- Sugar Beet Physiological Research Institute, Inner Mongolia Agricultural University, Hohhot, China
Constitutive photomorpogenic dwarf (CPD) is a pivotal enzyme gene for brassinolide (BR) synthesis and plays an important role in plant growth, including increasing plant biomass and plant height, elongating cells, and promoting xylem differentiation. However, little is known about the function of the CPD gene in sugar beet. In the current study, we isolated CPD from Beta vulgaris L. (BvCPD), which encodes protein localized in the nucleus, cell membrane, and cell wall. BvCPD was strongly expressed in parenchyma cells and vascular bundles. The transgenic sugar beet overexpressing BvCPD exhibited larger diameter than that of the wild type (WT), which mainly owing to the increased number and size of parenchyma cells, the enlarged lumen and area of vessel in the xylem. Additionally, overexpression of BvCPD increased the synthesis of endogenous BR, causing changes in the content of endogenous auxin (IAA) and gibberellin (GA) and accumulation of cellulose and lignin in cambium 1–4 rings of the taproot. These results suggest that BvCPD can promote the biosynthesis of endogenous BR, improve cell wall components, promote the development of parenchyma cells and vascular bundle, thereby playing an important role in promoting the growth and development of sugar beet taproot.
1 Introduction
Sugar beet (Beta vulgaris L.), a biennial plant in the Chenopodiaceae family, is the most important raw material in the production of sucrose (Ramesh et al., 2017). The fleshy taproot is the main harvest portion of the beet and is rich in sucrose. Sugar production occurs via the leaves through photosynthesis during the first year of growth. The sugar is continuously transferred from leaves to the taproots and stored in concentric rings of vascular tissue originating from the secondary cortex of early root development as well as in parenchyma cells that increase in number and become larger during growth (Rae et al., 2005). Therefore, understanding the regulatory mechanisms underlying taproot growth will enable the development new sugar beet cultivars with high yield and quality.
Brassinolide (BR) is an important plant hormone and plays an critical role in regulating many biological processes, including root development, cell division and elongation, vascular bundle differentiation, pollen fertility, photomorphology, and stress resistance processes (Wei and Li, 2016; Nolan et al., 2020; Chen et al., 2022). Specifically, the mechanism by which BR regulates root growth and lateral development has been extensively studied in a variety of plants, including Arabidopsis thaliana (Vukašinović et al., 2021), Oryza sativa L (Huang et al., 2022), Triticum aestivum L (Avalbaev et al., 2021; Xu et al., 2022), Solanum lycopersicum (Bai et al., 2021), Phyllostachys pubescens (Wang et al., 2019; Guo et al., 2020), Populus L (Tian et al., 2018; Feng et al., 2021), Eriobotrya japonica (Su et al., 2021), and Beta vulgaris (Zhang et al., 2017; Wang et al., 2021). For example, mutants associated with BR biosynthesis or signaling genes, such as Arabidopsis dwf7-1, bri1, and bin2, cause abnormal xylem differentiation ratios and defects in vascular bundle development compared with the wild type (WT), suggesting that BRs can regulate vascular structure and xylem formation. (Choe et al., 1999; Caño-Delgado et al., 2004). Exogenous spraying of BR promoted the development of parenchyma cell and secondary xylem in sugar beet taproot and regulated the expression of cell wall biosynthesis-related genes, BvXTH33, BvSHV3, BvCESA6, BvPARVUS, and BvCEL1, which affected the diameter of taproot (Wang et al., 2021). In Liriodendron, BR treatment significantly increased the length of fibers and vascular elements, and induced the biosynthesis of cellulose and hemicellulose (Hyunjung et al., 2014). This indicates that BR has a regulatory role in the biosynthesis and modification of secondary cell wall components and cell wall assembly during secondary xylem development in woody plants. Furthermore, BR is most abundant in actively growing tissues. Unlike other plant hormones, BR is not transported over long distances and acts only near the site of synthesis (Symons et al., 2008). Therefore, research on BR biosynthesis and its role in modulating root growth and development can provide a reference for regulating sugar beet taproot development using molecular biology or chemical control.
The biosynthesis of BR is controlled by multiple genes, most of which encode cytochrome P450 (CYP90 or CYP85), such as CPD, DWF4, DET2, and BR6OX2. The CPD gene encodes a C-3 oxidase and plays a vital role in multiple steps of BR synthesis. The CPD protein sequence of guayule rubber particles has all the functional domains of microsomal cytochrome P450 monooxygenase, including an N-terminal cell membrane anchoring sequence, a proline-rich region, and a region for oxygen and hemoglobin binding. Furthermore, the CPD gene has a specific sterol hydroxylase domain (Pan et al., 1995). AtCPD can catalyze early BR intermediates i.e., (22S)-22-hydroxy campesterol, (22R,23R)-22,23-dihydroxycampesterol, 6-deoxocathasterone, and 6-deoxoteasterone; however, it is most potent in the catalysis of (22S)-22-hydroxy campesterol (Ohnishi et al., 2012). CPD has the function and necessity for plant growth and is widely expressed in root, stalk, leaf, and inflorescence (Mathur et al., 1998; Wang et al., 2015; Vukašinović et al., 2021). The function of this gene has been extensively studied in a variety of plants. Arabidopsis cpd mutants exhibit a yellowing phenotype under dark conditions, with hypocotyls becoming shorter, no apical curved grooves, cotyledons opening, leaf primordia appearing, and photoinduced genes no longer being inhibited. Under light conditions, it exhibits a dwarf and male sterile phenotype (Miklós et al., 1996). OsCPD regulates the biosynthesis of BR in Oryza sativa L, resulting in enlarged leaf angles and increased grain size; furthermore, it also plays a significant and redundant role in maintaining plant structure (Zhan et al., 2022). Overexpression of the potato CPD gene strengthens the ability of seedlings to remove reactive oxygen species and reduce oxidative damage by increasing BR content, enhancing antioxidant enzyme activity and improving plant resistance to PEG-induced osmotic stress, thereby affecting plant growth. (Zhou et al., 2016). Overexpression of PeCPD enhances BR signaling, increase plant height and biomass, promote xylem differentiation, and reduced pith size, thereby improving wood quality (Feng et al., 2021). Moreover, PeCPD regulates BL synthesis, allowing BL to interact with growth hormones and partially replace the action of cytokinin to stimulate healing tissue formation (Tian et al., 2018). When CPD is knocked down, cell growth in the meristem and elongation regions of the roots is inhibited, leading to root shortening (Vukašinović et al., 2021). However, there is little research on the function of the CPD gene in the radial development of plant roots, which needs to be further explored.
Our previously published transcriptome sequencing results showed that the Bv_qyup gene in the CPD family is closely related to sugar beet taproot enlargement (Zhang et al., 2017). A total of 76 BvCPD family members were detected in a sugar beet database, clustered into 10 subgroups and distributed on 9 chromosomes. The Bv_qyup gene is located on chromosome 2 and encodes the CPD protein. Overexpression of this gene in Arabidopsis thaliana showed phenotypes of root elongation, increased root volume, enlarged leaves, and increased length of fruit pods, which may be important in improving plant development (Zhu et al., 2020). However, the sugar beet root belongs to a typical tritrophic structure, and it has not been reported how this gene plays a role in regulating taproot growth and development. Herein, we investigated in depth the tissue expression pattern of BvCPD (Bv_qyup) in sugar beet, constructed overexpression and RNAi lines using agrobacterium-mediated genetic transformation, and elucidated the mechanism of BvCPD in regulating the growth and development of sugar beet taproots through morphological, anatomical, and molecular biological studies. The research confirms for the first time that the function of CPD in sugar beets, providing candidate genes for genetic improvement of high yield and quality in sugar beet. Simultaneously, we created high yield planting resources for sugar beet and provided a scientific basis for chemical regulation of yield traits in sugar beets.
2 Materials and methods
2.1 Plant material
We used high-sugar homozygous sugar beet variety BS02, cultivated by the Institute of Sugar Beet Physiology, Inner Mongolia Agricultural University. The growth environment of the histoculture and artificial climate chamber was 22°C, 60% humidity, and 16/8 h light/dark cycles.
2.2 Subcellular localization of BvCPD
Coding sequences (CDS) of BvCPD were obtained by PCR amplification using gene-specific primers BvCPD-G. PCR products (1,468 bp) were digested with Kpn I and Sal I, then cloned into the pCAMBIA1300-35S-EGFP vector. The plasmids of the fusion vector and empty vector were extracted and transferred into agrobacterium tumefaciens GV3101 receptor cells to infect the onion epidermis.
Onion phosphorus stems were sterilized and washed in sterile water on an ultra-clean bench, cut lengthways, and the thicker inner layer of the middle scale-leaf was taken. Using a scalpel, we gently cut several 1 cm2 squares, tore off epidermal cells, and placed them in MS medium and incubated them at 25°C for 24 h. Then, we took 3.3 mg of gold powder and placed it in a centrifuge tube containing 55 µL of 40% PEG. We took 20 µl and add 5 μg of 35S-BvCPD-EGFP or 35S-EGFP plasmid DNA, 50 µL of 2.5 M CaCl2, 8 µL of 0.1 M Spermidine. This was fully mixed and centrifuged for 30 s, and the supernatant was discarded. The remained was rinsed once with 70% ethanol, and we then add 15 µL of absolute ethanol before vortex and mixing. Then, 15 μL of the mixture was placed on the carrier membrane to dry. The cultured onion epidermal cells were placed in a vacuum chamber for 30 h to rupture the membrane. The onion was incubated at 25°C in the dark for 48 h. In addition, onion epidermal cells were treated with 0.3 g/mL sucrose solution for plasmolysis and then imaged. Each onion epidermal cell was removed and placed on a slide for observation and image acquisition using a fluorescent microscope (OLYMPUS, U-RFL-T, Japan).
2.3 Fluorescence in situ hybridization in paraffin sections of sugar beet
Beet roots were removed, washed, cut crosswise, and immediately placed in fixative for 24 h. A vacuum pump was used to evacuate air; thereafter, dehydration by gradient alcohol was followed by wax immersion and embedding. The paraffin was sliced by a slicer, retrieved with slice spreaders, and baked at 62°C for 2 h. Sections were then placed in dewaxing clear solution I and II for 15 min each, and anhydrous ethanol I and II for 5 min each. The samples were dried spontaneously and then immersed in DEPC water. Proteinase K (20ug/ml; Servicebio; Article number: G1234) was added dropwise and digested at 37°C for 25 min. We then added pre-hybridization solution (Servicebio; Article number: G3016-3) and incubated at 37°C for 1 h before discarding the liquid. We added the probe CPD-ORF hybridization solution (Synthesized by Servicebio, China) to a concentration of 500 nM and hybridized overnight at 42°C. The probe information is presented in Supplementary Table S1. Next, the sections were washed with eluent and incubated with DAPI (4’,6-diamidino-2-phenylindole) for 8 min in the dark before mounting. Finally, the sections were placed under a fluorescent microscope (OLYMPUS, U-RFL-T, Japan) for observation and image acquisition.
