- 1Hainan Yazhou Bay Seed Lab, Sanya, Hainan, China
- 2Center for Advanced Bioindustry Technologies, and Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
Acid soil syndrome leads to severe yield reductions in various crops worldwide. In addition to low pH and proton stress, this syndrome includes deficiencies of essential salt-based ions, enrichment of toxic metals such as manganese (Mn) and aluminum (Al), and consequent phosphorus (P) fixation. Plants have evolved mechanisms to cope with soil acidity. In particular, STOP1 (Sensitive to proton rhizotoxicity 1) and its homologs are master transcription factors that have been intensively studied in low pH and Al resistance. Recent studies have identified additional functions of STOP1 in coping with other acid soil barriers: STOP1 regulates plant growth under phosphate (Pi) or potassium (K) limitation, promotes nitrate (NO3-) uptake, confers anoxic tolerance during flooding, and inhibits drought tolerance, suggesting that STOP1 functions as a node for multiple signaling pathways. STOP1 is evolutionarily conserved in a wide range of plant species. This review summarizes the central role of STOP1 and STOP1-like proteins in regulating coexisting stresses in acid soils, outlines the advances in the regulation of STOP1, and highlights the potential of STOP1 and STOP1-like proteins to improve crop production on acid soils.
Introduction
About 30% of the world’s ice-free land and 50% of the world’s potentially arable lands are acidic (characterized by pH<5.5) (von Uexküll and Mutert, 1995). Approximately 60% of the acid soils occur in tropical or subtropical regions (Kochian et al., 2004), where rainfall is high, leaching is intense, and the soil’s water-holding capacity is low. As a result, acid soils usually have many other factors besides low pH that can impair crop production (Delhaize and Ryan, 1995), including: (a) hypoxia stress caused by submergence and water-logging (Voesenek and Bailey-Serres, 2015); (b) deficiency of soluble basic cations of K, calcium (Ca), and magnesium (Mg) caused by leaching (Krug and Frink, 1983; von Uexküll and Mutert, 1995); (c) dissolving and enrichment of insoluble iron (Fe), Al, and Mn in oxides caused by low pH and hypoxic conditions (Kochian et al., 2004); (d) passivation and deficiency of Pi caused by the fixation of reactive toxic metals (Kochian et al., 2004; Zheng, 2010) together with (e) unbalanced nitrogen nutrition with predominantly ammonium (NH4+) rather than NO3- (Kidd and Proctor, 2001). These factors also accelerate the process of soil acidification.
Acid soils inhibit root elongation and function, affect root water and nutrient uptake, and suppress plant growth (Ma, 2007). As early responsive factors to environmental signals, transcription factors play an essential role in stress resistance. STOP1 is a critical Cys2His2-type zinc finger transcription factor for proton tolerance and Al resistance (Iuchi et al., 2007). Recent studies further demonstrated that STOP1 is involved in regulating nutrient homeostasis and multiple stress tolerance in acid soils. In the post-genomic era, molecular breeding and genetic engineering are effective measures to improve the stress resistance of various crop species. Identification and functional characterization of STOP1 offer promising results for overcoming acid soil syndrome (Iuchi et al., 2007). This review focuses on recent advances in the biological function and regulatory processes of STOP1, which highlights the application of STOP1 and STOP1-like proteins in improving crop resistance to acid soil syndrome.
Overview of STOP1 and STOP1-like proteins
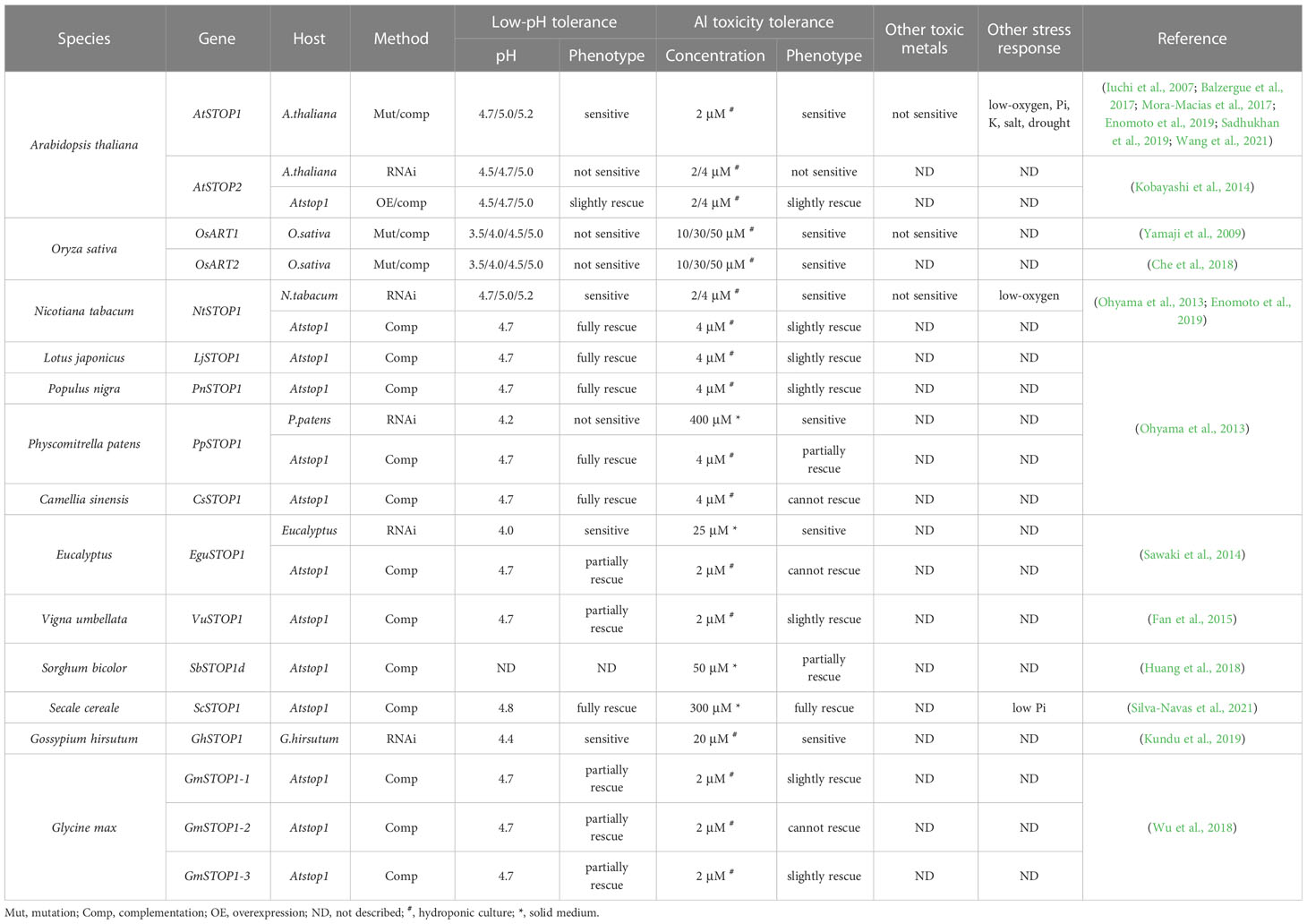
STOP1 is a C2H2 zinc finger transcription factor originally identified by forward genetics in Arabidopsis (Arabidopsis thaliana). The stop1 mutant was screened for its low pH sensitivity, and subsequent research showed that this mutant is also hypersensitive to Al stress (Iuchi et al., 2007). STOP1 localizes to the nucleus and up-regulates the expression of many genes involved in low pH tolerance and Al resistance (Sawaki et al., 2009). Recent studies revealed that STOP1 is essential for low-O2 (Enomoto et al., 2019), low-Pi (Balzergue et al., 2017; Mora-Macias et al., 2017), low-K (Wang et al., 2021), drought and salt tolerance (Sadhukhan et al., 2019) in Arabidopsis. These findings suggest that STOP1 functions as a central factor in modulating the response to coexisting environmental stresses in acid soils. STOP1 is evolutionarily conserved in a wide range of crops (Garcia-Oliveira et al., 2013; Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015; Huang et al., 2018; Wu et al., 2018; Kundu et al., 2019; Silva-Navas et al., 2021). Homologs of the Arabidopsis STOP1 (AtSTOP1) exist in wheat (Triticum aestivum), rice (Oryza sativa), soybean (Glycine max), tobacco (Nicotiana tabacum), sorghum (Sorghum bicolor), cotton (Gossypium hirsutum), rye (Secale cereale), and rice bean (Vigna umbellata), etc. Many plant species possess multiple STOP1-like proteins (Figure 1). Studies on the biological functions of STOP1-like proteins mainly focused on low pH tolerance and Al resistance. AtSTOP2, the paralog of AtSTOP1, is a physiologically minor isoform that activates the transcription of several AtSTOP1-regulated genes in Arabidopsis (Kobayashi et al., 2014). Knockdown of AtSTOP2 did not alter proton or Al sensitivity, but overexpression of AtSTOP2 partially rescued the low pH sensitivity of Atstop1 (Kobayashi et al., 2014). This is consistent with the fact that AtSTOP2 has lower expression and functions downstream of AtSTOP1, suggesting a possible unequal functional redundancy between them.
