
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 June 2023
Sec. Plant Proteomics and Protein Structural Biology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1198353
 Hailong Zhang1†
Hailong Zhang1† Jingwen Zhou1†
Jingwen Zhou1† Xiaoyue Kou1†
Xiaoyue Kou1† Yuqi Liu1
Yuqi Liu1 Xiaonan Zhao1,2
Xiaonan Zhao1,2 Guochen Qin3
Guochen Qin3 Mingyu Wang1
Mingyu Wang1 Guangtao Qian1
Guangtao Qian1 Wen Li1
Wen Li1 Yongshun Huang1
Yongshun Huang1 Xiaoting Wang1
Xiaoting Wang1 Zhenjie Zhao1
Zhenjie Zhao1 Shuang Li4
Shuang Li4 Xiaoqian Wu4
Xiaoqian Wu4 Lixi Jiang2
Lixi Jiang2 Xianzhong Feng5
Xianzhong Feng5 Jian-Kang Zhu6,7
Jian-Kang Zhu6,7 Lixin Li1*
Lixin Li1*SYP71, a plant-specific Qc-SNARE with multiple subcellular localization, is essential for symbiotic nitrogen fixation in nodules in Lotus, and is implicated in plant resistance to pathogenesis in rice, wheat and soybean. Arabidopsis SYP71 is proposed to participate in multiple membrane fusion steps during secretion. To date, the molecular mechanism underlying SYP71 regulation on plant development remains elusive. In this study, we clarified that AtSYP71 is essential for plant development and stress response, using techniques of cell biology, molecular biology, biochemistry, genetics, and transcriptomics. AtSYP71-knockout mutant atsyp71-1 was lethal at early development stage due to the failure of root elongation and albinism of the leaves. AtSYP71-knockdown mutants, atsyp71-2 and atsyp71-3, had short roots, delayed early development, and altered stress response. The cell wall structure and components changed significantly in atsyp71-2 due to disrupted cell wall biosynthesis and dynamics. Reactive oxygen species homeostasis and pH homeostasis were also collapsed in atsyp71-2. All these defects were likely resulted from blocked secretion pathway in the mutants. Strikingly, change of pH value significantly affected ROS homeostasis in atsyp71-2, suggesting interconnection between ROS and pH homeostasis. Furthermore, we identified AtSYP71 partners and propose that AtSYP71 forms distinct SNARE complexes to mediate multiple membrane fusion steps in secretory pathway. Our findings suggest that AtSYP71 plays an essential role in plant development and stress response via regulating pH homeostasis through secretory pathway.
Cell wall is an important feature of plant cells which provides cell shapes and strength (Keegstra, 2010). Cell wall is a complex extracellular matrix containing cellulose, hemicellulose, polysaccharides, lignin and a small amount of functional glycoproteins (Kumar and Turner, 2015), playing essential roles in cell morphogenesis, signal transduction, development, defense and stress tolerance (Fry, 2004; Liu et al., 2018). Plant cell wall is usually divided into two categories: primary cell wall (PCW) and secondary cell wall (SCW) (Keegstra, 2010). In Arabidopsis, the PCW consists of cellulose microfibrils cross-linked by xyloglucan (XyG), and embedded in pectin matrix (Gigli-Bisceglia et al., 2020), and the SCW consists of cellulose, hemicellulose, lignin, and xylans (Meents et al., 2018). XyG is the most enriched hemicellulose in PCWs (Julian and Zabotina, 2022). In addition, cell wall-specific proteins (e.g. extensins, expansins, hydroxyproline-/glycine-rich proteins and glycoproteins), polysaccharides and polyphenols are related to cell wall dynamics and response to environmental stimuli (Zhu, 2018). Lignin plays essential roles on plant growth and development and acts as a physical barrier against biotic and abiotic stress (Liu et al., 2018). Lignin is a heteropolymer of monolignols synthesized primarily through the general phenylpropanoid pathway (GPP), a main synthetic pathway for secondary metabolites e.g. flavonoids etc. (Geng et al., 2020). Lignin precursors are secreted by plasma membrane (PM)-localized transporters such as ATP binding cassette (ABC) transporters (Kaneda et al., 2010; Li and Chapple, 2010; Miao and Liu, 2010; Alejandro et al., 2012), and diffuse in both PCWs and SCWs for active lignification (Tobimatsu et al., 2013; Schuetz et al., 2014; Pandey et al., 2016; Geng et al., 2020). The monolignols polymerization is catalyzed by plant-specific class III peroxidases (PRXs) and laccases (Dixon and Barros, 2019). PRXs, the secretory peroxidases accumulated in the plant cell wall or the vacuole(Kidwai et al., 2020), are related to cell wall polymerization such as lignification, suberisation and cross-linking of cell-wall constituents, and consumption of hydrogen peroxide (H2O2), etc. (Meng et al., 2021). PRXs reduce H2O2 to oxidize monolignols (Meyer et al., 2009; Didi et al., 2015). Plant laccases, the multicopper oxidases, are necessary and nonredundant with PRXs for lignification during vascular development in Arabidopsis. Laccase 4 (LAC4)/11/17 are redundantly required for asymmetric lignification (Zhao et al., 2013). Unlike PRXs, laccases reduce O2 to H2O to oxidize monolignols without consuming reactive oxygen species (ROS) (Wang et al., 2015).
Flavonoids are broadly distributed in plants, and is essential for plant development and defense (Falcone Ferreyra et al., 2012; Sharma et al., 2019). In plant cells, flavonoids are mainly synthesized in the cytoplasmic face of the endoplasmic reticulum (ER) and then uploaded into the ER lumen (Zhao and Dixon, 2010; Falcone Ferreyra et al., 2012; Zhao, 2015). The PM-localized flavonoid transporters are mainly ABC transporters (Ferrer et al., 2008; McFarlane et al., 2010; Sharma et al., 2019), e.g. ABCG (ATP binding cassette sub-family G) protein. ABCG10 secrets isoflavonoids to the apoplast for plant defense (Klink et al., 2017), ABCG1 and ABCG16 are required for secretion of suberin and pollen wall synthetic materials (Zhao, 2015; Shanmugarajah et al., 2019; Liu et al., 2020; Dhara and Raichaudhuri, 2021). SNARE-mediated exocytosis pathways or nondirectional secretion are also proposed to mediate in the efflux of flavonoids into the extracellular space (Zhao and Dixon, 2010) Higher plants have developed a unique vesicle trafficking system to meet the needs of plant development and environmental adaption. Vesicle trafficking involves multiple organelles, and carries out material communication and signal transmission (Adnan et al., 2019). The Golgi functions as a vesicle transport hub, accepting ER-derived vesicles, and facilitating sorting of the cargo into various vesicles. Moreover, the Golgi is also a factory which holds protein post-translational modification (e.g. glycosylation) and substance synthesis (e.g. hemicellulose biosynthesis and assembly) (Bui et al., 2021). The protein glycosylation initiates in the ER lumen and is completed in the Golgi cisternae (Frankova and Fry, 2013). For example, glycosylation of arabinogalactan proteins (AGPs) is completed by Glycosyltransferases (GTs) at the Golgi (Frappaolo et al., 2020).The trans-Golgi network (TGN) is a sorting hub for both exocytosis and endocytosis (Heinze et al., 2020). At the TGN, hemicelluloses and secretory proteins are sorted into endosomes and targeted to the PM (Kang et al., 2011). The TGN-PM cycling pathway and vacuolar degradation pathways coordinately fine-tune homeostasis of the PM proteins, especially under environmental stimuli (Meents et al., 2018; Gigli-Bisceglia et al., 2020). Thus it can be seen that Golgi apparatus plays an important role in the secretion of cell wall biosynthetic materials and in stress response. The process of vesicle trafficking includes budding, directional movement, tethering, anchoring and membrane fusion (Cui et al., 2022), and each step is regulated by various factors, such as coatomer, SM, tether, SNARE and Rab proteins, which are highly conserved in yeast, mammals and plants (Singh and Jurgens, 2018). The soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins play an essential role on membrane fusion of arriving vesicles with the target membrane (Yoon and Munson, 2018; MartiniEre and Moreau, 2020). SNAREs usually contain an N-terminal region, a SNARE motif and a transmembrane domain (Fasshauer and Margittai, 2004). According to the conserved amino acids, glutamine or arginine residues in core of SNARE domain, SNARE proteins are divided into Q-SNAREs on the target membrane and R-SNAREs on the vesicle (Fasshauer et al., 1998). Q-SNAREs include Qa-, Qb- and Qc-SNARE, the three Q-SNAREs combine with R-SNARE to form trans-SNARE complex facilitating membrane fusion (Antonin et al., 2000). SYP71 is a plant-specific Qc-SNARE (Sanderfoot et al., 2000), with multiple subcellular localization of the ER, plasma membrane (Suwastika et al., 2008), endosome and cell plate (Bao et al., 2012; Klink et al., 2017). It is reported that SYP71 is essential for symbiotic nitrogen fixation in nodules in Lotus (Hakoyama et al., 2012), and participates in pathogen resistance in Glycine max (Klink et al., 2017), rice (Bao et al., 2012), wheat (Liu et al., 2016) and Arabidopsis (Wei et al., 2013). Arabidopsis SYP71 is proposed to localize at the PM, endosome and ER, and be involved in multiple membrane fusion steps during secretion (Tyrrell et al., 2007). Moreover, AtSYP71 coordinates with Qa-SNARE KNOLLE, Qb-SNARE NPSN11 and R-SNAREs VAMP721/722 to facilitate cell plate formation during cytokinesis (Suwastika et al., 2008; Bao et al., 2012; Yoon and Munson, 2018). However, the molecular mechanism underlying AtSYP71 regulatory roles on plant development is elusive.
In this study, we illustrate that AtSYP71 is essential for plant development and stress response. Homozygous mutant atsyp71-1 was seedling lethal, the knockdown mutants atsyp71-2 and atsyp71-3 exhibited severe early development defects, such as short roots and delayed bolting. Transcriptome analysis indicated that cell wall biosynthesis, metabolism and stress response were greatly affected in atsyp71-2. In response to transcriptomic data, the cell wall components and structures significantly altered, and ROS and pH homeostasis were disrupted in atsyp71-2. Furthermore, secretion was blocked in atsyp71-2. AtSYP71-interacting SNAREs were identified by pull down-LC-MS/MS analysis. The results suggest that AtSYP71 forms distinct SNARE complex at multiple steps in secretory pathway. Our findings suggest that AtSYP71 regulates pH homeostasis by controlling vesicle transport pathways, thereby affecting plant development and stress responses.
Arabidopsis thaliana ecotype Col-0 was used as wild type. T-DNA mutants were generated from Col-0. atsyp71-1 (GABI_367A08), atsyp71-2 (SALK_081547), atsyp71-3 (SALK_201897) and atsyp71-4 (SAIL_813_A05) were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University (https://abrc.osu.edu). Homozygous plants were isolated by PCR genotyping using the insertion-specific primers listed in Supplementary Table 3. The seeds were surface-sterilized and sown either in soil or on 1/2 Murashige and Skoog medium (PhytoTech) with 1% (w/v) sucrose and 0.8 or 1.2% agar. Plants were grown at 22°C under 16 h: 8 h/light: dark cycles, horizontally or vertically as needed. The root length was measures using ImageJ software.
Transgenic plants (Col-0) expressing myc (TAP)-tagged AtSYP71 were generated using a modified pNTAPa vector described by (Li et al., 2013).
To prepare antibodies against AtSYP71, a polypeptide (Cys-LPARIEAIPDGTA GGPKSTSAWTPSSTTSRPDIKFDSDGRFDDDYFQESN) was synthesized and conjugated to a Carrier protein KLH linked by an N-terminal Cys residue. The AtSYP71 peptide–KLH conjugates were injected into two rabbits to generate antibodies. The antibodies were subjected to ProteinA/G purification from the serum and ELISA detection. Polypeptide synthesis and antibody preparation were commissioned to GL Biochem (Shanghai) Ltd.
Immunoblot analysis was performed as described previously (Li et al., 2006). Antibodies were diluted as follows: anti-AtSYP71, 1:2,000. The secondary antibody was against rabbit IgG (ZB2301 and ZSGB-BIO), 1:5000. Immunoreactive signals were detected using a chemiluminescence detection system (LAS-4000 and FYJIFILM).
