Oono et al., 2020). However, in the literature, mutants defective in the synthesis of PHB have shown contrasting nitrogen fixation phenotypes, ranging from increased to decreased nitrogen fixation (Cevallos et al., 1996; Lodwig et al., 2005; Wang et al., 2007a; Wang et al., 2007b; Crang et al., 2021). Considering the redox balance and oxygen-limiting conditions that prevail in nodules, the accumulation of PHB or other carbon polymers may indeed be required for the persistence of bacteroids in nodules (Terpolilli et al., 2016; Schulte et al., 2021). There may therefore be a trade-off between rhizobial survival in the soil, persistence in nodules and nitrogen fixation efficiency (Ratcliff et al., 2008), but these effects might be species-specific and host-plant dependent (Chen et al., 2023).
Beyond the analysis of individual fitness components, an important question relates to the potential couplings or trade-offs between the different phenotypic traits. In particular, there is no selection of nitrogen fixation at the nodulation step (since nitrogen fixation occurs at the late stages of the interaction) (Daubech et al., 2017; Westhoek et al., 2017), but several lines of evidence indicate that co-evolutionary processes may lead to the selection of strains that are both competitive for nodulation and efficient for nitrogen fixation on a given host genotype (Burghardt et al., 2018; Younginger and Friesen, 2019; Batstone et al., 2020; Fields et al., 2023; Rahman et al., 2023). This can occur through partner-fidelity feedback, a positive feedback loop acting on the fitness of the two mutualistic partners as a result of repeated associations between these organisms (Sachs et al., 2004; Fujita et al., 2014).
Although most of the fitness measurements described above usually rely on traditional microbiological techniques [with potential new optimisations, e.g. (Mendoza-Suárez et al., 2020; Quides et al., 2023)], new approaches based on bacterial populations carrying short DNA tags, in combination with next generation sequencing, now enable to analyse rhizobial fitness in a high-throughput manner (Burghardt et al., 2018; Wheatley et al., 2020). These approaches are particularly powerful to gather integrative measures of fitness (e.g. encompassing all fitness components composing the entire life cycle) in genetically complex rhizobial populations, and open many opportunities to perform integrative fitness measurements in a variety of conditions (Burghardt, 2020; Burghardt et al., 2020; Burghardt et al., 2022).
Finally, it is worth remembering that measurements of bacterial fitness will likely be dependent on the abiotic and biotic environment during the experiment. Abiotic factors include experimental conditions (light, temperature, plant growth substrate…), and biotic factors include host plant genotype and microbial communities (composition and density), which is the focus of this review. In the next sections, we illustrate how microbial communities affect rhizobial fitness at the different steps of their life cycle (Figure 1; Table 1; Supplementary Table 1).
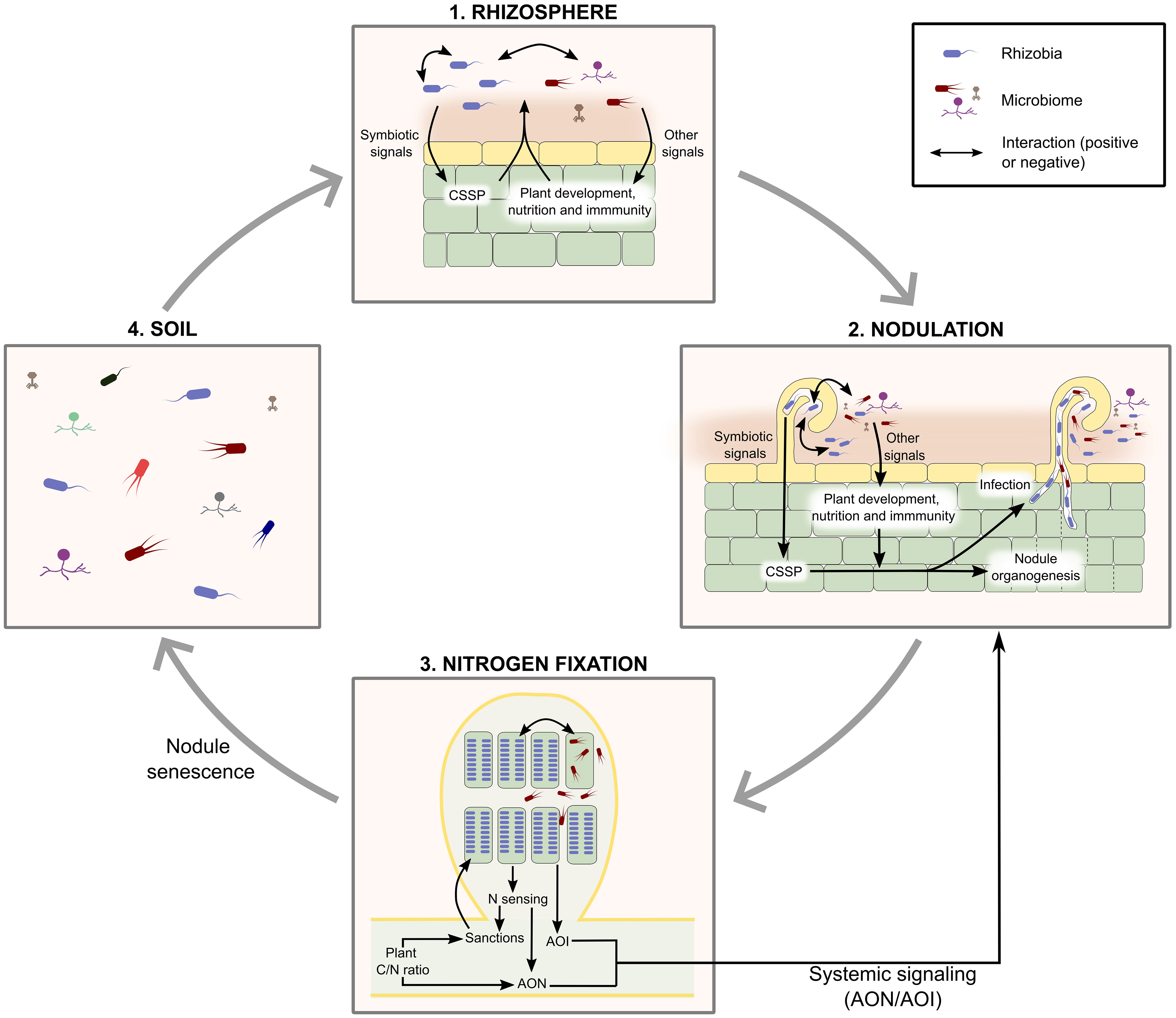
Figure 1 The life cycle of rhizobia and interactions with microbiome. A simplified view of the life cycle of rhizobia considers 4 successive steps: (1) colonisation of the rhizosphere, (2) initiation of nodulation, (3) nodule infection and nitrogen fixation, (4) nodule senescence and release of bacteria in the soil. Interactions with other microbes can occur at each step of the life cycle. (1) Rhizosphere: direct interactions (contact-dependent, or mediated by the secretion of beneficial or harmful chemical compounds) can occur as in any microbial community; indirectly, rhizobia and some members of the rhizosphere microbiome can modify the composition of their microbial community through plant-mediated effects: rhizobia and AMF through CSSP and non-rhizobia likely via other pathways linked to plant development, immunity, or nutrition. (2) Nodulation: rhizobia elicit plant symbiosis signaling pathways, more or less efficiently, which results in different levels of competitiveness. Other microbiome members can also induce physiological responses (for example through the secretion of plant hormones, or the modulation of hormone synthesis by the plant, or the synthesis of incompatible Nod Factors) that affect nodulation (positively or negatively, respectively). Direct physical or biochemical interactions between rhizobia and other microbes can also affect rhizobial nodulation success. (3) Nodule infection and nitrogen fixation: direct interactions can occur through the secretion of diffusible molecules (siderophores, antibiotics…) within nodules. Indirect interactions can result from the conditional sanctioning of rhizobia fixing less nitrogen than rhizobia present in other nodules. Of note, bacteria within nodules can influence the nodulation process (through AON, AOI, or the production of rhizobitoxin). AON, and probably conditional sanctioning, depend on the general physiological status of the plant (C/N ratio), which can be influenced by many biotic or abiotic factors. (4) Although we have not found any documented example (and are therefore not represented on this figure), it is also possible that in nodules indirect interactions between non-rhizobial endophytes and rhizobia occur, if nodule endophytes can activate plant signaling pathways that modify nodule physiology and rhizobial survival. Finally, although interactions likely occur during nodule senescence and bacterial release in the soil, we have not found any references on that topic. AOI, Auto-regulation of Infection; AON, Auto-regulation of Nodulation; CSSP, Common Symbiotic Signaling Pathway (induced by both AMF and rhizobium symbionts).
The rhizosphere, which is the soil surrounding and under the influence of the roots, is characterised by a high microbial biomass and a great diversity of tens of thousands of species (Berendsen et al., 2012; De La Fuente Cantó et al., 2020). It is composed of eukaryotic microorganisms (protozoa, fungi, oomycetes, yeasts and nematodes), prokaryotic microorganisms (bacteria and archaea) and viruses (Philippot et al., 2013). This complex microbial community is highly dynamic and depends on the soil chemical and biochemical characteristics, the plant genotypes, and the type of interactions between plants and microorganisms (mutualistic, pathogenic, saprophytic, or commensal) (Philippot et al., 2013; Trivedi et al., 2020). In particular, exudates released by the roots, such as the secretion of aromatic organic acids that are preferentially consumed by rhizosphere bacteria have a major effect on microbial abundance and composition (Bais et al., 2006; Shi et al., 2011; Zhalnina et al., 2018). Abiotic factors such as pH, salinity, the presence of biofertilizers, heavy metals and carbon resource availability are also strong factors influencing the rhizobiome (Ofek et al., 2006; Bellabarba et al., 2019). In addition, microbial interactions among community members have a fundamental effect on the rhizobiome composition (Hassani et al., 2018; Chepsergon and Moleleki, 2023). Network community analyses have found co-occurrence and exclusion patterns suggesting positive and negative interactions between microorganisms (Han et al., 2020). Several mechanisms can explain these patterns. For instance, competition between organisms sharing the same ecological niche and exploiting the same resources can lead to the exclusion of some strains (Hibbing et al., 2010; Ghoul and Mitri, 2016). The secretion of antibiotics or metabolites or even predation (i.e. killing and consuming the prey) by members of the community also influence the abundance of the other members (Granato et al., 2019; Peterson et al., 2020). On the other hand, cooperation by the production of public goods (Smith and Schuster, 2019) and cross-feeding between microorganisms (D’Souza et al., 2018; Jacoby and Kopriva, 2019) can maintain the co-occurrence of some species in the community. Plant-mediated indirect interactions can also occur when microbes induce a modification in the secretion of root exudates, which in turn affects other members of the microbiome (De La Fuente Cantó et al., 2020; Korenblum et al., 2020).
The root-associated rhizobial density varies in nature (Yan et al., 2014). It depends on both positive and negative interactions with other microorganisms. Among negative interactions, predation by protozoa or bacteria (Bdellovibrio or Myxococcus) can alter rhizobial populations (Danso et al., 1975; Keya and Alexander, 1975; Ramirez and Alexander, 1980). Soil bacteriophages, which have been shown to rapidly adapt to local bacterial host communities, can also reduce the rhizobial density in the rhizosphere (Van Cauwenberghe et al., 2021). Finally, the presence of antimicrobial compounds produced by other rhizosphere microorganisms such as antibiotics produced by Actinomycetes is likely to inhibit the growth of rhizobia (Patel, 1974; Pugashetti et al., 1982). However, certain rhizobia have the means to survive to these attacks and compete with other microorganisms in the soil. In an in vitro experiment, Pérez et al. (2014) showed that some strains of Sinorhizobium meliloti that produce the exopolysaccharide (EPS) galactoglucan are more resistant to predation by Myxococcus xanthus. Given that EPS play a crucial role in plant recognition and early steps of symbiosis, it is possible that in the rhizosphere, EPS production has a dual ecological advantage in reducing predation and promoting interactions with compatible host plants. The production of melanin was also shown to favour resistance against predation by Myxococcus xanthus (Contreras-Moreno et al., 2020), and a transcriptomic analysis identified several other putative defense mechanisms, such as the production of surface polysaccharides and membrane lipids, the activation of efflux pumps or the induction of iron uptake (Soto et al., 2023).
