- 1Section for Genetics and Evolutionary Biology (EVOGENE), Department of Biosciences, University of Oslo, Oslo, Norway
- 2Centre for Ecological and Evolutionary Synthesis (CEES), Department of Biosciences, University of Oslo, Oslo, Norway
Speciation involves reproductive isolation, which can occur by hybridization barriers acting in the endosperm of the developing seed. The nuclear endosperm is a nutrient sink, accumulating sugars from surrounding tissues, and undergoes coordinated cellularization, switching to serve as a nutrient source for the developing embryo. Tight regulation of cellularization is therefore vital for seed and embryonic development. Here we show that hybrid seeds from crosses between Arabidopsis thaliana as maternal contributor and A. arenosa or A. lyrata as pollen donors result in an endosperm based post-zygotic hybridization barrier that gives rise to a reduced seed germination rate. Hybrid seeds display opposite endosperm cellularization phenotypes, with late cellularization in crosses with A. arenosa and early cellularization in crosses with A. lyrata. Stage specific endosperm reporters display temporally ectopic expression in developing hybrid endosperm, in accordance with the early and late cellularization phenotypes, confirming a disturbance of the source-sink endosperm phase change. We demonstrate that the hybrid barrier is under the influence of abiotic factors, and show that a temperature gradient leads to diametrically opposed cellularization phenotype responses in hybrid endosperm with A. arenosa or A. lyrata as pollen donors. Furthermore, different A. thaliana accession genotypes also enhance or diminish seed viability in the two hybrid cross-types, emphasizing that both genetic and environmental cues control the hybridization barrier. We have identified an A. thaliana MADS-BOX type I family single locus that is required for diametrically opposed cellularization phenotype responses in hybrid endosperm. Loss of AGAMOUS-LIKE 35 significantly affects the germination rate of hybrid seeds in opposite directions when transmitted through the A. thaliana endosperm, and is suggested to be a locus that promotes cellularization as part of an endosperm based mechanism involved in post-zygotic hybrid barriers. The role of temperature in hybrid speciation and the identification of distinct loci in control of hybrid failure have great potential to aid the introduction of advantageous traits in breeding research and to support models to predict hybrid admixture in a changing global climate.
1 Introduction
Speciation is usually a continuous process towards increasing divergence and reproductive isolation between two lineages. Reproductive isolation can be obtained due to hybridization barriers, which act before fertilization (pre-zygotic) or after fertilization (post-zygotic) (Rieseberg and Willis, 2007; Widmer et al., 2009). A special case of post-zygotic hybridization barriers acts in the developing seed (Lafon-Placette and Köhler, 2016), resulting in developmental abnormality in the endosperm (Cooper and Brink, 1942). Seed death is accepted to be mainly due to failure of endosperm development since the embryo can be rescued in culture after microdissection (Sharma, 1999). Many studies have indicated endosperm deficiency to be the major cause of hybrid seed inviability (Brink and Cooper, 1947; Sukno et al., 1999; Dinu et al., 2005; Roy et al., 2011) and an endosperm-based hybridization barrier has been shown to be conserved across distinct species groups such as Arabidopsis (Lafon-Placette and Köhler, 2016), Capsella (Rebernig et al., 2015; Dziasek et al., 2021), rice (Ishikawa et al., 2011; Sekine et al., 2013; Zhang et al., 2016; Tonosaki et al., 2018; Wang et al., 2018), tomato (Florez-Rueda et al., 2016; Roth et al., 2019), monkeyflower (Oneal et al., 2016; Flores-Vergara et al., 2020; Kinser et al., 2021), and potato (Johnston and Hanneman, 1982; Cornejo et al., 2012). This suggests that the phenomenon represents a major mechanism of reproductive isolation in plants, however it is largely ignored in modern literature of speciation (Lafon-Placette and Köhler, 2016).
The endosperm is a triploid tissue that requires tight control of genome dosage (2:1 maternal:paternal ratio). Cellularization of the endosperm marks a transition in seed development, as up to this point, the endosperm functions as a nutrient sink. At this developmental time point, the endosperm concurrently switches from nutrient sink to source for the developing embryo (Lafon-Placette and Köhler, 2016), and manipulating the timing of endosperm cellularization through interploidy crosses arrests embryo development (Scott et al., 1998; Hehenberger et al., 2012). Similarly, hybridization between plant species was shown to result in embryo arrest due to endosperm cellularization failure (Haig and Westoby, 1988; Haig and Westoby, 1991; Comai et al., 2000; Bushell et al., 2003). Manipulating the ploidy of parents in interspecies crosses has also shown to improve the success of hybridization, demonstrating a requirement for genome balance in the endosperm (Comai et al., 2000; Bushell et al., 2003; Lafon-Placette et al., 2017).
The genus Arabidopsis has been widely used for studying evolutionary questions (Koenig and Weigel, 2015; Koch, 2019) including the effects of interspecific hybridization in controlled crossings (Chen et al., 1998; Comai et al., 2000; Nasrallah et al., 2000; Josefsson et al., 2006; Walia et al., 2009; Burkart-Waco et al., 2012; Burkart-Waco et al., 2013; Burkart-Waco et al., 2015; Bjerkan et al., 2020). When diploid A. thaliana is crossed to diploid A. arenosa, the endosperm shows late cellularization and high degree of seed abortion (Josefsson et al., 2006; Bjerkan et al., 2020). Furthermore, when A. arenosa is crossed as male to A. lyrata, the same phenotype can be seen with late endosperm cellularization and a very high seed lethality (Lafon-Placette et al., 2017). Interestingly, crossing A. lyrata as male to A. arenosa results in the opposite effect with early endosperm cellularization (Lafon-Placette et al., 2017).
The strength of the endosperm-based hybridization barrier can be influenced by accession specific genetic variation (Burkart-Waco et al., 2012; Burkart-Waco et al., 2013; Burkart-Waco et al., 2015; Bjerkan et al., 2020). In crosses between diploid A. thaliana to diploid A. arenosa, the choice of accessions in both species significantly acts to repress or enhance the endosperm barrier (Bjerkan et al., 2020; Burkart-Waco et al., 2012). The current knowledge on the effect of temperature in early seed development is limited (Paul et al., 2020), but an effect of temperature on hybrid seed development in reciprocal crosses of wheat and barley has previously been reported (Molnár-Láng and Sutka, 1994). The sensitivity of endosperm cellularization to heat stress during early endosperm development has been demonstrated in rice (Folsom et al., 2014) and type I MADS-box transcription factors (TFs) are deregulated during moderate heat stress (Chen et al., 2016). We have previously reported a temperature effect on endosperm based post-zygotic hybrid lethality of diploid species in the genus Arabidopsis (Bjerkan et al., 2020). This temperature effect was also shown using different accessions of both A. arenosa and A. thaliana, demonstrating a combinatorial effect of accessions and temperature (Bjerkan et al., 2020; Burkart-Waco et al., 2012).
Genomic imprinting is an epigenetic phenomenon, which infers parent-of-origin allele specific expression of maternally or paternally inherited alleles (Hornslien et al., 2019; Batista and Kohler, 2020). As proper endosperm development depends on a correct ratio of parental genomes, it is suggested that differences in genomic imprinting programs may be responsible for the evolution of sexual incompatibility in crosses between divergent individuals (Haig and Westoby, 1988; Haig and Westoby, 1991; Bushell et al., 2003; Schatlowski and Kohler, 2012). Alternatively, or additionally, epigenetic remodeling upon hybridization due to combination of diverged maternal and paternal siRNAs may lead to comprehensive failure of genomic imprinting and ectopic expression of transposons and imprinted genes (Martienssen, 2010; Ng et al., 2012). Recent evidence supports this emerging role of imprinted genes, and in Arabidopsis interspecies hybrids, paternally expressed genes (PEGs) shifted to be maternally expressed genes (MEGs) (Josefsson et al., 2006). Importantly, both PEGs and MEGs have been shown to erect hybridization barriers, and mutational loss of these genes has been reported to bypass hybridization barriers in interspecies crosses (Walia et al., 2009; Wolff et al., 2015). A major part of the MEGs and PEGs encodes proteins that activate pathways in the endosperm consistent with the prominent role of cellularization in seed survival.
Proper endosperm development in Arabidopsis is reliant on the FERTILIZATION INDEPENDENT SEED-Polycomb Repressive Complex 2 (FIS-PRC2) (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999; Köhler et al., 2003). FIS-PRC2 is important in endosperm development indirectly through the genes it regulates, which include several type I MADS-box TFs (Zhang et al., 2018). Deregulation of type I MADS-box TFs in interspecies crosses has been postulated to induce the endosperm-based hybridization barrier, but unfortunately most of these TFs have no clear function because of extensive genetic redundancy (De Bodt et al., 2003; Parřenicová et al., 2003; Walia et al., 2009; Bemer et al., 2010). One exception is AGAMOUS-LIKE 62 (AGL62), which is found to suppress cellularization of the endosperm in Arabidopsis (Kang et al., 2008). The clear function of AGL62 is further emphasized through its interaction with the FIS-PRC2 complex (Hehenberger et al., 2012), with mutation of FIE, MEA, FIS2 and MSI1 resulting in an ectopic proliferation of nuclear endosperm (Grossniklaus et al., 1998; Köhler et al., 2003; Guitton et al., 2004), whereas the agl62 mutant results in precocious cellularization (Kang et al., 2008). AGL62 mutation has also been shown to alleviate the hybridization barrier in the A. thaliana × A. arenosa cross, resulting in a higher germination rate (Walia et al., 2009; Bjerkan et al., 2020).
Here we report that hybrid seeds from crosses between A. thaliana mothers with A. arenosa or A. lyrata pollen donors result in diametrically opposed endosperm phenotypes, both giving rise to reduced seed germination rate, albeit caused by late cellularization in crosses with A. arenosa and early cellularization in crosses with A. lyrata. We demonstrate that the hybrid barriers are under the influence of abiotic factors, and show that a temperature gradient leads to opposed cellularization phenotype and seed viability in hybrid endosperm with A. arenosa or A. lyrata as pollen donors. In addition, A. thaliana accession genotypes also influence seed viability in the two hybrid cross-types in opposite directions. Using stage specific endosperm reporters, we demonstrate that the source-sink endosperm phase change is delayed or precocious in seeds of the two hybrids. Our data suggests an A. thaliana type I MADS-BOX family locus to act as a promoter of endosperm cellularization, affecting the germination rates of A. arenosa or A. lyrata hybrid seeds in opposite directions.
2 Materials and methods
2.1 Plant material and growth conditions
A. thaliana accessions (Col-0, C24, Ws-2 and Wa-1) and mutant lines were obtained from the Nottingham Arabidopsis Stock Center (NASC). The A. arenosa population MJ09-4 originates from Nízke Tatry Mts.; Pusté Pole (N 48.8855, E 20.2408) and the A. lyrata subsp. petraea population MJ09-11 originates from lower Austria; street from Pernitz to Pottenstein (N 47.9190, E 15.9755) (Jørgensen et al., 2011; Lafon-Placette et al., 2017; Bjerkan et al., 2020). Mutant lines agl35-1 (SALK_033801), agl40-1 (SALK_107011) and mea-9 (SAIL_724_E07) were in Col-0 accession background (Shirzadi et al., 2011; Kirkbride et al., 2019; Bjerkan et al., 2020). The agl35-1 T-DNA line was genotyped using primers AAACCAAAGTTTTGCCACTAAGAC, ATTTTTCAGTCAAGATTACCCACC and GCGTGGACCGCTTGCTGCAACTCTCTCAGG. Marker lines proAT5G09370>>H2A-GFP (EE-GFP) and proAT4G00220>>H2A-GFP (TE1-GFP) were in Col-0 accession background (van Ekelenburg et al., 2023).
Surface-sterilization of seeds was performed by treatment with 70% ethanol, bleach (20% Chlorine, 0.1% Tween20) and wash solution (0.001% Tween20) for 5 min each step (Lindsey et al., 2017). Sterilized seeds were transferred to petri dishes containing 0.5 MS growth-medium with 2% sucrose (Murashige and Skoog, 1962) and stratified at 4°C for 2 days (A. thaliana) or 10 days (A. lyrata, A. arenosa and hybrids) before germination at 22°C with a 16h/8h light/dark cycle. After two weeks, seedlings were transferred to soil and cultivated at 18°C (16h/8h light/dark cycle, 160 µmol/m2/sec, 60-65% humidity). A. arenosa strain MJ09-4 was previously demonstrated to be diploid (Bjerkan et al., 2020). A. lyrata and A. thaliana × A. lyrata individual hybrid plants were confirmed diploid by flow cytometry (Supplementary Datasheet S1).
