- 1Department of Southern Area Crop Science, National Institute of Crop Science, Rural Development Administration, Miryang, Republic of Korea
- 2Graduate School of Green-Bio Science and Crop Biotech Institute, Kyung Hee University, Yongin, Republic of Korea
- 3Division of Life Science, Plant Molecular Biology and Biotechnology Research Center, Gyeongsang National University, Jinju, Republic of Korea
- 4Extension Service Bureau, Rural Development Administration, Jeonju, Republic of Korea
- 5National Institute of Agricultural Science, Rural Development Administration, Jeonju, Republic of Korea
- 6National Institute of Crop Science, Rural Development Administration, Suwon, Republic of Korea
‘Seolgaeng’, an opaque-endosperm rice (Oryza sativa) mutant, is used to prepare high-quality dry-milled rice flour. The mutation causing its opaque-endosperm phenotype was unknown. Map-based cloning identified a missense mutation in the gene FRUCTOSE-6-PHOSPHATE 2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 2 (OsF2KP2) in Seolgaeng. Transfer DNA insertion and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-induced f2kp2 mutants exhibited opaque endosperm. Rice harbors another F2KP gene, OsF2KP1. CRISPR/Cas9-induced double mutants of OsF2KP1 and OsF2KP2 (f2kp-d) possessed more opaque endosperm compared to f2kp2 single mutants, whereas the endosperm of the f2kp1 single mutant was normal. Grain hardness and damaged starch content were significantly reduced in f2kp2 mutants compared to the wild type and f2kp1. Amylose content was lower than normal in f2kp2 mutants but not f2kp1. Grain hardness and amylose content were much lower in f2kp-d than in f2kp2. Starch polymerization analysis revealed altered amylopectin structure in f2kp2 and f2kp-d mutants. F2KP activity was lower in f2kp2 and much lower in the double mutants when compared to the wild types, but f2kp1 showed no significant difference. In coleoptiles, hypoxia induced OsF2KP2 expression but downregulated OsF2KP1. These results suggest that OsF2KP2 functions as the main F2KP isoform in endosperm experiencing hypoxia, but OsF2KP1 may partially compensate for the absence of OsF2KP2. We propose that F2KP has a crucial role in inorganic pyrophosphate-utilizing energy metabolism for starch biosynthesis in rice endosperm.
Introduction
Endosperm is the principal storage organ in cereal grains. Flour produced from rice (Oryza sativa) endosperm has many culinary uses, including as a gluten-free substitute for wheat flour, and is consumed in processed foods such as cakes, noodles, breads, and confections. Rice varieties vary in their suitability to produce high-quality flour via dry milling, which is a relatively simple and low-cost method. Because the main component of milled rice is starch, the functionality of rice flour largely depends on grain hardness and damaged starch content (Patindol et al., 2007).
In contrast to typical translucent rice endosperm, floury opaque endosperm has numerous air spaces that make it easy to mill and thus ideal for rice flour production. This type of endosperm results from the malfunction of amyloplastic starch biosynthetic enzymes, including starch synthases, starch branching enzymes, and starch debranching enzymes. The floury opaque phenotype can also be caused by defects in energy metabolic enzymes that contribute to adenosine triphosphate (ATP) production for starch biosynthesis (Nakamura, 2018; Lee and Jeon, 2020).
The central part of the rice endosperm is subject to hypoxia, which limits ATP production via mitochondrial oxidative respiration. The low-oxygen environment necessitates an alternative supply of ATP, derived in this case via the inorganic pyrophosphate (PPi)-dependent glycolytic pathway. The first committed step of glycolysis is catalyzed by PPi-dependent fructose-6-phosphate 1-phosphotransferase (PFP), which interconverts fructose-6-phosphate (F6P) and fructose-1,6-bisphosphate (F1,6BP). Rice PFP mutants possess a floury opaque endosperm (Duan et al., 2016; Chen et al., 2020). PFP activity is enhanced by fructose-2,6-bisphosphate (F2,6BP), which promotes PPi-dependent glycolytic metabolic flux (Stitt, 1990; Trevanion, 2002; Nielsen et al., 2004). The formation and degradation of F2,6BP is catalyzed by the bifunctional enzyme fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase (F2KP). Little is known about the role of F2KP in starch biosynthesis in the endosperm of crop plants, including rice.
The opaque-endosperm rice mutant variety ‘Seolgaeng’ was generated via N-methyl-N-nitrosourea treatment. Seolgaeng is commercially used to produce high-quality dry-milled rice flour that exhibits a uniformly fine particle size, with round starch granules and relatively little damaged starch (Kwak et al., 2017). The mutation causing the opaque-endosperm phenotype in Seolgaeng has not been identified. In this study, we discovered using map-based cloning that Seolgaeng harbors a mutation in an F2KP gene, Oryza sativa FRUCTOSE-6-PHOSPHATE 2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 2 (OsF2KP2). We showed that this mutation is responsible for the floury opaque-endosperm phenotype of Seolgaeng. Our results reveal the crucial role of F2KP in producing F2,6BP, an activator of PPi-dependent PFP, in rice endosperm during starch biosynthesis. In addition, we generated new rice mutants deficient in both F2KP isoforms, OsF2KP1 and OsF2KP2, via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing. The double mutants display improved rice flour characteristics and are more suitable for high-quality dry-milled flour production than Seolgaeng.
Materials and methods
Plant materials and growth conditions
Wild-type japonica rice (Oryza sativa) variety ‘Ilpumbyeo’ (Ilpum) and the opaque-endosperm mutant ‘Seolgaeng’, which was induced by N-methyl-N-nitrosourea treatment of ‘Ilpum’ (Kwak et al., 2017), were used in this study. Wild-type japonica rice variety ‘Saeilmi’ was used to produce the mapping population by crossing it with Seolgaeng. Wild-type japonica rice variety ‘Taichung’ and its respective transfer DNA (T-DNA) insertion mutant were used for analysis. Wild-type japonica rice variety ‘Dongjin’ was used to produce mutant alleles via CRISPR/Cas9-mediated genome editing. All plants were grown in the Living Modified Organism paddy fields at the Department of Southern Area Crop Science, Miryang, and Kyung Hee University, Yongin, Korea, under natural conditions during the summer.
Morphological and physicochemical analysis of rice grains
The ratio of amylose to amylopectin and total starch content were determined using an Amylose–amylopectin Assay kit (K-AMYL; Megazyme, Wicklow, Ireland) and a Total Starch Assay kit (K-TSTA; Megazyme, Wicklow, Ireland), respectively, according to the manufacturer’s instructions. Rice endosperm hardness was determined by measuring the pressure at the grain breakage point using a universal testing machine (Z 0.5, Zwick Roell, Ulm, Germany) (Kwak et al., 2017). To measure damaged starch content, 100 ± 10 mg of rice flour from each mutant and its respective wild type was tested using a Starch Damage Assay kit (K-SDAM; Megazyme, Wicklow, Ireland). For microscopy analysis of the opaque-endosperm phenotype, rice grains were observed on an LED illuminator. Scanning electron microscopy was carried out as described previously (Ryoo et al., 2007). Starch samples coated with gold were observed under a Stereoscan Leica Model 440 instrument (Leica Cambridge) at an accelerating voltage of 20 kV. The chain-length distribution of starch extracted from mature endosperm was determined according to a previously described high performance anion exchange chromatography method (Ryoo et al., 2007).
Map-based cloning
To develop a mapping population, Seolgaeng was crossed to Saeilmi to yield the line YR29150. The F2 individuals from self-fertilized YR29150 plants were used for phenotypic analysis of the endosperm. One of the progeny plants, YR29150-1, an opaque-endosperm line, was backcrossed to Saeilmi to produce a BC1F1 line named YR32422, which was further propagated to produce BC1F2 individuals. Kompetitive Allele-Specific PCR (KASP) markers were used to construct an initial map of the causal gene (Cheon et al., 2018). BC1F2 and BC1F3 individuals with wild-type endosperm were sequentially analyzed to narrow down the candidate region. To identify mutations in the target interval, genomic DNA sequencing was performed in the Seolgaeng and Ilpum backgrounds.
