
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 January 2024
Sec. Plant Systems and Synthetic Biology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1259347
 Xiang-Yu Liu1,2†
Xiang-Yu Liu1,2† Yi-Na Wang1,2†
Yi-Na Wang1,2† Jiang-Shun Du1,2†
Jiang-Shun Du1,2† Bi-Huan Chen1,2
Bi-Huan Chen1,2 Kun-Yi Liu2
Kun-Yi Liu2 Lei Feng1,2
Lei Feng1,2 Gui-Sheng Xiang1,2
Gui-Sheng Xiang1,2 Shuang-Yan Zhang1,2
Shuang-Yan Zhang1,2 Ying-Chun Lu2
Ying-Chun Lu2 Sheng-Chao Yang1,2
Sheng-Chao Yang1,2 Guang-Hui Zhang1,2*
Guang-Hui Zhang1,2* Bing Hao2,3*
Bing Hao2,3*Bergenin is a typical carbon glycoside and the primary active ingredient in antitussive drugs widely prescribed for central cough inhibition in China. The bergenin extraction industry relies on the medicinal plant species Bergenia purpurascens and Ardisia japonica as their resources. However, the bergenin biosynthetic pathway in plants remains elusive. In this study, we functionally characterized a shikimate dehydrogenase (SDH), two O-methyltransferases (OMTs), and a C-glycosyltransferase (CGT) involved in bergenin synthesis through bioinformatics analysis, heterologous expression, and enzymatic characterization. We found that BpSDH2 catalyzes the two-step dehydrogenation process of shikimic acid to form gallic acid (GA). BpOMT1 and AjOMT1 facilitate the methylation reaction at the 4-OH position of GA, resulting in the formation of 4-O-methyl gallic acid (4-O-Me-GA). AjCGT1 transfers a glucose moiety to C-2 to generate 2-Glucosyl-4-O-methyl gallic acid (2-Glucosyl-4-O-Me-GA). Bergenin production ultimately occurs in acidic conditions or via dehydration catalyzed by plant dehydratases following a ring-closure reaction. This study for the first time uncovered the biosynthetic pathway of bergenin, paving the way to rational production of bergenin in cell factories via synthetic biology strategies.
Carbon glycosides are a specific type of secondary plant metabolites (Franz and Grun, 1983). Based on the aromatic aglycone structure, carbon glycosides are classified into flavonoid C-glycosides, xanthone C-glycosides, chromone C-glycosides, anthrone C-glycosides, and C-glycosylated gallic acids (GAs) (Jihen et al., 2017; Wang et al., 2022). Carbon glycosides are formed through the combination of aglycones with sugars catalyzed by C-glycosyltransferases (CGTs), which can generate C-C bonds to endow acid and glycosidase hydrolysis tolerance (Mori et al., 2021; Bililign et al., 2005; Oualid and Artur, 2012; Chong et al., 2022). Owing to these advantages, C-glycoside drugs show remarkably stable drug absorption, molecular recognition, and drug metabolism (Vidal et al., 2013; Singh et al., 2017).
Bergenin is a GA C-glycoside that has selective central cough inhibition activity and is the main ingredient of the antitussive drugs Xuedansu Tablet and Capsule widely prescribed in China (Rohit et al., 2017). Bergenin reportedly has anti-inflammatory, anti-anxiety, anti-malaria, anti-cancer, anti-diabetes, anti-hepatotoxicity, immunomodulatory, and neuroprotective pharmacological activities (Rajesh et al., 2011; Liang et al., 2014; Gao et al., 2015; Jitender et al., 2017; Barai et al., 2019; Kumar et al., 2019; Shi et al., 2019; Xiang et al., 2020). Bergenin is widely distributed in higher plants and has been found in more than 90 plant species belonging to 37 genera in 20 families (Mehta et al., 2022). The Chinese Pharmacopoeia lists the roots of Bergenia purpurascens and Ardisia japonica as natural bergenin resources for the medicinal extraction industry (China Pharmacopoeia Committee, 2020). In both Bergenia purpurascens and Ardisia japonica, bergenin is distributed the whole plant, but predominantly accumulates and stored in the root (Li et al., 2009).
Despite the significant potential of bergenin, its medicinal value has been greatly compromised due to limited supply. In the last few years, the annual demand for dried bergenin root in the extraction industry has exceeded 2,000 tons (Lv et al., 2017). The excessive and uncontrolled harvesting over several decades has resulted in a significant depletion of natural resources, pushing them towards to exhaustion. Moreover, the cultivation of B. purpurascens and A. japonica has been challenging due to their preference for high altitudes, mountainous regions, and cold climate conditions. Resulted in an expanded gap in the bergenin raw material market and a gradual increase in prices. Furthermore, the total chemical synthesis of bergenin is insufficient to meet commercial demands (Parkan et al., 2014). Therefore, it is imperative to elucidate the biosynthetic pathways of bergenin and employ synthetic biology approaches for its large-scale production. In recent years, pathway elucidation of C-glycosides in plants has received wide interest. Researchers have mainly focused on the discovery and functional characterization of key CGTs involved in C-glycoside biosynthesis. At present, more than 50 functional CGTs, mostly involved in C-glycosyl flavonoid synthesis, have been identified in plants, including Oryza sativa, Zea mays, Fagopyrum esculentum, Gentiana triflora, Trollius chinensis, Glycyrrhiza glabra, Scutellaria baicalensis, and Dendrobium catenatum (Brazier-Hicks et al., 2009; Falcone et al., 2013; Nagatomo et al., 2014; Nobuhiro et al., 2015; He et al., 2019; Ren et al., 2020; Wang et al., 2020; Zhang et al., 2020). CGTs can be roughly divided into two functional types: the first type directly adds a glycone to the flavone aglycone to form C-glycosyl flavonoids, whereas the second type, which is currently the most reported, binds a sugar moiety to the open-ring form of the 2-hydroxyflavanone skeleton or its monosaccharide and subsequently undergoes cyclization dehydration to form C-glycosyl flavonoids (Wang et al., 2020; Chong et al., 2022).
To date, only a few CGTs involved in C-glycoside xanthone biosynthesis have been characterized. In Mangifera indica, MiCGT transfers UDP-glucose to maclurin to generate 3-C-glucosylmaclurin, which is subsequently cyclized by xanthone synthase to produce mangiferin (Chen et al., 2015; Chong et al., 2022). In contrast, in Hypericum perforatum, the xanthone skeleton is directly C-glycosylated to form C-glycoside xanthone, and N4CGT1 and N4CGT2 directly catalyze the C-4 glycosylation of norathyriol to form isomangiferin (Uchida et al., 2021). In Morus alba and Angelica decursiva, MaCGT and AbCGT, respectively, are involved in C-glycoside coumarin biosynthesis (Chen et al., 2021; Wang et al., 2022).
Bergenin is a C-glycosyl derivative of GA that is considered to have the simplest structure among C-glycosides that contain a lactone (Taneyama and Yoshida, 1979). Different from the cyclization mechanisms of flavonoid and xanthone C-glycosides, the cyclization step in bergenin synthesis occurs on the skeleton and the sugar moiety does not participate in the closure reaction (Franz and Grun, 1983; Chong et al., 2022). Under acidic conditions, the electrophilicity of the oxygen atom in the carboxyl group is enhanced, facilitating its susceptibility to attack by hydrogen atoms in alcohols and leading to the formation of ester intermediates. The protons were supplied by a 1M HCl solution herein. Subsequently, the oxygen atoms of the alcohol undergo electrophilic attacks and form new ester bonds with the carbon atoms in the carboxyl group within the intermediate stage of esterification. In the meantime, the oxygen atom in the original carboxyl group and the carbon atom in the ester intermediate form a new carbonyl group to form lactone. The involvement of the sugar moiety in the formation of the lactone underlies the uniqueness of this class of compounds. For GA C-glycosides, the sugar moiety participates in the cyclization reaction, and the lactone is formed through an esterification reaction between the carboxyl group of the skeleton and the C-2 hydroxyl group of the sugar moiety (Franz and Grun, 1983).
Bergenin is the best-known and major representative of C-glycosides. However, its complete biosynthetic pathway remains unclear. In this study, we for the first time elucidated the bergenin biosynthetic pathway. Four candidate genes encoding enzymes involved bergenin biosynthesis in B. purpurascens and A. japonica were characterized (Figure 1). The bergenin biosynthetic pathway starts from shikimic acid (SA), which is catalyzed by BpSDH2 to produce GA. BpOMT1 and AjOMT1 generate 4-O-methyl-GA (4-O-Me-GA) from GA. The novel AjCGT1 uses 4-O-Me-GA as a substrate to produce 2-Glucosyl-4-O-Me-GA. After intramolecular dehydration, the closure reaction occurs to form bergenin. The discovery of these enzymes has provided valuable insights into the biosynthetic pathways of bergenin, enabling subsequent efficient de novo synthesis of bergenin in cell factories.
