
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 August 2023
Sec. Plant Metabolism and Chemodiversity
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1243323
This article is part of the Research TopicPlant Natural Resins: From Formation Mechanism to Ecological SignificanceView all 8 articles
 Hao Zhang1†
Hao Zhang1† Xupo Ding1,2*†
Xupo Ding1,2*† Hao Wang1,2
Hao Wang1,2 Huiqin Chen1,2
Huiqin Chen1,2 Wenhua Dong1,2
Wenhua Dong1,2 Jiahong Zhu1,2
Jiahong Zhu1,2 Jian Wang3
Jian Wang3 Shiqing Peng1,2
Shiqing Peng1,2 Haofu Dai1,2
Haofu Dai1,2 Wenli Mei1,2*
Wenli Mei1,2*Introduction: Agarwood, the dark-brown resin produced by Aquilaria trees, has been widely used as incense, spice, perfume or traditional medicine and 2-(2-phenethyl) chromones (PECs) are the key markers responsible for agarwood formation. But the biosynthesis and regulatory mechanism of PECs were still not illuminated. The transcription factor of basic leucine zipper (bZIP) presented the pivotal regulatory roles in various secondary metabolites biosynthesis in plants, which might also contribute to regulate PECs biosynthesis. However, molecular evolution and function of bZIP are rarely reported in Malvales plants, especially in Aquilaria trees.
Methods and results: Here, 1,150 bZIPs were comprehensively identified from twelve Malvales and model species genomes and the evolutionary process were subsequently analyzed. Duplication types and collinearity indicated that bZIP is an ancient or conserved TF family and recent whole genome duplication drove its evolution. Interesting is that fewer bZIPs in A. sinensis than that species also experienced two genome duplication events in Malvales. 62 AsbZIPs were divided into 13 subfamilies and gene structures, conservative domains, motifs, cis-elements, and nearby genes of AsbZIPs were further characterized. Seven AsbZIPs in subfamily D were significantly regulated by ethylene and agarwood inducer. As the typical representation of subfamily D, AsbZIP14 and AsbZIP41 were localized in nuclear and potentially regulated PECs biosynthesis by activating or suppressing type III polyketide synthases (PKSs) genes expression via interaction with the AsPKS promoters.
Discussion: Our results provide a basis for molecular evolution of bZIP gene family in Malvales and facilitate the understanding the potential functions of AsbZIP in regulating 2-(2-phenethyl) chromone biosynthesis and agarwood formation.
Aquilaria sinensis (Lour.) Spreng. is a special member of Malvales (Thymelaeaceae family) since it is one of the resource plants for agarwood production. This resin is a valuable aromatic ingredient which was used as perfumes and incense in religious rituals and ceremonies for centuries. Nowadays, agarwood is a highly demanded and indispensable ingredient in the perfume industry, and is welcomed as craft productions (Ding et al., 2020). In addition, it is also used as a traditional medicine in Chinese therapies and Ayurveda (Persoon, 2008; Naef, 2011; Ding et al., 2020). High-quality agarwood represents a precious, expensive, and scarce resource since healthy agarwood trees only produce few agarwood in nature unless they were affected by injury, such as burning, gnawing insects, lightning strikes, or fungal infection (Ding et al., 2020). Fungal inoculum is the most effective method for the artificial induction of agarwood production compared with chemical methods and physical methods (Sangareswari et al., 2016; Chhipa et al., 2017). This suggests that microbes play an important role in agarwood formation by priming agarwood tree immunity. Plants produce diverse secondary metabolites against biotic or abiotic stress. The previous research suggested that agarwood is a mixture of defense chemicals from agarwood trees in response to environmental stresses, especially stimulated by microbes pressures (Mohamed et al., 2010; Chhipa et al., 2017).
Terpenes and 2-(2-phenethyl) chromones (PECs) are the vital biological components and the main contributor to agarwood aroma. Previous research also indicated that chromones and their derivatives in the acetone extract from the high-quality agarwood occupied up to 60%, which implies that the contents of chromones and their derivatives might relate to the quality of agarwood (Ishihara et al., 1993). Type III polyketide synthases (PKSs) are pivotal for synthesizing the core flavonoids and their derivatives in the plant kingdom. They might also produce the precursors of chromone or its derivatives. However, its regulatory mechanism is unknown in Aquilaria trees or during agarwood formation. Transcription factors (TFs) play crucial roles in functional gene activation and regulation in model plants and crops. However, the role of TFs in chromones and agarwood formation has gained little attention (Liao et al., 2018; Wang et al., 2022d).
Transcription factors are one of the largest functional classes of proteins in eukaryotic genomes. They regulate nearly all biological processes including growth and development, hormone response, and environmental stress responses in plants. They are also essential in plant secondary metabolite accumulation according to their ability to regulate multiple functional genes in the metabolic pathways and cellular processes (Broun, 2004; Weirauch and Hughes, 2011). Recent studies suggest that TFs might contribute to the interaction between pathogen and Aquilaria trees to generate agarwood (Liu et al., 2022a). Basic leucine zipper proteins (bZIPs) are a ubiquitous family of plant transcription factors that share a basic region composed of a conserved DNA-contacting structure at the N-terminus and a characteristic and unique leucine zipper structure required for TF dimerization (Kouzarides and Ziff, 1989). Most plant bZIPs that prefer to recognize cis regulatory elements have ACGT core sequences, such as G-box (CACGTG), C-box (GACGTC), and A-box (TACGTA) (Foster et al., 1994; Dröge-Laser et al., 2018; Qu et al., 2022). The bZIP gene family (and their dimers thereof) perform a plethora of functions in plant physiological processes (Jakoby et al., 2002; Corrêa et al., 2008; Weirauch and Hughes, 2011).
Previous research identified 78 bZIPs that were sorted into 13 groups based on the phylogenetic tree of the bZIP basic region and predictor of conserved motifs in Arabidopsis thaliana (Dröge-Laser et al., 2018). The functional and regulatory processes of each AtbZIP subfamily were further summarized and highlighted according to extensive experimental data. For example, most AtbZIPs in group D were involved in plant hormone response and increasing plant tolerance for pathogen and xenobiotic stress in the environment (Fonseca et al., 2022). In addition, bZIP helps to regulate flavonoid pathways in A. thaliana, soybean, Ginkgo biloba, apple, or grapevine (Loyola et al., 2016; An et al., 2018; Job et al., 2018; Lian et al., 2018; Zhao et al., 2020). However, most research has focused on bZIPs of subfamily H (also called HY5) or subfamily G interacting with chalcone synthase (CHS) to regulate flavonoid accumulation. The involvement of bZIP in type III PKS expression or chromone content is rarely reported, especially for subgroup D of bZIPs.
The bZIP family of TFs is widely distributed in the plant kingdom and their subfamilies are analyzed in detail in various plants, especially the staple oil or food crops (wheat, rice, maize, Olive, soybean), the model plant (Arabidopsis thaliana) and in horticultural crops (litchi, pear, walnut, pomegranate, Chinese jujube, and sweet potato) (Wei et al., 2012; Zhang et al., 2014; Dröge-Laser et al., 2018; Zhang et al., 2018a; Agarwal et al., 2019; Yang et al., 2019; Rong et al., 2020; Zhang et al., 2020b; Ma et al., 2021; Hou et al., 2022; Wang et al., 2022c; Zhang et al., 2022b). Moreover, the bZIP gene families in medicinal plants such as Carthamus tinctorius, Cannabis sativa, Andrographis paniculata, Isatis indigotica, and Salvia miltiorrhiza were studied in recent years (Zhang et al., 2018b; Li et al., 2020; Guan et al., 2022; Jiang et al., 2022; Lu et al., 2022). Interestingly, bZIPs were also involved in mediated or increased the accumulation of effective ingredients in medicinal plants (Hao et al., 2019). However, the research was limited to the genome of individual species and comprehensive analysis of bZIPs in the same order has received little attention. The differentiation of gene family in the same orders might elucidate species evolution and trait formation, especially for the identification of special paralogous genes. Besides A. sinensis, Malvales contains numerous species that contribute to economic, cultural, or ecological levels, such as cotton (Gossypium raimondii), cocoa (Theobroma cacao), durian (Durio zibethinus), jute (Corchorus capsularis and C. olitorius). Therefore, a comprehensive analysis of bZIPs in A. sinensis and other Malvales plants will provide the evolutionary history and regulatory mechanism of bZIPs in the differentiation of species from Malvales.
This study aimed to identify and compare the bZIP gene family in the A. sinensis genome and eight other Malvales species genomes (D. zibethinus, Hopea hainanensis, G. raimondii, T. cacao, C. capsularis, C. olitorius, Dipterocarpus turbinatus, and Hibiscus cannabinus) to obtain a better picture of the size and evolution of the bZIP family in Malvales. Three outgroup genomes (A. thaliana, Vitis vinifera, and Amborella trichopoda) were used to reveal the evolutionary and differentiation process of the bZIP gene family in Malvales. Sixty-two AsbZIP genes in the A. sinensis genome were subsequently characterized to illustrate their phylogenetic relationship, conservative domains and motifs, gene structures, chromosomal location, duplication modes, collinearity, cis-elements, and sequence alignments between A. sinensis and ten other species. Furthermore, the expression profiles of genes from bZIP subfamily D were investigated by treating A. sinensis stems with ethylene and agarwood-inducer. Eventually, two AsbZIPs genes in subfamily D (AsbZIP14 and AsbZIP41) that potentially participate in the regulation of chromone biosynthesis via activating or inhibiting the expression of AsPKS genes in A. sinensis were further investigated. These systematic results provide a theoretical basis for the further study of evolutionary processes of the bZIP gene family in Malvales and the roles of AsbZIP in agarwood formation and Aquilaria tree response to a wide range of environmental stresses.
Five-year-old A. sinensis trees were collected from the Institute of Tropical Bioscience and Biotechnology (110° 19′ 246 E, 19° 59′ 756 N), Haikou, China. A 5 mm diameter and 1 cm-deep hole was drilled into the A. sinensis tree stem at a 45-degree downward angle from the trunk. Then, the needle of infusion was inserted into the hole and the agarwood inducer was slowly and continuously dripped into the xylem of the stems. The xylem tissues around the hole were taken (500 mg per sample) at 0, 3, 6, and 9 days after treatment. Ethylene (ET) treatment involved wrapping the stems of A. sinensis trees with absorbent cotton moistened with 1% ET and then intertwining them with plastic wrap. The treated tissues of stems were harvested (500 mg per sample) at 0, 3, 6, 12, 24, and 48 h after treatment. The collected samples were immediately frozen in liquid nitrogen and stored at -80°C in the laboratory prior to subsequent studies.
The genome files of D. zibethinus, H. hainanensis, G. raimondii, T. cacao, C. capsularis, C. olitorius, D. turbinatus, H. cannabinus, V. vinifera, A. sinensis, and A. trichopoda were downloaded from JGI (https://phytozome-next.jgi.doe.gov/) and NCBI (Argout et al., 2011; Project et al., 2013; Guan et al., 2014; Zhou et al., 2017; Wang et al., 2019; Ding et al., 2020; Zhang et al., 2020a; Zhang et al., 2021; Wang et al., 2022b; Zuccolo et al., 2011).
The AsbZIPs in genomes were firstly identified by HMMER-3.1 with bZIP domains (PF00170 and PF12498) retrieving from the Pfam library (http://pfam.xfam.org). The homologous identification were used the seed sequences of all 78 AtbZIP proteins in A. thaliana downloaded from the TAIR library (https://www.arabidopsis.org/) to perform a blast search against the A. sinensis genome with e < 10-5 to obtain the protein sequences of AsbZIPs (Dröge-Laser et al., 2018). Then the non-redundant candidate database of AsbZIPs was obtained by combining the sequences from HMMER and homologous identification. The other elven non-redundant candidate databases were built by using the same method. Secondly, constructed the twelve non-redundant candidate databases as the final seed sequences and then aligned in each genome again to obtain the final candidates. These non-redundant candidates were subsequently screened by the NCBI-CDD library (https://www.ncbi.nlm.nih.gov/cdd), eggNOG-mapper library (http://eggnog-mapper.embl.de/), and Pfam database using the default parameters to verify the presence of conserved domains. The candidate proteins with no conserved domains of bZIP were removed and 62 genes were regarded as the final gene family of bZIP genes in A. sinensis.
The phylogenetic trees of species were initially constructed with r8s v 1.70 based on 302 single-copy genes from 12 species using OrthoFinder v 2.5.4. All bZIP protein sequences in 12 species were assembled as the final dataset for phylogenetic tree construction using FastTree v 2.1.11 after alignment with MAFFT v 7.310. The duplication and loss of bZIP genes in Marvales were analyzed using Notung v 2.9.13 based on two phylogenetic trees of species and bZIP protein sequences. The collinearity pairs of bZIP between A. sinensis and other ten species were detected using WGDI v 0.5.9 and visualized with JCVI v 0.8.4 (Goll et al., 2010). The protein alignments were translated into coding sequence alignments using an in-house Perl script of KaKs_calculator 3.0 for Ks (synonymous substitution rate) analysis. Ka (nonsynonymous substitution rate) and Ks values were then calculated based on the coding sequence alignments using the method of model averaging (MA) implemented in the KaKs_calculator 3.0 (Zhang, 2022). The divergence time was calculated with the formula T = Ks/2r. Ks being the synonymous substitutions per site and r represents the rate of divergence for nuclear genes from plants. The r was taken as 1.5×10-8 synonymous substitutions per site per year for dicotyledonous plants by evaluating chalcone synthase and alcohol dehydrogenase sequences using the fossil pollen data (Koch et al., 2000).
The amino acid sequences of AsbZIP were submitted to the online software of ExPASy (https://www.expasy.org/) to obtain their isoelectric point (pI) and molecular weight (MW) and the subcellular localizations were predicted by WoLF PSORT software (https://wolfpsort.hgc.jp/) (Gasteiger et al., 2003). The MEME program (https://meme-suite.org/meme/tools/meme) was used to identify the conserved motifs of AsbZIP protein sequences and NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd) was used to collect information about the domain composition of AsbZIPs (Bailey et al., 2009). The results were visualized with CFVisual v 2.1 software (https://github.com/ChenHuilong1223/CFVisual). The exon/intron structure of AsbZIP genes was analyzed by the GSDS program (http://gsds.gao-lab.org/index.php) from the CDS and genomic sequence data (Hu et al., 2015). The cis acting elements of AsbZIPs were predicted from 2 kb DNA sequences upstream of the AsbZIP translation sites using the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Elements that respond to adversity stress or phytohormones were selected and visualized using CFVisual v 2.1 software. The initial prediction and analyses of AsbZIP genes and nearby genes were presented by the online software (https://www.chiplot.online/). Detailed information regarding the nearby genes was obtained from gene annotations (GFF3 format) of the A. sinensis genome. The protein sequences of these genes were submitted to the Eggnog-mapper library (http://eggnog-mapper.embl.de/) and agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php) for gene classification and enrichment. The duplication events of the nearby genes were predicted by the script of duplicate_gene_classifer in MCscanX.