Isolation and production of Osf2kp mutants
To isolate the mutated DNA in the T-DNA insertional mutant f2kp2-2, the primer sets F2KP2-TF/F2KP2-TR and F2KP2-RB/F2KP2-TR (Supplementary Table S1) were used to amplify gene-specific and T-DNA-specific regions, respectively. To produce CRISPR/Cas9-mediated single knockout mutants for OsF2KP1 or OsF2KP2, the sequences 5′-AGACACCCCGTGCATCATTG-3′ and 5′-AGTTTGTGGAAGTAATGAGG-3′ were selected as targets, respectively, using the CRISPRdirect program, avoiding off-target effects (Naito et al., 2015). Single-strand oligos for each target were annealed, cloned into the entry vector pOs-sgRNA, and subcloned into the destination vector pH-Ubi-cas9-7 using the Gateway system (Miao et al., 2013). For the f2kp1 f2kp2 double mutant, tRNA-target-gRNA-tRNA-target-gRNA fragments were synthesized via Golden Gate assembly, with 5′-TACTGACATCAGGGATTCCC-3′ and 5′-TGACCTAGCTGCTAGTTGGA-3′ as the respective targets for each gene. The fragments were then cloned into the pRGEB32 vector, which was predigested with BsaI (Xie et al., 2015). The resulting vectors were introduced into wild-type japonica rice variety Dongjin by Agrobacterium (Agrobacterium tumefaciens)-mediated transformation (Jeon et al., 2000). PCR amplicons were generated using gene-specific primers (F2KP1C-seq-F and F2KP1C-seq-R for OsF2KP1 and F2KP2C-seq-F and F2KP2C-seq-R for OsF2KP2) and subjected to Sanger DNA sequencing to genotype the mutants (Supplementary Table S1). Finally, CRISPR/Cas9 transgene-free homozygous progeny plants were selected by performing PCR with the vector-specific primers pRGEB32-F and pRGEB32-R and used in subsequent experiments.
F2KP activity assay
F2KP activity in rice endosperm was determined by measuring the change in activity of PPi-dependent PFP from potato by F2,6BP, which was produced by F2KP, as described previously (Park et al., 2007) with slight modifications. The assay mixture (200 μL total volume) contained 25 mM HEPES (pH 7.5), 1 mM MgCl2, 1 mM ATP, 100 mM F-6-P, 1 mM NADH, 0.5 U aldolase, 1 U triose-phosphate isomerase, 1 U glycerophosphate dehydrogenase, and 20 μL crude extract from wild-type and mutant endosperms. A 40-mg sample of developing rice endosperms collected 14 days after pollination was extracted in 1 mL assay buffer. Relative F2KP activity represents the means of three independent experiments. The reaction was started by adding 50 μL 10-mM pyrophosphate. Positive associations between PPi-dependent PFP activity and F2,6BP concentration were determined using F2,6BP (Santa Cruz Biotechnology, Dallas, TX, USA).
Hypoxia treatment
For the hypoxia stress group, surface-sterilized rice seeds were germinated in water. Twenty seeds were transferred to a 50-mL Corning tube filled with 50 mL sterile water and covered with a lid. For the aerobic (control) group, 20 seeds in a 50-mL Corning tube without a lid were soaked in 1 mL sterile water. Every 12 h, the water was changed for both groups. After a 48-h incubation at a constant temperature of 20°C and a 12-h photoperiod with a uniform light intensity of 150 μmol photons m–2 s–1, the coleoptiles were harvested from each group for RNA extraction.
RNA extraction and reverse-transcription quantitative PCR analysis
Total RNA was extracted from the fully developed leaves of 10-week-old plants and coleoptiles of mock- and hypoxia-treated samples using RNAiso Plus (Takara, Japan) and reverse-transcribed into cDNA using a ReverTra Ace qPCR RT Master Mix cDNA Synthesis kit (Toyobo, Japan) according to the manufacturer’s instructions. To measure relative mRNA levels of OsF2KP1 and OsF2KP2, RT-qPCR was performed using Prime Q-Master Mix with SYBR Green I (GeNet Bio, Korea) and a Rotor-Gene 6000 real-time amplification instrument (Corbett Research, Australia). The F2KP1-qRT-F/R and F2KP2-qRT-F/R primer sets were used to examine OsF2KP1 and OsF2KP2 transcript levels, respectively. The resulting data were normalized to that of OsUBQ5 (Os01g0328400) using the primers UBQ5-qRT-F/R (Supplementary Table S1).
Results
Morphological and physicochemical analysis of Seolgaeng endosperm
Whereas its parental rice variety Ilpum develops a normal endosperm, Seolgaeng, a variety used to produce dry-milled rice flour, is a mutant with a transparent outer surface and an opaque endosperm (Kwak et al., 2017) (Figure 1A). Scanning electron microscopy of the endosperm revealed that Ilpum starch granules were polygonal and angular, whereas those of Seolgaeng were round, ellipsoidal, and irregular (Figure 1A). Seolgaeng showed significantly lower grain hardness and damaged starch content than Ilpum (Figures 1B, C). Amylose content was lower in Seolgaeng compared to Ilpum (Figure 1D). Moreover, analysis of the degree of polymerization (DP) of starch revealed that short-chain (DP 6–12) starch production was greater and intermediate-chain (DP 13–30) starch production was less in Seolgaeng compared to Ilpum (Figure 1F). These differences likely led to the more rounded starch granules in Seolgaeng. Total starch content was similar in Seolgaeng and Ilpum (Figure 1E).
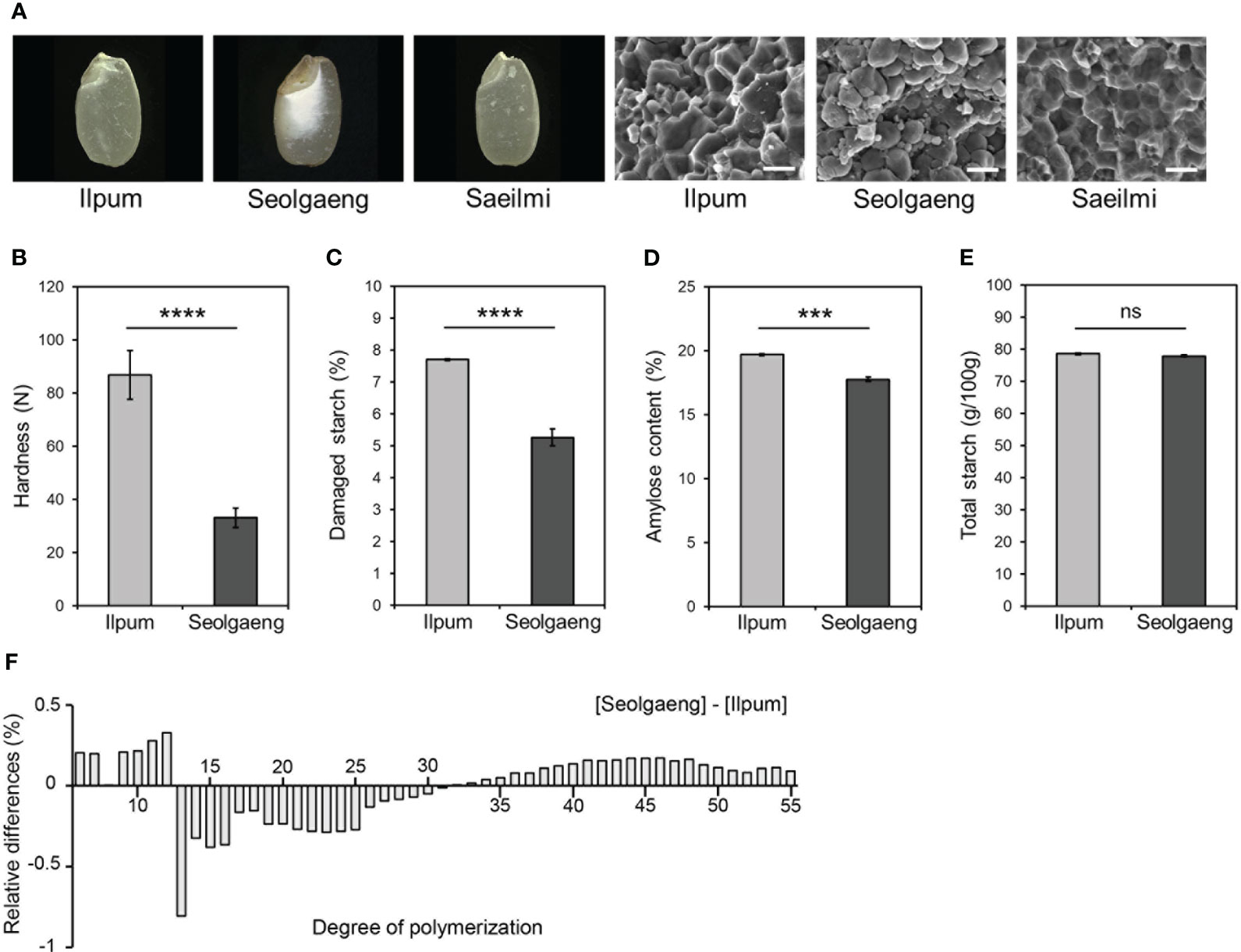
Figure 1 Morphological and physicochemical analysis of Seolgaeng. (A) Images of endosperm (left) and scanning electron micrographs of starch granules (right) from three different rice varieties. Scale bars, 50 μm. (B) Grain hardness. Data are means ± standard error of the mean (SEM) (n = 12). Student’s t-tests were used to detect significant differences. ****p < 0.0001. (C–F) Comparisons of the starch characteristics of Ilpum and Seolgaeng. (C) Damaged starch content. Data are means ± SEM (n = 9). ****p < 0.0001. (D) Amylose content. Data are means ± SEM (n = 4). ***p < 0.001. (E) Total starch content. Data are means ± SEM (n = 6). ns, not significant. (F) Amylopectin chain-length distribution. Relative differences are expressed as the percentage of peak areas of Seolgaeng subtracting Ilpum. Data are the means of three replicates.