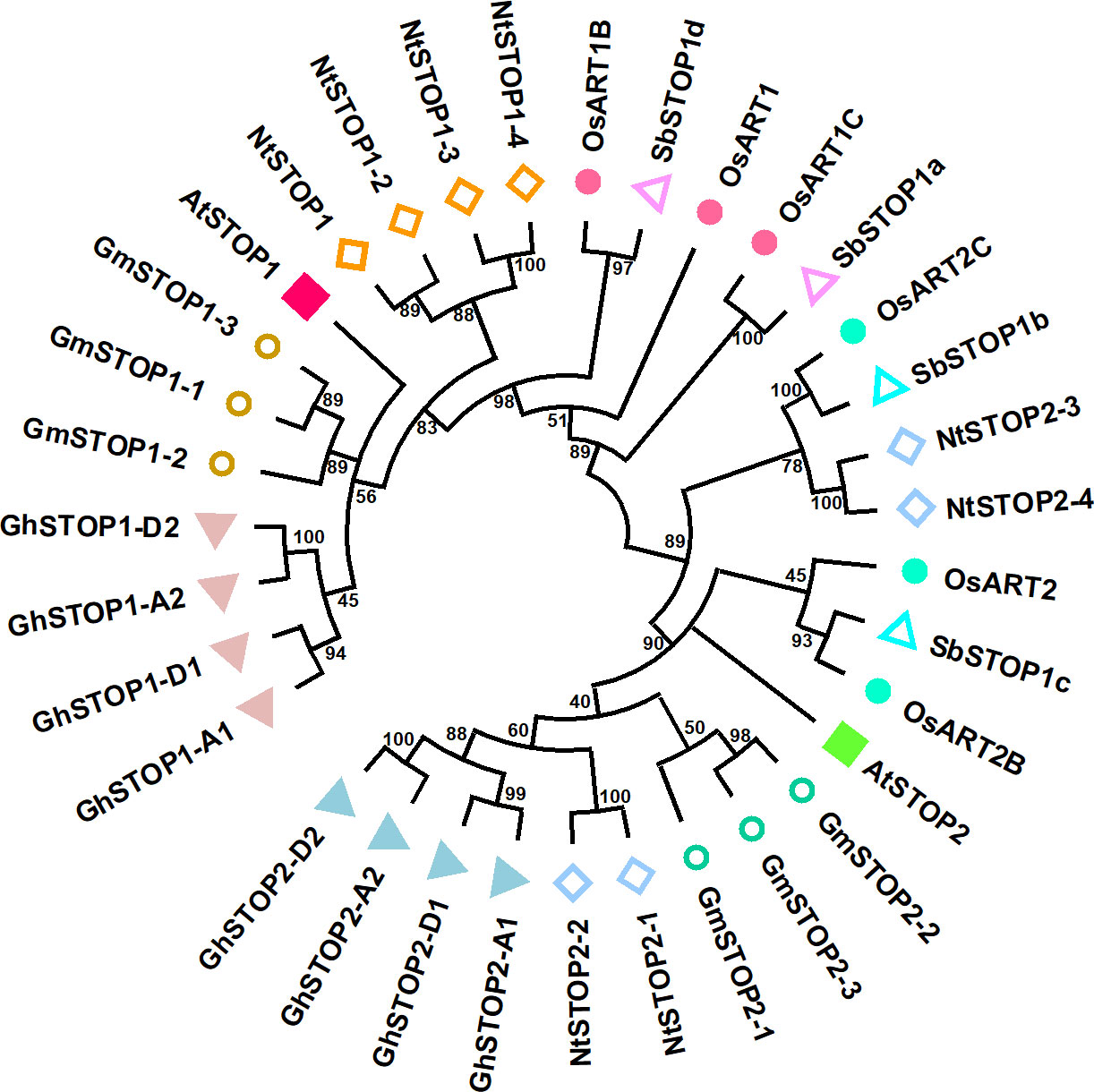
Figure 1 Phylogenetic analysis of STOP1-like proteins in representative crop species. The plant STOP1-like proteins analyzed include representatives from Arabidopsis (Arabidopsis thaliana Araport11 data: AtSTOP1, At1G34370; AtSTOP2, At5G22890), soybean (Glycine max Wm82 ISU-01 v2.1 data: GmSTOP1-1, Gm10G178500; GmSTOP1-2, Gm16G128700; GmSTOP1-3, Gm20G138900; GmSTOP2-1, Gm13G281700; GmSTOP2-2, Gm11G153300; GmSTOP2-3, Gm12G147100), upland cotton (Gossypium hirsutum v3.1 data: GhSTOP1/GhSTOP1-A1, GhA02G060700; GhSTOP1-A2, GhA09G117100; GhSTOP1-D1, GhD02G066100; GhSTOP1-D2, GhD09G114000; GhSTOP2-A1, GhA09G088500; GhSTOP2-D1, GhD09G088200; GhSTOP2-A2, GhA05G086600; GhSTOP2-D2, GhD05G087900), tobacco (Nicotiana tabacum v4.5 data: NtSTOP1/NtSTOP1-1, 0000159g0180; NtSTOP1-2, 0009001g0020; NtSTOP1-3, 0000303g0050; NtSTOP1-4, 0004461g0030; NtSTOP2-1, 0000173g0150; NtSTOP2-2 0028281g0010; NtSTOP2-3 0000083g0190; NtSTOP2-4, 0005475g0020), rice (Oryza sativa v7.0 data: OsART1, Os12g0170400; OsART1B, Os01g0871200; OsART1C, Os03g0838800; OsART2, Os04g0165200; OsART2B, Os08g0562300; OsART2C, Os02g0572900), sorghum (Sorghum bicolor v5.1 data:SbSTOP1a, Sb01G020200; SbSTOP1b, Sb04G188300; SbSTOP1c, Sb07G166000; SbSTOP1d, Sb 03G370700). Evolutionary relationships were inferred from amino acid sequences using the Neighbor-Joining method in MEGA11 (Tamura et al., 2021). The branching topology pattern of the condensed tree is shown under a 40% cut-off.
Similarly, OsART1 (Al resistance transcription factor 1), a STOP1 homolog in rice, was identified by mutant screening and map-based cloning (Yamaji et al., 2009). As a core transcription factor for Al resistance, OsART1 regulates many Al resistance genes through direct promoter binding and transcription activation (Tsutsui et al., 2011). Although not so sensitive as Osart1, the Osart2 mutants also showed reduced growth under Al3+ treatment (Che et al., 2018). However, unlike AtSTOP1, mutation of OsART1 or OsART2 in rice did not increase sensitivity to proton stress (Yamaji et al., 2009; Che et al., 2018). One possible reason is that OsART1 and OsART2 function redundantly with their homologs in regulating low pH tolerance; since rice has six copies of STOP1-like proteins, neither OsART1 nor OsART2 is the closest homolog to AtSTOP1 (Figure 1). Additionally, studies of STOP1 and STOP1-like proteins in other plant species (Table 1) showed they have slightly different roles in response to low pH and Al stress (Yamaji et al., 2009; Garcia-Oliveira et al., 2013; Ohyama et al., 2013; Kobayashi et al., 2014; Sawaki et al., 2014; Fan et al., 2015; Che et al., 2018; Daspute et al., 2018; Huang et al., 2018; Wu et al., 2018; Kundu et al., 2019; Silva-Navas et al., 2021). Therefore, future research is needed to evaluate the redundancy between multiple STOP1-like proteins in some plant species, especially in response to low pH, and to investigate whether functional preferences have evolved between them in dealing with specific stresses. The study of the functional preferences of STOP1-like proteins will benefit the extension of STOP1 and STOP1-like proteins to crop genetic improvement.
STOP1 and STOP1-like proteins mediated low pH tolerance
Proton stress is thought to be the proximal cause of poor plant growth in acid soils (Arnon and Johnson, 1942). The primary target of low pH toxicity might be related to the disturbance of the stability in the pectic polysaccharide network (Koyama et al., 2001). In Arabidopsis, AtSTOP1-regulated AtPGIPs (Polygalacturonase inhibitory proteins) inhibit pectin depolymerization in the root cell wall under acidic conditions, maintain the stability of the pectic polysaccharide network, and have a potential role in low pH tolerance (Spadoni et al., 2006).
The balance of cellular pH is influenced by proton transport across the membrane, H+-coupled ion transport, the production or degradation of organic acids, and the uptake and assimilation of nitrogen (Felle, 2001; Britto and Kronzucker, 2005; Reguera et al., 2015; Feng et al., 2020). Activation of several transporters by AtSTOP1 (Figure 2) has been reported to be critical for low pH tolerance in Arabidopsis. AtSTOP1 modulates the transcription of AtHAK5 (High-affinity K+ transporter 5) (Sawaki et al., 2009; Nakano et al., 2020), AtSULTR3;5 (Sulfate transporter 3;5) (Sawaki et al., 2009), H+-coupled high-affinity NO3- symporter gene AtNRT1.1 (Nitrate transporter 1.1) (Fang et al., 2016; Ye et al., 2021) and AtCIPK23 (CBL-interacting protein kinase 23) (Sawaki et al., 2009). AtCIPK23 additionally regulates the activity of AtHAK5 (Ragel et al., 2015; Wang et al., 2021), AtAKT1 (Arabidopsis K+ transporter 1) (Li et al., 2006; Xu et al., 2006), AtNRT1.1 (Liu and Tsay, 2003; Leran et al., 2015), and AtAMTs (Ammonium transporters) (Straub et al., 2017; Wang et al., 2021) through phosphorylation to influence ion uptake, overcome rhizosphere acidification and establish a favorable cellular pH. Besides, AtSTOP1 promotes the expression of AtTDT (tonoplast dicarboxylate transporter) to increase the concentration of dicarboxylate and, hence, enhance the capacity to produce OH- to regulate the pH homeostasis in the cytosol (Hurth et al., 2005).
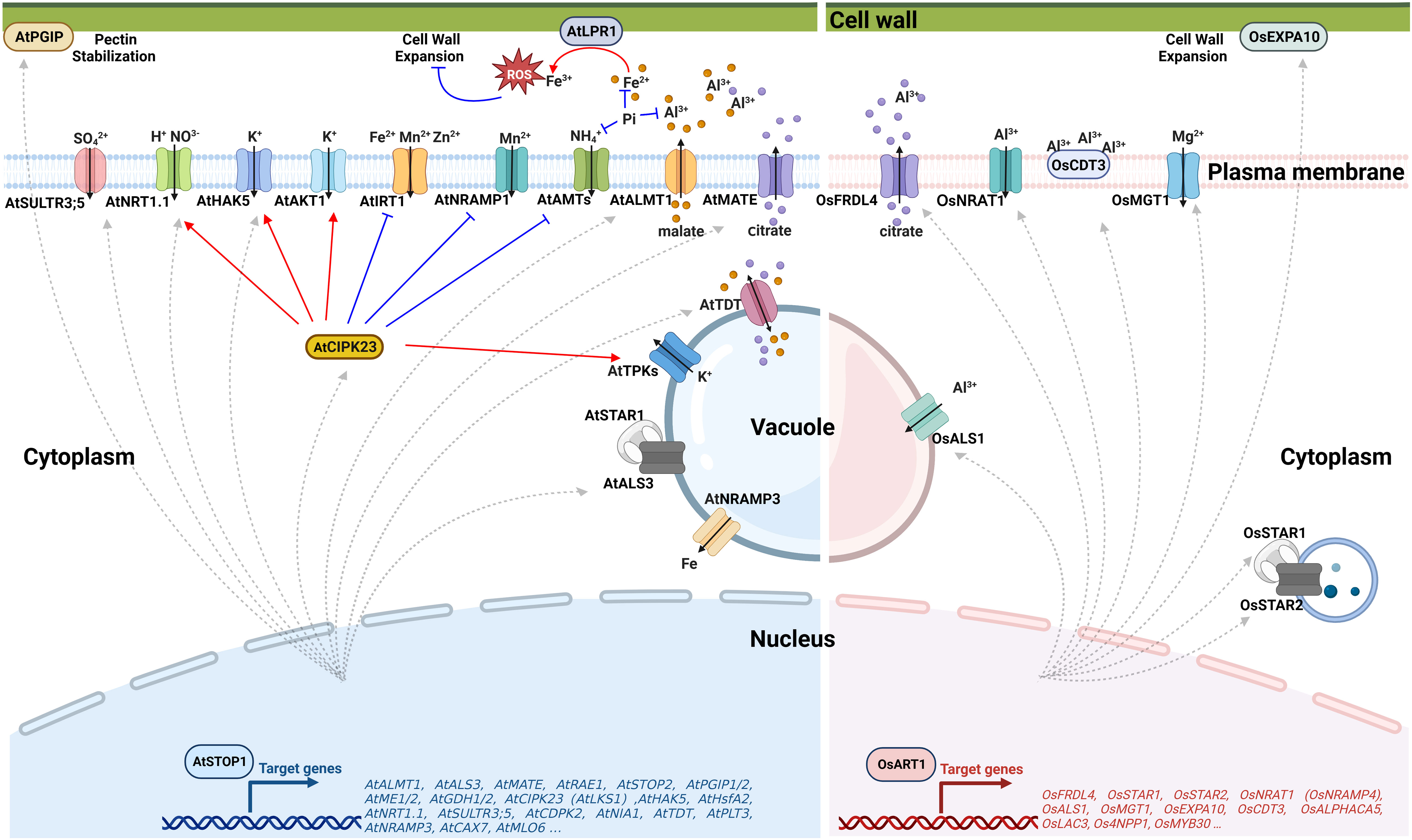
Figure 2 Schematic representation of STOP1/ART1 regulation of Al resistance and nutrient acquisition. Left panel: Under Al stress, AtSTOP1 regulates AtPGIPs to maintain cell walls stability. AtSTOP1 also up-regulates the organic acid transporter coding genes AtALMT1 and AtMATE to increase the secretion of malate and citrate, respectively, which chelate Al in the rhizosphere. For cellular nutrient management, AtSTOP1 enhances Pi bioavailability by regulating malate secretion and Fe-dependent remodeling of root system architecture. AtSTOP1 up-regulates the transcription of AtHAK5, AtNRT1.1, and AtSULTR3;5 to facilitate the uptake of K+, NO3-, and SO42-, respectively. In addition, AtSTOP1 regulates the activity of AtHAK5, AtNRT1.1, AtAKT1, AtIRT1, AtNRAMP1, AtTPKs, and AtAMTs through AtCIPK23. Right panel: In rice, OsART1 transcriptionally activates OsFRDL4 to exude citrate, positively regulates OsSTAR1/2 to maintain cell wall stability, and enhances the Al-induced expression of OsNRAT1 and OsALS1 to sequester Al into vacuoles for intracellular detoxification. OsART1 also promotes the expression of the cell wall loosening protein OsEXPA10 and the cysteine-rich peptide OsCDT3 to cope with Al stress. In addition, OsART1 confers Mg uptake through positive regulation of OsMGT1 transcription to alleviate the Al toxicity. ROS, Reactive oxygen species; STOP1, Sensitive to proton rhizotoxicity 1; ALMT1, Aluminum-activated malate transporter 1; MATE, Multidrug and toxic compound extrusion; TDT, Tonoplast dicarboxylate transporter; PGIPs, Polygalacturonase inhibitory proteins; NRAMP1/3, Natural resistance-associated macrophage protein 1/3; CIPK23/LKS1, CBL-interacting protein kinase 23/Low potassium sensitivity 1; SULTR3;5, Sulfate transporter 3;5; HAK5, High-affinity K+ transporter 5; NRT1.1, Nitrate transporter 1.1; AKT1, Arabidopsis K+ transporter 1; IRT1, Iron-regulated transporter 1; TPKs, Two-pore K+ channels; AMTs, Ammonium transporters; ART1, Al resistance transcription factor 1; FRDL4, Ferric reductase defective-like 4; STAR1/2, Sensitive to Al rhizotoxicity 1/2; EXPA10, Expansin-A10; NRAT1, NRAMP Al transporter 1; ALS1, Al-sensitive 1; MGT1, Magnesium transporter 1. Figure created using BioRender (https://biorender.com/).