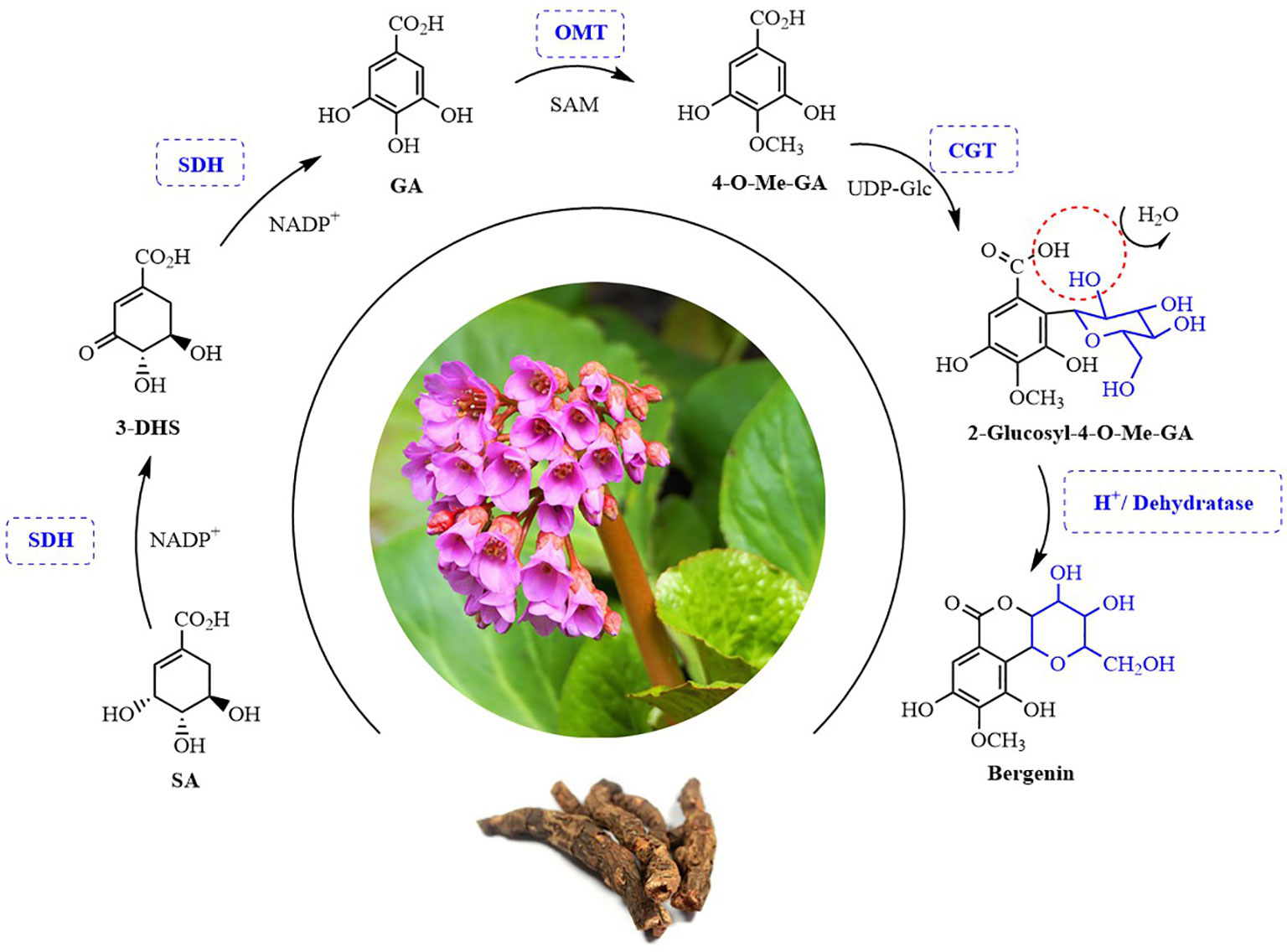
Figure 1 Possible biosynthesis pathway of bergenin in plants. SA, Shikimic acid; 3-DHS, 3-Dehydroshikimic acid; GA, Gallic acid; 4-O-Me-GA, 4-O-Methyl gallic acid. SDH, Shikimate dehydrogenases; OMT, O-methyltransferase; CGT, C-glycosyltransferases. SAM, S-adenosy-L-methionine; UDP-Glc, Uridine diphosphate-Glucose.
B. purpurascens was collected in Lijiang, Yunnan, China and A. japonica was collected in Nanning, Guangxi, China. root parts from healthy plants were collected and frozen at –80°C.
SA, 3-DHS, GA, NADP+, 4-O-Me-GA, S-adenosyl-l-methionine (SAM), UDP-glucose, bergenin, and protocatechuic acid were all purchased from Yuan Ye (Shanghai, China).
The tender plant parts were cut and frozen in liquid nitrogen. Total RNA was extracted using the HiPure HP Plant RNA Mini Kit (Magen, Guangzhou, China) and reverse-transcribed into first-strand cDNA using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (6210A; Takara, Beijing, China).
Transcriptome sequencing was performed at Gene Denovo Biotechnology (Guangzhou, China) using the Illumina HiSeq™2000 high-throughput sequencing platform. Raw data were processed by removing reads containing adaptors, reads with a proportion of N >10%, and low-quality reads. The remaining clean reads were subjected to de novo assembly using the transcriptome splicing software Trinity, with K-mer set to 25 (Grabherr et al., 2011).
The coding regions (CDS) of the unigenes obtained were predicted using the software tools BLASTX and ESTscan. First, the unigenes were compared to the Nr and Swiss-Prot databases in priority order using BLASTX, with an E-value threshold of 1 × 10–5 (Altschul et al., 1997). If a significant match was found in the higher-priority databases, further comparisons with lower-priority databases were omitted. This process resulted in the identification of CDS for the unigenes. The best matching result from the comparison was also utilized to determine the sequence direction of the unigenes. In cases where the unigenes could not be matched in either of the two databases, the CDS were predicted using the ESTscan software. Finally, the predicted CDS were translated into amino acid sequences, and only sequences with a length exceeding 70 amino acids were retained for subsequent in-depth analysis.
Based on searches using the transcriptome data and annotation results from protein databases obtained by local BLAST, key candidate genes encoding shikimate dehydrogenases, O-methyltransferase, and C-glycosyltransferase were obtained. The open reading frames (ORFs) and amino acid sequences of the SDHs, OMTs, and CGTs were identified using ORFfinder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). We used InterPro (www.ebi.ac.uk/Tools/InterProScan) to identify functional domains. SDH, OMT, and CGT amino acid sequences from other species were downloaded from the National Biotechnology Information Center database and compared using ClustalW (Supplementary Table 1). A maximum likelihood tree was constructed using the IQ-tree software, with 1,000 bootstrap replicates (Nguyen et al., 2015).
The SnapGene software was used to design specific primers containing homologous arms of the Pet28a vector for the candidate genes (Supplementary Table 2). Q5 Mix DNA polymerase (NEB, USA) was used to amplify the genes from cDNA using the following thermal cycling conditions: 98°C for 30 s, 35 cycles of 98°C for 15 s, 58°C for 30 s, and 72°C for 90 s, and finally, 72°C for 10 min. Successful target gene amplification was detected by agarose gel electrophoresis. Successfully amplified genes were recovered from the gel and purified using the EasyPure Quick Gel Extraction Kit (TransGen Biotech, China) and stored at –20°C.
The candidate genes encoding SDHs, OMTs, and CGTs were inserted into the BamHI site of pET28a(+) by homologous recombination using NEBuilder® HiFi DNA Assembly Master Mix (E2621) (NEB). The recombinant vector was transformed into Escherichia coli BL21 (DE3) cells and sequenced. Individual colonies carrying the correct gene expression vector were inoculated into Luria Broth culture medium supplemented with 50 μg/mL kanamycin and incubated at 37°C under shaking at 220 rpm/min. When the culture reached an optical density at 600 nm of 0.6–0.8, 0.1 mM isopropyl-β-d-1-thiogalactopyranoside was added and the culture was further incubated at 16°C under shaking at 180 rpm/min for 16 h to induce protein expression. Then, the E. coli cells were collected by centrifugation at 5,000 rpm and resuspended in binding buffer (50 mM Tris-HCl, 0.2 M NaCl, pH 8.0). The cell membranes were disrupted using an ultrasonic crusher (Scientz-IID, China). The proteins were purified using nickel affinity chromatography. After washing the column with 50 mL of washing buffer (20 mM Tris-HCl, 0.2 M NaCl, and 50 mM imidazole, pH 8.0), the protein was eluted with 10 mL of elution buffer (20 mM Tris-HCl, 0.2 M NaCl, and 250 mM imidazole, pH 8.0). The flow rate was 1 mL/min. Finally, the solution was concentrated using an ultrafiltration centrifuge tube (Merck KGaA, Darmstadt, Germany). Protein purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the protein concentration was determined using a protein quantification assay kit (TransGen Biotech, China).
For the functional characterization of SDH proteins, an analytical reaction was carried out in a 100-μL system containing 50 mM Tris-HCl (pH 8.0), 0.1 mM SA, 0.2 mM NADP+, and 20 μg purified enzyme (30°C, 2 h). The reaction was terminated by adding 100 μL of ice-cold HCl (1 M) (Huang et al., 2019). The mixture was centrifuged at 12,000 × g for 15 min and the supernatant was analyzed using an Agilent 1290 series ultrahigh-performance liquid chromatography (UHPLC) system (Agilent Technologies, Germany).
For the functional characterization of OMT proteins, an analytical reaction was carried out in a 100-μL system containing 50 mM Tris-HCl (pH 8.0), 0.1 mM GA, 0.1 mM SAM, and 20 μg purified enzyme (35°C, 2 h). The reaction was terminated by adding 100 μL of ice-cold HCl (1 M) (Mageroy et al., 2012). The mixture was centrifuged at 12,000 × g for 15 min the supernatant solution is analyzed by UHPLC.
CGT enzyme activity was assessed in a 100-μL reaction system containing 50 mM Tris-HCl (pH 8.0), 0.1 mM 4-O-Me-GA, 0.1 mM UDP-glucose, and 20 μg purified enzyme (32°C, 2 h). The reaction was terminated by adding 100 μL of ice-cold HCl (1 M) (Wang et al., 2020). The mixture was centrifuged at 12,000 × g for 15 min and the supernatant was analyzed by UHPLC system.
Ten microliters of supernatant were used for UHPLC analysis. The sample was separated on a Waters XBridge Shield RP18 column (4.6 mm × 250 mm, 5 μm). The mobile phase consisted of a 0.1% v/v formic acid aqueous solution (A) and acetonitrile (B). The gradient elution conditions were as follows: 0–8 min, 1%–5% B; 8–13 min, 5%–10% B; 13–20 min, 10%–20% B; 20–25 min, 20%–45% B; 25–35 min, 45%–90% B; 35–40 min, 90% B. The total run time was 40 min. The temperature of the chromatographic column was set to 30°C, and the flow rate was 0.6 mL/min. The detection wavelengths for GA, 4-O-Me-GA, and bergenin were 260 nm, 230 nm, and 270 nm, respectively.
The reaction products were detected using an Agilent 1290 UPLC Q-TOF UHPLC triple quadrupole MS spectrometer equipped with a heated electrospray ionization source. The MS conditions were as follows: electrospray ionization in negative ion mode, voltage: 3,500 V, fragmentation voltage: 135 V, taper hole voltage: 60V, radio frequency voltage: 700 V, Scanning range: 100–1,000 m/z, scanning mode: SRM. The samples were separated on a Waters XBridge Shield RP18 column (4.6 mm × 250 mm, 5 μm). The mobile phase consisted of a 0.1% v/v formic acid aqueous solution (A) and acetonitrile (B). The gradient elution conditions were as follows: 0–8 min, 1%–5% B; 8–13 min, 5%–10% B; 13–20 min, 10%–20% B; 20–25 min, 20%–45% B; 25–35 min, 45%–90% B; 35–38 min, 90% B; 38–45 min, 90%–100% B. The stripping time was 40 min. The temperature of the chromatographic column was set to 32°C, and the flow rate was 0.6 mL/min. The detection wavelengths of GA, 4-O-Me-GA, and bergenin were 260 nm, 230 nm, and 270 nm, respectively.
mRNA was extracted from the young roots of B. purpurascens and A. japonica, and cDNA libraries were prepared. The transcriptomes were sequenced and the data were assembled using the Trinity software (Table 1). We obtained 309,597,772 and 137,767,838 raw reads in total, and 303,534,544 and 136,379,226 clean reads with a Q20 ratio >97% after quality filtering. The clean data for B. purpurascens and A. japonica were 44.85 Gb and 20.38 Gb. De novo assembly of the clean reads using Trinity software yielded 102,974 and 768,428 unigenes with an N50 length of 1,588 bp and 1,889 bp for B. purpurascens and A. japonica, respectively. The average unigene length was 881 bp for B. purpurascens and 983 bp for A. japonica.