2.4 Vector construction and genetic transformation of sugar beet
Construction of BvCPD overexpression vector: The CDSs of BvCPD were acquired by PCR amplification using gene specific primers BvCPD-OE. PCR products (1,523 bp) were digested with Bam H I and Pst I, then cloned into the ToPopBoLn-TA vector. We selected the positive colonies with correct sequencing to extract the recombinant plasmid, cut BvCPD with Bam H I and Pst I double enzymes, recovered the digested product by agarose gel electrophoresis, and inserted the T4 ligase into the plant expression vector pCAMBIA1300-35S-X cut by the corresponding restriction enzyme. In this way, we successfully constructed a plant overexpression vector containing the complete CDS region of the BvCPD gene for genetic transformation into sugar beet.
Construction of BvCPD silencing vector: CDSs of BvCPD were acquired by PCR amplification using gene specific primers BvCPD-RP and BvCPD-RA. PCR products (334 bp) were digested with Bam H I and Spe I, Kpnl, and Sac I, then cloned into the pEASYR-Blunt Zero Cloning Vector. We selected the positive colonies with correct sequencing and extracted the recombinant plasmid using BamH I/Spe I double enzyme digestion for BvCPD forward fragments, and KpnI/SacI double enzyme digestion for BvCPD reverse fragments. After recovering the enzymatic cleavage products, T4 ligase was inserted into the plant expression vector pCAMBIA1300-35S-X after corresponding restriction endonuclease cleavage. In this way, we constructed a plant silencing expression vector containing both positive and negative fragments of the BvCPD gene.
Planting sterile seedlings: Seeds were washed for 30 min, treated with 0.1% HgCl2 solution for 10 min, flushed 3–4 times with sterile water, dried on filter paper for 5–10 min, and planted into a germination medium. Agrobacterium-mediated genetic transformation of sugar beet: We cut the sprouted beet cotyledonary node to form a wound, pre-cultured for 24 h, overnight cultured the agrobacterium containing the target gene to an OD value of 0.6–0.8, centrifuged and resuspend to 0.5 with suspension, and dipped the beet cotyledon node for 10 min. Excess agrobacterium was blotted dry on filter paper and incubated on a co-culture medium for 2–3 days in the dark. Inhibiting agrobacterium growth: After 2–3 days, we washed the cotyledonary nodes 3–4 times and transferred them to a suppressive medium for 5–7 days. Screening for positive plants: We transferred the sterile beet cotyledons after inhibition to a screening medium containing Temetin, and after 15–20 days transferred the screened beet resistant shoots to a differentiation medium for differentiation and expansion. Promoting root growth: When sugar beet seedlings reached 3–5 cm, they were transferred to rooting medium to induce root formation. Transplanting: After 1 month, the beet seedlings were differentiated based on thick roots and leaves topping out to the cap; we opened the cap and refined these seedlings for 3–5 days, transplanted them into seedling trays (nutrient soil: vermiculite = 3:1), covered the box, and cultivate in the dark for 2–3 days (Supplementary Figure S1). The composition of all the media used is presented in Supplementary Table S2.
2.5 Phenotypic determination of sugar beet
2.5.1 Phenotype
Morphological indicators of sugar beet were measured when transplanted transgenic sugar beet seedlings were incubated in an artificial climate chamber for 80 days. Three plants were taken from each line. Fresh weight per plant, leaf plexus weight, fresh leaf weight, petiole weight, root weight, taproot weight, and fibrous root weight were weighed on an electronic balance; plant height, leaf length, leaf width, root length, and taproot length were measured with a tape measure; the diameter of the taproot was measured with a vernier caliper.
2.5.2 Anatomic structure
Beet roots were cut to a thickness of 5 mm along the first leaf scar after 80 days of transplant culture and placed in FAA fixative for 24 h and dehydrated. Next, 12-μm thick transverse sections were obtained using a rotary slicer, stained with astra blue and saffron, and then fixed on paraffin (Maia et al., 2018). The samples were observed under a microscope and micrographs were taken. Images were analyzed using the CaseViewer software. We assessed the following histological parameters: ring spacing, area of parenchyma cells, number of parenchyma cell layers, area of xylem, and area of vessel lumen. The distance between the outermost xylem of two vascular forming layers was defined as the spacing between the rings of forming layers; the area of 10 adjacent parenchyma cells was selected to measure the area; the number of parenchyma cell layers between the rings of two forming layers was counted; the xylem area per unit area was counted using CaseViewer magnified to 47x field of view; and the area of the ductal pore lumen on the xylem of each forming layer was counted. All lines had three biological replicates.
2.6 Hormone determination
We sampled sugar beet roots by inserting a punch diagonally from the outside to the inside. Three plants from each line were taken. Plant tissue was ground under liquid nitrogen freezing conditions, and 0.1 g of fresh sample was mixed with 900 μL of 0.1% PBS buffer. After sufficient shaking, the tissue was centrifuged at 12,000 rpm for 30 min in a cryogenic freezing centrifuge. The supernatant was taken for hormone determination. BR, IAA, and GA contents were determined by enzyme-linked immunoassay (ELISA) according to the manufacturer’s instructions (Jiangsu Su Enzyme Science and Technology Co., Ltd.).
2.7 Gene expression identification by qRT–PCR
Total RNA was extracted according to the operating instructions of the TransZol Up Plus RNA Kit (ER501). RNA was reverse transcribed to complementary DNA (cDNA) using TransScript One-Step gDNA Removal and cDNA Synthesis Super Mix (AT311), and diluted 5-fold for use. Experiments were performed using the CFX96 Real-Time Quantitative PCR System (Bio-Rad, Hercules, CA, USA). The reaction system included 10 μL of PerfectStartTM Green qPCR SuperMix (AQ601), upstream and downstream primers (10 μM) of 0.4 μL each, 7.2 μL of nuclease-free water, and 2 μL of cDNA template. The PCR amplification procedure was 30 s at 94°C, followed by 5 s at 94°C, 15 s at 55°C, and 30 s at 72°C for a total of 45 cycles, with only one product amplified and detectable after each reaction for melt curve analysis. The 2-ΔΔCT method was used to calculate gene relative expression (Livak and Schmittgen, 2001). Three biological replicates and three technical replicates were taken for the experimental samples. Primer sequences are shown in Supplementary Table S3.
2.8 Cellulose and lignin quantification
After 120 days of transplantation of sugar beet, samples were taken from the vascular rings and inter-rings of the 1–4 layers of the taproot, cut into fine shreds to dry and then ground into powder. Determination of cellulose and lignin content was performed according to the manufacturer’s instructions (Jiangsu Addison Biotechnology Co., China).
2.9 Data analysis
Differences between means were assessed using SPSS (VER) at the 5% and 1% probability levels. Statistical plots of the data were produced using GraphPad Prism.
3 Results
3.1 BvCPD is localized in the nucleus, cell membrane, and cell wall
We used the onion epidermis as a model for subcellular localization of BvCPD proteins. The 35S-BvCPD-EGFP fusion expression vector and 35S-EGFP empty vector were genetically transformed into onion epidermal cells for transient expression by particle bombardment. Fluorescence microscopy observations showed that the control 35S-initiated EGFP protein signal was distributed throughout the onion epidermal cells (Figures 1A–C). The BvCPD protein was expressed in the nucleus, cell membrane, and cell wall (Figures 1D–I).
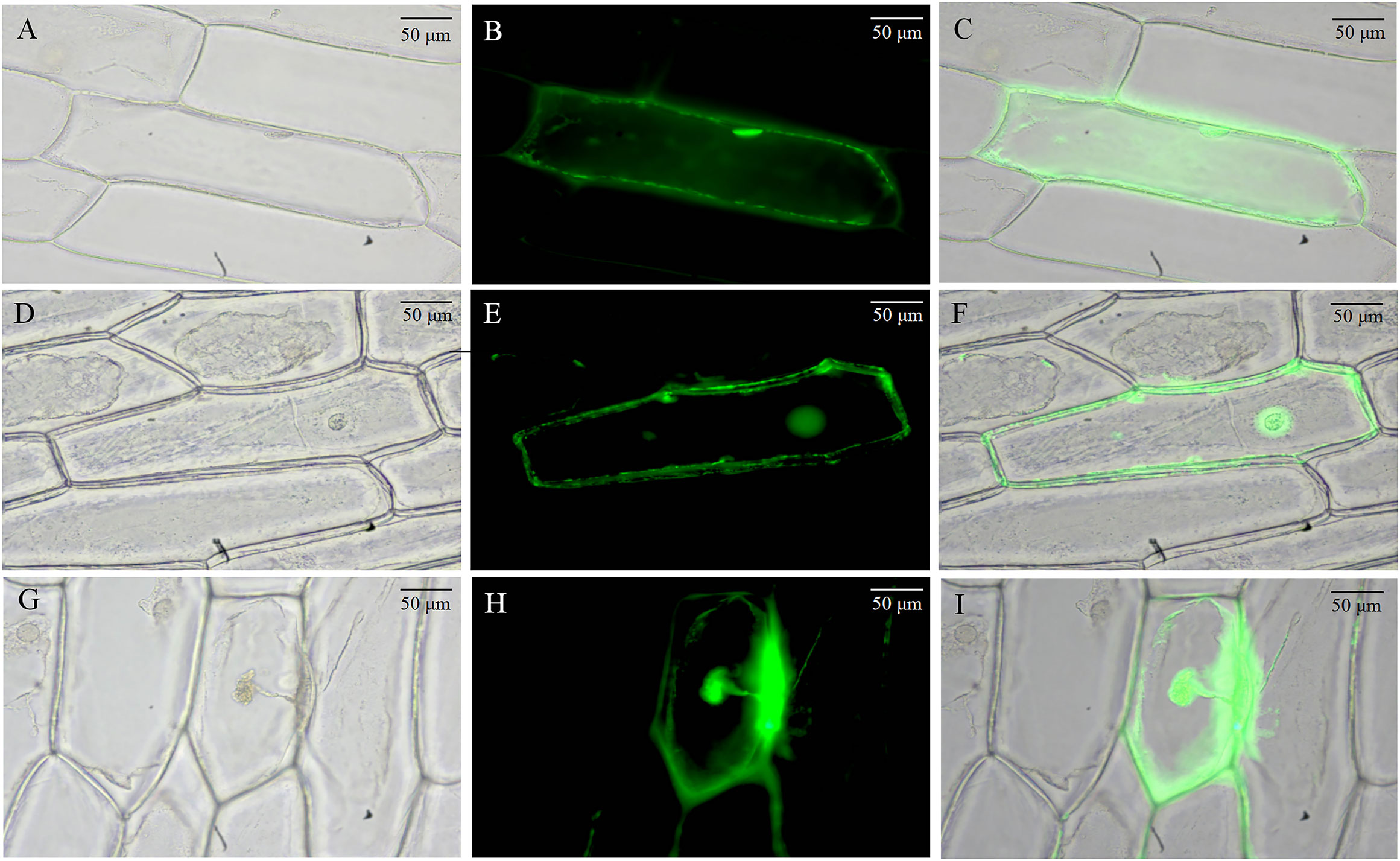
Figure 1 Subcellular localization of BvCPD in onion epidermis. (A–C) Subcellular localization of 35S-EGFP. (D–F) Subcellular localization of 35S-BvCPD-EGFP before plasmolysis. (G–I) Subcellular localization of 35S-BvCPD-EGFP after plasmolysis. Scale bars = 50 μm.
