- 1Institute of Cotton Research of Chinese Academy of Agricultural Sciences/Research Base, Anyang Institute of Technology, National Key Laboratory of Cotton Bio-breeding and Integrated Utilization, Anyang, Henan, China
- 2Engineering Research Centre of Cotton, Ministry of Education/College of Agriculture, Xinjiang Agricultural University, Urumqi, China
- 3Hunan Institute of Cotton Science, Changde, Hunan, China
A-galactosidases (AGALs), the oligosaccharide (RFO) catabolic genes of the raffinose family, play crucial roles in plant growth and development and in adversity stress. They can break down the non-reducing terminal galactose residues of glycolipids and sugar chains. In this study, the whole genome of AGALs was analyzed. Bioinformatics analysis was conducted to analyze members of the AGAL family in Gossypium hirsutum, Gossypium arboreum, Gossypium barbadense, and Gossypium raimondii. Meanwhile, RT-qPCR was carried out to analyze the expression patterns of AGAL family members in different tissues of terrestrial cotton. It was found that a series of environmental factors stimulated the expression of the GhAGAL3 gene. The function of GhAGAL3 was verified through virus-induced gene silencing (VIGS). As a result, GhAGAL3 gene silencing resulted in milder wilting of seedlings than the controls, and a significant increase in the raffinose content in cotton, indicating that GhAGAL3 responded to NaCl stress. The increase in raffinose content improved the tolerance of cotton. Findings in this study lay an important foundation for further research on the role of the GhAGAL3 gene family in the molecular mechanism of abiotic stress resistance in cotton.
1 Introduction
Plants are subjected to a variety of abiotic stresses in their growth process, including drought, salt, high temperature, and cold stresses. To enable the normal growth of plants under various stress conditions, plants have developed multiple signaling and regulatory pathways to withstand stress and thrive. Raffinose family oligosaccharides (RFOs), the essential substances mediating stress response in plants, play an important role in enhancing plant resistance to abiotic stresses (Li et al., 2011). Studies have reported that the expression of genes related to RFO metabolites is significantly upregulated under stress conditions such as low temperature, drought, and high salt. These findings suggest that RFOs may have a crucial effect on enhancing resistance to abiotic stresses (Joshi et al., 2021; Lee et al., 2021). In addition, studies have reported that RFOs can facilitate the elimination of reactive oxygen species (ROS) in plants experiencing stress (Ma et al., 2021). Raffinose can be transported to the chloroplast, protects the thylakoid, and maintains the stability of the photosynthetic system, PSII (Schneider and Keller, 2009). As ROS scavengers, both raffinose and galactose can trap hydroxyl radicals and reduce oxidative damage in plants under stressful conditions (Nishizawa-Yokoi et al., 2008). AGAL, the gene that breaks down RFOs in plants, has received little attention, but it is just as important as the gene that synthesizes RFOs.
AGAL is a ubiquitous enzyme in the plant kingdom (Wang et al., 2022). AGALs of Family 26 are of eukaryotic origin, whereas the AGALs of Family 37 is mainly of prokaryotic origin (Fialho Lda et al., 2008). A notable function of this enzyme is in the germination of seeds and tubers (Hughes et al., 2016). AGAL exhibits a high activity during seed maturation, during germination, and in the seedling stage, which can be attributed to hydration factors that result in the complete degradation of soluble sugars in the embryo axis and primarily in the cotyledons (Mittal and Sharma, 1991). AGAL is involved in numerous aspects of plant metabolism, including the hydrolysis of α-1,6 chains of cotton seed oligosaccharides during deforming. Downregulation of α-Gal gene expression in Petunia spp. results in increased whole plant frost resistance in non-domesticated and cold-domesticated plants. In contrast, overexpression of the α-Gal gene leads to the reduced endogenous raffinose and decreased frost resistance (Pennycooke et al., 2003). Osh69, a rice alkaline galactosidase, was found to be located in chloroplasts and to play a role in leaf senescence. It is upregulated in darkness and in response to injury (Lee et al., 2004). Meanwhile, the AGA3 protein was identified in cucumber chloroplasts and its expression was downregulated under cold stress. However, it was upregulated after temperature recovery. This strongly suggests that AGA3 plays an important role in the catabolism of chloroplast RFOs under cold stress. Notably, the alkaline environment of the chloroplast stroma is suitable for AGA3 to exert its catalytic action. AGA2, another alkaline galactosidase, is responsible for the catabolism of RFOs in the cytoplasm once the temperature returns to the normal level. RFOs accumulate in different subcellular compartments of cucumber leaves under cold stress, whereas upon the removal of the stress, they can be broken down in situ by various galactosidases (Sui et al., 2012; Gu et al., 2018). In the research on spinach in New Zealand, the expression of the TtAGAl1 gene was affected by various abiotic stresses. Among them, drought was found to be a particularly strong promoter of TtAGAl expression (Hara et al., 2008). Moreover, the expression of related genes in Vitis vinifera was studied under salt and drought stresses as well as and during seed development. It was found that the Vv-a-gal/SIP gene is differentially expressed under different osmotic stress conditions. Additionally, Vv-α-gal/SIP-specific transcripts were preferentially accumulated in salt-tolerant Vitis vinifera varieties (Daldoul et al., 2012). ZmAGA1 expression was enhanced in maize seedlings under cold and drought stress conditions, but not upon sodium chloride stress (Zhao et al., 2006). AGAL is an essential enzyme for plant growth and development, which plays a crucial role in plant development Although characterized AGAL has been studied in several species, it has not been reported in cotton. It is crucial to delve into the mechanisms underlying the salinity stress resistance in cotton.
Cotton is an economically important crop that is grown worldwide for the production of fiber and cottonseed oil. However, stress conditions often affect the growth and development of cotton, thereby reducing its yield. Different abiotic stresses may have diverse effects on cotton (Bawa et al., 2022). With the rapid development of sequencing technology, the genome sequences of Gossypium hirsutum, Gossypium arboreum, Gossypium barbadense, and Gossypium raimondii have been sequenced and resolved. In this study, the AGAL family of cotton was identified and characterized by bioinformatics analyses (Huang et al., 2022).The expression pattern of the GhAGALs gene was also analyzed, and the results showed that GhAGALs expression varied in different tissues. In addition, the AGAL gene plays a role in the function of Gossypium hirsutum and its specific site of action. Findings in this study provide a theoretical foundation for future studies on the utilization of the AGAL family in Gossypium hirsutum.
2 Experimental materials and methods
2.1 Identification of AGAL family members and construction of evolutionary tree
The protein sequences of four major cotton species Gossypium hirsutum, Gossypium arboreum, Gossypium barbadense, and Gossypium raimondii were downloaded from the cotton database Cotton FGD (https://cottonfgd.org), Gossypium hirsutum (ZJU), Gossypium arboreum (CRI), Gossypium barbadense (ZJU), Gossypium raimondii (JGI), the genome sequence, and the CDS sequence (Zhu et al., 2017). Additionally, protein sequences, genomes, and other biological information of five diploid species were downloaded using the online database JGI Phytozome v12.1. Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Vitis vinifera, and Zea mays (Wang et al., 2021). Furthermore, the hidden Markov model (HMM) file for the conserved domain (PF16499) was downloaded from the Pfam database (https://pfam.xfam.org/). Hidden Markov models were used with (PF16499) as the query file to search for candidate AGAL family genes in the genomes of cotton and other species. This search was conducted using HMMER (version 3.3.1) (http://www.hmmer.org/) and BLASTP. Manual removal of redundant sequences from HMMER (version 3.3.1) and BLASTP results for incomplete genes (Henderson et al., 1997). These genes were identified as members of the AGAL gene family. The identified AGAL family members were renamed GhAGAL1-GhAGAL15 based on their chromosomal positions (Zhang et al., 2021).
To clearly understand the evolutionary relationship of the AGAL gene, those identified AGAL gene family members were used to search for protein sequences of cotton and five diploid species: Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Vitis vinifera, and Zea mays. We employed using HMMER (version 3.3.1) and BLASTP for this purpose. Multiple sequence alignment was performed using MEGA 7.0 software (Cui et al., 2021b). Construction of intraspecific and interspecific evolutionary trees for AGAL using the online software ChiPlot (Xie et al., 2023) online tools to enhance the visualization of evolutionary trees (Kumar et al., 2016).
2.2 Chromosome localization in the AGAL family
Biological information concerning the location and structure of AGAL family members was extracted from the genome gff3 annotation files of four cotton species (Chen et al., 2018). Afterward, locations of the cotton AGAL family genes on the chromosome were analyzed and mapped with TBtools software (Chen et al., 2020; Cai et al., 2021), so as to reveal the coevolutionary relationships between the AGAL families of cotton species.
2.3 Gene structure and conserved motif analysis
Phylogenetic tree, gene structure, and conserved protein motif were triply mapped with MAST files, GhAGALs evolutionary tree NWK files, and gff3 files by adopting TBtools software.
2.4 Analysis of GhAGALs gene promoter elements and expression level analysis
To investigate the relationships of the AGAL family with hormones and stress, the upstream 2,000-bp sequence (Malik et al., 2020) of the start codon in the AGAL family was selected from the Gossypium hirsutum genome sequence by using the PlantCARE data database query system (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Afterward, cis-acting elements in the upstream regions of genes were identified, analyzed and later visualized with TBtools after screening (Chen et al., 2020).
2.5 Analysis of the tissue expression pattern of GhAGALs family genes
To analyze the expression of GhAGALs in various tissues, GhAGALs data were obtained from the online database of the Cotton Research Institute (http://grand.cricaas.com.cn/page/tools/expressionVisualization). In addition, representative organization values for root, stem, and leaf tissues were selected based on the characteristics of the GhAGALs family. The original data were analyzed to obtain the gene expression levels, which were subsequently visualized using a histogram.
2.6 Gene interaction network
The GhAGAL protein interaction network was analyzed based on the STRING database (https://string-db.org/) after taking Arabidopsis thaliana orthologs into consideration, so as to predict the interactions between GhAGAL family genes and other genes in cotton.
2.7 RNA isolation and quantitative reverse transcription-polymerase chain reaction of GhAGALs family genes
Gossypium hirsutum plants, provided by the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences (Zhong 9807), were grown in an incubator at 25°C.When the cotton grew to three leaves and one heart stage, the plants were subjected to the stress of 100 mM NaCl, and the leaves were collected at 0 h, 6 h, 12 h, and 24 h. RNA was then extracted using the Aidlab kit. cDNA was synthesized through reverse transcription using TransGen Biotech. Clear water-treated plants were used as the control. The procedure was as follows: 94°C for 30 s; 94°C for 5 s, 55°C for 15 s, and 72°C for 10 s for 45 cycles; followed by storage at 4°C (Supplementary Table S1). The relative expression of genes was calculated using the 2−ΔΔCt method, with actin being the internal reference (Chen et al., 2020).
