
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 August 2023
Sec. Plant Systematics and Evolution
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1228551
This article is part of the Research Topic Advances in the Study of Polyploid Evolution in Wild Populations View all 9 articles
Chrysanthemum indicum, a species native to Eastern Asia is well known as one of the progenitor species of the cultivated Chrysanthemum which is grown for its ornamental and medicinal value. Previous genomic studies on Chrysanthemum have largely ignored the dynamics of plastid genome (plastome) and mitochondria genome (mitogenome) evolution when analyzing this plant lineage. In this study, we sequenced and assembled the plastomes and mitogenomes of diploid and tetraploid C. indicum as well as the morphologically divergent variety C. indicum var. aromaticum. We used published data from 27 species with both plastome and mitogenome complete sequences to explore differences in sequence evolution between the organellar genomes. The size and structure of organellar genome between diploid and tetraploid C. indicum were generally similar but the tetraploid C. indicum and C. indicum var. aromaticum were found to contain unique sequences in the mitogenomes which also contained previously undescribed open reading frames (ORFs). Across Chrysanthemum mitogenome structure varied greatly but sequences transferred from plastomes in to the mitogenomes were conserved. Finally, differences observed between mitogenome and plastome gene trees may be the result of the difference in the rate of sequence evolution between genes in these two genomes. In total the findings presented here greatly expand the resources for studying Chrysanthemum organellar genome evolution with possible applications to conservation, breeding, and gene banking in the future.
Chrysanthemum indicum, a perennial herbaceous plant in the Asteraceae family, is widely used as a medicinal, ornamental, and food plant (Luo and Li, 2013). Previous studies have found that C. indicum possesses variable traits across the species range and also varies in ploidy level (Li et al., 2013; Klie et al., 2014; Qi et al., 2021), including diploid, tetraploid, and hexaploid (Si-Lan. et al., 1998; C. et al., 2004; El-Twab and Kondo, 2007; Li et al., 2013; Klie et al., 2014). For example C. indicum from coastal saline soils were noted to be dwarfed, while those found on Mount Lu (Jiangxi Province, China) were found to have a greater density of trichomes on their leaves than plants from the other regions (Luo and Li, 2013). Frequent gene flow has been reported between species in the Chrysanthemum genus (Si-Lan. et al., 1998; C. and S, 2002; C. et al., 2004; El-Twab and Kondo, 2007; Luo et al., 2016), as well as among different accessions within C. indicum (Li et al., 2013; Luo et al., 2016; Ma et al., 2020; Qi et al., 2021). In previous phylogenetic studies about Chrysanthemum genus, C. indicum were frequently found to be most closely related with C. zawadskii and C. nankingenese (C. et al., 2004). However, substantial gene flow has led to conflicting phylogenetic outcomes within the genus and among wild populations of C. indicum (C. et al., 2004; El-Twab and Kondo, 2007). Previous studies have suggested that tetraploid C. indicum originated from hybridization between diploid C. indicum and related species (Li et al., 2013), while other studies have found that some C. indicum polyploid populations were homoeologous polyploids (Li et al., 2014). Within C. indicum the variety C. indicum var. aromaticum was described to account for individuals that possess thicker leaf blades and glandular hairs (Luo and Li, 2013), but studies on this variety have also found that gene flow has occurred between it and other C. indicum lineages (Luo and Li, 2013; Yuan et al., 2022). Much of the research regarding C. indicum has been in its contribution to the Chinese cultivated Chrysanthemum which is an allohexaploid lineage used as a horticultural ornament (Si-Lan. et al., 1998; C. and S, 2002; C. et al., 2004; El-Twab and Kondo, 2007). The above-mentioned evidence for genetic admixture in Chrysanthemum has been derived from the nuclear loci with little known about how mitogenomes and plastomes have been sorted and evolved during episodes of admixture and increases in ploidy.
Sequence data from plant mitogenomes and plastomes has been widely used for plant phylogenetic research due to the presence of conserved nonrecombinant sequences that flank more rapidly evolving regions (Kelchner, 2002; Rogalski et al., 2015; Uncu et al., 2015). For example, the phylogeny of the three large families: Arecaceae, Moraceae, and Saxifragaceae, have been resolved with plastome data (Liu et al., 2020; Zhang et al., 2022; Yao et al., 2023). With the decreasing cost of high-throughput sequencing and the improvement of assembly techniques, an increasing number of studies are using complete organellar genomes instead of one or several loci to conduct phylogenetic and evolutionary studies in plants. Currently, 1852 plastomes (mostly from different varieties of the same species) and 62 mitogenomes of Asteraceae are available in GenBank. The smaller number of published mitogenomes is a result of the difficulty in assembling plant mitochondrial genomes (high frequency of rearrangements) using short read sequencing techniques (Uncu et al., 2015). For example, it is difficult to resolve complex mitogenome assembly results with different contig-repeat-contig combinations by using only short read sequencing data (Zhang et al., 2023). However, the issue of complete mitogenome assembly is gradually being addressed with the use of long-read sequencing methods. Although many organellar genomes have been published from the Asteraceae, most studies have been limited to phylogenetic research, without further exploring the evolutionary patterns within a species or how organellar genome evolution correlates with changes in nuclear genome ploidy (Xia et al., 2016; Kim et al., 2018; Won et al., 2018; Feng et al., 2020; Xia et al., 2021a; Xia et al., 2021b; Yu et al., 2021; Masuda et al., 2022).
In this study, we sequenced and assembled the complete mitogenomes and plastomes of diploid and tetraploid C. indicum, as well as C. indicum var. aromaticum, and compared them with those of 23 Asteraceae species for which public data was available for both organellar genomes. This is the first study to explore the evolutionary histories of varieties of C. indicum with different ploidy levels and phenotypic variations using organellar genomes as a basis for historical inference. We compared the differences of the sequence and structure of organellar genomes among different varieties of C. indicum, resolved phylogenetic relationships among different varieties of C. indicum, analyzed the diversity of simple sequence repeats (SSRs), and quantified the gene transfers from plastomes to mitogenomes.
By utilizing long PacBio HiFi reads, the mitogenomes of diploid (2x) and tetraploid (4x) C. indicum were assembled with published C. indicum mitogenome (NCBI accession number: MH716014.1) used to extract sequences from whole genome data. In addition the mitogenome of C. indicum var. aromaticum was also assembled with Illumina reads. The mitogenome of C. indicum (2x) was assembled into a single circular genome, and was used to resolve the mitogenome of C. indicum (4x) and C. indicum var. aromaticum with multiple linkage structures (Figure S1). The sizes of the three circular mitogenome were 192,408 bp with 45.51% GC content for C. indicum (2x), 193,563 bp with 45.43% GC content for C. indicum (4x), and 198,095 bp with 45.33% GC content for C. indicum var. aromaticum, similar with that in found in most Asteraceae species sequenced at present (Figure 1). All three of these newly assembled mitogenomes contain 51 unique genes, including 33 protein coding genes (PCGs) genes, three rRNA genes, and 16 tRNA, with trnS-GCU having two copies and trnM-CAU having three copies in C. indicum (2x) and C. indicum (4x), but five copies in C. indicum var. aromaticum (Table 1). Among the PCGs, eight genes contained introns, three of which (ccmFC, nad4 and rps3) contained one intron, and five others (cox2, nad1, nad2, nad5, and nad7) contained two or more introns. Among the three mitogenomes, all PCGs are single copy, except atp9 that has two copies in C. indicum var. aromaticum.