3.2 BvCPD genes are strongly expressed in sugar beet parenchyma cells and vascular bundles
mRNA fluorescence in situ hybridization results showed that BvCPD was mainly expressed in cell walls of inter-ring parenchymal cells and in the xylem of vascular bundles in root cross sections (Figures 2A–F). In leaf cross sections, BvCPD was mainly expressed in vascular bundles and mesophyll cells, with strong expression in the xylem of vascular bundles (Figures 2G–L). In petiole cross sections, BvCPD was expressed in the vascular bundle, with high expression in the xylem and low expression in the phloem (Figures 2M–Q). These results suggest that the BvCPD gene may play an essential role in the development of parenchymal cells and vascular bundles in sugar beet.
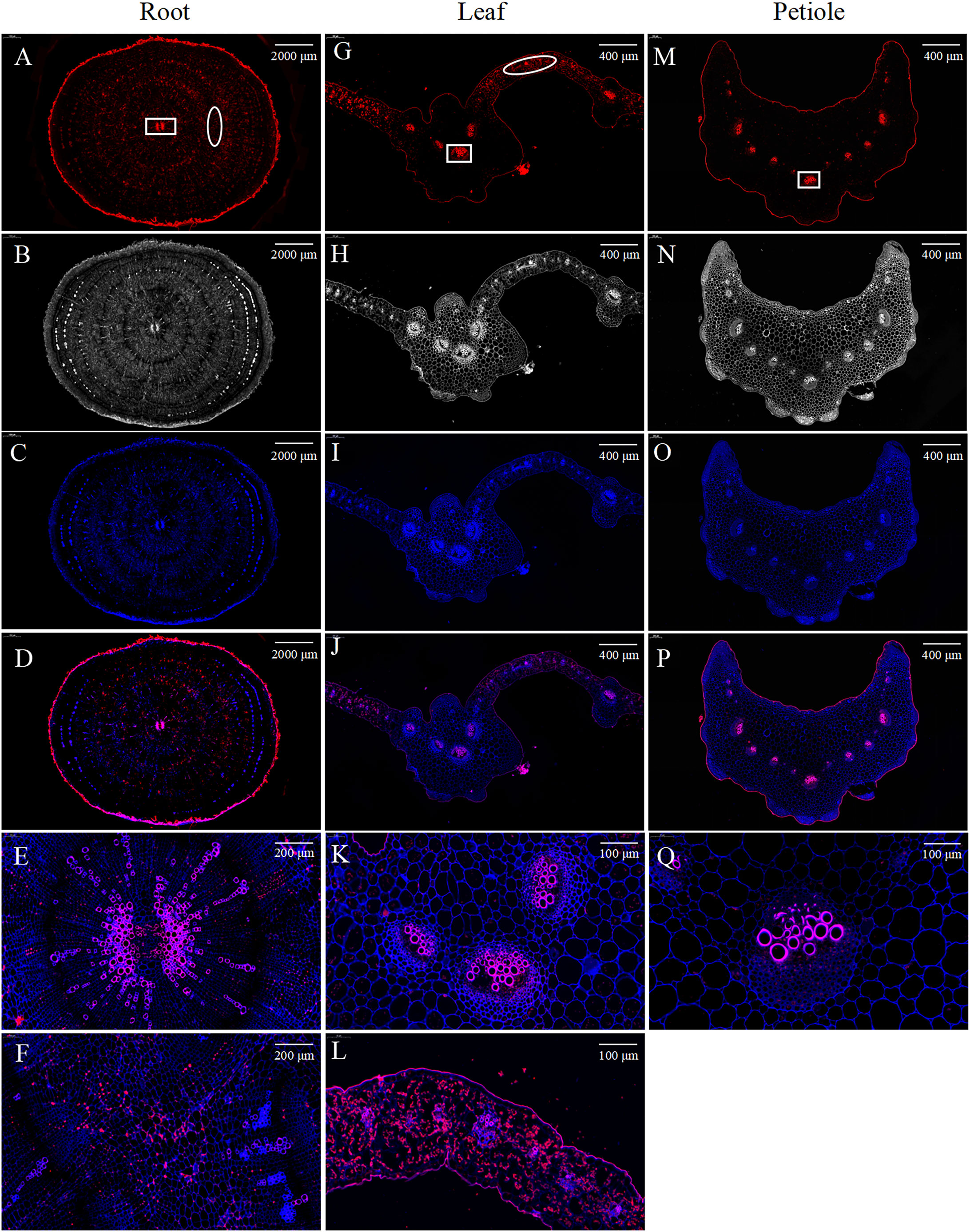
Figure 2 Fluorescence in situ hybridization of BvCPD in sugar beet. (A–D) Root. Scale bar = 2,000 μm (E) Enlarged area of root, denoted by the square in (A), representing the vascular bundle. Scale bar = 200 μm. (F) Enlarged area of root, denoted by the oval (A), representing parenchyma cells. Scale bar = 200 μm. (G–J) Leaf. Scale bar = 400 μm (K) enlarged area of leaf, denoted by the square in (G). Scale bar = 100 μm. (L) Enlarged area of leaf, denoted by the oval in (G). Scale bar = 100 μm. (M–P) Petiole. Scale bar = 400 μm. (Q) Enlarged area of petiole, denoted by the square in (M). Scale bar = 100 μm. (A, G, M) Red images showing BvCPD expression regions. (B, H, N) Blue images showing cell wall autofluorescence. (C, I, O) Bright field images. (D, J, P) Composite images.
3.3 Acquisition of BvCPD transgenic sugar beet plants
The optimized Agrobacterium tumefaciens genetic transformation system was used to impregnate cotyledon nodes of sugar beet. After screening for hygromycin (6 mg/L) resistance, 21 lines with hygromycin resistance were obtained, including 12 overexpression-resistant lines and 9 silenced-resistant lines. Polymerase chain reaction (PCR) with primer HYG-2 was performed using DNA from resistant plants as the template, and the expected target gene products (Figure 3A) were obtained in all the cases, confirming the insertion of exogenous genes in the genomes of these plants. The lines were subjected to mass expansion, rooting, and transplanting. Reverse transcription–polymerase chain reaction (RT–PCR) results showed that transcripts of the screening gene hygromycin (Hyg) were detected in all the lines of the transgenic sugar beet, confirming that the target gene was already stably expressed (Figure 3B). Quantitative real–time polymerase chain reaction (qRT–PCR) technology was used to detect the expression of BvCPD in transgenic lines. The results showed that compared with WT, the expression level of BvCPD was increased in overexpressed lines, but decreased in silent lines (Figures 3C, D). Due to the different insertion sites of target gene, the expression levels varied among different lines. The three lines with the highest differences in expression levels as compared to WT were selected from overexpressed and silent lines, and named OE1, OE2, OE3 and R1, R2, and R3, respectively, for subsequent research.
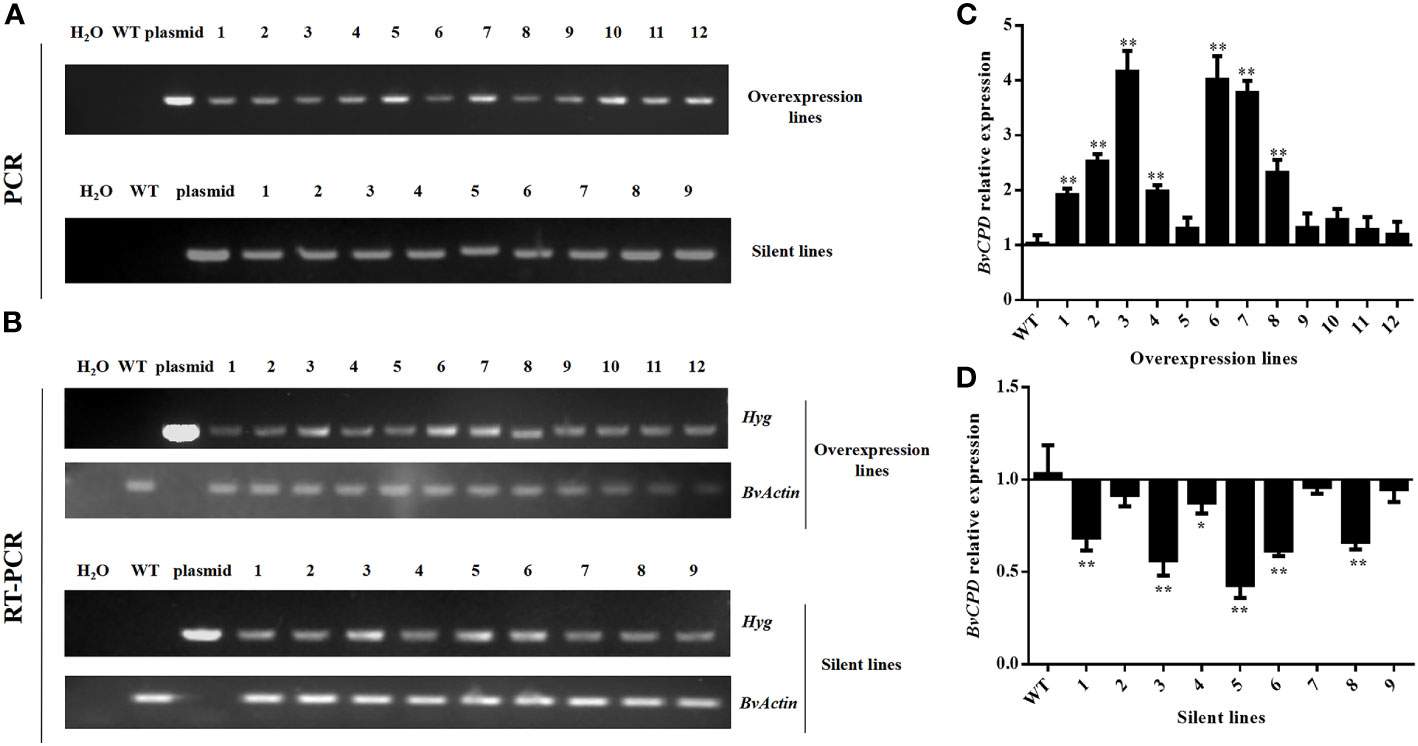
Figure 3 Identification of transgenic lines. (A) Electrophoresis plots of PCR products of the Hyg gene of the overexpressing and silent lines. (B) Electrophoresis plot of the RT–PCR products of the Hyg gene of the overexpressing and silent lines. (C) Relative expression of BvCPD in the overexpression lines. (D) Relative expression of BvCPD in silent lines. *Student’s-t test, P < 0.05; ** Student’s-t test, P < 0.01.
