
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 January 2024
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1263595
This article is part of the Research Topic The Interplay of Plant Biotic and Abiotic Stresses: Mechanisms and Management View all 7 articles
 Jing Li1
Jing Li1 Shukuan Guo1
Shukuan Guo1 Yin Min Htwe1
Yin Min Htwe1 Xiwei Sun1
Xiwei Sun1 Lixia Zhou1
Lixia Zhou1 Fangyuan Wang1
Fangyuan Wang1 Chunru Zeng1
Chunru Zeng1 Shuangyan Chen1,2
Shuangyan Chen1,2 Amjad Iqbal1,3
Amjad Iqbal1,3 Yaodong Yang1*
Yaodong Yang1*MYB transcription factors regulate the growth, development, and secondary metabolism of plant species. To investigate the origin of color variations in coconut pericarp, we identified and analyzed the MYB gene family present in coconut. According to the sequence of MYB genes in Arabidopsis thaliana, homologous MYB gene sequences were found in the whole genome database of coconut, the conserved sequence motifs within MYB proteins were analyzed by Motif Elicitation (MEME) tool, and the sequences without conservative structure were eliminated. Additionally, we employed RNA-seq technology to generate gene expression signatures of the R2R3-MYB genes across distinctive coconut parts exhibiting diverse colors. To validate these profiles, we conducted quantitative PCR (qPCR). Through comprehensive genome-wide screening, we successfully identified a collection of 179 MYB genes in coconut. Subsequent phylogenetic analysis categorized these 179 coconut MYB genes into 4-subfamilies: 124 R2R3-MYB, 4 3R-MYB types, 4 4R-MYB type, and 47 unknown types. Furthermore, these genes were further divided into 34 subgroups, with 28 of these subgroups successfully classified into known subfamilies found in Arabidopsis thaliana. By mapping the CnMYB genes onto the 16 chromosomes of the coconut genome, we unveiled a collinearity association between them. Moreover, a preservation of gene structure and motif distribution was observed across the CnMYB genes. Our research encompassed a thorough investigation of the R2R3-MYB genes present in the coconut genome, including the chromosomal localization, gene assembly, conserved regions, phylogenetic associations, and promoter cis-acting elements of the studied genes. Our findings revealed a collection of 12 R2R3-MYB candidate genes, namely CnMYB8, CnMYB15, CnMYB27, CnMYB28, CnMYB61, CnMYB63, CnMYB68, CnMYB94, CnMYB101, CnMYB150, CnMYB153, and CnMYB164. These genes showed differential expressions in diverse tissues and developmental stages of four coconut species, such as CnMYB68, CnMYB101, and CnMYB28 exhibited high expression in majority of tissues and coconut species, while CnMYB94 and CnMYB164 showed lower expression. These findings shed light on the crucial functional divergence of CnMYB genes across various coconut tissues, suggesting these genes as promising candidate genes for facilitating color development in this important crop.
All eukaryotic organisms contain the MYB transcription factor (TF) family, which is known for its extensive size and its ability to regulate a wide range of physiological processes in plants. These processes include environmental adaptation, hormone signal transduction, development, metabolic regulation, (Li et al., 2019; Li T. et al., 2023; Ma et al., 2023). The name MYB is derived from the conserved DNA-binding domain known as the MYB domain. The MYB proteins can be classified into different groups based on the repetitive units they possess: 1R-MYB (1- repeat), R2R3-MYB (2-repeats), R1R2R3-MYB (3-repeats), and 4R-MYB (4-repeats). Each repeat contains approximately 50-53 amino acids and encodes three α-helices, with the second and third helices forming a helix-turn-helix (HTH) structure (Du et al., 2012). 1R-MYB plays an important role in regulating plant transcription and maintaining chromosome structure. The R2R3-MYB family genes contain two conserved R2 and R3 repeat sequences in the MYB binding domain, as well as a regulatory domain (activation or inhibition function) in the C-terminus variable region. It has numerous members and diverse functions, widely participating in cell differentiation, secondary metabolism, environmental stress, and invasion of diseases and pests; The conserved domains of the 3R-MYB family genes are composed of R1, R2, and R3, which are mainly involved in the regulation of cell differentiation and cell cycle; The conserved domain of the 4R-MYB subfamily genes consists of four R1/R2 repeat sequences. The plant TF database (http://planttfdb.gaolab.org) contains a total of 22,032 MYB and 15,369 MYB-related sequences (Tian et al., 2020). The MYB family is abundant in numerous plant species, with specific examples including 197 AtMYB genes in Arabidopsis thaliana (Katiyar et al., 2012), 155 PaMYB genes in Petunia axillaris (Chen et al., 2021), 174 MrMYB genes in Myrica rubra (Cao et al., 2021), 133 DcMYB genes in Dendrobium catenatum (Zhang et al., 2021), and 159 EgMYB genes in Elaeis guineensis (Zhou et al., 2020). The R2R3-MYB subfamily is the most prevalent among plants, distinguished by the presence of two types of R domains located at the N-terminal end. These R2R3-MYB proteins commonly demonstrate transcriptional activation or repression capacity at their C-terminal end (Tolosa and Zhang, 2020). Because of these DNA-binding domain characteristics, the R2R3-MYB family was also divided into 25 subgroups in Arabidopsis (Stracke et al., 2001). In other plant species, the R2R3-MYB proteins can be classified into 25 or more classes (Li et al., 2020; Ding et al., 2023).
Pigments are significant secondary metabolites that play vital roles in photosynthesis and petal coloration. Extensive research has shown that MYB genes are involved in pigment formation within various plant parts, such as petals and peels (Chen et al., 2021; Yang et al., 2021). Anthocyanins are natural, water soluble pigments that play a vital role in plants. They contribute a variety of colors to reproductive organs and vegetative tissues, ranging principally from reddish to purplish or bluish shades (Li et al., 2022). The MYB genes in plants play a crucial role in the accumulation of anthocyanins and have a significant influence on the development of color in various organs such as floral parts, leaves, and pericarp (Mackon et al., 2021). Numerous studies have established the crucial role of R2R3-MYB transcription factor in directlyre gulating genes expression associated with anthocyanin biosynthesis. It serves as a key regulator in controlling anthocyanin production in various horticultural plants, including vegetables, fruits, and ornamentals (Duan et al., 2022; Wang et al., 2022; Jiang et al., 2023). The AaMYB2, an R2R3-MYB gene isolated by Li et al. (2016) from Anthurium andraeanum (Hort.), has been identified as a specific transcriptional regulator for anthocyanin production in spathes and leaves. Furthermore, recent studies have revealed that certain members of the R2R3-MYB family also act as principal regulators of carotenoid synthesis (Sagawa et al., 2016; Zhu et al., 2017; Ampomah-Dwamena et al., 2019; Li et al., 2020; Yin et al., 2022). Thus, the influence of R2R3-MYB genes extends beyond anthocyanin synthesis and encompasses the regulation of carotenoid production as well.
Coconut (Cocos nucifera L.) is an important fruit tree and woody oil crop that thrives in hot regions and possesses a unique quality trait- the color of its epicarp. Coconut peels can display a range of colors, including orange, brown, yellow, and green. Interestingly, the leaf stalk epidermis (LSE), sepal (SE), and flower spike branch (FSB) of different coconut species exhibit the same color as their respective peels (Figure 1). In contrast to many other fruits, the outer skin color of the coconut remains relatively stable throughout its development, without undergoing significant changes. Unlike some fruits that undergo a distinct transition process associated with maturity, coconut do not exhibit such a marked transformation. Availability of the comprehensive genome sequence of the coconut (Xiao et al., 2017) has provided a valuable resource for analyzing MYB genes across the entire genome.
