
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 30 August 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1247707
This article is part of the Research Topic Applications of Novel Gene Editing Tools and Approaches in Plants View all 10 articles
 Dan Yao1,2†
Dan Yao1,2† Junming Zhou1†
Junming Zhou1† Aijing Zhang3
Aijing Zhang3 Jiaxin Wang1
Jiaxin Wang1 Yixuan Liu1
Yixuan Liu1 Lixue Wang1
Lixue Wang1 Wenxuan Pi1
Wenxuan Pi1 Zihao Li1
Zihao Li1 Wenjun Yue1
Wenjun Yue1 Jinliang Cai1
Jinliang Cai1 Huijing Liu1
Huijing Liu1 Wenyuan Hao4*
Wenyuan Hao4* Xiangchun Qu2*
Xiangchun Qu2*Soybean [Glycine max (Linn.) Merr] is a source of plant-based proteins and an essential oilseed crop and industrial raw material. The increase in the demand for soybeans due to societal changes has coincided with the increase in the breeding of soybean varieties with enhanced traits. Earlier gene editing technologies involved zinc finger nucleases and transcription activator-like effector nucleases, but the third-generation gene editing technology uses clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9). The rapid development of CRISPR/Cas9 technology has made it one of the most effective, straightforward, affordable, and user-friendly technologies for targeted gene editing. This review summarizes the application of CRISPR/Cas9 technology in soybean molecular breeding. More specifically, it provides an overview of the genes that have been targeted, the type of editing that occurs, the mechanism of action, and the efficiency of gene editing. Furthermore, suggestions for enhancing and accelerating the molecular breeding of novel soybean varieties with ideal traits (e.g., high yield, high quality, and durable disease resistance) are included.
Soybean is a significant source of vegetable proteins for humans and an important oilseed crop, making it a commercially valuable plant (Zhang A, et al., 2023). More than 90% of the soybean plants cultivated in the three main soybean-producing countries (USA, Brazil, and Argentina) are genetically modified varieties generated using gene editing technology (Fang et al., 2023). In terms of sustainable food production, the demand for soybeans has continued to increase because of the scarcity of arable land. In the field of molecular breeding, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) has emerged as a commonly used third-generation gene editing technology (Nakamori, 2023). Thus, many new and desirable soybean traits have been developed using gene editing technology, which is currently a hot topic in scientific research (Osakabe and Osakabe, 2017; Chen et al., 2022; Zhou et al., 2023a).
In recent years, CRISPR/Cas9 gene editing technology has been used by plant molecular breeders to improve various plant traits (Ma et al., 2016; Zhang et al., 2017; Rao et al., 2022). Because it can simply, effectively, and precisely edit target genes responsible for specific characteristics, CRISPR/Cas9 has replaced previously used gene editing techniques (Zheng et al., 2021; Impens et al., 2022; Liu H. et al., 2022). Several crop traits, including yield, quality, stress tolerance, disease resistance, and herbicide resistance, can be improved using CRISPR/Cas9 systems. This can lead to the development of novel germplasm with superior traits as well as significant advancements in plant molecular breeding (Yin et al., 2017; Hussain et al., 2018; Wada et al., 2020; Gan and Ling, 2022; Qi et al., 2023).
The limitations of early genome editing methods included the inability to explore the relationships between several related genes (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013). These previous methods were mostly employed to edit individual genes. Because soybean is a paleotetraploid, it has many homologous and redundant genes, which makes the functional characterization of soybean genes challenging (Tran and Mochida, 2010; Du et al., 2023). The CRISPR/Cas9 system has recently been used to edit multiple genes in the soybean genome. This has considerably decreased the effects of redundant genes on the efficient editing of specific genes for breeding soybean varieties with desirable traits (Bao et al., 2020; Xu H. et al., 2020; Baek et al., 2022; Guan et al., 2022; Rasheed et al., 2022a).
This review describes the recent improvements in soybean traits via the application of the CRISPR/Cas9 gene editing technology. It also presents information regarding the target genes and their mechanism of action, while providing a brief overview of transformation efficiency and gene editing efficiency. Furthermore, suggestions for future CRISPR/Cas9 development and use in soybean molecular breeding programs are included.
There has recently been an increase in the use of CRISPR/Cas9 to edit genes in soybean, corn, wheat, rice, cotton, and other crops (Figure 1, Table 1). The creation of new soybean germplasm with many excellent traits using various transformation methods (e.g., Agrobacterium-mediated transformation) has laid the foundation for further improving CRISPR/Cas9 gene editing technology for soybean molecular breeding (Figure 2).
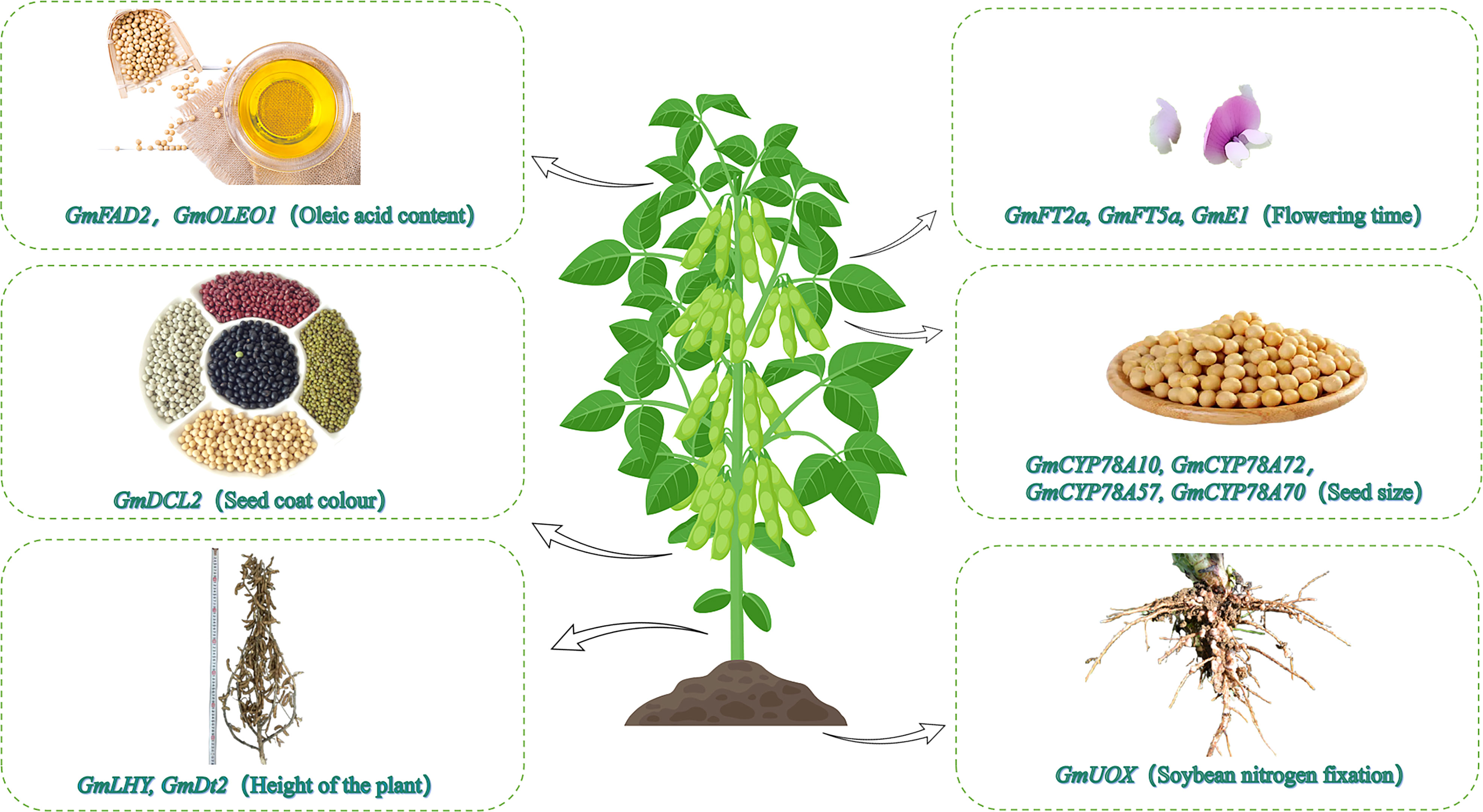
Figure 1 Utility of CRISPR/Cas9 for editing soybean functional genes. The CRISPR/Cas9 gene editing technology has recently been used to modify soybean genes affecting the oil content, photoperiodic flowering, seed coat color, seed size, plant height, and nodulation.
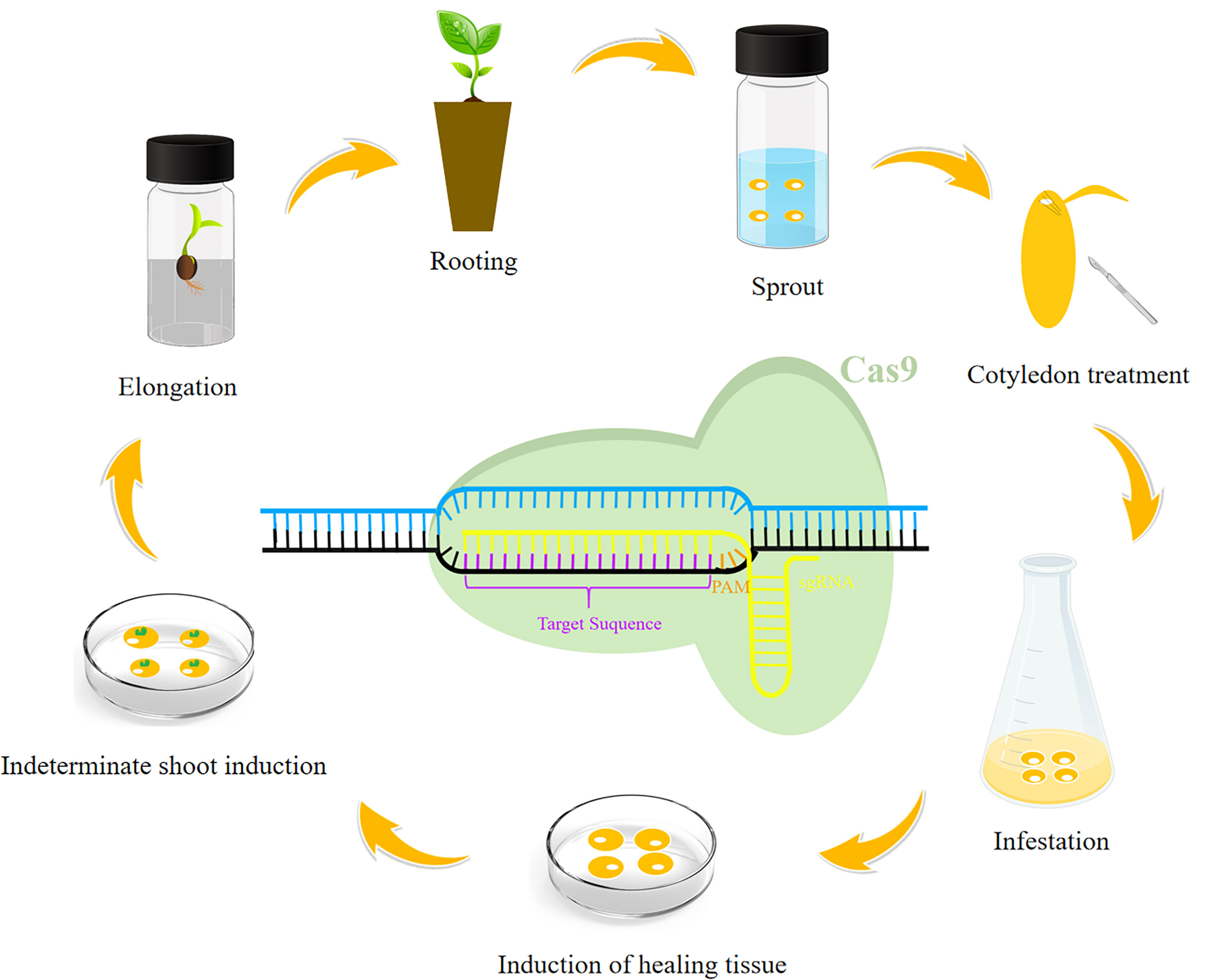
Figure 2 General soybean genetic transformation process. The following steps are generally included in the genetic transformation of soybean: sprouting, cotyledon treatment, infestation, induction of healing tissue, indeterminate shoot induction, elongation, and rooting. A schematic diagram is provided to show how the CRISPR/Cas9 system cleaves the target genomic segment.
