- College of Agriculture, Ibaraki University, Ibaraki, Japan
To examine the physiological change in the growth suppression and abortion of parthenocarpic cucumber fruit, the expression of candidate marker genes of sugar starvation in relation to growth activity was examined. Fruits that failed to start exponential growth seemed to eventually abort. Hexose concentration of fruits was low in growth-suppressed fruit and increased in normally growing fruit consistent with the vacuolization. The correlation matrix indicated that the transcript levels of the genes, except CsaV3_6G046050 and CsaV3_5G032930, had a highly significant negative correlation with the relative growth rate in fruit length and had highly significant mutual positive correlations, suggesting that the asparagine synthetase gene, Cucumis sativus putative CCCH-type zinc finger protein CsSEF1, C. sativus BTB/POZ domain-containing protein At1g63850-like, CsaV3_3G000800, CsaV3_3G041280, and CsaV3_7G032930 are good markers of sugar starvation in cucumber fruit. The expression of candidate marker genes together with the hexose analysis strongly suggests that severe sugar starvation is occurring in growth-suppressed fruit.
1 Introduction
Sugar starvation occurs in plant tissues under various stress conditions (Slocombe et al., 2004; Barros et al., 2017; van Rensburg et al., 2019). In Arabidopsis and other plants, hexokinase 1 (Moore et al., 2003), SnRK1, (Lu et al., 2007), trehalose 6-phosphate (Fichtner and Lunn, 2021), TOR (Dobrenel et al., 2016; Li et al., 2019; Rodriguez, 2019), mR156 (Buendia-Monreal and Gillmore, 2017) are known to be involved in sugar signaling. On the other hand, asparagine synthase gene is regarded to be a very good marker of sugar starvation (Rolland et al., 2006). This may mean that the expression of the genes related to sugar signaling is not necessarily a good marker of sugar starvation. Also, sugar starvation has been studied mainly in leaves. So, we looked for marker genes of sugar starvation in cucumber fruit in relation to the formation of malformed fruits. Using the subtraction method (Tazuke and Asayama, 2013; Tazuke et al., 2015), we identified three candidate genes that are upregulated by total defoliation including the asparagine synthetase (AS) gene,. The function of the remaining two genes remains unclear. However, the expression of Cucumis sativus putative CCCH-type zinc finger protein CsSEF1 (CsSEF1) and C. sativus BTB/POZ domain-containing protein At1g63850-like (CsFDI1) was markedly upregulated by total defoliation (Tazuke and Asayama, 2013; Tazuke et al., 2015), prolonged darkness (Tazuke and Asayama, 2013; Tazuke et al., 2015), salinity (Tazuke et al., 2017), removal of the ovary (Tazuke et al., 2017), and fruit harvest (Tazuke and Kinoshita, 2018), strongly suggesting that these genes are good markers of sugar starvation. In a preliminary experiment, the growth cessation of parthenocarpic cucumber fruit was accompanied by upregulation of the AS, CsSEF1, and CsFDI1 genes, suggesting the occurrence of sugar starvation during growth cessation or abortion of a fruit (Tazuke, unpublished data). In addition, we isolated five genes by RNA sequencing (RNA-seq) followed by real-time PCR (Tazuke et al., 2019) of total defoliated ‘Tokiwa’; the expression of PREDICTED: C. sativus homeobox-leucine zipper protein ATHB-40 (HB), PREDICTED: C. sativus peptide methionine sulfoxide reductase (PMS), PREDICTED: C. sativus NAC domain-containing protein 1 (NAC), PREDICTED: C. sativus NAC domain-containing protein 100 (NAC3), and PREDICTED: C. sativus hypothetical protein (uk4) was markedly upregulated by total defoliation, although the functions of these genes remain unclear (Tazuke et al., 2019). As described in Tazuke et al. (2019), varietal difference of the effect of total defoliation on the expression of the chosen 8 genes was examined. Cultivars used were as follows. Japanese type: ‘‘Tokiwa’, ‘High green 21’, and ‘Freedom house No. 2’; North-Chinese type: ‘Suyo’’; South-Chinese type: ‘Sagami hanshiro’;;Slicer:’Poinsett’; English greenhouse slicer: ‘Proloog RZ’; Beit Alpha ‘Khassib RZ’; Pickling: ‘Mogami’ together covering representative cucumber types of the world. When the fruit was about 9 cm long, total defoliation treatment was done at 9 a.m. and harvested 9 a.m. the next day. Although there was some varietal difference, in all cultivars, all the 8 genes were markedly upregulated (Tazuke et al., 2019). This suggested that the 8 genes may be good markers of sugar starvation for all the cucumber cultivars in the world.
Because microspore formation is sensitive to environmental conditions, such as moderate heat stress and low temperature (Picken, 1984; Sato et al., 2006; Mesihovic et al., 2016), parthenocarpy is a desired trait. Cucumber has naturally parthenocarpic cultivars. Many reports have shown that parthenocarpy is induced by plant hormones (Fu et al., 2008; Gorget et al., 2005; Serrani et al., 2010). Introduction of the indole-3-acetic acid (IAA)-synthesizing DefH9-iaaM gene in cucumber enhances parthenocarpy (Yin et al., 2006). In tomato, overexpression of the auxin (Aux) receptor Solanum lycopersicum Transport Inhibitor Response 1 (SlTIR1) gene (Ren et al., 2011) and suppression of expression of the S. lycopersicum Aux response factor 7 (SlARF7) gene (De Jong et al., 2009) and Aux/IAA SlIAA9 gene (Wang et al., 2005) induce parthenocarpy. Parthenocarpy is also induced by gibberellin, and it is suggested that Aux works upstream of gibberellin (Serrani et al., 2010; Hu et al., 2018). However, the direct target of gibberellin is not known (Shinozaki et al., 2020). Kusano et al. (2022) performed transcriptomic, hormonomic, and metabolomic analyses of well-established tomato parhenocarpic mutants.
In cucumber, the results of the search for the genes responsible for parthenocarpy were equivocal. This may be partly attributed to differences in the measurements of parthenocarpy because Lietzow et al. (2016) used early growth as the measure of the extent of parthenocarpy, Wu et al. (2016) regarded fruits that did not abort as parthenocarpic, and Sun et al. (2006) used fruit yield as the measure of parthenocarpy. Therefore, additional observations of fruit growth and abortion are necessary to address this issue.
Cucumber fruits that fail to set eventually abort (Hikosaka and Sugiyama, 2004; Hikosaka and Sugiyama, 2005). It has been suggested that fruit abortion is caused by the source–sink relationship, which triggers sugar shortage in fruit (Marcelis, 1993). Thus, the investigation of sugar starvation in parthenocarpic fruit will be informative in this context. In addition to a genetic approach, understanding the physiological response of fruit set/abortion will be informative.
In this study, we observed the expression of candidate marker genes of sugar starvation in association with growth cessation in parthenocarpic cucumber fruit in a moderately parthenocarpic cultivar, as well as with fruit growth and sugar concentration to assess the physiological responses occurring in fruit set and abortion.
2 Materials and methods
2.1 Plant cultivation and growth analysis of fruits
Cucumber (Cucumis sativus L.) cv. ‘Tokiwa’ was used. It is a moderately parthenocarpic cultivar, suited to the study of parthenocarpy responses. Plants were grown in a glasshouse. Single plants were grown in 15 L containers filled with vermiculite. The plants were watered every morning with half-strength Hoagland No. 2 solution. Tap water was used to prepare the solution. The plants were detopped, leaving about 10 leaves. The lateral shoots were pinched, leaving the first node. The female flowers on the first nodes were used for the experiment. In the evening, female flower buds in which the petals had become yellow were judged to flower the next day and were wrapped with a paper bag to prevent pollination. The bag was removed on the day following the flowering. In the spring of 2019, fruit length was measured every morning until the fruit grew to marketable size or became soft indicating fruit abortion. In the spring of 2021, fruit length was measured every 2 days in the morning from the day of anthesis until 0, 2, 4, 6, and 8 days after anthesis (DAA), when the fruit was harvested. The relative growth rate in length of a fruit (RGR % h–1) was estimated as follows:
where L0 and L1 are the lengths of the fruit at the beginning and end of the period of RGR measurement, respectively; and d0 and d1 are the dates of the beginning and end of the period, respectively. In 2019, one culture was done, and four cultures were done in 2021. In 2019, the growth curve was constructed until the fruits grew to marketable size or aborted. In 2021, the growth curve was constructed for fruits harvested on 8 DAA or for those that had aborted on 8 DAA or earlier combining the four cultures.