2.8 Construction of the GhAGAL3 recombinant vector and transformation by Agrobacterium tumefaciens
Firstly, we downloaded the CDS sequence of GhAGAL3 from the Cotton FGD database. Then, a 300-bp region was selected from the silent site to design VIGS primers for amplifying the target fragment. The pYL156 silencing vector was constructed by double digesting the pYL156 vector with two restriction enzymes, BamHI and SacI, using the In-Fusion technique. Next, the recombinant vector product was transfected into E. coli, PCR was performed using VIGS primers, and the resulting bands were sent to the testing company for sequencing. After obtaining the correct sequencing results, re-transformation with Agrobacterium tumefaciens was conducted. In brief, the LBA4404 bacteriophage carrying the following constructs, including control pYL156 (empty vector), pYL156: GhAGAL3, pYL156: PDS (positive control), and pYL192 (help vector), was inserted into the cotyledons of Zhong 9807. After 24 h of dark treatment, cotton was grown in an incubator at 25°C/16-h light and 23°C/8-h dark cycle conditions (Fan et al., 2022). When cotton grew to three leaves and one heart stage, lines injected with pYL156 and successfully silenced were immersed in the 100-mM NaCl solution, whereas control lines were immersed in ddH2O. After 36 h, phenotypic differences were observed among the various treatments and individual plants were sampled. Each sample weighed 0.1 g.
2.9 Determination of raffinose, D-galactose, and D-glucose contents
The leaves of control and NaCl-treated cotton seedlings were collected at each time point, immediately frozen in liquid nitrogen, and stored at −80°C to determine the raffinose content. The content of raffinose was detected using a plant raffinose ELISA kit, that of D-galactose content using the D-galactose content kit (Article number: ADS-W-TDX046). D-Glucose content test was performed using the D-glucose content (GOPOD oxidase method) kit (Article number: ADS-W-TDX002).
2.10 Determination of Pro and MDA contents
For determination of proline (Pro) and malondialdehyde (MDA) contents, the leaves of control and NaCl-treated cotton seedlings were collected at each time point and immediately frozen in liquid nitrogen and stored at −80°C to determine the content of Pro and MDA. The content of Pro was determined using the test kit from Nanjing Jiancheng Bioengineering Institute, the content of MDA was determined using the test kit from Beijing Solarbio Science & Technology Co., Ltd. (Liu et al., 2022; Meng et al., 2023).
2.11 Determination of chlorophyll content
Chlorophyll was extracted from 0.1 g of fresh cotyledon leaves by overnight immersion in an 85% (v/v) acetone solution, the supernatant was centrifuged at 4,000 rpm for 10 min and diluted with 85% acetone to the appropriate concentration, and absorbance values were measured by absorbance at 424.5, nm 644 nm, and 663 nm, with 85% acetone serving as a blank (Farooq et al., 2013).
2.12 Leaf DAB staining
First, the DAB working solution was made in accordance with the DAB stain kit’s instructions and kept chilled at 4°C. The leaves were removed from the staining solution and soaked in anhydrous ethanol. The ethanol can be changed several times during the decolorization process to ensure that the green color of the leaves disappears. Cotton samples were submerged in the staining solution and stained overnight in the dark. When the green tint had finally faded completely, ROS distribution and accumulation was seen.
2.13 Statistical analysis
A minimum of three biological replicates was deemed necessary for each dataset. Spass19.0 was used for histogram drawing, and GraphPad Prism 9.0 was used for data significance analysis.
3 Results
3.1 Identification of AGAL family members and construction of the evolutionary trees
To further understand the evolutionary relationships of the AGAL family in cotton, nine different species were identified in this study, namely, Gossypium hirsutum, Gossypium arboreum, Gossypium barbadense, Gossypium raimondii, Oryza sativa, Arabidopsis thaliana, Zea mays, Populus trichocarpa, and Vitis vinifera. According to our identification results, 15 genes were in Gossypium hirsutum, 15 in Gossypium barbadense, 7 in Gossypium raimondii, 7 in Gossypium arboreum, 9 in Oryza sativa, 7 in Vitis vinifera, 18 in Zea mays, 22 in Populus trichocarpa, and 4 in Arabidopsis thaliana. Using MEGA 7.0 software, the amino acid sequences were compared and analyzed by the maximum likelihood method, and a phylogenetic tree was constructed (Figure 1A). The classification of the genes within the species’ family can be clearly observed through the phylogenetic tree. The genes are divided into three subclasses: I, II, and III. Subclass I was further divided into two subfamilies, and subclass II and subclass III were also divided into two subfamilies. The (Figure 1B) shows that the genes of subclass I and subclass III families tend to converge, whereas subclass II had the lowest number of family genes. The closest relationship was seen between cotton and Zea mays. Thereafter, the evolutionary trees were compared, which showed that the number of genes in Gossypium hirsutum and Gossypium barbadense was higher than that in Vitis vinifera, Oryza sativa, and Arabidopsis thaliana (Rui et al., 2022). This suggested that cotton underwent massive amplification during the evolutionary process (Figure 1B).
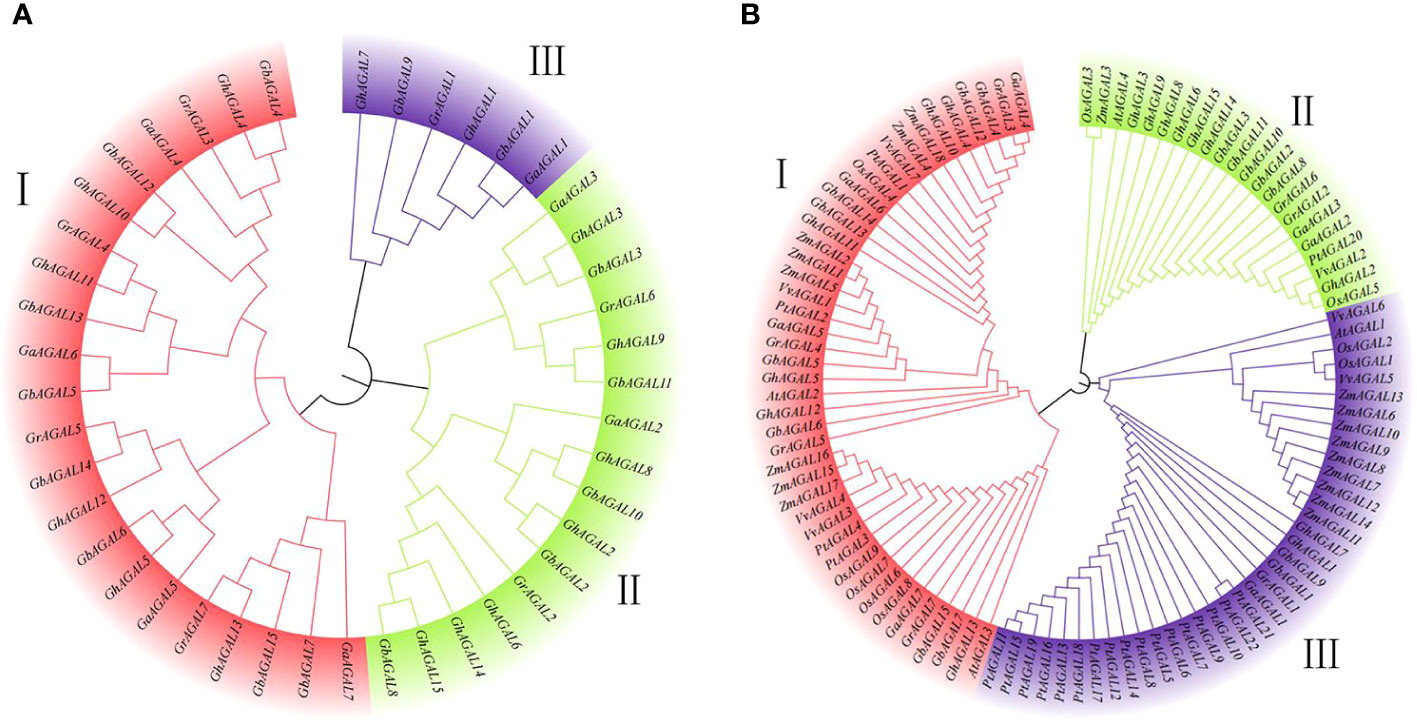
Figure 1 Phylogenetic tree constructed with MEGA7 by the neighbor-joining (NJ) method. (A) Evolutionary analysis of the AGAL family members of four cotton species, (B) Phylogenetic relationships of AGAL genes from four cotton species and five other plant species.
To further analyze the evolutionary relationships among the common ancestors of the four cotton species, their amino acid sequences were compared and analyzed with MEGA 7.0 software. In addition, the neighbor-joining method was applied in constructing the phylogenetic tree of the AGAL family (Figure 1B). As clearly observed from the phylogenetic tree, the species’ family genes were classified into three subclasses I, II, and III. Among the homologous genes of Gossypium hirsutum, the AGAL gene was subjected to stable selection across different cotton species. Meanwhile, there were only a few genes undergoing natural selection, as indicated by the conserved amino acid phenotype of the protein.
3.2 Chromosome distribution of the AGAL family genes in cotton
We further investigated the distribution of AGAL family genes on chromosomes. It was found that the genes were unevenly distributed among certain chromosomes or closely aligned regions (Figure 2). Among them, 14 GhAGALs family genes of cotton were located on 11 chromosomes and distributed on each of these chromosomes. However, GhAGAL15 did not appear in chromosome localization, possibly indicating that it was not located on any specific chromosome. Moreover, the 15 GbAGALs family genes in Gossypium barbadense were also localized on 11 chromosomes, with distribution on each of these chromosomes. The localization of the AGAL family genes on chromosomes was generally similar in both Gossypium barbadense and Gossypium hirsutum, suggesting a certain degree of similarity between these different species. Additionally, all the seven genes of evolutionary Gossypium arboreum and Gossypium raimondii were evenly distributed on each chromosome. Upon further analysis, most of the genes were highly conserved in their location on the chromosomes. For example, the positions of GhAGAL1-GbAGAL1-GaAGAL1-GrAGAL1 and GhAGAL2-GbAGAL2-GaAGAL2-GrAGAL2 on the chromosomes all show similar locations of gene distribution (Supplementary Table S2).
