
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 July 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1209384
This article is part of the Research Topic СRISPR tools, technology development, and application View all 9 articles
 Jinlian Yang1†
Jinlian Yang1† Yaoyu Fang1†
Yaoyu Fang1† Hu Wu1
Hu Wu1 Neng Zhao1
Neng Zhao1 Xinying Guo1
Xinying Guo1 Enerand Mackon1
Enerand Mackon1 Haowen Peng1
Haowen Peng1 Sheng Huang2
Sheng Huang2 Yongqiang He1
Yongqiang He1 Baoxiang Qin1
Baoxiang Qin1 Yaoguang Liu3
Yaoguang Liu3 Fang Liu1
Fang Liu1 Shengwu Chen4*
Shengwu Chen4* Rongbai Li1*
Rongbai Li1*Rice (Oryza sativa L.) is a staple food in many countries around the world, particularly in China. The production of rice is seriously affected by the bacterial leaf streak and rice blast, which can reduce rice yield or even cause it to fail to be harvested. In this study, susceptible material 58B was edited by CRISPR/Cas9, targeting a target of the Pi21 gene and a target of the effector-binding element (EBE) of the OsSULTR3;6 gene, and the mutants 58b were obtained by Agrobacterium-mediated method. The editing efficiency of the two targets in the T0 generation was higher than 90.09%, the homozygous mutants were successfully selected in the T0 generation, and the homozygous mutation rate of each target was higher than 26.67%. The expression of the edited pi21 and EBE of Ossultr3;6 was significantly reduced, and the expression of defense responsive genes was significantly upregulated after infected with rice blast. The lesion areas of rice blast and bacterial leaf streak were significantly reduced in 58b, and the resistance of both was effectively improved. Furthermore, the gene editing events did not affect the agronomic traits of rice. In this study, the resistance of 58b to rice blast and bacterial leaf streak was improved simultaneously. This study provides a reference for using Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 (CRISPR/Cas9) to accelerate the improvement of rice varieties and the development of new materials for rice breeding.
Rice (Oryza sativa L.) is one of the most important food crops in the world. Bacterial leaf streak and rice blast are two deadly diseases that can cause serious damage to rice (Ke et al., 2017). Bacterial leaf streak is a bacterial disease caused by Xanthomonas oryzae pv.oryzicola (Xoc) that mainly infects rice leaves through leaf stomata or wounds (Jiang et al., 2020). The genome similarity between Xoc and another Xanthomonas oryzae pv. oryzae (Xoo) is more than 90%, and they both lead to rice disease by introducing transcription activator–like effectors (TALEs) into plant cells to activate the expression of the susceptibility gene(Cox et al., 2017). TALE is a class of proteins unique to Xanthomonas species (Jens and Ulla, 2010; Boch et al., 2014). TALE can activate either the susceptibility gene (S) or the resistance gene (R) of the plant, thereby making the host susceptible or activating the defense mechanism of the pathogen (Boch et al., 2014). TALE contains a conserved central repeat region consisting of 34 amino acid repeats, an N-terminal region of the type III secretion system, and a C-terminal region containing transcriptional activation domains and nucleoplasm localization signals (Xu et al., 2022). So far, the targeting of TALE has been determined by the central repeat region, where each repeat unit recognizes a nucleotide through a specific degenerate codon, resulting in a contiguous DNA sequence [effector-binding element (EBE)] (Matthew and Adam, 2009; Jens and Ulla, 2010). Magnaporthe oryzae–caused rice blast is one of the most damaging diseases to rice, with a large damage area and severity (Liu et al., 2014). Rice blast can infect the leaves, stems, panicles, and roots of rice at various developmental stages (Wilson and Talbot, 2009), resulting in a significant decrease in rice yield (Ebbole, 2007). Following pathogen infection, rice plants activate the biosynthesis and signal transduction of various hormones that act as immune signals to activate host defense responses against pathogen invasion (David et al., 2013; Yang et al., 2013). Jasmonic acid (JA) enhances resistance to rice blast by activating defense-related genes and accumulating antimicrobial substances (Shiduku et al., 2014). It has been reported that the ubiquitin-proteasome system negatively regulates OsWRKY45 through periodic degradation in the absence of pathogen infection (Zhou et al., 2022) and salicylic acid (SA) signaling, whereas OsWKRY45-mediated defense can be activated in the presence of SA signaling or pathogen infection reactions (Matsushita et al., 2013).
Known as a powerful gene editing tool, CRISPR/Cas9 has been widely used in rice to improve yield and quality traits, enhance disease resistance, and create male sterile rice lines to accelerate the process of hybrid rice breeding. For example, Yamauchi et al. used CRISPR/Cas9 technology to knock out the RBOHH gene and demonstrated its role in reducing Reactive oxygen species (ROS) accumulation in rice roots (Yamauchi et al., 2017); Li et al. used CRISPR/Cas9 to knock out four rice yield genes Gn1a, DEP1, GS3, and IPA1 to assess their roles in rice yield (Li et al., 2016); Usman et al. (2020) improved the fragrance quality of rice by editing the Badh2 gene (Usman et al., 2020); Zhou et al., (2022) edited the Bsr-d1, Pi21, and ERF922 genes of LK638S and improved the resistance of LK638S to rice blast and bacterial blight (Zhou et al., 2022).
The evolution of Magnaporthe oryzae may lead to decreased or even completely lost rice blast resistance. Therefore, developing new rice lines with broad-spectrum resistance (BSR) to blast is necessary. However, bacterial leaf streak resistance is a quantitative trait controlled by multiple quantitative trait loci, and it is difficult to effectively select Xoc-resistant varieties by traditional breeding (Bossa-Castro et al., 2018; Xie et al., 2021). CRISPR/Cas9 gene editing technology has advantages in high efficiency, simple operation, affordable cost, and the ability to simultaneously edit multiple targets. CRISPR transgenic progeny can be screened for homozygous mutants without T-DNA insertion, reducing breeding time and labor costs significantly. In this study, CRISPR/Cas9 technology was used to edit the TALE-binding region of the susceptibility gene OsSULTR3;6 and the second exon of Pi21 gene in the high-quality indica maintainer line 58B, which simultaneously improved the resistance to rice blast and bacterial leaf streak. This work provides new and interesting breeding materials with both broad-spectrum blast resistance and bacterial leaf streak resistance.
In this experiment, the indica maintainer line 58B preserved in Guangxi University was selected as the recipient material. This material has high quality and yield but is susceptible to rice blast and bacterial leaf streak. All experimental materials were independently planted in a planting pool in the rice net room or planting pond of Guangxi University. The CRISPR/Cas9 gene editing system used in this experiment was provided by YL, the South China Agricuture University. Xoc GX01 used was from in Guangxi University; M. oryzae H322 was a strain of rice blast isolated and preserved in the experimental field of Guangxi University by HP’s laboratory. The list of primers used in the study is shown in Supplementary Table 1.
