
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 May 2024
Sec. Plant Systematics and Evolution
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1219354
This article is part of the Research Topic Orchid Phylogenomics, Trait Evolution and Diversification Dynamics View all 4 articles
The hyperdiverse orchid genus Bulbophyllum is the second largest genus of flowering plants and exhibits a pantropical distribution with a center of diversity in tropical Asia. The only Bulbophyllum section with a center of diversity in Australasia is sect. Adelopetalum. However, the phylogenetic placement, interspecific relationships, and spatio-temporal evolution of this section remain largely unclear. To infer broad-level relationships within Bulbophyllum, and interspecific relationships within sect. Adelopetalum, a genome skimming dataset was generated for 89 samples, which yielded 70 plastid coding regions and a nuclear ribosomal DNA cistron. For 18 additional samples, Sanger data from two plastid loci (matK and ycf1) and nuclear ITS were added using a supermatrix approach. The study provided new insights into broad-level relationships in Bulbophyllum, including phylogenetic evidence for the non-monophyly of sections Beccariana, Brachyantha, Brachypus, Cirrhopetaloides, Cirrhopetalum, Desmosanthes, Minutissima, Oxysepala, Polymeres, and Sestochilos. Section Adelopetalum and sect. Minutissima s.s. formed a highly supported clade that was resolved as a sister group to the remainder of the genus. Divergence time estimations based on a relaxed molecular clock model placed the origin of Bulbophyllum in the Early Oligocene (ca. 33.2 Ma) and sect. Adelopetalum in the Late Oligocene (ca. 23.6 Ma). Ancestral range estimations based on a BAYAREALIKE model identified the Australian continent as the ancestral area of the sect. Adelopetalum. The section underwent crown diversification from the mid-Miocene to the late Pleistocene, predominantly in continental Australia. At least two independent long-distance dispersal events were inferred eastward from the Australian continent to New Zealand and to New Caledonia from the early Pliocene onwards, likely mediated by predominantly westerly winds of the Southern hemisphere. Retraction and fragmentation of the eastern Australian rainforests from the early Miocene onwards are likely drivers of lineage divergence within sect. Adelopetalum facilitating allopatric speciation.
The hyperdiverse orchid genus Bulbophyllum Thouars (Epidendroideae) is the second largest genus of flowering plants, with more than 2,100 species, and exhibits exceptional morphological and ecological diversity (Frodin, 2004; Pridgeon et al., 2014; POWO, 2022). Species of this predominantly epiphytic genus occur in a wide range of tropical and subtropical habitats, from montane rainforests to dry deciduous forests, savanna woodlands, and rocky fields with shrubby vegetation (Pridgeon et al., 2014). Bulbophyllum is distributed pantropically, occupying all botanical continents defined by Brummitt et al. (2001), except Antarctica and Eurasia. The genus is most diverse on the botanical continent of tropical Asia (1562 species), also occurring on the botanical continents of Africa (305), temperate Asia (152), Southern America (88), the Pacific (49), Australasia (Australia and New Zealand; 35), and Northern America (7) (POWO, 2022). Centers of diversity are found in tropical Asia in the floristic regions of Malesia (667), Papuasia (656), and Afrotropics in the western Indian Ocean region, on the islands of Madagascar, and Mascarenes (218) (POWO, 2022).
The high number of species and complex patterns of morphological variation have presented significant challenges for resolving relationships in Bulbophyllum, which is reflected in the substantial taxonomic revisions proposed. Traditionally, the Bulbophyllinae Schltr. (tribe Dendrobieae Endl.) included the large genus Bulbophyllum along with smaller genera, such as Cirrhopetalum Lindl., Drymoda Lindl., Pedilochilus Schltr., Sunipia Buch.-Ham. ex Sm., and Trias Lindl. (Dressler, 1993; Garay et al., 1994; Szlachetko and Margonska, 2001). Recent revisions treat all genera within the subtribe Bulbophyllinae as a more broadly defined Bulbophyllum and recognize 97 sections within the genus (Pridgeon et al., 2014; Vermeulen et al., 2014). Molecular phylogenetic studies have largely focused on species from specific geographic regions, such as Madagascar and Mascarenes (Fischer et al., 2007; Gamisch et al., 2015), the Neotropics (Smidt et al., 2011; Smidt et al., 2013), and Peninsular Malaysia (Hosseini et al., 2012) or on taxonomic groups such as the Cirrhopetalum alliance (Hu et al., 2020), and few have taken a global perspective (e.g., Gamisch and Comes, 2019). These studies revealed a strong biogeographic pattern within the genus with four main clades that include species largely confined or endemic within one broader geographical area: 1) continental Africa, 2) Madagascar and the Mascarene Islands, 3) Southern America, or 4) Asia (Fischer et al., 2007; Smidt et al., 2011; Gamisch et al., 2015; Gamisch and Comes, 2019). The Southern American clade, the Madagascan clade, and the continental African clade together form a highly supported lineage (Fischer et al., 2007; Smidt et al., 2011; Smidt et al., 2013; Gamisch et al., 2015; Gamisch and Comes, 2019), in sister group position to the Asian clade (Fischer et al., 2007; Gamisch et al., 2015; Gamisch and Comes, 2019). Previous molecular phylogenetic studies have mainly elucidated relationships within Madagascan, continental African and Neotropical sections and within the Cirrhopetalum alliance (Fischer et al., 2007; Smidt et al., 2011; Gamisch et al., 2015; Hu et al., 2020). However, evolutionary relationships within the Asian clade, which also includes taxa from the Australasian and Pacific regions, are still poorly understood, and the monophyly of sections within this clade has remained largely untested within a phylogenetic framework.
The study of hyperdiverse groups such as Bulbophyllum requires a robust phylogenetic framework for assessing the monophyly of its infrageneric taxa. High-throughput sequencing approaches facilitate the establishment of such a framework phylogeny to clarify broader evolutionary relationships and assess the monophyly of infrageneric taxa and their trait evolution (Hassemer et al., 2019; Nargar et al., 2022; van Kleinwee et al., 2022). However, phylogenomic studies that provide insights into the broad-level evolutionary relationships within the Asian clade of Bulbophyllum are still lacking. This hampers progress in understanding the spatio-temporal diversification of evolutionary lineages and trait evolution within this highly diverse genus.
Within Bulbophyllum, section Adelopetalum has a unique distribution, being the only section with a center of diversity in Australia (Vermeulen, 1993; Brummitt et al., 2001; Pridgeon et al., 2014), and thus presents an interesting case study for range evolution within Bulbophyllum. This section comprises 12 tropical to temperate epi-lithophytic species (Figure 1). Nine species occur along Australia’s east coast in the montane forest communities of the Great Dividing Range, with one species (B. argyropus) found on Australian islands (Lord Howe Island and Norfolk Island). Two species (B. corythium and B. lingulatum) are endemic to the montane forests of New Caledonia, and one species (B. tuberculatum) is endemic to the lowland coastal forests of New Zealand. The section was circumscribed based on morphological affinities recognized among 10 species from Australia and New Caledonia previously assigned to the Bulbophyllum sections Desmosanthes, Racemosae, and Sestochilus (Dockrill, 1992; Vermeulen, 1993). Subsequent treatments identified two additional species within the Adelopetalum group, B. weinthalii and B. exiguum (Jones and Clements, 2002; Jones, 2020). Section Adelopetalum is characterized by thin creeping rhizomes adpressed to the host, anchored by filamentous roots with small pseudobulbs that are crowded to widely spaced, and a small single flat leaf arising from the apex of the pseudobulb. The inflorescence is single- to few-flowered, with small white, cream, or yellow flowers, sometimes with red or purple patterns. The petals are smaller than the sepals but similar in shape, with the bases of the lateral sepals fused to the column foot. The fleshly three-lobed labellum is firmly hinged to the apex of the column foot (Jones, 2020).

Figure 1 Morphological diversity within Bulbophyllum section Adelopetalum. (A) B. argyropus; (B) B. weinthalii; (C) B. bracteatum; (D) B. elisae; (E) B. exiguum; (F) B. lilianae; (G) B. lageniforme; (H) B. newportii. Photos M. Clements.
Previous cladistic analyses of the sect. Adelopetalum, based on morphological characters, resolved two main clades within the section, differentiated by the pseudobulb rib and ovary texture and the size and shape of the lower margin of the stelidia: the filiform column appendages typical for most Bulbophyllum (Vermeulen, 1993). Previous molecular phylogenetic studies based on the nuclear ribosomal ITS region (ITS1 + 5.8S + ITS2) have included two to three species from section Adelopetalum (Gamisch et al., 2015; Gamisch and Comes, 2019), placing them in an early diverging position within the Asian clade. However, the placement of sect. Adelopetalum remains unclear because of the lack of statistical support. Furthermore, phylogenetic relationships within sect. Adelopetalum and the evolution of its ancestral range are poorly understood and have not yet been investigated using molecular phylogenetic approaches.
This study aims to improve our understanding of the poorly understood evolutionary relationships among Asian and Australasian Bulbophyllum species and the spatio-temporal evolution of section Adelopetalum. The specific aims of this study were to 1) build a phylogenomic framework for Bulbophyllum with a focus on the Asian and Australasian species; 2) assess the monophyly and phylogenetic placement of sect. Adelopetalum within Bulbophyllum; 3) to infer interspecific relationships within sect. Adelopetalum; and 4) to reconstruct the range evolution of sect. Adelopetalum. The study sheds light on molecular systematics and evolution of Orchidaceae in the Australasian region over the Cenozoic.
