- 1Department of Biology Education, Pusan National University, Busan, Republic of Korea
- 2Department of Pharmacology and Neuroscience, Creighton University School of Medicine, Omaha, NE, United States
- 3Marine Industry Research Institute for East Sea Rim, Uljin, Republic of Korea
- 4Department of Systems Biotechnology, Chung-Ang University, Anseong, Republic of Korea
Candidate strains that contribute to drought resistance in plants have been previously screened using approximately 500 plant growth-promoting rhizobacteria (PGPR) obtained from Gotjawal, South Korea, to further understand PGPR associated with plant drought tolerance. In this study, a selected PGPR candidate, Flavobacterium sp. strain GJW24, was employed to enhance plant drought tolerance. GJW24 application to Arabidopsis increased its survival rate under drought stress and enhanced stomatal closure. Furthermore, GJW24 promoted Arabidopsis survival under salt stress, which is highly associated with drought stress. GJW24 ameliorated the drought/salt tolerance of Brassica as well as Arabidopsis, indicating that the drought-resistance characteristics of GJW24 could be applied to various plant species. Transcriptome sequencing revealed that GJW24 upregulated a large portion of drought- and drought-related stress-inducible genes in Arabidopsis. Moreover, Gene Ontology analysis revealed that GJW24-upregulated genes were highly related to the categories involved in root system architecture and development, which are connected to amelioration of plant drought resistance. The hyper-induction of many drought/salt-responsive genes by GJW24 in Arabidopsis and Brassica demonstrated that the drought/salt stress tolerance conferred by GJW24 might be achieved, at least in part, through regulating the expression of the corresponding genes. This study suggests that GJW24 can be utilized as a microbial agent to offset the detrimental effects of drought stress in plants.
Introduction
Various abiotic stresses including heat, cold, UV, drought, high salt, and heavy metals, adversely affect plant productivity (Zhu, 2002). However, plants have developed regulatory mechanisms via plant-microbe interactions as a strategy to overcome these stresses. Plant growth-promoting rhizobacteria (PGPR), which are indigenous to the plant rhizosphere, can play a positive role in plant growth via diverse mechanisms, including enhancement of abiotic stress tolerance (Vejan et al., 2016). When plants face abiotic stresses, PGPR provides plants with adaptation mechanisms such as the regulation of hormones such as indole-3-acetic acid (IAA), cytokinin (CK), gibberellic acid (GA), and abscisic acid (ABA); the production of 1-aminocyclopropane 1-carboxylic acid (ACC) deaminase; the production of antioxidants; the secretion of osmolytes; and the secretion of microbial exopolysaccharide (EPS) (Ma et al., 2020). Although a diverse range of bacteria belonging to Pseudomonas and Bacillus have been reported as PGPR, the study on PGPR responsible for abiotic stress tolerance in other groups has been relatively poor (Beneduzi et al., 2012).
Drought is one of the abiotic stresses determining plant productivity. It has several adverse effects during plant life cycle, such as the inhibition of photosynthesis, alteration of the respiration rate, disruption of metabolic processes, and improper ion uptake (Hussain et al., 2018). Therefore, to offset these adverse effects, studies on useful PGPR involved in improving plant drought tolerance have been rapidly conducted in recent years. Many studies have revealed that various PGPR ameliorate drought stress resistance in diverse plant species. IAA and GA from several PGPR promote nutrient acquisition and growth of plants, enabling plant survival under drought/salt stress conditions (Ma et al., 2020; Uzma et al., 2022). The treatment of CK-producing Bacillus subtilis has been shown to improve drought resistance in Platycladus orientalis (Liu et al., 2013). These PGPR-produced phytohormones regulate the formation of root hairs and lateral roots, leading to increased uptake of water and nutrients. PGPR, such as Bacillus licheniformis Rt4M10, Phyllobacterium brassicacearum STM196, and Azospirillum brasilense Sp 245, increase ABA content in plants, consequently ameliorating drought tolerance (Bresson et al., 2013; Cohen et al., 2015). Abiotic stress-induced ethylene inhibits plant growth; however, ACC deaminase (an ACC-degrading enzyme)-producing PGPR can mitigate the adverse effects of abiotic stressors and positively regulate plant growth (Lephatsi et al., 2021). Several reports have demonstrated that ACC deaminase-producing microbes confers enhanced tolerance to drought/salt stress in diverse plant species (Forni et al., 2017). Abiotic stress triggers the production of reactive oxygen species, which are highly toxic to macromolecules and lipids and result in oxidative damage in plant cells (You and Chan, 2015). Some PGPR are involved in regulating the activities of antioxidant enzymes and their gene expression, which mitigates the deleterious effects of abiotic stressors, including drought (Bharti et al., 2016; Batool et al., 2020). Plants utilize compatible osmolytes as osmoprotectants to stabilize macromolecules and membrane structure under drought/salt stress. PGPR-produced EPS is able to help keep the water potential of the rhizosphere high by binding soil particles, thereby ameliorating plant abiotic stress tolerance (Naseem et al., 2018). The existence of PGPR, which produces these osmolytes and EPS, has been widely reported, with a focus on its effect on enhancing resistance to drought/salt stress (Naseem et al., 2018; Khan and Bano, 2019; Ma et al., 2020). Collectively, these studies show that various PGPR strains play crucial roles in ameliorating plant drought stress resistance via diverse mechanisms.
In this research, we show that Flavobacterium sp. strain GJW24 improved drought and drought-related salt stress resistance in Arabidopsis and Brassica plants. The abiotic stress resistance phenotype conferred by GJW24 increases the possibility that GJW24 can be utilized as a biological material to positively regulate plant growth and yield under such unfavorable environments.
Materials and methods
Plant materials and culture conditions
Arabidopsis (A. thaliana) ecotype Col-0 and cabbage (Brassica campestris L. ssp. pekinensis, cultivar; ‘Chun Yeon Gold’) were used as the wild types in this study. The surfaces of the plant seeds were sterilized, and the seeds were grown on 1× MS plate supplemented with 0.8% bacto agar and 1% sucrose (pH 5.8). The seeds were kept for 7 d in a growth chamber at 23°C under a 16 h light/8 h dark cycle. Then, the resulting seedlings were moved to into a pot filled with horticultural soil mix (cocopeat 51.5%, peat moss 10%, vermiculite 13%, perlite 15%, zeolite10%, humic acid 0.1%, fertilizer 0.4%) and kept in a growth chamber at 23°C under a 16 h light/8 h dark cycle for further analysis.
Bacterial growth conditions and plant inoculation
The Flavobacterium sp. strain GJW24 was obtained from Gotjawal soil, Jeju Island, South Korea (Kim et al., 2018). GJW24 was incubated on R2A plates overnight at 28°C. The resulting microbes were adjusted to an OD600 of 0.02~0.03 using MS solution. Next, one milliliter of the bacterial suspension was added to the region around the growth areas of Arabidopsis and Brassica.
Assay for drought/salt stress resistance in Arabidopsis
Assays for drought and salt stress resistance in Arabidopsis were performed using the MS liquid (mock) or GJW24-treated plants. For the drought tolerance assay, 8-day-old plants were exposed to mock or GJW24. Seven days after treatment, they were exposed to water deficiency (drought stress) for 14 d, followed by re-watering for 4 d. The assays were conducted with four biological replicates. For each assay, at least ten plants were used for mock or GJW24 treatment. The assay for salt stress resistance was conducted by supplying 50 mL of water or 150 mM NaCl solution to the soil 7 d after mock or GJW24 treatment, three times for 6 d at 3-d intervals. Survival rates were monitored 3 d after the last day of NaCl treatment. The assays were conducted with six biological replicates. In four combinations of GJW24 and salt stress treatments for each assay, at least eight plants were used for each combination.