Target sites of Pi21 and OsSULTR3;6-EBE were selected by the CRISPR-Genome Editing (GE) (http://skl.scau.edu.cn/home). The target sites were introduced separately into the promoter and the Single guide RNA (sgRNA) using overlapping PCR. Subsequently, the promoter-target-sgRNA units were assembled into the CRISPR/Cas9 vector, following the method described by Zeng et al. (Zeng et al., 2018). The validated CRISPR/Cas9 plasmid was transformed into Agrobacterium tumefaciens EHA105, which was then used for rice transformation of the 58B variety (Supplementary Figure 1). Specific primer pairs Cas9-F/Cas9-R and HPT-F/HPT-R were used to confirm T0 transgenic-positive plants. Pi21-TF/Pi21-TR and OsSULTR3;6-TF/OsSULTR3;6-TR were used to amplify the genomic regions containing each target site, and the amplified products were followed for Sanger sequencing in T0 and T1 generations. The sequencing results were analyzed to determine the target mutation by an online tool DSDecodeM (http://skl.scau.edu.cn/dsdecode/) (Liu et al., 2015). Transgene-free plants were identified using the primer pairs Cas9-F/Cas9-R and HPT-F/HPT-R and determined by both showing negative amplification (Supplementary Figure 3). The genomic DNA was amplified with specific primers, and the amplified products were sequenced. The sequencing results were compared with the The Rice Annotation Project-Database (RAP-DB) sequence to analyze the off-target of each gene.
Magnaporthe oryzae H322 was used for inoculation in this experiment. The activated strain was transferred to oat medium for culture, placed in a 28°C incubator, and exposed to light for 24 h a day for 10 days to induce sporulation. Before inoculation, rice was selected with consistent growth to the three-leaf stage and the suspension prepared (the spore concentration was adjusted to 1 × 105/mL, the total number of spores is about 30 in 16 middle squares of the blood count plate). The seedlings were sprayed evenly with the suspension, three biological replicates per strain. After inoculation, a layer of black film was placed on the transparent plastic film for shading and treatment for 36 h. After the treatment, the black film was removed, and it was kept moist for 5 days to investigate the incidence and lesions.
At the tillering stage, bacterial leaf streak inoculation was performed on transgene-free homozygous mutant. A single colony was selected from the streak-activated GX01 strain on NA medium (5 g/L tryptone, 1 g/L beeg extract, 1 g/L yeast extract, 10 g/L sucrose, 17 g/L agar, PH 6.8–7.0), and put into 500 mL of NB medium, which was cultured at 28°C at 180 rpm for 2 days. The OD600 value of bacterial suspension ranged between 0.6 and 0.8. GX01 was inoculated by acupuncture at the tillering stage (6 weeks) of rice (Supplementary Figure 4). The disease infection was investigated 14 days after inoculation and photographed.
Total RNA was extracted from fresh leaves using the FastPure Universal Plant Total RNA Isolation Kit (catalog no. RC411, Vazyme). The extracted RNA reverse-transcribed into Complementary DNA (cDNA), and Quantitative real-time (qRT)-PCR was performed. The gene expression levels of pi21 and Ossultr3;6 were detected in the mutant and wild types.
Wild-type (58B) and mutant lines were planted in Guangxi University. Each line was planted in four rows with eight plants in each row. At the maturity stage, five plants were randomly selected to investigate the plant height, effective panicle number, panicle length, number of grains per panicle, and 1,000-grain weight. Then, the data were analyzed using Excel and IBM SPSS Statistics 20.
To generate Pi21 and OsSULTR3;6-EBE mutants, two sgRNAs that are in the second exon of the Pi21 gene (LOC_Os04g32850) and EBE in the promoter region of OsSULRT3;6 gene (LOC_Os01g52130) were designed (Figure 1A). Two sgRNAs were constructed into the CRISPR/Cas9 vertor (Figure 1B), and the vector plasmid was used to transform 58B rice by Agrobacterium tumefaciens EHA105. All 15 transgenic seedlings of 58b were positive plants with a transformation rate of 100% (Supplementary Figure 2). There was only one transgenic plant in target Pi21 that did not have a detected target mutation, with a frequency of 6.67%. The frequencies of heterozygous and biallelic mutations were also the same at 26.67%, and the frequency of homozygous mutations was as high as 40%. Only one transgenic plant in the OsSULTR3;6-EBE target was not mutated. There were five heterozygous and five biallelic plants with a frequency of 33.33%, respectively. There were four plants with homozygous mutations with a frequency of 26.67% (Table 1). The mutation types of Pi21 and OsSULTR3;6-EBE both include base insertions, substitutions, and deletions, including mainly base deletions (43.33% and 40%, respectively; Table 2).
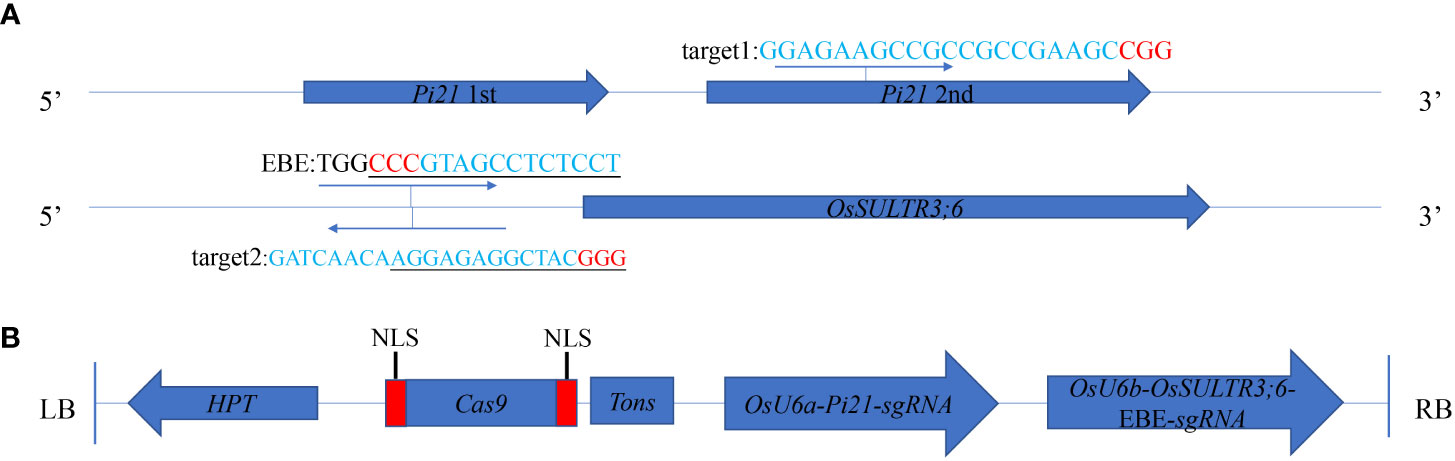
Figure 1 CRISPR/Cas9-mediated targeted mutagenesis of Pi21 and OsSULTR3;6-EBE. (A) Target sites of CRISPR/Cas9. One target was chosen in the in the second exon of the Pi21 gene; another target was chosen around effector-binding element (EBE) in the promoter region of OsSULRT3;6 gene; the PAM sequences were marked in red. (B) The expression CRISPR/Cas9 vector. OsU6a and OsU6b, rice promoter; HPT, hygromycin; NLS, nuclear localization signal; Tons, the terminator; LB and RB, left border and right border, respectively.