Fluorescent images were obtained using a point scanning confocal microscope (Leica TCS SP8). Confocal imaging was preset for GFP (Ex:488 nm, Em:500–550 nm) or for propidium iodide staining (Ex:543 nm, Em: 580–640 nm).
For the yeast two-hybrid assay, the fragments of AtSYP71 (cytosolic region), MIP2 (Sec39 domain) and MIP3 were amplified using cDNA obtained from seedlings with specific primers and ligated into pEASY-Blunt vector (TransGen, #CB101-01), respectively. After Sanger sequencing confirmation, the fragments were transferred into pGADT7 or pGBKT7 vectors, respectively. AtSYP81, AtSEC20 and MAG2 constructs were generated in our previous study (Li et al., 2006; Zhao et al., 2018). The paired constructs were introduced into Saccharomyces cerevisiae strain AH109 (Clontech) and selected on SD/-Leu/-Trp medium. The interactions were detected on SD/-Leu/-Trp/-His/-Ade medium.
Total RNA was extracted as described previously (Guan et al., 2021) using 9-day-old seedlings grown on 1/2MS medium horizontally. Reverse transcription Quantitative polymerase chain reaction (RT-qPCR) was performed according to the manufacturer’s instructions. The specific primers are listed in Supplementary Table 3.
Total RNA was extracted from 9-day-old seedlings grown on 1/2MS medium horizontally. Illumina cDNA libraries were constructed with the TruSeq RNA Sample Prep Kits v2 (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Sequencing of the cDNA libraries was performed by pair-end methousing an Illumina HISEQ-x10 with a 150-bp read length and a sequence depth ~20 million uniquely mapped reads. Three biological replicates per sample. The data presented in the study are deposited in the NCBI repository, the BioProject ID is PRJNA971388.
Reads were trimmed using the CLC Genomics Workbench 6.5.1 (CLC bio, Denmark) with the following parameters: ‘quality scores-0.005; trim ambiguous nucleotides-2; remove 5’ terminal nucleotides-1; remove 3’ terminal nucleotides-1; discard reads below length 25’. Trimmed reads were mapped using the RNA-seq mapping algorithm implemented in the CLC Genomics Workbench to the reference Arabidopsis thaliana genome (TAIR10) allowing only unique mapping (length fraction=1, similarity fraction=0.95). In order to estimate the influence of non-uniquely mapped reads on gene expression we also mapped reads using the same software and parameters as indicated above, but allowing multiple mapping (up to 10 hits). For each gene, total gene reads (TGR) was determined as the sum of all reads mapped to this gene. To avoid bias due to different library sizes, TGR values were normalized by a size factor as described in Anders and Huber (Anders and Huber, 2010).
DEGs were identified using the R package ‘DESeq’ (Anders and Huber, 2010). A false discovery rate (FDR) of 0.05 and a fold change of 2.0 were chosen as the threshold for significantly differential expression. The original transcriptomic data of atsyp71-2 vs Col-0 is in Supplementary Table 1.
Downregulated and upregulated DEGswere analyzed for GO and other annotation (as key words or protein domain) enrichment using the DAVID gene functional annotation tool https://david.ncifcrf.gov/, with an FDR value of 0.05 and a fold change of category representation of 2.0 as the threshold of significance (Huang da et al., 2009a; Huang da et al., 2009b).
KEGG enrichment analysis was performed using KOBAS (v2.0). Rich factor, P-value and the number of genes indicated the degree of KEGG enrichment. Rich factor means the ratio of enriched DEGs to all genes annotated in the pathway. Therefore, the higher the Rich factor value, the greater the enrichment of the pathway. Parameter setting: Corrected P-Value < 0.05.
The first segment of stems from ten-weeks-old plants were harvested and dried at 80°C to constant weight, then were ground and passed through a 40 mesh sieve. About 5 mg was put into a 10 mL glass test tube with 1 mL of glacial acetic acid containing 30% acetyl bromide and 40 uL of perchloric acid, sealed with a sealing film, fully mixed, then incubated in water bath at 80°C for 40 min, shaking every 10 min. After natural cooling, 1 mL of 2 mol/L NaOH and glacial acetic acid (equal volume) were added and fully mixed. Add 1,960 uL of acetic acid to 40 uL of supernatant and fully mixed. Then, absorbance of the mixture was determined by spectrophotometer at 280 nm wavelength versus the prepared blank.
The first segment of stems from ten-weeks-old plants were harvested and incubated overnight in 80% ethanol at 65°C. Dry materials were ball milled to fine powder. Cellulose content was determined by Anthrone sulfuric acid colorimetry using Cellulose determination kit (CLL-2-Y, Comin, Suzhou Keming Biotechnology Co., Ltd.) according to Xiao et al. (2016). D-Glc (Sigma) was used as a standard for calculation of cellulose content. Three repeats per sample.
The flavonoid content was determined by colorimetric assay (Kim et al., 2003). A 250 μL of standard solution of rutin at different concentrations or appropriately diluted samples were added to 10 ml volumetric flask containing 1 ml of distillate water, respectively, then, 75 μl of NaNO2 (5%) was added and mixed thoroughly. After 6 min of incubation, 75 μl of AlCl3 (10%) was added, fully mixed and incubated for 6 min, then 500 μl of NaOH (1N) was added. Immediately, the solution was diluted by adding 2.5 ml of methanol and mixed thoroughly. Absorbance of the mixture was determined by spectrophotometer at 506 nm wavelength versus the prepared blank. Total flavonoid compounds in plant were indicated as mg rutin equivalents (CE mg/ml). Three repeats per sample.
For determination of activities of ROS-scavenging enzyme, 0.2 g of roots from nine-day-old seedlings were ground in 1.6 ml of precooled 50 mM phosphate-buffered saline (PBS) buffer (pH7.8). The homogenate was centrifuged at 16,000 g for 20 min at 4°C, and the supernatant was used for the assays. Determination of activities of peroxidase (POD), superoxide dismutase (SOD) and superoxide catalase (CAT) were performed according to established protocols (Luo et al., 2020). Three biological replicates per sample.
0.1g dry powder of the first stem segments from ten-weeks-old Col-0 and atsyp71-2 plants were used for determination of cell wall monosaccharides according to the method reported by Liu et al. (Liu et al., 2022a).
The first stem segments from 10-weeks-old Col-0 and atsyp71-2 plants were fixed and made into paraffin slices (Wang et al., 2020), and subjected to immunofluorescence analysis according to previously reported method (Wang et al., 2016).
Pull-down assay was performed as described previously using a μMACS™ epitope tag protein isolation kit (Anti-c-myc MicroBeads, Miltenyi Biotec, Order No. 130-091-123). Two grams of ten-day-old seedlings were used for initiation. The eluates were used for Shotgun liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS analysis was mainly performed as described previously (Qin et al., 2017), with slight modifications.
To explore the biological function of AtSYP71 on plant growth and development, four T-DNA insertion mutants of AtSYP71, atsyp71-1 to atsyp71-4 were obtained. Genotyping and sequencing confirmed that T-DNA was inserted in 5’-UTR, different sites of the first intron region and the seventh intron of AtSYP71, respectively, in atsyp71-1 to atsyp71-4 (Figure 1A). RT-qPCR analysis indicated that AtSYP71 expression was eliminated in atsyp71-1, and was significantly downregulated in atsyp71-2 to atsyp71-4 (Figure 1B). Consequently, atsyp71-2 to atsyp71-4 were knockdown alleles, and atsyp71-1 was knockout allele. The complementation lines of atsyp71-1 (atsyp71-1 com) and atsyp71-2 (atsyp71-2 com) were generated by crossing atsyp71-1 (using heterozygous plant) and atsyp71-2 with pAtSYP71::GFP-AtSYP71-expressing wild type (Col-0) line, respectively. The complemented lines restored the AtSYP71 expression in atsyp71-1 and atsyp71-2 mutants (Figure 1B). p35S::TAP-AtSYP71 (AtSYP71 OE) conferred significantly increased AtSYP71 expression in wild-type background (Figure 1B). Comparison of the early development phenotypes of Col-0, atsyp71 mutants, complementation lines (atsyp71-1/-2 com) and AtSYP71 OE revealed that seven-day-old seedlings of atsyp71-2 and atsyp71-3 developed much shorter primary roots (about 67% decrease) and smaller shoots than Col-0, while, the root length and shoots of atsyp71-4 and AtSYP71 OE had no significance compared with that of Col-0 (Figures 1C, D). However, the development of the knockout allele atsyp71-1 was inhibited. After germination, roots of atsyp71-1 cannot elongate, cotyledons were albino or vitrified, and the true leaves were very small and turned yellow and vitrified (Figure 1E). And the so-called seedlings dead at about ten days after germination. The bolting of atsyp71-2 and atsyp71-3 plants was significantly delayed compared with that of Col-0 (Figure 1F). The phenotypes of atsyp71-1 com and atsyp71-2 com were restored (Figures 1C, D, F). These results suggest that AtSYP71 is essential for plant morphogenesis and early development.
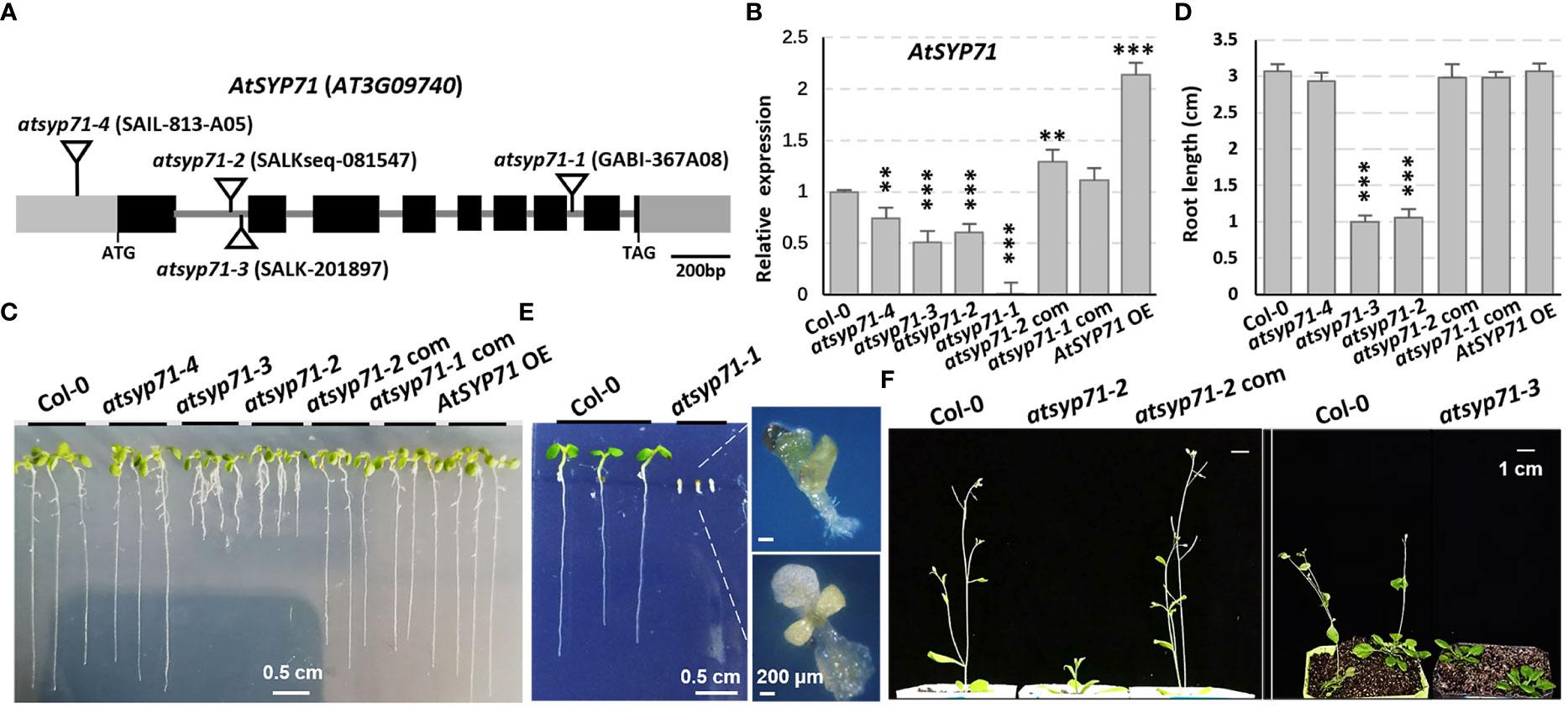
Figure 1 AtSYP71 is essential for plant morphogenesis and early development. (A) AtSYP71 gene structure diagram. Black boxes represent exons, gray lines represent introns, and gray boxes represent untranslated regions (UTRs). The triangles indicate the T-DNA insertion sites of the atsyp71 mutants. (B) Statistics of RT-qPCR detection of the relative expression levels of AtSYP71 in atsyp71 mutants, the atsyp71 complementation lines and AtSYP71 overexpression line. Three independent experiments per sample, two repeats per experiment. (C) Phenotype of seven-day-old seedlings of Col-0, atsyp71 mutants, atsyp71-2 com and AtSYP71 OE lines. (D) Statistics of primary root length of the displayed genotypes (n ≥ 44). Three biological replicates per sample. **P < 0.01; ***P < 0.001. Student’s t test. (E) Five-day-old seedlings of Col-0 and atsyp71-4 mutant. Magnified pictures highlighted growth defects of atsyp71-4. Bar, 0.5 cm. (F) Bolting of atsyp71-2 and atsyp71-3 was delayed. Four- (right panel) and five (left panel)-week-old plants of the displayed genotypes.
