- 1Joint International Research Laboratory of Agriculture and Agri-Product Safety of the Ministry of Education of China, Yangzhou University, Yangzhou, China
- 2Key Lab of Crop Genetics & Physiology of Jiangsu Province, Yangzhou University, Yangzhou, China
- 3Department of Crop Genetics and Breeding, College of Agriculture, Yangzhou University, Yangzhou, China
- 4Faculty of Forestry, University of Khartoum, Khartoum North, Sudan
Salt stress affects large cultivated areas worldwide, thus causing remarkable reductions in plant growth and yield. To reduce the negative effects of salt stress on plant growth and yield, plant hormones, nutrient absorption, and utilization, as well as developing salt-tolerant varieties and enhancing their morpho-physiological activities, are some integrative approaches to coping with the increasing incidence of salt stress. Numerous studies have been conducted to investigate the critical impacts of these integrative approaches on plant growth and yield. However, a comprehensive review of these integrative approaches, that regulate plant growth and yield under salt stress, is still in its early stages. The review focused on the major issues of nutrient absorption and utilization by plants, as well as the development of salt tolerance varieties under salt stress. In addition, we explained the effects of these integrative approaches on the crop’s growth and yield, illustrated the roles that phytohormones play in improving morpho-physiological activities, and identified some relevant genes involve in these integrative approaches when the plant is subjected to salt stress. The current review demonstrated that HA with K enhance plant morpho-physiological activities and soil properties. In addition, NRT and NPF genes family enhance nutrients uptake, NHX1, SOS1, TaNHX, AtNHX1, KDML, RD6, and SKC1, maintain ion homeostasis and membrane integrity to cope with the adverse effects of salt stress, and sd1/Rht1, AtNHX1, BnaMAX1s, ipal-1D, and sft improve the plant growth and yield in different plants. The primary purpose of this investigation is to provide a comprehensive review of the performance of various strategies under salt stress, which might assist in further interpreting the mechanisms that plants use to regulate plant growth and yield under salt stress.
Introduction
The world faces a tremendous challenge in crop production (López-Marqués et al., 2020). According to (FAO, 2017), the human population will increase to 10 billion, and the requirements for cereals and livestock production will exceed 60 percent (Springmann et al., 2018). Agriculture growth depends on productivity achieved through increased crop yields. However, the higher yield was only achieved during the green revolution period (López-Marqués et al., 2020). The percentage increase in yield has decreased after the green revolution.
Salt stress is one of the most important environmental factors, significantly decreasing crop growth and yield worldwide (Zörb et al., 2019; Negi et al., 2020). Salt stress affects approximately 20 percent of all agricultural lands and 33 percent of irrigated agricultural lands (FAO, 2017). The neutral salt concentrations of sodium chloride (NaCl) and sodium sulfate (Na2SO4) caused salt stress in soil (Van Zelm et al., 2020). The higher accumulation of NaCl in the soil depletes the water content and harms plants. It thus causes toxic effects from the sodium and chloride ions in plants (Ahmad et al., 2022b). To cope with the negative effects of salt stress’s, plants use various responses, such as regulating gene expression and stimulating hormones (Raza et al., 2022; Feng et al., 2023). During salt stress, different kinds of strategies can be used to increase plant growth and yields. Currently, researchers and growers realize the importance of identifying suitable cultivars (Jiang et al., 2022), nutrient absorption (Adil et al., 2022), the roles of hormones (Ahmad et al., 2022a), and gene identification (Tang et al., 2022) under salt stress (Adil et al., 2022; Jiang et al., 2022; Tang et al., 2022; Ahmad et al., 2022b).
Different studies have shown that the regulation of genes under salt stress is affected by numerous transcriptional cascades (Wu et al., 2019). Abscisic acid (ABA) and gibberellin (GA), both acting as endogenous signaling hormones and are essential regulators of salt stress (Pu et al., 2019; Ahmad et al., 2022b). WRKY genes in cotton respond to salt stress via ABA signaling and regulate the production of reactive oxygen species (ROS) in plant cells (Yan et al., 2014). In wheat, MYB genes respond to salt stress by regulating ion homeostasis in order to control osmotic pressure and lower ROS concentrations (Song et al., 2020). Understanding the molecular mechanism underlying salt resistance in plants is essential for improving crop quality and yield, and this can only be achieved by studying different salt-tolerant genes in plants (Wang et al., 2022).
Salt stress affects nutrient absorption and disturbs plant metabolic activities such as lipid and carbohydrate metabolism, which reduce crop growth and yield (Parida and Das, 2005; Zafar et al., 2022). In addition, salt stress affects the plant’s root system. It creates osmotic stress due to the elevated sodium (Na+), resulting in a water shortage in plant cells and thus affecting water potential (Ekinci et al., 2022). Due to the imbalances of nutrient availability in soil, salt stress thus causes ion toxicity in different plants (Ali et al., 2021).
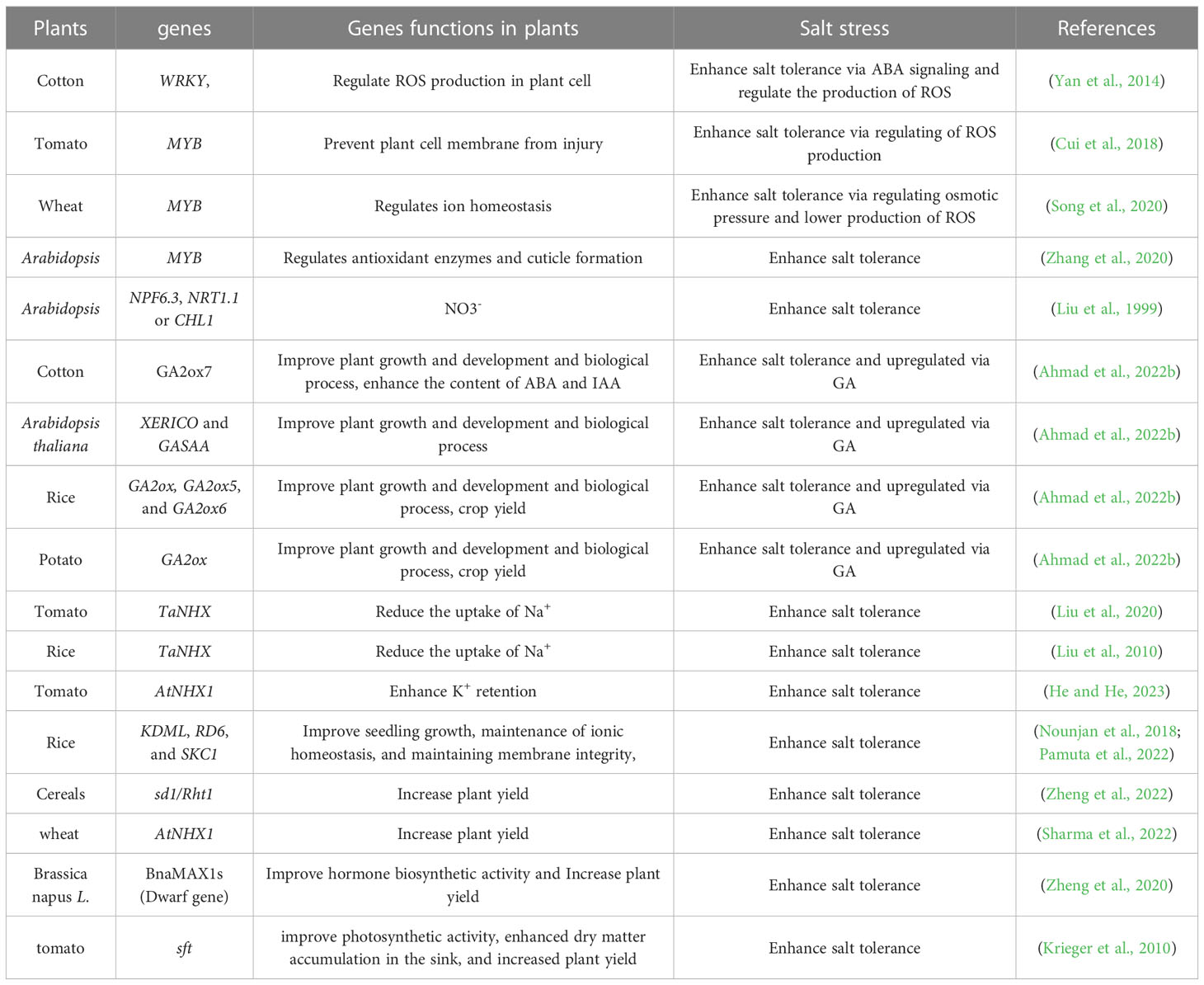
Introducing suitable cultivars and desirable genes, etc., has been widely investigated to improve crop growth and productivity under salt stress (Chattha et al., 2020). However, these approaches are time-consuming and costly. In order to improve the growth and yield of desirable cultivars, plant genomic editing with the CRISPR/Cas9 system is currently being used. Nevertheless, the regulation of genome-edited crops is still unknown, especially under abiotic stresses (Razzaq et al., 2019). Applying seed priming, nutrient management, and phyto-hormones to overcome the adverse effects of salt stress can suggest promising conclusions for various plants to improve yield (Ahmad et al., 2022a; Ahmad et al., 2022b; Figure 1). In this review, we examined how salt stress affects nutrient uptake and utilization in various crops under salt stress from the aspects of breeding salt-tolerant varieties, identifying salt-resistant genes, and using CRISPR-Cas9 tools for genome editing (Table 1).
