
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 December 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1255805
This article is part of the Research Topic Advances in Environmental Stress Biology and Important Agronomic Traits Improvement in Non-staple Crops View all 11 articles
 Yaya Hu1†
Yaya Hu1† Zhuoru Dai2†
Zhuoru Dai2† Jinan Huang1†
Jinan Huang1† Meikun Han1
Meikun Han1 Zhiwei Wang3
Zhiwei Wang3 Weijing Jiao1
Weijing Jiao1 Zhiyuan Gao1
Zhiyuan Gao1 Xinliang Liu4*
Xinliang Liu4* Lanfu Liu1*
Lanfu Liu1* Zhimin Ma1*
Zhimin Ma1*Plant glutamate receptor (GLR) homologs are crucial calcium channels that play an important role in plant development, signal transduction, and response to biotic and abiotic stresses. However, the GLR gene family has not yet been thoroughly and systematically studied in sweet potato. In this study, a total of 37 GLR genes were identified in the cultivated hexaploid sweet potato (Ipomoea batatas), and 32 GLR genes were discovered in each of the two diploid relatives (Ipomoea trifida and Ipomoea triloba) for the first time. Based on their evolutionary relationships to those of Arabidopsis, these GLRs were split into five subgroups. We then conducted comprehensive analysis to explore their physiological properties, protein interaction networks, promoter cis-elements, chromosomal placement, gene structure, and expression patterns. The results indicate that the homologous GLRs of the cultivated hexaploid sweet potato and its two relatives are different. These variations are reflected in their functions related to plant growth, hormonal crosstalk, development of tuberous roots, resistance to root rot, and responses to abiotic stress factors, all of which are governed by specific individual GLR genes. This study offers a comprehensive analysis of GLR genes in sweet potato and its two diploid relatives. It also provides a theoretical basis for future research into their regulatory mechanisms, significantly influencing the field of molecular breeding in sweet potatoes.
Glutamate, a ubiquitous amino acid, participates in various important chemical reactions in animals, plants, and microorganisms and plays an indispensable function in protein metabolism and signal transduction processes (Forde and Lea, 2007). Glutamate receptors (GLRs), including the ionotropic and metabotropic GLRs, were first discovered in animals. The ionotropic GLR of animal is a ligand-gated non-selective cation channel that regulates excitatory nerve signal transmission and guides neuronal development (Mayer, 2006; Zhu and Gouaux, 2017). Due to the existence of conserved signature domains, plant GLR structures are similar to those for animal ionotropic GLRs (Wudick et al., 2018a). Plant GLRs composed of receptor domains and four transmembrane helical domains are typical membrane proteins belonging to a class of amino acid-gated Ca2+ channels. When GLRs are activated by their corresponding ligands, they can mediate the transmembrane influx of Ca2+, thus activating Ca2+ signaling to regulate plant responses to stress and simultaneously impact their overall development and growth (Sobolevsky et al., 2009; Traynelis et al., 2010; Kong et al., 2015; Tian et al., 2020; Luan and Wang, 2021; Ahmed et al., 2023).
Plant GLRs have been studied for more than 25 years, during which significant progress has been made in understanding their structural and functional characteristics. At present, 20, 13, 24, 34, 29, 35, 36, 16, 43, 16, and 21 GLRs, have been identified in Arabidopsis thaliana (Chiu et al., 2002), Solanum lycopersicum (Aouini et al., 2012), Oryza sativa (Singh et al., 2014), Pyrus bretschneideri (Chen et al., 2016), Medicago truncatula (Philippe et al., 2019), Glycine max (Zeng et al., 2020), Gossypium hirsutum (Liu et al., 2021), Zea mays (Zhou et al., 2021), Saccharum (Zhang et al., 2022), Brassica rapa (Yang et al., 2022), and Erigeron breviscapus (Yan et al., 2022), respectively. Furthermore, genetic research has demonstrated that GLRs play a crucial role in regulating various plant developmental processes and responding to environmental stresses such as salt, drought, heat, wounding, and pathogen attacks. These developmental processes encompass seed germination (Kong et al., 2015), root development (Vincill et al., 2013; Singh et al., 2016), hypocotyl elongation and pollen tube growth (Michard et al., 2011; Wudick et al., 2018b), xylem growth (Chen et al., 2016), Ca2+ distribution (Kim et al., 2001), stomatal closure (Cho et al., 2009), nitrogen and carbon metabolism (Kang et al., 2004), and abscisic acid (ABA) synthesis. Tea transcriptome data showed that the homologs for GLR2.7 and GLR2.8 manifest an upregulation in their expression levels under salt stress (Wan et al., 2018). Following coldstress, AtGLR1.2 and AtGLR1.3 activate core binding factors/dehydration-responsive element binding protein 1 signaling pathway through endogenous jasmonic acid (JA) accumulation, which contributes towards enhancing cold tolerance (Hu et al., 2013; Zheng et al., 2018). Moreover, OsGLR3.4 is involved in brassinosteroid-mediated damage response from root to stem in rice. (Yu et al., 2022). GLRs can also improve the regeneration of plants after wounding and are crucial for establishing a balance between plant defense and regeneration following injury (Bian et al., 2022; Hernández-Coronado et al., 2022). As Ca2+ channels, plant GLRs can participate in disease-resistance responses by regulating Ca2+ signals. Kang et al. cloned an RsGLuR gene located on the plasma membrane from small radish, and found that overexpression of RsGLuR in Arabidopsis can improve resistance to the pathogen Botrytis cinema by triggering JA biosynthesis (Kang et al., 2006). However, there is no substantial volume of research on GLRs in sweet potato, in terms of their regulatory mechanisms and biological functions.
Sweet potato, Ipomoea batatas (L.) Lam. (2n = 6x = 90), is characterized by drought resistance, high and stable yields, strong adaptability, and rich nutrition, and is the best food recommended by the World Health Organization (Ma et al., 2012). It is both a food and cash crop with a wide range of uses. Specifically, it can be used as fresh food (Xie et al., 2018), starch processing (Zhou et al., 2020), food processing (Xiang et al., 2020), leafy vegetables (Su et al., 2018), and ornamental purposes (Meng and Lai, 2019). Accordingly, sweet potato has become an essential feed, food, and industrial raw material, and it is widely cultivated in over 100 regions and countries across the globe (Food and Agriculture Organization of the United Nations, 2021). The sweet potato genome is large and complex owing to its hexaploidy and high heterozygosity. In recent years, the assembly and reporting of the genomes for hexaploid sweet potato (Taizhong6) and its two diploid relatives (Ipomoea trifida, NCNSP0306, 2n=2x=30 and Ipomoea triloba, NCNSP0323, 2n=2x=30) have made it possible to analyze and identify the important gene family in sweet potato across the whole-genome level (Yang et al., 2017; Wu et al., 2018).
The cultivated hexaploid sweet potato and two of its diploid relatives (I. trifida and I. triloba) were used to screen and identify the GLR gene members in this study. The GLR genes were then analyzed for phylogenetic relationships, protein physicochemical properties, chromosomal localizations, gene structures, promoter cis-elements, protein interaction network, and expression patterns. The findings provide a basis for further understanding of the biological functions of GLRs and the future molecular breeding of sweet potatoes.
The whole-genome sequences for I. batatas, I. trifida, and I. triloba were obtained through Ipomoea Genome Hub (https://ipomoea-genome.org/) (viewed: 6 March 2023) and Sweetpotato Genomics Resource (http://sweetpotato.plantbiology.msu.edu/) (viewed: 6 March 2023). Three different screening techniques (Dai et al., 2022) were used at once to ensure that all members of GLR family were accurately identified.
The molecular weight, hydrophilicity, instability index, and theoretical isoelectric point for GLRs were computed through the Expert Protein Analysis System (ExPASy, https://www.expasy.org/) (viewed: 12 March 2023). Protein Subcellular Localization Prediction (PSORT, https://wolfpsort.hgc.jp/) (viewed: 12 March 2023) was employed to predict the subcellular localizations for GLRs.
IbGLRs, ItfGLRs, and ItbGLRs were separately mapped to the respective chromosomes for I. batatas, I. trifida, and I. triloba by Ipomoea Genome Hub (https://ipomoea-genome.org/) (viewed: 14 March 2023) and Sweetpotato Genomics Resource (http://sweetpotato.plantbiology.msu.edu/) (viewed: 14 March 2023). The Toolkit for Biologists integrating various biological data handling tools (TBtools) program (South China Agricultural University, Guangzhou, China) was used for visualization.
MEGA ClustalW 7.0 was used to phylogenetically analyze the amino acid sequences for GLRs of Arabidopsis, I. batatas, I. trifida, and I. triloba (Kumar et al., 2016). Bootstrapping was carried out with 1000 replicates, and Interactive Tree of Life (iTOL) software (http://itol.embl.de/) (viewed: 8 June 2023) was used to construct a phylogenetic tree.
To perform an in-depth study of GLRs’ conserved motifs, the Multiple Expectation Maximizations for Motif Elicitation (MEME) program (https://meme-suite.org/meme/) (viewed: 15 March 2023) was used. The maximum number of motifs that could be used has been set at five.
A database of Plant Cis-Acting Regulatory Element (PlantCARE, (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (viewed: 16 March 2023) predicted cis-elements within 1500 bp promoter region for GLRs (Lescot et al., 2002). Gene Structure Display Server (GSDS) 2.0 developed by Peking University, Beijing, China (http://gsds.gao-lab.org/) (viewed: 16 March 2023) and the TBtools software developed by South China Agricultural University, Guangzhou, China was used to obtain and visualize the exon-intron structures for GLRs, respectively.
Depending upon Arabidopsis orthologous proteins, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://www.string-db.org/) (viewed: 18 March 2023) predicted GLR protein interaction network. The Cytoscape version 3.2 was used to construct the network map (Kohl et al., 2011).
Transcriptome analysis and real-time quantitative PCR (qRT-PCR) was carried out at duration points (0 h, 36 h, 72 h, 120 h, and 10 d) following root rot induction using the underground stem of resistant variety Jishuzi203 and susceptible variety Jishuzi563. These plants were provided by the Institute of Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences (Shijiazhuang, China) and planted in the natural root rot disease nursery in Xiong'an New Area, China. qRT-PCR was conducted on a CFX Opus 384 Real-Time PCR system (Bio-Rad, USA) by the ChamQ Universal SYBR qPCR Master Mix (Vazyme, China). The relative expression level of the target gene was presented as fold change compared with the internal control using the 2 - Δct method, and data were analyzed with Duncan’s multiple range test (p < 0.05). Three biological replications were performed for each test. A gene of I. batatas ADP-ribosylation factor (ARF, GenBank JX177359) was used as an internal control. The specific primers used for the qRT-PCR analysis were listed in Supplementary Table S2. Based on related investigations (Zhang et al., 2017; Dong et al., 2019; Zhu et al., 2019) ribonucleic nucleic acid sequencing (RNA-seq) data for IbGLRs were collected. RNA-seq data for ItfGLRs and ItbGLRs were collected through Sweetpotato Genomics Resource (http://sweetpotato.plantbiology.msu.edu/) (viewed: 22 March 2023). The fragments per kilobase of exon per million mapped fragments (FPKM) method was used to calculate the GLRs expression levels. Tbtools was used to build the heat maps.
37 GLRs for sweet potato (named Ib) and 32 for each of two diploid relatives (named Itf and Itb) were identified through a combination of three methods used by Dai et al. (2022). The sequences from I. batatas were used for the analysis of the physicochemical features of GLRs (Table 1). The coding sequence length of IbGLRs varied from 1977 bp (IbGLR7) to 7326 bp (IbGLR36). The molecular weights of IbGLRs ranged from 73.627 to 270.105 kDa, the amino acid lengths extended from 658 to 2441 amino acids, and their isoelectric points varied from 5.49 (IbGLR32) to 9.31 (IbGLR30). Most IbGLRs were stable, only IbGLR2/10/11/13/14/15/21 and IbGLR30 were unstable (instability index > 40). 17 hydrophobic proteins (the grand average of hydropathy [GRAVY] score > 0) and 20 hydrophilic proteins (GRAVY score < 0) were identified for the IbGLR family. Subcellular localization prediction assessment demonstrated that except for IbGLR1 (located on chloroplast) and IbGLR12 (located on chloroplast and nucleus), the remaining IbGLRs were located on the plasma membrane.
Based on the physical locations of genes in the I. batatas, I. trifida, and I. triloba genomes, the chromosomal positions of GLRs in these crops were mapped. In I. batatas, IbGLRs were distributed unevenly on nine chromosomes. LG13 had the most IbGLR genes, with 11, followed by LG7 with seven. LG3, LG9, and LG12 had four IbGLRs. There were less than four IbGLRs on LG1, LG5, LG8, and LG15 (Figure 1A). In I. trifida, except for ItfGLR31 and ItfGLR32 located on Chr00 (not shown in Figure 1B), the other ItfGLRs were located on the same chromosomes (Chr02, Chr03, Chr05, Chr06, Chr07, Chr10, Chr11, Chr12, and Chr14) compared to those in I. triloba. Chr02 had the most ItfGLR genes, with 12, followed by Chr14 with four. There were less than four ItfGLRs on Chr03, Chr05, Chr06, Chr07, Chr10, Chr11, and Chr12 (Figure 1B). In I. triloba, Chr02 had the most ItbGLR genes, with 11, followed by Chr03 with six and Chr14 with five. There were less than four ItbGLRs on Chr05, Chr06 Chr07, Chr10, Chr11, and Chr12 (Figure 1C). Chr06, Chr10, Chr11, and Chr12 had the same numbers of GLRs in I. trifida and I. triloba. These findings imply that two diploid relatives differed in the proportion and distribution of GLRs on chromosomes in comparison to sweet potato.
