- 1College of Life Science and Engineering, Henan University of Urban Construction, Pingdingshan, China
- 2State Key Laboratory of Hybrid Rice, Department of Plant Sciences, College of Life Sciences, Wuhan University, Wuhan, China
Dirigent (DIR) proteins play essential roles in regulating plant growth and development, as well as enhancing resistance to abiotic and biotic stresses. However, the whole-genome identification and expression profiling analysis of DIR gene family in millet (Setaria italica (Si)) have not been systematically understood. In this study, we conducted genome-wide identification and expression analysis of the S. italica DIR gene family, including gene structures, conserved domains, evolutionary relationship, chromosomal locations, cis-elements, duplication events, gene collinearity and expression patterns. A total of 38 SiDIR members distributed on nine chromosomes were screened and identified. SiDIR family members in the same group showed higher sequence similarity. The phylogenetic tree divided the SiDIR proteins into six subfamilies: DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f, and DIR-g. According to the tertiary structure prediction, DIR proteins (like SiDIR7/8/9) themselves may form a trimer to exert function. The result of the syntenic analysis showed that tandem duplication may play the major driving force during the evolution of SiDIRs. RNA-seq data displayed higher expression of 16 SiDIR genes in root tissues, and this implied their potential functions during root development. The results of quantitative real-time PCR (RT-qPCR) assays revealed that SiDIR genes could respond to the stress of CaCl2, CdCl, NaCl, and PEG6000. This research shed light on the functions of SiDIRs in responding to abiotic stress and demonstrated their modulational potential during root development. In addition, the membrane localization of SiDIR7/19/22 was confirmed to be consistent with the forecast. The results above will provide a foundation for further and deeper investigation of DIRs.
1 Introduction
Gramineae family member of millet (Setaria italica) originated in China. The prolific yield and diverse ecological niches of millet are in sharp contrast to its small stature and short life cycle (Li and Brutnell, 2011). The remarkable drought tolerance and wide-ranging germplasm collection of millet have provided ideal model systems for the studies of C4 evolution, comparative grass genomics, and biofuel feedstocks (Doust et al., 2009; Brutnell et al., 2010).
Dirigent (DIR) proteins were first reported and isolated in Forsythia intermedia and were found to be a model for regioselective and stereoselective coupling during biological processes (Davin et al., 1997; Davin and Lewis, 2005). Since then, DIR proteins have been more and more cloned and studied in seed plants, including Thuja plicata, Schisandra chinensis (Kim et al., 2012), Pisum sativum (Seneviratne et al., 2015), Linum usitatissimum (Corbin et al., 2018), Glycine max (Li et al., 2017), Arabidopsis (Gasper et al., 2016; Yonekura-Sakakibara et al., 2021), rice (Duan et al., 2023), and cotton (Liu et al., 2021). According to the previous reports, DIR proteins in seed plants could be divided into six different subfamilies, named DIR-a and DIR-like groups (b/d, c, e, f, and g) (Ralph et al., 2007). The protein crystal structures of (+)-DIR (PsDRR206) and (−)-DIR (AtDIR6) have been obtained. The DIR protein is composed of eight reverse parallel β spiral structures connected in series β-barrel, which exists in the form of a trimer (Kim et al., 2015; Gasper et al., 2016).
The number of DIR and DIR-like genes is different in distinct plant species: 26 in Arabidopsis (Paniagua et al., 2017), 19 in Isatis indigotica (Li et al., 2014), and 55 in rice (Duan et al., 2023). Also, DIR genes are expressed variously in plant tissues including but not limited to leaves and roots (Paniagua et al., 2017). The promoter activity of DIR genes has been detected primarily in the vascular bundle of red cedar (Kim et al., 2002). Additionally, five conserved motifs (motifs I–V) have been identified within the DIR and DIR-like protein sequences in Arabidopsis, spruce, and rice (Ralph et al., 2007; Duan et al., 2023). It is worth mentioning that the DIR domain within ESB1 protein is indispensable for the lignin deposition of Casparian strips (Hosmani et al., 2013), and the N-glycosylation sites of Asn were reported to be representative amino acids of FiDIR1 (Burlat et al., 2001). Except for this, some DIR proteins were proposed to mediate the formation of gossypol in cotton (Effenberger et al., 2015; Effenberger et al., 2017; Lin et al., 2023). DIR proteins in leguminous plants showed dehydratase activity, although lacking a catalytic active center (Uchida et al., 2017).
The rapid progress rate of whole-genome sequencing projects also gives opportunities to resolve agricultural difficulties (Schnable et al., 2009; Li and Brutnell, 2011). Increasing findings suggest that DIRs also have various developmental regulation roles. For instance, the expression level of ScDIR genes was induced by PEG6000 and NaCl stresses (Guo et al., 2012). Most VrDIR genes varied their expression levels under high salt and drought stresses (Xu et al., 2021). The silencing of CaDIR7 reduced peppers’ tolerance to Phytophthora capsici and abiotic stress (Khan et al., 2018). Recent research showed that GhDIR5 mutation prevented gossypol formation in cotton (Guo et al., 2012). DIR members also function in regulating lignin biosynthesis to stand and defend against microorganisms and insects (Burlat et al., 2001; Hosmani et al., 2013). The resistance to Phytophthora sojae and the total lignan accumulation of GmDIR22 overexpressor were significantly enhanced (Li et al., 2017). After being infected by Fusarium solani, the transcriptional level of PsDRR266 was induced (Seneviratne et al., 2015). These similar benefits can be also found in GhDIR overexpression plants (Shi et al., 2012). Therefore, DIR proteins are necessary during the diverse biological and physiological processes of plants. The whole-genome identification of DIR protein is practicable for us to enable plant tolerance. However, an understanding of DIR proteins in S. italica is lacking.
In this study, genome-wide analysis of 38 DIR gene families in S. italica (SiDIRs) has been performed using bioinformatics methods. The evolutionary history was investigated through their phylogenetic relationships, sequence similarity, gene structure features, motif positions, tertiary structure, chromosome distributions, cis-elements, duplication events, and gene collinearity. The comprehensive expression patterns of SiDIRs in the five tissues of flag leaf, stem, root, panicle, and mesophyll unravel their important regulatory roles during millet development. Also, the active transcriptional responses of SiDIR genes to abiotic stress highlight the functional potential involved in these physiological processes. The finding of interacting proteins will provide candidate members that are involved in the adaptivity of millets to various environments. Additionally, the analysis of subcellular localization illustrates the potential function of SiDIRs in cell membranes. Our studies will provide useful theoretical support and new insights into exploring millet resistance and crop yield potential.
2 Results
2.1 Identification of DIR family members in S. italica
A total of 38 DIR and DIR-like members in S. italica were confirmed by HMMER and BLASTP methods. According to their chromosomal location, the 38 DIR genes were named SiDIR1 to SiDIR38. The protein sequence length of all the SiDIRs was less than 350 amino acids, ranging from 158 (SiDIR10) to 340 (SiDIR38); the molecular weight ranged from 16.8 (SiDIR8) to 34.9 (SiDIR7) kDa; the theoretical PI ranged from 4.79 (SiDIR38) to 9.82 (SiDIR9) (Supplemental Data 1). The predicted subcellular localization of most SiDIR proteins was in the cell membrane, while the remaining family members were located in the cell wall, chloroplast, mitochondrion, nucleus, etc. (Supplemental Data 1). The N-glycosylation sites of SiDIR protein sequences were also analyzed. It is interesting to find that 60% of proteins have the distribution of N-glycosylation sites (Supplemental Data 1). These results showed the divergence of millet DIR genes and hinted at their distinct functional potential.
2.2 Phylogenetic analysis and sequence similarity of SiDIRs
To analyze the homology of SiDIRs, we used ClustalW and found that the protein sequences within the same subfamily have high similarity (Figure 1A). For example, SiDIR7, SiDIR8, and SiDIR9 have high sequence similarity (78%–86%); SiDIR19, SiDIR20, SiDIR21, SiDIR23, SiDIR25, and SiDIR26 have high sequence similarity (72%–89%). Also, we found that individual amino acids within the conserved motifs have undergone specific mutations. The amino acid of SiDIR35 changed from “FG” to “FS”, and SiDIR2, SiDIR10, and SiDIR11 changed from “FG” to “LG”. In addition, the conservative amino acid of SiDIR2, SiDIR5, SiDIR8, SiDIR9, SiDIR10, and SiDIR11 changed from “VGRAQG” to “VARAQG” (Supplemental Data 2). Due to the differences in amino acid sequences, different DIR proteins may have diverse functions.
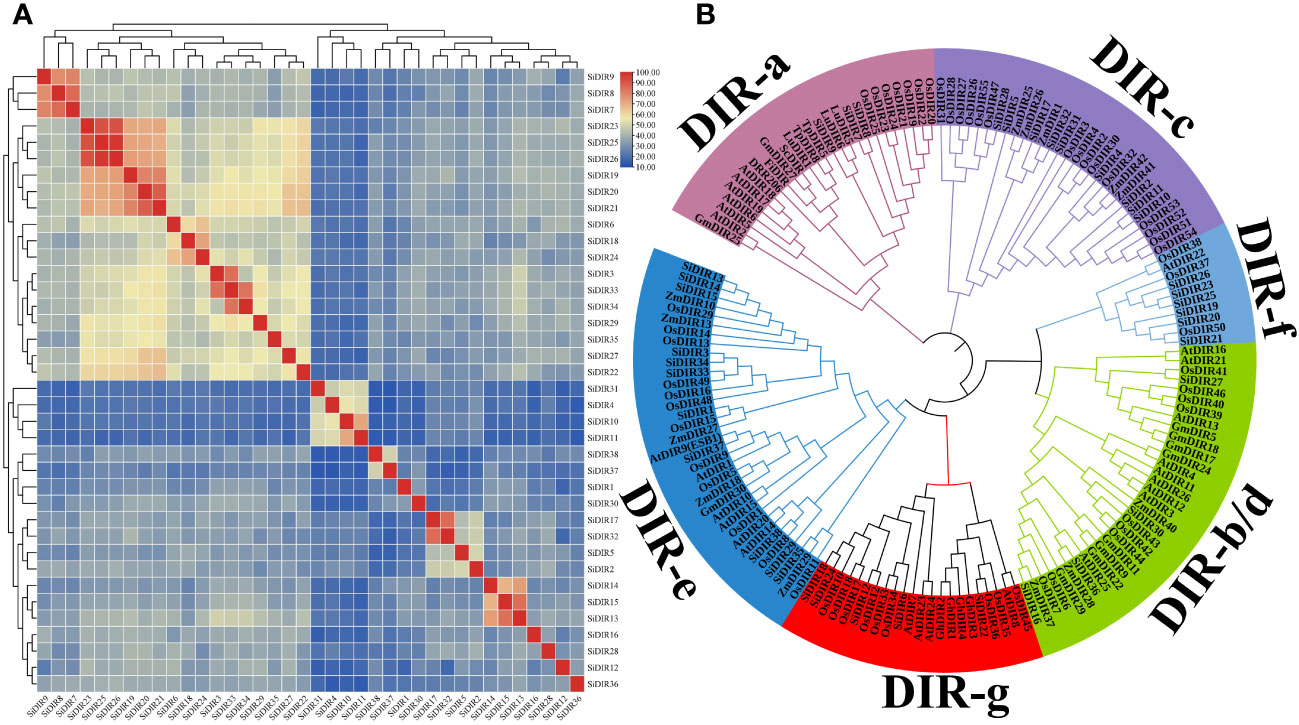
Figure 1 Sequence similarity analysis and the phylogenetic tree of SiDIRs. (A) The heatmap of SiDIR sequence similarity. The protein pairwise similarity matrix was obtained and visualized using TBtools software. The color indicates the similarity percentage, and the color scale values are shown on the upper right. (B) Phylogenetic tree of SiDIRs, AtDIRs, OsDIRs, ZmDIRs, GmDIRs, GhDIRs, LuDIRs, TpDIRs, FiDIR, and ScDIR. The evolutionary tree was constructed by neighbor-joining method (bootstrap values: 1,000 replicates). Pink purple, green, purple, blue, pale blue, and red represent the subgroup of DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f, and DIR-g, respectively.
