
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 June 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1215084
This article is part of the Research TopicIdentification of Genes Involved in Environmental Adaption and Yield Traits in Forage and Other Ecologically Important PlantsView all 10 articles
Abiotic stresses have deleterious effects on seed germination and seedling establishment, leading to significant crop yield losses. Adverse environmental conditions can cause the accumulation of methylglyoxal (MG) within plant cells, which can negatively impact plant growth and development. The glyoxalase system, which consists of the glutathione (GSH)-dependent enzymes glyoxalase I (GLX1) and glyoxalase II (GLX2), as well as the GSH-independent glyoxalase III (GLX3 or DJ-1), plays a crucial role in detoxifying MG. However, genome-wide analysis of glyoxalase genes has not been performed for one of the agricultural important species, oat (Avena sativa). This study identified a total of 26 AsGLX1 genes, including 8 genes encoding Ni2+-dependent GLX1s and 2 genes encoding Zn2+-dependent GLX1s. Additionally, 14 AsGLX2 genes were identified, of which 3 genes encoded proteins with both lactamase B and hydroxyacylglutathione hydrolase C-terminal domains and potential catalytic activity, and 15 AsGLX3 genes encoding proteins containing double DJ-1 domains. The domain architecture of the three gene families strongly correlates with the clades observed in the phylogenetic trees. The AsGLX1, AsGLX2, and AsGLX3 genes were evenly distributed in the A, C, and D subgenomes, and gene duplication of AsGLX1 and AsGLX3 genes resulted from tandem duplications. Besides the core cis-elements, hormone responsive elements dominated the promoter regions of the glyoxalase genes, and stress responsive elements were also frequently observed. The subcellular localization of glyoxalases was predicted to be primarily in the cytoplasm, chloroplasts, and mitochondria, with a few presents in the nucleus, which is consistent with their tissue-specific expression. The highest expression levels were observed in leaves and seeds, indicating that these genes may play important roles in maintaining leaf function and ensuring seed vigor. Moreover, based on in silico predication and expression pattern analysis, AsGLX1-7A, AsGLX2-5D, AsDJ-1-5D, AsGLX1-3D2, and AsGLX1-2A were suggested as promising candidate genes for improving stress resistance or seed vigor in oat. Overall, the identification and analysis of the glyoxalase gene families in this study can provide new strategies for improving oat stress resistance and seed vigor.
Reactive dicarbonyl compounds, including methylglyoxal (MG), glyoxal (GO), and 3-deoxyglucosone (3-DG), are the main toxic metabolites spontaneously produced by various metabolic pathways when plants are exposed to stress, and these compounds can hinder normal growth and development (Chinchansure et al., 2015; Ramu et al., 2020). MG, which is the most prevalent reactive dicarbonyl compound, is a by-product of nonenzymatic reactions through glycolysis and the Calvin cycle, and enzymatic pathways through proteins and fatty acid metabolism (Kaur et al., 2014; Takagi et al., 2014). High concentrations of MG can be cytotoxic to cells as they spontaneously form advanced glycation end-products (AGEs) when interacting with nucleic acids, proteins, and lipids (Kaur et al., 2014; Singla-Pareek et al., 2020). However, despite its potential cytotoxicity at high concentrations, MG also serves as a vital signaling molecule involved in various biological processes (Singla-Pareek et al., 2020). Therefore, the maintenance of MG homeostasis is crucial for the normal growth and development of plants.
Plants maintain MG homeostasis through the glyoxalase system and non-glyoxalase system. The former primarily comprises three enzymes, namely, glyoxalase I (GLX1, GLXI or GLYI), also known as lactoylglutathione lyase (EC 4.4.1.5); glyoxalase II (GLX2, GLXII or GLYII), also known as hydroxyacylglutathione hydrolase (EC 3.1.2.6); and glyoxalase III (GLX3, GLYIII or DJ-1)(Li, 2016). GSH spontaneously reacts with MG to form hemithioacetal (HTA), which is then converted to S-D-lactoylglutathione (SLG) by GLX1. GLX2 hydrolyses SLG to produce D-lactate and GSH, which is then recycled back into the system (Thornalley, 1990). The functions of these two GSH-dependent glyoxalases have been extensively explored in plants and reviewed in terms of MG detoxification, cell aging, signal transduction, cell division and differentiation, starch synthesis, pollination, nutrient response, and stress response (Singla-Pareek et al., 2020). Among these, stress response is considered their primary function (Kaur et al., 2014; Hasanuzzaman et al., 2017). The expression and activity of GLX1 and GLX2 vary significantly under various stress conditions, such as hypoxia, salt, drought, heat, cold, and heavy metal (Kaur et al., 2014; Sankaranarayanan et al., 2017). GLX3, a novel glyoxalase enzyme of the DJ-I protein family, directly catalyzes the conversion of MG to D-lactate (Ghosh et al., 2016). Only a few studies have investigated GLX3 in model plants and cereal crops, demonstrating its participation in stress responses (Jana et al., 2021; Kumar et al., 2021; Gambhir et al., 2023). Recent studies have shown that GLX1 and GLX2 are also involved in regulating seed vigor in Oryza sativa and Arabidopsis (Schmitz et al., 2017; Liu et al., 2022), but their role in seed development, storage, and germination is still poorly understood.
The identification and functional analysis of the glyoxalase gene family members have gained significant attention due to the diverse functions of glyoxalase enzymes. Although GLX1 and GLX2 genes have been genome-wide identified in model plants and important crops, such as Arabidopsis (Mustafiz et al., 2011), O. sativa (Mustafiz et al., 2011), Sorghum bicolor (Bhowal et al., 2020), Glycine max (Ghosh and Islam, 2016), and Brassica rapa (Yan et al., 2018), the genome-wide identification on GLX3 is limited (Li et al., 2019; Jana et al., 2021; Yan et al., 2023). Notably, both GLX1 and GLX2 genes in these species have multiple members with diverse subcellular localizations (Li, 2016). GLX1s are classified into two types: Ni2+-dependent and Zn2+-dependent (Schmitz et al., 2018), while GLX2s belong to the beta-lactamase protein family and have a binuclear metal center consisting of Fe3+, Zn2+, and Mn2+ (Schilling et al., 2003). GLX3s belong to the DJ-1/PfpI superfamily and do not require metal ions for their optimal activity (Ghosh et al., 2016). The identification and analysis of GLX1, GLX2, and GLX3 genes are crucial for comprehending the regulation of plant growth, development, and stress response.
Oat (A. sativa) is an important cereal and feed crop that has recently had its genome data released (Rasane et al., 2015; Kamal et al., 2022; Peng et al., 2022), but has not yet been extensively studied in terms of the GLX1, GLX2, and GLX3 genes. Therefore, the identification and expression analysis of these genes at the whole-genome level will help to rapidly advance stress-resistant breeding and seed vigor improvement in oat. In our previous research, we discovered a close association between the GSH-dependent glyoxalases and the AsA-GSH cycle with oat seed vigor, where GSH content serves as a potential marker for seed germination percentage (Sun et al., 2022b). We also conducted a genome-wide identification of the glutathione reductase (GR) genes involved in the AsA-GSH cycle and analyzed its expression pattern during seed germination under stress conditions (Sun et al., 2022a). The objective of this study is to identify the GLX1, GLX2, and GLX3 genes in oat at the whole-genome level, and to conduct chromosome mapping, phylogenetic analysis, synteny analysis, conserved domain analysis, cis-regulatory element and subcellular localization prediction, and tissue-specific analysis. In addition, the role of these members in seed germination under stress conditions will be analyzed using qPCR. This comprehensive analysis of the glyoxalase gene families in oat will significantly contribute to the genetic enhancement of stress resistance and seed vigor in this essential crop.
The HMM (Hidden Markov Model) files for the conserved domains of GLX1, GLX2, and GLX3 proteins, including conserved glyoxalase domain (PF00903), metallo-beta-lactamase domain (PF00753), and DJ-1/PfpI domain (PF01965), were obtained from the Pfam database (Mustafiz et al., 2011; Ghosh et al., 2016; Bhowal et al., 2020). The Simple HMM Search module in TBtools software was used to perform the alignment of HMM files to the oat genome (PepsiCo_OT3098_v2_genome, https://wheat.pw.usda.gov/GG3/) to initially identify oat GLX1, GLX2, and GLX3 genes. The protein sequences were then submitted to the Pfam website for individual confirmation of their conserved domains, and ultimately determine the members of the GLX1, GLX2, and GLX3 families.
The TBtools software was also used to visualize the chromosome distribution of oat GLX1, GLX2, and GLX3 genes (Chen et al., 2020). The nomenclature of oat GLX1, GLX2, and GLX3 genes followed the international wheat gene nomenclature rules (http://wheat.pw.usda.gov/ggpages/wgc/98/intro.htm), which could reflect the chromosomal locations of these genes in the oat subgenomes.
The identified AsGLX1, AsGLX2, and AsGLX3 members were aligned with glyoxalase members from Arabidopsis and O. sativa using ClustalX (v2.1) with default settings (Larkin et al., 2007). The protein sequences of O. sativa and Arabidopsis GLX1s, GLX2s, and GLX3s used for phylogenetic analysis were sourced from previously published genome-wide identification studies (Mustafiz et al., 2011; Ghosh et al., 2016). The phylogenetic tree was constructed using the neighbor-joining method in MEGA 6.0, with 10,000 bootstrap tests and support values expressed as percentages based on 1000 replications (Tamura et al., 2013). Gene duplication events were analyzed using the Multiple Collinearity Scan toolkit (MCScanX) with default parameters (Wang et al., 2012). To illustrate the interspecies syntenic relationships of GLX1, GLX2, and GLX3 genes between oat, Arabidopsis, and O. sativa, a synteny analysis plot was constructed using Dual Synteny Plotter for MC ScanX in TBtools (Chen et al., 2020).
The online program ExPaSy-ProtParam (https://web.expasy.org/protparam/) was utilized to analyze the physical and chemical properties of AsGLX1, AsGLX2, and AsGLX3 proteins, including the amino acid number (AA), molecular weight (MW), and theoretical isoelectric point (pI), instability index (II), negatively charged residues (NCRs), and positively charged residues (PCRs). Protein subcellular localization was predicted using online tools, including WoLF PSORT (https://wolfpsort.hgc.jp/), CELLov.2.5 (http://CELLO.life.nctu.edu.tw/), and Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
The TBtools software was used to analyze the protein domains of AsGLX1s, AsGLX2s, and AsGLX3s through HMM files alignment (Chen et al., 2020).
To comprehensively investigate the potential response of the AsGLX1, AsGLX2, and AsGLX3 genes to stress, we extracted a putative promoter region of 2 kb upstream of the genes from the oat genome sequence and identified cis-regulatory elements using the PlantCARE online tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). It may be helpful to further explore the transcriptional regulation mechanism of the glyoxalase genes and reveal its roles in plant development and stress response.