Assay for drought/salt stress resistance in Brassica
For the drought tolerance assay for Brassica, 8-day-old Brassica plants were exposed to MS liquid (mock) or GJW24. Seven days after treatment, the plants were exposed to water deficiency (drought stress) for 10 d, followed by re-watering for 6 d. The assays were conducted with four biological replicates. For each assay, at least six plants were used for mock or GJW24 treatment. The assay for salt stress resistance was performed by supplying 50 mL of water or 150 mM NaCl solution to the soil 7 d after mock or GJW24 treatment, eight times at 3-d intervals. Survival rates were monitored 1 d after the last day of NaCl treatment. For each assay, at least eleven plants were used for mock or GJW24 treatment.
Measurement of stomatal aperture
Eight-day-old Arabidopsis plants were exposed to mock or GJW24. Seven days after treatment, detached leaves from mock- or GJW24-treated plants were floated in stomatal opening solution containing 10 mM MES-KOH (pH 6.15), 50 mM KCl and 10 mM CaCl2 for 2.5 h in the light (Lee et al., 2013). Then, the samples were additionally incubated in the same buffer with 0 or 20 μM ABA for 2.5 h in the light. The stomatal apertures from the abaxial part of the treated leaves were measured using a microscope (DM750, Leica) and the ImageJ program, as described by Kim et al. (2020).
Measurement of chlorophyll concentration
The total chlorophyll contents of the mock- or GJW24-treated samples under salt stress were quantified as described by Woo et al. (2020). The extracts were prepared using 80% acetone, and were then maintained in the dark for 30 min. The assays were conducted with six biological replicates. The chlorophyll content was quantified using the supernatants from centrifugation, and determined as described by Woo et al. (2020).
RNA isolation and RT-qPCR analysis
Total RNA was extracted using TRIzol reagent (Ambion). From one microgram of total RNA, complementary DNA (cDNA) was synthesized using RevertAid™ reverse transcriptase (Fermentas). Quantitative real-time PCR (qRT-PCR) was performed as described by Woo et al. (2020). The qRT-PCR was performed using the Solg™ 2 × real-time PCR Smart Mix (SolGent) and analyzed with the Rotor-Gene Q system (Qiagen). The transcript amounts of each gene were monitored using the comparative cycle threshold (CT) method, which was normalized against the transcripts from the reference genes; Arabidopsis ACTIN2 or Brassica ACT7 (Schmittgen and Livak, 2008). The DNA sequences for RT-qPCR analysis are listed in Table S1. The qRT-PCR analyses were performed with at least six biological replicates.
Transcriptome sequencing analysis
For transcriptome sequencing analysis, eight-day-old Arabidopsis plants were exposed to mock or GJW24. Total RNAs were independently extracted from the entire Arabidopsis plants (8 plants for each treatment) with three biological replicates 7 d after mock or GJW24 treatment. The replicates were not pooled per treatment. The analysis was done by MacroGen Inc. (Korea). To evaluate total RNA integrity, samples were run on a TapeStation RNA Screentape (Agilent Technologies). The RNA library was generated using RNA with RNA integrity number higher than 7.0. A total RNA library (0.5 μg) for each sample was produced using the Illumina TruSeq Stranded Total RNA Library Prep Plant Kit (Illumina, Inc.). After removing the total ribosomal RNA, the messenger RNA was fragmented using divalent cations at elevated temperatures. The resulting fragmented mRNAs were converted into first-strand cDNA with random primers and SuperScript II reverse transcriptase (Invitrogen). After second-strand cDNA synthesis using DNA Polymerase I, RNase H and dUTP, the resulting cDNA fragments go through an end repair process, the addition of a single ‘A’ base, and then ligation of the adapters. After purification, the products were enriched using PCR to construct the final cDNA library. Libraries were quantified using KAPA Library Quantification kits for Illumina Sequencing platforms (Kapa Biosystems) and qualified using a TapeStation D1000 ScreenTape (Agilent Technologies). Paired-end (2×100 bp) sequencing of indexed libraries was conducted using an Illumina NovaSeq system (Illumina, Inc.), by MacroGen Inc. (Korea). After removing adapter sequences and trimming bases with poor base quality using Trimmomatic v0.38, the cleaned reads were aligned to TAIR 10.1 with HISAT v2.1.0 (Kim et al., 2015), based on the HISAT and Bowtie2 implementations. The reference genome sequence and the gene annotation data were retrieved from NCBI Genome assembly and NCBI RefSeq database, respectively. Aligned data were sorted and indexed with SAMtools v 1.9, and the transcripts were assembled and quantified with StringTie v2.1.3b (Pertea et al., 2015; Pertea et al., 2016). Gene-level and transcript-level quantification were calculated with raw read count, FPKM and TPM.
Differential gene expression analysis
Among total 38,338 genes, 21,834 genes with non-zero counts in all replicates at least one period group were retrieved during the QC step. The fold change and the statistical significance of DEGs was investigated by exactTest using edgeR (Robinson et al., 2010). Among the 21,834 genes analyzed, the expression of the transcript that satisfied with FC ≥ 2 and p < 0.05 at all comparisons was considered as significantly up-regulated genes. All p-values are adjusted using Benjamini-Hochberg algorithm to control false discovery rate. To investigate characteristic biological attributes of the genes upregulated by GJW24 treatment, Gene Ontology (GO) analysis was performed with the functional annotation tool on DAVID (https://david.ncifcrf.gov/tools.jsp), and the GJW24-upregulated genes were assigned to different categories for biological processes (BPs) and molecular functions (MFs). The inducibility of GJW24-upregulated genes under drought stress were checked using the database from the Arabidopsis eFP Browser (Winter et al., 2007).
Phylogenetic analysis
To generate a phylogenetic tree based on the 16s rRNA sequences, the 16s rRNA sequences of 17 Flavobacterium type strains, which exhibited the highest similarity to that of GJW24 based on a BLAST search, and 9 type strains from other genera in the family Flavobacteriaceae were obtained from ‘The List of Prokaryotic names with Standing in Nomenclature’ (LPSN) (https://lpsn.dsmz.de/text/approved-lists). The 16s rRNA sequences were aligned with ClustalW running in the MEGA11 program (Tamura et al., 2021). A rooted phylogenetic tree was generated with the MEGA11 using the maximum-likelihood method. The bootstrap method was performed to assess statistical significance of the tree using 1000 replicates.
Results and discussion
Identification of a Flavobacterium involved in drought tolerance process in Arabidopsis
Approximately 500 soil microbes, which included a unique microbial community, have previously been obtained from Gotjawal, Jeju Island (Kim et al., 2018; Woo et al., 2020). Through a preliminary screening process to select PGPR involved in drought stress resistance in Arabidopsis, several microbes whose applications resulted in the upregulation of drought resistance marker genes in Arabidopsis were identified (Woo et al., 2020). The 16S rRNA gene of the selected candidate exhibited the highest similarity to those of diverse Flavobacterium strains based on a BLAST search, showing that it belongs to the genus Flavobacterium. Therefore, the strain was named Flavobacterium sp. strain GJW24. The 16S rRNA gene sequence of GJW24 has been uploaded in GenBank under accession number OR272272. To examine how closely related GJW24 is to other Flavobacterium strains that have already been characterized, the 16s rRNA sequences of type strains from diverse Flavobacteria and other genera in the family Flavobacteriaceae were obtained from LPSN, and a phylogenetic tree based on the 16s rRNA sequences of GJW24 and the selected type strains was generated (Figure 1). Although the Flavobacterium genus has been reported as a PGPR group, studies on its biological contribution to the drought stress response in plants are highly limited (Gontia-Mishra et al., 2016; Kim et al., 2020); therefore, the biological function of GJW24 was further analyzed in this study.
