- 1Iowa State University, Department of Agronomy, Ames, IA, United States
- 2Agriculture Victoria, Agribio, La Trobe University, Melbourne, VIC, Australia
- 3Iowa State University, Department of Statistics, Ames, IA, United States
- 4Iowa State University, Department of Electrical and Computer Engineering, Ames, IA, United States
- 5Iowa State University, Roy J. Carver Department of Biochemistry, Biophysics, and Molecular Biology, Ames, IA, United States
Efforts to increase genetic gains in breeding programs of flowering plants depend on making genetic crosses. Time to flowering, which can take months to decades depending on the species, can be a limiting factor in such breeding programs. It has been proposed that the rate of genetic gain can be increased by reducing the time between generations by circumventing flowering through the in vitro induction of meiosis. In this review, we assess technologies and approaches that may offer a path towards meiosis induction, the largest current bottleneck for in vitro plant breeding. Studies in non-plant, eukaryotic organisms indicate that the in vitro switch from mitotic cell division to meiosis is inefficient and occurs at very low rates. Yet, this has been achieved with mammalian cells by the manipulation of a limited number of genes. Therefore, to experimentally identify factors that switch mitosis to meiosis in plants, it is necessary to develop a high-throughput system to evaluate a large number of candidate genes and treatments, each using large numbers of cells, few of which may gain the ability to induce meiosis.
1 Introduction to in vitro nurseries
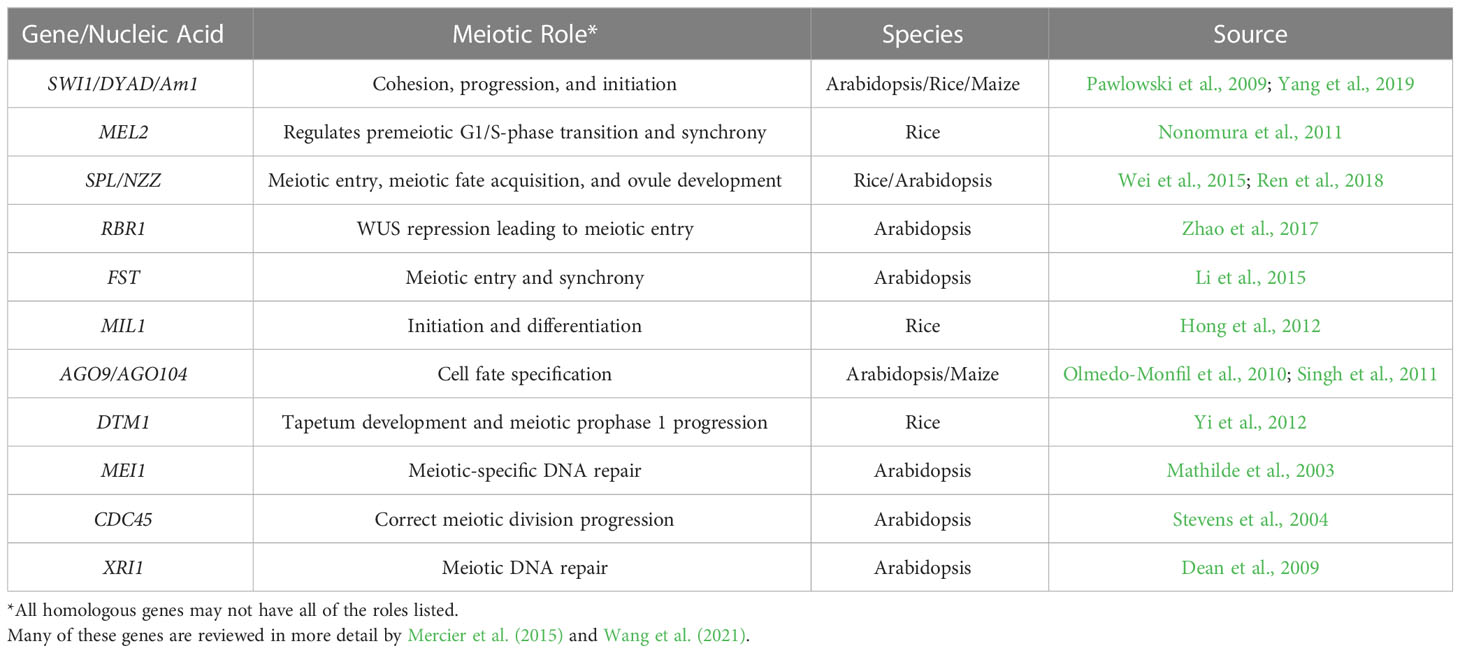
Globally the number of undernourished people is expected to increase to 840 million by 2030 (FAO et al, 2020). Even though we need to produce more food in the future, the current levels of food production are at risk as climate change has the potential to disrupt food availability (Brummer et al., 2011; De La Fuente et al., 2013; Brown et al., 2015; US Embassy and Consulate in Italy, 2022). Innovative breeding techniques to improve food security to increase genetic gains are needed. Genetic gain is associated with selection intensity, heritability, genetic variance, and the time needed for a breeding cycle (Figure 1; Li et al., 2018). Several breeding methods and technologies have been developed to increase genetic gain by reducing the time needed to complete a breeding cycle, and these include winter nurseries, doubled-haploids (Geiger, 2009; De La Fuente et al., 2013; Boerman et al., 2020), speed breeding (Watson et al., 2018; Jähne et al., 2020), gene editing or gene expression regulation (Gao et al., 2020; Pan et al., 2022; reviewed in Zhang et al., 2018), marker-assisted selection (Karunarathna et al., 2021; López-Malvar et al., 2021; Tibbs Cortes et al., 2021), genomic selection (Bhat et al., 2016; López-Malvar et al., 2021), phytohormonal induction of early flowering (Espinosa et al., 2017), and combination of doubled-haploids with other breeding strategies such as gene editing during haploid induction and genomic prediction (Kelliher et al., 2019; Wang et al., 2020). These technologies focus on reducing the number of generations needed to develop a line, or reducing the time to flowering and seed production, thereby allowing more generations per unit of time. Technologies such as speed breeding and hormone manipulation provide earlier flowering times but are only available for a limited number of crop species, (Iqbal et al., 2017; Watson et al., 2018; Jähne et al., 2020), and they still require that plants produce floral organs and gametes for sexual reproduction.
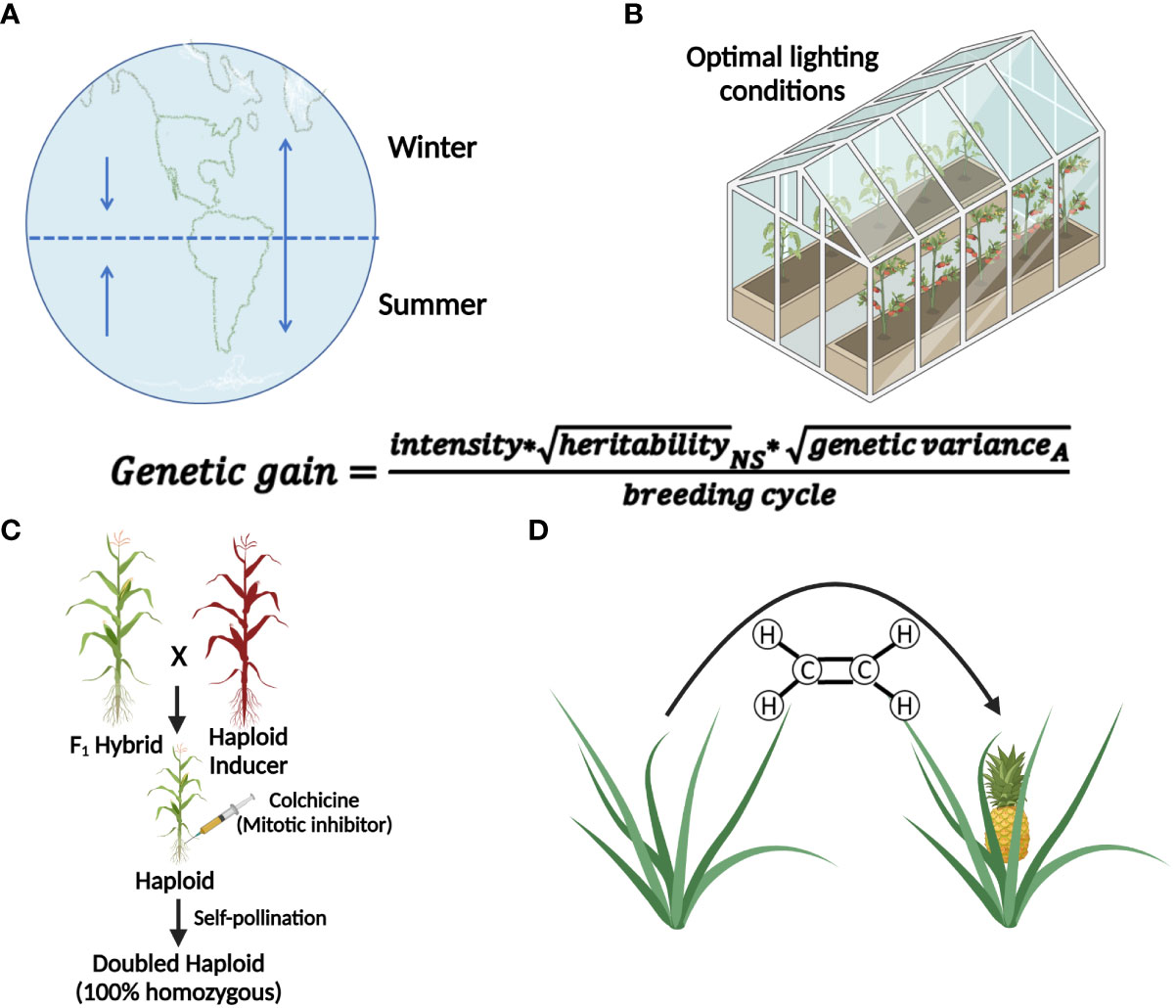
Figure 1 Methods that accelerate breeding cycles or generation time of plants. The genetic gain equation provides the basis for understanding how the use of each technique leads to genetic gain (A) The use of winter nurseries to capitalize on proximity to the equator or opposite hemispheric seasons, increase diurnal photoperiods, and warmer temperatures to increase the number of growing seasons per year. (B) Speed-breeding techniques that utilize optimal lighting conditions to induce early flowering to decrease breeding cycle times. (C) Doubled-haploid technology reduces the time needed to develop a homozygous line. (D) Use of chemicals (e.g. ethylene) to induce early flowering and fruit development, decreasing breeding cycle time. Created with BioRender.com.
