
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 August 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1219856
This article is part of the Research Topic Germline Development and Fertilization Mechanisms of Poaceae View all 4 articles
 Dezhou Wang1,2†
Dezhou Wang1,2† Jinghong Zuo1,2*†
Jinghong Zuo1,2*† Shan Liu1,2†
Shan Liu1,2† Weiwei Wang1,2
Weiwei Wang1,2 Qing Lu3
Qing Lu3 Xiaocong Hao1,2
Xiaocong Hao1,2 Zhaofeng Fang1,2
Zhaofeng Fang1,2 Ting Liang3
Ting Liang3 Yue Sun3
Yue Sun3 Chunman Guo1,2
Chunman Guo1,2 Changping Zhao1,2*
Changping Zhao1,2* Yimiao Tang1,2*
Yimiao Tang1,2*BRI1 EMS SUPPRESSOR1 (BES1) family members are crucial downstream regulators that positively mediate brassinosteroid signaling, playing vital roles in the regulation of plant stress responses and anther development in Arabidopsis. Importantly, the expression profiles of wheat (Triticum aestivum L.) BES1 genes have not been analyzed comprehensively and systematically in response to abiotic stress or during anther development. In this study, we identified 23 BES1-like genes in common wheat, which were unevenly distributed on 17 out of 21 wheat chromosomes. Phylogenetic analysis clustered the BES1 genes into four major clades; moreover, TaBES1-3A2, TaBES1-3B2 and TaBES1-3D2 belonged to the same clade as Arabidopsis BES1/BZR1 HOMOLOG3 (BEH3) and BEH4, which participate in anther development. The expression levels of 23 wheat BES1 genes were assessed using real-time quantitative PCR under various abiotic stress conditions (drought, salt, heat, and cold), and we found that most TaBES1-like genes were downregulated under abiotic stress, particularly during drought stress. We therefore used drought-tolerant and drought-sensitive wheat cultivars to explore TaBES1 expression patterns under drought stress. TaBES1-3B2 and TaBES1-3D2 expression was high in drought-tolerant cultivars but substantially repressed in drought-sensitive cultivars, while TaBES1-6D presented an opposite pattern. Among genes preferentially expressed in anthers, TaBES1-3B2 and TaBES1-3D2 expression was substantially downregulated in thermosensitive genic male-sterile wheat lines compared to common wheat cultivar under sterile conditions, while we detected no obvious differences under fertile conditions. This result suggests that TaBES1-3B2 and TaBES1-3D2 might not only play roles in regulating drought tolerance, but also participate in low temperature-induced male sterility.
The steroid phytohormone brassinosteroid (BR) plays an essential role in a broad range of plant growth and vital physiological processes (Li and Deng, 2005; Wang et al., 2012; Anwar et al., 2018; Feng et al., 2021; Li et al., 2021). BR-INSENSITIVE1 (BRI1)-EMS SUPPRESSOR1 (BES1) and BRASSINAZOLE RESISTANT1 (BZR1) are core transcription factors directly mediating BR signaling (Wang et al., 2012; Chen et al., 2019b; Cui et al., 2019; Cao et al., 2020; Kono and Yin, 2020; Su et al., 2021). A highly conserved DNA binding motif in the N-terminal of BES1-type proteins binds to E-boxes or to BR-response elements to regulate the transcription of target genes (Yin et al., 2005; Wang et al., 2012; Anwar et al., 2018; Su et al., 2021). Recent studies have revealed that BES1/BZR1 family members not only participate in BR signal transduction but also integrate multiple phytohormone signals to control tapetum development and regulate various stress responses (Yin et al., 2002; Wang et al., 2012; Jiang et al., 2015; Saha et al., 2015; Chen et al., 2019a; Chen et al., 2019b; Cui et al., 2019; Planas-Riverola et al., 2019; Setsungnern et al., 2020; Yan et al., 2020; Mecchia et al., 2021; Su et al., 2021; Korwin Krukowski et al., 2022; Sun et al., 2022; Yang et al., 2022).
BES1 and BZR1 possess a conserved BES1-type domain in their N termini and belong to the plant-specific BES1/BZR1 transcription factor family, which consists of eight members in Arabidopsis (Arabidopsis thaliana L.), 11 in maize (Zea mays L.), 16 in soybean (Glycine max L.), 22 in apple (Malus domestica L.), and nine in tomato (Solanum lycopersicum L.) (Wang et al., 2012; Lachowiec et al., 2018; Li et al., 2018; Li et al., 2019b; Li Y. et al., 2018; Cao et al., 2020; Feng et al., 2021; Su et al., 2021; Sun et al., 2022). The functions of the eight BES1/BZR1 family members in the model species Arabidopsis (BES1, BZR1, BEH1, BEH2, BEH3, BEH4, BMY2, and BMY4) have been well studied (Yin et al., 2005; Guo et al., 2009; Reinhold et al., 2011; Anwar et al., 2018; Lachowiec et al., 2018; Chen et al., 2019b; Otani et al., 2020). BES1 and BZR1 share high sequence similarity and perform essential functions in stress responses. The four BES1 homologs named BEH1, BEH2, BEH3, and BEH4 are partially redundant with BES1 and BZR1 (Chen et al., 2019a; Chen et al., 2019b). Although the single mutants bes1 and bzr1 do not exhibit typical BR mutant phenotypes, the quintuple mutant (bes1 bzr1 beh1 beh3 beh4/qui-1) is defective in tapetum and microsporocyte development (Chen et al., 2019a; Chen et al., 2019b). The other two members of the BES1/BZR1 family, the β-amylase proteins BMY2 and BMY4, are also associated with BR signaling, controlling shoot growth and development (Thalmann et al., 2019). Anthers of Arabidopsis mutants lacking function of BZR1, BES1, and their homologs are mostly loculeless, indicating that BES1 family members play indispensable roles during anther development (Chen et al., 2019a; Chen et al., 2019b).
Studies on the functions of BES1/BZR1 gene family members have recently been extended to other species. The protein encoded by the drought-responsive BES1 transcription factor gene TaBZR2 in wheat (Triticum aestivum L.) binds to the promoter region of TaGST1 (glutathione S-transferase 1) to activate its transcription, leading to the induction of drought responses (Cui et al., 2019). The liverwort (Marchantia polymorpha L.) BES1 member MpBES1 controls cell proliferation and differentiation (Mecchia et al., 2021). Studies on the nine tomato BES1 members revealed that SlBES1.5 (also named SlBZR1) plays a critical role in the BR-mediated regulation of tapetal cell degeneration and pollen development; overexpression of SlBZR1 increases pollen viability, while the pollen viability of SlBZR1 loss-of-function mutants decreased compared to that of the wild type (Yan et al., 2020). SlBZR1 also affects tomato anther development by directly binding to the promoter region of the gene SlRBOH1 (RESPIRATORY BURST OXIDASE HOMOLOG1), encoding an NADPH oxidase that produces reactive oxygen species (ROS), leading to tapetum degradation, a decline in pollen viability, decreased pollen germination, and reduced seed number (Yan et al., 2020). These results demonstrate that BES1 members have a profound influence on anther development.
Wheat is a major cereal crop worldwide. Globalization has turned crop productivity into a serious issue affecting food security; hence, improving wheat yield and quality under extreme weather conditions is of vital importance (Khan et al., 2020). Previous studies have shown that BES1/BZR1 family members participate in plant responses to environmental stress (Cui et al., 2019; Kono and Yin, 2020; Liao et al., 2020; Sun et al., 2020; Jia et al., 2021; Van Nguyen et al., 2021), with overexpression of the TaBZR2 positively regulating wheat drought stress (Cui et al., 2019). Hence, studies on the relationship of wheat BES1 family members with stress responses is very important. The proper development of functional pollen is closely related to wheat yield. Thermosensitive genic male sterile (TGMS) lines are defective in anther development under sterile environmental conditions (low temperature at the meiosis stage) but can reproduce under fertile conditions (moderate temperature during anther development), a characteristic that is widely used in hybrid wheat breeding to aid in wheat production (Bai et al., 2017; Xu et al., 2020). Previous research in Arabidopsis and tomato provides a rationale for the exploration of the role played by TaBES1 in TGMS lines during anther development in wheat (Chen et al., 2019a; Chen et al., 2019b). As a first step, the global identification of all wheat BES1 family members and the investigation of their biological functions will be helpful for identifying stress-related BES1-like genes in wheat to improve adaptation to stress (Ahad et al., 2021; Kesawat et al., 2021; Liu et al., 2021). Twenty TaBZR genes were recently identified, and the expression profiles of some were explored (Kesawat et al., 2021; Liu et al., 2021). However, the potential functions of TaBES1 members in male sterility remain unknown.
In this study, we performed a global survey of BES1 genes in common wheat and identified 23 TaBES1-like genes through comprehensive bioinformatics analysis. We also analyzed their expression patterns under drought, salt, cold, and heat stress conditions. In particular, we tested the expression levels of TaBES1 members in male sterile lines and common wheat cultivar under sterile and fertile conditions. In subsequent research, we are constructing knockout and overexpression transgene materials specifically targeting TaBES1-3B2 and TaBES1-3D2 genes, aiming to elucidate the precise functions of each BES1 gene. Moreover, these materials show significant potential for utilization in wheat breeding. Our study provides new insights into the functions of wheat BES1 members, which will be of vital importance for wheat molecular breeding and wheat production.
