
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 June 2023
Sec. Plant Symbiotic Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1211107
This article is part of the Research TopicPlant-Rhizobia Symbiosis and Nitrogen Fixation in LegumesView all 12 articles
Three PHO2-like genes encoding putative ubiquitin-conjugating E2 enzymes of Medicago truncatula were characterized for potential roles in phosphorous (P) homeostasis and symbiotic nitrogen fixation (SNF). All three genes, MtPHO2A, B and C, contain miR399-binding sites characteristic of PHO2 genes in other plant species. Distinct spatiotemporal expression patterns and responsiveness of gene expression to P- and N-deprivation in roots and shoots indicated potential roles, especially for MtPHO2B, in P and N homeostasis. Phenotypic analysis of pho2 mutants revealed that MtPHO2B is integral to Pi homeostasis, affecting Pi allocation during plant growth under nutrient-replete conditions, while MtPHO2C had a limited role in controlling Pi homeostasis. Genetic analysis also revealed a connection between Pi allocation, plant growth and SNF performance. Under N-limited, SNF conditions, Pi allocation to different organs was dependent on MtPHO2B and, to a lesser extent, MtPHO2C and MtPHO2A. MtPHO2A also affected Pi homeostasis associated with nodule formation. Thus, MtPHO2 genes play roles in systemic and localized, i.e., nodule, P homeostasis affecting SNF.
Nitrogen (N) and phosphorus (P) are essential macronutrients for plant growth and development. Low availability of these nutrients in most soils limits crop production necessitating the use of fertilizers to secure food production. Symbiotic nitrogen fixation (SNF) in legumes is the primary natural source of nitrogen in agroecosystems, although industrial nitrogen-fertilizers now provide most of the nitrogen for crop production. Use and loss to the environment of industrial N-fertilizer is not sustainable and more needs to be done to boost the use of legumes and N derived from SNF in agriculture to remedy this (Udvardi et al., 2021).
SNF results from a mutualistic symbiosis between soil bacteria, called rhizobia, and legumes during which the bacteria reduce atmospheric di-nitrogen into ammonia within specialized root organs called nodules. In exchange for ammonia provided to the plant, the bacteria receive carbon (C) in the form of organic acids and other nutrients, including P for instance (Udvardi & Poole, 2013). Complex regulatory networks have evolved to control acquisition and allocation of C, N, P and other essential nutrients for optimal growth, development and functioning of plant organs and the plant as a whole (Gautrat et al., 2021; Helliwell, 2022), although our understanding of these networks remains incomplete.
SNF is sensitive to environmental stress, including P-deficiency. Nodules contain relatively high concentrations of P especially in nucleic acids (plant and bacterial DNA and RNA), which underpin protein synthesis and high metabolic activity, including SNF (Sulieman et al., 2013; Cabeza et al., 2014). P limitation severely inhibits root nodule organogenesis and SNF (Hernandez et al., 2009). Maintenance of P homeostasis in nodules is considered a main adaptive strategy to maintain symbiotic performance under P-deficiency, although underlying mechanisms are poorly understood. (Cabeza et al., 2014; Nasr Esfahani et al., 2017).
In non-legumes, there is growing evidence for crosstalk between P and N regulation of nutrient acquisition, growth and metabolism (Hu et al., 2019; Medici et al., 2019; Ueda et al., 2020). In Arabidopsis and other species, P homeostasis is systemically regulated by the transcriptional activators Phosphate Starvation Response 1 (PHR1/PHL) (Rubio et al., 2001; Bustos et al., 2010), the negative regulator SPX-like (Puga et al., 2014; Wang et al., 2014), and tuned by the balance of specific microRNAs (miR399 and miR827) (Chiou et al., 2006; Lin et al., 2013) and long non-coding RNAs (IPS-like) (Franco-Zorrilla et al., 2007) in coordination with PHOSPHATE2 (PHO2) (Aung et al., 2006; Bari et al., 2006; Lin et al., 2008). PHO2 is a ubiquitin-conjugating (UBC) E2 enzyme involved in the degradation of multiple type of Pi transporters including members of the PHT1/PT (PHOSPHATE TRANSPORTER 1) protein family, PHOSPHATE 1 (PHO1) and PHF1 (PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR 1) (Bari et al., 2006; Liu et al., 2012; Cao et al., 2014; Park et al., 2014; Ouyang et al., 2016; Pacak et al., 2016). The PHO2-miR399-IPS1 and PHO2-NLA-miR827 regulatory modules (Franco-Zorrilla et al., 2007; Kant et al., 2011; Lin et al., 2013) function independently but cooperatively, regulating acquisition and root-to-shoot translocation of Pi in response to P and N availability, protecting aboveground organs from excessive Pi accumulation. The physiological role of PHO2 protein in maintaining whole-plant Pi-homeostasis has been described for rice (Cao et al., 2014), wheat (Ouyang et al., 2016) and Arabidopsis (Bari et al., 2006). Further, Arabidopsis PHO2 is considered a local and systemic integrator of N availability in phosphate systemic signaling (Medici et al., 2019).
Although the mechanisms of N and P crosstalk in legumes related to nodule development and SNF are largely unknown, there is evidence that systemic signaling pathways controlling N fixation and acquisition are linked to phosphate systemic signaling. Phosphorus deficiency influence rhizobial infection and nodulation through miR2111/Too Much Love (TML), PHR (Phosphate Starvation Response) - RICs (Rhizobium-induced CLE Peptides) - NARK (Nodulation Autoregulation Receptor Kinase) and through PHR depending on P homeostasis regulatory modules in legumes (Gautrat et al., 2021; Zhong et al., 2023 and references therein). Some Phosphate Transporter (PHT/PT) and PHOSPHATE1 (PHO1)-type P transporters, downstream targets of PHR transcription factors, have been assigned an important role in maintaining Pi homeostasis in nodules, supporting SNF (Qin et al., 2012; Chen et al., 2019; Nguyen et al., 2021). Likewise, two alfalfa PHO2 genes have been implicated in systemic P-homeostasis, although their roles during symbiosis have not been explored (Miller et al., 2022) A Medicago truncatula PHO2-like gene contributes to quantitative variation in nodulation in this species, but the underlying mechanism remains unknown (Curtin et al., 2017). Medicago truncatula has two other PHO2-like genes, although their roles, if any, in P-homeostasis and SNF also remain unknown (Miller et al., 2022).
Here, we explore the roles of the three Medicago truncatula PHO2-like genes in P homeostasis and SNF. Our results implicate PHO2 proteins in systemic Pi homeostasis and the support of SNF.
Medicago truncatula ecotype R108 (HM340) was used in all experiments as a wild-type control as this is the genetic background of the Tnt1, CRISPR/Cas9 and TALEN pho2 mutants. Offspring of the CRISPR/Cas9 line WPT210-9, described previously (Curtin et al. (2017), were screened to isolate homozygous pho2-ACRISPR and pho2-BCRISPR mutants. Similarly, offspring of TALEN line WPT52-4-8 (Cermak et al. (2017), were used to identify the homozygous mutant pho2-BTALEN. Offspring of self-pollinated plants from the WPT210-9 and WPT52-4-8 lines were genotyped by combining PCR amplification and NlaIV and HaeIII restriction enzyme digestion assays, respectively. Changes in genomic DNA were confirmed by Sanger sequencing of undigested PCR products of the homozygous mutant lines and wild-type plants. Sequence comparisons were performed using Geneious software.
The pho2-ATnt1 homozygous line was obtained from Tnt1 line NF12360 (Curtin et al., 2017). Tnt1 line NF16248 was used to isolate the homozygous exonic mutant allele pho2-CTnt1 by PCR genotyping. Tnt1-specific and gene-specific primers are listed in Supplementary Table 2. Pho2-ACRISPR, pho2-BCRISPR and pho2-CTnt1 were the mutant alleles selected for phenotypic analysis, while the pho2-ATnt1 and pho2-BTALEN mutant alleles were used to confirm phenotypes (Supplementary Figure 6).
Seeds were sterilized, scarified, and stratified as described before (Kryvoruchko et al., 2018). Seedlings with fully opened cotyledons and similar radicles were individually transferred into 2” x 7” plastic cones (Stuewe & Sons Inc.) containing a mixture (3:1, v/v) of sterilized Turface (calcined [illite] clay) and Vermiculite (Sun Gro Horticulture). Seedlings were fertilized with one-half-strength B&D solution (Broughton & Dilworth, 1971). Seven days after transplanting, seedlings were watered with full-strength modified B&D nutrient solution. Nitrogen was supplied as a 2:1 mixture of KNO3 and NH4NO3, while phosphorus was supplied as KH2PO4. The different treatments and final concentrations were “control” (8 mM N and 0.5 mM P), “reduced-P” (8 mM N and 20 μM P), “controlsym” (0.25 mM N and 0.5 mM P) and “reduced-Psym” (0.25 mM N and 20 μM P). K2SO4 was used to balance the potassium concentration in the reduced-P and reduced-Psym solutions. All other macro- and micro-nutrients of the B&D nutrient solution were provided as specified (Broughton & Dilworth, 1971).
For the non-symbiotic experiments, plants were watered with control or reduced-P nutrient solutions, while for the symbiotic experiments they were inoculated with 50 mL suspension (OD600 ∼0.02) of Sinorhizobium meliloti strain 1021 (Meade et al., 1982) in controlsym or reduced-Psym nutrient solutions. Plants were grown under controlled conditions of light (200 μmol m-2 s-1, 16h day/8h night), constant temperature of 22°C, and 40% relative humidity, and irrigated twice per week - once with the corresponding nutrient solution and once with B&D without N or P, to avoid accumulation of N or P. Plants were harvested four weeks after starting treatments. Each plant was removed from its cone and the root carefully washed with water to remove substrate while avoiding loss of roots and nodules. Independent plant tissues were frozen in liquid nitrogen and stored at −80°C for later use, or oven dried at 60°C and weighed. Total plant dry weight (DW) was the sum of the shoot and root dry weights.
The acetylene reduction assay (ARA) was carried out as previously described (Hardy et al., 1968). Briefly, four weeks after inoculation, entire root systems were transferred onto sterile Whatman paper strips placed inside 12-mL glass vials containing 2 mL of sterile distilled water. The tubes were sealed with rubber stoppers. Each tube contained roots from independent plants. Samples were incubated in dark in the presence of 10% (v/v) acetylene at 28°C for up to 16 h. Ethylene and acetylene concentrations were measured using an Agilent 7890A gas chromatograph (Agilent Technologies). Serial dilutions of a known quantity of ethylene were used to make standard curves of GC chromatogram peak area to calculate the amount of ethylene produced. The amount of ethylene produced was determined by measuring the area of the ethylene peak relative to background. Nitrogenase activity was calculated as the amount of ethylene produced per unit root dry weight. The number and biomass of the nodules was determined by detaching them from the roots.
Four weeks after treatments, roots, shoots, and nodules were collected into liquid nitrogen. After grinding in liquid nitrogen, total RNA was extracted using the TRIzol reagent (Life Technologies). Residual genomic DNA was removed using Turbo DNase I (Ambion). RNA was quantified using a Nanodrop Spectrophotometer ND-100 (NanoDrop Technologies). For qPCR, reverse transcription was carried out using SuperScript III Reverse Transcriptase (Invitrogen) and oligo(dT)20 primer, as describe previously. Transcript levels were normalized using the geometric mean of three housekeeping genes, MtPI4K (Medtr3g091400), MtPTB2 (Medtr3g090960), and MtUBC28 (Medtr7g116940), whose transcript levels were stable across all the samples analyzed (Kakar et al., 2008). Three biological replicates were included per gene. qPCR cycle threshold (Ct) values were analyzed using the ΔΔCt method (Livak & Schmittgen, 2001). Primer sequences used in this analysis are listed in Supplementary Table 2. Sequence alignments and the design of gene specific primers were performed using Geneious software.
