
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 07 November 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1256091
 Nitish Dave1
Nitish Dave1 Atif Iqbal1
Atif Iqbal1 Margi Patel2
Margi Patel2 Tarun Kant1*
Tarun Kant1* Virendra Kumar Yadav2*
Virendra Kumar Yadav2* Dipak Kumar Sahoo3*
Dipak Kumar Sahoo3* Ashish Patel1*
Ashish Patel1*Since ancient times, Azadirachta indica, or Neem, has been a well-known species of plant that produces a broad range of bioactive terpenoid chemicals that are involved in a variety of biological functions. Understanding the molecular mechanisms that are responsible for the biosynthesis and control of terpenoid synthesis is majorly dependent on successfully identifying the genes that are involved in their production. This review provides an overview of the recent developments concerning the identification of genes in A. indica that are responsible for the production of terpenoids. Numerous candidate genes encoding enzymes that are involved in the terpenoid biosynthesis pathway have been found through the use of transcriptomic and genomic techniques. These candidate genes include those that are responsible for the precursor synthesis, cyclization, and modification of terpenoid molecules. In addition, cutting-edge omics technologies, such as metabolomics and proteomics, have helped to shed light on the intricate regulatory networks that govern terpenoid biosynthesis. These networks are responsible for the production of terpenoids. The identification and characterization of genes involved in terpenoid biosynthesis in A. indica presents potential opportunities for genetic engineering and metabolic engineering strategies targeted at boosting terpenoid production as well as discovering novel bioactive chemicals.
Neem, or Azadirachta indica, belongs to the family Meliaceae. It is native to India and Burma and has since been introduced to several nations in Africa and North America. A. indica holds significant importance in traditional medicine systems, particularly in India and other parts of South Asia (Kumar and Navaratnam, 2013; Moga et al., 2018; Blum et al., 2019).
Various biological actions have been discovered in A. indica, and it has been investigated for its possible antibacterial, antiviral, antifungal, molluscicidal, and antihyperglycemic characteristics (Ufele et al., 2013; Ezeigwe et al., 2015; S. Abdelhady et al., 2015; Ashfaq et al., 2016; Joy Sinha et al., 2017; Osman Mohamed Ali et al., 2017; Altayb et al., 2022).
Furthermore, the SARS-CoV-2 (COVID-19) pandemic (Kalasariya et al., 2022) has lately posed a challenge to humanity, and different compounds have been investigated in silico to treat the disease. Docking investigations of A. indica molecules have also provided encouraging results for their inhibitory action against various illnesses such as SARS-COV-2, malaria, and dengue (Lavanya et al., 2015; Dwivedi et al., 2016; Khanal et al., 2019; Adegbola et al., 2021; Baildya et al., 2021).
The A. indica has been thoroughly studied for its secondary chemical compounds and for its potential application in the discovery and synthesis of triterpenes, which are among the most abundant and highly complex families of plant-derived natural products.
A predominant focus of research in A. indica is the presence of an important secondary metabolite compound known as azadirachtin, which is a triterpenoid class of limonoids. Azadirachtin, the principal insecticidal component contained in the kernel of Neem seeds, displays high bioactivity against different kinds of insects (Schmutterer, 1995; Schmutterer and Singh, 1995; Hummel et al., 2015).
Growing concerns about the potential negative impacts of chemical pesticides on human health, the environment, and non-target organisms have led to an increasing preference for alternative crop protection methods (Ajiboye et al., 2022). Consequently, there is a greater focus on the development and utilization of plant- or microbe-based biopesticides that are both bioactive and biodegradable. Azadirachtin-based pesticides are environmentally friendly, biodegradable, and non-toxic to wildlife, plants, and birds (Raizada et al., 2001; Kilani-Morakchi et al., 2021). Azadirachtin has shown very minimal toxicity to mammals and has great selectivity for its target organisms (Mordue (Luntz) et al., 2005; Amaral et al., 2019). Azadirachtin is the predominant compound responsible for controlling of insects in agriculture (Vacante and Bonsignore, 2018). Over the past three decades, there has been an increase in the utilization of Neem-based insecticides, primarily attributed to the discovery and isolation of azadirachtin, the key bioactive compound responsible for its insecticidal properties (Chaudhary et al., 2017; Pasquoto-Stigliani et al., 2017). Azadirachtin has been successfully commercialized, and it is still widely accepted as being the most effective botanical pesticide that is in use in agricultural production all around the world (Isman and Grieneisen, 2014; Chaudhary et al., 2017; Aribi et al., 2020).
The biosynthesis route of Neem, which is known to synthesize physiologically and economically relevant triterpenoids with extraordinarily complex carbon skeletons and diverse functional groups, is of great interest among researchers. The first successful synthesis of azadirachtin took 20 years to complete (Jauch, 2008; Veitch et al., 2008) and comprises 71 steps; however, the yield is merely 0.00015%, and thus the production of azadirachtin at the industrial scale is not feasible.
The recent identification and functional characterization of genes involved in the formation of these triterpenoid precursors, which are responsible for the synthesize of limonoids, was made possible by studies in transcriptomics and genomics. Thus, omics research offers a useful technique for examining the biosynthesis of secondary metabolites. While the biochemical constituents of Neem have been widely investigated, its genetic, molecular, and genomic resources are scarce.
The ability of Meliaceae plants to metabolize structurally diverse and physiologically relevant compounds is well established (Lin et al., 2022). The massive amount of literature available across several platforms makes it challenging to find information on each Neem metabolite. Azadirachtin, the most researched, has a challenging chemical structure that belongs to the tetranortriterpenoid class and is present in several forms, the most well-known of which are azadirachtin A, and azadirachtin B ((EFSA) et al., 2018; Fernandes et al., 2019).
The secondary metabolites found in various parts of the tree endow Neem with an array of biological capabilities. Azadirachtin, Azadirone, Gedunin, Nimbin, Salannin, and Vilasinin are some of the major metabolites known to exhibit substantial pesticidal and/or therapeutic properties (Dhar et al., 1998; Isman, 2006; Pravin Kumar et al., 2007; Boursier et al., 2011; Gupta and Diwan, 2017). Out of the several limonoids, azadirachtin accounts for most of its metabolite pool. Table 1 shows the structures and properties of a few secondary metabolites.