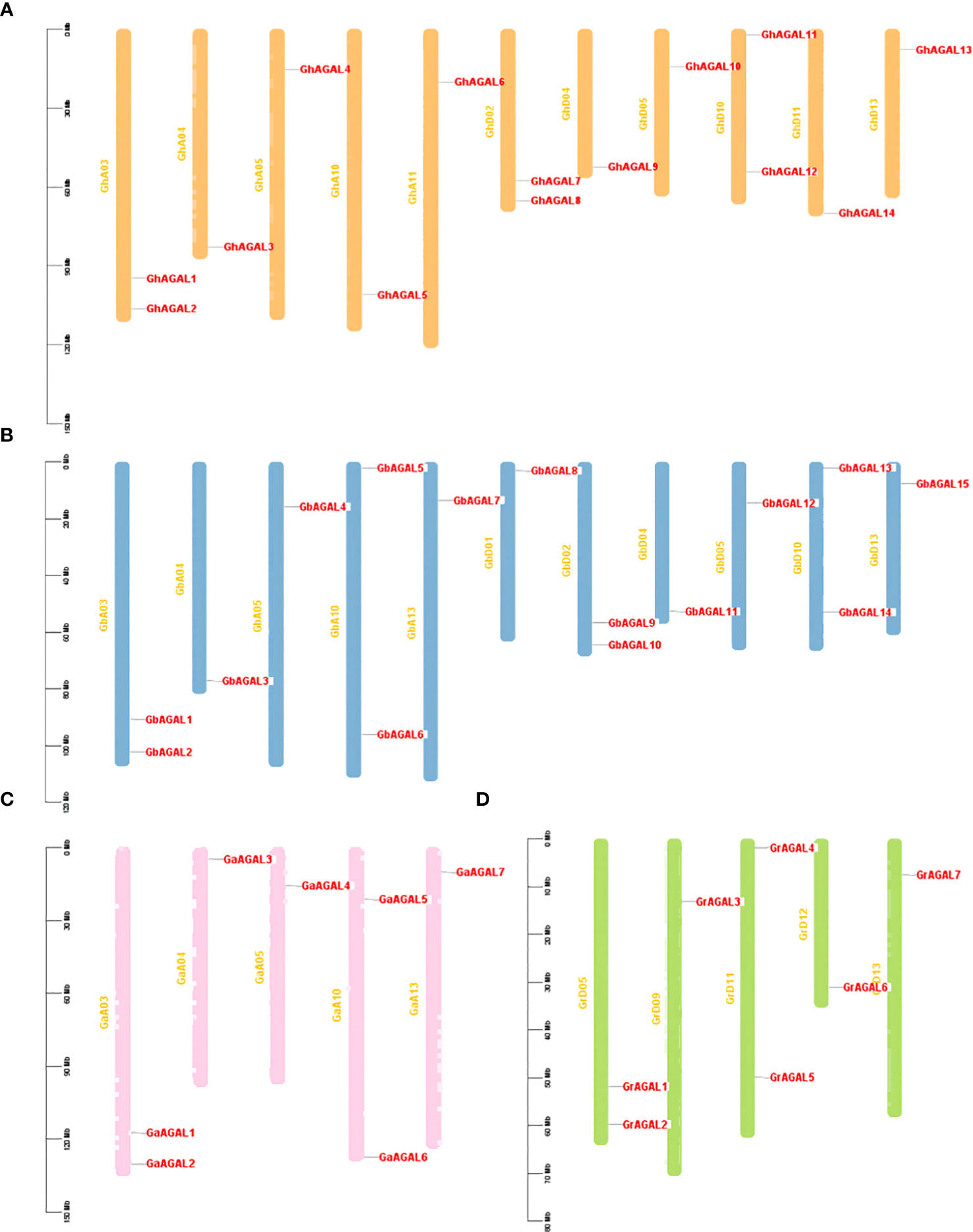
Figure 2 Chromosomal localization of AGAL Chromosome distribution of AGAL genes in four cotton species. (A) The chromosomes of Gossypium hirsutum, (B) The chromosomes of Gossypium barbadense. (C) The chromosomes of Gossypium arboreum. (D) The chromosomes of Gossypium raimondii.
3.3 Conserved protein motifs and gene structure analysis
Triadic joint analysis of four major cotton species was conducted by phylogenetic tree, gene structure, and conserved motifs (Figure 3). From the figure, it was seen that family members on the same evolutionary branch displayed a similar distribution of conserved motif, which suggested that the family members were functionally similar and structurally evolutionarily conserved (Bailey and Elkan, 1994).
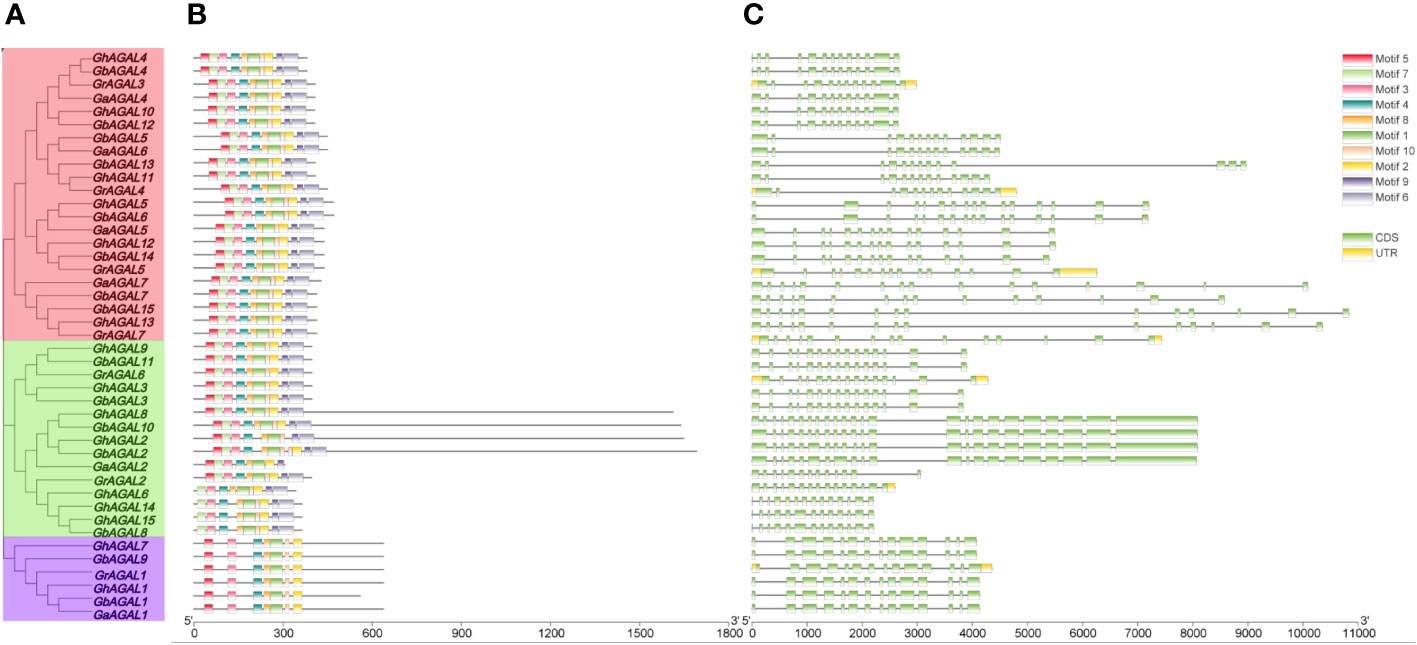
Figure 3 Conserved motifs and gene structure analysis of the GhAGAL family. (A) Phylogenetic tree of the GhAGAL family. (B) Conserved motifs of the GhAGAL family. (C) Gene structure of the GhAGAL family gene structure.
3.4 Analysis of cis-acting elements and expression levels in Gossypium hirsutum
To further investigate the mechanisms of the response of GhAGAL gene family members to abiotic stresses, relevant data were downloaded to analyze the differential expression of genes under cold, heat, salt, and PEG stresses. As observed from Figure 3, the expression of different genes changed to varying degrees under different stresses. Therefore, it was speculated that the GhAGAL gene family was involved in the regulation of abiotic stresses (Figure 3). In addition, members of the GhAGAL gene family also played a crucial role in the physiological and biochemical processes of plants. According to Figure 4, the GhAGAL gene family is involved in various environmental stimuli, including light response, methyl jasmone, low-temperature stress, abscisic acid, gibberellin, defense, and stress response (Hu et al., 2019) (Supplementary Table S3). Therefore, it inferred that the Gossypium hirsutum GhAGALs family primarily consists of phytohormones and cis-acting elements related to adversity. Afterward, the expression of 15 genes in the AGAL family of Gossypium hirsutum was analyzed under different abiotic stresses and the results were visualized in the form of heat maps (Chen et al., 2020). According to the results, under cold stress, GhAGAL4 and GhAGAL11, GhAGAL2, GhAGAL15, GhAGAL6, GhAGAL3, GhAGAL9, GhAGAL13, GhAGAL5, GhAGAL12, and GhAGAL11 were upregulated whereas GhAGAL1, GhAGAL2, GhAGAL3, GhAGAL8, and GhAGAL11 were downregulated. Under heat stress, GhAGAL2, GhAGAL3, GhAGAL5, GhAGAL8, GhAGAL9, GhAGAL12, GhAGAL11, and GhAGAL13 were upregulated. GhAGAL1, GhAGAL5, GhAGAL11, and GhAGAL13 were downregulated. Under PEG stress, the upregulated expressions of GhAGAL8, GhAGAL2, GhAGAL3, GhAGAL9, GhAGAL11, and GhAGAL1 were downregulated. Under NaCl stress, GhAGAL2, GhAGAL3, GhAGAL5, GhAGAL6, GhAGAL9, GhAGAL12, GhAGAL11, and GhAGAL1 were upregulated, whereas GhAGAL1, GhAGAL3, GhAGAL11, GhAGAL12, and GhAGAL13 were downregulated.
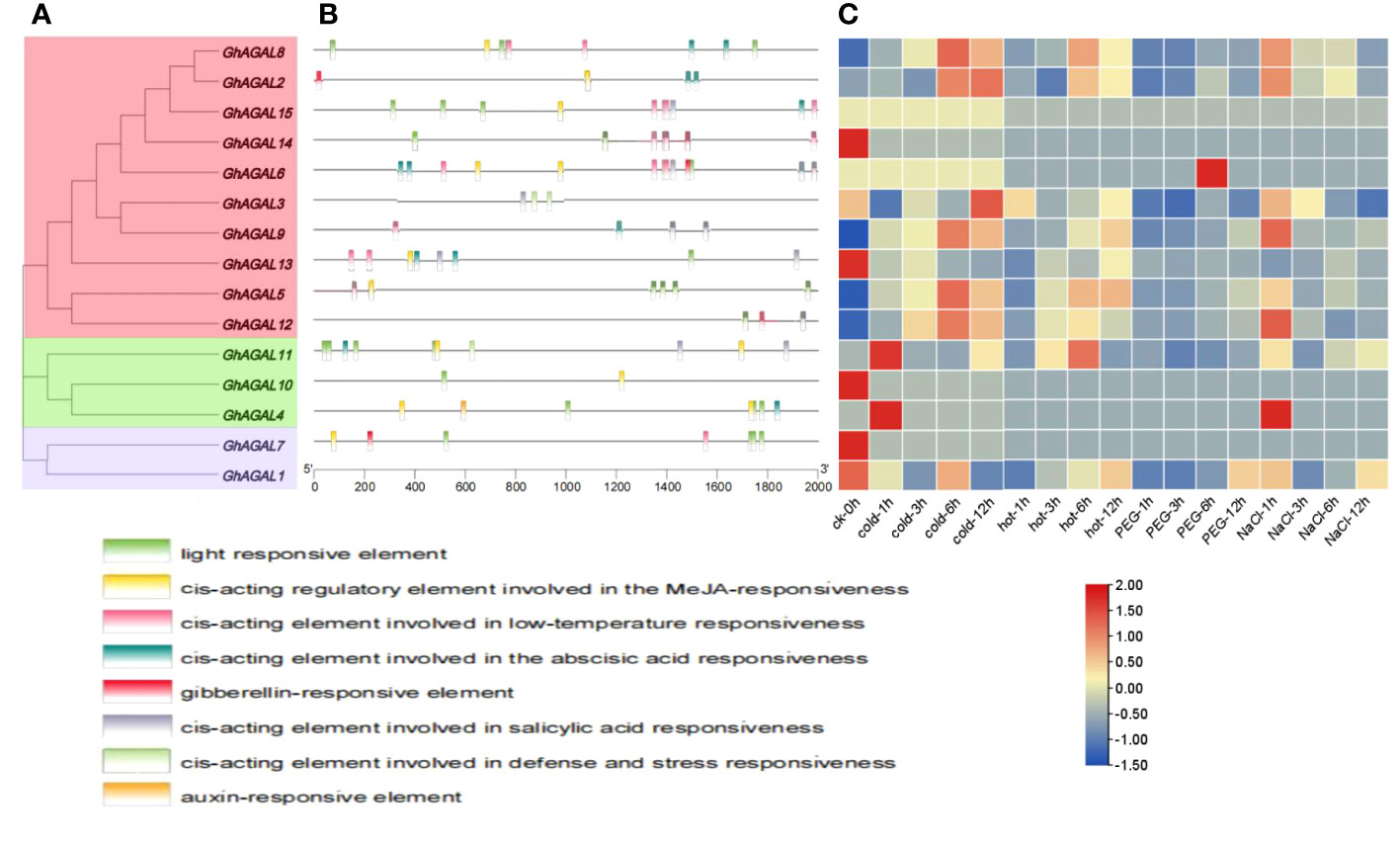
Figure 4 Analysis of cis-acting elements and differential expression patterns of the GhAGAL gene family members. (A) Phylogenetic tree of the GhAGAL gene family. (B) cis-Acting elements in the promoters of the GhAGAL gene family members. (C) Differential expression levels of GhAGAL gene family members in response to cold, heat, salt, and PEG stresses.