The genome assembly of Jemalong A17 (Mt4.0 v1) and Medicago R108 (v0.95) ecotypes from Phytozome and the Legume Information System (LIS) were consulted to retrieve DNA sequences and gene structures. The Integrative Genomics Viewer (IGV) software (https://igv.org) was used to visualize the original raw RNA-seq data used for the MtSSPdb (https://mtsspdb.zhaolab.org/database) (Boschiero et al., 2020) and confirm the 5’ and 3’ UTR regions as well as the expression profiles.
Precursor sequences of the pre-miR399s from several plant species were obtained from miRBase (www.mirbase.org) and used to identify Medicago truncatula miR399. The miR399 sequences and the potential miR399-binding sites (miR399BS) were validated using psRNATarget with default parameters (Dai et al., 2018). The consensus miR399 sequence, miR399 sequence logo, potential PHO-like ([G(G/T/A) (C/T/A)GTGG]; Mukatira et al., 2001) and P1BS (GnATATnC; Rubio et al., 2001) cis-regulatory elements were generated using Geneious software.
Protein sequences were extracted from Phytozome and NCBI Protein databases. The circular phylogenetic tree was constructed from a ClustalW multiple sequence alignment of the full‐length protein sequences in Geneious software using Juker-Cantor as the genetic distance model and the unweighted pair group method with arithmetic mean (UPGMA) as a tree build method with 500 replicates and 60% of support threshold. The gene IDs encoding each protein are described in Supplementary Table 1.
Soluble inorganic phosphate (Pi) was measured in four-week-old plants after treatments according to Miller et al. (2022), with minor modifications. Briefly, frozen tissue samples (leaves, roots and nodules) were ground. Deionized water was added to the homogenized samples, mixed, and centrifuged at 13,000 g for 3 minutes, and the clarified supernatant was transferred to a clean tube to quantify Pi content. Aliquots were diluted appropriately and mixed in 96 deep-well plates with HCl and malachite green reagent. After 15 minutes of incubation at room temperature, light absorbance was measured at 660 nm. The sample Pi concentration was determined by reference to a calibration curve using K2HPO4. Pi concentration was calculated based on fresh weight of samples. Measurements were performed in triplicate in three independent biological replicates.
Data on gene expression, biomass, Pi content, etc. were analyzed statistically for mean comparisons, between wild type and mutant alleles or between control and treatment conditions, by one-way analysis of variance (ANOVA) and t-tests (P<0.05). Statistical analyses and graphs were generated using GraphPad Prism software.
M. truncatula PHO2 genes were identified via BLASTP searches using the Phytozome and the Legume Information System (LIS) as databases and known plant PHO2 proteins as queries (Supplementary Table 1). Three separate genes were identified and named MtPHO2-A (Medtr4g020620), MtPHO2-B (Medtr2g013650) and MtPHO2-C (Medtr4g088835) (Figure 1A), keeping Medtr4g020620 as -A, the first PHO2 gene described in M. truncatula (Curtin et al., 2017). Note that Wang et al. (2017) described Medtr2g013650 as the only PHO2 gene in Medicago, which we named MtPHO2-B to distinguish it from the other two PHO2 genes we identified.
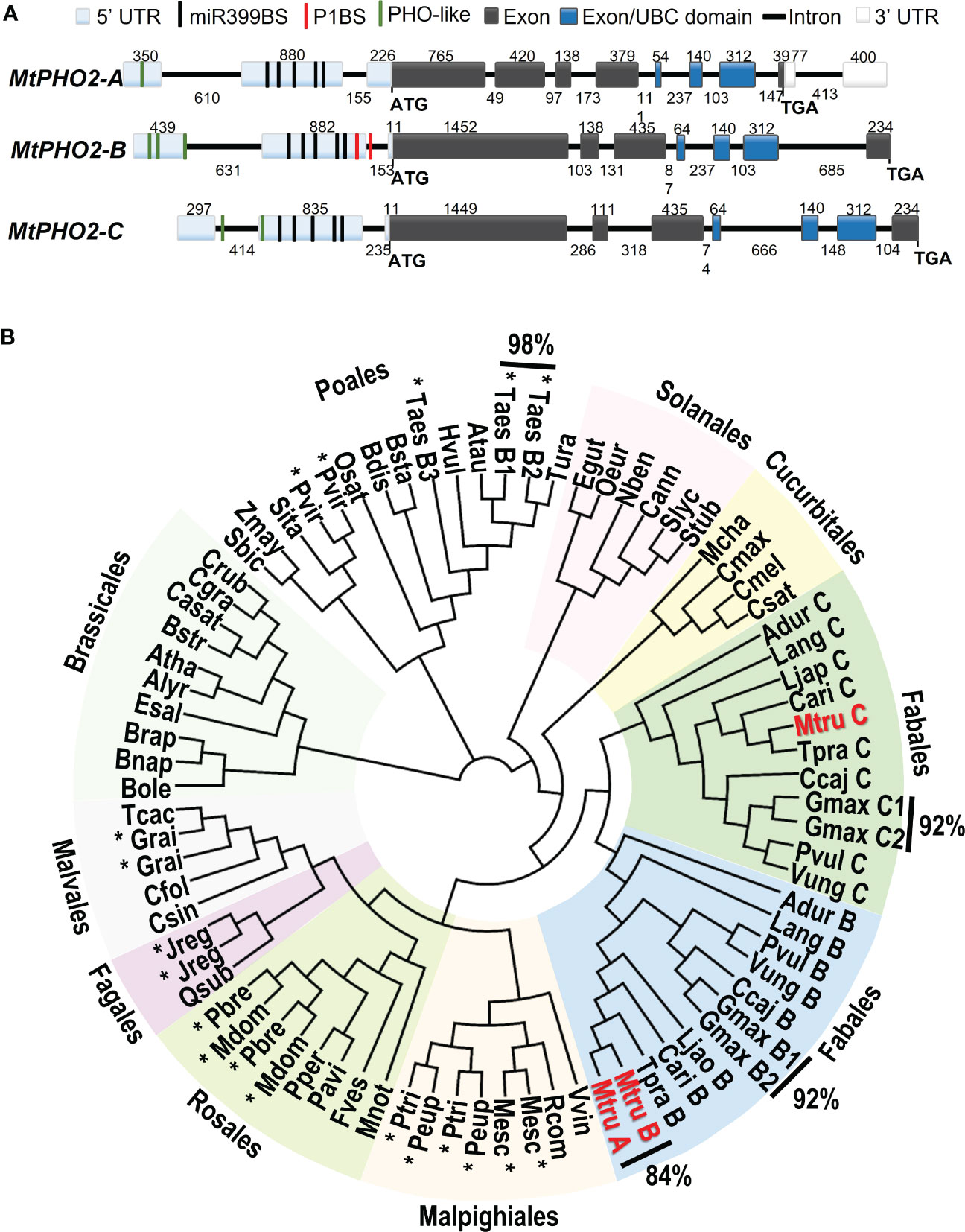
Figure 1 Gene structure and phylogenetic relationships of Medicago truncatula PHO2-like genes. (A) Gene structure of the three PHO2 genes present in the A17 Medicago truncatula genome (Mt4.0v1) and validated using RNA-seq data. Only splicing variant MtPHO2-A.2 is shown here (see Supplementary Figure 1A for other slicing variants). Exons are shown as grey or darker blue boxes, with the latter encoding the ubiquitin-conjugating (UBC) domain (see also Supplementary Figure 1B). UTRs are shaded light blue (5’ of the coding sequence) or white boxes (3’ of the coding sequence). The black, green and red lines depict the position within the 5’ UTR of the five potential miR399-binding sites (miR399BS), PHO-like elements, and the PHR1 binding sites (P1BS). Promoters and cis-regulatory motifs are detailed in Supplementary Figure 2. Gene structures are drawn to scale, and the associated numbers indicate sizes (numbers above exons and below introns). (B) Phylogenetic tree of the PHO2-like gene family in plants. The different orders are marked next to different colored backgrounds. The Medicago truncatula PHO2 genes are indicated in red (MtruA, MtPHO2-A; MtruB, MtPHO2-B; MtruC, MtPHO2-C). The * indicates gene duplication events outside of the order Fabales. Values outside of the Phylogenetic tree show the percentages of homology between MtPHO2A and B proteins as well as some other plant species. The protein sequences were extracted from Phytozome and NCBI Protein databases. The circular phylogenetic tree was constructed from a ClustalW alignment of the full‐length protein sequences in Geneious software using Juker-Cantor as the genetic distance model and UPGMA as a tree build method with 500 replicates and 60% of support threshold. The gene IDs encoding each protein are described in Supplementary Table S1.
Detailed in silico analysis of the annotated DNA sequences and visualization of raw RNA-seq data (Boschiero et al., 2020), with the Integrative Genomics Viewer (IGV) (Robinson et al., 2011), were used to validate gene structures. These three MtPHO2 genes shared the typical number (7-9) and arrangement of exons and regulatory elements in the proximal promoter region/5’-UTR (Figure 1A).
Analyses of the 5′-UTR of the three MtPHO2 genes showed five putative miR399-binding sites (miR399-BS) and PHO-like elements, while only tMtPHO2-B presented putative PHR1 binding sites (P1BS) in its 5′-UTR (Figure 1A; Supplementary Figure 2A). Using mature and stem-loop sequences of known miRNA399s obtained from miRbase (www.mirbase.org), up to 10 different miRNA399 species (miR399a to j) were identified in the M. truncatula genome. Sequence alignment distinguished up to 5 different variants of miR399 (Supplementary Figure 2B) that potentially could target the miR399BS identified within the 5′-UTR of the MtPHO2 genes. Depending on the miR399BS, mismatches were identified in the central or toward the 3′ ends of the miR399s sequences.
The MtPHO2-A gene was predicted to have three splicing isoforms (Supplementary Figure 1A). Amino acid sequence alignment and phylogenetic analysis of the 81 PHO2 proteins identified in the sequenced plant genomes revealed phylogenetic patterns, grouping into clades. Except for the Poales order, most of the PHO2 identified in the databases fell into different plant orders of Eudicots. Gene duplication events were identified in monocots and eudicots, with close evolutionary relationships (92-98% homology). In legumes (order Fabales), two distinct branches associated with the well-known genome duplication event (Cannon et al., 2006) were identified. MtPHO2-A and MtPHO2-B appeared clustered together showing 84% homology at the protein level, while MtPHO2-C clustered in a duplicated branch (Figure 1B) sharing 68% homology with MtPHO2-B and 60% with MtPHO2-A (data not shown).
A detailed analysis revealed that four of the five possible MtPHO2 protein variants (including the three MtPHO2-A variants) conserve the distinctive ubiquitin-conjugating catalytic (UBCc) domain at the C-terminus, including E3 ligase interaction residues and the E2 active site cysteine according to PROSITE database (https://prosite.expasy.org/). MtPHO2-A.3 protein variant was the only exception, lacking the UBCc domain (Supplementary Figure 1B).
We investigated the spatial-temporal expression patterns of MtPHO2 genes. Relative transcript levels, based on sequence fragments per kilobase of transcript per million reads mapped (FPKM) (MtSSPdb; https://mtsspdb.zhaolab.org/database), revealed that MtPHO2 genes were ubiquitously expressed in different organs and pod developmental stages, although their expression was consistently higher in the root. MtPHO2-B had the highest expression level in all the organs evaluated, followed by MtPHO2-C. MtPHO2-A expression was relatively low in all organs (Figure 2A). Similar expression differences between these genes were detected during nodulation, with only slight changes between 10- and 28-days post inoculation (dpi). Again, MtPHO2-A expression levels were very low compared to the other two MtPHO2 genes (Figure 3A).