CRISPR/Cas9 gene editing technology inevitably presents off-target phenomena, which will interfere with experimental results. To avoid off-target events as much as possible, the potential off-target sites were predicted through the CRISPR-GE website (Table 3), and specific primers were designed for amplification and sequencing to analyze the off-target rate of each target. Pi21 has three off-target sites located in the CDS region of the gene, namely, Os03g0349200, Os01g0184800, and Os09g0380300, and the highest off-target index reached 0.557. Os03g0349200 presumably encodes a cyclin-dependent kinase C-2 protein involved in the cell cycle; Os01g0184800 presumably encodes a photoconductive protein; and Os09g0380300 presumably encodes a cytochrome P450 family protein. The two off-target sites of OsSULTR3;6-EBE were in the CDS region of Os03g0209500 and Os11g0568600, respectively, and the other off-target sites were located in the non-coding region, with the highest off-target index of 0.405. The protein encoded by Os03g0209500 belongs to the zinc finger family of proteins, whereas Os11g0568600 encodes a protein containing a THUMP domain. The specific functions of these two proteins are unknown. Specific primers were designed for the abovementioned off-target sites in the CDS region of the gene, the DNA of the homozygous plants of the T0 generation was extracted for PCR amplification, and then the off-target situation was analyzed by sequencing. The results showed that no off-target events were detected in the two homozygous seedlings of 58b (Supplementary Figure 5).
The resistance to rice blast of a single Pi21 homozygous mutant with a base insertion in the T1 generation was identified in seedling and maturity rice blast, three biological replications (Table 4). The single Pi21 mutant was found to exhibit increased resistance to M. oryzae H322 at the seedling stage. To test whether enhanced blast resistance still existed at reproductive stage, the blast evaluation on rice panicles was conducted, and the mutants were transplanted in a field rice blast area in Wutang, Nanning. The results showed that percentage of diseased panicles in mutants were significantly lower than that in 58B (Figure 2).
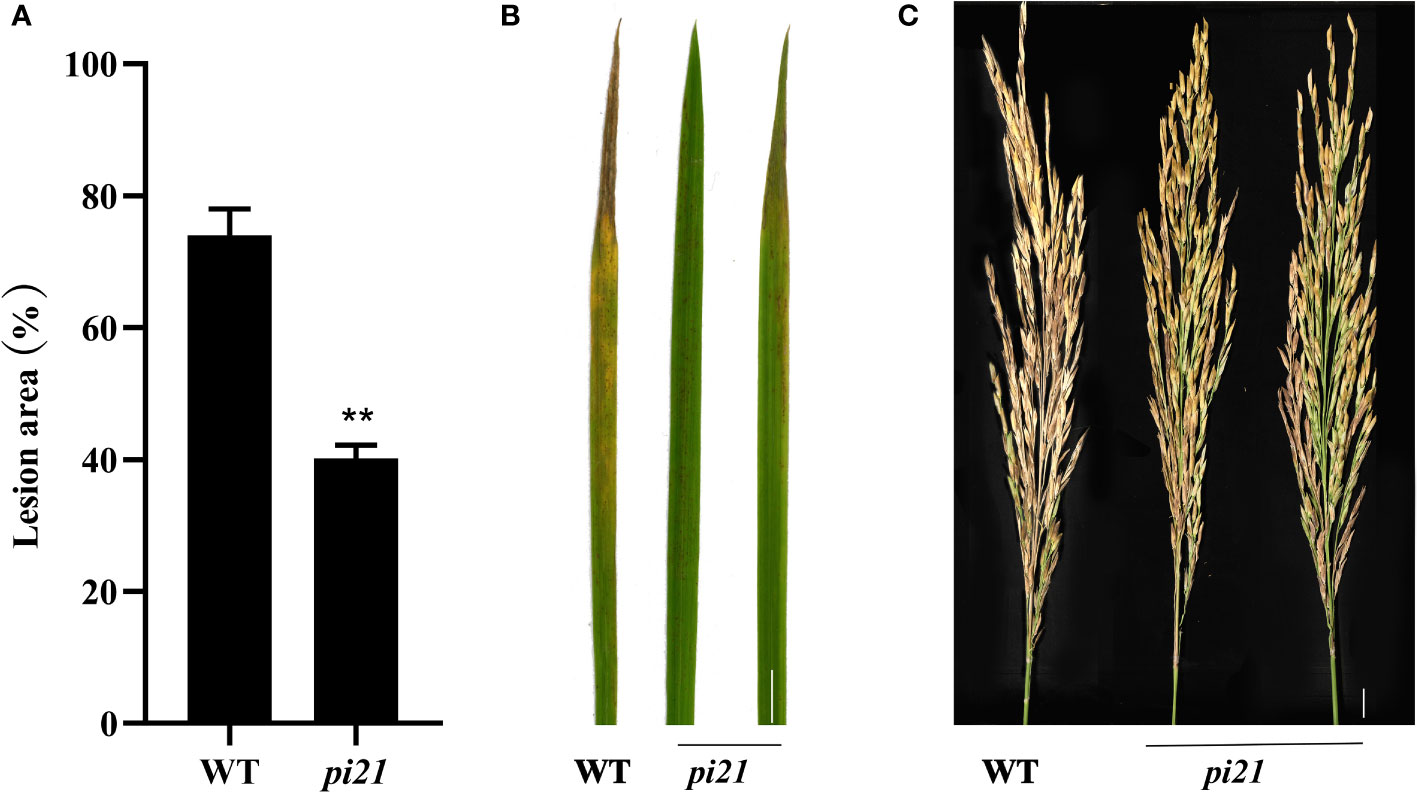
Figure 2 Enhanced blast resistance of the single Pi21 mutant lines. (A) The percentage of lesion areas of rice blast (n = 3 leaves). (B) Rice mutant lines and wild-type 58B were tested for resistance to M. oryzae at the seedling stage. (C) The resistance to M. oryzae rice mutant lines and wild-type 58B was tested at rice reproductive stage. t-test: **P < 0.01.
A single homozygous OsSULTR3;6-EBE mutant line with a 4-base deletion was selected for further analysis (Table 5). The mutants were inoculated Xoc strain GX01 using the acupuncture method at the tillering stage, three biological replications. At 15 days after inoculation, disease length was about 75% shorter on the Ossultr3;6-EBE mutant than that on 58B (Figure 3). The results demonstrate that disrupting OsSULTR3;6-EBE significantly reduces susceptibility to Xoc in rice.
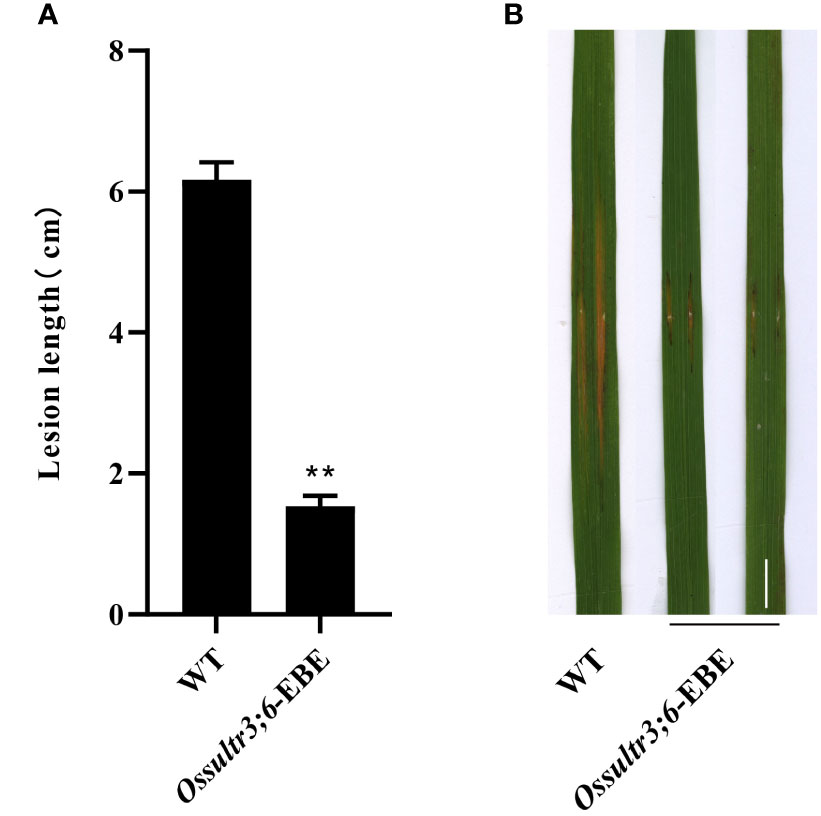
Figure 3 Enhanced bacterial leaf streak resistance of the single OsSULTR3;6-EBE mutant lines. (A) The percentage of lesion length of bacterial leaf streak (n = 3 leaves). (B) Rice mutant lines and wild-type 58B were tested for resistance to Xoc at the tillering stage. t-test: **P < 0.01.