The alignment of 140 full-length protein sequences (78 AtbZIPs and 62 AsbZIPs) was initially performed using MUSCLE with default parameters. A phylogenetic tree was constructed using the data with IQ-tree version 1.6.12 with the best model, VT+F+R7 (Minh et al., 2020). Analysis of the phylogenetic tree and the conserved motifs placed AsbZIPs into 13 groups (A-K, M and S) according to the classification method in A. thaliana and were visualized with Evolview 3.0 (http://www.evolgenius.info/) (Dröge-Laser et al., 2018).
The information about the physical locations of AsbZIP genes in 8 chromosomes was obtained from the annotations of 62 genes (GFF3 format) and the AsbZIP genes were renamed according to their chromosomal locations. The duplication types of bZIP1 genes in A. sinensis were analyzed by the script of the duplicate_gene_classifer in MCscanX and visualized by Circos v0.69 (Krzywinski et al., 2009). Collinearity analysis of AsbZIP was also detected using WGDI v 0.5.9 (Sun et al., 2022b).
The expression levels of 62 AsbZIP genes were obtained from previous transcriptome data and the log2 fragments per kilobase per million mapped reads (FPKM) values of AsbZIP genes were visualized by Heml 1.0 software (Ding et al., 2020; Li et al., 2021a). Total RNA was isolated from collected samples with the RNAprep Pure Plant Plus kit (Tiangen, China) and stored as previously described. First-strand cDNA was immediately synthesized using a FastKing gDNA Dispelling RT SuperMix kit (Tiangen, China) and was employed as the template in quantitative real-time PCR (RT-qPCR). Meanwhile, all seven genes in subfamily D were selected for RT-qPCR validation analysis and histone was used as a housekeeping gene (Kumeta and Ito, 2010). Primers were designed by the IDT PrimerQuest tool (https://sg.idtdna.com/pages) (Table S1). Quantitative real-time PCR was conducted using SuperReal PreMix Plus (SYBR Green) (Tiangen, China) with the CFX96™ Real-Time system (Bio-Rad, Hercules, CA, USA) using three biological replicates. The results were analyzed by the 2−∆∆Ct method (Ding et al., 2018; Liang et al., 2023).
The coding sequences of AsbZIP14 and AsbZIP41 were amplified using a Taq Plus DNA Polymerase kit (Tiangen, China) with primers listed in Table S2 in Escherichia coli DH5α cells. The PCR program was set as follows: 5 min at 95 °C for initial denaturation, followed by 35 cycles with 95 °C for 10 s, 58 °C for 20 s, and 72 °C for 20 s. Then, the coding segments were respectively ligated into the pNC-Cam1304-SubC vectors containing green fluorescent protein (GFP) gene to generate pAsbZIP14-GFP and pAsbZIP41-GFP plasmids. Agrobacterium tumefaciens GV3101 strains harboring pAsbZIP14-GFP, pAsbZIP41-GFP, or pNC-Cam1304-SubC plasmids were separately grown to OD600 = 0.8 in LB medium at 200 rpm and 28°C and infiltrated into the onion epidermis by Agrobacterium-mediated transformation. The transformed onion epidermis was incubated in darkness at 27°C for 70 h. The GFP fluorescent signal of these onion epidermal cells was monitored using a Leica confocal fluorescence microscope (Wetzlar, Germany) after the nuclei were stained with 20 μg/mL 4´, 6-diamidino-2- phenylindole (DAPI) for 5 min and washed three times with the saline (Qu et al., 2020; Li et al., 2021a).
The promoters of AsPKS3 (1770 bp), AsPKS4 (1770 bp), AsPKS6 (1590 bp), AsPKS8 (1380 bp), and AsPKS9 (1335 bp) were amplified by PCR (Table S2) based on the other six AsPKS promoters containing abundant tandem TA or high GC content. The PCR products were separately inserted into bait vector pHIS2 with a Ready-to-Use Seamless Cloning Kit (Bhbio, China) to generate pHIS-pAsPKS3, pHIS-pAsPKS4, pHIS-pAsPKS6, pHIS-pAsPKS8, and pHIS-pAsPKS9 constructs. Meanwhile, the ORF sequence of AsbZIP14 and AsbZIP41 were respectively fused into the pGAD7 vectors to form the prey construct pGAD-AsbZIP14 and pGAD-AsbZIP41. Each of the bait vectors and the prey construct was transformed into yeast strain Y187 and the cells were cultivated on nutrient deficiency medium (SD/-His/-Leu/-Trp, SD-TLH) supplemented with different amounts of 3-amino-1,2,4-triazole (3-AT) at 30°C. The interaction between AsbZIP protein and the promoters of these AsPKS genes were scored after 3 days.
The promoters of AsPKS3, AsPKS4, AsPKS6, AsPKS8, and AsPKS9 were respectively cloned into pGreenII-0800-LUC plasmid and the ORF of AsbZIP14 and AsbZIP41 respectively ligated into pGreenII 62-SK to form the reporter vectors and effector vector. The resulting constructs and the empty pGreenII 62-SK vector (control) were individually transformed into A. tumefaciens GV3101 competent cells with pSoup-p19. A. tumefaciens cells were resuspended in infiltration medium (10 mM MES, 10 mM MgCl2 and 160 mM acetosyringone, pH 5.7) to a final OD600 = 0.8. The mixed bacterial cells of the effecter (1 mL) and reporter (3 mL) were infiltrated into N. benthamiana leaf tissues. The LUC and REN activities were detected using a Dual-Luciferase® reporter assay system (Promega, USA) following the manufacturer’s instructions after the tobacco was infiltrated for 3 days (Li et al., 2021a). Experiments were performed using eight independent biological replicates and three technical replicates.
The seed proteins from the were non-redundant database of bZIP proteins from twelve species genomes used as a query to search against eleven species genomes to investigate the candidate bZIP genes in each species and uncover the evolutionary process of the bZIP gene family in Malvales. A total of 62 AsbZIP proteins were identified in A. sinensis genome and the results were verified by Pfam, CDD, and eggNOG databases (Table S3). The bZIP transcription factor gene family of three representative model plants and eight Malvales species was identified and verified using the same method. The number of bZIP genes in each genome were differed. A total of 251 DzbZIP genes were identified in D. zibethinus, followed by H. hainanensis (172), G. raimondii (120), D. turbinatus (107), H. cannabinus (103), A. thaliana (78), and V. vinifera (68). The three species without a recent whole genome duplication (WGD) event (T. cacao, C. capsularis, or C. olitorius) contained fewer bZIPs than A. sinensis. The species of A. trichopoda without WGD and whole genome triplication (WGT) only contained 44 bZIPs (Figure 1).
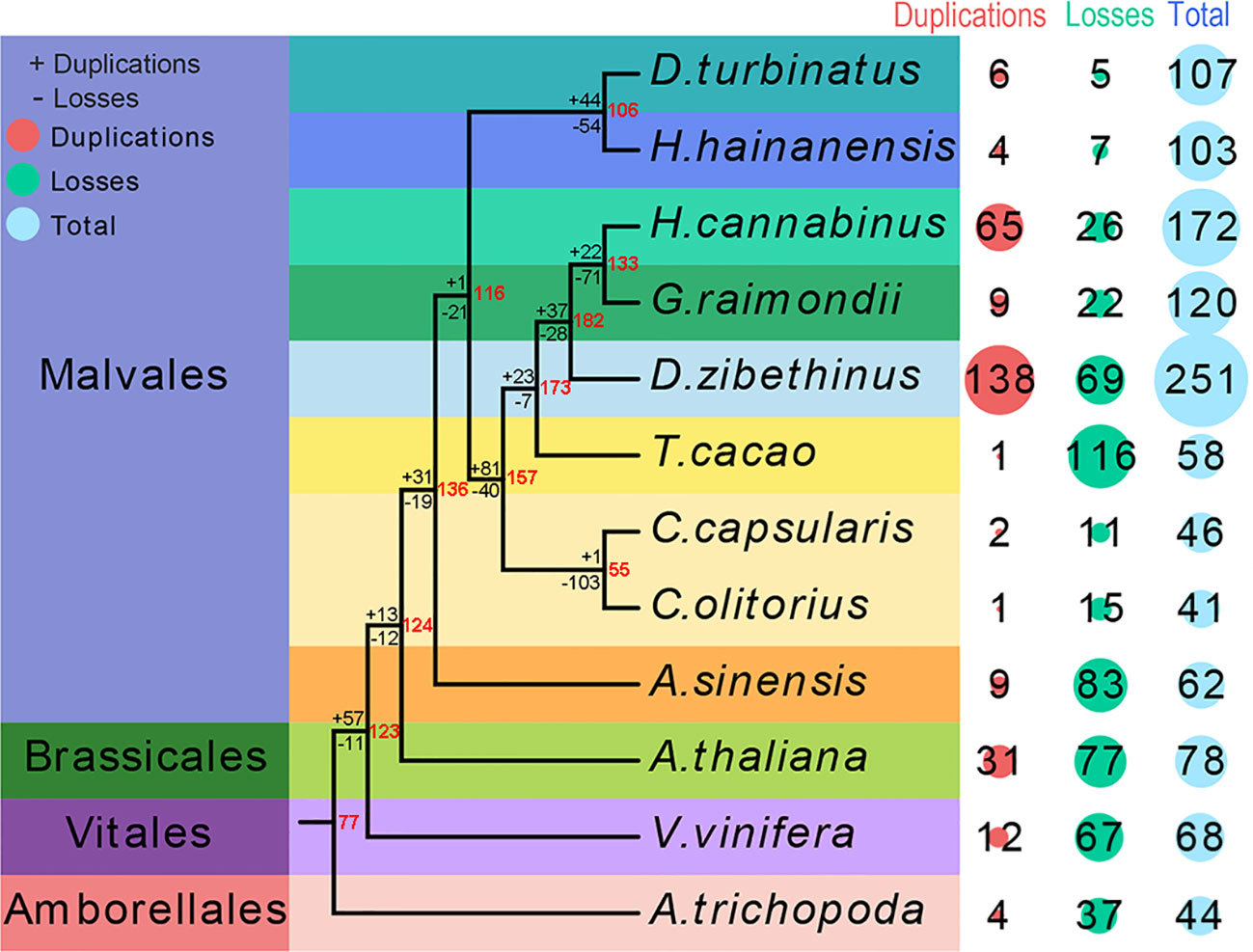
Figure 1 Quantitative distribution and duplication or loss analyses of bZIPs in nine Malvales plants and three model plants: Arabidopsis thaliana, Vitis vinifera, and Amborella trichopoda. The number of bZIP gene variations at different stages of plant evolution. The number of gene losses and duplications is indicated by “-” or “+” on each branch. The red number at node represents the number of bZIPs genes in ancestors species.
Subsequently, the phylogenies of these 12 species were reconstructed to reveal the evolutionary process of bZIP genes in Malvales using A. trichopoda as the outgroup species. The analysis of genes loss and duplication suggested that bZIPs gene family present rapid expansion in Malvales species experienced both WGD and WGT (Figure 1). Interestingly is that only 62 bZIPs genes in A. sinensis even this agarwood tree also undergone WGT and WGD. 77 ancestral genes were present in the lineage leading to the common ancestor of A. trichopoda and V. vinifera, and Malvales based on the duplication or losses of bZIP genes and the number of variations at different stages of evolution. Meanwhile, 57 ancestral genes were duplicated, and 11 ancestral genes were lost in the lineage leading to the common ancestor of A. thaliana, V. vinifera, and Malvales. Thirteen ancestral genes were duplicated, and 12 ancestral genes were lost for the branch of the common ancestor of A. thaliana and 9 Malvales species. The separation of A. sinensis from eight other Malvales species resulted in the duplication of 31 ancestral genes and the loss of 19 ancestral genes. This result suggests that the duplication or losses of bZIP gene families in each Malvales species are not uniform since the divergence. Nine out of sixty-two bZIP genes in A. sinensis originated from duplication, while 83 members were lost in species evolution or differentiation. In summary, more bZIP genes were lost than duplicated in A. trichopoda, A. thaliana, V. vinifera, and Malvales species except in D. zibethinus and H. cannabinus. This result implied that the bZIP gene families of D. zibethinus and H. cannabinus were significantly larger than that of other species (Figure 1).
The comparative synteny and collinearity maps of bZIP genes were constructed using eleven species (excluding A. trichopoda because it lacks a chromosome-level genome) to identify the orthologous bZIPs genes between A. sinensis and other species. There were 136, 105, 124, 114, 155, 68, 63, 59, 42, and 49 orthologous pairs between A. sinensis and the other ten species (D. zibethinus, H. hainanensis, G. raimondii, D. turbinatus, H. cannabinus, A. thaliana, T. cacao, V. vinifera, C. olitorius, and C. capsularis, respectively) (Figure 2A). Only AsbZIP09 and AsbZIP25 had no orthologous pairs with the other 11 species; this indicated that they might be unique to A. sinensis (Figure 2B). The bZIPs were more conserved in these species.