A total of 136 orchid samples, representing 114 species, were included in the study. A broad sampling was included from the Asian, Australasian, and Pacific regions, representing 41 sections, i.e., 60% of sections recognized from these regions in the most recent treatment of the group (Pridgeon et al., 2014). From the Australasian region, all Bulbophyllum species were sampled (Australia: 30, New Zealand: 2). For Bulbophyllum sect. Adelopetalum, 28 samples were included, representing all 12 species recognized in the section. Morphologically closely related sect. Minutissima was included with nine samples representing five species comprising all four Australasian and Pacific species and two (of ca. 19) tropical Asian species. Sampling of five representative species from the Afrotropical clade was informed by previous phylogenetic studies (Fischer et al., 2007; Smidt et al., 2011; Gamisch and Comes, 2019). Species names followed the accepted taxonomy of Plants of the World Online (POWO, 2022). Sectional taxonomy followed the Internet Orchid Species Photo Encyclopedia (IOSPE, 2022), except for B. exiguum, which was placed in section Adelopetalum, and B. wolfei, which was placed in section Polymeres based on Jones and Clements (2002) and Jones (2020). The outgroup comprised representatives of subtribe Dendrobineae which is sister to Bulbophyllinae, and tribes Arethuseae, Malaxideae, Nervilieae, Neottieae based on previous molecular phylogenetic studies (Górniak et al., 2010; Givnish et al., 2015; Serna-Sánchez et al., 2021). Details of the plant material studied, voucher information, and the number of loci included for each sample are provided in Supplementary Material S1, and a complete list of loci analyzed is provided in Supplementary Material S2.
Total genomic DNA was extracted from approximately 10 to 20 mg of silica-dried leaf material. Extractions were carried out with commercial extraction kits: Qiagen DNeasy plant kit (Venlo, Netherlands) and ChargeSwitch gDNA plant kit (Invitrogen, Carlsbad, USA), following the manufacturers’ protocols, or using the CTAB method (Doyle and Doyle, 1990), with modifications as described by Weising et al. (2005). Sequence data were generated using Sanger sequencing (46 samples) for the nuclear ribosomal ITS region (ITS1, 5.8s, ITS2) and two plastid genes (matK, ycf1) and shotgun high-throughput sequencing (89 samples) to extract 70 plastid coding sequences (CDS) and the nuclear ribosomal DNA cistron (Supplementary Material S2) for subsequent analyses. Libraries for high-throughput sequencing were constructed from 50 ng to 100 ng of total DNA using the TruSeq Nano DNA LT library preparation kit (Illumina, San Diego, USA) for an insert size of 350 base pairs (bp) and paired-end reads according to the manufacturer’s protocol. Libraries were multiplexed with 96 indices and DNA sequencing with 125 bp paired-end reads was carried out on an Illumina HiSeq 2500 platform at the Australian Genomic Research Facility, Melbourne (Australia).
For Sanger sequencing, amplification of ITS was carried out with primers 17F and 26SER (Sun et al., 1994), matK with primers 19F and 1326R (Gravendeel, et al., 2001; Cuénoud et al., 2002), and ycf1 with primers 3720F, intR, intF, and 5500R (Neubig et al., 2009). The PCR protocol is described in Supplementary Material S3. Sequencing reactions were carried out using amplification primers, and sequencing was conducted on an AB3730xl 96-capillary sequencer (Australian Genome Research Facility, Brisbane, Australia).
Sequences were assembled and edited in Geneious R10 (Kearse et al., 2012) using Geneious mapper. Illumina sequences were mapped to a reference set of plastid CDS extracted from Dendrobium catenatum (GenBank accession number KJ862886) and ycf68 from Anoectochilus roxburghii (GenBank accession number KP776980). Plastid coding regions were targeted in preference over whole plastomes to avoid challenges in aligning non-coding regions and to ensure homology across alignments that included representative sampling from the mega diverse genus Bulbophyllum. To build a closely related reference for the nuclear ribosomal ITS-ETS region, Illumina reads of Bulbophyllum sect. Adelopetalum representative B. boonjee (CNS_G07175) were first mapped to the ITS-ETS region of Corallorhiza trifida (JVF2676a). The B. boonjee Illumina reads were then mapped to the B. boonjee consensus sequence generated in the initial step to extend the region assembled to include the complete nuclear ribosomal DNA cistron (5´ETS, 18S, ITS1, 5.8S, ITS2, 28S, and 3´ETS). Illumina sequences for all the remaining samples were assembled against the B. boonjee nuclear ribosomal DNA cistron reference. Consensus sequences were generated with bases matching at least 60% of the total adjusted chromatogram quality, and a minimum coverage of ten reads. The quality of the assemblies was checked and edited manually where required. For Sanger sequencing, bidirectional reads were assembled using Geneious and manually edited. Additional sequences were sourced from DRYAD (https://doi.org/10.5061/dryad.n9r58) for Coelogyne flaccida (Givnish et al., 2015). DNA sequences were aligned using MAFFT v.7.222 (Katoh et al., 2002; Katoh et al., 2005) with default settings, visually inspected, and then concatenated into a nuclear and plastid supermatrix. The nuclear supermatrix included 136 accessions partitioned into coding and non-coding regions (alignment length: 6,341 bp, number of parsimony informative sites: 995 (16%)), and the plastid supermatrix included 130 accessions and 70 plastid coding regions, partitioned by gene and codon position (alignment length: 61,553 bp, number of parsimony informative sites: 5,789 (9%)). A plastid dataset comprising high-throughput sequencing data (excluding Sanger sequences) was analyzed separately, including 90 accessions and 70 plastid coding regions (alignment length: 61,553 bp, number of parsimony informative sites: 5,682 (9%)).
For divergence time estimations, the plastid supermatrix was reduced to one representative per species (indicated by an asterisk in Supplementary Material S1), comprising 111 accessions (alignment length: 60,984 bp, number of parsimony-informative sites: 5,755 (9%)). Edited ITS, matK and ycf1 generated from Sanger sequencing and raw Illumina reads used for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB72010 (https://www.ebi.ac.uk/ena/browser/view/PRJEB72010).
Phylogenetic relationships were inferred using maximum likelihood in IQ-TREE v. 1.6.12 (Nguyen et al., 2015). The best-fit partition scheme and nucleotide substitution model for each partition were determined using IQ-TREE’s ModelFinder (Kalyaanamoorthy et al., 2017) based on the Akaike information criterion (AIC) (Akaike, 1974). Nodal support was assessed based on 1,000 replicates of ultrafast bootstrap approximation, with clades receiving >95 ultrafast bootstrap support (UFBS) considered as well supported (Minh et al., 2013; Hoang et al., 2018).
The divergence times were estimated based on the plastid dataset in beast v. 2.4.8 (Bouckaert et al., 2014), applying the best-fit partition scheme and substitution model, as determined by IQ-TREE’s ModelFinder. We tested two molecular clock models: 1) strict clock (Zuckerkandl and Pauling, 1965) and 2) relaxed lognormal clock (Drummond et al., 2006), and two models of speciation and extinction: 1) Yule and 2) birth-death (Yule, 1925; Gernhard, 2008). Three secondary calibration points were used by applying priors with a normal distribution, mean ages, and 95% higher posterior density (HDP) intervals based on the results of a family wide molecular clock analysis by Chomicki et al. (2015). The root age was set at 55.02 Ma (HDP: 42.0–68.0). The next secondary calibration point was applied to the last common ancestor of Dendrobineae, Malaxideae, and Arethuseae and was set to 47.77 Ma (HDP: 36.4–59.1). Monophyly was constrained for this node, which is consistent with the relationships reconstructed in previous phylogenetic analyses (Chomicki et al., 2015; Givnish et al., 2015). The last secondary calibration was set at the stem node of Dendrobieae and Malaxideae with 38.68 Ma (HDP: 30.8–46.6). An additional calibration based on the fossil Dendrobium winikaphyllum (Conran et al., 2009) was applied to the stem node of the Australasian Dendrobium clade (D. macropus, D. cunninghamii, and D. muricatum), using a uniform distribution with an infinite maximum age and the minimum age constrained to 20.4 Ma, based on the minimum age of the strata containing the fossil (Mildenhall et al., 2014). Ten independent beast analyses were performed for 30 million MCMC generations, with trees sampled every 3 × 104 generations. To assess the convergence of independent runs and determine burn-in fractions, log files were assessed using Tracer v.1.7.1 (Rambaut and Drummond, 2007). Log and tree files from independent runs were combined in LogCombiner (from the beast package) with a cumulative burn-in fraction of 10%–31%, and the sampling frequency was set to generate at least 10,000 tree and log files (Drummond and Bouckaert, 2015). The combined log file was assessed in Tracer to ensure that the effective sample size for all parameters was above 200. Five additional independent beast runs were conducted for the final analysis using a relaxed lognormal clock with birth death speciation to achieve an effective sample size above 200 for the ucldmean parameter. A maximum clade credibility tree was generated using TreeAnnotator (beast package) with median node heights. To compare the clock and speciation models, the Akaiki information criterion using the MCMC app from the beast package v 2.6.2 was used to measure the AICM for the combined MCMC runs generated in the beast analysis for each model (Supplementary Material S4).
The species distributions was extracted from POWO (2022). Biogeographic areas were largely delineated based on the botanical continents defined by Brummitt et al. (2001). The subcontinental regions of Papuasia, Australia, and New Zealand allowed a more fine-scale resolution of the range evolution in section Adelopetalum (Brummitt et al., 2001). The following seven biogeographic areas were coded: A) Africa, B) temperate Asia, C) tropical Asia, D) Papuasia, E) Australia, F) New Zealand, and G) Pacific. Ancestral ranges were estimated using RASP v. 4.0 (Yu et al., 2015) with the BioGeoBEARS package (Matzke, 2013), based on the maximum clade credibility tree obtained from the beast analysis of the plastid supermatrix, pruned of the outgroups to Dendrobieae. Three models of range evolution were tested: the dispersal-extinction cladogenesis model (DEC) (Ree and Smith, 2008), a maximum likelihood version of Ronquist’s parsimony dispersal-vicariance (DIVA) (Ronquist, 1997; Ronquist, 2001), termed DIVALIKE (Matzke, 2013), and a simplified likelihood interpretation of the Bayesian “BayArea” program (Landis et al., 2013), known as BAYAREALIKE (Matzke, 2013). No constraints were applied to the dispersal direction, and the maximum number of ranges was set to five based on the maximum number of observed areas of extant species. Likelihood values were compared, and the model of best fit determined by the AIC score (Akaike, 1974) was used to infer the marginal probabilities of alternative ancestral ranges at each node in the phylogeny (Supplementary Material S5).