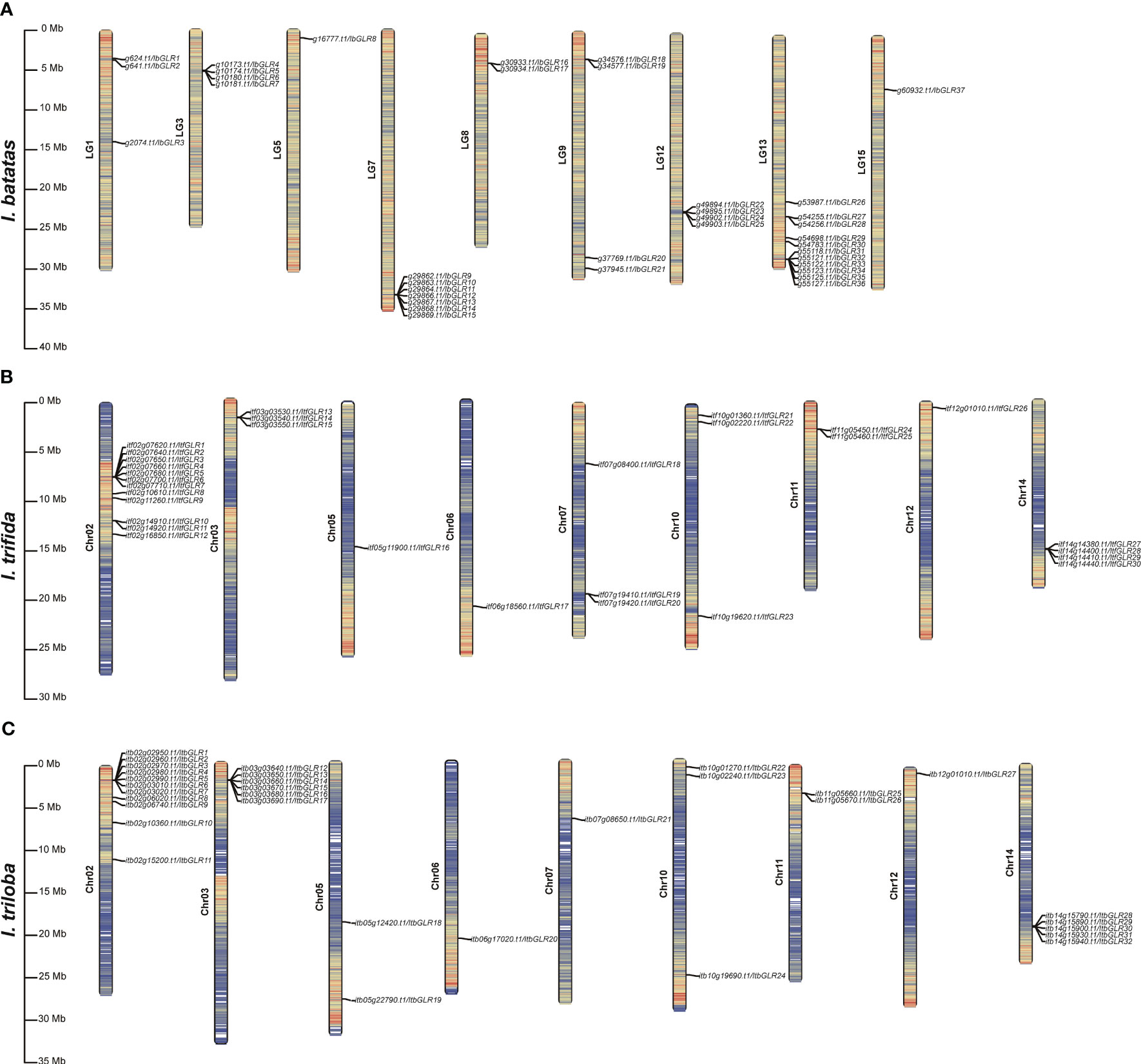
Figure 1 Chromosomal distribution and localization information of GLR genes in Ipomoea batatas (A), Ipomoea trifida (B), and Ipomoea triloba (C). The bar chart represents chromosomes, with chromosome numbers on the left side and gene names on the right side for bar chart. The black line on the right side for bar chart marks the position of each GLR gene on the chromosome and represents it in units of Mbp.
Phylogenetic trees were constructed to elucidate the evolutionary interactions between 121 different GLRs from Arabidopsis (20), I. batatas (37), I. trifida (32), and I. triloba (32) (Figure 2). The GLRs for different species were categorized according to evolutionary distances into different subgroups, including five (Groups I to V) of I. batatas, four (Groups I to III, and V) of I. trifida, five (Groups I to V) of I. triloba, and three (Groups I to III) of Arabidopsis. The specific distributions for GLRs were as follows (total: I. batatas, I. trifida, I. triloba, Arabidopsis): Group I (37: 11, 12, 10, 4), Group II (27: 6, 6, 6, 9), Group III (47: 16, 10, 14, 7), Group IV (2: 1, 0, 1, 0), and Group V (8: 3, 4, 1, 0) (Figure 2; Supplementary Table S1). All AtGLRs were found to have homologous proteins in I. batatas, I. trifida, and I. triloba, but IbGLR18/19/27/28, ItfGLR10/11/23/32, and ItbGLR10/24 showed no homology with Arabidopsis GLRs. Except for Groups II and IV, the numbers and types of GLRs distributed in the I. batatas subgroups were different from those in I. trifida, I. triloba, and Arabidopsis.
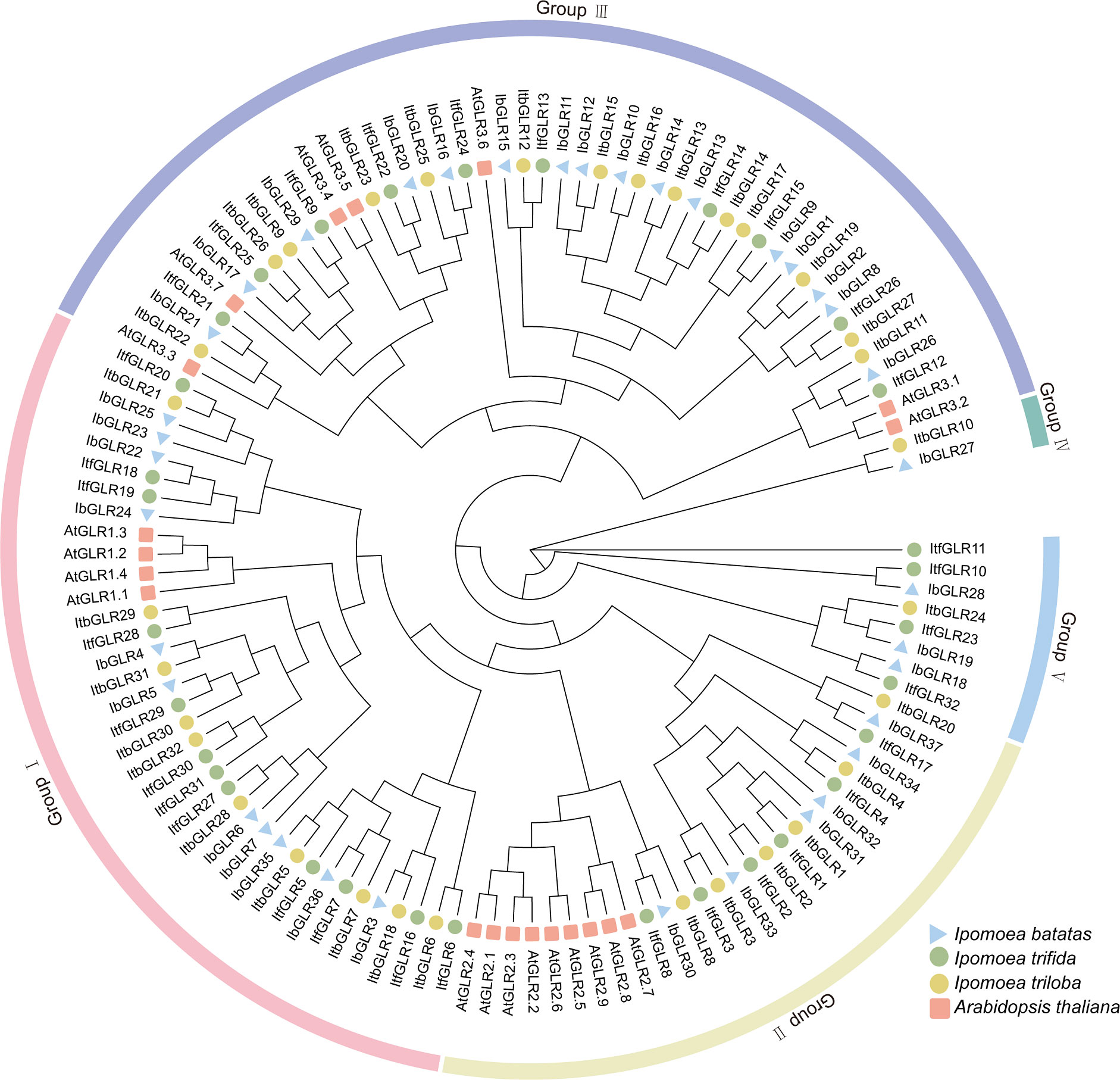
Figure 2 Phylogenetic relationship analysis of GLRs in Arabidopsis, Ipomoea batatas, Ipomoea trifida, and Ipomoea triloba. The 20 AtGLRs for Arabidopsis are represented by red squares. The 37 IbGLRs for I.batatas are represented by blue triangles. The 32 ItfGLRs for I. trifida are represented by green circles. The 32 ItbGLRs in I. triloba are represented by yellow circles.
The MEME website was used to analyze the sequence motifs of 37 IbGLRs, 32 ItfGLRs, and 32 ItbGLRs, and motif 1 to 5 were identified as five conserved motifs (Figure 3A). All GLR sequences were used to produce the sequences for the five most conserved motifs, shared between sweet potato and two diploid relatives (Supplementary Figure S1). Despite being substantially similar, the GLRs for I. batata, I. trifida together with I. triloba could have different numbers and types of conserved domains. For example, ItfGLR1 contained motifs 2 to 5, ItbGLR1 contained motifs 1 to 5, and IbGLR31 contained motifs 1 to 5. ItfGLR4 contained motifs 1 to 5, ItbGLR4 contained motifs 1 to 5, and IbGLR34 contained motifs 1 to 3 and 5. ItfGLR8 comprised motifs 1 to 5, ItbGLR8 contained motifs 3 to 5, and IbGLR30 contained motifs 1 to 5 (Figure 3A). Additionally, the receptor domains (atrial natriuretic factor [ANF]_receptor and periplasmic_binding_protein) and four transmembrane helix domains (Lig_chan domains) of GLRs are closely related to plant functions. Most GLRs of I. batata, I. trifida, and I. triloba (30 IbGLRs, 28 ItfGLRs, and 29 ItbGLRs) contained ANF_receptor and Lig_chan domains. However, ItbGLR31 only contained a Lig_chan domain. IbGLR7/18/23/28, ItfGLR32, and ItbGLR24 contained periplasmic_binding_protein_type1 and Lig_chan domains, and IbGLR6/22/24, ItfGLR18/19/24, and ItbGLR25 contained ANF_receptor, periplasmic_binding_protein_type2, and Lig_chan domains (Figure 3B).
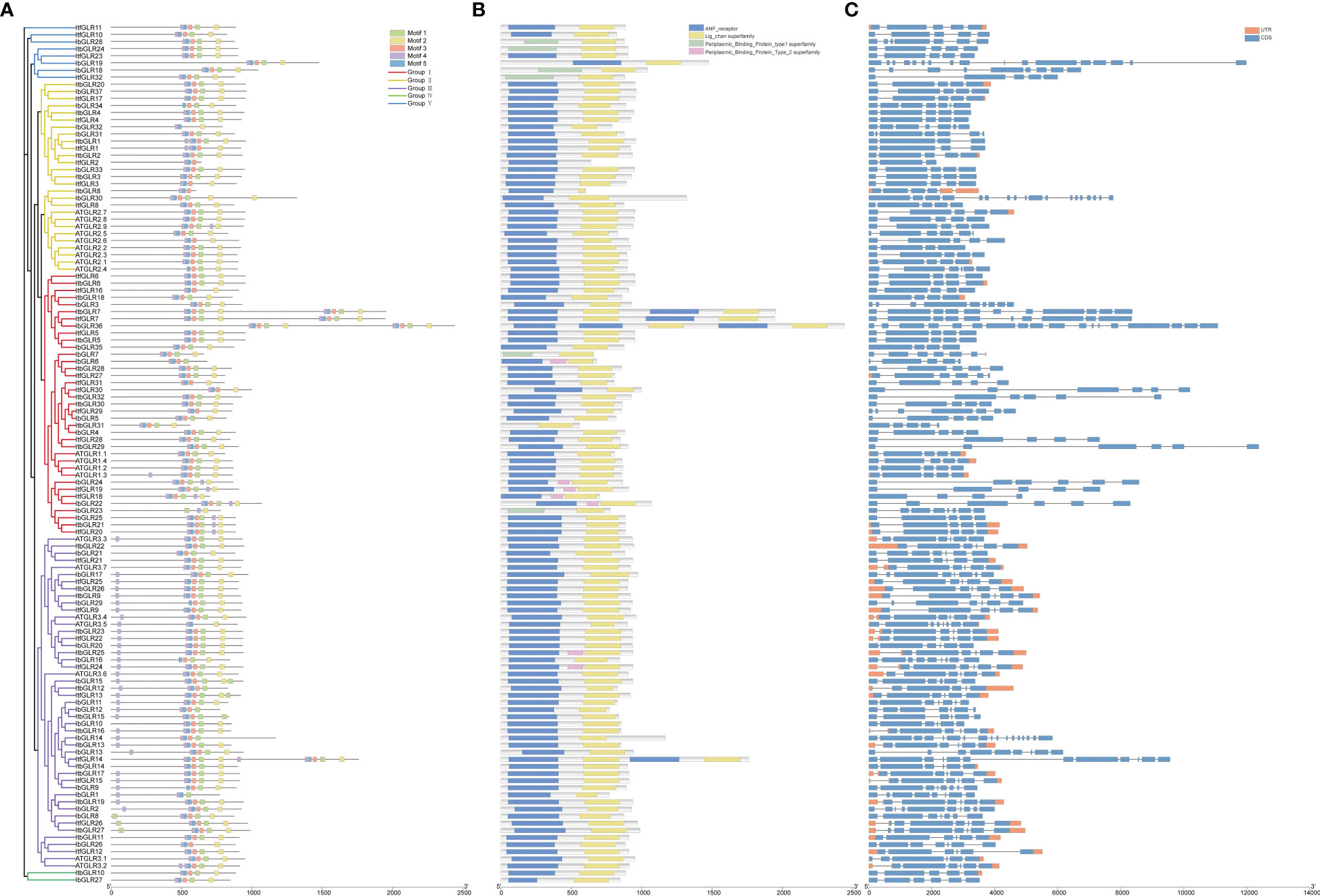
Figure 3 The exon–intron structural and conserved motifs analysis of GLR family in Ipomoea batatas, Ipomoea trifida, and Ipomoea triloba. (A) GLRs were distributed in five subgroups by phylogenetic tree (left), and motif 1-5 are shown in different colors (right). (B) The GLRs conserved domain structures. The blue boxes, yellow boxes, green boxes, and pink boxes represent the ANF_receptor domain, Lig_chan domain, periplasmic_binding_protein_type1 domain and periplasmic_binding_protein_type2 domain, respectively. (C) The GLRs exon-intron structures. The black lines, blue boxes, and orange boxes represent the introns, exons, and UTRs, respectively.