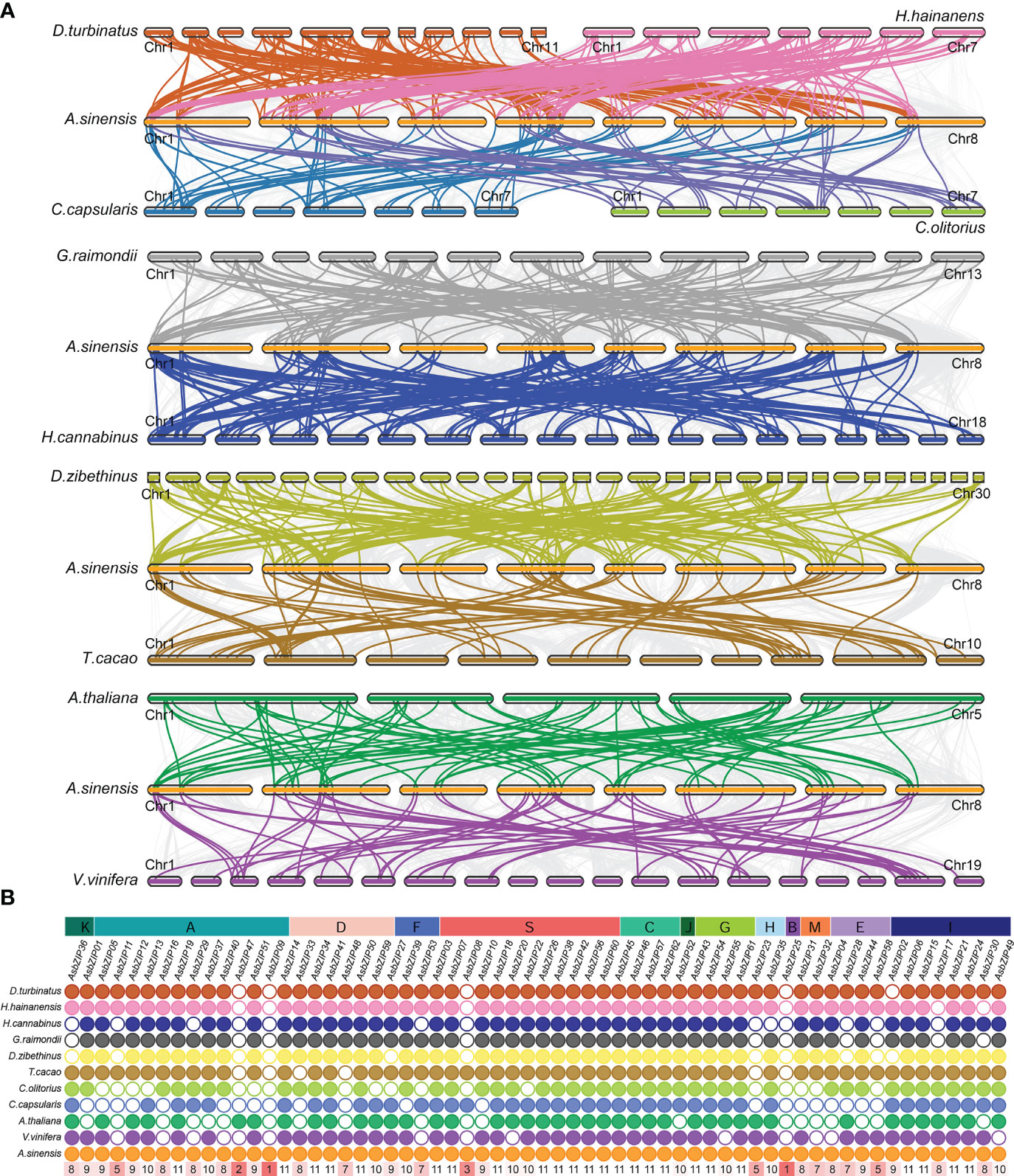
Figure 2 Synteny relationship of AsbZIPs with bZIPs from eight other Malvales plants, A. thaliana, and V.vinifera. (A) Synteny analyses of AsbZIPs and bZIPs from other species. (B) The statistics of collinear relationship. Gray lines in the background indicated the syntenic blocks within A. sinensis and other plant genomes. The highlighted lines indicate the AsbZIPs with their collinearity gene in different species. The number in red rectangle show the number of species holding the orthologous gene of this AsbZIP.
Nonsynonymous (Ka) and synonymous (Ks) values for bZIP gene pairs between A. sinensis and other eleven selected species were calculated to detect the driving force of the bZIP gene family evolutionary process (Figures 3A, B). The Ka/Ks value ranged from 0.04–1.18 and all of the Ka/Ks ratios were < 1 except for one pair (AsbZIP45 and AsbZIP46) (Figure 3C). All Ka/Ks values of bZIP gene pairs between A. sinensis and eleven other species were significantly below 1. This indicated that the bZIP genes were subjected to negative selection and high conservation in plant evolution. The Ka/Ks value of the collinearity pairs of AsbZIP45 and AsbZIP46 was above 1 within the A. sinensis genome, whereas the Ka/Ks values of 15 other paralogous AsbZIP gene pairs were below 1. This confirmed that the AsbZIP gene family primarily underwent negative selection in the subsequent evolution of A. sinensis.
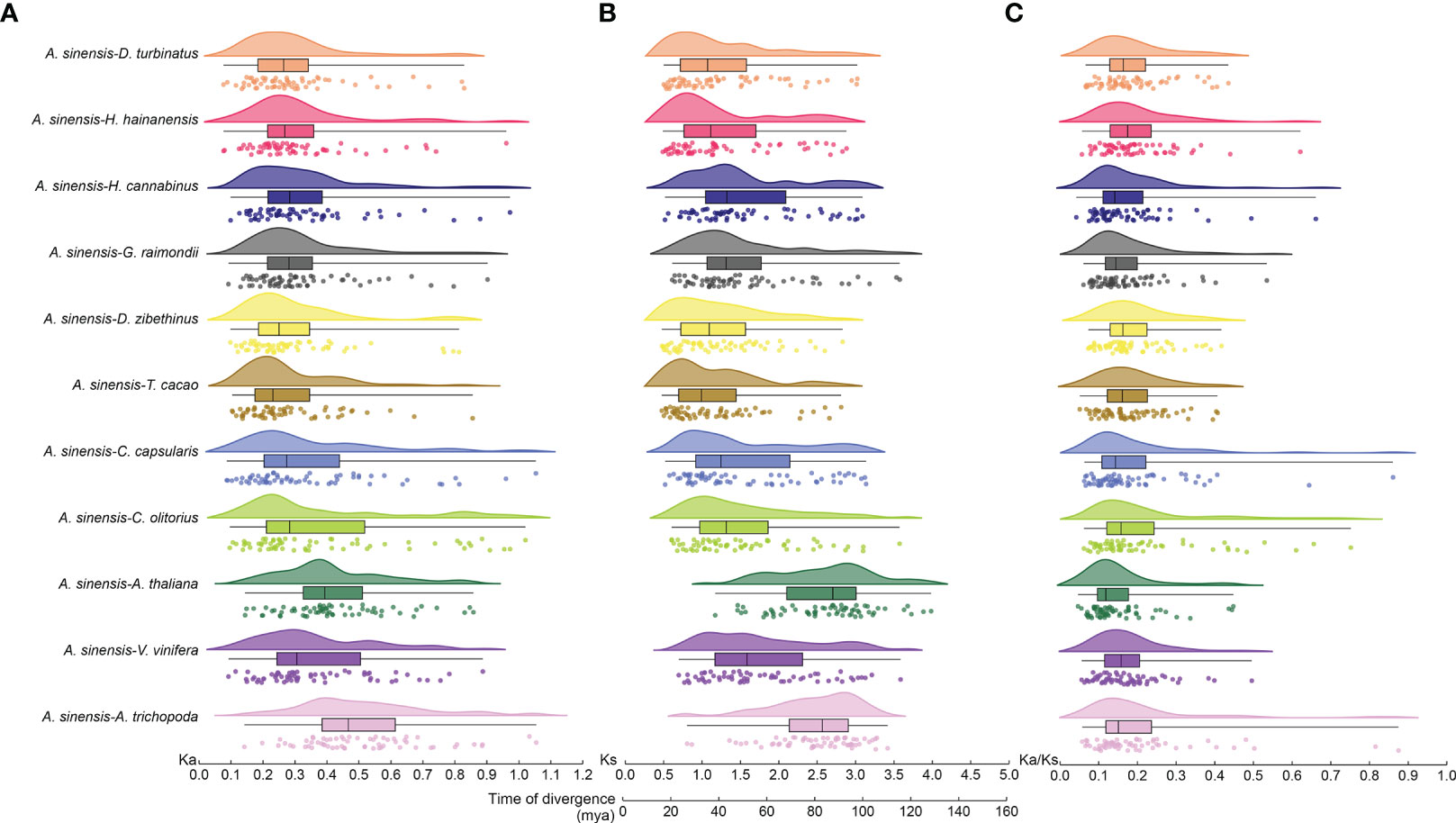
Figure 3 Ka, Ks, and Ka/Ks analysis, and divergence time of bZIP syntenic gene pairs in A. sinensis and other species. (A) Nonsynonymous (Ka) values. (B) Synonymous (Ks) values and divergence time. (C) The Ka/Ks ratio. Different species were plotted using different colors, while the same plant was drawn with the same color.
The locations of their means, variances, and peaks were statistically determined after using the normal distribution function to fit the Ks distribution. The Ks peaks from the bZIP gene families of different species were in different positions, while some species had similar Ks peaks and shared a similar evolutionary rate (Figure 3B). The Ks peak of bZIPs in A. trichopoda, A. thaliana, and A. sinensis was 3.08, 2.97, and 1.10, respectively. The Ks peak of bZIPs from H. hainanensis was similar to that of H. cannabinus and G. raimondii, while the bZIP Ks peak of D. turbinatus was similar to that of D. zibethinus and T. cacao. Besides, the Ks peak of bZIPs in V. vinifera resembled that of C. olitorius and C. capsularis. This might be caused by the retention of ancient WGD events and the mission of the recent WGD events in these three species.
The Ks values were further used to track the divergence time of these gene pairs for the 11 species to deduce the divergence time between bZIP genes of different species. The divergence time of bZIP genes in A. sinensis extended from 36.00–102.06 million years ago (Mya) except a duplication event of AsbZIP45 and AsbZIP46 occurred at 0.25 Mya, which was dividing during the recent WGD. The divergence time of bZIP genes in A. sinensis was approximately 26.69–122.32 (Mya) compared with eight other Malvales species. Meanwhile, the divergence time of bZIPs in A. sinensis occurred at ~ 48.18–134.95 Mya, ~ 33.53–122.66 Mya, and ~ 36.89–117.46 Mya, respectively compared with A. thaliana, V. vinifera, and A. trichopoda (Figure 3B). These results showed that the divergence times of bZIP genes in A. sinensis and other Malvales are closer compared with other species. These results suggested that the divergence of the common ancestor bZIP genes in these species occurred near the core eudicot-common hexaploidy (ECH) event at ~ 115–130 million years ago (Mya) and the bZIP genes were the ancient and conserved transcription factors in angiosperm evolution.
A joint analysis of bZIP genes in the A. sinensis genome was performed using Blastp and MCScanX to further explore the expansion mechanism of the bZIP gene family. The 62 AsbZIP genes were unevenly distributed on eight chromosomes of A. sinensis: Chr 8 contained only three bZIP genes, while Chr 2 and Chr 4 harbored 13 AsbZIPs each. In addition, Chr 1 and Chr 6 possessed 9 AsbZIP genes each, while Chr 7, Chr 3, and Chr 5 contained 6, 5, and 4 AsbZIP genes, respectively (Figure 4).
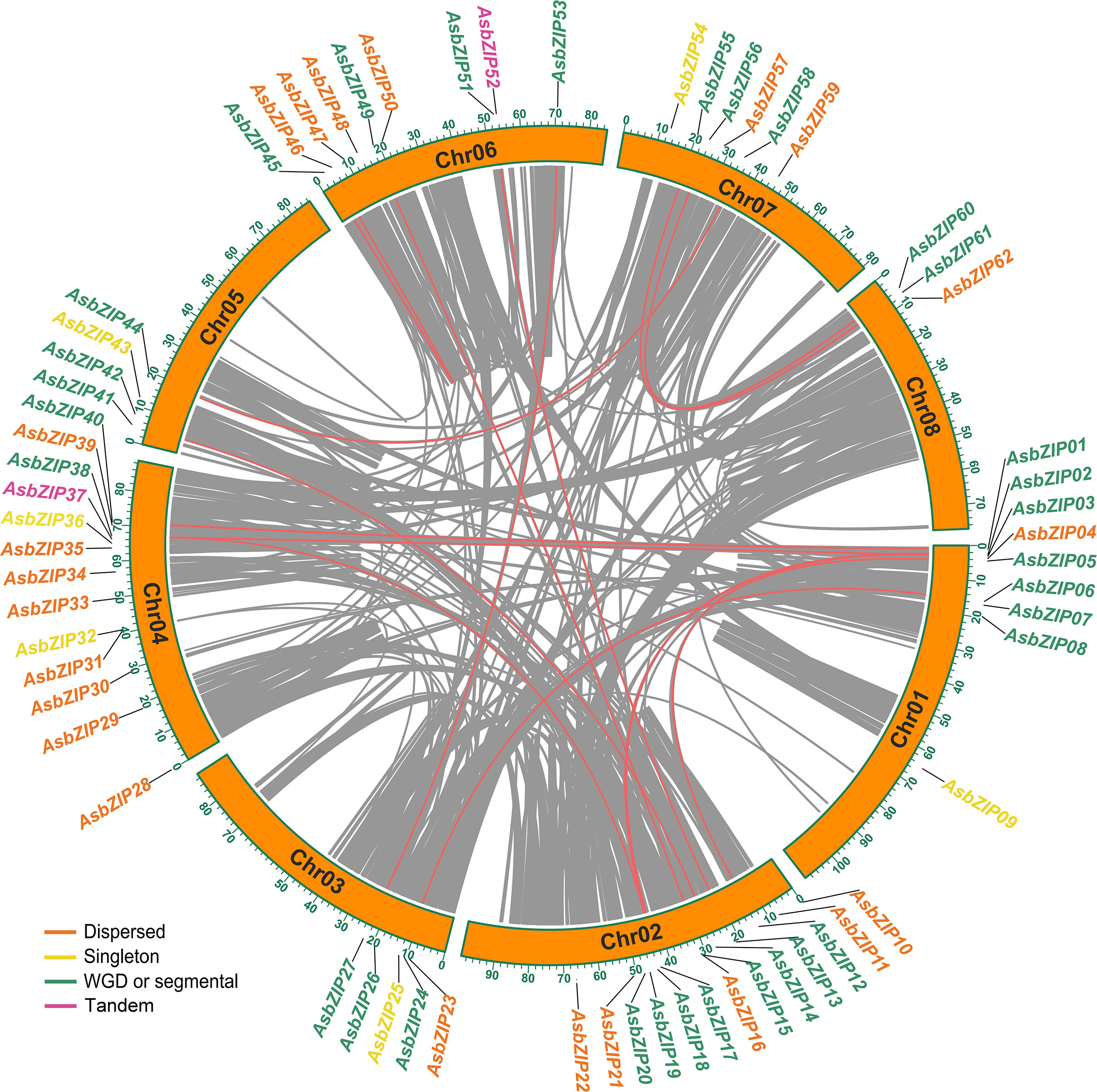
Figure 4 Chromosomal distribution and synteny analysis of AsbZIP genes. Gray lines represented syntenic blocks in the agarwood genome and red lines indicate syntenic AsbZIP gene pairs. Five gene duplication modes of AsbZIP gene names were highlighted with different colors in the external circles.