To explore the mechanism of AtSYP71 regulatory role on plant growth and development, we performed transcriptome analysis using roots of nine-day-old Col-0 and atsyp71-2 seedlings. The sequencing obtained 34,524 and 33,113 reliable clean reads above 100 bp in Col-0 and atsyp71-2, respectively. After comparison using tophat2 software, 87.68% and 86.59% of clean reads matched to the reference genome sequence, respectively. Statistical evaluation of sequencing quality value showed that base Q30% was 92.37% and 92.16%, respectively (Supplementary Figure 1A), indicating the high quality of transcriptome sequencing and reliable original data for subsequent analysis. Volcano map indicated the overall distribution of the differentially expressed genes (DEGs). A total of 165 DEGs were identified, of which 69 were up regulated, and 96 were down regulated (Supplementary Figure 1B). Gene Ontology (GO) analysis revealed that in Biological process (BP), the DEGs were enriched in Response to biotic and abiotic stimuli/stress, Defense responses (Figure 2A, red arrows), and Redox processes (yellow arrows), etc. In Cellular component (CC), DEGs were enriched in Cell wall and External encapsulating structure (blue arrows). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis indicates that DEGs were mainly enriched in Glucosinolate Biosynthesis, 2-Oxocarboxylic acid metabolism and Phenylpropanoid biosynthesis, Metabolic and Secondary metabolic processes, and Amino acids metabolism, etc (Figure 2B). These results suggest that down regulation of AtSYP71 disturbed plant response to stimuli and stress through redox processes and metabolism etc.
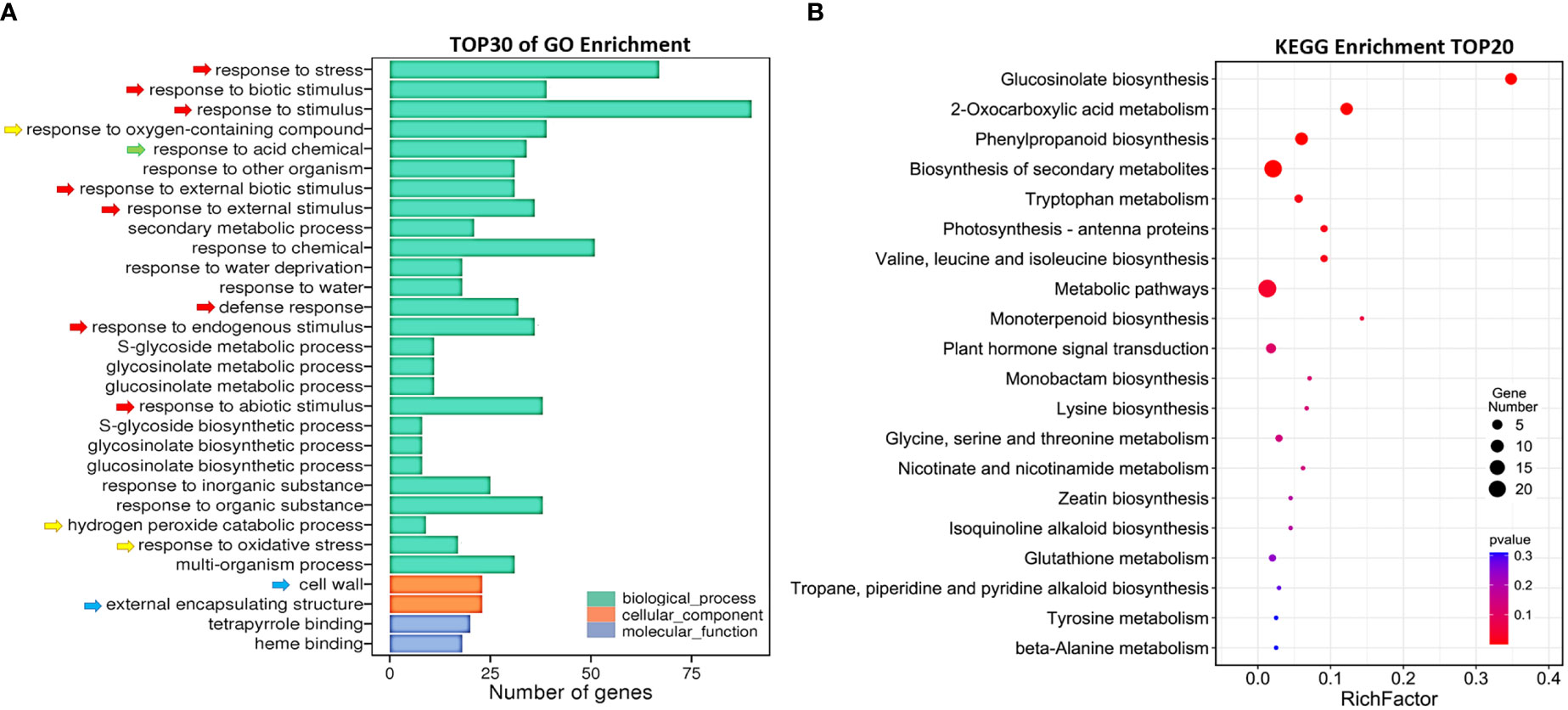
Figure 2 Comparative transcriptome analysis of DEGs in atsyp71-2. (A) Gene ontology (GO) enrichment analysis of DEGs. X axis represents DEG number. Y axis represents GO terms. (B) Top 20 pathways of KEGG enrichment analysis. X axis represents enrichment factors; Y axis represents pathways. The color bar indicates the P value, the circle size indicates DEG number.
Go enrichment analysis of the DEGs in Cellular component (GO:0005575, CC) indicated that the DEGs were enriched in the Extracellular region (GO:0005576) and External encapsulating structure (GO:0030312) containing Cell wall (GO:0005618) which includes Plant-type cell wall (GO:0009505) (Figure 3A, rectangles). The DEGs include five PRXs (class III peroxidases), three LTPs (lipid transfer proteins), three XTHs (xyloglucan endotransglucosylase/hydrolase), five EXPAs (expansins) and some other cell wall biosynthesis-related genes, and most of these genes were down-regulated (Figure 3B; Supplementary Table 2), suggesting that AtSYP71 is related to cell wall biosynthesis and dynamics.
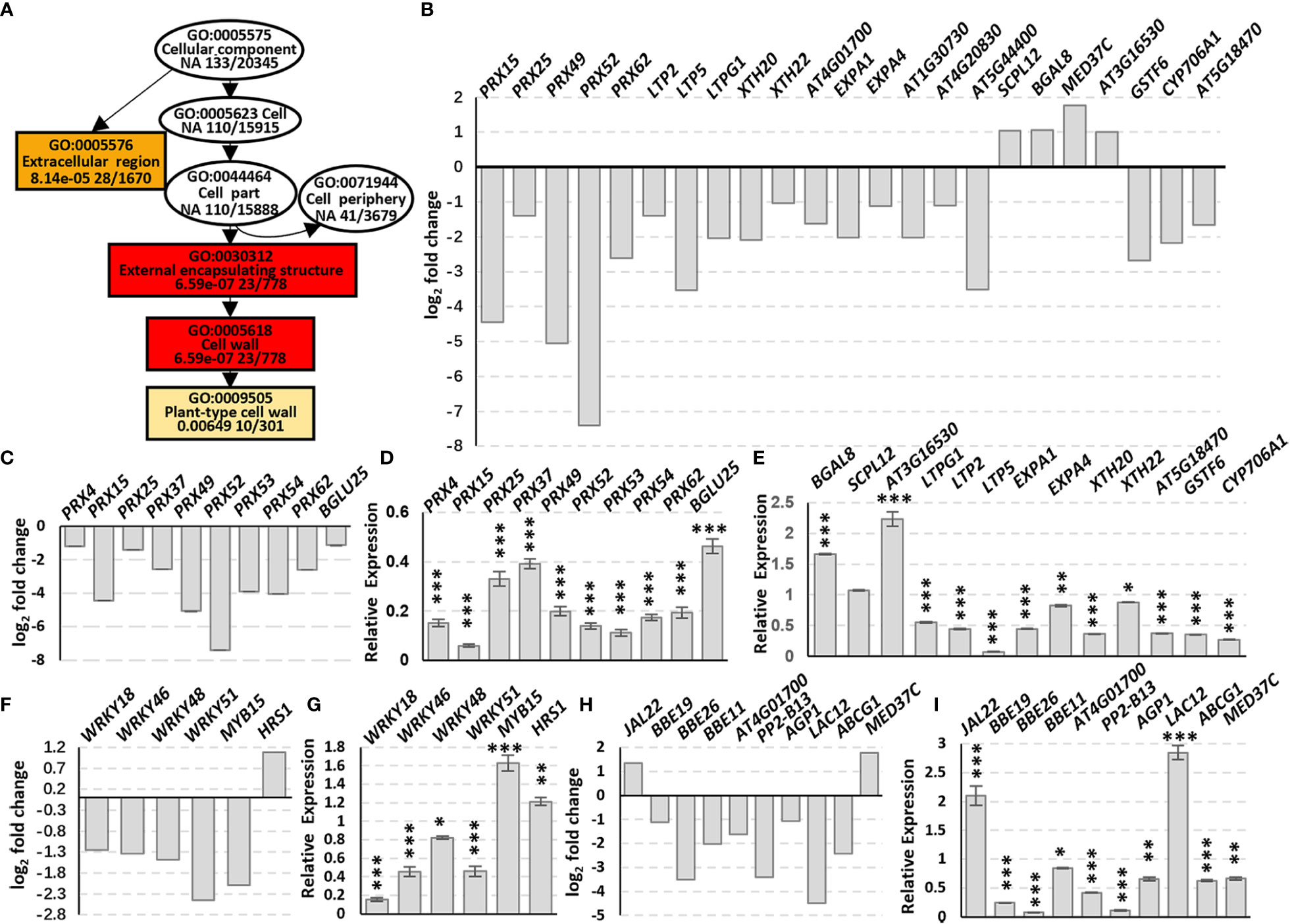
Figure 3 Changes in DEG Enrichment in atsyp71-2 mutant. (A) The enriched GO terms in Cellular component (GO:0005575) of DEGs in Col-0 and atsyp71-2 seedlings. Rectangles indicate the significant terms. The colors of rectangles and ovals represent the relative significances, ranging from red (the most significant, P < 0.0001), orange (the second significant, P < 0.001), light yellow (the third significant; P < 0.05) and white (no significance). (B) Transcriptomic analysis of DEGs in CC. (C) Statistics of RT-qPCR analysis of DEG expression levels in (B). (D, E) Transcriptomic analysis and statistics of RT-qPCR analysis of DEGs in Phenylpropanoid biosynthesis pathway. (F, G) Transcriptomic analysis and statistics of RT-qPCR analysis of some other DEGs related to cell wall biosynthesis. (H, I) Transcriptomic analysis and statistics of RT-qPCR detection of Transcription factors related to cell wall biosynthesis. Total RNA for RT-qPCR analysis were from roots of seven-day-old seedlings. All RT-qPCR Data are presented from three independent experiments performed with four technical replicates per sample. *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t-test. Abbreviations: ABCG1, ATP binding cassette sub-family G 1; AGP1, Arabinogalactan protein 1; BBE19/OGOX1, FAD-binding Berberine Bridge Enzyme 19/oligogalacturonide oxidase 1; BGAL8, beta-galactosidase 8; BGLU25, beta glucosidase 25; CYP706A1, cytochrome p450 family 706, subfamily A, polypeptide 1; EXPA1, expansin A1; GSTF6, glutathione s-transferase 6; JAL22, jacalin-related lectin 22; LAC12, LACCASE 12; LTP2 (lipid transfer protein 2); LTPG1 (glycosylphosphatidylinositol-anchored lipid protein transfer 1), PP2-B13, PHLOEM PROTEIN 2-B13; PRX, peroxidase; SCPL12, serine carboxypeptidase-like 12; XTH20, xyloglucan endotransglucosylase/hydrolase 20.
