
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 October 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1243030
Juglans mandshurica (Manchurian walnut) is a precious timber and woody grain and oil species in Northeast China. The heterodichogamous characteristic phenomenon resulted in the non-synchronous flowering and development of male and female flowers, which limited the mating and the yield and quality of fruits. LFY is a core gene in the flowering regulatory networks, which has been cloned in J. mandshurica, and the function has also been verified preliminarily. In this study, the JmLFY promoter sequence with different lengths of 5′-deletion (pLFY1-pLFY6) were cloned and conducted bioinformatics analysis, the promoter activities were analyzed by detecting their driving activity to GUS gene in the tobacco plants that transformed with different promoter sequence stably or transiently. After that, the interaction between JmSOC1 and JmLFY gene promoter was also analyzed via yeast single-hybrid. The results showed that the promoter sequence contains core cis-acting elements essential for eukaryotic promoters, hormone response elements, defense- and stress-responsive elements, flowering-related elements, etc. Transgenic tobacco plants with pLFY1 were obtained by Agrobacterium infection using the pCAMBIA1301 expression vector, and the GUS gene driven by the JmLFY promoter was detected to express in the leaf, stem, flower, and root of the transformed tobacco plant, which indicated that the obtained JmLFY promoter had driving activity. GUS histochemical staining and enzyme activity detection showed that promoter fragments with different lengths had promoter activity and could respond to the induction of long photoperiod, low temperature, salicylic acid (SA), IAA, GA3, and methyl jasmonate (MeJA). The core regulatory region of JmLFY gene promoter in J. mandshurica was between −657 bp and −1,904 bp. Point-to-point validation of yeast single-hybrid confirmed the interaction between JmSOC1 and JmLFY gene promoter, which indicated that JmLFY gene is the downstream target of JmSOC1. These results reveal relevant factors affecting JmLFY gene expression and clarify the molecular mechanism of JmLFY gene regulation in the flower developmental partially, which will provide a theoretical basis for regulating the flowering time by regulating JmLFY gene expression in J. mandshurica.
Flowering is an important process in the plant life cycle, which means the end of childhood life and the plants begin reproductive growth (Zhang and Liu, 2003; Zhang et al., 2019a; Zhang et al., 2019b). The transitions from vegetative growth to reproductive growth were affected by the combination of internal factors and external environmental factors (Aidyn et al., 2002; Zeng et al., 2018), such as age, photoperiod, vernalization, autonomous pathway, and gibberellin pathway (Martina et al., 2015; Jian et al., 2019; Jin et al., 2019; Liu et al., 2021). Thus, the flower development and flower bud differentiation processes in plants are regulated by complex gene regulatory networks (Wang et al., 2004).
LFY has a core position in the flowering regulatory networks (Gordon and Caroline, 2002; Liu et al., 2009; Zhao et al., 2020), which participate in several pathways mentioned above, and plays crucial roles in promoting the formation of floral primordia, maintaining floral meristem function and floral initiation, and preventing the reversal of floral meristem (Shannon and Meeks-Wagner, 1993; Mandel and Yanofsky, 1995; Yanofsky, 1995; Chen et al., 1997; Alvarez-Buylla et al., 2006; He et al., 2018). The overexpression of LFY genes promoted early flowering and supplemented the phenotypic defects of lfy mutant partially (Weigel et al., 1993; He et al., 2000; Ahearn et al., 2001; Peña et al., 2001; Cai et al., 2023), which demonstrates the important role of LFY genes in flowering regulation further.
Juglans mandshurica is a precious timber and woody grain and oil species in Northeast China, which has important economic, nutritional, and medicinal values. As a monoecious species, we found that the heterodichogamous characteristics are a common phenomenon in J. mandshurica in the previous investigation of the reproductive phenological characteristics of the species (Guo, 2020; Qin et al., 2021), which resulted in the non-synchronous flowering and development of male and female flowers and then limited the mating and the yield and quality of fruits. Therefore, it is necessary to solve the bottleneck problem of low fruit yield caused by the inconsistent development of male and female flowers. We have cloned the JmLFY from J. mandshurica successfully (Song, 2019; Liu et al., 2022), and the genetic transformation and function verification studies were also conducted by transformed JmLFY into Arabidopsis (Cai, 2022). Overexpression of JmLFY gene in Arabidopsis inhibited vegetative growth, promoted reproductive growth, and supplemented the phenotypic defects of lfy mutant partially.
Promoters are able to determine the expression level of genes and occupy an important role in the regulation of gene transcription. In this study, we cloned the promoter of JmLFY genes and predicted the cis-acting elements and active sites on promoters using online bioinformatics software. In order to identify factors affecting the expression of JmLFY genes, we constructed a series of plant expression vectors using different lengths of the promoter with 5′-deletion and verified the activities of the promoter. Further validation of interaction between JmSOC1 and the JmLFY promoter was also conducted by point-to-point validation of yeast single-hybrid. The results will reveal relevant factors affecting JmLFY gene expression and clarify the molecular mechanism of JmLFY gene regulation in the flower developmental partially, which will provide a theoretical basis for regulating the flowering time by regulating JmLFY gene expression in J. mandshurica.
DNA extraction from leaves of J. mandshurica was conducted as described by Song (2019). According to the obtained JmLFY gene sequence (GenBank Accession No.: KX364241), an upstream non-coding nucleotide sequence (NC_049909.1) from Juglans regia genomic DNA was selected for JmLFY promoter cloning (Table 1). PCR amplification (the reaction system included LA Taq Mix 12.5 µL, DNA 1 µL, LFY-P-F and LFY-P-R (10 µmol/L) 1 µL, and ddH2O 9.5 µL). The reaction procedure was as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s, 58°C for 1 min, 72°C for 2 min; and 72°C for 7 min; the product was detected by 1% agarose gel electrophoresis and then recovered using the MiniBEST Agarose Gel DNA Extraction Kit version 4.0 (Takara, Dalian, China). The recovered product was then ligated to the pMD-19-T Vector (Takara) according to the instructions. The recombined vector was then transformed into Escherichia coli DH5α competent cells, the positive clones selected by 50 mg/L of ampicillin (Amp) were used for PCR amplification and then sequenced, and the correct recombined vector confirmed by sequencing was named pMD19-T-pLFY.