In order to identify relationships among the AsbZIP genes and define the potential gene duplication events, paralogous information was examined and 16 paralogous AsbZIP gene pairs (27 AsbZIP gene and one other gene) located on different chromosomes were found in the A. sinensis genome. One paralogous gene (evm.model.Scaffold68.87) has no bZIP conserved domain and can be removed from AsbZIP gene family. This is an example of the fact that the structure of a gene may be changed during the duplication events. The percentage of paralogous AsbZIP genes was 43.5% (27 paralogous genes in 62 AsbZIPs). This indicated that duplications were probably conducive to the expansion of the AsbZIP family. Therefore, five gene duplication modes (WGD or segmental, tandem, proximal, singleton, and dispersed duplications) of 62 AsbZIP genes were examined to understand different gene duplication contributions to the expansion of AsbZIP gene family. The percentage of WGD or segmental AsbZIPs was 51.6% (32), followed by dispersed duplication (35.5%, 22), singleton (6, 9.7%), and tandem (2, 3.2%); proximal duplication was not detected (Table S4). Briefly, the origin of the AsbZIP gene family occurred by the WGD or dispersed duplication in A. sinensis.
The phylogenetic tree was constructed based on the protein sequences of 62 AsbZIPs and 78 AtbZIPs to explore the phylogenetic relationship and subfamily classification of AsbZIPs proteins (Figure 5). Sixty-two AsbZIPs were divided into 13 subfamilies (A-K, M, and S); this is the same as bZIPs in A. thaliana based on conserved domains and motifs (Figures 5, 6A). A and S are the largest subfamilies with 13 and 12 members, respectively, followed by the I subfamily (8), D subfamily (7), E subfamily (5), C subfamily (4), G subfamily (4), F subfamily (3), and H subfamily (2); meanwhile, the B, J, K, and M subfamilies contained only one member each.
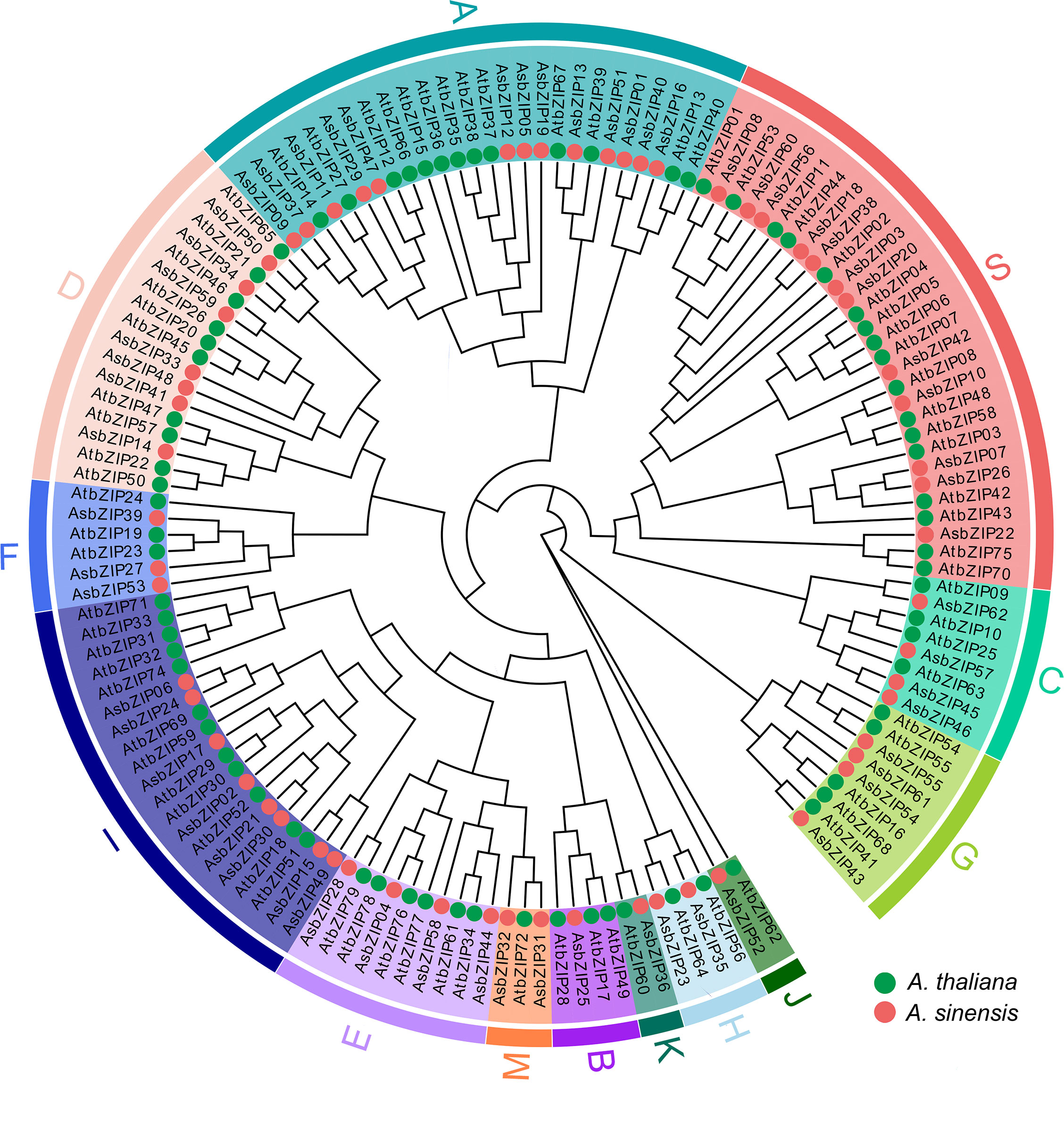
Figure 5 Phylogenetic tree of the bZIP gene family in A. sinensis and A. thaliana. Iqtree was used for model selection, followed by the selection of the best model (VT+F+R7) to generate the phylogenetic tree. The classification into 13 groups (A-K, M, and S) was marked by different colors.
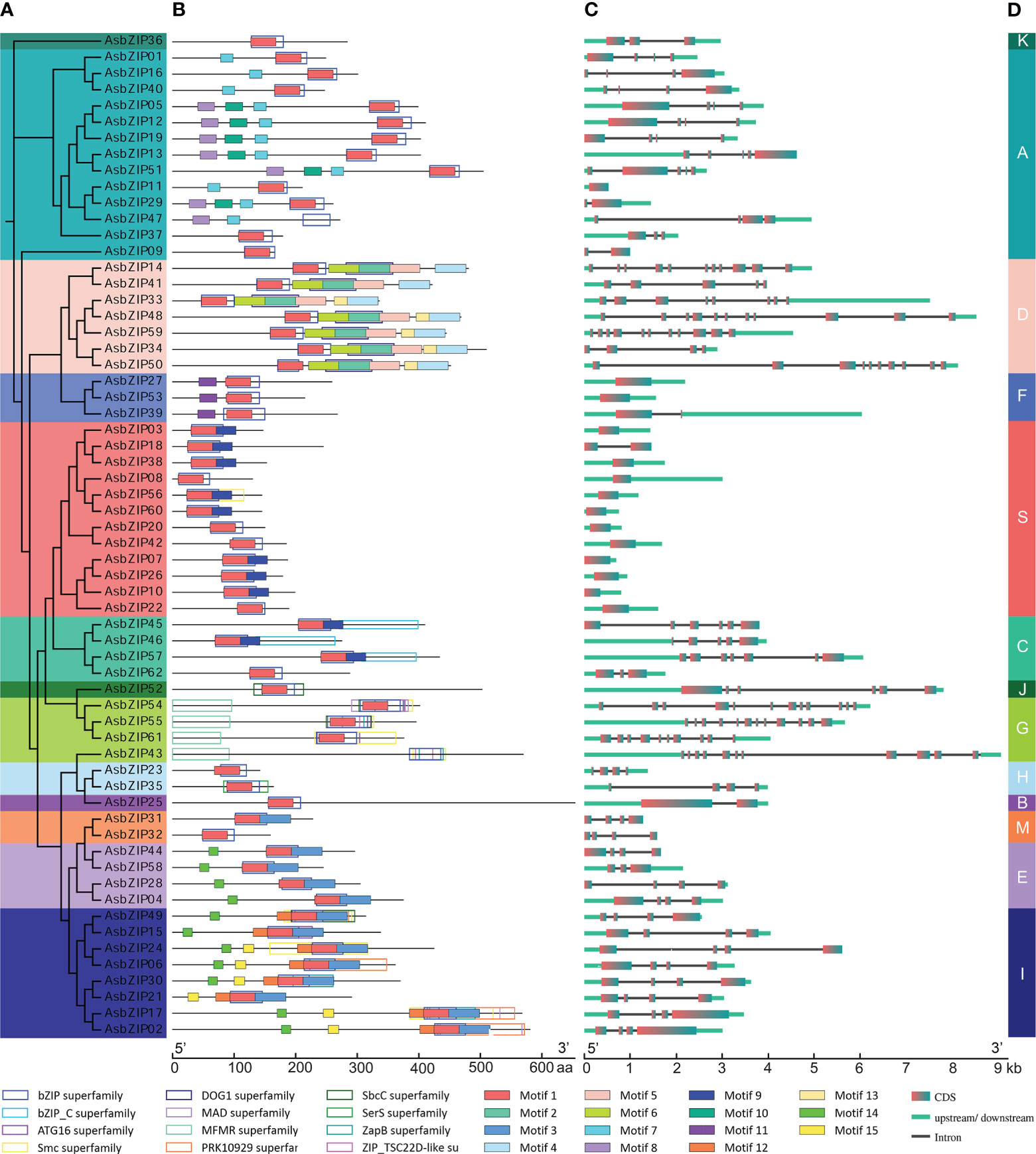
Figure 6 Phylogenetic relationship, conserved domains, motif pattern, and exon–intron structure of AsbZIP proteins and genes. (A) Phylogenetic analysis of AsbZIP proteins. (B) Functional domain and motif compositions of AsbZIP proteins. Fifteen motifs were indicated with rectangles using different fill colors. Twelve conserved domains were shown in rectangles with different outline colors, and their detailed information is provided in Table S6 and Figure S1. (C) Exon-intron structures of AsbZIP genes. (D) Organization of the AsbZIP family. Thirteen subfamilies are marked by different colors.
The lengths of AsbZIPs ranged from 130 AA (AsbZIP08) to 663 AA (AsbZIP25). The predicted molecular weight (MW) of the 62 AsbZIPs was between 15.26 kDa (AsbZIP08) to 72.34 kDa (AsbZIP25). All AsbZIP proteins were predicted to be hydrophilic and the isoelectric point (pI) was predicted to be between 4.77 (AsbZIP36) to 10.69 (AsbZIP37) (Table S5). Almost all AsbZIP proteins were predicted to localize to the nucleus, except for AsbZIP25 (endoplasmic reticulum) and AsbZIP32 (chloroplast) (Table S5).
Protein sequences of 62 AsbZIPs were analyzed using CDD and MEME for the detection of conserved domains and putative motifs to further confirm and understand the composition and gene structure of AsbZIPs (Figure 6B, Figure S1 and Table S6). All AsbZIP proteins almost contain motif 1 which has a specific N-X7-R/K motif that is regarded as a basic DNA-binding region of bZIP proteins, and an adjacent so-called leucine zipper (Figure 6B). The leucine zipper is a domain region that has characteristic heptad repeats of leucine (L) or related hydrophobic amino acids and is general associated with enabling dimerization (Vinson et al., 1989). AsbZIP43 and AsbZIP47 have no motif 1; however, it is hard to eliminate them from the bZIP family since each of them contains the same conserved bZIP superfamily domain.
Exon-intron organization was sequentially investigated to comprehensively understand the similarity and diversity of AsbZIP genes. The number of introns in the AsbZIPs ranged from 0 to 11 out of 13 groups of intron/exon structures (Figure 6C). The D and G subfamilies are the most intron-rich genes compared with the other subfamilies and contained 7–14 introns, except for AsbZIP34 and AsbZIP41 since they had incomplete gene structures. AsbZIP14 and AsbZIP41 of subfamily D had the closest relationship (Figure 6A), although they contained 10 and 4 introns, respectively (Figure 6C). This suggested that their expression and the regulatory mechanism were different. However, no intron was found in the S and F subfamilies except for AsbZIP18 and AsbZIP39 which contained one intron each. Furthermore, one member in the B subfamily (AsbZIP25) contained only one intron. Meanwhile, almost all of the eight other subfamilies presented 3-4 introns. These results indicated that AsbZIPs classified into the same subfamily shared a highly similar composition and position of introns/exons, but different subfamilies had large variations.
The functional diversity of the AsbZIP family might be initially predicted based on the experimentally tested function of Arabidopsis bZIP subfamilies and the diverse conserved domains of each subfamily. The plausible proposition is that the proteins categorized within the different groups tended to perform different functions according to phylogenetic tree analysis combined with structural diversity, motif prediction, and domain map of AsbZIPs (Figures 5, 6) (Hu et al., 2016; Unel et al., 2019; Rong et al., 2020).
The cis acting elements in the promoter of AsbZIP genes were analyzed using the Plantcare database. A total of 51 cis acting elements related to light response (30), plant development (7), hormone response (8), and related to environmental stress (6) were further analyzed (Figure S2). All of the AsbZIPs promoters contained light response element, which is the most abundant and kinds of element. Multiple-stress responsive elements are common in the promoters of all AsbZIPs, except for AsbZIP10 and AsbZIP44. Among them, 27, 54, 25, 27, and 18 AsbZIPs contained wound-responsive elements (WUN-motif), anaerobic induction elements, or enhancer-like element involved in anoxic specific inducibility (ARE and GC-motif), low-temperature responsive elements (LTRs), drought inducibility elements (MBS), and defense and stress-responsive elements (TC-rich repeats), respectively. We also found multiple-hormone responsive elements in the AsbZIP family, and these plant hormone response-related acting cis elements were distributed in almost all members except AsbZIP11 and AsbZIP49. Meanwhile, 26, 50, 39, 21, and 38 AsbZIPs contained salicylic acid (SA) cis elements (TCA-element), abscisic acid responsiveness cis elements (ABRE), methyl jasmonate (MeJA) cis elements (TGACG-motif and CGTCA-motif), auxin cis elements (TGA-element and AuxRR-core), and gibberellin cis elements (GARE-motif, TATC-box, and P-box), respectively. In addition, 46 members of the AsbZIP family contained plant growth and development-related cis elements. Namely, 5, 4, 10, 26, 15, and 24 AsbZIPs contained cis acting elements involved in cell cycle regulation (MSA-like), cis elements responding to the differentiation of palisade mesophyll cells (HD-Zip 1), circadian control cis elements (circadian), meristem expression cis elements (CAT-box), endosperm expression cis elements (GCN4_motif), and cis acting elements involved in zein metabolism regulation (O2-site), The promoters of AsbZIP07 harbored a cis element with an AACA_motif involved in the endosperm-specific negative expression. Together, these results indicated that AsbZIPs may respond to many different kinds of light, growth and development, and stress and hormone signaling. These results suggested that AsbZIP family genes might have various functions and each subfamily might play different roles in plant developmental events, and biotic and abiotic responses.
