
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 October 2023
Sec. Plant Systematics and Evolution
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1234148
This article is part of the Research TopicMechanisms Underlying Phenotypic Convergence in PlantsView all 5 articles
East Asia is the richest region of plant biodiversity in the northern temperate zone, and its radiation provides key insights for understanding rapid speciation, including evolutionary patterns and processes. However, it is challenging to investigate the recent evolutionary radiation among plants because of the lack of genetic divergence, phenotypic convergence, and interspecific gene flow. Epimedium sect. Diphyllon is a rarely studied plant lineage endemic to East Asia, especially highly diversified in its southern part. In this study, we report a robust phylogenomic analysis based on genotyping-by-sequencing data of this lineage. The results revealed a clear biogeographic pattern for Epimedium sect. Diphyllon with recognition into two major clades corresponding to the Sino–Himalayan and Sino–Japanese subkingdoms of East Asian Flora and rapid diversification of the extant species dated to the Pleistocene. Evolutionary radiation of Epimedium sect. Diphyllon is characterized by recent and predominant parallel evolution and atavism between the two subkingdom regions, with extensive reticulating hybridization within each region during the course of diversification in southern East Asia. A parallel-atavism-introgression hypothesis is referred to in explaining the radiation of plant diversity in southern East Asia, which represents a potential model for the rapid diversification of plants under global climate cooling in the late Tertiary. Our study advances our understanding of the evolutionary processes of plant radiation in East Asia as well as in other biodiversity hotspot regions.
East Asia (EA) is the richest region in the Northern Hemisphere in terms of the biogeographic diversification of flowering plants (Takhtajan, 1986; Wu and Wu, 1998) and has been considered a massive sanctuary for Tertiary relict flora and associated forest plants during the climatic fluctuations over the Neogene (Tiffney, 1985; Liu, 1988; Wen, 1999). EA has been a natural floristic region since Grisebach first discussed it in 1872 (Takhtajan, 1986; Wu, 1988), well known for its abundant endemic elements from more than 31 families and 600 genera of flowering plants (Good, 1964; Takhtajan, 1969; Takhtajan, 1986; Wu and Wu, 1998; Qian, 2002). The high species richness in EA can be attributed to secondary diversification due to habitat heterogeneity and a lower extinction rate in the Neogene, whereas the lower species diversity in North America and Europe is often explained by the hypothesis that more severe extinctions occurred as a consequence of global cooling in the late Eocene to Oligocene (Xiang et al., 2004; Wen et al., 2010). It has also been suggested as the ancestral area for most EA–North American disjunct lineages (Wen, 1999; Milne and Abbott, 2002). Biogeographic studies on flowering plants in the Northern Hemisphere have largely focused on the classic EA and North American disjunctions; however, few studies have explored the morphological and evolutionary patterns and mechanisms to explain the radiation and high diversity of EA flora (Wen, 1999; Donoghue et al., 2001).
Evolutionary radiation, a widely recognized mode of rapid species diversification, is a fascinating phenomenon for biologists. Recently, the mechanisms underlying evolutionary radiation have been extensively studied, and several lineages of organisms have been characterized by rapid evolutionary radiation, such as Darwin’s finches (Grant and Grant, 2002; Lamichhaney et al., 2015), African lake cichlids (Seehausen, 2006; Wagner et al., 2012) and representatives of Andean flora (Pennington et al., 2010; Hughes and Atchison, 2015). Introgression and hybridization are common in plant evolution, especially in radiation lineages (Ellstrand and Schierenbeck, 2000; Linder and Rieseberg, 2004; Vriesendorp and Bakker, 2005; Soltis and Soltis, 2009; Harrison and Larson, 2014; Vargas et al., 2017). Hybridization and introgression often lead to chromosomal rearrangements, whole-genome duplication, and differential gene expression; some of which play important roles in plant speciation and adaptive radiation (Linder and Rieseberg, 2004; Seehausen, 2004; Baack and Rieseberg, 2007; Alix et al., 2017). Interspecific hybridization has been estimated to have occurred in at least 25% of flowering plant species (Mallet, 2005; Mallet, 2007).
Epimedium sect. Diphyllon from Berberidaceae is an excellent model for studying species radiation with possible network evolution in southern EA. First, Epimedium sect. Diphyllon is the largest section within the morphologically unique genus of Epimedium characterized by spider-like flowers (Figure 1). This section encompasses over 80% of all Epimedium species, which is primarily distributed within southern EA from southwestern, central to southeastern China. The section has two main distribution centers of Hengduan Mountain region and south-central China, located on each side of the boundary between the Sino–Japanese Forest and the Sino–Himalayan Forest subkingdoms in the East Asian Flora (Wu and Wu, 1998), providing a good chance to test the possible link between floristic division and taxa phylogenetic relationships.
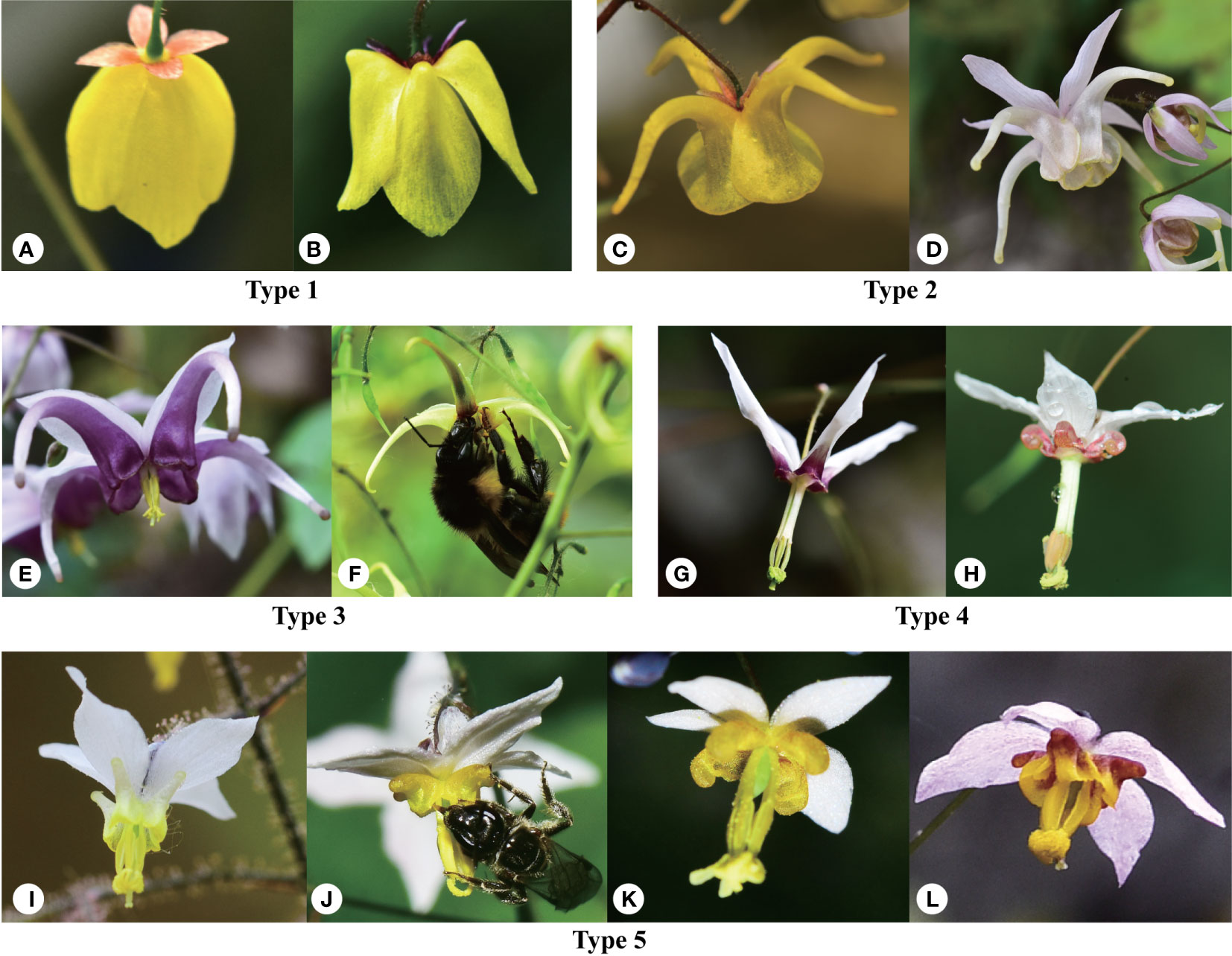
Figure 1 Morphological diversity of Epimedium sect. Diphyllon showing the five types of flowers and pollinators. Type 1: lamina flat, obovate, with a slight median nectariferous furrow but no spur or a saccate spur. (A) E. campanulatum, (B) E. ecalcaratum. Type 2: lamina rounded and conspicuous but expanded outwards at base into an elongated spur usually longer than the inner sepal. (C) E. davidii, (D) E. hunanense. Type 3: lamina reduced to a rim around the mouth of the elongated basally-swollen spur which is longer than the inner sepal. (E) E. aciminatum, (F) E. jingchengshanense. Type 4: lamina reduced to a rim around the mouth of the slipper-like cylindric spur, which is shorter than inner sepal. (G) E. fargesii, (H) E. dolichostemon. Type 5: lamina rand spur alike reduced, forming a minute pouch. (I) E. brevicornu, (J) E. stellulatum, (K) E. myrianthum, and (L) E. truncatum.
Furthermore, species delimitation and morphological variations in the speciose Epimedium sect. Diphyllon are extremely complicated owing to their diverse morphological traits, which vary widely in petal type, form, relative size of inner sepals and petals, and flower dimensions (Ying, 2002; Zhang et al., 2007; De Smet et al., 2012; Zhang et al., 2014b). Both nuclear and chloroplast data suggest that Epimedium sect. Diphyllon is a monophyletic lineage with a recent evolutionary history (Sun et al., 2005; Zhang et al., 2007; De Smet et al., 2012; Zhang et al., 2014b; Zhang et al., 2021). Traditionally, the section is classified into four series (Campanulatae, Davidianae, Dolichocerae, and Brachycerae) based on flower morphology (Stearn, 2002). However, phylogenetic studies have not confirmed the classification and phylogeny within the Epimedium sect. Diphyllon based on pollen types, flavonoid types, karyotypes, and molecular markers (Guo and Xiao, 1999; Sun et al., 2005; Sheng and Chen, 2007; Zhang et al., 2007; Guo et al., 2008; Zhang et al., 2008; Sheng et al., 2010; De Smet et al., 2012; Zhang et al., 2018). The backbone and species relationships in this section remain unclear, as evidenced by species from the same series that are not clustered into one clade (Zhang et al., 2014b). Despite recent phylogenetic analysis from the plastid genome sequences provided much better phylogenetic resolution within Epimedium sect. Diphyllon, the results could not provide a clear route for morphological evolution in Epimedium sect. Diphyllon (Guo et al., 2021). Further investigation of morphological diversity and evolution in EA is necessary, building on robust phylogenetic relationships from nuclear genomic data.
Third, possible hybridization or introgression among species has been frequently suggested for the Epimedium sect. Diphyllon. An outcrossing breeding system with strong self-incompatibility and high cross-compatibility between infraspecific and interspecific individuals is prevalent in Epimedium (Suzuki, 1984; Sheng et al., 2011). Observations of garden experiences have also indicated that all Epimedium species might be interfertile when brought together (Stearn, 2002; Sheng et al., 2011). Epimedium species rarely overlap in the different areas they occupy, and they seemingly evolved in geographical isolation although sympatric species are also common and may have produced several hybrid swarms. For example, the meeting of E. acuminatum and E. fangii on Mt. Omei resulted in the creation of a hybrid swarm: E.×omeiense (Stearn, 1995). The interspecies similarity of Epimedium is close to or even higher than the intraspecies similarity (Guo et al., 2021). Frequent gene flow among sympatric populations due to pollen or seed dispersal might be one of the reasons for the low interspecific differentiation among Epimedium species (Xu et al., 2007). We hypothesized that introgression and hybridization played an important role in evolution and triggered rapid species diversification and diverse phenotypic traits in our studied group.
