
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 June 2023
Sec. Plant Metabolism and Chemodiversity
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1213662
A correction has been applied to this article in:
Corrigendum: SbWRKY75- and SbWRKY41- mediated jasmonic acid signaling regulates baicalin biosynthesis
Introduction: Scutellaria baicalensis Georgi is a traditional Chinese medicinal plant with broad pharmacological activities whose main active ingredient is the flavonoid baicalin. Given its medicinal value and increasing market demand, it is essential to improve the plant’s baicalin content. Flavonoid biosynthesis is regulated by several phytohormones, primarily jasmonic acid (JA).
Methods: In this study, we conducted transcriptome deep sequencing analysis of S. baicalensis roots treated with methyl jasmonate for different durations (1, 3, or 7 hours). Leveraging weighted gene co-expression network analysis and transcriptome data, we identified candidate transcription factor genes involved in the regulation of baicalin biosynthesis. To validate the regulatory interactions, we performed functional assays such as yeast one-hybrid, electrophoretic mobility shift, and dual-luciferase assays.
Results: Our findings demonstrated that SbWRKY75 directly regulates the expression of the flavonoid biosynthetic gene SbCLL-7, whereas SbWRKY41 directly regulates the expression of two other flavonoid biosynthetic genes, SbF6H and SbUGT, thus regulating baicalin biosynthesis. We also obtained transgenic S.baicalensis plants by somatic embryo induction and determined that overexpressing SbWRKY75 increased baicalin content by 14%, while RNAi reduced it by 22%. Notably, SbWRKY41 indirectly regulated baicalin biosynthesis by modulating the expression of SbMYC2.1, SbJAZ3 and SbWRKY75.
Discussion: This study provides valuable insights into the molecular mechanisms underlying JA-mediated baicalin biosynthesis in S. baicalensis. Our results highlight the specific roles of transcription factors, namely SbWRKY75 and SbWRKY41, in the regulation of key biosynthetic genes. Understanding these regulatory mechanisms holds significant potential for developing targeted strategies to enhance baicalin content in S. baicalensis through genetic interventions.
Chinese skullcap (Scutellaria baicalensis Georgi), a plant used in traditional Chinese medicine, is renowned for its diverse pharmacological activities, such as heat-clearing, detoxification, diuresis, sore-throat relief, and anti-inflammatory effects, which are primarily attributed to its flavonoid constituents, including baicalein, baicalin, wogonin, and wogonoside (Zhao et al., 2016a). Zhao et al. discovered a specialized flavone biosynthetic pathway in S. baicalensis (Zhao et al., 2016b). This pathway encompasses the enzymatic conversion of cinnamoyl-CoA to cinnamoyl, subsequent catalytic transformation of pinocembrin to chrysin mediated by flavone synthase II (FNSII), and further conversions of chrysin into wogonin and baicalein (Zhao et al., 2019). The elucidation of this pathway establishes a crucial foundation for comprehending the biosynthesis of these essential flavonoid compounds in S. baicalensis. Given the medicinal value and increasing market demand of S. baicalensis, enhancing the content of active ingredients in this plant is of great economical and pharmacological importance. Previous studies have shown that the exogenous application of phytohormones, especially jasmonic acid (JA), stimulates the accumulation of baicalin in S. baicalensis (Xu et al., 2010). However, the mechanism by which JA increases baicalin biosynthesis is likely to involve multiple pathways and factors that are not well understood.
Baicalin biosynthesis and accumulation in S. baicalensis are known to be tightly regulated by various internal and external signals, although the underlying mechanisms remain largely unclear (Wang et al., 2022b). Among these signals, JA and its derivatives are plant stress hormones that play key roles in regulating flavonoid biosynthesis, root growth, and defense against pathogen infection and insect attack (Zhu et al., 2022). Studies on Arabidopsis (Arabidopsis thaliana) have revealed that JA regulates anthocyanin biosynthesis through the activation of transcription factors (TFs) (Shan et al., 2009). The TF MYC2 and JASMONATE-ZIM-DOMAIN PROTEIN (JAZ) repressors have emerged as the master regulators of most aspects of the JA signaling pathway in Arabidopsis (Kazan and Manners, 2013). Indeed, MYC2 is an important transcription factor that activates the expression of flavonoid biosynthetic genes by binding to specific E-box elements in their promoter regions or by forming complexes with other TFs (Van Moerkercke et al., 2019). In the absence of JA, JAZ repressors directly bind to MYC2 to form a repressive transcriptional complex. In the presence of JA, CORONATINE INSENSITIVE1 (COI1) recruits JAZ repressors, leading to their ubiquitination and subsequent degradation through the 26S proteasome, which releases MYC2 that induces JA responses (Chini et al., 2007; Thines et al., 2007).
The sophisticated transcriptional regulation of JA response in plants is mediated by various TF families, including APETALA2 (AP2)/ETHYLENE-RESPONSE FACTOR (ERF), basic helix-loop-helix (bHLH), MYB, and WRKY (Zhao et al., 2020; Huang et al., 2022; Liu et al., 2023; Yuan et al., 2023). Among these TF families, the WRKY family has emerged as a crucial player in the regulation of biosynthetic pathways for various bioactive compounds. Notably, studies conducted in S. baicalensis have provided evidence for a close association between WRKY TFs and the accumulation of anthocyanins (Zhang et al., 2022). Furthermore, the expression of NnWRKY40a and NnWRKY40b, two WRKY transcription factor genes cloned from lotus (Nelumbo nucifera), is significantly induced by JA and promotes BIA biosynthesis by modulating the expression of BIA biosynthetic genes (Li et al., 2022). Similarly, TcWRKY26 in Chinese yew (Taxus chinensis) directly binds to the W-box in the DBAT (10-deacetylbaccatin III-10β-O-acetyltransferase gene) promoter to activate its expression, with the TcWRKY26–TcJAV3 complex defining another JA signal transduction pathway that effectively regulates taxol biosynthesis (Chen et al., 2022). The diverse roles of WRKY family members in regulating the biosynthesis of different bioactive compounds indicate their significance in the JA response of plants.
In this study, we investigated the transcriptional regulation of baicalin biosynthesis in the roots of S. baicalensis following treatment with methyl jasmonate (MeJA). To this end, we collected root samples at four different time points and performed a transcriptome deep sequencing (RNA-seq) analysis, followed by weighted gene co-correlation network analysis (WGCNA) to construct a TF-gene regulatory network. By analyzing the regulatory network, we identified key TF genes likely to be involved in the JA-mediated regulation of baicalin biosynthesis in S. baicalensis. Our results shed light on the complex signaling and transcriptional cascade underlying the biosynthesis of this important flavone compound.
All plant materials were cultivated in a greenhouse under a controlled environment of 22°C with a 16-h light/8-h dark photoperiod. For JA response analysis, 1-month-old S. baicalensis seedlings were transferred to an aqueous solution containing 200 μM MeJA. The roots of the seedlings were collected at four time points, including 0, 1, 3, and 7 h of MeJA treatment.
Roots of S. baicalensis seedlings subjected to MeJA treatment for 0, 1, 3, and 7 h were used for transcriptome analysis. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. LC-Bio Technology Co., Ltd. (Hangzhou, China) generated the sequencing libraries and conducted sequencing. Reference transcriptome data are deposited in the Sequence Read Archive (SRA) under accession number SRA: PRJNA961700.
WGCNA was conducted using the OmicStudio tools available at https://www.omicstudio.cn/tool/WGCNA. A pairwise Pearson’s correlation matrix was constructed and transformed into a weighted matrix. To create a topological overlap matrix, a soft threshold of 0.85 was applied. Module-trait associations were estimated to identify the relationship between modules and various treatment stages, by calculating the correlation between the module eigengene and the flavone contents at each infection stage. This approach facilitated the identification of expression modules that were highly correlated with the flavone contents. The networks were visualized using Cytoscape software.
