- 1Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
- 2Plant Gene Expression Center, United States Department of Agriculture-Agricultural Research Service, Albany, CA, United States
CLAVATA3/ESR-related (CLE) peptides perform a variety of important functions in plant development and historically have been targeted during the domestication of existing crops. Pennycress (Thlaspi arvense) is an emerging biofuel crop currently undergoing domestication that offers novel monetary and environmental incentives as a winter cover crop during an otherwise fallow period of the corn/soybean farming rotation. Here we report the characterization of the CLE gene family in pennycress through homology comparison of the CLE motif with other dicot species by conducting a homology comparison and maximum likelihood phylogenetic analysis supplemented with manual annotation. Twenty-seven pennycress CLE genes were identified, and their expression analyzed through transcriptome profiling and RT-qPCR. Our study provides a genome-wide analysis of the CLE gene family in pennycress and carries significant value for accelerating the domestication of this crop through identification of potential key developmental regulatory genes.
1 Introduction
CLE genes encode a family of extracellular signaling peptides that are involved in numerous plant developmental processes. CLAVATA3 (CLV3) is the founding member of the CLE gene family in Arabidopsis thaliana and is a primary player in maintaining the shoot apical meristem: the pool of stem cells in the shoot tip that acts as a source of all cells for above ground organ development (Fletcher et al., 1999; Beauzamy et al., 2015; Fletcher, 2018). CLV3 activity as a key stem cell regulator comprises an ancient land plant function found in plants as early as bryophytes such as Physcomitrella patens (Whitewoods et al., 2018). CLV3 has an overlapping function in limiting shoot meristem maintenance with CLE16 and CLE17 (Dao et al., 2022) but acts oppositely to CLE40 (Schlegel et al., 2021). Additionally, the CLE19, CLE22 and CLE40 peptides each function in the root apical meristem, with CLE19 regulating root meristem size, CLE22 playing a role in meristem maintenance, and CLE40 regulating the distance of the quiescent center from the root tip (Casamitjana-Martıínez et al., 2003; Fiers et al., 2004; Stahl et al., 2009; Jun et al., 2010). CLE41 and CLE44 are involved in determining the division plane of cambium cells in the vascular root meristem (Ito et al., 2006; Hirakawa et al., 2008; Etchells and Turner, 2010). CLE1, CLE3, CLE4 and CLE7 are implicated in lateral root development under nitrogen deficiency (Araya et al., 2014). CLE8 is involved in seed development (Fiume and Fletcher, 2012) whereas CLE9/10 and CLE25 regulate stomate and vascular bundle development (Qian et al., 2018; Song et al., 2021). Further, CLE45 activity is implicated in phloem development (Song et al., 2021).
CLE genes are highly conserved in plants (Oelkers et al., 2008; Zhang et al., 2020). The full-length proteins comprise a conserved structure consisting of a signal peptide sequence near the amino-terminus of the peptide, followed by a variable domain that shows relatively low amino acid similarity between CLE proteins in a given plant species, and a CLE motif towards the carboxyl-terminus (Cock and McCormick, 2001; Strabala, 2008). The CLE motif is cleaved from the full-length protein and undergoes downstream post-translational modification to generate the mature, functional 12-13 amino acid peptide (Kim et al., 2017). The structure of CLE peptides among land plants is well conserved (Oelkers et al., 2008; Zhang et al., 2020), allowing for comprehensive phylogenetic comparison between Arabidopsis and other angiosperms.
The broad range of roles the CLE peptides play reflects potential uses in future domestication; for instance, meristem enlargement can lead to increased fruit size and seed number in crops such as maize, rice and tomato (Somssich et al., 2016). Domestication of crops such as tomato and maize was undertaken through significant changes in plant architecture traits that are mediated by shoot and floral meristem size such as flower number, fruit size, and fruit number. With meristem size being primarily regulated by the CLV3 pathway (Doebley et al., 2006; Bennetzen and Hake, 2009; Xu et al., 2015; Wu et al., 2018), understanding of the CLE family in new species can provide insight into its plant architecture and thereby provide a next step in its domestication effort by improving yield.
The midwestern United States primarily uses a rotation of corn and soybean as agricultural output. Fertilizer applied to these crops is subject to runoff into local waterways during the fallow winter period in between crop rotations; further, soil erodes from the empty fields. Cover crops can absorb excess nitrogen, preserve soil health, and offer a significant monetary incentive to growers after their primary harvest: providing an alternative to fallow winter fields (Isbell, 2009). Pennycress, a brassica closely related to Arabidopsis, has a short enough life cycle to be planted during this otherwise fallow period (Moser et al., 2009; Franzke et al., 2011; Sedbrook et al., 2014). Further, pennycress is cold hardy and its seeds can be harvested for oil (Warwick et al., 2002; Moser et al., 2009; Fan et al., 2013). Current efforts to promote pennycress as a cover crop have made it a candidate for use in the emerging biofuels industry (Phippen and Phippen, 2012). Pennycress is currently being implemented as an off-season rotation crop in the Midwest (Phippen et al., 2022) with previously generated varieties featuring various fatty acid profiles for biodiesel, jet fuel, and industrial fuel applications (Isbell et al., 2015; Esfahanian et al., 2021; Jarvis et al., 2021). As well, classic domestication traits such as early flowering and loss of seed shattering have also been engineered in pennycress to fit its planting into maize/soybean rotations while reducing loss of seed yield (Chopra et al., 2020).
We have undertaken a genome-wide analysis of the CLE gene family in pennycress to gain insight into their potential conservation in this emerging crop species and identify candidate genes for domestication efforts. We identify 27 CLE (TaCLE) genes in the pennycress genome. These genes are highly conserved between pennycress and Arabidopsis, although several sets of homologous CLE gene pairs in Arabidopsis are present as single copy genes in pennycress. Transcript profiling using various pennycress tissues shows that TaCLE genes are expressed in a variety of tissues during plant development; as well, some pennycress CLE genes show tissue expression profiles distinct from their Arabidopsis counterparts. Defining the pennycress CLE family members provides candidates for genetic engineering that can be undertaken to accelerate the domestication of this emerging biofuel crop.
2 Methods
2.1 Hidden markov modelling and sequence identification
HMMer 3.3.2 was used to identify amino acid sequences containing a CLE motif in the pennycress genome annotation version 1 (Dorn et al., 2015) using the hmmscan command and a CLE profile.hmm file generated by Oelkers et al., 2008. nblastn searches of the annotated genomic sequences of each of the 32 Arabidopsis CLE genes were then used to further identify and refine candidate sequences found during the HMM search. Finally, manual comparison of pennycress candidate peptides with Arabidopsis peptides and Sanger sequencing of pennycress cDNA amplified from whole seedling tissue using predicted TaCLE gene-specific primers (Supplementary Table 2) provided final consensus on identified sequences.
2.2 Multiple sequence alignment and conserved motif analysis
Multiple sequence alignment was performed on pennycress peptides using Clustal Omega and visualized using Jalview (Waterhouse et al., 2009; Madeira et al., 2022). Visualization of the 27 pennycress and 32 Arabidopsis peptide consensus sequences was undertaken using the publicly available weblogo application (Crooks et al., 2004) using default settings.
2.3 cDNA extraction and sequencing
MN106 seeds were sown on MS-Agar plates and seedlings were allowed to grow under long day (16 hour light: 8 hour dark) conditions at 22°C for three weeks after germination (Murashige and Skoog, 1962). RNA was extracted from 10 whole seedlings using a commercial RNeasy mini kit (Qiagen, 74004). Application of DNase I was used to destroy remaining genomic DNA according to the manufacturer’s protocol (Thermo Fisher Scientific, M0303S). 1 µg of extracted RNA was reverse transcribed using a commercial cDNA synthesis kit (Bio-Rad, 1708890). CLE gene sequences were amplified from 1 µl of 1/20th dilution of 1000 ng/µl MN106 cDNA using gene-specific primers (Supplementary Table 2) and the resulting DNA amplicons run on gels and extracted using the QIAquick gel extraction kit (Qiagen, 28706). Extracted DNA fragments were sequenced through Eurofins Genomics (Eurofins Genomics LLC).
2.4 Phylogenetic comparison and genomic organization
CLE genomic nucleotide sequences were aligned using Clustal Omega and the terminal ends were eliminated in Jalview alignment to align the signal peptide sequences as well as CLE motifs (Waterhouse et al., 2009; Madeira et al., 2022). One thousand phylogenetic maximum likelihood trees were generated using RAxML to generate sufficient bootstrap values and visualized using iTOL (Kozlov et al., 2019; Letunic and Bork, 2021). The following command was issued for RAxML generation:
raxmlHPC-PTHREADS-SSE3 -f a -x 1123 -p 2341 -#1000 -m GTRGAMMA -T 3 -s [Alignment File].fa -n [Output File].raxml
Genomic organization was visualized using the gene structure display server (Hu et al., 2015). Signal peptide analysis of full-length TaCLE amino acid sequences was undertaken using the SignalP web browser on the ‘Eukarya’ setting (Teufel et al., 2022).
