
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 November 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1266032
This article is part of the Research Topic Highlights from the 12th Plant Growth-Promoting Rhizobacteria Workshop View all 12 articles
By improving plant nutrition and alleviating abiotic and biotic stresses, plant growth-promoting bacteria (PGPB) can help to develop eco-friendly and sustainable agricultural practices. Besides climatic conditions, soil conditions, and microbe-microbe interactions, the host genotype influences the effectiveness of PGPB. Yet, most GWAS conducted to characterize the genetic architecture of response to PGPB are based on non-native interactions between a host plant and PGPB strains isolated from the belowground compartment of other plants. In this study, a GWAS was set up under in vitro conditions to describe the genetic architecture of the response of Arabidopsis thaliana to the PGPB Pseudomonas siliginis, by inoculating seeds of 162 natural accessions from the southwest of France with one strain isolated from the leaf compartment in the same geographical region. Strong genetic variation of plant growth response to this native PGPB was observed at a regional scale, with the strain having a positive effect on the vegetative growth of small plants and a negative effect on the vegetative growth of large plants. The polygenic genetic architecture underlying this negative trade-off showed suggestive signatures of local adaptation. The main eco-evolutionary relevant candidate genes are involved in seed and root development.
Plant-Growth-Promoting Bacteria (PGPB) are bacterial strains isolated from diverse environmental reservoirs with the potential to provide multiple benefits to food and non-food crops (Bashan, 1998; Glick, 2012; Santoyo et al., 2016; Ramakrishna et al., 2019; Tian et al., 2020). For instance, PGPB can promote plant growth by improving plant nutrition and alleviating abiotic stresses such as drought and salinity (Choudhary et al., 2016; Singh et al., 2018; Kumar et al., 2020; Mokrani et al., 2020; Santoyo et al., 2021; Gamalero and Glick, 2022; Gupta et al., 2022). PGPB can also promote plant health by participating in defense against pathogens and pests (Liu et al., 2017; Liu et al., 2020; Morelli et al., 2020; Ruiu, 2020; Noman et al., 2021; Santoyo et al., 2021). In addition to the potential of PGPB to increase crop yield, PGPB can contribute to reducing environmental degradation by participating in phytoremediation techniques for soil and water decontamination (de-Bashan et al., 2012; Glick, 2012; Santoyo et al., 2016; Martínez-Hidalgo et al., 2019; Riva et al., 2020).
PGPB represent therefore a unique opportunity to develop eco-friendly and sustainable agricultural practices, a lofty goal that is especially relevant in the context of current global changes (Glick, 2012; Santoyo et al., 2016; Ramakrishna et al., 2019). For instance, such practices include the biotechnological use of PGPB as biofertilizers or biocontrol agents (De Silva et al., 2019; Leila and El-Hafid, 2020; Ünüvar and Ünlü, 2022). However, the effectiveness of most PGPB is highly influenced by climatic conditions, soil conditions, and microbe-microbe interactions, thereby deeply affecting their use in a wide range of agricultural conditions (Fageria and Stone, 2006; Gaiero et al., 2013; Rilling et al., 2019; Canfora et al., 2021). Moreover, the effects of a PGPB can highly depend on the genotype of the host plant (Wintermans et al., 2016; Ramakrishna et al., 2019; Yassue et al., 2021; Ramírez-Sánchez et al., 2022; Schultz et al., 2022). There is, therefore, a growing interest in the potential of harnessing the beneficial effects of individual members of the microbiota through plant breeding (Bergelson et al., 2021; Gutierrez and Grillo, 2022; Nerva et al., 2022; Santoyo, 2022). This requires the identification of host genetic factors either by using artificial genetic variation or by exploiting natural genetic variation (Bergelson et al., 2021). The latter approach was adopted by setting up genome-wide association studies (GWAS) on diverse plants including the model plants Arabidopsis thaliana (Wintermans et al., 2016; Cotta et al., 2020; Plucani do Amaral et al., 2023) and Medicago truncatula (Stanton-Geddes et al., 2013) as well as diverse crops such as maize (Vidotti et al., 2019; Yassue et al., 2021; Yassue et al., 2023), soybean (Torkamaneh et al., 2020) and common bean (Kamfwa et al., 2015). These GWAS revealed a highly polygenic architecture of response to PGPB, with the identification of multiple Quantitative Trait Loci (QTLs) with small effects. The fine mapping of these QTLs revealed candidate genes involved in plant immunity (Kamfwa et al., 2015; Vidotti et al., 2019; Yassue et al., 2023), hormonal pathways (Vidotti et al., 2019; Cotta et al., 2020; Plucani do Amaral et al., 2023), nutrient uptake and provision (Stanton-Geddes et al., 2013; Curtin et al., 2017; Torkamaneh et al., 2020; Plucani do Amaral et al., 2023; Yassue et al., 2023) and plant development (Wintermans et al., 2016; Cotta et al., 2020), which is in line with the main pathways identified by analysis of mutants affecting microbiota structure in plants (Bergelson et al., 2021).
While informative, most of these GWAS were conducted with PGPB isolated from the belowground compartment of plants (e.g. roots and rhizosphere), thereby neglecting the close interplay between plant genetics and PGPB isolated from the phyllosphere (Copeland et al., 2015; Ahmed, 2017; Remus-Emsermann and Schlechter, 2018; Abadi et al., 2020; Chalot and Puschenreiter, 2021). In addition, the number of GWAS investigating the genetic architecture of plant response to native PGPB strains remains scarce, thereby impeding the discovery of genetic and molecular mechanisms that might have been selected during plant-PGPB co-evolution (Baltrus, 2017; Lyu et al., 2021). For instance, in the three GWAS conducted on A. thaliana, plants were inoculated with either the strain Pseudomonas simiae WCS417r isolated from the rhizosphere of wheat (Wintermans et al., 2016), the strain Bacillus pumilus TUAT-1 isolated from rice roots (Cotta et al., 2020) or the strain Azoarcus oleearius DQS-4T isolated from oil-contaminated soil in Taiwan (Faoro et al., 2017; Plucani do Amaral et al., 2023). Finally, to our knowledge, it is still unknown whether plant polymorphic genes involved in interactions with PGPB have been shaped by natural selection. Yet, identifying candidate genes presenting suggestive signatures of local adaptation might be a starting point to unravel eco-evolutionary relevant biological pathways involved in the responsiveness of plants to PGPB (Bergelson and Roux, 2010; Roux and Bergelson, 2016).
In this study, we set up a GWAS under in vitro conditions to describe the genetic architecture of the response of A. thaliana to the bacterial species Pseudomonas siliginis. P. siliginis has been identified as the 6th most abundant bacterial species in the leaf and root microbiota across 163 natural populations of A. thaliana located in the southwest of France (Bartoli et al., 2018). Based on a bacterial strain isolated from the rhizosphere of wheat, P. siliginis was recently described as a new species of the Pseudomonas genus (Girard et al., 2021). Since then, P. siliginis has been isolated from the phyllosphere of plants of the genus Flaveria (Murillo-Roos et al., 2022) and we recently isolated six strains of P. siliginis from the A. thaliana leaf compartment (Ramírez-Sánchez et al., 2022). Two and four of these strains showed a PGPB effect on A. thaliana when inoculated at the seed and seedling stages, respectively (Ramírez-Sánchez et al., 2022). By inoculating seeds of 162 whole-genome sequenced natural accessions from the southwest of France with one of these PGPB strains isolated in the same geographical region, we aimed at (i) estimating the extent of genetic variation of aboveground vegetative growth response to this strain at different time points, (ii) describing the underlying genetic architecture by combining a Bayesian hierarchical model with a local score approach that has been applied in diverse plant and animal species (Fariello et al., 2017; Bonhomme et al., 2019; Bonhomme et al., 2021; Aoun et al., 2020; Apuli et al., 2021; Libourel et al., 2021; Brachi et al., 2022; Demirjian et al., 2022; Andrews et al., 2023; Boitard et al., 2023; Demirjian et al., 2023; Frachon et al., 2023; Neto and Hancock, 2023; Roux et al., 2023), and (iii) evaluating the strength of selection acting on the candidate genes by testing whether the SNPs significantly associated with natural variation of the plant growth response to the strain overlapped significantly with suggestive signatures of local adaptation.
In this study, we used the OTU6_Psi_1 strain of P. siliginis that has been isolated from the rosette of one individual of A. thaliana collected in spring 2015 in the natural population ESPE-B located in the southwest of France (Bartoli et al., 2018; Ramírez-Sánchez et al., 2022). This strain was demonstrated to have a PGPB effect on A. thaliana under in vitro conditions when inoculated both at seed or seedling stage (Ramírez-Sánchez et al., 2022). In addition, we revealed a high genetic variation in response to this strain among seven A. thaliana accessions located in the southwest of France to be suppressed (Ramírez-Sánchez et al., 2022). Using single-molecule real-time long reads with a PacBio Sequel II system, a de novo genome sequence was obtained for the OTU6_Psi_1 strain, showing a single chromosome containing 5,458 genes (Ramírez-Sánchez et al., 2022).
Fifty-four populations (each represented by three accessions) were selected to represent the ecological and genetic diversity observed among a set of 168 natural populations of A. thaliana from southwest of France (Frachon et al., 2018; Frachon et al., 2019) (Supplementary Table S1). The seeds coming from the maternal plants were harvested in June 2015. To reduce differences in the maternal effects of the 162 seed lots (i.e. 54 populations × three accessions), one plant per accession per generation was grown as followed: (i) several seeds of each accession were sown on October 1st 2016 in 7 x 7 x 6 cm plastic pots (Soparco®) filled with damp standard culture soil (PROVEEN MOTTE 20, Soprimex®); (ii) seeds were stratified at 4°C for four days; (iii) pots were put on November 4th 2016 to a greenhouse at 22°C with a 16 hours photoperiod; (iv) seedlings were thinned to one on November 25th 2016; (v) seedlings were transferred to the INRAE campus of Auzeville field station (France) on December 5th 2016; (vi) when plants started to flower, they were moved to a greenhouse that reproduces outdoor conditions (no extra light or heating) but protects the plants from rain; (vii) aratubes (Arasystem®) were put on each plant to prevent cross-pollination between accessions; (viii) seeds were collected from late April to early May 2017 and conserved at 4°C.
The sterilization of the seeds was performed with chlorine gas as previously described (Ramírez-Sánchez et al., 2022) and the seeds were then kept at 4°C.
An experiment of 3,888 plants was set up using a split-plot design with two treatments (i.e. inoculation with OTU6_Psi_1 and mock treatment) nested within two blocks. Each ‘block × treatment’ interaction was represented by 24 48-well plates with each well filled with 700 µL of 0.5x MS medium (Murashige and Skoog medium), which contains 2.2 g of MS medium, 0.5 g of 2-(N-Morpholino)-ethanesulfonic acid, 6.0 g of plant tissue culture agar, 1 L of deionized water and a pH adjusted to the range of 5.7-5.8. The six wells of the last column of each plate were sown with seeds from the Col-0 reference accession to control for micro-environmental variation among plates. The 162 accessions were randomly assigned to the remaining columns of the 24 plates, resulting for each accession in a total of 12 replicates (two columns of six wells) in each treatment. The same randomization was done among treatments within a block but was modified between the two blocks.
