- Centre for Novel Agricultural Products (CNAP), Department of Biology, University of York, York, United Kingdom
Introduction: Climate change is likely to lead to not only increased global temperatures but also a more variable climate where unseasonal periods of heat stress are more prevalent. This has been evidenced by the observation of spring-time temperatures approaching 40°C in some of the main spring-wheat producing countries, such as the USA, in recent years. With an optimum growth temperature of around 20°C, wheat is particularly prone to damage by heat stress. A warming climate with increasingly common fluctuations in temperature therefore threatens wheat crops and subsequently the lives and livelihoods of billions of people who depend on the crop for food. To futureproof wheat against a variable climate, a better understanding of the response to early heat stress is required.
Methods: Here, we utilised DESeq2 to identify 7,827 genes which were differentially expressed in wheat landraces after early heat stress exposure. Candidate hub genes, which may regulate the transcriptional response to early heat stress, were identified via weighted gene co-expression network analysis (WGCNA), and validated by qRT-PCR.
Results: Two of the most promising candidate hub genes (TraesCS3B02G409300 and TraesCS1B02G384900) may downregulate the expression of genes involved in the drought, salinity, and cold responses—genes which are unlikely to be required under heat stress—as well as photosynthesis genes and stress hormone signalling repressors, respectively. We also suggest a role for a poorly characterised sHSP hub gene (TraesCS4D02G212300), as an activator of the heat stress response, potentially inducing the expression of a vast suite of heat shock proteins and transcription factors known to play key roles in the heat stress response.
Discussion: The present work represents an exploratory examination of the heat-induced transcriptional change in wheat landrace seedlings and identifies several candidate hub genes which may act as regulators of this response and, thus, may be targets for breeders in the production of thermotolerant wheat varieties.
Introduction
The damaging effect of heat stress exposure on Triticum aestivum L. (bread wheat) yields is well known, with reductions between 3% and 6% being observed for every degree increase above the crop’s optimal growth temperature of 20°C (Chowdhury and Wardlaw, 1978; Kobza and Edwards, 1987; Wardlaw et al., 1989; Nagai and Makino, 2009; Ray et al., 2013; Zhao et al., 2017; Tian et al., 2018), with such heat-induced yield losses being evidenced in recent field trials (Riaz et al., 2021; Roychowdhury et al., 2023; Wang et al., 2023). These kinds of yield losses are likely to become more common in the coming years as a result of climate change and global warming, as, according to Intergovernmental Panel on Climate Change (IPCC) predictions, an increase in global mean surface temperatures of between 0.3°C and 4.8°C, compared with the prior century, are expected by 2100 (Collins et al., 2013), whereas other models predict more rapid global temperature increases, with such levels being reached by the year 2060 (Wigley and Raper, 2001; Murphy et al., 2004; De Costa, 2011). This is particularly worrying considering that global wheat consumption in 2021/2022 reached almost 800 million metric tonnes and currently accounts for 20% of the globe’s annual calorie consumption (Pfeifer et al., 2014; Food and Agriculture Organization of the United Nations et al., 2018; United States Department of Agriculture - Foreign Agricultural Service, 2023), meaning the lives, and livelihoods, of billions around the world depend on the success of the yields of this single crop.
Not only are yearly average global temperatures rising, but seasonal temperature patterns are likely to shift over the coming years, with warmer springs already being increasingly reported; for example, 10 of the 13 springs to have occurred since 2010 make up the warmest springs ever recorded, with spring 2022 ranking sixth on this list (NOAA National Centers for Environmental Information, 2022a). This trend toward increasingly warm spring months saw much of Western, Southern, and Central Europe experience record temperatures in May 2022, whereas Southern states of the USA experienced their fourth warmest May since records began in 1895 (NOAA National Centers for Environmental Information, 2022a; NOAA National Centers for Environmental Information, 2022b). Not only are average temperatures during the spring months rising globally, but unseasonal periods of extreme temperatures are expected to occur more frequently as the climate continues to change (Easterling et al., 2000; Thornton et al., 2014; Haokip et al., 2020). Evidence of such events have been observed recently, with the USA, Italy, and Turkey all experiencing periods of elevated temperature, above 30°C, in May 2020 (NOAA National Centers for Environmental Information, 2020), whereas parts of the American Midwest, such as major spring-wheat-producing state Minnesota, saw temperatures reach 38°C (NOAA National Centers for Environmental Information, 2018).
This is a pressing issue for much of the Northern Hemisphere, in addition to South Asia and the Middle East, as, in many western countries, spring wheat is often sown during March and April. The springtime sowing of seeds in these regions means warmer spring months, and increasingly prevalent periods of extreme temperatures coincide with the early vegetative development of spring wheat crops in regions of high production, such as the USA, Canada, and the United Kingdom. These countries produced over 80 million tonnes of wheat combined in 2021 (Food and Agriculture Organization of the United Nations, 2023) and so play crucial roles in global food security. Therefore, it is essential that spring wheat crops in such countries are protected against the increasingly likely threat of heat stress during early development.
The first step toward achieving this is to gain a better understanding of both thermotolerance and the heat stress response during early vegetative development. Having previously identified a candidate master hub, and three validated genetic markers, for early basal thermotolerance (Barratt et al., 2023a), the present work builds on this previous experiment, this time aiming to understand the transcriptional response to early heat stress in spring habit wheat landraces and identify candidate hub genes, which may regulate this response using weighted gene co-expression network analysis (WGCNA). Together, these works provide a comprehensive examination of early heat stress exposure in bread wheat, generating insights into how these processes may be regulated transcriptionally, and identifying genes which may be responsible for this regulation.
A handful of studies have examined the effect of heat stress on the transcriptome of wheat during vegetative development (Qin et al., 2008; Liu et al., 2015; Jin et al., 2020); however, this type of analysis paired with subsequent network analysis is less common, despite this approach enabling the identification of a small number of promising candidate genes potentially playing large regulatory roles in the stress response, reducing the time spent laboriously screening all of the identified stress-responsive genes. Similar combined approaches have been used in other contexts, however, such as to identify regulators of thermotolerance during vegetative development in wheat (Girousse et al., 2018; Mishra et al., 2021); response to heat and cold stresses, and basal thermotolerance in rice (Wang et al., 2022; Zeng et al., 2022; Boulanger et al., 2023); response to combined heat, drought, and salinity stresses in Brachypodium (Shaar-Moshe et al., 2017); and drought stress response in sugarcane (Tang et al., 2023), whilst we have previously used this combined approach to study the drought stress response in wheat (Barratt et al., 2023b). However, there are no similarly exploratory examples of this approach being used to study the heat stress response in wheat, as yet. Although one study has utilised similar approaches to identify genes that may be regulated by preexisting candidate genes under heat stress, by examining the effect of heat stress exposure on knockout mutants and wild-type plants (Tian et al., 2022), our study represents the first exploratory example of WGCNA utilisation to identify candidate hub genes which may regulate the heat stress response during vegetative development in climate-adapted bread wheat landraces.
Materials and methods
Plant growth conditions
The seeds used in the present work were from plants which derived from at least three generations of selfing. There were 13 accessions previously shown to be distinctively tolerant or susceptible under heat stress (Barratt et al., 2023a; Supplementary Data Sheet 1) sown in Levington Advance Seed & Modular F2S compost mixed with Aggregate Industries Garside Sands 16/30 sand in an 80:20 ratio, which was treated with Calypso insecticide (Bayer Crop Science Ltd., 0.083 ml mixed with 100 ml water, applied to each litre of compost). The heat stress treatment used in the present work was identical to that used previously (Barratt et al., 2023a), as it was found to significantly disrupt plant growth, without being lethal. Plants were placed into a Percival AR-75L growth cabinet with 18-h day length and respective day/night temperatures of 22°C and 16°C until the three-leaf stage. At this point, four replicates of each accession were transferred to a separate Percival AR-75L growth cabinet and exposed to 35°C/30°C (day/night) for 14 days, with all other conditions being the same as in the control cabinet. Both control and heat-stressed plants were well-watered, ensuring that the compost was kept moist with daily watering. 2-cm leaf tissue samples taken at the three-leaf stage and after 14 days of heat stress exposure were frozen in liquid nitrogen and stored at −80°C prior to RNA extraction.
RNA extraction, sequencing, and mapping
Leaf tissue samples weighing less than 100 mg were used for total RNA extraction via the E.Z.N.A Plant RNA Kit (Omega Bio-Tek, GA, USA) including a DNase treatment, according to the manufacturer’s protocol. Both NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, MA, USA) and Qubit 4 Fluorometer (Life Technologies, CA, USA) were used for quantification of RNA concentration, whereas Agilent Technology 2100 Bioanalyzer (Agilent Technologies, CA, USA) was used to assess RNA quality. Samples were deemed to be acceptable for use in subsequent analysis if their RNA Integrity Number (RIN) value was >7. To help control the effect of the environment on the transcriptome, prior to sequencing, we pooled acceptable RNA from at least three replicate plants per accession, per condition (pre- or post-heat stress), whereas biological replication for each treatment was provided by the different accessions. Samples were stored at −80°C and shipped on dry ice to Novogene (Cambridge, United Kingdom) for sequencing using the Illumina NovaSeq 6000 platform (Illumina, CA, USA) with a 150-bp paired-end sequencing strategy. Raw reads were trimmed using Trimmomatic v0.39 (Bolger et al., 2014) by removing leading and trailing low-quality or N bases (below quality 3), minimum length 36 bp and sliding window 4:15. FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of the data, then Salmon v0.8.1 (Patro et al., 2017) was used to map trimmed reads to the Triticum aestivum reference genome v1.1 (IWGSC RefSeq v1.1, http://ftp.ensemblgenomes.org/pub/plants/release-46/fasta/triticum_aestivum/). Salmon transcripts per million (TPM), counts, and lengths were inputted into R (version 4.1.2.; R Core Team, 2021) using TxImport (Soneson et al., 2015) for further analysis.
Transcriptomic and differential expression analyses
There were 26 pooled RNA samples from 13 accessions (before and after heat stress, for 13 accessions) used for transcriptomic analysis. After importing transcriptome data into R using TxImport, the principal component analysis (PCA) function of DESeq2 (version; 1.36.0; Love et al., 2014) was first used to explore count data from RNA-Seq. Genes with fewer than 10 non-zero entries were then removed, leaving 75,732 genes for differential expression analysis (DEA; Supplementary Data Sheet 2). DEA was carried out using the DESeq2 package (version 1.36.0; Love et al., 2014) in R, whereby an additive model was used to identify genes differentially expressed between tolerant and susceptible accessions, as well as between pre- and post-stress samples. To make our selection of DEGs robust to replicate variability, we used the adaptive shrinkage function (ashr) to shrink effect sizes of genes with high dispersion values. Genes which showed a log2FC above/below 1.5/−1.5 and an FDR-adjusted p-value (Benjamini and Hochberg, 1995) below, or equal to, 0.05 for either of these comparisons were deemed to be tolerance or response differentially expressed genes (DEGs), respectively. The adaptive shrinkage function was employed in the ranking of genes to shrink the log fold-change estimates of genes with low counts or high dispersion (Stephens, 2017). Due to extremely low numbers of tolerance DEGs identified in the DEA, only response DEGs were studied further.
GO term enrichment analysis
To identify gene ontology (GO) terms significantly enriched amongst upregulated and downregulated response DEGs identified via DEA, GO enrichment analysis was conducted. An approach, used previously (Borrill et al., 2019; Andleeb et al., 2023), was adopted to transfer GO terms to the v1.1 annotation, from the IWGSC RefSeqv1.0 genome annotation, as GO terms are only available for the v1.0 annotation. The list of genes for which GO terms can be transferred can be found in Andleeb et al. (2023). IWGSC v1.0 GO terms were read into R using the base R function readRDS() for analysis, after being retrieved from https://opendata.earlham.ac.uk/wheat/under_license/toronto/Ramirez-Gonzalez_etal_2018-06025-Transcriptome-Landscape/data/TablesForExploration/FunctionalAnnotation.rds.
GO terms that upregulated and downregulated response DEGs are annotated with were then collated into two groups, before the agriGO Singular Enrichment Analysis tool (Du et al., 2010; Tian et al., 2017) was used to conduct a Fisher’s exact test on both groups of GO terms, with the GO terms of all genes included in DEA serving as background. 0.05 was the p-value threshold; Hochberg (FDR) was the multi-test adjustment method (Benjamini and Hochberg, 1995), and 5 was the minimum number of mapping entries threshold. Significantly enriched GO terms had an FDR-adjusted p-value < 0.05. AgriGO’s DAG Drawer tool was also used to generate DAG trees for significantly enriched GO terms.