2.5 Expression analysis
In silico transcription analysis was undertaken using a publicly available transcriptome dataset (Dorn et al., 2013) to generate a heatmap using ggplot (Wickham, 2016). Individual TaCLE gene expression within nine tissue types was profiled from a publicly available dataset (Nunn et al., 2022), using a minimum normalized read count cutoff of 10 or higher. For RT-qPCR, MN106 seedlings were grown on ½ MS-Agar plates and harvested three weeks after germination for RNA extraction using the Qiagen RNeasy Mini Kit. Genomic DNA was digested using NEB DNase I and the remaining RNA was reverse transcribed using the Bio-Rad iScript Reverse Transcription Supermix. Finally, qPCR was undertaken using the Bio-Rad Syber Green Master Mix. Differential expression analysis was performed manually taking Cq values according to the following formulae:
ΔCq = CqTarget – CqActin
ΔCq Expression = 2–ΔCq
and normalizing the ΔCq of the different tissue types to that of an ACTIN reference gene. A cutoff of a p-value < 0.05 from a one-tailed student’s t-test between null and experimental values was used to establish detectable expression. Three technical replicates were performed for each of three biological replicates.
3 Results
3.1 Identification and verification of CLE gene family in Thlaspi arvense
Extracting the predicted CLE peptides from the publicly available Thlaspi arvense version 1 genome using a downloadable FASTA file generated a searchable protein sequence list from which a Hidden Markov Model (HMM) search was conducted using HMMer 3.3.2 (Eddy, 2011; Dorn et al., 2015). Protein sequences were verified as members of the CLE family based on the presence of a conserved signaling peptide sequence as well as a characteristic CLE motif sequence (Cock and McCormick, 2001). Corresponding genomic and coding sequences were identified based on these peptides using a CLE HMM profile (Oelkers et al., 2008). Further manual characterization of pennycress CLE coding sequences was undertaken using tblastn of predicted pennycress CLE proteins against known Arabidopsis CLE genes. Pairwise sequence alignment after tblastn verified similarity of sequence and annotation of homologous coding sequence between Arabidopsis and pennycress CLE sequences. Finally, genomic DNA and cDNA were isolated from 3-week-old pennycress seedlings and sequenced to confirm the annotated sequences of the Thalaspi arvense CLE (TaCLE) genes. This work yielded 27 identified TaCLE genes as well as their respective genome annotation location on the pennycress chromosome scaffolds (Table 1). Pennycress CLE genes were named based on their overall amino acid sequence similarity to the corresponding Arabidopsis CLE gene. When a single TaCLE gene showed strong similarity to two Arabidopsis CLE genes across the full coding sequence, the TaCLE gene was named according to which Arabidopsis CLE gene showed the greatest degree of similarity in the CLE domain.
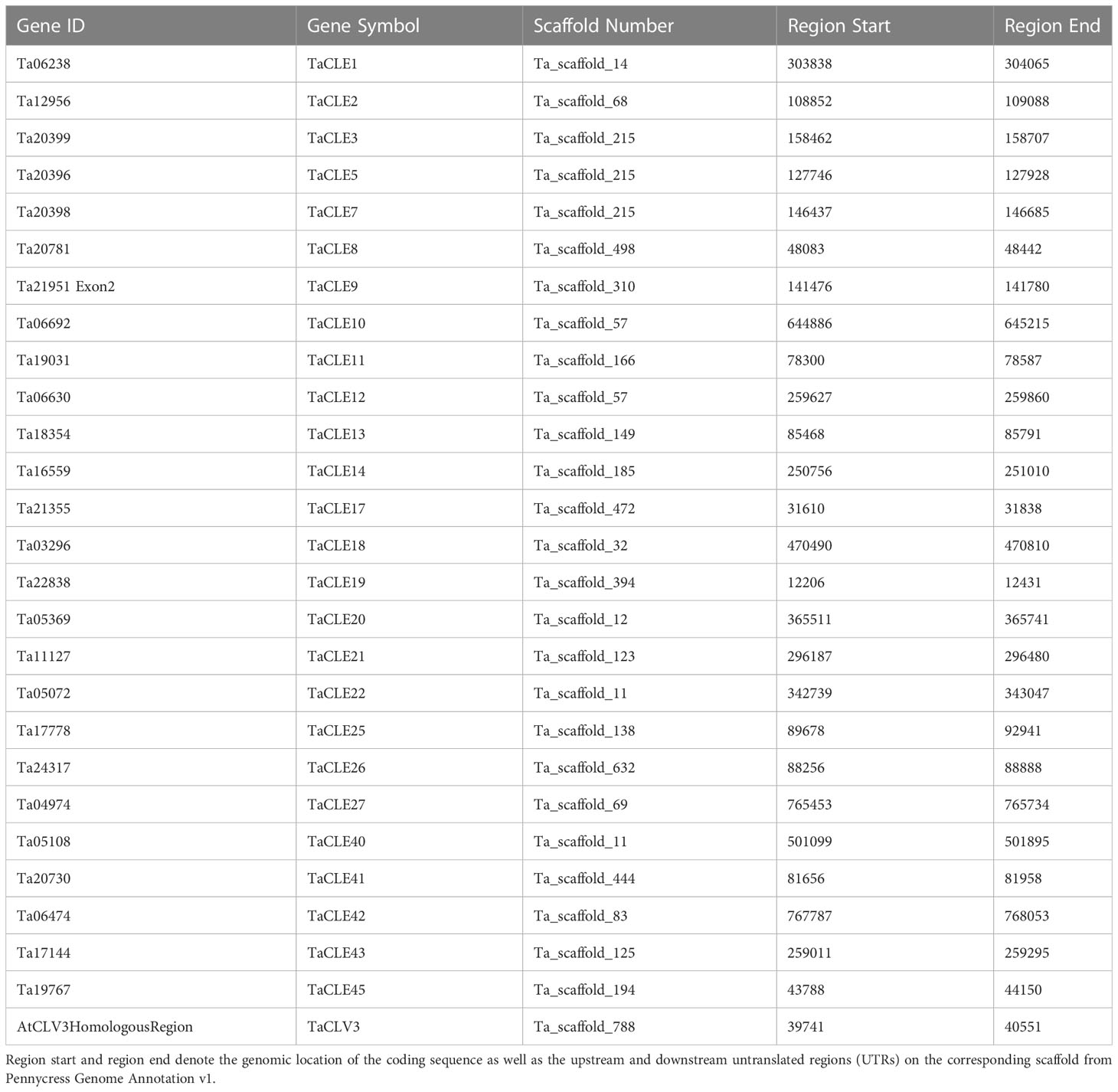
Table 1 Complete list of CLE genes identified in the pennycress genome. Gene Symbol refers to the given name based on the closest sister peptide in Arabidopsis.
Sequencing of amplified cDNA using TaCLE gene-specific primers confirmed the corrected coding sequence annotation and generated updated scaffold locations for the TaCLV3, TaCLE19, TaCLE25 and TaCLE40 loci (Table 1). TaCLV3 was not annotated in the Pennycress Genome Annotation v1 (Dorn et al., 2015) so we mapped it to Ta_scaffold_788 based on nucleotide sequence homology to the Arabidopsis CLV3 gene (Fletcher et al., 1999). The genomic sequence corresponding to TaCLE19 was originally annotated as the second exon of a larger gene with the Gene ID Ta21951. Our reannotation of the TaCLE25 coding sequence removes 25 base pairs (bp) from the 5’ end and 1 bp from the 3’ end of exon 1, as well as 28 bp from the 3’ end of exon 2, relative to the draft genome annotation (Figure S1). Finally, our reannotation of TaCLE40 identifies exon 1 as a 100 bp sequence that initiates 179 bp downstream of the exon 1 sequence annotated in the draft genome, and additionally omits 22 bp from the 3’ end of exon 2 (Figure S1). A recent release of an improved pennycress reference genome assembly representing ~97.5% of the estimated genome size (Nunn et al., 2022) correctly presented the sequences of the TaCLV3 and TaCLE40 genes, denoted TAV2_LOCUS13686 and TAV2_LOCUS22323, respectively, illustrating the limitations of the initial assembly built from shorter-read sequences.
A multiple sequence alignment of the full-length TaCLE proteins was constructed using Clustal Omega and Jalview (Waterhouse et al., 2009) to observe the conservation of amino acid sequences among the family members. TaCLE peptides display the characteristic conserved amino-terminal signal peptide sequence as well as the highly conserved 13 amino acid CLE motif located towards the C-terminus (Figure 1A). Similar to the full-length Arabidopsis CLE proteins (Cock and McCormick, 2001), the intervening variable domain presents little conservation between the different full-length CLE proteins in pennycress (Figure 1A).