3.5 Interaction network of GhAGAL proteins
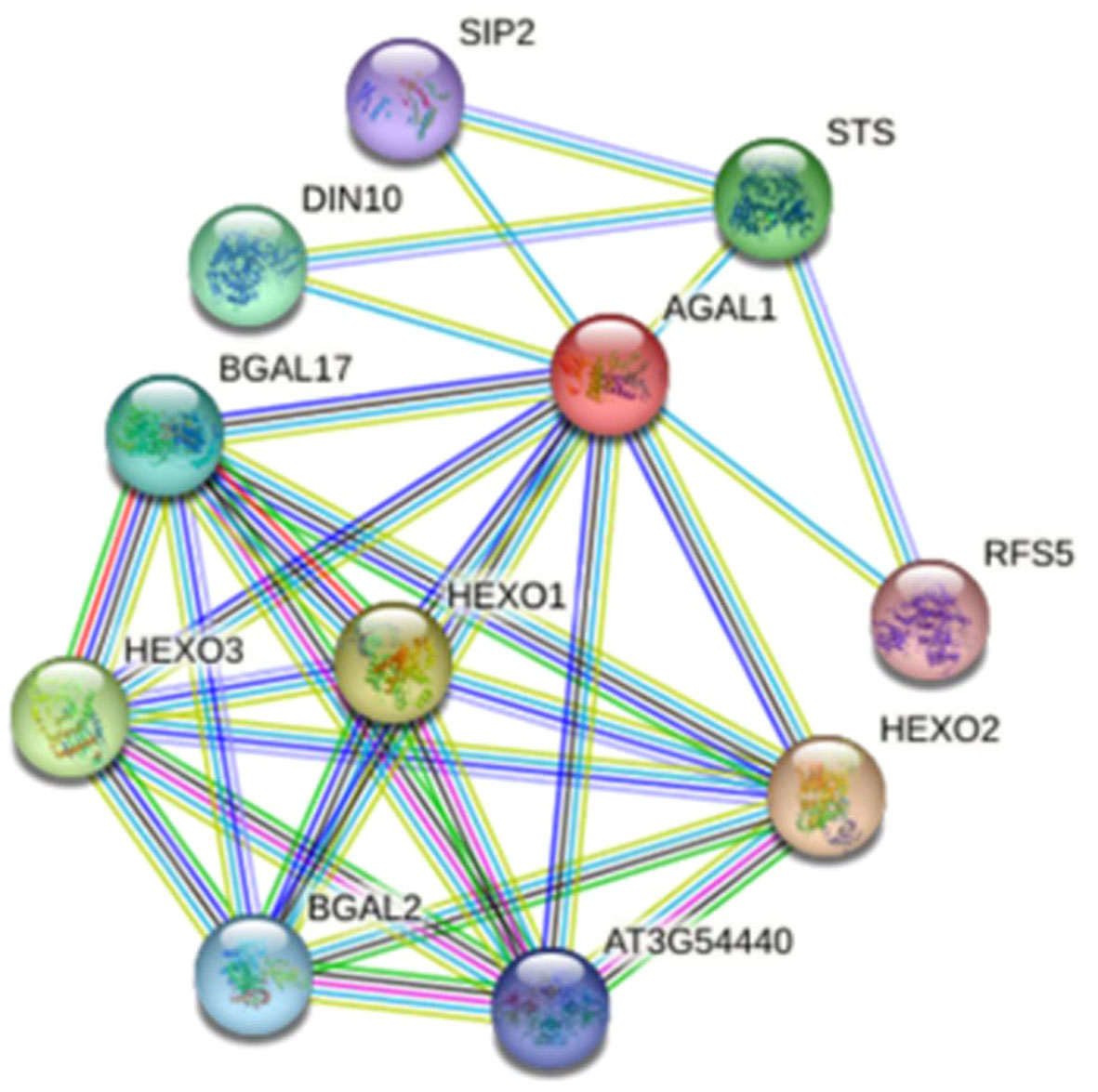
A protein interaction network map of GhAGAL was constructed based on the protein sequence of GhAGAL by adopting the online STRING database (https://string-db.org/) (Yu et al., 2009). In the predicted network, 10 proteins were predicted to interact with GhAGAL, including DIN10, STS, SIP2, RFS5, BGAL17, HEXO1, HEXO3, HEXO2, and BGAL2, (Figure 5). Afterward, the CottonFGD database was searched to identify the related genes that interact with GhAGAL. Among them, RFS5 interacted with GhAGAL and played a crucial role in mitigating salt stress in cotton. RFS5 not only helps scavenge free radicals but also enhances salt tolerance in cotton (Cui et al., 2021a). In this study, GhAGAL was identified as the decomposition gene of RFS5. Consequently, it was speculated that there might be a relationship between GhAGAL and RFS5.
3.6 Tissue-specific expression pattern of the GhAGALs gene and RT-qPCR analysis
To understand the expression of AGAL family genes in Gossypium hirsutum, the expression levels of 15 AGAL genes were analyzed in different tissues (roots, leaves, and stems) of Gossypium hirsutum (Figure 6). As a result, various genes exhibited significant differential expression within the same tissue, meanwhile the same also gene showed significant differential expression across different tissues. The expression pattern maps were created through clustering and analyzing differences in gene expression from a holistic perspective. These maps were divided into three categories, namely, high expression, no expression, and low expression. It was obviously observed from the graph that the relative expressions of GhAGAL3 and GhAGAL9 were the highest in leaves, whereas those of GhAGAL5, GhAGAL11, and GhAGAL12 were the highest. The relative expression levels of GhAGAL1, GhAGAL6, and GhAGAL8 were the highest in roots, where the relative expressions of GhAGAL2, GhAGAL4, GhAGAL14, and GhAGAL15 were expressed only in roots and not in stems and leaves. GhAGAL7 and GhAGAL13 are expressed in stems, but not in roots and leaves.
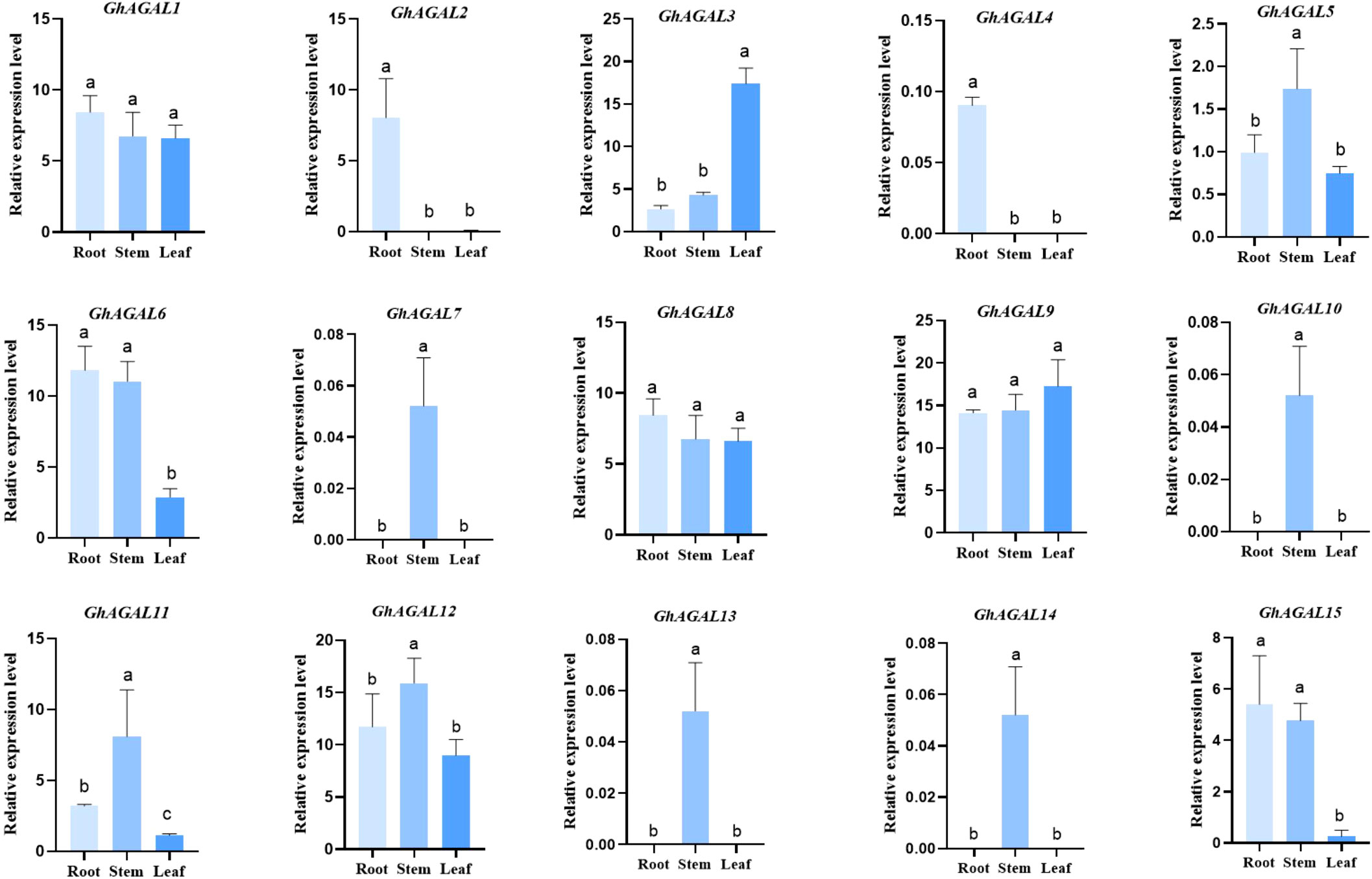
Figure 6 Specific expression of AGAL family genes in roots, stems, and leaves. Signifcance level α;=0.05: The resulting values are expressed in relative units. The error bar in the figure is the standard deviation (SD) of the three biological replicates in each treatment group.