Phenylpropanoid biosynthesis pathway is reported to be involved in cell wall biosynthesis (Lewis et al., 1987; Merali et al., 2007), and the enriched DEGs include nine PRXs and BGLU25, a β-D-glucosidase gene (Figure 3C). The five PRXs in CC were also included in this pathway. RT-qPCR analysis indicate that the changes of the DEGs in this pathway and CC were the same, and most of the DEGs had significances (Figures 3D, E). Among these factors, PRXs are involved in lignification, suberization, cross-linking of extensins, metabolism of reactive oxygen species (ROS), as well as cell wall dynamics, e.g. cell wall loosening and strengthening, etc. (Almagro et al., 2009; Francoz et al., 2015). XTH enzymes play a role in cell wall loosening through the modification of xyloglucan chains (Opazo et al., 2017). EXPAs are located in the cell wall and activated by low apoplastic pH (Cosgrove, 2005), and act on cell wall loosening resulting in cell elongation (Pacifici et al., 2018). Moreover, BGAL8 (beta-galactosidase) (Chandrasekar and van der Hoorn, 2016), LTPs (lipid transfer proteins) (Chae et al., 2009; Bard et al., 2016), BBE19/OGOX1 (FAD-binding Berberine Bridge Enzyme 19/oligogalacturonide oxidase 1) (Benedetti et al., 2018), ABCG1 (Shanmugarajah et al., 2019) and ATPP2 (Phloem protein 2)-B13 (Bobbili et al., 2018) participate in cell wall biosynthesis and dynamics, and GSTF6 (glutathione s-transferase 6), CYP706A1 (cytochrome p450 family 706, subfamily A, polypeptide 1), BGLU25 (beta glucosidase 25) and SCPL12 (serine carboxypeptidase-like 12) are involved in biosynthesis of flavonoids which is related to stress response (Xu et al., 2004; Su et al., 2011).
In addition to the DEGs in CC, expression of some transcription factors (TFs) related to cell wall dynamics/biosynthesis and stress response also altered significantly (Figure 3F). RT-qPCR analysis confirmed that except for MYB15, the changes of the TF genes were consistent with those of transcriptomic data, and most of them had significances (Figure 3G). It has been demonstrated that MYB15 plays a central role on pathogen-induced lignification (Kim et al., 2020a), and WRKY18 and WRKY53 coordinate with histone acetyltransferase1 (HAC1) to regulate responses to sugars, the structural components of cell wall (Rolland and Sheen, 2005; Chen et al., 2019). WRKY46, WRKY48 and WRKY51 are responsible for abiotic stress response, and HRS1 is involved in nitrogen saturation signaling (Xing et al., 2008; Gao et al., 2011; Chen et al., 2017; Li et al., 2021). To further confirm that the alteration of expression of these genes was resulted from knockdown of AtSYP71, we checked the expression of some of the genes randomly in atsyp71-3. The results showed the same trends of change (Supplementary Figure 2). The significant alteration of expression of these genes in atsyp71 mutants suggest that AtSYP71 regulates cell wall biosynthesis, dynamics and plant stress response.
Transcriptome analysis reveals that in CC, all DEGs were enriched in Cell wall and External encapsulating structure (Figure 2A, blue arrows), indicating AtSYP71 is closely related to cell wall homeostasis. Therefore, we first determined the contents of cell wall components in atsyp71-2 stems. The results indicate that the content of lignin in atsyp71-2 stems increased significantly, while, the content of cellulose didn’t change obviously (Figure 4A), but the content of flavonoids reduced significantly compared with that in Col-0 (Figure 4B). Determination of cell wall polysaccharides indicate that the contents of glucose and XyG increased significantly, of galactose decreased significantly, and of arabinose didn’t change significantly compared with that in Col-0 (Figure 4C). Immunofluorescence images indicate that the content of AGPs decreased significantly, and of XyG increased significantly which is consistent with the result of polysaccharides determination (Figure 4D; Supplementary Figure 3). To investigate the effects of changes in the component abundance on cell wall structure, we observed the cross section of stems. The images revealed that the stem diameter didn’t change obviously (Figures 4E, F), but the average xylem number and total area in atsyp71-2 increased significantly compared with that in Col-0 (Figures 4E, G). Moreover, the cell wall thickness of interfascicular fiber (IF) cells was significantly thicker than that of Col-0 (Figures 4H, I). It can be speculated that the increased xylem area and the thickness of IF cell wall may be the main reasons for the increase in lignin content. These results strongly suggest that AtSYP71 regulates cell wall biosynthesis and dynamics.
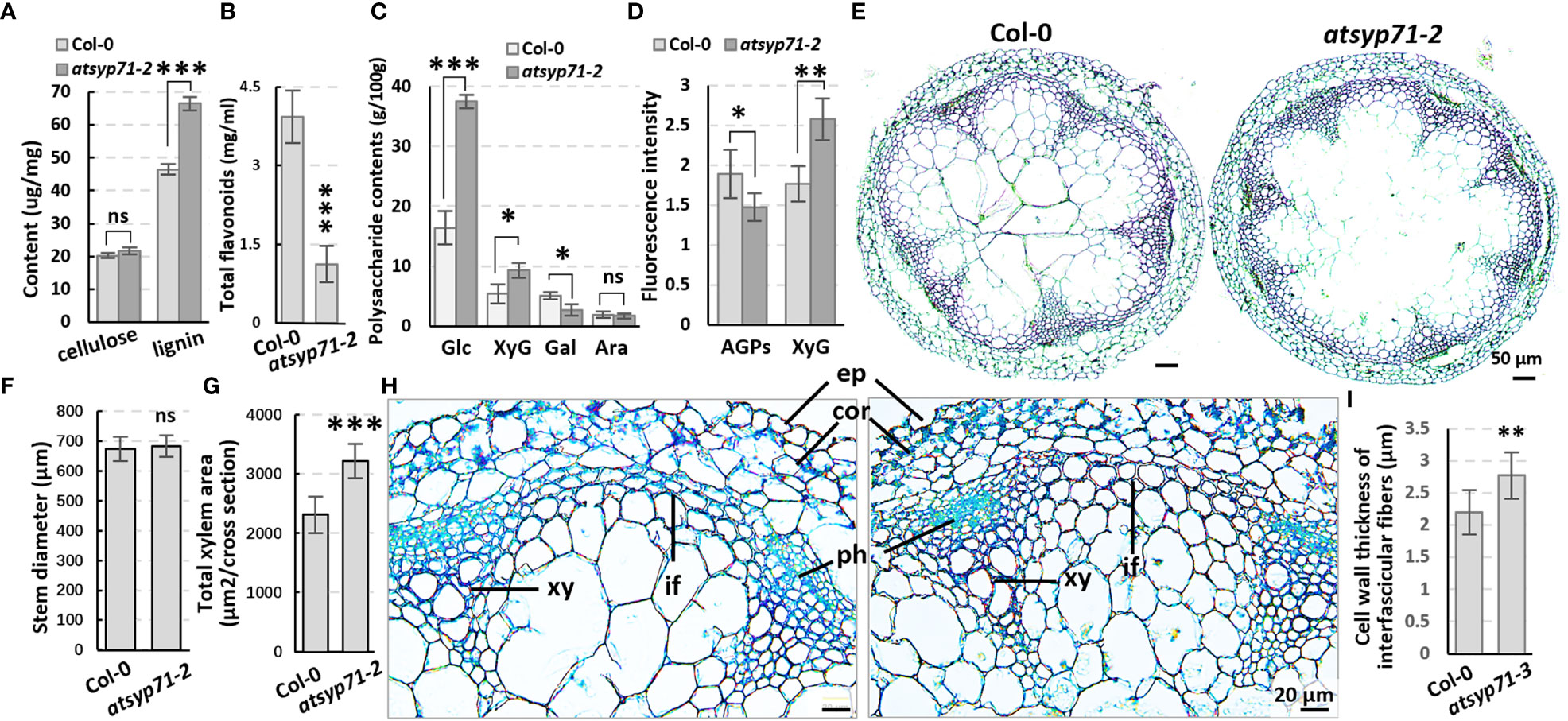
Figure 4 The cell wall components and structure altered in atsyp71-2 mutant. (A-C) Statistics of contents of lignin, cellulose (A), polysaccharides (B) and total flavonoids (C) in the first section of stems from ten-week-old plants. Three biological replicates per sample. (D) Statistics of fluorescence intensity of Immunolabeling of AGPs with anti-LM2 antibody and XXXG xyloglucan with anti-LM15 antibody, respectively.nCol-0 = 6, natsyp71-2 = 7. (E) The paraffin sections with Safranine and Fast Green double staining of stems from the same batch as those in (A). (F, G) Statistics of stem diameter (E) and total xylem area (F) shown representatively in (D). n≥8 stems. (H) Magnified images of the stem cross sections. (I) Statistics of cell wall thickness of interfascicular fiber cells. n≥50 cells. *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test. Ara, arabinose; Gal, galactose; Glu, glucose; XyG, xyloglucan; co, cortex; ep, epidermis; if, interfascicular fibers; ph, phloem; xy, xylem. ns, no significance.
Transcriptome analysis indicates that in BP, a large amount of the DEGs were enriched in processes of stimuli/stress response (Figure 2A, red arrows), indicating AtSYP71 is essential for plant stress adaption. Therefore, we investigated the redox status of atsyp71 mutants. The activities of antioxidases e.g. peroxidase (POD), superoxide dismutase (SOD) and superoxide catalase (CAT) significantly increased in atsyp71-2 (Figure 5A). The DAB and NBT staining results revealed that H2O2 and levels in atsyp71-2 decreased significantly compared with those in Col-0 (Figures 5B, C). These results indicate that ROS homeostasis was disturbed due to knockdown of AtSYP71. Then, we checked stress response of atsyp71-2. Under 75 μm H2O2 treatment, root length of Col-0 declined by about 13%, whereas that of atsyp71-2 and atsyp71-3 only declined by 2-5%. Under treatment of 0.1 μM methylviologen (MV, donor of ), root length of Col-0 declined dramatically by about 71%, however, that of atsyp71 mutants only declined by 17-19%. While, under 0.1 μM MV + 75 μm H2O2 treatment, root length of both Col-0 and atsyp71 mutants decreased more sharply, that of Col-0 declined by about 80%, and that of atsyp71 mutants declined by 41-43% (Figures 5D-F). These results indicate that the ROS homeostasis was seriously interrupted in atsyp71 mutants. Under 120 mM NaCl and 150 mM mannitol treatments, root length of Col-0 and AtSYP71 OE seedlings reduced significantly, but that of atsyp71-2 didn’t change obviously (Figures 5G-J). These results suggest that AtSYP71 modulates ROS homeostasis and affects plant stress response.