Genome-wide introgression signals are difficult to detect, especially in older interspecific hybridization events or rapidly diverged lineages (Soltis and Soltis, 2009). Robust phylogenetic inferences are required to understand mechanisms underlying species evolution. In plants, unfortunately, most groups that rapidly speciated exhibit a low resolution of interspecific relationships with a wide occurrence of polytomy and conflicting phylogenies due to a lack of informative loci, frequently accompanied by incomplete lineage sorting (ILS) and/or hybridization including introgression in the early reticulate history. Thus, more robust data and analyses are required to explore hybridization events and distinguish ILS from hybridization and introgression (Vargas et al., 2017).
Studying the evolutionary history of Epimedium sect. Diphyllon proves to be challenging because of its recent and rapid radiation, potential phenotypic convergence, and limited informative loci from nrDNA, cpDNA, and AFLP data (Sun et al., 2005; Zhang et al., 2007; De Smet et al., 2012; Zhang et al., 2014b; Zhang et al., 2021), which resulted in polytomies of the phylogenetic tree, making researches unable to generate strong phylogenetic hypotheses. Here, we applied the genotyping-by-sequencing (GBS) strategy to collect the genomic sequences of Epimedium sect. Diphyllon with the integration of morphological evidence; then, we employed concatenated, coalescent-based, biogeographic, and phylogenetic network methods to determine the underlying mechanism of phenotypic evolution and recent rapid radiation in the evolutionary history of Epimedium sect. Diphyllon in southern EA. Our study sheds light on the processes of plant radiation patterns and reveals possible mechanisms underlying the high level of plant diversity in this region.
We sampled 57 accessions for GBS sequencing, including 43 species and 2 putative new species representing the taxa from all four series of Epimedium sect. Diphyllon (Campanulatae: 2 species, Davidianae: 14 species, Dolichocerae: 14 species, and Brachycerae: 14 species), and one species from sect. Maceceras (E. koreanum) was used as an outgroup (Supplementary Table S1). Voucher specimens were deposited in the Herbarium of Beijing Normal University. Taxonomic systems and species identification were largely based on Stearn (2002) except for the seven recently published species that were recognized following the original literature (Guo et al., 2007; Zhang and Li, 2009; Zhang et al., 2014a; Zhang et al., 2015; Zhang et al., 2016).
Total genomic DNA was extracted from approximately 20 mg of fresh leaf tissue from each plant using a modified CTAB protocol (Doyle and Doyle, 1987; Cullings, 1992). DNA concentrations were quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific Inc., California, USA) with a dsDNA Broad-Range Assay Kit.
The GBS library was prepared from DNA samples using the PstI-HF restriction enzyme according to previously published protocols (Elshire et al., 2011; Escudero et al., 2014), with some modifications. For each sample, 500 ng of genomic DNA was combined with 0.6 pmol of a sample-specific barcode adapter and 0.6 pmol of a common adapter. Each sample was digested with four units of PstI-HF (New England Biolabs Inc., Massachusetts, USA) at 37°C overnight. The adapters were ligated using 400 units of T4 DNA Ligase (New England Biolabs Inc., Massachusetts, USA) at room temperature for 4 h. We combined 50 ng of each sample, and the pool was purified using Agencourt AMPure XP (Beckman Coulter, Inc., Indiana, USA). The DNA fragments were amplified for 15 PCR cycles starting from 35 ng of DNA using NEB 2x Taq MasterMix (New England Biolabs Inc., Massachusetts, USA). The PCR product was purified using different volume ratios of AMPure XP beads to sample, and fragment size distributions were assessed using a 2100 Bioanalyzer (Agilent Technologies, Inc., California, USA). The library with the optimal profile and concentration (AMPure to sample ratio: 0.8:1, concentration: 1.87 ng/μl, average size: 595 bp) was submitted to Majorbio Bio-Pharm Technology (Shanghai, China) for 100 bp HiSeq 2000 Illumina sequencing. Quality control of the sequencing results was performed using FastQC 0.11.3 (Andrews, 2010).
The assembly of GBS loci was performed using the ipyrad 0.9.30 pipeline (Eaton and Overcast, 2020), which allows for indel variation (commonly found in phylogenetic datasets) and is optimized for downstream phylogenetic analyses. We conducted a de novo assembly of the GBS data. To compare and verify the de novo assembly results, a second assembly was performed based on the reference genome by mapping the full-length transcriptome sequence of E. pseudowushanense (Pan et al., 2017). The following parameters were applied to assemble the complete loci and single nucleotide polymorphism (SNP) sequences using ipyrad: a maximum of 4 low-quality base calls, a minimum depth of 6 for statistical and majority rule base calling, a maximum clustering depth of 1,000 within samples, 0 mismatches allowed in barcodes, a 50 base-pair minimum length of reads after trimming, and a maximum of 10 SNPs per locus. A minimum sample coverage per locus was set to 43 (ca. 75% of the total) considering the balance between locus coverage and data resolution, which is much higher than those in Fernández-Mazuecos et al. (2020). Finally, we obtained four GBS datasets for downstream analysis: complete de novo sequences (Denovo), de novo SNPs (Denovo_SNP), complete reference-based sequences (Ref), and reference-based SNPs (Ref_SNP).
The gene trees of the four GBS datasets were analyzed using the maximum likelihood (ML) method, as implemented in both RAxML 8.2.10 (Stamatakis, 2014) and IQ-TREE v1.6.10 (Hoang et al., 2018). All datasets were analyzed without partitioning. A rapid bootstrap search (100 replicates), followed by a thorough ML search (-m GTRGAMMA), was implemented for the RAxML approach. The IQ-TREE analysis employed an ML search with the best-fit substitution model automatically selected, and branch support was assessed using an ultrafast bootstrap (Minh et al., 2013). The datasets were also inferred using Bayesian inference (BI), as implemented in ExaBayes 1.4.1 (Aberer et al., 2014) with the GTRGAMMA substitution model.
Species trees were inferred using the SVD quartets method (Chifman and Kubatko, 2014; Chifman and Kubatko, 2015). It is well known that gene trees, especially the rapid radiation group that frequently accompanies ILS, may result in obscure phylogenetic topologies. The SVD quartets method is run under a multispecies coalescent approach and accounts for the ILS. Analyses were performed in PAUP* 4 under multispecies coalescent conditions to evaluate all possible quartets. One hundred bootstrap replicates were conducted and the results were summarized in a 50% consensus tree.
Two morphometric datasets (vegetative and reproductive traits) of Epimedium sect. Diphyllon were assembled for morphological analysis (Supplementary Table S2). The characteristics of leaves covering 42 species and an unknown species representative of the four series were appraised based on dried, pressed specimens obtained from our field collection and herbarium. The selected characters included 11 variables, and the final morphometric data set included 737 samples (Supplementary Table S3). The characteristics of the flowers covering 41 species and one unknown species representative of the four series were assessed based on living plants obtained from our field collection; the selected characters included 11 variables, and the final morphometric dataset included 699 individuals (Supplementary Table S4).
To assess the reproductive and vegetative morphological affinities of the four series of Epimedium sect. Diphyllon discriminant function analysis (DFA) was performed to identify the main characteristics contributing to the morphological differentiation of the taxa. We used the basic package MASS (Venables and Ripley, 2002) of R v3.2.5 (R Development Core Team, 2016) for the DFA. The R package ggord (Beck, 2017) was used to represent the first two axes of the DFA (Supplementary Tables S5, S6) in a biplot with 95% confidence ellipses of concentration for the four series.
Morphometric and phylogenomic data were integrated to investigate the patterns of morphological evolution in Epimedium sect. Diphyllon. We used the phylomorphospace approach (Sidlauskas, 2008) implemented in the R package phytools (Revell, 2012) to reconstruct the evolutionary history of morphological diversification. A backbone input tree from the IQ-TREE analysis based on the de novo dataset was selected, and one individual from each taxon was used. The tree included 35 species from Epimedium sect. Diphyllon and one species from Epimedium sect. Macroceras. As previous AFLP (Zhang et al., 2014b) and cpDNA (Liu, 2021) results suggest that the sister lineage of Epimedium sect. Diphyllon is E. elatum from sect. Polyphyllon, we used the morphological traits of E. elatum rather than those of E. koreanum in the ancestral state analysis to estimate the ancestral character state of the section. The phylogenetic tree was then projected onto the multivariate morphospace defined by the first two principal components from the reproductive (Supplementary Tables S7, S8) and vegetative traits (Supplementary Tables S9, S10) to investigate the patterns of morphological evolution.
To investigate particular morphological characters and phenotypic convergence, we conducted ancestral state reconstructions (ASRs) in Mesquite 3.70 (Maddison and Maddison, 2021) using the same phylogenetic tree used for the morphospace analysis. As previous AFLP (Zhang et al., 2014b) and cpDNA (Liu, 2021) results suggest that the sister lineage of Epimedium sect. Diphyllon is E. elatum from sect. Polyphyllon, we used the morphological traits of E. elatum rather than those of E. koreanum in the ancestral state analysis to estimate the ancestral character state of the section. Four characters that represent reproductive variation across the section were analyzed: (1) flower size (small and large); (2) flower shape (Type 1: lamina flat, obovate, with a slight median nectariferous furrow without spur or a saccate spur; Type 2: lamina rounded and conspicuous but expanded outwards at the base into an elongated spur usually longer than the inner sepal; Type 3: lamina reduced to a rim around the mouth of the elongated basally swollen spur, which is longer than the inner sepal; Type 4: lamina reduced to a rim around the mouth of the slipper-like cylindrical spur, which is shorter than the inner sepal; Type 5: both lamina and spur are reduced, forming a minute pouch); (3) inner sepal color (white, yellow, red, and purple); (4) spur color (white, yellow, red, purple, and brown).
We estimated a time-calibrated tree using penalized likelihood as implemented in treePL (Smith and O’meara, 2012). When working with phylogenies obtained from a massive amount of data, such as those produced by GBS, treePL software is appropriate for estimating divergence times (Zheng and Wiens, 2015). The IQ-TREE tree obtained from the de novo dataset was used as input. The tree was pruned to include a single, arbitrarily chosen individual of each species. Calibration ages were based on the results of the dating analysis of the plastid genome (Guo et al., 2021). The root node between the outgroup and Epimedium sect. Diphyllon with a maximum age of 2.11 million years ago (Ma).
Inferences of reticulate events were run in the Julia package PhyloNetworks (Solís-Lemus et al., 2017) using the SNaQ method (Solís-Lemus and Ané, 2016), which performs maximum pseudolikelihood estimation of phylogenetic networks from a list of unrooted gene trees or a table of quartet concordance factors (CFs) of 4-taxon trees (quartets) under multispecies coalescence, incorporating ILS and reticulation events. The CFs were used to estimate a semi-direct species network with estimated reticulation events and γ-values (inheritance probabilities of ancestral contributions to hybridization events). This strategy accelerates the likelihood computation by considering subsets of quartets instead of simultaneously calculating the full likelihood across all taxa. SNP2CF (Olave and Meyer, 2020) was used in R to convert the unlinked SNP PHYLIP file of ipyrad into a CF table. Afterward, the phylogenetic networks were reconstructed using PhyloNetworks. The SNaQ function used the CF table and ran 50 independent analyses with the number of reticulation events (h) ranging from zero to nine, using the gene tree as the starting topology for each h value. The best-fit number of reticulation events was inferred by plotting the likelihood scores for each run and observing the point at which additional reticulations did not improve the pseudolikelihood score (Solís-Lemus et al., 2017). We then used the pseudoreplication approach in SNP2CF to create a CF table with credibility intervals that were used in the PhyloNetworks Bootsnaq command to conduct 100 bootstrap replications to estimate the support for the inferred inheritance probabilities.