To perform tissue-specific expression profiling of AsGLX1, AsGLX2, and AsGLX3 genes, oat (cv Challenger) samples were collected from seeds, roots, leaves, stems, florets, and lemmas. The seed samples were collected during the imbibition phase (0 h, 12 h, and 24 h) and the development phase (8, 15, and 30 days after flowering, DAF). At the flowering stage, young leaves, lemmas, and florets were collected, while old leaves were obtained from plants at 30 DAF. The roots used in the study were from 10-day-old plants. All seeds used in the study had a germination percentage of 100%.
Seed germination was conducted in a plant growth chamber using plastic petri dishes (11.5 cm × 11.5 cm) containing three layers of filter paper and 50 seeds under a 16-h dark and 8-h light cycle according to the guidelines of the International Seed Testing Association (ISTA, 2019). Normal seed imbibed in distilled water at 20°C was used as the control (CK), while salt, drought, and MG stress treatments employed solutions of 150 mM NaCl (Xu et al., 2021), 20% PEG6000 (Xie et al., 2021), and 10 mM MG (Hoque et al., 2012), respectively. Cold treatment was conducted by imbibing seeds in distilled water at 10°C. Seeds aged for 30 days were also imbibed in distilled water at 20°C. The seed aging treatment was based on the method described by Xia et al. (Xia et al., 2020). Seed samples were collected after 0 h, 6 h, 12 h, 24 h, 36 h, and 72 h of imbibition, with each treatment consisting of three biological replicates, and 20 seeds collected as one replicate.
The total RNA was extracted from oat tissue samples using the Quick RNA isolation Kit (Huayueyang Biotech Co., Ltd., China). The first-strand cDNA was synthesized from 1 μg of RNA using the EasyScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR Kit (TransGen Biotech, China). The qRT-PCR was performed on a CFX96 Real-Time System using 2×RealStar Fast SYBR qPCR Mix (Genstar, China), with AsEIF4A as the reference gene (Yang et al., 2020). The thermal cycle program was as follows: an initial step at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The relative expression level was determined using the 2-ΔΔCt method.
The comparison of the relative expression levels among different tissues was analyzed using ANOVA and a Duncan’s test in SPSS Statistics 22 and was visualized using GraphPad Prism version 8.0. The expression heatmaps for AsGLX1, AsGLX2, and AsGLX3 genes during seed germination under stress were created using TBtools. The significant expression changes were calculated using Student’s t test. The details of the primers used in the qRT-PCR assay are listed in Table S1.
Through whole genome alignment in oat, 26 AsGLX1 genes, 14 AsGLX2 genes, and 15 AsGLX3 genes were identified. Chromosomal mapping showed all 26 AsGLX1 genes were evenly distributed in the A, C, and D subgenomes, with 10 in the A subgenome and 8 in both the C and D subgenomes. AsGLX1 genes in chromosome set 1 had 9 genes, but only one gene on chromosome set 5. As for 14 AsGLX2 genes, they were also evenly distributed across the subgenomes, with 5 in both the A and C subgenomes, and 4 in the D subgenome. AsGLX2 genes were only present once on each chromosome. Regarding AsGLX3 genes, 5 genes were located in the A subgenome, 4 in the C subgenome, and 6 in the D subgenome. AsGLX3 genes were not present in chromosome sets 1 and 2, but were highly represented in chromosome sets 3 and 7, with two genes located on each of 3A, 3C, 3D, 7A, and 7D chromosomes. Additionally, some genes of the GLX1 and GLX3 families in oat are adjacent on chromosomes (Figure 1).
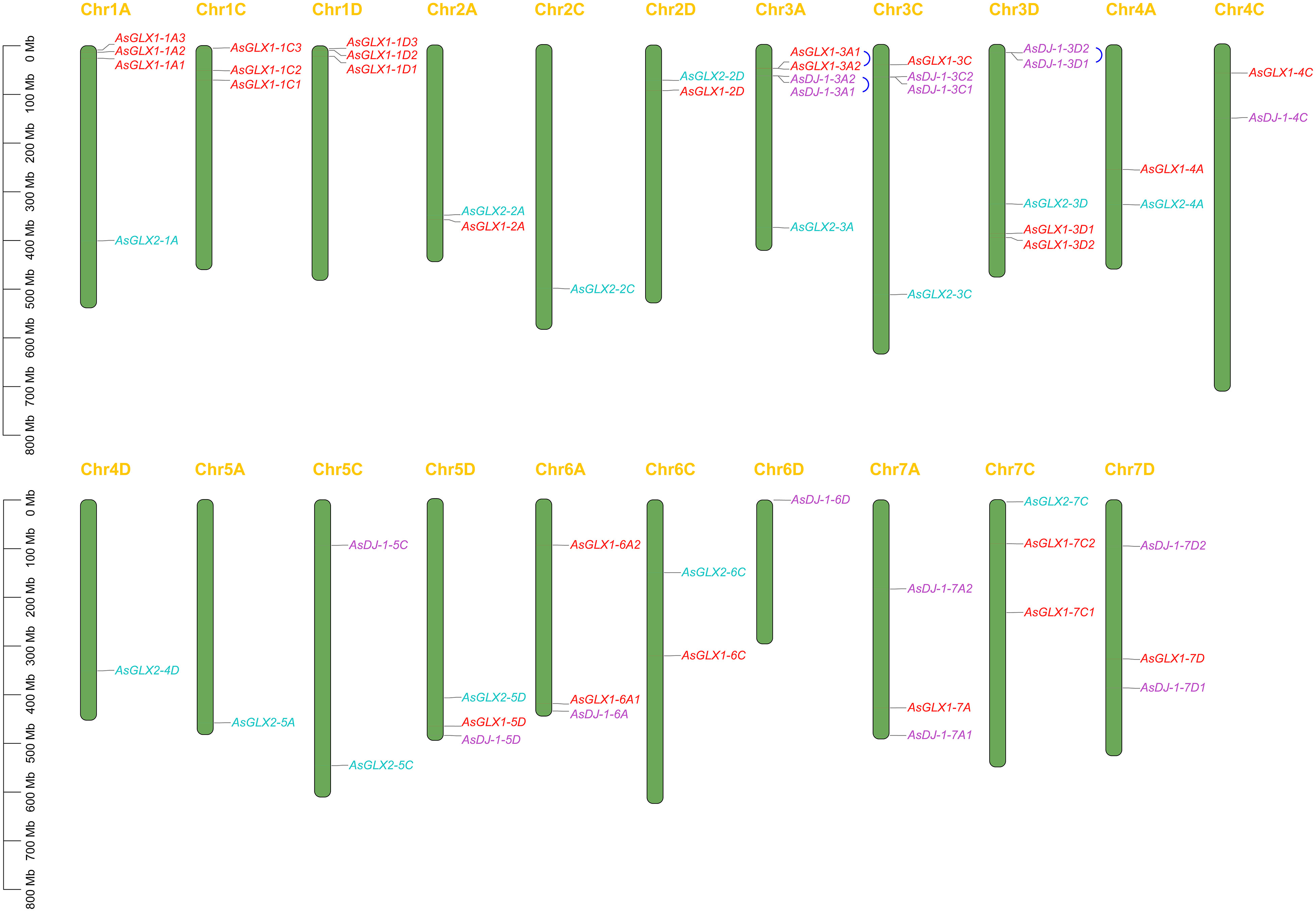
Figure 1 The chromosomal mapping of AsGLX1, AsGLX2, and AsGLX3 genes across oat chromosomes. The blue connecting lines represent tandem duplications. The chromosome number is indicated at the top of each chromosome and the scale is shown in megabases (Mb).
To elucidate the phylogenetic relationships among members of the oat GLX families, amino acid-based phylogenetic trees were constructed using the sequences of GLX1s, GLX2s, and GLX3s from oat, Arabidopsis, and O. sativa (Figure 2). Notably, the number of GLX1 members exceeded that of GLX2 and GLX3 in A. sativa, O. sativa, and Arabidopsis. The 26 AsGLX1s were categorized into two clades. Clade I consisted of 11 AsGLX1s, 4 AtGLX1s, and 5 OsGLX1s, while Clade II contained 16 AsGLX1s, 7 AtGLX1s, and 6 OsGLX1s (Figure 2A). The 14 AsGLX2s were classified into four clades, with Clade I and Clade II exclusively containing AsGLX2s, with 2 and 6 members, respectively. Clade III consisted of 1 AtGLX2, 1 OsGLX2s, and 3 AsGLX2s, while Clade IV comprised 3 AsGLX2s, 4 AtGLX2s, and 2 OsGLX2s (Figure 2B). Additionally, the 15 AsGLX3 (AsDJ-1) members were also categorized into four clades, with Clade I containing 3 AtDJ-1s, 2 OsDJ-1s, and 3 AsDJ-1s. Clade II included 1 OsDJ-1 and 3 AsDJ-1s, and Clade III consisted of 1 AtDJ-1s, 1 OsDJ-1s, and 3 AsDJ-1s. Clade IV has the largest number of AtDJ-1 members, including 2 AtDJ-1s, 2 OsDJ-1s, and 6 AsDJ-1s (Figure 2C).
Gene duplication analysis revealed that certain gene pairs of the GLX1 and GLX3 gene families, namely AsGLX1-3A1 and AsGLX1-3A2, AsDJ-1-3A1 and AsDJ-1-3A2, and AsDJ-1-3D1 and AsDJ-1-3D2, were the result of tandem duplications, and they were adjacent on the chromosomes. However, no evidence of segmental duplications was found among the genes of the GLX1, GLX2, and GLX3 families (Figure 1).
The synteny analysis between oat and rice, as well as Arabidopsis, revealed that there is no collinearity between oat and Arabidopsis in the glyoxalase gene families. However, collinearity was observed between oat and rice in the GLX1, GLX2, and GLX3 gene families. Specifically, 18 AsGLX1 genes showed collinearity with 8 O. sativa genes, 9 AsGLX2 genes showed collinearity with 4 O. sativa genes, and 8 AsDJ-1 genes showed collinearity with 3 O. sativa genes (Figures 3A–C). Among these collinear gene pairs, OsGLYI6 has the highest number of homologous genes in oat, with six homologous genes (AsGLX1-1A3, AsGLX1-1C3, AsGLX1-1D3, AsGLX1-3A1, AsGLX1-3C, and AsGLX1-3D1). The second highest is OsGLYI1, which has five oat homologous genes (AsGLX1-1A3, AsGLX1-1D3, AsGLX1-3A1, AsGLX1-3C, and AsGLX1-3D1). OsGLYII3 has three oat homologous genes including AsGLX2-5A, AsGLX2-5C, and AsGLX2-5D. Similarly, OsDJ-1A has three homologous genes (AsDJ-1-3A2, AsDJ-1-3C2, and AsDJ-1-3D2), and OsDJ-1E also has three oat homologous genes (AsDJ-1-4C, AsDJ-1-7A1, and AsDJ-1-7D1).