For the 162 natural accessions and Col-0, one seed was sown in each well. After the 7-day cold treatment, seeds were inoculated and placed in a phytotron (10 hours photoperiod, light intensity ~ 80 µmol m-2 s-1, 21°C, 50% hygrometry) with a daily plate randomization.
The OTU6_Psi_1 strain was grown from glycerol stock on solid medium in a 9 cm x 1.5 cm circle polystyrene Petri dish filled with TSA medium for one day. Colonies were diluted in 500 µl of sterile deionized water. 250 µl were then deposited in two new 9 cm x 1.5 cm circle polystyrene Petri dishes filled with TSA and spread with sterile beads. The two plates were incubated at 28°C overnight. Bacterial colonies were then resuspended and diluted in sterile deionized water to an OD600nm of 0.5 which corresponds to 3.90 x108 CFU/mL. Each seed was inoculated either with 5 µl of OTU6_Psi_1 (P. siliginis treatment) or 5 µl of sterile deionized water (mock treatment). Col-0 seeds that were sown in the last column of each 48-well plate were not inoculated. Plates were sealed with a micropore tape (3M Micropore Surgical x 9.14 m).
The germination date was recorded each day, between three and seven days after sowing. A picture of each 48-well plate was taken on 14, 21, and 28 days after inoculation (dai) using a photo-box designed in the lab and with a mobile camera (Samsung S6 16 Mpx). The vegetative growth was then visually scored for each plant using a scale established in Ramírez-Sánchez et al. (2022) and ranging from one (very small plant) to seven (well-grown plant). A total of 4,460 plants (3,888 plants of the 162 natural accessions and 572 Col-0 plants) were therefore phenotyped after inoculation at 14 dai, 21 dai and 28 dai.
We studied the genetic variation between the 54 natural populations of A. thaliana in response to OTU6_Psi_1 using the following mixed model (PROC MIXED procedure in SAS v. 9.4, SAS Institute Inc., Cary, NC, USA):
where Y corresponds to the score of plant development at a given dai; ‘µ’ is the overall mean of the phenotypic data; ‘Block’ accounts for differences in micro-environmental conditions between the two blocks; ‘Treatment’ corresponds to the mean effect of OTU6_Psi_1 in comparison with the mock treatment; ‘Population’ corresponds to the genetic differences among the 54 populations; ‘Accession(Population)’ corresponds to the mean genetic differences among accessions within populations; ‘Population*Treatment’ and ‘Accession(Population)*Treatment’ test if the rank among the 54 populations and the three accessions within populations differs among the two treatments, respectively; ‘Germ_date’ corresponds to the date of germination, ‘Score_dai_Col’ is a covariate that represents the mean value of the Col-0 plants for each plate and accounts for plate effects within a block; and ‘ϵ’ is the residual term. All factors were treated as fixed effects, as the levels of no factor were random samples from a population to which we intended to extrapolate. For calculating F-values, terms were tested over their appropriate denominators. As a split-plot design was set-up, the variance associated with ‘block × treatment’ was used as the error term to test ‘block’ and ‘treatment’ effects.
For each ‘treatment × dai’ combination, genotypic values of the 54 populations were estimated by calculating least-squares (LS) mean values of the term ‘Population’ in the following linear model (PROC MIXED procedure in SAS v. 9.4, SAS Institute Inc., Cary, NC, USA):
To calculate broad-sense heritability values (H²) of vegetative growth for each ‘treatment × dai’ combination, we first ran a linear model (PROC MIXED procedure in SAS v. 9.4, SAS Institute Inc., Cary, NC, USA):
We then ran the following model based on the residuals obtained from model 3:
The percentage of phenotypic variance explained by each term of Model 4 was estimated by the PROC VARCOMP procedure (REML method, SAS v. 9.4, SAS Institute Inc., Cary, NC, USA). H² values were then estimated as previously described (Lynch & Walsh, 1998; Huard-Chauveau et al., 2013) and using a formula adapted from Gallais (1990):
where ‘VF’ corresponds to the genetic variance among the 162 accessions, ‘VB’ is the variance associated with the ‘Block’ effect, ‘B’ is the number of blocks per treatment, ‘VR’ is the residual variance, and ‘N’ is the number of blocks.
At each dai, genotypic values of the 162 accessions were estimated by calculating LSmean values of the term ‘Accession’ of the following model (PROC MIXED procedure in SAS v. 9.4, SAS Institute Inc., Cary, NC, USA):
Based on genotypic values, we estimated the extent of plant growth response (PGR) to OTU6_Psi_1 for each population and each accession at 14 dai, 21 dai and 28 dai using the following formula:
Based on within-population genetic variation previously available for 168 natural populations of A. thaliana (Frachon et al., 2018), a Bayesian hierarchical model (Gautier, 2015) was applied to estimate the standardized allele frequencies corrected for the effect of population structure within each population for 1,638,649 SNPs across the genome (Frachon et al., 2018; Frachon et al., 2019). Standardized population allele frequencies were then retrieved for the 54 populations used in this work. Then, for each of the three PGR traits (i.e. PGR estimated at 14 dai, 21 dai and 28 dai), a genome scan was launched by estimating for each SNP the Spearman’s rho value and associated p-values between standardized allele frequencies and genotypic values obtained at the population level. Manhattan plots and quantile-quantile plots drawn on the p-values associated with Spearman’s rho values indicate an absence of an excess of low p-values. To better describe the genetic architecture associated with PGR, notably the identification of QTLs with small effects, we then implemented a local score approach on the set of p-values (Fariello et al., 2017). The local score allows detection of significant genomic segments by accumulating the statistical signals derived from multiple adjacent SNPs, thereby limiting the number of tests performed while utilizing all the available data (Fariello et al., 2017; Apuli et al., 2021). By following Bonhomme et al. (2019); Aoun et al. (2020); Libourel et al. (2021); Demirjian et al. (2022) and Demirjian et al. (2023), we then implemented a local score approach (with tuning parameter ξ = 2) on these p values. Finally, significant SNP-phenotype associations were found by estimating a chromosome-wide significance threshold for each chromosome (Bonhomme et al., 2019).
For each of the three PGR traits, the candidate genes underlying the QTLs were retrieved using a custom script written under the R environment (Libourel et al., 2021). The lists of the candidate genes were then submitted to the classification SuperViewer tool on the University of Toronto website (http://bar.utoronto.ca/ntools/cgibin/ntools_classification_superviewer.cgi) using the MapMan classification, to allow the identification of biological pathways significantly over-represented (P< 0.05).
To test whether the SNPs underlying the QTLs identified by BHM-LS (hereafter named top SNPs) have suggestive signatures of local adaptation, we followed the method previously described in Brachi et al. (2015) for each of the three PGR traits. We looked for an over-representation of the top SNPs in the extreme upper tail of the XtX distribution obtained for the set of 168 natural populations of A. thaliana (Frachon et al., 2018). For a given SNP, XtX is a measure of the variance of the standardized population allele frequencies, which results from a rescaling based on the covariance matrix of population allele frequencies (Gautier, 2015). The formula used to calculate the fold enrichment in suggestive signatures of local adaptation was:
Here n is the number of SNPs in the upper tail of the XtX distribution. Here, we considered XtX statistic values as suggestive of local adaptation if they were among the top 1% of genome-wide XtX statistic values (i.e. 16,386 SNPs). na is the number of top SNPs that were also in the upper tail of the XtX distribution. N is the total number of SNPs tested genome-wide and Na is the total number of top SNPs. Following the methodology described in Hancock et al. (2011), the statistical significance of enrichment was assessed by running 10,000 null circular permutations across the five chromosomes of A. thaliana.
The vegetative growth of the 54 natural populations of A. thaliana was on average significantly promoted by seed inoculation with the OTU6_Psi_1 strain of P. siliginis at 28 dai, but not at 14 dai and 21 dai (Table 1, Figure 1). At each time point of scoring, significant quantitative genetic variation was detected among populations as well as among accessions within populations across the two treatments (Table 1, Figure 1). Based on the 162 natural accessions, we detected significant and high broad-sense heritability (H²) values for each ‘treatment × time point of scoring’ combination (mock - 14 dai: H² = 0.80, P< 0.001; mock - 21 dai: H² = 0.79, P< 0.001; mock - 28 dai: H² = 0.82, P< 0.001; OTU6_Psi_1 - 14 dai: H² = 0.80, P< 0.001; OTU6_Psi_1 - 21 dai: H² = 0.80, P< 0.001; OTU6_Psi_1 - 28 dai: H² = 0.81, P< 0.001), suggesting that a large fraction of vegetative growth variation is explained by host genetic differences in our in vitro conditions. At each time point of scoring, we also detected a large and significant genetic variation among the 54 natural populations as well as among accessions within populations, for both the direction and the strength of the PGR to inoculation with the OTU6_Psi_1 strain (Table 1, Figure 2). Importantly, we observed a negative trade-off between the direction and the strength of the PGR to the OTU6_Psi_1 strain and the score of plant growth in absence of OTU6_Psi_1, with OTU6_Psi_1 having a positive effect on the vegetative growth of small plants and a negative effect on the vegetative growth of large plants (Figures 3A-C, Supplementary Table S2). This negative trade-off was observed at both the population and accession levels (Figures 3A-C, Supplementary Figure S1), thereby suggesting a phenomenon occurring among natural populations and within populations.
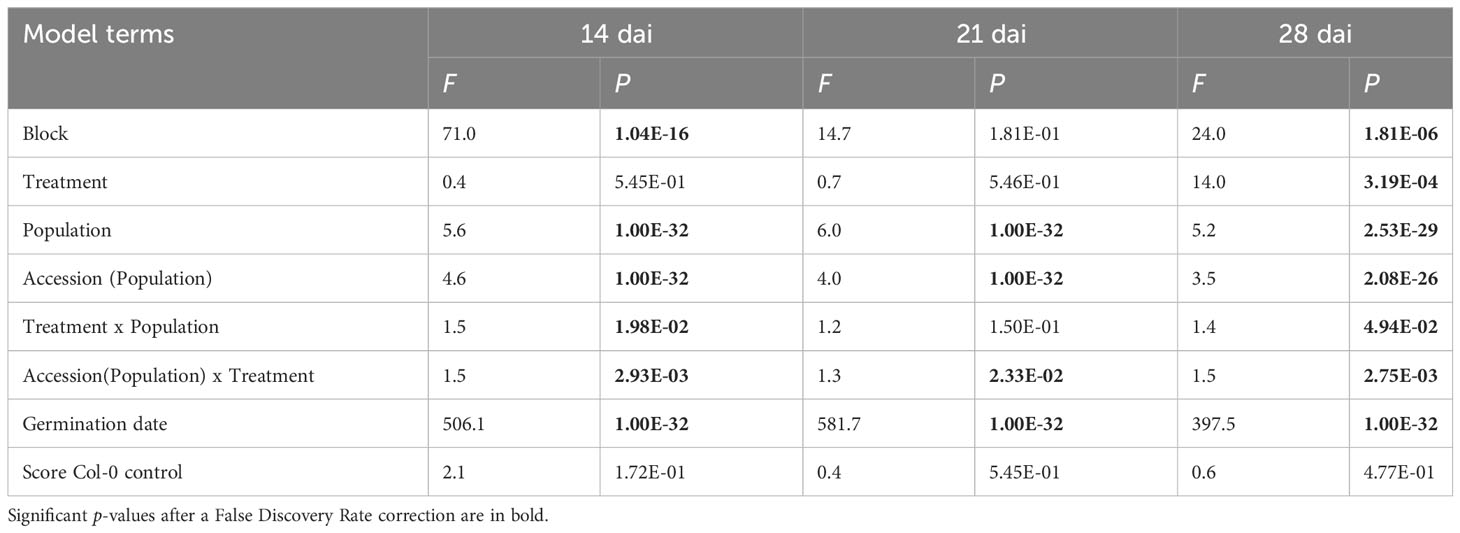
Table 1 Natural genetic variation of plant growth response to seed inoculation with the OTU6_Psi_1 strain at 14 dai, 21 dai and 28 dai.