Map-based cloning of the mutant gene in Seolgaeng
To identify the mutation causing the opaque-endosperm phenotype in Seolgaeng, a mapping population was developed from a genetic cross between the mutant Seolgaeng and wild-type Saeilmi (Figure 1A). The progeny of self-fertilized F2 plants showed a segregation ratio of normal:opaque endosperm of 115:35, which is approximately 3:1 (X2 = 0.222; P = 0.637). This ratio indicated that a single recessive mutation was responsible for the opaque-endosperm phenotype. We used 153 of 512 available KASP markers to construct an initial map of the causal mutation via recessive class analysis by analyzing 12 individuals with floury opaque endosperm. We then sequentially analyzed 86 BC1F2, 584 BC1F3, and 38 BC1F3 backcrossed individuals with wild-type endosperm to narrow down the candidate region. This analysis delineated the causal mutation to a ~600-kb region on chromosome 3 containing 112 annotated genes (Figure 2; Supplementary Table S2). Genomic DNA sequencing within this region in Seolgaeng and Ilpum detected two intergenic nucleotide changes as well as an A-to-G substitution in the sixth exon of Os03g0294200 in Seolgaeng. This gene encodes OsF2KP2, an enzyme that regulates F2,6BP levels (Park et al., 2007). The A-to-G substitution caused a missense mutation at position 266 of OsF2KP2, from tyrosine (Y) to cysteine (C) (Figure 2).
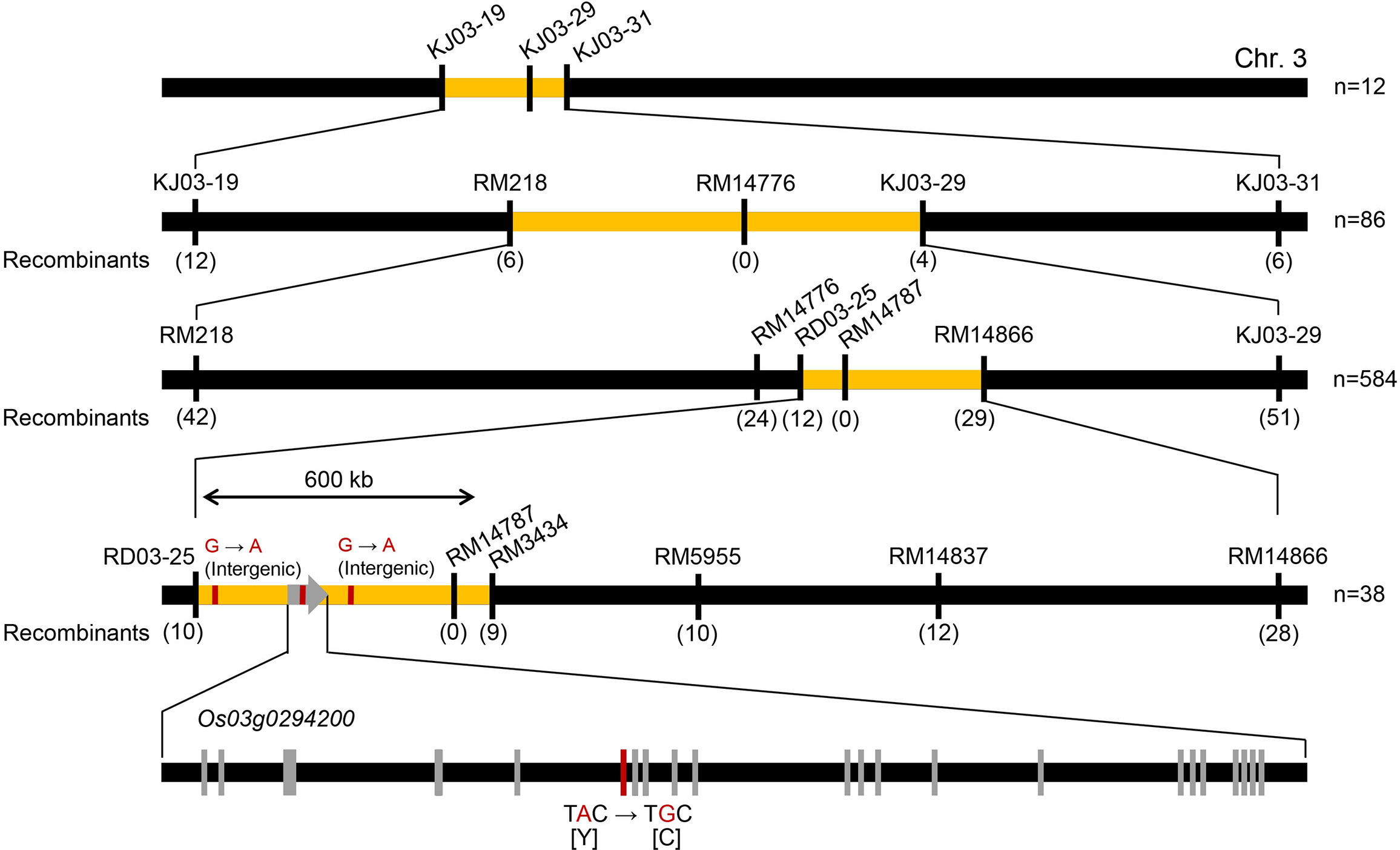
Figure 2 Map-based cloning of the mutated gene in Seolgaeng. The primary map constructed from 12 floury opaque-endosperm individuals located the causal mutation to a 6.6-Mb interval between markers KJ03-019 and KJ03-31 on chromosome 3. The first fine map, constructed from 86 wild-type endosperm individuals, narrowed the candidate region down to a 3.96-Mb interval between markers RM218 and KJ03-29. The second fine map, constructed from 584 wild-type endosperm individuals, identified a 1.6-Mb candidate region between markers RD03-25 and RM14866. The final map, constructed from 38 wild-type endosperm individuals, pinpointed the candidate region to a 600-kb interval between markers RD03-25 and RM3434. Finally, genomic DNA sequencing detected two variations in intergenic regions and one in the sixth exon of Os03g0294200: a nucleotide substitution from A in Ilpum to G in Seolgaeng, which caused an amino acid change at position 266 from tyrosine (Y) to cysteine (C).
We developed a cleaved amplified polymorphic sequence (CAPS) marker based on the A-to-G change in OsF2KP2 (Supplementary Table S3). The polymorphism associated with the CAPS marker co-segregated perfectly with the wild-type and floury opaque-endosperm phenotypes in F2 individuals from a cross between Seolgaeng and Saeilmi (Supplementary Figure S1A). In an analysis of 210 floury opaque-endosperm genetic resources, only 4 showed a Seolgaeng-specific allelic pattern, all of which were either Seolgaeng or Seolgaeng-derived lines (Supplementary Figure S1B). This result indicates that the mutation is unique to Seolgaeng.