Five upstream genes and five downstream genes around 62 AsbZIPs were further analyzed to investigate the potential effect of nearby genes on AsbZIPs. Sixty-one pseudo gene clusters were constructed ranging between 65.59–900.93 kb according to AsbZIP31 and AsbZIP32 tandem genes on the same chromosome (Figures S3, S4). A total of 671 genes (62 AsbZIPs and 609 nearby genes) were enriched into 21 COG categories (Figure S3). Transcription (103, 15.35%) was enriched (except in the largest unknown functional categories: 295, 43.96%), followed by posttranslational modification, protein turnover, and chaperones (37, 5.51%), signal transduction mechanisms (26, 3.87%), translation, ribosomal structure, and biogenesis (25, 3.73%), energy production and modification (21, 3.13%), and carbohydrate transport and metabolism (19, 2.83%) (Figure S3). Different from WGD or segmental had the greatest contributions to the expansion of AsbZIP gene family, more dispersed duplications (269, 40.09%) were discovered in nearby genes of AsbZIPs, followed by WGD or segmental (181, 26.97%), singleton (175, 26.08%), tandem (36, 5.37%) and proximal duplications (10, 1.49%) (Figure S4). These results suggested that nearly all genes distributed around AsbZIPs were none functional gene directly involved in secondary metabolism while they might provide regulatory function via protein interaction with DNA elements.
The expression levels of 62 AsbZIP genes were analyzed to further distinguish candidate AsbZIP genes related to PEC biosynthesis (Figure 7). The expression profiles of AsbZIP genes are significantly different during agarwood formation. Five AsbZIPs had no transcript abundance (AsbZIP09, 11, 31, 32, and 44), while the other 57 AsbZIPs were expressed in this process (32 were upregulated, while 25 were downregulated). Transcriptomic analysis showed that there 12 AsbZIP genes (03, 08, 14, 18, 35, 38, 41, 42, 49, 56, 60, and 62) were expressed at high levels and AsbZIP03 was significantly expressed. Both candidates of subgroup M (AsbZIP31 and 32) and all members of subgroup E had low expression except AsbZIP04. The expression of subfamily S AsbZIPs was starkly polarized on account of seven genes out of twelve AsbZIPs showing high expression levels compared with all AsbZIPs; however, the last five genes were expressed at relatively low levels. Subfamily D contained seven members, with four genes showing low expression during agarwood formation, while AsbZIP48 was consistently expressed. Meanwhile, AsbZIP14 and AsbZIP41 had high expression levels, this suggests that they might be involved in regulating type III PKS expression.
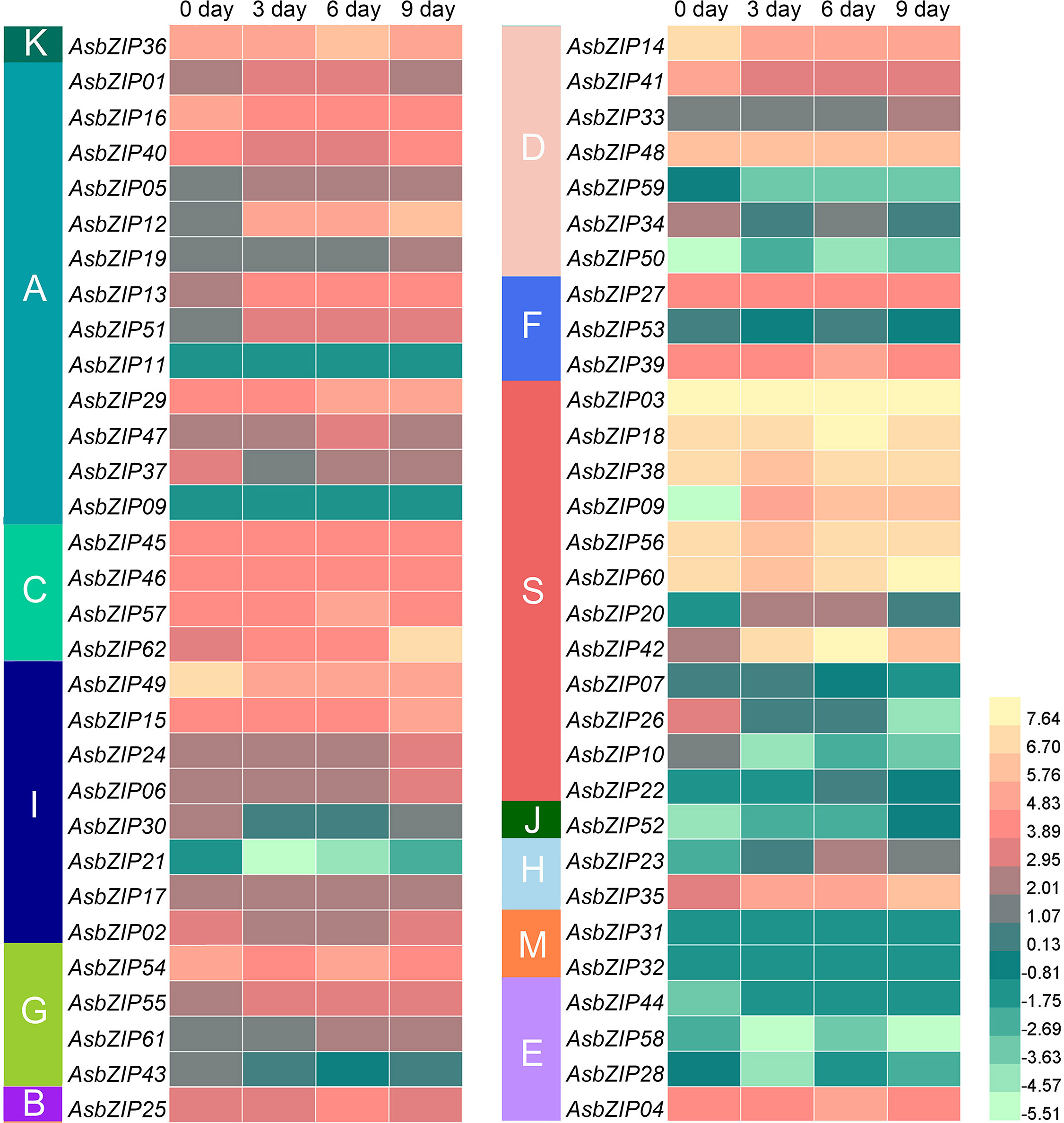
Figure 7 Expression profiles of AsbZIP genes in A. sinensis stems treated with agarwood-inducer. The 0 day, 3 day, 6 day, and 9 day tags indicated the time-points after the stems of A. sinensis were treated with the agarwood inducer. The RPKM (reads per kilobase of exon model per million mapped reads) were transformed to log2.
The expression levels of all AsbZIP genes from subfamily D were detected by qRT-PCR following ethylene (ET) and agarwood-inducer treatment to distinguish PEC biosynthesis-related AsbZIPs genes in subfamily D (Figure 8). Ethylene stimulus increased AsbZIP1 expression by 71,427.18-fold compared with the control at 3 h, followed by a decrease after 48 h, with a small peak at 24 h. The expression of AsbZIP41 and AsbZIP33 was also significantly induced by ET (Figure 8A). In comparison, the expression of AsbZIP14, AsbZIP41, and AsbZIP59 was significantly down-regulated under the agarwood-inducer treatment. The agarwood-inducer treatment caused a decrease in AsbZIP50 expression and reached the lowest level at six hours. This was followed by a considerable increase to its highest level at 48 hours (Figure 8B).
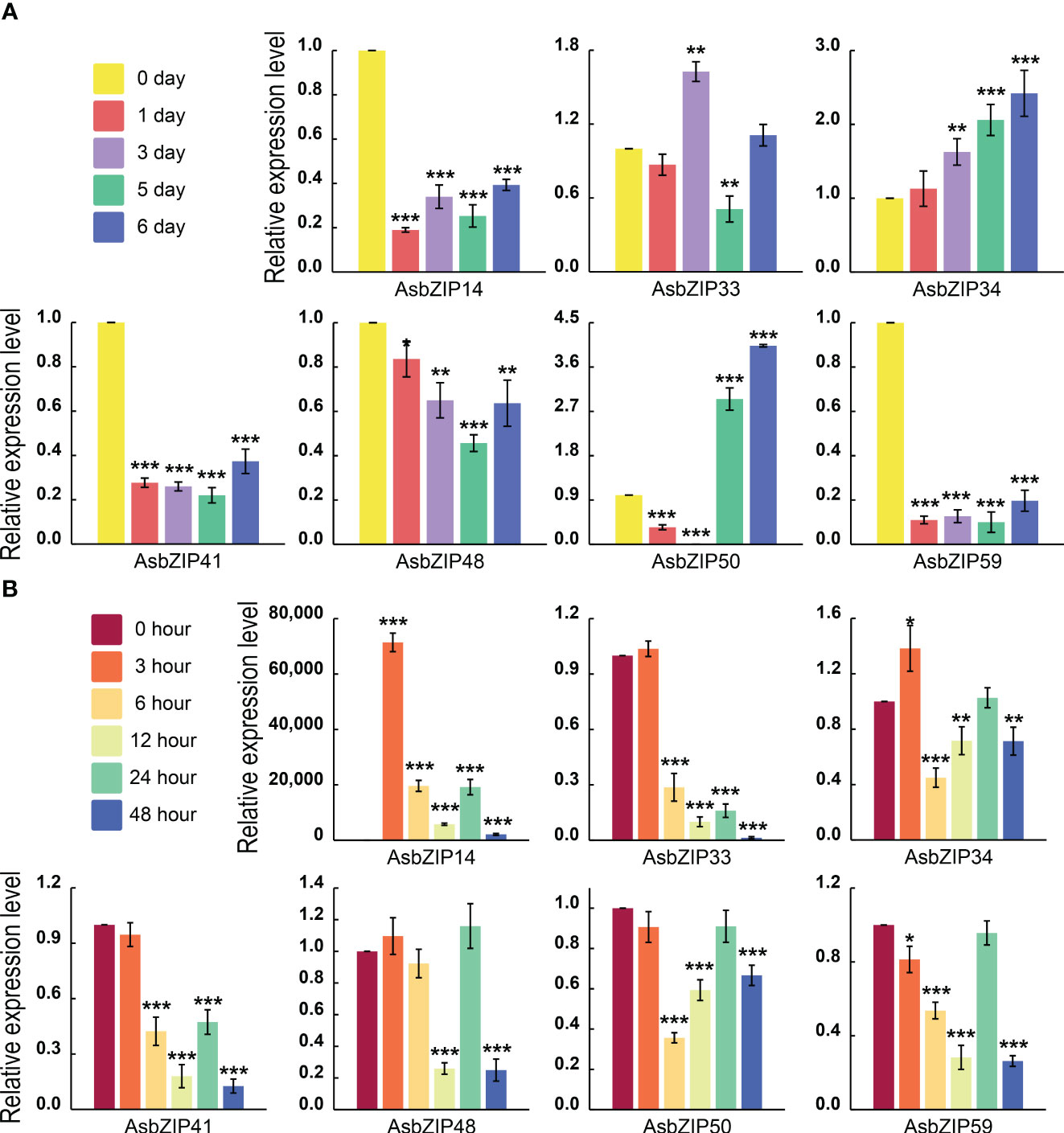
Figure 8 Relative expression of AsbZIP genes of subfamily D in A. sinensis stems after treatment with agarwood inducer and ethylene (ET). (A) Relative expression of AsbZIP genes of subfamily D in the A. sinensis stem after treatment with agarwood inducer at day 0, 1, 3, 5, and 6. (B) Relative expression of AsbZIP genes of subfamily D in A. sinensis stems after treatment with ET at 0, 3, 6, 12, 24, and 48 (h) qRT-PCR data from three independent biological replicates were shown using standard errors (SE) and AsHistone was used as the internal control. Asterisks represent significant differences (*p < 0.05; **p < 0.01; ***p < 0.001).
The subcellular location of AsbZIP14 and AsbZIP41 was analyzed to investigate their roles in regulating chromone biosynthesis by activating AsPKS expression. The GFP fluorescence produced by pAsbZIP14-GFP or pAsbZIP41-GFP was preponderantly located in the nucleus (Figure 9A). This demonstrated that AsbZIP14 and AsbZIP41 perform their regulatory function as TFs in the nucleus. Next, Yeast one-hybrid (Y1H) assay was used to verify the ability of AsbZIP14 and AsbZIP41 interacted with the promoter of AsPKS3, AsPKS4, AsPKS6, AsPKS8 and AsPKS9. The results of Y1H assays confirmed that AsbZIP14 physically interacted with the promoter of AsPKS3, AsPKS6, AsPKS8, and AsPKS9 and might regulate their transcription; however, it did not interact with the AsPKS4 promoter (Figure 9B). Meanwhile, AsbZIP41 presented a completely different function (Figure 9B). The dual-luciferase reporter gene assay (Dual-LUC) was conducted to better understand the AsbZIP14 and AsbZIP41 mode of action on AsPKS genes (Figure 9C). It further verified the binding of AsbZIP14 with the promoter regions of AsPKS3, AsPKS6, AsPKS8, and AsPKS9, respectively (Figure 9D). AsbZIP14 exhibited an 11.0-fold increase in the activity of the AsPKS3 promoter and a 9.7-fold increase in the activity of the AsPKS6 promoter. Activation of the AsPKS8 promoter was also suppressed by AsbZIP14 with a 1.6-fold increase. Meanwhile, the expression of AsPKS9 resulted in a more than 1.5 folds decrease in the luciferase activity controlled by AsbZIP14. The level of the luciferase activity controlled by AsbZIP41 and AsPKS4 promoters was suppressed more than 1.6 folds suggested that AsbZIP41 could transcriptionally downregulate AsPKS6. (Figure 9D). AsbZIP14 could activate the expression of AsPKS3, AsPKS6 and AsPKS8 whereas that could inhibit the expression of AsPKS9 based on Dual-LUC assays and Y1H assays. Besides, the AsPKS4 promoter was also mediated by AsbZIP41.