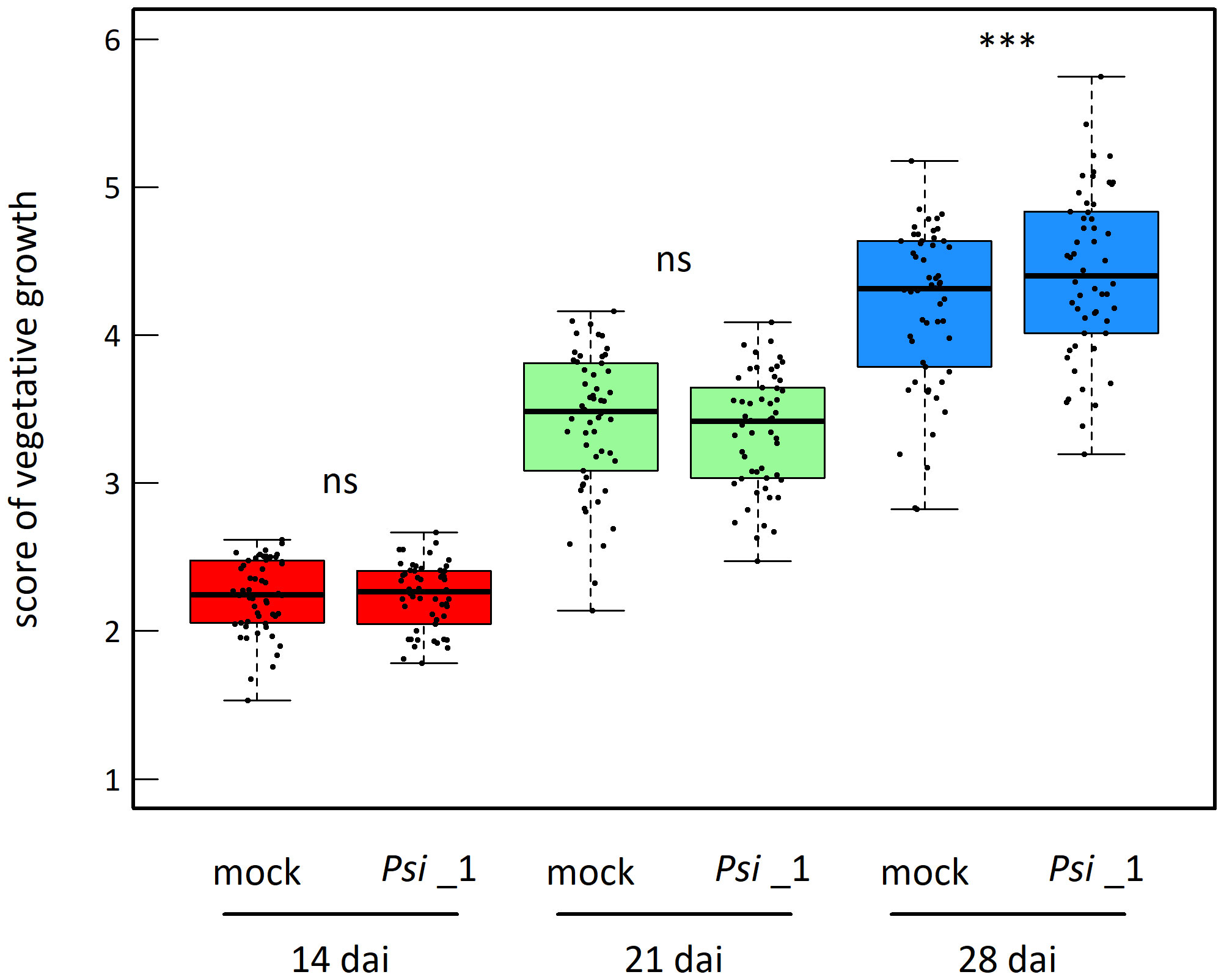
Figure 1 Mean genetic response to seed inoculation with the OTU6_Psi_1 strain at 14 dai, 21 dai and 28 dai. Each dot corresponds to the genotypic value of one of the 54 natural populations of A. thaliana. ns, non-significant, *** P< 0.001.
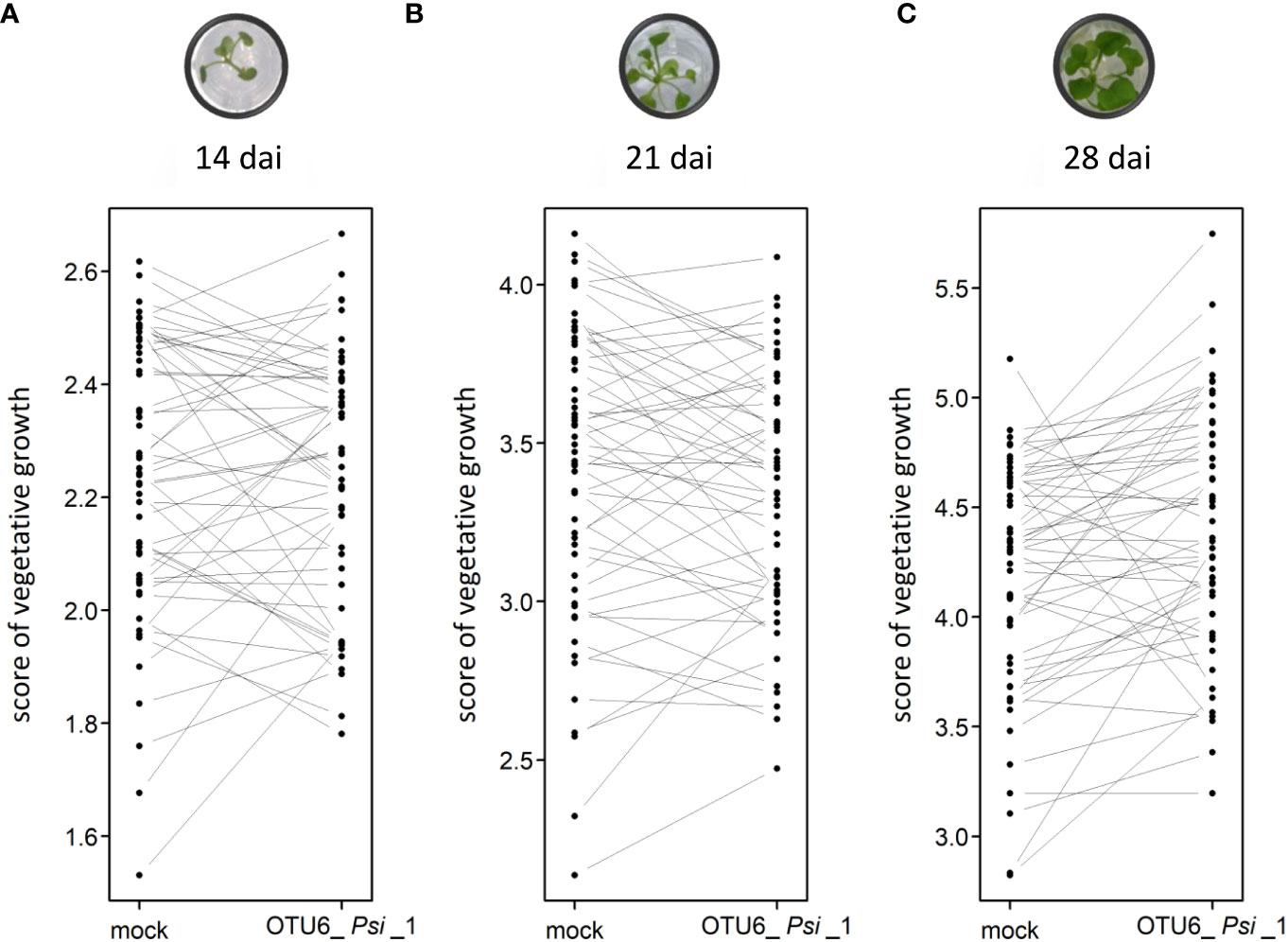
Figure 2 Interaction plots illustrating the genetic variation of response to the OTU6_Psi_1 strain at the population level at 14 dai (A), 21 dai (B) and 28 dai (C). Each dot corresponds to the genotypic value of one of the 54 populations of A thaliana. Each line corresponds to the response of one of the 54 populations to the inoculation with the OTU6_Psi_1 strain.