3.4 BvCPD can promote the growth of sugar beet
The shoot morphologies of the different transgenic lines and the wild type varied significantly. The morphological indicators of each leaf bush of the transgenic plants overexpressing BvCPD were significantly greater than those of WT, while those of silent lines were significantly smaller (Figures 4A–I). Compared with WT, the increase in leaf fresh weight of the overexpressed lines was greater than that of other indicators, increasing by 57.92%–152.24% (Figure 4G); leaf length and leaf width increased by 16.41%–35.29% and 14.71%–35.78%, respectively (Figures 4D, E). Petiole was significantly thickened (Figure 4M). Silent lines had a slender petiole (Figure 4M) and the largest reduction in petiole weight (decrease of 71.89%–90.08%; Figure 4H). Leaf length and leaf width decreased by 8.98%–33.75% and 30.39%–40.20%, respectively. In summary, the overexpression of BvCPD facilitates more vigorous growth of the shoots (Figures 4J–M).
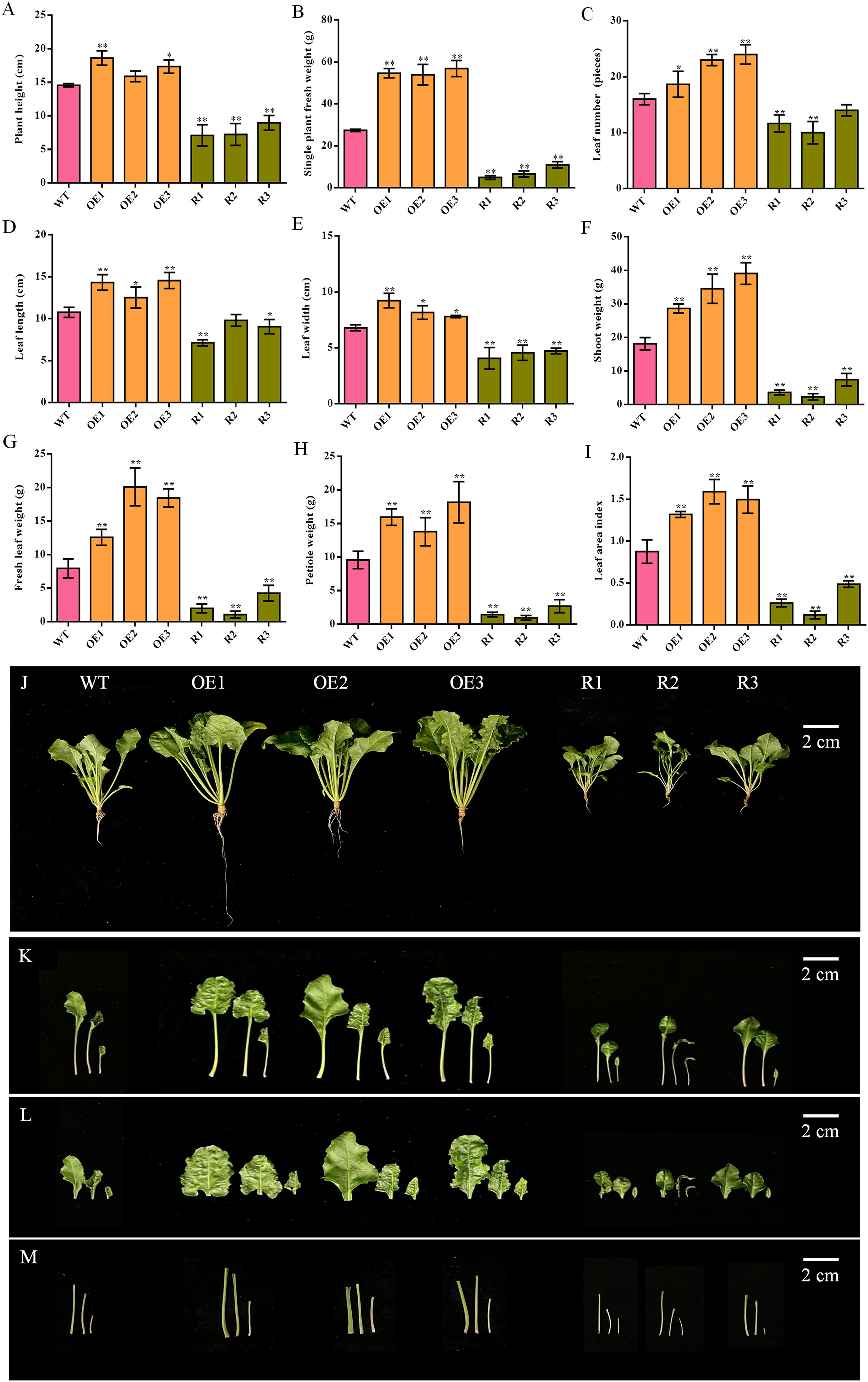
Figure 4 Phenotype determination of the aerial part between transgenic sugar beet and WT. (A) Plant height. (B) Single plant fresh weight. (C) Leaf number. (D) Leaf length. (E) Leaf width. (F) Shoot weight. (G) Fresh leaf weight. (H) Petiole weight. (I) Leaf area index. (J) Phenotype of each transgenic line and WT 1 month after transplanting. (K) Phenotype of the aerial part. (L) Phenotype of leaf. (M) Phenotype of petiole. *Student’s t-test, P < 0.05; **Student’s-t test, P < 0.01. Scale bars = 2 cm.
Comparing the root of sugar beet from various lines, all indicators were significantly increased in BvCPD overexpression lines and significantly decreased in silent lines, except for taproot length, for which there was no significant difference in most lines (Figures 5A–G). The taproot weight and lateral root weight of the overexpression lines were the most significantly different, increasing by 75.70%–166.01% and 178.16%–297.70%, respectively as compared to WT; silent lines decreased by 54.17%–85.89% and 50.57%–87.93%, respectively (Figures 5B, C). In addition, taproot diameters of overexpressed and silenced lines increased and decreased by 36.80%–77.58% and 41.27%–55.08%, respectively (Figures 5F, I). Circular numbers increased and decreased by 1–2 compared with WT (Figure 5G). In summary, overexpression of BvCPD promotes taproot development (Figures 5H, I) by increasing the circular number of the taproot formation layer and the taproot diameter.
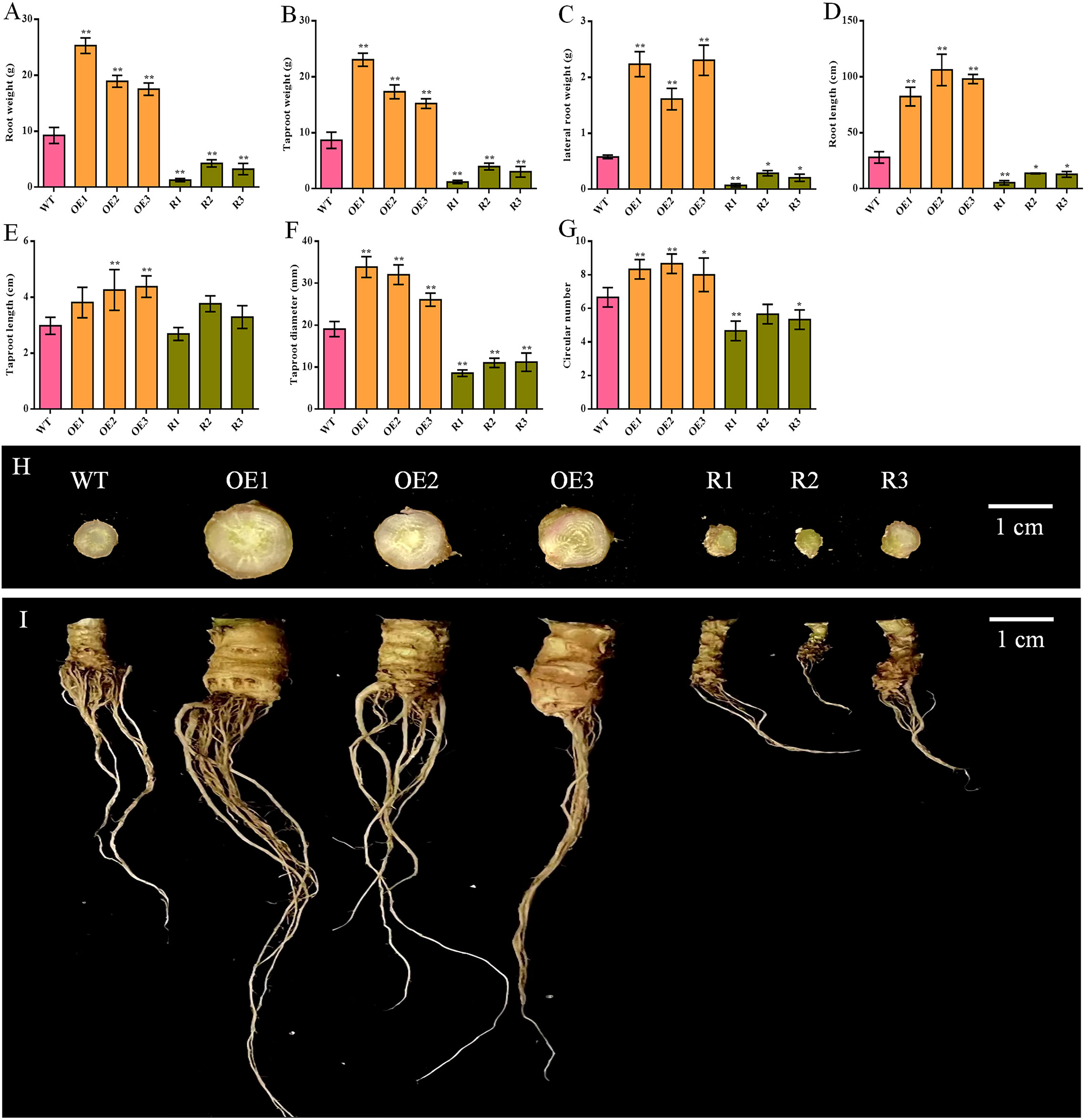
Figure 5 Phenotype determination of the underground parts of transgenic sugar beet and WT. (A) Root weight. (B) Taproot weight. (C) Fibrous root weight. (D) Root length. (E) Taproot length. (F) Taproot diameter. (G) Circular number. (H) Taproot cross-section of each transgenic line and WT. (I) Phenotype of the roots of transgenic lines and WT 1 month after transplanting. * Student’s-t test, P < 0.05; ** Student’s-t test, P < 0.01. Scale bars = 1 cm.
3.5 BvCPD expression is highest in the roots of transgenic sugar beet
qRT–PCR was used to detect the relative expression of BvCPD in the roots, petiole, and leaf of transgenic beets to clarify expression changes in various organs. The expressions of BvCPD in the root, petiole, and leaf of each overexpressed and silenced line were significantly higher and lower than those of WT, respectively. BvCPD expression in the roots of overexpressed lines was greater than those in the leaf and petiole (Figure 6A). Among the three overexpression lines, OE3 had the highest expression level in the root. R2 was the silent line with the lowest expression level of BvCPD in the root.
