- Department of Environmental Microbiology and Biotechnology, Nicolaus Copernicus University in Toruń, Toruń, Poland
Even though canola is one of the most important industrial crops worldwide, it has high nutrient requirements and is susceptible to pests and diseases. Therefore, natural methods are sought to support the development of these plants. One of those methods could be a plant growth–promoting rhizobacteria (PGPR) that have a beneficial effect on plant development. The aim of this study was a genomic comparison of two PGPR strains chosen based on their effect on canola growth: Peribacillus frigoritolerans 2RO30, which stimulated canola growth only in sterile conditions, and Pseudomonas sivasensis 2RO45, which promoted canola growth in both sterile and non-sterile conditions. First of all, six bacterial strains: RO33 (Pseudomonas sp.), RO37 (Pseudomonas poae), RO45 (Pseudomonas kairouanensis), 2RO30 (Peribacillus frigoritolerans), 2RO45 (Pseudomonas sivasensis), and 3RO30 (Pseudomonas migulae), demonstrating best PGP traits in vitro, were studied for their stimulating effect on canola growth under sterile conditions. P. frigoritolerans 2RO30 and P. sivasensis 2RO45 showed the best promoting effect, significantly improving chlorophyll content index (CCI) and roots length compared to the non-inoculated control and to other inoculated seedlings. Under non-sterile conditions, only P. sivasensis 2RO45 promoted the canola growth, significantly increasing CCI compared to the untreated control and to other inoculants. Genome comparison revealed that the genome of P. sivasensis 2RO45 was enriched with additional genes responsible for ACC deaminase (acdA), IAA (trpF, trpG), and siderophores production (fbpA, mbtH, and acrB) compared to 2RO30. Moreover, P. sivasensis 2RO45 showed antifungal effect against all the tested phytopathogens and harbored six more biosynthetic gene clusters (BGC), namely, syringomycin, pyoverdin, viscosin, arylpolyene, lankacidin C, and enterobactin, than P. frigoritolerans 2RO30. These BGCs are well known as antifungal agents; therefore, it can be assumed that these BGCs were responsible for the antifungal activity of P. sivasensis 2RO45 against all plant pathogens. This study is the first report describing P. sivasensis 2RO45 as a canola growth promoter, both under controlled and natural conditions, thus suggesting its application in improving canola yield, by improving nutrient availability, enhancing stress tolerance, and reducing environmental impact of farming practices.
1 Introduction
Plant growth–promoting rhizobacteria (PGPR) that colonize plant rhizosphere are group of bacteria exerting a beneficial effect on plant development (Adedeji et al., 2020). The application of these microorganisms can reduce the requirement of synthetic chemical fertilizers and pesticides, which is important for the development of sustainable agriculture (Patel et al., 2021). PGPR can promote plant growth directly or indirectly. Direct plant growth promotion includes mechanisms providing plants with phosphorus, nitrogen, iron, and indol-3-acetic-acid, while indirect promotion consists of preventing phytopathogens due to antimicrobial metabolites and extracellular enzymes (Chouyia et al., 2020; Wang et al., 2023). Many beneficial microorganisms, including certain bacteria and fungi, produce antibiotics as secondary metabolites, inhibiting the growth and development of phytopathogens by disrupting their cell walls, membranes, or metabolic processes. Beneficial microorganisms can outcompete phytopathogens for essential nutrients in the rhizosphere. Additionally, some microbial metabolites can induce systemic resistance in plants, making them more resistant to pathogenic attacks. By breaking down the cell walls of phytopathogens, bacterial enzymes such as chitinases and glucanases inhibit the growth and spread of pathogens. Meanwhile, extracellular proteases and lipases produced by beneficial microorganisms can hydrolyze proteins and lipids in the cell membranes (Kumari et al., 2019; de Andrade et al., 2023).
The genus Pseudomonas and Bacillus are well known as PGPRs and biocontrol agents due to their ability to solubilize phosphates and produce phytohormones or secondary metabolites such as hydrogen cyanide (HCN), siderophores, and lipopeptides (Sheoran et al., 2015; Lastochkina et al., 2019; Oni et al., 2022). It was reported that bio-inoculations of the three PGPR Bacillus strains with the ability to catabolize ACC, increased shoots, and roots length of canola plants (Kashyap et al., 2019). Whereas Pseudomonas sp. strain, which was able to produce siderophores and fix nitrogen, affected canola seeds germination and seedlings growth (Jamalzadeh et al., 2021). Ecological applications of Pseudomonas and Bacillus species as bioinoculants are crucial for maintaining food security (Sah et al., 2021; Etesami et al., 2023).
Peribacillus frigoritolerans is a rod-shaped, Gram-positive bacterium belonging to the family Bacillaceae and classified originally as Brevibacterium frigoritolerans (Montecillo and Bae, 2022). It is well documented that Brevibacterium frigoritolerans stimulates plant growth and suppresses diseases caused by phytopathogens. B. frigoritolerans promotes growth of wheat (Triticum aestivum L.) increasing roots, shoots, seedlings length, and plant biomass (Tara and Saharan, 2017), while Raufa et al. (2019) found this strain as biocontrol agent in suppression of maize (Zea mays L.) stalk rot caused by Fusarium moniliforme. Whereas, Pseudomonas sivasensis was isolated from farm fisheries in Turkey and described for the first time by Duman et al. (2020). Świątczak et al. (2023a) found that PGPR Pseudomonas sivasensis bacterization altered the taxonomic structure of bacterial and fungal communities by increasing the abundance of plant beneficial microorganisms and increasing metabolic activity and functional diversity of microbial communities in the canola rhizosphere.
In our study, we present new insights into the plant growth–promoting (PGP) properties of Pseudomonas sivasensis 2RO45, which promoted canola growth under both controlled and natural conditions. Moreover, PGP and biosynthetic cluster genes of Pseudomonas sivasensis 2RO45 were compared with other PGPR—Peribacillus frigoritolerans 2RO30 that promoted canola growth only in sterile conditions.
2 Materials and methods
2.1 Isolation of plant growth–promoting rhizobacteria
Canola (Brassica napus L. var. napus) plant samples collected from three following growth stages: vegetative (RO), flowering (2RO), and maturity (3RO) were sourced from farmland in Ostroda, Poland (53°41′38″N 19°57′58″E). To isolate bacteria from the rhizosphere, canola roots were washed with sterile distilled water and cut into small pieces (2 mm). Following the serial dilution process, each 10-fold diluent was spread onto nutrient agar (NA) (Biomaxima, Warsaw, Poland) plates with amphotericin B (40 µg/mL) as an antifungal agent and incubated for 3 days at 28°C. After incubation, the rhizospheric bacteria were enumerated and 50 colonies from each plant growth stage were isolated and stored in glycerol stocks at −80°C until further studies.
2.2 Assay of plant growth–promoting traits in vitro
2.2.1 Qualitative estimations
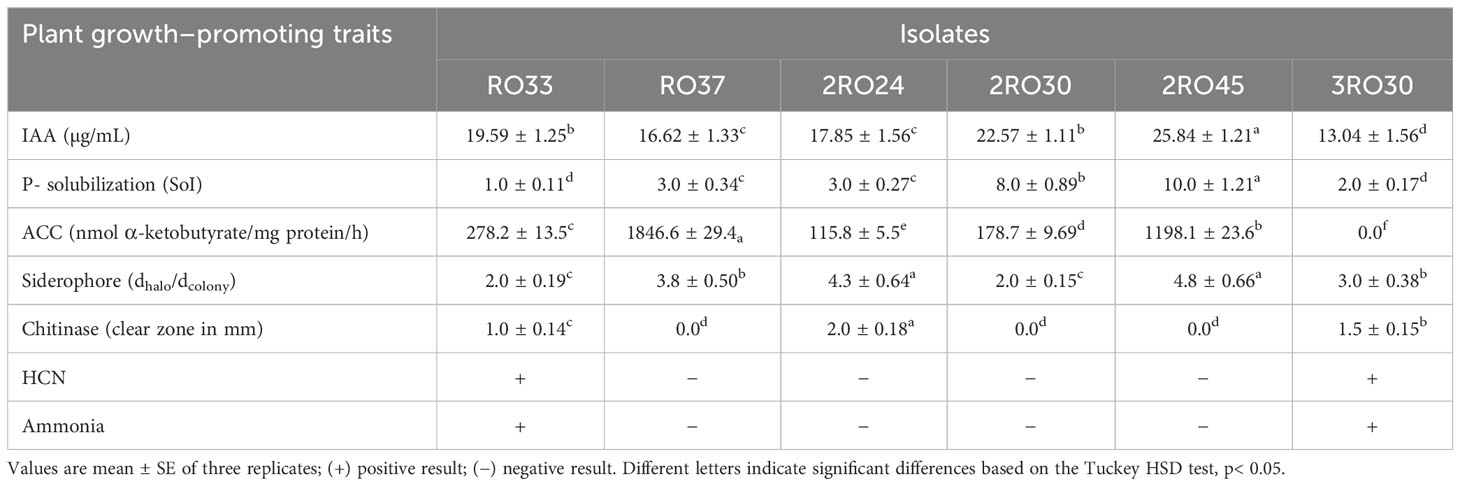
The rhizobacterial strains were qualitatively screened for PGP traits, namely, phosphates, siderophores, chitinases, hydrogen cyanide (HCN), and ammonia production. Phosphorus solubilization and siderophores sequestration abilities were assessed using Pikovskaya agar plates and Chrome Azurol Sulphonate (CAS) medium, respectively (Pikovskaya, 1948; Alexander and Zuberer, 1991). Pikovskaya agar plates were incubated at 26°C for 7 days for observation of a halo zone around the colonies. The solubilization index (SoI) was evaluated as the ratio of total diameter (colony + halo zone) to colony diameter (Luziatelli et al., 2019). The CAS plates were incubated for 4 days at 26°C for observation of an orange halo zone around the colonies. The siderophores index (SI) was calculated as the ratio of the halo zone diameter to colony diameter. For chitinase production, bacteria were inoculated on medium containing (g/L): peptone 1.0, FeSO4 × 7H2O 0.1, iron gluconate 0.1, yeast extract 0.1, colloidal chitin 7.0 g dry mass, and agar 15.0 (Swiontek Brzezinska et al., 2013). Plates were incubated for 14 days at 22°C to observe clearing zone around the colonies. The colloidal chitin was prepared with the Lingappa and Lockwood method (1962). HCN production was checked using NA plates supplemented with glycine (0.44%) according to Lorck (1948). A sterile filter paper (Whatman No. 1) was soaked in solution of picric acid (0.5%) and sodium carbonate (2%) and was kept on the plate lid. After 4 days of incubation at 28°C, color change of filter paper from yellow to brown was a positive indicator for HCN production. Ammonia production was assessed by inoculating bacteria in nutrient broth (Biomaxima, Warsaw, Poland) for 3 days at 26°C. After adding a Nessler reagent (0.5 mL), the development of orange color was considered as a positive result.