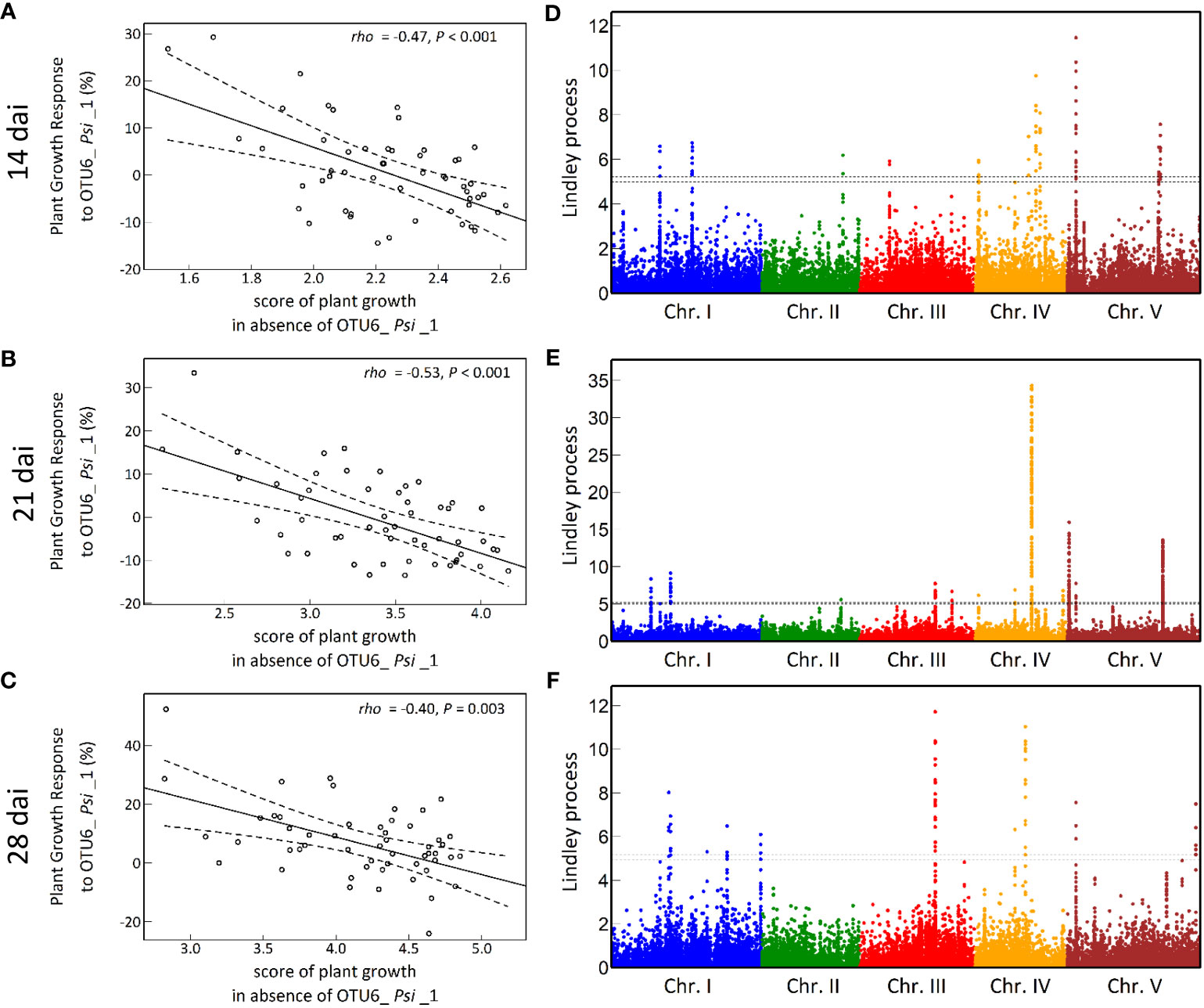
Figure 3 The genetic architecture of plant growth response (PGR) to the OTU6_Psi_1 strain. (A-C) Negative trade-offs at the population level between the level of PGR to OTU6_Psi_1 (expressed in percentage relative to the mock treatment) and the score of plant growth in absence of OTU6_Psi_1, at 14 dai, 21 dai and 28 dai. Each dot corresponds to the genotypic value of one of the 54 natural populations of A. thaliana. rho: correlation coefficient of Spearman between the response to OTU6_Psi_1 and the score of plant growth in absence of OTU6_Psi_1. P: p-value. The solid line corresponds to the fitted regression line, whereas the dashed lines delimit the band of 99% confidence intervals. (D-F) Manhattan plots of the Lindley process for PGR to OTU6_Psi_1 at 14 dai, 21 dai and 28 dai. The x-axis corresponds to the physical position of 1,638,649 SNPs on the five chromosomes. The dashed line indicates the chromosome-wide significance threshold.
A GWA mapping analysis combining a Bayesian hierarchical model with a local score approach (BHM-LS) revealed a polygenic architecture of PGR to the OTU6_Psi_1 strain, with the identification of a total of 570 top SNPs underlying a total of 43 QTLs, with 14, 18 and 11 QTLs detected at 14 dai, 21 dai and 28 dai, respectively (Figures 3D-F; Supplementary Tables S3, S4). The genetic architecture was highly dynamic over time, with five QTLs in common between the three time points of scoring (Figures 3D-F, Supplementary Table S4). Importantly, the top SNPs were significantly enriched in suggestive signatures of local adaptation across the genome of A. thaliana in southwest of France, with a fold enrichment (FE) that increases with the time of scoring (14 dai: FE = 2.4, P = 0.0763; 21 dai: FE = 3.5, P = 0.0236; 28 dai: FE = 5.2, P = 0.0092) (Supplementary Table S5). Relationships between PGR variation and allele frequencies of a top SNP presenting a suggestive signature of local adaptation are illustrated for each time point of scoring in Figure 4.

Figure 4 Illustration of the relationship between the level of plant growth response to the OTU6_Psi_1 strain (expressed in percent to the mock treatment) and the standardized allele frequencies of a top SNP presenting both one of the highest genotype-phenotype relationships and a signature of local adaptation at 14 dai (A), 21 dai (B) and 28 dai (C). rho: correlation coefficient of Spearman. P: p-value. The solid line corresponds to the fitted regression line, whereas the dashed lines delimit the band of 99% confidence intervals.
In line with the very short linkage disequilibrium of ~50 bp observed in French mapping populations of A. thaliana at the regional and local scales (Brachi et al., 2013; Frachon et al., 2017), the mean length of QTL intervals was rather small (~803 bp, quantile 5% ~ 47 bp, quantile 95% ~ 3.09 kb) (Supplementary Table S4), thereby allowing the fine mapping of candidate genes. Accordingly, the 43 detected QTLs overlapped with only 95 unique candidate genes, including 37, 50 and 25 unique candidate genes at 14 dai, 21 dai and 28 dai, respectively (Supplementary Table S4). In agreement with the dynamic genetic architecture between the three time points of scoring (Figure 3), only 14 candidate genes were common between two or three time points of scoring (Table 2, Figure 5). Interestingly, 4 out of the 14 candidate genes encode glycosyl transferases (GTs), including the three previously studied genes AtGALT31A, UGT76C4, and UGT76C5 (Geshi et al., 2013; Li et al., 2015; Poulsen et al., 2015; Liu et al., 2019). AtGALT31A, which encodes a ß–galactosyltransferase involved in the elongation of ß–1,6-galactan side chains on arabinogalactan proteins, is important for the progression of embryo development beyond the globular stage (Geshi et al., 2013). UGT76C4 and UGT76C5 are two nicotinate N-glycosyltransferases (Li et al., 2015; Liu et al., 2019), with a proposed physiological function for UGT76C4 in seed development and germination. While UGT76C4 is predominantly expressed in dry seeds, UGT76C5 was mainly detected in the root tissue of 7-day-old seedlings (Li et al., 2015). We also identified the AtCathB3 gene, encoding a cathepsin B-like protease, which is strongly induced during seed germination and early post-germination in A. thaliana (Iglesias-Fernández et al., 2014). Two other candidates, SBT3.5 and RLP48 (Receptor Like Protein 48), were previously characterized for their role in root growth and root hair development, respectively (Sénéchal et al., 2014; Stetter et al., 2015). The subtilisin-like serine protease SBT3.5 may play a role in the regulation of PME17 encoding a putative pectin methylesterase (PME) in A. thaliana roots (Sénéchal et al., 2014). First identified as a candidate gene in a GWAS on root hair traits in response to the scarce local phosphorus supply, RLP48 was then validated as being involved in root hair density (Stetter et al., 2015). Finally, an interesting candidate is JAZ11, a gene part of the jasmonate (JA)-zinc-finger inflorescence meristem (ZIM)-domain (JAZ) family (Liu et al., 2021). The jaz11 mutant exhibits JA-regulated root growth inhibition and increased susceptibility to Pseudomonas syringae pv. tomato (Pst) DC3000 (Liu et al., 2021).
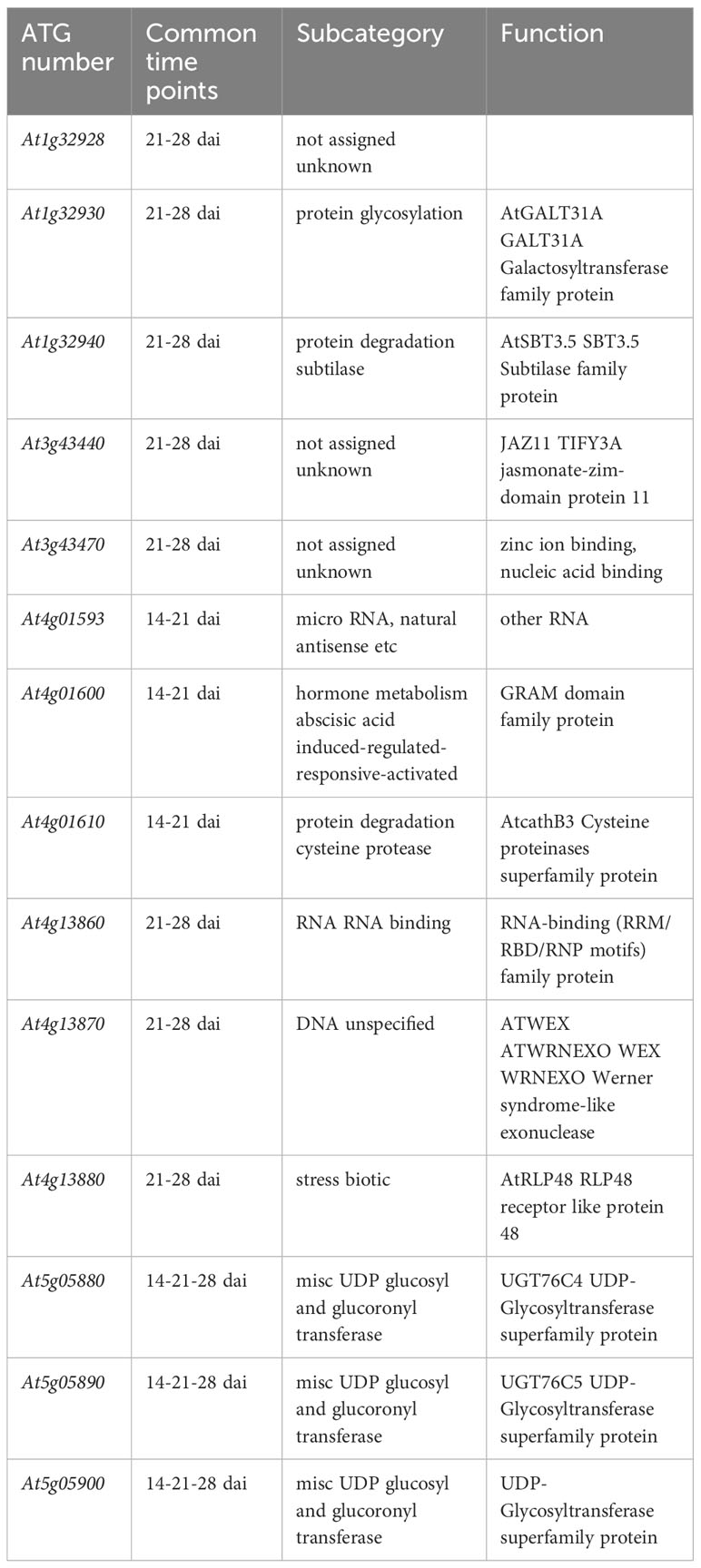
Table 2 List of the 14 candidate genes in response to the OTU6_Psi_1 strain and common between two or three time points of scoring.