Several enzymes involved in metabolic processes that generate or consume protons are also associated with low pH tolerance (Sawaki et al., 2009). Genes encoding malic enzymes (AtME1 and AtME2) that supply pyruvate in the biochemical pH-stat pathway and enzymes (AtGDH1 and AtGAD1) that reduce H+ by accumulating GABA in the GABA shunt pathway are transcriptionally regulated by AtSTOP1 (Sakano, 1998; Magneschi and Perata, 2009; Bown and Shelp, 2016). Furthermore, AtSTOP1 transcriptionally regulates its minor isoform AtSTOP2, which co-regulates a subset of AtSTOP1-regulated genes that confer low pH tolerance (Kobayashi et al., 2014).
Although low pH toxicity is the most direct abiotic stress in acid soils, limited studies are still insufficient to fully elucidate the molecular mechanisms by which plants, especially crops, respond to low pH. Further progress is needed to increase our understanding of the diversity and regulating mechanism of low pH resistance genes.
STOP1 and STOP1-like proteins mediated Al resistance
Al toxicity is one of the most critical factors limiting crop yield in acid soils (Kochian et al., 2015). In acid soils, Al3+ ions dissolved from clay minerals enter root cells within 30 minutes and rapidly inhibit root growth within an hour (Lazof et al., 1994; Delhaize and Ryan, 1995). Al-induced exudation of organic acid anions from the roots is the first barrier for plants to cope with Al toxicity. These organic acid anions chelate Al3+ to form non-toxic compounds, thereby inhibiting Al entry into the roots (Kochian et al., 2015). In Arabidopsis, AtSTOP1 transcriptionally regulates genes encoding organic acid transporters (Figure 2), including AtALMT1 (Aluminum-activated malate transporter 1) and AtMATE (Multidrug and toxic compound extrusion), to increase the secretion of malate and citrate, respectively (Hoekenga et al., 2006; Liu et al., 2009; Sawaki et al., 2009). These organic acids sequestrate toxic Al3+ in the rhizosphere, forming a non-toxic complex to reduce plant damage. Similarly, OsART1 activates the transcription of MATE family gene OsFRDL4 (Ferric reductase defective-like 4) to exude citrate to cope with Al stress in rice (Yokosho et al., 2011). Excess Al3+ can still break the barrier of organic acids. The root cell wall is the next site where Al directly contacts and interacts with the plant. The negatively charged groups of pectin and hemicellulose have a high affinity for Al3+ and can alleviate Al toxicity by reducing its entry into the root cell (Yang et al., 2008). However, the replacement of Ca2+ by Al3+ results in a thick and rigid cell wall. Too much Al bound to the cell wall also inhibits root growth and development (Tabuchi and Matsumoto, 2001).
Different strategies have evolved in plants to modify the cell wall in response to Al toxicity. In Arabidopsis, AtSTOP1 up-regulates AtPGIPs to strengthen the pectic polysaccharide network in the cell wall under Al stress (Agrahari et al., 2021). In sorghum, SbSTOP1 activates the transcription of a β-1,3-glucanase gene SbGLU1 to degrade callose and avoid cell wall rigidity (Gao et al., 2019). In rice, OsART1 up-regulates the expression of ABC (ATP binding cassette) transporters OsSTAR1 (Sensitive to Al rhizotoxicity 1) and OsSTAR2 to transport UDP-glucose for cell wall modification, which is required for Al detoxification (Huang et al., 2009). Direct inhibition of OsMYB30 transcription by OsART1 reduces 4-coumaric acid accumulation, preventing excess Al3+ from binding to the cell wall (Gao et al., 2022). OsART1 also promotes the expression of the cell wall loosening protein OsEXPA10 (Expansin-A10) under Al stress, which regulates cell elongation but contributes less to Al tolerance (Che et al., 2016). In addition to the cell wall, Al3+ can also be bound to the plasma membrane-anchored cysteine-rich peptide OsCDT3 (Cadmium tolerance 3). OsCDT3 is downstream of OsART1, and it binds Al3+ directly to prevent Al3+ from entering the root cell, thus alleviating Al toxicity (Xia et al., 2013).
Despite almost 90% of the soluble Al3+ in roots being tightly bound to the cell wall (Ma, 2007), a small proportion of toxic Al3+ entering the cell can still inhibit root growth (Lazof et al., 1996; Blancaflor et al., 1998; Yamamoto et al., 2002). Once Al enters the root cell, sequestration and storage of Al in the vacuole is an essential mechanism for detoxification (Figure 2). OsART1 positively regulates the Al-induced expression of OsNRAT1 (NRAMP Al transporter 1) and OsALS1 (Al-sensitive 1) (Yamaji et al., 2009). OsNRAT1 is localized to the plasma membrane, which takes up extracellular Al3+ to alleviate cell wall damage (Xia et al., 2010). OsALS1 is a half-size ABC transporter that sequesters the cytoplasmic Al into vacuoles for safe storage (Huang et al., 2012). OsNRAT1 may function cooperatively with OsALS1 to be involved in the intracellular detoxification of Al. In addition, OsART1-regulated OsMGT1 (Magnesium transporter 1) alleviates Al toxicity by increasing intracellular Mg concentration (Chen et al., 2012).
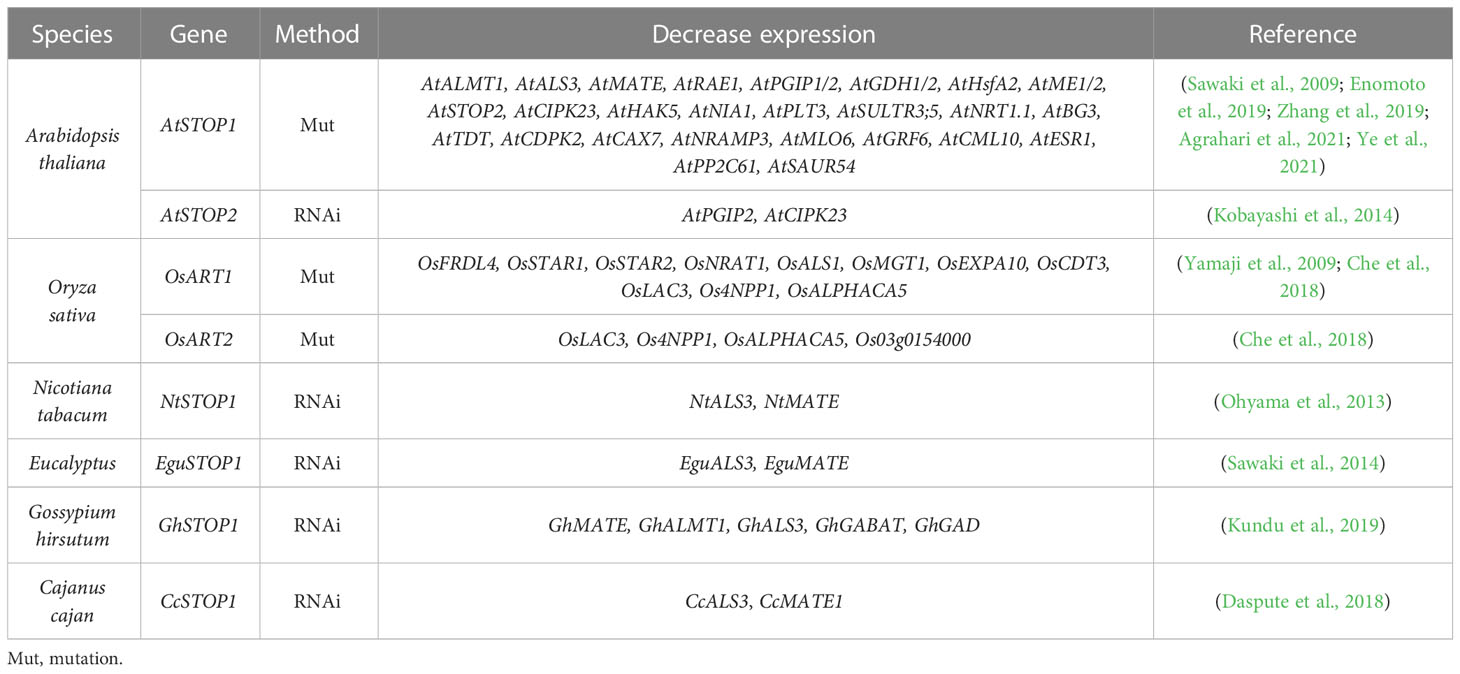
STOP1-like proteins have also been characterized in many plant species (Table 1) and show some functional differentiation in response to Al and low pH stress (Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015; Huang et al., 2018; Wu et al., 2018). As orthologous genes with similar functions, the transcription of OsSTAR1 and OsALS1 in rice is regulated by OsART1, whereas the expression of AtSTAR1 and AtALS1 in Arabidopsis is unaffected by AtSTOP1 (Larsen et al., 2007; Huang et al., 2010). This suggests that different living environments may affect the function of STOP1 and STOP1-like proteins by evolving their preferences for downstream genes. For example, in dryland crops, Al-mediated root exudation of organic acids plays a more important role in Al resistance, whereas rice lives in an aqueous environment that easily disrupts the organic acid barrier (Famoso et al., 2010). Thus, STOP1-like proteins in dryland crops may tend to activate Al resistance genes associated with organic acids secretion, while rice may rely more on cell wall modification and internal detoxification. In addition, overexpression of AtSTOP2 partially rescued Al resistance and low pH tolerance of Atstop1 by restoring the expression of AtSTOP1-regulated genes, including AtPGIP1/2, AtALS3, and AtMATE, but not AtALMT1 (Kobayashi et al., 2014). Whereas in rice, mutation of OsART2 did not affect the expression of previously identified OsART1-regulated genes (Che et al., 2018). This difference may be due to different experimental approaches, as the knockdown of AtSTOP2 reduced the expression of AtPGIP2 and AtCIPK23 but did not affect the transcription of AtPGIP1, AtALS3 and AtMATE (Kobayashi et al., 2014). This difference may also be related to rice having six STOP1-like genes that may compensate for each other, with additional copies increasing functional redundancy. In fact, according to RT-qPCR results (Che et al., 2018), some of the potential downstream Al resistance genes identified in Osart2 are also regulated by OsART1.
