
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 September 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1233996
This article is part of the Research Topic Crop Improvement in the Era of Next-Generation Sequencing View all 14 articles
 Harsh Raman1*
Harsh Raman1* Rosy Raman1
Rosy Raman1 Niharika Sharma2†
Niharika Sharma2† Xiaobo Cui3†
Xiaobo Cui3† Brett McVittie1†
Brett McVittie1† Yu Qiu1†
Yu Qiu1† Yuanyuan Zhang3
Yuanyuan Zhang3 Qiong Hu3
Qiong Hu3 Shengyi Liu3
Shengyi Liu3 Nelson Gororo4
Nelson Gororo4Pod shatter is a trait of agricultural relevance that ensures plants dehisce seeds in their native environment and has been subjected to domestication and selection for non-shattering types in several broadacre crops. However, pod shattering causes a significant yield reduction in canola (Brassica napus L.) crops. An interspecific breeding line BC95042 derived from a B. rapa/B. napus cross showed improved pod shatter resistance (up to 12-fold than a shatter-prone B. napus variety). To uncover the genetic basis and improve pod shatter resistance in new varieties, we analysed F2 and F2:3 derived populations from the cross between BC95042 and an advanced breeding line, BC95041, and genotyped with 15,498 DArTseq markers. Through genome scan, interval and inclusive composite interval mapping analyses, we identified seven quantitative trait loci (QTLs) associated with pod rupture energy, a measure for pod shatter resistance or pod strength, and they locate on A02, A03, A05, A09 and C01 chromosomes. Both parental lines contributed alleles for pod shatter resistance. We identified five pairs of significant epistatic QTLs for additive x additive, additive dominance and dominance x dominance interactions between A01/C01, A03/A07, A07/C03, A03/C03, and C01/C02 chromosomes for rupture energy. QTL effects on A03/A07 and A01/C01 were in the repulsion phase. Comparative mapping identified several candidate genes (AG, ABI3, ARF3, BP1, CEL6, FIL, FUL, GA2OX2, IND, LATE, LEUNIG, MAGL15, RPL, QRT2, RGA, SPT and TCP10) underlying main QTL and epistatic QTL interactions for pod shatter resistance. Three QTLs detected on A02, A03, and A09 were near the FUL (FRUITFULL) homologues BnaA03g39820D and BnaA09g05500D. Focusing on the FUL, we investigated putative motifs, sequence variants and the evolutionary rate of its homologues in 373 resequenced B. napus accessions of interest. BnaA09g05500D is subjected to purifying selection as it had a low Ka/Ks ratio compared to other FUL homologues in B. napus. This study provides a valuable resource for genetic improvement for yield through an understanding of the genetic mechanism controlling pod shatter resistance in Brassica species.
Plants have evolved vivid mechanisms for survival and fitness across various ecological niches. In the wild, plants dehisce their fruits and disperse seeds to ensure the multiplication and adaptation of their progenies and confront challenges posed by climatic and ecological vagaries. Seeds of the Brassicaceae family members are enclosed in a silique (pod), which consists of two congenitally fused carpels (valves); each is separated with a thin layer called a pseudo-septum or replum (Figure 1) (Bowman et al., 1999). Both valves and replum are differentiated with valve margins where pod dehiscence and seed abscission occur via pod drop and seed shattering, possibly by similar molecular mechanisms (Balanzà et al., 2016). Pod drop – a phenomenon where a whole fruit (silique) drops on the ground, is a common problem in some canola production regions, particularly Canada. As the pod matures physiologically, valves detach from the replum, resulting in pod dehiscence (Figure S1A) and the seeds attached to the replum with a funiculus fall to the ground (Figure S1C). Pod dehiscence occurs via the dehiscence zone formation at the valve margins by two layers: a lignification layer of 1-2 thick and rigid cells and the separation (also called abscission) layer of iso-diametrically shaped cells, separating the valve from the replum (Spence et al., 1996; Rajani and Sundaresan, 2001; Dinneny and Yanofsky, 2005). At maturity, cells in the separation layer degrade by polygalacturonase, cellulase, and mannanase enzymes (Ogawa et al., 2009). Shattering occurs when the abscission force becomes more significant than the binding force of the pod valve (Lee et al., 2017). External influences such as wind velocity, machinery, and high temperatures further escalate pod shattering in brassicas.
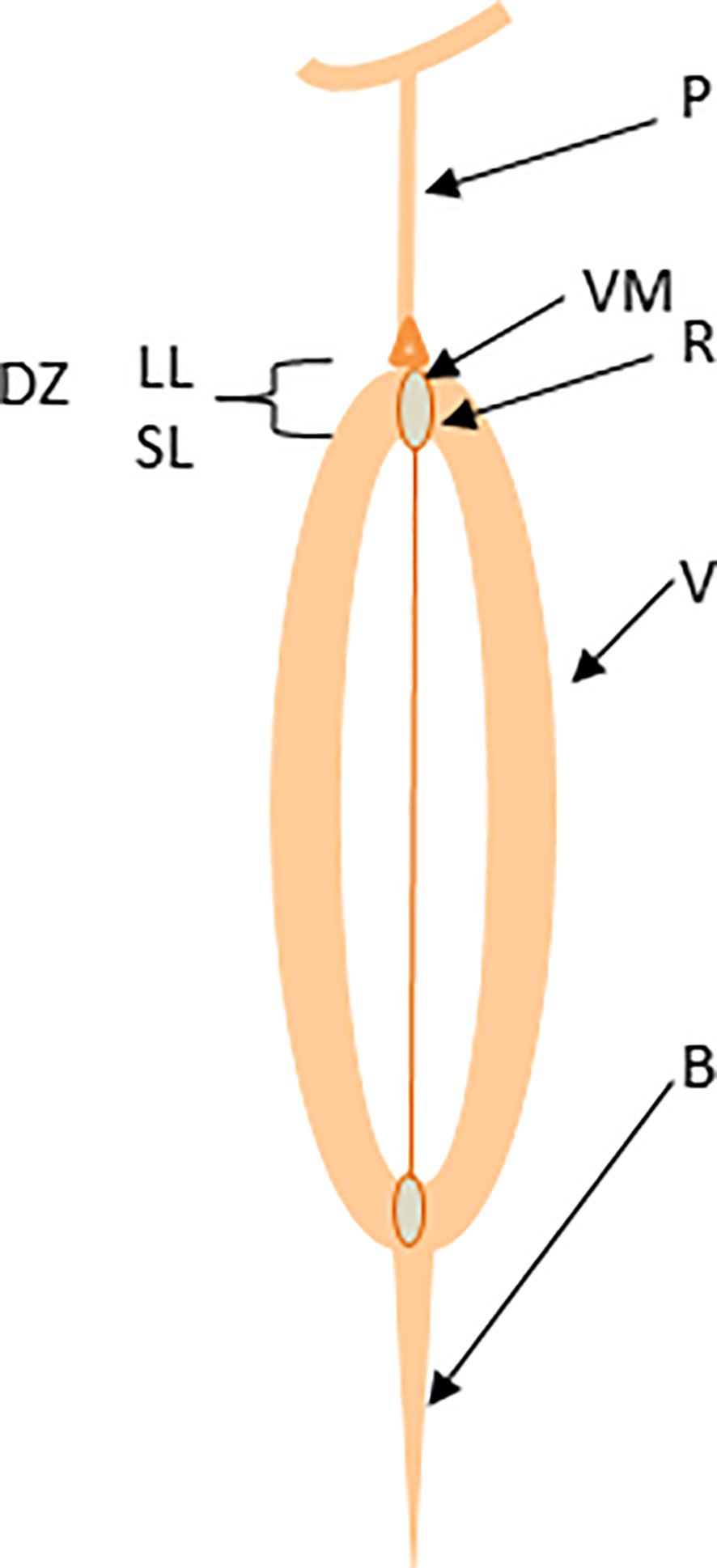
Figure 1 Schematic representation of canola pod structure. The valve (V), dehiscence zone (DZ), separation layer (SL), lignification layer (LL), valve margin (VM), replum (R), and beak (B).
Molecular mechanisms underlying pod dehiscence are well-dissected in a model plant, Arabidopsis thaliana - a distant relative of Brassica napus L. At least thirteen genes that are responsible for pod dehiscence in Arabidopsis have been identified, such as MADS-box genes: SHATTERPROOF1 (SHP1), SHATTERPROOF2 (SHP2) and FRUITFULL (FUL); Basic-loop-helix genes: INDEHISCENT (IND), ALCATRAZ (ALC) and SPATULA (SPT); REPLUMLESS (RPL) and APETALA2 (AP2), ARABIDOPSIS DEHISCENCE ZONE POLYGALACTUROSE1 (ADPG1), ADPG2, a C2H2 zinc finger transcription factors JAGGED (JAG) and BnLATE FLOWERING (BnLATE); NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), ENDO-BETA-MANNANASE7 (MAN7), and CELLULASE6 (Ferrandiz et al., 2000; Liiljegren et al., 2000; Rajani and Sundaresan, 2001; Roeder et al., 2003; Sorefan et al., 2009; He et al., 2018; Li et al., 2021). Different genes involved in auxin, gibberellin and cytokinin biosynthesis also regulate pod development and dehiscence (Sorefan et al., 2009; Arnaud et al., 2010; Marsch-Martínez et al., 2012).
Canola, the second most crucial oilseed crop after soybean, contributes about 13-16% of global vegetable oil production. The allotetraploid canola genome (2n = 4× = 38, genome AACC) originated about 7,500 years ago via ancient hybridisation events between two diploid progenitors Brassica species, B. rapa (2n = 2× = 20, AA genome) and B. oleracea (2n = 2× = 18, CC genome) (Chalhoub et al., 2014; Lu et al., 2019). However, seed shattering (commonly referred to as pod-shattering) is a universal constraint in canola production, and in the literature, none of the domesticated accessions of B. napus is reported to be ‘completely’ resistant to pod shattering. Generally, canola pods are highly sensitive to pre-mature shattering, significantly reducing yield. The seed loss varies from 8 to 70% across environments depending on genotypic attributes (canopy architecture, resistance to lodging and diseases), method of harvesting (windrow/direct heading), and time of harvesting (early, optimal time vs late) and environmental conditions at the time of harvest (MacLeod, 1981; Price et al., 1996; Child et al., 1998; Vera et al., 2007; de la Pasture, 2018). Shattered seeds grow in the field at a much higher rate (60x) than those sowed initially (Figure S1) and become a weed in the next crop; hence must be controlled (Wang et al., 2007).
To overcome pod-shattering, the majority of broadacre canola varieties are harvested by windrowing/swathing - a practice of cutting plants at physiological maturity (50 to 60% seed colour change from green to dark brown, red or black) and leaving them in the field before threshing with a combine harvester. This practice can also lead to significant losses from seed shattering, mainly when not accomplished at the ‘right’ time. The window for windrowing is often small and subjected to labour and combined harvester availability and congenial weather conditions. High temperatures, high-velocity winds, rainfall, and hailstorm events significantly impact canola seed yield and oil content. High yield is essential for meeting global demands for healthy vegetable oil, protein for animal feed, and canola growers for return on their investment.
Understanding the genetic determinants and novel alleles underlying this domestication trait would provide an improved genetics-based solution to reduce yield loss in B. napus. The functionality of some of Arabidopsis pod dehiscence genes has also been demonstrated in Brassica species via overexpression, RNAi, gene editing, and induced mutation studies (Ostergaard et al., 2006; Kord et al., 2015; Lawrenson et al., 2015; Braatz et al., 2018a; Braatz et al., 2018b; Stephenson et al., 2019; Li et al., 2021). Recently, it has also been shown that miR319-targeted TEOSINTE BRANCHED 1, CYCLOIDEA, and PROFEERATIN CELL NUCLEAR ANTIGEN BINDING FACTOR (TCPs) inhibit pod elongation and dehiscence via regulation of FUL expression in A. thaliana and B. napus (Cao et al., 2022). Although the network of pod dehiscence genes has been investigated in Arabidopsis, their expression level has not been fine-tuned in commercial canola varieties with genetic modification approaches, except in POD GURAD varieties where TILLING has been deployed only in the BASF canola breeding program (Laga et al., 2008). In fact, ectopic (over-) expression of FUL and SHP genes led to indehiscent pods due to the non-lignification of cells between the valve and replum and the absence of dehiscence zone formation (Ferrandiz et al., 2000; Liiljegren et al., 2000; Ostergaard et al., 2006).
Previous research has shown a limited range of genetic variation for pod shatter resistance in B. napus (Morgan et al., 2007; Raman et al., 2014). However, a wide range of genetic variation for pod shattering is observed in diploid and amphidiploid species of Brassica, such as B. rapa, B. juncea (2n = 4× = 36, AABB), and B. carinata (2n = 4× = 34, BBCC) (Kadkol et al., 1984; Kadkol et al., 1985; Raman et al., 2017). In a previous study, Raman et al. (2014) reported that pod shatter resistance could improve up to 12-fold in a shatter-prone variety of B. napus via the introgression of resistant alleles from B. rapa. To uncover the genetic basis underlying seed shattering in this interspecific source, we investigated an F2 mapping population and its F2:3 progenies derived from a cross between B. napus (BC95041) and B. rapa/B. napus (BC95042). We further identified epistatic quantitative trait loci (QTLs) for additive × additive, additive dominance, and dominance × dominance interactions. Candidate genes and their sequence variants in parental lines underlying QTL regions for pod shatter resistance were identified, which could regulate variation in pod shatter resistance.