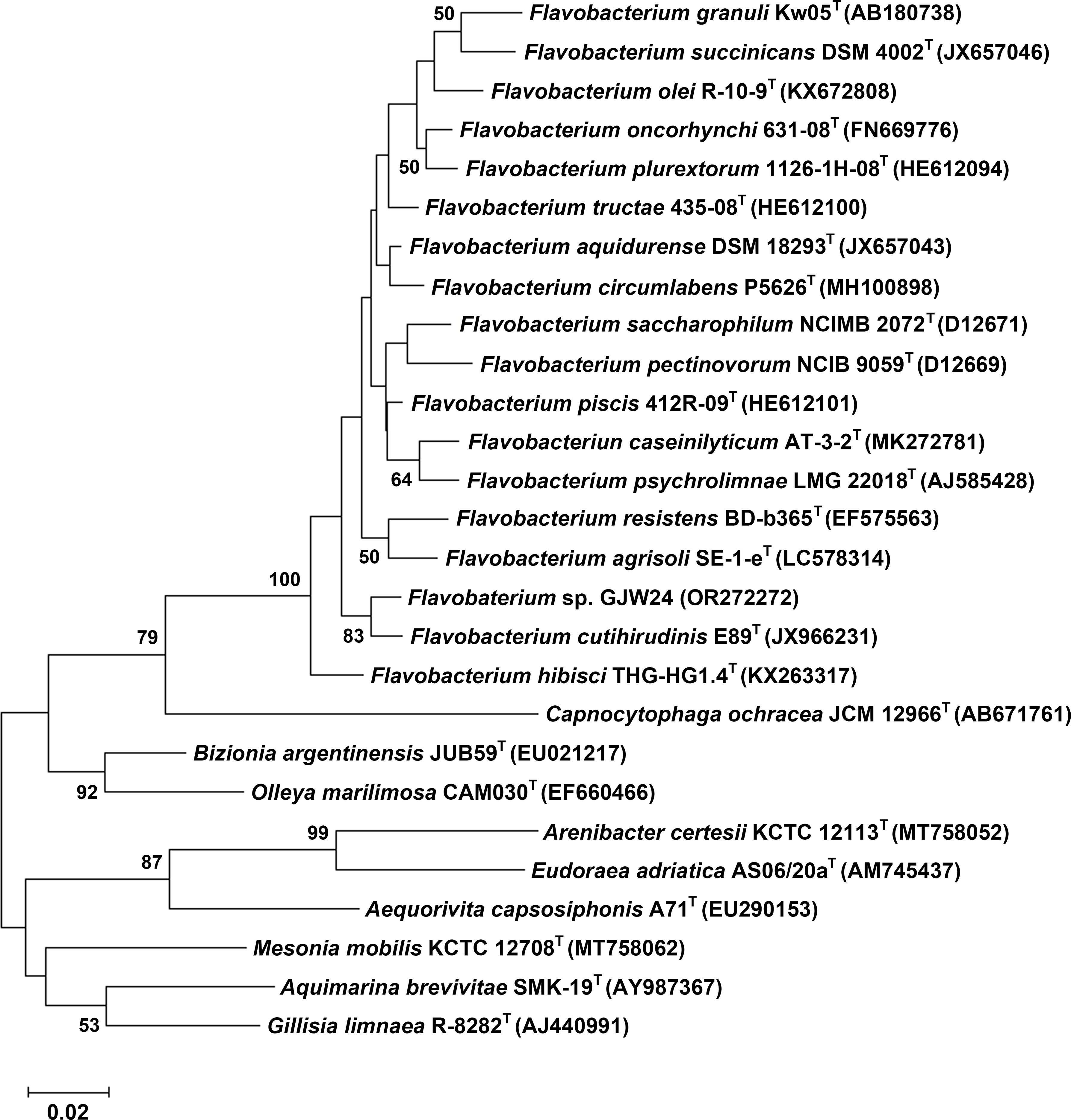
Figure 1 A phylogenetic tree among Flavobacterium sp. strain GJW24, diverse Flavobacterium type strains, and type strains from other genera in the family Flavobacteriaceae, based on their 16S rRNA gene sequences. The 16s rRNA sequences were obtained from LPSN and aligned with ClustalW running in the MEGA11 program. A rooted phylogenetic tree was generated with the MEGA11 using the maximum-likelihood method. The bootstrap method was performed to assess statistical significance of the tree using 1000 replicates. Bootstrap values (>50%) are indicated at the nodes.
Improved drought/salt stress resistance in Arabidopsis and Brassica following GJW24 application
To examine the potential functional role of GJW24 in the drought stress response, a phenotypic analysis of drought resistance in Arabidopsis was performed in the presence and absence of GJW24. The survival rate of GJW24-treated plants under drought condition was more than 2.5-fold higher than that of the mock-treated plants (Figure 2A). Given that tolerance against drought stress is largely achieved through the modulation of the stomatal aperture in plants, the effect of GJW24 on stomatal aperture regulation was further monitored. Although the difference of the stomatal apertures between mock-treated and GJW24-treated plants was not detected in the absence of ABA, the stomatal closure of GJW24-treated plants was significantly enhanced compared to mock-treated plants in the presence of ABA, indicating that GJW24 is involved in the promotion of ABA-mediated stomatal closure (Figure 2B). ABA is a crucial phytohormone that mediates drought tolerance by promoting stomatal closure and modulating drought-responsive genes (Takahashi et al., 2020). Thus, the enhancement of ABA-dependent stomatal closure in Arabidopsis by GJW24 (Figure 2B) shows that the regulation of the ABA signal transduction pathway is involved in the GJW24-mediated drought tolerance process. Collectively, these findings indicate that GJW24 improves tolerance against drought stress and that this process may be accomplished by minimizing water loss from transpiration, which results, at least in part, from the promotion of ABA-mediated stomatal closure. As salt stress increases endogenous ABA levels in plants, the plant salt stress response has been reported to largely overlap with the ABA-mediated drought stress response (Shinozaki and Yamaguchi-Shinozaki, 2007). Therefore, to confirm the GJW24-induced improvement in drought tolerance, the effect of GJW24 on salt stress resistance was examined. Similar to drought stress, the survival rate and chlorophyll content of GJW24-treated plants were approximately two- and three-fold higher than those of mock-treated ones under salt stress, respectively (Figure 2C), indicating that GJW24 application enhanced salt stress tolerance. To determine whether the GJW24-induced drought/salt stress resistance in Arabidopsis could be applied to other plants, the effect of GJW24 on abiotic stress resistance in Brassica, a crop most closely related to Arabidopsis, was investigated (Lagercrantz et al., 1996; Woo et al., 2020). Similar to its effect on Arabidopsis, GJW24 improved drought/salt stress tolerance in Brassica. Under drought and salt stress, the survival rate of GJW24-treated Brassica plants was more than three- and two-fold higher than that of mock-treated plants, respectively (Figures 2D, E). GJW24-induced improved drought/salt stress tolerance in Arabidopsis and Brassica increases the possibility that GJW24 may be utilized as a biological resource to facilitate plant growth under drought/salt stress condition.
