- School of Life Science and Engineering, Southwest Jiaotong University, Chengdu, Sichuan, China
Due to global climate change, drought is emerging as a major threat to plant growth and agricultural productivity. Abscisic acid (ABA) has been implicated in plant drought tolerance, however, its retarding effects on plant growth cannot be ignored. The reactions catalyzed by 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) proteins are critical steps within the isoprenoid biosynthesis in plants. Here, five DXS (CtDXS1-5) and two DXR (CtDXR1-2) genes were identified from Cassia tora genome. Based on multiple assays including the phylogeny, cis-acting element, expression pattern, and subcellular localization, CtDXS1 and CtDXR1 genes might be potential candidates controlling the isoprenoid biosynthesis. Intriguingly, CtDXS1 transgenic plants resulted in drought tolerance but retardant growth, while CtDXR1 transgenic plants exhibited both enhanced drought tolerance and increased growth. By comparison of β-carotene, chlorophyll, abscisic acid (ABA) and gibberellin 3 (GA3) contents in wild-type and transgenic plants, the absolute contents and (or) altered GA3/ABA levels were suggested to be responsible for the balance between drought tolerance and plant growth. The transcriptome of CtDXR1 transgenic plants suggested that the transcript levels of key genes, such as DXS, 9-cis-epoxycarotenoid dioxygenases (NCED), ent-kaurene synthase (KS) and etc, involved with chlorophyll, β-carotene, ABA and GA3 biosynthesis were induced and their contents increased accordingly. Collectively, the trade-off effect induced by CtDXR1 was associated with redesigning architecture in phytohormone homeostasis and thus was highlighted for future breeding purposes.
1 Introduction
Drought stress is one of the primary factors limiting the global geographic distribution of plants as well as their growth and productivity (Fang and Xiong, 2015). To counteract drought stress, plants have evolved a range of response mechanisms, one of which is the phytohormone-mediated pathway. Abscisic acid (ABA) is the main hormone that plants use in response to drought stress and plays a key role in plant growth and development (Lee and Luan, 2012). ABA together with stress response transcription factors, including DREBs and AREBs, trigger various physiological processes and metabolic changes that allow whole tissues to acquire drought tolerance (Muhammad Aslam et al., 2022). However, over the years, there has been increasing lines of evidence of delayed effects of ABA on normal plant growth (De Smet et al., 2003; Sreenivasulu et al., 2012). In Arabidopsis, the reduction in ABA content promoted root growth (Miao et al., 2021). Conversely, exogenous application of ABA inhibited lateral root emergence (De Smet et al., 2003), rhizoidal infection, and nodule formation (Xu et al., 2021). Similarly, overexpression of ABA-induced MAPKs resulted in seedlings and leaf senescence (Matsuoka et al., 2018). Overexpression of protein phosphatase 2C (PP2C), a negative regulator of ABA signaling, increased plant biomass production mainly due to accelerated inflorescence stem growth (Sugimoto et al., 2014). Thus, a key challenge for the development of next-generation agriculture is to balance agro-industrial needs (enhanced plant growth yield) and plant adaptation during the growing season (tolerance to abiotic stresses), and ABA antagonistic regulators have been further introduced to address this challenge (Zhang et al., 2022).
The plant hormone gibberellin (GA) is generally considered a negative regulator of ABA and mediates many physiological processes antagonistically with ABA (Shu et al., 2018). ABA and GA are 15- and 20-carbon plant hormones, respectively, and can be broadly attributed to isoprenoids. In addition to ABA and GA, isoprenoids also include photosynthetic pigments (chlorophyll and carotenoid), electron transporters (plastoquinone and ubiquinone), structural components of membranes (phytosterols), and anti-pathogen agents (phytoalexins), which play a role in nutrient accumulation, photosynthesis, plant growth and development and adaptation to environmental stresses. In addition to these plant-specific functions, many plants isoprenoids have been shown to be of industrial and therapeutic interest. The plant-produced tetraterpene β-carotene (provitamin A) is one of basic nutrients needed to maintain human health. Intriguingly, the terpenoids and sesquiterpenes produced by rhizosphere-inhabiting bacteria/fungi have been reported to potentially promote plant growth and stress tolerance (Tyagi et al., 2018). In plants, isoprenoid is synthesized mainly by sequential condensation and modification of two common building-blocks, isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMADP), via the mevalonic acid (MVA) pathway in the cytosol and the 2-methyl-d-erythritol-4-phosphate (MEP) pathway in the plastid. From the common intermediate, geranylgeranyl diphosphate (GGPP), the biosynthetic pathways of GA and ABA diverge (Rodríguez-Concepción and Boronat, 2015). Since GA and ABA share common upstream biosynthetic pathways and common intermediate, the genetic manipulation on the genes in the upstream biosynthetic pathways might simultaneously increase the GA and ABA contents. In the MEP pathway, 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) catalyze the first and second steps, respectively. In previous studies, DXS and DXR were considered as different regulators of fluxes during the formation of various isoprenoids (You et al., 2020) and their roles remained elusive. Specifically, both AtDXS and AtDXR have been suggested to be rate-limiting enzymes that increase carotenoid, chlorophyll, and ginkgolide content (Estévez et al., 2001; Carretero-Paulet et al., 2006; Kim et al., 2006), yet only DXS but not DXR caused increases in carotenoids and chlorophyll in Daucus carota or rice (Simpson et al., 2016; You et al., 2020), and increases in ABA and GA in poplars (Wei et al., 2019). In transgenic Arabidopsis plants with overexpressed DXS, the isoprenoid products were not markedly increased due to diversion of flux via 2-C-methylerythritol-2,4-cyclodiphosphate (MEcDP) (Wright et al., 2014). On the other hand, overexpression of the Lilium DXR gene has also been reported to increase monoterpene content in tobacco flowers (Zhang et al., 2018). In this way, plant DXS and DXR genes have been suggested to function in isoprenoid metabolism in a species- or organ-specific manner, highlighting the significance of elucidating the distinct roles of DXS and DXR genes in various plant species (You et al., 2020).
Cassia tora L. (Leguminosae), also known as Senna tora, is an important herb that grows mainly in Asia and Africa and has attracted attention for centuries due to its widespread use as a traditional medicine and food ingredient. Consumer awareness and knowledge of the health benefits of C. tora have greatly stimulated the marketing and consumption of C. tora products, which contain a variety of bioactive compounds such as betulinic acid, flavonoids, rotenoids, stigmasterol and β-carotene (Vats and Kamal, 2014; Vats, 2018; Górnaś et al., 2019). Besides, C. tora contains a large amount of anthraquinones whose part of backbone is biosynthesized via the MEP pathway (Liu et al., 2014). However, few studies on genes involved in MEP pathway in C. tora have been reported, mainly due to the lack of the C. tora genome. Recently, thorough genomic analysis and functional identification of candidate genes involved in the MEP pathway of C. tora became possible due to the availability of genomic data of C. tora (Kang et al., 2020). Therefore, the objectives of this study were (1) to perform an in-depth analysis of the DXS and DXR genes of C. tora (ASM1485142v1) by structural and functional annotation. Accordingly, the CtDXS1 and CtDXR1 genes, which may play a role in isoprenoid biosynthesis, were selected as candidates for further studies; (2) to investigate the different roles of the CtDXS1 and CtDXR1 genes in isoprenoid biosynthesis, plant growth, and drought tolerance; and (3) to put forward the potential molecular mechanism to achieve a trade-off between drought tolerance and plant growth. As a consequence, this report provides, for the first time, a basis for further investigation of the biological functions of DXS and DXR genes in C. tora, which can be used to develop genomic strategies for next-generation agriculture.
2 Results
2.1 Genome-wide identification of DXS and DXR genes in C. tora
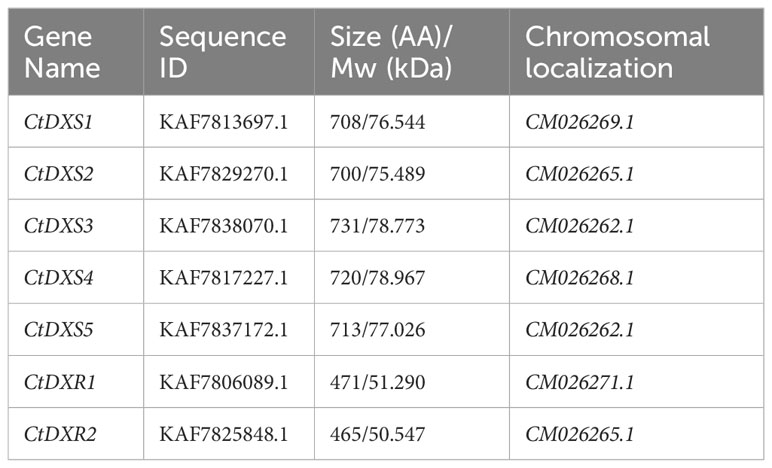
DXSs and DXRs are key regulators of the MEP pathway and are conserved in the plant kingdom (Zeng and Dehesh, 2021). The genomes of Arabidopsis, soybean, and Panicum virgatum encode 3, 10, and 11 DXS genes, respectively, whereas the genome of algae contains only one DXS gene copy (de Luna-Valdez et al., 2021). In Arabidopsis, rice and Coffea arabica, DXR is encoded by a single-copy gene (Silva et al., 2020). In this study, a genome-wide analysis of the C. tora genome was performed to identify DXS and DXR genes. There are two main methods for identifying DXS and DXR family genes. In the first approach, a BlastP search was performed in the C. tora genome database using hidden Markov model (HMM) profiles of known DXS and DXR sequences. In the second method, NCBI’s conserved domain database (CDD) was performed to validate DXS and DXR members. After removing redundant sequences, a total of five DXS and two DXR genes were identified in C. tora (Table 1). Gene names, sequence numbers, protein sizes, molecular weights and chromosomal locations are listed in Table 1.
The nucleotide sequences of five CtDXS genes were phylogenetically analyzed with the plant, algae, and bacteria DXS sequences. Consistent with previous reports (You et al., 2020; de Luna-Valdez et al., 2021), plant DXS was clustered into three major clades, namely, clade I including CtDXS1 and CtDXS5, clade II including CtDXS2 and CtDXS3, and clade III including CtDXS4. All DXS clades were monophyletic clusters with highly supported bootstrap (100, 86 and 100 BP, respectively). Furthermore, our phylogenetic results indicated that DXSs from the same species, such as Oryza sativa (abbreviated as Os in Figure 1), Medicago truncatula (abbreviated as Mt in Figure 1), and C. tora, were distributed in different clades, suggesting that the replication that gave rise to these different clades occurred prior to the divergence of these species. Members of clade I have been reported to play important roles in the biosynthesis of housekeeping and photosynthetic terpenoids such as chlorophylls and carotenoids, whose gene expression is dependent on light conditions (You et al., 2020). Members in clade II play a minor ecological role in the production of functional terpene metabolites, whereas clade III members are associated with postembryonic development in Arabidopsis and are not involved in primary metabolism and isoprenoid synthesis (de Luna-Valdez et al., 2021). In phylogenetic analysis (Figure 1A), CtDXS1 was closely related to legumes and had the highest sequence similarity to MtDXS1 (AJ430047.1), which encoded an enzyme with significant DXS activity, i.e., it catalyzed glyceraldehyde 3-phosphate and pyruvate to produce DXP (Walter et al., 2002), so it was subsequently selected to study its effect on isoprenoid biosynthesis. Based on the results of amino acid comparison and classification of conserved structural domains, the CtDXS1 protein belongs to the transketolase superfamily. For example, similar to other DXSs, CtDXS1 contains a TPP-binding domain (coenzyme of DXS) in motif 10 at the N-terminal end with the highly conserved sequences -GDG- and -LNDN- at the starting and end points, respectively (Figure S1). In addition, the His residue at position 93 (H93) in motif 5 at the N terminus of CtDXS1 showed a good match with His49 in Escherichia coli DXS, which has been reported to be involved in the catalytic action of DXS (Yang et al., 2012c). Invariant Asp294 residue in motif 7 was also found in the intermediate region of CtDXS1, and is thought to be specific for the enzymatic activity (Crowell et al., 2003). In addition, a DRAG structural domain, which is thought to play an important role, was found in motif 2 at its C-terminus, which starts and ends with the sequences -HCGS- and -PSD, respectively, similar to other plant DXSs. The highly conserved H499 was proposed to interact with glyceraldehyde 3-phosphate (Carretero-Paulet et al., 2013).