Exon-intron architectures were examined in order to gain a better understanding of structural variation among the GLRs (Figure 3C). The number of exons in the GLR genes varied from 3 to 18. In detail, GLRs of Group I contained 4 to 18 exons, GLRs of Group II contained 3 to 18 exons, GLRs of Group III contained 6 to 17 exons, GLRs of Group IV contained 5 to 6 exons, and GLRs of Group V carried 5 to 17 exons (Figure 3C). Additionally, exon-intron architecture for several homologous GLRs in I. batatas was identified as possibly different from the counterparts in I. trifida and I. triloba. Such as, IbGLR36 carried 18 exons, but ItfGLR7 and ItbGLR7, the corresponding homologous genes of IbGLR36 contained 10 and 11 exons in Group I, IbGLR30 contained 18 exons, but ItfGLR8 and ItbGLR8 both contained five exons in Group II, and IbGLR13 carried nine exons, but ItfGLR14 and ItbGLR14 contained 12 and six exons in Group III (Figure 3C). These findings suggest that sweet potato genome potentially underwent a lineage-specific differentiation event involving GLR gene family members.
A cis-element, such as the sequence upstream of GLRs, could play a significant role in plant development and stress responses. Hence, we used a 1500 bp DNA sequence upstream of IbGLRs for cis-element analysis in I. batatas. According to the predicted functions, such components were separated into five groups (core, hormone, developmental, light, and abiotic/biotic) (Figure 4). The CAAT-box and TATA-box core promoter elements were present in all 37 IbGLRs. There were 19 to 47 CAAT-box and 17 to 129 TATA-box core promoter elements in 37 IbGLRs. In addition to IbGLR35, other IbGLRs were found to have several hormone elements, such as the P-box for gibberellic acid (GA)-responsive elements, TCA for SA-responding elements, AuxRR-core together with TGA-element for indole-3-acetic acid (IAA)-responsive elements, CGTCA-motif and TGACG-motif for MeJA-responsive elements/ABRE for ABA-responsive elements (Figure 4).
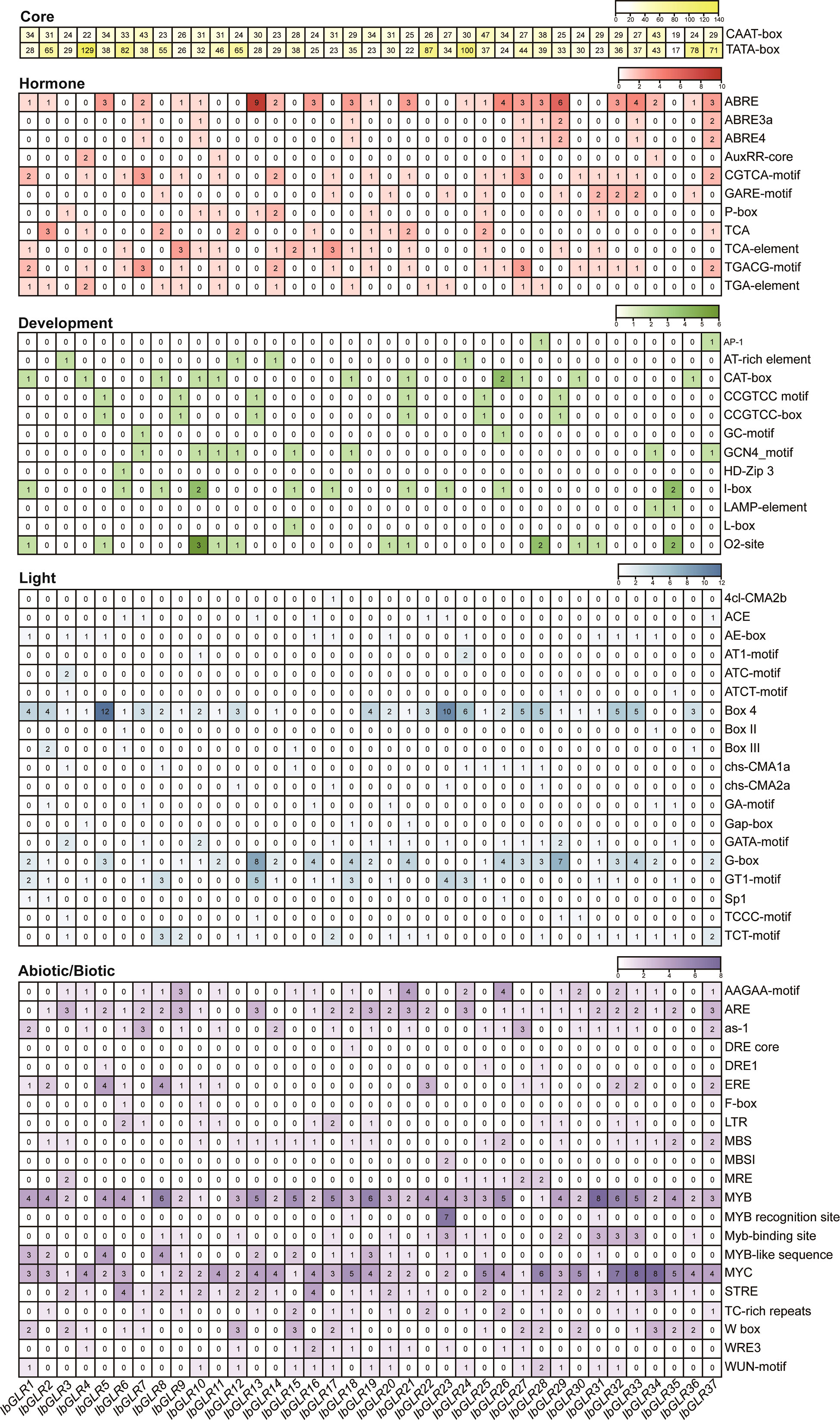
Figure 4 The cis-element analysis of IbGLRs in Ipomoea batatas. Yellow, red, green, blue, and purple represents core elements, hormone elements, development elements, light elements, and abiotic/biotic elements, respectively. For the same color, a darker color indicates a greater number for cis-elements.
For development-related elements, the CAT-box related to meristem formation (found in IbGLR1/4/8/10/11/18/21/26/27/30/36), O2-site, a zein metabolism-regulatory element (observed in IbGLR1/5/10/11/12/20/21/28/30/31/35), I-box responding to light (found in IbGLR1/6/8/10/15/17/21/23/26/35), and GCN4_motif participating in regulating seed-specific expression (found in IbGLR7/10/11/12/15/18/34/37) were abundant in promoters of IbGLRs. The promoter regions of IbGLR2/16/19/22/32/33 did not contain any development-related elements (Figure 4). Similarly, the promoters of 37 IbGLRs contained a number of light-responsive elements. IbGLRs were found to be rich in BOX4, G-boxes, GT1-motifs, and TCT-motifs. The ATC-motif was only present in IbGLR3, and 4cl-CMA2b only in IbGLR17 (Figure 4).
Moreover, several abiotic elements, the MYC, MYB, and DRE core responded to drought stress, LTR, MBS, and W box responded to salt stress and biotic elements, the WUN, WRE3, and W box motifs were found in most IbGLRs (Figure 4). This indicated that IbGLRs may have different roles under different stress conditions. The DRE core was only found in the promoter region of IbGLR18. The results demonstrate that IbGLRs take part in regulating the growth and development of plants, hormone crosstalk, and adaptation to abiotic and biotic stress in sweet potato.
An IbGLR interaction network was built through orthologous proteins from Arabidopsis to examine possible regulatory networking for IbGLRs (Figure 5). The prediction of protein interactions suggested that some IbGLRs (IbGLR2/5/7/12/18/20/25) could interact with other IbGLRs. Moreover, IbGLR7, IbGLR17, and IbGLR20 were determined to interact with CNGC18, which was reported to regulate germination of pollen grains and the growth of pollen tubes (Chang et al., 2007; Cao et al., 2016; Gu et al., 2017). IbGLR17, IbGLR20, and IbGLR25 were determined to interact with GF14, which participates in transport, growth, metabolism, and the drought stress reactions (Li et al., 2016; Hajibarat et al., 2022). IbGLR2, IbGLR7, and IbGLR17 were determined to interact with TPK1, which participates in potassium transportation, fruit ripening, and quality formation (Lu, 2011; Latza et al., 2013; Wang et al., 2018). IbGLR2/3/12/20 were determined to interact with TPC1, which mediates Ca2+ release (Moccia et al., 2021; Navarro-Retamal et al., 2021; Pottosin and Dobrovinskaya, 2022), and IbGLR2/3/5/6/7/12/17/18/20 and IbGLR25 were determined to interact with AGB1, which regulates salt stress tolerance, fungal and bacterial immunity, and plant growth (Takahashi et al., 2018; Yu and Assmann, 2018; Zhang et al., 2020; Qi et al., 2021; Afrin et al., 2022). These findings indicate that IbGLRs have a distinct function in mediating biotic and abiotic stress and influence plant development in sweet potatoes.
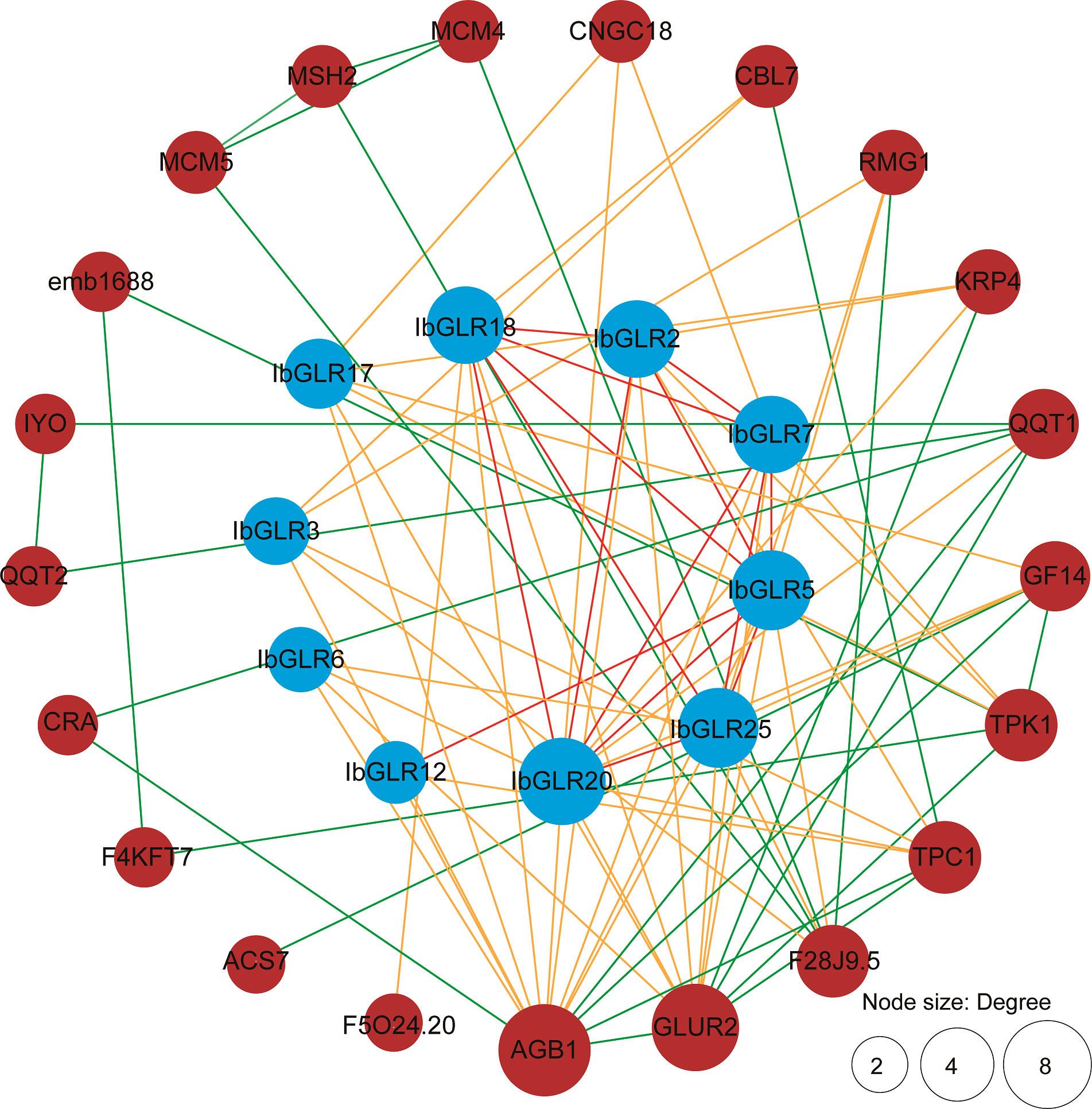
Figure 5 IbGLR functional interaction network analysis in Ipomoea batatas. The network nodes represent proteins. The number of interacting proteins is represented by node size. The lines represent the interactions between proteins. The interactions between different GLRs are represented by red lines. The interactions between GLRs and other proteins are represented by orange lines. The interactions between proteins other than GLR are represented by green lines.