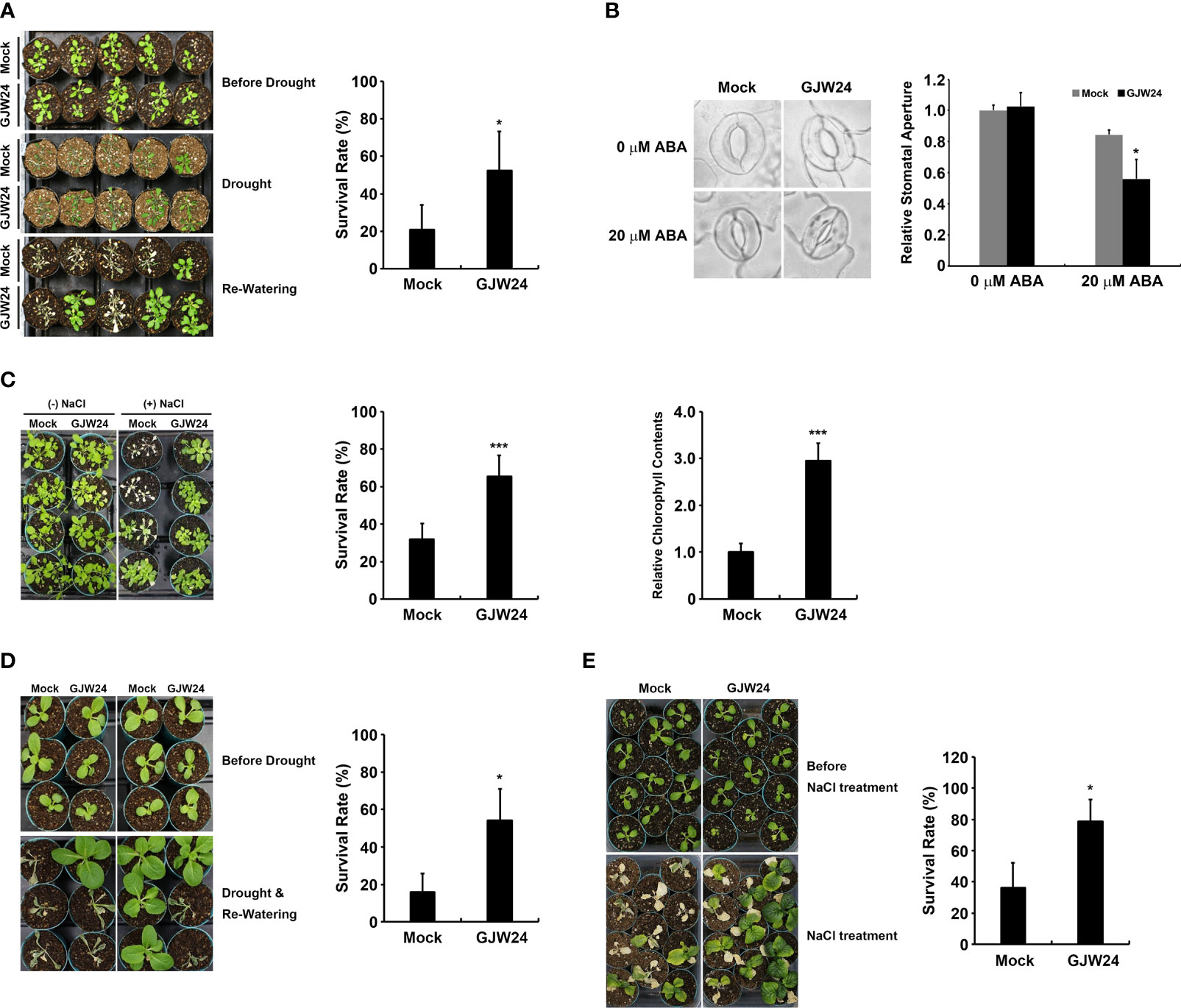
Figure 2 Phenotypic analysis of Arabidopsis and Brassica plants in response to drought and salt stressors in the presence and absence of GJW24. (A) Increased drought tolerance of Arabidopsis. Eight-day-old Arabidopsis plants were treated with mock or GJW24 and watered regularly for 1 week. The resulting plants were exposed to dehydration stress for 14 d, followed by re-watering for 4 (d) Error bars indicate the standard deviation (SD) of four biologically independent experiments. A Student’s t-test was conducted to identify statistically significant differences; *P < 0.05. (B) Stomatal movements of Arabidopsis with or without GJW24, in the absence or presence of ABA. Eight-day-old Arabidopsis plants were treated with mock or GJW24. Seven days after treatment, detached leaves from mock- or GJW24-treated plants were prepared to measure the stomatal apertures in the absence or presence of ABA. Error bars indicate the SD of three biologically independent experiments (at least 40 stomata for each biological sample); *P < 0.05. (C) Enhanced salt resistance in Arabidopsis following GJW24 application. Eight-day-old plants were treated with mock or GJW24. Salt stress resistance was observed by introducing 50 mL of water or 150 mM NaCl solution to the soil 7 d after GJW24 treatment, three times for 6 d at 3-d intervals. The survival rates (n = 6) and the chlorophyll contents (n = 6) under salt stress were determined 3 and 4 d after the final salt treatment, respectively; ***P < 0.001. (D) Improved drought resistance in Brassica following GJW24 application. Eight-day-old Brassica plants were treated with mock or GJW24 and watered regularly for 1 week. The resulting plants were subjected to dehydration stress for 10 d, followed by re-watering for 6 (d) Error bars indicate the SD of four biologically independent experiments; *P < 0.05. (E) Enhanced salt tolerance in Brassica following GJW24 application. Ten-day-old Brassica plants were treated with mock or GJW24. Salt stress tolerance was observed by introducing 50 mL of water or 150 mM NaCl solution to the soil 7 d after mock or GJW24 treatment, eight times at 3-d intervals. The survival rates under salt stress were determined 1 d after the final salt treatment (n = 6); ***P < 0.001.