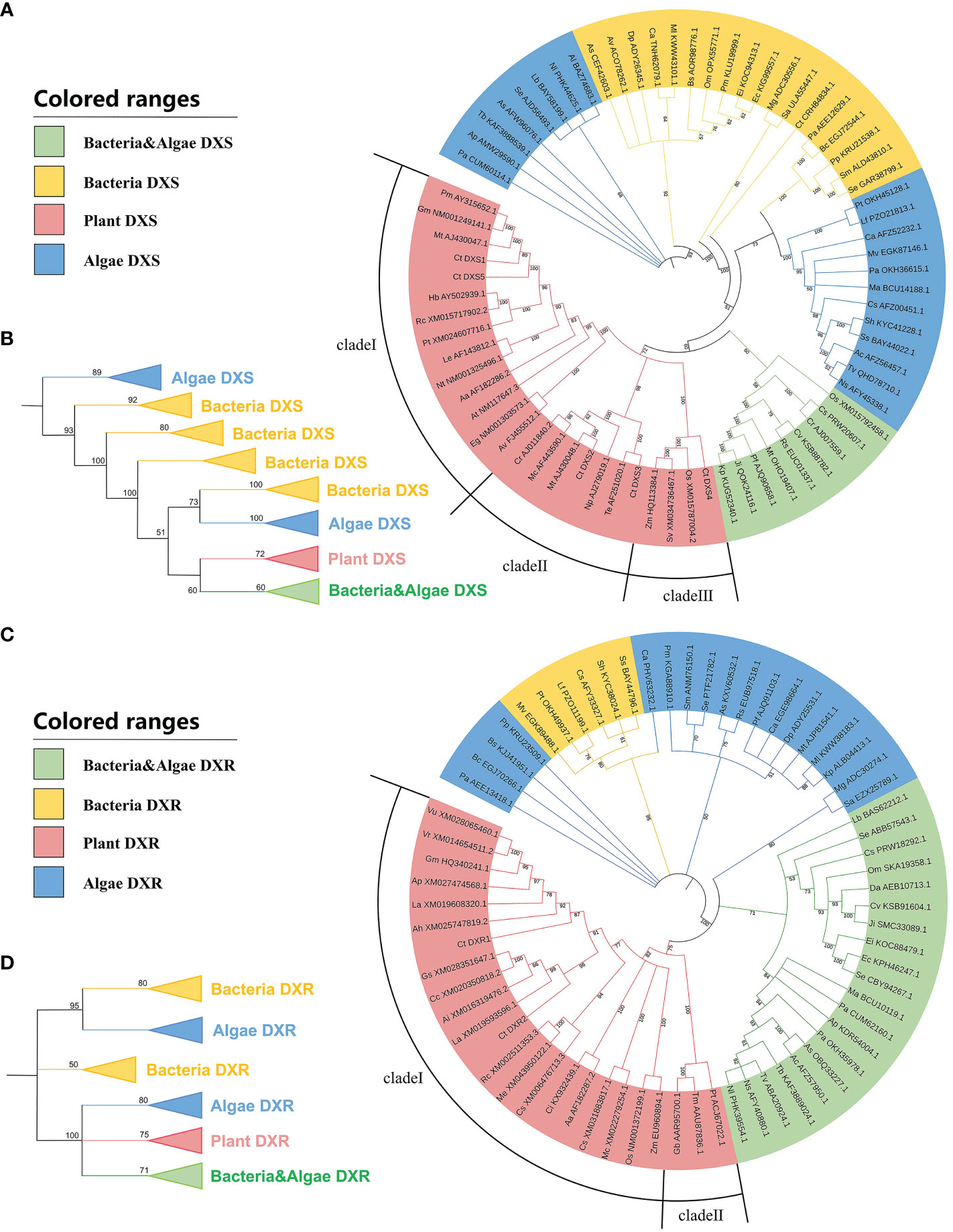
Figure 1 Phylogenetic analysis of the DXSs and DXRs. The bootstrap values were represented above the nodes. (A) NJ tree of DXSs from various organisms. (B) Topological structure of the NJ tree of DXSs. (C) NJ tree of DXRs from various organisms. (D) Topological structure of the NJ tree of DXRs.
C. tora DXRs included two members (KAF7806089.1 and KAF7825848.1) that were phylogenetically assigned to plant DXR clade I, which included the legumes Glycine max (abbreviated as Gm in Figure 1) and M. truncatula (Figure 1C). In respect to CtDXR2, CtDXR1 was more closely related to GmDXR1 (accession no HQ340241.1), which played a role in biosynthesis of isoprenoids, i.e. GmDXR1-overexpressing plants exhibited increasing isoprenoid content (Zhang et al., 2012). The enzyme consists of three distinct structural domains: an N-terminal NADPH-binding domain, a central linker domain and a C-terminal helical structural domain (Proteau, 2004). Since NADPH is involved in the catalytic reaction of DXR, the NADPH-binding structural domain is essential for the catalytic activity of DXR. Two highly conserved NADPH binding regions, GSTGS(I/V)GT and LAAGSN(V/I), are identified in motif 5 in the N-terminal structural domain of CtDXR1, while two substrate binding regions, LPADSEHSAI in motif 4 and NKGLEVIEAHY in motif 2, located in its central linker domain, are highly homologous to DXRs from other plants (Figure S2).
Furthermore, as an annual land plant, C. tora shows a symbiotic relationship with bacterial endophytes (Kumar et al., 2015). To confirm whether the CtDXS1 and CtDXR1 genes were indeed derived from C. tora, we performed a series of exercises. The first thing we did was to search for homologous regions of both genes using Blastn. The highest scoring sequences were DXS from Prosopis alba (XP_028781649.1), and DXR from Prosopis alba (XM_028922493.1), respectively. In addition, phylogenetic analysis using nucleotide sequences of DXSs and DXRs from plants and microorganisms demonstrated that CtDXS1 and CtDXR1 were clustered into plant clade (Figures 1B, D). Third, the average identity of CtDXS1 with plant DXSs was approximately 5% higher than that with microorganism DXSs. while the average identity of CtDXR1 with plant DXRs was approximately 9% higher than that with microorganism DXRs, respectively. The sequences of CtDXS1 and CtDXR1 were much more similar to those of other plants, while both were far from those of microorganisms. Therefore, it was inferred that CtDXS1 and CtDXR1 were genuine member from C. tora and not endophytes.
2.2 Cis-acting elements analysis of CtDXS and CtDXR genes
The identification of cis-acting elements may help to determine the molecular functions of CtDXS and CtDXR genes. Based on the corresponding gene sequences, cis-acting elements were obtained using PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) analysis. The results showed that the promoters of the CtDXS1, CtDXS3, CtDXS5, CtDXR1 and CtDXR2 genes have multiple cis-acting elements involved in stress responses and hormones (Figure 2A). These include ABRE (response to ABA, ACGTG), MYB (response to stress, GA and ABA, TAA CCA), CGTCA-motif (response to stress and MeJA, CGTCA), and TGACG-motif (response to stress and MeJA, TGACG). In contrast, the promoter of the CtDXS2 gene lacks the ABRE and TGACG-motif, and the promoter of the CtDXS4 gene lacks the TGACG-motif. Therefore, we can infer that these five genes with multiple response elements may be involved in plant responses to abiotic stresses.
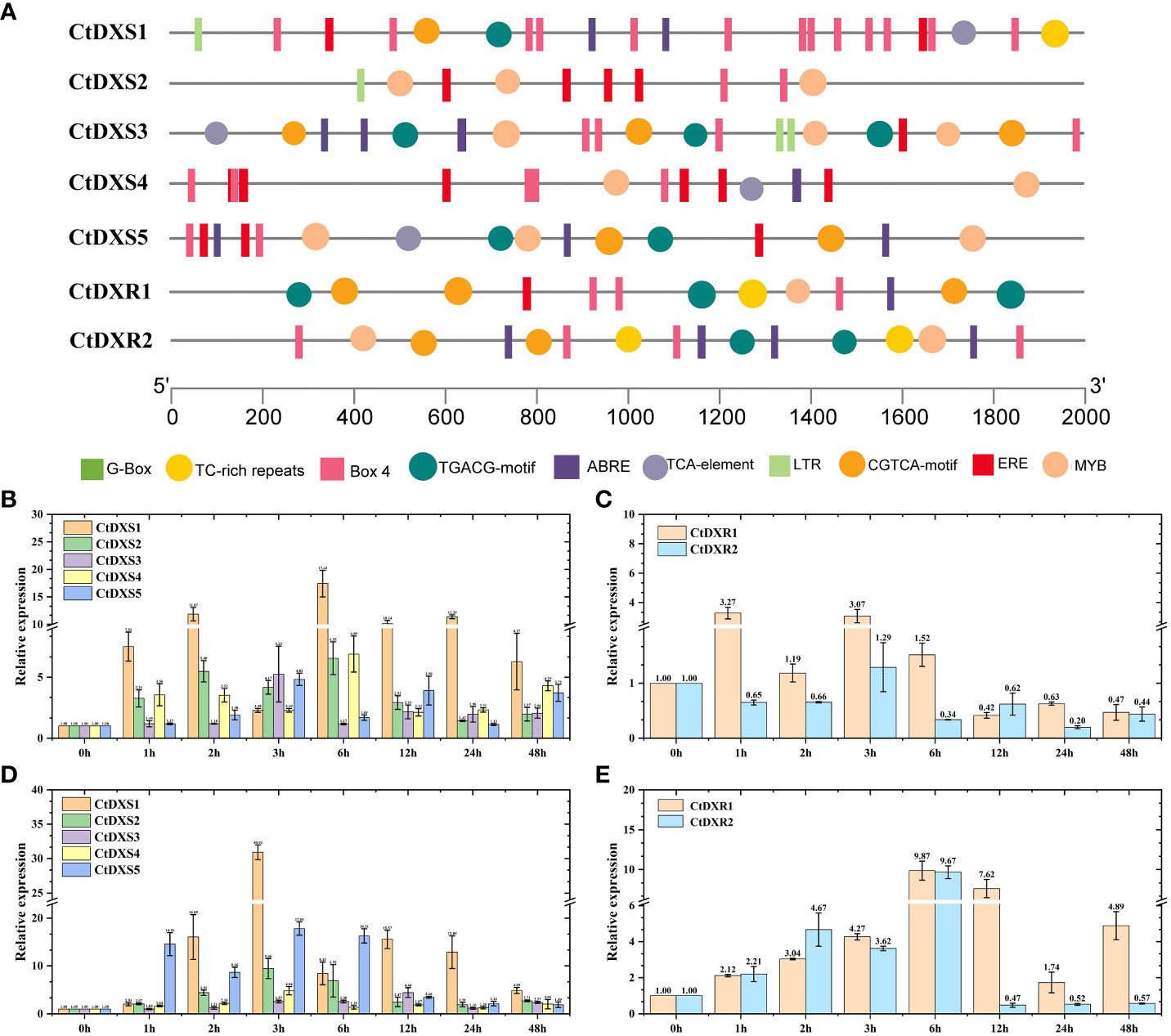
Figure 2 Cis-element analysis and expressional pattern of CtDXS and CtDXR genes. (A) Cis-acting elements of CtDXS and CtDXR genes. Real-time PCR of CtDXS (B) and CtDXR (C) genes under simulated drought treatment. Real-time PCR of CtDXS (D) and CtDXR (E) genes under ABA treatment. To normalize the relative expression level, Elongation factor 1a was used as an internal control.
2.3 The expressional pattern of CtDXS and CtDXR genes
As shown in Figures 2B–E, both CtDXS1 and CtDXR1 genes were sensitive under drought and ABA treatments. Under drought treatment, the expression level of CtDXS1 gene gradually increased and reached the highest expression level (17.42-fold) at 6 h (Figure 2B), whereas the expression level of CtDXR1 gene increased rapidly and reached the highest expression level at 1 h (3.27-fold), compared with the control without treatment, respectively (Figure 2C). Under ABA treatment, the CtDXS1 gene showed a similar trend to the drought treatment, reaching its highest expression level at 3 h (30.92-fold) (Figure 2D), while the highest expression level of the CtDXR1 (9.87-fold) gene occurred at 6 h (Figure 2E). The different expression patterns of the CtDXS1 and CtDXR1 genes imply that these two genes may play different roles in the drought and ABA responses. Compared with the CtDXS1 gene, the other four CtDXS genes were expressed at relatively lower levels under drought and ABA treatments, respectively. The CtDXR2 gene was also expressed at lower levels than CtDXR1 under drought and ABA treatment conditions, respectively. Therefore, based on the phylogenetic tree, cis-acting element analysis, and expression patterns under various stresses, the CtDXS1 and CtDXR1 genes are promising for drought response and were therefore selected for further functional identification.
2.4 Subcellular localization of CtDXS1 and CtDXR1 proteins
TARGETP showed a putative chloroplast transfer peptide in CtDXS1 with a high probability of 0.8976, whereas ChloroP predicted that CtDXS1 is not located in chloroplasts. Considering the subcellular localization of the MEP pathway in plants (Llamas et al., 2017; Zhang et al., 2019), it is a better prediction that CtDXS1 is located in chloroplasts. The predicted chloroplast transfer peptide at sites 46-47 of CtDXS1 has a conserved VX-A pattern cleavage site (for CtDXS1, X: C, Figure S1). Meanwhile, there was a putative chloroplast transit peptide at sites 48-49 (Figure S2) of CtDXR1 with a conserved CS-X patterned cleavage site (for CtDXR1, X: V), suggesting that CtDXR1 is preferentially located in chloroplasts.
To verify the predicted subcellular location of CtDXS1 and CtDXR1 proteins, pBI121 CtDXS1: GFP and pBI121 CtDXR1: GFP expression vectors were transiently transformed into N. benthamiana leaves, respectively. Under laser confocal scanning, chloroplasts and GFP showed red and green fluorescence, respectively. The subcellular localization of CtDXS1 and CtDXR1 proteins was able to be observed based on the combined fluorescence, where the visualized yellow fluorescence was evident, indicating that both CtDXS1 and CtDXR1 are located in chloroplasts (Figure 3), in which the MEP pathway is located.