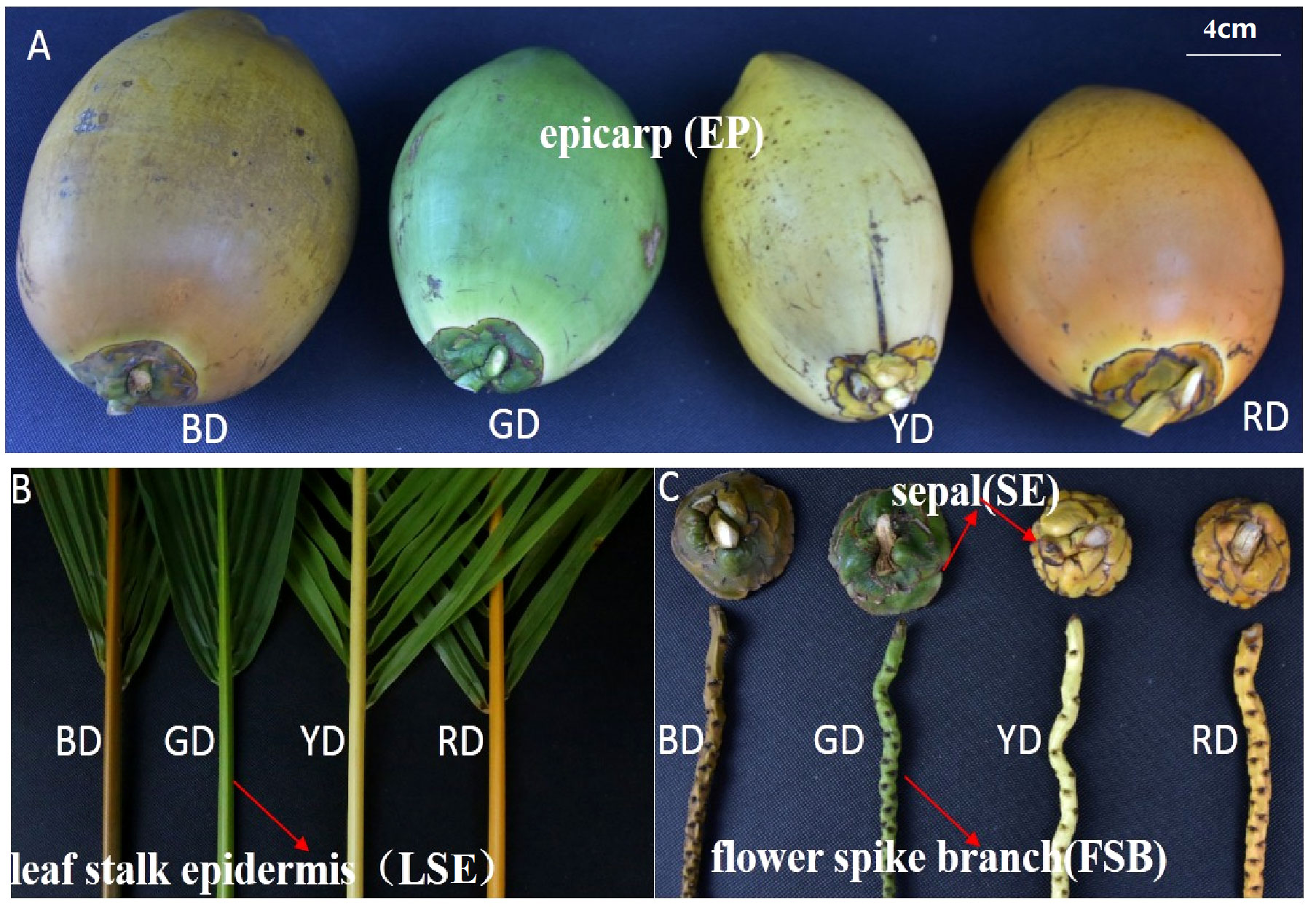
Figure 1 Different tissue parts of different coconut species. Leaf stalk epidermis (LSE), epicarp (EP), sepal (SE) and flower spike branch (FSB) of 7-month-old fruit of Red dwarf Coconut (RD), Yellow dwarf Coconut (YD), Brown dwarf Coconut (BD) and Green dwarf Coconut (GD).
Although several studies have established the role of MYBtranscription factors (TFs) in many biochemical and physiological processes in plants, a comprehensive identification and characterization of MYB TFs specific to coconuts is still lacking. Additionally, the expression patterns of MYB genes in different parts of the coconut are not yet fully understood. Notably, there has been no comprehensive systematic analysis of the entire array of MYB genes conducted in coconuts. Therefore, addressing this knowledge gap and conducting a comprehensive genomic analysis of the MYB gene family, with a specific emphasis on R2R3-MYB in coconuts, is crucial. The information provided by these results may help further analyze the function of the CnMYB gene and elucidate its secondary metabolic mechanism. This report aims to address this critical need by focusing on comprehensive genomic profiling and expression analysis of MYB genes in coconuts, with a specific emphasis on highlighting the R2R3-MYB subgroup.
The draft coconut genome served as the reference for obtaining the coconut MYB gene sequences (Xiao et al., 2017). A Hidden Markov Model (HMM) profile representing the MYB DNA-binding domain with accession (PF00249) was acquired from the Pfam protein family database (http://pfam.xfam.org/) (Finn et al., 2016). Subsequently, this profile was employed as a query (P < 0.001) for the identification of all potential CnMYB genes. In parallel, AtMYB gene sequences were utilized as query sequences to detect MYB genes within the coconut genome. The identified MYB genes containing conserved domains underwent further analysis, while those lacking the PF00249 conserved domain were excluded from the dataset. Amino acid sequences of Arabidopsis MYB proteins (AtMYBs) were obtained from the Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/). The BLAST searches were conducted against the coconut genome databases using Arabidopsis MYB protein sequences as queries to identify CnMYB gene families (Katiyar et al., 2012). In total, 179 CnMYB genes were identified from the coconut genome. Additional information regarding the number of amino acids, molecular weight (MW) and isoelectric point (pI) of each MYB protein of interest was gathered using the ExPASy proteomic website (https://web.expasy.org/compute_pi/). The CELLO tool was used to predict the intracellular distribution of all CnMYB genes.
A neighbor-joining (NJ) phylogenetic analysis was conducted by MEGA-X based on the alignment. Bootstrap analysis with 1000 replicates was performed to calculate the reliability of the NJ tree (Kumar et al., 2018). The necessary data, including mRNA sequences, CDS and gene annotation summaries of coconut MYB genes, were retrieved from Gigascience Database 2017 (http://dx.doi.org/10.5524/100347) (Supplementary Table 1). To confirm the structures of the coconut MYB genes, the mRNA sequences were aligned with the complete coconut genome sequence. The Gene Structure Display Server was then employed to analyze and determine the intron-exon organization of the coconut MYB genes. For the analysis of conserved sequence motifs within MYB proteins, the Motif Elicitation (MEME) tool (http://meme-suite.org/tools/meme) (Bailey et al., 2009) was utilized, and the results are presented in Supplementary Table 2. Additionally, the TBtools software (version 1.045) developed by Chen et al. (2020) was used to visualize the chromosomal localization of MYB genes in the coconut genome. This analysis utilized the annotated genomic data from the coconut genome database to accurately map the MYB genes onto the corresponding chromosomes.
Samples from four distinct species of coconuts, namely Red Dwarf (RD), Yellow Dwarf (YD), Brown Dwarf (BD), and Green Dwarf (GD), were carefully selected for analysis. The specific parts examined included the leaf stalk epidermis (LSE), epicarp (EP), sepal (SE), and flower spike branch (FSB) of 7-month-old fruits (Figure 1). Nine coconut fruits from the same fruit bunch were taken. Three coconut fruits were taken for collection of peel, sepals, floral branches and petioles separately and mixed them to form one biological replicate. In this way two different group were prepared to make another two biological replicate. These research samples were generously provided by the Coconut Research Institute (CRI), Chinese Academy of Tropical Agricultural Sciences (CATAS), located in Wenchang, Hainan, China. Following collection, all samples were immediately cryopreserved using liquid nitrogen and subsequently transferred to a freezer set at -80°C for future use.
