
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Plant Sci., 26 May 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1204896
This article is part of the Research TopicUtilization of Microbiome to Develop Disease Resistance in Crop Plants against PhytopathogensView all 5 articles
Editorial on the Research Topic
Utilization of microbiome to develop disease resistance in crop plants against phytopathogens
Sustainable agriculture is important for ensuring food security at an affordable cost. Agricultural practices heavily depend on agrochemicals, such as fertilizers, fungicides, and pesticides, to ensure optimal productivity and prevent losses due to pathogens and pests. However, with time, it has become clear that most commonly used agrochemicals have residual side effects on consumers, soil health, and the environment. Hence, developing newer environmentally friendly approaches to ensure crop productivity, with minimal usage of agrochemicals, is highly desirable. The potential of plant/rhizospheric soil-associated microbes to support the overall growth of plants and provide protection against phytopathogens has been actively studied in recent years (Busby et al., 2017; Liu et al., 2020). A large number of beneficial bacteria, including Bacillus, Pseudomonads and Burkholderia sp. and fungi, including Trichoderma sp. and Piriformospora indica, have been shown to have growth-promoting and disease-protection abilities (Hacquard et al., 2017). It is to be noted that billions of microbes live within or in very close association with plants; together, they influence the growth and immunity of the host plant. Moreover, due to intraspecific or intraspecific competitions, there is a constant flux in the population dynamics of the microbiota (Das et al., 2021). Hence it is important to understand the interactions among the plant-associated microbes to comprehend their ecological fitness under field conditions. The genome sequence of plant-associated beneficial microorganisms has unravelled that they are enriched in novel antimicrobial compounds and secondary metabolite-encoding biosynthetic gene clusters (BGCs), however, most of them remain to be characterised. Characterising the coding potential of plant-associated microbes will profusely contribute to the global bio-economy.