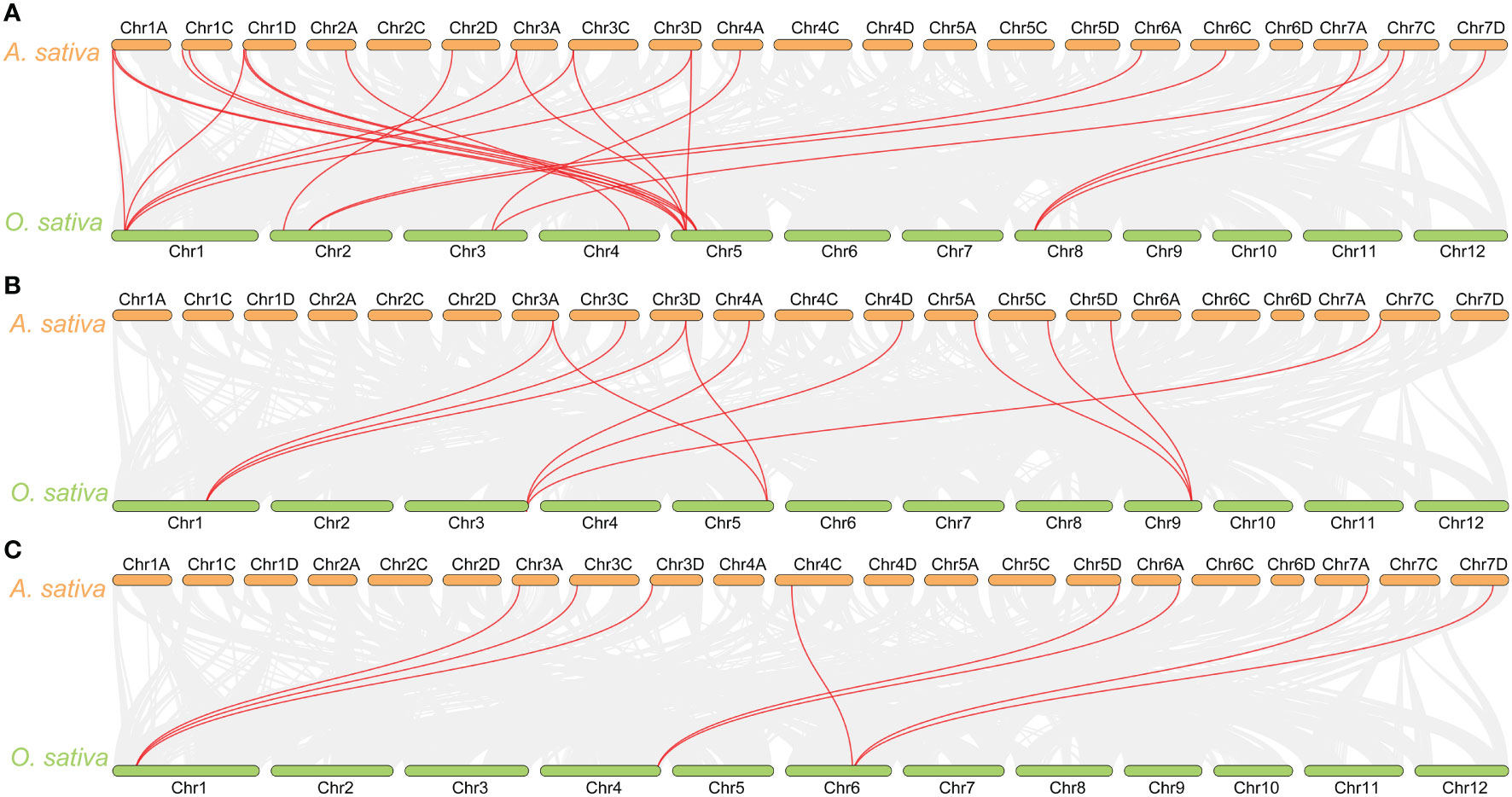
Figure 3 Synteny analysis of GLX1 (A), GLX2 (B), and GLX3 (C) genes between A. sativa and O. sativa. The gray lines show colinear blocks in the genomes of A. sativa and O. sativa, while the red line highlights the colinear GLX1, GLX2, and GLX3 gene pairs.
The online subcellular localization prediction of AsGLX1, AsGLX2, and AsGLX3 genes showed that these genes are primarily expressed in the cytoplasm, chloroplasts, and mitochondria (Table S2). Based on the predicted results of two or three online servers, 13 genes of the AsGLX1 family were found to be expressed in the cytoplasm, 17 in chloroplasts, and 4 in mitochondria. While, 5 genes of the AsGLX2 family were found to be expressed in the cytoplasm, 8 in chloroplasts, and 7 in mitochondria. Furthermore, 5 genes of the AsGLX3 family were found to be expressed in the cytoplasm, 13 in chloroplasts, and 3 in mitochondria. In addition, AsGLX1-2A was predicted by all three online servers to be expressed in the nucleus. Other members, including 6 AsGLX1s, 6 AsGLX2s, and 4 AsGLX3s, predicted by a single program to potentially be expressed in the nucleus, suggest that these members are important for maintaining nuclear stability.
Through the calculation of amino acid length, molecular weight, theoretical isoelectric points, instability index, and charged residues of AsGLX1, AsGLX2, and AsGLX3 proteins, it was found that among the 26 AsGLX1 members, the molecular weight ranged from 15.219 to 94.136 kDa, the amino acid length ranged from 140 AA to 834 AA, and the pI ranged from 4.81 to 8.98. Among them, 12 proteins had an instability index of less than 40, and 20 proteins were rich in negatively charged residues. For the 14 members of AsGLX2, the molecular weight ranged from 32.495 to 143.432 kDa, the amino acid length ranged from 297 AA to 1300 AA, and the pI ranged from 5.87 to 9.12. Among them, 4 proteins had an instability index of less than 40, and 11 proteins were rich in negatively charged residues. As for the 15 members of AsGLX3, the molecular weight ranged from 41.235 to 76.399 kDa, the amino acid length ranged from 395 AA to 723 AA, and the pI ranged from 4.90 to 9.26. Among them, 5 proteins had an instability index of less than 40, and 11 proteins were rich in negatively charged residues (Table S2).
Conserved domain analysis of glyoxalase families showed that all AsGLX1s possess conserved glyoxalase domain, AsGLX2s possess conserved metallo-β-lactamase domain, and AsGLX3s possess two conserved DJ-1/PfpI domains (Figure 4). Among AsGLX1 members, AsGLX1-1A1, AsGLX1-1C1, AsGLX1-1D1, AsGLX1-6C, AsGLX1-6A2, AsGLX1-7A, AsGLX1-7C1, and AsGLX1-7D possess two conserved glyoxalase domains, with the first domain consisting of 120 aa and the second domain consisting of 115 aa in AsGLX1-1A1, AsGLX1-1C1, AsGLX1-1D1, AsGLX1-6C, and AsGLX1-6A2, while the first domain is 121 aa and the second domain is 120 aa in AsGLX1-7A, AsGLX1-7C1, and AsGLX1-7D. These eight members, along with AtGLYI-3, AtGLYI-6, OsGLYI2, OsGLYI7, and OsGLYI11, belong to Clade I and are Ni2+-dependent GLX1s. AsGLX1-5D and AsGLX1-6A1 contain only one glyoxalase domain, with a length of 141 aa, and belong to the same branch in Clade I as AtGLYI-2 and OsGLYI8, indicating that they are Zn2+-dependent GLX1s. AsGLX1-1A2, AsGLX1-1D2, and AsGLX1-1C2, which have a domain length of 115 aa, belong to Clade I as well but are not Ni2+-dependent GLX1s. The 13 AsGLX1s with functional domains ranging from 108 to 295 aa in Clade II may be GLX1-like proteins. In addition, AsGLX1-1C3 has one reverse transcriptase domain (RVT_1), and AsGLX1-2A has one NADH dehydrogenase complex I subunit M domain (NdhM), which is closely related to its predicted chloroplast localization (Figures 2A, 4A).
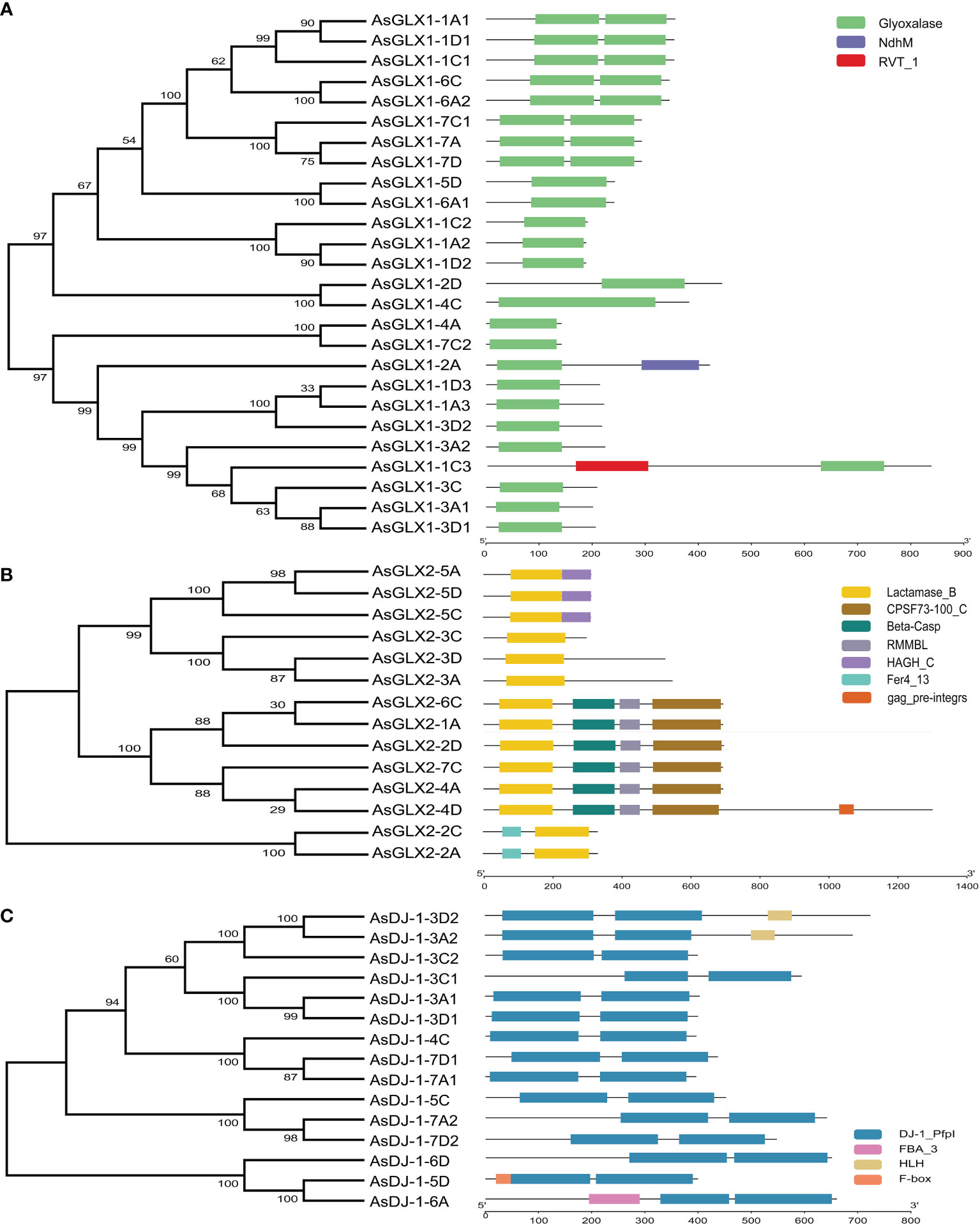
Figure 4 Schematic representation of domain architecture of GLX1s (A), GLX2s (B) and GLX3s (C) from oat.