IbGLR expression levels were examined using RNA-seq data in four typical I. batatas tissues (leaves, stems, fibrous roots, and tuberous roots), in order to investigate their potential biological functions in plant developmental processes (Figure 6A). Interestingly, different IbGLRs had distinct patterns of expression in the four tissues, with some IbGLRs showing tissue-specific expression. IbGLR6/13/26/32 and IbGLR35 had a high expression in leaves, whereas the expression of IbGLR1/2 and IbGLR4 was high in the stems. IbGLR8/12/18/19/21/23/25/28/33/34/36 and IbGLR37 highly expressed in fibrous roots, IbGLR29 and IbGLR31 highly expressed in tuberous roots, IbGLR3 and IbGLR27 highly expressed in leaves and fibrous roots, IbGLR11/14/15/16/17/20/22 and IbGLR24 highly expressed in leaves and stems, and IbGLR10 and IbGLR30 highly expressed in stems and fibrous roots. These findings imply that IbGLRs may have different roles in sweet potato tissue development.
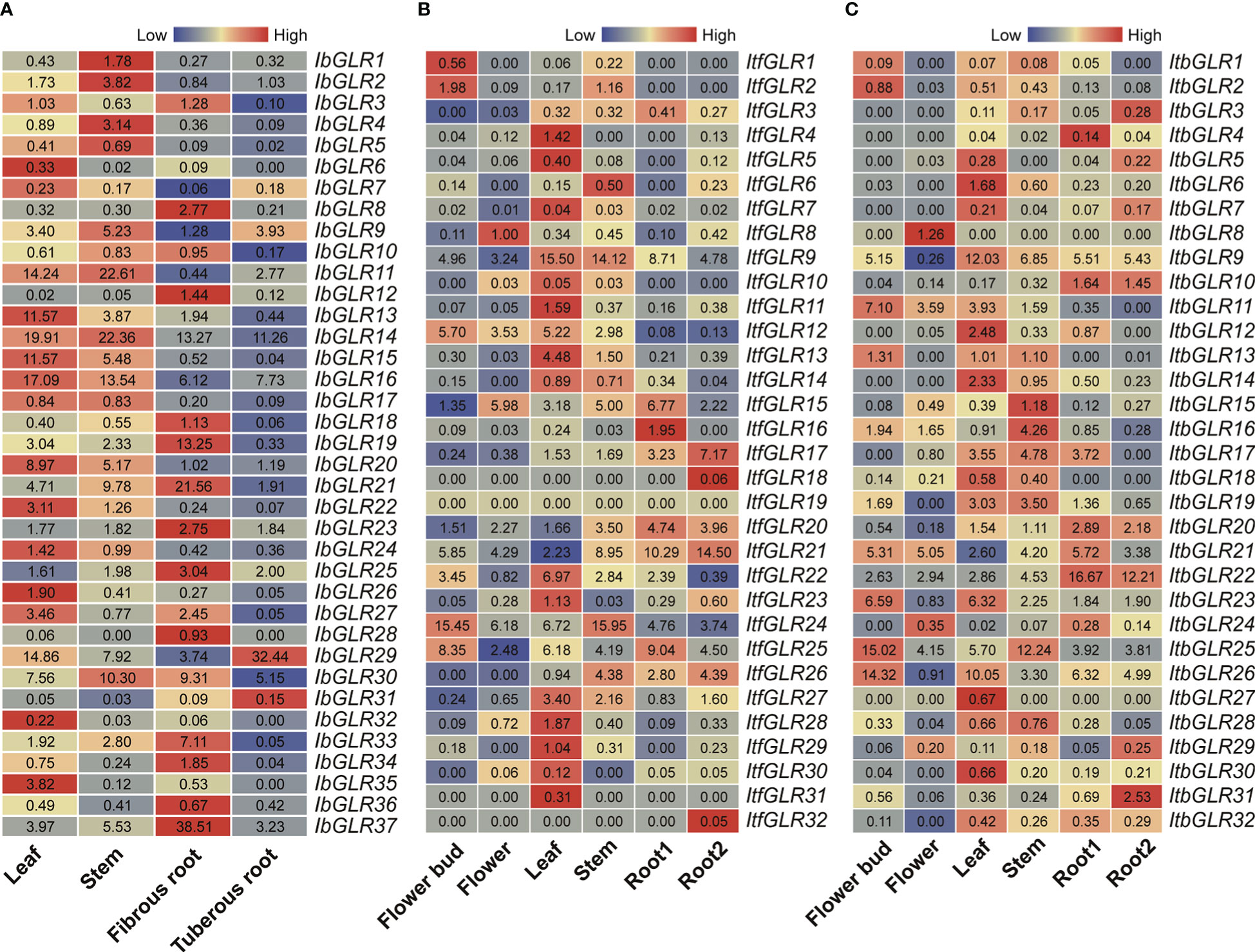
Figure 6 GLR expression study in different tissues for different cultivars. (A) Represents expression analysis for Ipomoea batatas based on the leaf, stem, fibrous root, and tuberous root. (B, C) Represent gene expression patterns for Ipomoea trifida and Ipomoea triloba based on the flower bud, flower, leaf, stem, root 1, and root 2. The FPKM values are displayed in the boxes. The color bar only represents the expression values of one GLR gene in different tissues.
Additionally, the expression patterns of ItfGLRs and ItbGLRs were studied via re-analyzing RNA-seq data published in six tissues (flower buds, flowers, leaves, stems, root 1, and root 2) (Wu et al., 2018) (Figures 6B, C). In I. trifida, ItfGLR9 was highly expressed in leaves (FPKM value = 15.50) and stems (FPKM value = 14.12). In I. triloba, ItbGLR8, and ItbGLR27 were only expressed in flowers and leaves, respectively, whereas the other ItbGLRs were expressed in different tissues. ItbGLR9 exhibited a high expression in leaves. ItbGLR22 was highly expressed in root 1 and root 2. While ItbGLR25 was highly expressed in stems and flower buds, ItbGLR26 exhibited a high expression in leaves and flower buds. These findings suggest that different GLR genes exhibit diverse growth regulatory roles in sweet potato and two diploid relatives, as well as different tissue expression patterns.
The expression patterns for IbGLRs in the roots of Xushu22 plants were studied at five developmental stages using RNA-seq data (Dong et al., 2019). The results indicated that among the 37 IbGLRs, 22 IbGLRs shared similar expression patterns, with a higher transcriptional level in fibrous roots (diameter of approximately 1 mm) than roots from other growth stages. Within this group, IbGLR21 had the highest level of expression (FPKM value = 16.38) (Figure 7). The expression of IbGLR29 was the highest (FPKM value = 31.83) in the initial tuberous root (diameter of approximately 1 cm), and the expression of IbGLR2 was the highest (FPKM value = 6.51) in tuberous roots (diameter of approximately 3 cm). Meanwhile, the expression of IbGLR14 was highest (FPKM value = 12.69) in tuberous roots (diameter of approximately 5 cm), and the expression of IbGLR16 was the highest (FPKM value = 11.31) in tuberous roots (diameter of approximately 10 cm), which was consistent with the tuberous root of approximately 5 cm in diameter. Overall, the expression patterns imply different contributions of IbGLRs in the root development of sweet potato.
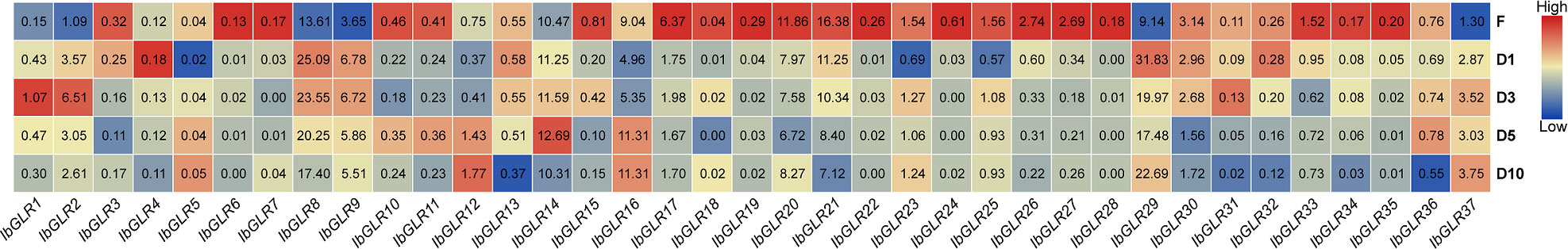
Figure 7 The expression analysis of IbGLRs at different stages of root development. F, D1, D3, D5 and D10 represent fibrous root (diameter of approximately 1 mm), initial tuberous root (diameter of approximately 1 cm), tuberous root (diameter of approximately 3 cm), tuberous root (diameter of approximately 5 cm), and tuberous root (diameter of approximately 10 cm), respectively. The FPKM values are displayed in the boxes. The color bar only represents the expression values of one GLR gene in different stages of root development.
To study the possible role of IbGLRs during interactions with the sweet potato root rot pathogen, we analyzed the expression levels of IbGLRs at different time points after root rot induction in Jishuzi563 (root rot-sensitive variety) and Jishuzi203 (root rot-resistant variety) using RNA-seq data (Figure 8). Subsequently, we further evaluated the expression levels of some IbGLRs using qRT-PCR (Supplementary Figure S2). Without root rot infection, the expression of IbGLR10/11/12/16/18 was repressed at 36 h, 72 h, 120 h, and 10 d compared with that at 0 h in Jishuzi563 and Jishuzi203. After root rot infection, only IbGLR18 expression was repressed in Jishuzi563 infected by the root rot but was induced in Jishuzi203 infected by the root rot. This result was consistent with qRT-PCR results. It has been speculated that IbGLR18 may participate in sweet potato resistance to root rot. In addition, the RNA-seq results showed that IbGLR14/26/37 expression levels were upregulated in Jishuzi563 and Jishuzi203 in response to root rot, and the expression levels in the resistant variety Jishuzi203 were higher than those in susceptible variety Jishuzi563, which was consistent with real-time quantitative results. qRT-PCR results showed that IbGLR14 manifested the highest level of expression at 120 h, IbGLR26 at 72 h, and IbGLR37 at 10 d. The outcome suggests that different IbGLRs may function differently in developing disease resistance at different stages during the interaction between sweet potato and root rot pathogens.
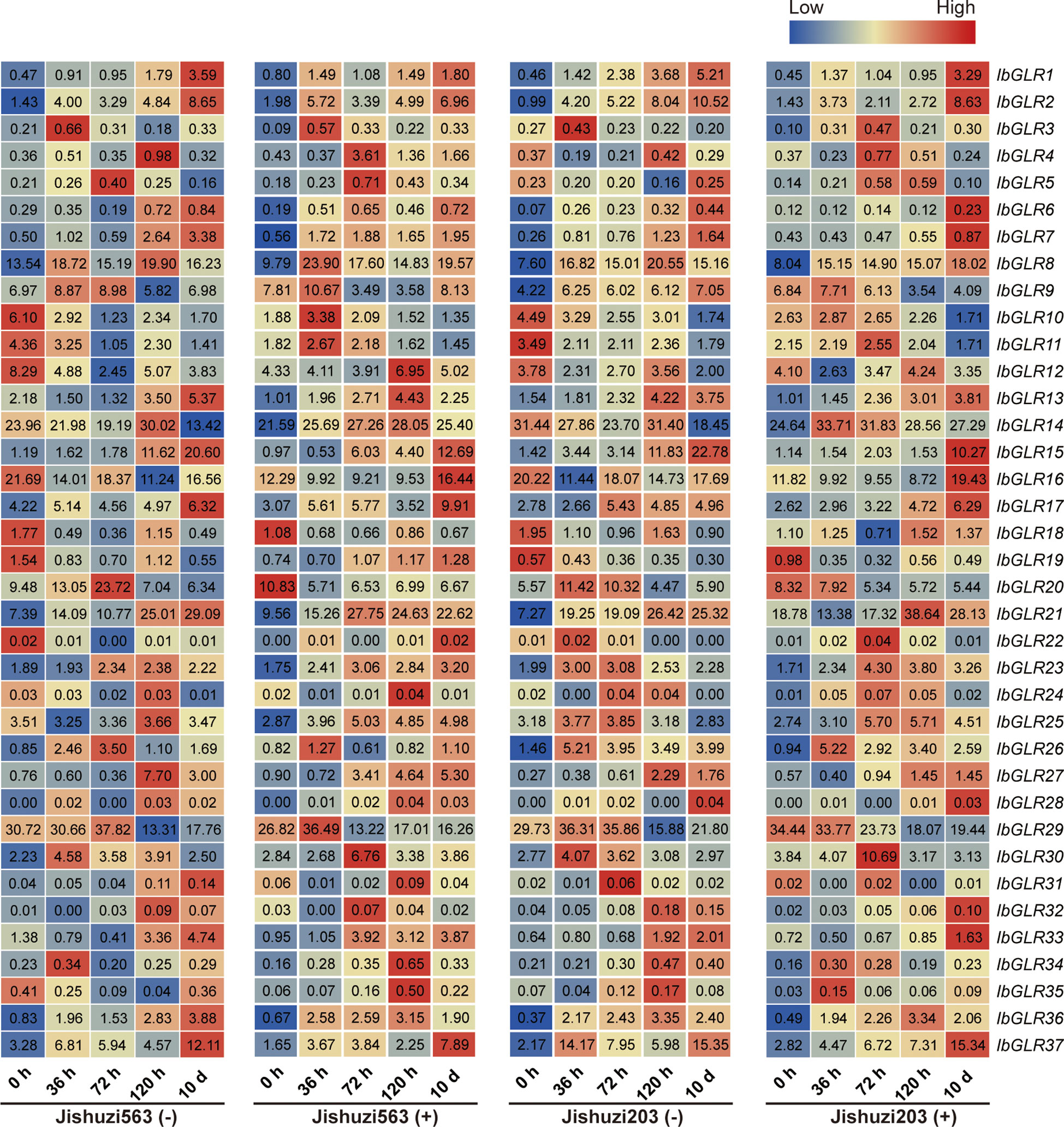
Figure 8 IbGLRs expression analysis in response to root rot. Jishuzi563(-): a root rot sensitive variety without root rot; Jishuzi563(+): a root rot sensitive variety with root rot; Jishuzi203(-): a root rot resistant variety without root rot; Jishuzi203(+): a root rot resistant variety with root rot. The FPKM values are displayed in the boxes. The color bar only represents the expression values of one GLR gene at different times after root rot induction in different varieties.