Network construction and module detection
A single co-expression network was constructed via the WGCNA R package (Langfelder and Horvath, 2008; Langfelder and Horvath, 2012), using TPM data provided by Salmon. No samples were removed after clustering, but 19,965 genes were removed due to too many zero values: leaving 87,580 genes from 26 samples (13 accessions, before and after heat stress exposure) for network construction. The blockwiseModules() function conducted blockwise network construction according to the function’s default parameters, except the following: network type = signed hybrid, maximum block size = 5000, soft threshold power = 8 (advised by the package’s authors for this number of samples, as no soft threshold power exceeded a reasonable scale-free topology fix index of 0.8), minimum module size = 30, merge cut height = 0.25. After module detection, edge and node files were created using the “exportNetworkToCytoscape()” function with a threshold of 0.1; filtering out weak connections between genes (nodes). Results of sample clustering, scale-free topology fit index as a function of the soft-thresholding power, and mean connectivity as a function of the soft-thresholding power can be found in Figure 1. Gene expression data after sample clustering and processing via WGCNA, and network construction data are available on GitHub: https://github.com/andreaharper/HarperLabScripts/.
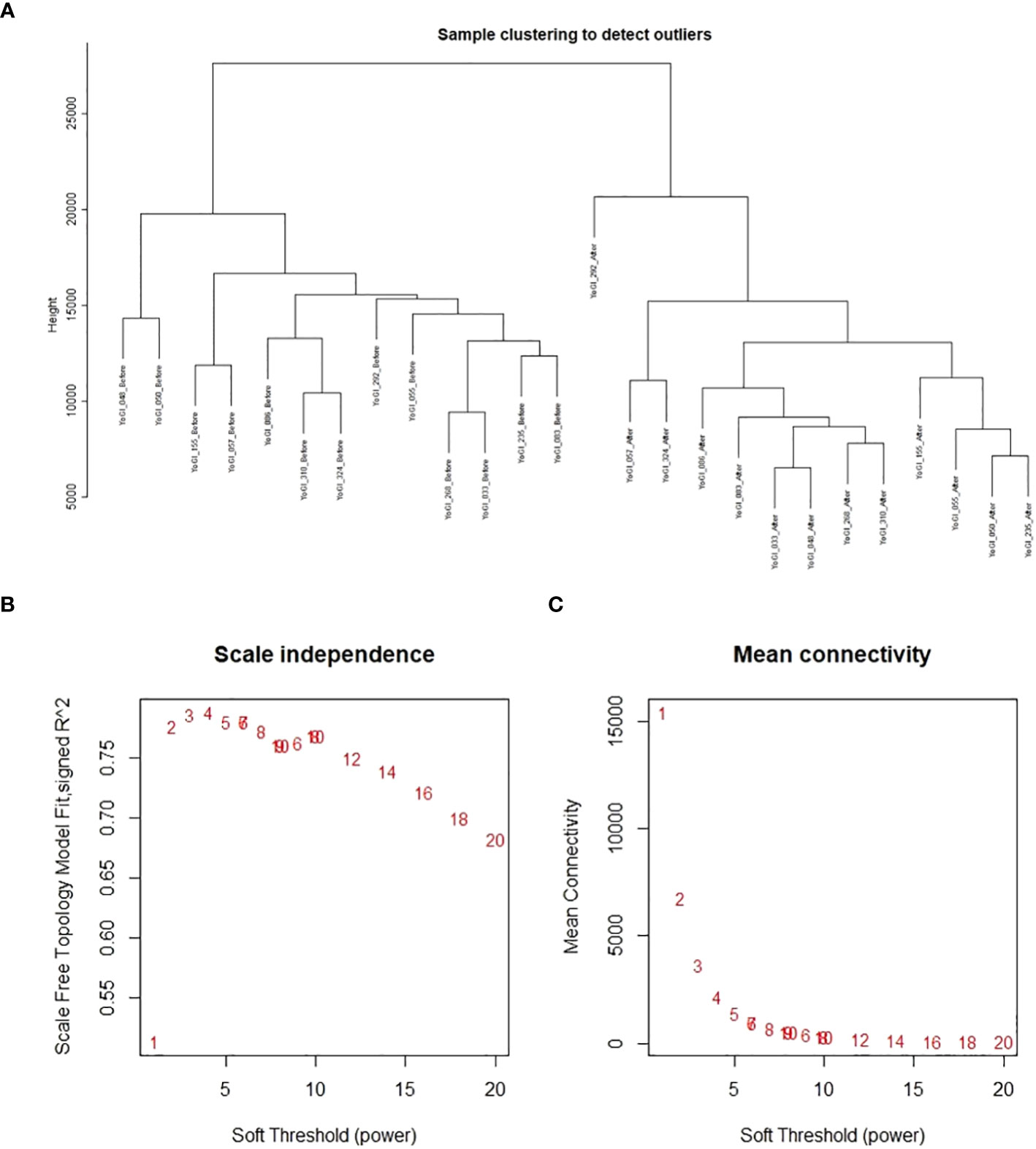
Figure 1 Analyses performed by WGCNA prior to co-expression network construction. Results of sample clustering (A), scale-free topology fit index as a function of the soft-thresholding power (B), and mean connectivity as a function of the soft-thresholding power (C).
Identifying stress-associated modules
To understand the likely functions of genes within each module, GO enrichment analysis was conducted using the same approach as outlined above. Here, however, GO terms associated with genes within a module were collated and submitted to the agriGO Singular Enrichment Analysis tool, with the GO terms of all genes included in the network serving as background. All other parameters were the same as described above.
As well as this, we also conducted DEG enrichment analysis to identify which modules in the co-expression network contained a significantly larger proportion of response DEGs than expected and thus may be particularly associated with the heat stress response. To test whether a module was significantly enriched in response DEGs (observed proportion of DEGs above 8.94%), a one-proportion Z-test was used. Modules were deemed to be significantly enriched in DEGs if p was < 0.05.
Network visualisation and hub identification
Degree (connection) scores were calculated for each gene, via either the Cytoscape network analyser tool (Assenov et al., 2008), after first visualising network modules in Cytoscape (version 3.9.1.; Shannon et al., 2003), or via counting the number of connections to and from each gene in the module’s WGCNA edge file using the table() function in R. The script used to calculate degree scores in R is available on GitHub (https://github.com/andreaharper/HarperLabScripts/). Visualisation and analysis in Cytoscape were used to identify hub genes in the majority of the modules; however, the edge counting method in R was used to calculate degree scores for genes in particularly large modules (containing more than ~2,000 genes), which often cannot easily be loaded, viewed, and analysed in Cytoscape. For the largest modules, the R package vroom (version 1.6.3.; https://vroom.r-lib.org) was used to read Cytoscape edge files into R for analysis.
We selected hub genes for further analysis based on their high degree scores, significant levels of differential expression, and annotated functions with potential regulatory roles. In cases where multiple genes within a module shared the highest degree score, or the highest degree-scoring genes were found not to be differentially expressed under heat stress, the highest-scoring DEG was deemed to be the hub gene. These well-connected DEGs were selected for further enquiry as they were deemed to be more likely to act as coordinators of the transcriptional response to heat stress than well-connected genes that were not differentially expressed. Where the putative function of the most well-connected DEG suggested no involvement in either the control of gene expression (be that directly as a transcription factor, or more indirectly via involvement in signalling pathways), or in the heat stress response/thermotolerance (for example, as a heat shock protein; HSP), other DEGs with similar degree scores, which were predicted to play such roles based on their annotation, were favoured as the candidate hub gene. If no such well-connected DEGs within a module were likely involved in such processes, the most well-connected DEG was deemed to be the module’s hub gene. Uncharacterised hub genes were studied further, as they represented novel candidates for potential regulators of the heat stress response. Orthologues of hub genes, and genes they were connected to, were identified via Ensembl Plants (Yates et al., 2022).
qRT-PCR
cDNA was obtained from the RNA extracted for each one of the biological replicates of the 13 landraces (4 biological replicates, 2 treatments) of the mRNA-Seq experiment. The reaction was carried out using the ImProm-II™ Reverse Transcription System (Promega) using the manufacturer’s instruction, 1 µl of Oligo(dT)16 (5 µM) (Eurogentec Ltd, Camberley, UK) and 1µl of each RNA sample. Quantification was performed with a Nanodrop 2000 and after that, the cDNA of the 4 biological replicates were pooled in equimolar concentrations. qRT-PCR was performed for the genes TraesCS1B02G384900, TraesCS3B02G409300 and TraesCS4D02G212300 and tubulin using the primers described in Supplementary Data Sheet 7 and the iTaq Universal SYBR Green Supermix (Bio-Rad), adding 200ng of cDNA and 0.1 µM of each primes. The qRT-PCR protocol was set on QuantStudio™ 7 Pro Real-Time PCR System (ThermoFisher) as follows: 95 °C for 4.5 min, 40 cycles of 95°C for 15s and 60°C for 15s. The melting curve was performed by initially heating in a 4.5°C/s ratio up to 95°C and maintaining for 10s reducing the temperature to 3.44°C/s up to 60°C and heating in a 0.15°C/s to 95°C kept for 10s with fluorescence measurement in the last step of the PCR and melting curve. The relative expression between after- and before-heat samples was calculated using the delta-delta Ct method using tubulin as the reference.
Results
Transcriptome sequencing and quantification
An average of 49,798,391 reads were obtained from each sample (minimum of 39,938,210 and maximum of 60,591,112) with an average of 92.5% of reads with Q30 and a GC content of 54.9. After trimming, an average of 2,888,493 reads were kept for each sample. An average of 71.5% of the trimmed reads mapped against the wheat genome. Raw data and Salmon outputs are publicly available in the Gene Ontology Repository (accession number: GSE23236). DESeq2 was used to variance-stabilize counts from all 26 samples, before the 500 most variable genes were assessed via principal component analysis (PCA; Figure 2A). PC1 and PC2, combined, explained 37% of the total variance, with clear distinction between samples taken before and after heat stress exposure being apparent on PC1 (which explained 27% of the observed variation). PC2 explained a smaller proportion of the total variation (10%) and provided some separation between samples, likely relating to variation in each accession’s geographical point of origin.
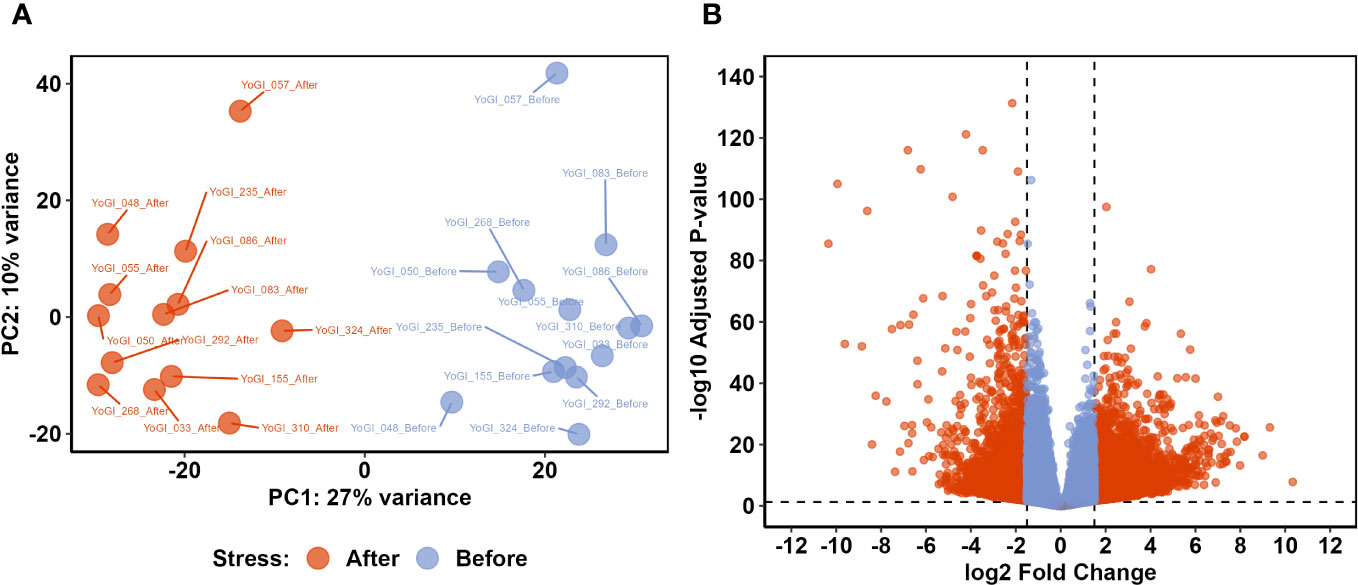
Figure 2 Comparative transcriptomic analysis identified a shift in the wheat transcriptome after exposure to early heat stress. Principal component analysis (PCA) of variance-stabilised counts from all 26 samples (A) showed clear separation between the two groups on PC1. Differential expression analysis identified 7,827 DEGs with significantly different expression before and after exposure to early heat stress (B). Dashed lines indicate DEG thresholds: vertical lines represent the log2FC thresholds of ±1.5, whereas horizontal lines represent the p-value threshold of 0.05. DEGs which met these criteria are beyond these threshold lines, coloured red.
Identification of DEGs and comparative transcriptomic analysis
To identify genes which may be involved in the heat stress response and basal thermotolerance, we employed DEA via DESeq2 (Love et al., 2014). The analysis identified 7,827 genes which were significantly differentially expressed before and after heat stress exposure (response DEGs; Supplementary Data Sheet 3), as well as 93 genes which were differentially expressed between tolerant and susceptible accessions (tolerance DEGs; Supplementary Data Sheet 3). Of the response DEGs, 5,384 were significantly upregulated after heat stress exposure, whereas 2,443 were significantly downregulated (Figure 2B). There were 41 tolerance DEGs expressed at significantly higher levels in tolerant accessions, whereas 52 tolerance DEGs were expressed more in susceptible accessions. The total number of tolerance DEGs was almost 100-fold less than the total number of response DEGs, and so response DEGs became the main point of inquiry in the subsequent analyses.