The wheat (Triticum aestivum) genome (wheat genome version IWGSCV1.1) and protein sequences were obtained from the Ensembl Plants website (http://plants.ensembl.org/index.html accessed on 11 March 2022). HMMER 3.0 (hidden Markov model, HMM) (http://hmmer.org/) software, BLASTP (https://blast.ncbi.nlm.nih.gov/) method and Pfam (http://pfam.xfam.org/) database searches were used to identify BES1 genes in Triticum aestivum, Aegilops tauschii, Triticum dicoccoides, Triticum turgidum, Triticum urartu, and Hordeum vulgare using reported Arabidopsis, rice, and maize BES1 genes as queries. BES1 proteins were verified using HMMER software (E value < 0.01) and the NCBI conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The HMM profile of the BES1 domain was downloaded from the Pfam (PF05687) and WheatOmics (http://wheatomics.sdau.edu.cn/tools/proteinfamily.html) databases. The physical and chemical properties of TaBES1 proteins from the longest transcripts were determined using the ExPASy website (http://web.ExPASy.org/translate/).
MEGA 10.0 software was used to reconstruct a phylogenetic tree of the TaBES1 family using the maximum-likelihood method with the full-length protein sequences. The BES1-like proteins from eight species were divided into subfamilies according to their clustering patterns. Each TaBES1 gene was mapped onto one of the 21 wheat chromosomes according to the IWGSC RefSeq v1.1 (‘Chinese Spring’) reference genome of the Triticeae Multi-omics Center (http://wheatomics.sdau.edu.cn/). The Ensembl database (http://plants.ensembl.org/biomart/martview/) was used to extract information about chromosome length, and a physical map was drawn using MG2C software v2.1 (http://mg2c.iask.in/mg2c-v2.1/).
Gene structures were visualized using the Gene Structure Display Server (http://gsds.gao-lab.org/). Conserved motifs of TaBES1 proteins were analyzed using MEME (http://meme-suite.org/index.html). TBtools software (https://github.com/CJ-Chen/TBtools/releases) was then used to analyze and visualize the TaBES1 gene structures and conserved motifs of TaBES1 proteins. A 2-kb upstream promoter region for each TaBES1 gene was retrieved from the Ensembl Plants website and then analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), finally the prediction of cis-regulatory elements was visualized using TBtools software. Alignment of BES1-domain sequences was performed using MEGA 10 software, and the sequence logo of this domain was created using the weblogo website (http://weblogo.berkeley.edu/).
Sequences of TaBES1 genes were uploaded to the psRNATarget website (https://www.zhaolab.org/psRNATarget/) to search for potential miRNA-mediated regulation based on known miRNAs and gene sequence information. The psRobot website (http://omicslab.genetics.ac.cn/) was used for the analysis of small RNA target predictions, and putative regulatory networks were visualized using Cytoscape software v3.8.2 (https://cytoscape.org/). Expression data based on transcriptome deep sequencing (RNA-seq) and microarray databases of T. aestivum ‘Chinese Spring’ from the WheatOmics database (http://wheatomics.sdau.edu.cn/). To analyze tissue-specific expression patterns, the coding sequence of each gene was submitted to the WheatOmics database to obtain expression data for roots, stems, leaves, spikes, anthers, and grain tissues. Transcripts per million (TPM) values for TaBES1 genes under drought, salt, cold, and heat stress conditions were also retrieved from the Triticeae Multi-omics Center database and visualized as heatmaps using TBtools software.
To investigate the tissue-specific expression profiles of TaBES1 genes, the following tissues were collected: root and shoot tissues at the seedling stage, leaves at the tilling stage, spikes and anthers at the full heading stage, and grain at the milking stage, from the wheat cultivar ‘Xiaobaimai’ (XBM), which serves as the original backbone parent wheat variety in northern China. As XBM is known for its sensitivity to abiotic stress at seedling stage (Li L. et al., 2018; Cao et al., 2021), its seeds were placed in Petri dishes containing two pieces of filter paper soaked with water at 23°C under a 16 h light/8 h dark photoperiod; germinated seeds were then transferred to plastic mesh grids at 28°C under a 16 h light/8-h dark photoperiod. Fourteen-day-old XBM seedlings were exposed to drought (25%, [w/v] polyethylene glycol 6000 [PEG-6000]), salt (250 mM NaCl), cold (4°C), or heat (40°C) for 0, 1, 2, 5, 10, or 24 h (Liu et al., 2015; Xu et al., 2020), and they were used for the gene expression analysis.
Seedlings from the cultivars ‘Taiyuan806’ (TY806), ‘Zhongyou9507’ (ZY9507), ‘Yumai8’ (YM8), ‘Xinong318’ (XN318), and ‘Zhongyin6’ (ZY6) were used to test drought resistance. TY806, XN318, and ZY6 were reported to be drought-tolerant, while XBM, ZY9507, and YM8 were considered drought-sensitive cultivars (Li L. et al., 2018; Cao et al., 2021). They were planted in pots containing a mixture of vermiculite and humus soil in equal volumes, and grown under appropriate watering conditions at 70% relative humidity and a temperature of 25°C, with a photoperiod of 16 hours of light and 8 hours of darkness in a climate incubator. When the seedlings reached the 2-leaf stage, watering was withheld for 2 weeks to induce drought stress. Additionally, PEG-induced drought stress was applied to these six wheat varieties. The seeds of the six wheat cultivars were germinated in a dark environment at a temperature of 23°C. After a two-day germination period, uniform seedlings were selected and transplanted into plastic containers filled with a hydroponic solution based on the Hoagland culture method. The growth conditions were maintained at a temperature of 28°C, with a photoperiod of 16 hours of light and 8 hours of darkness. Subsequently, ten-day-old seedlings with two leaves were subjected to PEG-induced drought stress (PEG-6000, 25% w/v) for 24 hours. To assess the impact of drought stress, the superoxide dismutase (SOD) activity, malondialdehyde (MDA) content, and peroxidase (POD) activity of the entire seedlings were measured. These measurements were conducted using the Total Superoxide Dismutase assay kit, Plant Malondialdehyde assay kit, and Peroxidase assay kit, respectively (Jiancheng Bioengineering Institute located in Nanjing, China). All assays were performed according to the manufacturer’s instructions. The expression levels of the other five wheat cultivars were assessed using 14-day-old seedlings under 0, 1, 2, 5, 10, and 24 hours of PEG treatment, which was the same as the treatment given to XBM. Finally, the seedlings of all six wheat cultivars under PEG treatment were used for gene expression analysis. Three independent experiments were performed for each line (Li L. et al., 2018; Cao et al., 2021)
Temperature was found to play a crucial role in controlling the fertility of TGMS lines, with moderate temperature promoting fertility and low temperature inducing sterility during the anther development (Liu et al., 2015; Xu et al., 2020). In this study, TGMS lines BS1453 and BS366 were grown in Beijing (BJ) (moderate temperature fertile environment) and Nanyang (NY) of Henan province (low temperature sterile environment). The common wheat cultivar ‘Jing411’ (J411) was used as a control and exhibited a fertile phenotype in both the BJ and NY environments. Anthers from BJ and NY environments were collected and frozen in liquid nitrogen immediately at each male reproductive development stage of central callose (S6), meiotic (S7), tetrad (S8), and young microspore (S9) (Browne et al., 2018; Xu et al., 2020). After confirming the anther development stages by microscope, anthers at the four developmental stages from BS1453, BS366 and J411 were performed for the following gene expression analysis by qPCR. The pollen starch was subjected to staining using a 10% (v/v) Lugol solution (Lee et al., 2016; Xu et al., 2020). The RNAs of 12 samples (anthers at S9 stage of BS1453, BS366 and J411 under fertile and sterile conditions, two biological replicates per stage of three cultivars) were subjected to 150 bp paried-end sequencing using Novaseq 6000 platform (Illumina), sequencing were performed according to the manufacturer’s standard protocol. Illumina RNA seq was constructed using the method of Külahoglu et al. (Browne et al., 2018; Xu et al., 2020). Each gene’s transcript profile was evaluated using fragments per kilobase of exon per million fragments mapped (FPKM).
Seedlings under various abiotic stress conditions and anthers from sterile and fertile conditions were collected and stored at -80°C. Total RNA was isolated from the samples using TRIzol reagent (Invitrogen) (Pandey et al., 2022). For qPCR, first-strand cDNA was synthesized using a Primer Script RT reagent Kit with gDNA Eraser (TaKaRa). The transcript levels were determined with ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China), and the qPCR were conducted in a CFXTM qPCR detection system (Bio-Rad, California, USA). Reaction systems were prepared in 20 μL volumes as follows: 10μL of 2μL Taq Pro Universal SYBR qPCR Master Mix, 6 μL of RNase-free water, 2 μL of cDNA, 1 μL of forward primer, and 1 μL of reverse primer. The primers were designed using Primer Premier 5.0. (Table S2). The qPCR reaction conditions were 94°C for 30 s, followed by 40 cycles of 95°C for 5s, 60°C for 30 s. Then a melting curve of 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s was used. TaACTIN gene (Gene ID: LOC542814) was performed as the reference gene and the relative gene expression levels were calculated using the 2-ΔΔCt method. Three biological replicates were performed for each treatment, with three technical replicates per sample.