According to Verpoorte and Alfermann (2000), there are three main classes of secondary metabolites that may be distinguished from one another based on their biosynthetic pathways. These classes include terpenoids, polyketides, and phenylpropanoids (Verpoorte and Alfermann, 2000). Two major metabolic pathways—mevalonate (MVA) and methylerythritol 4-phosphate/deoxyxylulose 5-phosphate (MEP)—are used by plants for synthesizing terpenoids (Shi et al., 2010).
In higher plants, the traditional mevalonate pathway mostly produces the precursors, which are essential for the synthesis of sesquiterpenes, triterpenes, and sterols in the cytosol and mitochondria, while the hemi-, mono-, sesqui-, and diterpenes are produced by the non-mevalonic acid pathway.
Although the azadirachtin biosynthesis in Neem is not well established, the initial step in triterpenoid biosynthesis involves the cyclization of 2,3-oxidosqualene, which is catalyzed by oxidosqualene cyclase (Figure 1). This cyclization reaction represents the primary diversification level in the biosynthesis of triterpenoids (Abe et al., 1993). In addition to this, tirucallol (C30 Triterpene), a steroid of triterpenoids, is a potential precursor of Neem azadirachtin biosynthesis (Ley et al., 1993; Johnson et al., 1996; Ley et al., 2008).
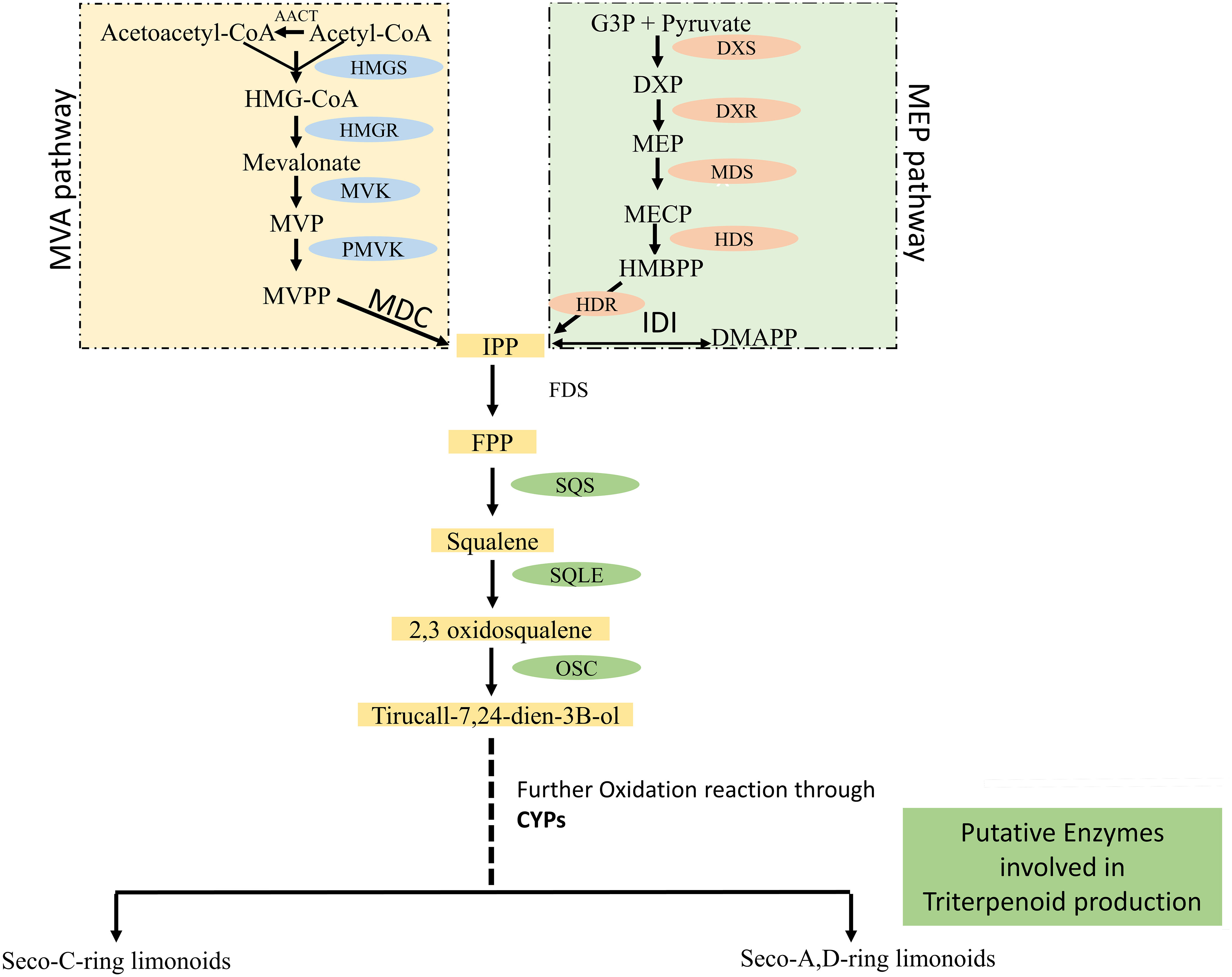
Figure 1 Overview of Triterpenoid Biosynthesis in A. indica. IPP and DMAPP are the fundamental building blocks for isoprenoid synthesis. They can combine in different ways to form larger molecules like FPP (farnesyl diphosphate). FPP can then be converted to squalene via the enzyme SQS (squalene synthase). Squalene undergoes an epoxidation reaction to form 2,3-oxidosqualene, catalyzed by the enzyme SQLE (squalene epoxidase). 2,3-oxidosqualene is then converted to different triterpenoids, such as tirucall-7,24-dien-3β-ol, through the action of the enzyme OSC (oxidosqualene cyclase). Further modifications, including oxidation reactions by various CYP (cytochrome P450) enzymes, can lead to the production of various seco-C-ring and seco-A,D-ring limonoids. (The comprehensive chemical structures for each compound have been provided in Supplementary Table 1).
In A. indica, two different levels of biochemical complexity are assumed to be involved in the production of azadirachtin from tirucallol (Hansen et al., 1994; Ley et al., 2008). Initially, a reduction of four atoms occurs in the lateral chain (Ley et al., 1993; Dewick, 2002), followed by the cyclization of the residual atoms to generate a furan ring. This leads to the formation of limonoids, namely azadirone and azadiradione. Following this, the C-ring undergoes an opening process, leading to the generation of C-seco-limonoids, namely nimbin, and salannin, and the third ring of apotirucallol is oxidized (Ley et al., 1993; Johnson et al., 1996; Puri, 1999; Ley et al., 2008). Additional rearrangements and oxidations are necessary to produce azadirachtin, which is classified as one of the most extensively oxidized triterpenoids (Aerts and Mordue, 1997).