During different soybean developmental stages, many genetic and biochemical processes control how soybean perceives and responds to abiotic stresses, including salinity and drought. One of the primary objectives of molecular breeding research is improving stress tolerance (Deshmukh et al., 2014; Amoanimaa-Dede et al., 2022; Cadavid et al., 2023). Osmotic stress in plant cells is typically caused by abiotic factors (e.g., drought or excessive salinity). Analyses of the sequences of the related genes revealed the regulatory effects of various plant cellular components, such as sensors, receptors, phytohormones, transcription factors, kinases, phosphatases, and microRNAs, on abiotic stress response-related pathways (Ramesh et al., 2019; Mangena, 2020; Staniak et al., 2023).
Water deficiency substantially restricts soybean growth and development, which can decrease the soybean yield by up to 40% (Khan, 2018). Thus, there is a critical need for exploring the mechanism underlying soybean drought resistance and generating new drought-resistant soybean germplasm (Ramlal et al., 2022). By deleting miR398c in soybean, Zhou et al. (2020) increased the expression of GmCSD1a/b, GmCSD2a/b/c, and GmCCS (relative to the corresponding levels in over-expression strains), thereby increasing the capacity to scavenge O2− (Zhou et al., 2020). In 2021, Xiao et al. identified 112 GmPLA family genes in the soybean genome and used CRISPR/Cas9 technology to knock out two homologous genes (GmpPLA-II epsilon and zeta). Knocking out one or both genes affected the root response to phosphorus deficiency, with some mutant lines exhibiting increased resistance to flooding and drought conditions (compared with the control) (Xiao et al., 2021). Additionally, in 2021, Yu et al. reported that the GmNF-YC14 deletion mutant created using CRISPR/Cas9 technology is more susceptible to drought stress than wild-type soybean, implying GmNF-YC14 may be useful for increasing soybean drought tolerance (Yu et al., 2021). By comparing the agronomic features of soybean plants over-expressing sHSP26 with those of soybean plants in which sHSP26 had been edited, Liu S. Y., et al. (2022) revealed that sHSP26 may considerably increase soybean drought tolerance and yield (Liu S. Y., et al., 2022). In 2022, Yang et al. edited the soybean transcription factor gene GmNAC12, which decreased the survival of the transgenic plants exposed to drought stress by at least 12%. They concluded that GmNAC12 is a key gene that positively regulates soybean tolerance to drought conditions (Yang C.F., et al., 2022).
Salinity can severely decrease the seed yield and quality of soybean, which is a salt-sensitive crop species (Phang et al., 2008; Cai et al., 2022; Feng et al., 2023). In addition to accelerating the development of salt-tolerant soybean varieties to increase grain yield, research on salt stress tolerance can also optimize the use of saline farmland (Chen et al., 2018; Jin et al., 2021). In 2021, Niu et al. clarified the effects of knocking down and over-expressing lncRNA77580 on the expression of nearby protein-coding genes linked to the soybean response to salt stress. Additionally, increases in the length of the DNA fragment deleted from lncRNA77580 via the application of CRISPR/Cas9 technology increased the changes in the expression of lncRNA77580 and nearby genes (Niu et al., 2021). By simultaneously targeting six GmAITR genes using a CRISPR/Cas9 system, Wang et al. (2021) produced a Cas9-free GmAITR3 and GmAITR6 double mutant and a GmAITR2 GmAITR3 GmAITR4 GmAITR5 GmAITR6 quintuple mutant. They determined that salt tolerance was more pronounced in the higher-order mutants, suggesting that mutating GmAITR genes can enhance soybean salt tolerance (Wang et al., 2021). Zhang M.H. et al. (2022) produced three soybean mutants in which GmSOS1 was edited and observed that Na+ accumulated significantly more in the mutants than in the control. Accordingly, this gene is essential for soybean salt tolerance because it helps maintain Na+ homeostasis (Zhang M.H., et al., 2022).
The adaptation of soybean to severe drought and salt stresses involves the activation of overlapping pathways at the morphological, physiological, and molecular levels. Drought tolerance and salt tolerance are polygenic traits (Chen et al., 2018; Kofsky et al., 2018; Mammadov et al., 2018). Additionally, the perception of stress and its effects on soybean growth or development are similar among the abiotic stress factors. In an earlier study by Du et al. (2018), soybean plants in which the transcription factor gene GmMYB118 was silenced were more susceptible to drought and saline conditions than soybean plants over-expressing GmMYB118. Moreover, the decreased production of minor heat shock proteins increased the resistance of plants to drought, cold, and salt stresses (Du et al., 2018). However, when Zhang M.H. et al. (2022) knocked out GmHsps_p23, which encodes a minor heat shock protein in soybean, the transgenic plants were highly susceptible to salt and drought conditions. Future research will need to focus on the use of several gene editors to simultaneously target and regulate the expression of functional genes mediating drought and salinity tolerance to produce novel soybean genotypes with superior traits (Zhang Y.Z., et al., 2022).
Tobacco ringspot virus, soybean dwarf virus, soybean vein necrosis virus, soybean mosaic virus (SMV), bean pod mottle virus, and alfalfa mosaic virus are only a few of the viruses that can infect soybean (Liu et al., 2016; Widyasari et al., 2020; Lin et al., 2022). Multiple viruses can simultaneously infect soybean plants, causing more harm than an infection by a single virus. Hence, the use of gene editing tools to target genes that control soybean disease resistance and improve disease resistance-related traits has become a major objective in soybean molecular breeding programs (Chang et al., 2016; Chandra et al., 2022; Zhao et al., 2023).
Several non-homologous end-joining and homology directed repair-mediated gene replacement mutants were produced by Fang et al. (2015), who targeted the soybean blast fungal pathogenicity gene Avr4/6. These mutants were more resistant to diseases caused by oomycetes than the controls (Fang and Tyler, 2016). Ochola et al. (2020) edited the usual effector genes of the soybean root pathogen Phytophthora sojae. They observed that disease resistance was affected by the Avr gene expression level in soybean (Ochola et al., 2020). In 2020, Ma et al. confirmed that GmLMM2 deficiencies increased the resistance to P. sojae by increasing tetrapyrrole biosynthesis, but decreased the chlorophyll content by disrupting tetrapyrrole biosynthesis. The elimination of GmLMM2 expression resulted in the appearance of necrotic regions in the growing leaves of the CRISPR/Cas9-edited mutants (Ma et al., 2020). Zhang P.P, et al. (2020) targeted GmF3H1, GmF3H2, and GmFNSII-1 in soybean plants (including the hairy roots) using a CRISPR/Cas9-mediated multiple gene editing system. They detected a significant increase in the isoflavone content and a significant decrease in the SMV coat protein content (approximately 33% decrease) in the mutants, indicating that the increased isoflavone content enhanced the leaf resistance to SMV (Zhang P.P., et al., 2020). Three crucial genes in the soybean Rsc4 gene family (Rsc4-1, Rsc4-2, and Rsc4-3) were modified by CRISPR/Cas9 in 2021 to alter soybean resistance to SMV (Yin et al., 2021). To investigate the effector gene Avr1b-1 in the soybean pathogen Blastomyces in terms of its function as well as the underlying mechanism. Gu et al. (2021) created target locus-specific knockout and knock-in mutants. All selected knockout mutants were virulent on plants expressing Rps1b, whereas the infection of plants lacking Rps1b was unaffected. When a sgRNA-resistant variant of Avr1b-1 was re-introduced into the Avr1b-1 locus of the mutants in which Avr1b was knocked out, the resulting knock-in transformants expressing Avr1b-1 were unable to infect soybean plants carrying Rps1b (Gu et al., 2021). Compared with the RNAi and over-expression strains, the soybean plants in which GmDRR1 was knocked down (in 2022) were considerably less resistant to Blastomyces infections (Yu et al., 2022). By altering the coding region of the soybean transcription factor gene GmTCP19L, Fan et al. (2022) produced a mutant with a 2 bp deletion. This mutant soybean germplasm resource exhibited increased susceptibility to blast molds (Fan et al., 2022).
Plants that are resistant to Rps gene products can perceive certain pathogen effectors encoded by Avr genes. By deleting Avr45a, Arsenault-Labrecque et al. (2022) produced novel soybean plants resistant to Rps8 (Arsenault-Labrecque et al., 2022). In 2022, Zhang et al. identified Glyma.07g110300 (LOC100775351) as a quantitative trait locus (QTL)-M marker gene encoding the UDP-glycosyltransferase (UGT) primarily responsible for soybean resistance to leaf-chewing insects. Using a CRISPR/Cas9 system, they enhanced the resistance of soybean to Helicoverpa armigera and Spodoptera litura via the following two mutation types: large fragment deletion and single base insertion. Zhang Y.X., et al. (2022) confirmed that GmUGT confers resistance to leaf-chewing insects by changing the flavonoid content and the expression of genes related to flavonoid biosynthesis and defense (Zhang Y.X., et al., 2022). By editing the soybean 14-3-3 gene (Glyma05g29080) via large fragment insertions and deletions and producing transgenic plants with increased susceptibility to hard tick infestations and decreased nodulation, Zhang Y.F., et al. (2023) showed Glyma05g29080 contributes to nodulation and defense responses (Zhang Y.F., et al., 2023). Using a CRISPR/Cas9 gene editing method, Liu et al. (2023b) silenced GmTAP1 in soybean, which resulted in increased resistance to P. sojae strains P231, P233, and P234. An analysis of reactive oxygen species revealed that a loss-of-function mutation to GmTAP1 does not substantially alter plant basal immunity (Liu T.F., et al., 2023).
The soybean cyst nematode (SCN) is responsible for the soybean disease associated with the largest economic losses (Bent, 2022). By altering two functional genes (Glyma.12G194800 and Glyma.16G154200) in the syntaxin family of SCN resistance genes, Dong et al. (2020) produced SCN-resistant soybean cultivars (Dong et al., 2020). In 2021, Butler et al. demonstrated that Glyma.15G191200 of cqSCN-006, which encodes gamma-SNAP, influences SCN resistance. Additionally, using CRISPR/Cas9 gene editing technology to disrupt the cqSCN-006 allele decreased the SCN resistance of the transgenic roots (Butler et al., 2021). In 2022, Zhang et al. mutated Glyma.07g110300 by introducing a CRISPR/Cas9 expression vector into the Tianlong 1 soybean variety to increase the resistance to S. litura and H. armigera (Zhang Y.X., et al., 2022).
Soybean is used as a source of food for animals, including humans (Medic et al., 2014). It has the highest protein content of any crop and is a significant source of edible oils (Gupta and Manjaya, 2022; Zaaboul et al., 2022; Song et al., 2023). In the past few years, several studies have employed CRISPR/Cas9 gene editing technology to enhance the protein and oleic acid contents of soybean.