To better understand the capacity of IbGLRs to combat abiotic stress, RNA-seq data from a salt-tolerant line (ND98) and a salt-sensitive variety (Lizixiang) were used to compare the expression patterns of IbGLRs under salt stress. Meanwhile, the RNA-seq data from Xu55-2, a drought-resistant cultivar, was studied under drought stress (Zhang et al., 2017; Zhu et al., 2019). Salt stress enhanced the expression of 13 IbGLRs in the ND98 line (the expression for IbGLR1/11/14/19/24/33/34 was highest at 12 h, while that of IbGLR10/18/22/30/35/37 was highest at 48 h). For these genes, IbGLR19 expression in salt sensitive variety Lizixiang was repressed by salt stress (Figure 9A). In Xu55-2, the expression of 26 IbGLRs was induced rapidly following drought stress and reached its maximum at the early stage (≤3 h). Meanwhile, IbGLR21 expression was repressed by polyethylene glycol (PEG) treatment, whereas IbGLR9/10/13/15/19/29/31/32 expression levels were induced (Figure 9B). Thus, it may be speculated that IbGLR19 is related to salt and drought tolerance in sweet potato.
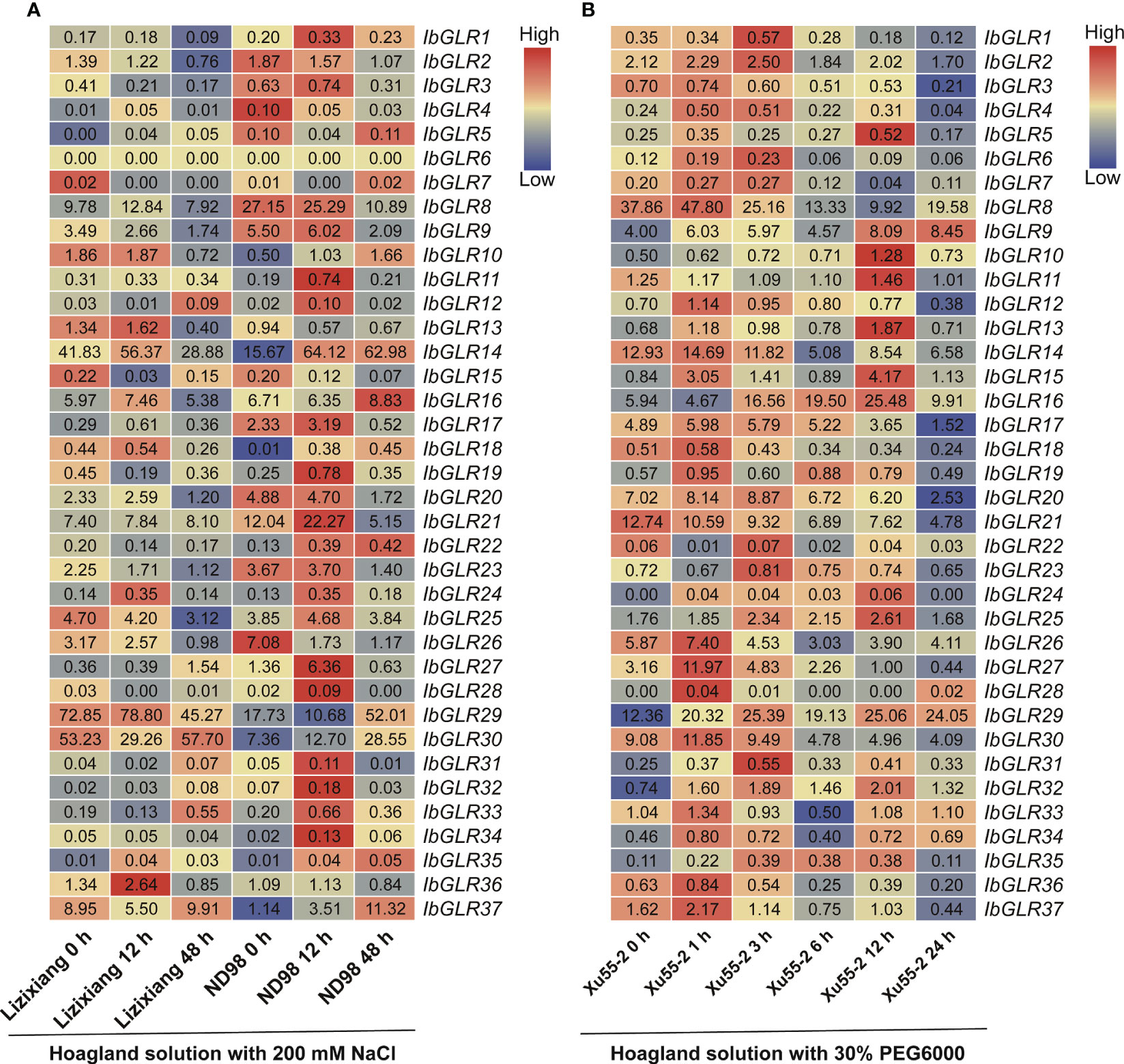
Figure 9 IbGLRs expression analysis in response to NaCl and PEG treatments. (A) Expression analysis for IbGLRs under NaCl treatment conditions in salt-sensitive variety Lizixiang and salt-tolerant line ND98. (B) Expression analysis for IbGLRs under PEG treatment conditions in drought-tolerant variety Xu55-2. The FPKM values are displayed in the boxes. The color bar only represents the expression values of one IbGLR in different treatments.
Additionally, the evaluation of ItfGLRs and ItbGLRs expression in I. trifida and I. triloba following drought and salt treatment was conducted using RNA-seq data (Wu et al., 2018). Expression levels of ItfGLR1/2/18/31 were not induced, whereas those for ItfGLR3/23/24 and ItbGLR3/8/10/11/21/31 were induced (Figure 10). These results indicate that there are differences in the expression of GLRs in sweet potato and two diploid relatives under drought and salt stress.
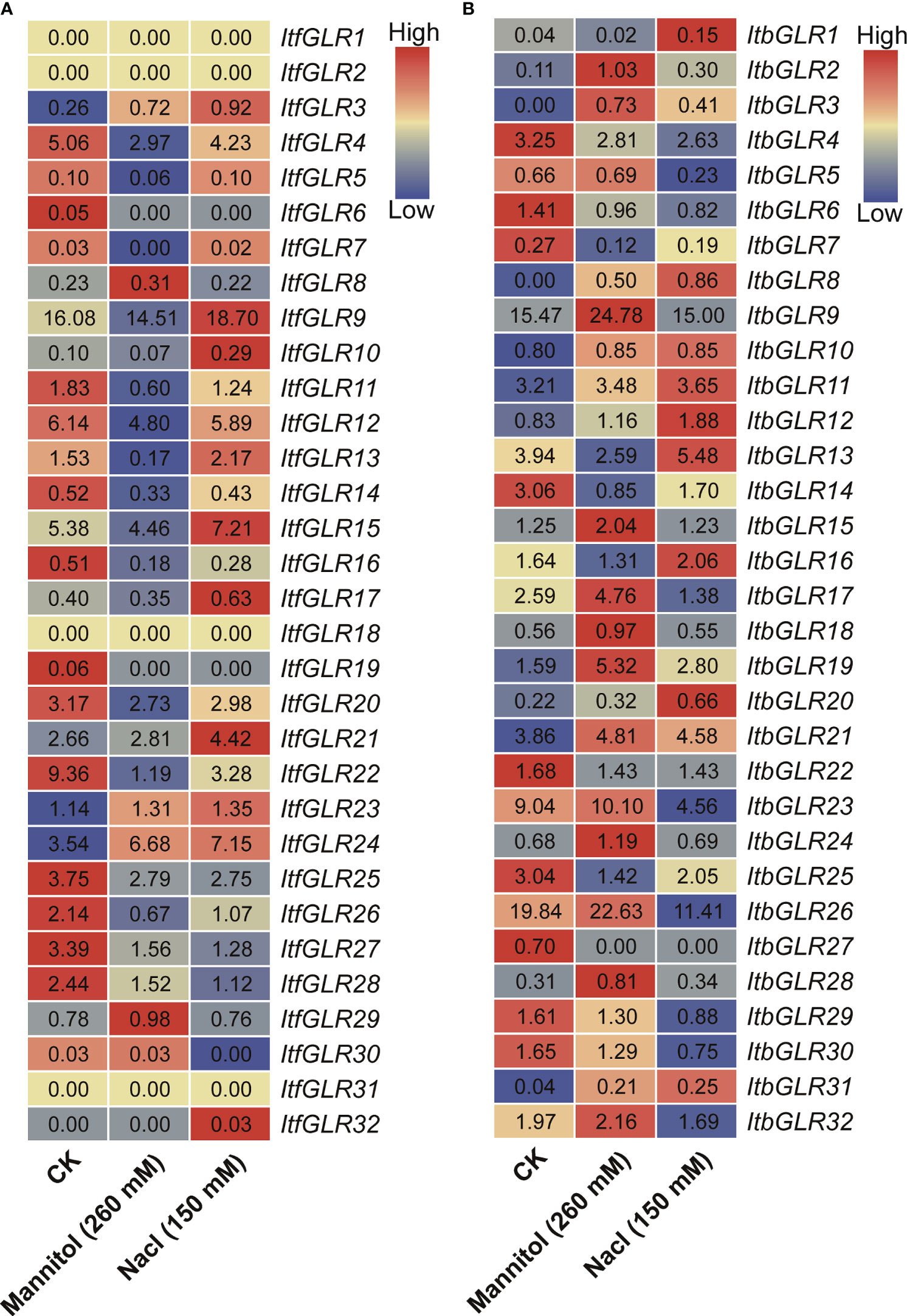
Figure 10 GLRs expression analysis in response to mannitol and NaCl stresses in Ipomoea trifida (A) and Ipomoea triloba (B). The FPKM values are displayed in the boxes. The color bar only represents the expression values of one GLR gene in different treatments.
Ultimately, an assessment of expression patterns for ItfGLRs and ItbGLRs under ABA, GA, and IAA treatments was carried out through RNA-seq data from I. trifida and I. triloba (Wu et al., 2018). In I. trifida, the expression of ItfGLR18 and ItfGLR31 was not induced by any of the hormones, whereas the expression levels of five ItfGLRs were induced by ABA, those of 11 ItfGLRs were induced by GA, and those of two ItfGLRs were induced by IAA. ItfGLR6 and ItfGLR30 expression was induced by all hormones, and ItfGLR1/5/7/8/9/11/13/15/16/20/23/25 and ItfGLR27 expression was suppressed by the three hormones (Figure 11A). In the case of I. triloba, expression levels of 15 ItbGLRs were induced by ABA, those of four ItbGLRs were induced by GA, and those of five ItbGLRs were induced by IAA. ItbGLR18 and ItbGLR19 expression was induced by all hormones, whereas ItbGLR5/6/7/13/23/25/26 and ItbGLR32 expression was repressed by all hormones (Figure 11B). The findings suggest that GLRs in the two diploid relatives participate in various hormonal pathways and take part in the crosstalk among different hormones.