Differences in downstream gene sets and regulatory preferences of STOP1-like proteins have been reported in different plants (Table 2). In the Atstop1 complementation assay, STOP1-like proteins showed slight differences in the activation of AtSTOP1 downstream genes (Table 3). This suggests there may be a functional differentiation of STOP1-like proteins in different species, or STOP1 partners in Arabidopsis not cooperating well with STOP1-like proteins. Therefore, it is necessary to carry out in vivo functional studies in these plants. Despite small differences in the activation of downstream genes by STOP1 and STOP1-like proteins, they remain central factors regulating Al resistance. Further dissection and engineering of STOP1 and STOP1-like proteins have great potential in improving Al resistance in acid soils.
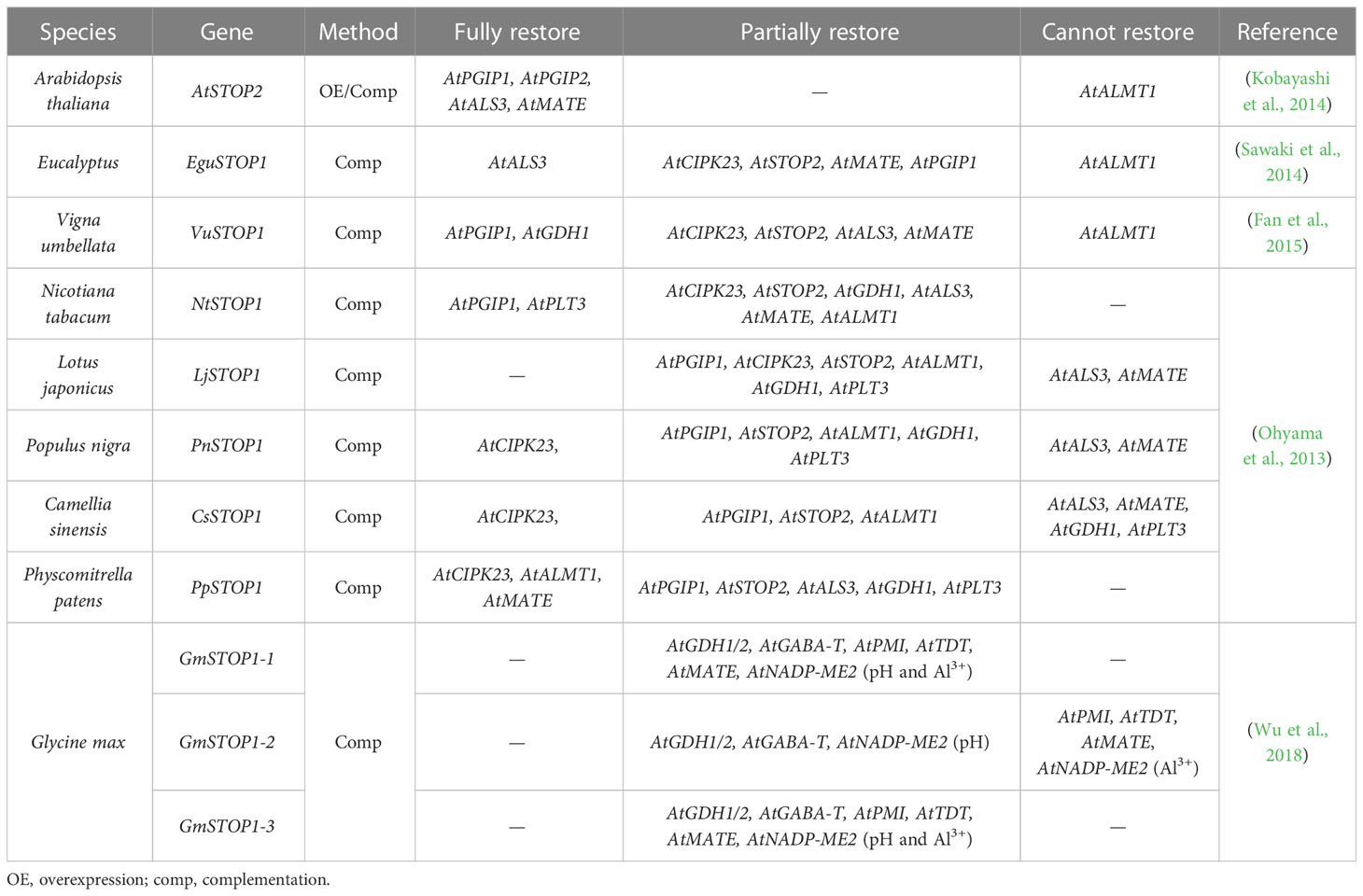
Table 3 Expression of STOP1 downstream genes in the Atstop1 mutant complemented with STOP1-like proteins.
STOP1 and STOP1-like proteins mediated nutrient homeostasis
Nutrient sensing and homeostasis are crucial for plants to adapt to the environment. Soil acidification begins with the loss of salt-based ions, so the acid soils are typically deficient in salt-based ions such as K+ and Mg2+ (von Uexküll and Mutert, 1995). AtSTOP1 contributes to K uptake by mediating the transcription of AtHAK5, which encodes a high-affinity K+ transporter (Sawaki et al., 2009). In addition, the AtCIPK23, downstream of AtSTOP1, together with AtCBL1/9 (Calcineurin B-like), enhances K+ uptake by phosphorylating the K+ transporters AtHAK5 and AtAKT1 (Li et al., 2006; Xu et al., 2006; Ragel et al., 2015; Wang et al., 2021). As a partner of AtCBL2/3, AtCIPK23 also regulates K homeostasis redundantly with AtCIPK3/9/26 through activating tonoplast AtTPK (Two-pore K+) channels that promote K+ remobilization, which plays a vital role in plant adaptation to K deficiency (Tang et al., 2020). Besides, the AtCBL2/3-CIPK3/9/23/26 module regulates vacuolar Mg storage, thereby influencing Mg homeostasis (Tang et al., 2015). In rice, OsART1 transcriptionally regulates the plasma membrane-localized Mg2+ transporter OsMGT1 to promote Mg uptake, especially under Al treatment, thereby increasing cellular Mg content and, on the other hand alleviating Al toxicity (Chen et al., 2012). Apart from the deficiency of salt-based ions, acid soils usually contain excessive levels of metal nutrients such as Fe2+, Mn2+, and Zn2+, which in excess cause phytotoxicity (Kochian et al., 2004). AtCIPK23 phosphorylates the broad-spectrum high-affinity metal transceptor AtIRT1 (Iron-regulated transporter 1) when excess non-iron metals are present and bound to the histidine-rich motif of AtIRT1, which subsequently recruits the E3 ubiquitin ligase AtIDF1, targeting AtIRT1 to the vacuole for its degradation, thus prevent non-iron metal toxicity (Dubeaux et al., 2018). Together with AtCBL1/9, AtCIPK23 interacts with and phosphorylates the high-affinity Mn2+ transporter NRAMP1 (Natural resistance-associated macrophage protein 1), promotes its clathrin-mediated endocytosis, reduces its plasma membrane distribution and improves plant tolerance to Mn toxicity (Zhang et al., 2023). In conclusion, AtSTOP1/OsART1 affects the uptake of metals, including K+ and Mg2+, by controlling the transcription of their transporters directly, and regulates the absorption of K+, Mg2+, Fe2+, Mn2+, and Zn2+ via CIPK23-mediated phosphorylation.
In addition to affecting the homeostasis of salt-based ions and toxic metals, AtSTOP1 regulates the uptake of non-metallic elements. Under Pi deficiency conditions, AtSTOP1 accumulates in the nucleus and enhances the expression of AtALMT1 and AtMATE1 to secrete malate and citrate, respectively. These organic acids desorb Pi from mineral surfaces and dissolve Pi from complexes of Al and Fe oxides, increasing bioavailable Pi concentrations in soil (Kochian et al., 2004). Furthermore, AtSTOP1-promoted malate secretion triggers ROS (reactive oxygen species) production and callose deposition in the presence of the ferroxidases AtLPR1 and AtLPR2 (Low phosphate root 1 and 2), thereby regulating root system architecture, repressing primary root elongation and stimulating lateral root development to efficiently utilize the low mobility Pi in the topsoil (Balzergue et al., 2017; Mora-Macias et al., 2017). During these processes, Fe and Al promote AtSTOP1 accumulation in the nucleus, possibly by inhibiting AtSTOP1 degradation (Godon et al., 2019). Low Pi-induced AtSTOP1 transcriptionally activates AtALS3, while AtALS3 interacts with AtSTAR1 to inhibit the nuclear accumulation of AtSTOP1 to prevent AtSTOP1 overactivation (Wang et al., 2019). Low Pi also promotes the expression of AtAMT1;1 and AtAMT1;2, inducing rhizosphere acidification through NH4+ uptake, which in turn promotes nuclear accumulation of AtSTOP1 to increase soil Pi availability (Tian et al., 2021). In response to the imbalance in nitrogen availability in acidic soils, AtSTOP1-induced AtCIPK23 inhibits the transport activity of AtAMTs through phosphorylation, which alleviates rhizosphere acidification and avoids NH4+ toxicity (Straub et al., 2017; Wang et al., 2021). AtSTOP1 also controls the NO3- uptake by activating AtNRT1.1 transcription directly, and AtCIPK23 activates AtNRT1.1 through phosphorylation (Ye et al., 2021). Besides, AtSTOP1 positively regulates the expression of AtSULTR3;5 in the root vasculature, and AtSULTR3;5 is localized to the plasma membrane for sulfate (SO42-) uptake and affects the transport of SO42- from root to shoot (Kataoka et al., 2004; Sawaki et al., 2009). Collectively, STOP1 promotes Pi uptake by regulating root system architecture and regulates the uptake of SO42-, NH4+, and NO3- by regulating their transporters. It is interesting to note that STOP1 functions as a center for nutrient management under deprivation conditions, controlling nutrient homeostasis other than stress tolerance.
