- 1Crop Improvement Section, Plant Breeding, ICAR-Indian Institute of Rice Research (ICAR-IIRR), Hyderabad, India
- 2Genetics and Plant Breeding, Sugarcane Research Station, Vuyyuru, India
- 3Crop Production Section, Agronomy, ICAR-Indian Institute of Rice Research (ICAR-IIRR), Hyderabad, India
- 4Crop Diversification and Genetics Section, International Center for Biosaline Agriculture (ICBA), Dubai, United Arab Emirates
Salt stress is the second most devastating abiotic stress after drought and limits rice production globally. Genetic enhancement of salinity tolerance is a promising and cost-effective approach to achieve yield gains in salt-affected areas. Breeding for salinity tolerance is challenging because of the genetic complexity of the response of rice plants to salt stress, as it is governed by minor genes with low heritability and high G × E interactions. The involvement of numerous physiological and biochemical factors further complicates this complexity. The intensive selection and breeding efforts targeted towards the improvement of yield in the green-revolution era inadvertently resulted in the gradual disappearance of the loci governing salinity tolerance and a significant reduction in genetic variability among cultivars. The limited utilization of genetic resources and narrow genetic base of improved cultivars have resulted in a plateau in response to salinity tolerance in modern cultivars. Wild species are an excellent genetic resource for broadening the genetic base of domesticated rice. Exploiting novel genes of underutilized wild rice relatives to restore salinity tolerance loci eliminated during domestication can result in significant genetic gain in rice cultivars. Wild species of rice, Oryza rufipogon and Oryza nivara, have been harnessed in the development of a few improved rice varieties like Jarava and Chinsura Nona 2. Furthermore, increased access to sequence information and enhanced knowledge about the genomics of salinity tolerance in wild relatives has provided an opportunity for the deployment of wild rice accessions in breeding programs, while overcoming the cross-incompatibility and linkage drag barriers witnessed in wild hybridization. Pre-breeding is another avenue for building material that are ready for utilization in breeding programs. Efforts should be directed towards systematic collection, evaluation, characterization, and deciphering salt tolerance mechanisms in wild rice introgression lines and deploying untapped novel loci to improve salinity tolerance in rice cultivars. This review highlights the potential of wild relatives of Oryza to enhance tolerance to salinity, track the progress of work, and provide a perspective for future research.
1 Introduction
Cultivated rice, which primarily includes Oryza sativa (Asian cultivated rice) and Oryza glaberrima (African cultivated rice), contains 22 wild species that are not cultivated (Solis et al., 2020). O. sativa is cultivated worldwide, whereas O. glaberrima, is predominantly grown in Africa. Wild rice ancestors have adapted to various geographically distinct habitats (Atwell et al., 2014). The 22 wild ancestors constituted the largest gene pool. Oryza species are highly variable and comprise 11 distinct genomes, including six diploids (AA, BB, CC, EE, FF, and GG) and five allotetraploids (BBCC, CCDD, KKLL, HHJJ, and HHKK) (Stein et al., 2018). They differ in morphological characteristics such as growth habit, plant height, flowering, leaf size, panicle size, and branching, awning, and seed size.
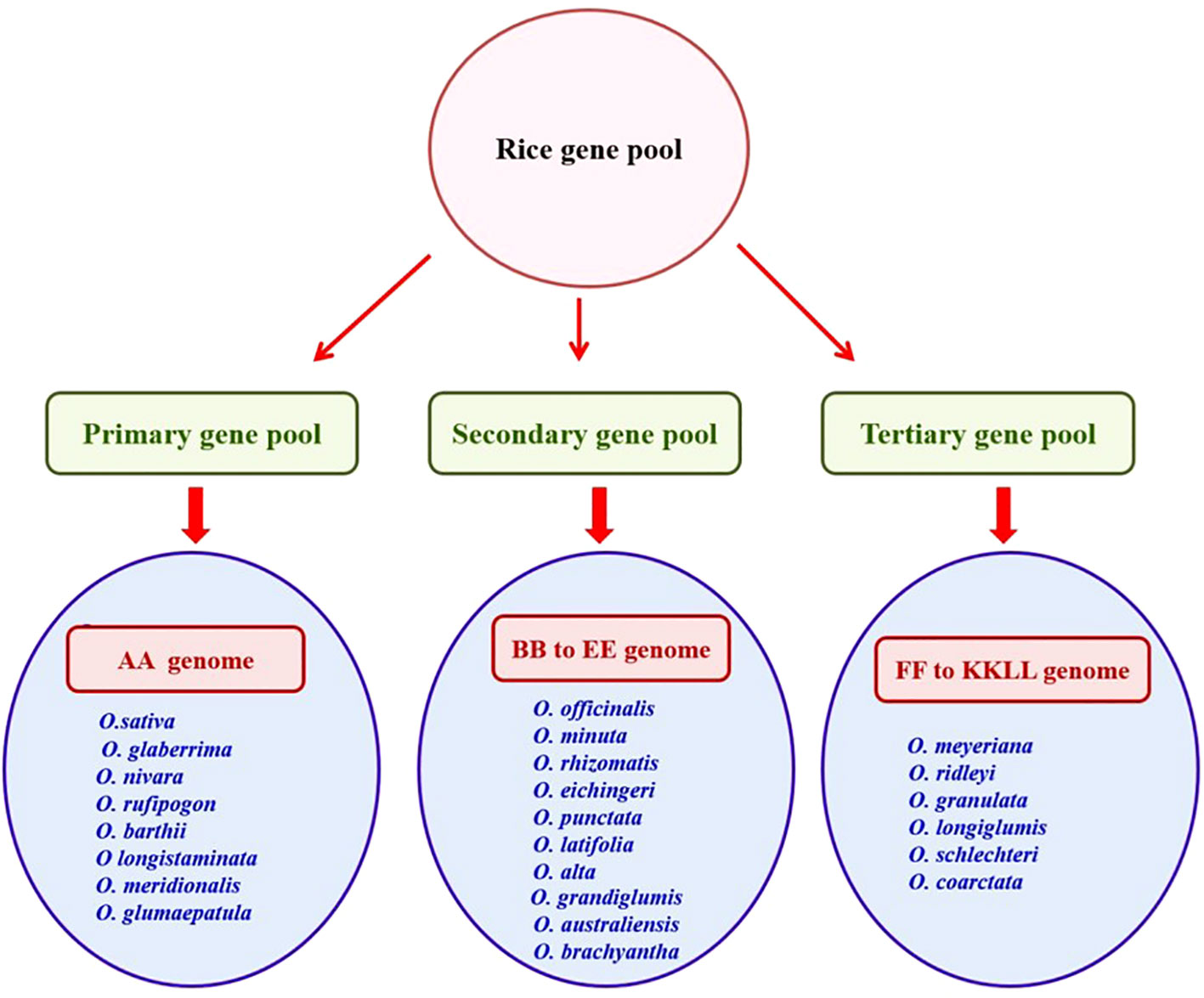
Wild relatives of rice are grouped into three gene pools (primary, secondary, and tertiary) based on their ease of hybridization with cultivated rice and phylogenetic relationships (Harlan and de Wet, 1971). The primary gene pool comprises the O. sativa complex (AA genome) with close relatives of cross-compatible rice and a secondary gene pool consisting of O. officinalis complex (BB to FF genomes) with less closely related species and a tertiary gene pool constituted with O. meyeriana complex, O. ridleyi complex, and O. schlechteria complex (GG to KKLL genomes) with more distant relatives of rice, necessitating embryo rescue, chromosome doubling, or bridging species to facilitate gene transfer (Solis et al., 2020) (Figure 1).
Studies have revealed that indigenous species of rice could potentially be used as genetic resources for abiotic stresses such as submergence (Luo et al., 2020; Sasayama et al., 2022) drought (Hamzelou et al., 2020; Anilkumar et al., 2023), heat (Prasanth et al., 2012; Khan et al., 2019; Jagadish et al., 2021), etc. However, few limited published studies have reported the significance of wild rice species in elevating salinity tolerance in cultivars (Solis et al., 2020).
Indeed, very few review articles have focused on the genetic enhancement of salinity tolerance in wild O. sativa species. Hence, in this manuscript, we have reviewed the impact of salinity effects in rice, the importance of wild rice species in breeding for salinity tolerance, characterization and mechanisms of tolerance in wild rice, deployment of wild rice genes in rice breeding, and challenges and opportunities for the incorporation of wild gene loci to develop salt-resilient rice cultivars.
2 Importance of wild species in rice breeding
Wild progenitors adapted to various changing climatic conditions display tremendous genetic diversity and carry genetic loci associated with unique agronomic and adaptive traits (Hellwig et al., 2021). The basic requirement of a successful crop breeding program is the availability and accessibility of genetic variability in target traits (Perrino and Perrino, 2020). When there is exhaustion in genetic variation among domesticated genotypes, breeders are left with access the diversity available in land races and wild ancestors as alternate sources of variability.
Asian cultivated rice has evolved from its wild ancestor Oryza rufipogon through human efforts (Wang et al., 1992). Similarly, the African rice O. glaberrima was domesticated from the African wild progenitor Oryza barthii. Erect growth, non-shattering, and increased grain number and weight were targeted during selective breeding. The intense selection and breeding that occurred during domestication could have probably resulted in the loss of genetic variability and the loci associated with stress tolerance in cultivars. O. sativa represents <20% of the total variation compared to that found in various wild species (Stein et al., 2018), and significant natural variability for salt tolerance still exists in wild species (Munns et al., 2016), which can be exploited to improve cultivated rice. There are certain wild species such as Oryza coarctata, a distantly related wild rice that thrives under extreme salinity (450 mM NaCl). However, the crossability barrier limits their utility.
Owing to tall plant stature, photosensitivity, poor grain type, low seed set, high grain shattering, and low yield potential, natural wild accessions are regarded as poor agronomic performers (Sanchez et al., 2013). Despite this, the recovery of widely adaptable cultivars to diverse challenging environments would be higher when wild relatives of rice are used in the crossing program (Jin et al., 2021). Insights into morphological and physiological responses to salt stress are crucial for exploiting stress tolerance through distant crosses. The extent to which wild rice progenitors demonstrate tolerance and the probable genetic defense strategies behind tolerance remains partially understood. Crossing barriers, linkage drag, and epistatic effects from unadapted wild genes when introgressed into elite cultivars may complicate mainstream breeding. However, advances in genomic tools and techniques, particularly embryo rescue techniques and deploying molecular marker-based advanced backcrosses, identification, selection, and incorporation of target quantitative trait loci (QTLs) into elite varieties, could successfully generate improved versions of cultivars (Dai et al., 2022).
The genetic wealth of rice diversity was systematically preserved in gene banks. As of December 2023 (URL-https://www.irri.org/international-rice-genebank), the International Rice Gene Bank Collection Information System (IRGCIS) of the International Rice Research Institute (IRRI) documented 1,32,000 accessions of both cultivated and wild rice. Nevertheless, only a few wild accessions have been evaluated using various salt-screening methods (Kumar et al., 2015). The primary gene pool consisting of cultivated rice species (O. sativa and O. glaberrima) and wild rice species (Oryza nivara, O. rufipogon, O. barthii, Oryza longistaminata, Oryza meridionalis, and Oryza glumaepatula) are analyzed for detecting tolerant wild accessions in limited research investigations. A large untapped genetic diversity is available in the secondary and tertiary gene pools, which are additional sources of new salinity tolerance genes. These gene pools can be used to introduce beneficial genes into superior rice varieties (Prusty et al., 2018; Solis et al., 2020).
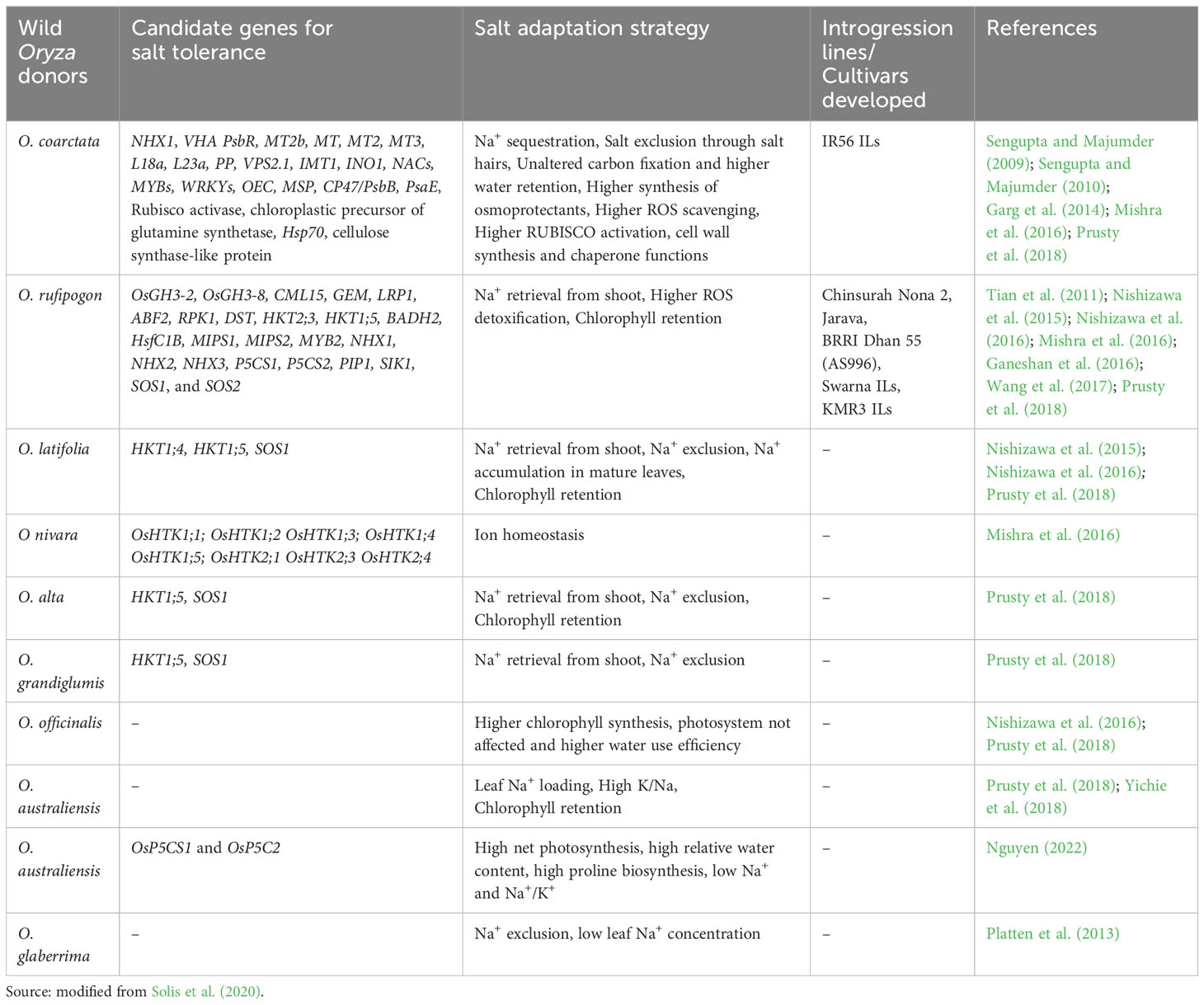
Over the past three decades, there has been substantial advancement in the genetic diversity analysis of wild rice resources and the selection of suitable donor parents. Following the standard Yoshida solution culture method (Gregorio et al., 1997) and the modified Yoshida solution culture method (Singh et al., 2010), several germplasm accessions were screened at the seedling stage to identify the donors. A few accessions of O. rufipogon and O. nivara that survived salinity levels as high as 12 dSm−1 (Habiba et al., 2015) were identified. O. rufipogon is considered the best donor among the closest wild relatives belonging to the AA genome, followed by the distant wild relatives O. coarctata, O. latifolia, and Oryza alta (Solis et al., 2020).