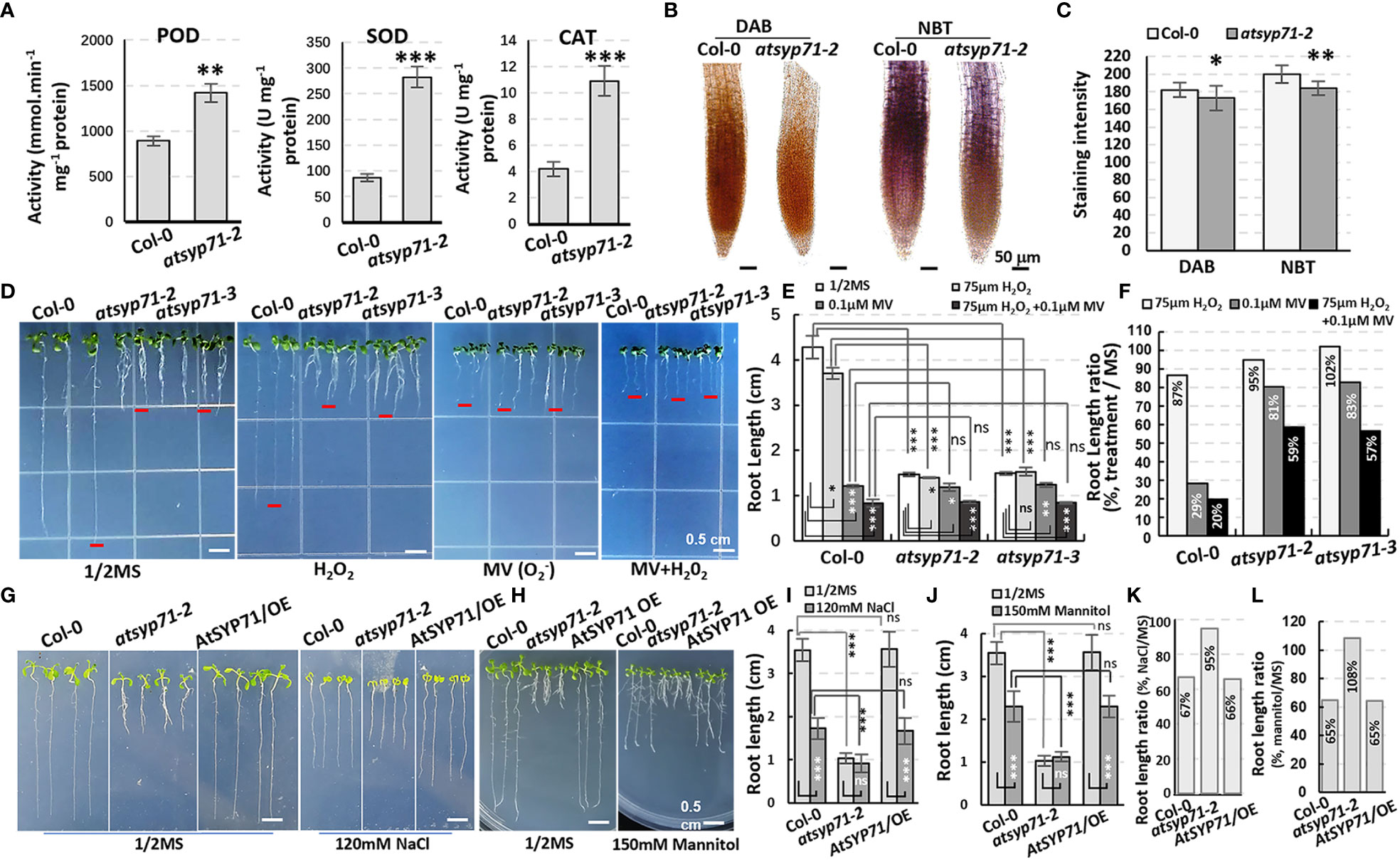
Figure 5 Alteration of ROS homeostasis and stress response in atsyp71 mutants. (A) Statistics of antioxidase activities in roots of nine-day-old Col-0 and atsyp71-2 seedlings. (B) DAB and NBT staining of roots of Col-0 and atsyp71-2 nine-day-old seedlings. (C) Statistics of DAB and NBT staining intensities measured by Photoshop 2019. (D) Treatments on Col-0 and atsyp71 nine-day-old seedlings with 75 μm H2O2, 0.1 μM MV, or 75 μm H2O2 + 0.1 μM MV. n≈50. Three biological replicates per sample. (E) Statistics of root length shown representatively in (D). (F) Statistics of ratio of root length in (E). (G) Seven-day-old seedlings under 120 mM NaCl treatment. (H) Nine-day-old seedlings under 150 mM mannitol treatment. (I, J) Statistics of root length in (G, H), respectively. n≈50. Three biological replicates per sample. (K, L) Statistics of ratio of root length in (I, J), respectively. ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test.
Since GO Enrichment analysis revealed a DEG enrichment in Response to acid chemical process (Figure 2A, green arrow), we further investigated growth of atsyp71 mutants under different pH value. In 1/2MS medium with pH7 condition, the root length of both Col-0 and atsyp71-2 increased significantly. Under pH8 condition, root growth of Col-0 largely recovered, however, root growth of atsyp71-2 increased more significantly than that at pH5.8, and at pH9, root growth of Col-0 was inhibited significantly, however, root length of atsyp71-2 was still significantly higher than that at pH5.8 (Figures 6A-C), means that atsyp71-2 root growth was better under alkaline conditions. Therefore, we speculated that atsyp71-2 roots probably secreted more acidic substances to the apoplast to alleviate the inhibition of root growth under alkali stress. To verify our speculation, bromocresol purple, a pH indicator, was used to detect the acidification of the medium growing atsyp71-2 mutants and Col-0. As expected, the medium around atsyp71-2 and atsyp71-3 roots showed higher acidification than that around Col-0 and AtSYP71 OE roots (Figure 6D). These results demonstrate that AtSYP71 may affect root pH homeostasis. Then, we added MES [2-(N-morpholino) ethanesulfonic acid], a broadly used Good’s buffer, to 1/2MS medium to observe its effects on root growth. To obtain the appropriate MES concentration for treatment, 0.1% and 0.5% MES were tested. It was found that at 0.5% MES condition, root length of atsyp71 mutants was recovered better that that at 0.1% MES condition (Supplementary Figure 4). Thus, 0.5% MES was used for subsequent experiments.
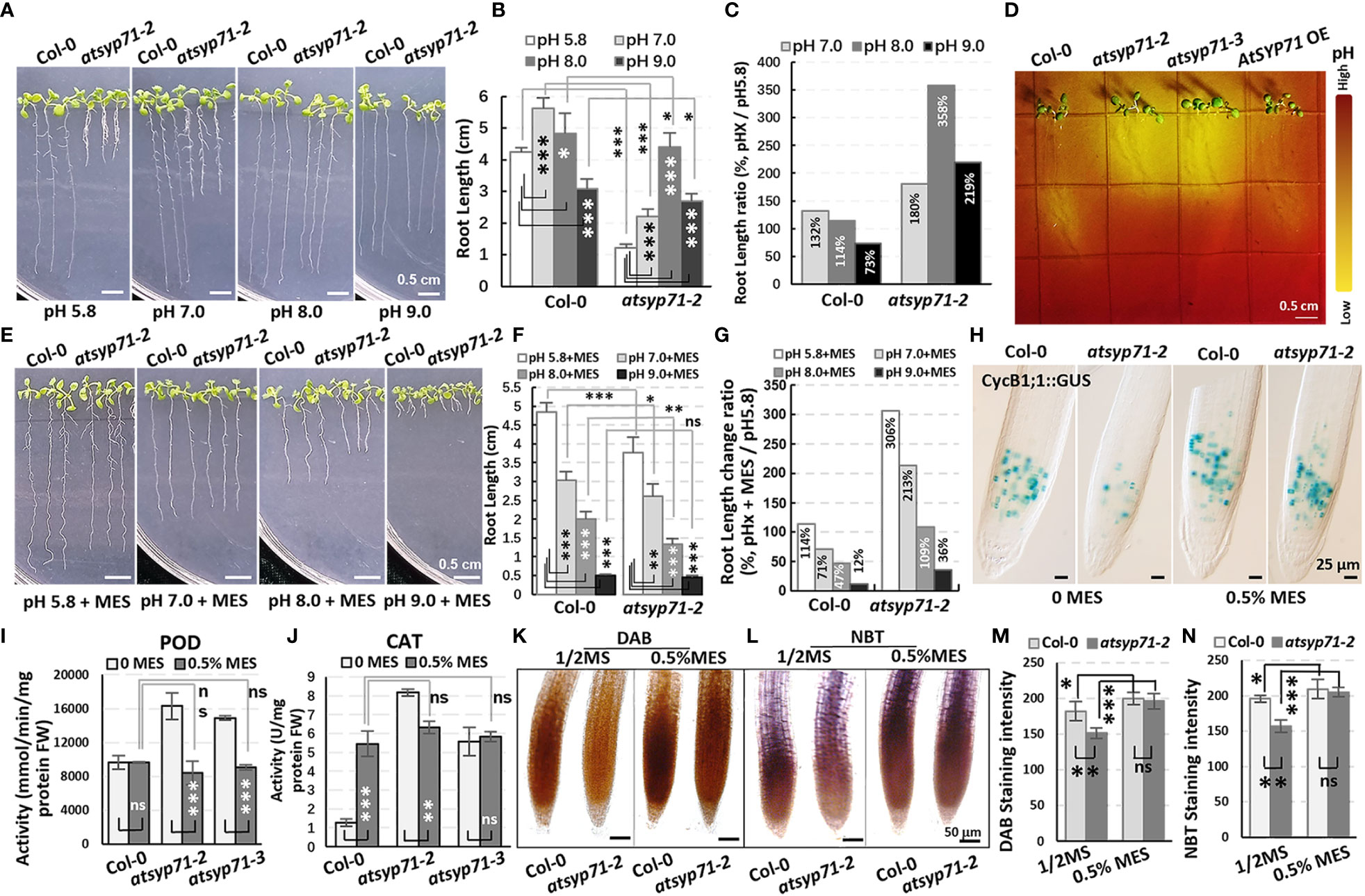
Figure 6 Alkali stress response altered in atsyp71 mutants. (A) Phenotype of nine-day-old seedlings grown on 1/2MS medium with different pH value. (B) Statistics of root length of seedlings in (A). (C) Statistics of ratio of root length in (B). (D) Visualization of root acidification of Col-0, atsyp71 mutants nd AtSYP71 OE seedlings using the pH indicator, bromocresol purple. Five-day-old seedlings grown on 1/2MS medium (pH5.8) were transferred to 1/2MS medium (pH 6.8) containing 0.006% (w/v) bromocresol purple, and photographed three-day after transfer. (E) Phenotype of nine-day-old seedlings grown on 1/2MS medium containing 0.5% MES with different pH value. (F) Statistics of root length of seedlings in (E). (G) Statistics of ratio of root length in (F). (H) Nomarski images of CycB1;1::GUS-expressing cells in roots of nine-day-old Col-0 and atsyp71-2 seedlings. (I, J) POD (H) and CAT (I) activities of nine-day-old seedlings grown on 1/2MS medium (pH5.8) with or without 0.5% MES. (K, L) DAB (K) and NBT (L) staining of nine-day-old seedlings grown on 1/2MS medium (pH5.8) with or without 0.5% MES. (M, N) Statistics of staining intensities in (K) and (L). ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001. Student’s t test.
