
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 June 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1207764
 Xu Jiang1,2
Xu Jiang1,2 Zhen Wang1
Zhen Wang1 Jing Feng3
Jing Feng3 Ziyi Du4
Ziyi Du4 Zhongjun Zhang1
Zhongjun Zhang1 Yibin Zhang5
Yibin Zhang5 Mingzhe Che1
Mingzhe Che1 Junda Ren6
Junda Ren6 Haiguang Wang1
Haiguang Wang1 Wei Quan7*
Wei Quan7*Wheat yield has been constrained by stripe rust disease globally. A wheat landrace (Qishanmai, QSM) consistently showed lower stripe rust severities in multiple year studies than susceptible check varieties including Suwon11 (SW) at the adult plant stage. To detect QTL for reducing the severity in QSM, 1218 recombinant inbred lines (RILs) were developed from SW × QSM. QTL detection was conducted firstly using 112 RILs selected for similarity in pheno-morphological characters. The 112 RILs were assessed for stripe rust severity at the 2nd leaf, 6th leaf and flag leaf stages under field and greenhouse conditions, and genotyping was done primarily with a single nucleotide polymorphism (SNP) array. On the basis of these phenotypic and genotypic data, a major QTL (QYr.cau-1DL) was detected on chromosome 1D at the 6th leaf and flag leaf stages. Further mapping was conducted by genotyping 1218 RILs using new simple sequence repeat (SSR) markers, which were developed by referring to the sequences of the wheat line Chinese Spring (IWGSC RefSeq v1.0). QYr.cau-1DL was mapped within a 0.5 cM (5.2 Mb) interval delimited by the SSR markers 1D-320.58 and 1D-325.79. These markers were applied to select for QYr.cau-1DL by screening F2 or BC4F2 plants of the wheat crosses RL6058 × QSM, Lantian10 × QSM and Yannong21 × QSM. F2:3 or BC4F2:3 families derived from the selected plants were assessed for stripe rust resistance in the fields of two locations and in a greenhouse. Wheat plants carrying the resistant marker haplotype in homozygous state for QYr.cau-1DL showed lower stripe rust severities (by 44% to 48%) than plants lacking this QTL. The trial of RL6058 (a carrier of Yr18) × QSM also indicated that QYr.cau-1DL had larger effect than Yr18 on reducing severity; they acted synergistically, yielding an elevated level of stripe rust resistance.
Wheat (Triticum aestivum L.) is one of the most important staple food crops globally (IWGSC, 2018). Wheat yields have been constrained in many countries/regions of the world by stripe/yellow rust disease caused by the obligate biotrophic fungus Puccinia striiformis Westend. f. sp. tritici Erikss. (Pst). Major Pst epidemics have been documented in certain geographical regions of, such as, Australia, China, Europe and the United States of America, causing regional yield losses up to 25% (Wellings, 2011).
Identification and deployment of genes and quantitative trait locus/loci (QTL) conferring resistance to stripe rust is a cost-effective strategy to manage this disease (Johnson, 1984; Line, 2002). Eighty four stripe rust resistance (Yr) genes have been formally named (McIntosh et al., 2020; Klymiuk et al., 2022) and more than 350 stripe rust resistance QTL have been documented (Rosewarne et al., 2013; Maccaferri et al., 2015; Jan et al., 2021). During the last two decades, new Pst races became prevalent or even predominant in Pst populations in some countries/regions, which are virulent to many of the Yr genes (Chen, 2005; Milus et al., 2008; Milus et al., 2009; Wellings, 2011; Hovmøller et al., 2016; Ali et al., 2017; Dong et al., 2017) and can also defeat the resistance conferred by certain QTL (Sorensen et al., 2014). Identification of new resistance genes/QTL is an essential work to enhance genetic diversity of resistance resources for sustainably controlling stripe rust.
Some of stripe rust resistance genes/QTL function at all plant growth stages, and some others confer adult plant resistance (APR) (Johnson, 1984; Line, 2002; Dong et al., 2017). The former are usually effective against a limited number of Pst races and typically encode NLR proteins (Dong et al., 2017; Javier and Keller, 2021), whereas the latter encode more diverse proteins (Fu et al., 2009; Krattinger et al., 2009; Moore et al., 2015; Dong et al., 2017). APR genes/QTL are often considered to be race non-specific, while exceptions were reported (Johnson, 1984; Johnson, 1992; Singh et al., 2011; Sorensen et al., 2014; Brown, 2015). Certain APR genes/QTL, such as Yr18, Yr29 and Yr46 (Rosewarne et al., 2013) and QYrst.wgp-6BS.1/Yr78 (Santra et al., 2008; Dong et al., 2017), have been demonstrated to confer durable resistance sensu Johnson (1984) and Line (2002). There are also reports where APR was detectable as early as at the 4th leaf stage (Ma and Singh, 1996; Quan et al., 2013; Segovia et al., 2014; Chhetri et al., 2016; Kanwal et al., 2021), being described as “mid-stage resistance” (Kanwal et al., 2021) and “juvenile stage” resistance (Segovia et al., 2014).
Effect size/magnitude of a QTL can be measured in percentage of phenotypic variance explained (PVE%) by the QTL. APR QTL may vary considerably in effect size with a PVE% value ranging from 1% to 88% (Rosewarne et al., 2013; Maccaferri et al., 2015; Jan et al., 2021). QTL with large effects, so-called “major QTL”, are usually more appreciated than minor ones in practical breeding programs (Rosewarne et al., 2013; Wang et al., 2019).
A wheat landrace (Qishanmai, QSM) was observed to possess APR to stripe rust in our previous multiple year studies (Supplementary Table 1), although it was susceptible at the seedling stage. The current study was carried out to (1) map QTL for stripe rust resistance in QSM, (2) screen for DNA markers closely flanking a mapped major QTL, (3) validate the major QTL across different environments and genetic backgrounds, and (4) examine combination effect of the mapped QTL with Yr18.
The mapping population comprising 1218 F8 recombinant inbred lines (RILs) from the wheat cross Suwon11 (SW) × QSM was developed as described in Zhang et al. (2023). Briefly, F2 was advanced to F6 using single seed descent method; seeds of the same RIL were pooled at each of the generations F7, F8, etc. The winter wheats QSM and SW have the accession numbers S-00402 and AUS-22519, respectively, at http://wheatpedigree.net/. SW is pheno-morphologically similar with QSM as well as is highly susceptible to stripe rust. The wheat variety Mingxian169 (MX) was used as susceptible check throughout the study. QTL mapping was performed firstly using 112 RILs and then the location of a QTL was refined using 1218 RILs.
Our pilot trials indicated that expression level of the stripe rust resistance in QSM varied with plant growth stages. Here, the resistance of wheat plants was examined at the 2nd leaf, 6th leaf and flag leaf stages. To minimize possible interference with disease recording by difference in growth progress among plants, 112 RILs were selected from the SW × QSM population to serve as test materials for preliminary mapping. The 112 RILs unfolded their main shoot leaves on the same day at each of the 3 plant growth stages and they are also very similar one another in plant height. These RILs together with their parents were evaluated for disease severity in the winter wheat cropping season from autumn 2018 to summer 2019 (abbreviated as “season 2019” hereafter) in the fields of southern Gansu province and Shandong province, and in a greenhouse. Each of the 3 locations was considered as an individual “environment”.
In southern Gansu, trial plots were located in Wushan county (34°42′15′′N, 104°40′08″E, and elevation 1650 m). The trial was carried out in the same ways as in Zhang et al. (2021). Briefly, each plot was constituted with a single 1-m-long row spaced 25 cm apart. A plot was sown with about 40 seeds from an individual RIL of SW × QSM. SW, QSM and the susceptible check wheat MX were grown after every 28 RIL rows. Each RIL was replicated in three plots in a randomized complete block layout. MX was also sown in the bordering areas surrounding the trial plots. As detailed in Zhang et al. (2017) and Wang et al. (2019), the geo-morphological and weather conditions are highly favorable for Pst to over-summer, and consequently, to infect the autumn-sown wheat seedlings naturally with no need for artificial inoculation. Pst hyphae in the infected wheat plants can over-winter, and then produce and release urediniospores that infect wheat plants in the forthcoming spring (Chen et al., 2009). To minimize a possible cold damage to the wheat plants that harbor Pst, the test plots were protected using plastic films in winter months. On drought days in autumn, spring and the early-summer, the test wheat plants were frequently atomized with water after sunset to facilitate Pst infection as we previously detailed (Wang et al., 2019; Zhang et al., 2021). Pst samples collected from infected MX plants were differentiated for their pathotypes applying the methods used in the USA (Wan and Chen, 2014), but choosing 17 Yr genes as differentials that have high differential ability in China (Supplementary Table 2). The identified pathotypes were collectively virulent against the genes 1, 6, 7, 8, 9, 10, 17, 24, 26, 27, 28, 29, 32, SP and Tr1, and avirulent to Yr5 and Yr15 (Supplementary Table 2). Disease severity was scored as detailed in Zhang et al. (2017) and Wang et al. (2019). Briefly, the score was in a percentage of infected leaf area (Peterson et al., 1948); stripe rust areas were visually averaged over all the flag leaves within a plot when severities on SW and MX flag leaves reached approximately 90%. Mean severity of three plot replicates for each RIL was used for QTL mapping.