To explore the effects of AGAL family genes on the plant tissue of Gossypium hirsutum. Quantitative reverse transcription-polymerase chain reaction (RT-qPCR) analysis was conducted, and a histogram was created to visualize the expression of 15 AGAL family genes in Gossypium hirsutum leaves under NaCl stress at 0 h (CK), 6 h, 12 h, and 24 h. As clearly observed from Figure 7, the expression level of GhAGAL1 increased at 6 h and 12 h but decreased at 24 h compared with the control (CK). GhAGAL2 expression decreased at 6 h, 12 h, and 24 h relative to the control group (CK), whereas GhAGAL3 expression continued to increase at 0 h, 6 h, 12 h, and 24 h, and GhAGAL4 expression decreased at 6 h, 12 h, and 24 h. The expression of GhAGAL5 increased at 6 h and 12 h but decreased significantly at 24 h. GhAGAL6 expression decreased at 6 h and then increased rapidly at 12 h compared with CK but decreased again at 24 h. GhAGAL7 expression increased at 6 h, 12 h, and 24 h but decreased again at 24 h after increasing at 12 h. GhAGAL8 expression decreased at 24 h compared with CK and increased at 0 h, 6 h, 12 h, and 24 h relative to CK. In addition, the expression of GhAGAL8 decreased compared with the control (CK). GhAGAL12 expression initially increased at 6 h and then slightly decreased at 12 h and later rapidly increased at 24 h compared with the control. GhAGAL13 expression did not show any significant change at 6 h compared with the control but then continued to decrease at 12 h and 24 h. GhAGAL14 expression decreased at 6 h, 12 h, and 24 h relative to the control, whereas GhAGAL15 expression was also decreased at 6 h, 12 h, and 24 h compared with the control. The expression of GhAGAL15 decreased significantly at 6 h, 12 h, and 24 h. It decreased significantly at 6 h compared with CK and then increased significantly at 12 h but decreased again at 24 h. GhAGAL3 expression continued to rise with time under NaCl stress (Figure 7). Therefore, it was screened for functional verification by combining the tissue specificity of the GhAGAL family and conducting RT-qPCR analysis in leaves.
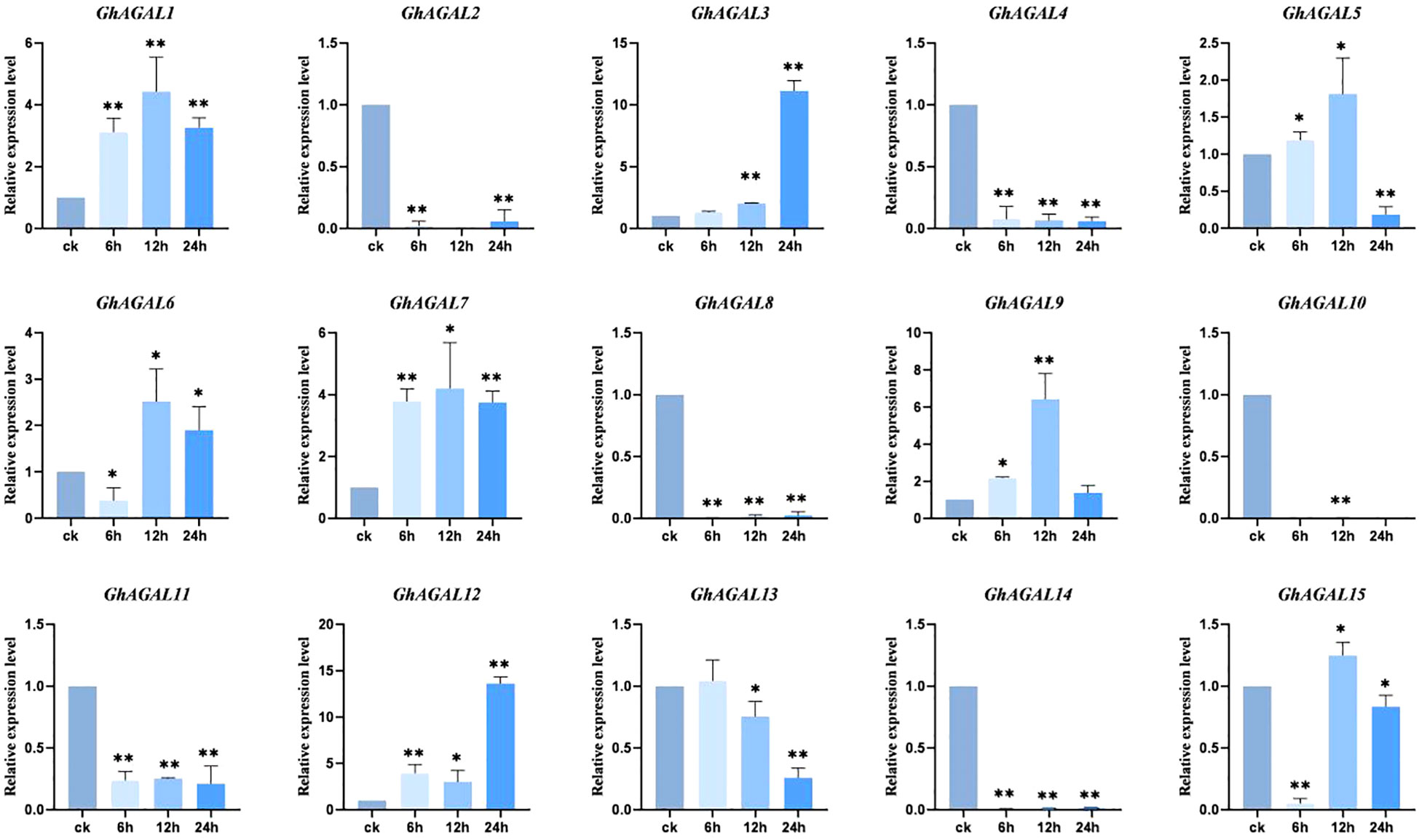
Figure 7 Expression of AGAL family genes in leaves at 0 h, 6 h, 12 h, and 24 h under NaCl stress. *0.01 < p < 0.05, **p < 0.01; the resulting mean values are presented as relative units.
3.7 Phenotype of cotton plants with GhAGAL3 gene silencing by VIGS under NaCl stress
To validate the above result, the genetic virus-mediated gene silencing technique was performed on the GhAGAL3 gene (Figure 8). When the cotton reached the three-leaf stage, PDS plants exhibited albinism. Then, plants injected with the pYL156 carrier and pYL156:GhAGAL3 were transferred to a tripod and further treated with 100 mM NaCl. As a result, the plant phenotype of pYL156 was more wilted than that of pYL156: GhAGAL3 at 36 h. The expression level of the plants was analyzed using RT-qPCR. The results showed that the expression level of GhAGAL3 plants was lower than that of pYL156 carriers, which further indicated the successful gene silencing. At the same time, the MDA content of GhAGAL3 gene-silenced plants decreased slightly after salt stress, whereas the Pro content increased significantly. In contrast, the MDA content of pYL156: GhAGAL3 was higher than that of pYL156 in the case of CK treatment. However, under NaCl stress, there was a slight downward trend in the MDA content of pYL156: GhAGAL3 compared with that of pYL156. This suggests that the degree of membrane lipolysis damage was also reduced, resulting in lesser damage to the plants. As revealed by DAB staining results, the leaves of pYL156 plants under NaCl stress were darker than those of GhAGAL3 plants. Based on the above results, after GhAGAL3 silencing, cotton maintained normal growth by regulating the contents of osmotic substances such as proline. Additionally, cotton scavenged ROS by improving the activities of antioxidant enzymes, thus enabling cotton to resist NaCl stress.
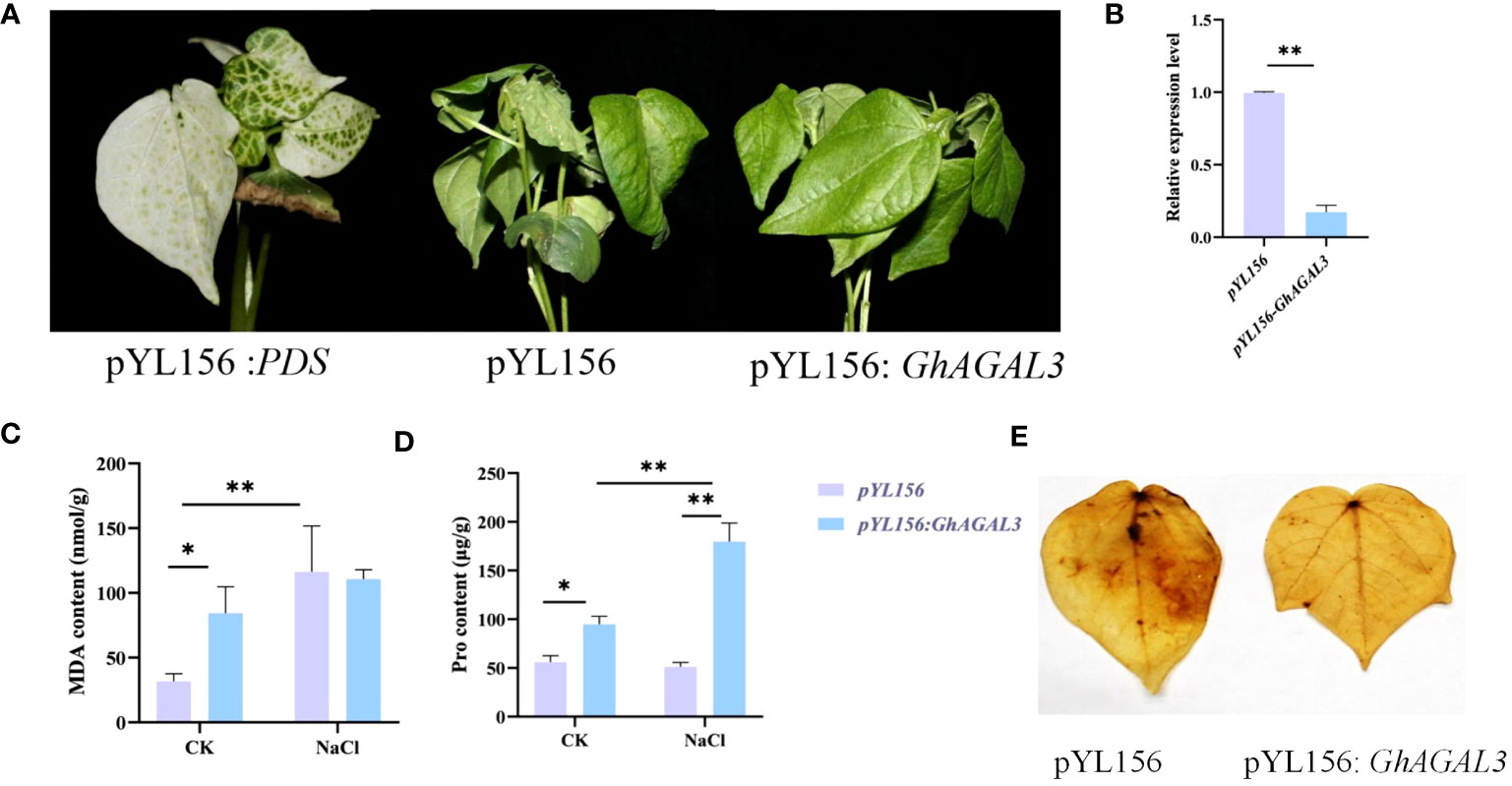
Figure 8 Phenotype of cotton leaves after virus infection and expression of GhAGAL3 under NaCl stress. (A) Phenotype of cotton after GhAGAL3 gene silencing under NaCl stress. pYL 156: PDS as a positive control, pYL156 was an empty vector as control, and pYL156 GhAGAL3 was the GhAGAL3-silenced lines. (B) Relative expression level of GhAGAL3 under NaCl stress. (C) MDA content of empty control and VIGS plants under normal growth and NaCl stress. (D) Pro content of empty control and VIGS plants under normal growth and NaCl stress. (E) DAB staining. *0.01 < p < 0.05, **p < 0.01; the resulting mean values are presented as relative units.