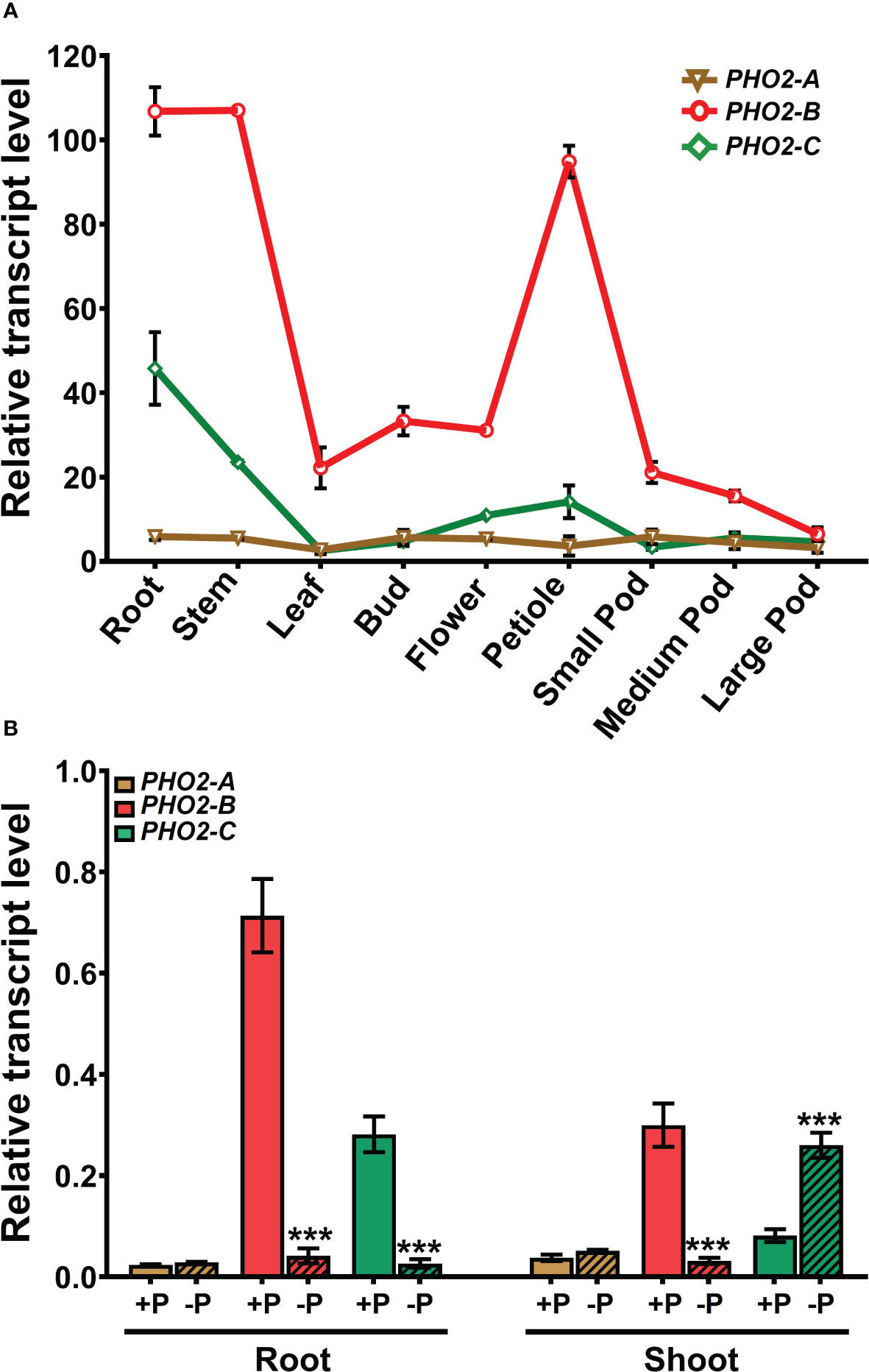
Figure 2 Expression profiles of Medicago truncatula PHO2-like genes in various organs and treatments.(A) Relative transcript levels of MtPHO2-A, B, C in different organs derived from RNA-seq data with numbers indicating fragments per kilobase of transcript per million reads mapped (FPKM) and represent the average of three biological replicates with standard errors. Further details are given in Boschiero et al. (2020) and the M. truncatula SSP database (MtSSPdb). (B) Relative transcript levels of MtPHO2-A, B and C, determined by qRT-PCR analysis of roots and shoots of four-week-old plants grown under optimal P-nutrition (+P, 0.5 mM Pi) and limiting-P (-P, 20 μM Pi) conditions, as shown in Figure 4 and as described in the methodology. PCR primer sequences are presented in Supplementary Table S2. Data shown are the mean and SEM of three independent experiments (n=6). For each replicate, transcript levels were normalized against two housekeeping genes, MtPTB2 and MtPDF2; asterisks indicate significant differences between the optimal and reduced P (*p < 0.05, **p < 0.01, ***p < 0.001) calculated using two-tailed Student’s t-tests.
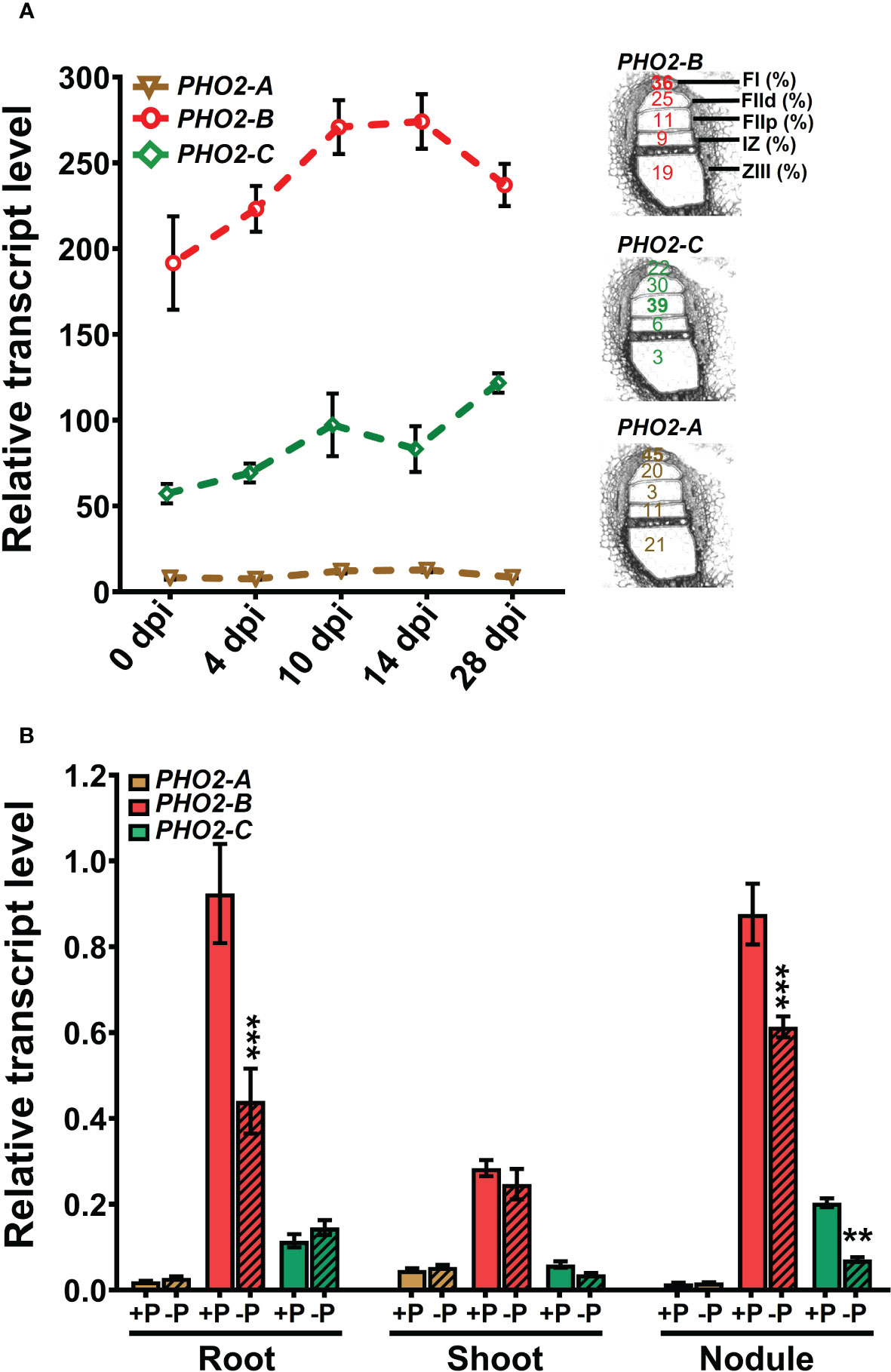
Figure 3 Expression profiles of Medicago truncatula PHO2 genes in various organs and treatments during nodulation. (A) Relative transcript levels of MtPHO2-A, B and C in nodules over time. Left, RNA-seq data expressed as fragments per kilobase of transcript per million reads mapped (FPKM) represent the average of three biological replicates with standard errors. Further details are given in the MtSSPdb (Boschiero et al. (2020). Right: scheme of the five laser micro-dissected regions of M. truncatula nodules, containing the percentage of normalized counts per region for each PHO2 gene, according to Roux et al. (2014). (B) Relative transcript levels, quantified by qRT-PCR, of MtPHO2-A, B and C genes in four-week-old roots, shoots and nodules of plants grown under symbiotic nitrogen fixation conditions (0.5 mM N and inoculated with S. meliloti strain Sm1021) with optimal-P (+P, 0.5 mM P) or reduced-P (-P, 20 μM P), under the same conditions as those shown in Figures 5–8. PCR primer sequences are presented in Supplementary Table 2. Data are the mean and SEM of three independent experiments (n=6 for root and shoot, n=18 for nodules). For each replicate, transcript levels were normalized against two housekeeping genes (MtPTB2 and MtPDF2); asterisks indicate significant differences between optimal- and reduced-P, calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
Expression profiles determined by qPCR confirmed that in the absence of nutritional deficits, MtPHO2-B was the primary PHO2 transcript in roots and shoots, followed by MtPHO2-C and MtPHO2-A (Figure 2B). P-limitation resulted in lower transcript levels of MtPHO2-B and MtPHO2-C in roots. In shoots, only MtPHO2-B was down-regulated under P-limitation while MtPHO2-C was up-regulated (Figure 2B; Supplementary Figure 3A). N-limitation resulted in up-regulation of MtPHO2-B and MtPHO2-C in roots and shoots, but no significant change in MtPHO2-A transcript levels (Supplementary Figure 3B).
Expression profiles of the three MtPHO2 genes in symbiotic, nitrogen-fixing plants were similar to those of non-symbiotic plants, with MtPHO2-B exhibiting the highest transcript levels in roots, shoots, and nodules, followed by MtPHO2-C and MtPHO2-A (Figure 3B; Supplementary Figure 3A). P-limitation under symbiotic conditions down-regulated MtPHO2-B in roots and nodules but not shoots. MtPHO2-B and MtPHO2-C, but not MtPHO2-A, were down-regulated in nodules in response to P-limitation (Figure 3B).
To explore the function of the MtPHO2 proteins, homozygous pho2-ACRISPR, pho2-BCRISPR, pho2-ATnt1, pho2-BTALEN and pho2-CTnt1 mutants were used (Supplementary Figure 4). Under optimal, nutrient-replete conditions, lack of MtPHO2-B or MtPHO2-C reduced root and shoot growth, especially in the case of the pho2-BCRISPR mutant (Figures 4A, B). Measurement of Pi accumulation in roots, young leaves and fully expanded mature leaves revealed that only the pho2-BCRISPR mutant had significantly higher Pi accumulation in mature leaves, compared to the WT, while the pho2-CTnt1 mutant showed a slight but significant reduction in Pi content in such leaves (Figure 4C). Plants of each mutant line were also grown in rich soil (Metro-Mix) in a greenhouse under optimal nutritional conditions for analysis of late developmental phenotypes and seed replication. All three mutants grew less than WT plants, although the pho2-BCRISPR mutant was the most severely affected, followed by pho2-CTnt1 and pho2-ACRISPR mutants (Supplementary Figure 5). Stunted growth was accompanied by a decrease in seed production in both the pho2-BCRISPR and pho2-CTnt1 mutants, especially the former, which also displayed symptoms of necrosis in its mature leaves (Supplementary Figure 5).