Under pH5.8 with 0.5% MES condition, the root length of both Col-0 and atsyp71-2 increased significantly compared with that at pH5.8, while, the root elongation of atsyp71-2 (by 306%) was more significant than that of Col-0 (by 114%). At pH7 and pH8 with 0.5% MES, the root growth of Col-0 was inhibited and the root length was significantly shorter than that at pH5.8 (by 71% and 47%, respectively), whereas root length of atsyp71-2 was still significantly longer than that at pH5.8 (by 216% and 109%, respectively). And under pH9 with 0.5% MES, root growth of both Col-0 and atsyp71-2 was inhibited seriously (Figures 6E-G). To study root apical meristem (RAM) activity under adding MES condition, we observed the mitotic marker CycB1;1::GUS which was introduced into atsyp71-2 by crossing. The GUS staining results revealed that under pH5.8, the RAM activity in atsyp71-2 roots was seriously inhibited compared with that in Col-0, but when added 0.5% MES, the RAM activity in atsyp71-2 was restored and was close to the level in Col-0 (Figure 6H). These results suggest that AtSYP71 regulates root development via modulating pH homeostasis. To figure out whether pH homeostasis is connected with ROS homeostasis, we checked ROS level in atsyp71 roots under MES condition. First, we determinated antioxidase activities of the seedlings grown on 1/2MS medium (pH5.8) with or without 0.5% MES. At pH5.8 with 0.5% MES, the POD activities of Col-0 did not change significantly, whereas that of atsyp71 mutants decreased significantly compared with that at pH5.8. There was no significant difference in POD activities between Col-0 and atsyp71 mutants after adding 0.5% MES (Figure 6I). On the other hand, in the presence of 0.5% MES, the CAT activities of Col-0 increased significantly, while, that of atsyp71-2 reduced significantly compared with that without MES. And there was no significant difference in CAT activities between Col-0 and atsyp71 mutants after adding 0.5% MES (Figure 6J). The DAB and NBT staining revealed that under 0.5% MES condition, H2O2 and levels in both Col-0 and atsyp71-2 increased significantly, but there was little difference between Col-0 and atsyp71-2 (Figures 6K-N). These results indicate that MES buffered the excess H+ produced by atsyp71-2 roots and consequently restored the ROS level, suggesting that AtSYP71 affects ROS homeostasis and root development via regulating pH homeostasis.
Since cell wall biosynthesis and acid substance secretion are regulated by secretion pathway, we observed the localization of the secretory marker SecGFP (Batoko et al., 2000; Leucci et al., 2007), in atsyp71-2 root cells. Confocal images revealed that in atsyp71-2 root cells, SecGFP displayed apparent cytoplasmic localization which was absent in Col-0 (Figure 7A, arrows), indicating that part of SecGFP was blocked in secretion pathway, suggesting that AtSYP71 plays crucial role on secretion.
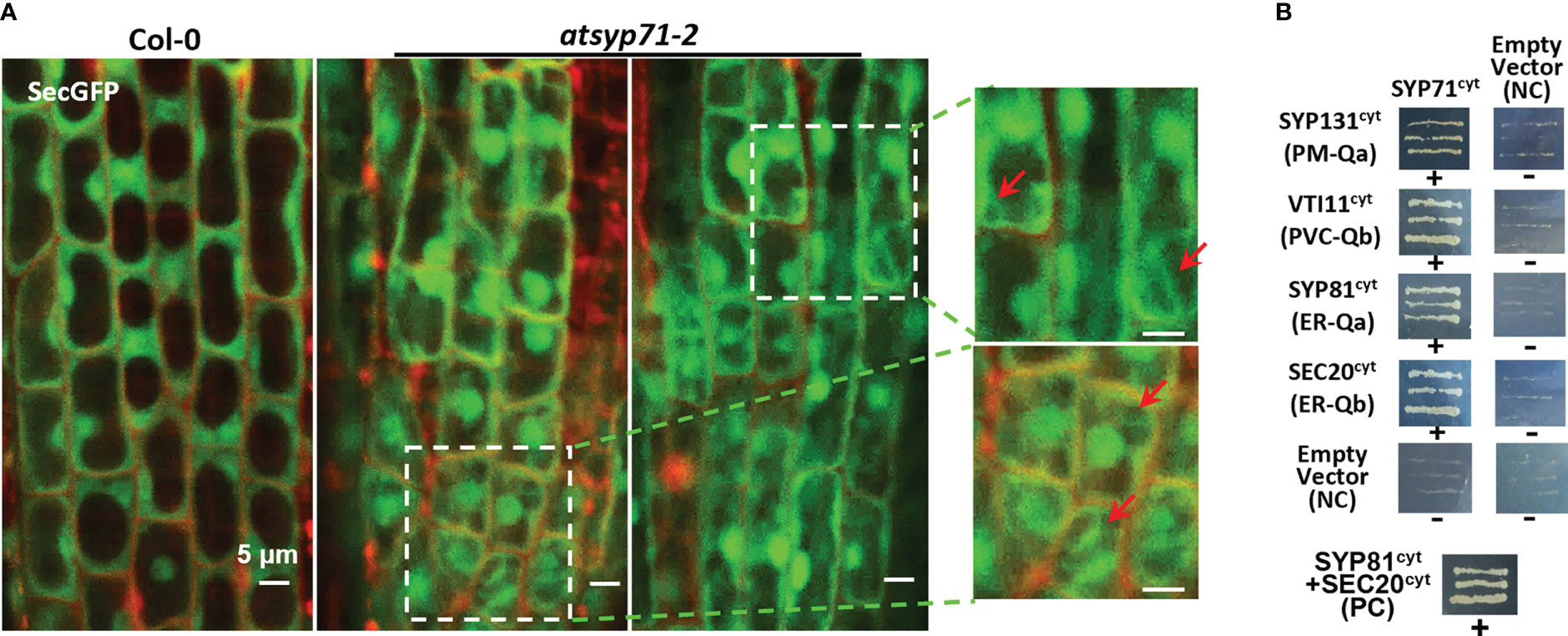
Figure 7 AtSYP71 regulated secretion. (A) Confocal images of SecGFP in Col-0 and atsyp71-2 with PI staining. The magnified images on the right are the part with white boxes. Arrows indicate the cytoplasmic localization of Sec-GFP. (B) Yeast two hybrid analysis of AtSYP71 interactors. Yeast strain AH109 was transformed with the paired constructs as shown. Transformants were streaked onto SD/_Leu/_Trp/_His/_Ade medium. AtSYP81/pGADT7 vs AtSec20/pGBKT7 served as a positive control. Each construct and its corresponding empty vector were used as negative controls. cyt, cytoplasmic fragment; FL, full length. +, has an interaction; -, no interaction.
Then, we identified AtSYP71- associating proteins. AtSYP71 is reported to localize on the ER, endosome, plasma membrane and cell plate (Alexandersson et al., 2004; Marmagne et al., 2004; Mongrand et al., 2004; Uemura et al., 2004; Morel et al., 2006), suggesting its multiple functions. During cytokinesis, AtSYP71 associates with Qa-SNARE KNOLLE, Qb-SNARE NPSN11, and R-SNARE VAMP721/722 to form a tetrameric SNARE complex in cell plate (El Kasmi et al., 2013). To further identify AtSYP71 partners, we generated myc-AtSYP71/atsyp71-2 lines by crossing myc-AtSYP71 (AtSYP71OE) line with atsyp71-2 mutant. Immunoblot analysis indicates recovery of AtSYP71 protein level and root length compared with atsyp71-2 (Supplementary Figure 5), implying that myc-AtSYP71 proteins were functional. Then, we performed pull down of myc-AtSYP71 and identified PM-localized Qa-SNARE SYP131, SYP121 and SYP122, PM-Qb-SNARE NPSN11 and NPSN12, prevacuolar compartment (PVC)-localized Qb-SNARE VTI11 and SYP22 (Table 1). Among these factors, SYP121/SYP122, NPSN11/NPSN12, and SYP22 are demonstrated to be co-immunoprecipitated with AtSYP71 (Fujiwara et al., 2014). For the rest unclarified factors, VTI11 and SYP131, we performed yeast two hybrid (Y2H) analysis. As expected, Y2H results confirmed the interactions of AtSYP71 with VTI11 and SYP131, respectively (Figure 7B). AtSYP71 is also localized at the ER, but the pull down products did not include ER-localized SNAREs, suggesting the interaction of AtSYP71 with the ER-SNARE was minor. Therefore, we performed pull down-LC-MS/MS analysis using myc-tagged ER-Qa-SNARE AtSYP81-overexpressing plants. The elution products include AtSYP71, and ER-Qb-SNARE SEC20, as well as the PM- and PVC-localized AtSYP71 partner proteins (Table 2). Y2H confirmed the interactions of AtSYP71 with AtSYP81 and SEC20, respectively (Figure 7B). These results suggest that during interphase, AtSYP71 may form distinct SNARE complexes with different set of SNAREs to mediate membrane fusion of multiple steps in secretory or vacuole-targeting pathways, respectively.
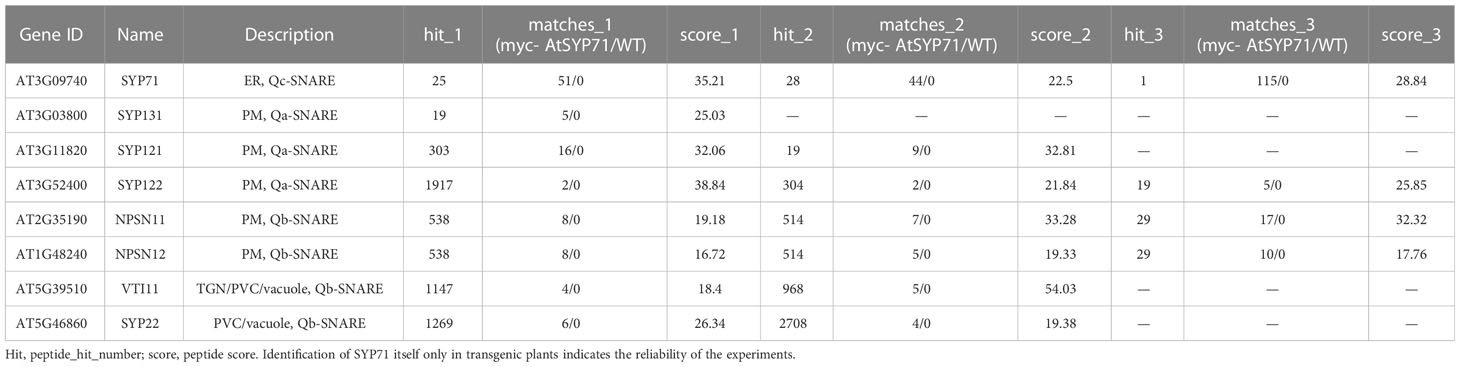
Table 1 Pull down-LC-MS/MS analysis identified AtSYP71-associated proteins related to vesicle transport using myc-AtSYP71-overexpressing plants.
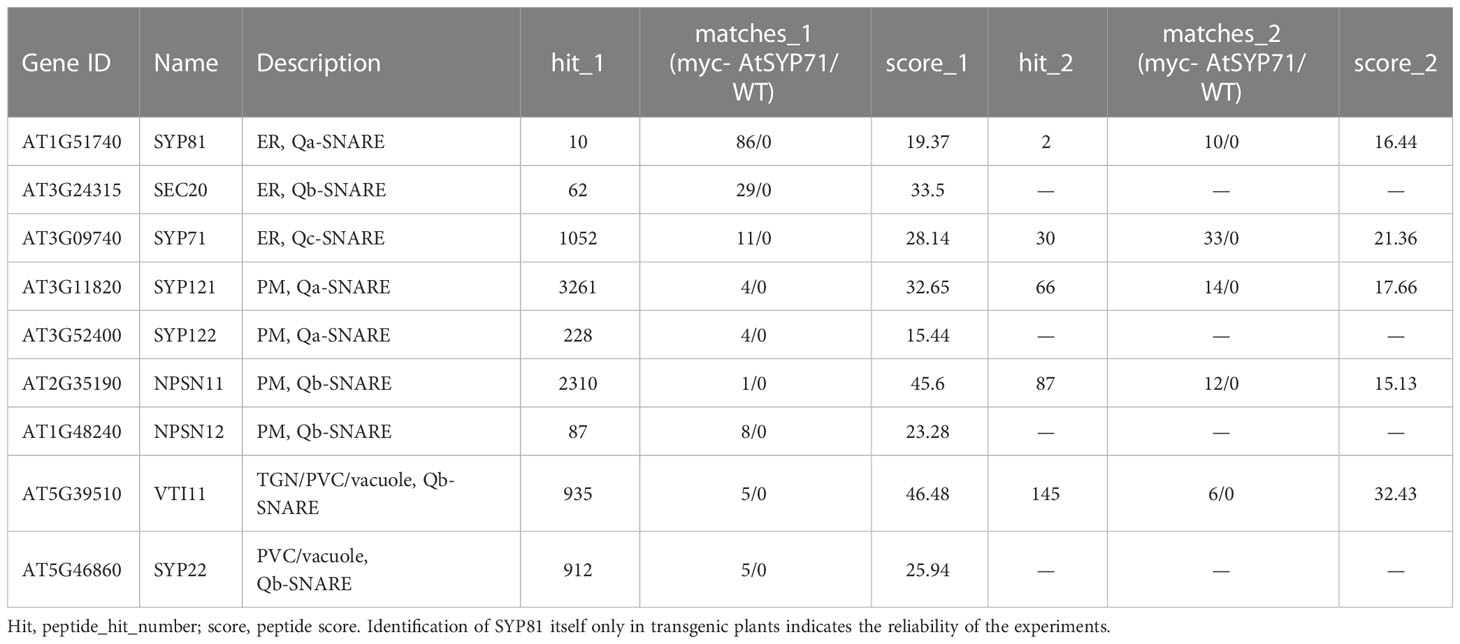
Table 2 Pull down-LC-MS/MS analysis identified AtSYP81-associated proteins which are related to vesicle trafficking using myc-AtSYP81-overexpressing plants.