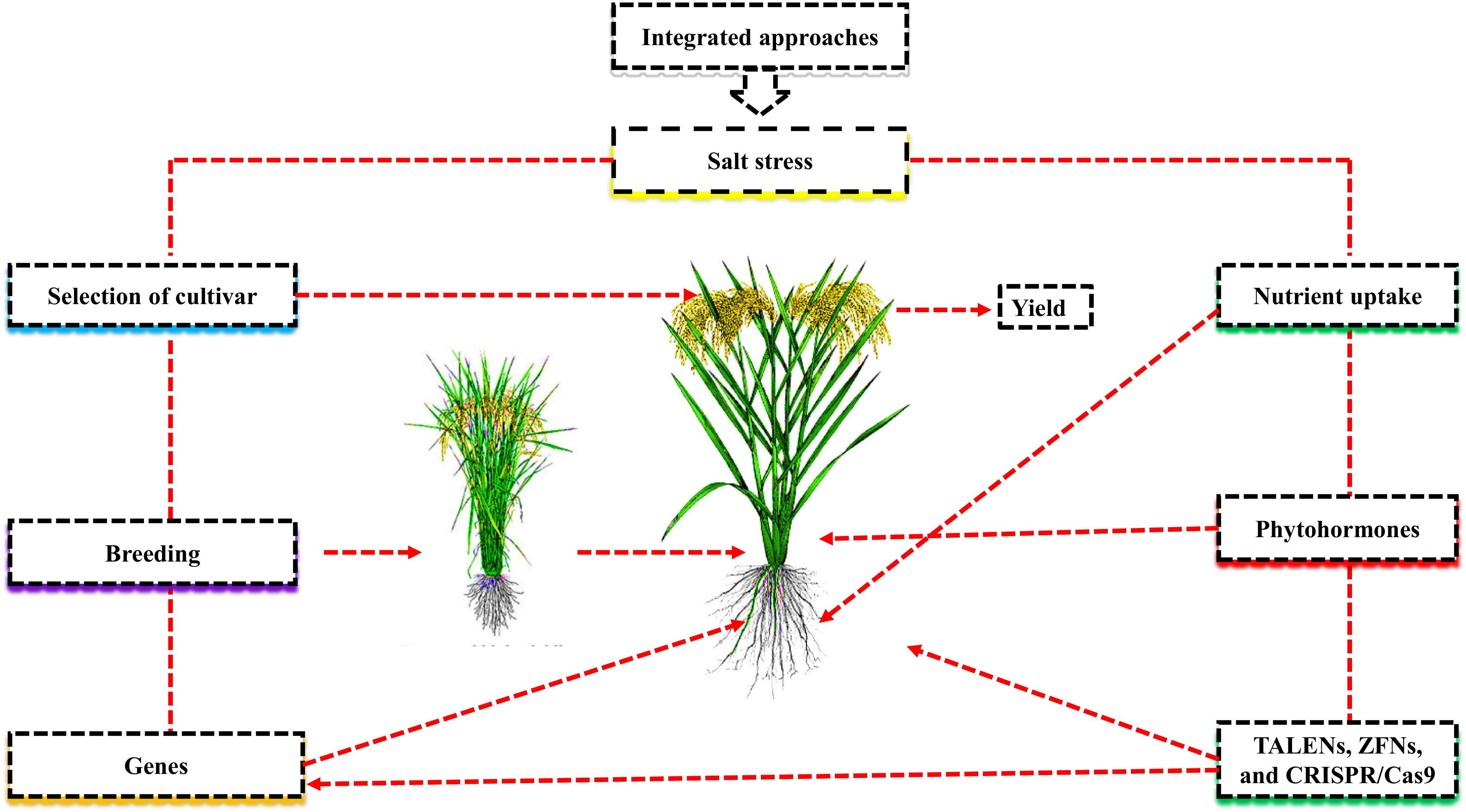
Figure 1 Integrated approach mitigate the negative effects of salt stress and improve plant growth and yield.
Different approaches for increasing plant growth and yield
Effects of salt stress on crop growth and development
Salt stress is one of the major abiotic stresses that causes seed dormancy and reduces normal plant growth and yield. Soluble salt at higher concentrations causes both osmotic and ionic stresses in soil, which lead to secondary stresses such as nutritional imbalance and oxidative stress. In the root zone, the higher concentrations of salts cause higher osmotic pressure in the soil compared to plant cells, which reduces the capability of plants to uptake water and nutrients (El-Mageed et al., 2022). When plants are subjected to high salt stress, the soil solution becomes more hyper-osmotic and causes the root cells to lose water, resulting in plant senescence or wilting (Verma et al., 2022). Osmotic stress, caused by a lack of water in plant tissues, primarily reduces leaf growth and causes a reduction in shoots and reproductive growth. (Chourasia et al., 2022). Secondary stress, such as oxidative stress, mainly occurs due to the higher production of ROS, which contributes to the primary effects of salt stress described above (Kaur et al., 2023). In addition, the overproduction of ROS in plants increases the fluidity and permeability of cell membranes and degrades functional and structural proteins under salt stress (Sharma et al., 2023).
Salt stress and genes involve in plant’s nutrient uptake
Due to salt-induced osmotic stress, plant growth and nutrient uptake decreased under salt stress. (Abbas et al., 2022). The uptake of nutrients by roots and the efficiency of photosynthesis by leaves are sources of plant development. Plant cells accumulate salt-affected soil ions (Na+, and Cl-) that inhibit nitrogen (N), phosphorus (P), and potassium (K) uptake and photosynthesis (Zhang et al., 2011; Ait-El-Mokhtar et al., 2020; Figure 2). Many plant organelles, including mitochondria, chloroplasts, and peroxisomes, produce more reactive oxygen species (ROS) when subjected to higher salt stress, such as hydroxyl radical (OH), superoxide (O2-), and hydrogen peroxide (H2O2). Proteins, lipids, nucleic acids, and cellular membranes were all negatively affected due to the higher accumulation of ROS in plants (Zulfiqar et al., 2019). Plants protect themselves against ROS by producing different antioxidant enzymes such as catalase, ascorbate peroxidase, and superoxide dismutase (Ahmad et al., 2022a).
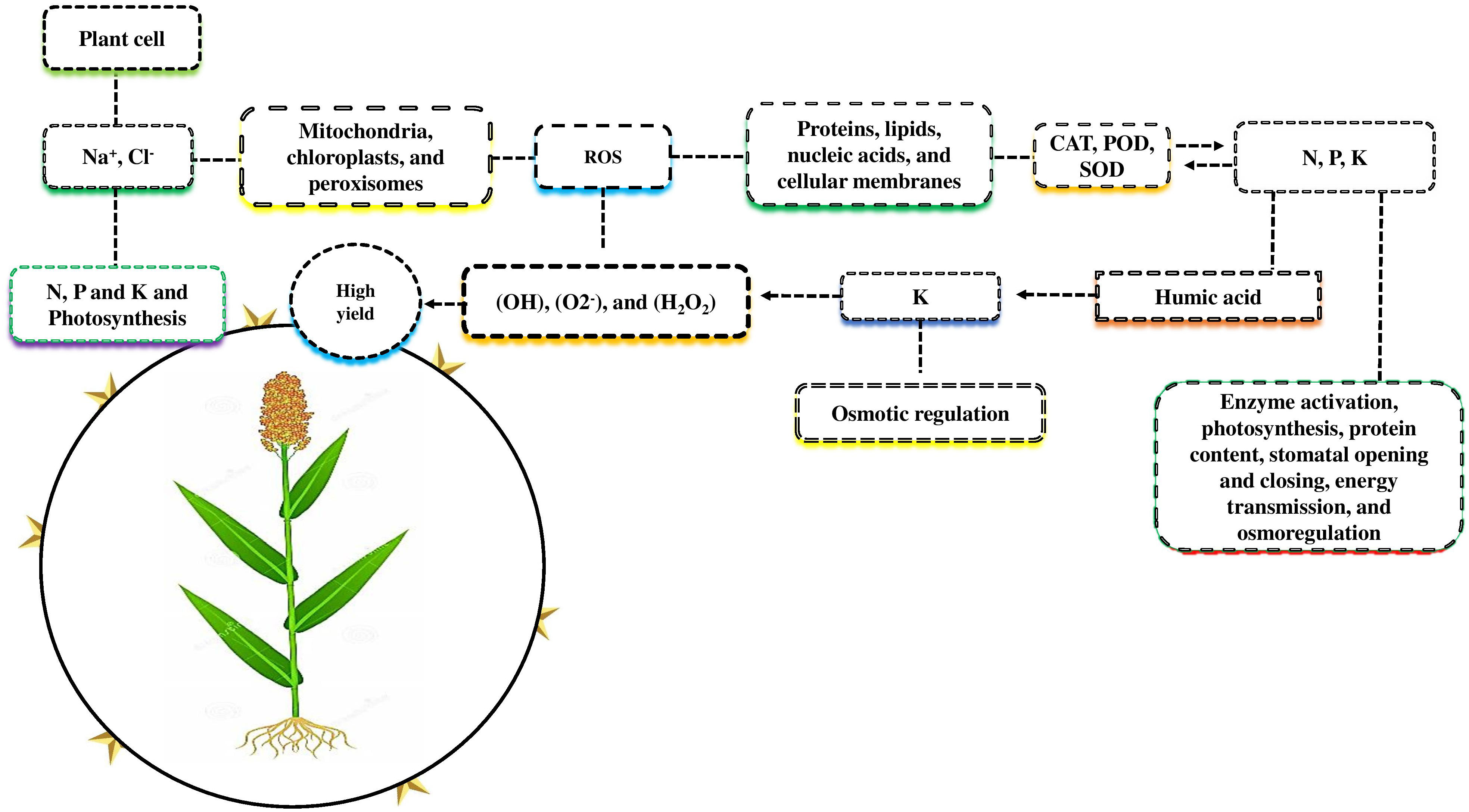
Figure 2 Nutrients mitigate the adverse effects of salt stress. Plant cells accumulate salt-effected ions such as Na+ and Cl- that reduce N, P, and K uptake and photosynthetic activity. Plants organelles such as mitochondria, chloroplast, and peroxisome are exposed to higher salt stress, including the hydroxyl radical (OH), superoxide (O2-), and hydrogen peroxide (H2O2), produce more reactive oxygen species (ROS), which negatively affect protein, lipids nucleic acid, and cellular damage. For self-defense, plants produce different antioxidant enzymes, such as CAT, POD, and SOD, to eliminate the negative effects of salt stress. Macronutrients N, P and K play a major role in enzyme activation photosynthesis, protein content, stomatal opening and closing, energy transmission, and osmoregulation under salt stress. K play a vital role in osmotic regulation compared to N and P. The combined application of K with humic acid counters the negative effects of ROS and improves the morph-physiological activity and yield of various crops.