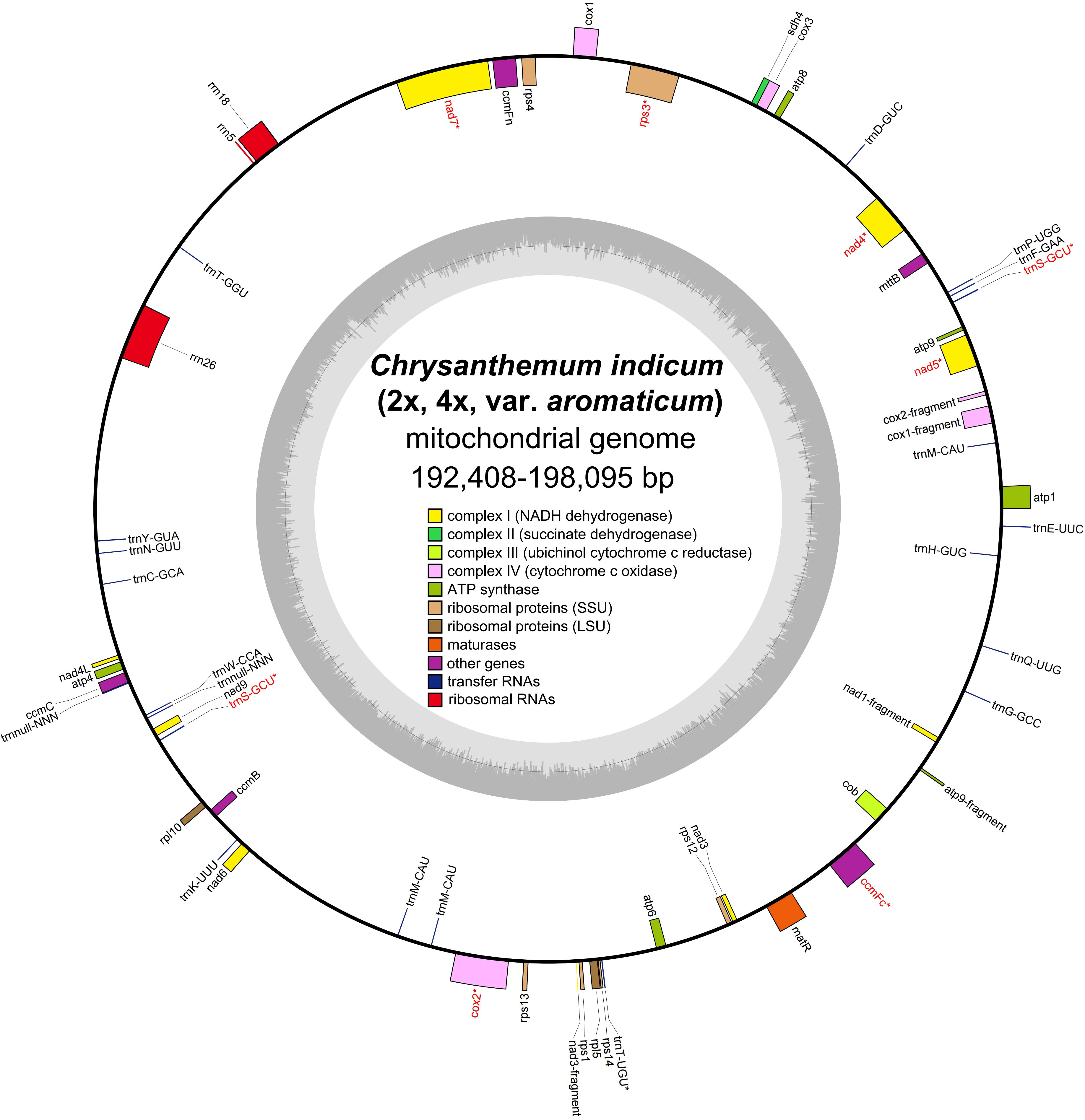
Figure 1 Mitogenome structure from three C. indicum accessions in this study. * indicates gene containing intron (s).
All three newly assembled chloroplast genomes maintain the classical dumbbell-shaped genome structure. The plastome sizes of the three samples were very similar at 151,033 bp for C. indicum (2x), 151,036 bp for C. indicum (4x) and 151,053 bp for C. indicum var. aromaticum with the same GC content at 37.47% for all these plastomes (Figure S2). All three of these newly assembled plastomes of Chrysanthemum contain 109 unique genes, including 80 PCGs, 25 tRNAs, and four rRNAs with the IR region containing six protein coding genes, eight tRNAs and four rRNAs, with 13 PCGs containing introns, and two of these (rps12 and ycf3) have more than one intron. Overall, compared to mitogenomes, these three plastomes differ only slightly in a few regions and are generally conserved in sequence similarity.
To better understand organellar genome evolution in Chrysanthemum, the three mitogenomes from Chrysanthemum indicum assembled in this study were used for genome-wide collinear comparisons with published mitogenomes of four Chrysanthemum species C. indicum (MH716014), C. boreale (NC_039757), C. makinoi (OU343227), and C. zawadskii (ON053202) (Figure 2). The four accessions from C. indicum have a more similar mitogenome structure in the genome-wide alignment results for sequence lengths greater than 1 Kb, except in C. indicum (MH716014) which did not have the 22.6 Kb and 23.7 Kb inversions found in the other three species (Figure 2). Between the four Chrysanthemum species mitogenomes highly divergent rearrangements were found pointing to a process of rapid structural turnover between species (Figure 2). Among the three newly assembled mitogenomes, C. indicum (4x) has a unique sequence of 2.8 Kb compared to C. indicum (2x), on which two ORFs encoding proteins of 496aa and 72aa in length were predicted. In C. indicum var. aromaticum two unique sequences of length 3.3 Kb and 5.6 Kb were found with ORFs encoding proteins of 307aa, 165aa, and 73aa in length predicted on the 3.3 Kb sequence. On the 5.6 Kb sequence ORFs encoding proteins 383 aa, 346 aa, 94 aa, 72 aa, and 71 aa in length, and two trnM-CAU were predicted (Table S2). Comparative sequence analysis of the plastomes corresponding to seven Chrysanthemum accessions showed that the plastome sequences were conserved within the genus (Figure S3). In addition, to understand the structural differences of mitogenomes within cultivated C. indicum, the three newly assembled mitogenomes were compared separately for intra-genomic collinearity, and it was found that these mitogenomes all contain few repetitive sequences, which explains the relatively small size of these mitogenomes. The C. indicum (2x) and C. indicum (4x) mitogenomes were more similar in intra-genomic collinearity patterns and showed similarity between their genome structure (Figure S4). In general, the mitogenomes showed large structural differences between species within Chrysanthemum and greater synteny within species while the plastomes were structurally conserved across all samples.