In addition, studies have shown that type VI secretion systems (Bladergroen et al., 2003; De Campos et al., 2017) as well as the production of phages (Schwinghamer and Brockwell, 1978; Joglekar et al., 2023) or bacteriocins (Hirsch, 1979; Triplett and Barta, 1987; Oresnik et al., 1999) in rhizobia can play an important role in the direct inhibition of bacterial competitors. Among antagonistic molecules produced by rhizobia, diffusible quorum sensing molecules, in particular Acyl Homoserine Lactones (AHLs), can inhibit the growth of other rhizobial strains by activating LuxR-type regulators in the neighbouring strains and inducing genes leading to growth arrest (Schripsema et al., 1996; Wilkinson et al., 2002). The production of these antimicrobial compounds by rhizobia offers a competitive advantage in nutrient-limited environments but also for the establishment of symbiosis, by inhibiting other nodulating bacteria. A recent in vitro study suggests that both facilitative and inhibitory interactions exist even between rhizobial genotypes of the same species (Fields et al., 2022).
Besides negative interactions, cases of cooperation between rhizobia and other microorganisms are frequently found. An example of positive interaction was recently observed in co-cultures of the rhizobium Rhizobium etli and the yeast Saccharomyces cerevisiae. This interaction led to the formation of mixed biofilms in which the growth of the rhizobium was promoted (Andrade-Domínguez et al., 2021). This study has shown that this synergism is an effect of commensal interactions, where the rhizobia benefit from compounds secreted by the yeast such as dicarboxylic acids that bacteria use for their nutrition, and sophoroside, an antimicrobial compound detoxified by R. etli, which can shape the composition of the microbial community associated with this yeast. In the rhizosphere, nutritional interdependencies and reciprocal exchange of metabolites between microorganisms are commonly found (Jacoby and Kopriva, 2019). For example, a feeding of rhizobia by Actinobacteria capable of hydrolyzing cellulose has been evidenced (Silva et al., 2019). Other synergistic interactions were observed between Mesorhizobia and Actinobacteria (Vo et al., 2021), and between Sinorhizobia and bacteria of the Bacillus cereus group (Han et al., 2020), which enhance the growth of rhizobia by mechanisms that have not yet been elucidated.
Finally, recent research has led to the discovery that plant functional genes, and in particular symbiotic signaling genes in legumes, are involved in modulating the structure of the root-associated microbiota and the interactions among these communities (Zgadzaj et al., 2016; Thiergart et al., 2019; Liu et al., 2023). Legumes can establish mutualistic interactions with rhizobia and arbuscular mycorrizal fungi (AMF) that induce symbiotic signaling pathways involving specific components but also sharing some common components (Oldroyd, 2013). Disruption of both common and specific components of the symbiotic pathways affects the relative abundance of many bacterial and fungal taxa in the rhizosphere. For example, Lotus japonicus mutants defective in rhizobial NF perception (nfr5 mutants) or in rhizobium infection (nin mutants) associate with a different bacterial community than the wild-type Lotus japonicus accession, with several bacterial orders such as Flavobacteriales, Myxococcales, Pseudomonadales, Rhizobiales, and Sphingomonadales being depleted in the roots of mutant plant genotypes (Zgadzaj et al., 2016; Wippel et al., 2021). Furthermore, inactivation of both root nodule and arbuscular mycorrhizal symbiosis pathways (symrk and ccamk mutants) was surprisingly associated with the root microbiota network structure (increase in both network connectivity and degree centrality) suggesting a higher number of interactions between community members in the absence of both symbioses (Thiergart et al., 2019). Interestingly, the activation of the specific mycorrhizal symbiosis pathway in peanut was shown to promote the accumulation of rhizobia in the rhizosphere and stimulate nodulation (Wang et al., 2021). Altogether, these studies showed a broad role of plant symbiotic genes in shaping the structure of microbial communities on legume roots or rhizosphere and the interactions between members of these communities.
The success of nodulation by rhizobia depends not only on the rhizobial density near the root but also on a process of selection mediated by the plant. In nature, legumes are usually nodulated by a diversity of rhizobial strains that produce NF with compatible structure. However, among compatible strains, even from the same species complex, some are more competitive than others to form nodules (Elliott et al., 2009; Melkonian et al., 2014; Batstone et al., 2017; Boivin et al., 2020; Boivin et al., 2021). This preferential association is dependent on both the plant genotype, the bacterial genotypes and the interaction between the plant and bacterial genotypes (Burghardt et al., 2017; Boivin et al., 2021; Fagorzi et al., 2021). On the bacterial side, using classical genetics or large-scale transposon insertion mutant libraries, several studies have shown that hundreds of genes are involved in the competitive ability of rhizobia to form nodules (Pobigaylo et al., 2008; Wheatley et al., 2020; Mendoza-Suárez et al., 2021). Beyond NF biosynthesis and regulatory genes, genes involved in motility and chemotaxis, or other functions that help bacteria to migrate near the roots (Caetano-Anollés et al., 1988; Wheatley et al., 2020; Ji et al., 2023), and in surface polysaccharide biosynthesis (Pobigaylo et al., 2008; Janczarek et al., 2009) were shown to be important for competitive nodule formation. Also, the bacterial capacity to metabolise components found in root exudates such as the homoserine or mimosine (Soedarjo and Borthakur, 1998; Vanderlinde et al., 2014), particular sugars such as erythritol (Yost et al., 2006; Wheatley et al., 2020), rhamnose (Oresnik et al., 1998), myo-inositol (Fry et al., 2001) or amino acids such as proline (Jimenez-Zurdo, 1995) contribute to nodulation competitiveness. In addition, genes involved in purine biosynthesis (Xie et al., 2009; Wheatley et al., 2020) and nitrogen metabolism (Wheatley et al., 2020) play a role in competitive nodule occupancy. Although in nature strains that exhibit high competitiveness for nodulation often demonstrate high nitrogen fixation efficiency as well (Burghardt et al., 2018; Fields et al., 2023; Rahman et al., 2023), the capacity to fix atmospheric nitrogen itself does not contribute to nodulation competitiveness (Amarger, 1981; Hahn and Studer, 1986; Daubech et al., 2017; Westhoek et al., 2017; Bourion et al., 2018). Interestingly, co-inoculation of highly competitive and efficient nitrogen-fixing strains with another rhizobial strain can lead either to higher plant benefits than in single inoculation or, on the contrary, to a decrease in both nodulation and plant benefits, indicating the existence of positive and negative competitive interferences between the rhizobial strains (Heath and Tiffin, 2007; Montoya et al., 2023; Rahman et al., 2023). Other interactions negatively affecting nodulation initiation were described by Gano-Cohen et al. (2016), who showed that the presence of non-nodulating Bradyrhizobium reduced the number of nodules formed by nodulating Bradyrhizobium on the legume Acmispon strigosus, probably by directly competing for attachment sites on the root. In another study, a phenomenon called “competitive nodulation blocking” was observed in the presence of some rhizobial strains that produce high levels of NF of incompatible structure, thus inhibiting nodulation by other compatible symbionts (Hogg et al., 2002). The presence of plant pathogens can also inhibit nodule formation as shown under laboratory conditions by Benezech et al. (2021) using the Medicago-Sinorhizobium-Ralstonia model system.
On the other hand, direct cooperations between rhizobial strains are possible through the production of extracellular diffusible molecules that can functionally complement each other. For example, a Mesorhizobium loti strain deficient in NF production (R7A nodA mutant) and another one deficient in exopolysaccharide production (R7A exoU mutant) are both unable to nodulate Lotus japonicus but can complement each other to form functional nodules. In that case, 80% of nodules contained both strains (Kelly et al., 2013). Similar results were obtained with nod and exo mutants of Sinorhizobium meliloti nodulating alfalfa (Klein et al., 1988; Kapp et al., 1990). A more recent study showed that complementation of rhizobium EPS deficient strain can be done by non rhizobial nodule endophytic bacteria (Zgadzaj et al., 2015). Another example of cooperation has been observed between the Phaseolus vulgaris symbiont R. etli and a commensal rhizobium R. fabae. In this cooperation, R. fabae produces AHLs, quorum sensing molecules that are perceived by the CinR regulator of R. etli leading to changes in gene expression and optimisation of nodulation (Miao et al., 2018).
In addition, the presence of plant growth-promoting rhizobacteria (PGPR) in the soil can indirectly increase the nodulation efficiency of rhizobia by producing or decreasing the amounts of phytohormones known to interfere either positively or negatively with the nodulation process (Alemneh et al., 2020). For example, the production of indole-3-acetic acid (IAA), a plant hormone of the auxin class, by bacteria of the genera Azospirillum, Micromonosperma, Pseudomonas or Bacillus, increases nodule formation by rhizobia on legumes (see (Alemneh et al., 2020) for a review on bacteria able to synthesise IAA). Indeed, these molecules are known to increase lateral root growth (Casimiro et al., 2001), potentially providing more infection sites for rhizobia. IAA may also promote nodulation through induction of a rhizobium infection signaling pathway in root hairs (Breakspear et al., 2014). In addition, many PGPRs, such as bacteria of the genera Serratia, Arthrobacter or Pseudomonas, are able to produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase which sequesters and cleaves the plant ethylene precursor ACC, reducing levels of ethylene, a negative regulator of nodule formation, and thus promoting nodulation (see (Alemneh et al., 2020) for a review on bacteria able to synthesise ACC deaminases). This process is even more important under stressful conditions, such as salinity stress, known to increase the levels of plant-produced ethylene (Ahmad et al., 2011). Finally, some PGPR can increase the availability of nutrients for rhizobia and can stimulate their growth in vitro (Le et al., 2016; Vo et al., 2021). An example of cooperation between Streptomyces and Rhizobium sp. was described by Tokala et al. (2002), where iron- and molybdate-sequestering siderophores produced by Streptomyces provide these metals to the rhizobium strain at early and late symbiotic stages, increasing both nodulation and nitrogen fixation.
Rhizobia can also interact with eukaryotic microorganisms such as fungi. A large number of studies have shown the positive effect of AMF on nodulation (Antunes and Goss, 2005; Barea et al., 2005; Pang et al., 2023; Tsikou et al., 2023). Phosphate supply by the fungus seems to be the main factor improving the rhizobium-legume symbiosis, although other factors such as the modulation of plant hormone levels by fungi, as described for PGPRs, also contribute to increase nodule number and plant biomass. Nevertheless, a meta-analysis showed that the synergistic effect between AMF and rhizobia is mostly observed in perennial plants but was not observed in annual plants (Primieri et al., 2022). These results suggest that the effect of the tripartite interaction between legumes, AMF and rhizobia can be influenced by the life history of the host plant. Additionally, these effects can vary based on the co-inoculated species, suggesting a particular compatibility between the symbionts is necessary to obtain combined positive effects in plants (Xavier and Germida, 2003), and on abiotic conditions (Afkhami et al., 2020). For example, under limited light, co-inoculation of rhizobia and AMF can lead to a reduction in plant growth, likely due to the important allocation of carbohydrates to the symbionts (Ballhorn et al., 2016). Finally, fungal mycelia form dispersion networks, which may facilitate root invasion. This was observed for a Bradyrhizobium strain, which initiates nodulation on peanut by a crack entry process and uses mycelia produced by a biotrophic fungus, Phomopsis liquidambaris, to migrate to legume rhizosphere and reach infection sites (Zhang et al., 2020).
Nodules represent a specific niche for rhizobia, in which they are largely shielded from competition with other soil bacteria. However, this isolation is not strict. The presence of non-rhizobial strains in nodules has been reported for a long time, but the recent advent of large-scale sequencing experiments provided some more complete and systematic descriptions of these 'nodule-associated bacteria' (NAB). These findings open the possibility that rhizobia may interact directly with other non-rhizobia within nodules.
Reported NAB belong to many different genera of Alpha-, Gamma- or Beta- proteobacteria (the latter now been included as a subset of Gamma-proteobacteria (Parks et al., 2018)) as well as to the Actinobacteria and Firmicutes phyla (Martínez-Hidalgo and Hirsch, 2017). NAB tend to have lower densities than rhizobia within nodules (Etesami, 2022), but their precise localisation within nodules is often unknown. Numerous studies have shown that NAB can affect the number of nodules formed by rhizobia and/or plant growth. In fact, many NAB seem to possess plant-growth promoting traits, making them good candidate as inoculants together with selected rhizobial strains (Martínez-Hidalgo and Hirsch, 2017; Velázquez et al., 2017; da Silva et al., 2023). Yet, there is little information currently available on the activity and biological relevance of these NAB within nodules.