Different mineral nutrients and organic amendments have been widely used to improve salt resistance and nutrient uptake in different kinds of crop species. Among these nutrients, N, P, and K are essential for plants as they are involved in different physiological and biochemical processes in plant growth and yield (Ahmad et al., 2022a). Previous studies demonstrated that macronutrients are required for various plant cellular processes, such as enzyme activation, photosynthesis, protein content, stomatal opening and closing, energy transmission, and osmoregulation under salt stress (Ashraf et al., 2013; Taha et al., 2020). K is suggested to be more efficient for osmotic regulation as compared to N and P under salt stress (Abbas et al., 2022). A key factor in plant salt resistance is the higher uptake of potassium over sodium (Zrig et al., 2021). Abbas et al. (2022) demonstrated that the application of K with humic acid (HA) enhanced different physiological and biochemical activities, such as nutrient uptake, water relations, stomatal conductance, and enzymes activation to counter the adverse effects of ROS (Abbas et al., 2022). HA, are organic compounds that are necessary for improving soil properties and plant growth (Ampong et al., 2022). The corresponding findings were supported by (Ali et al., 2019), who demonstrated that K with HA, increased salt resistance in sorghum by increasing its nutrient uptake and antioxidant activities, and reducing ROS production. The current review showed that K in combination with HA improved different morpho-physiological activities in plants. The application of HA with N and P or other molecules, such as fulvic acid (FA), which enhances soil qualities and plant growth in different crops under salt stress, is still not well understood.
Plant nutrient use efficiency and yield can be increased by identifying critical genes (Kumar et al., 2021). Both the NRT and NPF gene families have been recently discovered to be involved in nitrate uptake and transport throughout the plant (Fan et al., 2017; Figure 3). There are two types of transport systems involved in NO3- uptake: low-affinity transport systems (LATS) and high-affinity transport systems (HATS) (Faure et al., 2021). Many NRT family genes have been identified as having high affinity, whereas NPF are thought to function as the primary components of the LATS for NO3- at high concentrations (Li et al., 2021). Previous studies showed that some RNT and NPF family genes are involved in the dual-transport system; for example, in Arabidopsis, the RNT and NPF genes such as NPF6.3, NRT1.1 or CHL1, was identified in both high- and low-affinity nitrate uptakes (Liu et al., 1999). The NPF6.3 gene transport a variety of substrates, including protein concentration, dipeptides, chloride, glucosinolates, and plant hormones such as gibberellins (GAs), jasmonates (JAs), indole-3-acetic acid (IAA), and abscisic acid (ABA) (Fan et al., 2017; Chao et al., 2021). In Arabidopsis, the gene families NRT and NPF were crucial for nitrate uptake and transport to other parts of the plant. However, the remarkable performance of the NPF6.3 gene under salt stress, as well as the identification of NRT and NPF low-to-high affinities genes in other plant species, remain unclear.
Figure 3 Different genes are involved in plant nutrient uptake and enhanced plant growth and yield under salt stress. Two gene families, NRT and NPF, enhance the uptake of NO3- via two transportation methods: low-affinity transport systems (LATS) and high-affinity transport systems (HATS). RNT genes are especially involved in HATS, while NPF is involved in LATS. Some genes, such as NPF6.3 and NRT1.1/CHL1, are involved in the dual-transport system. The genes NPF6.3 transport a variety of substrates such as protein, dipeptides, chloride, glucosinolates, gibberellins (GAs), jasmonate (JAs), indole-3-acetic acid (IAA), abscisic acid (ABA) to the various parts of the plants.
Effects of salt stress on physiological and biochemical activities
Nevertheless, internally, plants have developed comprehensive resistance systems to cope with the adverse effects of ROS (Wang et al., 2023). SOD, CAT, POD, GR, and APX are ROS-scavenging and mediate in the reaction cycle of antioxidant chemicals, including ascorbic acid (AsA) and glutathione (GSH) (Apel and Hirt, 2004; Dietz et al., 2006; Türkan and Demiral, 2009).
Several studies demonstrated that salt stress reduces the physiological activities of wheat, barley, maize, sunflower, rice, tomatoes, and beets (Turan et al., 2009; Shahbaz et al., 2011; Shiyab et al., 2013; Umnajkitikorn et al., 2013; Zeeshan et al., 2020; He et al., 2022). In wheat and barley, the negative effects of salt stress may be due to a reduction in stomatal conductance or the excessive production of ROS in plants, which can enhance oxygen-induced cellular damage. Similarly, Netondo et al. (2004) in sorghum showed that changes in stomatal conductance and the concentration of cellular CO2 were positively correlated during salt stress, showing that stomatal conductance was a key factor that played an important role in plant net photosynthesis (Netondo et al., 2004). It has been confirmed that stomatal conductance plays a crucial role in net photosynthesis in wheat, barley, and sorghum, but the detailed mechanism in various species is still unknown.
Genes improve plant physiological and biochemical activities under salt stress
Genes play an important role in reducing abiotic stresses in plants by facilitating their growth and development, nutrient uptake, and carrying them from one part of the plant to another. Salt-related genes, such as SKC1, CDPK, and MAPK pathways, overlay-sensitive (SOS) pathways, PAL and CHS, actively maintain response to salt stress in plants (Pamuta et al., 2022). SOS1 and NHX1 genes encode antiporter Na+/H+; however, SOS1 is located on the plant plasma membrane (Du et al., 2023). The SOS1 gene regulates the transport of Na+ from roots to the shoots of the plant (Liu et al., 2019). The TaNHX gene enhance plant tolerance against salt stress due to less uptake of Na+ and its translocation to the shoots in tomatoes and rice (Liu et al., 2010). The performance of the protein, NHX has been widely investigated in various crops such as tomatoes, rice, and cotton (Liu et al., 2010; Gouiaa and Khoudi, 2015). Though, in tomatoes, AtNHX1 gene overexpression improved K+ retention in cells under higher salt stress (He and He, 2023). Similarly, in transgenic tobacco, the expression of the TNHXS1-IRES-TVP1 bicistronic transcriptional unit led to an increase in the accumulation of K+ and a decrease in N+ concentration in leaf tissue (Gouiaa et al., 2012). An increase in antioxidant activities such as SOD, POD, and CAT prevents ROS accumulation and reduces cellular damage in plants (Ahmad et al., 2022a). In rice KDML, RD6, and SKC1 genes reduced the adverse effect of ROS and increase seedling growth due to the higher antioxidant enzymes activities under salt stress (Pamuta et al., 2022). The possible results might be due to the SKC1 genes responsible for the maintenance of ionic homeostasis, maintaining membrane integrity, and coping against salt-induced damages such as ROS detoxification (Nounjan et al., 2018; El Moukhtari et al., 2020). Hence, genes alleviated the negative effects of salt stress by improving physiological and biochemical activities of the plants. However, these gene’s roles and underlying mechanisms in various crops remain unclear under salt stress.