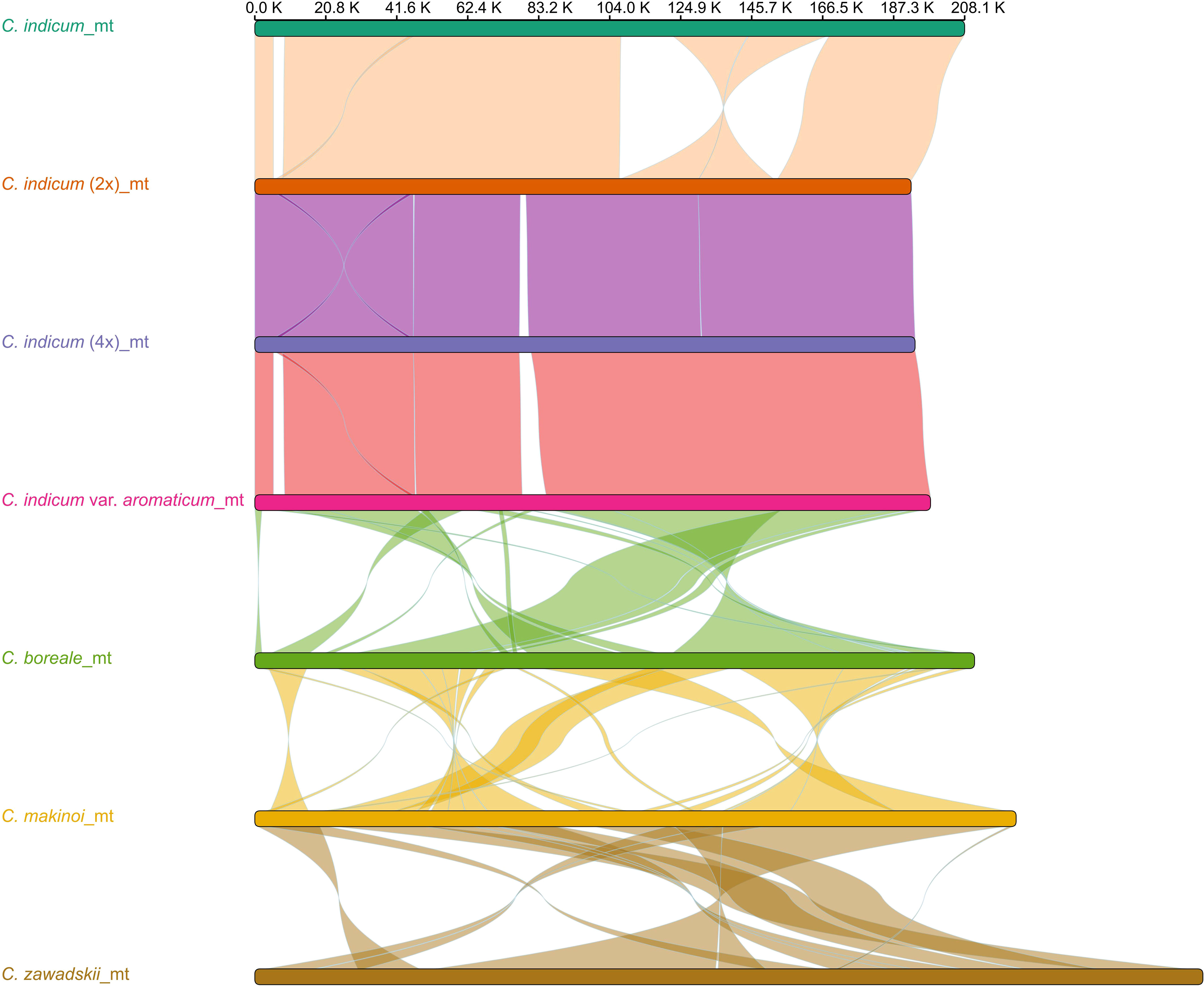
Figure 2 Synteny between Chrysanthemum mitogenomes. Mitogenomes from seven accessions with sequence rearrangements longer than 1 Kb between two pairs shown.
To further understand the differences between organellar genomes of Chrysanthemum repeat sequences were annotated and compared among seven Chrysanthemum accessions (Figure 3). The Chrysanthemum organellar genomes contained repeat types Complement (C), Forward (F), Palindromic (P), and Reverse (R) at different ratios, with repeats less than 50 bp being the most common (Figures 3A, B). In all accessions, the number of repeats of the F and P type were dominant in both genomes but in plastomes the repeat types were more evenly distributed especially among the shorter repeat sequences (Figures 3A, B). The abundance of F and P type repeats in the mitogenome is correlated with larger mitogenomes. To better understand the dynamics of repetitive sequences in the organelles of the Chrysanthemum plastome and mitogenome, SSR sequences of seven accessions were analyzed (Figures 3C, D). A total of 23, 24, 25, 23, 25, 39, and 29 SSRs were detected in the mitogenomes of C. indicum (2x), C. indicum (4x), C. indicum var. aromaticum, C. indicum, C. boreale, C. makinoi, and C. zawadskii (Figure 3C), respectively, and a total of 47, 48, 47, 42, 43, 40, and 45 SSRs were detected in the plastomes of C. indicum (2x), C. indicum (4x), C. indicum var. aromaticum, C. indicum, C. boreale, C. makinoi, and C. zawadskii (Figure 3D), respectively. The number of SSRs in plastomes was greater than that in the corresponding mitogenomes (Figures 3C, D). The number of A/T SSRs in the plastomes and mitogenomes far exceeded the combined number of all other types of SSRs while the mitogenome of C. makinoi had a much higher number of A/T repeats than the other accessions (Figures 3C, D). The mitogenomes of all four accessions from C. indicum did not contain AG/CT SSRs, but C. indicum (4x) possessed a unique ACAGAT/ATCTGT SSR, and C. boreale possessed a unique AAGGCT/AGCCTT SSR (Figure 3C). In the plastomes the number and type of SSRs are more conserved, except for C. boreale and C. zawadskii that did not contain any C/G SSRs, all other accessions contained four C/G SSR loci (Figure 3D).
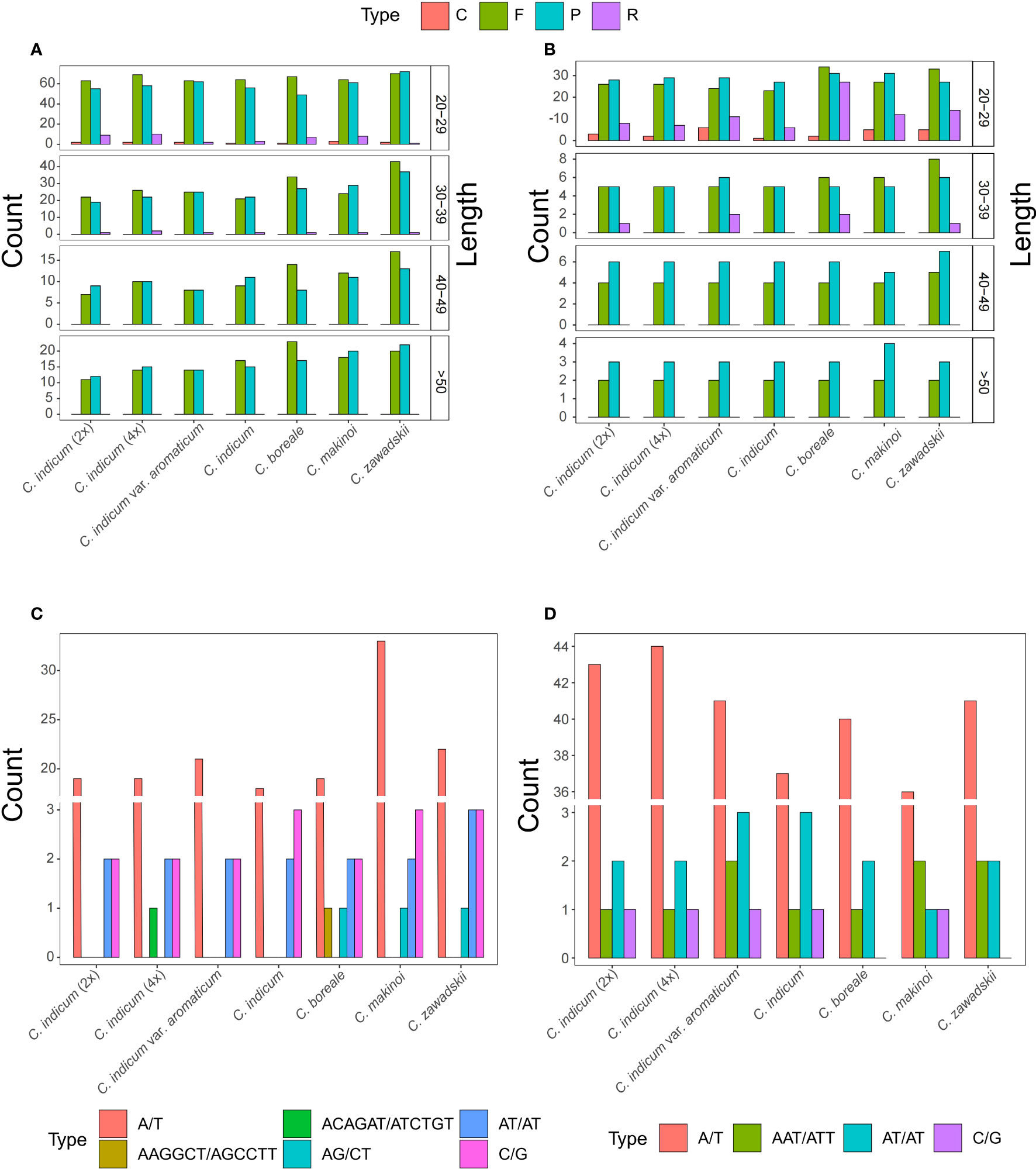
Figure 3 Repeat sequences in Chrysanthemum organellar genomes. (A) Mitogenome repeats identified with REPuter include F (forward), R (reversed), C (complement) and P (palindromic); (B) Plastome repeats identified with REPuter include F, P, C, and R repeats; (C) The SSRs identified from Chrysanthemum mitogenomes; (D) The SSRs identified from Chrysanthemum plastomes.