STOP1 and STOP1-like proteins regulate other stress responses
Approximately 60% of the acid soil occurs in rainfed areas of the tropics or subtropics (Kochian et al., 2004), where plants are sometimes submerged in low (hypoxia) or no oxygen (anoxia) conditions, which are an important abiotic constraint on lowland yields (Zeigler and Puckridge, 1995). In Arabidopsis, AtSTOP1 transcription is induced by low oxygen. Subsequently, it contributes to low-oxygen tolerance by activating the transcription of AtGDH1/2 and AtHsfA2 (Heat shock factor A2), and a conserved mechanism that NtSTOP1 involved in hypoxia tolerance has also been identified in tobacco (Enomoto et al., 2019). Furthermore, AtSTOP1 enhances salt tolerance by transcriptionally regulating several salt tolerance genes, including AtCIPK23, which negatively regulates drought resistance by maintaining K+ transport to maintain stomatal opening (Sadhukhan et al., 2019). AtCIPK23 also directly phosphorylates and activates the S-type anion channels AtSLAC1 (Slow anion channel associated 1) and AtSLAH3 (SLAC1 homolog 3) to regulate the stomatal aperture (Maierhofer et al., 2014). Additionally, AtSTOP1-mediated pH tolerance is involved in the root response of plant-fungal communication between Arabidopsis and Trichoderma (Pelagio-Flores et al., 2017).
The STOP1 homolog in pineapple (Ananas comosus) shows a diurnal oscillation expression coinciding with the oscillation of malate concentration in leaves and may be the key circadian oscillator regulating CAM metabolism (Sharma et al., 2017). Mutation of AtSTOP1 influences the transcript levels of many genes, including AtPLT3 (Probable polyol transporter 3), AtCDPK2 (Calcium-dependent protein kinase 2), AtCAX7 (Calcium exchanger 7), AtNRAMP3, AtNIA1 (Nitrate reductase 1), AtMLO6 (Mildew resistance locus O 6), AtGRF6 (Growth regulating factor 6), AtCML10 (Calmodulin like 10), AtESR1 (Enhancer of shoot regeneration 1), AtPP2C61 (Protein phosphatase 2C 61) and AtSAUR54 (Small auxin upregulated RNA 54) (Sawaki et al., 2009; Sadhukhan et al., 2019). DAP-seq data also showed that AtSTOP1 binds to the promoters of AtPLT3, AtCDPK2, AtCAX7, AtNRAMP3, AtNIA1 and AtMLO6 (O'Malley et al., 2016). Further studies and more evidence are needed to clarify whether STOP1 participates in other biological processes through these downstream genes.
Regulation of STOP1 and STOP1-like proteins
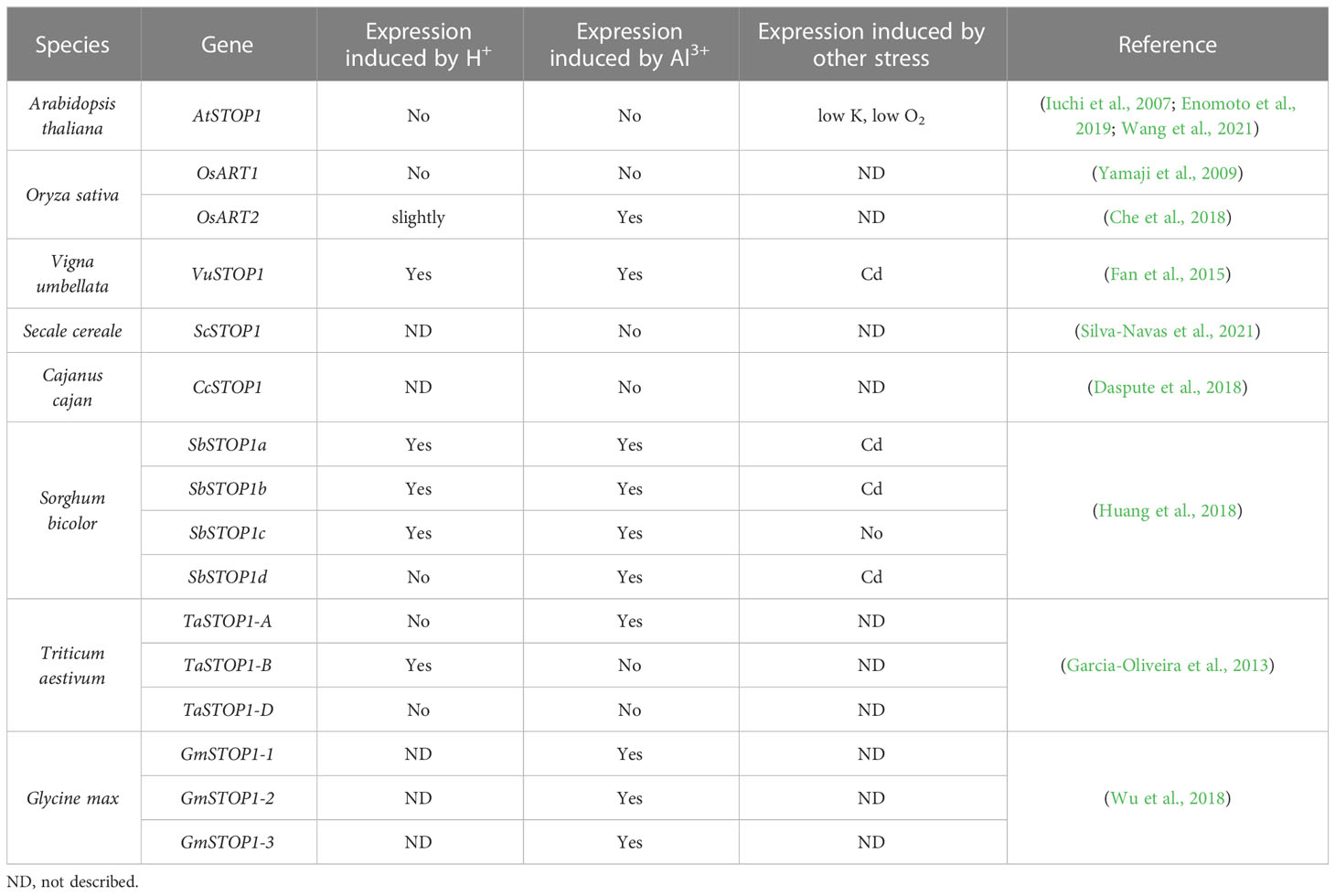
As a master transcription factor, the activity and protein levels of STOP1 and STOP1-like proteins are regulated by complex mechanisms at multiple levels, including transcriptional regulation, post-transcriptional regulation, and post-translational modifications. Transcript levels of AtSTOP1 and OsART1 are not affected by low pH or Al stress, but low K+ and hypoxic stress induce AtSTOP1 transcription in Arabidopsis (Enomoto et al., 2019; Wang et al., 2021). In some plant species, such as sorghum and rice bean, there exists transcriptional regulation of STOP1-like genes in response to low pH, Al stress, or cadmium (Cd) stress (Table 4). In addition, the mRNA level of the STOP1 homolog in pineapple showed a more than 3- fold diurnal oscillation within a day, which coincided with the oscillation of malate concentration in leaves (Sharma et al., 2017). Although the transcription of AtSTOP1 in Arabidopsis is not affected by low pH or Al stress, AtSTOP1 is required for the expression of downstream genes such as AtALMT1 under Al stress (Iuchi et al., 2007). This suggests the existence of some post-transcriptional regulatory mechanism that promotes the expression of downstream genes by activating AtSTOP1 under Al treatment. Recent studies revealed the mechanisms of RAE (Regulation of AtALMT1 expression) genes in regulating STOP1 at the post-transcriptional level (Figure 3). The nuclear membrane localized THO/TREX complex processes AtSTOP1 mRNA with two key members, AtRAE2/AtTEX1 and AtRAE3/AtHPR1. Mutation of AtTEX1 or AtHPR1 decreases the protein level of AtSTOP1 in roots. AtHPR1 affects AtSTOP1 mRNA export from the nucleus, while AtTEX1 does not (Guo et al., 2020; Zhu et al., 2021).
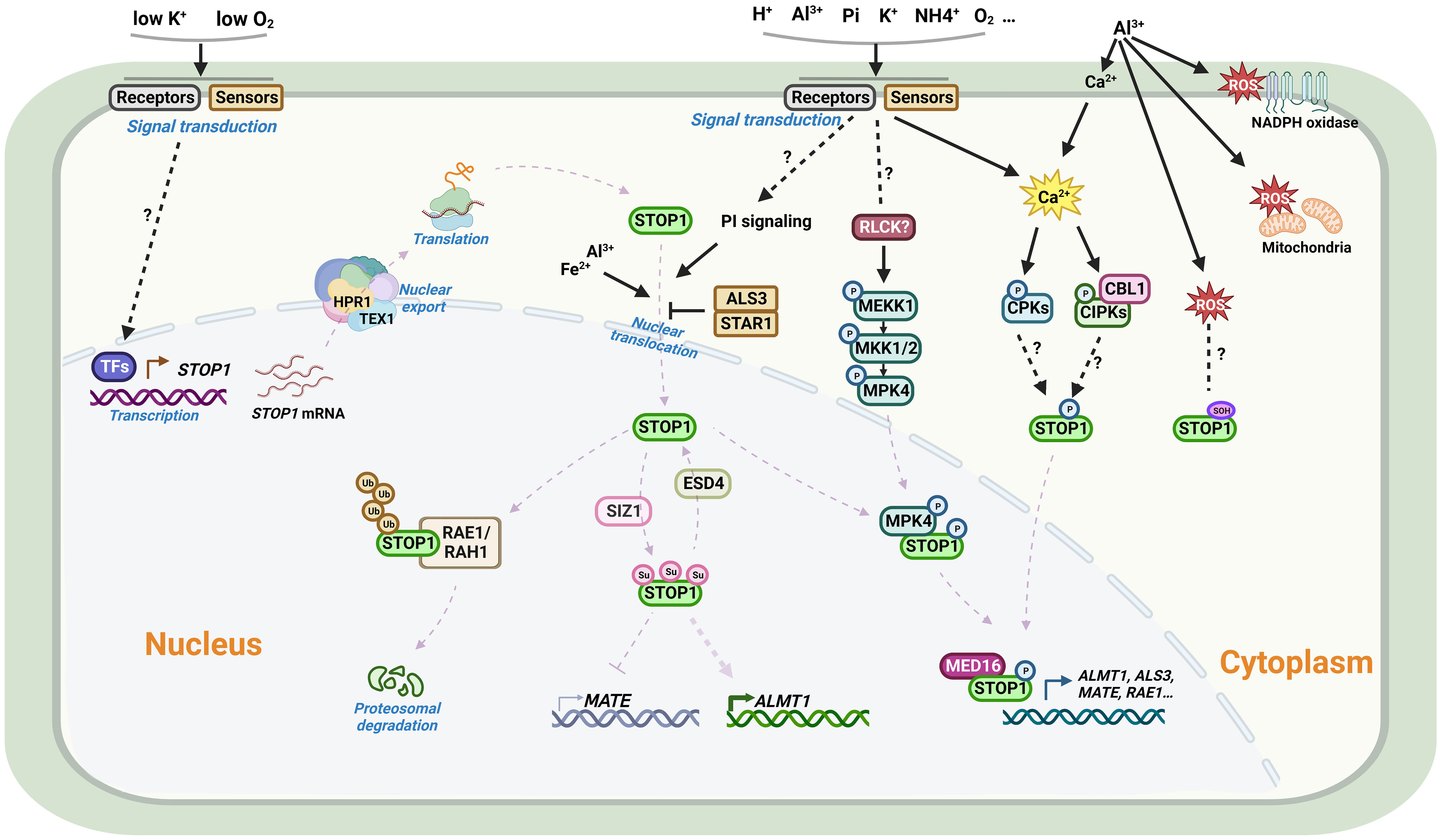
Figure 3 Schematic representation of the regulation of STOP1 in Arabidopsis. From upper left: Under most conditions, STOP1 is constitutively expressed in plants. Plants can sense hypoxic stress and low K+ signals to activate STOP1 transcription. The mRNA of STOP1 is processed and transported by the THO/TREX complex on the nuclear envelope. RAE3/HPR1 and RAE2/TEX1 are two key members of the complex, in which HPR1, but not TEX1, affects STOP1 mRNA export from the nucleus. For the STOP1 protein, in addition to the PI signaling pathway, Al and Fe promote STOP1 nuclear accumulation, whereas ALS3/STAR1 inhibits nuclear STOP1 accumulation. The MEKK1-MKK1/2-MPK4 cascade plays an important role in Al signaling. Al activates the kinase activity of MPK4, which interacts with and phosphorylates STOP1, thereby stabilizing STOP1 by reducing its interaction with RAE1. Al3+ releases Ca2+ from the cell wall via cation exchange, Ca2+ signaling followed by signal transduction may promote STOP1 phosphorylation via CBL/CIPK or CPKs signaling pathways, and Al-induced ROS accumulation may modulate the oxidation of cysteine residues on STOP1. In the nucleus, the F-box proteins RAE1 and RAH1 are components of the SCF-type E3 ligase complex that ubiquitinates STOP1 and facilitates its 26S proteasomal degradation. The SUMO E3 ligase SIZ1 and the SUMO protease RAE5/ESD4 are involved in the SUMOylation and de-SUMOylation modifications of STOP1, regulating its stability and altering its association with different target gene promoters. STOP1 interacts with MED16 to co-activate the transcription of downstream genes. Al, aluminum; Fe, iron; K, Potassium; Pi, Phosphate; Ca, Calcium; H+, Proton; NH4+, Ammonium; O2, Oxygen; ROS, Reactive oxygen species; TF, Transcription factor; STOP1, Sensitive to proton rhizotoxicity 1; HPR1, Hyper-Recombination 1; TEX1, Transcription-Export 1; CBL, Calcineurin B-like; CIPK, CBL-interacting protein kinase; CPK, Calcium-dependent protein kinase; MEKK1, MAPK/ERK kinase kinase 1; MKK1/2, MAP kinase kinase 1/2; MPK4, MAP kinase 4; RLCKs, Receptor-like cytoplasmic kinases; PI, phosphatidylinositol; RAE1, Regulation of AtALMT1 expression; RAH1, RAE1 homolog 1; SUMO, Small ubiquitin-related modifier; SIZ1, SAP and MIZ1 domain-containing ligase 1; ESD4, Early in short days 4; ALS3, Al-sensitive 3; STAR1, Sensitive to Al rhizotoxicity 1; MED16, Mediator 16. Figure created using BioRender (https://biorender.com/).