Effects of salt stress on crop yield and yield-related genes
The world population is expected to increase by 34 percent in 2050, and the requirements for food production are expected to increase by 34 percent to meet the demand for cereals (FAO, 2017). This growth in cereal productivity will need to occur in a world with higher salt stress, where regular higher salt stress negatively affect plant yield (Adil et al., 2022). Hence, to improve the yield of cereals, increase the current germplasm’s yield and improve yield stability through enhanced tolerance to salt stress (Hu and Schmidhalter, 2023). It has been demonstrated that better management of land and the introduction of new genotypes through genetic engineering and breeding programs can lead to advances in yield. Recently, various approaches, traditional and state-of-the-art amelioration, have been put forward to improve plant yield (Panagea et al., 2016). Plant breeding has played a paramount role in maintaining food security, leading to increased plant productivity over the past few decades. For salt reclamation, breeding is considered one of the most efficient strategy for improving plant tolerance against salt stress (Ashraf and Munns, 2022). Despite much research documented in understanding the response of plants to salt stress, the breeding of salt-resistant genotypes remains slow, with limited progress in plants (Asif et al., 2018; Kotula et al., 2020). Compared to the slow breeding progress, to enhance the salt tolerance of barley and wheat, salt-resistant genotypes have been introduced and commercialized in various Asian countries via conventional breeding methods (Ismail and Horie, 2017). The improved rice genotypes development through this breeding method increase grain yield production in fields under salt-affected areas by 0.5 to more than 2 tons per hectare (Singh et al., 2016). Nevertheless, the development of these varieties took 5-10 years of rigorous evaluation of many breeding lines with high process costs; hence, various approaches, such as genome-based and marker-assisted breeding, are becoming more promising and attractive (Thomson et al., 2012). Different studies have shown that yield of transgenic wheat and barley genotypes with salt resistance can be developed by manipulating the expression of introducing genes or native genes (Hu and Schmidhalter, 2023). The green revolution gene, sd1/Rht1, significantly increased plant’s yield when it was successfully adopted in cereals (Zheng et al., 2022). In addition, the highly expressed AtNHX1 gene in transgenic wheat lines, a gene encoding an Arabidopsis vacuolar Na+/H+ antiporter, showed a higher grain yield in saline field (Sharma et al., 2022). A few dwarf genes, such as BnaMAX1s, are involved in hormone biosynthetic activities and increased plant yield in brassica napus L. (Zheng et al., 2020). Similarly, rice over-dominance ipa1-1D and tomato sft genes increased plant photosynthetic activities, enhanced dry matter accumulation in sink, and increased yield (Jiao et al., 2010; Krieger et al., 2010). Nevertheless, the identified genes against salt resistance have not yet been transferred into relevant commercial genotypes nor used to generate a salt-resistant plant (Asif et al., 2018). The reason is that quantitative trait loci (QTLs) or genes have only been checked in controlled growth conditions with short periods of salt stress, which do not reflect realistic field conditions (Singh et al., 2021). Therefore, in the future, more field trials are required to measure the value of these genes and dwarf-related genes in breeding to achieve higher yield under salt stress. Moreover, clustered regularly interspaced short palindromic repeats (CRISPR-Cas9), transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases (ZFNs) are techniques that easily modify genetic loci or multiple homologous genes.
Practices of alleviating salt stress on crops
Different technologies are used for gene modification
Genome editing technologies, which can change the target genes of the plant genome, is increasingly preferred for use in different fields, including crop breeding and plant science. Genome-editing technologies characterize crop improvement and gene function (Xia et al., 2021). The leading three technologies used in genome editing such as TALENs, ZFNs, and CRISPR/Cas9. TALENs and ZFNs are time-consuming and require a lengthy protocol to gain the specific target (Liu et al., 2021). Compared with TALENs and ZFNs, the CRISPR-Cas9 techniques are more convenient, easy to design, and vigorous (Chennakesavulu et al., 2021). CRISPER-Cas9 is a simple toolkit that is easy to design because of the involvement of only single-guided RNA (sgRNA) and the cas9 protein compared to TALENs and ZFNs (Razzaq et al., 2019). Additionally, the procedure involved in TALENs and ZFNs is complicated because they need protein engineering for their construction (Razzaq et al., 2019). Due to this obstacle, the tools of TALENs and ZFNs in plants have been limited (Jinek et al., 2012). In several plants, continuous innovation for efficient genome editing has expanded the application of CRISPR-Cas9 and is rapidly becoming a promising tool for gene modifications (Shan et al., 2013; Zhang et al., 2019). CRISPR-Cas9 (CRISPR-associated) is a prokaryotic adaptive immune system that binds and cleaves foreign nucleic acids (Brouns et al., 2008). The type II CRISPR system most frequently used is composed of two components, such as Cas9 nuclease and an artificial single guide RNA (sgRNA) (Jinek et al., 2012). CRISPR-Cas9 plays a vital role in improving plant quality and yield. Plant yield is a complicated, multigenic, and quantitative characteristic affected by various features. The CRISPR-Cas9 technique has proven to be effective in increasing plant yield. The CRISPR-Cas9 genome editing technique is only important for gene knock-in and knock-out, not for the base version (Chennakesavulu et al., 2021). The corresponding findings of (Li et al., 2017; Zhang et al., 2018) revealed that genes negatively regulate yield traits such as tiller number by OSAAP3, and grain size by OsGRF, used CRISPR-Cas9 to knock out multiple genome yield-related genes, including Hd2, Hd4, and Hd5 (Li et al., 2017; Zhang et al., 2018). Recently, 30 genotypes of “Green Revolution miracle rice” were investigated through genome sequencing, and 57 genes controlling yield-relevant traits were knocked out via the CRIPR-Cas9 technique (Huang et al., 2018). Phenotyping results showed that many genes identified during screening were crucial to regulate yield-related traits in rice. However, more studies need further investigation to identify various genes in other crops via CRISPR-Cas9.
Abiotic stress tolerance through CRISPR-Cas9-mediated genome editing has been documented in Arabidopsis, wheat, rice, tomatoes, barley, and sorghum (Gobena et al., 2017; Wang et al., 2017; Sánchez-León et al., 2018; Lawrenson and Harwood, 2019; Liu et al., 2019; Tran et al., 2021). In tomatoes, the functional domains of hybrid proline-rice protein 1 (S1HyPRP1), a negative regulator of salt stress resistance, were disrupted using a CRIPR-Cas9 mediated multiple genome editing approaches (Tran et al., 2021). Further investigation showed that the precise elimination of S1HyPRP1 functional domains in tomatoes led to higher salt tolerance during all growth stages (Tran et al., 2021). The Slmapk3 editing gene, via CRISPR-Cas9, exhibited lesser ROS, higher enzyme activities, lower membrane damage, and reduced severe plant welting under heat stress (Yu et al., 2019). Several studies have been conducted about the role of CRISPR-Cas9 in genome editing that enhanced different plant’s growth and yield, but further research is needed to investigate the performance of these modified genes via CRISPR-Cas9 techniques in different crops under salt stress.
Selection of suitable cultivars under salt stress
Different strategies assist in the improvement of suitable cultivars under salt stress
Salt resistance relies on the selection of suitable genotypes and their families (Figure 4). Investigating the comparison or examining the physiological mechanism of salt stress in some cultivars that belong to the same family can provide more knowledge of the discriminative growth pattern of plant and salt resistance levels in both cultivars (Shahzad et al., 2021). Hence, due to the lack of examination of physiological mechanisms concerning salt stress in genotypes, it is critical to determine the negative impact of salt stress on crop growth and yield. Adapting cultivars to salt stress involves complex biochemical, physiological, and molecular mechanisms, which are still in an early phase (Denaxa et al., 2022). To reduce the negative effects of salt stress on crop growth and development, various strategies, like seed priming and foliar application, should be used to understand plant morpho-physiological and biochemical activities (Figure 4). Seed priming is when various crop seeds are soaked with one or more growth regulators at an appropriate level before they are sown.