To further elucidate the evolutionary relationship of SiDIRs, a phylogenetic tree containing 38 SiDIRs, 26 AtDIRs, 55 OsDIRs, 13 ZmDIRs, 11 GmDIRs, 4 GhDIRs, 3 LuDIRs, 2 TpDIRs, 1 FiDIR, and 1 ScDIR were constructed (Figure 1B). The DIR members could be grouped into DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f, and DIR-g. DIR-e (blue in Figure 1B) contained the largest number at 11 SiDIRs, while DIR-a (pink purple in Figure 1B) contained the smallest number at three SiDIRs. DIR-b/d (green in Figure 1B), DIR-c (purple in Figure 1B), DIR-f (pale blue in Figure 1B), and DIR-g (red in Figure 1B) contained four, nine, six, and five SiDIRs, respectively. Interestingly, no AtDIR member was found in the subgroup of DIR-c.
2.3 Analysis of gene structures, protein domains, and conserved motifs
We used NCBI-CDD and MEME databases to analyze the gene structure and the distribution of conserved motifs, respectively. We found that the gene structure and conserved motifs of SiDIRs were similar within the same subgroups (Figures 2A–D). For example, SiDIR4, SiDIR7, SiDIR8, SiDIR9, and SiDIR38 contained five to six identical conserved motifs (Figure 2B). Except for the dirigent domains, the DIR family also contains three other specific domains including jacatin, dirigent superfamily domain, and tudor_AtPTM-like domain (Figure 2C). Some DIR-c subfamily members contain an N-terminal DIR domain and C-terminal end of a jacalin-related lectin (JRL) domain (Figure 2C), which show high specificity to bind mono- or oligo-saccharides (Kittur et al., 2007; Huwa et al., 2021; Huwa et al., 2022). Among the 38 SiDIRs, four members contained the jacatin domain, two members contained the dirigent superfamily domain, and no member contained the tudor_AtPTM-like domain (Figure 2C). Additionally, nine members (SiDIR17, SiDIR32, SiDIR31, SiDIR37, SiDIR10, SiDIR28, SiDIR5, SiDIR2, and SiDIR11) contained introns ranging from one to three, which is higher than that of the other DIR members. Most of the DIR and DIR-like family members contained exons ranging from two to three (Figure 2D). The differences in gene structure or conserved motifs might be due to various biological functions of SiDIRs.
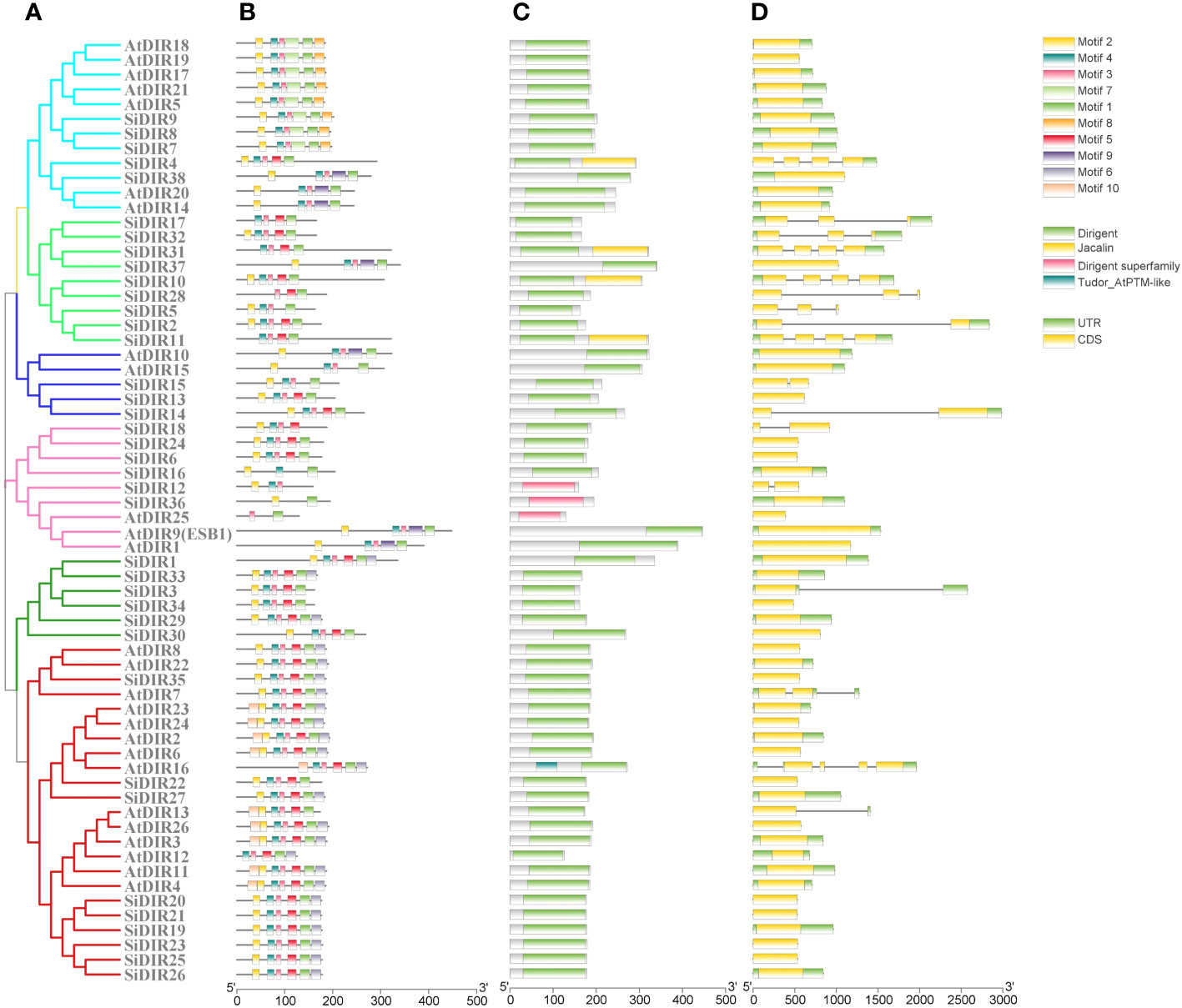
Figure 2 Analysis of gene structure, protein domains, and conserved motifs of SiDIRs. (A) Phylogenetic tree of SiDIRs and AtDIRs. (B) The conserved motifs of SiDIRs that were predicted by MEME. (C) The conserved domains were predicted and analyzed by NCBI-CDD. (D) Exo-intron distribution of SiDIR genes. This picture was visualized using TBtools software.
2.4 Amino acid alignment and the tertiary structure prediction of SiDIR7/8/9
FiDIR1 and DRR206 participate in the formation of (+)-pinoresinol (Pickel et al., 2010; Seneviratne et al., 2015). AtDIR6 and LuDIR5/LuDIR6 are able to form (−)-pinoresinol (Dalisay et al., 2015; Gasper et al., 2016). Based on the results, SiDIR7, SiDIR8, SiDIR9, TpDIR5/8, LuDIR1/5/6, AtDIR5/6, FiDIR, and ScDIR belonged to the same DIR-a subfamily (Figure 1). We aligned the amino acids and found that some sites were highly conserved, such as alanine (A) located in β3 (Figure 3A). Phenylalanine (F) sites located in the β4 structure were similar, except for their mutation to isoleucine (I) in SiDIR9 (Figure 3A). Also, the hydrophilic amino acid “Y” located in the β4 structure mutated into unhydrophobic amino acid “F” (phenylalanine), while the hydrophobic amino acid “I” located in β5 structure mutated to hydrophobic amino acid “L” (leucine) (Figure 3A).