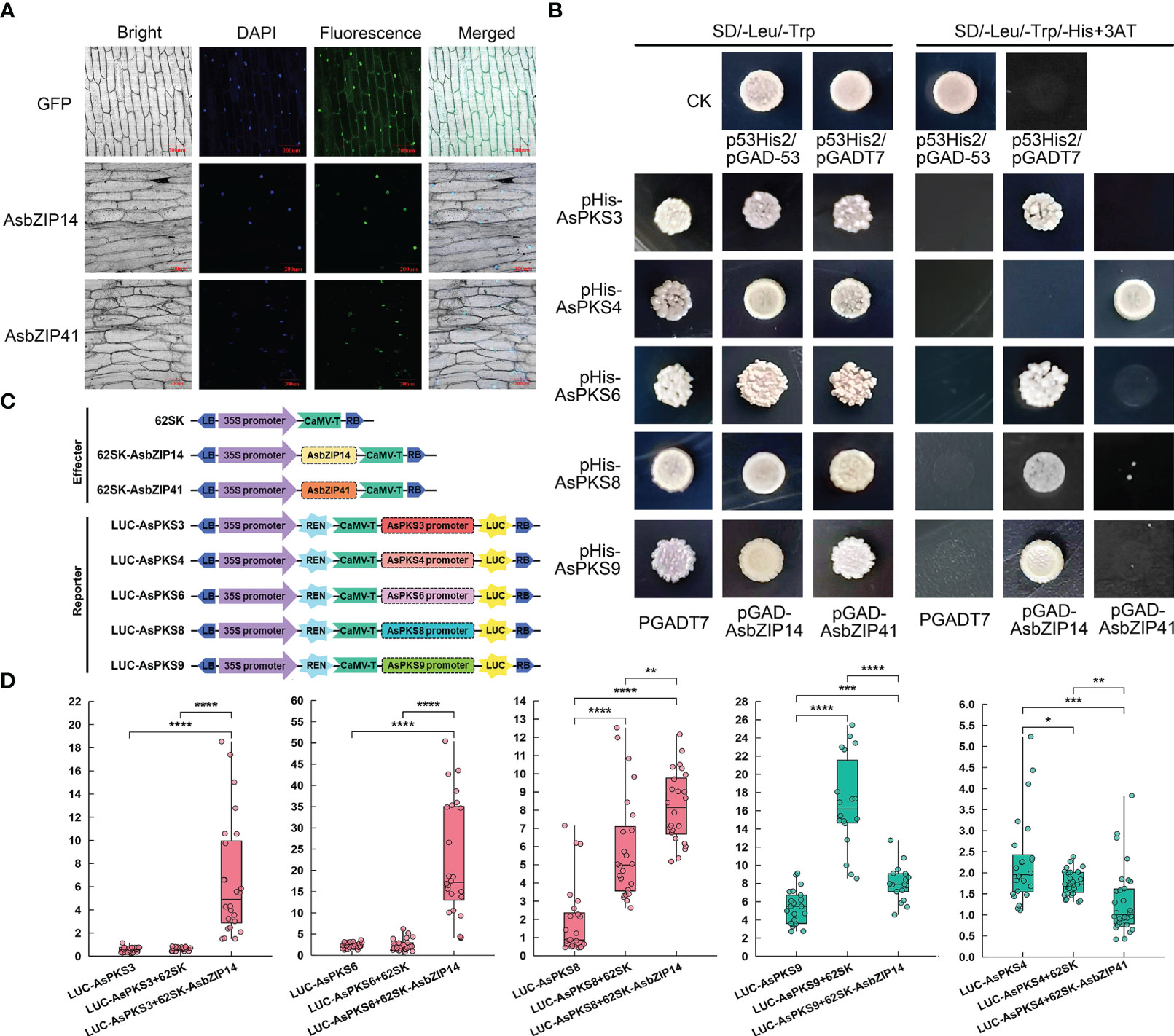
Figure 9 Promoter activation of AsPKS3, AsPKS4, AsPKS6, AsPKS8, and AsPKS9 by AsbZIP14 and AsbZIP41. (A) Nuclear localization of AsbZIP14 and AsbZIP41 in onion epidermal cells. (B) Yeast one-hybrid analysis of AsbZIP14 and AsbZIP41 proteins binding to the promoters of five AsPKSs. Assays were repeated three times. (C) Schematic diagrams of the effecter and reporter plasmids used in Dual-LUC assays. REN, Renilla luciferase internal reference gene. LUC, firefly luciferase reporter gene. (D) Dual-luciferase (Dual-LUC) assays showed that AsbZIP14 or AsbZIP41 regulated the transcriptional activation of the promoters of five AsPKSs. The suppression and promotion effect were colored green and red, respectively. The activities of LUC and REN were determined 3 days after infiltration. The data were indicated by the ratio of LUC to REN and the units of y-asix are 1×e-2.. Data represent the mean ± SE of eight biological replicates and three technical replicates. Significant differences were tested by analysis of variance (ANOVA: *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001).
There is an increasingly systematic and comprehensive identification of the bZIP gene family of different plants with high-quality plant genomes in recent years. Nevertheless, the exhaustive analyses and functional characterization of bZIP families in Malvales plants are unavailable. This work provided a detailed and systemic analysis of AsbZIP genes in A. sinensis and estimated bZIP candidates of eight other Malvales plants (cocoa, C. capsularis, C. olitorius, cotton, durian, D. turbinatus, H. hainanensis, and kenaf) with A. thaliana, grape, and A. trichopoda used as the references.
The bZIPs are ancient and conserved transcription factors in plant evolution; however, their numbers in each plant are different and diverse (Weirauch and Hughes, 2011; Dröge-Laser et al., 2018). In this study, 1150 bZIP candidate genes were identified in eight Malvales species, a model plant and two species at the key nodes of plant evolution. This included, 62, 78, 120, 251, 58, 46, 41, 107, 103, 172, 68, and 44 bZIP genes in agarwood tree, A. thaliana, cotton, durian, cocoa, C. capsularis, C. olitorius, D. turbinatus, H. hainanensis, kenaf, grape, and A. trichopoda, respectively. Polyploidization might always contribute to the gene functions differentiation and number change (Song et al., 2018; Meng et al., 2021; Hou et al., 2022). Duplications and gene losses in this process are the important driving forces for shaping bZIP families The differences in bZIP genes in numerous species might result from gene duplication or losses to various degrees (Figure 1). The ancestry of all angiosperms suffered from an ancestral whole genome triplication (WGT), followed by an additional recent WGD event that affected agarwood tree, cotton, durian, D. turbinatus, H. hainanensis, and H. cannabinus (Vanneste et al., 2014). In contrast, no additional recent polyploidization event was found in cocoa, C. capsularis, C. olitorius, grape, and A. trichopoda (Figure 3B) (Argout et al., 2011; Guan et al., 2014; Zheng et al., 2015; Canaguier et al., 2017; Lian et al., 2018; Wang et al., 2019; Meng et al., 2021; Zhang et al., 2021; Wang et al., 2022b). Interestingly, the number of bZIP genes in A. sinensis was similar to that of cocoa and grape which only experienced the angiosperms-common hexaploidy, but was noticeably less than Malvales experienced additional WGD. The distinct result might be caused by A. sinensis is the relative ancient species in Malvales. Based on these results, we speculate that more AsbZIP candidates were lost during the evolution of A. sinensis compared with other analyzed Malvales species.
The divergence time analysis agreed that the ancient ancestor genes of bZIPs in these species occurred during the core eudicot-common hexaploidy (ECH); therefore, bZIPs were an evolutionarily ancient gene family for angiosperms (Figure 3B). In addition, collinear analysis of agarwood tree, Arabidopsis, grape, cotton, cocoa, C. capsularis, C. olitorius, durian, D. turbinatus, H. hainanensis, and kenaf showed the evolutionary conservation of the bZIP family between Malvales and non-Malvales plants (Figure 2). These results were consistent with previous analyses of bZIPs in green plants. The observed purifying selection of the AsbZIP gene family is supported by the fact that most Ka/Ks values of paralogous AsbZIP gene pairs were below 1 (Figure 3C). Moreover, analysis of AsbZIP gene duplication modes showed that bZIP gene expansion was mainly derived from WGD or segmental duplication (Figure 4).
The basic region of bZIP protein consists of a DNA-binding region that is an invariant N-X7-R/K motif with asparagine (N) and basic (R/K) residues with exact spacing(Dröge-Laser et al., 2018). However, sites that contained variations in the N and R/K residues were also considered bZIP genes. For instance, the bZIP 76-bZIP79 in subfamily E of A. thaliana harbors a deletion or even lacks the N residue (Corrêa et al., 2008; Dröge-Laser et al., 2018). In this study, the six AsbZIPs (AsbZIP04, AsbZIP09, AsbZIP28, AsbZIP32, AsbZIP43, and AsbZIP51) identified with this method contained the conservative bZIP domain and variations in the N or R/K residues (Figure S1). The close relationship between these six AsbZIPs with the typical bZIPs in A. sinensis or A. thaliana suggests that more details should be considered to identify the bZIP gene family in other plants.
Enormous functional analysis of bZIPs was generated in the model plant, A. thaliana. Most of the results determined a prototypic model to define the subfamily classification and predict the function of bZIP genes in each subfamily (Zheng et al., 2015; Duan et al., 2022; Zhang et al., 2022a). Sixty-two AsbZIPs from the agarwood tree genome were sorted into 13 groups similar to bZIP in A. thaliana according to the phylogenetic tree and the conserved motif analysis compared with previously identified and classified gene families (Figure 6) (Dröge-Laser et al., 2018). The group-specific functional and regulatory properties of AsbZIPs were predicated according to the exhaustive summary of bZIPs in A. thaliana and multitudinous angiosperm. Specifically, most bZIP members in subfamily A directly bind to abscisic acid responsive cis-elements (ABRE) and are involved in the abscisic acid pathway to counteract water deficit, floral transition control, and seed development (Banerjee and Roychoudhury, 2017; Shu et al., 2018; Collin et al., 2020). Group B and K members were involved in regulating stress response in the endoplasmic reticulum (Kim et al., 2018; Ruberti et al., 2018). The bZIP members of subgroup C preferentially heterodimerize with group S1 members and form the C/S1-bZIP network that always functions as signaling hub genes to manage plant energy; this leads to species survival from environmental stress and metabolic adaptation (Weltmeier et al., 2009; Pedrotti et al., 2018; Li et al., 2020). Moreover, group S always comprises many bZIP members to benefit plant survival following various abiotic and biotic stresses, including salt stress, cold stress, bicarbonate alkaline stress, extended darkness, and pathogen defense responses (Fu and Dong, 2013; Weiste et al., 2017; Wu et al., 2018; Shine et al., 2019; Wang et al., 2022a). Subfamily D bZIP members (also known as TGA factors) control various important signaling molecules of the phytohormone transduction pathway. They physically interact with genes that are crucial participants of plant innate pathogen responses and are regulators in systemic acquired resistance; this is a broad-spectrum immunity triggered by a prior local pathogen infection at the whole plant level (Dröge-Laser et al., 2018). For example, AtTGA3 mediates the interaction between SA and cytokinin and prevents pathogen damage (Choi et al., 2010); TGA2, TGA5, and TGA6 evoke necrotrophic pathogen defense responses activated by jasmonic acid (JA) and ET in A. thaliana (Zander et al., 2010); while TGA1 and TGA4 induce defense mechanisms against bacterial pathogens in A. thaliana (Wang and Fobert, 2013). AcTGA01, AcTGA06, and AcTGA07 are responsive to hormones at different levels and increase their resistance to kiwifruit canker caused by pathogens (Liu et al., 2022b). Taken together, the AsbZIP members of the same subgroup possess similar adjacent phylogenetic relationships, type of conserved domains and motifs, and exon-intron patterns; this may contribute to proteins with similar functions. The exact regulating function of AsbZIPs involved in agarwood tree was unmapped; however, preliminary results indicated that AsbZIPs participated in agarwood formation and plant hormone response (Figures 7, 8), especially for subfamily D of AsbZIPs.
Chromones and sesquiterpenes are the principal constituents of agarwood (Li et al., 2021b). Chromones and their derivatives have various important biological activities and contribute to the balsamic, long-acting, characteristically pleasant fragrance of agarwood (Naef, 2011; Li et al., 2021b). The abundance of PECs is an important indicator of high-quality agarwood. Dynamic changes of the predicted precursors and chromones during agarwood formation indicated that type III PKSs may be responsible for the biosynthesis of PECs and PKSs also contribute to the biosynthesis of flavonoids, which are high similarity to the backbone structures of chromones (Liao et al., 2018). Type III PKSs were the key enzymes in the formation of the C6–C5–C6 scaffold of chromones and in chromone biosynthesis (Li et al., 2021b; Wang et al., 2022d). Transcription factors regulate the synthesis of primary and special secondary metabolites in plants. For example, the TFs of ERF, MYC, bHLH, and WRKY could regulate sesquiterpene biosynthesis as the positive or negative regulator in A. sinensis (Li et al., 2021a). Nevertheless, the regulatory role of TFs involved in chromone biosynthesis is unknown.