As a paradigm shift, Murray et al. (2013) and De La Fuente et al. (2013) suggested the concept of a cell-based in vitro breeding system (called in vitro nurseries; IVNs). In IVNs, breeding cycle time could be substantially reduced by enabling rapid cell-level breeding cycles, without the need for flowering. Somatic tissue from parental lines could be cultured and challenged to induce haploid cells after recombination without gametophyte development (will be referred to as artificial gametes throughout the text), these cells can then be fused artificially to develop sexual pairing in vitro. In addition to time, IVNs will significantly reduce required field space and avoid exposure to environmental risks in field settings. The benefits of IVNs are of particular interest for species with a long generation time. Some woody species do not produce flowers for more than 30 years (Hackett, 1985). For example, poplar trees provide many ecosystem services such as phytoremediation, substrate for biofuels, and other bioproducts (Zalesny et al., 2020), but can take 10 years to flower (Hsu et al., 2006). Coffee trees do not flower until the second year, and it is not until the third year that they reach maturity (Santos et al., 2019). Even in annual crops such as maize, where two generations per year are routinely completed using winter nurseries, more generations per year would increase the annual genetic gain significantly.
Successful implementation of IVNs will require systematically overcoming a variety of bottlenecks. We anticipate three distinct phases (Figure 2). Phase I addresses the main bottleneck: meiosis induction (or meiosis-like recombination followed by reductional division) from somatic vegetative tissue, which currently is unavailable in plants. We assume that there will be a need to evaluate a potentially large number of genes, external treatments, and their combinations before identifying a path for meiosis induction in plants. Thus, an assay for high-throughput and low-cost screening of candidate genes or treatments for meiosis induction is needed. The other two phases are based on the successful development of protocols for meiosis induction. Phase II addresses artificial gamete formation and identification. Identifying and isolating these artificial gametes in a mixture consisting primarily of somatic cells will be critical for manipulation in the next phase. Phase III includes the assessment of induced artificial gametes that carry favorable alleles using genomic selection methods. This can only be done after artificial gametes have been isolated and developed into cell lines. Only then, can a sample of cells from each line be sacrificed for DNA isolation and genotyping for genomic selection. Further, this phase includes the fusion of selected artificial gametes to generate diploid cells, as a starting point for the next generation in IVNs. In this review, we will assess the concept of a cell-based in vitro breeding system, which circumvents the need for flowering (De la Fuente et al., 2013; Murray et al., 2013). The overall objective of this paper is to investigate the feasibility of Phase I through the (i) identification of bottlenecks and uncertainties (ii) while proposing possible solutions, and thus (iii) providing a starting point for the development of IVN technologies.
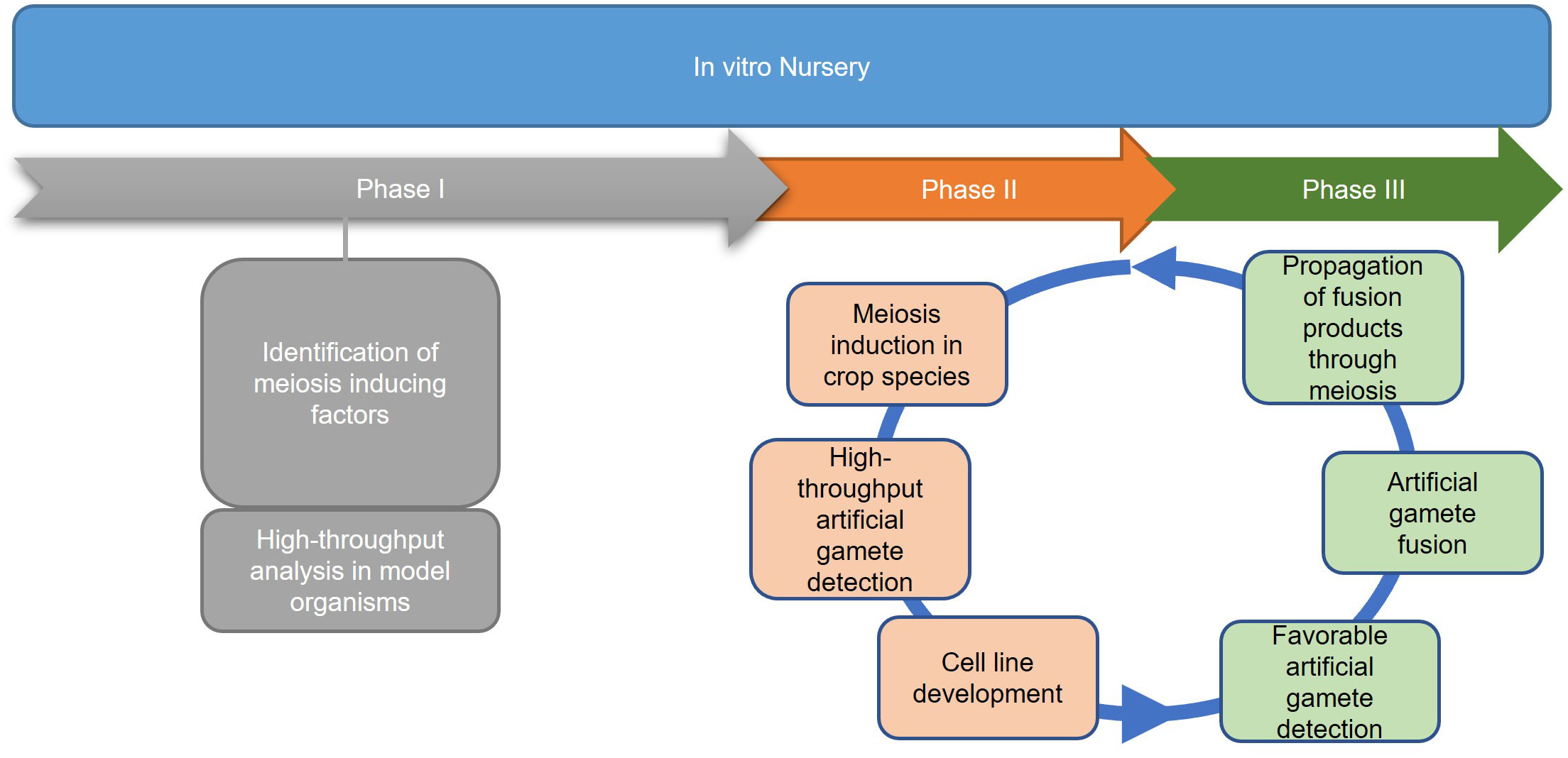
Figure 2 Three proposed phases of an in vitro nursery. Phase I emphasizes the identification of meiosis-inducing factors as a major obstacle in establishing IVNs. Identification of the meiosis-inducing factors needs a quantitative, high throughput assay to detect meiosis. Phase II involves the development of the nursery in the context of a specific crop species, using the meiosis-inducing factors identified in Phase I and developing haploid cell lines for the selection of desirable traits. Phase III identifies haploid cell lines that are expressing desirable traits by the use of markers and traditional breeding tools. After diploidization, the products are propagated by mitosis for further cycling through Phases II and III.
2 Eukaryotic meiosis
2.1 Meiosis in plants
Sex is a fundamental process shared among eukaryotes (Colnaghi et al., 2020), with meiosis being a key step to generating variation by recombining genomes. Meiosis consists of DNA replication followed by two divisions that reduce the genome size by half (Mercier et al., 2015). During meiosis, chromosomes recombine via crossovers (COs), a mechanism to reshuffle genes and respective physically linked alleles on a chromosome (Mercier et al., 2015). The major obstacle in establishing IVNs in plants is the inability to induce meiosis outside of the male or female reproductive cell structures of the flower. To be practical for IVNs, in vitro meiosis induction has to be based on a limited number of factors to enable a practical, routine application for artificial gamete formation.
In contrast to the predetermined germline of animals, the transition from vegetative to reproductive growth in plants occurs later in development where archesporial cells are generated from primordia, beginning the plant germline (Zhou et al., 2017). Differences between plant and human germline development have been outlined in Figure 3. In angiosperms, gametogenesis is a highly conserved process and occurs within specialized tissues of the anther and the ovule. The production of gametes proceeds in two steps: sporogenesis, followed by gametogenesis. In the anther, hypodermal archesporial cells divide to produce outer primary parietal cells which become somatic tissues, and inner primary sporogenous cells which will then divide mitotically to become microspore mother cells that undergo meiosis (Lora and Hormaza, 2021). Ovule initiation arises from the medial meristem tissue within the carpel. Immediately following ovule initiation three distinct regions arise: funiculus, chalaza, and nucellus. The nucellus gives rise to the megaspore mother cell which undergoes meiosis to generate four megaspores, three of which degrade leaving a single functional megaspore (Lora and Hormaza, 2021).
Figure 3 A schematic comparison of germline development in humans vs. plants. Blastocyst formation takes approximately five days (Popovic et al., 2021), after which germline in humans develops and can be detected as early as the beginning of gastrulation (Wen and Tang, 2019). Three sets of arrows indicate additional developmental processes that were not discussed. In plants, germline development is much later in a plant’s life cycle, occurring during flowering. Somatic hypodermal cells divide to develop an archesporial cell, which is considered the origin of the germline (Zhou et al., 2017). Archesporial cells will then form spore mother cells which undergo meiosis to develop gametes. Created with BioRender.com.
2.2 Controlling meiosis in other eukaryotes
Since the underlying evolutionary path of sexual reproduction is thought to have evolved only once in eukaryotes (Goodenough and Heitman, 2014), there is insight to gain from other non-plant, eukaryotic species. For example, Medrano et al. (2016) found that human somatic cells can be converted into germline-like cells with the ectopic expression of six genes. These genes, PRDM1, PRDM14, LIN28A, DAZL, VASA, and SYCP3, have conserved regions in many plant species and the products have been shown to have important functions in such processes such as the repression of transposable elements, nucleic acid binding, and stem cell maintenance in human meiotic processes (Bateman, 2019; Howe et al., 2020). About 1% of these germline-like cells were able to complete meiosis (Medrano et al., 2016). Further, overexpression of human CD61 (integrin-β3) in canine adipose-derived mesenchymal stem cells led to the upregulation of markers for primordial germ-like cells (Fang et al., 2017). Vernet et al. (2020) suggested that the exogenous application of retinoic acid may force meiosis induction in mice. In vitro mouse studies of spermatogonia overexpressing telomerase catalytic component, mTERT, resulted in cells that could be induced to undergo meiosis in vitro, with the application of stem cell factor (Feng et al., 2002; Griswold, 2005; Riou et al., 2005). This outcome suggests that sex cells can be developed without structure-specific nurse cells, which is encouraging in the case of IVNs. In S. cerevisiae, antisense transcription was found to control meiotic cell entry by regulating IME4 (Initiator of Meiosis 4), an RNA methyltransferase (Hongay et al., 2006). This gene is also expressed in the testes and ovaries of Drosophila (Hongay and Orr-Weaver, 2011). In Arabidopsis thaliana, MTA (mRNA adenosine methylase), which is a homolog of IME4, was found to be essential for embryogenesis (Zhong et al., 2008). In addition to IME4, nutritional stress can also induce meiosis in yeast (Mata et al., 2002). Taken together, such studies in other eukaryotic species suggest that meiosis or meiotic precursors can be artificially induced and that this may also be achievable with plants.