For the 57 samples sequenced for GBS, we obtained a total of 895 million reads with 15.7 million reads per sample on average. The final data from de novo assembly contained 88135 parsimony-informative sites with a total length of 820547 bp (73.0% missing data) and 10331306 bp (73.5% missing data) for the Denovo_SNP and Denovo datasets, respectively; for the reference-based data there were 49034 parsimony-informative sites with a total length of 931802 bp (76.4% missing data) and 9039893 (77.4% missing data) for the Ref_SNP and Ref datasets, respectively.
Based on the four GBS datasets, phylogenetic analyses using the RAxML, IQ-TREE, and ExaBayes approaches produced 12 similar topologies with solid support (Supplementary Figures S1A-C, S2A-C, S3A-C, S4A-C). A phylogenetic tree based on the IQ-TREE method using the de novo GBS dataset with fully resolved and well-supported interspecific relationships was presented in Figure 2. All species of Epimedium sect. Diphyllon were grouped into two clades—a west clade and an east clade—except for E. brevicornu, which was discovered as a basal lineage of Epimedium sect. Diphyllon (Supplementary Figure 2, Supplementary Figures S1A-C, S2B, S3A, C), or a sister lineage to either the west or (Supplementary Figures S2A, B, S3B, S4A-C) east clades (Supplementary Figure S1C).
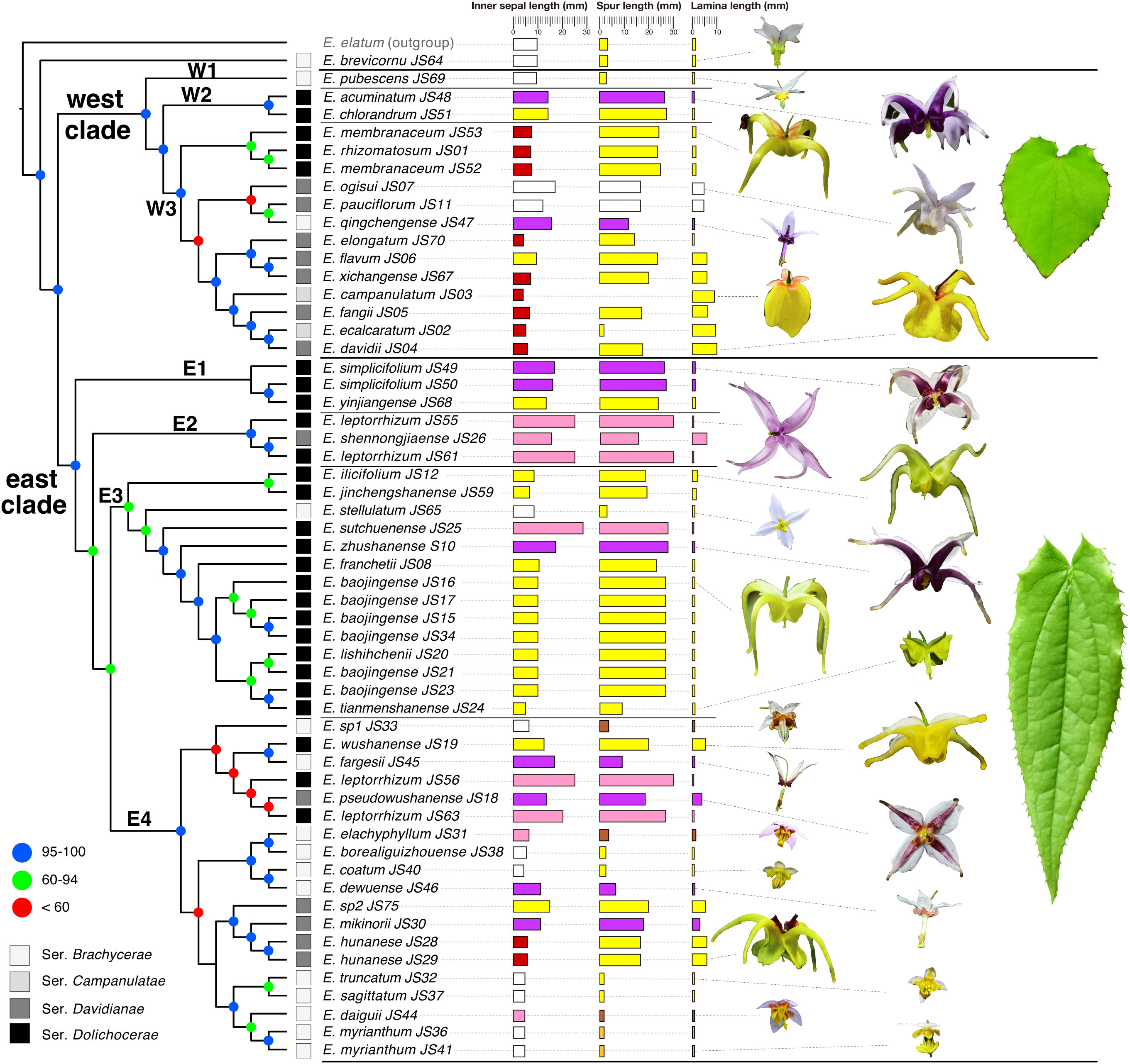
Figure 2 Phylogenetic relationships and morphological diversity of Epimedium sect. Diphyllon in the east Asia. Phylogenetic tree obtained from the IQ-TREE method based on the denovo GBS dataset, and node support is indicated with colored circles. The four series of sect. Diphyllon is indicated by different colored quadrate. Morphological traits are shown for the taxa, including mean values of inner sepal length, spur length, and lamina length. Flowers and leaves of sect. Diphyllon are shown.
Seven subclades with strong support were identified using concatenated approaches. The west clade was divided into three subclades (W1, W2, and W3), with E. pubescens as the first diverging lineage (W1), and the remaining species were grouped into two subclades (W2 and W3) (Figure 2, Supplementary Figures S1A-C, S2A-C, S3A-C, S4A-C).
The east clade was divided into four subclades (E1, E2, E3, and E4) (Figure 2; Supplementary Figures S1A-C, S2A-C, S3A-C, S4A-C). E1 contains two species (E. yinjiangense and E. simplicifolium) and is supported as a distinct evolutionary lineage by both Denovo and Denovo_SNP datasets (Figure 2, Supplementary Figures S1A-C, S2A-C). However, the two reference-based datasets present different topologies, with E. jinchengshanense nested within them (Supplementary Figures S3A-C, S4A-C). E2 includes two species (E. leptorrhizum and E. shennongjiaense), and all topologies support it as a monophyletic lineage (Figure 2, Supplementary Figures S1A-C, S2A-C, S3A-C, S4A-C). E3 is recovered as a monophyletic lineage from the Denovo and Denovo_SNP datasets (Figure 2, Supplementary Figures S1A-C, S2A-C), but paraphyletic from the reference-based datasets (Supplementary Figure S3A-C, S4A-C). Finally, the remaining species in E4, such as E. fargesii, E. hunanense, and E. myrianthum, are supported as monophyletic lineages across all topologies (Figure 2; Supplementary Figures S1A-C, S2A-C, S3A-C, S4A-C).
SVD quartet analyses produced topologies similar to those of the concatenated gene trees (Supplementary Figures S1D, S2D, S3D & S4D). Two slightly different species-level topologies were generated from the Denovo and Denovo_SNP datasets (Supplementary Figures S1D, S2D). In the first topology, an individual of E. leptorrhizum_JS63 from Suiyang, Guizhou, was found to be closely related to E. jinchengshanense, as it was recovered as a sister lineage. In the second topology, a cluster of E. wushanense and E. fargesii was found to be a sister lineage to E3 (Supplementary Figures S1D, S2D). The SVD species tree generated from the Ref dataset supports E. brevicornu as the basal species in Epimedium sect. Diphyllon, with E. jinchengshanense as a sister species to E1 (Supplementary Figure S3D). The SVD species tree generated from the Ref_SNP dataset was similar to the Ref tree at the subgroup level; however, E. brevicornu is suggested as the basal species in the east clade (Supplementary Figure S4D).
Morphometric analyses were performed based on 11 vegetative traits from 737 individuals of 42 taxa and one unknown species from Epimedium sect. Diphyllon. The results showed that the individuals from the four series identified by Stearn (2002) were mixed together (Figure 3A). Biplots of sites and variables depicted differences between clusters, in particular vegetative morphometric characters for leaf texture, the ratio of middle leaflet length to width, and sawtooth density. The first two LD coordinates (explaining 98.12% of the variance) depicted a complex morphospace (Figure 3A). The results are based on 11 reproductive characters from 699 individuals of 42 taxa and one unknown species from Epimedium sect. Diphyllon revealed that the morphometric differentiation is consistent with the taxonomic treatment by Stearn (2002) (Figure 3B). Biplots of sites and variables depicted differences between clusters, in particular, flower morphometric characters for lamina presence or absence, inner sepal color, spur color, and spur reflection. The first two LD coordinates (explaining 97.62% of the variance) depicted a complex morphospace (Figure 3B).
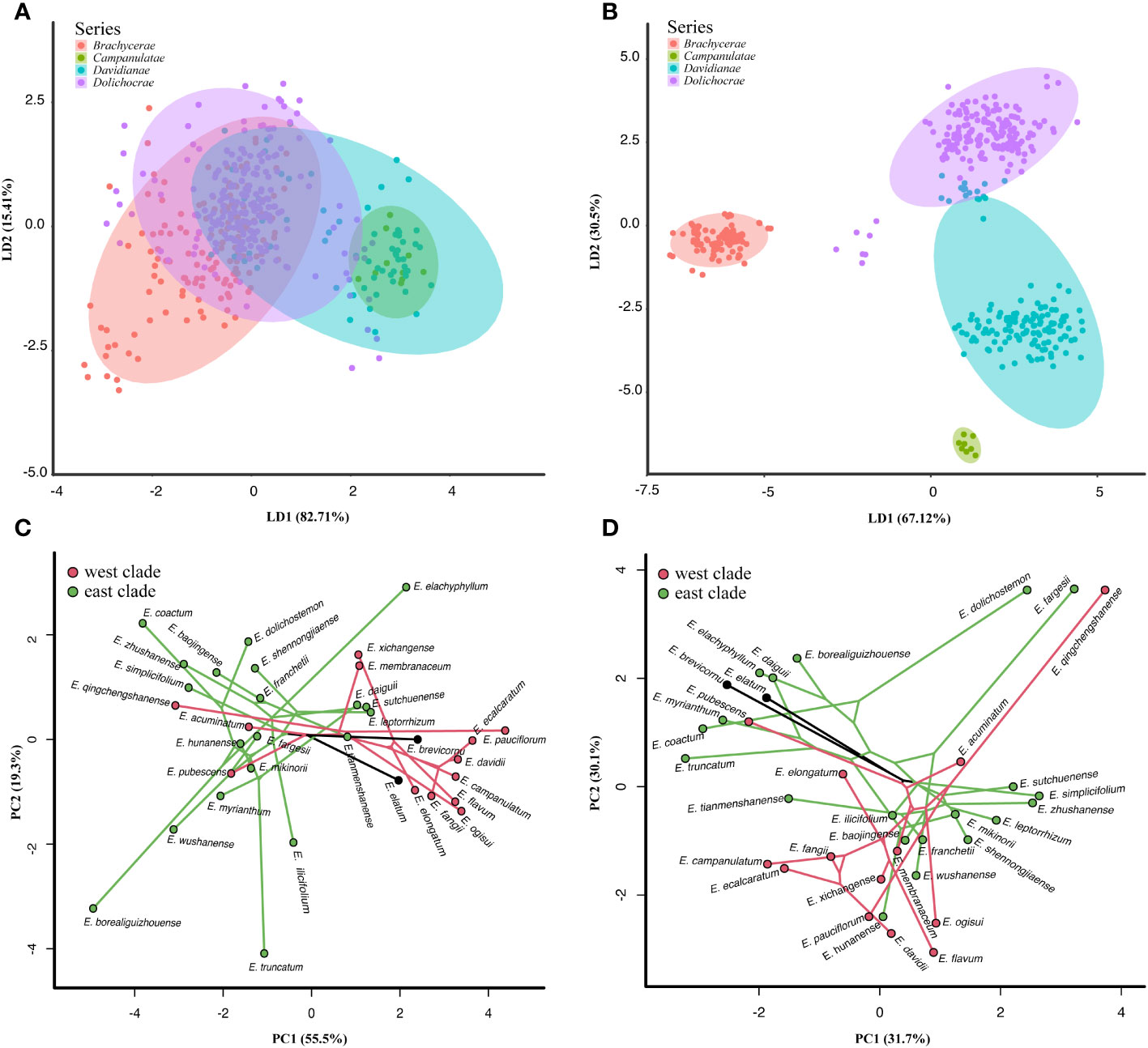
Figure 3 Morphometric analyses of Epimedium sect. Diphyllon. (A) Morphometric analyses on DFA of the vegetative traits (737 individuals, 42 taxa and one unknown species). (B) Morphometric analyses based on DFA of the reproductive traits (699 individuals, 41 taxa one unknown species). (C) Phylomorphospace of vegetative traits defined by a backbone tree with 35 species from IQ-TREE analysis based on the denovo GBS dataset. Each species is represented by the mean values. (D) Phylomorphospace of reproductive traits defined by a backbone tree with 35 species from IQ-TREE analysis based on the denovo GBS dataset. Each species is represented by the mean values of the coordinates.