At the ICAR-National Institute of Plant Biotechnology, India, 800 accessions of O. nivara and O. rufipogon collected across ecologies were evaluated (Singh et al., 2018). The accession NKSWR 173 recorded high seedling stage salt tolerance (150 mM NaCl) (Mishra et al., 2016). A survey of 22 accessions of wild species screened under high salinity (240 mM NaCl) in a hydroponic system revealed seven accessions, Oryza minuta, Oryza grandiglumis, Oryza latifolia, O. alta, Oryza rhizomatis, O. coarctata, and Oryza eichingeri, with higher levels of tissue tolerance and chlorophyll preservation in leaves as compared to the donor parents for salinity tolerance(Pokkali, Nona Bokra, and FL478) and salt-sensitive checks (IR29 and IR75862-206-2-8-3) (Prusty et al., 2018). Mondal et al. (2018) and Mishra et al. (2022) studied the halophytic wild species O. coarctata abundant in Indian coastal regions as a source of salinity tolerance genes.
In the past two decades, genome sequencing technology has improved the utilization of genetic variation in wild Oryza species for crop improvement. Efforts to sequence wild rice genomes began in 2003 with the establishment of the International Oryza Map Alignment Project (IOMAP). It has provided an in-depth characterization of wild rice genomes to discover and exploit genes/genomic regions governing diverse traits for their transfer into cultivated rice (Wing et al., 2005). Stein et al. (2018) sequenced genomes of seven wild species (O. rufipogon, O. nivara, O. barthii, O. glumaepatula, O. meridionalis, O. punctata, and L. perrieri) and deposited at National Center for Biotechnology Information (NCBI), Bethesda, Maryland and provided an array of diversity panels. The genome of O. coarctata has been sequenced at ICAR-NIPB, India, which would be a valuable addition to the I-OMAP project and would supplement the current genomic resources of wild and cultivated species (Mondal et al., 2018). This provides insight into the genetic makeup of O. coarctata and broadens the gene pool for the enhancement of cultivated rice. Multiple studies have employed whole genome sequencing and resequencing information from cultivated and wild rice population species to identify genomic regions harboring improved agronomic traits as well as adaptive traits, such as resistance to biotic stresses and tolerance to abiotic stresses, including salinity tolerance (Huang et al., 2012; Xu et al., 2012).
3 Impact of salt stress on rice
Rice is the main nutritional source for more than half of the world’s population (Zhou et al., 2020). Globally, rice is the third-largest cereal grown in 162 million hectares, with an annual production of 755 million tons (FAO, 2022). Soil salinity and alkalinity are the leading abiotic stressors in coastal and inland areas, followed by drought (Islam et al., 2021). Even moderate salt stress has been reported to reduce the rice yield by 68% (Naheed et al., 2007). The changing global climate is expected to cause a significant increase in soil salinization owing to inadequate rainfall, high evaporation rates, and seawater intrusion (Cheng et al., 2020).
Salt-affected soils are broadly classified as sodic–alkaline, saline, or saline–sodic soils (Eynard et al., 2005). In alkaline soils, carbonates and bicarbonates of sodium and magnesium are the most prevalent anions, whereas chlorides and sulfates of sodium and magnesium are frequently found in saline soils. Sodic soils have pH >8.5, the electrical conductivity of saturation extract (ECe) <4 dS m−1 and exchangeable sodium percentage (ESP) >15. Saline soils near the coastal regions, ECe >4 dS m−1, pH < 8.5, and ESP <15. Saline sodic soils exhibit saline and sodic characteristics, such as variable pH, ECe ≥4 dS m−1, and ESP ≥15%.
Plants are categorized into halophytes and glycophytes based on their responses to salt stress, (Flowers and Colmer, 2008; Mishra and Tanna, 2017). Halophytes thrive in highly saline soils. Most of the crops are salt-sensitive glycophytes. Rice has a threshold salt concentration of >30 mM NaCl (ECe = 3 dS m−1) (USDA et al., 2008). A decrease in yield of 12% was reported for each unit, exceeding the specified threshold (Reddy et al., 2014). Excessive salinity causes osmotic stress and ion toxicity in crop plants. Osmotic pressure reduces soil osmotic potential and causes decreased water uptake, further inhibiting stomatal opening, photosynthesis, elongation, and cell proliferation. This results in a slower growth. Ionic stress causes rapid accumulation of toxic Na+ and Cl−, disrupting metabolic processes, resulting in early senescence and reduced stomatal conductance, leading to decreased photosynthesis, biomass, and poor yield (Yu et al., 2017). Salinity stress, apart from reducing germination, causes whitening of affected leaf tips, leaf rolling, stunted plant growth, patchy appearance in the field, reduced tillering, delayed panicle emergence, length and the number of panicles, reduces pollen viability, spikelet fertility, spikelet number/panicle, and ultimately grain yield, and in cases of increased severity, results in the death of the rice plant (Figure 2).

Figure 2 Rice genotypes showing field symptoms of alkalinity stress grown at Chandra Shekhar Azad University of Agriculture & Technology (CSAUAT), Kanpur, India.
Plants adapt to high salt through various physiological and biochemical defense systems namely high initial seedling vigor, early maturity, sodium exclusion, sodium compartmentalization in vacuoles of roots and older leaves, osmotic adjustment, control of Reactive Oxygen Species (ROS) through production of antioxidants, and programmed cell death.
During the domestication of cultivated rice from local landraces and traditional varieties, selection pressure for productivity traits favored a few genes at the expense of many others. Hence, domesticated varieties have less genetic diversity for other non-selected traits, such as biotic and abiotic stresses, including salinity tolerance, compared to wild species and landraces. This implies that the cultivated gene pool has a narrow genetic base; hence, further gain in salinity tolerance is difficult to achieve. Consequently, the developed cultivars were found to be salt sensitive or moderately tolerant to salinity. Interestingly, wild rice gene pools possess extensive genetic diversity as they grow in undisturbed natural habitats. This diversity can enrich the cultivated gene pool with higher salt tolerance through introgression of wild genes. Salt stress profoundly affects various morphological, physiological, and biochemical processes in rice plants (Table 1).
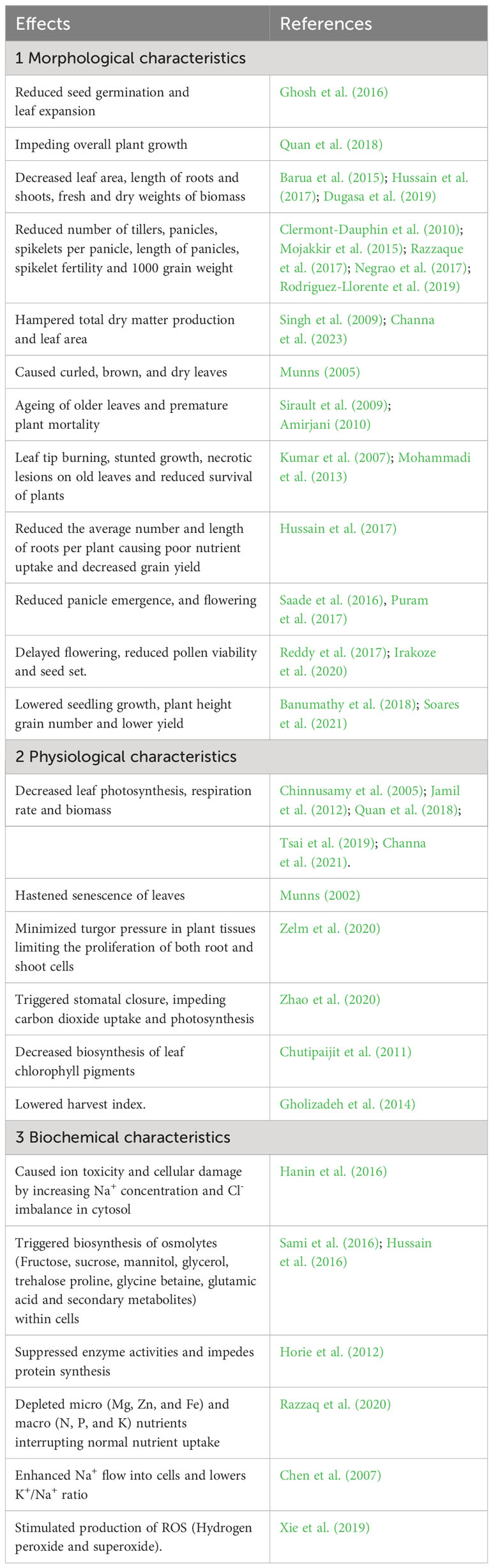
Table 1 Effects of salt stress on morphological, physiological, and biochemical characteristics in rice.
4 Defense strategies of salt tolerance in rice
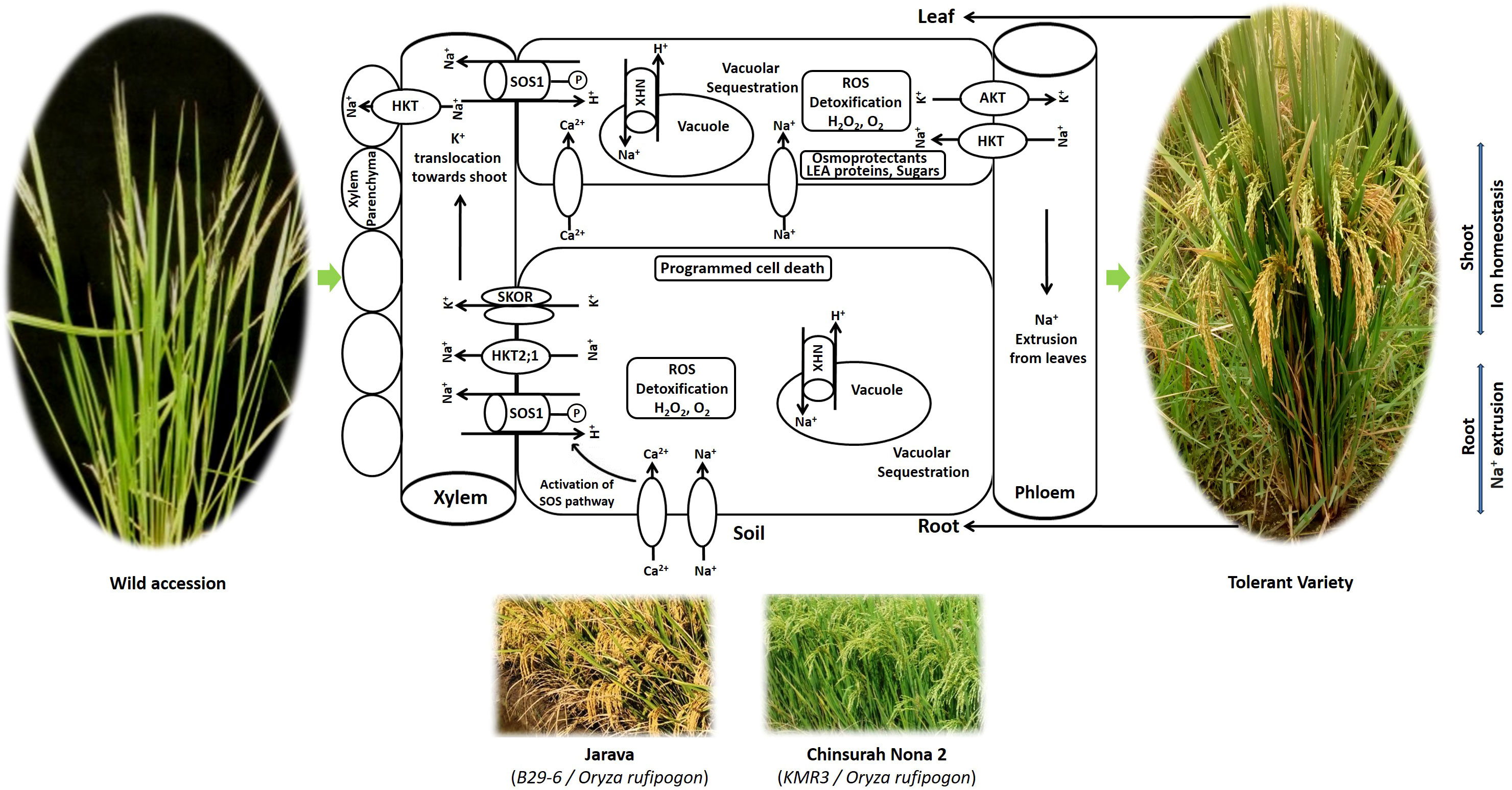
Plants have evolved protective mechanisms at the cellular, organellar, and whole plant levels to recover from salinity stress. Rice cultivars exhibit genetic variation in their adaptive strategies against salinity (Khan et al., 2020). The crop has developed a series of adaptive mechanisms, including (i) osmotic adjustment, (ii) compartmentation and ion homeostasis, (iii) antioxidant defense, and (iv) programmed cell death (Figure 3).