2.2.2 Quantitative estimations
2.2.2.1 Indole acetic acid production
Bacterial indole acetic acid (IAA) may increase roots length and surface, allowing the plant better uptake of soil nutrients and water, which, in turn, can stimulate plant growth (Gilbert et al., 2018).
IAA production was quantified using medium (g/L): peptone 5.0; yeast extract 3.0; and L-tryptophan 1.0 following the modified method of Ehmann (1977). After 4 days of incubation at 28°C, culture suspension was centrifuged at 10080g for 10 min. Centrifuged culture suspension supernatant was mixed with Salkowski reagent (2% 0.5 FeCl3 in 35% HClO4) and incubated for 30 min in the dark. Intensity of the color was measured at 530 nm using Hitachi U-2500 spectrophotometer.
2.2.2.2 ACC deaminase activity
PGP bacteria that express ACC deaminase activity may increase roots length and facilitate adaptation and survival of plants (del Carmen Orozco-Mosqueda et al., 2020).
Quantitative estimation of ACC was carried out according to the modified method described by Honma and Shimomura (1978). Bacterial cultures were inoculated in nutrient broth medium (Biomaxima, Warsaw, Poland), following incubation at 30°C for 24h in a rotary shaker. Culture suspension was centrifuged at 6000g for 10 min (4°C) and Dworkin and Foster (DF) salts minimal medium (5 mL) was added to the pellet (Dworkin and Foster, 1958). Culture suspension was centrifuged again at 6000g for 10 min (4°C) and DF salts minimal medium (5 mL) with 0.5 M ACC (30 μL) were added to the pellet. After incubation at 30°C for 24h, culture suspension was centrifuged at 4032g for 10 min (4°C) and bacterial cells were washed with 0.1M Tris-HCl (5 mL; pH 7.6). After centrifugation at 10000g for 5 min, 0.1 M Tris-HCl (600 μL; pH 8.5), toluene (30 μL), and 0.5 M ACC substrate (20 μL) were added to the pellet following incubation at 30°C for 30 min. 0.56 M HCl (1 ml) was added and culture suspension was centrifuged at 10000g for 5 min. Centrifuged culture suspension supernatant (1 mL) was mixed with 0.56 M HCl (800 μL) and 0.2% 2,4-dinitrophenylhydrazine (300 μL). After a 30-min long incubation at 30°C, 2 M NaCl (2 mL) was added and the absorbance was determined at 540 nm using Hitachi U-2500 spectrophotometer. The ACC deaminase was expressed in terms of nanomoles of α-ketobutyrate produced per milligram protein per hour. Protein content was estimated according Bradford (1976) method.
2.3 Greenhouse experiment in sterile and non-steriled conditions
2.3.1 Seed sterilization and inoculum preparation
The winter rapeseed of the Areti variety was selected for the study. It is a hybrid variety characterized by outstanding health and a very high yield potential. Sterilization of canola seeds was performed by disinfection in 1% NaOCl for 30 min and by washing 3 times in sterile distilled water following the method of Rudolph et al. (2015). Fresh bacterial cultures were inoculated in LB broth and incubated for 2 days at 26°C in a shaker incubator. The sterilized seeds were resuspended in bacterial suspension (10 mL of 108 CFU/mL) supplemented with 0.05 g carboxymethyl cellulose (CMC) and agitated for 30 min. The control was the seeds resuspended in 10 mL of nutrient broth without bacterial inoculum and supplemented with 0.5% CMC (Rudolph et al., 2015).
2.3.2 Seed treatment
Four canola seeds were sown per pot in eight replicates for each treatment. Seeds resuspended in six bacterial suspension (RO33, RO37, RO45, 2RO30, 2RO45, and 3RO30) were germinated in a sterile sand and vermiculite (1:1). Seeds with 2RO30, 2RO45 strains and their consortium were additionally sown in non-sterile soil taken from the field in Ostroda, Poland (53°41′38″N 19°57′58″E). Canola seedlings were maintained in a day–night cycle of 16h light (100 μmol/m2/s) and a temperature of 22°C. The plants were moistened with an equal amount of water and were harvested after 44 days (Świątczak et al., 2023b).
2.3.3 Compatibility assay
For non-sterile greenhouse experiment, in-vitro antagonism compatibility test of two strains: Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 was performed. Each isolates were resuspended in sterile water (108 CFU/mL) and 100 µL of the tested microorganism were spread onto NA plates. A sterile filter paper disc (5 mm in diameter) was placed onto NA plate containing the spread bacteria and 10 µL of bacterial suspension was inoculated on the paper disc. Plates were incubated at 28°C for 48h. When a clear zone of inhibition around the disc was observed, microorganisms were considered incompatible. When inhibition zone was not observed, they were classified as compatible (Widyantoro, 2019; Tabacchioni et al., 2021). Each experiment was performed in triplicate.
2.3.4 Canola parameters determination
Chlorophyll content index (CCI) was measured using a chlorophyll meter CCM-200plus (Opti-Sciences, Hudson, USA). Other canola growth parameters – length of roots, shoots and epicotyl were measured after washing the plant roots with distilled water. Photosynthetic area of leaves was calculated using DigiShape 1.3 software (Moraczewski, 2005). The plant roots, shoots, epicotyl, petioles and leaves were dried at 85°C for 48 hours and dry weight of these plant parts was determined. Furthermore, two following indexes were calculated: specific leaf area (SLA) and leaf weight ratio (LWR). The SLA index was calculated as follows: SLA = assimilation area [cm2]/leaves dry biomass [g], while LWR index was evaluated as follows: LWR = leaves dry biomass [g]/total plant dry biomass [g] (Piernik et al., 2017).
2.3.5 Soil physicochemical analyses
The physical and chemical parameters of the non-sterile soil were determined at District Chemical and Agricultural Station (Bydgoszcz, Poland). The physicochemical analyses such as pH, phosphorus, potassium, magnesium, ammonium nitrogen, nitrate nitrogen, and organic carbon were evaluated according to PN-ISO10390, 1997; PN-R-04020:1994/Az1, 2004; PN-R-04022, 1996; PN-R-04022:1996/Az1, 2002; PN-R-04028, 1997, and PN-ISO14235, 2003 standards, respectively.
2.4 Statistical analysis
The statistical analysis of the data was performed using Past3 (version 3.25). To determine significant differences between treatments, one-way analysis of variance (ANOVA) was applied, followed by Tuckey post-hoc test. ANOVA assumptions were checked using Shapiro–Wilk test for normality and Levene’s test for homogeneity of variances. When not normal distribution occurred, test for equal medians—Mann–Whitney was performed.