To understand the likely functionalities of the genes differentially expressed under heat stress, and to examine the differences in gene functionalities between these groups, we conducted GO enrichment analysis on the two DEG groups (Supplementary Figure 1; Supplementary Data Sheet 4). We found that GO terms related to DNA damage and replication [for example; “DNA integrity checkpoint” (GO:0031570), “DNA damage checkpoint” (GO:0000077), and “DNA replication” (GO:0006260)] were significantly enriched amongst upregulated DEGs, as was the term “protein refolding” (GO:0042026). We also found an abundance of terms related to cell wall processes [for example; “Cell wall assembly” (GO:0070726) and “Plant-type cell wall organization or biogenesis” (GO:0071669)], as well terms related to both cellulose [for example; “Cellulose microfibril organization” (GO:0010215) and “Cellulose biosynthetic process” (GO:0030244)] and lignin [for example; “Lignin metabolic process” (GO:0009808) and “Phenylpropanoid metabolic process” (9.7e-05)] synthesis and organization.
However, amongst downregulated DEGs, terms related to photosynthesis were significantly enriched, for example: “Photosynthesis” (GO:0015979), “Photosynthesis, light reaction” (GO:0019684) and “Photosynthetic electron transport in photosystem II” (GO:0009772). Terms related to the drought response were also significantly enriched, for example: “Response to water” (GO:0009415) and “Trehalose biosynthetic process” (GO:0005992), as were terms related to the general stress response, for example: “Response to stress” (GO:0006950), “Response to oxidative stress” (GO:0006979) and “Response to stimulus” (GO:0050896). Similarly, terms potentially related to the salinity response, for example: “Ion transport” (GO:0006811), “Cation transport” (GO:0006812), “Ion homeostasis” (GO:0050801) and “Sodium ion transport” (GO:0006814) were also significantly enriched amongst downregulated DEGs.
Identifying stress-associated modules
The co-expression network was consisted of 73 modules (Supplementary Data Sheet 5), housing 87,580 genes. Modules within the co-expression network ranged in size from 36 to 26,420 genes, whereas mean and median module size were 1,120 and 310 genes, respectively.
Modules significantly enriched in the “response to heat” (GO:0006951), “response to temperature stimulus” (GO:0009266) or “response to stress” (GO:0006950) GO terms likely contain genes involved in the response to heat stress. We found that 11 modules were significantly enriched in these, or other stress-associated, GO terms (Table 1), with the turquoise and yellow modules being significantly enriched, specifically, in the “response to heat” GO term. Although it was enriched in the “response to water” (GO:0009415) GO term, the black module may also contain genes involved in responding to elevated temperatures, as drought stress often occurs simultaneously with heat stress. The most significantly enriched GO term, and any significantly enriched stress-associated GO terms, in each module can be seen in Supplementary Data Sheet 6.
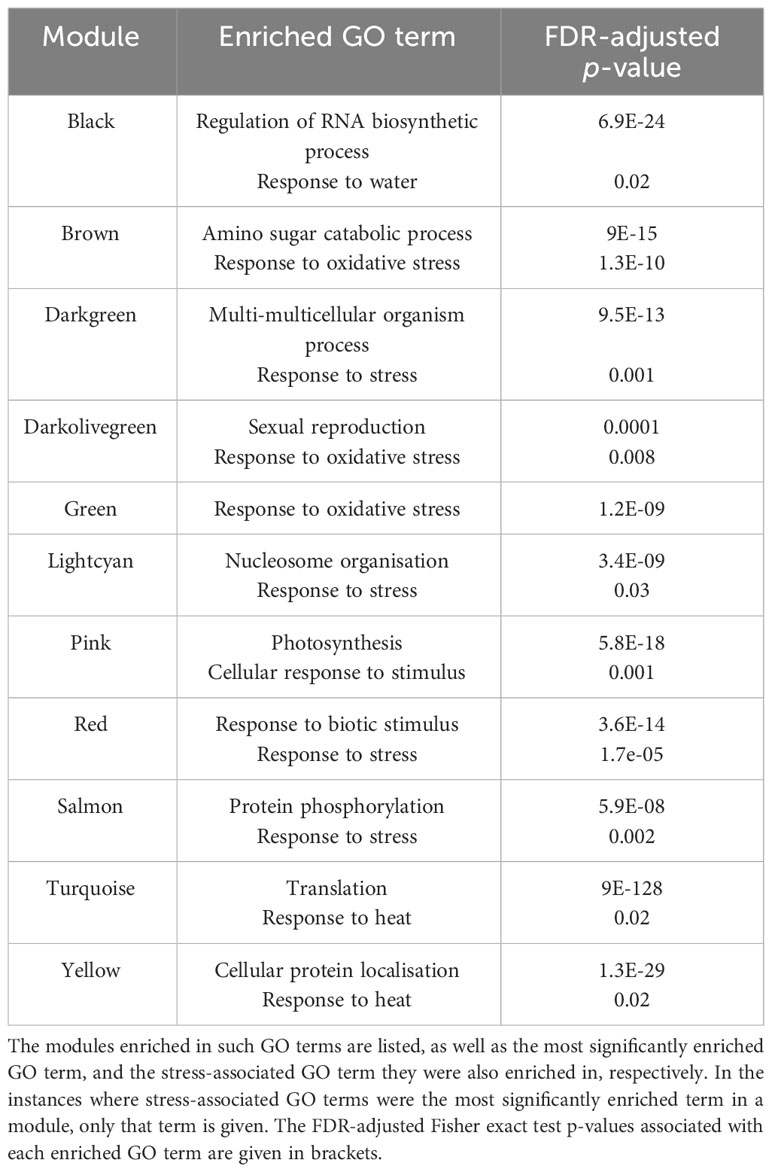
Table 1 A total of 11 modules significantly enriched in GO terms related to the stress response, according to GO enrichment analysis by the AgriGO v2.0 Singular Enrichment Analysis tool (Du et al., 2010; Tian et al., 2017).
To further explore which modules may be associated with the heat stress response, we aimed to identify modules significantly enriched in response DEGs, via DEG enrichment analysis. We found that 11 modules were significantly enriched in response DEGs (p < 0.05), with a one-proportion Z-test identifying that the proportion of DEGs in these modules was significantly greater than the expected proportion of 8.94% (Table 2). Amongst these modules, the observed proportions of DEGs ranged from 11.4% (turquoise) to 39.9% (pink).
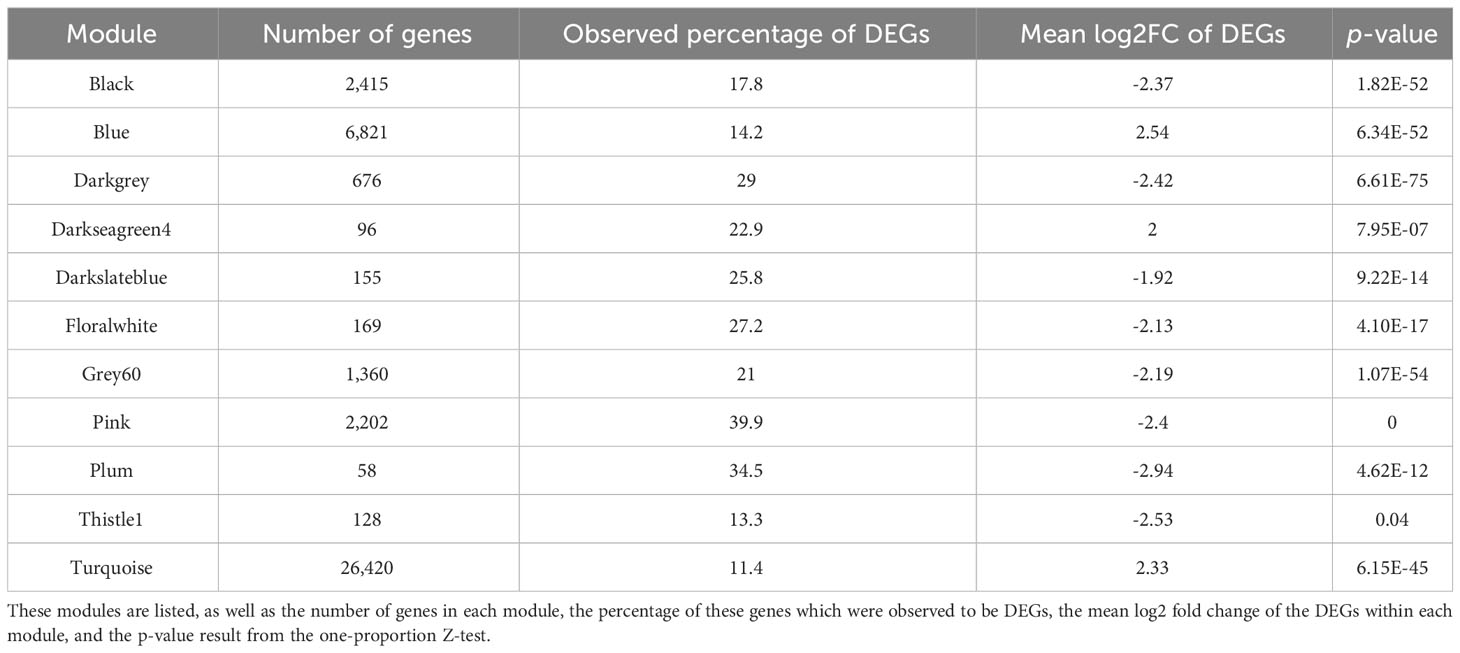
Table 2 A total of 11 modules were significantly enriched in DEGs, as they contained a significantly higher proportion of DEGs than expected should the total number have been distributed across modules according to their size (8.94%).
Hub gene identification
Within these stress-associated modules, determined either due to an enrichment of stress-associated GO terms (Table 1) or an enrichment of DEGs (Table 2), well-connected DEGs were identified as hub genes which may act to coordinate the transcriptional response to early heat stress. These hub genes are seemingly involved in a range of processes, from thermotolerance, to stress hormone signalling and photosynthesis (Table 3). However, three of these hub genes, in particular (TraesCS1B02G384900, TraesCS3B02G409300, and TraesCS4D02G212300; Figure 3), were deemed to be the most promising candidate hub genes, not only potentially regulating the transcriptional heat stress response (like the other hub genes) but also the physiological heat stress response—thanks to their own likely function, and the likely functions of the genes they are connected to in their respective modules. Both TraesCS1B02G384900 and TraesCS3B02G409300 may determine the expression of potentially superfluous genes, as well as the expression of stress hormone signalling repressors and photosynthesis genes, respectively. TraesCS4D02G212300, however, may coordinate the expression of a vast suite of heat shock proteins (HSPs), small heat shock proteins (sHSPs), and stress-responsive transcription factors.
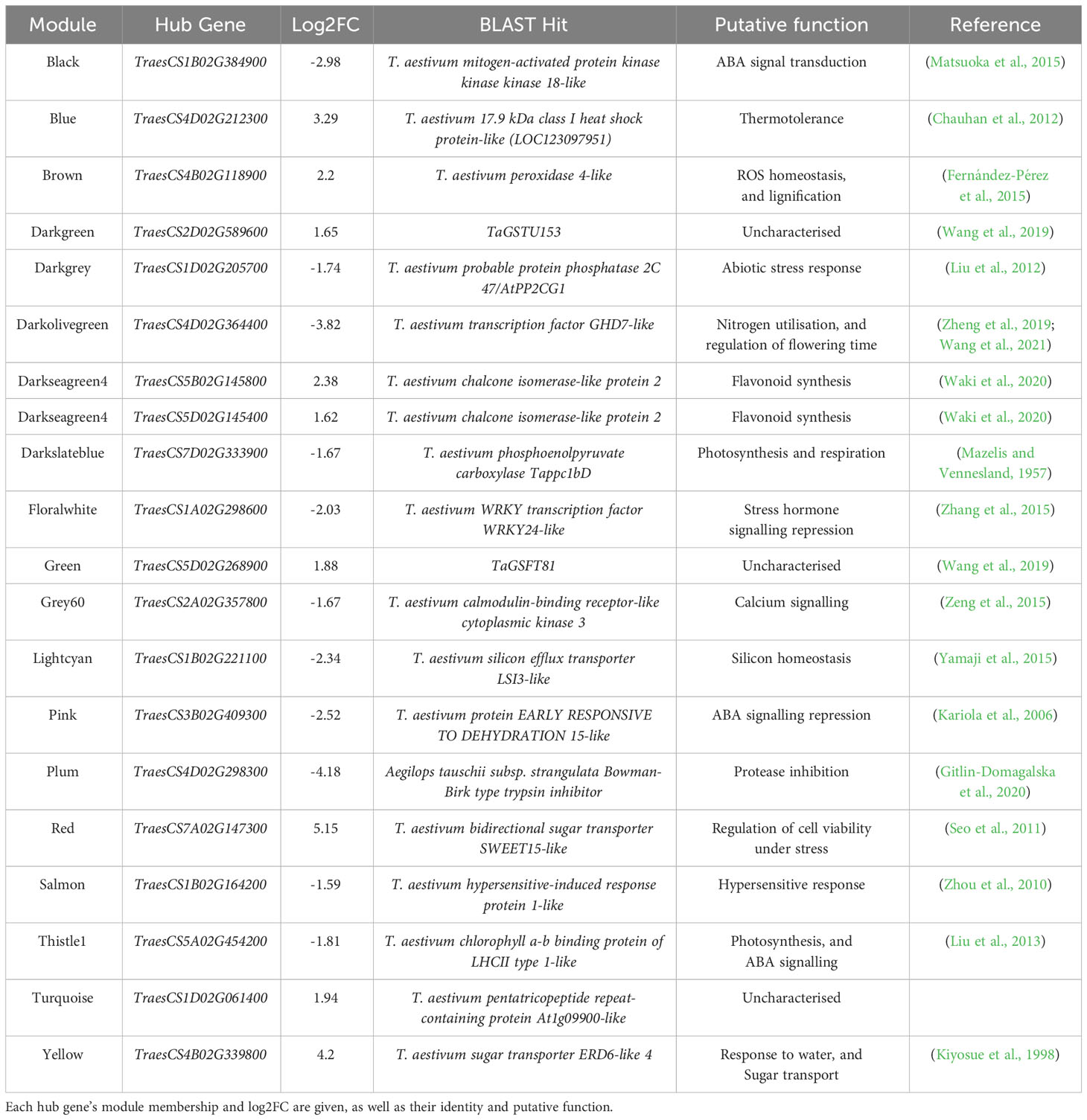
Table 3 Hub genes identified in stress-associated modules may be strong candidates for regulators of the heat stress response, based on their high number of connections to other genes within stress-associated modules.