Two homozygous pi21/Ossultr3;6-EBE double mutant plants with different editing types in the T1 generation (pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11) were used to identify the resistance (Table 6). pi21/Ossultr3;6-EBE-7 produced 21-bp deletion at Pi21, resulting in seven–amino acid “PEKPPPK” deletion in the Pi21 protein; 1-bp insertion at OsSULTR3;6-EBE did not cause a sequence change in the OsSULTR3;6 gene and OsSULTR3;6 protein. pi21/Ossultr3;6-EBE-11 produced 1-bp insertion at Pi21 and caused the frameshift in the Pi21 coding region, generating the premature translation termination codon; pi21/Ossultr3;6-EBE-11 produced 33-bp deletion at OsSULTR3;6-EBE but did not directly affect the OsSULTR3;6 gene and OsSULTR3;6 protein (Figure 4).
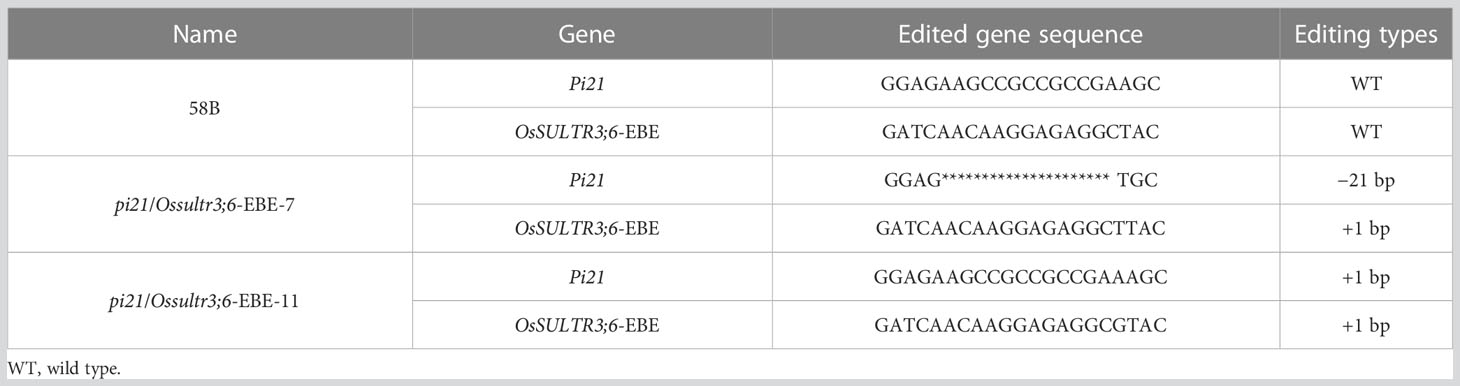
Table 6 sgRNA sequence and mutations at the target site of Pi21/OsSULTR3;6-EBE in the T1 homozygous mutant.
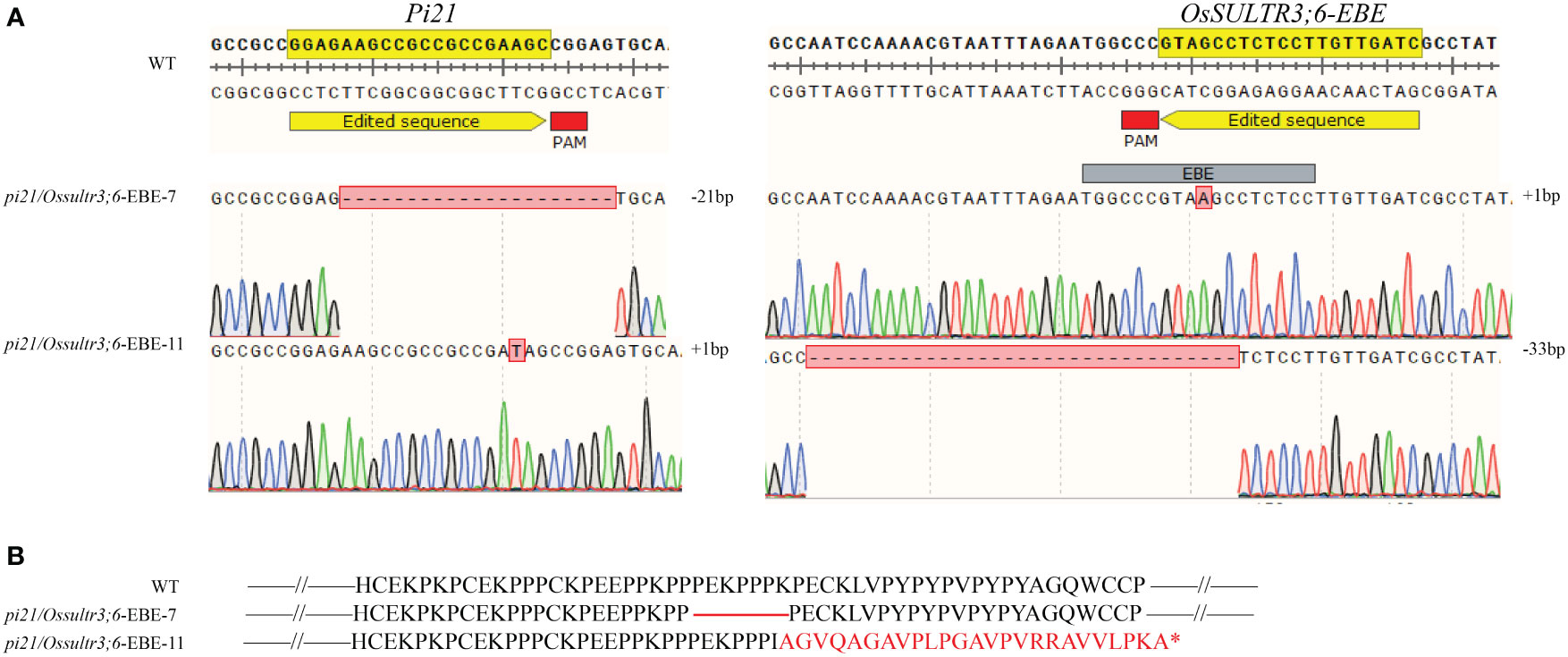
Figure 4 pi21/Ossultr3;6-EBE double mutation plants with different editing types (A) The mutated sequences of Pi21 and OsSULTR3;6. The number of base deletion and insertion is shown by the mark of minus (−) and plus (+). (B) Amino acid variations of the Pi21 protein in the mutant. The red line indicates the missing protein. *indicates the termination of translation.