2.5 Identification of isolates based on 16S rRNA gene
Six bacterial isolates were identified based on 16S rRNA gene sequence according to Kalwasińska et al. (2020). Total genomic DNA was extracted using GeneMATRIX Bacterial and Yeast Genomic DNA Purification Kit (EURx, Gdańsk, Poland), following the manufacturer’s protocol. For polymerase chain reaction (PCR) amplification, the following 20 µL of reaction total volume was used: Taq DNA polymerase, 0.2 mM dNTP mixture, Polbuffer B with 1.5 mM MgCl2, 0.25µM of 27 F (5-AGAGTTTGATCCTGGCTCAG-3) and 1492 R (5-TACGGTTACCTTGTTACGACTT-3) primers, and 1 µL of genomic DNA. The PCR conditions were: initial denaturation (95°C for 3 min), 30 cycles of amplification: denaturation (95°C for 30s), annealing (52°C for 20s), extension (72°C for 1 min 40s) and final extension (72°C for 5 min). PCR amplicons were checked in 1% (w/v) agarose gel stained with Midori Green DNA Stain. Sequencing of PCR products was performed using Big Dye Terminator v 3.1 Cycle Sequencing Kit according to manufacturer’s instruction. Capillary electrophoresis was performed by the Sequencing and Oligonucleotides Synthesis Laboratory, IBB (Warsaw, Poland). Nucleotide sequences of RO33, RO37, RO45, 2RO30, 2RO45, and 3RO30 were submitted to GenBank under the accession numbers MW599360, MW599361, MW599362, MW599363, MW599366, and MW599367, respectively. The taxonomy of bacterial isolates were determined using the EzBioCloud database (Yoon et al., 2017).
2.6 Genome sequencing, prediction of PGP genes and biosynthetic gene cluster
Genomic DNA of Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 was extracted and whole genome was sequenced on Illumina HiSeq using 250 bp paired-end protocol at the University of Birmingham (UK). Library construction was prepared with the use of Nextera XT Library Prep kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions with the following modifications: PCR elongation time was increased from 30 s to 1 min, and 2 ng of DNA was used as an input instead of 1 ng. The library preparation and quantification of DNA were done with a Microlab STAR automated liquid handling system (Hamilton, Bonaduz, Switzerland). Quantification of pooled libraries was performed using the Kapa Biosystems Library Quantification Kit for Illumina on a Roche light cycler 96 qPCR machine. Reads were adapter trimmed using Trimmomatic 0.30 with a sliding window quality cutoff of Q15 (Bolger et al., 2014). De-novo assembly was performed on samples using SPAdes version 3.7 (Bankevich et al., 2012), and contigs were annotated using Prokka 1.11 (Seemann, 2014). Further annotation was performed using NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP). The genomic sequences are available in DDBJ/ENA/GenBank under the accession numbers JAOAQM000000000 and JAOAQN000000000, respectively, for Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45.
The prediction of PGP genes was obtained from KEGG pathway analysis (https://www.kegg.jp/kegg/mapper/reconstruct.html), while secondary metabolite biosynthesis gene clusters were determined by antiSMASH (version 6.0) (https://antismash.secondarymetabolites.org).
2.7 Assay of plant pathogens growth inhibition in vitro
Bacterial isolates were screened for inhibition against fungal pathogens: Alternaria alternata 783, Botrytis cinerea 873, Fusarium culmorum 2333, Fusarium oxysporum 872, Fusarium solani 25, Phytophthora cactorum 1925, and Phytophthora megasperma 404, which can affect canola plants (Fernandez, 2007; Krasnow and Hausbeck, 2015; Al-Lami et al., 2019; Romero et al., 2008; Monnier et al., 2018; Ismaiel et al., 2021; Yu et al., 2023). Plant pathogenic fungi were obtained from the Plant Pathogenic Bank of the Institute of Plant Protection in Poznan (Poland). Fungi were cultivated on PDA plates (Biomaxima, Warsaw, Poland) for 5 days at 26°C, while bacteria were cultivated on PDA for 24h at 26°C. After incubation, mycelium agar discs (5 mm in diameter) were inserted onto PDA plate containing spread bacteria. Cultures were incubated for 7 days at 26°C and the diameter of the fungal mycelium was estimated. Pure cultures of each fungus were used as a control. Each experiment was performed in triplicate. The fungal growth inhibition zone was estimated by the following formula:
where C is the diameter of fungi colony in the control plate, B is the diameter of fungi colony that grew in the presence of rhizobacteria (Wonglom et al., 2019).
3 Results
3.1 Assay of plant growth–promoting traits in vitro
The total rhizobacterial population was counted from different plant growth stages: the vegetative, flowering, and maturity. This analysis showed that the total bacterial load was the highest in the flowering stage followed by a maturity and vegetative stage (Supplementary Table S1). Fifty canola rhizospheric bacteria were isolated from the three plant growth stages and were tested for PGP traits including IAA, phosphate, ACC deaminase, siderophores, chitinases, HCN, and ammonia production. Ammonia and HCN production by bacterial strains can also have a positive effect on plant growth, for example by elongation of plant roots and shoots (Bhattacharyya et al., 2020). The plant growth promotion assay in vitro showed that rhizobacterial strains isolated from different growth stages exhibited various PGP traits (Supplementary Figure S1). Six isolates possessed the best PGP properties (Table 1). All of the strains were able to produce IAA, sequester siderophores and solubilize phosphates. Among these strains 2RO45 was the most effective regarding all three PGP. RO37 demonstrated the highest ACC deaminase activity. Three of the isolates: RO33, RO45, and 3RO30 were able to produce chitinases. Moreover, RO33 and 3RO30 strains were HCN and ammonia producers.
3.2 Greenhouse experiment in sterile and non-steriled conditions
Six bacterial strains identified as Pseudomonas sp. RO33, Pseudomonas poae RO37, Pseudomonas kairouanensis RO45, Peribacillus frigoritolerans 2RO30, Pseudomonas sivasensis 2RO45, and Pseudomonas migulae 3RO30 were used in greenhouse experiment to evaluate their in-vivo ability to stimulate canola growth under sterile conditions (Figure 1). The results showed that P. frigoritolerans 2RO30 and P. sivasensis 2RO45 significantly increased CCI and roots length compared to the non-inoculated control and to seedlings inoculated with RO33, RO37, RO45, and 3RO30. In addition, these isolates significantly improved shoots length and epicotyl weight compared to the untreated control. Seedlings inoculated with P. frigoritolerans 2RO30 showed significantly higher mean value of epicotyl length compared to RO37, RO45, and 3RO30, while for P. sivasensis 2RO45, this parameter was higher compared to RO37 and 3RO30 strains. Inoculated canola with 2RO30 and 2RO45 had significantly higher shoots weight compared to non-inoculated plants and those inoculated with RO33, RO37, and RO45 strains. Moreover, P. sivasensis 2RO45 induced leaves weight compared to the control and to RO33, RO37, RO45, and 3RO30 treatments while, for petioles weight, significant increase was observed compared to seedlings inoculated with RO33, RO45, and 3RO30, but not to the control. The PGP effect of P. frigoritolerans 2RO30 and P. sivasensis 2RO45 under sterile conditions is visible in the Figure 2.
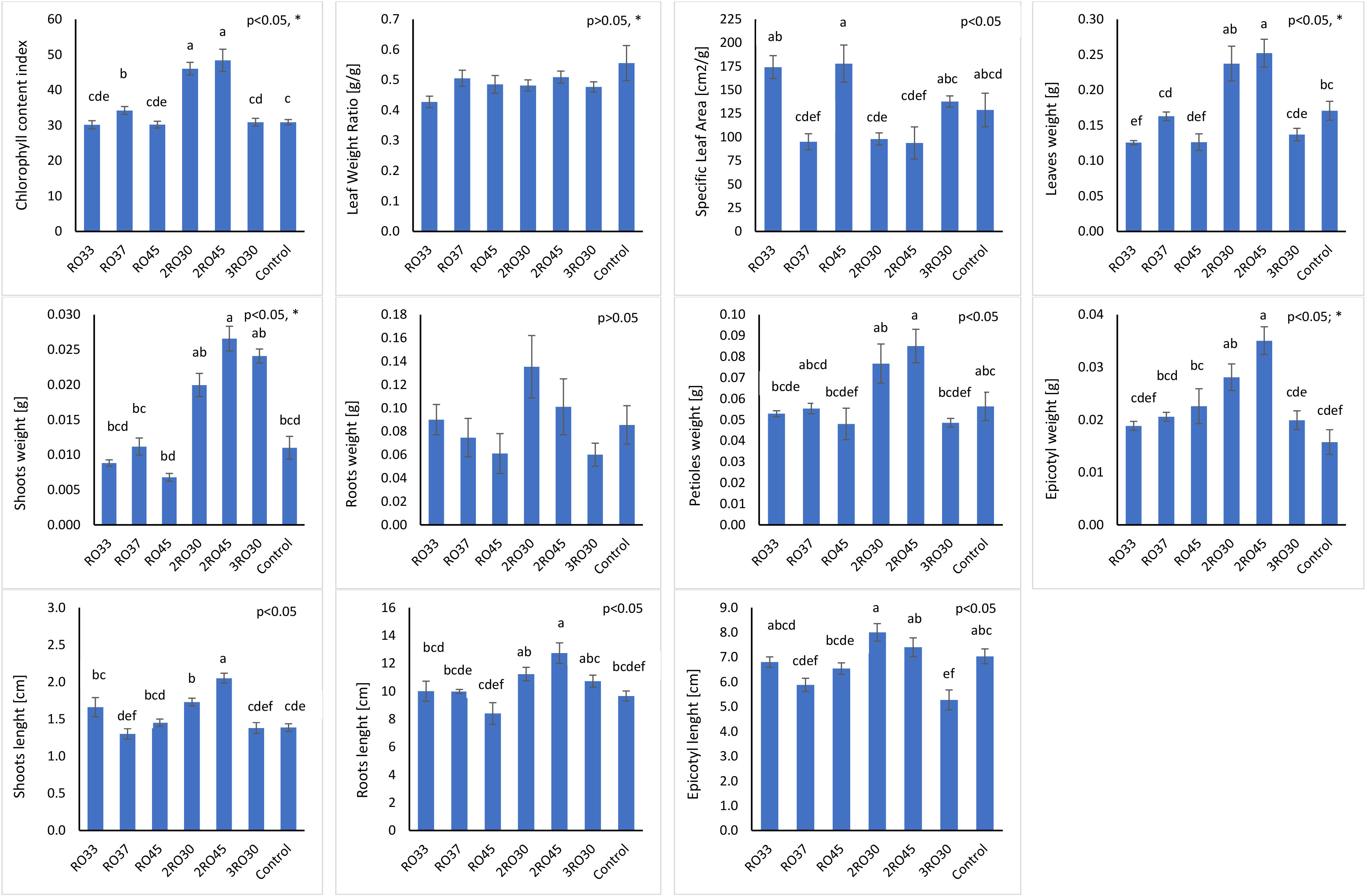
Figure 1 Plant growth parameters in sterile soil. Different letters indicate significant differences based on Tukey test as a post hoc, p< 0.05; *not normal distribution (test for equal medians: Mann–Whitney).