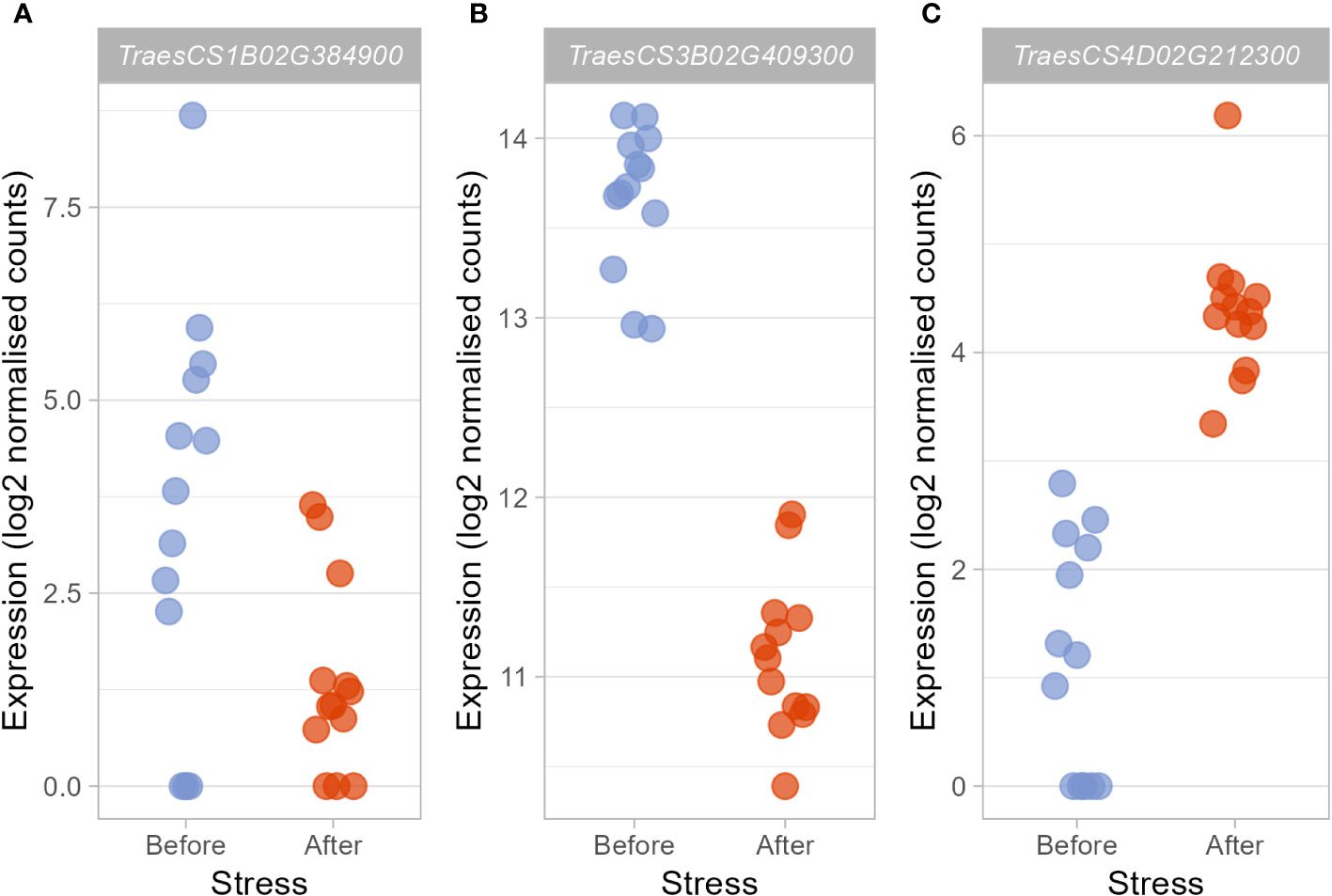
Figure 3 Candidate hub genes which may regulate the heat stress response were differentially expressed after heat stress exposure. Those hub genes deemed to be particularly promising based on their membership within stress-associated modules, their putative function, and the putative functions of the DEGs they were connected to showed varying responses to heat stress. TraesCS1B02G384900 (A) and TraesCS3B02G409300 (B) were significantly downregulated (log2FC = −2.98 and −2.52, respectively), whereas expression of TraesCS4D02G212300 (C) was significantly upregulated (log2FC = 3.29).
qRT PCR validation
qRT-PCR confirmed the patterns of expression for TraesCS3B02G409300 (t-test, t(24)=5.09, p<0.0001, n=26) and TraesCS4D02G212300 (t-test, t(19.83)=-6.56, p<0.0001, n=26), which were found to be down- and up-regulated respectively after heat stress, supporting the role of these genes in activating the early heat stress response. However we were not able to confirm the down-regulation of TraesCS1B02G384900 by qRT-PCR (t-test, t(23.59, p=0.98, n=26).
Discussion
Heat stress causes widespread changes in the wheat transcriptome
In the present work, we demonstrate that the expression profiles of almost 8,000 genes in the spring wheat transcriptome are significantly altered by exposure to early heat stress; 5,384 and 2,443 genes being significantly upregulated and downregulated, respectively. Amongst these groups of DEGs, genes with different functionalities were significantly enriched. Perhaps predictably given their importance as part of the heat stress response, genes involved in protein refolding were enriched amongst upregulated DEGs (Wang et al., 2004; Kotak et al., 2007; Al-Whaibi, 2011; Mogk et al., 2018; Tian et al., 2021). As well as disrupting protein homeostasis, periods of elevated temperature will also cause single- and double-stranded breaks in DNA, whilst also halting the progression of the replication fork (Velichko et al., 2012; Kantidze et al., 2016; Han et al., 2021). The need to protect cells against such heat-induced DNA damage is a key part of the heat stress response, shown previously to increase thermotolerance in Arabidopsis (Han et al., 2020) and evidenced by the enrichment of GO terms related to DNA replication and repair amongst upregulated DEGs. Similarly, we observed the enrichment of GO terms related to cell wall processes and lignin biosynthesis—likely evidence of the cell wall remodelling known to occur in plants during periods of high temperature (Yang et al., 2006; Lima et al., 2013; Le Gall et al., 2015; Wu et al., 2018; Pinski et al., 2021), with lignin synthesis being identified as an important thermotolerance mechanism in rice (Cai et al., 2020).
Amongst downregulated DEGs, however, we saw the significant enrichment of many terms related to photosynthesis, and photosystem II (PSII) in particular. PSII is particularly vulnerable to damage by heat stress (Yamamoto, 2016; Wang et al., 2018; Hu et al., 2020); therefore, the abundance of these genes amongst downregulated DEGs suggests a partial shutdown of PSII and, thus, a reduced photosynthetic rate under heat stress. Interestingly, we saw the enrichment of terms related to the general stress response amongst downregulated DEGs, suggesting these genes play no role in the tailored heat stress response. Perhaps related to this, we also found that terms related to the response to drought and salinity were enriched amongst downregulated DEGs, including the orthologue of AtPP2CG1, which responds to abscisic acid and positively regulates salt stress tolerance in Arabidopsis (Liu et al., 2012). It may be possible, therefore, that these genes are downregulated to increase transcriptional capacity for genes involved directly in the heat stress response. Similar widespread downregulation of drought- and salinity-responsive genes under heat stress has not been extensively described previously in similar works in wheat (Qin et al., 2008; Rangan et al., 2020; Azameti et al., 2022; Lee et al., 2022). Nor was a comparable shift seen when we examined the effect of early drought stress on the wheat transcriptome (Barratt et al., 2023b); for example, only 161 (2.99%) of the 5,384 upregulated heat-responsive DEGs identified in the present work were also downregulated under early drought stress in our previous work, whereas almost double this number of upregulated drought DEGs (321) were downregulated in the present work after heat stress exposure.
This perhaps speaks to the similarity of the different abiotic stresses, as although drought, salinity, freezing, and heat stresses all cause damage to protein structure, functionality, and cell membrane stability, there exist stress-specific cellular environments under drought, salinity, and freezing stresses that are not observed in well-watered plants exposed to high temperatures—such as desiccation, ion imbalance, and ice crystal formation. It is those genes involved in responding to these specialised cellular environments, therefore, that are likely to be superfluous under heat stress and, subsequently, are also likely to be downregulated. The same cannot be said for many of the genes involved in responding to heat stress, however, as these genes are largely involved in mitigating the effects common amongst all the abiotic stresses, particularly damage to proteins and membranes. The relatively large crossover potential of such genes in responding to different abiotic stresses, as a result of the similarities in cellular damage caused by these stresses, is evidenced by the fact that 22% (1,184 of 5,384) of the upregulated DEGs identified in the present work were also upregulated after exposure to early drought stress in our previous work (Barratt et al., 2023b), and by the observations of key heat stress-responsive gene families, such as HSPs, acting to enhance tolerance to drought and salinity stresses, as well as heat, in other species (Gao et al., 2012; Li et al., 2016; Zhai et al., 2016; Guo et al., 2020; Jiang et al., 2020; Rahman et al., 2022; Do et al., 2023).
TaMAPKKK18-like and TaERD15-like may coordinate a transcriptional shift away from growth and the response to abiotic stresses other than heat
Given that the downregulation of genes likely involved in responding to abiotic stresses other than heat seems to be a substantial constituent of the transcriptional heat stress response in wheat landraces, it was interesting that two of the hub genes identified in the co-expression network were connected to a large number of such downregulated DEGs in their respective modules, with one itself likely playing a role in the cold stress response and cold tolerance. The black module contained almost twice as many DEGs as expected (expected number = 216, observed number = 429, p = 1.82E-52) and was enriched in the “Response to abiotic stimulus” and “Response to water” GO terms (FDR-adjusted p = 0.004 and 0.019, respectively). TraesCS1B02G384900, TaMAPKKK18-like, was downregulated under heat stress (log2FC = -2.98, Figure 3A) and identified as a hub gene within the black module, being connected to 428 DEGs (100% of the remaining DEGs in the module). Meanwhile, the pink module was identified as particularly associated with the heat stress response as it was significantly enriched in DEGs (expected number = 197, observed number = 879, p = 0), whilst also being enriched in the GO terms “photosynthesis” and “cellular response to stimulus” (p = 1.5E-33 and 0.001, respectively). The hub gene in the module was TraesCS3B02G409300, Triticum aestivum EARLY RESPONSIVE TO DEHYDRATION 15-like (TaERD15-like). TaERD15-like was also found to be downregulated under heat stress (−2.52, Figure 3B), and was connected to 845 of the 879 remaining DEGs in the module (96%)—all of which were also downregulated.