This Research Topic brings four important articles that exemplify and emphasise the utility of plant-associated microbes in sustainable agriculture (Figure 1). Lee et al. report that a gram-positive bacterium, Bacillus velezensis GH1-13, helps manage brown patch disease of cool-season turfgrass caused by a necrotrophic fungal pathogen, Rhizoctonia solani. The study demonstrates that the application of GH1-13 helps reduce the usage of azoxystrobin fungicide to 50%, as GH1-13 + 50% of azoxystrobin provided a similar level of protection to that observed in the case of 100% of azoxystrobin treatment, even under field conditions. In addition, they have developed a qRT-PCR-based assay to precisely detect the GH1-13. The analysis reflected that the bacterium could stably cohabit and survive in the rhizosphere suggesting its use as a stable biocontrol agent. This highlights that the focus should be on developing more straightforward approaches to quantify the ecological fitness of the microbes under field conditions.
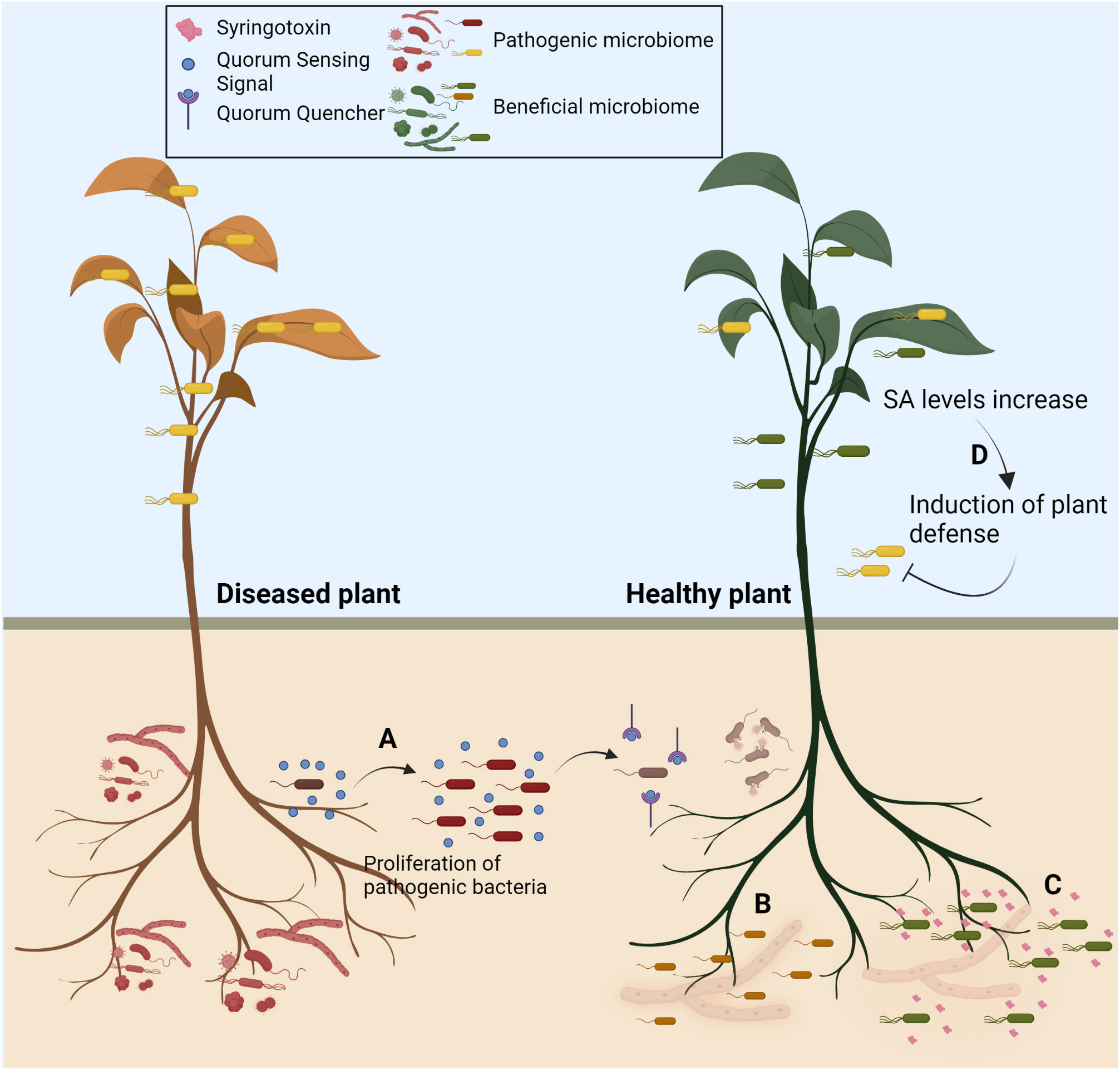
Figure 1 Strategies to deploy microbiome for crop improvement. Recent studies have highlighted that plant-associated microbes and the microbiome can be harnessed for crop protection. The articles in the research topic exemplify (A) Use of quorum quenching to disrupt the quorum sensing of the pathogenic bacteria to prevent disease (B) Use of antagonistic bacteria to protect plants from fungal diseases (C) Some novel secondary metabolites secreted from the plant-associated bacteria can antagonise fungal pathogens (D) Microbiome-mediated priming of Salicylic acid-mediated plant defence to prevent subsequent attacks from pathogenic microorganisms. Created with BioRender.com.
Ferrarini et al. have characterised the role of the biosynthetic gene clusters (BGCs) associated with the synthesis of cyclic lipopeptides (CLPs) in a phytopathogenic bacterium Pseudomonas fuscovaginae UPB264 that causes sheath brown rot disease in rice. The authors have obtained a single-contig genome of the UPB264 and identified that the bacterium encodes novel CLPs, asplenin, in addition to the previously reported CLPs, syringotoxin and fuscopeptin. Using the mutant strains, fuscopeptin and syringotoxin are important for the bacterium causing rice sheath rot disease. In contrast, asplenin and syringotoxin are important for bacterial swarming motility and antifungal activity. Syringotoxin has been reported to have antifungal activity against sheath blight pathogen Rhizoctonia solani. The study emphasises the characterisation of putative BGCs is an important area of research, which would lead to the identification of novel bioactive molecules for potential exploitation for plant growth promotion and disease control.