2.2 Crosses, temperature- and germination assays
A. thaliana plants were emasculated 2 days before pollination. Crossed plants were placed at experimental growth temperature until silique maturity/harvesting. For each cross combination, 4-8 different individual plants were used as pollinators and 15-85 siliques (biological replicates) were harvested individually (Supplementary Datasheet S2). After short-term storage at 4°C seeds from induvidual siliques were surface-sterilized ON using chlorine gas (Lindsey et al., 2017). All seeds from the harvested individual siliques were counted and planted on individual 0.5 MS growth-medium containing petri-dishes and scored for germination by counting protrusions through the seed coat after 10 days at 22°C growth conditions. On day 20, germinated seedlings were checked for A. thaliana accidental self-pollination (formation of floral shoots without vernalization). These rare events occurred at a frequency less than 1% and self-plants were removed from the analyses if present.
2.3 Statistics
R-studio (version 2023.03.1 + 446) (R Core Team, 2023) was used for data analyses. Plots were generated using the ggplot2 (Wickham, 2016) and dplyr (Wickham et al., 2023) packages. For statistical analyses the car package (Fox and Weisberg, 2019) and ggpubr package (Kassambara, 2023) were employed. To assess the homogeneity of variance for the germination assays we conducted Levene’s test (Levene, 1960). If the null hypothesis was rejected, the Welch’s t-test (Welch, 1947) was used for statistical analyses. In all other cases statistical analyses were performed using Wilcoxon rank sum test (Mann and Whitney, 1947).
2.4 Microscopy
Feulgen stained seeds were harvested at 6 days after pollination (DAP), stained using Schiff’s reagent (Sigma-Aldrich S5133), fixed and embedded in LR White (London Resin) (Braselton et al., 1996). Imaging was performed using an Andor DragonFly spinning disc confocal microscope with a Zyla4.2 sCMOS 2048x2048 camera attachment and excitation 488 nm/emission 500 to 600 nm. Seeds were scored for embryo and endosperm developmental stage and the number of endosperm nuclei. Endosperm nuclei counts were assigned an endosperm division value (EDV), which estimates the number of divisions to reach the corresponding number of endosperm nuclei (Ungru et al., 2008). Mean EDV was calculated using the formula: 2x = mean number of nuclei, where x is the EDV (x = LOG(mean number of nuclei)/LOG(2)).
Crosses with EE-GFP and TE1-GFP markers were imaged using the Andor DragonFly as described. Whole-mount imaging of seeds was performed using an Axioplan2 imaging microscope after 24 h/4°C incubation in 8:2:1 (w/v/v) chloral hydrate:water:glycerol (Grini et al., 2002). Mature dry seeds were imaged on a 1.5 x 2.5 cm grid using a Leica Z16apoA microscope connected to a Nikon D90 camera. Seed size and circularity were measured by converting images to black and white and then using the ImageJ “Threshold” and “Analyze particles” functions (https://imagej.nih.gov/ij/).
3 Results
3.1 Antagonistic effects on endosperm barriers in hybrid seeds from A. lyrata and A. arenosa crossed to A. thaliana
In order to compare the success of interspecific hybrids of A. thaliana crossed with A. lyrata or A. arenosa, we first observed the seed-set. Hybrid crosses with A. lyrata fathers had significantly reduced seed set per silique compared to crosses with A. arenosa fathers or A. thaliana (Col-0) self crosses (Supplementary Figures S1A, B). The lower seed-set correlates with failure of pollen tube burst after entering the female gametophyte (Supplementary Figure S1C), a pre-zygotic barrier previously described (Escobar-Restrepo et al., 2007). Interestingly, pollen tube burst failure can also be observed in crosses between A. thaliana and A. arenosa, but to a lower degree, correlating with the observed seed-set frequency (Supplementary Figures S1A, C). Differences in seed set between the two hybrid crosses thus have a gametophytic pre-zygotic base, and therefore do not affect the postzygotic hybridization barrier in these crosses.
A. thaliana self seeds and A. thaliana × A. arenosa or A. thaliana × A. lyrata hybrid seeds were comparable in size at 6 DAP (Figure 1A), but most A. thaliana × A. lyrata hybrid embryos were at an earlier developmental stage (Figure 1B). Large variation in embryonic stages including developmental arrest was observed in 12 DAP whole-mount chloral hydrate cleared hybrid seeds (Supplementary Figure S2A). A time series of Feulgen stained A. thaliana × A. lyrata hybrid seeds further identified variation in endosperm cellularization, starting at 3 DAP and resulting in developmental arrest at the globular embryo stage (Supplementary Figure S2B). The frequency of A. thaliana × A. lyrata early cellularization was higher than for A. thaliana self, and in strong contrast to the late cellularization in hybrid seeds from A. thaliana crossed with A. arenosa [Figure 1B; (Josefsson et al., 2006; Bjerkan et al., 2020)]. In order to compare the observed cellularization phenotype with nuclear proliferation in the syncytial endosperm, we investigated the number of nuclei in hybrid endosperm. The endosperm nuclei-number was significantly lower in A. thaliana × A. lyrata compared to A. thaliana × A. arenosa hybrid seeds, suggesting a reduction in endosperm proliferation rate (Figure 1C). Nonetheless, germination rates of hybrid seeds from crosses with A. lyrata were significantly higher than crosses with A. arenosa (Figure 1D; Supplementary Datasheet S2), indicating that the endosperm hybrid barrier is influenced in a diametrically opposed manner, both in terms of the endosperm cellularization phenotype and the effective output measured as the ability of hybrid seeds to germinate.
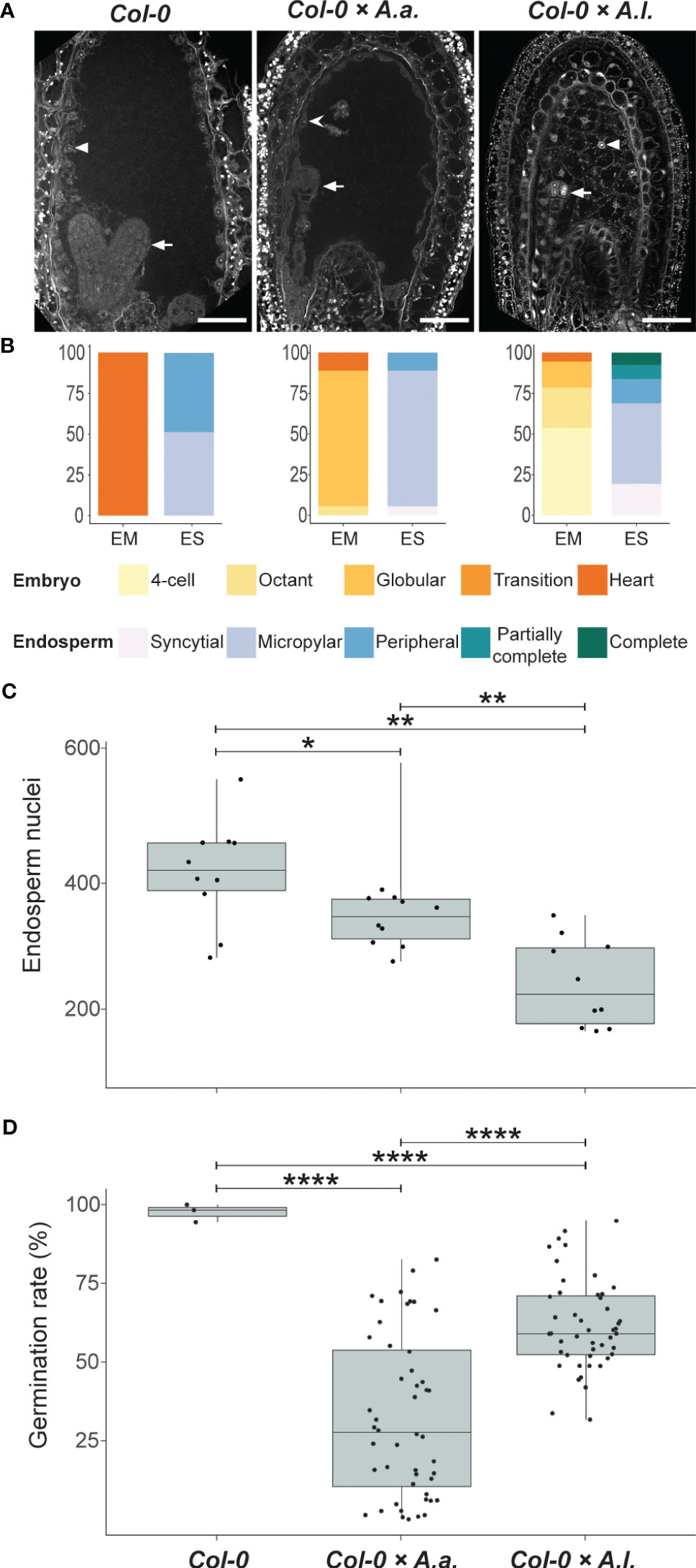
Figure 1 Antagonistic endosperm phenotypes in hybrid seeds. Seeds from crossing A. arenosa (A.a.) or A. lyrata (A.l.) as pollen donor to A. thaliana (Col-0) have antagonistic endosperm phenotypes and display opposing seed germination rates. (A) Confocal images of Feulgen-stained Col-0 and hybrid seeds 6 days after pollination (DAP) emphasizing endosperm cellularization. Open arrowhead points to syncytial endosperm nuclei, closed arrowheads point to cellularized endosperm nuclei and full arrows point to the embryo. In Col-0 self cellularization occurs at the 6 DAP embryo heart stage whereas in A.a. hybrid seeds the endosperm is mainly syncytial at 6 DAP. In A.l. hybrid seeds precocious endosperm cellularization is observed 6 DAP already at the early globular stage. All crosses are female × male. Scale bar = 50 μm. (B) Relative frequencies of embryo and endosperm stages in seeds from the same crosses as above. Col-0, n = 37; Col-0 × A.a., n = 18; Col-0 × A.l., n = 93; EM, embryo stages; ES, endosperm stages. (C) Number of endosperm nuclei in seeds from the same crosses as above, n = 10. Significance is indicated for the comparisons between all genotypes (Wilcoxon rank-sum test: *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001). (D) Germination rates in seeds from the same crosses as above. Biological replicates (siliques): Col-0, n = 4; Col-0 × A.a., n = 48; Col-0 × A.l., n = 48. Significance is indicated for the comparisons between all genotypes (Welch’s t-test: *P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001).
3.2 Endosperm phase change and cellularization in hybrids are not synchronized with embryo development
Compared to A. thaliana, viable seeds resulting from crosses between A. thaliana mothers and A. arenosa or A. lyrata fathers appear to develop along a slower embryo and endosperm developmental path. This suggests that as long as endosperm cellularization and embryo development are synchronized, seed viability is not impacted. In A. lyrata × A. arenosa hybrid seeds with unsynchronized endosperm and embryo development, the embryo can be rescued in vitro, indicating that the barrier is caused by lack of nutrient support to the growing embryo (Lafon-Placette and Köhler, 2016).
To investigate synchronization of embryo and endosperm in A. thaliana (Col-0) hybrid seeds with A. arenosa or A. lyrata fathers, we used genetic markers of endosperm development that are expressed before and after endosperm cellularization in A. thaliana (van Ekelenburg et al., 2023). In A. thaliana the EE-GFP genetic marker is expressed after fertilization and up to endosperm cellularization at 6-7 DAP (van Ekelenburg et al., 2023) and not expressed after cellularization (Figures 2, S3). In A. thaliana × A. arenosa hybrid seeds, expression of the EE-GFP marker was observed for the full duration of the time series (15 DAP) and no visible downregulation could be observed (Figures 2, S3). This expression pattern supports the observation that A. thaliana × A. arenosa hybrid seeds fail to initiate or have delayed endosperm cellularization (Figure 1) as described previously (Josefsson et al., 2006; Bjerkan et al., 2020). In contrast, when crossed to A. lyrata, expression of the EE-GFP marker decreased from 5 DAP and only a low frequency of seeds expressed the marker at cellularization around 9 DAP (Figures 2, S3). Taking the developmental delay in the A. thaliana × A. lyrata hybrid into account, the EE-GFP marker is prematurely terminated (Figure 2B), in accordance with the early cellularization phenotype (Figure 1). In A. thaliana self seeds, downregulation of the EE-GFP marker coincides with endosperm cellularization (van Ekelenburg et al., 2023). In hybrid seeds from A. arenosa or A. lyrata fathers, continued expression or premature EE-GFP downregulation, respectively, (Figure 2B) coincides with diametrically opposed cellularization phenotypes both indicating that the endosperm phase change and cellularization is not synchronized with embryo development, leading to embryo and seed failure.