An interspecific line derived B. rapa/B. napus with the highest pod rupture energy (RE), BC95042 (shatter resistant with high RE (Raman et al., 2014)) was crossed with the advanced breeding lines of B. napus, BLN3303 (BC95041, maternal parent, shatter prone with low RE). This study utilised an F2 population comprising 203 individuals generated from the self-pollination of a single F1 cross from BC95041/BC94042. Each F2 line was selfed to generate an F2:3 population for confirming phenotypes.
The two parental lines and their F2 population of 203 plants were grown in 2021 in white plastic pots (Garden City Plastics, NSW, Australia)) under birdcage conditions at the Wagga Wagga Agricultural Institute, New South Wales, Australia. The cultivation of canola plants followed standard management practices. Plants were watered thrice per week, fertilised weekly using in-line liquid fertilisers, and protected from blackleg and sclerotinia diseases by applications of Prosaro® 420 SC and Aviator fungicides (Bayer Crop Sciences, Australia) and aphids using chemicals recommended in Australia. Day to flowering was recorded daily for each F2 plant. To avoid outcrossing and get pure F3 progenies, all F2 plants were bagged with perforated pollination bags before flower initiation, leaving the primary stem out for the natural pod development for shatter testing. Ten pods were collected from each line at maturity (BBCH scale 95) in the 50 mL plastic tubes containing a silica sachet, as detailed in our previous study (Raman et al., 2014). Pods were desiccated in a dehydrator (G. T. D. Pty. Ltd., Australia) at 40°C for 48 hours to reduce variation due to moisture content and further tested for variation in pod rupture energy. For validation, 40 F2:3 families (20 high rupture energy and 20 low rupture energy) and parents were grown in pots in 2016 under birdcage conditions and tested with a pendulum test described earlier (Raman et al., 2014). The phenotypic means for each genotype were used for further genetic analysis. A pair-wise correlation between rupture energy and pod length in F2 and F2:3 populations was calculated. The rupture energy of five pods of each F2 plant was averaged and used for QTL analysis.
Young leaf tissue of the field-grown plants was collected from each line in a 96-well format. The tissue was frozen immediately and kept at - 80°C until used for DNA isolation. Tissue was ground in liquid nitrogen and extracted for DNA using a method described by Raman et al. (2005). DNA concentration was determined by a Qubit fluorometer and Qubit dsDNA broad-range assay kit according to the manufacturer’s recommendation. DNA quality was checked on the Tris-Acetate-EDTA buffered 0.8% agarose gel. The F2 population and parental lines were genotyped with the genotyping-by-sequencing-based DArTseq marker approach (Raman et al., 2014) using the HiSeq 2500 system (Illumina, USA) at the DArT P/L, University of Canberra, Bruce, Australia. We considered only high-quality DArTseq markers, which included SNPs (single nucleotide polymorphism) and in-sillco presence-absence markers, having BLAST alignments (E-value: 5e-5) and minimum sequence identity of 90% with the reference B. napus cv. Darmor-bzh v 4.1.
The linkage map of the F2 population was constructed using DArT P/L’s OCD MAPPING program (Petroli et al., 2012), as described previously (Raman et al., 2017). The association between markers and rupture energy was tested using linear marker regression, Fisher’s exact test, and the X2 test. We applied the additive, dominant and recessive models and full scan permutation with 1000 iterations for the genome scan. Haplotype blocks (HB) were detected using 0.98 upper confidence and 0.7 lower bound recombination value at threshold 0.01, Expectation maximization algorithm (EM) iteration 1,000 and EM convergence tolerance value of 0.00010 (Gabriel et al., 2002). P values for haplotyping association test were determined using 10,000 iterated permutations in the SVS package (Golden Helix, Bozeman, USA). We used binary data of contrasting 141 F2 phenotypes for resistance or sensitivity to shattering (Table S5a) for haplotype analysis. Manhattan plots were generated in the SVS package (Golden Helix, Bozeman, USA).
QTL mapping was performed by single interval mapping (IM), inclusive composite interval mapping (ICIM-ADD) of additive and dominant QTL, and inclusive composite interval mapping of epistatic QTL (ICIM-EPI) functions implemented in the QTL IciMapping v4.1 (www.isbreeding.net). The threshold logarithm of odds (LOD) value was determined by a permutation test involving 1,000 runs at a significance level of P = 0.05. Threshold P values for ICIM and IM for rupture energy were 3.07 and 3.25, respectively. While for pod length, threshold P values for ICIM and IM are 2.66 and 1.78, respectively. QTLs having LOD values more than the estimated threshold were declared as significant. LOD score greater than 2.5 but less than estimated threshold P values were termed suggestive QTL. The phenotypic variance explained (% PVE) and the additive effects of QTLs were directly derived from the QTL analysis outputs files. For digenic epistatic QTL interactions, LOD threshold values for each trait were estimated after 1,000 permutations using a type I error = 0.05. Epistatic effect QTLs were analysed using ICIM-EPI at the threshold LOD 4.87. Favorable parental alleles that enhance the trait expression were identified using an additive effect’s direction (+ and -ve).
The physical map positions of significant markers associated with pod shatter resistance were obtained using the reference B. napus cv Darmor-bzh genome by BlastN (Altschul et al., 1990) searches, as detailed in Raman et al. (2014). We also used the BnaOmics platform (https://bnaomics.ocri-genomics.net/) that integrates pan-genome and multi-omics data of B. napus (Cui et al., 2023) to search candidate genes. The only single top hit with the cutt-of E value of 1E-5 was considered for identifying syntenic region underlying candidate genes. B. napus annotated genes which were mapped within the marker intervals with ICIM/ICIM-EPI, were assumed candidate genes. The candidates that map within 500 kb from the significant markers identified with genome scan approaches were also identified. Genes involved in the pod shatter trait of Arabidopsis (Table S14) were used to search the corresponding copies in B. napus, with an e-value of 1e-10.
Arabidopsis thaliana genic and protein sequences of AT5G60910 from the Arabidopsis Information Resource (TAIR) were used to search the homologues in B. napus using TBLASTN and BLASTP (B. napus cv. Darmor-bzh genome, versions 4.1; http://www.genoscope.cns.fr, and the pan-genome) (Cui et al., 2023).
We used the Geneious tree builder pipeline to generate a Neighbour-Joining phylogenetic tree of DNA sequences from B. rapa, B. oleracea and B. napus for FUL (Figure 2) and FUL-Like genes (Figure S5). Sequences were aligned with global alignment with free end gaps, Blosum62 cost matrix, and Jukes-Cantor genetic distance model, implemented in the Geneious prime package (https://www.geneious.com). A. thaliana FUL gene was used as an outgroup to verify functional divergence. The synonymous substitution rate (Ks), non-synonymous substitution rate (Ka), and Ka/Ks ratio were calculated with SNPGenie (https://github.com/chasewnelson/SNPGenie).
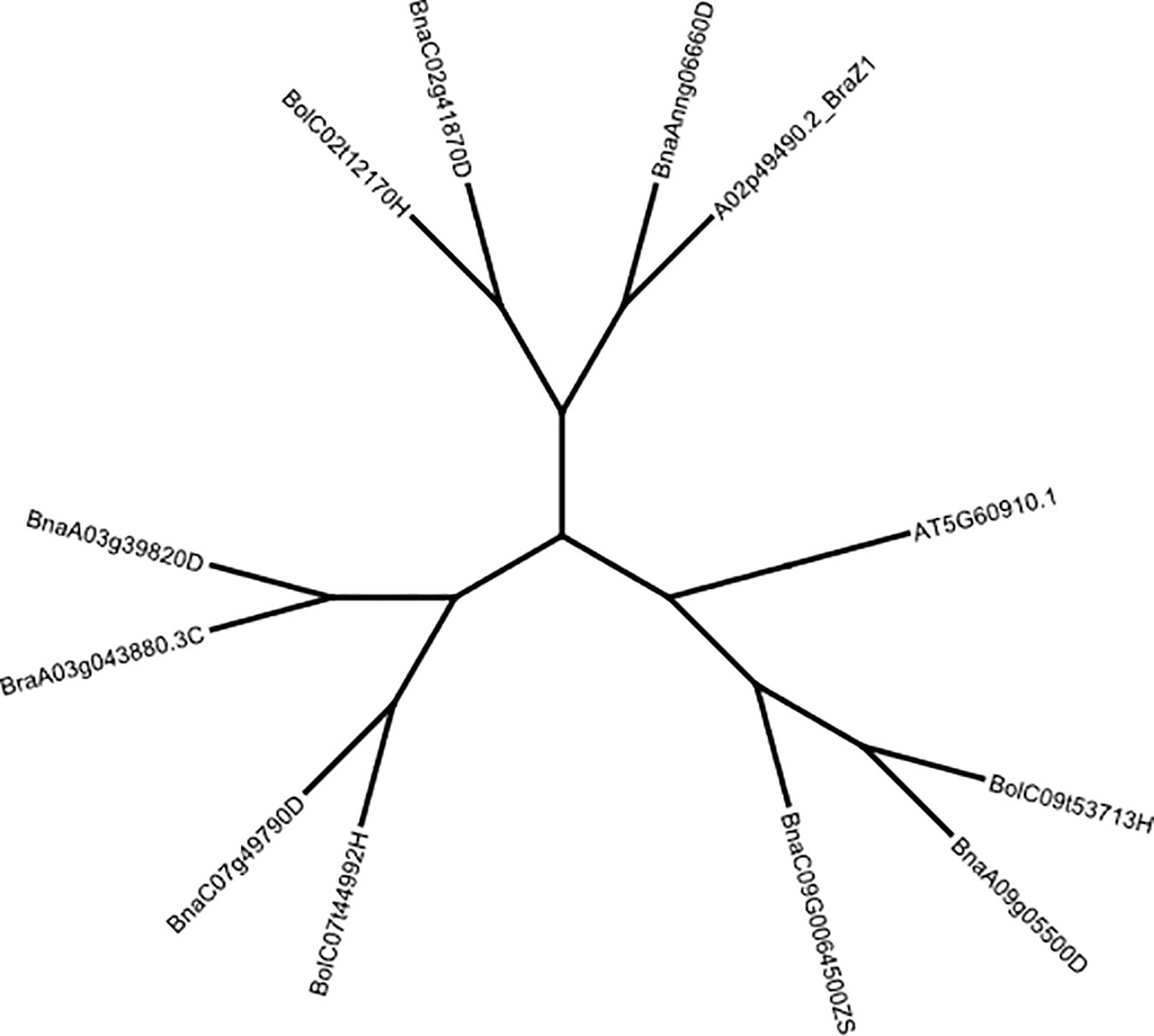
Figure 2 Neighbour-joining tree showing the grouping of B. rapa, B oleracea and B. napus FUL copies using Jukes-Cantor distance and Blosum62 cost matrices implemented in Geneious Prime. The FUL gene of A. thaliana (AT5G60910, TAIR) was used as an outgroup. Multiple sequence alignments were also carried-out for the FUL homologues. FUL protein sequences were retrieved from the BRAD database (www.brassicadb.cn, Accessed 18 April 2023).
The intron-exon distribution of FUL genes was obtained from genome annotation files from the online resources described above and confirmed using sequence analysis with AtFUL. Multiple sequence alignment of protein sequences was performed with ClustalX 2.0 (http://www.custal.org/clustal2/) and implemented in the BioEdit package to visualise functional variation in the FUL genes. Conserved domains in the FUL were predicted using the NCBI Conserved Domain Database (http://www.ncbi.nih.gov/cdd) at E-value <0.001. Analysis of 5Kb upstream sequences of five FUL homologues for locating known motifs in the cis-acting regulatory elements was conducted using SIGNALSCAN program in Plant cis-Regulatory DNA Elements (PLACE, https://www.dna.affrc.go.jp/PLACE/?action=newplace). The number of motifs identified for each type were counted, and their roles were described (https://www.dna.affrc.go.jp/PLACE/place_seq.shtml). Also, the same dataset (5Kb upstream sequences of FUL homologues) was investigated for the presence of any novel motifs (sequence pattern that repeatedly occurs in a group of related protein or DNA sequences) using MEME (Multiple EM for Motif Elicitation, https://meme-suite.org/meme/tools/meme).
Anatomical features of valve margins from pods of parental lines were collected 35 to 40 days after anthesis. Hand sections were prepared from the middle of the pod, where the replum was narrow. Fresh sections were observed for autofluorescence using a fluorescence microscope. Photographs were taken using a Zeiss Axiphot microscope fitted with a Sony Cyber-shot digital camera.
We evaluated 203 F2 lines derived from a cross between the B. napus line BLN3343-C00402 (maternal parent, NBGIP accession BC95041, shattering type) and interspecific line BC95042 (paternal parent derived from B. rapa/B. napus, resistant to pod shattering, Raman et al., 2014) using the pendulum test to investigate the genetic inheritance and genetic determinants underlying pod-shattering resistance. Herein, we implemented the pendulum test to detect genetic variation in rupture energy - a measure of pod strength/resistance to shattering (Kadkol et al., 1984; Kadkol et al., 1986; Liu et al., 1994; Raman et al., 2014). The interspecific line, BC95042, required a higher level of force to break up the pod and release seed; therefore, it had a higher value for rupture energy than the maternal line BC95041.