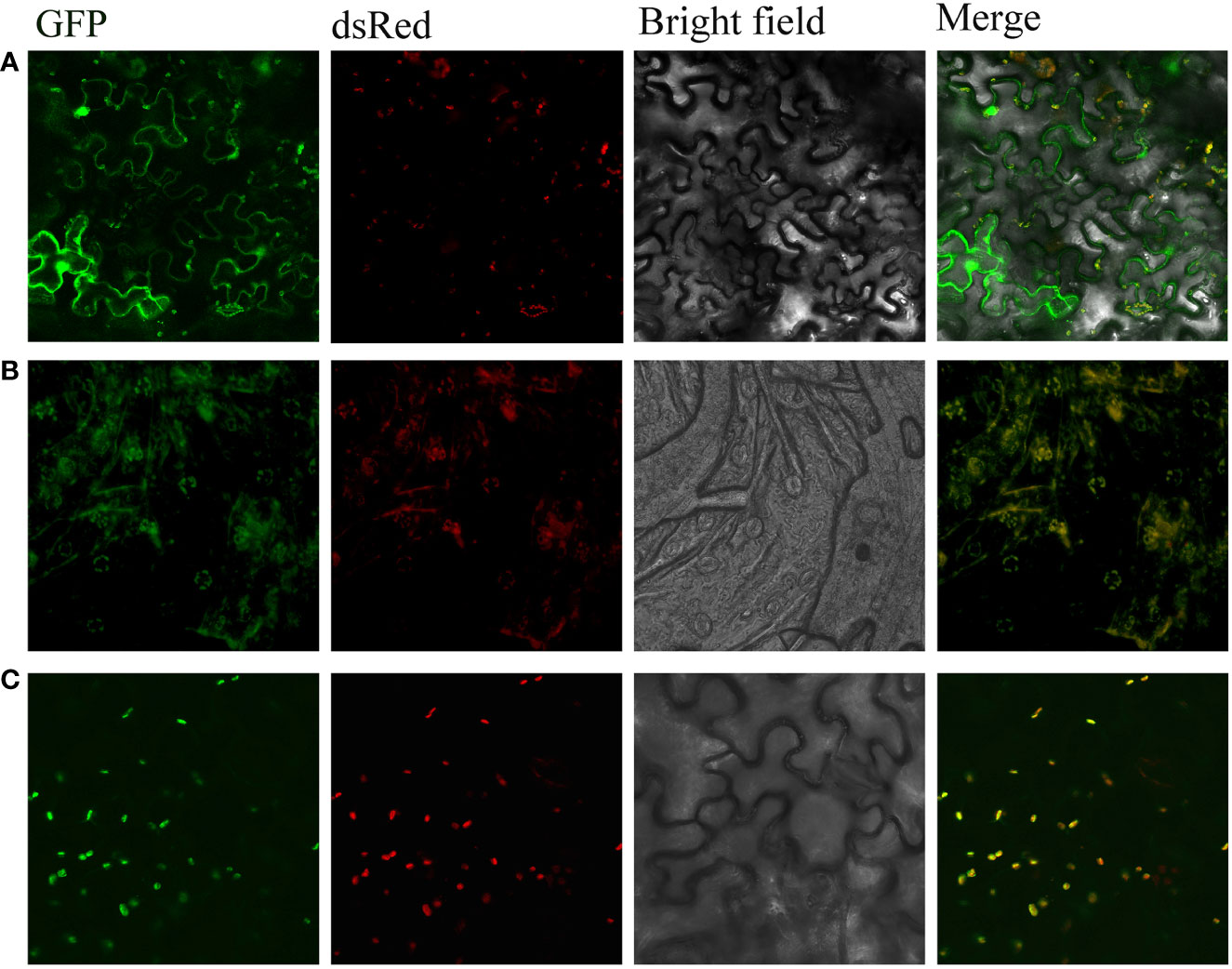
Figure 3 CtDXS1 and CtDXR1 were located in chloroplast. (A) Expression of 35S: GFP, (B) Expression of 35S: CtDXS1-GFP, (C) Expression of 35S: CtDXR1-GFP in N. benthamiana is localized in yellow dots around the chloroplast, indicating the subcellular location of the fusion proteins. Free DsRed marks the chloroplast. Free GFP serves as a localization control.
2.5 Production of transgenic plants
DXS and DXR catalyze the first and second steps of the MEP pathway and are considered to be the rate-limiting enzymes for isoprenoid production in bacteria and plants (Estévez et al., 2001; Liao et al., 2023). To verify the role of these two genes in influencing the level of plastid isoprenoids in plants, recombinant plasmids containing CtDXS1 and CtDXR1 ORFs, respectively, controlled at their 5` ends by the CaMV 35S promoter, were constructed. Both recombinant plasmids were introduced into N. benthamiana by Agrobacterium-mediated transformation, respectively. Transgenic plants were selected on media containing kanamycin. Transgenic plants exhibited a green phenotype similar to that of wild-type plants. The T2 lines were analyzed for gene expression, isoprenoid content and phenotype.
To determine the expression of CtDXS1 and CtDXR1, reverse-transcription PCR (RT-PCR) analysis was performed on 20-day-old seedlings of transgenic and wild-type plants using cDNA as template. As shown in Figure S3, CtDXS1 and CtDXR1 were substantially expressed in CtDXS1 transgenic plants and CtDXR1 transgenic plants, respectively, while CtDXS1 and CtDXR1 were not detected in wild-type plants.
2.6 CtDXS1 and CtDXR1 transgenic plants increased isoprenoid contents
To determine the amounts of isoprenoids, four kinds of plastidic isoprenoids, such as chlorophyll, β-carotene, ABA and GA3 were selected. The β-carotene in CtDXS1 transgenic plants was 1.10 ± 0.10-fold higher than that in wild-type plants, while plants overexpressing CtDXR1 gene presented a 2.29 ± 0.12-fold increase of β-carotene relative to that in the wild-type plants (p<0.01) (Figure S4A). As shown in Figure S4B, total chlorophyll content in CtDXS1 transgenic plants achieved a 1.34 ± 0.03-fold increase, compared to the wild-type levels (p<0.01). In CtDXR1 transgenic plants, a total of chlorophyll content was observed to be a 1.46 ± 0.03-fold increase in respect to that in wild-type plants (p<0.01). It would be of particular interest to explore the effect of altering the expression of CtDXS1 and CtDXR1 on GA levels. Since it has been shown that GA3 plays a central role in plant development (Jaudal et al., 2018), its level was used as a marker of GA levels in this study. As shown in Figure S4C, for those CtDXR1 transgenic plants, the levels of GA3 were detected to be 3.53 ± 0.09-fold higher than the wild-type levels. In CtDXS1 transgenic plants, lower levels of GA3 were observed to be 6.56 ± 0.2% of the wild-type levels. Since a major part of ABA biosynthesis is carried out in the plastid from β-carotene, overexpression of CtDXS1 and CtDXR1 may affect the levels of this phytohormone. It was observed that CtDXS1-overexpressing plants accumulated a 4.32 ± 1.19-fold increase of ABA content compared to wild-type levels (p<0.01). In CtDXR1-overexpressing plants, the ABA content was 3.77 ± 0.84-fold higher than the wild-type levels (p<0.01) (Figure S4D).
2.7 GA3 and ABA played role in plant growth and drought tolerance
In order to determine whether two isoprenoid-type phytohormones, GA3 and ABA, played role in plant growth and drought tolerance, the GA3 and ABA were exogenously applied to wild-type plants, respectively. The phenotypes were recorded after treatment of 7 days. As shown in Figure S5 and Table S1, exogenous GA3 application improved plant growth since the plants treated by GA3 exhibited larger leaf size (1.51 cm2, p<0.05), number of lateral roots (14.2, p<0.05), fresh weight (111.4 mg, p<0.05) and length of taproots (96 mm) in respect to control plants with (0.90 cm2, 9.8, 86.08 mg and 90.6 mm), respectively. These key results support the critical importance of GA3 in explaining the growth phenotype, but also imply the importance of phytohormone ratio for plant growth.
Meanwhile, exogenous ABA application enhanced drought tolerance of wild-type plants. After drought treatment for 7 days, the plants treated by ABA had a higher value of height (71 mm; p<0.05), number of lateral roots (9.4), length of taproots (58.6 mm, p<0.05), fresh weight (36.26 mg, p<0.05), but similar number of leaves (7) compared to control plants with (56.8 mm, 8.2, 44.4 mm, 26.98 mg and 6.8), respectively (Figure S5, Table S2).
2.8 CtDXS1 and CtDXR1 transgenic plants increased defense-related enzymes activities
To elucidate the plant tolerance induced by ABA accumulation, further biochemical analyses were performed. The activities of phenylalanine ammonia lyase (PAL), peroxidase (POD) and superoxide dismutase (SOD) were measured using 30-day-old leaves of wild-type and transgenic plants. As a result, transgenic plants showed enhanced activities of PAL, POD and SOD.
As shown in Figure S6, total PAL activity was increased in CtDXS1 transgenic plants, up to 1.51 ± 0.03-fold higher than wild-type levels. In CtDXR1 transgenic plants, a total PAL activity was observed to be a 3.12 ± 0.10-fold increase relative to wild-type levels (p<0.05).
Overexpression of CtDXS1 and CtDXR1 genes also resulted in increased POD activity. Relative to wild-type levels, CtDXS1 transgenic plants showed a 2.23 ± 0.31-fold increase of total POD activity. Plants overexpressing CtDXR1 contained a 4.66 ± 0.54-fold higher total POD activity than wild-type levels (p<0.05).
As shown in Figure S6, CtDXS1 transgenic plants had SOD levels of a 1.25 ± 0.02-fold increase compared to wild-type levels (p<0.01). For those transgenic plants that accumulated CtDXR1, SOD activity was 1.59 ± 0.03-fold higher than that in wild-type plants (p<0.001).
2.9 Phenotypes of transgenic plants
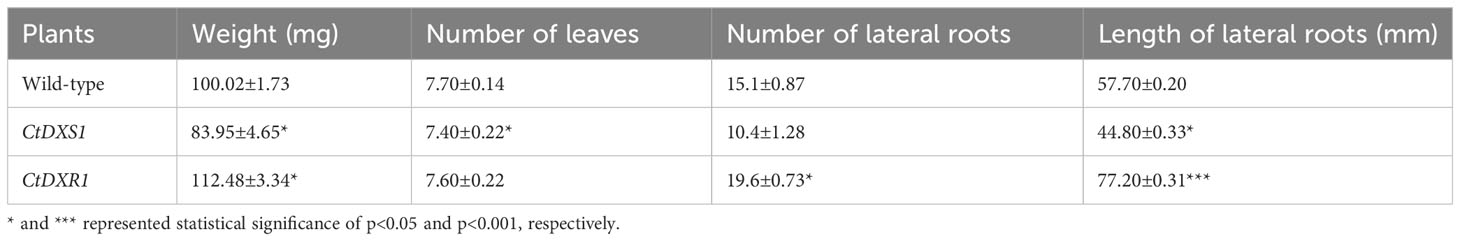
To determine whether the observed changes in isoprenoid levels affect plant morphology, the plants of wild-type, CtDXS1, and CtDXR1 transgenic plants were grown and their phenotypes were compared under normal conditions. No significant differences in general plant morphology were observed between our transgenic plants and the wild-type control, except for biomass. To explore this phenotype more closely, the biomass of the transgenic plants was estimated at the seedling stage (20 days) by leaf size, number and length of lateral roots. It was observed (Figure 4A, Table 2) that the weight of wild-type plants had 100.02 mg, while those of CtDXS1 and CtDXR1 transgenic plants were 83.95 mg (p<0.05) and 112.48 mg (p<0.05), respectively (Figure 4A, Table 2). The CtDXR1 transgenic plants had a greater number of lateral roots (19.6, p<0.05), while wild-type and CtDXS1 transgenic plants had fewer number of 15.1 and 10.4, respectively. In addition, the highest length of total lateral roots was observed of 77.20 mm (p<0.001) in CtDXR1 transgenic plants, while those in wild-type and CtDXS1 transgenic plants were relatively lower of 57.70 and 44.80 mm (p<0.05), respectively.

Figure 4 Phenotypes of the CtDXS1 transgenic, CtDXR1 transgenic and wild-type plants. (A) The 20-day-old seedlings of CtDXS1 transgenic, CtDXR1 transgenic and wild-type plants cultivated on culture medium under normal conditions; (B) The 20-day-old seedlings of CtDXS1-transgenic, CtDXR1-transgenic and wild-type plants cultivated on culture medium under drought stress; (C) The 60-day-old seedlings of CtDXS1 transgenic, CtDXR1 transgenic and wild-type plants cultivated in soil mixture under normal conditions; (D) The 60-day-old seedlings of CtDXS1 transgenic, CtDXR1 transgenic and wild-type plants cultivated in soil mixture under drought stress.
Intriguingly, both CtDXS1 and CtDXR1 transgenic plants exhibited higher drought tolerance than wild-type plants at seedling stage. After drought stress induced by 200 mM mannitol for 10 days, the leaves of the transgenic plants remained green and unfold, while those of some wild-type seedlings curled up, implying that the wild-type plants may be in a dehydrated state (Figure 4B). Compared with wild-type plants, the CtDXS1 and CtDXR1 transgenic plants showed greater height and fresh weight, and higher number of lateral root and leaf, respectively (Table 3). Therefore, in our definition both CtDXS1 and CtDXR1 transgenic plants may be able to improve tolerance to drought stress.
Meanwhile, to investigate the drought tolerance of transgenic and wild-type plants at their mature stages, 30-day-old seedlings were cultivated in pots containing a soil mixture (nutrient soil:vermiculite:peat, 1.5:1:1, V/V/V) and grown at 25°C, 16 h of light and 8 h of darkness. After 30 days, the plants under normal and drought conditions, respectively, were used to estimate the height of plants, the number of leaves, the number and length of lateral roots, average lateral and longitudinal lengths of leaves. We observed that the phenotypes of the potted plants showed similar tendencies to those of the plants on the culture medium. Under normal conditions, CtDXR1 transgenic plants showed the highest height (12.8 cm, p<0.001) and leaf area (1453.38 mm2), while CtDXS1 transgenic and wild-type exhibited similar height and leaf area (Figure 4C, Table 4). Under drought stress, the transgenic plants essentially exhibited greater drought tolerance than the wild-type plants. The CtDXR1 transgenic plants exhibited the highest height (12.4 cm, p<0.01), lateral root length (59.5 mm, p<0.001), number of leaves (10, p<0.01) and largest leaf areas (789.13 mm2, p<0.001), respectively, while CtDXS1 transgenic plants had higher height (11.4 cm, p<0.05), lateral root length (53.75 mm, p<0.001), number of leaves (8.75) and larger leaf areas (488.75 mm2) (Figure 4D, Table 4). Thus, our results suggested that transgenic plants were able to simultaneously improve drought tolerance in young and more mature plants.