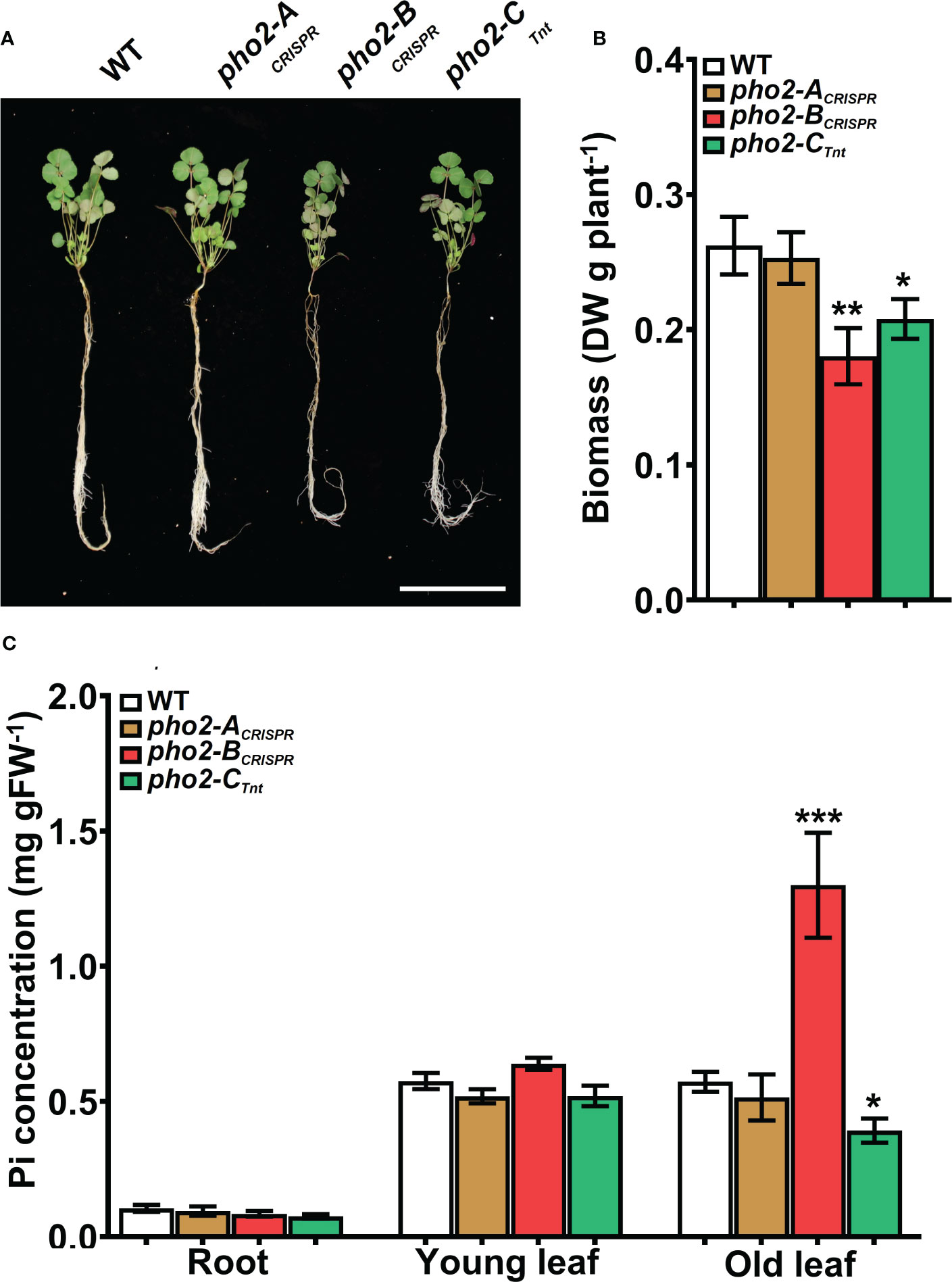
Figure 4 Performance of the pho2 mutants under optimal nutritional conditions. (A) Representative four-week-old plants growth under optimal nutritional conditions, including 0.5 mM Pi. Scale bar = 10 cm. (B) Plant dry weight. (C) Free phosphate (Pi) concentration in roots, young and old leaves. Data shown are the mean and SEM of three independent experiments (n=5 per experiment). Asterisks indicate significant differences between the wild type and the mutants calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
All three mutants, pho2-ACRISPR, pho2-BCRISPR and pho2-CTnt1 exhibited reduced growth and biomass under optimal symbiotic conditions, including high Pi (0.5 mM; Figure 5). Again, mature leaves of the pho2-BCRISPR mutant, but not the other mutants, exhibited necrotic symptoms (Figures 5A, B). Measurements of organ Pi content revealed hyper-accumulation of Pi in young and especially mature leaves of the pho2-BCRISPR, which was mirrored by a drastic decrease in Pi content of its roots. Pi also accumulated in older leaves of both the pho2-ACRISPR and pho2-CTnt1 mutants relative to the WT (Figure 5C).
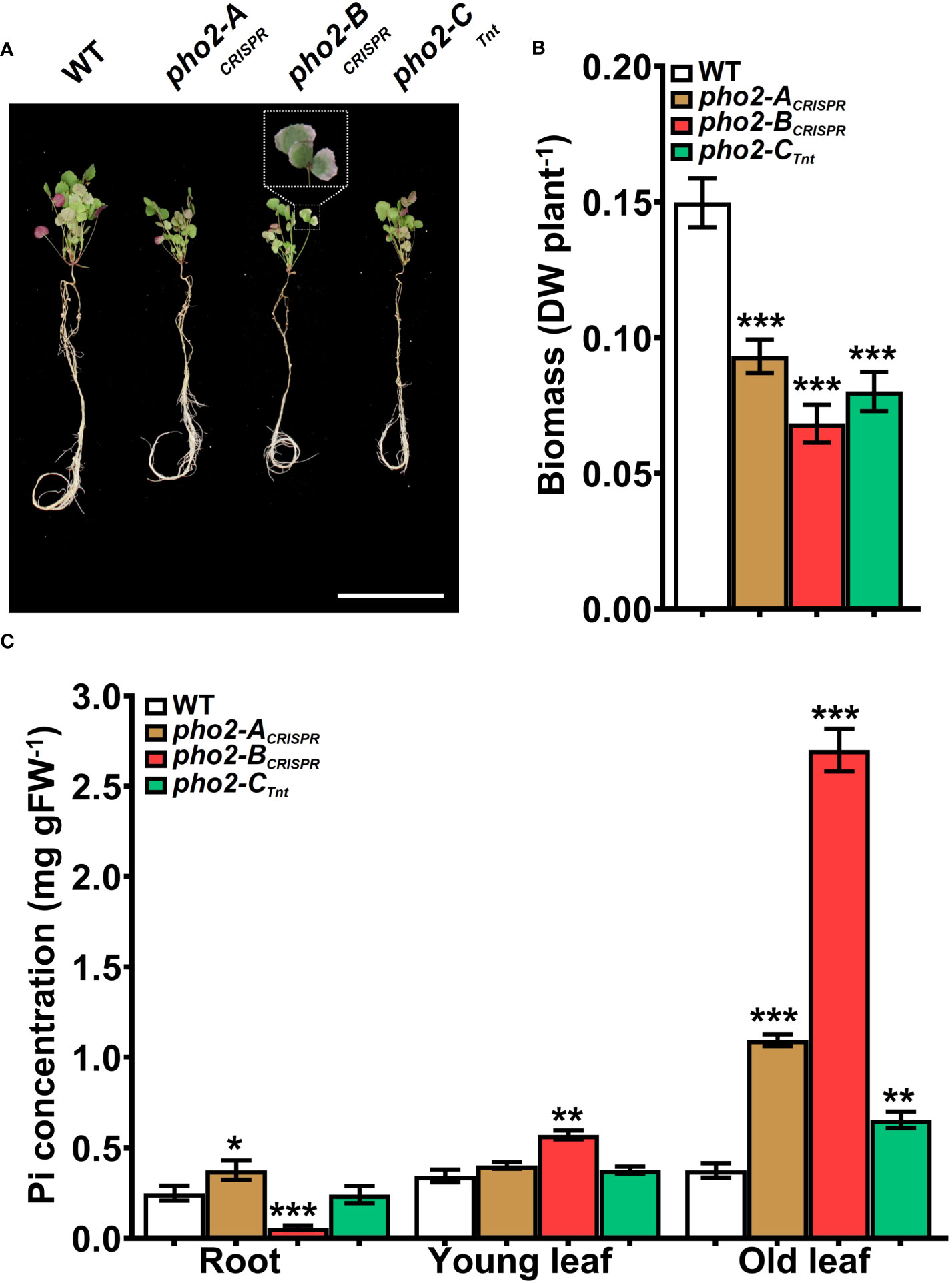
Figure 5 Performance of the Mtpho2 mutants under symbiotic nitrogen fixation conditions. Plants were inoculated with S. meliloti strain Sm1021 and given optimal-P (0.5 mM Pi). (A) Representative four-week-old plants. Scale bar = 10 cm. (B) Plant dry weight. (C) Free phosphate (Pi) concentration in roots, young and old leaves. Data shown are the mean and SEM of three independent experiments (n=5/experiment). Asterisks indicate significant differences between the wild type and the mutants calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
Traits related to SNF were differentially affected in the three mutants. The pho2-ACRISPR mutant showed a reduced number of nodules compared to the WT, but with similar biomass and nitrogenase activity (Figures 6A-E). The pho2-BCRISPR mutant was affected in all the traits evaluated, with reduced number and biomass of nodules, as well as nitrogen fixation capacity. Likewise, pho2-CTnt1 exhibited reduced nodule number, biomass and nitrogen fixation (Figures 6A-E). Loss of gene function had a variable effect on nodule Pi concentration, with the pho2-BCRISPR accumulating less, pho2-ACRISPR accumulating more, and pho2-CTnt1 accumulating the same concentration as the WT (Figure 6B).