4.1 Osmotic adjustment by accretion of osmolytes
Plants have less access to water and nutrients under osmotic stress. To save water and minimize transpiration, which causes Na+ ion inflow from the roots to the shoots, plants respond by closing their stomata (Flowers and Flowers, 2005). Osmotic stress builds up Na+ in the leaves and reduces growth (Afrasyab et al., 2010). Plants respond to salinity-induced osmotic stress by synthesizing osmoprotectants in the cytoplasm, such as quaternary amino acid derivatives (proline and glycine betaine), sugars (glucose, fructose, and trehalose), sugar alcohols (glycerol and methylated inositols), and late embryogenesis-abundant (LEA) proteins to maintain high cytosolic osmotic adjustment (Horie et al., 2012). The proline synthesis genes OsP5CS1 and OsP5CR enhance proline biosynthesis and salt tolerance (Sripinyowanich et al., 2013). Glycine betaine, encoded by OsCMO and OsBADH1, accumulates in rice when exposed to high salt (Tang et al., 2014). OsSWEET13 and OsSWEET1 regulate sugar homeostasis in rice under saline conditions (Mathan et al., 2020). Salt and drought tolerance was considerably increased by the LEA genes OsLEA3-2, OsLEA4, OsLEA5, and OsEm1 (Duan and Cai, 2012).
4.2 Compartmentation and ion homeostasis or tissue tolerance
Tolerant plants minimize the initial entry of Na+, restrict Na+ movement in the xylem, induce outflow of Na+ in the soil, enhance the absorption of K+, and regulate the Na+/K+ ratio to protect the leaves from ion toxicity (Munns and Tester, 2008). In rice, plasma membrane-based histidine kinase transporters (HKTs) modulate the accumulation of Na+ in the cytosol. They either absorb sodium from the soil solution or remove sodium from the xylem sap and load Na+ into the phloem sap to reduce sodium accumulation in leaves,when K+ is limited (Campbell et al., 2017). In the Saltol region of FL478, a Pokkali derivative and a sodium transporter gene, OsHKT1;5, mediate Na+ exclusion in rice (Hauser and Horie, 2010).
The capacity of a tissue to function normally while containing a high internal concentration of Na+ is known as tissue tolerance. It involves the sequestration of excessive Na+ from the cytoplasm into the vacuoles of non-functional older leaves and leaf sheaths, and enzymatic detoxification of reactive oxygen species. Selective uptake of Na+ into vacuoles is performed by four vacuolar Na+/H+ transporters (OsNHX1, OsNHX2, OsNHX3, and OsNHX4). Thus, plants maintain a high tissue K+/Na+ ratio, which prevents cytosolic Na+ toxicity (Wu et al., 2020). Excess Na+ is pumped out from the root xylem by a salt overly sensitive 1 (SOS1) transporter, the plasma membrane Na+/H+ antiporter, and OsNHX1 and OsSOS1 (Kumar et al., 2013) and OsTPC1 (Kurusu et al., 2012), which contribute to ion homeostasis. Tissue tolerance also involves the biosynthesis of compatible solutes and the formation of enzymes, such as superoxide dismutase and catalase, which detoxify ROS and conserve cell size and turgor (Munns et al., 2016).
4.3 Defense against oxidate damage through detoxification of reactive oxygen species
Excessive reactive oxygen species (ROS) are formed in chloroplasts, peroxisomes, and mitochondria under salt stress. This causes oxidative damage to lipids, proteins in cell membranes, enzymes, and nucleic acids, and death in plants (Arif et al., 2020). Rice plants overproduce enzymatic antioxidants, viz., glutathione peroxidase, superoxide dismutase, catalase, and ascorbate peroxidase, and non-enzymatic antioxidants, such as proline, glycine betaine, glutathione, and polyphenols, to protect the cell against oxidative damage (Kim et al., 2017; Meng et al., 2018).
4.4 Programmed cell death
Plants adopt PCD to destroy excess or injured cells and prevent sodium influx into the shoots and roots. Reports have suggested that the upregulation of genes linked to PCD in rice increases salinity tolerance (Hoang et al., 2015). Cell death in rice roots during salinity stress was found to occur in a highly regulated manner (Liu et al., 2019).
Other tolerance mechanisms manifest through early vigorous growth to avoid salt toxicity and enhanced stomatal closure in rice (Kumar et al., 2013). The osmotic tolerance phase has not been much researched in rice compared to ionic stress. Tolerant rice accessions possess only a few of these mechanisms. Pooling the superior genes associated with adaptive mechanisms governing low Na+ uptake, high K+ uptake, Na+ sequestration, reduced transpiration, and synthesis of osmolytes to develop highly tolerant elite cultivars is urgently needed.
5 Overview of morphological, physiological, genetic, and molecular mechanisms underlying salt tolerance in wild rice
Wild species have evolved stronger morphological, physiological, genetic, and molecular adaptive mechanisms than cultivars have. They exhibit sodium secretion through special structures such as salt glands, improved ion homeostasis in roots and shoots, increased osmolyte biosynthesis, strong tissue tolerance, greater detoxification of ROS, and enhanced osmotic tolerance compared to cultivars. Several wild rice donors with increased tolerance to salinity have been, i.e., O. coarctata (Majee et al., 2004; Sengupta and Majumder, 2010; Garg et al., 2014), O. punctata, O. officinalis (Prusty et al., 2018), and O. rufipogon (Tian et al., 2011; Zhou et al., 2016).
Nakamura et al. (2002) reported higher seedling survival and high photosynthetic rates in O. latifolia and O. rufipogon up to 300 mM NaCl concentration than cultivated salt tolerant SR26-B and salt sensitive IR28. O. coarctata, earlier known as Porteresia coarctata, an allotetraploid wild rice halophyte with KKLL genome (2n = 4x = 48), is the only distant rice which could withstand extreme salinity (500 mM to 650 mM NaCl) as reported by Sengupta et al. (2008). It is a unique wild rice that grows in mangroves along coastal belts. It exhibits multiple defense mechanisms of tolerance and mainly adopts a salt excretion strategy to reduce the high salt load in photosynthetic tissues (Sengupta and Majumder, 2010), thereby retaining a high photosynthetic rate. It contains characteristic hairs known as trichomes on the upper surface of the leaves, through which sodium and chloride salts are excreted (Prusty et al., 2018). It deploys vacuolar Na+ compartmentation mediated by Na+ transporters (OcNHX1, OcSOS1, OcHKT1;4, and OcHKT1;5) to maintain a low cytosolic Na+/K+ ratio in the leaf mesophyll, even though it continues to grow in saline water. Despite high salinity, it sustains a low water content (Senthilkumar et al., 2005). Additionally, it possesses a greater Na+ and K+ retention capacity in shoots than in roots and eliminates ROS through enzymatic processes (Sengupta and Majumder, 2009).
A study conducted by Platten et al. (2013) using 103 O. sativa and 12 O. glaberrima accessions indicated the operation of ion homeostasis mechanisms in O. glaberrima through salt accumulation in leaves, which was independent of OsHKT1;5, as observed in O. sativa. Pushpalatha et al. (2013) evaluated 15 O. nivara and O. rufipogon introgression lines in the background of KMR3 and Swarna cultivars at various salinity levels (0 mM to 200 mM NaCl) during germination, vegetative, and reproductive growth stages and study revealed a Na+ exclusion mechanism and osmoprotection by proline synthesis in KMR3 ILs (K463 and K478), vacuolar sodium sequestration, and consistent chlorophyll content in Swarna ILs (S166, S3-1, S14, and S75) in response to salt stress.
Nishizawa et al. (2015), (Nishizawa et al., 2016) reported similar constant photosynthetic rates and sodium accumulation in O. officinalis and O. latifolia accessions. However, a relationship between stomatal conductance and net photosynthetic rate could not be demonstrated in O. latifolia compared to O. officinalis, indicating the possibility of the presence of a new set of salinity tolerance loci. O. rufipogon and O. nivara have been reported to possess a pool of genes that maintain K+ homeostasis, Na+ exclusion, and sodium compartmentalization (Ganeshan et al., 2016; Tian et al., 2017). Another wild relative, O. australiensis recorded proline accumulation, low Na+ content, and a low Na+/K+ ratio in the shoots and roots (Yichie et al., 2018).
Prusty et al. (2018) investigated 22 wild species in a hydroponic experiment along with four cultivated tolerant checks (Nona Bokra, Pokkali, and FL478) and two sensitive checks (IR 29 and IR 75862-206-2-8-3) under high saline stress (240 mM NaCl). Two wild species, O. latifolia and O. alta, survived up to 26 days to 33 days, and O. coarctata grew without deleterious effects. Interestingly, O. eichingeri, O. minuta, and O. coarctata accumulated high Na+ content in their roots and had low oxidative damage. Gene expression studies suggested the involvement of the OsHKT1;4 transporter gene for mediating Na+ exclusion in leaves, but Na compartmentation occurred independently of the tonoplast-localized OsNHX1 transporter gene. Shahzad et al. (2022) attributed the tolerance in wild rice genotypes of O. alta and O. barthii, to a greater tissue tolerance mechanism. Orthologous alleles of the stress-responsive VOZ gene from wild species, viz., O. brachyantha, O. longistaminata, and O. nivara, could act as potential donors for salinity stress improvement (Ganie et al., 2020). Nan et al. (2020) identified activation of OrWRKY genes in O. rufipogon under salt stress.
Nguyen (2022) investigated physiological traits governing salt tolerance in 18 Australian wild rice accessions of O. australiensis, O. rufipogon, and O. meridionalis under high salinity (200 mM NaCl) together with three cultivars, namely IR 29, salt sensitive and Pokkali, salt tolerant. O. australiensis accessions displayed high net photosynthesis, high relative water content, and low Na+ and Na+/K+ in the shoots and roots. Gene expression analysis revealed upregulation of proline synthesis genes OsP5CS1 and OsP5C2, and downregulation of the proline degradation gene OsProDH. Thus, osmoregulation and ion homeostasis are the key tolerance mechanisms in O. australiensis accessions. Solis et al. (2022) demonstrated higher Na+ uptake and reduced Na+ effluxes in O. alta, O. latifolia, and O. coarctata. The expression of NHX1 and SOS1/NHX7 genes that govern tissue tolerance triggered by salt stress. Detailed information on the candidate genes and their mechanisms are listed in Table 2.
Adaptive mechanisms may occur singly or in combination, depending on the species and growth stage of the crop. Different accessions of the same wild Oryza species could have a distinct genetic basis for phenotypic expression. With better knowledge of the mechanisms in wild species, appropriate breeding methods could be formulated to enhance yield under salt stress than what is currently obtained in studies involving O. sativa donors.
6 Approaches to improve salt tolerance in rice
Tolerance to salinity is a complex polygenic trait linked to several morphological and physiological traits, with huge environmental influence and poor heritability (Gregorio and Senadhira, 1993; Flowers, 2004). Based on an earlier standard hydroponic system of screening (Gregorio et al., 1997) and the modified Yoshida culture-based method (Singh et al., 2010), there have been several investigations in screening the germplasm of O. sativa subspecies indica and japonica (Negrao et al., 2011) to identify donors. In rice, three major strategies, namely conventional breeding, molecular breeding, and genetic engineering, were employed to generate superior salinity-tolerant cultivars.
6.1 Traditional breeding
Breeders have employed introduction, hybridization, pedigree selection, bulk method, modified bulk pedigree, recurrent selection, backcross method, and induced mutations to adapt to salt stress. Early breeding efforts have focused on improving locally domesticated landraces using pure line selection. Notable among them are Pokkali, Nona Bokra, Bhura Rata, and Kalarata. Globally, Pokkali is the most extensively utilized donor because it maintains a low shoot Na+/K+ ratio with tissue tolerance under high salinity. Unfortunately, linkage drag contribute to poor yield and grain quality, and the late maturity of landraces is often brought into new cultivars (Solis et al., 2020).
Approximately 101 salinity-tolerant rice cultivars developed using conventional breeding techniques have been developed worldwide (Singh et al., 2021). However, many of them are only moderately tolerant to salinity and not during all growth stages of the crop. The complex inheritance, pleiotropy, and high G × E interactions of salt-tolerance traits hinder traditional breeding efforts (Cohen and Leach, 2019). It is vital to breed rice varieties that can withstand high salt levels without compromising the yield. Under these conditions, the marker-assisted introgression approach shows more promise in the rapid development of tolerant cultivars and in lowering the risk of unwanted linkage drag with the negative traits of wild species and landraces. This has increased the need for the integration of molecular breeding techniques into the breeding process (Ismail et al., 2007; Thomson et al., 2010).