Figure 2 Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 effect on canola growth compared to untreated control under sterile conditions.
Based on growth promotion effects under sterile conditions P. frigoritolerans 2RO30 and P. sivasensis 2RO45 were selected for greenhouse experiment in non-sterile soil. Additionally, as compatibility between these strains was not observed, the consortium of the isolates was used for the experiment (Figure 3).
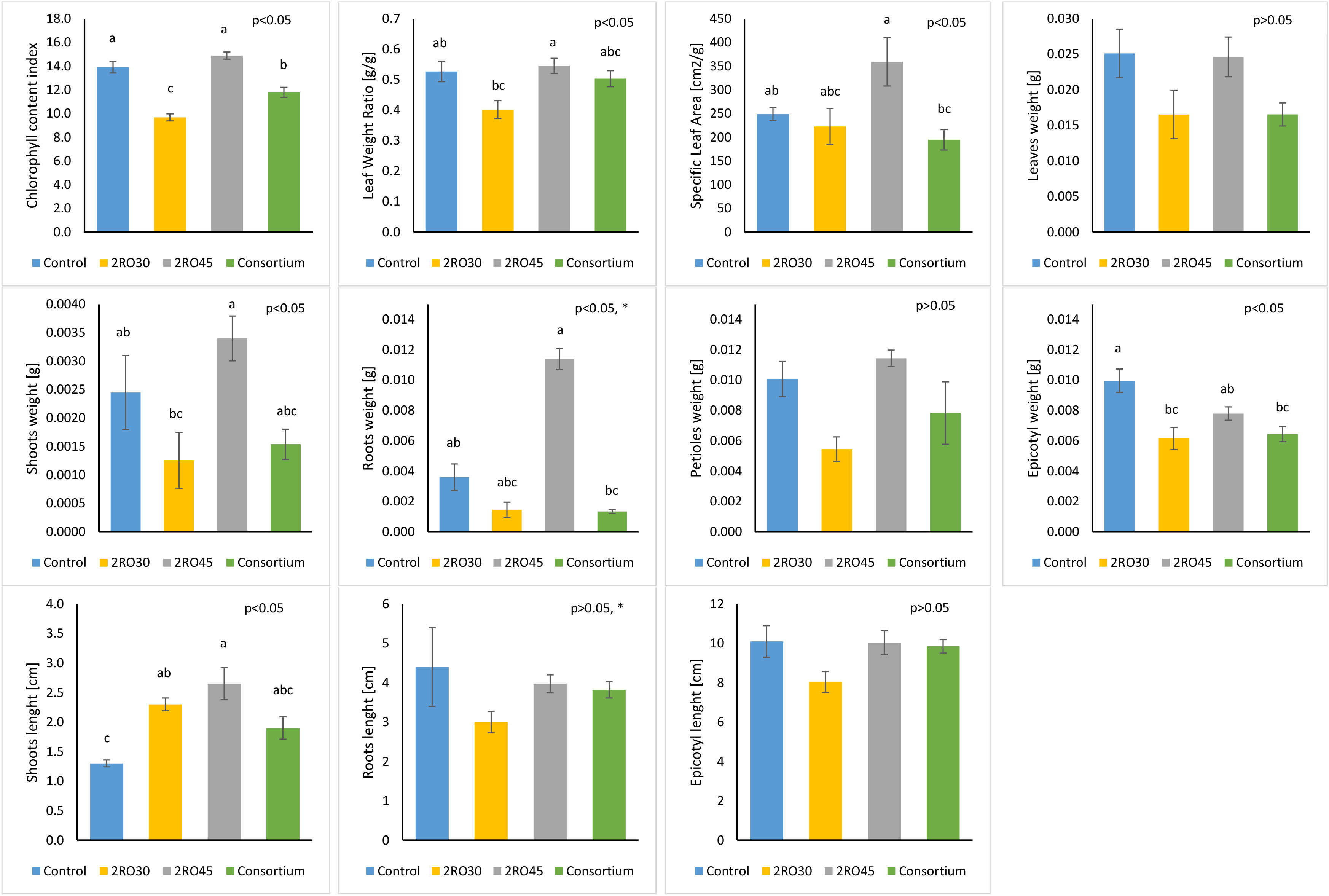
Figure 3 Plant growth parameters in non-sterile soil. Different letters indicate significant differences based on Tukey test as a post hoc, p< 0.05; *not normal distribution (test for equal medians: Mann–Whitney).
Our findings demonstrated that P. sivasensis 2RO45 significantly improved CCI compared to the untreated control and other inoculant treatments. Moreover, 2RO45 strain significantly increased shoots length compared to the non-inoculated control, but not to the other inoculated seedlings. Seedlings inoculated with the strain showed significantly higher SLA index and weight of roots compared to consortium but not to the control. Significant differences in LWR index and shoots weight between P. frigoritolerans 2RO30 and P. sivasensis 2RO45 were observed with higher mean values for P. sivasensis 2RO45. Figure 4 showed the PGP effect of P. sivasensis 2RO45 compared to untreated control under non-sterile conditions.
The physicochemical analysis of the non-sterile soil (Supplementary Table S2) recorded potassium content of 350 mg/kg, followed by phosphorus content of 310 mg/kg, and magnesium content of 91 mg/kg. The ammonium nitrogen, nitrate nitrogen, and organic carbon contents were 6.57 mg/kg, 79.13 mg/kg, and 1.65%, respectively. The soil had a slightly acidic pH.
3.3 Genome sequencing, prediction of PGP genes, and biosynthetic gene clusters
Annotation of P. frigoritolerans 2RO30 and P. sivasensis 2RO45 genomes identified 5,435 and 5,797 coding genes, respectively, and 120 and 77 pseudogenes, respectively. In total, 1,877,684 and 2,803,949 reads were obtained from the whole-genome sequencing of 2RO30 and 2RO45 and assembled into 85 and 92 contigs with GC content of 40.19% and 59.63%, respectively (Supplementary Table S3).