In plants, the exchange of genetic material between cellular compartments (nucleus, mitochondria, and chloroplasts) is often referred to as Intracellular Gene Transfer (IGT) (Bergthorsson et al., 2003; Aubin et al., 2021). Characterizing these IGT events is key to understanding the genetics and evolution of organellar genomes. We did this in Chrysanthemum, by quantifying the sequence transfers from plastomes to mitogenomes (Figure 4). Each accession had a total of 8-10 (from 5220-9176 bp in total length) fragments from a plastomic origin integrated into 8-12 (5212-9164 bp) locations in the mitogenome. The least number (8) of transferred sequences was in C. indicum (2x) and the most (10) in C. makinoi. The length of the transferred fragments ranged from the shortest at 79 bp to the longest of 2556 bp (Table 2; Table S1). Correlation analyses indicated that the number of plastome transferred sequences and the size of the corresponding mitogenomes were positively correlated, as the number and length of plastome transfer sequences was greater for accessions with large mitogenomes (Figure S5). The transferred sequences across the seven accessions were conserved in respect to both size and sequence of origin. Specifically all accessions contained seven completely transferred gene sequences (petL, petG, trnM-CAU, trnH-GUG, trnN-GUU, trnW-CCA, and trnP-UGG) and five partially transferred gene sequences (16S rRNA, rps11, ycf2, psaB, and psaA) (Table 2). Interestingly, C. indicum var. aromaticum, C. boreale, and C. makinoi have partially transferred sequences of the 23S rRNA gene, but the lengths of these fragments vary from 135 bp in C. indicum var. aromaticum to 357 bp and 443 bp in C. makinoi, and 238 bp in C. boreale (Table S1). These results suggest that the plastomic fragments were transferred to the mitogenome predating speciation, and that shared transfers specifically between C. indicum var. aromaticum, and other Chrysanthemum species may be the result of past hybridization, incomplete lineage sorting, or retention of ancestral transfers lost in other C. indicum species.
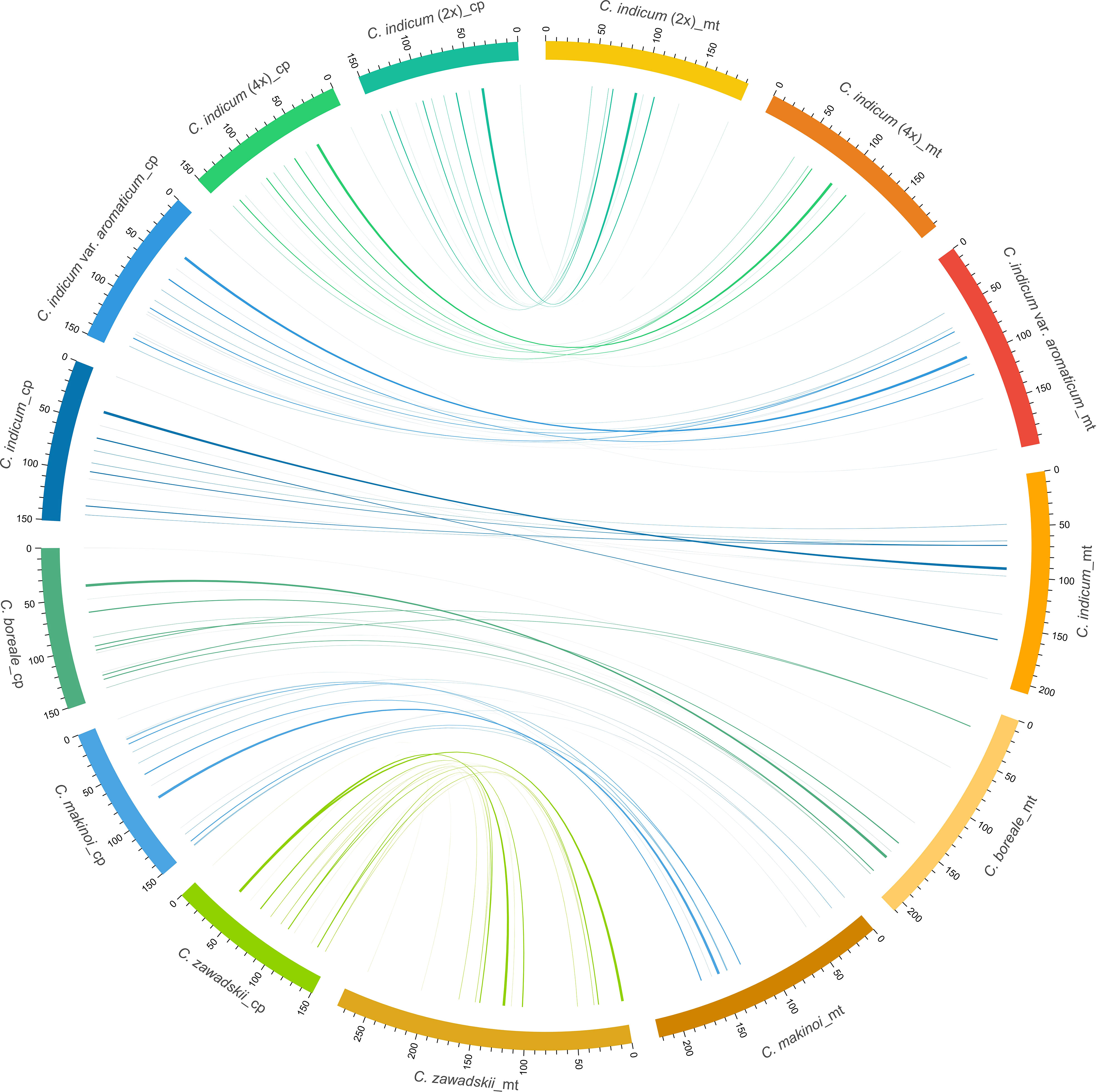
Figure 4 Schematic diagram of homologous sequences identified among the two organellar genomes of seven C. indicum accessions. Plastomes are on the left and corresponding mitogenomes are on the right. Lengths are given in Kb.
A maximum likelihood (ML) phylogenetic tree of 26 Asteraceae species (Platycodon grandifiorus was set as outgroup) with complete mitogenome and plastome data was constructed based on 80 plastome genes and 32 mitogenome genes, respectively (Figure 5). In our study, the phylogenetic topology at tribe level was generally consistent with previous research (Zhang et al., 2021). However, the internal phylogenetic relationships within the Anthemideae were somewhat different. On the mitogenome tree, C. boreale grouped with Artemisia giraldii, while on plastome tree, it grouped with Chrysanthemum. This may be due to the fact that the four mitogenome genes (atp9, mttb, nad2 and nad6) in C. boreale show greater similarity with that in A. giraldii, especially the atp9 gene which was identical between the two species. In a previous study, it was suggested that C. boreale may be an early diverging species within Chrysanthemum, and thus may have retained more ancestral characters (El-Twab and Kondo, 2007). From the phylogenetic tree all four genomes of C. indicum clustered together in both plastome and mitogenome analyses. The phylogenetic analysis also suggested that the maternal donor of the tetraploid C. indicum may be the diploid C. indicum, while C. indicum var. aromaticum appears to have undergone some degree of divergence from the other C. indicum.