To identify the blast resistance of the pi21 in homozygous mutant, pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11 were collected and inoculated M. oryzae H322 at the three-leaf stage by spraying. After 7 days, the area of lesions was counted, and the resistance to rice blast at the seedling stage was identified. Simultaneously, pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11 were also planted in Wutang, Nanning, where the rice blast naturally occurred, and the rice blast resistance was identified after the rice was mature. The results showed that, compared with the wild type, the pi21/Ossultr3;6-EBE homozygous mutant had significantly decreased lesion area, indicating that knockout of the pi21 gene significantly improved the rice blast resistance at seedling and mature stages (Figures 5A–C). To identify bacterial leaf streak resistance of homozygous mutants of pi21/Ossultr3;6-EBE, acupuncture was used to inoculate Xoc GX01, and the lesion length was measured and analyzed 15 days after inoculation. The results showed that, compared with the wild type, the length of the lesions of the pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11 lines was significantly reduced, indicating that the knockout of the OsSULTR3;6-EBE had significantly improved resistance (Figures 5D, E).
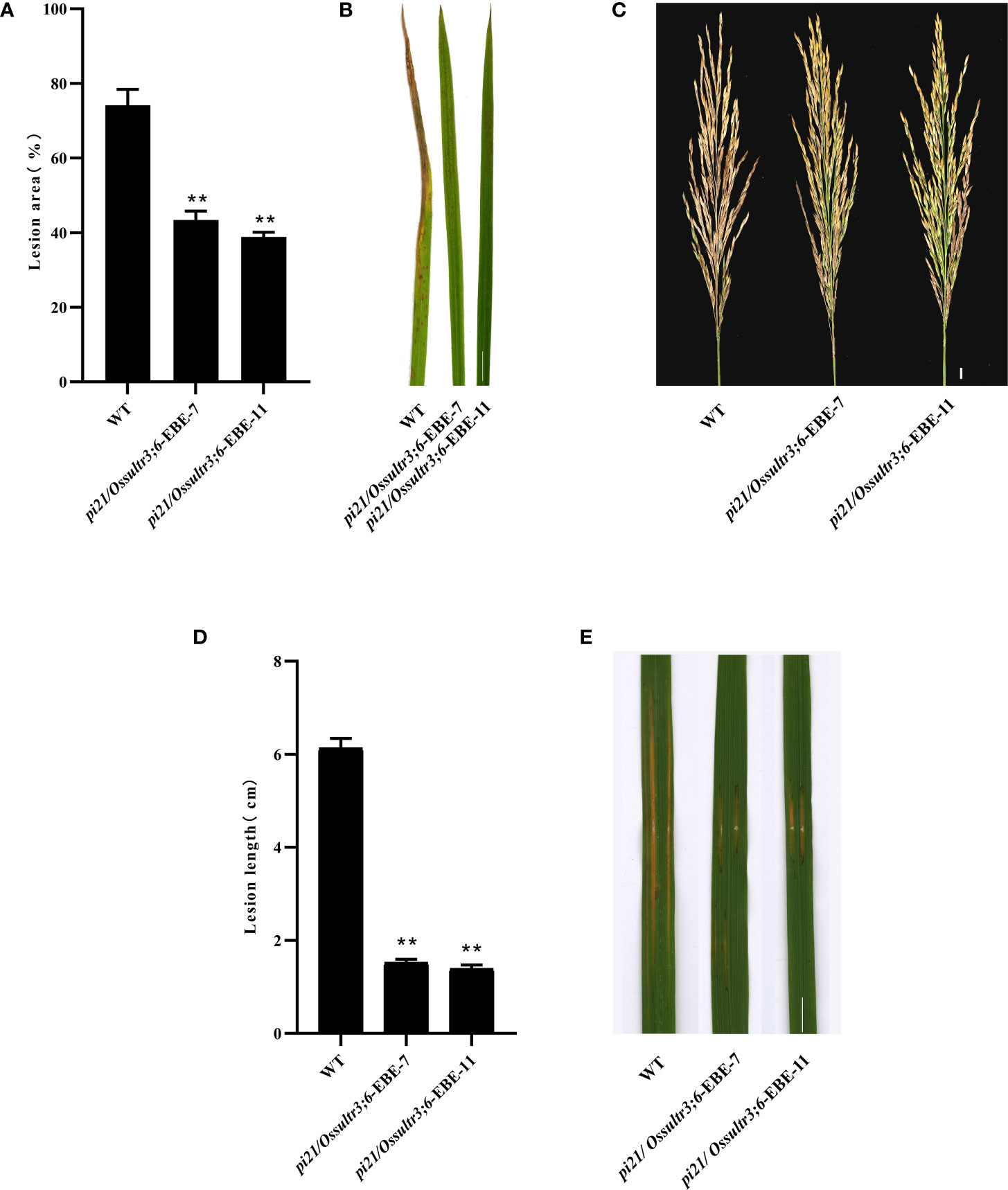
Figure 5 Bacterial leaf streak and rice blast resistance were enhanced in mutants. (A) The percentage of lesion areas of rice blast. (B) Rice mutant lines and wild-type 58B were tested for resistance to M. oryzae at the seedling stage. (C) Rice mutant lines and wild-type 58B were tested for resistance to M. oryzae at the mature stage. (D) The percentage of lesion length of bacterial leaf streak. (E) Rice mutant lines and wild-type 58B were tested for resistance to Xoc at the tillering stage. t-test: **P < 0.01.
As susceptibility genes, the Pi21 and OsSULTR3;6 genes play an important role in the invasion of pathogens and promote rice disease through transcription and translation into corresponding proteins, and their expression levels have a huge impact on the degree of disease. To further study the effect of pi21 and Ossultr3;6 genes mutations on rice plant susceptibility, the expression levels of pi21 and Ossultr3;6 in the knockout mutants were detected. The wild type was used as the control, and OsActin was used as the internal reference gene for qRT-PCR detection. The results showed that the expression of the pi21 gene of pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11 plants was significantly decreased by compared with that of the wild type (Figure 6A). The Ossultr3;6 gene expression levels of pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11 were decreased, respectively, compared with that of the wild type (Figure 6B). The result showed that the pi21 gene can significantly reduce its expression and improve the resistance to rice blast; after editing EBE, it can prevent the TALE secreted by Xoc from binding with EBE, thereby significantly reducing the expression of Ossultr3;6 gene and achieving disease resistance.