2.3 Plant genetic factors involved in meiosis induction
In plants, a limited number of genetic factors have been identified to play a role in meiosis induction or early meiotic processes by studying aberrant phenotypes presented by mutant alleles. Table 1 summarizes the genes that have been found in previous studies. Maize AMEIOTIC 1 (AM1) is required for meiotic progression while it is also likely required for meiosis initiation as premeiotic cells with am1 mutations led to mitosis instead of meiosis (Pawlowski et al., 2009). The SWI(SWITCH1)/DYAD protein is a putative homolog of AM1 in Arabidopsis, but its role appears to be more important in early meiosis instead of initiation. Consistent with this, SWI/DYAD maintains chromatid cohesion during meiosis as a WINGS APART-LIKE antagonist (Pawlowski et al., 2009; Yang et al., 2019). Evidence for the role of FEHLSTART (FST), a basic helix-loop-helix protein, in meiosis is shown with early meiotic entry in Arabidopsis mutants, and these mutants also show meiotic asynchrony (Li et al., 2015). KRP4, KRP6, and KRP 7 (KIP-RELATED PROTEIN 4,5,6) as well as RETINOBLASTOMA RELATED1 (RBR1), prevent the formation of supernumerary meiocytes from forming next to an already existing meiocyte while the repression of WUSCHEL (WUS) by RBR1 allows entry into meiosis (Zhao et al., 2017). An RNA-helicase (RH17) was found to play a role in reproduction as supernumerary reproductive cell lineages developed at a rate of over 20% in lines that were heterozygous for an rh17 mutant allele (Stein et al., 2021). Evidence for potential clues in phase change induction is further supported by the Mitosis instead of Meiosis phenotype in rice and Arabidopsis, where mutations in only three genes prevent meiotic cell entry and instead meiocytes in the gametophyte undergo mitosis (Mieulet et al., 2016). This phenotype can be developed with triple mutations in REC8 and OSD1 in combinations with either SPO11 or PRD1,2,3 mutants in Arabidopsis and with the combination of REC8, PAIR1, and OSD1 mutations in rice (Mieulet et al., 2016). These previous studies lay a strong foundation on which we can build an understanding of meiosis induction in plants.
2.4 Plant hormonal and environmental factors
Hormonal cues from surrounding somatic tissue in the developing gametophyte also affect meiotic processes. Auxin signaling is likely to provide cues for the differentiation of egg cells vs. synergid cells in the egg apparatus (Sun et al., 2021). Auxin and brassinosteroids are important factors in meiocyte development as peak expression in biosynthesis and signaling is found in meiotic anthers (Dhaka et al., 2020). In addition, an auxin gradient appears to play a role in male germ cell development (Zheng et al., 2021). Cytokinin is shown to play a role in meiotic processes as well. For example, cytokinin histidine kinase receptors, AHK2, AHK3, and CRE1, are attributed with the ability to sense environmental cytokinin to create a kinase cascade, while triple knockouts of these three genes, result in cytokinin unresponsive plants (Inoue et al., 2001; Higuchi et al., 2004; Cheng et al, 2013). Loss of function with these cytokinin receptors results in female gametophytic lethality but can be recovered via TDNA complement insertions (Higuchi et al, 2004; Cheng et al, 2013). Environmental conditions such as hypoxia and oxidation-reduction have also been shown to induce meiotic fate (Kelliher and Walbot, 2012). Mutations in genes associated with redox reactions, like MSCA1 (maize), MIL1 (rice), ROXY1, and ROXY2 (Arabidopsis), led to fertility disruptions (Xing and Zachgo, 2008; Hong et al., 2012; Kelliher & Walbot, 2012). Further, a switch from apomeiosis to meiosis occurs with increased oxidative stress treatment in Boechera premeiotic ovules (Mateo de Arias et al., 2020). In Arabidopsis, retinal was determined to be an endogenous metabolite that plays a role in root organogenesis and root clock functions (Dickinson et al., 2021). Interestingly, TEMPERATURE INDUCED LIPOCALIN (TIL) acts as a retinal binder in plants with protective functions in heat stress, light stress, and oxidative stress (Chi et al., 2009; Boca et al., 2014; Dickinson et al., 2021). Evidence for a stress-mediated switch between meiosis and apomeiosis has been demonstrated (Mateo de Arias et al., 2020), and since retinoic acid may force meiosis induction in mice (Vernet et al., 2020), the closely related retinal may function in stress response in plants, and possibly be of interest to explore for meiotic induction. Moreover, the number of candidate genes and factors for meiosis induction has grown substantially in the past years. However, an efficient test system is needed to determine the relevance of candidate factors in meiosis initiation.
3 Tools for meiotic factor testing
3.1 Cell-based system
Detailed analyses of specific genetic factors and growth hormones provide a great starting place to begin testing factors as meiotic induction candidates but low induction rates in mammals (Medrano et al., 2016) suggest that a high-throughput system is required to evaluate these candidates.
High-throughput, single-cell culture systems, such as protoplasts, may provide a robust approach to detecting the rare meiotic events induced by multiple factors. Protoplasts are spherical-shaped cells that are devoid of the cell wall, removed by enzymatic digestion, and can provide totipotent homogeneous populations of cells useful for plant genetic improvement studies in some species (Davey et al., 2005; Eeckhaut et al, 2013; Sahab et al., 2019). An important factor for the viability of protoplasts is maintaining osmotic stabilization to prevent cell lysis after cell wall removal (Marx, 2016; Reed and Bargmann, 2021). Protoplast-based platforms can allow for the hybridization of different species via protoplast fusion and plant regeneration (Melchers et al., 1978). Moreover, protoplasts can enable the exploration of signal transduction and metabolic pathways (Sheen, 2001), cell type-specific functions (Petersson et al., 2015; Denyer et al., 2019), and determine the subcellular localization, transport, and interactions of intracellular proteins (Goodman et al., 2004; Zhang et al., 2011). Cellular division and subsequent regeneration from protoplasts have been reported in numerous species with varying levels of efficiency (Nagata and Takebe, 1971; Xu et al., 1982; Shillito et al., 1989; Kiełkowska and Adamus, 2012; Chupeau et al., 2013; Jeong et al., 2021). However, recalcitrance to protoplast regeneration has also been observed across many species and is particularly challenging in monocotyledonous species (Hahne et al., 1989; Xu et al., 2022). There is a range of factors that can influence establishing reliable protoplast transient assays and regeneration protocols (reviewed in Reed and Bargmann, 2021).
3.2 In vitro meiosis induction testing system using single cells
Using protoplasts allows many cells to be analyzed at one time while also providing the potential to be collected and used in downstream IVN experiments in addition to simple ploidy analysis. With these single cells, two options have been considered for high-throughput screening of meiosis induction. First, protoplasts can be isolated and then challenged to undergo cellular division. Division would then be followed by a meiotic induction treatment from which dividing cells can be reisolated for ploidy-state analysis. This option can be laborious but provides a means for the analysis of introduced genetic factors (Yoo et al., 2007). The second option can utilize dividing callus, which can be treated with meiotic induction factors followed by protoplast isolation for ploidy-state analysis. This system is potentially less laborious and enables efficient testing of exogenous factors, but the assessment of genetic elements would rely on an efficient transformation system. Both options, however, require callus formation as a result of cellular division of which cell lines could be maintained for analysis and subsequent selection. These protoplast-based approaches would also benefit from culture suspension as multiple factors could be tested while easily moving aliquots of cells for processing and systematic treatment application.
Efficient delivery of genetic elements and the induction and detection of meiosis may be difficult to establish in protoplasts, as cell survival, fitness, and division can be impacted by the product of transgenes and mutagenesis. To test genetic factors, DNA delivery into the cells is required. Conventionally, transgenic plants can be generated via the delivery of DNA-encoding gene constructs via microprojectile bombardment or Agrobacterium-mediated transformation (Cunningham et al, 2018). Other types of transformation methods can utilize nanoparticles (Cunningham et al, 2018; Mao et al., 2019), electroporation, microinjection, PEG-mediated direct delivery in protoplasts (Yoo et al, 2007), and viral-vectors (Catoni et al., 2018; reviewed by Abrahamian et al., 2020). Alternatively, DNA-free CRISPR/Cas genome editing systems can deploy ribonucleoproteins to cells using similar DNA delivery approaches for targeted mutagenesis of genes or regulatory regions to modulate the expression of genes (Woo et al, 2015; Liu et al., 2020; Ma et al., 2020; Zhang et al., 2021). In protoplasts, transcriptional regulation was also an effective means to control gene expression and could be multiplexed (Pan et al., 2021). Given the potential difficulties of genetic element testing in protoplasts, however, protoplasts derived from callus may be highly suited for evaluating chemical factors which can be simply applied in culture media. For instance, chemical factors have been applied to callus cultures to test cell cycle regulation and ploidy increase (Wan et al., 1989; Elmaghrabi et al., 2017), while hormones and stress factors added to callus culture media have been evaluated to increase metabolite production (Beygi et al., 2021). Interestingly, chemical agents can be assessed and deployed to reduce chromosome number, somewhat like haploids or meiosis-like reductions. For example, decades ago a chloramphenicol antibiotic treatment was shown to reduce chromosomes to a haploid state in root cells of barley seedlings (Yoshida and Yamaguchi, 1973). Caffeine treatments have been shown to induce somatic meiosis-like reductions in Vicia root tips (Chen et al., 2000). Meiosis-like reductions have also been observed in somatic embryogenic callus cultures of Arabidopsis (Yihua et al., 2001) and non-embryogenic carrot cell culture lines that were considered to be permanently expressed in a meiotic or sporogenous tissue state (Ronchi et al., 1992). These studies indicate the potential for screening and deploying chemical agents on explant tissue sources and in in vitro culture for meiosis induction in an IVN system. However, the reliable and efficient development of such approaches likely requires extensive work and validation that may vary across different species (Yan et al., 2017).