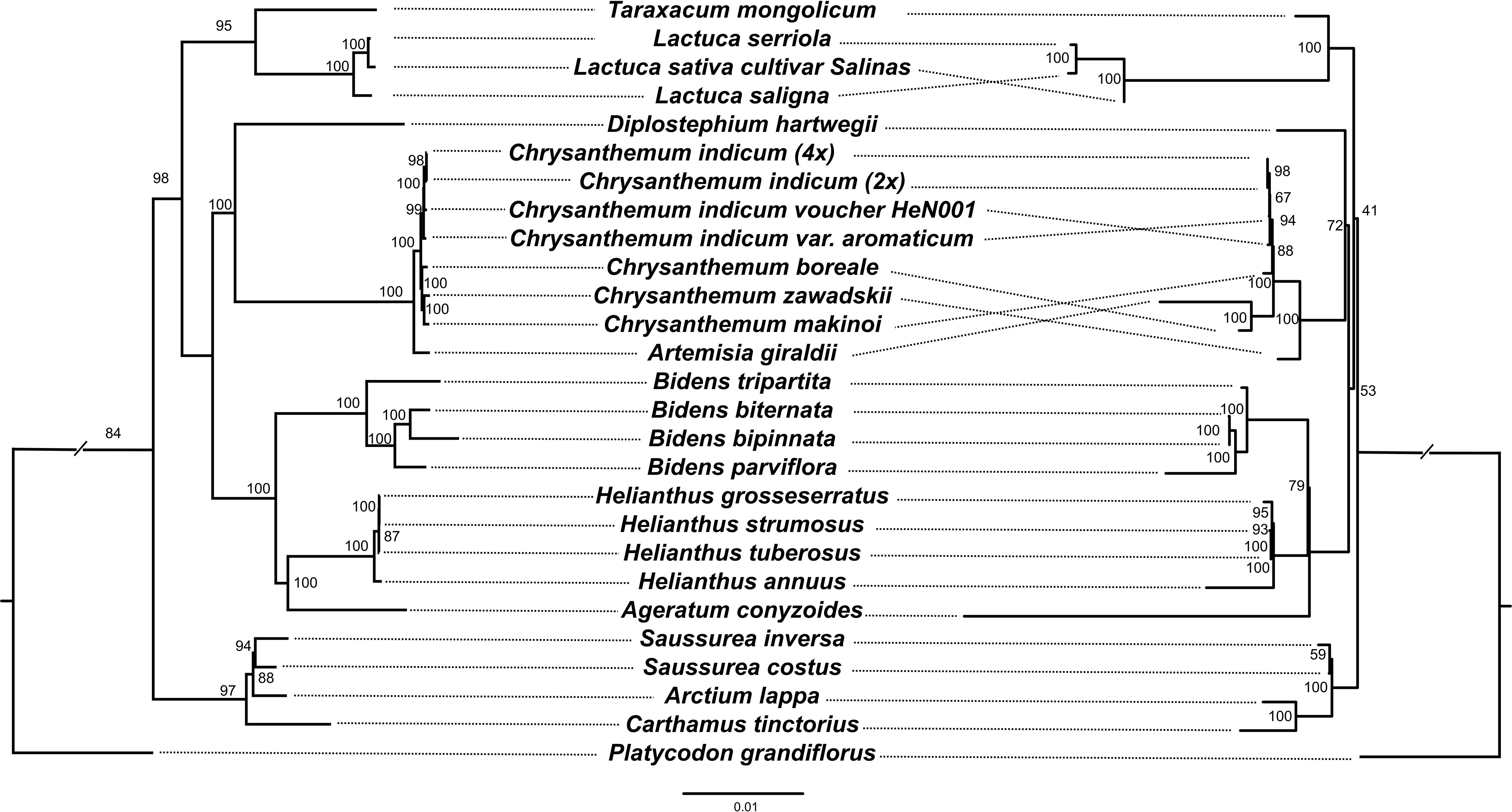
Figure 5 Comparison of ML trees between different gene sets. Left. ML tree based on 80 plastid genes. Right. ML tree based on 32 mitochondrial genes. Numbers at nodes are bootstrap values.
With the set model parameters, we obtained two time tree result based on plastome estimation and mitogenome estimation (Figures S6, S7). From the results, the 95% intervals obtained for plastomes are more convergent, while those for mitogenomes are broader, which may be related to the mutation rate and the number of mutation sites in mitochondria (Figure S6). Here we take the plastome results as the basis. From the results, we learnt that the C. indicum was born at 1.43 Mya, and C. indicum var. aromaticum and C. indicum were separated at 0.81 Mya; while tetraploid C. indicum and diploid C. indicum were separated at 0.38 Mya (Figure S7).
Plant mitogenomes have undergone tremendous structural changes during evolution, which have resulted in a near 200-fold variation in mitogenome size among plants (from ~66 Kb of Viscum scurruloideum to ~12 Mb of Larix sibirica) and frequent rearrangements with low gene density, in contrast to plant plastomes which are conserved in genome size from 115-165 Kb and containing a dumbbell-shaped genome structure (Putintseva et al., 2020; Wu et al., 2020). Owing to the release of a large amount of plant whole genome sequencing data, an increasing number of mitogenomes have been assembled and subjected to comparative studies, in which many mitogenomes have been found to exhibit a reticulated complex genomic structure. The diversity of many mitochondrial genomic structures is thought to be the result of recombination mediated by repetitive sequences (Wu and Sloan, 2019). However, the three mitogenomes of C. indicum newly assembled in this study show a simple monocyclic structure, along with a low number and percentage (0.9%-2.2%) of repetitive sequences, which may account for the structure and relatively small size at just over 40 Kb longer than their respective plastomes. In a comparison among four accessions of C. indicum, it was found that the mitogenome structure and sequence within species is relatively conserved, in contrast to large differences observed among mitogenomes within cultivated sorghum (Zhang et al., 2023). Meanwhile, only a 2.8 Kb segment of the mitogenome differed between accessions of C. indicum of different ploidy levels, two ORFs encoding proteins 496aa and 72aa in length were predicted on this segment, and the functions of these two ORFs have yet to be investigated by mitochondrial gene editing techniques (Kazama et al., 2019). However, when comparing among four different species in Chrysanthemum, it was found that the mitogenomes differed not only in size, but also in structure, mainly in genome sequence order. In conclusion, the relationship between mitogenome sequence and structural variation within Chrysanthemum and what effect this has on molecular function needs to be further investigated from a greater number of samples. What factors are driving structural conservation within C. indicum and divergence between species should also be investigated in greater depth.
IGT occurs frequently and continuously during nucleoplasmic coevolution between different genomes in plant cells (Zhang et al., 2020). In this study, we assembled the mitogenomes and plastomes of three C. indicum accessions and examined the sequence transfers from plastome to mitogenome in combination with four published organellar genomes of species in Chrysanthemum. It was found that the number and the size of these transferred sequences within Chrysanthemum showed a positive correlation with the size of the mitogenome. The transferred genes were conserved among the accessions, and the size of the transferred fragments containing these genes was also generally conserved. All samples contained seven completely transferred gene sequences from the plastome (petL, petG, trnM-CAU, trnH-GUG, trnN-GUU, trnW-CCA, and trnP-UGG) and five partially transferred genes (16S rRNA, rps11, ycf2, psaB, and psaA). These results suggest that these sequence transfers occurred prior to the divergence of Chrysanthemum species and have been retained through time albeit in different locations of the genome given rapid turnover in structure from recombination. Interestingly, all four accessions of C. indicum have the same transferred gene, except for C. indicum var. aromaticum that has an additional 135 bp transferred sequence containing a partially transferred sequence of the 23S rRNA gene. This transferred fragment shared between C. indicum var. aromaticum and the other species in Chrysanthemum may suggest that this lineage is early diverging or that introgression or incomplete lineage sorting may have been involved in the evolution of this variety.
