
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 13 July 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1223861
This article is part of the Research Topic Quality of Ornamental Crops: Effect of Genotype, Preharvest, and Improved Production Chains on Quality Attributes of Ornamental Crops, volume II View all 11 articles
 Silvia Farinati
Silvia Farinati Samela Draga
Samela Draga Angelo Betto
Angelo Betto Fabio Palumbo
Fabio Palumbo Alessandro Vannozzi
Alessandro Vannozzi Margherita Lucchin
Margherita Lucchin Gianni Barcaccia*
Gianni Barcaccia*Plant male sterility (MS) represents the inability of the plant to generate functional anthers, pollen, or male gametes. Developing MS lines represents one of the most important challenges in plant breeding programs, since the establishment of MS lines is a major goal in F1 hybrid production. For these reasons, MS lines have been developed in several species of economic interest, particularly in horticultural crops and ornamental plants. Over the years, MS has been accomplished through many different techniques ranging from approaches based on cross-mediated conventional breeding methods, to advanced devices based on knowledge of genetics and genomics to the most advanced molecular technologies based on genome editing (GE). GE methods, in particular gene knockout mediated by CRISPR/Cas-related tools, have resulted in flexible and successful strategic ideas used to alter the function of key genes, regulating numerous biological processes including MS. These precision breeding technologies are less time-consuming and can accelerate the creation of new genetic variability with the accumulation of favorable alleles, able to dramatically change the biological process and resulting in a potential efficiency of cultivar development bypassing sexual crosses. The main goal of this manuscript is to provide a general overview of insights and advances into plant male sterility, focusing the attention on the recent new breeding GE-based applications capable of inducing MS by targeting specific nuclear genic loci. A summary of the mechanisms underlying the recent CRISPR technology and relative success applications are described for the main crop and ornamental species. The future challenges and new potential applications of CRISPR/Cas systems in MS mutant production and other potential opportunities will be discussed, as generating CRISPR-edited DNA-free by transient transformation system and transgenerational gene editing for introducing desirable alleles and for precision breeding strategies.
Plant male sterility (MS) refers to the inability of the plant to generate functional anthers, pollen, or male gametes, although female fertility remains unaffected (Kaul, 1988). Therefore, male sterile plants cannot undergo self-pollination, but they can be fertilized by male fertile plants. The establishment of MS lines is a major goal in F1 hybrid production and marketing because by disabling self-fertilization, it is possible to facilitate the exploitation of heterosis in predominantly autogamous species (Longin et al., 2012; Kim and Zhang, 2018; Li et al., 2022; Ramlal et al., 2022). In the past, the main way to avoid considerable shares of progeny derived from self-pollination, even in species with predominantly allogamous fertilization, was to perform physical emasculation with chemical, mechanical or even manual methods. The main advantage in the use of MS lines is the reduction of costs, time and energy related to these emasculation procedures (Colombo and Galmarini, 2017). For these reasons, MS lines have been developed in several species of economic interest (Abbas et al., 2021; Wang et al., 2023), particularly in horticultural crops and ornamental plants (Yamagishi and Bhat, 2014; Barcaccia et al., 2016; Khan and Isshiki, 2016; Jindal et al., 2019; Singh and Khar, 2021). MS exhibits, in most cases, Mendelian inheritance, which is controlled either by the coordinated action of nuclear and cytoplasmatic genes or exclusively by nuclear genes (Kaul, 1988). The first scenario, defined as cytoplasmic male sterility (CMS), or three-line breeding system, relies on loci localized within the mitochondrial genome (Rogers and Edwardson, 1952; Chen and Liu, 2014). Cytoplasmic maternal inheritance causes all progeny derived from plants carrying the S locus (sterile) cytoplasm to inherit the male sterility trait (Budar and Pelletier, 2001; Yamagishi and Bhat, 2014; Xu et al., 2022). This condition can be overcome by nuclear genes that are functional in dominant conditions (Jindal et al., 2019); they are defined as restorers of fertility (Rf) and can suppress or downregulate the CMS genes and revert male sterility (Schnable and Wise, 1998; Ning et al., 2020). In contrast, genic male sterility (GMS), also reported as nuclear male sterility (NMS), or two-lines breeding system, is generally controlled by single nuclear genes, mostly by recessive alleles (ms) (Colombo and Galmarini, 2017; Manjunathagowda, 2021).
Although genes involved in MS have not yet been characterized in many species, the complex molecular mechanisms at the bases of GMS and CMS have been studied in the most important crops at the transcriptomic, biochemical and epigenetic levels (Fan et al., 2016; Li et al., 2019; Liu et al., 2022; Nie et al., 2023). Potential limitations in the application of MS systems in agricultural species reside first in the availability of MS resources, in the difficulty encountered when introgressing the trait into commercial varieties (e.g., linkage drag issues), and in the maintenance of the MS lines. Furthermore, considering that MS is detectable only during flowering stages, selecting plants characterized by GMS systems may be a challenge for preventing self-pollination. In addition, MS can be influenced by environmental conditions, resulting in instability and being a major issue for conducting crosses. Nevertheless, external conditions have been successfully exploited in rice and wheat, manipulating temperature or photoperiod to guarantee alternating cross-pollination or self-pollination (environmental genic male sterility – EGMS) (Zhou et al., 2012; Meng et al., 2016).
Developing MS lines therefore represents one of the most important challenges in plant breeding programs. Over the years, MS has been accomplished through many different techniques ranging from cross-mediated breeding to advanced methods based on knowledge of genetics and genomics to the most advanced molecular technologies based on genome editing (GE). If MS sources have not been found in the species of interest or if their transfer to the productive varieties is hindered by technical limitations, MS can also be induced by mutagenesis with chemical or physical agents (Hawkes et al., 2011). At present, the ability to precisely recognize and edit DNA sequences can have a significant impact on functional genomics and crop advancement studies. The recent development of GE-based technologies has provided researchers with powerful tools not only for decoding gene functions but also for improving or introducing new plant traits. This progress offers an increasing number of approaches considered revolutionary in molecular biology since it allows modifications at genomic loci in a precise and efficient manner (Malzahn et al., 2017). GE methods, being less time-consuming, can accelerate the creation of new genetic variability with the accumulation of favorable alleles, able to dramatically change the biological process and resulting in a potential efficiency of cultivar development bypassing sexual crosses (Chen and Gao, 2014; Gao, 2015; Arora and Narula, 2017; Scheben et al., 2017). Furthermore, since the development of a new commercial male-sterile line using traditional breeding systems usually takes several years or decades, these modern genetic engineering techniques can reduce dramatically the breeding time (Zhou et al., 2016).
Starting from these assumptions, the aim of this manuscript is to provide a general overview of insights and advances into plant male sterility, first providing a brief description of conventional breeding programs, and then focus attention on the recent new breeding GE-based applications capable of inducing MS by targeting specific nuclear genic loci. In particular, a summary of the mechanisms underlying the recent CRISPR technology and relative success applications will be described for the main crop and ornamental species. Finally, we discuss the future challenges and potential opportunities of such technologies for introducing desirable alleles and improving many traits for precision breeding strategies.
The selection of improved varieties through conventional breeding primarily relies on phenotypic observations and the breeder’s experience. For planning a promising plant breeding program, the following association establishment between phenotype and relative genotype results is fundamental (Chen and Lubberstedt, 2010). As reported in the Introduction section, MS is an important trait for different purposes, primarily for the production of hybrid seeds. The development and propagation process of an F1 hybrid obtained through an MS system involves multiline maintenance strategies (Khan and Isshiki, 2016; Kim and Zhang, 2018; Xu et al., 2022; Scariolo et al., 2023). Various agronomic strategies can be used to introduce the MS trait in a commercial line of interest through conventional breeding approaches: after identifying naturally occurring male sterility within a species, the MS trait may be transferred to elite germplasm by cross pollination (Yamagishi and Bhat, 2014; Bruns, 2017; Zheng et al., 2020). Briefly, as schematically reported in Figure 1, CMS bases on a three-line system that includes, in addition to the MS line, a fertility restorer line and a maintainer line (Chen and Liu, 2014). Conversely, the use of the Mendelian recessive genes of GMS requires the discrimination of male fertile and sterile progeny prior to anthesis to ensure the maintenance of the MS line. This task can be challenging unless functional molecular markers associated with the MS locus are available (Wu et al., 2016). EGMS could overcome this problem by altering specific environmental conditions to make MS lines either male fertile or sterile (Sun et al., 2022).
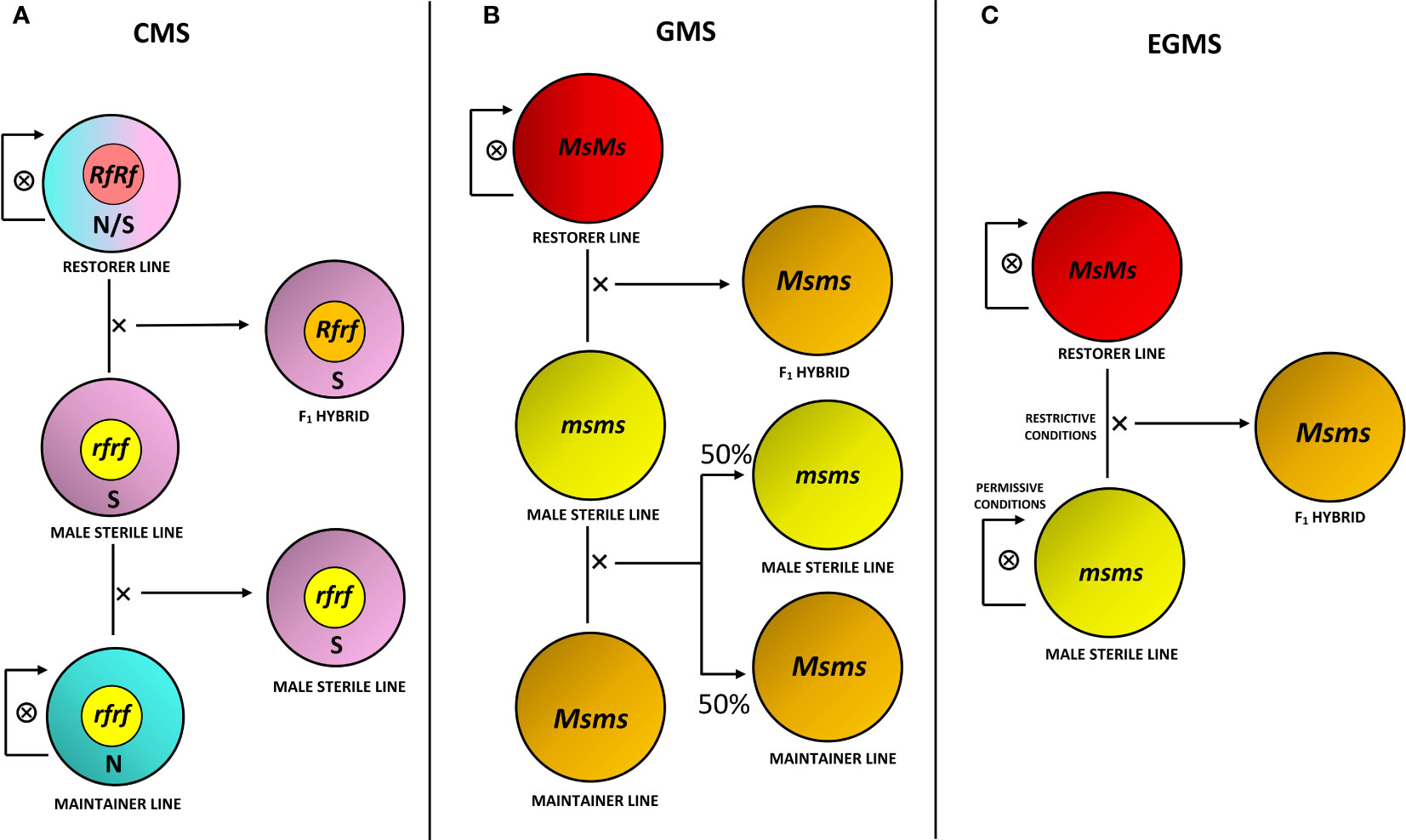
Figure 1 Production and maintenance strategies for MS systems. (A) Three-line system for cytoplasmic male sterility (CMS), involving an MS line with sterile cytoplasm (S) and restorer gene in recessive homozygous condition (rfrf), crossed with a maintainer line carrying normal fertile cytoplasm (N) and rf alleles for its maintenance, in addition to a fertility restorer line with N or S cytoplasm without distinction and restorer alleles in dominant homozygous, hence functional, condition (RfRf), crossed with the MS line for F1 hybrid production. The F1 hybrid consequently brings S cytoplasm and is heterozygous for the restorer gene (Rfrf), hence male fertility. Maintainer and restorer lines are self-pollinated for their maintenance. (B) Three-line system for genic male sterility (GMS), involving a recessive homozygous MS line for the MS gene (msms), crossed with a heterozygous maintainer line (Msms) for maintenance, producing half recessive homozygous and half heterozygous progeny, in addition to a dominant homozygous restorer line (MsMs), crossed with the MS line for heterozygous and fertile F1 hybrid production. (C) Two-line system for environmental genic male sterility (EGMS), involving a recessive homozygous MS line, kept in permissive environmental conditions (i.e., low temperatures or short-day photoperiod) in order to make it become male fertile and self-pollinate for its maintenance, while kept in restrictive conditions (i.e., high temperatures or long-day photoperiod) to make it male sterile and to cross it with a dominant homozygous restorer line for F1 hybrid production.
The identification of functional molecular markers linked to specific traits can be of primary importance to allow future selection programs mediating marker-assisted selection (MAS), which identifies the following mapping gene responsible for the observed phenotype (Page and Grossniklaus, 2002; Schneeberger, 2014; Aklilu, 2021). The use of MAS results in fact in a useful predictive tool for the identification of male sterile genotypes, mapping markers closely associated with the MS locus (Mackenzie, 2012). The complexity and long times of these breeding strategies make the exploration of the molecular mechanisms a key feature to improve productivity and other traits of interest (Bohra et al., 2016; Van Ginkel and Ortiz, 2018; Yu et al., 2021a). On the basis of these articulated schemes of MS maintenance, defining the conditions in which the markers can be predictive molecular tools of genotype is a key point of each program. Furthermore, starting from the knowledge of a well-characterized mechanism of the MS system in model species, substantial genetic resources can be used for the discovery of homologous ms-related genes in other species (Leino et al., 2003; Fernandez Gomez and Wilson, 2014; Morales et al., 2022).
Four different models have been described to explain how CMS can produce male sterility Chen and Liu (2014):
i) Cytotoxicity model: the proteins encoded by the CMS genes directly cause the death of the cells involved. At the basis of the mechanisms by which this occurs it has been hypothesized that there is mitochondrial dysfunction, but a well-defined model has not yet been developed, lacking molecular evidence of cytotoxicity itself. As a result, a simple explanation for CMS in these systems is that the CMS proteins cause mitochondrial malfunction in the anthers’ sporophytic or gametophytic cells, resulting in male abortion (Levings, 1993).
ii) Lack of energy model: the cellular respiration process is altered. In fact, CMS proteins can act as dysfunctional homologues of parts of complexes forming the electron transport chain, or changing proton gradients critical to the cellular respiration process, resulting in no ATP production. The molecular evidence supports the concept that some CMS are caused by an energy deficit in growing anthers, which demand more energy (Sabar et al., 2003; Wang et al., 2013).
iii) Asynchronous programmed cell death (PCD) model: PCD is induced in tapetum cells earlier than its normal course. It is implemented through the release of cytochrome C, a protein complex of the electron transport chain, and by increasing the production of reactive oxygen compounds (ROS, from Reacting Oxygen Species). By starting the autolysis before the pollen is mature, the tapetum cannot continue to nourish it and this does not complete its development. Plant male gametophytes form in anthers through cooperative contacts between sporophytic (anther wall) and gametophytic (microspore) cells, as well as correct PCD-controlled cellular degeneration of the tapetum, the deepest cell layer of the anther wall tissue (Mah, 2005).
iv) Retrograde regulation model: some CMS proteins are able to regulate the expression of nuclear genes, including some involved in the processes for correct reproduction. For example, they can disturb the formation activity of the stamens, in place of which carpels or petals develop. Or they can nullify the action of fertility restorative genes, when they are in their recessive allelic/haplotype form (Linke et al., 2003).