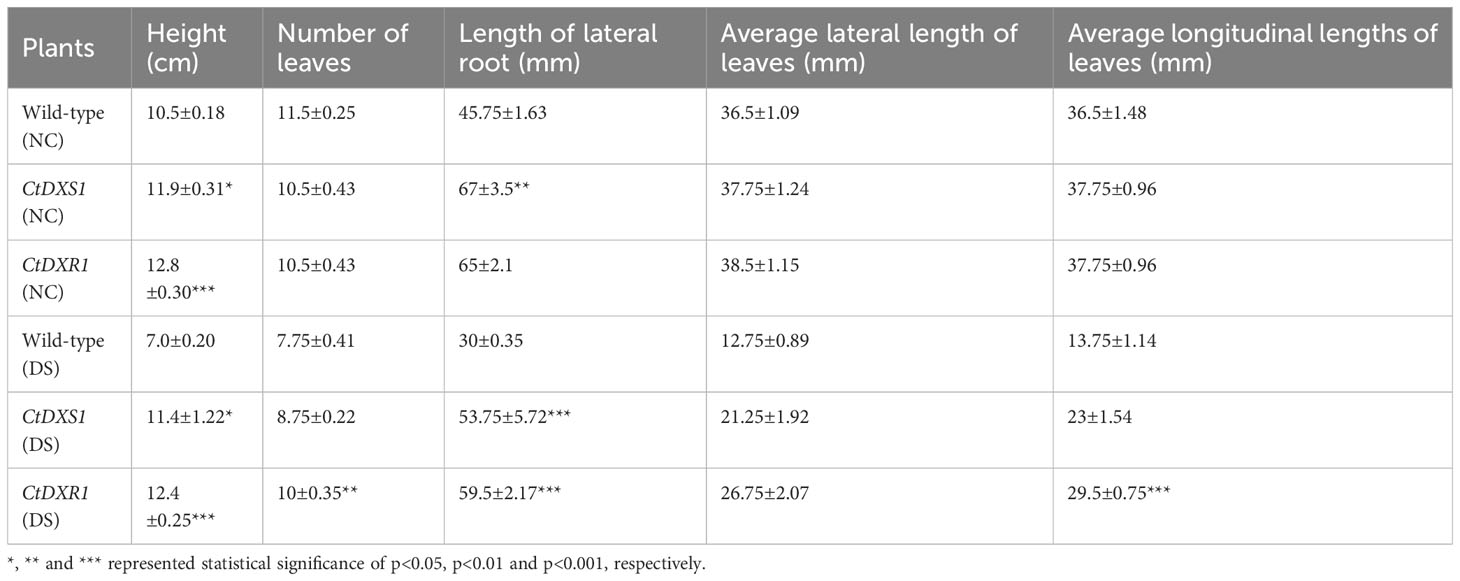
Table 4 Phenotypic statistics of seedlings cultivated in soil mixture under normal conditions (NC) and drought stress (DS), respectively.
2.10 Transcriptomic alteration in CtDXR1 transgenic plants
Since CtDXR1 transgenic plants strike a balance between plant growth and drought tolerance, RNA-seq analysis was performed to reveal transcriptional changes in CtDXR1 transgenic plants. Overall, wild-type and CtDXR1 transgenic plants produced 44.63 (6.67 Gb) and 41.45 (6.21 Gb) clean reads, respectively. In total, 96.35% of clean reads with Q30 rate of at least 94.15% were mapped to the N. benthamiana genome, indicating that the transcriptome data was reliable. A total of 4,333 genes were up-regulated and 3,340 genes were down-regulated in the DEGs, with fold changes greater than 2 and Q-values less than 0.05. Gene ontology (GO) was further investigated and the top 20 GO terms of biological processes (BP), cellular components (CC) and molecular functions (MF) in the up-regulated genes were analyzed (Figure 5). Among the up-regulated genes, “response to abscisic acid”, “response to water deprivation” and “regulation of salicylic acid biosynthetic process” in biological process, as well as “calcium ion binding” and “calmodulin binding” in molecular functions indicated that the enhanced drought tolerance of CtDXR1 transgenic plants might be involved with ABA-related pathway. The “xyloglucan metabolic process”, “cell wall biogenesis”, “cell wall organization” and “response to auxin” in biological process, “apoplast” in cellular component, as well as “carbohydrate binding” and “xyloglucan: xyloglucosyl transferase activity” in molecular functions were also consistent with the enhanced growth of CtDXR1 transgenic plants. Photosynthesis is the most affected pathway of biological processes, cellular components, and molecular functions among the regulated genes, which is consistent with the increase in chlorophyll content.
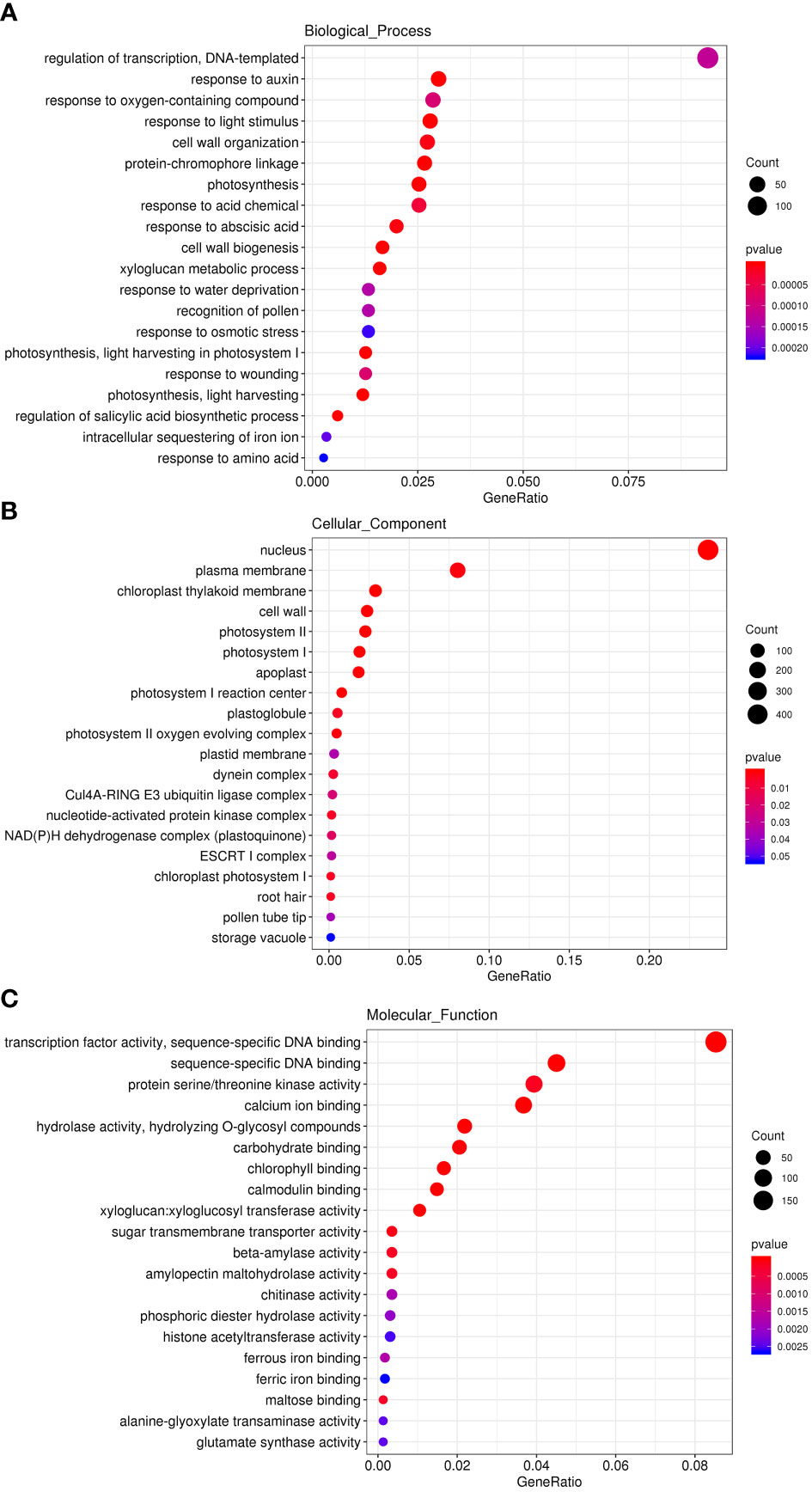
Figure 5 GO analyses of up-regulated genes in top 20 GO terms of Biological Process (A), Cellular Component (B), and Molecular Function (C), respectively.
2.11 Transcriptional alterations of genes responsible for isoprenoid biosynthesis
Previously, up-regulation of several genes involved in carotenoid biosynthesis was observed in LCYB1-overexpressing tobacco lines (Moreno et al., 2020). Therefore, the effect of CtDXR1 on the expression of genes related to isoprenoid metabolism was analyzed in detail using N. benthamiana, which is closely related to tobacco (Figure 6A, Table S3). In total, 10 genes had increased transcript levels, including one DXS(DXS2), three geranylgeranyl diphosphate synthases (GGPPS3, GGPPS10, and GGPPS13), two 9-cis-epoxycarotenoid dioxygenases (NCED10 and NCED11), one ent-kaurene synthase (KS5), one phytoene synthase (PSY1), and two carotenoid cleavage dioxygenases (CCD3 and CCD4). And the expression levels of eight genes were down-regulated, including two GGPPSs (GGPP2 and GGPP5), one isopentenyl diphosphate isomerase (IDI5), one gibberellin 20-oxidase (GA20ox10), one 15-cis-zeta-carotene isomerase (Z-ISO1), one beta-carotene hydroxylase (HYD1), one neoxanthin synthase (NSY1) and one CCD (CCD2) (Figure 6B, Table S3).
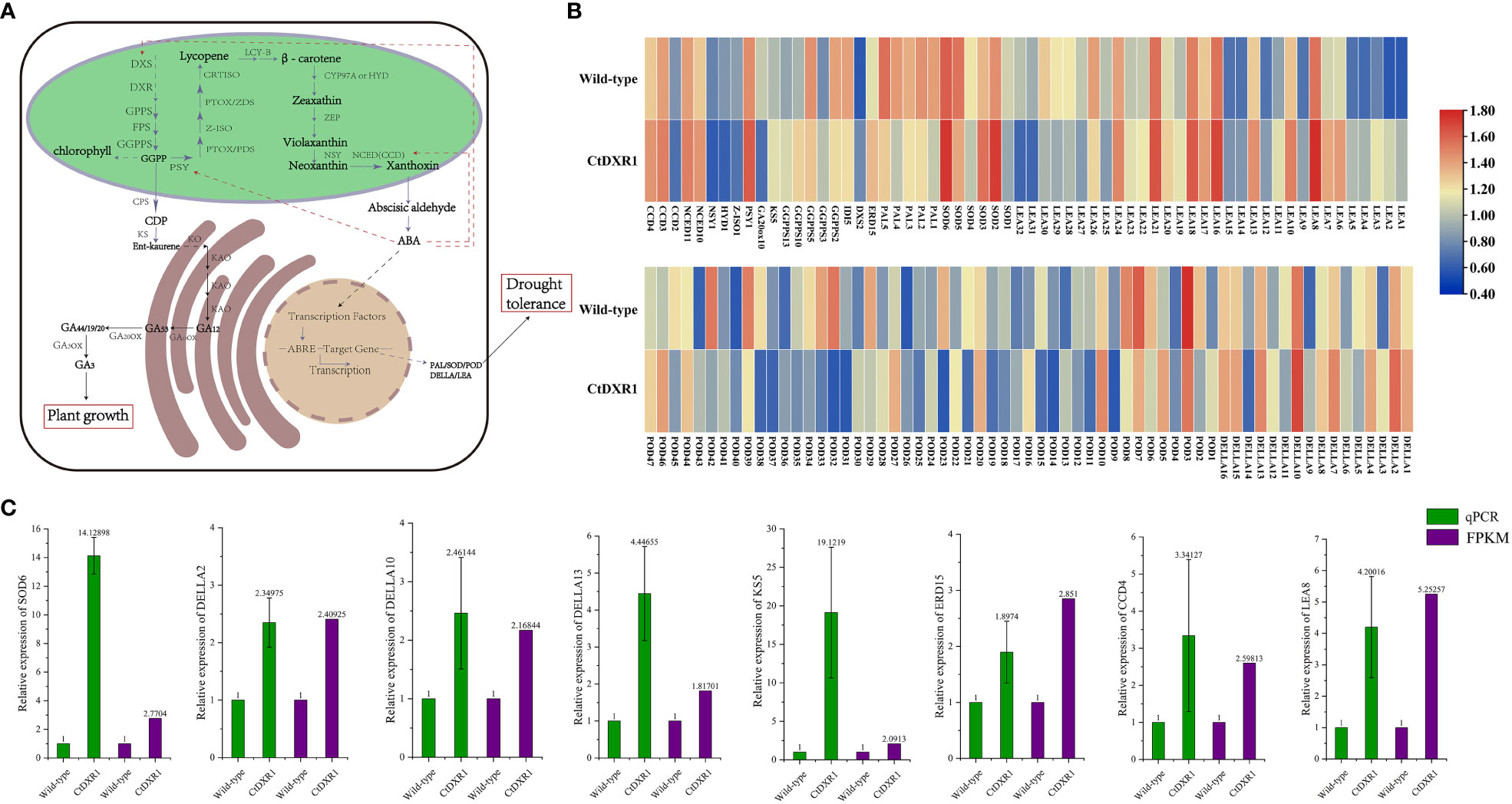
Figure 6 Isoprenoids biosynthesis and downstream signal transduction pathway. (A) Isoprenoids biosynthesis pathway is represented (adapted from Diretto et al., 2020 and Liu et al., 2015), with the differential expressed genes highlighted in heatmaps (B). The real-time PCR results of partial differential expressed genes were represented in (C). Error bars represent the standard error of the mean.
In the present study, many downstream genes regulated by ABA were significantly enhanced (Figure 6B, Table S4), such as three DELLA proteins (DELLA2, 10 and 13), one Late embryogenesis abundant protein (LEA8), one SOD (SOD6), and an early response to dehydration (ERD15), whose expression was also confirmed by real-time PCR (Figure 6C, the nucleotide sequence and primers of each gene are listed in Tables S5, S7, respectively). In addition, cis-acting element analysis on six potential target genes in the N. benthamiana genome was performed using the sequence of the 2-kb segment upstream of ATG. It was found that the promoters of all six genes contain ABRE and (or) MYB, which are involved in ABA-dependent pathways under drought and salt stress (Figure S7, Table S6) (Yang et al., 2011). In addition, the G-boxes in the DELLA2, DELLA13, ERD15 and LEA8 promoters are also associated with the ABA-dependent pathway (Yang et al., 2012a). Therefore, the increased transcript levels of SOD were consistent with increased SOD activity in CtDXR1 transgenic plants.