AsGLX2-5A, AsGLX2-5C, and AsGLX2-5D belong to Clade IV in the GLX2 family, which are characterized by the presence of both a lactamase B domain (148 aa) and a HAGH_C domain (hydroxyacylglutathione hydrolase C-terminal) with 84 aa. These three members share a conserved metal ion-binding site (THHHYDH). Clade III of the GLX2 family comprises members that contain the lactamase B domain with169 aa, while the lactamase B domain of 6 AsGLX2s in Clade II is 154 aa. AsGLX2s of Clade I, including AsGLX2-2C and AsGLX2-2A, have lactamase B domains that are 156 aa and 158 aa in length, respectively. Addition, the active site of AsGLX2-5A, AsGLX2-5C, AsGLX2-5D, AsGLX2-3A, AsGLX2-3C, AsGLX2-3D, AsGLX2-2C, and AsGLX2-2A all feature the GHT residue, which is essential for catalytic activity. In addition to the lactamase B domain and the HAGH_C domain, some AsGLX2s also contain other domains such as the Pre-mRNA 3’-end-processing endonuclease polyadenylation factor C-term domain (CPSF73-100_C), Beta-Casp domain, Zn-dependent metallo-hydrolase RNA specificity domain (RMMBL), GAG-pre-integrase domain, and 4Fe-4S single cluster domain of Ferredoxin I (Fer4_13) (Figures 2B, 4B).
Although all AsDJ-1s have two DJ-1/PfpI domains, there are differences in the amino acid length that connects the two domains among these members. The number of amino acids between the two domains in Clade I members of the AsDJ-1 family is shorter, ranging from 11 to 14 aa, while the number of amino acids between the two domains in members of the other three Clades is longer, ranging from 39 to 41 aa, except for AsDJ-1-3C2, which has 15 aa. Additionally, AsDJ-1-3A2 and AsDJ-1-3D2 each have one Helix-loop-helix DNA binding domain (HLH), AsDJ-1-6A has one F-box associated domain (FBA_3), and AsDJ-1-5D has one F-box domain (F-box) (Figures 2C, 4C).
The analysis of the upstream 2 kb promoter regions of the AsGLX1, AsGLX2, and AsGLX3 genes showed that the basic eukaryotic promoter elements CAAT-box and TATA-box are widely distributed in the promoter regions of these genes. Other cis-regulatory elements mainly related to hormone responsiveness, anaerobic induction, defense and stress responsiveness, light responsiveness, endosperm expression, MYB binding sites, meristem expression, and seed-specific regulation (Figure 5). The hormone responsive elements mainly included ABA, GA, MeJA, SA, and IAA responsiveness. The stress responsiveness was mainly related to low temperature, anaerobic induction, and defense and stress responsiveness. Apart from core elements, hormone responsive elements are the most abundant in the AsGLX1, AsGLX2, and AsGLX3 promoters, accounting for 47%, 46%, and 53% of all elements, respectively (Table S3). Light responsive elements are also present at high proportions in the AsGLX1, AsGLX2, and AsGLX3 gene promoters, accounting for 32%, 24%, and 23%, respectively. Among hormone responsive elements, ABA responsive elements are the most abundant in the AsGLX1 gene promoters, accounting for about 50%, while MeJA responsive elements are the most abundant in the AsGLX2 and AsGLX3 gene promoters, accounting for 46% and 55%, respectively. Elements associated with adverse conditions are present in descending order of number in AsGLX1, AsGLX2, and AsGLX3 gene promoters, namely anaerobic induction, low-temperature responsiveness, and defense and stress responsiveness (Table S3). Each of the three gene families contains three genes with seed-specific regulatory elements in their promoter regions, namely AsGLX1-1A3, AsGLX1-3A1, AsGLX1-4C, AsGLX2-2A, AsGLX2-3A, AsGLX2-4D, AsDJ-1-3A1, AsDJ-1-3D1, and AsDJ-1-3D2. In addition, MYB binding site elements are also widely distributed in the promoter of genes in the three gene families, particularly in AsGLX2, where each gene member’s promoter region contains MYB binding site elements (Figure 5).
Three members from each of the AsGLX1, AsGLX2, and AsGLX3 gene families were randomly selected in different clades with distinct protein domain compositions and cis-element compositions to investigate their tissue-specific expression. AsGLX1-1A1 and AsGLX2-2D showed high expression levels in leaves, particularly in old leaves, and were predicted to be localized in chloroplasts, indicating their importance in leaf function and development (Figures 6A, D, Table S2). AsGLX1-7A and AsGLX1-3D2 exhibited the highest expression levels in dry seeds, with increasing expression levels during seed development, indicating their potential roles in seed development and dehydration (Figures 6B, C). AsGLX2-3C was highly expressed in germinated seeds and roots, but had low expression levels in leaves, which is consistent with its predicted mitochondrial localization (Figure 6E, Table S2). AsDJ-1-3D2, AsDJ-1-4C, and AsDJ-1-5D showed high expression levels in leaves, glumes, dry seeds, and developing seeds, with expression levels gradually increasing during seed development (Figures 6G–I). Overall, AsGLX1, AsGLX2, and AsGLX3 genes exhibited relatively high expression levels in leaves and seeds, suggesting their potential roles in maintaining leaf function and seed vigor.
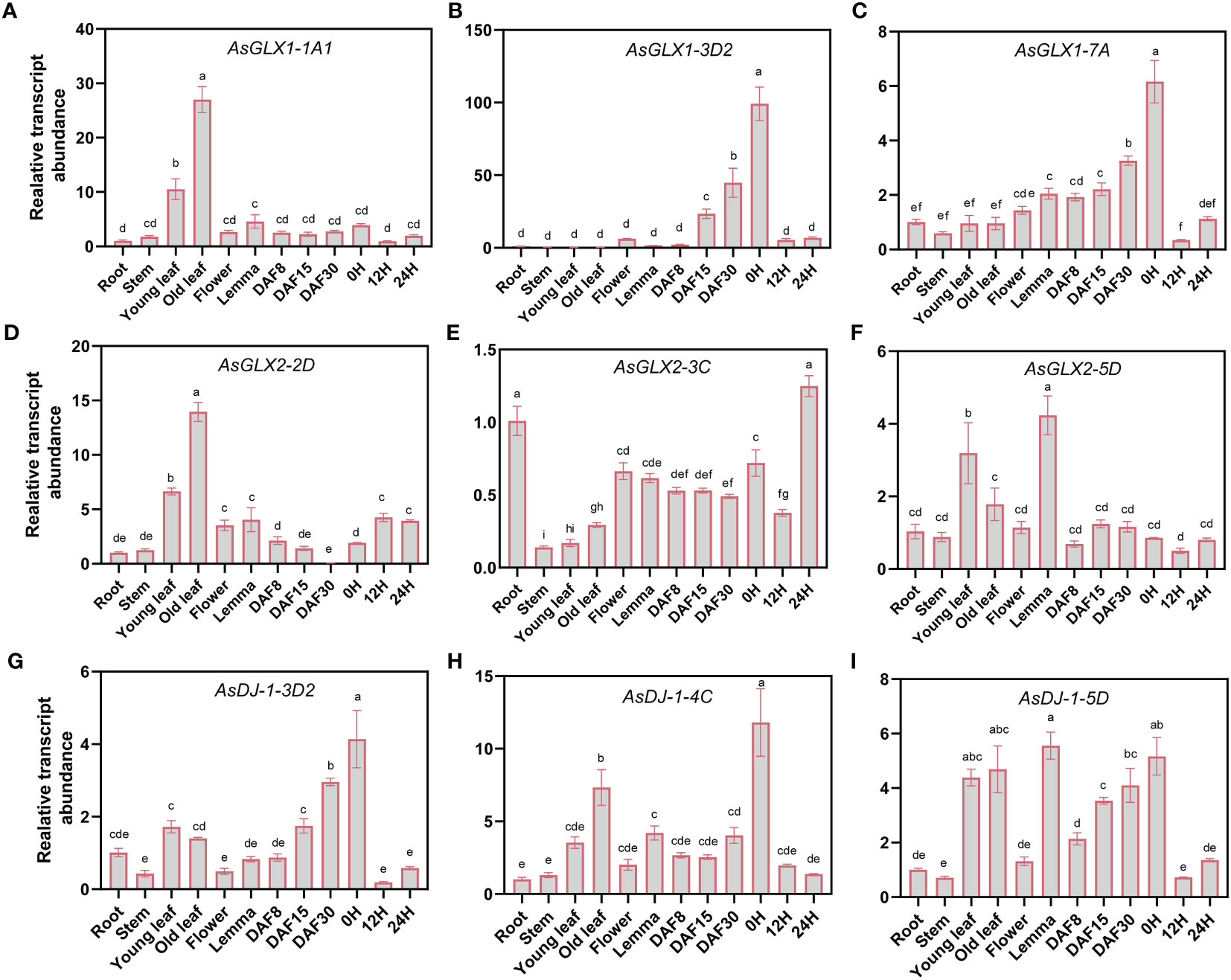
Figure 6 Expression profiling of AsGLX1, AsGLX2, and AsGLX3 genes in different tissues of oat by qPCR. The relative expression was calculated using root as reference. Seeds imbibed for 0 h, 12 h, and 24 h in germination phases were marked as 0 H, 12 H, and 24 H, respectively. Developing seeds at 8, 15, and 30 days after flowering were marked as DAF8, DAF15, and DAF30, respectively. The lower-case letters (A–I) represent statistical significance among the samples and the vertical bars represent the ± SEM for three replicates. The mean values sharing different letters, obtained from Duncan test, are different significantly at p < 0.05 level.
The expression patterns of AsGLX1-1A1 and AsGLX1-7A during seed germination showed a similar trend of first decreasing and then increasing, while AsGLX1-3D2 exhibited a distinct trend of gradual decrease during germination. After aging treatment, the expression levels of AsGLX1-1A1 and AsGLX1-7A in seeds significantly decreased, while the expression level of AsGLX1-3D2 showed no significant changes (Figure 7, Supplementary Table 4).