The F2 population derived from a single F1 plant showed a continuous distribution of rupture energy scores, ranging from 2.32 mJ to 17.76 mJ) (Figure 3A). We observe that both pod valves separate length-wise (vertically) under field conditions (Figure S1A). This shattering pattern differs from pod drop, which often occurs in related species of Brassica, such as Raphanus raphanistrum subsp. sativus (L.) (Figure S1B). Microscopic analysis revealed that the dehiscence zone is well-differentiated in shatter-prone parental lines of the mapping population BC95041 compared to pod shatter-resistant parental lines (BC95042). Interspecific line BC95042 required high energy to rupture the pod (threshing) than the shatter-prone line BC95041 (Figure 3A). In the resistant parental line, there was less lignification of cells near the dehiscence zone and a less conspicuous distinction between lignified and separation layer from the replum compared to shatter-prone lines (Figures 3B, C). These observations suggest that the pod shatter resistance genes play an essential role in the dehiscence zone differentiating and subsequent seed dispersal (Liiljegren et al., 2000). To verify the rupture energy scores of the F2 lines, we raised a subset of 40 F2:3 progenies representing extreme phenotypes (the top 20 and bottom 20 F2 lines based on their pod energy scores) under natural field conditions. A positive correlation (r = 0.7) between the rupture energy scores of F2 plants and their F2:3 progenies (Figure 3D) indicates that rupture energy scores are reliable and suitable for genetic analysis.
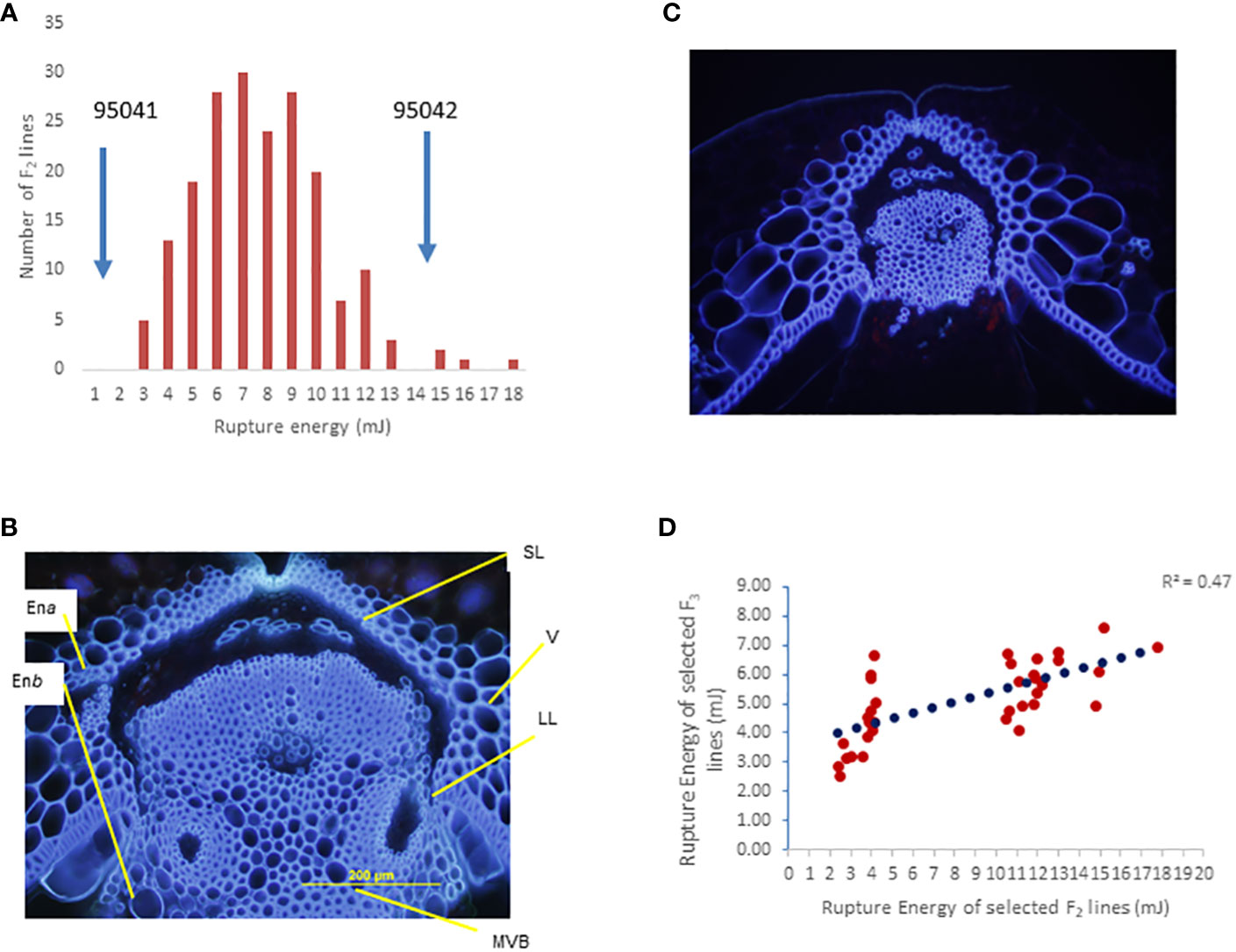
Figure 3 Genetic analysis of the BC95041 (shatter prone)/BC95042 (shatter resistant) F2 population for pod shatter resistance. (A) Frequency distribution of rupture energy (RE) scores in a segregation population containing 179 individuals. Solid arrows indicate the average RE scores of the maternal line BC95041 and the paternal interspecific line 95042. (B) Cross section of developing pods showing well-developed dehiscence zone/abscission layer (DZ) in BC95041, whereas BC95042 shows limited DZ differentiation. The dehiscence zone: DZ, valve: V, the main vascular bundle of replum: MVB, the two endocarp layers, endocarp a: Ena, and endocarp b: Enb are shown. (C) Arrowheads indicate a lack of complete cell separation in the pod shatter-prone line. (D) Relationship `of pod rupture energy scores between F2 and F3 individuals.
Using the DArTseq technology (Raman et al., 2014), a total of 26,002 high-quality SNPs (single nucleotide polymorphism) and in-sillco presence-absence markers, which showed (i) polymorphism between the parents and (ii) segregation in a mapping population, were used. We constructed a genetic linkage map spanning a total length of 2117.53 cM, with an average interval of 7.32 cM. The length of the chromosomes (linkage groups) ranged from 22.25 (C02) to 179.81 cM (A09). The marker density of the linkage groups ranged from 3.61 (A02) to 10.15 (A10). On average, 80.51% of markers were anchored to the 19 linkage groups, representing the An and Cn subgenomes of the reference B. napus cv. Darmor-bzh genome (Table 1). Using a genetic framework map based on 15,498 DArTseq markers (Table S1), we identified and located the significant QTLs conferring resistance to pod shatter on the B. napus genome. Different algorithms were used to identify robust associations for breeding use. Linear regression analysis using an additive model revealed that the top 99 markers mapped on chromosomes A01, A05, A09, C03 and C04 have a significant association (LOD ≥3.00) with resistance to pod shatter (Figure 4A, Table S2A). Of them, the top 16 markers were localised on A09 within 4.59 to 21.47 cM, and in-silico DArTseq marker 3101411 showed the most significant association (-log10P = 5.16) with resistance to pod shatter (Supplementary Table S2B). This marker showed a complete linkage with 15 other markers (Table S2C). Haplotype-based association test was conducted to detect the association between observed variations of pod shatter and marker haplotypes rather than single SNPs using the SVS package. We detected 677 haplotype blocks (HB, Supplementary Table S3A) following parameters described by Gabriel et al. (2002). Two markers in HB 303 on A09 detected the most significant association for pod shatter resistance with logistic regression (Table S3B). Haplotype trend regression revealed that HB308 (delimited with 3105829|F|0-8:C>G-8:C>G, 5121480|F|0-11:T>C-11:T>C, 3074795|F|0-19:G>T-19:G>T, 5050199|F|0-8:T>C-8:T>C markers, followed by HB309 with 3146480|F|0-46:A>G-46:A>G was the most significantly associated with pod rupture energy in the BC95041/BC95042 population (Table S4).
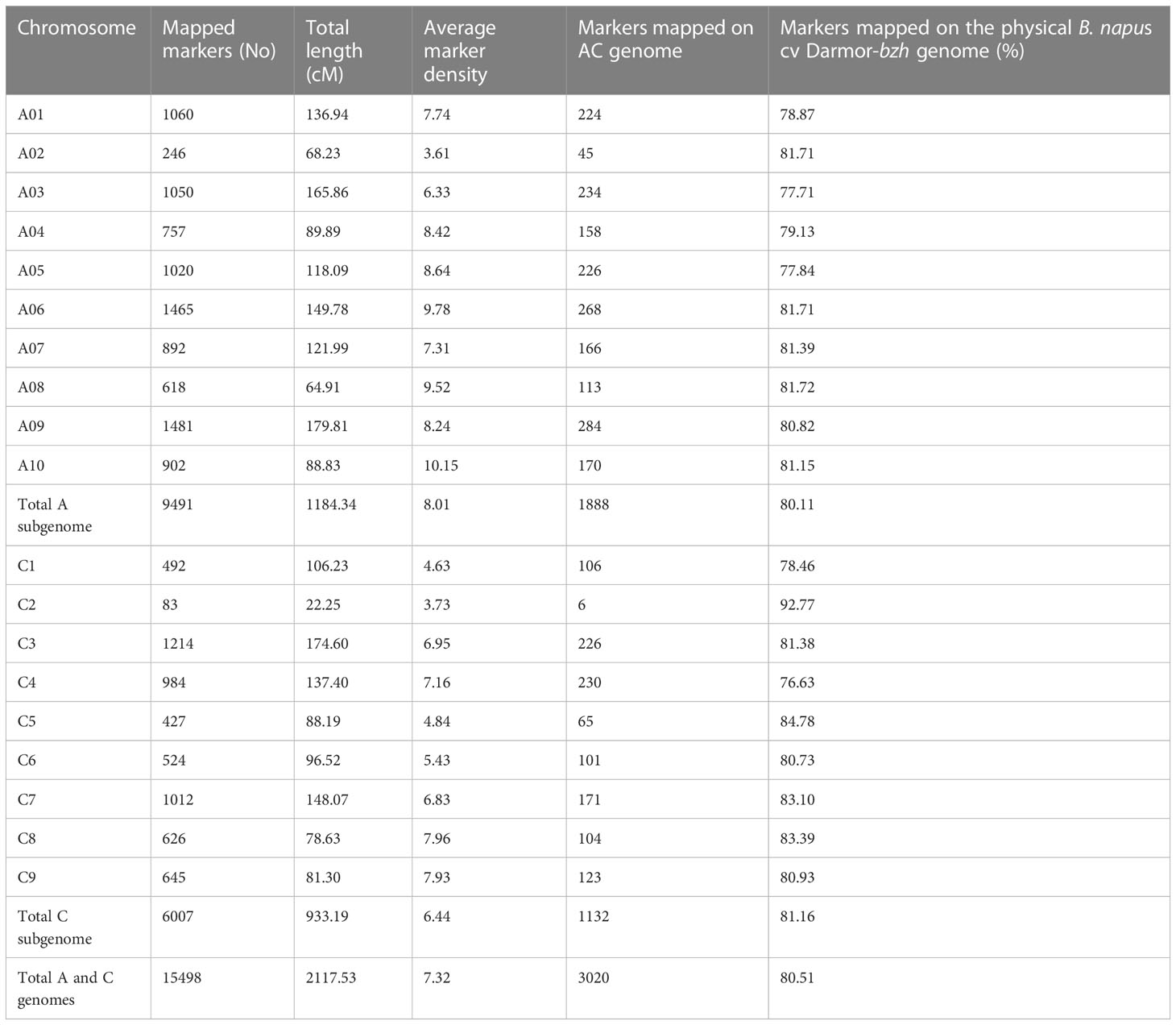
Table 1 Linkage map showing genetic distance, distribution and distance (cM) of DArTseq markers in the F2 population from BC95041/BC95042.