6.2 Molecular breeding
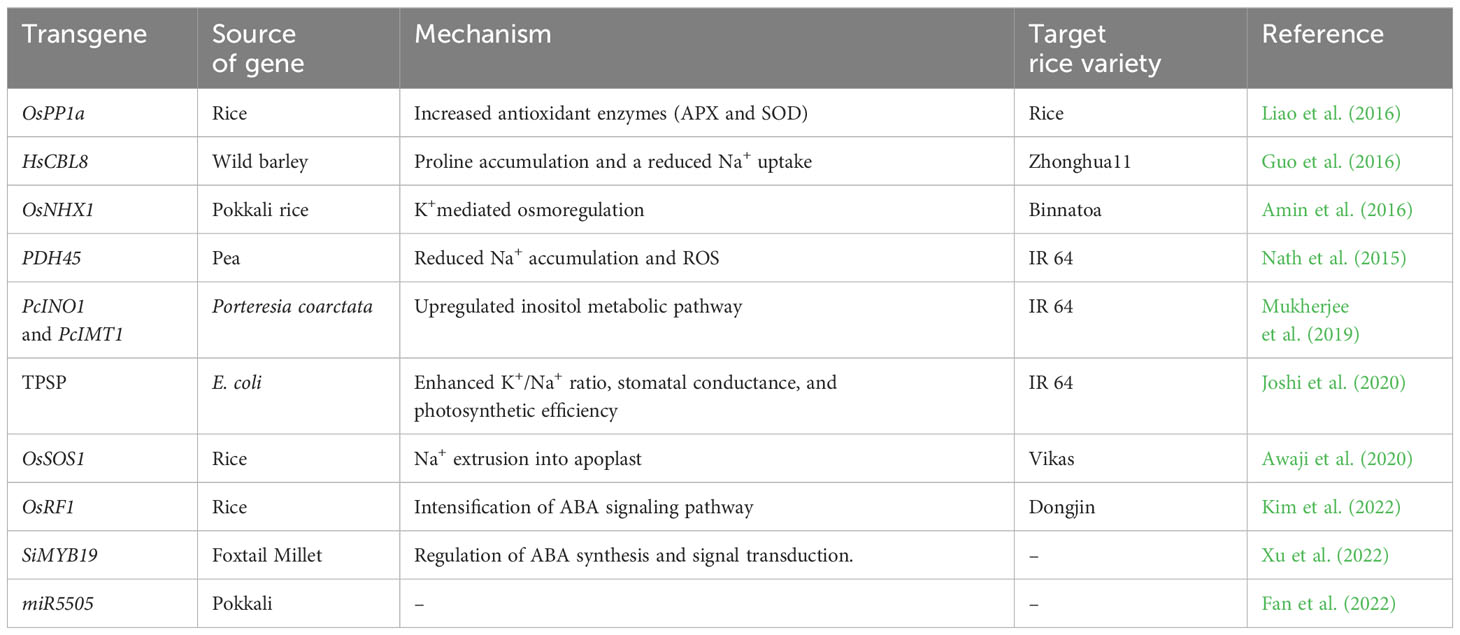
With the advent of DNA-based markers, approximately 1,000 QTLs for salt tolerance in rice have been mapped (Prakash et al., 2022), and several candidate genes for salinity tolerance have been identified (Supplementary Table 1). The genetic regions corresponding to these QTLs and genes have been implicated in various molecular and physiological processes. The major landmark in salinity tolerance breeding was the detection of a Saltol QTL for shoot K+/Na+ homeostasis on chromosome 1 (Gregorio, 1997) in an RIL designated as FL478 (IR66496-3R-78-1-1) derived from Pokkali/IR29. In the Saltol region, the SKC1 gene of the Nona Bokra landrace controlling K+ concentration in shoots was identified and later cloned as OsHKT1;5, which encodes a plasma membrane Na+ transporter that regulates Na+ partitioning between roots and shoots (Ren et al., 2005).
Saltol QTL have been successfully transferred to many popular varieties through marker-aided back cross breeding (MABB) in India, Bangladesh, Russia, and Vietnam. (Padmavathi et al., 2023). The tightly linked markers within the Saltol QTL region (AP3206, RM8094, and RM3412), flanking markers, i.e., RM1287 and RM10694, RM493 and RM10793 enabled its successful transfer (Thomson et al., 2010) in the genetic background of BT7 (Linh et al., 2012), AS996 (Huyen et al., 2012), Bacthom 7 (Vu et al., 2012) in Vietnam, BRRI Dhan 49 (Hoque et al., 2015) in Bangladesh, Novator (Usatov et al., 2015) in Russia; ADT43 (Geetha et al., 2017), PB 1121 (Babu et al., 2017), Pusa Basmati 1 (Singh et al., 2018), Yukinko-mai (Rana et al., 2019), Pusa44 and Sarjoo 52 (Krishnamurthy et al., 2020), Pusa Basmati 1509 (Yadav et al., 2020), Aiswarya (Nair and Shylaraj, 2021) and Improved Samba Mahsuri (Rekha et al., 2022) in India. These MAS-derived varieties are already available to farmers for cultivation purposes.
Numerous QTLs for salinity tolerance in rice have been identified through biparental mapping populations. However, this approach may only partially reveal the genetic diversity of traits. A genome-wide association study (GWAS) facilitates the detection of a wide array of QTLs, thereby revealing a more extensive genetic diversity of the trait than bi-parental populations. Due to the higher recombination rate of the genome of natural genotypes, GWAS is employed for high resolution rapid mapping of genome-wide SNPs associated with morphological, physiological, photosynthetic and yield and its component traits under salinity such as K+/Na+ ratio, salt injury score, Na+ and K+ content of root and shoot, Na+ sheath: blade ratio, seedling length, fresh and dry weight of shoots and roots, chlorophyll and water content, number of panicles, filled grains and grain yield in seedling stage using hydroponics (Batayeva et al., 2018; Lekklar et al., 2019; Rohila et al., 2019; Zhang et al., 2020; Yuan et al., 2020; Le et al., 2021; Nayyeripasand et al., 2021; Kim and Suk-Man, 2023) and during the reproductive stage (Theerawitaya et al., 2020; Warraich et al., 2020; Chen et al., 2022).
The enrichment of MABB-derived salt-tolerant cultivars is constrained to bring major advancement, as it can only correct the deficiency of popular varieties, for example, salt sensitivity retaining the recurrent parent genome rather than creating highly heterotic salt-tolerant varieties. Wild rice has not been exploited much compared to cultivated rice. With the advent of next-generation sequencing (NGS) techniques, the available sequence information of genomes of seven wild species, viz., O. rufipogon, O. nivara, O. barthii, O. glumaepatula, O. meridionalis, O. punctata, and L. perrieri (Stein et al., 2018), provides opportunities to detect new genes and novel functional markers for incorporation into cultivars.
6.3 Genetic engineering
Genetic engineering is a promising approach for trait transfer to overcome hybridization barriers. This approach for increasing salinity tolerance centers on manipulating genes encoding the synthesis of compatible osmotica, antioxidants, sodium/potassium transport proteins, and transcription factors underlying salt tolerance mechanisms, focusing on cultivated rice (Table 3). Despite the improvement in transgenic rice possessing the reported genes produced under glasshouse conditions, they have hardly reached farmers’ fields for commercial cultivation (Kotula et al., 2020). Transgenic methods have focused only on altering individual genes and a single tolerance mechanism that hinders salinity tolerance improvement. Hence further research is needed to harness the potential of these wild sources.
6.4 Genome editing
The technique permits editing of the target locus, knockout, and allele exchange in the genome, and culminating in the development of transgene-free edited plants. In rice, the CRISPR/Cas9 gene editing method has been effectively used to edit the OsRR22 gene, which encodes a transcription factor that controls signaling and cytokinin metabolism in plants, thereby improving salt stress tolerance in rice (Zhang et al., 2019). The mutated salt-tolerant gene (OsDST A), which was developed using CRISPR/Cas9, has been reported to increase tolerance to salt stress by decreasing stomata and increasing leaf water retention in the MTU1010 rice variety (Santosh Kumar et al., 2020). In another study, CRISPR/Cas9 mediated mutagenesis of the BEARI transcription factor enhanced the tolerance to excessive salts by controlling ion transport (Teng et al., 2022). Improvement in salt tolerance with decreased salt build-up has been achieved by editing the OsNAC3 gene in rice (Zhang et al., 2021). Mutants with CRISPR/Cas9-mediated OsmiR535 knockout exhibit increased NaCl tolerance (Yue et al., 2020). The CRISPR/Cas9 mediated mutagenesis of the rice gene BG3, which promotes the transport of cytokinin hormones, revealed increased salinity tolerance (Yin et al., 2020). The CRISPR/Cas technique can be applied to investigate the wild rice gene pool and address challenges associated with linkage drag during the introgression of target wild genes into high-yielding backgrounds.
7 Deployment of wild rice relatives in breeding for salt tolerance
Wild rice species are treasure troves with various beneficial traits linked to yield, quality, and tolerance/resistance to abiotic and biotic stresses. Breeders often neglect the utilization of wild rice species for two main reasons. First, it is difficult to ensure gene flow from wild rice into the cultivated gene pool because of cross incompatibility, sterility, or non-viability in F1 or backcrosses, restricted genetic recombination between wild and elite genomes, and linkage drag from wild rice. However, these genetic complications could be resolved with MABB, ensuring precise gene introgression with selections to minimize unwanted linkage drag and backcrosses, as compared to the traditional approach (Warburton et al., 2017). Second, when wild rice is grown outside their native habitat, either it is poorly acclimated or the expression of beneficial alleles is concealed. Ultimately, the performance of the derived introgression lines (ILs) is inferior.
Novel genes from the AA genome containing wild species in the primary gene pool could be easily transferred into domesticated rice through the traditional back-cross method. However, distant crosses between O. sativa and genetically remote wild species of the secondary and tertiary gene pools are difficult to achieve due to cross incompatibility and embryo abortion and/or degeneration. However, embryo rescue techniques can overcome these hurdles by producing distant fertile interspecific hybrids (Jena, 2010). Intergeneric hybrids between Porteresia coarctata, distant rice relatives, and O. sativa were produced with limited success by adopting vegetative multiplication of rescued hybrid embryos (Jena, 1994). Salt-tolerant genes can also be incorporated from O. porteresia into O. sativa through bridge crossing with O. australiensis (Latha et al., 2004; Mammadov et al., 2018).
With the latest advancements in NGS technology and high-throughput phenotyping, the historical natural genetic variation for salt tolerance present in a panel of wild rice accessions can easily be captured following GWAS. This is a potential strategy for mapping salt tolerance genes/QTLs, particularly in wild rice, where cross-incompatibility complicates the generation of mapping populations. The use of genetically distant wild species in the development of improved cultivars with superior trait performance has been greatly facilitated by the identification of candidate genes, GWAS, and development of introgression lines through MABB (Singh et al., 2021; Dai et al., 2022).
Transgenic approaches using cloned genes from wild species offer a solution to the issue of direct introgression of wild rice genes. Most transgenic methods have focused on single genes and/or one type of salt-tolerance mechanism in cultivars. However, genetic engineering utilizing cloned genes governing antioxidants, osmolytes, and ion transporters, photosynthesis enhancement, and yield simultaneously could potentially lead to the creation of more resilient rice varieties capable of thriving under high-salinity conditions.
The double haploid technique (DH) is another potential method for generating new homozygous salt-tolerant lines from distant crosses involving wild species in a single generation from heterozygous parents, which would otherwise require several generations of selfing to achieve near homozygosity in conventional breeding.
Genome editing is an emerging strategy for speeding up the development of advanced breeding lines, wherein a specific site in a target wild gene is edited directly by insertion, deletion, or alteration of existing nucleotide/s in the genomic segment of commercial varieties developed through distant hybridization. It corrects the deficiency, as in the present case, of salt sensitivity of the cultivars by avoiding laborious conventional or MABB and the incorporation of large introgressed genomic regions of wild species. Speed breeding is another promising approach for utilizing wild progenitors in breeding that manipulates the photoperiod and temperature to rapidly generate multiple generations within a year. In scenarios where precise genome editing is not an option, speed breeding plays a pivotal role in accelerating generation advancement, enabling selection against undesirable traits inherited from wild species, and stabilizing the genetic background of newly developed cultivars.
Although extensive studies have not been carried out in rice using wild species for genome editing and transgenic approaches, a considerable amount of research has been conducted on QTL mapping and introgression of QTLs/candidate genes in agronomically superior cultures.
7.1 QTLs/genes in wild species
As the QTLs/genes from indigenous wild relatives cannot be utilized directly in breeding populations, pre-breeding strategies could be followed to identify and transfer wild rice genes into an intermediate breeding material that can easily be hybridized with modern elite varieties. Although wild rice is not as widely exploited as cultivated rice, it contains positive alleles, genes, and QTLs associated with salt tolerance. These effects have been documented using advanced backcross populations and introgression lines. Due to the large natural genetic diversity of wild species, QTLs associated with novel tolerance mechanisms could be prime candidates for improving salt tolerance (Stein et al., 2018). A few studies, as detailed below, have attempted to discover natural alleles governing salt tolerance in wild rice donors.
A collection of 87 ILs derived from Teqing/O. rufipogon was screened for salinity tolerance during seedling stage by Tian et al. (2011). They detected 15 QTLs, 13 containing O. rufipogon alleles that conferred higher tolerance in the Teqing background. These alleles enhanced relative root, shoot, and total dry weight at three loci (qRRW10, qRSW10, and qRTW10) on chromosome 10 in salt-tolerant ILs.
In a set of 285 ILs derived from 93-11/O. rufipogon, Wang et al. (2017) identified 10 QTLs for salt tolerance traits on chromosomes 1, 5, 7, 9 to 12 at seedling stage. They observed that qST7 on chromosome 7 coincided with qRRW7, qRSW7, and qRTW7 in O. rufipogon for salt tolerance reported by Tian et al. (2011). They also found that qST10 on chromosome 10 shared a similar QTL hotspot as qRRW10, qRSW10, and qRTW10 reported by Tian et al. (2011). Four candidate genes linked to salinity tolerance namely LRP1 (LOC_Os05g32070), acetyltransferase (LOC_Os05g31254), GRAM domain containing protein (GEM, LOC_Os10g34730) and calmodulin-related calcium sensor protein (OsCML15, LOC_Os05g31620) were recorded in an O. rufipogon derived salt-tolerant IL 9L136. It is perceived that the accumulation of enzymatic antioxidants such as peroxidase, catalase and superoxide dismutase in 9L136 served as a probable antioxidant defense mechanism.