One of the important attributes of beneficial microbes is their ability to induce host immune responses and prevent disease establishment by phytopathogens. It is important to understand the defence-priming ability of the beneficial microbes to utilise them for disease control. Li et al. report that a Priestia megaterium strain JR48, isolated from the vegetable field, has a plant-growth-promoting and biocontrol effect. The data reflected that bacterial Treatment not only enhances the Chinese cabbage’s growth but also prevents the plants from black rot disease caused by Xanthomonas campestris pv. campestris 8004 (Xcc) infection. Notably, induced expression of various defence-related genes, including pathogenesis-related (PR) genes, enhanced callose deposition, salicylic acid (SA) accumulation, and hydrogen peroxide, were observed in the JR48-treated plants. Overall, the study emphasizes the importance of salicylic acid (SA)- mediated defence in the biocontrol ability of JR48.
Notably, plant-associated microbes continuously interact with the host plants and other co-having microbes in a hostile environment. They have intricate ability to communicate with each other using quorum sensing molecules. Extensive progress has been made in understanding the quorum-sensing mechanism and exploring whether using quorum-quenching molecules could prevent phytopathogens from establishing disease. Zhu et al. provide the current understanding in this field and elaborate on how quorum quenching is effective in countering pathogen growth and attenuating virulence in crop plants. The quorum quenchers either function through competitive inhibition of the active site or physical degradation of the signalling molecules. They are primarily considered to have fewer side effects on non-target micro-organisms. An additional advantage of using quorum quenchers is that, unlike antibiotics, for which there is selection pressure in bacteria to develop resistance, it is challenging to build tolerance against quorum quenchers. The review not only elaborates on various applications of quorum quenchers in agriculture but also discusses their exploitation in controlling aquacultural pathogens, human pathogens, and biofouling mitigation. The article advocates that it is important to investigate quorum quenching molecules’ side effects on non-target organisms, particularly whether they can alter the colonisation of beneficial microorganisms. The special issue on utilising microbiome for controlling phytopathogens provide novel approaches for biocontrol of diseases.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
GJ acknowledges Swarna Jayanti fellowship from SERB, Ministry of Science and Technology, Govt. of India, research funding from Department of Biotechnology (DBT), Govt. of India, and NIPGR core research grant. The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
We acknowledge the reviewers, guest editors and editorial team members for their valuable contributions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Busby, P. E., Soman, C., Wagner, M. R., Friesen, M. L., Kremer, J., Bennett, A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PloS Biol. 15, 1–14. doi: 10.1371/journal.pbio.2001793
Das, J., Yadav, S. K., Ghosh, S., Tyagi, K., Magotra, A., Krishnan, A., et al. (2021). Enzymatic and non-enzymatic functional attributes of plant microbiome. Curr. Opin. Biotechnol. 69, 162–171. doi: 10.1016/j.copbio.2020.12.021
Hacquard, S., Spaepen, S., Garrido-Oter, R., Schulze-Lefert, P. (2017). Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589. doi: 10.1146/annurev-phyto-080516-035623
Keywords: microbiome, phytopathogen, biocontrol, antagonists, disease suppresion
Citation: Ghosh S and Jha G (2023) Editorial: Utilization of microbiome to develop disease resistance in crop plants against phytopathogens. Front. Plant Sci. 14:1204896. doi: 10.3389/fpls.2023.1204896
Received: 12 April 2023; Accepted: 02 May 2023;
Published: 26 May 2023.
Edited and Reviewed by:
Prem Lal Kashyap, Indian Institute of Wheat and Barley Research (ICAR), IndiaCopyright © 2023 Ghosh and Jha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Srayan Ghosh, c3JheWFuZ2hvc2g5QGdtYWlsLmNvbQ==; Gopaljee Jha, am1zZ29wYWxAbmlwZ3IuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.