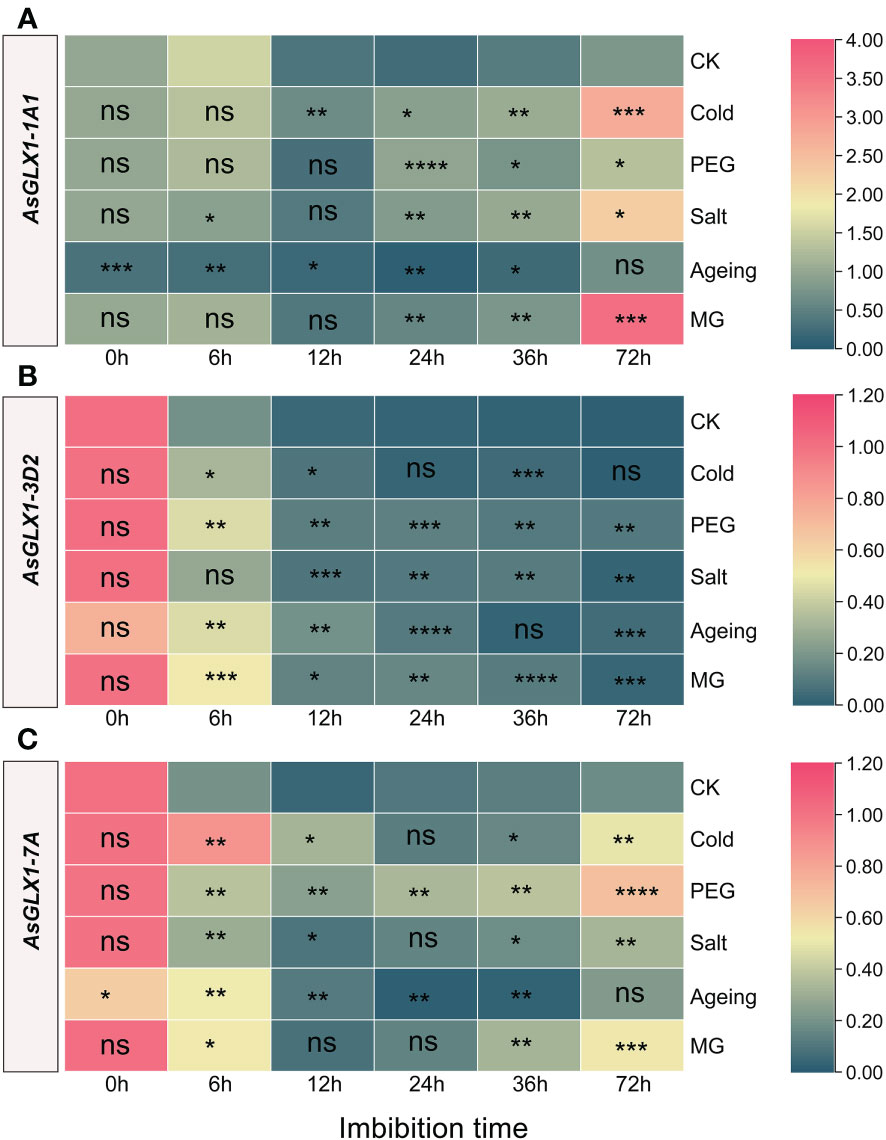
Figure 7 Expression profiling of AsGLX1-1A1 (A), AsGLX1-3D2 (B) and AsGLX1-7A (C) during seed germination of oat under stress by qPCR. The relative expression was calculated using dry seed (0 h) of CK as reference. The significant change in comparison to CK has been calculated using Student’s t test. *indicates a significant difference at p < 0.05; **indicates a significant difference at p < 0.01; ***indicates a significant difference at p < 0.001; ****indicates a significant difference at p < 0.0001; ns represents not significant.
Compared to the control, AsGLX1-1A1 showed significant downregulation during 0-36 h imbibition of aged seed, and it showed relatively small changes during the early imbibition stage (0-12 h) under cold, PEG, salt, and MG treatments. However, after 24 h of treatment, AsGLX1-1A1 expression was significantly induced, particularly under salt, cold, and MG treatments at 72 h of imbibition (Figure 7A). AsGLX1-3D2 exhibited relatively small changes under cold treatment, with no significant difference between the control and treatments at 24 h and 72 h. AsGLX1-3D2 was upregulated at most imbibition time points during PEG and MG treatments, and in aged seeds, indicating its detoxification role during seed germination under PEG and MG stress and after aging treatment (Figure 7B). AsGLX1-7A was significantly induced during imbibition for 0-72 h under PEG treatment, with the most significant upregulation occurring at 72 h. However, the expression level of AsGLX1-7A in aged dry seeds significantly decreased, being significantly higher than the control during early imbibition (6-12 h), but significantly lower than or not significantly different from the control at 24-72 h of imbibition, suggesting a potential role during the early stages of germination in aged seeds. In addition, AsGLX1-7A was significantly induced under cold, salt, and MG treatments at all imbibition time points except for 24 h under cold and salt treatments or 36 h under MG treatment (Figure 7C). Overall, AsGLX1 genes exhibit specificity in response to different stresses, and AsGLX1-3D2 and AsGLX1-7A may play important detoxification roles during seed germination under stress conditions.
The response of the three AsGLX2 genes to stress treatments is relatively smaller compared to the tested AsGLX1 genes. Following aging treatment, both AsGLX2-3C and AsGLX2-5D exhibited significant downregulation, while AsGLX2-2D2 showed no significant difference compared to the CK (Figure 8, Supplementary Table 4).
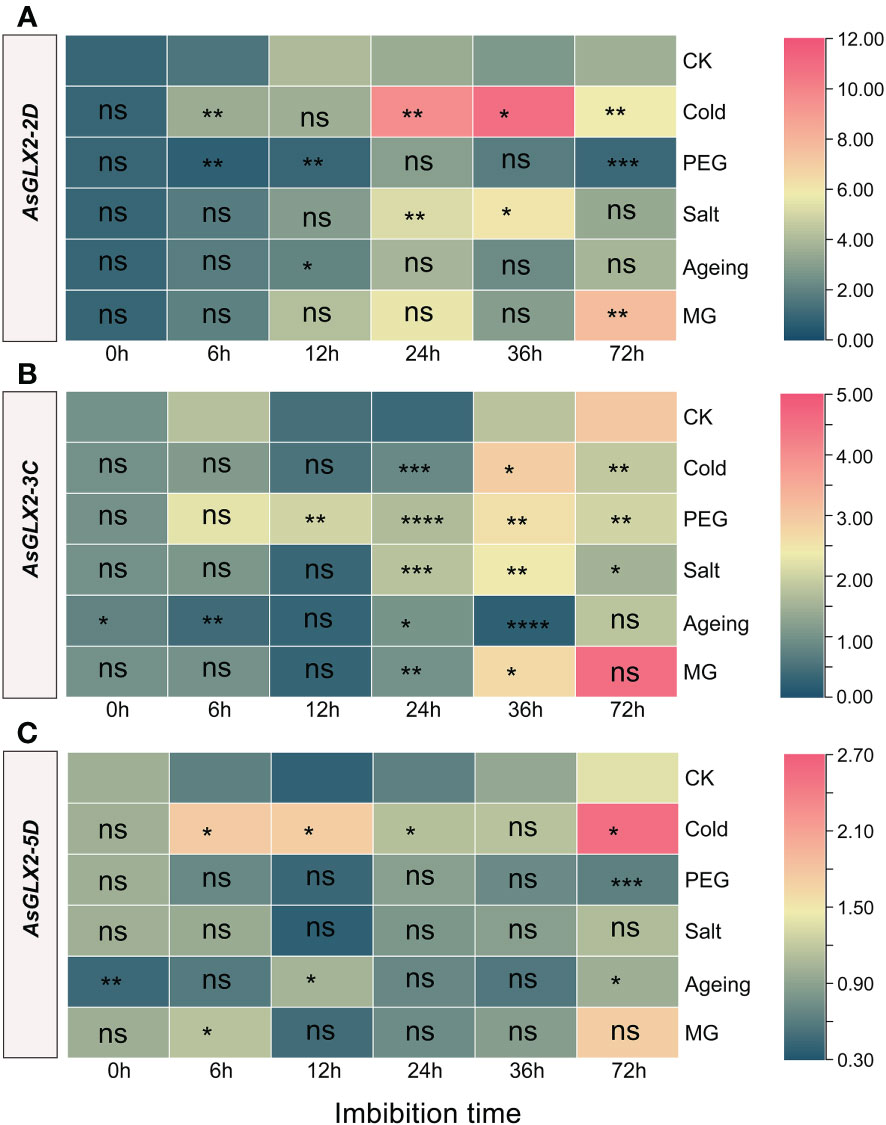
Figure 8 Expression profiling of AsGLX2-2D (A), AsGLX2-3C (B), AsGLX2-5D (C) during seed germination of oat under stress by qPCR. The relative expression was calculated using dry seed (0 h) of CK as reference. The significant change in comparison to CK has been calculated using Student’s t test. *indicates a significant difference at p < 0.05; **indicates a significant difference at p < 0.01; *** indicates a significant difference at p < 0.001; ****indicates a significant difference at p < 0.0001; ns represents not significant.
AsGLX2-2D2 was significantly up-regulated during imbibition under cold treatment, except for 12 h, and showed marked response at 24 h and 36 h. In aged seeds, AsGLX2-2D2 showed almost no significant response. AsGLX2-2D2 was up-regulated at 24 h and 36 h of imbibition under salt stress, while it was up-regulated at 72 h under MG treatment (Figure 8A). AsGLX2-3C showed a weak response to different treatments in the early stage of imbibition, and was only significantly up-regulated under PEG treatment for 12 h and significantly down-regulated at 6 h of imbibition in aged seeds. During 24-36 h of imbibition, AsGLX2-3C was up-regulated under various treatments, except for significant down-regulation at 36 h of imbibition in aged seeds. At 72 h of imbibition, the expression of AsGLX2-3C showed no significant difference compared to the control under various treatments (Figure 8B). AsGLX2-5D also showed a strong response under cold treatment, and was up-regulated at all-time points except for 36 h of imbibition. Under PEG and salt treatments, AsGLX2-5D showed no significant difference in expression level compared to the control at most time points, except for significant down-regulation at 72 h under PEG treatment. AsGLX2-5D was significantly up-regulated more than 2-fold at 12 h of imbibition in aged seeds, and its expression level was significantly higher than the control at 6 h of imbibition under MG treatment (Figure 8C). Overall, AsGLX2 genes showed a more significant response to cold treatment, indicating their important role in maintaining seed vigor or promoting seed germination under cold stress.
The AsDJ-1 genes exhibit a diverse response to various stresses during seed imbibition, with AsDJ-1-5D displaying a more pronounced response to stress, whereas the responses of AsDJ-1-4C and AsDJ-1-3D2 are comparatively minor. Following aging treatment, all three tested AsDJ-1 genes show a substantial downregulation (Figure 9, Supplementary Table 4).