GJW24 increases upregulation of a large portion of drought/salt/osmotic stress−inducible genes
To understand the action mechanism of GJW24 in drought/salt stress resistance in Arabidopsis, transcriptome sequencing analysis of Arabidopsis was performed in the presence and absence of GJW24. Among the 21,834 genes analyzed, the expression of 251 showed at least a two-fold increase in the GJW24-treated samples compared to the mock-treated samples (Tables 1, S2). To investigate characteristic biological attributes of the genes upregulated by GJW24 treatment, Gene Ontology (GO) analysis was done with the functional annotation tool on DAVID (https://david.ncifcrf.gov/tools.jsp), and the GJW24-upregulated genes were assigned to different categories for biological processes (BPs) (Figure 3A and Table S3) and molecular functions (MFs) (Figure 3B and Table S4). For BP, the genes were highly related to categories involved in root system architecture and development. Furthermore, GO terms related to ‘response to stress’ were notably enriched. To investigate whether the GJW24-conferred drought resistance phenotype in Arabidopsis is functionally associated with the altered expression patterns observed in GJW24-treated plants, the inducibility of GJW24-upregulated genes under drought stress was monitored using the database from the Arabidopsis eFP Browser (Winter et al., 2007). Based on information from the Arabidopsis eFP Browser, for each gene, the expression values from drought-treated samples (approximately 10% fresh weight loss) at various time points (0.25, 0.5, 1, 3, 6, 12 or 24 h) were divided by the values from untreated samples at the same time points. The maximum fold increase for each gene was then retrieved. Of the total 251 GJW24-specific genes, 100 (39.8%) exhibited increased expression patterns by more than twofold by drought stress, indicating that the application of GJW24 upregulated several drought-inducible genes (Tables 1, S2). Given that drought stress responses in plants largely overlaps with the responses to salt and osmotic stressors (Zhu, 2002), the inducibility of the GJW24-upregulated genes to salt and osmotic stress was also investigated based on information from the Arabidopsis eFP Browser. For each gene, the expression values from 150 mM NaCl-treated samples (salt) at various time points (0.5, 1, 3, 6, 12 or 24 h) or 300 mM mannitol-treated samples (osmotic) at various time points (0.5, 1, 3, 6, 12 or 24 h) were divided by the values from untreated samples at the same time points. The maximum fold increase for each gene was then obtained. Similar to drought-inducible genes, 115 (45.8%) and 110 (43.8%) of the total 251 GJW24-specific genes exhibited upregulated expression of more than twofold by salt and osmotic stress, respectively (Tables 1, S2). Moreover, 47.9% (70 genes) of the 146 genes induced by drought, salt, or osmotic stress among the GJW24-upregulated genes were commonly upregulated by these three stressors (Figure 3C). Collectively, these findings indicate that GJW24 upregulated a large portion of drought- and drought-related stress-inducible genes in Arabidopsis. Previous researches have shown that drought resistance traits are functionally associated with the upregulation of drought-inducible genes in plants (Kim et al., 2020; Woo et al., 2020); therefore, GJW24-induced enhanced drought/salt stress tolerance may result from the overall enhancement of the upregulation of genes induced by drought and drought-related stressors. The GO analysis results revealed that the GJW24-upregulated genes were highly assigned to the categories related to root development. Several PGPR have been demonstrated to modify root system architecture and modulate root development (Vacheron et al., 2013; Zamioudis et al., 2013; Zhou et al., 2016), which supports our suggestion that GJW24 can play a role as a PGPR. Furthermore, many reports indicate that several PGPR involved in root development and growth contribute to enhanced plant drought stress tolerance (Cohen et al., 2015; Vurukonda et al., 2016; Jochum et al., 2019; Woo et al., 2020; Zia et al., 2021), showing that the GJW24-induced improvement of drought resistance may be associated with its possible role in root system architecture and development for efficient water uptake.
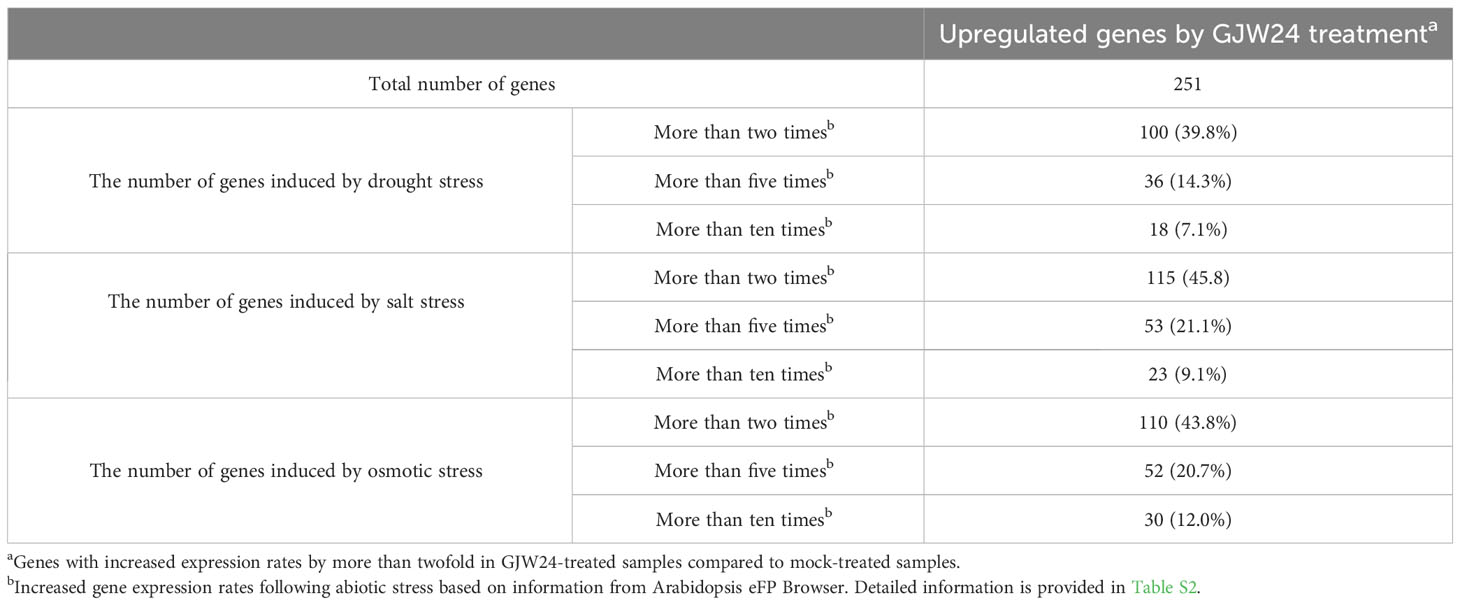
Table 1 Changes in expression patterns of abiotic stress-inducible genes following GJW24 application.
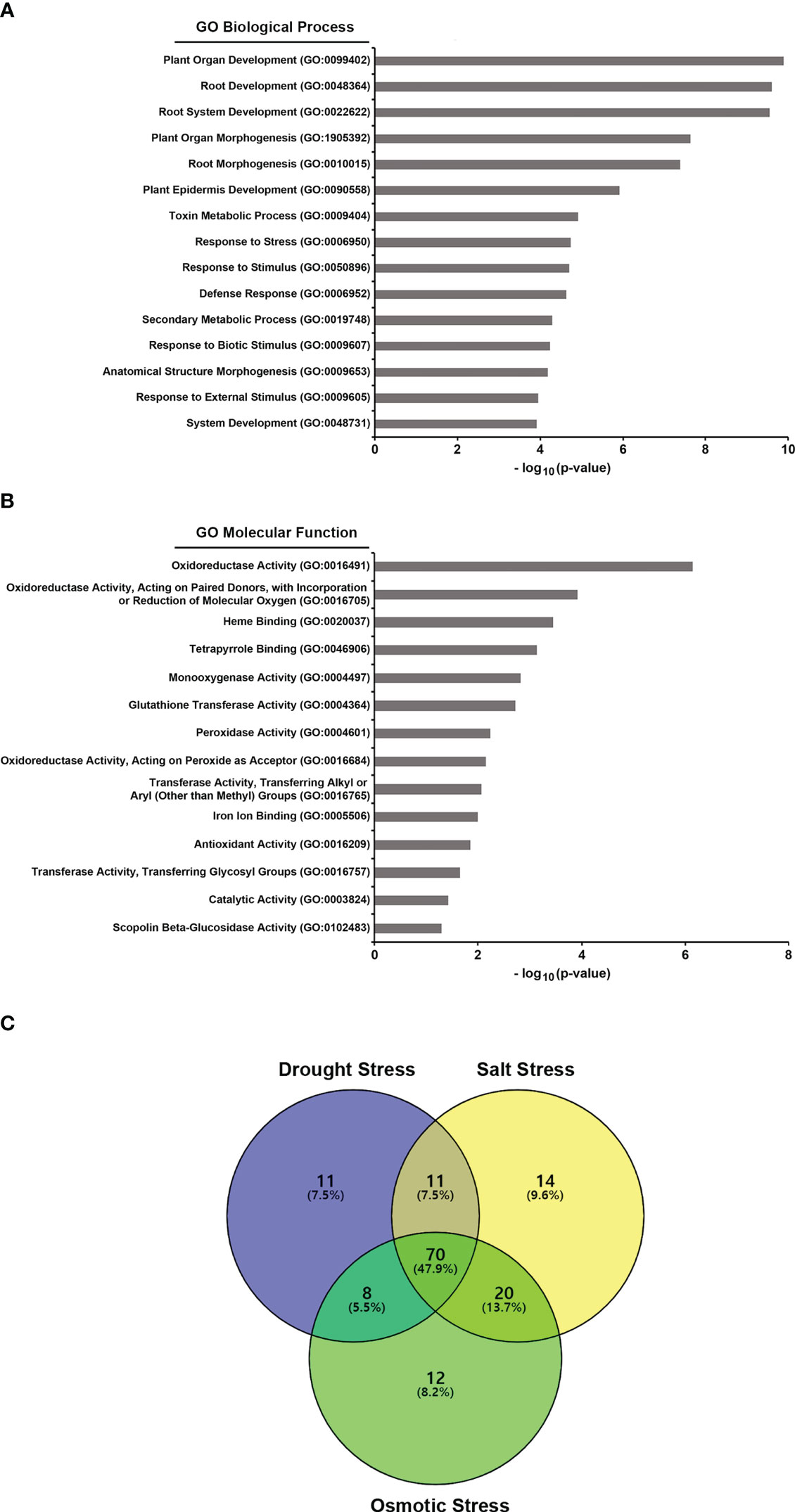
Figure 3 Gene ontology (GO) analysis of the GJW24-upregulated genes and Venn diagram of the genes induced by drought and drought-related stressors among the GJW24-upregulated genes. (A, B) GO analysis of biological processes (A) and molecular functions (B) for GJW24-upregulated genes. GO terms were identified by the functional annotation tool on DAVID. (C) Three-set Venn diagram of the overlap of drought stress-, salt stress- and osmotic stress-inducible genes in the GJW24-upregulated gene expression datasets. Among 251 genes that showed increased gene expression by at least twofold in the GJW24-treated samples, drought stress- (100 genes), salt stress- (115 genes), and osmotic stress-inducible genes (110 genes), which exhibited more than two-fold increases by each stressor (based on Arabidopsis eFP Browser) were retrieved and used generate Venn diagram. Courtesy: Oliveros, J.C. (2007-2015) Venny. An interactive tool for comparing lists with Venn diagrams: http://bioinfogp.cnb.csic.es/tools/venny/index.html.