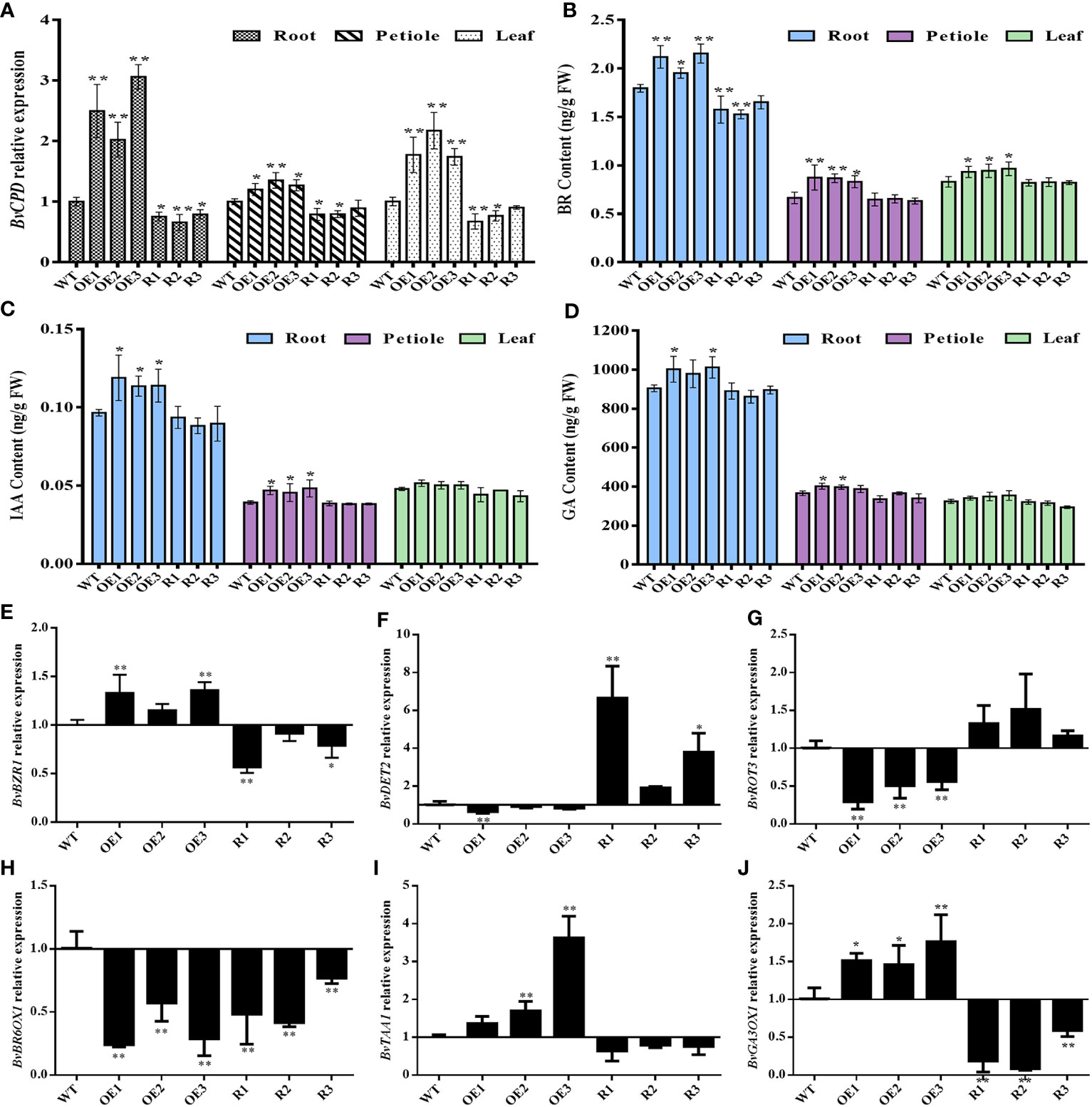
Figure 6 Determination of gene expression levels and hormone content in different transgenic lines and WT. (A) Relative expression of BvCPD in the root, petiole, and leaf of transgenic type and WT sugar beet. (B–D) BR, IAA, and GA content in root, petiole, and leaf. (E–J) Relative expression levels of BvBZR1, BvDET2, BvROT3, BvBR6OX1, BvTAA1, and BvGA3OX1 in the root. * Student’s-t test, P < 0.05; ** Student’s-t test, P < 0.01.
3.6 BvCPD increases BR content and sugar beet root growth & development by cooperating with IAA and GA
To verify whether BvCPD plays a role in sugar beet BR synthesis, transgenic sugar beet BR content was measured. The BR contents in the root, petiole, and leaf of each overexpression line were significantly higher than those of WT, while for silent lines they were significantly lower, especially in the root (Figure 6B). Hormones IAA and GA, which are related to BR, interact to promote plant growth and development (Li et al., 2021). Therefore, endogenous IAA and GA in various organs were also analyzed. The variation trend of IAA and GA content in transgenic lines was consistent with BR, but the difference in BR content was more significant compared with WT (Figures 6C, D). This indicates that BvCPD can increase the BR content. Endogenous BR may play a leading role in the simultaneous mobilization of IAA and GA, which synergistically controls the growth and development of sugar beet, especially the root.
To analyze expression changes of genes related to BR, IAA, and GA synthesis and regulation in sugar beet, real-time fluorescence quantitative analysis was used to determine the brassinazole-resistant (BvBZR), de-etiolated2 (BvDET2), rotundifolia3 (BvROT3), BR-6-oxidase (BvBR6OX1), L-Tryptophan aminotransferase1 (BvTAA1), and gibberellin 3-oxidase1 (BvGA3OX1) genes. The expression of the BR regulatory gene BvBZR1 was significantly upregulated in BvCPD overexpression lines, while the expression of BR synthesis genes BvDET2 and BvROT3 was downregulated. In silent lines, the expression of BvBZR1 was downregulated, while BvDET2 and BvROT3 were upregulated. In contrast, the expression level of BvBR6OX1 was downregulated in both overexpressed and silenced lines. BvTAA1 and BvGA3OX1, as IAA and GA related synthetase genes, were upregulated in overexpressed lines, and downregulated in each silent line (Figures 6E–J). The results indicate that BvCPD is involved in the regulation of BR, IAA, and GA biosynthesis.
3.7 BvCPD promotes parenchyma cell enlargement, xylem development, and ductal pore lumen area
To assess how taproot structure affects taproot diameter, we examined 1–4 rings of each transgenic line taproot and WT (Figures 7A, B). Compared with WT, the ring spacing of the 1–2 formation layer of taproot was significantly increased in BvCPD overexpression lines. Ring spacing of OE1 is greatest in overexpression lines (2,275.39 μm). In contrast, there was an extremely significant reduction in ring spacing for silent lines, with the ring spacing of R3 being the smallest (587.84 μm; Figure 7C). Parenchymal cells between the 1–2 cambial rings of BvCPD overexpressed beet taproot were significantly larger than that of WT. The parenchymal cell area of OE2 was the largest in the overexpression lines (32,097.844 μm2). Parenchymal cells of RNAi silencing lines were significantly smaller than that of WT. RNAi silenced line R1 had the smallest parenchymal cell area (7,209.375 μm2; Figure 7D). Moreover, the amount of cell layers in all overexpressed lines increased significantly. The cell layer with the highest number was OE3, reaching 36 layers, which is 12 layers more than WT. The silence lines were significantly reduced, and the number of layers in R3 was half that of WT (Figure 7E). Similar results were found for the 2–3 and 3–4 cambium rings. In summary, BvCPD increases the size of parenchyma cells and number of layers in the first and second formation layer rings, thereby increasing the diameter of beet taproot (Figures 7F–K).
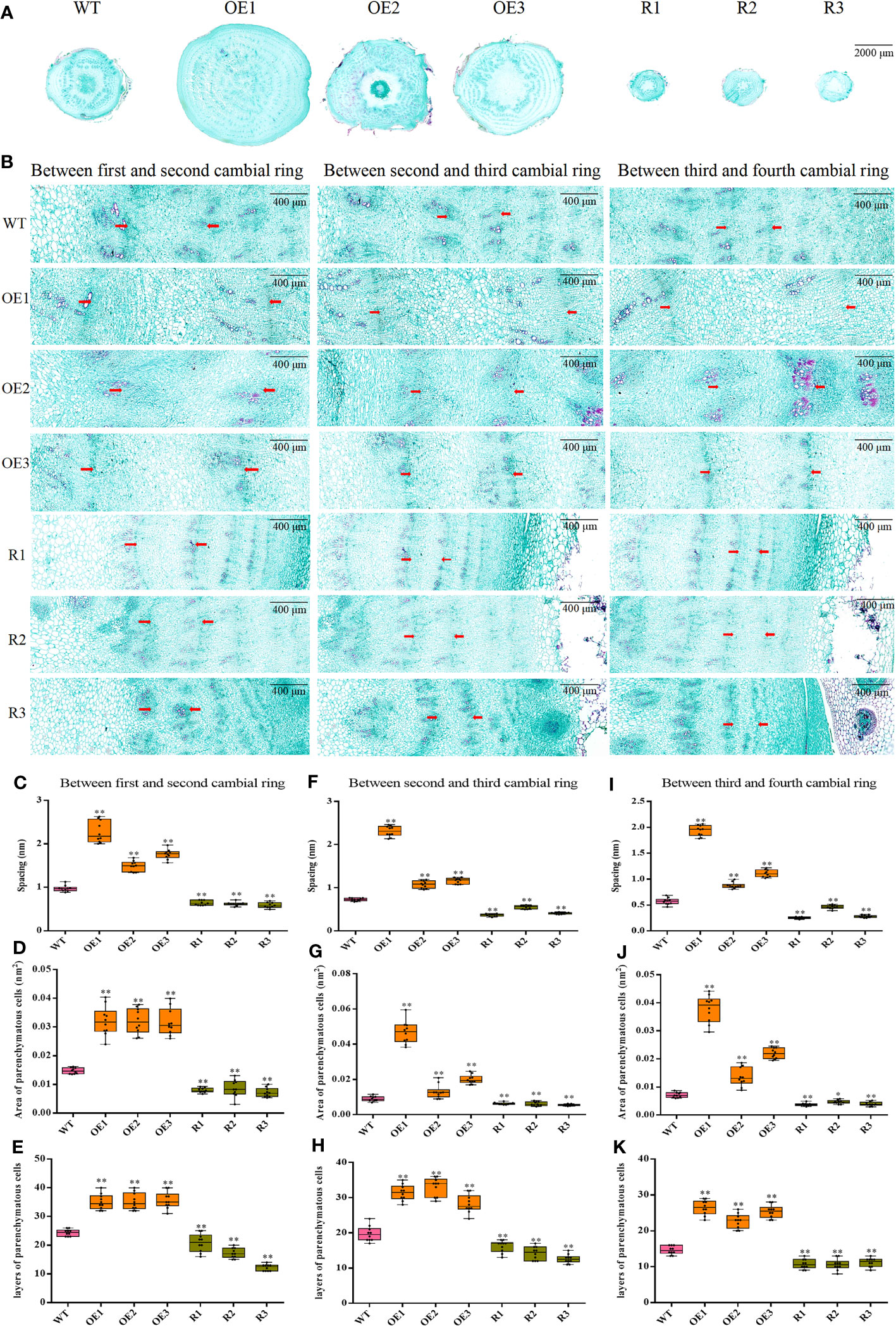
Figure 7 Determination of ring spacing, parenchyma cell size, and cell layer number in the first four rings. (A) Cross-section of wild type and transgenic sugar beet taproot. Scale bar = 2,000 μm. (B) First four ring-forming layers of different lines. Scale bar = 400 μm. (C–E) Spacing, area of parenchyma cells, and layers of parenchyma cells between the first and second cambial rings. (F–H) Spacing, area of parenchyma cells, and layers of parenchyma cells between the second and third cambial rings. (I–K) Spacing, area of parenchyma cells, and layers of parenchyma cells between the third and fourth cambial rings. * Student’s-t test, P < 0.05; ** Student’s-t test, P < 0.01.