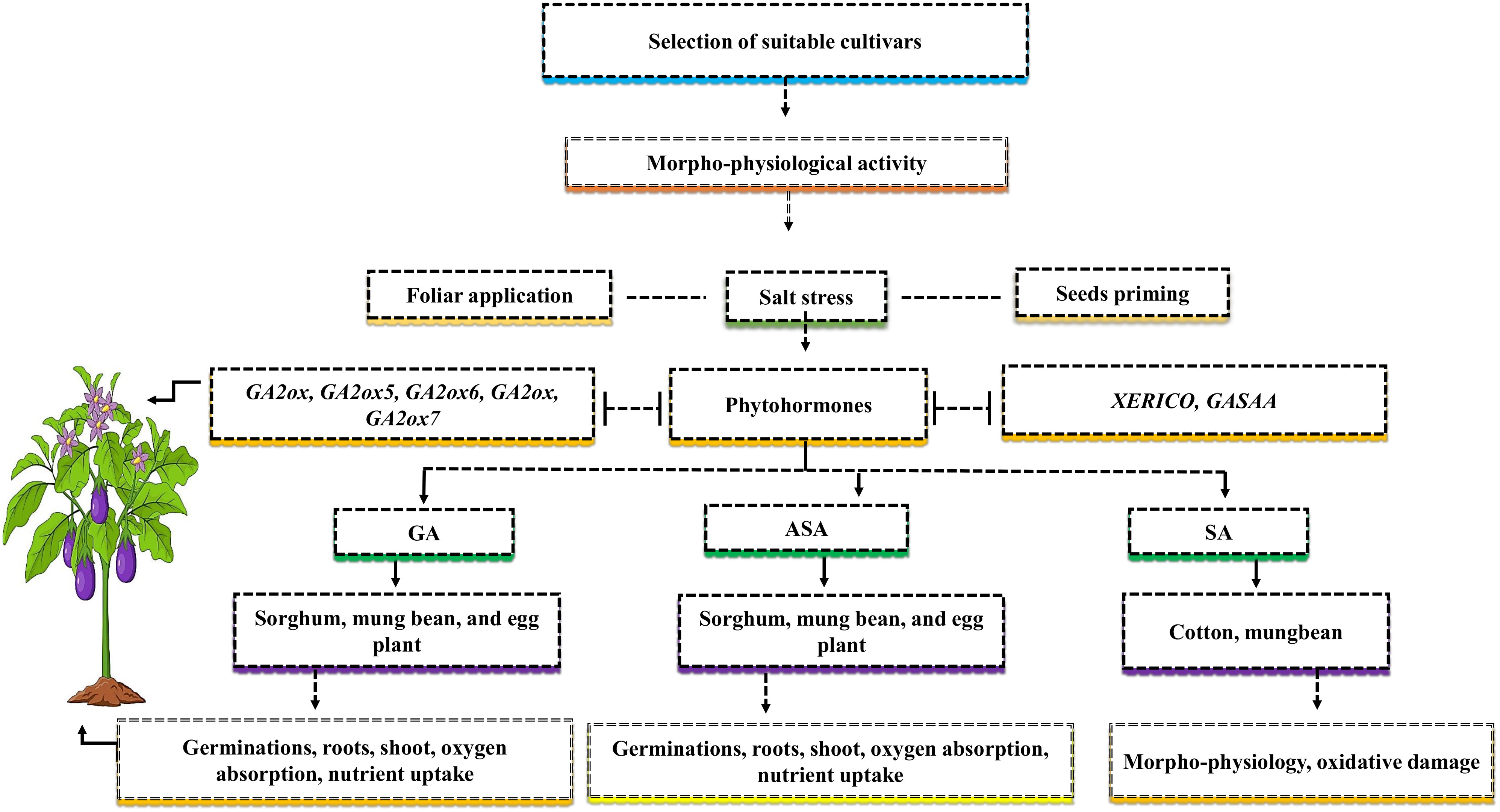
Figure 4 The selection of suitable cultivars can mitigate the negative effects of salt stress. For higher yield, it’s important to know the morph-physiological activity of the cultivars. The phyto-hormones such as gibberellins (GAs), ascorbate (ASA), and salicylic acid (SA) can mitigate the adverse effects of salt stress via two application methods such as foliar and seed priming. The application of GAs improves germinations, roots, shoot, oxygen absorption, and nutrient uptake in sorghum, mung bean, and eggplant, ASA improves germinations, roots, shoot, oxygen absorption, nutrient uptake in sorghum, mung bean, and eggplant, and SA improves morpho-physiology activities, and oxidative damage in cotton and mung bean. The genes such as GA2ox, GA2ox5, GA2ox6, GA2ox, GA2ox7, XERICO and GASAA are involved in endogenous phyto-hormones and increase plant yield.
In contrast, the foliar application is the treatment of one or more growth regulators applied at an appropriate level in a liquid form directly to the leaves. Previous studies demonstrated that NaCl reduced germination percentage in three beans and two sorghum cultivars (Hasanuzzaman et al., 2020). The reduction in germination percentage might be caused by increased NaCl osmotic pressure, which slows down the water imbibition, germination and metabolism processes of the seeds of sorghum and beans (Xie et al., 2019). The application of ASA enhanced the germination percentage of sorghum and eggplant under the higher level of NaCl. The treatment reduced the adverse effects of salt stress, improved oxygen absorption, and improved the transportation of nutrients from cotyledon to embryos (Irfan et al., 2021). Plant roots and shoots are considered essential parameters for salt stress because roots come into contact with the soil and absorb water from it and transfer it to the shoots. For instance, the reports of Xie et al. (2019) on various plants and Hussien Ibrahim et al. (2020) on sorghum showed that tissue alteration under salt stress caused a significant decrease in the seedling growth characteristics (Xie et al., 2019; Hussien Ibrahim et al., 2020). The application of exogenous ASA increased sorghum seedling growth characteristic while mitigating the negative effects of salt stress (Mohammed Ibrahim Elsiddig et al., 2022). Similar results were supported by Mittal et al. (2018) in Brassica rapa L., who demonstrated that seed treatment with ASA before sowing has dramatically improved seedling growth characteristics and protects the plants roots and shoots form altering (Mittal et al., 2018). The improved seedling growth characteristic might be due to the ascorbic acid antioxidant action and or increased cell enlargement within the apical meristem of seedlings (Wang et al., 2019).
Moreover, seed priming with suitable amount of GA can protect against seed deterioration and mitigate the adverse effects of salt stress, such as ion toxicity, osmotic stress, and an imbalance of nutrients uptake (Ahmad et al., 2022b). Seeds soaked with GA application facilitate germination and increased seedling length in rice and sorghum, as GA stimulated cell division and cell elongation (Saudi, 2017; Shihab and Hamza, 2020). Similarly, SA applications can enhance plant tolerance to salt stress in different crops (Khan et al., 2015). Such as, the reasonable concentration of SA in stressed plants via seeds soaking before sowing and after germination via spraying or adding to the nutrient solution can improve the morpho-physiological activities of cotton seedlings under salt stress (Ahmad et al., 2022a). SA improved mung beans growth characteristics and photosynthetic activities, and at the same time, oxidative damage from salt stress was reduced (Ahanger et al., 2019). The seed priming and foliar application of ASA, GA, and SA play an important role in enhancing salt tolerance mechanism and promoting the germination of Medicago sativa L. Brassica juncea L. and cotton (Ahmad et al., 2022b). Further studies are required to close the knowledge gap regarding the application of ASA, GA, and SA to other growth attributes, such as the physiological and biochemical characteristics of various plants via other molecular techniques.
Additionally, to improve plant growth and yield with the help of the selection of suitable cultivars, it is crucial to understand the genes in plants that are responsive to phyto-hormones. In response to salt stress, the genes GA2ox7 in cotton, XERICO and GASAA in Arabidopsis thaliana, GA2ox, GA2ox5, and GA2ox6 in rice, and GA2ox in potatoes were upregulated by GAs (Ahmad et al., 2022b). These genes support the growth and development of plants and are involved in a number of biological processes. The GA2ox7 genes enhance the content of abscisic acid (ABA) and indole acetic acid (IAA) in cotton (Ahmad et al., 2022b). The expression of GA2ox6 genes in rice transgenic plants increased the grain yield by 10-30% during abiotic stresses (Lo et al., 2017). These genes are involved in plant endogenous hormones, but their signaling and transduction pathways in different species under salt stress are still not being clearly understood.
Conclusions and future recommendations
The impact of salt stress on plant growth is considered a significant threat to agricultural productivity. Salt stress mainly reduces the plant’s growth through physiology and the imbalance of ion homeostasis, which can alter gene expression. Therefore, to enhance the yield of plants under salt stress conditions, it is crucial to understand the integrative approaches. Salt stress is a highly complicated process, and coping with the negative effects of this stress is still poorly understood.
The application of HA with K+ improves morpho-physiological activities and soil properties. However, its performance with N, P, and other molecules such as FA, are far from clear.
The current study investigated that phyto-hormones such as ASA, SA, and GA improve the growth attributes in several plants. However, the detailed mechanism of these phyto-hormones in physiological and biochemical activities in different plants under salt stress is still in an early phase.
Stomatal conductance is paramount in net photosynthesis activities in several crops, such as wheat, barley, and sorghum. However, elucidating the detailed mechanism of stomatal conductance under salt stress in various crops is still unknown.
Gene families such as NRT and NPF uptake nitrate and translocate it to other parts of the plants. However, the performance of these genes family, especially the NPF6.3, NRT1.1/CHL1 under salt stress in different crops is still unknown.
It has been confirmed that various genes, including NHX1, SOS1, TaNHX, AtNHX1, KDML, RD6, and SKC1, maintain ion homeostasis and membrane integrity to cope with salt-induced damage in different plants. However, these genes’ performance and underlying mechanisms in several crops remain unknown under salt stress.
Different genes, such as sd1/Rht1, AtNHX1, BnaMAX1s, ipal-1D, and sft, improved the growth and yield of various plants. However, the identification of these genes against salt tolerance has not yet been transferred into the relevant commercial genotypes or used to generate salt-resistant plants.
CRISPER-Cas9 successfully knocks out various genes such as OSAAP3, OsGRF4, OsGS3, TaGW2, TaGASR7, Hd2, HD4, and Hd5, which negatively regulate the yield traits of rice. In contrast, the genes modified by CRISPR-Cas9-modified genes Slmapk3 enhanced enzyme activities, reduced plant wilting, and increased plant yield under heat stress. However, further studies require to investigated to knock in or knock out different genes via CRISPR-Cas9 under salt stress.
Author contributions
IA: Visualized the idea, participated in the writing (review and editing), and arranged the contents and draft of the original manuscript. GSZ and GLZ: Acquired funding and contributed to the reviewing and editing of the manuscript. JL, YZ and MY: Assisted in the collection of literatures. ES and MS: Polished the manuscript and eliminated grammatical errors. All authors contributed to the article and approved the submitted version.