GJW24-mediated expression pattern of several drought- and drought-related stress-responsive genes in Arabidopsis and Brassica
The GJW24 treatment led to an overall enhancement of the upregulation of drought stress-inducible genes (Table 1). To confirm the list of GJW24-upregulated genes based on the information from the transcriptome sequencing analysis and their drought responsiveness using the Arabidopsis eFP Browser, several genes whose expression was increased by more than two folds by both GJW24 and drought stress were retrieved from Table S2, and their expression patterns were validated using qPCR (Figure 4A). The four selected genes, MYB74, CRK36, SQP2, and YSL7, showed similar GJW24- and drought-inducible patterns to those shown in the transcriptome sequencing analysis and the Arabidopsis eFP Browser database, supporting the reliability of the gene expression profiling results related to both GJW24- and drought-inducible genes (Figure 4A). Furthermore, GJW24 enhanced the upregulation of the selected genes under drought stress (Figure 4A). The expression levels of PERK13, SLAH1, PGL4, YUC9, and ACS4, which were selected from the list of GJW24- and salt-up-regulated genes, were also increased by GJW24 under salt stress (Figure 4B). Next, to determine whether the GJW24-induced drought- and salt-stress resistant phenotypes in Brassica were functionally connected to the expression of the corresponding stress-responsive genes, the expression patterns of drought- or salt-inducible genes in Brassica were examined with or without GJW24 treatment under stress conditions. GJW24 increased the expression of selected drought− (BrEXLB1, BrDREB2A, BrTIFY3a, BraCSD3 and Bra009300) and salt−inducible genes (BrRD29B, Bra034402, BrSR3, BrLAS and Bra005748) in Brassica plants, similar to Arabidopsis (Figures 4C, D) (Yadav et al., 2015; Saha et al., 2016; Li et al., 2018; Yoon et al., 2018; Alam et al., 2019; Jin et al., 2019; Verma et al., 2019; Muthusamy et al., 2020). Collectively, these gene expression results indicate that GJW24 was involved in the upregulation of various drought- and salt-inducible genes in Brassica and Arabidopsis, suggesting that the modulation of drought and salt stress resistance by GJW24 may be achieved, at least in part, by regulating the expression of genes associated with drought and salt resistance in both plant species.
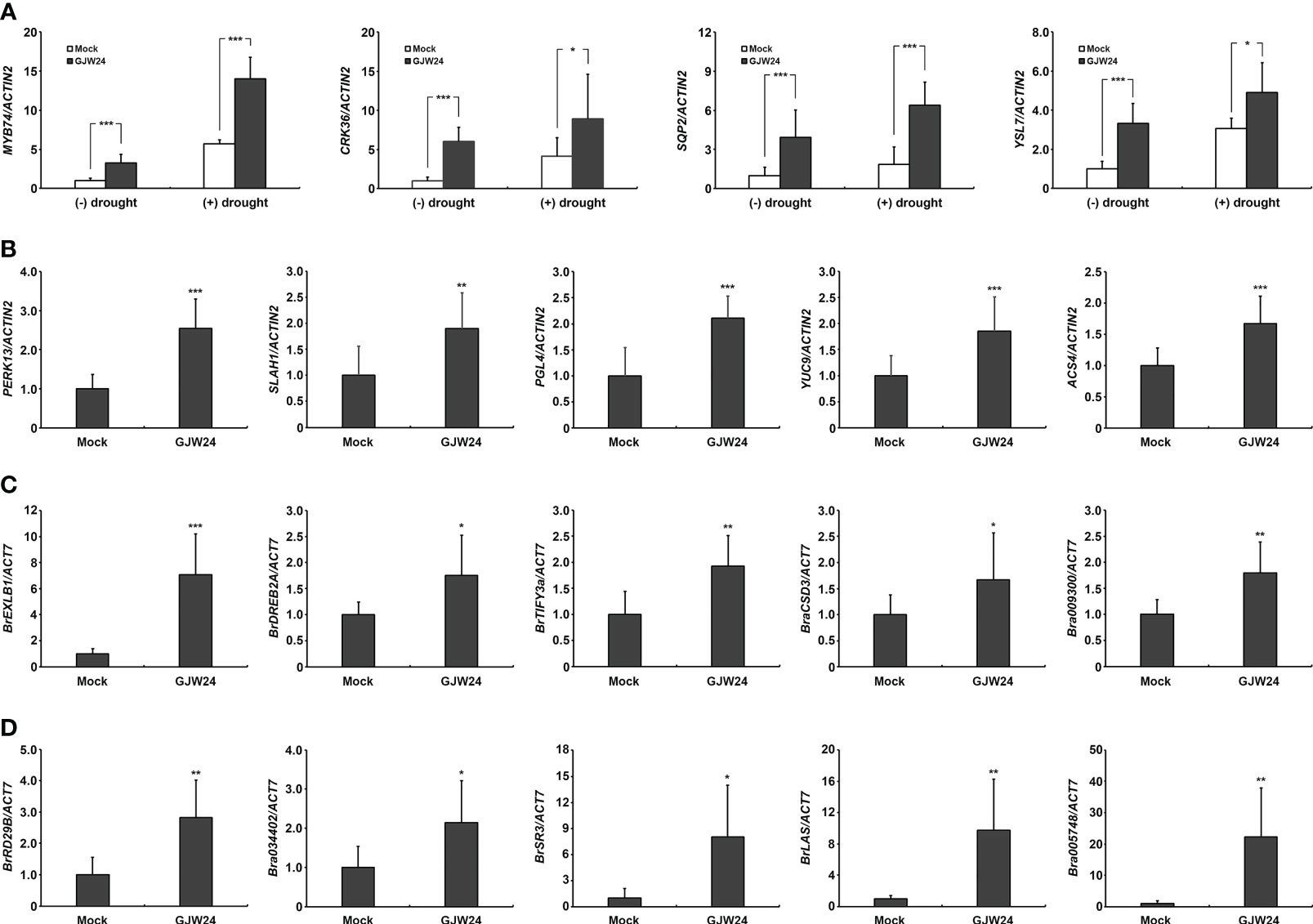
Figure 4 Induction of several drought- and salt-inducible genes in response to GJW24 treatment in Arabidopsis and Brassica. (A) Validation of the GJW24- and drought-upregulated genes from transcriptome sequencing analysis and Arabidopsis eFP Browser. Eight-day-old Arabidopsis seedlings were exposed to mock or GJW24. After 7 d, they were taken out of the pots in which they were growing. After checking the fresh weight, they were placed on filter paper until approximately 20% of their total fresh weight was lost or maintained without dehydration. The average expression value of each gene from mock-treated samples without drought stress was set to 1.0. Values indicate means ± SD (n ≥ 6). A Student’s t-test was conducted to identify statistically significant differences; *P < 0.05; ***P < 0.001. (B) Upregulation of various salt-inducible genes in Arabidopsis by GJW24 treatment under salt stress. Eight-day-old Arabidopsis seedlings were exposed to mock or GJW24. The salt stress resistance assay was conducted by adding 50 mL of 200 mM NaCl solution or water to the soil 7 d after mock or GJW24 treatment, twice at 3-d intervals for 3 (d) The samples were obtained 3 d after the last day of NaCl treatment. The average expression value of each gene from mock-treated samples was set to 1.0. Values indicate means ± SD (n ≥ 6); **P < 0.01; ***P < 0.001. (C) Upregulation of various drought-inducible genes in Brassica by GJW24 treatment under drought stress. Eight-day-old Brassica seedlings were treated with mock or GJW24. After 7 d, they were taken out of the pots in which they were growing. After checking the fresh weight, they were placed on filter paper until approximately 20% of their total fresh weight was lost. Values indicate means ± SD (n ≥ 6); *P < 0.05; **P < 0.01; ***P < 0.001. (D) Upregulation of various salt-inducible genes in Brassica by GJW24 treatment under salt stress. Eight-day-old Brassica seedlings were treated with mock or GJW24. The salt stress resistance assay was conducted by adding 50 mL of water or 300 mM NaCl solution to the soil 7 d after mock or GJW24 treatment, twice at a 3-day interval for 3 (d) The samples were obtained 3 d after the last day of NaCl treatment. Values indicate means ± SD (n ≥ 6); *P < 0.05; **P < 0.01.
Conclusion