The RNA extraction was carried out by the MRIP method (Method for RNA isolation from Palm) (Xiao et al., 2012) and has been improved. The protocol of the RNA extraction method according to the Iqbal et al. (2019). The quality of the extracted RNA (i.e., degradation and contamination) was assessed using 1% agarose gels. Additionally, the integrity of the RNA was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States), while the concentration was measured using a Nanodrop Spectrophotometer (IMPLEN, CA, United States). Sequencing of the samples was performed on the BGISEQ-MGI2000 platform instrument at BGI Genomics (Shenzhen 518083, China) with three biological replicates for each sample. To ensure high-quality data, the raw sequencing reads were processed to obtain clean reads. This involved excluding reads with adapters, reads with unknown bases more than 5% and low-quality base ratios more than 20% were filtered using SOAPnuke (version 1.4.0) (Chen et al., 2018). The resulting clean reads were stored in FASTQ format. Subsequently, the data were aligned to the reference genome using HISAT (v2.1.0) (Kim et al., 2015), and then matched with the assembled unique genes using Bowtie2 (v2.2.5) (Langmead and Salzberg, 2012). RNA-seq by expectation maximization (RSEM) (version 1.2.8) was utilized to calculate the expression levels of the genes (Li and Dewey, 2011). For functional annotation, the assembled unigenes were annotated using databases such as KEGG and GO, and transcription factors were predicted as well. Differential gene analysis within groups was conducted using DESeq with the conditions of Fold Change ≥ 2 and adjusted P-value ≤ 0.001 (Wang et al., 2010).
To conduct a comprehensive analysis of coconut MYB genes, transcriptoe datasets from various coconut tissues, including LSE, EP, SE and FSB, were utilized. The expression levels of genes were quantified using Reads Per Kilobase Million (RPKM) values, which were further Log2 transformed to facilitate comparative analysis. To visualize the expression patterns of 12 R2R3-MYB genes, a heatmap was generated using TBtools.
Forward and reverse primers for qPCR analysis were designed by National Center for Biotechnology Information (NCBI) Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast) with melting temperatures of 55-60°C, primer length 19-25bp, GC content 50-60% and amplicon size of 80-200bp. To prevent amplification of non-target gDNA, the primers were designed to span intronic regions. The properties of each primer were evaluated using the PCR Primer Stats software. The primers used for qPCR analysis can be found in Supplementary Table 4. The reference gene CnACTIN was used as an internal control (Xia et al., 2014).The Quick and Reliable RNA Extraction Method (QRREM) was employed to isolate total RNA from the epicarp of coconut fruit, following the protocol of Iqbal et al. (2019). Subsequently, the isolated RNA underwent quality and quantity assessment through agarose gel electrophoresis and Nanodrop spectrophotometer analysis. For reverse transcription, 1 μg of RNA was used with the MightyScript first-strand cDNA synthesis kit following the manufacturer’s instructions. The qPCR reactions were conducted using the 2 × SYBR Green qPCR ProMix in 96-well optical plates on a Mastercycler ep realplex4 machine. The qPCR reactions were carried out with a total reaction volume of 10 μL, consisting of amplification at 95°C for 5 s, 55°C for 15 s, and 68°C for 20 s. The melting stage involved heating from 60°C to 95°C for 20 min. Each experiment was conducted with biological and technical triplicates. The fold change in expression level for each sample was calculated by normalizing the CT value relative to a reference gene, using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
The experiments were conducted in triplicate (n=3) to ensure the reliability of the findings. Mean values, accompanied by standard errors of the mean, were used to present the data. SAS software (SAS Inc., Cary, NC, USA) was employed to perform ANOVA (analysis of variance) and DMRT (Duncan’s Test) with a significance level set at p < 0.05 and p < 0.01 to assess the significance among various treatments. Correlation coefficients were calculated based on the mean values.
A comprehensive analysis of MYB genes in the coconut genome revealed a total of 179 MYB genes, which wereeffectively profiled from the coconut genome. According to the numbering in the coconut genome, it is arranged from small to large and positioned as CnMYB01 to CnMYB179. Detailed information about each of these profiled MYB genes as shown in Supplementary Tables 1 and 2. The length of the peptide chains in the CnMYB proteins varied from 61 to 1150 amino acids, as shown in Supplementary Table 1. Additionally, the predicted proteins exhibited a molecular weight range of 7.8 to 128.1 and isoelectric points ranging from 4.09 to 10.11, as indicated in Supplementary Table 1. Regarding the intracellular distribution of the CnMYB putative proteins the vast majority was predicted to be localized in the nucleus. Based on the analysis of the N-terminal aa region, 154 CnMYBs were predicted to be localized in the nucleus whereas CnMYB02 would be localized in the cytoplasm and CnMYB03 and CnMYB132 both in the cytoplasm and nucleus (Supplementary Table 1).
Through a search using DNA sequence annotations, we observed that 179 identified coconut MYB genes were distributed across 16 chromosomes. Among these genes, 148 genes exhibited uneven distribution patterns across the chromosomes (Figure 2). More specifically, our analysis revealed that 11 CnMYB genes were located on chromosome 1, 15 genes on chromosome 2, 11 genes on chromosome 3, 12 genes on chromosome 4, 10 genes on chromosome 5, 12 genes on chromosome 6, 11 genes on chromosome 7, 10 genes on chromosome 8, 12 on chromosome 9 held, 8 genes on chromosome 10 held, 4 genes on chromosome 11, 9 genes on chromosome 12, 13 genes on chromosome 13, 11 genes on chromosome 14, 2 genes on chromosome 15, and 3 genes on chromosome 16 (Figure 2). Additionally, 29 CnMYB genes were assigned to chromosomes that could not be determined. Notably, chromosome 2 contained the highest number of CnMYB genes (15), while chromosome 4, 6 and 9 each contain 12 CnMYB genes. In contrast, chromosome 15 had the lowest number of MYB genes (2) (Figure 2).
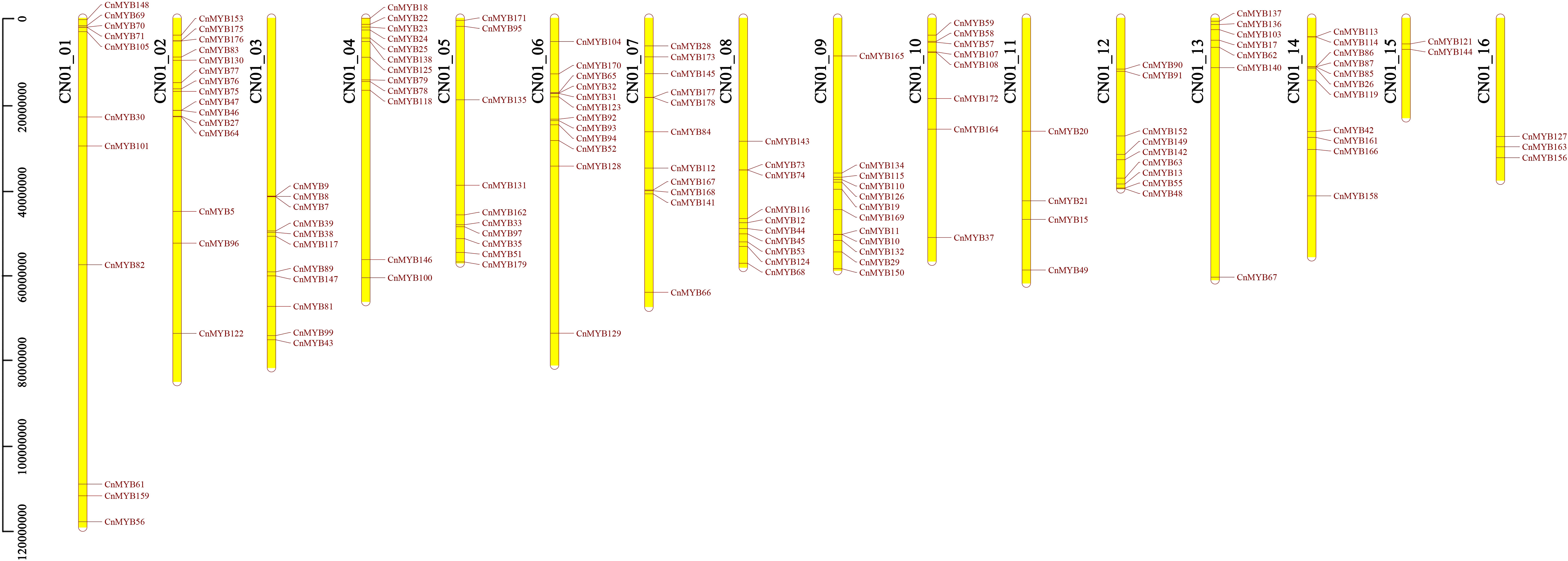
Figure 2 CnMYB genes distribution across 16 chromosomes of coconut genome. The scale represents the length of coconut chromosomes.