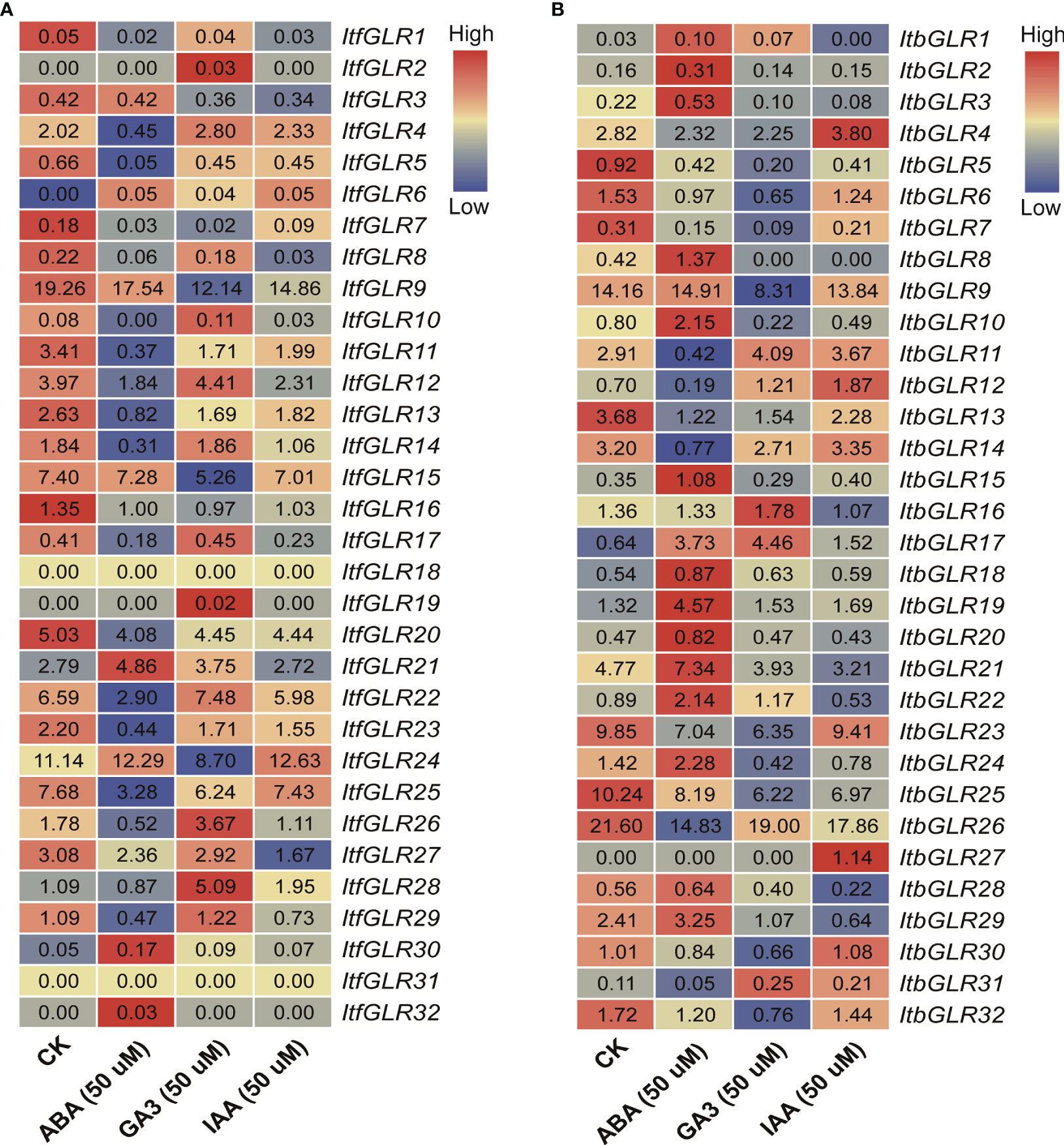
Figure 11 GLRs expression analysis in response to abscisic acid (ABA), gibberellin (GA), and indolent-3-acetic acid (IAA) treatments in Ipomoea trifida (A) and Ipomoea triloba (B). The FPKM values are displayed in the boxes. The color bar only represents the expression values of one GLR gene in different treatments.
Additionally, the pattern of expression for ItfGLRs and ItbGLRs was analyzed utilizing I. trifida and I. triloba RNA-seq data, respectively, at 10/4°C (day/night) and 35/35°C (day/night) treatment (Wu et al., 2018). In I. trifida, ItfGLR4/8/17/23/27 and ItfGLR28 expression was induced by 10/4°C (day/night), whereas ItfGLR16 expression was induced by 35/35°C (day/night) temperatures comparison with the control levels (Figure 12A). In I. triloba, ItbGLR3/8/20/24/29 and ItbGLR32 expression was induced by 10/4°C (day/night) and ItfGLR31 expression was induced by 35/35°C (day/night) compared with control levels temperatures (Figure 12B). These results show that different GLRs participate in the responses of I. trifida and I. triloba to different temperature stresses.
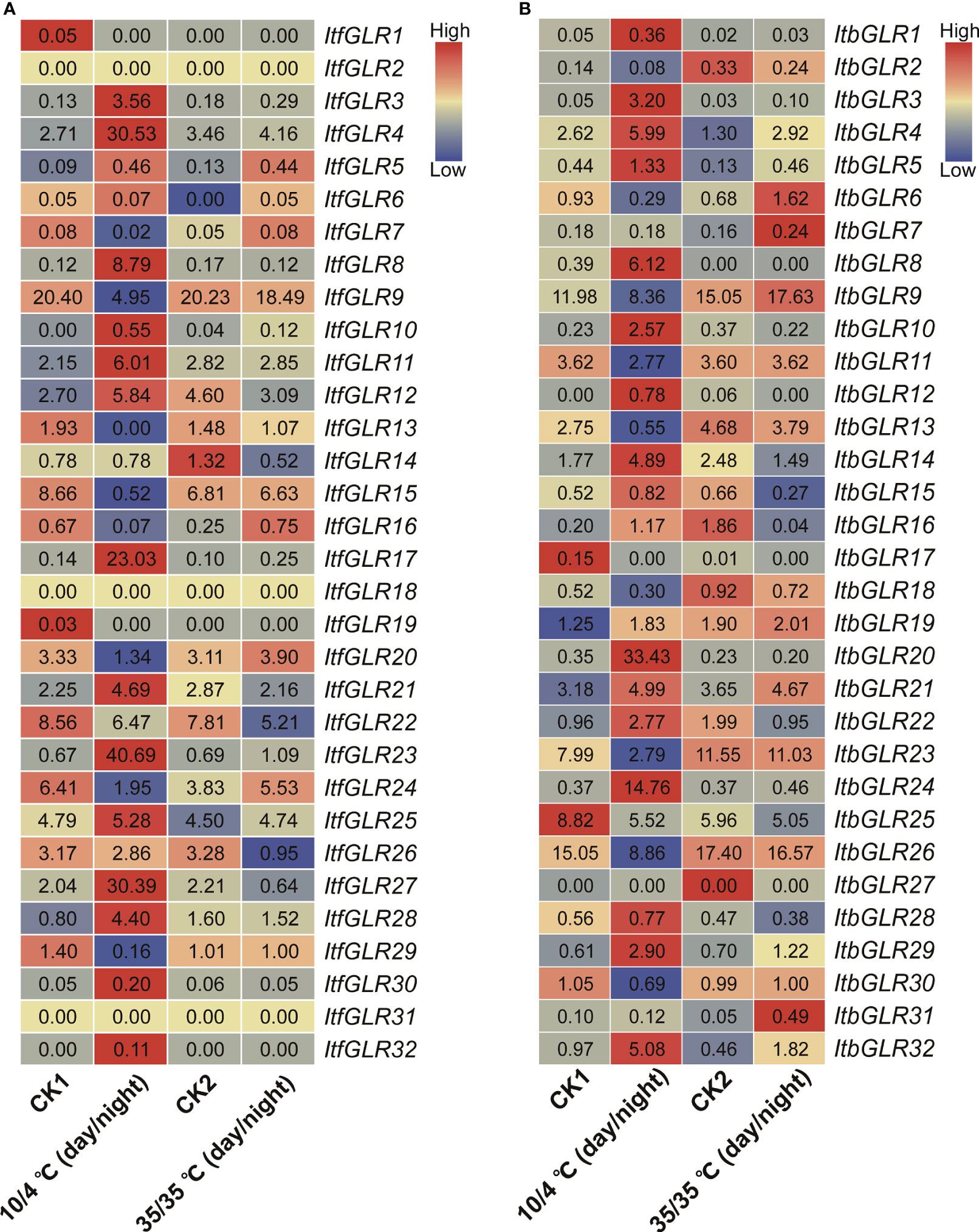
Figure 12 GLRs expression analysis under 10/4°C (day/night) and 35/35°C (day/night) treatments in Ipomoea trifida (A) and Ipomoea triloba (B). CK1, cold control; CK2, heat control. FPKM values are shown in the boxes. The color bar only represents the expression values of one GLR gene in different treatments.
Plant GLRs are mainly located on vacuolar and plasma membranes (Davenport, 2002). They are potential candidates for the plasma membrane-level regulation for calcium influx (Mäser et al., 2001) and could be amino acid sensors in plants (Tapken et al., 2013). Arabidopsis, rice, and other model plants have been investigated for the functions of GLRs, however, until now, there have been no reports on the identification of the GLR gene family members in sweet potato. Because of the complicated genetic background of cultivated sweet potato, research on the sweet potato gene family is limited in the cultivated variety, mostly focusing on its two diploid relatives (Chen et al., 2019a; Chen et al., 2019b; Li et al., 2019; Lu et al., 2019; Wan et al., 2020; Zhu et al., 2020; Huang et al., 2021; Dai et al., 2022; Nie et al., 2023). Through this investigation, we conducted an in-depth study of the characteristics of GLR genes and compared their expression patterns in response to different types of biotic and abiotic stresses using genomic sequences for the hexaploid sweet potato and two diploid relatives. The genome-wide study of GLR genes provides vital guidance for the further studying their functions.
Overall, I. trifida and I. triloba had the same number of GLRs (32 ItfGLRs and 32 ItbGLRs) identified, however they were less numerous than those (37 IbGLRs) in I. batatas. This is consistent with the fact that I. trifida and I. triloba are diploid and have a close genetic relationship. The evolution and differentiation of chromosomes were revealed by genomic alignment (Mukherjee et al., 2018). I. batatas, I. trifida, and I. triloba differed in terms of the distribution and proportion of GLRs in individual chromosomes. GLRs were located on nine chromosomes for I. batatas and I. triloba, however, GLRs were located on ten chromosomes for I. trifida. If different members of the same family are located within the same or adjacent intergenic regions, they have tandem repeat relationships (Freeling, 2009). Based on this standard, eight, six, and four tandem repeats were identified in I. batatas, I. trifida, and I. triloba, respectively (Figure 1). This indicates that tandem repeats might be one of the reasons for the amplification of GLR genes in I. batatas, I. trifida, and I. triloba.
This research is based on the GLRs for I. batatas, I. trifida, and I. triloba, which were segregated into five groups (Groups I to V), four groups (Groups I to III and V), and five groups (Groups I to V), respectively. The fourth and fifth subgroups consisted of 10 GLRs that were absent in Arabidopsis. The type and number of GLRs in various subgroups for sweet potato and two diploid relatives are different from those in Arabidopsis. These findings imply that the genomes may have experienced lineage-specific differentiation for GLR gene family.
Introns are important components of eukaryotic protein-coding genes, eliminated during messenger RNA precursor molecule splicing. They are characterized by their clear organization and abundance in eukaryotic genes (Rogozin et al., 2005; Mukherjee et al., 2018). Because of the presence of introns, gene expression in eukaryotic cells is much more complex than that in prokaryotic cells. Under stressful conditions, introns assume a key role in regulating cell growth (Morgan et al., 2019). Herein, in comparison with I. trifida and I. triloba, I. batatas had distinct exon-intron patterns for some homologous GLRs (Figure 3C). For example, IbGLR36, which was expressed in fibrous roots, contained 17 introns, whereas its homologous genes ItfGLR7 and ItbGLR7 contained nine and ten introns, respectively, and were expressed in leaves. Moreover, IbGLR30, which was expressed in the stem, contained 17 introns, whereas ItfGLR8 and ItbGLR8 contained four and five introns, respectively, and were expressed in flowers (Figures 3C, 6). The corresponding differences in exon–intron structure between sweet potato and its two diploid relatives might lead to different functions for GLRs in various aspects of plant growth and development (Pang et al., 2018; Ma et al., 2019; Ma J et al., 2020).
Plant GLRs are actively involved in hormone biosynthesis and signal transduction concerning the regulation of plant growth and responses to stress (Weiland et al., 2015). PpGLR1 is involved in ABA-mediated growth regulation in Physcomitrium patens (Wang et al., 2022). Meanwhile, RsGluR could serve as a defensive mechanism against pathogen infection by triggering MeJA biosynthesis (Kang et al., 2006). In this work, the promoters of the IbGLRs contained 11 hormone-responsive elements, 12 developmental elements, 19 light-responsive elements, and 21 abiotic/biotic-response elements. In addition to IbGLR35, other IbGLRs comprised at least one hormone element (Figure 4). IbGLR3 only contained the P-box of the GA-responsive elements, whereas its homologous gene ItbGLR18 was determined to be induced by ABA. IbGLR4 was found to contain the TGACG-motif and CGTCA-motif for MeJA-responsive elements, TCA for SA-responsive elements, and the AuxRR-core together with TGA-element for IAA-responsive elements, whereas its homologous gene, ItbGLR31, was determined to be induced by GA and IAA. IbGLR21 contained ABRE for ABA-responsive elements, the TGACG-motif together with CGTCA-motif for MeJA-responsive elements, and TCA for SA-responsive elements, whereas its homologous genes, ItfGLR21 and ItbGLR22, were both determined to be induced by ABA and GA. IbGLR30 carried the TGACG-motif and CGTCA-motif for MeJA-responsive elements, whereas its homologous gene, ItbGLR8, was determined to be induced by ABA (Figures 4, 11). Such results demonstrate GLRs involvement in the interplay of several hormones and the participation of homologous GLR genes in numerous hormone pathways in sweet potato and two diploid relatives. Further research is needed to fully understand how GLRs control hormonal crosstalk.
GLR plays an indispensable role in the response of plants to different biological stresses (He et al., 2016). Liu and colleagues discovered a point mutation for GhGLR4.8 exon that can increase the resistance of upland cotton to Fusarium wilt, while knocking out the GhGLR4.8 lead the defense ability of cotton cell wall against G. hirsutum Fov race 7 to weaken (Liu et al., 2021). AtGLR3.3 also has a valuable function in the defense response induced by Pseudomonas syringae pv tomato DC3000. An AtGLR3.3 mutant was shown to exhibit high sensitivity to DC3000, and the response of defense genes was reduced in mutant lines induced by pathogenic bacteria (Li et al., 2013). Sweet potato root rot infection usually begins at the underground stem and tip or middle of fibrous roots. Severely diseased plants do not produce tuberous roots, whereas slightly diseased plants produce tuberous roots with diseased spots (Ma Z et al., 2020).