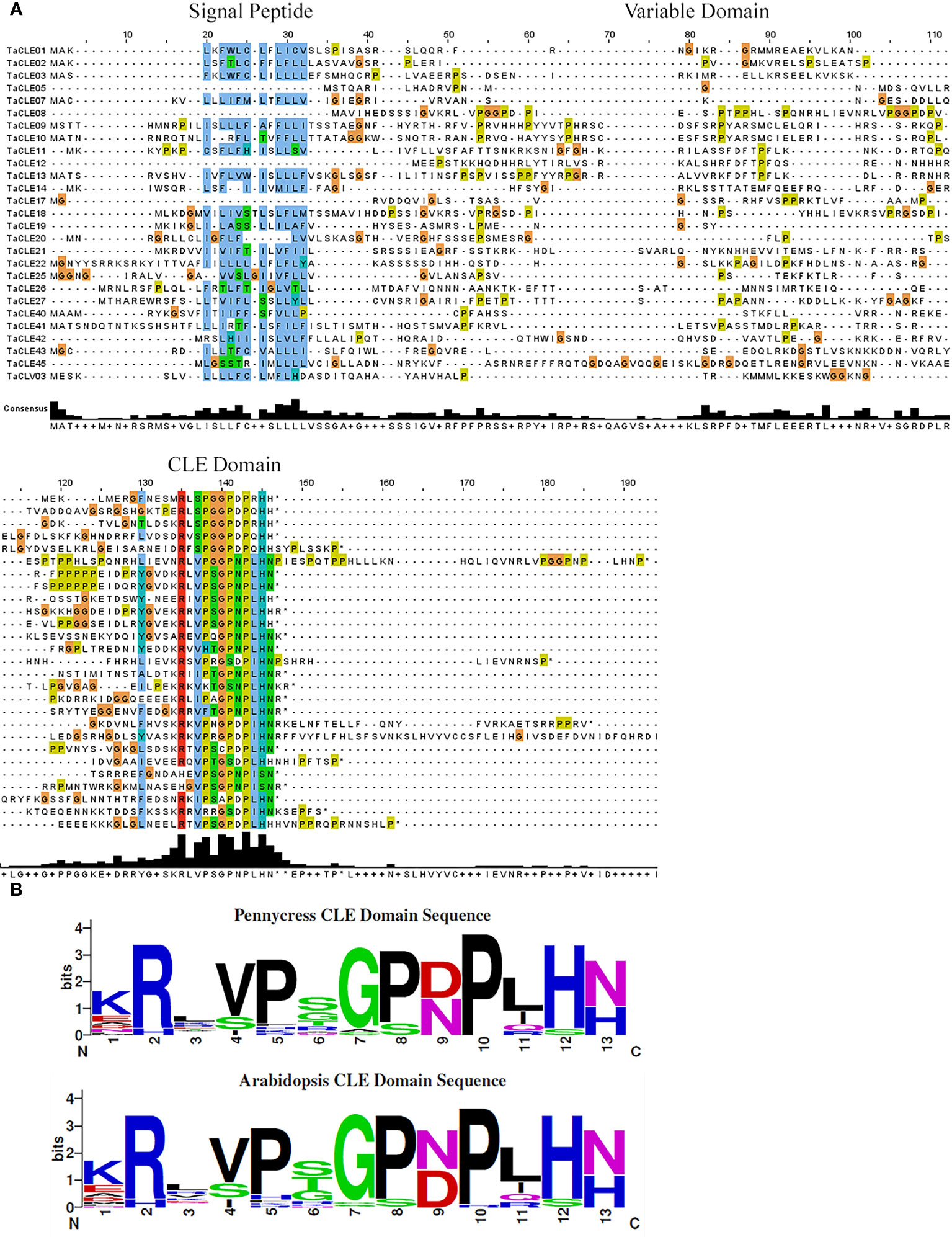
Figure 1 Multiple sequence alignment of the TaCLE proteins and consensus sequence for the CLE domains of pennycress and Arabidopsis. (A) Multiple sequence alignment of all 27 TaCLE proteins with the signal peptide, variable domain and CLE domain regions denoted. Colored boxes indicate conserved amino acids, and an amino acid consensus plot is shown below the CLE peptide sequences. (B) Weblogo plots of the pennycress and Arabidopsis CLE domain amino acid consensus sequences.
All 32 Arabidopsis CLE proteins contain an amino-terminal hydrophobic region that is predicted to act either as a signal peptide or a signal anchor sequence, directing the CLE peptide to the extracellular space (Sharma et al., 2003). Interestingly, the TaCLE5, TaCLE8, TaCLE12 and TaCLE17 proteins seem to lack a conserved signal peptide based on the amino acid sequence alignment (Figure 1A). We confirmed this prediction by examining their full-length amino acid sequences using SignalP 6.0, a machine learning model that detects all five known signal peptide types (Nielsen et al., 2019; Teufel et al., 2022). SignalP 6.0 failed to predict a signal peptide in any of these four pennycress CLE proteins (Table S1). Experimental analysis of these proteins therefore will be required to determine their subcellular localization.
The CLE domain amino acid consensus between pennycress and Arabidopsis proteins is highly similar (Figure 1B). The most striking difference lies at (N/D)9 with aspartate being slightly more prevalent in pennycress when compared to Arabidopsis. Divergence also exists at the sixth residue with a prevalence of(S/G/T)6 in pennycress compared to (S/T/G)6 in Arabidopsis; however, other sites display high levels of conservation (Figure 1B). CLE peptides undergo various post-translational modifications to gain functional potence (Kondo et al., 2006; Ohyama et al., 2009). Proline8, the site required for glycosylation, is highly conserved among the pennycress CLE domains (Figure 1B), which is consistent with our understanding of the importance of glycosylation at Pro8 of CLV3 to facilitate its ability to restrict stem cell activity (Shinohara and Matsubayashi, 2013). While experimental evidence will be necessary to assess the biochemical activities of the pennycress CLE peptides, conservation of the functional residue suggests overall conservation of function.
3.2 Phylogenetic analysis and genome organization of TaCLE genes
We next determined the general nucleotide sequence similarity of the pennycress to the Arabidopsis CLE genes through a maximum likelihood phylogenetic analysis. A Hidden Markov Model, BLAST comparison, and manual homology modeling of genomic nucleotide sequences followed by maximum likelihood tree generation shows a near one-to-one correspondence between pennycress CLE genes and their relatives in Arabidopsis (Figure 2). An exception occurs with the TaCLE8 and TaCLE18 genes, which are named based on the TaCLE8 CLE motif having greatest similarity to that of AtCLE8 and the TaCLE18 CLE motif having greatest similarity to that of AtCLE18. Our phylogenetic analysis shows that the AtCLE8 and AtCLE18 genomic nucleotide sequences are more similar to one another than either is to the TaCLE8 or TaCLE18 sequences (Figure 2); however, due to the low bootstrap values we are unable to draw this conclusion definitively. Interestingly, the gene pairs AtCLE3/4, AtCLE5/6, AtCLE16/17 and AtCLE41/44 that are predicted to be the result of duplication events in Arabidopsis (Sharma et al., 2003) each have one homologue in pennycress, denoted TaCLE3, TaCLE5, TaCLE17, and TaCLE41 respectively based on which of the two Arabidopsis homologues has the greatest similarity to the pennycress CLE domain (Table 1; Figure 2). Overall, the bootstrap values for the various branches of the tree are reproducibly high (>50), demonstrating that the pennycress CLE genes are reproducibly different from one another while maintaining a largely one-to-one relationship with their sister Arabidopsis genes.