The protein level of AtSTOP1 increases after Al treatment, while the E3 ubiquitin ligase AtRAE1 interacts with and ubiquitinates AtSTOP1, promoting its 26S proteasomal degradation (Zhang et al., 2019). As a paralog of AtRAE1, AtRAH1 (RAE1 homolog 1) plays an unequally redundant role in regulating AtSTOP1 stability (Fang et al., 2021b). AtSTOP1 in turn promotes the transcription of AtRAE1/AtRAH1, forming a negative feedback loop to prevent excessive AtSTOP1 accumulation and over-activation of Al resistance (Zhang et al., 2019; Fang et al., 2021b). Accumulation of AtSTOP1 under Al treatment may result from protein modifications that prevent AtSTOP1 from interacting with AtRAE1/AtRAH1 or inhibit its ubiquitination. Given that inhibitors of PI (phosphatidylinositol) signaling blocked nuclear localization of AtSTOP1 under Al stress, other factors may contribute to the accumulation of AtSTOP1. Under low Pi conditions, Fe and Al-promoted AtSTOP1 accumulation is higher in the als3 mutants, although the mechanism by which AtALS3/AtSTAR1 inhibits AtSTOP1 accumulation remains unclear (Godon et al., 2019; Wang et al., 2019). In tomato, SlRAE1 is also involved in the ubiquitination and degradation of SlSTOP1, and SlSZP1 (STOP1-interacting zinc finger protein 1) interacts with SlSTOP1 to protect it from degradation by SlRAE1 (Zhang et al., 2022).
Reversible protein phosphorylation affects AtSTOP1-regulated AtALMT1 transcription and malate secretion (Kobayashi et al., 2007). In rye, a conserved phosphorylatable serine site in ScSTOP1 is vital for activating ScALMT1 transcription (Silva-Navas et al., 2021). Mutation of AtCBL1 results in reduced expression of AtALMT1, demonstrating that Ca2+ signaling may be involved in AtSTOP1 phosphorylation via CBL–CIPK networks (Ligaba-Osena et al., 2017). In addition, the AtMEKK1-AtMKK1/2-AtMPK4 cascade plays a role in AtSTOP1 phosphorylation. Al exposure causes AtMPK4 to phosphorylate AtSTOP1, reducing its interaction with AtRAE1 and thus contributing to the stabilization and accumulation of AtSTOP1 (Zhou et al., 2023). Furthermore, SUMOylation modifications stabilize AtSTOP1, and blocking the SUMOylation of AtSTOP1 reduces AtSTOP1 accumulation. Mutations in the SUMO protease AtRAE5/AtESD4 affect the de-SUMOylation of AtSTOP1 and alter its association with the promoters of different target genes (Fang et al., 2020). Consistently, mutations in the SUMO E3 ligase AtSIZ1 reduce SUMOylation of AtSTOP1 and decrease the protein levels of AtSTOP1 (Fang et al., 2021a; Xu et al., 2021).
In the nucleus, AtSTOP1 interacts with AtMED16 (Mediator 16), a component of the transcriptional co-activation complex, and co-regulates the expression of several downstream genes (Raya-Gonzalez et al., 2021). While AtBZR1 (Brassinazole resistant 1) competitively inhibits the activation of AtALMT1 expression by AtSTOP1 (Liu et al., 2022). In sorghum, SbSTOP1d is self-interacting and interacts with SbSTOP1b, suggesting that SbSTOP1s may function as homodimers or heterodimers (Huang et al., 2018). The ability that STOP1 to regulate the expression of different genes under different stress conditions suggests that these environmental signals may activate or modify STOP1 in various manners, or there are different kinds of proteins that interact with STOP1 and regulate its promoter binding preferences. Further studies are required to elucidate how these interacting proteins affect the function of STOP1 and STOP1-like proteins, and whether STOP1 and STOP1-like proteins undergo different modifications in response to different environments.
Concluding remarks and future perspectives
In addition to low pH and proton stress, acidic soils often have many other coexisting factors that impair crop yields. With significant advances in our understanding of the acid soil syndrome, researchers are becoming increasingly aware that although tolerance to low pH is a prerequisite for plant growth in acid soils, STOP1 tolerance is not limited to proton tolerance and Al resistance, but also includes enhanced bioavailability of Pi and other nutrients, as well as tolerance to other limiting factors. As a core transcription factor to cope with acid soil syndrome, STOP1 is a node for the cross-talk of multiple environmental signals. The phenomenon that STOP1 determines many different traits is a classic case of pleiotropy. An evolutionarily effective strategy is using a limited number of genes to perform more functions through combinations of transcriptional regulation, mRNA processing, protein modification, and protein-protein interaction. Because STOP1 has a role in resistance or tolerance to many different stresses, applying STOP1 or STOP1-like proteins in agricultural production is expected to improve crop resistance to acid soil syndrome.
In this review, we summarized the biological functions of STOP1 and STOP1-like proteins, especially in the context of the various constraints of acid soil syndrome. We hope this will provide researchers with insights into exploiting STOP1 and STOP1-like proteins, related signaling components and regulatory networks through molecular breeding and biotechnology to improve crop tolerance to acid soil syndrome, especially those plants that are not well adapted to acid soils, such as alfalfa and soybean. Genome editing is a powerful tool for improving crop varieties. By knocking in cis-elements or high-throughput editing at the STOP1 promoter region (Shen et al., 2023; Tian et al., 2023), STOP1 expression can be environmentally induced or constitutively enhanced. The STOP1 protein also can be stabilized by point substitution through base editing of phosphorylation sites (Tian et al., 2022). In addition, the strength of the STOP1 effect can be fine-tuned by modulating potential regulators of STOP1.
Although many regulators of STOP1 under Al stress have been identified, STOP1 and STOP1-like proteins are also regulated by multiple stress signals on acidic soils. Future studies of the regulatory mechanisms of STOP1 may identify more upstream components in the signaling pathway, determine how different receptors sense different upstream signals, and investigate how downstream genes are precisely regulated by STOP1 or STOP1-like proteins. It appears that there are ‘too many’ genes regulated by STOP1 or STOP1-like proteins, and it will also be possible to classify the main process in which STOP1 is involved and to target specific downstream genes to improve a particular trait. Although the function of STOP1 and STOP1-like proteins in Al resistance has been extensively studied in many plant species, it is unclear whether STOP1 and STOP1-like proteins play conserved roles in other biological processes. Further studies are needed to investigate whether other processes regulated by STOP1 are conserved when plants adapt to different living environments. In addition, STOP1 regulates many stress-responsive genes but has a limited effect on certain downstream genes. For some specific genes, transcriptional regulation is not the dominant mode of regulation. Overall, STOP1 contributes more to acid soil tolerance. The other functions of STOP1 are more likely to play a supporting role in dealing with acid soil syndrome. Therefore, we believe that genetic engineering of STOP1 and its homologs is preferred for crops to counteract acidic soils.
Author contributions
XL conceived the ideas and wrote the draft. YT reviewed and edited the manuscript and the figures. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the China Postdoctoral Science Foundation (No. BX20220098 and No. 2022M720973) and the Hainan Yazhou Bay Seed Laboratory (B22C1000P).