In Shandong, trial plots were located in Tai’an district (36°18′09′′N, 117°13′05″E, and elevation 90 m). The trial was done in the same ways as mentioned above with some exceptions, i.e., spreader (MX) plants were grown in rows adjacent and perpendicular to the test plot rows. Artificial inoculation is needed in this location. The inoculums were the mixed urediniospores of the Chinese Pst races CYR32 and CYR34, which have been prevalent in China for multiple years (Han et al., 2015; Wu et al., 2016; Bai et al., 2018). These races are collectively virulent against the Yr genes 1, 2, 3a, 4a, 4b, 6, 7, 8, 9, 10, 17, 23, 24, 25, 26, 27, 28, 29, 31, 43, 44, A, Alba, Cle, CV1, CV2, CV3, Exp2, Gaby, Res, SD, SO, SP, SpP and Su, and avirulent to Yr5 and Yr15 (Wan et al., 2004; Li et al., 2006; Chen et al., 2009; Wu et al., 2016). Initial urediniospores of the races were kindly provided by Chen et al. (2009) and Dr. Ruiming Lin (Institute of Plant Protection, Chinese Academy of Agricultural Science). Propagation of urediniospores and inoculation were performed as detailed in Wang et al. (2019) and Zhang et al. (2021). Briefly, the urediniospores were increased on MX plants grown in a greenhouse, and fresh urediniospores were harvested just before they were needed for inoculating the field trial plants. At the tillering to stem elongation stages (mid-March to early April) of the field test plants, a clear afternoon was chosen for inoculation. The MX spreader plants were sprayed with water solution containing well suspended fresh Pst urediniospores (in a mean density of about 75,800 urediniospores mL-1) and 0.04% Tween 20. Efforts were made to distribute spore dewdrops evenly over wheat leaf surfaces as fine as possible with minimized coalescence; the plants were then protected with plastic films for 16 h. Such an inoculation was repeated three times in the period from mid-March to early April. Three weeks after inoculation, fresh Pst urediniospores released from the spreader plants were readily observed. Beginning from this time until last disease recording, the test plants were regularly sprayed with water after sunset. Disease severities were assessed in the ways as mentioned above.
In greenhouse, trial was done in the same ways as described above with some exceptions, i.e., each test plot (20 cm × 20 cm) was sown with 9 seeds from one of the RILs/parents in a drill manner. When their 2nd leaves unfolded completely, the wheat seedlings were inoculated with Pst in the same ways as described above. The inoculated seedlings were incubated for 18 h at 6°C to 12°C. The greenhouse temperature was then controlled to a range between 5°C and 25°C with natural daylight supplemented artificially to 16 h at 11000 lux until disease recording was completed. Such a procedure involving inoculation, incubation and greenhouse condition control was repeated when the main shoots’ 6th leaves of the wheat plants unfolded completely (concurrent with 1 to 4 tillers depending on plants). Disease severity was recorded on the inoculated 2nd and 6th leaves, respectively, in the same ways as mentioned above.
Genotyping and QTL mapping were conducted in the same ways as in Zhang et al. (2023). Briefly, DNA samples were prepared from a single F8 plant of each of the 112 SW × QSM RILs as well as SW and QSM with the CTAB method in Saghaimaroof et al. (1984). These plants were subjected to a genotypic test applying a 90 K wheat SNP array (Wang et al., 2014) at the CapitalBio Technology (Beijing, China; http://www.capitalbiotech.com). After excluding heterozygous data points, 10026 polymorphic SNPs between SW and QSM were identified from the remaining data. To improve quality, filtering was conducted by removing the SNPs with an allele frequency of lower than 0.35 or higher than 0.65, or with missing scores higher than 5%; 8637 SNPs with high quality were identified. Redundant SNPs were filtered out applying the software in Li et al. (2007) and manually; the retained 1578 unique SNP markers were subjected to a linkage analysis applying JoinMap 4.0 (Stam, 1993). Genetic distance (in cM) was calculated using Kosambi function. Each linkage group was assigned to a specific chromosome by aligning the sequences containing the SNPs to the chromosomal survey sequence map (IWGSC, 2014; IWGSC, 2018).
However, we observed that the SNP map contained 32 large gaps (>20 cM). To reduce such gaps, 988 SSR markers were developed on the basis of the sequence IWGSC RefSeq v1.0 (IWGSC, 2018). These markers were designated with chromosome name plus numbers indicating coordinates in Mb in the IWGSC RefSeq v1.0; for instance, “1D-322.55” denotes an SSR marker physically positioned at 322.55 Mb on chromosome 1D. PCR reaction was carried out as we previously described (Zhang et al., 2023). Briefly, each reaction tube included 4.65 μL ddH2O, 10 × PCR buffer (Mg2+ plus, 1.0 μL), DNA template (25–55 ng/μL, 2.0 μL), forward and reverse primers (both in 10 μM, 1.0 μL each primer), dNTPs (10 mM, 0.25 μL), and Taq DNA polymerase (5 unit/μL, 0.1 μL). PCR amplification was performed following Roder et al. (1998), i.e., started with 94°C for 3 min, then 45 cycles at 94°C for 1 min, 60°C for 1 min and 72°C for 2 min, and ended with 72°C for 10 min. In cases where this cycling failed to produce amplicons, a touchdown program was used, i.e., started with 94°C for 5 min; then 10 cycles at 94°C for 30 s, 60°C for 30 s (touchdown with -0.5°C each cycle), and 72°C for 30 s; followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and ended with 72°C for 10 min. PCR products were differentiated using 6% denaturing polyacrylamide gels and stained with AgNO3 (Bassam et al., 1991). Thirty-two of the 988 markers were selected to genotype the 112 RILs. Sequences and details of the 32 PCR markers including the PCR profiles were presented in Supplementary Table 3.
The composite interval mapping (CIM) method in Windows QTL Cartographer 2.5 (Wang et al., 2010) was chosen to do QTL mapping. A threshold LOD value was determined for each of the trials applying the permutation program that was run repeatedly for 1000 times using α = 0.05 as the type I error rate. The threshold value varied with different trials, ranging from 2.7 to 3.5. For the sake of simplicity, 3.5 was used as the threshold for all trials. CIM control parameters were set with the model 6 (standard model), a window size of 10 cM, and the forward and backward regression at a criteria probability of 0.05 for both “into” and “out”. The walking speed was at 0.5 cM. Effect size/magnitude of a QTL was expressed in phenotypic variance explained (PVE) by the QTL.
A major QTL, detected using the aforementioned 112 RILs, was further mapped using all the 1218 RILs and hundreds of new SSR markers that were developed and named as mentioned above. These markers were screened firstly against SW and QSM and then against SW × QSM RILs. It was found that two (1D-298.34 and 1D-356.22) (Supplementary Table 3) of these markers delimited the detected major QTL to a relatively short interval. From the 1218 RILs, 92 recombinants were identified between 1D-298.34 and 1D-356.22. The 92 RILs were assessed for stripe rust resistance in season 2020 in the fields of southern Gansu and Shandong, and in a greenhouse as described above.
To validate a QTL mapped in SW × QSM population, three wheat populations were used, namely, 30 BC4F2:3 families selected from Lantian10 (LT) × QSM, 30 BC4F2:3 families from Yannong21 (YN) × QSM, and 80 F2:3 families from RL6058 × QSM. Such selections were conducted using DNA markers that tag the mapped QTL as detailed in Supplementary File 1. LT and YN were commercial cultivars, respectively, in southern Gansu and Shandong with susceptibility to stripe rust. RL6058 carries Yr18, and thus RL6058 × QSM was also used for examining interaction between the detected QTL and Yr18. The plants of LT × QSM, YN × QSM, and RL6058 × QSM were tested in season 2022 for stripe rust resistance in, respectively, southern Gansu, Shandong, and a greenhouse. The trials were conducted basically in the same ways as described above, but seeds were sown individually 20 cm apart. Greenhouse conditions were the same as mentioned above. Disease severities were assessed on flag leaves of individual plants in all the three locations. Infection types were also recorded in southern Gansu based on a 1 to 9 scale following Line and Qayoum (1992).
Disease data were analyzed applying SAS/STAT version 9.3 (SAS Institute Inc., Cary, NC, USA). Analyses of variance, heritability and correlation for RIL disease data were performed with PROC GLM, PROC VARCOMP and PROC CORR, respectively. Disease means between different groups of BC4F2:3 or F2:3 plants were compared based on ANOVA and Tukey test at α = 0.05.
Stripe rust severities were equal to or higher than 90% on the susceptible parent SW at all stages and on the APR parent QSM at the 2nd leaf stage (Figure 1; Supplementary File 2 and Supplementary Table 4). At the 6th leaf and flag leaf stages, however, QSM showed obviously lower severities than SW, confirming our previous observation that QSM possesses APR. The disease severities of the 112 RILs ranged broadly from 5% to 100% (Figures 1A–C) and varied significantly among the RILs at the 6th leaf and flag leaf stages (Table 1), compared to the narrow range (Figure 1D) and insignificant variance at the 2nd leaf stage (Table 1). Correlation coefficients for RIL disease severity were significant between different plant growth stages and among different test environments (Table 2) with a heritability being 0.89. These data illustrate that the APR was genetically segregating in a quantitative manner and the resistance began to express at the 6th leaf stage.