By comparing the proteomes of CMS and fertile lines, some CMS causative proteins, such as URF13 of maize CMS-T (Forde et al., 1978) and truncated COX2 of sugar beet CMS-G (Ducos et al., 2001), were discovered in other CMS systems. The CMS candidate genes were found in a few cases, such as radish CMS-Ogu (Bonhomme et al., 1991) and wheat alloplasmic CMS-AP (Rathburn and Hedgcoth, 1991), by analyzing the mitochondrial DNAs of segregating somatic hybrids (cybrids) produced from protoplast fusion between CMS-carrying lines and normal fertile lines. However, owing of the difficulties in acquiring cybrids and the uncertainty of recombination events between the mitochondrial DNAs of the fusion lines, this strategy is ineffective for most crops. Several methodologies can be used to identify CMS candidate genes. The most common approach is to look for changes in mitochondrial gene organization and/or mitochondrial transcriptome or proteome differences in CMS cytoplasm lines with and without the Rf gene(s). Nuclear Rf genes perform their action at different levels, involving various steps of protein synthesis or cellular metabolism (Chen and Liu, 2014).
Since in the case of CMS, 100% of offspring individuals will be MS, the use of molecular markers results a strategic key if identified and mapped in association with Rf genes. Table 1 shows main examples of the important crop classes for which mapped and retrievable information regarding reproducible, codominant molecular markers linked to Rf genes, offering fast and reliable detection tools to select, by MAS, parental lines for production of the desired progeny (Jordan et al., 2010; Yan et al., 2017).
In addition to major cereal crops and species, a great interest in mapping CMS-related loci has also been reflected in ornamental plants such as sunflower and petunia (Bentolila et al., 1998; Gentzbittel et al., 1999; Feng and Jan, 2008; Yue et al., 2010; Liu et al., 2012; Qi et al., 2012; Liu et al., 2013; Talukder et al., 2019).
Several molecular mechanisms underlie GMS in different species, in many of them genes coding transcription factors are capable of modifying the expression of genes involved in reproductive processes. The result is disturbance of gamete formation, due to failure of homologous chromosome separation in meiotic anaphase I and delayed of programmed cell death in tapetum (Jeong et al., 2014). In particular, several nuclear genes have been found responsible for MS, causing arrest of microspore development. Furthermore, as mentioned in Introduction section, the male sterility can also depend on environmental conditions, and in this case the GMS is define as EGMS. Temperature-sensitive genic male sterile (TGMS) and photoperiod-sensitive genic male sterile (PGMS) lines were developed especially in cereals crops like rice and wheat. TGMS lines are sterile at high temperatures and fertile at low temperatures, while PGMS lines can either be sterile when the day is longer than the night and fertile when it is shorter, or vice versa. In China, EGMS lines occupy 20% of the area dedicated to the cultivation of hybrid rice (Li et al., 2007). Also in this case, the molecular markers result strategic keys if identified and mapped in association with the ms locus, as testified in past (Barcaccia et al., 2016). However, to date, in more than 610 species of flowering plants the MS trait is under investigation, and specifically in the past few decades, at least 40 GMS genes have been identified by MAS and characterized in model Arabidopsis and rice (Chen and Liu, 2014; Singh et al., 2019; Wan et al., 2019). As similarly reported in Tables 1, 2 shows main examples of the several crop classes for which mapped and retrievable information regarding reproducible, codominant molecular markers linked to nuclear male sterility genes. Molecular markers such as SSR, RFLP, SCAR, and SNP were fully employed for mapping male sterility genes, while insertion−deletion (InDel), target region amplification polymorphism (TRAP), sequence-related amplified polymorphism (SRAP), high resolution melting (HRM), and conserved orthologous set (COS) markers were sporadically used among these research studies. However, the data availability of mapped genes was correlated with species of great agronomic and economic importance: studies on cereals and other horticulture crop were prevalent, resulting in numerous mapped markers and associated genes involved in both MS systems, which offer open access for hybrid production using male-sterile lines (Li et al., 2007; Rout et al., 2021; Morales et al., 2022). These insights were interesting because proper MAS application could offer competitive phenotypes for market demand and contribute to reducing production costs, which is also fundamental for ornamental plant companies.
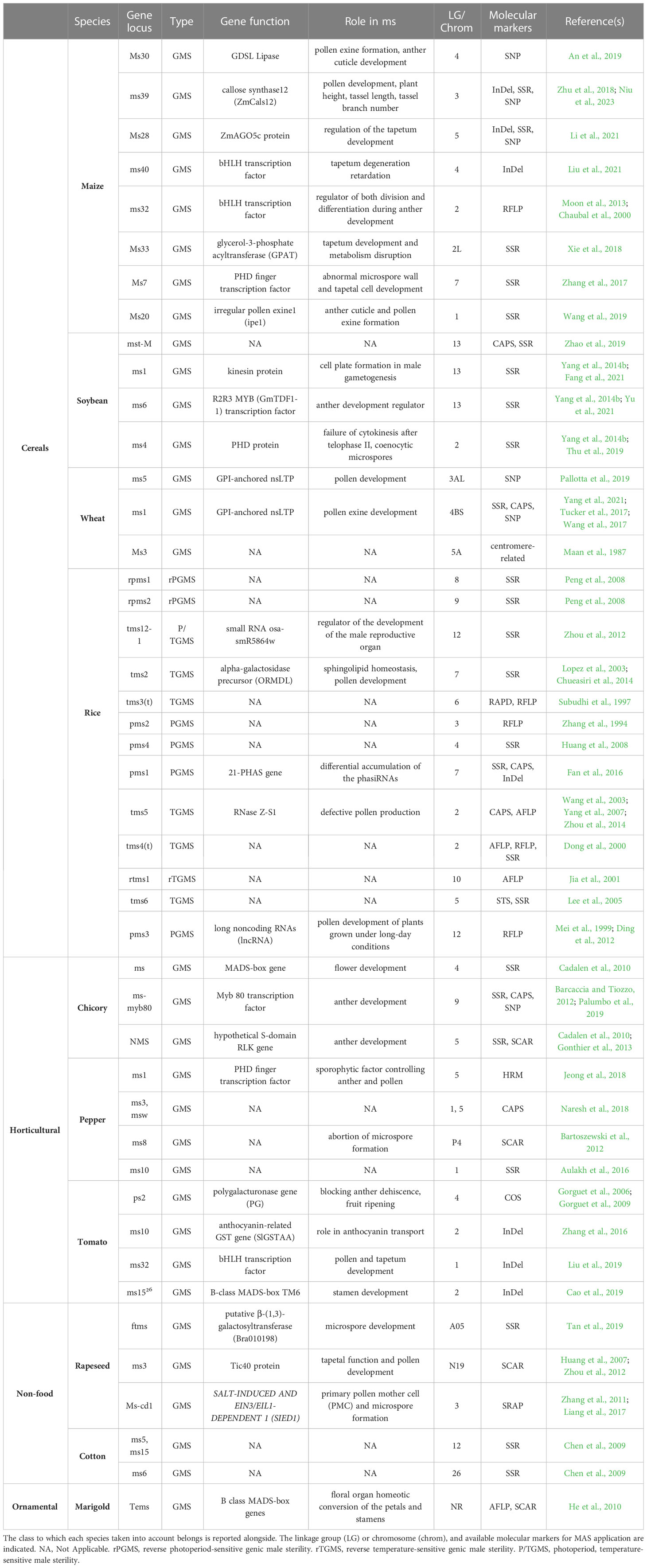
Table 2 Male sterility-related molecular markers mapped to relative genes associated with the GMS trait.
Novel GE technologies have been intensively developed through diverse biological systems depending on sequence-specific nucleases (SSNs). Upon induction, all SSNs may detect a specific DNA fragment and cause double-stranded breaks (DSBs), repaired by two endogenous repair machinery of plant. Initially, ZFNs (zinc-finger nucleases) and TALENs (transcription activator-like effector nucleases) were the two systems primarily employed in genome editing techniques (Smith et al., 2006; Petolino, 2015; Malzahn et al., 2017). However, the difficulties of array and vector design in each of these methods, as well as the time-consuming work necessary to construct vectors for each new DNA sequence target, have hampered their widespread usage for plant genome editing. In contrast, with subsequent scientific breakthroughs, CRISPR/Cas-based genome editing systems (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein) have been increasingly employed in the last decade (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2015). Their use is constantly expanding in numerous applications, resulting in a wider array of editing tools developed for several purposes (Figure 2). CRISPR/Cas-based systems are considered more robust and simpler for targeting gene editing since they present a significant advancement over previous systems, such as the simplicity and versatility in vector design and construction for subsequent plant transformation (Chen et al., 2019; Bhat et al., 2020; Nadakuduti and Enciso-Rodriguez, 2020; Zhu et al., 2020). An increasing number of studies attest to the expanded applications of Cas9 nuclease for editing beyond double strand breaks, and the accompanying benefits of those systems have resulted in quick, widespread acceptance for editing applications in a diverse range of plant species. Cas9-related nuclease, if associated with an RNA guide (single guide RNA, sgRNA), is able to identify a special site PAM (Protospacer Adjacent Motif) in the host DNA and cut the target sequence recognized, mediated by the complement to which the sgRNA binds (Mojica et al., 2009; Jinek et al., 2012), inducing the development of modified and improved forms of Cas9 and Cas9-like nucleases. In addition to Cas9, other related enzymes (Cas12a, CasΦ, and Cms1), derived from other CRISPR systems, have been implemented since they are potentially useful for editing approaches, each with slightly different capabilities to recognize and modify PAM sites (Zetsche et al., 2015; Begemann et al., 2017; Gao et al., 2017; Li et al., 2018; Pausch et al., 2020).
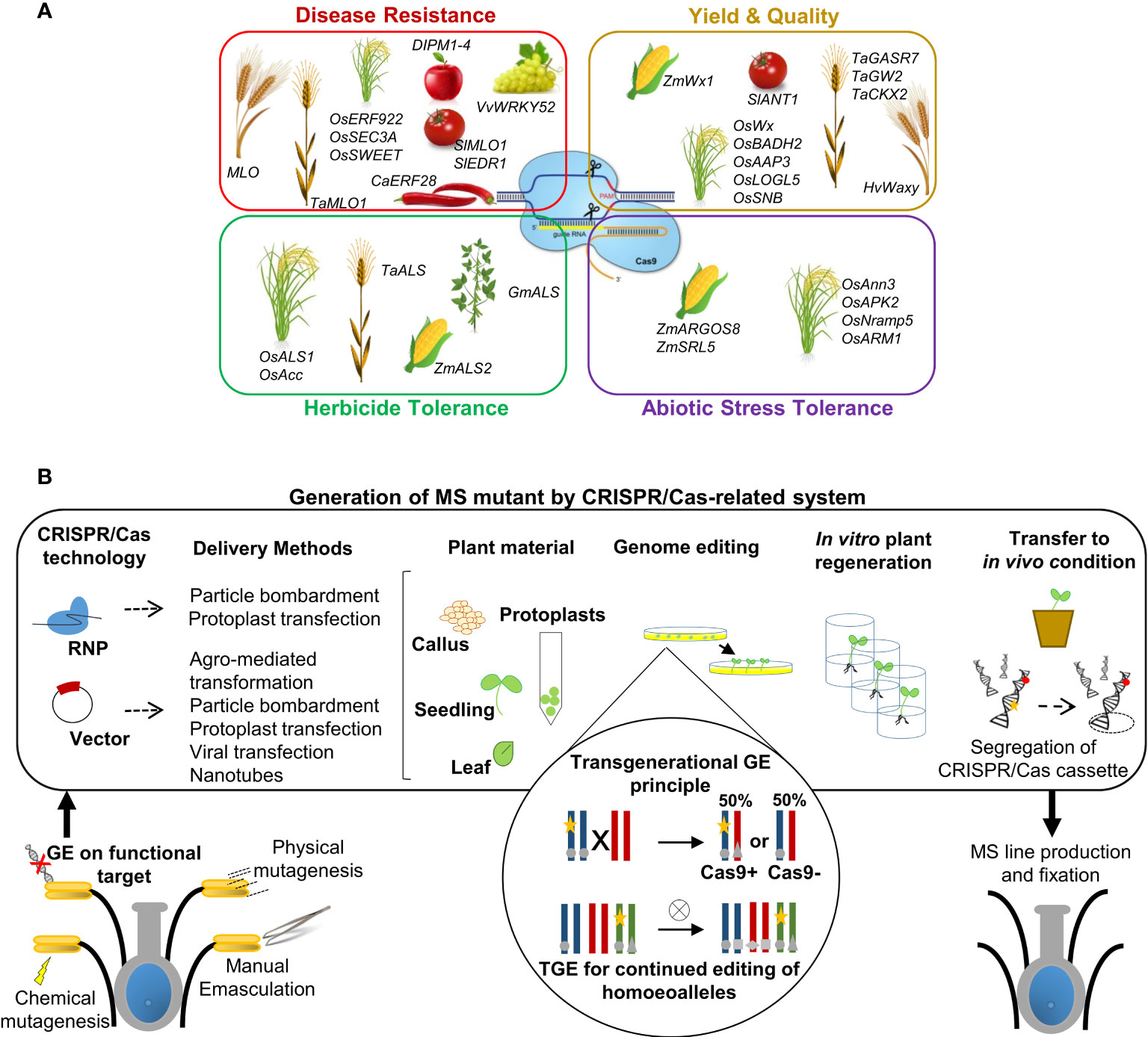
Figure 2 GE applications in precision plant breeding approaches. (A) Schematic representation of the main applications of GE for crop improvement through CRISPR/Cas and related systems. Examples of genes modified for improving specific traits are listed in each specific box for various reference crops. (B) Simplified representation of the workflow for MS generation mediated by CRISPR/Cas technology applied to target genes. In summary, gene editing is induced by transient or stable expression of a Cas nuclease and gRNA through the transformation/transfection of the ribonucleoprotein (RNP) complex or DNA vector. Both CRISPR machinery reagents can be delivered into plant cells using biolistic transformation or other methods, according to suggested transformation/transfection protocols related to species, plant tissues and the methodological approach followed. Such events can lead to the generation of edited whole plants. The transgene locus is usually heterozygous in the first generation of transgenic plants (T0). Afterwards, elimination of the CRISPR/Cas cassette transgene (yellow star) by genetic segregation, according to Mendelian genetics, occurred to obtain transgene-free material. Transgene-free and edited T1 plants can be identified by PCR-based genotyping. The transgenerational GE principle and potential applications in plants are highlighted schematically in circles: transgenic plants represented as a chromosome pair are hemizygous for a CRISPR/Cas9-containing T-DNA locus (yellow stars) and edited in both alleles (grey circles). When crossed with a WT, the resulting progeny either lacks the T-DNA and inherits a single edited allele or inherits the T-DNA, resulting in (transgenerational) editing of the inherited WT allele. TGE for continued editing of homoeoalleles in polyploids species: a transgenic line may have edits only in a subset of homoeoalleles at the homologous chromosomes. After self-crossing and selecting plants that inherited the T-DNA, all homoeoalleles may now be edited. The details can be found within the text.
