- 1Department of Plant Biology, University of Georgia, Athens, GA, United States
- 2Department of Plant & Environmental Science, Clemson University, Florence, SC, United States
Introgression can produce novel genetic variation in organisms that hybridize. Sympatric species pairs in the carnivorous plant genus Sarracenia L. frequently hybridize, and all known hybrids are fertile. Despite being a desirable system for studying the evolutionary consequences of hybridization, the extent to which introgression occurs in the genus is limited to a few species in only two field sites. Previous phylogenomic analysis of Sarracenia estimated a highly resolved species tree from 199 nuclear genes, but revealed a plastid genome that is highly discordant with the species tree. Such cytonuclear discordance could be caused by chloroplast introgression (i.e. chloroplast capture) or incomplete lineage sorting (ILS). To better understand the extent to which introgression is occurring in Sarracenia, the chloroplast capture and ILS hypotheses were formally evaluated. Plastomes were assembled de-novo from sequencing reads generated from 17 individuals in addition to reads obtained from the previous study. Assemblies of 14 whole plastomes were generated and annotated, and the remaining fragmented assemblies were scaffolded to these whole-plastome assemblies. Coding sequence from 79 homologous genes were aligned and concatenated for maximum-likelihood phylogeny estimation. The plastome tree is extremely discordant with the published species tree. Plastome trees were simulated under the coalescent and tree distance from the species tree was calculated to generate a null distribution of discordance that is expected under ILS alone. A t-test rejected the null hypothesis that ILS could cause the level of discordance seen in the plastome tree, suggesting that chloroplast capture must be invoked to explain the discordance. Due to the extreme level of discordance in the plastome tree, it is likely that chloroplast capture has been common in the evolutionary history of Sarracenia.
1 Introduction
Evolutionary biologists have long been interested in hybridization as a process that generates biodiversity. Hybridization leading to introgression introduces genetic information to a species, which increases genetic variation for selection to act on and provides opportunity for adaptive evolution (Pease et al., 2016; Grant and Grant, 2019; Meier et al., 2019). Organisms that readily hybridize may be subject to these evolutionary forces. However, the formation of hybrids does not imply that introgression (transfer of genome segments between hybridizing species) is occurring, as hybrids must reproduce with the parental population and introgressed alleles must survive in the face of natural selection and genetic drift. Identifying the extent to which hybridizing taxa are exchanging genetic material sheds light on the processes that generate and maintain variation within them.
Sarracenia L. is a genus of 8-11 species of carnivorous plants native to North America. It is one of the three extant genera in the family Sarraceniaceae, with species forming tube shaped traps adapted to catch and digest insects. Due to this unique adaptation they are commonly called pitcher plants, although pitcher-shaped carnivorous leaves have evolved convergently in at least two other lineages (Nepenthes L., Cephalotus Labill.) (Albert et al., 1992). Most Sarracenia species occur sympatrically with at least one other species, and all species pairs can produce fertile hybrids (Bell, 1952). Hybrids between sympatric species are frequently observed in nature (Bell, 1952), and population genetics studies using a few microsatellite loci have shown evidence of gene-flow between species at some sites and not at others (Furches et al., 2013; Rentsch and Holland, 2020). The forces maintaining species boundaries are not well known, but it is possible that outbreeding depression is contributing to species coherence in the face of hybridization. Sarracenia hybrids exhibit intermediate pitcher morphology which may decrease prey capture efficacy. Another possible factor contributing to the maintenance of species boundaries is asynchronous flowering phenology (Bell, 1952).
Sarracenia diverged from the rest of Sarraceniaceae an estimated 23 MYA, with most of the diversification within Sarracenia occurring between 1-3 MYA (Ellison et al., 2012). Given the rapid speciation, significant gene tree discordance is expected due to incomplete lineage sorting (ILS) (Degnan and Rosenberg, 2009). Despite this, Stephens et al. (2015) estimated a multi-species coalescent phylogeny using 199 nuclear genes that resolved most of the species relationships with high support. This study also presented a plastome tree that was highly discordant with the nuclear tree; no species was reciprocally monophyletic. Cytonuclear discordance such as this can be the result of ILS or introgression of the plastid genome, otherwise referred to as chloroplast capture.
Although the plastome phylogeny estimated in Stephens et al. (2015) is relatively well supported, the analysis was limited by the recovery of only 42kbp of plastome sequence limited to the long single copy and short single copy regions of the plastome. To confirm that the extreme cytonuclear discordance observed in the Stephens et al. (2015) phylogenies was not an artifact of a lack of data, we reassembled plastomes from those sequencing reads using an alternative assembly pipeline to recover more sequence. Seventeen additional accessions are added to this analysis. The cause of cytonuclear discordance is formally assessed using a coalescent based simulation approach to distinguish between ILS and chloroplast capture. Additionally, whole plastomes are assembled and gene content evolution is assessed within the context of carnivory.