In Lotus, NAB can enter nodules by co-colonising infection threads formed by the compatible rhizobium Mesorhizobium loti (Zgadzaj et al., 2015). A fluorescently labeled non-nodulating and non-fixing strain Rhizobium mesosinicum KAW12 was then shown to colonise both inter- and intra-cellular nodule spaces, when co-inoculated with M. loti. Colonisation was dependent on the symbiotic genetic program of the host plant, activated by the compatible NF from M. loti. Crosbie et al. (2022) later identified an antagonistic interaction between another intracellular nodule commensal (Pseudomonas sp.) and an ineffective Rhizobium strain. Indeed, Pseudomonas strains were detected in L. japonicus nodules formed by an efficient M. loti strain, but not in nodules formed by the non-fixing Rhizobium sp. BW8-2. Co-inoculating one of these Pseudomonas strains with Rhizobium sp. BW8-2 reduced the number of nodules formed by the latter strain, suggesting a negative effect of Pseudomonas on Rhizobium during the early stages of symbiosis. This effect was observed on Lotus japonicus but not on L. burnetii, showing that the host plant plays a role in the mediation of this rhizobium-NAB interaction.
Hansen et al. (2020) were also able to isolate cultivable NAB from Medicago sativa nodules formed by S. meliloti. These strains, representative of a simplified nodule microbial community established after three cycles of in planta selection, were used to perform functional studies on synthetic communities, by measuring the rate of nodule colonisation of each strain when co-inoculated with other members of this community (but always including S. meliloti). Both cooperative and antagonistic interactions between strains were identified in this system. Several interesting results emerge from this study. First, while two strains (Pseudomonas sp. and Paenibacillus sp.) showed a mutually beneficial effect on the rate of nodule colonisation, Paenibacillus sp. proliferation within nodules was reduced in the presence of Pseudomonas sp., showing that the type of interaction can be dependent on the phase of the life cycle. Second, all endophytes were able to antagonise S. meliloti in in vitro conditions and reduced nodule formation. However, co-inoculation of the entire community with S. meliloti reduced neither nodule formation nor plant growth, suggesting that higher-order interactions modify the effect of these NAB on S. meliloti. Finally, spatial metabolic analyses indicate that some antagonistic interactions might be mediated by the production of antibiotics by one of the nodule commensal strains.
Overall, these three studies gathered compelling evidence that rhizobia-NAB and rhizobia-plant-NAB interactions do occur within nodules. Future investigations on the precise effects of NAB on rhizobial fitness and nitrogen fixation will undoubtedly complete our understanding of these fascinating aspects of rhizobia biology.
Direct interactions between rhizobia and other bacteria may also occur at longer distance. A subset of rhizobial species produce rhizopines, a class of inositol-derived molecules synthesised in bacteroids (Murphy et al., 1995). Rhizopines are believed to be catabolised by rhizobia that are in the rhizosphere or infection threads and carry the corresponding degradation operons. In agreement with this hypothesis, rhizopine-catabolising strains have a fitness advantage over non-catabolizing strains during symbiosis (Gordon et al., 1996). However, this advantage was observed at early symbiotic stages, and it is therefore unclear if it arises from the ability to catabolise rhizopines produced by bacteroids, or other compounds produced by plants or bacteria in the rhizosphere. Nevertheless, the ability to catabolise rhizopines was also found in non-rhizobia (Gardener and De Bruijn, 1998), suggesting that other rhizospheric bacteria or possibly NAB may benefit from rhizopine produced by nodule bacteria and that rhizopines may represent a ‘public good’ for rhizospheric bacteria (although the capacity to catabolise it is restricted to a relatively small subset of bacteria). This consideration opens interesting evolutionary questions that have been addressed by mathematical modeling (Simms and Bever, 1998).
Indirect long-distance interactions also occur between rhizobia present within nodules and rhizobia located in the rhizosphere. The process of auto-regulation of infection (AOI) initiated by nodule rhizobia regulates root-hair infection by rhizospheric bacteria (Tian et al., 2012). This phenomenon involves complex signal exchanges between plants and nodule bacteria (Garnerone et al., 2018; Sorroche et al., 2019; Sorroche et al., 2020). However, the relevance of AOI for rhizobial fitness when plants are exposed to different rhizobial or non-rhizobial strains remains to be investigated. In addition, infection and nitrogen fixation by rhizobia have strong effects on plant physiology, and can lead to the production of systemic signals that modulate rhizobial fitness throughout the plant (Lepetit and Brouquisse, 2023). For example, the auto-regulation of nodulation (AON) allows the plant to block the nodulation process when its needs in nitrogen are covered by the functioning nodules (Ferguson et al., 2019). Long-distance interactions are also known to occur between rhizospheric microorganisms and nodule bacteria (Ruiz-Lozano et al., 2001; Tokala et al., 2002). In soybean, symbiosis with the AMF Glomus mosseae protects nodules from drought-induced senescence (Ruiz-Lozano et al., 2001). The authors observed that AMF reduced the effect of drought on several markers associated with nodule senescence, such as the decreased nitrogen fixation activity and the early appearance of oxidative damages. It is likely that this protective effect also translates into better rhizobial fitness.
Finally, indirect interactions between rhizobia present in different nodules have recently been reported when plants are colonised by several different rhizobial strains. While it is now well established that rhizobial persistence within nodules depends on their rate of nitrogen fixation (see section 2), Whesthoek et al. (2021) showed that this effect can be modulated depending on the symbiotic effectiveness of the diverse strains nodulating the same host plant. Using a strain that fixes reduced amounts of nitrogen (‘intermediate fixer’), the authors showed that bacterial load within mature nodules colonised by this strain is lower when the plant is co-inoculated with an efficient strain than with a non-fixing strain. These results show that plants are able to impose 'conditional sanctions' by comparing nitrogen output from the different nodules and responding accordingly in ways that affect rhizobial survival within nodules. This response seems to involve differential transport of sugars and di-carboxylic acids to the nodules, but the precise molecular mechanisms involved in the sensing of differential nitrogen fixation levels and the establishment of conditional sanctions remain to be discovered.
All the examples mentioned so far in this review (and also reviewed by others; (Barea et al., 2005; Afkhami et al., 2020; Checcucci and Marchetti, 2020; Burghardt and diCenzo, 2023)) show that rhizobial fitness can be modified by the presence of other microorganisms at the different steps of the life cycle. Therefore, microbial communities will change the selective pressures acting on rhizobial populations, and thus their evolutionary trajectories. Below we discuss two processes by which the biotic environment can affect rhizobial evolution: eco-evolutionary feedbacks and the alteration of the mechanisms of evolution. For more general perspectives on the evolution of rhizobia, we refer interested readers to the following recent reviews (Andrews et al., 2018; Burghardt, 2020; Denison and Muller, 2022; Remigi et al., 2016; Masson-Boivin and Sachs, 2018; Tian and Young, 2019; Tang and Capela, 2020; Provorov et al., 2022; Wardell et al., 2022; Liu S, et al., 2023).
Eco-evolutionary feedbacks refer to the interplay between the modification of selective pressures by ecological mechanisms (e.g. biotic interactions) and the modification of ecological interactions by evolutionary mechanisms (Schoener, 2011; Hendry, 2017; Ware et al., 2019). Eco-evolutionary feedbacks are expected to be particularly relevant when the time scales of ecological and evolutionary processes overlap, which is typically the case in microbial populations (Fields and Friman, 2022). In the case of rhizobia, the presence of a complex microbiome can shape not only the evolution of the interactions between rhizobia and other microbiome members, but also interactions between rhizobia and their host plant. This seems be the case for example with the emergence of phage-resistant rhizobia that survive better in the rhizosphere but show a decreased symbiotic performances on their host plant (Kleczkowska, 1950; Stacey et al., 1982; Handelsman et al., 1984; Mishra et al., 2004; Anand et al., 2012). In another example of eco-evolutionary feedback, the co-culture of R. etli with yeasts leads to the rapid appearance of bacterial phenotypic variants (Andrade-Domínguez et al., 2014). These variants carry diverse mutations that allow R. etli to grow in the presence of orotic acid, a metabolite secreted by yeasts and that inhibits the growth of wild-type bacteria. These variants then further evolved to become yeast antagonists, thus completely changing the nature of the interaction between these organisms. Although it is unknown whether these two strains interact with one another in their natural environment, this work nicely unravels how the presence of yeasts can rapidly affect the phenotypic and genetic composition of rhizobial populations.
Experimental evolution can also be used to study how plant-rhizobia interactions can change in a variety of ecological contexts (Remigi et al., 2019). Doin De Moura et al. (2023) analysed an evolution experiment designed to turn a plant pathogenic bacterium into a legume symbiont, following the artificial transfer of a symbiotic plasmid into the plant pathogenic ancestor (Doin de Moura et al., 2020). In the course of this experiment, multiple sub-populations of bacteria emerge and compete in the rhizosphere. Evolved bacteria (that are still not fixing nitrogen at the end of the experiment) predominantly improved their nodulation competitiveness. These results show that, in emerging rhizobia with poor symbiotic phenotypes, the selective bottleneck at the nodulation step exerts a strong selective pressure on bacterial populations.
Batstone et al. (2020) performed serial nodulation cycles on five lines of M. truncatula using a mix of two rhizobial strains as starting inoculum: one effective and one ineffective nitrogen-fixing strain. During this experiment, the effective strain increased in frequency in almost all lines, and became even more beneficial to the plant lines on which they evolved. An interesting follow-up of this experiment would be to evolve strains alone or in co-inoculation, to assess the effect of competition between multiple rhizobial strains on their respective evolutionary trajectories.
The presence of a complex microbial community can also modify the basic evolutionary mechanisms acting on a focal species, such as the rates of mutations and of horizontal gene transfer (HGT). Bacterial mutation rates are plastic (Ferenci, 2019; Matic, 2019; Pribis et al., 2022), and can be influenced by various stresses, nutrient availability (Maharjan and Ferenci, 2017), or cell density (Krašovec et al., 2017). Interestingly, in E. coli, the modulation of mutation rate by cell density depends on the quorum-sensing gene luxS, indicating a ‘social-dependence’ of mutation rate in this strain (Krašovec et al., 2014). Given that microbial communities can constitute stressful environments for bacteria (Hibbing et al., 2010; LeRoux et al., 2015; Traxler and Kolter, 2015; Granato et al., 2019), it is plausible that mutation rates of rhizobia might be affected by other microorganisms. In addition, many symbiotic plasmids also carry stress-inducible error-prone DNA polymerases (Remigi et al., 2014). After transferring one of these plasmids (from the rhizobium Cupriavidus taiwanensis) into a non-symbiotic bacterium, the expression of error-prone DNA polymerases in stressful environments (the rhizosphere) induces transient bursts in mutation rate and accelerates adaptation to the symbiotic lifestyle. Antagonistic interactions between microbiome members can also lead to the selection of constitutive hypermutator strains (e.g., that are defective in DNA repair systems). This has been documented for phage-bacteria interactions where lytic phages impose strong mortality in bacterial populations. Second-order selection of constitutive hypermutators can thus occur, as these strains will have a higher chance to become resistant to phages (Chevallereau et al., 2022).
HGT is another major driver of rhizobia evolution. Evidently, dense and diverse microbial communities, like those found in the rhizosphere, facilitate HGT due to both the elevated transfer rates and the extensive pool of genes that could potentially be transferred (Brito, 2021; Moura De Sousa et al., 2023; Sánchez-Salazar et al., 2023). Yet additional mechanisms exist that further increase HGT in rhizobia. The transfer rate of symbiotic integrative and conjugative elements (ICE) from Azorhizobium caulinodans to other rhizobia is elevated in the rhizosphere of several plants compared to empty soil (Ling et al., 2016). The perception of flavonoid compounds by a transcription regulator of the LysR family controls the expression of the ICE integrase, together with two other genes located on this symbiotic island (Ling et al., 2016; Li et al., 2022). Recently, the transfer of symbiotic plasmids from R. etli CFN42 to other rhizobia and NAB was also detected in bean nodules (Bañuelos‐Vazquez et al., 2019; Bañuelos-Vazquez et al., 2020). These examples show that symbiosis genes are particularly prone to be transferred in the presence of host plants. Indeed, HGT of symbiosis genes has been extensively documented by phylogenetic studies (Remigi et al., 2016; Andrews et al., 2018; Wardell et al., 2022). In the field, non-symbiotic strains that are well adapted to the abiotic environment can become efficient symbionts by acquiring symbiotic ICE from ‘elite’ rhizobial inoculants that are not adapted to the soil conditions (Sullivan et al., 1995; Nandasena et al., 2007; Hill et al., 2021; Colombi et al., 2023). Moreover, the frequent re-arrangements and recombinations between symbiotic genes from different strains create a dynamic equilibrium where less cooperative strains regularly appear as well as strains with new or improved symbiotic abilities, illustrating how genetic diversity of rhizobial strains fuels the evolvability of symbiosis (Weisberg et al., 2022). Altogether, these studies demonstrate that incorporating the whole microbial communities will be crucial to understand the rate and patterns of rhizobial evolution.