Utilizing bioinformatics tools such as HMMER analysis and searches with the BLASTP tool and the Pfam database, we identified 59 putative BES1-like genes from the genomes of common wheat and its relatives. Of these genes, 23 were in Triticum aestivum, four in red wild einkorn wheat (Triticum urartu L.), 13 in wild emmer wheat (Triticum dicoccoides L.), 6 in Tausch’s goatgrass (Aegilops tauschii L.), and thirteen in durum wheat (Triticum turgidum L.), (Table S1). According to the gene nomenclature, we named the 23 wheat BES1-like genes based on their chromosomal locations and homoeologous genes was named in the same number with different alphabet letters corresponding to their respective subgenomes (Table S1). TaBES1 genes were unevenly distributed across 17 of 21 wheat chromosomes (Figure S1). Each chromosome harbored one TaBES1 gene except chromosomes 3A, 3B, 3D, 6A, and 6B, which each carried two genes. No TaBES1 gene mapped to chromosomes 1B, 1D, 5A, or 5D. The 23 TaBES1 genes were almost evenly distributed across the A (8), B (8), and D (6) subgenomes. As listed in Table S1, the average amino acid length of the putative TaBES1 proteins was 365 amino acids (aa), ranging from 178 to 686 aa, and TaBES1-4D was the longest protein (686 aa). The predicted molecular weights of TaBES1 proteins ranged from 19.27 kDa (TaBES1-3D1) to 75.47 kDa (TaBES1-4D); all TaBES1 proteins were hydrophobic.
We constructed an unrooted phylogenetic tree using the full-length sequences for the 59 BES1 proteins from common wheat and its relatives identified in this study and those of reported BES1 proteins from Arabidopsis (eight), rice (six), and maize (eleven) (Table S3). The tree formed four major clades (I-IV) based on the phylogenetic relationship between each BES1-like protein and Arabidopsis BES1 members (Figure 1). TaBES1-4B, TaBES1-4D, TaBES1-6A1, TaBES1-6B1, and TaBES1-Un were closely related to AtBMY2 and AtBMY4, which clustered in clade I. TaBES1-3A2, TaBES1-3B2, and TaBES1-3D2 formed a tight clade with Arabidopsis BEH3 and BEH4 within clade II. Clade III contained TaBES1-6A2, TaBES1-6B2, TaBES1-6D, TaBES1-7A, TaBES1-7B, and TaBES1-7D, with no Arabidopsis member. Finally, TaBES1-1A, TaBES1-2A, TaBES1-2B, TaBES1-2D, TaBES1-3A1, TaBES1-3B1, TaBES1-3D1, TaBES1-4A, and TaBES1-5B were closely related to Arabidopsis tapetum development associated proteins BEH1, BEH2, BES1, and BZR1, which all belonged to the largest clade IV.
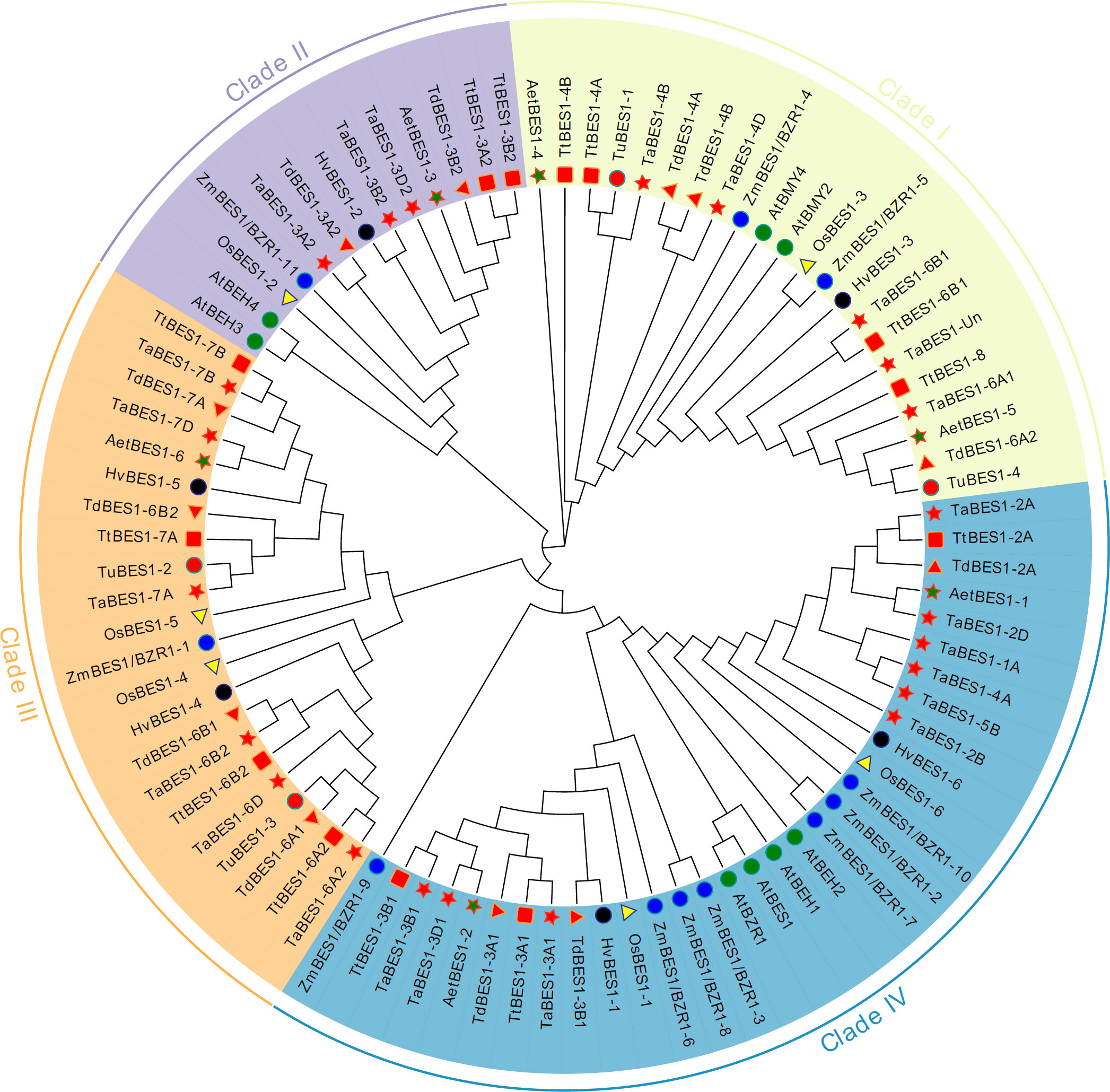
Figure 1 Phylogenetic analysis of BES1 proteins. The maximum-likelihood tree was constructed based on the BES1 proteins from Arabidopsis (At), rice (Os), maize (Zm), wheat (Ta) and its relatives (Triticum Urartu, Tu; Triticum dicoccoides, Td; Triticum turgidum, Tt; and Aegilops tauschii, Aet) via Mega 10 software.
We analyzed the exon-intron structure of all wheat BES1-like genes by aligning their predicted coding sequences to the wheat genome. TaBES1 genes from the same subclade shared a common gene structure (Figure S2A). Most wheat BES1 genes contained one intron and two exons; TaBES1-4A had two introns and three exons, while TaBES1-4B, TaBES1-4D, TaBES1-6A1, TaBES1-6B1, and TaBES1-Un belonging to clade I had more than seven introns. The intron and exon number of clade I was far greater than that of the other clades.
We also examined cis-regulatory elements within 2-kb fragments of promoter sequence upstream of each TaBES1 gene. As shown in Figure S2B, we identified cis-acting elements related to phytohormone signaling, including methyl jasmonate, abscisic acid, salicylic acid, gibberellin, and auxin. We also detected several abiotic stress responsive cis-acting elements, such as those associated with wounding, drought, cold, and light.
Proteins belonging to the same subfamily and sharing similar motif compositions are likely to share similar functions, prompting us to characterize functional domains in each TaBES1 protein (Li et al., 2019b). Motif analysis highlighted the high level of conservation of TaBES1 proteins, with motif 1 being shared by all TaBES1 proteins except TaBES1-5B (Figure S2C). Moreover, we detected motif 2 in all proteins except TaBES1-6A1, while motif 5 was present in all proteins except TaBES1-3A1, TaBES1-3B1, and TaBES1-3D1. Closely related family members in the phylogenetic tree shared a common motif composition (Li et al., 2019b); we identified motif 10 in members from subclades I, II, and III, while motifs 3, 8, and 9 only existed within subclade I members (Figure S2C).
The BES1-type domain is conserved across plant species. An analysis of conserved amino acid residues among wheat BES1-type domains produced results similar to comparable analyses in Arabidopsis, maize, tomato, and other species, illustrating the high conservation of the N terminus, while the C terminus of TaBES1 members was less conserved (Figure S3A). BES1 proteins contain a putative nuclear localization sequence, a highly conserved N-terminal region, a BIN2 (BR-INSENSITIVE 2) phosphorylation domain, a PEST motif, and a C-terminal structure (Cao et al., 2020). Similar to other species, sequence logo analysis showed that the N-terminal BES1-type domain was highly conserved between Arabidopsis and wheat BES1 family members (Figure S3B).