The exon-intron organization of the 179 CnMYB genes was examined using the Gene Structure Display Server program, as illustrated in Figure 3. The analysis revealed that the majority MYB genes exhibited varying numbers of introns, ranging from 0 to 15 (Figure 3). Among the 179 CnMYB genes, CnMYB166 had the highest number of introns (15), followed by CnMYB80 and CnMYB117 with 11 introns each. Additionally, CnMYB81 and CnMYB151 contained 10 introns, while CnMYB130 had 9 introns. Furthermore, CnMYB12 had 7 introns, and CnMYB145 had 6 introns. Notably, 28 genes, including CnMYB1, CnMYB3, CnMYB4, CnMYB15, CnMYB25, CnMYB31, CnMYB32, CnMYB34, CnMYB44, CnMYB46, CnMYB63, CnMYB65, CnMYB73, CnMYB74, CnMYB85, CnMYB86, CnMYB87, CnMYB93, CnMYB97, CnMYB100, CnMYB101, CnMYB125, CnMYB139, CnMYB143, CnMYB158, CnMYB173, CnMYB174, and CnMYB177, did not contain any introns. Furthermore, three genes (CnMYB80, CnMYB117 and CnMYB166) had the maximum number of exons (≥12). Most of the MYB genes exhibited a small number of introns, typically ranging from 0 to 3, suggesting a conserved pattern of intron distribution within the MYB genes. The disparity observed in the CnMYB gene composition suggests noteworthy deviation within the coconut genome. Additionally, we performed protein motif profiling to assess the variation in the CnMYB gene family of coconut. Our findings revealed the presence of 10 preserved motifs across all 179 identified MYB genes, as depicted in Figures 4, 5 and Supplementary Table 3.

Figure 5 A total of ten conserved motifs distribution in CnMYB genes. Each motif is represented by a number (1-10) in the colored box. Sequence logos of amino acid residues of 10 conserved motifs of CnMYB proteins are also shown.
To construct a phylogeny of MYB proteins, a maximum likelihood (ML) method was employed, using 179 MYB proteins sequences from coconut and 154 MYB proteins sequences from Arabidopsis thaliana (Figure 6). The resulting dendrogram classified the MYB genes into 34 distinct subgroups, denoted as S1-S26, C1-C6, 3R-MYB, and 4R-MYB (Figure 6). These subgroups represent four main types of MYB protein families: 4R-MYB, 3R-MYB, R2R3-MYB, and the coconut-specific subgroups (Figure 6). Out of the 34 subgroups, 29 included proteins from both coconut and Arabidopsis, while the remaining five were specific to either coconut (S9, S12 and S15) or Arabidopsis (C1 and C2). Similar species-specific subgroups of MYBs have been found in other plant species, such as Solanum tuberosum (Li Y. et al., 2020), Casuarina equisetifolia (Wang et al., 2021) and Petunia (Chen et al., 2021). Within the 3R-MYB subfamily, we identified four CnMYB genes (CnMYB18, CnMYB81, CnMYB130, CnMYB139) and ten AtMYB genes. Similarly, the 4R-MYB subfamily consisted of four CnMYB genes (CnMYB70, CnMYB71, CnMYB151, CnMYB166) and four AtMYB genes. The C1 to C6 subfamily included forty-seven CnMYB genes and thirteen AtMYB genes. The remaining 124 CnMYB genes and 127 AtMYB genes were associated with the R2R3-MYB family. The R2R3-MYB family was further divided into subgroups S1-S26, with varying numbers of CnMYB and AtMYB genes. Notably, subgroups S3 and S11 comprised only one CnMYB member each, making them the smallest groups, while subgroup S14 comprised 15 members, making it the largest group. Interestingly, no CnMYB genes were found in subgroups S9, S12, or S15, suggesting potential gene loss during coconut genome evolution or acquisition in Arabidopsis evolution. The presence of more CnMYB genes than AtMYB genes in certain subgroups indicates functional distinction of MYB genes among various plant species. These phylogenetic findings suggest that MYB genes clustered within the same set may share preserved functions, which should be further investigated through experimental approaches.
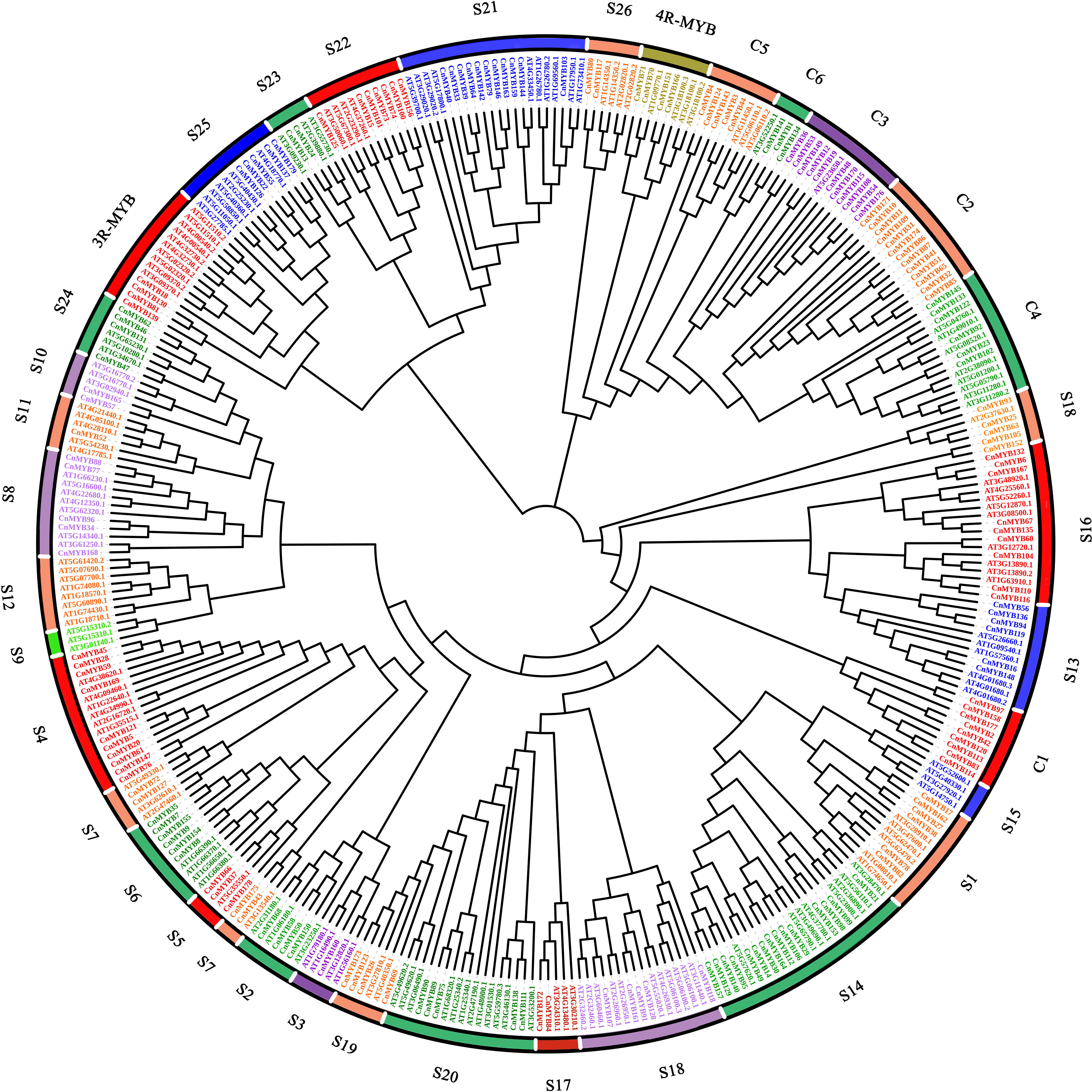
Figure 6 Phylogenetic analysis of 179 CnMYB (coconut) and 130 AtMYB (Arabidopsis) genes. A maximum likelihood (ML) phylogenetic tree of 309 MYB genes of two plants was constructed using MEGA 6.06 software with protein sequences.