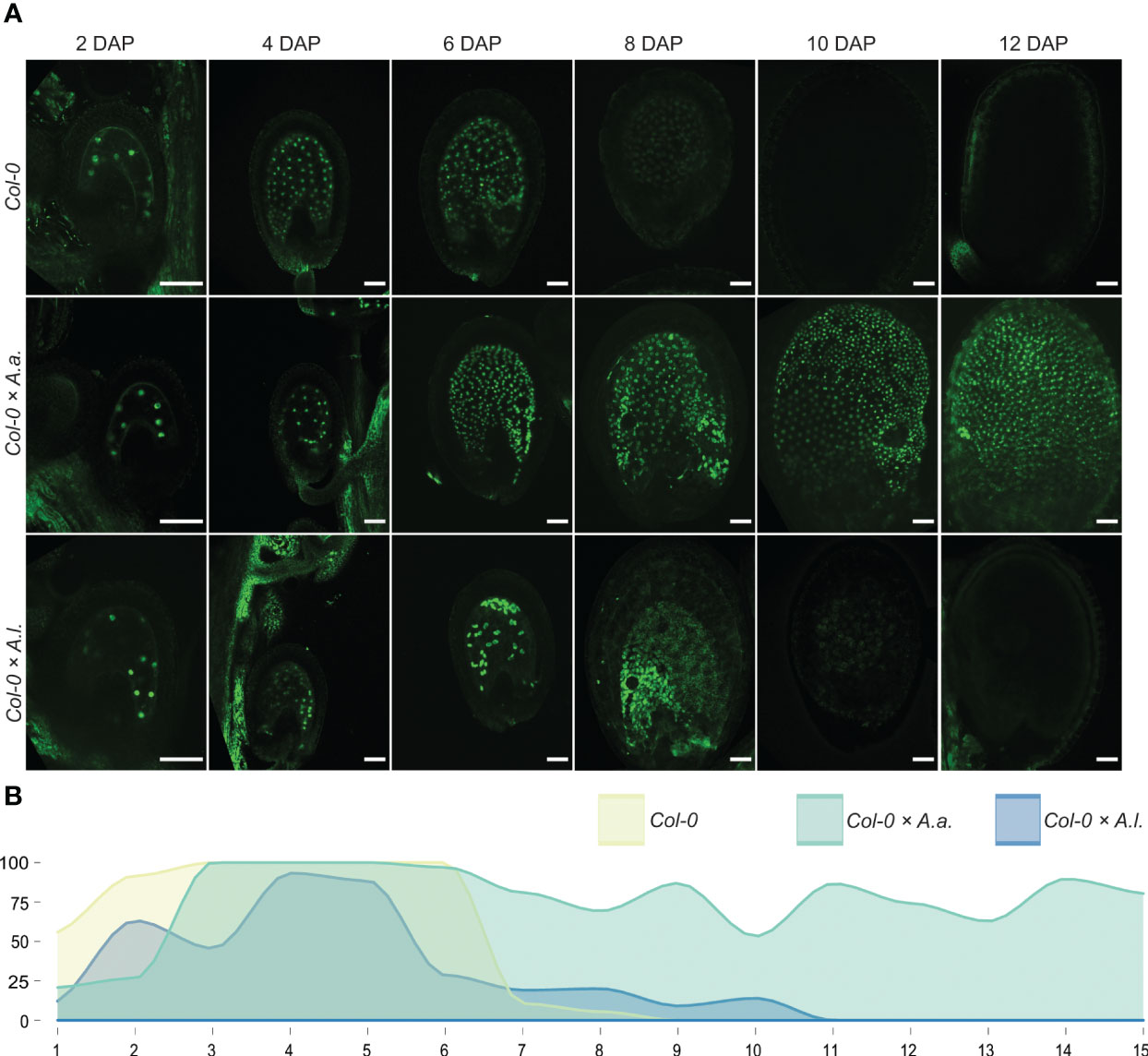
Figure 2 Early endosperm marker confirms aberrant cellularization timing in hybrid seeds. (A) Expression patterns of proAT5G09370>>H2A-GFP (EE-GFP) in seeds of A. thaliana (Col-0) and in hybrid seeds from crossing A. arenosa (A.a.)/A. lyrata (A.l.) as pollen donor to Col-0 at 2, 4, 6, 8, 10 and 12 days after pollination (DAP). Scale bar = 50 µm. (B) Percentage of hybrid seeds expressing EE-GFP at 1-15 DAP. A. thaliana (Col-0) self crosses and hybrid crosses are indicated with different colors.
In A. thaliana, the TE1-GFP genetic marker is expressed after cellularization at 6-7 DAP (van Ekelenburg et al., 2023) and until seed maturation at 17-19 DAP (Figures 3, S4). Delayed activation of marker expression was observed at low frequency in the A. thaliana × A. arenosa hybrid seeds with expression from 9 to 18 DAP (Figures 3, S4). Interestingly, in A. thaliana × A. lyrata hybrid seeds the TE1-GFP marker expressed prematurely from before 6 DAP globular stage seeds lasting until 18 DAP (Figures 3, S4), indicating premature endosperm phase change initiation and supporting the observed precocious endosperm cellularization phenotype (Figure 1).
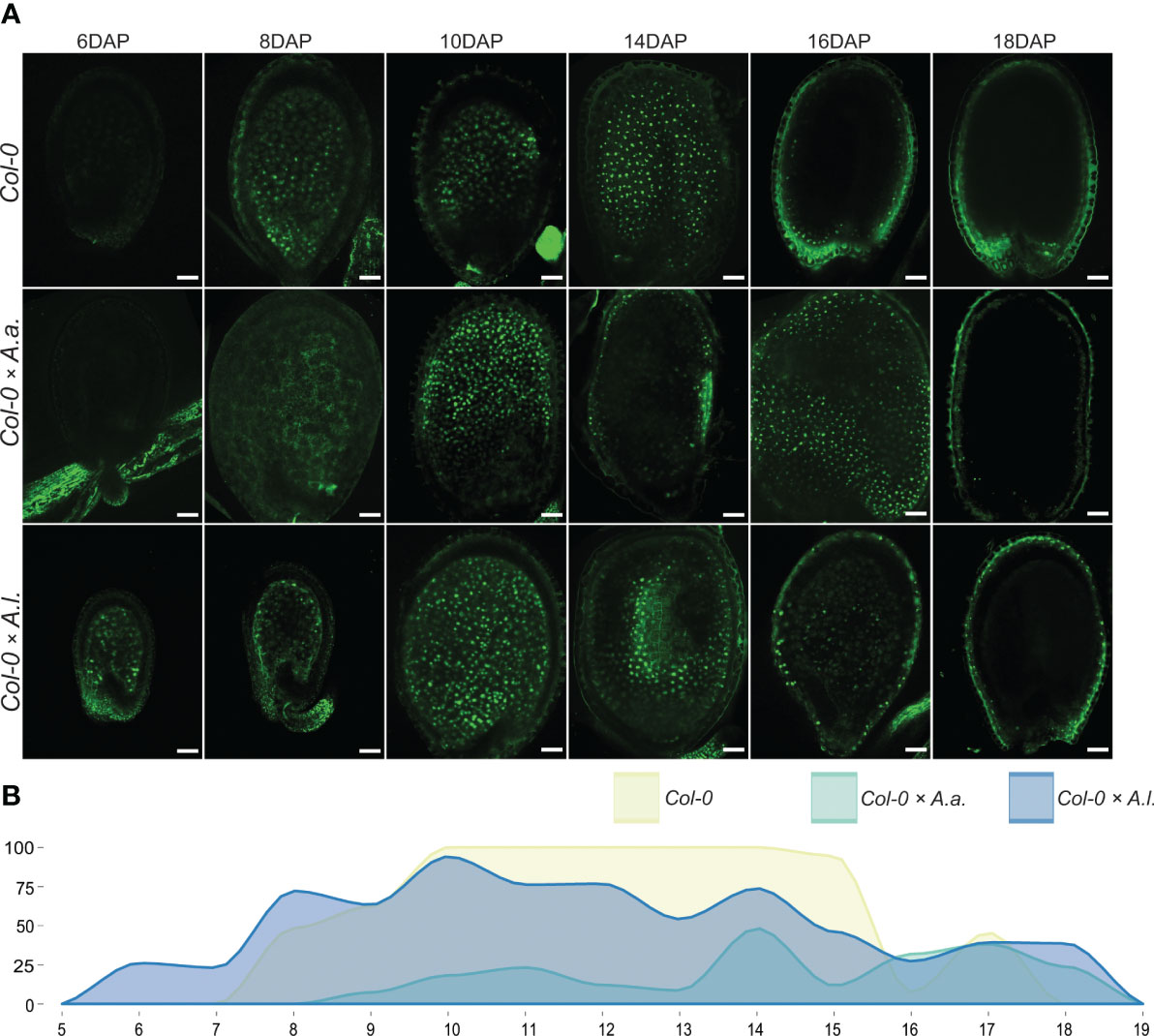
Figure 3 Total endosperm marker confirms aberrant cellularization timing in hybrid seeds. (A) Expression patterns of proAT4G00220>>H2A-GFP (TE1-GFP) in seeds of A. thaliana (Col-0) and in hybrid seeds from crossing A. arenosa (A.a.)/A. lyrata (A.l.) as pollen donor to A. thaliana (Col-0) at 6, 8, 10, 14, 16 and 18 days after pollination (DAP). Scale bar = 50 µm. (B) Percentage of hybrid seeds expressing TE1-GFP at 5-19 DAP. A. thaliana (Col-0) self crosses and hybrid crosses are indicated with different colors.
In A. thaliana seeds the decrease of EE-GFP expression and increase of TE1-GFP expression is strictly coordinated, with limited overlap. This is contrasted by the EE-GFP and TE1-GFP expression in seeds from hybrid crosses (Figure 4). Since the marker transgenes are expressed from the maternal A. thaliana genomes, the only difference in these crosses is the paternal contribution. For A. thaliana × A. arenosa an overlap in expression of the markers was observed from 9 DAP, caused by the prolonged expression of the EE-GFP marker (Figure 4). In A. thaliana × A. lyrata overlapping expression was observed from 6 to 10 DAP (Figure 4), showing a shift in TE1-GFP expression towards earlier developmental stages, although the A. thaliana × A. lyrata hybrid seeds develop slower compared to both A. thaliana selfed and A. thaliana × A. arenosa seeds (Figure 1).