2.2 Fruit harvest
In the spring culture of 2021, fruits were harvested for gene expression and sugar analyses separately at 9 a.m. on 0, 2, 4, 6, and 8 DAA immediately after the length measurement. The harvested fruit was immediately weighed, placed in a jar filled with liquid nitrogen, and transferred to the laboratory. Samples for RNA extraction were stored at –80°C, if necessary, whereas samples for sugar extraction were stored at –30°C.
2.3 RNA extraction and reverse transcription
The frozen sample was powdered with a mortar and pestle and total RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). DNA was removed with the RNase-Free DNase Set (QIAGEN). For reverse transcription, PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa, Kusatsu, Japan) was used. 2 mL total RNA was mixed with 2 mL Buffer, 0.5 mL enzyme, 0.5 mL oligo dT primer0.5 mL random 6 mersand and 4.5 mL RNase Free Water in a PCR tube and incubated at 37°C for 15 minutes and incubated at 85°C for 5 seconds to inactivate the enzyme then kept at 4°C with Eppendorf 5331 Mastercycler Gradient Thermal Cycler (Eppendorf Co. Ltd., Tokyo, Japan) and stored at –30°C.
2.4 RNA-seq and real-time PCR
The result of RNA-seq without biological replication (conducted by Hokkaido System Science Co. Ltd.) is shown in Table S1. Genes whose expression was enhanced by defoliation larger than 32 times are listed in Tazuke et al. (2019). The abbreviations of the name of genes are based on the description in GenBank. Expression of these genes were analyzed by real-time PCR, whose result is shown in Tazuke et al. (2019). From this result, 8 genes who showed large enhancement were selected as shown in Table 1. The primers used in this experiment were designed using Primer3web (https://primer3.ut.ee/). Selected reverse transcripts were amplified by PCR. Aliquots of the PCR products were subjected to electrophoresis to verify the specificity of the primers. The aliquots of the PCR products were subjected to a series of 10-fold dilutions with EASY Dilution (for real-time PCR) solution (TaKaRa) and used to produce the calibration curve. C. sativus clone CU36H1 actin was used as the internal standard. Two μL reverse transcript solution was mixed with 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 12 μL Premix Ex Taq™ (Perfect Real Time; TaKaRa) and 4 μL sterilized water on a PCR plate. The PCR plate was set in a LightCycler® 96 System (Roche, Basel, Switzerland). The cycle condition was 40 cycles of 95°C 5 seconds and 60°C 30 seconds.
2.5 Post harvest expression of the chosen 8 genes
Cucumber cultivar ‘Tokiwa’ was grown in a glasshouse in Spring of 2020 as described above. At 9 a.m., about 9 cm long pollinated young fruits were put into water vapor saturated boxes and transferred to the labolatory and the boxes were incubated at 20°C or 30°C. After 6, 12 and 24 hours of incubation, total RNA was extracted and analyzed by real-time PCR. For 0 hour, fruits harvested in the glasshouse was immediately put in a jar containing liquid nitrogen and transferred to the laboratory. Every treatment was triplicated.
2.6 Sugar analyses
The stored frozen samples were powdered with a mortar and pestle, homogenized with 80% ethanol, extracted for 15 min at 80°C, and filled up to 100 mL. Aliquots of 1 mL were dried with a rotary evaporator and redissolved in 1 mL water, and the concentrations of sucrose, glucose, and fructose were analyzed enzymatically with an F-kit (JK International Co. Ltd., Tokyo, Japan). Glucose and fructose are the major sugars in cucumber fruit (Handley et al., 1983). Here, we refer to the sum of the concentration of glucose and fructose as the hexose concentration. In cucumber fruit, sugar starvation must be hexose starvation.
2.7 Statistical analyses
The statistical analyses were performed using R (R Core Team, 2022). Correlation matrix was calculated by cor() function. P-value was calculated by cor.test() function. Multiple comparison was performed using TukeyHSD() and aov() functions. The data were log-transformed if appropriate. For the test of difference between average and constant, pt() function was used.
3 Results
3.1 Fruit growth
Fruit on the day of anthesis is shown in Figure 1A. Fruit at 3 DAA is shown in Figure 1B, at which time point they had not begun exponential growth, which is a typical symptom of growth suppression of parthenocarpic fruit. The growth of fruit based on length is shown in Figures 2A, D. RGR in fruit length is shown in Figures 2B, E. In the growth analysis of 2021 (Figures 2A, B), the growth of most fruit could be classified into those that eventually aborted and those that did not abort. Among the 41 fruits examined, 18 aborted yielding an abortion rate of 43.9%. The date of fruit abortion was concentrated on 6 DAA and 8 DAA (Figure 2C). In the growth analysis of 2019, the date of onset of exponential growth was variable (Figures 2D, E). Most of the fruits belonged to one of two classes: those that grew to marketable size and those that eventually aborted. Fruits with low growth activity eventually aborted, but the date of abortion was variable (Figure 2F). In the 41 examined fruits, 23 aborted yielding an abortion rate of 56.1%. These abortion rates verified that ‘Tokiwa’ is an intermediately parthenocarpic cultivar, which confirms that it is suited for analysis of the parthenocarpic traits.

Figure 1 (A) Flower on the day of anthesis of parthenocarpic cucumber fruit. (B) Fruit 3 days after anthesis of parthenocarpic cucumber fruit.