Functional unigene annotation was based on sequence similarity searches against the Nr (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/), SwissProt (https://www.uniprot.org/), KEGG (http://www.kegg.jp/), and KOG (https://www.ncbi.nlm.nih.gov/KOG/) databases. The genes with the highest sequence similarity to the unigene sequences were identified, and functional annotations for the unigenes were obtained based on these results (Supplementary Table 3). A total of 44,726 (43.43%) and 37,356 (48.61%) unigenes were thus annotated for B. purpurascens and A. japonica, respectively.
GA is produced from SA by SDH using NADP+ as a hydride acceptor after a two-step dehydrogenation reaction via 3,5-dihydroxydehydro-SA. Subsequently, 3,5-dihydroxybenzoic acid isomerized to form gallic acid (Muir et al., 2011). This pathway has been confirmed in multiple plants (Bontpart et al., 2016; Huang et al., 2019; Tahara et al., 2020). To identify the genes involved in catalyzing the two-step dehydrogenation of SA to produce gallic acid in B. purpurascens and A. japonica, a phylogenetic tree was constructed using the identified 14 SDHs as references along with 2 SDH proteins from B. purpurascens and 2 SDH proteins found in A. japonica (Figure 2). All sequences information used in the phylogenetic analysis is listed in Supplementary Table 3. BpSDH2 and AjSDH2 clustered together with VvSDH3, CsDQD/SDHc, and EcDQD/SDH3, and the corresponding proteins showed a 89.83% amino acid sequence similarity (Supplementary Figure 6). Since the functionality of SA two-step dehydrogenation to produce GA has been individually confirmed in Vitis vinifera, Camellia sinensis, and Eucalyptus camaldulensis for VvSDH3, CsDQD/SDHc, and EcDQD/SDH3, respectively, we hypothesize that BpSDH2 and AjSDH2 might also have similar functions.
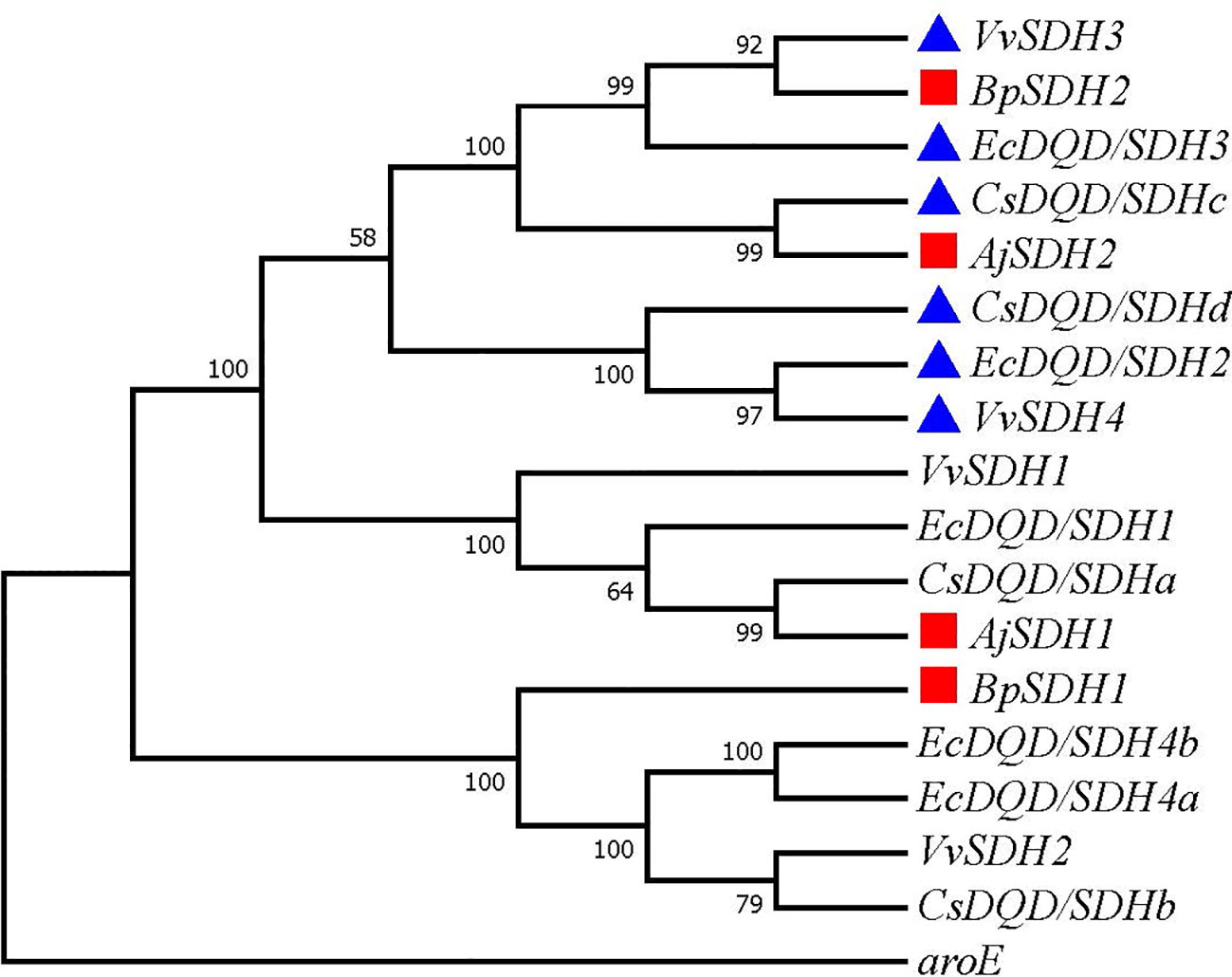
Figure 2 Phylogenetic tree of SDHs involved in GA biosynthesis. A phylogenetic tree was constructed based on the amino acid sequences containing the shikimate dehydrogenases domain from B. purpurascens, A. japonica, and other species. The red box represents SDH found in B. purpurascens, A. japonica, while the blue box represents reported SDH with GA-producing function. All other species’ SDH sequences were obtained from the NCBI database (Supplementary Table S3).
Upon expression in E. coli, only BpSDH2 was obtained in the supernatant and could be used for enzyme assays (Supplementary Figure 1A). We first conducted an enzymatic activity assay on the total protein extract of BpSDH2 using SA as the substrate and NADP+ as the hydride acceptor. Two peaks were observed in the chromatogram of the protein extract, whereas there were no corresponding peaks observed in the control (Figure 3). The retention times of the peaks matched those of 3-dehydro-SA (3-DHS) and GA standards. The spectrum of BpSDH2 showed LC-MS/MS fragmentation ions of 3-DHS at m/z 171 [M-H]−, m/z 127 [M-H-44]−, and GA at m/z 169 [M-H]−, m/z 125 [M-H-44]−, which were consistent with those of the 3-DHS and GA standards (Supplementary Figures 2, 3). These results indicated that the recombinant BpSDH2 protein could catalyze the dehydrogenation of SA in two consecutive steps to produce 3-DHS and GA. HPLC results after the enzymatic assay of BpSDH2 showed a new characteristic peak that was different from those of 3-DHS and GA (Figure 3). GA can also be formed via the synthetic pathway of protocatechuic acid (PCA) or 3,4,5-trihydroxycinnamic acid (Vladimir et al., 2003; Choubey et al., 2015). LC-MS/MS results confirmed that the presence of an unexpected peak with a mass of 153 (m/z, [M-H]−) at 21.12 min, which corresponded to the PCA standard (Supplementary Figures 2, 3).
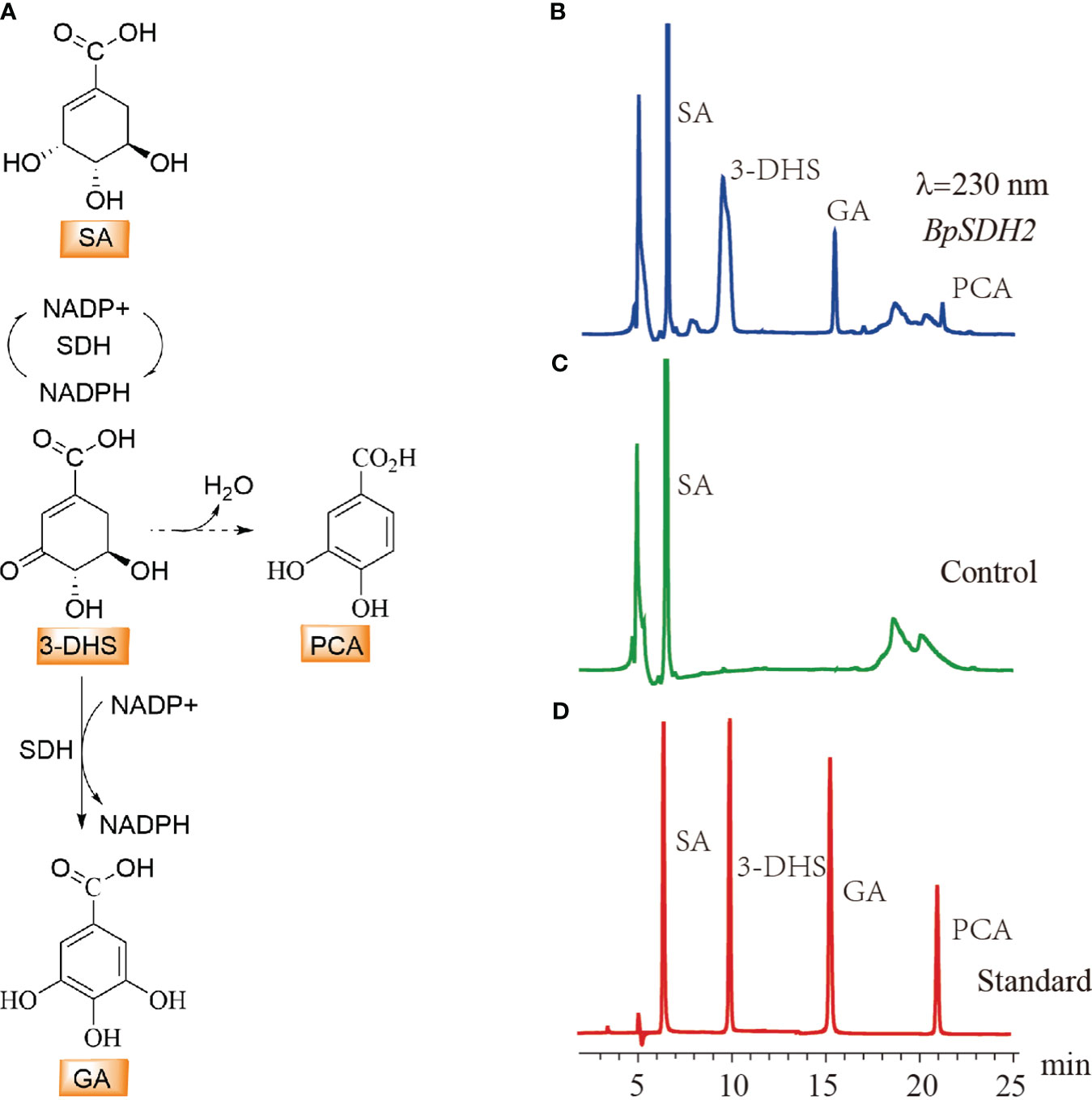
Figure 3 HPLC detection of substrate activity for shikimic acid with BpSDH1 protein. (A) The biosynthetic pathway for the conversion of shikimic acid to gallic acid, as well as the possible pathway for the synthesis of protocatechuic acid. (B) In vitro enzyme activity assay products of shikimic acid with BpSDH2 and NADP+. (C) The reaction of inactivated BpSDH2 protein with shikimic acid and NADP+ serves as a negative control. (D) The retention times of SA, 3-DHS, GA, and PCA.