To investigate the potential regulation of TaBES1 genes by small regulatory RNAs, we uploaded all 23 TaBES1 sequences to the wheat miRNA database; which predicted that eight wheat miRNAs have the capacity to regulate TaBES1 transcript levels, based on their high sequence complementarity (Figure 2). Most TaBES1 genes were regulated by only one miRNA, although TaBES1-2A, TaBES1-2D, TaBES1-3B2, TaBES1-6A2, TaBES1-6B2, and TaBES1-6D were each regulated by two miRNAs. Notably, Tae-miRNA9780 targeted the transcripts of five TaBES1 genes (TaBES1-2A, TaBES1-2D, TaBES1-6A2, TaBES1-6B2, and TaBES1-6D), suggesting its importance during BR signal transduction. The miR171, miR1119, miR9657, miR9776, and miR9780 families have been reported to play roles in plant stress responses, with the miR171, miR9657, and miR9677 families being also associated with anther development (Bai et al., 2017; Shi et al., 2018; Sun et al., 2018; Li et al., 2019a; Zeeshan et al., 2021; Zhang et al., 2022).
We obtained initial tissue expression data from the WheatOmics database. The expression levels of TaBES1 genes varied across tissues and developmental stages (Figure S4). Expression levels of nine of the 23 TaBES1 family members were extremely low during all developmental stages and in all tissues. By contrast, TaBES1-3A2, TaBES1-3B2, and TaBES1-3D2 on chromosome 3 showed higher expression levels during the whole life cycle (Figure S4). We confirmed the expression patterns of TaBES1 genes in roots, stems, leaves, spikes, anthers, and seed from XBM by qPCR, which revealed that expression levels in root tissues are relatively higher than those in other tissues (Figure S5). TaBES1-1A belongs to the same subgroup as SlBZR1, which also contains Arabidopsis BES1, BZR1, and BEH1 involved in anther development (Figure 1). According to the wheat-expression databases and qPCR results, TaBES1-3A2, TaBES1-3B2, TaBES1-3D2, TaBES1-4B, and TaBES1-4D exhibited relatively higher expression levels in anthers than in other tissues and were therefore considered anther-preferential genes (Figure S5).
To determine how TaBES1 transcription responds to abiotic stresses, we downloaded expression data for TaBES1 genes in plants exposed to drought, salt, cold, or heat from the WheatOmics database; most TaBES1 genes were substantially downregulated under these abiotic stress conditions (Figure S6). To validate the expression patterns of TaBES1 genes, we conducted qPCR using total RNA extracted from fourteen-day-old XBM seedlings subjected to drought, salt, cold, and heat abiotic stresses for 0, 1, 2, 5, 10, 24 h. Finally, we observed a high degree of congruence between qPCR and RNA-seq data, with the transcript levels of most genes being affected by stress treatments visualized as heatmaps using TBtools software with log scale base as 2 and log with as 1 (Figure 3). Drought stress considerably downregulated the expression levels of most TaBES1 genes, with the exception of TaBES1-2A, TaBES1-6D, and TaBES1-7A, which were upregulated under the same conditions (Figure 3A). In response to salt stress, the expression levels of all family members tended to decline, except TaBES1-3A2, which was considerably upregulated (Figure 3B). The expression levels of almost all TaBES1 genes were very low upon exposure to cold stress, while TaBES1-3D1 and TaBES1-6D exhibited an upregulation after low-temperature treatment (Figure 3C). The expression of TaBES1-3A2, TaBES1-6B1, TaBES1-6D, TaBES1-7D, and TaBES1-Un was induced at the beginning of heat treatment, while that of the other TaBES1 genes was substantially suppressed (Figure 3D).
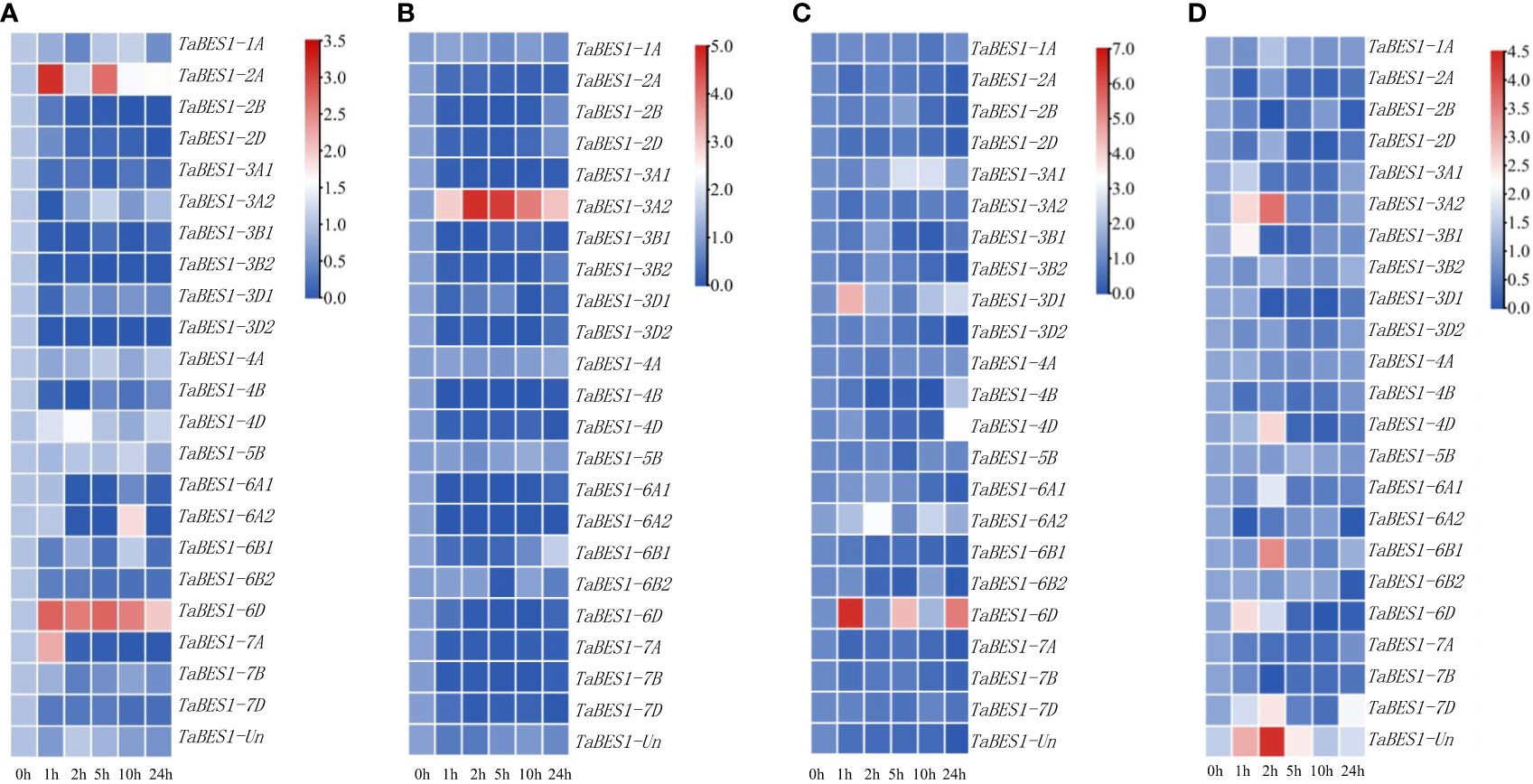
Figure 3 Expression profiles of TaBES1s under abiotic stresses: (A) drought (PEG6000), (B) salt, (C) cold and (D) heat stress conditions using qPCR. A heatmap of the relative expression of all the TaBES1 genes. The color key (blue to red) represent the relative gene expression values as fold change. For each gene, the 0 h treatment expression value was set as 1.0. Relative expression of each TaBES1 gene was normalized to TaACTIN.
To further elucidate the drought resistance level of each wheat cultivar at the seedling stage, we conducted both water-withhold treatment and PEG-induced drought stress on the six cultivars. The experimental design included a well-watered control group and a drought stress group where water was withheld, validating the previously reported levels of drought resistance. Specifically, TY806, XN318, and ZY6 were found to exhibit drought tolerance, while XBM, ZY9507, and YM8 were identified as drought-sensitive cultivars (Li L. et al., 2018; Cao et al., 2021) (Figure S8A). These findings were further confirmed by the application of PEG treatment, which resulted in significantly higher levels of SOD values in the three drought-resistant cultivars compared to the sensitive cultivars. Similarly, POD values exhibited a similar trend to that of the SOD values. Furthermore, the MDA values in the three drought-resistant cultivars were significantly lower than those observed in the sensitive cultivars (Figure S8B). Based on these significant findings, the aforementioned three cultivars (TY806, XN318, ZY6) were classified as drought-resistant, whereas the remaining three cultivars (YM8, ZY9507, XBM) were categorized as drought-sensitive.