S. baicalensis transcription factor genes were identified through a comparative analysis of transcripts assembled from RNA-seq data against the PlantTFDB database using the BlastX algorithm. Candidate SbMYC2 genes were further selected by BLAST using the AtMYC2 sequence as query. Additionally, the hidden Markov model (HMM) profiles PF06200 (TIFY domain) and PF09425 (Jas motif) of the JAZ family were retrieved from the Pfam database, and the transcriptome sequence was subjected to hmmsearch analysis to identify candidate JAZ genes. Finally, a phylogenetic tree was constructed with all candidate TFs using MEGA7 software to facilitate visualization and interpretation of the results.
The coding region of SbWRKY75 was cloned into the pHB vector to generate the pHB : SbWRKY75 overexpression vector. A fragment of the SbWRKY75 coding sequence (bp 9–293) was selected for RNA interference (RNAi) and cloned into the intermediate vector pENTR. The target fragment was then recombined into the pHELLSGATE vector via LR Clonase to obtain the RNAi interference construct pHELLSGATE : SbWRKY75 (Figure S2). The pHB : SbWRKY75 and pHELLSGATE : SbWRKY75 clones were subsequently transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101.
The seeds were washed with running water for a duration of 30 minutes, followed by a cold storage period of 72 hours at 4°C. Subsequently, the seeds were immersed in 70% ethanol for 30 seconds and rinsed thrice with sterile water to ensure surface sterilization. A 1% sodium hypochlorite treatment was then administered for 15 minutes, followed by three additional rinses with sterile water. The surface-sterilized seeds were then inoculated onto Murashige and Skoog (MS) medium to facilitate the cultivation of sterile seedlings. Leaf and stem segments were excised from S. baicalensis plantlets and used as explants to induce somatic embryogenesis. The explants were immersed in Agrobacterium suspension (OD=0.6) with uniform shaking in the dark for 10 min. After being blotted dry onto sterile tissue paper, the explants were transferred to MS medium with 10.0 mg/L 1-naphthaleneacetic acid. The inoculated explants were selected on selective medium containing 200 mg/L timentin and cultivated at 25°C in the dark to induce somatic embryogenesis. After the formation of somatic embryos, they were subjected to an alternating light/dark cycle of 16 hours of light and 8 hours of darkness under a light intensity of 2,000 lux. The seedlings were then transferred to 1/2 MS medium to induce root formation. The rooted seedlings were transferred to flower discs containing substrate for further cultivation. Genomic DNA was extracted from the young leaves of transgenic S. baicalensis seedlings for confirmation of transformation.
The promoter fragments of baicalin biosynthesis pathway genes were cloned into the pLacZ vector as bait. The coding sequences of SbWRKY75, SbWRKY41, and SbRAP2.1 were recombined into the pB42AD vector as prey. The primer sequences are listed in Table S1. Different combinations of vectors were co-transformed into yeast strain EGY48. Transformants were selected on synthetic defined (SD) medium lacking tryptophan and leucine. Positive yeast colonies were grown for 3 days at 30°C, collected, and resuspended in sterile water, before being spotted onto an X-gal medium plate, which was then incubated at 30°C for 24 h. Negative controls were established using the empty pLacZ and pB42AD plasmids.
The coding sequences of SbWRKY75, SbWRKY41, and SbRAP2.1 were cloned into the pET32a expression vector to produce His-tagged fusion proteins. The sequences of the primers used for cloning are provided in Table S1. The resulting plasmids were transformed into E. coli BL21 cells, and the production of recombinant proteins was induced by adding 0.1 mM IPTG at 18°C for 12 h. The bacterial cells pelleted by centrifugation and lysed using a low-temperature ultra-high-pressure continuous flow cell disrupter (JNBIO). The supernatant was purified using Ni-NTA resin (Qiagen, Cat. No. 30210), and the bound proteins were eluted with 200 mM imidazole and subsequently concentrated using a Millipore (Merck, Cat. No. UFC903024) ultrafiltration device.
To investigate whether SbWRKY75, SbWRKY41, and SbRAP2.1 can bind to the promoters of their target genes, electrophoretic mobility shift assays (EMSAs) were conducted. The probes used in EMSA were labeled with digoxigenin; their sequences are listed in Table S1. The EMSA reactions were performed using a DIG Gel Shift Kit, 2nd Generation, following the manufacturer’s instructions (Roche). The reaction products were loaded onto a 5% polyacrylamide gel and electrophoresed at 80 V for 90 min. The DNA-protein complexes were transferred to a nylon membrane for 60 min at 300 mA and crosslinked at 120 MJ. Probe detection was carried out according to the protocol provided by Roche.
The coding sequences of SbWRKY75 and SbWRKY41 without a stop codon were individually cloned into the fusion YFP vector pHB through one-step cloning. The resulting constructs were transformed into Agrobacterium strain GV3101 and infiltrated into Nicotiana benthamiana leaves using needleless syringes. YFP fluorescence in lower epidermal cells was observed 48 h to 72 h after Agrobacterium infiltration using a confocal microscope (Leica TCS SP8). Primers used are listed in Table S1.
To perform the dual-luciferase reporter gene assay (dual-LUC), the promoter sequences of SbCLL-7, SbF6H, and SbUGT were cloned individually into pGreen II 0800 by one-step cloning (Lv et al., 2019). The resulting constructs were transformed into Agrobacterium strain GV3101. N. benthamiana leaves were infiltrated with the Agrobacterium cultures using needleless syringes. The leaves were collected 48 h after infiltration for dual-LUC assays, which were performed using a Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega). Primers used are listed in Table S1.
The quantification of baicalin, baicalein, wogonoside, and wogonin was carried out using an Agilent 6410 Triple Quad LC/MS system. Root samples of S. baicalensis were collected and homogenized to obtain a fine powder, which was then subjected to ultrasonic extraction with 70% (v/v) ethanol at 60°C for 1 h. The obtained extracts were centrifuged at 10,000 g at 4°C for 15 min and subsequently filtered through a Millipore Millex-HV filter (Merck Millipore, Billerica, MA, USA). To determine the flavone content, an absolute quantification method with external standards was employed. Calibration curves were constructed using standard solutions of baicalin, baicalein, wogonoside, and wogonin at known concentrations. The analytical parameters of the Agilent 6410 Triple Quad LC/MS were as follows: column, Waters T3 column (2.1 mm × 100 mm, 3 μm); oven temperature, 350°C. The mobile phase contained D: water (containing 0.1% [v/v] formic acid); C: acetonitrile; flow rate, 0.3 mL/min; column temperature, 50°C; and injection volume, 5 μL.
Total RNA was extracted from the roots of control S. baicalensis seedlings and those of seedlings treated with 200 µM MeJA for 0, 1, 3, or 7 h using TRIzol Reagent. Total RNA samples was then reverse-transcribed to first-strand cDNA using PrimeScript II RTase (Takara cat. no. 6110A). Primer pairs were designed using Primer Premier 5 software. Quantitative PCR was performed on a QuantStudio 3 System following the manufacturer’s protocol. The relative expression levels of target genes were calculated using the 2–ΔΔCt method with SbACT7 as the reference gene. The primer sequences are available in Table S1.
All boxplots, bar graphs, and connecting lines were generated using Origin 2018 Software. The error bars in the figures represent the standard deviation of the means obtained from three independent biological replicates.
To investigate the effects of MeJA treatment on baicalin biosynthesis in S. baicalensis roots, we treated plants with an aqueous MeJA solution for 0, 1, 3, or 7 h (Figure 1A). We determined that the levels of baicalein, baicalin, wogonin, and wogonoside change rapidly over the course of the treatment (Figure 1B). Specifically, the levels of baicalein, baicalin, wogonin, and wogonoside significantly decreased by 54%, 96%, 31% and 97%, respectively, after 1 h of MeJA treatment compared to the 0-h samples. However, after 7 h of MeJA exposure, the levels of these compounds had increased to reach 95% (baicalein), 19% (baicalin), 113% (wogonin), and 13% (wogonoside) of their levels in the 0-h samples. These results suggest that the treatment resulted in a substantial decrease in flavone glycoside content, followed by a slow recovery. Conversely, the decrease in glycoside content was modest and this essentially returned to pre-treatment levels after 7 h of MeJA treatment. Therefore, MeJA treatment has a significant influence on the flavone contents of S. baicalensis roots.