We conducted an analysis of the expression profiles of 179 CnMYB genes in four distinct tissues of coconut, including LSE, EP, SE and FSB using RNA-seq data from a database. The results were visualized using a heatmap, which revealed the expression patterns of 12 R2R3-MYB genes in coconut tissues (Figure 7, Supplementary Table 5). Among these genes, three (CnMYB15, CnMYB68, CnMYB101) exhibited the highest expression across all four coconut tissues (Figure 7). Conversely, genes with the lowest expression levels were predominantly observed in LSE, specifically CnMYB94 and CnMYB164 (Figure 7). Notably, genes with higher expression levels were primarily observed in the EP tissues of all four coconut species (Figure 7). Furthermore, the expression of CnMYB8 was found to be higher in all four tissues of the Green Dwarf (GD) species compared to other coconut varieties.
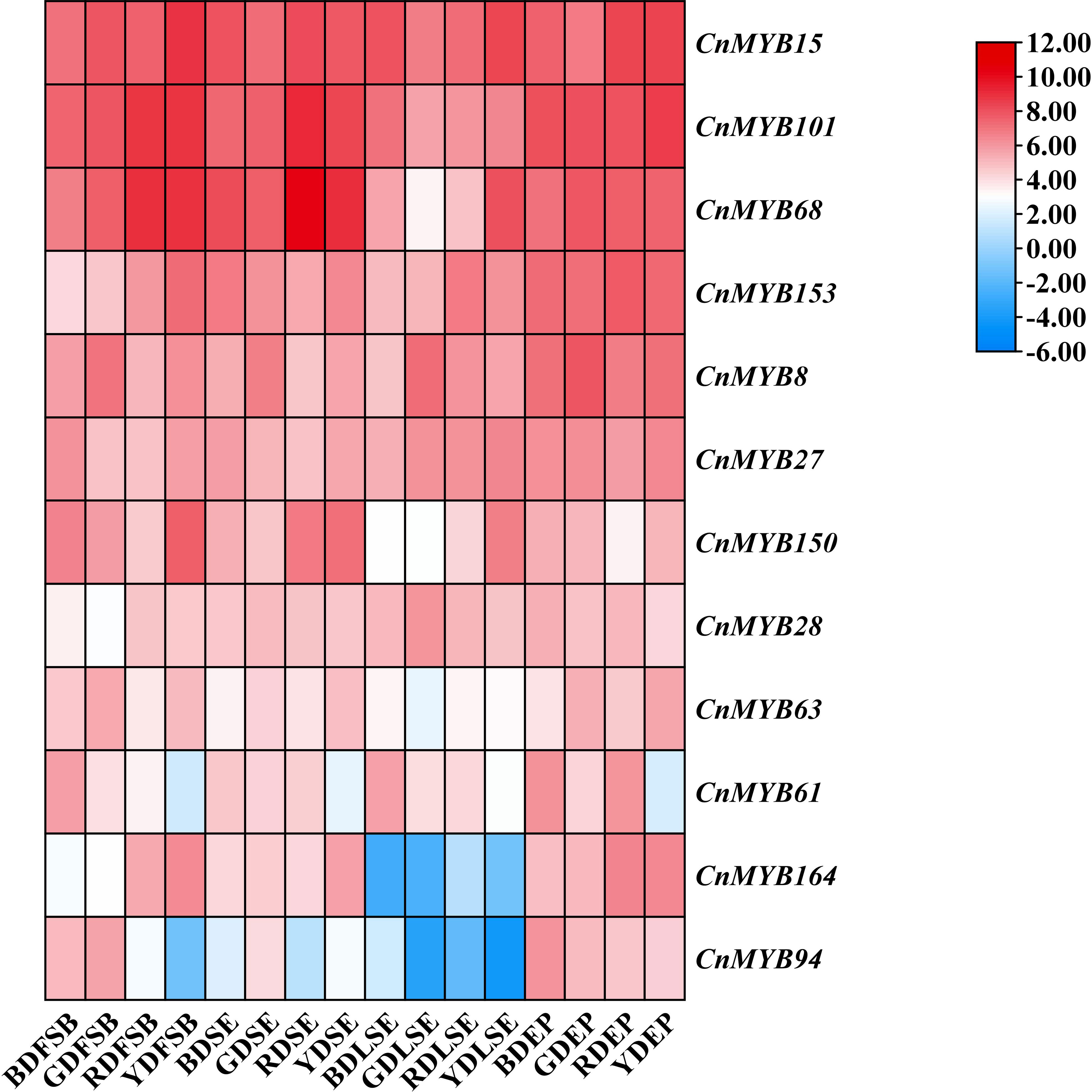
Figure 7 Heat map of candidate genes expression in different tissues of different coconut species. Note: Log2FPKM value were sued to construct the heat map with clustering.
Moreover, we performed qPCR analysis to investigate the expression patterns of 12 candidate MYB genes (CnMYB8, CnMYB15, CnMYB27, CnMYB28, CnMYB61, CnMYB63, CnMYB68, CnMYB94, CnMYB101, CnMYB150, CnMYB153, CnMYB164) during different stages of fruit development (4M, 7M, and 10M). Among these genes, CnMYB28, CnMYB68, and CnMYB101 displayed higher expression levels compared to the other genes, especially CnMYB68 (Figure 8). Notably, the expression level of CnMYB68 in the 10-month-old fruit peel of Red Dwarf (RD), Yellow Dwarf (YD), and Brown Dwarf (BD) coconuts was significantly higher than in other stages and Green Dwarf (GD) coconut (P<0.05). Additionally, the expression levels of CnMYB101 in RD, YD, and BD were higher than in GD, with the highest expression level observed in the 4-month-old coconut peel of RD, YD, and BD (Figure 8). Conversely, the expressions of CnMYB8, CnMYB15, CnMYB27, CnMYB63, CnMYB150, CnMYB153, and CnMYB164 were relatively lower compared to the other genes.
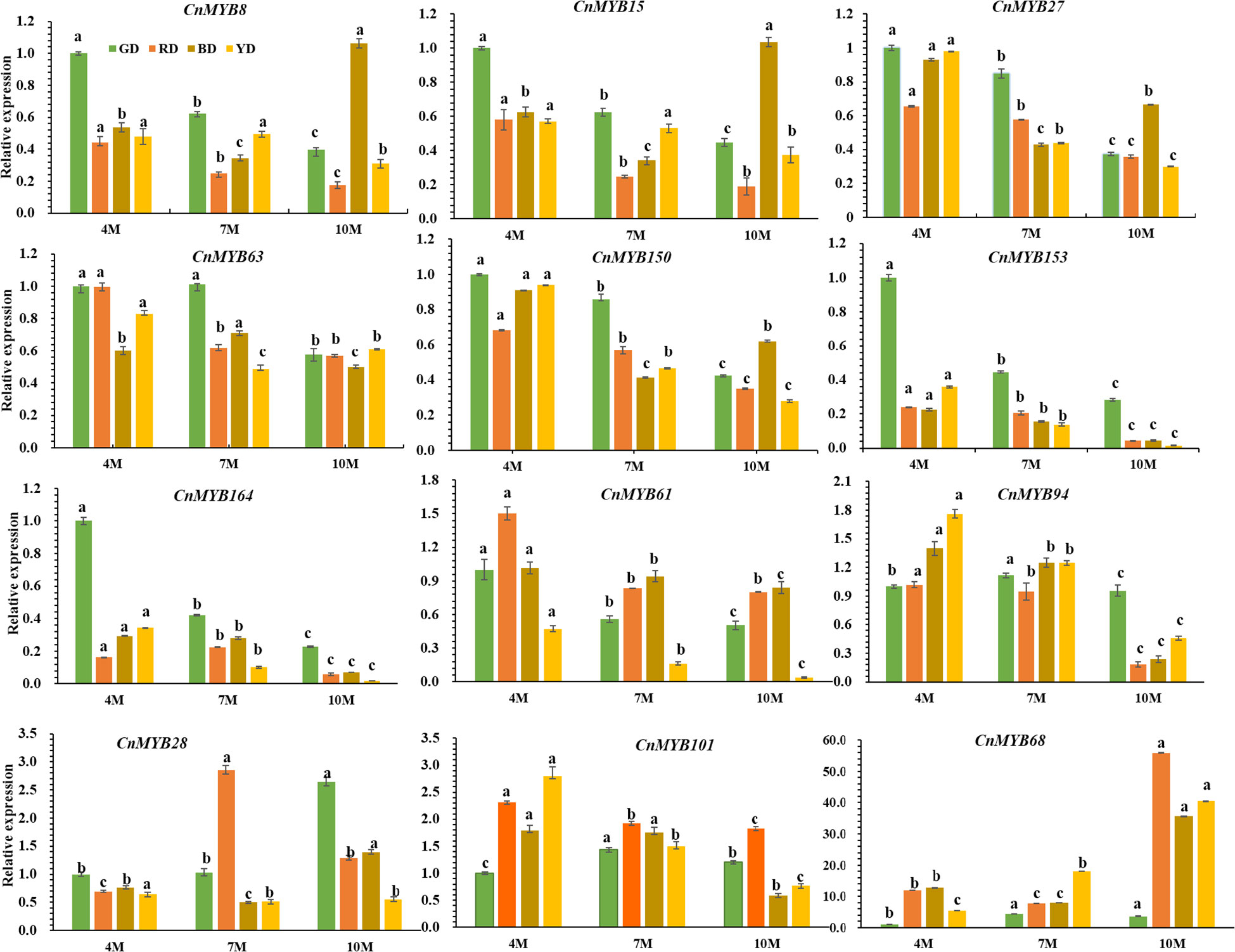
Figure 8 Relative expression levels of CnMYB8, CnMYB15, CnMYB27, CnMYB28, CnMYB61, CnMYB63, CnMYB68, CnMYB94, CnMYB101, CnMYB150, CnMYB153, CnMYB164 genes in the peel of different coconut varieties at different developmental stages, using CnActin as an internal reference gene. Data represent the mean ± SD of three independent experiments. Different lowercase letters indicate significant differences according to the least significant difference test (LSD) at p < 0.05.