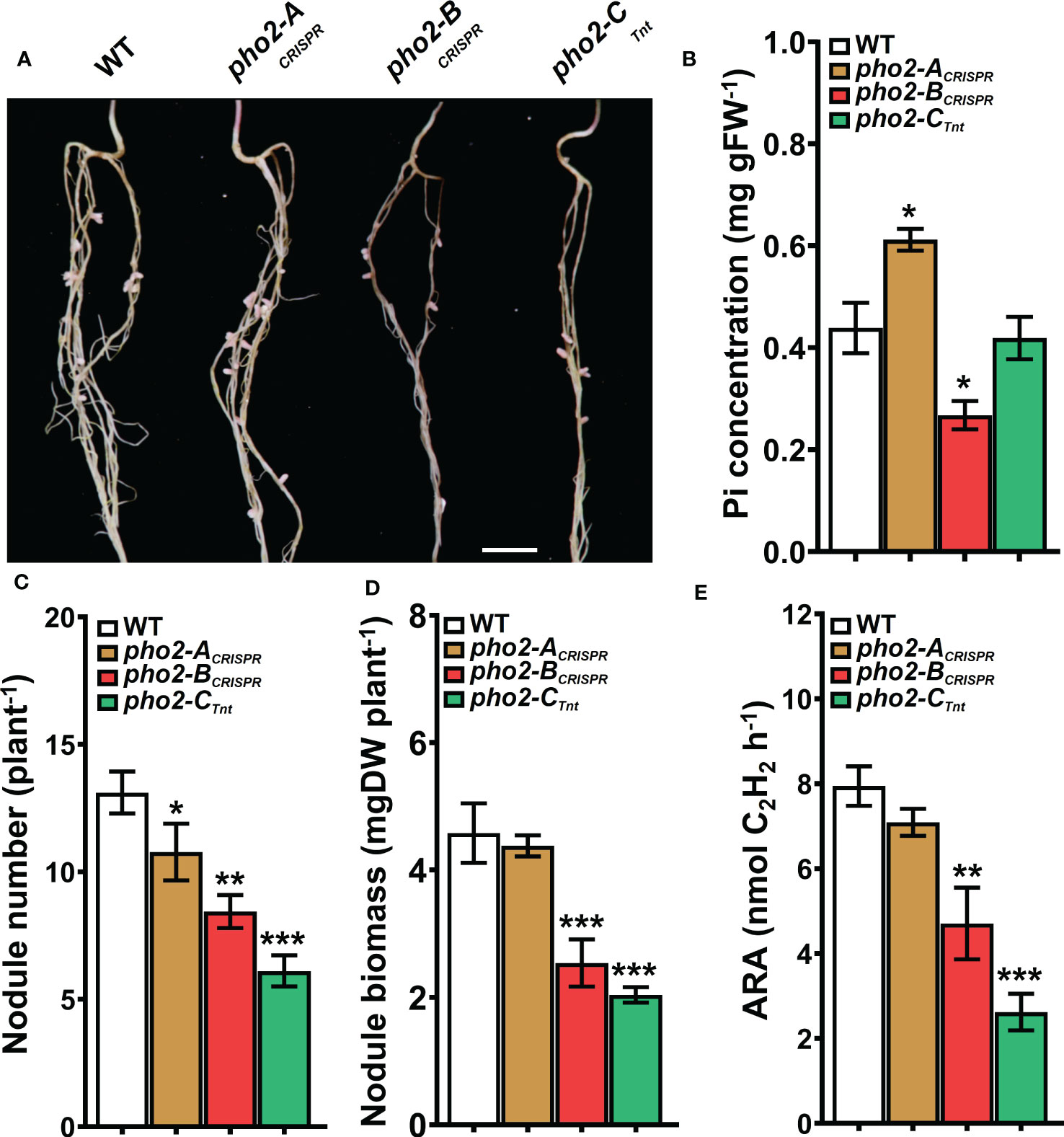
Figure 6 Symbiotic phenotypes of the Mtpho2 mutants with optimal-P. (A) Nodulated roots at 21 dpi with S. meliloti strain Sm1021. Scale bar = 1 cm. (B) Free phosphate (Pi) concentration in nodules (C) Average nodule number. (D) Average nodule biomass. E) Acetylene reduction assay (ARA) on whole nodulated roots. Data shown are the mean and SEM of three independent experiments (n=5/experiment). Asterisks indicate significant differences between the wild type and mutants calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
P-limitation reduced WT plant growth, biomass, nodule number and biomass, nitrogen fixation, and Pi concentration in all organs relative to P-replete plants (compare Figures 7, 8 with Figures 5, 6, respectively). Even so, pho2-BCRISPR and pho2-CTnt1 mutants were smaller than the WT under P-limiting conditions (Figures 7A, B). The reduced size of these two mutants was accompanied by a moderate but significant accumulation of Pi in mature leaves compared to the WT (Figure 7C). pho2-BCRISPR and pho2-CTnt1 mutants also exhibited defects in symbiotic traits, including reduced nodule biomass and nitrogen fixation (Figures 8C-E). Interestingly, reduced nodule biomass resulted from reduced nodule number of pho2-CTnt1 but not pho2-BCRISPR, which produced the same number of nodules as the wild-type, albeit smaller (Figure 8).
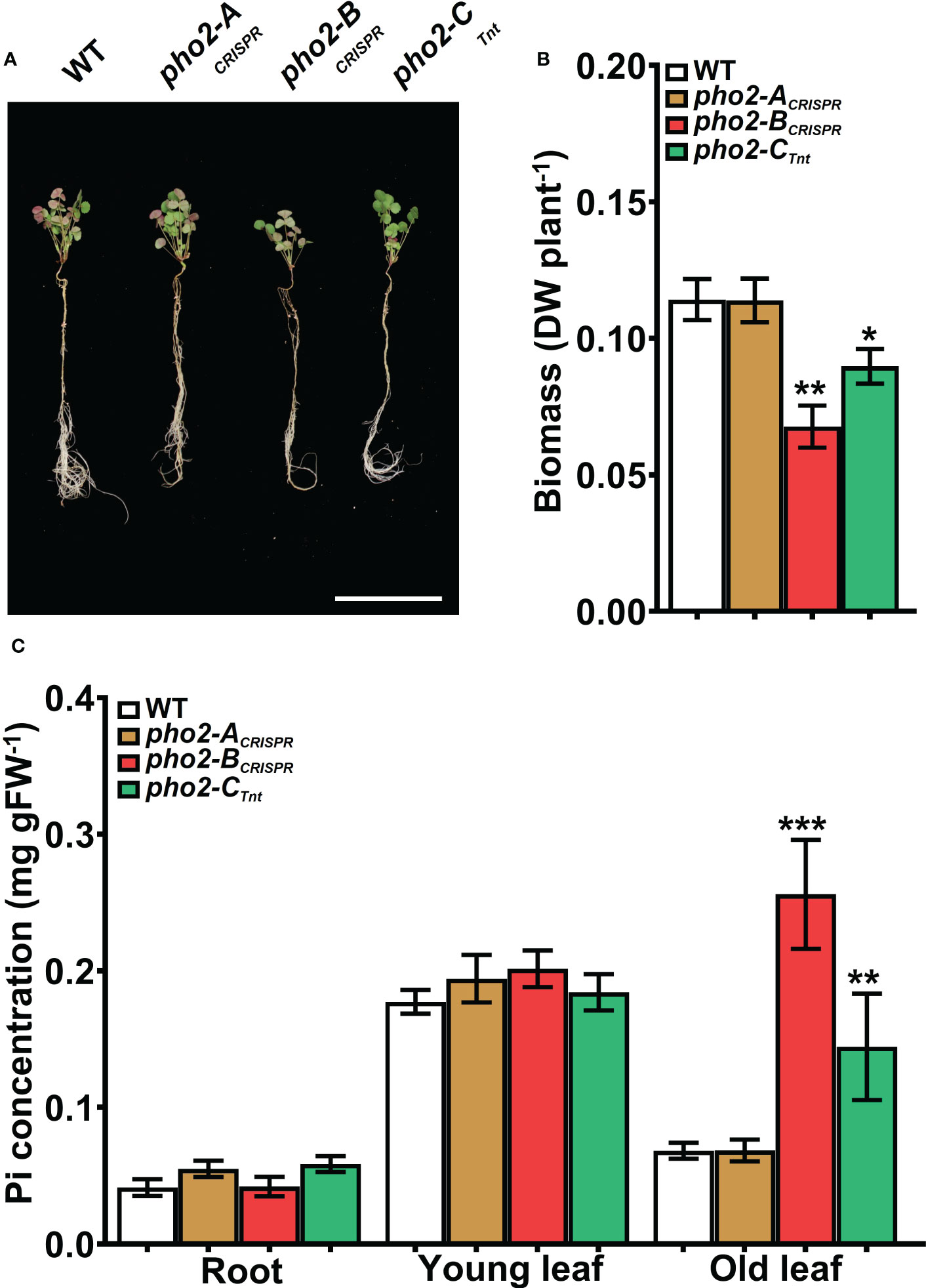
Figure 7 Performance of the Mtpho2 mutants under symbiotic nitrogen fixation conditions. Plants were inoculated with S. meliloti strain Sm1021 and given reduced-P (20 μM Pi). (A) Representative four-week-old plants. Scale bar = 10 cm. (B) Plant dry weight. (C) Free phosphate (Pi) concentration in roots, young and old leaves. Data shown are the mean and SEM of three independent experiments (n=5/experiment). Asterisks indicate significant differences between the wild type and the mutants calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).

Figure 8 Symbiotic phenotypes of the Mtpho2 mutants with reduced-P. (A) Nodulated roots at 21 dpi with S. meliloti strain Sm1021. Scale bar = 1 cm. (B) Free phosphate (Pi) concentration in nodules (C) Average nodule number. (D) Average nodule biomass. (E) Acetylene reduction activity (ARA) of whole nodulated roots. Data shown are the mean and SEM of three independent experiments (n=5/experiment). Asterisks indicate significant differences between the wild type and mutants calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
To better understand the role of PHO2 homologs in Pi homeostasis, we measured the expression levels of eight MtPT/PHT1-like, five MtPHO1-like, three nitrogen limitation adaptation (NLA) and three PHR1-like genes in roots and shoots of WT and mutant plants (Supplementary Table 2). Quantification of expression levels by qPCR revealed that all MtPT/PHT1-like and MtPHO1-like transporters were induced by P-limitation (data not shown). In the absence of nutritional deficits, MtPT5, MtPT3 and MtPT13 transporters were highly upregulated in roots and/or shoots of pho2-BCRISPR plants compared to the WT plants, whereas MtPT5 was slightly upregulated in shoots of pho2-ACRISPR plants (Figure 9A). Under symbiotic conditions without P limitation, the three mutant alleles presented deregulation of the expression levels of MtPT/PHT1-like transporters. The expression levels of MtPT5, MtPT3, MtPT6 and MtPHO1;3 transporters were deregulated in roots and/or shoots of pho2-BCRISPR plants compared to the WT plants. These transporters, except for MtPHO1;3, also showed deregulation in their expression levels in pho2-ACRISPR plants, but more moderate than those in pho2-ACRISPR plants. Similarly, MtPT5, MtPT6 and MtPHO1;3 transporters were deregulated in roots and/or shoots of pho2-CCRISPR plants compared to the WT plants. Although MtPT6 was upregulated at different levels in roots and shoots of all the three mutants, it was the only transporter upregulated in the nodule of the pho2-ACRISPR mutant compared with the WT plants (Figure 9B).