Figure 1 Co-expression network analysis and transcription factor regulation of baicalin biosynthesis in S. baicalensis with MeJA treatment. (A) WGCNA-based clustering analysis of samples and construction of the co-expression modules. Each module is indicated by different colors in the row; (B) Heatmap representation of the correlation between WGCNA modules and flavone contents. Darker red represents a high positive correlation; darker blue indicates a high negative correlation; (C) Analysis and visualization of the correlation between the top 30% transcription factor genes from the ‘Mebrown’ module and baicalin biosynthesis genes; (D) Heatmap representation of the expression levels of transcription factor gene families. The color shows the number of differentially expressed genes (DEGs) in a particular gene family. The asterisks indicate the top five transcription factor gene families with high numbers of DEGs between groups.
We examined the expression of various genes known to encode enzymes involved in baicalin biosynthesis in S. baicalensis (Figure 1C). These genes consisted of SbPAL-1 (Phenylalanine ammonia-lyase 1), SbPAL-2, SbPAL-3 (Park et al., 2012), SbCLL-7 (Cinnamate-CoA ligase 7), SbCHS-2 (Chalcone synthase 2), SbCHI (Chalcone isomerase), SbFNSII-2 (Flavone synthase II-2), SbF6H (Flavonoid 6-hydroxylase) (Zhao et al., 2016b), SbUGT (UDP-Glucosyl transferase) (Nagashima et al., 2000) and SbGUS (BETA-D-Glucuronidase) (Ikegami et al., 1995). We observed significant changes in the expression levels of these genes in seedling roots upon treatment with exogenous MeJA, in agreement with the observed alterations in flavone contents described above. These findings suggest that the MeJA-induced regulation of flavone biosynthesis in S. baicalensis is mediated through transcriptional modulation of various key biosynthetic genes in this pathway.
We employed the WGCNA R package to identify modules of co-expressed genes in response to MeJA treatment. With a soft minimal threshold for Pearson’s correlation coefficient of 0.85, we identified 15 co-expression modules, as defined on differential expression of their constituent genes, with each module being indicated by a different color (Figure 2A, Table S2). We clustered the consensus module eigengenes and visualized them to analyze module-phenotype correlations. Among the different gene expression modules, we found that the ‘MEturquoise’ module exhibited a strong and positive correlation with both baicalin and wogonoside contents, suggesting that the genes within this module play important roles in regulating the biosynthesis of these two compounds. Additionally, the ‘MEbrown’ module showed a strong and positive correlation specifically with wogonin content, indicating that the genes within this module may be involved in the specific biosynthesis of wogonin (Figure 2B). KEGG pathway enrichment analysis revealed that the ‘MEturquoise’ module does not contain genes directly involved in the phenylpropanoid pathway. Therefore, we focused on the ‘MEbrown’ module for subsequent analysis (Figure S1). We selected genes encoding baicalin biosynthetic enzymes and TFs within the top 30% genes from the ‘MEbrown’ module as candidate genes and identified key transcription factor genes regulating baicalin biosynthesis through correlation analysis (Figure 2C).
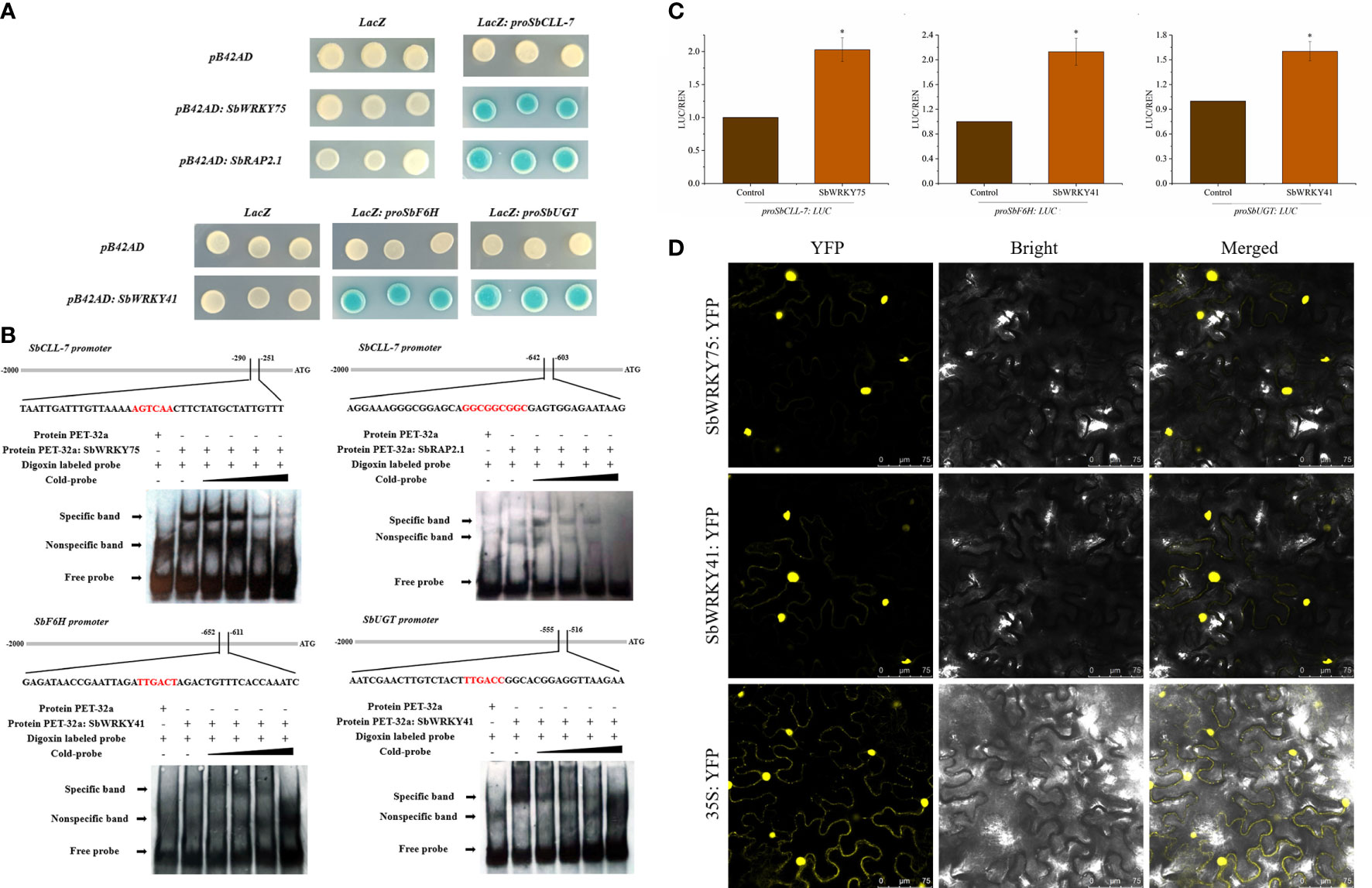
Figure 2 Detection of binding between the SbWRKY75, SbWRKY41 and SbRAP2.1 and their target promoters by Y1H assay (A) and EMSA (B); (C) Dual-LUC assay showing that SbWRKY75 and SbWRKY41 activate the transcription of SbCLL-7 and SbF6H, SbUGT in N. benthamiana. The YFP effector construct was used as a negative control, and the LUC/REN ratios for YFP were set to 1; (D) Subcellular localization of SbWRKY75 and SbWRKY41 in N. benthamiana leaves.
To investigate the TFs involved in regulating baicalin biosynthesis during MeJA processing, we looked at the number of differentially expressed TF genes in different TF gene families at different time points. We observed that the bHLH, ERF, MYB, NAC, and WRKY families are represented by the highest numbers of differentially expressed genes, with the proportion of differentially expressed TF genes in the ERF and WRKY families being particularly high (Figure 2D). Notably, TFs in the WRKY and ERF families have been shown to play important roles in flavonoid biosynthesis (Lloyd et al., 2017; Wu et al., 2022). Therefore, we chose five WRKY TF genes and four ERF TF genes with a p<0.01, as determined by Pearson’s correlation analysis with baicalin biosynthesis pathway genes, from the top 30% central TF genes in the ‘MEbrown’ module defined by WGCNA analysis. We investigated the function of these selected TF genes below.