We observed different topologies between phylogenetic analysis based on plastome and mitogenome sequences. Overall, the results obtained from chloroplasts are consistent with previous studies based on nuclear genes, while mitochondria tend to exhibit conflicting resolutions compared with nuclear gene results in adjacent clades (Zhang et al., 2021). This may be due to the slow mutation rate in sequences that have been observed in plant mitogenomes (Duff and Nickrent, 1999; Xi et al., 2013; Grewe et al., 2014). Additionally, the high frequency of RNA editing sites in mitochondrial genes imposes difficulties for mitochondrial gene tree construction (Drouin et al., 2008; Knoop, 2011). For example, in angiosperms, the number of editing sites in the mitogenome often ranges from 200 to 500, while in the plastome, there are only 30 to 50 editing sites (Malek et al., 1996; Knoop, 2011; Zhang et al., 2021). Furthermore, the editing sites in the mitochondrial genome are not only evident in the coding region, but also in tRNA, introns, and 5’ and 3’ untranslated regions (Malek et al., 1996; Oldenkott et al., 2014). Our study supports the view that mitochondrial genes may not be appropriate for plant phylogenetic inferences at some levels of taxonomic organization (Malek et al., 1996; Drouin et al., 2008; Zhang et al., 2021).
The current research used highly accurate long-read HiFi sequencing data to assemble mitogenomes from diploid and tetraploid C. indicum and short read sequencing data for C. indicum var. aromaticum using assembly software GSAT. We compared the differences among mitogenomes from different species within Chrysanthemum and different accessions of C. indicum using the newly assembled and previously published organellar genomes, and found that between species large structural differences are present, but within C. indicum species the structure is more conserved between accessions of different ploids. Meanwhile the plastome is structurally conserved across Asteraceae. Transferred genes from plastome to mitogenome showed high levels of sequence conservation within the accessions of different ploids from C. indicum species. In addition, we resolved phylogenetic relationships using plastome and mitogenome CDSs of 27 Asteraceae accessions, with Platycodon grandiflorus as an outgroup and found conflicting resolutions between these gene sets potentially because of the extremely slow rates of evolution among mitogenome CDSs.
The samples collected for this study were all from the Chrysanthemum indicum germplasm base of China Resources Sanjiu Medical & Pharmaceutical Co., Ltd in Yangxin County, Huangshi City, Hubei Province. The total genomic DNA was extracted using a Cetyltrimethylammonium Bromide (CTAB) method (Arseneau et al., 2017). The same DNA sample was used for Illumina sequencing and HiFi sequencing by using a Hiseq Xten PE150 sequencing platform (Emerman et al., 2017) and PacBio Sequel II sequencing platform respectively (Eid et al., 2009).
Mitogenomes from three Chrysanthemum accessions were assembled with short read (Illumina) or long-read sequencing (PacBio HiFi) data. Specifically, for the diploid- and tetraploid-Chrysanthemum, first, a total of three Gb of reads were randomly extracted from the PacBio HiFi data by using SeqKit v2.1.0 (Shen et al., 2016). Then the published mitogenome of C. indicum (NCBI accession number: MH716014.1) was used as a reference to extract mitochondrial reads by using minimap2 with the parameter settings’-cx map-hifi -H’ (Li, 2018). Flye-meta v2.8.3-b1695 (Kolmogorov et al., 2020) was used to assembly the mitogenomes with PacBio HiFi reads, with the master circle conformation mitogenome of C. indicum (2x) and the complex mitogenome conformation of C. indicum (4x) with multiple connections obtained. Next the reference and C. indicum (2x) mitogenomes were mapped to C. indicum (4x) to simplify it to a single circle in Bandage (Wick et al., 2015). For C. indicum var. aromaticum with short read data, a total of five Gb reads were randomly extracted. GSAT was used to assembly the mitogenome with the pipeline ‘graphShort’, then simplified the genome in the same method as above (He et al., 2022). The three single circles were annotated by GeSeq (Tillich et al., 2017)with the reference mitogenome of C. indicum (MH716014), and the trans-spliced genes checked manually.
A similar pipeline as mitogenome assembly was used to assemble the plastome of diploid- and tetraploid-Chrysanthemum accessions. Given the high copy number of plastome sequences within a cell, one Gb of HiFi reads were randomly extracted for assembly. For C. indicum var. aromaticum the plastome was extracted from the de novo assembly results of GSAT from above. The plastomes were annotated by GeSeq (Tillich et al., 2017; Qu et al., 2019) and PGA software, using the reference plastome of C. indicum (NC_020320).
The program Blastn v2.11.0+ with parameter settings ‘-evalue 1e-6’ was used to identify syntenic sequences between the seven accessions from Chrysanthemum (Camacho et al., 2009). Syntenic sequences longer than 1 Kb were visualized with NGenomeSyn v1.41 (He et al., 2023).
The repeat types, F (forward), P (palindrome), R (reverse), and C (complement) of dispersed repeat sequences in the seven Chrysanthemum accessions organelles were detected using REPuter with parameter settings ‘-c -f -p -r -l 20 -h 3 -best 300’ (Kurtz et al., 2001). The MISA software was used to identify simple sequence repeats (SSRs) with 10, 6, 5, 5, 5, and 5 repeat units set as minimum thresholds for mono-, di-, tri-, tetra-, penta-, and hexa-motifs respectively (Beier et al., 2017).The results were visualized with ggplot2 (Villanueva and Chen, 2019).
To determine the plastid-derived sequences within Chrysanthemum, the seven mitogenomes were searched against their corresponding plastomes by Blastn v2.11.0+ with parameter settings ‘-evalue 1e-6’. Then the plastid-derived sequences were annotated. The location of plastid-derived sequences in both the mitogenome (destination) and plastome (origin) were visualized with Circos v0.69-9 (Krzywinski et al., 2009). The plastid-derived sequences in mitogenomes were annotated by GeSeq (Tillich et al., 2017)and PGA software, using the reference plastome of C. indicum (NC_020320).
To analyze the phylogenetic relationships among Asteraceae, the plastomes and mitogenomes of C. indicum (2x), C. indicum (4x), C. indicum var. aromaticum, and 23 other Asteraceae were used, with Platycodon. grandiflorus (Campanulaceae) as an outgroup. The CDSs of 80 chloroplast protein-coding genes and 32 mitochondrial protein-coding genes were extracted, concatenated, and aligned using MAFFT v7.490 (Katoh et al., 2002) with poorly aligned sections trimmed with TrimAL v1.4 (Capella-Gutiérrez et al., 2009). These datasets were then used to conduct two separate phylogenetic analysis using IQ-TREE v2.0 with 1000 ultrafast bootstrap replicates to assess branch support based on the auto-selected best-fit model ‘TVM + F + R2’, with FigTree v1.4.3 used for tree visualization (Minh et al., 2020).
All gene pairs alignments were converted into PAML format using EasyCodeML (Yang, 2007; Gao et al., 2019). All the sequences pairs was merge in one file. Molecular divergence times were estimated by placing soft boundaries on the split nodes using records from the timetree database (http://timetree.org/). For divergence time markers we used two nodes, one for the divergence of the outgroup from the Asteraceae, set at 67.7-93.0 Mya, and the other divergence time for the node represented by Arctium lappa from the node represented by Chrysanthemum indicum, set at 36.6-45.1 Mya. The MCMCtree module of PAML software was implemented to estimate the divergence time of each node (Dos Reis, 2022). Tracer was used to evaluate whether the results of MCMCtree have converged. The result of divergence time tree results are visualized using ggtree package (Xu et al., 2022).
The mitogenome and plastome sequences supporting the conclusions of this article are available in GenBank (https://www.ncbi.nlm.nih.gov/) repository, accession numbers: NC_067879, NC_042378, NC_042756, NC_042406, NC_034354, MH716014, NC_039757, ON053202, OU343227.1, NC_064134, NC_062671, NC_062672, NC_060635, NC_062670, NC_051989, NC_051990, NC_058585, NC_023337, NC_053927, NC_066407, NC_059793, NC_058644, NC_066808, and NC_035958 for mitogenome, MK905238, MH375874, OK128342, MZ127827, NC_060634, MW691204, NC_058915, KX822074, NC_037388, NC_020320, OU343226.1, MK604174, KX063880, NC_007977, MT302568, MT302567, MG696658, NC_066756, DQ383816, NC_066758, NC_067730, NC_050977, NC_031396, and NC_035624 for plastome. The data presented in the study are deposited in the GenBank repository, accession number OQ835564 -OQ835569 and the Genome Sequence Archive (Wang et al., 2017) in the National Genomics Data Center (Partners, 2023), Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences repository, accession number CRA011215.