We further investigated the roles of TaBES1 members in response to drought stress by measuring their expression profiles in drought-tolerant and drought-sensitive cultivars. Wheat cultivar XBM is stress-sensitive at the seedling stage (Li L. et al., 2018; Cao et al., 2021); we determined that most TaBES1 genes are considerably suppressed in response to drought stress (Figure 3A). By contrast, the expression patterns of TaBES1-2A, TaBES1-3B2, TaBES1-3D2, TaBES1-6B1, TaBES1-6D, and TaBES1-7D were different in the drought-tolerant cultivar TY806 from those in XBM (Figure S7). We observed similar expression profiles for these six genes in three drought-sensitive cultivars (XBM, ZY9507, and YM8) and three drought-tolerant cultivars (TY806, XN318, and ZY6) at the seedling stage (Li L. et al., 2018) (Figures 4A, B).In general, we detected no obvious commonality in the expression patterns of TaBES1-2A, TaBES1-6B1, or TaBES1-7A between drought-tolerant and -sensitive cultivars (Figure S8C). The relative expression levels of TaBES1-3B2 in drought-tolerant cultivars showed a substantial elevation compared to those measured in drought-sensitive cultivars (Figure 4C). TaBES1-3B2 is the homolog of the positive drought regulator gene TaBZR2 (TaBES1-3D2), which positively regulates the drought stress response(Cui et al., 2019). Thus, we hypothesize that TaBES1-3B2 might have a role in drought signaling, and its function should be investigated in more detail future experiments. The expression patterns of TaBES1-6D were different among drought-sensitive and drought-tolerant cultivars; TaBES1-6D expression was greatly upregulated in drought-sensitive cultivars but downregulated in drought-tolerant cultivars. We therefore proposed that TaBES1-6D is a negative regulator in the response to drought stress.
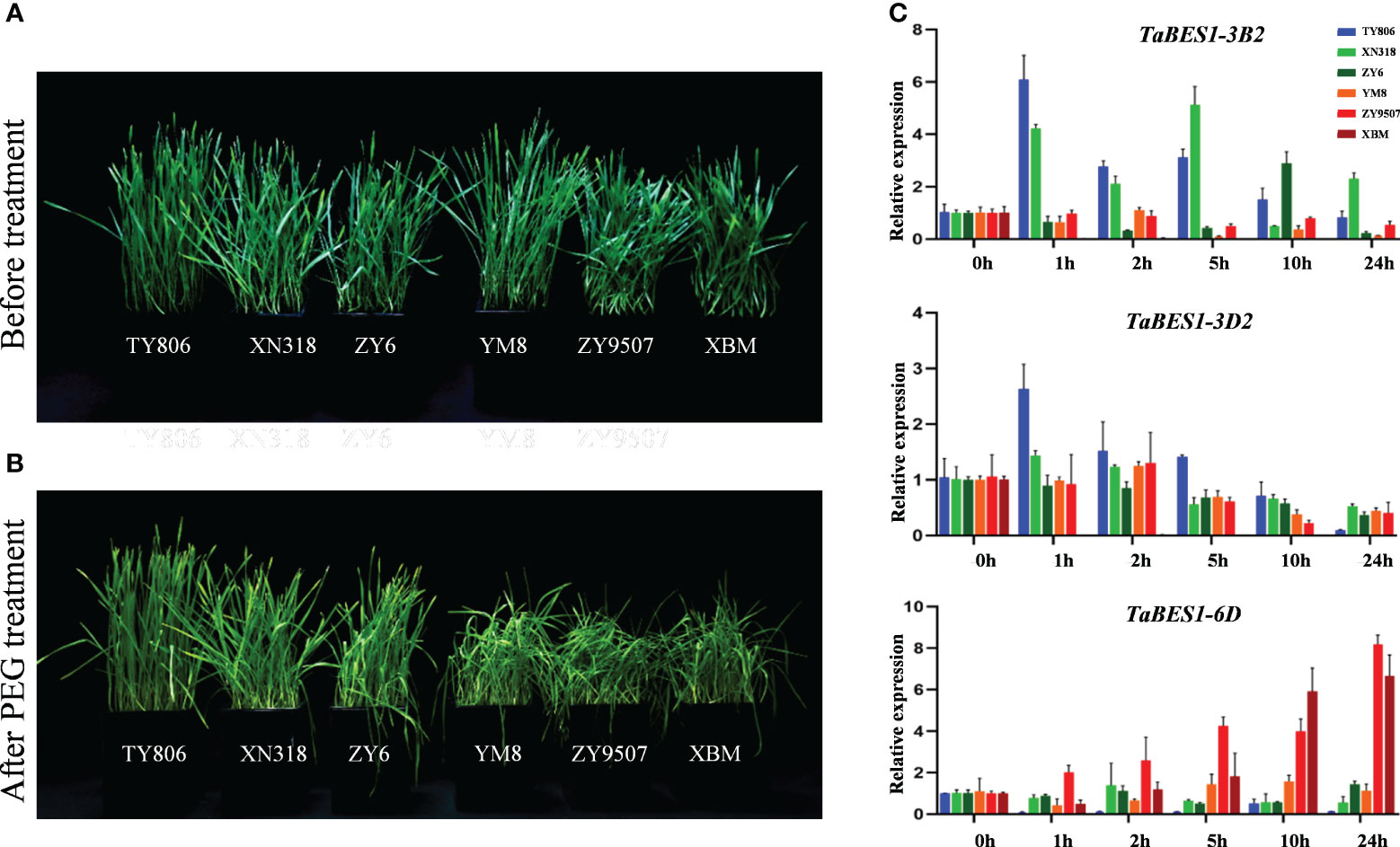
Figure 4 (A) Phenotypes of drought tolerant (TY806, XN318, ZY6) and sensitive cultivars (YM8, ZY9507, XBM) at the seedling stage under well-watered control conditions and (B) PEG-induced stress conditions (PEG-6000, 25% w/v). (C) The expression patterns of TaBES1-3B2, TaBES1-3D2 and TaBES1-6D under 0, 1, 2, 5, 10, 24 h PEG treatment in drought-tolerant (TY806, XN318, and ZY6) and drought-sensitive cultivars (XBM, ZY9507, and YM8). For each gene, the 0 h treatment expression value was set as 1.0. Relative expression of each TaBES1 gene was normalized to TaACTIN.
Previous studies have reported that BES1 members play roles in anther development (Chen et al., 2019a; Chen et al., 2019b). Therefore, besides the responses to different abiotic stresses, we also explored the potential functions of anther-preferential TaBES1 genes. Using data from the wheat expression database and qPCR results, we determined the expression levels of anther-preferential wheat BES1 genes (TaBES1-1A, TaBES1-3A2, TaBES1-3B2, TaBES1-3D2, TaBES1-4B, and TaBES1-4D) in sterile lines and a common wheat cultivar at the S6~S9 anther developmental stages (Browne et al., 2018), which are extremely important for regulation of anther fertility (Xu et al., 2020). In the case of anthers of TGMS lines under sterile conditions (NY), the sensitive period of anther development aligns with a low-temperature environment, leading to significant inhibition in the development of TGMS anthers and a low seed setting rate. Conversely, under fertile conditions (BJ), anther development is delayed compared to the sterile conditions, occurring in a more favorable moderate-temperature environment, resulting in normal seed setting (Li et al., 2006; Bai et al., 2017; Xu et al., 2020) (Figure S9A). Under moderate temperature fertile conditions, the pollen morphology of TGMS lines was similar to the common wheat cultivar, with almost full filled starch; under sterile (low temperature) conditions, however, the pollen of TGMS lines nearly no starch-filled, while the common wheat cultivar pollen was still comparable to that seen under fertile conditions (Figures 5A, S9A).