We cloned the promoter sequence of several baicalin biosynthesis pathway genes into the pLacZ vector. To identify TFs potentially binding to these promoter sequences, we conducted a yeast one-hybrid (Y1H) assay using the above five WRKY TF genes and four ERF TF genes. As indicated by the blue color of the corresponding yeast colonies harboring the relevant constructs, we determined that SbWRKY75 (Sb01t34690) bound to the SbCLL-7 promoter, while SbWRKY41 (Sb06t15420) bound to the SbF6H and SbUGT promoters, and SbRAP2.1 (Sb08t10780) bound to the SbCLL-7 promoter (Figure 3A).
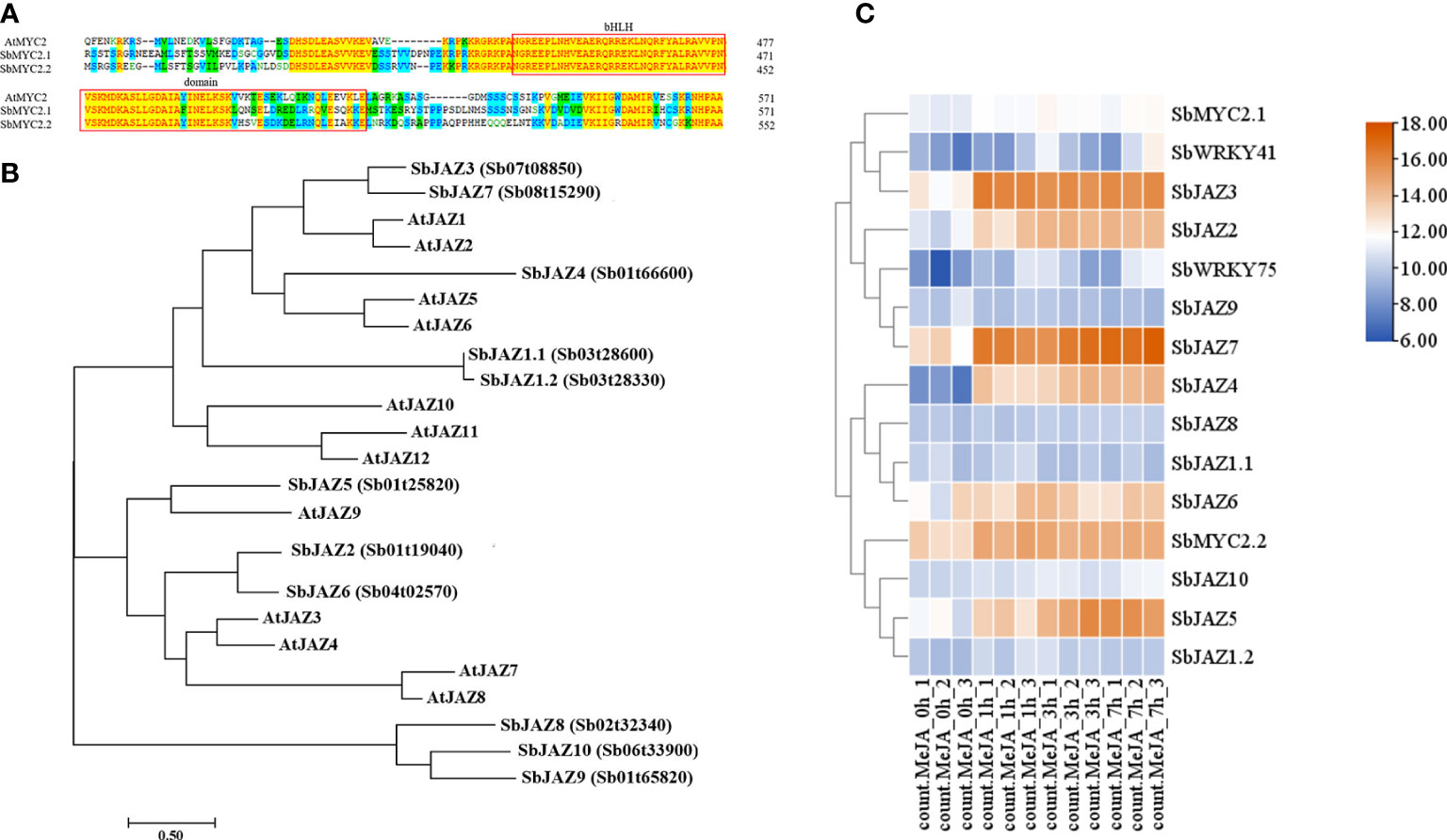
Figure 3 (A) Sequence alignment of SbMYC2 and AtMYC2; (B) Phylogenetic analysis of SbJAZ and AtJAZ proteins; (C) Heatmap representation showing the expression levels of SbMYC2s, SbJAZs, SbWRKY75 and SbWRKY41 in control S. baicalensis roots and after MeJA treatment. Each value was transformed using a Log2 conversion.
We validated the above results by conducting an electrophoretic mobility shift assay (EMSA) and a dual-luciferase (LUC) assay. To this end, we produced recombinant SbWRKY75 and SbRAP2.1 fused to a thioredoxin tag and incubated the purified proteins with digoxigenin-labeled probes for the SbCLL-7 promoter. We detected specific binding of both recombinant proteins to labeled probes, as evidenced by the lower mobility of the probes from the corresponding target genes (Figure 3B). Importantly, this lower mobility was gradually competed by incubation with unlabeled probe (Figure 3B). In the dual-LUC assay, using the reporter constructs proSbCLL-7:LUC, proSbF6H:LUC, and proSbUGT : LUC, we observed that co-expression of SbWRKY75 with the proSbCLL-7:LUC reporter construct resulted in a 103% increase in the LUC/REN ratio (firefly to Renilla luciferase activity). Additionally, co-expression of SbWRKY41 with the proSbF6H:LUC and proSbUGT : LUC reporter constructs resulted in a 113% increase and a 60% increase in the LUC/REN ratio, respectively (Figure 3C).
We also generated constructs consisting of the SbWRKY75 or SbWRKY41 coding sequence cloned in-frame with the sequence of yellow fluorescent protein (YFP) and driven by the cauliflower mosaic virus (CaMV) 35S promoter to establish the subcellular localization of these two TFs. We detected YFP fluorescence for SbWRKY75:YFP and SbWRKY41:YFP mainly in the nucleus of N. benthamiana leaf cells, indicating that SbWRKY75 and SbWRKY41 localize to the nucleus (Figure 3D).
To investigate the effect of SbWRKY75 on the baicalin biosynthesis pathway in S. baicalensis, we transiently overexpressed SbWRKY75 via Agrobacterium-mediated transient transformation. We detected a significant 73% increase in SbWRKY75 transcript levels in the roots of these transiently overexpressing plants (Figure 4A). Conversely, we measured about a 43% decrease in SbWRKY75 transcript levels in S. baicalensis plants transiently expressing an RNAi construct specific for SbWRKY75 (Figure 4B). We then analyzed the relative expression levels of ten baicalin biosynthesis genes in these SbWRKY75 overexpression and RNAi plants. The expression levels of SbCLL-7, SbPAL-1, SbPAL-2, SbCHS-2, SbFNSII-2, SbF6H, and SbUGT were significantly upregulated in the SbWRKY75 overexpression seedlings, showing approximately 0.9-, 1.1-, 1.77-, 0.45-, 7.70-, 1.64-, and 0.17-fold increases, respectively. Conversely, the expression levels of SbCLL-7, SbPAL-1, SbCHS-2, SbCHI, SbUGT, and SbGUS were approximately 84%, 34%, 21%, 59%, 48%, and 84% lower in the SbWRKY75 RNAi seedlings compared to wild-type S. baicalensis seedlings.