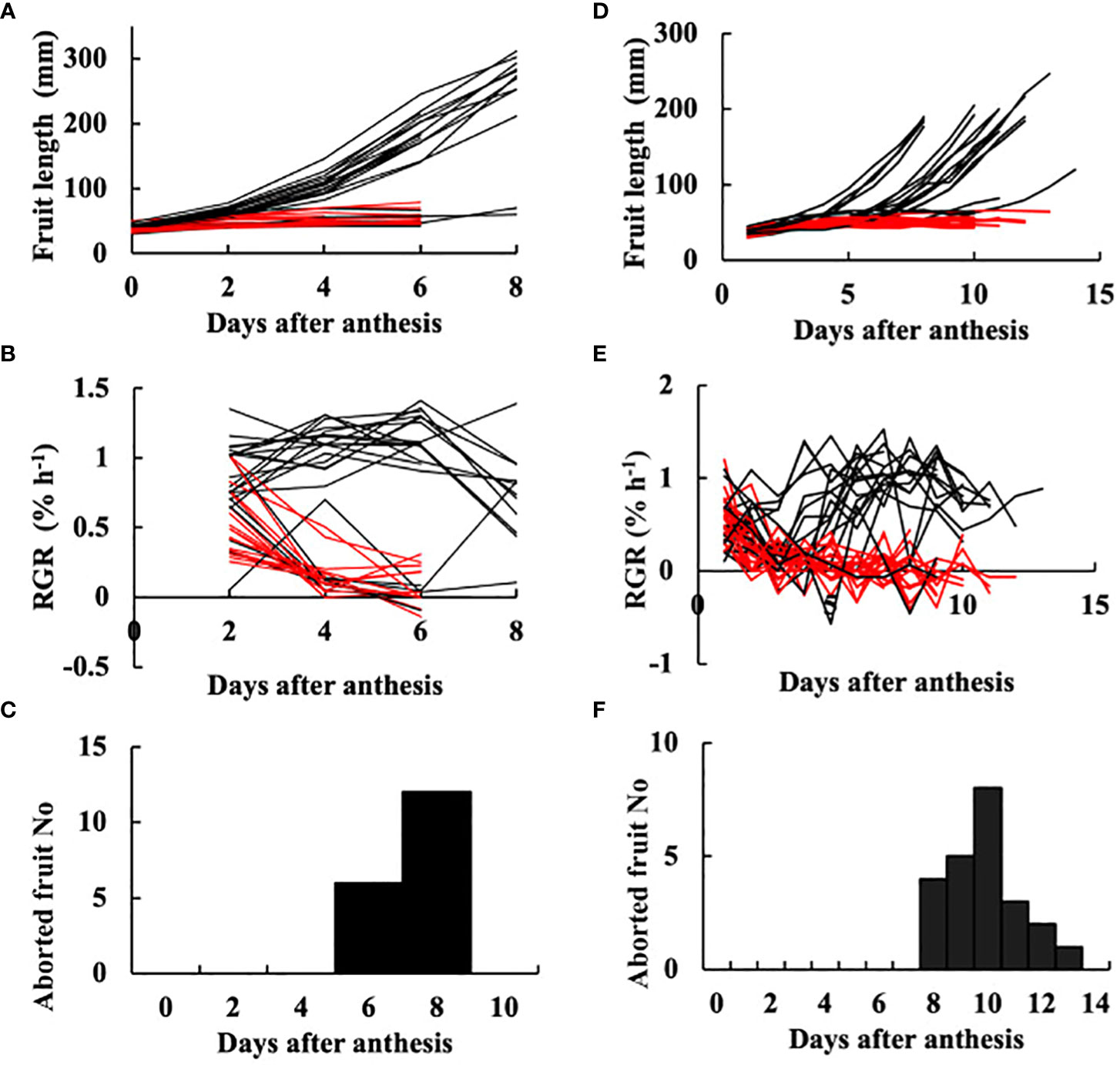
Figure 2 Growth of parthenocarpic cucumber fruit. (A–C) Experiment in 2021. (D–F) Experiment in 2019. (A, D) Growth in length plotted against days after anthesis (DAA). Each line corresponds to an individual fruit. (B, E) Relative growth rate (RGR) in fruit length. Each line corresponds to an individual fruit. (C, F) Histogram of the date of fruit abortion. (A, B, D, E) Red lines indicate individual fruits that eventually aborted. Black lines indicate fruits that did not abort.
3.2 Evaluation of the expression of genes related to sugar signaling
3.2.1 Hexokinase 1
In Arabidopsis, hexokinase 1 (HXK1) is a well-known sugar sensor. So, we sought in the Table S1 for the keyword “hexokinase 1”. One hit was obtained: Csa2G000870. Because the RNA-seq was analyzed using Chinese Long ver. 2, the gene code is different from that of Chinese long ver. 3. The enhancement of the expression was less than 2.
3.2.2 SnRK1
Luo et al. (2022) have done genome-wide analysis of SnRK gene family in cucumber. They found CsaV3_6G006250 was homologous to AtSnRK1. This gene corresponds to Csa6G077450 in Chinese Long ver. 2. However, this gene was not found in Table S1. So, Arabidopsis protein sequence AT3G01090’s homolog was sought in Chinese Long ver. 2 database. Csa6G077450 had E-value 0, but this gene could not be found in Table S1. Csa4G269780 had E-value e-92. This gene’s expression enhancement was about 2.
3.2.3 TOR
The protein sequence of AtTORC1: AT1G50030’s homolog was sought by BLAST in Chinese long ver. 2 database. Csa7G070760 was obtained. Its enhancement of expression was about 2.
3.3 Gene expression of fruits without pollination
Transcript levels plotted against fruit fresh weight are shown in Figure 3. At 0 DAA, the transcript levels of all eight genes were very small. The increase in transcript levels was restricted to very small fruits, suggesting that sugar starvation only occurred in growth-suppressed fruit. An exception was HB in which some fruits showed high transcript levels at a relatively high fresh weight.
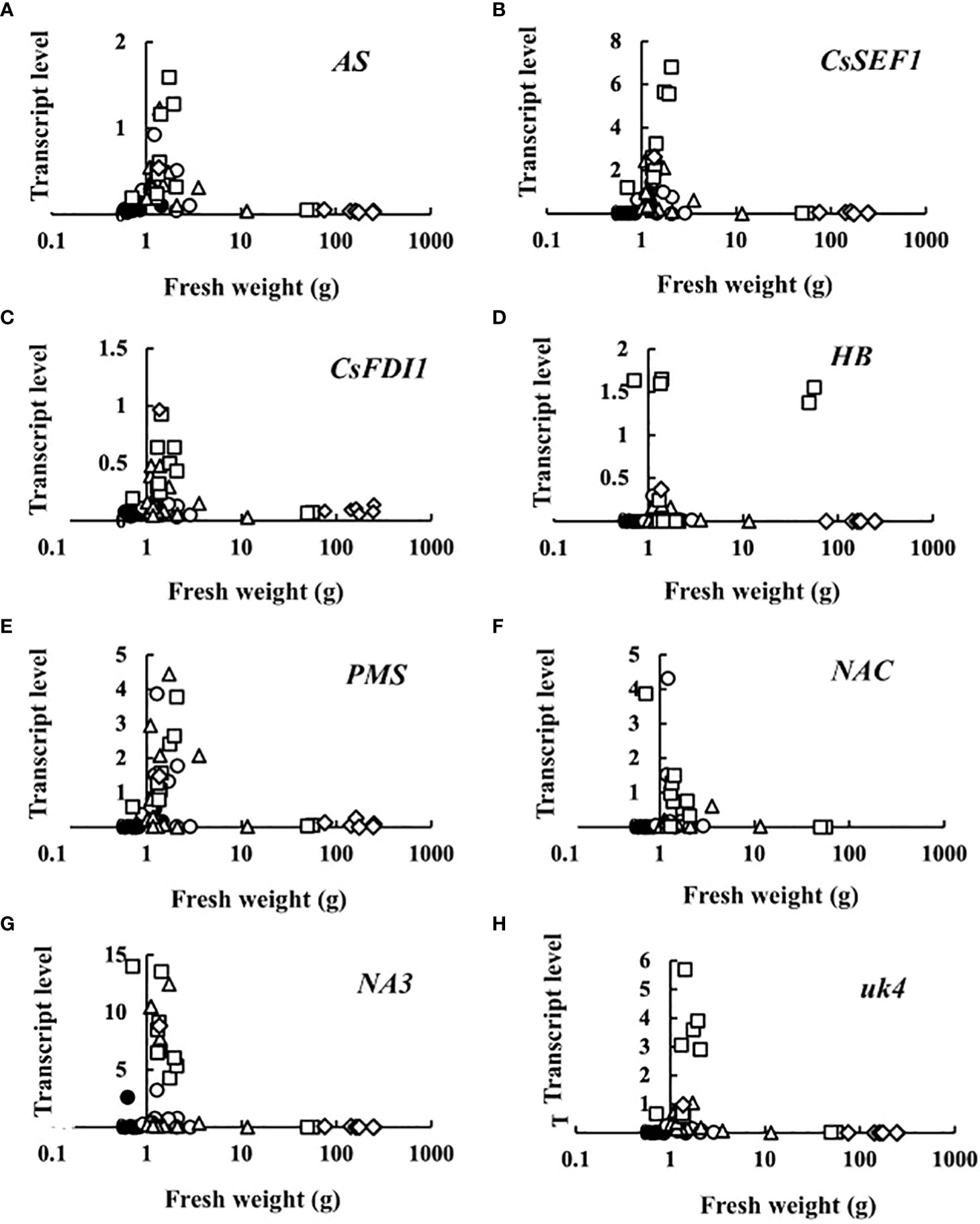
Figure 3 Relationship between fruit fresh weight and transcript levels of (A) asparagine synthetase (AS), (B) Cucumis sativus putative CCCH-type zinc finger protein CsSEF1 (CsSEF1), (C) C. sativus BTB/POZ domain-containing protein At1g63850-like (CsFDI1), (D) C. sativus homeobox-leucine zipper protein ATHB-40 (HB), (E) C. sativus peptide methionine sulfoxide reductase (PMS), (F) C. sativus NAC domain-containing protein 1 (NAC), (G) C. sativus NAC domain-containing protein 100 (NA3), and (H) C. sativus hypothetical protein (uk4) of parthenocarpic cucumber fruit. •: The day of anthesis, ○: 2 days after anthesis (DAA), △: 4 DAA, □: 6 DAA, ◇: 8 DAA. Description of the genes is given in Table 1.