Functional validation of the Osf2kp2 mutation
To determine whether the mutation in OsF2KP2 is responsible for the opaque endosperm of Seolgaeng, we obtained the T-DNA insertional mutant f2kp2-2 (variety Taichung) and named the Seolgaeng allele f2kp2-1 (Figure 3A). The expression levels of OsF2KP2 were reduced in Seolgaeng and f2kp2-2 compared to the respective wild types (Supplementary Figure S2A). The progeny of homozygous f2kp2-2 mutant plants exhibited opaque endosperm (Figure 3C). We created two CRISPR/Cas9-mediated deletion mutants (variety Dongjin) with frameshift mutations in OsF2KP2: f2kp2-3 and f2kp2-4 (Figure 3A). OsF2KP2 transcript levels were reduced in f2kp2 mutants (Supplementary Figure S2A). The progeny of all homozygous mutants showed opaque endosperm (Figure 3C). These results demonstrate that the mutation of OsF2KP2 leads to opaque endosperm.
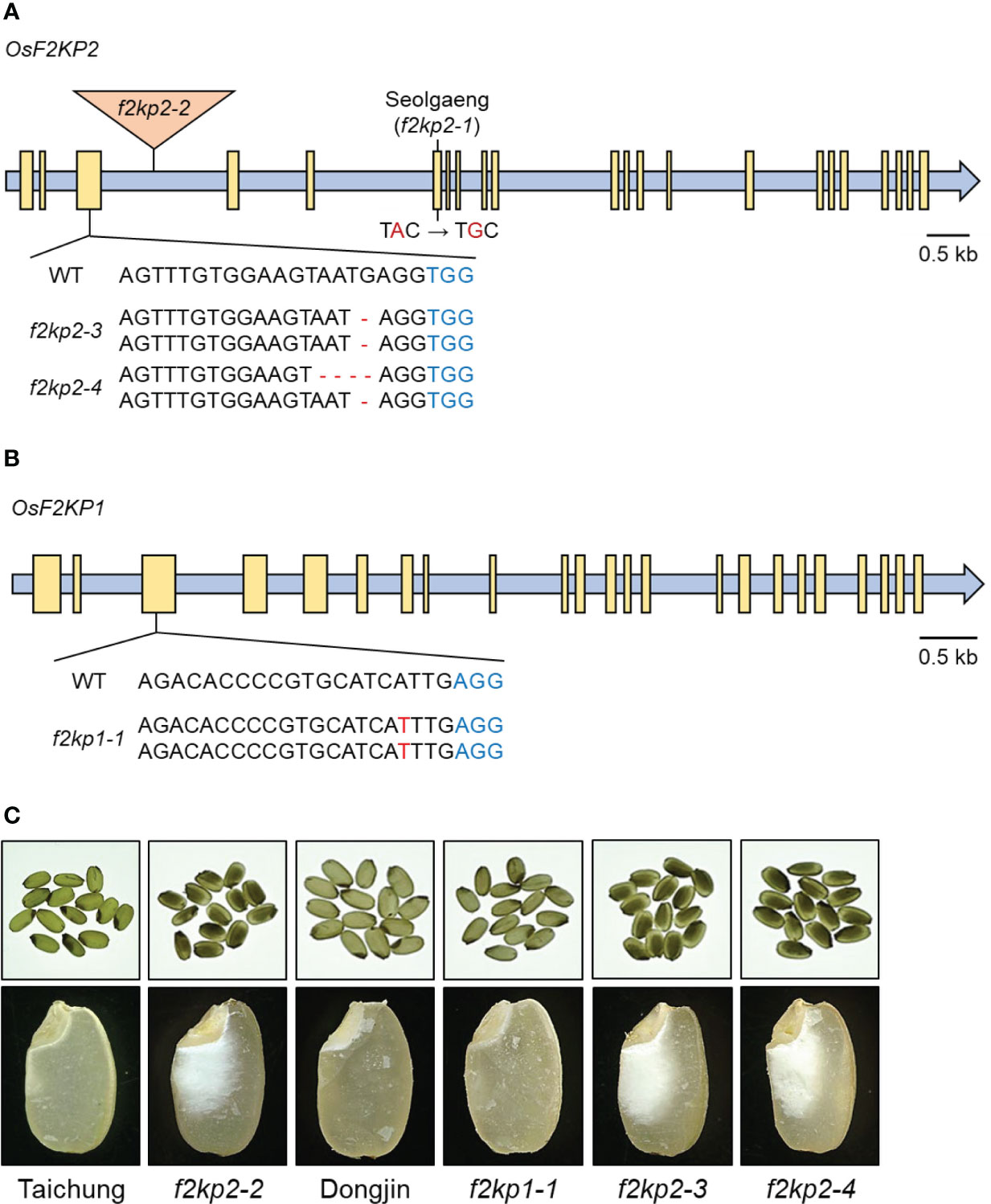
Figure 3 Production of f2kp1 and f2kp2 mutants and analysis of their endosperm phenotypes. (A) Production of T-DNA insertional and CRISPR/Cas9-edited f2kp2 mutants. Blue indicates protospacer adjacent motifs and red indicates deletions and substitutions. Triangle, T-DNA insertion site; yellow boxes, exons. (B) Production of the CRISPR/Cas9-edited f2kp1 mutant. Protospacer adjacent motifs and insertion mutations are indicated in blue and red, respectively. Yellow boxes, exons. (C) Opaque-endosperm phenotypes (top) and longitudinal endosperm sections (bottom) of f2kp mutants compared to the respective wild types.
Production and characterization of f2kp1 f2kp2 double mutants
The rice genome contains a second F2KP isoform, OsF2KP1 (Os05g0164100). Whereas OsF2KP1 transcripts were abundant in leaves, OsF2KP2 transcripts were relatively abundant in the endosperm (Park et al., 2007). We generated an f2kp1 mutant (variety Dongjin) via CRISPR/Cas9-mediated genome editing: f2kp1-1 carried a homozygous T insertion in the third exon of OsF2KP1, introducing a frameshift (Figure 3B). The expression level of OsF2KP1 was reduced in f2kp1-1 (Supplementary Figure S2B). Phenotypic analysis revealed that f2kp1-1 endosperm appeared normal (Figure 3C).
We also generated f2kp1 f2kp2 double mutants (f2kp-d; variety Dongjin) via CRISPR/Cas9-mediated genome editing. Double mutants f2kp-d1 and f2kp-d2 carried the same homozygous A deletion in OsF2KP1 and distinct biallelic mutations in OsF2KP2, introducing frameshifts in both genes (Figure 4A). RT-qPCR analysis confirmed that the expression levels of both OsF2KP1 and OsF2KP2 were reduced in f2kp-d mutants (Supplementary Figure S2C). The endosperms of f2kp-d were more opaque than those of f2kp2 (Figures 3C, 4B).