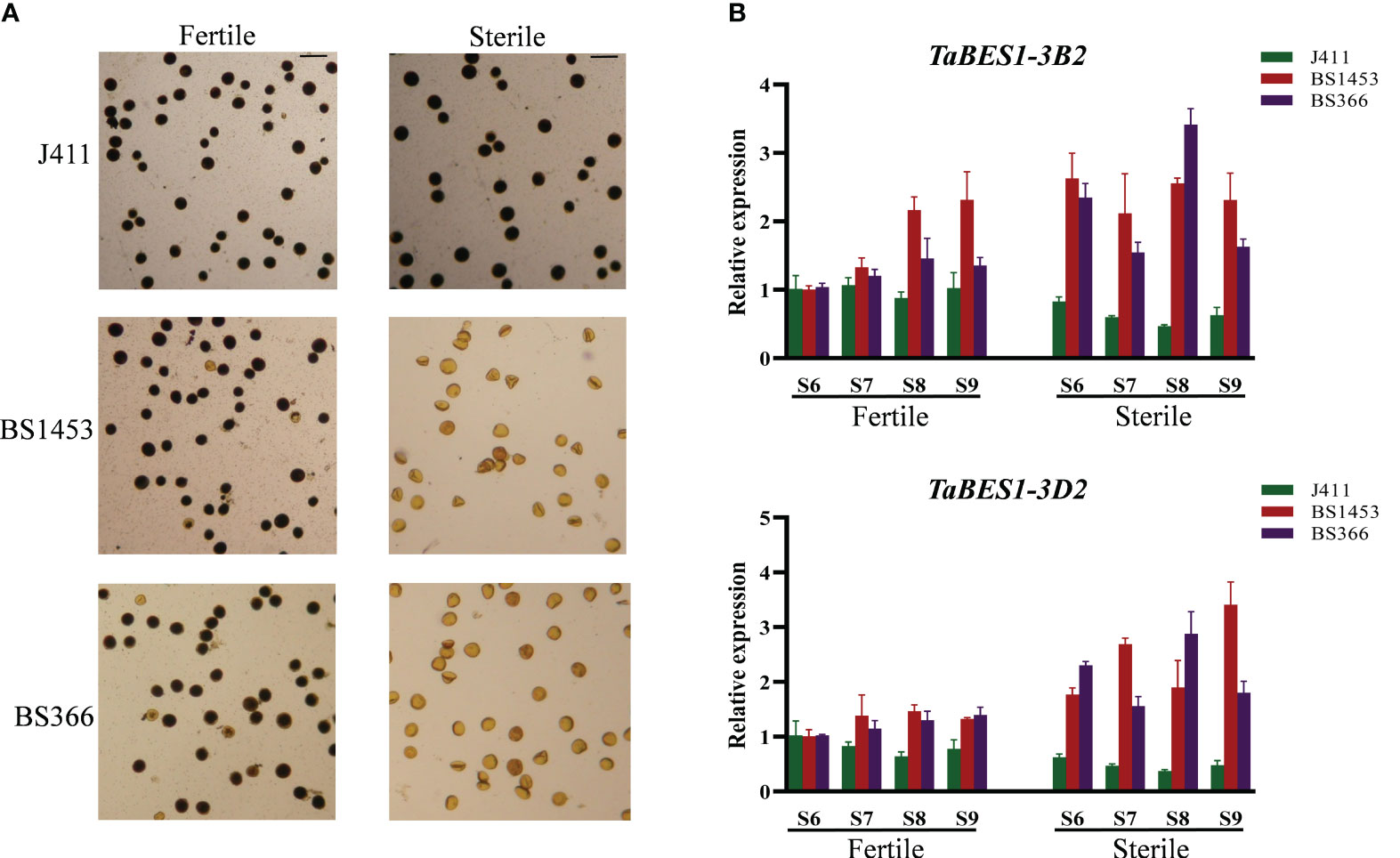
Figure 5 (A) Morphology of pollen grains stained with Lugol’s iodine in TGMS lines (BS1453 and BS366) and common wheat (J411) in the moderate temperature fertile conditions and low temperature sterile conditions. Bars=100 μm. (B) The expression patterns of TaBES1-3B2 and TaBES1-3D2 in TGMS lines (BS1453 and BS366) and common wheat cultivar (J411) in moderate temperature fertile conditions and low temperature sterile conditions. For each gene, the S6 stage under fertile condition expression value was set as 1.0. Relative expression of each TaBES1 gene was normalized to TaACTIN.
In general, no substantial changes were detected in expression levels between the TGMS lines and the common wheat cultivar under fertile conditions (Figure 5B); however, we detected substantial differences between the TGMS lines and the common wheat cultivar under sterile (low temperature) conditions (Figure 5B), especially for the expression patterns of TaBES1-3B2 and TaBES1-3D2 (Figure 5B). Indeed, TaBES1-3B2 expression showed no considerable difference in the common wheat cultivar between sterile and fertile conditions merely with a slight decline under sterile condition, whereas in the TGMS lines, TaBES1-3B2 expression was higher under sterile conditions as compared to fertile conditions. Notably, the expression pattern of TaBES1-3D2 was similar to that of TaBES1-3B2, showing a considerable rise in expression levels in the TGMS lines under low-temperature conditions. Then the transcriptome analysis was further performed to investigate the expression patterns of TaBES1s in TGMS lines and common wheat cultivar (Figure S10), and a high degree of congruence was observed between qPCR and RNA-seq results. Generally, in the common cultivar, TaBES1-3B2 and TaBES1-3D2 expression showed no considerable difference under fertile and sterile conditions; however, under sterile conditions, the transcript levels of TaBES1-3B2 and TaBES1-3D2 were substantially higher than the fertile conditions in TGMS lines.
BRs are necessary throughout the plant life cycle. BES1 family members are widely distributed in plants and play a vital role in BR signaling associated with various abiotic stress responses, cell proliferation and differentiation, seed germination, tapetal cell degeneration, and pollen development (Chen et al., 2019a; Chen et al., 2019b; Yan et al., 2020). Common wheat (Triticum aestivum) is an allohexaploid plant, with Triticum urartu as the A genome donor, Aegilops speltoides as the B genome donor, and Aegilops tauschii as the D genome donor. It is thought that T. urartu (AA) first hybridized with A. speltoides (BB) to form wild emmer wheat (Triticum dicoccoides; AABB); T. dicoccoides (AABB) then hybridized with A. tauschii (DD) to produce common wheat (AABBDD) (Pont et al., 2019). We analyzed the BES1 gene family in common wheat and its relatives, and found that gene number was largely positively associated with species ploidy level. Wheat appears to contain more BES1 genes than rice, maize, or other graminaceous crops, and the 23 wheat BES1 genes are almost evenly distributed across the A, B, and D subgenomes. Among wheat and its relatives, the largest number of BES1-like genes were found mapped to group-3 chromosomes, group-6 chromosomes. Interestingly, the number of exons on group-3 chromosomes was generally two, moreover, the synteny analysis showed that the genes on group-3 chromosomes might be relatively conserved over long evolutionary periods among plant species (Figure S11).
Studies on BES1 family members have been reported in many species; however, knowledge on the functions of wheat BES1 transcription factors is very limited. Phylogenetic analysis revealed that the 23 wheat BES1 family members can be classified into four subgroups. Relatively conservative TaBES1-3A2, TaBES1-3B2, and TaBES1-3D2 belonged to the same clade as Arabidopsis BEH3 and BEH4, which participate in anther development. Predictions from the wheat-expression database and qPCR results showed that all TaBES1 genes were highly expressed in roots, with TaBES1-1A, TaBES1-3A2, TaBES1-3B2, TaBES1-3D2, TaBES1-4B, and TaBES1-4D displaying relatively high expression levels in anthers. Correspondingly, the transcriptome data also showed that TaBES1-3A2, TaBES1-3B2, TaBES1-3D2, with relatively higher expression levels among all the TaBES1-like genes.
BES1 proteins play crucial roles in plant adaptation to environmental stress (Li and Deng, 2005; Anwar et al., 2018; Cui et al., 2019; Liao et al., 2020; Sun et al., 2020); moreover, most TaBES1 genes displayed root-specific expression. Hence we examined the expression profiles of TaBES1 genes in response to various stress conditions. The expression of wheat BES1 genes was induced by drought, salt, cold, and heat treatments, with most TaBES1 genes exhibiting downregulated expression after stress treatment. Six TaBES1 genes showed different expression patterns between drought-tolerant and -sensitive cultivars; we confirmed their potential functions in drought stress responses by determining their expression levels in three drought-tolerant lines and three drought-sensitive lines. We identified TaBES1-3B2, TaBES1-3D2, and TaBES1-6D as drought-related genes. The expression levels of TaBES1-3D2 were high in three drought-tolerant cultivars, especially in the early stages of drought stress, while TaBES1-3D2 expression declined in drought-sensitive lines during stress. Our study therefore confirmed the function of TaBZR2/TaBES1-3D2 in wheat drought tolerance (Cui et al., 2019). The TaBES1-3D2 homolog on chromosome 3B is TaBES1-3B2, which has a potential function in drought-tolerance; however, another homologous gene, TaBES1-3A2, appears to have no obvious relation to drought tolerance.
Previous studies have shown that the miRNA171 family plays roles in responses to biotic and abiotic stresses (Hwang et al., 2011); moreover, tae-miR171a is predicted to enhance salt tolerance based on its substantial downregulation seen in RNA-seq datasets (Zeeshan et al., 2021). tae-miR171a might also mediate the male-sterile signaling pathway, possibly participating in anther development, since tae-miR171a is downregulated in wheat male-sterile lines (Bai et al., 2017). The miRNA1119 family is associated with drought stress, with tae-miR1119 being predicted to play critical roles in regulating wheat drought tolerance (Shi et al., 2018). tae-miR9657 may regulate salt tolerance by enhancing the expression of MYB-related transcription factors (Zeeshan et al., 2021); RNA-seq analysis also indicated that tae-miR9657a-3p is highly abundant in wheat male-sterile lines (Bai et al., 2017). Li et al. and Han et al. both reported that tae-miR9677b levels are lower in male-sterile lines compared to fertile lines (Li et al., 2019a), which might suggest a function in anther development. The gene regulated by miR9776 encoding lipoxygenase plays essential roles in lipid metabolism and confers the osmotic stress response (Zhao et al., 2020), while tae-miR9780 might be involved in the response to abiotic stress (Zhang et al., 2022). Moreover, RNA-seq analysis demonstrated that tae-miR171a displays differential abundance in the wheat TGMS line under both sterile and fertile conditions (Bai et al., 2017); tae-miR171a is believed to be involved in the regulation of wheat male sterility. TaBES1-3B2 was predicted to be a target gene of tae-miR171a, suggesting a potential role in male sterility. Whereas, the potential functions those miRNAs might have on the regulation of drought stress and anther development still needed further experiments to be proved.