Transcriptome and qRT-PCR analysis showed that IbGLR18 expression was induced in Jishuzi203 (root rot-resistant variety) and repressed in Jishuzi563 (root rot-sensitive variety) after root rot infection. In addition, the IbGLR18 expression in fibrous roots was higher in comparison to that in other tissues (leaf, stem, and tuberous root). Furthermore, IbGLR18 expression was also higher in the fibrous root (diameter of root 1 mm) than that in tuberous roots (diameter of root 1/3/5/10 cm) in the five developmental stages of the Xushu22 root. Therefore, we speculate that IbGLR18 might be involved in resistance to root rot and the formation of fibrous roots in sweet potato, but its function should be verified in the future studies.
Usually, tuberous roots are the main harvesting tissues for hexaploid sweet potato. I. trifida and I. triloba are not capable of forming tuberous roots (Wu et al., 2018). The expression for IbGLR29 in tuberous roots was higher than that in other tissues (leaf, stem, and fibrous roots) of sweet potato, and its homolog genes ItfGLR9 and ItbGLR9 were highly expressed in the leaf. Further, in the five developmental stages of Xushu22 roots, the expression of IbGLR29 in tuberous roots (root diameter of 1/3/5/10 cm) was high in comparison to that in fibrous roots (diameter of approximately 1 mm). IbGLR29 may therefore be involved in the generation of tuberous roots in sweet potato.
GLRs play a crucial role in mediating plants responses to abiotic environmental stress (He et al., 2016). The degree of expression of ZmGLR2.3/3.1 demonstrated a significant increase after drought stress (Zhou et al., 2021). In this work, MYB and MYC were found to respond to drought stress, MBS to salt stress, and LTR to cryogenic stress, and these were identified in the IbGLR19 promoter, which was rapidly expressed after drought stress and reached its expression maximum at the early stage (1 h). Moreover, its expression was upregulated by NaCl treatment in ND98 (salt-tolerant line) and downregulated in Lizixiang (salt-sensitive variety, Figures 4, 9). ItfGLR23, the homolog gene of IbGLR19, was induced by both drought and salt treatments in I. trifida (Figure 10). These results suggest that IbGLR19 may contribute to abiotic stress. Additionally, I. trifida and I. triloba may be utilized for searching and identifying functional genes, particularly those that provide tolerance or resistance to biotic and abiotic stresses, which may have been lost during the domestication of cultivated sweet potato (Cao et al., 2016). In I. trifida and I. triloba, several genes (ItfGLR8 and its homologous gene ItbGLR8, ItfGLR17 and its homologous gene ItbGLR20, ItfGLR23 and its homologous gene ItbGLR24, and ItfGLR28 and its homologous gene ItbGLR29) exhibited the same expression patterns and were induced under 10/4°C (day/night) (Figure 12). ItfGLR3 and its homologous gene ItbGLR3 were induced by drought and salt treatments (Figure 10). In summary, the GLRs induced in I. trifida and I. triloba could serve as candidate genes for enhancing the abiotic stress resistance of sweet potato.
The original contributions presented in the study are publicly available. This data can be found here: NCBI SRA (http://www.ncbi.nlm.nih.gov/Traces/sra), accession numbers SAMN10755180, SAMN10755181, SAMN10755182, SAMN10755183, SAMN10755184, SAMN10755185, SAMN10755186, SAMN10755187, SAMN10755188, SAMN10755189, SAMN10755190, SAMN10755191, SAMN10755192, SAMN10755193, SAMN10755194 and SRP092215.
YH: Writing – original draft, Writing – review & editing. ZD: Writing – original draft. JH: Writing – original draft. MH: Writing – original draft. ZW: Writing – original draft. WJ: Writing – original draft. ZG: Writing – original draft. XL: Writing – review & editing. LL: Writing – review & editing. ZM: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32301957), the China Agriculture Research System (CARS-10-Sweetpotato), the Basic Research Funds of Hebei Academy of Agriculture and Forestry Sciences (2021060207), the Key Research and Development Project of Hebei Province (22322911D), the Modern Agriculture Industrial Technology System Innovation Team Project of Hebei Province (HBCT2023060202), and the HAAFS Science and Technology Innovation Special Project (2022KJCXZX-LYS-12). The funding bodies were not involved in the design of the study, in the collection, analysis and interpretation of data or in writing the manuscript.
We are grateful to Huan Zhang (College of Agronomy and Biotechnology, China Agricultural University) for her suggestions on our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1255805/full#supplementary-material
Afrin, T., Costello, C. N., Monella, A. N., Kørner, C. J., Pajerowska-Mukhtar, K. M. (2022). The interplay of GTP-binding protein AGB1 with ER stress sensors IRE1a and IRE1b modulates Arabidopsis unfolded protein response and bacterial immunity. Plant Signal Behav. 17, 2018857. doi: 10.1080/15592324.2021.2018857
Ahmed, I., Kumar, A., Bheri, M., Srivastava, A. K., Pandey, G. K. (2023). Glutamate receptor like channels: Emerging players in calcium mediated signaling in plants. Int. J. Biol. Macromol. 234, 123522. doi: 10.1016/j.ijbiomac.2023.123522
Aouini, A., Matsukura, C., Ezura, H., Asamizu, E. (2012). Characterisation of 13 glutamate receptor-like genes encoded in the tomato genome by structure, phylogeny and expression profiles. Gene 493, 36–43. doi: 10.1016/j.gene.2011.11.037
Bian, C., Demirer, G. S., Brady, S. M. (2022). GLRs: mediating a defense-regeneration tradeoff in plants. Dev. Cell 57, 417–418. doi: 10.1016/j.devcel.2022.02.007
Cao, Q., Li, A., Chen, J., Sun, Y., Tang, J., Zhang, A., et al. (2016). Transcriptome sequencing of the sweet potato progenitor (Ipomoea Trifida (HBK) G. Don.) and discovery of drought tolerance genes. Trop. Plant Biol. 9, 63–72. doi: 10.1007/s12042-016-9162-7
Chang, F., Yan, A., Zhao, L., Wu, W., Yang, Z. (2007). A putative calcium-permeable cyclic nucleotide-gated channel, CNGC18, regulates polarized pollen tube growth. J. Integr. Plant Biol. 49, 1261–1270. doi: 10.1111/j.1672-9072.2007.00524.x
Chen, J., Jing, Y., Zhang, X., Li, L., Wang, P., Zhang, S., et al. (2016). Evolutionary and expression analysis provides evidence for the plant glutamate-like receptors family is involved in woody growth-related function. Sci. Rep. 6, 32013. doi: 10.1038/srep32013
Chen, Y., Zhu, P., Wu, S., Lu, Y., Sun, J., Cao, Q., et al. (2019a). Identification and expression analysis of GRAS transcription factors in the wild relative of sweet potato Ipomoea trifida. BMC Genomics 20, 911. doi: 10.1186/s12864-019-6316-7
Chen, Y., Zhu, P., Zhang, L., Wu, S., Ma, D., Cao, Q., et al. (2019b). Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet. 20, 41. doi: 10.1186/s12863-019-0743-y
Chiu, J. C., Brenner, E. D., DeSalle, R., Nitabach, M. N., Holmes, T. C., Coruzzi, G. M. (2002). Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol. Biol. Evol. 19, 1066–1082. doi: 10.1093/oxfordjournals.molbev.a004165
Cho, D., Kim, S. A., Murata, Y., Lee, S., Jae, S. K., Nam, H. G., et al. (2009). De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J. 58, 437–449. doi: 10.1111/j.1365-313X.2009.03789.x
Dai, Z., Yan, P., He, S., Jia, L., Wang, Y., Liu, Q., et al. (2022). Genome-wide identification and expression analysis of SWEET family genes in sweet potato and its two diploid relatives. Int. J. Mol. Sci. 23, 15848. doi: 10.3390/ijms232415848
Dong, T., Zhu, M., Yu, J., Han, R., Tang, C., Xu, T., et al. (2019). RNA-Seq and iTRAQ reveal multiple pathways involved in storage root formation and development in sweet potato (Ipomoea batatas L.). BMC Plant Biol. 19, 136. doi: 10.1186/s12870-019-1731-0
Food and Agriculture Organization of the United Nations. (2021). Sweetpotato. Available at: https://www.fao.org/faostat/zh/#search/Sweet%20potatoes (Accessed March 15, 2023).
Forde, B. G., Lea, P. J. (2007). Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 58, 2339–2358. doi: 10.1093/jxb/erm121
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Gu, L. L., Gao, Q. F., Wang, Y. F. (2017). Cyclic nucleotide-gated channel 18 functions as an essential Ca2+ channel for pollen germination and pollen tube growth in Arabidopsis. Plant Signal. Behav. 12, e1197999. doi: 10.1080/15592324.2016.1197999
Hajibarat, Z., Saidi, A., Hajibarat, Z. (2022). Genome-wide identification of 14-3-3 gene family and characterization of their expression in developmental stages of Solanum tuberosum under multiple biotic and abiotic stress conditions. Funct. Integr. Genomics 22, 1377–1390. doi: 10.1007/s10142-022-00895-z
He, M., Sun, Y., Chen, X., Shi, D., Li, D., Chen, Y., et al. (2016). Current research advances on glutamate receptors (GLRs) in plants. Acta Bot. Sin. 51, 827–840. doi: 10.11983/CBB15212
Hernández-Coronado, M., Araujo, P. C. D., Ip, P. L., Nunes, C. O., Rahni, R., Wudick, M. M., et al. (2022). Plant glutamate receptors mediate a bet-hedging strategy between regeneration and defense. Dev. Cell 57, 451–465.e6. doi: 10.1016/j.devcel.2022.01.013
Hu, Y. R., Jiang, L. Q., Wang, F., Yu, D. Q. (2013). Jasmonate regulates the inducer of CBF expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Huang, Z., Wang, Z., Li, X., He, S., Liu, Q., Zhai, H., et al. (2021). Genome-wide identification and expression analysis of JAZ family involved in hormone and abiotic stress in sweet potato and its two diploid relatives. Int. J. Mol. Sci. 22, 9786. doi: 10.3390/ijms22189786
Kang, J., Mehta, S., Turano, F. J. (2004). The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 45, 1380–1389. doi: 10.1093/pcp/pch159
Kang, S., Kim, H. B., Lee, H., Choi, J. Y., Heu, S., Oh, C. J., et al. (2006). Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Mol. Cells 21, 418–427.
Kim, S. A., Kwak, J. M., Jae, S. K., Wang, M. H., Nam, H. G. (2001). Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol. 42, 74–84. doi: 10.1093/pcp/pce008
Kohl, M., Wiese, S., Warscheid, B. (2011). Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303. doi: 10.1007/978-1-60761-987-1_18
Kong, D., Ju, C., Parihar, A., Kim, S., Cho, D., Kwak, J. M. (2015). Arabidopsis glutamate receptor homolog 3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol. 167, 1630–1642. doi: 10.1104/pp.114.251298
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Latza, A., Mehlmerb, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 6, 1274–1289. doi: 10.1093/mp/sss158
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, F., Wang, J., Ma, C., Zhao, Y., Wang, Y., Hasi, A., et al. (2013). Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol. 162, 1497–1509. doi: 10.1104/pp.113.217208
Li, T., Sun, Y., Ruan, Y., Xu, L., Hu, Y., Hao, Z., et al. (2016). Potential role of D-myo-inositol-3-phosphate synthase and 14-3-3 genes in the crosstalk between Zea mays and Rhizophagus intraradices under drought stress. Mycorrhiza 26, 879–893. doi: 10.1007/s00572-016-0723-2
Li, Y., Zhang, L., Zhu, P., Cao, Q., Sun, J., Li, Z., et al. (2019). Genome-wide identification, characterisation and functional evaluation of WRKY genes in the sweet potato wild ancestor Ipomoea trifida (H.B.K.) G. Don. under abiotic stresses. BMC Genet. 20, 90. doi: 10.1186/s12863-019-0789-x
Liu, S., Zhang, X., Xiao, S., Ma, J., Shi, W., Qin, T., et al. (2021). A single-nucleotide mutation in a glutamate receptor-like gene confers resistance to Fusarium wilt in Gossypium hirsutum. Adv. Sci. 8, 2002723. doi: 10.1002/advs.202002723
Lu, L. (2011). In silico cloning and bioinformatic analysis of TPK1 Gene in tobacco. Sci. Agric. Sin. 44, 28–35. doi: 10.3864/j.issn.0578-1752.2011.01.004
Lu, Y., Sun, J., Yang, Z., Zhao, C., Zhu, M., Ma, D., et al. (2019). Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative Ipomoea trifida. Gene 686, 177–186. doi: 10.1016/j.gene.2018.11.044
Luan, S., Wang, C. (2021). Calcium signaling mechanisms across kingdoms. Annu. Rev. Cell Dev. Biol. 37, 311–340. doi: 10.1146/annurev-cellbio-120219-035210
Ma, J., Deng, S., Jia, Z., Sang, Z., Zhu, Z., Zhou, C., et al. (2020). Conservation and divergence of ancestral AGAMOUS/SEEDSTICK subfamily genes from the basal angiosperm Magnolia wufengensis. Tree Physiol. 40, 90–107. doi: 10.1093/treephys/tpz091
Ma, Z., Gao, W., Liu, L., Liu, M., Zhao, N., Han, M., et al. (2020). Identification of QTL for resistance to root rot in sweetpotato (Ipomoea batatas (L.) Lam) with SSR linkage maps. BMC Genomics 21, 366. doi: 10.1186/s12864-020-06775-9
Ma, D. F., Li, Q., Cao, Q. H., Niu, F. X., Xie, Y. P., Tang, J., et al. (2012). Development and prospect of sweetpotato industry and its technologies in China. Jiangsu J. Agric. Sci. 28, 969–973.