Using germinal root transformation technology, Li et al. altered the soybean seed storage protein-encoding genes Glyma.20g148400, Glyma.03g163500, and Glyma.19g164900 to increase soybean seed protein contents (Li et al., 2019a). By simultaneously modifying the soybean genes GmFAD2-1A and GmFAD2-1B, Do et al. (2019) managed to increase the oleic acid content by more than 80%, while also decreasing the linoleic acid level by 1.3%–1.7% (Do et al., 2019). Zhang et al. (2019) silenced the soybean phospholipase D1-encoding gene, which increased the oil content and germination rate of the mutant seeds (compared with the wild-type seeds) at high temperatures and high humidity levels (Zhang et al., 2019). In 2021, Qu et al. analyzed the oleic acid contents of soybean plants over-expressing Gm15G117700 and soybean plants in which the gene was edited; the oleic acid content increased in the gene-edited plants by 3.49% (Qu et al., 2021). Zhou et al. (2023a) recently edited five important enzyme-encoding genes in the GmFAD2 family and analyzed the associated effects on soybean oil synthesis. Editing GmFAD2-1A increased the oleic acid content by 91.49% (Zhou et al., 2023a). In another recent study, Li et al. (2023) edited two target genes by altering the conserved PAP2 structural domain-encoding sequences of GmPDCT1 and GmPDCT2. The decrease in phosphatidylcholine-derived diacylglycerol contents via the knockdown of GmPDCT prevented the entry of phosphatidylcholine-modified polyunsaturated fatty acids into the triacylglycerol biosynthesis pathway (Li et al., 2023b).
In addition to increasing the protein and oleic acid contents, researchers have attempted to enhance other soybean characteristics. Phytic acid (PA) is an anti-nutrient in grains that prevents humans from absorbing trace minerals (e.g., iron and zinc). In soybean, GmIPK1 encodes an enzyme that converts inositol 1,3,4,5,6-pentaphosphate to inositol 1,2,3,4,5,6-hexaphosphate (Alkarawi and Zotz, 2014; Sarkhel and Roy, 2022). Using the CRISPR/Cas9 system, Song et al. (2022) edited the GmIPK1 gene and sgRNA to introduce mutations to create soybean lines with low PA levels. The decreased PA levels in the T2 generation mutant seeds were not accompanied by defective growth or seed development (Song et al., 2022).
Flavor is an important soybean quality-related attribute. Accordingly, CRISPR/Cas9 technology has been exploited to develop soybean germplasm with superior flavor-related traits (Fernandez-Marin et al., 2014). Because soybean proteins are allergens, decreasing the abundance of allergenic proteins will likely increase the utility of soybean as a source of protein (e.g., in processed food) (Cordle, 2004; L'Hocine and Boye, 2007; Gharibzahedi et al., 2022; Gracio et al., 2023). In 2020, Sugano et al. simultaneously targeted and edited GmBd28k and GmBd30K to eliminate two allergenic proteins in the Japanese soybean cultivars Enrei and Kariyutaka (Sugano et al., 2020). Soybean flavor and quality are influenced by three lipoxygenases (LOX1, LOX2, and LOX3). By editing three genes in the soybean GmLox family (GmLox1, GmLox2, and GmLox3), Wang J., et al. (2020) improved the edibility of soybean oil and protein products. Editing these genes decreased soybean odors (Wang J., et al., 2020). The raffinose oligosaccharide (RFO) family members are the main soluble carbohydrates in soybean seeds, but they are anti-nutritional seed components because they typically cause gas and indigestion, while also decreasing energy efficiency (Salvi et al., 2022). In 2021, Le et al. decreased the soybean seed RFO content by knocking down two galactinol synthase-encoding genes, namely GmGOLS1A and its homolog GmGOLS1B (Le et al., 2020). To decrease the RFO content in mature seeds, Cao et al. (2022) used a CRISPR/Cas9 multi-gene editing method to delete the RS2 and RS3 genes in soybean and cottonseed (Cao et al., 2022). Qian et al. (2022) mutated GmBADH2 and confirmed this gene contributes to soybean odors (Qian et al., 2022). In addition, Bai et al. (2022) used CRISPR/Cas9 gene editing technology to produce two multi-gene mutants, one lacking the 7S subunit and the other lacking the 11S subunit. Both of these mutations enhanced the flavor of soybean meal (Bai et al., 2022).
One of the key factors influencing the development of high-yielding soybean cultivars is the appropriate regulation of plant structural features (e.g., plant height, number of nodes, number of pods, internode length, number of branches, and number of grains) (Hu and Wiatrak, 2012; Kuzbakova et al., 2022). In recent years, soybean phenotype-related genes have been edited using CRISPR/Cas9 gene editing technology to produce soybean germplasm with a variety of improved features.
Using the CRISPR/Cas9 system, Bao et al. (2019) mutated four SPL9 family genes that encode SQUAMOSA promoter-binding protein-like (SPL) transcription factors. The higher-order mutant plants with different combinations of mutations had more nodes and branches on the main stem (compared with the control plants), resulting in varying numbers of nodes per plant (Bao et al., 2019). In 2019, Cheng et al. used four gRNAs to alter four late elongated hypocotyl (LHY)-encoding GmLHY genes in soybean. Phenotypic analyses showed that the quadruple mutant plants had relatively short internodes and exhibited dwarfism (Cheng et al., 2019). In the Tianlong 9 variety, Jia et al. (2020) knocked out two copies of the soybean DCL2 gene, which altered the color of the soybean seed coat from yellow to brown (Jia et al., 2020). To increase soybean production, Cai et al. (2021) modified the low-latitude spring soybean variety Huachun 6 using a CRISPR/Cas9 multi-gene editing technique. Specifically, they targeted GmJAG, which affects the number of seeds per pod (Cai et al., 2021). In 2022, Mu et al. targeted six GmBIC genes in soybean using CRISPR/Cas9 technology. The single, double, and quadruple mutants were shorter than normal (Mu et al., 2022). In another recent study, Zhong et al. (2022) edited the soybean GmHdz4 gene, which increased the total root length, root surface area, and number of root tips (compared with the mutant lines over-expressing GmHdz4) (Zhong et al., 2022). Furthermore, Zhang Z. et al. (2023) silenced the soybean GmNSS gene, which resulted in the production of abnormally small seeds. (Zhang Z. et al., 2023).
Abscisic acid is an essential phytohormone that controls various processes related to plant growth, development, and stress responses (Nguyen et al., 2023). Using a CRISPR/Cas9 system, Zhang Z. H. et al. (2022) mutated GmPYL17, GmPYL18, and GmPYL19. Compared with the wild-type plants, the mutants were taller, had more branches, and were less sensitive to abscisic acid during the seed germination stage (Zhang Z. H. et al., 2022).
The shattering of soybean pods can significantly decrease yield. By altering the GmPDH gene family in soybean variety Huachun 6, Zhang Z. et al. (2022) showed that the PDH1 mutation dramatically increases pod shatter resistance without modifying other important agronomic parameters (Zhang Z. et al., 2022).
Rhizobia can produce a symbiotic nitrogen-fixation system with legumes that increases plant output without damaging the local ecosystem (Chakraborty et al., 2022; Hawkins and Oresnik, 2022). More than 65% of the nitrogen fixation is due to the symbiotic interaction between rhizobia and legumes (Fields et al., 2021; Jimenez-Guerrero et al., 2022). Soybean converts free nitrogen in the air to chemosynthetic nitrogen that can be absorbed and used by the plant via nitrogen-fixing nodules. This process yields soybean seeds with a high protein content, thereby increasing the nutritional value of soybean (Dadnia, 2011; Meng et al., 2015; Igiehon et al., 2021).
Xu et al. (2021) promoted soybean nodulation by using CRISPR/Cas9 technology to knock down miR9c (Xu et al., 2021). By deleting the soybean RFG193 gene, Fan et al. (2020) generated transgenic plants with mature nitrogen-fixing nodules on purple or red roots, which produced anthocyanins, whereas nodules were undetectable on the non-transgenic roots (Fan et al., 2020). In 2021, Yang et al. reported that a loss-of-function mutation to GmHSP17.9 significantly affects soybean plant growth and seed yield through the associated changes to the number of root nodules, nodule fresh weight, nitrogenase activity, poly-hydroxybutyrate vesicles, and urea and total nitrogen contents (Yang Z.W., et al., 2022). Nguyen et al. (2021) silenced GmUOX in a soybean mutant, which exhibited nitrogen deficit atrophy and early nodule senescence as revealed by decreased nitrogenase (acetylene reduction) activities in the nodules, a greenish-white hue inside the nodules, and a decreased root protein output (Nguyen et al., 2021). Gao et al. (2021) investigated the role of the PIN protein during the nitrogen fixation by soybean nodules. More specifically, they produced a triple mutant (GmPIN1-abc family) (Gao et al., 2021). The modification of the soybean Rfg1 allele by Fan et al. (2022) revealed Rfg1 mediates the resistance to Sinorhizobium fredii and Bradyrhizobium japonicum strains, leading to broad-spectrum resistance to nodulation in transgenic plants (Fan et al., 2017). After knocking down GmNN1, Li et al. (2022) detected yellowing leaves as well as decreased nitrogen contents and decreased nodulation (compared with the wild-type control plants) (Li et al., 2022). By silencing GmNAC039 and GmNAC018 as well as the four target genes GmCYP35, GmCYP37, GmCYP39, and GmCYP4, Yu et al. (2023) showed that the transcription factors encoded by GmNAC039 and GmNAC018 directly increase the expression of GmCYP genes to induce root tumor senescence (Yu et al., 2023).
Because soybean is a short-day (SD) plant, it blooms more quickly during SD conditions than during long-day (LD) conditions (Weller and Ortega, 2015; Lin et al., 2021; Xia et al., 2021). Modulating the blooming time and minimizing the sensitivity to sunshine duration through molecular breeding can increase soybean adaptability and production by mitigating photoperiodic responses (Zhang L.X. et al., 2020; Zhang M. et al., 2022; Du et al., 2023).
Cai et al. (2018a) edited the soybean genes GmFT2a and GmFT9a and discovered that both mutants in the T2 generation exhibited a late-blooming phenotype (Cai et al., 2018a). Using a double sgRNA design and CRISPR/Cas9 technology, Cai et al. (2018b) deleted specific DNA fragments in GmFT2a (Glyma16g26660) and GmFT5a (Glyma16g04830). The homozygous GmFT2a mutants (1,618 bp deletion) in the T2 generation flowered late (Cai et al., 2018b). Two QTL regions that respectively included GmFT2a and GmFT5a were identified by Cai et al. (2020b). They were linked to various genetic effects on flowering during various photoperiods. Under LD and SD conditions, the flowering times of transgenic plants over-expressing GmFT2a or GmFT5a, GmFT2a mutants, GmFT5a mutants, and GmFT2a and GmFT5a double mutants were examined. There was no overlap between GmFT2a and GmFT5a, which cooperatively control the blooming time, but GmFT2a has a greater effect than GmFT5a under SD conditions, while GmFT5a has a greater effect than GmFT2a under LD conditions (Cai et al., 2020a). Wang L. W. et al. (2020) mapped QTLs and identified GmPRR37 as a functional gene encoding a regulator of soybean flowering. A natural mutation to GmPRR37 results in early flowering, thereby enabling the cultivation of soybean plants at high latitudes (Wang L. W., et al., 2020). Li et al. (2020) used CRISPR/Cas9 technology to knock out GmPRR3b. The resulting soybean mutant exhibited retarded growth and a delayed transition to the flowering stage (Li et al., 2020). In 2020, Chen et al. modified the soybean GmAP1 gene in a quadruple mutant. The observed increase in plant height was associated with delayed flowering, altered flower shapes, and increases in the number of nodes and the internode length. In contrast, under SD conditions, the over-expression of GmAP1 led to early flowering and dwarfism (Chen et al., 2020). Li et al. (2021) edited four LNK2 genes using a CRISPR/Cas9 system to produce a quadruple mutant lacking transgenes. This mutant flowered earlier than the wild-type control under LD conditions. In addition, the LNK2 transcript level was lower in the quadruple mutant than in the wild-type plants (Li et al., 2021). Zhao et al. (2022) mutated GmPHYA or GmPHYB using CRISPR/Cas9 technology. The phenotypic changes due to the mutations to GmPHYA2 and GmPHYA3, which have redundant and additive roles in seedling responses to daylight, indicated GmPHYB1 is primarily responsible for daylight-induced photomorphogenesis (Zhao et al., 2022). In 2022, Zhai et al. suggested that GmMDE and GmFT2a/GmFT5a contribute to a positive feedback regulatory loop that promotes flowering in soybean. Knocking down the soybean E1 gene induces GmMDE expression. Moreover, the over-expression of GmMDE06 increases the expression of GmFT2a and GmFT5a, which regulate flowering (Zhai et al., 2022). In 2023, Wan et al. investigated the relationship between the dominant E1 gene and photoperiodic regulation via the CRISPR/Cas9-mediated targeted mutation of E1 in soybean variety Tianlong 1. Four mutations were introduced into the E1 coding region. The significant structural changes in the generated mutants included the commencement of terminal flowering, the creation of distinct stems, and a decrease in the number of branches (Wan et al., 2022).