Compared to chemical fertilizers or pesticides, which cause environmental pollution, the use of environmentally friendly PGPR may contribute to improved plant growth in a more desirable manner. Here, GJW24 was selected from Gotjawal soil as a useful PGPR that increased the drought resistance of Arabidopsis and Brassica. Although the potential value of GJW24 as a biological agent in agriculture has been demonstrated in this study, the detailed mechanism by which GJW24 enhances plant drought tolerance still requires further investigation. Given that various secondary metabolites and phytohormones (IAA, cytokinin, ABA, and GA) produced by PGPR have been reported to aid the adaptation of plants to abiotic stress conditions (Ma et al., 2020), predicting the list of secondary metabolites and phytohormones that GJW24 can produce from the whole genome sequencing of GJW24 may be useful for a better understanding of the function of GJW24. The finding that several auxin- and ABA-responsive genes are included in the list of GJW24-upregulated Arabidopsis genes increases the possibility that GJW24 may utilize the regulation of plant hormone signaling as a strategy to confer drought tolerance in plants (Table S2).
Data availability statement
The data presented in this study are deposited in the NCBI repository (https://www.ncbi.nlm.nih.gov/), accession number OR272272.
Author contributions
HK: Conceptualization, Data curation, Funding acquisition, Investigation, Validation, Writing – review & editing, Formal Analysis. O-GW: Data curation, Formal Analysis, Investigation, Validation, Writing – review & editing, Writing – original draft. JBK: Data curation, Formal Analysis, Investigation, Validation, Writing – review & editing. S-YY: Data curation, Validation, Writing – review & editing. J-SK: Writing – review & editing, Resources. WJS: Writing – review & editing, Data curation, Formal Analysis. J-YH: Data curation, Formal Analysis, Writing – review & editing. J-HL: Data curation, Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Validation, Writing – original draft.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF- 2020R1A6A3A01100030 to HK and NRF-2021R1F1A1045505 to J-HL), by the LB692 Nebraska Biomedical Research Development Fund (to J-YH), and by the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (916007021HD040 to J-HL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1257137/full#supplementary-material
References
Alam, I., Cui, D. L., Batool, K., Yang, Y. Q., Lu, Y. H. (2019). Comprehensive genomic survey, characterization and expression analysis of the HECT gene family in Brassica rapa L. and Brassica oleracea L. Genes (Basel) 10, 400. doi: 10.3390/genes10050400
Batool, T., Ali, S., Seleiman, M. F., Naveed, N. H., Ali, A., Ahmed, K., et al. (2020). Plant growth promoting rhizobacteria alleviates drought stress in potato in response to suppressive oxidative stress and antioxidant enzymes activities. Sci. Rep. 10, 16975. doi: 10.1038/s41598-020-73489-z
Beneduzi, A., Ambrosini, A., Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Bharti, N., Pandey, S. S., Barnawal, D., Patel, V. K., Kalra, A. (2016). Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6, 34768. doi: 10.1038/srep34768
Bresson, J., Varoquaux, F., Bontpart, T., Touraine, B., Vile, D. (2013). The PGPR strain Phyllobacterium brassicacearum STM196 induces a reproductive delay and physiological changes that result in improved drought tolerance in Arabidopsis. New Phytol. 200, 558–569. doi: 10.1111/nph.12383
Cohen, A. C., Bottini, R., Pontin, M., Berli, F. J., Moreno, D., Boccanlandro, H., et al. (2015). Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant 153, 79–90. doi: 10.1111/ppl.12221
Forni, C., Duca, D., Glick, B. R. (2017). Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 410, 335–356. doi: 10.1007/s11104-016-3007-x
Gontia-Mishra, I., Sapre, S., Sharma, A., Tiwari, S. (2016). Amelioration of drought tolerance in wheat by the interaction of plant growth-promoting rhizobacteria. Plant Biol. (Stuttg) 18, 992–1000. doi: 10.1111/plb.12505
Hussain, H. A., Hussain, S., Khaliq, A., Ashraf, U., Anjum, S. A., Men, S., et al. (2018). Chilling and drought stresses in crop plants: Implications, cross talk, and potential management opportunities. Front. Plant Sci. 9, 393. doi: 10.3389/fpls.2018.00393
Jin, L., Ouyang, N., Huang, Y., Liu, C., Ruan, Y. (2019). Genome-wide analysis of sulfotransferase genes and their responses to abiotic stresses in Chinese cabbage (Brassica rapa L.). PloS One 14, e0221422. doi: 10.1371/journal.pone.0221422
Jochum, M. D., McWilliams, K. L., Borrego, E. J., Kolomiets, M. V., Niu, G., Pierson, E. A., et al. (2019). Bioprospecting plant growth-promoting rhizobacteria that mitigate drought stress in grasses. Front. Microbiol. 10, 2106. doi: 10.3389/fmicb.2019.02106
Khan, N., Bano, A. (2019). Exopolysaccharide producing rhizobacteria and their impact on growth and drought tolerance of wheat grown under rainfed conditions. PLoS One 14, e0222302. doi: 10.1371/journal.pone.0222302
Kim, J. S., Kim, D. S., Lee, K. C., Lee, J. S., King, G. M., Kang, S. (2018). Microbial community structure and functional potential of lava-formed Gotjawal soils in Jeju, Korea. PLoS One 13, e0204761. doi: 10.1371/journal.pone.0204761
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, J. E., Woo, O. G., Bae, Y., Keum, H. L., Chung, S., Sul, W. J., et al. (2020). Enhanced drought and salt stress tolerance in Arabidopsis by Flavobacterium crocinum HYN0056T. J. Plant Biol. 63, 63–71. doi: 10.1007/s12374-020-09236-8
Lagercrantz, U., Putterill, J., Coupland, G., Lydiate, D. (1996). Comparative mapping in Arabidopsis and Brassica, fine scale genome collinearity and congruence of genes controlling flowering time. Plant J. 9, 13–20. doi: 10.1046/j.1365-313X.1996.09010013.x