2 Materials and methods
2.1 Sequence data
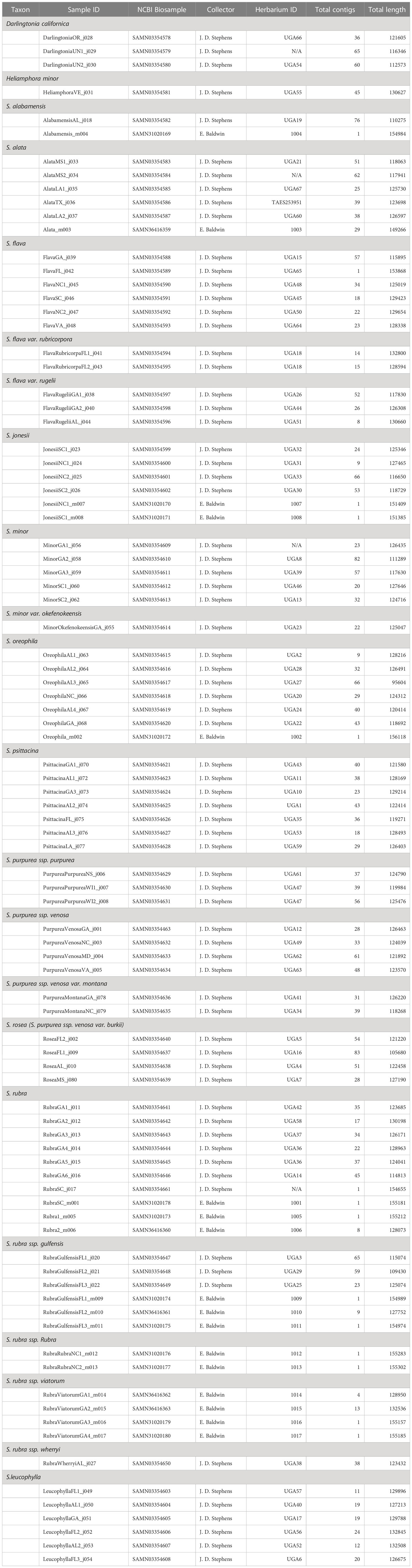
Leaf tissue was obtained from 17 individuals in total: 11 accessions were obtained from the Atlanta Botanical Garden’s living conservation collection (S. oreophila, S. jonesii, S. alata, S. alabamensis and S. rubra) and six accessions were obtained from two field sites (S. rubra subsp. rubra and S. rubra subsp. viatorum). DNA was extracted from silica dried samples using the Qiagen DNeasy Plant Mini Kit. Library prep was performed using the Kapa Biosystems HyperPlus Kit using iTru adapters (Glenn et al., 2019). Libraries were pooled at equal concentrations and enriched for putative single-copy orthologs enrichment using the Angiosperms353 bait set (Johnson et al., 2019). The enriched pool was sequenced on an Illumina NextSeq 500 at the Georgia Genomics and Bioinformatics Core using a High Output 300 cycle flow cell generating 150bp paired-end reads.
In addition, sequencing reads from Stephens et al. (2015) were downloaded from NCBI Short Read Archive. The Stephens et al. data set includes 71 accessions of Sarracenia and 4 accessions of outgroups in Sarraceniaceae (Heliamphora minor and Darlingtonia californica).
2.2 Plastome assembly
All raw reads were trimmed using Trimmomatic (v. 0.39) (Bolger et al., 2014). Both the new data set and the data set obtained from Stephens et al. were sequenced from libraries enriched for targeted nuclear loci. However, the majority of the reads from both data sets are off-target. Stephens et al. reported an average of 1.6% of reads on target, and analysis of the new data set revealed that less than 1% of the reads were on target. The large proportion of off-target reads enable the assembly of the plastome.
Initial de-novo plastome assembly was attempted with GetOrganelle (v. 1.7.5.2) (Jin et al., 2020). GetOrganelle often produced two assembly versions differing only in the orientation of the short single copy regions (SSC). SSC orientation was determined by aligning assemblies to the reference plastome (Clethra L. delavayi, Genbank accession NC_041129) using MUMmer (v. 4.0.0) (Kurtz et al., 2004), and only the assemblies with concordant SSC orientation were retained.
GetOrganelle did not generate complete de-novo plastome assemblies from every sample. In these cases, the following reference-based pipeline was used. Reads were aligned to one of the complete Sarracenia plastome assemblies using BWA (v. 0.7.17) (Li et al., 2009). The aligned reads were then extracted and assembled de-novo using SPAdes (Bankevich et al., 2012). Afin (https://github.com/afinit/afin) was used to extend the resulting contigs and fuse any contigs with significant overlap. At this stage, assemblies were either mostly complete (1-3 contigs consisting of the large single copy region (LSC), short single copy regions (SSC), and one IR), or they were more fragmented. The mostly complete assemblies were manually pasted together. The IR boundaries were verified by mapping reads to the assemblies and identifying the coordinate where half of the reads spanned the IR and LSC and the other half spanned the IR and SSC.
2.3 Plastome annotation
Complete plastome assemblies were annotated using PGA (Qu et al., 2019). Fragmented assemblies were aligned to one of the complete, PGA annotated plastomes using the Minimap2 (v. 2.17) (Li, 2018) plugin in Geneious. The “transfer annotation” function was used before generating a consensus sequence.
2.4 Alignment and phylogeny estimation
Coding sequences (CDS) from 79 plastid genes were extracted from the annotated assemblies and aligned with MAFFT (v. 7.470) (Katoh and Standley, 2013). All resulting gene alignments were concatenated. Regions of the concatenated alignment that were poorly aligned or had gaps in 50% or more of the samples were filtered out of the gene alignments using Gblocks (v. 0.91b) (Castresana, 2000). A maximum-likelihood phylogeny was estimated from the concatenated gene alignments using IQ-Tree (v. 2.0.6) (Nguyen et al., 2015). 1000 bootstrap replicates were performed using UFBoot (Minh et al., 2013). The GTR + F + R4 substitution model was used.
2.5 Plastome tree simulations
To differentiate between incomplete lineage sorting (ILS) and chloroplast capture, a tree simulation approach similar to Folk et al., 2017 (Folk et al., 2016) was used. Plastome trees under ILS were simulated using the dendropy python package (v. 4.5.2) (Sukumaran and Holder, 2010) with the species tree from Stephens et al. (2015) as a guide tree. Since plastomes are effectively haploid and inherited uniparentally, plastomes have one quarter of the effective population size of diploid nuclear loci. Since the guide tree used for these simulations was estimated exclusively using nuclear loci, its branch lengths were scaled by four to account for the effective population size differential between plastomes and nuclear loci. A distribution of tree discordance under the null hypothesis of ILS was generated by calculating a tree distance metric [information-based generalized Robinson-Foulds distance (Smith, 2020)] between 1000 simulated trees and the species tree. Then the distance between the empirical plastome tree from this study and the species tree was calculated and compared to the null distribution. Since the empirical plastome tree has samples that are not in the Stephens et al. (2015) species tree, those tips were dropped from the plastome tree to enable calculating distance.