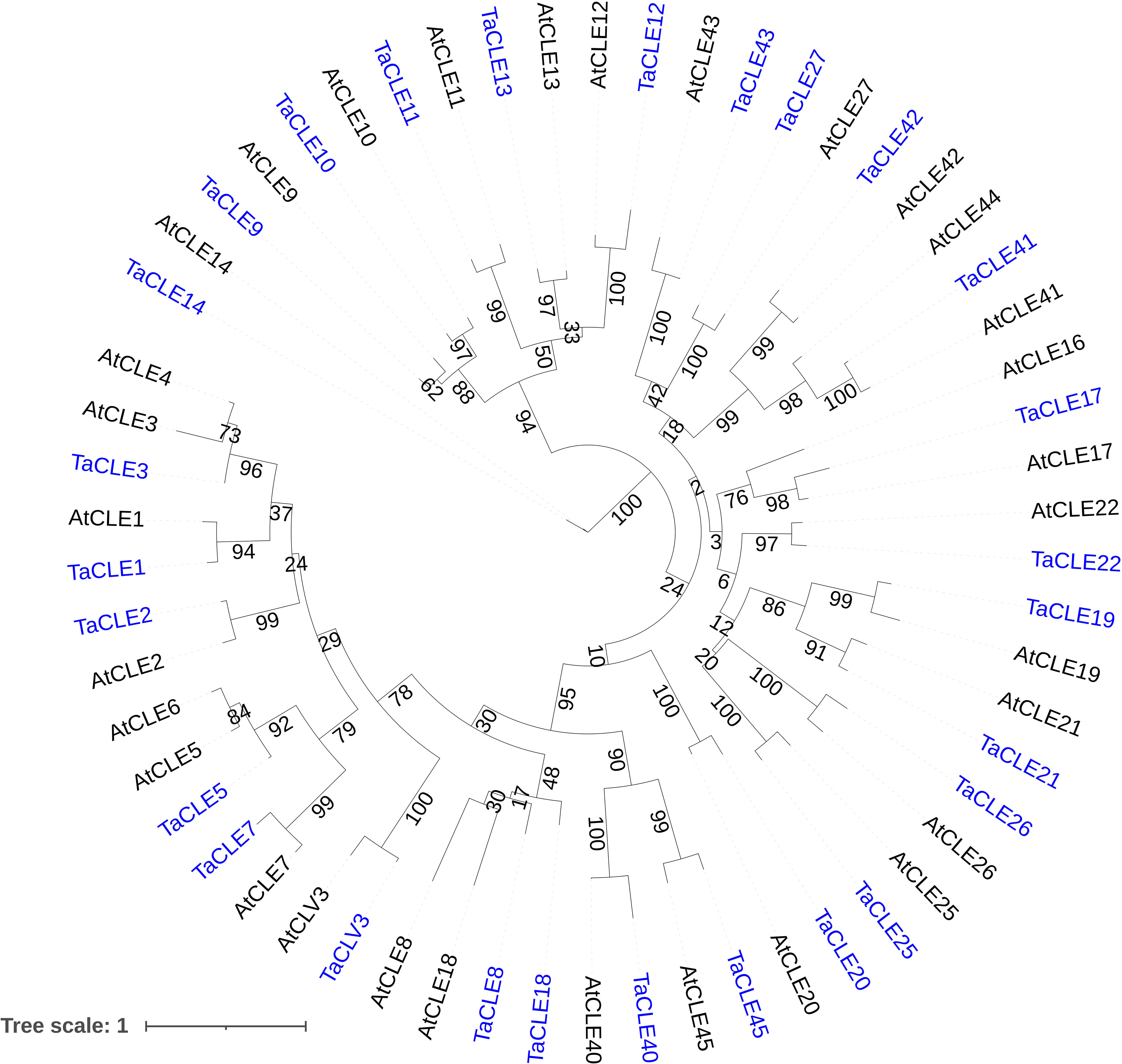
Figure 2 Phylogenetic analysis of pennycress and Arabidopsis CLE peptides. Maximum likelihood tree generated from alignment of all pennycress (blue) and Arabidopsis (black) CLE domains with 1000 bootstrap replicates. Bootstrap values from 1-100 are displayed at each branch.
Examination of the genomic organization of the TaCLE loci indicates that the genes predominantly lack introns and that the length of the genomic sequence is on the order of 200-700 base pairs (Figure 3A). In Arabidopsis, both the CLV3 and CLE40 loci consist of three exons and two introns (Cock and McCormick, 2001), and these gene structures are conserved in their pennycress counterparts. Yet whereas AtCLE16, AtCLE17, AtCLE19 and AtCLE25 consist of a single exon, the TaCLE17 and TaCLE19 loci both feature a small intron and the TaCLE25 locus features a much longer intron (Figure 3A), suggesting the regulation of these three genes may differ from that of their Arabidopsis counterparts.
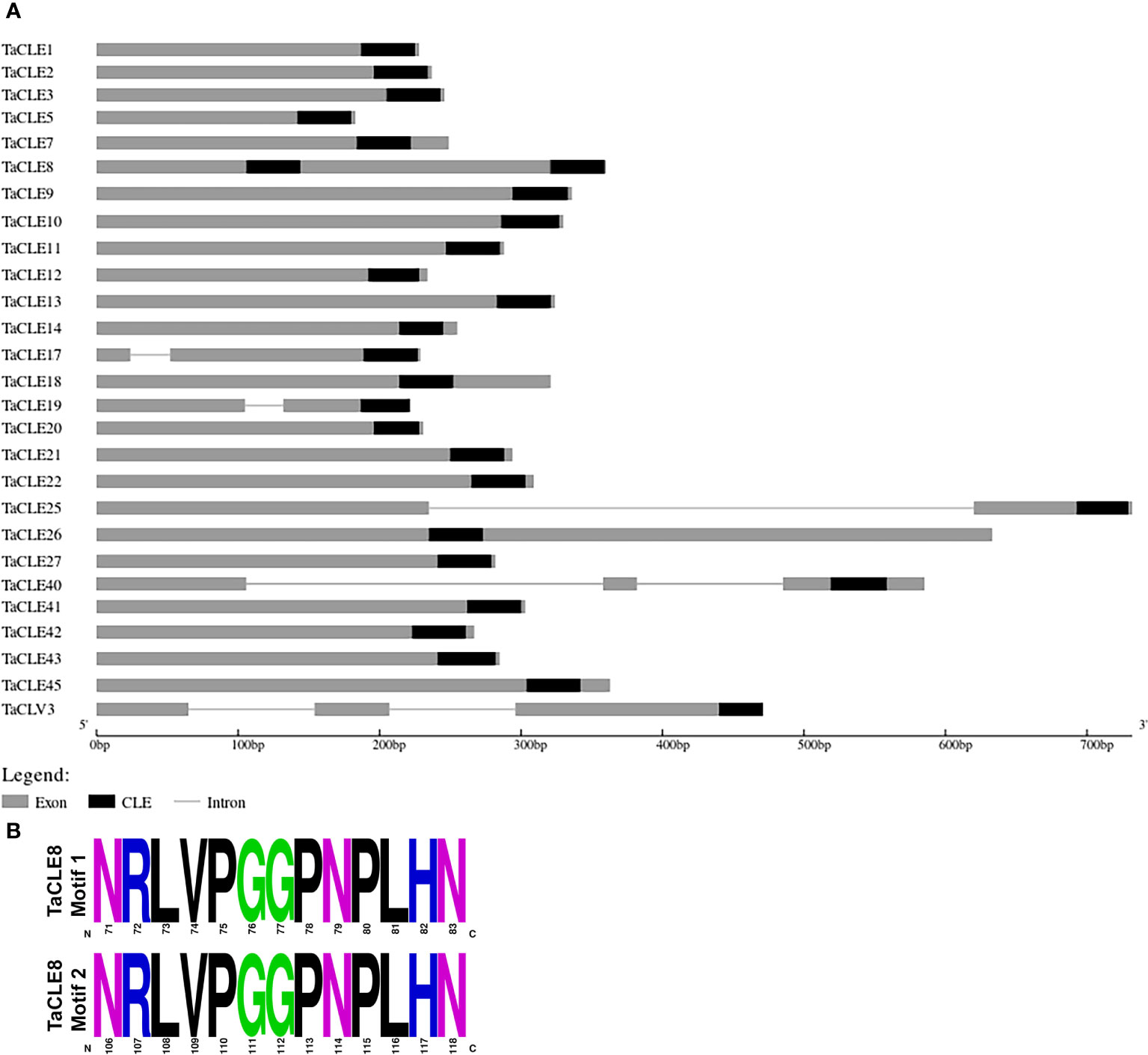
Figure 3 Genomic organization of the TaCLE genes and sequence of the duplicate TaCLE8 CLE domains. (A) TaCLE genome structure showing exons as grey boxes, introns as lines, and CLE motifs as black boxes. (B) Amino acid sequences of the two TaCLE8 CLE domains.
Although the 32 Arabidopsis CLE genes each contain a single CLE motif, some CLE genes in other plants contain multiple CLE motifs (Gao and Guo, 2012). Among the 27 identified CLE genes in pennycress, the TaCLE8 gene encodes two separate CLE motifs, one located in the center and the other towards the C terminus of the protein (Figure 3A). The CLE domains of TaCLE8 are identical in amino acid sequence (Figure 3B), suggesting they may have identical functions.
3.3 TaCLE gene expression analysis
Next, we analyzed the expression profiles of the pennycress CLE genes, as understanding gene transcription levels and patterns can provide insight into their involvement in various aspects of growth and development. We first used a publicly available transcriptome database (Dorn et al., 2013) to examine TaCLE gene expression in four broad tissue types: flowers, inflorescences, roots, and vegetative rosettes. Four main expression groups could be observed, which we demarcated Groups I-IV based on their differential expression patterns (Figure S2). The Group I genes TaCLE5, TaCLE10, and TaCLE17 display relatively high transcript levels across all four tissue types when compared to the other TaCLE genes. Group II consists of 12 TaCLE genes with low or undetectable levels of expression in the four tissues. Group III features 8 genes with detectable mRNA expression in one or more of the tissues, but with far lower expression levels than either Group I or IV genes. Finally, the Group IV genes TaCLE1 and TaCLE11 show comparatively high expression in roots and rosettes relative to the Group I genes that display moderately high expression across all four tissue types. This analysis demonstrates that nearly half of the TaCLE genes are transcribed in one or more of the major pennycress tissue types.