TaMAPKKK18-like’s orthologue (identified via Ensembl Plants; Yates et al., 2022) in Arabidopsis, AtMAPKKK18, is a key part of ABA-mediated signal transduction, as it acts to phosphorylate proteins in an ABA-dependent manner (Matsuoka et al., 2015). This kinase activity can determine leaf senescence, growth, and stomatal dynamics, as overexpression of the gene led to smaller plants and increased leaf senescence of rosette leaves (Matsuoka et al., 2015), whereas knockout mutants showed more vigorous root growth, as well as increased stomatal aperture (Mitula et al., 2015)—suggesting a link with water use, and subsequently, the drought response. TaERD15-like’s orthologue in rice, OsERD15, is known to be both expressed more in cold-tolerant varieties and also to be induced during cold stress exposure (Sperotto et al., 2018; Rativa et al., 2020). Rice and wheat ERD15 proteins are relatively poorly characterised; however, in Arabidopsis, they are known to be integral players in the response to abiotic stress, mainly drought and cold, as they act as negative regulators of ABA signalling (Kariola et al., 2006; Aalto et al., 2012). In addition to a likely role responding to cold stress, which would be unrequired under high temperatures, a potential duty repressing ABA signalling may also explain the downregulation of the hub gene here, due to the key roles ABA plays during the heat stress response, including increasing antioxidant activity and sucrose metabolism, as well as upregulating the expression of HSPs and Hsfs (Li et al., 2020).
The likely involvement of both genes in ABA signal transduction, therefore, perhaps explains their identification as hub genes within their respective modules—as the expression of many genes in the wheat transcriptome will respond to this integral signal. TaMAPKKK18-like, however, may also be able to have far-reaching effects on gene expression thanks to connections to a suite of heat-responsive transcription factors and signalling proteins: 14% of the DEGs TaMAPKKK18-like was connected to in the module were transcription factors (from gene families such as MYB, WRKY, DREB, ERF, and Hsf), whereas TaMAPKKK18-like was also connected to 17 differentially expressed JAZ proteins—key repressors of JA-signalling and JA-induced gene expression (Santner and Estelle, 2007; Kazan and Manners, 2012; Wager and Browse, 2012; Sasaki-Sekimoto et al., 2014).
Within their respective modules, TaMAPKKK18-like and TaERD15-like were connected to a large number of DEGs which appear to be involved in responding to abiotic stresses other than heat. For instance, connected to TaMAPKKK18-like in the black module were as follows: homoeologues TraesCS1A02G423800 (−2.25) and TraesCS1B02G455900 (−2.51), Triticum aestivum late embryogenesis abundant 14-A-like genes, whose orthologue in Arabidopsis increased salt tolerance when overexpressed (Jia et al., 2014); TraesCS6B02G268100 (−3.57) and its homoeologue TraesCS6D02G238200 (−4.4), Triticum aestivum AP2 domain CBF (CBFI), which are likely involved in the cold response (Medina et al., 1999); TraesCS6D02G332500 (−2.58), Triticum aestivum cold-shock CS120, which is also likely involved in the response to cold stress thanks to shared sequence identity with regions of cold-response genes in Arabidopsis, such as AtRAB18 (Lång and Palva, 1992; Lang et al., 1994; Mantyla et al., 1995; Puhakainen et al., 2004); and TraesCS1D02G263200 (−1.87), Triticum aestivum ERF019-like, which encodes an ethylene-responsive transcription factor whose orthologue in Arabidopsis improves drought tolerance and water use, through reduced stomatal aperture and transpiration, when overexpressed (Scarpeci et al., 2017). Similarly, TaERD15-like was connected to TraesCS4B02G332700 (−2.32), TraesCS4B02G332800 (−2.68), TraesCS4D02G329500 (−2.21), and TraesCS5A02G503800 (−2.91)—copies of Triticum aestivum ABA-inducible PHV A1-like, also known as HVA1 or WCOR615. The barley gene, HVA1, has been found to increase drought and salinity tolerance when overexpressed in rice and wheat (Xu et al., 1996; Sivamani et al., 2000; Rohila et al., 2002; Chandra Babu et al., 2004; Bahieldin et al., 2005; Chen et al., 2015). TaERD15-like was also connected to TraesCS5A02G503900 (−1.58), Triticum aestivum cold-responsive LEA/RAB-related COR (Wrab17.1), another COR protein which has been shown to respond to ABA and cold stress (Tsuda et al., 2000), and may play a role in the biotic stress response (Gaoshan et al., 2018).
Despite both appearing to be involved in determining the downregulation of superfluous drought- and cold-responsive genes under early heat stress, TaMAPKKK18-like and TaERD15-like may also play key roles in the regulation of other genes involved in separate processes. For instance, reduced expression of TaMAPKKK18-like under heat stress may also activate stress hormone signalling, thanks to the co-downregulation of several ABA and JA signalling repressors such as TraesCS7A02G201200 (−5.43), TaTIFY 11e-like, encoding a likely repressor of jasmonate responses due to its membership in the JAZ family (Santner and Estelle, 2007; Kazan and Manners, 2012; Wager and Browse, 2012; Sasaki-Sekimoto et al., 2014); and TraesCS3A02G347500 (−2.66), Triticum aestivum WRKY24-like, whose orthologue in rice is a negative regulator of GA and ABA signalling (Zhang et al., 2015). The downregulation of such signalling genes, therefore, may allow key stress hormones to accumulate in plant tissue under heat stress and act as part of the heat stress response (Li et al., 2020).
Similarly, TaERD15-like is itself a likely repressor of ABA signalling downregulated under early heat stress; however, we found that it was connected to a large number of genes which seemingly play roles in photosynthesis, largely as part of PSII—the most heat-labile part of the photosynthetic apparatus (Yamamoto, 2016; Wang et al., 2018; Hu et al., 2020). Nine of these downregulated genes, TraesCS1A02G403300 (−2.79), TraesCS1B02G432700 (−2.73), TraesCS1D02G411300 (−2.66), TraesCS2A02G204800 (−1.87), TraesCS2B02G220100 (−1.53), TraesCS5B02G463100 (−3.06), TraesCS5D02G329200 (−2.61), TraesCS5D02G464900 (−3.9), TraesCS7D02G276300 (−2.53), encode Chlorophyll a-b binding proteins, which form antenna complexes in PSII and act to absorb sunlight, transferring excitation energy to PSII to power photosynthetic electron transport (Jansson, 1994; Jansson, 1999). Under intense heat stress, PSII light-harvesting complexes fall off of thylakoid membranes, subsequently reducing the efficiency of electron transfer, which results in reduced photosynthesis (Janka et al., 2013; Mathur et al., 2014; Hu et al., 2020). There were 16 other DEGs connected to the hub that were constituent parts of the heat-labile PSII reaction centre, all of which were also downregulated under heat stress. The downregulation of these genes, as well as the nine Chlorophyll a-b binding protein genes, suggests inactivation of PSII under heat stress. As well as being connected to genes which are part of PSII, TaERD15-like was also connected to TraesCS4A02G177500 (−2.15), Triticum aestivum ribulose bisphosphate carboxylase/oxygenase activase A, chloroplastic-like—otherwise known as TaRca2 (Caruana et al., 2022). The TaRca2 isoforms are the most heat-labile of the Rubisco activase proteins; meaning, during periods of heat stress, less functional protein is available to remodel the active site of Rubisco to release tightly bound inhibitors, leading to a reduced photosynthetic rate (Salvucci et al., 1985; Bhat et al., 2017; Degen et al., 2021).
The damaging effect of heat stress exposure on PSII activity is well known (Yamamoto, 2016; Wang et al., 2018; Hu et al., 2020); however, here we see evidence that TaERD15-like may be playing a central role in this inactivation, as it was connected to a large number of downregulated PSII genes. Downregulation of TaERD15-like, therefore, may be a preventative tactic taken by plants to limit the build-up of damaged photosynthesis proteins under heat stress—a wise tactic considering that this is often toxic to cells (McClellan et al., 2005; Gil et al., 2017).
sHSP hub gene may promote the heat stress response via upregulation of thermoprotectants and stress-responsive transcription factors
The blue module was enriched in DEGs (expected number = 610, observed number = 966, p = 6.34E-52), but not in any GO terms related to the stress response. The most well-connected gene in this module, TraesCS4D02G212300, Triticum aestivum 17.9 kDa class I heat shock protein-like (LOC123097951), was also upregulated under heat stress (3.29, Figure 3C). The small HSP (sHSP) hub gene also shares remarkable sequence identity (97%) with TaHSP26; a sHSP located in the chloroplast, whose expression is induced by heat stress exposure (Chauhan et al., 2012; Khurana et al., 2013). TaHSP26 has been known to be involved in thermotolerance for over two decades with Joshi et al. (1997) finding that the gene was expressed in thermotolerant recombinant inbred lines, but not susceptible ones. More recently, further evidence for the gene’s role in increasing thermotolerance has been provided, as when the gene was expressed in Arabidopsis, PSII activity, photosynthetic pigment production, biomass, and seed yield under heat stress were all higher than that of WT plants (Chauhan et al., 2012). The present work suggests that the similar gene, and fellow sHSP, TraesCS4D02G212300 (herein referred to as the “sHSP hub gene”), may act as a key coordinator of the response to early heat stress.
The sHSP hub was connected to 952 of the 966 remaining DEGs in the module, 60 of which were annotated as HSPs. For example, TraesCS1D02G284000 (2.99), TaHSP70d, is a known thermotolerance gene (Hu et al., 2018); TraesCS2A02G033700 (2.44), Triticum aestivum heat shock protein 90-1, shows sequence identity (76%) to AtHSP81.4, whereas TraesCS1A02G340100 (4.87), and homeologues TraesCS3B02G308100 (5.82) and TraesCS3D02G273600 (3.6), encode Triticum aestivum chaperone protein ClpB1-like and share sequence identity (71%, 72%, and 72%, respectively) with AtHSP101 (ClpB1)—a gene whose expression is known to respond to heat stress, and whose protein aids protein refolding under high temperatures, and facilitates the deaggregation of toxic ubiquitylated protein aggregates via interaction with the proteasome (Queitsch et al., 2000; Hong and Vierling, 2001; Tonsor et al., 2008; McLoughlin et al., 2019). The sHSP hub gene was also connected to six genes: TraesCS1B02G294300 (4.02), TraesCS1A02G285000 (2.93), TraesCS3B02G390700 (4.32), TraesCS3D02G351900 (3.85), TraesCS3D02G352400 (2.0), and TraesCS4A02G098600 (3.58), which share 78% sequence identity with AtHSC70-1—a repressor of thermotolerance in Arabidopsis (Tiwari et al., 2020). The upregulation of these six genes, and our previous observation that increased expression of a wheat orthologue of AtHSC70-1 can be used as a marker for increased thermotolerance (Barratt et al., 2023a), suggests these genes may play positive roles in both thermotolerance and the heat stress response in T. aestivum. The majority of the 60 HSPs connected to the hub were fellow sHSPs, a group of proteins known to delay formation of harmful protein aggregates under heat stress and enhance thermotolerance in a wide variety of plant species, such as rice, maize, and poplar (Murakami et al., 2004; Kim et al., 2012; Sun et al., 2012; Zhou et al., 2012; Chen et al., 2014; Merino et al., 2014; Tian et al., 2021). The upregulation of these genes under heat stress in wheat corroborates these observations and suggests that the hub gene may act as a regulator of these crucial protective genes.
The sHSP hub was also connected to a large group of transcription factors known to play key roles in abiotic stress responses in wheat and other species. Expression of homoeologues TraesCS7A02G270100 (2.05) and TraesCS7B02G168300 (2.08), TaHsfB2-3 and TaHsfB2-4, respectively, respond to heat stress treatment (Duan et al., 2019), and these genes belong to a family of transcription factors which act to determine the expression of many other stress-responsive gene family members as part of abiotic stress responses, particularly HSPs (Guo et al., 2016). TraesCS3A02G281900 (1.62), Triticum aestivum probable WRKY transcription factor 65, shows some sequence identity (74%) to a small region of its Arabidopsis orthologue, AtWRKY65—a gene known to increase thermotolerance and repress thermomorphogenesis when acting alongside its homologues (Qin et al., 2022). However, TraesCS2D02G414300 (2.72), Triticum aestivum ethylene-responsive transcription factor ERF105-like, may act as part of other stress responses. The gene’s orthologue in Arabidopsis is involved in promoting freezing tolerance and cold acclimation (Bolt et al., 2017), suggesting an action as part of the generalised stress response in wheat, as opposed to a tailored response to cold stress. Similarly, the hub is connected to other genes which have previously been described as playing roles in response to stresses other than heat, but their upregulation, and connection to the hub gene, in the present work suggests they may act as part of the general stress response: TraesCS4B02G176700 (2.03), TaWRKY19, has been shown to regulate abiotic stress tolerance when overexpressed in Arabidopsis—leading to increased salt, drought, and freezing tolerance, likely via the upregulation of stress-responsive genes such as DREB2A, RD29A, RD29B, and Cor6.6 (Niu et al., 2012), whereas Aegilops tauschii subsp. strangulata ethylene-responsive transcription factor 1-like (TraesCS4B02G200200; 1.8) has been shown to prevent disease progression and regulate the expression of genes involved in the biotic stress response (Lorenzo et al., 2003). Likewise, TraesCS5D02G148800 (4.58), TaNAC29, increases salt and drought tolerance when overexpressed in Arabidopsis (Huang et al., 2015), whereas overexpression of the Arabidopsis namesake of TraesCS5A02G510100 (2.86) Triticum aestivum zinc finger protein CONSTANS-LIKE 4-like, AtCOL4, led to reduced ABA sensitivity and increased salinity tolerance (Min et al., 2015). The upregulation of such genes in the present work suggests a shared role as part of the general stress response, unlike the downregulated drought-, salinity-, and cold-responsive genes connected to TaMAPKKK18-like and TaERD15-like which likely act as part of the tailored response to these stresses. The hub’s connection to this suite of upregulated transcription factors, as well as its connection to 60 fellow upregulated HSPs and sHSPs, suggests a new function for the poorly characterised sHSP hub gene as a potential activator of the heat stress response.