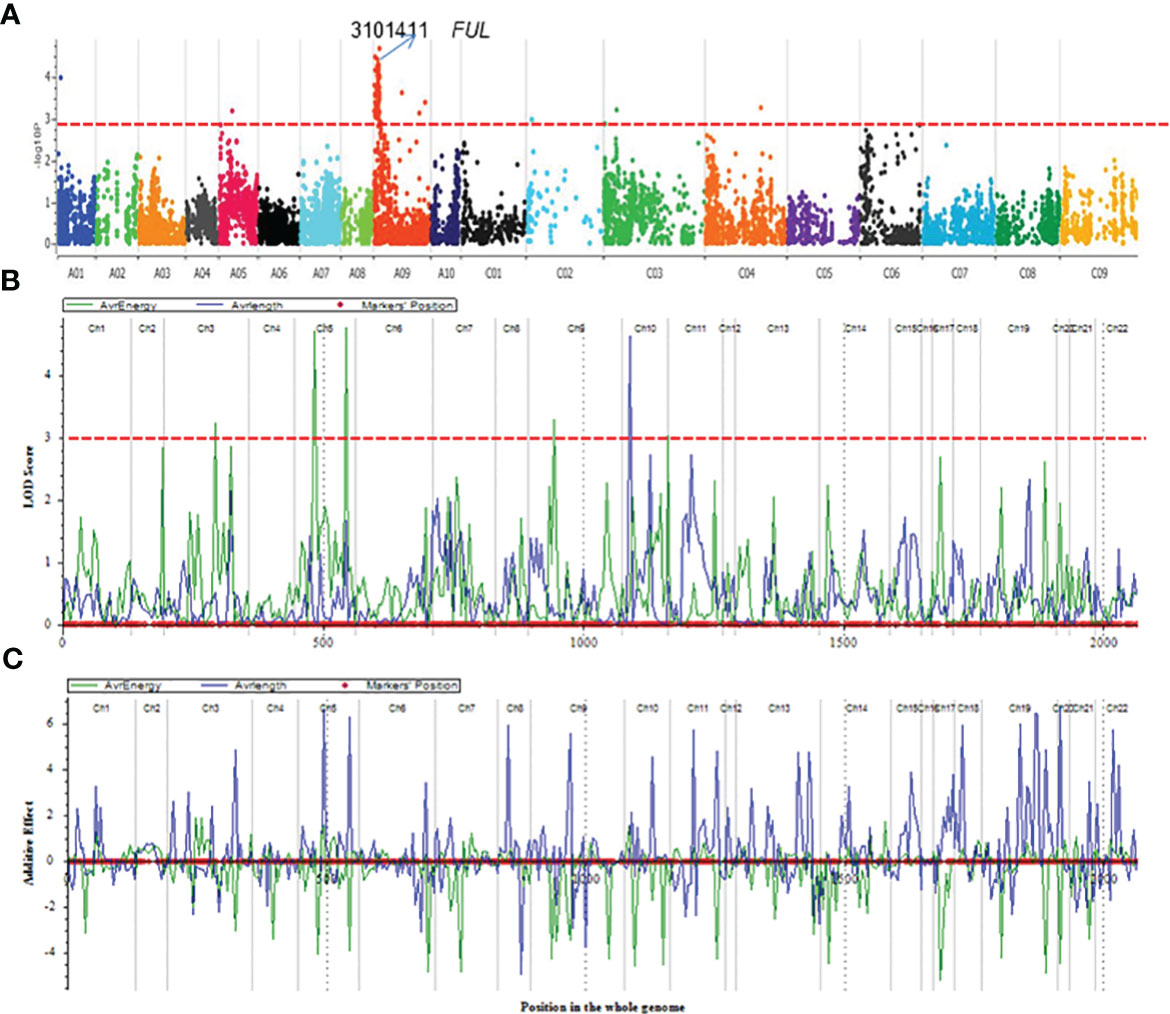
Figure 4 QTL mapping for pod shatter resistance measured as rupture energy (mj) by the pendulum test and pod length in the F2:3 population derived from BC95041/BC95042. (A) Manhattan plots showing genomic regions associated with resistance to pod shatter: significant regions are labelled. (B) Gene scan showing a single QTL on chromosome A09 for pod shatter resistance in an F2 population derived from a cross between BC95041 and BC95042. Significant QTL having a LOD score of 4 are shown by the dashed line (in blue colour). Pod shatter resistance was evaluated under birdcage conditions at Wagga Wagga, Australia and tested for rupture energy using a pendulum. (C) Allelic effects estimated by CIM approach. Linkage groups: Ch1-Ch10 relate to chromosomes A1-A10, Ch11-Ch14 to C01-C04; Ch15-16 to C05, Ch17-18 to C06, Ch19 to C07, Ch20-21 to C08 and Ch22 to C09.
We further detected QTLs associated with rupture energy and pod length using the simple interval mapping (IM) and composite interval mapping (CIM) approaches using the ICIM package. Five to seven significant QTLs for rupture energy were detected on chromosomes A03, A05 and A09 and C01 with IM and CIM (Table 2, Figure 4B). Three consistent QTLs were localised to the same genomic regions on chromosomes A02 and A05 across the analytical methods (Table 2). LOD scores of QTLs ranged from 2.8 to 4.77 and accounted proportion of variance explained (PVE) from 6.29% to 20.80% (Table 2). QTLs displayed both additive and dominant effects. Both parental lines contributed alleles for pod shatter resistance (Figure 4C). However, the interspecific paternal line BC95042 showed higher allelic effects (more than 2 folds) than the maternal B. napus line BC95041.
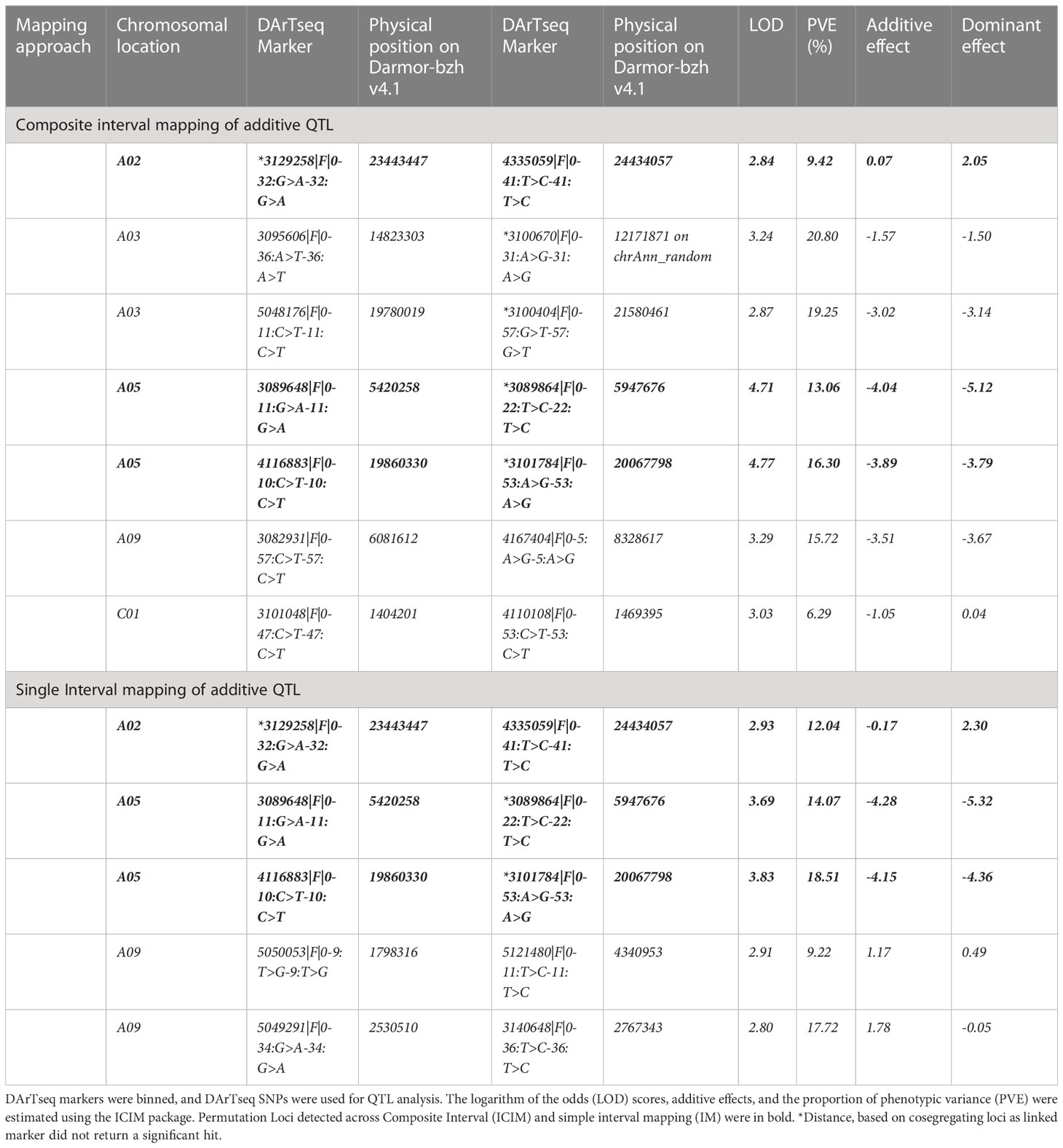
Table 2 Quantitative Trait Loci (QTLs) associated with pod shatter resistance measured as average rupture energy with the pendulum test.
To investigate the major genetic determinants controlling rupture energy, we binned pod shatter variation scores into two discrete categories, resistant (1, rupture energy: 2.32 to 6.94 mJ) and susceptible (0, rupture energy: 7.0 to 17.76 mJ) phenotypes, in conjunction with the seven highly significant markers (Supplementary Table S5A) and performed haplotype analysis to determine trait-marker association. The chi-squared analysis supported the presence of a single shatter resistance gene in BnF2 (χ2 3:1 = 0.17, with 1 degree of freedom, Two-tailed P value = 0.90). The HB 309 (defined by 15 SNPs: 3146480|F|0-46:A>G-46:A>G, 3096696|F|0-28:T>C-28:T>C, 3101752|F|0-29:C>T-29:C>T, 3159673|F|0-15:T>G-15:T>G, 5818650|F|0-5:C>T-5:C>T, 7250077|F|0-9:G>A-9:G>A, 3076890|F|0-52:A>T-52:A>T, 3079266|F|0-41:T>A-41:T>A, 3113543|F|0-40:A>C-40:A>C, 5121412|F|0-9:A>G-9:A>G, 5120748|F|0-29:G>C-29:G>C, 7249512|F|0-32:T>C-32:T>C, 3076528|F|0-55:T>C-55:T>C, 3077272|F|0-18:C>T-18:C>T, 3081487|F|0-26:C>T-26:C>T) revealed the most significant marker association with pod shatter resistance (χ2 -log10P: 9.99, Supplementary Table S5B) on chromosome A09. No other significant association was detected on B. napus chromosomes. Significantly associated markers detected on A09 showed collinearity between genetic and physical maps (Figure S2A). Different analytic methods revealed at least one significant locus on chromosome A09 that conditions variation in pod shatter resistance in the BC95041/BC95042 population. Mendelisation of quantitative variation revealed the limitation of identifying significant QTLs for trait variation (Tables 2, 3).
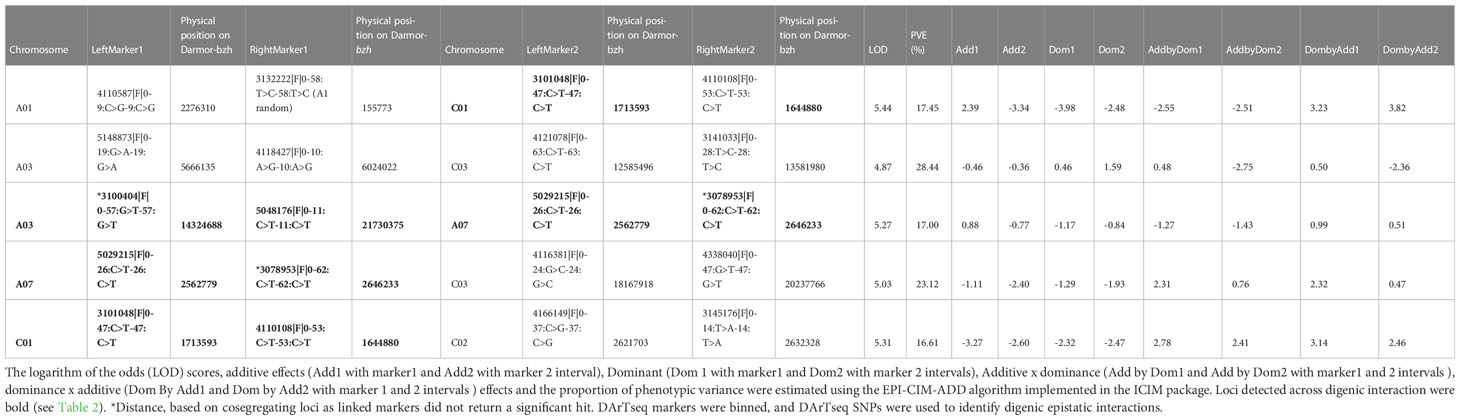
Table 3 Epistatic Quantitative Trait Loci (QTL) associated with pod shatter resistance measured as average rupture energy with the pendulum test.
Previous studies showed pod shatter resistance, measured as a random impact test, correlates with pod length (Cui, 2013). To determine whether pod length variation relates to pod shattering tested with pendulum test in the F2 population from BC95041/BC95042, we mapped QTLs associated with pod length on A02, A05, A07, A08, A10, C02 and C05 (Supplementary Table S6A). Simple interval mapping identified three significant QTLs on chromosomes A05, A07, A10, and C02, whereas composite interval mapping identified two QTLs on A10 and C01 (Supplementary Table S7A). None of the QTLs associated with pod length was collocated with QTLs for rupture energy, suggesting that pod length is genetically not associated with rupture energy (Table 2, Supplementary Table S7A). This was further substantiated by the lack of phenotypic correlation between pod length and shatter resistance scores (r = 0.01, Figure S2B).
Using a threshold estimated by permutation test at P = 0.05, 1,000 iteration (4.87), five pairs of significant epistatic QTLs for rupture energy were detected on A01/C01, A03/A07, A03/C03, A07/C03, and C01/C02 and revealed effects for additive × additive, additive × dominance and dominance × dominance interactions (Figure 5, Table 3). These EPI-QTLs accounted for 16.61% to 28.44% of PVE. Both parental alleles contributed to the epistasis in the intercross population. Additive marker effects between A03 and A07 chromosomes and A01 and C01 were in the repulsion phase. Epistatic QTLs for pod length were identified on chromosomes; A03/C07, A03/A05, A05/A08 and A05/A09, A05/C01, A05/C03, A09/C08, A09/A10, A10/C03 and A10/C08 at threshold 5 (Table S7B). However, using the threshold permutation test value estimated using 1,000 iterations, we did not identify any significant epistatic QTL for pod length.