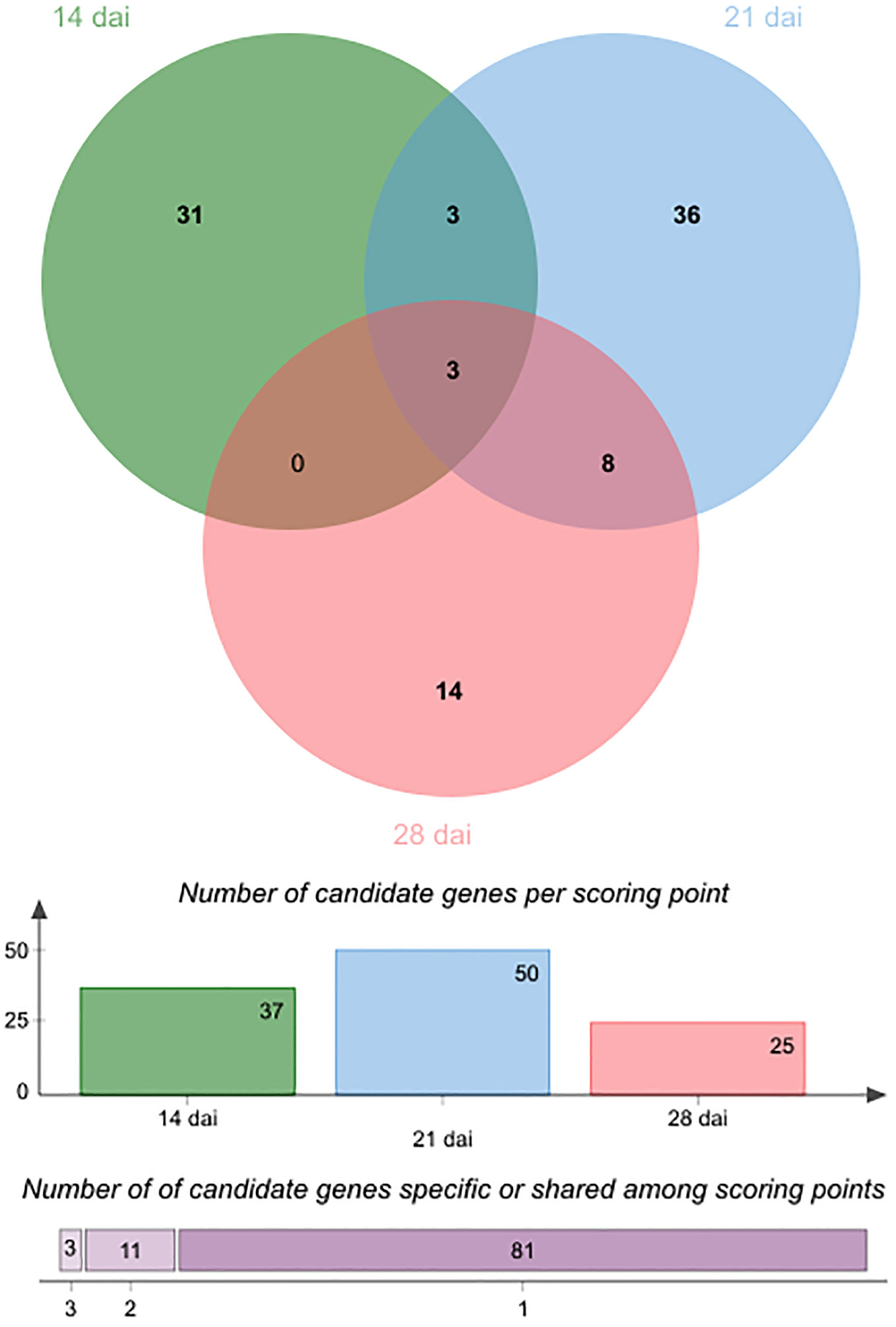
Figure 5 Venn diagram illustrating the number of specific and common candidate genes of plant growth response to the OTU6_Psi_1 strain between the three different time points of scoring. Colored bars indicate the number of candidate genes for each time point. The horizontally stacked bar plot indicates the number of candidate genes specific to one time point of scoring or common between two or three time points of scoring.
We also looked for biological processes significantly over-represented compared to the overall class frequency in the A. thaliana MapMan annotation. This allowed us to identify relevant candidate genes of PGR to the OTU6_Psi_1 strain. Based on the lists of unique candidate genes found for each time point of scoring, we detected one and two significantly over-represented biological processes at 14 dai and 21 dai, respectively. The over-represented biological process at 14 dai corresponds to the ‘cell wall modification’ class, whereas the over-represented biological processes at 21 dai correspond to the ‘co-factor and vitamin metabolism’ and ‘nucleotide metabolism’ classes. No significantly over-represented biological processes were detected at 28 dai. The three enriched classes contain seven candidate genes (Supplementary Table S4). For the ‘cell wall modification’ class, EXP17 encodes an expansin, a non-hydrolytic cell wall-loosening protein, which was suggested to participate in cell separation to promote lateral roots (LRs) emergence via the overlaying tissues of the primary root. Overexpression and silencing of EXP17 in A. thaliana increased and delayed the density of emerged LRs in the presence of auxin, respectively (Lee and Kim, 2013). The two other cell wall remodeling genes encode the xyloglucan endotransglucosylases/hydrolases XTH14 and XTH23 (Maris et al., 2009; Xu et al., 2020). XTH23 is involved in LR development under salt stress (Xu et al., 2020). The ‘co-factor and vitamin metabolism’ class contains two genes. Pyridoxine synthase 1 (PDX1.1) is part of a specific pathway involved in the biosynthesis of vitamin B6 (pyridoxal 5’-phosphate) in higher plants, which acts as a coenzyme for many metabolic enzymes but also as a potent antioxidant (Tambasco-Studart et al., 2005). Strikingly, pdx1 knockout mutants are impaired in root growth and early seedling development and are hypersensitive to osmotic and oxidative stresses (Chen and Xiong, 2005; Boycheva et al., 2015). AtFMN/FHy encodes a bi-functional enzyme involved in the metabolism of vitamin B2 (riboflavin) (Maruta et al., 2012). The ‘nucleotide metabolism’ class also contains two genes. AtNUDX2 is part of an A. thaliana Nudix (nucleoside diphosphates linked to some moiety X) hydrolase family of 28 genes. AtNUDX2 encodes an ADP-ribose pyrophosphatase that confers enhanced tolerance of oxidative stress in A. thaliana (Ogawa et al., 2009; Maruta et al., 2012). AMK2 encodes an adenosine monophosphate kinase that has a role in the architecture of chloroplasts (Lange et al., 2008).
Despite the phyllosphere representing 60% of the total biomass on Earth and concentrating 1026 bacteria (Vorholt, 2012), most GWAS carried out with non-pathogenic bacteria have focused on symbiotic bacteria or non-symbiotic PGPB isolated from the belowground compartment of plants (Stanton-Geddes et al., 2013; Wintermans et al., 2016; Curtin et al., 2017; Vidotti et al., 2019; Cotta et al., 2020; Torkamaneh et al., 2020). In this study, extensive genetic variation was observed among 162 natural accessions of A. thaliana in response to one strain of the native PGPB P. siliginis, which is an abundant and prevalent bacterial species in the leaf and root compartments of natural populations of A. thaliana located in the southwest of France (Ramírez-Sánchez et al., 2022). Because other strains of P. siliginis have been isolated from the leaf compartment of A. thaliana and characterized at the genomic level (Ramírez-Sánchez et al., 2022), it would be interesting to test whether the level of genetic variation of PGR and the underlying genetic architecture are similar among P. siliginis strains when inoculated at the seed stage. In addition, because P. siliginis and other Pseudomonas species with a PGPB effect, such as Pseudomonas moraviensis (Ramírez-Sánchez et al., 2022), belong to the subgroup Pseudomonas koreensis (Girard et al., 2021), it would be informative to check whether the genetics of PGR to P. siliginis extends to other phylogenetically close Pseudomonas species. Finally, in agreement with seed coating as an efficient way of introducing PGPB to seedlings (Ma, 2019; de Souza et al., 2020), we set up our GWAS by inoculating P. siliginis on seeds. Because the strength of the PGPB effect of P. siliginis on A. thaliana can depend on the developmental stage of the plants (Ramírez-Sánchez et al., 2022), it would be complementary to set up a GWAS by inoculating P. siliginis at the seedling stage.
Importantly, we observed a strong negative trade-off between plant growth in absence of P. siliginis and PGR to the OTU6_Psi_1 strain. To our knowledge, such a negative trade-off has not been reported in the literature. Identifying the mechanisms underlying this negative trade-off deserves further investigation. For instance, these mechanisms might rely on differences in seed size and physiology among the 162 accessions tested in this study. Beyond identifying the mechanisms, such a negative trade-off should promote the maintenance of genetic diversity at the underlying candidate genes, with the selection of growth-inductor responsive A. thaliana genotypes in presence of P. siliginis and the selection of growth-inhibitor responsive A. thaliana genotypes in absence of P. siliginis. Because the trade-off was observed both at the among-population and within-population levels, it suggests that the dynamics of A. thaliana - PGPB interactions should be studied at the metapopulation level rather than at the population level, as previously evidenced by studies on natural plant pathosystems such as Plantago lanceolata -Podosphaera plantaginis (Laine, 2005; Thrall et al., 2012) and A. thaliana - Pseudomonas syringae (Karasov et al., 2014).
Theoretical predictions suggest that the temporal regulation of QTLs often drives phenotypic changes in ontogenetic time, typically time-to-event or time-to-failure traits such as flowering time or death time (Johannes, 2007). Accordingly, previous GWAS performed on plant response to pathogens revealed temporal patterns in the detection of QTLs along the infection stages, with association peaks being detected only either at the earlier or at the later stages of infection (Aoun et al., 2017; Bartoli and Roux, 2017; Aoun et al., 2020; Demirjian et al., 2022; Demirjian et al., 2023). For instance, the atypical meiotic cyclin SOLO DANCERS gene was functionally validated in A. thaliana as conferring susceptibility to the bacterial pathogen Ralstonia solanacearum but only at the early stages of the infection (Aoun et al., 2020). Another A. thaliana gene, BWS1 (bacterial wilt susceptibility 1), was also revealed by GWAS as a susceptibility factor with a temporal dynamic in response to R. solanacearum (Demirjian et al., 2023). In this study, a similar dynamic in the detection of QTLs was observed for PGR to the OTU6_Psi_1 strain, suggesting that the PGPB effects conferred by P. siliginis depend on the time specificity of the genetic effects of A. thaliana. Importantly, in line with the mean PGPB effect of the strain OTU6_Psi_1 that increases over time, the enrichment in suggestive signatures of local adaptation of the candidate genes also increases over time, thereby highlighting the eco-evolutionary relevance of this native A. thaliana - P. siliginis interactions, similarly to the native interactions between A. thaliana and the bacterial pathogen P. syringae (Karasov et al., 2014; Roux and Bergelson, 2016). Our population genomics approach for identifying suggestive signatures of local adaptation across the genome of A. thaliana allows taking into account both the effect of selective processes at all life stages of A. thaliana while controlling for the effect of local demographic history (Frachon et al., 2017; Roux et al., 2023). This indirect approach is based on the calculation of the XtX statistics, analogous to FST but explicitly corrected for the covariance matrix of allele frequencies among populations (Gautier, 2015), and has been used to identify candidate genes associated with suggestive signatures of local adaptation in diverse species such as the European white oak (Leroy et al., 2020), the Aleppo pine Pinus halepensis (Ruiz Daniels et al., 2019), Caenorhabditis elegans (Crombie et al., 2022), the white-footed mice Peromyscus leucopus (Harris and Munshi-South, 2017) and the fungal wheat pathogen Zymoseptoria tritici (Hartmann et al., 2018). The PGPB effect observed on vegetative growth should therefore translate to fitness proxies such as total seed production in natural conditions, but remains to be tested. However, although a direct approach for testing local adaptation is based on setting up field experiments, in particular reciprocal field experiments, some fitness components such as germination rate can be hard to estimate under natural conditions (Savolainen et al., 2013). In addition, we must caution that estimating the effect of an adaptive allele on fitness proxies such as total seed production does not always predict the fate of the evolution of the frequency of this allele, as previously demonstrated for alleles conferring herbicide resistance (Roux et al., 2004; Roux et al., 2005; Roux et al., 2006; Vila-Aiub et al., 2011).