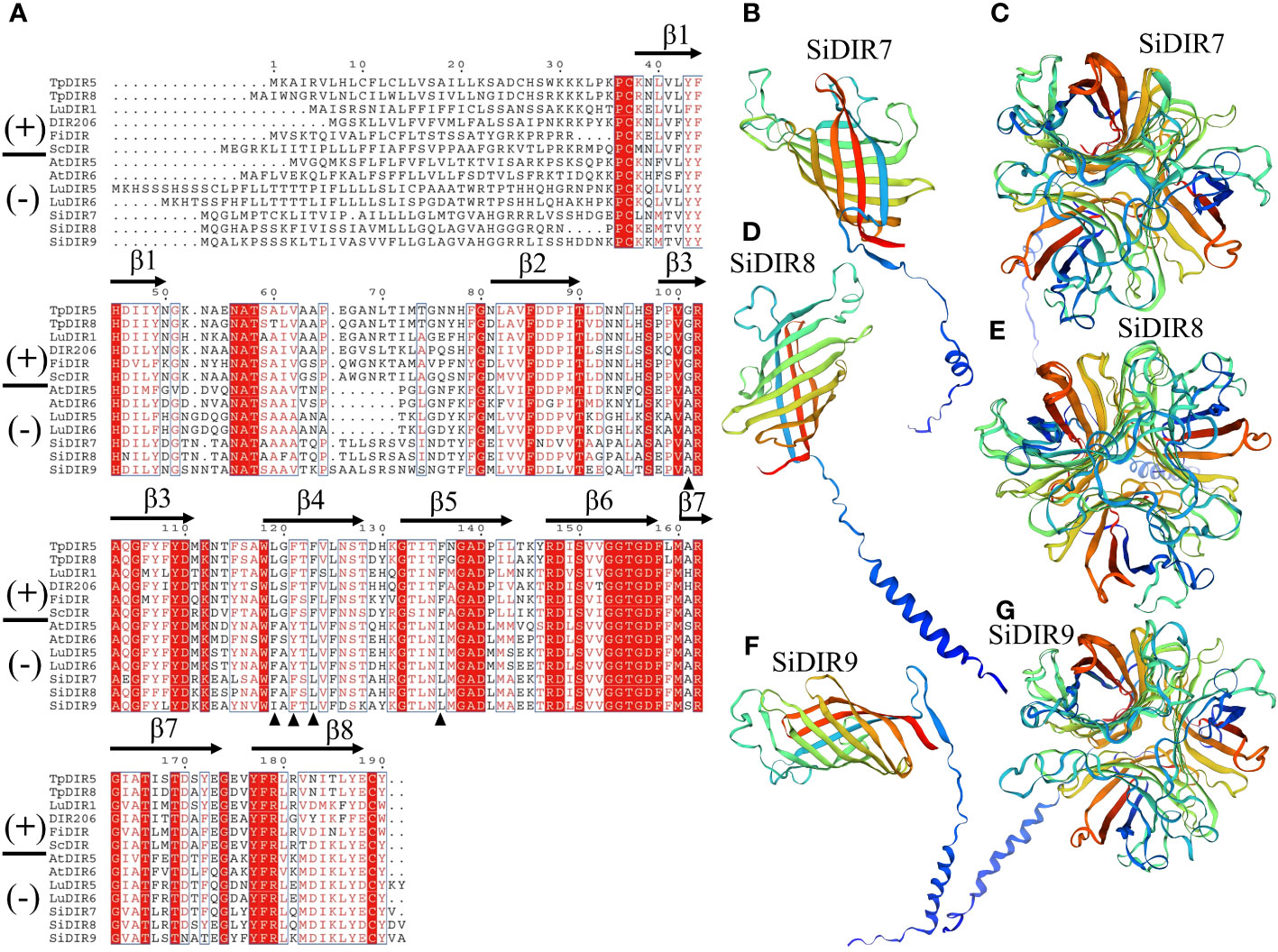
Figure 3 Amino acid alignment and the tertiary structure prediction of SiDIR7, SiDIR8, and SiDIR9. (A) The sequence alignment of SiDIR7, SiDIR8, and SiDIR9 with (−)- and (+)-DIRs. The black triangle indicates residues that are differently conserved in (+)- and (−)-DIRs. (B–G) Predicted tertiary structures of SiDIR proteins.
It has been known that the tertiary structure of the AtDIR6 trimer is an eight-stranded antiparallel β-barrel with spatially well-separated cavities for substrate binding. The binding cavity is composed of two lobes, and each of the two lobes is lined with a set of hydrophilic and potentially catalytic residues that are conserved in (+)- and (−)-pinoresinol-forming DIRs (Gasper et al., 2016). A further forecast showed that the tertiary structure of SiDIR7, SiDIR8, and SiDIR9 consisted of eight-stranded antiparallel β-barrels (Figures 3B, D, F). SiDIR7/8/9 were able to form trimers by themselves through homologous modeling prediction (Figures 3C, E, G). These suggested that SiDIR7, SiDIR8, and SiDIR9 may be able to direct the formation of (−)-pinoresinol. Meanwhile, the different locations of hydrophobic cavities and diverse substrate binding sites (Figures 3B–G) also potentially revealed their different functions.
2.5 Duplication event analysis of SiDIR genes
According to genome annotation, we analyzed the distribution of 38 SiDIR genes. The 38 SiDIR genes were randomly distributed on nine chromosomes (Chr) (Figure 4). Both Chr3 and Chr4 contain three members (~7.89%), while Chr9 contains two members (~5.26%). Compared to Chr5 (1 gene, ~2.63%), Chr8 contained the largest number of SiDIR family (15 genes, ~39.47%), which appears in the form of gene clusters (Figure 4). Although there was no distribution on the first and sixth chromosomes, most SiDIR genes were distributed on the ends of the chromosomes.
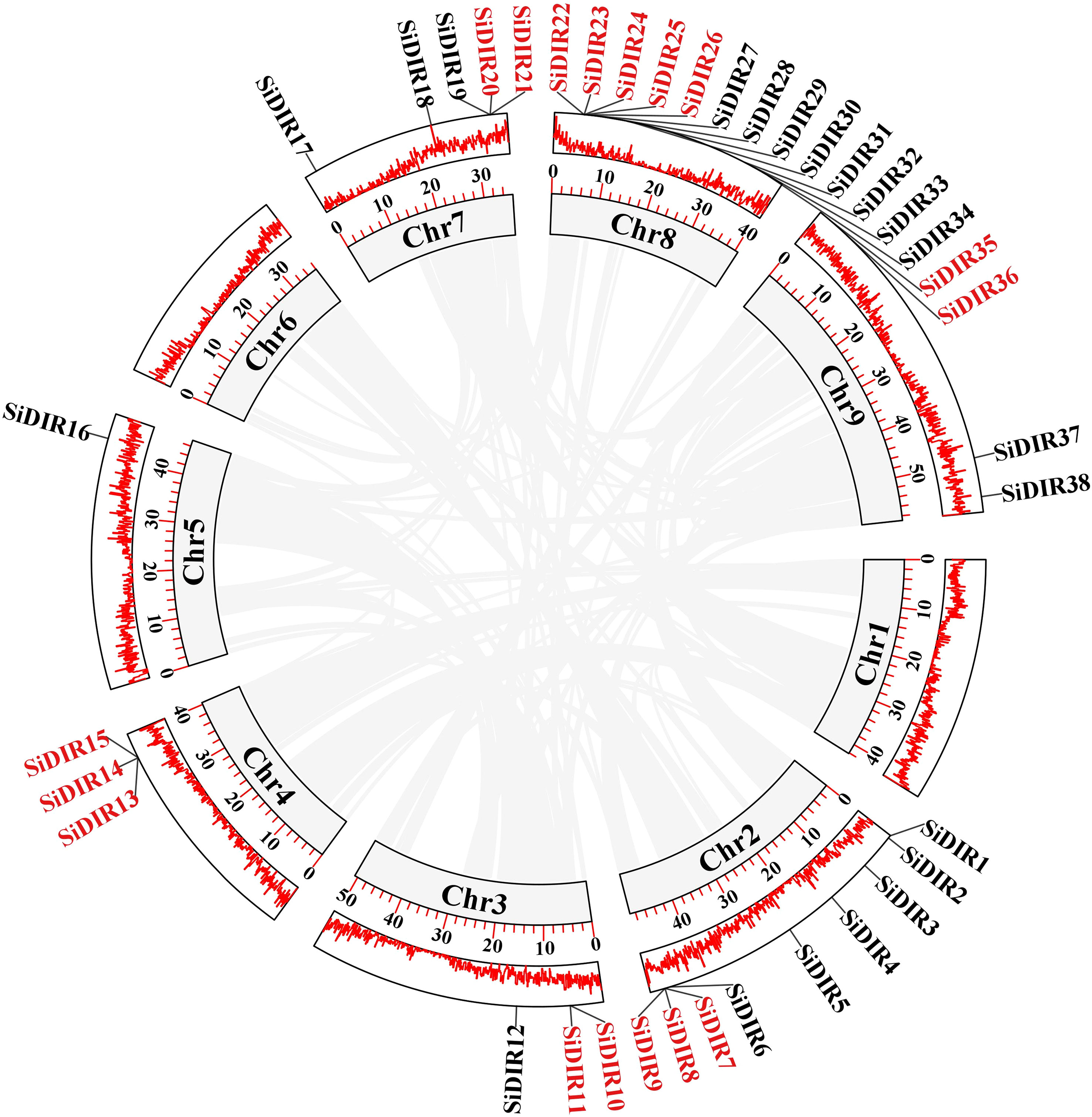
Figure 4 The gene location and duplication events of SiDIR genes. The tandem duplicated genes are indicated in red color.
Tandem duplication (TD) and whole-genome duplication (WGD)/segmental duplication (SD) of genes drive the evolution and expansion of gene family (Weidenbach et al., 2016; Zhang et al., 2022; Guo et al., 2023). We analyzed the duplication events of SiDIR genes and found 10 tandem duplication events involving 17 SiDIR genes on Chr2, 3, 4, 7, and 8 (Figure 4; Supplemental Data 3). The genes of SiDIR7/8/9, SiDIR10/11, SiDIR13/15, SiDIR20/21, and SiDIR25/26 in tandem duplication events were from the same subgroup (Figure 4), indicating the accuracy of the group division of the phylogenetic tree. However, no genome duplication (WGD)/SD events were found. These results revealed that TD events contribute largely to expanding SiDIR gene family.
Meanwhile, we counted the Ka, Ks, and Ka/Ks ratios of the duplication gene pairs using DNASP to analyze the evolutionary selection of duplication pairs in SiDIR gene family. We found that the Ka/Ks ratios of most gene pairs were less than 1 (Supplemental Data 3), implying that these SiDIRs have undergone negative selection. Only two gene pairs, SiDIR13 and SiDIR14, and SiDIR23 and SiDIR24 (Supplemental Data 3) had Ka/Ks ratios greater than 1, indicating that SiDIR13 and SiDIR14, and SiDIR23 and SiDIR24 may undergo positive selection and that they are important for the evolution of millets.
2.6 Collinearity analysis of SiDIRs
To investigate more deeply the evolution mechanisms of SiDIR genes, 12, 16, 14, and 8 ortholog DIR gene pairs were identified when compared millets with Arabidopsis thaliana, Oryza sativa, Zea mays, and G. max, respectively (Figure 5; Supplemental Data 4). Interestingly, we found that some SiDIR genes were identified to be associated with at least three homologous gene pairs, such as SiDIR6 to AT4G11180.1, AT4G23690.1 and AT5G42500.1; SiDIR37 to AT1G65870.1, AT2G28670.1, AT2G39430.1 and AT3G55230.1; SiDIR3 to Glyma.07G157100.1, Glyma.08G258300.1, Glyma.18G282600.1 and Glyma.18G207700.1 (Figure 5; Supplemental Data 4). Strikingly, some collinear gene pairs identified between Si and Os, and Zm were not found between Si and Os, and Zm were not found between Si and At, and Gm, such as SiDIR7/LOC_Os07g44250.1, and SiDIR7/Zm00001d022270 (Figure 5; Supplemental Data 4). Some collinear gene pairs identified between Si and At were not found between Si and Os, Gm and Zm, such as SiDIR10/AT3G16450.1. We speculated that these orthologous genes may exert vital roles during the evolutionary process of DIRs.