We assumed that 4-O-Me-GA is formed via methylation of GA at the 4-OH position by OMT. While, the phylogenetic tree contained the reported 7 OMTs in plants and OMTs identified in B. purpurascens and A. japonica was constructed with ODOMT from Oesophagostomum dentatum (Figure 4). BpOMT1 and AjOMT1 clustered together with the catechol OMT gene (CTOMT1) from Solanum lycopersicum, and the corresponding amino acid sequences were highly homologous. CTOMT1 not only shows catechol OMT catalytic activity, but also methylates other substrates with structures similar to that of catechol, such as protocatechuic aldehyde, pyrogallol, and caffeic acid. Considering the structural similarity between GA and these compounds, we hypothesize that BpOMT1 and AjOMT1 catalyze the methylation at 4-OH of GA.
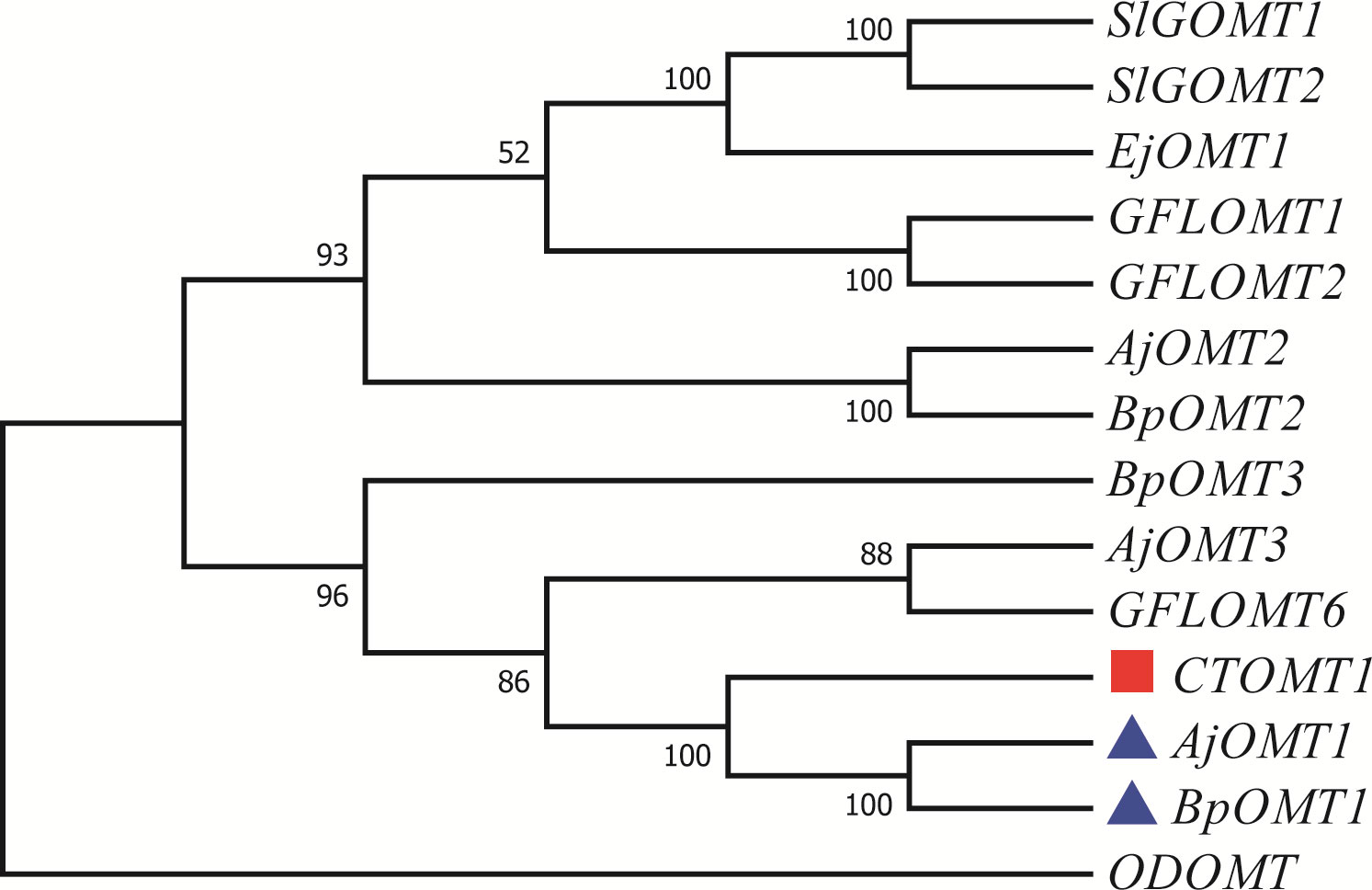
Figure 4 Phylogenetic tree of OMTs involved in 4-O-Me-GA biosynthesis. A phylogenetic tree was constructed based on the amino acid sequences containing the O-methyltransferase domain from B. purpurascens, A. japonica, and other species. The blue boxes represent the candidate OMTs identified from B. purpurascens, A. japonica, which are potential enzymes involved in catalyzing the conversion of GA to 4-O-Me-GA. The OMTs depicted in red boxes serve as the main reference sequences, as they have been previously reported to have the methylating catalytic function on structures similar to GA. All other species’ OMTs were obtained from the NCBI database (Supplementary Table S3).
The candidate genes involved in 4-O-Me-GA biosynthesis were heterologously expressed in E. coli, and the results showed that both BpOMT1 and AjOMT1 were successfully expressed (Supplementary Figures 1B, C). The purified proteins were mixed with the substrate GA and the methyl donor SAM and incubated at a constant temperature of 32°C for the enzymatic reaction. HPLC analysis showed a new peak in the enzymatic reaction products of both BpOMT1 and AjOMT1 with the same retention time as that of the 4-O-Me-GA standard (Figure 5). The enzymatic products of BpOMT1 and AjOMT1 were confirmed by LC-MS/MS analysis. The spectra of BpOMT1 and AjOMT1 products showed LC-MS/MS fragmentation ions of 4-O-Me-GA product at m/z 183 [M-H]−, m/z 168 [M-H-15]−, and m/z 124 [M-H-59]−, which were consistent with those of the 4-O-Me-GA standard (Supplementary Figure 4). These results demonstrated that BpOMT1 and AjOMT1 can catalyze the methylation of 4-OH-GA to produce 4-O-Me-GA.
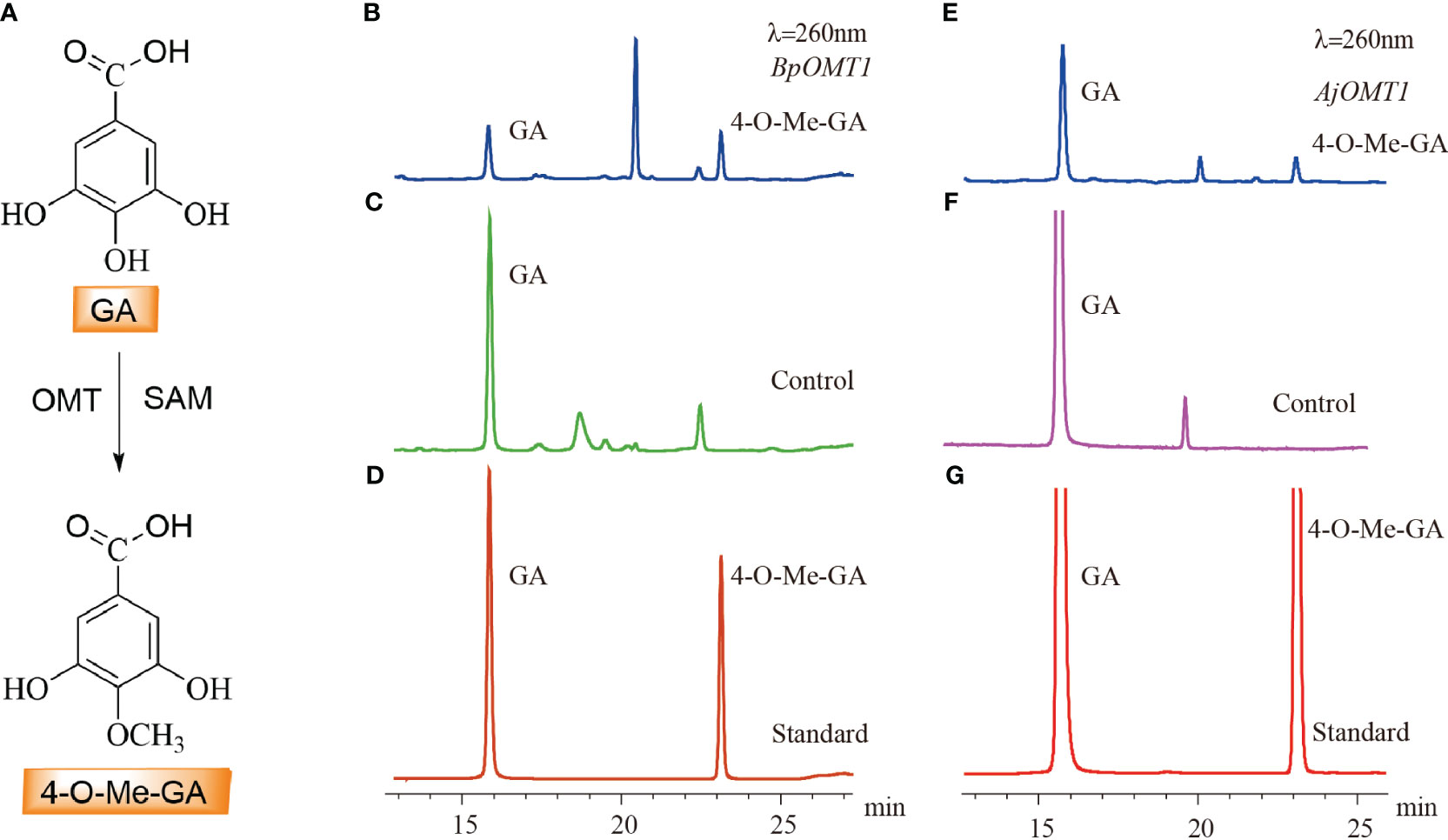
Figure 5 HPLC detection of the C-4 methylation activity of OMT protein on GA. (A) The biosynthetic pathway of 4-O-Me-GA through the C-4 methylation of GA. (B) The enzymatic activity assay of GA with BpOMT1 and SAM in vitro. (C) Inactivated BpOMT1 protein was used as a negative control in the reaction with GA and SAM. (D) The retention times of GA and 4-O-Me-GA standard. (E) The enzymatic activity assay of GA with AjOMT1 and SAM in vitro. (F) The reaction of inactivated AjOMT1 protein with GA and SAM serves as a negative control. (G) The retention time of authentic standard of GA and 4-O-Me-GA.