The maximum likelihood phylogeny inferred from the 70 loci plastid supermatrix provided strong support for the monophyly of Bulbophyllum and its sister group relationship to Dendrobium (Figure 2). Sections Adelopetalum and Minutissima s.s. formed a highly supported clade, referred to as the Adelopetalum/Minutissima clade, which was resolved in the sister group position to the remainder of the genus (ultrafast bootstrap support/UFBS 98) (Figures 2, 3). Within the Adelopetalum/Minutissima clade, all the Adelopetalum species and B. pygmaeum (sect. Minutissima) formed a highly supported lineage (UFBS 100) termed the Adelopetalum clade. Within the Adelopetalum clade several highly supported groups were resolved: 1) the argyropus clade consisting of B. argyropus, B. corythium and B. tuberculatum (UFBS 100), reconstructed in a highly supported sister group relationship to B. weinthalii (UFBS 100); 2) the bracteatum clade, including B. boonjee, B. bracteatum, and B. elisae (UFBS 99); and 3) the newportii clade comprised of B. exiguum, B. lageniforme, B. lilianae, B. lingulatum, and B. newportii (UFBS 100). Relationships among the B. pygmaeum, argyropus clade + B. weinthalii, bracteatum, and newportii clades received weak support. The sister to the Adelopetalum clade was the highly supported Minutissimum clade comprising three species of sect. Minutissima (B. globuliforme, B. keekee, and B. minutissimum), including the type species of the section (UFBS 100) (Figure 2). Section Minutissima was identified as polyphyletic, with sect. Minutissima species placed within the Adelopetalum clade (B. pygmaeum) and Asian clade (B. mucronatum and B. moniliforme). Within the Asian clade, sections Beccariana, Brachyantha, Brachypus, Cirrhopetaloides, Cirrhopetalum, Desmosanthes, Oxysepala, Polymeres, and Sestochilos were identified as polyphyletic or paraphyletic. The phylogenetic relationships described here based on the plastid supermatrix (Figures 1, 3) are supported by reconstructions based on the 70 gene plastid dataset (Supplementary Material S6).

Figure 2 Maximum likelihood phylogenetic reconstruction of Bulbophyllum based on the supermatrix of 70 plastid coding regions in inset a. with Bulbophyllum sections Adelopetalum and Minutissima s.s. in detail. Ultrafast bootstrap values are given adjacent to nodes. Australian species are shown with an asterisk.

Figure 3 Maximum likelihood phylogenetic reconstruction of Bulbophyllum based on the supermatrix of 70 plastid coding regions in inset a. with Afrotropical and Asian Bulbophyllum clades in detail. Ultrafast bootstrap values are given adjacent to nodes. Australian species are shown with an asterisk.
Our analyses showed that sect. Adelopetalum does not share a close relationship with other Australasian Bulbophyllum species, such as those in sect. Brachypus (B. nematopodum), sect. Brachystachyae (B. evasum), sect. Cirrhopetalum (B. longiflorum), sect. Ephippium (B. gracillimum), sect. Monanthes (B. macphersonii), sect. Oxysepala (B. gadgarrense, B. grandimesense, B. lamingtonense, B. lewisense, B. schillerianum, B. shepherdii, B. wadsworthii, B. windsorense), sect. Polymeres (B. bowkettiae, B. johnsonii, B. radicans, and B. wolfei), and sect. Sestochilus (B. baileyi). Australian species from each of these sections were placed at nine different positions within the Asian clade. Australian species from sect. Polymeres formed a highly supported clade (UFBS 100). Australian species from sect. Oxysepala formed a moderately supported clade (UFBS 91) and together with the type species of section Oxysepala from Papuasia (B. cladistinum) formed a close relationship with the Australian representative of section Monanthes (B. macphersonii) (UFBS 100).
The maximum likelihood phylogeny based on the nuclear ribosomal DNA cistron was resolved with overall lower support compared to analyses based on the 70 plastid loci supermatrix (Figures 4, 5). Relationships among the outgroup taxa were concordant with the plastid phylogeny, and Bulbophyllum was resolved with maximum support. Within Bulbophyllum, the Afrotropical (UFBS 97), Asian (UFBS 100), and Adelopetalum/Minutissima (UFBS 100) clades were resolved with high to maximum support; however, their relationships were poorly supported. Within the Adelopetalum/Minutissima clade, the highly supported clades revealed in the plastid phylogeny were also reconstructed based on the nuclear dataset (argyropus clade (UFBS 100), bracteatum clade (UFBS 93), minutissimum clade (UFBS 86), and newportii clade (UFBS 83), however relationships among these remained poorly supported. Similar to reconstructions based on the plastid phylogeny: relationships among the argyropus, bracteatum and newportii clades, B. pygmaeum and B. weinthalii were poorly supported; sections Beccariana, Brachyantha, Brachypus, Cirrhopetaloides, Cirrhopetalum, Desmosanthes, Minutissima, Oxysepala, Polymeres, and Sestochilos were identified as polyphyletic or paraphyletic; and Australian species from sect. Brachystachyae, sect. Cirrhopetalum, sect. Monanthes, sect. Oxysepala, sect. Brachypus, sect. Polymeres, sect. Ephippium, sect. Stenochilus were placed in nine clades across the Asian clade.
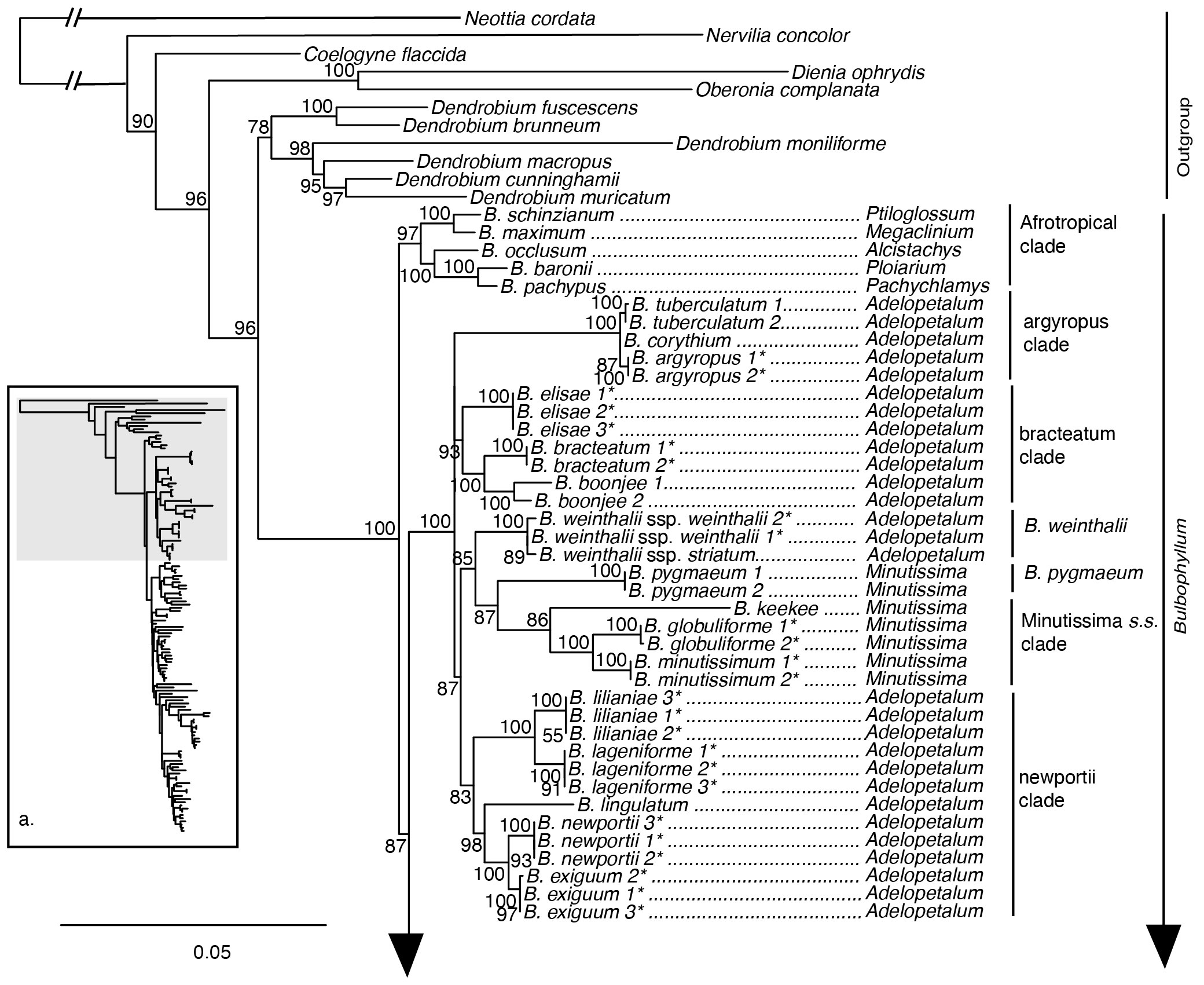
Figure 4 Maximum likelihood phylogenetic reconstruction of Bulbophyllum based on the nuclear ribosomal DNA cistron (5’ETS, 18s, ITS1, 5.8s, ITS2, 28s, 3’ETS) in inset a. with Bulbophyllum sections Adelopetalum and Minutissima in detail. Ultrafast bootstrap values are given adjacent to nodes. Australian species are shown with an asterisk.