The rhizobium-legume symbiosis has long been a model system to study both the molecular and eco-evolutionary aspects of host-microbe interactions. In the recent years, we have witnessed an increased interest in incorporating the effect of complex microbial communities on the functioning of this symbiosis, and numerous interactions with a multitude of microbes have already been detected throughout the rhizobial life cycle in both laboratory and field conditions (Table 1).
However, certain stages of the rhizobial life cycle have been understudied. For instance, interactions within the nodule microbiome, although much less complex than the rhizosphere microbiome, still lack a comprehensive understanding. Similarly, interactions of bacteria released from senescent nodules with soil communities remain largely unknown. In addition, research has predominantly focused on pairwise interactions so far, while in nature, interspecies interactions occur in multispecies communities. In such complex ecosystems, higher-order interactions can profoundly impact the fitness of a focal strain (Levine et al., 2017). Another set of open questions relates to the genetic bases of inter-microbial interactions. Once we start to uncover the molecular mechanisms involved in these interactions and to describe the existing natural genetic variations in the life history traits, it will be particularly interesting to further test whether there are genetic trade-offs or couplings between these different traits, including those involved in the interaction with the host plant. In parallel, assessing the eco-evolutionary dynamics of these interactions with laboratory evolution or field experiments will provide complementary information on the selective pressures acting on rhizobial populations.
Therefore, we have probably only scratched the surface of the extent to which rhizobial-microbial interactions contribute to the establishment and functioning of legume-rhizobia symbioses. Additional discoveries are expected as research progresses on this topic, concerning both the mechanisms that mediate these interactions and their ecological and evolutionary consequences. A future challenge will be to integrate all these findings in a coherent framework, and use this knowledge to improve agricultural and ecosystem services that can be obtained from rhizobia. An ambitious goal would be to be able to design synthetic communities where microbial interactions enhance rhizobial fitness and symbiotic services, and mitigate the effects of plant pathogens. This will require cooperation between several disciplines (molecular microbiology, ecology and evolutionary biology, plant physiology, agronomy…) and cross-fertilisation between laboratory and field experiments.
MGA: Writing – original draft, Writing – review & editing. BR: Writing – original draft, Writing – review & editing. DC: Writing – original draft, Writing – review & editing. PR: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is funded by the French National Research Agency (ANR-21-CE02-0019-01), the “Laboratoires d’Excellence (LABEX)” TULIP (ANR-10-LABX-41), and the “École Universitaire de Recherche (EUR)” TULIP-GS (ANR-18-EURE-0019).
We apologise to colleagues whose work could not be cited to keep the review concise.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1277262/full#supplementary-material
Afkhami, M. E., Almeida, B. K., Hernandez, D. J., Kiesewetter, K. N., Revillini, D. P. (2020). Tripartite mutualisms as models for understanding plant–microbial interactions. Curr. Opin. Plant Biol. 56, 28–36. doi: 10.1016/j.pbi.2020.02.003
Ahmad, M., Zahir, Z. A., Asghar, H. N., Asghar, M. (2011). Inducing salt tolerance in mung bean through coinoculation with rhizobia and plant-growth-promoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 57, 578–589. doi: 10.1139/w11-044
Alemneh, A. A., Zhou, Y., Ryder, M. H., Denton, M. D. (2020). Mechanisms in plant growth-promoting rhizobacteria that enhance legume–rhizobial symbioses. J. Appl. Microbiol. 129, 1133–1156. doi: 10.1111/jam.14754
Amarger, N. (1981). Competition for nodule formation between effective and ineffective strains of Rhizobium meliloti. Soil Biol. Biochem. 13, 475–480. doi: 10.1016/0038-0717(81)90037-7
Anand, A., Jaiswal, S. K., Dhar, B., Vaishampayan, A. (2012). Surviving and thriving in terms of symbiotic performance of antibiotic and phage-resistant mutants of Bradyrhizobium of soybean [Glycine max (L.) Merrill]. Curr. Microbiol. 65, 390–397. doi: 10.1007/s00284-012-0166-8
Andrade-Domínguez, A., Salazar, E., Del Carmen Vargas-Lagunas, M., Kolter, R., Encarnación, S. (2014). Eco-evolutionary feedbacks drive species interactions. ISME J. 8, 1041–1054. doi: 10.1038/ismej.2013.208
Andrade-Domínguez, A., Trejo-Hernández, A., Vargas-Lagunas, C., Encarnación-Guevara, S. (2021). Phenotypic plasticity and a new small molecule are involved in a fungal-bacterial interaction. Sci. Rep. 11, 19219. doi: 10.1038/s41598-021-98474-y
Andrews, M., De Meyer, S., James, E., Stępkowski, T., Hodge, S., Simon, M., et al. (2018). Horizontal transfer of symbiosis genes within and between rhizobial genera: Occurrence and importance. Genes 9, 321. doi: 10.3390/genes9070321
Antunes, P. M., Goss, M. J. (2005). “Communication in the tripartite symbiosis formed by arbuscular mycorrhizal fungi, rhizobia and legume plants: A review,” in Roots and Soil Management: Interactions between Roots and the Soil. Eds. Zobel, R. W., Wright, S. F. (Madison, WI, USA: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America), 199–222. doi: 10.2134/agronmonogr48.c11
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Ballhorn, D. J., Schädler, M., Elias, J. D., Millar, J. A., Kautz, S. (2016). Friend or foe — Light availability determines the relationship between mycorrhizal fungi, rhizobia and Lima bean (Phaseolus lunatus L.). PloS One 11, e0154116. doi: 10.1371/journal.pone.0154116
Bañuelos-Vazquez, L. A., Cazares, D., Rodríguez, S., Cervantes-De la Luz, L., Sánchez-López, R., Castellani, L. G., et al. (2020). Transfer of the symbiotic plasmid of Rhizobium etli CFN42 to endophytic bacteria inside nodules. Front. Microbiol. 11, 1752. doi: 10.3389/fmicb.2020.01752
Bañuelos-Vazquez, L. A., Torres Tejerizo, G., Cervantes-De La Luz, L., Girard, L., Romero, D., Brom, S. (2019). Conjugative transfer between Rhizobium etli endosymbionts inside the root nodule. Environ. Microbiol. 21, 3430–3441. doi: 10.1111/1462-2920.14645
Barea, J.-M., Pozo, M. J., Azcón, R., Azcón-Aguilar, C. (2005). Microbial co-operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. doi: 10.1093/jxb/eri197
Batstone, R. T., Burghardt, L. T., Heath, K. D. (2022). Phenotypic and genomic signatures of interspecies cooperation and conflict in naturally occurring isolates of a model plant symbiont. Proc. R. Soc B Biol. Sci. 289, 20220477. doi: 10.1098/rspb.2022.0477
Batstone, R. T., Dutton, E. M., Wang, D., Yang, M., Frederickson, M. E. (2017). The evolution of symbiont preference traits in the model legume Medicago truncatula. New Phytol. 213, 1850–1861. doi: 10.1111/nph.14308
Batstone, R. T., O’Brien, A. M., Harrison, T. L., Frederickson, M. E. (2020). Experimental evolution makes microbes more cooperative with their local host genotype. Science 370, 476–478. doi: 10.1126/science.abb7222
Bellabarba, A., Fagorzi, C., diCenzo, G. C., Pini, F., Viti, C., Checcucci, A. (2019). Deciphering the symbiotic plant microbiome: Translating the most recent discoveries on rhizobia for the improvement of agricultural practices in metal-contaminated and high saline lands. Agronomy 9, 529. doi: 10.3390/agronomy9090529
Benezech, C., Le Scornet, A., Gourion, B. (2021). Medicago - Sinorhizobium - Ralstonia: A model system to investigate pathogen-triggered inhibition of nodulation. Mol. Plant-Microbe Interact. 34, 499–503. doi: 10.1094/MPMI-11-20-0319-SC
Berendsen, R. L., Pieterse, C. M. J., Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bergersen, F. J., Gibson, A. H., Licis, I. (1995). Growth and N2-fixation of soybeans inoculated with strains of Bradyrhizobium japonicum differing in energetic efficiency and PHB utilization. Soil Biol. Biochem. 27, 611–616. doi: 10.1016/0038-0717(95)98639-6
Berrabah, F., Ratet, P., Gourion, B. (2015). Multiple steps control immunity during the intracellular accommodation of rhizobia. J. Exp. Bot. 66, 1977–1985. doi: 10.1093/jxb/eru545
Bladergroen, M. R., Badelt, K., Spaink, H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 16, 53–64. doi: 10.1094/MPMI.2003.16.1.53
Boivin, S., Ait Lahmidi, N., Sherlock, D., Bonhomme, M., Dijon, D., Heulin-Gotty, K., et al. (2020). Host-specific competitiveness to form nodules in Rhizobium leguminosarum symbiovar viciae. New Phytol. 226, 555–568. doi: 10.1111/nph.16392
Boivin, S., Lepetit, M. (2020). “Partner preference in the legume-rhizobia symbiosis and impact on legume inoculation strategies,” in Advances in Botanical Research. Eds. Frendo, P., Frugier, F., Masson-Boivin, C. (Cambridge, MA, USA: Academic Press), 323–348. doi: 10.1016/bs.abr.2019.09.016
Boivin, S., Mahé, F., Debellé, F., Pervent, M., Tancelin, M., Tauzin, M., et al. (2021). Genetic variation in host-specific competitiveness of the symbiont Rhizobium leguminosarum symbiovar viciae. Front. Plant Sci. 12, 719987. doi: 10.3389/fpls.2021.719987
Bourion, V., Heulin-Gotty, K., Aubert, V., Tisseyre, P., Chabert-Martinello, M., Pervent, M., et al. (2018). Co-inoculation of a pea core-collection with diverse rhizobial strains shows competitiveness for nodulation and efficiency of nitrogen fixation are distinct traits in the interaction. Front. Plant Sci. 8, 2249. doi: 10.3389/fpls.2017.02249
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2014). The root hair “Infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/tpc.114.133496
Brito, I. L. (2021). Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 19, 442–453. doi: 10.1038/s41579-021-00534-7
Burghardt, L. T. (2020). Evolving together, evolving apart: Measuring the fitness of rhizobial bacteria in and out of symbiosis with leguminous plants. New Phytol. 228, 28–34. doi: 10.1111/nph.16045
Burghardt, L. T., diCenzo, G. C. (2023). The evolutionary ecology of rhizobia: Multiple facets of competition before, during, and after symbiosis with legumes. Curr. Opin. Microbiol. 72, 102281. doi: 10.1016/j.mib.2023.102281
Burghardt, L. T., Epstein, B., Guhlin, J., Nelson, M. S., Taylor, M. R., Young, N. D., et al. (2018). Select and resequence reveals relative fitness of bacteria in symbiotic and free-living environments. Proc. Natl. Acad. Sci. U.S.A. 115, 2425–2430. doi: 10.1073/pnas.1714246115
Burghardt, L. T., Epstein, B., Hoge, M., Trujillo, D. I., Tiffin, P. (2022). Host-associated rhizobial fitness: Dependence on nitrogen, density, community complexity, and legume genotype. Appl. Environ. Microbiol. 88, e00526–e00522. doi: 10.1128/aem.00526-22