3 Results
3.1 Plastome assemblies
Fourteen complete, circularized plastomes have been assembled and annotated including the following Sarracenia species: S. jonesii, S. alabamensis, S. oreophila, S. rubra subsp. gulfensis, S. rubra subsp. rubra, and S. rubra subsp. viatorum. Average assembly statistics for the all assemblies are shown in Table 1. The assembly pipeline for fragmented assemblies recovered an average of 114kbp of plastome sequence, almost tripling the 42kbp recovered in Stephens et al. (2015). The use of different references is one potential factor explaining this difference; this study used a complete Sarracenia plastome (Ericales) as a reference whereas Stephens et al. (2015) used a plastome from Vitis vinifera (Vitales). Eighty protein-coding genes were extracted from assemblies, and sequences were aligned for all samples, and alignments were concatenated for the phylogeny estimation.
3.2 Pseudogenization of plastome encoded genes
All complete Sarracenia plastomes include some pseudogenized plastome-encoded genes. With the exception of ndhB and ndhE, all ndh genes either have been pseudogenized due to premature stop codons or large deletions (Figure 1). Similarly, all samples contain a premature stop codon within the rps12 gene.
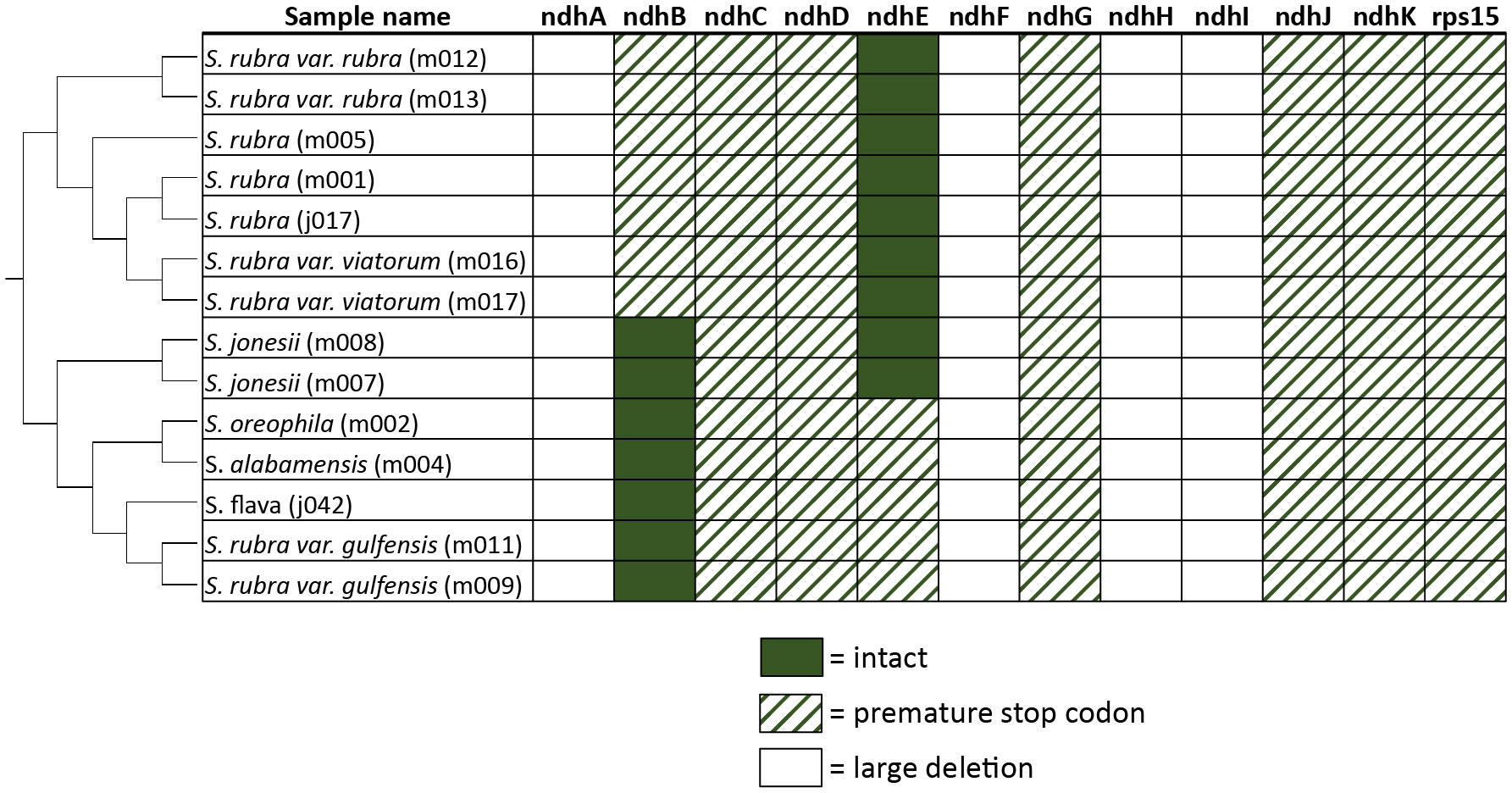
Figure 1 Status of ndh genes in all complete plastome assemblies. Filled cells represent intact genes, dashed cells represent premature stop codons, and blank cells represent genes with large deletions. Plastome tree trimmed from these samples is shown on the left.
3.3 Plastid phylogeny
Consistent with Stephens et al. (2015), no species were found to exhibit monophyly of their plastomes, and the plastid tree is highly incongruent with the published species tree (Figure 2). Support values across the backbone of the tree are all greater than 70, and most internal nodes are highly supported as well (Figure 2). Branch lengths within Sarracenia are generally very short in comparison to the outgroups. An exception is the split at the base of the Sarracenia clade. This branch splits Sarracenia into two distinct plastid lineages. These main lineages are arbitrarily termed clade A and clade B (Figure 2). Clade B contains all sampled individuals of minor, oreophila, jonesii, and purpurea var. montana, and clade A contains all sampled individuals of alata and purpurea (excluding var. montana). All other species are split across these two main lineages (flava, psittacina, rubra, and leucophylla).