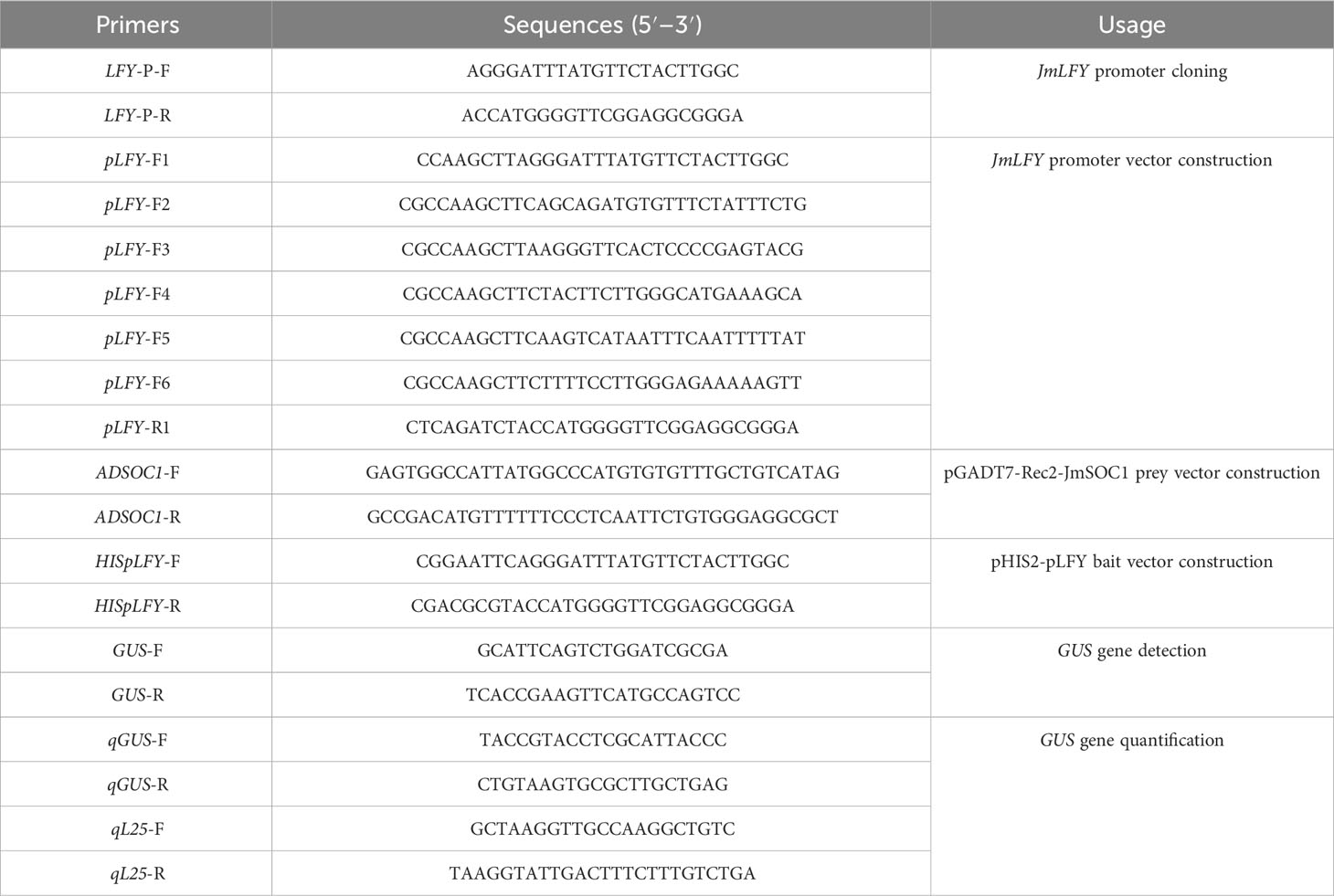
Table 1 Primer sequences for cloning and functional analysis of JmLFY promoter of Juglans mandshurica.
The obtained promoter sequence was aligned by DNAMAN6.0, and then regulatory elements were predicted and analyzed using online analysis software Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (https://www.dna.affrc.go.jp/PLACE/).
According to the predicted cis-acting element positions on the promoter, the plasmid DNA of pMD19-T-pLFY was extracted and amplified using six primers, which could amplify different lengths of JmLFY promoter fragments by designing 5′-deletion primers (Table 1, Figure 1A). The PCR products were named pLFY1 to pLFY6. The recovered PCR products and pCAMBIA1301 vector were then dual-digested with HindIII and BglII rapid endonuclease and ligated using T4 DNA Ligase. The recombined vector was then transformed into E. coli DH5α competent cells, and then the positive clones selected by 50 mg/L of kanamycin (Kan) were used for PCR amplification and then sequenced; the correct recombined vectors confirmed by sequencing were named pCAMBIA1301-pLFY1 to pCAMBIA1301-pLFY6, which were abbreviated respectively as p1301-pLFY1 to p1301-pLFY6 hereafter.
Figure 1 GUS histochemical staining and enzyme activity determination of transiently transformed Nicotiana benthamiana plants with different JmLFY promoter fragments. (A) Schematic illustration of different lengths of 5’-deletion primers for JmLFY promoter. (B) GUS histochemical staining of positive control (injected Agrobacterium with pCAMBIA1301 empty vector, i), negative control (injected Agrobacterium without vector, ii), p1301- pLFY1 (iii), p1301-pLFY2 (iv), p1301-pLFY3 (v), p1301-pLFY4 (vi), pA1301-pLFY5 (vii), and p1301-pLFY6 (viii). (C) GUS enzyme activity determination with different JmLFY promoter fragments. The different uppercase letters above the error bars mean significant difference at α = 0.01.
Agrobacterium EHA105 strain was cultured on YEP solid medium for 36 h at 28°C, and a single clone was selected and incubated in liquid YEP medium containing 50 mg/L of rifampin (Rif) with 200-rpm shaking frequency at 28°C till OD600 reached 0.5–0.6. The liquid was then centrifuged for 5 min at 4,000 g at 4°C, the supernatant was discarded, and the precipitate was resuspended in 1 mL of precooled CaCl2. The Agrobacterium EHA105 competent cells were added in 500 μL of 50% sterilized glycerol, then divided into 100 μL, frozen in liquid nitrogen, and stored in a −80°C freezer.
Plasmid DNA extraction of the recombinant vectors (p1301-pLFY1 to p1301-pLFY6) was conducted, mixed with Agrobacterium EHA105 competent cells on ice for 30 min, then frozen in liquid nitrogen for 5 min, and transferred to 37°C water bath for 5 min. The transformed products were cultured in liquid YEP medium overnight at 28°C with 200-rpm shaking frequency and then centrifuged for 1 min at 4,000 g, and most of the supernatant was discarded. The remaining approximately 100 μL was used for resuspending the precipitate, then cultured on YEP solid medium containing 50 mg/L of Kan and 50 mg/L of Rif, inverted petri dishes, and incubated at 28°C for 2–3 days. The single clone was selected and incubated in a liquid YEP medium containing 50 mg/L of Kan and 50 mg/L of Rif with 200-rpm shaking frequency at 28°C. The solution with propagated transformation products was detected by PCR amplification and 1% agarose gel electrophoresis.
The verified transformed products were coated on a solid YEP medium containing 50 mg/L of Kan and 50 mg/L of Rif. A single clone was selected and incubated in 50 mL of liquid YEP medium containing 50 mg/L of Kan and 50 mg/L of Rif till OD600 reached 0.5–0.6. The liquid was then centrifuged for 10 min at 5,000 g, and the supernatant was discarded. The precipitate was resuspended with MS medium [containing 30 g/L of sucrose and 100 μM of acetosyringone (AS)] till OD600 reached 0.5–0.6, which was used as an infection solution.
The prepared p1301-pLFY1 infection solution was then used for infected leaves of aseptic Nicotiana benthamiana seedlings for 30–45 days. The leaves were cut into 0.5–1 cm2 and soaked in the infection solution for 8–10 min. The soaked leaves were co-cultured on MS + 1 mg/L 6-BA + 0.1 mg/L NAA at 25°C for 3 days, then washed successively with sterile water containing ceftazidime (Cef) 1,000 mg/L (2 min) and 500 mg/L (1 min), and then washed with sterile water for three times. The sterile leaves were transferred successively on selection medium for callus induction (MS + 1 mg/L 6-BA + 0.1 mg/L NAA + 500 mg/L Cef + 10 mg/L hygromycin (Hyg), 3 weeks), differentiation (MS + 0.5 mg/L 6-BA + 0.05 mg/L NAA + 500 mg/L Cef + 10 mg/L Hyg, 1 week), elongation (MS + 0.2 mg/L 6-BA + 0.02 mg/L NAA + 500 mg/L Cef + 10 mg/L Hyg, 1 week), and rooting (MS + 0.02 mg/L NAA + 500 mg/L Cef + 10 mg/L Hyg, 2 weeks). The rooted tobacco seedlings were transferred into pots and covered with plastic cups with high light transmittance for 2–3 days, and then the covers were removed.