MYB transcription factors have been extensively studied and recognized as one of the largest families of transcription factors in plants. Among all MYB subgroups, the R2R3-MYB protein comprises the highest number of members that can play a crucial role in various aspects of the secondary metabolism in plants (Song et al., 2021; Zhang et al., 2021). The R2R3-MYB family can control the diverse biological processes such as plant metabolism, growth and development (Li et al., 2023; Zhang et al., 2023). Additionally, it regulates plant responses to hormones and various types of stresses (Zhao et al., 2023). Several genomic-scale investigations have been concluded to identify MYB family members in various plant genomes (Li Y. et al., 2020; Liu et al., 2021; Zhao et al., 2023). The genomic identification of R2R3-MYB transcription factors has been conducted in various plant species, revealing their significant presence in plant genomes. For instance, extensive sequencing efforts have led to the identification of 138 R2R3-AtMYBs in Arabidopsis thaliana (Katiyar et al., 2012), 99 R2R3-DcMYB in Dendrobium catenatum (Zhang et al., 2021), 393 R2R3-TaMYBs in Triticum aestivum (Wei et al., 2020), 111 R2R3-StMYB in Solanum tuberosum (Li Y. et al., 2020) and 174 R2R3-NtMYB in Nicotiana tabacum (Yang et al., 2022). However, little is known about the MYB gene family in Cocos nucifera.
In this study, we identified 179 MYB members in Cocos nucifera through bioinformatics analysis, and characterized their phylogenetic relationships with A. thaliana MYBs. Our compresinsive analysis encompassed phylogenetic characteristics, physical and chemical properties, gene structure, chromosome location, tissue-specific expression patterns, and expression profiles during coconut fruit development stages. Our findings revealed that the MYB domain and other motifs of CnMYBs exhibit a high degree of conservation with those of Arabidopsis AtMYBs. Nonetheless, we also observed divergence between CnMYBs and AtMYBs, indicating a combination of conservation and diversity within plant MYBs.
Specifically focusing on the R2R3-MYB family, our analysis identified 124 members within the coconut genome. Phylogenetic analysis categorized these members into 26 subfamilies, with unified subfamily members displaying similar motifs and gene structures (Figure 6). The oil palm genome was found to contain 152 R2R3-MYB family members, which were classified into 23 subfamilies (Zhou et al., 2020). Furthermore, the protein sequences of the 124 coconut R2R3-MYB family members exhitited varying lengths, ranging from 89 (CnMYB123) to 555 (CnMYB118) amino acids, with an average length of 314 amino acids. Their molecular masses also varied, spanning from 10.37 (CnMYB123) to 59.79 kDa (CnMYB118), with an average molecular mass of 34.84 kDa. Theoretical isoelectric points ranged from 4.28 (CnMYB90) to 10.06 (CnMYB107), with an average molecular mass of 6.99 (Supplementary Table 1). In a separate investigation involving 124 longan R2R3-MYB family members, their protein sequences ranged from 134 to 661 amino acids, with an average length of 307 amino acids. The molecular masses ranged from 15.69 to 775.81 kDa, with an average of 34.68 kDa and theoretical isoelectric points ranged from 4.91 to 10.6 (Lv et al., 2023). Based on the findings of a previous study conducted by Zhang et al. 2021, our study provides further support for the hypothesis that the MYB gene family exhibits a significant level of evolutionary conservation among various plant species.
The pattern of gene structure is a useful tool for studying the evolutionary associations within a gene family. In our study, we identified 124 R2R3-CnMYB genes, which exhibited a range of exon numbers from 1 to 16 (Figure 3). Most of the R2R3-CnMYBs, similar to those found in other plant species, consisted of three exons and two introns (Liu et al., 2014; Chen et al., 2022; Du et al., 2022). It is worth noting that within the same subfamily, the exon/intron patterns of R2R3- CnMYB genes showed remarkable, with the majority of genes having no more than two introns. This finding aligns with previous research that also observed the presence of a maximum of two introns in most plant R2R3-MYB genes (Sabir et al., 2022; Yin et al., 2022). Furthermore, we analyzed the motif compositions of CnMYB proteins. Our findings revealed that the majority of MYB proteins contained motifs 1, 2, 3, 4, 5, and 6 (Figure 4), while most MYB proteins contained motifs 1, 3, 5, and 8 in Dendrobium catenatum (Zhang et al., 2021). The majority of MYB genes, included several motifs (1, 2, 3, 4, and 5) in Prunus avium (Sabir et al., 2022). These motifs were conserved within specific subgroups, and proteins in the same subgroup that share these motifs likely (Qin et al., 2023).
Prior research has implicated R2R3-MYB genes in essential functions in the anthocyanins, carotenoids and flavonoid biosynthesis in a variety of plants. For instance, in red-fleshed apple, MdMYB22 and MdMYB12 have been identified as key regulatorsof proanthocyanidins and flavonols biosynthesis (Wang et al., 2017). Moreover, PpMYB10 has been found to primarily control anthocyanin biosynthesis in the exocarp of Prunus persica (Ravaglia et al., 2013), while MdMYB1 is a major regulator of anthocyanin biosynthesis in red-skinned fruit (Zhang et al., 2019). In pear fruit, PbMYB12b has been found to positively regulate flavonol biosynthesis by enhancing the expression of PbCHSb and PbFLS (Zhai et al., 2019). In wolfberry, Lba11g0183 and Lba02g01219 have been identified as candidate genes involved in carotenoid biosynthesis (Yin et al., 2022). Additionally, in Elaeis guineensis, the VIR gene, encoding R2R3-MYB-like transcription factor with homology to Lilium LhMYB12 and similarity to Arabidopsis PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), regulate the heterogeneity of red and yellow fruit color (Singh et al., 2014). Genetic variations in the VIRESCENS gene have been associated with the conspicuousness of fruit colors in palms, indicating potential selection by frugivores (Wang et al., 2022). Notably, when comparing the VIRESCENS gene sequences of oil palm and date palms, no matching R2R3-MYB gene was found in coconut. In coconut, the VIRESCENS gene appears to be disrupted by the insertion of a highly repetitive sequence spanning 100 kb (Wang et al., 2022).