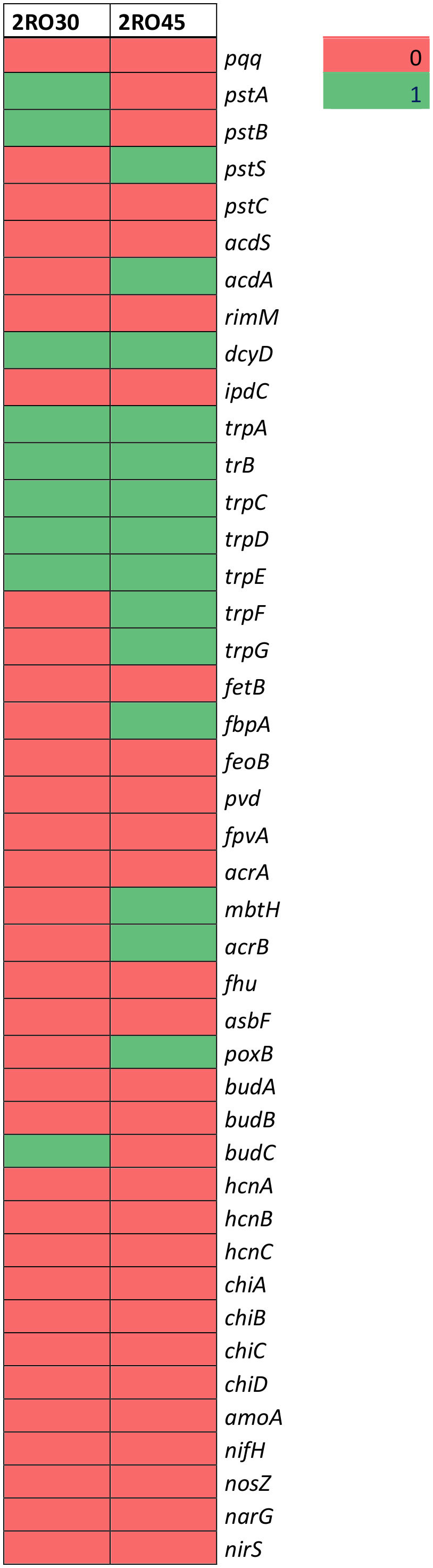
Whole genome sequencing of the P. frigoritolerans 2RO30 and P. sivasensis 2RO45 revealed differences in identified PGP genes between strains (Figure 5 and Supplementary Table S4). 2RO30 genome harbored pstA, and pstB, while 2RO45 genome coded for pstS, genes, which are responsible for phosphate transport. The genomes of 2RO30 and 2RO45 included genes involving in ACC deaminase (dcyD) and IAA (trpA,B,C,D,E) production. However, additional genes contributing to ACC deaminase (acdA) and IAA (trpF, trpG) were detected in P. sivasensis 2RO45 genome. Among 2RO30 and 2RO45 genomes, only 2RO45 genome contained genes involved in siderophores sequestration (fbpA, mbtH, and acrB). Moreover, genes related to acetoin and butanediol synthesis were found in 2RO30 and 2RO45 genomes. 2RO30 genome coded for budC gene, while 2RO45 coded for poxB.
The analysis of biosynthetic gene clusters (BGC) revealed the presence of betalactone (fengycin) in both P. frigoritolerans 2RO30 and P. sivasensis 2RO45 genomes. Moreover, four additional type of genes, including nonribosomal peptide synthetases (NRPS) (viscosin, syringomycin, and pyoverdin), terpene (enterobactin), arylpolyene (APE Vf), and redox-cofactor (lankacidin C) were detected in 2RO45 genome. The abovementioned BGCs of P. frigoritolerans 2RO30 and P. sivasensis 2RO45 are present in the Figure 6 and Supplementary Table S5.
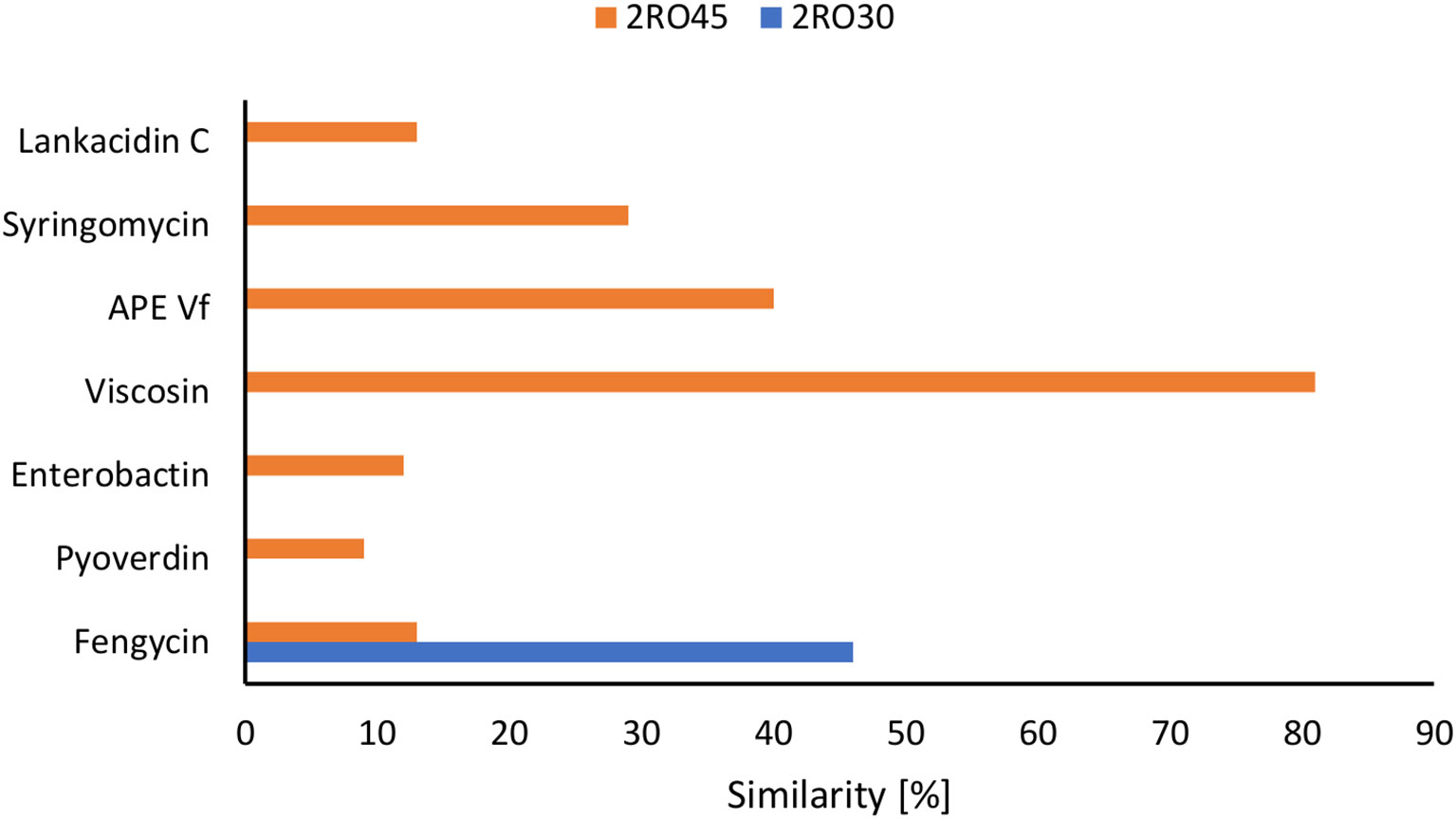
Figure 6 Detection of the secondary metabolite genes in Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 genomes.
3.4 Assay of plant pathogens growth inhibition in vitro
The phytopathogens inhibition assay revealed that P. sivasensis 2RO45 had antifungal effect against all tested fungi. 2RO45 strain inhibited the mycelial growth of six plant pathogens (A. alternata, B. cinerea, F. culmorum, F. oxysporum, F. solani, and P. cactorum) with the inhibition rate of more than 10%. Whereas, P. frigoritolerans 2RO30 antagonistic effect in the rate greater than or equal to 10% was observed only against B. cinerea and F. oxysporum (Figure 7).
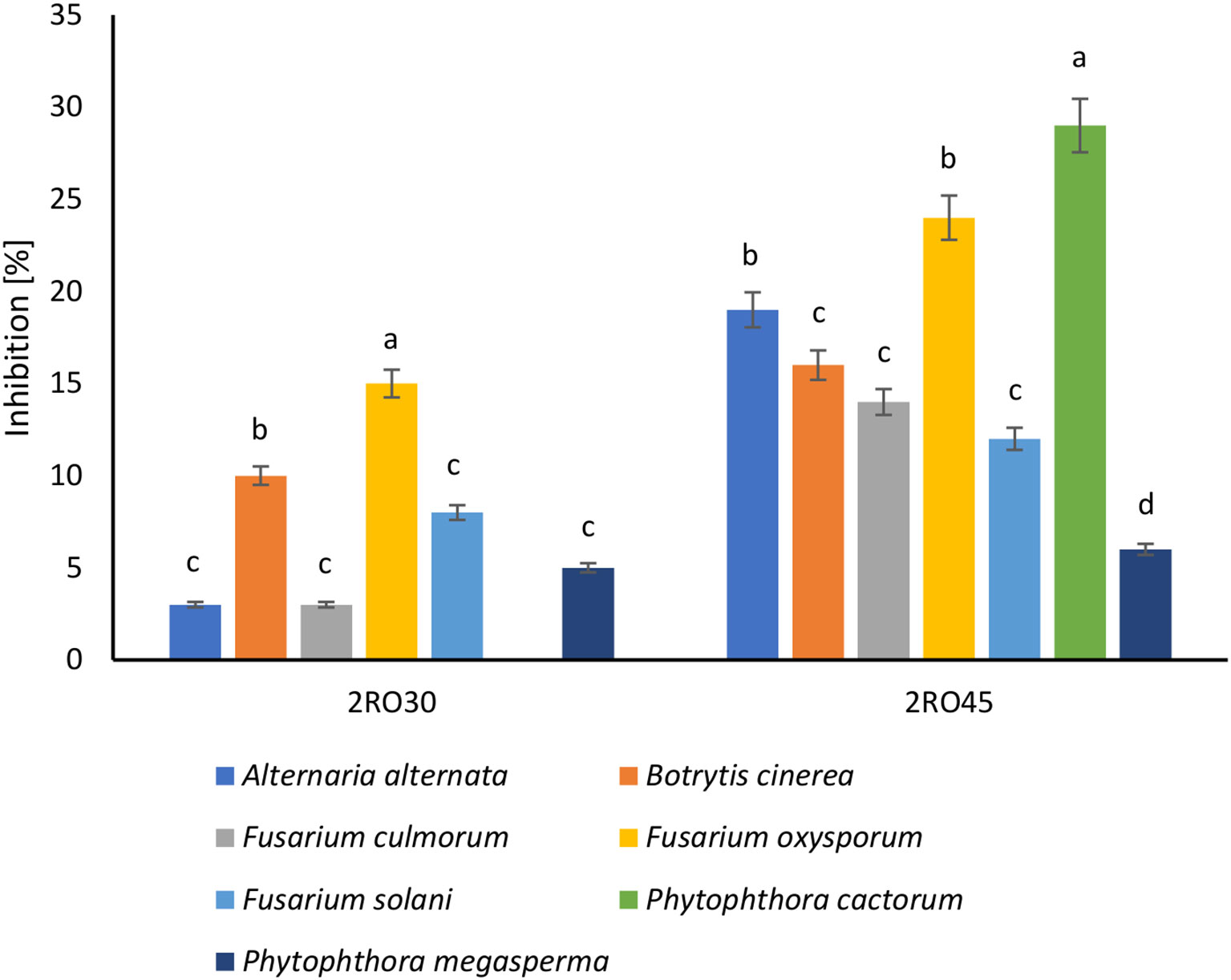
Figure 7 Inhibition of plant pathogens by Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45. Different letters indicate significant differences based on the Tuckey HSD test, p< 0.05.