3 Discussion
3.1 Identification of DXS and DXR members in the C. tora genome
Since isoprenoids play a key protective role and are nutritionally important, the regulation of their production has been a target for agricultural improvement. In this study, four different isoprenoids were derived from the intermediate molecule GGPP, which is produced by condensation of IDP and DMADP units. DXS and/or DXR enzymes have all been shown to promote increased MEP-derived products when overproduced in different crop systems, including rice (You et al., 2020), carrot (Simpson et al., 2016) and barley (Walter et al., 2000). However, the relationship between drought tolerance and changes in plant isoprenoid-type hormones is still less clear. This is fundamentally different from plants transformed with genes encoding the CtDXS1 and CtDXR1 enzymes. In this study, a comprehensive genome-wide analysis of the DXS and DXR genes of C. tora was first done, and five DXS and two DXR genes were identified. The expansion of the DXS family was not limited to specific lineages, as these examples included both dicotyledonous and monocotyledonous plant species. In some cases, this expansion of the gene family was associated with genome-wide replication events that occurred in some of these species (like Glycine max) (Wang et al., 2021). In contrast, this is consistent with previous report (Silva et al., 2020) that DXR is a low-copy gene, with no more than two DXR genes in more than 96% of land plants, with the exception of G. max (three copies) and Kalanchoe laxiflora (four copies).
All published phylogenetic analyses of the DXS family agree on the existence of these three distinct groups, but they differ in the number of genes in the plant lineage. For example, the DXS2 group disappears in all crucifers, DXS3 in some Euphorbiaceae, and DXS1 is apparently absent in Triticum aestivum and Arabidopsis halleri (de Luna-Valdez et al., 2021). Consistent with previous report (You et al., 2020), our phylogeny shows that DXS genes from land plants clustered into three monophyletic clades (Figure 1) with high bootstrap support. Our phylogeny showed that these three clades included members from the same species (Medicago truncatula and Oryza sativa) (Figure 1), suggesting that the replication that gave rise to the different clades occurred prior to the divergence of these groups. More importantly, CtDXS1 and CtDXS5 were included in clade I involved in essential isoprenoid biosynthesis. The phylogenetic tree of DXR showed a more complex division than DXS (Figure 1). In the NJ tree, angiosperms were observed to be clustered in clade I, where legumes also appeared to be a strongly supported monophyletic group (91 BP). In clade II, gymnosperms were monophyletic and sister to angiosperms, similar to previous result (Tong et al., 2015). Since the two CtDXR genes were located in a monophyletic group and were sister to each other, it appeared that the duplication of the two CtDXR genes occurred after the divergence of C. tora.
Conservative cis-acting element assays indicated that the promoters of both CtDXS1 and CtDXR1 genes contained drought-responsive and ABA-responsive elements, suggesting that both genes might play a role in drought tolerance. This hypothesis was initially verified by real-time PCR, and the mRNA levels of both genes were increased under simulated drought and ABA treatments (Figures 2B-E). In general, genes with altered expression levels under environmental stress are thought to play a role in the stress response. Considering the phylogeny, cis-acting elements, expression pattern, and subcellular location detection, it was very likely that CtDXS1 or CtDXR1 genes influenced the biosynthesis of isoprenoid in plants.
3.2 The isoprenoid contents were responsible for the phenotypes of transgenic plants
In the next steps, four important plastidic isoprenoids, such as β-carotene, chlorophyll, GA3 and ABA, were analyzed. These isoprenoids derive via three pathways from the common GGPP intermediate, which is formed by condensation of IDP and DMADP units. β-carotene and ABA are downstream products of phytoene, a C40 intermediate molecule, via GGPP (Barja and Rodríguez-Concepción, 2021). Chlorophyll is a downstream product of phytyl pyrophosphate (phytyl-pp) via GGPP (Barja and Rodríguez-Concepción, 2021). GA constitutes a large family of diterpenoids that act as plant hormones and are involved in many developmental processes. These compounds are formed by the conversion of GGPP to ent-kaurene, and these initial steps are catalyzed by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) (Su et al., 2016).
β-carotene, ABA and chlorophyll contents increased at different levels in CtDXS1 and CtDXR1 transgenic plants, except that CtDXR1 transgenic plants had the highest GA3, while CtDXS1 transgenic plants had the lowest GA3 content. It was particularly interesting that the relative change in ABA was greater than the observed change in total β-carotene. As shown in Figures S4A, D, the relative increase in β-carotene was more limited than the relative increase in ABA. Similar result was reported by Estévez et al. (2001) who found changes in ABA contents were greater than those observed for total carotenoids in DXS-overexpressing Arabidopsis. The reason for this may be attributed in part to the highly regulated and complex carotenoid biosynthetic pathway (Banerjee and Sharkey, 2014). For example, pro-vitamin A, which plays a central role in human growth, reproduction, immunity, and vision, is also derived from β-carotene (Spiegler et al., 2012). It is known that vitamin A and ABA are isolated from a common β-carotene precursor, so an increase in a particular product may not be reflected in the total isoprenoid levels of the transgenic plant. Also, ABA can be hydrolyzed back from its glucose-conjugated form to the active form by β-glucosidase (Xu et al., 2012). Furthermore, for a specific product, there are additional restrictive reaction steps, for example, 9-cis-epoxycarotenoid dioxygenase (NCED) catalyzes the formation of xanthoxin from neoxanthin and plays a key role in ABA biosynthesis (Martínez-Andújar et al., 2020). Based on the following comparative transcriptome, two NCED genes showed higher expression in CtDXR1 transgenic plants than wild-type plants (Figure 6B, Table S3). Therefore, these results suggested that CtDXS1 and CtDXR1 genes might influence the accumulation of ABA in direct and (or) indirect way.
This altered isoprenoid accumulation would directly result in the observed protection against drought stress, (or) alternatively alter plant growth. To distinguish between these possibilities, plants were grown under normal conditions (plants without drought stress) and were exposed to drought stress, respectively. Under normal conditions, transgenic plants exhibited no significant differences in morphology, but significantly higher plant growth was observed in CtDXR1 transgenic plants (Figure 4A). The increase in chlorophyll content (1.46-fold higher than that in wild-type plants) was initially thought to play a role in the large phenotype of the CtDXR1 transgenic plants. However, CtDXS1 transgenic plants also had a 1.34-fold increase of chlorophyll content compared to wild-type plants but exhibited a relatively dwarf phenotype under normal conditions (Figure 4A). It was possible that the chlorophyll levels in the transgenic plants were not sufficient to produce these phenotypes in N. benthamiana. It was considered that a more detailed analysis of these plants could provide additional information about chlorophyll regulation and function in plants. Therefore, it was hypothesized that the main effect on growth was attributed to the alternation of phytohormonal biosynthesis, particularly that of GA3 and ABA. It is well known that GA3 and ABA antagonistically mediate many physiological processes. Some phenotypic characteristics of CtDXR1 transgenic plants were similar to those with increased GA3 content, such as accumulation of biomass, increase in plant length and expansion of leaf area. Meanwhile, characteristic phenotypes such as dwarf growth and reduced biomass were observed in CtDXS1 transgenic plants (Figure 4A, Table 2). Since the ratio between different phytohormones is more important than the absolute content (Moreno et al., 2020; Kaur et al., 2021), it is possible that the higher GA3/ABA ratio contributed to the strong increase in biomass (the ratio of GA3/ABA in CtDXR1 transgenic plants was approximately 1:1 compared to 1:6.6 in CtDXS1 transgenic plants).
Under drought stress conditions, both CtDXS1 and CtDXR1 transgenic plants exhibited significant drought tolerance (Figure 4). Drought stress led to an increased production of ROS, which could severely damage photosynthetic apparatus and pigments such as chlorophyll (Guadagno et al., 2017). Since ROS can be efficiently quenched by ABA-induced antioxidant and protective systems (such as PAL, SOD and POD), the abundance of ABA may be a major factor for chlorophyll protection in the transgenic lines. In addition, β-carotene can also reduce ROS production directly and/or indirectly, and hence is proposed to play role in drought tolerance (Moreno et al., 2020; Yu et al., 2020). However, the role of β-carotene in drought tolerance might be slight in this study, since the β-carotene content in CtDXS1 transgenic plants was only 1.10-fold increase compared with that in wild-type plants (Figure S4A). The additional results obtained in Figure S5 provided proof that two isoprenoid-type phytohormones, GA3 and ABA, played substantial role in plant growth and drought tolerance. Considering that both CtDXS1 and CtDXR1 transgenic plants showed similar trends of increased ABA (Figure S4D), ABA might act alone or be of more significance than β-carotene under drought stress.
3.3 The comparative transcriptome on CtDXR1 transgenic and wild-type plants
Since overexpression of DXS and DXR genes resulted in changes in isoprenoid abundance, it is possible that both DXS and DXR are rate-limiting enzymes in the MEP pathway. However, the CtDXS1 and CtDXR1 transgenic plants showed opposite trend of changes in GA3 levels, and hence it was likely that other rate-limiting enzymes for each specific isoprenoid exist. Data from other research groups (Do et al., 2016) support the hypothesis that GA levels are increased or decreased by regulating enzyme levels involved in the later GA biosynthetic steps. Interestingly, in addition to enhanced drought tolerance, accelerated plant growth was observed in CtDXR1 transgenic plants and resulted in higher biomass accumulation. Thus, CtDXR1 expression in crops might provide a suitable strategy to achieve a trade-off between plant growth and drought tolerance under field conditions. However, to explore its potential in crops, a better understanding of the molecular basis of its phenotype is essential, so a comparative transcriptome on CtDXR1 transgenic plants and wild-type plants was performed. The results revealed that several genes involved in isoprenoid biosynthesis were up-regulated, including DXS, KS, and NCED, with KS being a potential regulator of GA biosynthesis (Do et al., 2016), while DXS and NCED are key genes involved in ABA biosynthesis (Thompson et al., 2007; Arias et al., 2022), suggesting that the remodeled architecture of genes involved in isoprenoid biosynthesis might contribute to the trade-off between plant hormone content. In soybean and Chinese cabbage, mutations in KS resulted in reduced plant height, shorter internodes, and non-headed phenotype (Li et al., 2018; Gao et al., 2020), whereas a series of dwarf phenotypes in GA-deficient mutants reverted to wild type by heterologous overexpression of KS (Otsuka et al., 2004), thus supporting the conclusion that increased GA levels might largely explain the developmental phenotypes observed in CtDXR1 transgenic plants. Previous studies have shown that DXS and NCED in tobacco could induce high accumulation of ABA (Thompson et al., 2007; Arias et al., 2022), leading to the drought-resistant phenotype observed in CtDXR1 transgenic plants.
Interestingly, the observed changes in gene expression (Figure 6) may be partly a direct consequence of altered phytohormonal levels. Previously, it was reported that enhanced ABA and (or) GAs contents altered the expression of genes involved in isoprenoid biosynthesis (Lv et al., 2017; Chen et al., 2022). For example, PSY expression in Arabidopsis roots (Meier et al., 2011), NCEDs expression in peach (Tian et al., 2020), DXS expression in S. miltiorrhiza (Yang et al., 2012b), and GGPPS expression in Osmanthus fragrans (Liu et al., 2020) were induced after treatment with ABA, respectively. Furthermore, GAs had a feedback effect on GA3ox expression in watermelon (Middleton et al., 2012), but no feedback effect on GA20ox expression in Gibberella fujikuroi (Tudzynski et al., 2002) and KS expression in sunflower (Pugliesi et al., 2011). Furthermore, characterization of the up-regulated GGPPSs (GGPPS1, GGPPS4 and GGPPS5), NCEDs (NCED1 and NCED2), PSY and DXS promoters revealed 16, 10, nine and two ABREs, respectively (Table S6). Although it was expected that GA response elements might be present in the promoters of KS, no GA regulatory elements were revealed. Therefore, the higher ABA content in CtDXR1 transgenic plants might be mainly responsible for the feedback effect through which carotenoid content was increased.
In addition, it was previously reported that increased ABA content altered the expression of downstream stress-responsive genes (Yang et al., 2020). In our study, enhanced expression levels of DELLA, LEA, SOD and ERD were observed in transgenic plants. ABA promotes accumulation of DELLA protein, which contributes to remodeling growth and development of Arabidopsis seedlings (Guo et al., 2014). LEA is a key regulator of plant water loss control, and its overexpression exhibits enhanced drought tolerance (Lim et al., 2018). SOD plays a key role in protecting plants from oxidative stress, and its expression level is induced by ABA (Yu et al., 2020). ERD is rapidly activated in response to ABA and drought stress (Kariola et al., 2006). These up-regulated genes implicated that plants have evolved multi-level mechanisms for drought tolerance. Enzymatic analyses have shown that the enhanced drought tolerance of transgenic plants through ABA accumulation appears to be associated with increased activity of defense-related enzymes such as PAL, SOD, and POD. These enzymes are usually associated with induced tolerance of plants to environmental stresses (Yu et al., 2020; Qin et al., 2022). This hypothesis is consistent with previous analyses that the application of ABA together with the pathogen Alternaria solani induced higher expression of PAL and POD activities and defense genes (Song et al., 2011). However, this hypothesis contradicts previous analyses, which showed limited increase in POD activity in sitiens plants of tomatoes treated with ABA pulses prior to infection (Asselbergh et al., 2008). This result may be due to the diversity of each member of the multigene family. Based on the transcriptome data and real-time PCR results presented herein, the up-regulation of mRNA levels of defense-related genes might contribute to the increase in their activities.