Genomic studies in Neem have focused on sequencing and analyzing the complete set of genes and genomic elements present in the species. The first draught genome was published by a team led by Krishnan (Krishnan et al., 2012). The investigators reported diverse insights from the genome of A. indica. The researchers identified genes such as Terpene Synthase 21 (TPS21), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (lytB), 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (ispH), 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (ispE), Geranylgeranyl diphosphate synthase (GGPS), Farnesyl diphosphate synthase (FDPS), squalene synthase (FDFT1), and Squalene epoxidase (SQLE) that are involved in terpenoid production and are also associated with steroid biosynthesis pathways. These genes were observed to be more abundant in Neem compared to Arabidopsis thaliana, Oryza sativa, Citrus sinensis, and Vitis vinifera. According to their report, it was found that the genome of A. indica is characterized by a high AT content, a low abundance of repetitive DNA sequences, and a mean gene length of 1.69Kb. Additionally, A. indica was observed to be phylogenetically related to Citrus sinensis (Krishnan et al., 2012).
However, in another study, the genome published by Kuravadi and their group reported the presence of about 87 megabases (Mb) of repetitive DNA sequences in the Neem genome, accounting for approximately 33% of the total genome size. This percentage is higher than what was previously reported, suggesting a significant presence of repetitive elements in the Neem genome. Furthermore, the study identified molecular markers such as SSRs (Simple Sequence Repeats), SNPs (Single Nucleotide Polymorphisms), and InDels (insertions and deletions) within the Neem genome. These markers can serve as genetic signposts, allowing researchers to identify and study elite Neem genotypes with desirable traits. The genome was also compared with the citrus genome, which revealed extensive syntenic blocks between Neem and citrus chromosomes, indicating genetic relatedness (Kuravadi et al., 2015).
A recent study conducted by Du and their groups successfully reported the genome of A. indica at a chromosome-scale level. The assembled genome had a size of approximately 281Mb, covering around 73.2% of the estimated total genome size. This achievement of chromosome-scale assembly provides a comprehensive understanding of the Neem genome. Moreover, they reported that the Neem genome exhibited a high level of heterozygosity (0.896%), indicating significant genetic diversity within the species. They also reported that A. indica possesses a higher number of terpene-related gene clusters compared to other species, and chromosome 13 played a central role in the evolution of terpenoid biosynthesis in A. indica. They observed that a lot of genes linked to terpenes were clustered on this chromosome. This shows that chromosome 13 may have gone through certain types of evolution that led to the accumulation and organization of genes related to terpenes in the Neem genome (Du et al., 2022).
According to Du and their team, there are 70 terpene synthase (TPS) genes and 355 cytochrome P450 (CYP) genes that were responsible for terpenoid biosynthesis. The abundance of TPS genes in A. indica was consistent with other terpenoid-rich plant species. Notably, the A. indica TPS and CYP genes were mostly found in the terpene-related groups on chromosome 13, which further suggests that they might be involved in the biosynthesis of azadirachtin (Du et al., 2022). Table 2 shows a comparison of the different genomes of A. Indica.
Transcriptomic studies have been performed to examine gene expression patterns and identify differentially expressed genes in various tissues and under different conditions (Figure 2). RNA-seq technology has been utilized to analyze the transcriptomes of A. indica leaves, flowers, seeds, and other tissues, providing valuable information on gene expression dynamics and regulatory networks.
Krishnan and their group published the first draught genome and Transcriptome from various parts of A. indica. They conducted phylogenetic studies that confirmed the taxonomic closeness between Neem and citrus, which also belong to the same order. Also, Neem was found to be related to Melia species, which is another plant that has terpenoid chemicals. This suggests that these chemicals are made in the Meliaceae family by a similar evolutionary process (Krishnan et al., 2012; Krishnan et al., 2016).
The tissue-specific variation was also identified in triterpenoids (Pandreka et al., 2015). Their findings indicated that the mature seed kernel and pericarp of A. indica during the early stages contained the highest levels of triterpenoids. Furthermore, as compared to other tissues, the kernel contained a diverse range of triterpenoids, particularly C-seco triterpenoids. They identified and functionally characterized the genes which are involved in the initial steps of isoprenoid biosynthesis, such as AiGDS, AiFDS, and AiSQS. They also examined the levels of 15 triterpenoids in various Neem tissues, including flowers, leaves, stem, bark, and different developmental stages of pericarp and kernel. Using solvent partition for extraction and UPLC-ESI(+)-HRMS for analysis, they observed that the concentration of these triterpenoids varied among the tissues. Notably, kernel displayed the highest triterpenoid content. This experimental aspect was aimed at corelates with the omics data with actual triterpenoid levels in various tissues. In another study conducted by (Bhambhani et al., 2017), various developmental stages of the fruit (FS1, FS2, FS3, FS4) and leaves of A. indica were sampled from a five-year-old tree. Upon conducting a phytochemical analysis focused on tetranortriterpenoids, several observations were made. Azadirachtin displayed a fruit-specific accumulation, reaching its peak in the FS3 stage. While nimbin was present in the leaves, it accumulated more significantly in the fruit stages. Notably, both azadirachtin and salannin were absent in the leaf tissue. Furthermore, only a trace amount of nimbin was found in the leaves, underscoring the observation that the fruit, particularly the FS3 stage, is richer in these phytochemicals compared to the leaves (Bhambhani et al., 2017).
An important enzyme was functionally characterized in a significant study by a team led by Hodgson. The researchers characterized the tirucalla-7,24-dien-3β-ol synthase, which is an Oxidosqualene Cyclase (OSCs), from three distinct plant species: A. indica, Melia azedarach, and Citrus sinensis. They also identified three cytochrome P450 (CYP) sequences, namely AiCYP71BQ5, AiCYP72A721, and AiCYP88A108, which showed high co-expression with AiOSC1. The study suggested that certain Cytochrome P450 enzymes (CYPs) could potentially be responsible for oxidizing the tirucalla-7,24-dien-3β-ol scaffold generated by AiOSC1. Furthermore, it was observed that AiOSC1 showed the highest expression in the fruit, aligning with a previous report that highlighted elevated levels of ring-intact limonoids, such as azadiradione and epoxyazadiradione, in the fruit of A. indica (Hodgson et al., 2019). A Group led by Pandreka, also cloned and functionally characterized tirucalla-7,24-dien-3β-ol synthase (AiTTS1), an enzyme responsible for the synthesis of tirucalla-7,24-dien-3β-ol. Additionally, they cloned and characterized squalene epoxidase (AiSQE1), cycloartenol synthase (AiCAS), and two cytochrome P450 reductases. Through comparative tissue expression analysis, the researchers also identified genes involved in terpenoid synthesis and found higher levels of expression for AiFDS (farnesyl diphosphate synthase), AiSQS (squalene synthase), AiSQE3 (squalene epoxidases), and AiTTS1 (triterpene synthases) in the kernel (Pandreka et al., 2021).