This approach is defined as a precision-type plant breeding technology, and it is currently being utilized to change the characteristics of various plants, including important crops, as well as to produce new germplasm resources. (Gaillochet et al., 2021). The applications have been primarily focused on traits related to stress tolerance, disease resistance, quality improvement, and higher yields with minimal input (Liu et al., 2022) (Figure 2A). In particular, the CRISPR/Cas system has been widely employed to edit plant genomes to modify genes in various ways, e.g., gene knockout, gene knock-in, gene regulation, base editing, and prime editing (Zhang et al., 2021a). Gene knockout/-in and base and/or prime editing mediated by CRISPR/Cas-related tools have resulted in flexible and successful strategic ideas used to alter the function of key genes and their associated networks, regulating complicated crop traits (Gaillochet et al., 2021). The successes of CRISPR/Cas application in crop improvement have been reviewed in many papers, which are focused on the improvement of crop resistance to fungi, bacteria, and viruses, typically mediating targeting susceptibility systems to increase resistance (Wang et al., 2014; Malnoy et al., 2016; Wang et al., 2016; Nekrasov et al., 2017; Borrelli et al., 2018; Ma et al., 2018; Pu et al., 2018; Wang et al., 2018b; Dong and Ronald, 2019; Oliva et al., 2019; Mishra et al., 2021), resistance to an assortment of herbicides (Li et al., 2015; Chen et al., 2017; Zong et al., 2018; Zhang et al., 2019; Kuang et al., 2020), and abiotic stresses such as drought, salinity, high temperatures, and soil pollution (Lou et al., 2017; Nieves-Cordones et al., 2017; Shi et al., 2017; Tang et al., 2017; Pan et al., 2020). In particular, the main purposes of food crop improvement research using GE technology are to raise yield (e.g., grain size/weight/number per panicle) and crop quality traits determined by internal (e.g., contents of nutrients and bioactive substances) and external (e.g., size, color, and texture) factors related to a response to adverse surrounding environments (Shan et al., 2015; Zhang et al., 2016b; Lou et al., 2017; Dahan-Meir et al., 2018; Wang et al., 2018c; Chen et al., 2019; Ma et al., 2019; Voss-Fels et al., 2019; Wang et al., 2020; Zhu et al., 2020; Liu et al., 2021b). To simplify the overview of this complicated scenario, Figure 2A summarizes the main traits that can be enhanced by genome editing, with a list of example genes edited by the CRISPR/Cas system to improve related plant species.
In addition to stress response, and traits related to quality and yield, CRISPR/Cas-based technology offers a new strategic tool to affect other crop traits associated to fertility/sterility (Figure 2B). As described in section 2, researchers have employed several strategies for integrating MS traits into genomes of interest using information and methods arising from conventional breeding approaches, with the aim to guarantee high varietal purity breeding and to have better offspring in terms of uniformity, yield, and stress tolerance (Bao et al., 2022). Thanks to important forward genetic tools, as mutagenesis approaches and TILLING populations, it has been possible to discover and investigate new candidate genes controlling male sterility. Furthermore, the increasing number of transcriptomic and proteomic studies in recent decades, mostly on crop species, has allowed us to characterize an emergent number of genes with different roles in the development of male reproductive organs and consequently with a putative role in MS induction. If the role of these genes is confirmed as influencer of MS trait, they could be potential targets for subsequent gene editing strategies (Carroll, 2011; Li et al., 2012). The elucidation of molecular processes regulating anther and pollen development has increased the identification and characterization of new putative candidate male-sterility genes (MSGs) in several species, allowing the development and effective use of numerous biotechnology-based male-sterility systems for crop hybrid breeding (Perez-Prat and Van Lookeren Campagne, 2002; Whitford et al., 2013; Wu et al., 2016; Zhang et al., 2019). As reported in more detail in following sections, CRISPR/Cas technology is resulted a novel, rapid and alternative method for the generation of MS lines through target gene editing, both in food crops (monocots and horticultural dicots) and in the increasing ornamental sector, implementing also the knowhow underlying male sterility in plants (Cong et al., 2013; Wang et al., 2018a).
The CRISPR/Cas system, especially based on Cas9, has been successfully applied for the generation of male sterile lines in important worldwide food crops (Barman et al., 2019; Okada et al., 2019). Studies in main crops, such as rice, soybean, maize, and tomato, have reported that pooled CRISPR/Cas9 methods can result in valid strategies to generate a population of mutants for the MS trait (Jacobs et al., 2017; Meng et al., 2017; Liu et al., 2019; Liu et al., 2020). The application of CRISPR/Cas technologies for generating mutants with a male sterile phenotype is an effective tool, mainly mediating a knock-out approach towards target GMS genes with nuclear origin since, compared to cytoplasmic male sterile lines, it is much easier and more useful to produce hybrid seeds (Qi et al., 2020).
Generally, MS mutants result from mutations in target genes involved in microsporogenesis and/or microgametogenesis. Meiosis-related, tapetum-specific and transcription regulatory genes, such as eme1/exs, tpd1, ams and ms1, have been elucidated as key candidate genes involved in these biological processes (Canales et al., 2002; Zhao et al., 2002; Yang et al., 2003). Furthermore, many Arabidopsis transcription factors (TFs) genes, such as MYB103, DYT1, TDF1, AMS, bHLH10, bHLH89 and bHLH91, have been investigated as direct controllers of pollen development (Sorensen et al., 2003; Zhang et al., 2006; Zhang et al., 2007; Zhu et al., 2008; Zhu et al., 2015; Pan et al., 2020). The molecular and functional information has been then easily transferred from models to crops, as reported for Arabidopsis and rice, in which two analogous pathways regulating pollen and tapetum development have been identified in previous research (Fu et al., 2014; Jeong et al., 2014; Zhu et al., 2015; Mishra et al., 2018). In tomato, two homologous genes have been identified as regulators of tapetum and pollen formation. The first, SlMS10 (Solyc02g079810_ms1035) gene, encoding a basic helix-loop-helix TF (bHLH) and homologue to AtDYT1 and OsUDT1, carries both PCD and meiosis alteration in the tapetum during microsporogenesis (Jeong et al., 2014). Its editing has confirmed that SlMS10 is a possible good target candidate for male sterility induction since its knockout mediated by the CRISPR/Cas9 system conferred a male sterility phenotype (Jung et al., 2020). Recently, Liu and colleagues demonstrated that the creation of a mutation in ms1035 by CRISPR/Cas9 technology in association with its linkage marker genes led to marker use for creating mutants exhibiting complete male sterility and recognition during the early developmental stage, confirming promising application possibilities in the production of hybrid seeds (Liu et al., 2021a). The second, Solyc01g081100, homologous to the AtbHLH10/89/90 and OsEAT1 genes, is a candidate gene for the male sterile 32 (ms32) mutant, a locus affecting tapetum and pollen development, and for this reason, it is suggested as a good target for gene editing to quickly develop such lines of interest (Liu et al., 2019). Furthermore, knockout by CRISPR/Cas9 of the SlAMS gene, encoding another basic helix-loop-helix (bHLH) TF, caused downregulation leading to abnormal pollen development, which in turn decreased pollen viability and subsequently generated a male-sterile phenotype (Bao et al., 2022). Recently, other tomato CRISPR/Cas9-edited lines with male sterility phenotypes were obtained by knock-out of SlPHD_ms1 (Solyc04g008420), encoding a PHD-type TF involved in pollen formation and tapetum development, suggesting a key role for SlPHD in male sterility and aiding research into the regulatory processes of pollen and tapetum growth in tomato (Gökdemir et al., 2022). With analogous purposes, CRISPR/Cas technology was also applied in other horticultural crops, as demonstrated in cucurbit species. For example, the knockout of eIF4E by CRISPR/Cas9 in melon highlighted for the first time the association between eIF4E editing and the development of male sterility (Pechar et al., 2022). In watermelon (Citrullus lanatus L.), knockout of ClATM1 by CRISPR/Cas9 causes male sterility, confirming its self-regulatory activity and providing new insights into the molecular mechanism underlying anther development (Zhang et al., 2021b).
In monocots, several CRISPR/Cas systems for producing MS have been reported as successful applications in precision breeding. An improved CRISPR/Cas9 system was driven by the TaU3 RNA polymerase III U3 promoter, and three homologous alleles expressing the wheat redox enzyme NO POLLEN 1 (NP1) were altered to produce totally male-sterile wheat mutants (Li et al., 2020). Furthermore, with recent molecular identification of the Ms1 gene and exploiting strategies related to transgenerational gene editing (see below section 5.2), it has been possible to extend the use of the CRISPR/Cas9 system to generate Ms1 knockout wheat lines that exhibit male sterility in the first generation, demonstrating the utility of the CRISPR/Cas9 system for the rapid generation of nuclear male sterility in hexaploid species like wheat (Okada et al., 2019). Chen and colleagues created a CRISPR/Cas9 vector in maize to target the male sterility gene 8 (Ms8). The resulting mutant was male-sterile, which was compatible with Mendelian genetic rules and was stably acquired by subsequent generations (Chen et al., 2018b). Furthermore, editing ZmMTL (ZmPLA1) with the CRISPR/Cas9 system has produced maternal haploid inducers with powerful haploid identification markers useful for breeding doubled-haploid crops, such as maize itself (Dong et al., 2018). Additionally, ZmMS26, a known nuclear fertility gene (Loukides et al., 1995; Djukanovic et al., 2013) that is conserved in other monocots, like rice, wheat, and sorghum (Cigan et al., 2017), was subjected to precision editing: targeted mutagenesis of MS26 utilizing the modified I-CreI homing endonuclease or CRISPR/Cas9 resulted in the generation of new ms26 male sterile lines (Djukanovic et al., 2013; Svitashev et al., 2015; Qi et al., 2020). In rice, gene knockout by CRISPR/Cas9 of the OsHXK5 gene resulted in male sterility, contributing to demonstration that OsHXK5 contributes to a large portion of the hexokinase activity necessary for the starch utilization pathway during pollen germination and tube growth, as well as for starch biosynthesis during pollen maturation (Lee et al., 2020).
The success of CRISPR/Cas technology application has also been provided in EGMS conditions, or rather the ability to switch from fertile to sterile conditions and vice versa, by adjusting environmental variables such as temperature and photoperiod. Great progress has been recently achieved in the understanding of PGMS or TGMS traits in cereal crops, and several genes controlling P/TGMS traits have been investigated and transferred, mediating conventional breeding and/or biotechnological transformation, in specific lines on which more than 30% of cereal hybrid production depends in China (Ding et al., 2012; Zhou et al., 2012; Zhang et al., 2013; Huang et al., 2014; Zhou et al., 2014). Several studies elucidated the molecular genetic mechanisms at the base of EMGS, confirming also the interesting role assumed by phasiRNAs (phased small-interfering RNAs) generated by long-noncoding RNAs. In rice, for example, the phasiRNAs originated from PMS1T locus regulates PSMS in rice (Fan et al., 2016). Especially in rice, in the last few years, many genes influencing PGMS or TGMS traits have been discovered and cloned, and several reports describe different CRISPR/Cas-based approaches to obtain photo- and thermosensitive male-sterile lines. For example, a simple and efficient rice TGMS cultivation system using CRISPR/Cas9 editing technology was proposed to knock out the TMS5 (thermosensitive genic male-sterile 5) gene target, with great value in new commercial “transgene free” TGMS rice lines (Zhou et al., 2016). TMS5 is a nuclear recessive gene that controls the TGMS trait and extensively used in two-line hybrid rice breeding. It was the first spontaneously mutated Oryza sativa ssp. indica, identified more than 30 years ago, and encodes an RNase ZS1 endonuclease, able to degrade the temperature-sensitive ubiquitin fusion ribosomal protein L40 (UbL40) mRNA (Zhou et al., 2014). A study found that when plants were grown under a high temperature regime, several tms5 mutants developed in a background of the japonica type showed a high degree (85.3%) of pollen sterility (Zhou et al., 2016), confirming that targeted modification of TMS5 by the CRISPR/Cas9 system is a successful approach to develop TGMS lines for hybrid rice production. Huang et al. targeted the TMS5 gene, producing a mutant that was entirely male-sterile at high temperatures but male-fertile at low temperatures, with a pollen fertility transition temperature fixed at 28°C. (Huang et al., 2014). Recent studies revealed the molecular mechanism of tms5 leading to male sterility in rice to easily obtain excellent TGMS lines (Fang et al., 2022) and potentially applicable in other crops. CRISPR/Cas9-engineered mutation of TMS5 also resulted in the formation of thermosensitive male sterility in maize (Li et al., 2017). In addition, Li et al. altered the carbon starvation CSA gene in pollen grains of the rice variety ‘Kongyu 131’ and found that the csa mutant had a male-sterile phenotype in short-day and a male-fertile phenotype in long-day conditions. (i.e., photosensitive nuclear male sterile mutant) (Li et al., 2016), whereas in tomato, Shen and colleagues generated photosensitive/thermosensitive male-sterile lines by using CRISPR/Cas9 modifying the genic male-sterile 2-2 (PTGMS2-2) gene (Liu et al., 2019).
As described in the previous sections, site-specific genome editing by CRISPR/Cas9 technology is becoming a progressively more successful tool for functional, basic and applied plant research because it can generate a high rate of mutation while being relatively easy to use (Zhang et al., 2013; Lowder et al., 2015; Ma et al., 2015; Wang et al., 2016). Numerous methods have been used to create CRISPR-edited plants devoid of CRISPR constructs and other transgenes because the lack of any transgenes in gene-edited plants is a requirement for the commercialization of any CRISPR-edited plants with stable valuable traits. For public approval, gene elimination or bypassing alien elements to edit endogenous genes is fundamental and could be a strategic approach, even if transgenic intermediates are transiently necessary (Figure 2B). The main different strategies useful to avoid the maintenance of transgene integration have been deeply described by He and Zhao, 2020 (He and Zhao, 2020). Commonly, after CRISPR-mediated mutagenesis, the Cas9 gene and associated DNA sequences are eliminated through genetic segregation, which frequently allays public concerns about genetically modified individuals. The biggest advantage of the method is that it could allow the selection of plants that no longer contain the T-DNA sequence, producing plant materials not containing any foreign DNA even though they were produced using transgenic technology mediating stable transformation methodologies. However, the fact that many commercial crop varieties are polyploid, heterozygous, or asexually reproduced complicates these efforts. Many commercial cultivars’ genome complexity, long juvenile phase, and/or self-incompatibility limit the development of CRISPR-mediated transgenic crops since backcrossing is required to remove the CRISPR transgene.
In the past, plant transient transformation technology has been widely used as an alternative approach to facilitate rapid and efficient gene function analysis (Sheen, 2001; Chen et al., 2006). Using transient transformation methods, such as particle bombardment (Romano et al., 2003), transient transformation by Agrobacterium sp. (Cui et al., 2017) and polyethylene glycol (PEG)-mediated protoplast transfection (Cankar et al., 2022), excellent results in plant research have been achieved. Among these, the protoplast transient expression system has played a relevant role in genomics and proteomics research, resulting in a potential, rapid, and convenient technique for testing new technologies, such as GE approaches. In general, transient expression methods for protoplasts have been designed for many crop species, including monocots, dicots, herbaceous and woody species, such as rice (Yang et al., 2014a), barley (Bai et al., 2014), corn (Cao et al., 2014), apple (Maddumage et al., 2002), and grapevine (Zhao et al., 2016). These findings demonstrate the possibility and feasibility of utilizing protoplasts for CRISPR-mediated gene editing, particularly in species with a protracted juvenile phase, heterozygosity, or asexual propagation. Likewise, this strategy could represent the most feasible way to directly apply CRISPR-mediated DNA-free genome editing technologies for improving traits and increasing commercial value, as already experimentally confirmed for food and non-food crops, such as strawberry (Martin-Pizarro et al., 2019; Wilson et al., 2019), potato (Gonzalez et al., 2019; Nicolia et al., 2021; Zhao et al., 2021), lettuce (Woo et al., 2015), chicory (De Bruyn et al., 2020; Cankar et al., 2022), Nicotiana tabacum (Lin et al., 2018; Hsu et al., 2019) and Brassica oleracea (Lee et al., 2020; Hsu et al., 2021), and ornamental species, as petunia (Yu et al., 2021b). For these reasons, protoplast transient expression systems represent a promising and valid approach for generating CRISPR-edited DNA-free plant material and MS mutant production (Figure 2B). Numerous studies describe the different gene modification methods using transient expression of the Cas protein and associated sgRNA, mediating the main delivery methods into somatic plant cells, which may be done either as DNA vectors, through Agrobacterium infiltration (Chen et al., 2018a), or as ribonucleoprotein (RNP), using biolistic delivery (Liang et al., 2018), nanotubes (Demirer et al., 2019), virus transfection (Ellison et al., 2020), PEG-calcium (PEG–Ca2+) (Toda et al., 2019). Because there is no foreign DNA present during transfection, direct transfection of the RNP complex eliminates the risk of plasmid DNA insertions into the plant genome (Andersson et al., 2018). Genome editing is realizable utilizing protoplasts without the insertion of foreign CRISPR DNA and without the necessity for hybridization, introgression, or back-crossing of progeny in the T0 generation. Furthermore, protoplasts are single cells that are edited before the first cell division: new plants grow from a single modified protoplast, ensuring that all cells share the same genetic background and that edited alleles are passed down to the next generation However, RNP-mediated genome editing has been employed successfully in many plant species, targeting genes with agronomic interest, involved disease resistance (Malnoy et al., 2016), in grain yield (Toda et al., 2019), nutritional composition (Andersson et al., 2018), and male fertility (Svitashev et al., 2016). MS induction, using an analogous approach, has been successfully achieved only in maize. Svitashev and colleagues demonstrated the success of their research, in which two male fertility nuclear genes (MS26 and MS45) were targeted by purified Cas9 protein preassembled with in vitro transcribed gRNAs, demonstrating DNA-free genome editing in a major crop species using biolistically delivered Cas9–gRNA RNPs on immature embryos and subsequent plant regeneration (Svitashev et al., 2016). These positive results suggest the potential of applying similar methodologies in other large crops to increase the number of examples of male sterile lines CRISPR-edited DNA-free by transient transformation system by RNP complex.