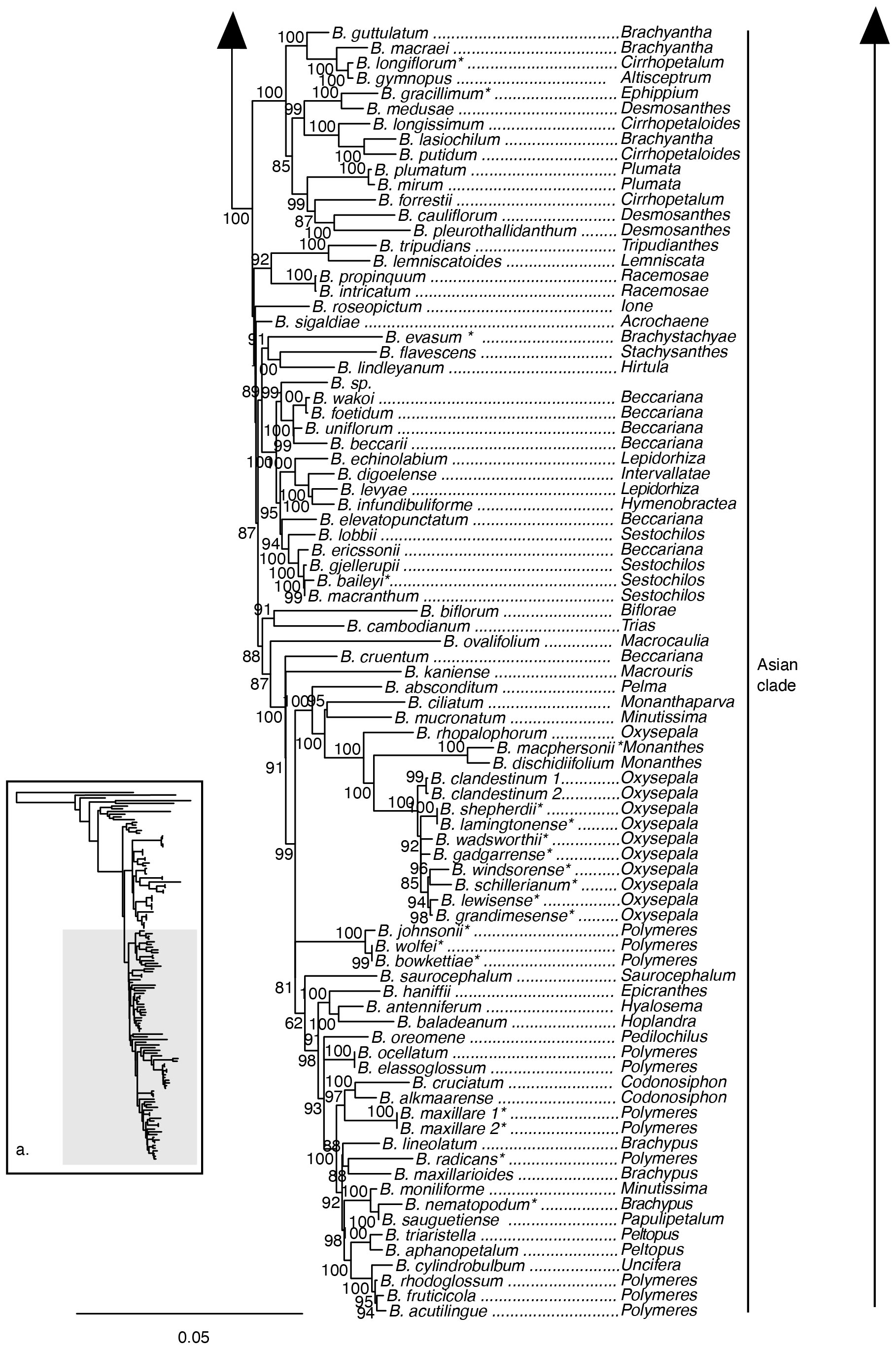
Figure 5 Maximum likelihood phylogenetic reconstruction of Bulbophyllum based on nuclear ribosomal DNA cistron (5’ETS, 18s, ITS1, 5.8s, ITS2, 28s, 3’ETS) in inset a. with Afrotropical and Asian Bulbophyllum clades in detail. Ultrafast bootstrap values are given adjacent to nodes. Australian species are shown with an asterisk.
The divergence time analysis based on a relaxed lognormal clock and birth death prior to speciation and extinction, which was identified as the model of best fit based on the Akaike information criterion (Supplementary Material S4), is presented here (Figure 6), with the Asian and Afrotropical clades collapsed and the complete chronogram provided in Supplementary Material S7. The divergence time analysis based on the plastid dataset was well-resolved and highly supported (Figure 6, Supplementary Material S7). The divergence between Bulbophyllum and Dendrobium was estimated to have occurred during the Early Oligocene, ca. 33.2 Ma (95% highest posterior probability density, HPD: 27.7–39.0). The crown of Bulbophyllum, constituting the divergence of the Adelopetalum/Minutissima clade from the remainder of the genus, was dated to the late Oligocene, ca. 24.9 Ma (HPD: 20.1–30.7). The divergence between the Asian clade and the Afrotropical clade was estimated to have occurred during the Late Oligocene, ca. 24.3 Ma (HPD: 19.4–29.8) and diversification within the Asian clade was estimated to have occurred from the mid Miocene 21.4 Ma (HPD: 17.2–26.4 Ma) and the Afrotropical clade from 16.0 Ma (HPD: 10.4–21.7). The crown age of the Adelopetalum/Minutissima clade was dated to the late Oligocene, ca. 23.6 Ma (HPD: 18.6–29.1), with a split of the Minutissimum clade from the Adelopetalum clade. The crown age of the Adelopetalum clade was dated to mid-Miocene, ca. 15.3 Ma (HPD: 10.6–21.2). The stem branches of major lineages within the Adelopetalum clade were estimated to have diversified during the mid-Miocene; the bracteatum clade was dated to ca. 14.5 Ma (HPD: 10.0–20.4); the lineage giving rise to B. weinthalii to ca. 12.1Ma (6.9–17.6); the argyropus clade to ca. 12.1 Ma (6.9–17.6); and the newportii clade to ca. 14.9 Ma (HPD 10.3–20.7). Diversification among species within these lineages occurred from the mid-Miocene onwards, with the most recent divergence identified during the late Pleistocene among B. argyropus, B. corythium, and B. tuberculatum.
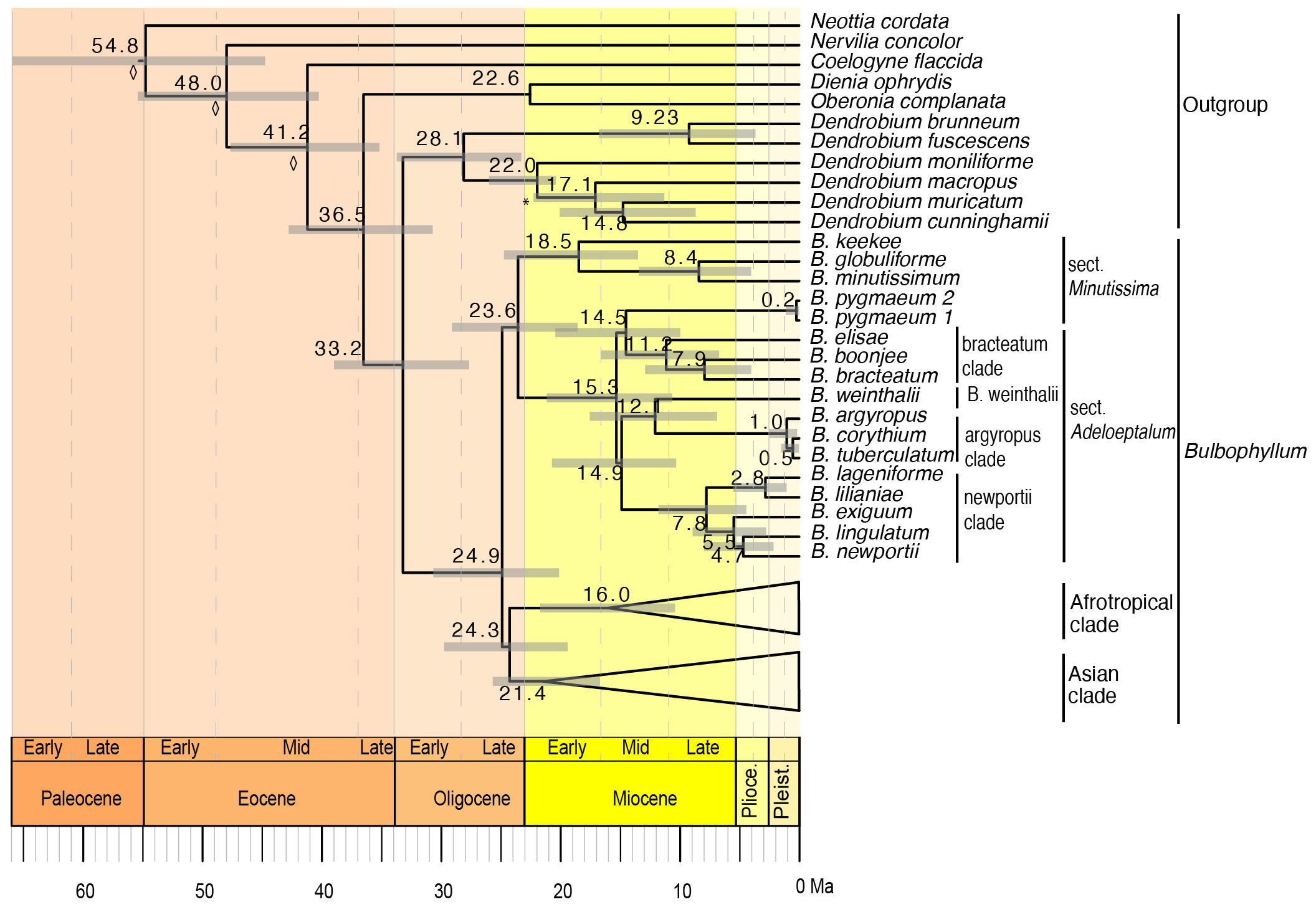
Figure 6 Maximum clade credibility chronogram for Bulbophyllum sect. Adelopetalum based on 70 plastid coding sequences, relaxed lognormal clock and birth death prior. Divergence dates and 95% highest posterior density values are indicated adjacent to nodes. Gray bars indicate 95% highest posterior density. The asterisk denotes the node constrained with a fossil calibration point; the diamond shape denotes nodes that were constrained by secondary calibration points.