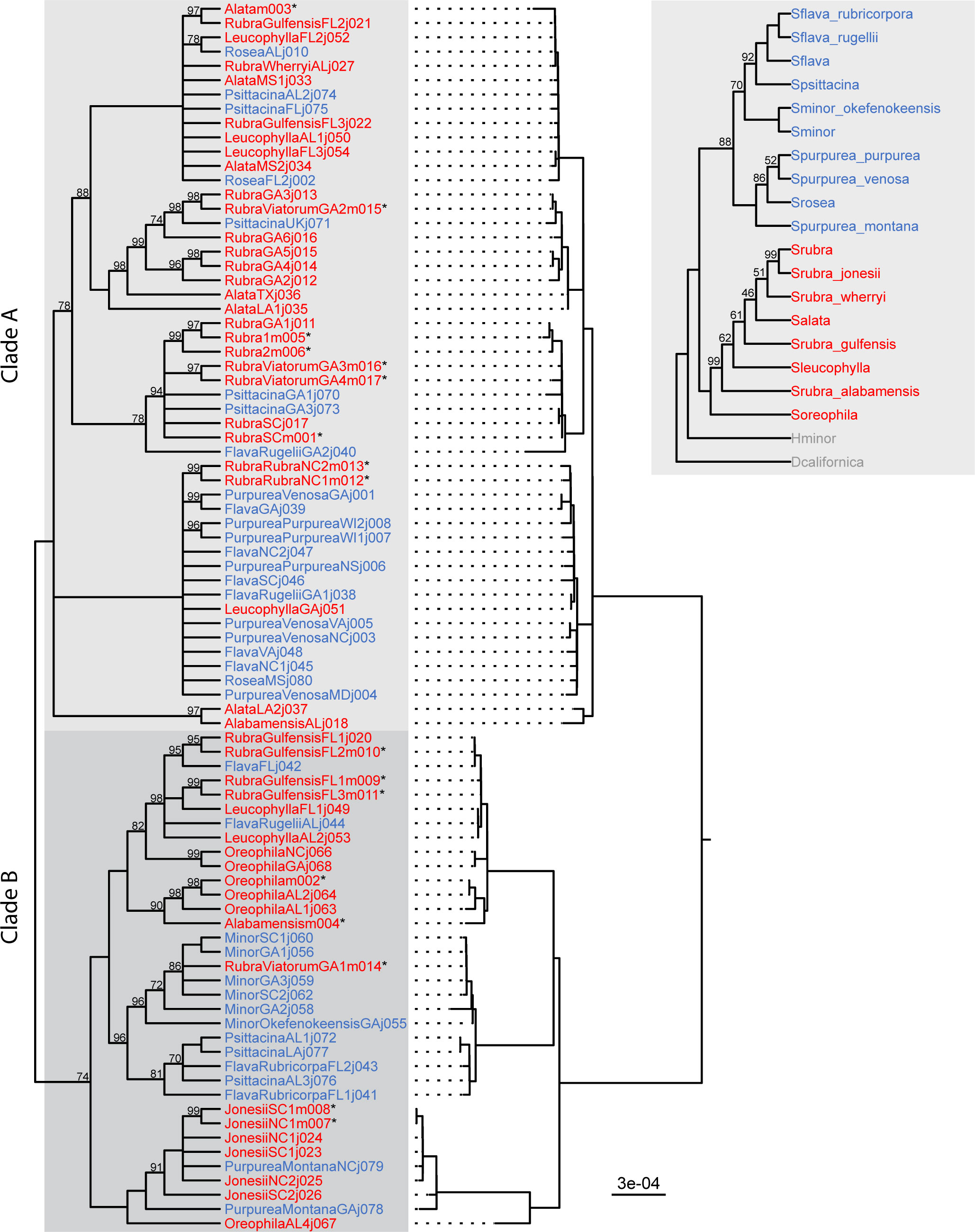
Figure 2 Maximum likelihood plastome cladogram (left) and phylogram (center) and species tree (inset, top right) from Stephens et al. (2015). Nodes on the cladogram with bootstrap values less than 70 are collapsed. Uncollapsed cladogram nodes with bootstrap values less than 100 are labelled. Tip names are either red or blue based on which of the two major clades the species belongs to in the species tree. Asterisks next to tip labels indicate samples that were newly sequenced for this study.