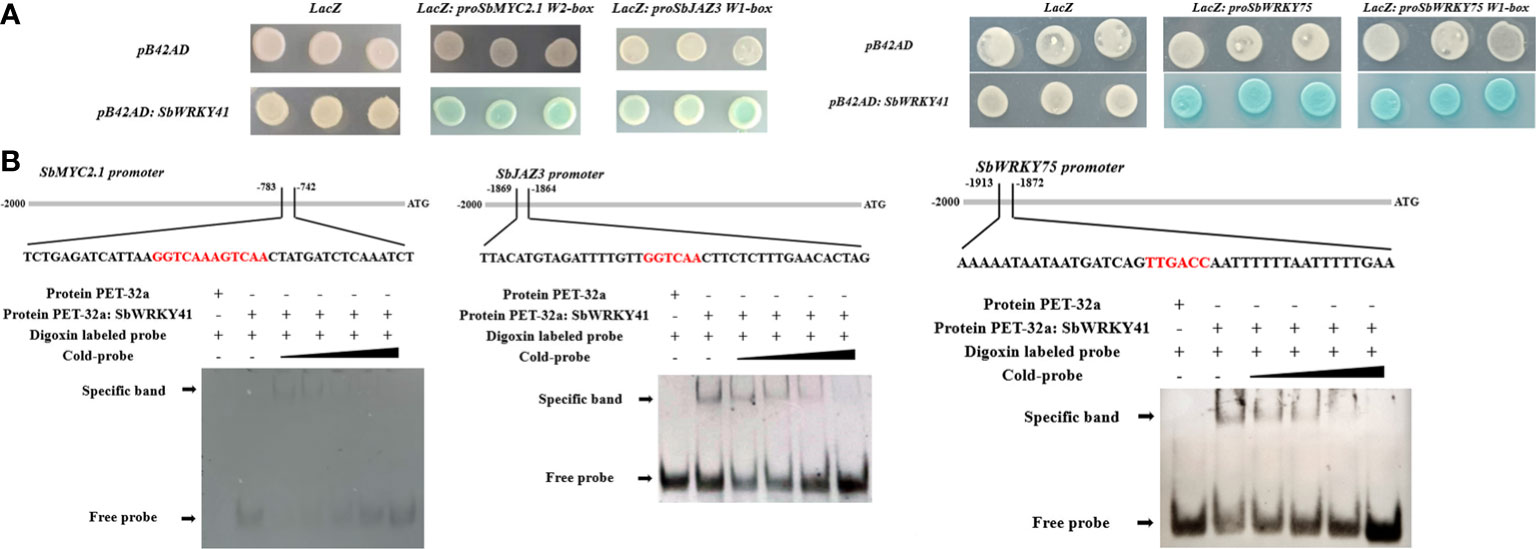
Figure 4 Y1H (A) and EMSA (B) results showing the binding of SbWRKY41 to the W2-box of the SbMYC2.1 promoter, the W1-box of the SbJAZ3 promoter, and the W1-box of the SbWRKY75 promoter.
We also measured baicalin contents in the roots of SbWRKY75 overexpression and RNAi seedlings (Figure 4C). Compared to those in wild-type S. baicalensis seedlings, we detected baicalin and wogonoside levels that were higher by 14% and 13%, respectively, in SbWRKY75 overexpression seedlings, whereas the levels of these compounds decreased by 22% and 19% in the SbWRKY75-RNAi seedlings. These results further support a role for SbWRKY75 in regulating baicalin accumulation.
The altered expression of baicalin biosynthesis genes in MeJA-treated roots suggested that transcriptional regulation contributes to JA-induced baicalin biosynthesis. MYC TFs and JAZ proteins are critical regulators of the JA signaling pathway, making them potential targets for investigation in S. baicalensis (Li et al., 2017). Analysis of SbMYC2.1 and SbMYC2.2 revealed the presence of conserved domains shared with their homologs in Arabidopsis (Figure 5A). In addition, we identified three and one W-boxes in their promoters, respectively, raising the possibility that their transcription may be regulated via direct binding by WRKY TFs. Moreover, using HMMsearch, we identified 11 members of the JAZ family in S. baicalensis and reconstructed a phylogenetic tree together with JAZ proteins from Arabidopsis (Figure 5B). This analysis revealed that SbJAZ3 is similar to SbJAZ7, AtJAZ1, and AtJAZ2, while SbJAZ4 was more similar to AtJAZ5 and AtJAZ6. Additionally, SbJAZ5 clustered together with AtJAZ9, suggesting that these TFs may exert similar functions in plants. However, SbJAZ8, SbJAZ9, and SbJAZ10 were grouped together in a distinct branch, indicating that they may have undergone unique changes in their evolution and have different biological functions.
Turning to our RNA-seq analysis of gene expression in the roots of S. baicalensis following MeJA treatment, we observed similar upregulation of gene expression patterns after MeJA treatment for SbWRKY75, SbJAZ9, SbJAZ7, SbWRKY41, SbMYC2.1, and SbJAZ3 (Figure 5C), indicating that SbWRKY75 and SbWRKY41 are necessary for JA-induced baicalin biosynthesis and suggesting that they may regulate the transcription of both SbMYC2s and SbJAZs.
We investigated the binding of SbWRKY75 to the promoters of SbJAZ7 and SbJAZ9, and of SbWRKY41 to the promoters of SbMYC2.1 and SbJAZ3 using a Y1H assay. We tested various promoter fragments, finding that SbWRKY41 binds to the W2-box motif in the SbMYC2.1 promoter and to the W1-box motif in the SbJAZ3 promoter (Figure 6A). To confirm this interaction, we purified full-length recombinant SbWRKY41 and performed an EMSA using digoxigenin-labeled fragments containing the W2-box motif from the SbMYC2.1 promoter and the W1-box from the SbJAZ3 promoter motif as probes. We determined that SbWRKY41 binds to the W-box motifs of the SbMYC2.1 and SbJAZ3 promoters; importantly, binding decreased when unlabeled probes were added as competitors (Figure 6B). This finding indicates that SbWRKY41 specifically binds to the W-box motifs in the SbMYC2.1 and SbJAZ3 promoters.
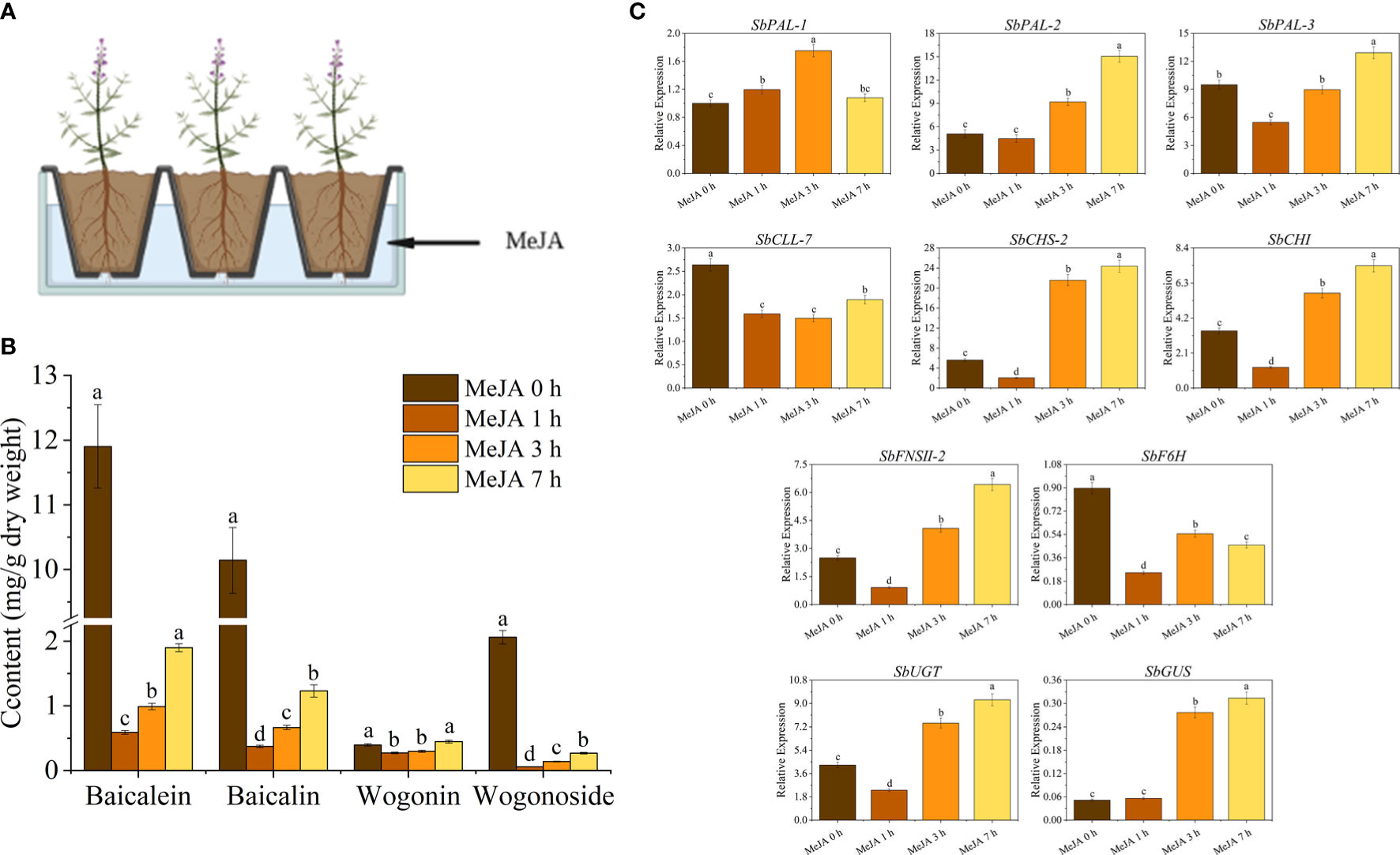
Figure 6 MeJA treatment influenced flavone production and baicalein biosynthesis genes in roots of S. baicalensis seedlings. (A) Schematic diagram of MeJA treatment of the roots of 1-month-old S. baicalensis seedlings; (B) Changes in flavonoid contents in roots of S. baicalensis seedlings treated with MeJA of 0, 1, 3, and 7 h; (C) RT-qPCR analysis of the expression of baicalein biosynthetic pathway-related genes in response to MeJA treatment.