Model testing of the three biogeographic models (DEC, DIVALIKE, BAYAREALIKE) using the Akaike information criterion identified the BAYAREALIKE model as the model of best fit for ancestral range estimation (Supplementary Material S5). The ancestral ranges estimated using the BAYAREALIKE model are presented here, with the Asian and Afrotropical clades collapsed (Figure 7). The complete chronogram is provided in Supplementary Material S8, the marginal probabilities for ancestral ranges at all nodes in Supplementary Material S9, and the node IDs in Supplementary Material S10.
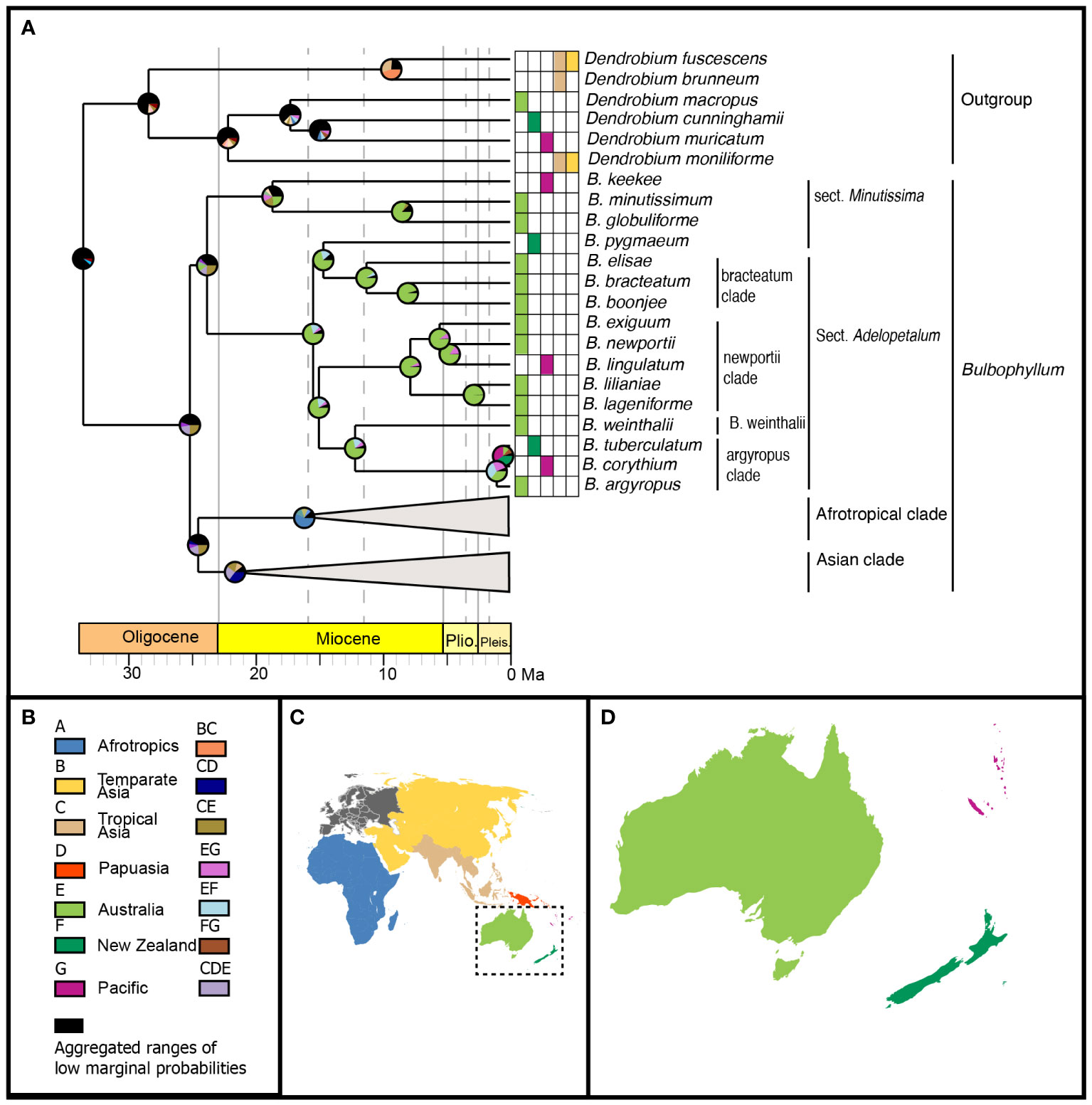
Figure 7 Range evolution of Bulbophyllum sect. Adelopetalum. (A) ancestral area reconstruction based on the BAYAREALIKE model with species extant distributions shown within the grid and pie charts at internal nodes representing marginal probabilities for alternative ancestral areas; (B) legend of color coded geographic regions and shared ancestral areas; (C) world map of color coded geographic regions delineated in the biogeographic analysis; (D) detail of Australasian color coded geographic regions.
Australia was reconstructed as the most likely ancestral range for the most recent common ancestor (MRCA) of the Adelopetalum clade (range probability (RP) 82) and all nodes within this lineage (RP 72–99), except for the argyropus clade (Figure 7). Range shifts from Australia were inferred from the early Pliocene to the Pacific region (New Caledonia) in the newportii clade, which gave rise to B. lingulatum. Range shifts were also inferred from Australia to the Pacific region (New Caledonia) and New Zealand, either in the lineage giving rise to MRCA of the argyropus clade or subsequently within this lineage. Three alternative ancestral ranges were reconstructed for the MRCA of the argyropus clade: Australia (RP 36) or widespread distributions, including Australia and New Zealand (RP 33) or Australia and New Caledonia (RP 26). Two alternative ranges were also reconstructed for MRCA of B. corythium and B. tuberculatum: New Zealand (RP 41) and New Caledonia (RP 34). Considering these alternative scenarios, range shifts within the argyropus clade were estimated to have occurred sometime between the mid-Miocene and the late Pliocene (12.1 Ma–0.5 Ma). The ancestral range of the MRCA of the Adelopetalum/Minutissima clade and Bulbophyllum remains unresolved in the ancestral range reconstruction. For the MRCA of the Adelopetalum/Minutissima clade, a wide ancestral range including Australia and tropical Asia received the highest relative probability (RP 26), while alternative ranges included a wide distribution across Australia, tropical Asia and Papuasia (RP 13) or Australia (RP 11).
This study provided a broad plastid phylogenetic framework for Asian and Australasian sections of Bulbophyllum and revealed a close relationship between sections Adelopetalum and Minutissima s.s., that together form a highly supported early diverging lineage within the genus (Figures 2, 3). Relationships based on 70 plastid genes support a sister group relationship between the Adelopetalum/Minutissima clade and the remainder of the genus (Asian + Afrotropical clades). Within the Adelopetalum/Minutissima clade, analyses based on our 70 plastid loci supermatrix showed a dichotomous split between the highly supported Minutissima s.s. and Adelopetalum clades. Species were reconstructed in each of these clades according to their sectional placement, except for the New Zealand endemic B. pygmaeum (sect. Minutissima), which was nested within the Adelopetalum clade, rendering the section Adelopetalum paraphyletic. Section Minutissima was identified as polyphyletic with the Australian (B. minutissimum (sect. type), B. globuliforme), and Pacific species (B. keekee) placed in the Minutissima clade and New Zealand species (B. pygmaeum) in the Adelopetalum clade, while the Asian species, B. mucronatum and B. moniliforme were resolved within the Asian clade. Section Minutissima has undergone numerous taxonomic changes with treatments ranging from a narrower circumscription recognizing species from Australia (Jones and Clements, 2001), to broader classifications, including 23 species from Thailand, Indonesia, Australia, New Zealand, New Caledonia, and New Guinea (Pridgeon et al., 2014). Our phylogenetic analysis based on plastid and nuclear markers did not reveal a close relationship between the sect. Minutissima species from the Australasian/Pacific region and Asian species B. mucronatum and B. moniliforme. Rather, in our analyses the Australasian species fell within the Adelopetalum/Minutissima clade while the Asian species were nested within the Asian clade. These results support the morphological studies that differentiate sect. Minutissima species from Australasia and Asia (Jones and Clements, 2001) and indicate minute pseudobulbs are a trait that has evolved more than once independently in the genus. Further studies, including dense sampling and ancestral character reconstruction, are required to assess the evolution of morphological traits and their phylogenetic utility across the Asian clade.
Within the section Adelopetalum, phylogenetic analyses supported current species concepts, except for species within the argyropus clade, which exhibited shallow genetic differentiation (Figures 2, 4). Previous taxonomic treatments of this morphologically similar group have delineated up to three species: B. argyropus (Australia’s east coast and offshore islands: Lord Howe Island, and Norfolk Island), B. corythium (New Caledonia), and B. tuberculatum (New Zealand) (Halle, 1981; Vermeulen, 1993; Jones and Clements, 2002). Divergence dating analyses showed that this group represents a relatively recent radiation, reconstructing divergence among species during the Pleistocene. Further studies are required to clarify species delimitation and dispersal patterns utilising population-level sampling and genomic techniques suited to resolving relationships among recently diverged lineages, such as reduced representation high-throughput approaches like ddRAD, DArT or target sequence capture methods (Sansaloni et al., 2011; Peterson et al., 2012; Weitemier et al., 2014; Folk et al., 2015; Bagley et al., 2020; Schmidt-Lebuhn et al., 2022).