To discover structural changes in cambial rings, we used CaseViewer to calculate the area of the xylem on the first, second, and third lamellar rings of beet taproot, and the size of the lumen area of the vessel on the xylem. The size of the xylem per unit area decreased sequentially from the first to the third layer for both the BvCPD transgenic and WT lines (Figure 8A). In each formation ring, the area of xylem in BvCPD overexpression lines was significantly larger than that of WT, while the development of xylem was inhibited and the area was significantly smaller in RNAi silencing lines (Figure 8B). It is worth noting that the reduction of the 1, 2, and 3 formative rings in the overexpression lines is not as significant as the WT. This indicates that BvCPD promotes the development of xylem, and the 2 and 3 rings are the most significant.
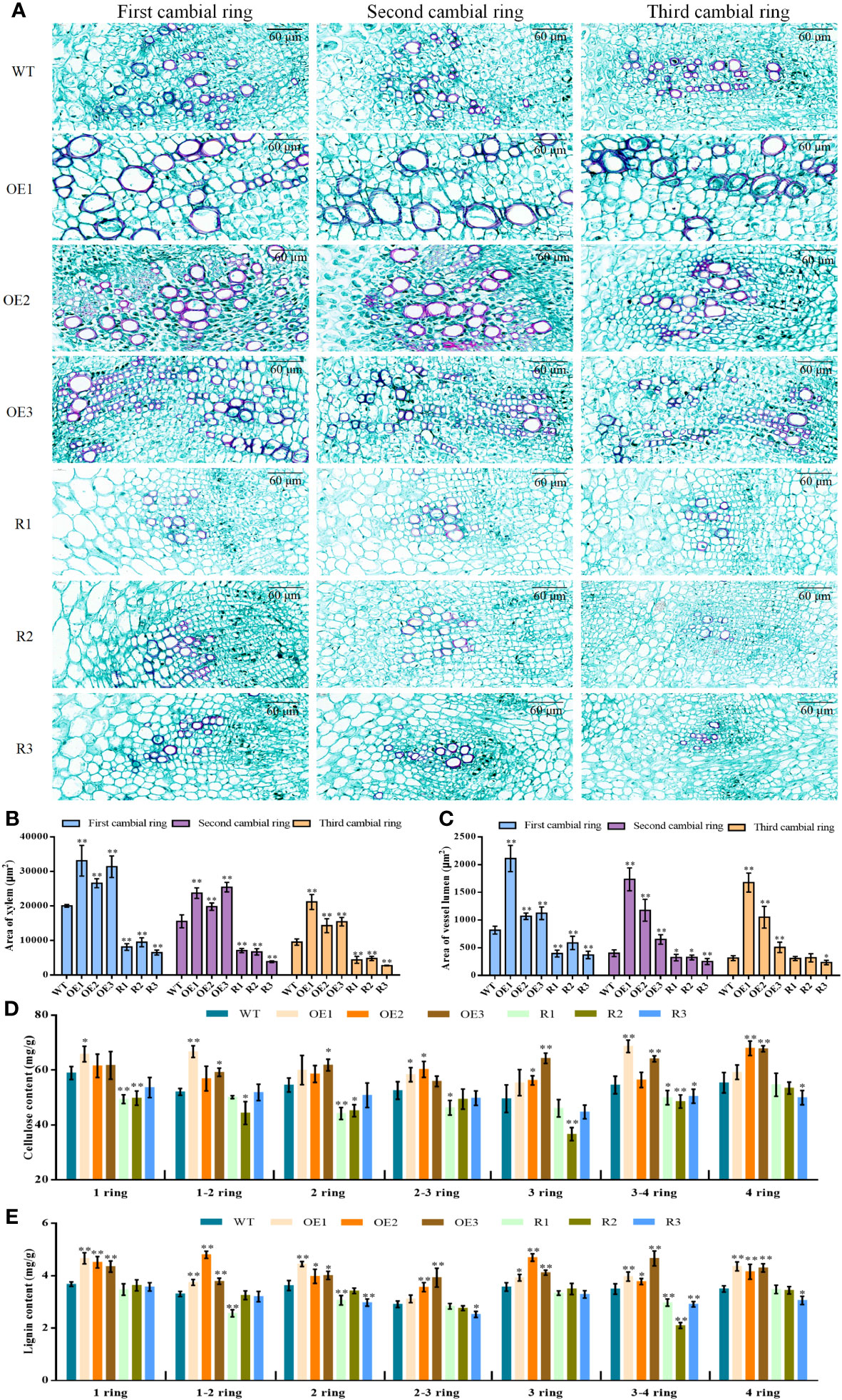
Figure 8 Xylem structure of the first, second, and third formative layers per unit area for each line. (A) Xylem structure of the first, second, and third formative layers. (B) Area of xylem in taproot 1,2,3 cambium. (C) Area of vessel lumen in taproot 1,2,3 cambium. (D) Cellulose content in taproot 1–4 cambium. (E) Lignin content in taproot 1–4 cambium. * Student’s-t test, P < 0.05; ** Student’s-t test, P < 0.01. Scale bars = 60 μm.
From microscopic observations, in the first, second, and third formative layers, ducts of the transgenic overexpression lines were all thicker compared with those of WT, while those of RNAi lines were slightly thinner compared with those of WT (Figure 8A). The vessel lumen areas of BvCPD overexpression lines were significantly higher compared with those of WT on the first, second, and third formative rings, while those of silenced lines were significantly lower compared with those of WT. There were highly significant differences for the first formative ring, but the differences gradually decreased in the second and third rings (Figure 8C). BvCPD overexpression lines had increased ductal pore lumen areas on the xylem of rings 1, 2, and 3, and the effect became more pronounced as the number of rings increased. The effect was more pronounced in the first and second formative loops of RNAi-interfering lines but weakened as the number of loops increased. In summary, overexpression of BvCPD was able to promote ductal development.
To assess whether BvCPD affects the composition of parenchyma cells and xylem in sugar beet taproot, cellulose and lignin contents in the 1–4 rings of each transgenic lines were measured. Cellulose and lignin contents were higher in overexpression lines than in WT (Figures 8D, E), but lower in silent lines compared with WT. This result shows that BvCPD affected the content of cellulose and lignin in 1–4 rings.
4 Discussion
C-3 oxidase, 5 α reductase, C-22 hydroxylase, C-23 hydroxylase, and C-6 oxidase are key enzymes in the BR synthesis pathway. CPD plays an important role as a C-3 oxidase in multiple steps of the BR synthesis pathway. Our results of subcellular localization revealed that BvCPD acts on the nucleus, cell membrane and cell wall. It was expressed in the taproot, petiole, and leaf of sugar beet, and was strongly expressed in the vascular bundles of various organs and parenchyma cells of taproot. This is slightly different from the other research. PeCPD of poplar was expressed in the endoplasmic reticulum in addition to being localized in the cell membrane (Wu et al., 2013). OsCPD1 and OsCPD2 of rice were both expressed in the endoplasmic reticulum (Zhan et al., 2022). Furthermore, AtCPD was strongly expression restricted to procambial cells of Arabidopsis roots when expressed under its natural promoter (Vukašinović et al., 2021). This indicates that the role of CPD in different plants also varies. The results of this study suggest that the expression of BvCPD in the nucleus, cell membrane, and cell wall may regulate cell wall relaxation, enhance the relative stability of the internal environment of cells, affect the development and differentiation of vascular bundles and parenchyma cells, and thus promote the expansion of sugar beet taproot. This is similar to the function of the poplar PeCPD gene, which regulates vascular development growth in Arabidopsis thaliana (Wu et al., 2013). Overexpression of BvCPD in sugar beet promotes BR synthesis, especially in roots. Unlike IAA and GA, BR lacks long-distance transport and acts only near the site of synthesis (Symons et al., 2008). Widespread expression of BvCPD in various tissues and organs may be more effective in maintaining local BR biosynthesis or gradients for optimal growth and development. The CPD gene in cotton and soybean both rescue root development, leaf expansion, and plant conformation in cpd Arabidopsis mutants (Wang et al., 2015; Liu et al., 2021). This was verified based on the observations in sugar beet BvCPD overexpressed and silenced plants.
BvCPD affected the morphological growth of sugar beet, especially the radial growth of the taproot. Different overexpression lines were taller than WT plants, with wide and long leaves, stout petioles, well-developed root systems, and clusters of lateral roots. Silent lines were shorter than WT, with short and narrow leaves and weak petioles. Further, individual lines also showed severe leaf curling and tiny root systems with almost no lateral roots. Notably, we found that BvCPD did not have a significant effect on the length of the taproot; however, it had a significant impact on the diameter of the taproot. When the Arabidopsis bri1 mutant was disturbed by BR signaling, both radial and lateral growth were reduced, but the reduction in radial growth was less than that in lateral growth (Fridman et al., 2021). This suggests that BvCPD plays an important role in regulating the radial growth of sugar beet roots. Low concentrations of BR induce lateral root formation, but for lateral root formation, BR and growth hormone have a synergistic effect—a process that may be partially mediated by phospholipase A (Bao et al., 2004). BvCPD is significantly upregulated during the taproot growth and sucrose accumulation stages in beet taproot (Zhang et al., 2017), indicating that BvCPD is associated with beetroot development. The evidence that BR autonomously promotes root growth in Arabidopsis thaliana also supports the idea that CPD can regulate plant root growth (Müssig et al., 2003). The most significant differences in the relative expression of BvCPD and content of BR were found in the root (as compared with the petiole and leaf of transgenic sugar beet), with corresponding changes in levels of IAA and GA. BvCPD increased the endogenous content of BR and enhanced BR signaling in sugar beet while downregulating other synthetic genes such as BvDET2, BvROT3, and BvBR6OX1. This is in general agreement with the results of Wu et al. (2013) and Zheng et al. (2018).