In another study, Wang et al. (2020) used a novel hybrid-sequencing approach using Illumina HiSeq and Pacific Biosciences, and they identified five different types of genes potentially involved in azadirachtin biosynthesis. They identified 22 unigenes encoding enzymes, including the oxidosqualene cyclase (OSC), alcohol dehydrogenase (ADH), cytochrome P450 (CYP450), acyltransferase (ACT), and esterase (EST). Table 3 shows the comparisons of different attempts taken by various researchers (Wang et al., 2020).
Omics is a potent tool for identifying essential genes for significant traits, clarifying physiological event mechanisms, and revealing unknown metabolic pathways. A whole genome sequence provides a complete overview of how the functional elements of the genome are structurally organized. These structural elements carry the knowledge of the evolutionary history of an organism (Subramanian et al., 2020).
In addition to genomes, transcriptomes have also been shown to be essential in deciphering the molecular mechanisms and metabolic pathways underpinning a wide range of biological functions. High-throughput sequencing technologies such as RNA sequencing (RNA-seq) are utilized to create extensive transcriptome atlases, and these technologies also contribute to a better knowledge of the functional components that make up the genome of any species (Jiang et al., 2015).
Considering the advantages of omics technology can aid in identifying the unexplored pathways across different species, and the integration of transcriptome data with other omics approaches, such as proteomics and metabolomics, can provide a more comprehensive understanding of the biology of any given species (Yan et al., 2022). By correlating gene expression with protein abundance and metabolite levels, researchers can unravel the complex interactions and regulatory networks underlying physiology and biochemistry.
In the case of A. indica, the biosynthetic pathway for triterpenoid production was not well studied until the publication of the first genome and transcriptome of A. indica. These studies helped in identifying the repeat elements, the nucleotide composition of nucleotides, and expression profiles of initial genes involved in terpenoid production in different tissues of Neem. A. indica was the first Meliaceae family member to be sequenced genome-wide (Krishnan et al., 2012).
The Relative expression of HMGR (HMG-CoA reductase) was higher when compared to the MEP pathway, confirming that the Mevalonate pathway might contributes to the isoprene units of triterpenoids. The distribution of limonoids varies across different tissues reported (Pandreka et al., 2015; Aarthy et al., 2018).
Some of the critical enzyme like oxidosqualene cyclase (AiOSC1) involved in the pathway for the synthesis of triterpenoid was reported by (Hodgson et al., 2019), but they failed to mention triterpene synthase (TTSs), including tirucalla-7, 24-dien-3β-ol synthase, which were functionally characterized by different researchers (Pandreka, 2018; Thulasiram et al., 2018). These Triterpene Synthases (TTSs) work on 2,3-oxidosqualene to create cyclic compounds, which is the crucial step for steroid and triterpenoid biosynthesis (Volkman, 2005). Later, Triterpene synthase (TTS1), was also functionally characterized by a team led by Pandreka (Pandreka et al., 2021).
These clusters of genes mentioned by the various authors from 2012 to 2022 have paved the way for the identification of genes responsible for the production of industrially and medically important triterpenoids in A. indica. The integration of omics, bioinformatics, and genetic engineering technologies holds a great deal of promise for expanding our understanding of the process and locating any missing links in the chain of events that led to the creation of azadirachtin (Kumar et al., 2021). Although there aren’t any well-established genes involved in the numerous processes leading from tirucallol to azadirachtin, utilizing these methodologies can nevertheless yield really helpful insights.
Researchers are able to conduct an exhaustive investigation of the genetic and molecular components that are associated with a pathway by making use of omics tools such as genomics, transcriptomics, and proteomics. Tools and techniques from the field of bioinformatics can be used to assist in the processing and interpretation of huge amounts of biological data, which can in turn facilitate the discovery of candidate genes and probable enzymes involved in this class of triterpenoids.
The discovery of the missing link in the biosynthesis of azadirachtin in A. indica presents a significant difficulty because the biosynthesis of azadirachtin involves a complicated network of processes, with metabolites serving as both substrates and products and gene products acting as enzymes that catalyze the appropriate reactions. Once the underlying molecular mechanisms are understood, new opportunities arise for altering and optimizing the production of azadirachtin, which may result in increased yields or novel applications in the pharma and agriculture industries.
A. indica is classified as a high-value medicinal tree and a great source for azadirachtin, which is known for its insecticidal activity. Along with these molecules, A. indica also synthesizes several other biologically active compounds with demonstrated pharmacological attributes. Despite extensive research on A. indica’s enzymatic pathway machinery, the downstream genes responsible for azadirachtin production have not yet been identified. This knowledge gap highlights the need for in-depth studies that employ an integrated omics approach. By combining various omics technologies, such as genomics, transcriptomics, proteomics, and metabolomics, researchers can gain a comprehensive understanding of the biosynthesis of different alkaloids and their analogs present in A. indica. Identifying and isolating these compounds is crucial for their industrial and pharmacological applications. With the help of omics tools, the biosynthesis pathways of several bioactive substances in A. indica can be mapped out and can provide more useful genetic information about A. indica. The potential of omics research in A. indica appears to be promising, presenting prospects for enhancing our understanding of Neem’s biosynthesis pathways, identifying new bioactive compounds, evaluating genetic variability, expediting functional gene discovery, directing breeding endeavors, and implementing systems biology methodologies. These advancements can have significant implications for the development of Neem-based applications in the medical, agricultural, and industrial sectors.
ND: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing- original draft. AI: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing- original draft. MP: Data curation, Formal analysis, Investigation, Methodology, Software, Writing- review & editing. TK: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Writing- original draft. VY: Conceptualization, Project administration, Supervision, Visualization, Writing- original draft, Writing- review & editing. DS: Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision, Writing- original draft. AP: Conceptualization, Project administration, Supervision, Visualization, Writing- original draft, Writing- review & editing.