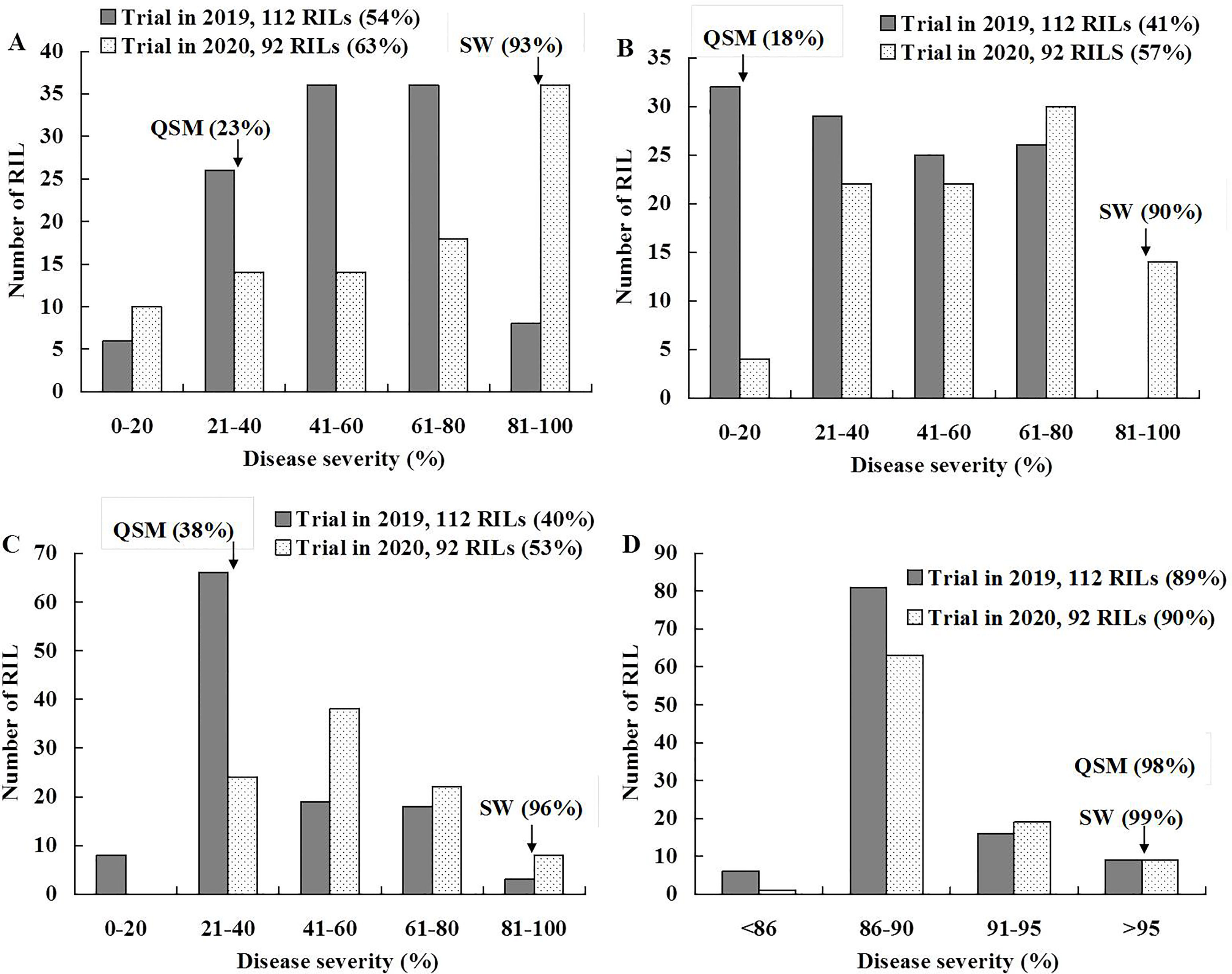
Figure 1 Frequency distribution of recombinant inbred lines (RILs) of Suwon11 (SW) × Qishanmai (QSM) for stripe rust severity. Disease was recorded on flag leaves in the fields of southern Gansu province (A) and Shandong province (B), and on 6th leaves (C) and 2nd leaves (D) in greenhouses. Mean severities are presented in parentheses.
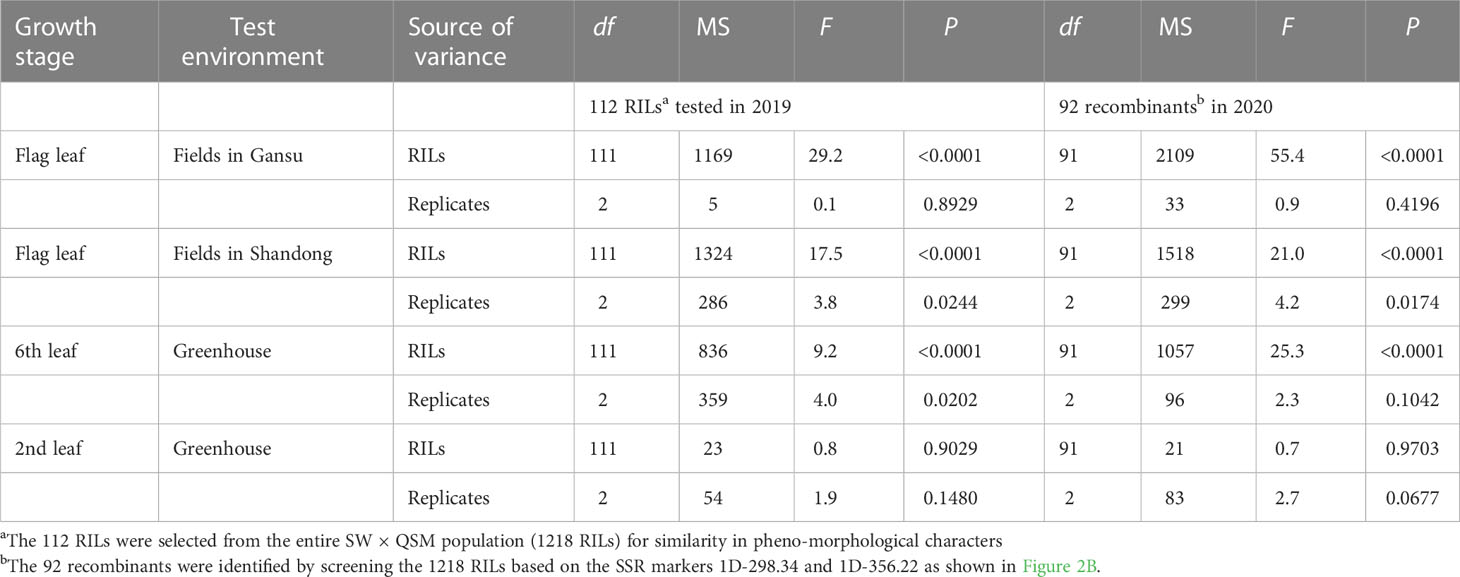
Table 1 Analysis of variance on disease severity of the recombinant inbred lines (RILs) of Suwon11 (SW) × Qishanmai (QSM).
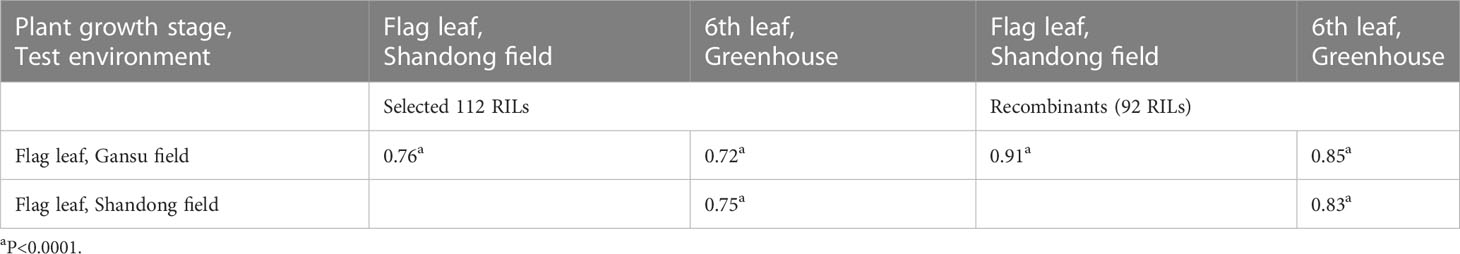
Table 2 Correlation coefficients for disease severity of recombinant inbred lines (RILs) of Suwon11 × Qishanmai among different plant growth stages and test environments.
The genetic map of the 112 RILs covers all 21 chromosomes with 1610 (1578 SNP and 32 SSR) markers and spans 3656.4 cM (Supplementary Table 5). The mean distance between markers was 2.3 cM with a maximum of 19.2 cM and a minimum of 0.1 cM. Based on this map and disease severity data of the 112 RILs, a QTL (QYr.cau-1DL) was detected on chromosome arm 1DL (Figure 2A). QYr.cau-1DL showed a LOD peak value from 12.4 to 23.4 that exceeded the threshold (3.5) at the 6th leaf and flag leaf stages in all trials (Table 3); at the 2nd leaf stage, however, LOD value was lower than 3.5 (Figure 2A). QYr.cau-1DL was flanked within a 5.7 cM interval by the SNP markers IWB15913 and IWB2332 (Figure 2A). IWB12282 was located at the chromosome position where LOD peak of QYr.cau-1DL occurred. Particularly, we could identify an SSR marker (1D-322.55) that was also located at the peak (Figure 2A). QYr.cau-1DL illustrated a PVE value of 35% at the 6th leaf stage and 48% to 55% at the flag leaf stage (Table 3); the stripe rust resistant allele was contributed by QSM.