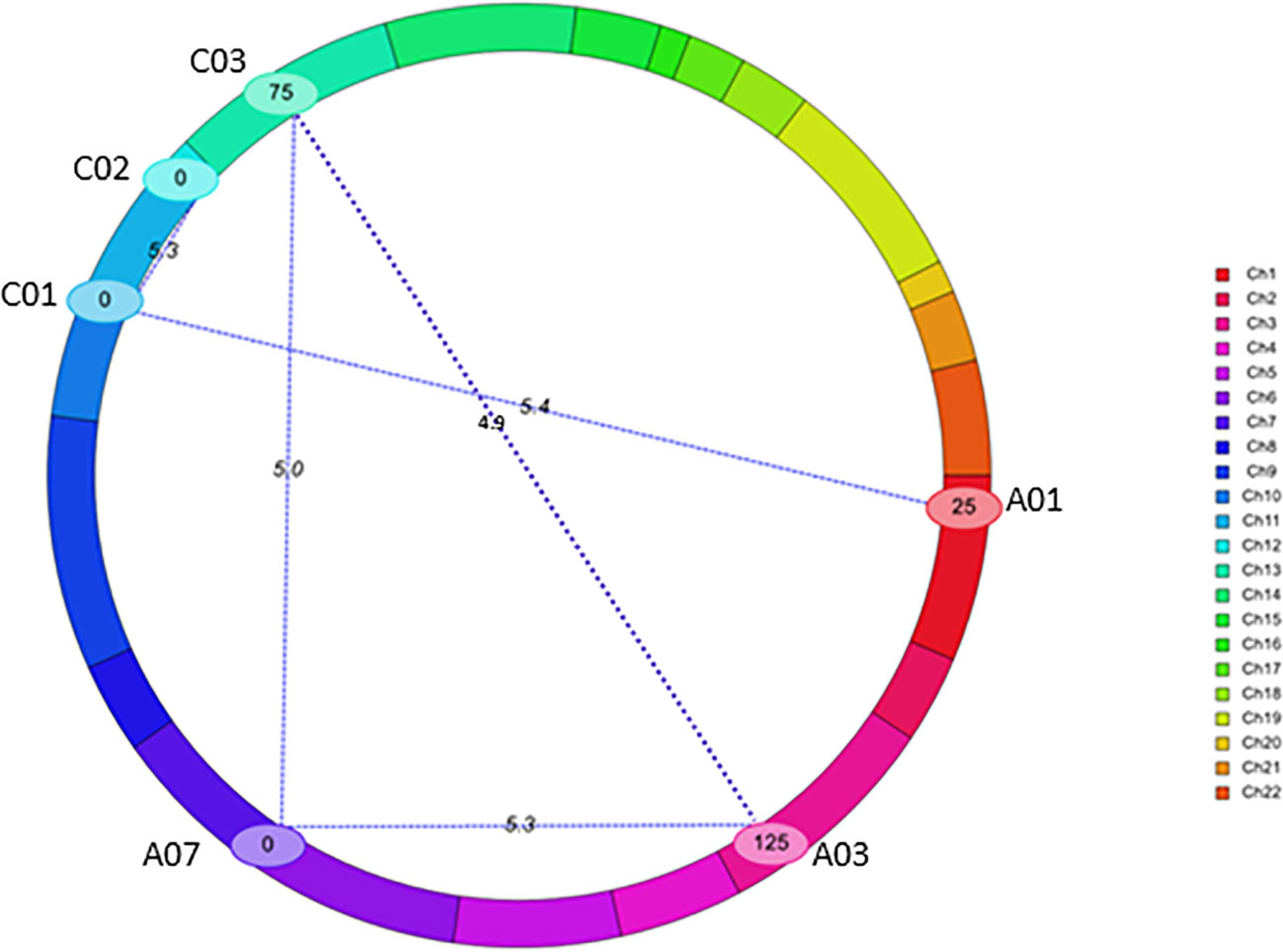
Figure 5 QTL interactions that showed epistatic effects for pod shatter resistance in the F2 population from BC95041/BC95042. The epistatic interaction was identified using the CIM-EPI approach in the ICIM package. Different linkage groups relating to B. napus chromosomes are shown (Ch1-Ch10 relate to chromosomes A1-A10, Ch11-Ch14 to C01-C04; Ch15-16 to C05, Ch17-18 to C06, Ch19 to C07, Ch20-21 to C08 and Ch22 to C09) in different colours. Interactions are shown with blue lines. Chromosomes that showed significant interactions are labelled.
We searched for the physical location of significant markers flanking QTLs for main effects and epistatic interactions (Tables 2, 3) using the B. napus cv. Darmor-bzh reference genome v4.1 (Supplementary Tables S8A, B). Annotated genes mapped with the QTLs marker intervals and the homologues of priori genes involved in pod shattering of A. thaliana. were inspected. Annotated genes in the reference assemblies located within QTL intervals in reference assemblies were prioritized as candidates for pod shatter resistance. The highly significant marker 3101411 associated with pod shatter resistance on A09 was mapped to the reference sequence of C08, and other cosegregating markers with 3101411 that were located at the same locus on the genetic map (16.45 cM) were mapped to the 2,177,920 to 2,443,302 bp of the Darmor-bzh v4.1 reference sequence (Supplementary Table S2C). Comparative analysis identified several candidate genes, including AP2, ABI3, ARF, BP1, CEL6, CESA3, FIL, FUL, GA2OX2, IAA31, IND, LAC4, LEUNIG, KNOTTED, MAGL15, PG1, RPL, QRT2, RGA, SPL and TCP10) underlying main QTL and epistatic QTL interactions for pod shatter resistance. Three copies of the FUL gene underlie the QTLs for pod shatter resistance on chromosomes A02, A03 and A09 (Table 2, Supplementary Table S10). Marker 3129258|F|0-32:G>A-32:G>A was located 63.6 kb from BnaAnng06660D homologue of FUL on A02 (Supplementary Table S9A). The A03 QTL delimited with 5048176|F|0-11:C>T-11:C>T was mapped ~116kb apart from the FUL homolog (BnaA03g39820D), accounting for 19.25% % PVE. QTL on chromosome A09 delimited with 5121480|F|0-11:T>C-11:T>C marker (19.25% of the total PVE) was located near the FUL gene (~248Kb, BnaA09g05500D, Table 2). Therefore, FUL may contribute to genetic variation in pod shatter resistance in the population used herein. To check whether there are candidate genes that could not be retrieved based on a single reference (Darmor-bzh versions v4.1/10) genome assembly, we utilised the BnaOmics platform that integrates pan-genome of 26 B. napus reference genomes and re-sequencing data of 2,885 accessions (Cui et al., 2023). At least two FUL copies of A02 and A03 were located in the pan-genome (Table S10).
FUL is a MADS-box transcription factor that is shown to be a part of a complex regulatory network that controls floral meristem identity, shoot maturation, floral transition, cell proliferation in pod valves and cell differentiation by limiting the dehiscence zone formation in A. thaliana, B. napus and B. juncea (Gu et al., 1998; Ferrandiz et al., 2000; Rajani and Sundaresan, 2001; Ostergaard et al., 2006). TBLASTN and reciprocal BLASTP searches against Arabidopsis proteins confirmed that the FUL (AGL8, AT5G60910) clade includes five homologues in B. napus on chromosomes Ann_random (BnaAnng06660D, A02 in the pan-genome), A03 (BnaA03g39820D), A09 (BnaA09g05500D), C02 (BnaC02g41870D) and C07 (BnaC07g49790D) detected in both reference genome assemblies v4.1 and 10 (Figures S3, S5). However, seven homologues were annotated in the B. napus pan-genome gene assembly on A02, A03, A09, C02, C07 and C09 chromosomes and validated for the presence of MADS-box domain-containing protein with a K-box coil and the MEF2 DNA-binding/dimerisation regions (Table S10B). FUL homologues of B. napus: BnaA03g39820D, BnaA09g0550D and BnaAnn06660D were clustered into distinct clades with B. rapa and BnaC02g41870D and BnaC07g49790D with B. oleracea clade, as expected (Figure 2). FUL homologue of B. oleracea (LOC10631378) showed grouping with BnaA09g0550D. Since we identified several QTLs that map near to MADS-box transcription factors such as AGAMOUS (AG), APETALA and AG-LIKE transcription factors could also regulate FUL expression throughout vegetative and reproductive phases during the plant development; we performed phylogenetic analysis using the Bayesian clustering method. This analysis differentiated AG, FUL (AGL8), SHP1 (AGL1), SHP2 (AGL5), and AGL3/SEPALLATA4 (SEP4) clades (Figure S5).
To date, BnaA09g05500D is the only FUL orthologue of A. thaliana and its closely related MADS-box gene in Sinapis alba: MADSB, which is shown to be involved in pod dehiscence via gene expression studies (Ferrandiz et al., 2000; Liiljegren et al., 2000; Chandler et al., 2005). Therefore, we further investigated its gene structure, evolution rate, and sequence variants using a dataset of 373 resequenced B. napus accessions utilised in the Australian National Brassica germplasm improvement program for gene discovery projects (Table S10). To determine the gene structure of the FUL homologues in B. napus, we used AtFUL (AT5G60910, TAIR). Sequence analysis of BnaA09g0550D) revealed that it encodes a 726 bp transcript with a 242 amino acid protein and comprises 8 exons and 7 introns (http://www.genoscopegen.cns.fr/brassicanapus/cgi-bin/geneView?src=colza;name=BnaA09g55330D) (Figure S3). The size of the first intron (intron 1) varied from 861 (A02) to 2462 (C02) bp, in contrast to some plant species, such as tomato and the wild D-genome progenitor of bread wheat, Aegilops tauschii (Takumi et al., 2009; Maheepala et al., 2019). The parental lines of the mapping population from BC95041 and BC95042 revealed 364 polymorphic SNPs and deletions NCBI, Banklt accession ID 2735083, (Table S11), and two non-synonymous variants were identified in exon 1 (c.A25G:p.K9E) and 7 (c.G616T:p.A206S). There were five non-synonymous SNV in exon 1 (c.G166A:p.E56K, c.G155A:p.G52D, c.G139A:p.V47I and c.A25G:p.K9E) and exon 7 (c.G616T:p.A206S) of BnaA09g05500D. Among all 373 accessions, up to 578 variants were detected in FUL homologues in B. napus; the majority (~50%) occurred in the intergenic region, followed by intronic regions (Table S11). Sequence variants were detected in the exonic and upstream sequence of FUL homologues, ranging from 19 to 36 and 11-99, respectively. We also identified splice variants for BnaA09g05500D (1 variant) and BnaAnng06660D gene (2 variants).
We performed selection pressure analysis to determine the evolution rate as the ratio of Ka/Ks of FUL copies. Our results show that BnaA09g05500D copy on chromosome A09 had purifying selection (<0.1) followed by copies on C02, suggesting conserved function compared to BnaA03g39820D and BnaC07g49790D on A03 and C07, respectively (Supplementary Table S10C).
Analysis of 5 kb upstream regions of five FUL homologues with the SIGNALSCAN program within the PLACE database (https://www.dna.affrc.go.jp/PLACE/?action=newplace) revealed several motifs found in plant cis-acting regulatory DNA elements. The search identified 183 motifs, ranging from 127 in BnaC02g41870D to 145 in BnaA03g39820D). Of these 183 motifs, 91 common motifs were present in all five homologues, while 25 were unique to one of them. The duplication frequency of these common motifs in all five genes is depicted in Figure 6A, and the numbers are given in Table S12. Among the common motifs, DOFCOREZM is the most abundant one, with duplication frequency of 66 to 98 in the 5 Kb upstream region of FUL homologues, followed by CACTFTPPCA1, GT1CONSENSUS GATABOX and CAATBOX1. The FUL gene is shown to bind to a specific CArG box, with the consensus sequence CC(A/T)6GG (de Folter and GC, 2006). In B. napus, 2 to 20 CArG motifs (CARGCW8GAT and CARGATCONSENSUS) were found in the upstream sequence of FUL homologs. We identified CArG consensus sequence (CCWWWWWWGG) in BnaAnng06660D and BnaC07g49790D only, whereas a variant of CArG motif with a more extended A/T-rich core (CWWWWWWWWG) is found in upstream sequences of all five FUL homologues (Figure 6B). There were 14 motifs (ABRELATERD1, ACGTATERD1, ACGTABREMOTIFA2OSEM, CBFHV, DRECRTCOREAT, LTRECOREATCOR15, MYB1AT, MYB2AT, MYBATRD22, MYBCORE, MYB2CONSENSUSAT, MYCCONSENSUSAT, MYCATERD1 and MYCATRD22) detected in the dataset which are associated with water stress or dehydration. Consistent with previous studies, we also found auxin response elements (GGTCCCATGMSAUR, AUXREPSIAA4, AUXRETGA1GMGH3, ARFAT, SURECOREATSULTR11 and CATATGGMSAUR) in our upstream sequences dataset. Among these motifs, SURECOREATSULTR11 and CATATGGMSAUR were found in the upstream sequences of all five genes, whereas GGTCCCATGMSAUR and AUXREPSIAA4 were unique to the upstream sequence of BnaA03g39820D (Table S12). Furthermore, seven motifs (WRKY71OS, PYRIMIDINEBOXOSRAMY1A, PYRIMIDINEBOXHVEPB1, GAREAT, MYBGAHV, GADOWNAT and GARE2OSREP1) were associated with gibberellin signalling pathway. The chromosome A09 FUL copy also had the maximum number (14) of SAUR (Small Auxin-Up RNA, CATATGGMSAUR) motifs, implicated in auxin responsiveness (Xu and Guilfoyle, 1997). Copy number variation and distribution of motifs in the upstream regulatory region of FUL may account for natural variation in gene expression and regulation of valve growth by interacting with other genes involved in valve margin differentiation, such as SHP1, SHP2, IND and ALC. IND also forms auxin minimum by coordinating auxin efflux in separation layer cells (Sorefan et al., 2009). We also found the GTGANTG10 motif (with duplication frequency 28-43), which shows homology to pectate lyase (Rogers et al., 2001).