3.8 Changes of raffinose content under NaCl stress
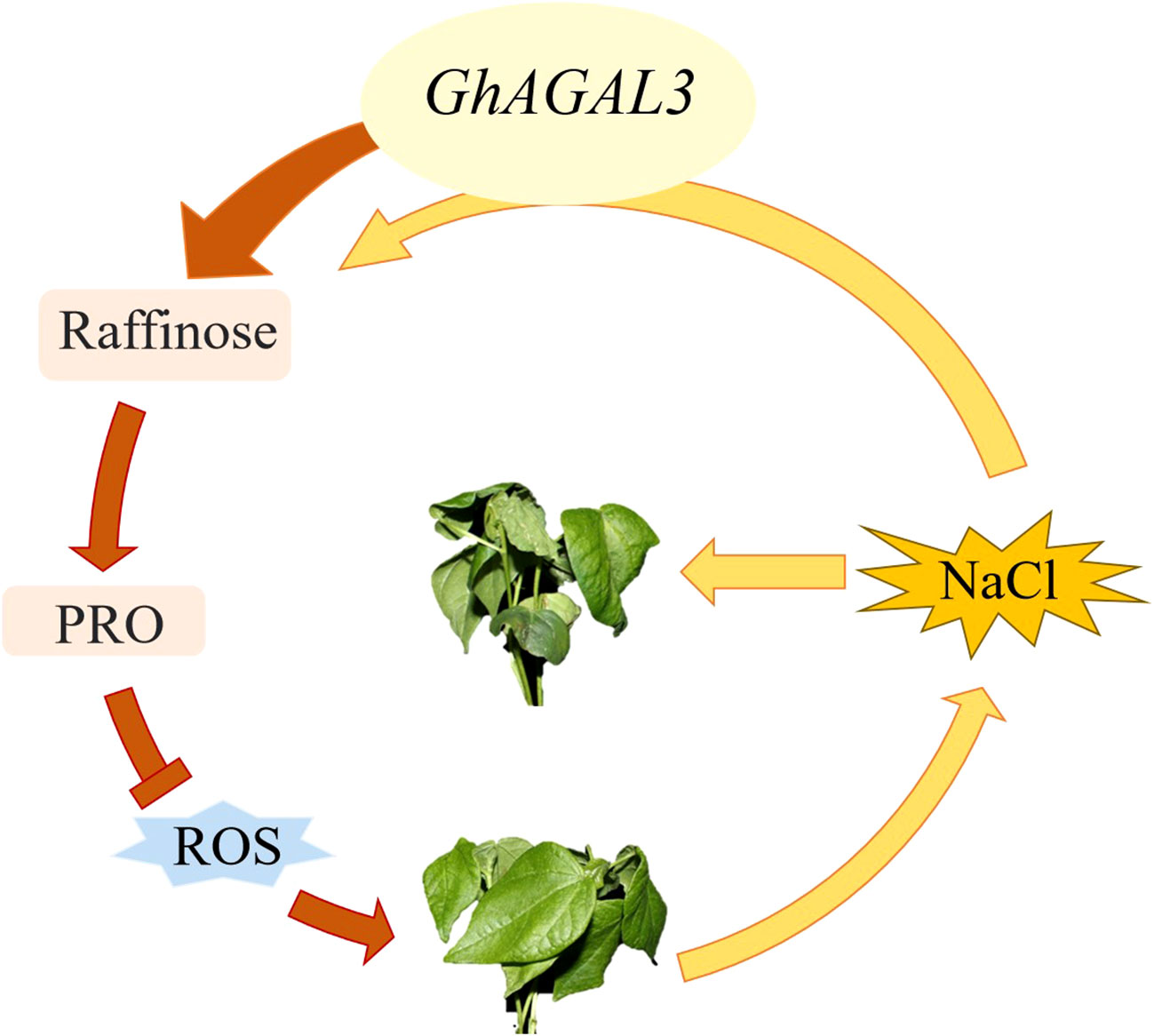
To further investigate the potential relationship between raffinose and salt tolerance in cotton, the raffinose content was measured in both control and salt-treated plants before and after treatment (Figure 9). According to our results, the content of raffinose increased after silencing GhAGAL3. Meanwhile, the levels of D-glucose and D-galactose, which are the downstream metabolic substances controlled by GhAGAL3, were determined, respectively. As a result, the levels of D-glucose and D-galactose decreased under salt stress. the chlorophyll content of GhAGAL3 plant increased significantly compared with that of the control plant pYL156. These results indicated that salt stress was alleviated by increasing the raffinose content.
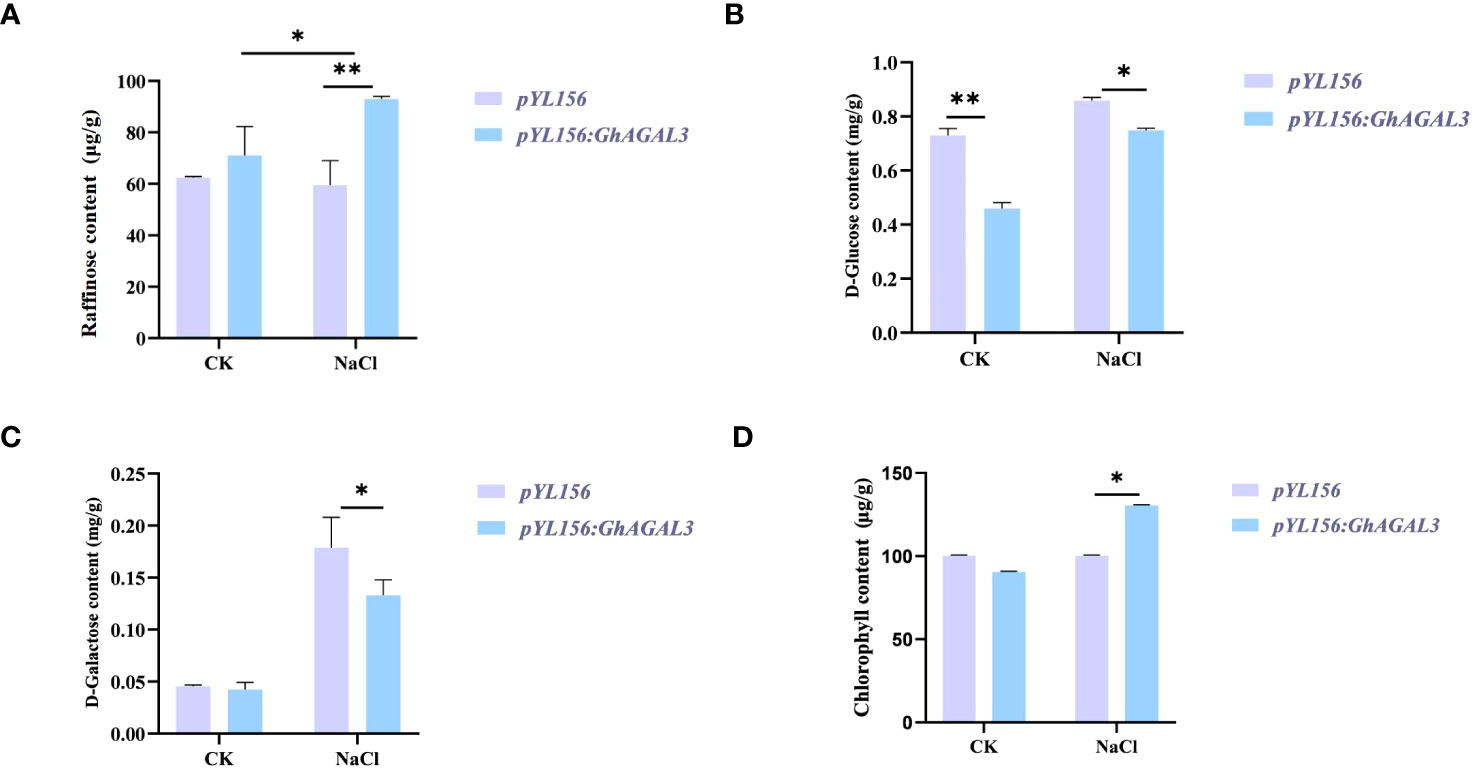
Figure 9 Determination of raffinose, D-glucose, D-galactose, and chlorophyll content after GhAGAL3 silencing. (A) Raffinose content of empty control and VIGS plants under normal growth and NaCl stress. (B) D-glucose content of empty control and VIGS plants under normal growth and NaCl stress. (C) D-galactose content of empty control and VIGS plants under normal growth and NaCl stress. (D) content of empty control and VIGS plants under normal growth and NaCl stress. *0.01 < p < 0.05, **p < 0.01; the resulting mean values were presented as relative units.