Wheat lines overexpressing TaBZR2/TaBES1-3D2 exhibit improved drought tolerance, while TaBZR2/TaBES1-3D2 RNA interference (RNAi) lines display a drought-sensitive phenotype (Cui et al., 2019). TaBES1-3B2 and TaBES1-3D2 share high sequence identity, suggesting a similar function in the drought stress response. Moreover, our small RNA target prediction analysis revealed that TaBES1-3B2 is a predicted target gene of tae-miR171a, which confers drought memory to wheat plants (Hwang et al., 2011; Yue et al., 2022); this result suggests that TaBES1-3B2 might be involved in drought stress adaptation. TaBES1-6D exhibited the expression pattern opposite to that of TaBES1-3B2 and TaBES1-3D2, suggesting it is a negative regulator of drought response. TaBES1-6D was predicted to be the target gene of tae-miR9776 and tae-miR9780, which are both associated with plant osmotic stress signaling pathways (Zeeshan et al., 2021; Zhang et al., 2022), indicating that TaBES1-6D is probably related to stress responses.
Besides their regulation of drought responses, BES1 members participate in anther developmental in several species; for example, tomato SlBZR1 directly binds to the SlRBOH1 promoter, leading to pollen defects. While TGMS lines also display a pollen defect phenotype, they greatly benefit wheat hybrid breeding and production. Hence, we explored the potential contribution of TaBES1 genes to TGMS during anther developmental stages. We determined the expression profiles of six genes preferentially expressed in anthers (TaBES1-1A, TaBES1-3A2, TaBES1-3B2, TaBES1-3D2, TaBES1-4B and TaBES1-4D) in sterile lines and common wheat cultivar, as well as under fertile and sterile conditions. The expression levels of TaBES1-3B2 and TaBES1-3D2 showed large differences in sterile lines compared to the common wheat cultivar. Under fertile conditions, we detected no substantial change between sterile lines and common wheat cultivar. However, under the sterile, low-temperature environment, sterile lines displayed considerably higher expression levels of TaBES1-3B2 and TaBES1-3D2, while those of common wheat cultivar greatly decreased. RNA-seq analysis revealed the expression profiles of TaBES1-3B2 and TaBES1-3D2 were basically congruent with qPCR analysis. No substantial changes were observed between sterile lines and common wheat cultivar under fertile conditions, while the expression levels of TaBES1-3B2 gene was highly increased under sterile conditions in TGMS lines comparing with common wheat cultivar. The phylogenetic analysis illustrated that TaBES1-3B2 and TaBES1-3D2 were closely to Arabidopsis tapetum and microsporocyte development proteins BEH3 and BEH4. In addition, the synteny analysis revealed that AtBEH3 represented the orthologous gene of TaBES1-3A2, TaBES1-3B2 and TaBES1-3D2. Moreover, both the qPCR analysis and transcriptome results indicated TaBES1-3B2 and TaBES1-3D2 exhibited high expression levels in the TGMS lines under sterile conditions, which implies a plausible role of these genes in low temperature-induced wheat male sterility. Collectively, our investigation provides a fundamental understanding of wheat male sterility and molecular breeding. Nonetheless, the precise functions and mechanisms of these genes in male sterility require further exploration through the construction of gene knocking-out lines in common wheat.
Previous studies have demonstrated the functions of BES1 proteins in response to various stresses and their roles in anther development in Arabidopsis. The current study confirmed the potential roles of BES1 members under abiotic stress conditions and during anther development in common wheat. We identified 23 TaBES1 family members, and their expression profiles indicated that TaBES1-3B2, TaBES1-3D2, and TaBES1-6D might be drought-related genes. Furthermore, TaBES1-3B2 and TaBES1-3D2 might play roles in low temperature-induced male sterility signaling pathways. Taken together, these results suggest that TaBES1-3B2 and TaBES1-3D2 are not only involved in drought tolerance, but also possibly participate in the regulation of low temperature-induced male sterility. These results further our understanding of the molecular mechanisms underlying TaBES1 members in regulating plant growth and development.
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) repository, accession number SAMN35358019-SAMN35358030 and the BioProject ID PRJNA976038.
YT designed the experiment, SL and JZ carried out the experiment, JZ wrote the paper, DW contributed to data analysis. SL, WW, XH, ZF, TL, QL, YS, CG, and CZ participated in field trials. All authors contributed to the article and approved the submitted version.
This research was supported by Collaborative Innovation Center of Genomics Breeding of BAAFS (KJCX201907-2), Foundation for Youths of BAAFS (QNJJ202316), and Beijing Natural Science Foundation Project (6222011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1219856/full#supplementary-material
Ahad, A., Aslam, R., Gul, A., Amir, R., Munir, F., Batool, T. S., et al. (2021). Genome-wide analysis of bZIP, BBR, and BZR transcription factors in Triticum aestivum. PloS One 16, e0259404. doi: 10.1371/journal.pone.0272533
Anwar, A., Liu, Y., Dong, R., Bai, L., Yu, X., Li, Y. (2018). The physiological and molecular mechanism of brassinosteroid in response to stress: a review. Biol. Res. 51, 46. doi: 10.1186/s40659-018-0195-2
Bai, J. F., Wang, Y. K., Wang, P., Duan, W. J., Yuan, S. H., Sun, H., et al. (2017). Uncovering male fertility transition responsive mirna in a wheat photo-thermosensitive genic male sterile line by deep sequencing and degradome analysis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01370
Browne, R. G., Iacuone, S., Li, S. F., Dolferus, R., Parish, R. W. (2018). Anther morphological development and stage determination in Triticum aestivum. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00228
Cao, X., Khaliq, A., Lu, S., Xie, M., Ma, Z., Mao, J., et al. (2020). Genome-wide identification and characterization of the BES1 gene family in apple (Malus domestica). Plant Biol. 22, 723–733. doi: 10.1111/plb.13109
Cao, Z., Wu, X., Wang, D., Liu, S., Yang, H., Hao, X., et al. (2021). Cloning and expression analysis of wheat multiprotein bridging factor gene TaBF1c. J. Triticeae Crops 41, 391–400. doi: 10.7606/jissn.1009-1041.2021.04.01
Chen, L. G., Gao, Z., Zhao, Z., Liu, X., Li, Y., Zhang, Y., et al. (2019a). BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant 12, 1408–1415. doi: 10.1016/j.molp.2019.06.006
Chen, W., Lv, M., Wang, Y., Wang, P. A., Cui, Y., Li, M., et al. (2019b). BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 10, 4164. doi: 10.1038/s41467-019-12118-4
Cui, X. Y., Gao, Y., Guo, J., Yu, T. F., Zheng, W. J., Liu, Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Feng, Z., Shi, H., Lv, M., Ma, Y., Li, J. (2021). Protein farnesylation negatively regulates brassinosteroid signaling via reducing BES1 stability in Arabidopsis thaliana. J. Integr. Plant Biol. 63, 1353–1366. doi: 10.1111/jipb.13093
Guo, H., Ye, H., Li, L., Yin, Y. (2009). A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal. Behav. 4, 784–786. doi: 10.1073/pnas.0812346106
Hwang, E. W., Shin, S. J., Yu, B. K., Byun, M. O., Kwon, H. B. (2011). miR171 family members are involved in drought response in Solanum tuberosum. J. Plant Biol. 54, 43–48. doi: 10.1007/s12374-010-9141-8
Jia, C., Zhao, S., Bao, T., Zhao, P., Peng, K., Guo, Q., et al. (2021). Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 302, 110719. doi: 10.1016/j.plantsci.2020.110719
Jiang, J., Zhang, C., Wang, X. (2015). A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 27, 361–374. doi: 10.1105/tpc.114.133678
Kesawat, M. S., Kherawat, B. S., Singh, A., Dey, P., Kabi, M., Debnath, D., et al. (2021). Genome-wide identification and characterization of the Brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 22, 8743. doi: 10.3390/ijms22168743
Khan, M. K., Pandey, A., Athar, T., Choudhary, S., Deval, R., Gezgin, S., et al. (2020). Fusarium head blight in wheat: contemporary status and molecular approaches. 3 Biotech. 10, 172. doi: 10.1007/s13205-020-2158-x
Kono, A., Yin, Y. (2020). Updates on BES1/BZR1 regulatory networks coordinating plant growth and stress responses. Front. Plant Sci. 11, 617162. doi: 10.3389/fpls.2020.617162
Korwin Krukowski, P., Visentin, I., Russo, G., Minerdi, D., Bendahmane, A., Schubert, A., et al. (2022). Transcriptome analysis points to BES1 as a transducer of strigolactone effects on drought memory in Arabidopsis thaliana. Plant Cell Physiol 63, 1873–1889. doi: 10.1093/pcp/pcac058