Acknowledgments
We thank Dr. Yi-Fang Zhu from the College of Agriculture and Biotechnology (CAB), Zhejiang University, for critical reading of the manuscript and valuable suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agrahari, R. K., Enomoto, T., Ito, H., Nakano, Y., Yanase, E., Watanabe, T., et al. (2021). Expression GWAS of PGIP1 identifies STOP1-dependent and STOP1-independent regulation of PGIP1 in aluminum stress signaling in Arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.774687
Arnon, D. I., Johnson, C. M. (1942). Influence of hydrogen ion concentration on the growth of higher plants under controlled conditions. Plant Physiol. 17 (4), 525–539. doi: 10.1104/pp.17.4.525
Balzergue, C., Dartevelle, T., Godon, C., Laugier, E., Meisrimler, C., Teulon, J. M., et al. (2017). Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 8, 15300. doi: 10.1038/ncomms15300
Blancaflor, E. B., Jones, D. L., Gilroy, S. (1998). Alterations in the cytoskeleton accompany aluminum-induced growth inhibition and morphological changes in primary roots of maize. Plant Physiol. 118 (1), 159–172. doi: 10.1104/pp.118.1.159
Bown, A. W., Shelp, B. J. (2016). Plant GABA: not just a metabolite. Trends Plant Sci. 21 (10), 811–813. doi: 10.1016/j.tplants.2016.08.001
Britto, D. T., Kronzucker, H. J. (2005). Nitrogen acquisition, PEP carboxylase, and cellular pH homeostasis: new views on old paradigms. Plant Cell Environ. 28 (11), 1396–1409. doi: 10.1111/j.1365-3040.2005.01372.x
Che, J., Tsutsui, T., Yokosho, K., Yamaji, N., Ma, J. F. (2018). Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 220 (1), 209–218. doi: 10.1111/nph.15252
Che, J., Yamaji, N., Shen, R. F., Ma, J. F. (2016). An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. 88 (1), 132–142. doi: 10.1111/tpj.13237
Chen, Z. C., Yamaji, N., Motoyama, R., Nagamura, Y., Ma, J. F. (2012). Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 159 (4), 1624–1633. doi: 10.1104/pp.112.199778
Daspute, A. A., Kobayashi, Y., Panda, S. K., Fakrudin, B., Kobayashi, Y., Tokizawa, M., et al. (2018). Characterization of CcSTOP1; a C2H2-type transcription factor regulates Al tolerance gene in pigeonpea. Planta 247 (1), 201–214. doi: 10.1007/s00425-017-2777-6
Delhaize, E., Ryan, P. R. (1995). Aluminum toxicity and tolerance in plants. Plant Physiol. 107 (2), 315–321. doi: 10.1104/pp.107.2.315
Dubeaux, G., Neveu, J., Zelazny, E., Vert, G. (2018). Metal sensing by the transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69 (6), 953–64 e5. doi: 10.1016/j.molcel.2018.02.009
Enomoto, T., Tokizawa, M., Ito, H., Iuchi, S., Kobayashi, M., Yamamoto, Y. Y., et al. (2019). STOP1 regulates the expression of HsfA2 and GDHs that are critical for low-oxygen tolerance in Arabidopsis. J. Exp. Bot. 70 (12), 3297–3311. doi: 10.1093/jxb/erz124
Famoso, A. N., Clark, R. T., Shaff, J. E., Craft, E., McCouch, S. R., Kochian, L. V. (2010). Development of a novel aluminum tolerance phenotyping platform used for comparisons of cereal aluminum tolerance and investigations into rice aluminum tolerance mechanisms. Plant Physiol. 153 (4), 1678–1691. doi: 10.1104/pp.110.156794
Fan, W., Lou, H. Q., Gong, Y. L., Liu, M. Y., Cao, M. J., Liu, Y., et al. (2015). Characterization of an inducible C2H2 -type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 208 (2), 456–468. doi: 10.1111/nph.13456
Fang, X. Z., Tian, W. H., Liu, X. X., Lin, X. Y., Jin, C. W., Zheng, S. J. (2016). Alleviation of proton toxicity by nitrate uptake specifically depends on nitrate transporter 1.1 in Arabidopsis. New Phytol. 211 (1), 149–158. doi: 10.1111/nph.13892
Fang, Q., Zhang, J., Yang, D. L., Huang, C. F. (2021a). The SUMO E3 ligase SIZ1 partially regulates STOP1 SUMOylation and stability in Arabidopsis thaliana. Plant Signal Behav. 16 (5), 1899487. doi: 10.1080/15592324.2021.1899487
Fang, Q., Zhang, J., Zhang, Y., Fan, N., van den Burg, H. A., Huang, C. F. (2020). Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 32 (12), 3921–3938. doi: 10.1105/tpc.20.00687
Fang, Q., Zhou, F., Zhang, Y., Singh, S., Huang, C. F. (2021b). Degradation of STOP1 mediated by the f-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant J. 106 (2), 493–506. doi: 10.1111/tpj.15181
Felle, H. H. (2001). pH: signal and messenger in plant cells. Plant Biol. 3 (6), 577–591. doi: 10.1055/s-2001-19372
Feng, H., Fan, X., Miller, A. J., Xu, G. (2020). Plant nitrogen uptake and assimilation: regulation of cellular pH homeostasis. J. Exp. Bot. 71 (15), 4380–4392. doi: 10.1093/jxb/eraa150
Gao, L. J., Liu, X. P., Gao, K. K., Cui, M. Q., Zhu, H. H., Li, G. X., et al. (2023). ART1 and putrescine contribute to rice aluminum resistance via OsMYB30 in cell wall modification. J. Integr. Plant Biol. 65 (4), 934–949. doi: 10.1111/jipb.13429
Gao, J., Yan, S., Yu, H., Zhan, M., Guan, K., Wang, Y., et al. (2019). Sweet sorghum (Sorghum bicolor L.) SbSTOP1 activates the transcription of a β-1,3-glucanase gene to reduce callose deposition under Al toxicity: a novel pathway for Al tolerance in plants. Biosci. Biotechnol. Biochem. 83 (3), 446–455. doi: 10.1080/09168451.2018.1540290
Garcia-Oliveira, A. L., Benito, C., Prieto, P., de Andrade Menezes, R., Rodrigues-Pousada, C., Guedes-Pinto, H., et al. (2013). Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and proton H+ toxicity in bread wheat (Triticum aestivum L.). BMC Plant Biol. 13, 134. doi: 10.1186/1471-2229-13-134
Godon, C., Mercier, C., Wang, X., David, P., Richaud, P., Nussaume, L., et al. (2019). Under phosphate starvation conditions, Fe and Al trigger accumulation of the transcription factor STOP1 in the nucleus of Arabidopsis root cells. Plant J. 99 (5), 937–949. doi: 10.1111/tpj.14374
Guo, J., Zhang, Y., Gao, H., Li, S., Wang, Z. Y., Huang, C. F. (2020). Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol. 228 (1), 179–193. doi: 10.1111/nph.16658
Hoekenga, O. A., Maron, L. G., Pineros, M. A., Cancado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 (25), 9738–9743. doi: 10.1073/pnas.0602868103
Huang, S., Gao, J., You, J., Liang, Y., Guan, K., Yan, S., et al. (2018). Identification of STOP1-like proteins associated with aluminum tolerance in sweet sorghum (Sorghum bicolor L.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00258
Huang, C. F., Yamaji, N., Chen, Z., Ma, J. F. (2012). A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 69 (5), 857–867. doi: 10.1111/j.1365-313X.2011.04837.x
Huang, C. F., Yamaji, N., Ma, J. F. (2010). Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 153 (4), 1669–1677. doi: 10.1104/pp.110.155028
Huang, C. F., Yamaji, N., Mitani, N., Yano, M., Nagamura, Y., Ma, J. F. (2009). A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21 (2), 655–667. doi: 10.1105/tpc.108.064543
Hurth, M. A., Suh, S. J., Kretzschmar, T., Geis, T., Bregante, M., Gambale, F., et al. (2005). Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 137 (3), 901–910. doi: 10.1104/pp.104.058453
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Kobayashi, Y., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A. 104 (23), 9900–9905. doi: 10.1073/pnas.0700117104
Kataoka, T., Hayashi, N., Yamaya, T., Takahashi, H. (2004). Root-to-shoot transport of sulfate in Arabidopsis. evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 136 (4), 4198–4204. doi: 10.1104/pp.104.045625
Kidd, P. S., Proctor, J. (2001). Why plants grow poorly on very acid soils: are ecologists missing the obvious? J. Exp. Bot. 52 (357), 791–799. doi: 10.1093/jexbot/52.357.791
Kobayashi, Y., Hoekenga, O. A., Itoh, H., Nakashima, M., Saito, S., Shaff, J. E., et al. (2007). Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145 (3), 843–852. doi: 10.1104/pp.107.102335
Kobayashi, Y., Ohyama, Y., Kobayashi, Y., Ito, H., Iuchi, S., Fujita, M., et al. (2014). STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 7 (2), 311–322. doi: 10.1093/mp/sst116
Kochian, L. V., Hoekenga, O. A., Pineros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kochian, L. V., Pineros, M. A., Liu, J., Magalhaes, J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598. doi: 10.1146/annurev-arplant-043014-114822
Koyama, H., Toda, T., Hara, T. (2001). Brief exposure to low-pH stress causes irreversible damage to the growing root in Arabidopsis thaliana: pectin-Ca interaction may play an important role in proton rhizotoxicity. J. Exp. Bot. 52 (355), 361–368. doi: 10.1093/jexbot/52.355.361
Krug, E. C., Frink, C. R. (1983). Acid rain on acid soil: a new perspective. Science 221 (4610), 520–525. doi: 10.1126/science.221.4610.520
Kundu, A., Das, S., Basu, S., Kobayashi, Y., Kobayashi, Y., Koyama, H., et al. (2019). GhSTOP1, a C2H2 type zinc finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. (Stuttg) 21 (1), 35–44. doi: 10.1111/plb.12895
Larsen, P. B., Cancel, J., Rounds, M., Ochoa, V. (2007). Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225 (6), 1447–1458. doi: 10.1007/s00425-006-0452-4
Lazof, D. B., Goldsmith, J. G., Rufty, T. W., Linton, R. W. (1994). Rapid uptake of aluminum into cells of intact soybean root tips (a microanalytical study using secondary ion mass spectrometry). Plant Physiol. 106 (3), 1107–1114. doi: 10.1104/pp.106.3.1107
Lazof, D. B., Goldsmith, J. G., Rufty, T. W., Linton, R. W. (1996). The early entry of Al into cells of intact soybean roots (a comparison of three developmental root regions using secondary ion mass spectrometry imaging). Plant Physiol. 112 (3), 1289–1300. doi: 10.1104/pp.112.3.1289
Leran, S., Edel, K. H., Pervent, M., Hashimoto, K., Corratge-Faillie, C., Offenborn, J. N., et al. (2015). Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal 8 (375), ra43. doi: 10.1126/scisignal.aaa4829
Li, L., Kim, B. G., Cheong, Y. H., Pandey, G. K., Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 (33), 12625–12630. doi: 10.1073/pnas.0605129103
Ligaba-Osena, A., Fei, Z., Liu, J., Xu, Y., Shaff, J., Lee, S. C., et al. (2017). Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis. New Phytol. 214 (2), 830–841. doi: 10.1111/nph.14420
Liu, T., Deng, S., Zhang, C., Yang, X., Shi, L., Xu, F., et al. (2023). Brassinosteroid signaling regulates phosphate starvation-induced malate secretion in plants. J. Integr. Plant Biol. 65 (5), 1099–1112. doi: 10.1111/jipb.13443
Liu, J., Magalhaes, J. V., Shaff, J., Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57 (3), 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Liu, K. H., Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22 (5), 1005–1013. doi: 10.1093/emboj/cdg118