Although the role of sHSPs, and TaHSP26 in particular, in acquired thermotolerance and the response to heat stress is widely accepted to be the prevention of protein misfolding and aggregation of heat labile proteins, work by Guihur et al. (2020) suggests that this group of proteins may also act to regulate the activity of signalling proteins, which, in turn, improves thermotolerance by impeding processes such as cell death (Guihur et al., 2020). It may be possible, therefore, that the sHSP hub gene identified in the present work regulates the heat-responsive expression of the genes it is connected to in the co-expression network via effects on the activity of these signalling proteins; however, further work is required to determine the exact mechanism by which the sHSP hub gene may indeed regulate the expression of these genes in response to heat stress.
In the present work, we have demonstrated that early heat stress exposure causes large shifts in the wheat transcriptome, with almost 8,000 response DEGs being identified, whereas the likely functionalities of genes being upregulated and downregulated suggests a shift away from growth and development, to stress response and damage mitigation. We also observed the widespread downregulation of genes potentially involved in responding to other abiotic stresses, likely due to the fact that the cellular conditions these genes respond to are not present under heat stress. We then paired these data with the co-expression network to identify heat-associated modules, within which were several promising candidates which may act as regulators of the transcriptional and physiological early heat stress response. Downregulation of two of the most promising candidates under early heat stress (TaMAPKKK18-like and TaERD15-like) may act to downregulate the expression of these superfluous stress-responsive genes, whereas a sHSP hub gene may activate the expression of HSPs and fellow sHSPs, as well as transcription factors known to play key roles in various abiotic stress responses, including the response to heat. This work, therefore, represents a vital step toward the creation of more thermotolerant wheat varieties, and provides key new insights into the transcriptional response of wheat to early heat stress, as well as candidate genes which may regulate this response.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI gene expression omnibus (GEO) under accession number GSE232367.
Author contributions
LB and AH conceived and planned the project. LB performed plant growth experiments and RNA extraction. SFO performed transcriptome data mapping and QPCR. LB conducted transcriptomic analyses. LB and SFO wrote the manuscript, and all authors reviewed and edited it. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by UK Biotechnology and Biological Sciences Research Council White Rose Doctorial Training Partnerships (DTP) in Mechanistic Biology (BB/M011151/1). The authors thank the Germplasm Resource Unit at John Innes Centre, UK (Grant number BBS/E/J/000PR8000) for providing germplasm and sharing the genotype and genetic map information, CIMMYT, Mexico, and the Crop Research Institute, Czechia, for providing additional germplasm.
Acknowledgments
We thank the horticultural staff at the University of York for their help in maintaining and growing plants, as well as Sally James and Lesley Gilbert from the Technology Facility at the University of York for their assistance in assessing RNA quality prior to sequencing, and Isaac Reynolds for his advice and assistance during transcriptomic analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1252885/full#supplementary-material
Supplementary Figure 1 | Gene Ontology enrichment analysis of DEGs in graphical format: (A) Up-regulated genes – Biological Process; (B) Up-regulated genes – Cellular Component; (C) Up-regulated genes – Molecular Function; (D) Down-regulated genes – Biological Process; (E) Down-regulated genes – Cellular Component; (F) Down-regulated genes – Molecular Function.
References
Aalto, M. K., Helenius, E., Kariola, T., Pennanen, V., Heino, P., Hõrak, H., et al. (2012). ERD15–an attenuator of plant ABA responses and stomatal aperture. Plant Sci. 182, 19–28. doi: 10.1016/j.plantsci.2011.08.009
Al-Whaibi, M. H. (2011). Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 23, 139–150. doi: 10.1016/j.jksus.2010.06.022
Andleeb, T., Knight, E., Borrill, P. (2023). Wheat NAM genes regulate the majority of early monocarpic senescence transcriptional changes including nitrogen remobilization genes. G3 13, jkac275. doi: 10.1093/g3journal/jkac275
Assenov, Y., Ramírez, F., Schelhorn, S.-E., Lengauer, T., Albrecht, M. (2008). Computing topological parameters of biological networks. Bioinformatics 24, 282–284. doi: 10.1093/bioinformatics/btm554
Azameti, M. K., Ranjan, A., Singh, P. K., Gaikwad, K., Singh, A. K., Dalal, M., et al. (2022). Transcriptome profiling reveals the genes and pathways involved in thermo-tolerance in wheat (Triticum aestivum L.) genotype Raj 3765. Sci. Rep. 12, 14831. doi: 10.1038/s41598-022-18625-7
Bahieldin, A., Mahfouz, H. T., Eissa, H. F., Saleh, O. M., Ramadan, A. M., Ahmed, I. A., et al. (2005). Field evaluation of transgenic wheat plants stably expressing the HVA1 gene for drought tolerance. Physiol. Plant 123, 421–427. doi: 10.1111/j.1399-3054.2005.00470.x
Barratt, L. J., He, Z., Fellgett, A., Wang, L., McQueen Mason, S., Bancroft, I., et al. (2023a). Co-expression network analysis of diverse wheat landraces reveals marker of early thermotolerance and candidate master-regulator of thermotolerance genes. Plant J. 115 (3), 614–626. doi: 10.1111/tpj.16248
Barratt, L. J., Reynolds, I. J., Franco Ortega, S., Harper, A. L. (2023b). Transcriptomic and co-expression network analyses on diverse wheat landraces identifies candidate master regulators of the response to early drought. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1212559
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B Stat. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bhat, J. Y., Miličić, G., Thieulin-Pardo, G., Bracher, A., Maxwell, A., Ciniawsky, S., et al. (2017). Mechanism of enzyme repair by the AAA+ Chaperone rubisco activase. Mol. Cell 67, 744–756.e6. doi: 10.1016/j.molcel.2017.07.004
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolt, S., Zuther, E., Zintl, S., Hincha, D. K., Schmülling, T. (2017). ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 40, 108–120. doi: 10.1111/pce.12838
Borrill, P., Harrington, S. A., Simmonds, J., Uauy, C. (2019). Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling. Plant Physiol. 180, 1740–1755. doi: 10.1104/pp.19.00380
Boulanger, H. G., Guo, W., Monteiro, L., de, F. R., Calixto, C. P. G. (2023). Co-expression network of heat-response transcripts: A glimpse into how splicing factors impact rice basal thermotolerance. Front. Mol. Biosci. 10. doi: 10.3389/fmolb.2023.1122201
Cai, Z., He, F., Feng, X., Liang, T., Wang, H., Ding, S., et al. (2020). Transcriptomic analysis reveals important roles of lignin and flavonoid biosynthetic pathways in rice thermotolerance during reproductive stage. Front. Genet. 11. doi: 10.3389/fgene.2020.562937
Caruana, L., Orr, D. J., Carmo-Silva, E. (2022). Rubiscosome gene expression is balanced across the hexaploid wheat genome. Photosynth. Res. 152, 1–11. doi: 10.1007/s11120-022-00897-9
Chandra Babu, R., Zhang, J., Blum, A., David Ho, T.-H., Wu, R., Nguyen, H. T. (2004). HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci. 166, 855–862. doi: 10.1016/j.plantsci.2003.11.023
Chauhan, H., Khurana, N., Nijhavan, A., Khurana, J. P., Khurana, P. (2012). The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 35, 1912–1931. doi: 10.1111/j.1365-3040.2012.02525.x
Chen, X., Lin, S., Liu, Q., Huang, J., Zhang, W., Lin, J., et al. (2014). Expression and interaction of small heat shock proteins (sHsps) in rice in response to heat stress. Biochim. Biophys. Acta 1844, 818–828. doi: 10.1016/j.bbapap.2014.02.010
Chen, Y.-S., Lo, S.-F., Sun, P.-K., Lu, C.-A., Ho, T.-H. D., Yu, S.-M. (2015). A late embryogenesis abundant protein HVA1 regulated by an inducible promoter enhances root growth and abiotic stress tolerance in rice without yield penalty. Plant Biotechnol. J. 13, 105–116. doi: 10.1111/pbi.12241
Chowdhury, S. I., Wardlaw, I. F. (1978). The effect of temperature on kernel development in cereals. Aust. J. Agric. Res. 29, 205–223. doi: 10.1071/ar9780205
Collins, M., Knutti, R., Arblaster, J., Dufresne, J.-L., Fichefet, T., Friedlingstein, P., et al. (2013). “Long-term climate change: projections, commitments and irreversibility,” in Climate change 2013-The physical science basis: Contribution of working group I to the fifth assessment report of the intergovernmental panel on climate change (New York NY USA: Cambridge University Press), 1029–1136. Available at: https://research.monash.edu/en/publications/long-term-climate-change-projections-commitments-and-irreversibil.
De Costa, W. A. J. M. (2011). A review of the possible impacts of climate change on forests in the humid tropics. J. Natl. Sci. Found. Sri Lanka 39, 281. doi: 10.4038/jnsfsr.v39i4.3879
Degen, G. E., Orr, D. J., Carmo-Silva, E. (2021). Heat-induced changes in the abundance of wheat Rubisco activase isoforms. New Phytol. 229, 1298–1311. doi: 10.1111/nph.16937
Do, J.-M., Kim, H.-J., Shin, S.-Y., Park, S.-I., Kim, J.-J., Yoon, H.-S. (2023). OsHSP 17.9, a small heat shock protein, confers improved productivity and tolerance to high temperature and salinity in a natural paddy field in transgenic rice plants. Collect. FAO Agric. 13, 931. doi: 10.3390/agriculture13050931
Du, Z., Zhou, X., Ling, Y., Zhang, Z., Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. doi: 10.1093/nar/gkq310
Duan, S., Liu, B., Zhang, Y., Li, G., Guo, X. (2019). Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genomics 20, 257. doi: 10.1186/s12864-019-5617-1
Easterling, D. R., Meehl, G. A., Parmesan, C., Changnon, S. A., Karl, T. R., Mearns, L. O. (2000). Climate extremes: observations, modeling, and impacts. Science 289, 2068–2074. doi: 10.1126/science.289.5487.2068
Fernández-Pérez, F., Vivar, T., Pomar, F., Pedreño, M. A., Novo-Uzal, E. (2015). Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J. Plant Physiol. 175, 86–94. doi: 10.1016/j.jplph.2014.11.006
Food and Agriculture Organization of the United Nations. (2023). FAOSTAT: Crops and livestock products. Available at: https://www.fao.org/faostat/en/#data/QCL (Accessed March 31, 2023).
Food and Agriculture Organization of the United Nations, United Nations International Children’s Emergency Fund, World Health Organization, World Food Programme, International Fund for Agriculture Development (2018). The State of Food Security and Nutrition in the World 2018: Building climate resilience for food security and nutrition (Rome: Food & Agriculture Org). Available at: https://play.google.com/store/books/details?id=LC9uDwAAQBAJ.
Gao, C., Jiang, B., Wang, Y., Liu, G., Yang, C. (2012). Overexpression of a heat shock protein (ThHSP18.3) from Tamarix hispida confers stress tolerance to yeast. Mol. Biol. Rep. 39, 4889–4897. doi: 10.1007/s11033-011-1284-2
Gaoshan, Y., Na, L., Dong, D., Shuaishuai, W., Peng, L., Shengfang, H., et al. (2018). Functional characterization of the Wrab17 gene in the interaction process between wheat and Puccinia triticina. Plant Physiol. Biochem. 133, 100–106. doi: 10.1016/j.plaphy.2018.10.018
Gil, K.-E., Kim, W.-Y., Lee, H.-J., Faisal, M., Saquib, Q., Alatar, A. A., et al. (2017). ZEITLUPE contributes to a thermoresponsive protein quality control system in Arabidopsis. Plant Cell 29, 2882–2894. doi: 10.1105/tpc.17.00612
Girousse, C., Roche, J., Guerin, C., Le Gouis, J., Balzegue, S., Mouzeyar, S., et al. (2018). Coexpression network and phenotypic analysis identify metabolic pathways associated with the effect of warming on grain yield components in wheat. PloS One 13, e0199434. doi: 10.1371/journal.pone.0199434
Gitlin-Domagalska, A., Maciejewska, A., Dębowski, D. (2020). Bowman-birk inhibitors: insights into family of multifunctional proteins and peptides with potential therapeutical applications. Pharmaceuticals 13, 421. doi: 10.3390/ph13120421
Guihur, A., Fauvet, B., Finka, A., Quadroni, M., Goloubinoff, P. (2020). Quantitative proteomic analysis to capture the role of heat-accumulated proteins in moss plant acquired thermotolerance. Plant Cell Environ. 44, 2117–2133. doi: 10.1111/pce.13975
Guo, L.-M., Li, J., He, J., Liu, H., Zhang, H.-M. (2020). A class I cytosolic HSP20 of rice enhances heat and salt tolerance in different organisms. Sci. Rep. 10, 1383. doi: 10.1038/s41598-020-58395-8
Guo, M., Liu, J.-H., Ma, X., Luo, D.-X., Gong, Z.-H., Lu, M.-H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00114
Han, S.-H., Kim, J. Y., Lee, J.-H., Park, C.-M. (2021). Safeguarding genome integrity under heat stress in plants. J. Exp. Bot. 72 (21), 7421–7435. doi: 10.1093/jxb/erab355
Han, S.-H., Park, Y.-J., Park, C.-M. (2020). HOS1 activates DNA repair systems to enhance plant thermotolerance. Nat. Plants 6, 1439–1446. doi: 10.1038/s41477-020-00809-6
Haokip, S. W., Shankar, K., Lalrinngheta, J. (2020). Climate change and its impact on fruit crops. J. Pharmacogn. Phytochem. 9, 435–438.