Of the 21 candidate genes highlighted, i.e. 14 genes in common between two or three scoring time points and seven genes of the three enriched biological processes, more than half of them are potentially involved in cell wall proliferation during seed and root development. Among these candidates, we observed both primary (i.e. expansins) and secondary (i.e. endoglycosylase/hydrolases) wall-loosening factors that are key players in cell wall structuring. For instance, expansins are cell wall-loosening proteins that directly induce cell wall extension by breaking non-covalent bonds between cellulose micro-fibrils and associated matrix polysaccharides in the cell wall (Lee and Kim, 2013). This study highlights EXP17 and xyloglucan endotransglucosylases/hydrolases, which facilitate LR emergence (Maris et al., 2009; Lee and Kim, 2013; Xu et al., 2020). XTH isoenzymes also strengthen the side-walls and cell walls of root hairs in the root differentiation zone after the completion of cell expansion (Maris et al., 2009). In addition, we identified GTs that catalyze protein glycosylation, a major post-translational modification of proteins, which significantly affects protein folding, conformation, distribution, stability and activity (Poulsen et al., 2015). Specifically, we identified a galactosyltransferase (Geshi et al., 2013) and three UGTs (UDP-glycosyltransferases), which are described to glycosylate various phytohormones and metabolites in response to biotic and abiotic stress in plants (Li et al., 2015; Rehman et al., 2018). Finally, studying PDX1 revealed that vitamin B6 is essential for root development and stress tolerance (Chen and Xiong, 2005; Boycheva et al., 2015). Other candidates are involved in both growth development and plant defense. For instance, the candidate gene SBT3.5 may have a direct or indirect role in root and/or root hair development, particularly via the processing of PME7 in planta (Sénéchal et al., 2014), as PME are ubiquitous cell wall enzymes involved in important developmental processes (Micheli, 2001). Beyond the role of SBT3.5 in root development, another subtilase, SBT3.3, plays a role in immune priming during plant-pathogen interactions (Ramírez et al., 2013). In addition, the candidate gene JAZ11 inhibits A. thaliana hypersensitivity to the key phytohormone JA and represses susceptibility to Pst DC3000 (Liu et al., 2021).
The next step to understand the genetic and molecular mechanisms underlying the adaptive negative trade-off in response to P. siliginis observed in this study would be to phenotype (i) the mutant lines of the candidate genes for PGR of both leaves and roots, and (ii) the ability of the OTU6_Psi_1 strain to multiply in the leaf and root compartment of seedlings. In addition, it would be of particular interest to study the expression profiles with the spatial and subcellular localization of the candidate genes after inoculation with the OTU6_Psi_1 strain. Finally, exploiting the haplotypic diversity of the candidate genes among the 162 natural accessions of A. thaliana used in this study may help to identify the polymorphisms that have been selected in nature to respond to the PGPB P. siliginis.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
DR-S: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Data curation, Software, Visualization. CG-V: Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. FR: Data curation, Formal Analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Visualization. FV: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. DR-S was funded by a Ph.D. fellowship from CONACyT. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 951444-PATHOCOM). This study was performed at the LIPME belonging to the Laboratoire d’Excellence (LABEX) entitled TULIP (ANR-10-LABX-41).
We are grateful to Rémi Duflos for his assistance during the statistical analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1266032/full#supplementary-material
Abadi, V. A. J. M., Sepehri, M., Rahmani, H. A., Zarei, M., Ronaghi, A., Taghavi, S. M., et al. (2020). Role of dominant phyllosphere bacteria with plant growth–promoting characteristics on growth and nutrition of maize (Zea mays L.). J. Soil Sci. Plant Nutr. 20, 2348–2363. doi: 10.1007/s42729-020-00302-1
Ahmed, A. (2017). Phyllospheric plant growth promoting bacteria. J. Bacteriol. Mycol. 5, 215–216. doi: 10.15406/jbmoa.2017.05.00119
Andrews, K. R., Seaborn, T., Egan, J. P., Fagnan, M. W., New, D. D., Chen, Z., et al. (2023). Whole genome resequencing identifies local adaptation associated with environmental variation for redband trout. Mol. Ecol. 32, 800–818. doi: 10.1111/mec.16810
Aoun, N., Desaint, H., Boyrie, L., Bonhomme, M., Deslandes, L., Berthomé, R., et al. (2020). A complex network of additive and epistatic quantitative trait loci underlies natural variation of Arabidopsis thaliana quantitative disease resistance to Ralstonia solanacearum under heat stress. Mol. Plant Pathol. 21, 1405–1420. doi: 10.1111/mpp.12964
Aoun, N., Tauleigne, L., Lonjon., F., Deslandes, L., Vailleau, F., Roux, F., et al. (2017). Quantitative disease resistance under elevated temperature: genetic basis of new resistance mechanisms to Ralstonia solanacearum. Front. Plant Sci. 8, 1387. doi: 10.3389/fpls.2017.01387
Apuli, R.-P., Richards, T., Rendon-Anaya, M., Karacic, A., Rönnberg-Wästljung, A.-C., Ingvarsson, P. K. (2021). The genetic basis of adaptation in phenology in an introduced population on Black Cottonwood (Populus trichocarpa, Torr. & Gray). BMC Plant Biol. 21, 317. doi: 10.1186/s12870-021-03103-5
Baltrus, D. A. (2017). Adaptation, specialization, and coevolution within phytobiomes. Curr. Opin. Plant Biol. 38, 109–116. doi: 10.1016/j.pbi.2017.04.023
Bartoli, C., Frachon, L., Barret, M., Rigal, M., Huard-Chauveau, C., Mayjonade, B., et al. (2018). In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 12, 2024–2038. doi: 10.1038/s41396-018-0152-7
Bartoli, C., Roux, F. (2017). Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front. Plant Sci. 8, 763. doi: 10.3389/fpls.2017.00763
Bashan, Y. (1998). Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 16, 729–770. doi: 10.1016/S0734-9750(98)00003-2
Bergelson, J., Brachi, B., Roux, F., Vailleau, F. (2021). Assessing the potential to harness the microbiome through plant genetics. Curr. Opin. Biotechnol. 70, 167–173. doi: 10.1016/j.copbio.2021.05.007
Bergelson, J., Roux, F. (2010). Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat. Rev. Genet. 11, 867–879. doi: 10.1038/nrg2896
Boitard, S., Liaubet, L., Paris, C., Fève, K., Dehais, P., Bouquet, A., et al. (2023). Whole-genome sequencing of cryopreserved resources from French Large White pigs at two distinct sampling times reveals strong signatures of convergent and divergent selection between the dam and sire lines. Genet. Sel. Evol. 55, 13. doi: 10.1186/s12711-023-00789-z
Bonhomme, M., Bensmihen, S., André, O., Amblard, E., Garcia, M., Maillet, F., et al. (2021). Distinct genetic basis for root responses to lipo-chitooligosaccharide signal molecules from different microbial origins. J. Exp. Bot. 72, 3821–3834. doi: 10.1093/jxb/erab096
Bonhomme, M., Fariello, M. I., Navier, H., Hajri, A., Badis, Y., Miteul, H., et al. (2019). A local score approach improves GWAS resolution and detects minor QTL: application to Medicago truncatula quantitative disease resistance to multiple Aphanomyces euteiches isolates. Heredity 123, 517–531. doi: 10.1038/s41437-019-0235-x
Boycheva, S., Dominguez, A., Rolcik, J., Boller, T., Fitzpatrick, T. B. (2015). Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis. Plant Physiol. 167, 102–117. doi: 10.1104/pp.114.247767
Brachi, B., Filiault, D., Whitehurst, H., Darme, P., Le Gars, P., Le Mentec, M., et al. (2022). Plant genetic effects on microbial hubs impact host fitness in repeated field trials. Proc. Natl. Acad. Sci. 119, e2201285119. doi: 10.1073/pnas.2201285119
Brachi, B., Meyer, C. G., Villoutreix, R., Platt, A., Morton, T. C., Roux, F., et al. (2015). Coselected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc. Natl. Acad. Sci. 112, 4032–4037. doi: 10.1073/pnas.1421416112
Brachi, B., Villoutreix, R., Faure, N., Hautekèete, N., Piquot, Y., Pauwels, M., et al. (2013). Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Mol. Ecol. 22, 4222–4240. doi: 10.1111/mec.12396
Canfora, L., Costa, C., Pallottino, F., Mocali, S. (2021). Trends in soil microbial inoculants research: A science mapping approach to unravel strengths and weaknesses of their application. Agriculture 11, 158. doi: 10.3390/agriculture11020158
Chalot, M., Puschenreiter, M. (2021). Exploring plant rhizosphere, phyllosphere and endosphere microbial communities to improve the management of polluted sites. Front. Microbiol. 12, 763566. doi: 10.3389/fmicb.2021.763566
Chen, H., Xiong, L. (2005). Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 44, 396–408. doi: 10.1111/j.1365-313X.2005.02538.x
Choudhary, D. K., Kasotia, A., Jain, S., Vaishnav, A., Kumari, S., Sharma, K. P., et al. (2016). Bacterial-mediated tolerance and resistance to plants under abiotic and biotic stresses. J. Plant Growth Regul. 35, 276–300. doi: 10.1007/s00344-015-9521-x
Copeland, J. K., Yuan, L., Layeghifard, M., Wang, P. W., Guttman, D. S. (2015). Seasonal community succession of the phyllosphere microbiome. Mol. Plant-Microbe Interact. 28, 274–285. doi: 10.1094/MPMI-10-14-0331-FI
Cotta, M. S., do Amaral, F. P., Cruz, L. M., Wassem, R., de Oliveira Pedrosa, F., Yokoyama, T., et al. (2020). Genome-wide association studies reveal important candidate genes for the Bacillus pumilus TUAT-1 - Arabidopsis thaliana interaction. BioRxiv. doi: 10.1101/2020.05.26.117002
Crombie, T. A., Battlay, P., Tanny, R. E., Evans, K. S., Buchanan, C. M., Cook, D. E., et al. (2022). Local adaptation and spatiotemporal patterns of genetic diversity revealed by repeated sampling of Caenorhabditis elegans across the Hawaiian Islands. Mol. Ecol. 31, 2327–2347. doi: 10.1111/mec.16400
Curtin, S. J., Tiffin, P., Guhlin, J., Trujillo, D. I., Burghart, L. T., Atkins, P., et al. (2017). Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 173, 921–931. doi: 10.1104/pp.16.01923
de-Bashan, L. E., Hernandez, J. P., Bashan, Y. (2012). The potential contribution of plant growth-promoting bacteria to reduce environmental degradation – A comprehensive evaluation. Appl. Soil Ecol. 61, 171–189. doi: 10.1016/j.apsoil.2011.09.003
Demirjian, C., Razavi, N., Desaint, H., Lonjon, F., Genin, S., Roux, F., et al. (2022). Study of natural diversity in response to a key pathogenicity regulator of Ralstonia solanacearum reveals new susceptibility genes in Arabidopsis thaliana. Mol. Plant Pathol. 23, 321–338. doi: 10.1111/mpp.13135
Demirjian, C., Razavi, N., Yu, G., Mayjonade, B., Zhang, L., Lonjon, F., et al. (2023). An atypical NLR gene confers bacterial wilt susceptibility in Arabidopsis. Plant Commun. 4, 100607. doi: 10.1016/j.xplc.2023.100607
De Silva, N. I., Brooks, S., Lumyong, S., Hyde, K. D. (2019). Use of endophytes as biocontrol agents. Fungal Biol. Rev. 33, 133–148. doi: 10.1016/j.fbr.2018.10.001
de Souza, N. L., Rocha, S. S., Narezzi, N. T., Tiepo, A. N., de Oliveira, A. L. M., Oliveira, H. C., et al. (2020). Differential impacts of plant growth-promoting bacteria (PGPB) on seeds of neotropical tree species with contrasting tolerance to shade. Trees 34, 121–132. doi: 10.1007/s00468-019-01902-w
Fageria, N. K., Stone, L. F. (2006). Physical, chemical, and biological changes in the rhizosphere and nutrient availability. J. Plant Nutr. 29, 1327–1356. doi: 10.1080/01904160600767682
Faoro, H., Rene Menegazzo, R., Battistoni, F., Gyaneshwar, P., do Amaral, F. P., Taulé, C., et al. (2017). The oil-contaminated soil diazotroph Azoarcus olearius DQS-4T is genetically and phenotypically similar to the model grass endophyte Azoarcus sp. BH72. Environ. Microbiol. Rep. 9, 223–238. doi: 10.1111/1758-2229.12502
Fariello, M.-I., Boitard, S., Mercier, S., Robelin, D., Faraut, T., Arnould, C., et al. (2017). Accounting for linkage disequilibrium in genome scans for selection without individual genotypes: The local score approach. Mol. Ecol. 26, 3700–3714. doi: 10.1111/mec.14141
Frachon, L., Arrigo, L., Rusman, A., Poveda, L., Qi, W., Scopece, G., et al. (2023). Putative signals of generalist plant species adaptation to local plant communities and abiotic factors. Mol. Biol. Evol. 40, msad036. doi: 10.1093/molbev/msad036
Frachon, L., Bartoli, C., Carrère, S., Bouchez, O., Chaubet, A., Gautier, M., et al. (2018). A genomic map of climate adaptation in Arabidopsis thaliana at a micro-geographic scale. Front. Plant Sci. 9, 967. doi: 10.3389/fpls.2018.00967
Frachon, L., Libourel, C., Villoutreix, R., Carrère, S., Glorieux, C., Huard-Chauveau, C., et al. (2017). Intermediate degrees of synergistic pleiotropy drive adaptive evolution in ecological time. Nat. Ecol. Evol. 1, 1551–1561. doi: 10.1038/s41559-017-0297-1
Frachon, L., Mayjonade, B., Bartoli, C., Hautekèete, N. C., Roux, F. (2019). Adaptation to plant communities across the genome of Arabidopsis thaliana. Mol. Biol. Evol. 36, 1442–1456. doi: 10.1093/molbev/msz078
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., Dunfield, K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750. doi: 10.3732/ajb.1200572
Gallais, A. (1990). Théorie de la sélection en amélioration des plantes. Eds. Gallais, A., Bannerot, H. (Paris, France: Masson), 588.