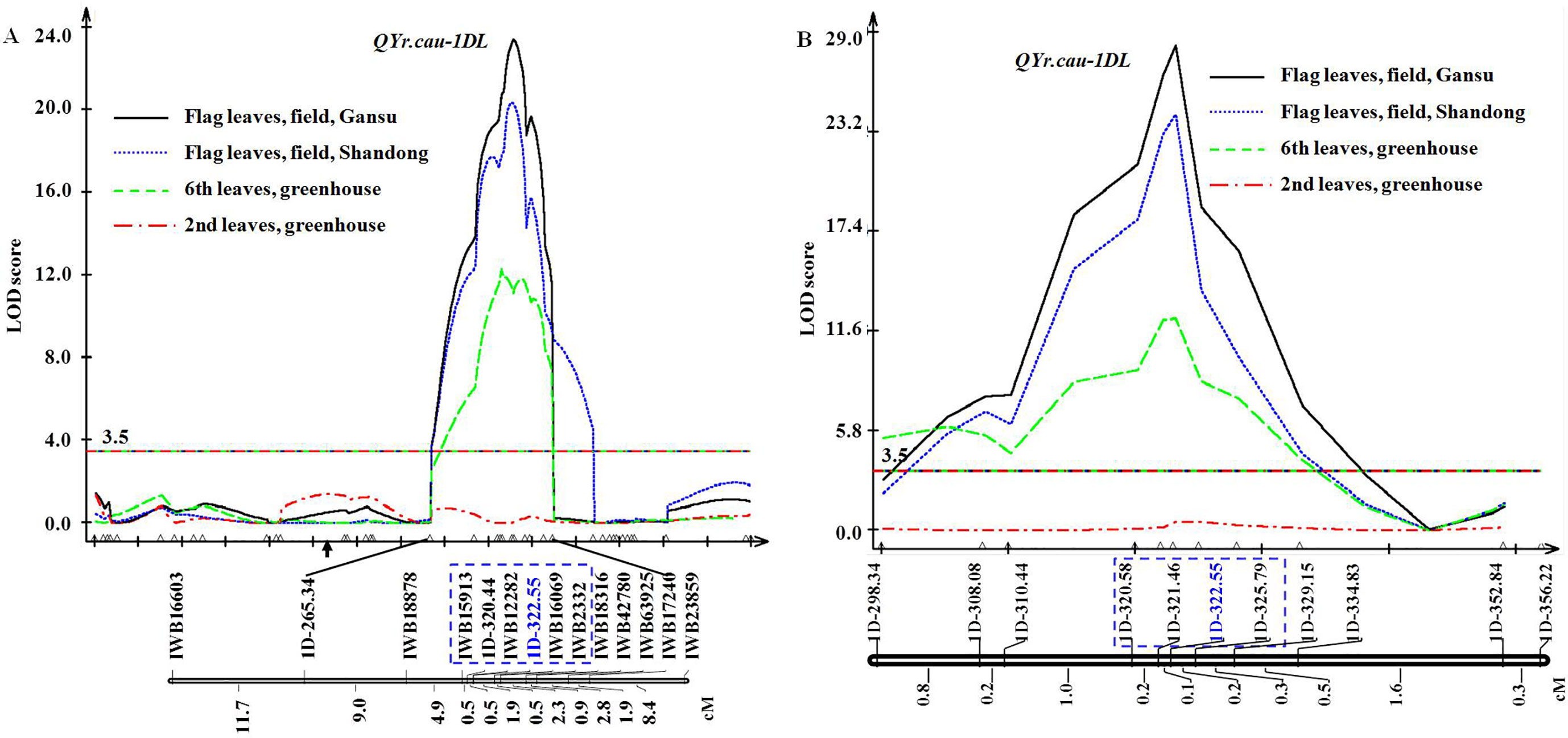
Figure 2 Genetic map of QYr.cau-1DL region in chromosome arm 1DL and logarithm of the odds (LOD) curves. (A) Location of QYr.cau-1DL mapped using 112 recombinant inbred lines (RILs). (B) Location of QYr.cau-1DL refined based on the map of 1218 RILs. Marker names and intervals (in cM of Kosambi) between adjacent markers are shown along the chromosome orientated with the telomere of 1DL to the right. Arrow suggests approximate position of the centromere. Horizontal lines indicate the threshold LOD of 3.5. Each of the small triangles along the x axis represents a marker used for QTL mapping. For reading ease, only certain marker names are displayed, while all marker names with their positions are presented in Supplementary Table 5.
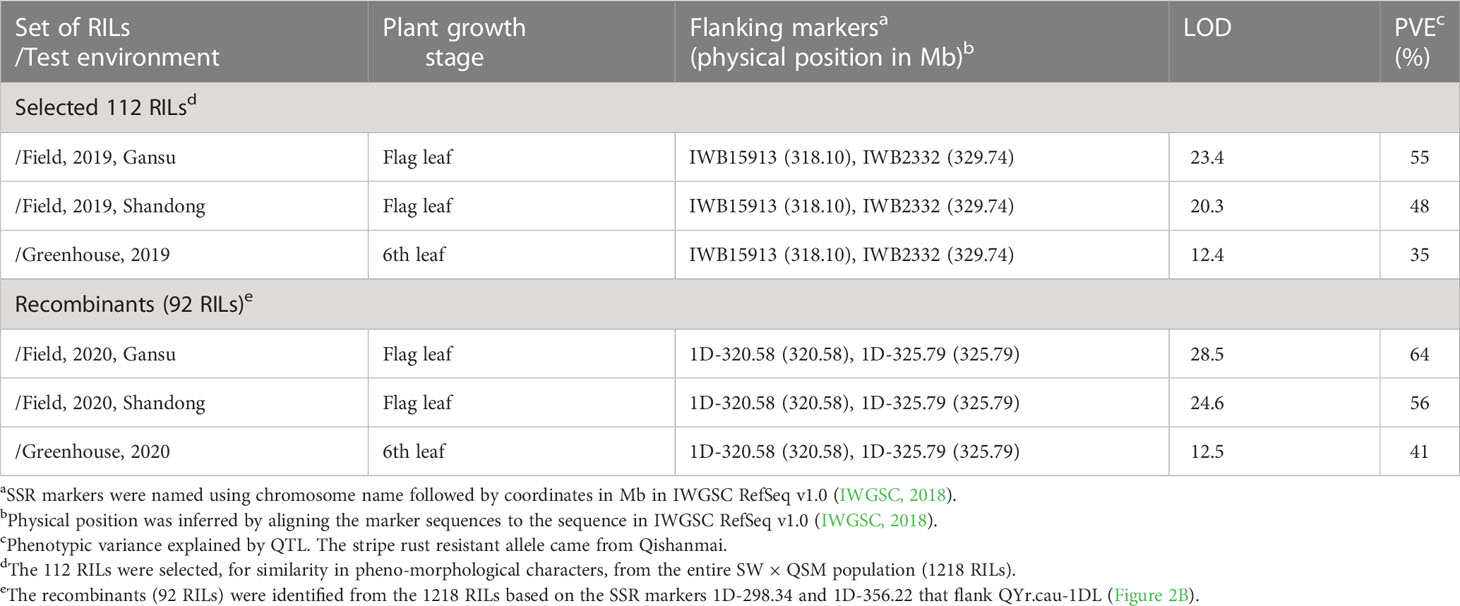
Table 3 A QTL (QYr.cau-1DL) on chromosome arm 1DL for stripe rust resistance in the recombinant inbred line (RIL) mapping population of Suwon11 (SW) × Qishanmai (QSM), detected in different environments.
Additional QTL were mapped on chromosomes 3A, 5A, 6A and 7A with PVE values being lower than 11%. At two of the four loci, resistant alleles came from QSM. These minor QTL will be further examined in future. Only QYr.cau-1DL is addressed here.
The genetic map (Figure 2A) constructed using 112 RILs provided an SSR marker (1D-322.55) that was considered as a tag for QYr.cau-1DL. Around the position of 1D-322.55 in the reference physical assembly (IWGSC, 2018), 607 new SSR markers were developed and used firstly to check QSM and SW. The identified 189 polymorphic markers were then applied to screen 20 RILs, sampled randomly from the 1218 ones, for checking marker segregation and quality. From the 189 markers, 18 ones of high quality were selected to genotype all 1218 RILs; 10 of the 18 markers (Supplementary Table 3) were mapped around 1D-322.55 (Figure 2B). Genetic distance between the most proximal marker (1D-298.34) and the most distal marker (1D-356.22) was 5.2 cM. Ninety-two out of the 1218 RILs were identified to be recombinants (Supplementary Table 6) between 1D-298.34 and 1D-356.22. Disease data of the 92 RILs consolidated the disease results of the 112 RILs as shown in Figure 1 and Tables 1, 2.
Based on the disease severity data of the 92 RILs (Supplementary Table 6) and the genetic map of 1218 RILs (Figure 2B), QYr.cau-1DL was delimited, by 1D-320.58 and 1D-325.79 (Figure 2B), to a 0.5 cM genetic interval that was equivalent to a physical distance of 5,207,169 bp on 1D, spanning 320,584,561–325,791,730 bp in the reference map (IWGSC, 2018).
PCR amplicon of QSM DNA was a 232 bp fragment at 1D-320.58 and a 290 bp fragment at 1D-325.79 (Figure 3); combination of the two fragments constitutes the marker haplotype for the resistant allele of QYr.cau-1DL. RL6058 and SW showed a susceptible haplotype, and LT and YN showed another susceptible haplotype. QSM had a 523 bp amplicon that represents the susceptible allele of Yr18, while the 751 bp amplicon of RL6058 represents the resistant allele (Figure 3).