A RIL mapping population from a cross between salt sensitive cultivar, Ningjing16 and salt tolerant Dongxiang wild rice, O. rufipogon was used by Quan et al. (2018) to map 9 QTLs for salt tolerance at the seedling stage (qST) on chromosomes 1, 3, 4, 5, 6, 8, and 10. They reported that qST6, a major QTL influencing survival rate, accounted for 19.3% of the phenotypic variance and showed additive effects. They indicated protein kinases, MYB and zinc finger transcriptional factors and SKC1, HKT1;5 transporters and HAK6 as the potential genes within the QTL region. They hypothesized that ion homeostasis and kinase signalling pathways were the possible mechanisms of salt tolerance.
Wairich et al. (2021) conducted a research experiment to examine the effect of salinity stress on two different populations of interspecific introgression from O. sativa × O. meridionalis and O. sativa × O. rufipogon crosses. They identified three potential QTLs on chromosomes 1, 3, and 5 in O. sativa cv. Curinga O. meridionalis (Ng. acc. W2112) ILs, and 19 QTLs on chromosomes 1, 4, and 7 in O. sativa cv. Curinga O. rufipogon accession (IRGC 105491) population for various vegetation indices under salinity. The study demonstrated that introgression line (IL) CR47 of O. sativa/O. rufipogon cross had a tissue tolerance mechanism, while IL CM6, derived from O. sativa/O. meridionalis, had a higher Na+/K+ ratio in roots to cope with salinity.
Kumari et al. (2021) subjected back cross progenies derived from IR64 and O. nivara accession NKSWR 173 to screening against salinity stress. They genotyped a set of 74 BC1F2 families for the presence of seedling stage (qSES1.1 and qSES3.2) and reproductive-stage salt-tolerant QTLs (qSTY11.1). Four backcrossed families displayed enhanced tolerance, as determined by phenotypic performance and QTL peak markers, during both stages of growth.
In another study employing backcross inbred lines (BILs) from the cross 9311 and an African wild rice, O. longistaminata, 18 QTLs conferring salt tolerance were found (Yuan et al., 2022), and one QTL each for salt injury score (qSIS2), the water content of seedlings under salt treatment (qWCSST2), and the relative water content of seedlings (qRWCS2) colocalized on chromosome 2. Sequence and expression analyses suggest that MH02t0466900, encoding cytochrome P450 86B1, may contribute to ion homeostasis.
In a study involving 117 DHs derived from F1s of Savitri and Pokkali, Samantaray et al. (2021) identified four candidate genes for salinity tolerance, namely LOC_Os01g09550, LOC_Os01g09560, LOC_Os12g06560, and LOC_Os12g06570 during germination stage.
7.2 Wild rice derived salt-tolerant elite lines/cultivars
Elite rice lines with salinity tolerance have been successfully developed by harnessing the genetic variability in a limited number of wild rice species (Chatterjee et al., 2006; Tian et al., 2011; Ganeshan et al., 2016; Wang et al., 2017; Quan et al., 2018; Kumari et al., 2021; Wairich et al., 2021).
The wild species O. rufipogon (2n = 24, AA) is frequently used to breed salt-tolerant lines because of its close evolutionary proximity and high compatibility with O. sativa (2n = 24, AA) (Londo et al., 2006). The potential salt-tolerant introgression line, YIL 16, is a derivative of Teqing/O. rufipogon has been reported by Tian et al. (2011). It has three O. rufipogon genomic regions that hold qRRW3 and qSTS2, which are responsible for the relative root dry weight and salt tolerance score, respectively. With its enhanced salt tolerance, YIL 16 could be directly utilized or suggested for breeding programs. Ganeshan et al. (2016) reported salinity tolerance genes in ILs derived from the crosses O. sativa with O. rufipogon or O. nivara.
Four candidate genes (LOC_Os05g31254, LOC_Os05g31620, LOC_Os05g32070, and LOC_Os10g34730) were identified in the QTL regions (qRW10, qRSW10, and qRTW10) of an introgression line (9L 136) according to a study carried out by Wang et al. (2017). This line was developed from a cross between a Chinese O. rufipogon accession and the O. sativa cultivar 93-1. The researchers hypothesized that the overexpression of these genes could improve salt tolerance in rice varieties. Furthermore, the potential of O. rufipogon QTLs to boost the antioxidant system in domesticated varieties could help them tolerate salt-induced oxidative stress through marker-assisted introgression.
Quan et al. (2018) employed an accession of wild rice, O. rufipogon, recovered DJ15, a salt-tolerant introgression line from O. rufipogon and O. sativa (Ningjing16) cross. Subsequently, six high-yielding salt-tolerant RILs developed between NIL DJ15/Koshihikari possessing both qST1.2DJ15 and qST6DJ15 were identified with an improved seed set.
Despite the availability of 22 potential wild species and two cultivated species (O. sativa and O. glaberrima), only two salt-tolerant varieties have been developed to date. Jarava and Chinsura Nona 2 were both developed using O. rufipogon donors. Jarava is a coastal saline-tolerant rice cultivar bred by ICAR-IIRR, Hyderabad, India, through distant hybridization between O. rufipogon and O. sativa. In 2005, it was released and notified by the Central Sub-Committee on Crop Standards, Notification and Release of Varieties for cultivation in West Bengal, Andaman & Nicobar, Puducherry states of India due to its superior yield under saline soils (Gazette of India notification No: S.O.1566 E dated 11-05-2005). It is a long-duration (143 days to 145 days) variety possessing short bold grains with 4.5 t/ha grain yield (Figure 4).
Another coastal saline tolerant cultivar, Chinsura Nona 2 (Gosaba 6), a medium duration (130 days to 135 days) variety with bold grains, originated from the KMR3 × O. rufipogon cross developed at ICAR-IIRR, Hyderabad in collaboration with Rice Research Station, Chinsurah, West Bengal, India (Thummala et al., 2022). It recorded 5.56 t/ha of grain yield under salinity; and was released and notified by the State Variety Release Committee (SVRC) for commercial cultivation in the West Bengal state of India (Gazette of India notification No S.O. 3220 E dated 5.9.2019). Thus, wild rice genes contributed immensely to the enhanced salt tolerance in the improved cultivars.
Employing genetic engineering, the MIPS coding gene from the most distant rice ancestor, O. coarctata, PcINO1, encoding L-myoinositol 1-phosphate synthase, was introgressed into the cultivated rice variety, Pusa Basmati-1, which displayed increased salt tolerance (Chatterjee et al., 2006).
8 Conclusions
Salinity, particularly during the reproductive stage, is a major abiotic stress factor that drastically reduces rice crop productivity. If not properly addressed, it poses a severe threat to global food security. Breeding for salinity tolerance is challenging due to its polygenic control, interaction with the environment, intricate physiological and metabolic processes, and growth stage. Despite the complexity, a moderate headway has been achieved with the development and release of approximately 101 salt-tolerant rice varieties using traditional global breeding methods.
The development of salt-tolerant cultivars has been hindered by the scarcity of genetic resources among domesticated cultivars. Therefore, future research should prioritize broadening the genetic base of modern cultivars by utilizing the salinity-adaptive genes present in unexplored wild species from secondary and tertiary gene pools. Prior to this endeavor, it is necessary to systematically explore and characterize various unexploited accessions of wild species for different salt stress defense mechanisms and identify genes for incorporation into breeding programs.
Obstacles associated with crossing and unwanted linkage drag are common during distant hybridization with wild species. This deficiency can be corrected by adopting the MABB method, which facilitates the accurate and accelerated introgression of target candidate genes/QTLs in popular cultivars. It is vital to reinforce pre-breeding programs that employ wild species in their breeding schemes worldwide. Continued research is needed to pinpoint more genomic resources in terms of salt-responsive QTLs and molecular markers and to profile the genes and their upstream regulatory regions in potential wild relatives. This will facilitate an in-depth understanding of the mechanisms that are effective in breeding tolerant varieties.
Exotic candidate genes have the potential to generate transgenic plants. The accurate pyramiding of genes responsible for different mechanisms of salt tolerance from divergent wild genetic sources into elite cultivars is needed for an hour to increase salt tolerance. Progress in transgenic and genome-editing techniques has paved the way for more possibilities to utilize and investigate valuable genes from wild rice to enhance salinity tolerance in cultivated rice.
The specific salt tolerance mechanisms in halophytes have yet to be fully elucidated. A comprehensive analysis of the process by which plants recognize salt stress and the crosstalk between different genes and pathways that are involved in regulating salt tolerance mechanisms is still needed. Concerted efforts are necessary to preserve both ex situ and in situ wild rice species for integration into salinity breeding programs.
Author contributions
GP: Conceptualization, Writing – original draft, Writing – review & editing. UB: Writing – review & editing. KR: Writing – review & editing. DB: Writing – review & editing. MA: Data curation, Writing – review & editing. RKS: Writing – review & editing. RMS: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1253726/full#supplementary-material
References
Afrasyab, R., James Richard, A., Kazem, P., Rana., M. (2010). Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 37, 255–263. doi: 10.1071/FP09148
Amin, U. S. M., Biswas, S., Elias, S. M., Razzaque, S., Haque, T., Malo, R., et al. (2016). Enhanced Salt Tolerance Conferred by the Complete 2.3 kb cDNA of the Rice Vacuolar Na+/H+ Antiporter Gene Compared to 1.9 kb Coding Region with 5′ UTR in Transgenic Lines of Rice. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00014
Amirjani, M. (2010). Effect of salinity stress on growth, mineral composition, proline content, antioxidant enzymes of soybean. Am. J. Plant Physiol. 5 (6), 350–360. doi: 10.3923/ajpp.2010.350.360
Anilkumar, C., Sah, R. P., Beena, R., TP, M. A., Kumar, A., Behera, S., et al. (2023). Conventional and contemporary approaches for drought tolerance rice breeding: Progress and prospects. Plant Breed 130, 330–345. doi: 10.1111/pbr.13119
Arif, Y., Singh, P., Siddiqui, H., Bajguz, A., Hayat, S. (2020). Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem 156, 64–77. doi: 10.1016/j.plaphy.2020.08.042
Atwell, B. J., Wang, H., Scafaro, A. P. (2014). Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 215-216, 48–58. doi: 10.1016/j.plantsci.2013.10.007
Awaji, S. M., Hanjagi, P. S., BN, P., VR, S. (2020). Overexpression of plasma membrane Na+/H+ antiporter OsSOS1 gene improves salt tolerance in transgenic rice plants. Oryza 57 (4), 277–287. doi: 10.35709/ory.2020.57.4.3
Babu, N. N., Krishnan, S. G., Vinod, K. K., Krishnamurthy, S. L., Singh, V. K., Singh, M. P., et al. (2017). Marker Aided Incorporation of Saltol, a Major QTL Associated with Seedling Stage Salt Tolerance, into Oryza sativa ‘Pusa Basmati 1121’. Front. Plant Sci. 8, 41. doi: 10.3389/fpls.2017.00041
Banumathy, S., Veni, K., Anandhababu, R., Arunachalam, P., Raveendran, M., Vanniarajan, C. (2018). Character association and stress indices for yield components in Saltol introgressed backcross inbred lines of rice (Oryza sativa L.). Indian J. Agric. Res. 52 (1), 28–33. doi: 10.18805/IJARe.A-4926
Barua, R., De Ocampo, M., Egdane, J., Ismail, A. M., Mondal, S. (2015). Phenotyping rice (Oryza sativa L.) genotypes for physiological traits associated with tolerance of salinity at seedling stage. Sci. Agric. 12, 156–162. doi: 10.15192/PSCP.SA.2015.12.3.156162
Batayeva, D., Labaco, B., Ye, C., Li, X., Usenbekov, B., Rysbekova, A., et al. (2018). Genome-wide association study of seedling stage salinity tolerance in temperate japonica rice germplasm. BMC Genet. 19 (1), 18. doi: 10.1186/s12863-017-0590-7
Campbell, M. T., Bandillo, N., Al Shiblawi, F. R. A., Sharma, S., Liu, K., Du, Q., et al. (2017). Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PloS Genet. 13 (6), e1006823. doi: 10.1371/journal.pgen.1006823
Channa, G. S., Mahar, A. R., Memon, A. H., Bhagat, M. A., Saand, M. A., Mirbahar, A. A., et al. (2021). Effect of salinity on emergence and early growth stages of aromatic and non-aromatic Rice (Oryza sativa L.) genotypes. Biol. Sci.-PJSIR 64, 64–74. doi: 10.52763/PJSIR.BIOL.SCI.64.1.2021.64.74
Channa, G. S., Mahar, A. R., Rajpar, I., Mirani, A. A., Bhatti, S. M., Bhagat, M. A., et al. (2023). Evaluation of non-aromatic rice (Oryza sativa L.) cultivars for NaCl tolerance under hydroponic conditions. Sarhad J. Agriculture. 39 (3), 573–580. doi: 10.17582/journal.sja/2023/39.3.573.580
Chatterjee, A., Goswami, L., Maitra, S., Ghosh Dastidar, K., Ray, S., Lahiri Majumder, A. (2006). Introgression of a novel salt-tolerant L-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Lett. 580 (16), 3980–3988. doi: 10.1016/j.febslet.2006.06.033
Chen, G., Hu, K., Zhao, J., Guo, F., Shan, W., Jiang, Q., et al. (2022). Genome-wide association analysis for salt–induced phenotypic and physiologic responses in rice at seedling and reproductive stages. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.822618
Chen, Z., Pottosin, I. I., Cuin, T. A., Fuglsang, A. T., Tester, M., Jha, D., et al. (2007). Root plasma membrane transporters controlling K+/na+ Homeostasis in salt-stressed barley. Plant Physiol. 145 (4), 1714–1725. doi: 10.1104/pp.107.110262
Cheng, L., Abraham, J., Zhu, J., Trenberth, K. E., Fasullo, J., Boyer, T., et al. (2020). Record setting ocean warmth continued in 2019. Adv. Atmos Sci. 37, 137–142. doi: 10.1007/s00376-020-9283-7
Chinnusamy, V., Jagendorf, A., Zhu, J. K. (2005). Understanding and improving salt tolerance in plants. Crop Sci. 45 (2), 437. doi: 10.2135/cropsci2005.0437
Chutipaijit, S., Suriyan, C. H., Sompornpailin, K. (2011). High contents of proline and anthocyanin increase protective response to salinity in Oryza sativa L. spp. indica. Aust. J. Crop Sci. 5, 1191–1198.