Gamalero, E., Glick, B. R. (2022). Recent advances in bacterial amelioration of plant drought and salt stress. Biology 11, 437. doi: 10.3390/biology11030437
Gautier, M. (2015). Genome-wide scan for adaptive divergence and association with population-specific covariates. Genetics 201, 1555–1579. doi: 10.1534/genetics.115.181453
Geshi, N., Johansen, J. N., Dilokpimol, A., Rolland, A., Belcram, K., Verger, S., et al. (2013). A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 76, 128–137. doi: 10.1111/tpj.12281
Girard, L., Lood, C., Höfte, M., Vandamme, P., Rokni-Zadeh, H., van Noort, V., et al. (2021). The ever-expanding Pseudomonas genus: Description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 9, 1766. doi: 10.3390/microorganisms9081766
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012, 963401. doi: 10.6064/2012/963401
Gupta, A., Mishra, R., Rai, S., Bano, A., Pathak, N., Fujita, M., et al. (2022). Mechanistic insights of plant growth promoting bacteria mediated drought and salt stress tolerance in plants for sustainable agriculture. Int. J. Mol. Sci. 23, 3741. doi: 10.3390/ijms23073741
Gutierrez, A., Grillo, M. A. (2022). Effects of domestication on plant-microbiome interactions. Plant Cell Physiol. 63, 1654–1666. doi: 10.1093/pcp/pcac108
Hancock, A. M., Brachi, B., Faure, N., Horton, M. W., Jarymowycz, L. B., Sperone, F. G., et al. (2011). Adaptation to climate across the Arabidopsis thaliana genome. Science 334, 83–86. doi: 10.1126/science.1209244
Harris, S. E., Munshi-South, J. (2017). Signatures of positive selection and local adaptation to urbanization in white-footed mice (Peromyscus leucopus). Mol. Ecol. 26, 6336–6350. doi: 10.1111/mec.14369
Hartmann, F. E., McDonald, B. A., Croll, D. (2018). Genome-wide evidence for divergent selection between populations of a major agricultural pathogen. Mol. Ecol. 27, 2725–2741. doi: 10.1111/mec.14711
Huard-Chauveau, C., Perchepied, L., Debieu, M., Rivas, S., Kroj, T., Kars, I., et al. (2013). An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PloS Genet. 9, e1003766. doi: 10.1371/journal.pgen.1003766
Iglesias-Fernández, R., Wozny, D., Iriondo-de Hond, M., Oñate-Sánchez, L., Carbonero, P., Barrero-Sicili,a, C. (2014). The AtCathB3 gene, encoding a cathepsin B-like protease, is expressed during germination of Arabidopsis thaliana and transcriptionally repressed by the basic leucine zipper protein GBF1. J. Exp. Bot. 65, 2009–2021. doi: 10.1093/jxb/eru055
Johannes, F. (2007). Mapping temporally varying quantitative trait loci in time-to-failure experiments. Genetics 175, 855–865. doi: 10.1534/genetics.106.059808
Kamfwa, K., Cichy, K. A., Kelly, J. D. (2015). Genome-wide association analysis of symbiotic nitrogen fixation in common bean. Theor. Appl. Genet. 128, 1999–2017. doi: 10.1007/s00122-015-2562-5
Karasov, T. L., Kniskern, J. M., Gao, L., DeYoung, B. J., Ding, J., Dubiella, U., et al. (2014). The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512, 436–440. doi: 10.1038/nature13439
Kumar, A., Singh, S., Gaurav, A. K., Srivastava, S., Verma, J. P. (2020). Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 11, 1216. doi: 10.3389/fmicb.2020.01216
Laine, A. L. (2005). Spatial scale of local adaptation in a plant-pathogen metapopulation. J. Evol. Biol. 18, 930–938. doi: 10.1111/j.1420-9101.2005.00933.x
Lange, P. R., Geserick, C., Tischendorf, G., Zrenner, R. (2008). Functions of chloroplastic adenylate kinases in Arabidopsis. Plant Physiol. 146, 492–504. doi: 10.1104/pp.107.114702
Lee, H. W., Kim, J. (2013). EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 54, 1600–1611. doi: 10.1093/pcp/pct105
Leila, B., El-Hafid, N. (2020). “Biofertilizers and biopesticides: Microbes for sustainable agriculture,” in Advances in plant microbiome and sustainable agriculture. Eds. Yadav, A. N., Rastegari, A. A., Yadav, N., Kour, D. (Singapore: Springer), 257–279.
Leroy, T., Louvet, J.-M., Lalanne, C., Le Provost, G., Labadie, K., Aury, J.-M., et al. (2020). Adaptive introgression as a driver of local adaptation to climate in European white oaks. New Phytol. 226, 1171–1182. doi: 10.1111/nph.16095
Li, W., Zhang, F., Chang, Y., Zhao, T., Schranz, M. E., Wang, G. (2015). Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27, 1907–1924. doi: 10.1105/tpc.15.00223
Libourel, C., Baron, E., Lenglet, J., Amsellem, L., Roby, D., Roux, F. (2021). The genomic architecture of competitive response of Arabidopsis thaliana is highly flexible among plurispecific neighborhoods. Front. Plant Sci. 12, 741122. doi: 10.3389/fpls.2021.741122
Liu, H., Brettell, L. E., Singh, B. (2020). Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 25, 841–844. doi: 10.1016/j.tplants.2020.06.003
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8, 2552. doi: 10.3389/fmicb.2017.02552
Liu, B., Seong, K., Pang, S., Song, J., Gao, H., Wang, C., et al. (2021). Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 231, 1525–1545. doi: 10.1111/nph.17477
Liu, L., Zhang, F., Li, G., Wang, G. (2019). Qualitative and quantitative NAD+ metabolomics lead to discovery of multiple functional nicotinate N-glycosyltransferase in Arabidopsis. Front. Plant Sci. 10, 1164. doi: 10.3389/fpls.2019.01164
Lynch, M., Walsh, B. (1998). Genetics and analysis of quantitative traits (Sunderland MA: Sinauer Associates, Inc), 980.