Lachowiec, J., Mason, G. A., Schultz, K., Queitsch, C. (2018). Redundancy, feedback, and robustness in the Arabidopsis thaliana BZR/BEH gene family. Front. Genet. 9. doi: 10.3389/fgene.2018.00523
Lee, S. K., Eom, J. S., Hwang, S. K., Shin, D., An, G., Okita, T. W., et al. (2016). Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J. Exp. Bot. 67, 5557–5569. doi: 10.1093/jxb/erw324
Li, L., Deng, X. W. (2005). It runs in the family: regulation of brassinosteroid signaling by the BZR1-BES1 class of transcription factors. Trends Plant Sci. 10, 266–268. doi: 10.1016/j.tplants.2005.04.002
Li, Q., Guo, L., Wang, H., Zhang, Y., Fan, C., Shen, Y. (2019b). In silico genome-wide identification and comprehensive characterization of the BES1 gene family in soybean. Heliyon 5, e01868. doi: 10.1016/j.heliyon.2019.e01868
Li, H. X., Guo, J. L., Zhang, C. Y., Zheng, W. J., Song, Y. L., Wang, Y. (2019a). Identification of differentially expressed mirnas between a wheat K-type cytoplasmic male sterility line and its near-isogenic restorer line. Plant Cell Physiol. 60, 1604–1618. doi: 10.1093/pcp/pcz065
Li, Y., He, L., Li, J., Chen, J., Liu, C. (2018). Genome-wide identification, characterization, and expression profiling of the Legume BZR transcription factor gene family. Front. Plant Sci. 9, 1332. doi: 10.3389/fpls.2018.01332
Li, L., Mao, X., Wang, J., Chang, X., Liu, Y., Jing, R. (2018). Drought tolerance evaluation of wheat germplasm resources. Acta Agronomica Sin. 44, 988–999. doi: 10.3724/SP.J.1006.2018.00988
Li, Q., Xu, F., Chen, Z., Teng, Z., Sun, K., Li, X., et al. (2021). Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat. Plants 7, 1108–1118. doi: 10.1038/s41477-021-00959-1
Li, Y. F., Zhao, C. P., Zhang, F. T., Sun, H., Sun, D. F. (2006). Fertility alteration in the photo-thermo-sensitive male sterile line BS20 of wheat (Triticum aestivum L.). Euphytica. 151, 207–213. doi: 10.1007/s10681-006-9141-4
Liao, K., Peng, Y. J., Yuan, L. B., Dai, Y. S., Chen, Q. F., Yu, L. J., et al. (2020). Brassinosteroids antagonize jasmonate-activated plant defense responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol. 182, 1066–1082. doi: 10.3389/fpls.2018.01332
Liu, D., Cui, Y., Zhao, Z., Li, S., Liang, D., Wang, C., et al. (2021). Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genomics 22, 682. doi: 10.1186/s12864-021-08002-5
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15, 152. doi: 10.1186/s12870-015-0511-8
Mecchia, M. A., Garcia-Hourquet, M., Lozano-Elena, F., Planas-Riverola, A., Blasco-Escamez, D., Marques-Bueno, M., et al. (2021). The BES1/BZR1-family transcription factor MpBES1 regulates cell division and differentiation in Marchantia polymorpha. Curr. Biol. 31, 4860–4869 e4868. doi: 10.1016/j.cub.2021.08.050
Otani, Y., Tomonaga, Y., Tokushige, K., Kamimura, M., Sasaki, A., Nakamura, Y., et al. (2020). Expression profiles of four BES1/BZR1 homologous genes encoding bHLH transcription factors in Arabidopsis. J. Pestic. Sci. 45, 95–104. doi: 10.1584/jpestics.D20-001
Pandey, A., Khan, M. K., Hamurcu, M., Brestic, M., Topal, A., Gezgin, S. (2022). Insight into the root transcriptome of a boron-tolerant Triticum zhukovskyi genotype grown under boron toxicity. Agronomy 12, 2421. doi: 10.3390/agronomy12102421
Planas-Riverola, A., Gupta, A., Betegon-Putze, I., Bosch, N., Ibanes, M., Cano-Delgado, A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development 146, dev151894. doi: 10.1242/dev.151894
Pont, C., Leroy, T., Seidel, M., Tondelli, A., Duchemin, W., Armisen, D., et al. (2019). Tracing the ancestry of modern bread wheats. Nat. Genet. 51, 905–911. doi: 10.1038/s41588-019-0393-z
Reinhold, H., Soyk, S., Simkova, K., Hostettler, C., Marafino, J., Mainiero, S., et al. (2011). β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. Plant Cell 23, 1391–1403. doi: 10.1105/tpc.110.081950
Saha, G., Park, J. I., Jung, H. J., Ahmed, N. U., Kayum, M. A., Kang, J. G., et al. (2015). Molecular characterization of BZR transcription factor family and abiotic stress induced expression profiling in Brassica rapa. Plant Physiol. Biochem. 92, 92–104. doi: 10.1016/j.plaphy.2015.04.013
Setsungnern, A., Munoz, P., Perez-Llorca, M., Muller, M., Thiravetyan, P., Munne-Bosch, S. (2020). A defect in BRI1-EMS-SUPPRESSOR 1 (bes1)-mediated brassinosteroid signaling increases photoinhibition and photo-oxidative stress during heat stress in Arabidopsis. Plant Sci. 296, 110470. doi: 10.1016/j.plantsci.2020.110470
Shi, G. Q., Fu, J. Y., Rong, L. J., Zhang, P. Y., Guo, C. J., Xiao, K. (2018). TaMIR1119, a miRNA family member of wheat (Triticum aestivum), is essential in the regulation of plant drought tolerance. J. Integr. Agr. 17, 2369–2378. doi: 10.1016/S2095-3119(17)61879-3
Su, D., Xiang, W., Wen, L., Lu, W., Shi, Y., Liu, Y., et al. (2021). Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 21, 161. doi: 10.1186/s12870-021-02933-7
Sun, Z., Liu, X., Zhu, W., Lin, H., Chen, X., Li, Y., et al. (2022). Molecular traits and functional exploration of BES1 gene family in plants. Int. J. Mol. Sci. 23, 4242. doi: 10.3390/ijms23084242
Sun, L., Sun, G., Shi, C., Sun, D. (2018). Transcriptome analysis reveals new microRNAs-mediated pathway involved in anther development in male sterile wheat. BMC Genomics 19, 333. doi: 10.1186/s12864-018-4727-5
Sun, F., Yu, H., Qu, J., Cao, Y., Ding, L., Feng, W., et al. (2020). Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int. J. Mol. Sci. 21, 996. doi: 10.3390/ijms21030996
Thalmann, M., Coiro, M., Meier, T., Wicker, T., Zeeman, S. C., Santelia, D. (2019). The evolution of functional complexity within the β-amylase gene family in land plants. BMC Evol. Biol. 19, 66. doi: 10.1186/s12862-019-1395-2
Van Nguyen, T., Park, C. R., Lee, K. H., Lee, S., Kim, C. S. (2021). BES1/BZR1 Homolog 3 cooperates with E3 ligase AtRZF1 to regulate osmotic stress and brassinosteroid responses in Arabidopsis. J. Exp. Bot. 72, 636–653. doi: 10.1093/jxb/eraa458
Wang, Z. Y., Bai, M. Y., Oh, E., Zhu, J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Xu, L., Wang, D., Liu, S., Fang, Z., Su, S., Guo, C., et al. (2020). Comprehensive atlas of wheat (Triticum aestivum L.) AUXIN RESPONSE FACTOR expression during male reproductive development and abiotic stress. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.586144
Yan, M. Y., Xie, D. L., Cao, J. J., Xia, X. J., Shi, K., Zhou, Y. H., et al. (2020). Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J. 102, 931–947. doi: 10.1111/tpj.14672
Yang, J., Wu, Y., Li, L., Li, C. (2022). Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 173, 1–13. doi: 10.1016/j.plaphy.2022.01.019
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Yin, Y., Wang, Z. Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/s0092-8674(02)00721-3
Yue, H., Zhang, H., Su, N., Sun, X., Zhao, Q., Weining, S., et al. (2022). Integrate small rna and degradome sequencing to reveal drought memory response in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 23, 5917. doi: 10.3390/ijms23115917
Zeeshan, M., Qiu, C. W., Naz, S., Cao, F., Wu, F. (2021). Genome-wide discovery of mirnas with differential expression patterns in responses to salinity in the two contrasting wheat cultivars. Int. J. Mol. Sci. 22. doi: 10.3390/ijms222212556
Zhang, L., Wang, K., Han, Y., Yan, L., Zheng, Y., Bi, Z., et al. (2022). Genome-wide analysis of the VQ motif-containing gene family and expression profiles during phytohormones and abiotic stresses in wheat (Triticum aestivum L.). BMC Genomics 23, 292. doi: 10.1186/s12864-022-08519-3
Keywords: BRI1 EMS SUPPRESSOR (BES1), expression pattern, abiotic stresses, anther development, thermosensitive genic male sterile
Citation: Wang D, Zuo J, Liu S, Wang W, Lu Q, Hao X, Fang Z, Liang T, Sun Y, Guo C, Zhao C and Tang Y (2023) BRI1 EMS SUPPRESSOR1 genes regulate abiotic stress and anther development in wheat (Triticum aestivum L.). Front. Plant Sci. 14:1219856. doi: 10.3389/fpls.2023.1219856
Received: 09 May 2023; Accepted: 14 July 2023;
Published: 09 August 2023.
Edited by:
Liang-Zi Zhou, Shandong Agricultural University, ChinaReviewed by:
Mohd. Kamran Khan, Selçuk University, TürkiyeCopyright © 2023 Wang, Zuo, Liu, Wang, Lu, Hao, Fang, Liang, Sun, Guo, Zhao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinghong Zuo, enVvamluZ2hvbmcxMDFAMTYzLmNvbQ==; Changping Zhao, emhhb2NoYW5ncGluZ0BiYWFmcy5uZXQuY24=; Yimiao Tang, dGFuZ3lpbWlhb0BiYWFmcy5uZXQuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.