The prepared p1301-pLFY1 to p1301-pLFY6 infection solutions were transiently transformed into tobacco leaves by injecting using a 1-mL disposable injector. The injected plants were bagged in the dark for 2–3 days and then transferred to light.
Considering that the JmLFY promoter contains the light-responsive elements, hormone response elements, and low-temperature responsive elements, the transiently transformed tobacco plants were then treated with photoperiod (16-h light/8-h dark and 8-h light/16-h dark), temperature (4°C and 25°C), and hormone (salicylic acid (SA), ABA, IAA, GA3, and methyl jasmonate (MeJA), with H2O as control) spraying to verify the function of the core regulatory region and the cis-acting elements of the JmLFY promoter. The photoperiod and low-temperature treatments were conducted in transiently transformed tobacco plants with p1301-pLFY1, p1301-pLFY2, p1301-pLFY3, p1301-pLFY4, p1301-pLFY5, and p1301-pLFY6, and the hormone treatments were conducted only in transiently transformed tobacco plants with p1301-pLFY1. After 24 h of treatments, the GUS histochemical staining and GUS enzyme activity determination were performed.
RNA was extracted from the roots, stems, leaves, and flowers of transgenic plants, which were reverse transcribed into cDNA. GUS gene expression in these organs was then detected by qRT-PCR with tobacco L25 gene as the reference gene, and the primers are listed in Table 1. Furthermore, in order to examine the transcriptional activity of GUS genes driven by the JmLFY promoter in different organs in the transgenic plants, GUS histochemical staining was performed using a GUS staining kit (Beijing Coolaber Technology Co., Ltd., Beijing, China).
For the transient transformation tobacco plants, GUS histochemical staining and enzyme activity determination (GUS enzymatic activity assay kit, Beijing Coolaber Technology Co., Ltd.) of punched leaves were performed.
Based on the JmSOC1 sequence information and the restriction endonuclease SmaI site and its flanking sequences of the pGADT7-Rec2 vector, the upstream and downstream primers ADSOC1-F and ADSOC1-R were designed (Table 1). The pMD19-T-JmSOC1 target fragment was then amplified by PCR using plasmid DNA extracted from pMD19-T-JmSOC1 (conserved by Key Laboratory of Forest Tree Genetics and Breeding of Liaoning Province) as a template. Plasmid of yeast vector pGADT7-Rec 2 was extracted and digested using the restriction endonuclease SmaI; the products were recovered, purified, and then ligated with pMD19-T-JmSOC1 target fragment using NovoRec plus One step PCR Cloning Kit. The ligated products were then transformed into E. coli Top10 competent cells, and the bacteria solution was detected by PCR and sequenced to identify the recombinant vector. The correct vector was used as a prey vector for yeast one-hybrid and named pGADT7-SOC1.
According to the JmLFY promoter sequence and the recognition site of the pHIS2 restriction endonuclease, the EcoRI and MluI restriction sites were introduced at the 5′ end of the upstream and downstream primers of the JmLFY promoter. The primers HISpLFY-F and HISpLFY-R with the EcoRI and MluI restriction sites were designed (Table 1) and used for JmLFY promoter amplification. The amplified JmLFY promoter was recovered, purified, dual-digested by EcoRI and MluI, and then ligated to pHIS2 linear vector fragments that were also dual-digested by EcoRI and MluI using T4 DNA Ligase. The ligated products were then transformed into E. coli Top10 competent cell, and the bacteria solution was detected by PCR and sequenced to identify the recombinant vector. The correct vector was used as a bait vector for yeast one-hybrid and named pHIS2-LFY.
The positive control (pGADT7-rec2-p53 and pHIS2-p53), negative control (pGADT7-Rec2 and pHIS2-p53), self-activation assay (pGADT7-Rec2 and pHIS2-LFY), and interaction assay (pGADT7-SOC1 and pHIS2-LFY) were co-transformed into yeast competent cell Y187, and the yeast solution was then coated on DDO medium (SD/-Leu/-Trp) and TDO medium (SD/-His/-Leu/-Trp) with various concentrations of 3-AT (60 mM, 90 mM, 150 mM, and 200 mM). The cultures were placed upside down and incubated at 30°C for 2–4 days.
A 2,170-bp sequence was obtained and compared to the cDNA of JmLFY gene and the upstream sequences of LFY in J. regia published by the National Center for Biotechnology Information (NCBI). A total of 55 bases with 95% similarity at the 3′ end of the obtained sequence overlapped with the 5′ end cDNA of JmLFY gene, which indicated that we obtained the upstream sequence of JmLFY. Compared with the remaining 2,115 bp to upstream sequences of LFY in J. regia, over 90% similarity suggested that we obtained the promoter sequence of JmLFY gene successfully (SRR24958161, Figure S1).
The LFY promoter regulatory element analysis showed that various functional cis-acting elements exist on JmLFY gene promoter, including the core cis-acting elements essential for eukaryotic promoters, such as TATA-box, CAAT-box; light-responsive elements, such as Box 4, G-Box, GT1-motif; hormone response elements, such as abscisic acid response element ABRE, MeJA response element CGTCA-motif and TGACG-motif, ethylene-responsive ERE, gibberellin-responsive TATC-box, cis-acting element involved in salicylic acid-responsive TCA, and auxin response element TGA, etc.; stress-responsive elements, such as low temperature-responsive cis-acting element (LTR), MYB binding site involved in drought inducibility (MBS) and light responsiveness (MRE), cis-acting element involved in defense and stress responsiveness (TC-rich repeats), MYB response elements associated with drought, salt and abscisic acid response, MYC associated with drought and abscisic acid response; flowering-related elements, such as the cis-acting element POLLEN1LELAT52 that is specifically expressed by pollen, the late pollen gene initiator element GTGANTG10, and the binding site CArG-box motif for flowering-related proteins; and other elements, such as anaerobic-induced regulatory element ARE, cis-regulatory element involved in regulation of zein metabolism O2-site, damage response element WRE3, etc.; and some elements of unknown function (Table S1, Figure S1). These results suggested that the expression of LFY gene may be regulated by several factors.
To identify the core regulatory regions of the JmLFY promoter, a total six of expression vectors (p1301-pLFY1 to p1301-pLFY6) with different lengths of 5′-deletion fragments were constructed successfully. The expression vector p1301-pLFY1 was transformed into N. benthamiana, and regenerated plants were obtained via callus induction, differentiation, elongation, rooting, and acclimatization (Figure 2), and six transgenic tobacco plants were determined by PCR with GUS gene universal primer and pLFY 1-specific primer (Figure S2).