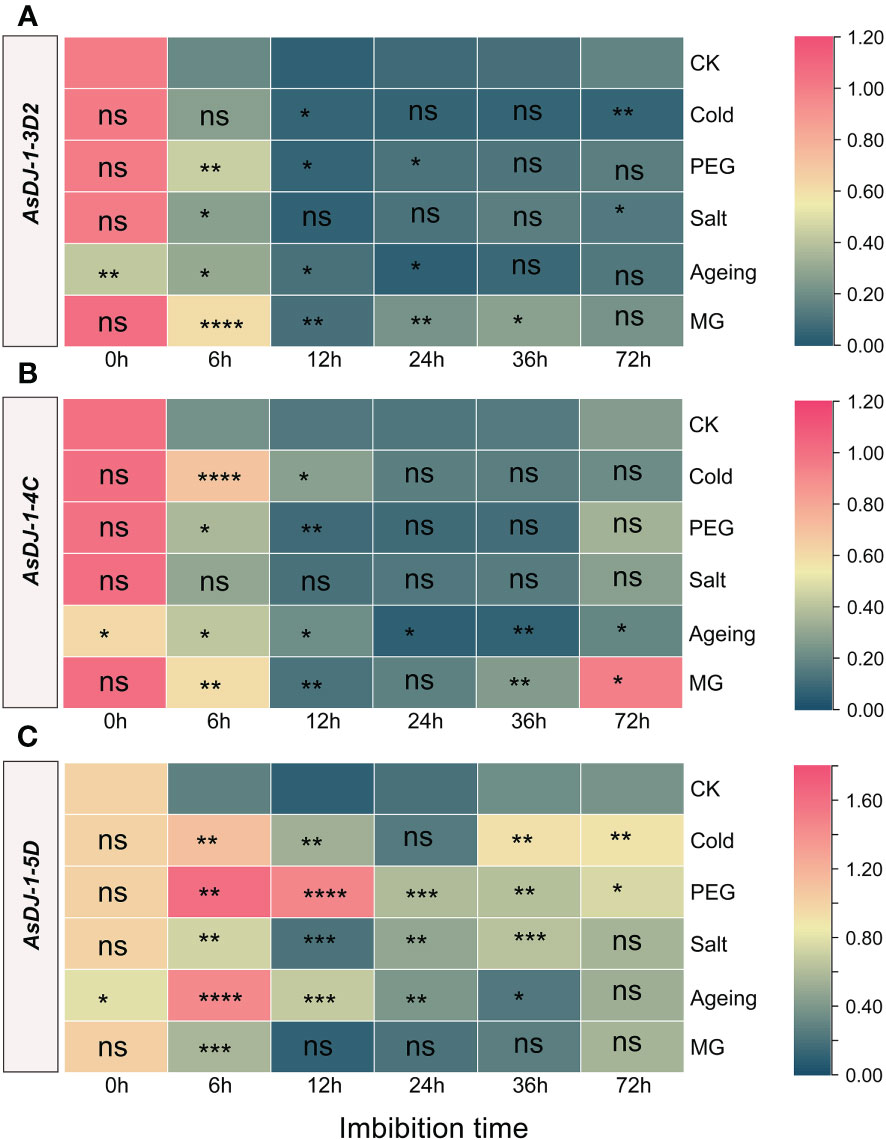
Figure 9 Expression profiling of AsDJ-1-3D2 (A), AsDJ-1-4C (B), and AsDJ-1-5D (C) during seed germination of oat under stress by qPCR. The relative expression was calculated using dry seed (0 h) of CK as reference. The significant change in comparison to CK has been calculated using Student’s t test. *indicates a significant difference at p < 0.05; **indicates a significant difference at p < 0.01; ***indicates a significant difference at p < 0.001; ****indicates a significant difference at p < 0.0001; ns represents not significant.
AsDJ-1-3D2 showed the most significant response under MG treatment, with significant upregulation compared to the control at 6-36 h, and the most significant response occurred at 6 h. Under cold treatment, AsDJ-1-3D2 was significantly upregulated at 12 h and significantly downregulated at 72 h. It was significantly upregulated at 6-24 h under PEG treatment, and showed a significant response at 12 h and 72 h of imbibition under salt treatment. In aged seeds, it was significantly upregulated during the early imbibition stage (6-12 h) (Figure 9A). AsDJ-1-4C did not show any significant response under salt treatment. Under cold and PEG treatments, it showed a significant response during the early imbibition stage, and cold treatment significantly induced its expression at 6 h of imbibition. In aged seeds, the expression level of AsDJ-1-4C was significantly upregulated during the early imbibition stage (6-12 h), and then showed a significant decrease. Moreover, under MG treatment, its expression level was significantly upregulated at 6 h, 36 h, and 72 h of imbibition (Figure 9B). AsDJ-1-5D was significantly upregulated during the imbibition under PEG treatment, especially in the early imbibition stage (6-12 h). There was no significant response under cold treatment at 24 h and under salt treatment at 72 h. At other time points, both cold and salt treatments significantly induced the expression of AsDJ-1-5D. During the imbibition of aged seeds, AsDJ-1-5D was significantly upregulated during 6-24 h, with the most significant response observed at 6 h, with an increase of more than 5 folds. Under MG treatment, its expression level was only significantly upregulated at 6 h (Figure 9C). Overall, AsDJ-1-5D has a relatively strong role in detoxification during the early germination stage of aged seeds and under PEG treatment.
The glyoxalases participate in various biological processes in plants (Singla-Pareek et al., 2020), such as stress response (Kaur et al., 2014), seed germination (Schmitz et al., 2017), plant senescence (Singla-Pareek et al., 2009), nutrient regulation (Borysiuk et al., 2022), signal transduction (Sankaranarayanan et al., 2017), starch synthesis (You et al., 2019), and pollen development (Sankaranarayanan et al., 2015). Their primary role in plants is to detoxify MG, which is spontaneously produced in plants and significantly accumulates under abiotic stresses such as salinity, drought, heavy metals, and low temperature, thus hindering plant growth and development (Kaur et al., 2014; Hasanuzzaman et al., 2017; Sankaranarayanan et al., 2017). Higher plants often contain multiple members of the GLX1, GLX2, and GLX3 gene families, and different members exhibit variations in their subcellular localization, expression patterns, and functional roles (Ghosh and Islam, 2016; Schmitz et al., 2017; Singla-Pareek et al., 2020). Therefore, a comprehensive genome-wide identification of glyoxalase gene families and understanding their chromosome distribution, evolutionary relationships, conserved domains, cis-regulatory elements, and gene expression patterns is crucial for exploring the functional diversity of glyoxalase genes and improving plant stress resistance.
The GSH-dependent pathway is the main pathway for clearing MG by GLX1 and GLX2 enzymes, which has led previous studies on plant glyoxalases to focus mainly on these two enzymes. However, the GSH-independent GLX3 has only recently received attention. Genome-wide identification and analysis of GLX1 and GLX2 families have been completed in in various plants. In Arabidopsis, a total of 11 GLX1 genes and 5 GLX2 genes were identified, while 11 GLX1 genes and 3 GLX2 genes were found in O. sativa (Mustafiz et al., 2011). In G. max, 24 GLX1 genes and 12 GLX2 genes were identified (Ghosh and Islam, 2016). In S. bicolor, 15 GLX1 genes and 6 GLX2 genes were found (Bhowal et al., 2020). However, the extensive whole-genome identification of glyoxalase families has ignored GLX3 genes, and only a few plants, such as Medicago truncatula and V. vinifera, have been systematically identified for the GLX1, GLX2, and GLX3 families (Ghosh, 2017; Li et al., 2019). In addition, recent studies have also individually identified GLX3s in some plant species. For example, 217 GLX3s were obtained by using AtDJ-1d (AT3g02720) to compare the Swiss-Prot database, including 8 Oryza species, 2 Triticum species, Hordeum vulgare, Zea mays, S. bicolor, Setaria italica, and Brachypodium distachyon. Among them, 12 GLX3s were identified in O. sativa, which were encoded by 6 genes and strongly induced by MG (Ghosh et al., 2016). To gain insight into the evolutionary patterns of GLX3, the evolution of GLX3s across prokaryotes and eukaryotes were studied, and 183 GLX3s belonging to 69 species was used for evolutionary analysis in plants (Kumar et al., 2021). In summary, the study of plant GLX3 have become a focus and hotspot, and systematic analysis of GLX1, GLX2, and GLX3 members will play an important role in plant improvement.
A total of 26 AsGLX1 genes, 14 AsGLX2 genes, and 15 AsGLX3 genes were identified in oat. The number of AsGLX1 and AsGLX2 genes identified in Arabidopsis, O. sativa, G. max, S. bicolor, and B. rapa was lower than that in oat, which may be related to genome duplication events during evolution (Ghosh and Islam, 2016; Yan et al., 2018; Bhowal et al., 2020; Peng et al., 2022). In the secondary branches of the evolutionary tree of the GLX1, GLX2, and GLX3, there were two or three oat homologous genes, originating from two or three oat subgenomes (A, C, and D), corresponding to one Arabidopsis glyoxalase gene. Oat has diploid and tetraploid ancestors, and genome duplication may be the main reason for the high number of homologous genes in oat (Peng et al., 2022). Some clades contained only GLX members from O. sativa and oat, such as OsGLYI11, AsGLX1-7A, AsGLX1-C1, and AsGLX1-D in GLX1, as well as OsDJ-1A, AsDJ-1-3A1, AsDJ-1-3C1, and AsDJ-1-3D1 in GLX3, indicating that the three families evolved asynchronously in the O. sativa, oat, and Arabidopsis genomes. Furthermore, the gene duplication events analysis of AsGLX1, AsGLX2, and AsGLX3 genes showed that only AsGLX1 and AsGLX3 gene families had tandem duplications, while none of the three families had segmental duplications. In contrast, in O. sativa, Arabidopsis, B. napus and G. max, gene duplications of GLX1 and GLX2 genes were caused by the segmental duplication, rather than tandem duplication (Mustafiz et al., 2011; Ghosh and Islam, 2016; Yan et al., 2023). Therefore, there are significant differences in gene duplication events that occur in the glyoxalase families in different plant species.
The AsGLX1 family members, AsGLX1-1A1, AsGLX1-1C1, AsGLX1-1D1, AsGLX1-6C, AsGLX1-6A2, AsGLX1-7A, AsGLX1-7C1, and AsGLX1-7D, contain two conserved glyoxalase domains and are clustered with the Ni2+-dependent GLX1 members in Arabidopsis, indicating that they are Ni2+-dependent AsGLX1s. AsGLX1-5D and AsGLX1-6A1 contain a single glyoxalase domain of 141 amino acids in length and are clustered with Zn2+-dependent GLX1s in Arabidopsis and O. sativa, indicating that they are Zn2+-dependent AsGLX1s (Ghosh and Islam, 2016; Bhowal et al., 2020). The activity of GLX1 depends on divalent metal ions, and early studies suggested that the type of divalent ion required for GLX1 activity varies between prokaryotes and eukaryotes. GLXI in humans and yeast is Zn2+-dependent, while GLX1 in E. coli requires Ni2+ for optimal activity (He et al., 2000). However, studies on plants have identified two types of divalent ion-dependent GLX1s. For instance, in Arabidopsis, AtGLYI2 is dependent on Zn2+, whereas AtGLYI3 and AtGLYI6 are dependent on Ni2+. Most studies on the function of plant GLX1s have focused on these two types, with less research on other GLX1-like proteins. In O. sativa, OsGLYII-1 has ethylmalonic encephalopathy-1 activity, which can be activated by Ca2+, and OsGLYII-2 has a binuclear zinc/iron center at its active site that is crucial for its activity. AsGLX2-5A, AsGLX2-5C, and AsGLX2-5D have both lactamase B and HAGH_C domains, all of which contain conserved THHHYDH metal ion-binding sites and GHT activity sites, indicating that these members encode for putative functionally active AsGLX2 enzymes (Ghosh and Islam, 2016; Bhowal et al., 2020; Singla-Pareek et al., 2020). GLX2 enzymes with these two domains, including the THHHYDH and GHT sites, have been identified in other plants such as sorghum (SbGLYII-3 and SbGLYII-4) and grape (VvGLYII-like1 and VvGLYII-like2)(Ghosh and Islam, 2016; Li et al., 2019).