Previous cladistic analyses based on morphological traits of sect. Adelopetalum differentiated two main clades within the section, one comprising B. lageniforme, B. lilianae, B. lingulatum, and B. newportii, and the other uniting B. argyropus, B. bracteatum, and B. elisae (Vermeulen, 1993). Phylogenetic relationships based on plastid and nuclear markers found strong to moderate support for the first clade recovered in the cladistic analysis, corresponding to the newportii clade in the present analyses (Figures 1, 4). The second group found in the cladistic analysis included three species placed in the phylogenetic analyses within either the bracteatum clade (B. bracteatum, B. elisae) or the argyropus clade (B. argyropus). However, the relationship between these two lineages remains unclear owing to low support. The sister group relationship between B. bracteatum and B. argyropus recovered in the cladistic analysis was not supported by phylogenetic reconstructions based on molecular data, suggesting that character states shared by these species, such as the number of veins in petals and number of pollinia, may be homoplasious. Further studies using ancestral character reconstruction are required to test the phylogenetic utility of the morphological traits used in prior studies.
While plastid phylogenomics has clarified the major clades and interspecific relationships within sect. Adelopetalum and broad-level relationships within Bulbophyllum, further studies are required. Non-monophyletic sections identified in the present study (e.g., sections Beccariana, Brachyantha, Brachypus, Cirrhopetaloides, Cirrhopetalum, Desmosanthes, Minutissima, and Polymeres) (Figure 3) and in previous molecular phylogenetic studies (Fischer et al., 2007; Smidt et al., 2011; Pridgeon et al., 2014; Hu et al., 2020) highlight the need for further taxonomic revision of Bulbophyllum. Further studies are required with an expanded sampling of diverse Asian and Pacific taxa to increase our understanding of evolutionary relationships and assess sectional classification in more detail. Phylogenetic relationships reconstructed from the nuclear ribosomal DNA cistron were not strongly supported, which is in line with previous molecular studies based on ITS (Gamisch et al., 2015; Gamisch and Comes, 2019; Hu et al., 2020). Approaches yielding a higher number of nuclear markers, such as target sequence capture, provide an opportunity to improve our understanding of evolutionary relationships in future studies. While assembling datasets with comprehensive species coverage within mega diverse groups, such as Bulbophyllum, remains a challenge, the present study provides an example of the use of a broad phylogenetic framework with targeted sampling within a section to test the monophyly and phylogenetic placement of groups of interest.
Our divergence time analysis and ancestral range estimations showed that Bulbophyllum sect. Adelopetalum represents an Australasian lineage that originated on the Australian continent during the late Oligocene to early Miocene (Figures 6, 7). The Australian ancestral range is largely conserved within the lineage, indicating that diversification among species has predominantly occurred on the Australian continent. The conservation of ancestral range observed within sect. Adelopetalum is consistent with previous phylogenetic analyses of Bulbophyllum that have shown a strong biogeographic signal among clades which are largely confined to biogeographic regions such as Madagascar, continental Africa and South America (Fischer et al., 2007; Smidt et al., 2011; Gamisch et al., 2015; Gamisch and Comes, 2019). The evolution of Bulbophyllum during the early Oligocene occurred subsequent to the breakup of Gondwana (Matthews et al., 2016; Zahirovic et al., 2016), indicating long-distance dispersal (LDD) in the evolution of biogeographical lineages within the genus (van den Berg, 2003; Smidt et al., 2011; Gamisch et al., 2015; Gamisch and Comes, 2019). Nevertheless, the conservation of ancestral ranges observed within the Adelopetalum lineage in this study and strong biogeographic signals among clades identified in previous studies indicate that LDD with successful establishment and persistence have been relatively infrequent within Bulbophyllum. Although the minute wind-dispersed seeds of orchids have a high dispersal potential, their successful establishment in a new area is limited by several factors, such as the presence of mycorrhizal partners necessary for germination and development, a suitable host or substrate and microclimatic conditions, and the availability of pollinators (Arditti and Ghani, 2000; Jersáková and Malinová, 2007; McCormick et al., 2012). Our results are consistent with those of previous studies that have identified in situ diversification as the dominant biogeographic process, despite evidence for LDD, and provides further support for the hypothesis that the complex requirements for successful establishment, rather than dispersal limitations, play an important role in constraining the geographic distribution of orchids (Givnish et al., 2016; Pérez-Escobar et al., 2017).
Our phylogenetic analyses further resolved the interspecific relationships in sect. Adelopetalum (Figure 2). Divergence time estimation showed that divergence among species occurred mainly during the Miocene and Pliocene (Figure 6), during a period of extensive changes in the distribution of forest vegetation on the Australian continent in response to drastic climatic changes. During the early Miocene, Australian vegetation diversified in response to the aridification of the Australian continent and the abrupt shift to a cool-dry climate during the mid-Miocene resulted in considerable fragmentation of rainforest habitats (Martin, 2006; Byrne et al., 2011). Bulbophyllum sect. Adelopetalum comprises epiphytic species that occur in mesic forest habitats; thus, diversification and fragmentation of these habitats were likely drivers of allopatric lineage divergence within this group. Sister group relationships were identified between two species pairs with disjunct distributions in Australia’s northern wet tropical rainforests and southeastern rainforests (B. boonjee/B. bracteatum and B. newportii/B. exiguum). These relationships support the hypothesis that the diversification and fragmentation of forest habitats in Australia have been important drivers of lineage divergence in Australia’s mesic biome (Byrne et al., 2011; Simpson et al., 2018).
While the ancestral range was predominantly conserved within the Adelopetalum lineage, range expansion events were inferred from continental Australia across the Coral and Tasman Seas to New Caledonia in the lineage giving rise to B. lingulatum and to New Zealand and New Caledonia in the argyropus clade (Figure 7). New Caledonia and New Zealand each have a long history of isolation from Australia that predates the evolution of Bulbophyllum, indicating that the colonization of these islands by Bulbophyllum species has been via LDD (Matthews et al., 2016). It remains unclear whether LDD to New Zealand and New Caledonia in the argyropus clade occurred from the early Miocene in the lineage, giving rise to the MRCA of the group or subsequently within this clade during the late Pleistocene; thus, the spatiotemporal evolution of this lineage requires further study.
The pattern of eastward dispersal observed in range shifts from Australia, across the Coral and Tasman Seas, is consistent with dispersal patterns inferred in other angiosperms, including Abrotanella, Dendrobium, Dracophyllum, Korthalsella, Leucopogon, Northofagus, Oreobolus, Pterostylis, Rytidosperma, and Veronica (Linder, 1999; Molvray et al., 1999; Lavarack et al., 2000; Swenson et al., 2001; Wagstaff et al., 2002; Chacón et al., 2006; Wagstaff et al., 2006; Albach and Meudt, 2010; Wagstaff et al., 2010; Puente-Lelièvre et al., 2013; Nargar et al., 2022). The bias towards eastward dispersal observed within section Adelopetalum among other plant groups may be facilitated by the predominant westerly winds occurring in the southern hemisphere that were initiated after the rifting of Australia and South America from Antarctica during the Eocene (Sanmartín et al., 2007).
This study provides an important phylogenomic framework for the mega genus Bulbophyllum, facilitating studies on trait and range evolution within the genus. Several Asian sections were resolved as paraphyletic, warranting taxonomic revision. Our plastid phylogenomic analyses revealed an early diverging lineage within Bulbophyllum composed of sect. Adelopetalum and sect. Minutissima s.s. For Bulbophyllum sect. Adelopetalum, this study reconstructed its origin in the early Oligocene and identified the Australian continent as the ancestral range. Species diversification within the section occurred predominantly on the Australian continent, with the fragmentation of mesic habitats during the Miocene identified as likely drivers of allopatric lineage divergence. Multiple independent long-distance dispersal events were inferred from the Australian continent eastward to the islands of New Zealand and New Caledonia.
The data used for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB72010 (https://www.ebi.ac.uk/ena/browser/view/PRJEB72010).
Conceptualization: LS, KN, and MC; Data curation: LS and MC; Formal Analysis: LS and HO; Funding acquisition: LS, KN, DC, and MC; Investigation and Methodology: LS; Supervision: KN, DM, and MC; Writing—original draft: LS; Writing—review and editing: KN, HO, DM, and MC. All authors contributed to the article and approved the submitted version.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Australian Biological Resources Study (BBR 210-34), the Hermon Slade Foundation (HSF 16-04), and the Australian Orchid Foundation (AOF 320/17; AOF 325.18.). The authors acknowledge the contribution of Bioplatforms Australia (enabled by NCRIS) in the generation of data used in this publication.