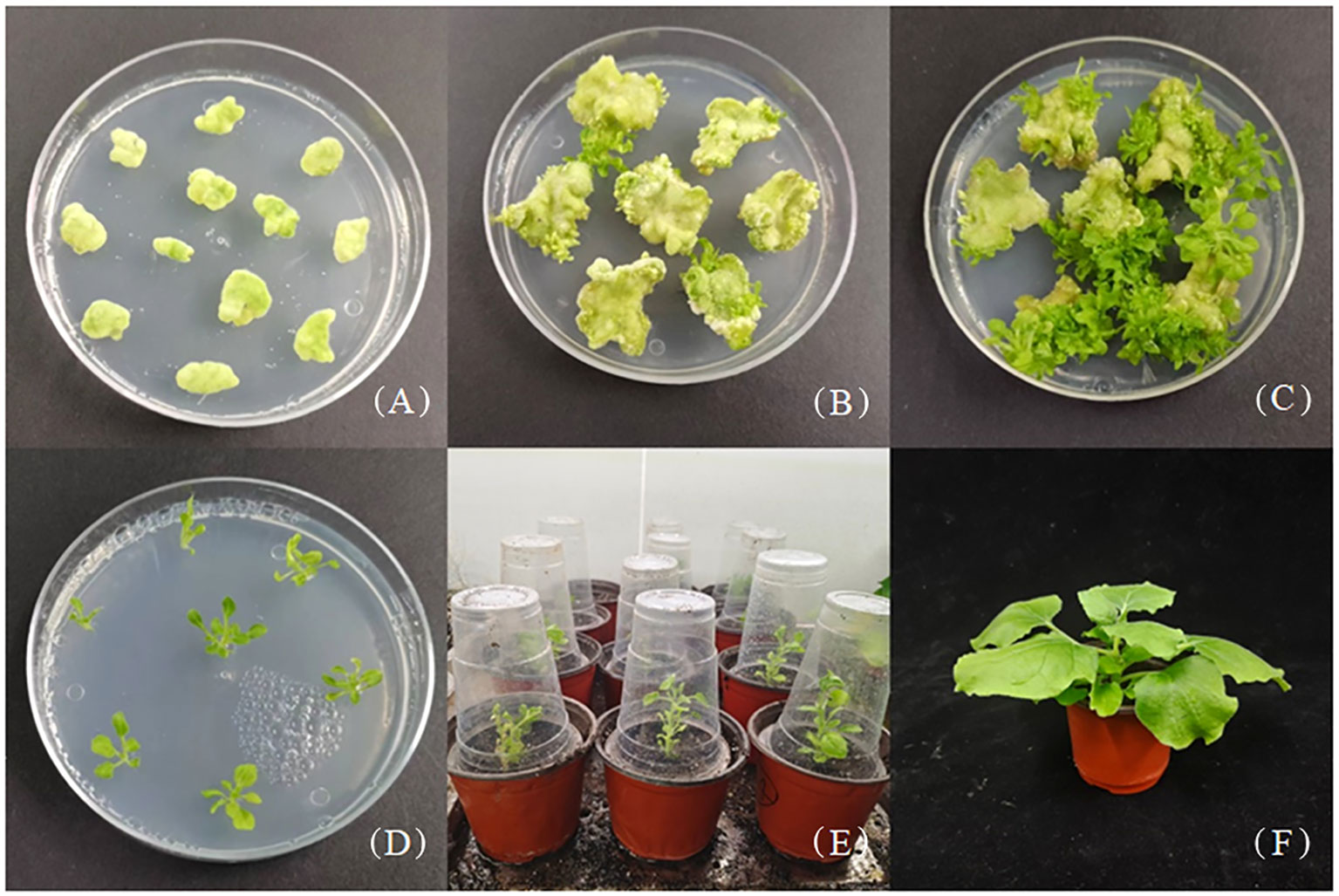
Figure 2 Genetic transformation of Nicotiana benthamiana with p1301-pLFY1. (A) Callus induction. (B, C) Callus differentiation and adventitious shoot elongation. (D) Rooting of adventitious shoots. (E) Transplanting. (F) Transgenic plant.
GUS histochemical staining in different organs of transgenic tobacco showed that JmLFY gene promoter drove GUS gene transcriptional activity in all detected organs, which stained the deepest in the leaf and the slightest in the root (Figure 3A). qRT-PCR was also performed on roots, stems, leaves, and flowers of the transgenic tobacco, which showed similar results as GUS histochemical staining. The expression of GUS gene was the highest in the leaf of transgenic tobacco and the lowest in the root (Figure 3B).
Figure 3 GUS histochemical staining and qRT-PCR analysis of GUS gene in different organs of transgenic Nicotiana benthamiana. (A) Root, stem, leaf, and flower of wild type (upper) and transgenic (lower) tobacco. (B) qRT-PCR analysis of GUS gene in root, stem, leaf, and flower of transgenic N. benthamiana. The different uppercase letters above the error bars mean significant difference at α = 0.01.
GUS staining showed that the positive control (injected Agrobacterium with pCAMBIA1301 empty vector) was stained the deepest, and the leaves of negative control (injected Agrobacterium without vector) were not stained. The other leaves from transiently transformed plants were stained except for p1301-pLFY6, of which p1301-pLFY1 and p1301-pLFY2 were stained deeply, followed by p1301-pLFY3 and p1301-pLFY 4, and p1301-pLFY5 was stained slightly (Figure 1B).
GUS enzyme activity determination confirmed the histochemical staining results further (Figure 1C), which showed the highest activity of positive control, followed by p1301-pLFY2, p1301-pLFY1, p1301-pLFY3, p1301-pLFY4, p1301-pLFY5, and p1301-pLFY6 successively.
GUS histochemical staining of transiently transformed plants treated by 2 days of long photoperiod (16-h light/8-h dark) and short photoperiod (8-h light/16-h dark) showed that transiently transformed plants with different JmLFY promoter fragments were stained in different levels, which decrease progressively from p1301-pLFY1 to p1301-pLFY6 (Figure 4A). However, all the leaves from plants treated by a long photoperiod were stained more strongly than those treated by a short photoperiod (Figure 4A). GUS enzyme activity determination showed similar results to GUS histochemical staining (Figure 4B).
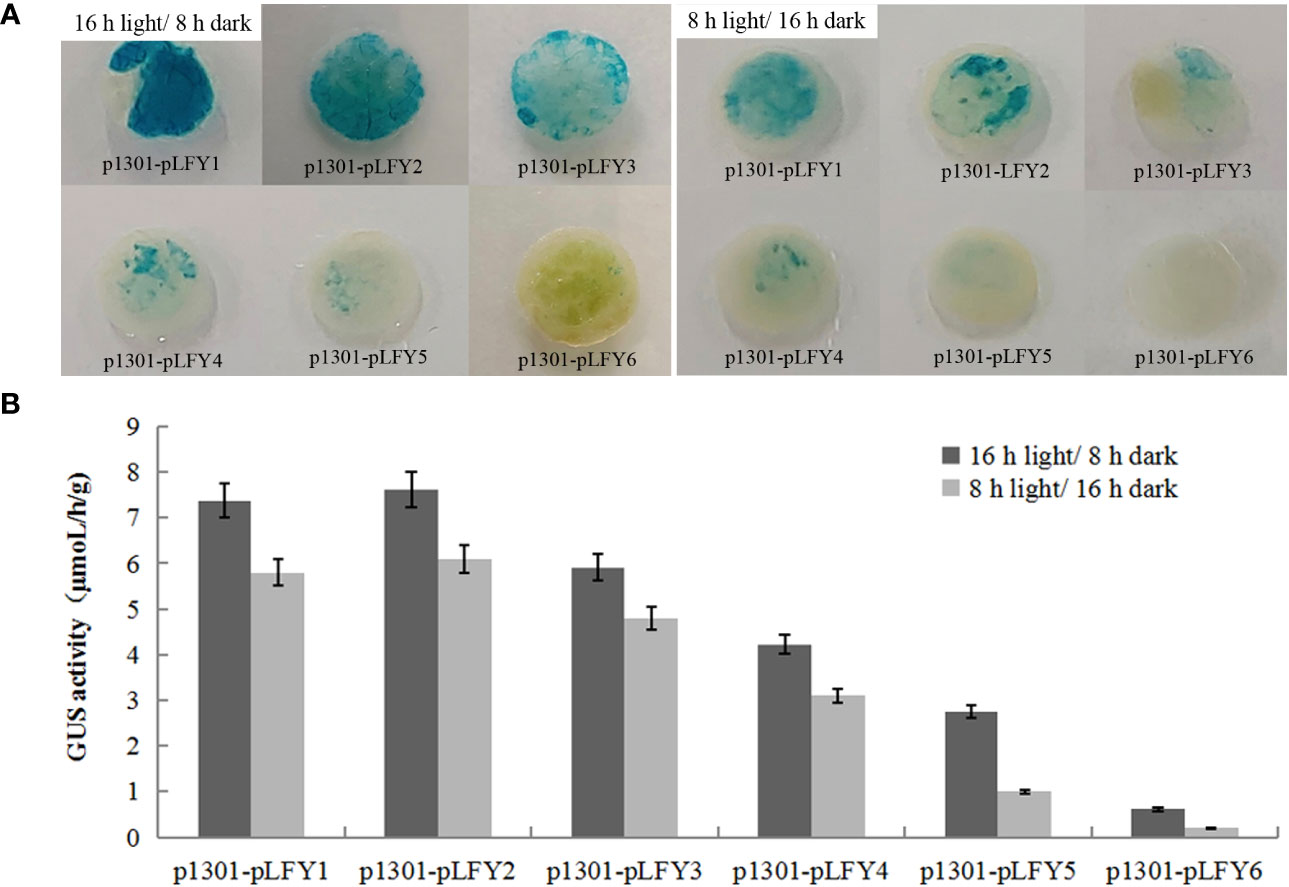
Figure 4 GUS histochemical staining (A) and enzyme activity determination (B) of transient transformed Nicotiana benthamiana plants with different JmLFY promoter fragments under long (16-h light/8-h dark) and short (8-h light/16-h dark) photoperiod.