Subcellular localization prediction analysis revealed that AsGLX1s are mainly expressed in the cytoplasm, chloroplasts, and mitochondria, AsGLX2s are mainly expressed in chloroplasts and mitochondria, and AsGLX3s are mainly expressed in chloroplasts. Chloroplasts and mitochondria are organelles responsible for photosynthesis and respiration, respectively, and are metabolically active. They are not only the main source of ROS, but also the potential organelles for MG production (Hasanuzzaman et al., 2017). Excessive MG has been found to inhibit photosynthesis and disrupt mitochondrial function (Kaur et al., 2016). Under stress conditions, chloroplast and mitochondrial components are often hotspots for glycation (Tripathi et al., 2023). The expression and activity of glyoxalase enzymes in chloroplasts and mitochondria are crucial for protecting the photosynthetic system and mitochondrial function. In addition, there are also some members that function in the nucleus, which may be important for protecting the nucleus and maintaining DNA and RNA stability. For example, in sorghum, SbGLYI-8/8.1 proteins were also found to harbor putative nuclear localization signals and therefore, may catalyze the conversion of nuclear MG to SLG (Bhowal et al., 2020). And OsGLYI-8 is located in the nucleus and can alleviate DNA damage caused by MG in the nucleus (Kaur et al., 2017).
Through tissue-specific expression analysis of some members, it was found that their expression patterns were consistent with subcellular localization predictions. For example, AsGLX1-7A was predicted to be expressed in the cytoplasm and was found to be highly expressed in developing seeds and dry seeds. AsGLX2-3C was predicted to be primarily expressed in mitochondria and was found to be highly expressed in roots, flowers, dry seeds, and seeds imbibed for 24 h. AsDJ-1-4C was predicted to be expressed in chloroplasts and cytoplasm and was found to be highly expressed in dry seeds and leaves. Overall, most of the tested members were expressed in leaves and dry seeds, indicating their important roles in maintaining leaf function and seed vigor. AsGLX1-3D2 is exclusively expressed in developing and dry seeds, indicating that its function may be seed-specific, such as enhancing seed tolerance to stress during seed maturation and dehydration. In Arabidopsis and O. sativa, AtGLYI8, OsGLYI3, and OsGLYI10 were also found to be highly expressed in developing seeds (Mustafiz et al., 2011). Glyoxalases have been suggested play an important role in seed development, seed germination, and seed vigor regulation. In Arabidopsis, a cytosolic GLXI3 isoform works on the elimination of toxic reactive carbonyl species during germination and seedling establishment (Schmitz et al., 2017). And the lack of AtGLYI2 resulted in severe inhibition of seed germination under MG treatment, and the growth of seedlings was also limited under salt stress (Liu et al., 2022). In rice, OsGLYI7 participates in starch synthesis in the endosperm, and its mutant had significantly reduced starch content and altered expression of starch synthesis genes (You et al., 2019). While, OsGLYI3 is specifically expressed in rice seeds and contributes to seed longevity and salt stress tolerance (Liu et al., 2022). AsGLX2-3C is highly expressed in roots and flowers, showing a significant difference from other members, and may play a detoxifying role during root and flower development. In B. napus, GLX1 is required for pollination and is targeted by the self-incompatibility system (Sankaranarayanan et al., 2015). Previous studies have mainly neglected the role of the glyoxalases in seeds. The diverse expression patterns and subcellular localization of GLX1, GLX2, and GLX3 genes provide a basis for their functional diversity in plants, and their important roles in seeds should be given more attention in the future.
The expression patterns of glyoxalase gene families, especially GLX1 and GLX2 genes, under stresses have been extensively studied. It has proven GLX1 and GLX2 transcript levels and enzyme activities could be induced by various adverse conditions (Kaur et al., 2014; Sankaranarayanan et al., 2017). But their expression patterns during seed germination have been rarely reported. However, from seed sowing to seedling establishment stage, crops often suffer from stress such as drought, low temperature, or salinity, which can lead to a significant reduction in crop yield. Therefore, analyzing the expression patterns of GLX1, GLX2, and GLX3 genes during seed germination under stress conditions is crucial for understanding their role in regulating seed vigor and coping with stress. In this study, it was found that the GLX1 genes AsGLX1-3D2 and AsGLX1-7A may play an important role in detoxification during seed germination under stress conditions. They were significantly induced by cold, drought, salt, MG, and aging treatments in the early stages of germination (6 h and 12 h). AsGLX2-5D was upregulated in the early stages of germination by cold, MG, and aging induction, while AsGLX2-3C was induced by various stresses in the later stages (24h-72h) of imbibition, indicating that these genes may play a detoxification role during seed germination under stress conditions, but with specificity in terms of stress time and types. GLX1 and GLX2 transgenic plants often exhibit increased stress resistance, while their function-deficient mutants exhibit reduced stress resistance (Kaur et al., 2014; Kaur et al., 2016; Sankaranarayanan et al., 2017). For instance, in Arabidopsis, complementation of AtGLYI2 and the rice homolog OsGLYI8 significantly enhanced stress resistance in the atglyI-2 mutant (Kaur et al., 2017). Overexpression of OsGLYII3 in tobacco significantly improved plant resistance to MG and NaCl (Singla-Pareek et al., 2003). Overexpression of OsGLYII2 in E. coli and tobacco resulted in a significant increase in resistance to both MG and salt stresses, along with improvements in plant photosynthetic performance and a decrease in oxidative damage (Ghosh et al., 2014). The expression analyzed GLX3 members, AsDJ-1-3D2, AsDJ-1-4C, and AsDJ-1-5 AsDJ-1-5D, show significant responses to stress during the early stages of seed imbibition, with AsDJ-1-5 AsDJ-1-5D being significantly upregulated throughout the imbibition process under stress conditions such as cold, drought, salt, and aging. In both E. arundinaceus and commercial sugarcane hybrids, Gly III respond to drought and salt stress like Gly II and Gly I, but the expression level of Gly III is higher under stress (Manoj et al., 2019). Additionally, the overexpression of EaGly III in sorghum confers significant improvements in drought and salt stress resistance, as evidenced by increased levels of proline and soluble sugars, enhanced photosynthetic and antioxidant abilities, and decreased lipid peroxidation in transgenic lines (Mohanan et al., 2020; Mohanan et al., 2021). Although studies on GLX3 genes in plants are limited, they all indicate the great potential of GLX3 in combating unfavorable conditions.
In this study, it was found that AsGLX1-7A has two glyoxalase domains and is a Ni2+-dependent GLX1, located in the cytoplasm, and has a high gene expression level in seeds. AsGLX2-5D has THHHYDH metal ion and GHT active sites and may play a protective role in mitochondria, which are important for respiration and stress responses during seed germination. AsDJ-1-5D has two conserved DJ-1 domains and a high gene expression level in seeds. The three genes showed significant responses to various stresses and encode for putative functionally active glyoxalase enzymes. In addition, AsGLX1-3D2 is seed-specifically expressed, and AsGLX1-2A may function in the protection of nucleic acid stability under stress conditions. These members of the glyoxalase gene families can be focused on in future research as potential candidate genes for oat stress resistance breeding and seed vigor improvement.
The identification of oat genes involved in stress resistance and seed vigor regulation represents a crucial endeavor in oat molecular breeding, with significant implications for crop yield and germplasm conservation. This study found that oat has more glyoxalase genes than most other plant species due to genome duplication events and tandem duplications during evolution. These genes are generally regulated by hormones and respond to adverse conditions. Their diverse tissue expression patterns and subcellular localizations indicate their functional diversity in plants, especially in leaf development and seed vigor formation. AsGLX1-3D2 was specifically expressed in seeds, and AsGLX1-2A may play an important role in alleviating nucleic acid glycation. In addition, AsGLX1-7A has potential Ni2+-dependent GLX1 activity, and AsDJ-1-5D has double DJ-1 domains, both of which can significantly respond to cold, drought, salt, MG and aging treatments. AsGLX2-5D has potential GLX2 activity and can respond to cold, aging, and MG stress. These highlighted genes are promising candidates for further investigation into oat stress resistance or seed vigor regulation. However, the specific functions of other members of AsGLX1, AsGLX2, and AsGLX3 families require further investigation. Nonetheless, such research may lead to the development of new strategies for breeding oats with improved stress tolerance and seed vigor through the utilization of glyoxalase genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
PM conceived and designed the experiment. MS and SS performed the experiments and analyzed the data. ZJ, CO, JW, HZ, and WM contributed to the experiment. MS and SS wrote the paper, and ML and PM revised the paper. All authors contributed to the article and approved the submitted version.
This research was supported by R&D of seed coating technology for key pasture grasses in the Qinghai-Tibet Plateau (SJCZFY2022-10) and CARS (CARS-34).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1215084/full#supplementary-material
Bhowal, B., Singla-Pareek, S. L., Sopory, S. K., Kaur, C. (2020). From methylglyoxal to pyruvate: a genome-wide study for the identification of glyoxalases and d-lactate dehydrogenases in Sorghum bicolor. BMC Genomics 21, 145. doi: 10.1186/s12864-020-6547-7
Borysiuk, K., Ostaszewska-Bugajska, M., Kryzheuskaya, K., Gardeström, P., Szal, B. (2022). Glyoxalase I activity affects Arabidopsis sensitivity to ammonium nutrition. Plant Cell Rep. 41, 2393–2413. doi: 10.1007/s00299-022-02931-5
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chinchansure, A. A., Korwar, A. M., Kulkarni, M. J., Joshi, S. P. (2015). Recent development of plant products with anti-glycation activity: a review. RSC Adv. 5, 31113–31138. doi: 10.1039/C4RA14211J
Gambhir, P., Singh, V., Raghuvanshi, U., Parida, A. P., Pareek, A., Roychowdhury, A., et al. (2023). A glutathione-independent DJ-1/PfpI domain-containing tomato glyoxalaseIII2, SlGLYIII2, confers enhanced tolerance under salt and osmotic stresses. Plant Cell Environ. 46, 518–548. doi: 10.1111/pce.14493
Ghosh, A. (2017). Genome-wide identification of glyoxalase genes in medicago truncatula and their expression profiling in response to various developmental and environmental stimuli. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00836
Ghosh, A., Islam, T. (2016). Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 16, 87. doi: 10.1186/s12870-016-0773-9
Ghosh, A., Kushwaha, H. R., Hasan, M. R., Pareek, A., Sopory, S. K., Singla-Pareek, S. L. (2016). Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 6, 18358. doi: 10.1038/srep18358
Ghosh, A., Pareek, A., Sopory, S. K., Singla-Pareek, S. L. (2014). A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 80, 93–105. doi: 10.1111/tpj.12621
Hasanuzzaman, M., Nahar, K., Hossain, M., Mahmud, J. A., Rahman, A., Inafuku, M., et al. (2017). Coordinated actions of glyoxalase and antioxidant defense systems in conferring abiotic stress tolerance in plants. Int. J. Mol. Sci. 18, 200. doi: 10.3390/ijms18010200
He, M. M., Clugston, S. L., Honek, J. F., Matthews, B. W. (2000). Determination of the structure of escherichia coli glyoxalase I suggests a structural basis for differential metal activation. Biochemistry 39, 8719–8727. doi: 10.1021/bi000856g
Hoque, T. S., Uraji, M., Tuya, A., Nakamura, Y., Murata, Y. (2012). Methylglyoxal inhibits seed germination and root elongation and up-regulates transcription of stress-responsive genes in ABA-dependent pathway in arabidopsis. Plant Biol. (Stuttg) 14, 854–858. doi: 10.1111/j.1438-8677.2012.00607.x
ISTA (2019). International rules for seed testing. Bassersdorf: International Seed Testing Association.