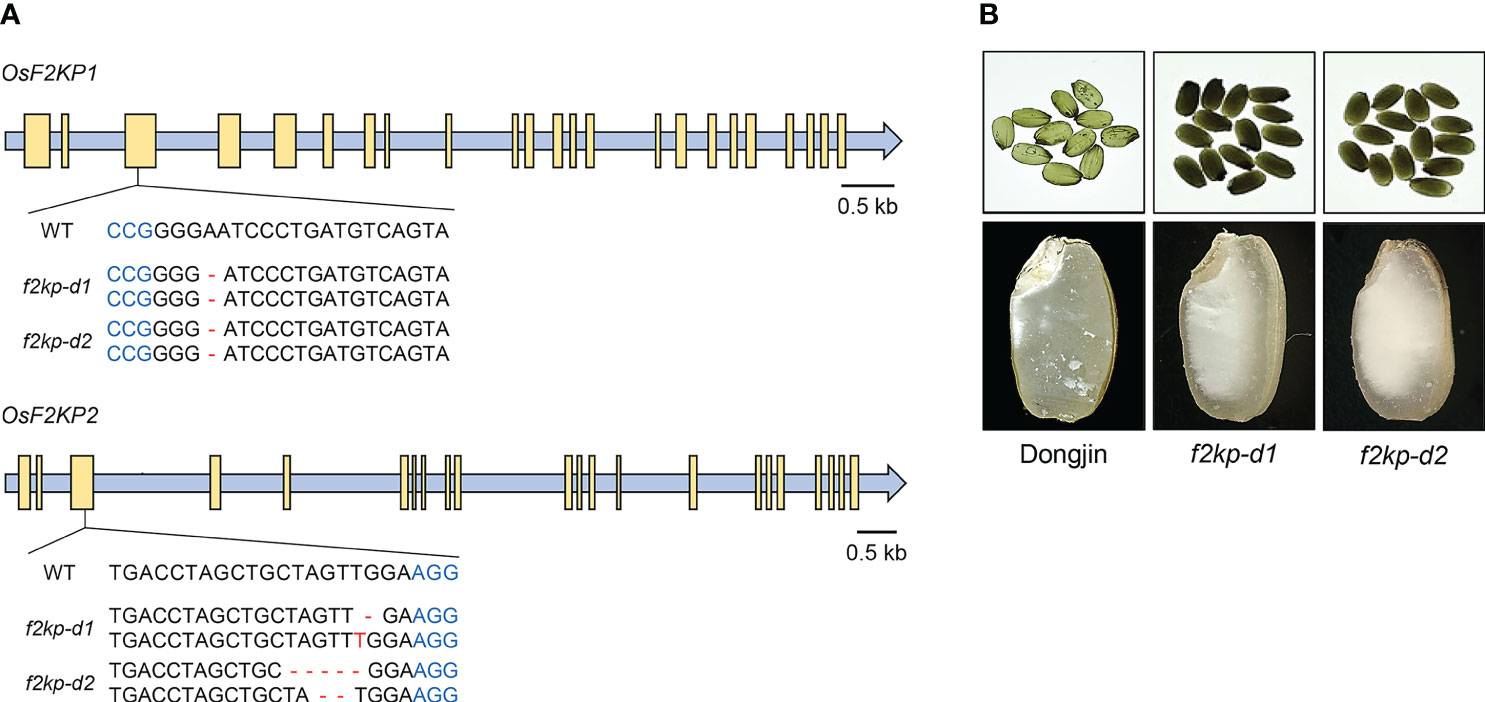
Figure 4 Production of f2kp double mutants (f2kp-d) and analysis of their endosperm phenotypes. (A) Schematic representations of f2kp-d mutants generated by CRISPR/Cas9-mediated genome editing. Protospacer adjacent motifs and insertion or deletion (–) mutations are indicated in blue and red, respectively. Yellow boxes, exons. (B) Opaque-endosperm phenotypes (top) and longitudinal sections of endosperm (bottom) of f2kp-d mutants compared to wild type (Dongjin).
Grain hardness and damaged starch content were significantly reduced in f2kp2 and f2kp-d compared to wild-type and f2kp1 grains (Figures 5A, B). Grain hardness was much lower in f2kp-d than in f2kp2, which is consistent with the severe opaqueness of the double mutant grains (Figure 4B). Amylose content was lower in f2kp2 and much lower in f2kp-d compared to the wild types, but f2kp1-1 showed no significant difference (Figure 5C). Analysis of the DP of starch revealed that, as in Seolgaeng, short-chain starch production was higher and intermediate-chain starch production was lower in f2kp2 and f2kp-d compared to the respective wild types (Figures 1F, 5E). Total starch content did not differ among the f2kp2 mutants compared to the respective wild types but decreased by 2.9% and 2.4% in f2kp-d1 and f2kp-d2, respectively (Figure 5D). These results indicate that changes in amylose content and amylopectin structure resulted in an abnormal starch granule shape in the mutants (Jeon et al., 2010).
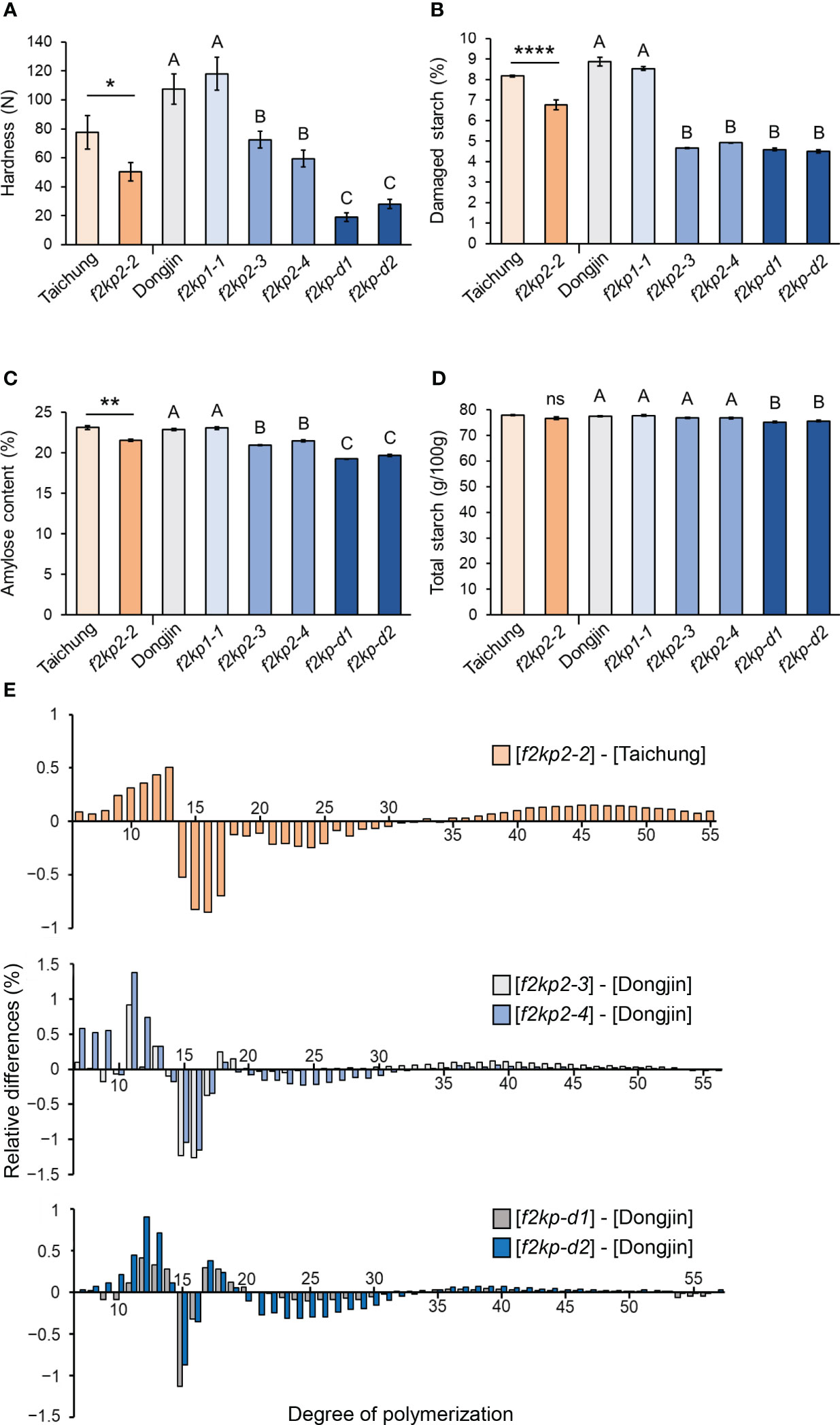
Figure 5 Physicochemical analysis of f2kp mutant grains. (A) Grain hardness. Data are means ± SEM (n = 12). Student’s t-tests were used to detect significant differences. *p < 0.05. Different uppercase letters indicate significant differences (p < 0.05). (B) Damaged starch content. Data are means ± SEM (n = 9). ****p < 0.0001. Different uppercase letters indicate significant differences (p < 0.05). (C) Amylose content. Data are means ± SEM (n = 4). **p < 0.01. Different uppercase letters indicate significant differences (p < 0.05). (D) Total starch content. Data are means ± SEM (n = 6). ns, not significant. Different uppercase letters indicate significant differences (p < 0.05). (E) Amylopectin chain-length distribution. Relative differences are expressed as the percentage of peak areas of mutants subtracting their respective wild types. Data are means of three replications.
Biochemical analysis of F2KP activity
Given that the Y residue at position 266 of OsF2KP2 is conserved in F2KP isoforms from various plant species (Supplementary Figure S3), its missense mutation in Seolgaeng likely affects F2KP activity in the endosperm. Enzyme-coupled analysis of F2KP activity in developing endosperms collected 14 days after pollination (when starch biosynthesis is active) revealed that F2KP activity was ~55% lower in Seolgaeng compared to Ilpum (Figure 6). F2KP activity was lower in all f2kp2 mutants and much lower in the double mutants compared to their respective wild types, but f2kp1-1 showed no significant difference (Figure 6). The severity of endosperm opaqueness of the mutants was strongly associated with a reduction in F2KP activity in the endosperm (Figures 1A, 3C, 4B). These results support the idea that reduced F2KP activity leads to opaque endosperm development.