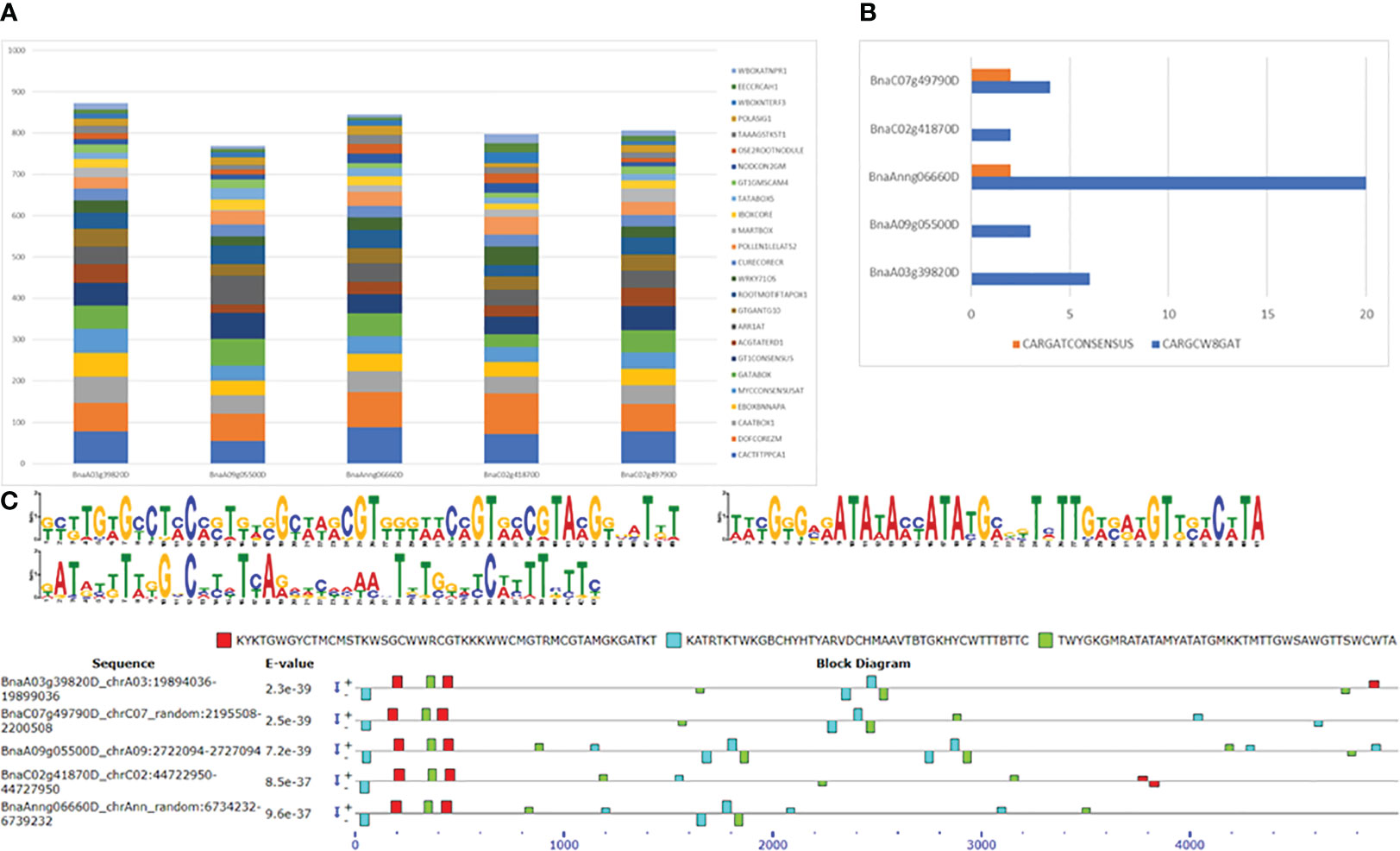
Figure 6 Motif identification in 5 Kb upstream sequences of five FUL homologues in B. napus. (A) Duplication frequency of 25 most abundant motifs. (B) CArG motifs and their frequency of occurrence. (C) Three novel motifs found using MEME and their occurrence in the sequence. + and – indicates the motif occurrence on sense and antisense strands.
We also discovered three unknown motifs in the 5 Kb upstream sequences of all five FUL homologues. The first motif KYKTGWGYCTMCMSTKWSGCWWRCGTKKKWWCMGTRMCGTAMGKGATKT (GCGTGTGCCTCCCCTGTCGCAAGCGTGGGAACCGTGCCGTACGGGATGT) is potentially located within first 500bp upstream, whereas the second motif KATRTKTWKGBCHYHTYARVDCHMAAVTBTGKHYCWTTTBTTC (GATGCGTTGGCCCCCTCAGCGCCCAACTGTGGCCCATTTCTTC) and the third motif TWYGKGMRATATAMYATATGMKKTMTTGWSAWGTTSWCWTA (TACGGGCGATATACCATATGCGGTCTTGACAAGTTCACATA) are randomly dispersed with no particular pattern detected in their occurrence with respect to positions (Figure 6C). Also, the first motif is mainly detected on the sense strand, whereas the second and third motifs are comparatively present on both sense and antisense strands.
Seed shattering is a massive issue in commercial canola production worldwide, underpinning growers’ profitability. Pod shatter-resistant varieties suitable for direct harvesting with combines are essential to reduce (i) reliance on windrowing, (ii) yield losses, (iii) inputs cost (labour and fuel for windrowing and controlling rogues in subsequent crops), (iv) carbon emissions occurred while windrowing followed by threshing with combine harvesters, and to improve (v) gross margins of farmers (return on the investment).
Herein, we investigated the genetic basis of pod shatter resistance in an interspecific derivative of B. rapa/B. napus. In this study, we used the pendulum test to describe the genetic variation for pod shatter resistance in a quantitative manner and understand its underlying genetic and anatomical bases. Previously, several methods, such as the number of seeds lost from pods, the number of seedlings germinated, the random impact test, and the pendulum test, have been used to determine genetic variation for pod shatter resistance in Brassica species (Morgan et al., 1998). There were 6.23-fold differences in pod shatter resistance between parental lines, suggesting that the interspecific source, BC95042, could be used to improve resistance to pod shatter.
Genetic analysis showed that pod shatter resistance is due to seven QTLs located on A02, A03, A05, A09 and C01 chromosomes in an F2 population derived from a cross between BC95041 and BC95042 (Table 2). With linear marker regression, HTR, IM, and CIM algorithms, we repeatedly detected four QTLs for pod shatter resistance on A02, A05 and A09, suggesting these QTLs are reliable for research and development activities such as introducing appropriate favourable alleles into canola varieties. Using different mapping algorithms with robust statistical power ensured the identification of significant marker-trait associations by reducing false positives to make genetic gains in canola breeding programs. Previous genetic mapping studies identified QTLs for pod shatter resistance in B. rapa (Mongkolporn et al., 2003; Bagheri et al., 2012), B. juncea (Kaur et al., 2020) and B. napus (Hu et al., 2012; Wen et al., 2013; Raman et al., 2014; Liu et al., 2016). Some of the QTLs were located in similar genetic positions on B. napus genome, which were detected in earlier studies (Table S9). However, there were no overlapping QTL regions across populations of Chinese origin. For example, Liu et al. (2016)) reported six significant QTLs for pod shatter resistance in a B. napus GWAS panel and two structured biparental populations on A01, A06, A07, A09, C02, and C05 chromosomes. Two QTLs on A06 and A09 were repeatedly detected across environments and mapping panels. QTL on A09 delimited with an Illumina SNP marker, Bn-A09-p30171993, was mapped near the SHP1 gene (A09_random chromosome on the 4.1 Darmor-bzh assembly). However, SHP1 and Bn-A09-p30171993 were located at the distal end of the A09 chromosome (Darmor-bzh version 10). However, this study identified three QTLs on chromosomes A02, A03 and A09 that significantly contributed to pod shatter resistance, accounting for 9.42% and 19.25% of the total PVE, respectively, and map near the FUL homologues (BnaAnng06660D, BnaA03g39820D and BnaA09g05500D, Table 1). These QTLs were not detected in other B. napus populations (Wen et al., 2013; Liu et al., 2016). We could not compare the map position of 13 QTLs for pod shatter resistance, measured by improved random impact method on A01, A04, A07, A08, C05, and C08 (Wen et al., 2013) as they were not mapped on any physical map of B. napus. Our study did not detect any QTL on A06 for pod shatter resistance located near the GIBBERELLEIN-3-OXIDASE1 gene in B. napus populations of Chinese origin (Liu et al., 2016). Most QTLs on A01, C02, and C05 were not closely mapped. These observations hint that selection for pod-shattering may have occurred at several independent loci and shaped the genomic architecture of pod-shatter resistance during cultivation and selective breeding in B. napus. This hypothesis is supported by independent seed-shattering QTLs (on A03, A09, this study) and the absence of the SHP1 and TCP8 genes, as shown in earlier studies (Liu et al., 2016; Liu et al., 2020; Chu et al., 2022). During domestication, Brassica species may have acquired several shattering resistance mechanisms to reach the desirable level of shattering resistance, suitable for manual harvesting, probably under humid climates, e.g., Europe and Wuhan. However, the resistance level is insufficient for hot and dry climates, e.g., Australia.
The PVE (6.29 to 20.80%) and additive effects from both parental lines (-4.28 to 1.78) that we identified in this study were consistent with most of the published B. napus studies revealing a small to moderate proportion of genotypic variation (4.01 to 28.9%) in pod shatter resistance (Wen et al., 2013; Raman et al., 2014; Liu et al., 2016; Liu et al., 2020). A recent study shows a major gene (i.e. TCP8 on C09) effect on pod shatter resistance via a lignified-layer bridge in a B. napus population (Chu et al., 2022). Our digenic interaction analysis showed five epistatic QTL interactions between chromosomes (A01-C01, A03-A07, A07-C03, A03-C03, and C01-C02). The positive epistatic effect of additive x additive suggested that the two epistatic loci (e.g. A03/C03, A07/C03, and C01/C02) with homozygous/heterozygous alleles from the same parent could increase the pod shatter resistance. However, the positive additive × dominance epistatic effect indicated that BC95042 could increase the pod shatter resistance. Breeding programs must consider additive and additive x additive epistatic interactions to improve resistance to pod shatter.
Based on the physical location of linked markers associated with pod shatter resistance, we prioritized AG, ABI3, ARF3, BP1, CEL6, FIL, FUL, GA2OX2, IND, LATE, LEUNIG, MAGL15, RPL, QRT2, RGA, SPT and TCP10, as candidate genes for pod shatter resistance (Table S9). The mechanisms and genetic factors involved in pod dehiscence have been investigated in A. thaliana and its closely related Brassica species. MADX-box transcription factors encoding FUL, SHP1, and SHP2 are the major players that control fruit patterning, lignin deposition, and pod dehiscence in Arabidopsis (Gu et al., 1998; Liiljegren et al., 2000). FUL negatively regulates SHP and IND expression in the valve margin and APETALA 1 in the outer whorl of the flower (Ferrandiz et al., 2000; Kaufmann et al., 2010). FUL and BEL-subfamily homeodomain gene RPL also negatively regulate SHP expression in the valve margin (Roeder et al., 2003). The floral homeotic gene AP2 also negatively regulates the expression of SHP, RPL, and IND genes and the expansion of replum and lignified layers (Ripoll et al., 2011). SHP1 and SHP2, which act redundantly, regulate the expression of basic helix-loop-helix (bHLH) genes: ALC, IND, and SPATULA (SPT). SHP1/2 and IND cause pod dehiscence by promoting cell proliferation and are involved in the differentiation of the lignification and separation layers in the stripes of the valve margin, whereas ALC and SPT are involved in forming the separation layer (Rajani and Sundaresan, 2001; Liljegren et al., 2004; Lewis et al., 2006; Groszmann et al., 2011). IND activates the expression of ALC and SPT but also promotes its own heterodimerisation with them through DELLA protein degradation (Girin et al., 2010; Girin et al., 2011). Finally, ALC and SPT are able to repress IND expression (Lenser and Theissen, 2013). IND regulates gibberellin levels through the GA3 Oxidase 1/GA4 gene (Arnaud et al., 2010; Kay et al., 2013). FIL, YABBY and JAG can control the expression patterns of FUL and SHP in the valve and valve margins ( (Dinneny and Yanofsky, 2005; Mühlhausen et al., 2013). We also identified downstream genes such as BETA-1-4 GLUCANASE (CELLULASE6), ENDO-POLYGALACTURONASE (RDPG1, QRT2), MAN7, NST1/3 and other MADS family transcription factors like SEPALLATA3, AGL15, SEP4, associated with pod shatter resistance in the mapping population. These genes are implicated in pod dehiscence in A. thaliana and B. napus (Jiang et al., 2016; Li et al., 2021). Di Marzo et al. (2022), found that the expression of α-XYLOSIDASE1 (XYL1) is directly regulated in developing seeds and fruit by the MADS-box transcription factor SEEDSTICK (STK). They demonstrated that XYL1 complement the stk smaller seed phenotype, confirming the importance of cell wall modulation in shaping organs. Some priori genes for pod shatter resistance were localised more than 1Mb from significant QTL regions. Small populations with low-density markers cannot resolve recombination between markers and candidate genes (Raman et al., 2016). However, the homologs of pod shatter resistance genes that map further apart from significantly associated markers on other chromosomes could regulate genetic variation in pod shatter resistance. Further research is required to substantiate this hypothesis. We identified sequence variants between the parental lines of the mapping population and other elite lines of B. napus. Further studies are required to establish the role of sequence variants in pod shatter resistance genes and their functional role via gene expression and gene editing approaches. Overall, our data on genetic mapping and putative candidate/priority genes suggest the complex network involved in pod shatter resistance in B. napus germplasm, broadly consistent with A. thaliana (Figure 7), as reiterated earlier (Stephenson et al., 2019). This observation is consistent with the high syntenic relationships between B. napus and A. thaliana (Parkin et al., 2005).