3.3 High-throughput fluorescence analysis
A potentially efficient method to detect and quantify meiosis induction with callus-derived protoplasts is through the use of a transgenic, bi-fluorescent system to track chromosomal segregation after meiotic cell division. By utilizing a dual marker system, an assay to detect the induction of meiosis can be achieved based on the presence or absence of fluorescence signals in cells through fluorescence-activated cell sorting (FACS) instruments (Bargmann and Birnbaum, 2010; Borges et al., 2012; Ortiz-Ramírez et al., 2018). DNA content analysis using FACS or flow cytometry can also be used to determine artificial gametes (haploid) and somatic cells (diploid), but traditional stains such as 4’,6-diamidino-2-phenylindole and propidium iodide inefficiently pass through intact cell membranes (Wallberg et al., 2016), while there are other commercially available stains for DNA analysis of live cells, these will have to be controlled for and considered when testing meiotic candidates. These factors present difficulties for further downstream uses in IVN’s and are the basis for the fluorescent system development suggestion. In the proposed fluorescent system, two different fluorescent single-copy reporter genes such as RFP or GFP can be integrated into the genome either in allelic or non-allelic positions (Figure 4) and detected without the need for DNA stains.
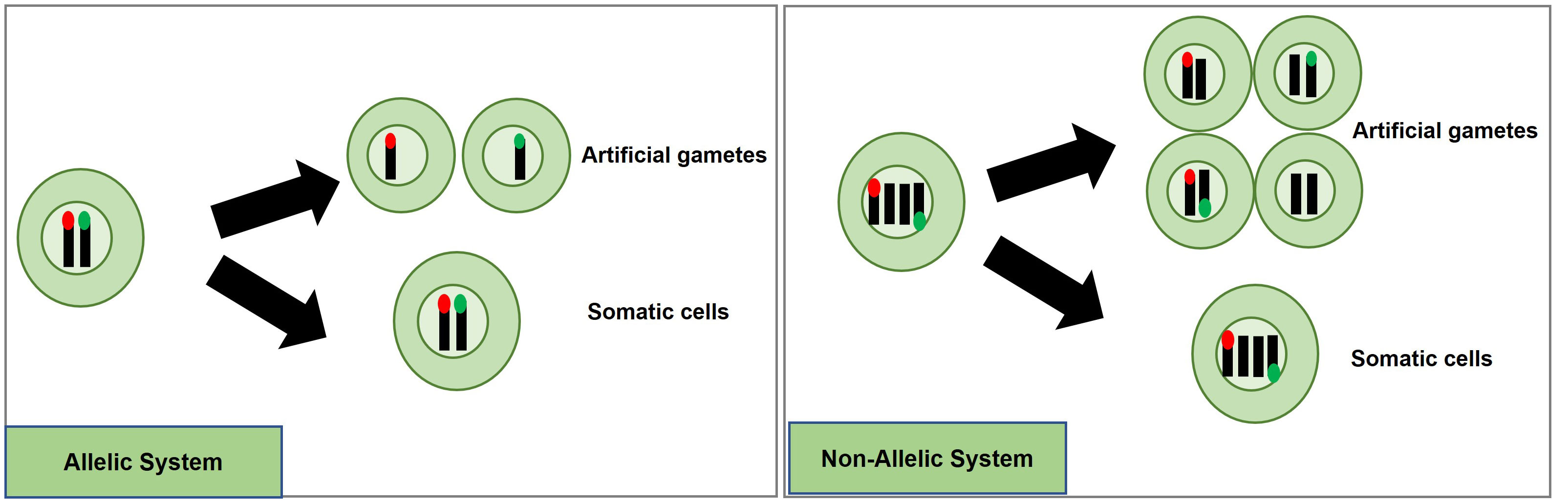
Figure 4 Proposed high-throughput meiosis induction detection tools using bi-fluorescent single cells to track chromosomal segregation. The left panel depicts an allelic system where chromosomal segregation can be identified 1:1. The right panel depicts a non-allelic system where chromosomal segregation can be detected with 50% less efficiency. Red and green dots represent different fluorescent markers on a chromosome, arrows indicate a treatment, and cells right of the arrows are the potential products after treatment, either artificial gametes (if meiosis was induced) or somatic cells (if meiosis was not induced).
Generating a genotype with two different markers in allelic positions is more complex than for non-allelic markers. For example, one way to obtain different marker genes in allelic positions would be to establish a homozygous, fluorescent marker line, and use gene-editing techniques to replace this marker with another fluorescent marker in the allelic position. Low rates for homology-directed repair (HDR) in plants have prevented such targeted knock-ins from being efficiently accomplished. Recent developments, however, have provided more efficient approaches with HDR rates being reported at levels as high as 6.3% (Sun et al., 2023), 3.2% (Wang et al., 2023), 9.1% (Miki et al., 2018); and targeted T-DNA integration via Agrobacterium-mediated transformation in rice ranging from 4 to 5.3% (Lee et al., 2019). Moreover, constructs encoding a Cas9-VirD2 fusion have succeeded in improving HDR-mediated integration in rice transformation as well (Ali et al., 2020).
An informative marker tool is possible by using the resulting F1 progeny from a cross between parents carrying different fluorescent markers in allelic positions. Haploid cells would express only one fluorescent marker while diploid cells would express both (Figure 4). Repressor/activator systems such as the Q-system from Neurospora crassa may provide another option to track chromosomal segregation, as the presence of a repressor in a diploid containing an activator would prevent the expression of a marker, but in a haploid, the marker would be expressed due to the absence of the repressor. The opposite effect could also be obtained using only a transcriptional activator in the Q-system. Transcriptional activation also controls the Gal4/UAS system where the presence of Gal4 would lead to marker expression in a diploid while a haploid would be repressed. Both systems have been established as molecular tools in plant systems (Waki et al., 2013; Persad et al., 2020). These systems would still require allelic positioning to be relevant in IVNs, in addition to overcoming false identification with leaky signaling. These site-specific allelic placements of reporter genes could also be achieved using recombinases such as the Cre-lox and FLP-FRT gene-stacking system (Nandy et al., 2015).
Alternatively, a non-allelic bi-fluorescent reporter system may provide a readily available alternative to this process. The non-allelic, hybrid line would only require establishing, two single-locus fluorescent marker lines that would be crossed, and the resulting F1, which would carry both markers, can be used for testing meiosis-inducing factors. However, this system has a decreased efficiency caused by a 50% reduction of “informative” artificial gametes, expressing only a single fluorophore compared to the allelic system. However, the speed of development of marker lines provides a relevant strategy for Phase I (Figure 4).
3.4 RNA sequencing and fusion technologies
Technological advances in RNA sequencing may also contribute to determining meiosis-induction factors in plants. Single-cell RNA sequencing (scRNA-seq) has proven to be an efficient and cost-effective approach to analyzing multiple tissue types in response to treatment (Shin et al., 2019; Shulse et al., 2019). This technology could be used as an assay to assess candidate meiotic induction factors and provide expression data. scRNA-seq with barcoding permits the sequencing of multiple samples through multiplexing, which allows for simultaneous evaluation of multiple treatments and factors (Macosko et al., 2015; Shin et al., 2019; Shulse et al., 2019). Nelms and Walbot (2019) demonstrated the use of this tool to determine gene expression profiles during different stages of meiosis development. Comparative studies of germline and somatic cells have provided insight into differential gene expression in the meiosis of plants (Dukowic-Schulze et al., 2014a; Nelms and Walbot, 2019; Barakate et al., 2021; Dukowic-Schulze et al., 2014b). The availability of reference genes for meiotic processes also provides an opportunity for factors to be tested using quantitative PCR (Ji et al., 2014; Nelms and Walbot, 2019; Garrido et al., 2020). Therefore, these molecular tools could be used to determine the onset of artificial meiosis induction. It has also been considered, that meiosis induction could be assessed by fluorescent fusions with meiosis-specific genes such as the PROREC8:REC8:GFP line developed by Prusicki et al. (2019). These methods would provide evidence for specific steps in meiosis and could be scaled for high-throughput investigation. Given this, however, fluorescent-based markers may provide many benefits to tracking and assessing meiosis induction, especially with up-scaling and cost reduction using available commercial instruments, as laborious nucleic acid isolation would not be required.
3.5 Statistical approaches for the detection of rare events
The complexity of datasets and inherent variance expected among biological samples would require the optimization of robust statistical analysis methods to detect and discriminate artificial gametes at low meiotic induction rates. The method would rely on analyzing a multitude of data points and determining which factor(s), if any, play a role in meiosis induction. The limit of detection must be possible with induction rates as low as 1%, based on in vitro meiosis induction rates found in human cells (Medrano et al., 2016). Hence, large totipotent protoplast populations that can be analyzed are preferred.
The two bi-fluorescent systems outlined above can use flow cytometry or FACS to detect the different cell populations that show different fluorescence signals. Analysis of fluorescence values can be done using the popular method of “gating” (Adan et al., 2017). Gating is a technique where regions of fluorescence are manually selected to identify events, in our case artificial gametes and diploid cells. Figure 5 depicts a theoretical gating approach, where a balance between accuracy of cell identification and the number of cells identified must be reached. Cells containing both fluorescent markers (i.e., diploid cells), would show similar fluorescence signals for both markers (i.e., population near the middle of the plot), and cells containing only one of the fluorescent markers (i.e., haploid cells), would predominantly show fluorescence of one of the two markers (i.e., population near either of the two axes). Borges et al. (2012) used a similar FACS method to successfully sort nuclei tagged with either RFP in vegetative nuclei or GFP in sperm nuclei from intact bi-fluorescent pollen, obtaining purity rates as high as 99%. Gating is a potential solution to fluorescent cell discrimination, but the subjectivity in gating may induce unwarranted biases in the follow-up statistical analysis to determine the difference among multiple treatments. Alternative methods for gating could be support vector machines (SVM) or clustering. SVM (Cortes and Vapnik, 1995; Lee et al., 2012) is a supervised machine learning technique that learns from labeled training data and creates hyper-planes that separate the artificial gametes from the diploid cells. Clustering is an unsupervised learning (Lo et al., 2008) for automated gating of flow cytometry data that can estimate the cluster means, covariance matrices, and proportions of each cell type. All these data science tools have non-zero probabilities of misclassification, that is, classifying an artificial gamete as diploid and vice versa, and these misclassification probabilities affect the power of the statistical analyses. Thus, the number of cells needs to be adjusted to account for the loss in power due to misclassifications. Figure 6 shows a chi-squared test’s power curves under possible misclassifications by SVM for a range of meiosis induction percentages. The number of cells required to detect, for example, a 1% meiosis induction rate with at least 80% power is around 13,000, which is well within the limit of flow cytometry. Further, these results suggest that pooling samples after meiosis induction treatment may provide an efficient approach to testing multiple factors to reduce flow cytometry costs. Pools of interest can then be analyzed more in-depth to identify the factor responsible for artificial gamete induction.