The influence of light exposure time on flavonoid and anthocyanin biosynthesis in fruits has been extensively studied by Premathilake et al. (2020). It was found that exposing pear fruit to light for a long duration up-regulated the expression of the R2R3-MYB DNA-binding protein PpMYB17 (Premathilake et al., 2020), resulting in higher anthocyanin biosynthesis (Alabd et al., 2022). In our study, we observed that the epicarp covered by sepals exhibited a lighter or even whiter color, which may be attributed to the obstruction of other coconut fruits within the clusters, which can result in variations in the overall pericarp color. It is speculated that the uneven coloration is influenced by light obstruction (Supplementary Figure 1). We also identified higher expression levels of CnMYB68, CnMYB101, and CnMYB28 in various tissues and developmental stages across the four coconut species, as evident from both transcriptome data and quantitative data. However, further experiments are required to confirm their functional roles.
This study successfully identified 179 MYB genes in the coconut genome through a comprehensive genome-wide screening approach. A thorough investigation was conducted into the genomic architecture, genetic lineages, chromosomal localization, gene replication events, preserved motifs, and expression patterns across different tissues. The expression pattern analysis of CnMYBs in various coconut tissues revealed their constitutive expression with significant functional differentiation. Additionally, qPCR testing of 12 selected CnMYB genes demonstrated their diverse expression patterns. These findings provide valuable insights into the essential functional divergence observed among CnMYB genes across diverse coconut tissues, establishing them as potential candidate genes responsible for color development in this important crop. Furthermore, this research offers a comprehensive understanding of the MYB gene family in coconut, laying a strong foundation for future explorations into the functional roles and evolutionary dynamics of MYB genes in coconut.
The data presented in the study are deposited in the NCBI repository, accession number: PRJNA374600. The transcriptome data used in this article is attached in Supplementary Table 5.
YY: Writing – review & editing. JL: Data curation, Writing – original draft, Writing – review & editing. SG: Data curation, Writing – review & editing. YMH: Writing – review & editing. XS: Writing – review & editing. LZ: Software, Writing – review & editing. FW: Writing – review & editing. CZ: Writing – review & editing. SC: Writing – review & editing. AI: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We would like to express our gratitude for the financial assistance received from the National Natural Science Foundation of China. (Grant No. 32071805) and the fundamental Scientific Research Funds for Chinese Academy of Tropical Agriculture Sciences (CATAS-Nos. 1630152023012). We would like to extend our heartfelt appreciation to the National Germplasm Nursery of Tropical Palm and the Scientific Observation and Experiment Station of Tropical Oil Crops, Ministry of Agriculture and Rural Affairs, P.R. China, for generously providing the coconut fruits utilized in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1263595/full#supplementary-material
Supplementary Figure 1 | Coloring of different coconut varieties.
Alabd, A., Ahmad, M., Zhang, X., Gao, Y., Peng, L., Zhang, L., et al. (2022). Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Horticulture Res. 9, uhac199. doi: 10.1093/hr/uhac199
Ampomah-Dwamena, C., Thrimawithana, A. H., Dejnoprat, S., Lewis, D., Espley, R. V., Allan, A. C. (2019). A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytologist. 221, 309–325. doi: 10.1111/nph.15362
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Web Server issue), W202–W208. doi: 10.1093/nar/gkp335
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Science. 12. doi: 10.3389/fpls.2021.691384
Chen, D., Chen, H., Dai, G., Zhang, H., Liu, Y., Shen, W., et al. (2022). Genome-wide identification of R2R3-MYB gene family and association with anthocyanin biosynthesis in Brassica species. BMC Genomics 23 (1), 441. doi: 10.1186/s12864-022-08666-7
Chen, Y., Chen, Y., Shi, C., Huang, Z., Zhang, Y., Li, S., et al. (2018). SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7, gix120. doi: 10.1093/gigascience/gix120
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., He, W., Guo, X., Pan, J. (2021). Genome-wide identification, classification and expression analysis of the MYB transcription factor family in Petunia. Int. J. Mol. Sci. 22, 4838. doi: 10.3390/ijms22094838
Ding, Y., Yang, Q., Waheed, A., Zhao, M., Liu, X., Kahar, G., et al. (2023). Genome-wide characterization and functional identification of MYB genes in Malus sieversii infected by Valsa Mali. Front. Plant science. 14. doi: 10.3389/fpls.2023.1112681
Du, H., Yang, S. S., Liang, Z., Feng, B. R., Liu, L., Huang, Y. B., et al. (2012). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12, 106. doi: 10.1186/1471-2229-12-106
Du, J., Zhang, Q., Hou, S., Chen, J., Meng, J., Wang, C., et al. (2022). Genome-wide identification and analysis of the R2R3-MYB gene family in theobroma cacao. Genes 13 (9), 1572. doi: 10.3390/genes13091572
Duan, A.-Q., Tan, S.-S., Deng, Y.-J., Xu, Z.-S., Xiong, A.-S. (2022). Genome-wide identification and evolution analysis of R2R3-MYB gene family reveals S6 subfamily R2R3-MYB transcription factors involved in anthocyanin biosynthesis in carrot. Int. J. Mol. Sci. 23, 11859. doi: 10.3390/ijms231911859
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 (D1), D279–D285. doi: 10.1093/nar/gkv1344
Iqbal, A., Yang, Y., Qadri, R., Wu, Y., Li, J., Shah, F., et al. (2019). QRREM method for the isolation of high-quality RNA from the complex matrices of coconut. Bioscience Rep. 39, BSR20181163. doi: 10.1042/BSR20181163
Jiang, L., Yue, M., Liu, Y., Zhang, N., Lin, Y., Zhang, Y., et al. (2023). A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × ananassa). Plant Biotechnol. J. 21 (6), 1140–1158. doi: 10.1111/pbi.14024
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13, 1–19. doi: 10.1186/1471-2164-13-544
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 1–16. doi: 10.1186/1471-2105-12-323
Li, J., Han, G., Sun, C., Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signaling Behavior. 14, 1613131. doi: 10.1080/15592324.2019.1613131
Li, Y., Lin-Wang, K., Liu, Z., Allan, A. C., Qin, S., Zhang, J., et al. (2020). Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. macromolecules. 148, 817–832. doi: 10.1016/j.ijbiomac.2020.01.167
Li, C., Qiu, J., Yang, G., Huang, S., Yin, J. (2016). Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.). Plant Cell Rep. 35, 2151–2165. doi: 10.1007/s00299-016-2025-8
Li, J., Xu, S., Mei, Y., Gu, Y., Sun, M., Zhang, W., et al. (2023). Genomic-wide identification and expression analysis of R2R3-MYB transcription factors related to flavonol biosynthesis in Morando officials. BMC Plant Biol. 23 (1), 381. doi: 10.1186/s12870-023-04394-6
Li, C., Yu, W., Xu, J., Lu, X., Liu, Y. (2022). Anthocyanin biosynthesis bnduced by MYB transcription factors in plants. Int. J. Mol. Sci. 23 (19), 11701. doi: 10.3390/ijms231911701
Li, T., Zhang, S., Li, Y., Zhang, L., Song, W., Chen, C., et al. (2023). Simultaneous promotion of salt tolerance and phenolic acid biosynthesis in Salvia miltiorrhiza via overexpression of Arabidopsis MYB12. Int. J. Mol. Sci. 24 (21), 15506. doi: 10.3390/ijms242115506
Li, B. J., Zheng, B. Q., Wang, J. Y., Tsai, W.-C., Lu, H. C., Zou, L. H., et al. (2020). New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 3, 89. doi: 10.1038/s42003-020-0821-8