The enrichment of W-box motifs in WRKY promoters suggests that WRKY TFs can regulate their own expression or cross-regulate with other WRKY TFs (Maleck et al., 2000). To investigate the potential regulatory relationship between SbWRKY75 and SbWRKY41, we conducted Y1H and EMSA experiments. We established that SbWRKY41 can modulate the expression of SbWRKY75 and may function upstream of SbWRKY75 (Figure 6B).
S. baicalensis has been a staple of traditional Chinese medicine for over two millennia. JA has been shown to play a role in enhancing flavone production in a number of medicinal plants, including S. baicalensis (Zhou et al., 2012; Horbowicz et al., 2021; Yao et al., 2022). For instance, treating three-week-old cultured roots of S. baicalensis with MeJA resulted in a doubling of total flavone content in roots, with greater increases seen for the aglycones baicalein and wogonin, which increased 230% and 330%, respectively (Kuzovkina et al., 2005). However, the molecular mechanism underlying JA-induced baicalin production and expression of the relevant biosynthesis pathway genes has remained elusive. In this study, we discovered that JA-activated SbWRKY75 regulates SbCLL-7 transcription, while SbWRKY41 regulates SbF6H and SbUGT transcription. Additionally, we uncovered the involvement of SbWRKY41 in JA-regulated baicalin biosynthesis through its transcriptional regulation of SbMYC2.1, SbJAZ3, and SbWRKY75 (Figure 7).
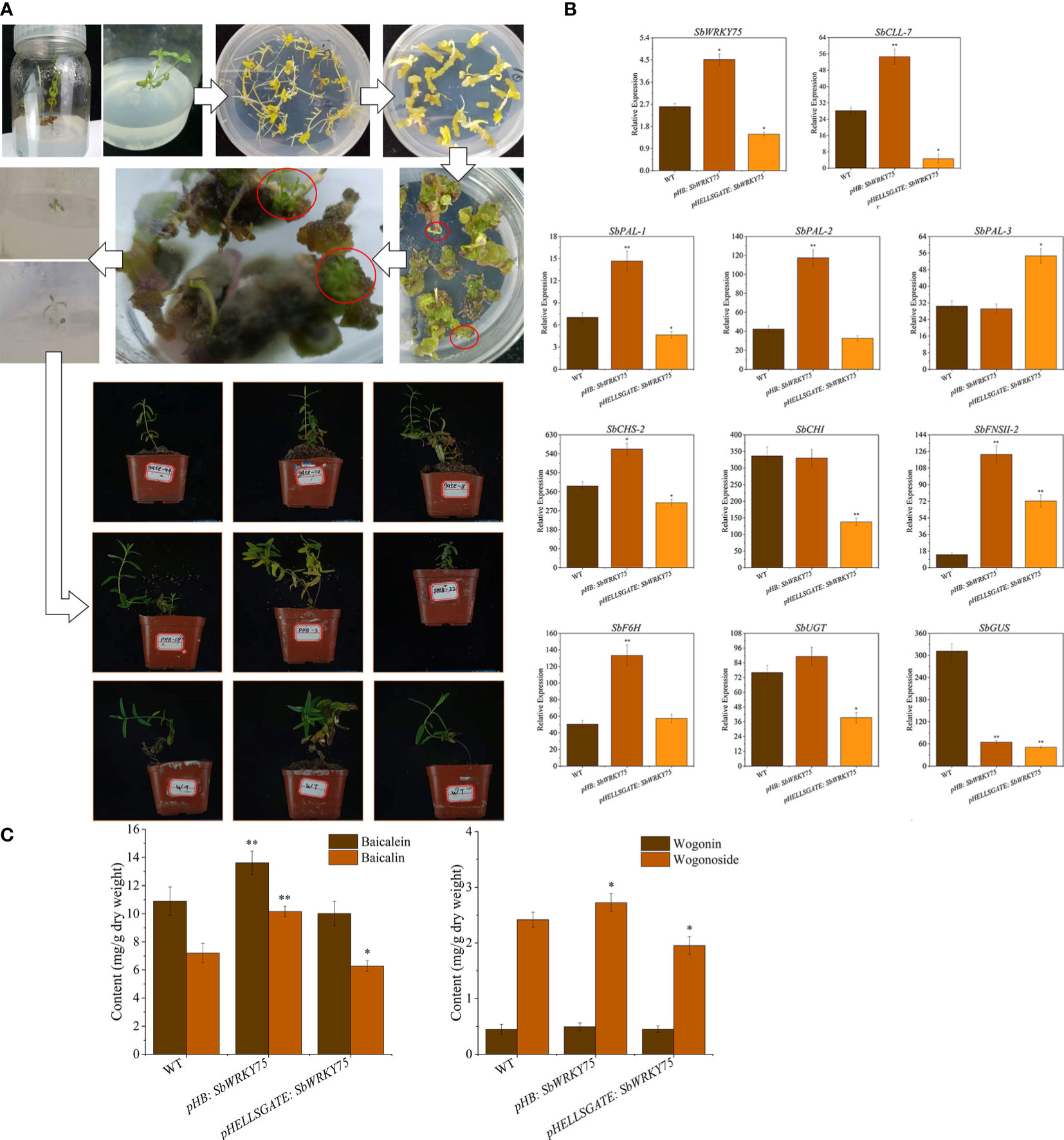
Figure 7 Transgenic S. baicalensis seedlings were obtained via somatic embryogenesis. (A) Schematic overview of somatic embryogenesis in S. baicalensis; (B) Relative expression levels of flavonoid biosynthetic genes and (C) flavonoid contents in transgenic S. baicalensis overexpressing (pHB) or silenced (pHELLSGATE) for SbWRKY75.
The construction of gene regulatory networks is a powerful approach for identifying TF-gene interactions in diverse biological contexts, and WGCNA has demonstrated superior performance in detecting functionally associated gene pairs through examination of pairwise correlations in gene expression patterns (Kuang et al., 2021). In this study, we performed a transcriptome analysis of control S. baicalensis roots and of roots exposed to MeJA treatment for 1, 3, or 7 h to establish a co-regulated gene network associated with baicalin biosynthesis. Through this analysis, we identified nine TF genes and their corresponding regulatory pathways responsible for baicalin biosynthesis. We confirmed the validity of the network through Y1H assays, EMSAs, and dual-LUC assays, which demonstrated that SbWRKY75, SbWRKY41, and SbRAP2.1 bound to the promoters of their respective target genes.