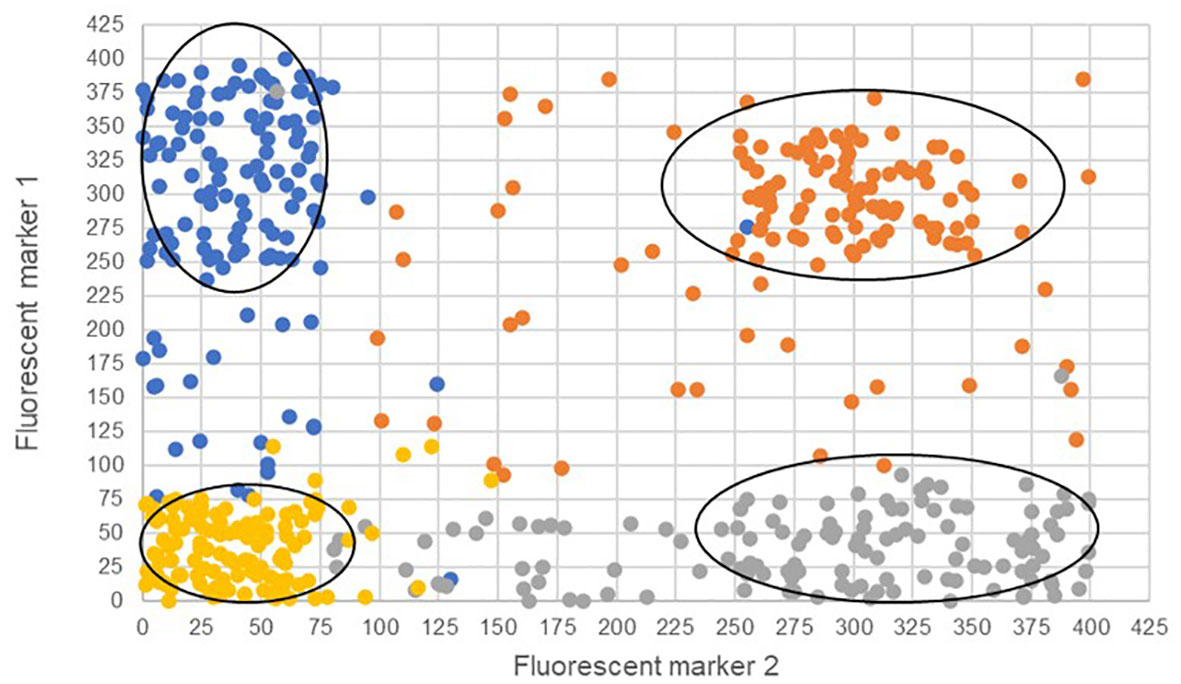
Figure 5 Four theoretical cell populations were produced to simulate mock flow cytometry analysis data with theoretical gating classifications (ovals) for either only fluorescent marker 1 (blue), only fluorescent marker 2 (gray), both fluorescent markers (orange), or the absence of fluorescent markers (gold). The ovals represent potential gating for individual populations. Misclassified cells are depicted as those that have fallen outside the gating ovals or those that have an incorrect fluorescence classification and are a different color than others in the same population.
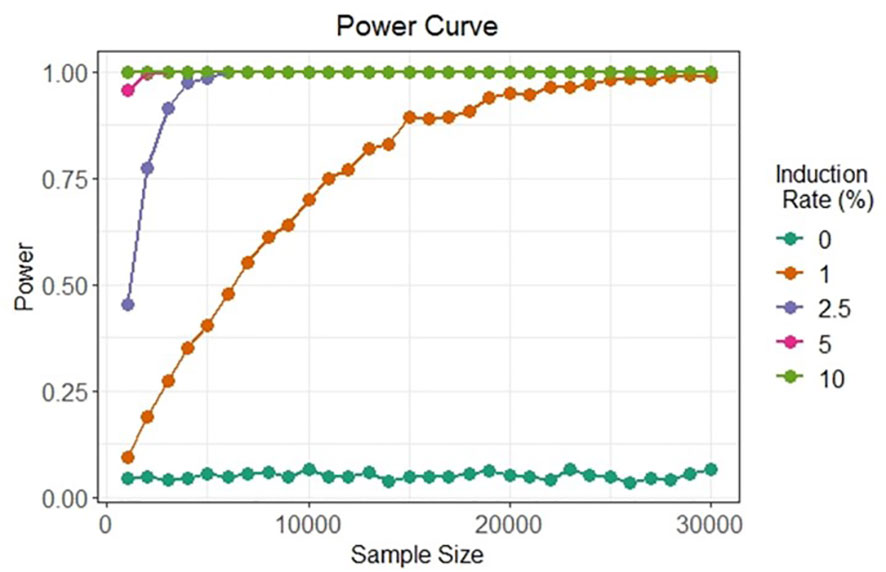
Figure 6 Simulated in silico power for statistical testing of fluorescent cell detection. Each curve denotes the power of the chi-squared test to detect artificial gametes at four different rates of meiosis induction upon the evaluation of different cell sample sizes under possible misclassification by an SVM classifier.
In summary, detection of the few induced gametic cells in a large population of predominantly diploid cells must be supported by a robust statistical framework to provide confidence in factors that result in artificial gametes at low rates, considerations such as this help to define testing procedures and technical limits.
4 Discussion
The development of IVNs could greatly benefit plant breeding as a new tool to increase genetic gain. The conservation of meiotic processes in eukaryotes provides evidence of the potential to develop a universal system to induce meiosis for all plant species in vitro with only minimal changes to culture conditions. To make progress in IVNs, however, a cost-efficient, high-throughput detection tool must be developed for detecting artificial gametes, which is supported by a robust statistical framework. Such a tool would allow the evaluation of a large number of factors as potential inducers of meiosis. Additionally, as custom molecules for targeted biological processes such as the PROTAC system (reviewed in Békés et al., 2022) become more widely available, opportunities to target genetic factors may be tested more efficiently without needing genetic transformation.
There is much to learn from natural phenomena such as apomixis and parthenogenesis, which may provide insights into approaches that can be reversed in order to induce meiosis. Gene activation technologies paired with increased gene editing capabilities have promise in plant meiosis induction, especially in reversing the effect of a knock-out (Pan et al., 2021; Pan et al., 2022). Additionally, while environmental factors and hormone signaling show clear effects on reproduction, systematic testing of these factors will need to be well-thought out as these factors usually have global consequences on plants. By using liquid based culture systems, factors can be applied easier and in a more uniform fashion, which may further increase the scale of a meiosis induction screening system.
For detection we have proposed a protoplast system and while protoplasts can be isolated easily and in large numbers, which is amenable to the detection systems discussed in this article, protoplast regeneration can be species-dependent and recalcitrant (Hahne et al., 1989; Xu et al., 2022). There may be other technologies that provide different benefits to such a system and should also be explored. Additionally, as new cytometric and microfluidic technologies such as impedance flow cytometry (Heidmann et al., 2016) continue to improve, DNA stains and fluorescent markers may no longer be needed as cells could be detected, quantified, and sorted for downstream manipulation using label-free approaches.
5 Concluding remarks and future directions
IVN’s have the potential to change cultivar development in big ways as they can increase genetic gain by decreasing breeding cycle time while also being kept in controlled laboratory conditions. In this review, we have assessed the bottleneck that we believe to be the most limiting at the current state, meiosis induction, but in order to implement and scale IVN’s to efficient sizes, other bottlenecks will need to be overcome. These include the induction and detection of meiosis in crop species, artificial gamete selection, fusion, and subsequent propagation. These bottlenecks will be addressed in subsequent review articles as considerable research is needed. By fully understanding the gaps in knowledge in IVNs, solutions can be more efficiently explored and shared. Progress in single cell analyses, transformation, and sequencing technologies will continue to push IVN’s from ideas to reality.
Author contributions
TC drafted and compiled manuscript. DI, SS, PK contributed to writing and editing manuscript. SA finalized figures and editing. SD and EB, contributed to writing and figure development. SH and LD contributed to draft writing. Additionally, BN and TL contributed to draft development and editing. All authors contributed to the article and approved the submitted version.
Funding
We want to thank the Iowa State University Plant Sciences Institute, RF Baker Center for Plant Breeding, and KJ Frey Chair in Agronomy for their generous support. This article is also a product of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, Project No. IOW03717 (S.D.), and Project No. IOW04714(T.L.) which is supported by USDA/NIFA and State of Iowa funds. This work was also supported by the Foundation for Food & Agriculture Research under award number CA19-SS-0000000128 (T.L.). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the Foundation for Food & Agriculture Research or the U.S. Department of Agriculture.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrahamian, P., Hammond, R. W., Hammond, J. (2020). Plant virus-derived vectors: applications in agricultural and medical biotechnology. Annu. Rev. Virol. 7 (1), 513–535. doi: 10.1146/annurev-virology-010720-054958
Adan, A., Alizada, G., Kiraz, Y., Baran, Y., Nalbant, A. (2017). Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 37 (2), 163–176. doi: 10.3109/07388551.2015.1128876
Ali, Z., Shami, A., Sedeek, K., Kamel, R., Alhabsi, A., Tehseen, M., et al. (2020). Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun. Biol. 3 (1), 44. doi: 10.1038/s42003-020-0768-9
Barakate, A., Orr, J., Schreiber, M., Colas, I., Lewandowska, D., McCallum, N., et al. (2021). Barley anther and meiocyte transcriptome dynamics in meiotic prophase I. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.619404
Bargmann, B. O. R., Birnbaum, K. D. (2010). Fluorescence activated cell sorting of plant protoplasts. J. Visualized Experiments 36, 1673. doi: 10.3791/1673
Bateman, A. (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47 (D1), D506–D515. doi: 10.1093/nar/gky1049
Békés, M., Langley, D. R., Crews, C. M. (2022). PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discovery 21 (3), 181–200. doi: 10.1038/s41573-021-00371-6
Beygi, Z., Nezamzadeh, Z., Rabiei, M., Mirakhorli, N. (2021). Enhanced accumulation of trigonelline by elicitation and osmotic stresses in fenugreek callus culture. Plant Cell Tissue Organ Culture 147 (1), 169–174. doi: 10.1007/s11240-021-02055-w
Bhat, J. A., Ali, S., Salgotra, R. K., Mir, Z. A., Dutta, S., Jadon, V., et al. (2016). Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 7. doi: 10.3389/fgene.2016.00221
Boca, S., Koestler, F., Ksas, B., Chevalier, A., Leymarie, J., Fekete, A., et al. (2014). Arabidopsis lipocalins AtCHL and AtTIL have distinct but overlapping functions essential for lipid protection and seed longevity. Plant Cell Environ. 37 (2), 368–381. doi: 10.1111/pce.12159
Boerman, N. A., Frei, U. K., Lübberstedt, T. (2020). Impact of spontaneous haploid genome doubling in maize breeding. Plants 9 (3), 369. doi: 10.3390/plants9030369
Borges, F., Gardner, R., Lopes, T., Calarco, J. P., Boavida, L. C., Slotkin, R. K., et al. (2012). FACS-based purification of arabidopsis microspores, sperm cells and vegetative nuclei. Plant Methods 8 (1), 44. doi: 10.1186/1746-4811-8-44
Brown, M., Antle, J., Backlund, P., Carr, E., Easterling, W. E., Walsh, M. K., et al. (2015). Climate change, global food security and the U.S. food system. Available at: http://www.usda.gov/oce/climate_change/FoodSecurity2015Assessment/FullAssessment.pdf.