Lyu, D., Msimbira, L. A., Nazari, M., Antar, M., Pagé, A., Shah, A., et al. (2021). The coevolution of plants and microbes underpins sustainable agriculture. Microorganisms 9, 1036. doi: 10.3390/microorganisms9051036
Ma, Y. (2019). Seed coating with beneficial microorganisms for precision agriculture. Biotechnol. Adv. 37, 107423. doi: 10.1016/j.biotechadv.2019.107423
Maris, A., Suslov, D., Fry, S. C., Verbelen, J. P., Vissenberg, K. (2009). Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 60, 3959–3972. doi: 10.1093/jxb/erp229
Martínez-Hidalgo, P., Maymon, M., Pule-Meulenberg, F., Hirsch, A. M. (2019). Engineering root microbiomes for healthier crops and soils using beneficial, environmentally safe bacteria. Can. J. Microbiol. 65, 91–104. doi: 10.1139/cjm-2018-0315
Maruta, T., Yoshimoto, T., Ito, D., Ogawa, T., Tamoi, M., Yoshimura, K., et al. (2012). An Arabidopsis FAD pyrophosphohydrolase, AtNUDX23, is involved in flavin homeostasis. Plant Cell Physiol. 53, 1106–1116. doi: 10.1093/pcp/pcs054
Micheli, F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419. doi: 10.1016/S1360-1385(01)02045-3
Mokrani, S., Nabti, E. H., Cruz, C. (2020). Current advances in Plant Growth Promoting Bacteria alleviating salt stress for sustainable agriculture. Appl. Sci. 10, 7025. doi: 10.3390/app10207025
Morelli, M., Bahar, O., Papadopoulou, K. K., Hopkins, D. L., Obradović, A. (2020). Role of endophytes in plant health and defense against pathogens. Front. Plant Sci. 11, 1312. doi: 10.3389/fpls.2020.01312
Murillo-Roos, M., Abdullah, H. S. M., Debbar, M., Ueberschaar, N., Agler, M. T. (2022). Cross-feeding niches among commensal leaf bacteria are shaped by the interaction of strain-level diversity and resource availability. ISME 16, 2280–2289. doi: 10.1038/s41396-022-01271-2
Nerva, L., Sandrini, M., Moffa, L., Velasco, R., Balestrini, R., Chitarra, W. (2022). Breeding toward improved ecological plant-microbiome interactions. Trends Plant Sci. 27, 1134–1143. doi: 10.1016/j.tplants.2022.06.004
Neto, C., Hancock, A. (2023). Genetic architecture of flowering time differs between populations with contrasting demographic and selective histories. Mol. Biol. Evol. 40, msad185. doi: 10.1093/molbev/msad185
Noman, M., Ahmed, T., Ijaz, U., Shahid, M., Azizullah, Li, D., et al. (2021). Plant-microbiome crosstalk: Dawning from composition and assembly of microbial community to improvement of disease resilience in plants. Int. J. Mol. Sci. 22, 6852. doi: 10.3390/ijms22136852
Ogawa, T., Ishikawa, K., Harada, K., Fukusaki, E., Yoshimura, K., Shigeoka, S. (2009). Overexpression of an ADP-ribose pyrophosphatase, AtNUDX2, confers enhanced tolerance to oxidative stress in Arabidopsis plants. Plant J. 57, 289–301. doi: 10.1111/j.1365-313X.2008.03686.x
Plucani do Amaral, F., Wang, J., Williams, J., Tuleski, T. R., Joshi, T., Ferreira, M. A. R., et al. (2023). Mapping genetic variation in Arabidopsis in response to plant growth-promoting bacterium Azoarcus olearius DQS-4T. Microorganisms 11, 331. doi: 10.3390/microorganisms11020331
Poulsen, C. P., Dilokpimol, A., Geshi, N. (2015). Arabinogalactan biosynthesis: Implication of AtGALT29A enzyme activity regulated by phosphorylation and co-localized enzymes for nucleotide sugar metabolism in the compartments outside of the Golgi apparatus. Plant Signal. Behav. 10, e984524. doi: 10.4161/15592324.2014.984524
Ramakrishna, W., Yadav, R., Li, K. (2019). Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 138, 10–18. doi: 10.1016/j.apsoil.2019.02.019
Ramírez, V., López, A., Mauch-Mani, B., Gil, M. J., Vera, P. (2013). An extracellular subtilase switch for immune priming in Arabidopsis. PloS Pathog. 9, e1003445. doi: 10.1371/journal.ppat.1003445
Ramírez-Sánchez, D., Gibelin-Viala, C., Mayjonade, B., Duflos, R., Belmonte, E., Pailler, V., et al. (2022). Investigating genetic diversity within the most abundant and prevalent non-pathogenic leaf-associated bacteria interacting with Arabidopsis thaliana in natural habitats. Front. Microbiol. 13, 984832. doi: 10.3389/fmicb.2022.984832
Rehman, H. M., Nawaz, M. A., Shah, Z. H., Ludwig-Müller, J., Chung, G., Ahmad, M. Q., et al. (2018). Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 8, 1875. doi: 10.1038/s41598-018-19535-3
Remus-Emsermann, M. N. P., Schlechter, R. O. (2018). Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 218, 1327–1333. doi: 10.1111/nph.15054
Rilling, J. I., Acuña, J. J., Nannipieri, P., Cassan, F., Maruyama, F., Jorquera, M. A. (2019). Current opinion and perspectives on the methods for tracking and monitoring plant growth−promoting bacteria. Soil Biol. Biochem. 130, 205–219. doi: 10.1016/j.soilbio.2018.12.012
Riva, V., Riva, F., Vergani, L., Crotti, E., Borin, S., Mapelli, F. (2020). Microbial assisted phytodepuration for water reclamation: Environmental benefits and threats. Chemosphere 241, 124843. doi: 10.1016/j.chemosphere.2019.124843
Roux, F., Bergelson, J. (2016). The genetics underlying natural variation in the biotic interactions of Arabidopsis thaliana: The challenges of linking evolutionary genetics and community ecology. Curr. Top. Dev. Biol. 119, 111–156. doi: 10.1016/bs.ctdb.2016.03.001
Roux, F., Camilleri, C., Bérard, A., Reboud, X. (2005). Multigenerational versus single generation studies to estimate herbicide resistance fitness cost in Arabidopsis thaliana. Evolution 59, 2264–2269. doi: 10.1111/j.0014-3820.2005.tb00934.x
Roux, F., Frachon, L., Bartoli, C. (2023). The genetic architecture of adaptation to leaf and root bacterial microbiota in Arabidopsis thaliana. Mol. Biol. Evol. 40, msad093. doi: 10.1093/molbev/msad093
Roux, F., Gasquez, J., Reboud, X. (2004). The dominance of the herbicide resistance coist in several Arabidopsis thaliana mutant lines. Genetics 166, 449–460. doi: 10.1534/genetics.166.1.449
Roux, F., Giancola, S., Durand, S., Reboud, X. (2006). Bulding of an experimental cline with Arabidopsis thaliana to estimate herbicide fitness cost. Genetics 173, 1023–0131. doi: 10.1534/genetics.104.036541
Ruiu, L. (2020). Plant-growth-promoting bacteria (PGPB) against insects and other agricultural pests. Agronomy 10, 861. doi: 10.3390/agronomy10060861
Ruiz Daniels, R., Taylor, R. S., Gonzalez-Martinez, S. C., Vendramin, G. G., Fady, B., Oddou-Muratorio, S., et al. (2019). Looking for local adaptation: convergent microevolution in Aleppo pine (Pinus halepensis). Genes 10, 673. doi: 10.3390/genes10090673
Santoyo, G. (2022). How plants recruit their microbiome? New insights into beneficial interactions. J. Adv. Res. 40, 45–58. doi: 10.1016/j.jare.2021.11.020
Santoyo, G., Gamalero, E., Glick, B. R. (2021). Mycorrhizal-bacterial amelioration of plant abiotic and biotic stress. Front. Sustain. Food Syst. 5, 672881. doi: 10.3389/fsufs.2021.672881
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda Mdel, C., Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Savolainen, O., Lascoux, M., Merilä, J. (2013). Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820. doi: 10.1038/nrg3522
Schultz, C. R., Brantley, K. M., Wallace, J. G. (2022). The role of genetic variation in Zea mays response to beneficial endophytes. Plant Growth Regul. 98, 167–177. doi: 10.1007/s10725-022-00842-9
Sénéchal, F., Graff, L., Surcouf, O., Marcelo, P., Rayon, C., Bouton, S., et al. (2014). Arabidopsis PECTIN METHYLESTERASE17 is co-expressed with and processed by SBT3.5, a subtilisin-like serine protease. Ann. Bot. 114, 1161–1175. doi: 10.1093/aob/mcu035
Singh, V. K., Singh, A. K., Singh, P. P., Kumar, A. (2018). Interaction of plant growth promoting bacteria with tomato under abiotic stress: A review. Agric. Ecosyst. Environ. 267, 129–140. doi: 10.1016/j.agee.2018.08.020
Stanton-Geddes, J., Paape, T., Epstein, B., Briskine, R., Yoder, J., Mudge, J., et al. (2013). Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PloS One 8, e65688. doi: 10.1371/journal.pone.0065688
Stetter, M. G., Schmid, K., Ludewig, U. (2015). Uncovering genes and ploidy involved in the high diversity in root hair density, length and response to local scarce phosphate in Arabidopsis thaliana. PloS One 10, e0120604. doi: 10.1371/journal.pone.0120604
Tambasco-Studart, M., Titiz, O., Raschle, T., Forster, G., Amrhein, N., Fitzpatrick, T. B. (2005). Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. 102, 13687–13692. doi: 10.1073/pnas.0506228102
Thrall, P. H., Laine, A. L., Ravensdale, M., Nemri, A., Dodds, P. N., Barrett, L. G., et al. (2012). Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol. Lett. 15, 425–435. doi: 10.1111/j.1461-0248.2012.01749.x
Tian, L., Lin, X., Tian, J., Ji, L., Chen, Y., Tran, L. S. P., et al. (2020). Research advances of beneficial microbiota associated with crop plants. Int. J. Mol. Sci. 21, 1792. doi: 10.3390/ijms21051792
Torkamaneh, D., Chalifour, F. P., Beauchamp, C. J., Agrama, H., Boahen, S., Maaroufi, H., et al. (2020). Genome-wide association analyses reveal the genetic basis of biomass accumulation under symbiotic nitrogen fixation in African soybean. Theor. Appl. Genet. 133, 665–676. doi: 10.1007/s00122-019-03499-7
Ünüvar, Ö.C., Ünlü, E. S. (2022). “From hologenomes to biofertilizers in wheat production,” in Ancient wheats. Eds. Zencirci, N., Ulukan, H., Baloch, F. S., Mansoor, S., Rasheed, A. (Cham: Springer), 181–196.
Vidotti, M. S., Lyra, D. H., Morosini, J. S., Granato, Í.S.C., Quecine, M. C., Azevedo, J. L., et al. (2019). Additive and heterozygous (dis)advantage GWAS models reveal candidate genes involved in the genotypic variation of maize hybrids to Azospirillum brasilense. PloS One 14, e0222788. doi: 10.1371/journal.pone.0222788
Vila-Aiub, M. M., Neve, P., Roux, F. (2011). A unified approach to the estimation and interpretation of resistance costs in plants. Heredity 107, 386–394. doi: 10.1038/hdy.2011.29
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Wintermans, P. C. A., Bakker, P. A. H. M., Pieterse, C. M. J. (2016). Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. 90, 623–634. doi: 10.1007/s11103-016-0442-2
Xu, P., Fang, S., Chen, H., Cai, W. (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 104, 59–75. doi: 10.1111/tpj.14905
Yassue, R. M., Carvalho, H. F., Gevartosky, R., Sabadin, F., de Souza, P. H., Bonatelli, M. L., et al. (2021). On the genetic architecture in a public tropical maize panel of the symbiosis between corn and plant growth-promoting bacteria aiming to improve plant resilience. Mol. Breed. 41, 63. doi: 10.1007/s11032-021-01257-6
Yassue, R. M., Galli, G., Chen, C. P. J., Fritsche-Neto, R., Morota, G. (2023). Genome-wide association analysis of hyperspectral reflectance data to dissect the genetic architecture of growth-related traits in maize under plant growth-promoting bacteria inoculation. Plant Direct 7, e492. doi: 10.1002/pld3.492
Keywords: PGPB, Pseudomonas siliginis, seed inoculation, vegetative growth, negative tradeoff, GWA mapping
Citation: Ramírez-Sánchez D, Gibelin-Viala C, Roux F and Vailleau F (2023) Genetic architecture of the response of Arabidopsis thaliana to a native plant-growth-promoting bacterial strain. Front. Plant Sci. 14:1266032. doi: 10.3389/fpls.2023.1266032
Received: 24 July 2023; Accepted: 23 October 2023;
Published: 09 November 2023.
Edited by:
Louise Mary Nelson, University of British Columbia, CanadaReviewed by:
Joseph Edwards, The University of Texas at Austin, United StatesCopyright © 2023 Ramírez-Sánchez, Gibelin-Viala, Roux and Vailleau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabienne Vailleau, ZmFiaWVubmUudmFpbGxlYXVAaW5yYWUuZnI=
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.