Burghardt, L. T., Guhlin, J., Chun, C. L., Liu, J., Sadowsky, M. J., Stupar, R. M., et al. (2017). Transcriptomic basis of genome by genome variation in a legume-rhizobia mutualism. Mol. Ecol. 26, 6122–6135. doi: 10.1111/mec.14285
Burghardt, L. T., Trujillo, D. I., Epstein, B., Tiffin, P., Young, N. D. (2020). A select and resequence approach reveals strain-specific effects of Medicago nodule-specific PLAT-domain genes. Plant Physiol. 182, 463–471. doi: 10.1104/pp.19.00831
Caetano-Anollés, G., Wall, L. G., De Micheli, A. T., Macchi, E. M., Bauer, W. D., Favelukes, G. (1988). Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol. 86, 1228–1235. doi: 10.1104/pp.86.4.1228
Carrión, V. J., Cordovez, V., Tyc, O., Etalo, D. W., De Bruijn, I., De Jager, V. C. L., et al. (2018). Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 12, 2307–2321. doi: 10.1038/s41396-018-0186-x
Casimiro, I., Marchant, A., Bhalerao, R. P., Beeckman, T., Dhooge, S., Swarup, R., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. doi: 10.1105/tpc.13.4.843
Cevallos, M. A., Encarnación, S., Leija, A., Mora, Y., Mora, J. (1996). Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-beta-hydroxybutyrate. J. Bacteriol. 178, 1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996
Checcucci, A., Azzarello, E., Bazzicalupo, M., Galardini, M., Lagomarsino, A., Mancuso, S., et al. (2016). Mixed nodule infection in Sinorhizobium meliloti–Medicago sativa symbiosis suggest the presence of cheating behavior. Front. Plant Sci. 7, 835. doi: 10.3389/fpls.2016.00835
Checcucci, A., Marchetti, M. (2020). The rhizosphere talk show: The rhizobia on stage. Front. Agron. 2, 591494. doi: 10.3389/fagro.2020.591494
Chen, W. F., Meng, X. F., Jiao, Y. S., Tian, C. F., Sui, X. H., Jiao, J., et al. (2023). Bacteroid development, transcriptome, and symbiotic nitrogen-fixing comparison of Bradyrhizobium arachidis in nodules of peanut (Arachis hypogaea) and medicinal legume Sophora flavescens. Microbiol. Spectr. 11, e01079–e01022. doi: 10.1128/spectrum.01079-22
Chepsergon, J., Moleleki, L. N. (2023). Rhizosphere bacterial interactions and impact on plant health. Curr. Opin. Microbiol. 73, 102297. doi: 10.1016/j.mib.2023.102297
Chevallereau, A., Pons, B. J., Van Houte, S., Westra, E. R. (2022). Interactions between bacterial and phage communities in natural environments. Nat. Rev. Microbiol. 20, 49–62. doi: 10.1038/s41579-021-00602-y
Colombi, E., Hill, Y., Lines, R., Sullivan, J. T., Kohlmeier, M. G., Christophersen, C. T., et al. (2023). Population genomics of Australian indigenous Mesorhizobium reveals diverse nonsymbiotic genospecies capable of nitrogen-fixing symbioses following horizontal gene transfer. Microb. Genomics 9. doi: 10.1099/mgen.0.000918
Compton, K. K., Scharf, B. E. (2021). Rhizobial chemoattractants, the taste and preferences of legume symbionts. Front. Plant Sci. 12, 686465. doi: 10.3389/fpls.2021.686465
Contreras-Moreno, F. J., Muñoz-Dorado, J., García-Tomsig, N. I., Martínez-Navajas, G., Pérez, J., Moraleda-Muñoz, A. (2020). Copper and melanin play a role in Myxococcus xanthus predation on Sinorhizobium meliloti. Front. Microbiol. 11, 94. doi: 10.3389/fmicb.2020.00094
Coyte, K. Z., Rakoff-Nahoum, S. (2019). Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29, R538–R544. doi: 10.1016/j.cub.2019.04.017
Crang, N., Borah, K., James, E. K., Jorrín, B., Green, P., Tkacz, A., et al. (2021). Role and regulation of poly-3-hydroxybutyrate in nitrogen fixation in Azorhizobium caulinodans. Mol. Plant-Microbe Interact. 34, 1390–1398. doi: 10.1094/MPMI-06-21-0138-R
Crook, M. B., Lindsay, D. P., Biggs, M. B., Bentley, J. S., Price, J. C., Clement, S. C., et al. (2012). Rhizobial plasmids that cause impaired symbiotic nitrogen fixation and enhanced host invasion. Mol. Plant-Microbe Interact. 25, 1026–1033. doi: 10.1094/MPMI-02-12-0052-R
Crosbie, D. B., Mahmoudi, M., Radl, V., Brachmann, A., Schloter, M., Kemen, E., et al. (2022). Microbiome profiling reveals that Pseudomonas antagonises parasitic nodule colonisation of cheater rhizobia in Lotus. New Phytol. 234, 242–255. doi: 10.1111/nph.17988
Czernic, P., Gully, D., Cartieaux, F., Moulin, L., Guefrachi, I., Patrel, D., et al. (2015). Convergent evolution of endosymbiont differentiation in Dalbergioid and inverted repeat-lacking clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 169, 1254–1265. doi: 10.1104/pp.15.00584
Danso, S. K. A., Keya, S. O., Alexander, M. (1975). Protozoa and the decline of Rhizobium populations added to soil. Can. J. Microbiol. 21, 884–895. doi: 10.1139/m75-131
da Silva, T. R., Rodrigues, R. T., Jovino, R. S., Carvalho, J. R. D. S., Leite, J., Hoffman, A., et al. (2023). Not just passengers, but co-pilots! Non-rhizobial nodule-associated bacteria promote cowpea growth and symbiosis with (brady)rhizobia. J. Appl. Microbiol. 134, lxac013. doi: 10.1093/jambio/lxac013
Daubech, B., Remigi, P., Doin De Moura, G., Marchetti, M., Pouzet, C., Auriac, M.-C., et al. (2017). Spatio-temporal control of mutualism in legumes helps spread symbiotic nitrogen fixation. eLife 6, e28683. doi: 10.7554/eLife.28683
De Campos, S. B., Lardi, M., Gandolfi, A., Eberl, L., Pessi, G. (2017). Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Front. Microbiol. 8, 2473. doi: 10.3389/fmicb.2017.02473
De Faria, S. M., Ringelberg, J. J., Gross, E., Koenen, E. J. M., Cardoso, D., Ametsitsi, G. K. D., et al. (2022). The innovation of the symbiosome has enhanced the evolutionary stability of nitrogen fixation in legumes. New Phytol. 235, 2365–2377. doi: 10.1111/nph.18321
De La Fuente Cantó, C., Simonin, M., King, E., Moulin, L., Bennett, M. J., Castrillo, G., et al. (2020). An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant J. 103, 951–964. doi: 10.1111/tpj.14781
Denison, R. F., Kiers, E. T. (2011). Life histories of symbiotic rhizobia and mycorrhizal fungi. Curr. Biol. 21, R775–R785. doi: 10.1016/j.cub.2011.06.018
Denison, R. F., Muller, K. E. (2022). An evolutionary perspective on increasing net benefits to crops from symbiotic microbes. Evol. Appl. 15, 1490–1504. doi: 10.1111/eva.13384
Doin De Moura, G. G., Mouffok, S., Gaudu, N., Cazalé, A.-C., Milhes, M., Bulach, T., et al. (2023). A selective bottleneck during host entry drives the evolution of new legume symbionts. Mol. Biol. Evol. 40, msad116. doi: 10.1093/molbev/msad116
Doin de Moura, G. G., Remigi, P., Masson-Boivin, C., Capela, D. (2020). Experimental evolution of legume symbionts: What have we learnt? Genes 11, 339. doi: 10.3390/genes11030339
D’Souza, G., Shitut, S., Preussger, D., Yousif, G., WasChina, S., Kost, C. (2018). Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488. doi: 10.1039/C8NP00009C
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e14. doi: 10.1016/j.cell.2018.10.020
Elliott, G. N., Chou, J.-H., Chen, W.-M., Bloemberg, G. V., Bontemps, C., Martínez-Romero, E., et al. (2009). Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 11, 762–778. doi: 10.1111/j.1462-2920.2008.01799.x
Epstein, B., Burghardt, L. T., Heath, K. D., Grillo, M. A., Kostanecki, A., Hämälä, T., et al. (2023). Combining GWAS and population genomic analyses to characterize coevolution in a legume-rhizobia symbiosis. Mol. Ecol. 32, 3798–3811. doi: 10.1111/mec.16602
Etesami, H. (2022). Root nodules of legumes: A suitable ecological niche for isolating non-rhizobial bacteria with biotechnological potential in agriculture. Curr. Res. Biotechnol. 4, 78–86. doi: 10.1016/j.crbiot.2022.01.003
Fagorzi, C., Bacci, G., Huang, R., Cangioli, L., Checcucci, A., Fini, M., et al. (2021). Nonadditive transcriptomic signatures of genotype-by-genotype interactions during the initiation of plant-rhizobium symbiosis. mSystems 6, e00974–e00920. doi: 10.1128/mSystems.00974-20
Ferenci, T. (2019). Irregularities in genetic variation and mutation rates with environmental stresses. Environ. Microbiol. 21, 3979–3988. doi: 10.1111/1462-2920.14822
Ferguson, B. J., Mens, C., Hastwell, A. H., Zhang, M., Su, H., Jones, C. H., et al. (2019). Legume nodulation: The host controls the party. Plant Cell Environ. 42, 41–51. doi: 10.1111/pce.13348
Fields, B., Friman, V.-P. (2022). Microbial eco-evolutionary dynamics in the plant rhizosphere. Curr. Opin. Microbiol. 68, 102153. doi: 10.1016/j.mib.2022.102153
Fields, B., Moeskjær, S., Deakin, W. J., Moffat, E. K., Roulund, N., Andersen, S. U., et al. (2023). Rhizobium nodule diversity and composition are influenced by clover host selection and local growth conditions. Mol. Ecol. 32, 4259–4277. doi: 10.1111/mec.17028
Fields, B., Moffat, E. K., Harrison, E., Andersen, S. U., Young, J. P. W., Friman, V.-P. (2022). Genetic variation is associated with differences in facilitative and competitive interactions in the Rhizobium leguminosarum species complex. Environ. Microbiol. 24, 3463–3485. doi: 10.1111/1462-2920.15720
Frederickson, M. E. (2020). No selection for cheating in a natural meta-population of rhizobia. Ecol. Lett. 23, 409–411. doi: 10.1111/ele.13293
Friesen, M. L. (2012). Widespread fitness alignment in the legume-rhizobium symbiosis. New Phytol. 194, 1096–1111. doi: 10.1111/j.1469-8137.2012.04099.x
Fry, J., Wood, M., Poole, P. S. (2001). Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant-Microbe Interact. 14, 1016–1025. doi: 10.1094/MPMI.2001.14.8.1016
Fujita, H., Aoki, S., Kawaguchi, M. (2014). Evolutionary dynamics of nitrogen fixation in the legume–rhizobia symbiosis. PloS One 9, e93670. doi: 10.1371/journal.pone.0093670
Gano-Cohen, K. A., Stokes, P. J., Blanton, M. A., Wendlandt, C. E., Hollowell, A. C., Regus, J. U., et al. (2016). Nonnodulating Bradyrhizobium spp. modulate the benefits of legume-rhizobium mutualism. Appl. Environ. Microbiol. 82, 5259–5268. doi: 10.1128/AEM.01116-16
Gano‐Cohen, K. A., Wendlandt, C. E., Stokes, P. J., Blanton, M. A., Quides, K. W., Zomorrodian, A., et al. (2019). Interspecific conflict and the evolution of ineffective rhizobia. Ecol. Lett. 22, 914–924. doi: 10.1111/ele.13247
Gardener, B. B. M., De Bruijn, F. J. (1998). Detection and isolation of novel rhizopine-catabolizing bacteria from the environment. Appl. Environ. Microbiol. 64, 4944–4949. doi: 10.1128/AEM.64.12.4944-4949.1998
Garnerone, A.-M., Sorroche, F., Zou, L., Mathieu-Demazière, C., Tian, C. F., Masson-Boivin, C., et al. (2018). NsrA, a predicted β-Barrel outer membrane protein involved in plant signal perception and the control of secondary infection in Sinorhizobium meliloti. J. Bacteriol. 200, e00019-18. doi: 10.1128/JB.00019-18
Ghoul, M., Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Gordon, D. M., Ryder, M. H., Heinrich, K., Murphy, P. J. (1996). An experimental test of the rhizopine concept in Rhizobium meliloti. Appl. Environ. Microbiol. 62, 3991–3996. doi: 10.1128/aem.62.11.3991-3996.1996