3.3.1 Southern Appalachian species
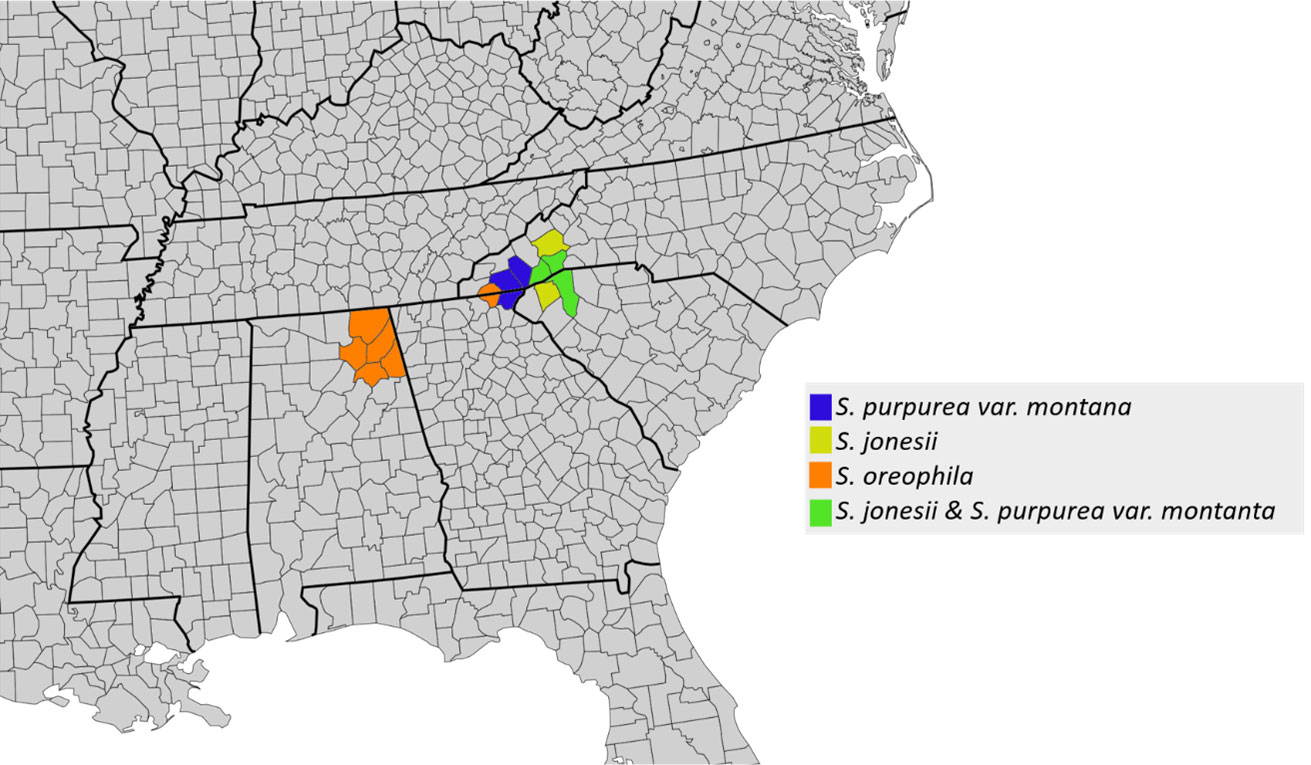
S. purpurea var. montana and S. jonesii form a clade. Both taxa have distributions restricted to a small area in the southern Appalachian Mountains (Figure 3) and hybridize at sympatric sites. The only other species found in the southern Appalachians is S. oreophila, although it is not sympatric with S. jonesii or S. purpurea var. montana, but may have been historically (McPherson and Schnell, 2011). Two S. oreophila accessions from Alabama are sister to the Appalachian clade, and the other S. oreophila accessions are placed in a clade sister to this.
3.3.2 Sarracenia flava, S. minor, and S. psittacina
S. flava, S. minor, and S. psittacina form a clade sister to S. purpurea on the species tree, however the placement of these species on the plastid tree is not congruent. All S. minor accessions are placed within clade B sister to the clade containing S. oreophila, S. jonesii, and S. purpurea var. montana. Some S. flava and S. psittacina accessions from the Gulf coastal plain are also placed in the S. minor clade, despite all S. minor accessions in this study originating from the Atlantic coastal plain. This could indicate either ancient introgression or retention of plastome diversity from the ancestor of these three species. S. flava and S. psittacina are scattered across the chloroplast phylogeny; both species have accessions found in clades A and B. In S. flava, all Gulf coastal plain accessions are found in clade B and all Atlantic coastal plain accessions are found in clade A.
3.3.3 Sarracenia purpurea complex
With the exception of S. purpurea var. montana, all S. purpurea accessions (including S. rosea) are placed in clade A. There is no discernible pattern to their placement within this lineage. This is surprising given the vast geographic range represented by these taxa; the individuals sampled for this study originate from throughout their distribution from Mississippi to Nova Scotia. Only S. purpurea subsp. purpurea is found north of Maryland, so the relatedness of plastomes between this taxon and other species are unlikely to be the result of recent introgression.
3.4 Plastome phylogeny simulations
The tree distance metric that was used ranges from 0 (an identical tree) to 1 (the most distal tree). The plastome trees simulated under the pure coalescent model have distances from the species tree ranging from 0.29 to 0.56, while the distance from the empirical plastome tree is 0.73 (Figure 4). A T-test using the distribution of simulated plastome tree distances as the null distribution gives a p-value of >2.2e-16, rejecting the null hypothesis of ILS causing the discordance alone.