Clermont-Dauphin, C., Suwannang, N., Grunberger, O., Claude, H., Maeght, J. I. (2010). Yield of rice under water and soil salinity risks in farmers’ fields in northeast Thailand. Field Crop Res. 118, 289–296. doi: 10.1016/j.fcr.2010.06.009
Cohen, S. P., Leach, J. E. (2019). Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 9 (1), 6273. doi: 10.1038/s41598-019-42731-8
Dai, L., Li, P., Li, Q., Leng, Y., Zeng, D., Qian, Q. (2022). Integrated multi-omics perspective to strengthen the understanding of salt tolerance in rice. Int. J. Mol. Sci. 23 (9), 5236. doi: 10.3390/ijms23095236
Duan, J., Cai, W. (2012). OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PloS One 7 (9), e45117. doi: 10.1371/journal.pone.0045117
Dugasa, M. T., Cao, F., Ibrahim, W., Wu, F. (2019). Differences 990 in physiological and biochemical characteristics in response to single and combined drought and salinity stresses between wheat genotypes differing in salt tolerance. Physiol. Plant 165 (2), 134–143. doi: 10.1111/ppl.12743
Eynard, A., Lal, R., Wiebe, K. (2005). Crop response in salt-affected soils. J.Sustain Agric. 165 (2), 134–143. doi: 10.1300/j064v27n01_03
Fan, Y., Xie, J., Zhang, F. (2022). Overexpression of miR5505 enhanced drought and salt resistance in rice (Oryza sativa). doi: 10.1101/2022.01.13.476146
FAO, IFAD, UNICEF, WFP and WHO (2022). The State of Food Security and Nutrition in the World 2022. Repurposing food and agricultural policies to make healthy diets more affordable (Rome: FAO). doi: 10.4060/cc0639en
Flowers, T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55 (396), 307–319. doi: 10.1093/jxb/erh003
Flowers, T. J., Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179 (4), 945–963. doi: 10.1111/j.1469-8137.2008.02531.xv
Flowers, T., Flowers, S. (2005). Why does salinity pose such a difficult problem for plant breeders. Agric. Water Manage. 78 (1-2), 15–24. doi: 10.1016/j.agwat.2005.04.015
Ganeshan, P., Jain, A., Brajendra, P., Rao, A. R., Sreenu, K., Mishra, P., et al. (2016). Identification of salt tolerant rice lines among interspecific BILs developed by crossing Oryza sativa × O. rufipogon and O. sativa × O. nivara. Australian Journal of Crop Science 10, 220–228.
Ganie, S. A. (2020). RNA chaperones: Potential candidates for engineering salt tolerance in rice. Crop Science. 60, 530–540. doi: 10.1002/csc2.20134
Garg, R., Verma, M., Agrawal, S., Shankar, R., Majee, M., Jain, M. (2014). Deep Transcriptome Sequencing of Wild Halophyte Rice, Porteresia coarctata, Provides Novel Insights into the Salinity and Submergence Tolerance Factors. DNA Res. 21 (1), 69–84. doi: 10.1093/dnares/dst042
Geetha, S., Vasuki, A., Selvam, P., Jagadeesh, S. R., Krishnamurthy, S. L., Palanichamy, M., et al. (2017). Development of sodicity tolerant rice varieties through marker assisted backcross breeding. Electron. J. Plant Breed. 8 (4), 1013–1031. doi: 10.5958/0975-928X.2017.00151.X
Gholizadeh, A., Dehghania, H., Dvorakb, J. (2014). Determination of the most effective traits on wheat yield under saline stress. Agric. Adv. 3, 103–110. doi: 10.14196/aa.v3i4.1304
Ghosh, B., Ali Md, N., Gantait, S. (2016). Response of rice under salinity stress: A review update. Rice Research: Open Access 4 (2), 167. doi: 10.4172/2375-4338.1000167
Gregorio, G. B. (1997). Tagging salinity tolerance genes in rice using amplified fragment length polymorphism (AFLP) (Los Baños, Philippines: IRRI, Philippines).
Gregorio, G. B., Senadhira, D. (1993). Genetic analysis of salinity tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 86-86 (2-3), 333–338. doi: 10.1007/bf00222098
Gregorio, G. B., Senadhira, D., Mendoza, R. D. (1997). Screening rice for salinity tolerance, vol 22, IRRI discussion paper series. International Rice research Institute (Manila), 30.
Guo, W., Chen, T., Hussain, N., Zhang, G., Jiang, L. (2016). Characterization of salinity tolerance of transgenic rice lines harboring hsCBL8 of wild barley (Hordeum spontanum) line from qinghai-tibet plateau. Front.Plant Sci. 7. doi: 10.3389/fpls.2016.01678
Habiba, U., Abedin, M. A., Hassan, A. W. R., Shaw, R. (2015). Food security and risk reduction in Bangladesh (Tokyo: Springer).
Hamzelou, S., Kamath, K. S., Masoomi-Aladizgeh, F., Johnsen, M. M., Atwell, B. J., Haynes, P. A. (2020). Wild and cultivated species of rice have distinctive proteomic responses to drought. Int. J. Mol. Sci. 21 (17), 5980. doi: 10.3390/ijms21175980
Hanin, M., Ebel, C., Ngom, M., Laplaze, L., Masmoudi, K. (2016). New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01787
Harlan, J. R., de Wet, J. M. J. (1971). Toward a rational classification of cultivated plants. Taxon 20 (4), 509. doi: 10.2307/1218252
Hauser, F., Horie, T. (2010). A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant, Cell and Environment 33, 552–565. doi: 10.1111/j.1365-3040.2009.02056.x
Hellwig, T., Abbo, S., Sherman, A., Ophir, R. (2021). Prospects for the natural distribution of crop wild-relatives with limited adaptability: The case of the wild pea, Pisum fulvum. Plant Sci. 310, 110957. doi: 10.1016/j.plantsci.2021.110957
Hoang, T. M. L., Moghaddam, L., Williams, B., Khanna, H., Dale, J., Mundree, S. G. (2015). Development of salinity tolerance in rice by constitutive overexpression of genes involved in the regulation of programmed cell death. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00175
Hoque, A. B. M. Z., Haque, M. A., Sarker, M. R. A., Rahman, M. A. (2015). Marker-assisted introgression of Saltol locus into genetic background of BRRI Dhan-49. Int.J. Biosci. 6, 71–80. doi: 10.12692/ijb/6.12.71-80
Horie, T., Karahara, I., Katsuhara, M. (2012). Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 5, 11. doi: 10.1186/1939-8433-5-11
Hussain, M. I., Lyra, D. A., Farooq, M., Nikoloudakis, N., Khalid, N. (2016). Salt and drought stresses in safflower: a review. Agron. Sustain. Dev. 36 (1), 4. doi: 10.1007/s13593-015-0344-8
Huang, X., Kurata, N., Wei, X., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490 (7421), 497–501. doi: 10.1038/nature11532
Hussain, S., Zhang, J. H., Zhong, C., Zhu, L., Cao, X., Yu, S., et al. (2017). Effects of salt stress on rice growth, development characteristics, and the regulating ways: A review. J. Integr. Agric. 16 (11), 2357–2374. doi: 10.1016/S2095-3119(16)61608-8
Huyen, L. T. N., Cuc, L. M., Ismail, A. M., Ham, L. H. (2012). Introgression the salinity tolerance QTLs saltol into AS996, the elite rice variety of Vietnam. Am. J. Plant Sci. 03, 981–987. doi: 10.4236/ajps.2012.37116
Irakoze, W., Prodjinoto, H., Nijimbere, S., Rufyikiri, G., Lutts, S. (2020). NaCl and na2SO4 salinities have different impact on photosynthesis and yield-related parameters in rice (Oryza sativa L.). Agron 10 (6), 864. doi: 10.3390/agronomy10060864
Islam, M. A., Lobry de Bruyn, L., Warwick, N. W. M., Koech, R. (2021). Salinity-affected threshold yield loss: A signal of adaptation tipping points for salinity management of dry season rice cultivation in the coastal areas of Bangladesh. J. Environ. Manage. 288, 112413. doi: 10.1016/j.jenvman.2021.112413
Ismail, A. M., Heuer, S., Thomson, M. J., Wissuwa, M. (2007). Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol. Biol. 65 (4), 547–570. doi: 10.1007/s11103-007-9215-2
Jagadish, S. V. K., Way, D. A., Sharkey, T. D. (2021). Scaling plant responses to high temperature from cell to ecosystem. Plant Cell Environ. 44, 1987–1991. doi: 10.1111/pce.14082
Jamil, M., Bashir, S., Anwar, S., Bibi, A., Bangash, A., Ullah, F., et al. (2012). Effect of salinity on physiological and biochemical characteristics of different varieties of rice. Pak. J. Bot. 44, 7–13.
Jena, K. (1994). Production of intergeneric hybrid between Oryza sativa L. and Porteresia coarctata T. Curr. Sci. 67, 744–746.
Jena, K. K. (2010). The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa. Breed. Sci. 60, 518–523. doi: 10.1270/jsbbs.60.518
Jin, S., Lin, Q., Luo, Y., Zhu, Z., Liu, G., Li, Y., et al. (2021). Genome-wide specificity of prime editors in plants. Nat. Biotechnol 39, 1292–1299. doi: 10.1038/s41587-021-00891-x
Joshi, R., Sahoo, K. K., Singh, A. K., Anwar, K., Gautam, R. K., Krishnamurthy, S. L., et al. (2020). Engineering trehalose biosynthesis pathway improves yield potential in rice under drought, saline and sodic conditions. J. Exp. Bot 71 (2), 653–668. doi: 10.1093/jxb/erz462
Khan, S., Anwar, S., Ashraf, M. Y., Khaliq, B., Sun, M., Hussain, S., et al. (2019). Mechanisms and adaptation strategies to improve heat tolerance in rice. A review. Plants 8 (11), 508. doi: 10.3390/plants8110508
Khan, M. A., Asaf, S., Khan, A. L., Adhikari, A., Jan, R., Ali, S., et al. (2020). Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. (Stuttgart Germany) 22 (5), 850–862. doi: 10.1111/plb.13124
Kim, M. J., Kim, H. J., Pak, J. H., Cho, H. S., Choi, H. K., Jung, H. W., et al. (2017). Overexpression of AtSZF2 from Arabidopsis showed enhanced tolerance to salt stress in soybean. Plant Breed. Biotechnol. 5, 1–15. doi: 10.9787/PBB.2017.5.1.001
Kim, S., Park, S.-I., Kwon, H., Cho, M. H., Kim, B.-G., Chung, J. H., et al. (2022). The Rice Abscisic Acid-Responsive RING Finger E3 Ligase OsRF1 Targets OsPP2C09 for Degradation and Confers Drought and Salinity Tolerance in Rice. Front. Plant Sci. 12, 797940. doi: 10.3389/fpls.2021.797940
Kim, T. H., Suk-Man, K. (2023). Identification of candidate genes for salt tolerance at the seedling stage using integrated genome-wide association study and transcriptome analysis in rice. Plants 12 (6), 1401. doi: 10.3390/plants12061401
Kotula, L., Garcia, P., Zorb, C., Colmer, T. D., Flowers, T. J. (2020). Improving crop salt tolerance using transgenic approaches: an update and physiological analysis. Plant Cell Environ 43 (12), 2932–2956. doi: 10.1111/pce.13865
Krishnamurthy, S. L., Pundir, P., Warraich, A. S., Rathor, S., Lokeshkumar, B. M., Singh, N. K., et al. (2020). Introgressed saltol QTL lines improves the salinity tolerance in rice at seedling stage. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00833
Kumar, K., Kumar, M., Kim, S. R., Ryu, H., Cho, Y. G. (2013). Insights into genomics of salt stress response in rice. Rice 6 (1), 27. doi: 10.1186/1939-8433-6-27
Kumar, N., Kumar, N., Shukla, A., Shankhdhar, S. C., Shankhdhar, D. (2015). Impact of terminal heat stress on pollen viability and yield attributes of rice (Oryza sativa L.). Cereal Res. Commun. 43 (4), 616–626. doi: 10.1556/0806.43.2015.023
Kumar, V., Shriram, V., Jawali, N., Shitole, M. G. (2007). Differential response of indica rice genotypes to NaCl stress in relation to physiological and biochemical parameters. Arch. Agron. Soil Sci. 53 (5), 581–592. doi: 10.1080/03650340701576800
Kumari, S., Singh, B., Singh, S. K., Satya, D., Singh, S., Tripathy, K., et al. (2021). Exploring novel QTLs among backcross lines for salinity tolerance in rice. Indian J. Agric. Sci. 91 (3), 426–429. doi: 10.56093/ijas.v91i3.112517
Kurusu, T., Hamada, H., Koyano, T., Kuchitsu, K. (2012). Intracellular localization and physiological function of a rice Ca2+ permeable channel OsTPC1. Plant Sign. Beh. 7, 1428–1430. doi: 10.4161/psb.22086
Latha, R., Chaluvadi, S., Subramaniam, H., Palanisami, E., Swaminathan, M. (2004). Approaches to breeding for salinity tolerance – a case study on Porteresia coarctata. Ann. Appl. Biol. 144, 177–184. doi: 10.1111/j.1744-7348.2004.tb00331.x
Le, T. D., Gathignol, F., Vu, H. T., Nguyen, K. L., Tran, L. H., Vu, H. T. T., Dinh, T. X., et al. (2021). Genome-wide association mapping of salinity tolerance at the seedling stage in a panel of Vietnamese landraces reveals new valuable QTLs for salinity stress tolerance breeding in rice. Plants 10 (6), 1088. doi: 10.3390/plants10061088
Lekklar, C., Pongpanich, M., Suriya-arunroj, D., Chinpongpanich, A., Tsai, H., Comai, L., et al. (2019). Genome-wide association study for salinity tolerance at the flowering stage in a panel of rice accessions from Thailand. BMC Genom 20 (1), 76. doi: 10.1186/s12864-018-5317-2
Liao, Y. D., Lin, K. H., Chen, C. C., Chiang, C. M. (2016). Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 36 (3), 22. doi: 10.1007/s11032-016-0446-2
Linh, L. H., Linh, T. H., Xuan, T. D., Ham, L. H., Ismail, A. M., Khanh, T. D. (2012). Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the red river delta of Vietnam. Int. J. Plant Genomics, 1–9. doi: 10.1155/2012/949038
Liu, C., Chen, K., Zhao, X., Wang, X., Shen, C., Zhu, Y., et al. (2019). Identification of genes for salt tolerance and yield-related traits in rice plants grown hydroponically and under saline field conditions by genome-wide association study. Rice 12 (1), 88. doi: 10.1186/s12284-019-0349-z
Luo, Q., Nakazawa, M., Sasayama, D., Fukayama, H., Hatanaka, T., Azuma, T. (2020). Responses to complete submergence in African rice (Oryza glaberrima). J.Crop Res. 65, 37–41. doi: 10.18964/jcr.65.0_37
Majee, M., Maitra, S., Dastidar, K. G., Pattnaik, S., Chatterjee, A., Hait, N. C., et al. (2004). A Novel Salt-tolerant l-myo-Inositol-1-phosphate Synthase from Porteresia coarctata (Roxb.) Tateoka, a Halophytic Wild Rice. J. Biol. Chem. 279 (27), 28539–28552. doi: 10.1074/jbc.m310138200
Mammadov, J., Buyyarapu, R., Guttikonda, S. K., Parliament, K., Abdurakhmonov, I. Y., Kumpatla, S. P. (2018). Wild relatives of maize, rice, cotton, and soybean: treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00886
Mathan, J., Singh, A., Ranjan, A. (2020). Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant 171 (4), 620–637. doi: 10.1111/ppl.13210
Meng, X., Zhou, J., Sui, N. (2018). Mechanisms of salt tolerance in halophytes: current understanding and recent advances. Open Life Sci. 13 (1), 149–154. doi: 10.1515/biol-2018-0020
Mishra, S., Chowrasia, S., Mondal, T. (2022). Dhani (Oryza coarctata): A wild relative of rice is a potential source of coastal salinity tolerance genes suitable for rice breeding (Transforming Coastal Zone for Sustainable Food and Income Security). doi: 10.1007/978-3-030-95618-9_2
Mishra, S., Singh, B., Panda, K., Singh, B. P., Singh, N., Misra, P., et al. (2016). Association of SNP haplotypes of HKT family genes with salt tolerance in Indian wild rice germplasm. Rice 9 (1), 15. doi: 10.1186/s12284-016-0083-8
Mishra, A., Tanna, B. (2017). Halophytes: potential resources for salt stress tolerance genes and promoters. Front. Plant Sci. 8 (829). doi: 10.3389/fpls.2017.00829
Mohammadi, R., Mendioro, M. S., Diaz, G. Q., Gregorio, G. B., Singh, R. K. (2013). Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice (Oryza sativa L.). J. Genet. 92 (3), 433–443. doi: 10.1007/s12041-013-0285-4
Mojakkir, A. M., Tareq, M. Z., Mottalib, M. A., Hoque, A. B. M. Z., Hossain, M. A. (2015). Effect of salinity at reproductive stage in rice. Int. J. Bus Soc. Sci. 3, 7–12.