We showed that SbWRKY75 activated transcription from the SbCLL-7 promoter, while SbWRKY41 activated that from two baicalin biosynthesis pathway genes. Our investigation indicated that the overexpression and RNA interference of SbWRKY75 in transiently transformed seedlings significantly influenced the expression of baicalin biosynthetic genes and the accumulation of baicalin. Similarly, TcWRKY1 is a TF encoded by a gene whose expression is induced by MeJA and enhances the expression of DBAT when overexpressed in Taxus chinensis suspension cells. Moreover, RNAi-mediated knockdown of TcWRKY1 transcript levels decreases DBAT transcript levels, thereby decreasing paclitaxel biosynthesis (Li et al., 2013). We established here that SbWRKY75 acts as a positive regulator of baicalin biosynthesis by modulating the expression of genes involved in its biosynthetic pathway. SbWRKY75 overexpression may affect the expression of SbCLL-7 or modulate other signaling pathways, thereby either directly or indirectly influencing the expression of baicalin biosynthesis pathway genes. However, as plants seek to maintain a balance between growth, development, and secondary metabolite biosynthesis, negative feedback regulation can inhibit gene expression when the concentration of secondary metabolites is too high. Thus, some baicalin pathway genes in S. baicalensis may experience a more modest decrease or change in their expression levels. Previous studies have demonstrated that WRKY TFs are closely associated with anthocyanin accumulation in S. baicalensis (Zhang et al., 2022). Our findings suggest that both SbWRKY75 and SbWRKY41 transcriptionally activate the expression of baicalin biosynthesis genes, thereby positively regulating baicalin accumulation.
In addition, we demonstrated that SbWRKY41 also regulates the expression of SbMYC2.1 and SbJAZ3. MYC2 and JAZ proteins are key regulators of the JA signaling pathway, interacting with TFs that control plant secondary metabolism (Li et al., 2022). JA induces the expression of WRKY TF genes and regulates the activity and stability of their encoded proteins through various modifications, including phosphorylation and acetylation (Mao et al., 2007; Liao et al., 2016). Furthermore, several WRKY TFs have been implicated in the JA signaling pathway, such as WRKY40a in peppers (Capsicum annuum), which directly regulates the expression of CaJAZ8 to modulate JA signaling (Raffeiner et al., 2022), and GhMYC2, which serves as a link between the WRKY–MAPK cascade and flavonoid biosynthesis in upland cotton (Gossypium hirsutum) (Wang et al., 2022a). Given the interaction between SbWRKY41, SbMYC2.1, and SbJAZ3, we speculate that JA-responsive SbWRKY41 also regulates baicalin biosynthesis via the JA signaling pathway in S. baicalensis.
Both the Y1H assay and EMSA revealed potential interactions between SbWRKY41 and SbWRKY75, suggesting that these two TFs may form a regulatory network to optimize the signal transduction and cooperatively regulate baicalin biosynthesis. In plants, WRKY TFs interact and cross-regulate one another, and many possess self-regulatory feedback (Turck et al., 2004). For instance, WRKY6 can inhibit its own activity by binding to the W-box in the promoter of its encoding gene and that of WRKY42 in Arabidopsis (Robatzek and Somssich, 2002). Similar observations have been reported for CaWRKY6 and CaWRKY40 in pepper, with CaWRKY6 showing functional and expression similarities with CaWRKY40 (Cai et al., 2015). Based on the regulation of SbWRKY75 by SbWRKY41, it is reasonable to speculate that SbWRKY41 and SbWRKY75 may functionally interact and form a transcriptional network to achieve optimal coordination and fine-tuning of baicalin biosynthesis.
Based on the above findings, we propose a mechanistic model outlining the possible mode of action of SbWRKY75 and SbWRKY41 in regulating the biosynthesis of baicalin in S. baicalensis. In the presence of bioactive JAs, SbWRKY75 and SbWRKY41 are activated, with SbWRKY41 in turn regulating the expression of SbWRKY75. Additionally, SbWRKY41 participates in the JA signaling pathway by modulating the expression of SbMYC2.1 and SbJAZ3. By binding to the W-box cis-elements present in the promoters of baicalin biosynthetic genes, SbWRKY75 and SbWRKY41 activate the transcription of these genes. Our findings provide evidence for a positive regulatory role for SbWRKY75 and SbWRKY41 in baicalin biosynthesis in S. baicalensis, thereby offering a promising strategy for enhancing the production of baicalin through genetic engineering of specific TF genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SF, ZL, KC and WC conceived and designed the experiments. SF, CZ and SQ performed the experiments. SF wrote the paper. YX, ZL, KC and WC revised the manuscript. All authors contributed to the article and approved the submitted version.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (81830109), National Key Research and Development Program of China (2022YFC3501700), the Shanghai local Science and Technology Development Fund Program guided by the Central Government (YDZX20203100002948) and Young Elite Scientists Sponsorship Program by Cast (2021-QNRC1-02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1213662/full#supplementary-material
Cai, H. Y., Yang, S., Yan, Y., Xiao, Z. L., Cheng, J. B., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66 (11), 3163–3174. doi: 10.1093/jxb/erv125
Chen, L., Wu, L., Yang, L., Yu, H. Y., Huang, P. L., Wang, Y. H., et al. (2022). TcJAV3-TcWRKY26 cascade is a missing link in the jasmonate-activated expression of taxol biosynthesis gene DBAT in taxus chinensis. Int. J. Mol. Sci. 23 (21), 13194. doi: 10.3390/ijms232113194
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 (7154), 666–66+. doi: 10.1038/nature06006
Horbowicz, M., Wiczkowski, W., Goraj-Koniarska, J., Miyamoto, K., Ueda, J., Saniewski, M. (2021). Effect of methyl jasmonate on the terpene trilactones, flavonoids, and phenolic acids in ginkgo biloba l. leaves: relevance to leaf senescence. Molecules 26 (15), 4682. doi: 10.3390/molecules26154682
Huang, L. J., Zhang, J. Y., Lin, Z., Yu, P. Y., Lu, M. Z., Li, N. (2022). The AP2/ERF transcription factor ORA59 regulates ethylene-induced phytoalexin synthesis through modulation of an acyltransferase gene expression. J. Cell. Physiol. doi: 10.1002/jcp.30935
Ikegami, F., Matsunae, K., Hisamitsu, M., Kurihara, T., Yamamoto, T., Murakoshi, I. (1995). PURIFICATION AND PROPERTIES OF a PLANT BETA-D-GLUCURONIDASE FROM SCUTELLARIA ROOF. Biol. Pharm. Bull. 18 (11), 1531–1534. doi: 10.1248/bpb.18.1531
Kazan, K., Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6 (3), 686–703. doi: 10.1093/mp/sss128
Kuang, J. F., Wu, C. J., Guo, Y. F., Walther, D., Shan, W., Chen, J. Y., et al. (2021). Deciphering transcriptional regulators of banana fruit ripening by regulatory network analysis. Plant Biotechnol. J. 19 (3), 477–489. doi: 10.1111/pbi.13477
Kuzovkina, I. N., Guseva, A. V., Kovacs, D., Szoke, E., Vdovitchenko, M. Y. (2005). Flavones in genetically transformed scutellaria baicalensis roots and induction of their synthesis by elicitation with methyl jasmonate. Russian J. Plant Physiol. 52 (1), 77–82. doi: 10.1007/s11183-005-0012-y