We assumed that 4-O-Me-GA is glycosylated with UDP-glucose at the C-2 position by CGT, and a lactone ring is formed to produce bergenin after intramolecular dehydration catalyzed by a dehydratase. Based on the conserved motif of UDP-glycosyltransferases (UGTs; PSPG BOX), 43 UGTs were screened from the transcriptome data of B. purpurascens and A. japonica. A phylogenetic tree was constructed with the 43 screened and characterized CGTs (Figure 6).
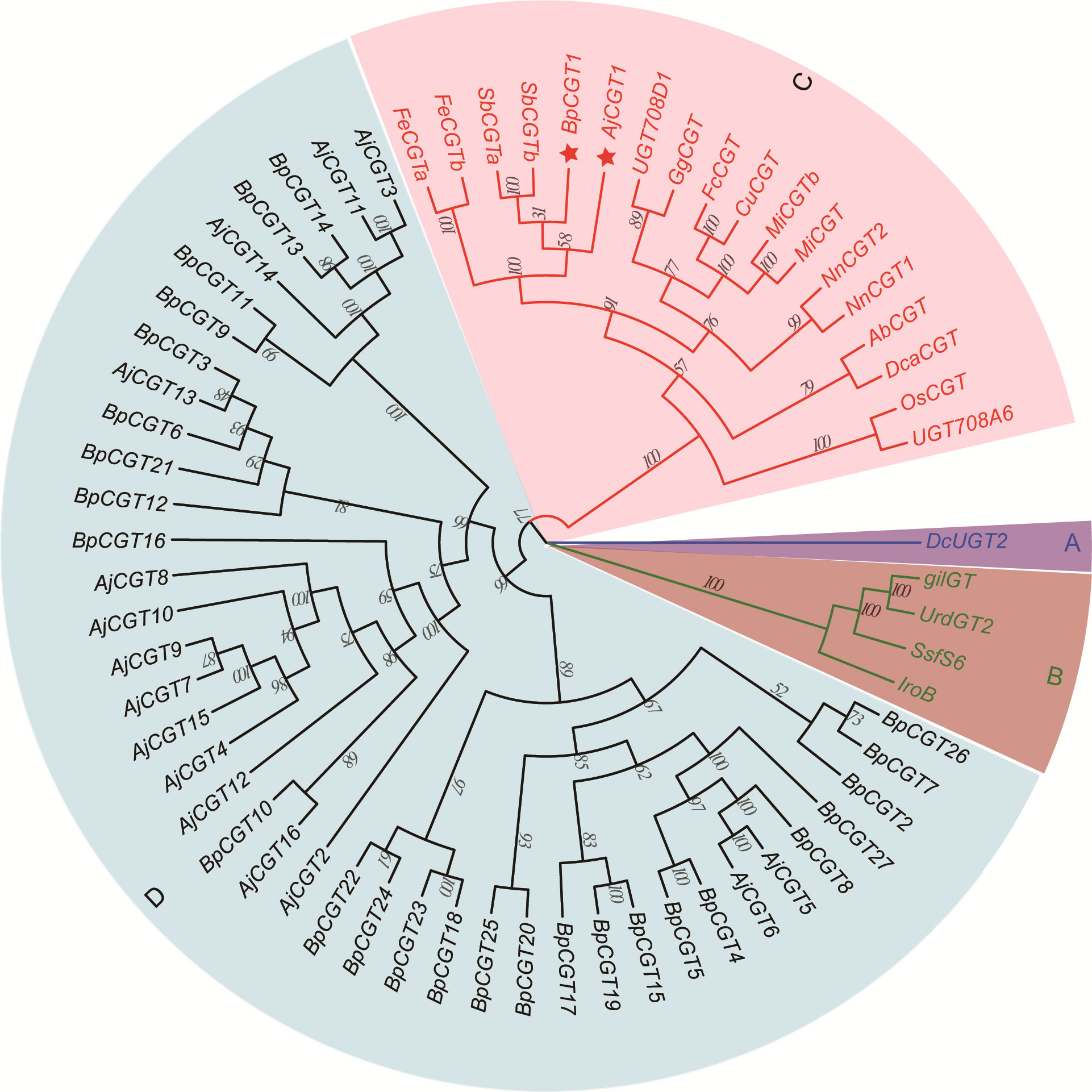
Figure 6 Phylogenetic tree of CGTs involved in bergenin biosynthesis. The sector (A) highlights the CGT sequences of glycosyltransferases found in animals, while sector (B) highlights those found in bacteria. Sector (C) represents the reported CGTs with glycosyltransferase function in plants. Sector (D) represents the UGTs identified in B. purpurascens and A. japonica by transcriptomics. The red pentagrams represent the candidate CGTs identified in B. purpurascens and A. japonica. The CGTs of other species in sectors A, B, and C were obtained from the NCBI database (Supplementary Table S3).
The genes were clearly divided into four groups, including animal, bacterial, and plant UGTs and those of B. purpurascens and A. japonica. Among all UGTs identified in B. purpurascens and A. japonica, only BpCGT1 and AjCGT1 clustered together with the reported plant CGTs in the phylogenetic tree. The corresponding amino acid sequences displayed a high degree of similarity to these plant CGTs. Based on these results, it can be inferred that BpCGT1 and AjCGT1 may possess CGT activity to catalyze the substrate 4-O-Me-GA to produce bergenin.
Both BpCGT1 and AjCGT1 were expressed in E. coli to assess their catalytic activity. Both BpCGT1 and AjCGT1 were successfully expressed and purified (Supplementary Figures 1D, E). The purified proteins were incubated with 4-O-Me-GA and UDP-glucose, and the products were treated with H+ before HPLC analysis. Bergenin production was not detected in the BpCGT1 assay, whereas for AjCGT1, a new peak appeared with the same retention time as the bergenin standard (Figure 7). The spectrum of AjCGT1 products showed LC-MS/MS fragmentation ions of bergenin at m/z 327 [M-H]−, m/z 312 [M-H-15]−, m/z 249 [M-H-78]−, and m/z 234 [M-H-93]−, which were consistent with those of the bergenin standard (Supplementary Figure 5). This result indicated that AjCGT1 can catalyze the glycosylation of 4-O-Me-GA at the C-2 position, which is followed by cyclization by a dehydratase or under acidic conditions to form bergenin.
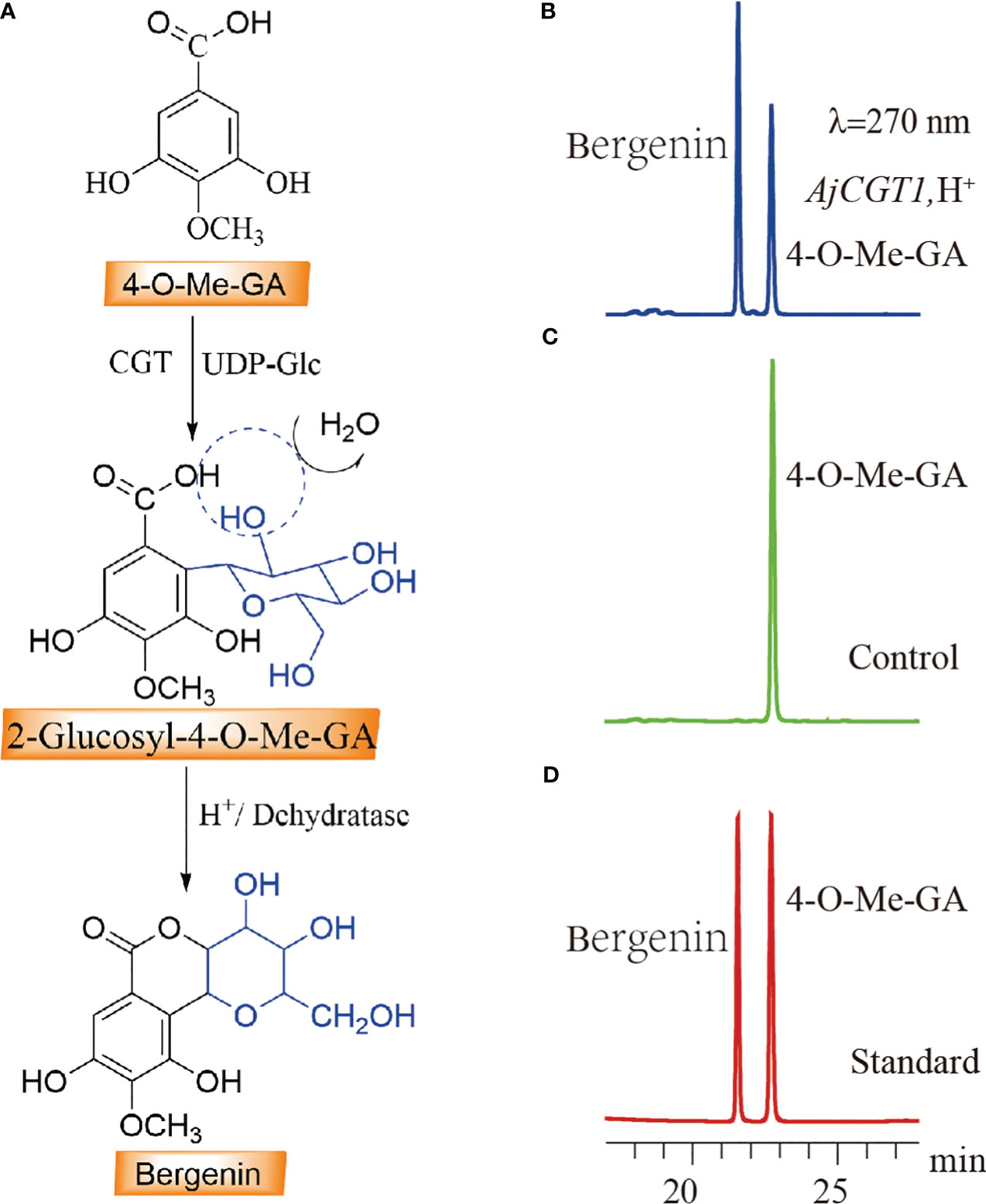
Figure 7 HPLC detection of the C-2 glycosylation activity of CGT protein on 4-O-Me-GA. (A) The biosynthetic pathway of bergenin through the C-2 glycosylation of 4-O-Me-GA. (B) The enzymatic activity assay of 4-O-Me-GA with AjCGT1 and UDP-Glc in vitro. (C) Inactivated AjCGT1 protein was used as a negative control in the reaction with 4-O-Me-GA and UDP-Glc. (D) The retention times of the standard compounds 4-O-Me-GA and bergenin.