3.4 Potential molecular mechanism on trade-off between plant growth and drought tolerance
Derived from these results, a molecular model was proposed to explain the compromised phenotype observed in CtDXR1 transgenic plants. First, overexpression of CtDXR1 promoted the accumulation of isoprenoids, such as β-carotene, chlorophyll, GA3, and ABA. Second, in CtDXR1 transgenic plants, isoprenoid biosynthesis was remodeled at the transcriptional level (such as enhanced levels of DXS, KS, GA20ox, and NCED, or silencing of GGPPS), possibly due to a feedback effect. In addition, the remodeling effect might also be due to altered levels of ABA and GA3. Third, under normal conditions, higher GA3 levels or higher GA3/ABA ratios promoted the growth of CtDXR1 transgenic plants. Fourth, enhanced ABA levels and/or β-carotene directly or indirectly protected plants in response to drought stress. In addition, ABA induced the expression of defense-related genes (such as DELLA, LEA, SOD and PAL) (Lim et al., 2018; Yu et al., 2020), which subsequently reduced the damage caused by drought, leading to improved drought tolerance (Lim et al., 2018; Wang et al., 2020; Yu et al., 2020; Qin et al., 2022).
Although the phenotypes of CtDXS1 and CtDXR1 transgenic plants could be largely linked to altered phytohormonal levels, it remained difficult to understand why expression of CtDXS1 and CtDXR1 genes from C. tora, but not other DXS and DXR genes from Arabidopsis, P. trichocarpa, would result in significant isoprenoid accumulation and developmental phenotypes (Simpson et al., 2016; Wei et al., 2019). One possibility is a species-specific effect, namely that similar trends in phytohormonal accumulation may not occur even with other DXS and DXR genes. Another possible explanation may lie in the different biochemical properties of the enzymes. In C. tora, CtDXS1 and CtDXR1 operate mainly in the leaves (Figure 2A, Figure 2B), where large amounts of chlorophyll accumulate. Thus, CtDXS1 and CtDXR1 may be less susceptible to feedback inhibition by isoprenoid products and may have a stronger positive effect on isoprenoid biosynthesis. According to our proposed mechanism, this discordance might be related to the balance of various phytohormones, and the expression structure of key genes involved in phytohormonal biosynthesis could be evaluated in the future.
4 Conclusion
In this study, five DXS genes CtDXS1-5 and two DXR genes CtDXR1-2, which are encoded by small multigene families, were identified for the first time from the genome of C. tora. Among them, CtDXS1 and CtDXR1 were proposed to play a potential role in plant isoprenoid biosynthesis and drought tolerance, and such hypothesis was verified by the following transgenic experiments. CtDXS1 and CtDXR1 transgenic plants substantially influenced the isoprenoid accumulation and exhibited enhanced drought tolerance. In respect to CtDXS1 transgenic plants, CtDXR1 transgenic plants exhibited trade-off effect between plant growth and drought tolerance, which expands their role in future agricultural operations. Furthermore, this paper provided a molecular mechanism for the trade-off between plant growth and drought tolerance involving the remodeled structure of GA3/ABA biosynthesis. Overall, our study expanded the family of plant DXSs and DXRs and enhanced the understanding of the molecular mechanisms underlying the crosstalk between various plant hormones and the trade-off between plant growth and drought tolerance.
5 Materials and methods
5.1 Identification of DXS and DXR genes in Cassia tora
To obtain the DXS and DXR genes, all protein sequences of Cassia tora were downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) and made available as a local protein database. The complete HMM maps of the DXS and DXR protein domains (PF02670 and PF02779) were also downloaded from PFam (http://pfam.sanger.ac.uk/) to identify DXS and DXR members separately. BLASTP searches were performed using HMM profiles from the Local Protein Data Bank that had 50% identity and an E-value ≤ 0.001. In the second approach, candidate was refined by CDD classification (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) sequences to remove proteins without the conserved DXS (PLN02582) and DXR (PLN02696) structural domains, respectively (Marchler-Bauer et al., 2005). The corresponding results were used as queries to search again for DXS and DXR in the C. tora protein database. In addition, HMMER and BLAST hits were compared and the results were parsed by an in-house PERL script. The open reading frame was generated by the ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.htML). Phylogenetic tree construction was performed with MEGA 6.0 program (Song et al., 2022). The motif organization of protein sequences was identified using the MEME analysis at the website (http://meme.nbcr.net/meme/meme.html) (Pan et al., 2019).
5.2 Plant materials, total RNA isolation and cDNA preparation
The plant materials preparation, total RNA isolation and cDNA preparation were carried out based on the method reported by Liu et al. (2015). C. tora was grown in a greenhouse in city of Chengdu (30°41′47.04″N, 104°3′12.6″E) at 25°C, 50% humidity, and 16 h of light/8 h of darkness. Seeds of C. tora were collected on the 30th day after flowering in summer (09/2020), frozen in liquid N2 and stored at -80°C for RNA isolation. Total RNA was isolated using RNA plant-plus reagent (Omega, USA). First-strand cDNA of C. tora was synthesized with oligo dT-AP primers using M-MLV reverse transcriptase (TaKaRa, Japan).
5.3 Expression pattern analysis of CtDXS and CtDXR genes under different treatments
To analyze the expression levels of CtDXS and CtDXR genes under different stresses, sterile seedlings (7 days after germination) cultured on Murashige and Skoog (MS) solid medium were collected at 25°C with 16 h of light/8 h of darkness. For simulated drought and ABA treatments, seedlings were placed on MS medium supplemented with mannitol (200 mM) and ABA (30 μg mL-1) and cultured for 0, 1, 2, 3, 6, 12, 24, and 48 h, respectively. Total RNA was extracted and cDNA synthesis was performed following the same method as Liu et al. (2015). Gene-specific primer pairs for real-time PCR of CtDXS (CtDXS1-qF to CtDXS5-qF, CtDXS1-qR to CtDXS5-qR in Table S7), CtDXR (CtDXR1-qF, CtDXR2-qF, CtDXR1-qR and CtDXR2-qR in Table S7) genes and the reference gene Elongation factor 1a (EF1α2-qF and EF1α2-qR in Table S7) were designed using Primer Premier 5.0 on the basis of full-length cDNA (Liu et al., 2015). Real-time PCR was performed using SYBR Premix Ex Taq II (Takara, Japan) and LightCycler 96 system (Roche Diagnostics, Germany) based on 2 μL cDNA template in 20 μL reaction buffer using gene-specific primer pairs under the following conditions: pre-denaturation at 95°C for 1 min, followed by 40 cycles of 10 s at 95°C, 10 s at 60°C, and 20 s at 72°C. The expression levels of CtDXS and CtDXR genes were analyzed using ΔΔCT-method in LightCycler 96 software, respectively.
5.4 Subcellular location of CtDXS1 and CtDXR1
Firstly, primers CtDXS-1F and CtDXS-1R containing EcoRI and BamHI site, respectively, were designed to amplify the corresponding full-length CDS region of CtDXS1 (without the stop codon). CtDXR-1F and CtDXR-1R containing EcoRI and BamHI, respectively, were designed to amplify the corresponding cDNAs of the full-length CDS region of CtDXR1 (without the stop codon), respectively. Secondly, the CtDXS1 fragment was introduced into the vector carrying the GFP fragment. Similarly, the CtDXR1 fragment was ligated into the vector pBI121 carrying the GFP fragment to construct the vector pBI121 CtDXR1: GFP. Agrobacterium tumefaciens GV3101 carrying the pBI121 CtDXS1: GFP and pBI121 CtDXR1: GFP vectors were infiltrated into the young leaves of 4-week-old Nicotiana benthamiana plants, respectively. Leaves of N. benthamiana were examined four days post infiltration (dpi) using a Fluoview FV10i confocal laser scanning microscope as described previously (Qin et al., 2022). Fluorescence signals were detected using the following excitation and emission wavelengths: GFP (488 nm/510 nm) and chloroplasts (640 nm/675 nm). Images were processed with Image J software.
5.5 Generation of CtDXS1 and CtDXR1-transgenic plants
To analyze the functional roles of CtDXS1 and CtDXR1 genes in transgenic plants, CtDXS1 and CtDXR1 genes were overexpressed into wild-type N. benthamiana, respectively. First, the ORF of CtDXS1 was amplified by PCR using primers CtDXS-2F and CtDXS-2R containing the BamHI and SnaBI sites, respectively. The ORF of CtDXR1 was amplified by PCR using primers CtDXR-2F and CtDXR-2R containing the XbaI and SmaI sites, respectively. Subsequently, the PCR products were cloned into the pBI121 binary vector under the control of CaMV 35S promoter. The recombinant vectors pBI-CtDXS1 and pBI-CtDXR1 were transformed into A. tumefaciens strain GV3101 and then introduced into N. benthamiana by Agrobacterium-mediated transformation, respectively. Transgenic T1 generation plants were generated and only lines showing kanamycin (50mg/L) and rifampicin (50mg/L) resistance were selected for further studies. DNA-based molecular screening of CtDXS1 and CtDXR1 transgenic plants was performed using primer pair PCR (CtDXS-2F and CtDXS-2R, CtDXR-2F and CtDXR-2R), respectively. Meanwhile, the expression of CtDXS1 and CtDXR1 in transgenic T2 plants was analyzed by RT-PCR (CtDXS-3F and CtDXS-3R, CtDXR-3F and CtDXR-3R), respectively.
5.6 Determination of β-carotene, ABA, GA3 and chlorophyll contents
The β-carotene, ABA, GA3, and chlorophyll contents of 30-day transgenic and wild-type seedlings were determined as described (Estévez et al., 2001), respectively. Total β-carotene was extracted from fresh sample (500 mg) with 20 mL of cooled standard solution and incubated for 16 h at 4°C. The β-carotene content was analyzed by HPLC using a Hypersil C18 column (4.6 mm × 150 mm) with a column temperature of 30°C. The mobile phase consisted of solvents A (0.02% butylated hydroxytoluene in methanol) and B (tetrahydrofuran). The ratio of solvent A to B in the mobile phase was set to 9:1. The elution conditions were set to a flow rate of 1 mL min-1, with a detection wavelength of 448 nm. The determination of each plant was carried out with three individual samples as replicates.
To determine the total chlorophyll content, 100 mg of fresh sample was extracted with 95% ethanol. Spectrophotometric quantification was performed on a Shimadzu UV-2200 spectrophotometer. To quantify the ABA and GA3 content of transgenic and wild-type plants, 500 mg of fresh sample was homogenized in 80% ethanol and incubated for 16 h at 4°C with continuous shaking. The supernatant was collected after centrifugation (10,000 × g, 15 min) and extracted with ethyl acetate. The supernatant was then dried and dissolved in 1 mL of 80% methanol, and filtered through a membrane filter (0.45 μm, 13 mm, Millipore). Finally, a mixed solution composed of ABA and GA3 was used as standard for the simultaneous analysis of ABA and GA3 by HPLC using a Hypersil C18 column (4.6 mm × 150 mm) with a column temperature of 35°C. The mobile phase consisted of methanol: 0.1% phosphoric acid (7:13, V/V) with an elution rate of 0.8 mL min-1. The wavelength for detection of ABA and GA3 was set to 210 nm. The determination of each plant was carried out with three individual samples as replicates.
5.7 Assays of PAL, SOD and POD activities
Leaf tissue (500 mg of fresh weight) was thoroughly ground in liquid nitrogen. To prepare the PAL sample, the leaf powder was homogenized with 3 mL of 100 mM ice-cold borate buffer (pH 8.8) containing 1% (w/v) polyvinylpyrrolidone (PVPP) and 0.1 mM EDTANa2 (Qin et al., 2022), but SOD and POD samples were prepared by homogenizing the leaf powder with 50 mM sodium phosphate buffer (pH 7.8) containing 1% (w/v) PVPP and 0.1 mM EDTANa2. The homogenates were centrifuged at 12,000 rpm for 20 min and the supernatant (crude extract) was used as the source of enzymes. All steps were carried out at 4°C.
PAL activity was determined according to the method of Qin et al. (2022) with some modifications. Briefly, 0.14 mL of crude extract was incubated with 3 mL of reaction buffer (100 mM boric acid (pH 8.8), 200 μM L-phenylalanine) for 30 min at 30°C. The reaction was terminated by adding 1 mL of 6N hydrochloric acid and the absorbance was measured at 290 nm using a spectrophotometer. The specific activity was expressed in units per gram of fresh weight. The reaction mixture consisted of 0.3 mL of crude extract and 3 mL of reaction solution (2% of guaiacol, 30% of H2O2, 100 mM sodium phosphate buffer (pH 7.8)), incubated for 3 min at 30°C and stopped by the addition of 10 mL of 6N hydrochloric acid. POD activity was measured as an increase in absorbance at 470 nm. SOD activity was measured using a commercial SOD assay kit (ab65354, Abcam). Reaction reagents were prepared according to the manufacturer’s instructions. After incubation at 37°C for 20 min, absorbance was measured at 450 nm. Each test was performed in triplicate.
5.8 Transcriptome on the wild-type and CtDXR1 transgenic plants
20-day-old CtDXR1 transgenic and wild-type seedlings were collected, frozen in liquid nitrogen, and subsequently stored at -80°C until use. Total RNA from independent transgenic and wild-type lines was extracted. RNA-seq was performed by Biomarker Technologies (http://www.Biomarker.com.cn/) using the Illumina HiSeq™ 4000 system (Xiang et al., 2018). Clean reads were obtained after removing low-quality reads [more than 10% of bases with Q values <30 or more than 5% of Ns (where N represents ambiguous bases in the reads) from the original reads]. The clean reads were mapped to the reference genome of N. benthamiana “V1.0.1” (https://solgenomics.net/) using HISAT2 (Musich et al., 2021). StringTie was used for the normalization of exon reads (Pertea et al., 2015), reported as FPKM, and the number of gene reads was calculated using htseq-count (Florea et al., 2013). The IDEG6 package was used to find DEGs (Romualdi et al., 2003). According to the Benjamini-Hochberg formula, only those genes with a fold change (FC) ratio ≥ 2 and a corrected P-value < 0.05 were considered as DEGs. GO and KEGG pathway enrichment analysis of DEGs was performed using the R package (Kanehisa et al., 2008), which is based on hypergeometric distribution.