The author(s) declare no financial support was received for the research, authorship, and/or publication of this article.
The authors are grateful to Genetics and Tree Improvement Division, Arid Forest Research Institute, Jodhpur, India and Department of Life Sciences, Hemchandracharya North Gujarat University, Patan, Gujarat, India for providing the laboratory facilities to carry out the research work and CAMPA Authority, MoEFCC, New Delhi for financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1256091/full#supplementary-material
Aarthy, T., Mulani, F. A., Pandreka, A., Kumar, A., Nandikol, S. S., Haldar, S., et al. (2018). Tracing the biosynthetic origin of limonoids and their functional groups through stable isotope labeling and inhibition in neem tree (Azadirachta indica) cell suspension. BMC Plant Biol. 18 (1). doi: 10.1186/s12870-018-1447-6
Abe, I., Rohmer, M., Prestwich, G. D. (1993). Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 93, 2189–2206. doi: 10.1021/cr00022a009
Adegbola, P. I., Semire, B., Fadahunsi, O. S., Adegoke, A. E. (2021). Molecular docking and ADMET studies of Allium cepa, Azadirachta indica and Xylopia aethiopica isolates as potential anti-viral drugs for Covid-19. VirusDisease 32 (1), 85–97. doi: 10.1007/s13337-021-00682-7
Aerts, R. J., Mordue, A. J. (1997). Feeding deterrence and toxicity of neem triterpenoids. J of Chem Ecol 23, 2117–2132. doi: 10.1023/B:JOEC.0000006433.14030.04
Ajiboye, T. O., Oladoye, P. O., Olanrewaju, C. A., Akinsola, G. O. (2022). Organophosphorus pesticides: Impacts, detection and removal strategies. Environ. Nanotechnology Monit. Manage. 17, 100655. doi: 10.1016/j.enmm.2022.100655
Akihisa, T., Zhang, J., Manosroi, A., Kikuchi, T., Manosroi, J., Abe, M. (2021). “Chapter 2 - Limonoids and other secondary metabolites of Azadirachta indica (neem) and Azadirachta indica var. siamensis (Siamese neem), and their bioactivities,” in Studies in Natural Products Chemistry, vol. 68 . Ed. Rahman, A.-u. (Amsterdam, The Netherlands: Elsevier), 29–65. doi: 10.1016/B978-0-12-819485-0.00013-X
Altayb, H. N., Yassin, N. F., Hosawi, S., Kazmi, I. (2022). In-vitro and in-silico antibacterial activity of Azadirachta indica (Neem), methanolic extract, and identification of Beta.d-Mannofuranoside as a promising antibacterial agent. BMC Plant Biol. 22 (1). doi: 10.1186/s12870-022-03650-5
Alzohairy, M. A. (2016). Therapeutics role of azadirachta indica (Neem) and their active constituents in diseases prevention and treatment. Evidence-Based Complementary Altern. Med. 2016. doi: 10.1155/2016/7382506
Amaral, K. D., Gandra, L. C., de Oliveira, M. A., de Souza, D. J., Della Lucia, T. M. C. (2019). Effect of azadirachtin on mortality and immune response of leaf-cutting ants. Ecotoxicology 28 (10), 1190–1197. doi: 10.1007/s10646-019-02124-z
Aribi, N., Denis, B., Kilani-Morakchi, S., Joly, D. (2020). Azadirachtin, a natural pesticide with multiple effects. Medecine/Sciences 36 (1), 44–49. doi: 10.1051/medsci/2019268
Ashfaq, U. A., Jalil, A., ul Qamar, M. T. (2016). Antiviral phytochemicals identification from Azadirachta indica leaves against HCV NS3 protease: an in silico approach. Natural Product Res. 30 (16), 1866–1869. doi: 10.1080/14786419.2015.1075527
Baildya, N., Khan, A. A., Ghosh, N. N., Dutta, T., Chattopadhyay, A. P. (2021). Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: An insight from molecular docking and MD-simulation studies. J. Mol. Structure 1227. doi: 10.1016/j.molstruc.2020.129390
Bartelsmeier, I., Kilian, M., ten Broeke, C. J. M., Mertens, D., Dicke, M. (2022). Local and systemic effect of azadirachtin on host choice and feeding activity of Macrosiphum rosae on rose plants. Arthropod-Plant Interact. 16 (2), 191–204. doi: 10.1007/s11829-022-09889-x
Bhambhani, S., Lakhwani, D., Gupta, P., Pandey, A., Dhar, Y. V., Kumar Bag, S., et al. (2017). Transcriptome and metabolite analyses in Azadirachta indica: Identification of genes involved in biosynthesis of bioactive triterpenoids. Sci. Rep. 7 (1). doi: 10.1038/s41598-017-05291-3
Blum, F. C., Singh, J., Merrell, D. S. (2019). In vitro activity of neem (Azadirachta indica) oil extract against Helicobacter pylori. J. Ethnopharmacology 232, 236–243. doi: 10.1016/j.jep.2018.12.025
Boursier, C. M., Bosco, D., Coulibaly, A., Negre, M. (2011). Are traditional neem extract preparations as efficient as a commercial formulation of azadirachtin A? Crop Prot. 30 (3), 318–322. doi: 10.1016/j.cropro.2010.11.022
Brandt, G. E. L., Schmidt, M. D., Prisinzano, T. E., Blagg, B. S. J. (2008). Gedunin, a novel Hsp90 inhibitor: semisynthesis of derivatives and preliminary structure–activity relationships. J. Medicinal Chem. 51 (20), 6495–6502. doi: 10.1021/jm8007486
Chaudhary, S., Kanwar, R. K., Sehgal, A., Cahill, D. M., Barrow, C. J., Sehgal, R., et al. (2017). Progress on Azadirachta indica based biopesticides in replacing synthetic toxic pesticides. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00610
Dhar, R., Zhang, K., Talwar, G. P., Garg, S., Kumar, N. (1998). Inhibition of the growth and development of asexual and sexual stages of drug-sensitive and resistant strains of the human malaria parasite Plasmodium falciparum by Neem (Azadirachta indica) fractions. J. Ethnopharmacology 61 (1), 31–39. doi: 10.1016/S0378-8741(98)00012-9
Drijfhout, F. P., David Morgan, E. (2010). “4.11 - terrestrial natural products as antifeedants,” in Comprehensive Natural Products II. Eds. (Ben) Liu, H.-W., Mander, L. (Netherlands: Elsevier), 457–501. doi: 10.1016/B978-008045382-8.00103-9
Du, Y., Song, W., Yin, Z., Wu, S., Liu, J., Wang, N., et al. (2022). Genomic analysis based on chromosome-level genome assembly reveals an expansion of terpene biosynthesis of Azadirachta indica. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.853861
Dwivedi, V., Tripathi, I., Mishra, S. (2016). In silico evaluation of inhibitory potential of triterpenoids from Azadirachta indica against therapeutic target of dengue virus, NS2B-NS3 protease. J. Vector Borne Dis. 53 (2), 156–161.