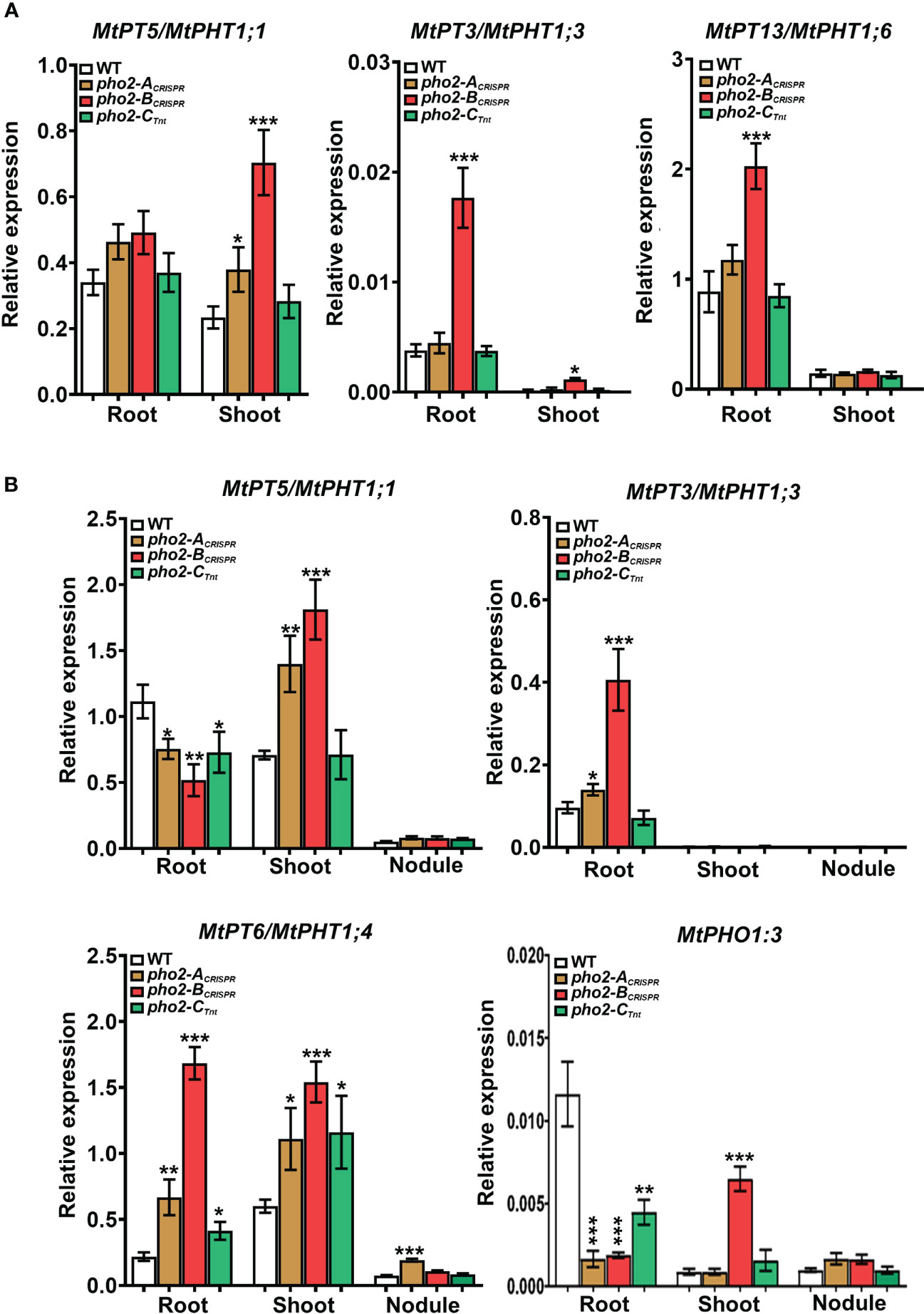
Figure 9 Relative expression levels, quantified by qPCR, of MtPHT1-like genes in various experimental conditions and organs (A) Optimal-P. (B) Symbiotic nitrogen fixation with optimal-P. PCR primers used are included in Supplementary Table S2. Data shown are the mean and SEM of three independent experiments. Plants were grown under the same conditions as those shown on Figures 4–8. For each replica, copy numbers were normalized using the mean average of two housekeeping genes (MtPTB2 and MtPDF2); asterisks indicate significant differences between the WT and mutants in each tissue of the same condition calculated using two-tailed Student’s t-tests (*p < 0.05, **p < 0.01, ***p < 0.001). Double nomenclature (PT/PHT) due to Breuillin-Sessoms et al., 2015. Plant Cell. 27(4):1352-66.999.
PHOSPHATE2 (PHO2) genes encode PHO2/Ubiquitin-Conjugating E2 24 (UBC24) proteins. The role of PHO2 proteins in regulating inorganic phosphate (Pi) homeostasis and Pi translocation and remobilization has been illuminated using mutants of Arabidopsis (Bari et al., 2006), rice (Cao et al., 2014), common wheat (Ouyang et al., 2016), and alfalfa (Miller et al., 2022), and transient silencing in barley (Pacak et al., 2010; Pacak et al., 2016) and citrus (Wang et al., 2020). The regulatory role of PHO2 in Pi homeostasis is based on its joint action with E3 ligases to polyubiquitinate and degrade Pi transporters, thus controlling Pi translocation, accumulation and/or remobilization in plant tissues when P is not limiting. Under P limitation, turnover of Pi transporters is prevented by undermining PHO2 protein production via microRNA399 (miR399)-guidedPHO2 mRNA degradation, orchestrated by the transcription factor PHOSPHATE STARVATION RESPONSE 1 (PHR1) (Aung et al., 2006; Bari et al., 2006).
Although some species, e.g. Arabidopsis and rice (Bari et al., 2006; Cao et al., 2014), have a single PHO2 gene, others have more than one, which begs the question of their roles in P-homoeostasis or other plant processes. For instance, autotetraploid alfalfa has two PHO2 genes, with multiple haplotypes of each. However, only one of the two alfalfa PHO2 proteins seems to have a role in P-homeostasis (Miller et al., 2022).
Here, we used sequence and phylogenomic analysis to identify three different PHO2 genes in the Medicago truncatula, encoding up to five possible PHO2 proteins, including splice variants (Figure 1; Supplementary Figure 1). In addition to having three PHO2 genes, the presence of splicing isoforms adds an extra level of complexity that may have biological significance. Alternate splicing to produce different functional proteins is a tightly regulated process essential for plants development and adaptation to environment (Huertas et al., 2019). In fact, PHO2 transcript splicing is regulated by P stress in rice (Secco et al., 2013), while Arabidopsis produces a shorter spliced form of its single PHO2 gene transcript that does not contain miR399 binding sites, to escape from miR399 mediated target degradation (Scheible et al., 2023). Two splice variants of different length have been reported for PHO2 in barley (Pacak et al., 2016) but have not been characterized. Interestingly, detailed study of splicing events in tomato under P stress revealed none in SlPHO2 (Tian et al., 2021), suggesting that alternative splicing of PHO2 transcript may not occur in all species and/or conditions. It is interesting that one of the Medicago splice variants, MtPHO2-A.3, which lacks its UBC catalytic domain sequences (Supplementary Figure 1), has a counterpart in rice (Dong et al., 2018). Further work is required to understand what role, if any, such a protein plays in Medicago and rice.
Transcript expression levels of the PHO2 genes are modulated by nutritional status, being normally repressed in roots and shoots by P limitation (Bari et al., 2006; Hackenberg et al., 2013; Cao et al., 2014; Ouyang et al., 2016; Wang et al., 2017; Miller et al., 2022; Santoro et al., 2022). Recent reports in Arabidopsis, tomato and barley revealed that PHO2 genes also can alter their expression as a function of N levels, indicating their integrative role between P and N homeostasis (de Souza Campos et al., 2019; Medici et al., 2019; Marro et al., 2022). Remarkably, some PHO2 genes, however, have different time-, intensity-, and genotype-dependent transcriptomic responses under P limitation (de Souza Campos et al., 2019; Gamir et al., 2020), indicating complex spatial-temporal Pi starvation signaling. Modulation of PHO2 transcripts requires recognition of PHO2 mRNAs by miR399s, regulated in turn by the transcription factor PHR1. Thus, the presence of five different miR399-BS conserved in the 5′-UTR of the three MtPHO2 genes underscores their likely role in P regulation (Figure 1A; Supplementary Figure 2). The high identity between the 10 Medicago truncatula miR399s and miR399-BSs on their 5′-UTR (Supplementary Figure 2) suggests their potential to cleave MtPHO2 mRNAs. Although we did not validate miR399 cleavage, the high homology of the Medicago miR399 and miR399-BS sequences with those of other plant species already validated in vitro (Aung et al., 2006; Bari et al., 2006; Hackenberg et al., 2013; Xu et al., 2013; Miller et al., 2022) or by transient expression (Ouyang et al., 2016), strongly suggest their cleavage capabilities. Indeed, regulation of expression of MtPHO2-B by a miR399 has already demonstrated indirectly through the mutation of the Medicago PDIL1 (Wang et al., 2017), a Pi deficiency-responsive At4-like lncRNA that suppresses the effect of miR399 by acting as mimics of PHO2 (Franco-Zorrilla et al., 2007). Interestingly, MtPHO2-B gene also contain a PHR1-binding DNA sequence (P1BS) motif. To our knowledge, this is the first description of the P1BS motif in a PHO2 gene, suggesting direct PHR1-dependent regulation in addition of the mediated regulation by the various miR399s. The presence of miR399s variants and cis-regulatory elements (Figure 1A; Supplementary Figure 2) could lead to different post-transcriptional regulations that could explain the different levels of expression and/or transcriptomic responses between the MtPHO2-B and MtPHO2-C genes to the P nutritional status in the plant (Figures 2, 4), suggesting then a differential role for both genes. Likewise, MtPHO2-B, and to a much lesser extent MtPHO2-C, are induced by N limitation in roots and shoots (Supplementary Figure 3), suggesting a similar regulation dependent on the N status of the plant.
The impaired growth observed under control conditions by the lack of the protein MtPHO2-B can directly be associated with un unbalanced Pi homeostasis due to Pi hyperaccumulation in old mature leaves (Figure 4). This is the result of a deficiency PHO2-dependent degradation of Pi transporters, altering Pi movement between tissues and organs and preventing Pi from being used properly by the plant (Huang et al., 2013; Park et al., 2014). Taken together, our results identify MtPHO2-B as the functional ortholog of the characterized PHO2 proteins (Aung et al., 2006; Bari et al., 2006; Cao et al., 2014; Ouyang et al., 2016; Miller et al., 2022), integral to Pi homeostasis in the absence of nutritional deficits. Similarly, the reduced plant growth in the absence of the protein MtPHO2-C can also be associated with an imbalanced P homeostasis. However, since pho2 mutants are described as Pi hyperaccumulators (Aung et al., 2006; Bari et al., 2006) and the M. truncatula mutant has a marginal, albeit significant, reduction in Pi content in old leaves (Figure 4), our results implies that MtPHO2-C is not a functional ortholog of the characterized PHO2, although it does appear to have a role in Pi homeostasis. The proposed secondary role controlling Pi homeostasis is in line with results recently described for the alfalfa homologous PHO2-C, where mutations of the different haplotypes lead to limited Pi hyperaccumulation possibility because to the inability of miR399s to cleave MtPHO2-C mRNAs (Miller et al., 2022). Demonstrating the role of the MtPHO2-C proteins in Pi homeostasis, regardless of any putative role in plant development (Supplementary Figure 5), will require further experiments. Likewise, the reduced expression levels and lack of transcriptomic response to P limitation suggest a limited role in controlling Pi homeostasis, or compensation by the other two MtPHO2 proteins. This is partially supported by the absence of phenotypes in the mutant alleles during the early stages of development but does not rule out a long-term role in regulating certain levels of Pi necessary to support plant growth (Supplementary Figure 5). In fact, Medicago truncatula is more sensitive to elevated Pi content compared with most other leguminous and non-leguminous plant species (Sulieman et al., 2013), so it is possible to reason that it also requires a finely tuned Pi homeostasis, controlled by multiple PHO2 activities.