Ma, J. F. (2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol 264, 225–252. doi: 10.1016/S0074-7696(07)64005-4
Magneschi, L., Perata, P. (2009). Rice germination and seedling growth in the absence of oxygen. Ann. Bot. 103 (2), 181–196. doi: 10.1093/aob/mcn121
Maierhofer, T., Diekmann, M., Offenborn, J. N., Lind, C., Bauer, H., Hashimoto, K., et al. (2014). Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal 7 (342), ra86. doi: 10.1126/scisignal.2005703
Mora-Macias, J., Ojeda-Rivera, J. O., Gutierrez-Alanis, D., Yong-Villalobos, L., Oropeza-Aburto, A., Raya-Gonzalez, J., et al. (2017). Malate-dependent fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc. Natl. Acad. Sci. U.S.A. 114 (17), E3563–E3E72. doi: 10.1073/pnas.1701952114
Nakano, Y., Kusunoki, K., Hoekenga, O. A., Tanaka, K., Iuchi, S., Sakata, Y., et al. (2020). Genome-wide association study and genomic prediction elucidate the distinct genetic architecture of aluminum and proton tolerance in Arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00405
O'Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165 (5), 1280–1292. doi: 10.1016/j.cell.2016.04.038
Ohyama, Y., Ito, H., Kobayashi, Y., Ikka, T., Morita, A., Kobayashi, M., et al. (2013). Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 162 (4), 1937–1946. doi: 10.1104/pp.113.218958
Pelagio-Flores, R., Esparza-Reynoso, S., Garnica-Vergara, A., Lopez-Bucio, J., Herrera-Estrella, A. (2017). Trichoderma-induced acidification is an early trigger for changes in Arabidopsis root growth and determines fungal phytostimulation. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00822
Ragel, P., Rodenas, R., Garcia-Martin, E., Andres, Z., Villalta, I., Nieves-Cordones, M., et al. (2015). The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 169 (4), 2863–2873. doi: 10.1104/pp.15.01401
Raya-Gonzalez, J., Ojeda-Rivera, J. O., Mora-Macias, J., Oropeza-Aburto, A., Ruiz-Herrera, L. F., Lopez-Bucio, J., et al. (2021). MEDIATOR16 orchestrates local and systemic responses to phosphate scarcity in Arabidopsis roots. New Phytol. 229 (3), 1278–1288. doi: 10.1111/nph.16989
Reguera, M., Bassil, E., Tajima, H., Wimmer, M., Chanoca, A., Otegui, M. S., et al. (2015). pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell 27 (4), 1200–1217. doi: 10.1105/tpc.114.135699
Sadhukhan, A., Enomoto, T., Kobayashi, Y., Watanabe, T., Iuchi, S., Kobayashi, M., et al. (2019). Sensitive to proton rhizotoxicity1 regulates salt and drought tolerance of Arabidopsis thaliana through transcriptional regulation of CIPK23. Plant Cell Physiol. 60 (9), 2113–2126. doi: 10.1093/pcp/pcz120
Sakano, K. (1998). Revision of biochemical pH-stat: involvement of alternative pathway metabolisms. Plant Cell Physiol. 39 (5), 467–473. doi: 10.1093/oxfordjournals.pcp.a029393
Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., et al. (2009). STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150 (1), 281–294. doi: 10.1104/pp.108.134700
Sawaki, Y., Kobayashi, Y., Kihara-Doi, T., Nishikubo, N., Kawazu, T., Kobayashi, M., et al. (2014). Identification of a STOP1-like protein in eucalyptus that regulates transcription of Al tolerance genes. Plant Sci. 223, 8–15. doi: 10.1016/j.plantsci.2014.02.011
Sharma, A., Wai, C. M., Ming, R., Yu, Q. (2017). Diurnal cycling transcription factors of pineapple revealed by genome-wide annotation and global transcriptomic analysis. Genome Biol. Evol. 9 (9), 2170–2190. doi: 10.1093/gbe/evx161
Shen, R., Yao, Q., Zhong, D., Zhang, X., Li, X., Cao, X., et al. (2023). Targeted insertion of regulatory elements enables translational enhancement in rice. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1134209
Silva-Navas, J., Salvador, N., Del Pozo, J. C., Benito, C., Gallego, F. J. (2021). The rye transcription factor ScSTOP1 regulates the tolerance to aluminum by activating the ALMT1 transporter. Plant Sci. 310, 110951. doi: 10.1016/j.plantsci.2021.110951
Spadoni, S., Zabotina, O., Di Matteo, A., Mikkelsen, J. D., Cervone, F., De Lorenzo, G., et al. (2006). Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 141 (2), 557–564. doi: 10.1104/pp.106.076950
Straub, T., Ludewig, U., Neuhauser, B. (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 29 (2), 409–422. doi: 10.1105/tpc.16.00806
Tabuchi, A., Matsumoto, H. (2001). Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol. Plant 112 (3), 353–358. doi: 10.1034/j.1399-3054.2001.1120308.x
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Tang, R. J., Zhao, F. G., Garcia, V. J., Kleist, T. J., Yang, L., Zhang, H. X., et al. (2015). Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112 (10), 3134–3139. doi: 10.1073/pnas.1420944112
Tang, R.J., Zhao, F.G., Yang, Y., Wang, C., Li, K., Kleist, T. J., et al. (2020). A calcium signalling network activates vacuolar K+ remobilization to enable plant adaptation to low-K environments. Nat. Plants 6 (4), 384–393. doi: 10.1038/s41477-020-0621-7
Tian, Y., Shen, R., Li, Z., Yao, Q., Zhang, X., Zhong, D., et al. (2022). Efficient C to G editing in rice using an optimized base editor. Plant Biotechnol. J. 20 (7), 1238–1240. doi: 10.1111/pbi.13841
Tian, W. H., Ye, J. Y., Cui, M. Q., Chang, J. B., Liu, Y., Li, G. X., et al. (2021). A transcription factor STOP1-centered pathway coordinates ammonium and phosphate acquisition in. Arabidopsis. Mol. Plant 14 (9), 1554–1568. doi: 10.1016/j.molp.2021.06.024
Tian, Y., Zhong, D., Li, X., Shen, R., Han, H., Dai, Y., et al. (2023). High-throughput genome editing in rice with a virus-based surrogate system. J. Integr. Plant Biol. 65 (3), 646–655. doi: 10.1111/jipb.13381
Tsutsui, T., Yamaji, N., Feng Ma, J. (2011). Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 156 (2), 925–931. doi: 10.1104/pp.111.175802
Voesenek, L., Bailey-Serres, J. (2015). Flood adaptive traits and processes: an overview. New Phytol. 206 (1), 57–73. doi: 10.1111/nph.13209
von Uexküll, H. R., Mutert, E. (1995). Global extent, development and economic impact of acid soils. Plant Soil 171 (1), 1–15. doi: 10.1007/bf00009558
Wang, Z. F., Mi, T. W., Gao, Y. Q., Feng, H. Q., Wu, W. H., Wang, Y. (2021). STOP1 regulates LKS1 transcription and coordinates K+/NH4+ balance in Arabidopsis response to low-K+ stress. Int. J. Mol. Sci. 23 (1), 383. doi: 10.3390/ijms23010383
Wang, X., Wang, Z., Zheng, Z., Dong, J., Song, L., Sui, L., et al. (2019). Genetic dissection of Fe-dependent signaling in root developmental responses to phosphate deficiency. Plant Physiol. 179 (1), 300–316. doi: 10.1104/pp.18.00907
Wu, W., Lin, Y., Chen, Q., Peng, W., Peng, J., Tian, J., et al. (2018). Functional conservation and divergence of soybean GmSTOP1 members in proton and aluminum tolerance. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00570
Xia, J., Yamaji, N., Kasai, T., Ma, J. F. (2010). Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. U.S.A. 107 (43), 18381–18385. doi: 10.1073/pnas.1004949107
Xia, J., Yamaji, N., Ma, J. F. (2013). A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J. 76 (2), 345–355. doi: 10.1111/tpj.12296
Xu, J., Li, H.-D., Chen, L.-Q., Wang, Y., Liu, L.-L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in. Arabidopsis. Cell 125 (7), 1347–1360. doi: 10.1016/j.cell.2006.06.011
Xu, J., Zhu, J., Liu, J., Wang, J., Ding, Z., Tian, H. (2021). SIZ1 negatively regulates aluminum resistance by mediating the STOP1-ALMT1 pathway in Arabidopsis. J. Integr. Plant Biol. 63 (6), 1147–1160. doi: 10.1111/jipb.13091
Yamaji, N., Huang, C. F., Nagao, S., Yano, M., Sato, Y., Nagamura, Y., et al. (2009). A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21 (10), 3339–3349. doi: 10.1105/tpc.109.070771
Yamamoto, Y., Kobayashi, Y., Devi, S. R., Rikiishi, S., Matsumoto, H. (2002). Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 128 (1), 63–72. doi: 10.1104/pp.010417
Yang, J. L., Li, Y. Y., Zhang, Y. J., Zhang, S. S., Wu, Y. R., Wu, P., et al. (2008). Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 146 (2), 602–611. doi: 10.1104/pp.107.111989
Ye, J. Y., Tian, W. H., Zhou, M., Zhu, Q. Y., Du, W. X., Zhu, Y. X., et al. (2021). STOP1 activates NRT1.1-mediated nitrate uptake to create a favorable rhizospheric pH for plant adaptation to acidity. Plant Cell 33 (12), 3658–3674. doi: 10.1093/plcell/koab226
Yokosho, K., Yamaji, N., Ma, J. F. (2011). An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J. 68 (6), 1061–1069. doi: 10.1111/j.1365-313X.2011.04757.x
Zeigler, R. S., Puckridge, D. W. (1995). Improving sustainable productivity in rice-based rainfed lowland systems of south and southeast Asia. GeoJournal 35 (3), 307–324. doi: 10.1007/bf00989138
Zhang, L., Dong, D., Wang, J., Wang, Z., Zhang, J., Bai, R. Y., et al. (2022). A zinc finger protein SlSZP1 protects SlSTOP1 from SlRAE1-mediated degradation to modulate aluminum resistance. New Phytol. 236 (1), 165–181. doi: 10.1111/nph.18336
Zhang, Y., Zhang, J., Guo, J., Zhou, F., Singh, S., Xu, X., et al. (2019). F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116 (1), 319–327. doi: 10.1073/pnas.1814426116
Zhang, Z., Fu, D., Xie, D., Wang, Z., Zhao, Y., Ma, X., et al. (2023). CBL1/9-CIPK23-NRAMP1 axis regulates manganese toxicity. New Phytol. doi: 10.1111/nph.18968
Zheng, S. J. (2010). Crop production on acidic soils: overcoming aluminium toxicity and phosphorus deficiency. Ann. Bot. 106 (1), 183–184. doi: 10.1093/aob/mcq134
Zhou, F., Singh, S., Zhang, J., Fang, Q., Li, C., Wang, J., et al. (2023). The MEKK1-MKK1/2-MPK4 cascade phosphorylates and stabilizes STOP1 to confer aluminum resistance in Arabidopsis. Mol. Plant 16 (2), 337–353. doi: 10.1016/j.molp.2022.11.010
Keywords: STOP1 transcription factor, STOP1-like proteins, acid soil syndrome, aluminum toxicity, proton toxicity, plant nutrient, regulatory network
Citation: Li X and Tian Y (2023) STOP1 and STOP1-like proteins, key transcription factors to cope with acid soil syndrome. Front. Plant Sci. 14:1200139. doi: 10.3389/fpls.2023.1200139
Received: 04 April 2023; Accepted: 25 May 2023;
Published: 21 June 2023.
Edited by:
Weiqiang Li, RIKEN, JapanReviewed by:
Sheliang Wang, Huazhong Agricultural University, ChinaSatomi Kanno, Nagoya University, Japan
Copyright © 2023 Li and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifu Tian, dGlhbnlpZnVAY2Fhcy5jbg==
 Xinbo Li
Xinbo Li Yifu Tian
Yifu Tian