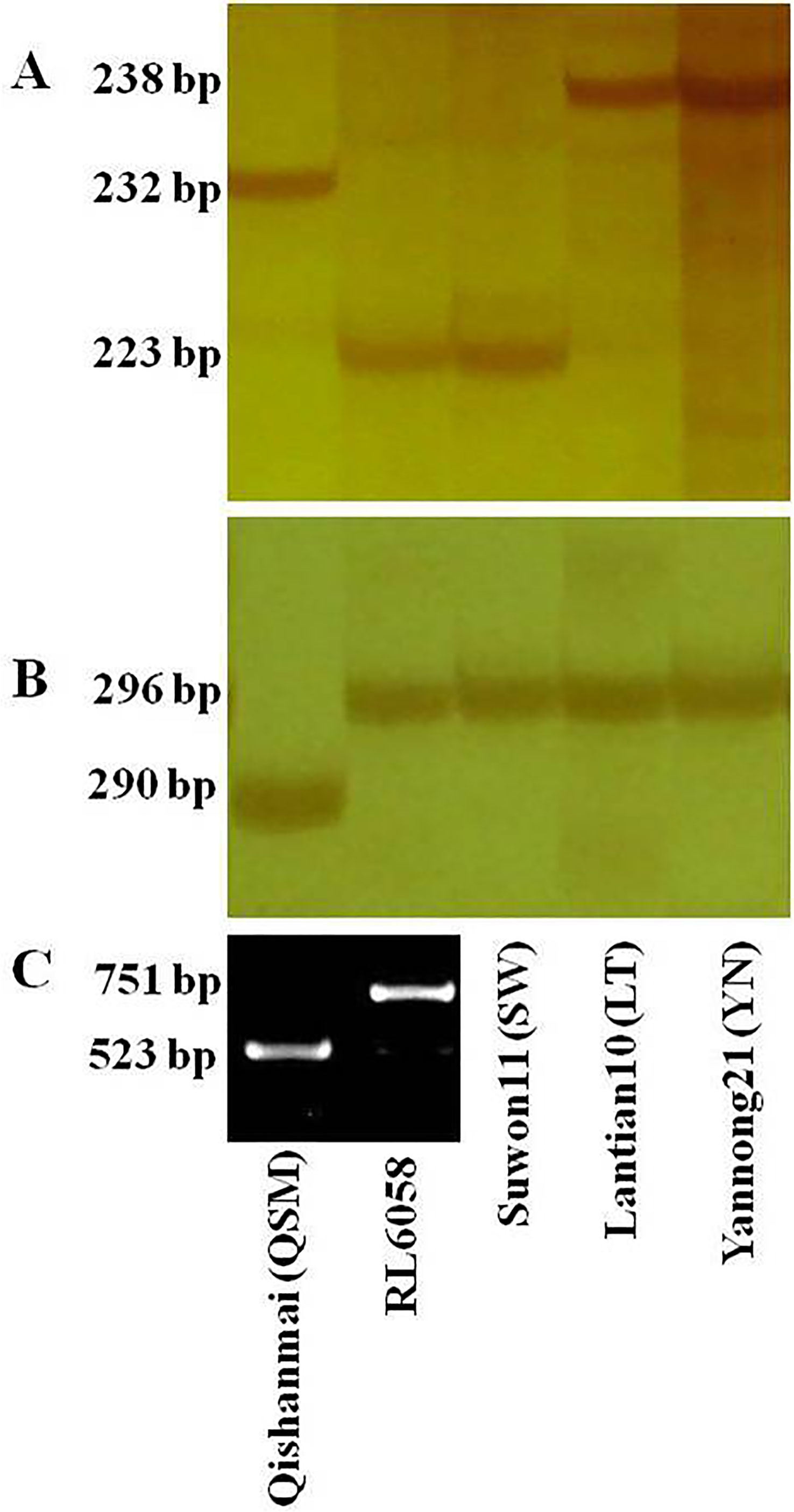
Figure 3 DNA fragments produced from the marker loci 1D-320.58 (A) and 1D-325.79 (B) for QYr.cau-1DL, and cssfr5 (C) for Yr18 by PCR amplification with DNA templates from the five tested wheat lines. DNA fragments were separated using polyacrylamide gels for (A, B), while using agrose gel for (C). Primer sequences are presented in Supplementary Table 3.
The R-plants and S-plants of BC4F2:3 families selected from LT × QSM (Supplementary File 1) had a homozygous resistant haplotype at QYr.cau-1DL and a homozygous susceptible haplotype, respectively. The R-plants showed a significantly lower mean disease severity (by 46%) than the S-plants (Figure 4A; Supplementary Table 7). Significant difference was also observed for infection type (Figure 4B) between the R- and S-plants. Correlation between disease severity and infection type was positive and significant (r = 0.96, P < 0.0001). Likewise, comparison between the R- and S-plants selected from YN × QSM (Supplementary File 1) showed a lower severity (by 44%) in the former than that in the latter (Figure 4A; Supplementary Table 8).
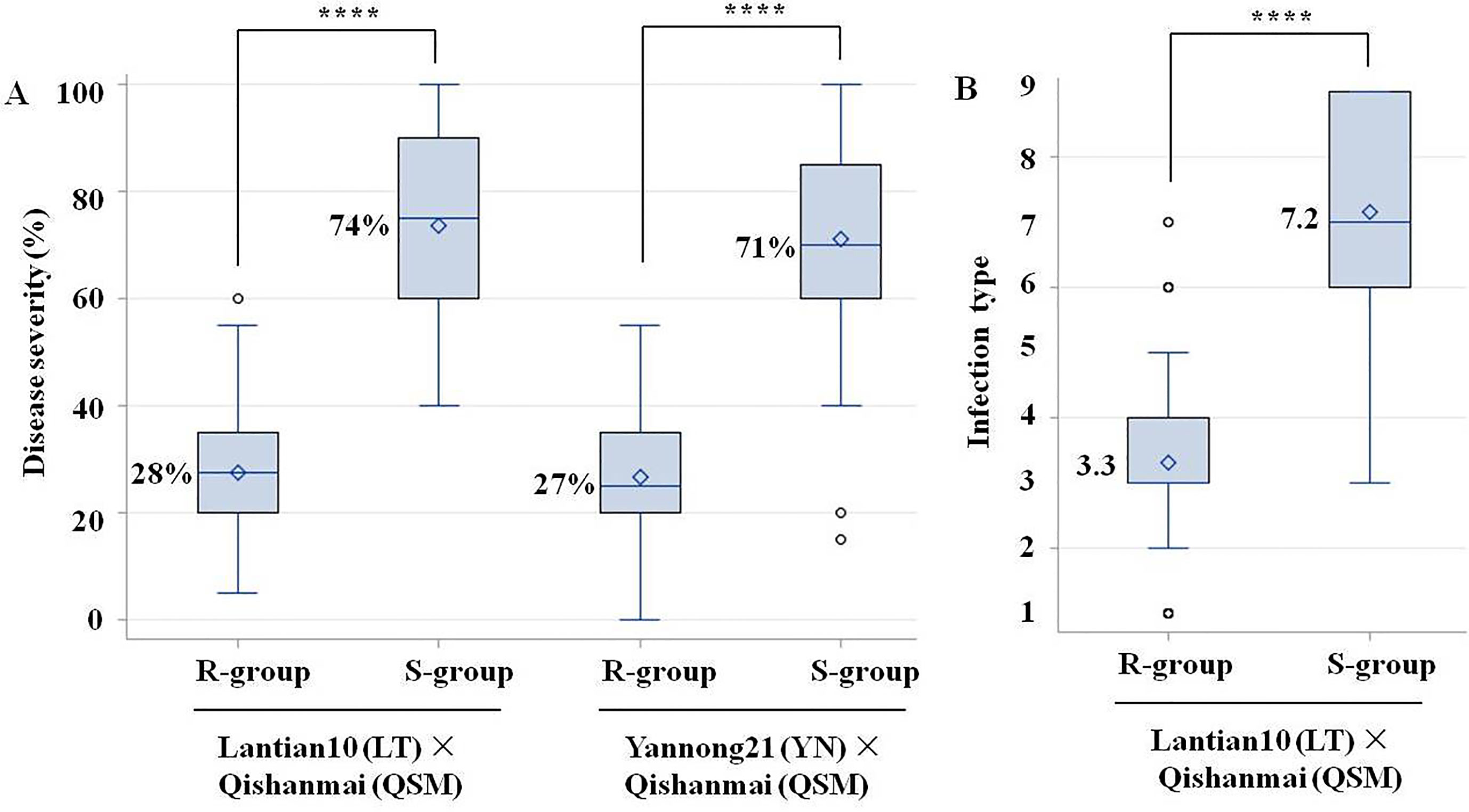
Figure 4 Boxplots showing effects of QYr.cau-1DL (represented by the flanking markers 1D-320.58 and 1D-325.79) on reducing stripe rust severity (A) and infection type (B) in Gansu field for LT × QSM and Shandong field for YN × QSM. R-plants carried the marker haplotype of stripe rust resistance in homozygous state at QYr.cau.-1DL and S-plants carried the haplotype of susceptibility in homozygous state. **** Indicates significant difference at α = 0.0001 based on an ANOVA and Tukey test. Small diamond and solid line within a box indicate the mean and median disease value, respectively. The top and bottom edges of a box illustrate the 75th and 25th percentiles, respectively. The whiskers outside a box extend to the extreme data points, and small circles denote outliers.
The four groups of F2:3 plants selected from RL6058 × QSM, namely, QYr.cau-1DL+Yr18, QYr.cau-1DL alone, Yr18 alone, and None (Supplementary File 1; Figure 5), were significantly different in disease severity (Figure 5; Supplementary Table 9). The lowest (13%) and highest (75%) severities were observed in the first group (i.e., QYr.cau-1DL+Yr18) and in the last group (None; susceptible control), respectively. The group carrying QYr.cau-1DL alone displayed a lower severity (by 14%) than the group carrying Yr18 alone, and a further lower severity (by 49%) than the susceptible control group (Figure 5).