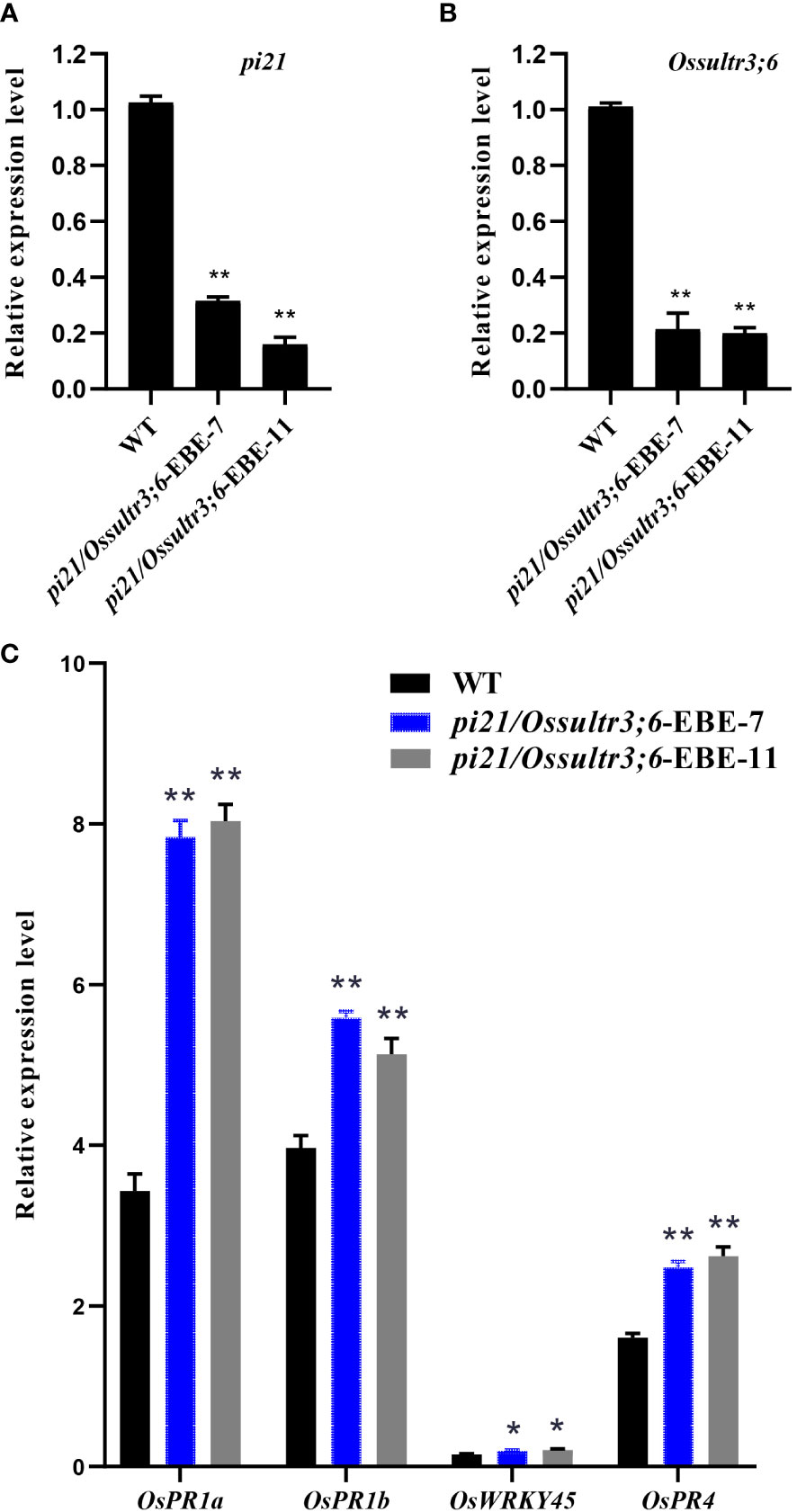
Figure 6 Expression analysis of pi21, Ossultr3;6 genes and defense responsive genes. (A) Relative expression of pi21. (B) Relative expression of ossultr3;6. (C) Relative expression level of defense responsive genes after inoculation of rice blast. t-test: *P < 0.05 and **P < 0.01.
After the pathogenic bacteria infect rice, the plant will initiate an immune response through the hormone signaling pathway. To study the role of the pi21 in the pathogenic infection of rice, the leaves of the inoculated site were taken 24 h after inoculation, and the extracted RNA was used to detect the expression of SA signaling pathway marker genes OsPR1a, OsPR1b, OsWRKY45, and JA signaling pathway marker gene OsPR4. The results showed that the gene expression levels of the SA signaling pathway and the JA signaling pathway were significantly increased after rice blast inoculation (Figure 6C), which may be the reason why pi21 showed disease resistance.
To study the effects of Pi21 and OsSULTR3;6 gene editing on the main agronomic traits of rice, the homozygous mutants without foreign genes in the T2 generation were chosen and planted in separate planting ponds. Their main agronomic traits were then statistically analyzed under the same growing conditions. pi21/Ossultr3;6-EBE-7 and pi21/Ossultr3;6-EBE-11 lines with both Pi21 and OsSULTR3;6-EBE mutations did not differ significantly from the wild type in plant height, 1,000-grain weight, panicle length, number of grains per panicle, or effective panicle number, This results indicated that the Pi21 and OsSULTR3;6-EBE mutation did not affect the main agronomic traits (Figure 7).
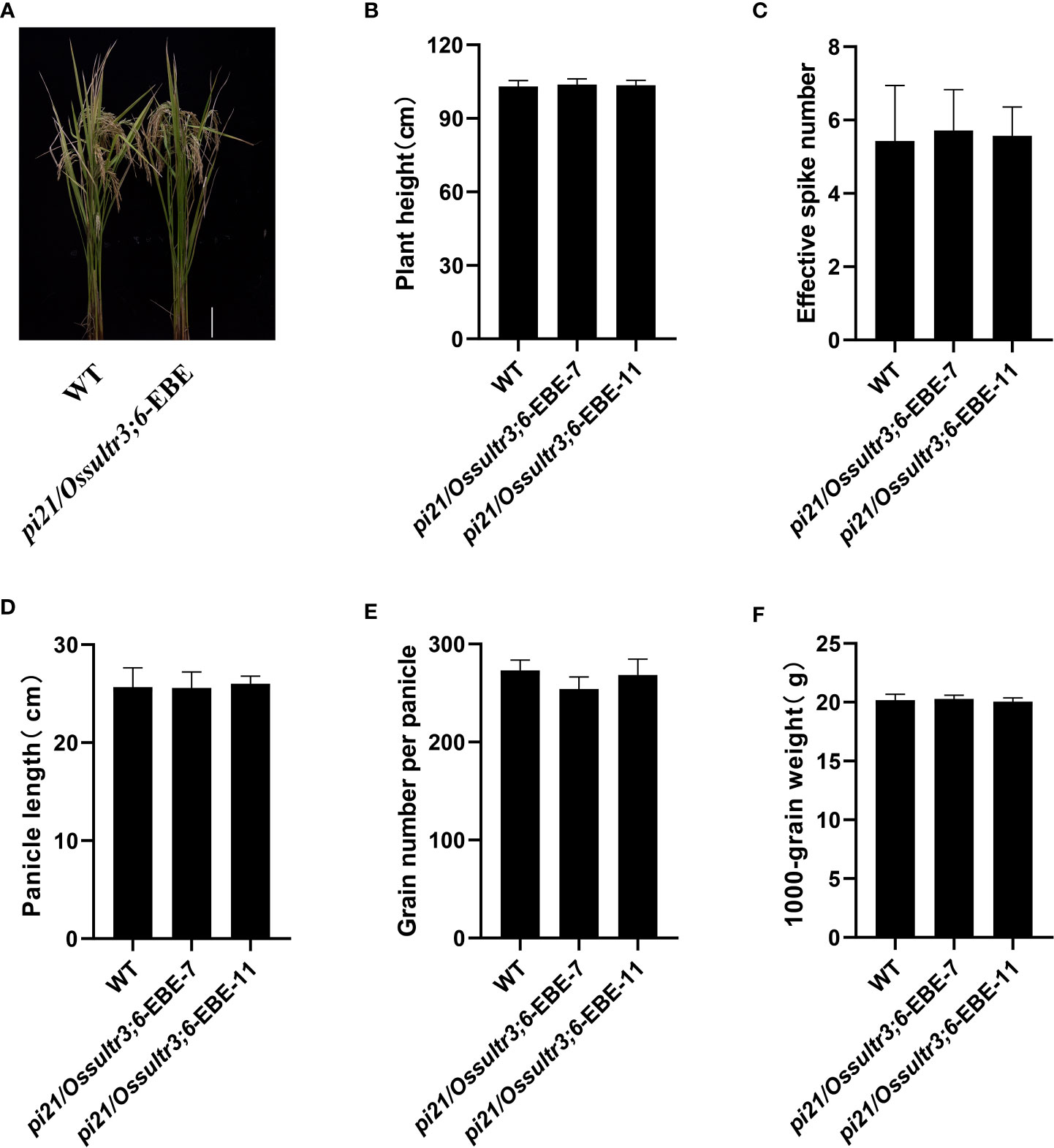
Figure 7 The agronomic traits of the mutants and the wild type (WT). (A) Phenotypes of pi21/Ossultr3;6-EBE mutant lines. (B) Plant height. (C) Effective spike number. (D) Panicle length. (E) Grain number per panicle. (F) 1,000-grain weight.