4 Discussion
Due to global climate change, the abiotic environment has become one of the most crucial factors influencing plant growth and development. Salt stress, in particular, has a direct impact on crop growth, development, and yield (Vaughan et al., 2018). AGAL is a hydrolyzing enzyme related to raffinose, which has an irreplaceable role in plant growth and development, as well as in response to adversity (Tsaniklidis et al., 2016). Studies on the AGAL gene family have revealed that AGAL exists not only in cotton but also in other evolutionary branches of species, including Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Vitis vinifera, and Zea mays. To be specific, 15 AGAL genes were identified in Gossypium hirsutum, 15 in Gossypium barbadense, 7 in Gossypium raimondii, 7 in Gossypium arboreum, 9 in Oryza sativa, 7 in Vitis vinifera, 18 in Zea mays, 22 in Populus trichocarpa, and 4 in Arabidopsis thaliana (Chrost et al., 2007). There were 44 AGAL genes identified in the four major cotton species, with the same number being identified in Vitis vinifera, Gossypium arboreum, and Gossypium raimondii. Therefore, it was speculated that amplification might occur during the evolutionary process. Although less studied in plant AGAL, four members have been identified in Arabidopsis thaliana, but none of them has been reported in cotton as of now. At the same time, the constructed phylogenetic tree showed that cotton was closely related to Zea mays and distantly related to Vitis vinifera. In addition, AGAL genes tended to be conserved throughout the cotton genome. AGAL plays an important role in plant growth, development, and response to adversity stress (Zhou et al., 2012; Han et al., 2015).
From the perspectives of chromosome localization and motif structure analysis, it was found that the gene structure of the AGAL family was highly conserved. In addition, the structure of the GhAGAL gene family members was highly consistent with that of the GbAGAL gene family members both of which evolve from diploid to tetraploid.
The expression of AGAL genes is regulated by various environmental factors, including light, phytohormones, and adversity stress. The present study revealed the patterns of response to salt, PEG, cold, and heat stresses, as well as the tissue-specific expression of the AGAL gene family. Notably, the cis-acting element has an important function when plants are subjected to abiotic stresses(Yadav et al., 2011). According to our results, cis-acting elements responded and generated excitons to regulate gene expression. A large number of hormone response elements, such as salicylic acid, jasmonic acid, and abscisic acid, are present in the promoter of GhAGALs. In this study, corresponding cis-acting elements of salicylic acid, jasmonic acid, abscisic acid, and other plant hormones exerted a crucial effect plant adaptation to abiotic stresses. When cotton is exposed to abiotic stress, it undergoes a variety of physiological and biochemical responses to mitigate the adverse effects of the stress (Zhang et al., 2022). Abiotic stresses including drought, cold, heat, and salt can reduce cotton yield. This study provided the first comprehensive analysis of the AGAL gene family in four cotton species. Finally, to verify the function of the GhAGAL3 gene, we silenced it through the VIGS experiment. The results showed that silencing GhAGAL3 reduced the salt tolerance of cotton.
Studies have reported that the metabolism of RFOs is a complex regulatory network in plants, and numerous associated enzymes are involved in the accumulation of RFOs (Sui et al., 2012). The first step in the catabolism of raffinose is the hydrolysis mediated by α-Gal, which produces sucrose and galactose (Peng et al., 2014). RFOs are distributed in plants and exert a protective effect in response to a wide range of abiotic stresses (Sonali et al., 2015). Overexpression of the α-Gal gene decreases the level of raffinose, which in turn reduces the cold tolerance of plants (Salvi et al., 2022). In the meantime, low temperature induced an increase in fructose content and a decrease in sucrose content within cucumber stems (Dos Santos et al., 2011). Moreover, the alkaline α-Gal activity of stem samples was lower than that of control samples after low temperature stress. Thus, α-Gal plays an important role in inhibiting glucose metabolism.
According to the expression patterns of AGAL gene in different tissues under NaCl stress and under different abiotic stresses, GhAGAL3 was selected for a functional verification study. After silencing GhAGAL3 gene, the wilting degree of seedlings decreased relative to the negative control. It was inferred that GhAGAL3 might play an important role in responding to NaCl stress (Figure 10). Under NaCl stress, the expression of GhAGAL3 decreased and the raffinose content increased. Additionally, the proline content of GhAGAL3 significantly increased. Typically, the proline content in plants reflects the extent of their resistance to stress. Plants accumulate proline under stressful conditions. The higher the proline content, the stronger the resistance (Xie et al., 2011). When plants accumulate an excessive amount of reactive oxygen species, lipid peroxidation occurs, leading to the production of free radicals. Under such circumstances, plants are affected by oxidative stress, and as a result, cells cannot function normally. Lipid peroxidation is the most serious injury process in an organic body. As revealed by the DAB staining in this study, the leaves of pYL156 plants treated with NaCl were darker than those of GhAGAL3, which might be ascribed oxidative stress. Brown spots were clearly visible on cotton leaves under stress, and the color of veins was darker. This suggested the significant buildup deepening, indicating the accumulation of ROS in cotton leaves after the stress treatment (Montillet et al., 2005). Therefore, it was concluded that increasing the raffinose content in cotton seedlings effectively eliminated hydroxyl free radicals and reduced the salt stress-induced damage to cotton seedlings. To enhance the salt stress tolerance of cotton to salt stress, raffinose is used as an osmotic regulator in previous studies to alleviate salt stress and eliminate hydroxyl radicals from the body. ROS production in plants is reduced under stress. (Nishizawa et al., 2019). In some studies, the mechanism by which CsGolS4 enhances drought resistance is illustrated, and whether CsGolS4 improves oxidative damage caused by low temperature and drought stresses is examined by measuring the activities of reactive oxygen enzymes (Ma et al., 2021). Various environmental stresses induce the accumulation of ROS whereas an excessive amount of ROS can cause cell damage, including lipid peroxidation damage (Van den Ende and Valluru, 2008). Collectively, these studies suggest that RFOs play an important role in enhancing stress resistance in plants.
5 Conclusion
According to the findings in this study, raffinose played a vital role in salt stress. The phenotype and raffinose content of cotton leaves under salt stress were investigated in this study. AGAL genes were discovered in cotton, and 7, 15, 15, and 7 AGAL genes were found in Gossypium arboreum, Gossypium barbadense, Gossypium hirsutum, and Gossypium raimondii, respectively. The AGAL genes were divided into three branches based on the phylogenetic tree, gene structure, and motifs. The AGAL family was engaged in a variety of abiotic stressors. Raffinose played a crucial role in salt stress. Therefore, silencing GhAGAL3 resulted in a more favorable phenotype under NaCl stress. Results in this study can lay a theoretical foundation for further research on the link between GhAGAL3 and NaCl stress.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
WC, YC, YH, LZ, and RC: Designed the experiments, methodology, experiment, analysis of data, writing-original draft preparation, writing-review and editing. XYL: Methodology, experiment. HH: Methodology. YF and YZ: Experiment. XF, KN, and TJ: Experiment. YM and ML: Experiment. MH and YL: Experiment. XKL, XC, and DW: Methodology. LZ, LG, JW, and SW: Methodology. QC: Writing-review and editing. WY: Conceived and designed the experiments, supervision. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences, China Agriculture Research System of MOF and MARA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1246677/full#supplementary-material
References
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Intenational Conf. On Intelligent Syst. Mol. Biol. 2 (1), 28–36.
Bawa, G., Liu, Z., Zhou, Y., Fan, S., Ma, Q., Tissue, D. T., et al. (2022). Cotton proteomics: Dissecting the stress response mechanisms in cotton. Front. Plant Sci. 13, 1035801. doi: 10.1186/s12864-021-07622-1
Cai, K., Liu, H., Chen, S., Liu, Y., Zhao, X., Chen, S. (2021). Genome-wide identification and analysis of class III peroxidases in Betula pendula. BMC Genomics 22 (1), 314. doi: 10.1186/s12864-021-07622-1
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Si, G. Y., Zhao, G., Abdullah, M., Guo, N., Li, D. H., et al. (2018). Genomic comparison of the P-ATPase gene family in four cotton species and their expression patterns in Gossypium hirsutum. Molecules 23 (5), 1092. doi: 10.3390/molecules23051092
Chrost, B., Kolukisaoglu, U., Schulz, B., Krupinska, K. (2007). An α-galactosidase with an essential function during leaf development. Planta 225, 311–320. doi: 10.1007/s00425-006-0350-9
Cui, R., Lu, X., Chen, X., Malik, W. A., Wang, D., Wang, J., et al. (2021a). A novel raffinose biological pathway is observed by symbionts of cotton≡Verticillium dahliae to improve salt tolerance genetically on cotton. J. Agron. Crop Sci. 207 (6), 956–969. doi: 10.1111/jac.12556
Cui, R., Wang, X., Malik, W. A., Lu, X., Chen, X., Wang, D., et al. (2021b). Genome-wide identification and expression analysis of Raffinose synthetase family in cotton. BMC Bioinf. 22 (1), 356. doi: 10.1186/s12859-021-04276-4
Daldoul, S., Toumi, I., Reustle, G. M., Krczal, G., Ghorbel, A., Mliki, A., et al. (2012). Molecular cloning and characterisation of a cDNA encoding a putative alkaline alpha-galactosidase from grapevine (Vitis vinifera L.) that is differentially expressed under osmotic stress. Acta physiologiae plantarum 34, 891–903. doi: 10.1007/s11738-011-0887-5
Dos Santos, T. B., Budzinski, I. G., Marur, C. J., Petkowicz, C. L., Pereira, L. F., Vieira, L. G. (2011). Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol. Biochem. 49 (4), 441–448. doi: 10.1016/j.plaphy.2011.01.023
Fan, Y., Zhang, Y., Rui, C., Zhang, H., Xu, N., Wang, J., et al. (2022). Molecular structures and functional exploration of NDA family genes respond tolerant to alkaline stress in Gossypium hirsutum L. Biol. Res. 55 (1), 4. doi: 10.1186/s40659-022-00372-8
Farooq, M. A., Ali, S., Hameed, A., Ishaque, W., Mahmood, K., Iqbal, Z. (2013). Alleviation of cadmium toxicity by silicon is related to elevated photosynthesis, antioxidant enzymes; suppressed cadmium uptake and oxidative stress in cotton. Ecotoxicology Environ. Saf. 96, 242–249. doi: 10.1016/j.ecoenv.2013.07.006
Fialho Lda, S., Guimaraes, V. M., Callegari, C. M., Reis, A. P., Barbosa, D. S., Borges, E. E., et al. (2008). Characterization and biotechnological application of an acid alpha-galactosidase from Tachigali multijuga Benth. seeds. Phytochemistry 69 (14), 2579–2585. doi: 10.1016/j.phytochem.2008.08.017
Gu, H., Lu, M., Zhang, Z., Xu, J., Cao, W., Miao, M. (2018). Metabolic process of raffinose family oligosaccharides during cold stress and recovery in cucumber leaves. J. Plant Physiol. 224-225, 112–120. doi: 10.1016/j.jplph.2018.03.012
Han, Q., Li, T., Zhang, L., Yan, J., Dirk, L.M., Downie, B., et al. (2015). Functional analysis of the 5′-regulatory region of the maize alkaline alpha-galactosidase1 gene. Plant Mol. Biol. Reporter 33, 1361–1370.