The transcript levels plotted against RGR are shown in Figure 4. Because RGR could not be calculated for 0 DAA fruits, the relationship between RGR and transcript levels was examined at 2, 4, 6, and 8 DAA. The transcript levels were mostly close to zero but tended to become high in fruits whose RGRs became close to zero. The correlation matrix of RGR and transcript levels are shown in Table 2. All of the correlation coefficients, except those for HB and NAC, were highly significant. For HB, the absolute values of the correlation coefficient were very low and insignificant. For NAC, correlation coefficients were insignificant or poorly significant. Negative correlation coefficients between RGR and transcript levels indicate that transcript levels become higher at lower RGRs. Highly significant positive correlation coefficients between transcript levels of AS, CsSEF1, CsFDI1, PMS, NA3, and uk4 indicate that these genes are responding similarly to fruit growth.
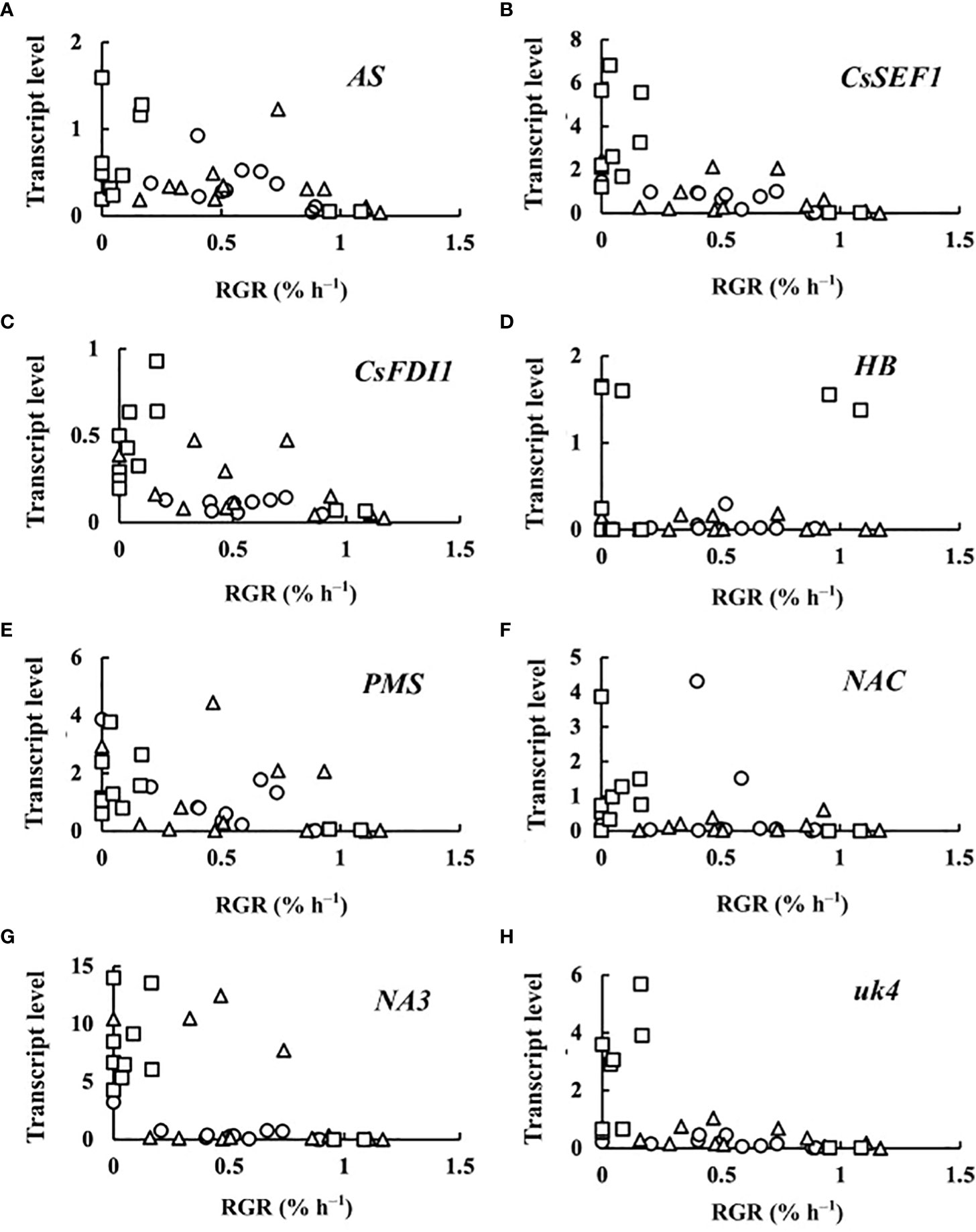
Figure 4 Relationship between relative growth rate (RGR) in fruit length and transcript levels of (A) asparagine synthetase (AS), (B) Cucumis sativus putative CCCH-type zinc finger protein CsSEF1 (CsSEF1), (C) C. sativus BTB/POZ domain-containing protein At1g63850-like (CsFDI1), (D) C. sativus homeobox-leucine zipper protein ATHB-40 (HB), (E) C. sativus peptide methionine sulfoxide reductase (PMS), (F) C. sativus NAC domain-containing protein 1 (NAC), (G) C. sativus NAC domain-containing protein 100 (NA3), and (H) C. sativus hypothetical protein (uk4) of parthenocarpic cucumber fruit. ○: 2 days after anthesis (DAA), △: 4 DAA, □: 6 DAA, ◇: 8 DAA. Description of the genes is given in Table 1.