Currently, rice breeding should consider not only high yield but also high rice quality. Rice blast and bacterial leaf streak are the main diseases that seriously harm the yield and quality of rice (Liu et al., 2014; Asibi et al., 2019). With the rapid development of CRISPR/Cas9-mediated genomic technologies, modifying S genes to produce new varieties with BSR has become feasible for many crops (Xu et al., 2019). For example, the CRISPR/Cas9 knockout mutant of the rice S gene Pi21 is resistant to M. oryzae (Nawaz et al., 2020). 58B is an Indica rice variety independently bred by our laboratory for many years. 58B originated from yexiang B/IR58025B//43B/DP15; among them, yexiang B and 43B are indica rice maintainer line from China. IR58025B is derived from the International Rice Research Institute indica rice maintainer line. DP15 is a common wild rice (O. rufipogon Griff). It has outstanding advantages, high rice quality, first grade rice, and good taste. It has short stalks, strong tillering power, narrow leaves, upright, good plant shape, strong compatibility, strong hybrid advantage, and good productivity. However, it is susceptible to rice blast and bacterial leaf streak, which affects yield and quality.
To address this issue, in the study, CRISPR/Cas9 technology was used to precisely edit both the BSR rice blast gene Pi21 and the promoter region EBE of the susceptibility gene OsSULRT3;6, which produced to the single pi21 mutants, single Ossultr3;6-EBE mutants, and pi21/Ossultr3;6-EBE double mutants. After inoculation of M. oryzae and Xoc, the results showed that single mutants of the S gene Pi21 or OsSULTR3;6-EBE can enhance resistance to M. oryzae or Xoc, and this is consistent with the previous results (Nawaz et al., 2020; Xu et al., 2020). Noteworthy, when both Pi21 and OsSULTR3;6-EBE were edited, the pi21/Ossultr3;6-EBE double mutant has resistance against both M. oryzae and Xoc. Similar to the resistance conferred by the single mutants, no superimposed or attenuated effects were found. The mutant that was planted in the rice blast–infected field also demonstrated heightened resistance against rice blast. However, the current shortage of M. oryzae or Xoc strains in the laboratory hinders the ability to determine whether the pi21/Ossultr3;6-EBE double mutants exhibit BSR. This crucial investigation will need to be conducted in the future.
Disruption of the S gene can cause other effects including reduced growth, low fertility, and reduced tolerance to other stresses (Zaidi et al., 2018). For example, knockdown of the S gene OsSWEET11 in rice significantly reduced the sucrose concentration in the embryo sac of the mutants, resulting in seed germination deficiency (Ma et al., 2017). The Xa13 gene controls not only disease resistance but also reproductive growth in rice. If its expression is suppressed while enhancing resistance to strain PX099A, then it can also cause pollen abortion in rice and reduce the fruiting rate (Yang et al., 2018). To eliminate this side effect, Li used CRISPR/Cas9 to edit the Xa13 promoter to obtain Xoo resistant rice with normal fertility (Li et al., 2011). Xoc introduces TALE into the plant cell through the type III secretion system, which recognizes and binds EBEs in the promoter region of the host susceptibility gene, activating the transcriptional expression of the susceptibility gene and making the host susceptible to disease (Hui et al., 2019). At present, there are still few studies on the genes corresponding to TALE in Xoc. The sulfate transporter protein gene OsSULTR3;6 is a susceptible gene, which can be bound by Tal2g, one TALE of Xoc, and cause bacterial leaf streak in host rice (Cernadas et al., 2014). By modifying the Tal2g-binding region (EBE) of the OsSULTR3;6 gene, we were able to obtain homozygous mutants that had a 1-bp base insertion and a 33-bp base deletion within the EBE region. It is important to note that the actual sequence of the OsSULTR3;6 gene itself remained unchanged. The bacterial leaf streak resistance was identified by acupuncture method, and both mutant strains showed significantly lower spot length and higher resistance than wild-type 58B, indicating that editing the EBE region of the susceptibility gene could effectively improve the resistance of rice to bacterial leaf streak, and this is consistent with the previous results (Xu et al., 2020). In the study, higher levels of SA signaling related genes OsPRla, OsPRlb, OsWRKY45 and JA signaling related gene OsPR4 were detected in mutants than in wild-type plants when the plants were infected with M. oryzae (Figure 6). These results suggest that the enhanced resistance of the mutant to rice blast may be associated with the activation of SA and JA signaling transduction genes. In the study, five key agronomic traits were evaluated in the field for the pi21/Ossutr3;6-EBE double mutants and observed no significant differences in these traits between the mutant and WT plants.
In conclusion, our work provides a rapid and effective approach to breed rice varieties resistant to rice blast and bacterial leaf streak, which could significantly accelerate the breeding of rice varieties with multiple disease resistance.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
RL, SC, and FL designed and supervised the research. JY, YF, and HW performed most experiments. JY, YF, and XG analyzed date. JY, YF, NZ, EM, and RL wrote the paper. HP, SH, YH, BQ, YL, SC, and RL provided resources. All authors contributed to the article and approved the submitted version.
This study was supported by the State Key Laboratory for the Conservation and Utilization of Subtropical Agro bioresources (No. SKLWSA‐a201914 and No. SKLCUSA-b202203). This research was funded by the Guangxi Zhuang Autonomous Region Science and Technology Department, grant numbers AA17204070, AB16380066, and AB16380093.