Granato, E. T., Meiller-Legrand, T. A., Foster, K. R. (2019). The evolution and ecology of bacterial warfare. Curr. Biol. 29, R521–R537. doi: 10.1016/j.cub.2019.04.024
Gresshoff, P. M., Rolfe, B. G. (1978). Viability of Rhizobium bacteroids isolated from soybean nodule protoplasts. Planta 142, 329–333. doi: 10.1007/BF00385085
Hahn, M., Studer, D. (1986). Competitiveness of a nif – Bradyrhizobium japonicum mutant against the wild-type strain. FEMS Microbiol. Lett. 33, 143–148. doi: 10.1111/j.1574-6968.1986.tb01228.x
Han, Q., Ma, Q., Chen, Y., Tian, B., Xu, L., Bai, Y., et al. (2020). Variation in rhizosphere microbial communities and its association with the symbiotic efficiency of rhizobia in soybean. ISME J. 14, 1915–1928. doi: 10.1038/s41396-020-0648-9
Handelsman, J., Ugalde, R. A., Brill, W. J. (1984). Rhizobium meliloti competitiveness and the alfalfa agglutinin. J. Bacteriol. 157, 703–707. doi: 10.1128/jb.157.3.703-707.1984
Hansen, B. L., Pessotti, R., de, C., Fischer, M. S., Collins, A., El-Hifnawi, L., et al. (2020). Cooperation, competition, and specialized metabolism in a simplified root nodule microbiome. mBio 11, e01917–e01920. doi: 10.1128/mBio.01917-20
Hassani, M. A., Durán, P., Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6, 58. doi: 10.1186/s40168-018-0445-0
Heath, K. D., Tiffin, P. (2007). Context dependence in the coevolution of plant and rhizobial mutualists. Proc. R. Soc B Biol. Sci. 274, 1905–1912. doi: 10.1098/rspb.2007.0495
Hendry, A. P. (2017). Eco-evolutionary dynamics (Princeton: Princeton University Press). doi: 10.1515/9781400883080
Hibbing, M. E., Fuqua, C., Parsek, M. R., Peterson, S. B. (2010). Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hill, Y., Colombi, E., Bonello, E., Haskett, T., Ramsay, J., O’Hara, G., et al. (2021). Evolution of diverse effective N2-fixing microsymbionts of Cicer arietinum following horizontal transfer of the Mesorhizobium ciceri CC1192 symbiosis integrative and conjugative element. Appl. Environ. Microbiol. 87, e02558–e02520. doi: 10.1128/AEM.02558-20
Hirsch, P. R. (1979). Plasmid-determined bacteriocin production by Rhizobium leguminosarum. J. Gen. Microbiol. 113, 219–228. doi: 10.1099/00221287-113-2-219
Hogg, B., Davies, A. E., Wilson, K. E., Bisseling, T., Downie, J. A. (2002). Competitive nodulation blocking of cv. Afghanistan pea is related to high levels of nodulation factors made by some strains of Rhizobium leguminosarum bv. viciae. Mol. Plant-Microbe Interact. 15, 60–68. doi: 10.1094/MPMI.2002.15.1.60
Jacoby, R. P., Kopriva, S. (2019). Metabolic niches in the rhizosphere microbiome: New tools and approaches to analyse metabolic mechanisms of plant–microbe nutrient exchange. J. Exp. Bot. 70, 1087–1094. doi: 10.1093/jxb/ery438
Janczarek, M., Jaroszuk-Ściseł, J., Skorupska, A. (2009). Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie Van Leeuwenhoek 96, 471–486. doi: 10.1007/s10482-009-9362-3
Ji, Y.-Y., Zhang, B., Zhang, P., Chen, L.-C., Si, Y.-W., Wan, X.-Y., et al. (2023). Rhizobial migration toward roots mediated by FadL-ExoFQP modulation of extracellular long-chain AHLs. ISME J. 17, 417–431. doi: 10.1038/s41396-023-01357-5
Jimenez-Zurdo, J. I. (1995). Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol. Plant Microbe Interact. 8, 492. doi: 10.1094/MPMI-8-0492
Joglekar, P., Ferrell, B. D., Jarvis, T., Haramoto, K., Place, N., Dums, J. T., et al. (2023). Spontaneously produced lysogenic phages are an important component of the soybean Bradyrhizobium mobilome. mBio 14, e0029523. doi: 10.1128/mbio.00295-23
Kapp, D., Niehaus, K., Quandt, J., Muller, P., Puhler, A. (1990). Cooperative action of Rhizobium meliloti nodulation and infection mutants during the process of forming mixed infected alfalfa nodules. Plant Cell 2, 139–151. doi: 10.1105/tpc.2.2.139
Kelly, S. J., Muszyński, A., Kawaharada, Y., Hubber, A. M., Sullivan, J. T., Sandal, N., et al. (2013). Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol. Plant-Microbe Interact. 26, 319–329. doi: 10.1094/MPMI-09-12-0227-R
Kern, L., Abdeen, S. K., Kolodziejczyk, A. A., Elinav, E. (2021). Commensal inter-bacterial interactions shaping the microbiota. Curr. Opin. Microbiol. 63, 158–171. doi: 10.1016/j.mib.2021.07.011
Keya, S. O., Alexander, M. (1975). Regulation of parasitism by host density: The Bdellovibrio-Rhizobium interrelationship. Soil Biol. Biochem. 7, 231–237. doi: 10.1016/0038-0717(75)90044-9
Kiers, E. T., Ratcliff, W. C., Denison, R. F. (2013). Single-strain inoculation may create spurious correlations between legume fitness and rhizobial fitness. New Phytol. 198, 4–6. doi: 10.1111/nph.12015
Kiers, E. T., Rousseau, R. A., Denison, R. F. (2006). Measured sanctions: Legume hosts detect quantitative variation in rhizobium cooperation and punish accordingly. Evol. Ecol. Res. 8, 1077–1086.
Kiers, E. T., Rousseau, R. A., West, S. A., Denison, R. F. (2003). Host sanctions and the legume-rhizobium mutualism. Nature 425, 78–81. doi: 10.1038/nature01931
Kimbrel, J. A., Thomas, W. J., Jiang, Y., Creason, A. L., Thireault, C. A., Sachs, J. L., et al. (2013). Mutualistic co-evolution of type III effector genes in Sinorhizobium fredii and Bradyrhizobium japonicum. PloS Pathog. 9, e1003204. doi: 10.1371/journal.ppat.1003204
Kleczkowska, J. (1950). A study of phage-resistant mutants of Rhizobium trifolii. J. Gen. Microbiol. 4, 298–310. doi: 10.1099/00221287-4-3-298
Klein, S., Hirsch, A. M., Smith, C. A., Signer, E. R. (1988). Interaction of nod and exo Rhizobium meliloti in alfalfa nodulation. Mol. Plant-Microbe Interact. 1, 94–100. doi: 10.1094/mpmi-1-094
Konopka, A. (2009). What is microbial community ecology? ISME J. 3, 1223–1230. doi: 10.1038/ismej.2009.88
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Krašovec, R., Belavkin, R. V., Aston, J. A. D., Channon, A., Aston, E., Rash, B. M., et al. (2014). Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell–cell interactions. Nat. Commun. 5, 3742. doi: 10.1038/ncomms4742
Krašovec, R., Richards, H., Gifford, D. R., Hatcher, C., Faulkner, K. J., Belavkin, R. V., et al. (2017). Spontaneous mutation rate is a plastic trait associated with population density across domains of life. PloS Biol. 15, e2002731. doi: 10.1371/journal.pbio.2002731
Le, X. H., Franco, C. M. M., Ballard, R. A., Drew, E. A. (2016). Isolation and characterisation of endophytic actinobacteria and their effect on the early growth and nodulation of lucerne (Medicago sativa L.). Plant Soil 405, 13–24. doi: 10.1007/s11104-015-2652-9
Lepetit, M., Brouquisse, R. (2023). Control of the rhizobium–legume symbiosis by the plant nitrogen demand is tightly integrated at the whole plant level and requires inter-organ systemic signaling. Front. Plant Sci. 14, 1114840. doi: 10.3389/fpls.2023.1114840
LeRoux, M., Peterson, S. B., Mougous, J. D. (2015). Bacterial danger sensing. J. Mol. Biol. 427, 3744–3753. doi: 10.1016/j.jmb.2015.09.018
Levine, J. M., Bascompte, J., Adler, P. B., Allesina, S. (2017). Beyond pairwise mechanisms of species coexistence in complex communities. Nature 546, 56–64. doi: 10.1038/nature22898
Li, M., Chen, Q., Wu, C., Li, Y., Wang, S., Chen, X. (2022). A novel module promotes horizontal gene transfer in Azorhizobium caulinodans ORS571. Genes 13, 1895. doi: 10.3390/genes13101895
Ling, J., Wang, H., Wu, P., Li, T., Tang, Y., Naseer, N., et al. (2016). Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc. Natl. Acad. Sci. U.S.A. 113, 13875–13880. doi: 10.1073/pnas.1615121113
Liu, Q., Cheng, L., Nian, H., Jin, J., Lian, T. (2023). Linking plant functional genes to rhizosphere microbes: A review. Plant Biotechnol. J. 21, 902–917. doi: 10.1111/pbi.13950
Liu, S., Jiao, J., Tian, C.-F. (2023). Adaptive evolution of rhizobial symbiosis beyond horizontal gene transfer: From genome innovation to regulation reconstruction. Genes 14, 274. doi: 10.3390/genes14020274
Lodwig, E. M., Leonard, M., Marroqui, S., Wheeler, T. R., Findlay, K., Downie, J. A., et al. (2005). Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids. Mol. Plant-Microbe Interact. 18, 67–74. doi: 10.1094/MPMI-18-0067
Lodwig, E., Poole, P. (2003). Metabolism of rhizobium bacteroids. Crit. Rev. Plant Sci. 22, 37–78. doi: 10.1080/713610850
Maharjan, R. P., Ferenci, T. (2017). A shifting mutational landscape in 6 nutritional states: Stress-induced mutagenesis as a series of distinct stress input–mutation output relationships. PloS Biol. 15, e2001477. doi: 10.1371/journal.pbio.2001477
Marchetti, M., Catrice, O., Batut, J., Masson-Boivin, C. (2011). Cupriavidus Taiwanensis bacteroids in Mimosa pudica indeterminate nodules are not terminally differentiated. Appl. Environ. Microbiol. 77, 2161–2164. doi: 10.1128/aem.02358-10
Martínez-Hidalgo, P., Hirsch, A. M. (2017). The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes J. 1, 70–82. doi: 10.1094/PBIOMES-12-16-0019-RVW
Masson-Boivin, C., Giraud, E., Perret, X., Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Masson-Boivin, C., Sachs, J. L. (2018). Symbiotic nitrogen fixation by rhizobia - the roots of a success story. Curr. Opin. Plant Biol. 44, 7–15. doi: 10.1016/j.pbi.2017.12.001
Matic, I. (2019). Mutation rate heterogeneity increases odds of survival in unpredictable environments. Mol. Cell 75, 421–425. doi: 10.1016/j.molcel.2019.06.029
Melkonian, R., Moulin, L., Béna, G., Tisseyre, P., Chaintreuil, C., Heulin, K., et al. (2014). The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype: Competition for nodulation in α- and β-rhizobia. Environ. Microbiol. 16, 2099–2111. doi: 10.1111/1462-2920.12286
Mendoza-Suárez, M., Andersen, S. U., Poole, P. S., Sánchez-Cañizares, C. (2021). Competition, nodule occupancy, and persistence of inoculant strains: Key factors in the rhizobium-legume symbioses. Front. Plant Sci. 12, 690567. doi: 10.3389/fpls.2021.690567
Mendoza-Suárez, M. A., Geddes, B. A., Sánchez-Cañizares, C., Ramírez-González, R. H., Kirchhelle, C., Jorrin, B., et al. (2020). Optimizing Rhizobium-legume symbioses by simultaneous measurement of rhizobial competitiveness and N2 fixation in nodules. Proc. Natl. Acad. Sci. U.S.A. 117, 9822–9831. doi: 10.1073/pnas.1921225117
Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium –legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235. doi: 10.1073/pnas.0600912103
Miao, J., Zhang, N., Liu, H., Wang, H., Zhong, Z., Zhu, J. (2018). Soil commensal rhizobia promote Rhizobium etli nodulation efficiency through CinR-mediated quorum sensing. Arch. Microbiol. 200, 685–694. doi: 10.1007/s00203-018-1478-2
Mishra, A., Dhar, B., Singh, R. M. (2004). Nodulation competitiveness between contrasting phage phenotypes of pigeonpea rhizobial strains. Indian J. Exp. Biol. 42, 611–615.