5.9 Statistical analysis
All experiments had at least three biological replicates, so the results are expressed as mean ± standard error. For statistical analysis, SPSS software version 19.00 (SPSS, Chicago, IL, United States) was used. Any statistically significant differences between the two groups were determined using independent t-tests. p-values < 0.05 were considered statistically significant.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HL: Writing – original draft. CT: Data curation, Writing – original draft. HQ: Writing – original draft. RJ: Writing – review & editing. QZ: Writing – review & editing. SH: Writing – review & editing. GT: Writing – review & editing. CY: Writing – review & editing. JZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was partially funded by Sichuan Science and Technology Program (No. 2018SZ0061 and 2021ZHFP0170), Sichuan Administration of TCM program (No. 2021MS116), Innovation Program for Technology of Chengdu City (2022-YF05-01357-SN), Fundamental Research Funds for the Central Universities (No. 2682022ZTPY053).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1270396/full#supplementary-material
Supplementary Figure 1 | Motifs organization and sequence logos in DXS proteins produced by MEME analysis. The DXSs in the motif analysis included AAQ84169 (Pueraria montana var. lobata), AMJ39459 (Bixa orellana), AEM42997 (Siraitia grosvenorii), AAS99588 (Elaei guineensis) and FJ455512 (Amomum Villosum), respectively. (A) motif organization of DXS proteins. The motifs are shown as different colored boxes. (B) the sequence logos of important motifs in DXS proteins. The height of the letter represents the degree of conservation at each position. The residue positions in the motifs are presented as the numbers on the x-axis, while the number on the y-axis represents the content measured in bits. The conserved His93, Asp294 and H499 residues for DXS catalysis were indicated with triangles, respectively. The TPP-binding domain was marked with the horizontal line and the pyridine binding DRAG domain is indicated with the double horizontal line on the top.
Supplementary Figure 2 | Motifs organization and sequence logos in DXR proteins produced by MEME analysis. The DXRs in the alignment included AFP43697 (Rhaphiolepis bibas), BAF98290 (Hevea brasiliensis), QGZ18966 (Pinus massoniana) and ABJ80680 (Salvia miltiorrhiza), respectively. The NADPH-binding domain was marked with the horizontal line and the substrate binding motifs is indicated with the double horizontal line on the bottom, respectively.
Supplementary Figure 3 | Expression of CtDXS1 and CtDXR1 in transgenic and wild-type plants. (A) RT-PCR results of CtDXS1 gene in CtDXS1 transgenic and wild-type plants, respectively. The primers (CtDXS-3F and CtDXS-3R) were used in this experiment; (B) RT-PCR results of CtDXR1 gene in CtDXR1 transgenic and wild-type plants, respectively. The primers (CtDXR-3F and CtDXR-3R) were used in this experiment.
Supplementary Figure 4 | The β-carotene (A), chlorophyll (B), gibberellins (GA3) (C) and abscisic acid (ABA) (D) contents in transgenic and wild-type plants. Bars represent standard deviation of six independent replicates.
Supplementary Figure 5 | The wild-type N. benthamiana plants treated by exogenous GA3 and ABA, respectively. For GA3 treatment, 14-day-old seedlings cultivated in 1/2 MS medium containing 50 μM GA3 were used as GA3 experiment group (A), while those cultivated in 1/2 MS medium not containing GA3 were used as GA3 control group (B). For ABA treatment, 14-day-old seedlings cultivated in 1/2 MS medium containing 100 μM ABA were used as ABA experiment group (C), while those cultivated in 1/2 MS medium not containing ABA were used as ABA control group (D).
Supplementary Figure 6 | PAL, POD and SOD activities in CtDXS1 transgenic (A), CtDXR1 transgenic (B) and wild-type plants, respectively.
Supplementary Figure 7 | Cis-acting element analysis of potential downstream genes of ABA.
References
Arias, D., Ortega, A., González-Calquin, C., Quiroz, L. F., Moreno-Romero, J., Martínez-García, J. F., et al. (2022). Development and carotenoid synthesis in dark-grown carrot taproots require PHYTOCHROME RAPIDLY REGULATED1. Plant Physiol. 189 (3), 1450–1465. doi: 10.1093/plphys/kiac097
Asselbergh, B., Achuo, A. E., Höfte, M., Van Gijsegem, F. (2008). Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol. Plant Pathol. 9, 11–24. doi: 10.1111/j.1364-3703.2007.00437.x
Banerjee, A., Sharkey, T. D. (2014). Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod Rep. 31 (8), 1043–1055. doi: 10.1039/c3np70124g
Barja, M. V., Rodríguez-Concepción, M. (2021). Plant geranylgeranyl diphosphate synthases: every (gene) family has a story. aBIOTECH 2, 289–298. doi: 10.1007/s42994-021-00050-5
Carretero-Paulet, L., Cairó, A., Botella-Pavía, P., Besumbes, O., Campos, N., Boronat, A., et al. (2006). Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol. Biol. 62 (4-5), 683–695. doi: 10.1007/s11103-006-9051-9
Carretero-Paulet, L., Cairó, A., Talavera, D., Saura, A., Imperial, S., Rodríguez-Concepción, M., et al. (2013). Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase like protein in Arabidopsis thaliana. Gene 524 (1), 40–53. doi: 10.1016/j.gene.2012.10.071
Chen, C., Chen, H., Chen, Y., Yang, W., Li, M., Sun, B., et al. (2022). Joint metabolome and transcriptome analysis of the effects of exogenous GA3 on endogenous hormones in sweet cherry and mining of potential regulatory genes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1041068
Crowell, D. N., Packard, C. E., Pierson, C. A., Giner, J. L., Downes, B. P., Chary, S. N. (2003). Identification of an allele of CLA1 associated with variegation in Arabidopsis thaliana. Physiol. Plant 118 (1), 29–37. doi: 10.1034/j.1399-3054.2003.00063.x
de Luna-Valdez, L., Chenge-Espinosa, M., Hernández-Muñoz, A., Cordoba, E., López-Leal, G., Castillo-Ramírez, S., et al. (2021). Reassessing the evolution of the 1-deoxy-D-xylulose 5-phosphate synthase family suggests a possible novel function for the DXS class 3 proteins. Plant Sci. 310, 110960. doi: 10.1016/j.plantsci.2021.110960
De Smet, I., Signora, L., Beeckman, T., Inzé, D., Foyer, C. H., Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33, 543–555. doi: 10.1046/j.1365-313x.2003.01652.x
Diretto, G., Frusciante, S., Fabbri, C., Schauer, N., Busta, L., Wang, Z., et al. (2020). Manipulation of β-carotene levels in tomato fruits results in increased ABA content and extended shelf life. Plant Biotechnol. J. 18 (5), 1185–1199. doi: 10.1111/pbi.13283
Do, P. T., De Tar, J. R., Lee, H., Folta, M. K., Zhang, Z. J. (2016). Expression of ZmGA20ox cDNA alters plant morphology and increases biomass production of switchgrass (Panicum virgatum L.). Plant Biotechnol. J. 14 (7), 1532–1540. doi: 10.1111/pbi.12514
Estévez, J. M., Cantero, A., Reindl, A., Reichler, S., León, P. (2001). 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276 (25), 22901–22909. doi: 10.1074/jbc.M100854200
Fang, Y., Xiong, L. (2015). General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 72 (4), 673–689. doi: 10.1007/s00018-014-1767-0
Florea, L., Song, L., Salzberg, S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2, 188. doi: 10.12688/f1000research.2-188.v2
Gao, Y., Huang, S., Qu, G., Fu, W., Zhang, M., Liu, Z., et al. (2020). The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Res. 7 (1), 178. doi: 10.1038/s41438-020-00399-6
Górnaś, P., Czubinski, J., Rudzińska, M., Grygier, A., Ying, Q., Chakradhari, S., et al. (2019). Selected uncommon Legumes as a source of essential fatty acids, tocopherols, tocotrienols, sterols, carotenoids, and squalene. Plant Foods Hum. Nutr. 74 (1), 91–98. doi: 10.1007/s11130-018-0706-x
Guadagno, C. R., Ewers, B. E., Speckman, H. N., Aston, T. L., Huhn, B. J., DeVore, S. B., et al. (2017). Dead or alive? Using membrane failure and chlorophyll a fluorescence to predict plant mortality from drought. Plant Physiol. 175 (1), 223–234. doi: 10.1104/pp.16.00581
Guo, W., Cong, Y., Hussain, N., Wang, Y., Liu, Z., Jiang, L., et al. (2014). The remodeling of seedling development in response to long-term magnesium toxicity and regulation by ABA-DELLA signaling in Arabidopsis. Plant Cell Physiol. 55 (10), 1713–1726. doi: 10.1093/pcp/pcu102
Jaudal, M., Zhang, L., Che, C., Li, G., Tang, Y., Wen, J., et al. (2018). A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J. Exp. Bot. 69 (20), 4867–4880. doi: 10.1093/jxb/ery284
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36 (Database issue), 480–484. doi: 10.1093/nar/gkm882
Kang, S. H., Pandey, R. P., Lee, C. M., Sim, J. S., Jeong, J. T., Choi, B. S., et al. (2020). Genome-enabled discovery of anthraquinone biosynthesis in Senna tora. Nat. Commun. 11 (1), 5875. doi: 10.1038/s41467-020-19681-1
Kariola, T., Brader, G., Helenius, E., Li, J., Heino, P., Palva, E. T. (2006). EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 142 (4), 1559–1573. doi: 10.1104/pp.106.086223
Kaur, H., Ozga, J. A., Reinecke, D. M. (2021). Balancing of hormonal biosynthesis and catabolism pathways, a strategy to ameliorate the negative effects of heat stress on reproductive growth. Plant Cell Environ. 44 (5), 1486–1503. doi: 10.1111/pce.13820
Kim, S. M., Kuzuyama, T., Chang, Y. J., Song, K. S., Kim, S. U. (2006). Identification of class 2 1-deoxy-D-xylulose 5-phosphate synthase and 1-deoxy-D-xylulose 5-phosphate reductoisomerase genes from Ginkgo biloba and their transcription in embryo culture with respect to ginkgolide biosynthesis. Planta Med. 72 (3), 234–240. doi: 10.1055/s-2005-916180
Kumar, V., Kumar, A., Pandey, K. D., Roy, B. K. (2015). Isolation and characterization of bacterial endophytes from the roots of Cassia tora L. Ann. Microbiol. 65, 1391–1399. doi: 10.1007/s13213-014-0977-x
Lee, S. C., Luan, S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 35 (1), 53–60. doi: 10.1111/j.1365-3040.2011.02426.x
Li, Z. F., Guo, Y., Ou, L., Hong, H., Wang, J., Liu, Z. X., et al. (2018). Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 131 (5), 1001–1016. doi: 10.1007/s00122-017-3044-8
Liao, H., Quan, H., Huang, B., Ji, H., Zhang, T., Chen, J., et al. (2023). Integrated transcriptomic and metabolomic analysis reveals the molecular basis of tissue-specific accumulation of bioactive steroidal alkaloids in Fritillaria unibracteata. Phytochemistry 214, 113831. doi: 10.1016/j.phytochem.2023.113831
Lim, J., Lim, C. W., Lee, S. C. (2018). The pepper late embryogenesis abundant protein, CaDIL1, positively regulates drought tolerance and ABA signaling. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01301
Liu, Y., Dong, B., Zhang, C., Yang, L., Wang, Y., Zhao, H. (2020). Effects of exogenous abscisic acid (ABA) on carotenoids and petal color in Osmanthus fragrans 'Yanhonggui'. Plants 9 (4), 454. doi: 10.3390/plants9040454
Liu, Z., Song, T., Zhu, Q., Wang, W., Zhou, J., Liao, H. (2014). De novo assembly and analysis of Cassia obtusifolia seed transcriptome to identify genes involved in the biosynthesis of active metabolites. Biosci. Biotechnol. Biochem. 78 (5), 791–799. doi: 10.1080/09168451.2014.905182
Liu, Z., Zhu, Q., Li, J., Yu, J., Li, Y., Huang, X., et al. (2015). Selection and evaluation of reference genes for expression analysis of Cassi. Biosci. Biotechnol. Biochem. 79 (11), 1818–1826. doi: 10.1080/09168451.2015.1052771
Llamas, E., Pulido, P., Rodríguez-Concepción, M. (2017). Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PloS Genet. 13 (9), e1007022. doi: 10.1371/journal.pgen.1007022
Lv, Z., Zhang, L., Tang, K. (2017). New insights into artemisinin regulation. Plant Signaling Behav. 12 (10), e1366398. doi: 10.1080/15592324.2017.1366398
Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., et al. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33 (Database issue), D192–D196. doi: 10.1093/nar/gki069
Martínez-Andújar, C., Martínez-Pérez, A., Ferrández-Ayela, A., Albacete, A., Martínez-Melgarejo, P. A., Dodd, I. C., et al. (2020). Impact of overexpression of 9-cis-epoxycarotenoid dioxygenase on growth and gene expression under salinity stress. Plant Sci. 295, 110268. doi: 10.1016/j.plantsci.2019.110268