Because soybean is a self-pollinated plant that has small flower organs, artificial cross-breeding is both difficult and ineffective (Li et al., 2019b; Chen G. M., et al., 2021). Furthermore, differences in flowering times among varieties originating from various geographical regions frequently further restrict the exchange of genes, resulting in a limited genetic base for soybean breeding and genetic modifications (Li et al., 2019b). Accordingly, methods for increasing the genetic diversity of soybean varieties are needed (Bohra et al., 2016). In particular, for sexually reproducing crops, male sterility is a crucial precondition for hybrid seed generation and crop reproduction (Jiang et al., 2011; Yang et al., 2014). Male sterile lines can increase the quality of hybrids, lower the cost of hybrid seed production, and even broaden the utility of hybrids. The scarcity of adequate male sterile lines has limited the commercial use of soybean accessions (Li et al., 2016; Ramlal et al., 2022).
To create stable male sterile soybean lines, Chen et al. (2021) targeted AMS homologs using CRISPR/Cas9 technology. Although editing GmAMS2 failed to produce a male sterile line, editing GmAMS1 yielded plants with a male sterile phenotype. GmAMS1 contributes to the development of pollen walls as well as the regulation of soybean tapetum degeneration (Chen et al., 2021a). Jiang et al. (2021) modified Glyma.13G114200 using a CRISPR/Cas9 system; the phenotypes of two gene-edited lines were consistent with the male sterility of the MS1 mutant (Jiang et al., 2021). By eliminating GmSPL2b, Ding et al. (2023) decreased the heat tolerance of a soybean cytoplasmic male sterility-based recovery line during flowering (Ding et al., 2023).
Compared with Cas9, the CRISPR family member Cas12a is more practical and effective. Hence, CRISPR/Cas12a can effectively edit multiple genes because of the specific way that CRISPR RNA (crRNA) functions (Bandyopadhyay et al., 2020; Paul and Montoya, 2020; Zhou et al., 2023b). In 2017, Jiang et al. used CRISPR/Cas12a to achieve editing in the soybean FAD2 gene for the first time (Jiang et al., 2017). In addition, large chromosomal segments of the target genome were deleted by Duan et al. (2021) using CRISPR/Cas12a, with an editing efficiency of 91.7% (Duan et al., 2021). In 2023, Liang et al. produced CRISPR/Cas12a-edited soybeans in just 45 days, with transformation and gene editing efficiencies of 30% and 50%, respectively (Liang et al., 2023). To produce gene-edited soybeans with better traits, CRISPR/Cas12a-based multi-gene editing methods will increasingly be used to modify the soybean genome.
Because they enable the replacement of a single base via RNA editing without introducing DNA double-strand breaks or requiring donor templates, base editor tools created using the CRISPR/Cas9 system are especially useful for plant molecular breeding (Molla et al., 2021; Yang et al., 2021; Hua et al., 2022). A CRISPR/Cas9-mediated base editing tool was designed by Cai et al. (2020a) to alter individual bases in the soybean genome. A base editor was developed by combining Cas9n (D10A), rat cytosine deaminase (APOBEC1), and a uracil glycosylase inhibitor. This base editor was then cloned into the pTF101.1 vector. The targeted genes were GmFT2a and GmFT4a, which were under the control of the 2× CaMV 35S promoter. There were two types of base substitutions (C to T and C to G), both of which occurred within the target sequence (Cai et al., 2020a). Single nucleotide polymorphisms, which influence phenotypic diversity and are linked to many significant agronomic parameters, are abundant in the soybean genome. Future genetic improvement and breeding of soybean can greatly benefit from the application of base editing technology (Bharat et al., 2020; Xu R. F., et al., 2020).
Because of increases in the global population and living standards, CRISPR/Cas9 technology must be exploited to quickly develop high-yielding, high-quality soybean varieties (Khan et al., 2018; Zhang and Showalter, 2020). Field tests of high-oleic soybean varieties produced using CRISPR/Cas9 gene editing technology in the US have produced positive results, with potential implications for soybean molecular breeding. There have been considerable advances in the molecular breeding of soybean since the development of CRISPR/Cas9 gene editing technology, which has decreased concerns about the safety of products made from genetically modified soybeans, leading to the gradual acceptance of genetically modified crops. The CRISPR/Cas9 system, which continues to be refined and enhanced, has largely outperformed the older technologies involving zinc finger nucleases and transcription activator-like effector nucleases in terms of gene editing efficiency and convenience (Samanta et al., 2016; Demirci et al., 2018; Farooq et al., 2018). Researchers will use CRISPR/Cas9 gene editing systems to develop soybean lines with improved features as more functional soybean genes are identified and characterized.
However, there are certain limitations to the utility of CRISPR/Cas9 for soybean breeding. Unanswered questions include the following: (i) How can genome editing tools be efficiently delivered to soybean plants? (ii) How can the functional redundancy in gene families be rapidly and precisely determined? (iii) How can the editing of multiple genes be exploited to modify various traits? (iv) How can base editing, prime editing, and government regulations regarding genome-edited crops further increase the effectiveness of gene editing? Despite encouraging results, many obstacles must be overcome before CRISPR/Cas9 can be widely used for soybean breeding.
Additionally, numerous sgRNAs for different plant genomes have been assembled into CRISPR editing vectors. Moreover, sgRNA pooling techniques have made it possible to mutate multiple genes. The diversity in the sequences that PAM can detect has increased, leading to improved gene editing, because of the creation of Cas9 homologs, such as StCas9 and SaCas9, for plant molecular breeding. The highly efficient editing of plant genomes has been achieved using the nCas9-mediated single-base editing system, while the saturation mutagenesis of plant genomes and optimal gene editing efficiencies have been attained via the two-base editing method. The CRISPR/Cas9 gene editing method will be applied to soybean molecular breeding more effectively, conveniently, and broadly in the future, thereby facilitating increasingly precise molecular breeding and accelerating soybean molecular breeding.
DY and JZ performed the manuscript writing; AZ, JW, YL, LW, WP, ZL summarized the literature reports; WY and JC carried out the production of pictures; HL performed the organization of the table; WH and XQ reviewed and proofread the manuscript. All authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Key Research and Development Program of Science and the Technology of Jilin Province (No. 20210202006NC), and the Key Laboratory of Crop Genetic Resources and Germplasm Creation in Northeast China, Ministry of Agriculture and Rural Affairs (KYJF2023DX015-3).
We thank Liwen Bianji (Edanz) (www.liwenbianji.cn ) for editing the English text of a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alkarawi, H. H., Zotz, G. (2014). Phytic acid in green leaves. Plant Biol. 16, 697–701. doi: 10.1111/plb.12136
Amoanimaa-Dede, H., Su, C. T., Yeboah, A., Zhou, H., Zheng, D., Zhu, H. (2022). Growth regulators promote soybean productivity: a re1 view. Peerj 10, e12556. doi: 10.7717/peerj.12556
Arsenault-Labrecque, G., Santhanam, P., Asselin, Y., Cinget, B., Lebreton, A., Labbé, C., et al. (2022). Rxlr effector gene avr3a from Phytophthora Sojae is recognized by rps8 in soybean. Mol. Plant Pathol. 23, 693–706. doi: 10.1111/mpp.13190
Baek, D., Chun, H. J., Kim, M. C. (2022). Genome editing provides a valuable biological toolkit for soybean improvement. Plant Biotechnol. Rep. 16, 357–368. doi: 10.1007/s11816-022-00778-6
Bai, M. Y., Yuan, C. C., Kuang, H. Q., Sun, Q., Hu, X. C., Cui, L. N., et al. (2022). Combination of two multiplex genome-edited soybean varieties enables customization of protein functional properties. Mol. Plant 15, 1081–1083. doi: 10.1016/j.molp.2022.05.011
Bandyopadhyay, A., Kancharla, N., Javalkote, V. S., Dasgupta, S., Brutnell, T. P. (2020). CRISPR/Cas12a (cpf1): a versatile tool in the plant genome editing toolbox for agricultural advancement. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.584151
Bao, A. L., Chen, H. F., Chen, L. M., Chen, S. L., Hao, Q. N., Guo, W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of gmspl9 genes alters plant architecture in soybean. BMC Plant Biol. 19 (1), 131. doi: 10.1186/s12870-019-1746-6
Bao, A., Tran, L. P., Cao, D. (2020). CRISPR/Cas9-based gene editing in soybean. Methods Mol. Biol. (Clifton N.J.) 2107, 349–364. doi: 10.1007/978-1-0716-0235-5_19
Bent, A. F. (2022). Exploring soybean resistance to soybean cyst nematode. Annu. Rev. Phytopathol. 60, 379–409. doi: 10.1146/annurev-phyto-020620-120823