(EFSA), E. F. S. A, Arena, M., Auteri, D., Barmaz, S., Brancato, A., Brocca, D., et al. (2018). Peer review of the pesticide risk assessment of the active substance azadirachtin (Margosa extract). EFSA J. (EFSA) Eur. Food Saf. Authority 16 (9), e05234. doi: 10.2903/j.efsa.2018.5234
Ezeigwe, O. C., Ononamadu, C. J., Enemchukwu, B. N., Umeoguaju, U. F., Okoro, J. C. (2015). Antidiabetic and antidiabetogenic properties of the aqueous extracts of Azadirachta indica leaves on alloxan induced diabetic wistar rats. Int. J. Biosci. (IJB) 7 (2), 116–126. doi: 10.12692/ijb/7.2.116-126
Fan, M. X., Chen, G. L., Guo, M. Q. (2022). Potential antioxidative components in azadirachta indica revealed by bio-affinity ultrafiltration with SOD and XOD. Antioxidants 11 (4). doi: 10.3390/antiox11040658
Fernandes, S. R., Barreiros, L., Oliveira, R. F., Cruz, A., Prudêncio, C., Oliveira, A. I., et al. (2019). Chemistry, bioactivities, extraction and analysis of azadirachtin: State-of-the-art. Fitoterapia 134, 141–150. doi: 10.1016/j.fitote.2019.02.006
Garg, H. S., Bhakuni, D. S. (1984). Salannolide, a meliacin from Azadirachta indica. Phytochemistry 23 (10), 2383–2385. doi: 10.1016/S0031-9422(00)80562-2
Gupta, P., Diwan, B. (2017). Bacterial Exopolysaccharide mediated heavy metal removal: A Review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 13, 58–71. doi: 10.1016/j.btre.2016.12.006
Hansen, D. J., Cuomo, J., Khan, M., Gallagher, R. T., Ellenberger, W. P. (1994). Advances in neem and azadirachtin chemistry and bioactivity. ACS Symposium Series. Am. Chem. Soc. 551, 103–129. doi: 10.1021/bk-1994-0551.ch008
Hodgson, H., de la Peña, R., Stephenson, M. J., Thimmappa, R., Vincent, J. L., Sattely, E. S., et al. (2019). Identification of key enzymes responsible for protolimonoid biosynthesis in plants: Opening the door to azadirachtin production. Proc. Natl. Acad. Sci. United States America 116 (34), 17096–17104. doi: 10.1073/pnas.1906083116
Hummel, H. E., Langner, S. S., Hein, D. F., Sanguanpong, U., Schmutterer, H. (2015). Unusually versatile plant genus Azadirachta with many useful and so far incompletely exploited properties for agriculture, medicine and industry. Acta Fytotechnica Zootechnica 18 (Special Issue), 169–175. doi: 10.15414/afz.2015.18.si.169-175
Isman, M. B. (2006). Botanical insecticides, deterrents, and repellents in modern agriculture and an increasingly regulated world. Annu. Rev. Entomology 51 (1), 45–66. doi: 10.1146/annurev.ento.51.110104.151146
Isman, M. B., Grieneisen, M. L. (2014). Botanical insecticide research: Many publications, limited useful data. Trends Plant Sci. 19 (3), 140–145. doi: 10.1016/j.tplants.2013.11.005
Jauch, J. (2008). Total synthesis of azadirachtin - Finally completed after 22 years. Angewandte Chemie - Int. Edition 47 (1), 34–37. doi: 10.1002/anie.200703814
Jiang, Z., Zhou, X., Li, R., Michal, J. J., Zhang, S., Dodson, M. V., et al. (2015). Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell. Mol. Life Sci. 72 (18), 3425–3439. doi: 10.1007/s00018-015-1934-y
Johnson, S., David Morgan, E., Peiris, C. N. (1996). Development of the major triterpenoids and oil in the fruit and seeds of neem (Azadirachta indica). Ann. Bot. Company 78, 383–388. doi: 10.1006/anbo.1996.0133
Joy Sinha, D., Nandha, K. D. S., Jaiswal, N., Vasudeva, A., Prabha Tyagi, S., Pratap Singh, U. (2017). Antibacterial Effect of Azadirachta indica (Neem) or Curcuma longa (Turmeric) against Enterococcus faecalis Compared with That of 5% Sodium Hypochlorite or 2% Chlorhexidine in vitro. Bull. Tokyo Dental Coll. 58 (2), 103–109. doi: 10.2209/tdcpublication.2015-0029
Juin, S. K., Pushpakumar, S., Tyagi, S. C., Sen, U. (2022). Glucosidase inhibitor, Nimbidiol ameliorates renal fibrosis and dysfunction in type-1 diabetes. Sci. Rep. 12 (1), 21707. doi: 10.1038/s41598-022-25848-1
Kalasariya, H. S., Patel, N. B., Gacem, A., Alsufyani, T., Reece, L. M., Yadav, V. K., et al. (2022). Marine alga ulva fasciata-derived molecules for the potential treatment of SARS-CoV-2: an in silico approach. Mar. Drugs 20 (9), 586. doi: 10.3390/md20090586