Our findings corroborate that GA serves as the primary substrate for bergenin biosynthesis and plays a crucial role in the production of various secondary metabolites in plants (Franz and Grun, 1983; Petra et al., 2001). GA has been extensively studied for more than half a century (Dewick and Haslam, 1969). Isotope labeling experiments have revealed that it is primarily formed through direct dehydrogenation of SA (Ishikura et al., 1984). Crude extracts of birch leaves have been found to undergo a reaction with 3-DHS and NADP+, resulting in the production of GA. This finding provides further evidence that the intermediate 3-DHS in the shikimate pathway serves as a precursor for GA biosynthesis in plants (Vladimir et al., 2003). SDH is a multifunctional enzyme that also catalyzes the reversible reduction of 3-dehydroshikimate to shikimate (Muir et al., 2011), thereby playing a crucial role in the biosynthesis of aromatic compounds. Recent studies have validated these findings for VvSDH3 and VvSDH4 in grapes, CsDQD/SDHc and CsDQD/SDHd in tea plants, and EcDQD/SDH2 and EcDQD/SDH3 in Eucalyptus camaldulensis (Bontpart et al., 2016; Huang et al., 2019; Tahara et al., 2020). In this study, we discovered that BpSDH1 is capable of catalyzing two consecutive dehydrogenation reactions on SA to produce 3-dehydro-SA and GA, providing further evidence for the derivation of GA via the SA pathway in plants (Werner et al., 1997; Roland et al., 2004). In addition, we observed an anomalous phenomenon in the reaction products, wherein PCA was detected alongside 3-DHS and GA. PCA is primarily generated through the dehydration of 3-DHS, a process that has been extensively researched in microorganisms (Elsemore and Ornston, 1995; Muir et al., 2011; Brückner et al., 2018; Kim et al., 2020). However, it remains to be verified whether PCA in this reaction is formed through the dehydration of 3-DHS or other mechanisms.
OMTs play a crucial role in plants by catalyzing the transfer of a methyl group from SAM to hydroxyl groups on receptor molecules (Ibrahim et al., 1998; Joseph et al., 2003; David, 2005). They are actively involved in the biosynthesis of diverse polyphenolic compounds. Although numerous OMTs involved in the methylation of polyphenols have been identified, such as SlGOMT1 in Silene latifolia and EjOMT1 in Eriobotrya japonica, which catalyze the formation of veratrole from guaiacol (Gupta et al., 2012; Takao et al., 2016). Guaiacol is produced through the methylation of catechol by CTOMT1 in tomato (Mageroy et al., 2012). SaOMT2 in Sorbus aucuparia has exhibited some activity towards caffeic acid and 5-hydroxyferulic acid (Khalil et al., 2015); however, no enzyme capable of methylating GA had been identified to date. In this study, we discovered that BpOMT1 and AjOMT1 both exhibit O-methyltransferase activity towards the 4-OH position of GA, resulting in the formation of 4-O-Me-GA upon the addition of SAM as a methyl donor. Moreover, CTOMT1 and SaOMT2 exhibit substrate promiscuity towards compounds that share structural similarity with GA, which is in line with the homology results from the phylogenetic tree analysis (Mageroy et al., 2012; Khalil et al., 2015).
Since the first experimental validation of a CGT (OsCGT, UGT72B1) in Oryza sativa ssp. indica, numerous key CGTs involved in the synthesis of C-glycosyl flavonoids, xanthone C-glycosides, and coumarin C-glycosides have been verified in plants (Brazier-Hicks et al., 2009; Wang et al., 2020; Chen et al., 2021; Uchida et al., 2021). The formation process of these C-glycosides can be roughly divided into two steps. First, a CGT catalyzes the C-glycosylation reaction on the open-ring conformation of 2-hydroxyflavanone or benzophenones (Negi et al., 2013; Chen et al., 2015; Feng et al., 2021; Sun et al., 2022). Subsequently, the products undergo cyclization in the presence of acidic conditions and other enzymes to yield C-glycosides. Our study revealed the existence of AjCGT1, a C-glycosyltransferase capable of catalyzing the biosynthesis of C-glycosylated GAs in plants, marking the first time such an enzyme has been identified. AjCGT1 possesses a unique characteristic as it promotes C-2 glycosylation of 4-O-Me-GA. Under acidic conditions, the -OH group of glucose on 2-Glucosyl-4-O-Me-GA undergoes esterification dehydration with the -COOH group of 4-O-Me-GA, ultimately resulting in the production of bergenin. This is not only the first confirmed CGT involved in the biosynthesis of C-glycosylated GAs, but also the first demonstration that the -OH group of glucose participates in the formation of C-glycosides. Further, our findings serve as a valuable reference for future exploration of other types of CGTs.
In recent years, GA biosynthesis has been accomplished in microorganisms, primarily via modifications to the 3-DHS and chorismate pathways (Xinxiao et al., 2021). Introduction of 3-DHS dehydratase (AroZ) and 4-hydroxybenzoic acid ester hydroxylase (PobA Y385F) in engineered yeast under fed-batch conditions resulted in the production of 20 g/L GA (Kambourakis et al., 2000). GA production of 440.53 mg/L was achieved by introducing the more efficient double mutant Y385F/T294A, which continuously hydroxylates 4-HBA to GA (Chen et al., 2017). S. cerevisiae overexpressing upstream genes of the SA pathway and AroZ and PobA Y385F produced 682 mg/L GA (Brückner et al., 2018). The introduction of chorismate lyase (UbiC) effectively increased the flux of 4-HBA and promoted the synthesis of downstream products (Shang et al., 2020). Based on these findings, the UbiC and PobAY385F/T294A genes will be optimized and integrated with BpSDH1, BpOMT1 or AjOMT1, and AjCGT1 into engineered yeast. In conclusion, this study uncovered the catalytic functions of an SDH (BpSDH1), two OMTs (BpOMT1 and AjOMT1), and a CGT (AjCGT1) involved in the bergenin biosynthetic pathway in B. purpurascens and A. japonica. The molecular mechanism of bergenin biosynthesis was elucidated for the first time, paving the way to de novo biosynthesis in engineered yeast.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI accession numbers: PRJNA1008861 and PRJNA1008855.
X-YL: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Y-NW: Data curation, Investigation, Validation, Writing – review & editing. J-SD: Data curation, Investigation, Writing – original draft. B-HC: Formal analysis, Supervision, Software, Writing – original draft. K-YL: Data curation, Supervision, Writing – review & editing. LF: Investigation, Methodology, Project administration, Writing – review & editing. G-SX: Conceptualization, Data curation, Formal analysis, Software, Writing – original draft. S-YZ: Formal analysis, Visualization, Writing – review & editing. Y-CL: Supervision, Writing – review & editing. S-CY: Funding acquisition, Supervision, Writing – review & editing. G-HZ: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing. BH: Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Fundamental Research Project of Yunnan (202101AS070037, 202301BD070001-100, 202201AT070274), Yunnan Characteristic Plant Extraction Laboratory (2022YKZY001), The Major Science and Technique Programs in Yunnan Province (202102AA310048, 202102AE090042-01, 202102AA310045-01), National Natural Science Foundation of China (Grant No. 82160727).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1259347/full#supplementary-material
Supplementary Figure 1 | SDS-PAGE analysis of protein expression for BpSDH2, BpOMT1, AjOMT1, BpCGT1, and AjCGT1. (A–E) respectively represent the SDS-PAGE analysis results of the protein expressions of BpSDH2, BpOMT1, AjOMT1, BpCGT1, and AjCGT1. M represented the Direct-load™ Color Prestained Protein Marker; 1 represented the electrophoretic results of SDS-PAGE protein in 250 mM imidazole flow penetrating solution.
Supplementary Figure 2 | The extracted ion chromatogram (EICs) of compounds with molecular weights of 154, 170, and 172. (A) The extracted ion chromatogram (EICs) of the protease activity extract of BpSDH2 protein. (B) The extracted ion chromatogram (EICs) of standard PCA, 3-DHS, and GA.
Supplementary Figure 3 | The identification of reaction products and standards by LC-MS/MS analysis. (A, C, E) represent characteristic ion peaks with molecular weights of 172, 170, and 154, respectively, in the enzyme activity extract of BpSDH2. (B, D, F) represent the characteristic ion peaks of the standard with molecular weights of 172 (3-DHS), 170 (GA), and 154 (PCA), respectively.
Supplementary Figure 4 | The identification of reaction products and 4-O-Me-GA standard of OMT protein by LC-MS/MS analysis. (A) The extracted ion chromatogram (EICs) of the enzyme activity extracts of BpOMT1 and AJOMT1 with a molecular weight of 184, as well as the extracted ion chromatogram (EICs) of the standard 4-O-Me-GA. (B) The characteristic ion peak of the standard 4-O-Me-GA with a molecular weight of 184. (C) The characteristic ion peak with a molecular weight of 184 in the enzyme activity extract of BpOMT1. (D) The characteristic ion peak with a molecular weight of 184 in the enzyme activity extract of AjOMT1.