Bharat, S. S., Li, S. Y., Li, J. Y., Yan, L., Xia, L. Q. (2020). Base editing in plants: current status and challenges. Crop J. 8, 384–395. doi: 10.1016/j.cj.2019.10.002
Bohra, A., Jha, U. C., Adhimoolam, P., Bisht, D., Singh, N. P. (2016). Cytoplasmic male sterility (cms) in hybrid breeding in field crops. Plant Cell Rep. 35, 967–993. doi: 10.1007/s00299-016-1949-3
Butler, K. J., Fliege, C., Zapotocny, R., Diers, B., Hudson, M., Bent, A. F., et al. (2021). Soybean cyst nematode resistance quantitative trait locus cqscn-006 alters the expression of a gamma-snap protein. Mol. Plant-Microbe Interact. 34, 1433–1445. doi: 10.1094/MPMI-07-21-0163-R
Cadavid, I. C., Balbinott, N., Margis, R. (2023). Beyond transcription factors: more regulatory layers affecting soybean gene expression under abiotic stress. Genet. Mol. Biol. 46, e20220166. doi: 10.1590/1678-4685-GMB-2022-0166
Cai, Y. P., Chen, L., Liu, X. J., Guo, C., Sun, S., Wu, C. X., et al. (2018a). CRISPR/Cas9-mediated targeted mutagenesis of gmft2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. doi: 10.1111/pbi.12758
Cai, Y. P., Chen, L., Sun, S., Wu, C. X., Yao, W. W., Jiang, B. J., et al. (2018b). CRISPR/Cas9-mediated deletion of large genomic fragments in soybean. Int. J. Mol. Sci. 19 (12), 3835. doi: 10.3390/ijms19123835
Cai, Y. P., Chen, L., Zhang, Y., Yuan, S., Su, Q., Sun, S., et al. (2020a). Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 18, 1996–1998. doi: 10.1111/pbi.13386
Cai, X. X., Jia, B. W., Sun, M. Z., Sun, X. L. (2022). Insights into the regulation of wild soybean tolerance to salt-alkaline stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1002302
Cai, Y. P., Wang, L. W., Chen, L., Wu, T. T., Liu, L. P., Sun, S., et al. (2020b). Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18, 298–309. doi: 10.1111/pbi.13199
Cai, Z. D., Xian, P. Q., Cheng, Y. B., Ma, Q. B., Lian, T. X., Nian, H., et al. (2021). CRISPR/Cas9-mediated gene editing of GmJAGGED1 increased yield in the low-latitude soybean variety huachun 6. Plant Biotechnol. J. 19, 1898–1900. doi: 10.1111/pbi.13673
Cao, L., Wang, Z. R., Ma, H. Y., Liu, T. F., Ji, J., Duan, K. X., et al. (2022). Multiplex CRISPR/Cas9-mediated raffinose synthase gene editing reduces raffinose family oligosaccharides in soybean. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1048967
Chakraborty, S., Valdes-Lopez, O., Stonoha-Arther, C., Ane, J. M. (2022). Transcription factors controlling the rhizobium-legume symbiosis: integrating infection, organogenesis and the abiotic environment. Plant Cell Physiol. 63, 1326–1343. doi: 10.1093/pcp/pcac063
Chandra, S., Choudhary, M., Bagaria, P. K., Nataraj, V., Kumawat, G., Choudhary, J. R., et al. (2022). Progress and prospectus in genetics and genomics of phytophthora root and stem rot resistance in soybean (Glycine max L.). Front. Genet. 13, 939182. doi: 10.3389/fgene.2022.939182
Chang, H. X., Lipka, A. E., Domier, L. L., Hartman, G. L. (2016). Characterization of disease resistance loci in the usda soybean germplasm collection using genome-wide association studies. Phytopathology 106, 1139–1151. doi: 10.1094/PHYTO-01-16-0042-FI
Chen, Y. Z., Fu, M. C., Li, H., Wang, L. G., Liu, R. Z., Liu, Z. J., et al. (2021b). High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) Generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 19, 424–426. doi: 10.1111/pbi.13507
Chen, J. L., Li, S. Y., He, Y. B., Li, J. Y., Xia, L. Q. (2022). An update on precision genome editing by homology-directed repair in plants. Plant Physiol. 188, 1780–1794. doi: 10.1093/plphys/kiac037
Chen, H. T., Liu, X. Q., Zhang, H. M., Yuan, X. X., Gu, H. P., Cui, X. Y., et al. (2018). Advances in salinity tolerance of soybean: genetic diversity, heredity, and gene identification contribute to improving salinity tolerance. J. Integr. Agric. 17, 2215–2221. doi: 10.1016/S2095-3119(17)61864-1
Chen, L. Y., Nan, H. Y., Kong, L. P., Yue, L., Yang, H., Zhao, Q. S., et al. (2020). Soybeanap1homologs control flowering time and plant height. J. Integr. Plant Biol. 62, 1868–1879. doi: 10.1111/jipb.12988
Chen, X., Yang, S. X., Zhang, Y. H., Zhu, X. B., Yang, X. J., Zhang, C. B., et al. (2021a). Generation of male-sterile soybean lines with the CRISPR/Cas9 system. Crop J. 9, 1270–1277. doi: 10.1016/j.cj.2021.05.003
Chen, H. M., Ye, R., Liang, Y., Zhang, S. C., Liu, X. L., Sun, C. J., et al. (2023). Generation of low-cadmium rice germplasms via knockout of oslcd using CRISPR/Cas9. J. Environ. Sci. 126, 138–152. doi: 10.1016/j.jes.2022.05.047
Chen, C., Zhao, Y. Q., Tabor, G., Nian, H. Q., Phillips, J., Wolters, P., et al. (2023). A leucine-rich repeat receptor kinase gene confers quantitative susceptibility to maize southern leaf blight. New Phytol. 238, 1182–1197. doi: 10.1111/nph.18781
Chen, G. M., Zhou, Y. Z., Kishchenko, O., Stepanenko, A., Jatayev, S., Zhang, D. B., et al. (2021). Gene editing to facilitate hybrid crop production. Biotechnol. Adv. 46, 107676. doi: 10.1016/j.biotechadv.2020.107676
Cheng, Q., Dong, L. D., Su, T., Li, T. Y., Gan, Z. R., Nan, H. Y., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of gmlhy genes alters plant height and internode length in soybean. BMC Plant Biol. 19 (1), 562. doi: 10.1186/s12870-019-2145-8
Chin, H. S., Wu, Y. P., Hour, A. L., Hong, C. Y., Lin, Y. R. (2016). Genetic and evolutionary analysis of purple leaf sheath in rice. Rice 9 (1), 8. doi: 10.1186/s12284-016-0080-y
Cordle, C. T. (2004). Soy protein allergy: incidence and relative severity. J. Nutr. 134, 1213S–1219S. doi: 10.1093/jn/134.5.1213S
Dadnia, M. R. (2011). Effect of different formulations of rhizobium japonicum on yield and yield attributes of soybean (Glycine max L.). Res. On Crops 12, 413–416.
Demirci, Y., Zhang, B. H., Unver, T. (2018). CRISPR/Cas9: an rna-guided highly precise synthetic tool for plant genome editing. J. Cell. Physiol. 233, 1844–1859. doi: 10.1002/jcp.25970
Deshmukh, R., Sonah, H., Patil, G., Chen, W., Prince, S., Mutava, R., et al. (2014). Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 5, 244. doi: 10.3389/fpls.2014.00244
Ding, X. L., Guo, J. F., Lv, M. L., Wang, H. J., Sheng, Y., Liu, Y., et al. (2023). The mi R156b-GmSPL2b module mediates male fertility regulation of cytoplasmic male sterility-based restorer line under high-temperature stress in soybean. Plant Biotechnol. J. 21 (8), 1542–1559. doi: 10.1111/pbi.14056
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L., Stacey, G., Gillman, J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and alpha-linolenic acid phenotype in soybean. BMC Plant Biol. 19 (1), 311. doi: 10.1186/s12870-019-1906-8
Dong, J., Zielinski, R. E., Hudson, M. E. (2020). T-SNAREs bind the Rhg1 alpha-SNAP and mediate soybean cyst nematode resistance. Plant J. 104, 318–331. doi: 10.1111/tpj.14923
Du, H. P., Fang, C., Li, Y. R., Kong, F. J., Liu, B. H. (2023). Understandings and future challenges in soybean functional genomics and molecular breeding. J. Integr. Plant Biol. 65, 468–495. doi: 10.1111/jipb.13433
Du, Y. T., Zhao, M. J., Wang, C. T., Gao, Y., Wang, Y. X., Liu, Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18 (1), 320. doi: 10.1186/s12870-018-1551-7
Duan, N. N., Tang, S. Q., Zeng, B. T., Hu, Z. Q., Hu, Q., Wu, L. Q., et al. (2021). An episomal CRISPR/Cas12a system for mediating efficient gene editing. Life-Basel 11 (11), 1262. doi: 10.3390/life11111262
Errum, A., Rehman, N., Uzair, M., Inam, S., Ali, G. M., Khan, M. R., et al. (2023). CRISPR/Cas9 editing of wheat PPD-1 gene homoeologs alters spike architecture and grain morphometric traits. Funct. Integr. Genomics 23 (1), 66. doi: 10.1007/s10142-023-00989-2
Fan, Y. L., Liu, J., Lyu, S. H., Wang, Q., Yang, S. M., Zhu, H. Y., et al. (2017). The soybean Rfg1 gene restricts nodulation by sinorhizobium fredii usda193. Front. Plant Sci. 8, 1548. doi: 10.3389/fpls.2017.01548
Fan, Y., Wang, X., Li, H., et al. (2020). Anthocyanin, a novel and user-friendly reporter for convenient, non-destructive, low cost, directly visual selection of transgenic hairy roots in the study of rhizobia-legume symbiosis. Plant Methods. 16, 94. doi: 10.1186/s13007-020-00638-w
Fan, S. J., Zhang, Z., Song, Y., Zhang, J., Wang, P. W. (2022). CRISPR/Cas9-mediated targeted mutagenesis of GmTCP19L increasing susceptibility to Phytophthora Sojae in soybean. PloS One 17 (6), e0267502. doi: 10.1371/journal.pone.0267502
Fang, X. L., Sun, Y. Y., Li, J. H., Li, M. A., Zhang, C. B. (2023). Male sterility and hybrid breeding in soybean. Mol. Breed. 43 (6), 47. doi: 10.1007/s11032-023-01390-4
Fang, Y. F., Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora Sojae using CRISPR/Cas9. Mol. Plant Pathol. 17, 127–139. doi: 10.1111/mpp.12318
Farooq, R., Hussain, K., Nazir, S., Javed, M. R., Masood, N. (2018). CRISPR/Cas9; A robust technology for producing genetically engineered plants. Cell. Mol. Biol. 64, 31–38. doi: 10.14715/cmb/2018.64.14.6
Feng, C., Gao, H. T., Zhou, Y. G., Jing, Y., Li, S. Q., Yan, Z., et al. (2023). Unfolding molecular switches for salt stress resilience in soybean: recent advances and prospects for salt-tolerant smart plant production. Front. Plant Sci. 14, 1162014. doi: 10.3389/fpls.2023.1162014