WZ and SZ conceived the study. HH and ZW assembled and annotated the mitogenome and carried out the comparative analysis. LX, QM, and MW analyzed the structural and sequence of plastid genomes. HH, ZW, SZ, LT, and WZ discussed the results. HH, ZW, SZ, and LT wrote the manuscript. WZ and SZ revised the paper. All authors approved the final manuscript.
This project was funded by the Science, Technology, and Innovation Commission of Shenzhen Municipality (RCYX20200714114538196), and it was also supported by the National Natural Science Foundation of China (31970244 and 32170238), and the Funding through the Elite Young Scientists Program of the Chinese Academy of Agricultural Sciences was also given to Zhiqiang Wu. In addition, this project was supported by Science Technology and Innovation Commission of Shenzhen Municipality of China (ZDSYS 20200811142605017), Innovation Team and Talents Cultivation Program of the National Administration of Traditional Chinese Medicine (ZYYCXTD-D-202005) and molecular marker-assisted selection of new varieties of Chrysanthemum indicum (R21017).
Authors HL, LX, QM, and MW were employed by the company China Resources Sanjiu Medical & Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1228551/full#supplementary-material
Arseneau, J. R., Steeves, R., Laflamme, M. (2017). Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour 17, 686–693. doi: 10.1111/1755-0998.12616
Aubin, E., El Baidouri, M., Panaud, O. (2021). Horizontal gene transfers in plants. Life (Basel) 11, 857. doi: 10.3390/life11080857
Beier, S., Thiel, T., Münch, T., Scholz, U., Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Bergthorsson, U., Adams, K. L., Thomason, B., Palmer, J. D. (2003). Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201. doi: 10.1038/nature01743
C., W., J., C., and A., J. M. (2004). Molecular evolution and phylogeny of florist's chrysanthemum and related species. J. Beijing Forestry Univ., 91–96.
C., Z., S, D. (2002). AFLP analysis of some plants of the genus Chrysanthemum. J. Beijing Forestry Univ. 24, 72–76.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10, 421. doi: 10.1186/1471-2105-10-421
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Dos Reis, M. (2022). Dating microbial evolution with MCMCtree. Methods Mol. Biol. 2569, 3–22. doi: 10.1007/978-1-0716-2691-7_1
Drouin, G., Daoud, H., Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 827–831. doi: 10.1016/j.ympev.2008.09.009
Duff, R. J., Nickrent, D. L. (1999). Phylogenetic relationships of land plants using mitochondrial small-subunit rDNA sequences. Am. J. Bot. 86, 372–386. doi: 10.2307/2656759
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
El-Twab, M. H. A., Kondo, K. J. C. B. (2007). FISH physical mapping of 5S rDNA and telomere sequence repeats identified a peculiar chromosome mapping and mutation in Leucanthemella linearis and Nipponanthemum nipponicum in Chrysanthemum sensu lato. Chromosome Bot. 2, 11–17. doi: 10.3199/iscb.2.11
Emerman, A. B., Bowman, S. K., Barry, A., Henig, N., Patel, K. M., Gardner, A. F., et al. (2017). NEBNext direct: A novel, rapid, hybridization-based approach for the capture and library conversion of genomic regions of interest. Curr. Protoc. Mol. Biol. 119, 7.30.31–37.30.24. doi: 10.1002/cpmb.39
Feng, H., Xu, H., Chen, J., Zheng, H., Ye, Y., Liu, Y. (2020). Characterization of the complete chloroplast genome of Chrysanthemum oreastrum (Asteraceae) as a traditional medicinal plant to China. Mitochondrial DNA Part B 5, 1172–1173. doi: 10.1080/23802359.2020.1731341
Gao, F., Chen, C., Arab, D. A., Du, Z., He, Y., Ho, S. Y. W. (2019). EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Grewe, F., Edger, P. P., Keren, I., Sultan, L., Pires, J. C., Ostersetzer-BIran, O., et al. (2014). Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion 19 Pt B, 135–143. doi: 10.1016/j.mito.2014.05.008
He, W., Xiang, K., Chen, C., Wang, J., Wu, Z. (2022). Master graph: an essential integrated assembly model for the plant mitogenome based on a graph-based framework. Brief. Bioinform. 24, bbac522. doi: 10.1093/bib/bbac522
He, W., Yang, J., Jing, Y., Xu, L., Yu, K., Fang, X. (2023). NGenomeSyn: an easy-to-use and flexible tool for publication-ready visualization of syntenic relationships across multiple genomes. Bioinformatics 39, btad121. doi: 10.1093/bioinformatics/btad121
Katoh, K., Misawa, K., Kuma, K., Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kazama, T., Okuno, M., Watari, Y., Yanase, S., Koizuka, C., Tsuruta, Y., et al. (2019). Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat. Plants 5, 722–730. doi: 10.1038/s41477-019-0459-z
Kelchner, S. A. (2002). Group II introns as phylogenetic tools: structure, function, and evolutionary constraints. Am. J. Bot. 89, 1651–1669. doi: 10.3732/ajb.89.10.1651
Kim, J. S., Lee, W., Pak, J. H. (2018). The complete plastid genome sequence of Chrysanthemum lucidum (Asteraceae): an endemic species of Ulleung Island of Korea. Mitochondrial DNA. Part B Resour. 3, 476–477. doi: 10.1080/23802359.2018.1463147
Klie, M., Schie, S., Linde, M., Debener, T. (2014). The type of ploidy of chrysanthemum is not black or white: a comparison of a molecular approach to published cytological methods. Front. Plant Sci. 5, 479. doi: 10.3389/fpls.2014.00479
Knoop, V. (2011). When you can't trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 68, 567–586. doi: 10.1007/s00018-010-0538-9
Kolmogorov, M., Bickhart, D. M., Behsaz, B., Gurevich, A., Rayko, M., Shin, S. B., et al. (2020). metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 17, 1103–1110. doi: 10.1038/s41592-020-00971-x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, J., Wan, Q., Abbott, R. J., Rao, G.-Y. (2013). Geographical distribution of cytotypes in theChrysanthemum indicumcomplex as evidenced by ploidy level and genome-size variation. J. Syst. Evol. 51, 196–204. doi: 10.1111/j.1759-6831.2012.00241.x
Li, J., Wan, Q., Guo, Y. P., Abbott, R. J., Rao, G. Y. (2014). Should I stay or should I go: biogeographic and evolutionary history of a polyploid complex (Chrysanthemum indicum complex) in response to Pleistocene climate change in China. New Phytol. 201, 1031–1044. doi: 10.1111/nph.12585
Liu, L. X., Du, Y. X., Folk, R. A., Wang, S. Y., Soltis, D. E., Shang, F. D., et al. (2020). Plastome evolution in saxifragaceae and multiple plastid capture events involving heuchera and tiarella. Front. Plant Sci. 11, 361. doi: 10.3389/fpls.2020.00361
Luo, C., Chen, D., Cheng, X., Zhao, H., Huang, C. (2016). Genome size estimations in Chrysanthemum and correlations with molecular phylogenies. Genet. Resour. Crop Evol. 64, 1451–1463. doi: 10.1007/s10722-016-0448-2
Ma, Y. P., Zhao, L., Zhang, W. J., Zhang, Y. H., Xing, X., Duan, X. X., et al. (2020). Origins of cultivars of Chrysanthemum—Evidence from the chloroplast genome and nuclear LFY gene. J. Syst. Evol. 58, 925–944. doi: 10.1111/jse.12682
Malek, O., Lättig, K., Hiesel, R., Brennicke, A., Knoop, V. (1996). RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J. 15, 1403–1411. doi: 10.1002/j.1460-2075.1996.tb00482.x
Masuda, Y., Nakano, M., Kusaba, M. (2022). The complete sequence of the chloroplast genome of Chrysanthemum rupestre, a diploid disciform capitula species of Chrysanthemum. Mitochondrial DNA. Part B Resour. 7, 603–605. doi: 10.1080/23802359.2022.2057252
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Oldenkott, B., Yamaguchi, K., Tsuji-Tsukinoki, S., Knie, N., Knoop, V. (2014). Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20, 1499–1506. doi: 10.1261/rna.045575.114
Partners C.-N.M.a (2023). Database resources of the national genomics data center, China national center for bioinformation in 2023. Nucleic Acids Res. 51, D18–d28. doi: 10.1093/nar/gkac1073
Putintseva, Y. A., Bondar, E. I., Simonov, E. P., Sharov, V. V., Oreshkova, N. V., Kuzmin, D. A., et al. (2020). Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genom. 21, 654. doi: 10.1186/s12864-020-07061-4
Qi, S., Twyford, A. D., Ding, J. Y., Borrell, J. S., Wang, L. Z., Ma, Y. P., et al. (2021). Natural interploidy hybridization among the key taxa involved in the origin of horticultural chrysanthemums. J. Syst. Evol. 60, 1281–1290. doi: 10.1111/jse.12810
Qu, X.-J., Moore, M. J., Li, D.-Z., Yi, T.-S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 50. doi: 10.1186/s13007-019-0435-7
Rogalski, M., do Nascimento Vieira, L., Fraga, H. P., Guerra, M. P. (2015). Plastid genomics in horticultural species: importance and applications for plant population genetics, evolution, and biotechnology. Front. Plant Sci. 6, 586. doi: 10.3389/fpls.2015.00586
Shen, W., Le, S., Li, Y., Hu, F. (2016). SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PloS One 11, e0163962. doi: 10.1371/journal.pone.0163962
Si-Lan., D., Jun-Yu., C., Wen-Bin., L. (1998). Application of RAPD analysis in the study on the origin of chinese cultivated chrysanthemum. J. Integr. Plant Biol. 40, 11.