Supplementary Figure 5 | The identification of products of CGT protein and bergenin standard by LC-MS/MS analysis. (A) The extracted ion chromatogram (EICs) of the enzyme activity extracts of AJCGT1 with a molecular weight of 346 under conditions in the presence of methanol. (B) The MS fragments of the enzyme activity extracts of AJCGT1 with a molecular weight of 346 under conditions in the presence of methanol. (C) The characteristic ion peak with a molecular weight of 346 in the enzyme activity extract of AjCGT1 under conditions in the presence of methanol. (D) The extracted ion chromatogram (EICs) of the enzyme activity extracts of AJCGT1 with a molecular weight of 328 under conditions in the presence of hydrochloric acid, as well as the extracted ion chromatogram (EICs) of the standard bergenin. (E) The characteristic ion peak of the standard bergenin with a molecular weight of 328. (F) The characteristic ion peak with a molecular weight of 328 in the enzyme activity extract of AjCGT1 under conditions in the presence of hydrochloric acid.
Supplementary Figure 6 | Alignment of amino acid sequences of shikimate dehydrogenase BpSDH2, AjSDH2, VvSDH3, CsDQD/SDHc, and EcDQD/SDH3. Amino acid sequences alignment were performed using DNAMAN 8.0.
CGT, C-glycosyltransferases; OMT, O-methyltransferases; SDH, Shikimate dehydrogenases; SAM, S-adenosyl-L-methionine; UDP-Glu, Uridine diphosphate glucose; 4-O-Me-GA, 4-O-methyl gallic acid; SA, Shikimic acid; 3-DHS, 3-Dehydroshikimic acid; 2-Glucosyl-4-O-Me-GA, 2-Glucosyl-4-O-methyl gallic acid; PCA, Protocatechuic acid; PCA, Non-Redundant Protein Sequence Database; Nr, Non-Redundant Protein Sequence Database; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Barai, P., Raval, N., Acharya, S., Borisa, A., Bhatt, H., Acharya, N. (2019). Neuroprotective effects of bergenin in alzheimer’s disease: investigation through molecular docking, in vitro and in vivo studies. Behav. Brain Res. 356, 18–40. doi: 10.1016/j.bbr.2018.08.010
Bililign, T., Griffith, B. R., Thorson, J. S. (2005). Structure, activity, synthesis and biosynthesis of aryl-C-glycosides. Natural Product Rep. 22 (6), 742–760. doi: 10.1039/b407364a
Bontpart, T., Marlin, T., Vialet, S., Guiraud, J., Pinasseau, L., Meudec, E., et al. (2016). Two shikimate dehydrogenases, VVSDH3 and VVSDH4, are involved in gallic acid biosynthesis in grapevine. J. Exp. Botany. 67 (11), 3537–3550. doi: 10.1093/jxb/erw184
Brazier-Hicks, M., Evans, K. M., Gershater, M. C., Puschmann, H., Steel, P. G., Edwards, R. (2009). The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 284 (27), 17926–17934. doi: 10.1074/jbc.M109.009258
Brückner, C., Oreb, M., Kunze, G., Boles, E., Tripp, J. (2018). An expanded enzyme toolbox for production of cis, cis-muconic acid and other shikimate pathway derivatives in saccharomyces cerevisiae. FEMS Yeast Res. 18 (2), 1–12. doi: 10.1093/femsyr/foy017
Chen, D., Chen, R., Wang, R., Li, J., Xie, K., Bian, C., et al. (2015). Probing the catalytic promiscuity of a regio- and stereospecific C-glycosyltransferase from mangifera indica. Angew Chem. Int. Ed Engl. 54, 12678–12682. doi: 10.1002/anie.201506505
Chen, D., Fan, S., Yang, Z., Dai, J. (2021). Biocatalytic application of a membrane-bound coumarin C-glucosyltransferase in the synthesis of coumarin and benzofuran C-glucosides. Advanced Synthesis Catalysis. 363, 5072–5078. doi: 10.1002/adsc.202100041
Chen, Z., Shen, X., Wang, J., Wang, J., Yuan, Q., Yan, Y. (2017). Rational engineering of p-hydroxybenzoate hydroxylase to enable efficient gallic acid synthesis via a novel artificial biosynthetic pathway. Biotechnol. Bioengineering. 114 (11), 2571–2580. doi: 10.1002/bit.26364
China Pharmacopoeia Committee (2020). Chinese Pharmacopoeia, Volume I, PRC Vol. 226 (Beijing: China Medical Science and Technology Press), 386–387.
Chong, Y., Lee, S. W., Ahn, J. H. (2022). Phenolic C-glycoside synthesis using microbial systems. Curr. Opin. Biotechnol. 78, 102827. doi: 10.1016/j.copbio.2022.102827
Choubey, S., Varughese, L. R., Kumar, V., Beniwal, V. (2015). Medicinal importance of gallic acid and its ester derivatives: a patent review. Pharm. Patent Analyst. 4 (4), 305–315. doi: 10.4155/ppa.15.14
David, R. G. (2005). Evolution of flavors and scents. Annu. Rev. Plant Biol. 56, 301–325. doi: 10.1146/annurev.arplant.56.032604.144128
Dewick, P. M., Haslam, E. (1969). Phenol biosynthesis in higher plants. Gallic acid. Biochem. J. 113 (3), 537–542. doi: 10.1042/bj1130537
Elsemore, D. A., Ornston, L. N. (1995). Unusual ancestry of dehydratases associated with quinate catabolism in acinetobacter calcoaceticus. J. Bacteriology. 177 (20), 5971–5978. doi: 10.1128/jb.177.20.5971-5978.1995
Falcone, F. M. L., Rodriguez, E., Casas, M. I., Labadie, G., Grotewold, E., Casati, P. (2013). Identification of a bifunctional maize C- and O-glucosyltransferase. J. Biol. Chem. 288 (44), 31678–31688. doi: 10.1074/jbc.M113.510040
Feng, C. Y., Li, S. S., Taguchi, G., Wu, Q., Yin, D. D., Gu, Z. Y., et al. (2021). Enzymatic basis for stepwise C-glycosylation in the formation of flavonoid di-C-glycosides in sacred lotus (Nelumbo nucifera Gaertn.). Plant J. 106 (2), 351–365. doi: 10.1111/tpj.15168
Franz, G., Grun, M. (1983). Chemistry, occurrence and biosynthesis of C-glycosyl compounds in plants. Planta Med. 47, 131–140. doi: 10.1055/s-2007-969972
Gao, X., Guo, M., Zhang, Z., Wang, T., Cao, Y., Zhang, N. (2015). Bergenin plays an anti-inflammatory role via the modulation of mapk and NF-κB signaling pathways in a mouse model of lps-induced mastitis. Inflammation 38 (3), 1142–1150. doi: 10.1007/s10753-014-0079-8
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Gupta, A. K., Akhtar, T. A., Widmer, A., Pichersky, E., Schiestl, F. P. (2012). Identification of white campion (Silene latifolia) guaiacol O-methyltransferase involved in the biosynthesis of veratrole, a key volatile for pollinator attraction. BMC Plant Biol. 12, 158. doi: 10.1186/1471-2229-12-158
He, J. B., Zhao, P., Hu, Z. M., Liu, S., Kuang, Y., Zhang, M., et al. (2019). Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angew Chem. Int. Ed Engl. 58, 11513–11520. doi: 10.1002/anie.201905505
uang, K., Li, M., Liu, Y., Zhu, M., Zhao, G., Zhou, Y., et al. (2019). Functional analysis of 3-dehydroquinate dehydratase/shikimate dehydrogenases involved in shikimate pathway in Camellia sinensis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01268
Ibrahim, R. K., Bruneau, A., Bantignies, B. (1998). Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol. Biol. 36 (1), 1–10. doi: 10.1023/a:1005939803300
Ishikura, N., Hayashida, S., Tazaki, K. K. U. J. (1984). Biosynthesis of gallic and ellagic acids with 14C-labeled compounds in acer and rhus leaves. Botanical Magazine Tokyo (Japan). 97, 355–367. doi: 10.1007/BF02488668
Jihen, A., Pierre, L., Richard, D. (2017). Enzymatic synthesis of glycosides: from natural O- and N-glycosides to rare C- and S-glycosides. Beilstein J. Organic Chem. 13, 1857–1865. doi: 10.3762/bjoc.13.180
Jitender, S., Ashwani, K., Anupam, S. (2017). Antianxiety activity guided isolation and characterization of bergenin from caesalpinia digyna rottler roots. J. Ethnopharmacol. 195, 182–187. doi: 10.1016/j.jep.2016.11.016
Joseph, P. N., Richard, A. D., Eran, P., Chloe, Z., Jean-Luc, F. (2003). Chapter two structural, functional, and evolutionary basis for methylation of plant small molecules. Recent Adv. Phytochemistry. 37, 37–58. doi: 10.1016/S0079-9920(03)80017-5
Kambourakis, S., Draths, K. M., Frost, J. W. (2000). Synthesis of gallic acid and pyrogallol from glucose: replacing natural product isolation with microbial catalysis. J. Am. Chem. Society. 122 (37), 8–17. doi: 10.1021/ja000853r
Khalil, M. N. A., Brandt, W., Beuerle, T., Reckwell, D., Groeneveld, J., Hänsch, R., et al. (2015). O-methyltransferases involved in biphenyl and dibenzofuran biosynthesis. Plant Journal: For Cell Mol. Biol. 83 (2), 263–276. doi: 10.1111/tpj.12885
Kim, H., Kim, S. Y., Sim, G. Y., Ahn, J. H. (2020). Synthesis of 4-hydroxybenzoic acid derivatives in Escherichia coli. J. Agric. Food Chem. 68, 9743–9749. doi: 10.1021/acs.jafc.0c03149
Kumar, S., Sharma, C., Kaushik, S. R., Kulshreshtha, A., Chaturvedi, S., Nanda, R. K., et al. (2019). The phytochemical bergenin as an adjunct immunotherapy for tuberculosis in mice. J. Biol. Chem. 294 (21), 8555–8563. doi: 10.1074/jbc.RA119.008005
Li, P. P., Yang, S. C., Zeng, Y. H. (2009). Research progress on the medicinal plant resources of Rhodiola crenulata. Chin. Traditional Herbal Drugs 40 (09), 1500–1505. Available at: https://kns.cnki.net/kcms2/article/abstract?v=sSXGFc3NEDJ2qPJ0ZzBsw_b0xDbAiePNkhnSXVOsnyXERvMRzoF4i8xO9qy2EoNLrk_OjHnksR1UYpFKWxhPkNA9DNM_54vl7dBhPRaBFOftheOKZ__gqw==&uniplatform=NZKPT&language=gb.