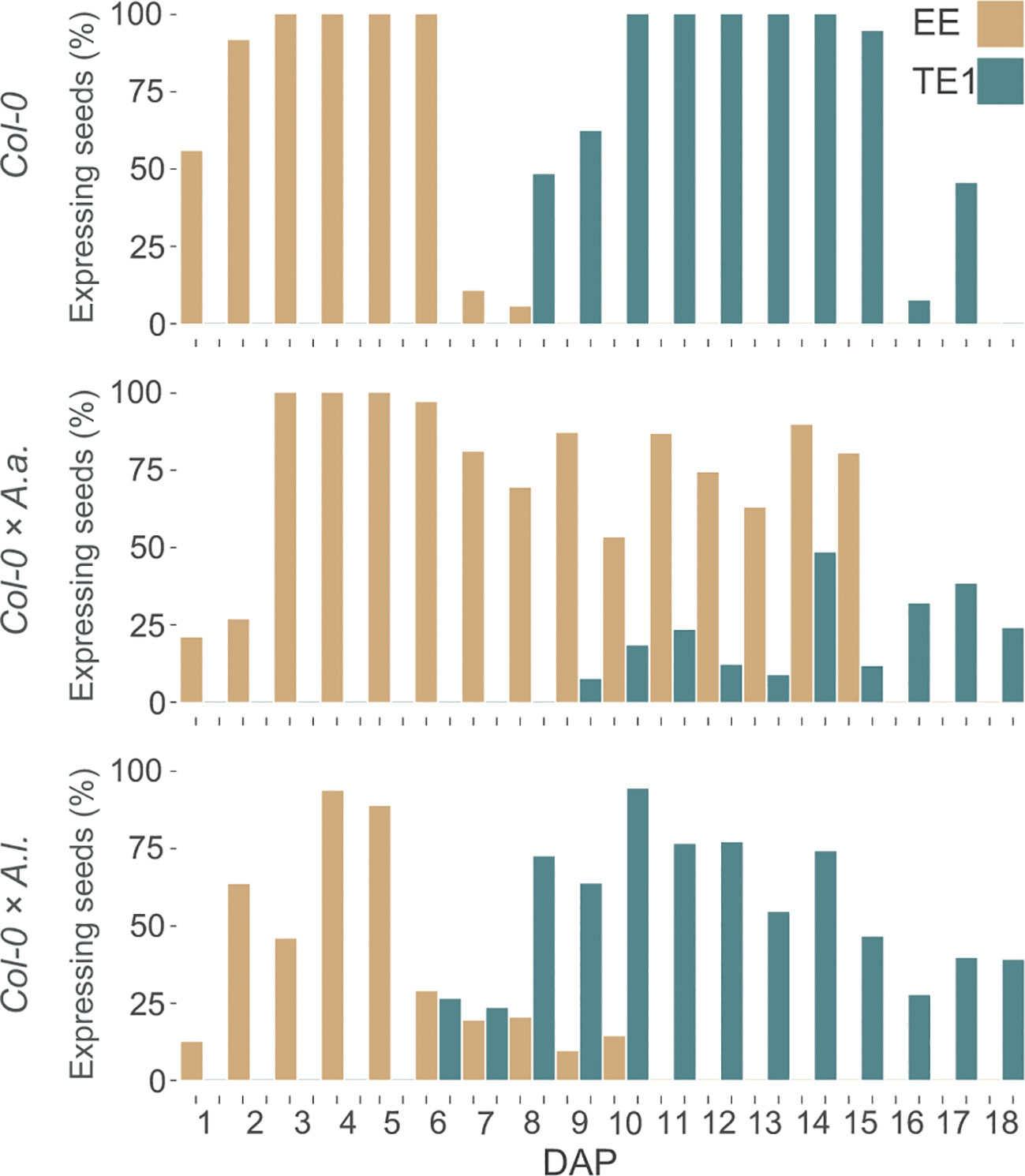
Figure 4 Overlapping expression of early and late endosperm markers in hybrid seeds. Percentage of seeds expressing proAT5G09370>>H2A-GFP (EE-GFP) and proAT4G00220>>H2A-GFP (TE1-GFP) at 1-18 days after pollination (DAP) in A. thaliana (Col-0) self-crosses and from crossing A. arenosa (A.a.) or A. lyrata (A.l.) as pollen donor to A. thaliana (Col-0). EE-GFP expression in the Col-0 × A. arenosa hybrid seeds were not documented after 15 DAP.
3.3 Temperature alters the hybrid barrier strength in diametrically opposed directions
Lowering of temperature from 22°C to 18°C ameliorates the germination efficiency of the hybrid seeds from A. thaliana mothers crossed to A. arenosa fathers (Bjerkan et al., 2020). In order to investigate if the phenotypically contrasting hybrid barrier observed in seeds from A. lyrata fathers was affected by temperature in a corresponding way, we performed crosses between A. thaliana (Col-0) mothers and A. lyrata or A. arenosa fathers at 4°C temperature windows ranging from 14°C to 26°C. Interestingly, the germination rate of A. thaliana × A. arenosa hybrid seeds was significantly enhanced by progressive lowering of the temperature, and contrasted by the germination rate of A. thaliana × A. lyrata hybrid seeds that was significantly enhanced by progressive increase of the temperature (Figure 5; Supplementary Datasheet S2). In the temperature window, the effects on the hybrid barriers followed a close to linear, but opposed reaction norm. We applied more extreme temperatures previously reported to be within the normal, and not stress inducing, growth-range of A. thaliana (Lloyd et al., 2018), but a further enhancement could not be obtained (Supplementary Figure S5).
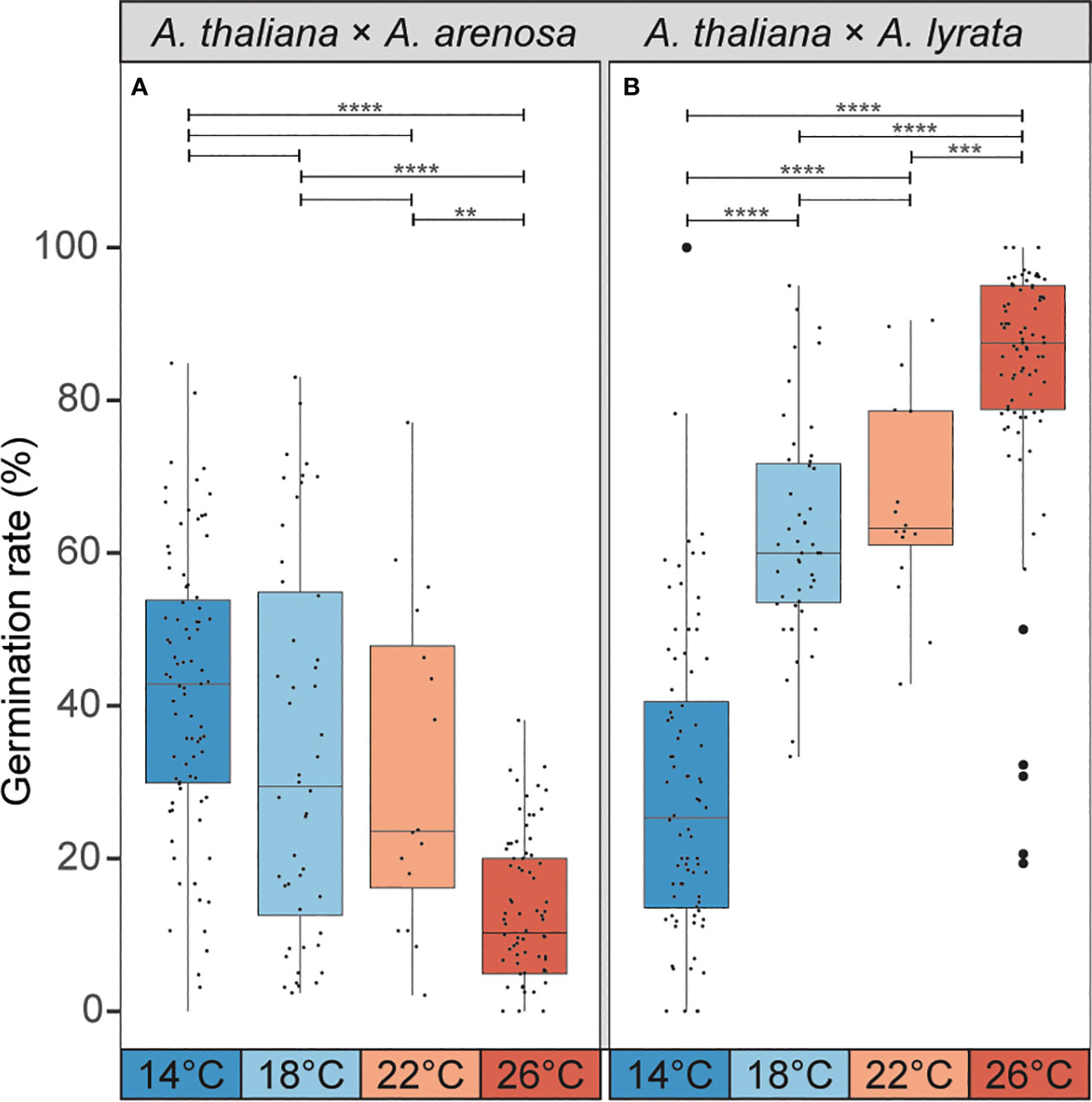
Figure 5 Temperature during seed development affects germination rate of hybrid seeds. (A) Germination rate of A. thaliana (Col-0) × A. arenosa hybrid seeds from crosses performed at 14°C, 18°C, 22°C and 26°C. Germination rate decreases with increasing temperature. Biological replicates (siliques): 14°C, n = 83 (3243 seeds); 18°C, n = 48 (2610 seeds); 22°C, n = 16 (796 seeds); 26°C, n = 78 (2713 seeds). (B) Germination rate of A. thaliana (Col-0) × A. lyrata hybrid seeds at 14°C, 18°C, 22°C and 26°C. Germination rate increases with increasing temperature. Biological replicates (siliques): 14°C, n = 84 (1676 seeds); 18°C, n = 47 (1540 seeds); 22°C, n = 16 (533 seeds); 26°C, n = 85 (2043 seeds). Box plot contains scattered data points representing germination rates observed per silique. Outliers are plotted as large data points. Significance is indicated for the comparisons between all temperatures (Welch’s t-test: **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
3.4 Accessions of A. thaliana influence A. arenosa and A. lyrata hybrid barriers antagonistically
Different accessions of A. thaliana affect the strength of the hybrid barrier when crossed to A. arenosa (Burkart-Waco et al., 2012; Bjerkan et al., 2020). Having demonstrated an antagonistic temperature effect on the hybrid barrier when A. arenosa or A. lyrata are crossed to A. thaliana mothers, we next investigated if different A. thaliana accessions influence the hybrid barrier in similar or opposite manner. We performed hybrid crosses with A. arenosa and A. lyrata fathers to the diploid A. thaliana accessions Col-0, C24 and Ws-2. Additionally, the tetraploid accession Wa-1 was used as a control, as tetraploid A. thaliana crossed to diploid A. arenosa has been shown to increase hybrid seed survival (Josefsson et al., 2006).
Compared to the Col-0 accession crosses, the C24 accession enhanced seed survival significantly when crossed to A. arenosa, contrasted by the A. lyrata hybrid where the germination rate declined highly significantly compared to the Col-0 cross (Figure 6; p ≤ 0.0001). For Ws-2, the germination rate was severely reduced in the A. arenosa hybrid cross, contrasted by moderately high (though lower than for Col-0) germination rate in the A. lyrata hybrid cross (Figure 6, Supplementary Datasheet S2).
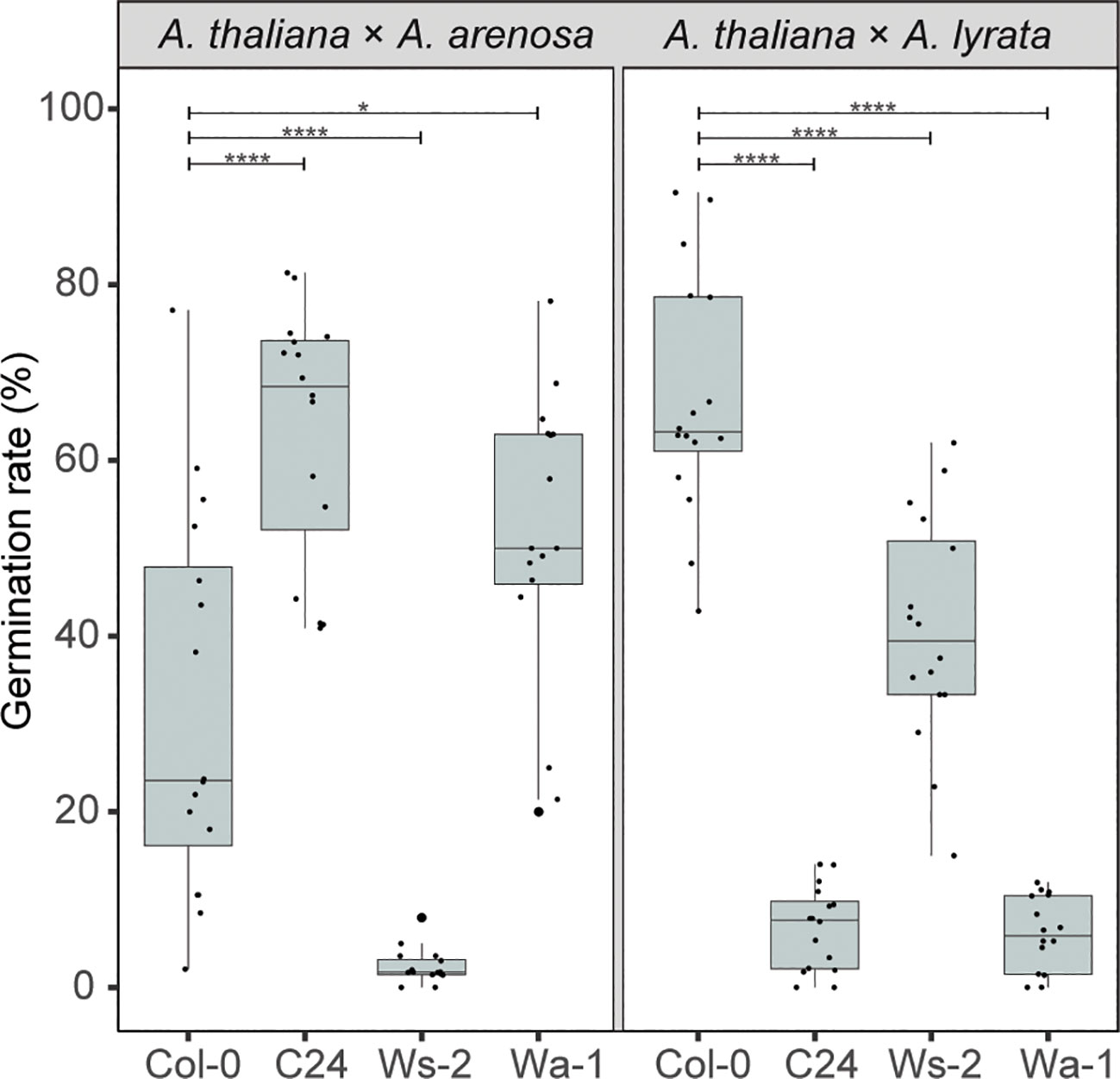
Figure 6 A. thaliana accessions affect hybrid barriers antagonistically. Germination rate of seeds from crossing A. arenosa (A.a.)/A. lyrata (A.l.) as pollen donor to A. thaliana (Col-0/C24/Ws-2/Wa-1) at 22°C. Biological replicates (siliques): Col-0 × A.a., n = 16 (533 seeds); C24 × A.a., n = 16 (805 seeds); Ws-2 × A.a., n = 16 (947 seeds); Wa-1 × A.a., n = 16 (805 seeds); Col-0 × A.l., n = 16 (533 seeds); C24 × A.l., n = 16 (821 seeds); Ws-2 × A.l., n = 16 (572 seeds); Wa-1 × A.l., n = 16 (759 seeds). Box plot contains scattered data points representing germination rates observed per silique. Outliers are plotted as large data points. Significance is indicated for comparisons between Col-0 × A.a./Col-0 × A.l. and crosses involving other A. thaliana accessions (Welch’s t-test; *P ≤ 0.05; ****P ≤ 0.0001).