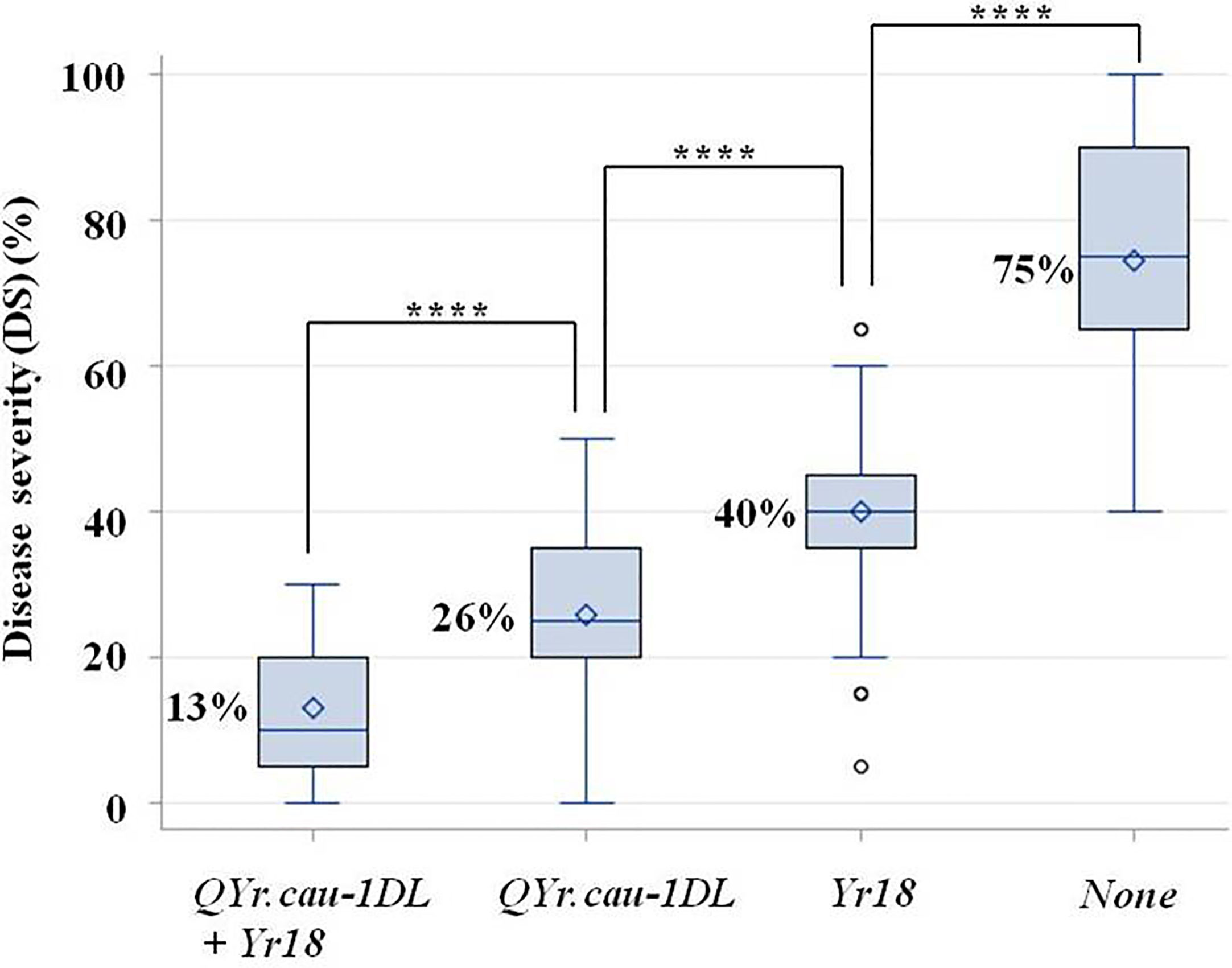
Figure 5 Boxplots showing the effects of QTL combinations on stripe rust severity in RL6058 × Qishanmai F2:3 population in a greenhouse. The x-axis defines four F2:3 family groups with different QTL combinations, i.e., presence of QYr.cau-1DL plus Yr18, QYr.cau-1DL alone, Yr18 alone, and None (neither QYr.cau-1DL nor Yr18), respectively. Refer to the legend of Figure 4 for descriptions of **** and box.
Wheat landraces have been considered to possess a higher level of genetic diversity than modern cultivars (Newton et al., 2010; Marone et al., 2021). A number of new stripe rust resistance genes/QTL have been found from wheat landraces as exemplified by those in the Watkins Collection (Bansal et al., 2011) and the Vavilov Collection (Jambuthenne et al., 2022). Likewise, the QYr.cau-1DL identified here from the landrace QSM constitutes an example from China, where there is a long wheat cultivation history with considerably diverse landraces (Zhou et al., 2018).
It is notable that QYr.cau-1DL was significantly effective in all the 9 field and greenhouse trials and in all the 4 wheat crosses SW × QSM, LT × QSM, YN × QSM and RL6058 × QSM (Table 3; Figures 2, 4, 5). The plants selected from RL6058 × QSM have a genetic background of spring habit (Supplementary File 1), contrasting to the winter/facultative habit of the other 3 crosses. The semi-dwarf wheat cultivar LT (95 cm) and dwarf YN (75 cm) were substantially different each other in a number of pheno-morphological traits (data not shown) and adapt to highland (Gansu province, elevation 1650 m) and lowland (Shandong province, 90 m) ecosystems, respectively. The trial plots in Gansu were separated by 1100 km from those in Shandong. These data indicate that QYr.cau-1DL can consistently act in greatly varied circumstances.
QYr.cau-1DL is located on chromosome 1D where 30 stripe rust resistance QTL were previously detected (Figure 6; Supplementary Table 10). These QTL were physically projected by aligning the sequences of their representing marker/s to 1D sequence in the IWGSC RefSeq v1.0 (Supplementary Table 10). QYr.cau-1DL was projected to a 5.2 Mb region, spanning 320.58–325.79 Mb. There is a distance of >60 Mb between QYr.cau-1DL and the other 30 QTL (Figure 6). Yr25 was also located on 1D by Calonnec and Johnson (1998) using Chinese Spring monosomic lines, but it could not be physically positioned due to lack of DNA marker. Our observations illustrated that the wheat variety Reichersberg 42 (a carrier of Yr25) was highly susceptible (Supplementary File 2) in terms of both infection type and disease severity. Therefore, it is highly likely that QYr.cau-1DL is a new QTL for resistance to stripe rust.
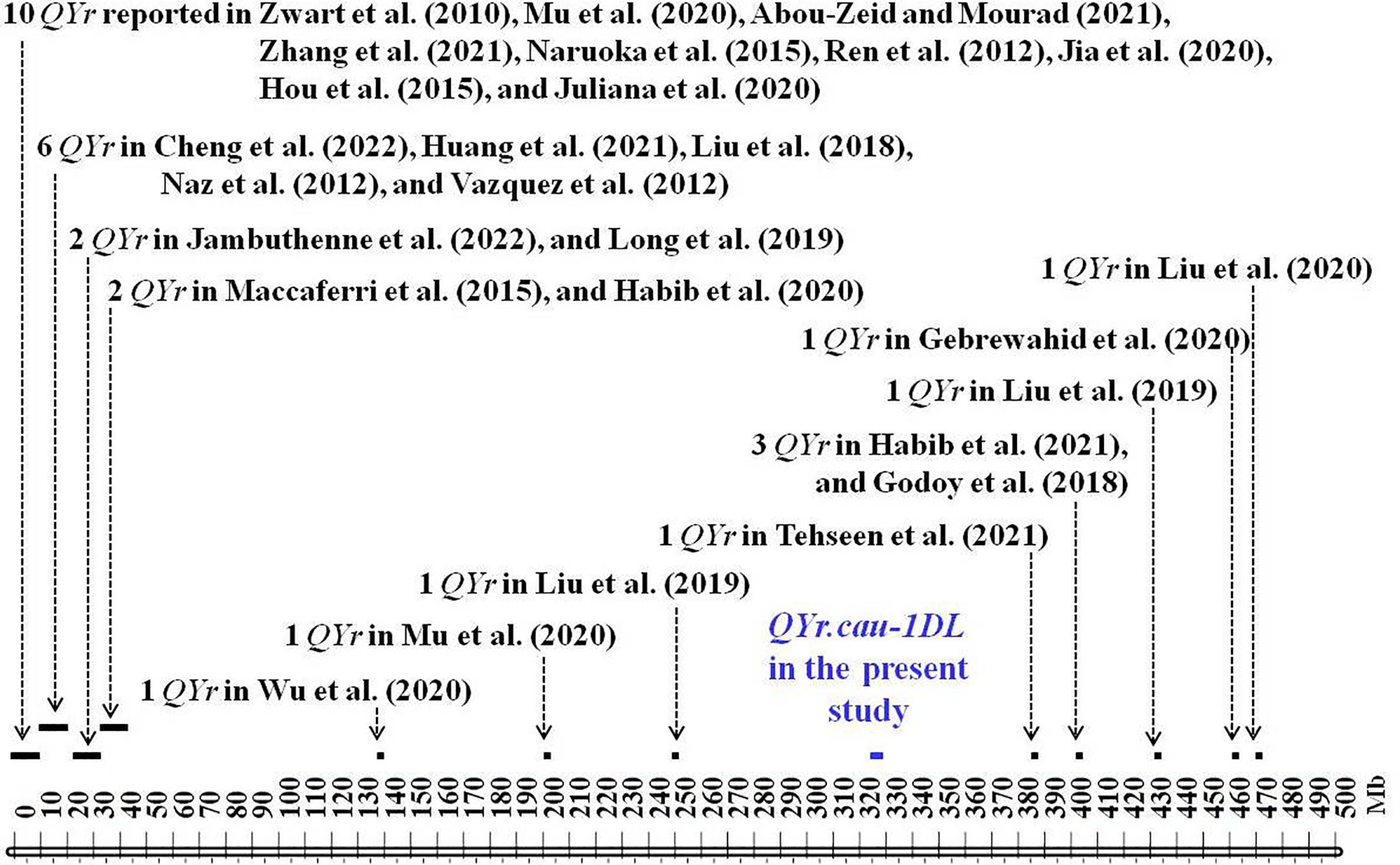
Figure 6 Physical positions of QYr.cau-1DL and the previously reported QTL for resistance to stripe rust on chromosome 1D (10-Mb tick size map) inferred by aligning marker sequences of these QTL to the 1D sequences in the IWGSC RefSeq v1.0 (IWGSC, 2018). Arrows indicate the approximate positions of previously reported QTL. Refer to Supplementary Table 10 for further information and a list of full references.