Combined with phylogenetic and morphometric data, the phylomorphospace revealed distinct morphological patterns in the east and west clades within Epimedium sect. Diphyllon. The phylomorphospace, based on vegetative data, indicates the ancestral leaf shape of Epimedium sect. Diphyllon appears to have broadly ovate leaflets (Figure 3C). Epimedium brevicornu, the basal species of Epimedium sect. Diphyllon, has maintained vegetative traits that closely resemble those of the common ancestor of the Epimedium sect. Diphyllon. In contrast, the other species underwent significant morphological changes in multiple directions. In the phylomorphospace plot, the species of the west clade broadly occurred on the right side of the diagonal and displayed broad ovate to ovate leaflets (e.g., E. davidii, E. elongatum, and E. xichangense). In contrast, the species of the east clade are mostly distributed on the left side of the diagonal and displayed ovate-to-lanceolate leaflets. Additionally, convergence in vegetative traits is suggested by the close coordinates in different phylogenetic clades, as seen in E. pubescens/E. myrianthum with ovate leaflets.
The phylomorphospace defined by the reproductive traits (Figure 3D) suggests the floral ancestor state of Epimedium sect. Diphyllon appears to be a small flower with an inconspicuous spur and distinct lamina. This basic floral shape was retained by E. brevicornu, E. pubescens, and other small flowering species (Type 5). However, other species have undergone vast morphological shifts in different directions, even in sister-species pairs, including E. acuminatum/E. chlorandrum (purple corolla vs. yellow corolla), E. membranaceum/E. pauciflorum (yellow corolla, petals without lamina vs. white corolla, petals with lamina), E. elongatum/E. xichangense (petals without lamina vs. petals with lamina) and E. fangii/E. ecalcaratum (petals with spur vs. flat petals), E.leptorrhizum/E.shennongjiaense (petals without lamina vs. petals with lamina), E. wushanense/E. fargesii (large flowers, short stamens vs. small flowers, prolonged stamens), E. hunanense/E. mikinorii (corolla yellow and significant lamina vs. corolla purple and medium-sized lamina), and E. coatum/E. dolichostemon (short stamen vs. prolonged stamen). The mapping of phylogenetic relationships onto geographic distributions revealed pairs of morphologically divergent sister species found in close proximity to one another. In contrast, convergence is suggested by the neighborhood in the morphospace of non-closely related species pairs (Figure 3D). Large flowers, yellow-dominated corollas, short sepals, large nectar spurs, and significant laminae have evolved independently in E. hunanense and E. davidii (Type 1). Overall, semblable floral morphologies, large flowers, broadly obovate inner sepals, and large nectar spurs without lamina have evolved independently in E. simplicifolium and E. acuminatum (Type 2). The floral traits of large flowers, yellow corollas, large nectar spurs, and those without lamina have evolved independently in E. membranaceum and E. baojinense (Type 2).
MP reconstructions facilitated the unambiguous inference of multiple shifts in flower size, flower type, inner sepal color, and spur color (Figure 4). The ancestral flowers were small (Figure 4A). The character evolution of the five flower types also suggests that the small flowers of Types 4 and 5 were in the ancestral state (Figure 4B). Large flowers and long spurs have independently evolved multiple times in both the west and east clades. This phenomenon was also observed in flower color (Figures 4C, D). The plesiomorphic state should be white, with diversification into yellow, red, pink to purple flower colors (Figure 4C). For petals or spurs, yellow color was the ancestral state; therefore, flower colors have evolved multiple times from yellow to purple and white in the west clade and from yellow to purple, red, and brown in the east clade (Figure 4D).
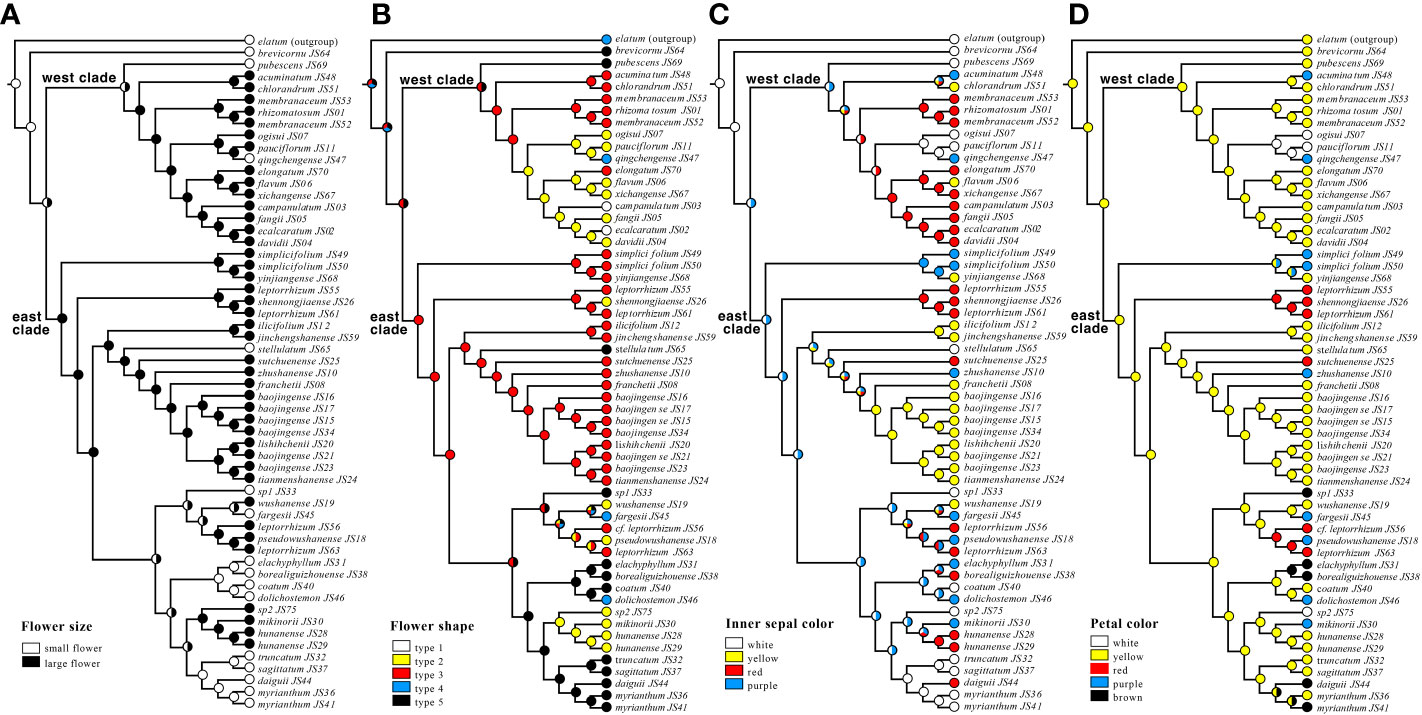
Figure 4 Patterns of morphological evolution of the Epimedium sect. Diphyllon. (A) Ancestral states were reconstructed from flower size, (B) flower shape, (C) inner sepal color, (D) petal color.
Time estimation suggests that the extant species of Epimedium sect. Diphyllon diverged in the late Pleistocene at about 0.84 (0.57–1.25) Ma with E. brevicornu as the first split lineage (Figure 5). The divergence age between the east and west clades was estimated to be 0.77 (0.43–1.16) Ma (Figure 5).
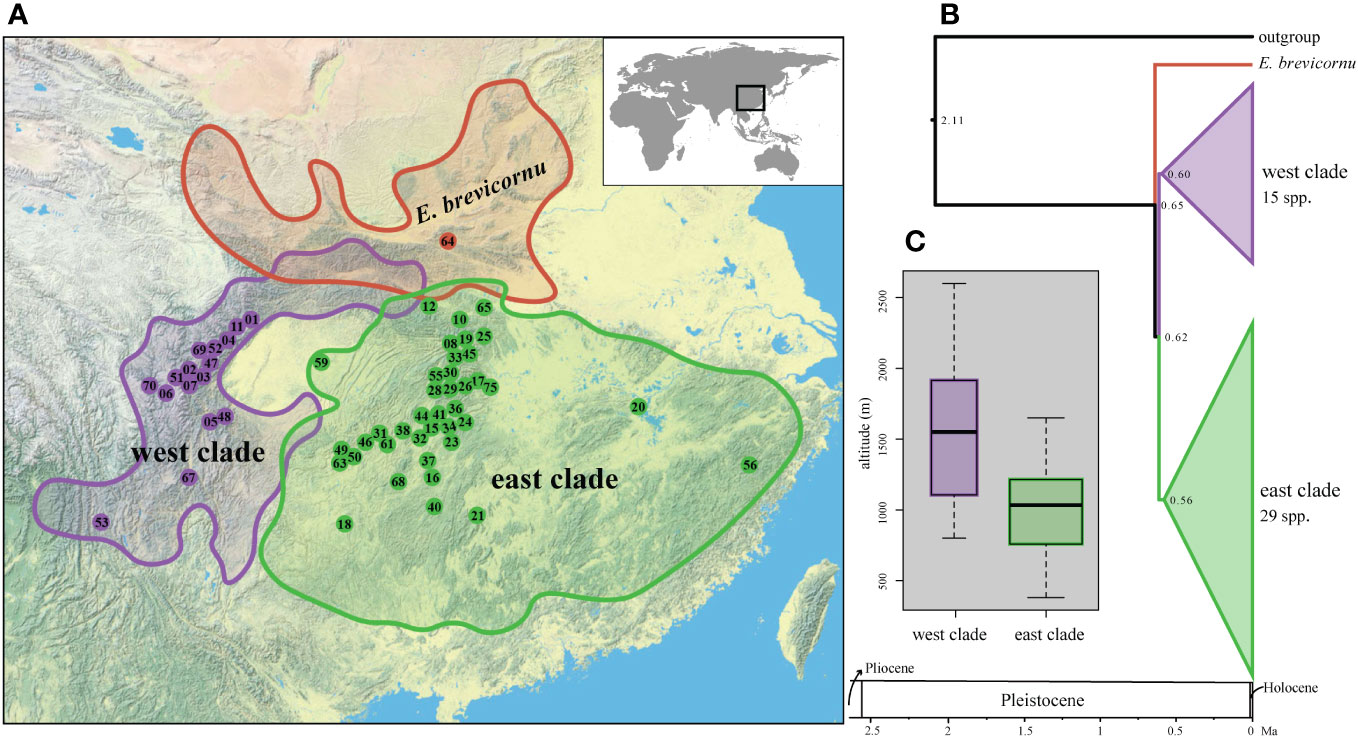
Figure 5 Phylogenetic and spatiotemporal context of the Epimedium sect. Diphyllon radiation in EA. (A) Geographic distribution of three lineages within Epimedium sect. Diphyllon. (B) Divergence times estimated using treePL approach from IQ-TREE analysis based on denovo dataset. (C) Boxplot of altitude difference between west clade and east clade.