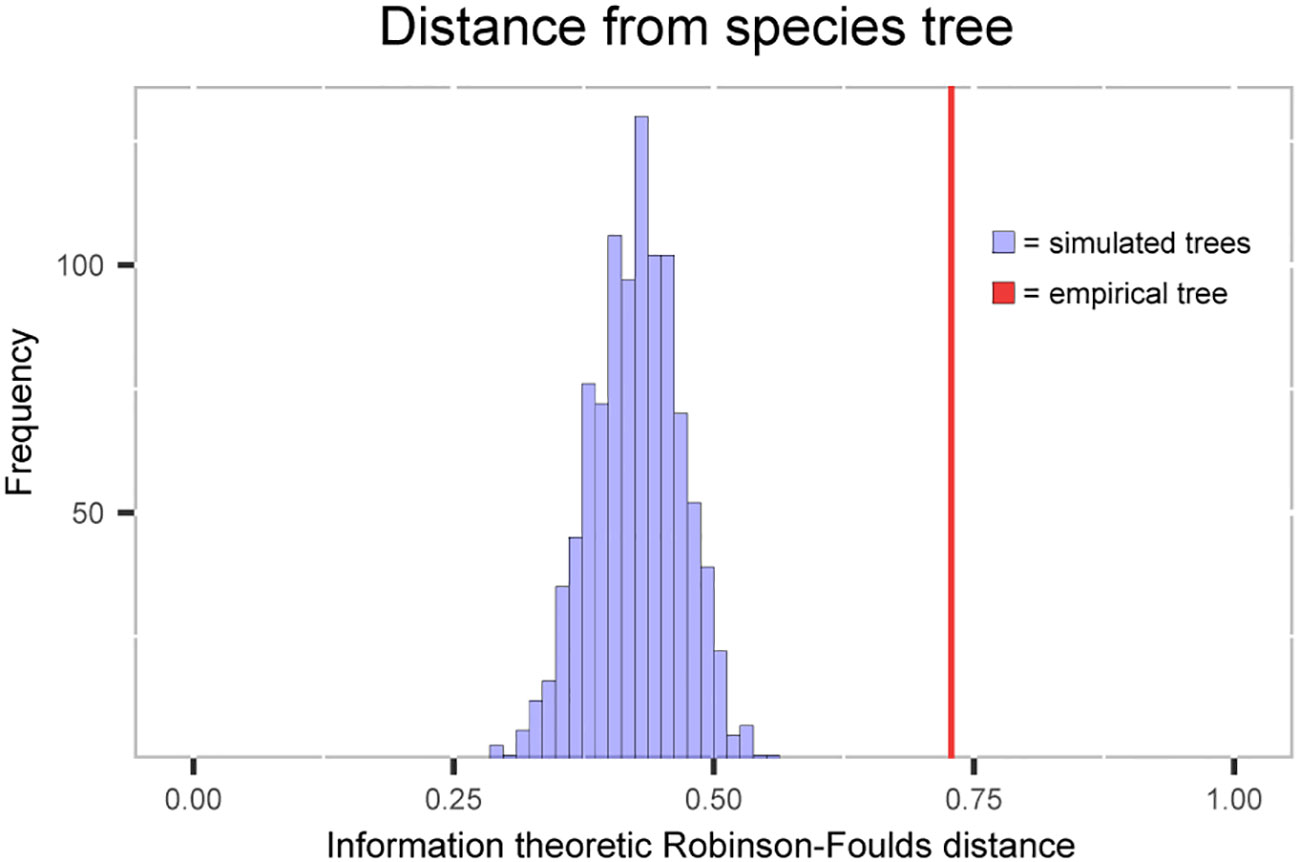
Figure 4 Histogram of information-based generalized Robinson-Foulds distance between the simulated plastome trees and the species tree. Red line shows the distance between empirically estimated plastome tree and the species tree.
4 Discussion
4.1 Pseudogenization of ndh genes
Independent pseudogenization or complete loss of ndh genes has been shown in many plant lineages, including holoparasitic, hemiparastitic, and carnivorous plant lineages (Barrett et al., 2014; Lin et al., 2017; Cao et al., 2019; Gruzdev et al., 2019; Nevill et al., 2019). Functional ndh genes are rarely found in non-photosynthetic parasitic plants, and the loss of ndh genes is strongly correlated with the transition to heterotrophy in parasitic plant lineages (Wicke et al., 2016). Since plastid encoded ndh genes are thought to optimize photosynthetic chemistry in fluctuating or stressful environments [reviewed in (Sabater, 2021)], the loss of ndh genes in parasitic lineages that are no longer fully dependent on photosynthesis as a source of carbon is unsurprising. In carnivorous plants, however, evidence for significant heterotrophic uptake of carbon is limited (Rischer et al., 2002), and a transition to full heterotrophy seems unlikely, so this line of reasoning does not explain the independent pseudogenization of functional ndh genes across carnivorous plant lineages. It is possible that the acquisition of organic nitrogen has an interaction with photosynthetic chemistry that relaxes the need for ndh. As Nevill et al. (2019) noted, organic nitrogen acquisition bypasses the need to assimilate nitrate using photosynthetically-derived reductant. Alternatively, the pseudogenization of ndh genes in parasitic plants and carnivorous plants could be due to unrelated mechanisms. The pseudogenization of almost all of the ndh genes across the genus Sarracenia shown here provides further evidence that carnivorous plants do not require these genes. Sequencing of full plastomes from other carnivorous species would reveal if the pseudogenization of ndh occurs early in carnivorous plant evolution.
4.2 Cytonuclear discordance
The plastome phylogeny in this study shows a similarly extreme level of discordance with the species tree as that of the Stephens et al. (2015) plastome phylogeny. That study ascribed the discordance to a combination of chloroplast capture and a lack of informative polymorphisms in the chloroplast sequence. A third source of discordance, ILS, is considered here. A lack of informative polymorphisms is not an issue here, as almost all the plastome coding sequences are used and the resulting phylogeny has high bootstrap values across the spine, suggesting that there is sufficient evidence that major clades within the tree are correct.
To distinguish between the two remaining sources of discordance, plastome phylogenies under ILS were simulated. The simulated phylogenies showed much lower levels of discordance with the species tree than the empirically estimated plastome. To simulate the plastome phylogenies, the branch lengths of the guide tree were multiplied by four due to the assumption that the chloroplast is inherited matrilineally in Sarracenia like most seed plants (Mogensen, 1996). Since this assumption hasn’t been empirically proven and biparental inheritance of the chloroplast is possible, simulations with branch lengths multiplied by two were performed and show similar results (Supplemental Data).
There is ample signal of introgression in the plastome, but Stephens et al. (2015) reported no evidence of gene flow in the nuclear data. A search through the nuclear gene trees revealed that none of the trees had a similar topology to the plastome tree, but some trees did exhibit a high degree of discordance with the species tree, possibly due to occasional nuclear gene introgression. Cytonuclear discordance is commonly observed and is attributed to introgression in plant and animal systems (Rieseberg and Soltis, 1991; Berthier et al., 2006; Gernandt et al., 2018), including several instances where there is limited signal for introgression in nuclear data (Winkler et al., 2013; Good et al., 2015; Folk et al., 2016; Rose et al., 2020). However, the mechanism for organellar introgressions without accompanying nuclear loci is poorly understood (Rieseberg and Soltis, 1991; Folk et al., 2018). Sarracenia is a genus where hybridization is common and thus some level of nuclear introgression might be expected. The extreme level of chloroplast capture and lack of signal for nuclear gene flow in Sarracenia illustrates the comparative ease of introgression of organelles over nuclear loci.