Yr18 is an important resistance resource since it confers durable resistance (Dyck et al., 1966; Krattinger et al., 2009). RL6058 carries Yr18 (Lagudah et al., 2009) and thus RL6058 × QSM F2:3 families were segregating at both QYr.cau-1DL and Yr18. This provides an opportunity for comparing effects of the two QTL in the same genetic background and also for examining their interaction. QYr.cau-1DL displayed a larger effect on reducing stripe rust severity than Yr18 (Figure 5). Rosewarne et al. (2013) indicated that “Yr29 may be less effective against stripe in the presence of Yr18”; similar interaction between Yr18 and Yr46 was addressed by Lagudah (2011). Different from these situations, combination of QYr.cau-1DL with Yr18 can elevate the level of resistance (Figure 5) without unfavorable interaction. The RL6058 × QSM results also illustrate that QYr.cau-1DL can be effectively selected based on the resistant marker haplotype, i.e., a 232 bp amplicon at 1D-320.58 combined with a 290 bp amplicon at 1D-325.79 (Figure 3). The 751 bp amplicon, a fragment of the cloned Yr18 gene, is diagnostic of the resistant allele at Yr18 (Krattinger et al., 2009; Lagudah et al., 2009). Therefore, QYr.cau-1DL and Yr18 can be utilized simultaneously in breeding programs with marker-assisted selection.
We previously described (Zhang et al., 2017; Wang et al., 2019) that southern Gansu is a Pst hotspot within the near-Himalayan region. This region has been hypothesized to be a part of Pst diversity center (Ali et al., 2014). We also described some supporting data (Zhang et al., 2017; Wang et al., 2019). In southern Gansu and northwestern Sichuan of China, for instance, the geo-morphological and weather conditions are highly favorable for Pst to over-summer, over-winter and survive year-round, leading to a brooder of new Pst virulence races (Wan et al., 2004; Chen et al., 2009). Pst population can reproduce sexually on alternate hosts with naturally occurring recombinants being observed (Mboup et al., 2009; Ali et al., 2010; Duan et al., 2010; Zhao et al., 2016). Since 1940s when the first survey on Pst races was conducted in China (Chen et al., 2009), extraordinarily diverse virulent variants have been identified (Wan et al., 2004; Chen et al., 2009; Bai et al., 2018; Li et al., 2023), and nearly every of Pst races first appeared in the hotspot. Resistance genes, as exemplified by Yr9 and Yr26, also first became ineffective there (Wan et al., 2004; Chen et al., 2009; Han et al., 2015; Bai et al., 2018). We consider that any race-specific stripe rust resistance gene/QTL might be defeated by Pst variants in such a hotspot within limited years. But, during the 36 years from 1987 to 2022, stripe rust severities never reached 25% on QSM (the donor of QYr.cau-1DL), relative to >85% on the susceptible check wheat MX (Supplementary Table 1). Another fact is that QSM was cultivated on approximately 43000 hectares annually in the 1940s and early 1950s in southern Gansu (Zhang, 1995), namely, this landrace was ever widely grown. Therefore, QYr.cau-1DL might be associated with a form of durable resistance sensu Johnson (1984).
An APR QTL (QYr.cau-1DL) identified in the present study has larger effects on reducing stripe rust severity than the well-known gene Yr18; synergistic interaction between the two QTL can yield an elevated level of resistance. QYr.cau-1DL can stably function over substantially diverse genetic backgrounds and highly different agro-geographical environments. Thus, it should have a potential for application in wheat breeding across wide circumstances. The SSR markers 1D-320.58 and 1D-325.79 can be used to assist selection for QYr.cau-1DL in breeding processes following parental polymorphism checks.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The authors declare that the experiments comply with the current laws of the country.
XJ and WQ conceived the study. JF, ZZ, and WQ constructed the wheat populations. XJ, ZW, ZD, ZZ, WQ, YZ, MC, and JR prepared inoculums, carried out disease trials, and genotyped wheat lines using SSR markers. XJ, ZW, and ZZ designed SSR primers and constructed linkage groups. XJ and ZZ analyzed disease data. HW contributed to inoculums preparation and supervised XJ in learning related to this study. XJ drafted the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by National Natural Science Foundation of China (32001496; 31871923; 32101702), National Key Research and Development Program of China (2020YFD0900102), and Beijing Academy of Agriculture and Forestry Sciences Sci-Tech Innovation Capacity Building Program (KJCX20200115).
We thank Bai, Z., Mou, X., Lin, Y., Wang, J. and Wang, X. for their assistance in conducting field and greenhouse trials.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1207764/full#supplementary-material
Ali, S., Gladieux, P., Leconte, M., Gautier, A., Justesen, A. F., Hovmøller, M. S., et al. (2014). Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici. PloS Pathog. 10, e1003903. doi: 10.1371/journal.ppat.1003903
Ali, S., Leconte, M., Walker, A. S., Enjalbert, J., de Vallavieille-Pope, C. (2010). Reduction in the sex ability of worldwide clonal populations of Puccinia striiformis f.sp. tritici. Fungal Genet. Biol. 47, 828–838. doi: 10.1016/j.fgb.2010.07.002
Ali, S., Rodriguez-Algaba, J., Thach, T., Sorensen, C. K., Hansen, J. G., Lassen, P., et al. (2017). Yellow rust epidemics worldwide were caused by pathogen caces from divergent genetic lineages. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01057
Bai, B. B., Liu, T. G., Liu, B., Gao, L., Chen, W. Q. (2018). High relative parasitic fitness of G22 derivatives is associated with the epidemic potential of wheat stripe rust in China. Plant Dis. 102, 483–487. doi: 10.1094/pdis-04-17-0511-sr
Bansal, U. K., Forrest, K. L., Hayden, M. J., Miah, H., Singh, D., Bariana, H. S. (2011). Characterisation of a new stripe rust resistance gene Yr47 and its genetic association with the leaf rust resistance gene Lr52. Theor. Appl. Genet. 122, 1461–1466. doi: 10.1007/s00122-011-1545-4
Bassam, B. J., Caetanoanolles, G., Gresshoff, P. M. (1991). Fast and sensitive silver staining of DNA in polyacrylamide gels. Analytical Biochem. 196, 80–83. doi: 10.1016/0003-2697(91)90120-i
Brown, J. K. M. (2015). Durable resistance of crops to disease: a Darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539. doi: 10.1146/annurev-phyto-102313-045914
Calonnec, A., Johnson, R. (1998). Chromosomal location of genes for resistance to Puccinia striiformis in the wheat line TP1295 selected from the cross of soissonais-desprez with lemhi. Eur. J. Plant Pathol. 104, 835–847. doi: 10.1023/a:1008660904975
Chen, X. M. (2005). Epidemiology and control of stripe rust Puccinia striiformis f. sp tritici on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Chen, W. Q., Wu, L. R., Liu, T. G., Xu, S. C., Jin, S. L., Peng, Y. L., et al. (2009). Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 93, 1093–1101. doi: 10.1094/pdis-93-11-1093
Chhetri, M., Bariana, H., Kandiah, P., Bansal, U. (2016). Yr58: a new stripe rust resistance gene and its interaction with Yr46 for enhanced resistance. Phytopathology 106, 1530–1534. doi: 10.1094/phyto-04-16-0182-r
Dong, Z. Z., Hegarty, J. M., Zhang, J. L., Zhang, W. J., Chao, S. M., Chen, X. M., et al. (2017). Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor. Appl. Genet. 130, 2127–2137. doi: 10.1007/s00122-017-2946-9
Duan, X., Tellier, A., Wan, A., Leconte, M., de Vallavielle-Pope, C., Enjalbert, J. (2010). Puccinia striiformis f.sp tritici presents high diversity and recombination in the over-summering zone of gansu, China. Mycologia 102, 44–53. doi: 10.3852/08-098
Dyck, P. L., Samborsk., Dj, Anderson (1966). Inheritance of adult plant leaf rust resistance derived from the common wheat varieties exchange and frontana. Can. J. Genet. Cytology 8, 665–671. doi: 10.1139/g66-082
Fu, D. L., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X. M., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Han, D. J., Wang, Q. L., Chen, X. M., Zeng, Q. D., Wu, J. H., Xue, W. B., et al. (2015). Emerging Yr26-virulent races of Puccinia striiformis f. sp. tritici are threatening wheat production in the sichuan basin, China. Plant Dis. 99, 754–760. doi: 10.1094/pdis-08-14-0865-re
Hovmøller, M. S., Walter, S., Bayles, R. A., Hubbard, A., Flath, K., Sommerfeldt, N., et al. (2016). Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant Pathol. 65, 402–411. doi: 10.1111/ppa.12433
IWGSC (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. doi: 10.1126/science.1251788
IWGSC. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Jambuthenne, D. T., Riaz, A., Athiyannan, N., Alahmad, S., Ng, W. L., Ziems, L., et al. (2022). Mining the vavilov wheat diversity panel for new sources of adult plant resistance to stripe rust. Theor. Appl. Genet. 135, 1355–1373. doi: 10.1007/s00122-022-04037-8
Jan, I., Saripalli, G., Kumar, K., Kumar, A., Singh, R., Batra, R., et al. (2021). Meta-QTLs and candidate genes for stripe rust resistance in wheat. Sci. Rep. 11, 22923. doi: 10.1038/s41598-021-02049-w
Javier, S. M., Keller, B. (2021). NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr. Opin. Plant Biol. 62, 102053. doi: 10.1016/j.pbi.2021.102053
Johnson, R. (1984). A critical analysis of durable resistance. Annu. Rev. Phytopathol. 22, 309–330. doi: 10.1146/annurev.py.22.090184.001521
Johnson, R. (1992). Past, present and future opportunities in breeding for disease resistance, with examples from wheat. Euphytica 63, 3–22. doi: 10.1007/bf00023908
Kanwal, M., Qureshi, N., Gessese, M., Forrest, K., Babu, P., Bariana, H., et al. (2021). An adult plant stripe rust resistance gene maps on chromosome 7A of Australian wheat cultivar axe. Theor. Appl. Genet. 134, 2213–2220. doi: 10.1007/s00122-021-03818-x
Klymiuk, V., Chawla, H. S., Wiebe, K., Ens, J., Fatiukha, A., Govta, L., et al. (2022). Discovery of stripe rust resistance with incomplete dominance in wild emmer wheat using bulked segregant analysis sequencing. Commun. Biol. 5, 826. doi: 10.1038/s42003-022-03773-3
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lagudah, E. S. (2011). Molecular genetics of race non-specific rust resistance in wheat. Euphytica 179, 81–91. doi: 10.1007/s10681-010-0336-3
Lagudah, E. S., Krattinger, S. G., Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., Spielmeyer, W., et al. (2009). Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 119, 889–898. doi: 10.1007/s00122-009-1097-z
Li, Y., Dai, J., Zhang, T., Wang, B., Zhang, S., Wang, C., et al. (2023). Genomic analysis, trajectory tracking, and field surveys reveal sources and long-distance dispersal routes of wheat stripe rust pathogen in China. Plant Commun. 4, 100563. doi: 10.1016/j.xplc.2023.100563
Li, Z. F., Xia, X. C., Zhou, X. C., Niu, Y. C., He, Z. H., Zhang, Y., et al. (2006). Seedling and slow rusting resistance to stripe rust in Chinese common wheats. Plant Dis. 90, 1302–1312. doi: 10.1094/pd-90-1302
Li, H. H., Ye, G. Y., Wang, J. K. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374. doi: 10.1534/genetics.106.066811
Line, R. F. (2002). Stripe rust of wheat and barley in north America: a retrospective historical review. Annu. Rev. Phytopathol. 40, 75–118. doi: 10.1146/annurev.phyto.40.020102.111645
Line, R. F., Qayoum, A. (1992). “Virulence aggressiveness, evolution, and distribution of races of puccinia striiformis (the cause of stripe rust of wheat) in north americ –1987,” in U.S. department of agriculture technical bulletin no. 1788, the national technical information service (Pullman, WA, USA: Springfield), p44.