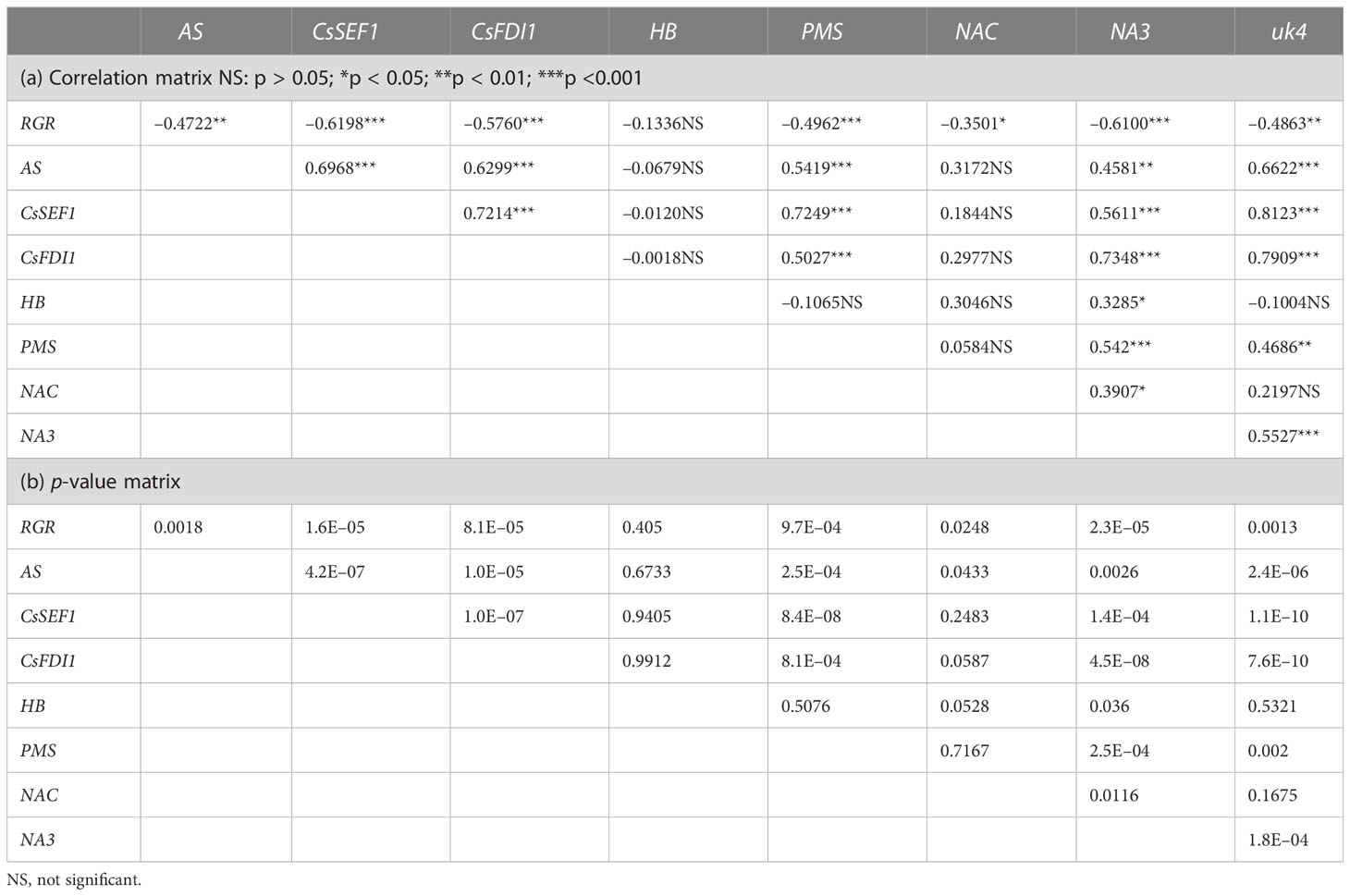
Table 2 Correlation matrix of relative growth rate (RGR) in fruit length and transcript levels of candidate marker genes of fruit sugar starvation in cucumber.
3.4 Sugar levels
In all fruits, the sucrose concentration was very low, and did not differ much from zero (p = 0.044). The sum of the concentrations of glucose and fructose was regarded as the hexose concentration. The relationships between fruit fresh weight or RGR and hexose concentration at 0, 2, 4, 6, and 8 DAA are shown in Figure 5. At 0 DAA, the hexose concentration was very low but the average was significantly larger than zero (p = 1.18E–05). In fruits whose growth was suppressed, as seen from the low values of fresh weight and RGR, hexose concentration was very low. In fruits that grew normally, hexose concentration was high. The correlation coefficient between fruit fresh weight and hexose concentration was 0.5336 and the p-value was 9.4E–05 (n = 48). The correlation coefficient between RGR and hexose concentration was 0.5476, and the p-value was 0.00012 (n = 37). The relationship between hexose concentration and RGR was remarkable. There was a clear transition from low to high hexose concentration around RGR = 0.5% h–1 (Figure 5B). Under salinity, fruit RGR declined to 0.5% h–1 (Tazuke, 1997). So, the average of all data of 0 DAA and the average of data for which RGR was lower than 0.5% h–1 after 2 DAA were plotted. Two fruits at 8 DAA that had low RGR but had relatively high fresh weight were excluded. Averages of hexose concentration of fruits whose RGR was larger than 0.5% h–1 were also plotted (Figure 6). Hexose concentration of growth-suppressed fruits decreased up to 6 DAA toward zero.
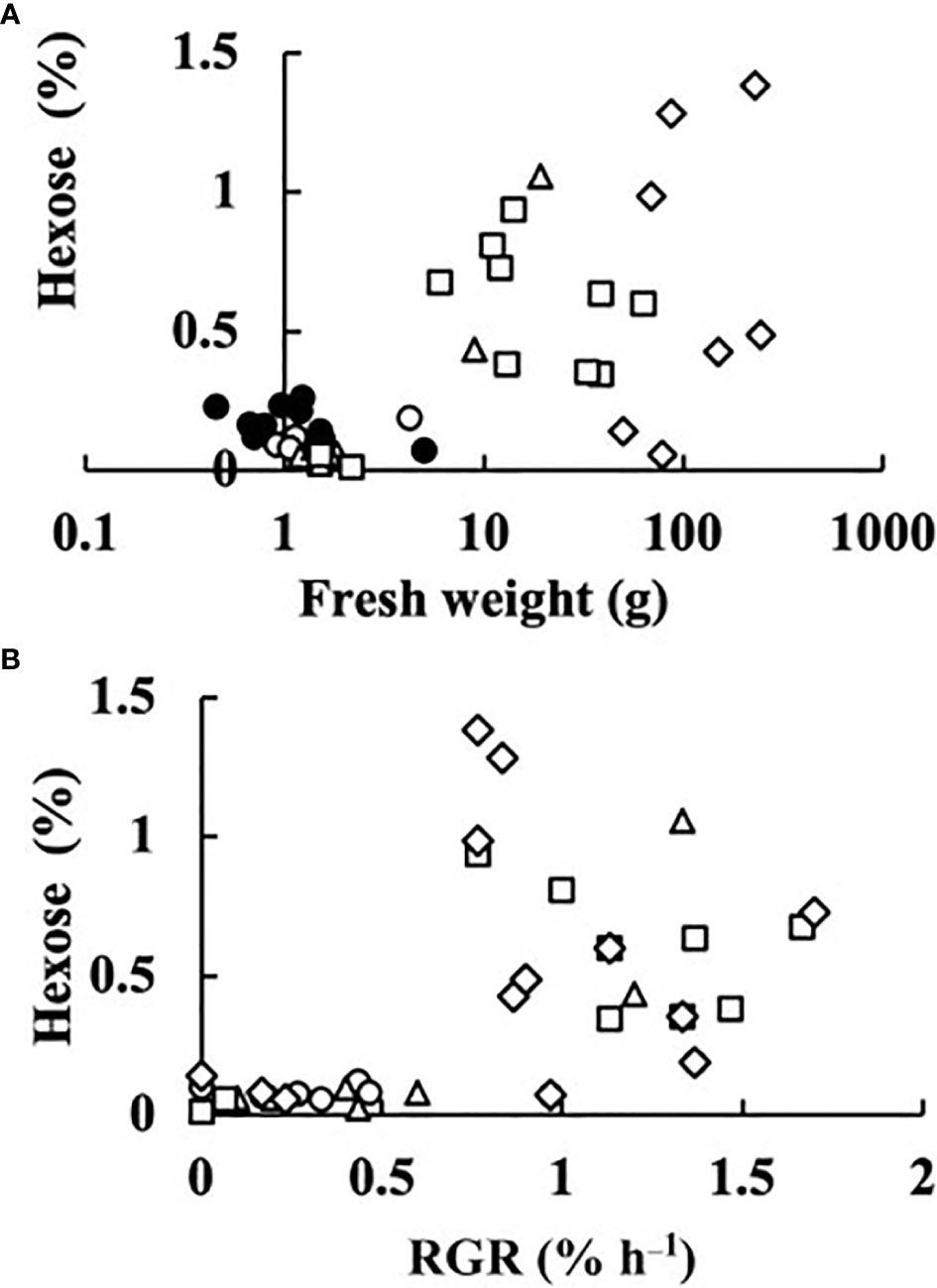
Figure 5 Dependence of hexose (glucose + fructose) concentration of parthenocarpic cucumber fruits on growth parameters. •: The day of anthesis, ○: 2 days after anthesis (DAA), △: 4 DAA, □: 6 DAA, ◇: 8 DAA. (A) Fruit fresh weight. (B) Relative growth rate (RGR) in fruit length.