Agarwood is the resin mixed with diverse secondary metabolites produced by wounded agarwood trees (Rasool and Mohamed, 2016; Naziz et al., 2019). Plant hormones directly or indirectly regulate signaling networks by inducing or repressing defense genes; this results in plants adapting to a wide range of environmental stresses. Ethylene is an essential and ubiquitous plant hormone. Plants typically increase ET levels when they suffer from various environmental stresses and ET always elicits plant defense responses (Fatma et al., 2022). Ethylene and jasmonic acid are essential signaling molecules for wound-induced activation of secondary metabolism by modulating reactive oxygen species levels in carrot tissue (Jacobo-Velázquez et al., 2015); ethylene is also a vital signaling molecule in the synthesis of ginsenoside and catechins (Rahimi et al., 2015; Ke et al., 2018). The cultured shoots of agarwood trees treated with MeJA promote the production of sesquiterpene and a chromone derivative of agarwood in A. sinensis (Faizal et al., 2021). 2-(2-phenethyl) chromones and their derivatives were induced by various exogenous phytohormones in the calli of agarwood tree, such as MeJA, SA, and abscisic acid (Dong et al., 2018). The involvement of subfamily D bZIPs in plant response to hormone and pathogen infection implied that AsbZIPs might also participate in the defense response of A. sinensis resulting in agarwood formation (Mohamed et al., 2010; Chhipa et al., 2017). Two AsbZIP genes of subfamily D (AsbZIP14 and AsbZIP41) were highly expressed and significantly downregulated following treatment with a liquid mixture used in agarwood formation (Figure 8). This suggested that AsbZIP14 and AsbZIP41 might participate in agarwood formation. Ethylene responds to bZIP factors in group D to active pathogen defense and it also is a signaling molecule that induces the expression of subgroup D bZIP genes (Tucker et al., 2002; Zander et al., 2014). Furthermore, AsbZIP14 expression was induced 71,427.18 times following ET treatment, while the expression of other subgroup DAsbZIPs have little change according to qRT-PCR assays (Figure 8A). These results suggested that ASbZIP14 and AsbZIP41 may be the candidate genes for Aquilaria trees responding to pathogen challenge and regulating the production of key components in agarwood.
bZIP transcription factors could activate chalcone synthase (CHS), a kind of plant type III PKS involved in flavonoid biosynthesis and plant development or plant signaling transduction in response to pathogens and other stresses. The G/HBF-1 (bZIP) in soybean interacts with CHS promoters to improve disease resistance (Dröge-Laser et al., 1997). GmbZIP45 overexpression specifically activated CHS8 transcription and plant defense against pathogens in soybean protoplasts (Gonçalves et al., 2020). HY5 (bZIPs of subgroup H), a light, and a UV-B radiation response factor acted as a transcriptional activator of CHS participating in flavonoid accumulation in A. thaliana (Stracke et al., 2010). VvibZIPC22 activates the promoters of the CHS gene and controls flavonol biosynthesis (Malacarne et al., 2016). Some type III PKS are involved in chromones or their precursor; however, the roles of bZIP in chromone biosynthesis via regulating type III PKS expression is poorly studied. This study determined whether AsbZIP14 and AsbZIP41 from subfamily D bind with the promoters of AsPKS3, AsPKS4, AsPKS6, AsPKS8, and AsPKS9. AsbZIP14 and AsbZIP41 are a pair of paralogous subfamily D AsbZIP genes sharing the most adjacent phylogenetic relationship and having the same number and type of conserved domains and motifs (Figure 6). They regulate AsPKS expression in different manners, and both were generated from WGD (Figure 4). AsbZIP14 positively activated the expression of AsPKS3, AsPKS6, and AsPKS8, and negatively regulated AsPKS9 expression. AsbZIP14 did not interact with the promoters of AsPKS4. Interestingly, AsbZIP41 positively regulated AsPKS4 by binding its promoters, but cannot activate AsPKS3, AsPKS6, AsPKS8, and AsPKS9 (Figure 9D). The results furtherly indicated that paralogous AsbZIPs from the same subgroup with high sequence homology regulate different target-genes of the same family by varying degrees. This results also demonstrated that bZIPs regulate type III PKS expression to promote or suppress flavonoid and chromone generation.
Systemic genome-wide analysis of 1150 bZIPs identified from eight Mavales and three model species indicated that bZIPs are an ancient and conserved gene family in the plant kingdom and the recent WGD is the driving force in the evolution of the bZIP gene family. The bZIP gene family in A. sinensis underwent purifying selection in species generation and WGD or segmental duplication resulted in AsbZIP expansion. Sixty-two AsbZIPs were divided into 13 subfamilies and showed diverse expression profiles during agarwood formation. AsbZIP14 and AsbZIP41 from subgroup D had the closest phylogenetic relationship and responded to ET stimulation and agarwood inducer. AsbZIP14 could interacted with the promoters of AsPKS3, AsPKS6, AsPKS8 and positively regulated their expression. Meanwhile, AsbZIP14 could negatively regulate AsPKS9 expression. AsbZIP41 negatively regulated AsPKS4 expression and could not interact with the other four AsPKS. This genome-wide analysis of the bZIP gene family provided a basis to further investigate the evolutionary process of bZIPs in the plant kingdom and their regulatory mechanisms of type III AsPKS in A. sinensis. Furthermore, our study also provided potential genetic resources for improving the yield and quantity of agarwood.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XD and WM conceived the experiments. HZ and XD carried out the experiments with the help of HW, HC, WD, JZ, and HD. SP and JW contributed the plant materials and data analysis. HZ and XD wrote the manuscript and WM edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31870668 and 32171824), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630052020003) and the Earmarked Fund of China Agriculture Research System (CARS-21).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1243323/full#supplementary-material
Supplementary Figure 1 | Motif logos of AsbZIPs.
Supplementary Figure 2 | Analysis of cis elements in the promoters of AsbZIP genes.
Supplementary Figure 3 | COG enrichment and classification of AsbZIPs and their nearby genes within gene clusters.
Supplementary Figure 4 | Gene duplicated AsbZIPs and their nearby genes within gene clusters.
Agarwal, P., Baranwal, V. K., Khurana, P. (2019). Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 9 (1), 1–18. doi: 10.1038/s41598-019-40659-7
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., Hao, Y. J. (2018). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 41 (11), 2678–2692. doi: 10.1111/pce.13393
Argout, X., Salse, J., Aury, J.-M., Guiltinan, M. J., Droc, G., Gouzy, J., et al. (2011). The genome of Theobroma cacao. Nat. Genet. 43 (2), 101–108. doi: 10.1038/ng.736
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (suppl_2), W202–W208. doi: 10.1093/nar/gkp335
Banerjee, A., Roychoudhury, A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254 (1), 3–16. doi: 10.1007/s00709-015-0920-4
Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7 (2), 202–209. doi: 10.1016/j.pbi.2004.01.013
Canaguier, A., Grimplet, J., Di Gaspero, G., Scalabrin, S., Duchêne, E., Choisne, N., et al. (2017). A new version of the grapevine reference genome assembly (12X. v2) and of its annotation (VCost. v3). Genomics Data 14, 56. doi: 10.1016/j.gdata.2017.09.002
Chhipa, H., Chowdhary, K., Kaushik, N. (2017). Artificial production of agarwood oil in Aquilaria sp. by fungi: a review. Phytochem. Rev. 16 (5), 835–860. doi: 10.1007/s11101-017-9492-6
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K.-H., Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19 (2), 284–295. doi: 10.1016/j.devcel.2010.07.011
Collin, A., Daszkowska-Golec, A., Kurowska, M., Szarejko, I. (2020). Barley ABI5 (Abscisic Acid INSENSITIVE 5) is involved in abscisic acid-dependent drought response. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01138
Corrêa, L. G. G., Riaño-Pachón, D. M., Schrago, C. G., Vicentini dos Santos, R., Mueller-Roeber, B., Vincentz, M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PloS One 3 (8), e2944. doi: 10.1371/journal.pone.0002944
Ding, X., Mei, W., Huang, S., Wang, H., Zhu, J., Hu, W., et al. (2018). Genome survey sequencing for the characterization of genetic background of Dracaena Cambodiana and its defense response during dragon’s blood formation. PloS One 13, e0209258. doi: 10.1371/journal.pone.0209258
Ding, X., Mei, W., Lin, Q., Wang, H., Wang, J., Peng, S., et al. (2020). Genome sequence of the agarwood tree Aquilaria sinensis (Lour.) Spreng: the first chromosome-level draft genome in the Thymelaeceae family. GigaScience 9 (3), giaa013. doi: 10.1093/gigascience/giaa013
Dong, X., Gao, B., Feng, Y., Liu, X., Wang, J., Wang, J., et al. (2018). Production of 2-(2-phenylethyl) chromones in Aquilaria sinensis calli under different treatments. Plant Cell Tiss. Org. 135 (1), 53–62. doi: 10.1007/s11240-018-1442-5
Dröge-Laser, W., Kaiser, A., Lindsay, W. P., Halkier, B. A., Loake, G. J., Doerner, P., et al. (1997). Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 16 (4), 726–738. doi: 10.1093/emboj/16.4.726
Dröge-Laser, W., Snoek, B. L., Snel, B., Weiste, C. (2018). The Arabidopsis bZIP transcription factor family—an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Duan, L., Mo, Z., Fan, Y., Li, K., Yang, M., Li, D., et al. (2022). Genome-wide identification and expression analysis of the bZIP transcription factor family genes in response to abiotic stress in Nicotiana tabacum L. BMC Genomics 23 (1), 1–17. doi: 10.1186/s12864-022-08547-z
Faizal, A., Esyanti, R. R., Adn’ain, N., Rahmani, S., Azar, A. W. P., Turjaman, M. (2021). Methyl jasmonate and crude extracts of Fusarium solani elicit agarwood compounds in shoot culture of Aquilaria malaccensis Lamk. Heliyon 7 (4), e06725. doi: 10.1016/j.heliyon.2021.e06725
Fatma, M., Asgher, M., Iqbal, N., Rasheed, F., Sehar, Z., Sofo, A., et al. (2022). Ethylene signaling under stressful environments: Analyzing collaborative knowledge. Plants 11 (17), 2211. doi: 10.3390/plants11172211
Fonseca, A., Urzúa, T., Jelenska, J., Sbarbaro, C., Seguel, A., Duarte, Y., et al. (2022). The TGA transcription factors from clade II negatively regulate the salicylic acid accumulation in Arabidopsis. Int. J. Mol. Scl. 23 (19), 11631. doi: 10.3390/ijms231911631
Foster, R., Izawa, T., Chua, N. H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8 (2), 192–200. doi: 10.1096/fasebj.8.2.8119490
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31 13), 3784–3788. doi: 10.1093/nar/gkg563
Goll, J., Rusch, D. B., Tanenbaum, D. M., Thiagarajan, M., Li, K., Methé, B. A., et al. (2010). METAREP: JCVI metagenomics reports—an open source tool for high-performance comparative metagenomics. Bioinformatics 26 (20), 2631–2632. doi: 10.1093/bioinformatics/btq455
Gonçalves, A. B., Fontes, P. P., Dadalto, S. P., de Souza, G. B., Marcelino-Guimarães, F. C., Alves, M. S., et al. (2020). GmbZIP45 binds to H-box cis-element in vitro and overexpression in soybean protoplasts induces the expression of CHS8 gene. Physiol. Mol. Plant P. 112, 101556. doi: 10.1016/j.pmpp.2020.101556
Guan, X., Nah, G., Song, Q., Udall, J. A., Stelly, D. M., Chen, Z. J. (2014). Transcriptome analysis of extant cotton progenitors revealed tetraploidization and identified genome-specific single nucleotide polymorphism in diploid and allotetraploid cotton. BMC Res. Notes 7 (1), 1–10. doi: 10.1186/1756-0500-7-493
Guan, R., Xu, S., Lu, Z., Su, L., Zhang, L., Sun, W., et al. (2022). Genomic characterization of bZIP transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Int. J. Biol. Macromol. 223, 1619–1631. doi: 10.1016/j.ijbiomac.2022.10.283
Hao, X., Zhong, Y., Nützmann, H.-W., Fu, X., Yan, T., Shen, Q., et al. (2019). Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 60 (8), 1747–1760. doi: 10.1093/pcp/pcz084
Hou, H., Kong, X., Zhou, Y., Yin, C., Jiang, Y., Qu, H., et al. (2022). Genome-wide identification and characterization of bZIP transcription factors in relation to litchi (Litchi chinensis Sonn.) fruit ripening and postharvest storage. Int. J. Biol. Macromol. 222, 2176–2189. doi: 10.1016/j.ijbiomac.2022.09.292
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, W., Yang, H., Yan, Y., Wei, Y., Tie, W., Ding, Z., et al. (2016). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6 (1), 1–12. doi: 10.1038/srep22783
Ishihara, M., Tsuneya, T., Uneyama, K. (1993). Components of the volatile concentrate of agarwood. J. Essent. Oil Res. 5 (3), 283–289. doi: 10.1080/10412905.1993.9698221
Jacobo-Velázquez, D. A., González-Agüero, M., Cisneros-Zevallos, L. (2015). Cross-talk between signaling pathways: the link between plant secondary metabolite production and wounding stress response. Sci. Rep. 5 (1), 1–10. doi: 10.1038/srep08608
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 (3), 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jiang, M., Wang, Z., Ren, W., Yan, S., Xing, N., Zhang, Z., et al. (2022). Identification of the bZIP gene family and regulation of metabolites under salt stress in lsatis indigotica. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1011616
Job, N., Yadukrishnan, P., Bursch, K., Datta, S., Johansson, H. (2018). Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 176 (4), 2963–2976. doi: 10.1104/pp.17.00856
Ke, S.-W., Chen, G.-H., Chen, C.-T., Tzen, J. T., Yang, C.-Y. (2018). Ethylene signaling modulates contents of catechin and ability of antioxidant in Camellia sinensis. Bot. Stud. 59 (1), 1–8. doi: 10.1186/s40529-018-0226-x
Kim, J.-S., Yamaguchi-Shinozaki, K., Shinozaki, K. (2018). ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176 (3), 2221–2230. doi: 10.1104/pp.17.01414
Koch, M. A., Haubold, B., Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 (10), 1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248
Kouzarides, T., Ziff, E. (1989). Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature 340 (6243), 568–571. doi: 10.1038/340568a0
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kumeta, Y., Ito, M. (2010). Characterization of δ-guaiene synthases from cultured cells of Aquilaria, responsible for the formation of the sesquiterpenes in agarwood. Plant Physiol. 154 (4), 1998–2007. doi: 10.1104/pp.110.161828