We further took advantage of a larger-scale expression atlas to profile TaCLE gene expression in nine distinct pennycress tissue types (Nunn et al., 2022). TaCLE1 is primarily expressed in 1-week-old roots and shoots with a subsequent increase in expression in seed pods (Figure 4A). TaCLE5 is primarily expressed in shoots, rosette leaves, and inflorescences (Figure 4B). TaCLE7 is predominantly expressed in green seeds (Figure 4C) whereas TaCLE10 and TaCLE11 are more highly expressed in 1-week-old shoots and young green siliques (Figures 4D, E). TaCLE12 is expressed in open flowers, young green siliques and green seeds (Figure 4F). The expression pattern of TaCLE14 appears to be restricted to green seeds (Figure 4G). TaCLE17 features broad expression, particularly in the developing rosette leaves, inflorescences and reproductive tissues (Figure 4H). TaCLE18 and TaCLE20 are primarily expressed in 1-week-old shoots and rosette leaves while TaCLE20 is also expressed in green seeds (Figures 4I, J). Finally, the CLE genes TaCLE41 and TaCLE43 are both expressed in 1-week-old shoots, with also expressed in young green siliques and TaCLE43 in green seeds (Figures 4K, L). Among the pennycress CLE genes TaCLE1 is the most highly expressed, but others such as TaCLE5, TaCLE7, TaCLE18, TaCLE20, TaCLE41, and TaCLE43 show low levels of expression (Figure S3). In contrast, the extremely high expression of TaCLE14 in green seeds is likely to be an artefact of sequencing as it is orders of magnitude greater than those of the other TaCLE genes in either published transcriptome dataset (Figure 4; Figure S2).
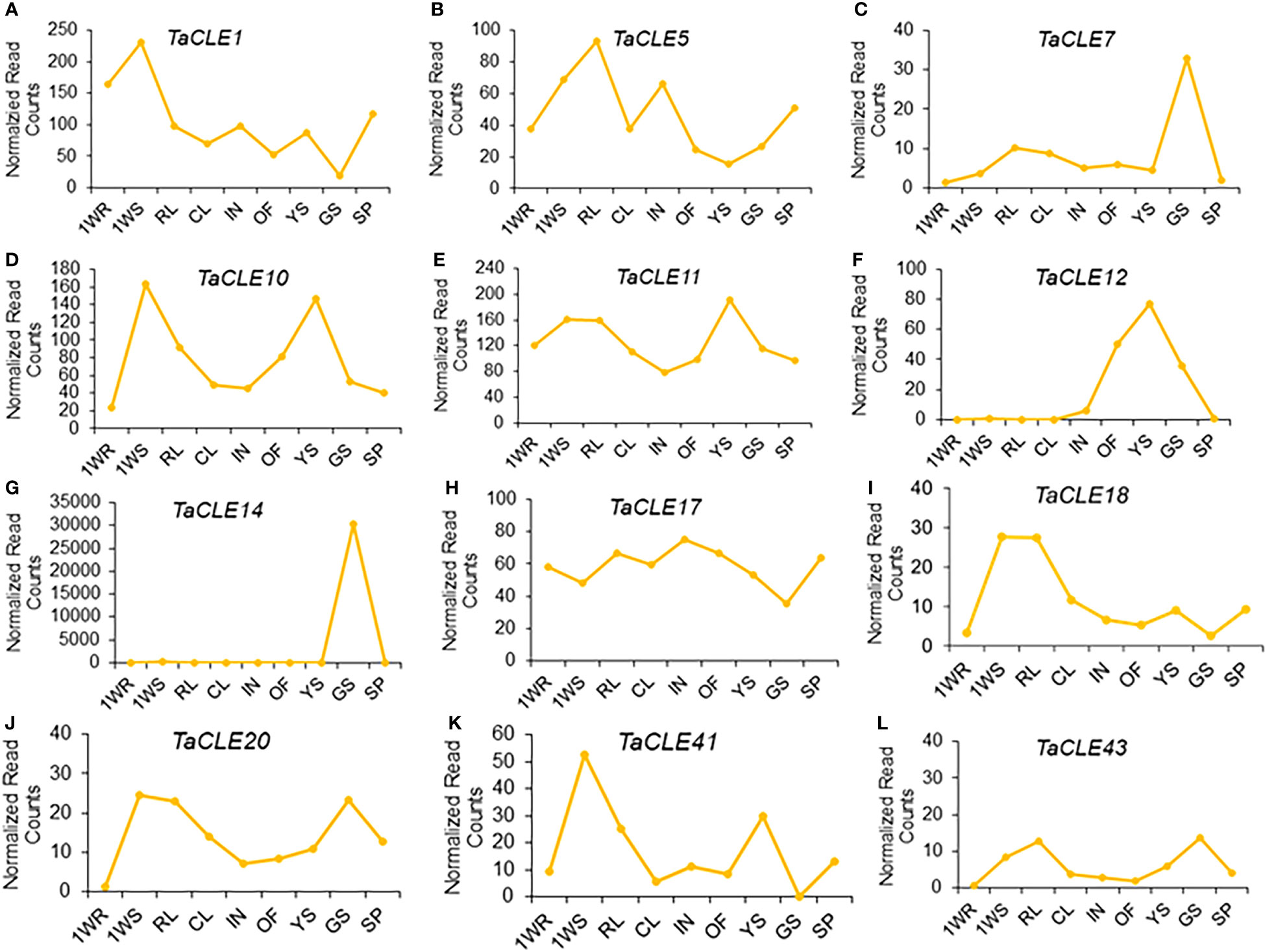
Figure 4 Expression profiles of TaCLE genes in various pennycress tissues. mRNA expression levels (normalized read counts) of the (A) TaCLE1 (B) TaCLE5 (C) TaCLE7 (D) TaCLE10 (E) TaCLE11 (F) TaCLE12 (G) TaCLE14 (H) TaCLE17 (I) TaCLE18 (J) TaCLE20 (K) TaCLE41 and (L) TaCLE43 genes. 1WR, roots from 1-week-old seedlings; 1WS, shoots from 1-week-old seedlings; RL, rosette leaves; CL, cauline leaves; IN, inflorescences; OF, open florescences; YS, young green siliques; GS, green seeds; SP, seed pods.
In total, 13 out of the 27 pennycress CLE family members display measurable expression in the published transcriptome datasets. To elucidate the expression of the other 14 TaCLE genes, we queried development stages not previously examined by extracting RNA from three-week-old whole seedlings as well as dissected shoots and dissected roots, and then undertaking reverse transcription-quantitative polymerase chain reaction (RT-qPCR) to quantify TaCLE gene transcript levels. Most of the CLE genes are found to be significantly expressed using a one-tailed Student’s t-test against a negative control in at least one tissue type, except for TaCLE3 and TaCLE21 which do not feature significant levels of expression in these tissues (Figure 5). Importantly, quantitative PCR also confirmed the expression of our annotated versions of the TaCLV3, TaCLE19, TaCLE25 and TaCLE40 genes. Due to the differences between our putative sequence for TaCLE40 and the annotated sequence in the draft genome with exon 1 being 179 base pairs apart, we designed one primer for our sequence and one for the draft sequence. Only our primer was successful in cDNA fragment amplification (Figure S2; Table S2).