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Uncu, A. O., Uncu, A. T., Celık, İ., Doganlar, S. (2015). Frary A. A primer to molecular phylogenetic analysis in plants. CRC. Crit. Rev. Plant Sci. 34, 454–468. doi: 10.1080/07352689.2015.1047712
Villanueva, R. A., Chen, Z. (2019). ggplot2: elegant graphics for data analysis (2nd ed.). Measurement: Interdiscip. Res. Perspect. 17, 160–167. doi: 10.1080/15366367.2019.1565254
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T., et al. (2017). GSA: genome sequence archive. Genom. Proteom. Bioinf. 15, 14–18. doi: 10.1016/j.gpb.2017.01.001
Wick, R. R., Schultz, M. B., Zobel, J., Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Won, S. Y., Jung, J. A., Kim, J. S. (2018). The complete chloroplast genome of Chrysanthemum boreale (Asteraceae). Mitochondrial DNA. Part B Resour. 3, 549–550. doi: 10.1080/23802359.2018.1468225
Wu, Z., Liao, X., Zhang, X., Tembrock, L., Broz, A. (2020). Genomic architectural variation of plant mitochondria – a review of multichromosomal structuring. J. Syst. Evol. 60, 160–168. doi: 10.1111/jse.12655
Wu, Z., Sloan, D. B. (2019). Recombination and intraspecific polymorphism for the presence and absence of entire chromosomes in mitochondrial genomes. Heredity 122, 647–659. doi: 10.1038/s41437-018-0153-3
Xi, Z., Wang, Y., Bradley, R. K., Sugumaran, M., Marx, C. J., Rest, J. S., et al. (2013). Massive mitochondrial gene transfer in a parasitic flowering plant clade. PloS Genet. 9, e1003265. doi: 10.1371/journal.pgen.1003265
Xia, Y., Hu, Z., Li, X., Wang, P., Zhang, X., Li, Q., et al. (2016). The complete chloroplast genome sequence of Chrysanthemum indicum. Mitochondrial DNA. Part A DNA mapping sequencing Anal. 27, 4668–4669. doi: 10.3109/19401736.2015.1106494
Xia, H., Xu, Y., Liao, B., Huang, Z., Zhou, H., Wang, F. (2021a). Complete chloroplast genome sequence of a Chinese traditional cultivar in Chrysanthemum, Chrysanthemum morifolium 'Anhuishiliuye'. Mitochondrial DNA. Part B Resour. 6, 1281–1282. doi: 10.1080/23802359.2020.1866451
Xia, H., Xu, Y., Zhang, J., Huang, Z., Luo, H., Ye, Z., et al. (2021b). Complete chloroplast genome sequence of a Dutch cultivar of Chrysanthemum, Chrysanthemum morifolium 'Orizaba' (Asteraceae). Mitochondrial DNA. Part B Resour. 6, 1937–1938. doi: 10.1080/23802359.2021.1936672
Xu, S., Li, L., Luo, X., Chen, M., Tang, W., Zhan, L., et al. (2022). Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 1, e56. doi: 10.1002/imt2.56
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yao, G., Zhang, Y. Q., Barrett, C., Xue, B., Bellot, S., Baker, W. J., et al. (2023). A plastid phylogenomic framework for the palm family (Arecaceae). BMC Biol. 21, 50. doi: 10.1186/s12915-023-01544-y
Yu, J., Xia, M., Xu, H., Han, S., Zhang, F. (2021). The complete chloroplast genome sequence of Neopallasia pectinata (Asteraceae). Mitochondrial DNA. Part B Resour. 6, 430–431. doi: 10.1080/23802359.2020.1870893
Yuan, R., Wang, X., Zhang, J., Sen, L., Dong, G., Liu, Y. (2022). Evaluation of genetic variation in natural populations of chrysanthemum indicum var. Aromaticum and screening of core germplasm. World Sci. Technol. - Modernising Chin. Med. 24, 1325–1334. doi: 10.1097/CM9.0000000000002199
Zhang, G. J., Dong, R., Lan, L. N., Li, S. F., Gao, W. J., Niu, H. X. (2020). Nuclear integrants of organellar DNA contribute to genome structure and evolution in plants. Int. J. Mol. Sci. 21, 707. doi: 10.3390/ijms21030707
Zhang, C., Huang, C. H., Liu, M., Hu, Y., Panero, J. L., Luebert, F., et al. (2021). Phylotranscriptomic insights into Asteraceae diversity, polyploidy, and morphological innovation. J. Integr. Plant Biol. 63, 1273–1293. doi: 10.1111/jipb.13078
Zhang, S., Wang, J., He, W., Kan, S., Liao, X., Jordan, D. R., et al. (2023). Variation in mitogenome structural conformation in wild and cultivated lineages of sorghum corresponds with domestication history and plastome evolution. BMC Plant Biol. 23, 91. doi: 10.1186/s12870-023-04104-2
Keywords: Chrysanthemum indicum, Asteraceae, organelle genome, intragenomic gene transfers, phylogenetic analysis, tetraploid, variants
Citation: Liu H, Hou Z, Xu L, Ma Q, Wei M, Tembrock LR, Zhang S and Wu Z (2023) Comparative analysis of organellar genomes between diploid and tetraploid Chrysanthemum indicum with its relatives. Front. Plant Sci. 14:1228551. doi: 10.3389/fpls.2023.1228551
Received: 25 May 2023; Accepted: 28 July 2023;
Published: 18 August 2023.
Edited by:
Muhammad Awais Farooq, Hebei Agricultural University, ChinaReviewed by:
Yun-peng Du, Beijing Academy of Agricultural and Forestry Sciences, ChinaCopyright © 2023 Liu, Hou, Xu, Ma, Wei, Tembrock, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Wu, d3V6aGlxaWFuZ0BjYWFzLmNu; Shuo Zhang, c3poYW5nQHdlYm1haWwuaHphdS5lZHUuY24=
† These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.