Because CRISPR/Cas9 expression cassettes and target sites are distributed throughout the genome, segregation and deletion of CRISPR/Cas9 cassettes is conceivable through subsequent selfing or crossing (Figure 2B). However, in crops with a high level of genome complexity, highly heterozygous, polyploid genomes, and usually propagated vegetatively, this is not easily achieved. Specifically, efficient propagation and stacking of first-generation mutations becomes increasingly difficult or nearly impossible with polyploidy.
Numerous new strategies have been developed to extend the CRISPR toolbox, and many of these new schemes could also take advantage from transgenerational gene editing (TGE)-based strategies, defined as the continued ability of Cas9 to edit also after cross: this means that if the Cas9 nuclease is still active, after cross it will encounter a new WT allele, which can be edited to create independent alleles. TGE has been utilized for a variety of applications, some of which are not always defined as TGE, such as the editing new alleles in polyploid crops, the creating allelic variation, and the editing target genes in refractory genetic backgrounds (Impens et al., 2022). Mutations are frequently found only in a fraction of the homoeoalleles targeted by the same sgRNA in polyploid crops such as hexaploid common wheat (Triticum aestivum) and tetraploid cotton (Gossypium hirsutum) (Wang et al., 2018a; Wang et al., 2018b; Wang et al., 2018c). While expressing CRISPR/Cas9 for more than one generation during TGE promotes on-target homoeoallele editing, it does not always boost off-targeting.
On the basis of TGE, with the purpose of accelerating the understanding of MS and ensuring speedy improvement, a new approach (Ramadan et al., 2021) was tested for example in cotton system, in which the use of pooled sgRNAs targeting single or duplicated genes belonging to different families provided a large number of intentional mutants that would help us know male sterility in cotton itself. Furthermore, this strategy ensured a rapid characterization of the key genes which may influence fertility in cotton, with important consequences for cotton future genetic improvement (Ramadan et al., 2021). Furthermore, as previously mentioned, a TGE-based methodology was implemented to facilitate the ongoing modification of homoeoalleles in species like hexaploid wheat, which is not easily amenable to conventional mutagenesis techniques. In this approach, a transgenic line may exhibit modifications in only a subset of homoeoalleles. However, through self-crossing and careful selection of plants inheriting the T-DNA, it becomes possible to modify all homoeoalleles. Singh and colleagues proposed an effective utilization of the CRISPR/Cas system and next-generation sequencing for mutant analysis in wheat. They successfully established the role of TaMs26 in wheat pollen generation by combining mutations in TaMs26 from the A-, B-, and D-genomes through crossing, resulting in the development of male sterile plants (Singh et al., 2017). Orthologous Ms26 mutations in rice and sorghum plants, as in maize, confer a recessive male sterile phenotype, and restoration of fertility in these mutant sorghum plants was achieved by a copy of maize Ms26 (Cigan et al., 2017). Afterwards, with recent molecular identification of the male fertility Ms1 gene, it has been possible to extend the use of the CRISPR/Cas9 system to generate Ms1 knockout wheat lines with male sterility in the first generation, demonstrating the potential of the CRISPR/Cas9 system for the fast generation of GMS in hexaploid wheat (Singh et al., 2018; Okada et al., 2019).
This evidence on transgenerational gene editing activity demonstrates that TGE can contribute to novel variation in the offspring of CRISPR/Cas9-expressing plants, and that Cas9-inducible trait can be transferred by crossing the plants expressing the gene editing constructs with the lines of interest.
The interest in obtaining MS lines by molecular precision breeding mediated by the CRISPR/Cas system has been generally described as a fundamental step for the production of F1 hybrids in horticultural crops. In contrast, in ornamental plant research, this aspect has not been deeply investigated to date, despite an increasing number of studies on potential CRISPR/Cas system applications in precision breeding in ornamental plants being continually tested for improving several traits. In fact, in ornamental species, where traits such as high heterozygosity, large genomes, high chromosome numbers, polyploidy, long life cycles, self-sterility, or the inability to produce seeds frequently limit the applicability of conventional breeding methods, genome editing approaches are particularly desirable (Azadi et al., 2016; Sharma and Messar, 2017). Furthermore, obtaining nontransgenic first-generation altered plants and permitting the development of foreign DNA-free editing approaches would be extremely beneficial in such instances. However, the potential of using such methodologies in ornamental species breeding is dependent on information on the availability of efficient transformation and regeneration protocols, as well as the structure of plant genomes and function of genes. In recent decades genome sequencing technology played a significant role, allowing site-specific mutagenesis approaches on several key genes controlling traits of high interest and suggesting that CRISPR/Cas9-induced mutagenesis is effective also in ornamental sector (Zhang et al., 2016a; Kishi-Kaboshi et al., 2017; Yan et al., 2019; Yu et al., 2021b). In fact, it has been successfully employed to create gene knockouts and induce genetic alterations in ornamental Petunia inflate and Petunia hybrid (Subburaj et al., 2016; Zhang et al., 2016a; Sun and Kao, 2018; Yu et al., 2021b; Xu et al., 2022), Chrysanthemum morifolium (Kishi-Kaboshi et al., 2017), Dendrobium officinale (Kui et al., 2017), Ipomoea nil (Watanabe et al., 2017), Lilium longiflorum and Lilium pumilum (Yan et al., 2019), and Phalaenopsis equestris (Tong et al., 2020). In particular, in polyploid species, such as chrysanthemum, the possibility of mutating multiple copies of a target gene has been indirectly shown, as demonstrated in other polyploid crops, e.g., hexaploid wheat (Wang et al., 2014; Mekapogu et al., 2022).
Regarding MS induction, the production of male-sterile ornamental plants is of great interest for many purposes, such as facilitating hybrid seed production, eliminating pollen allergens (i.e., gene escape), reduce the need for deadheading to extend the flowering period, redirect resources from seeds to vegetative growth and increase flower longevity and self-life (Garcia-Sogo et al., 2010). In past decades, the production of engineered male sterile plants by canonical transgenesis approaches was documented in ornamental Kalanchoe blossfeldiana through the directed expression of the ribonuclease Barnase gene under control of the PsEND1 promoter, which determines tissue-specific expression of the Barnase gene in anther tissues (epidermis, endothecium, middle layer, connective). The Barnase gene affected normal anther development, inducing the ablation of specific tissues at early stages of anther development with a consequent lack of pollen at anthesis in transgenic flowers (Garcia-Sogo et al., 2010). The use of this technology was especially useful to produce environmentally friendly transgenic ornamentals carrying new traits, as this modification would prevent gene flow between the genetically modified plants and related species (Roque et al., 2007; Gardner et al., 2009). A similar approach was used to efficiently create male sterile versions of existing Pelargonium spp. cultivars, which represent one of the most popular garden plants around the world, have considerable economic importance in the market of ornamental plants. Using a cotransformation protocol, two new traits were introduced in P. zonale, one to produce long-life plants by inducing the IPT gene during plant senescence and the other to produce male sterile plants without pollen (Garcia-Sogo et al., 2012). With similar molecular strategies and related purposes, male sterility was induced in Chrysanthemums spp. In this specific case, since many wild chrysanthemum relatives in the Compositae family are cross-compatible with chrysanthemum cultivars, to reduce the possibility of transgene flow into wild relatives, a male sterility trait using the mutated ethylene receptor gene Cm-ETR1/H69A was introduced into chrysanthemum cultivars (Shinoyama et al., 2012). Recently, thanks to the release of whole genome sequence information (Hirakawa et al., 2019), Shinoyama et al. (Shinoyama et al., 2020) reported an important example of MS induction in Chrysanthemums spp. by a genome editing approach targeting the CmDMC1 gene through the use of TALENS technology to knock out all six identified CmDMC1 genes. Two chrysanthemum cultivars with the TALEN expression vector resulted in the development of lines with disruption of all CmDMC1 loci, successfully inducing male and female sterility (Shinoyama et al., 2020). The interest in creating MS lines in ornamental species, together with the positive results obtained in some of them, supports the idea of implementing CRISPR/Cas-based technologies as a potential tool for genetic improvement in floricultural research.
Conventional breeding approaches still depend on breeders choosing materials based on phenotypic analyses. Breeders and scientists choose purposefully different parents to produce crop varieties that combine the desired characteristics of both parents. However, the usefulness of traditional breeding methods may be restricted to complex traits. To complement traditional breeding techniques, molecular breeders have developed and applied GE technologies, which should supplement rather than replace traditional breeding methods. Generally, two major criteria should be considered while assessing the applicability and future development of GE technology. Firstly, the development of cost-effective, low-risk, and efficient transformation systems that align with agricultural requirements is crucial in expanding the utilization of this molecular techniques. Secondly, the regulatory practices implemented by governments play a pivotal role. Currently, there is a global debate around whether CRISPR-edited lines should undergo similar regulations as conventional genetically modified (GM) plants, or if they should be allowed to enter the market without regulation once the CRISPR-cassette mediating segregating cycles have been removed (Chen and Gao, 2014; Voytas and Gao, 2014; Gao, 2015).
This review aims to emphasize that these approaches could make available potential and alternative methods for many breeding purposes. Several examples report that CRISPR/Cas technology has thus far been proven to be successful in genome editing of numerous food and non-food crops, as well as ornamental plants, whose genomes have been efficiently modified to induce genetic variability, resulting in a strong tool in plant genetics and precision breeding. The use of these modification tools, in comparison to their adaptability and final use, has provided a remarkable breakthrough in biological applications thanks to a growing number of accessible genome sequencing data related to the reduction in sequencing costs. In this intricate scenario, this review provides an overview of recent successes for MS induction based on GE applications, accelerating and lowering the cost of male sterility induction by targeting known candidate functional loci. The following development of male sterility, especially in food crops, has been greatly investigated for seed hybrid production. Conditional MS mutants, for example, created through genome editing, are particularly useful in major crops such as rice and maize, opening the possibility for applying the idea to many other crops Interestingly an increasing number of new additional applications of GE technology for MS producing have been reported, especially in promising ornamental species, in which the final goal of obtaining a MS ornamental species arises from the need to have allergenic free plant material. Furthermore, because only a few nucleotides are changed to modify the genome, the new improved methods based on simultaneous editing of gene sequences could be an important starting point for the development of new elite varieties by utilizing efficient and specific modifications at genomic loci, offering advantages over GM crops. Many of these new tools also benefit from TGE-based methods for editing additional alleles in polyploid species. Additionally, according to the studies mentioned above, the transient expression of the CRISPR/Cas cassette, and in particular the direct transfection of the RNP complex, exhibits a number of benefits compared to DNA plasmid delivery, followed by stable integration. With a transient approach, we have a DNA-free transfer, eliminating the possibility of unintended recombinant DNA insertion into the plant genome, a bypass of the cell’s transcriptional and translational machinery, with an immediate activity of the RNP complex per single cell, and finally a quick breakdown of complexes after delivery, which lowers the incidence of mosaicism effects. For these reasons, they are regarded as the most innovative and the new frontier of precision plant breeding programs, and this is the strategic direction that breeding could take in the future, supporting the idea that these approaches could be the new strategic assisted evolution technology towards reproductive systems, with potential to form new varieties.
Conceptualization, SF and GB. Investigation and resources, SF, SD, and AB. Data curation, SF and FP. Writing—original draft preparation, SF, SD, and AB. Writing—review and editing, SF, FP, and AV. Visualization, SF, FP, AV, and GB. Supervision, ML and GB. Project administration, GB. Funding Acquisition, GB. All authors contributed to the article and approved the submitted version.
This study was carried out within the research contract signed by the Gruppo Padana S.S. company (Paese, TV, Italy), and Department of Agronomy, Food, Natural resources, Animals and Environment (DAFNAE), University of Padua (Italy), within action IV.5_GREEN, PON 2014–2021.
The authors would like to thank the company Gruppo Padana S.S. (Referent person Marco Gazzola) for funding part of the PhD program of AB and the RTDA research project of SF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbas, A., Yu, P., Sun, L., Yang, Z., Chen, D., Cheng, S., et al. (2021). Exploiting genic Male sterility in rice: from molecular dissection to breeding applications. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.629314
Ahmadikhah, A., Karlov, G. I. (2006). Molecular mapping of the fertility-restoration gene Rf4 for WA-cytoplasmic males terility in rice. Plant Breed 125, 363–367. doi: 10.1111/j.1439-0523.2006.01246.x
Aklilu, E. (2021). Review on forward and reverse genetics in plant breeding. All Life 14, 127–135. doi: 10.1080/26895293.2021.1888810
An, X., Dong, Z., Tian, Y., Xie, K., Wu, S., Zhu, T., et al. (2019). ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant. 12(3), 343–359. doi: 10.1016/j.molp.2019.01.011
Andersson, M., Turesson, H., Olsson, N., Falt, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant 164, 378–384. doi: 10.1111/ppl.12731
Arora, L., Narula, A. (2017). Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01932
Aulakh, P. S., Dhaliwal, M. S., Jindal, S. K., Schafleitner, R., Singh, K. (2016). Mapping of male sterility gene ms10 in chilli pepper (Capsicum annuum l.). Plant Breed 135, 531–535. doi: 10.1111/pbr.12389
Azadi, P., Bagheri, H., Nalousi, A. M., Nazari, F., Chandler, S. F. (2016). Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol. Adv. 34, 1073–1090. doi: 10.1016/j.biotechadv.2016.06.006
Bai, Y., Han, N., Wu, J. X., Yang, Y. N., Wang, J. H., Zhu, M. Y., et al. (2014). A transient gene expression system using barley protoplasts to evaluate microRNAs for post-transcriptional regulation of their target genes. Plant Cell Tiss Org 119, 211–219. doi: 10.1007/s11240-014-0527-z
Bao, H., Ding, Y., Yang, F., Zhang, J., Xie, J., Zhao, C., et al. (2022). Gene silencing, knockout and over-expression of a transcription factor ABORTED MICROSPORES (SlAMS) strongly affects pollen viability in tomato (Solanum lycopersicum). BMC Genomics 23, 346. doi: 10.1186/s12864-022-08549-x
Barcaccia, G., Tiozzo, C. S. (2012). New Male Sterile Cichorium Spp. mutant, parts or derivatives, where male sterility is due to a recessive nuclear mutation linked to a polymorphic molecular marker, useful for producing F1 hybrids of cichorium spp. EU Patent No. WO2012163389-A1.