4 Discussion
PGPR can promote plant growth by direct and indirect mechanisms, which can be active simultaneously or independently at different plant growth stages (Kumar et al., 2012). In our study, the bacterial community was enumerated from three canola growth stages: vegetative, flowering and maturity. The results showed that the total bacterial load was the highest in the flowering stage followed by a maturity and vegetative stage. It can be explained by the fact that, when a plant grows, rhizospheric bacteria enter the plant system at vegetative stage and multiply during flowering stage but again start declining at maturity stage, for example, due to water stress, nutrient deficiency or free radical formation (Marag and Suman, 2018). Our results are in agreement with other studies. Marag and Suman (2018) enumerated endophytic population from different maize growth stages and found that the maximum bacterial loads were at the flowering stage. However, it is not known whether there is any association between the isolation of bacteria from different growth stages and their plant growth promoting (PGP) characteristics. Our study showed that rhizobacterial strains isolated from different periodic growth stages exhibited various PGP traits. However, a higher number of strains with the following PGP activities: IAA, ACC, phosphates, siderophores, and ammonia production were isolated from the flowering stage. This could be explained by much higher bacterial loads at the flowering stage than at the vegetative and maturity stages.
In addition, our results showed that two strains isolated from flowering stage growth: P. frigoritolerans 2RO30 and P. sivasensis 2RO45 were the most effective in stimulating canola growth in sterile conditions. According to Pérez-Montaño et al. (2014) when PGPR promote plant growth under sterile conditions, their effect on plant development should be evaluated also under non-sterile conditions. Therefore, to demonstrate the plant growth promotion ability of P. frigoritolerans 2RO30 and P. sivasensis 2RO45, a greenhouse experiment in non-sterile conditions was performed. In addition to testing the PGP effect of P. frigoritolerans 2RO30 and P. sivasensis 2RO45, a consortium of these two strains was used in the non-sterile experiment. Swiontek Brzezinska et al. (2022) showed that co-inoculation of three rhizobacterial strains from different taxa (Pseudomonas, Sphingobacterium and Microbacterium) enhanced canola growth in comparison to single-inoculant treatments. However, our results showed that canola growth parameters increased only when plants were inoculated with P. sivasensis 2RO45. Interestingly, P. frigoritolerans 2RO30, which promoted plant growth under sterile conditions, did not promote canola under non-sterile conditions. Beneficial effects of PGPR on plant development are highly variable under natural conditions due to the native rhizosphere microbial communities’ presence (Pacheco da Silva et al., 2022). According to Bhattacharyya et al. (2018), certain indigenous microbes might negatively affect the function and survival of introduced PGPR. Our results could be explained by that P. frigoritolerans 2RO30 failed in competition with soil natural microbiota and therefore it did not promote in non-sterile conditions. Moreover, it could be associated with differences in PGP characteristics in vitro and PGP genes between P. frigoritolerans 2RO30 and P. sivasensis 2RO45 strains.
High IAA production and phosphates solubilization features responsible for promoting canola growth under sterile conditions have been confirmed by the presence of appropriate genes in the genomes of these strains. However, P. sivasensis 2RO45 coded for many additional genes related to ACC and IAA production, such as acdA and trpF,G, which could have contributed to better canola growth promotion both in sterile and non-sterile soil. PGPR that produce IAA may increase root biomass, allowing the plant better access to soil nutrients and water uptake, which in turn can stimulate plant growth (Gilbert et al., 2018). Whereas, ACC deaminase can facilitate adaptation and survival of plants and increase roots length, thereby promote plant growth (del Carmen Orozco-Mosqueda et al., 2020). Świątczak et al. (2023c) found that the presence of trpA,B,C,D,E,F,S and pqq, pstAB genes in Brevibacillus laterosporus K75 genome and its high IAA and phosphates activity in vitro contributed to maize growth promotion under sterile and non-sterile conditions.
Apart from the genes mentioned above, 2RO45 genome contained fbpA, mbtH, acrB genes involved in siderophores production. BGC analysis revealed the presence of siderophore–enterobactin in its genome. Moreover, P. sivasensis 2RO45 showed the highest siderophores production in the PGP assay in vitro. It was reported that siderophores are responsible for plant growth promotion indirectly by inhibition of plant pathogens. However, siderophores can also stimulate plant species directly by enhancing iron (Fe) uptake in plants (dos Santos et al., 2020). Therefore, we assumed that high siderophores activity in vitro and the presence of many genes related to siderophores production in comparison to P. frigoritolerans 2RO30 are also responsible for P. sivasensis 2RO45 PGP effect under non-sterile conditions.
Moreover, P. sivasensis 2RO45 inhibited the mycelial growth of all fungi and with higher inhibition rates than P. frigoritolerans 2RO30. These observations are confirmed by genome mining and BGC analysis where P. sivasensis 2RO45 harbored six more secondary metabolite cluster genes than P. frigoritolerans 2RO30. The founded NRPS: syringomycin, pyoverdine, and viscosin are well known as antifungal agents (Bensaci et al., 2011; Alsohim et al., 2014; Sass et al., 2021). Fengycin is a lipopeptide producing by Bacillus amyloliquefaciens PPL, which contributes to antifungal activity against Fusarium oxysporum (Kang et al., 2020). Arylpolyene from Vibrio fischeri ES114 (APE Vf) was found in many Pseudomonas species (Gaete et al., 2022; Han et al., 2022) and according to Dutta et al. (2020) this lipid influences the antifungal effect of P. fluorescens NBC275. Strain Streptomyces rochei FS18 producing polyketide antibiotic–lankacidin and its derivatives, showed significant antifungal effect against Aspergillus niger (Cao et al., 2015). It can be assumed that these BGCs were responsible for the antifungal activity of P. sivasensis 2RO45 against plant pathogens, and if it has more possibilities of defense against pathogens, it promotes plants more effectively.