In the tetraploid A. thaliana Wa-1 to diploid A. arenosa hybrid cross, the seed germination rate was enhanced, as previously reported (Josefsson et al., 2006). In contrast, in the Wa-1 to A. lyrata hybrid cross, the germination rate was significantly decreased compared to the Col-0 cross (Figure 6). Notably, the effect on hybrid seed viability was higher with the diploid C24 accession than with the tetraploid Wa-1 accession.
Crosses performed in parallel at 18°C and 22°C demonstrated the same trends at both temperatures (Supplementary Figure S6, Supplementary Datasheet S2). Although large differences in the barrier strength was observed, as measured by germination and seed viability, no obvious correlation could be found between seed survival and seed size and circularity (Supplementary Figure S7).
3.5 A. thaliana accession effects are not readily explained by endosperm cellularization phenotype
To investigate if the endosperm phenotype reflects the influence of accessions on hybrid seed viability, we inspected Feulgen stained 6 DAP hybrid seeds by confocal microscopy. We scored the number of endosperm nuclei in A. thaliana accessions and accession hybrids with A. arenosa and A. lyrata at 18°C and 22°C. The endosperm division value [EDV; (Ungru et al., 2008)] was generally higher when A. arenosa was involved (Figure 7; Supplementary Datasheet S3). No obvious correlation between the number of endosperm nuclei and hybrid seed viability was found (Supplementary Figure S8), however a significant correlation between endosperm proliferation rates and growth temperature could be observed in all crosses except A. thaliana C24 x A. lyrata. The latter hybrid cross did indeed exhibit very low germination rates (Figure 6), however similar low germination rates were found in A. thaliana Ws-2 x A. arenosa hybrid seeds but here accompanied by a high endosperm proliferation rate (Supplementary Figure S8, Supplementary Datasheet S3).
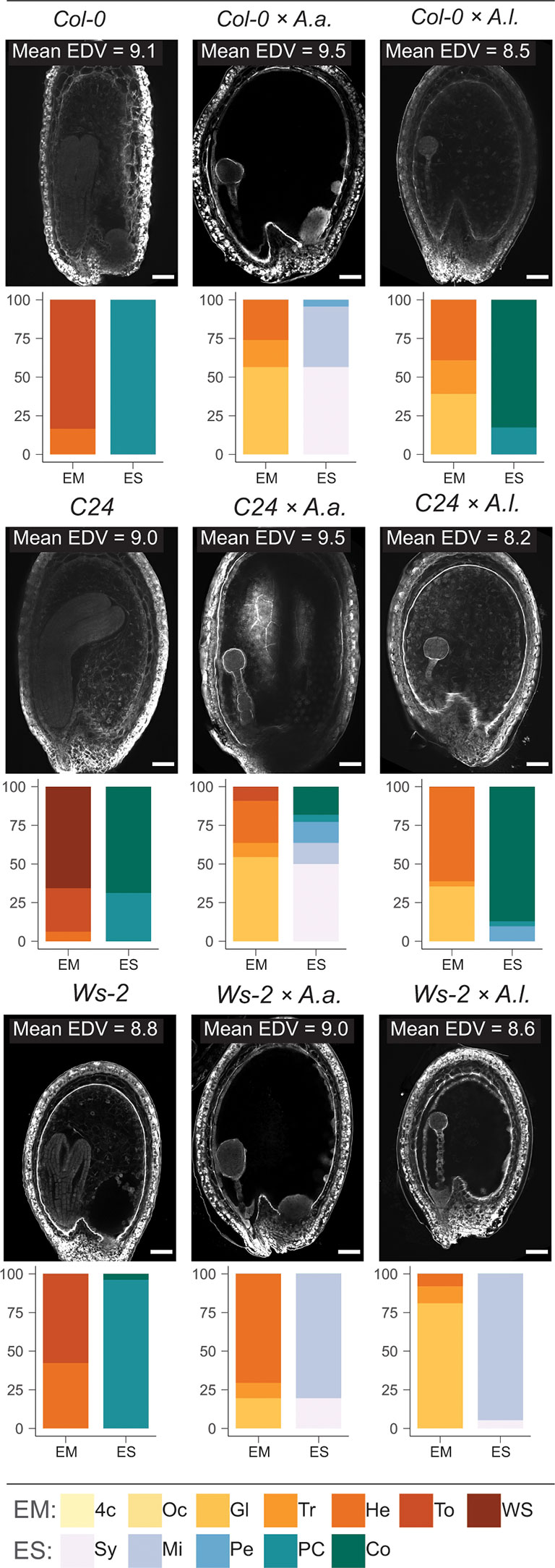
Figure 7 Effect of temperature, accession and hybridization on endosperm development. Confocal images showing endosperm cellularization of 6 DAP Feulgen-stained seeds from crossing A. arenosa (A.a.) or A. lyrata (A.l.) as pollen donor to A. thaliana (accession Col-0 or C24 or Ws-2) at 22°C. Scale bar = 50 μm. Mean endosperm division value (EDV) is shown within each image, nEDV = 10 seeds. Beneath each image quantification of the described embryo and endosperm stages is shown as bar charts: Col-0, n = 36; Col-0 × A.a., n = 23; Col-0 × A.l., n = 23; C24, n = 32; C24 × A.a., n = 22; C24 × A.l., n = 31; WS-2, n = 26; WS-2 × A.a., n = 51; WS-2 × A.l., n = 37. Embryo stages (EM): 4c, 4-cell; Oc, Octant; Gl, Globular; Tr, Transition; He, Heart; To, Torpedo; WS, Walking stick; Endosperm cellularization stages (ES): Sy, Syncytial endosperm; Mi, Micropylar endosperm cellularization; Pe, Peripheral endosperm cellularization; PC, Partially complete endosperm cellularization; Co, Complete endosperm cellularization.
Seed phenotypes were scored for defined stages of embryo and endosperm development. A. thaliana accession self crosses at 22°C displayed embryo stages in the late heart to walking stick stage, with C24 exhibiting the fastest embryonic development. Endosperm cellularization was partly complete, though fully completed in most A. thaliana C24 self-seeds (Figure 7; Supplementary Datasheet S3). In A. thaliana accession x A. arenosa hybrid seeds, embryo development ranged from globular to transition stages. The endosperm was mainly syncytial or had initiated cellularization in the micropylar endosperm. In the C24 cross almost half of the seeds exhibited advanced cellularization stages and also complete cellularization. In this cross, higher germination rate was correlated with temporally correct timing of endosperm cellularization (Figures 6, 7).
In A. thaliana accession x A. lyrata hybrid seeds, embryonic stages ranged from globular to heart, where the C24 accession displayed a majority of heart stages, and Ws-2 a majority of globular stages. In the Col-0 and C24 accession hybrids, near uniform complete endosperm cellularization was observed, contrasted by early peripheral cellularization in Ws-2 (Figure 7). In the case of the latter hybrid cross, embryo development and endosperm cellularization appeared to be synchronized, leading to higher seed viability (Figure 6). However, large differences in seed viability between C24 (low) and Col-0 (high) hybrid crosses (Figure 6) were not reflected by the endosperm cellularization phenotype as both crosses had mostly fully cellularized endosperm and appeared to be in a similar embryonic stage (Figure 7). A. thaliana accession hybrid crosses at 18°C exhibited a similar pattern (Supplementary Figure S9). Major significant differences in seed viability between C24 and Ws-2 (low vs medium-high; Supplementary Figure S6) were not reflected by endosperm cellularization as both accession hybrids exhibited mostly micropylar endosperm (Supplementary Figure S9). We conclude that the effect of using different accessions in the hybrid crosses can not readily be explained by a direct effect on the endosperm cellularization phenotype alone and that a more complex interaction between different genotypes occur.
3.6 Mutation of the MADS-box transcription factor AGL35 influences A. arenosa and A. lyrata hybrid barriers antagonistically
Deregulation of type I MADS-box TFs has been correlated with endosperm-based hybridization barriers (Walia et al., 2009), and many of these TFs are epigenetically regulated by the so-called FIS-PRC2 and the histone methyltransferase MEDEA (MEA) (Zhang et al., 2018). Mutation of MEA results in ectopic proliferation of endosperm nuclei and delayed cellularization (Grossniklaus et al., 1998; Köhler et al., 2003; Guitton et al., 2004) and we therefore investigated if endosperm overproliferation in A. thaliana mea mutant mothers crossed to A. arenosa or A. lyrata enhances or alleviates the hybrid barriers, respectively.
Heterozygous self-crossed A. thaliana mea mutants resulted in a reduced germination rate of 60% meaning that 80% of seeds carrying the mutant maternal allele failed to germinate due to delayed endosperm cellularization (Supplementary Figure S10). Crossing A. thaliana mea to A. arenosa or A. lyrata resulted in a significant decrease in seed survival compared to Col-0 crosses (Supplementary Figure S10, Supplementary Datasheet S2). Reduced germination rate in both crosses corresponded to an additive effect of the reduced germination of the heterozygous A. thaliana mea mutation. Compared to expected values, single gene mutation of mea could not bypass the A. thaliana x A. lyrata species barrier, nor enhance the A. thaliana x A. arenosa barrier (Supplementary Figure S10, Supplementary Datasheet S2).
We studied the effect of single candidate genes regulated by FIS-PRC2 (Zhang et al., 2018). The mutant agl35-1 in the Col-0 background was previously shown to strengthen the barrier when A. thaliana was crossed to A. arenosa (Bjerkan et al., 2020) and AGL35 was upregulated in the same hybrid cross (Walia et al., 2009). AGL40 is similarly expressed in the endosperm (Zhang et al., 2018), upregulated in hybrids (Walia et al., 2009) and mutant seeds have reduced seed size (Kirkbride et al., 2019). To investigate if mutation of single candidate genes could produce opposed effects on the hybrid barrier when crossing A. thaliana mothers to A. lyrata or A. arenosa, as observed when changing temperature or A. thaliana accession (Figures 5, 6), we performed A. lyrata and A. arenosa crosses to A. thaliana agl35-1 and agl40-1 and scored seed germination.
Interestingly, in crosses where agl35-1 was crossed to A. arenosa or A. lyrata a highly significant decrease or increase in germination rate was observed, respectively, compared to wild type Col-0 crosses (Figure 8A). Single mutation of AGL35 affected the hybrid barrier strength in diametrically opposed directions, as in crosses to A. lyrata the germination rate was significantly enhanced, in contrast to A. thaliana x A. arenosa crosses where the germination rate was significantly reduced (Figure 8A). Mutation of AGL40 crossed to A. arenosa did not significantly affect germination rate, but in crosses to A. lyrata germination rate was significantly reduced. Col-0 crossed to A. lyrata displayed reduced seed size (Figures S2, S7) due to early endosperm cellularization (Figure 1), and thus the mutation of AGL40 may increase the frequency of early cellularization.