Funding
This work was partially funded by China National Key Research and Development Program (2022YFE0113400, 2018YFE0108100), the Natural Science Foundation of Jiangsu Province of China (BK20221371), and the Rural Revitalization Program of Xinghua City, additionally the key disciplines of higher education in Jiangsu Province.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, G., Rehman, S., Siddiqui, M. H., Ali, H. M., Farooq, M. A., Chen, Y. (2022). Potassium and humic acid synergistically increase salt tolerance and nutrient uptake in contrasting wheat genotypes through ionic homeostasis and activation of antioxidant enzymes. Plants 11, 263. doi: 10.3390/plants11030263
Adil, M., Bashir, S., Bashir, S., Aslam, Z., Ahmad, N., Younas, T., et al. (2022). Zinc oxide nanoparticles improved chlorophyll contents, physical parameters, and wheat yield under salt stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.932861
Ahanger, M. A., Aziz, U., Alsahli, A. A., Alyemeni, M. N., Ahmad, P. (2019). Influence of exogenous salicylic acid and nitric oxide on growth, photosynthesis, and ascorbate-glutathione cycle in salt stressed vigna angularis. Biomolecules 10, 42. doi: 10.3390/biom10010042
Ahmad, I., Zhu, G., Zhou, G., Song, X., Hussein Ibrahim, M. E., Ibrahim Salih, E. G. (2022a). Effect of n on growth, antioxidant capacity, and chlorophyll content of sorghum. Agronomy 12, 501. doi: 10.3390/agronomy12020501
Ahmad, I., Zhu, G., Zhou, G., Song, X., Hussein Ibrahim, M. E., Ibrahim Salih, E. G., et al. (2022b). Pivotal role of phytohormones and their responsive genes in plant growth and their signaling and transduction pathway under salt stress in cotton. Int. J. Mol. Sci. 23, 7339. doi: 10.3390/ijms23137339
Ait-El-Mokhtar, M., Baslam, M., Ben-Laouane, R., Anli, M., Boutasknit, A., Mitsui, T., et al. (2020). Alleviation of detrimental effects of salt stress on date palm (Phoenix dactylifera l.) by the application of arbuscular mycorrhizal fungi and/or compost. Front. Sustain. 4. doi: 10.3389/fsufs.2020.00131
Ali, A. Y. A., Ibrahim, M. E. H., Zhou, G., Nimir, N. E. A., Elsiddig, A. M. I., Jiao, X., et al. (2021). Gibberellic acid and nitrogen efficiently protect early seedlings growth stage from salt stress damage in sorghum. Sci. Rep. 11, 6672. doi: 10.1038/s41598-021-84713-9
Ali, A. Y. A., Ibrahim, M. E. H., Zhou, G., Nimir, N. E. A., Jiao, X., Zhu, G., et al. (2019). Ameliorative effects of jasmonic acid and humic acid on antioxidant enzymes and salt tolerance of forage sorghum under salinity conditions. Agron. J. . 111, 3099–3108. doi: 10.2134/agronj2019.05.0347
Ampong, K., Thilakaranthna, M. S., Gorim, L. Y. (2022). Understanding the role of humic acids on crop performance and soil health. Front. Agron. 4. doi: 10.3389/fagro.2022.848621
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Ashraf, M., Munns, R. (2022). Evolution of approaches to increase the salt tolerance of crops. Crit. Rev. Plant Sci. 41, 128–160. doi: 10.1080/07352689.2022.2065136
Ashraf, M. Y., Rafique, N., Ashraf, M., Azhar, N., Marchand, M. (2013). Effect of supplemental potassium (K+) on growth, physiological and biochemical attributes of wheat grown under saline conditions. J. Plant Nutr. 36, 443–458. doi: 10.1080/01904167.2012.748065
Asif, M. A., Pearson, A. S., Schilling, R. K., Roy, S. J. (2018). Opportunities for developing salt-tolerant wheat and barley varieties. Annu. Rev. Plant Biol. 157-218. doi: 10.1002/9781119312994.apr0681
Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Chao, H., He, J., Cai, Q., Zhao, W., Fu, H., Hua, Y., et al. (2021). The expression characteristics of NPF genes and their response to vernalization and nitrogen deficiency in rapeseed. Int. J. Mol. Sci. 22, 4944. doi: 10.3390/ijms22094944
Chattha, M. U., Hassan, M. U., Khan, I., Chattha, M. B., Aamer, M., Nawaz, M., et al. (2020). Impact of planting methods on biomass production, chemical composition and methane yield of sorghum cultivars. Pak. J. Agric. Sci. 57. doi: 10.21162/PAKJAS/20.7112
Chennakesavulu, K., Singh, H., Trivedi, P. K., Jain, M., Yadav, S. R. (2021). State-of-the-art in CRISPR technology and engineering drought, salinity, and thermo-tolerant crop plants. Plant Cell Rep., 1–17. doi: 10.1007/s00299-021-02681-w
Chourasia, K. N., More, S. J., Kumar, A., Kumar, D., Singh, B., Bhardwaj, V., et al. (2022). Salinity responses and tolerance mechanisms in underground vegetable crops: an integrative review. Planta 255, 68. doi: 10.1007/s00425-022-03845-y
Cui, J., Jiang, N., Zhou, X., Hou, X., Yang, G., Meng, J., et al. (2018). Tomato MYB49 enhances resistance to phytophthora infestans and tolerance to water deficit and salt stress. Planta 248, 1487–1503. doi: 10.1007/s00425-018-2987-6
Denaxa, N.-K., Nomikou, A., Malamos, N., Liveri, E., Roussos, P. A., Papasotiropoulos, V. (2022). Salinity effect on plant growth parameters and fruit bioactive compounds of two strawberry cultivars, coupled with environmental conditions monitoring. Agronomy 12, 2279. doi: 10.3390/agronomy12102279
Dietz, K.-J., Jacob, S., Oelze, M.-L., Laxa, M., Tognetti, V., De Miranda, S. M. N., et al. (2006). The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 57, 1697–1709. doi: 10.1093/jxb/erj160
Du, N., Xue, L., Xue, D., Dong, X., Yang, Q., Jahan, M. S., et al. (2023). The transcription factor SlNAP1 increases salt tolerance by modulating ion homeostasis and ROS metabolism in Solanum lycopersicum. Gene 849, 146906. doi: 10.1016/j.gene.2022.146906
Ekinci, M., Turan, M., Yildirim, E. (2022). Biochar mitigates salt stress by regulating nutrient uptake and antioxidant activity, alleviating the oxidative stress and abscisic acid content in cabbage seedlings. Turk. J. Agric. For. 46, 28–37. doi: 10.3906/tar-2104-81
El-Mageed, T., Mekdad, A. A., Rady, M. O., Abdelbaky, A. S., Saudy, H. S., Shaaban, A. (2022). Physio-biochemical and agronomic changes of two sugar beet cultivars grown in saline soil as influenced by potassium fertilizer. J. Soil Sci. Plant Nutr. 22, 3636–3654. doi: 10.1007/s42729-022-
El Moukhtari, A., Cabassa-Hourton, C., Farissi, M., Savouré, A. (2020). How does proline treatment promote salt stress tolerance during crop plant development? Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01127
Fan, X., Naz, M., Fan, X., Xuan, W., Miller, A. J., Xu, G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68, 2463–2475. doi: 10.1093/jxb/erx011
FAO, F. (2017). “Agriculture organization of the United Nations the future of food and agriculture,” in Trends and challenges (Rome: FAO). doi: 10.1093/jxb/erx011
Faure, S., Malagoli, P., Lainé, P., Le Deunff, E., Ledily, F., Ourry, A., et al. (2021). Modelling nitrogen uptake in winter oilseed rape by using influx kinetics of nitrate transport systems.