Brummer, C. E., Barber, W. T., Collier, S. M., Cox, T. S., Johnson, R., Murray, S. C., et al. (2011). Plant breeding for harmony between agriculture and the environment. Front. Ecol. Environ. 9 (10), 561–568. doi: 10.1890/100225
Catoni, M., Noris, E., Vaira, A. M., Jonesman, T., Matić, S., Soleimani, R., et al. (2018). Virus-mediated export of chromosomal DNA in plants. Nat. Commun. 9 (1), 5308. doi: 10.1038/s41467-018-07775-w
Chen, Y., Zhang, L., Zhou, Y., Geng, Y., Chen, Z. (2000). Inducing somatic meiosis-like reduction at high frequency by caffeine in root-tip cells of vicia faba. Mutat. Res. - Fundam. Mol. Mech. Mutagenesis 452 (1), 67–72. doi: 10.1016/S0027-5107(00)00045-2
Cheng, C. Y., Mathews, D. E., Schaller, G. E., Kieber, J. J. (2013). Cytokinin-dependent specification of the functional megaspore in the arabidopsis female gametophyte. Plant J. 73 (6), 929–940. doi: 10.1111/tpj.12084
Chi, W. T., Fung, R. W. M., Liu, H. C., Hsu, C. C., Charng, Y. Y. (2009). Temperature-induced lipocalin is required for basal and acquired thermotolerance in arabidopsis. Plant Cell Environ. 32 (7), 917–927. doi: 10.1111/j.1365-3040.2009.01972.x
Chupeau, M. C., Granier, F., Pichon, O., Renou, J. P., Gaudin, V., Chupeaua, Y. (2013). Characterization of the early events leading to totipotency in an arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25 (7), 2444–2463. doi: 10.1105/tpc.113.109538
Colnaghi, M., Lane, N., Pomiankowski, A. (2020). Genome expansion in early eukaryotes drove the transition from lateral gene transfer to meiotic sex. ELife 9, 1–16. doi: 10.7554/ELIFE.58873
Cortes, C., Vapnik, V. (1995). Support-vector networks. Mach. Learn. 20 (3), 273–297. doi: 10.1007/bf00994018
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L., Landry, M. P. (2018). Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36 (9), 882–897. doi: 10.1016/j.tibtech.2018.03.009
Davey, M. R., Anthony, P., Power, J. B., Lowe, K. C. (2005). Plant protoplasts: status and biotechnological perspectives. Biotechnol. Adv. 23 (2), 131–171. doi: 10.1016/j.biotechadv.2004.09.008
Dean, P. J., Siwiec, T., Waterworth, W. M., Schlögelhofer, P., Armstrong, S. J., West, C. E. (2009). A novel ATM-dependent X-ray-inducible gene is essential for both plant meiosis and gametogenesis. Plant J. 58 (5), 791–802. doi: 10.1111/j.1365-313X.2009.03814.x
De La Fuente, G. N., Frei, U. K., Lübberstedt, T. (2013). Accelerating plant breeding. Trends Plant Sci. 18 (12), 667–672. doi: 10.1016/j.tplants.2013.09.001
Denyer, T., Ma, X., Klesen, S., Scacchi, E., Nieselt, K., Timmermans, M. C. P. (2019). Spatiotemporal developmental trajectories in the arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48 (6), 840–852.e5. doi: 10.1016/j.devcel.2019.02.022
Dhaka, N., Krishnan, K., Kandpal, M., Vashisht, I., Pal, M., Sharma, M. K., et al. (2020). Transcriptional trajectories of anther development provide candidates for engineering male fertility in sorghum. Sci. Rep. 10 (1), 897. doi: 10.1038/s41598-020-57717-0
Dickinson, A. J., Zhang, J., Luciano, M., Wachsman, G., Sandoval, E., Schnermann, M., et al. (2021). A plant lipocalin promotes retinal-mediated oscillatory lateral root initiation. Science 373 (6562), 1532–1536. doi: 10.1126/science.abf7461
Dukowic-Schulze, S., Harris, A., Li, J., Sundararajan, A., Mudge, J., Retzel, E. F., et al. (2014a). Comparative transcriptomics of early meiosis in arabidopsis and maize. J. Genet. Genomics 41 (3), 139–152. doi: 10.1016/j.jgg.2013.11.007
Dukowic-Schulze, S., Sundararajan, A., Mudge, J., Ramaraj, T., Farmer, A. D., Wang, M., et al. (2014b). The transcriptome landscape of early maize meiosis. BMC Plant Biol. 14 (1), 118. doi: 10.1186/1471-2229-14-118
Eeckhaut, T., Lakshmanan, P. S., Deryckere, D., Van Bockstaele, E., Van Huylenbroeck, J. (2013). Progress in plant protoplast research. Planta 238 (6), 991–1003. doi: 10.1007/s00425-013-1936-7
Elmaghrabi, A. M., Rogers, H. J., Francis, D., Ochatt, S. J. (2017). Peg induces high expression of the cell cycle checkpoint gene WEE1 in embryogenic callus of medicago truncatula: potential link between cell cycle checkpoint regulation and osmotic stress. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01479
Espinosa, M.E.Á., Moreira, R. O., Lima, A. A., Ságio, S. A., Barreto, H. G., Luiz, S. L. P., et al. (2017). Early histological, hormonal, and molecular changes during pineapple (Ananas comosus (L.) Merrill) artificial flowering induction. J. Plant Physiol. 209, 11–19. doi: 10.1016/j.jplph.2016.11.009
Fang, J., Wei, Y., Lv, C., Peng, S., Zhao, S., Hua, J. (2017). CD61 promotes the differentiation of canine ADMSCs into PGC-like cells through modulation of TGF-β signaling. Sci. Rep. 7 (1), 43851. doi: 10.1038/srep43851
FAO, IFAD, UNICEF, WFP, WHO. (2020). The state of food security and nutrition in the world 2020. (Rome: FAO). doi: 10.4060/ca9692en
Feng, L. X., Chen, Y., Dettin, L., Reijo Pera, R. A., Herr, J. C., Goldberg, E., et al. (2002). Generation and in vitro differentiation of a spermatogonial cell line. Science 297 (5580), 392–395. doi: 10.1126/science.1073162
Gao, H., Gadlage, M. J., Lafitte, H. R., Lenderts, B., Yang, M., Schroder, M., et al. (2020). Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 38 (5), 579–581. doi: 10.1038/s41587-020-0444-0
Garrido, J., Aguilar, M., Prieto, P. (2020). Identification and validation of reference genes for RT-qPCR normalization in wheat meiosis. Sci. Rep. 10 (1), 2726. doi: 10.1038/s41598-020-59580-5
Geiger, H. H. (2009). Doubled Haploids. In: Bennetzen, J. L., Hake, S. (eds) Handbook of Maize. Springer, New York, NY. doi: 10.1007/978-0-387-77863-1_32
Goodenough, U., Heitman, J. (2014). Origins of eukaryotic sexual reproduction. Cold Spring Harb Perspect. Biol. 6 (3). doi: 10.1101/cshperspect.a016154
Goodman, C. D., Casati, P., Walbot, V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in zea mays. Plant Cell 16 (7), 1812–1826. doi: 10.1105/tpc.022574
Griswold, M. D. (2005). Perspective on the Function of Sertoli Cells. Sertoli Cell Biol. 15–18. doi: 10.1016/B978-012647751-1/50003-9
Hackett, W. P. (2011). Juvenility, Maturation, and Rejuvenation in Woody Plants. In Horticultural Reviews, Janick, J. (Ed.). doi: 10.1002/9781118060735.ch3
Hahne, B., Fleck, J., Hahne, G. (1989). Colony formation from mesophyll protoplasts of a cereal, oat. Proc. Natl. Acad. Sci. 86 (16), 6157–6160. doi: 10.1073/pnas.86.16.6157
Heidmann, I., Schade-Kampmann, G., Lambalk, J., Ottiger, M., Di Berardino, M. (2016). Impedance flow cytometry: a novel technique in pollen analysis. PloS One 11 (11), e0165531. doi: 10.1371/journal.pone.0165531
Higuchi, M., Pischke, M. S., Mähönen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. United States America 101 (23), 8821–8826. doi: 10.1073/pnas.0402887101
Hong, L., Tang, D., Zhu, K., Wang, K., Li, M., Cheng, Z. (2012). Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24 (2), 577–588. doi: 10.1105/tpc.111.093740
Hongay, C. F., Grisafi, P. L., Galitski, T., Fink, G. R. (2006). Antisense transcription controls cell fate in saccharomyces cerevisiae. Cell 127 (4), 735–745. doi: 10.1016/j.cell.2006.09.038
Hongay, C. F., Orr-Weaver, T. L. (2011). Drosophila inducer of MEiosis 4 (IME4) is required for notch signaling during oogenesis. Proc. Natl. Acad. Sci. United States America 108 (36), 14855–14860. doi: 10.1073/pnas.1111577108
Howe, K. L., Contreras-Moreira, B., De Silva, N., Maslen, G., Akanni, W., Allen, J., et al. (2020). Ensembl genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 48 (D1), D689–D695. doi: 10.1093/nar/gkz890
Hsu, C. Y., Liu, Y., Luthe, D. S., Yuceer, C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18 (8), 1856–1861. doi: 10.1105/tpc.106.041038
Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., et al. (2001). Identification of CRE1 as a cytokinin receptor from arabidopsis. Nature 409 (6823), 1060–1063. doi: 10.1038/35059117
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., Khan, M. I. R. (2017). Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00475
Jähne, F., Hahn, V., Würschum, T., Leiser, W. L. (2020). Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 133 (8), 2335–2342. doi: 10.1007/s00122-020-03601-4
Jeong, Y. Y., Lee, H. Y., Kim, S. W., Noh, Y. S., Seo, P. J. (2021). Optimization of protoplast regeneration in the model plant arabidopsis thaliana. Plant Methods 17 (1), 21. doi: 10.1186/s13007-021-00720-x
Ji, Y., Tu, P., Wang, K., Gao, F., Yang, W., Zhu, Y., et al. (2014). Defining reference genes for quantitative real-time PCR analysis of anther development in rice. Acta Biochim. Biophys. Sin. 46 (4), 305–312. doi: 10.1093/abbs/gmu002
Karunarathna, K. H. T., Mewan, K. M., Weerasena, O. V. D. S. J., Perera, S. A. C. N., Edirisinghe, E. N. U. (2021). A functional molecular marker for detecting blister blight disease resistance in tea (Camellia sinensis l.). Plant Cell Rep. 40 (2), 351–359. doi: 10.1007/s00299-020-02637-6
Kelliher, T., Starr, D., Su, X., Tang, G., Chen, Z., Carter, J., et al. (2019). One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 37 (3), 287–292. doi: 10.1038/s41587-019-0038-x
Kelliher, T., Walbot, V. (2012). Hypoxia triggers meiotic fate acquisition in maize. Science 337 (6092), 345–348. doi: 10.1126/science.1220080
Kiełkowska, A., Adamus, A. (2012). An alginate-layer technique for culture of brassica oleracea l. protoplasts. Vitro Cell. Dev. Biol. - Plant 48 (2), 265–273. doi: 10.1007/s11627-012-9431-6
Lee, K., Eggenberger, A. L., Banakar, R., McCaw, M. E., Zhu, H., Main, M., et al. (2019). CRISPR/Cas9-mediated targeted T-DNA integration in rice. Plant Mol. Biol. 99 (4–5), 317–328. doi: 10.1007/s11103-018-00819-1
Lee, G., Stoolman, L., Scott, C. (2012). Transfer learning for auto-gating of flow cytometry data Vol. 27. Eds. Guyon, I., Dror, G., Lemaire, V., Taylor, G., Silver, D. (JMLR(workshop), 155–166. Available at: http://www.clopinet.com/isabelle/Projects/ICML2011/slides/lee11.pdf.