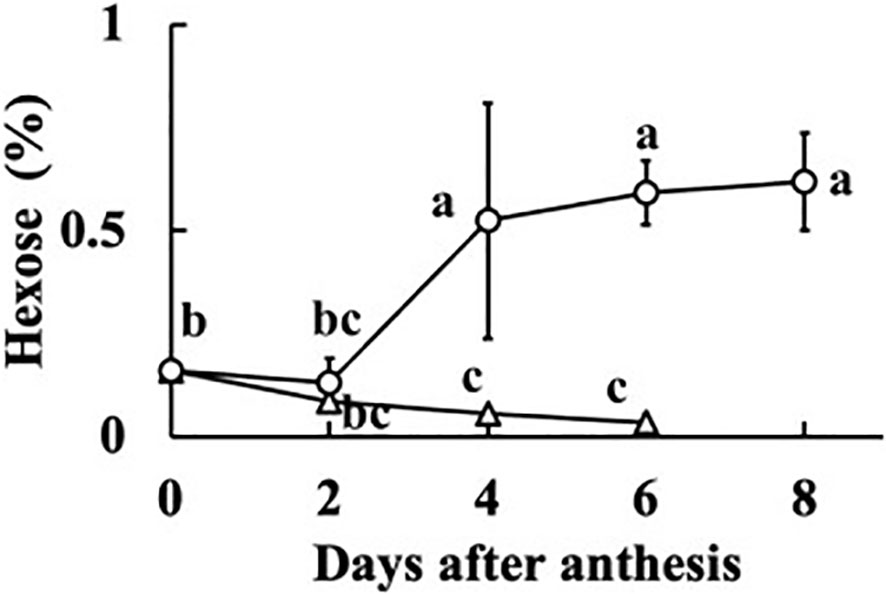
Figure 6 Change in hexose concentration of growth-suppressed (relative growth rate [RGR] < 0.5% h–1) and not growth-suppressed (RGR > 0.5% h–1) parthenocarpic cucumber fruit. Vertical bars represent the standard error of the mean. Different letters indicate 5% significant difference.
3.5 Postharvest gene expression
The expression of the 8 genes increased up to 12 hours after harvest. However, in many genes especially at 20°C, the expression decreased at 24 hours after harvest. The enhancement of the expression of each gene compared to that at the harvest was similar to total defoliation treatment (Figure 7).
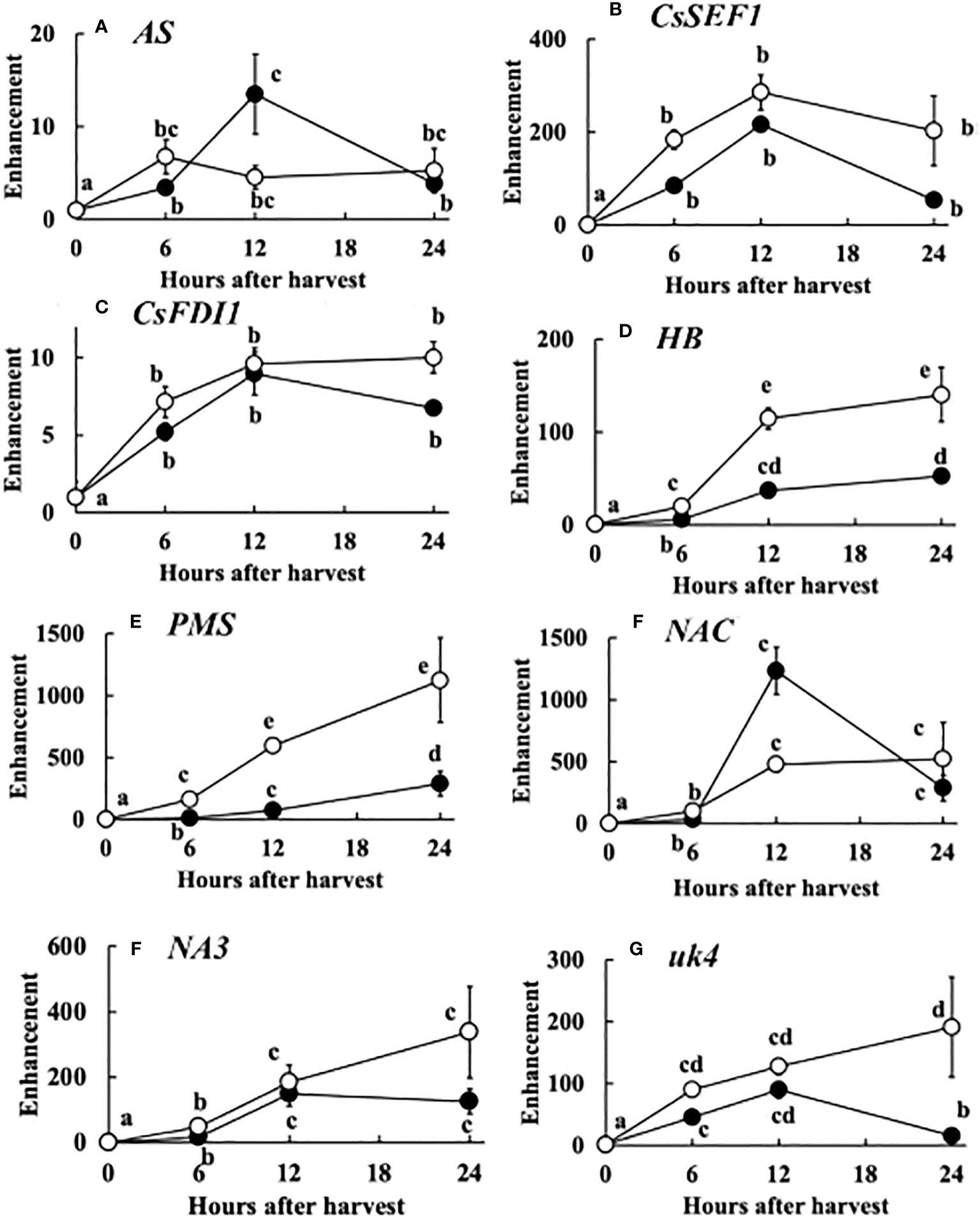
Figure 7 Post-harvest change in the expression of 8 genes, i.e. (A) AS, (B) CsSEF1, (C) CsFDI1, (D) HB, (E) PMS, (F) NAC, (G) NA3, (H) uk4. The meanings of the abbreviations are shown in Table 1. Harvested cucumber fruits were incubated at 20°C (closed circles) or 30°C (open circles). Vertical bars are SE of means. Different letters indicate 5% significant difference.
4 Discussion
As seen from RNA-seq data (Table S1), the enhancement of the expression of well-known sugar signaling genes, HXK1, SnRK1 and TOR was not much larger than 2. This may mean that these genes are constantly expressed and deliver the signal to the genes at lower cascade. Therefore, these genes are not suited for the marker of sugar starvation.
Hexose concentration was low at 0 DAA and increased in fruits with high RGR, reflecting exponential growth. This is consistent with vacuolization with the accumulation of hexose in the vacuole in normally growing fruit (Shinozaki et al., 2020). The hexose concentration at 0 DAA was very low but was significantly higher than zero. From this stage, fruit can grow normally, meaning that this low hexose concentration may be sufficient. At this stage, the vacuole may not have developed much, and this hexose concentration may therefore reflect that of cytosol. Hexose concentration in growth-suppressed fruit decreased until 6 DAA toward zero. In about 9 cm cucumber fruit, total defoliation and prolonged darkness enhanced the expression of AS and CsSEF1, but hexose concentration was not much affected by the treatment (Tazuke and Asayama, 2013), possibly because most hexose was in the vacuole. On the other hand, the close-to-zero hexose concentration seen in growth-suppressed parthenocarpic fruit may reflect little vacuolization. In this condition, explanation of the expression of candidate marker genes will be very influential. Sucrose concentration in all of the fruits examined was very close to zero. In most plants that translocate sucrose, a sucrose concentration gradient is often supposed to determine the sink activity (Ho, 1988; Ho, 1996). However, the main translocated sugars are stachyose and raffinose in cucumber (Turgeon and Wolf, 2009; Dai et al., 2023). Whether the gradient of these sugars is related to sink activity in cucumber fruit remains to be investigated.