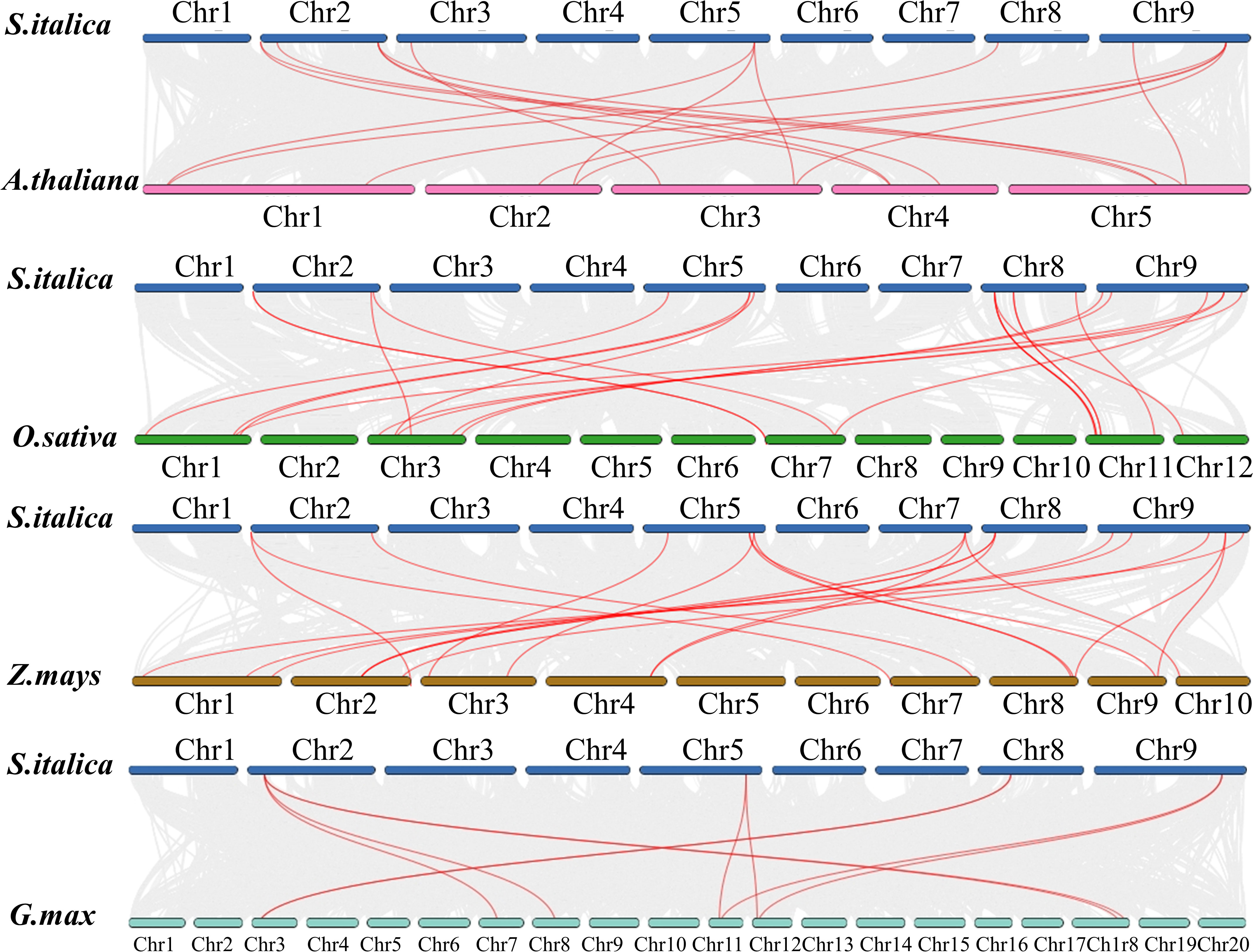
Figure 5 Syntenic analysis between SiDIRs and the DIR genes of Arabidopsis thaliana, Oryza sativa, Zea mays, and Glycine max. The collinear blocks are shown by gray lines, while the syntenic DIR homologous gene pairs are highlighted by red lines. “Chr1–20” means the chromosome number.
2.7 cis-Element analysis of SiDIRs
To predict the functions and regulatory mechanisms of SiDIR genes, the cis-elements within their promoters were analyzed. A total of 23 different cis-elements in the promoter of SiDIRs were detected. These include cis-elements involved in phytohormone response (ABRE: abscisic acid, AuxRR-core: auxin, GARE-motif: gibberellin, CGTCA-motif: methyl jasmonate, and TCA-element: salicylic acid), light response, developmental regulation (circadian control, endosperm, flavonoid biosynthesis, and meristem), and environmental stress (defense and stress, drought, and low temperature) (Figure 6). In plants, these cis-elements regulate various signaling pathways, hinting at the complicated regulatory function of SiDIRs.
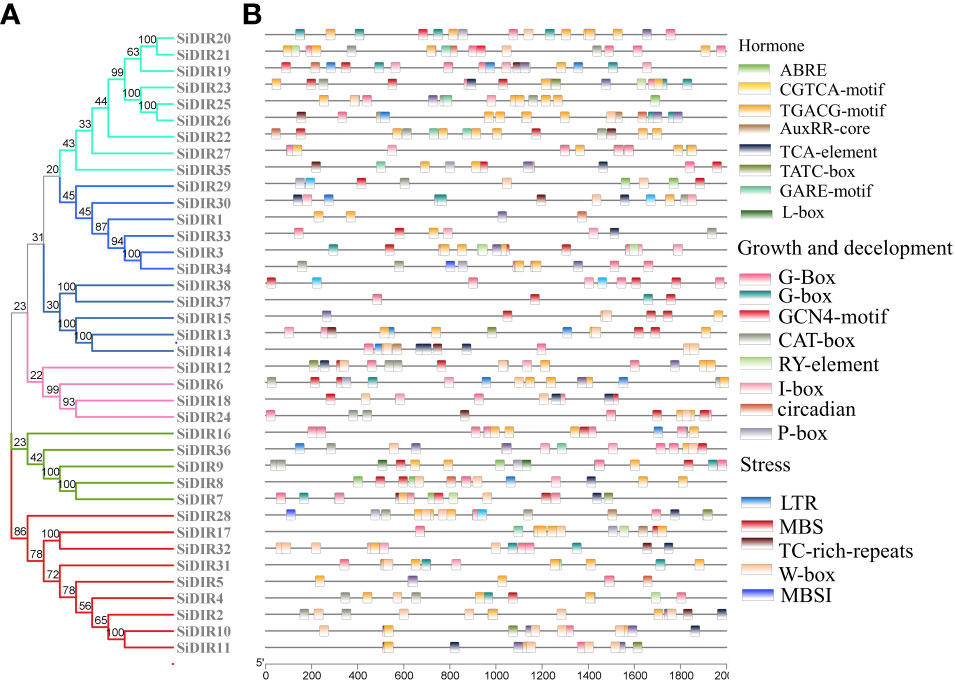
Figure 6 cis-Element analysis of SiDIRs. (A) Phylogenetic tree of SiDIRs. (B) cis-Element analysis of SiDIR genes; the colorful boxes represent different cis-elements.
2.8 Interaction network of SiDIR proteins
In order to further clarify the functions and regulatory pathways among DIR family, a protein–protein interaction network of AtDIR family was analyzed and predicted by STRING software (Figure S1). Nearly all of the AtDIR proteins could interact with other members. AtDIR9 (ESB1), At4g13580, and At2g39430 were central to the interaction network. At4g13580 might interact with AtDIR9 (ESB1), At2g39430, At5g42500, At3g24020, At4g11190, and AtDIR6. Similarly, At4g11190 may also interact with At3g24020 and AtDIR6. The interaction protein numbers of At3g13650, At1g22900, and At5g42510 might be the least. Interestingly, the interaction between At5g42510 and At2g39430 might be strong (Figure S1). According to the protein similarity, we inferred that SiDIR family members might have a similar protein–protein interaction network to that of AtDIRs.
2.9 The expression analysis of SiDIRs in five different tissues
To gain insights into the functions of SiDIRs, the expression patterns of SiDIRs were analyzed based on RNA‐seq data. A total of 16 SiDIR genes including SiDIR1, SiDIR10, SiDIR11, SiDIR17, SiDIR19, SiDIR20, SiDIR21, SiDIR22, SiDIR23, SiDIR24, SiDIR26, SiDIR27, SiDIR31, SiDIR36, SiDIR37, and SiDIR38 expressed higher in root than other tissues (Figure 7; Supplemental Data 5). Unlike the expression of SiDIR12, which was detected mainly in the stem and panicle, that of SiDIR3 was expressed mainly in the stem and root (Figure 7; Supplemental Data 5). SiDIR10, SiDIR11, SiDIR19, and SiDIR31 are expressed only in root. Additionally, there were no transcripts of SiDIR5, SiDIR7, SiDIR9, SiDIR18, SiDIR28, SiDIR29, and SiDIR32 being detected in the organs of flag leaf, stem, root, panicle, and mesophyll (Figure 7; Supplemental Data 5). These results hinted at the essential regulatory function of SiDIRs in plant development.
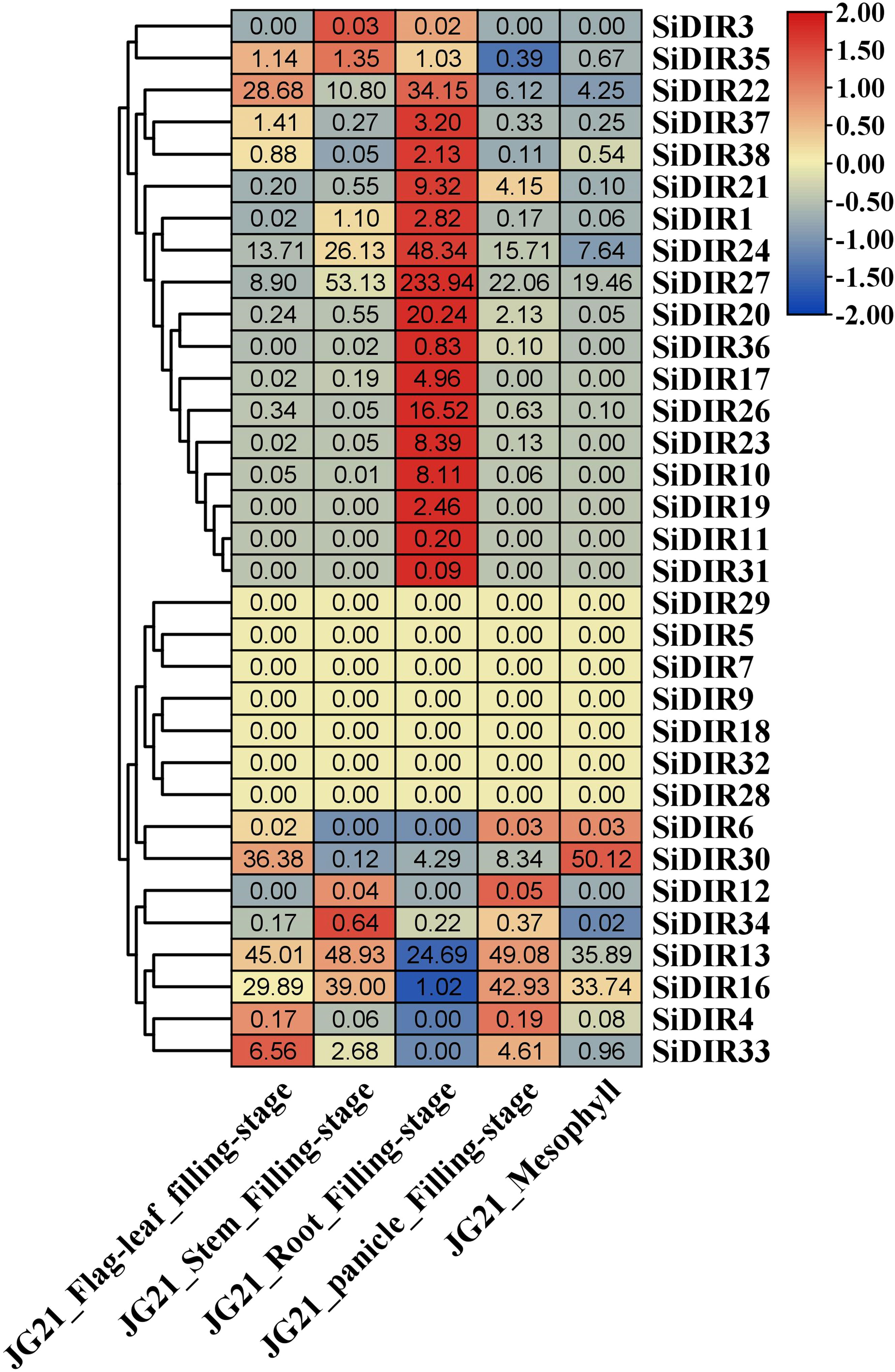
Figure 7 The heatmap of the expression profiles of SiDIRs in five tissues (Flag leaf, Stem, Root, Panicle, and Mesophyll). TPM values of SiDIRs were transformed by log2, and the heatmap was created by the software of TBtools.