The authors wish to acknowledge the use of Next-Generation Sequencing services and facilities at the Australian Genomic Research Facilities (AGRF). Plant material was collected with the permission of the KuKu Nyungkal traditional owners, Queensland Parks and Wildlife (permit numbers: WISP11258812 and WITK11258712). We thank B. Gray, D.L. Jones, G. McCraith, M. Harrison, M.T. Mathieson, B.P. Molloy, L. Roberts, and P.D. Ziesing for contributing plant material to the study. We thank Guillaume Chomicki for providing files used for secondary calibrations in our divergence dating analyses. A preprint of the manuscript was released online (Simpson et al., 2021; doi: https://doi.org/10.1101/2022.07.24.500920).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1219354/full#supplementary-material
Akaike, H. (1974). A new look at the statistical model identification. IEEE Trans. Automatic Control 19 (6), 716–723. doi: 10.1109/TAC.1974.1100705
Albach, D. C., Meudt, H. M. (2010). Phylogeny of Veronica in the Southern and Northern Hemispheres based on plastid, nuclear ribosomal and nuclear low-copy DNA. Mol. Phylogenet. Evol. 54 (2), 457–471. doi: 10.1016/j.ympev.2009.09.030
Arditti, J., Ghani, A. K. A. (2000). Tansley Review No. 110: Numerical and physical properties of orchid seeds and their biological implications. New Phytol. 145 (3), 367–421. doi: 10.1046/j.1469-8137.2000.00587.x
Bagley, J. C., Uribe-Convers, S., Carlsen, M. M., Muchhala, N. (2020). Utility of targeted sequence capture for phylogenomics in rapid, recent angiosperm radiations: Neotropical Burmeistera bellflowers as a case study. Mol. Phylogenet. Evol. 152, 106769. doi: 10.1016/j.ympev.2020.106769
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: A software platform for bayesian evolutionary analysis. PloS Comput. Biol 10(4), e1003537. doi: 10.1371/journal.pcbi.1003537
Brummitt, R. K., Pando, F., Hollis, S., Brummitt, N. A. (2001). World geographical scheme for recording plant distributions. Pittsburg: International working group on taxonomic databases for plant sciences (TDWG). Vol. 951, p. 952.
Byrne, M., Steane, D. A., Joseph, L., Yeates, D. K., Jordan, G. J., Crayn, D., et al. (2011). Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. J. Biogeography 38 (9), 1635–1656. doi: 10.1111/j.1365-2699.2011.02535.x
Chacón, J., Madriñán, S., Chase, M. W., Bruhl, J. J. (2006). Molecular phylogenetics of Oreobolus (Cyperaceae) and the origin and diversification of the American species. TAXON 55 (2), 359–366. doi: 10.2307/25065583
Chomicki, G., Bidel, L. P. R., Feng, M., Coiro, M., Zhang, X., Wang, Y., et al. (2015). The velamen protects photosynthetic orchid roots against UV-B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytol. 205 (3), 1330–1341. doi: 10.1111/nph.13106
Conran, J. G., Bannister, J. M., Lee, D. E. (2009). Earliest orchid macrofossils: Early Miocene Dendrobium and Earina (Orchidaceae: Epidendroideae) from New Zealand. Am. J. Bot. 96 (2), 466–474. doi: 10.3732/ajb.0800269
Cuénoud, P., Savolainen, V., Chatrou, L. W., Powell, M., Grayer, R. J., Chase, M. W. (2002). Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 89 (1), 132–144. doi: 10.3732/ajb.89.1.132
Dockrill, A. W. (1992). Australian indigenous orchids: the epiphytes, the tropical terrestrial species Surrey Beatty & Sons in assoication with the Society for Ground Australian (Sydney, NSW, Australia: Surrey Beatty & Sons in assoication with the Society for Ground Australian Plants—NSW).
Dressler, R. L. (1993). Phylogeny and classification of the orchid family (Cambridge, England: Cambridge University Press).
Drummond, A. J., Bouckaert, R. R. (2015). Bayesian evolutionary analysis with BEAST. (Cambridge, England: Cambridge University Press).
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PloS Biol. 4 (5), 699–710. doi: 10.1371/journal.pbio.0040088
Fischer, G. A., Gravendeel, B., Sieder, A., Andriantiana, J., Heiselmayer, P., Cribb, P. J., et al (2007). Evolution of resupination in Malagasy species of Bulbophyllum (Orchidaceae). Mol. Phylogenet. Evol. 45 (1), 358–376. doi: 10.1016/j.ympev.2007.06.023
Folk, R. A., Mandel, J. R., Freudenstein, J. v. (2015). A protocol for targeted enrichment of intron-containing sequence markers for recent radiations: A phylogenomic example from Heuchera (Saxifragaceae). Appl. Plant Sci. 3 (8), 1500039. doi: 10.3732/apps.1500039
Frodin, D. G. (2004). History and concepts of big plant genera. TAXON 53 (3), 753–776. doi: 10.2307/4135449
Gamisch, A., Comes, H. P. (2019). Clade-age-dependent diversification under high species turnover shapes species richness disparities among tropical rainforest lineages of Bulbophyllum (Orchidaceae). BMC Evolutionary Biol. 19 (1), 1–16. doi: 10.1186/s12862-019-1416-1
Gamisch, A., Fischer, G. A., Comes, H. P. (2015). Multiple independent origins of auto-pollination in tropical orchids (Bulbophyllum) in light of the hypothesis of selfing as an evolutionary dead end. BMC Evolutionary Biol. 15 (1), 1–18. doi: 10.1186/s12862-015-0471-5
Garay, L. A., Hamer, F., Siegerist, E. S. (1994). The genus Cirrhopetalum and the genera of the Bulbophyllum alliance. Nordic J. Bot. 14 (6), 609–646. doi: 10.1111/j.1756-1051.1994.tb01080.x
Gernhard, T. (2008). The conditioned reconstructed process. J. Theor. Biol. 253 (4), 769–778. doi: 10.1016/j.jtbi.2008.04.005
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B: Biol. Sci. 282 (1814), 20151553. doi: 10.1098/rspb.2015.1553
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2016). Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeography 43 (10), 1905–1916. doi: 10.1111/jbi.12854
Górniak, M., Paun, O., Chase, M. W. (2010). Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: Congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 56 (2), 784–795. doi: 10.1016/j.ympev.2010.03.003
Gravendeel, B., Chase, M. W., de Vogel, D. F., Roos, M. C., Mes, H. M., Bachmann, K. (2001). Molecular phylogeny of Coelogyne (Epidendroideae; Orchidaceae) based on plastid RFLPs, matK, and nuclear ribosomal ITS sequences: evidence for polyphyly. Am. J. Bot. 88 (10), 1915–1927.
Hassemer, G., Bruun-Lund, S., Shipunov, A. B., Briggs, B. G., Meudt, H. M., Rønsted, N. (2019). The application of high-throughput sequencing for taxonomy: The case of Plantago subg. Plantago (Plantaginaceae). Mol. Phylogenet. Evol. 138, 156–173. doi: 10.1016/j.ympev.2019.05.013
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35 (2), 518–522. doi: 10.1093/molbev/msx281
Hosseini, S., Go, R., Dadkhah, K., Nuruddin, A. A. (2012). Studies on maturase K sequences and systematic classification of Bulbophyllum in Peninsular Malaysia. Pakistan J. Bot. 44 (6), 2047–2054.
Hu, A. Q., Gale, S. W., Liu, Z. J., Suddee, S., Hsu, T. C., Fischer, G. A., et al. (2020). Molecular phylogenetics and floral evolution of the Cirrhopetalum alliance (Bulbophyllum, Orchidaceae): Evolutionary transitions and phylogenetic signal variation. Mol. Phylogenet. Evol. 143 (10668), 9. doi: 10.1016/j.ympev.2019.106689
IOSPE (2022) Internet Orchid Species Photo Encyclopedia. Available at: http://www.orchidspecies.com/.
Jersáková, J., Malinová, T. (2007). Spatial aspects of seed dispersal and seedling recruitment in orchids. New Phytol. 176, 237–241. doi: 10.1111/j.1469-8137.2007.02223.x
Jones, D. L. (2020). A complete guide to native orchids of Australia (Sydney, NSW, Australia: Reed New Holland).
Jones, D. L., Clements, M. A. (2001). Oncophyllum, a new genus of Orchidaceae from Australia. Orchadian 13, 420–425.
Jones, D. L., Clements, M. A. (2002). Nomenclatural changes in the Australian and New Zealand bulbophyllinae and eriinae (Orchidaceae). Orchadian 13, 498–501.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Kuma, K., Toh, H., Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Katoh, K., Misawa, K., Kuma, K. I., Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 (14), 3059–3066. doi: 10.1093/nar/gkf436
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Landis, M. J., Matzke, N. J., Moore, B. R., Huelsenbeck, J. P. (2013). Bayesian analysis of biogeography when the number of areas is large. Systematic Biol. 62 (6), 789–804. doi: 10.1093/sysbio/syt040
Lavarack, B., Harris, W., Stocker, G. (2000). Dendrobium and its relatives (Portland, Oregon, United States: Timber Press).
Linder, H. P. (1999). Rytidosperma vickeryae - A new danthonioid grass from Kosciuszko (New South Wales, Australia): Morphology, phylogeny and biogeography. Aust. Systematic Bot. 12 (5), 743–755. doi: 10.1071/SB97046
Martin, H. A. (2006). Cenozoic climatic change and the development of the arid vegetation in Australia. J. Arid Environments 66 (3 SPEC. ISS.), 533–563. doi: 10.1016/j.jaridenv.2006.01.009
Matthews, K. J., Maloney, K. T., Zahirovic, S., Williams, S. E., Seton, M., Müller, R. D. (2016). Global plate boundary evolution and kinematics since the late Paleozoic. Global Planetary Change 146, 226–250. doi: 10.1016/j.gloplacha.2016.10.002
Matzke, N. J. (2013). BioGeoBEARS: BioGeography with Bayesian (and likelihood) evolutionary analysis in R Scripts (R package, version 0.2).