For AS, CsSEF1, and CsFDI1, correlation of their transcript levels with each other was positive and highly significant, and with RGR, the correlation was negative and highly significant. In these three genes, the upregulation of the genes was mainly seen on 6 DAA. Because the expression of these three genes was tested in many experiments (Tazuke and Asayama, 2013; Tazuke et al., 2015; Tazuke et al., 2017; Tazuke and Kinoshita, 2018; Tazuke et al., 2019), it is highly likely that they are good markers of sugar starvation, although the gene function is known only in AS. Thus, in fruits whose RGR became close to zero, it is most probable that severe sugar starvation is occurring. The upregulation of candidate sugar starvation genes together with the reduction of hexose concentration close to zero strongly suggests that the candidate genes are genuine sugar starvation markers. In an experiment removing the ovary, it was suggested that stopping of sugar import (loss of sink activity) together with the remaining growth activity causes sugar starvation (Tazuke et al., 2017). Thus, loss of sink activity may occur in slow growing fruits, and it is possible to hypothesize that sugar starvation caused by loss of sink activity triggers fruit abortion. In the growth analysis of 2019, DAA of the onset of exponential growth was variable (Figures 2D, E), which indicates that the use of initial growth rate as the indicator of parthenocarpy (Lietzow et al., 2016) is inappropriate. The source–sink relationship and environmental factors reportedly affect the onset of exponential fruit growth (Marcelis, 1993; Hikosaka and Sugiyama, 2004; Hikosaka and Sugiyama, 2005). Because the suppression of fruit growth and fruit abortion that follows are affected by the source–sink relationship and environmental factors, the use of fruit abortion as the indicator of parthenocarpy (Wu et al., 2016) may lead to confusion, which is also the case in the use of fruit yield (Sun et al., 2006).
For newly identified genes by RNA-seq (Tazuke et al., 2019), PMS, NA3, and uk4 showed a highly significant negative correlation with RGR and a highly significant positive correlation with the transcript levels of AS, CsSEF1, and CsFDI1. They also showed a highly significant positive correlation among their transcript levels. Thus, it is likely that PMS, NA3, and uk4 are also good marker genes of sugar starvation. For the other two genes, HB and NAC, correlation matrix elements were mostly insignificant. In HB, the relationship with fruit fresh weight was abnormal. In NAC, outliers of the relationship between RGR and transcript levels were seen, which might have caused the insignificant correlation. The lack of correlation in HB and NAC is interesting. Both HB and NAC responded to total defoliation as examined by RNA-seq and real-time PCR (Tazuke et al., 2019). It is therefore possible that they respond to some signal under total defoliation other than sugar starvation. The investigation of HB and NAC may reveal unidentified physiological responses under total defoliation.
The post-harvest gene expression was similar to that of total defoliation except enhancement was much larger in post-harvest (Figure 7). This suggest that drastic sugar starvation is occurring after harvest. Therefore, these 8 genes can be good tools for the study of post-harvest physiology.
In sum, hexose measurement and expression of candidate marker genes together conclusively suggest that severe sugar starvation is occurring in growth-suppressed parthenocarpic cucumber fruit. As suggested by Shinozaki et al. (2020), hexose accumulation and vacuolization may be critical to the fruit set. How the sink activity is regulated to import sugars needs further investigations. The effects of total defoliation, parthenocarpy and post-harvest on the expression of the 6 genes were similar, but there were some differences. These differences may suggest the physiological status of fruits under these treatments. Prolonged failure of the onset of fruit growth eventually cause fruit abortion. Sugar starvation triggers autophagy (van Rensburg et al., 2019). So, it is possible that fruit abortion is caused by autophagy. Six out of eight candidate genes were suggested to be valid. These genes may be sensitive sensors of sugar starvation even in well-vacuolated tissue and can contribute to the study of stress physiology. Analyses of expression from the gene promoters may contribute to clarifying the signal transduction mechanism under sugar starvation. The remaining two genes seemed to respond to different stimuli.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
AT planned and supervised the experiment, MA helped in molecular techniques and joined discussion. TK helped in statistical analyses. All authors contributed to the article and approved the submitted version.
Funding
We were funded from Ministry of Education.
Acknowledgments
We thank Kaori Murakami and Ryusei Matsumura for helping in experiment. We thank Tsutomu Kuboyama for joining the discussion.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1241267/full#supplementary-material
Supplementary Table 1 | Analysis of RNA-seq data.
References
Barros, J. A. S., Cavalcanti, J. H. F., Medeiros, D. B., Nunes-Nesi, A., Avin-Wittenberg, T., Fernie, A. R., et al. (2017). Autophagy deficiency compromise alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiol. 175, 62–76. doi: 10.1104/pp.16.01576
Buendia-Monreal, M., Gillmore, C. S. (2017). Convergent repression of miR156 by sugar and the CDK8 module of Arabidopsis mediator. Dev. Biol. 423, 19–23. doi: 10.1016/j.ydbio.2017.01.007
Dai, H., Zhang, W., Hua, B., Zhu, Z., Zhang, J., Zhang, Z., et al. (2023). Cucumber STACHYOSE SYNTHASE is regulated by its cis-antisense RNA asCsSTS to balance source–sink carbon partitioning. Plant Cell 35, 435–452. doi: 10.1093/plcell/koac317
De Jong, M., Walters-Arts, M., Feron, R., Mariani, C., Vriezen, W. H. (2009). The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, 160–170. doi: 10.1111/j.1365-313X.2008.03671.x
Dobrenel, T., Caldana, C., HHanson, J., Robaglia, C., Vincentz, M., Veit B. and Meyer, C. (2016). TOR signaling and nutrient sensing. Ann. Rev. Plant Biol. 67, 261–285. doi: 10.1146/annurev-arplant-043014-114648
Fichtner, F., Lunn, J. E. (2021). The role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Ann. Rev. Plant Biol. 72, 737–760. doi: 10.1146/annurev-arplant-050718-095929
Fu, F. Q., Mao, W. H., Zhou, Y. H., Asami, T., Yu, J. Q. (2008). A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 59, 2299–2308. doi: 10.1093/jxb/ern093
Gorget, B., van Heusden, A. W., Lindhout, P. (2005). Parthenocarpic fruit development in tomato. Plant Biol. 7, 131–139.