Feng, D., Gao, Q., Liu, J., Tang, J., Hua, Z., Sun, X. (2023). Categories of exogenous substances and their effect on alleviation of plant salt stress. Eur. J. Agron. 142, 126656. doi: 10.1016/j.eja.2022.126656
Gobena, D., Shimels, M., Rich, P. J., Ruyter-Spira, C., Bouwmeester, H., Kanuganti, S., et al. (2017). Mutation in sorghum low germination stimulant 1 alters strigolactones and causes striga resistance. Proc. Natl. Acad. Sci. 114, 4471–4476. doi: 10.1073/pnas.1618965114
Gouiaa, S., Khoudi, H. (2015). Co-Expression of vacuolar Na+/H+ antiporter and h+-pyrophosphatase with an IRES-mediated dicistronic vector improves salinity tolerance and enhances potassium biofortification of tomato. Phytochemistry 117, 537–546. doi: 10.1016/j.phytochem.2015.05.016
Gouiaa, S., Khoudi, H., Leidi, E. O., Pardo, J. M., Masmoudi, K. (2012). Expression of wheat Na+/H+ antiporter TNHXS1 and h+-pyrophosphatase TVP1 genes in tobacco from a bicistronic transcriptional unit improves salt tolerance. Plant Mol. Biol. 79, 137–155. doi: 10.1007/s11103-012-9901-6
Hasanuzzaman, M., Bhuyan, M. B., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9, 681. doi: 10.3390/antiox9080681
He, H., He, L.-F. (2023). “Advances in potato breeding for abiotic stress tolerance,” in Smart plant breeding for vegetable crops in post-genomics era (Springer), 383–407. doi: 10.1007/978-981-19-5367-5
He, H., Zhou, W., Lü, H., Liang, B. (2022). Growth, leaf morphological and physiological adaptability of leaf beet (Beta vulgaris var. cicla) to salt stress: a soil culture experiment. Agronomy 12, 1393. doi: 10.3390/agronomy12061393
Hu, Y., Schmidhalter, U. (2023). Opportunity and challenges of phenotyping plant salt tolerance. Trends Plant Sci. doi: 10.1016/j.tplants.2022.12.010
Huang, J., Li, J., Zhou, J., Wang, L., Yang, S., Hurst, L. D., et al. (2018). Identifying a large number of high-yield genes in rice by pedigree analysis, whole-genome sequencing, and CRISPR-Cas9 gene knockout. Proc. Natl. Acad. Sci. 115, E7559–E7567. doi: 10.1073/pnas.1806110115
Hussien Ibrahim, M. E., Adam Ali, A. Y., Zhou, G., Ibrahim Elsiddig, A. M., Zhu, G., Ahmed Nimir, N. E., et al. (2020). Biochar application affects forage sorghum under salinity stress. C Chil. J. Agric. Res. 80, 317–325. doi: 10.4067/S0718-58392020000300317
Irfan, M., Jan, G., Jan, F. G., Murad, W. (2021). Influence of ascorbic acid against salt stress on the germination, biochemicals and micronutrients concentrations of Solanum melongena (L.). doi: 10.20944/preprints202101.0278.v1
Ismail, A. M., Horie, T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
Jiang, X., Zhang, W., Fernie, A. R., Wen, W. (2022). Combining novel technologies with interdisciplinary basic research to enhance horticultural crops. Plant J. 109, 35–46. doi: 10.1111/tpj.15553
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. science 337, 816–821. doi: 10.1126/science.1225829
Kaur, M., Tak, Y., Bhatia, S., Kaur, H. (2023). “Phenolics biosynthesis, targets, and signaling pathways in ameliorating oxidative stress in plants,” in Plant phenolics in abiotic stress management (Springer), 149–171. doi: 10.1007/978-981-19-6426-8_8
Khan, M. I. R., Fatma, M., Per, T. S., Anjum, N. A., Khan, N. A. (2015). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00462
Kotula, L., Garcia Caparros, P., Zörb, C., Colmer, T. D., Flowers, T. J. (2020). Improving crop salt tolerance using transgenic approaches: an update and physiological analysis. Plant Cell Environ. 43, 2932–2956. doi: 10.1111/pce.13865
Krieger, U., Lippman, Z. B., Zamir, D. (2010). The flowering gene single flower truss drives heterosis for yield in tomato. Nat. Genet. 42, 459–463. doi: 10.1038/ng.550
Kumar, S., Kumar, S., Mohapatra, T. (2021). Interaction between macro and micro-nutrients in plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.665583
Lawrenson, T., Harwood, W. A. (2019). Creating targeted gene knockouts in barley using CRISPR/Cas9. Barley: Methods Protoc., 217–232. doi: 10.1007/978-1-4939-8944-7_14
Li, M., Tian, H., Gao, Y. (2021). A genome-wide analysis of NPF and NRT2 transporter gene families in bread wheat provides new insights into the distribution, function, regulation and evolution of nitrate transporters. Plant Soil 465, 47–63. doi: 10.1007/s11104-021-04927-8
Li, X., Zhou, W., Ren, Y., Tian, X., Lv, T., Wang, Z., et al. (2017). High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genomics 44, 175–178. doi: 10.1016/j.jgg.2017.02.001
Liu, Y., Gao, Y., Gao, Y., Zhang, Q. (2019). Targeted deletion of floral development genes in Arabidopsis with CRISPR/Cas9 using the RNA endoribonuclease Csy4 processing system. Hortic. Res. 6. doi: 10.1038/s41438-019-0179-
Liu, K.-H., Huang, C.-Y., Tsay, Y.-F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.1105/tpc.11.5.865
Liu, G., Lin, Q., Jin, S., Gao, C. (2021). The CRISPR-cas toolbox and gene editing technologies. Mol. Cell. doi: 10.1016/j.molcel.2021.12.002
Liu, Y., Shi, Y., Zhu, N., Zhong, S., Bouzayen, M., Li, Z. (2020). SlGRAS4 mediates a novel regulatory pathway promoting chilling tolerance in tomato. Plant Biotechnol. J. 18, 1620–1633. doi: 10.1111/pbi.13328
Liu, S., Zheng, L., Xue, Y., Zhang, Q., Wang, L., Shou, H. (2010). Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. J. Plant Biol. 53, 444–452. doi: 10.1007/s12374-010-9135-6
Lo, S. F., Ho, T. H. D., Liu, Y. L., Jiang, M. J., Hsieh, K. T., Chen, K. T., et al. (2017). Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice. Plant Biotechnol. J. 15, 850–864. doi: 10.1111/pbi.12681
López-Marqués, R. L., Nørrevang, A. F., Ache, P., Moog, M., Visintainer, D., Wendt, T., et al. (2020). Prospects for the accelerated improvement of the resilient crop quinoa. J. Exp. Bot. 71, 5333–5347. doi: 10.1093/jxb/eraa285
Mittal, N., Thakur, S., Verma, H., Kaur, A. (2018). Interactive effect of salinity and ascorbic acid on Brassica rapa l. plants. G.J.B.B 7, 27–29.
Mohammed Ibrahim Elsiddig, A., Zhou, G., Nimir, N. E. A., Yousif Adam Ali, A. (2022). Effect of exogenous ascorbic acid on two sorghum varieties under different types of salt stress. Chil. J. Agric. Res. 82, 10–20. doi: 10.4067/S0718-58392022000100010
Negi, P., Pandey, M., Dorn, K. M., Nikam, A. A., Devarumath, R. M., Srivastava, A. K., et al. (2020). Transcriptional reprogramming and enhanced photosynthesis drive inducible salt tolerance in sugarcane mutant line M4209. J. Exp. Bot. 71, 6159–6173. doi: 10.1093/jxb/eraa339
Netondo, G. W., Onyango, J. C., Beck, E. (2004). Sorghum and salinity: II. gas exchange and chlorophyll fluorescence of sorghum under salt stress. Crop Sci. 44, 806–811. doi: 10.2135/cropsci2004.8060
Nounjan, N., Chansongkrow, P., Charoensawan, V., Siangliw, J. L., Toojinda, T., Chadchawan, S., et al. (2018). High performance of photosynthesis and osmotic adjustment are associated with salt tolerance ability in rice carrying drought tolerance QTL: physiological and co-expression network analysis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01135
Pamuta, D., Siangliw, M., Sanitchon, J., Pengrat, J., Siangliw, J. L., Toojinda, T., et al. (2022). Physio-biochemical traits in improved ‘KDML105’jasmine rice lines containing drought and salt tolerance gene under drought and salt stress. Chil. J. Agric. Res. 82, 97–110. doi: 10.4067/S0718-58392022000100097
Panagea, I., Daliakopoulos, I., Tsanis, I., Schwilch, G. (2016). Evaluation of promising technologies for soil salinity amelioration in timpaki (Crete): a participatory approach. Solid Earth 7, 177–190. doi: 10.5194/se-7-177-2016
Parida, A. K., Das, A. B. (2005). Salt tolerance and salinity effects on plants: a review. Ecotoxicol. Environ. Saf. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Pu, Y., Liu, L., Wu, J., Zhao, Y., Bai, J., Ma, L., et al. (2019). Transcriptome profile analysis of winter rapeseed (Brassica napus l.) in response to freezing stress, reveal potentially connected events to freezing stress. Int. J. Mol. Sci. 20, 2771. doi: 10.3390/ijms20112771
Raza, A., Tabassum, J., Fakhar, A. Z., Sharif, R., Chen, H., Zhang, C., et al. (2022). Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol., 1–28. doi: 10.1080/07388551.2022.2093695
Razzaq, A., Saleem, F., Kanwal, M., Mustafa, G., Yousaf, S., Imran Arshad, H. M., et al. (2019). Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci. 20, 4045. doi: 10.3390/ijms20164045
Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi: 10.1111/pbi.12837
Saudi, A. H. (2017). Effect of seed size, plant growth regulators and some chemical materials on germination charactristics and seedling vigour of rice (Oryza sativa l.) seeds. D.A.S.J 9, 91–106.