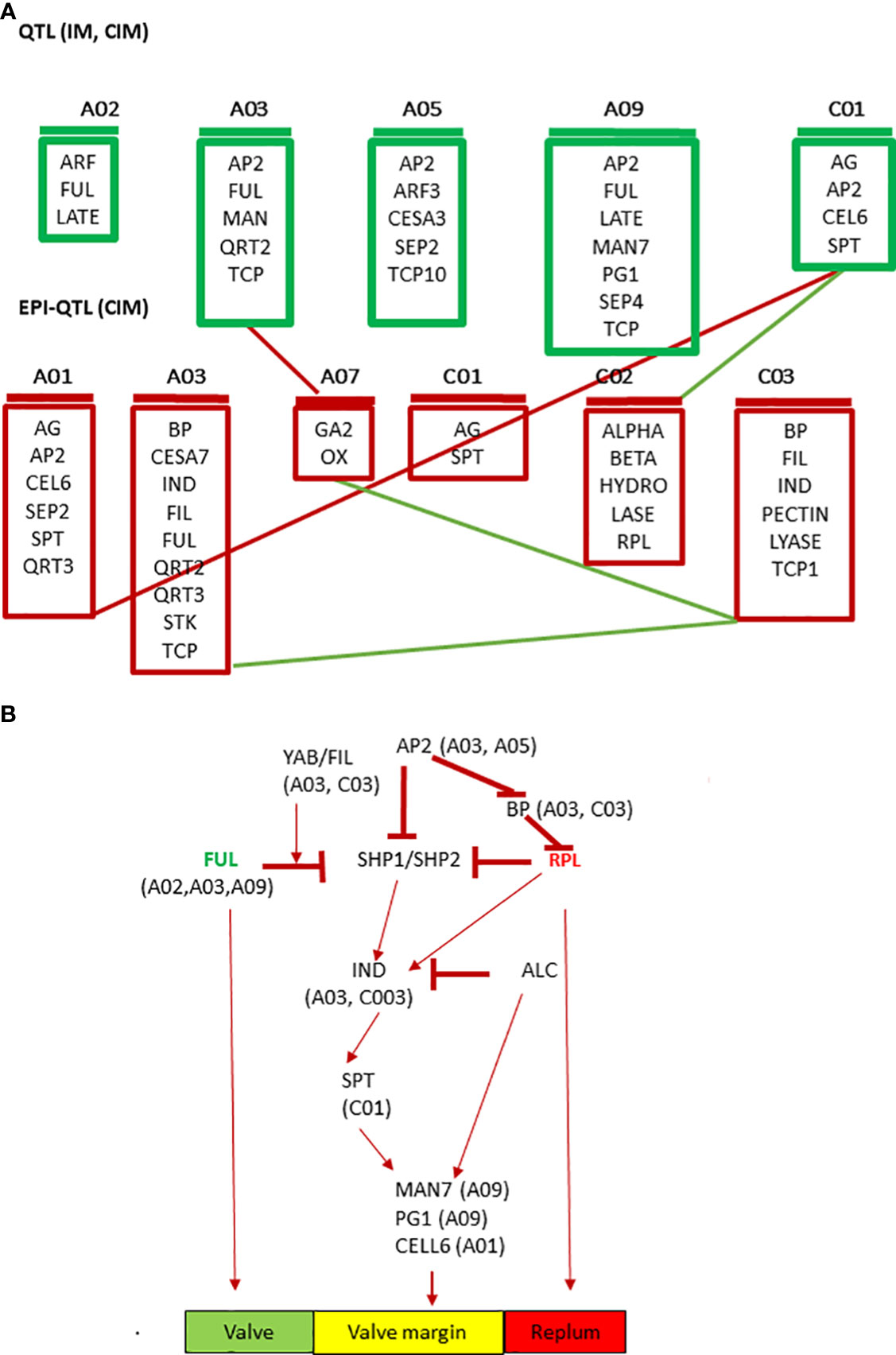
Figure 7 Prioritized candidate genes underlying QTL for pod shatter resistance using the simple interval, composite interval mapping, and epistatic-composite interval mapping algorithms implemented in the ICIM package. (A) Cartoon showing QTL with main effects (IM, CIM) and epistatic interactions (epi-QTL) along with their chromosomal location and (B) Extrapolated QTL-based candidate genes involved in pod shatter resistance network in B. napus. The green colour indicates valve, the yellow colour valve margin identity-related genes, and the orange colour indicates the replum.
In summary, we constructed the genetic framework map and identified seven genomic regions associated with pod rupture energy on A02, A03, A05, A09, and C01 chromosomes in an F2 population derived from the BC95041/BC95042 line developed from B. rapa/B. napus. In addition, five pairs of significant epistatic QTL interactions for rupture energy between A01/C01, A03/A07, A07/C03, A03/C03, and C01/C02 chromosomes. Overall, our results showed that independent QTLs (on A02, A03, A05, A09 and C01 chromosomes) and interactive QTLs (on A01/C01, A03/A07, A07/C03, A03/C03, and C01/C02) contribute to genetic variation in pod shatter resistance. Epistatic QTL interactions possibly reflect the regulatory network (repressor and activators) involved in pod dehiscence in A. thaliana. Several QTL regions were mapped near the candidate genes (AG, ABI3, ARF3, BP1, CEL6, FIL, FUL, GA2OX2, IND, LATE, LEUNIG, MAGL15, RPL, QRT2, RGA, SPT, and TCP10) which are involved in pod dehiscence, primarily in Arabidopsis. We described putative cis-acting motifs and sequence variants in genic and promoter regions of FUL homologues in 373 B. napus accessions. This study provides a valuable resource for gene discovery, the molecular mechanism underlying pod shatter resistance and yield improvement in Brassica species. DNA markers could accelerate the use of QTL in the Brassica breeding programs for marker-assisted selection, backcross, and genomic selection pipelines.
This study found that the interspecific line, BC94052 has superior alleles for resistance to pod shatter. Our genetic mapping suggests pod shatter resistance is due to multiple loci; three QTLs map to the A02, A03 and A09 chromosomes near FUL homologues. Our research provides a valuable genetic resource for improving pod shatter resistance in canola and for future studies on understanding molecular mechanisms underlying pod shatter resistance.
The datasets presented in this study can be found in online repositories. The Illumima sequence data of FULL genes can be found in NCBI Banklt accesssion 2735083.
The authors declare that the experiments comply with the current laws of the country in which they were performed and comply with ethical standards.
HR and RR designed the research and analyzed the data. RR developed the mapping population and conducted the experiments. YQ and BM assisted in phenotyping and performed pod anatomy and DNA extractions. NS, XC, YZ, QH, HR, and SL aligned DArTseq data with the reference genomes and analysed the dataset. NG provided the seeds of an interspecific line. HR wrote the first draft, NS contributed to the sections and all authors approved the final draft of the manuscript.
We thank the GRDC and NSW DPI for supporting this research under the DAN00208 and the Key Research Project of Hubei province (No.2021EHB026).
We thank Ms. Hannah Roe and Mr. John Bromfield for the pendulum testing of F2 and F2:3 families and Dr Gururaj Kadkol and Greg Buzza for the discussion. HR thanks Ms. Charmaine Carlisle, Charles Sturt University, Wagga Wagga, for making the microscope available.
NG works for Nuseed Pty Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1233996/full#supplementary-material
Altschul, S., Gish, W., Miller, W., Myers, E., Lipman, D. (1990). Basic local alignment search tool. J. Mole Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T. A., Lawrenson, T., et al. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24 (19), 2127–2132. doi: 10.1101/gad.593410
Bagheri, H., El-Soda, M., van Oorschot, I., Hanhart, C., Bonnema, G., Jansen - van den Bosch, T., et al. (2012). Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Front. Plant Sci. 3 (183). doi: 10.3389/fpls.2012.00183
Balanzà, V., Roig-Villanova, I., Di Marzo, M., Masiero, S., Colombo, L. (2016). Seed abscission and fruit dehiscence required for seed dispersal rely on similar genetic networks. Development 143 (18), 3372–3381. doi: 10.1242/dev.135202
Bowman, J. L., Baum, S. F., Eshed, Y., Putterill, J., Alvarez, J. (1999). Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 45, 155–205. doi: 10.1016/S0070-2153(08)60316-6
Braatz, J., Harloff, H. J., Emrani, N., Elisha, C., Heepe, L., Gorb, S. N., et al. (2018a). The effect of INDEHISCENT point mutations on silique shatter resistance in oilseed rape (Brassica napus). Theor. Appl. Genet. 131 (4), 959–971. doi: 10.1007/s00122-018-3051-4
Braatz, J., Harloff, H.-J., Jung, C. (2018b). EMS-induced point mutations in ALCATRAZ homoeologs increase silique shatter resistance of oilseed rape (Brassica napus). Euphytica 214 (2), 29. doi: 10.1007/s10681-018-2113-7
Cao, B., Wang, H., Bai, J., Wang, X., Li, X., Zhang, Y., et al. (2022). miR319-regulated TCP3 modulates silique development associated with seed shattering in brassicaceae. Cells 11 (19). doi: 10.3390/cells11193096
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chandler, J., Corbesier, L., Spielmann, P., Dettendorfer, J., Stahl, D., Apel, K., et al. (2005). Modulating flowering time and prevention of pod shatter in oilseed rape. Mol. Breed. 15 (1), 87–94. doi: 10.1007/s11032-004-2735-4
Child, R., Chauvaux, N., John, K., Ulvskov, P., Onckelen, H. (1998). Ethylene biosynthesis in oilseed rape pods in relation to pod shatter. J. Exp. Bot. 49, 829–838. doi: 10.1093/jxb/49.322.829
Chu, W., Liu, J., Cheng, H., Li, C., Fu, L., Wang, W., et al. (2022). A lignified-layer bridge controlled by a single recessive gene is associated with high pod-shatter resistance in. Brassica napus L. Crop J. 10 (3), 638–646. doi: 10.1016/j.cj.2021.09.005
Cui, J., Mei, D., Li, Y., Liu, J., Fu, L., Peng, P., et al (2013). Genetic contribution of silique related traits to silique shatter resistance of Brassica napus L. Chin. J. Oil Crop Sci. 35 (5), 461. doi: 10.7505/j.issn.1007-9084.2013.05.001
Cui, X., Hu, M., Yao, S., Zhang, Y., Tang, M., Liu, L., et al. (2023). BnaOmics: a comprehensive platform combining pan-genome and multi-omics data of Brassica napus. Plant Commun., 100609. doi: 10.1016/j.xplc.2023.100609
de Folter, S., GC, A. (2006). trans meets cis in MADS science. Trends Plant Sci. 11, 224–231. doi: 10.1016/j.tplants.2006.03.008
de la Pasture, L. (2018). Pod shatter—shattering implications for pod pop. Crop Production Magazine. Available at: https://www.cpm-magazine.co.uk/technical/pod-shatter-shattering-implications-pod-pop/ (Accessed 25 March 2023).
Di Marzo, M., Viana, V. E., Banfi, C., Cassina, V., Corti, R., Herrera-Ubaldo, H., et al. (2022). Cell wall modifications by α-XYLOSIDASE1 are required for control of seed and fruit size in Arabidopsis. J. Exp. Bot. 73 (5), 1499–1515. doi: 10.1093/jxb/erab514
Dinneny, J. R., Yanofsky, M. F. (2005). Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. BioEssays 27 (1), 42–49. doi: 10.1002/bies.20165
Ferrandiz, C., Liljegren, S. J., Yanofsky, M. F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289 (5478), 436–438.
Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstiel, B., et al. (2002). The structure of haplotype blocks in the human genome. Science 296 (5576), 2225–2229. doi: 10.1126/science.1069424
Girin, T., Paicu, T., Stephenson, P., Fuentes, S., Körner, E., O’Brien, M., et al. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in arabidopsis. Plant Cell 23 (10), 3641–3653. doi: 10.1105/tpc.111.090944
Girin, T., Stephenson, P., Goldsack, C. M. P., Kempin, S. A., Perez, A., Pires, N., et al. (2010). Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. Plant J. 63 (2), 329–338. doi: 10.1111/j.1365-313X.2010.04244.x
Groszmann, M., Paicu, T., Alvarez, J. P., Swain, S. M., Smyth, D. R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68 (5), 816–829. doi: 10.1111/j.1365-313X.2011.04732.x
Gu, Q., Ferrándiz, C., Yanofsky, M. F., Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125 (8), 1509–1517. doi: 10.1242/dev.125.8.1509
He, H., Bai, M., Tong, P., Hu, Y., Yang, M., Wu, H. (2018). CELLULASE6 and MANNANASE7 affect cell differentiation and silique dehiscence. Plant Physiol. 176 (3), 2186–2201. doi: 10.1104/pp.17.01494
Hu, Z., Hua, W., Huang, S., Yang, H., Zhan, G., Wang, X., et al. (2012). Discovery of pod shatter-resistant associated SNPs by deep sequencing of a representative library followed by bulk segregant analysis in rapeseed. PloS One 7 (4), e34253. doi: 10.1371/journal.pone.0034253
Jiang, X., He, H., Wang, T., Wang, X., Wu, H. (2016). Gene expression profile analysis indicate SEPALLATA3 and AGL15 potentially involved in arabidopsis silique dehiscence by regulating glycosyl hydrolase. J. Plant Biol. 59 (2), 133–142. doi: 10.1007/s12374-016-0567-5
Kadkol, G., Halloran, G., Macmillan, R. (1985). Evaluation of Brassica genotypes for resistance to shatter. II. Variation in siliqua strength within and between accessions. Euphytica 34, 915–924. doi: 10.1007/BF00035431
Kadkol, G., Halloran, G., MacMillan, R. (1986). Inheritance of siliqua strength in Brassica campestris L. I. Studies of F2 and backcross populations. Can. J. Genetical Cytology 28, 365–373. doi: 10.1139/g86-054
Kadkol, G. P., Macmillan, R. H., Burrow, R. P., Halloran, G. M. (1984). Evaluation of Brassica genotypes for resistance to shatter. I. Development of a laboratory test. Euphytica 33 (1), 63–73. doi: 10.1007/BF00022751
Kaufmann, K., Wellmer, F., Muiño, J. M., Ferrier, T., Wuest, S. E., Kumar, V., et al. (2010). Orchestration of floral initiation by APETALA1. Science 328 (5974), 85–89. doi: 10.1126/science.118524
Kaur, J., Akhatar, J., Goyal, A., Kaur, N., Kaur, S., Mittal, M., et al. (2020). Genome wide association mapping and candidate gene analysis for pod shatter resistance in Brassica juncea and its progenitor species. Mol. Biol. Rep. 47 (4), 2963–2974. doi: 10.1007/s11033-020-05384-9
Kay, P., Groszmann, M., Ross, J. J., Parish, R. W., Swain, S. M. (2013). Modifications of a conserved regulatory network involving INDEHISCENT controls multiple aspects of reproductive tissue development in Arabidopsis. New Phytol. 197 (1), 73–87. doi: 10.1111/j.1469-8137.2012.04373.x
Kord, H., Shakib, A. M., Daneshvar, M. H., Azadi, P., Bayat, V., Mashayekhi, M., et al. (2015). RNAi-mediated down-regulation of SHATTERPROOF gene in transgenic oilseed rape. 3 Biotech. 5 (3), 271–277. doi: 10.1007/s13205-014-0226-9
Laga, B., den Boer, B., Lambert, B. (2008). Brassica plant comprising a mutant indehiscent allele (Patent US8809635B2, Bayer CropScience NV). Available at: patents.google.com/patent/US9475849B2/en.
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Ostergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 258. doi: 10.1186/s13059-015-0826-7
Lee, J. S., Kim, K. R., Ha, B.-K., Kang, S. (2017). Identification of SNPs tightly linked to the QTL for pod shattering in soybean. Mol. Breed. 37 (4), 54. doi: 10.1007/s11032-017-0656-2
Lenser, T., Theissen, G. (2013). Conservation of fruit dehiscence pathways between Lepidium campestre and Arabidopsis thaliana sheds light on the regulation of INDEHISCENT. Plant J. 76, 545–556. doi: 10.1111/tpj.12321
Lewis, M. W., Leslie, M. E., Liljegren, S. J. (2006). Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 9 (1), 59–65. doi: 10.1016/j.pbi.2005.11.009
Li, Y. L., Yu, Y. K., Zhu, K. M., Ding, L. N., Wang, Z., Yang, Y. H., et al. (2021). Down-regulation of MANNANASE7 gene in B. napus L. enhances silique dehiscence-resistance. Plant Cell Rep. 40 (2), 361–374. doi: 10.1007/s00299-020-02638-5
Liiljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liljegren, S. J., Roeder, A. H. K., Kempin, S. A., Gremski, K., Østergaard, L., Guimil, S., et al. (2004). Control of fruit patterning in arabidopsis by INDEHISCENT. Cell 116 (6), 843–853. doi: 10.1016/S0092-8674(04)00217-X
Liu, X. Y., Macmillan, R. H., Burrow, R. P., Kadkol, G. P., Halloran, G. M. (1994). Pendulum test for evaluation of rupture strength of seed pods. J. Texture Stud. 25, 179–189. doi: 10.1111/j.1745-4603.1994.tb01325.x
Liu, J., Wang, J., Wang, H., Wang, W., Zhou, R., Mei, D., et al. (2016). Multigenic control of pod shattering resistance in chinese rapeseed germplasm revealed by genome-wide association and linkage analyses. Front. Plant Sci. 7 (1058). doi: 10.3389/fpls.2016.01058
Liu, J., Zhou, R., Wang, W., Wang, H., Qiu, Y., Raman, R., et al. (2020). A copia like-retrotransposon insertion in the upstream region of SHATTERPROOF 1 gene, BnSHP1.A9 is associated with quantitative variation in pod shattering resistance in oilseed rape. J. Exp. Bot. 71 (18), 5402–5413. doi: 10.1093/jxb/eraa281
Lu, K., Wei, L. J., Li, X. L., Wang, Y. T., Wu, J., Liu, M., et al. (2019). Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 10. doi: 10.1038/s41467-019-09134-9
MacLeod, J. (1981). Harvesting in oilseed rape (Cambridge: Cambridge Agricultural Publishing), 107–120.
Maheepala, D. C., Emerling, C. A., Rajewski, A., Macon, J., Strahl, M., Pabón-Mora, N., et al. (2019). Evolution and diversification of FRUITFULL genes in solanaceae. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00043
Marsch-Martínez, N., Ramos-Cruz, D., Irepan Reyes-Olalde, J., Lozano-Sotomayor, P., Zúñiga-Mayo, V. M., de Folter, S. (2012). The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 72 (2), 222–234. doi: 10.1111/j.1365-313X.2012.05062.x
Mongkolporn, O., Kadkol, G. P., Pang, C. K., Taylor, P. W. J. (2003). Identification of RAPD markers linked to recessive genes conferring siliqua shatter resistance in Brassica rapa. Plant Breed. 122, 479–484. doi: 10.1046/j.0179-9541.2003.00910.x
Morgan, C., Bavage, A., Bancroft, I., Bruce, D., Child, R., Chinoy, C., et al. (2007). Using novel variation in Brassica species to reduce agricultural inputs and improve agronomy of oilseed rape—a case study in pod shatter resistance. Plant Genet. Resour. 1 (1), 59–65. doi: 10.1079/PGR200311
Morgan, C. L., Bruce, D. M., Child, R., Ladbrooke, Z. L., Arthur, A. E. (1998). Genetic variation for pod shatter resistance among lines of oilseed rape developed from synthetic B. napus. Field Crops Res. 58, 153–165. doi: 10.1016/S0378-4290(98)00099-9
Mühlhausen, A., Lenser, T., Mummenhoff, K., Theißen, G. (2013). Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. Plant J. 73 (5), 824–835. doi: 10.1111/tpj.12079
Ogawa, M., Kay, P., Wilson, S., Swain, S. M. (2009). Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21 (1), 216–233. doi: 10.1105/tpc.108.063768
Ostergaard, L., Kempin, S. A., Bies, D., Klee, H. J., Yanofsky, M. F. (2006). Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol. J. 4 (1), 45–51. doi: 10.1111/j.1467-7652.2005.00156.x
Parkin, I. A. P., Gulden, S. M., Sharpe, A. G., Lukens, L., Trick, M., Osborn, T. C., et al. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171 (2), 765–781. doi: 10.1534/genetics.105.042093
Petroli, C. D., Sansaloni, C. P., Carling, J., Steane, D. A., Vaillancourt, R. E., Myburg, A. A., et al. (2012). Genomic characterisation of DArT markers based on high-density linkage analysis and physical mapping to the eucalyptus genome. PloS One 7 (9), e44684. doi: 10.1371/journal.pone.0044684
Price, J. S., Hobson, R. N., Neale, M. A., Bruce, D. M. (1996). Seed losses in commercial harvesting of oilseed rape. J. Agric. Eng. Res. 65 (3), 183–191. doi: 10.1006/jaer.1996.0091
Rajani, S., Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11 (24), 1914–1922. doi: 10.1016/S0960-9822(01)00593-0
Raman, R., Qiu, Y., Coombes, N., Song, J., Kilian, A., Raman, H. (2017). Molecular diversity analysis and genetic mapping of pod shatter resistance loci in Brassica carinata L. Front. Plant Science. 8 (1765). doi: 10.3389/fpls.2017.01765
Raman, H., Raman, R., Coombes, N., Song, J., Prangnell, R., Bandaranayake, C., et al. (2016). Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 39 (6), 1228–1239. doi: 10.1111/pce.12644
Raman, R., Raman, H., Johnstone, K., Lisle, C., Smith, A., Martin, P., et al. (2005). Genetic and in silico comparative mapping of the polyphenol oxidase gene in bread wheat (Triticum aestivum L.). Funct. Integrated Genomics 5, 185–200. doi: 10.1007/s10142-005-0144-3
Raman, H., Raman, R., Kilian, A., Detering, F., Carling, J., Coombes, N., et al. (2014). Genome-wide delineation of natural variation for pod shatter resistance in B. napus. PloS One 9 (7), e101673. doi: 10.1371/journal.pone.0101673
Ripoll, J. J., Roeder, A. H., Ditta, G. S., Yanofsky, M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138 (23), 5167–5176. doi: 10.1242/dev.073031
Roeder, A. H. K., Ferrándiz, C., Yanofsky, M. F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the arabidopsis fruit. Curr. Biol. 13 (18), 1630–1635. doi: 10.1016/j.cub.2003.08.027
Rogers, H. J., Bate, N., Combe, J., Sullivan, J., Sweetman, J., Swan, C., et al. (2001). Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol. Biol. 45 (5), 577–585. doi: 10.1023/A:1010695226241
Sorefan, K., Girin, T., Liljegren, S. J., Ljung, K., Robles, P., Galvan-Ampudia, C. S., et al. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459, 583–586. doi: 10.1038/nature07875
Spence, J., Vercher, Y., Gates, P., Harris, N. (1996). ‘Pod shatter’ in Arabidopsis thaliana, Brassica napus and B. juncea. J. Microscopy 181 (2), 195–203. doi: 10.1046/j.1365-2818.1996.111391.x
Stephenson, P., Stacey, N., Brüser, M., Pullen, N., Ilyas, M., O’Neill, C., et al. (2019). The power of model-to-crop translation illustrated by reducing seed loss from pod shatter in oilseed rape. Plant Reprod. 32 (4), 331–340. doi: 10.1007/s00497-019-00374-9
Takumi, S., Nishioka, E., Morihiro, H., Kawahara, T., Matuoka, Y. (2009). Natural variation of morphological traits in wild wheat progenitor Aegilops tauschii Coss. Breed. Sci. 59, 579–588. doi: 10.1270/jsbbs.59.579
Vera, C. L., Downey, R. K., Woods, S. M., Raney, J. P., McGregor, D. I., Elliott, R. H., et al. (2007). Yield and quality of canola seed as affected by stage of maturity at swathing. Can. J. Plant Sci. 87 (1), 13–26. doi: 10.4141/P05-077
Wang, R., Ripley, V. L., Rakow, G. (2007). Pod shatter resistance evaluation in cultivars and breeding lines of Brassica napus, B. juncea and Sinapis alba. Plant Breed. 126 (6), 588–595.
Wen, Y. C., Zhang, S. F., Yi, B., Wen, J., Wang, J. P., Zhu, J. C., et al. (2013). Identification of QTLs involved in pod-shatter resistance in Brassica napus L. Crop Pasture Sci. 63 (12), 1082–1089.
Keywords: pod shattering, domestication, genetic mapping, canola, genetic analysis, sequence variation
Citation: Raman H, Raman R, Sharma N, Cui X, McVittie B, Qiu Y, Zhang Y, Hu Q, Liu S and Gororo N (2023) Novel quantitative trait loci from an interspecific Brassica rapa derivative improve pod shatter resistance in Brassica napus. Front. Plant Sci. 14:1233996. doi: 10.3389/fpls.2023.1233996
Received: 03 June 2023; Accepted: 31 July 2023;
Published: 06 September 2023.
Edited by:
Umesh K. Reddy, West Virginia State University, United StatesReviewed by:
Javed Akhatar, Punjab Agricultural University, IndiaCopyright © 2023 Raman, Raman, Sharma, Cui, McVittie, Qiu, Zhang, Hu, Liu and Gororo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Harsh Raman, aGFyc2gucmFtYW5AZHBpLm5zdy5nb3YuYXU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.