We conducted SNaQ analysis using 16 representative species to reconstruct a phylogenetic network within Epimedium sect. Diphyllon in EA. Based on the log pseudo-likelihood scores, the optimal network model was identified as h=5 (Supplementary Figure S5). The phylogenetic network generated by SNaQ showed topologies similar to those of the concatenated and coalescent analyses, supporting the differentiation between the west and east clades. This network illustrates the rapid radiation of Epimedium sect. Diphyllon species with four hybridizing edges: a) E. davidii is a hybrid species between E. ogisui (γ = 0.414) and E. campanulatum (γ = 0.586); b) E. membranaceum shows minor genetic introgression from the common ancestor of the west clade (γ = 0.0363); c) E. stellulatum has ca. 30% genomic exchange from E. brevicornu (γ = 0.294); and d) the E. myrianthum–E. hunanense lineage has an introgressed origin between E. baojinense (γ =0.372) and E. pseudowushanense (γ =0.628) (Figure 6).
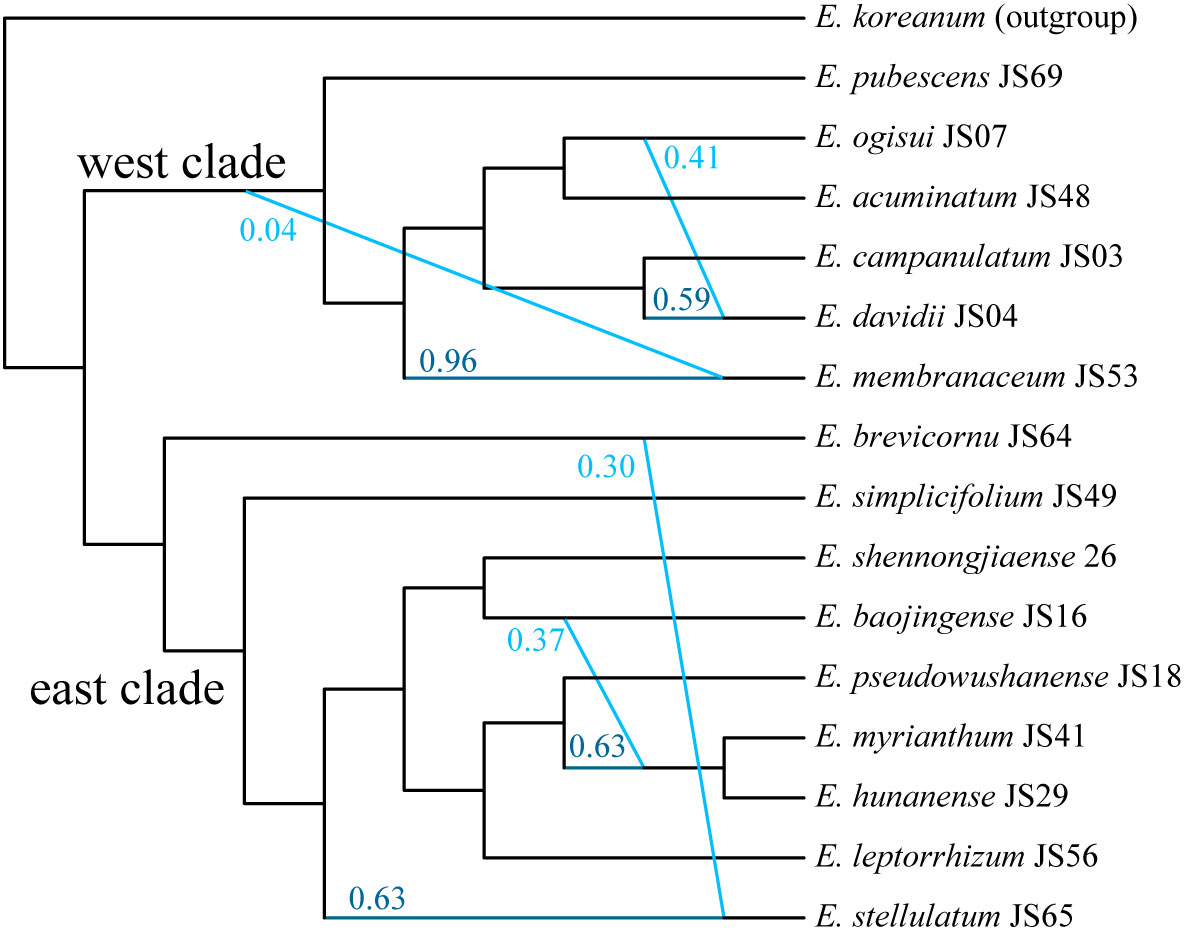
Figure 6 Phylogenetic network of Epimedium sect. Diphyllon estimated from SNaQ analysis with hmax = 5.
Phylogenetic relationships among Epimedium sect. Diphyllon remains unclear despite the use of numerous conventional molecular markers, such as AFLP, or plastid genome data (Sun et al., 2005; Zhang et al., 2007; De Smet et al., 2012; Zhang et al., 2014b; Guo et al., 2021). Phylogenomic approaches can help reconstruct speciation patterns and processes in detail, particularly in cases where conventional phylogenetic markers fail, as demonstrated for Galapagos giant daisies (Fernández-Mazuecos et al., 2020). Techniques based on high-throughput genomic sequencing, such as GBS, can generate genome-wide phylogenetic markers, providing unparalleled opportunities to test competing evolutionary hypotheses (Elshire et al., 2011; Qi et al., 2015; Fernández-Mazuecos et al., 2018). After generating a GBS library and testing both de novo and reference-based sequence dataset assemblies with multiple analytical analyses (Supplementary Figures S1-S4), the optimal combination produced a phylogenetic tree with fully resolved and well-supported interspecific relationships (Figure 2). Trees based on coalescent methods generally had lower support values, although the evolutionary patterns derived from both approaches were largely consistent (Supplementary Figures S1D, S2D, S3D & S4D).
Our phylogenomic analyses based on the GBS data revealed a much clearer evolutionary relationship for Epimedium sect. Diphyllon than before, with average node supports of 93.86%/1/90.34% from ML/BI/SVD analyses, respectively (Figure 2; Supplementary Figures S1-S4, Supplementary Table S11). Except for the uncertain placement of E. brevicornu, all accessions were grouped into two well-supported clades (Figure 2; Supplementary Figures S1-S4). All the results of our phylogenetic trees showed that E. brevicornu is an early diverging lineage, but its phylogenetic position was unstable. Specifically, E. brevicornu was placed as a sister species to the remaining species of Epimedium sect. Diphyllon or a sister species to either the west or east clades (Figure 2; Supplementary Figures S1-S4). In a recent study by Guo et al. (2021), however, E. brevicornu HBXY is deeply nested in the west clade based on chloroplast genomic data. This species is characterized by small flowers, which may indicate the ancestral status and distribution of the Epimedium sect. Diphyllon.
The results show a comparable divergence pattern to the plastid genome phylogeny of Guo et al. (2021), albeit with relatively lower resolution in the chloroplast results. In line with the chloroplast genome findings of Guo et al. (2021), our nuclear genomic results also identify two groups. However, our nuclear genomic analysis offers a more detailed resolution of species relationships within each subgroup. Notably, most of the analyses strongly support the identification of three subgroups within the west clade and four subgroups within the east clade (Figure 2; Supplementary Figures S1-S4).
Except for E. brevicornu, topological differences were also detected between previous chloroplast and current GBS datasets for the other taxa within the section. Epimedium pubuscens is deeply nested within the west clade in the chloroplast results (Guo et al., 2021); however, it is consistently recognized as the first lineage of the west clade in the current nuclear genomic results (Figure 2; Supplementary Figures S1-S4). Epimedium stellulatum SXNZ in Guo et al. (2021) is placed in the west clade; however, our collection from Hubei belonged to the east clade (Figure 2; Supplementary Figures S1-S4). Epimedium pauciflorum SCMS belongs to the east clade in the study of Guo et al. (2021), while our collection from Sichuan, Maoxian, belonged to the west clade (Figure 2; Supplementary Figures S1-S4). These phylogenetic conflicts between different genomic data might have resulted from the low resolution of the chloroplast sequences or from hybridization or introgression between them. Further tests with more data are needed.
A morphology-based taxonomic treatment by Stearn (2002) was rejected by our team and chloroplast results (Guo et al., 2021) that the four series defined by flower characters are not monophyletic (Figure 2; Supplementary Figures S1-S4). Epimedium sect. Diphyllon diverged into two clades, and species belonging to different series were mixed together in each clade (Figure 2; Supplementary Figures S1-S4). The west clade includes all four series: Campanulatae, Davidianae, Dolichocerae, and Brachycerae, whereas the east clade consists of Davidianae, Dolichocerae, and Brachycerae series (Figure 2; Supplementary Figures S1-S4). The taxonomic treatment was largely based on reproductive rather than vegetative characteristics (Stearn, 2002), which was also supported by the morphological analyses (Figure 3B). The vegetative data did not corroborate the taxonomic treatments when the four series were mixed together (Figure 3A). In contrast, the reproductive data provided evidence in favor of the taxonomic treatments that distinguished the species into four series (Figure 3B).
Despite the incongruence between morphological and phylogenetic relationships, there is strong evidence of geographic patterns within Epimedium sect. Diphyllon. The two main clades within Epimedium sect. Diphyllon broadly corresponds to the biogeographic distribution of southern EA (Figure 5). The west clade is composed of species mostly from western China, whereas those of the east clade are largely from eastern China. The division of the two clades into distinct biogeographic regions roughly matched the EA floristic division between the Sino–Japanese Forest and the Sino–Himalayan Forest subkingdoms as well as the environmental division between southwestern and eastern China (Wu and Wu, 1998), suggesting a close link between phylogenetic relationships, floristic evolution, and geographical changes in EA (Zhang et al., 2007). Geological differences, together with the formation and development of the Asian monsoon, may have been the main factors driving the evolution of EA flora (Zhang et al., 2007), as indicated by the divergence in Epimedium sect. Diphyllon. Moreover, these phytogeographical units are in agreement with the infraspecific relationships in Spiraea japonica, with two clades corresponding to the eastern and western divisions (Zhang et al., 2006). Major genetic subdivisions between the Sino–Himalayan and Sino–Japanese Forest subkingdoms have also been found in other plant taxa, such as Ainsliaea (Mitsui et al., 2008), Cardiocrinum (Lu et al., 2017), and across the East China Sea between Southeast China and Japan, such as Kalopanax septemlobus (Sakaguchi et al., 2012) and Euptelea (Cao et al., 2016).
Interestingly, we observed a morphological differentiation trend in leaf traits between the two clades (Figure 3A). The phylomorphospace results revealed that species of the west clade usually have membranous and broadly ovate leaflets, whereas those of the east clade have leathery, ovate, narrowly ovate, and lanceolate leaflets (Figure 3A). The phylogenetic separation of the two clades is related to the patterns of morphological differentiation in the two clades of Epimedium sect. Diphyllon. It appears that climatic and geological differences played important roles in the divergence of Epimedium sect. Diphyllon and probably initiated the leaf divergence of the two groups within southern EA (Cao et al., 2016).