4.2.1 Geographic patterns of plastome introgression
Although the lack of monophyletic species in the plastome tree makes it difficult to interpret specific instances of plastome introgression, a handful of such instances can be elucidated using geographic context. For example, all accessions of S. purpurea var. montana and S. jonesii, two taxa restricted to a small region in the southern Appalachians, form a well-supported clade within clade B. Given that all other S. purpurea accessions are placed in clade A, it is likely that a plastome derived from S. jonesii was introgressed into S. purpurea var. montana. Similarly, an accession of S. rubra that was sampled from the Georgia fall line near S. minor populations is placed within the S. minor clade. Again, we hypothesize this to be an instance of S. minor plastome being introgressed into S. rubra. More generally, the weak species clustering in the plastome tree implies a long history of interspecific exchange of cytoplasmic genomes in Sarracenia.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/; PRJNA884359.
Author contributions
EB, MM, JL-M contributed to study design. EB performed all data analysis and wrote initial manuscript draft. MM collected samples and generated data. MM, JL-M contributed to refinement of manuscript and approved final submission. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by an National Science Foundation grant (DEB 2110875) to JL-M.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1237749/full#supplementary-material
Supplementary Figure 1 | png - Histograms of information-based generalized Robinson-Foulds distance between the simulated plastome trees and the species tree. Panel titles indicate how the branches of the guide tree were scaled when plastome trees were simulated. Red line shows the distance between empirically estimated plastome tree and the species tree.
alignment.fasta
Concatenated gene alignments used to estimate plastome tree.
Plastome.newick
Plastome tree in newick format.
Simtrees.1.newick
Plastome trees simulated with unscaled guide tree.
Simtrees.2.newick
Plastome trees simulated with guide tree branch lengths scaled by two.
Simtrees.4.newick
Plastome trees simulated with guide tree branch lengths scaled by four.
Fragmented_assemblies.zip
Zipped folder containing all fragmented assemblies in fasta format.
References
Albert, V. A., Williams, S. E., Chase, M. W. (1992). Carnivorous plants: phylogeny and structural evolution. Science 257 (5076), 1491–1495. doi: 10.1126/science.1523408
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. doi: 10.1089/cmb.2012.0021
Barrett, C. F., Freudenstein, J. V., Li, J., Mayfield-Jones, D. R., Perez, L., Pires, J. C., et al. (2014). Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31 (12), 3095–3112. doi: 10.1093/molbev/msu252
Bell, C. R. (1952). Natural hybrids in the genus sarracenia: I. Hist. Distribut. Taxonomy. J. Elisha Mitchell Sci. Soc. 68 (1), 55–80. Available at: https://www.jstor.org/stable/24334305.
Berthier, P., Excoffier, L., Ruedi, M. (2006). Recurrent replacement of mtDNA and cryptic hybridization between two sibling bat species Myotis myotis and Myotis blythii. Proc. R. Soc. B: Biol. Sci. 273 (1605), 3101–3123. doi: 10.1098/rspb.2006.3680
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Cao, M., Li, Z., Dai, X., Wu, X., Li, Y., Wu, S. (2019). The complete plastid genome of carnivorous pitcher plant Cephalotus follicularis. Mitochondrial DNA Part B. 4 (1), 2025–2027. doi: 10.1080/23802359.2019.1617054
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 (4), 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Degnan, J. H., Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24 (6), 332–340. doi: 10.1016/j.tree.2009.01.009
Ellison, A. M., Butler, E. D., Hicks, E. J., Naczi, R. F. C., Calie, P. J., Bell, C. D., et al. (2012). Phylogeny and biogeography of the carnivorous plant family Sarraceniaceae. PloS One 7 (6), e39291–e3929e. doi: 10.1371/journal.pone.0039291
Folk, R. A., Mandel, J. R., Freudenstein, J. V. (2016). Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Systemat. Biol. 66 (3), 320–337. doi: 10.1093/sysbio/syw083
Folk, R. A., Soltis, P. S., Soltis, D. E., Guralnick, R. (2018). New prospects in the detection and comparative analysis of hybridization in the tree of life. Am. J. Botany. 105 (3), 364–375. doi: 10.1002/ajb2.1018
Furches, M. S., Small, R. L., Furches, A. (2013). Hybridization leads to interspecific gene flow in Sarracenia (Sarraceniaceae). Am. J. Bot. 100 (10), 2085–2091. doi: 10.3732/ajb.1300038
Gernandt, D. S., Aguirre Dugua, X., Vázquez-Lobo, A., Willyard, A., Moreno Letelier, A., Pérez de la Rosa, J. A., et al. (2018). Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. Am. J. Bot. 105 (4), 711–725. doi: 10.1002/ajb2.1052
Glenn, T. C., Nilsen, R. A., Kieran, T. J., Sanders, J. G., Bayona-Vásquez, N. J., Finger, J. W., et al. (2019). Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ 7, e7755. doi: 10.7717/peerj.7755
Good, J. M., Vanderpool, D., Keeble, S., Bi, K. (2015). Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks. Evolution 69 (8), 1961–1972. doi: 10.1111/evo.12712
Grant, P. R., Grant, B. R. (2019). Hybridization increases population variation during adaptive radiation. Proc. Natl. Acad. Sci. 116 (46), 23216–23224. doi: 10.1073/pnas.1913534116
Gruzdev, E. V., Kadnikov, V. V., Beletsky, A. V., Kochieva, E. Z., Mardanov, A. V., Skryabin, K. G., et al. (2019). Plastid Genomes of Carnivorous Plants Drosera rotundifolia and Nepenthes × ventrata Reveal Evolutionary Patterns Resembling Those Observed in Parasitic Plants. Int. J. Mol. Sci. 20 (17), 4107. doi: 10.3390/ijms20174107
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., dePamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21 (1), 241. doi: 10.1186/s13059-020-02154-5
Johnson, M. G., Pokorny, L., Dodsworth, S., Botigué, L. R., Cowan, R. S., Devault, A., et al. (2019). A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol. 68 (4), 594–606. doi: 10.1093/sysbio/syy086
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5 (2), R12. doi: 10.1186/gb-2004-5-2-r12
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34 (18), 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi: 10.1093/bioinformatics/btp352
Lin, C.-S., Chen, J. J. W., Chiu, C.-C., Hsiao, H. C. W., Yang, C.-J., Jin, X.-H., et al. (2017). Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90 (5), 994–1006. doi: 10.1111/tpj.13525
McPherson, S., Schnell, D. E. (2011). Sarraceniaceae of North America: redfern natural history productions.