5 Conclusions
Pseudomonas sivasensis 2RO45 and Peribacillus frigoritolerans 2RO30 possessing the highest IAA production and phosphates solubilization, showed the best promoting effect on canola growth under sterile conditions. Under non-sterile conditions, only Pseudomonas sivasensis 2RO45 promoted canola growth, which can be associated with the presence of additional genes responsible for ACC deaminase (acdA), IAA (trpF, trpG), and siderophores production (fbpA, mbtH, and acrB) in its genome. Our study is the first report describing Pseudomonas sivasensis 2RO45 as a plant growth promoter, both under controlled and natural conditions, thus suggesting its application in improving canola yield, by improving nutrient availability, enhancing stress tolerance, and reducing the environmental impact of farming practices. Its application aligns with the principles of environmentally friendly and economically viable agriculture. However, more research about how this strain competes with native soil microbes is necessary.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JŚ wrote the original draft, made plant growth – promoting assays in vitro and in vivo, conducted statistical data and genome analysis, and prepared the manuscript editorially; AK coordinated the study, and checked the validity of the original draft; MS designed and coordinated the study, checked the validity of the original draft. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by funds provided by Nicolaus Copernicus University (Torun, Poland) to maintain research potential and Excellence Initiative Research University competition for scientific groups - BIOdegradable PACKaging materials research group.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1233237/full#supplementary-material
References
Adedeji, A. A., Häggblom, M. M., Babalola, O. O. (2020). Sustainable agriculture in Africa: Plant growth-promoting rhizobacteria (PGPR) to the rescue. Sci. Afr. 9, e00492. doi: 10.1016/j.sciaf.2020.e00492
Alexander, D. B., Zuberer, D. A. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12, 39–45. doi: 10.1007/BF00369386
Al-Lami, H. F. D., You, M. P., Barbetti, M. J. (2019). Incidence, pathogenicity and diversity of Alternaria spp. associated with Alternaria leaf spot of canola (Brassica napus) in Australia. Plant Pathol. 68, 492–503. doi: 10.1111/ppa.12955
Alsohim, A. S., Taylor, T. B., Barrett, G. A., Gallie, J., Zhang, X. X., Altamirano-Junqueira, A. E., et al. (2014). The biosurfactant viscosin produced by Pseudomonas fluorescens SBW 25 aids spreading motility and plant growth promotion. Environ. Microbiol. 16, 2267–2281. doi: 10.1111/1462-2920.12469
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bensaci, M. F., Gurnev, P. A., Bezrukov, S. M., Takemoto, J. Y. (2011). Fungicidal activities and mechanisms of action of Pseudomonas syringae pv. syringae lipodepsipeptide syringopeptins 22A and 25A. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00216
Bhattacharyya, C., Banerjee, S., Acharya, U., Mitra, A., Mallick, I., Haldar, A., et al. (2020). Evaluation of plant growth promotion properties and induction of antioxidative defense mechanism by tea rhizobacteria of Darjeeling, India. Sci. Rep. 10, 15536. doi: 10.1038/s41598-020-72439-z
Bhattacharyya, D., Duta, S., Yu, S. M., Jeong, S. C., Lee, Y. H. (2018). Taxonomic and functional changes of bacterial communities in the rhizosphere of kimchi cabbage after seed bacterization with Proteus vulgaris JBLS202. Plant Pathol. J. 34, 286. doi: 10.5423/PPJ.OA.03.2018.0047
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1006/abio.1976.9999
Cao, Z., Yoshida, R., Kinashi, H., Arakawa, K. (2015). Blockage of the early step of lankacidin biosynthesis caused a large production of pentamycin, citreodiol and epi-citreodiol in Streptomyces rochei. J. Antibiot. 68, 328–333. doi: 10.1038/ja.2014.160
Chouyia, F. E., Romano, I., Fechtali, T., Fagnano, M., Fiorentino, N., Visconti, D., et al. (2020). P-Solubilizing Streptomyces roseocinereus MS1B15 with multiple plant growth-promoting traits enhance barley development and regulate rhizosphere microbial population. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01137
de Andrade, L. A., Santos, C. H. B., Frezarin, E. T., Sales, L. R., Rigobelo, E. C. (2023). Plant growth-promoting rhizobacteria for sustainable agricultural production. Microorganisms 11, 1088. doi: 10.3390/microorganisms11041088
del Carmen Orozco-Mosqueda, M., Glick, B. R., Santoyo, G. (2020). ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 235, 126439. doi: 10.1016/j.micres.2020.126439
dos Santos, R. M., Diaz, P. A. E., Lobo, L. L. B., Rigobelo, E. C. (2020). Use of plant growth-promoting rhizobacteria in maize and sugarcane: characteristics and applications. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.00136
Duman, M., Mulet, M., Saticioglu, I. B., Altun, S., Gomila, M., Lalucat, J., et al. (2020). Pseudomonas sivasensis sp. nov. isolated from farm fisheries in Turkey. Syst. Appl. Microbiol. 43, 126103. doi: 10.1016/j.syapm.2020.126103
Dutta, S., Yu, S. M., Lee, Y. H. (2020). Assessment of the contribution of antagonistic secondary metabolites to the antifungal and biocontrol activities of Pseudomonas fluorescens NBC275. Plant Pathol. J. 36, 491. doi: 10.5423/PPJ.FT.08.2020.0149
Dworkin, M., Foster, J. (1958). Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 75, 592–603. doi: 10.1128/jb.75.5.592-603.1958
Ehmann, A. (1977). The Van Urk-Salkowski reagent—a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr A 132, 267–276. doi: 10.1016/S0021-9673(00)89300-0
Etesami, H., Jeong, B. R., Glick, B. R. (2023). Potential use of Bacillus spp. as an effective biostimulant against abiotic stresses in crops—A review. Curr. Res. Biotechnol. 5, 100128. doi: 10.1016/j.crbiot.2023.100128
Fernandez, M. R. (2007). Fusarium populations in roots of oilseed and pulse crops grown in eastern Saskatchewan. Can. J. Plant Sci. 87, 945–952. doi: 10.4141/P06-145
Gaete, A., Andreani-Gerard, C., Maldonado, J. E., Muñoz-Torres, P. A., Sepúlveda-Chavera, G. F., González, M. (2022). Bioprospecting of plant growth-promoting traits of Pseudomonas sp. strain C3 isolated from the atacama desert: Molecular and culture-based analysis. Diversity 14, 388. doi: 10.3390/d14050388
Gilbert, S., Xu, J., Acosta, K., Poulev, A., Lebeis, S., Lam, E. (2018). Bacterial production of indole related compounds reveals their role in association between duckweeds and endophytes. Front. Chem. 6. doi: 10.3389/fchem.2018.00265
Han, P., Liu, T., Zheng, Y., Song, R., Nan, T., Yang, X., et al. (2022). A Mycorrhizal bacteria strain isolated from Polyporus umbellatus exhibits broad-spectrum antifungal activity. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.954160
Honma, M., Shimomura, T. (1978). Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42, 1825–1831. doi: 10.1080/00021369.1978.10863261
Ismaiel, M. I., Samar, S., Ali, I. N., EL-Korashy, M. A. (2021). Effectiveness of some plant extracts as safe control means against damping-off and root rot diseases in canola plants. J. Appl. Plant Prot. 10, 21–27. doi: 10.21608/JAPP.2021.222792
Jamalzadeh, A., Darvishnia, M., Khodakaramian, G., Bazgir, E., Zafari, D. (2021). Genetic diversity and plant growth-promoting activity of the dominant bacteria from canola plants in Western Iran. Egypt. J. Biol. Pest Control 31, 1–9. doi: 10.1186/s41938-021-00442-1
Kalwasińska, A., Krawiec, A., Deja-Sikora, E., Gołębiewski, M., Kosobucki, P., Swiontek Brzezinska, M., et al. (2020). Microbial diversity in deep-subsurface hot brines of Northwest Poland: From community structure to isolate characteristics. Appl. Environ. Microbiol. 86, e00252-20. doi: 10.1128/AEM.00252-20
Kang, B. R., Park, J. S., Jung, W. J. (2020). Antifungal evaluation of fengycin isoforms isolated from Bacillus amyloliquefaciens PPL against Fusarium oxysporum f. sp. lycopersici. Microb. Pathog. 149, 104509. doi: 10.1016/j.micpath.2020.104509
Kashyap, B. K., Solanki, M. K., Pandey, A. K., Prabha, S., Kumar, P., Kumari, B. (2019). Bacillus as plant growth promoting rhizobacteria (PGPR): a promising green agriculture technology. Plant Health Under Biotic Stress: Volume 2: Microbial Interactions 219–236.
Krasnow, C. S., Hausbeck, M. K. (2015). Pathogenicity of Phytophthora capsici to Brassica vegetable crops and biofumigation cover crops (Brassica spp.). Plant Dis. 99, 1721–1726. doi: 10.1094/PDIS-03-15-0271-RE
Kumar, A., Kumar, A., Devi, S., Patil, S., Payal, C., Negi, S. (2012). Isolation, screening and characterization of bacteria from Rhizospheric soils for different plant growth promotion (PGP) activities: an in vitro study. Recent. Res. Sci. Technol. 4, 1–5.
Kumari, B., Mallick, M. A., Solanki, M. K., Solanki, A. C., Hora, A., Guo, W. (2019). “Plant growth promoting rhizobacteria (PGPR): modern prospects for sustainable agriculture Plant health under biotic stress. Eds. Ansari, R., Mahmood, I. (Singapore: Springer). doi: 10.1007/978-981-13-6040-4_6
Lastochkina, O., Seifikalhor, M., Aliniaeifard, S., Baymiev, A., Pusenkova, L., Garipova, S., et al. (2019). Bacillus spp.: efficient biotic strategy to control postharvest diseases of fruits and vegetables. Plants 8, 97. doi: 10.3390/plants8040097
Lingappa, Y., Lockwood, J. (1962). Chitin media for selective isolation and culture of actinomycetes. Phytopathology 52, 317–323.