GUS histochemical staining of transiently transformed plants treated at normal temperature (25°C) and low temperature (4°C) showed that transiently transformed plants with different JmLFY promoter fragments were stained at different levels (Figure 5A). However, leaves from plants treated with low temperatures were stained more strongly than those treated with normal temperatures except for p1301-pLFY5 and p1301-pLFY6 (Figure 5A). GUS enzyme activity determination showed similar results to GUS histochemical staining (Figure 5B).
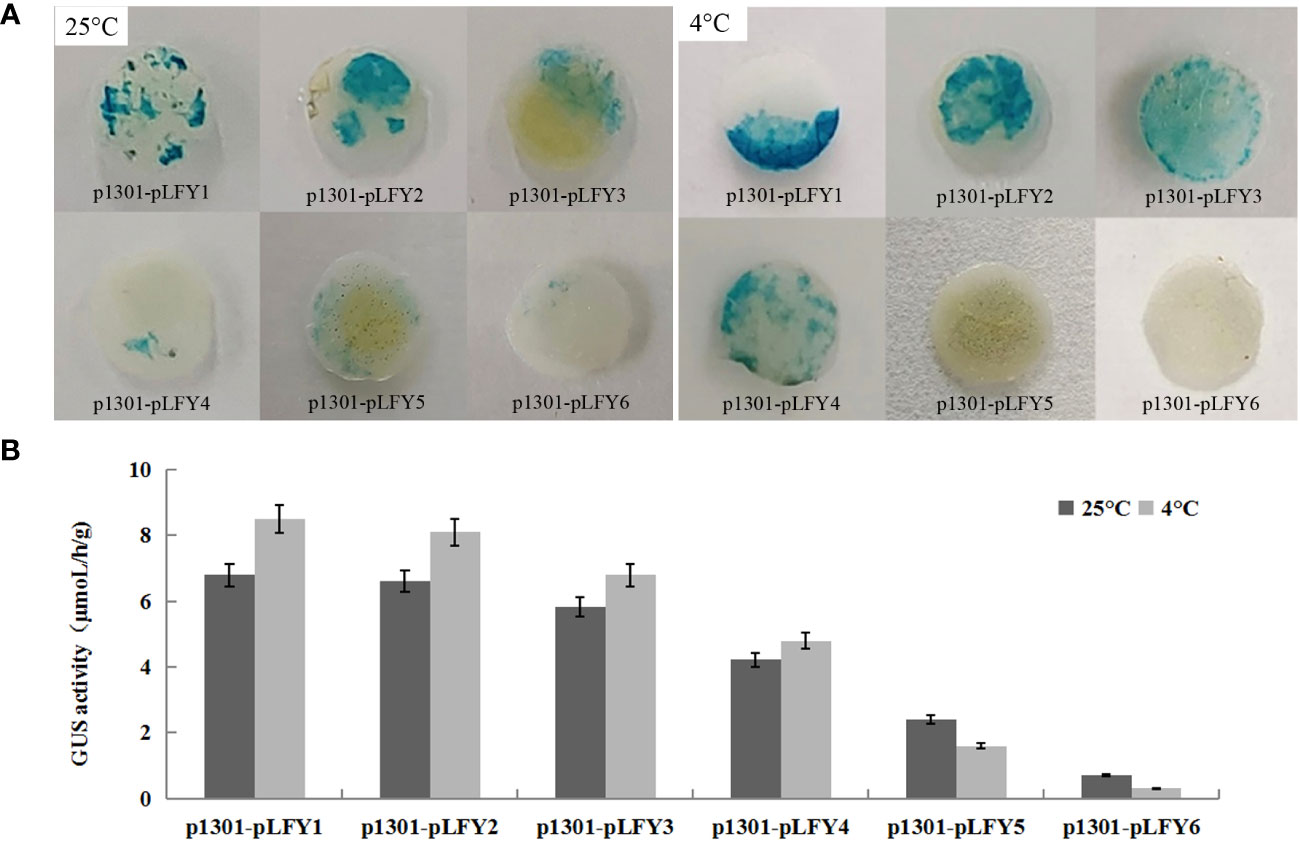
Figure 5 GUS histochemical staining (A) and enzyme activity determination (B) of transiently transformed Nicotiana benthamiana plants with different JmLFY promoter fragments under 25°C and 4°C.
GUS histochemical staining and enzyme activity determination of transiently transformed plants treated by hormones showed that the treated transiently transformed plants with p1301-pLFY1 had higher activities than those treated by H2O (control), except for ABA, which showed lower activities than control (Figure 6). These results indicated that the p1301-pLFY1 promoter fragment responded to the induction of SA, IAA, GA3, and MeJA, and the treatments of SA, IAA, GA3, and MeJA increased the activities of JmLFY1 promoter.