We speculated that overexpression of BvCPD would also mimic the high level of BR in sugar beet and produce feedback inhibition of BR-related synthase genes, thereby maintaining normal levels of Campestanol in the plant (Simona et al., 2002). Studies on Arabidopsis showed that the expression of AtDWF4, AtCPD, AtROT3, and AtCYP90D1 is negatively regulated by the BR signaling transcription factor AtBZR1/2 (He et al., 2005). Exogenous BR treatment inhibits the expression of AtDWF4, AtCPD, AtROT3, and AtCYP90D1 through AtBZR1/2 (Goda et al., 2004; Ohnishi et al., 2006). This study found a similar BR negative feedback regulation mechanism in sugar beets. BvBZR1 expression was upregulated to balance increased BR content in vivo, and BvBZR1 reduced the expression of BR synthesis-related enzyme genes BvDET2 and BvROT3 through negative feedback regulation. BvBR6OX1 is located downstream of BvCPD in the synthesis pathway, and it catalyzes the CS to BL transition. Unlike the two BR synthesis genes described above, BvBR6OX1 was downregulated in expression in both the overexpression and RNAi-silenced lines compared with WT. We hypothesized that in the overexpression lines, as in the other synthetic genes, negative feedback regulation represses BvBR6OX1 gene expression, owing to increased BR content. However, silent lines may not be affected by changes in BR content, but only by the disrupted expression of BvCPD, which generates less substrate after catalysis, ultimately leading to significant downregulation of BvBR6OX1. The GA biosynthetic pathway is linked to the BR signaling pathway via BZR1 and DELLA (Li et al., 2012). In addition, earlier studies reported that BRs could induce the expression of GAs synthesis genes (Bouquin et al., 2001). In this study, enhanced BR signaling caused upregulated expression of BvTAA1 and BvGA3OX1, stimulating the synthesis of IAA and GA. All three hormones were significantly increased in overexpressed sugar beet taproot. BR, IAA, and GA may act synergistically to affect sugar beet root growth and lead to taproot expansion. However, the specific regulatory mechanisms still need to be elucidated.
BvCPD promotes the division and expansion of parenchyma cells between the rings of the sugar beet taproot; additionally, it also promotes the development of the xylem of the vascular bundle and the lumen of the vessel. The synergy of the two factors makes the sugar beet taproot develop rapidly and increases the diameter of the taproot. In this study, BvCPD caused changes in endogenous hormones to produce changes in sugar beet root morphology. Therefore, cross-sectional taproot paraffin sections were made for the transgenic lines and WT. The first four rings on the taproot were observed. The ring spacing, number of cell layers, size of parenchyma cells, xylem area, and area of vessel lumen of BvCPD overexpressing lines were significantly larger than those of WT, whereas all indicators of silent lines were smaller. This indicates that BvCPD promotes BR synthesis and may exert synergistic effects with IAA and GA. This increases the size and number of parenchyma cells and the spacing between rings. HAT7 and GTL1 are regulatory factors induced by BR along cortical trajectories, controlling cell extension (Nolan et al., 2023). Exogenous BR treatment can induce the expression of cell Cyclin D3 to regulate cell division in roots (González-García et al., 2011). After enhanced BR signal in cotton, the transcription factor GhBES1.4 regulates the downstream target gene GhKRP6, causing it to interact with the G-type CDK protein GhCDKG that affects cell cycle splicing, ultimately regulating cell proliferation (Gu et al., 2023). We speculate that the increase or decrease in BR content caused by BvCPD in transgenic sugar beet strains will also regulate such genes, affecting the expansion of parenchyma cells in the taproot.
Moreover, BR accelerates the transport of water and nutrients by promoting the development of the xylem and ducts. Therefore, the beet experiences strong physiological metabolism that accelerates growth and development. The reason is that BR can promote cell wall growth by regulating genes associated with cell wall biosynthesis, such as BvXTH33, BvSHV3, BvCESA6, BvPARVUS, and BvCEL1, and it can also promote secondary xylem development (Wang et al., 2021). The BR biosynthetic mutant cpd, the BR receptor mutant bri1, and the BR signaling mutant bin2 all produced fewer vascular bundles. In contrast, overexpression of the BRI1 and BR signaling genes in plants results in more vascular bundles (Ibañes et al., 2009). Overexpression of BvCPD between and on different rings also promotes the accumulation of cellulose and lignin, the main components of the cell wall, which provides a reliable basis for the development of vascular bundles. The relative expression of PbrMYB24 in pear flesh is significantly positively correlated with lignin and cellulose content. PbrMYB169 and PbrNSC can activate the promoter of PbrMYB24, enhancing gene expression (Xue et al., 2023). The LAC4 and LAC17 genes can also affect the content of lignin, especially the silencing of LAC17, which specifically affects the deposition of G–lignin units in fibers (Serge et al., 2011). Therefore, future research should focus on the mechanism of action of BvCPD in the growth of sugar beet taproot, and identify genes that are regulated to cause changes in cellulose and lignin content.
In summary, CPD plays a major role in root development. Overexpression of BvCPD in taproot increases BR levels and upregulates BvBZR1 expression. BvBZR1 regulates BvTAA1 and BvGA3OX1 to increase IAA and GA contents. We speculate that the three hormones act synergistically to induce changes in parenchyma cells and vascular bundles to promote taproot expansion (Figure 9). Reduced BvCPD expression restricts taproot expansion. This study provides direct genetic evidence that BvCPD is a functional gene for BR synthase in sugar beet and affects the lateral root growth of beet. Our results provide a theoretical basis for sugar beet root expansion study.
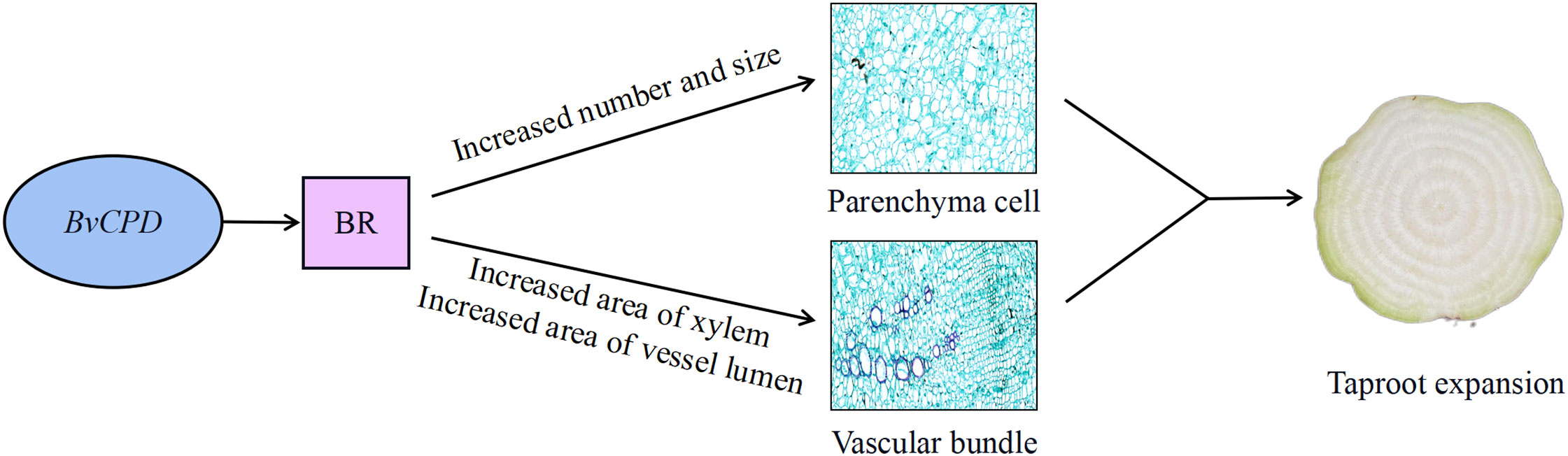
Figure 9 Hypothetical model of BvCPD roles in taproot expansion of sugar beet. Overexpression of BvCPD in taproot increases BR levels and induces changes in parenchyma cells and vascular bundles to promote taproot expansion. Arrows indicate positive effects.
5 Conclusions
BvCPD promotes the synthesis of BR and causes changes in endogenous IAA and GA contents. This increases taproot diameter by regulating the number and size of parenchyma cells and areas of the xylem and vessel lumen of vascular bundles, leading to the expansion of sugar beet taproot. This is the first report to identify the function of CPD in sugar beet and provides a basis for further studies on the mechanism of action of CPD in sugar beet.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding authors.
Author contributions
XG: Data curation, Writing – original draft, Writing – review & editing. YL: Writing – review & editing. NL: Writing – review & editing. GL: Writing – review & editing. YS: Writing – review & editing. SZ: Project administration, Writing – review & editing.
Funding
This work was supported by the following grants: the Inner Mongolia Autonomous Region ‘‘the open competition mechanism to select the best candidates’’ project (2022JBGS0029), the National Natural Science Foundation of China (31760416), the Postgraduate research funding innovation project (DC2100002433), and the Basic Research Funds for Universities (RZ2200001154).