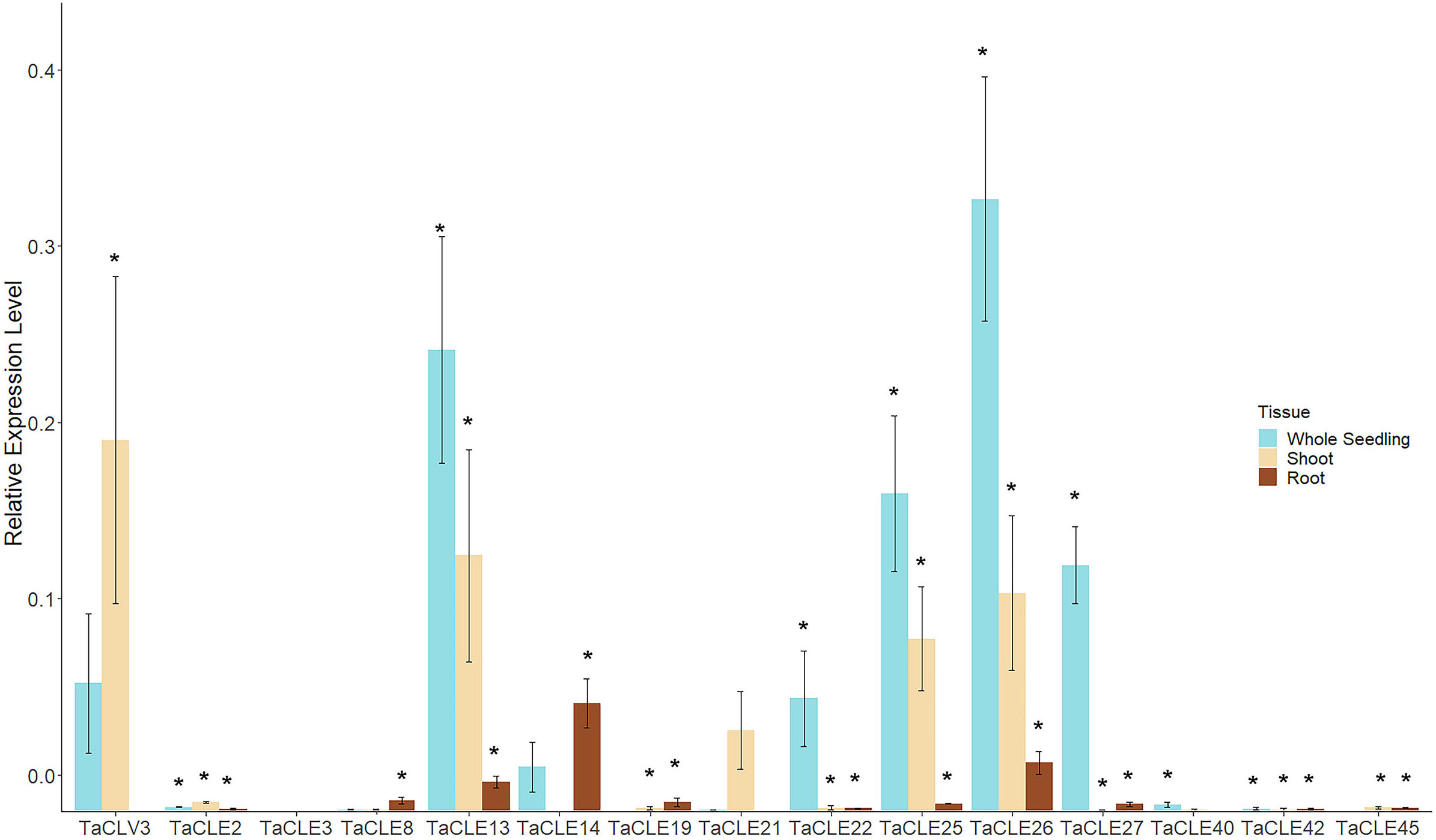
Figure 5 Expression profiles of TaCLE genes in 3-week-old pennycress tissues. Relative mRNA expression levels (mean ± SE) of TaCLE genes in whole seedling, shoot and root tissues. Asterisks (*) denote significance of p < 0.05 when compared via a Student’s one-tailed t-test against the null value.
Transcripts from the TaCLE2, TaCLE19, TaCLE42 and TaCLE45 genes are expressed at low but detectable levels in all three tissues analyzed (Figure 5). TaCLV3 displays higher expression in shoot than whole seedling tissue but is barely detectable in root tissue (Figure 5). TaCLE13, TaCLE25 and TaCLE26 show similar patterns of expression, with their transcript levels highest in whole seedlings, lower in shoot tissue and very low in root tissue. TaCLE22, TaCLE27 and TaCLE40 transcripts are detected in whole seedling but not shoot or root tissue, suggesting they may be expressed in leaves. TaCLE8 and TaCLE14 appear to be expressed at low levels in root tissue, whereas TaCLE21 transcripts are not detected at statistically significant levels in seedling, shoot or root tissues. In sum, we provide evidence for the expression under normal growth conditions of 25 of the 27 pennycress CLE genes.
4 Discussion
The CLE genes form an evolutionarily-conserved signaling protein family that control numerous aspects of plant growth and development. Some of these genes regulate molecular pathways that may be exploited to accelerate the domestication of orphan or emerging crops such as the biofuel crop pennycress. For example, the CLV3-mediated meristem maintenance pathway that has been targeted to enhance fruit and seed yield during the domestication of tomato, rice and maize (Fletcher, 2018) may be engineered to improve the low seed yield of pennycress plants (Sedbrook et al., 2014). Likewise, targeting the pennycress homolog of CLE8 could enlarge the size of the oilseeds to improve the harvest for biofuel applications, whereas manipulation of the pennycress CLE1-7 genes could enhance shoot regeneration and thus improve transformation efficiency (McGinn et al., 2019).
We have therefore undertaken an investigation into the CLE peptide family in pennycress through the identification of 27 TaCLE genes. Overall, the TaCLE genes display similar genetic organization and sequence composition to their Arabidopsis counterparts. We detected expression of 13 of the TaCLE genes in publicly available transcriptome datasets, and expression of a further 12 genes in our RT-qPCR dataset from seedling tissues. Because CLE peptides have previously been implicated in crucial developmental pathways, this study provides a solid foundation for genetic engineering of pennycress architecture, a key step in its wider adoption as a cost-effective biofuels crop.
Pennycress features a similar number of CLE peptides as other Brassica species, such as 29 in Brassica rapa, 32 in Brassica oleracea, and 32 in Arabidopsis thaliana (Xie et al., 2022). Full-length pennycress CLE proteins display a similar structure to CLE proteins in other species, featuring a signal peptide, variable domain, and CLE domain (Figure 1). Interestingly, SignalP predicts no signal peptide in TaCLE5, TaCLE8, TaCLE12 and TaCLE17, indicating either that these proteins are not exported or they lack a predictable signal peptide export sequence (Table S1). The TaCLE8 protein is also distinct in featuring two identical CLE domains (Figure 3B), indicating a possible duplication of this motif. Overall, the CLE domain consensus sequence shows greater conservation between Arabidopsis and pennycress (Figure 1B) than between Arabidopsis and Brassica rapa, Brassica napus or Brassica oleracea (Xie et al., 2022). This high level of sequence conservation is suggestive of conserved roles for CLE family members between Arabidopsis and pennycress.
Phylogenetic analysis and hidden Markov modelling showcase a near one-to-one correspondence between Arabidopsis and pennycress CLE genes, although notably the Arabidopsis gene pairs AtCLE3/4, AtCLE5/6, AtCLE16/17, and AtCLE41/44 have only one copy in pennycress (Figure 1A; Figure 3). Yet AtCLE9/10, another predicted gene duplication in Arabidopsis, features both homologues in pennycress. These divergences suggest that either these CLE genes were duplicated in a common ancestor of Arabidopsis and pennycress and then subsequently some were lost in pennycress, or the genes have undergone duplication following the divergence between these two closely related Brassica species. Further phylogenetic analysis of the CLE family in closer relatives of pennycress could help to distinguish between these hypotheses.
Generally, the pennycress CLE gene structures are similar to those of the Arabidopsis CLE genes (Figure 3A). Most TaCLE loci consist of a single exon, although TaCLE40 and TaCLV3, like AtCLE40 and AtCLV3, consist of two introns and three exons. Yet unlike their Arabidopsis counterparts, the TaCLE17, TaCLE19 and TaCLE25 genes each contain a single intron, suggesting differences may exist in their transcription regulation. The overall similarity in gene structure is demonstrative of the relatedness between these two species.
To gain insight into the potential biological functions of the pennycress CLE genes, we examined their transcription profiles using in silico expression analysis as well as RT-qPCR. We found evidence for the expression of 25 TaCLE genes within the various tissues profiled (Figures 4, 5, S2). However, we were unable to detect measurable TaCLE3 and TaCLE21 expression across the datasets sampled, and several TaCLE gene transcripts, including TaCLE42, TaCLE43 and TaCLE45 were detected at extremely low levels. This is not unexpected because many CLE genes in various species are expressed at very low levels, making them difficult to parse in transcriptomics data (Goad et al., 2017). Further, some CLE genes are restricted to small subsets of tissues: for example, CLV3 expression is restricted to the central zone of the Arabidopsis shoot and floral meristem (Fletcher et al., 1999). These attributes can make accurately quantifying transcript levels within bulk tissues such as leaves or roots difficult. Future analysis using transcriptional reporter lines can more accurately pinpoint when and where the TaCLE genes are expressed.
Importantly, absolute or relative gene expression values are not necessarily reflective of the importance of a given gene in growth and development. CLE genes that are lowly expressed may still carry out important functions. Further work will clarify the expression of the entire TaCLE gene family in tissues not yet assayed, such as floral organs and lateral roots, and determine the precise gene expression patterns within tissues.
Like the Arabidopsis CLE genes (Jun et al., 2010), the majority of TaCLE genes are detected in multiple tissues during pennycress development (Figures 4, 5; Figures S2, S3). In addition, all pennycress tissues examined express multiple TaCLE genes. We observe that a number of CLE genes including TaCLE1, TaCLE5, TaCLE11, and TaCLE17 are broadly expressed in pennycress, whereas others such as TaCLE12 and TaCLE18 show more tissue-restricted expression patterns.
Many TaCLE genes are expressed in pennycress roots, as is the case in Arabidopsis (Jun et al., 2010). In root tissue from 1-week-old seedlings, TaCLE1, TaCLE5, TaCLE9, TaCLE11, and TaCLE17 are all expressed, as are TaCLE10 and TaCLE41 at very low levels (Figures 4, S2). In root tissue from 3-week-old seedings, TaCLE8 and TaCLE14 are notably expressed whereas TaCLE2, TaCLE13, TaCLE19, TaCLE22, TaCLE25, TaCLE26, TaCLE27, TaCLE42 and TaCLE45 are all expressed at low yet measurable levels (Figure 5).