Barcaccia, G., Ghedina, A., Lucchin, M. (2016). Current advances in genomics and breeding of leaf chicory (Cichorium intybus l.). Agriculture 6, 6–50. doi: 10.3390/agriculture6040050
Barman, H. N., Sheng, Z., Fiaz, S., Zhong, M., Wu, Y., Cai, Y., et al. (2019). Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 19, 109. doi: 10.1186/s12870-019-1715-0
Bartoszewski, G., Waszczak, C., Gawronski, P., Stepien, I., Bolibok-Bragoszewska, H., Palloix, A., et al. (2012). Mapping of the ms8 male sterility gene in sweet pepper (Capsicum annuum l.) on the chromosome P4 using PCR-based markers useful for breeding programmes. Euphytica 186, 453–461. doi: 10.1007/s10681-012-0637-9
Begemann, M., Gray, B., January, E., Singer, A., Kesler, D., He, Y., et al. (2017). Characterization and validation of a novel group of type V, class 2 nucleases for in vivo genome editing. bioRxiv, 192799. doi: 10.1101/192799
Bentolila, S., Zethof, J., Gerats, T., Hanson, M. R. (1998). Locating the petunia Rf gene on a 650-kb DNA fragment. Theor. Appl. Genet. 96, 980–988. doi: 10.1007/s001220050829
Bhat, M. A., Bhat, M. A., Kumar, V., Wani, I. A., Bashir, H., Shah, A. A., et al. (2020). The era of editing plant genomes using CRISPR/Cas: a critical appraisal. J. Biotechnol. 324, 34–60. doi: 10.1016/j.jbiotec.2020.09.013
Bohra, A., Jha, U. C., Adhimoolam, P., Bisht, D., Singh, N. P. (2016). Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep. 35, 967–993. doi: 10.1007/s00299-016-1949-3
Bonhomme, S., Budar, F., Férault, M., et al. (1991). A 2.5 kb NcoI fragment of ogura radish mitochondrial DNA is correlated with cytoplasmic male-sterility in brassica cybrids. Curr. Genet. 19, 121–127. doi: 10.1007/BF00326293
Borrelli, V. M. G., Brambilla, V., Rogowsky, P., Marocco, A., Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01245
Bruns, H. A. (2017). Southern corn leaf blight: a story worth retelling. Agron. J. 109, 1218–1224. doi: 10.2134/agronj2017.01.0006
Budar, F., Pelletier, G. (2001). Male Sterility in plants: occurrence, determinism, significance and use. C R Acad. Sci. III 324, 543–550. doi: 10.1016/s0764-4469(01)01324-5
Cadalen, T., Morchen, M., Blassiau, C., Clabaut, A., Scheer, I., Hilbert, J. L., et al. (2010). Development of SSR markers and construction of a consensus genetic map for chicory (Cichorium intybus l.). Mol. Breed. 25, 699–722. doi: 10.1007/s11032-009-9369-5
Canales, C., Bhatt, A. M., Scott, R., Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in arabidopsis. Curr. Biol. 12, 1718–1727. doi: 10.1016/s0960-9822(02)01151-x
Cankar, K., Hakkert, J. C., Sevenier, R., Campo, E., Schipper, B., Papastolopoulou, C., et al. (2022). CRISPR/Cas9 targeted inactivation of the kauniolide synthase in chicory results in accumulation of costunolide and its conjugates in taproots. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.940003
Cao, X., Liu, X., Wang, X., Yang, M., Van Giang, T., Wang, J., et al. (2019). B-class MADS-box TM6 is a candidate gene for tomato male sterile-15(26). Theor. Appl. Genet. 132, 2125–2135. doi: 10.1007/s00122-019-03342-z
Cao, J. M., Yao, D. M., Lin, F., Jiang, M. Y. (2014). PEG-mediated transient gene expression and silencing system in maize mesophyll protoplasts: a valuable tool for signal transduction study in maize. Acta Physiologiae Plantarum 36, 1271–1281. doi: 10.1007/s11738-014-1508-x
Carroll, D. (2011). Genome engineering with zinc-finger nucleases. Genetics 188, 773–782. doi: 10.1534/genetics.111.131433
Chaubal, R., Zanella, C., Trimnell, M. R., Fox, T. W., Albertsen, M. C., Bedinger, P. (2000). Two male‐sterile mutants of Zea mays (Poaceae) with an extra cell division in the anther wall. Am. J. Bot. 87(8), 1193–1201. doi: 10.2307/2656657
Chen, D., Ding, Y., Guo, W., Zhang, T. (2009). Molecular mapping of genic male-sterile genesms15,ms5andms6in tetraploid cotton. Plant Breed 128, 193–198. doi: 10.1111/j.1439-0523.2008.01562.x
Chen, K., Gao, C. (2014). Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 33, 575–583. doi: 10.1007/s00299-013-1539-6
Chen, L., Li, W., Katin-Grazzini, L., Ding, J., Gu, X., Li, Y., et al. (2018a). A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 5, 13. doi: 10.1038/s41438-018-0023-4
Chen, L., Liu, Y. G. (2014). Male Sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Chen, Y., Lubberstedt, T. (2010). Molecular basis of trait correlations. Trends Plant Sci. 15, 454–461. doi: 10.1016/j.tplants.2010.05.004
Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M. E., Umemura, K., Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/j.1364-3703.2006.00346.x
Chen, Y., Wang, Z., Ni, H., Xu, Y., Chen, Q., Jiang, L. (2017). CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in arabidopsis. Sci. China Life Sci. 60, 520–523. doi: 10.1007/s11427-017-9021-5
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, R., Xu, Q., Liu, Y., Zhang, J., Ren, D., Wang, G., et al. (2018b). Generation of transgene-free maize Male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01180
Cheng, C., Nie, H., Li, H., Adjibolosoo, D., Li, B., Jiang, K., et al. (2023). Identification of fertility restoration candidate genes from a restorer line R186 for Gossypium harknessii cytoplasmic male sterile cotton. BMC Plant Biol. 23(1):175. doi: 10.1186/s12870-023-04185-z
Chueasiri, C., Chunthong, K., Pitnjam, K., Chakhonkaen, S., Sangarwut, N., et al. (2014). Rice ORMDL controls sphingolipid homeostasis affecting fertility resulting from abnormal pollen development. PloS One 9 (9), e106386. doi: 10.1371/journal.pone.0106386
Cigan, A. M., Singh, M., Benn, G., Feigenbutz, L., Kumar, M., Cho, M. J., et al. (2017). Targeted mutagenesis of a conserved anther-expressed P450 gene confers male sterility in monocots. Plant Biotechnol. J. 15, 379–389. doi: 10.1111/pbi.12633
Colombo, N., Galmarini, C. R. (2017). The use of genetic, manual and chemical methods to control pollination in vegetable hybrid seed production: a review. Plant Breed 136, 287–299. doi: 10.1111/pbr.12473
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cui, M. Y., Wei, W., Gao, K., Xie, Y. G., Guo, Y., Feng, J. Y. (2017). A rapid and efficient agrobacterium-mediated transient gene expression system for strawberry leaves and the study of disease resistance proteins. Plant Cell Tiss Org 131, 233–246. doi: 10.1007/s11240-017-1279-3
Dahan-Meir, T., Filler-Hayut, S., Melamed-Bessudo, C., Bocobza, S., Czosnek, H., Aharoni, A., et al. (2018). Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95, 5–16. doi: 10.1111/tpj.13932
De Bruyn, C., Ruttink, T., Eeckhaut, T., Jacobs, T., De Keyser, E., Goossens, A., et al. (2020). Establishment of CRISPR/Cas9 genome editing in witloof (Cichorium intybus var. foliosum). Front. Genome Ed 2. doi: 10.3389/fgeed.2020.604876
Demirer, G. S., Zhang, H., Goh, N. S., Gonzalez-Grandio, E., Landry, M. P. (2019). Carbon nanotube-mediated DNA delivery without transgene integration in intact plants. Nat. Protoc. 14, 2954–2971. doi: 10.1038/s41596-019-0208-9
Dewey, R. E., Timothy, D. H., Levings, C. S., 3rd. (1991). Chimeric mitochondrial genes expressed in the c male-sterile cytoplasm of maize. Curr. Genet. 20 (6), 475–482. doi: 10.1007/BF00334775
Dill, C. L., Wise, R. P., Schnable, P. S. (1997). Rf8 and rf* mediate unique T-urf13-transcript accumulation, revealing a conserved motif associated with RNA processing and restoration of pollen fertility in T-cytoplasm maize. Genetics 147, 1367–1379. doi: 10.1093/genetics/147.3.1367
Ding, J., Lu, Q., Ouyang, Y., Mao, H., Zhang, P., Yao, J., et al. (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. 109, 2654–2659. doi: 10.1073/pnas.1121374109
Djukanovic, V., Smith, J., Lowe, K., Yang, M., Gao, H., Jones, S., et al. (2013). Male-Sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J. 76, 888–899. doi: 10.1111/tpj.12335
Dong, L., Li, L., Liu, C., Liu, C., Geng, S., Li, X., et al. (2018). Genome editing and double-fluorescence proteins enable robust maternal haploid induction and identification in maize. Mol. Plant 11, 1214–1217. doi: 10.1016/j.molp.2018.06.011
Dong, O. X., Ronald, P. C. (2019). Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. 180, 26–38. doi: 10.1104/pp.18.01224
Dong, N. V., Subudhi, P. K., Luong, P. N., Quang, V. D., Quy, T. D., Zheng, H. G., et al. (2000). Molecular mapping of a rice gene conditioning thermosensitive genic male sterility using AFLP, RFLP and SSR techniques. Theor. Appl. Genet. 100, 727–734. doi: 10.1007/s001220051345
Ducos, E., Touzet, P., Boutry, M. (2001). The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes. Plant J. 26 (2), 171–180. doi: 10.1046/j.1365-313x.2001.01017.x
Ellison, E. E., Nagalakshmi, U., Gamo, M. E., Huang, P. J., Dinesh-Kumar, S., Voytas, D. F. (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 6, 620–624. doi: 10.1038/s41477-020-0670-y
Fan, Y., Yang, J., Mathioni, S. M., Yu, J., Shen, J., Yang, X., et al. (2016). PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA 113, 15144–15149. doi: 10.1073/pnas.1619159114
Fang, Y., Yang, J., Guo, X., Qin, Y., Zhou, H., Liao, S., et al. (2022). CRISPR/Cas9-induced mutagenesis of TMS5 confers thermosensitive genic Male sterility by influencing protein expression in rice (Oryza sativa l.). Int. J. Mol. Sci. 23 (15), 8354. doi: 10.3390/ijms23158354
Fang, X., Sun, X., Yang, X., Li, Q., Lin, C., Xu, J., et al. (2021). MS1 is essential for male fertility by regulating the microsporocyte cell plate expansion in soybean. Sci. China Life Sci. 64, 1533–1545. doi: 10.1007/s11427-021-1973-0
Feng, J., Jan, C. C. (2008). Introgression and molecular tagging of Rf (4), a new male fertility restoration gene from wild sunflower helianthus maximiliani l. Theor. Appl. Genet. 117, 241–249. doi: 10.1007/s00122-008-0769-4
Fernandez Gomez, J., Wilson, Z. A. (2014). A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12, 765–777. doi: 10.1111/pbi.12181
Forde, B. G., Oliver, R. J., Leaver, C. J. (1978). Variation in mitochondrial translation products associated with male-sterile cytoplasms in maize. Proc. Natl. Acad. Sci. USA 75 (8), 3841–3845. doi: 10.1073/pnas.75.8.3841
Fu, Z., Yu, J., Cheng, X., Zong, X., Xu, J., Chen, M., et al. (2014). The rice basic helix-Loop-Helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26, 1512–1524. doi: 10.1105/tpc.114.123745
Fujii, S., Toriyama, K. (2009). Suppressed expression of retrograde-regulated Male sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc. Natl. Acad. Sci. U.S.A. 106, 9513–9518. doi: 10.1073/pnas.0901860106
Gaillochet, C., Develtere, W., Jacobs, T. B. (2021). CRISPR screens in plants: approaches, guidelines, and future prospects. Plant Cell 33, 794–813. doi: 10.1093/plcell/koab099
Gao, C. X. (2015). Genome editing in crops: from bench to field. Natl. Sci. Rev. 2, 13–15. doi: 10.1093/nsr/nwu054
Gao, L., Cox, D. B. T., Yan, W. X., Manteiga, J. C., Schneider, M. W., Yamano, T., et al. (2017). Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 35, 789–792. doi: 10.1038/nbt.3900
Garcia-Sogo, B., Pineda, B., Castelblanque, L., Anton, T., Medina, M., Roque, E., et al. (2010). Efficient transformation of kalanchoe blossfeldiana and production of male-sterile plants by engineered anther ablation. Plant Cell Rep. 29, 61–77. doi: 10.1007/s00299-009-0798-8
Garcia-Sogo, B., Pineda, B., Roque, E., Anton, T., Atares, A., Borja, M., et al. (2012). Production of engineered long-life and male sterile pelargonium plants. BMC Plant Biol. 12, 156. doi: 10.1186/1471-2229-12-156
Gardner, N., Felsheim, R., Smith, A. G. (2009). Production of male- and female-sterile plants through reproductive tissue ablation. J. Plant Physiol. 166, 871–881. doi: 10.1016/j.jplph.2008.10.002
Gentzbittel, L., Mestries, E., Mouzeyar, S., Mazeyrat, F., Badaoui, S., Vear, F., et al. (1999). A composite map of expressed sequences and phenotypic traits of the sunflower (Helianthus annuus l,) genome. Theor. Appl. Genet. 99, 218–234. doi: 10.1007/s001220051228
Geyer, M., Bund, A., Albrecht, T., Hartl, L., Mohler, V. (2016). Distribution of the fertility-restoring gene Rf3 in common and spelt wheat determined by an informative SNP marker. Mol. Breed. 36, 167. doi: 10.1007/s11032-016-0592-6
Gökdemir, G., Seçgin, Z., Uluisik, S., Kavas, M. (2022). CRISPR/Cas9 knock-out of SlPHD_MS1 (Solyc04g008420) gene results in complete male sterility in tomato. Plant Growth Regul. 98, 329–341. doi: 10.1007/s10725-022-00869-y
Gonthier, L., Blassiau, C., Morchen, M., Cadalen, T., Poiret, M., Hendriks, T., et al. (2013). High-density genetic maps for loci involved in nuclear male sterility (NMS1) and sporophytic self-incompatibility (S-locus) in chicory (Cichorium intybus l., asteraceae). Theor. Appl. Genet. 126, 2103–2121. doi: 10.1007/s00122-013-2122-9
Gonzalez, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Falt, A. S., et al. (2019). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01649
Gorguet, B., Schipper, D., Van Heusden, A. W., Lindhout, P. (2006). High-resolution fine mapping of ps-2, a mutated gene conferring functional male sterility in tomato due to non-dehiscent anthers. Theor. Appl. Genet. 113, 1437–1448. doi: 10.1007/s00122-006-0389-9
Gorguet, B., Schipper, D., van Lammeren, A., Visser, R. G., van Heusden, A. W. (2009). Ps-2, the gene responsible for functional sterility in tomato, due to non-dehiscent anthers, is the result of a mutation in a novel polygalacturonase gene. Theor. Appl. Genet. 118, 1199–1209. doi: 10.1007/s00122-009-0974-9
Hawkes, T., Pline-Srnic, W., Dale, R., Friend, E., Hollinshead, T., Howe, P., et al. (2011). D-glufosinate as a male sterility agent for hybrid seed production. Plant Biotechnol. J. 9, 301–314. doi: 10.1111/j.1467-7652.2010.00549.x
He, Y. H., Ning, G. G., Sun, Y. L., Hu, Y., Zhao, X. Y., Bao, M. Z. (2010). Cytological and mapping analysis of a novel male sterile type resulting from spontaneous floral organ homeotic conversion in marigold (Tagetes erecta l.). Mol. Breed. 26, 19–29. doi: 10.1007/s11032-009-9372-x
He, Y., Zhao, Y. (2020). Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH 1, 88–96. doi: 10.1007/s42994-019-00013-x
Hirakawa, H., Sumitomo, K., Hisamatsu, T., Nagano, S., Shirasawa, K., Higuchi, Y., et al. (2019). De novo whole-genome assembly in chrysanthemum seticuspe, a model species of chrysanthemums, and its application to genetic and gene discovery analysis. DNA Res. 26, 195–203. doi: 10.1093/dnares/dsy048