Montiel, J., Szűcs, A., Boboescu, I. Z., Gherman, V. D., Kondorosi, É., Kereszt, A. (2016). Terminal bacteroid differentiation is associated with variable morphological changes in legume species belonging to the inverted repeat-lacking clade. Mol. Plant-Microbe Interact. 29, 210–219. doi: 10.1094/MPMI-09-15-0213-R
Montoya, A. P., Wendlandt, C. E., Benedict, A. B., Roberts, M., Piovia-Scott, J., Griffitts, J. S., et al. (2023). Hosts winnow symbionts with multiple layers of absolute and conditional discrimination mechanisms. Proc. R. Soc B Biol. Sci. 290, 20222153. doi: 10.1098/rspb.2022.2153
Moura De Sousa, J., Lourenço, M., Gordo, I. (2023). Horizontal gene transfer among host-associated microbes. Cell Host Microbe 31, 513–527. doi: 10.1016/j.chom.2023.03.017
Muller, K. E., Denison, R. F. (2018). Resource acquisition and allocation traits in symbiotic rhizobia with implications for life-history outside of legume hosts. R. Soc Open Sci. 5, 181124. doi: 10.1098/rsos.181124
Müller-Santos, M., Koskimäki, J. J., Alves, L. P. S., de Souza, E. M., Jendrossek, D., Pirttilä, A. M. (2021). The protective role of PHB and its degradation products against stress situations in bacteria. FEMS Microbiol. Rev. 45, fuaa058. doi: 10.1093/femsre/fuaa058
Murphy, P. J., Wexler, W., Grzemski, W., Rao, J. P., Gordon, D. (1995). Rhizopines—Their role in symbiosis and competition. Soil Biol. Biochem. 27, 525–529. doi: 10.1016/0038-0717(95)98627-Z
Nandasena, K. G., O’Hara, G. W., Tiwari, R. P., Sezmiş, E., Howieson, J. G. (2007). In situ lateral transfer of symbiosis islands results in rapid evolution of diverse competitive strains of mesorhizobia suboptimal in symbiotic nitrogen fixation on the pasture legume Biserrula pelecinus L. Environ. Microbiol. 9, 2496–2511. doi: 10.1111/j.1462-2920.2007.01368.x
Ofek, M., Ruppel, S., Waisel, Y. (2006). “Effects of salinity on rhizosphere bacterial communities associated with different root types of Vicia faba L,” in Biosaline Agriculture and Salinity Tolerance in Plants. Eds. Öztürk, M., Waisel, Y., Khan, M. A., Görk, G. (Basel: Birkhäuser Basel), 1–13. doi: 10.1007/3-7643-7610-4_1
Oldroyd, G. E. D. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oono, R., Anderson, C. G., Denison, R. F. (2011). Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc. R. Soc B Biol. Sci. 278, 2698–2703. doi: 10.1098/rspb.2010.2193
Oono, R., Muller, K. E., Ho, R., Jimenez Salinas, A., Denison, R. F. (2020). How do less-expensive nitrogen alternatives affect legume sanctions on rhizobia? Ecol. Evol. 10, 10645–10656. doi: 10.1002/ece3.6718
Oresnik, I. J., Pacarynuk, L. A., O’Brien, S. A. P., Yost, C. K., Hynes, M. F. (1998). Plasmid-encoded catabolic genes in Rhizobium leguminosarum bv. trifolii: Evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant-Microbe Interact. 11, 1175–1185. doi: 10.1094/MPMI.1998.11.12.1175
Oresnik, I. J., Twelker, S., Hynes, M. F. (1999). Cloning and characterization of a Rhizobium leguminosarum gene encoding a bacteriocin with similarities to RTX toxins. Appl. Environ. Microbiol. 65, 2833–2840. doi: 10.1128/AEM.65.7.2833-2840.1999
Orr, H. A. (2009). Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 10, 531–539. doi: 10.1038/nrg2603
Pang, J., Ryan, M. H., Wen, Z., Lambers, H., Liu, Y., Zhang, Y., et al. (2023). Enhanced nodulation and phosphorus acquisition from sparingly-soluble iron phosphate upon treatment with arbuscular mycorrhizal fungi in chickpea. Physiol. Plant 175, e13873. doi: 10.1111/ppl.13873
Parks, D. H., ChuvoChina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P.-A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Patel, J. J. (1974). Antagonism of actinomycetes against rhizobia. Plant Soil 41, 395–402. doi: 10.1007/BF00017266
Pérez, J., Jiménez-Zurdo, J. I., Martínez-Abarca, F., Millán, V., Shimkets, L. J., Muñoz-Dorado, J. (2014). Rhizobial galactoglucan determines the predatory pattern of Myxococcus xanthus and protects Sinorhizobium meliloti from predation. Environ. Microbiol. 16, 2341–2350. doi: 10.1111/1462-2920.12477
Peterson, S. B., Bertolli, S. K., Mougous, J. D. (2020). The central role of interbacterial antagonism in bacterial life. Curr. Biol. 30, R1203–R1214. doi: 10.1016/j.cub.2020.06.103
Philippot, L., Raaijmakers, J. M., Lemanceau, P., van der Putten, W. H. (2013). Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pierce, E. C., Dutton, R. J. (2022). Putting microbial interactions back into community contexts. Curr. Opin. Microbiol. 65, 56–63. doi: 10.1016/j.mib.2021.10.008
Pobigaylo, N., Szymczak, S., Nattkemper, T. W., Becker, A. (2008). Identification of genes relevant to symbiosis and competitiveness in Sinorhizobium meliloti using signature-tagged mutants. Mol. Plant-Microbe Interact. 21, 219–231. doi: 10.1094/MPMI-21-2-0219
Poole, P., Ramachandran, V., Terpolilli, J. (2018). Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303. doi: 10.1038/nrmicro.2017.171
Porter, S. S., Simms, E. L. (2014). Selection for cheating across disparate environments in the legume-rhizobium mutualism. Ecol. Lett. 17, 1121–1129. doi: 10.1111/ele.12318
Pribis, J. P., Zhai, Y., Hastings, P. J., Rosenberg, S. M. (2022). Stress-induced mutagenesis, gambler cells, and stealth targeting antibiotic-induced evolution. mBio 13, e01074–e01022. doi: 10.1128/mbio.01074-22
Primieri, S., Magnoli, S. M., Koffel, T., Stürmer, S. L., Bever, J. D. (2022). Perennial, but not annual legumes synergistically benefit from infection with arbuscular mycorrhizal fungi and rhizobia: a meta-analysis. New Phytol. 233, 505–514. doi: 10.1111/nph.17787
Provorov, N. A., Andronov, E. E., Kimeklis, A. K., Onishchuk, O. P., Igolkina, A. A., Karasev, E. S. (2022). Microevolution, speciation and macroevolution in rhizobia: Genomic mechanisms and selective patterns. Front. Plant Sci. 13, 1036943. doi: 10.3389/fpls.2022.1026943
Pugashetti, B. K., Angle, J. S., Wagner, G. H. (1982). Soil microorganisms antagonistic towards Rhizobium japonicum. Soil Biol. Biochem. 14, 45–49. doi: 10.1016/0038-0717(82)90075-X
Quides, K. W., Lee, Y., Hur, T., Atamian, H. S. (2023). Evaluation of qPCR to detect shifts in population composition of the rhizobial symbiont Mesorhizobium japonicum during serial in planta transfers. Biology 12, 277. doi: 10.3390/biology12020277
Quides, K. W., Stomackin, G. M., Lee, H.-H., Chang, J. H., Sachs, J. L. (2017). Lotus japonicus alters in planta fitness of Mesorhizobium loti dependent on symbiotic nitrogen fixation. PloS One 12, e0185568. doi: 10.1371/journal.pone.0185568
Rahman, A., Manci, M., Nadon, C., Perez, I. A., Farsamin, W. F., Lampe, M. T., et al. (2023). Competitive interference among rhizobia reduces benefits to hosts. Curr. Biol. 33, 2988–3001.e4. doi: 10.1016/j.cub.2023.06.081
Ramirez, C., Alexander, M. (1980). Evidence suggesting protozoan predation on rhizobium associated with germinating seeds and in the rhizosphere of beans (Phaseolus vulgaris L.). Appl. Environ. Microbiol. 40, 492–499. doi: 10.1128/aem.40.3.492-499.1980
Ratcliff, W. C., Kadam, S. V., Denison, R. F. (2008). Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia: PHB increases rhizobium fitness. FEMS Microbiol. Ecol. 65, 391–399. doi: 10.1111/j.1574-6941.2008.00544.x
Ratcliff, W. C., Underbakke, K., Denison, R. F. (2011). Measuring the fitness of symbiotic rhizobia. Symbiosis 55, 85–90. doi: 10.1007/s13199-011-0150-2
Regus, J. U., Quides, K. W., O’Neill, M. R., Suzuki, R., Savory, E. A., Chang, J. H., et al. (2017). Cell autonomous sanctions in legumes target ineffective rhizobia in nodules with mixed infections. Am. J. Bot. 104, 1299–1312. doi: 10.3732/ajb.1700165
Remigi, P., Capela, D., Clerissi, C., Tasse, L., Torchet, R., Bouchez, O., et al. (2014). Transient hypermutagenesis accelerates the evolution of legume endosymbionts following horizontal gene transfer. PloS Biol. 12, e1001942. doi: 10.1371/journal.pbio.1001942
Remigi, P., Masson-Boivin, C., Rocha, E. P. C. (2019). Experimental evolution as a tool to investigate natural processes and molecular functions. Trends Microbiol. 27, 623–634. doi: 10.1016/j.tim.2019.02.003
Remigi, P., Zhu, J., Young, J. P. W., Masson-Boivin, C. (2016). Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol. 24, 63–75. doi: 10.1016/j.tim.2015.10.007
Ruiz-Lozano, J. M., Collados, C., Barea, J. M., Azcón, R. (2001). Arbuscular mycorrhizal symbiosis can alleviate drought-induced nodule senescence in soybean plants. New Phytol. 151, 493–502. doi: 10.1046/j.0028-646x.2001.00196.x
Sachs, J. L., Mueller, U. G., Wilcox, T. P., Bull, J. J. (2004). The evolution of cooperation. Q. Rev. Biol. 79, 135–160. doi: 10.1086/383541
Sánchez-Salazar, A. M., Taparia, T., Olesen, A. K., Acuña, J. J., Sørensen, S. J., Jorquera, M. A. (2023). An overview of plasmid transfer in the plant microbiome. Plasmid 127, 102695. doi: 10.1016/j.plasmid.2023.102695
Sauviac, L., Rémy, A., Huault, E., Dalmasso, M., Kazmierczak, T., Jardinaud, M., et al. (2022). A dual legume-rhizobium transcriptome of symbiotic nodule senescence reveals coordinated plant and bacterial responses. Plant Cell Environ. 45, 3100–3121. doi: 10.1111/pce.14389
Schoener, T. W. (2011). The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science 331, 426–429. doi: 10.1126/science.1193954
Schripsema, J., de Rudder, K. E., van Vliet, T. B., Lankhorst, P. P., de Vroom, E., Kijne, J. W., et al. (1996). Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-L-homoserine lactone molecules, known as autoinducers and as quorum sensing co-transcription factors. J. Bacteriol. 178, 366–371. doi: 10.1128/jb.178.2.366-371.1996
Schulte, C. C. M., Borah, K., Wheatley, R. M., Terpolilli, J. J., Saalbach, G., Crang, N., et al. (2021). Metabolic control of nitrogen fixation in rhizobium-legume symbioses. Sci. Adv. 7, eabh2433. doi: 10.1126/sciadv.abh2433
Schwinghamer, E. A., Brockwell, J. (1978). Competitive advantage of bacteriocin and phage-producing strains of Rhizobium trifolii in mixed culture. Soil Biol. Biochem. 10, 383–387. doi: 10.1016/0038-0717(78)90062-7
Shi, S., Richardson, A. E., O’Callaghan, M., DeAngelis, K. M., Jones, E. E., Stewart, A., et al. (2011). Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 77, 600–610. doi: 10.1111/j.1574-6941.2011.01150.x
Silva, V. M. A., Martins, C. M., Cavalcante, F. G., Ramos, K. A., Silva, L. L. D., Menezes, F. G. R. D., et al. (2019). Cross-feeding among soil bacterial populations: Selection and characterization of potential bio-inoculants. J. Agric. Sci. 11, 23. doi: 10.5539/jas.v11n5p23
Simms, E. L., Bever, J. D. (1998). Evolutionary dynamics of rhizopine within spatially structured rhizobium populations. Proc. R. Soc Lond. B Biol. Sci. 265, 1713–1719. doi: 10.1098/rspb.1998.0493
Smith, P., Schuster, M. (2019). Public goods and cheating in microbes. Curr. Biol. 29, R442–R447. doi: 10.1016/j.cub.2019.03.001