In shoot tissue from 1-week-old seedlings, the TaCLE1, TaCLE5, TaCLE10, TaCLE11, TaCLE17, TaCLE18, TaCLE20 and TaCLE41 genes are all expressed (Figure 4). Among these, neither TaCLE18 nor TaCLE20 is expressed in roots, indicating a potential role in above-ground tissue development. TaCLE41, being primarily expressed in young root and shoot tissues reflects potential vascular and root roles like those of its Arabidopsis sister (Ito et al., 2006; Hirakawa et al., 2008; Etchells and Turner, 2010). Further, TaCLV3, TaCLE2, TaCLE13, TaCLE25, TaCLE26, TaCLE42 and TaCLE45 are all expressed in 3-week shoot tissue, although the transcript levels of the latter two genes are very low (Figure 5). TaCLV3 expression in shoot tissue suggests a potentially similar role to Arabidopsis CLV3 in maintaining the shoot apical meristem (Clark et al., 1995; Fletcher, 1999), as its expression is moderate in whole seedling tissue but increased when the tissue is narrowed down to just the shoot while being absent in roots.
Pennycress rosette and rosette leaf tissues also express numerous CLE genes. TaCLE1, TaCLE5, TaCLE7, TaCLE9, TaCLE10, TaCLE11, TaCLE17, TaCLE18, TaCLE20, TaCLE41 and TaCLE43 transcripts are all detectable in rosette and rosette leaf tissue (Figure S2). Additionally, TaCLE22, TaCLE27 and TaCLE40 display higher mRNA expression levels in whole seedlings than in either shoots or roots (Figure 5), suggesting that all of these genes may function during the vegetative phase.
During the reproductive phase, we detect expression of TaCLE1, TaCLE5, TaCLE7, TaCLE9, TaCLE10, TaCLE11, TaCLE12 and TaCLE17 in inflorescences (Figure S2; Figure 4), as well as TaCLE18, TaCLE20 and TaCLE41 in inflorescences and open florescences (Figure 4). The same set of genes is expressed in young green siliques, with TaCLE10, TaCLE11, TaCLE12 and TaCLE41 showing higher levels of transcription in siliques than in other reproductive tissues.
Seed yield is intrinsically tied with seed weight, as seed weight increases results in an increased harvest index, the seed weight divided by the dry shoot mass. However, little is understood about the role of CLE genes in seed development beyond the involvement of AtCLE8 in embryo and endosperm size regulation (Fiume and Fletcher, 2012). Our transcriptome data indicate that at least 8 TaCLE genes are expressed in green seeds: TaCLE5, TaCLE7, TaCLE10, TaCLE11, TaCLE12, TaCLE17, TaCLE20 and TaCLE43 (Figure 4), excluding TaCLE14 for which the extremely high level of expression is likely to be an artifact. Interestingly, although AtCLE5, AtCLE7 and AtCLE12 gene expression is absent in Arabidopsis seed tissue (Klepikova et al., 2016), their pennycress homologs TaCLE5, TaCLE7 and TaCLE12 are all expressed in green seeds (Figure 4). Conversely, AtCLE17 and TaCLE17 are both expressed in seed tissue whereas AtCLE16 transcription is notably absent. The similarity in expression pattern, as well as the greater amino acid similarity in the CLE motif (Figure 2), provide further suggestion that the single copy TaCLE17 gene is more closely related to AtCLE17 than its sister gene AtCLE16. Understanding the functions of CLE genes expressed in pennycress seeds could provide a path forward for domestication while also furthering understanding of the family’s role in this crucial step of plant reproduction.
Members of the CLAVATA3/ESR-related peptide family regulate many important biological processes in plants and have played a role in the domestication of diverse crop species. Our study presents the sequence information, conservation, and expression analysis of this family in the emerging biofuel crop pennycress. Using a combination of evolution and transcriptomics analysis, we have provided a solid foundation for future domestication efforts by elucidating potential candidates for future genetic engineering efforts. Such domestication efforts can provide a significant financial incentive that could accelerate the adoption of pennycress as a cover crop to maintain soil health by preventing runoff and erosion during an otherwise fallow planting period. Also, a deeper understanding of this family across different species can provide further insight into the role of CLE genes in plant growth and development.
Data availability statement
The data presented in this study can be found online in the supplemental material at https://www.frontiersin.org/articles/10.3389/fpls.2023.1240342/full#supplementary-material. Genomic data used in this study have previously been deposited in the NCBI Sequence Read Archive under accession number SRP033211 (Dorn et al., 2015). Expression data has previously been deposited in the ENA Sequence Read Archive under accession number PRJEB46635 (Nunn et al., 2022).
Author contributions
LH and JF conceived and designed the study. LH conducted the experiments and acquired the data. LH and JF analyzed the data. LH wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the United States Department of Agriculture (CRIS 2030-21000-048-00D).
Acknowledgments
We thank Ratan Chopra and M. David Marks at the University of Minnesota for MN106 seeds and information about pennycress, and Liu Hong, Dean Pettinga, and Citlali Fonseca for comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1240342/full#supplementary-material
References
Araya, T., Miyamoto, M., Wibowo, J., Suzuki, A., Kojima, S., Tsuchiya, Y. N., et al. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. 111 (5), 2029–2034. doi: 10.1073/pnas.1319953111
Beauzamy, L., Louveaux, M., Hamant, O., Boudaoud, A. (2015). Mechanically, the shoot apical meristem of Arabidopsis behaves like a shell inflated by a pressure of about 1 MPa. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01038
Bennetzen, J. L., Hake, S. C. (2009). Handbook of maize: its biology (NY: Springer New York). doi: 10.1007/978-0-387-79418-1
Casamitjana-Martıínez, E., Hofhuis, H. F., Xu, J., Liu, C.-M., Heidstra, R., Scheres, B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13 (16), 1435–1441. doi: 10.1016/S0960-9822(03)00533-5
Chopra, R., Johnson, E. B., Emenecker, R., Cahoon, E. B., Lyons, J., Kliebenstein, D. J., et al. (2020). Identification and stacking of crucial traits required for the domestication of pennycress. Nat. Food 1 (1), 84–91. doi: 10.1038/s43016-019-0007-z
Clark, S. E., Running, M. P., Meyerowitz, E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 (7), 2057–2067. doi: 10.1242/dev.121.7.2057
Cock, J. M., McCormick, S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126 (3), 939–942. doi: 10.1104/pp.126.3.939
Crooks, G. E., Hon, G., Chandonia, J.-M., Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14 (6), 1188–1190. doi: 10.1101/gr.849004
Dao, T. Q., Weksler, N., Liu, H., Leiboff, S., Fletcher, J. C. (2022). Interactive CLV3, CLE16, and CLE17 signaling mediates stem cell homeostasis in the Arabidopsis shoot apical meristem. Development 149(19), dev200787. doi: 10.1242/dev.200787
Doebley, J. F., Gaut, B. S., Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127 (7), 1309–1321. doi: 10.1016/j.cell.2006.12.006
Dorn, K. M., Fankhauser, J. D., Wyse, D. L., Marks, M. D. (2013). De novo assembly of the pennycress (Thlaspi arvense) transcriptome provides tools for the development of a winter cover crop and biodiesel feedstock. Plant J. 75 (6), 1028–1038. doi: 10.1111/tpj.12267
Dorn, K. M., Fankhauser, J. D., Wyse, D. L., Marks, M. D. (2015). A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Research: Int. J. Rapid Publ. Rep. Genes Genomes 22 (2), 121–131. doi: 10.1093/dnares/dsu045
Eddy, S. R. (2011). Accelerated profile HMM searches. PloS Comput. Biol. 7 (10), e1002195. doi: 10.1371/journal.pcbi.1002195
Esfahanian, M., Nazarenus, T. J., Freund, M. M., McIntosh, G., Phippen, W. B., Phippen, M. E., et al. (2021). Generating pennycress (Thlaspi arvense) seed triacylglycerols and acetyl-triacylglycerols containing medium-chain fatty acids. Front. Energy Res. 9. doi: 10.3389/fenrg.2021.620118
Etchells, J. P., Turner, S. R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137 (5), 767–774. doi: 10.1242/dev.044941
Fan, J., Shonnard, D. R., Kalnes, T. N., Johnsen, P. B., Rao, S. (2013). A life cycle assessment of pennycress (Thlaspi arvense L.) -derived jet fuel and diesel. Biomass and Bioenergy. 55, 87–100. doi: 10.1016/j.biombioe.2012.12.040
Fiers, M., Hause, G., Boutilier, K., Casamitjana-Martinez, E., Weijers, D., Offringa, R., et al. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327 (1), 37–49. doi: 10.1016/j.gene.2003.11.014