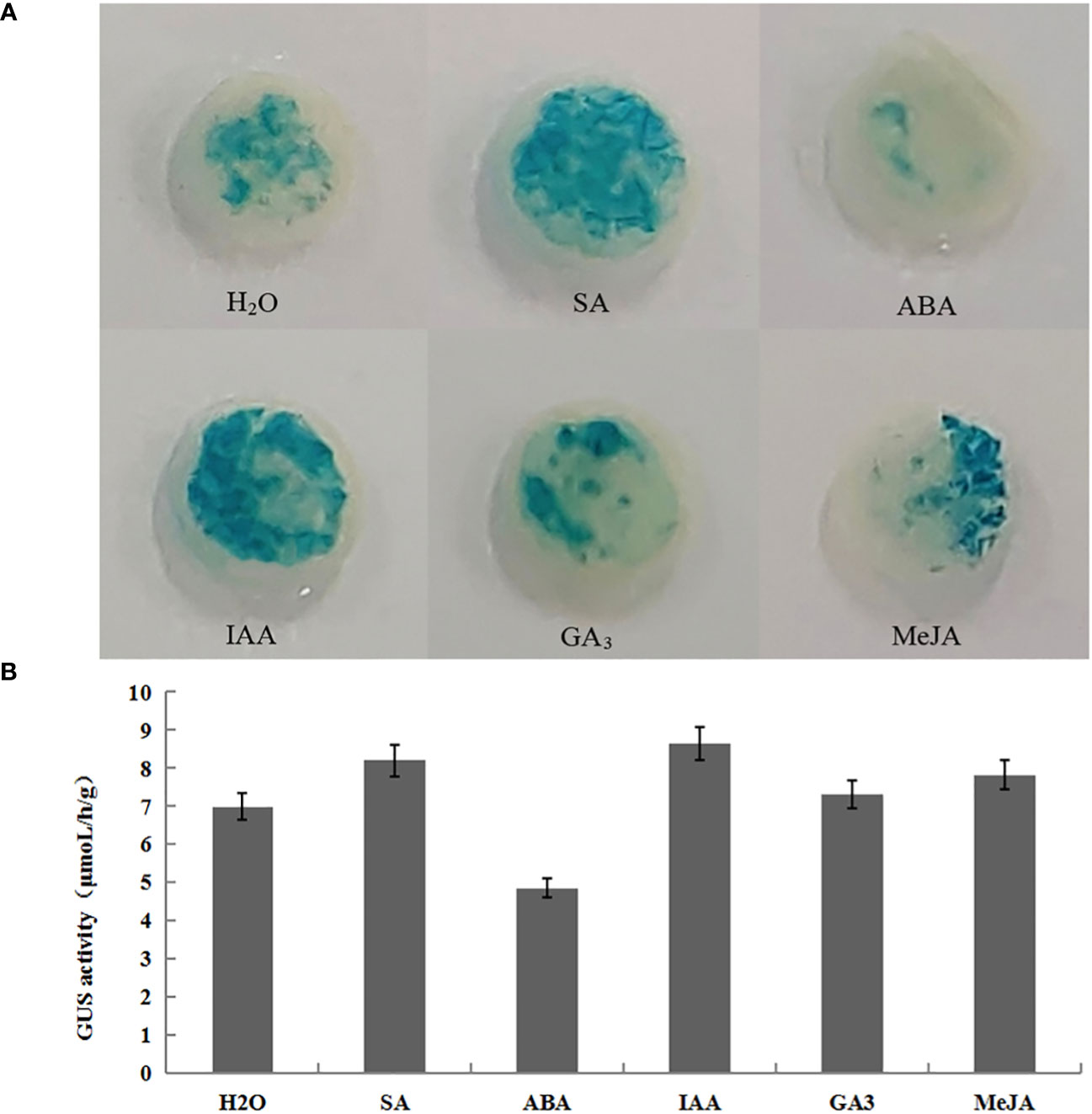
Figure 6 GUS histochemical staining (A) and enzyme activity determination (B) of transiently transformed Nicotiana benthamiana plants with p1301-pLFY1 by spraying different hormones.
Point-to-point validation by yeast one-hybrid of JMSOC1 and the JmLFY promoter showed that all co-transformed yeast plasmid grew well on the DDO medium, which suggested that the co-transformations are successful and without obvious toxic effect on the Y187 yeast strain (Figure 7). Self-activation decreased followed by increasing concentration of 3-AT on TDO medium, which was inhibited completely when 3-AT concentration was over 90 mM. The strains of positive control (pGADT7-Rec2 + pHIS2-p53) and the interaction assay (pGADT7-Rec2 + pHIS2-LFY) grew well on TDO medium supplied with 60 mM and 90 mM, which indicated the interaction between JmSOC1 and the JmLFY promoter.
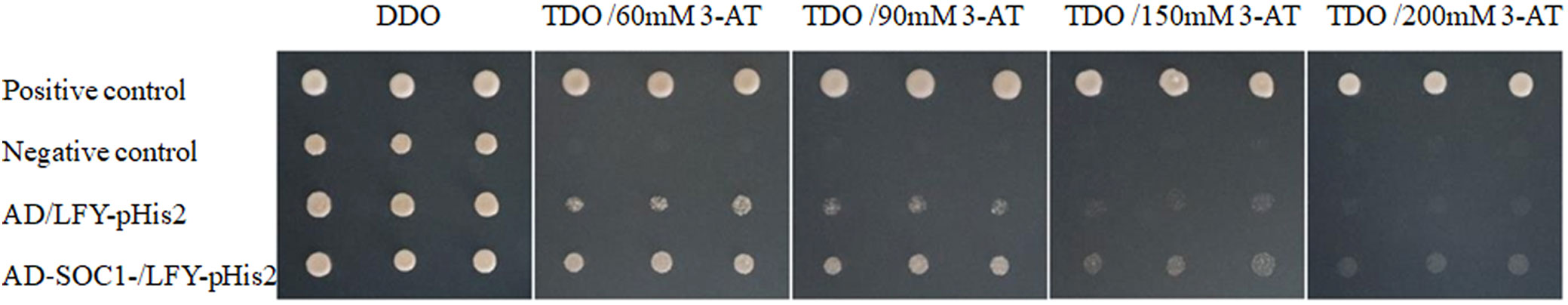
Figure 7 Point-to-point verification of yeast one-hybrid for co-transforming yeast plasmid. DDO and TDO mean Synthetic Dropout Media (SD) with Double Dropout Supplements (DDO, SD/-Leu/-Trp) and Triple Dropout Supplements (TDO, SD/-His/-Leu/-Trp).
J. mandshurica is a monoecious species with heterodichogamous characteristics. The non-synchronous flowering and development of male and female flowers limited the mating and the yield and quality of fruits of the species. LFY has a core position in the flowering regulatory networks in plants, and the overexpression of JmLFY gene has been confirmed to promote flowering approximately 7 days in advance and supplement the phenotypic defects of lfy mutant partially in Arabidopsis (Cai et al., 2023). In this study, we cloned the JmLFY promoter of 2,115 bp in length, which has more than 90% homologous sequence aligned with the J. regia promoter. GUS histochemical staining and expression analysis showed that the JmLFY promoter drove the expression of GUS gene in the leaves, stems, flowers, and roots of transgenic tobacco plants, which indicated that the LFY promoter had driving activity. Similar results were also reported in Populus tomentosa LFY promoter (Li et al., 2012a).
Bioinformatics analysis of the obtained sequence revealed that the JmLFY promoter contains the core cis-acting element essential to the eukaryotic promoter, such as TATA-box and CAAT-box, which are consistent with the basic structural characteristics of the promoter (Molina and Grotewold, 2005). In addition, some flowering-related elements, such as POLLEN1LELAT52, GTGANTG10, and CArG-box motif, were also reported in the LFY promoter of Dimocarpus longan (Xu et al., 2011).
To further determine the cis-elements and their function on the JmLFY promoter, several promoter fragments with different lengths of 5′-deletion were cloned and transformed transiently into tobacco plants. Decreased GUS activities followed by the reduction of JmLFY promoter sequences (Figures 4–6) indicated that the driving capacities of promoter fragments decreased by reduction of length, which might be caused by the decreasing numbers of responsive elements in the JmLFY promoter fragments. The higher activity of p1301-pLFY2 than p1301-pLFY1 suggested that a negative regulation region might exist in −1,904 bp to −2,115 bp of the JmLFY promoter, which needs to be studied further.
Light-responsive elements, such as Box 4, G-Box, and GT1-motif, which have been reported in LFY promoters in J. regia and Carya cathayensis (Sun et al., 2017), were also detected on the JmLFY promoter, which indicated that the promoters might be induced by light. In this study, long photoperiod (16-h light/8-h dark) enhanced the driving capacity of all the JmLFY promoters and confirmed that the light-responsive elements on the JmLFY promoter are more sensitive to the long photoperiod than the short photoperiod. The differences in GUS staining and enzyme activity among different-length promoter fragments might be related to the deletion of light-responsive elements in different promoter fragments.