Flower morphology is a critical feature that can be used to distinguish Epimedium species (Stearn, 2002). Based on the size relationship between the petals and inner sepals, the genus is divided into two groups (Stearn, 1993a; Stearn, 1993b). The small group usually consists of flat, slightly saccate, saccate, and short spurs with petals shorter than the inner sepals and < 1 cm in diameter, whereas the large-flowered group has long spurs with petals longer than the inner sepals and > 1 cm in diameter (Figure 1). Southern EA is the distribution and differentiation center of Epimedium sect. Diphyllon with both small and large flowers (Ying, 2002). The plesiomorphic state should be a relatively small flower, inner sepal larger than the petal, and petal with a short spur (Ying, 2002). Our data support the general hypothesis that the ancestral flower size should be small (Figure 4A) although a previous ancestral character reconstruction analysis suggests that a large flower with a long spur (large-flowered group) is the plesiomorphic state (Guo et al., 2021).
Stearn (2002) identified five types of flower shapes of Epimedium (Figure 1). Character evolution also suggests that the small flowers of types 4 and 5 are ancestral states (Figure 4B). The flowers independently evolved mainly in length and size, from small to large and back to small, in both the east and west clades (Figure 4B). It seems that parallel evolution is dominant in Epimedium sect. Diphyllon, as evidenced by the evolution of large flowers and long spurs in both west and east clades.
The evolution of spur length through pollinator shifts has been examined using phylogenetic evidence in recent studies (Whittall and Hodges, 2007; Boberg et al., 2014). Guo et al. (2021) suggested that the diversification of petal shape likely affected reproductive success and drove the evolution of Epimedium, leading to the evolution of the long spur to respond to pollinator shifts. Effective pollinators, such as bumblebees (Bombus spp.) and nectar-foraging bees, have been observed to be associated with species that have long spurs (Suzuki, 1984; Li et al., 2009). Bumblebees are a group of social bee species that form structured colonies, and the genus is composed of numerous species found widely in the Northern Hemisphere. The parallel evolution of large flowers with long spurs in both the west and east clades may be explained by the presence of similar bumblebees around flowers with long spurs in both regions.
A similar situation was also determined in the evolution of flower color (Figures 4C, D). The plesiomorphic state for flower color should be white, with diversification into yellow, red, and purple in inner sepals (Figure 4C). For petals or spurs, yellow is the ancestral state with a parallel shift from yellow to purple and white in the west clade or purple, red, and brown in the east clade (Figure 4D). Parallel evolution is common in plants and animals, and the repeated and similar large-scale morphological evolutionary trends of distinct lineages suggest that adaptation through natural selection (functional constraints) is the major cause of parallel evolution, which is a common phenomenon in extinct and extant lineages (Monnet et al., 2011). For example, enlarged flower heads have evolved repeatedly in Artemisia, which is considered helpful in attracting pollinators better (Tkach et al., 2008).
In biology, atavism is the modification of a biological structure, whereby an ancestral genetic trait reappears after having been lost through evolutionary changes in previous generations (Hall, 1984). Atavism probably occurred multiple times in both clades of southern EA Epimedium. Type 1, with flat petals, was assumed to be the ancestral type, which evolved into four other types: slightly saccate, saccate, short spur, and long spur (Figure 1). However, our results suggest that species with flat petals were deeply nested within the west clade (Figure 4B). The ancestral state for the petal shape of Epimedium should be flat without shifting to a saccate-like shape or spur (Stearn, 2002; Ying, 2002). The morphological traits of its sister genus Vancouveria and other genera of the family support this hypothesis although most Epimedium species have petals with short or long spurs (Zhang et al., 2007). Although there are very few species with flat petals, the phylogenetic results suggest that they are deeply nested within the tree, and character evolution analyses suggest that they have shifted back from species with spurs. The ancestral state of Epimedium sect. Diphyllon should be small flowers; however, a species with small flowers with short spurs have also evolved independently and multiple times in both clades (Figure 4).
The origin of Epimedium sect. Diphyllon dates back to c. 2.11 Ma in the early Quaternary (Zhang et al., 2007; Guo et al., 2021), and the divergence of the crown of Epimedium sect. Diphyllon is estimated to have taken place in the late Pleistocene epoch (Figure 5). It appears that the radiation of Epimedium sect. Diphyllon with parallel and atavism evolution of flowers is very recent, which is consistent with the relatively recent geological history of EA. The uplift of the Qinghai–Tibetan Plateau to its current altitude in southwest China, with allopatric differentiation of plants between southwest and east China, has been suggested to have a relatively recent history (Wen et al., 2014). Meta-analyses of seed plants from EA also suggest that the EA flora might be relatively young, with most of its clades originating from the Miocene (Chen et al., 2017).
Epimedium species occupy different areas in the wild and have evolved in geographical isolation. However, several species have overlapping ranges, which may result in hybridization (Stearn, 1995). We found strong evidence of extensive reticulation events during the diversification of Epimedium sect. Diphyllon in southern EA from the discordant phylogenetic relationships among gene trees, species trees (Supplementary Figures S1-S4), and especially, the SNaQ analysis (Figure 6). For example, the SNaQ results revealed that E. davidii is a hybrid species resulting from the interbreeding of E. ogisui (γ = 0.414) and E. campanulatum (γ = 0.586). The distribution of E. davidii overlapped with that of both E. ogisui and E. campanulatum in western China. Epimedium davidii exhibits intermediate characteristics; for example, its inner sepals (red, small, and lanceolate) and petal color (yellow) resemble those of E. campanulatum, while its petal form (with an elongated spur and obvious lamina) is similar to that of E. ogisui. Epimedium stellulatum, which has approximately 30% genomic exchange from E. brevicornu (γ = 0.294), has small flowers and broadly ovate leaflets that closely resemble those of E. brevicornu. It appears that E. hunanense, a species related to E. baojinense and E. pseudowushanense, exhibits intermediate traits between the two parental species. Specifically, its red-colored inner sepals and conspicuous lamina are similar to those of E. pseudowushanense, while its yellow-colored petals are similar to those of E. baojinense.
Evidence of extensive introgression has been found throughout the evolutionary history of Epimedium sect. Diphyllon (Figure 6) and reticulate evolution played significant roles in shaping the existing diversity in EA. Numerous cases have invaluably contributed to our understanding of the evolution of plant diversity in certain areas, with an increasing number of natural hybridization or introgression events have been identified in crops and wild species with genomic analyses recently (Rieseberg et al., 2007; Eaton and Ree, 2013; Penjor et al., 2016; Yang et al., 2016; Vargas et al., 2017; Morales-Briones et al., 2018; Lee-Yaw et al., 2019; Reichelt et al., 2021; Rose et al., 2021; Wang et al., 2021). For instance, significant interspecific introgression of Pedicularis sect. Cyathophora was identified using restriction site-associated DNA sequencing data (Eaton and Ree, 2013). Using the targeted enrichment approach to capture hundreds of nuclear loci and nearly complete plastome sequences, ancient and recent hybridization events in the genus Lachemilla were identified based on significant phylogenetic discordance and the results of species network analyses (Morales-Briones et al., 2018). Using phylogenomic data to explore the evolutionary history of the genus Polemonium showed that the cytoplasmic-nuclear discordance in this recently radiated genus was caused by both ILS and reticulate evolution (Rose et al., 2021). Other examples, such as Hawaii silversword, East Asian kiwifruit, and Iberian toadflax, have suggested that hybridization results in rapid speciation (Barrier et al., 1999; Liu et al., 2017; Fernández-Mazuecos et al., 2018).
The rampant introgressions in Epimedium sect. Diphyllon were probably caused by the lack of reproductive isolation. A series of experimental crosses provided strong evidence for an outbreeding system and a weak internal barrier to hybridization among the Epimedium species studied (Sheng et al., 2011). This is not surprising, considering that an outcrossing breeding system with strong self-incompatibility but high cross-compatibility between infraspecific and interspecific individuals is prevalent in Epimedium (Suzuki, 1983; Sheng et al., 2011). Interestingly, introgression events were mostly detected within the east or west clades, rarely between species from the two groups (Figure 6). Furthermore, hybridization events promote morphological diversity among species and obscure their evolutionary relationships, resulting in the extremely high diversity and complicated evolution of this lineage. The influence of hybridization on speciation and adaptive evolution should be further investigated.
Evolutionary radiations have long fascinated biologists since the publication of “On the Origin of Species” by Charles Darwin. EA is a natural floristic region with the highest biodiversity of flowering plants in the northern temperate zone. We conducted robust phylogenomic analyses to untangle the evolutionary patterns and processes of the barrenwort plant (Epimedium) during the course of its radiation mainly in southern EA. A parallel-atavism-introgression hypothesis (Figure 7) was referred to in explaining the radiation of plant diversity in southern EA, which provides a potential answer to the fundamental biological question of plant radiation on Earth. Moreover, massive genomic sequencing provided an essential framework for investigating evolutionary and ecological processes in the complex natural laboratories of EA, thereby advancing our understanding of plant radiation in biodiversity hotspots.
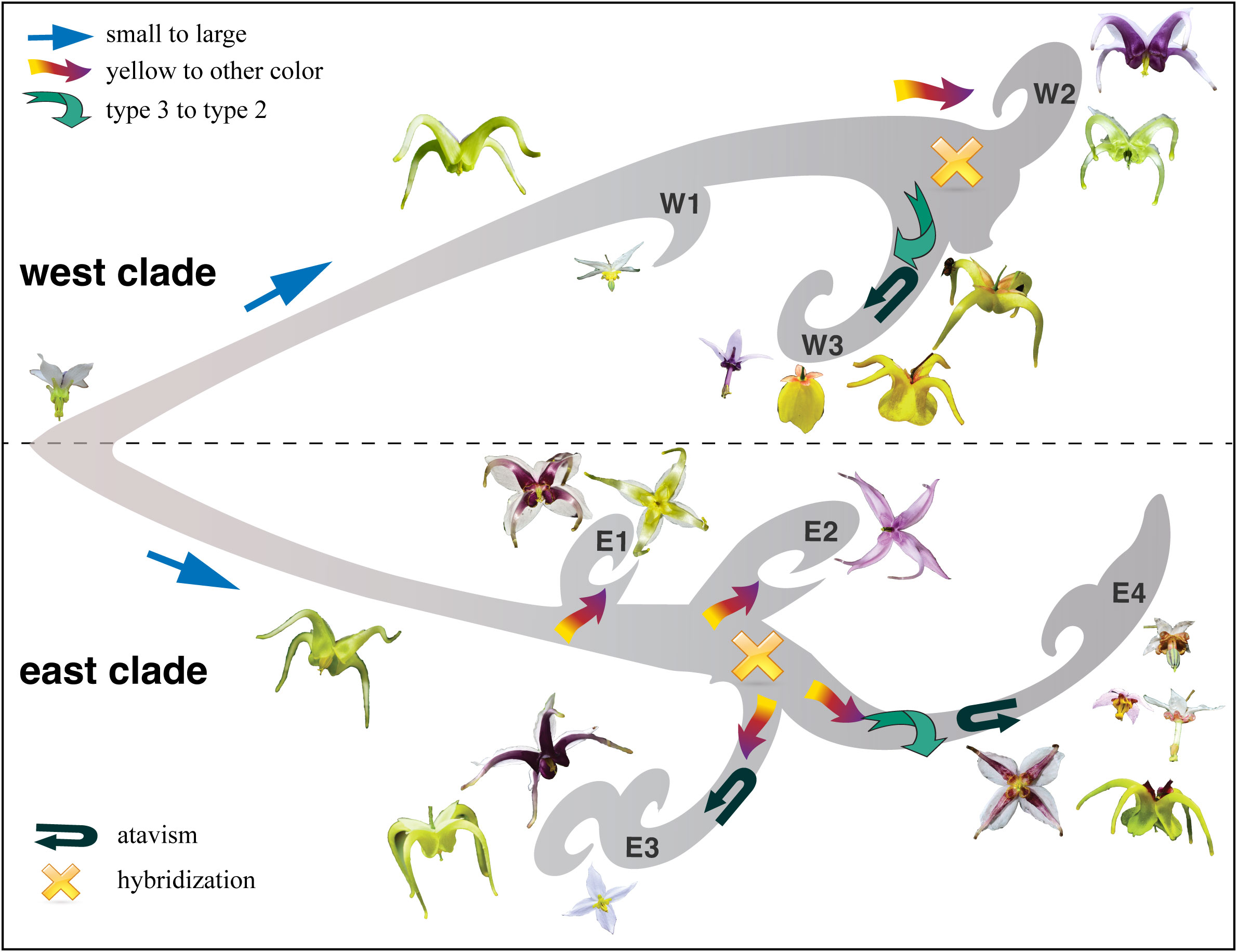
Figure 7 A schematic diagram showing the parallel-atavism-introgression pattern of Epimedium sect. Diphyllon radiated in southern EA.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Z-LN, CZ, B-LG, and Q-RL designed the research. CZ collected the samples. CZ and B-LG identified all plant samples. Z-LN, CZ, RM, and YM performed experiments and lab work. Z-LN, CZ, and RM analyzed the data. All authors contributed to the article and approved the submitted version.