Fiume, E., Fletcher, J. C. (2012). Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24 (3), 1000–1012. doi: 10.1105/tpc.111.094839
Fletcher, J. C., Brand, U, Running, M. P., Simon, R., Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 (5409), 1911–1914. doi: 10.1126/science.283.5409.1911
Fletcher, J. C. (2018). The CLV-WUS stem cell signaling pathway: A roadmap to crop yield optimization. Plants 7 (4), 87. doi: 10.3390/plants7040087
Franzke, A., Lysak, M. A., Al-Shehbaz, I. A., Koch, M. A., Mummenhoff, K. (2011). Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 16 (2), 108–116. doi: 10.1016/j.tplants.2010.11.005
Gao, X., Guo, Y. (2012). CLE peptides in plants: proteolytic processing, structure-activity relationship, and ligand-receptor interaction. J. Integr. Plant Biol. 54 (10), 738–745. doi: 10.1111/j.1744-7909.2012.01154.x
Goad, D. M., Zhu, C., Kellogg, E. A. (2017). Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol. 216 (2), 605–616. doi: 10.1111/nph.14348
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. 105 (39), 15208–15213. doi: 10.1073/pnas.0808444105
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Isbell, T. A. (2009). US effort in the development of new crops (Lesquerella, Pennycress Coriander and Cuphea). Oléagineux Corps Gras Lipides 16 (4-5–6), 4-5–4-6. doi: 10.1051/ocl.2009.0269
Isbell, T. A., Evangelista, R., Glenn, S. E., Devore, D. A., Moser, B. R., Cermak, S. C., et al. (2015). Enrichment of erucic acid from pennycress (Thlaspi arvense L.) seed oil. Ind. Crops Products 66, 188–193. doi: 10.1016/j.indcrop.2014.12.050
Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., Dohmae, N., et al. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313 (5788), 842–845. doi: 10.1126/science.1128436
Jarvis, B. A., Romsdahl, T. B., McGinn, M. G., Nazarenus, T. J., Cahoon, E. B., Chapman, K. D., et al. (2021). CRISPR/Cas9-Induced fad2 and rod1 Mutations Stacked With fae1 Confer High Oleic Acid Seed Oil in Pennycress (Thlaspi arvense L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.652319
Jun, J., Fiume, E., Roeder, A. H. K., Meng, L., Sharma, V. K., Osmont, K. S., et al. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154 (4), 1721–1736. doi: 10.1104/pp.110.163683
Kim, H.-J., Wu, C.-Y., Yu, H.-M., Sheen, J., Lee, H. (2017). Dual CLAVATA3 peptides in Arabidopsis shoot stem cell signaling. J. Plant Biol. 60 (5), 506–512. doi: 10.1007/s12374-017-0083-2
Klepikova, A. V., Kasianov, A. S., Gerasimov, E. S., Logacheva, M. D., Penin, A. A. (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88 (6), 1058–1070. doi: 10.1111/tpj.13312
Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., et al. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 (5788), 845–848. doi: 10.1126/science.1128439
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35 (21), 4453–4455. doi: 10.1093/bioinformatics/btz305
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res., 50, W276-279. doi: 10.1093/nar/gkac240
McGinn, M., Phippen, W. B., Chopra, R., Bansal, S., Jarvis, B. A., Phippen, M. E., et al. (2019). Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnol. J. 17 (4), 776–788. doi: 10.1111/pbi.13014
Moser, B. R., Shah, S. N., Winkler-Moser, J. K., Vaughn, S. F., Evangelista, R. L. (2009). Composition and physical properties of cress (Lepidium sativum L.) and field pennycress (Thlaspi arvense L.) oils. Ind. Crops Products 30 (2), 199–205. doi: 10.1016/j.indcrop.2009.03.007
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15 (3), 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nielsen, H., Tsirigos, K. D., Brunak, S., von Heijne, G. (2019). A brief history of protein sorting prediction. Protein J. 38 (3), 200–216. doi: 10.1007/s10930-019-09838-3
Nunn, A., Rodríguez-Arévalo, I., Tandukar, Z., Frels, K., Contreras-Garrido, A., Carbonell-Bejerano, P., et al. (2022). Chromosome-level Thlaspi arvense genome provides new tools for translational research and for a newly domesticated cash cover crop of the cooler climates. Plant Biotechnol. J. 20 (5), 944–963. doi: 10.1111/pbi.13775
Oelkers, K., Goffard, N., Weiller, G. F., Gresshoff, P. M., Mathesius, U., Frickey, T. (2008). Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8, 1. doi: 10.1186/1471-2229-8-1
Ohyama, K., Shinohara, H., Ogawa-Ohnishi, M., Matsubayashi, Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5 (8), 8. doi: 10.1038/nchembio.182
Phippen, W. B., Phippen, M. E. (2012). Soybean seed yield and quality as a response to field pennycress residue. Crop Sci. 52 (6), 2767–2773. doi: 10.2135/cropsci2012.03.0192
Phippen, W. B., Rhykerd, R., Sedbrook, J. C., Handel, C., Csonka, S. (2022). From farm to flight: coverCress as a low carbon intensity cash cover crop for sustainable aviation fuel production A review of progress towards commercialization. Front. Energy Res. 10. doi: 10.3389/fenrg.2022.793776
Qian, P., Song, W., Yokoo, T., Minobe, A., Wang, G., Ishida, T., et al. (2018). The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nat. Plants 4 (12), 12. doi: 10.1038/s41477-018-0317-4
Schlegel, J., Denay, G., Wink, R., Pinto, K. G., Stahl, Y., Schmid, J., et al. (2021). Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways. ELife 10, e70934. doi: 10.7554/eLife.70934
Sedbrook, J. C., Phippen, W. B., Marks, M. D. (2014). New approaches to facilitate rapid domestication of a wild plant to an oilseed crop: Example pennycress (Thlaspi arvense L.). Plant Sci. 227, 122–132. doi: 10.1016/j.plantsci.2014.07.008
Sharma, V. K., Ramirez, J., Fletcher, J. C. (2003). The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 51 (3), 415–425. doi: 10.1023/A:1022038932376
Shinohara, H., Matsubayashi, Y. (2013). Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54 (3), 369–374. doi: 10.1093/pcp/pcs174
Somssich, M., Je, B. I., Simon, R., Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143 (18), 3238–3248. doi: 10.1242/dev.133645
Song, X.-F., Hou, X.-L., Liu, C.-M. (2021). CLE peptides: Critical regulators for stem cell maintenance in plants. Planta 255 (1), 5. doi: 10.1007/s00425-021-03791-1
Stahl, Y., Wink, R. H., Ingram, G. C., Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19 (11), 909–914. doi: 10.1016/j.cub.2009.03.060
Strabala, T. J. (2008). CLE genes in plant development. Plant Signaling Behav. 3 (7), 457–459. doi: 10.4161/psb.3.7.5602
Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gíslason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40 (7), 7. doi: 10.1038/s41587-021-01156-3
Warwick, S. I., Francis, A., Susko, D. J. (2002). The biology of Canadian weeds. 9. Thlaspi arvense L. (updated). Can. J. Plant Sci. 82 (4), 803–823. doi: 10.4141/P01-159
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., Barton, G. J. (2009). Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25 (9), 1189–1191. doi: 10.1093/bioinformatics/btp033
Whitewoods, C. D., Cammarata, J., Nemec Venza, Z., Sang, S., Crook, A. D., Aoyama, T., et al. (2018). CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr. Biol. 28 (15), 2365–2376.e5. doi: 10.1016/j.cub.2018.05.068
Wu, Q., Xu, F., Jackson, D. (2018). All together now, a magical mystery tour of the maize shoot meristem. Curr. Opin. Plant Biol. 45, 26–35. doi: 10.1016/j.pbi.2018.04.010
Xie, M., Zhao, C., Song, M., Xiang, Y., Tong, C. (2022). Genome-wide identification and comparative analysis of CLE family in rapeseed and its diploid progenitors. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.998082
Xu, C., Liberatore, K. L., MacAlister, C. A., Huang, Z., Chu, Y.-H., Jiang, K., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47 (7), 7. doi: 10.1038/ng.3309
Keywords: pennycress, phylogenetics, expression, CLE, development, biofuel
Citation: Hagelthorn L and Fletcher JC (2023) The CLAVATA3/ESR-related peptide family in the biofuel crop pennycress. Front. Plant Sci. 14:1240342. doi: 10.3389/fpls.2023.1240342
Received: 14 June 2023; Accepted: 18 July 2023;
Published: 04 August 2023.
Edited by:
Li Lei, Berkeley Lab (DOE), United StatesReviewed by:
Xin Li, North Carolina State University, United StatesChanghao Li, Texas A and M University, United States
Haifei Hu, Guangdong Academy of Agricultural Sciences (GDAAS), China
Copyright © 2023 Hagelthorn and Fletcher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer C. Fletcher, amZsZXRjaGVyQGJlcmtlbGV5LmVkdQ==
 Lynne Hagelthorn
Lynne Hagelthorn Jennifer C. Fletcher
Jennifer C. Fletcher