Ma, H., Singh, R. P. (1996). Expression of adult resistance to stripe rust at different growth stages of wheat. Plant Dis. 80, 375–379. doi: 10.1094/pd-80-0375
Maccaferri, M., Zhang, J., Bulli, P., Abate, Z., Chao, S., Cantu, D., et al. (2015). A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum l.). G3-Genes Genomes Genet. 5, 449–465. doi: 10.1534./g3.114.014563
Marone, D., Russo, M. A., Mores, A., Ficco, D. B. M., Laido, G., Mastrangelo, A. M., et al. (2021). Importance of landraces in cereal breeding for stress tolerance. Plants-Basel 10, 1267. doi: 10.3390/plants10071267
Mboup, M., Leconte, M., Gautier, A., Wan, A. M., Chen, W., de Vallavieille-Pope, C., et al. (2009). Evidence of genetic recombination in wheat yellow rust populations of a Chinese oversummering area. Fungal Genet. Biol. 46, 299–307. doi: 10.1016/j.fgb.2008.12.007
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., Raupp, W. J. (2020). Catalogue of gene symbols for wheat: 2020 supplement. 13th Int. Wheat Genet. Symposium 0711.
Milus, E. A., Kristensen, K., Hovmøller, M. S. (2008). Increased aggressiveness of Puccinia striiformis f. sp. tritici at least partially explains recent stripe rust epidemics. Phytopathology 98, S107–S107.
Milus, E. A., Kristensen, K., Hovmøller, M. S. (2009). Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99, 89–94. doi: 10.1094/phyto-99-1-0089
Moore, J. W., Herrera-Foessel, S., Lan, C. X., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Newton, A. C., Akar, T., Baresel, J. P., Bebeli, P. J., Bettencourt, E., Bladenopoulos, K. V., et al. (2010). Cereal landraces for sustainable agriculture. a review. Agron. Sustain. Dev. 30, 237–269. doi: 10.1051/agro/2009032
Peterson, R. F., Campbell, A. B., Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity of leaves and stems of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Quan, W., Hou, G. L., Chen, J., Du, Z. Y., Lin, F., Guo, Y., et al. (2013). Mapping of QTL lengthening the latent period of Puccinia striiformis in winter wheat at the tillering growth stage. Eur. J. Plant Pathol. 136, 715–727. doi: 10.1007/s10658-013-0201-z
Roder, M. S., Korzun, V., Wendehake, K., Plaschke, J., Tixier, M. H., Leroy, P., et al. (1998). A microsatellite map of wheat. Genetics 149, 2007–2023. doi: 10.1093/genetics/149.4.2007
Rosewarne, G. M., Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Lan, C. X., He, Z. H. (2013). Quantitative trait loci of stripe rust resistance in wheat. Theor. Appl. Genet. 126, 2427–2449. doi: 10.1007/s00122-013-2159-9
Saghaimaroof, M. A., Soliman, K. M., Jorgensen, R. A., Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. United States America-Biological Sci. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Santra, D. K., Chen, X. M., Santra, M., Campbell, K. G., Kidwell, K. K. (2008). Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum l.) cultivar 'Stephens'. Theor. Appl. Genet. 117, 793–802. doi: 10.1007/s00122-008-0820-5
Segovia, V., Hubbard, A., Craze, M., Bowden, S., Wallington, E., Bryant, R., et al. (2014). Yr36 confers partial resistance at temperatures below 18 degrees to UK isolates of Puccinia striiformis. Phytopathology 104, 871–878. doi: 10.1094/phyto-10-13-0295-r
Singh, R. P., Huerta-Espino, J., Bhavani, S., Herrera-Foessel, S. A., Singh, D., Singh, P. K., et al. (2011). Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179, 175–186. doi: 10.1007/s10681-010-0322-9
Sorensen, C. K., Hovmøller, M. S., Leconte, M. (2014). Dedryver, fNew races of Puccinia striiformis found in Europe reveal race specificity of long-term effective adult plant resistance in wheat. Vallavieille-Pope C. Phytopathology 104, 1042–1051. doi: 10.1094/phyto-12-13-0337-r
Stam, P. (1993). Construction of integrated genetic linkage maps by means of a new computer package: JOINMAP. Plant J. 3, 739–744. doi: 10.1111/j.1365-313X.1993.00739.x
Wan, A. M., Chen, X. M. (2014). Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the united states in 2010. Plant Dis. 98, 1534–1542. doi: 10.1094/pdis-01-14-0071-re
Wan, A. M., Zhao, Z. H., Chen, X. M., He, Z. H., Jin, S. L., Jia, Q. Z., et al. (2004). Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 88, 896–904. doi: 10.1094/pdis.2004.88.8.896
Wang, S., Basten, J. C., Zeng, Z. B. (2010). “Windows QTL cartographer 2.5,” in Department of statistics (Raleigh, NC: North Carolina State University). Available at: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm.
Wang, Z., Ren, J. D., Du, Z. Y., Che, M. Z., Zhang, Y. B., Quan, W., et al. (2019). Identification of a major QTL on chromosome arm 2AL for reducing yellow rust severity from a Chinese wheat landrace with evidence for durable resistance. Theor. Appl. Genet. 132, 457–471. doi: 10.1007/s00122-018-3232-1
Wang, S. C., Wong, D. B., Forrest, K., Allen, A., Chao, S. M., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wellings, C. R. (2011). Global status of stripe rust: a review of historical and current threats. Euphytica 179, 129–141. doi: 10.1007/s10681-011-0360-y
Wu, J. H., Wang, Q. L., Chen, X. M., Wang, M. J., Mu, J. M., Lv, X. N., et al. (2016). Stripe rust resistance in wheat breeding lines developed for central shaanxi, an overwintering region for Puccinia striiformis f. sp tritici in China. Can. J. Plant Pathol. 38, 317–324. doi: 10.1080/07060661.2016.1206039
Zhang, Z. J. (2023). Evidence of durable resistance in nine Chinese landraces and one Italian cultivar of Triticum aestivum to Puccinia striiformis. Eur. J. Plant Pathol. 101, 405–409. doi: 10.1007/BF01874854
Zhang, X., Jiang, X., Zhang, Y., Ren, J., Feng, J., Quan, W. (2023). Identification of QTL for reducing loss of grain yield under salt stress conditions in bi-parental populations derived from wheat landrace hongmangmai. Theor. Appl. Genet. 136, 49. doi: 10.1007/s00122-023-04290-5
Zhang, Y., Wang, Z., Quan, W., Zhang, X., Feng, J., Ren, J., et al. (2021). Mapping of a QTL with major effect on reducing leaf rust severity at the adult plant growth stage on chromosome 2BL in wheat landrace hongmazha. Theor. Appl. Genet. 134, 1363–1376. doi: 10.1007/s00122-021-03776-4
Zhang, H. Y., Wang, Z., Ren, J. D., Du, Z. Y., Quan, W., Zhang, Y. B., et al. (2017). A QTL with major effect on reducing stripe rust severity detected from a Chinese wheat landrace. Plant Dis. 101, 1533–1539. doi: 10.1094/pdis-08-16-1131-re
Zhao, J., Wang, M. N., Chen, X. M., Kang, Z. S. (2016). Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Annu. Rev. Phytopathol. 54, 207–228. doi: 10.1146/annurev-phyto-080615-095851
Keywords: wheat landrace, stripe rust, disease resistance, QTL mapping, marker-assisted selection, QTL interaction
Citation: Jiang X, Wang Z, Feng J, Du Z, Zhang Z, Zhang Y, Che M, Ren J, Wang H and Quan W (2023) Mapping and validation of a novel major QTL for resistance to stripe rust in four wheat populations derived from landrace Qishanmai. Front. Plant Sci. 14:1207764. doi: 10.3389/fpls.2023.1207764
Received: 18 April 2023; Accepted: 30 May 2023;
Published: 16 June 2023.
Edited by:
Satish Kumar, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Deepak Baranwal, Bihar Agricultural University Sabour, IndiaCopyright © 2023 Jiang, Wang, Feng, Du, Zhang, Zhang, Che, Ren, Wang and Quan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Quan, cXVhbndlaV9iYWFmc0BzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.