Symbiotic nitrogen fixation (SNF) is a complex series of physical and chemical interactions built upon the trading of reduced carbon (C) from a legume for reduced N from the compatible symbiont. Atmospheric nitrogen (N2), once fixed in the root nodules, is partially transported to the aerial parts of the plant supporting multiple metabolic processes (Roy et al., 2020). The intensive C turnover during nodule development and N2 fixation are energy-consuming processes, requiring a large amount of P (Cabeza et al., 2014), as well as a limited presence of mineral N to promote nodule development without actually inhibiting N2 fixation (Ferguson et al., 2019). Under this condition of mineral N deprivation, most of the N available for plant use is obtained from the N2 fixation, so plants need a fine adjustment of Pi homeostasis in the nodule coordinated with the whole-plant Pi homeostasis (Sulieman & Tran, 2015; Sulieman et al., 2019). Such level of coordination in Medicago truncatula seems to be provided by the three MtPHO2 proteins, although the protein PHO2-B has a leading role in maintaining Pi homeostasis, as evidenced by the huge Pi hyperaccumulation not only in old leaves but also in young leaves, while it locally disappears from the roots (Figure 5C) and nodules (Figure 6B) in the pho2-B mutant. To our knowledge, this is the first time that a pho2 mutant imbalances Pi throughout the whole plant, including nodules. The abnormal distribution of Pi impacts nodule formation and development, causing the reduction in N2 fixation activity (Figures 6C, D) (Hernandez et al., 2009), impairing plant growth (Figures 5A, B). Thus, although plants preferentially distribute available Pi to the nodules to maintain symbiotic N2 fixation (Cabeza et al., 2014; Sulieman & Tran, 2015), our results confirm that nodule Pi homeostasis is subordinated to the whole-plant Pi homeostasis. Nevertheless, Pi homeostasis under SNF is not regulated by a single MtPHO2 gene. Even if at a lower scale than MtPHO2-B, the proteins MtPHO2-C and MtPHO2-A are also necessary to maintain whole-plant Pi homeostasis, the latter controlling Pi accumulation in roots and old leaves (Figure 5), and specially regulating Pi content for the formation of nodules (Figures 6B, D). The fact that neither the diploid nor the tetraploid alfalfa genome contains an orthologous MtPHO2-A gene while it is present in the wild relative Medicago ruthenica (Wang et al., 2021; Miller et al., 2022) and in Medicago truncatula genomes, evidence the evolutionary importance of this gene controlling quantitative variation for nodule formation (Stanton-Geddes et al., 2013; Curtin et al., 2017). Therefore, we can speculate that MtPHO2-A gene diverged from MtPHO2-B (Figure 1B) to support a finely tuned Pi homeostasis, especially in root nodules.
Symbiotic nitrogen fixation under limited mineral P is severely impacted by reducing the formation and the development of nodules, reducing the fixed N2 to be used by the plant (Figure 8) (Sulieman et al., 2013). In these conditions, MtPHO2-A protein does not seem to play a relevant role in the whole-plant Pi homeostasis while MtPHO2-B and MtPHO2-C proteins continue to exert some Pi regulatory capacity (Figure 7), necessary to regulate SNF traits (Figure 8). This, which could be somewhat unexpected since the role of PHO2 proteins, associated with the regulation of Pi when it is in excess, is necessary to coordinate N and P homeostasis (Liese et al., 2017; Medici et al., 2019) of the nodules to use Pi efficiently according to N availability. However, the reduced SNF due to P limitation also involves a lower degree of N2 assimilation, and therefore it is expected that plants exhibit local and/or systemic N-limited-related responses. As mentioned earliest, MtPHO2-B and MtPHO2-C genes are both induced by N deficiency (Supplementary Figure 3), but their tissue-dependent transcriptomic responses to mineral N and P limitation suggest common functions within the nodule, but also some type of speciation between tissues (Figure 3B). In tomato, for example, Pi starvation signaling depends on the plant’s N status, such that the downregulation of SlPHO2 by Pi deficiency happens only under optimal N conditions, whereas it can be upregulated by N deficiency only under optimal P conditions (Marro et al., 2022). Our results point here, indicating that both proteins MtPHO2-B and MtPHO2-C have a similar role, coordinating whole-plant P and N homeostasis to sustain plant growth.
Overall, our results confirm that under mineral N deprivation, the fine adjustment of Pi homeostasis is primarily due to the integral action of MtPHO2-B, assisted by the action of MtPHO2-C and MtPHO2-A. The latter also seems to have a specific role regulating nodule Pi homeostasis associated to nodule formation.
Tissue hyperaccumulation of Pi in pho2 mutants is known to result in local Pi limitation in other plant tissues and organs. To compensate for this local Pi limitation, these plants alter the expression levels of PHO1-like and PT/PHT1-like transporters, components of the P starvation response (PSR) (Aung et al., 2006; Bari et al., 2006; Hu et al., 2011; Liu et al., 2012; Huang et al., 2013; Ouyang et al., 2016). In fact, some of the PT/PHT1 transporters targeted by the PHO2-E3 complex to be degraded can increase their mRNA levels in the pho2 mutants (Huang et al., 2013; Lin et al., 2013). Under non-limiting nutritional conditions, only the lack of MtPHO2-B protein resulted in a higher expression of MtPT5, MtPT3 and MtPT13 in roots and/or shoots compared to WT (Figure 9A), affirming that PHO2-B is the main regulator of Pi homeostasis in Medicago truncatula. Although it is not possible to know if these changes are directly or indirectly linked to MtPHO2-B activity, the fact that there are no differences in the others individual mutants, points at these Pi transporters as PHO2-E3 targets for future experiments in Medicago truncatula.
The different patterns of Pi hyperaccumulation also lead to transcriptomic responses of the PSR components during SNF. Interestingly, although the genes MtPHO2-B and MtPHO2-C are the more abundant MtPHO2 genes within the nodules (Figure 3) (Roux et al., 2014), the lack of these individual proteins did not alter the expression of any of the MtPHO1-like and MtPT/PHT1-like genes in the nodules, but in roots and/or shoots (Figure 9B), supporting their role in maintaining whole-plant Pi homeostasis instead of nodule Pi homeostasis. These results contrast with the fact that some of the PHO1-like and PT/PHT1-like transporters, expressed between the root vascular system and vascular bundles of nodules, are crucial to maintain the Pi reallocation and homeostasis in young and mature nodules (Qin et al., 2012; Chen et al., 2019; Lu et al., 2020; Nguyen et al., 2021). On the contrary, the lack of the protein MtPHO2-A altered the expression of MtPT6/PHT1;4 in nodules (Figure 9B), one of the two PT/PHT1-like transporters that are induced during nodulation according to the MtSSPdb (Boschiero et al., 2020; https://mtsspdb.zhaolab.org/atlas-internal), which support the unique role of MtPHO2-A in maintaining Pi homeostasis in the nodule. New studies are needed to determine the role of MtPT6-like in SNF and the different variants of the MtPHO2-A protein, and a much more specific approach is required to determine the ubiquitination targets of the MtPHO2-E3 complex during SNF without P limitation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
RH: conceptualization, methodology, investigation, formal analysis – tables and figures, writing - original draft, review and editing final manuscript; IT-J: methodology (RNA isolation and qPCR reactions); SJC: methodology (provided the biological material to isolate some of the mutant alleles) and editing; WS: supervision and editing; MU: review and editing final manuscript, supervision and funding. All authors contributed to the article and approved the submitted version.
This work was supported by the Noble Research Institute LLC. Noble Research Institute was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication. SJC was supported by the U.S. Department of Agriculture, Agricultural Research Service. Mention of any trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. USDA is an equal opportunity provider and employer, and all agency services are available without discrimination.
Authors RH, IT-J, WS and MU are/were employed by the Noble Research Institute, LLC.
All the authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1211107/full#supplementary-material
Aung, K., Lin, S. I., Wu, C. C., Huang, Y. T., Su, C. L., Chiou, T. J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141 (3), 1000–1011. doi: 10.1104/pp.106.078063
Bari, R., Datt Pant, B., Stitt, M., Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141 (3), 988–999. doi: 10.1104/pp.106.079707
Boschiero, C., Dai, X., Lundquist, P. K., Roy, S., Christian de Bang, T., Zhang, S., et al. (2020). MtSSPdb: the medicago truncatula small secreted peptide database. Plant Physiol. 183 (1), 399–413. doi: 10.1104/pp.19.01088
Breuillin-Sessoms, F, Floss, DS, Gomez, SK, Pumplin, N, Ding, Y, Levesque-Tremblay, V, et al. (2015). Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2; 3. The Plant Cell. 27 (4):1352–66. https://doi.org/10.1105/tpc.114.131144
Broughton, W. J., Dilworth, M. J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125 (4), 1075–1080. doi: 10.1042/bj1251075
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Perez-Perez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in arabidopsis. PloS Genet. 6 (9), e1001102. doi: 10.1371/journal.pgen.1001102
Cabeza, R. A., Liese, R., Lingner, A., von Stieglitz, I., Neumann, J., Salinas-Riester, G., et al. (2014). RNA-Seq transcriptome profiling reveals that medicago truncatula nodules acclimate N(2) fixation before emerging p deficiency reaches the nodules. J. Exp. Bot. 65 (20), 6035–6048. doi: 10.1093/jxb/eru341
Cannon, S. B., Sterck, L., Rombauts, S., Sato, S., Cheung, F., Gouzy, J., et al. (2006). Legume genome evolution viewed through the medicago truncatula and lotus japonicus genomes. Proc. Natl. Acad. Sci. U.S.A. 103 (40), 14959–14964. doi: 10.1073/pnas.0603228103
Cao, Y., Yan, Y., Zhang, F., Wang, H. D., Gu, M., Wu, X. N., et al. (2014). Fine characterization of OsPHO2 knockout mutants reveals its key role in pi utilization in rice. J. Plant Physiol. 171 (3-4), 340–348. doi: 10.1016/j.jplph.2013.07.010
Cermak, T., Curtin, S. J., Gil-Humanes, J., Cegan, R., Kono, T. J. Y., Konecna, E., et al. (2017). A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29 (6), 1196–1217. doi: 10.1105/tpc.16.00922
Chen, L., Qin, L., Zhou, L., Li, X., Chen, Z., Sun, L., et al. (2019). A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N(2) fixation and yield in soybean. New Phytol. 221 (4), 2013–2025. doi: 10.1111/nph.15541
Chiou, T. J., Aung, K., Lin, S. I., Wu, C. C., Chiang, S. F., Su, C. L. (2006). Regulation of phosphate homeostasis by MicroRNA in arabidopsis. Plant Cell 18 (2), 412–421. doi: 10.1105/tpc.105.038943
Curtin, S. J., Tiffin, P., Guhlin, J., Trujillo, D. I., Burghart, L. T., Atkins, P., et al. (2017). Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 173 (2), 921–931. doi: 10.1104/pp.16.01923
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 Release). Nucleic Acids Res. 46 (W1), W49–W54. doi: 10.1093/nar/gky316
de Souza Campos, P. M., Cornejo, P., Rial, C., Borie, F., Varela, R. M., Seguel, A., et al. (2019). Phosphate acquisition efficiency in wheat is related to root:shoot ratio, strigolactone levels, and PHO2 regulation. J. Exp. Bot. 70 (20), 5631–5642. doi: 10.1093/jxb/erz349
Dong, C., He, F., Berkowitz, O., Liu, J., Cao, P., Tang, M., et al. (2018). Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 30 (10), 2267–2285. doi: 10.1105/tpc.18.00051