Low temperature enhanced the driving capacity of all the JmLFY promoters except for p1301-pLFY5 and p1301-pLFY6, which showed higher activities under normal temperature than low temperature. However, the location of low temperature-responsive cis-acting element (LTR, −2,016 bp in Table S1) was contained by all the promoter fragments, suggesting that other sites that responded to low temperature might exist in the −657-bp to −2,115-bp fragments of the JmLFY promoter. In C. cathayensis, the LFY promoter expression also increased after being treated with low temperature and light (Li, 2012b), which is consistent with our research. However, the specific mechanism that low temperature and long photoperiod promoted the expression of LFY promoter was still unknown, which is worth studying further.
In Arabidopsis, some hormone response elements existed on LFY gene promoter, which responded to the induction of auxin (Nobutoshi et al., 2016). Similarly, a series of hormone response elements such as ABRE, MeJA response element, CGTCA-motif, TGACG-motif, ERE, P-box, TCA-element, and TGA-element (Table S1) were also detected on the JmLFY promoter. Exogenous spraying of SA, IAA, GA3, and MeJA to the transiently transformed plants with p1301-pLFY1 increased GUS activities than that treated by H2O (control), and exogenous spraying of ABA decreased GUS activities (Figure 6), which indicated the positive regulation of SA, IAA, GA3, and MeJA and the negative regulation of ABA. In N. benthamiana and rice, Ferredoxin 1 (FD1) promoter activity was repressed when applying exogenous ABA. The result was explained as that the accumulation of ABA stimulates the expression of ABI5 (ABSCISIC ACID-INSENSITIVE 5), an ABA-responsive transcriptional factor, which negatively regulates FD1 by binding to ABRE motifs in the NbFD1 promoter (Cui et al., 2021). Thus, we speculated that the inhibition of JmLFY promoter activity by ABA might also be due to a similar causation because two ARBEs existed on the JmLFY promoter. Currently, both positive and negative effects of endogenous or exogenous ABA on plant flowering have been reported. For example, ABI4 and ABI5 activated transcription of the flowering repressor gene FLOWERING LOCUS C (FLC) by binding the FLC promoter directly and then repressed the floral transition (Wang et al., 2013; Xiong et al., 2019). Exogenous application of ABA delayed flowering in plants (Wang et al., 2013). These results indicate the negative effects of ABA on floral transition. On the contrary, endogenous ABA upregulated FLOWERING LOCUS T (FT) expression in Arabidopsis thaliana, and root application of exogenous ABA in soil accelerated Arabidopsis flowering (Riboni et al., 2016), which indicated the positive effects of ABA on floral transition. In J. mandshurica, our previous study showed that the endogenous ABA content increased gradually with the differentiation of flower buds (Qin et al., 2022); however, no further studies were conducted. Thus, it is necessary to study the specific effects of ABA on the flowering of J. mandshurica.
It was found that the conserved binding domain of MADS-box on the SOC1 could specifically bind to the DNA sequence containing CArG-box, thus regulating the expression of the downstream target LFY gene (Kaufmann et al., 2005). However, a missense mutation in the MADS box of SOC1 could not bind to the LFY promoter and then suppressed the flowering promotion function. Similarly, the LFY promoter without CArG-box could not be bound with MADS-box on the SOC1 in Gossypium hirsutum (Li et al., 2013). In this study, the bioinformatics analysis of the JmLFY promoter showed that two CArG-box domains (CAATATATAG, −1,868 bp, and CCTTTATAGG, −1,919 bp; Table S1, Figure S1) were present on the promoter sequence. In order to prove the interaction between JmSOC1 and JmLFY, the pGADT7-Rec2-JmSOC1 prey vector and the pHIS2-pLFY bait vector were constructed for yeast one-hybrid point-to-point verification. The results showed that JmSOC1 interacted with JmLFY gene promoter. However, whether the JmSOC1 can affect the transcriptional activation of the JmLFY promoter in vivo needs to be studied in the future. Similar results were also confirmed by the CHIP test of SOC1 and LFY promoter in Arabidopsis (Lee et al., 2008; Liu et al., 2008).
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
LZ and CL contributed to the conception and design of the study. JF, TD, MZ, and JW organized the database. JF and TD performed the statistical analysis. LZ and JF wrote the first draft of the manuscript. CL revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the 13th Five-Year National Key Research and Development Plan of China, grant number 2017YFD060060; the Key Research Project of Liaoning Provincial Department of Education, grant number LSNZD201905; and Applied Basic Research Project of Liaoning Provincial Department of Science and Technology, grant number 2022JH2/101300170.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1243030/full#supplementary-material
Ahearn, K., Johnson, H., Detlef, W., Ry, W. (2001). NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristem initiation and floral structure. Plant Cell Physiol. 10, 1130–1139. doi: 10.1093/pcp/pce143
Aidyn, M., Frédéric, C., George, C. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14, 111–130. doi: 10.1105/tpc.001362
Alvarez-Buylla, E. R., Garcia-Ponce, B., Garay-Arroyo, A. (2006). Unique and redundant functional domains of APETALA1 and CAULI FLOWER, two recently duplicated Arabidopsis thaliana floral MADS-genes. J. Exp. Bot. 57, 3099–3107. doi: 10.1093/jxb/erl081
Cai, J. (2022). Cloning of flowering related genes and functional verification of JmLFY gene in Juglans mandshurica Maxim (Shenyang: Shenyang Agricultural University).
Cai, J., Jia, R., Jiang, Y., Fu, J., Dong, T., Deng, J., et al. (2023). Functional verification of the JmLFY gene associated with the flowering of Juglans mandshurica Maxim. Peer J. 11, e14938. doi: 10.7717/peerj.14938
Chen, L., Cheng, J. C., Castle, L., Sung, Z. R. (1997). EMF genes regulate Arabidopsis inflorescence development. Plant Cell 9, 2011–2024. doi: 10.1105/tpc.9.11.2011
Cui, W., Wang, S., Han, K., Zheng, E., Ji, M., Chen, B., et al. (2021). Ferredoxin 1 is downregulated by the accumulation of abscisic acid in an ABI5-dependent manner to facilitate rice stripe virus infection in Nicotiana benthamiana and rice. Plant J. 107, 1183–1197. doi: 10.1111/tpj.15377
Gordon, G. S., Caroline, D. (2002). Arabidopsis, the rosetta stone of flowering time. Science 296, 285–289. doi: 10.1126/science.296.5566.285
Guo, C. (2020). Preliminary study on reproductive biology of hermaphroditic and heterozygotic sorbus in Juglans mandshurica (Shenyang: Shenyang Agricultural University).
He, C., Cai, Y., Dong, L., Wang, Y. (2018). Screening and expression analysis of related genes based on transcriptome sequencing of ginkgo flower buds at three differentiation stages. Acta Hortic. Sin. 45, 1479–1490. doi: 10.16420/j.issn.0513-353x.2018-0023
He, Z., Zhu, Q., Dabi, T., Li, D., Weigel, D., Lamb, C. (2000). Transformation of rice with the Arabidopsis floral regulator LEAFY causes early heading. Transgenic Res. 9, 223–227. doi: 10.1023/a:1008992719010
Jian, H., Zhang, A., Ma, J., Wang, T., Yang, B., Shuang, L., et al. (2019). Joint QTL mapping and transcriptome sequencing analysis reveal candidate flowering time genes in Brassica napus L. BMC Genomics 20, 4–14. doi: 10.1186/s12864-018-5356-8
Jin, Q., Zhao, Y., Zhang, Z., Ma, Y. (2019). Cloning and structural analysis of the LEAFY homologous gene in Leucanthemella linearis. Mol. Plant Breed. 17, 396–399. doi: 10.13271/j.mpb.017.000396
Kaufmann, K., Melzer, R., Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Lee, J., Oh, M., Park, H., Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 55, 832–843. doi: 10.1111/j.1365-313X.2008.03552.x
Li, Z. (2012b). Cloning and functional analysis of CcLFY gene promoter in Pecan carya (Lin’an: Zhejiang A&F University).