Handley, L. W., Pharr, D. M., McFeeters, R. F. (1983). Carbohydrate changes during maturation of cucumber fruit: implications for sugar metabolism and transport. Plant Physiol. 72, 498–502. doi: 10.1104/pp.72.2.498
Hikosaka, S., Sugiyama, N. (2004). Characteristics of flower and fruit development of multi-pistillate type cucumbers. J. Hortic. Sci. Biotech. 79, 219–222. doi: 10.1080/14620316.2004.11511751
Hikosaka, S., Sugiyama, N. (2005). Effect of fruit-load on growth patterns of fruit at the middle nodes of gynoecious-type cucumbers. J. Hortic. Sci. Biotech. 80, 130–134. doi: 10.1080/14620316.2005.11511903
Ho, L. C. (1988). Metabolism and compartmentation of imported sugars in sink organs in relation to sink strength. Ann. Rev. Plant Physiol. Plant Mol. Biol. 39, 355–378. doi: 10.1146/annurev.pp.39.060188.002035
Ho, L. C. (1996). The mechanism and assimilate partitioning and carbohydrate compartmentation in fruit in relation to the quality and yield of tomato. J. Exp. Bot. 47, 1239–1243. doi: 10.1093/jxb/47.Special_Issue.1239
Hu, J., Israeli., A., Ori, N., Sun, T.-P. (2018). The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 30, 1716–1728. doi: 10.1105/tpc.18.00363
Kusano, M., Worarad, K., Fukushima, A., Kamiya, K., Mitani, Y., Okazaki, Y., et al. (2022). Transcriptomic, hormonomic and metabolomic analyses highlighted the common modules related to photosynthesis, sugar metabolism and cell division in parthenocarpic tomato fruits during early fruit set. Cells 11, 1420. doi: 10.3390/cells11091420
Li, B., Wang, Y., Zhang, Y., Tian, W., Chong, K., Jang, J.-C., et al. (2019). PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis. Nucleic Acids Res. 47, 5001–5015. doi: 10.1093/nar/gkz191
Lietzow, C. D., Zhu, H., Pandey, S., Havey, M. J. (2016). QTL mapping of parthenocarpic fruit set in North American processing cucumber. Theor. Appl. Genet. 129, 2387–2401. doi: 10.1007/s00122-016-2778-z
Lu, C.-A., Lin, C.-C., Lee, K.-W., Chen, J.-L., Huang, L.-F., Ho, S.-L., et al. (2007). The SnRK1A protein kinase plays a key role in sugar signaling during germination and seeding growth of rice. Plant Cell 19, 2484–2499. doi: 10.1105/tpc.105.037887
Luo, Y., Niu, Y., Gao, R., Wang, C., Liao, W. (2022). Genome-Wide identification and expression analysis of SnRK gene family under abiotic stress in cucumber (Cucumis sativus L.). Agronomy 12, 1550. doi: 10.3390/agronomy12071550
Marcelis, L. F. M. (1993). Effect of assimilate supply on the growth of individual cucumber fruits. Physiol. Plant 87, 313–320. doi: 10.1111/j.1399-3054.1993.tb01736.x
Mesihovic, A., Iannacone, R., Firon, N., Fragkostefanakis, S. (2016). Heat stress regimes for the investigation of pollen thermotolerance in crop plants. Plant Reprod. 29, 93–105. doi: 10.1007/s00497-016-0281-y
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.-H., Liu, Y.-X., et al. (2003). Role of the Arabidopsis glucose sensor HKX1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi: 10.1126/science.1080585
Picken, A. J. F. (1984). A review of pollination and fruit set in the tomato (Lycopersicon esculentum Mill.). J. Hort Sci. doi: 10.1080/00221589.1984.11515163
R Core Team (2022). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Ren, Z., Li, Z., Mao, Q., Yang, Y., Deng, W., Hao, Y. (2011). The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. J. Exp. Bot. 62, 2815–2826. doi: 10.1093/jxb/erq455
Rodriguez, M., Parola, R., Andreola, S., Pereyra, C. (2019). TOR and SnRK1 signaling pathways in plant response to abiotic stresses: do they always act according to the “yin–yang” model? Plant Sci. 288. doi: 10.1016/j.plantsci.2019.110220
Rolland, F., Baena-Gonzalez, E., Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Ann. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Sato, S., Kamiyama, M., Iwata, T., Makita, N., Furukawa, H., Ikeda, H. (2006). Moderate increase of mean daily temperature adversely affects fruit set of Lycopersicon esculentum by disrupting specific physiological processes in male reproductive development. Ann. Bot. 97, 731–738. doi: 10.1093/aob/mcl037
Serrani, J. C., Carrera, E., Ruiz-Rivero, O., Gallego-Giraldo, L., Peres, L. E. P., Garcia-Martinez, J. L. (2010). Inhibition of auxin transport from the ovary or from the apical shoot induces parthenocarpic fruit-set in tomato mediated by gibberellins. Plant Physiol. 153, 851–862. doi: 10.1104/pp.110.155424
Shinozaki, Y., Beauvoit, B. P., Takahara, M., Hao, S., Ezura, K., Andrieu, M.-H., et al. (2020). Fruit setting rewires central metabolism via gibberellin cascades. PNAS 117, 23970–23981. doi: 10.1073/pnas.2011859117
Slocombe, S. P., Beaudoin, F., Donaghy, P. G., Hardie, D. G., Dickinson, J. R., Halford, N. G. (2004). SNF-related protein kinase (SnRK1) phosphorylates class I heat shock protein. Plant Physiol. Biochem. 42, 111–116. doi: 10.1016/j.plaphy.2003.11.009
Sun, Z., Lower, R. L., Staub, J. E. (2006). Variance component analysis of parthenocarpy in elite U.S. processing type cucumber (Cucumis sativus L.) lines. Euphytica 14, 331–339. doi: 10.1007/s10681-005-9041-z
Tazuke, A. (1997). Growth of cucumber fruit as affected by the addition of NaCl to nutrient solution. J. Japan Soc Hortic. Sci. 66, 519–526. doi: 10.2503/jjshs.66.519
Tazuke, A., Asayama, M. (2013). Expression of CsSEF1 gene encoding putative CCCH zinc finger protein is induced by defoliation and prolonged darkness in cucumber fruit. Planta 237, 681–691. doi: 10.1007/s00425-012-1787-7
Tazuke, A., Kanisawa., K., Kinoshita, T., Asayama, M. (2019). Varietal differences in the effects of total defoliation on the expression of candidate marker genes of sugar starvation in cucumber fruit. Hortic. Res. 18, 157–165. doi: 10.2503/hrj.18.157
Tazuke, A., Kinoshita, T. (2018). Analysis of the decline in respiration rate of cucumber fruits on vines under prolonged darkness, total defoliation, cutting, and heat-girdling of the peduncle. Hortic. Res. 17, 55–59. doi: 10.2503/hrj.17.55
Tazuke, A., Kinoshita, T., Asayama, M. (2015). Expression of the BTB/POZ domain-containing protein Atg63850-like gene CsFDI1 is enhanced by sugar starvation in cucumber fruit. Acta Physiol. Plant 37, 15. doi: 10.1007/s11738-014-1766-7
Tazuke, A., Kinoshita, T., Asayama, M. (2017). The expression of a candidate cucumber fruit sugar starvation marker gene CsSEF1 is enhanced in malformed fruit induced by salinity. Physiol. Mol. Biol. Plants 23, 565–570. doi: 10.1007/s12298-017-0452-9
Turgeon, R., Wolf, S. (2009). Phloem transport: cellular pathways and molecular trafficking. Ann. Rev. Plant Biol. 60, 207–221. doi: 10.1146/annurev.arplant.043008.092045
van Rensburg, H. C. J., Van den Ende, W., Signorelli, S. (2019). Autophagy in plants: both a puppet and a puppet master of sugars. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00014
Wang, H., Jones, B., Li, Z., Frasse, P., Delalande, C., Regad, F., et al. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17, 2676–2692. doi: 10.1105/tpc.105.033415
Wu, Z., Zhang, T., Li, L., Xu, J., Qin, X., Zhang, T., et al. (2016). Identification of a stable major-effect QTL (Parth 2.1) controlling parthenocarpy in cucumber and associated candidate gene analysis via genome re-sequencing. BMC Plant Biol. 16, 182. doi: 10.1186/s12870-016-0873-6
Keywords: asparagine synthetase, CsSEF1, fruit abortion, fruit growth suppression, RNA-seq
Citation: Tazuke A, Kinoshita T and Asayama M (2023) Expression of candidate marker genes of sugar starvation is upregulated in growth-suppressed parthenocarpic cucumber fruit. Novel gene markers for sugar starvation in growth-suppressed cucumber fruit. Front. Plant Sci. 14:1241267. doi: 10.3389/fpls.2023.1241267
Received: 16 June 2023; Accepted: 27 July 2023;
Published: 18 August 2023.
Edited by:
Neftali Ochoa-Alejo, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoReviewed by:
Minmin Miao, Yangzhou University, ChinaSagar Datir, Naoroji Godrej Centre for Plant Research (NGCPR), India
Copyright © 2023 Tazuke, Kinoshita and Asayama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akio Tazuke, dGF6dWtlQHZjLmliYXJha2kuYWMuanA=
 Akio Tazuke
Akio Tazuke Tsuguki Kinoshita
Tsuguki Kinoshita Munehiko Asayama
Munehiko Asayama