2.10 Expression analysis of SiDIRs under CaCl2, NaCl, CdCl, and PEG6000 treatments
To further characterize the SiDIR genes in response to abiotic stresses, the millets were treated with CaCl2, NaCl, CdCl, and PEG6000, and the transcriptional analysis of six SiDIRs in roots was carried out. After treatment with 20 mM of CaCl2, the transcriptional level of SiDIR19 and SiDIR36 reached the maximum at 24 h, while SiDIR10 and SiDIR20 reached the maximum at 48 h (Figure 8A). Specifically, the expression level of SiDIR10 (at 48 h) and SiDIR36 (at 24 h) was approximately eight times higher than that of control. Unlike this, the relative expression of SiDIR22 and SiDIR27 was downregulated (Figure 8A). For 1 mM CdCl treatment, the transcription level of SiDIR10/19/20 was upregulated, while SiDIR22/27 was significantly downregulated. However, the expression induction of SiDIR36 was not obvious under CdCl treatment (Figure 8B). Upon treatment with 150 mM of NaCl, the expression of SiDIR10/19/22/27/36 was upregulated at 24 h and 48 h, and SiDIR20 was upregulated only at the time point of 48 h (Figure 8C). In addition, the expression levels of SiDIR19/20/22/27/36 were upregulated after being treated with 10% PEG6000 for 24 h, and SiDIR10 was not upregulated until being treated for 48 h (Figure 8D). These results showed that different SiDIR genes with different expression patterns may function differently during millet growth.
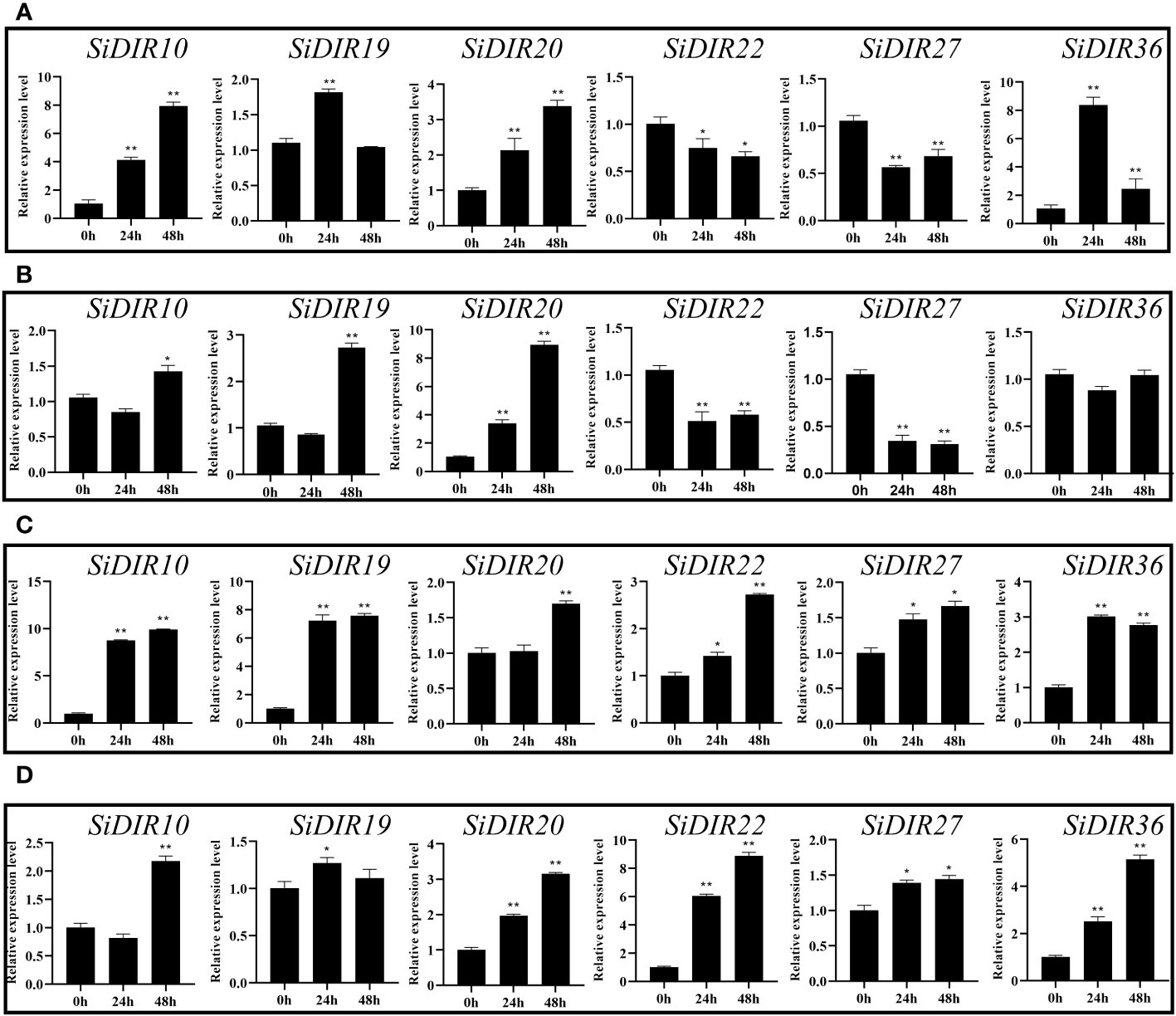
Figure 8 Expression patterns of six SiDIR genes under abiotic stresses. (A) 20 mM CaCl2 treatment, (B) 1 mM CdCl treatment, (C) 150 mM NaCl treatment, and (D) 10% PEG6000 treatment. The expression levels were calculated and shown as means ± SDs (n = 3). Statistically significant differences were analyzed by Student’s t-test (* p < 0.05, ** p < 0.01).
2.11 Gene co-expression analysis
Co-expression analysis can help find genes that were closely co-regulated during the physiological process. Based on the MDSI database (Li et al., 2023), we constructed co-expression networks centered on the SiDIR10, SiDIR19, SiDIR20, SiDIR22, SiDIR27, and SiDIR36. As shown in Figure 9, we obtained a total of six co-expression networks. Among them, the network centered on SiDIR19 is the largest (including 21 genes). In contrast, the network centered on SiDIR20 and SiDIR27 is the smallest (including one gene).
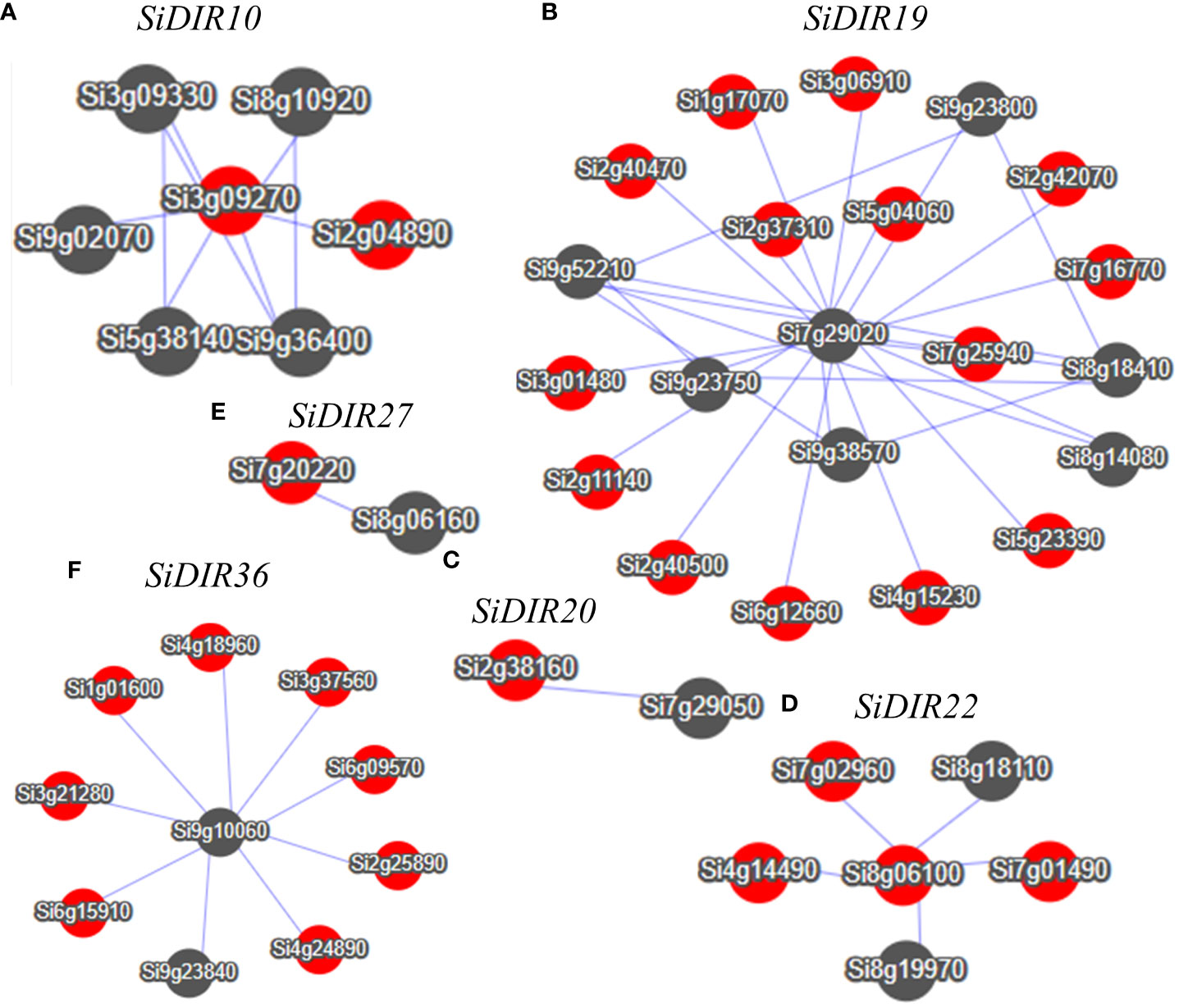
Figure 9 Co-expression network of SiDIR10 (A), SiDIR19 (B), SiDIR20 (C), SiDIR22 (D), SiDIR27 (E), and SiDIR36 (F). Dots represent genes, and lines indicate that they have co-expression relationship.