Jana, G. A., Krishnamurthy, P., Kumar, P. P., Yaish, M. W. (2021). Functional characterization and expression profiling of glyoxalase III genes in date palm grown under abiotic stresses. Physiol. Plant 172, 780–794. doi: 10.1111/ppl.13239
Kamal, N., Tsardakas Renhuldt, N., Bentzer, J., Gundlach, H., Haberer, G., Juhász, A., et al. (2022). The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 606, 113–119. doi: 10.1038/s41586-022-04732-y
Kaur, C., Sharma, S., Singla-Pareek, S. L., Sopory, S. K. (2016). Methylglyoxal detoxification in plants: role of glyoxalase pathway. Ind. J. Plant Physiol. 21, 377–390. doi: 10.1007/s40502-016-0260-1
Kaur, C., Singla-Pareek, S. L., Sopory, S. K. (2014). Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant Sci. 33, 429–456. doi: 10.1080/07352689.2014.904147
Kaur, C., Tripathi, A. K., Nutan, K. K., Sharma, S., Ghosh, A., Tripathi, J. K., et al. (2017). A nuclear-localized rice glyoxalase I enzyme, OsGLYI-8, functions in the detoxification of methylglyoxal in the nucleus. Plant J. 89, 565–576. doi: 10.1111/tpj.13407
Kumar, B., Kaur, C., Pareek, A., Sopory, S. K., Singla-Pareek, S. L. (2021). Tracing the evolution of plant glyoxalase III enzymes for structural and functional divergence. Antioxid. (Basel) 10, 648. doi: 10.3390/antiox10050648
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, Z.-G. (2016). Methylglyoxal and glyoxalase system in plants: old players, new concepts. Bot. Rev. 82, 183–203. doi: 10.1007/s12229-016-9167-9
Li, T., Cheng, X., Wang, Y., Yin, X., Li, Z., Liu, R., et al. (2019). Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera l.) and their expression profiling in response to downy mildew infection. BMC Genomics 20, 362. doi: 10.1186/s12864-019-5733-y
Liu, S., Liu, W., Lai, J., Liu, Q., Zhang, W., Chen, Z., et al. (2022). OsGLYI3, a glyoxalase gene expressed in rice seed, contributes to seed longevity and salt stress tolerance. Plant Physiol. Biochem. 183, 85–95. doi: 10.1016/j.plaphy.2022.04.028
Manoj, V. M., Anunanthini, P., Swathik, P. C., Dharshini, S., Ashwin Narayan, J., Manickavasagam, M., et al. (2019). Comparative analysis of glyoxalase pathway genes in Erianthus arundinaceus and commercial sugarcane hybrid under salinity and drought conditions. BMC Genomics 19, 986. doi: 10.1186/s12864-018-5349-7
Mohanan, M. V., Pushpanathan, A., Padmanabhan, S., Sasikumar, T., Jayanarayanan, A. N., Selvarajan, D., et al. (2021). Overexpression of glyoxalase III gene in transgenic sugarcane confers enhanced performance under salinity stress. J. Plant Res. 134, 1083–1094. doi: 10.1007/s10265-021-01300-9
Mohanan, M. V., Pushpanathan, A., Sasikumar, S. P. T., Selvarajan, D., Jayanarayanan, A. N., Arun, K. R., et al. (2020). Ectopic expression of DJ-1/PfpI domain containing Erianthus arundinaceus glyoxalase III (EaGly III) enhances drought tolerance in sugarcane. Plant Cell Rep. 39, 1581–1594. doi: 10.1007/s00299-020-02585-1
Mustafiz, A., Singh, A. K., Pareek, A., Sopory, S. K., Singla-Pareek, S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11, 293–305. doi: 10.1007/s10142-010-0203-2
Peng, Y., Yan, H., Guo, L., Deng, C., Wang, C., Wang, Y., et al. (2022). Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 54, 1248–1258. doi: 10.1038/s41588-022-01127-7
Ramu, V. S., Preethi, V., Nisarga, K. N., Srivastava, K. R., Sheshshayee, M. S., Mysore, K. S., et al. (2020). Carbonyl cytotoxicity affects plant cellular processes and detoxifying enzymes scavenge these compounds to improve stress tolerance. J. Agric. Food Chem. 68, 6237–6247. doi: 10.1021/acs.jafc.0c02005
Rasane, P., Jha, A., Sabikhi, L., Kumar, A., Unnikrishnan, V. S. (2015). Nutritional advantages of oats and opportunities for its processing as value added foods - a review. J. Food Sci. Technol. 52, 662–675. doi: 10.1007/s13197-013-1072-1
Sankaranarayanan, S., Jamshed, M., Kumar, A., Skori, L., Scandola, S., Wang, T., et al. (2017). Glyoxalase goes green: the expanding roles of glyoxalase in plants. Int. J. Mol. Sci. 18, 898. doi: 10.3390/ijms18040898
Sankaranarayanan, S., Jamshed, M., Samuel, M. A. (2015). Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 1, 1–7. doi: 10.1038/nplants.2015.185
Schilling, O., Wenzel, N., Naylor, M., Vogel, A., Crowder, M., Makaroff, C., et al. (2003). Flexible metal binding of the metallo-β-lactamase domain: glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry 42, 11777–11786. doi: 10.1021/bi034672o
Schmitz, J., Dittmar, I. C., Brockmann, J. D., Schmidt, M., Hüdig, M., Rossoni, A. W., et al. (2017). Defense against reactive carbonyl species involves at least three subcellular compartments where individual components of the system respond to cellular sugar status. Plant Cell 29, 3234–3254. doi: 10.1105/tpc.17.00258
Schmitz, J., Rossoni, A. W., Maurino, V. G. (2018). Dissecting the physiological function of plant glyoxalase I and glyoxalase I-like proteins. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01618
Singla-Pareek, S. L., Kaur, C., Kumar, B., Pareek, A., Sopory, S. K. (2020). Reassessing plant glyoxalases: large family and expanding functions. New Phytol. 227, 714–721. doi: 10.1111/nph.16576
Singla-Pareek, S. L., Reddy, M. K., Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. U S A. 100, 14672–14677. doi: 10.1073/pnas.2034667100
Singla-Pareek, S. L., Yadav, S. K., Sopory, A. M., Sopory, S. K. (2009). Role of the glyoxalase pathway in delaying plant senescence under stress conditions. SEB Exp. Biol. Ser. 62, 171–185.
Sun, M., Sun, S., Jia, Z., Ma, W., Mao, C., Ou, C., et al. (2022a). Genome-wide analysis and expression profiling of glutathione reductase gene family in oat (Avena sativa) indicate their responses to abiotic stress during seed imbibition. Int. J. Mol. Sci. 23, 11650. doi: 10.3390/ijms231911650
Sun, M., Sun, S., Mao, C., Zhang, H., Ou, C., Jia, Z., et al. (2022b). Dynamic responses of antioxidant and glyoxalase systems to seed aging based on full-length transcriptome in oat (Avena sativa l.). Antioxidants 11, 395. doi: 10.3390/antiox11020395
Takagi, D., Inoue, H., Odawara, M., Shimakawa, G., Miyake, C. (2014). The Calvin cycle inevitably produces sugar-derived reactive carbonyl methylglyoxal during photosynthesis: a potential cause of plant diabetes. Plant Cell Physiol. 55, 333–340. doi: 10.1093/pcp/pcu007
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thornalley, P. J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269, 1. doi: 10.1042/bj2690001
Tripathi, D., Oldenburg, D. J., Bendich, A. J. (2023). Oxidative and glycation damage to mitochondrial DNA and plastid DNA during plant development. Antioxidants 12, 891. doi: 10.3390/antiox12040891
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Xia, F., Cheng, H., Chen, L., Zhu, H., Mao, P., Wang, M. (2020). Influence of exogenous ascorbic acid and glutathione priming on mitochondrial structural and functional systems to alleviate aging damage in oat seeds. BMC Plant Biol. 20, 104. doi: 10.1186/s12870-020-2321-x
Xie, H., Li, M., Chen, Y., Zhou, Q., Liu, W., Liang, G., et al. (2021). Important physiological changes due to drought stress on oat. Front. Ecol. Evol. 9. doi: 10.3389/fevo.2021.644726
Xu, Z., Chen, X., Lu, X., Zhao, B., Yang, Y., Liu, J. (2021). Integrative analysis of transcriptome and metabolome reveal mechanism of tolerance to salt stress in oat (Avena sativa l.). Plant Physiol. Biochem. 160, 315–328. doi: 10.1016/j.plaphy.2021.01.027
Yan, G., Xiao, X., Wang, N., Zhang, F., Gao, G., Xu, K., et al. (2018). Genome-wide analysis and expression profiles of glyoxalase gene families in Chinese cabbage (Brassica rapa l). PloS One 13, e0191159. doi: 10.1371/journal.pone.0191159
Yan, G., Zhang, M., Guan, W., Zhang, F., Dai, W., Yuan, L., et al. (2023). Genome-wide identification and functional characterization of stress related glyoxalase genes in Brassica napus l. Int. J. Mol. Sci. 24, 2130. doi: 10.3390/ijms24032130
Yang, Z., Wang, K., Aziz, U., Zhao, C., Zhang, M. (2020). Evaluation of duplicated reference genes for quantitative real-time PCR analysis in genome unknown hexaploid oat (Avena sativa l.). Plant Methods 16, 138. doi: 10.1186/s13007-020-00679-1
Keywords: Avena sativa, glyoxalase, seed germination, abiotic stress, expression profiling
Citation: Sun M, Sun S, Jia Z, Zhang H, Ou C, Ma W, Wang J, Li M and Mao P (2023) Genome-wide analysis and expression profiling of glyoxalase gene families in oat (Avena sativa) indicate their responses to abiotic stress during seed germination. Front. Plant Sci. 14:1215084. doi: 10.3389/fpls.2023.1215084
Received: 01 May 2023; Accepted: 31 May 2023;
Published: 15 June 2023.
Edited by:
Qiang Guo, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Guangyan Feng, Sichuan Agricultural University, ChinaCopyright © 2023 Sun, Sun, Jia, Zhang, Ou, Ma, Wang, Li and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peisheng Mao, bWFvcHNAY2F1LmVkdS5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.