This study was supported in part by grants from the Natural Sciences Foundation of China (31570211, 31770213), the Natural Sciences Foundation of Hunan Province (2019JJ40232), and the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-031).
We would like to thank the Smithsonian Institution High Performance Computing Cluster (SI/HPC, “Hydra”: https://doi.org/10.25572/SIHPC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1234148/full#supplementary-material
Aberer, A. J., Kobert, K., Stamatakis, A. (2014). ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol. Biol. Evol. 31, 2553–2556. doi: 10.1093/molbev/msu236
Alix, K., Gérard, P. R., Schwarzacher, T., Heslop-Harrison, J. (2017). Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann. Bot. 120, 183–194. doi: 10.1093/aob/mcx079
Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Baack, E. J., Rieseberg, L. H. (2007). A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17, 513–518. doi: 10.1016/j.gde.2007.09.001
Barrier, M., Baldwin, B. G., Robichaux, R. H., Purugganan, M. D. (1999). Interspecific hybrid ancestry of a plant adaptive radiation: allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Mol. Biol. Evol. 16, 1105–1113. doi: 10.1093/oxfordjournals.molbev.a026200
Beck, M. (2017) ggord: Ordination Plots with ggplot2. Available at: https://fawda123.github.io/ggord/.
Boberg, E., Alexandersson, R., Jonsson, M., Maad, J., Ågren, J., Nilsson, L. A. (2014). Pollinator shifts and the evolution of spur length in the moth-pollinated orchid Platanthera bifolia. Ann. Bot. 113, 267–275. doi: 10.1093/aob/mct217
Cao, Y. N., Comes, H. P., Sakaguchi, S., Chen, L. Y., Qiu, Y. X. (2016). Evolution of East Asia’s Arcto-tertiary relict Euptelea (Eupteleaceae) shaped by late Neogene vicariance and quaternary climate change. BMC Evolutionary Biol. 16, 1–17. doi: 10.1186/s12862-016-0636-x
Chen, Y. S., Deng, T., Zhou, Z., Sun, H. (2017). Is the East Asian flora ancient or not? Natl. Sci. Rev. 5, 920–932. doi: 10.1093/nsr/nwx156
Chifman, J., Kubatko, L. (2014). Quartet inference from SNP data under the coalescent model. Bioinformatics 30, 3317–3324. doi: 10.1093/bioinformatics/btu530
Chifman, J., Kubatko, L. (2015). Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes, site-specific rate variation, and invariable sites. J. Theor. Biol. 374, 35–47. doi: 10.1016/j.jtbi.2015.03.006
Cullings, K. (1992). Design and testing of a plant-specific PCR primer for ecological and evolutionary studies. Mol. Ecol. 1, 233–240. doi: 10.1111/j.1365-294X.1992.tb00182.x
De Smet, Y., Goetghebeur, P., Wanke, S., Asselman, P., Samain, M.-S. (2012). Additional evidence for recent divergence of Chinese Epimedium (Berberidaceae) derived from AFLP, chloroplast and nuclear data supplemented with characterisation of leaflet pubescence. Plant Ecol. Evol. 145, 73–87. doi: 10.5091/plecevo.2012.646
Donoghue, M. J., Bell, C. D., Li, J. (2001). Phylogenetic patterns in Northern Hemisphere plant geography. Int. J. Plant Sci. 162, S41–S52. doi: 10.1086/323278
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19, 11–15.
Eaton, D. A., Overcast, I. (2020). ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592–2594. doi: 10.1093/bioinformatics/btz966
Eaton, D. A., Ree, R. H. (2013). Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae). Systematic Biol. 62, 689–706. doi: 10.1093/sysbio/syt032
Ellstrand, N. C., Schierenbeck, K. A. (2000). Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl. Acad. Sci. 97, 7043–7050. doi: 10.1073/pnas.97.13.7043
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379. doi: 10.1371/journal.pone.0019379
Escudero, M., Eaton, D. A., Hahn, M., Hipp, A. L. (2014). Genotyping-by-sequencing as a tool to infer phylogeny and ancestral hybridization: a case study in Carex (Cyperaceae). Mol. Phylogenet. Evol. 79, 359–367. doi: 10.1016/j.ympev.2014.06.026
Fernández-Mazuecos, M., Mellers, G., Vigalondo, B., Sáez, L., Vargas, P., Glover, B. J. (2018). Resolving recent plant radiations: power and robustness of genotyping-by-sequencing. Systematic Biol. 67, 250–268. doi: 10.1093/sysbio/syx062
Fernández-Mazuecos, M., Vargas, P., Mccauley, R. A., Monjas, D., Otero, A., Chaves, J. A., et al. (2020). The radiation of Darwin’s giant daisies in the Galápagos Islands. Curr. Biol. 30, 4989–4998. e4987. doi: 10.1016/j.cub.2020.09.019
Grant, P. R., Grant, B. R. (2002). Unpredictable evolution in a 30-year study of Darwin's finches. Science 296, 707–711. doi: 10.1126/science.107031
Guo, B. L., He, S. Z., Zhong, G. Y., Xiao, P. G. (2007). Two new species of Epimedium (Berberidaceae) from China. J. Systematics Evol. 45, 813–821. doi: 10.1360/aps06138
Guo, B. L., Pei, L. K., Xiao, P. G. (2008). Further research on taxonomic significance of flavonoids in Epimedium (Berberidaceae). J. Systematics Evol. 46, 874–885. doi: 10.3724/SP.J.1002.2008.06172
Guo, B. L., Xiao, P. G. (1999). The flavonoids in Epimedium L. and their taxonomic significance. J. Systematics Evol. 37, 228–243.
Guo, M., Pang, X., Xu, Y., Jiang, W., Liao, B., Yu, J., et al. (2021). Plastid genome data provide new insights into the phylogeny and evolution of the genus Epimedium. J. Advanced Res. 36, 175–185. doi: 10.1016/j.jare.2021.06.020
Hall, B. K. (1984). Developmental mechanisms underlying the formation of atavisms. Biol. Rev. 59, 89–122. doi: 10.1111/j.1469-185X.1984.tb00402.x
Harrison, R. G., Larson, E. L. (2014). Hybridization, introgression, and the nature of species boundaries. J. Heredity 105, 795–809. doi: 10.1093/jhered/esu033
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hughes, C. E., Atchison, G. W. (2015). The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytol. 207, 275–282. doi: 10.1111/nph.13230
Lamichhaney, S., Berglund, J., Almén, M. S., Maqbool, K., Grabherr, M., Martinez-Barrio, A., et al. (2015). Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375. doi: 10.1038/nature14181
Lee-Yaw, J. A., Grassa, C. J., Joly, S., Andrew, R. L., Rieseberg, L. H. (2019). An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytol. 221, 515–526. doi: 10.1111/nph.15386
Li, Y. X., Quan, Q. M., Sun, G. L. (2009). Effect of floral morphology on fruit set in Epimedium sagittatum (Berberidaceae). Plant systematics Evol. 279, 51–58. doi: 10.1007/s00606-009-0145-0
Linder, C. R., Rieseberg, L. H. (2004). Reconstructing patterns of reticulate evolution in plants. Am. J. Bot. 91, 1700–1708. doi: 10.3732/ajb.91.10.1700
Liu, T. S. (1988). Floristic relationships between eastern Asia and eastern North America (Taipei: National Taiwan University).
Liu, X. (2021). Phylogenetic study of Epimedium genus based on chloroplast genome and rDNA sequences (Beijing: Peking Union Medical College).
Liu, Y., Li, D., Zhang, Q., Song, C., Zhong, C., Zhang, X., et al. (2017). Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 215, 877–890. doi: 10.1111/nph.14607
Lu, R. S., Li, P., Qiu, Y. X. (2017). The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02054
Maddison, W. P., Maddison, D. R. (2021) Mesquite: a modular system for evolutionary analysis. Available at: http://www.mesquiteproject.org.
Mallet, J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. doi: 10.1016/j.tree.2005.02.010
Milne, R. I., Abbott, R. J. (2002). The origin and evolution of Tertiary relict floras. Adv. Botanical Res. 38, 281–314. doi: 10.1016/S0065-2296(02)38033-9
Minh, B. Q., Nguyen, M., Von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mitsui, Y., Chen, S. T., Zhou, Z. K., Peng, C. I., Deng, Y. F., Setoguchi, H. (2008). Phylogeny and biogeography of the genus Ainsliaea (Asteraceae) in the Sino-Japanese region based on nuclear rDNA and plastid DNA sequence data. Ann. Bot. 101, 111–124. doi: 10.1093/aob/mcm267
Monnet, C., De Baets, K., Klug, C. (2011). Parallel evolution controlled by adaptation and covariation in ammonoid cephalopods. BMC evolutionary Biol. 11, 115. doi: 10.1186/1471-2148-11-115
Morales-Briones, D. F., Liston, A., Tank, D. C. (2018). Phylogenomic analyses reveal a deep history of hybridization and polyploidy in the Neotropical genus Lachemilla (Rosaceae). New Phytol. 218, 1668–1684. doi: 10.1111/nph.15099
Olave, M., Meyer, A. (2020). Implementing large genomic single nucleotide polymorphism data sets in phylogenetic network reconstructions: a case study of particularly rapid radiations of cichlid fish. Systematic Biol. 69, 848–862. doi: 10.1093/sysbio/syaa005
Pan, J., Chen, H., Guo, B., Liu, C. (2017). Understanding the molecular mechanisms underlying the effects of light intensity on flavonoid production by RNA-seq analysis in Epimedium pseudowushanense BL Guo. PloS One 12, e0182348. doi: 10.1371/journal.pone.0182348
Penjor, T., Mimura, T., Kotoda, N., Matsumoto, R., Nagano, A. J., Honjo, M. N., et al. (2016). RAD-Seq analysis of typical and minor Citrus accessions, including Bhutanese varieties. Breed. Sci. 66, 797–807. doi: 10.1270/jsbbs.16059
Pennington, R. T., Lavin, M., Särkinen, T., Lewis, G. P., Klitgaard, B. B., Hughes, C. E. (2010). Contrasting plant diversification histories within the Andean biodiversity hotspot. Proc. Natl. Acad. Sci. 107, 13783–13787. doi: 10.1073/pnas.1001317107
Qi, Z. C., Yu, Y., Liu, X., Pais, A., Ranney, T., Whetten, R., et al. (2015). Phylogenomics of polyploid Fothergilla (Hamamelidaceae) by RAD-tag based GBS—insights into species origin and effects of software pipelines. J. Systematics Evol. 53, 432–447. doi: 10.1111/jse.12176
Qian, H. (2002). Environmental determinants of latitudinal diversity gradient in North America mammals: species age, range size and species richness. Ecol. Lett. 5, 693–699. doi: 10.1111/geb.13075
R Development Core Team. (2016). R: A language and environment for statistical computing. (Vienna: R foundation for statistical computing).