Li, J., Dukowic-Schulze, S., Lindquist, I. E., Farmer, A. D., Kelly, B., Li, T., et al. (2015). The plant-specific protein FEHLSTART controls male meiotic entry, initializing meiotic synchronization in arabidopsis. Plant J. 84 (4), 659–671. doi: 10.1111/tpj.13026
Li, H., Rasheed, A., Hickey, L. T., He, Z. (2018). Fast-forwarding genetic gain. Trends Plant Sci. 23 (3), 184–186. doi: 10.1016/j.tplants.2018.01.007
Liu, W., Rudis, M. R., Cheplick, M. H., Millwood, R. J., Yang, J. P., Ondzighi-Assoume, C. A., et al. (2020). Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 39 (2), 245–257. doi: 10.1007/s00299-019-02488-w
Lo, K., Brinkman, R. R., Gottardo, R. (2008). Automated gating of flow cytometry data via robust model-based clustering. Cytometry Part A 73 (4), 321–332. doi: 10.1002/cyto.a.20531
López-Malvar, A., Butron, A., Malvar, R. A., McQueen-Mason, S. J., Faas, L., Gómez, L. D., et al. (2021). Association mapping for maize stover yield and saccharification efficiency using a multiparent advanced generation intercross (MAGIC) population. Sci. Rep. 11 (1), 3425. doi: 10.1038/s41598-021-83107-1
Lora, J., Hormaza, J. I. (2021). Crosstalk between the sporophyte and the gametophyte during anther and ovule development in angiosperms BT - progress in botany, Vol. 82. Eds. Cánovas, F. M., Lüttge, U., Risueño, M.-C., Pretzsch, H., (Cham: Springer) 113–129. doi: 10.1007/124_2020_50
Ma, X., Zhang, X., Liu, H., Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 6 (7), 773–779. doi: 10.1038/s41477-020-0704-5
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161 (5), 1202–1214. doi: 10.1016/j.cell.2015.05.002
Mao, Y., Botella, J. R., Liu, Y., Zhu, J. K. (2019). Gene editing in plants: progress and challenges. Natl. Sci. Rev. 6 (3), 421–437. doi: 10.1093/nsr/nwz005
Marx, V. (2016). Plants: a tool box of cell-based assays. Nat. Methods 13 (7), 551–554. doi: 10.1038/nmeth.3900
Mata, J., Lyne, R., Burns, G., Bähler, J. (2002). The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32 (1), 143–147. doi: 10.1038/ng951
Mathilde, G., Ghislaine, G., Daniel, V., Georges, P. (2003). The arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 35 (4), 465–475. doi: 10.1046/j.1365-313X.2003.01820.x
Mateo de Arias, M., Gao, L., Sherwood, D. A., Dwivedi, K. K., Price, B. J., Jamison, M., et al. (2020). Whether gametophytes are reduced or unreduced in angiosperms might be determined metabolically. Genes 11 (12), 1449. doi: 10.3390/genes11121449
Medrano, J. V., Martínez-Arroyo, A. M., Míguez, J. M., Moreno, I., Martínez, S., Quiñonero, A., et al. (2016). Human somatic cells subjected to genetic induction with six germ line-related factors display meiotic germ cell-like features. Sci. Rep. 6, 24956. doi: 10.1038/srep24956
Melchers, G., Sacristán, M. D., Holder, A. A. (1978). Somatic hybrid plants of potato and tomato regenerated from fused protoplasts. Carlsberg Res. Commun. 43 (4), 203–218. doi: 10.1007/BF02906548
Mercier, R., Mézard, C., Jenczewski, E., Macaisne, N., Grelon, M. (2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66 (1), 297–327. doi: 10.1146/annurev-arplant-050213-035923
Mieulet, D., Jolivet, S., Rivard, M., Cromer, L., Vernet, A., Mayonove, P., et al. (2016). Turning rice meiosis into mitosis. Cell Res. 26 (11), 1242–1254. doi: 10.1038/cr.2016.117
Miki, D., Zhang, W., Zeng, W., Feng, Z., Zhu, J. K. (2018). CRISPR/Cas9-mediated gene targeting in arabidopsis using sequential transformation. Nat. Commun. 9 (1), 1967. doi: 10.1038/s41467-018-04416-0
Murray, S. C., Eckhoff, P., Wood, L., Paterson, A. H. (2013). A proposal to use gamete cycling in vitro to improve crops and livestock. Nat. Biotechnol. 31 (10), 877–880. doi: 10.1038/nbt.2707
Nagata, T., Takebe, I. (1971). Plating of isolated tobacco mesophyll protoplasts on agar medium. Planta 99 (1), 12–20. doi: 10.1007/BF00392116
Nandy, S., Zhao, S., Pathak, B. P., Manoharan, M., Srivastava, V. (2015). Gene stacking in plant cell using recombinases for gene integration and nucleases for marker gene deletion. BMC Biotechnol. 15 (1), 93. doi: 10.1186/s12896-015-0212-2
Nelms, B., Walbot, V. (2019). Defining the developmental program leading to meiosis in maize. Science 364 (6435), 52–56. doi: 10.1126/science.aav6428
Nonomura, K. I., Eiguchi, M., Nakano, M., Takashima, K., Komeda, N., Fukuchi, S., et al. (2011). A novel RNA-recognition-motif protein is required for premeiotic G1/s-phase transition in rice (Oryza sativa l.). PloS Genet. 7 (1), e1001265. doi: 10.1371/journal.pgen.1001265
Olmedo-Monfil, V., Durán-Figueroa, N., Arteaga-Vázquez, M., Demesa-Arévalo, E., Autran, D., Grimanelli, D., et al. (2010). Control of female gamete formation by a small RNA pathway in arabidopsis. Nature 464 (7288), 628–632. doi: 10.1038/nature08828
Ortiz-Ramírez, C., Arevalo, E. D., Xu, X., Jackson, D. P., Birnbaum, K. D. (2018). An efficient cell sorting protocol for maize protoplasts. Curr. Protoc. Plant Biol. 3 (3), e20072. doi: 10.1002/cppb.20072
Pan, C., Li, G., Malzahn, A. A., Cheng, Y., Leyson, B., Sretenovic, S., et al. (2022). Boosting plant genome editing with a versatile CRISPR-combo system. Nat. Plants 8 (5), 513–525. doi: 10.1038/s41477-022-01151-9
Pan, C., Wu, X., Markel, K., Malzahn, A. A., Kundagrami, N., Sretenovic, S., et al. (2021). CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 7 (7), 942–953. doi: 10.1038/s41477-021-00953-7
Pawlowski, W. P., Wang, C. J. R., Golubovskaya, I. N., Szymaniak, J. M., Shi, L., Hamant, O., et al. (2009). Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc. Natl. Acad. Sci. United States America 106 (9), 3603–3608. doi: 10.1073/pnas.0810115106
Persad, R., Reuter, D. N., Dice, L. T., Nguyen, M. A., Rigoulot, S. B., Layton, J. S., et al. (2020). The q-system as a synthetic transcriptional regulator in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00245
Petersson, S. V., Lindén, P., Moritz, T., Ljung, K. (2015). Cell-type specific metabolic profiling of arabidopsis thaliana protoplasts as a tool for plant systems biology. Metabolomics 11 (6), 1679–1689. doi: 10.1007/s11306-015-0814-7
Popovic, M., Azpiroz, F., Chuva de Sousa Lopes, S. M. (2021). Engineered models of the human embryo. Nat. Biotechnol. 39 (8), 918–920. doi: 10.1038/s41587-021-01004-4
Prusicki, M. A., Keizer, E. M., Van Rosmalen, R. P., Komaki, S., Seifert, F., Müller, K., et al. (2019). Live cell imaging of meiosis in arabidopsis thaliana. ELife 8, e42834. doi: 10.7554/eLife.42834
Reed, K. M., Bargmann, B. O. R. (2021). Protoplast regeneration and its use in new plant breeding technologies. Front. Genome Editing 3. doi: 10.3389/fgeed.2021.734951
Ren, L., Tang, D., Zhao, T., Zhang, F., Liu, C., Xue, Z., et al. (2018). OsSPL regulates meiotic fate acquisition in rice. New Phytol. 218 (2), 789–803. doi: 10.1111/nph.15017
Riou, L., Bastos, H., Lassalle, B., Coureuil, M., Testart, J., Boussin, F. D., et al. (2005). The telomerase activity of adult mouse testis resides in the spermatogonial α6-integrin-positive side population enriched in germinal stem cells. Endocrinology 146 (9), 3926–3932. doi: 10.1210/en.2005-0502
Ronchi, V. N., Giorgetti, L., Tonelli, M., Martini, G. (1992). Ploidy reduction and genome segregation in cultured carrot cell lines. II. somatic meiosis. Plant Cell Tissue Organ Culture 30 (2), 115–119. doi: 10.1007/BF00034304
Sahab, S., Hayden, M. J., Mason, J., Spangenberg, G. (2019). Mesophyll protoplasts and PEG-mediated transfections: Transient assays and generation of stable transgenic canola plants. In Kumar, S., Barone, P., Smith, M. (Eds.). Methods Molecular Biol. 1864, 131–152. Springer New York. doi: 10.1007/978-1-4939-8778-8_10
Santos, T., Shuler, J., Guimarães, R., Farah, A. (2019). CHAPTER 1. Introduction to Coffee Plant and Genetics: Production, Quality and Chemistry. (1–25). doi: 10.1039/9781782622437-00001
Sheen, J. (2001). Signal transduction in maize and arabidopsis mesophyll protoplasts. Plant Physiol. 127 (4), 1466–1475. doi: 10.1104/pp.010820
Shillito, R. D., Carswell, G. K., Johnson, C. M., Dimaio, J. J., Harms, C. T. (1989). Regeneration of fertile plants from protoplasts of elite inbred maize. Bio/Technology 7 (6), 581–587. doi: 10.1038/nbt0689-581
Shin, D., Lee, W., Lee, J. H., Bang, D. (2019). Multiplexed single-cell RNA-seq via transient barcoding for simultaneous expression profiling of various drug perturbations. Sci. Adv. 5 (5), eaav2249. doi: 10.1126/sciadv.aav2249
Shulse, C. N., Cole, B. J., Ciobanu, D., Lin, J., Yoshinaga, Y., Gouran, M., et al. (2019). High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep. 27 (7), 2241–2247.e4. doi: 10.1016/j.celrep.2019.04.054
Singh, M., Goel, S., Meeley, R. B., Dantec, C., Parrinello, H., Michaud, C., et al. (2011). Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23 (2), 443–458. doi: 10.1105/tpc.110.079020
Stein, R. E., Nauerth, B. H., Binmöller, L., Zühl, L., Loreth, A., Reinert, M., et al. (2021). RH17 restricts reproductive fate and represses autonomous seed coat development in sexual arabidopsis. Dev. (Cambridge) 148 (19), dev198739. doi: 10.1242/dev.198739
Stevens, R., Grelon, M., Vezon, D., Oh, J., Meyer, P., Perennes, C., et al. (2004). A CDC45 homolog in arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16 (1), 99–113. doi: 10.1105/tpc.016865
Sun, C., Lei, Y., Li, B., Gao, Q., Li, Y., Cao, W., et al. (2023). Precise integration of large DNA sequences in plant genomes using PrimeRoot editors. Nat. Biotechnol. doi: 10.1038/s41587-023-01769-w
Sun, Y., Wang, X., Pan, L., Xie, F., Dai, B., Sun, M., et al. (2021). Plant egg cell fate determination depends on its exact position in female gametophyte. Proc. Natl. Acad. Sci. United States America 118 (8), e2017488118. doi: 10.1073/pnas.2017488118
Tibbs Cortes, L., Zhang, Z., Yu, J. (2021). Status and prospects of genome-wide association studies in plants. Plant Genome 14 (1), e20077. doi: 10.1002/tpg2.20077
US Embassy and Consulate in Italy (2022). “How climate change affects the food crisis,” in US Embassies and consulates in Italy. Available at: https://it.usembassy.gov/how-climate-change-affects-the-food-crisis/.