McCormick, M. K., Lee Taylor, D., Juhaszova, K., Burnett, R. K., Jr., Whigham, D. F., O’neill, J. P. (2012). Limitations on orchid recruitment: not a simple picture. Mol. Ecol. 21 (6), 1511–1523. doi: 10.1111/j.1365-294X.2012.05468.x
Mildenhall, D. C., Kennedy, E. M., Lee, D. E., Kaulfuss, U., Bannister, J. M., Fox, B., et al. (2014). Palynology of the early Miocene Foulden Maar, Otago, New Zealand: Diversity following destruction. Rev. Palaeobotany Palynology 204, 27–42. doi: 10.1016/j.revpalbo.2014.02.003
Minh, B. Q., Nguyen, M. A. T., von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30 (5), 1188–1195. doi: 10.1093/molbev/mst024
Molvray, M., Kores, P. J., Chase, M. W. (1999). Phylogenetic relationships within Korthalsella (Viscaceae) based on nuclear ITS and plastid trnL-F sequence data. Am. J. Bot. 86 (2), 249–260. doi: 10.2307/2656940
Nargar, K., O’Hara, K., Mertin, A., Bent, S., Nauheimer, L., Simpson, L., et al. (2022). Evolutionary relationships and range evolution of greenhood orchids (subtribe Pterostylidinae): insights from plastid phylogenomics. Fronteirs Plant Sci. 14, 10631. doi: 10.3389/fpls.2022.912089
Neubig, K. M., Whitten, W. M., Carlsward, B. S., Blanco, M. A., Endara, L., Williams, N. H., et al. (2009). Phylogenetic utility of ycf1 in orchids: A plastid gene more variable than matK. Plant Systematics Evol. 277 (1–2), 75–84. doi: 10.1007/s00606-008-0105-0
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Pérez-Escobar, O. A., Chomicki, G., Condamine, F. L., Karremans, A. P., Bogarín, D., Matzke, N. J., et al. (2017). Recent origin and rapid speciation of Neotropical orchids in the world’s richest plant biodiversity hotspot. New Phytol. 215 (2), 891–905. doi: 10.1111/nph.14629
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S., Hoekstra, H. E. (2012). Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PloS One 7, 5. doi: 10.1371/journal.pone.0037135
POWO (2022) Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Available at: http://www.plantsoftheworldonline.org/.
Pridgeon, A. M., Cribb, P. J., Chase, M. W., Rasmussen, F. N. (2014). Genera Orchidacearum Volume 6 Epidendroideae (Oxford, England: Oxford University Press).
Puente-Lelièvre, C., Harrington, M. G., Brown, E. A., Kuzmina, M., Crayn, D. M. (2013). Cenozoic extinction and recolonization in the New Zealand flora: The case of the fleshy-fruited epacrids (Styphelieae, Styphelioideae, Ericaceae). Mol. Phylogenet. Evol. 66 (1), 203–214. doi: 10.1016/j.ympev.2012.09.027
Rambaut, A., Drummond, A. J. (2007) Tracer v.1.5. Available at: http://beast.bio.ed.ac.uk/Tracer.
Ree, R. H., Smith, S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biol. 57 (1), 4–14. doi: 10.1080/10635150701883881
Ronquist, F. (1997). Dispersal-vicariance analysis: A new approach to the quantification of historical biogeography. Systematic Biol. 46 (1), 195–203. doi: 10.1093/sysbio/46.1.195
Ronquist, F. (2001). DIVA version 1.2. Computer program for MacOS and Win32. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University. Available at: https://sourceforge.net/projects/diva/.
Sanmartín, I., Wanntorp, L., Winkworth, R. C. (2007). West Wind Drift revisited: testing for directional dispersal in the Southern Hemisphere using event-based tree fitting. J. Biogeography 34 (3), 398–416. doi: 10.1111/j.1365-2699.2006.01655.x
Sansaloni, C., Petroli, C., Jaccoud, D., Carling, J., Detering, F., Grattapaglia, D., et al. (2011). Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 5 (S7), 1–2. doi: 10.1186/1753-6561-5-s7-p54
Schmidt-Lebuhn, A. N., Egli, D., Grealy, A., Nicholls, J. A., Zwick, A., Dymock, J. J., et al. (2022). Genetic data confirm the presence of Senecio Madagascariensis in New Zealand. New Z. J. Bot., 1–13. doi: 10.1080/0028825X.2022.2148544
Serna-Sánchez, M. A., Pérez-Escobar, O. A., Bogarín, D., Torres-Jimenez, M. F., Alvarez-Yela, A. C., Arcila-Galvis, J. E., et al. (2021). Plastid phylogenomics resolves ambiguous relationships within the orchid family and provides a solid timeframe for biogeography and macroevolution. Sci. Rep. 11 (1), 1–11. doi: 10.1038/s41598-021-83664-5
Simpson, L., Clements, M. A., Crayn, D. M., Nargar, K. (2018). Evolution in Australia’s mesic biome under past and future climates: Insights from a phylogenetic study of the Australian Rock Orchids (Dendrobium speciosum complex, Orchidaceae). Mol. Phylogenet. Evol. 118, 32–46. doi: 10.1016/j.ympev.2017.09.004
Simpson, L., Clements, M. A., Orel, H. K., Crayn, D. M., Nargar, K. (2021). Plastid phylogenomics clarifies broad-level relationships in Bulbophyllum (Orchidaceae) and provides insights into range evolution of Australasian section Adelopetalum. bioRxiv 2022.07.24.500920. doi: 10.1101/2022.07.24.500920
Smidt, E. C., Borba, E. L., Gravendeel, B., Fischer, G. A., Berg, C. (2011). Molecular phylogeny of the Neotropical sections of Bulbophyllum (Orchidaceae) using nuclear and plastid spacers. Taxon 60, 1050–1064. doi: 10.1002/tax.604009
Smidt, E., de, C., Gallo, L. W., Scatena, V. L. (2013). Leaf anatomical and molecular studies in Bulbophyllum section Micranthae (Orchidaceae) and their implications for systematics. Braz. J. Bot. 36 (1), 75–82. doi: 10.1007/s40415-013-0008-3
Sun, Y., Skinner, D. Z., Liang, G. H., Hulbert, S. H. (1994). Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor. Appl. Genet. 89 (1), 26–32. doi: 10.1007/BF00226978
Swenson, U., Backlund, A., McLoughlin, S., Hill, R. S. (2001). Nothofagus biogeography revisited with special emphasis on the enigmatic distribution of subgenus Brassospora in New Caledonia. Cladistics 17, 28–47. doi: 10.1111/j.1096-0031.2001.tb00109.x
Szlachetko, D. L., Margonska, H. B. (2001). Genera et species Orchidalium. 3. Polish Botanical J. 46 (2), 113–121.
van den Berg, C. (2003). Estudos em sistemática molecular na família Orchidaceae (Feira de Santana, Bahia, Brazil: Universidade Estadual de Feira de Santana). Thesis presented for obtaining the title of Full Professor.
van Kleinwee, I., Larridon, I., Shah, T., Bauters, K., Asselman, P., Goetghebeur, P., et al. (2022). Plastid phylogenomics of the Sansevieria Clade of Dracaena (Asparagaceae) resolves a recent radiation. Mol. Phylogenet. Evol. 169, 107404. doi: 10.1016/j.ympev.2022.107404
Vermeulen, J. J. (1993). A taxonomic revision of Bulbophyllum sections Adelopetalum, Lepanthanthe, Macrouris, Pelma, Peltopus, and Unicifera (Orchidaceae). Orchid Monogr. 7 (1), 1–191.
Vermeulen, J. J., Schuiteman, A., de Vogel, E. F. (2014). Nomenclatural changes in Bulbophyllum (Orchidaceae; epidendroideae). Phytotaxa 166, 101–113. doi: 10.11646/phytotaxa.166.2.1
Wagstaff, S. J., Bayly, M. J., Garnock-Jones, P. J., Albach, D. C. (2002). Classification, origin, and diversification of the New Zealand hebes (Scrophulariaceae). Ann. Missouri Botanical Garden 89 (1), 38–63. doi: 10.2307/3298656
Wagstaff, S. J., Breitwieser, I., Swenson, U. (2006). Origin and relationships of the Austral genus Abrotanella (Asteraceae) inferred from DNA sequences. Taxon 55, 95–106. doi: 10.2307/25065531
Wagstaff, S. J., Dawson, M. I., Venter, S., Munzinger, J., Crayn, D. M., Steane, D. A., et al. (2010). Origin, diversification, and classification of the australasian genus Dracophyllum (Richeeae, Ericaceae). Ann. Missouri Botanical Garden 97 (2), 235–258. doi: 10.3417/2008130
Weising, K., Nybom, H., Pfenninger, M., Wolff, K., Kahl, G. (2005). DNA fingerprinting in plants: principles, methods, and applications. 2nd. ed (Boca Raton, Florida, USA: CRC Press).
Weitemier, K., Straub, S. C., Cronn, R. C., Fishbein, M., Schmickl, R., McDonnell, A., et al. (2014). Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2 (9), 1400042. doi: 10.3732/apps.1400042
Yu, Y., Harris, A. J., Blair, C., He, X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Yule, G. U. (1925). ). II.—A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis, F. R. S. Philos. Trans. R. Soc. London. Ser. B Containing Papers Biol. Character 213 (402–410), 21–87. doi: 10.1098/rstb.1925.0002
Zahirovic, S., Matthews, K. J., Flament, N., M”uller, R. D., Hill, K. C., Seton, M., et al. (2016). Tectonic evolution and deep mantle structure of the eastern Tethys since the latest Jurassic. Earth-Science Rev. 162, 293–337. doi: 10.1016/j.earscirev.2016.09.005
Keywords: ancestral area reconstruction, Australasia, Bulbophyllinae, Bulbophyllum, divergence time estimation, high-throughput sequencing, Orchidaceae, phylogenomics
Citation: Simpson L, Clements MA, Orel HK, Crayn DM and Nargar K (2024) Plastid phylogenomics clarifies broad-level relationships in Bulbophyllum (Orchidaceae) and provides insights into range evolution of Australasian section Adelopetalum. Front. Plant Sci. 14:1219354. doi: 10.3389/fpls.2023.1219354
Received: 09 May 2023; Accepted: 13 November 2023;
Published: 24 May 2024.
Edited by:
Mi-Jeong Yoo, Clarkson University, United StatesReviewed by:
Rob Smissen, Manaaki Whenua Landcare Research, New ZealandCopyright © 2024 Simpson, Clements, Orel, Crayn and Nargar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lalita Simpson, bGFsaXRhLnNpbXBzb24xQGpjdS5lZHUuYXU=; Katharina Nargar, S2F0aGFyaW5hLk5hcmdhckBjc2lyby5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.