Liu, J., Wang, J., Wang, M., Zhao, J., Zheng, Y., Zhang, T., et al. (2021). Genome-wide analysis of the R2R3-MYB gene family in Fragaria× Ananassa and its function identification during anthocyanins biosynthesis in pink-flowered Strawberry. Front. Plant Science. 12. doi: 10.3389/fpls.2021.702160
Liu, C., Wang, X., Xu, Y., Deng, X., Xu, Q. (2014). Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol. Biol. Rep. 41 (10), 6769–6785. doi: 10.1007/s11033-014-3563-1
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lv, X., Tian, S., Huang, S., Wei, J., Han, D., Li, J., et al. (2023). Genome-wide identification of the longan R2R3-MYB gene family and its role in primary and lateral root. BMC Plant Biol. 23 (1), 448. doi: 10.1186/s12870-023-04464-9
Ma, R., Liu, B., Geng, X., Ding, X., Yan, N., Sun, X., et al. (2023). Biological function and stress response mechanism of MYB transcription factor family genes. J. Plant Growth Regulation. 42, 83–95. doi: 10.1007/s00344-021-10557-2
Mackon, E., Jeazet Dongho Epse Mackon, G. C., Ma, Y., Haneef Kashif, M., Ali, N., Usman, B., et al. (2021). Recent insights into anthocyanin pigmentation, synthesis, trafficking, and regulatory mechanisms in rice (Oryza sativa L.) caryopsis. Biomolecules 11, 394. doi: 10.3390/biom11030394
Premathilake, A. T., Ni, J., Bai, S., Tao, R., Ahmad, M., Teng, Y. (2020). R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 252, 1–16. doi: 10.1007/s00425-020-03473-4
Qin, S., Wei, F., Liang, Y., Tang, D., Lin, Q., Miao, J., et al. (2023). Genome-wide analysis of the R2R3-MYB gene family in Spatholobus suberectus and identification of its function in flavonoid biosynthesis. Front. Plant science. 14. doi: 10.3389/fpls.2023.1219019
Ravaglia, D., Espley, R. V., Henry-Kirk, R. A., Andreotti, C., Ziosi, V., Hellens, R. P., et al. (2013). Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 13, 1–14. doi: 10.1186/1471-2229-13-68
Sabir, I. A., Manzoor, M. A., Shah, I. H., Liu, X., Zahid, M. S., Jiu, S., et al. (2022). MYB transcription factor family in sweet cherry (Prunus avium L.): genome-wide investigation, evolution, structure, characterization and expression patterns. BMC Plant Biol. 22 (1), 2. doi: 10.1186/s12870-021-03374-y
Sagawa, J. M., Stanley, L. E., LaFountain, A. M., Frank, H. A., Liu, C., Yuan, Y. W. (2016). An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytologist. 209, 1049–1057. doi: 10.1111/nph.13647
Singh, R., Low, E.-T. L., Ooi, L. C.-L., Ong-Abdullah, M., Nookiah, R., Ting, N.-C., et al. (2014). The oil palm VIRESCENS gene controls fruit colour and encodes a R2R3-MYB. Nat. Commun. 5, 4106. doi: 10.1038/ncomms5106
Song, X., Yang, Q., Liu, Y., Li, J., Chang, X., Xian, L., et al. (2021). Genome-wide identification of Pistacia R2R3-MYB gene family and function characterization of PcMYB113 during autumn leaf coloration in Pistacia chinensis. Int. J. Biol. macromolecules. 192, 16–27. doi: 10.1016/j.ijbiomac.2021.09.092
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 (5), 447–456. doi: 10.1016/s1369-5266(00)00199-0
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Tolosa, L. N., Zhang, Z. (2020). The role of major transcription factors in Solanaceous food crops under different stress conditions: Current and future perspectives. Plants 9, 56. doi: 10.3390/plants9010056
Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26 (1), 136–138. doi: 10.1093/bioinformatics/btp612
Wang, R., Mao, C., Ming, F. (2022). PeMYB4L interacts with PeMYC4 to regulate anthocyanin biosynthesis in Phalaenopsis orchid. Plant Sci. 324, 111423. doi: 10.1016/j.plantsci.2022.111423
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wang, Y., Zhang, Y., Fan, C., Wei, Y., Meng, J., Li, Z., et al. (2021). Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 21 (1), 328. doi: 10.1186/s12870-021-03083-6
Wei, Q., Chen, R., Wei, X., Liu, Y., Zhao, S., Yin, X., et al. (2020). Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genomics 21 (1), 792. doi: 10.1186/s12864-020-07175-9
Xia, W., Liu, Z., Yang, Y., Xiao, Y., Mason, A., Zhao, S., et al. (2014). Selection of reference genes for quantitative real-time PCR in Cocos nucifera during abiotic stress. NRC Res. Press 34 (2), 525–535. doi: 10.1139/cjb-2013-0212
Xiao, Y., Xu, P., Fan, H., Baudouin, L., Xia, W., Bocs, S., et al. (2017). The genome draft of coconut (Cocos nucifera). Gigascience 6 (11), 1–11. doi: 10.1093/gigascience/gix095
Xiao, Y., Yang, Y., Cao, H., Fan, H., Ma, Z., Lei, X., et al. (2012). Efficient isolation of high quality rna from tropical palms for rna-seq analysis. Plant Omics 5, 584. doi: 10.1094/PDIS-07-12-0630-PDN
Yang, L., Liu, H., Hao, Z., Zong, Y., Xia, H., Shen, Y., et al. (2021). Genome-wide identification and expression analysis of R2R3-MYB family genes associated with petal pigment synthesis in Liriodendron. Int. J. Mol. Sci. 22, 11291. doi: 10.3390/ijms222011291
Yang, J., Zhang, B., Gu, G., Yuan, J., Shen, S., Jin, L., et al. (2022). Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.). BMC Genomics 23 (1), 432. doi: 10.1186/s12864-022-08658-7
Yin, Y., Guo, C., Shi, H., Zhao, J., Ma, F., An, W., et al. (2022). Genome-wide comparative analysis of the R2R3-MYB gene family in five solanaceae species and identification of members regulating carotenoid biosynthesis in wolfberry. Int. J. Mol. Sci. 23, 2259. doi: 10.3390/ijms23042259
Zhai, R., Zhao, Y., Wu, M., Yang, J., Li, X., Liu, H., et al. (2019). The MYB transcription factor PbMYB12b positively regulates flavonol biosynthesis in pear fruit. BMC Plant Biol. 19, 1–11. doi: 10.1186/s12870-019-1687-0
Zhang, T., Cui, Z., Li, Y., Kang, Y., Song, X., Wang, J., et al. (2021). Genome-Wide identification and expression analysis of MYB transcription factor superfamily in Dendrobium catenatum. Front. Genet. 12, 1–13. doi: 10.3389/fgene.2021.714696
Zhang, L., Hu, J., Han, X., Li, J., Gao, Y., Richards, C. M., et al. (2019). A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 101494. doi: 10.1038/s41467-019-09518-x
Zhang, L., Xu, Y., Li, Y., Zheng, S., Zhao, Z., Chen, M., et al. (2023). Transcription factor CsMYB77 negatively regulates fruit ripening and fruit size in citrus. Plant Physiol., kiad592. doi: 10.1093/plphys/kiad592
Zhao, D., Gao, F., Guan, P., Gao, J., Guo, Z., Guo, J., et al. (2023). Identification and analysis of differentially expressed trihelix genes in maize (Zea mays) under abiotic stresses. PeerJ 11, e15312. doi: 10.7717/peerj.15312
Zhou, L. X., Yarra, R., Jin, L. F., Cao, H. X. (2020). Genome-wide identification and expression analysis of MYB gene family in oil palm (Elaeis guineensis Jacq.) under abiotic stress conditions. Environ. Exp. Bot. 180, 104245. doi: 10.1016/j.envexpbot.2020.104245
Zhu, F., Luo, T., Liu, C., Wang, Y., Yang, H., Yang, W., et al. (2017). An R2R3-MYB transcription factor represses the transformation of α-and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 216, 178–192. doi: 10.1111/nph.14684
Keywords: MYBs, genome-wide analysis, color, RNA-seq, qPCR, coconut
Citation: Li J, Guo S, Min Htwe Y, Sun X, Zhou L, Wang F, Zeng C, Chen S, Iqbal A and Yang Y (2024) Genome-wide identification, classification and expression analysis of MYB gene family in coconut (Cocos nucifera L.). Front. Plant Sci. 14:1263595. doi: 10.3389/fpls.2023.1263595
Received: 20 July 2023; Accepted: 21 December 2023;
Published: 15 January 2024.
Edited by:
Aliki Kapazoglou, Hellenic Agricultural Organization -DEMETER (ELGO-DIMITRA), GreeceReviewed by:
Dongchao Ji, Shandong University of Technology, ChinaCopyright © 2024 Li, Guo, Min Htwe, Sun, Zhou, Wang, Zeng, Chen, Iqbal and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaodong Yang, yyang@catas.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.