Li, H., Chen, Z., Li, Y., Wang, J., An, X. (2012a). Cloning and transient expression analysis of PtLFY gene promoter from Populus tomentosa. Chin. J. Bioengineering 32, 41–46. doi: 10.13523/j.cb.20120408
Li, J., Fan, S. L., Song, M. Z., Pang, C. Y., Wei, H. L., Li, W., et al. (2013). Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L. Plant Cell Rep. 32, 1675–1686. doi: 10.1007/s00299-013-1479-1
Liu, C., Chen, H., Er, H. L., Soo, H. M., Kumar, P. P., Han, J. H., et al. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–1491. doi: 10.1242/dev.020255
Liu, C., Lu, X., Song, J., Zhou, Q., Sun, X., Deng, J., et al. (2022). Molecular cloning, expression analysis and subcellular localization of LEAFY in Juglans mandshurica Maxim. Pol. J. Environ. Stud. 31, 1713–1725. doi: 10.15244/pjoes/142381
Liu, C., Thong, Z. H., Yu, H. (2009). Coming into bloom: the specification of floral meristems. Development 136, 3379–3391. doi: 10.1242/dev.033076
Liu, X., Zhu, Z., Liu, C., Zhang, L., Wei, H., Tang, W. (2021). Cloning and expression pattern of LEAFY gene in Actinidia eriantha Benth. J. Hefei Univ. Technol. 44, 706–710. doi: 10.3969/j.issn.1003-5060.2021.05.023
Mandel, M. A., Yanofsky, M. F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. doi: 10.1038/377522a0
Martina, B., Nadine, D., Christian, J. (2015). Flowering time regulation in crops-what did we learn from Arabidopsis. Curr. Opin. Biotech. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Molina, C., Grotewold, E. (2005). Genome wide analysis of Arabidopsis core promoters. BMC Genomics 6, 25. doi: 10.1086/1471-2164-6-25
Nobutoshi, Y., Woong, J. C., Staci, N. W., Krizek, B. A., Wagner, D. (2016). Aintegumenta And Aintegumenta-Like6/Plethora3 Induce Leafy expression in response to auxin to promote the onset of flower formation in Arabidopsis. Plant Physiol. 170, 283–293. doi: 10.1104/pp.15.00969
Peña, L., Martin-Trillo, M., Juarez, J., José, A., Luis, P. (2001). Constitutive expressing of Arabidopsis LEAFY or APETALA genes in citrus reduces their generation time. Nat. Biotechnol. 19, 263–267. doi: 10.1038/85719
Qin, B., Cai, J., Fu, J., Liu, C., Zhang, L. (2022). Dynamic changes of endogenous hormones in different developmental organs of Juglans mandshurica. Mol. Plant Breed. 20, 5130–5137. doi: 10.13271/j.mpb.020.005130
Qin, B., Lu, X., Sun, X., Cui, J., Deng, J., Zhang, L. (2021). Transcriptome-based analysis of the hormone regulation mechanism of gender differentiation in Juglans mandshurica Maxim. Peer J. 9, e12328. doi: 10.7717/PEERJ.12328
Riboni, M., Robustelli, T. A., Galbiati, M., Tonelli, C., Conti, L. (2016). ABA-dependent control of GIGANTEA signalling enables drought escape via upregulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 67, 6309–6322. doi: 10.1093/jxb/erw384
Shannon, S., Meeks-Wagner, D. R. (1993). Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 5, 639–655. doi: 10.1105/TPC.5.6.639
Song, J. (2019). Cloning and expression analysis of the flowering related gene LFY in Juglans mandshurica (Shenyang: Shenyang Agricultural University).
Sun, Z. C., Li, Z., Huang, J. Q., Zheng, B. S., Zhang, L. S., Wang, Z. J. (2017). Genome-wide comparative analysis of LEAFY promoter sequence in angiosperms. Physiol. Mol. Biol. Pla. 23, 23–33. doi: 10.1007/s12298-016-0393-8
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64, 675–684. doi: 10.1093/jxb/ers361
Wang, L., Liang, H., Pang, J., Zhu, M. (2004). Regulation network and biological roles of LEAFY in Arabidopsis thaliana in floral development. Hereditas 01, 137–142. doi: 10.1088/1009-0630/6/5/011
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F., Meyerowitz, E. M. (1993). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. doi: 10.1016/0092-8674(92)90295-n
Xiong, F., Ren, J. J., Yu, Q., Wang, Y. Y., Lu, C. C., Kong, L. J., et al. (2019). AtU2AF65b functions in abscisic acid mediated flowering viaregulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. 223, 277–292. doi: 10.1111/nph.15756
Xu, J., Zhu, N., Wen, C., Zeng, L. (2011). Cloning and sequence analysis of LEAFY gene promoter from longan (Dimocarpus longan). J. Fruit Sci. 28, 689–693. doi: 10.13925/j.cnki.gsxb.2011.04.031
Yanofsky, M. F. (1995). Floral meristems to floral organs: genes controlling early events in Arabidopsis flower development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 167–188. doi: 10.1146/annurev.pp.46.060195.001123
Zeng, X., Liu, H., Du, H., Wang, S., Yang, W., Chi, Y., et al. (2018). Soybean MADS-box gene GmAGL1 promotes flowering via the photoperiod pathway. BMC Genomics 19, 2–17. doi: 10.1186/s12864-017-4402-2
Zhang, L., Guo, C., Qin, B., Lu, X., Yang, Y., Qi, Y., et al. (2019a). Flowering phenology and pollen viability of Juglans mandshurica. J. Northeast Forestry Univ. 47, 4–8. doi: 10.13759/j.cnki.dlxb.2019.05.002
Zhang, Y., Liu, Q. (2003). Proceedings on molecular mechanism of plant flower development. Bull. Bot. 5, 589–601. doi: 10.3969/j.issn.1674-3466.2003.05.011
Zhang, L., Qin, B., Guo, C., Lu, X., Yang, Y., Qi, Y., et al. (2019b). Study on changes of material metabolism during flower bud formation of Juglans mandshurica. J. Shenyang Agric. Univ. 50, 280–288. doi: 10.3969/j.issn.1000-1700.2019.03.004
Keywords: Manchurian walnut, JmLFY, promoter, functional analysis, genetic transformation, transient transformation, yeast one hybrid
Citation: Zhang L, Fu J, Dong T, Zhang M, Wu J and Liu C (2023) Promoter cloning and activities analysis of JmLFY, a key gene for flowering in Juglans mandshurica. Front. Plant Sci. 14:1243030. doi: 10.3389/fpls.2023.1243030
Received: 20 June 2023; Accepted: 25 September 2023;
Published: 12 October 2023.
Edited by:
Qingzhang Du, Beijing Forestry University, ChinaCopyright © 2023 Zhang, Fu, Dong, Zhang, Wu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunping Liu, bGl1Y2h1bnBpbmcyMDE5QHN5YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.