Shahbaz, M., Ashraf, M., Akram, N. A., Hanif, A., Hameed, S., Joham, S., et al. (2011). Salt-induced modulation in growth, photosynthetic capacity, proline content and ion accumulation in sunflower (Helianthus annuus l.). Acta Physiol. Plant 33, 1113–1122. doi: 10.1007/s11738-010-0639-y
Shahzad, B., Rehman, A., Tanveer, M., Wang, L., Park, S. K., Ali, A. (2021). Salt stress in brassica: effects, tolerance mechanisms, and management. J. Plant Growth Regul., 1–15. doi: 10.1007/s00344-021-10338-x
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Sharma, K., Kumar, M., Chandran, D. (2023). “Ameliorative effects of phenolics in oxidative stress management in plants,” in Plant phenolics in abiotic stress management (Springer), 369–390. doi: 10.1007/978-981-19-6426-8_17
Sharma, H., Sharma, A., Sidhu, S., Upadhyay, S. K. (2022). “Na+/H+ antiporter (NHX) and salt stress tolerance,” in Cation transporters in plants (Elsevier), 99–113. doi: 10.1016/B978-0-323-85790-1.00008-7
Shihab, M. O., Hamza, J. H. (2020). Seed priming of sorghum cultivars by gibberellic and salicylic acids to improve seedling growth under irrigation with saline water. J. Plant Nutr. 43, 1951–1967. doi: 10.1080/01904167.2020.1766066
Shiyab, S. M., Shatnawi, M. A., Shibli, R. A., Al Smeirat, N. G., Ayad, J., Akash, M. W. (2013). Growth, nutrient acquisition, and physiological responses of hydroponic grown tomato to sodium chloride salt induced stress. J. Plant Nutr. 36, 665–676. doi: 10.1080/01904167.2012.754037
Singh, A., Jones, S., Ganapathysubramanian, B., Sarkar, S., Mueller, D., Sandhu, K., et al. (2021). Challenges and opportunities in machine-augmented plant stress phenotyping. Trends Plant Sci. 26, 53–69. doi: 10.1016/j.tplants.2020.07.010
Singh, Y., Mishra, V., Singh, S., Sharma, D., Singh, D., Singh, U., et al. (2016). Productivity of sodic soils can be enhanced through the use of salt tolerant rice varieties and proper agronomic practices. Field Crops Res. 190, 82–90. doi: 10.1016/j.fcr.2016.02.007
Song, Y., Yang, W., Fan, H., Zhang, X., Sui, N. (2020). TaMYB86B encodes a R2R3-type MYB transcription factor and enhances salt tolerance in wheat. Plant Sci. 300, 110624. doi: 10.1016/j.plantsci.2020.110624
Springmann, M., Clark, M., Mason-D’croz, D., Wiebe, K., Bodirsky, B. L., Lassaletta, L., et al. (2018). Options for keeping the food system within environmental limits. Nature 562, 519–525. doi: 10.1038/s41586-018-0594-0
Taha, R., Seleiman, M., Alotaibi, M., Alhammad, B. A., Rady, M., Mahdi, A. (2020). Exogenous potassium treatments elevate salt tolerance and performances of Glycine max l. by boosting antioxidant defense system under actual saline field conditions. Agronomy 10, 1741. doi: 10.3390/agronomy10111741
Tang, Y., Du, G., Xiang, J., Hu, C., Li, X., Wang, W., et al. (2022). Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea l.). Genomics 114, 171–184. doi: 10.1016/j.ygeno.2021.12.015
Thomson, M. J., Zhao, K., Wright, M., Mcnally, K. L., Rey, J., Tung, C.-W., et al. (2012). High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol. Breed. 29, 875–886. doi: 10.1007/s11032-011-9663-x
Tran, M. T., Doan, D. T. H., Kim, J., Song, Y. J., Sung, Y. W., Das, S., et al. (2021). CRISPR/Cas9-based precise excision of SlHyPRP1 domain (s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 40, 999–1011. doi: 10.1007/s00299-020-02622-z
Turan, M. A., Elkarim, A. H. A., Taban, N., Taban, S. (2009). Effect of salt stress on growth, stomatal resistance, proline and chlorophyll concentrations on maize plant. Afr. J. Agric. Res. 4, 893–897.
Türkan, I., Demiral, T. (2009). Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 67, 2–9. doi: 10.1016/j.envexpbot.2009.05.008
Umnajkitikorn, K., Faiyue, B., Saengnil, K. (2013). Enhancing antioxidant properties of germinated Thai rice (Oryza sativa l.) cv. kum doi saket with salinity. Rice Res. Open Access. doi: 10.4172/jrr.1000103
Van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Verma, T., Bhardwaj, S., Singh, J., Kapoor, D., Prasad, R. (2022). Triacontanol as a versatile plant growth regulator in overcoming negative effects of salt stress. J. Agric. Food Inf. 100351. doi: 10.1016/j.jafr.2022.100351
Wang, M., Li, G., Feng, Z., Liu, Y., Yuan, X., Uscola, M. (2023). A wider spectrum of avoidance and tolerance mechanisms explained ozone sensitivity of two white poplar ploidy levels. Ann. Bot. mcad019. doi: 10.1093/aob/mcad019
Wang, M., Mao, Y., Lu, Y., Tao, X., Zhu, J. K. (2017). Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 10, 1011–1013.
Wang, W., Pang, J., Zhang, F., Sun, L., Yang, L., Fu, T., et al. (2022). Salt−responsive transcriptome analysis of canola roots reveals candidate genes involved in the key metabolic pathway in response to salt stress. Sci. Rep. 12, 1666. doi: 10.1038/s41598-022-05700-2
Wang, Y.-H., Zhang, G., Chen, Y., Gao, J., Sun, Y.-R., Sun, M. F., et al. (2019). Exogenous application of gibberellic acid and ascorbic acid improved tolerance of okra seedlings to NaCl stress. Acta Physiol. Plant 41, 1–10. doi: 10.1007/s11738-019-2869-y
Wu, D., Liang, Z., Yan, T., Xu, Y., Xuan, L., Tang, J., et al. (2019). Whole-genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol. Plant 12, 30–43. doi: 10.1016/j.molp.2018.11.007
Xia, X., Cheng, X., Li, R., Yao, J., Li, Z., Cheng, Y. (2021). Advances in application of genome editing in tomato and recent development of genome editing technology. Theor. Appl. Genet. 134, 2727–2747. doi: 10.1007/s00122-021-03874-3
Xie, X., He, Z., Chen, N., Tang, Z., Wang, Q., Cai, Y. (2019). The roles of environmental factors in regulation of oxidative stress in plant. BioMed. Res. Int. 2019. doi: 10.1155/2019/9732325
Yan, H., Jia, H., Chen, X., Hao, L., An, H., Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yu, W., Wang, L., Zhao, R., Sheng, J., Zhang, S., Li, R., et al. (2019). Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 19, 1–13. doi: 10.1186/s12870-019-1939-z
Zafar, S., Perveen, S., Kamran Khan, M., Shaheen, M. R., Hussain, R., Sarwar, N., et al. (2022). Effect of zinc nanoparticles seed priming and foliar application on the growth and physio-biochemical indices of spinach (Spinacia oleracea l.) under salt stress. PloS One 17, e0263194. doi: 10.1371/journal.pone.0263194
Zeeshan, M., Lu, M., Sehar, S., Holford, P., Wu, F. (2020). Comparison of biochemical, anatomical, morphological, and physiological responses to salinity stress in wheat and barley genotypes deferring in salinity tolerance. Agronomy 10, 127. doi: 10.3390/agronomy10010127
Zhang, Z., Hua, L., Gupta, A., Tricoli, D., Edwards, K. J., Yang, B., et al. (2019). Development of an agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 17, 1623–1635. doi: 10.1111/pbi.13088
Zhang, Y., Li, D., Zhang, D., Zhao, X., Cao, X., Dong, L., et al. (2018). Analysis of the functions of Ta GW 2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. doi: 10.1111/tpj.13903
Zhang, P., Wang, R., Yang, X., Ju, Q., Li, W., Lü, S., et al. (2020). The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 43, 1925–1943. doi: 10.1111/pce.13784
Zhang, H., Wu, X., Li, G., Qin, P. (2011). Interactions between arbuscular mycorrhizal fungi and phosphate-solubilizing fungus (Mortierella sp.) and their effects on kostelelzkya virginica growth and enzyme activities of rhizosphere and bulk soils at different salinities. Biol. Fertil. Soils 47, 543–554. doi: 10.1007/s00374-011-0563-3
Zheng, M., Terzaghi, W., Wang, H., Hua, W. (2022). Integrated strategies for increasing rapeseed yield. Trends Plant Sci. doi: 10.1016/j.tplants.2022.03.008
Zheng, M., Zhang, L., Tang, M., Liu, J., Liu, H., Yang, H., et al. (2020). Knockout of two bna MAX 1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus l.). Plant Biotechnol. J. 18, 644–654. doi: 10.1111/pbi.13228
Zörb, C., Geilfus, C. M., Dietz, K. J. (2019). Salinity and crop yield. Plant Biol. 21, 31–38. doi: 10.1111/plb.12884
Zrig, A., Abdelgawad, H., Touneckti, T., Mohamed, H. B., Hamouda, F., Khemira, H. (2021). Potassium and calcium improve salt tolerance of thymus vulgaris by activating the antioxidant systems. Sci. Hortic. 277, 109812. doi: 10.1016/j.scienta.2020.109812
Keywords: salt stress, morpho-physiological and biochemical activity, nutrient uptake, CRSIPER-Cas9, genes, yield
Citation: Ahmad I, Zhu G, Zhou G, Younas MU, Suliman MSE, Liu J, Zhu Ym and Salih EGI (2023) Integrated approaches for increasing plant yield under salt stress. Front. Plant Sci. 14:1215343. doi: 10.3389/fpls.2023.1215343
Received: 01 May 2023; Accepted: 28 June 2023;
Published: 18 July 2023.
Edited by:
Muhammad Nawaz, Khwaja Fareed University of Engineering and Information Technology (KFUEIT), PakistanReviewed by:
Ravinder Kumar, Central Potato Research Institute (ICAR), IndiaDepeng Wang, Linyi University, China
Shahbaz Atta Tung, Pir Mehr Ali Shah Arid Agriculture University, Pakistan
Copyright © 2023 Ahmad, Zhu, Zhou, Younas, Suliman, Liu, Zhu and Salih. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guisheng Zhou, Z3N6aG91QHl6dS5lZHUuY24=; Guanglong Zhu, Zy56aHVAeXp1LmVkdS5jbg==
 Irshad Ahmad
Irshad Ahmad Guanglong Zhu1*
Guanglong Zhu1* Guisheng Zhou
Guisheng Zhou