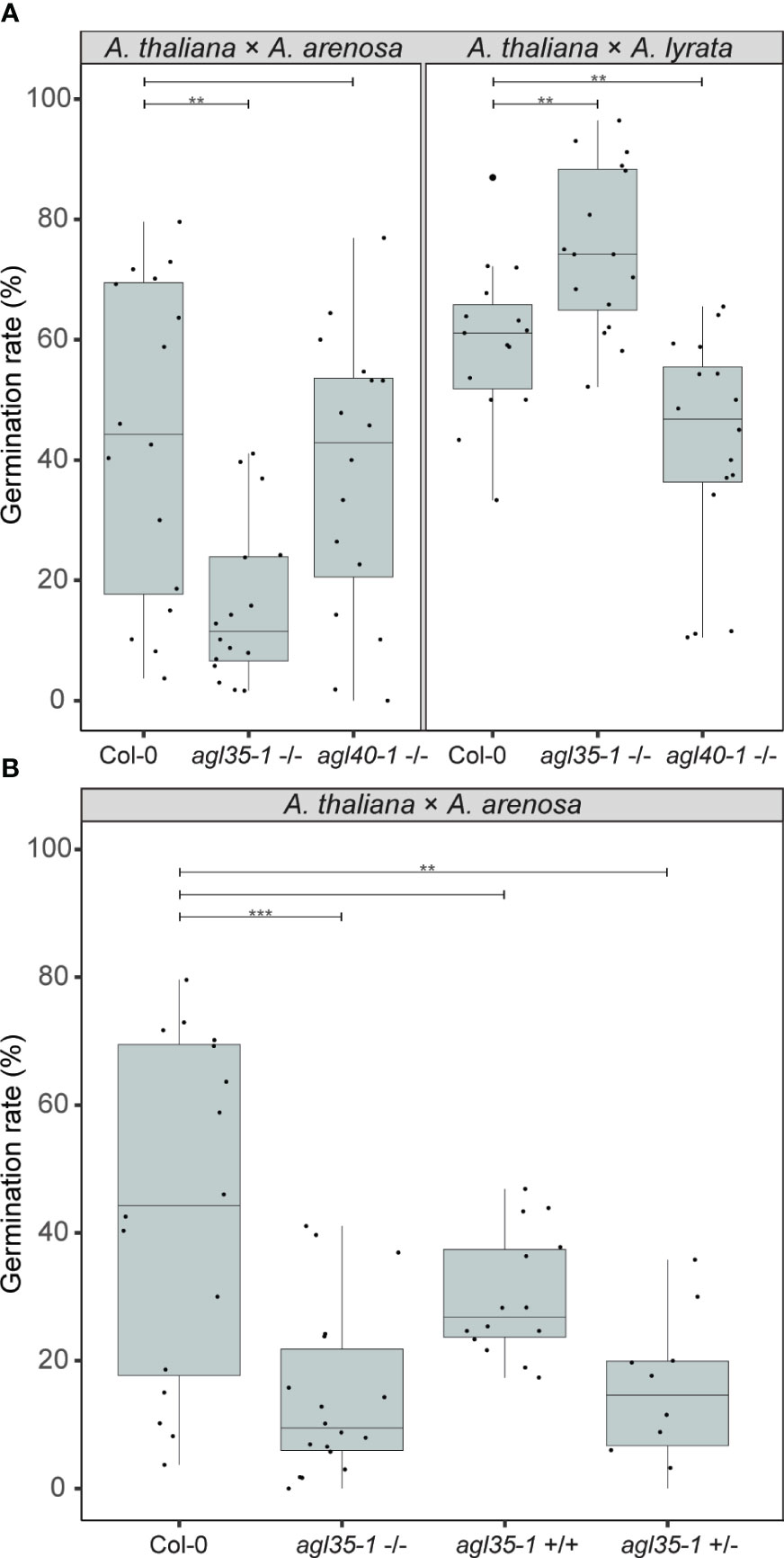
Figure 8 Genetic parameters influence the hybrid barrier. (A) Germination rate of seeds from crossing A. arenosa (A.a.)/A. lyrata (A.l.) as pollen donors to A. thaliana (Col-0), single mutants agl35-1-/- and agl40-1-/- at 18°C. Biological replicates (siliques): Col-0 × A.a., n = 16 (833 seeds); agl35-1-/- × A.a., n = 16 (904 seeds); agl40-1-/- × A.a., n = 16 (879 seeds); Col-0 × A.l., n = 16 (457 seeds); agl35-1-/- × A.l., n = 16 (509 seeds); agl40-1-/- × A.l., n = 16 (525 seeds). (B) Germination rate of seeds from crossing A. arenosa (A.a.) as pollen donor to A. thaliana (Col-0), homozygous agl35-1-/-, twice introgressed agl35-/- into Col-0 (agl35+/+), and heterozygous agl35-1 +/- at 18°C. Biological replicates (siliques): Col-0 × A.a., n = 16 (833 seeds); agl35-1-/- × A.a., n = 18 (1064 seeds); agl35+/+ × A.a., n = 14 (1562 seeds); agl35-1 +/- × A.a., n = 10 (525 seeds). Box plot contains scattered data points representing germination rates observed per silique. Outliers are plotted as large data points. Significance is indicated for the comparisons between Col-0 crosses and the mutant crosses (Welch’s t-test; **P ≤ 0.01; ***P ≤ 0.001).
To test if cryptic genetic variation in the agl35-1 mutant line could account for the observed phenotype, heterozygous agl35-1 was introgressed twice to Col-0 and segregating progeny of selfed heterozygotes were crossed to A. arenosa. The segregating agl35-1 A. thaliana mothers were genotyped for the agl35-1 insert and germination rate of hybrid seeds was scored (Figure 8B). Importantly, the germination rate of segregating wildtype plants (agl35+/+) was not significantly different from wildtype (Col-0) when crossed to A. arenosa. Both homozygous (agl35-1 -/-) and heterozygous agl35-1 +/- plants crossed to A. arenosa displayed significantly lower germination rate than Col-0 crossed to A. arenosa, indicating that the increased strength of the hybrid barrier was caused by mutation of AGL35. Furthermore, the observation that the strength of the hybrid barrier can be caused by heterozygous (agl35-1 +/-) plants crossed to A. arenosa indicates that the observed phenotype is caused by genetic interaction occurring in the fertilization products, the embryo or the endosperm.
4 Discussion
Recent advances in elucidating molecular mechanisms and genetic networks in hybrid endosperm lethality suggest that imprinted genes and genetic variation in the hybrid parents are important factors that can enhance or repress the frequency of the endosperm-based barrier (Bushell et al., 2003; Josefsson et al., 2006; Walia et al., 2009; Burkart-Waco et al., 2012; Burkart-Waco et al., 2013; Kradolfer et al., 2013; Schatlowski et al., 2014; Burkart-Waco et al., 2015; Rebernig et al., 2015; Wolff et al., 2015; Bjerkan et al., 2020). However, the mechanistic role of these factors and the interplay of genetic networks is largely unknown. For instance, ploidy can bypass the endosperm-based post-zygotic barrier, but a general role of ploidy cannot be attributed since ploidy plays a different role in maternal and paternal cross settings (Lafon-Placette et al., 2017). Furthermore, the role of gene dosage and genomic imprinting is supported by reports suggesting that mutation of MEGs and PEGs can overcome the endosperm-based post-zygotic barrier (Dilkes et al., 2008; Walia et al., 2009; Wolff et al., 2015; Borges et al., 2018), but a general role of imprinted genes cannot be defined since only some imprinted genes appear to have this effect (Walia et al., 2009; Burkart-Waco et al., 2015; Rebernig et al., 2015; Wolff et al., 2015). Since different accessions of the parental individuals in a hybrid cross can partly bypass the endosperm-based post-zygotic barrier without any change in ploidy (Walia et al., 2009; Wolff et al., 2015), the role of maternal and paternal genomes cannot be generalized.
Here we demonstrate that interspecies hybrid seeds from crossing A. lyrata or A. arenosa as the paternal parent to A. thaliana mothers show antagonistic endosperm cellularization phenotypes, with late cellularization in crosses with A. arenosa and early cellularization in crosses with A. lyrata. In both cases, cellularization failure results in an endosperm-based hybrid barrier and reduced viability of germinating seeds. This compares to previous studies where timing of endosperm cellularization is influenced by the paternal species in reciprocal A. arenosa and A. lyrata interspecies crosses and in crosses within the genus Capsella (Rebernig et al., 2015; Lafon-Placette et al., 2017). Intriguingly, we find that a temperature gradient leads to diametrically opposed cellularization phenotype responses in hybrid endosperm with A. arenosa or A. lyrata as pollen donors. In addition, A. thaliana accession genotypes also influence hybrid seed viability in opposite directions. To this end, we demonstrate that single gene mutation in A. thaliana MADS-box TF AGL35 independently can affect the germination rates of A. arenosa or A. lyrata hybrid seeds in opposite directions.
4.1 Ectopic timing of endosperm developmental phase change
The endosperm genetic markers EE-GFP and TE1-GFP mark syncytial endosperm development before cellularization and cellular endosperm stages, respectively (van Ekelenburg et al., 2023). We show that timing of the developmental phase change connected to endosperm cellularization is disturbed in hybrid seeds. In the Col-0 × A. arenosa hybrid seeds that fail to cellularize, the EE-GFP marker continues to be expressed throughout endosperm development, indicating phase change failure. In the Col-0 × A. lyrata hybrid seeds, characterized by early cellularization, the developmental time point of TE1-GFP expression indicates occurrence of a premature phase change. These findings support that not only the timing of endosperm cellularization is affected in these developing hybrid seeds, but also the developmental timing of the genetic network associated with endosperm phase change and maturation occurring at cellularization. In accordance with the incomplete endosperm hybridization barrier, prolonged expression of EE-GFP in Col-0 × A. arenosa seeds and precocious expression of TE1-GFP in Col-0 × A. lyrata seeds were not observed in all individual seeds. This suggests that gene regulation associated with the endosperm phase change within each hybrid seed varies and potentially is affected by genetic or epigenetic variation that modulates threshold levels for gene activation or repression.
In our system the sole difference between the two hybrids is the paternal parent, indicating a trans-acting mechanism, where differential expression from A. lyrata and A. arenosa genomes regulates the genetic markers expressed from the A. thaliana genome. Supporting this hypothesis, paternal transmission of mutants in NUCLEAR RNA POLYMERASE D1 (NRPD1) can bypass the cellularization phenotype in paternal excess inter-ploidy crosses (Erdmann et al., 2017; Martinez et al., 2018). NRPD1 is a main component in the the RNA-directed DNA methylation (RdDM) pathway, resulting in small RNA directed gene regulation by de novo DNA methylation (Law et al., 2013; Kirkbride et al., 2019) and could be a potential trans-acting regulatory mechanism (Erdmann et al., 2017). Future experiments to identify transcriptional differences from the parental genomes in hybrid seeds may point at key genes and mechanisms responsible for ectopic timing of the endosperm developmental phase change.
4.2 Temperature and accession affect the viability of hybrid seeds
We found that by increasing the temperature from 14°C to 26°C, Col-0 × A. arenosa seeds display significantly decreased germination rates (more than 30%). The same temperature range has an opposite effect in Col-0 × A. lyrata seeds resulting in increased germination rates (more than 50%). This demonstrates a temperature dependent genetic mechanism that acts antagonistically when A. thaliana is crossed to A. arenosa or A. lyrata and produces diametrically opposed cellularization phenotype responses in the hybrid endosperm.
Interestingly, in Brassica oleracea, temperature affects abscisic acid (ABA) levels specifically in the endosperm and cooler temperatures obstruct the breakdown of ABA in the desiccating endosperm (Chen et al., 2021). This is consistent with a recent report demonstrating that A. thaliana inter-ploidy uncellularized endosperm induced by paternal excess is correlated with increased ABA levels, suggesting that endosperm cellularization is connected to dehydration responses in the developing embryo (Xu et al., 2023). ABA catabolism in response to temperature may therefore be a potent mechanism to explain the temperature influence on the hybrid barrier when A. thaliana is crossed to A. arenosa. In a similar manner, we further speculate that precocious cellularization in crosses with A. lyrata may be associated with a similar mechanism that triggers ABA breakdown, but this needs further investigation.