Hsu, C. T., Cheng, Y. J., Yuan, Y. H., Hung, W. F., Cheng, Q. W., Wu, F. H., et al. (2019). Application of Cas12a and nCas9-activation-induced cytidine deaminase for genome editing and as a non-sexual strategy to generate homozygous/multiplex edited plants in the allotetraploid genome of tobacco. Plant Mol. Biol. 101, 355–371. doi: 10.1007/s11103-019-00907-w
Hsu, C.-T., Yuan, Y.-H., Lin, Y.-C., Lin, S., Cheng, Q.-W., Wu, F.-H., et al. (2021). Efficient and economical targeted insertion in plant genomes via protoplast regeneration. bioRxiv 2021, 3.09.434087. doi: 10.1101/2021.03.09.434087
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122. doi: 10.1105/tpc.111.093211
Huang, Z., Chen, Y., Yi, B., Xiao, L., Ma, C., Tu, J., et al. (2007). Fine mapping of the recessive genic male sterility gene (Bnms3) in brassica napus l. Theor. Appl. Genet. 115, 113–118. doi: 10.1007/s00122-007-0547-8
Huang, T. Y., Wang, Z., Hu, Y. G., Shi, S. P., Peng, T., Chu, X. D., et al. (2008). Genetic analysis and primary mapping of pms4, a photoperiod-sensitive genic male sterility gene in rice (Oryza sativa). Rice Sci. 15 (2), 153–156. doi: 10.1016/S1672-6308(08)60035-9
Huang, J. Z., E, Z. G., Zhang, H. L., Shu, Q. Y. (2014). Workable male sterility systems for hybrid rice: genetics, biochemistry, molecular biology, and utilization. Rice (NY) 7, 13. doi: 10.1186/s12284-014-0013-6
Igarashi, K., Kazama, T., Toriyama, K. (2016). A gene encoding pentatricopeptide repeat protein partially restores fertility in RT98-type cytoplasmic male-sterile rice. Plant Cell Physiol. 57 (10), 2187–2193. doi: 10.1093/pcp/pcw135
Impens, L., Jacobs, T. B., Nelissen, H., Inze, D., Pauwels, L. (2022). Mini-review: transgenerational CRISPR/Cas9 gene editing in plants. Front. Genome Ed 4. doi: 10.3389/fgeed.2022.825042
Itabashi, E., Iwata, N., Fujii, S., Kazama, T., Toriyama, K. (2011). The fertility restorer gene, Rf2, for lead rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J. 65, 359–367. doi: 10.1111/j.1365-313X.2010.04427.x
Jacobs, T. B., Zhang, N., Patel, D., Martin, G. B. (2017). Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol. 174, 2023–2037. doi: 10.1104/pp.17.00489
Jean, M., Brown, G. G., Landry, B. S. (1997). Genetic mapping of nuclear fertility restorer genes for the 'Polima' cytoplasmic male sterility in canola (Brassica napus l.) using DNA markers. Theor. Appl. Genet. 95, 321–328. doi: 10.1007/s001220050566
Jeong, K., Choi, D., Lee, J. (2018). Fine mapping of the genic male-sterile ms (1) gene in capsicum annuum l. Theor. Appl. Genet. 131, 183–191. doi: 10.1007/s00122-017-2995-0
Jeong, H. J., Kang, J. H., Zhao, M., Kwon, J. K., Choi, H. S., Bae, J. H., et al. (2014). Tomato Male sterile 1035 is essential for pollen development and meiosis in anthers. J. Exp. Bot. 65, 6693–6709. doi: 10.1093/jxb/eru389
Jia, J. H., Zhang, D. S., Li, C. Y., Qu, X. P., Wang, S. W., Chamarerk, V., et al. (2001). Molecular mapping of the reverse thermo-sensitive genic male-sterile gene (rtms1) in rice. Theor. Appl. Genet. 103, 607–612. doi: 10.1007/Pl00002916
Jindal, S., Dhaliwal, M., Meena, O. (2019). Molecular advancements in male sterility systems of capsicum: a review. Plant Breed 139, 42–64. doi: 10.1111/pbr.12757
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jo, Y. D., Ha, Y., Lee, J. H., Park, M., Bergsma, A. C., Choi, H. I., et al. (2016). Fine mapping of restorer-of-fertility in pepper (Capsicum annuum l.) identified a candidate gene encoding a pentatricopeptide repeat (PPR)-containing protein. Theor. Appl. Genet. 129, 2003–2017. doi: 10.1007/s00122-016-2755-6
Jordan, D. R., Mace, E. S., Henzell, R. G., Klein, P. E., Klein, R. R. (2010). Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum [Sorghum bicolor (L.) moench]. Theor. Appl. Genet. 120, 1279–1287. doi: 10.1007/s00122-009-1255-3
Jung, Y. J., Kim, D. H., Lee, H. J., Nam, K. H., Bae, S., Nou, I. S., et al. (2020). Knockout of SlMS10 gene (Solyc02g079810) encoding bHLH transcription factor using CRISPR/Cas9 system confers Male sterility phenotype in tomato. Plants 9 (9), 1198. doi: 10.3390/plants9091189
Kang, M. C., Kang, H. J., Jung, S. Y., Lee, H. Y., Kang, M. Y., Jo, Y. D., et al. (2022). The unstable restorer-of-fertility locus in pepper (Capsicum annuum. l) is delimited to a genomic region containing PPR genes. Theor. Appl. Genet. 135, 1923–1937. doi: 10.1007/s00122-022-04084-1
Kaul, M. L. H. (1988). Male Sterility in higher plants (Springer Science & Business Media). doi: 10.1007/978-3-642-83139-3
Khan, M., Isshiki, S. (2016). Cytoplasmic Male sterility in eggplant. Horticulture J. 85, 1–7. doi: 10.2503/hortj.MI-IR03
Kim, Y. J., Zhang, D. (2018). Molecular control of Male fertility for crop hybrid breeding. Trends Plant Sci. 23, 53–65. doi: 10.1016/j.tplants.2017.10.001
Kishi-Kaboshi, M., Aida, R., Sasaki, K. (2017). Generation of gene-edited chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol. 58, 216–226. doi: 10.1093/pcp/pcw222
Komori, T., Ohta, S., Murai, N., Takakura, Y., Kuraya, Y., Suzuki, S., et al. (2004). Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa l.). Plant J. 37, 315–325. doi: 10.1046/j.1365-313x.2003.01961.x
Kuang, Y., Li, S., Ren, B., Yan, F., Spetz, C., Li, X., et al. (2020). Base-Editing-Mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant 13, 565–572. doi: 10.1016/j.molp.2020.01.010
Kui, L., Chen, H., Zhang, W., He, S., Xiong, Z., Zhang, Y., et al. (2017). Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, dendrobium officinale. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02036
Lee, D. S., Chen, L. J., Suh, H. S. (2005). Genetic characterization and fine mapping of a novel thermo-sensitive genic male-sterile gene tms6 in rice (Oryza sativa l.). Theor. Appl. Genet. 111, 1271–1277. doi: 10.1007/s00122-005-0044-x
Lee, S. K., Kim, H., Cho, J. I., Nguyen, C. D., Moon, S., Park, J. E., et al. (2020). Deficiency of rice hexokinase HXK5 impairs synthesis and utilization of starch in pollen grains and causes male sterility. J. Exp. Bot. 71, 116–125. doi: 10.1093/jxb/erz436
Leino, M., Teixeira, R., Landgren, M., Glimelius, K. (2003). Brassica napus lines with rearranged arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor. Appl. Genet. 106, 1156–1163. doi: 10.1007/s00122-002-1167-y
Levings, C. S. . S. 3rd. (1993). Thoughts on Cytoplasmic Male Sterility in cms-T Maize. Plant Cell. 5 (10), 1285–1290. doi: 10.1105/tpc.5.10.1285
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, Z., Liu, Z. B., Xing, A., Moon, B. P., Koellhoffer, J. P., Huang, L., et al. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. doi: 10.1104/pp.15.00783
Li, J. J., Nadeem, M., Sun, G. L., Wang, X. B., Qiu, L. J. (2019). Male Sterility in soybean: occurrence, molecular basis and utilization. Plant Breed 138, 659–676. doi: 10.1111/pbr.12751
Li, J. F., Norville, J. E., Aach, J., Mccormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in arabidopsis and nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, Z., Sun, J., Hirsch, C. (2022). Understanding environmental modulation of heterosis. Plant Breed. Rev 45, 219–239. doi: 10.1002/9781119874157.ch4
Li, J., Wang, Z., He, G., Ma, L., Deng, X. W. (2020). CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J. Genet. Genomics 47, 263–272. doi: 10.1016/j.jgg.2020.05.004
Li, S. Q., Yang, D. C., Zhu, Y. G. (2007). Characterization and use of male sterility in hybrid rice breeding. J. Integr. Plant Biol. 49, 791–804. doi: 10.1111/j.1744-7909.2007.00513.x
Li, Q., Zhang, D., Chen, M., Liang, W., Wei, J., Qi, Y., et al. (2016). Development of japonica photo-sensitive genic Male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genomics 43, 415–419. doi: 10.1016/j.jgg.2016.04.011
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., Liu, J., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genomics 44, 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, S., Zhang, X., Wang, W., Guo, X., Wu, Z., Du, W., et al. (2018). Expanding the scope of CRISPR/Cpf1-mediated genome editing in rice. Mol. Plant 11, 995–998. doi: 10.1016/j.molp.2018.03.009
Li, Y., Huang, Y., Pan, L., Zhao, Y., Huang, W., Jin, W. (2021). Male sterile 28 encodes an ARGONAUTE family protein essential for male fertility in maize. Chromosome Res. 29, 189–201. doi: 10.1007/s10577-021-09653-6
Liang, Z., Chen, K., Zhang, Y., Liu, J., Yin, K., Qiu, J. L., et al. (2018). Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430. doi: 10.1038/nprot.2017.145
Liang, J., Ma, Y., Wu, J., Cheng, F., Liu, B., Wang, X. (2017). Map-based cloning of the dominant genic male sterile Ms-cd1 gene in cabbage (Brassica oleracea). Theor. Appl. Genet. 130, 71–79. doi: 10.1007/s00122-016-2792-1
Lin, C. S., Hsu, C. T., Yang, L. H., Lee, L. Y., Fu, J. Y., Cheng, Q. W., et al. (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. doi: 10.1111/pbi.12870
Linke, B., Nothnagel, T., Börner, T. (2003). Flower development in carrot CMS plants: mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS. Plant J. 34 (1), 27–37. doi: 10.1046/j.1365-313x.2003.01703.x
Liu, H., Chen, W., Li, Y., Sun, L., Chai, Y., Chen, H., et al. (2022). CRISPR/Cas9 technology and its utility for crop improvement. Int. J. Mol. Sci. 23 (18), 10442. doi: 10.3390/ijms231810442
Liu, H. J., Jian, L., Xu, J., Zhang, Q., Zhang, M., Jin, M., et al. (2020). High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 32, 1397–1413. doi: 10.1105/tpc.19.00934
Liu, X. Q., Xu, X., Tan, Y. P., Li, S. Q., Hu, J., Huang, J. Y., et al. (2004). Inheritance and molecular mapping of two fertility-restoring loci for honglian gametophytic cytoplasmic male sterility in rice (Oryza sativa l.). Mol. Gen. Genomics 271, 586–594. doi: 10.1007/s00438-004-1005-9
Liu, Z., Mulpuri, S., Feng, J., Vick, B., Jan, C. (2012). Molecular mapping of the Rf 3 fertility restoration gene to facilitate its utilization in breeding confection sunflower. Mol. Breed. 29, 275–284. doi: 10.1007/s11032-011-9563-0
Liu, Z., Wang, D., Feng, J., Seiler, G. J., Cai, X., Jan, C. C. (2013). Diversifying sunflower germplasm by integration and mapping of a novel male fertility restoration gene. Genetics 193, 727–737. doi: 10.1534/genetics.112.146092
Liu, J. W., Wang, S. F., Wang, H., Luo, B. T., Cai, Y. Y., Li, X. D., et al. (2021a). Rapid generation of tomato male-sterile lines with a marker use for hybrid seed production by CRISPR/Cas9 system. Mol. Breed. 41, 25. doi: 10.1007/s11032-021-01215-2
Liu, X., Yue, Y., Gu, Z., Huang, Q., Pan, Z., Zhao, Z., et al. (2021). The characterization and candidate gene isolation for a novel male-sterile mutant ms40 in maize. Plant Cell Rep. 40 (10), 1957–1970. doi: 10.1007/s00299-021-02762-w
Liu, X., Yang, M., Liu, X., Wei, K., Cao, X., Wang, X., et al. (2019). A putative bHLH transcription factor is a candidate gene for male sterile 32, a locus affecting pollen and tapetum development in tomato. Hortic. Res. 6, 88. doi: 10.1038/s41438-019-0170-2
Liu, Q., Yang, F., Zhang, J., Liu, H., Rahman, S., Islam, S., et al. (2021b). Application of CRISPR/Cas9 in crop quality improvement. Int. J. Mol. Sci. 22 (8), 4206. doi: 10.3390/ijms22084206
Longin, C. F., Muhleisen, J., Maurer, H. P., Zhang, H., Gowda, M., Reif, J. C. (2012). Hybrid breeding in autogamous cereals. Theor. Appl. Genet. 125, 1087–1096. doi: 10.1007/s00122-012-1967-7
Lopez, M. T., Toojinda, T., Vanavichit, A., Tragoonrung, S. (2003). Microsatellite markers flanking the tms2 gene facilitated tropical TGMS rice line development. Crop Sci. 43, 2267–2271. doi: 10.2135/cropsci2003.2267
Lou, D., Wang, H., Liang, G., Yu, D. (2017). OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00993
Loukides, C., Broadwater, A., Bedinger, P. (1995). Two new male-sterile mutants ofZea mays(Poaceae)with abnormal tapetal cell morphology. Am. J. Bot. 82, 1017–1023. doi: 10.1002/j.1537-2197.1995.tb11566.x
Lowder, L. G., Zhang, D., Baltes, N. J., Paul, J. W., Tang, X., Zheng, X., et al. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. doi: 10.1104/pp.15.00636
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Ma, J., Chen, J., Wang, M., Ren, Y., Wang, S., Lei, C., et al. (2018). Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 69, 1051–1064. doi: 10.1093/jxb/erx458
Ma, X., Feng, F., Zhang, Y., Elesawi, I. E., Xu, K., Li, T., et al. (2019). A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PloS Genet. 15, e1008191. doi: 10.1371/journal.pgen.1008191
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Maan, S. S., Carlson, K. M., Williams, N. D., Yang, T. (1987). Chromosomal arm location and gene-centromere distance of a dominant gene for Male sterility in wheat. Crop Sci. 27, 494–500. doi: 10.2135/cropsci1987.0011183X002700030013x
Mackenzie, S. (2012). “Male Sterility and hybrid seed production,” in Plant biotechnology and agriculture. Eds. Altman, A., Hasegawa, P. M. (San Diego: Academic Press). doi: 10.1016/b978-0-12-381466-1.00012-2
Maddumage, R., Fung, R. M. W., Weir, I., Ding, H., Simons, J. L., Allan, A. C. (2002). Efficient transient transformation of suspension culture-derived apple protoplasts. Plant Cell Tiss Org 70, 77–82. doi: 10.1023/A:1016073611902
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-Free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01904
Malzahn, A., Lowder, L., Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21. doi: 10.1186/s13578-017-0148-4
Manjunathagowda, D. C. (2021). Perspective and application of molecular markers linked to the cytoplasm types and male-fertility restorer locus in onion (Allium cepa). Plant Breed 140, 732–744. doi: 10.1111/pbr.12948
Martin-Pizarro, C., Trivino, J. C., Pose, D. (2019). Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70, 885–895. doi: 10.1093/jxb/ery400
Mei, M. H., Dai, X. K., Xu, C. G., Zhang, Q. F. (1999). Mapping and genetic analysis of the genes for photoperiod-sensitive genic male sterility in rice using the original mutant nongken 58S. Crop Sci. 39, 1711–1715. doi: 10.2135/cropsci1999.3961711x
Mekapogu, M., Kwon, O., Song, H., Jung, J. (2022). Towards the improvement of ornamental attributes in chrysanthemum: recent progress in biotechnological advances. Int. J. Mol. Sci. 23 (20), 12284. doi: 10.3390/ijms232012284