As shown in Supplemental Data 6, the co-expressed gene network centered on SiDIR10 is significantly enriched in the carbohydrate metabolic process, oxidation-reduction process, ion binding, response to cadmium ion, and oxidation-reduction process. The network centered around SiDIR19 showed significant enrichment in disease resistance, sterol biosynthetic process, malate transport, metal ion binding, response to oxidative stress, cell wall biogenesis, cell wall organization process, response to salt stress, response to abscisic acid, phosphate starvation, and response to stimulus process. Meanwhile, the network centered on SiDIR36 showed significant enrichment in response to water deprivation, ion binding, lignin catabolic process, cell wall biogenesis, and carbohydrate metabolic process. Overall, these findings present an interesting phenomenon that warrants further investigations.
2.12 Subcellular localization of SiDIR7/19/22
To deeply analyze the function of SiDIRs, the fusion expression vectors of 35S-SiDIR7-YFP, 35S-SiDIR19-YFP, and 35S-SiDIR22-YFP were constructed, with an empty vector of 35S-YFP used as a negative control and DAPI fluorescence signal used to indicate the nucleus. These vectors were transferred into the leaves of Nicotiana tabacum L., and the fluorescence was observed by ×20 laser confocal microscopy. The negative YFP signal was expressed in the membrane and nucleus, while the fluorescence signals of 35S-SiDIR7-YFP, 35S-SiDIR19-YFP, and 35S-SiDIR22-YFP were mainly expressed in the cell membrane (Figure 10). This was coherent with the predicted analysis (Supplemental Data 1).
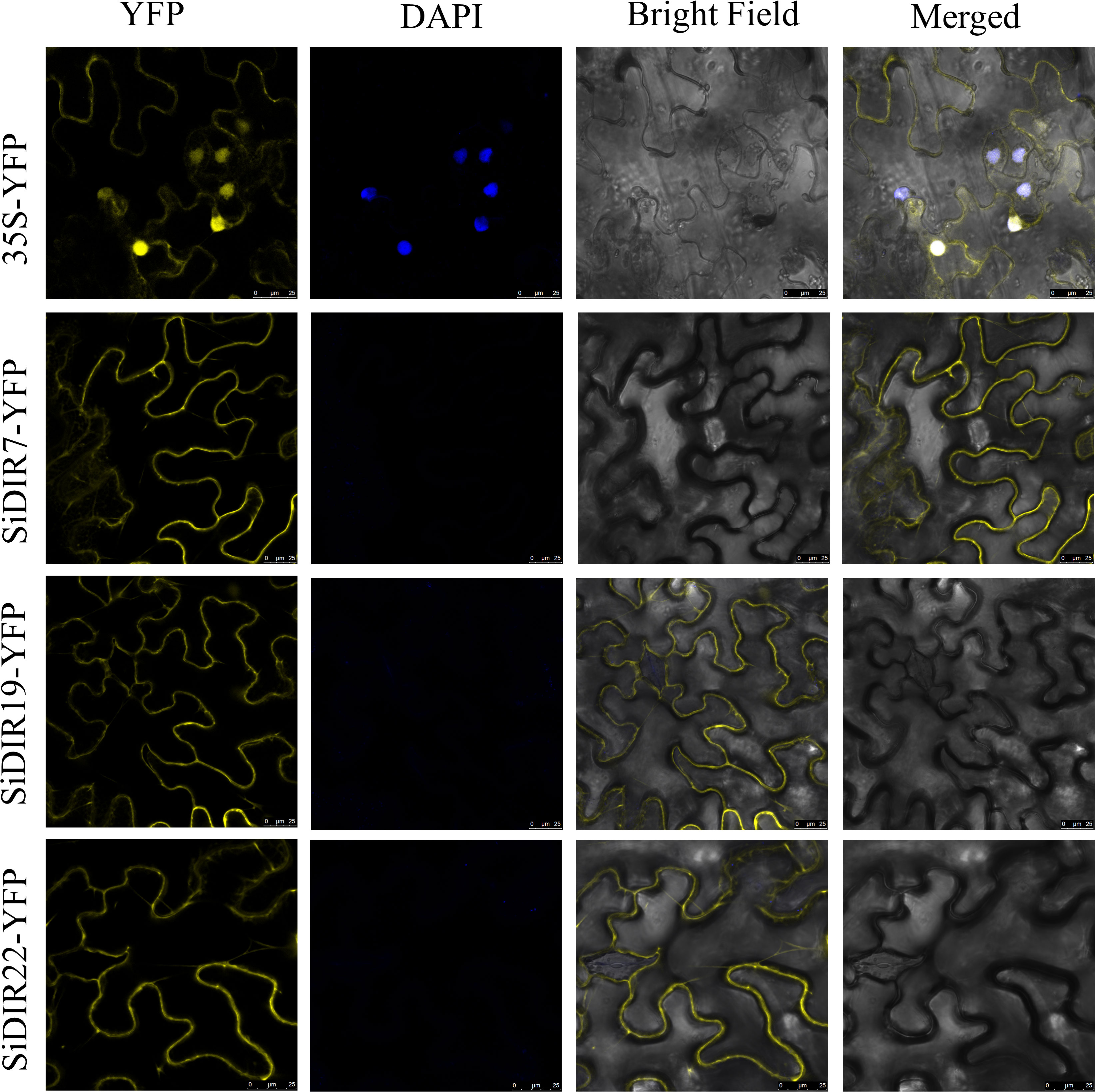
Figure 10 Subcellular localization analysis of SiDIR7/19/22 in Nicotiana tabacum L. leaves. Membrane localization of SiDIR7/19/22 was observed and confirmed by ×20 laser confocal microscopy, and 35S-YFP was the negative control. The blue fluorescence signal of DAPI indicated the nucleus. Scale bars: 25 µm.
3 Discussion
Dirigent proteins are ubiquitous in all vascular plants, such as ferns, gymnosperms, and angiosperms (Davin and Lewis, 2000; Ralph et al., 2007; Li et al., 2014). Although DIR genes have been identified in many species including rice, cotton, I. indigotica, and pepper (Li et al., 2014; Paniagua et al., 2017; Khan et al., 2018; Liu et al., 2021; Duan et al., 2023), SiDIR gene family has not been comprehensively analyzed. In this study, we completely identified 38 DIR proteins that belonged to six groups in S. italica, investigated their evolutionary events of TD duplication, and explored their functional potential during root development and expression diversity when responding to various abiotic stresses.
Phylogenetic analysis suggested that DIR proteins of millet, Arabidopsis, rice, soybean, maize, cotton, etc., were divided into DIR-a, DIR-b/d, DIR-c, DIR-e, DIR-f, and DIR-g subgroups (Figure 1), which was consistent with the previous studies (Ralph et al., 2007; Khan et al., 2018; Duan et al., 2023). Specifically, the subgroup DIR-e contained the most DIRs (Figure 1). Members in the same subgroups have higher sequence similarity (Figure 1). Our conserved domain research showed that the dirigent superfamily domain only appeared in AtDIR25, SiDIR12, and SiDIR36 (Figure 2C). The gene structure analysis displayed the range of the introns from one to four, while the exons were from one to five (Figure 2D). This may be revealed that DIR genes gain or lose exons or introns during the process of chromosomal rearrangements.
A further amino acid alignment analysis (Figure 3A) displayed and confirmed the classification of SiDIRs (Figure 1). It has been shown that DIR-a subfamily members contain the well-characterized 8–8′-linked-lignan forming dirigent proteins. For example, AtDIR6 and LuDIR5/6 participate in the formation of (−)-pinoresinol (Dalisay et al., 2015; Gasper et al., 2016). SiDIR7, SiDIR8, and SiDIR9 were closely related to LuDIR5/6 (Figure 1B). SiDIR7, SiDIR8, and SiDIR9 contained the necessary conserved residues of alanine (A), phenylalanine (F), and leucine (L) that could form (−)-pinoresinol (Figure 3A). This analysis indicates that SiDIR7, SiDIR8, and SiDIR9 might be involved in the formation of (−)-pinoresinol. However, whether SiDIR7/8/9 influences crop resistance to biotic stress, like GmDIR22 or GhDIR1 by promoting lignan accumulation, deserves further investigations (Shi et al., 2012; Li et al., 2017). Except for this, the predicted tertiary structure of SiDIR7/8/9 (Figures 3B–G) was similar to that of AtDIR6 (Gasper et al., 2016), which illustrated the possibility of SiDIR7, SiDIR8, and SiDIR9 to form trimers, respectively. In addition, homologous proteins might have similar functions in different species. AtESB1 was necessary for the formation of the Casparian strip in roots (Hosmani et al., 2013). SiDIR37 was closely classified into the same subgroup as AtESB1 (Figure 1B), suggesting that SiDIR37 is potentially involved in Casparian strip formation. Also, impaired lignin deposition resulted in a defective CS barrier in the mutants of ZmESBL, thus increasing Na+ transport and salt sensitivity (Wang et al., 2022). Our future work will be focused on exploring the function of SiDIRs when moderating salt resistance.
The number of SiDIR genes (Supplemental Data 1) was more than that in Arabidopsis (Paniagua et al., 2017), pepper (Khan et al., 2018), and I. indigotica (Li et al., 2014). Conversely, it was less than that in rice, Gossypium barbadense, and Gossypium hirsutum, in which TD and SD/WGD may be the main driving force behind DIR gene family expansion (Liu et al., 2021; Duan et al., 2023). According to our analysis of gene duplication events and collinearity analysis, a total of 17 TD SiDIR genes were identified, although no SD/WGD was found (Figures 4, 5). This revealed that tandem duplication may play an important role in expanding SiDIR gene family. Also, the number of tandem duplication genes (Figure 4) is considerable in the subgroups of DIR-f (Figure 1B), while the tandem duplication of the DIR-b/d group contributed to the expansion in pepper, cotton, spruce, and flax (Corbin et al., 2018; Khan et al., 2018; Liu et al., 2021; Duan et al., 2023). Based on the evolutionary functions of tandem duplication (Hanada et al., 2008), it is reasonable to infer that the rapid expansion of the DIR-f subfamily may be the adaptive evolution of millet. Interestingly, SiDIR genes were distributed unevenly on the chromosomes (except for chromosomes 1 and 6), and only SiDIR16 was located on chromosome 5 (Figure 4). Because of the DIR duplication gene pairs of cotton and other plants (Liu et al., 2021), it is not surprising to find that the Ka/Ks ratios of most SiDIR duplication gene pairs were under 1 (Supplemental Data 3, 4), which illustrated that the duplication gene pairs in millet were under purifying selection.