Notably, the effect of using different A. thaliana accessions in the hybrid crosses is larger than the temperature effect (close to 70% difference), and even larger than the interploidy effect. While the Col-0 and Ws-2 A. thaliana accessions resulted in a generally higher germination rate when hybridized with A. lyrata compared to A. arenosa, the C24 accession had the opposite effect. The way the accessions affected hybrid seed viability in opposite directions may point at a similar mechanism as observed in the temperature experiment. However, our results do not readily explain the observed germination rates by cellularization phenotype alone, and further investigations are required to resolve these observations.
Crossing tetraploid A. thaliana Wa-1 to diploid A. arenosa increases hybrid seed survival (Josefsson et al., 2006). In addition, ploidy affects the strength of the hybrid barrier in crosses between A. arenosa and A. lyrata, where higher ploidy in A. lyrata increases the hybrid seed survival rate, while higher ploidy in A. arenosa causes total seed lethality (Lafon-Placette et al., 2017). Our data show that a similar effect is found using the diploid accession C24, suggesting that C24 may have a higher effective ploidy and endosperm balance number [EBN; (Johnston and Hanneman, 1982)] compared to Col-0 and Ws-2. This corresponds well with the hypothesis that A. lyrata has a lower EBN compared to A. arenosa (Lafon-Placette and Köhler, 2016), explaining why crosses with C24 or Wa-1 decrease seed viability in the A. lyrata hybrid. However, it does not explain why A. lyrata crosses with the diploid C24 is more detrimental than crosses with the tetraploid Wa-1, suggesting that accession genotypes, in addition to ploidy, has an effect on the endosperm-based hybridization barrier.
4.3 AGL35 influences endosperm cellularization in hybrid seeds
AGAMOUS-LIKE (AGL) type I MADS-box TFs are highly expressed in the seed, specifically during endosperm cellularization (Bemer et al., 2010; Zhang et al., 2018; Bjerkan et al., 2020). Their importance in the endosperm-based hybridization barrier has been suggested by several studies (Josefsson et al., 2006; Walia et al., 2009; Bjerkan et al., 2020) and it has been hypothesized that timing of endosperm cellularization requires a stoichiometric balance between members of different MADS-box protein complexes (Batista et al., 2019). In this study we demonstrate that mutation in A. thaliana AGL35 has a highly significant and opposite effect on the hybrid barrier phenotype when crossed to A. lyrata and A. arenosa, respectively. AGL35 is bi-allelicly expressed in the chalazal endosperm (Bemer et al., 2010; Bjerkan et al., 2020) and upregulated in crosses between A. thaliana and A. arenosa compared with compatible crosses (Walia et al., 2009). Our results indicate that AGL35 is involved in the transition from syncytial to a cellularized endosperm, and may function as a promoter of cellularization, as mutant crosses to A. arenosa result in lower seed survival, while mutant crosses to A. lyrata result in increased survival compared to Col-0 crosses. Interestingly, a massive-multiplexed yeast two-hybrid study identified interaction between AGL62 and AGL35 (Trigg et al., 2017). These AGL TFs have seemingly antagonistic functions as AGL62 is a suppressor of endosperm cellularization (Kang et al., 2008). Paternal excess interploidy crosses cause increased AGL62 expression, correlated with endosperm cellularization failure (Erilova et al., 2009). AGL62 is also a direct target of the FIS PRC2 complex (Hehenberger et al., 2012) whereas we could see no direct effect on the A. arenosa or A. lyrata hybrid with A. thaliana by mutation of FIS PRC2. The antagonistic effects of single gene mutation of AGL35 is intriguing, and we speculate that expression differences between A. arenosa and A. lyrata in the hybrid endosperm may account for our observations but future investigation of this interaction and the role of AGL35 in regulation of endosperm-based hybridization barriers is required.
4.4 Conclusions
The findings in this study introduce a rigorous model system for the dissection of the influence of abiotic and genetic parameters in hybrid admixture, and have a large potential to support breeding and climate research. Further examination and usage of these approaches could help pinpoint genes, networks or gene dosage balances that are involved in overcoming the endosperm-based hybridization barrier. Species previously thought to be unable to hybridize due to postzygotic seed lethality may be able to do so given favorable conditions, and a similar effect could also apply to interploidy hybrids.
Currently, it is not known if the temperature effect on hybridization success is mediated by the same genetic network that is operated by changes in ploidy or genetic variation. Phenotypically, the temperature effect restores defects in timing of cellularization, but it is not known if the trigger is upstream or downstream of the causative genetic network. Elucidation of the genetic, epigenetic and mechanistic basis for this cross talk between the genic and environmental factors is therefore essential for our understanding of the plasticity of endosperm-based hybridization barriers.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files. Further inquiries can be directed to the corresponding author/s.
Author contributions
PG, RA, KB, and AB designed the research; RA, IM, and KB performed the experiments; PG, RA, KB, and AB analyzed and discussed the data; PG, RA, KB, and AB wrote the article; All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Norwegian Research Council (FRIPRO grants no. 276053 and 262247) to PG and AB. RA and IM were supported by the Norwegian Ministry of Education.
Acknowledgment
We thank the Laboratory of Flow Cytometry, Institute of Botany, Academy of Sciences (Czech Republic) for assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1229060/full#supplementary-material
Supplementary Data Sheet 1 | Flow Cytometry.
Supplementary Data Sheet 2 | Germination assays.
Supplementary Data Sheet 3 | Number of nuclei in crosses
Supplementary Data Sheet 4 | Feulgen phenotype observations
References
Batista, R. A., Kohler, C. (2020). Genomic imprinting in plants-revisiting existing models. Genes Dev. 34, 24–36. doi: 10.1101/gad.332924.119
Batista, R. A., Moreno-Romero, J., Qiu, Y., van Boven, J., Santos-González, J., Figueiredo, D. D., et al. (2019). The MADS-box transcription factor PHERES1 controls imprinting in the endosperm by binding to domesticated transposons. Elife 8, e50541. doi: 10.7554/eLife.50541.sa2
Bemer, M., Heijmans, K., Airoldi, C., Davies, B., Angenent, G. C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154, 287–300. doi: 10.1104/pp.110.160770
Bjerkan, K. N., Hornslien, K. S., Johannessen, I. M., Krabberød, A. K., van Ekelenburg, Y. S., Kalantarian, M., et al. (2020). Genetic variation and temperature affects hybrid barriers during interspecific hybridization. Plant J. 101, 122–140. doi: 10.1111/tpj.14523
Borges, F., Parent, J.-S., van Ex, F., Wolff, P., Martínez, G., Köhler, C., et al. (2018). Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50, 186–192. doi: 10.1038/s41588-017-0032-5
Braselton, J. P., Wilkinson, M. J., Clulow, S. A. (1996). Feulgen staining of intact plant tissues for confocal microscopy. Biotech. Histochem 71, 84–87. doi: 10.3109/10520299609117139
Brink, R. A., Cooper, D. C. (1947). The endosperm in seed development. Bot. Rev. 13, 479–541. doi: 10.1007/BF02861549
Burkart-Waco, D., Josefsson, C., Dilkes, B., Kozloff, N., Torjek, O., Meyer, R., et al. (2012). Hybrid incompatibility in Arabidopsis is determined by a multiple-locus genetic network. Plant Physiol. 158, 801–812. doi: 10.1104/pp.111.188706
Burkart-Waco, D., Ngo, K., Dilkes, B., Josefsson, C., Comai, L. (2013). Early disruption of maternal-zygotic interaction and activation of defense-like responses in Arabidopsis interspecific crosses. Plant Cell 25, 2037–2055. doi: 10.1105/tpc.112.108258
Burkart-Waco, D., Ngo, K., Lieberman, M., Comai, L. (2015). Perturbation of parentally biased gene expression during interspecific hybridization. PloS One 10, e0117293. doi: 10.1371/journal.pone.0117293
Bushell, C., Spielman, M., Scott, R. J. (2003). The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15, 1430–1442. doi: 10.1105/tpc.010496
Chen, C., Begcy, K., Liu, K., Folsom, J. J., Wang, Z., Zhang, C., et al. (2016). Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 171, 606–622. doi: 10.1104/pp.15.01992
Chen, Z. J., Comai, L., Pikaard, C. S. (1998). Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. U. S. A 95, 14891–14896. doi: 10.1073/pnas.95.25.14891
Chen, X., Yoong, F.-Y., O’Neill, C. M., Penfield, S. (2021). Temperature during seed maturation controls seed vigour through ABA breakdown in the endosperm and causes a passive effect on DOG1 mRNA levels during entry into quiescence. New Phytol. 232, 1311–1322. doi: 10.1111/nph.17646
Comai, L., Tyagi, A. P., Winter, K., Holmes-Davis, R., Reynolds, S. H., Stevens, Y., et al. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12, 1551–1568. doi: 10.1105/tpc.12.9.1551
Cooper, D. C., Brink, R. A. (1942). The endosperm as a barrier to interspecific hybridization in flowering plants. Science 95, 75–76. doi: 10.1126/science.95.2455.75
Cornejo, P., Camadro, E. L., Masuelli, R. W. (2012). Molecular bases of the postzygotic barriers in interspecific crosses between the wild potato species Solanum acaule and Solanum commersonii. Genome 55, 605–614. doi: 10.1139/g2012-047
De Bodt, S., Raes, J., Van de Peer, Y., Theißen, G. (2003). And then there were many: MADS goes genomic. Trends Plant Sci. 8, 475–483. doi: 10.1016/j.tplants.2003.09.006
Dilkes, B. P., Spielman, M., Weizbauer, R., Watson, B., Burkart-Waco, D., Scott, R. J., et al. (2008). The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PloS Biol. 6, 2707–2720. doi: 10.1371/journal.pbio.0060308
Dinu, I. I., Hayes, R. J., Kynast, R. G., Phillips, R. L., Thill, C. A. (2005). Novel inter-series hybrids in Solanum, section Petota. Theor. Appl. Genet. 110, 403–415. doi: 10.1007/s00122-004-1782-x
Dziasek, K., Simon, L., Lafon-Placette, C., Laenen, B., Wärdig, C., Santos-González, J., et al. (2021). Hybrid seed incompatibility in Capsella is connected to chromatin condensation defects in the endosperm. PloS Genet. 17, e1009370. doi: 10.1371/journal.pgen.1009370
Erdmann, R. M., Satyaki, P. R. V., Klosinska, M., Gehring, M. (2017). A small RNA pathway mediates allelic dosage in endosperm. Cell Rep. 21, 3364–3372. doi: 10.1016/j.celrep.2017.11.078
Erilova, A., Brownfield, L., Exner, V., Rosa, M., Twell, D., Mittelsten Scheid, O., et al. (2009). Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PloS Genet. 5, e1000663. doi: 10.1371/journal.pgen.1000663
Escobar-Restrepo, J.-M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W.-C., et al. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660. doi: 10.1126/science.1143562
Flores-Vergara, M. A., Oneal, E., Costa, M., Villarino, G., Roberts, C., De Luis Balaguer, M. A., et al. (2020). Developmental analysis of Mimulus seed transcriptomes reveals functional gene expression clusters and four imprinted, endosperm-expressed genes. Front. Plant Sci. 11, 132. doi: 10.3389/fpls.2020.00132
Florez-Rueda, A. M., Paris, M., Schmidt, A., Widmer, A., Grossniklaus, U., Städler, T. (2016). Genomic imprinting in the endosperm is systematically perturbed in abortive hybrid tomato seeds. Mol. Biol. Evol. 33, 2935–2946. doi: 10.1093/molbev/msw175
Folsom, J. J., Begcy, K., Hao, X., Wang, D., Walia, H. (2014). Rice Fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165, 238–248. doi: 10.1104/pp.113.232413
Fox, J., Weisberg, S. (2019). An R companion to applied regression, 3rd ed. (Thousand Oaks, CA: Sage Publication Inc.).