Melonek, J., Duarte, J., Martin, J., Beuf, L., Murigneux, A., Varenne, P., et al. (2021). The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat. Commun. 12, 1036. doi: 10.1038/s41467-021-21225-0
Meng, L., Liu, Z., Zhang, L., Hu, G., Song, X. (2016). Cytological characterization of a thermo-sensitive cytoplasmic male-sterile wheat line having K-type cytoplasm of aegilops kotschyi. Breed Sci. 66, 752–761. doi: 10.1270/jsbbs.16039
Meng, X., Yu, H., Zhang, Y., Zhuang, F., Song, X., Gao, S., et al. (2017). Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 10, 1238–1241. doi: 10.1016/j.molp.2017.06.006
Mishra, R., Joshi, R. K., Zhao, K. (2018). Genome editing in rice: recent advances, challenges, and future implications. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01361
Mishra, R., Mohanty, J. N., Mahanty, B., Joshi, R. K. (2021). A single transcript CRISPR/Cas9 mediated mutagenesis of CaERF28 confers anthracnose resistance in chilli pepper (Capsicum annuum l.). Planta 254, 5. doi: 10.1007/s00425-021-03660-x
Mojica, F. J. M., Diez-Villasenor, C., Garcia-Martinez, J., Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol. (Reading) 155, 733–740. doi: 10.1099/mic.0.023960-0
Moon, J., Skibbe, D., Timofejeva, L., Wang, C. J.R., Kelliher, T., Kremling, K., et al. (2013). Regulation of cell divisions and differentiation by MALE STERILITY 32 is required for anther development in maize. Plant J. 76 (4), 592–602. doi: 10.1111/tpj.12318
Morales, K. Y., Bridgeland, A. H., Hake, K. D., Udall, J. A., Thomson, M. J., Yu, J. Z. (2022). Homology-based identification of candidate genes for male sterility editing in upland cotton (Gossypium hirsutum l.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1006264
Nadakuduti, S. S., Enciso-Rodriguez, F. (2020). Advances in genome editing with CRISPR systems and transformation technologies for plant DNA manipulation. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.637159
Naresh, P., Lin, S. W., Lin, C. Y., Wang, Y. W., Schafleitner, R., Kilian, A., et al. (2018). Molecular markers associated to two non-allelic genic Male sterility genes in peppers (Capsicum annuum l.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01343
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., Kamoun, S. (2013). Targeted mutagenesis in the model plant nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. doi: 10.1038/s41598-017-00578-x
Nicolia, A., Fält, A., Hofvander, P., Andersson, M. (2021). “Protoplast-based method for genome editing in tetraploid potato,” in Crop breeding: genetic improvement methods. Ed. Tripodi, P. (New York, NY: Springer US). doi: 10.1007/978-1-0716-1201-9_12
Nie, H., Cheng, C., Kong, J., Li, H., Hua, J. (2023). Plant non-coding RNAs function in pollen development and male sterility. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1109941
Nieves-Cordones, M., Mohamed, S., Tanoi, K., Kobayashi, N. I., Takagi, K., Vernet, A., et al. (2017). Production of low-cs(+) rice plants by inactivation of the k(+) transporter OsHAK1 with the CRISPR-cas system. Plant J. 92, 43–56. doi: 10.1111/tpj.13632
Ning, L., Wang, H., Li, D., Li, Y., Chen, K., Chao, H., et al. (2020). Genome-wide identification of the restorer-of-fertility-like (RFL) gene family in brassica napus and expression analysis in Shaan2A cytoplasmic male sterility. BMC Genomics 21, 765. doi: 10.1186/s12864-020-07163-z
Niu, Q., Shi, Z., Zhang, P., Su, S., Jiang, B., Liu, X., et al. (2023). ZmMS39 encodes a callose synthase essential for male fertility in maize (Zea mays L.). Crop J. 11 (2), 394–404. doi: 10.1016/j.cj.2022.08.012
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17, 1905–1913. doi: 10.1111/pbi.13106
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Page, D. R., Grossniklaus, U. (2002). The art and design of genetic screens: arabidopsis thaliana. Nat. Rev. Genet. 3, 124–136. doi: 10.1038/nrg730
Pallotta, M. A., Warner, P., Kouidri, A., Tucker, E. J., Baes, M., Suchecki, R., et al. (2019). Wheat ms5 male-sterility is induced by recessive homoeologous a and d genome non-specific lipid transfer proteins. Plant J. 99, 673–685. doi: 10.1111/tpj.14350
Palumbo, F., Qi, P., Pinto, V. B., Devos, K. M., Barcaccia, G. (2019). Construction of the first SNP-based linkage map using genotyping-by-Sequencing and mapping of the Male-sterility gene in leaf chicory. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00276
Pan, Z., Liu, M., Zhao, H., Tan, Z., Liang, K., Sun, Q., et al. (2020). ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J. Integr. Plant Biol. 62, 1895–1909. doi: 10.1111/jipb.12982
Pausch, P., Al-Shayeb, B., Bisom-Rapp, E., Tsuchida, C. A., Li, Z., Cress, B. F., et al. (2020). CRISPR-CasPhi from huge phages is a hypercompact genome editor. Science 369, 333–337. doi: 10.1126/science.abb1400
Pechar, G. S., Donaire, L., Gosalvez, B., Garcia-Almodovar, C., Sanchez-Pina, M. A., Truniger, V., et al. (2022). Editing melon eIF4E associates with virus resistance and male sterility. Plant Biotechnol. J. 20, 2006–2022. doi: 10.1111/pbi.13885
Peng, H. F., Zhang, Z. F., Wu, B., et al. (2008). Molecular mapping of two reverse photoperiod-sensitive genic male sterility genes (rpms1 and rpms2) in rice (Oryza sativa l.). Theor. Appl. Genet. 118, 77–83. doi: 10.1007/s00122-008-0877-1
Perez-Prat, E., Van Lookeren Campagne, M. M. (2002). Hybrid seed production and the challenge of propagating male-sterile plants. Trends Plant Sci. 7, 199–203. doi: 10.1016/s1360-1385(02)02252-5
Petolino, J. F. (2015). Genome editing in plants via designed zinc finger nucleases. In Vitro Cell Dev. Biol. Plant 51, 1–8. doi: 10.1007/s11627-015-9663-3
Pu, Y., Liu, C., Li, J., Aerzu, G., Hu, Y., Liu, X. (2018). Different SlU6 promoters cloning and establishment of CRISPR/Cas9 mediated gene editing system in tomato. Scientia Agricultura Sin. 51, 315–326. doi: 10.3864/.issn.0578-1752.2018.02.011
Qi, L. L., Seiler, G. J., Vick, B. A., Gulya, T. J. (2012). Genetics and mapping of the R(1)(1) gene conferring resistance to recently emerged rust races, tightly linked to male fertility restoration, in sunflower (Helianthus annuus l.). Theor. Appl. Genet. 125, 921–932. doi: 10.1007/s00122-012-1883-x
Qi, X., Zhang, C., Zhu, J., Liu, C., Huang, C., Li, X., et al. (2020). Genome editing enables next-generation hybrid seed production technology. Mol. Plant 13, 1262–1269. doi: 10.1016/j.molp.2020.06.003
Ramadan, M., Alariqi, M., Ma, Y., Li, Y., Liu, Z., Zhang, R., et al. (2021). Efficient CRISPR/Cas9 mediated pooled-sgRNAs assembly accelerates targeting multiple genes related to male sterility in cotton. Plant Methods 17, 16. doi: 10.1186/s13007-021-00712-x
Ramlal, A., Nautiyal, A., Baweja, P., Kumar Mahto, R., Mehta, S., Pujari Mallikarunja, B., et al. (2022). Harnessing heterosis and male sterility in soybean [Glycine max (L.) merrill]: a critical revisit. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.981768
Rathburn, H. B., Hedgcoth, C. (1991). A chimeric open reading frame in the 5' flanking region of coxI mitochondrial DNA from cytoplasmic male-sterile wheat. Plant Mol. Biol. 16 (5), 909–912. doi: 10.1007/BF00015083
Rogers, J., Edwardson, J. (1952). The utilization of cytoplasmic Male-sterile inbreds in the production of corn hybrids. Agron. J. 44, 8–13. doi: 10.2134/agronj1952.00021962004400010004x
Romano, A., Raemakers, K., Bernardi, J., Visser, R., Mooibroek, H. (2003). Transgene organisation in potato after particle bombardment-mediated (co-)transformation using plasmids and gene cassettes. Transgenic Res. 12, 461–473. doi: 10.1023/a:1024267906219
Roque, E., Gomez, M. D., Ellul, P., Wallbraun, M., Madueno, F., Beltran, J. P., et al. (2007). The PsEND1 promoter: a novel tool to produce genetically engineered male-sterile plants by early anther ablation. Plant Cell Rep. 26, 313–325. doi: 10.1007/s00299-006-0237-z
Rout, D., Jena, D., Singh, V., Kumar, M., Arsode, P., Singh, P., et al. (2021). “Hybrid rice research: current status and prospects,” in Recent advances in rice research. Ed. Mahmood-UR- Rahman, A. (Rijeka: IntechOpen). doi: 10.5772/intechopen.93668
Sabar, M., Gagliardi, D., Balk, J., Leaver, C. J. (2003). ORFB is a subunit of F1F(O)-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 4 (4), 381–386. doi: 10.1038/sj.embor.embor800
Scariolo, F., Palumbo, F., Farinati, S., Barcaccia, G. (2023). Pipeline to design inbred lines and F1 hybrids of leaf chicory (Radicchio) using Male sterility and genotyping-by-Sequencing. Plants 12 (6), 1242. doi: 10.3390/plants12061242
Scheben, A., Wolter, F., Batley, J., Puchta, H., Edwards, D. (2017). Towards CRISPR/Cas crops - bringing together genomics and genome editing. New Phytol. 216, 682–698. doi: 10.1111/nph.14702
Schnable, P. S., Wise, R. P. (1994). Recovery of heritable, transposon-induced, mutant alleles of the rf 2 nuclear restorer of T-cytoplasm maize. Genetics 136, 1171–1185. doi: 10.1093/genetics/136.3.1171
Schnable, P. S., Wise, R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. doi: 10.1016/S1360-1385(98)01235-7
Schneeberger, K. (2014). Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15, 662–676. doi: 10.1038/nrg3745
Shahinnia, F., Geyer, M., Block, A., Mohler, V., Hartl, L. (2020). Identification of Rf9, a gene contributing to the genetic complexity of fertility restoration in hybrid wheat. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.577475
Shan, Q., Zhang, Y., Chen, K., Zhang, K., Gao, C. (2015). Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 13, 791–800. doi: 10.1111/pbi.12312
Sharma, R., Messar, Y. (2017). Transgenics in ornamental crops: creating novelties in economically important cut flowers. Curr. Sci. 113, 43–52. doi: 10.18520/cs/v113/i01/43-52
Sheen, J. (2001). Signal transduction in maize and arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. doi: 10.1104/pp.010820
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shinoyama, H., Ichikawa, H., Nishizawa-Yokoi, A., Skaptsov, M., Toki, S. (2020). Simultaneous TALEN-mediated knockout of chrysanthemum DMC1 genes confers male and female sterility. Sci. Rep. 10, 16165. doi: 10.1038/s41598-020-72356-1
Shinoyama, H., Sano, T., Saito, M., Ezura, H., Aida, R., Nomura, Y., et al. (2012). Induction of male sterility in transgenic chrysanthemums (Chrysanthemum morifolium ramat.) by expression of a mutated ethylene receptor gene, Cm-ETR1/H69A, and the stability of this sterility at varying growth temperatures. Mol. Breed. 29, 285–295. doi: 10.1007/s11032-010-9546-6
Singh, S., Dey, S. S., Bhatia, R., Kumar, R., Behera, T. K. (2019). Current understanding of male sterility systems in vegetable brassicas and their exploitation in hybrid breeding. Plant Reprod. 32, 231–256. doi: 10.1007/s00497-019-00371-y
Singh, H., Khar, A. (2021). Perspectives of onion hybrid breeding in India: an overview. Indian J. Agric. Sci. 91, 1426–1432. doi: 10.56093/ijas.v91i10.117404
Singh, M., Kumar, M., Thilges, K., Cho, M.-J., Cigan, A. M. (2017). MS26/CYP704B is required for anther and pollen wall development in bread wheat (Triticum aestivum l.) and combining mutations in all three homeologs causes male sterility. PloS One 12 (5), e0177632. doi: 10.1371/journal.pone.0177632
Singh, M., Kumar, M., Albertsen, M. C., Young, J. K., Cigan, A. M. (2018). Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum l.). Plant Mol. Biol. 97, 371–383. doi: 10.1007/s11103-018-0749-2
Sisco, P. H. (1991). Duplications complicate genetic mapping of Rf4, a restorer gene for cms-c cytoplasmic male sterility in corn. Crop Sci. 31, 1263–1266. doi: 10.2135/cropsci1991.0011183X003100050036x
Smith, J., Grizot, S., Arnould, S., Duclert, A., Epinat, J. C., Chames, P., et al. (2006). A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34, e149. doi: 10.1093/nar/gkl720
Sorensen, A. M., Krober, S., Unte, U. S., Huijser, P., Dekker, K., Saedler, H. (2003). The arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423. doi: 10.1046/j.1365-313x.2003.01644.x
Subburaj, S., Chung, S. J., Lee, C., Ryu, S. M., Kim, D. H., Kim, J. S., et al. (2016). Site-directed mutagenesis in petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi: 10.1007/s00299-016-1937-7
Subudhi, P. K., Borkakati, R. P., Virmani, S. S., Huang, N. (1997). Molecular mapping of a thermosensitive genetic male sterility gene in rice using bulked segregant analysis. Genome 40 (2), 188–194. doi: 10.1139/g97-027
Sun, L., Kao, T. H. (2018). CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of s-locus f-box protein-containing SCF complexes in recognition of non-self s-RNases during cross-compatible pollination in self-incompatible petunia inflata. Plant Reprod. 31, 129–143. doi: 10.1007/s00497-017-0314-1
Sun, Y., Zhang, Y., Jia, S., Lin, C., Zhang, J., Yan, H., et al. (2022). Identification of a candidate restorer-of-fertility gene Rf3 encoding a pentatricopeptide repeat protein for the cytoplasmic Male sterility in soybean. Int. J. Mol. Sci. 23 (10), 5388. doi: 10.3390/ijms23105388
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. doi: 10.1038/ncomms13274
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Talukder, Z. I., Ma, G. J., Hulke, B. S., Jan, C. C., Qi, L. L. (2019). Linkage mapping and genome-wide association studies of the Rf gene cluster in sunflower (Helianthus annuus l.) and their distribution in world sunflower collections. Front. Gen. 10. doi: 10.3389/fgene.2019.00216
Tan, C., Liu, Z., Huang, S., Feng, H. (2019). Mapping of the male sterile mutant gene ftms in brassica rapa l. ssp. pekinensis via BSR-seq combined with whole-genome resequencing. Theor. Appl. Genet. 132, 355–370. doi: 10.1007/s00122-018-3223-2
Tang, H., Luo, D., Zhou, D., Zhang, Q., Tian, D., Zheng, X., et al. (2014). The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol. Plant 7, 1497–1500. doi: 10.1093/mp/ssu047
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low cd-accumulating indica rice without compromising yield. Sci. Rep. 7, 14438. doi: 10.1038/s41598-017-14832-9
Thu, S.W., Rai, K.M., Sandhu, D., Rajangam, A., Balasubramanian, V. K., Palmer, R. G., et al. (2019). Mutation in a PHD-finger protein MS4 causes male sterility in soybean. BMC Plant Biol. 19, 378. doi: 10.1186/s12870-019-1979-4