Reichelt, N., Wen, J., Pätzold, C., Appelhans, M. S. (2021). Target enrichment improves phylogenetic resolution in the genus Zanthoxylum (Rutaceae) and indicates both incomplete lineage sorting and hybridization events. Ann. Bot. 128, 497–510. doi: 10.1093/aob/mcab092
Revell, L. J. (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. doi: 10.1111/j.2041-210X.2011.00169.x
Rieseberg, L. H., Kim, S. C., Randell, R. A., Whitney, K. D., Gross, B. L., Lexer, C., et al. (2007). Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129, 149–165. doi: 10.1007/s10709-006-9011-y
Rose, J. P., Toledo, C. A., Lemmon, E. M., Lemmon, A. R., Sytsma, K. J. (2021). Out of sight, out of mind: widespread nuclear and plastid-nuclear discordance in the flowering plant genus Polemonium (Polemoniaceae) suggests widespread historical gene flow despite limited nuclear signal. Systematic Biol. 70, 162–180. doi: 10.1093/sysbio/syaa049
Sakaguchi, S., Qiu, Y. X., Liu, Y. H., Qi, X. S., Kim, S. H., Han, J. Y., et al. (2012). Climate oscillation during the Quaternary associated with landscape heterogeneity promoted allopatric lineage divergence of a temperate tree Kalopanax septemlobus (Araliaceae) in East Asia. Mol. Ecol. 21, 3823–3838. doi: 10.1111/j.1365-294X.2012.05652.x
Seehausen, O. (2004). Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207. doi: 10.1016/j.tree.2004.01.003
Seehausen, O. (2006). African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B: Biol. Sci. 273, 1987–1998. doi: 10.1098/rspb.2006.3539
Sheng, M. Y., Chen, Q. F. (2007). Karyotype analysis of six Epimedium species native to Guizhou, China. Guihaia 27, 440–443. doi: 10.1016/S1872-2075(07)60055-7
Sheng, M. Y., Chen, Q. F., Wang, L. J., Tian, X. J. (2011). Hybridization among Epimedium (Berberidaceae) species native to China. Scientia Hortic. 128, 342–351. doi: 10.1016/j.scienta.2011.01.020
Sheng, M. Y., Wang, L. J., Tian, X. J. (2010). Karyomorphology of eighteen species of genus Epimedium (Berberidaceae) and its phylogenetic implications. Genet. Resour. Crop Evol. 57, 1165–1176. doi: 10.1007/s10722-010-9556-6
Sidlauskas, B. (2008). Continuous and arrested morphological diversification in sister clades of characiform fishes: a phylomorphospace approach. Evolution: Int. J. Organic Evol. 62, 3135–3156. doi: 10.1111/j.1558-5646.2008.00519.x
Smith, S. A., O’meara, B. C. (2012). treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28, 2689–2690. doi: 10.1093/bioinformatics/bts492
Solís-Lemus, C., Ané, C. (2016). Inferring phylogenetic networks with maximum pseudolikelihood under incomplete lineage sorting. PLoS Genet. 12, e1005896. doi: 10.1371/journal.pgen.1005896
Solís-Lemus, C., Bastide, P., Ané, C. (2017). PhyloNetworks: a package for phylogenetic networks. Mol. Biol. Evol. 34, 3292–3298. doi: 10.1093/molbev/msx235
Soltis, P. S., Soltis, D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588. doi: 10.1146/annurev.arplant.043008.092039
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stearn, W. T. (1993a). New large-flowered Chinese species of Epimedium (Berberidaceae). Kew Magazine 10, 178–184.
Stearn, W. T. (1993b). The small-flowered Chinese species of Epimedium (Berberidaceae). Kew Bull. 48 (4), 807–813. doi: 10.2307/4118862
Stearn, W. T. (1995). New Chinese taxa of epimedium (Berberidaceae) from Sichuan. Curtis's Botanical Magazine 12, 15–25.
Stearn, W. T. (2002). Genus Epimedium and other herbaceous Berberidaceae (London: The Royal Botanic Gardens, Kew).
Sun, Y., Fung, K. P., Leung, P. C., Shaw, P. C. (2005). A phylogenetic analysis of Epimedium (Berberidaceae) based on nuclear ribosomal DNA sequences. Mol. Phylogenet. Evol. 35, 287–291. doi: 10.1016/j.ympev.2004.12.014
Suzuki, K. (1983). Breeding system and crossability in Japanese Epimedium (Berberidaceae). botanical magazine = Shokubutsu-gaku-zasshi 96, 343–350. doi: 10.1007/BF02488178
Suzuki, K. (1984). Pollination system and its significance on isolation and hybridization in Japanese Epimedium (Berberidaceae). botanical magazine= Shokubutsu-gaku-zasshi 97, 381–396. doi: 10.1007/BF02488670
Takhtajan, A. (1969). Flowering plants: Origin and dispersal (Edinburgh and London: Oliver and Boyd).
Takhtajan, A. L. (1986). Floristic regions of the world (Berkeley, California: University of California press).
Tiffney, B. H. (1985). Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arnold Arboretum 66, 73–94.
Tkach, N. V., Hoffmann, M. H., Röser, M., Korobkov, A. A., Von Hagen, K. B. (2008). Parallel evolutionary patterns in multiple lineages of arctic Artemisia L. (Asteraceae). Evolution 62, 184–198. doi: 10.1111/j.1558-5646.2007.00270.x
Vargas, O. M., Ortiz, E. M., Simpson, B. B. (2017). Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 214, 1736–1750. doi: 10.1111/nph.14530
Vriesendorp, B., Bakker, F. T. (2005). Reconstructing patterns of reticulate evolution in angiosperms: what can we do? Taxon 54, 593–604. doi: 10.2307/25065417
Wagner, C. E., Harmon, L. J., Seehausen, O. (2012). Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487, 366–369. doi: 10.1038/nature11144
Wang, H. X., Morales-Briones, D. F., Moore, M. J., Wen, J., Wang, H. F. (2021). A phylogenomic perspective on gene tree conflict and character evolution in Caprifoliaceae using target enrichment data, with Zabelioideae recognized as a new subfamily. J. Systematics Evol. 59, 897–914. doi: 10.1111/jse.12745
Wen, J. (1999). Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Systematics 30, 421–455. doi: 10.1146/annurev.ecolsys.30.1.421
Wen, J., Ickert-Bond, S., Nie, Z. L., Li, R. (2010). “Timing and modes of evolution of eastern Asian-North American biogeographic disjunctions in seed plants,” in Darwin’s heritage today: Proceedings of the Darwin 2010 Beijing international conference. Eds. Long, M., Gu, H., Zhou, Z. (Beijing: Higher Education Press), 252–269.
Wen, J., Zhang, J. Q., Nie, Z. L., Zhong, Y., Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 5. doi: 10.3389/fgene.2014.00004
Whittall, J. B., Hodges, S. A. (2007). Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447, 706–709. doi: 10.1038/nature05857
Wu, Z. Y. (1988). Hengduan Mountains: A natural museum of seed plants in China (Beijing: Science Press).
Wu, Z. Y., Wu, S. G. (1998). “A proposal for a new floristic kingdom (realm) - the E. Asiatic kingdom, its delineation and characteristics,” in Floristic characteristics and diversity of East Asian plants. Eds. Zhang, A. L., Wu, S. G. (Beijing: China Higher Education Press), 3–42.
Xiang, Q. Y., Zhang, W. H., Ricklefs, R. E., Qian, H., Chen, Z. D., Wen, J., et al. (2004). Regional differences in rates of plant speciation and molecularevolution: a comparison between eastern Asia and eastern North America. Evolution 58, 2175–2184. doi: 10.1111/j.0014-3820.2004.tb01596.x
Xu, Y., Li, Z., Wang, Y., Huang, H. (2007). Allozyme diversity and population genetic structure of three medicinal Epimedium species from Hubei. J. Genet. Genomics 34, 56–71. doi: 10.1016/S1673-8527(07)60007-1
Yang, H., Wei, C. L., Liu, H. W., Wu, J. L., Li, Z. G., Zhang, L., et al. (2016). Genetic divergence between Camellia sinensis and its wild relatives revealed via genome-wide SNPs from RAD sequencing. PLoS One 11, e0151424. doi: 10.1371/journal.pone.0151424
Ying, T. S. (2002). Petal evolution and distribution patterns of Epimedium Linn.(Berberidaceae). J. Systematics Evol. 40, 481. doi: 10.1088/1009-1963/11/5/313
Zhang, Y. J., Dang, H. S., Li, J. Q., Wang, Y. (2014a). The Epimedium wushanense (Berberidaceae) species complex, with one new species from Sichuan, China. Phytotaxa 172, 039–045. doi: 10.11646/phytotaxa.172.1.5
Zhang, Y. J., Dang, H. S., Meng, A. P., Li, J. Q., Li, X. D. (2008). Karyomorphology of Epimedium (Berberidaceae) and its phylo-genetic implications. Caryologia 61, 283–293. doi: 10.1080/00087114.2008.10589640
Zhang, D. G., Deng, T., King, C. K., Zhang, J. W., Nie, Z. L., Sun, H. (2015). Epimedium tianmenshanensis (Berberidaceae), a new species from central China. Phytotaxa 222, 33–43. doi: 10.11646/phytotaxa.222.1.3
Zhang, Z., Fan, L., Yang, J., Hao, X., Gu, Z. (2006). Alkaloid polymorphism and ITS sequence variation in the Spiraea japonica complex (Rosaceae) in China: traces of the biological effects of the Himalaya-Tibet Plateau uplift. Am. J. Bot. 93, 762–769. doi: 10.3732/ajb.93.5.762
Zhang, Y. J., Li, J. Q. (2009). A new species of Epimedium (Berberidaceae) from Hubei, China. Novon: A J. Botanical Nomenclature 19, 567–569. doi: 10.3417/2007161
Zhang, Y. J., Li, J. Q., Wang, Y., Liang, Q. (2021). Taxonomy of Epimedium (Berberidaceae) with special reference to chinese species. Chin. Herbal Medicines 14, 20–35. doi: 10.1016/j.chmed.2021.12.001
Zhang, C., Peng, Z. B., Nie, Z. L., Zhang, D. G., Li, K. G., Meng, Y. (2018). Chromosome numbers and karyotypes of 18 species of Epimedium sect. Diphyllon (Berberidaceae) from central China. Caryologia 71, 428–437. doi: 10.1080/00087114.2018.1499482
Zhang, M. L., Uhink, C. H., Kadereit, J. W. (2007). Phylogeny and biogeography of Epimedium/Vancouveria (Berberidaceae): Western North American-East Asian disjunctions, the origin of European mountain plant taxa, and East Asian species diversity. Systematic Bot. 32, 81–92. doi: 10.1600/036364407780360265
Zhang, Y. J., Yang, L. L., Chen, J. J., Sun, W., Wang, Y. (2014b). Taxonomic and phylogenetic analysis of Epimedium L. based on amplified fragment length polymorphisms. Scientia Hortic. 170, 284–292. doi: 10.1016/j.scienta.2014.02.025
Zhang, Y. J., Zhang, S. J., Dang, H. S., Zheng, B., Li, J.-Q., Wang, Y. (2016). Epimedium xichangense (Berberidaceae), a new species from Sichuan, China. Phytotaxa 263, 286–290. doi: 10.11646/phytotaxa.263.3.9
Keywords: East Asia, evolutionary radiation, atavism, introgression, parallel evolution, Epimedium, Epimedium sect. Diphyllon
Citation: Zhang C, Meng R, Meng Y, Guo B-L, Liu Q-R and Nie Z-L (2023) Parallel evolution, atavism, and extensive introgression explain the radiation of Epimedium sect. Diphyllon (Berberidaceae) in southern East Asia. Front. Plant Sci. 14:1234148. doi: 10.3389/fpls.2023.1234148
Received: 03 June 2023; Accepted: 29 September 2023;
Published: 17 October 2023.
Edited by:
Qiu-Yun(Jenny) Xiang, North Carolina State University, United StatesCopyright © 2023 Zhang, Meng, Meng, Guo, Liu and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan-Ru Liu, bGl1cXVhbnJ1QGJudS5lZHUuY24=; Ze-Long Nie, bmllekBqc3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.