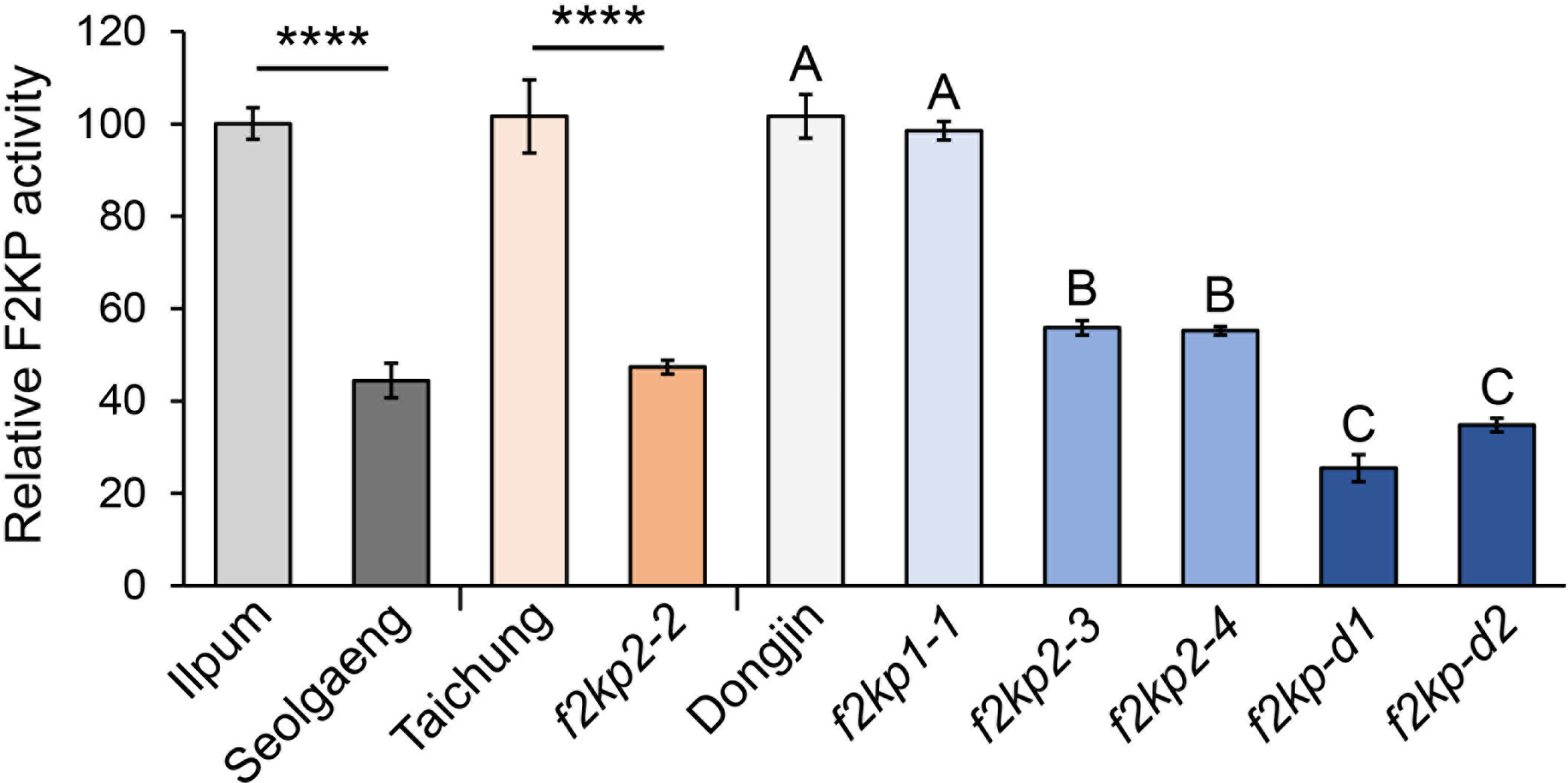
Figure 6 Relative F2KP activity in f2kp mutant endosperms. Data are means ± SEM (n = 3). Significant differences were analyzed by Student’s t-tests. ****p < 0.0001. Different uppercase letters indicate significant differences (p < 0.05).
Expression analysis of OsF2KP genes under hypoxia
PPi-utilizing enzymes can play a crucial role in starch biosynthesis in rice endosperm exposed to hypoxia, a condition in which mitochondrial respiration is limited (Lee and Jeon, 2020). To examine the expression of OsF2KP1 and OsF2KP2 in response to hypoxia, we performed RT-qPCR using cDNA produced from total RNA extracted from mock- and hypoxia-treated coleoptile samples. OsF2KP2 was strongly upregulated under hypoxic conditions, whereas OsF2KP1 was downregulated (Figure 7). These results are consistent with the finding that OsF2KP2 is the main isoform functioning in the central part of the endosperm during hypoxia.
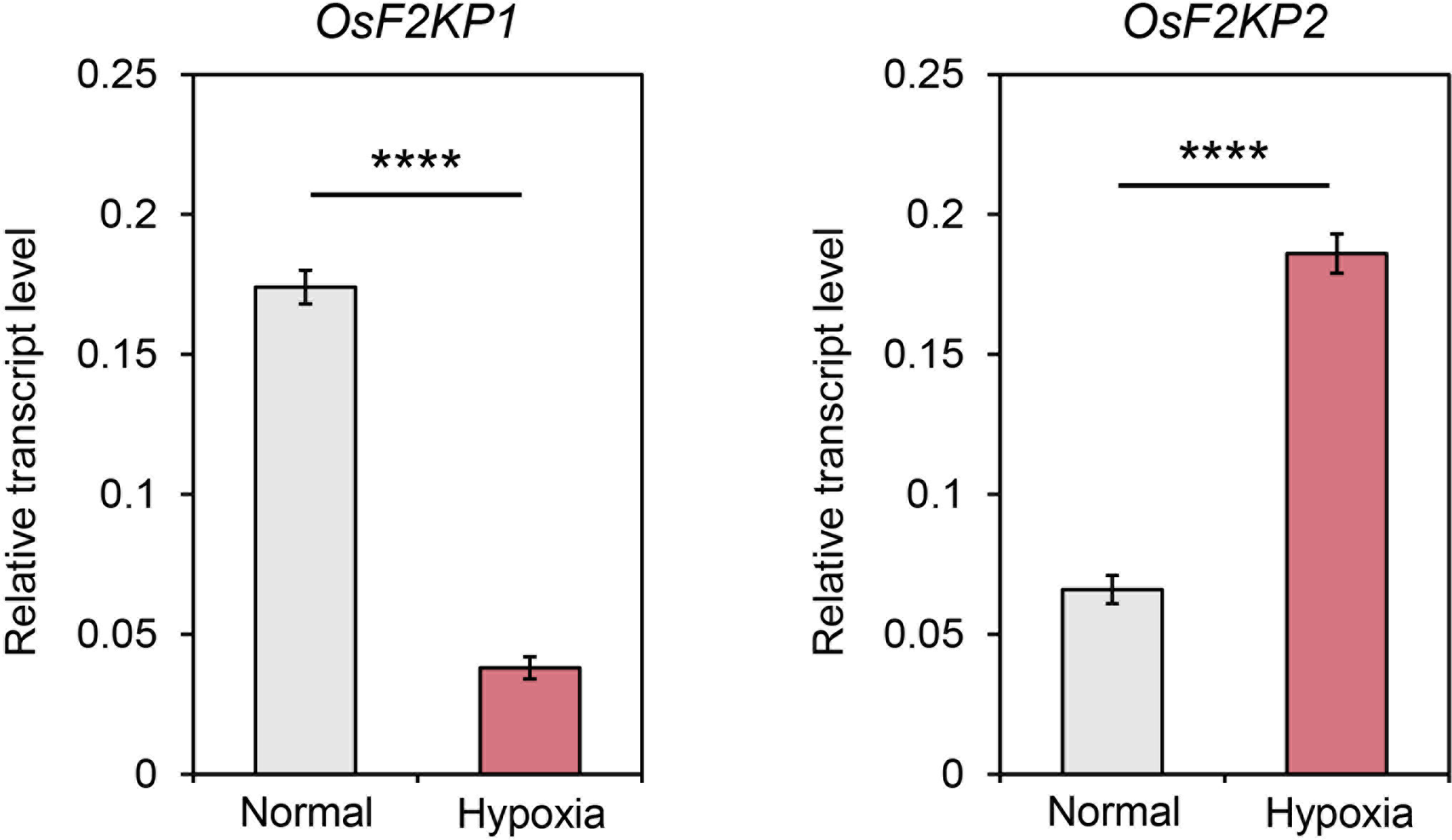
Figure 7 Expression analysis of OsF2KP1 and OsF2KP2 in rice coleoptiles under hypoxic conditions. Relative expression was normalized using OsUBQ5. Error bars indicate SEM (n = 4). Significant differences were analyzed by Student’s t-tests. ****p < 0.0001.