Acknowledgments
We would like to thank Dr. Wei Wang and Dr. Binjie Ma for providing valuable advice on the experimental methods of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1271329/full#supplementary-material
References
Avalbaev, A., Yuldashev, R., Fedorova, K., Petrova, N., Fedina, E., Gilmanova, R., et al. (2021). 24-epibrassinolide-induced growth promotion of wheat seedlings is associated with the changes in proteome and tyrosine phosphoproteome. Plant Biol. 23, 456–463. doi: 10.1111/plb.13233
Bai, C., Zheng, Y., Watkins, C. B., Fu, A., Ma, L., Gao, H. W., et al. (2021). Revealing the specific regulations of brassinolide on tomato fruit chilling injury by integrated multi-omics. Front. Nutr. 8. doi: 10.3389/FNUT.2021.769715
Bao, F., Shen, J., Brady, S. R., Muday, G. K., Asami, T., Yang, Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134, 1624–1631. doi: 10.1104/pp.103.036897
Bouquin, T., Meier, C., Foster, R., Nielsen, M. E., Mundy, J. (2001). Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 127, 450–458. doi: 10.1104/pp.010173
Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351. doi: 10.1242/dev.01403
Chen, X., Xue, H., Zhu, L., Wang, H., Long, H., Zhao, J., et al. (2022). ERF49 mediates brassinosteroid regulation of heat stress tolerance in Arabidopsis thaliana. BMC Biol. 20, 254–254. doi: 10.1186/S12915-022-01455-4
Choe, S., Noguchi, T., Fujioka, S., Takatsuto, S., Tissier, C. P., Gregory, B. D., et al. (1999). The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 11, 207–221. doi: 10.1105/tpc.11.2.207
Feng, Q., Ou, Y., Han, Y., De, D. V. R., Wang, L., Zhang, Q., et al. (2021). The brassinosteroid biosynthesis enzyme gene PeCPD improves plant growth and salt tolerance in Populus tomentosa. Ind. Crops Products 162, 113218. doi: 10.1016/J.INDCROP.2020.113218
Fridman, Y., Strauss, S., Horev, G., Ackerman, L., Reiner, B., Lane, B., Smith, RS., et al. (2021). The root meristem is shaped by brassinosteroid control of cell geometry. Nature Plants. 7, 1475–1484. doi: 10.1038/S41477-021-01014-9
Goda, H., Sawa, S., Asami, T., Fujioka, S., Shimada, Y., Yoshida, S. (2004). Comprehensive comparison brassinosteroid-regulated of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134, 1555–1573. doi: 10.1104/pp.103.034736
González-García, M. P., Vilarrasa-Blasi, J., Zhiponova, M., Divol, F., Mora-García, S., Russinova, E., et al. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859. doi: 10.1242/dev.057331
Gu, Y., Zhang, J., Liu, L., Qanmber, G., Liu, Z., Xing, K., et al. (2023). Cell cycle-dependent kinase inhibitor GhKRP6, a direct target of GhBES1.4, participates in BR regulation of cell expansion in cotton. Plant J. doi: 10.1111/tpj.16353
Guo, Z., Zhang, Z., Yang, X., Yin, K., Chen, Y., Zhang, Z., et al. (2020). PSBR1, encoding a mitochondrial protein, is regulated by brassinosteroid in moso bamboo (Phyllostachys edulis. Plant Mol. Biol. 103, 1–12. doi: 10.1007/s11103-020-00975-3
He, J.-X., Gendron, J., Sun, Y., Gampala, S., Gendron, N., Sun, C., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
Huang, Y., Dong, H., Mou, C., Wang, P., Hao, Q., Zhang, M., et al. (2022). Ribonuclease H-like gene SMALL GRAIN2 regulates grain size in rice through brassinosteroid signaling pathway. J. Integr. Plant Biol. 64, 1883–1900. doi: 10.1111/jipb.13333
Hyunjung, J., Jihye, D., Soo-Jeong, S., Joon, W. C., Young, I. C., Wook, K., et al. (2014). Exogenously applied 24- epi brassinolide reduces lignification and alters cell wall carbohydrate biosynthesis in the secondary xylem of Liriodendron tulipifera. Phytochemistry 101, 40–51. doi: 10.1016/j.phytochem.2014.02.003
Ibañes, M., Fàbregas, N., Chory, J., Caño, D. A. I. (2009). Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. U. S. A. 106, 13630–13635. doi: 10.1073/pnas.0906416106
Li, S., Liu, Z., Chen, G., Qanmber, G., Lu, L., Zhang, J., et al. (2021). Identification and analysis of ghEXO gene family indicated that ghEXO7_At promotes plant growth and development through brassinosteroid signaling in cotton (Gossypium hirsutum L.). Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.719889
Li, Q.-F., Wang, C., Jiang, L., Li, S., Sun, S. S. M., He, J.-X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5, ra72. doi: 10.1126/scisignal.2002908
Liu, L., Xie, Z., Lu, L., Qanmber, G., Chen, G., Li, S., et al. (2021). Identification of BR biosynthesis genes in cotton reveals that GhCPD-3 restores BR biosynthesis and mediates plant growth and development. Planta 254, 75. doi: 10.1007/S00425-021-03727-9
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real- time quantitative PCR and the 2-ΔΔCTMethod. METHODS 25, 402–408. doi: 10.1006/meth.2001.1262
Maia, C. F., Silva, B. R. S., Lobato, A. K. S. (2018). Brassinosteroids positively modulate growth, physiological, biochemical and anatomical evidence using two tomato genotypes contrasting to dwarfism. J. Plant Growth Regulation 37, 1099–1112. doi: 10.1007/s00344-018-9802-2
Mathur, J., Molnár, G., Fujioka, S., Takatsuto, S., Sakurai, A., Yokota, T., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14, 593–602. doi: 10.1046/j.1365-313X.1998.00158.x
Miklós, S., Kinga, N., Zsuzsanna, K.-K., Jaideep, M., Annette, K., Thomas, A., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Müssig, C., Shin, G.-H., Altmann, T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133, 1261–1271. doi: 10.1104/pp.103.028662
Nolan, T. M., Vukašinović, N., Hsu, C. W., Zhang, J., Vanhoutte, I., Shahan, R., et al. (2023). Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science 379, eadf4721. doi: 10.1126/SCIENCE.ADF4721
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids, multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 32, 295–318. doi: 10.1105/tpc.19.00335
Ohnishi, T., Godza, B., Watanabe, B., Fujioka, S., Hategan, L., Ide, K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287, 31551–31560. doi: 10.1074/jbc.M112.392720
Ohnishi, T., Szatmari, A., Watanabe, B., Fujita, S., Bancos, S., Koncz, C., et al. (2006). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell. 18, 3275–3288. doi: 10.1105/tpc.106.045443
Pan, Z., Durst, F., Werck, R. D., Gardner, H. W., Camara, B., Cornish, K., et al. (1995). The major protein of guayule rubber particles is a cytochrome P450. Characterization based on cDNA cloning and spectroscopic analysis of the solubilized enzyme and its reaction products. J. Biol. Chem. 270, 8487–8494. doi: 10.1074/JBC.270.15.8487
Rae, A. L., Perroux, J. M., Grof, C. P. L. (2005). Sucrose partitioning between vascular bundles and storage parenchyma in the sugarcane stem, a potential role for the ShSUT1 sucrose transporter. Planta 220, 817–825. doi: 10.1007/s00425-004-1399-y
Ramesh, D., Ketemaw, S., Abiyu, K. B. (2017). Production of Beet Sugar and Bio-ethanol from Sugar beet and it Bagasse, A Review. Int. J. Eng. Trends Technol. (IJETT) 43, 222–233. doi: 10.14445/22315381/IJETT-V43P237
Serge, B., Nathalie, D. C., Brigitte, P., Przemyslaw, B., Laurent, C., Phillipe, L., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 23, 1124–1137. doi: 10.1105/tpc.110.082792
Simona, B., Takahito, N., Tatsuro, S., Gergely, M., Bishop, G. J., Csaba, K., et al. (2002). Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513. doi: 10.1104.005439/pp
Su, W., Shao, Z., Wang, M., Gan, X., Yang, X., Lin, S. (2021). EjBZR1 represses fruit enlargement by binding to the EjCYP90 promoter in loquat. Hortic. Res. 8, 152–152. doi: 10.1038/S41438-021-00586-Z
Symons, G. M., Ross, J. J., Jager, C. E., Reid, J. B. (2008). Brassinosteroid transport. J. Exp. Botany 59, 17–24. doi: 10.1093/jxb/erm098
Tian, X., Xuan, L., Liu, B., Hu, T., Wang, C., Wang, X. (2018). Effects of heterologous expression of Populus euphratica brassinosteroids biosynthetic enzyme genes CPD (PeCPD) and DWF4 (PeDWF4) on tissue dedifferentiation and growth of Arabidopsis thaliana seedlings. Plant Cell Tissue Organ Culture (PCTOC) 132, 111–121. doi: 10.1007/s11240-017-1316-2
Vukašinović, N., Wang, Y., Vanhoutte, I., Fendrych, M., Guo, B., Kvasnica, M., et al. (2021). Local brassinosteroid biosynthesis enables optimal root growth. Nat. Plants 7, 619–632. doi: 10.1038/S41477-021-00917-X
Wang, T., Li, Q., Lou, S., Yang, Y., Peng, L., Lin, Z., et al. (2019). GSK3/shaggy-like kinase 1 ubiquitously regulates cell growth from Arabidopsis to Moso bamboo (Phyllostachys edulis). Plant Sci. 283, 290–300. doi: 10.1016/j.plantsci.2019.03.015
Wang, W., Sun, Y., Li, G., Zhang, S. (2021). Brassinosteroids promote parenchyma cell and secondary xylem development in sugar beet (Beta vulgaris L.) root. Plant Direct. 5, e340. doi: 10.1002/PLD3.340
Wang, M., Xu, X., Zhang, X., Sun, S., Wu, C., Hou, W., et al. (2015). Functional analysis of GmCPDs and investigation of their roles in flowering. PloS One 10, e0118476. doi: 10.1371/journal.pone.0118476
Wei, Z., Li, J. (2016). Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 9, 86–100. doi: 10.1016/j.molp.2015.12.003
Wu, H., Xu, D., Si, J., Lian, G., Qin, Y., Chen, X., et al. (2013). Cloning,characterization,and functional analysis of Populus euphratica CPD(PeCPD),a key brassinosteroid(BR) biosynthesis gene. Chin. Soc. Cell Biol. 260, 252.
Xu, H., Sun, H., Dong, J., Ma, C., Li, J., Li, Z., et al. (2022). The brassinosteroid biosynthesis gene TaD11-2A controls grain size and its elite haplotype improves wheat grain yields. Theor. Appl. Genet. 135, 2907–2923. doi: 10.1007/S00122-022-04158-0
Xue, Y., Shan, Y., Yao, J., Wang, R., Xu, S., Liu, D., et al. (2023). The transcription factor PbrMYB24 regulates lignin and cellulose biosynthesis in stone cells of pear fruits. Plant Physiol. 192, 1997–2014. doi: 10.1093/PLPHYS/KIAD200
Zhan, H., Lu, M., Luo, Q., Tan, F., Zhao, Z., Liu, M., et al. (2022). OsCPD1 and OsCPD2 are functional brassinosteroid biosynthesis genes in rice. Plant Sci. 325, 111482–111482. doi: 10.1016/J.PLANTSCI.2022.111482
Zhang, Y.-F., Li, G.-L., Wang, X.-F., Sun, Y.-Q., Zhang, S.-Y. (2017). Transcriptomic profiling of taproot growth and sucrose accumulation in sugar beet (Beta vulgaris L.) at different developmental stages. PloS One 12, e0175454. doi: 10.1371/journal.pone.0175454
Zheng, L., Zhao, C., Mao, J., Song, C., Ma, J., Zhang, D., et al. (2018). Genome-wide identification and expression analysis of brassinosteroid biosynthesis and metabolism genes regulating apple tree shoot and lateral root growth. J. Plant Physiol. 231, 68–85. doi: 10.1016/j.jplph.2018.09.002
Zhou, X., Zhang, N., Yang, J., Si, H. (2016). Functional analysis of potato CPD gene, A rate-limiting enzyme in brassinosteroid biosynthesis under polyethylene glycol-induced osmotic stress. Crop Sci. 56, 2675–2687. doi: 10.2135/cropsci2016.01.0067
Keywords: brassinosteroid, BvCPD gene, sugar beet, transgenic plant, parenchyma cell, vascular bundle development
Citation: Guo X, Li Y, Li N, Li G, Sun Y and Zhang S (2023) BvCPD promotes parenchyma cell and vascular bundle development in sugar beet (Beta vulgaris L.) taproot. Front. Plant Sci. 14:1271329. doi: 10.3389/fpls.2023.1271329
Received: 02 August 2023; Accepted: 28 August 2023;
Published: 12 September 2023.
Edited by:
Wenyi Wang, South China Agricultural University, Guangzhou, ChinaReviewed by:
Lei Liu, Jilin Agriculture University, Changchun, ChinaYuguang Wang, Heilongjiang University, Harbin, China
Copyright © 2023 Guo, Li, Li, Li, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoying Zhang, c3l6aDM2QGFsaXl1bi5jb20=; Yaqing Sun, c3lhcWluZ0BpbWF1LmVkdS5jbg==
 Xiaotong Guo
Xiaotong Guo Yue Li
Yue Li Ningning Li
Ningning Li Guolong Li
Guolong Li Yaqing Sun
Yaqing Sun Shaoying Zhang
Shaoying Zhang