Author SC was employed by the company Guangxi Lvhai Seed Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1209384/full#supplementary-material
Asibi, A., Chai, Q., Coulter, J. (2019). Rice blast: a disease with implications for global food security. Agronomy 9 (8), 451. doi: 10.3390/agronomy9080451
Boch, J., Bonas, U., Lahaye, T. (2014). TAL effectors- pathogen strategies and plant resistance engineering. New Phytol. 204 (4), 823–832. doi: 10.1111/nph.13015
Bossa-Castro, A., Tekete, C., Raghavan, C., Delorean, E., Dereeper, A., Dagno, K., et al. (2018). Allelic variation for broad-spectrum resistance and susceptibility to bacterial pathogens identified in a rice MAGIC population. Plant Biotechnol. J. 16 (9), 1559–1568. doi: 10.1111/pbi.12895
Cernadas, R., Doyle, E., Nino-Liu, D., Wilkins, K., Bancroft, T., Wang, L., et al. (2014). Code-assisted discovery of tal effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PloS Pathog. 10 (4). doi: 10.1371/journal.ppat.1004126
Cox, K., Meng, F., Wilkins, K., Li, F., Wang, P., Booher, N., et al. (2017). TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 24 (8), 15588. doi: 10.1038/ncomms15588
David, D., Godelieve, G., Monica, H. (2013). Hormone defense networking in rice: tales from a different world. Trends Plant Sci. 18 (10), 555–565. doi: 10.1016/j.tplants.2013.07.002
Ebbole, D. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45, 437–456. doi: 10.1146/annurev.phyto.45.062806.094346
Hui, S., Shi, Y., Tian, J., Wang, L., Li, Y., Wang, S., et al. (2019). TAL-carrying bacterial pathogens trap host nuclear import receptors for facilitation of infection of rice. Mol. Plant Pathol. 20 (4), 519–532. doi: 10.1111/mpp.12772
Hummel, A., Doyle, E., Bogdanove, A. (2012). Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195 (4), 883–893. doi: 10.1111/j.1469-8137.2012.04216.x
Jens, B., Ulla, B. (2010). Xanthomonas avrbs3 family-type III effectors:discovery and function. Annu. Rev. Phytopathol. 48 (1), 419–436. doi: 10.1146/annurev-phyto-080508-081936
Jiang, N., Yan, J., Liang, Y., Shi, Y., He, Z., Wu, Y., et al. (2020). Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa l.)–an updated review. Rice 13, 3. doi: 10.1186/s12284-019-0358-y
Ke, Y., Deng, H., Wang, S. (2017). Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 90 (4), 738–748. doi: 10.1111/tpj.13438
Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00377
Li, C., Wei, J., Lin, Y., Chen, H. (2011). Gene silencing using the recessive rice bacterial blight resistance gene xa13 as a new paradigm in plant breeding. Plant Cell Rep. 31 (5), 851–862. doi: 10.1007/s00299-011-1206-8
Li, W., Zhu, Z., Chen, M., Yin, J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170 (1), 114–126.e15. doi: 10.1016/j.cell.2017.06.008
Liu, W., Liu, J., Triplett, L., Leach, J., Wang, G. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. doi: 10.1146/annurev-phyto-102313-045926
Liu, , Xie, W. X., Ma, X., Li, J., Chen, J., Liu, Y.-G., et al. (2015). DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Molecular Plant. 8 (9), 1431–1433. doi: 10.1016/j.molp.2015.05.009
Ma, L., Zhang, D., Miao, Q., Yang, J., Xuan, Y., Hu, Y. (2017). Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant And Cell Physiol. 58 (5), 863–873. doi: 10.1093/pcp/pcx040
Matsushita, A., Inoue, H., Goto, S., Nakayama, A., Sugano, S., Hayashi, N., et al. (2013). Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 73 (2), 302–313. doi: 10.1111/tpj.12035
Matthew, J., Adam, J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326 (5959), 1501. doi: 10.1126/science.1178817
Nawaz, G., Usman, B., Peng, H., Zhao, N., Yuan, R., Liu, Y., et al. (2020). Knockout of pi21 by CRISPR/Cas9 and itraq-based proteomic analysis of mutants revealed new insights into M. oryzae resistance in elite rice line. Genes 11 (7), 735. doi: 10.3390/genes11070735
Shiduku, T., Seika, M., Daisuke, T., Shoko, Y., Keiichiro, T., Yuya, U. (2014). Isolation of jasmonate-induced sesquiterpene synthase of rice: product of which has an antifungal activity against Magnaporthe oryzae. J. Plant Physiol. 171 (8), 625–632. doi: 10.1016/j.jplph.2014.01.007
Usman, B., Nawaz, G., Zhao, N., Liao, S., Qin, B., Liu, F., et al. (2021). Programmed editing of rice (Oryza sativa l.) OsSPL16 gene using CRISPR/Cas9 improves grain yield by modulating the expression of pyruvate enzymes and cell cycle proteins. Int. J. Mol. Sci. 22 (1), 249. doi: 10.3390/ijms22010249
Usman, B., Nawaz, G., Zhao, N., Liu, Y., Li, R. (2020). Generation of high yielding and fragrant rice (Oryza sativa l.) lines by crispr/cas9 targeted mutagenesis of three homoeologs of cytochrome P450 gene family and OsBADH2 and transcriptome and proteome profiling of revealed changes triggered by mutations. Plants 9 (6), 788. doi: 10.3390/plants9060788
Wilson, R., Talbot, N. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nature reviews. Microbiology 7 (3), 185–195. doi: 10.1038/nrmicro2032
Xie, X., Zheng, Y., Lu, L., Yuan, J., Hu, J., Bu, S., et al. (2021). Genome-wide association study of QTLs conferring resistance to bacterial leaf streak in rice. Plants 10 (10), 2039. doi: 10.3390/plants10102039
Xu, X., Li, Y., Xu, Z., Yan, J., Wang, Y., Wang, Y., et al. (2022). TALE-induced immunity against the bacterial blight pathogen Xanthomonas oryzae pv. oryzae in rice. Phytopathol. Res. 4 (1), 47. doi: 10.1186/s42483-022-00153-x
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable tale-binding elements of multiple susceptibility genes in rice. Mol. Plant 12 (11), 1434–1446. doi: 10.1016/j.molp.2019.08.006
Xu, X., Xu, Z., Li, Z., Zakria, M., Zou, L., Chen, G. (2020). Increasing resistance to bacterial leaf streak in rice by editing the promoter of susceptibility gene OsSULRT3;6. Plant Biotechnol. J. 19 (6), 1101–1103. doi: 10.1111/pbi.13602
Yamauchi, T., Yoshioka, M., Fukazawa, A., Mori, H., Nishizawa, N., Tsutsumi, N., et al. (2017). An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29 (4), 775–790. doi: 10.1105/tpc.16.00976
Yang, J., Luo, D., Yang, B., Formmer, W., Emo, J. (2018). SWEET11 and 15 as key players in seed filling in rice. New Phytol. 218 (2), 604–615. doi: 10.1111/nph.15004
Yang, D., Yang, Y., He, Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6 (3), 675–685. doi: 10.1093/mp/sst056
Zaidi, S., Mukhtar, M., Mansoor (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends In Biotechnol. 36 (9), 898–906. doi: 10.1016/j.tibtech.2018.04.005
Zeng, D., Ma, X., Xie, X., Zhu, Q., Liu, Y. (2018). A protocol for CRISPR/Cas9-based multi-gene editing and sequence decoding of mutant sites in plants (in Chinese). Sci. Sin. Vitae 48, 783–794. doi: 10.1360/N052018-00069
Keywords: rice, CRISPR/Cas9, Pi21, OsSULTR3;6, rice blast, bacterial leaf streak
Citation: Yang J, Fang Y, Wu H, Zhao N, Guo X, Mackon E, Peng H, Huang S, He Y, Qin B, Liu Y, Liu F, Chen S and Li R (2023) Improvement of resistance to rice blast and bacterial leaf streak by CRISPR/Cas9-mediated mutagenesis of Pi21 and OsSULTR3;6 in rice (Oryza sativa L.). Front. Plant Sci. 14:1209384. doi: 10.3389/fpls.2023.1209384
Received: 20 April 2023; Accepted: 27 June 2023;
Published: 17 July 2023.
Edited by:
Suprasanna Penna, Amity University, Mumbai, IndiaReviewed by:
Subhasis Karmakar, National Rice Research Institute (ICAR), IndiaCopyright © 2023 Yang, Fang, Wu, Zhao, Guo, Mackon, Peng, Huang, He, Qin, Liu, Liu, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongbai Li, bGlyb25nYmFpQDEyNi5jb20=; Shengwu Chen, NjkyNzkyMDYzQHFxLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.