Kaur, S., Sharma, P., Bains, A., Chawla, P., Sridhar, K., Sharma, M., et al. (2022). Antimicrobial and anti-inflammatory activity of low-energy assisted nanohydrogel of Azadirachta indica oil. Gels 8 (7). doi: 10.3390/gels8070434
Khanal, P., Mandar, B. K., Magadum, P., Patil, B. M., Hullatti, K. K. (2019). In silico Docking Study of Limonoids from Azadirachta indica with pfpk5: A Novel Target for Plasmodium falciparum. Indian J. Pharm. Sci. 81 (2), 326–332. doi: 10.36468/pharmaceutical-sciences.514
Kilani-Morakchi, S., Morakchi-Goudjil, H., Sifi, K. (2021). Azadirachtin-based insecticide: overview, risk assessments, and future directions. Front. Agron. 3. doi: 10.3389/fagro.2021.676208
Koul, O., Singh, G., Singh, R., Singh, J., Daniewski, W. M., Berlozecki, S. (2004). Bioefficacy and mode-of-action of some limonoids of salannin group fromAzadirachta indica A. Juss and their role in a multicomponent system against lepidopteran larvae. J. Biosci. 29 (4), 409–416. doi: 10.1007/BF02712112
Krishnan, N. M., Jain, P., Gupta, S., Hariharan, A. K., Panda, B. (2016). An improved genome assembly of Azadirachta indica A. Juss. G3: Genes Genomes Genet. 6 (7), 1835–1840. doi: 10.1534/g3.116.030056
Krishnan, N. M., Pattnaik, S., Jain, P., Gaur, P., Choudhary, R., Vaidyanathan, S., et al. (2012). A draft of the genome and four transcriptomes of a medicinal and pesticidal angiosperm Azadirachta indica. BMC Genomics 13 (1). doi: 10.1186/1471-2164-13-464
Kumar, R., Mehta, S., Pathak, S. R. (2018). “Chapter 4 - Bioactive constituents of neem,” in Synthesis of Medicinal Agents from Plants. Eds. Tewari, A., Tiwari, S. (Netherlands: Elsevier), 75–103. doi: 10.1016/B978-0-08-102071-5.00004-0
Kumar, V. S., Navaratnam, V. (2013). Neem (Azadirachta indica): Prehistory to contemporary medicinal uses to humankind. Asian Pacific J. Trop. Biomedicine 3 (7), 505–514. doi: 10.1016/S2221-1691(13)60105-7
Kumar, R., Sharma, V., Suresh, S., Ramrao, D. P., Veershetty, A., Kumar, S., et al. (2021). Understanding Omics Driven Plant Improvement and de novo Crop Domestication: Some Examples. Front. Genet. 12. doi: 10.3389/fgene.2021.637141
Kuravadi, N. A., Yenagi, V., Rangiah, K., Mahesh, H. B., Rajamani, A., Shirke, M. D., et al. (2015). Comprehensive analyses of genomes, transcriptomes and metabolites of neem tree. PeerJ 2015 (8). doi: 10.7717/peerj.1066
Lavanya, P., Ramaiah, S., Anbarasu, A. (2015). Computational analysis reveal inhibitory action of nimbin against dengue viral envelope protein. VirusDisease 26 (4), 243–254. doi: 10.1007/s13337-015-0280-x
Ley, S. V., Abad-Somovilla, A., Anderson, J. C., Ayats, C., Bänteli, R., Beckmann, E., et al. (2008). The synthesis of azadirachtin: A potent insect antifeedant. Chem. – A Eur. J. 14 (34), 10683–10704. doi: 10.1002/chem.200801103
Ley, S. V., Denholm, A. A., Wood, A. (1993). The chemistry of azadirachtin. Natural Product Rep. 10 (2), 109–157. doi: 10.1039/NP9931000109
Lin, M., Bi, X., Zhou, L., Huang, J. (2022). Insecticidal triterpenes in meliaceae: plant species, molecules, and activities: part II (Cipadessa, melia). Int. J. Mol. Sci. 23 (10). doi: 10.3390/ijms23105329
Moga, M. A., Bălan, A., Anastasiu, C. V., Dimienescu, O. G., Neculoiu, C. D., Gavriş, C. (2018). An overview on the anticancer activity of azadirachta indica (Neem) in gynecological cancers. Int. J. Mol. Sci. 19 (12). doi: 10.3390/ijms19123898
Mordue (Luntz), A. J., Morgan, E. D., Nisbet, A. J. (2005). “6.4 - azadirachtin, a natural product in insect control,” in Comprehensive Molecular Insect Science. Ed. Gilbert, L. I. (Netherlands: Elsevier), 117–135. doi: 10.1016/B0-44-451924-6/00077-6
Osman Mohamed Ali, E., Shakil, N. A., Rana, V. S., Sarkar, D. J., Majumder, S., Kaushik, P., et al. (2017). Antifungal activity of nano emulsions of neem and citronella oils against phytopathogenic fungi, Rhizoctonia solani and Sclerotium rolfsii. Ind. Crops Products 108, 379–387. doi: 10.1016/j.indcrop.2017.06.061
Pandreka, A. (2018). De novo Sequencing and Analysis of Transcriptome from Azadirachta indica to Characterize the Genes Involved in Limonoid Biosynthesis (India: CSIR).