The diverse cis-elements of SiDIRs (Figure 6) could partially explain the diversified function of DIRs during plant development and plant defense against biotic and abiotic stresses (Baxter et al., 2009; Hosmani et al., 2013; Yang et al., 2021; Yonekura-Sakakibara et al., 2021; Wang et al., 2022).
It has been reported that 60% of the AtDIR genes show higher expression levels in roots compared with other organs (Paniagua et al., 2017). In millet, we found that approximately half of SiDIR genes similarly displayed higher expression levels in the root tissues (Figure 7). The expression pattern of SiDIRs is similar to that of most of the OsDIRs (Duan et al., 2023). In contrast, only a part of VrDIR genes was highly expressed in roots (Xu et al., 2021), implying the functional conservation and divergence of some DIR genes in different species. Considering universal DIR genes vary their number greatly in vascular plants, we supposed that DIRs are possibly the key family for aquatic plants to land.
Plants deal with abiotic stress to adapt to the circumstances and keep growing. Previous studies have confirmed that the expression of DIRs could respond to salt stress (Paniagua et al., 2017; Khan et al., 2018; Wang et al., 2022). For example, AtESB1 is involved in regulating the concentration of Na, S, K, As, Fe, Ca, Mn, and Zn in shoots (Baxter et al., 2009); VrDIRs are required in salt stress adjustment (Xu et al., 2021); the expression of ScDIR genes was induced by NaCl and PEG treatments (Guo et al., 2012); the transcription levels of CaDIR4/7/12 were significantly regulated by NaCl or mannitol treatment (Khan et al., 2018). We found that the expression levels of SiDIR10/19/20/22/27/36 could be induced by NaCl treatment (Figure 8). This hinted at the possibility of SiDIR10/19/20/22/27/36 to be potential candidate genes coping with salt stress.
Lignin, deposited mostly in the secondary cell walls of vascular plants, contributes to water transport and plant stress responses (Hu et al., 2019; Oliveira et al., 2020; Dabravolski and Isayenkov, 2023). Combining the results in Supplemental Data 6, the co-expression network of SiDIR36 illustrates its possible involvement in responding to salt or osmotic stresses by regulating lignin deposited in the cell walls. Unlike other family members, the expression levels of SiDIR22/27 were downregulated when treated with CaCl2 and CdCl (Figure 8). We speculated that SiDIR22 and SiDIR27 may play synergistic regulation roles. Additionally, the co-expression network centered around SiDIR27 exhibited significant enrichment in response to cytokinin and auxin-activated signaling pathways (Figure 9, Supplemental Data 6), indicating that SiDIR27 may play a role in responding to salt stresses through plant hormone signal transduction. These results clarify the distinct and diverse function of DIRs under abiotic stresses. Meanwhile, the membrane localization of SiDIRs (Figure 10) was consistent with the prediction (Supplemental Data 1) and further revealed their potential vital roles during plant growth and development.
4 Materials and methods
4.1 Plant materials and treatments
Yugu 1 was used as the experimental material in this study. Millets were grown in a greenhouse in Wuhan, Hubei Province, China. Millets were grown in Hoagland nutrient solution (Li et al., 2022). For CaCl2, NaCl, CdCl, and PEG6000 treatments, millet seedlings were grown in Hoagland solution for 10 days and then treated with 20 mM of CaCl2, 150 mM of NaCl, 1 mM of CdCl, and 10% PEG6000, respectively. Plant roots were collected after treatment for 0 h, 24 h, and 48 h. Three biological replicates were carried out for each treatment.
4.2 Data sources and identification of DIRs in different species
The genome data of A. thaliana, S. italica, Z. mays, G. max, and O. sativa spp. japonica were downloaded from Ensembl Plants (http://plants.ensembl.org/index.html/). The AtDIR protein sequence was downloaded from TAIR (https://www.Arabidopsis.org/). The hidden Markov Model (HMM) file of the dirigent domain (PF03018) was downloaded as reported previously (Duan et al., 2023). HMMER 3.0 (E-value ≤ 1e−5, similarity > 50%) was used to search the DIR protein from the S. italica protein database. Further, based on the BLASTP method, we searched SiDIR protein sequences using AtDIR protein sequences (E-value ≤ 1e−5, similarity > 50%). All candidate DIR protein sequences were used to verify the DIR domain as analyzed previously (Duan et al., 2023). The longest transcript was obtained using the R package seqfinder (https://github.com/yueliu1115/seqfinder).
4.3 Phylogenetic analysis of SiDIRs and AtDIRs
The phylogeny tree of identified SiDIRs and DIRs of rice, Arabidopsis, maize, soybean, cotton, etc., were constructed using the neighbor-joining (NJ) method of MEGA7.0 (bootstrap: 1,000 replications) (Kumar et al., 2016). The website of iTOL (Interactive Tree of Life, https://itol.embl.de/) was used to enhance the evolutionary tree.
4.4 Sequence alignment and the tertiary structure prediction of SiDIR7/8/9
The sequence alignment of DIR proteins was carried out by ClustalW, and ESPript 3.0 (https://espript.ibcp.fr/ESPript/ESPript/) was used to illustrate the conserved residues. For the prediction of tertiary structure, the protein sequences were input and analyzed through homologous modeling in SWISS-MODEL (https://swissmodel.expasy.org/).
4.5 Gene structure and conserved motif analysis
The conserved motifs of S. italica DIR proteins were determined by MEME (http://meme-suite.org/) with a conserved motif number of 10. The gene structure information was acquired from GFF data. The conserved domains were obtained from NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and were subsequently visualized using TBtools software (Chen et al., 2020).
4.6 Gene duplication events and the analysis of Ka/Ks ratios
Segmental and tandem duplications were detected by MCScanX (Wang et al., 2012). The non-synonymous (Ka)/synonymous (Ks) ratios of duplication gene pairs were calculated using TBtools software. TBtools software was used to visualize the duplication events (Chen et al., 2020). The divergence time of all duplicate gene pairs was estimated as previously (Deng et al., 2019).
4.7 Expression pattern analysis of SiDIRs using RNA-seq
Gene expression level data of different tissues were downloaded from MDSi: Multi-omics Database for S. italica (http://foxtail-millet.biocloud.net/page/tools/expressionVisualization) (Yang et al., 2020; Li et al., 2023). The heatmap was generated by TBtools software (Chen et al., 2020).
4.8 RNA extraction and quantitative real-time PCR
The primer 5.0 software was used to design specific primers for SiDIR genes in this study (Supplemental Data 7). Total RNA was extracted using the KKFast Plant RNApure Kit (ZOMANBIO, ZP405K-2). The cDNA was synthesized by PrimerScript™ IV 1st strand cDNA Synthesis Mix (TaKaRa, Mountain View, CA, USA; 6215A). The quantitative real-time PCR (RT-qPCR) system program was performed according to the previous research (Duan et al., 2023). The gene expression was analyzed by the 2−ΔΔCT method as used in a previous study (Gong et al., 2022).
4.9 Subcellular localization analysis
The CDS sequence of SiDIR7/19/22 was cloned from the cDNA of S. italica by the primers of SiDIR7-F/R, SiDIR19-F/R, and SiDIR22-F/R, respectively. The vector of 35S-YFP was digested with BamHI. The amplified products were then inserted into the linearized carrier of 35S-YFP by the kit of ClonExpress® MultiS One Step Cloning (Vazyme, Nanjing, China; C113) and verified by DNA sequencing. These four vectors were transformed into Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures harboring each construct were resuspended and mixed before being infiltrated into Nicotiana benthamiana leaves as described previously (Gong et al., 2022). After 48 h, the fluorescence signals were detected using a Leica TCS SP8 confocal microscope, and images were captured by LAS‐X software (Leica, Wetzlar, Germany).
5 Conclusions
In summary, 38 SiDIR gene family members were identified. We investigated the important role of SiDIR genes through the analysis of gene structure, evolutionary history, tertiary structure, cis-elements, stress responses, protein interaction, co-expression network, subcellular localization, and potential function. This study provides significant evidence and profound insights into the functional diversity of DIR proteins. Millet, a model for C4 photosynthesis, is one of the most traditional staple foods and the most economical and important source of energy for humans. This research may lay the foundation and pave a new way for improving the abiotic tolerance and agronomic traits of millet.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
LG, BL, and TZ conceived the idea. LG and BX wrote the first draft. LG and BX corrected the paper to the present form. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Doctoral research initiation fund, grant no. K-Q2023022.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1243806/full#supplementary-material
Supplementary Figure 1 | Predicted protein-protein interaction network for SiDIR based on their orthologs of AtDIR.
Supplementary Data Sheet 2 | Multi sequence alignment within the conserved motifs of SiDIRs.
Supplementary Data Sheet 7 | Primer sequences used in this study.