Toda, E., Koiso, N., Takebayashi, A., Ichikawa, M., Kiba, T., Osakabe, K., et al. (2019). An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 5, 363–368. doi: 10.1038/s41477-019-0386-z
Tucker, E.J., Baumann, U., Kouidri, A., Suchecki, R., Baes, M., Garcia, M., et al. (2017). Molecular identification of the wheat male fertility gene Ms1 and its prospects for hybrid breeding. Nat. Commun. 8, 869. doi: 10.1038/s41467-017-00945-2
Tong, C. G., Wu, F. H., Yuan, Y. H., Chen, Y. R., Lin, C. S. (2020). High-efficiency CRISPR/Cas-based editing of phalaenopsis orchid MADS genes. Plant Biotechnol. J. 18, 889–891. doi: 10.1111/pbi.13264
Van Ginkel, M., Ortiz, R. (2018). Cross the best with the best, and select the best: HELP in breeding selfing crops. Crop Sci. 58, 17–30. doi: 10.2135/cropsci2017.05.0270
Voss-Fels, K. P., Stahl, A., Hickey, L. T. (2019). Q&A: modern crop breeding for future food security. BMC Biol. 17, 18. doi: 10.1186/s12915-019-0638-4
Voytas, D. F., Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PloS Biol. 12, e1001877. doi: 10.1371/journal.pbio.1001877
Wan, X., Wu, S., Li, Z., Dong, Z., An, X., Ma, B., et al. (2019). Maize genic Male-sterility genes and their applications in hybrid breeding: progress and perspectives. Mol. Plant 12, 321–342. doi: 10.1016/j.molp.2019.01.014
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, K., Gao, F., Ji, Y., Liu, Y., Dan, Z., Yang, P., et al. (2013). ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in honglian cytoplasmic male sterile rice. New Phytol. 2, 408–418. doi: 10.1111/nph.12180
Wang, W., Pan, Q., He, F., Akhunova, A., Chao, S., Trick, H., et al. (2018b). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 1, 65–74. doi: 10.1089/crispr.2017.0010
Wang, D., Samsulrizal, N. H., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2019). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557. doi: 10.1104/pp.18.01187
Wang, W., Simmonds, J., Pan, Q., Davidson, D., He, F., Battal, A., et al. (2018c). Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 131, 2463–2475. doi: 10.1007/s00122-018-3166-7
Wang, F., Stewart, J. M., Zhang, J. (2007). Molecular markers linked to the Rf2 fertility restorer gene in cotton. Genome 50, 818–824. doi: 10.1139/g07-061
Wang, C., Wang, G., Gao, Y., Lu, G., Habben, J. E., Mao, G., et al. (2020). A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol. Biol. 102, 373–388. doi: 10.1007/s11103-019-00952-5
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PloS One 11, e0154027. doi: 10.1371/journal.pone.0154027
Wang, D., Wang, Y., Zhang, L., Yang, Y., Wu, Q., Hu, G., et al. (2023). Integrated transcriptomic and proteomic analysis of a cytoplasmic male sterility line and associated maintainer line in soybean. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1098125
Wang, Y. G., Xing, Q. H., Deng, Q. Y., Liang, F. S., Yuan, L. P., Weng, M. L., et al. (2003). Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5. Theor. Appl. Genet. 107, 917–921. doi: 10.1007/s00122-003-1327-8
Wang, P., Zhang, J., Sun, L., Ma, Y., Xu, J., Liang, S., et al. (2018a). High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16, 137–150. doi: 10.1111/pbi.12755
Wang, Z., Li, J., Chen, S., Heng, Y., Chen, Z., Yang, J., et al. (2017). Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. U.S.A. 114 (47), 12614–12619. doi: 10.1073/pnas.1715570114
Watanabe, K., Kobayashi, A., Endo, M., Sage-Ono, K., Toki, S., Ono, M. (2017). CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-b) locus in the Japanese morning glory ipomoea (Pharbitis) nil. Sci. Rep. 7, 10028. doi: 10.1038/s41598-017-10715-1
Whitford, R., Fleury, D., Reif, J. C., Garcia, M., Okada, T., Korzun, V., et al. (2013). Hybrid breeding in wheat: technologies to improve hybrid wheat seed production. J. Exp. Bot. 64, 5411–5428. doi: 10.1093/jxb/ert333
Wilson, F. M., Harrison, K., Armitage, A. D., Simkin, A. J., Harrison, R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 45. doi: 10.1186/s13007-019-0428-6
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-Free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wu, J., Zhang, M., Zhang, X., Guo, L., Qi, T., Wang, H., et al. (2017). Development of InDel markers for the restorer gene Rf1 and assessment of their utility for marker-assisted selection in cotton. Euphytica. 213, 1–8. doi: 10.1007/s10681-017-2043-9
Wu, Y., Fox, T. W., Trimnell, M. R., Wang, L., Xu, R. J., Cigan, A. M., et al. (2016). Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 14, 1046–1054. doi: 10.1111/pbi.12477
Xie, K., Wu, S., Li, Z., Zhou, Y., Zhang, D., Dong, Z., et al. (2018). Map-based cloning and characterization of Zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor. Appl. Genet. 131, 1363–1378. doi: 10.1007/s00122-018-3083-9
Xu, F., Yang, X., Zhao, N., Hu, Z., Mackenzie, S. A., Zhang, M., et al. (2022). Exploiting sterility and fertility variation in cytoplasmic male sterile vegetable crops. Hortic. Res. 9, uhab039. doi: 10.1093/hr/uhab039
Yamagishi, H., Bhat, S. R. (2014). Cytoplasmic male sterility in brassicaceae crops. Breed Sci. 64, 38–47. doi: 10.1270/jsbbs.64.38
Yan, G., Liu, H., Wang, H., Lu, Z., Wang, Y., Mullan, D., et al. (2017). Accelerated generation of selfed pure line plants for gene identification and crop breeding. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01786
Yan, R., Wang, Z., Ren, Y., Li, H., Liu, N., Sun, H. (2019). Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in lilium pumilum DC. fisch. and lilium longiflorum white heaven. Int. J. Mol. Sci. 20 (12), 2920. doi: 10.3390/ijms20122920
Yang, J. W., Fu, J. X., Li, J., Cheng, X. L., Li, F., Dong, J. F., et al. (2014a). A novel Co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein-protein interactions in rice. Plant Mol. Biol. Rep. 32, 153–161. doi: 10.1007/s11105-013-0633-9
Yang, W., Li, Y., Sun, L., Shoaib, M., Sun, J., Wang, D., et al. (2021). Genetic mapping of ms1s, a recessive gene for Male sterility in common wheat. Int. J. Mol. Sci. 22 (16), 8541. doi: 10.3390/ijms22168541
Yang, Q., Liang, C., Zhuang, W., Li, J., Deng, H., Deng, Q., et al. (2007). Characterization and identification of the candidate gene of rice thermo-sensitive genic male sterile gene tms5 by mapping. Planta 225, 321–330. doi: 10.1007/s00425-006-0353-6
Yang, Y., Speth, B. D., Boonyoo, N., Baumert, E., Atkinson, T. R., Palmer, R. G., et al. (2014b). Molecular mapping of three male-sterile, female-fertile mutants and generation of a comprehensive map of all known male sterility genes in soybean. Genome 57, 155–160. doi: 10.1139/gen-2014-0018
Yang, S. L., Xie, L. F., Mao, H. Z., Puah, C. S., Yang, W. C., Jiang, L., et al. (2003). Tapetum determinant1 is required for cell specialization in the arabidopsis anther. Plant Cell 15, 2792–2804. doi: 10.1105/tpc.016618
Yu, D., Gu, X., Zhang, S., Dong, S., Miao, H., Gebretsadik, K., et al. (2021a). Molecular basis of heterosis and related breeding strategies reveal its importance in vegetable breeding. Hortic. Res. 8, 120. doi: 10.1038/s41438-021-00552-9
Yu, J., Zhao, G., Li, W., Zhang, Y., Wang, P., Fu, A., et al. (2021). A single nucleotide polymorphism in an R2R3 MYB transcription factor gene triggers the male sterility in soybean ms6 (Ames1). Theor. Appl. Genet. 134, 3661–3674. doi: 10.1007/s00122-021-03920-0
Yu, J., Tu, L., Subburaj, S., Bae, S., Lee, G. J. (2021b). Simultaneous targeting of duplicated genes in petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Rep. 40, 1037–1045. doi: 10.1007/s00299-020-02593-1
Yue, B., Vick, B. A., Cai, X., Hu, J. (2010). Genetic mapping for the Rf1 (fertility restoration) gene in sunflower (Helianthus annuus l.) by SSR and TRAP markers. Plant Breed 129, 24–28. doi: 10.1111/j.1439-0523.2009.01661.x
Zabala, G., Gabay-Laughnan, S., Laughnan, J. R. (1997). The nuclear gene Rf3 affects the expression of the mitochondrial chimeric sequence r implicated in s-type male sterility in maize. Genetics. 147 (2), 847–860. doi: 10.1093/genetics/147.2.847
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, G., Lu, Y., Bharaj, T. S., et al. (1997). Mapping of the Rf-3 nuclear fertility-restoring gene for WA cytoplasmic male sterility in rice using RAPD and RFLP markers. Theoret. Appl. Genet. 94, 27–33. doi: 10.1007/s001220050377
Zhang, R., Chang, J., Li, J., Lan, G., Xuan, C., Li, H., et al. (2021b). Disruption of the bHLH transcription factor abnormal tapetum 1 causes male sterility in watermelon. Hortic. Res. 8, 258. doi: 10.1038/s41438-021-00695-9
Zhang, Q., Shen, B. C., Dai, X. K., Mei, M. L., Maroof, M. A. S., Li, Z. B. (1994). Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc. Natl. Acad. Sci. U.S.A. 18 (91), 8675–8679. doi: 10.1073/pnas.91.18.8675
Zhang, L. Y., Huang, Z. J., Wang, X. X., Gao, J. C., Guo, Y. M., Du, Y. C., et al. (2016b). Fine mapping and molecular marker development of anthocyanin absent, a seedling morphological marker for the selection of male sterile 10 in tomato. Mol. Breed. 36, 107. doi: 10.1007/s11032-016-0531-6
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi: 10.1038/ncomms12617
Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., et al. (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 5, 480–485. doi: 10.1038/s41477-019-0405-0
Zhang, W., Sun, Y., Timofejeva, L., Chen, C., Grossniklaus, U., Ma, H. (2006). Regulation of arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133, 3085–3095. doi: 10.1242/dev.02463
Zhang, X., Wu, J., Zhang, H., Ma, Y., Guo, A., Wang, X. (2011). Fine mapping of a male sterility gene MS-cd1 in brassica oleracea. Theor. Appl. Genet. 123, 231–238. doi: 10.1007/s00122-011-1579-7
Zhang, H., Che, J., Ge, Y., et al. (2017). Ability of Rf5 and Rf6 to restore fertility of chinsurah boro II-type cytoplasmic Male sterile oryza sativa (ssp. japonica) lines. Rice 10, 2. doi: 10.1186/s12284-017-0142-9
Zhang, H., Xu, C., He, Y., Zong, J., Yang, X., Si, H., et al. (2013). Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc. Natl. Acad. Sci. U.S.A. 110, 76–81. doi: 10.1073/pnas.1213041110
Zhang, B., Yang, X., Yang, C., Li, M., Guo, Y. (2016a). Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci. Rep. 6, 20315. doi: 10.1038/srep20315
Zhang, D., Zhang, Z., Unver, T., Zhang, B. (2021a). CRISPR/Cas: a powerful tool for gene function study and crop improvement. J. Adv. Res. 29, 207–221. doi: 10.1016/j.jare.2020.10.003
Zhang, Z. B., Zhu, J., Gao, J. F., Wang, C., Li, H., Li, H., et al. (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in arabidopsis. Plant J. 52, 528–538. doi: 10.1111/j.1365-313X.2007.03254.x
Zhao, X., Jayarathna, S., Turesson, H., Falt, A. S., Nestor, G., Gonzalez, M. N., et al. (2021). Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci. Rep. 11, 4311. doi: 10.1038/s41598-021-83462-z
Zhao, F. L., Li, Y. J., Hu, Y., Gao, Y. R., Zang, X. W., Ding, Q., et al. (2016). A highly efficient grapevine mesophyll protoplast system for transient gene expression and the study of disease resistance proteins. Plant Cell Tiss Org 125, 43–57. doi: 10.1007/s11240-015-0928-7
Zhao, Q., Tong, Y., Yang, C., Yang, Y., Zhang, M. (2019). Identification and mapping of a new soybean Male-sterile gene, mst-m. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00094
Zhao, D. Z., Wang, G. F., Speal, B., Ma, H. (2002). The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the arabidopsis anther. Gene Dev. 16, 2021–2031. doi: 10.1101/gad.997902
Zheng, X., He, L., Liu, Y., Mao, Y., Wang, C., Zhao, B., et al. (2020). A study of male fertility control in medicago truncatula uncovers an evolutionarily conserved recruitment of two tapetal bHLH subfamilies in plant sexual reproduction. New Phytol. 228, 1115–1133. doi: 10.1111/nph.16770
Zhou, H., He, M., Li, J., Chen, L., Huang, Z., Zheng, S., et al. (2016). Development of commercial thermo-sensitive genic Male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 6, 37395. doi: 10.1038/srep37395
Zhou, H., Liu, Q., Li, J., Jiang, D., Zhou, L., Wu, P., et al. (2012). Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22, 649–660. doi: 10.1038/cr.2012.28
Zhou, Z., Dun, X., Xia, S., Shi, D., Qin, M., Yi, B., et al. (2012). BnMs3 is required for tapetal differentiation and degradation, microspore separation, and pollen-wall biosynthesis in brassica napus. J. Experiment. Bot. 63 (5), 2041–2058. doi: 10.1093/jxb/err405
Zhou, H., Zhou, M., Yang, Y., Li, J., Zhu, L., Jiang, D., et al. (2014). RNase Z(S1) processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat. Commun. 5, 4884. doi: 10.1038/ncomms5884
Zhu, J., Chen, H., Li, H., Gao, J. F., Jiang, H., Wang, C., et al. (2008). Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in arabidopsis. Plant J. 55, 266–277. doi: 10.1111/j.1365-313X.2008.03500.x
Zhu, H., Li, C., Gao, C. (2020). Applications of CRISPR-cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21 (11), 661–677. doi: 10.1038/s41580-020-00288-9
Zhu, E., You, C., Wang, S., Cui, J., Niu, B., Wang, Y., et al. (2015). The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for arabidopsis anther development and transcriptome. Plant J. 83, 976–990. doi: 10.1111/tpj.12942
Zhu, Y., Shi, Z., Li, S., Liu, H., Liu, F., Niu, Q., et al. (2018). Fine mapping of the novel male-sterile mutant gene ms39 in maize originated from outer space flight. Mol. Breed. 38, 1–12. doi: 10.1007/s11032-018-0878-y
Keywords: male sterility, precision breeding, genome editing, CRISPR/Cas system, DNA free, transgenerational gene editing, food crops, ornamental species
Citation: Farinati S, Draga S, Betto A, Palumbo F, Vannozzi A, Lucchin M and Barcaccia G (2023) Current insights and advances into plant male sterility: new precision breeding technology based on genome editing applications. Front. Plant Sci. 14:1223861. doi: 10.3389/fpls.2023.1223861
Received: 16 May 2023; Accepted: 20 June 2023;
Published: 13 July 2023.
Edited by:
Azahara Carmen Martin, John Innes Centre, United KingdomReviewed by:
Sebastien Belanger, Donald Danforth Plant Science Center, United StatesCopyright © 2023 Farinati, Draga, Betto, Palumbo, Vannozzi, Lucchin and Barcaccia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianni Barcaccia, Z2lhbm5pLmJhcmNhY2NpYUB1bmlwZC5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.