Grini, P. E., Jürgens, G., Hülskamp, M. (2002). Embryo and endosperm development is disrupted in the female gametophytic capulet mutants of Arabidopsis. Genetics 162, 1911–1925. doi: 10.1093/genetics/162.4.1911
Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M. A., Gagliano, W. B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280, 446 LP–446450. doi: 10.1126/science.280.5362.446
Guitton, A.-E., Page, D. R., Chambrier, P., Lionnet, C., Faure, J.-E., Grossniklaus, U., et al. (2004). Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131, 2971–2981. doi: 10.1242/dev.01168
Haig, D., Westoby, M. (1988). On limits to seed production. Am. Nat. 131, 757–759. doi: 10.1086/284817
Haig, D., Westoby, M. (1991). Genomic imprinting in endosperm: Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc Lond. B Biol. Sci. 333, 1–13. doi: 10.1098/rstb.1991.0057
Hehenberger, E., Kradolfer, D., Köhler, C. (2012). Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039. doi: 10.1242/dev.077057
Hornslien, K. S., Miller, J. R., Grini, P. E. (2019). Regulation of parent-of-origin allelic expression in the endosperm. Plant Physiol. 180, 1498–1519. doi: 10.1104/pp.19.00320
Ishikawa, R., Ohnishi, T., Kinoshita, Y., Eiguchi, M., Kurata, N., Kinoshita, T. (2011). Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 65, 798–806. doi: 10.1111/j.1365-313X.2010.04466.x
Johnston, S. A., Hanneman, R. E. (1982). Manipulations of endosperm balance number overcome crossing barriers between diploid Solanum species. Science 217, 446–448. doi: 10.1126/science.217.4558.446
Jørgensen, M. H., Ehrich, D., Schmickl, R., Koch, M. A., Brysting, A. K. (2011). Interspecific and interploidal gene flow in Central European Arabidopsis (Brassicaceae). BMC Evol. Biol. 11, 1–13. doi: 10.1186/1471-2148-11-346
Josefsson, C., Dilkes, B., Comai, L. (2006). Parent-dependent loss of gene silencing during interspecies hybridization. Curr. Biol. 16, 1322–1328. doi: 10.1016/j.cub.2006.05.045
Kang, I.-H., Steffen, J. G., Portereiko, M. F., Lloyd, A., Drews, G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20, 635–647. doi: 10.1105/tpc.107.055137
Kassambara, A. (2023) ggpubr: “ggplot2” based publication ready plots. R package version 0.6.0. Available at: https://rpkgs.datanovia.com/ggpubr/.
Kinser, T. J., Smith, R. D., Lawrence, A. H., Cooley, A. M., Vallejo-Marín, M., Conradi Smith, G. D., et al. (2021). Endosperm-based incompatibilities in hybrid monkeyflowers. Plant Cell 33, 2235–2257. doi: 10.1093/plcell/koab117
Kirkbride, R. C., Lu, J., Zhang, C., Mosher, R. A., Baulcombe, D. C., Chen, Z. J. (2019). Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 116, 2761–2766. doi: 10.1073/pnas.1807621116
Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., et al. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A 96, 4186–4191. doi: 10.1073/pnas.96.7.4186
Koch, M. A. (2019). The plant model system Arabidopsis set in an evolutionary, systematic, and spatio-temporal context. J. Exp. Bot. 70, 55–67. doi: 10.1093/jxb/ery340
Koenig, D., Weigel, D. (2015). Beyond the thale: comparative genomics and genetics of Arabidopsis relatives. Nat. Rev. Genet. 16, 285–298. doi: 10.1038/nrg3883
Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., Gruissem, W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814. doi: 10.1093/emboj/cdg444
Kradolfer, D., Wolff, P., Jiang, H., Siretskiy, A., Köhler, C. (2013). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26, 525–535. doi: 10.1016/j.devcel.2013.08.006
Lafon-Placette, C., Johannessen, I. M., Hornslien, K. S., Ali, M. F., Bjerkan, K. N., Bramsiepe, J., et al. (2017). Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrata and Arabidopsis arenosa in Central Europe. Proc. Natl. Acad. Sci. U. S. A 114, E1027–E1035. doi: 10.1073/pnas.1615123114
Lafon-Placette, C., Köhler, C. (2016). Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol. Ecol. 25, 2620–2629. doi: 10.1111/mec.13552
Law, JA, Du, J, Hale, CJ, Feng, S, Krajewski, K, Palanca, AMS, et al (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389.
Levene, H. (1960). Robust tests for equality of variances. In I. Olkin (Ed.), Contributions to probability and statistics (Palo Alto, CA: Stanford University Press).
Lindsey, B. E., III, AU-Rivero, L., AU-Calhoun, C. S., AU-Grotewold, E., AU-Brkljacic, J. (2017). Standardized method for high-throughput sterilization of Arabidopsis seeds. J. Vis. Exp., 2017 (128), e56587. doi: 10.3791/56587
Lloyd, A., Morgan, C., H Franklin, F. C., Bomblies, K. (2018). Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208, 1409–1420. doi: 10.1534/genetics.117.300588
Luo, M., Bilodeau, P., Koltunow, A., Dennis, E. S., Peacock, W. J., Chaudhury, A. M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A 96, 296–301. doi: 10.1073/pnas.96.1.296
Mann, H. B., Whitney, D. R. (1947). On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60. doi: 10.1214/aoms/1177730491
Martienssen, R. A. (2010). Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol. 186, 46–53. doi: 10.1111/j.1469-8137.2010.03193.x
Martinez, G., Wolff, P., Wang, Z., Moreno-Romero, J., Santos-González, J., Conze, L. L., et al. (2018). Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet. 50, 193–198. doi: 10.1038/s41588-017-0033-4
Molnár-Láng, M., Sutka, J. (1994). The effect of temperature on seed set and embryo development in reciprocal crosses of wheat and barley. Euphytica 78, 53–58. doi: 10.1007/BF00021397
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nasrallah, M. E., Yogeeswaran, K., Snyder, S., Nasrallah, J. B. (2000). Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol. 124, 1605–1614. doi: 10.1104/pp.124.4.1605
Ng, D. W., Lu, J., Chen, Z. J. (2012). Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr. Opin. Plant Biol. 15, 154–161. doi: 10.1016/j.pbi.2012.01.007
Oneal, E., Willis, J. H., Franks, R. G. (2016). Disruption of endosperm development is a major cause of hybrid seed inviability between Mimulus guttatus and Mimulus nudatus. New Phytol. 210, 1107–1120. doi: 10.1111/nph.13842
Parřenicová, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Paul, P., Dhatt, B. K., Sandhu, J., Hussain, W., Irvin, L., Morota, G., et al. (2020). Divergent phenotypic response of rice accessions to transient heat stress during early seed development. Plant Direct 4, e00196. doi: 10.1002/pld3.196
R Core Team (2023). R: A language and environment for statistical computing (R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Rebernig, C. A., Lafon-Placette, C., Hatorangan, M. R., Slotte, T., Köhler, C. (2015). Non-reciprocal interspecies hybridization barriers in the Capsella genus are established in the endosperm. PloS Genet. 11, e1005295. doi: 10.1371/journal.pgen.1005295
Rieseberg, L. H., Willis, J. H. (2007). Plant speciation. Science 317, 910–914. doi: 10.1126/science.1137729
Roth, M., Florez-Rueda, A. M., Städler, T. (2019). Differences in effective ploidy drive genome-wide endosperm expression polarization and seed failure in wild tomato hybrids. Genetics 212, 141–152. doi: 10.1534/genetics.119.302056
Roy, A. K., Malaviya, D. R., Kaushal, P. (2011). Generation of interspecific hybrids of Trifolium using embryo rescue. Methods Mol. Biol. 710:141–151. doi: 10.1007/978-1-61737-988-8_12
Schatlowski, N., Kohler, C. (2012). Tearing down barriers: understanding the molecular mechanisms of interploidy hybridizations. J. Exp. Bot. 63, 6059–6067. doi: 10.1093/jxb/ers288
Schatlowski, N., Wolff, P., Santos-Gonzalez, J., Schoft, V., Siretskiy, A., Scott, R., et al. (2014). Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell 26, 3556–3568. doi: 10.1105/tpc.114.130120
Scott, R. J., Spielman, M., Bailey, J., Dickinson, H. G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341. doi: 10.1242/dev.125.17.3329
Sekine, D., Ohnishi, T., Furuumi, H., Ono, A., Yamada, T., Kurata, N., et al. (2013). Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J. 76, 792–799. doi: 10.1111/tpj.12333
Sharma, H. C. (1999). Embryo rescue following wide crosses. Methods Mol. Biol. 111, 293–307. doi: 10.1385/1-59259-583-9:293
Shirzadi, R., Andersen, E. D., Bjerkan, K. N., Gloeckle, B. M., Heese, M., Ungru, A., et al. (2011). Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PloS Genet. 7, e1001303. doi: 10.1371/journal.pgen.1001303
Sukno, S., Ruso, J., Jan, C. C., Melero-Vara, J. M., Fernandez-Martinez, J. M. (1999). Interspecific hybridization between sunflower and wild perennial Helianthus species via embryo rescue. Euphytica 106, 69–78. doi: 10.1023/A:1003524822284
Tonosaki, K., Sekine, D., Ohnishi, T., Ono, A., Furuumi, H., Kurata, N., et al. (2018). Overcoming the species hybridization barrier by ploidy manipulation in the genus Oryza. Plant J. 93, 534–544. doi: 10.1111/tpj.13803
Trigg, S. A., Garza, R. M., MacWilliams, A., Nery, J. R., Bartlett, A., Castanon, R., et al. (2017). CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 14, 819–825. doi: 10.1038/nmeth.4343
Ungru, A., Nowack, M. K., Reymond, M., Shirzadi, R., Kumar, M., Biewers, S., et al. (2008). Natural variation in the degree of autonomous endosperm formation reveals independence and constraints of embryo growth during seed development in Arabidopsis thaliana. Genetics 179, 829–841. doi: 10.1534/genetics.107.084889
van Ekelenburg, Y. S., Hornslien, K. S., Van Hautegem, T., Fendrych, M., Van Isterdael, G., Bjerkan, K. N., et al. (2023). Spatial and temporal regulation of parent-of-origin allelic expression in the endosperm. Plant Physiol. 191, 986–1001. doi: 10.1093/plphys/kiac520
Walia, H., Josefsson, C., Dilkes, B., Kirkbride, R., Harada, J., Comai, L. (2009). Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19, 1128–1132. doi: 10.1016/j.cub.2009.05.068
Wang, L., Yuan, J., Ma, Y., Jiao, W., Ye, W., Yang, D.-L., et al. (2018). Rice interploidy crosses disrupt epigenetic regulation, gene expression, and seed development. Mol. Plant 11, 300–314. doi: 10.1016/j.molp.2017.12.006
Welch, B. L. (1947). The generalisation of student’s problems when several different population variances are involved. Biometrika 34, 28–35. doi: 10.1093/biomet/34.1-2.28
Wickham, H., François, R., Henry, L., Müller, K., Vaughan, D. (2023) dplyr: a grammar of data manipulation. Available at: https://dplyr.tidyverse.orghttps://github.com/tidyverse/dplyr.
Widmer, A., Lexer, C., Cozzolino, S. (2009). Evolution of reproductive isolation in plants. Heredity 102, 31–38. doi: 10.1038/hdy.2008.69
Wolff, P., Jiang, H., Wang, G., Santos-Gonzalez, J., Köhler, C. (2015). Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. Elife 4, e10074. doi: 10.7554/eLife.10074.020
Xu, W., Sato, H., Bente, H., Santos-González, J., Köhler, C. (2023). Endosperm cellularization failure induces a dehydration-stress response leading to embryo arrest. Plant Cell 35, 874–888. doi: 10.1093/plcell/koac337
Zhang, H.-Y., Luo, M., Johnson, S. D., Zhu, X.-W., Liu, L., Huang, F., et al. (2016). Parental genome imbalance causes post-zygotic seed lethality and deregulates imprinting in rice. Rice 9, 1–12. doi: 10.1186/s12284-016-0115-4
Keywords: Arabidopsis thaliana, A. arenosa, A. lyrata, hybrid barrier, temperature, endosperm, seed development
Citation: Bjerkan KN, Alling RM, Myking IV, Brysting AK and Grini PE (2023) Genetic and environmental manipulation of Arabidopsis hybridization barriers uncovers antagonistic functions in endosperm cellularization. Front. Plant Sci. 14:1229060. doi: 10.3389/fpls.2023.1229060
Received: 26 May 2023; Accepted: 12 July 2023;
Published: 02 August 2023.
Edited by:
Dongfang Wang, Spelman College, United StatesReviewed by:
Jugou Liao, Yunnan University, ChinaFilipe Borges, INRA Centre Versailles-Grignon, France
Copyright © 2023 Bjerkan, Alling, Myking, Brysting and Grini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul E. Grini, cGF1bC5ncmluaUBpYnYudWlvLm5v
†These authors have contributed equally to this work
 Katrine N. Bjerkan1,2†
Katrine N. Bjerkan1,2† Renate M. Alling
Renate M. Alling Anne K. Brysting
Anne K. Brysting Paul E. Grini
Paul E. Grini