Liang, J., Li, Y., Liu, X., Huang, Y., Shen, Y., Wang, J., et al. (2014). In vivo and in vitro antimalarial activity of bergenin. BioMed. Rep. 2 (2), 260–264. doi: 10.3892/br.2013.207
Lv, X. L., Zhang, C. Y., Shen, L. Y., Guan, Y., Feng, Y. H., Shi, J. S., et al. (2017). Requirement and development of Bergenia. Chin. Agric. Sci. Bulletin. 33 (22), 53–57. Available at: https://kns.cnki.net/kcms2/article/abstract?v=sSXGFc3NEDLmbYHSYVSmUvkmIdYcwC97fy3mI2A0F_FSq8ihLjoHJ6wzFuFPWpS7RihoVT3O3eAeQo__asb0VXkEaAujtR6tMaLOXmHR3ylh3eGhQTNe3y41OScjfJM&uniplatform=NZKPT&language=gb.
Mageroy, M. H., Tieman, D. M., Floystad, A., Taylor, M. G., Klee, H. J. (2012). A solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant Journal: For Cell Mol. Biol. 69 (6), 1043–1051. doi: 10.1111/j.1365-313X.2011.04854.x
Mehta, S., Kadian, V., Dalal, S., Dalal, P., Kumar, S., Garg, M., et al. (2022). A fresh look on bergenin: vision of its novel drug delivery systems and pharmacological activities. Future Pharmacol. 2 (1), 64–91. doi: 10.3390/futurepharmacol2010006
Mori, T., Kumano, T., He, H., Watanabe, S., Senda, M., Moriya, T., et al. (2021). C-glycoside metabolism in the gut and in nature: identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes. Nat. Commun. 12 (1), 6294. doi: 10.1038/s41467-021-26585-1
Muir, R. M., Ibáñez, A. M., Uratsu, S. L., Ingham, E. S., Leslie, C. A., McGranahan, G. H., et al. (2011). Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 75 (6), 555–565. doi: 10.1007/s11103-011-9739-3
Nagatomo, Y., Usui, S., Ito, T., Kato, A., Shimosaka, M., Taguchi, G. (2014). Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (buckwheat) cotyledon. Plant Journal: For Cell Mol. Biol. 80 (3), 437–448. doi: 10.1111/tpj.12645
Negi, J. S., Bisht, V. K., Singh, P., Rawat, M. S. M., Joshi, G. P. (2013). Naturally occurring xanthones: chemistry and biology. J. Appl. Chem. 50 (1), 78–91. doi: 10.1155/2013/621459
Nguyen, L., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evolution. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Nobuhiro, S., Yuzo, N., Eri, Y., Fumi, T., Takashi, N., Hideyuki, T., et al. (2015). Identification of the glucosyltransferase that mediates direct flavone C-glucosylation in Gentiana triflora. FEBS Letters. 589 (1), 182–187. doi: 10.1016/j.febslet.2014.11.045
Oualid, T., Artur, M. S. S. (2012). Advances in C-glycosylflavonoid research. Curr. Organic Chem. 16 (7), 859–896. doi: 10.2174/138527212800194791
Parkan, K., Pohl, R., Kotora, M. (2014). Cross-coupling reaction of saccharide-based alkenyl boronic acids with aryl halides: the synthesis of bergenin. Chemistry 20 (15), 4414–4419. doi: 10.1002/chem.201304304
Petra, G., Ruth, N., Gerhard, S., Georg, G. G. (2001). Biosynthesis and subcellular distribution of hydrolyzable tannins. Phytochemistry 57 (6), 915–927. doi: 10.1016/s0031-9422(01)00099-1
Rajesh, K., Dinesh, K. P., Satyendra, K. P., Damiki, L., Sairam, K., S., H. (2011). Type 2 antidiabetic activity of bergenin from the roots of caesalpinia digyna rottler. Fitoterapia 83 (2), 395–401. doi: 10.1016/j.fitote.2011.12.008
Ren, Z., Ji, X., Jiao, Z., Luo, Y., Zhang, G. Q., Tao, S., et al. (2020). Functional analysis of a novel C-glycosyltransferase in the orchid Dendrobium catenatum. Hortic. Res. 7, 111. doi: 10.1038/s41438-020-0330-4
Rohit, S., Vikas, K., Sonali, S. B., Ram, A. V. (2017). Synthesis, pH dependent, plasma and enzymatic stability of bergenin prodrugs for potential use against rheumatoid arthritis. Bioorgan. Med. Chem. 25 (20), 5513–5521. doi: 10.1016/j.bmc.2017.08.011
Roland, A. W., Andreas, R., Christine, S., Adelbert, B., Hanns-Ludwig, S., Wolfgang, E. (2004). Biosynthesis of gallic acid in Rhus typhina: discrimination between alternative pathways from natural oxygen isotope abundance. Phytochemistry 65 (20), 2809–2813. doi: 10.1016/j.phytochem.2004.08.020
Shang, Y., Wei, W., Zhang, P., Ye, B. (2020). Engineering Yarrowia lipolytica for enhanced production of arbutin. J. Agric. Food Chem. 68 (5), 1364–1372. doi: 10.1021/acs.jafc.9b07151
Shi, X., Xu, M., Luo, K., Huang, W., Yu, H., Zhou, T. (2019). Anticancer activity of bergenin against cervical cancer cells involves apoptosis, cell cycle arrest, inhibition of cell migration and the STAT3 signalling pathway. Exp. Ther. Med. 17 (5), 3525–3352. doi: 10.3892/etm.2019.7380
Singh, R., Kumar, V., Bharate, S. S., Vishwakarma, R. A. (2017). Synthesis, pH dependent, plasma and enzymatic stability of bergenin prodrugs for potential use against rheumatoid arthritis. Bioorganic Medicinal Chem. 25, 5513–5521. doi: 10.1016/j.bmc.2017.08.011
Sun, X., Xue, X., Wang, X., Zhang, C., Zheng, D., Song, W., et al. (2022). Natural variation of zmcgt1 is responsible for isoorientin accumulation in maize silk. Plant J. 109 (1), 64–76. doi: 10.1111/tpj.15549
Tahara, K., Nishiguchi, M., Funke, E., Miyazawa, S. I., Miyama, T., Milkowski, C. (2020). Dehydroquinate dehydratase/shikimate dehydrogenases involved in gallate biosynthesis of the aluminum-tolerant tree species Eucalyptus camaldulensis. Planta 253 (1), 3. doi: 10.1007/s00425-020-03516-w
Takao, K., Mami, K., Takumi, F., Hideyuki, S., Tomohiko, T., Kenji, M. (2016). Characterization of an O-methyltransferase specific to guaiacol-type benzenoids from the flowers of loquat (Eriobotrya japonica). J. Bioscience Bioengineering. 122 (6), 679–684. doi: 10.1016/j.jbiosc.2016.06.012
Taneyama, M., Yoshida, S. (1979). Studies on C-glycosides in higher plants. II. Incorporation of 14C-glucose into bergenin and arbutin in Saxifraga stolonifera. Botanical Magazine 92, 69–73. doi: 10.1007/BF0248830
Uchida, K., Akashi, T., Hirai, M. Y. (2021). Identification and characterization of glycosyltransferases catalyzing direct xanthone 4-C-glycosylation in Hypericum perforatum. FEBS Lett. 595, 2608–2615. doi: 10.1002/1873-3468.14179
Vidal, P., Roldos, V., Fernandez-Alonso, M. C., Vauzeilles, B., Bleriot, Y., Cañada, J., et al. (2013). Conformational selection in glycomimetics: human galectin-1 only recognizes syn-Psi-type conformations of beta-1,3-linked lactose and its C-glycosyl derivative. Chemistry 19, 14581–14590. doi: 10.1002/chem.201301244
Vladimir, O., Juha-Pekka, S., Svetlana, O., Erkki, H., Kalevi, P. (2003). Gallic acid and hydrolysable tannins are formed in birch leaves from an intermediate compound of the shikimate pathway. Biochem. Systematics Ecology. 31 (1), 3–16. doi: 10.1016/S0305-1978(02)00081-9
Wang, Z., He, Y., Liao, L., Zhang, Y., Zhao, Y., Xiao, Y., et al. (2022). Forming coumarin C-glycosides via biocatalysis: characterization of a C-glycosyltransferase from Angelica decursiva. Biochem. Biophys. Res. Commun. 614, 85–91. doi: 10.1016/j.bbrc.2022.05.008
Wang, Z. L., Gao, H. M., Wang, S., Zhang, M., Chen, K., Zhang, Y. Q., et al. (2020). Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc. Natl. Acad. Sci. U. S. A. 117, 30816–30823. doi: 10.1073/pnas.2012745117
Werner, I., Bacher, A., Eisenreich, W. (1997). Retrobiosynthetic nmr studies with 13C-labeled glucose. Formation of gallic acid in plants and fungi. J. Biol. Chem. 272 (41), 25474–25482. doi: 10.1074/jbc.272.41.25474
Xiang, S., Chen, K., Xu, L., Wang, T., Guo, C. (2020). Bergenin exerts hepatoprotective effects by inhibiting the release of inflammatory factors, apoptosis and autophagy via the ppar-gamma pathway. Drug Des. Devel Ther. 14, 129–143. doi: 10.2147/DDDT.S229063
Xinxiao, S., Xianglai, L., Xiaolin, S., Jia, W., Qipeng, Y. (2021). Recent advances in microbial production of phenolic compounds. Chin. J. Chem. Engineering. 30 (02), 54–61. doi: 10.1016/j.cjche.2020.09.001
Keywords: biosynthetic pathway, bergenin, carbon glycosides, C-glycosyltransferase, O-methyltransferases
Citation: Liu X-Y, Wang Y-N, Du J-S, Chen B-H, Liu K-Y, Feng L, Xiang G-S, Zhang S-Y, Lu Y-C, Yang S-C, Zhang G-H and Hao B (2024) Biosynthetic pathway of prescription bergenin from Bergenia purpurascens and Ardisia japonica. Front. Plant Sci. 14:1259347. doi: 10.3389/fpls.2023.1259347
Received: 15 July 2023; Accepted: 14 December 2023;
Published: 04 January 2024.
Edited by:
Kristina Haslinger, University of Groningen, NetherlandsReviewed by:
Rufeng Wang, Shanghai University of Traditional Chinese Medicine, ChinaCopyright © 2024 Liu, Wang, Du, Chen, Liu, Feng, Xiang, Zhang, Lu, Yang, Zhang and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Hao, QmluZy5IYW9AaG90bWFpbC5jb20=; Guang-Hui Zhang, emdoNzMxMDczMTBAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.