Ferguson, B. J., Mens, C., Hastwell, A. H., Zhang, M., Su, H., Jones, C. H., et al. (2019). Legume nodulation: the host controls the party. Plant Cell Environ. 42 (1), 41–51. doi: 10.1111/pce.13348
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 (8), 1033–1037. doi: 10.1038/ng2079
Gamir, J., Torres-Vera, R., Rial, C., Berrio, E., de Souza Campos, P. M., Varela, R. M., et al. (2020). Exogenous strigolactones impact metabolic profiles and phosphate starvation signalling in roots. Plant Cell Environ. 43 (7), 1655–1668. doi: 10.1111/pce.13760
Gautrat, P., Laffont, C., Frugier, F., Ruffel, S. (2021). Nitrogen systemic signaling: from symbiotic nodulation to root acquisition. Trends Plant Sci. 26 (4), 392–406. doi: 10.1016/j.tplants.2020.11.009
Hackenberg, M., Shi, B.-J., Gustafson, P., Langridge, P. (2013). Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol. 13 (1), 1–17. doi: 10.1186/1471-2229-13-214
Hardy, R. W., Holsten, R. D., Jackson, E. K., Burns, R. C. (1968). The acetylene-ethylene assay for n(2) fixation: laboratory and field evaluation. Plant Physiol. 43 (8), 1185–1207. doi: 10.1104/pp.43.8.1185
Helliwell, K. E. (2022). Emerging trends in nitrogen and phosphorus signalling in photosynthetic eukaryotes. Trends Plant Sci. 28 (3), 208–2111. doi: 10.1016/j.tplants.2022.10.004
Hernandez, G., Valdes-Lopez, O., Ramirez, M., Goffard, N., Weiller, G., Aparicio-Fabre, R., et al. (2009). Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 151 (3), 1221–1238. doi: 10.1104/pp.109.143842
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate–NRT1. 1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5 (4), 401–413. doi: 10.1038/s41477-019-0384-1
Hu, B., Zhu, C., Li, F., Tang, J., Wang, Y., Lin, A., et al. (2011). LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156 (3), 1101–1115. doi: 10.1104/pp.110.170209
Huang, T. K., Han, C. L., Lin, S. I., Chen, Y. J., Tsai, Y. C., Chen, Y. R., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in arabidopsis roots. Plant Cell 25 (10), 4044–4060. doi: 10.1105/tpc.113.115998
Huertas, R., Catalá, R., Jiménez-Gómez, J. M., Mar Castellano, M., Crevillén, P., Piñeiro, M., et al. (2019). Arabidopsis SME1 regulates plant development and response to abiotic stress by determining spliceosome activity specificity. Plant Cell 31 (2), 537–554. doi: 10.1105/tpc.18.00689
Kakar, K., Wandrey, M., Czechowski, T., Gaertner, T., Scheible, W. R., Stitt, M., et al. (2008). A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods Dec;4(1):1–2. doi.org/10.1186/1746-4811-4-18
Kant, S., Peng, M., Rothstein, S. J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PloS Genet. 7 (3), e1002021. doi: 10.1371/journal.pgen.1002021
Kryvoruchko, I. S., Routray, P., Sinharoy, S., Torres-Jerez, I., Tejada-Jiménez, M., Finney, L. A., et al. (2018). An iron-activated citrate transporter, MtMATE67, is required for symbiotic nitrogen fixation. Plant Physiol. 176 (3), 2315–2329. doi: 10.1104/pp.17.01538
Liese, R., Schulze, J., Cabeza, R. A. (2017). Nitrate application or p deficiency induce a decline in medicago truncatula N(2)-fixation by similar changes in the nodule transcriptome. Sci. Rep. 7, 46264. doi: 10.1038/srep46264
Lin, S.-I., Chiang, S.-F., Lin, W.-Y., Chen, J.-W., Tseng, C.-Y., Wu, P.-C., et al. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147 (2), 732–746. doi: 10.1104/pp.108.116269
Lin, W. Y., Huang, T. K., Chiou, T. J. (2013). Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in arabidopsis. Plant Cell 25 (10), 4061–4074. doi: 10.1105/tpc.113.116012
Liu, T. Y., Huang, T. K., Tseng, C. Y., Lai, Y. S., Lin, S. I., Lin, W. Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in arabidopsis. Plant Cell 24 (5), 2168–2183. doi: 10.1105/tpc.112.096636
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lu, M., Cheng, Z., Zhang, X. M., Huang, P., Fan, C., Yu, G., et al. (2020). Spatial divergence of PHR-PHT1 modules maintains phosphorus homeostasis in soybean nodules. Plant Physiol. 184 (1), 236–250. doi: 10.1104/pp.19.01209
Marro, N., Lidoy, J., Chico, M. A., Rial, C., Garcia, J., Varela, R. M., et al. (2022). Strigolactones: new players in the nitrogen-phosphorus signalling interplay. Plant Cell Environ. 45 (2), 512–527. doi: 10.1111/pce.14212
Meade, H. M., Long, S. R., Ruvkun, G. B., Brown, S. E., Ausubel, F. M. (1982). Physical and genetic characterization of symbiotic and auxotrophic mutants of rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149 (1), 114–122. doi: 10.1128/jb.149.1.114-122.1982
Medici, A., Szponarski, W., Dangeville, P., Safi, A., Dissanayake, I. M., Saenchai, C., et al. (2019). Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 31 (5), 1171–1184. doi: 10.1105/tpc.18.00656
Miller, S. S., Dornbusch, M. R., Farmer, A. D., Huertas, R., Gutierrez-Gonzalez, J. J., Young, N. D., et al. (2022). Alfalfa (Medicago sativa l.) pho2 mutant plants hyperaccumulate phosphate. G3 (Bethesda) 12 (6). doi: 10.1093/g3journal/jkac096
Mukatira, U. T., Liu, C., Varadarajan, D. K., Raghothama, K. G. (2001). Negative regulation of phosphate starvation-induced genes. Plant Physiol. 127 (4), 1854–1862. doi: 10.1104/pp.010876
Nasr Esfahani, M., Inoue, K., Chu, H. D., Nguyen, K. H., Van Ha, C., Watanabe, Y., et al. (2017). Comparative transcriptome analysis of nodules of two mesorhizobium-chickpea associations with differential symbiotic efficiency under phosphate deficiency. Plant J. 91 (5), 911–926. doi: 10.1111/tpj.13616
Nguyen, N. N. T., Clua, J., Vetal, P. V., Vuarambon, D. J., De Bellis, D., Pervent, M., et al. (2021). PHO1 family members transport phosphate from infected nodule cells to bacteroids in medicago truncatula. Plant Physiol. 185 (1), 196–209. doi: 10.1093/plphys/kiaa016
Ouyang, X., Hong, X., Zhao, X., Zhang, W., He, X., Ma, W., et al. (2016). Knock out of the PHOSPHATE 2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci. Rep. 6, 29850. doi: 10.1038/srep29850
Pacak, A., Barciszewska-Pacak, M., Swida-Barteczka, A., Kruszka, K., Sega, P., Milanowska, K., et al. (2016). Heat stress affects pi-related genes expression and inorganic phosphate Deposition/Accumulation in barley. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00926
Pacak, A., Geisler, K., Jorgensen, B., Barciszewska-Pacak, M., Nilsson, L., Nielsen, T. H., et al. (2010). Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in brachypodium distachyon and oat. Plant Methods 6, 26. doi: 10.1186/1746-4811-6-26
Park, B. S., Seo, J. S., Chua, N. H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of arabidopsis phosphate homeostasis. Plant Cell 26 (1), 454–464. doi: 10.1105/tpc.113.120311
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in arabidopsis. Proc. Natl. Acad. Sci. 111 (41), 14947–14952. doi: 10.1073/pnas.140465411
Qin, L., Zhao, J., Tian, J., Chen, L., Sun, Z., Guo, Y., et al. (2012). The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 159 (4), 1634–1643. doi: 10.1104/pp.112.199786
Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29 (1), 24–26. doi: 10.1038/nbt.1754
Roux, B., Rodde, N., Jardinaud, M. F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77 (6), 817–837. doi: 10.1111/tpj.12442
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32 (1), 15–41. doi: 10.1105/tpc.19.00279
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 (16), 2122–2133. doi: 10.1101/gad.204401
Santoro, V., Schiavon, M., Visentin, I., Martin, M., Said-Pullicino, D., Cardinale, F., et al. (2022). Tomato plant responses induced by sparingly available inorganic and organic phosphorus forms are modulated by strigolactones. Plant Soil 474 (1-2), 355–372. doi: 10.1007/s11104-022-05337-0
Scheible, W. R., Pant, P., Pant, B. D., Krom, N., Allen, R. D., Mysore, K. S. (2023). Elucidating the unknown transcriptional responses and PHR1 mediated biotic and abiotic stress tolerance during phosphorus-limitation. J. Exp. Bot. 74(6):2083–2111 doi: 10.1093/jxb/erad009
Secco, D., Jabnoune, M., Walker, H., Shou, H., Wu, P., Poirier, Y., et al. (2013). Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25 (11), 4285–4304. doi: 10.1105/tpc.113.117325
Stanton-Geddes, J., Paape, T., Epstein, B., Briskine, R., Yoder, J., Mudge, J., et al. (2013). Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in medicago truncatula. PloS One 8 (5), e65688. doi: 10.1371/journal.pone.0065688
Sulieman, S., Ha, C. V., Schulze, J., Tran, L. S. (2013). Growth and nodulation of symbiotic medicago truncatula at different levels of phosphorus availability. J. Exp. Bot. 64 (10), 2701–2712. doi: 10.1093/jxb/ert122
Sulieman, S., Kusano, M., Ha, C. V., Watanabe, Y., Abdalla, M. A., Abdelrahman, M., et al. (2019). Divergent metabolic adjustments in nodules are indispensable for efficient N(2) fixation of soybean under phosphate stress. Plant Sci. 289, 110249. doi: 10.1016/j.plantsci.2019.110249
Sulieman, S., Tran, L.-S. P. (2015). Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 239, 36–43. doi: 10.1016/j.plantsci.2015.06.018
Tian, P., Lin, Z., Lin, D., Dong, S., Huang, J., Huang, T. (2021). The pattern of DNA methylation alteration, and its association with the changes of gene expression and alternative splicing during phosphate starvation in tomato. Plant J. 108 (3), 841–858. doi: 10.1111/tpj.15486
Udvardi, M., Below, F. E., Castellano, M. J., Eagle, A. J., Giller, K. E., Ladha, J. K., et al. (2021). A research road map for responsible use of agricultural nitrogen. 5, 660155 doi: 10.3389/fsufs.2021.660155
Udvardi, M., Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 63 (64), 781–805. doi: 10.1146/annurev-arplant-050312-120235
Ueda, Y., Kiba, T., Yanagisawa, S. (2020). Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of SPX genes. Plant J. 102 (3), 448–466. doi: 10.1111/tpj.14637
Wang, R., Fang, Y. N., Wu, X. M., Qing, M., Li, C. C., Xie, K. D., et al. (2020). The miR399-CsUBC24 module regulates reproductive development and Male fertility in citrus. Plant Physiol. 183 (4), 1681–1695. doi: 10.1104/pp.20.00129
Wang, T., Ren, L., Li, C., Zhang, D., Zhang, X., Zhou, G., et al. (2021). The genome of a wild medicago species provides insights into the tolerant mechanisms of legume forage to environmental stress. BMC Biol. 19 (1), 1–17. doi: 10.1186/s12915-021-01033-0
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. 111 (41), 14953–14958. doi: 10.1073/pnas.1404680111
Wang, T., Zhao, M., Zhang, X., Liu, M., Yang, C., Chen, Y., et al. (2017). Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant medicago truncatula. J. Exp. Bot. 68 (21-22), 5937–5948. doi: 10.1093/jxb/erx384
Xu, F., Liu, Q., Chen, L., Kuang, J., Walk, T., Wang, J., et al. (2013). Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14, 66. doi: 10.1186/1471-2164-14-66
Keywords: phosphorus, symbiotic nitrogen fixation (SNF), Medicago truncatula, PHO2, nitrogen
Citation: Huertas R, Torres-Jerez I, Curtin SJ, Scheible W and Udvardi M (2023) Medicago truncatula PHO2 genes have distinct roles in phosphorus homeostasis and symbiotic nitrogen fixation. Front. Plant Sci. 14:1211107. doi: 10.3389/fpls.2023.1211107
Received: 24 April 2023; Accepted: 22 May 2023;
Published: 13 June 2023.
Edited by:
Chang Fu Tian, China Agricultural University, ChinaReviewed by:
Pascal Ratet, UMR9213 Institut des Sciences des Plantes de Paris Saclay (IPS2), FranceCopyright © 2023 Huertas, Torres-Jerez, Curtin, Scheible and Udvardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Udvardi, bS51ZHZhcmRpQHVxLmVkdS5hdQ==; Raul Huertas, cmF1bC5odWVydGFzQGh1dHRvbi5hYy51aw==
†Present address: Raul Huertas, Environmental and Biochemical Sciences, The James Hutton Institute, Dundee, United Kingdom
Ivone Torres-Jerez, Institute for Agricultural Biosciences, Oklahoma State University, Ardmore, OK, United States
Michael Udvardi, Queensland Alliance for Agriculture and Food Innovation, University of Queensland, Brisbane, QLD, Australia
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.