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, W., Chen, H.-Q., Wang, H., Mei, W.-L., Dai, H.-F. (2021b). Natural products in agarwood and Aquilaria plants: chemistry, biological activities and biosynthesis. Nat. Prod. Rep. 38 (3), 528–565. doi: 10.1039/D0NP00042F
Li, H., Li, L., ShangGuan, G., Jia, C., Deng, S., NOman, M., et al. (2020). Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci. Rep. 10 (1), 1–15. doi: 10.1038/s41598-020-72390-z
Li, R.-S., Zhu, J.-H., Guo, D., Li, H.-L., Wang, Y., Ding, X.-P., et al. (2021a). Genome-wide identification and expression analysis of terpene synthase gene family in Aquilaria sinensis. Plant Physiol. Bioch. 164, 185–194. doi: 10.1016/j.plaphy.2021.04.028
Lian, H., Xu, P., He, S., Wu, J., Pan, J., Wang, W., et al. (2018). Photoexcited CRYPTOCHROME 1 interacts directly with G-protein β subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol. Plant 11 (10), 1248–1263. doi: 10.1016/j.molp.2018.08.004
Liang, Y.-E., Zhang, H., Zhu, J., Wang, H., Mei, W., Jiang, B., et al. (2023). Transcriptomic analysis reveals the involvement of flavonoids synthesis genes and transcription factors in dracaena Cambodiana response to ultraviolet-B radiation. Forests 14 (5), 979. doi: 10.3390/f14050979
Liao, G., Dong, W.-H., Yang, J.-L., Li, W., Wang, J., Mei, W.-L., et al. (2018). Monitoring the chemical profile in agarwood formation within one year and speculating on the biosynthesis of 2-(2-phenylethyl) chromones. Molecules 23 (6), 1261. doi: 10.3390/molecules23061261
Liu, J., Li, T., Chen, T., Gao, J., Zhang, X., Jiang, C., et al. (2022a). Integrating Multiple Omics Identifies Phaeoacremonium rubrigenum acting as Aquilaria sinensis marker fungus to promote agarwood sesquiterpene accumulation by inducing plant host phosphorylation. Microbiol. Spectr. 10 (4), e02722–e02721. doi: 10.1128/spectrum.02722-21
Liu, W., Zhao, C., Liu, L., Huang, D., Ma, C., Li, R., et al. (2022b). Genome-wide identification of the TGA gene family in kiwifruit (Actinidia chinensis spp.) and revealing its roles in response to Pseudomonas syringae pv. actinidiae (Psa) infection. Int. J. Biol. Macromol. 222, 101–113. doi: 10.1016/j.ijbiomac.2022.09.154
Loyola, R., Herrera, D., Mas, A., Wong, D. C. J., Höll, J., Cavallini, E., et al. (2016). The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67 (18), 5429–5445. doi: 10.1093/jxb/erw307
Lu, M., Meng, X.-X., Zhang, Y.-M., Zhu, X.-W., Li, J., Chen, W.-Q., et al. (2022). Genome-Wide Identification and Expression Profiles of bZIP Genes in Cannabis sativa L. Cannabis Cannabinoid. 7 (6), 882–895. doi: 10.1089/can.2021.0153
Ma, M., Chen, Q., Dong, H., Zhang, S., Huang, X. (2021). Genome-wide identification and expression analysis of the bZIP transcription factors, and functional analysis in response to drought and cold stresses in pear (Pyrus breschneideri). BMC Plant Biol. 21 (1), 1–19. doi: 10.1186/s12870-021-03356-0
Malacarne, G., Coller, E., Czemmel, S., Vrhovsek, U., Engelen, K., Goremykin, V., et al. (2016). The grapevine VvibZIPC22 transcription factor is involved in the regulation of flavonoid biosynthesis. J. Exp. Bot. 67 (11), 3509–3522. doi: 10.1093/jxb/erw181
Meng, F., Chu, T., Tang, Q., Chen, W. (2021). A tetraploidization event shaped the Aquilaria sinensis genome and contributed to the ability of sesquiterpenes synthesis. BMC Genomics 22 (1), 1–12. doi: 10.1186/s12864-021-07965-9
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa015
Mohamed, R., Jong, P., Zali, M. (2010). Fungal diversity in wounded stems of Aquilaria malaccensis. Fungal Divers. 43 (1), 67–74. doi: 10.1007/s13225-010-0039-z
Naef, R. (2011). The volatile and semi-volatile constituents of agarwood, the infected heartwood of Aquilaria species: a review. Flavour Frag. J. 26 (2), 73–87. doi: 10.1002/ffj.2034
Naziz, P. S., Das, R., Sen, S. (2019). The scent of stress: Evidence from the unique fragrance of agarwood. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00840
Pedrotti, L., Weiste, C., Nägele, T., Wolf, E., Lorenzin, F., Dietrich, K., et al. (2018). Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 30 (2), 495–509. doi: 10.1105/tpc.17.00414
Persoon, G. A. (2008). “Growing ‘the wood of the gods’: agarwood production in Southeast Asia”. in Smallholder tree growing for Rural Development and Environmental Services. Eds. Snelder, D. J., Lasco, R. D. (Dordrecht, NL: Springer Press), 245–262. doi: 10.1007/978-1-4020-8261-0_12
Project, A. G., Albert, V. A., Barbazuk, W. B., dePamphilis, C. W., Der, J. P., Leebens-Mack, J., et al. (2013). The Amborella genome and the evolution of flowering plants. Science 342 (6165), 1241089. doi: 10.1126/science.1241089
Qu, L., Li, H.-L., Guo, D., Wang, Y., Zhu, J.-H., Yin, L.-Y., et al. (2020). HbWRKY27, a group IIe WRKY transcription factor, positively regulates HbFPS1 expression in Hevea brasiliensis. Sci. Rep. 10 (1), 1–8. doi: 10.1038/s41598-020-77805-5
Qu, D., Wu, F., Zhao, X., Zhu, D., Gu, L., Yang, L., et al. (2022). A bZIP transcription factor VabZIP12 from blueberry induced by dark septate endocyte improving the salt tolerance of transgenic Arabidopsis. Plant Sci. 315, 111135. doi: 10.1016/j.plantsci.2021.111135
Rahimi, S., Kim, Y.-J., Yang, D.-C. (2015). Production of ginseng saponins: elicitation strategy and signal transductions. Appl. Microbiol. Biot. 99 (17), 6987–6996. doi: 10.1007/s00253-015-6806-8
Rasool, S., Mohamed, R. (2016). “Understanding agarwood formation and its challenges. in: Agarwood science behind the fragrance” Ed. Mohamed, R. (Singapore, SG: Springer Press), 39–56. doi: 10.1007/978-981-10-0833-7_3
Rong, S., Wu, Z., Cheng, Z., Zhang, S., Liu, H., Huang, Q. (2020). Genome-wide identification, evolutionary patterns, and expression analysis of bZIP gene family in olive (Olea europaea L.). Genes 11 (5), 510. doi: 10.3390/genes11050510
Ruberti, C., Lai, Y., Brandizzi, F. (2018). Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J. 93 (1), 155–165. doi: 10.1111/tpj.13768
Sangareswari, M., Parthiban, K. T., Kanna, S. U., Karthiba, L., Saravanakumar, D. (2016). Fungal microbes associated with agarwood formation. Am. J. Plant Sci. 7 (10), 1445–1452. doi: 10.4236/ajps.2016.710138
Shine, M., Xiao, X., Kachroo, P., Kachroo, A. (2019). Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 279, 81–86. doi: 10.1016/j.plantsci.2018.01.001
Shu, K., Chen, F., Zhou, W., Luo, X., Dai, Y., Shuai, H., et al. (2018). ABI4 regulates the floral transition independently of ABI5 and ABI3. Mol. Biol. Rep. 45 (6), 2727–2731. doi: 10.1007/s11033-018-4290-9
Song, X., Ma, X., Li, C., Hu, J., Yang, Q., Wang, T. (2018). Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genomics 19 (1), 1–15. doi: 10.1186/s12864-018-4744-4
Stracke, R., FAVORY, J. J., Gruber, H., Bartelniewoehner, L., Bartels, S., Binkert, M., et al. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33 (1), 88–103. doi: 10.1111/j.1365-3040.2009.02061.x
Sun, P., Jiao, B., Yang, Y., Shan, L., Li, T., Li, X., et al. (2022b). WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant 15 (12), 1841–1851. doi: 10.1016/j.molp.2022.10.018
Tucker, M. L., Whitelaw, C. A., Lyssenko, N. N., Nath, P. (2002). Functional analysis of regulatory elements in the gene promoter for an abscission-specific cellulase from bean and isolation, expression, and binding affinity of three TGA-type basic leucine zipper transcription factors. Plant Physiol. 130 (3), 1487–1496. doi: 10.1104/pp.007971
Unel, N. M., Cetin, F., Karaca, Y., Celik Altunoglu, Y., Baloglu, M. C. (2019). Comparative identification, characterization, and expression analysis of bZIP gene family members in watermelon and melon genomes. Plant Growth Regul. 87 (2), 227–243. doi: 10.1007/s10725-018-0465-6
Vanneste, K., Baele, G., Maere, S., Van de Peer, Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res. 24 (8), 1334–1347. doi: 10.1101/gr.168997.113
Vinson, C. R., Sigler, P. B., McKnight, S. L. (1989). Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246 (4932), 911–916. doi: 10.1126/science.2683088
Wang, L., Fobert, P. R. (2013). Arabidopsis clade I TGA factors regulate apoplastic defences against the bacterial pathogen Pseudomonas syringae through endoplasmic reticulum-based processes. PloS One 8 (9), e77378. doi: 10.1371/journal.pone.0077378
Wang, X.-H., Gao, B.-W., Nakashima, Y., Mori, T., Zhang, Z.-X., Kodama, T., et al. (2022d). Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl) chromones in agarwood. Nat. Commun. 13 (1), 1–12. doi: 10.1038/s41467-022-27971-z
Wang, S., Liang, H., Wang, H., Li, L., Xu, Y., Liu, Y., et al. (2022b). The chromosome-scale genomes of Dipterocarpus turbinatus and Hopea hainanensis (Dipterocarpaceae) provide insights into fragrant oleoresin biosynthesis and hardwood formation. Plant Biotechnol. J. 20 (3), 538–553. doi: 10.1111/pbi.13735
Wang, J., Yuan, J., Yu, J., Meng, F., Sun, P., Li, Y., et al. (2019). Recursive Paleohexaploidization shaped the durian genome. Plant Physiol. 179 (1), 209–219. doi: 10.1104/pp.18.00921
Wang, S., Zhang, X., Li, B., Zhao, X., Shen, Y., Yuan, Z. (2022c). Genome-wide identification and characterization of bZIP gene family and cloning of candidate genes for anthocyanin biosynthesis in pomegranate (Punica granatum). BMC Plant Biol. 22 (1), 1–18. doi: 10.1186/s12870-022-03560-6
Wang, H., Zhang, Y., Norris, A., Jiang, C.-Z. (2022a). S1-bZIP transcription factors play important roles in the regulation of fruit quality and stress response. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.802802
Wei, K., Chen, J., Wang, Y., Chen, Y., Chen, S., Lin, Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19 (6), 463–476. doi: 10.1093/dnares/dss026
Weirauch, M. T., Hughes, T. (2011). A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution, A handbook of transcription factors (Springer), 25–73.
Weiste, C., Pedrotti, L., Selvanayagam, J., Muralidhara, P., Fröschel, C., Novák, O., et al. (2017). The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PloS Genet. 13 (2), e1006607. doi: 10.1371/journal.pgen.1006607
Weltmeier, F., Rahmani, F., Ehlert, A., Dietrich, K., Schütze, K., Wang, X., et al. (2009). Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 69 (1), 107–119. doi: 10.1007/s11103-008-9410-9
Wu, S., Zhu, P., Jia, B., Yang, J., Shen, Y., Cai, X., et al. (2018). A Glycine soja group S2 bZIP transcription factor GsbZIP67 conferred bicarbonate alkaline tolerance in Medicago sativa. BMC Plant Biol. 18 (1), 1–10. doi: 10.1186/s12870-018-1466-3
Yang, Z., Sun, J., Chen, Y., Zhu, P., Zhang, L., Wu, S., et al. (2019). Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet. 20 (1), 1–18. doi: 10.1186/s12863-019-0743-y
Zander, M., La Camera, S., Lamotte, O., Métraux, J. P., Gatz, C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61 (2), 200–210. doi: 10.1111/j.1365-313X.2009.04044.x
Zander, M., Thurow, C., Gatz, C. (2014). TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 165 (4), 1671–1683. doi: 10.1104/pp.114.243360
Zhang, Z. (2022). KaKs_Calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinf. 20 (3), 536–540. doi: 10.1016/j.gpb.2021.12.002
Zhang, Y., Gao, W., Li, H., Wang, Y., Li, D., Xue, C., et al. (2020b). Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genet. 21 (1), 1–14. doi: 10.1186/s12864-020-06890-7
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018a). Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genet. 19 (1), 1–14. doi: 10.1186/s12864-018-4511-6
Zhang, L., Ma, X., Zhang, X., Xu, Y., Ibrahim, A. K., Yao, J., et al. (2021). Reference genomes of the two cultivated jute species. Plant Biotechnol. J. 19 (11), 2235–2248. doi: 10.1111/pbi.13652
Zhang, Z., Quan, S., Niu, J., Guo, C., Kang, C., Liu, J., et al. (2022b). Genome-Wide identification, classification, expression and duplication analysis of bZIP family genes in Juglans regia L. Int. J. Mol. Sci. 23 (11), 5961. doi: 10.3390/ijms23115961
Zhang, Y., Xu, Z., Ji, A., Luo, H., Song, J. (2018b). Genomic survey of bZIP transcription factor genes related to tanshinone biosynthesis in Salvia miltiorrhiza. Acta Pharm. Sin. B 8 (2), 295–305. doi: 10.1016/j.apsb.2017.09.002
Zhang, L., Xu, Y., Zhang, X., Ma, X., Zhang, L., Liao, Z., et al. (2020a). The genome of kenaf (Hibiscus cannabinus L.) provides insights into bast fibre and leaf shape biogenesis. Plant Biotechnol. J. 18 (8), 1796–1809. doi: 10.1111/pbi.13341
Zhang, Q., Zhang, W. J., Yin, Z. G., Li, W. J., Xia, C.-Y., Sun, H.-Y., et al. (2022a). Genome-wide identification reveals the potential functions of the bZIP gene family in common bean (Phaseolus vulgaris) in response to salt stress during the sprouting stage. J. Plant Growth Regul. 41 (8), 3075–3090. doi: 10.1007/s00344-021-10497-x
Zhang, Y., Zhou, J., Wang, L. (2014). Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. 13 (2), 3025–3036. doi: 10.4238/2014.april.16.11
Zhao, B., Wang, L., Pang, S., Jia, Z., Wang, L., Li, W., et al. (2020). UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind. Crops Prod. 151, 112483. doi: 10.1016/j.indcrop.2020.112483
Zheng, C., Santos Muñoz, D., Albert, V. A., Sankoff, D. (2015). Syntenic block overlap multiplicities with a panel of reference genomes provide a signature of ancient polyploidization events. BMC Genomics 16 (10), 1–6. doi: 10.1186/1471-2164-16-S10-S8
Zhou, Y., Massonnet, M., Sanjak, J. S., Cantu, D., Gaut, B. S. (2017). Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. P. Natl. Acad. Sci. 114 (44), 11715–11720. doi: 10.1073/pnas.1709257114
Keywords: bZIP, agarwood, Malvales, evolution, transcriptional regulation, chromone biosynthesis
Citation: Zhang H, Ding X, Wang H, Chen H, Dong W, Zhu J, Wang J, Peng S, Dai H and Mei W (2023) Systematic evolution of bZIP transcription factors in Malvales and functional exploration of AsbZIP14 and AsbZIP41 in Aquilaria sinensis. Front. Plant Sci. 14:1243323. doi: 10.3389/fpls.2023.1243323
Received: 20 June 2023; Accepted: 24 July 2023;
Published: 30 August 2023.
Edited by:
Bo Sun, Sichuan Agricultural University, ChinaReviewed by:
Baosheng Liao, Guangzhou University of Chinese Medicine, ChinaCopyright © 2023 Zhang, Ding, Wang, Chen, Dong, Zhu, Wang, Peng, Dai and Mei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xupo Ding, eHVwb2RpbmdAaG90bWFpbC5jb20=; Wenli Mei, bWVpd2VubGlAaXRiYi5vcmcuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.