Li, J., Li, Y., Dang, M. J., Li, S., Chen, S. M., Liu, R. Z., et al. (2022). Jasmonate-responsive transcription factors NnWRKY70a and NnWRKY70b positively regulate benzylisoquinoline alkaloid biosynthesis in lotus (Nelumbo nucifera). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.862915
Li, T., Xu, Y. X., Zhang, L. C., Ji, Y. L., Tan, D. M., Yuan, H., et al. (2017). The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29 (6), 1316–1334. doi: 10.1105/tpc.17.00349
Li, S., Zhang, P., Zhang, M., Fu, C., Yu, L. (2013). Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in taxus chinensis. Plant Biol. 15 (1), 19–26. doi: 10.1111/j.1438-8677.2012.00611.x
Liao, C. J., Lai, Z. B., Lee, S., Yun, D. J., Mengiste, T. (2016). Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 28 (7), 1662–1681. doi: 10.1105/tpc.16.00105
Liu, H., Li, L., Fu, X. Q., Li, Y. P., Chen, T. T., Qin, W., et al. (2023). AaMYB108 is the core factor integrating light and jasmonic acid signaling to regulate artemisinin biosynthesis in artemisia annua. New Phytol. 237 (6), 2224–2237. doi: 10.1111/nph.18702
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and Co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58 (9), 1431–1441. doi: 10.1093/pcp/pcx075
Lv, Z. Y., Guo, Z. Y., Zhang, L. D., Zhang, F. Y., Jiang, W. M., Shen, Q., et al. (2019). Interaction of bZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in artemisia annua. J. Exp. Bot. 70 (15), 3969–3979. doi: 10.1093/jxb/erz166
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K. A., et al. (2000). The transcriptome of arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26 (4), 403–410. doi: 10.1038/82521
Mao, P., Duan, M. R., Wei, C. H., Li, Y. (2007). WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol. 48 (6), 833–842. doi: 10.1093/pcp/pcm058
Nagashima, S., Hirotani, M., Yoshikawa, T. (2000). Purification and characterization of UDP-glucuronate: baicalein 7-o-glucuronosyltransferase from scutellaria baicalensis georgi. cell suspension cultures. Phytochemistry 53 (5), 533–538. doi: 10.1016/s0031-9422(99)00593-2
Park, N. I., Xu, H., Li, X., Kim, Y. S., Lee, M. Y., Park, S. U. (2012). Overexpression of phenylalanine ammonia-lyase improves flavones production in transgenic hairy root cultures of scutellaria baicalensis. Process Biochem. 47 (12), 2575–2580. doi: 10.1016/j.procbio.2012.09.026
Raffeiner, M., Ustun, S., Guerra, T., Spinti, D., Fitzner, M., Sonnewald, S., et al. (2022). The xanthomonas type-III effector XopS stabilizes CaWRKY40a to regulate defense responses and stomatal immunity in pepper (Capsicum annuum). Plant Cell 34 (5), 1684–1708. doi: 10.1093/plcell/koac032
Robatzek, S., Somssich, I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 (9), 1139–1149. doi: 10.1101/gad.222702
Shan, X. Y., Zhang, Y. S., Peng, W., Wang, Z. L., Xie, D. X. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in arabidopsis. J. Exp. Bot. 60 (13), 3849–3860. doi: 10.1093/jxb/erp223
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G. H., et al. (2007). JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 448 (7154), 661–U662. doi: 10.1038/nature05960
Turck, F., Zhou, A., Somssich, I. E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell 16 (10), 2573–2585. doi: 10.1105/tpc.104.024810
Van Moerkercke, A., Duncan, O., Zander, M., Simura, J., Broda, M., Vanden Bossche, R., et al. (2019). A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. United States America 116 (46), 23345–23356. doi: 10.1073/pnas.1911758116
Wang, L. J., Guo, D. Z., Zhao, G. D., Wang, J. Y., Zhang, S. X., Wang, C., et al. (2022a). Group IIc WRKY transcription factors regulate cotton resistance to fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytol. 236 (1), 249–265. doi: 10.1111/nph.18329
Wang, W. T., Hu, S. Y., Yang, J., Zhang, C. J., Zhang, T., Wang, D. H., et al. (2022b). A novel R2R3-MYB transcription factor SbMYB12 positively regulates baicalin biosynthesis in scutellaria baicalensis georgi. Int. J. Mol. Sci. 23 (24), 15452. doi: 10.3390/ijms232415452
Wu, Y., Li, X., Zhang, J. N., Zhao, H. Q., Tan, S. L., Xu, W. H., et al. (2022). ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1042084
Xu, H., Park, N. I., Li, X. H., Kim, Y. K., Lee, S. Y., Park, S. U. (2010). Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in scutellaria baicalensis. Bioresource Technol. 101 (24), 9715–9722. doi: 10.1016/j.biortech.2010.07.083
Yao, H., Wang, F., Bi, Q., Liu, H. L., Liu, L., Xiao, G. H., et al. (2022). Combined analysis of pharmaceutical active ingredients and transcriptomes of glycyrrhiza uralensis under PEG6000-induced drought stress revealed glycyrrhizic acid and flavonoids accumulation via JA-mediated signaling. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.920172
Yuan, M. Y., Shu, G. P., Zhou, J. H., He, P., Xiang, L., Yang, C. X., et al. (2023). AabHLH113 integrates jasmonic acid and abscisic acid signaling to positively regulate artemisinin biosynthesis in artemisia annua. New Phytol. 237 (3), 885–899. doi: 10.1111/nph.18567
Zhang, C. J., Wang, W. T., Wang, D. H., Hu, S. Y., Zhang, Q., Wang, Z. Z., et al. (2022). Genome-wide identification and characterization of the WRKY gene family in scutellaria baicalensis georgi under diverse abiotic stress. Int. J. Mol. Sci. 23 (8), 4225. doi: 10.3390/ijms23084225
Zhao, Q., Chen, X. Y., Martin, C. (2016a). Scutellaria baicalensis, the golden herb from the garden of Chinese medicinal plants. Sci. Bull. 61 (18), 1391–1398. doi: 10.1007/s11434-016-1136-5
Zhao, Q., Yang, J., Cui, M.-Y., Liu, J., Fang, Y., Yan, M., et al. (2019). The reference genome sequence of scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol. Plant 12 (7), 935–950. doi: 10.1016/j.molp.2019.04.002
Zhao, L., Zhang, W., Song, Q., Xuan, Y., Li, K., Cheng, L., et al. (2020). A WRKY transcription factor, TaWRKY40-d, promotes leaf senescence associated with jasmonic acid and abscisic acid pathways in wheat. Plant Biol. 22 (6), 1072–1085. doi: 10.1111/plb.13155
Zhao, Q., Zhang, Y., Wang, G., Hill, L., Weng, J. K., Chen, X. Y., et al. (2016b). A specialized flavone biosynthetic pathway has evolved in the medicinal plant, scutellaria baicalensis. Sci. Adv. 2 (4), e1501780. doi: 10.1126/sciadv.1501780
Zhou, J., Fang, L., Li, X., Guo, L. P., Huang, L. Q. (2012). Jasmonic acid (JA) acts as a signal molecule in LaCl3-induced baicalin synthesis in scutellaria baicalensis seedlings. Biol. Trace Element Res. 148 (3), 392–395. doi: 10.1007/s12011-012-9379-8
Keywords: JA, Baicalin, Scutellaria baicalensis Georgi, SbWRKY75, SbWRKY41
Citation: Fang S, Zhang C, Qiu S, Xiao Y, Chen K, Lv Z and Chen W (2023) SbWRKY75- and SbWRKY41-mediated jasmonic acid signaling regulates baicalin biosynthesis. Front. Plant Sci. 14:1213662. doi: 10.3389/fpls.2023.1213662
Received: 28 April 2023; Accepted: 05 June 2023;
Published: 21 June 2023.
Edited by:
Xu Lu, China Pharmaceutical University, ChinaReviewed by:
Xiaolong Hao, Zhejiang Chinese Medical University, ChinaCopyright © 2023 Fang, Zhang, Qiu, Xiao, Chen, Lv and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Xiao, eGlhb3lpbmd0Y21Ac2h1dGNtLmVkdS5jbg==; Kaixian Chen, a3hjaGVuQHNpbW0uYWMuY24=; Zongyou Lv, em9uZ3lvdWx2QDE2My5jb20=; Wansheng Chen, Y2hlbndhbnNoZW5nQHNodXRjbS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.