Lorck, H. (1948). Production of hydrocyanic acid by bacteria. Physiol. Plant 1, 142–146. doi: 10.1111/j.1399-3054.1948.tb07118.x
Luziatelli, F., Ficca, A. G., Colla, G., Baldassarre Švecová, E., Ruzzi, M. (2019). Foliar application of vegetal-derived bioactive compounds stimulates the growth of beneficial bacteria and enhances microbiome biodiversity in lettuce. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00060
Marag, P. S., Suman, A. (2018). Growth stage and tissue specific colonization of endophytic bacteria having plant growth promoting traits in hybrid and composite maize (Zea mays L.). Microbiol. Res. 214, 101–113. doi: 10.1016/j.micres.2018.05.016
Monnier, N., Furlan, A., Botcazon, C., Dahi, A., Mongelard, G., Cordelier, S., et al. (2018). Rhamnolipids from Pseudomonas aeruginosa are elicitors triggering Brassica napus protection against Botrytis cinerea without physiological disorders. Front. Plant Sci. 9, 1170. doi: 10.3389/fpls.2018.01170
Montecillo, J. A. V., Bae, H. (2022). Reclassification of Brevibacterium frigoritolerans as Peribacillus frigoritolerans comb. nov. based on phylogenomics and multiple molecular synapomorphies. Int. J. Syst. Evol. Microbiol. 72, 5389. doi: 10.1099/ijsem.0.005389
Moraczewski, I. (2005). digiShape. Automatic morphometry program (polish), Version 1.3 (Cortex Nova, Bydgoszcz, PL).
Oni, F. E., Esmaeel, Q., Onyeka, J. T., Adeleke, R., Jacquard, C., Clement, C., et al. (2022). Pseudomonas lipopeptide-mediated biocontrol: Chemotaxonomy and biological activity. Molecules 27, 372. doi: 10.3390/molecules27020372
Pacheco da Silva, M. L., Moen, F. S., Liles, M. R., Feng, Y., Sanz-Saez, A. (2022). The response to inoculation with PGPR plus orange peel amendment on soybean is cultivar and environment dependent. Plants 11, 1138. doi: 10.3390/plants11091138
Patel, J. S., Kumar, G., Bajpai, R., Teli, B., Rashid, M., Sarma, B. K. (2021). “PGPR formulations and application in the management of pulse crop health,” in In biofertilizers (Cambridge, UK: Woodhead Publishing), 239–251.
Pérez-Montaño, F., Alías-Villegas, C., Bellogín, R. A., Del Cerro, P., Espuny, M. R., Jiménez-Guerrero, I., et al. (2014). Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol. Res. 169, 325–336. doi: 10.1016/j.micres.2013.09.011
Piernik, A., Hrynkiewicz, K., Wojciechowska, A., Szymańska, S., Lis, M. I., Muscolo, A. (2017). Effect of halotolerant endophytic bacteria isolated from Salicornia europaea L. on the growth of fodder beet (Beta vulgaris L.) under salt stress. Arch. Agron. Soil Sci. 63, 1404–1418. doi: 10.1080/03650340.2017.1286329
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17, 362–370.
PN-ISO14235 (2003). Determination of organic carbon content by oxidation with dichromate (VI) in sulphuric acid medium [in Polish].
PN-R-04020:1994/Az1 (2004). Chemical and agricultural analysis of soil - Determination of available magnesium content [in Polish].
PN-R-04022 (1996). Chemical and agricultural analysis of soil - Determination of available potassium in mineral soils [in Polish].
PN-R-04022:1996/Az1 (2002). Soil chemical and agricultural analysis -Determination of available potassium in mineral soils [in Polish].
PN-R-04028 (1997). Soil chemical and agricultural analysis, sampling and determination of nitrogen and ammonium ions in mineral soils [in Polish].
Raufa, B., Mazhar, R., Javed, A., Tehmeena, M., Shehzad, M., Tariq, S., et al. (2019). Biocontrol potential of Bacillus gibsonii and Brevibacterium frigoritolerans in suppression of Fusarium stalk rot of maize: a sustainable approach. Asian J. Agric. Biol. 7, 320–333.
Romero, E., Zurera, C., Porras, M., Barrau, C., Romero, F. (2008). Comparative study of different Brassica species to control Phytophthora cactorum and Verticillium dahlie. J. Phytopathol. 90, S2.
Rudolph, N., Labuschagne, N., Aveling, T. A. S. (2015). The effect of plant growth promoting rhizobacteria on seed germination and seedling growth of maize. Seed Sci. Technol. 43, 507–518. doi: 10.15258/sst.2015.43.3.04
Sah, S., Krishnani, S., Singh, R. (2021). Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. CRMICR 2, 100084. doi: 10.1016/j.crmicr.2021.100084
Sass, G., Nazik, H., Chatterjee, P., Shrestha, P., Groleau, M. C., Déziel, E., et al. (2021). Altered Pseudomonas strategies to inhibit surface Aspergillus colonies. Front. Cell. Infect. Microbiol. 839. doi: 10.3389/fcimb.2021.734296
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sheoran, N., Nadakkakath, A. V., Munjal, V., Kundu, A., Subaharan, K., Venugopal, V., et al. (2015). Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds. Microbiol. Res. 173, 66–78. doi: 10.1016/j.micres.2015.02.001
Świątczak, J., Kalwasińska, A., Felföldi, T., Swiontek Brzezinska, M. (2023b). Bacillus paralicheniformis 2R5 and its impact on canola growth and N-cycle genes in the rhizosphere. FEMS Microbiol. Ecol. 99, fiad093. doi: 10.1093/femsec/fiad093
Świątczak, J., Kalwasińska, A., Szabó, A., Swiontek Brzezinska, M. (2023a). Pseudomonas sivasensis 2RO45 inoculation alters the taxonomic structure and functioning of the canola rhizosphere microbial community. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1168907
Świątczak, J., Kalwasińska, A., Wojciechowska, A., Swiontek Brzezinska, M. (2023c). Physiological properties and genomic insights into the plant growth-promoting rhizobacterium Brevibacillus laterosporus K75 isolated from maize rhizosphere. J. Sci. Food Agric. 103, 1432–1441. doi: 10.1002/jsfa.12238
Swiontek Brzezinska, M., Świątczak, J., Wojciechowska, A., Burkowska-But, A., Kalwasińska, A. (2022). Consortium of plant growth-promoting rhizobacteria enhances oilseed rape (Brassica napus L.) growth under normal and saline conditions. Arch. Microbiol. 204, 1–14. doi: 10.1007/s00203-022-03018-1
Swiontek Brzezinska, M., Jankiewicz, U., Walczak, M. (2013). Biodegradation of chitinous substances and chitinase production by the soil actinomycete Streptomyces rimosus. Int. Biodeter. Biodegr. 84, 104–110. doi: 10.1016/j.ibiod.2012.05.038
Tabacchioni, S., Passato, S., Ambrosino, P., Huang, L., Caldara, M., Cantale, C., et al. (2021). Identification of beneficial microbial consortia and bioactive compounds with potential as plant biostimulants for a sustainable agriculture. Microorganisms 9, 426. doi: 10.3390/microorganisms9020426
Tara, N., Saharan, B. S. (2017). Plant growth promoting traits shown by bacteria Brevibacterium frigrotolerans SMA23 isolated from Aloe vera rhizosphere. Agric. Sci. Digest. 37, 226–231. doi: 10.18805/asd.v37i03.9223
Wang, Z., Solanki, M. K., Kumar, A., Solanki, A. C., Pang, F., Ba, Z. X., et al. (2023). Promoting plant resilience against stress by engineering root microenvironment with Streptomyces inoculants. Microbiol. Res. 277, 127509. doi: 10.1016/j.micres.2023.127509
Widyantoro, A. (2019). Antagonism and compatibility of biofertilizer bacteria toward Fusarium oxysporum f. sp. cubense. Asian J. Agric. Biol. 7, 263–268.
Wonglom, P., Daengsuwan, W., Ito, S. I., Sunpapao, A. (2019). Biological control of Sclerotium fruit rot of snake fruit and stem rot of lettuce by Trichoderma sp. T76-12/2 and the mechanisms involved. Physiol. Mol. Plant Pathol. 107, 1–7. doi: 10.1016/j.pmpp.2019.04.007
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Keywords: PGPR, canola, sterile and non-sterile conditions, genome analysis, Peribacillus frigoritolerans, Pseudomonas sivasensis
Citation: Świątczak J, Kalwasińska A and Brzezinska MS (2024) Plant growth–promoting rhizobacteria: Peribacillus frigoritolerans 2RO30 and Pseudomonas sivasensis 2RO45 for their effect on canola growth under controlled as well as natural conditions. Front. Plant Sci. 14:1233237. doi: 10.3389/fpls.2023.1233237
Received: 02 June 2023; Accepted: 15 December 2023;
Published: 08 January 2024.
Edited by:
Eduardo V. Soares, Instituto Superior de Engenharia do Porto (ISEP), PortugalReviewed by:
Aditi Kundu, Indian Council of Agricultural Research, IndiaManoj Kumar Solanki, University of Silesia in Katowice, Poland
Copyright © 2024 Świątczak, Kalwasińska and Brzezinska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Świątczak, am9hbm5hc3dpYXRjemFrQGRva3RvcmFudC51bWsucGw=; Maria Swiontek Brzezinska, c3dpb250ZWtAdW1rLnBs
 Joanna Świątczak
Joanna Świątczak Agnieszka Kalwasińska
Agnieszka Kalwasińska Maria Swiontek Brzezinska*
Maria Swiontek Brzezinska*