Hong, S. W., Vierling, E. (2001). Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J. 27, 25–35. doi: 10.1046/j.1365-313x.2001.01066.x
Hu, S., Ding, Y., Zhu, C. (2020). Sensitivity and responses of chloroplasts to heat stress in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00375
Hu, X.-J., Chen, D., Lynne Mclntyre, C., Fernanda Dreccer, M., Zhang, Z.-B., Drenth, J., et al. (2018). Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 41, 79–98. doi: 10.1111/pce.12957
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15, 268. doi: 10.1186/s12870-015-0644-9
Janka, E., Körner, O., Rosenqvist, E., Ottosen, C.-O. (2013). High temperature stress monitoring and detection using chlorophyll a fluorescence and infrared thermography in chrysanthemum (Dendranthema grandiflora). Plant Physiol. Biochem. 67, 87–94. doi: 10.1016/j.plaphy.2013.02.025
Jansson, S. (1994). The light-harvesting chlorophyll ab-binding proteins. Biochim. Biophys. Acta (BBA) Bioenergetics 1184, 1–19. doi: 10.1016/0005-2728(94)90148-1
Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis/IT>. Trends Plant Sci. 4, 236–240. doi: 10.1016/s1360-1385(99)01419-3
Jia, F., Qi, S., Li, H., Liu, P., Li, P., Wu, C., et al. (2014). Overexpression of Late Embryogenesis Abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 454, 505–511. doi: 10.1016/j.bbrc.2014.10.136
Jiang, C., Bi, Y., Li, M., Zhang, R., Feng, S., Ming, F. (2020). A small heat shock protein gene (RcHSP17.8) from Chinese rose confers resistance to various abiotic stresses in transgenic tobacco. Plant Cell Tissue Organ Cult. 141, 407–415. doi: 10.1007/s11240-020-01798-2
Jin, J., Yang, L., Fan, D., Liu, X., Hao, Q. (2020). Comparative transcriptome analysis uncovers different heat stress responses in heat-resistant and heat-sensitive jujube cultivars. PloS One 15, e0235763. doi: 10.1371/journal.pone.0235763
Joshi, C. P., Klueva, N. Y., Morrow, K. J., Nguyen, H. T. (1997). Expression of a unique plastid-localized heat-shock protein is genetically linked to acquired thermotolerance in wheat. Theor. Appl. Genet. 95, 834–841. doi: 10.1007/s001220050633
Kantidze, O. L., Velichko, A. K., Luzhin, A. V., Razin, S. V. (2016). Heat stress-induced DNA damage. Acta Naturae 8, 75–78. doi: 10.32607/20758251-2016-8-2-75-78
Kariola, T., Brader, G., Helenius, E., Li, J., Heino, P., Tapio Palva, E. (2006). EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 142, 1559–1573. doi: 10.1104/pp.106.086223
Kazan, K., Manners, J. M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31. doi: 10.1016/j.tplants.2011.10.006
Khurana, N., Chauhan, H., Khurana, P. (2013). Wheat chloroplast targeted sHSP26 promoter confers heat and abiotic stress inducible expression in transgenic Arabidopsis Plants. PloS One 8, e54418. doi: 10.1371/journal.pone.0054418
Kim, K.-H., Alam, I., Kim, Y.-G., Sharmin, S. A., Lee, K.-W., Lee, S.-H., et al. (2012). Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol. Lett. 34, 371–377. doi: 10.1007/s10529-011-0769-3
Kiyosue, T., Abe, H., Yamaguchi-Shinozaki, K., Shinozaki, K. (1998). ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter1The nucleotide sequence reported in this paper has been submitted to DDBJ with the accession number of D89051.1. Biochim. Biophys. Acta (BBA) Biomembranes 1370, 187–191. doi: 10.1016/S0005-2736(98)00007-8
Kobza, J., Edwards, G. E. (1987). Influences of leaf temperature on photosynthetic carbon metabolism in wheat. Plant Physiol. 83, 69–74. doi: 10.1104/pp.83.1.69
Kotak, S., Larkindale, J., Lee, U., von Koskull-Döring, P., Vierling, E., Scharf, K.-D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. doi: 10.1016/j.pbi.2007.04.011
Lang, V., Mantyla, E., Welin, B., Sundberg, B., Palva, E. T. (1994). Alterations in Water Status, Endogenous Abscisic Acid Content, and Expression of rab18 Gene during the Development of Freezing Tolerance in Arabidopsis thaliana. Plant Physiol. 104, 1341–1349. doi: 10.1104/pp.104.4.1341
Lång, V., Palva, E. T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. doi: 10.1007/BF00027165
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Langfelder, P., Horvath, S. (2012). Fast R functions for robust correlations and hierarchical clustering. J. Stat. Software 46, 1–17. doi: 10.18637/jss.v046.i11
Lee, M. H., Kim, K.-M., Sang, W.-G., Kang, C.-S., Choi, C. (2022). Comparison of gene expression changes in three wheat varieties with different susceptibilities to heat stress using RNA-Seq analysis. Int. J. Mol. Sci. 23, 10734. doi: 10.3390/ijms231810734
Le Gall, H., Philippe, F., Domon, J.-M., Gillet, F., Pelloux, J., Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. doi: 10.3390/plants4010112
Li, N., Euring, D., Cha, J. Y., Lin, Z., Lu, M., Huang, L.-J., et al. (2020). Plant hormone-mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.627969
Li, J., Zhang, J., Jia, H., Li, Y., Xu, X., Wang, L., et al. (2016). The Populus trichocarpa PtHSP17.8 involved in heat and salt stress tolerances. Plant Cell Rep. 35, 1587–1599. doi: 10.1007/s00299-016-1973-3
Lima, R. B., dos Santos, T. B., Vieira, L. G. E., Ferrarese, M., de, L. L., Ferrarese-Filho, O., et al. (2013). Heat stress causes alterations in the cell-wall polymers and anatomy of coffee leaves (Coffea arabica L.). Carbohydr. Polym. 93, 135–143. doi: 10.1016/j.carbpol.2012.05.015
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15, 1–20. doi: 10.1186/s12870-015-0511-8
Liu, R., Xu, Y.-H., Jiang, S.-C., Lu, K., Lu, Y.-F., Feng, X.-J., et al. (2013). Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 64, 5443–5456. doi: 10.1093/jxb/ert307
Liu, X., Zhu, Y., Zhai, H., Cai, H., Ji, W., Luo, X., et al. (2012). AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem. Biophys. Res. Commun. 422, 710–715. doi: 10.1016/j.bbrc.2012.05.064
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Mantyla, E., Lang, V., Palva, E. T. (1995). Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 107, 141–148. doi: 10.1104/pp.107.1.141
Mathur, S., Agrawal, D., Jajoo, A. (2014). Photosynthesis: response to high temperature stress. J. Photochem. Photobiol. B 137, 116–126. doi: 10.1016/j.jphotobiol.2014.01.010
Matsuoka, D., Yasufuku, T., Furuya, T., Nanmori, T. (2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87, 565–575. doi: 10.1007/s11103-015-0295-0
Mazelis, M., Vennesland, B. (1957). Carbon dioxide fixation into oxalacetate in higher plants. Plant Physiol. 32, 591–600. doi: 10.1104/pp.32.6.591
McClellan, A. J., Tam, S., Kaganovich, D., Frydman, J. (2005). Protein quality control: chaperones culling corrupt conformations. Nat. Cell Biol. 7, 736–741. doi: 10.1038/ncb0805-736
McLoughlin, F., Kim, M., Marshall, R. S., Vierstra, R. D., Vierling, E. (2019). HSP101 interacts with the proteasome and promotes the clearance of ubiquitylated protein aggregates. Plant Physiol. 180, 1829–1847. doi: 10.1104/pp.19.00263
Medina, J., Bargues, M., Terol, J., Pérez-Alonso, M., Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–470. doi: 10.1104/pp.119.2.463
Merino, I., Contreras, A., Jing, Z.-P., Gallardo, F., Cánovas, F. M., Gómez, L. (2014). Plantation forestry under global warming: hybrid poplars with improved thermotolerance provide new insights on the in vivo function of small heat shock protein chaperones. Plant Physiol. 164, 978–991. doi: 10.1104/pp.113.225730
Min, J.-H., Chung, J.-S., Lee, K.-H., Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 57, 313–324. doi: 10.1111/jipb.12246
Mishra, D. C., Arora, D., Kumar, R. R., Goswami, S., Varshney, S., Budhlakoti, N., et al. (2021). Weighted gene co-expression analysis for identification of key genes regulating heat stress in wheat. Cereal Res. Commun. 49, 73–81. doi: 10.1007/s42976-020-00072-7
Mitula, F., Tajdel, M., Cieśla, A., Kasprowicz-Maluśki, A., Kulik, A., Babula-Skowrońska, D., et al. (2015). Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 56, 2351–2367. doi: 10.1093/pcp/pcv146
Mogk, A., Bukau, B., Kampinga, H. H. (2018). Cellular handling of protein aggregates by disaggregation machines. Mol. Cell 69, 214–226. doi: 10.1016/j.molcel.2018.01.004
Murakami, T., Matsuba, S., Funatsuki, H., Kawaguchi, K., Saruyama, H., Tanida, M., et al. (2004). Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol. Breed. 13, 165–175. doi: 10.1023/B:MOLB.0000018764.30795.c1
Murphy, J. M., Sexton, D. M. H., Barnett, D. N., Jones, G. S., Webb, M. J., Collins, M., et al. (2004). Quantification of modelling uncertainties in a large ensemble of climate change simulations. Nature 430, 768–772. doi: 10.1038/nature02771
Nagai, T., Makino, A. (2009). Differences between rice and wheat in temperature responses of photosynthesis and plant growth. Plant Cell Physiol. 50, 744–755. doi: 10.1093/pcp/pcp029
Niu, C.-F., Wei, W., Zhou, Q.-Y., Tian, A.-G., Hao, Y.-J., Zhang, W.-K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
NOAA National Centers for Environmental Information. (2018). Monthly National Climate Report for May 2018. Available at: www.ncei.noaa.gov/access/monitoring/monthly-report/national/201805 (Accessed March 30, 2023).
NOAA National Centers for Environmental Information. (2020). Monthly Global Climate Report for May 2020. Available at: https://www.ncei.noaa.gov/access/monitoring/monthly-report/global/202005 (Accessed March 30, 2023).
NOAA National Centers for Environmental Information. (2022a). Monthly Global Climate Report for May 2022. Available at: https://www.ncei.noaa.gov/access/monitoring/monthly-report/global/202205 (Accessed March 20, 2023).