Fernandez-Marin, B., Milla, R., Martin-Robles, N., Arc, E., Kranner, I., Becerril, J. M., et al. (2014). Side-effects of domestication: cultivated legume seeds contain similar tocopherols and fatty acids but less carotenoids than their wild counterparts. BMC Plant Biol. 14, 1599. doi: 10.1186/s12870-014-0385-1
Fields, B., Moffat, E. K., Friman, V. P., Harrison, E. (2021). The impact of intra- specific diversity in the rhizobia-legume symbiosis. Microbiology-Sgm 167 (4), 001051. doi: 10.1099/mic.0.001051
Gan, W. C., Ling, A. P. K. (2022). CRISPR/Cas9 in plant biotechnology: applications and challenges. Biotechnologia 103, 81–93. doi: 10.5114/bta.2022.113919
Gao, Z., Chen, Z. W., Cui, Y. Y., Ke, M. Y., Xu, H. F., Xu, Q. Z., et al. (2021). Gmpin-dependent polar auxin transport is involved in soybean nodule development. Plant Cell 33, 2981–3003. doi: 10.1093/plcell/koab183
Gharibzahedi, S., Smith, B., Altintas, Z. (2022). Bioactive and health-promoting properties of enzymatic hydrolysates of legume proteins: a review. Crit. Rev. Food Sci. Nutr 2022, 1–31. doi: 10.1080/10408398.2022.2124399
Gracio, M., Oliveira, S., Lima, A., Ferreira, R. B. (2023). Rubisco as a protein source for potential food applications: a review. Food Chem. 419, 135993. doi: 10.1016/j.foodchem.2023.135993
Gu, B., Shao, G. D., Gao, W. X., Miao, J. Q., Wang, Q. H., Liu, X. L., et al. (2021). Transcriptional variability associated with CRISPR-mediated gene replacements at the Phytophthora Sojae avr1b-1 locus. Front. Microbiol. 12, 645331. doi: 10.3389/fmicb.2021.645331
Guan, J. N., Xie, Z. M., Rasheed, A., Wang, T. C., Qian, Z., Zhang, Z., et al. (2022). CRISPR/Cas9 applications for improvement of soybeans, current scenarios, and future perspectives. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 50, 12678. doi: 10.15835/nbha50212678
Gupta, S. K., Manjaya, J. G. (2022). Advances in improvement of soybean seed composition traits using genetic, genomic and biotechnological approaches. Euphytica 99 (2022), 218. doi: 10.1007/s10681-022-03046-4
Hawkins, J. P., Oresnik, I. J. (2022). The rhizobium-legume symbiosis: co-opting successful stress management. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.796045
Hu, M. X., Wiatrak, P. (2012). Effect of planting date on soybean growth, yield, and grain quality: review. Agron. J. 104, 785–790. doi: 10.2134/agronj2011.0382
Hua, K., Han, P. J., Zhu, J. K. (2022). Improvement of base editors and prime editors advances precision genome engineering in plants. Plant Physiol. 188, 1795–1810. doi: 10.1093/plphys/kiab591
Huang, Q. L., Lin, B. R., Cao, Y. Q., Zhang, Y., Song, H. D., Huang, C. H., et al. (2023). CRISPR/Cas9-mediated mutagenesis of the susceptibility gene OsHPP04 in rice confers enhanced resistance to rice root-knot nematode. Front. Plant Sci. 14, 1134653. doi: 10.3389/fpls.2023.1134653
Hussain, B., Lucas, S. J., Budak, H. (2018). CRISPR/Cas9 in plants: at play in the genome and at work for crop improvement. Briefings Funct. Genomics 17, 319–328. doi: 10.1093/bfgp/ely016
Igiehon, N. O., Babalola, O. O., Cheseto, X., Torto, B. (2021). Effects of rhizobia and arbuscular mycorrhizal fungi on yield, size distribution and fatty acid of soybean seeds grown under drought stress. Microbiol Res. 242, 126640. doi: 10.1016/j.micres.2020.126640
Impens, L., Jacobs, T. B., Nelissen, H., Inze, D., Pauwels, L. (2022). Mini-review: transgenerational CRISPR/Cas9 gene editing in plants. Front. Genome Editing 4. doi: 10.3389/fgeed.2022.825042
Jia, J. B., Ji, R. H., Li, Z. W., Yu, Y. M., Nakano, M., Long, Y. P., et al. (2020). Soybean dicer-like2 regulates seed coat color via production of primary 22-nucleotide small interfering rnas from long inverted repeats. Plant Cell 32, 3662–3673. doi: 10.1105/tpc.20.00562
Jiang, B. J., Chen, L., Yang, C. Y., Wu, T. T., Yuan, S., Wu, C. X., et al. (2021). The cloning and CRISPR/Cas9-mediated mutagenesis of a male sterility gene ms1 of soybean. Plant Biotechnol. J. 19, 1098–1100. doi: 10.1111/pbi.13601
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., Weeks, D. P., et al. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi: 10.1111/pbi.12663
Jiang, W., Yang, S. P., Yu, D. Y., Gai, J. Y. (2011). Cloning and characterization of a novel gene gmmf1 in soybean (Glycine max L. Merr.). Agric. Sci. China 10, 1834–1841. doi: 10.1016/S1671-2927(11)60183-1
Jimenez-Guerrero, I., Medina, C., Vinardell, J. M., Ollero, F. J., Lopez-Baena, F. J. (2022). The rhizobial type 3 secretion system: the dr. Jekyll and mr. Hyde in the rhizobium-legume symbiosis. Int. J. Mol. Sci. 23 (19), 11089. doi: 10.3390/ijms231911089
Jin, J., Wang, J. F., Li, K. K., Wang, S. W., Qin, J., Zhang, G. H., et al. (2021). Integrated physiological, transcriptomic, and metabolomic analyses revealed molecular mechanism for salt resistance in soybean roots. Int. J. Mol. Sci. 22 (23), 12848. doi: 10.3390/ijms222312848
Karmacharya, A., Li, D., Leng, Y., Shi, G. S., Liu, Z., Yang, S. M., et al. (2023). Targeting disease susceptibility genes in wheat through wide hybridization with maize expressing cas9 and guide rna. Mol. Plant-Microbe Interact. Mpmi. doi: 10.1094/MPMI-01-23-0004-SC
Khan, M. A. (2018). Achievements and prospects of molecular breeding for drought tolerance in soybean [Glycine max (L.) MERR.]. Genetika-Belgrade 50, 1095–1109. doi: 10.2298/GENSR1803095K
Khan, M., Khan, S. U., Muhammad, A., Hu, L. M., Yang, Y., Fan, C. C., et al. (2018). Induced mutation and epigenetics modification in plants for crop improvement by targeting CRISPR/Cas9 technology. J. Cell. Physiol. 233, 4578–4594. doi: 10.1002/jcp.26299
Kofsky, J., Zhang, H. Y., Song, B. H. (2018). The untapped genetic reservoir: the past, current, and future applications of the wild soybean (glycine soja). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00949
Kong, K. K., Xu, M. G., Xu, Z. Y., Lv, W. H., Lv, P. Y., Begum, N., et al. (2023). Dysfunction of GmVPS8a causes compact plant architecture in soybean. Plant Sci. 331, 111677. doi: 10.1016/j.plantsci.2023.111677
Kuzbakova, M., Khassanova, G., Oshergina, I., Ten, E., Jatayev, S., Yerzhebayeva, R., et al. (2022). Height to first pod: a review of genetic and breeding approaches to improve combine harvesting in legume crops. Front. Plant Sci. 13, 948099. doi: 10.3389/fpls.2022.948099
L'Hocine, L., Boye, J. I. (2007). Allergenicity of soybean: new developments in identification of allergenic proteins, cross-reactivities and hypoallergenization technologies. Crit. Rev. Food Sci. Nutr. 47, 127–143. doi: 10.1080/10408390600626487
Le, H., Nguyen, N. H., Ta, D. T., Le, T., Bui, T. P., Le, N. T., et al. (2020). CRISPR/Cas9-mediated knockout of galactinol synthase-encoding genes reduces raffinose family oligosaccharide levels in soybean seeds. Front. Plant Sci. 11, 612942. doi: 10.3389/fpls.2020.612942
Li, X., Cai, Q., Yu, T., Li, S. J., Li, S. A., Li, Y. L., et al. (2023a). ZmG6PDH1 in glucose-6-phosphate dehydrogenase family enhances cold stress tolerance in maize. Front. Plant Sci. 14, 1116237. doi: 10.3389/fpls.2023.1116237
Li, Z. B., Cheng, Q., Gan, Z. R., Hou, Z. H., Zhang, Y. H., Li, Y. L., et al. (2021). Multiplex CRISPR/Cas9-mediated knockout of soybean LNK2 advances flowering time. Crop J. 9, 767–776. doi: 10.1016/j.cj.2020.09.005
Li, J. J., Ding, X. L., Han, S. H., He, T. T., Zhang, H., Yang, L. S., et al. (2016). Differential proteomics analysis to identify proteins and pathways associated with male sterility of soybean using itraq-based strategy. J. Proteomics 138, 72–82. doi: 10.1016/j.jprot.2016.02.017
Li, C., Li, Y. H., Li, Y. F., Lu, H. F., Hong, H. L., Tian, Y., et al. (2020). A domestication-associated gene gmprr3b regulates the circadian clock and flowering time in soybean. Mol. Plant 13, 745–759. doi: 10.1016/j.molp.2020.01.014
Li, J. J., Nadeem, M., Sun, G. L., Wang, X. B., Qiu, L. J. (2019b). Male sterility in soybean: occurrence, molecular basis and utilization. Plant Breed. 138, 659–676. doi: 10.1111/pbr.12751
Li, C., Nguyen, V., Liu, J., Fu, W., Chen, C., Yu, K. F., et al. (2019a). Mutagenesis of seed storage protein genes in soybean using CRISPR/Cas9. BMC Res. Notes 12, 176. doi: 10.1186/s13104-019-4207-2
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D. D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in arabidopsis and nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, X. X., Zhou, H. W., Cheng, L., Ma, N. N., Cui, B. F., Wang, W. F., et al. (2022). Shoot-to-root translocated GmNN1/FT2a triggers nodulation and regulates soybean nitrogen nutrition. PloS Biol. 20 (8), e3001739. doi: 10.1371/journal.pbio.3001739
Li, H. B., Zhou, R. N., Liu, P. Y., Yang, M. L., Xin, D. W., Liu, C. Y., et al. (2023b). Design of high-monounsaturated fatty acid soybean seed oil using gmpdcts knockout via a CRISPR/Cas9 system. Plant Biotechnol. J. 21 (7), 1317–1319. doi: 10.1111/pbi.14060
Liang, D., Liu, Y., Li, C., Wen, Q., Xu, J., Geng, L., et al. (2023). CRISPR/Lbcas12a-mediated genome editing in soybean. Eds. Yang, B., Harwood, W., Que., Q. (New York, NY: Springer US), 39–52.