Matsuoka, D., Soga, K., Yasufuku, T., Nanmori, T. (2018). Control of plant growth and development by overexpressing MAP3K17, an ABA-inducible MAP3K, in Arabidopsis. Plant Biotechnol-NAR 35 (2), 171–176. doi: 10.5511/plantbiotechnology.18.0412a
Meier, S., Tzfadia, O., Vallabhaneni, R., Gehring, C., Wurtzel, E. T. (2011). A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst. Biol. 5, 77. doi: 10.1186/1752-0509-5-77
Miao, R., Yuan, W., Wang, Y., Garcia-Maquilon, I., Dang, X., Li, Y., et al. (2021). Low ABA concentration promotes root growth and hydrotropism through relief of ABA INSENSITIVE 1-mediated inhibition of plasma membrane H+-ATPase 2. Sci. Adv. 7 (12), eabd4113. doi: 10.1126/sciadv.abd4113
Middleton, A. M., Úbeda-Tomás, S., Griffiths, J., Holman, T., Hedden, P., Thomas, S. G., et al. (2012). Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc. Natl. Acad. Sci. U. S. A 109 (19), 7571–7576. doi: 10.1073/pnas.1113666109
Moreno, J. C., Mi, J., Agrawal, S., Kössler, S., Turečková, V., Tarkowská, D., et al. (2020). Expression of a carotenogenic gene allows faster biomass production by redesigning plant architecture and improving photosynthetic efficiency in tobacco. Plant J. 103 (6), 1967–1984. doi: 10.1111/tpj.14909
Muhammad Aslam, M., Waseem, M., Jakada, B. H., Okal, E. J., Lei, Z., Saqib, H. S. A., et al. (2022). Mechanisms of abscisic acid-mediated drought stress responses in plants. Int. J. Mol. Sci. 23 (3), 1084. doi: 10.3390/ijms23031084
Musich, R., Cadle-Davidson, L., Osier, M. V. (2021). Comparison of short-read sequence aligners indicates strengths and weaknesses for biologists to consider. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.657240
Otsuka, M., Kenmoku, H., Ogawa, M., Okada, K., Mitsuhashi, W., Sassa, T., et al. (2004). Emission of ent-kaurene, a diterpenoid hydrocarbon precursor for gibberellins, into the headspace from plants. Plant Cell Physiol. 45 (9), 1129–1138. doi: 10.1093/pcp/pch149
Pan, X., Li, Y., Pan, G., Yang, A. (2019). Bioinformatics study of 1-deoxy-D-xylulose-5-phosphate synthase (DXS) genes in Solanaceae. Mol. Biol. Rep. 46 (5), 5175–5184. doi: 10.1007/s11033-019-04975-5
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Proteau, P. J. (2004). 1-deoxy-D-xylulose 5-phosphate reductoisomerase: an overview. Bioorg. Chem. 32 (6), 483–493. doi: 10.1016/j.bioorg.2004.08.004
Pugliesi, C., Fambrini, M., Salvini, M. (2011). Molecular cloning and expression profile analysis of three sunflower (Helianthus annuus) diterpene synthase genes. Biochem. Genet. 49 (1-2), 46–62. doi: 10.1007/s10528-010-9384-6
Qin, Y., Li, Q., An, Q., Li, D., Huang, S., Zhao, Y., et al. (2022). A phenylalanine ammonia lyase from Fritillaria unibracteata promotes drought tolerance by regulating lignin biosynthesis and SA signaling pathway. Int. J. Biol. Macromol 213, 574–588. doi: 10.1016/j.ijbiomac.2022.05.161
Rodríguez-Concepción, M., Boronat, A. (2015). Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 25, 17–22. doi: 10.1016/j.pbi.2015.04.001
Romualdi, C., Bortoluzzi, S., D'Alessi, F., Danieli, G. A. (2003). IDEG6: A web tool for detection of differentially expressed genes inmultiple tag sampling experiments. Physiol. Genomics 12 (2), 159–162. doi: 10.1152/physiolgenomics.00096.2002
Shu, K., Zhou, W., Chen, F., Luo, X., Yang, W. (2018). Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00416
Silva, N., Ivamoto-Suzuki, S. T., Camargo, P. O., Rosa, R. S., Pereira, L. F. P., Domingues, D. S. (2020). Low-copy genes in terpenoid metabolism: the evolution and expression of MVK and DXR genes in angiosperms. Plants 9 (4), 525. doi: 10.3390/plants9040525
Simpson, K., Quiroz, L. F., Rodríguez-Concepción, M., Stange, C. R. (2016). Differential contribution of the first two enzymes of the MEP pathway to the supply of metabolic precursors for carotenoid and chlorophyll biosynthesis in carrot (Daucus carota). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01344
Song, S., Chen, A., Zhu, J., Yan, Z., An, Q., Zhou, J., et al. (2022). Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism. Int. J. Biol. Macromol 194, 317–330. doi: 10.1016/j.ijbiomac.2021.11.135
Song, W., Ma, X., Tan, H., Zhou, J. (2011). Abscisic acid enhances resistance to Alternaria solani in tomato seedlings. Plant Physiol. Biochem. 49, 693–700. doi: 10.1016/j.plaphy.2011.03.018
Spiegler, E., Kim, Y. K., Wassef, L., Shete, V., Quadro, L. (2012). Maternal-fetal transfer and metabolism of vitamin A and its precursor β-carotene in the developing tissues. Biochim. Biophys. Acta 1821 (1), 88–98. doi: 10.1016/j.bbalip.2011.05.003
Sreenivasulu, N., Harshavardhan, V. T., Govind, G., Seiler, C., Kohli, A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506 (2), 265–273. doi: 10.1016/j.gene.2012.06.076
Su, P., Tong, Y., Cheng, Q., Hu, Y., Zhang, M., Yang, J., et al. (2016). Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci. Rep. 6, 23057. doi: 10.1038/srep23057
Sugimoto, H., Kondo, S., Tanaka, T., Imamura, C., Muramoto, N., Hattori, E., et al. (2014). Overexpression of a novel Arabidopsis PP2C isoform, AtPP2CF1, enhances plant biomass production by increasing inflorescence stem growth. J. Exp. Bot. 65 (18), 5385–5400. doi: 10.1093/jxb/eru297
Thompson, A. J., Mulholland, B. J., Jackson, A. C., McKee, J. M., Hilton, H. W., Symonds, R. C., et al. (2007). Regulation and manipulation of ABA biosynthesis in roots. Plant Cell Environ. 30 (1), 67–78. doi: 10.1111/j.1365-3040.2006.01606.x
Tian, W., Huang, D., Geng, B., Zhang, Q., Feng, J., Zhu, S. (2020). Regulation of the biosynthesis of endogenous nitric oxide and abscisic acid in stored peaches by exogenous nitric oxide and abscisic acid. J. Sci. Food Agric. 100 (5), 2136–2144. doi: 10.1002/jsfa.10237
Tong, Y., Su, P., Zhao, Y., Zhang, M., Wang, X., Liu, Y., et al. (2015). Molecular cloning and characterization of DXS and DXR genes in the terpenoid biosynthetic pathway of Tripterygium wilfordii. Int. J. Mol. Sci. 16 (10), 25516–25535. doi: 10.3390/ijms161025516
Tudzynski, B., Rojas, M. C., Gaskin, P., Hedden, P. (2002). The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J. Biol. Chem. 277 (24), 21246–21253. doi: 10.1074/jbc.M201651200
Tyagi, S., Mulla, S. I., Lee, K. J., Chae, J. C., Shukla, P. (2018). VOCs-mediated hormonal signaling and crosstalk with plant growth promoting microbes. Crit. Rev. Biotech. 38 (8), 1277–1296. doi: 10.1080/07388551.2018.1472551
Vats, S. (2018). Larvicidal activity and in vitro regulation of rotenoids from Cassia tora L. 3 Biotech. 8 (1), 13. doi: 10.1007/s13205-017-1038-5
Vats, S., Kamal, R. (2014). Identification of flavonoids and antioxidant potential of Cassia tora L. Am. J. Drug Discovery Dev. 4, 50–57. doi: 10.3923/ajdd.2014.50.57
Walter, M. H., Fester, T., Strack, D. (2000). Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate pathway of isoprenoid biosynthesis correlated with accumulation of the 'yellow pigment' and other apocarotenoids. Plant J. 21 (6), 571–578. doi: 10.1046/j.1365-313x.2000.00708.x
Walter, M. H., Hans, J., Strack, D. (2002). Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 31 (3), 243–254. doi: 10.1046/j.1365-313x.2002.01352.x
Wang, L., Gao, Y., Wang, S., Zhang, Q., Yang, S. (2021). Genome-wide identification of PME genes, evolution and expression analyses in soybean (Glycine max L.). BMC Plant Biol. 21 (1), 578. doi: 10.1186/s12870-021-03355-1
Wang, Z., Liu, L., Cheng, C., Ren, Z., Xu, S., Li, X. (2020). GAI functions in the plant response to dehydration stress in Arabidopsis thaliana. Int. J. Mol. Sci. 21 (3), 819. doi: 10.3390/ijms21030819
Wei, H., Movahedi, A., Xu, C., Sun, W., Almasi Zadeh Yaghuti, A., Wang, P., et al. (2019). Overexpression of PtDXS enhances stress resistance in poplars. Int. J. Mol. Sci. 20 (7), E1669. doi: 10.3390/ijms20071669
Wright, L. P., Rohwer, J. M., Ghirardo, A., Hammerbacher, A., Ortiz-Alcaide, M., Raguschke, B., et al. (2014). Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant Physiol. 165 (4), 1488–1504. doi: 10.1104/pp.114.245191
Xiang, M., Zhang, X., Deng, Y., Li, Y., Yu, J., Zhu, J., et al. (2018). Comparative transcriptome analysis provides insights of anti-insect molecular mechanism of Cassia obtusifolia trypsin inhibitor against Pieris rapae. Arch. Insect Biochem. Physiol. 97 (1), e21427. doi: 10.1002/arch.21427
Xu, Z. Y., Lee, K. H., Dong, T., Jeong, J. C., Jin, J. B., Kanno, Y., et al. (2012). A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24 (5), 2184–2199. doi: 10.1105/tpc.112.095935
Xu, S., Song, S., Dong, X., Wang, X., Wu, J., Ren, Z., et al. (2021). GmbZIP1 negatively regulates ABA-induced inhibition of nodulation by targeting GmENOD40-1 in soybean. BMC Plant Biol. 21 (1), 35. doi: 10.1186/s12870-020-02810-9
Yang, A., Dai, X., Zhang, W. H. (2012a). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63 (7), 2541–2556. doi: 10.1093/jxb/err431
Yang, Y., Guo, Y., Zhong, J., Zhang, T., Li, D., Ba, T., et al. (2020). Root physiological traits and transcriptome analyses reveal that root zone water retention confers drought tolerance to Opisthopappus taihangensis. Sci. Rep. 10 (1), 2627. doi: 10.1038/s41598-020-59399-0
Yang, D., Ma, P., Liang, X., Wei, Z., Liang, Z., Liu, Y., et al. (2012b). PEG and ABA trigger methyl jasmonate accumulation to induce the MEP pathway and increase tanshinone production in Salvia miltiorrhiza hairy roots. Physiol. Plant 146 (2), 173–183. doi: 10.1111/j.1399-3054.2012.01603.x
Yang, J., Adhikari, M. N., Liu, H., Xu, H., He, G., Zhan, R., et al. (2012c). Characterization and functional analysis of the genes encoding 1-deoxy-D-xylulose-5-phosphate reductoisomerase and 1-deoxy-D-xylulose-5-phosphate synthase, the two enzymes in the MEP pathway, from Amomum villosum Lour. Mol. Bio. Rep. 39 (8), 8287–8296. doi: 10.1007/s11033-012-1676-y
Yang, X., Yang, Y. N., Xue, L. J., Zou, M. J., Liu, J. Y., Chen, F., et al. (2011). Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 156 (3), 1397–1409. doi: 10.1104/pp.111.173427
You, M. K., Lee, Y. J., Kim, J. K., Baek, S. A., Jeon, Y. A., Lim, S. H., et al. (2020). The organ-specific differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, in carotenoid metabolism in Oryza sativa leaves and seeds. BMC Plant Biol. 20 (1), 167. doi: 10.1186/s12870-020-02357-9
Yu, J., Cang, J., Lu, Q., Fan, B., Xu, Q., Li, W., et al. (2020). ABA enhanced cold tolerance of wheat 'dn1' via increasing ROS scavenging system. Plant Signaling Behav. 15 (8), 1780403. doi: 10.1080/15592324.2020.1780403
Zeng, L., Dehesh, K. (2021). The eukaryotic MEP-pathway genes are evolutionarily conserved and originated from Chlaymidia and cyanobacteria. BMC Genomics 22 (1), 137. doi: 10.1186/s12864-021-07448-x
Zhang, M., Li, K., Liu, J., Yu, D. Y. (2012). Identification and differential expression of two isogenes encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase in Glycine max. Plant Biotechnol. Rep. 6, 363–371. doi: 10.1007/s11816-012-0233-4
Zhang, T., Sun, M., Guo, Y., Shi, X., Yang, Y., Chen, J., et al. (2018). Overexpression of LiDXS and LiDXR from Lily (Lilium 'Siberia') enhances the terpenoid content in tobacco flowers. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.0090
Zhang, H., Zhang, L., Ji, Y., Jing, Y., Li, L., Chen, Y., et al. (2022). Arabidopsis SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot. 73 (1), 182–196. doi: 10.1093/jxb/erab391
Keywords: Cassia tora, Leguminosae, isoprenoids, drought, trade-off, 1-deoxy-D-xylulose-5-phosphate synthase, 1-deoxy-D-xylulose-5-phosphate reductoisomerase
Citation: Tian C, Quan H, Jiang R, Zheng Q, Huang S, Tan G, Yan C, Zhou J and Liao H (2023) Differential roles of Cassia tora 1-deoxy-D-xylulose-5-phosphate synthase and 1-deoxy-D-xylulose-5-phosphate reductoisomerase in trade-off between plant growth and drought tolerance. Front. Plant Sci. 14:1270396. doi: 10.3389/fpls.2023.1270396
Received: 31 July 2023; Accepted: 06 October 2023;
Published: 20 October 2023.
Edited by:
Mohsin Tanveer, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Tian, Quan, Jiang, Zheng, Huang, Tan, Yan, Zhou and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiayu Zhou, c3BpbmV6aG91QGhvbWUuc3dqdHUuZWR1LmNu; Hai Liao, ZGRsaWFvaGFpQGhvbWUuc3dqdHUuZWR1LmNu
†These authors have contributed equally to this work
 Chunyao Tian†
Chunyao Tian† Hai Liao
Hai Liao