Lee, S. C., Lim, C. W., Lan, W., He, K., Luan, S. (2013). ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 6, 528–538. doi: 10.1093/mp/sss078
Lephatsi, M. M., Meyer, V., Piater, L. A., Dubery, I. A., Tugizimana, F. (2021). Plant responses to abiotic stresses and rhizobacterial biostimulants: metabolomics and epigenetics perspectives. Metabolites 11, 457. doi: 10.3390/metabo11070457
Li, P., Zhang, B., Su, T., Li, P., Xin, X., Wang, W., et al. (2018). BrLAS, a GRAS transcription factor from Brassica rapa, is involved in drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 1792. doi: 10.3389/fpls.2018.01792
Liu, F., Xing, S., Ma, H., Du, Z., Ma, B. (2013). Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl. Microbiol. Biotechnol. 97, 9155–9164. doi: 10.1007/s00253-013-5193-2
Ma, Y., Dias, M. C., Freitas, H. (2020). Drought and salinity stress responses and microbe-induced tolerance in plants. Front. Plant Sci. 11, 591911. doi: 10.3389/fpls.2020.591911
Muthusamy, M., Kim, J. Y., Yoon, E. K., Kim, J. A., Lee, S. I. (2020). BrEXLB1, a Brassica rapa Expansin-Like B1 gene is associated with root development, drought stress response, and seed germination. Genes (Basel) 11, 404. doi: 10.3390/genes11040404
Naseem, H., Ahsan, M., Shahid, M. A., Khan, N. (2018). Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J. Basic Microbiol. 58, 1009–1022. doi: 10.1002/jobm.201800309
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Saha, G., Park, J. I., Kayum, M. A., Nou, I. S. (2016). A genome-wide analysis reveals stress and hormone responsive patterns of TIFY family genes in Brassica rapa. Front. Plant Sci. 7, 936. doi: 10.3389/fpls.2016.00936
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shinozaki, K., Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Takahashi, F., Kuromori, T., Urano, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (2020). Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant Sci. 11, 556972. doi: 10.3389/fpls.2020.556972
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Uzma, M., Iqbal, A., Hasnain, S. (2022). Drought tolerance induction and growth promotion by indole acetic acid producing Pseudomonas aeruginosa in Vigna radiata. PLoS One 17, e0262932. doi: 10.1371/journal.pone.0262932
Vacheron, J., Desbrosses, G., Bouffaud, M. L., Touraine, B., Moënne-Loccoz, Y., Muller, D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4, 356. doi: 10.3389/fpls.2013.00356
Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., Nasrulhaq Boyce, A. (2016). Role of plant growth promoting rhizobacteria in agricultural sustainability-a review. Molecules 21, 573. doi: 10.3390/molecules21050573
Verma, D., Lakhanpal, N., Singh, K. (2019). Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genomics 20, 227. doi: 10.1186/s12864-019-5593-5
Vurukonda, S. S., Vardharajula, S., Shrivastava, M., SkZ, A. (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184, 13–24. doi: 10.1016/j.micres.2015.12.003
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., Provart, N. J. (2007). An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. doi: 10.1371/journal.pone.0000718
Woo, O. G., Kim, H., Kim, J. S., Keum, H. L., Lee, K. C., Sul, W. J., et al. (2020). Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem. 148, 359–367. doi: 10.1016/j.plaphy.2020.01.032
Yadav, D., Ahmed, I., Kirti, P. B. (2015). Genome-wide identification and expression profiling of annexins in Brassica rapa and their phylogenetic sequence comparison with B. juncea and A. thaliana annexins. Plant Gene 4, 109–124. doi: 10.1016/j.plgene.2015.10.001
Yoon, E. K., Krishnamurthy, P., Kim, J. A., Jeong, M. J., Lee, S. I. (2018). Genome-wide characterization of Brassica rapa genes encoding serine/arginine-rich proteins: expression and alternative splicing events by abiotic stresses. J. Plant Biol. 61, 198–209. doi: 10.1007/s12374-017-0391-6
You, J., Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6, 1092. doi: 10.3389/fpls.2015.01092
Zamioudis, C., Mastranesti, P., Dhonukshe, P., Blilou, I., Pieterse, C. M. (2013). Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 162, 304–318. doi: 10.1104/pp.112.212597
Zhou, C., Ma, Z., Zhu, L., Xiao, X., Xie, Y., Zhu, J., et al. (2016). Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. Int. J. Mol. Sci. 17, 976. doi: 10.3390/ijms17060976
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Keywords: GJW24, Flavobacterium, plant growth-promoting rhizobacteria, drought tolerance, salt stress tolerance
Citation: Kim H, Woo O-G, Kim JB, Yoon S-Y, Kim J-S, Sul WJ, Hwang J-Y and Lee J-H (2023) Flavobacterium sp. strain GJW24 ameliorates drought resistance in Arabidopsis and Brassica. Front. Plant Sci. 14:1257137. doi: 10.3389/fpls.2023.1257137
Received: 12 July 2023; Accepted: 06 September 2023;
Published: 13 October 2023.
Edited by:
Diogo Neves Proença, University of Coimbra, PortugalReviewed by:
Raul Riesco Jarrin, University of Salamanca, SpainSangeeta Paul, Indian Agricultural Research Institute (ICAR), India
Eduardo Valencia-Cantero, Michoacana University of San Nicolás de Hidalgo, Mexico
Copyright © 2023 Kim, Woo, Kim, Yoon, Kim, Sul, Hwang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Hoon Lee, amhsZWU3MkBwdXNhbi5hYy5rcg==
†These authors have contributed equally to this work
 Hani Kim1†
Hani Kim1† Og-Geum Woo
Og-Geum Woo Jong-Shik Kim
Jong-Shik Kim Woo Jun Sul
Woo Jun Sul Jee-Yeon Hwang
Jee-Yeon Hwang Jae-Hoon Lee
Jae-Hoon Lee