References
Baxter, I., Hosmani, P. S., Rus, A., Lahner, B., Borevitz, J. O., Muthukumar, B., et al. (2009). Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PloS Genet. 5, e1000492. doi: 10.1371/journal.pgen.1000492
Brutnell, T. P., Wang, L., Swartwood, K., Goldschmidt, A., Jackson, D., Zhu, X. G., et al. (2010). Setaria viridis: A model for C4 photosynthesis. Plant Cell 22, 2537–2544. doi: 10.1105/tpc.110.075309
Burlat, V., Kwon, M., Davin, L. B., Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897. doi: 10.1016/S0031-9422(01)00117-0
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Corbin, C., Drouet, S., Markulin, L., Auguin, D., Lainé, É., Davin, L. B., et al. (2018). A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: from gene identification and evolution to differential regulation. Plant Mol. Biol. 97, 73–101. doi: 10.1007/s11103-018-0725-x
Dabravolski, S. A., Isayenkov, S. V. (2023). The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 14, 1118313. doi: 10.3389/fpls.2023.1118313
Dalisay, D. S., Kim, K. W., Lee, C., Yang, H., Rübel, O., Bowen, B. P., et al. (2015). Dirigent protein-mediated lignan and cyanogenic glucoside formation in flax seed: integrated omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 78, 1231–1242. doi: 10.1021/acs.jnatprod.5b00023
Davin, L. B., Lewis, N. G. (2000). Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123, 453–462. doi: 10.1104/pp.123.2.453
Davin, L. B., Lewis, N. G. (2005). Lignin primary structures and dirigent sites. Curr. Opin. Biotechnol. 16, 407–415. doi: 10.1016/j.copbio.2005.06.011
Davin, L. B., Wang, H. B., Crowell, A. L., Bedgar, D. L., Martin, D. M., Sarkanen, S., et al. (1997). Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–367. doi: 10.1126/science.275.5298.362
Deng, X., An, B., Zhong, H., Yang, J., Kong, W., Li, Y. (2019). A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 10, 239. doi: 10.3390/genes10030239
Doust, A. N., Kellogg, E. A., Devos, K. M., Bennetzen, J. L. (2009). Foxtail Millet: a sequence-driven grass model system. Plant Physiol. 149, 137–141. doi: 10.1104/pp.108.129627
Duan, W., Xue, B., He, Y., Liao, S., Li, X., Li, X., et al. (2023). Genome-wide identification and expression pattern analysis of dirigent members in the Genus Oryza. Int. J. Mol. Sci. 24, 7189. doi: 10.3390/ijms24087189
Effenberger, I., Harport, M., Pfannstiel, J., Klaiber, I., Schaller, A. (2017). Expression in Pichia pastoris and characterization of two novel dirigent proteins for atropselective formation of gossypol. Appl. Microbiol. Biotechnol. 101, 2021–2032. doi: 10.1007/s00253-016-7997-3
Effenberger, I., Zhang, B., Li, L., Wang, Q., Liu, Y., Klaiber, I., et al. (2015). Dirigent proteins from cotton (Gossypium sp.) for the atropselective synthesis of gossypol. Angewandte Chemie 54, 14660–14663. doi: 10.1002/anie.201507543
Gasper, R., Effenberger, I., Kolesinski, P., Terlecka, B., Hofmann, E., Schaller, A. (2016). Dirigent protein mode of action revealed by the crystal structure of AtDIR6. Plant Physiol. 172, 2165–2175. doi: 10.1104/pp.16.01281
Gong, L., Liao, S., Duan, W., Liu, Y., Zhu, D., Zhou, X., et al. (2022). OsCPL3 is involved in brassinosteroid signaling by regulating OsGSK2 stability. J. Integr. Plant Biol. 64, 1560–1574. doi: 10.1111/jipb.13311
Guo, L., Lu, S., Liu, T., Nai, G., Ren, J., Gou, H., et al. (2023). Genome-wide identification and abiotic stress response analysis of PP2C gene family in Woodland and pineapple strawberries. Int. J. Mol. Sci. 24, 4049. doi: 10.3390/ijms24044049
Guo, J. L., Xu, L. P., Fang, J. P., Su, Y. C., Fu, H. Y., Que, Y. X., et al. (2012). A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 31, 1801–1812. doi: 10.1007/s00299-012-1293-1
Hanada, K., Zou, C., Lehti-Shiu, M. D., Shinozaki, K., Shiu, S. H. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003. doi: 10.1104/pp.108.122457
Hosmani, P. S., Kamiya, T., Danku, J., Naseer, S., Geldner, N., Guerinot, M. L., et al. (2013). Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. 110, 14498–14503. doi: 10.1073/pnas.1308412110
Hu, P., Zhang, K., Yang, C. (2019). BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol. 179, 700–717. doi: 10.1104/pp.18.01167
Huwa, N., Weiergräber, O. H., Fejzagić, A. V., Kirsch, C., Schaffrath, U., Classen, T. (2022). The crystal structure of the defense conferring rice protein OsJAC1 reveals a carbohydrate binding site on the dirigent-like domain. Biomolecules 12, 1126. doi: 10.3390/biom12081126
Huwa, N., Weiergräber, O. H., Kirsch, C., Schaffrath, U., Classen, T. (2021). Biochemical and initial structural characterization of the monocot chimeric jacalin OsJAC1. Int. J. Mol. Sci. 22, 5639. doi: 10.3390/ijms22115639
Khan, A., Li, R. J., Sun, J. T., Ma, F., Zhang, H. X., Jin, J. H., et al. (2018). Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 8, 5500. doi: 10.1038/s41598-018-23761-0
Kim, M. K., Jeon, J. H., Davin, L. B., Lewis, N. G. (2002). Monolignol radical–radical coupling networks in western red cedar and Arabidopsis and their evolutionary implications. Phytochemistry 61, 311–322. doi: 10.1016/S0031-9422(02)00261-3
Kim, K. W., Moinuddin, S. G. A., Atwell, K. M., Costa, M. A., Davin, L. B., Lewis, N. G. (2012). Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J. Biol. Chem. 287, 33957–33972. doi: 10.1074/jbc.M112.387423
Kim, K. W., Smith, C. A., Daily, M. D., Cort, J. R., Davin, L. B., Lewis, N. G. (2015). Trimeric structure of (+)-pinoresinol-forming dirigent protein at 1.95 Å resolution with three isolated active sites. J. Biol. Chem. 290, 1308–1318. doi: 10.1074/jbc.M114.611780
Kittur, F. S., Lalgondar, M., Yu, H. Y., Bevan, D. R., Esen, A. (2007). Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. J. Biol. Chem. 282, 7299–7311. doi: 10.1074/jbc.M607417200
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, P., Brutnell, T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J. Exp. Bot. 62, 3031–3037. doi: 10.1093/jxb/err096
Li, Q., Chen, J., Xiao, Y., Di, P., Zhang, L., Chen, W. (2014). The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance. BMC Genomics 15, 388. doi: 10.1186/1471-2164-15-388
Li, X. K., Hou, S. Y., Feng, M. M., Xia, R., Li, J. W., Tang, S., et al. (2023). MDSi: multi-omics database for setaria italica. BMC Plant Biol. 27, 223. doi: 10.1186/s12870-023-04238-3
Li, Y., Yu, S., Zhang, Q., Wang, Z., Liu, M., Zhang, A., et al. (2022). Genome-wide identification and characterization of the cct gene family in foxtail millet (Setaria italica) response to diurnal rhythm and abiotic stress. Genes 13, 1829. doi: 10.3390/genes13101829
Li, N., Zhao, M., Liu, T., Dong, L., Cheng, Q., Wu, J., et al. (2017). A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to phytophthora sojae. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01185
Lin, J. L., Fang, X., Li, J. X., Chen, Z. W., Wu, W. K., Guo, X. X., et al. (2023). Dirigent gene editing of gossypol enantiomers for toxicity-depleted cotton seeds. Nat. Plants 9, 605–615. doi: 10.1038/s41477-023-01376-2
Liu, Z., Wang, X., Sun, Z., Zhang, Y., Meng, C., Chen, B., et al. (2021). Evolution, expression and functional analysis of cultivated allotetraploid cotton. DIR genes. BMC Plant Biol. 21, 89. doi: 10.1186/s12870-021-02859-0
Oliveira, D. M., Mota, T. R., Salatta, F. V., Sinzker, R. C., Končitíková, R., Kopečný, D., et al. (2020). Cell wall remodeling under salt stress: insights into changes in polysaccharides, feruloylation, lignification, and phenolic metabolism in maize. Plant Cell Environ. 43, 2172–2191. doi: 10.1111/pce.13805
Paniagua, C., Bilkova, A., Jackson, P., Dabravolski, S., Riber, W., Didi, V., et al. (2017). Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 68, 3287–3301. doi: 10.1093/jxb/erx141
Pickel, B., Constantin, M. A., Pfannstiel, J., Conrad, J., Beifuss, U., Schaller, A. (2010). An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angewandte Chemie. Int. Ed. English 49, 202–204. doi: 10.1002/anie.200904622
Ralph, S. G., Jancsik, S., Bohlmann, J. (2007). Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.). Phytochemistry 68, 1975–1991. doi: 10.1016/j.phytochem.2007.04.042
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Seneviratne, H. K., Dalisay, D. S., Kim, K. W., Moinuddin, S. G. A., Yang, H., Hartshorn, C. M., et al. (2015). Non-host disease resistance response in pea (Pisum sativum) pods: Biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 113, 140–148. doi: 10.1016/j.phytochem.2014.10.013
Shi, H., Liu, Z., Zhu, L., Zhang, C., Chen, Y., Zhou, Y., et al. (2012). Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. (Shanghai) 44, 555–564. doi: 10.1093/abbs/gms035
Uchida, K., Akashi, T., Aoki, T. (2017). The missing link in leguminous pterocarpan biosynthesis is a dirigent domain-containing protein with isoflavanol dehydratase activity. Plant Cell Physiol. 58, 398–408. doi: 10.1093/pcp/pcw213
Wang, Y., Cao, Y., Liang, X., Zhuang, J., Wang, X., Qin, F., et al. (2022). A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat. Commun. 13, 2222. doi: 10.1038/s41467-022-29809-0
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Weidenbach, D., Esch, L., Möller, C., Hensel, G., Kumlehn, J., Höfle, C., et al. (2016). Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular poaceae-specific proteins. Mol. Plant 9, 514–527. doi: 10.1016/j.molp.2015.12.009
Xu, W., Liu, T., Zhang, H., Zhu, H. (2021). Mungbean dirigent gene subfamilies and their expression profiles under salt and drought stresses. Front. Genet. 12, 658148. doi: 10.3389/fgene.2021.658148
Yang, Z., Zhang, H., Li, X., Shen, H., Gao, J., Hou, S., et al. (2020). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants 6, 1167–1178. doi: 10.1038/s41477-020-0747-7
Yang, X., Zhong, S., Zhang, Q., Ren, Y., Sun, C., Chen, F. (2021). A loss-of-function of the dirigent gene TaDIR-B1 improves resistance to Fusarium crown rot in wheat. Plant Biotechnol. J. 19, 866–888. doi: 10.1111/pbi.13554
Yonekura-Sakakibara, K., Yamamura, M., Matsuda, F., Ono, E., Nakabayashi, R., Sugawara, S., et al. (2021). Seed-coat protective neolignans are produced by the dirigent protein AtDP1 and the laccase AtLAC5 in. Arabidopsis. Plant Cell 33, 129–152. doi: 10.1093/plcell/koaa014
Keywords: dirigent gene family, evolution, expression analysis, stress responses, Setaria italica
Citation: Gong L, Li B, Zhu T and Xue B (2023) Genome-wide identification and expression profiling analysis of DIR gene family in Setaria italica. Front. Plant Sci. 14:1243806. doi: 10.3389/fpls.2023.1243806
Received: 21 June 2023; Accepted: 28 August 2023;
Published: 20 September 2023.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Didla Ratna Babu, Acharya N. G. Ranga Agricultural University, IndiaSiyu Hou, Shanxi Agricultural University, China
Dan Liu, Tianjin Academy of Agricultural Sciences, China
Copyright © 2023 Gong, Li, Zhu and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luping Gong, Z2xwQGhuY2ouZWR1LmNu; Baoping Xue, eHVlYmFvcGluZ0B3aHUuZWR1LmNu
 Luping Gong
Luping Gong Bingbing Li1
Bingbing Li1 Baoping Xue
Baoping Xue