AtSYP71 has an unusual multiple localization on the ER, endosome, PM and cell plate, the compartments in the secretory pathways (Suwastika et al., 2008; El Kasmi et al., 2013). It is clarified that AtSYP71 binds with Qa-KNOLLE, Qb-SNARE NPSN11 and R-SNAREs VAMP721 and VAMP722 to form a SNARE complex regulating cytokinesis (El Kasmi et al., 2013). AtSYP71 is also demonstrated to be co-immunoprecipitated with SYP121/SYP122, NPSN11/NPSN12, and SYP22 (Fujiwara et al., 2014). In this study, we found that in addition to the mentioned factors, AtSYP71 interacted with PVC-localized SNARE, VTI11, suggesting that AtSYP71 may be also involved in vacuole-targeting pathway. Considering importance of AtSYP71 functions, deficiency of AtSYP71 will definitely seriously affect AtSYP71-dependent vesicle trafficking and organelle functions. Lethality of the knock-out mutant atsyp71-1 (Figure 1C) resembled the phenotype of syp71amiR and npsn11 syp71amiR, the cytokinesis defective mutants (El Kasmi et al., 2013). The failure of morphogenesis may be due to the blocked delivery of materials required for cell plate formation, implying essential role of AtSYP71 in secretion and development in Arabidopsis. On the contrary, Ljsyp71 mutants grow similarly to wild-type plant when supplied with combined nitrogen. OsSYP71 and TaSYP71 deficient mutants didn’t show obvious phenotypes under normal growth conditions (Bao et al., 2012; El Kasmi et al., 2013). These suggest that LjSYP71, OsSYP71 and TaSYP71 are not essential for plant growth and development. This probably because that SYP71 orthologues in different species gain functional division during evolutionary.
The knock-down mutants, atsyp71-2 and atsyp71-3, also displayed developmental defects. While, the short root phenotype was rescued by alkali pH and 0.5% MES which buffers H+ (Figure 6). The acidification of atsyp71-2 and atsyp71-3 rhizosphere indicate that the pH homeostasis was affected, suggesting that homeostasis of PM-localized ion channels and other PM-residents may also be disturbed. For example, SLAH3 mainly inhibit the inward-rectifying K+ channel KAT1 by protein-protein interaction and consequently prevent stomatal opening (Zhang et al., 2016). CIPK3 regulates K+ homeostasis through activating vacuolar K+ efflux to the cytoplasm (Tang et al., 2020). Transcriptional levels of these two genes altered significantly in atsyp71-2 (Supplementary Figure 6A), suggesting AtSYP71 effects on ion homeostasis. The fundamental reason of unbalanced homeostasis in the mutants was the block of secretory and cycling pathways which maintain homeostasis of PM-localized ion channels and other residents, as well as exocytosis of cell wall biosynthetic materials which led to changes of cell wall structure and components (Figure 4; Supplementary Figure 3).
Knockdown of AtSYP71 definitely impacted the organelle functions. The ER and the Golgi apparatus are responsible for protein glycosylation. Thus, post transcriptional modification of the glycoproteins, e.g. laccases, PRXs and AGPs, were likely disrupted in atsyp71 mutants. Transcriptome analysis indicate that the expression of PRX4/15/25/37/49/52/53/54/62, LAC12 and AGP1 genes decreased significantly (Figure 3), and the protein level of AGPs were also reduced significantly in atsyp71-2 (Figure 4D). Furthermore, the expression of UDP-glucosyltransferase, UGT74E2 and AT2G18560, the cell wall biosynthesis regulators, were also altered significantly (Supplementary Figure 6B). These changes validate effects of AtSYP71 on the regulatory machinery of cell wall biosynthesis. In addition, expression of many cell wall dynamics regulatory genes were also affected in atsyp71-2. For example, EXPA proteins are responsible for cell wall extension and induce stress relaxation and extension of cell wall in a pH-dependent manner (Cosgrove, 2000). XTH proteins catalyze molecular grafting and/or hydrolysis of cell wall xyloglucans (Jiang et al., 2020), and their function are pH-dependent (Cosgrove, 2000). LTP2 plays a role in maintaining the integrity of the cuticle-cell wall interface (Julke and Ludwig-Muller, 2015). PP2-B13, is a phloem protein 2 (PP2)-like protein. PP2 is a component of the phloem protein bodies, directly bind with the chitin cell wall and play important roles in defense and wound healing (Beneteau et al., 2010). BGLU25 belongs to beta-glucosidase family which is involved in cellulose degradation (Roepke and Bozzo, 2015; Han et al., 2020). Expression of these genes altered significantly (Figure 3), suggesting AtSYP71 regulatory role on cell wall dynamics. Among the genes, EXPA and XTH proteins function in a pH-dependent manner (Cosgrove, 2000; Jiang et al., 2020), and excessive acidification of atsyp71 mutants likely destroyed the optimal working pH of these kinds of proteins and resulted in defects on cell elongation.
Abiotic stresses induce high level of ROS production. The ROS scavenging system includes enzymatic and non-enzymatic antioxidant (You and Chan, 2015). The enzymatic antioxidants include SOD, CAT, POD, APX, glutathione peroxidase (GPX), glutathione-S-transferase (GST) etc. which reduce and H2O2 (Gill and Tuteja, 2010; Nadarajah, 2020). SODs have diverse forms containing distinct metal ions such as Fe, Mn or Cu/Zn, and are distributed in the apoplast (Kim et al., 2008), cytosol, chloroplasts, mitochondria and peroxisomes (Pilon et al., 2011). Respiratory burst oxidase homologues (RBOHs), the plant NADPH oxidases (NOXs), are localized on the PM and convert oxygen to in the apoplast (Segal, 2016). Under stress conditions, /H2O2 production occurs in cytoplasmic vesicles derived from the PM or ER (Leshem et al., 2006), and RBOHD is internalized into PM-derived vesicles (Hao et al., 2014). Oxidation also occurs inside the ER lumen to promote protein folding depending on disulfide bonds formation by a FAD-containing ER oxidase (ERO) and protein disulfide isomerases (PDIs) (Bulleid, 2012). The homeostasis of the organelles determines ROS homeostasis, thus, dysfunction of the ER and PM in atsyp71-2 definitely affected ROS homeostasis (Figure 5).
Apoplastic H2O2 influences PRX-mediated lignification and cross-linking of cell wall polymers (Marjamaa et al., 2009; Shigeto et al., 2015). In atsyp71-2, in addition to PRXs, expression of many redox-related genes altered significantly, such as AOC3 (endothelial amine oxidase), POX1 (proline oxidase family FAD-linked oxidoreductase), AT4G20830 and AT5G44400 (FAD linked oxidase homologues), LAC1 (multicopper oxidase) and AT1G31710 (Copper amine oxidase homologue) (Supplementary Figure 6B). The changes in these enzyme activities may be one of the reason of unbalanced ROS homeostasis. And the significantly increased lignin content was probably the consequence of the unbalanced ROS homeostasis in atsyp71-2.
The non-enzymatic antioxidants contain ascorbic acids, α-tocopherol, flavonoids, phenolic compounds, glutathione, carotenoids and lipids, which mitigate oxidative damage by their antioxidant activities through utilization of H2O2 (Duan et al., 2012; Kim et al., 2020b). Flavonoids distribute widely in plants and are one of the most bioactive plant secondary metabolites. Flavonoids are synthesized on the ER surface and uploaded into the ER lumen, and subsequently transported to the vacuole or is secreted to the apoplast (Zhao and Dixon, 2010; Falcone Ferreyra et al., 2012; Zhao, 2015). The PM-localized ABCG transporters mediated flavonoid efflux (Banasiak et al., 2013; Liu et al., 2022b). In atsyp71-2, expression of ABCG1 decreased significantly (Figure 3); in response, content of flavonoids decreased significantly (Figure 4B). Dysfunction of the ER and PM in atsyp71-2 definitely affected flavonoids production and secretion, which subsequently affected stress response of the mutants.
The original contributions presented in the study are publicly available. This data can be found here: The National Center for Biotechnology Information with Bioproject ID PRJNA971388 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA971388.
LL conceived and supervised the project. LL, HZ, JZ and XK designed the experiments. HZ, JZ, XK, YL, XZ, MW, WL, YH, XTW, GTQ and ZZ generated materials and performed the experiments. GCQ performed the LC-MS/MS analysis. SL and XQW determinated the cell wall polysaccharides. LL, HZ, JZ and XK drafted the manuscript. LJ, XF and J-KZ cooperated and supervised the project.
This work was supported by the National Natural Science Foundation of China (32170279) and Fundamental Research Funds for the Central Universities (2572019CT03). The funders have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
We would like to thank Prof. Xugang Li (Shandong Agricultural University) for donation of SecGFP marker line.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1198353/full#supplementary-material
Supplementary Figure 1 | Overview of transcriptome analysis.
Supplementary Figure 2 | Statistics of RT-qPCR analysis of DEG expression levels in atsyp71-3.
Supplementary Figure 3 | Detection of contents of AGPs and XyG.
Supplementary Figure 4 | Effects of different MES concentration on seedling development.
Supplementary Figure 5 | myc-AtSYP71 protein has functionality.
Supplementary Figure 6 | Transcriptomic analysis of ion channels (A) and redox-related genes (B).
Supplementary Table 1 | Original transcriptomic data of atsyp71-2 vs Col-0.
Supplementary Table 2 | Information of DEGs enriched in cell wall biosynthesis- and dynamics-related pathways.
Supplementary Table 3 | Primer information used in this study.