Lin, F., Chhapekar, S. S., Vieira, C. C., Da Silva, M. P., Rojas, A., Lee, D., et al. (2022). Breeding for disease resistance in soybean: a global perspective. Theor. Appl. Genet. 135, 3773–3872. doi: 10.1007/s00122-022-04101-3
Lin, X. Y., Liu, B. H., Weller, J. L., Abe, J., Kong, F. J. (2021). Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994. doi: 10.1111/jipb.13021
Liu, H., Chen, W. D., Li, Y. S., Sun, L., Chai, Y. H., Chen, H. X., et al. (2022). CRISPR/Cas9 technology and its utility for crop improvement. Int. J. Mol. Sci. 23 (18), 10442. doi: 10.3390/ijms231810442
Liu, J. Z., Fang, Y., Pang, H. X. (2016). The current status of the soybean-soybean mosaic virus (SMV) pathosystem. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01906
Liu, T. F., Ji, J., Cheng, Y. Y., Zhang, S. C., Wang, Z. R., Duan, K. X., et al. (2023). CRISPR/Cas9-mediated editing of GmTAP1 confers enhanced resistance to Phytophthora Sojae in soybean. J. Integr. Plant Biol. 65 (7), 1609–1612. doi: 10.1111/jipb.13476
Liu, S. Y., Liu, J. F., Zhang, Y. Z., Jiang, Y. S., Hu, S. W., Shi, A. D., et al. (2022). Cloning of the soybean sHSP26 gene and analysis of its drought resistance. Phyton-Internat J. Exp. Bot. 91, 1465–1482. doi: 10.32604/phyton.2022.018836
Liu, D., Yang, H. M., Zhang, Z. H., Chen, Q., Guo, W. L., Rossi, V., et al. (2023). An elite ?-Gliadin allele improves end-use quality in wheat. New Phytol. 239, 87–101. doi: 10.1111/nph.18722
Ma, J. J., Yang, S. X., Wang, D. M., Tang, K. Q., Feng, X. X., Feng, X. Z., et al. (2020). Genetic mapping of a light-dependent lesion mimic mutant reveals the function of coproporphyrinogen III oxidase homolog in soybean. Front. Plant Sci. 11, 557. doi: 10.3389/fpls.2020.00557
Ma, X. L., Zhu, Q. L., Chen, Y. L., Liu, Y. G. (2016). CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant 9, 961–974. doi: 10.1016/j.molp.2016.04.009
Mammadov, J., Buyyarapu, R., Guttikonda, S. K., Parliament, K., Abdurakhmonov, I. Y., Kumpatla, S. P., et al. (2018). Wild relatives of maize, rice, cotton, and soybean: treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 9, 886. doi: 10.3389/fpls.2018.00886
Mangena, P. (2020). Phytocystatins and their potential application in the development of drought tolerance plants in soybeans (Glycine max L.). Protein Pept. Lett. 27, 135–144. doi: 10.2174/0929866526666191014125453
Medic, J., Atkinson, C., Hurburgh, C. R. (2014). Current knowledge in soybean composition. J. Am. Oil Chem. Soc. 91, 363–384. doi: 10.1007/s11746-013-2407-9
Meng, L. B., Zhang, A. Y., Wang, F., Han, X. G., Wang, D. J., Li, S. M., et al. (2015). Arbuscular mycorrhizal fungi and rhizobium facilitate nitrogen uptake and transfer in soybean/maize intercropping system. Front. Plant Sci. 6, 339. doi: 10.3389/fpls.2015.00339
Molla, K. A., Sretenovic, S., Bansal, K. C., Qi, Y. P. (2021). Precise plant genome editing using base editors and prime editors. Nat. Plants 7, 1166–1187. doi: 10.1038/s41477-021-00991-1
Mu, R. L., Lyu, X., Ji, R. H., Liu, J., Zhao, T., Li, H. Y., et al. (2022). Gmbics modulate low blue light-induced stem elongation in soybean. Front. Plant Sci. 13, 803122. doi: 10.3389/fpls.2022.803122
Nakamori, T. (2023). Research on the deliciousness of processed soybean current state and future prospects of soybean breeding ii. J. Japanese Soc. Food Sci. Technology-Nippon Shokuhin Kagaku Kogaku Kaishi 70, 43–45. doi: 10.3136/nskkk.NSKKK-D-22-00008
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J., Kamoun, S. (2013). Targeted mutagenesis in the model plant nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Nguyen, C. X., Dohnalkova, A., Hancock, C. N., Kirk, K. R., Stacey, G., Stacey, M. G., et al. (2021). Critical role for uricase and xanthine dehydrogenase in soybean nitrogen fixation and nodule development. Plant Genome. 16 (2), e20171. doi: 10.1002/tpg2.20172
Nguyen, C. H., Yan, D. W., Nambara, E. (2023). Persistence of abscisic acid analogs in plants: chemical control of plant growth and physiology. Genes 14 (5), 1078. doi: 10.3390/genes14051078
Niu, F. J., Jiang, Q. Y., Sun, X. J., Hu, Z., Wang, L. X., Zhang, H. (2021). Large DNA fragment deletion in lncrna77580 regulates neighboring gene expression in soybean (Glycine max). Funct. Plant Biol. 48, 1139–1147. doi: 10.1071/FP20400
Niu, Q. K., Shi, Z. W., Zhang, P., Su, S., Jiang, B., Liu, X. W., et al. (2023). ZmMS39 encodes a callose synthase essential for male fertility in maize (Zea mays l.). Crop J. 11, 394–404. doi: 10.1016/j.cj.2022.08.012
Ochola, S., Huang, J., Ali, H., Shu, H. D., Shen, D. Y., Qiu, M., et al. (2020). Editing of an effector gene promoter sequence impacts plant-phytophthora interaction. J. Integr. Plant Biol. 62, 378–392. doi: 10.1111/jipb.12883
Osakabe, Y., Osakabe, K. (2017). Genome Editing to Improve Abiotic Stress Responses in Plants. Prog Mol Biol Transl Sci 149 (2017), 99–109. doi: 10.1016/bs.pmbts.2017.03.007
Paul, B., Montoya, G. (2020). CRISPR/Cas12a: functional overview and applications. Biomed. J. 43, 8–17. doi: 10.1016/j.bj.2019.10.005
Phang, T. H., Shao, G. H., Lam, H. M. (2008). Salt tolerance in soybean. J. Integr. Plant Biol. 50, 1196–1212. doi: 10.1111/j.1744-7909.2008.00760.x
Qi, Q. Y., Hu, B. C., Jiang, W. Y., Wang, Y. X., Yan, J. J., Ma, F. W., et al. (2023). Advances in plant epigenome editing research and its application in plants. Int. J. Mol. Sci. 24 (4), 3442. doi: 10.3390/ijms24043442
Qian, L. L., Jin, H. X., Yang, Q. H., Zhu, L. M., Yu, X. M., Fu, X. J., et al. (2022). A sequence variation in GmBADH2 enhances soybean aroma and is a functional marker for improving soybean flavor. Int. J. Mol. Sci. 23 (8), 4116. doi: 10.3390/ijms23084116
Qu, S., Jiao, Y. L., Abraham, L., Wang, P. W. (2021). Correlation analysis of new soybean [Glycine max (L.) Merr] gene Gm15g117700 with oleic acid. Phyton-Internat J. Exp. Bot. 90, 1177–1192. doi: 10.32604/phyton.2021.015206
Ramesh, S. V., Govindasamy, V., Rajesh, M. K., Sabana, A. A., Praveen, S. (2019). Stress-responsive mirnaome of Glycine max (L.) Merrill: molecular insights and way forward. Planta 249, 1267–1284. doi: 10.1007/s00425-019-03114-5
Ramlal, A., Nautiyal, A., Baweja, P., Mahto, R. K., Mehta, S., Mallikarunja, B. P., et al. (2022). Harnessing heterosis and male sterility in soybean [Glycine max (L.) Merrill]: a critical revisit. Front. Plant Sci. 13, 9817683. doi: 10.3389/fpls.2022.981768
Rangari, S. K., Sudha, M. K., Kaur, H., Uppal, N., Singh, G., Vikal, Y., et al. (2023). DNA-free genome editing for ZmPLA1 gene via targeting immature embryos in tropical maize. GM Crops Food-Biotechnol. Agric. Food 2023, 1–7. doi: 10.1080/21645698.2023.2197303
Rao, Y. C., Yang, X., Pan, C. Y., Wang, C., Wang, K. J. (2022). Advance of clustered regularly interspaced short palindromic repeats-Cas9 system and its application in crop improvement. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.839001
Rasheed, A., Mahmood, A., Maqbool, R., Albaqami, M., Sher, A., Sattar, A., et al. (2022a). Key insights to develop drought-resilient soybean: a review. J. King Saud Univ. Sci. 34, 102089. doi: 10.1016/j.jksus.2022.102089
Rasheed, A., Raza, A., Jie, H. D., Mahmood, A., Ma, Y. S., Zhao, L., et al. (2022b). Molecular tools and their applications in developing salt-tolerant soybean (Glycine max L.) Cultivars. Bioengineering-Basel 9 (10), 495. doi: 10.3390/bioengineering9100495
Salvi, P., Varshney, V., Majee, M. (2022). Raffinose family oligosaccharides (rfos): role in seed vigor and longevity. Biosci. Rep. 42 (10), BSR20220198. doi: 10.1042/BSR20220198
Samanta, M. K., Dey, A., Gayen, S. (2016). CRISPR/Cas9: an advanced tool for editing plant genomes. Transgenic Res. 25, 561–573. doi: 10.1007/s11248-016-9953-5
Sarkhel, S., Roy, A. (2022). Phytic acid and its reduction in pulse matrix: structure-function relationship owing to bioavailability enhancement of micronutrients. J. Food Process Eng. 45 (2022). doi: 10.1111/jfpe.14030
Schneider, H. M., Lor, V. S., Zhang, X., Saengwilai, P., Hanlon, M. T., Klein, S. P., et al. (2023). Transcription factor bHLH121 regulates root cortical aerenchyma formation in maize. Proc. Natl. Acad. Sci. United States America 120, e2075299176. doi: 10.1073/pnas.2219668120
Shan, Q. W., Wang, Y. P., Li, J., Zhang, Y., Chen, K. L., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR/Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Song, J. H., Shin, G., Kim, H. J., Lee, S. B., Moon, J. Y., Jeong, J. C., et al. (2022). Mutation of GmIPK1 gene using CRISPR/Cas9 reduced phytic acid content in soybean seeds. Int. J. Mol. Sci. 23 (18), 10583. doi: 10.3390/ijms231810583
Song, H., Taylor, D. C., Zhang, M. (2023). Bioengineering of soybean oil and its impact on agronomic traits. Int. J. Mol. Sci. 24 (3), 2256. doi: 10.3390/ijms24032256
Staniak, M., Szpunar-Krok, E., Kocira, A. (2023). Responses of soybean to selected abiotic stresses-photoperiod, temperature and water. Agriculture-Basel 13 (1), 146. doi: 10.3390/agriculture13010146
Sugano, S., Hirose, A., Kanazashi, Y., Adachi, K., Hibara, M., Itoh, T., et al. (2020). Simultaneous induction of mutant alleles of two allergenic genes in soybean by using site-directed mutagenesis. BMC Plant Biol. 20 (1), 513. doi: 10.1186/s12870-020-02708-6
Sun, J., Bie, X. M., Chu, X. L., Wang, N., Zhang, X. S., Gao, X. Q., et al. (2023). Genome-edited TaTFL1-5 mutation decreases tiller and spikelet numbers in common wheat. Front. Plant Sci. 14, 1142779. doi: 10.3389/fpls.2023.1142779
Tian, Y. H., Zhou, Y., Gao, G. J., Zhang, Q. L., Li, Y. H., Lou, G. M., et al. (2023). Creation of two-line fragrant glutinous hybrid rice by editing the wx and OsBADH2 genes via the CRISPR/Cas9 system. Int. J. Mol. Sci. 24 (1), 849. doi: 10.3390/ijms24010849
Tran, L., Mochida, K. (2010). Functional genomics of soybean for improvement of productivity in adverse conditions. Funct. Integr. Genomics 10, 447–462. doi: 10.1007/s10142-010-0178-z
Wada, N., Ueta, R., Osakabe, Y., Osakabe, K. (2020). Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 20 (1), 234. doi: 10.1186/s12870-020-02385-5
Wan, Z., Liu, Y. X., Guo, D. D., Fan, R., Liu, Y., Xu, K., et al. (2022). CRISPR/Cas9-mediated targeted mutation of the E1 decreases photoperiod sensitivity, alters stem growth habits, and decreases branch number in soybean. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1066820
Wang, J., Kuang, H. Q., Zhang, Z. H., Yang, Y. Q., Yan, L., Zhang, M. C., et al. (2020). Generation of seed lipoxygenase-free soybean using CRISPR/Cas9. Crop J. 8, 432–439. doi: 10.1016/j.cj.2019.08.008
Wang, L. W., Sun, S., Wu, T. T., Liu, L. P., Sun, X. G., Cai, Y. P., et al. (2020). Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 18, 1869–1881. doi: 10.1111/pbi.13346