Hara, M., Tokunaga, K., Kuboi, T. (2008). Isolation of a drought-responsive alkaline α-galactosidase gene from New Zealand spinach. Plant Biotechnol. 25 (5), 497–501. doi: 10.5511/plantbiotechnology.25.497
Henderson, J., Salzberg, S., Fasman, K. H. (1997). Finding genes in DNA with a hidden markov model. J. Comput. Biol. 4 (2), 127–141. doi: 10.1089/cmb.1997.4.127
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51 (4), 739–748.
Huang, H., He, Y., Cui, A., Sun, L., Han, M., Wang, J., et al. (2022). Genome-wide identification of GAD family genes suggests GhGAD6 functionally respond to Cd2+ stress in cotton. Front. Genet. 13. doi: 10.3389/fgene.2022.965058
Hughes, D., Boyd, S., Giraldo, P., Gonzalez, D., Holida, M., Goker-Alpan, O., et al. (2016). Novel treatment for Fabry disease: IV administration of plant derived alpha-gal-A enzyme safety and efficacy interim report. Mol. Genet. Metab. 2 (117), S59. doi: 10.1016/j.ymgme.2015.12.296
Joshi, J., Hasnain, G., Logue, T., Lynch, M., Wu, S., Guan, J. C., et al. (2021). A core metabolome response of maize leaves subjected to long-duration abiotic stresses. Metabolites 11 (11), 797. doi: 10.3390/metabo11110797
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Lee, C., Chung, C. T., Hong, W. J., Lee, Y. S., Lee, J. H., Koh, H. J., et al. (2021). Transcriptional changes in the developing rice seeds under salt stress suggest targets for manipulating seed quality. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.748273
Lee, R.-H., Lin, M.-C., Chen, S.-C. (2004). A novel alkaline α-galactosidase gene is involved in rice leaf senescence. Plant Mol. Biol. 55, 281–295. doi: 10.1007/s11103-004-0641-0
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, H. W., Zang, B. S., Deng, X. W., Wang, X. P. (2011). Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234 (5), 1007–1018. doi: 10.1007/s00425-011-1458-0
Liu, X., Cui, Y., Kang, R., Zhang, H., Huang, H., Lei, Y., et al. (2022). GhAAO2 was observed responding to NaHCO(3) stress in cotton compared to AAO family genes. BMC Plant Biol. 22 (1), 603. doi: 10.1186/s12870-022-03999-7
Ma, S., Lv, J., Li, X., Ji, T., Gao, L. (2021). Galactinol synthase gene 4 (CsGolS4) increases cold and drought tolerance in Cucumis sativus L. by inducing RFO accumulation and ROS scavenging. Environ. Exp. Bot. 185, 104406. doi: 10.1016/j.envexpbot.2021.104406
Malik, W. A., Wang, X., Wang, X., Shu, N., Cui, R., Chen, X., et al. (2020). Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int. J. Biol. Macromolecules 153 (prepublish), 492. doi: 10.1016/j.ijbiomac.2020.03.021
Meng, Y., Cui, Y., Peng, F., Guo, L., Cui, R., Xu, N., et al. (2023). GhCYS2 governs the tolerance against cadmium stress by regulating cell viability and photosynthesis in cotton. Ecotoxicology Environ. Saf. 263, 115386. doi: 10.1016/j.ecoenv.2023.115386
Mittal, Y., Sharma, C. B. (1991). Development of α-galactosidase isoenzymes in chickpea seeds. Plant Sci. 77 (2), 185–190. doi: 10.1016/0168-9452(91)90087-O
Montillet, J.-L., Chamnongpol, S., Rusterucci, C., Dat, J., van de Cotte, B., Agnel, J.-P., et al. (2005). Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 138 (3), 1516–1526. doi: 10.1104/pp.105.059907
Nishizawa, A., Yabuta, Y., Shigeoka, S. (2019). Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147 (3), 1251–1263. doi: 10.1104/pp.108.122465
Nishizawa-Yokoi, A., Yabuta, Y., Shigeoka, S. (2008). The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signaling Behav. 3 (11), 1016–1018. doi: 10.4161/psb.6738
Peng, Z., He, S., Gong, W., Sun, J., Pan, Z., Xu, F., et al (2014). Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genomics 15 (1), 1–28.
Pennycooke, J. C., Jones, M. L., Stushnoff, C. (2003). Down-regulating alpha-galactosidase enhances freezing tolerance in transgenic petunia. Plant Physiol. 133 (2), 901–909. doi: 10.1104/pp.103.024554
Rui, C., Chen, X., Xu, N., Wang, J., Zhang, H., Li, S., et al. (2022). Identification and structure analysis of KCS family genes suggest their reponding to regulate fiber development in long-staple cotton under salt-alkaline stress. Front. Genet. 13, 812449. doi: 10.3389/fgene.2022.812449
Salvi, P., Varshney, V., Majee, M. (2022). Raffinose family oligosaccharides (RFOs): role in seed vigor and longevity. Bioscience Rep. 42 (10), BSR20220198. doi: 10.1042/bsr20220198
Schneider, T., Keller, F. (2009). Raffinose in chloroplasts is synthesized in the cytosol and transported across the chloroplast envelope. Plant Cell Physiol. 50 (12), 2174–2182. doi: 10.1093/pcp/pcp151
Sonali, S., Sritama, M., Papri, B., Majumder, A. L. (2015). Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 6 (656), 656. doi: 10.3389/fpls.2015.00656
Sui, X. L., Meng, F. Z., Wang, H. Y., Wei, Y. X., Li, R. F., Wang, Z. Y., et al. (2012). Molecular cloning, characteristics and low temperature response of raffinose synthase gene in Cucumis sativus L. J. Plant Physiol. 169 (18), 1883–1891. doi: 10.1016/j.jplph.2012.07.019
Tsaniklidis, G., Benovias, A., Delis, C., Aivalakis, G. (2016). Acidic alpha galactosidase during the maturation and cold storage of cherry tomatoes. Acta Physiologiae Plantarum 38 (2), 57. doi: 10.1007/s11738-016-2075-0
Van den Ende, W., Valluru, R. (2008). Sucrose, sucrosyl oligosaccharides, and oxidative stress: scavenging and salvaging? J. Exp. Bot. 60 (1), 9–18. doi: 10.1093/jxb/ern297
Vaughan, M. M., Block, A., Christensen, S. A., Allen, L. H., Schmelz, E. A. (2018). The effects of climate change associated abiotic stresses on maize phytochemical defenses. Phytochem. Rev. 17 (1), 37–49. doi: 10.1007/s11101-017-9508-2
Wang, J., Zhang, Y., Xu, N., Zhang, H., Fan, Y., Rui, C., et al. (2021). Genome-wide identification of CK gene family suggests functional expression pattern against Cd2+ stress in Gossypium hirsutum L. Int. J. Biol. Macromolecules 188, 272–282. doi: 10.1016/j.ijbiomac.2021.07.190
Wang, Y., Wang, C., Chen, Y., Cui, M., Wang, Q., Guo, P. (2022). Heterologous expression of a thermostable α-galactosidase from parageobacillus thermoglucosidasius isolated from the lignocellulolytic microbial consortium TMC7. J. Microbiol. Biotechnol. 32 (6), 749–760. doi: 10.4014/jmb.2201.01022
Xie, H., Yang, L., Li, Z. G. (2011). The roles of proline in the formation of plant tolerance to abiotic stress. Biotechnol. Bull. 51 (2), 23–25.
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., Wang, H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51 (W1), W587–w592. doi: 10.1093/nar/gkad359
Yadav, N. R., Taunk, J., Rani, A., Aneja, B., Yadav, R. C. (2011). Role of transcription factors in abiotic stress tolerance in crop plants. Climate Change Plant Abiotic Stress Tolerance 25 (3), 2433–2442. doi: 10.5504/BBEQ.2011.0072
Yu, M., Lin, S., Bei, F., Zhao, D., Zheng, Y., Sheng, J. (2009). The effect of MeJA on ethylene biosynthesis and induced disease resistance to Botrytis cinerea in tomato. Postharvest Biol. Technol. 54 (3), 153–158. doi: 10.1016/j.postharvbio.2009.07.001
Zhang, H., Mao, L., Xin, M., Xing, H., Zhang, Y., Wu, J., et al. (2022). Overexpression of GhABF3 increases cotton(Gossypium hirsutum L.) tolerance to salt and drought. BMC Plant Biol. 22 (1), 1–15. doi: 10.1186/s12870-022-03705-7
Zhang, H., Zhang, Y., Xu, N., Rui, C., Fan, Y., Wang, J., et al. (2021). Genome-wide expression analysis of phospholipase A1 (PLA1) gene family suggests phospholipase A1-32 gene responding to abiotic stresses in cotton. Int. J. Biol. Macromolecules 192, 1058–1074. doi: 10.1016/j.ijbiomac.2021.10.038
Zhao, T.-Y., Corum Iii, J. W., Mullen, J., Meeley, R. B., Helentjaris, T., Martin, D., et al. (2006). An alkaline α-galactosidase transcript is present in maize seeds and cultured embryo cells, and accumulates during stress. Seed Sci. Res. 16 (2), 107–121. doi: 10.1093/glycob/cws121
Zhou, M. L., Zhang, Q., Zhou, M., Sun, Z. M., Zhu, X. M., Shao, J. R., et al. (2012). Genome-wide identification of genes involved in raffinose metabolism in maize. Glycobiology 22 (12), 1775–1785. doi: 10.1093/glycob/cws121
Keywords: α-galactosidase (AGALs), Gossypium hirsutum, raffinose family oligosaccharides (RFOs), abiotic stresses, functional verification
Citation: Chen W, Cui Y, He Y, Zhao L, Cui R, Liu X, Huang H, Zhang Y, Fan Y, Feng X, Ni K, Jiang T, Han M, Lei Y, Liu M, Meng Y, Chen X, Lu X, Wang D, Wang J, Wang S, Guo L, Chen Q and Ye W (2023) Raffinose degradation-related gene GhAGAL3 was screened out responding to salinity stress through expression patterns of GhAGALs family genes. Front. Plant Sci. 14:1246677. doi: 10.3389/fpls.2023.1246677
Received: 04 August 2023; Accepted: 27 November 2023;
Published: 19 December 2023.
Edited by:
Jian Chen, Jiangsu University, ChinaReviewed by:
Muhammad Waheed Riaz, Zhejiang Agriculture and Forestry University, ChinaAbdul Rehman, Zhengzhou University, China
Copyright © 2023 Chen, Cui, He, Zhao, Cui, Liu, Huang, Zhang, Fan, Feng, Ni, Jiang, Han, Lei, Liu, Meng, Chen, Lu, Wang, Wang, Wang, Guo, Chen and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wuwei Ye, eWV3MTU4QDE2My5jb20=; Quanjia Chen, Y2hxamlhQDEyNi5jb20=
†These authors have contributed equally to this work
 Wenhua Chen1,2†
Wenhua Chen1,2† Yupeng Cui
Yupeng Cui Yuexin Zhang
Yuexin Zhang Xiugui Chen
Xiugui Chen Xuke Lu
Xuke Lu Delong Wang
Delong Wang Quanjia Chen
Quanjia Chen Wuwei Ye
Wuwei Ye