Pandreka, A., Chaya, P. S., Kumar, A., Aarthy, T., Mulani, F. A., Bhagyashree, D. D., et al. (2021). Limonoid biosynthesis 3: Functional characterization of crucial genes involved in neem limonoid biosynthesis. Phytochemistry 184 (34). doi: 10.1016/j.phytochem.2021.112669
Pandreka, A., Dandekar, D. S., Haldar, S., Uttara, V., Vijayshree, S. G., Mulani, F. A., et al. (2015). Triterpenoid profiling and functional characterization of the initial genes involved in isoprenoid biosynthesis in neem (Azadirachta indica). BMC Plant Biol. 15 (1). doi: 10.1186/s12870-015-0593-3
Pasquoto-Stigliani, T., Campos, E. V. R., Oliveira, J. L., Silva, C. M. G., Bilesky-José, N., Guilger, M., et al. (2017). Nanocapsules containing neem (Azadirachta indica) oil: development, characterization, and toxicity evaluation. Sci. Rep. 7 (1). doi: 10.1038/s41598-017-06092-4
Ponnusamy, S., Haldar, S., Mulani, F., Zinjarde, S., Thulasiram, H., RaviKumar, A. (2015). Gedunin and azadiradione: Human pancreatic alpha-amylase inhibiting limonoids from neem (Azadirachta indica) as anti-diabetic agents. PloS One 10 (10). doi: 10.1371/journal.pone.0140113
Pravin Kumar, R., Manoj, M. N., Kush, A., Annadurai, R. S. (2007). In silico approach of azadirachtin binding with actins. Insect Biochem. Mol. Biol. 37 (6), 635–640. doi: 10.1016/j.ibmb.2007.03.010
Raizada, R. B., Srivastava, M. K., Kaushal, R. A., Singh, R. P. (2001). Azadirachtin, a neem biopesticide: subchronic toxicity assessment in rats. Food Chem. Toxicol. 39 (5), 477–483. doi: 10.1016/S0278-6915(00)00153-8
Rao, P. S., Subramanayam, G., Sridhar, P. R. (2019). Quercetin 3-galactoside from Azadirachta indica. J. Adv. Mol. Biol. 3 (1). doi: 10.22606/jamb.2019.31001
S. Abdelhady, M. I., Bader, A., Shaheen, U., El-Malah, Y., Abourehab, M. A. S., Barghash, M. F. (2015). Azadirachta indica as a source for antioxidant and cytotoxic polyphenolic compounds. Biosci. Biotechnol. Res. Asia 12 (2), 1209–1222. doi: 10.13005/bbra/1774
Sarkar, L., Oko, L., Gupta, S., Bubak, A. N., Das, B., Gupta, P., et al. (2022). Azadirachta indica A. Juss bark extract and its Nimbin isomers restrict β-coronaviral infection and replication. Virology 569, 13–28. doi: 10.1016/j.virol.2022.01.002
Sarkar, S., Singh, R. P., Bhattacharya, G. (2021). Exploring the role of Azadirachta indica (neem) and its active compounds in the regulation of biological pathways: an update on molecular approach. 3 Biotech. 11 (4), 178. doi: 10.1007/s13205-021-02745-4
Schmutterer, H. (1995). The neem tree: Azadirachta indica A. Juss. and other meliaceous plants: sources of unique natural products for integrated pest management, medicine, industry, and other purposes (Weinheim (Germany): VCH).
Schmutterer, H., Singh, R. P. (1995). “Effects on viruses and organisms: sections 3.9.3 - 3.10,” in The neem tree. (New Jersey, USA: WILEY). 326–366. doi: 10.1002/3527603980.ch3i
Shi, L., Ren, A., Mu, D., Zhao, M. (2010). Current progress in the study on biosynthesis and regulation of ganoderic acids. Appl. Microbiol. Biotechnol. 88 (6), 1243–1251. doi: 10.1007/s00253-010-2871-1
Shrirangasami, S., Murugaragavan, R., Rakesh, S., Ramesh, P. (2020). Chemistry behind in neem (Azadirachta indica) as medicinal value to living forms-A review. J. Pharmacognosy Phytochem. 9 (6), 467–469. doi: 10.22271/phyto.2020.v9.i6g.12936
Subramanian, I., Verma, S., Kumar, S., Jere, A., Anamika, K. (2020). Multi-omics data integration, interpretation, and its application. Bioinf. Biol. Insights 14, 1177932219899051. doi: 10.1177/1177932219899051
Thulasiram, H. V., Pandreka, A., Patil, C. S. (2018). cDNA sequences encoding enzymes of limonoid biosynthesis, co-expression of said enzymes and mutant of the same (Patent No. 201811047388IN) (India: Indian Patent).
Ufele, A. N., Nnajidenma, U. P., Ebenebe, C. I., Mogbo, T. C., Aziagba, B. O., Akunne, C. E. (2013). The effect of Azadirachta indica (Neem) leaf extract on longevity of snails (Achatina achatina). Int. Res. J. Biol. Sci. 2 (1), 61–63.
Vacante, V., Bonsignore, C. P. (2018). “Natural enemies and pest control,” in Handbook of pest management in organic farming (UK: CABI Publishing), 60–77. doi: 10.1079/9781780644998.0060
Veitch, G. E., Boyer, A., Ley, S. V. (2008). “The azadirachtin story,” in Angewandte Chemie - International Edition (New Jersey, USA: WILEY), vol. 47. , 9402–9429. doi: 10.1002/anie.200802675
Verpoorte, R., Alfermann, A. W. (2000). Metabolic engineering of plant secondary metabolism (Netherlands: Kluwer Academic Publishers).
Volkman, J. K. (2005). Sterols and other triterpenoids: Source specificity and evolution of biosynthetic pathways. Organic Geochemistry 36 (2), 139–159. doi: 10.1016/j.orggeochem.2004.06.013
Wang, H., Wang, N., Huo, Y. (2020). Multi-tissue transcriptome analysis using hybrid-sequencing reveals potential genes and biological pathways associated with azadirachtin A biosynthesis in neem (azadirachta indica). BMC Genomics 21 (1). doi: 10.1186/s12864-020-07124-6
Wang, S., Zhang, H., Li, X., Zhang, J. (2016). Gene expression profiling analysis reveals a crucial gene regulating metabolism in adventitious roots of neem (Azadirachta indica). RSC Adv. 6 (115), 114889–114898. doi: 10.1039/C6RA20494E
Wylie, M. R., Merrell, D. S. (2022). The antimicrobial potential of the neem tree Azadirachta indica. Front. Pharmacol. 13. doi: 10.3389/fphar.2022.891535
Yan, S., Bhawal, R., Yin, Z., Thannhauser, T. W., Zhang, S. (2022). Recent advances in proteomics and metabolomics in plants. Mol. Horticulture 2 (1), 17. doi: 10.1186/s43897-022-00038-9
Keywords: terpenoid biosynthesis, transcriptomics, proteomics, genetic engineering, metabolic engineering
Citation: Dave N, Iqbal A, Patel M, Kant T, Yadav VK, Sahoo DK and Patel A (2023) Deciphering the key pathway for triterpenoid biosynthesis in Azadirachta indica A. Juss.: a comprehensive review of omics studies in nature’s pharmacy. Front. Plant Sci. 14:1256091. doi: 10.3389/fpls.2023.1256091
Received: 10 July 2023; Accepted: 25 October 2023;
Published: 07 November 2023.
Edited by:
Peng Wang, Jiangsu Province and Chinese Academy of Sciences, ChinaReviewed by:
Nan Lu, Chinese Academy of Forestry, ChinaCopyright © 2023 Dave, Iqbal, Patel, Kant, Yadav, Sahoo and Patel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tarun Kant, dGFydW5rYW50QGljZnJlLm9yZw==; Virendra Kumar Yadav, eWFkYXZhOTRAZ21haWwuY29t; Dipak Kumar Sahoo, ZHNhaG9vQGlhc3RhdGUuZWR1; Ashish Patel, dW5pLmFzaGlzaEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.