NOAA National Centers for Environmental Information. (2022b). Monthly National Climate Report for May 2022. Available at: https://www.ncei.noaa.gov/access/monitoring/monthly-report/national/202205 (Accessed March 30, 2023).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Pfeifer, M., Kugler, K. G., Sandve, S. R., Zhan, B., Rudi, H., Hvidsten, T. R., et al. (2014). Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345, 1250091. doi: 10.1126/science.1250091
Pinski, A., Betekhtin, A., Skupien-Rabian, B., Jankowska, U., Jamet, E., Hasterok, R. (2021). Changes in the cell wall proteome of leaves in response to high temperature stress in Brachypodium distachyon. Int. J. Mol. Sci. 22, 6750. doi: 10.3390/ijms22136750
Puhakainen, T., Hess, M. W., Mäkelä, P., Svensson, J., Heino, P., Palva, E. T. (2004). Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol. 54, 743–753. doi: 10.1023/B:PLAN.0000040903.66496.a4
Qin, W., Wang, N., Yin, Q., Li, H., Wu, A.-M., Qin, G. (2022). Activation tagging identifies WRKY14 as a repressor of plant thermomorphogenesis in Arabidopsis. Mol. Plant 15, 1725–1743. doi: 10.1016/j.molp.2022.09.018
Qin, D., Wu, H., Peng, H., Yao, Y., Ni, Z., Li, Z., et al. (2008). Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genomics 9, 432. doi: 10.1186/1471-2164-9-432
Queitsch, C., Hong, S.-W., Vierling, E., Lindquist, S. (2000). Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12, 479–492. doi: 10.2307/3871063
Rahman, M. A., Woo, J. H., Song, Y., Lee, S.-H., Hasan, M. M., Azad, M. A. K., et al. (2022). Heat shock proteins and antioxidant genes involved in heat combined with drought stress responses in perennial rye grass. Life 12, 1426. doi: 10.3390/life12091426
Rangan, P., Furtado, A., Henry, R. (2020). Transcriptome profiling of wheat genotypes under heat stress during grain-filling. J. Cereal Sci. 91, 102895. doi: 10.1016/j.jcs.2019.102895
Rativa, A. G. S., Junior, A.T. de A., Friedrich, D. da S., Gastmann, R., Lamb, T. I., Silva, A. D. S., et al. (2020). Root responses of contrasting rice genotypes to low temperature stress. J. Plant Physiol. 255, 153307. doi: 10.1016/j.jplph.2020.153307
Ray, D. K., Mueller, N. D., West, P. C., Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PloS One 8, e66428. doi: 10.1371/journal.pone.0066428
R Core Team (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Riaz, M. W., Yang, L., Yousaf, M. I., Sami, A., Mei, X. D., Shah, L., et al. (2021). Effects of heat stress on growth, physiology of plants, yield and grain quality of different spring wheat (Triticum aestivum L.) genotypes. Sustain. Sci. Pract. Policy 13, 2972. doi: 10.3390/su13052972
Rohila, J. S., Jain, R. K., Wu, R. (2002). Genetic improvement of Basmati rice for salt and drought tolerance by regulated expression of a barley Hva1 cDNA. Plant Sci. 163, 525–532. doi: 10.1016/S0168-9452(02)00155-3
Roychowdhury, R., Zilberman, O., Chandrasekhar, K., Curzon, A. Y., Nashef, K., Abbo, S., et al. (2023). Pre-anthesis spike growth dynamics and its association to yield components among elite bread wheat cultivars (Triticum aestivum L. spp.) under Mediterranean climate. Field Crops Res. 298, 108948. doi: 10.1016/j.fcr.2023.108948
Salvucci, M. E., Portis, A. R., Ogren, W. L. (1985). A soluble chloroplast protein catalyzes ribulosebisphosphate carboxylase/oxygenase activation in vivo. Photosynth. Res. 7, 193–201. doi: 10.1007/BF00037012
Santner, A., Estelle, M. (2007). The JAZ proteins link jasmonate perception with transcriptional changes. Plant Cell 19, 3839–3842. doi: 10.1105/tpc.107.056960
Sasaki-Sekimoto, Y., Saito, H., Masuda, S., Shirasu, K., Ohta, H. (2014). Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal. Behav. 9, e27639. doi: 10.4161/psb.27639
Scarpeci, T. E., Frea, V. S., Zanor, M. I., Valle, E. M. (2017). Overexpression of AtERF019 delays plant growth and senescence, and improves drought tolerance in Arabidopsis. J. Exp. Bot. 68, 673–685. doi: 10.1093/jxb/erw429
Seo, P. J., Park, J.-M., Kang, S. K., Kim, S.-G., Park, C.-M. (2011). An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233, 189–200. doi: 10.1007/s00425-010-1293-8
Shaar-Moshe, L., Blumwald, E., Peleg, Z. (2017). Unique physiological and transcriptional shifts under combinations of salinity, drought, and heat. Plant Physiol. 174, 421–434. doi: 10.1104/pp.17.00030
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sivamani, E., Bahieldin1, A., Wraith, J. M., Al-Niemi, T., Dyer, W. E., Ho, T. D., et al. (2000). Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Plant Sci. 155, 1–9. doi: 10.1016/s0168-9452(99)00247-2
Soneson, C., Love, M. I., Robinson, M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. doi: 10.12688/f1000research.7563.2
Sperotto, R. A., de Araújo Junior, A. T., Adamski, J. M., Cargnelutti, D., Ricachenevsky, F. K., de Oliveira, B.-H. N., et al. (2018). Deep RNAseq indicates protective mechanisms of cold-tolerant indica rice plants during early vegetative stage. Plant Cell Rep. 37, 347–375. doi: 10.1007/s00299-017-2234-9
Stephens, M. (2017). False discovery rates: a new deal. Biostatistics 18, 275–294. doi: 10.1093/biostatistics/kxw041
Sun, L., Liu, Y., Kong, X., Zhang, D., Pan, J., Zhou, Y., et al. (2012). ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 31, 1473–1484. doi: 10.1007/s00299-012-1262-8
Tang, Y., Li, J., Song, Q., Cheng, Q., Tan, Q., Zhou, Q., et al. (2023). Transcriptome and WGCNA reveal hub genes in sugarcane tiller seedlings in response to drought stress. Sci. Rep. 13, 12823. doi: 10.1038/s41598-023-40006-x
Thornton, P. K., Ericksen, P. J., Herrero, M., Challinor, A. J. (2014). Climate variability and vulnerability to climate change: a review. Glob. Change Biol. 20, 3313–3328. doi: 10.1111/gcb.12581
Tian, F., Hu, X.-L., Yao, T., Yang, X., Chen, J.-G., Lu, M.-Z., et al. (2021). Recent advances in the roles of HSFs and HSPs in heat stress response in woody plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.704905
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Tian, X., Qin, Z., Zhao, Y., Wen, J., Lan, T., Zhang, L., et al. (2022). Stress granule-associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat (Triticum aestivum). New Phytol. 233, 1719–1731. doi: 10.1111/nph.17865
Tian, B., Talukder, S. K., Fu, J., Fritz, A. K., Trick, H. N. (2018). Expression of a rice soluble starch synthase gene in transgenic wheat improves the grain yield under heat stress conditions. In Vitro Cell. Dev. Biol. Plant 54, 216–227. doi: 10.1007/s11627-018-9893-2
Tiwari, L. D., Khungar, L., Grover, A. (2020). AtHsc70-1 negatively regulates the basal heat tolerance in Arabidopsis thaliana through affecting the activity of HsfAs and Hsp101. Plant J. 103, 2069–2083. doi: 10.1111/tpj.14883
Tonsor, S. J., Scott, C., Boumaza, I., Liss, T. R., Brodsky, J. L., Vierling, E. (2008). Heat shock protein 101 effects in A. thaliana: genetic variation, fitness and pleiotropy in controlled temperature conditions. Mol. Ecol. 17, 1614–1626. doi: 10.1111/j.1365-294X.2008.03690.x
Tsuda, K., Tsvetanov, S., Takumi, S., Mori, N., Atanassov, A., Nakamura, C. (2000). New members of a cold-responsive group-3 Lea/Rab-related Cor gene family from common wheat (Triticum aestivum L.). Genes Genet. Syst. 75, 179–188. doi: 10.1266/ggs.75.179
United States Department of Agriculture - Foreign Agricultural Service. (2023). Grain: World Markets and Trade. Available at: https://usda.library.cornell.edu/concern/publications/zs25x844t?locale=en#release-items (Accessed March 29, 2023).
Velichko, A. K., Petrova, N. V., Kantidze, O. L., Razin, S. V. (2012). Dual effect of heat shock on DNA replication and genome integrity. Mol. Biol. Cell 23, 3450–3460. doi: 10.1091/mbc.E11-12-1009
Wager, A., Browse, J. (2012). Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00041
Waki, T., Mameda, R., Nakano, T., Yamada, S., Terashita, M., Ito, K., et al. (2020). A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 11, 870. doi: 10.1038/s41467-020-14558-9
Wang, Q.-L., Chen, J.-H., He, N.-Y., Guo, F.-Q. (2018). Metabolic reprogramming in chloroplasts under heat stress in plants. Int. J. Mol. Sci. 19, 849. doi: 10.3390/ijms19030849
Wang, R., Ma, J., Zhang, Q., Wu, C., Zhao, H., Wu, Y., et al. (2019). Genome-wide identification and expression profiling of glutathione transferase gene family under multiple stresses and hormone treatments in wheat (Triticum aestivum L.). BMC Genomics 20, 986. doi: 10.1186/s12864-019-6374-x
Wang, Q., Su, Q., Nian, J., Zhang, J., Guo, M., Dong, G., et al. (2021). The Ghd7 transcription factor represses ARE1 expression to enhance nitrogen utilization and grain yield in rice. Mol. Plant 14, 1012–1023. doi: 10.1016/j.molp.2021.04.012
Wang, W., Vinocur, B., Shoseyov, O., Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, Y., Wang, Y., Liu, X., Zhou, J., Deng, H., Zhang, G., et al. (2022). WGCNA analysis identifies the hub genes related to heat stress in seedling of rice (Oryza sativa L.). Genes 13, 1020. doi: 10.3390/genes13061020
Wang, X., Zhang, J., Mao, W., Guan, P., Wang, Y., Chen, Y., et al. (2023). Association mapping identifies loci and candidate genes for grain-related traits in spring wheat in response to heat stress. Plant Sci. 331, 111676. doi: 10.1016/j.plantsci.2023.111676
Wardlaw, I. F., Dawson, I. A., Munibi, P., Fewster, R. (1989). The tolerance of wheat to high temperatures during reproductive growth. I. Survey procedures and general response patterns. Aust. J. Agric. Res. 40, 1–13. doi: 10.1071/ar9890001
Wigley, T. M., Raper, S. C. (2001). Interpretation of high projections for global-mean warming. Science 293, 451–454. doi: 10.1126/science.1061604
Wu, H.-C., Bulgakov, V. P., Jinn, T.-L. (2018). Pectin methylesterases: cell wall remodeling proteins are required for plant response to heat stress. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01612
Xu, D., Duan, X., Wang, B., Hong, B., Ho, T., Wu, R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257. doi: 10.1104/pp.110.1.249
Yamaji, N., Sakurai, G., Mitani-Ueno, N., Ma, J. F. (2015). Orchestration of three transporters and distinct vascular structures in node for intervascular transfer of silicon in rice. Proc. Natl. Acad. Sci. U. S. A. 112, 11401–11406. doi: 10.1073/pnas.1508987112
Yamamoto, Y. (2016). Quality control of photosystem II: the mechanisms for avoidance and tolerance of light and heat stresses are closely linked to membrane fluidity of the thylakoids. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01136
Yang, K. A., Lim, C. J., Hong, J. K., Park, C. Y., Cheong, Y. H., Chung, W. S., et al. (2006). Identification of cell wall genes modified by a permissive high temperature in Chinese cabbage. Plant Sci. 171, 175–182. doi: 10.1016/j.plantsci.2006.03.013
Yates, A. D., Allen, J., Amode, R. M., Azov, A. G., Barba, M., Becerra, A., et al. (2022). Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 50, D996–D1003. doi: 10.1093/nar/gkab1007
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00600
Zeng, Z., Zhang, S., Li, W., Chen, B., Li, W. (2022). Gene-coexpression network analysis identifies specific modules and hub genes related to cold stress in rice. BMC Genomics 23, 251. doi: 10.1186/s12864-022-08438-3
Zhai, M., Sun, Y., Jia, C., Peng, S., Liu, Z., Yang, G. (2016). Over-expression of JrsHSP17.3 gene from Juglans regia confer the tolerance to abnormal temperature and NaCl stresses. J. Plant Biol. 59, 549–558. doi: 10.1007/s12374-015-0507-9
Zhang, L., Gu, L., Ringler, P., Smith, S., Rushton, P. J., Shen, Q. J. (2015). Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 236, 214–222. doi: 10.1016/j.plantsci.2015.04.014
Zhao, C., Liu, B., Piao, S., Wang, X., Lobell, D. B., Huang, Y., et al. (2017). Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U. S. A. 114, 9326–9331. doi: 10.1073/pnas.1701762114
Zheng, T., Sun, J., Zhou, S., Chen, S., Lu, J., Cui, S., et al. (2019). Post-transcriptional regulation of Ghd7 protein stability by phytochrome and OsGI in photoperiodic control of flowering in rice. New Phytol. 224, 306–320. doi: 10.1111/nph.16010
Zhou, Y., Chen, H., Chu, P., Li, Y., Tan, B., Ding, Y., et al. (2012). NnHSP17.5, a cytosolic class II small heat shock protein gene from Nelumbo nucifera, contributes to seed germination vigor and seedling thermotolerance in transgenic Arabidopsis. Plant Cell Rep. 31, 379–389. doi: 10.1007/s00299-011-1173-0
Keywords: heat, transcriptomics, network analysis, Triticum aestivum, landrace, hub gene
Citation: Barratt LJ, Franco Ortega S and Harper AL (2024) Identification of candidate regulators of the response to early heat stress in climate-adapted wheat landraces via transcriptomic and co-expression network analyses. Front. Plant Sci. 14:1252885. doi: 10.3389/fpls.2023.1252885
Received: 04 July 2023; Accepted: 29 November 2023;
Published: 03 January 2024.
Edited by:
Ligeng Ma, Capital Normal University, ChinaReviewed by:
Rajib Roychowdhury, Volcani Center, IsraelAnthony Guihur, Université de Lausanne, Switzerland
Prachi Pandey, National Institute of Plant Genome Research (NIPGR), India
Copyright © 2024 Barratt, Franco Ortega and Harper. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea L. Harper, YW5kcmVhLmhhcnBlckB5b3JrLmFjLnVr
 Liam J. Barratt
Liam J. Barratt Sara Franco Ortega
Sara Franco Ortega Andrea L. Harper
Andrea L. Harper