Discussion
Impaired F2KP2 function causes floury opaque endosperm in Seolgaeng
Our study provides several lines of evidence that mutations in OsF2KP2 lead to the development of floury opaque endosperm in Seolgaeng. First, map-based cloning identified a missense mutation in the OsF2KP2 amino acid sequence in Seolgaeng at a residue conserved in F2KPs across plant species. Second, the T-DNA insertional mutant line f2kp2-2 and additional f2kp2 mutants created via CRISPR/Cas9-mediated genome editing all showed a similar floury opaque-endosperm phenotype. Third, all f2kp2 mutants showed reduced grain hardness and damaged starch content, low amylose content, and similar changes in amylopectin structure compared to their respective wild types. Last, a biochemical F2KP activity assay confirmed an association between reduced F2KP enzyme activity and the development of floury opaque endosperm. All these data suggest that reduced F2KP function triggers the development of abnormal floury opaque endosperm. Thus, our study demonstrates the crucial function of F2KP in rice endosperm development.
F2KP functions in PPi-utilizing energy metabolism during endosperm development
Rice endosperm is exposed to a hypoxic environment that generates abundant PPi as a byproduct of ADP-Glc pyrophosphorylase in the cytosol. PPi is an alternative energy currency that integrates carbon metabolism from sucrose breakdown with starch biosynthesis in the endosperm (Lee and Jeon, 2020). In this model, PPi-dependent glycolysis via PFP and cytosolic pyruvate, orthophosphate dikinase (cPPDK), coupled with the non-cyclic mode of the tricarboxylic acid cycle via cytosolic alanine aminotransferase (cAlaAT) and cytosolic malate dehydrogenase (cMDH), can sustain glycolytic flux, which produces adequate ATP levels in the endosperm (Figure 8). The malfunction of these enzymes, namely PFP (Duan et al., 2016; Chen et al., 2020), cPPDK (Kang et al., 2005; Lappe et al., 2018; Zhang et al., 2018), cAlaAT (Yang et al., 2015; Zhong et al., 2019), and cMDH (Teng et al., 2019), results in opaque endosperm.
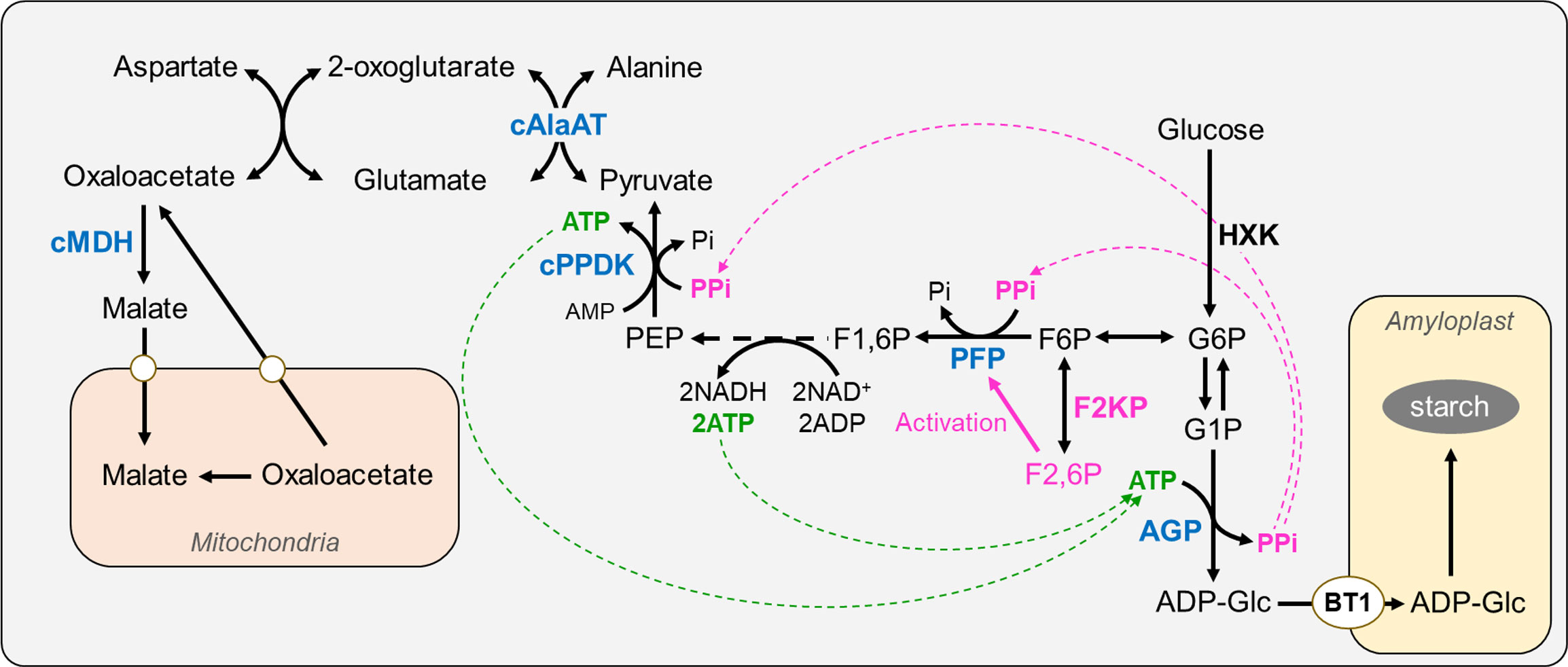
Figure 8 Proposed role of F2KP in rice endosperm starch biosynthesis. The F2KP-related pathway is highlighted in pink. F2KP generates F2,6BP, promoting synthesis of glycolysis-committed F1,6BP, which enters glycolysis. Reduced F2,6BP levels caused by OsF2KP2 deficiency restrict PFP activity, reducing the energy supply for starch biosynthesis. ADP-Glc, ADP-glucose; AGP, ADP-Glc pyrophosphorylase; cAlaAT, cytosolic alanine aminotransferase; BT1, BRITTLE 1 (ADP-glucose transporter); cPPDK, cytosolic pyruvate, orthophosphate dikinase; F1,6P, fructose-1-6-bisphosphate; F2,6P, fructose-2-6-bisphosphate; F2KP, fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase; F6P, fructose-6-phosphate; G1P, glucose-1-phosphate; G6P, glucose-6-phosphate; HXK, hexokinase; cMDH, cytosolic malate dehydrogenase; PEP, phosphoenolpyruvate; PFP, PPi-dependent fructose-6-phosphate 1-phosphotransferase.
The current study reveals a crucial function of F2KP during endosperm development. F2KP generates F2,6BP, thereby promoting glycolysis-committed F1,6BP production. F2,6BP activates PFP to convert F6P into F1,6BP, which enters glycolysis. Therefore, reduced F2,6BP levels caused by OsF2KP2 deficiency likely restrict PFP activity, reducing the energy supply for starch biosynthesis (Figure 8) and resulting in the development of floury opaque endosperm suitable for dry-milled rice flour.