Ma, R., Song, W., Wang, F., Cao, A., Xie, S., Chen, X., et al. (2019). A Cotton (Gossypium hirsutum) Myo-Inositol-1-Phosphate Synthase (GhMIPS1D) gene promotes root cell elongation in Arabidopsis. Int. J. Mol. Sci. 20, 1224. doi: 10.3390/ijms20051224
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. doi: 10.1104/pp.126.4.1646
Mayer, M. L. (2006). Glutamate receptors at atomic resolution. Nature 440, 456–462. doi: 10.1038/nature04709
Meng, Y. S., Lai, Q. X. (2019). Application and prospects of ornamental sweetpotato. J. Zhejiang Agric. Sci. 60, 2181–2184,2244. doi: 10.16178/j.issn.0528-9017.20191206
Michard, E., Lima, P. T., Borges, F., Silva, A. C., Portes, M. T., Carvalho, J. E., et al. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332, 434–437. doi: 10.1126/science.1201101
Moccia, F., Zuccolo, E., Nezza, F. D., Pellavio, G., Faris, P. S., Negri, S., et al. (2021). Nicotinic acid adenine dinucleotide phosphate activates two-pore channel TPC1 to mediate lysosomal Ca2+ release in endothelial colony-forming cells. J. Cell Physiol. 236, 688–705. doi: 10.1002/jcp.29896
Morgan, J. T., Fink, G. R., Bartel, D. P. (2019). Excised linear introns regulate growth in yeast. Nature 565, 606–611. doi: 10.1038/s41586-018-0828-1
Mukherjee, D., Saha, D., Acharya, D., Mukherjee, A., Chakraborty, S., Ghosh, T. C. (2018). The role of introns in the conservation of the metabolic genes of Arabidopsis thaliana. Genomics 110, 310–317. doi: 10.1016/j.ygeno.2017.12.003
Navarro-Retamal, C., Schott-Verdugo, S., Gohlke, H., Dreyer, I. (2021). Computational analyses of the AtTPC1 (Arabidopsis Two-Pore Channel 1) permeation pathway. Int. J. Mol. Sci. 22, 10345. doi: 10.3390/ijms221910345
Nie, N., Huo, J., Sun, S., Zuo, Z., Chen, Y., Liu, Q., et al. (2023). Genome-wide characterization of the PIFs family in sweet potato and functional identification of IbPIF3.1 under drought and Fusarium wilt stresses. Int. J. Mol. Sci. 24, 4092. doi: 10.3390/ijms24044092
Pang, X., Wei, Y., Cheng, Y., Pan, L., Ye, Q., Wang, R., et al. (2018). The tryptophan decarboxylase in Solanum lycopersicum. Molecules 23, 998. doi: 10.3390/molecules23050998
Philippe, F., Verdu, I., Morère-Le Paven, M. C., Limami, A. M., Planchet, E. (2019). Involvement of Medicago truncatula glutamate receptor-like channels in nitric oxide production under short-term water deficit stress. J. Plant Physiol. 236, 1–6. doi: 10.1016/j.jplph.2019.02.010
Pottosin, I., Dobrovinskaya, O. (2022). Major vacuolar TPC1 channel in stress signaling: what matters, K+, Ca2+ conductance or an ion−flux independent mechanism? Stress Biol. 2, 31. doi: 10.1007/s44154-022-00055-0
Qi, X., Tang, W., Li, W., He, Z., Xu, W., Fan, Z., et al. (2021). Arabidopsis G-Protein β subunit AGB1 negatively regulates DNA binding of MYB62, a suppressor in the gibberellin pathway. Int. J. Mol. Sci. 22, 8270. doi: 10.3390/ijms22158270
Rogozin, I. B., Sverdlov, A. V., Babenko, V. N., Koonin, E. V. (2005). Analysis of evolution of exon-intron structure of eukaryotic genes. Brief. Bioinform. 6, 118–134. doi: 10.1093/bib/6.2.118
Singh, S. K., Chien, C. T., Chang, I. F. (2016). The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J. Exp. Bot. 67, 1853–1869. doi: 10.1093/jxb/erv576
Singh, A., Kanwar, P., Yadav, A. K., Mishra, M., Jha, S. K., Baranwal, V., et al. (2014). Genome-wide expressional and functional analysis of calcium transport elements during abiotic stress and development in rice. FEBS J. 281, 894–915. doi: 10.1111/febs.12656
Sobolevsky, A. I., Rosconi, M. P., Gouaux, E. (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756. doi: 10.1038/nature08624
Su, Y. J., Dong, L. X., Wang, J., Dai, X. B., Zhang, A., Zhao, D. L., et al. (2018). Genetic diversity in vegetable and ornamental sweetpotato germplasm. J. Plant Genet. Res. 19, 57–64. doi: 10.13430/j.cnki.jpgr.2018.01.007
Takahashi, T., Murano, T., Ishikawa, A. (2018). SOBIR1 and AGB1 independently contribute to nonhost resistance to Pyricularia oryzae (syn. Magnaporthe oryzae) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 82, 1922–1930. doi: 10.1080/09168451.2018.1498727
Tapken, D., Anschütz, U., Liu, L. H., Huelsken, T., Seebohm, G., Becker, D., et al. (2013). A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci. Signal. 6, ra47. doi: 10.1126/scisignal.2003762
Tian, W., Wang, C., Gao, Q., Li, L., Luan, S. (2020). Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 6, 750–759. doi: 10.1038/s41477-020-0667-6
Traynelis, S. F., Wollmuth, L. P., McBain, C. J., Menniti, F. S., Vance, K. M., Ogden, K. K., et al. (2010). Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62, 405–496. doi: 10.1124/pr.109.002451
Vincill, E. D., Clarin, A. E., Molenda, J. N., Spalding, E. P. (2013). Interacting glutamate receptor-like proteins in phloem regulate lateral root initiation in Arabidopsis. Plant Cell 25, 1304–1313. doi: 10.1105/tpc.113.110668
Wan, R., Liu, J., Yang, Z., Zhu, P., Cao, Q., Xu, T. (2020). Genome-wide identification, characterisation and expression profile analysis of DEAD-box family genes in sweet potato wild ancestor Ipomoea trifida under abiotic stresses. Genes Genom. 42, 325–335. doi: 10.1007/s13258-019-00910-x
Wan, S., Wang, W., Zhou, T., Zhang, Y., Chen, J., Xiao, B., et al. (2018). Transcriptomic analysis reveals the molecular mechanisms of Camellia sinensis in response to salt stress. Plant Growth Regul. 84, 481–492. doi: 10.1007/s10725-017-0354-4
Wang, S., Song, M., Guo, J., Huang, Y., Zhang, F., Cheng, X., et al. (2018). The potassium channel FaTPK1 plays a critical role in fruit quality formation in strawberry (Fragaria × ananassa). Plant Biotechnol. J. 16, 737–748. doi: 10.1111/pbi.12824
Wang, Y., Yu, D., Zhao, H., Jiang, L., Gao, L., Song, Y., et al. (2022). A glutamate receptor-like gene is involved in ABA-mediated growth control in Physcomitrium (Physcomitrella) patens. Plant Signal. Behav. 17, e2145057. doi: 10.1080/15592324.2022.2145057
Weiland, M., Mancuso, S., Baluska, F. (2015). Signalling via glutamate and GLRs in Arabidopsis thaliana. Funct. Plant Biol. 43, 1–25. doi: 10.1071/FP15109
Wu, S., Lau, K. H., Cao, Q., Hamilton, J. P., Sun, H., Zhou, C., et al. (2018). Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 9, 4580. doi: 10.1038/s41467-018-06983-8
Wudick, M. M., Michard, E., Nunes, C. O., Feijó, J. A. (2018a). Comparing plant and animal glutamate receptors: common traits but different fates? J. Exp. Bot. 69, 4151–4163. doi: 10.1093/jxb/ery153
Wudick, M. M., Portes, M., Michard, T. E., Rosas-Santiago, P., Lizzio, M. A., Nunes, C. O., et al. (2018b). CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 360, 533–536. doi: 10.1126/science.aar6464
Xiang, C., Shen, S. F., Ji, Z. X., Li, B., Wu, L. H. (2020). Pedigree and quality traits of Zheshu sweetpotato varieties for table use and food processing use. J. Nucl. Agric. Sci. 34, 36–44. doi: 10.11869/j.issn.100-8551.2020.01.0036
Xie, Y. Z., Guo, X. D., Jia, Z. D., Ma, P. Y., Bian, X. F., Yu, Y. (2018). Progresses and prospects on edible sweetpotato breeding in China. Jiangsu J. Agric. Sci. 34, 1419–1424. doi: 10.3969/j.issn.1000-4440.2018.06.030
Yan, M., Chen, M., Zhang, W., Hu, Y., Lu, Y., He, S., et al. (2022). Identification and expression characteristics analysis of GLRs gene family of Erigeron breviscapus. Mol. Plant Breed. 20, 2842–2853. doi: 10.13271/j.mpb.020.002842
Yang, J., Moeinzadeh, M. H., Kuhl, H., Helmuth, J., Xiao, P., Haas, S., et al. (2017). Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 3, 696–703. doi: 10.1038/s41477-017-0002-z
Yang, L., Zhao, Y., Wu, X., Zhang, Y., Fu, Y., Duan, Q., et al. (2022). Genome wide identification and expression analysis of BraGLRs reveal their potential roles in abiotic stress tolerance and sexual reproduction. Cells 11, 3729. doi: 10.3390/cells11233729
Yu, Y., Assmann, S. M. (2018). Inter-relationships between the heterotrimeric Gβ subunit AGB1, the receptor-like kinase FERONIA, and RALF1 in salinity response. Plant Cell Environ. 41, 2475–2489. doi: 10.1111/pce.13370
Yu, B., Wu, Q., Li, X., Zeng, R., Min, Q., Huang, J. (2022). GLUTAMATE RECEPTOR-like gene OsGLR3.4 is required for plant growth and systemic wound signaling in rice (Oryza sativa). New Phytol. 233, 1238–1256. doi: 10.1111/nph.17859
Zeng, H., Zhao, B., Wu, H., Zhu, Y., Chen, H. (2020). Comprehensive in silico characterization and expression profiling of nine gene families associated with calcium transport in soybean. Agronomy 10, 1539. doi: 10.3390/agronomy10101539
Zhang, J., Cui, T., Su, Y., Zang, S., Zhao, Z., Zhang, C., et al. (2022). Genome-wide identification, characterization, and expression analysis of glutamate receptor-like gene (GLR) family in sugarcane. Plants (Basel). 11, 2440. doi: 10.3390/plants11182440
Zhang, T., Xu, P., Wang, W., Wang, S., Caruana, J. C., Yang, H., et al. (2020). Corrigendum: Arabidopsis G-protein β subunit AGB1 interacts with BES1 to regulate brassinosteroid signaling and cell elongation. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01122
Zhang, H., Zhang, Q., Zhai, H., Li, Y., Wang, X., Liu, Q., et al. (2017). Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci. Rep. 7, 40819. doi: 10.1038/srep40819
Zheng, Y., Luo, L. D., Wei, J. J., Chen, Q., Yang, Y. P., Hu, X. Y., et al. (2018). The glutamate receptors AtGLR1.2 and AtGLR1.3 increase cold tolerance by regulating jasmonate signaling in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 506, 895–900. doi: 10.1016/j.bbrc.2021.06.061
Zhou, Z. L., Tang, J., Cao, Q. H., Zhao, D. L., Zhang, A. (2020). Formation laws of quality characters in starch sweetpotato cultivars and its correlation with main agronomic characters. Jiangsu J. Agric. Sci. 36, 277–283. doi: 10.3969/j.issn.1000-4440.2020.02.004
Zhou, S., Zhang, L., Lv, Q., Huang, J. (2021). Identification and analysis of GLR family genes in maize. J. Maize Sci. 29, 35–42. doi: 10.13597/j.cnki.maize.science.20210206
Zhu, P., Dong, T., Xu, T., Kang, H. (2020). Identification, characterisation and expression analysis of MADS-box genes in sweetpotato wild relative Ipomoea trifida. Acta Physiol. Plant 42, 163. doi: 10.1007/s11738-020-03153-6
Zhu, S., Gouaux, E. (2017). Structure and symmetry inform gating principles of ionotropic glutamate receptors. Neuropharmacology 112, 11–15. doi: 10.1016/j.neuropharm.2016.08.034
Keywords: glutamate receptor, tissue-specific expression, root rot stress, abiotic stress, sweet potato, Ipomoea trifida, Ipomoea triloba
Citation: Hu Y, Dai Z, Huang J, Han M, Wang Z, Jiao W, Gao Z, Liu X, Liu L and Ma Z (2023) Genome-wide identification and expression analysis of the glutamate receptor gene family in sweet potato and its two diploid relatives. Front. Plant Sci. 14:1255805. doi: 10.3389/fpls.2023.1255805
Received: 09 July 2023; Accepted: 06 November 2023;
Published: 21 December 2023.
Edited by:
Meng Kou, Xuzhou Institute of Agricultural Sciences in Jiangsu Xuhuai District, ChinaReviewed by:
Lei Hua, Peking University, ChinaCopyright © 2023 Hu, Dai, Huang, Han, Wang, Jiao, Gao, Liu, Liu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhimin Ma, c3AxMzc4NTE4MTI1OUAxNjMuY29t; Lanfu Liu, bGxmNjcxMkAxNjMuY29t; Xinliang Liu, bGl1eGlubGlhbmcwNTU4QHNpbmEuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.