Mondal, T. K., Rawal, H. C., Chowrasia, S., Varshney, D., Panda, A. K., Mazumdar, A., et al. (2018). Draft genome sequence of first monocot-halophytic species Oryza coarctata reveals stress-specific genes. Sci. Rep. 8 (1), 13698. doi: 10.1038/s41598-018-31518-y
Mukherjee, R., Mukherjee, A., Bandyopadhyay, S., Mukherjee, S., Sengupta, S., Ray, S., et al. (2019). Selective manipulation of the inositol metabolic pathway for induction of salt-tolerance in indica rice variety. Sci. Rep. 9 (1), 5358. doi: 10.1038/s41598-019-41809-7
Munns, R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167 (3), 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Munns, R., James, R. A., Gilliham, M., Flowers, T. J., Colmer, T. D. (2016). Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Funct. Plant Biol. 43 (12), 1103–1113. doi: 10.1071/FP16187
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Naheed, G., Shahbaz, M., latif, A., Rha, E. S. (2007). Alleviation of the adverse effects of salt stress on rice (Oryza sativa L.) by phosphorus applied through rooting medium: growth and gas exchange characteristics. Pak. J. Bot. 39 (3), 729–737.
Nair, M. M., Shylaraj, K. S. (2021). Introgression of dual abiotic stress tolerance QTLs (Saltol QTL and Sub1 gene) into Rice (Oryza sativa L.) variety Aiswarya through marker assisted backcross breeding. Physiol. Mol. Biol. Plants 27 (3), 497–514.1215. doi: 10.1007/s12298-020-00893-0
Nakamura, I., Murayama, S., Tobita, S., Bong, B. B., Yanagihara, S., Ishimine, et al. (2002). Effect of naCl on the photosynthesis, water relations and free proline accumulation in the wild oryza species. Plant Prod Sci. 5, 305–310. doi: 10.1626/pps.5.305
Nan, H., Li, W., Lin, Y. L., Gao, L. Z. (2020). Genome-wide analysis of WRKY genes and their response to salt stress in the wild progenitor of Asian cultivated rice, oryza rufipogon. Front. Genet. 11. doi: 10.3389/fgene.2020.00359
Nath, M., Yadav, S., Sahoo, R., Passricha, N., Tuteja, R., Tuteja, N. (2015). PDH45 transgenic rice maintain cell viability through lower accumulation of Na+, ROS and calcium homeostasis in roots under salinity stress. J. Plant Physiol. 191, 1–11. doi: 10.1016/j.jplph.2015.11.008
Nayyeripasand, L., Garoosi, G. A., Ahmadikhah, A. (2021). Genome-wide association study (GWAS) to identify salt-tolerance QTLs carrying novel candidate genes in rice during early vegetative stage. Rice 14 (1), 9. doi: 10.1186/s12284-020-00433-0
Negrao, S., Courtois, B., Ahmadi, N., Abreu, I., Saibo, N., Oliveira, M. M. (2011). Recent updates on salinity stress in rice: from physiological to molecular responses. Crit. Rev. Plant Sci. 30 (4), 329–377. doi: 10.1080/07352689.2011.587725
Negrao, S., Schmöckel, S. M., Tester, M. (2017). Evaluating physiological responses of plants to salinity stress. Ann. Bot. 119 (1), 1–11. doi: 10.1093/aob/mcw191
Nguyen, T. T. H. (2022). Investigating Australian wild rice for improvement of salinity stress tolerance in cultivated rice Oryza sativa L.) (Brisbane, Australia: Queensland University of Technology).
Nishizawa, Y., Nakamura, I., Hossain, M., Akamine, H., Zheng, S. H. (2015). Dry Matter Production and Photosynthetic Ability of Oryza officinalis Wall ex Watt under Salinity Stress Condition. Japanese J. Crop Sci. 84, 49–55. doi: 10.1626/jcs.84.49
Nishizawa, Y., Nakamura, I., Tamaki, M., Imura, Y., Hossain, M. A., Zheng, S. H. (2016). Change in O2 evolution rate and protein property in the leaf blade of Oryza officinalis wall ex watt in response to salinity. Japanese J. Crop Sci. 85, 411–420. doi: 10.1626/jcs.85.411
Padmavathi, G., Singh, R. K., Arun, M. N., Umakanth, B., Prasad, G. ,. S. V., Muralidharan, K. (2023). “Genetic enhancement for salt tolerance in rice,” in Molecular and physiological insights into plant stress. Ed. Chen, J.-T. (Intech Open), 312–356.
Perrino, E. V., Perrino, P. (2020). Crop wild relatives: know how past and present to improve future research, conservation and utilization strategies, especially in Italy: a review. Genet. Resour. Crop Evol. 67 (5), 1067–1105. doi: 10.1007/s10722-020-00930-7
Platten, J. D., Egdane, J. A., Ismail, A. M. (2013). Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism? BMC Plant Biol. 13 (1), 32. doi: 10.1186/1471-2229-13-32
Prakash, N. R., Lokeshkumar, B. M., Rathor, S., Warraich, A. S., Yadav, S., Vinaykumar, N. M., et al. (2022). Meta-analysis and validation of genomic loci governing seedling and reproductive stage salinity tolerance in rice. Physiol. Plant 174, e13629. doi: 10.1111/ppl.13629
Prasanth, V. V., Chakravarthi, D., Vishnukiran, T., Rao, Y. V., Madhusmita, P., Mangrauthia, S. K., et al. (2012). Evaluation of rice germplasm and introgression lines for heat tolerance. Ann Biol. Res. 3, 5060–5068.
Prusty, M. R., Kim, S. R., Vinarao, R., Entila, F., Egdane, J., Diaz, M. G. Q., et al. (2018). Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00417
Puram, V. R. R., Ontoy, J., Linscombe, S., Subudhi, P. K. (2017). Genetic Dissection of Seedling Stage Salinity Tolerance in Rice Using Introgression Lines of a Salt Tolerant Landrace Nona Bokra. J. Heredity 108 (6), 658–670. doi: 10.1093/jhered/esx067
Pushpalatha, G., Subrahmanyam, D., Sreenu, K., Ram, T., Subbarao, L. V., Parmar, B., et al. (2013). Effect of salt stress on seedling growth and antioxidant enzymes in two contrasting rice introgression lines. Indian J. Plant Physiol. 18 (4), 360–366. doi: 10.1007/s40502-014-0061-3
Quan, R., Wang, J., Hui, J., Bai, H., Lyu, X., Zhu, Y., et al. (2018). Improvement of salt tolerance using wild rice genes. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02269
Rana, M. M., Takamatsu, T., Baslam, M., Kaneko, K., Itoh, K., Harada, N., et al. (2019). Salt tolerance improvement in rice through efficient SNP marker-assisted selection coupled with speed-breeding. Int. J. Mol. Sci. 20 (10), 2585. doi: 10.3390/ijms20102585
Razzaq, A., Ali, A., Safdar, L. B., Zafar, M. M., Rui, Y., Shakeel, A., et al. (2020). Salt stress induces physiochemical alterations in rice grain composition and quality. J. Food Sci. 85 (1), 14–20. doi: 10.1111/1750-3841.14983
Razzaque, S., Haque, T., Elias, S. M., Rahman, M. S., Biswas, S., Schwartz, S., et al. (2017). Reproductive stage physiological and transcriptional responses to salinity stress in reciprocal populations derived from tolerant (Horkuch) and susceptible (IR29) rice. Sci. Rep. 7 (1), 46138. doi: 10.1038/srep46138
Reddy, A. M., Francies, R. M., Rasool, S. N., Reddy, V. R. P. (2014). Breeding for tolerance stress triggered by salinity in rice. Int. J. Appl. Biol. Pharm. 5, 167–176.
Reddy, I. N. B. L., Kim, S. M., Kim, B. K., Yoon, I. S., Kwon, T. R. (2017). Identification of rice accessions associated with K +/na + Ratio and salt tolerance based on physiological and molecular responses. Rice Sci. 24 (6), 360–364. doi: 10.1016/j.rsci.2017.10.002
Rekha, G., Abhilash Kumar, V., Gokulan, C. G., Koushik, M. B.V.N., Laxmi Prasanna, B., Kulkarni, S., et al. (2022). DRR dhan 58, a seedling stage salinity tolerant NIL of improved Samba Mahsuri shows superior performance in multi-location trials. Rice 15 (1), 45. doi: 10.1186/s12284-022-00591-3
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 (10), 1141–1146. doi: 10.1038/ng1643
Rodriguez-Llorente, I., Pajuelo, E., Navarro-Torre, S., Mesa-Marín, J., Caviedes, M. (2019). Bacterial endophytes from halophytes: how do they help plants to alleviate salt stress? In: Kumar, M., Etesami, H., Kumar, V. (eds). Saline Soil-based Agriculture by Halotolerant Microorganisms (Singapore: Springer) doi: 10.1007/978-981-13-8335-9_6
Rohila, J. S., Edwards, J. D., Tran, G. D., Jackson, A. K., McClung, A. M. (2019). Identification of superior alleles for seedling stage salt tolerance in the USDA rice mini-core collection. Plants 8 (11), 472. doi: 10.3390/plants8110472
Saade, S., Maurer, A., Shahid, M., Oakey, H., Schmockel, S. M., Negrão, S., et al. (2016). Yield related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci. Rep. 6 (1), 32586. doi: 10.1038/srep32586
Samantaray, S., Ali, J., Nicolas, K.L., Katara, J.L., Verma, R.S., Parameswaran, C., et al. (2021). Doubled Haploids in Rice Improvement: approaches, Applications, and Future Prospects. Rice Improvement. doi: 10.1007/978-3-030-66530-2_12
Sami, F., Yusuf, M., Faizan, M., Faraz, A., Hayat, S. (2016). Role of sugars under abiotic stress. Plant Physiol. Biochem. 109, 54–61. doi: 10.1016/j.plaphy.2016.09.005
Sanchez, P. L., Wing, R. A., Brar, D. S. (2013). “The wild relative of rice: genomes and genomics,” in Genetics and genomics of rice. Plant genetics and genomics: crops and models, vol. 5 . Eds. Zhang, Q., Wing, R. (New York, NY: Springer). doi: 10.1007/978-1-4614-7903-1_2
Santosh Kumar, V. V., Verma, R. K., Yadav, S. K., Yadav, P., Watts, A., Rao, M. V., et al. (2020). CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants. 26 (6), 1099–1110. doi: 10.1007/s12298-020-00819-w
Sasayama, D., Niikawa, M., Hatanaka, T., Fukayama, H., Azuma, T. (2022). Adaptive responses to flooding in wild rice species with various genomes other than AA. Plant Prod. Sci. 25, 350–358. doi: 10.1080/1343943X.2022.2073896
Sengupta, S., Majumder, A. L. (2009). Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: a physiological and proteomic approach. Planta 229 (4), 911–929. doi: 10.1007/s00425-008-0878-y
Sengupta, S., Majumder, A. ,. L. (2010). Porteresia coarctata (Roxb.) Tateoka, a wild rice: A potential model for studying salt-stress biology in rice. Plant Cell Environ. 33, 526–542. doi: 10.1111/j.1365-3040.2009.02054.x
Sengupta, S., Patra, B., Ray, S., Majumder, A. L. (2008). Inositol methyl tranferase from a halophytic wild rice, Porteresia coarctata Roxb. (Tateoka): regulation of pinitol synthesis under abiotic stress. Plant Cell Environ. 31 (10), 1442–1459. doi: 10.1111/j.1365-3040.2008.01850.x
Senthilkumar, P., Jithesh, M. N., Parani, M., Rajalakshmi, S., Praseetha, K., Parida, A. (2005). Salt stress effects on the accumulation of vacuolarHC-ATPase subunit c transcripts in wild rice, Porteresia coarctata (Roxb.) Tateoka. Curr. Sci. 89, 1386–1394.