Wang, T. Y., Xun, H. W., Wang, W., Ding, X. Y., Tian, H. A., Hussain, S., et al. (2021). Mutation of gmaitr genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front. Plant Sci. 12, 779598. doi: 10.3389/fpls.2021.779598
Weller, J. L., Ortega, R. (2015). Genetic control of flowering time in legumes. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00207
Widyasari, K., Alazem, M., Kim, K. H. (2020). Soybean resistance to soybean mosaic virus. Plants-Basel 9 (2), 219. doi: 10.3390/plants9020219
Wu, Y., Xiao, N., Cai, Y., Yang, Q., Yu, L., Chen, Z., et al. (2023). CRISPR/Cas9-mediated editing of the oshppd 3' UTR confers enhanced resistance to hppd-inhibiting herbicides in rice. Plant Commun. 2023, 100605. doi: 10.1016/j.xplc.2023.100605
Xia, Z. J., Zhai, H., Wu, H. Y., Xu, K., Watanabe, S., Harada, K. (2021). The synchronized efforts to decipher the molecular basis for soybean maturity loci E1, E2, and E3 that regulate flowering and maturity. Front. Plant Sci. 12, 632754. doi: 10.3389/fpls.2021.632754
Xiao, Y. T., Karikari, B., Wang, L., Chang, F. G., Zhao, T. J. (2021). Structure characterization and potential role of soybean phospholipases a multigene family in response to multiple abiotic stress uncovered by CRISPR/Cas9 technology. Environ. Exp. Bot. 188, 104521. doi: 10.1016/j.envexpbot.2021.104521
Xu, R. F., Li, J., Liu, X. S., Shan, T. F., Qin, R. Y., Wei, P. C. (2020). Development of plant prime-editing systems for precise genome editing. Plant Commun. 1 (3), 100043. doi: 10.1016/j.xplc.2020.100043
Xu, H. Y., Li, Y. J., Zhang, K. F., Li, M. J., Fu, S. Y., Tian, Y. Z., et al. (2021). miR169c-NFYA-C-ENOD40 modulates nitrogen inhibitory effects in soybean nodulation. New Phytol. 229, 3377–3392. doi: 10.1111/nph.17115
Xu, H., Zhang, L. X., Zhang, K., Ran, Y. D. (2020). Progresses, challenges, and prospects of genome editing in soybean (Glycine max). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.571138
Yang, Z. W., Du, H., Xing, X. Z., Li, W. L., Kong, Y. B., Li, X. H., et al. (2022). A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J. 20, 103–115. doi: 10.1111/pbi.13698
Yang, C. F., Huang, Y. Z., Lv, P. Y., Antwi-Boasiako, A., Begum, N., Zhao, T. J., et al. (2022). Nac transcription factor GmNAC12 improved drought stress tolerance in soybean. Int. J. Mol. Sci. 23 (19), 12029. doi: 10.3390/ijms231912029
Yang, Y., Speth, B. D., Boonyoo, N., Baumert, E., Atkinson, T. R., Palmer, R. G., et al. (2014). Molecular mapping of three male-sterile, female-fertile mutants and generation of a comprehensive map of all known male sterility genes in soybean. Genome 57, 155–160. doi: 10.1139/gen-2014-0018
Yang, L., Tang, J., Ma, X. L., Lin, Y., Ma, G. R., Shan, M. H., et al. (2021). Progression and application of CRISPR/Cas genomic editors. Methods 194, 65–74. doi: 10.1016/j.ymeth.2021.03.013
Yin, K. Q., Gao, C. X., Qiu, J. L. (2017). Progress and prospects in plant genome editing. Nat. Plants 3, 17107. doi: 10.1038/nplants.2017.107
Yin, J. L., Wang, L. Q., Jin, T. T., Nie, Y., Liu, H., Qiu, Y. L., et al. (2021). A cell wall-localized nlr confers resistance to soybean mosaic virus by recognizing viral- encoded cylindrical inclusion protein. Mol. Plant 14, 1881–1900. doi: 10.1016/j.molp.2021.07.013
Yu, T. F., Liu, Y., Fu, J. D., Ma, J., Fang, Z. W., Chen, J., et al. (2021). The nf-y-pyr module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 19, 2589–2605. doi: 10.1111/pbi.13684
Yu, H. X., Xiao, A. F., Wu, J. S., Li, H. X., Duan, Y., Chen, Q. S., et al. (2023). GmNAC039 and GmNAC018 activate the expression of cysteine protease genes to promote soybean nodule senescence. Plant Cell. 35 (8), 2929–2951. doi: 10.1093/plcell/koad129
Yu, G. L., Zou, J. N., Wang, J. H., Zhu, R. S., Qi, Z. M., Jiang, H. W., et al. (2022). A soybean nac homolog contributes to resistance to Phytophthora Sojae mediated by dirigent proteins. Crop J. 10, 332–341. doi: 10.1016/j.cj.2021.08.009
Zaaboul, F., Zhao, Q. L., Xu, Y. J., Liu, Y. F. (2022). Soybean oil bodies: a review on composition, properties, food applications, and future research aspects. Food Hydrocolloids 124, 107296. doi: 10.1016/j.foodhyd.2021.107296
Zhai, H., Wan, Z., Jiao, S., Zhou, J. W., Xu, K., Nan, H. Y., et al. (2022). Gmmde genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 189, 1021–1036. doi: 10.1093/plphys/kiac092
Zhang, G. Y., Bahn, S. C., Wang, G. L., Zhang, Y. R., Chen, B. B., Zhang, Y. L., et al. (2019). PLDα1-knockdown soybean seeds display higher unsaturated glycerolipid contents and seed vigor in high temperature and humidity environments. Biotechnol. Biofuels 12, 9. doi: 10.1186/s13068-018-1340-4
Zhang, Y. F., Blahut-Beatty, L., Zheng, S. Q., Clough, S. J., Simmonds, D. H. (2023). The role of a soybean 14-3-3 gene (Glyma05g29080) on white mold resistance and nodulation investigations using CRISPR/Cas9 editing and rna silencing. Mol. Plant-Microbe Interact. 36, 159–164. doi: 10.1094/MPMI-07-22-0157-R
Zhang, M. H., Cao, J. F., Zhang, T. X., Xu, T., Yang, L. Y., Li, X. Y., et al. (2022). A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.870695
Zhang, P. P., Du, H. Y., Wang, J., Pu, Y. X., Yang, C. Y., Yan, R. J., et al. (2020). Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 18, 1384–1395. doi: 10.1111/pbi.13302
Zhang, Y. X., Guo, W., Chen, L. M., Shen, X. J., Yang, H. L., Fang, Y. S., et al. (2022). CRISPR/Cas9-mediated targeted mutagenesis of GmUGT enhanced soybean resistance against leaf-chewing insects through flavonoids biosynthesis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.802716
Zhang, Y. Z., Li, G. L., Hu, S. W., Liu, J. F., Jiang, Y. S., Liu, S. Y., et al. (2022). Cloning and drought resistance analysis of soybean GmHsps_p23-like gene. Phyton-Internat J. Exp. Bot. 91, 1183–1198. doi: 10.32604/phyton.2022.018853
Zhang, A., Li, Y., Wang, L., Wang, J., Liu, Y., Luan, X., et al. (2023). Analysis of LncRNA44234-associated cerna network reveals oil metabolism in soybean. J. Agric. Food Chem. 71 (25), 9815–9825. doi: 10.1021/acs.jafc.3c00993
Zhang, L. X., Liu, W., Tsegaw, M., Xu, X., Qi, Y. P., Sapey, E., et al. (2020). Principles and practices of the photo-thermal adaptability improvement in soybean. J. Integr. Agric. 19, 295–310. doi: 10.1016/S2095-3119(19)62850-9
Zhang, M., Liu, S. L., Wang, Z., Yuan, Y. Q., Zhang, Z. F., Liang, Q. J., et al. (2022). Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 20, 256–282. doi: 10.1111/pbi.13682
Zhang, Y. L., Ma, X. L., Xie, X. R., Liu, Y. G. (2017). CRISPR/Cas9-based genome editing in plants. Prog. Mol. Biol. Transl. Sci. 149, 133–150. doi: 10.1016/bs.pmbts.2017.03.008
Zhang, Y., Showalter, A. M. (2020). CRISPR/Cas9 genome editing technology: a valuable tool for understanding plant cell wall biosynthesis and function. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.589517
Zhang, Z. H., Wang, W. P., Ali, S., Luo, X., Xie, L. A. (2022). CRISPR/Cas9-mediated multiple knockouts in abscisic acid receptor genes reduced the sensitivity to aba during soybean seed germination. Int. J. Mol. Sci. 23 (24), 16173. doi: 10.3390/ijms232416173
Zhang, Z., Wang, J., Kuang, H., Hou, Z., Gong, P., Bai, M., et al. (2022). Elimination of an unfavorable allele conferring pod shattering in an elite soybean cultivar by CRISPR/Cas9. Abiotech 3, 110–114. doi: 10.1007/s42994-022-00071-8
Zhang, M., Wei, H. L., Hao, P. B., Wu, A. M., Ma, Q., Zhang, J. J., et al. (2021). GhGPAT12/25 are essential for the formation of anther cuticle and pollen exine in cotton (Gossypium hirsutum L.). Front. Plant Sci. 12, 667739. doi: 10.3389/fpls.2021.667739
Zhang, J., Wu, P., Li, N., Xu, X. L., Wang, S. X., Chang, S. Y., et al. (2023). A male-sterile mutant with necrosis-like dark spots on anthers was generated in cotton. Front. Plant Sci. 13, 1102196. doi: 10.3389/fpls.2022.1102196
Zhang, R. Z., Zhang, S. J., Li, J. H., Gao, J., Song, G. Q., Li, W., et al. (2023). CRISPR/Cas9-targeted mutagenesis of TaDCL4, TaDCL5 and TaRDR6 induces male sterility in common wheat. Plant Biotechnol. J. 21, 839–853. doi: 10.1111/pbi.14000
Zhao, R. H., Kang, I. J., Lee, S. W. (2023). Current status and future prospects in genomic research and breeding for resistance to xanthomonas citri pv. Glycines in soybean. Agronomy-Basel 13 (2), 490. doi: 10.3390/agronomy13020490
Zhao, F., Lyu, X., Ji, R. H., Liu, J., Zhao, T., Li, H. Y., et al. (2022). CRISPR/Cas9-engineered mutation to identify the roles of phytochromes in regulating photomorphogenesis and flowering time in soybean. Crop J. 10, 1654–1664. doi: 10.1016/j.cj.2022.03.008
Zheng, Y. Z., Li, Q. Q., Ye, M. W., Chen, A., Wang, H. Y. (2021). Applications of CRISPR/Cas9-based genome editing in the plant biology. Turkish J. Bot. 45, 253–268. doi: 10.3906/bot-2103-50
Zhong, X. B., Hong, W., Shu, Y., Li, J. F., Liu, L. L., Chen, X. Y., et al. (2022). CRISPR/Cas9 mediated gene-editing of GmHDZ4 transcription factor enhances drought tolerance in soybean (Glycine max [L.] Merr.). Front. Plant Sci. 13, 988505. doi: 10.3389/fpls.2022.988505
Zhou, J. M., Li, Z. Y., Li, Y., Zhao, Q. Z., Luan, X. C., Wang, L. X., et al. (2023a). Effects of different gene editing modes of CRISPR/Cas9 on soybean fatty acid anabolic metabolism based on GmFAD2 family. Int. J. Mol. Sci. 24 (5), 4769. doi: 10.3390/ijms24054769
Zhou, Y. G., Liu, W. C., Li, X. W., Sun, D. Q., Xu, K. H., Feng, C., et al. (2020). Integration of srna, degradome, transcriptome analysis and functional investigation reveals gma-miR398c negatively regulates drought tolerance via GmCSDs and GmCCS in transgenic arabidopsis and soybean. BMC Plant Biol. 20 (1), 190. doi: 10.1186/s12870-020-02370-y
Zhou, J. M., Luan, X. C., Liu, Y. X., Wang, L. X., Wang, J. X., Yang, S. N., et al. (2023b). Strategies and methods for improving the efficiency of CRISPR/Cas9 gene editing in plant molecular breeding. Plants-Basel 12 (7), 1478. doi: 10.3390/plants12071478
Keywords: CRISPR/Cas9, soybean, molecular breeding, gene editing, application
Citation: Yao D, Zhou J, Zhang A, Wang J, Liu Y, Wang L, Pi W, Li Z, Yue W, Cai J, Liu H, Hao W and Qu X (2023) Advances in CRISPR/Cas9-based research related to soybean [Glycine max (Linn.) Merr] molecular breeding. Front. Plant Sci. 14:1247707. doi: 10.3389/fpls.2023.1247707
Received: 26 June 2023; Accepted: 28 July 2023;
Published: 30 August 2023.
Edited by:
Tong Zhang, South China Agricultural University, ChinaReviewed by:
Yi Xu, Nanjing Agricultural University, ChinaCopyright © 2023 Yao, Zhou, Zhang, Wang, Liu, Wang, Pi, Li, Yue, Cai, Liu, Hao and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangchun Qu, amxzbmt5cXhjQDEyNi5jb20=; Wenyuan Hao, d2VueXVhbl9oQDE2My5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.