Soedarjo, M., Borthakur, D. (1998). Mimosine, a toxin produced by the tree-legume leucaena provides a nodulation competition advantage to mimosine-degrading rhizobium strains. Soil Biol. Biochem. 30, 1605–1613. doi: 10.1016/S0038-0717(97)00180-6
Sorroche, F., Morales, V., Mouffok, S., Pichereaux, C., Garnerone, A. M., Zou, L., et al. (2020). The ex planta signal activity of a Medicago ribosomal uL2 protein suggests a moonlighting role in controlling secondary rhizobial infection. PloS One 15, e0235446. doi: 10.1371/journal.pone.0235446
Sorroche, F., Walch, M., Zou, L., Rengel, D., Maillet, F., Gibelin-Viala, C., et al. (2019). Endosymbiotic Sinorhizobium meliloti modulate Medicago root susceptibility to secondary infection via ethylene. New Phytol. 223, 1505–1515. doi: 10.1111/nph.15883
Soto, M. J., Pérez, J., Muñoz-Dorado, J., Contreras-Moreno, F. J., Moraleda-Muñoz, A. (2023). Transcriptomic response of Sinorhizobium meliloti to the predatory attack of Myxococcus xanthus. Front. Microbiol. 14, 1213659. doi: 10.3389/fmicb.2023.1213659
Sprent, J. I. (2009). Legume nodulation: a global perspective. 1st ed (Oxford, UK: Wiley-Blackwell). doi: 10.1002/9781444316384
Stacey, G., Paau, A. S., Noel, K. D., Maier, R. J., Silver, L. E., Brill, W. J. (1982). Mutants of Rhizobium japonicum defective in nodulation. Arch. Microbiol. 132, 219–224. doi: 10.1007/BF00407954
Sullivan, J. T., Patrick, H. N., Lowther, W. L., Scott, D. B., Ronson, C. W. (1995). Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. U.S.A. 92, 8985–8989. doi: 10.1073/pnas.92.19.8985
Tang, M., Capela, D. (2020). Rhizobium diversity in the light of evolution, in: Advances in Botanical Research. Eds. Frendo, P., Frugier, F., Masson-Boivin, C.. (Cambridge, MA, USA: Academic Press), 251–288. doi: 10.1016/bs.abr.2019.09.006
Tavernier, P., Portais, J., Nava, S., Courtois, J., Courtois, B., Barbotin, J. (1997). Exopolysaccharide and poly-(beta)-hydroxybutyrate coproduction in two Rhizobium meliloti strains. Appl. Environ. Microbiol. 63, 21–26. doi: 10.1128/aem.63.1.21-26.1997
Terpolilli, J. J., Masakapalli, S. K., Karunakaran, R., Webb, I. U. C., Green, R., Watmough, N. J., et al. (2016). Lipogenesis and redox balance in nitrogen-fixing pea bacteroids. J. Bacteriol. 198, 2864–2875. doi: 10.1128/JB.00451-16
Thiergart, T., Zgadzaj, R., Bozsóki, Z., Garrido-Oter, R., Radutoiu, S., Schulze-Lefert, P. (2019). Lotus japonicus symbiosis genes impact microbial interactions between symbionts and multikingdom commensal communities. mBio 10, e01833–e01819. doi: 10.1128/mBio.01833-19
Tian, C. F., Garnerone, A.-M., Mathieu-Demazière, C., Masson-Boivin, C., Batut, J. (2012). Plant-activated bacterial receptor adenylate cyclases modulate epidermal infection in the Sinorhizobium meliloti–Medicago symbiosis. Proc. Natl. Acad. Sci. U.S.A. 109, 6751–6756. doi: 10.1073/pnas.1120260109
Tian, C. F., Young, J. P. W. (2019). Genomics and evolution of rhizobia, in: Ecology and Evolution of Rhizobia. Springer Singapore Singapore pp, 103–119. doi: 10.1007/978-981-32-9555-1_4
Timmers, A. C. J., Soupène, E., Auriac, M.-C., De Billy, F., Vasse, J., Boistard, P., et al. (2000). Saprophytic intracellular rhizobia in alfalfa nodules. Mol. Plant-Microbe Interact. 13, 1204–1213. doi: 10.1094/MPMI.2000.13.11.1204
Tokala, R. K., Strap, J. L., Jung, C. M., Crawford, D. L., Salove, M. H., Deobald, L. A., et al. (2002). Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl. Environ. Microbiol. 68, 2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002
Trainer, M. A., Charles, T. C. (2006). The role of PHB metabolism in the symbiosis of rhizobia with legumes. Appl. Microbiol. Biotechnol. 71, 377–386. doi: 10.1007/s00253-006-0354-1
Traxler, M. F., Kolter, R. (2015). Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970. doi: 10.1039/C5NP00013K
Triplett, E. W., Barta, T. M. (1987). Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by Rhizobium leguminosarum bv. trifolii strain T24 on clover. Plant Physiol. 85, 335–342. doi: 10.1104/pp.85.2.335
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Tsikou, D., Nikolaou, C. N., Tsiknia, M., Papadopoulou, K. K., Ehaliotis, C. (2023). Interplay between rhizobial nodulation and arbuscular mycorrhizal fungal colonization in Lotus japonicus roots. J. Appl. Microbiol. 134, lxac010. doi: 10.1093/jambio/lxac010
Van Cauwenberghe, J., Santamaría, R. I., Bustos, P., Juárez, S., Ducci, M. A., Figueroa Fleming, T., et al. (2021). Spatial patterns in phage-Rhizobium coevolutionary interactions across regions of common bean domestication. ISME J. 15, 2092–2106. doi: 10.1038/s41396-021-00907-z
Vanderlinde, E. M., Hynes, M. F., Yost, C. K. (2014). Homoserine catabolism by Rhizobium leguminosarum bv. viciae 3841 requires a plasmid-borne gene cluster that also affects competitiveness for nodulation. Environ. Microbiol. 16, 205–217. doi: 10.1111/1462-2920.12196
Vasi, F., Travisano, M., Lenski, R. E. (1994). Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. Am. Nat. 144, 432–456. doi: 10.1086/285685
Velázquez, E., Carro, L., Flores-Félix, J. D., Martínez-Hidalgo, P., Menéndez, E., Ramírez-Bahena, M.-H., et al. (2017). “The legume nodule microbiome: A source of plant growth-promoting bacteria,” in Probiotics and Plant Health. Eds. Kumar, V., Kumar, M., Sharma, S., Prasad, R. (Singapore: Springer Singapore), 41–70. doi: 10.1007/978-981-10-3473-2_3
Vo, Q. A. T., Ballard, R. A., Barnett, S. J., Franco, C. M. M. (2021). Isolation and characterisation of endophytic actinobacteria and their effect on the growth and nodulation of chickpea (Cicer arietinum). Plant Soil 466, 357–371. doi: 10.1007/s11104-021-05008-6
Walker, L., Lagunas, B., Gifford, M. L. (2020). Determinants of host range specificity in legume-rhizobia symbiosis. Front. Microbiol. 11, 585749. doi: 10.3389/fmicb.2020.585749
Wang, X., Feng, H., Wang, Y., Wang, M., Xie, X., Chang, H., et al. (2021). Mycorrhizal symbiosis modulates the rhizosphere microbiota to promote rhizobia–legume symbiosis. Mol. Plant 14, 503–516. doi: 10.1016/j.molp.2020.12.002
Wang, C., Saldanha, M., Sheng, X., Shelswell, K. J., Walsh, K. T., Sobral, B. W. S., et al. (2007a). Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 153, 388–398. doi: 10.1099/mic.0.29214-0
Wang, C., Sheng, X., Equi, R. C., Trainer, M. A., Charles, T. C., Sobral, B. W. S. (2007b). Influence of the poly-3-hydroxybutyrate (PHB) granule-associated proteins (PhaP1 and PhaP2) on PHB accumulation and symbiotic nitrogen fixation in Sinorhizobium meliloti Rm1021. J. Bacteriol. 189, 9050–9056. doi: 10.1128/JB.01190-07
Wardell, G. E., Hynes, M. F., Young, P. J., Harrison, E. (2022). Why are rhizobial symbiosis genes mobile? Philos. Trans. R. Soc B Biol. Sci. 377, 20200471. doi: 10.1098/rstb.2020.0471
Ware, I. M., Fitzpatrick, C. R., Senthilnathan, A., Bayliss, S. L. J., Beals, K. K., Mueller, L. O., et al. (2019). Feedbacks link ecosystem ecology and evolution across spatial and temporal scales: Empirical evidence and future directions. Funct. Ecol. 33, 31–42. doi: 10.1111/1365-2435.13267
Weisberg, A. J., Rahman, A., Backus, D., Tyavanagimatt, P., Chang, J. H., Sachs, J. L. (2022). Pangenome evolution reconciles robustness and instability of rhizobial symbiosis. mBio 13, e00074–e00022. doi: 10.1128/mbio.00074-22
Wendlandt, C. E., Roberts, M., Nguyen, K. T., Graham, M. L., Lopez, Z., Helliwell, E. E., et al. (2022). Negotiating mutualism: A locus for exploitation by rhizobia has a broad effect size distribution and context-dependent effects on legume hosts. J. Evol. Biol. 35, 844–854. doi: 10.1111/jeb.14011
Westhoek, A., Clark, L. J., Culbert, M., Dalchau, N., Griffiths, M., Jorrin, B., et al. (2021). Conditional sanctioning in a legume-Rhizobium mutualism. Proc. Natl. Acad. Sci. U.S.A. 118, e2025760118. doi: 10.1073/pnas.2025760118
Westhoek, A., Field, E., Rehling, F., Mulley, G., Webb, I., Poole, P. S., et al. (2017). Policing the legume-Rhizobium symbiosis: a critical test of partner choice. Sci. Rep. 7, 1419. doi: 10.1038/s41598-017-01634-2
Wheatley, R. M., Ford, B. L., Li, L., Aroney, S. T. N., Knights, H. E., Ledermann, R., et al. (2020). Lifestyle adaptations of Rhizobium from rhizosphere to symbiosis. Proc. Natl. Acad. Sci. U.S.A. 117, 23823–23834. doi: 10.1073/pnas.2009094117
Wippel, K., Tao, K., Niu, Y., Zgadzaj, R., Kiel, N., Guan, R., et al. (2021). Host preference and invasiveness of commensal bacteria in the Lotus and Arabidopsis root microbiota. Nat. Microbiol. 6, 1150–1162. doi: 10.1038/s41564-021-00941-9
Wilkinson, A., Danino, V., Wisniewski-Dyé, F., Lithgow, J. K., Downie, J. A. (2002). N -acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 184, 4510–4519. doi: 10.1128/JB.184.16.4510-4519.2002
Xavier, L. J. C., Germida, J. J., et al. (2003). Selective interactions between arbuscular mycorrhizal fungi and Rhizobium leguminosarum bv. viceae enhance pea yield and nutrition. Biol. Fertil. Soils. 37, 261–267. doi: 10.1007/s00374-003-0605-6
Xie, B., Chen, D., Cheng, G., Ying, Z., Xie, F., Li, Y., et al. (2009). Effects of the purL gene expression level on the competitive nodulation ability of Sinorhizobium fredii. Curr. Microbiol. 59, 193–198. doi: 10.1007/s00284-009-9420-0
Yan, J., Han, X. Z., Ji, Z. J., Li, Y., Wang, E. T., Xie, Z. H., et al. (2014). Abundance and diversity of soybean-nodulating rhizobia in black soil are impacted by land use and crop management. Appl. Environ. Microbiol. 80, 5394–5402. doi: 10.1128/AEM.01135-14
Yost, C. K., Rath, A. M., Noel, T. C., Hynes, M. F. (2006). Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiol. 152, 2061–2074. doi: 10.1099/mic.0.28938-0
Younginger, B. S., Friesen, M. L. (2019). Connecting signals and benefits through partner choice in plant–microbe interactions. FEMS Microbiol. Lett. 366, fnz217. doi: 10.1093/femsle/fnz217
Zgadzaj, R., Garrido-Oter, R., Jensen, D. B., Koprivova, A., Schulze-Lefert, P., Radutoiu, S. (2016). Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 113, E7996–E8005. doi: 10.1073/pnas.1616564113
Zgadzaj, R., James, E. K., Kelly, S., Kawaharada, Y., De Jonge, N., Jensen, D. B., et al. (2015). A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PloS Genet. 11, e1005280. doi: 10.1371/journal.pgen.1005280
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Keywords: rhizobia, symbiosis, nitrogen fixation, microbial communities, microbe-microbe interactions, eco-evolutionary dynamics
Citation: Granada Agudelo M, Ruiz B, Capela D and Remigi P (2023) The role of microbial interactions on rhizobial fitness. Front. Plant Sci. 14:1277262. doi: 10.3389/fpls.2023.1277262
Received: 14 August 2023; Accepted: 22 September 2023;
Published: 09 October 2023.
Edited by:
Chang Fu Tian, China Agricultural University, ChinaReviewed by:
Oswaldo Valdes-Lopez, National Autonomous University of Mexico, MexicoCopyright © 2023 Granada Agudelo, Ruiz, Capela and Remigi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippe Remigi, cGhpbGlwcGUucmVtaWdpQGlucmFlLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.