Shahzad, B., Yun, P., Shabala, L., Zhou, M., Sellamuthu, G., Venkataraman, G., et al. (2022). Unravelling the physiological basis of salinity stress tolerance in cultivated and wild rice species. Funct. Plant Biol. 49 (4), 351–364. doi: 10.1071/FP21336
Singh, R. K., Redoña, E., Refuerzo, L. (2009). Varietal improvement for abiotic stress tolerance in crop plants: Special reference to salinity in rice. In: Abiotic Stress adaptation plants. Springer pp, 387–415. doi: 10.1007/978-90-481-3112-9_18
Singh, R. K., Kota, S., Flowers, T. J. (2021). Salt tolerance in rice: seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet 34 (11), 3495–3533. doi: 10.1007/s00122-021-03890-3
Singh, R. K., Redona, E., Refuerzo, L. (2010). Varietal improvement for abiotic stress tolerance in crop plants: special reference to salinity in rice. Abiotic Stress Adaptation Plants, 387–415. doi: 10.1007/978-90-481-3112-9_18
Singh, V., Singh, A. K., Mohapatra, T., Krishnan, S. ,. G., Ellur, R. K. (2018). Pusa Basmati 1121 – a rice variety with exceptional kernel elongation and volume expansion after cooking. Rice 11 (1), 19. doi: 10.1186/s12284-018-0213-6
Sirault, X. R., James, R. A., Furbank, R. T. (2009). A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct. Plant Biol. 36 (11), 970. doi: 10.1071/fp09182
Soares, F. A., Chiangmai, P. N., Laosutthipong, C. (2021). Effect of salinity on agronomic characteristics of three rice varieties (Oryza sativa L.) at tillering stage. RSU Int. Res. Conf., 584–592. doi: 10.14458/RSU.res.2021.142
Solis, C. A., Yong, M. T., Vinarao, R., Jena, K., Holford, P., Shabala, L., et al. (2020). Back to the wild: on a quest for donors toward salinity tolerant rice. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00323
Solis, C. A., Yong, M.-T., Zhou, M., Venkataraman, G., Shabala, L., Holford, P., et al. (2022). Evolutionary Significance of NHX Family and NHX1 in Salinity Stress Adaptation in the Genus Oryza. Int. J. Mol. Sci. 23 (4), 2092. doi: 10.3390/ijms23042092
Sripinyowanich, S., Klomsakul, P., Boonburapong, B., Bangyeekhun, T., Asami, T., Gu, H., et al. (2013). Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): The role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ. Exp. Bot. 86, 94–105. doi: 10.1016/j.envexpbot.2010.01.0
Stein, J. C., Yu, Y., Copetti, D., Zwickl, D. J., Zhang, L., Zhang, C., et al. (2018). Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50 (2), 285–296. doi: 10.1038/s41588-018-0040-0
Tang, W., Sun, J., Liu, J., Liu, F., Yan, J., Gou, X., et al. (2014). RNAi-directed downregulation of betaine aldehyde dehydrogenase 1 (OsBADH1) results in decreased stress tolerance and increased oxidative markers without affecting glycine betaine biosynthesis in rice (Oryza sativa). Plant Mol. Biol. 86 (4-5), 443–454. doi: 10.1007/s11103-014-0239-0
Teng, Y., LV, M., Zhang, X., Cai, M., Chen, T. (2022). BEAR1, a bHLH transcription factor, controls salt response genes to regulate rice salt response. J. Plant Biol. 65, 217–230. doi: 10.1007/s12374-022-09347-4
Theerawitaya, C., Samphumphuang, T., Tisarum, R., Siangliw, M., Cha-um, S., Takabe, T., et al. (2020). Expression level of Na+ homeostasis-related genes and salt-tolerant abilities in backcross introgression lines of rice crop under salt stress at reproductive stage. Protoplasma 257 (6), 1595–1606. doi: 10.1007/s00709-020-01533-w
Thomson, M. J., Ocampo, M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., et al. (2010). Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice 3 (2-3), 148–160. doi: 10.1007/s12284-010-9053-8
Thummala, S. R., Guttikonda, H., Tiwari, S., Ramanan, R., Baisakh, N., Sarla, N., et al. (2022). Whole-Genome sequencing of KMR3 and oryza rufipogon-Derived introgression line IL50-13 (Chinsurah nona 2/Gosaba 6) identifies candidate genes for high yield and salinity tolerance in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.810373
Tian, L., Tan, L., Liu, F., Cai, H., Sun, C. (2011). Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon. J. Genet. Genomics 38 (12), 593–601. doi: 10.1016/j.jgg.2011.11.005
Tian, Z., Li, J., He, X., Jia, X., Yang, F., Wang, Z. (2017). Grain Yield, Dry Weight and Phosphorus Accumulation and Translocation in Two Rice (Oryza sativa L.) Varieties as Affected by Salt-Alkali and Phosphorus. Sustainability. 9 (8), 1461. doi: 10.3390/su9081461
Tsai, Y. C., Chen, K. C., Cheng, T. S., Lee, C., Lin, S. H., Tung, C. W. (2019). Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 19 (1), 403. doi: 10.1186/s12870-019-1983-8
Usatov, A. V., Alabushev, A. V., Kostylev, P. I., Azarin, K. V., Makarenko, M. S., Usatova, O. A. (2015). Introgression the saltol QTL into the elite rice variety of Russia by marker-assisted selection. Am. J. Agric. Biol. Sci. 10 (4), 165–169. doi: 10.3844/ajabssp.2015.165.169
USDA-ARS. (2008). Research databases. Bibliography on salt tolerance (Riverside, CA: Salinity Lab. US Dep Agric Res Serv). Available at: http://www.ars.usda.gov/Services/docs.htm?docid=8908. George E, Brown Jr.
Vu, H. T. T., Le, D. D., Ismail, A. M., Le, H. H. (2012). Marker-assisted backcrossing (MABC) for improved salinity tolerance in rice (Oryza sativa L.) to cope with climate change in Vietnam. Aust. J. Crop Sci. 6 (12), 1649–1654.
Wairich, A., Wember, L. S., Gassama, L. J., Wu, L., Murugaiyan, V., Ricachenevsky, F. K., et al. (2021). Salt resistance of interspecific crosses of domesticated and wild rice species. j. Plant Nutr. Soil Sc. 184 (4), 492–507. doi: 10.1002/jpln.202100068
Wang, S., Cao, M., Ma, X., Chen, W., Zhao, J., Sun, C., et al. (2017). Integrated RNA Sequencing and QTL Mapping to Identify Candidate Genes from Oryza rufipogon Associated with Salt Tolerance at the Seedling Stage. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01427
Wang, Z. Y., Second, G., Tanksley, S. D. (1992). Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor. Appl. Genet. 83, 565–5581. doi: 10.1007/BF00226900
Warburton, M. L., Rauf, S., Marek, L., Hussain, M., Ogunola, O., de Jesus Sanchez Gonzalez, J. (2017). The use of crop wild relatives in maize and sunflower breeding. Crop Sci. 57 (3), 1227. doi: 10.2135/cropsci2016.10.0855
Warraich, A. S., Krishnamurthy, S. L., Sooch, B. S., Vinaykumar, N. M., Dushyanthkumar, B. M., Bose, J., et al. (2020). Rice GWAS reveals key genomic regions essential for salinity tolerance at reproductive stage. Acta Physiol. Plant 42 (8). doi: 10.1007/s11738-020-03123-y
Wing, R. A., Ammiraju, J. S., Luo, M., Kim, H., Yu, Y., Kudrna, D., et al. (2005). The oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 59 (1), 53–62. doi: 10.1007/s11103-004-6237-x
Wu, F., Yang, J., Yu, D., Xu, P. (2020). Identification and Validation a Major QTL from “Sea Rice 86” Seedlings Conferred Salt Tolerance. Agronomy. 10 (3), 410. doi: 10.3390/agronomy10030410
Xie, X., He, Z., Chen, N., Tang, Z., Wang, Q., Cai, Y. (2019). The roles of environmental factors in regulation of oxidative stress in plant. BioMed. Res. Int. 2019, 1–11. doi: 10.1155/2019/9732325
Xu, X., Liu, X., Ge, S., Jensen, J. D., Hu, F., Li, X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30, 105–111. doi: 10.1038/nbt.2050
Xu, C., Luo, M., Su, X., Yan, J., Shi, H., Yan, H., et al. (2022). SiMYB19 from foxtail millet (Setaria italica)Confers transgenic rice tolerance to high salt stress in the field. Int. J.Mol. Sci. 23, 756. doi: 10.3390/ijms23020756
Yadav, A. K., Kumar, A., Grover, N., Ellur, R. K., Krishnan, S. G., Bollinedi, H., et al. (2020). Marker aided introgression of “Saltol”, a major QTL for seedling stage salinity tolerance into an elite Basmati rice variety “Pusa Basmati 1509. Sci.Rep 10 (1), 13877. doi: 10.1038/s41598-020-70664-0
Yichie, Y., Brien, C., Berger, B., Roberts, T. H., Atwell, B. J. (2018). Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. Rice 11 (1), 66. doi: 10.1186/s12284-018-0257-7
Yin, W., Xiao, Y., Niu, M., Meng, W., Li, L., Zhang, X., et al. (2020). ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell. 32, 2292–2306. doi: 10.1105/tpc.19.00542
Yu, J., Zao, W., He, Q., Kim, T. S., Park, Y. J. (2017). Genome-wide association study and gene set analysis for understanding candidate genes involved in salt tolerance at the rice seedling stage. Mol. Genet. Genom. 292 (6), 1391–1403. doi: 10.1007/s00438-017-1354-9
Yuan, J., Wang, X., Zhao, Y., Khan, N. U., Zhao, Z., Zhao, Y., et al. (2020). Genetic basis and identification of candidate genes for salt tolerance in rice by GWAS. Sci. Rep. 10, 9958. doi: 10.1038/s41598-020-66604-7
Yuan, L., Zhang, L., Wei, X., Wang, R., Li, N., Chen, G., et al. (2022). Quantitative trait locus mapping of salt tolerance in wild rice oryza longistaminata. Int. J. Mol. Sci. 23 (4), 2379. doi: 10.3390/ijms23042379
Yue, E., Cao, H., Liu, B. (2020). OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in oryza sativa. Plants 9 (10), 1337. doi: 10.3390/plants9101337
Zelm, V. E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Ann. Rev. Plant Biol. 71 (1), 403–433. doi: 10.1146/annurev-arplant-050718-100005
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed 39 (3), 47. doi: 10.1007/s11032-019-0954-y
Zhang, X., Long, Y., Chen, X., Zhang, B., Xin, Y., Li, L., et al. (2021). A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 21 (1), 546. doi: 10.1186/s12870-021-03333-7
Zhang, Y., Ponce, K. S., Meng, L., Chakraborty, P., Zhao, Q., Guo, L., et al. (2020). QTL identification for salt tolerance related traits at the seedling stage in indica rice using a multi-parent advanced generation intercross (MAGIC) population. Plant Growth Regul. 92, 365–373. doi: 10.1007/s10725-020-00644-x
Zhao, C., Zhang, H., Song, C., Zhu, J. K., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 1 (1), 100017. doi: 10.1016/j.xinn.2020.100017
Zhou, Y., Chebotarov, D., Kudrna, D., Llaca, V., Lee, S., Rajasekar, S., et al. (2020). A platinum standard pan-genome resource that represents the population structure of Asian rice. Sci. Data 7 (1), 113. doi: 10.1038/s41597-020-0438-2
Keywords: rice, salt tolerance, mechanisms, wild gene pool, traditional breeding, MAS, transgenics, genomics
Citation: Padmavathi G, Bangale U, Rao K N, Balakrishnan D, Arun M N, Singh RK and Sundaram RM (2024) Progress and prospects in harnessing wild relatives for genetic enhancement of salt tolerance in rice. Front. Plant Sci. 14:1253726. doi: 10.3389/fpls.2023.1253726
Received: 06 July 2023; Accepted: 13 December 2023;
Published: 31 January 2024.
Edited by:
Vijay Gahlaut, Chandigarh University, IndiaReviewed by:
Reetika Mahajan, University of Toledo, United StatesDebabrata Panda, Central University of Orissa, Koraput, India
Copyright © 2024 Padmavathi, Bangale, Rao, Balakrishnan, Arun, Singh and Sundaram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guntupalli Padmavathi, cGFkbWFndW50dXBhbGxpNkBnbWFpbC5jb20=
 Guntupalli Padmavathi
Guntupalli Padmavathi Umakanth Bangale
Umakanth Bangale K. Nagendra Rao2
K. Nagendra Rao2 Divya Balakrishnan
Divya Balakrishnan Rakesh Kumar Singh
Rakesh Kumar Singh