The f2kp1 f2kp2 double mutant is an ideal candidate rice variety for high-quality dry-milled rice flour production
Of the two F2KP isoforms, OsF2KP2 is predominant in rice endosperm (Park et al., 2007). Our expression analysis of hypoxia-treated rice coleoptiles suggests that OsF2KP2 might also be upregulated in the hypoxic central region of rice endosperm. These results are consistent with the floury opaque-endosperm phenotype of all f2kp2 mutants. Here, we demonstrated that the central region and outer surface of endosperm were more opaque in f2kp1 f2kp2 double mutant (f2kp-d) grains than in the f2kp2 single mutant. The endosperm of f2kp1 was indistinguishable from that of wild type. These data suggest that OsF2KP2 functions as the main F2KP isoform in the endosperm, whereas OsF2KP1 may partially compensate for the loss of OsF2KP2. It remains to be determined whether OsF2KP2 is specifically expressed in the hypoxic central region of the endosperm and OsF2KP1 in the outer surface.
f2kp-d showed reduced grain hardness and amylose content, two favorable characteristics for high-quality dry-milled rice flour. Under normal growth conditions in paddy fields in the summer, f2kp-d did not exhibit any marked vegetative growth phenotypes (Supplementary Figure S4). However, it will be necessary to observe the growth of the double mutants under stress conditions in the future. Thus, perhaps f2kp-d could be used to breed a new rice variety suitable for dry-milled rice flour production.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
J-WK, S-KL, S-HS, DS, J-HC, J-YL, J-MK, HJ, H-MP, and E-KA performed the experiments. J-MK, S-KL, S-HS, J-HL, and J-SJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation (2021M3E5E6025387 and 2023R1A2C1003142) and the Rural Development Administration (PJ01690101 and PJ01603004).
Acknowledgments
We thank Professor Li-Jia Qu (Peking University, China) for providing the CRISPR/Cas9 vectors pOs-sgRNA and pH-Ubi-cas9-7.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1231914/full#supplementary-material
References
Chen, C., He, B., Liu, X., Ma, X., Liu, Y., Yao, H., et al. (2020). Pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP1) regulates starch biosynthesis and seed development via heterotetramer formation in rice (Oryza sativa L.). Plant Biotechnol. J. 18, 83–95. doi: 10.1111/pbi.13173
Cheon, K. S., Baek, J., Cho, Y. I., Jeong, Y. M., Lee, Y. Y., Oh, J., et al. (2018). Single Nucleotide Polymorphism (SNP) discovery and Kompetitive Allele-Specific PCR (KASP) marker development with Korean Japonica rice varieties. Plant Breed. Biotechnol. 6, 391–403. doi: 10.9787/PBB.2018.6.4.391
Duan, E., Wang, Y., Liu, L., Zhu, J., Zhong, M., Zhang, H., et al. (2016). Pyrophosphate: fructose-6-phosphate 1-phosphotransferase (PFP) regulates carbon metabolism during grain filling in rice. Plant Cell Rep. 35, 1321–1331. doi: 10.1007/s00299-016-1964-4
Jeon, J.-S., Lee, S., Jung, K.-H., Jun, S.-H., Jeong, D.-H., Lee, J., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570. doi: 10.1046/j.1365-313x.2000.00767.x
Jeon, J.-S., Ryoo, N., Hahn, T.-R., Walia, H., Nakamura, Y. (2010). Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 48, 383–392. doi: 10.1016/j.plaphy.2010.03.006
Kang, H.-G., Park, S., Matsuoka, M., An, G. (2005). White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 42, 901–911. doi: 10.1111/j.1365-313X.2005.02423.x
Kwak, J., Yoon, M. R., Lee, J. S., Lee, J. H., Ko, S., Tai, T. H., et al. (2017). Morphological and starch characteristics of the Japonica rice mutant variety Seolgaeng for dry-milled flour. Food Sci. Biotechnol. 26, 43–48. doi: 10.1007/s10068-017-0006-5
Lappe, R. R., Baier, J. W., Boehlein, S. K., Huffman, R., Lin, Q., Wattebled, F., et al. (2018). Functions of maize genes encoding pyruvate phosphate dikinase in developing endosperm. Proc. Natl. Acad. Sci. U. S. A. 115, E24–E33. doi: 10.1073/pnas.1715668115
Lee, S., Jeon, J. (2020). Crucial role of inorganic pyrophosphate in integrating carbon metabolism from sucrose breakdown to starch synthesis in rice endosperm. Plant Sci. 298, 110572. doi: 10.1016/j.plantsci.2020.110572
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Naito, Y., Hino, K., Bono, H., Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nakamura, Y. (2018). Rice starch biotechnology: Rice endosperm as a model of cereal endosperms. Starch - Stärke 70, 1600375. doi: 10.1002/star.201600375
Nielsen, T. H., Rung, J. H., Villadsen, D. (2004). Fructose-2,6-bisphosphate: a traffic signal in plant metabolism. Trends Plant Sci. 9, 556–563. doi: 10.1016/j.tplants.2004.09.004
Park, S., Cho, M.-H., Bhoo, S. H., Jeon, J.-S., Kwon, Y.-K., Hahn, T.-R. (2007). Altered sucrose synthesis in rice plants with reduced activity of fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase. J. Plant Biol. 50, 38–43. doi: 10.1007/BF03030598
Patindol, J. A., Gonzalez, B. C., Wang, Y. J., McClung, A. M. (2007). Starch fine structure and physicochemical properties of specialty rice for canning. J. Cereal Sci. 45, 209–218. doi: 10.1016/j.jcs.2006.08.004
Ryoo, N., Yu, C., Park, C. S., Baik, M. Y., Park, I. M., Cho, M. H., et al. (2007). Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 26, 1083–1095. doi: 10.1007/s00299-007-0309-8
Stitt, M. (1990). Fructose 2,6-bisphosphate as a regulatory molecule in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 153–185. doi: 10.1146/annurev.pp.41.060190.001101
Teng, X., Zhong, M., Zhu, X., Wang, C., Ren, Y., Wang, Y., et al. (2019). FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synthesis and seed development in rice. Plant Biotechnol. J. 17, 1914–1927. doi: 10.1111/pbi.13108
Trevanion, S. (2002). Regulation of sucrose and starch synthesis in wheat (Triticum aestivum L.) leaves: role of fructose 2,6-bisphosphate. Planta 215, 653–665. doi: 10.1007/s00425-002-0792-7
Xie, K., Minkenberg, B., Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U. S. A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Yang, J., Kim, S.-R., Lee, S.-K., Choi, H., Jeon, J.-S., An, G. (2015). Alanine aminotransferase 1 (OsAlaAT1) plays an essential role in the regulation of starch storage in rice endosperm. Plant Sci. 240, 79–89. doi: 10.1016/j.plantsci.2015.07.027
Zhang, L., Zhao, L., Lin, L., Zhao, L., Liu, Q., Wei, C. (2018). A novel mutation of OsPPDKB, encoding pyruvate orthophosphate dikinase, affects metabolism and structure of starch in the rice endosperm. Int. J. Mol. Sci. 19, 2268. doi: 10.3390/ijms19082268
Keywords: endosperm, floury opaque mutant, fructose-6-phosphate 2-kinase/fructose-2, 6-bisphosphatase, rice flour, Seolgaeng, starch
Citation: Kang J-W, Lee S-K, Shim S-H, Shin D, Cho J-H, Lee J-Y, Ko J-m, Ji H, Park H-M, Ahn E-K, Lee J-H and Jeon J-S (2023) Dry-milled flour rice ‘Seolgaeng’ harbors a mutated fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase2. Front. Plant Sci. 14:1231914. doi: 10.3389/fpls.2023.1231914
Received: 31 May 2023; Accepted: 26 July 2023;
Published: 10 August 2023.
Edited by:
Antonio Costa De Oliveira, Federal University of Pelotas, BrazilReviewed by:
Robert Henry, The University of Queensland, AustraliaHongchun Xiong, Chinese Academy of Agricultural Sciences, China
Copyright © 2023 Kang, Lee, Shim, Shin, Cho, Lee, Ko, Ji, Park, Ahn, Lee and Jeon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jong-Seong Jeon, amplb25Aa2h1LmFjLmty; Jong-Hee Lee, Y2NyaWxqaEBrb3JlYS5rcg==
†These authors have contributed equally to this work
 Ju-Won Kang
Ju-Won Kang Sang-Kyu Lee
Sang-Kyu Lee Su-Hyeon Shim
Su-Hyeon Shim Dongjin Shin
Dongjin Shin Jun-Hyeon Cho1
Jun-Hyeon Cho1 Jong-Hee Lee
Jong-Hee Lee Jong-Seong Jeon
Jong-Seong Jeon