Meier, J. I., Stelkens, R. B., Joyce, D. A., Mwaiko, S., Phiri, N., Schliewen, U. K., et al. (2019). The coincidence of ecological opportunity with hybridization explains rapid adaptive radiation in Lake Mweru cichlid fishes. Nat. Commun. 10 (1), 5391. doi: 10.1038/s41467-019-13278-z
Minh, B. Q., Nguyen, M. A., von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30 (5), 1188–1195. doi: 10.1093/molbev/mst024
Mogensen, H. L. (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Botany. 83 (3), 383–404. doi: 10.1002/j.1537-2197.1996.tb12718.x
Nevill, P. G., Howell, K. A., Cross, A. T., Williams, A. V., Zhong, X., Tonti-Filippini, J., et al. (2019). Plastome-wide rearrangements and gene losses in carnivorous droseraceae. Genome Biol. Evol. 11 (2), 472–485. doi: 10.1093/gbe/evz005
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Pease, J. B., Haak, D. C., Hahn, M. W., Moyle, L. C. (2016). Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PloS Biol. 14 (2), e1002379. doi: 10.1371/journal.pbio.1002379
Qu, X. J., Moore, M. J., Li, D. Z., Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 50. doi: 10.1186/s13007-019-0435-7
Rentsch, J. D., Holland, R. C. (2020). Population Genetic Structure and Natural Establishment of Hybrids Between Sarracenia flava and Sarracenia minor in Francis Marion National Forest. Castanea 85 (1), 108–21, 14. doi: 10.2179/0008-7475.85.1.108
Rieseberg, L. H., Soltis, D. (1991). Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends Plants 5, 65–84. Available at: https://www.researchgate.net/publication/262005952_Phylogenetic_consequences_of_cytoplasmic_gene_flow_in_plants.
Rischer, H., Hamm, A., Bringmann, G. (2002). Nepenthes insignis uses a C2-portion of the carbon skeleton of l-alanine acquired via its carnivorous organs, to build up the allelochemical plumbagin. Phytochemistry 59 (6), 603–609. doi: 10.1016/S0031-9422(02)00003-1
Rose, J. P., Toledo, C. A. P., Lemmon, E. M., Lemmon, A. R., Sytsma, K. J. (2020). Out of sight, out of mind: widespread nuclear and plastid-nuclear discordance in the flowering plant genus polemonium (Polemoniaceae) suggests widespread historical gene flow despite limited nuclear signal. Systemat. Biol. 70 (1), 162–180. doi: 10.1093/sysbio/syaa049
Sabater, B. (2021). On the edge of dispensability, the chloroplast ndh genes. Int. J. Mol. Sci. 22 (22), 12505. doi: 10.3390/ijms222212505
Smith, M. R. (2020). Information theoretic generalized Robinson–Foulds metrics for comparing phylogenetic trees. Bioinformatics 36 (20), 5007–5013. doi: 10.1093/bioinformatics/btaa614
Stephens, J. D., Rogers, W. L., Heyduk, K., Cruse-Sanders, J. M., Determann, R. O., Glenn, T. C., et al. (2015). Resolving phylogenetic relationships of the recently radiated carnivorous plant genus Sarracenia using target enrichment. Mol. Phylogenet. Evol. 85, 76–87. doi: 10.1016/j.ympev.2015.01.015
Sukumaran, J., Holder, M. T. (2010). DendroPy: a Python library for phylogenetic computing. Bioinformatics 26 (12), 1569–1571. doi: 10.1093/bioinformatics/btq228
Wicke, S., Müller, K. F., dePamphilis, C. W., Quandt, D., Bellot, S., Schneeweiss, G. M. (2016). Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. U. S. A. 113 (32), 9045–9050. doi: 10.1073/pnas.1607576113
Winkler, M., Tribsch, A., Schneeweiss, G. M., Brodbeck, S., Gugerli, F., Holderegger, R., et al. (2013). Strong nuclear differentiation contrasts with widespread sharing of plastid DNA haplotypes across taxa in European purple saxifrages (Saxifraga section Porphyrion subsection Oppositifoliae). Botan. J. Linn. Soc. 173 (4), 622–636. doi: 10.1111/boj.12104
Keywords: hybridization, chloroplast capture, gene flow, carnivorous plant, Sarracenia, phylogenomics, plastome
Citation: Baldwin E, McNair M and Leebens-Mack J (2023) Rampant chloroplast capture in Sarracenia revealed by plastome phylogeny. Front. Plant Sci. 14:1237749. doi: 10.3389/fpls.2023.1237749
Received: 09 June 2023; Accepted: 20 July 2023;
Published: 14 August 2023.
Edited by:
Jeremie Benjamin Fant, Chicago Botanic Garden, United StatesReviewed by:
Michael J. Moore, Oberlin College, United StatesJosué Barrera-Redondo, Max Planck Society, Germany
Copyright © 2023 Baldwin, McNair and Leebens-Mack. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ethan Baldwin, ZXRoYW4uYmFsZHdpbkB1Z2EuZWR1
 Ethan Baldwin
Ethan Baldwin Mason McNair2
Mason McNair2 Jim Leebens-Mack
Jim Leebens-Mack