Vernet, N., Condrea, D., Mayere, C., Féret, B., Klopfenstein, M., Magnant, W., et al. (2020). Meiosis occurs normally in the fetal ovary of mice lacking all retinoic acid receptors. Sci. Adv. 6 (21), eaaz1139. doi: 10.1126/sciadv.aaz1139
Waki, T., Miyashima, S., Nakanishi, M., Ikeda, Y., Hashimoto, T., Nakajima, K. (2013). A GAL4-based targeted activation tagging system in arabidopsis thaliana. Plant J. 73 (3), 357–367. doi: 10.1111/tpj.12049
Wallberg, F., Tenev, T., Meier, P. (2016). Analysis of Apoptosis and Necroptosis by Fluorescence-Activated Cell Sorting. Cold Spring Harbor Protocols 2016 (4), pdb.prot087387. doi: 10.1101/pdb.prot087387
Wan, Y., Petolino, J. F., Widholm, J. M. (1989). Efficient production of doubled haploid plants through colchicine treatment of anther-derived maize callus. Theor. And Appl. Genet. 77 (6), 889–892. doi: 10.1007/BF00268344
Wang, N., Ryan, L., Sardesai, N., Wu, E., Lenderts, B., Lowe, K., et al. (2023). Leaf transformation for efficient random integration and targeted genome modification in maize and sorghum. Nat. Plants 9 (2), 255–270. doi: 10.1038/s41477-022-01338-0
Wang, Y., Van Rengs, W. M. J., Zaidan, M. W. A. M., Underwood, C. J. (2021). Meiosis in crops: from genes to genomes. J. Exp. Bot. 72 (18), 6091–6109. doi: 10.1093/jxb/erab217
Wang, N., Wang, H., Zhang, A., Liu, Y., Yu, D., Hao, Z., et al. (2020). Genomic prediction across years in a maize doubled haploid breeding program to accelerate early-stage testcross testing. Theor. Appl. Genet. 133 (10), 2869–2879. doi: 10.1007/s00122-020-03638-5
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4 (1), 23–29. doi: 10.1038/s41477-017-0083-8
Wei, B., Zhang, J., Pang, C., Yu, H., Guo, D., Jiang, H., et al. (2015). The molecular mechanism of SPOROCYTELESS/NOZZLE in controlling arabidopsis ovule development. Cell Res. 25 (1), 121–134. doi: 10.1038/cr.2014.145
Wen, L., Tang, F. (2019). Human germline cell development: from the perspective of single-cell sequencing. Mol. Cell 76 (2), 320–328. doi: 10.1016/j.molcel.2019.08.025
Woo, J. W., Kim, J., Kwon, S., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-Free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33 (11), 1162–1164. doi: 10.1038/nbt.3389
Xing, S., Zachgo, S. (2008). ROXY1 and ROXY2, two arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53 (5), 790–801. doi: 10.1111/j.1365-313X.2007.03375.x
Xu, Z. H., Davey, M. R., Cocking, E. C. (1982). Plant regeneration from root protoplasts of brassica. Plant Sci. Lett. 24 (1), 117–121. doi: 10.1016/0304-4211(82)90016-5
Xu, Y., Li, R., Luo, H., Wang, Z., Li, M. W., Lam, H. M., et al. (2022). Protoplasts: small cells with big roles in plant biology. Trends Plant Sci. 27 (8), 828–829. doi: 10.1016/j.tplants.2022.03.010
Yan, G., Liu, H., Wang, H., Lu, Z., Wang, Y., Mullan, D., et al. (2017). Accelerated generation of selfed pure line plants for gene identification and crop breeding. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01786
Yang, C., Hamamura, Y., Sofroni, K., Böwer, F., Stolze, S. C., Nakagami, H., et al. (2019). SWITCH 1/DYAD is a WINGS APART-LIKE antagonist that maintains sister chromatid cohesion in meiosis. Nat. Commun. 10 (1), 1755. doi: 10.1038/s41467-019-09759-w
Yi, J., Kim, S. R., Lee, D. Y., Moon, S., Lee, Y. S., Jung, K. H., et al. (2012). The rice gene DEFECTIVE TAPETUM and MEIOCYTES 1 (DTM1) is required for early tapetum development and meiosis. Plant J. 70 (2), 256–270. doi: 10.1111/j.1365-313X.2011.04864.x
Yihua, C., Lihua, Z., Yuxuan, G., Zhenghua, C. (2001). Meiosis-like reduction during somatic embryogenesis of arabidopsis thaliana. In Vitro Cell. Dev. Biol. Plant 37 (5), 654–657. Available at: https://www.jstor.org/stable/4293529.
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 (7), 1565–1572. doi: 10.1038/nprot.2007.199
Yoshida, H., Yamaguchi, H. (1973). Arrangement and association of somatic chromosomes induced by chloramphenicol in barley. Chromosoma 43 (4), 399–407. doi: 10.1007/BF00406746
Zalesny, R. S., Zhu, J. Y., Headlee, W. L., Gleisner, R., Pilipović, A., Van Acker, J., et al. (2020). Ecosystem services, physiology, and biofuels recalcitrance of poplars grown for landfill phytoremediation. Plants 9 (10), 1–26. doi: 10.3390/plants9101357
Zhang, Y., Iaffaldano, B., Qi, Y. (2021). CRISPR ribonucleoprotein-mediated genetic engineering in plants. Plant Commun. 2 (2), 100168. doi: 10.1016/j.xplc.2021.100168
Zhang, Y., Massel, K., Godwin, I. D., Gao, C. (2018). Applications and potential of genome editing in crop improvement. Genome Biol. 19 (1), 210. doi: 10.1186/s13059-018-1586-y
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7 (1), 30. doi: 10.1186/1746-4811-7-30
Zhao, X., Bramsiepe, J., Van Durme, M., Komaki, S., Prusicki, M. A., Maruyama, D., et al. (2017). RETINOBLASTOMA RELATED1 mediates germline entry in arabidopsis. Science 356 (6336), eaaf6532. doi: 10.1126/science.aaf6532
Zheng, Y., Wang, D., Ye, S., Chen, W., Li, G., Xu, Z., et al. (2021). Auxin guides germ-cell specification in arabidopsis anthers. Proc. Natl. Acad. Sci. United States America 118 (22), e2101492118. doi: 10.1073/pnas.2101492118
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20 (5), 1278–1288. doi: 10.1105/tpc.108.058883
Keywords: meiosis induction, in vitro biology, in vitro nurseries, high-throughput detection, plant breeding and biotechnology
Citation: Cook TM, Isenegger D, Dutta S, Sahab S, Kay P, Aboobucker SI, Biswas E, Heerschap S, Nikolau BJ, Dong L and Lübberstedt T (2023) Overcoming roadblocks for in vitro nurseries in plants: induction of meiosis. Front. Plant Sci. 14:1204813. doi: 10.3389/fpls.2023.1204813
Received: 12 April 2023; Accepted: 17 May 2023;
Published: 02 June 2023.
Edited by:
Manohar Chakrabarti, The University of Texas Rio Grande Valley, United StatesReviewed by:
Yiping Qi, University of Maryland, College Park, United StatesDhananjay K. Pandey, Amity University, Jharkhand, India
Copyright © 2023 Cook, Isenegger, Dutta, Sahab, Kay, Aboobucker, Biswas, Heerschap, Nikolau, Dong and Lübberstedt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Lübberstedt, dGhvbWFzbEBpYXN0YXRlLmVkdQ==
 Tanner M. Cook
Tanner M. Cook Daniel Isenegger
Daniel Isenegger Somak Dutta
Somak Dutta Sareena Sahab
Sareena Sahab Pippa Kay
Pippa Kay Siddique I. Aboobucker
Siddique I. Aboobucker Eva Biswas3
Eva Biswas3 Basil J. Nikolau
Basil J. Nikolau Liang Dong
Liang Dong Thomas Lübberstedt
Thomas Lübberstedt