Adnan, M., Islam, W., Zhang, J., Zheng, W., Lu, G. D. (2019). Diverse role of SNARE protein Sec22 in vesicle trafficking, membrane fusion, and autophagy. Cells 8, (4). doi: 10.3390/cells8040337
Alejandro, S., Lee, Y., Tohge, T., Sudre, D., Osorio, S., Park, J., et al. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22 (13), 1207–1212. doi: 10.1016/j.cub.2012.04.064
Alexandersson, E., Saalbach, G., Larsson, C., Kjellbom, P. (2004). Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45 (11), 1543–1556. doi: 10.1093/pcp/pch209
Almagro, L., Gomez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barcelo, A., Pedreno, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60 (2), 377–390. doi: 10.1093/jxb/ern277
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11 (10), R106. doi: 10.1186/gb-2010-11-10-r106
Antonin, W., Riedel, D., von Mollard, G. F. (2000). The SNARE Vti1a-beta is localized to small synaptic vesicles and participates in a novel SNARE complex. J. Neurosci. 20 (15), 5724–5732. doi: 10.1523/jneurosci.20-15-05724.2000
Banasiak, J., Biala, W., Staszków, A., Swarcewicz, B., Kepczynska, E., Figlerowicz, M., et al. (2013). A medicago truncatula ABC transporter belonging to subfamily G modulates the level of isoflavonoids. J. Exp. Bot. 64 (4), 1005–1015. doi: 10.1093/jxb/ers380
Bao, Y. M., Sun, S. J., Li, M., Li, L., Cao, W. L., Luo, J., et al. (2012). Overexpression of the qc-SNARE gene OsSYP71 enhances tolerance to oxidative stress and resistance to rice blast in rice (Oryza sativa l.). Gene 504 (2), 238–244. doi: 10.1016/j.gene.2012.05.011
Bard, G. C., Zottich, U., Souza, T. A., Ribeiro, S. F., Dias, G. B., Pireda, S., et al. (2016). Purification, biochemical characterization, and antimicrobial activity of a new lipid transfer protein from Coffea canephora seeds. Genet. Mol. Res.: GMR 15 (4). doi: 10.4238/gmr15048859
Batoko, H., Zheng, H. Q., Hawes, C., Moore, I. (2000). A Rab1 GTPase is required for transport between the. Plant Cell 12, 17. doi: 10.1104/pp.112.200782
Benedetti, M., Verrascina, I., Pontiggia, D., Locci, F., Mattei, B., De Lorenzo, G., et al. (2018). Four Arabidopsis berberine bridge enzyme-like proteins are specific oxidases that inactivate the elicitor-active oligogalacturonides. Plant J. 94 (2), 260–273. doi: 10.1111/tpj.13852
Beneteau, J., Renard, D., Marché, L., Douville, E., Lavenant, L., Rahbé, Y., et al. (2010). Binding properties of the n-acetylglucosamine and high-mannose n-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 153 (3), 1345–1361. doi: 10.1104/pp.110.153882
Bobbili, K. B., Pohlentz, G., Narahari, A., Sharma, K., Surolia, A., Mormann, M., et al. (2018). Coccinia indica agglutinin, a 17kDa PP2 like phloem lectin: Affinity purification, primary structure and formation of self-assembled filaments. Int. J. Biol. Macromol. 108, 1227–1236. doi: 10.1016/j.ijbiomac.2017.11.024
Bui, S., Mejia, I., Díaz, B., Wang, Y. (2021). Adaptation of the golgi apparatus in cancer cell invasion and metastasis. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.806482
Bulleid, N. J. (2012). Disulfide bond formation in the mammalian endoplasmic reticulum. Cold Spring Harbor Perspect. Biol. 4, (11). doi: 10.1101/cshperspect.a013219
Chae, K., Kieslich, C. A., Morikis, D., Kim, S. C., Lord, E. M. (2009). A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell. 21 (12), 3902–3914. doi: 10.1105/tpc.109.070854
Chandrasekar, B., van der Hoorn, R. A. (2016). Beta galactosidases in Arabidopsis and tomato - a mini review. Biochem. Soc. Trans. 44 (1), 150–158. doi: 10.1042/bst20150217
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 29 (6), 1425–1439. doi: 10.1105/tpc.17.00364
Chen, Q., Xu, X., Xu, D., Zhang, H., Zhang, C., Li, G. (2019). WRKY18 and WRKY53 coordinate with HISTONE ACETYLTRANSFERASE1 to regulate rapid responses to sugar. Plant Physiol. 180 (4), 2212–2226. doi: 10.1104/pp.19.00511
Cosgrove, D. J. (2000). Loosening of plant cell walls by expansins. Nature 407 (6802), 321–326. doi: 10.1038/35030000
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 (11), 850–861. doi: 10.1038/nrm1746
Cui, L., Li, H., Xi, Y., Hu, Q., Liu, H., Fan, J., et al. (2022). Vesicle trafficking and vesicle fusion: mechanisms, biological functions, and their implications for potential disease therapy. Mol. biomed. 3 (1), 29. doi: 10.1186/s43556-022-00090-3
Dhara, A., Raichaudhuri, A. (2021). ABCG transporter proteins with beneficial activity on plants. Phytochemistry 184, 112663. doi: 10.1016/j.phytochem.2021.112663
Didi, V., Jackson, P., Hejatko, J. (2015). Hormonal regulation of secondary cell wall formation. J. Exp. Bot. 66 (16), 5015–5027. doi: 10.1093/jxb/erv222
Dixon, R. A., Barros, J. (2019). Lignin biosynthesis: old roads revisited and new roads explored. Open Biol. 9 (12), 190215. doi: 10.1098/rsob.190215
Duan, J., Zhang, M., Zhang, H., Xiong, H., Liu, P., Ali, J., et al. (2012). OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa l.). Plant Sci. 196, 143–151. doi: 10.1016/j.plantsci.2012.08.003
El Kasmi, F., Krause, C., Hiller, U., Stierhof, Y. D., Mayer, U., Conner, L., et al. (2013). SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell. 24 (10), 1593–1601. doi: 10.1091/mbc.E13-02-0074
Falcone Ferreyra, M. L., Rius, S. P., Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00222
Fasshauer, D., Margittai, M. (2004). A transient n-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem. 279 (9), 7613–7621. doi: 10.1074/jbc.M312064200
Fasshauer, D., Sutton, R. B., Brunger, A. T., Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as q- and r-SNAREs. Proc. Natl. Acad. Sci. U S A 95 (26), 15781–15786. doi: 10.1073/pnas.95.26.15781
Ferrer, J. L., Austin, M. B., Stewart, C., Jr., Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46 (3), 356–370. doi: 10.1016/j.plaphy.2007.12.009
Francoz, E., Ranocha, P., Nguyen-Kim, H., Jamet, E., Burlat, V., Dunand, C. (2015). Roles of cell wall peroxidases in plant development. Phytochemistry 112, 15–21. doi: 10.1016/j.phytochem.2014.07.020
Frankova, L., Fry, S. C. (2013). Biochemistry and physiological roles of enzymes that 'cut and paste' plant cell-wall polysaccharides. J. Exp. Bot. 64 (12), 3519–3550. doi: 10.1093/jxb/ert201
Frappaolo, A., Karimpour-Ghahnavieh, A., Sechi, S., Giansanti, M. G. (2020). The close relationship between the golgi trafficking machinery and protein glycosylation. Cells 9, (12). doi: 10.3390/cells9122652
Fry, S. C. (2004). Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol. 161 (3), 641–675. doi: 10.1111/j.1469-8137.2004.00980.x
Fujiwara, M., Uemura, T., Ebine, K., Nishimori, Y., Ueda, T., Nakano, A., et al. (2014). Interactomics of qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 55 (4), 781–789. doi: 10.1093/pcp/pcu038
Gao, Q. M., Venugopal, S., Navarre, D., Kachroo, A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155 (1), 464–476. doi: 10.1104/pp.110.166876
Geng, P., Zhang, S., Liu, J., Zhao, C., Wu, J., Cao, Y., et al. (2020). MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 182 (3), 1272–1283. doi: 10.1104/pp.19.01070
Gigli-Bisceglia, N., Engelsdorf, T., Hamann, T. (2020). Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell Mol. Life Sci. 77 (11), 2049–2077. doi: 10.1007/s00018-019-03388-8
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 (12), 909–930. doi: 10.1016/j.plaphy.2010.08.016
Guan, L., Yang, S., Li, S., Liu, Y., Liu, Y., Yang, Y., et al. (2021). AtSEC22 regulates cell morphogenesis via affecting cytoskeleton organization and stabilities. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.635732
Hakoyama, T., Oi, R., Hazuma, K., Suga, E., Adachi, Y., Kobayashi, M., et al. (2012). The SNARE protein SYP71 expressed in vascular tissues is involved in symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Physiol. 160 (2), 897–905. doi: 10.1104/pp.112.200782
Han, Y., Watanabe, S., Shimada, H., Sakamoto, A. (2020). Dynamics of the leaf endoplasmic reticulum modulate beta-glucosidase-mediated stress-activated ABA production from its glucosyl ester. J. Exp. Bot. 71 (6), 2058–2071. doi: 10.1093/jxb/erz528
Hao, H., Fan, L., Chen, T., Li, R., Li, X., He, Q., et al. (2014). Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell. 26 (4), 1729–1745. doi: 10.1105/tpc.113.122358
Heinze, L., Freimuth, N., Rößling, A. K., Hahnke, R., Riebschläger, S., Fröhlich, A., et al. (2020). EPSIN1 and MTV1 define functionally overlapping but molecularly distinct trans-golgi network subdomains in Arabidopsis. Proc. Natl. Acad. Sci. U S A 117 (41), 25880–25889. doi: 10.1073/pnas.2004822117
Huang da, W., Sherman, B. T., Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 (1), 1–13. doi: 10.1093/nar/gkn923
Huang da, W., Sherman, B. T., Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 (1), 44–57. doi: 10.1038/nprot.2008.211
Jiang, Y., Li, Y., Lu, C., Tang, Y., Jiang, X., Gai, Y. (2020). Isolation and characterization of populus xyloglucan endotransglycosylase/hydrolase (XTH) involved in osmotic stress responses. Int. J. Biol. Macromol. 155, 1277–1287. doi: 10.1016/j.ijbiomac.2019.11.099
Julian, J. D., Zabotina, O. A. (2022). Xyloglucan biosynthesis: from genes to proteins and their functions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.920494
Julke, S., Ludwig-Muller, J. (2015). Response of Arabidopsis thaliana roots with altered lipid transfer protein (LTP) gene expression to the clubroot disease and salt stress. Plants (Basel) 5, (1). doi: 10.3390/plants5010002
Kaneda, M., Rensing, K., Samuels, L. (2010). Secondary cell wall deposition in developing secondary xylem of poplar. J. Integr. Plant Biol. 52 (2), 234–243. doi: 10.1111/j.1744-7909.2010.00925.x
Kang, B. H., Nielsen, E., Preuss, M. L., Mastronarde, D., Staehelin, L. A. (2011). Electron tomography of RabA4b- and PI-4Kbeta1-labeled trans golgi network compartments in Arabidopsis. Traffic 12 (3), 313–329. doi: 10.1111/j.1600-0854.2010.01146.x
Kidwai, M., Ahmad, I. Z., Chakrabarty, D. (2020). Class III peroxidase: an indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 39 (11), 1381–1393. doi: 10.1007/s00299-020-02588-y
Kim, D. O., Chun, O. K., Kim, Y. J., Moon, H. Y., Lee, C. Y. (2003). Quantification of polyphenolics and their antioxidant capacity in fresh plums. J. Agric. Food Chem. 51 (22), 6509–6515. doi: 10.1021/jf0343074
Kim, Y., Chung, Y. S., Lee, E., Tripathi, P., Heo, S., Kim, K. H. (2020b). Root response to drought stress in rice (Oryza sativa l.). Int. J. Mol. Sci. 21, (4). doi: 10.3390/ijms21041513
Kim, H. J., Kato, N., Kim, S., Triplett, B. (2008). Cu/Zn superoxide dismutases in developing cotton fibers: evidence for an extracellular form. Planta 228 (2), 281–292. doi: 10.1007/s00425-008-0734-0
Kim, S. H., Lam, P. Y., Lee, M. H., Jeon, H. S., Tobimatsu, Y., Park, O. K. (2020a). The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.583153
Klink, V. P., Sharma, K., Pant, S. R., McNeece, B., Niraula, P., Lawrence, G. W. (2017). Components of the SNARE-containing regulon are co-regulated in root cells undergoing defense. Plant Signal Behav. 12 (2), e1274481. doi: 10.1080/15592324.2016.1274481
Kumar, M., Turner, S. (2015). Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112, 91–99. doi: 10.1016/j.phytochem.2014.07.009
Leshem, Y., Melamed-Book, N., Cagnac, O., Ronen, G., Nishri, Y., Solomon, M., et al. (2006). Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. U. S. A. 103 (47), 18008–18013. doi: 10.1073/pnas.0604421103
Leucci, M. R., Di Sansebastiano, G. P., Gigante, M., Dalessandro, G., Piro, G. (2007). Secretion marker proteins and cell-wall polysaccharides move through different secretory pathways. Planta 225 (4), 1001–1017. doi: 10.1007/s00425-006-0407-9
Lewis, N. G., Yamamoto, E., Wooten, J. B., Just, G., Ohashi, H., Towers, G. H. (1987). Monitoring biosynthesis of wheat cell-wall phenylpropanoids in situ. Sci. (New York NY) 237 (4820), 1344–1346. doi: 10.1126/science.237.4820.1344
Li, X., Chapple, C. (2010). Understanding lignification: challenges beyond monolignol biosynthesis. Plant Physiol. 154 (2), 449–452. doi: 10.1104/pp.110.162842
Li, L., Shimada, T., Takahashi, H., Koumoto, Y., Shirakawa, M., Takagi, J., et al. (2013). MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. Plant J. 76 (5), 781–791. doi: 10.1111/tpj.12347
Li, L., Shimada, T., Takahashi, H., Ueda, H., Fukao, Y., Kondo, M., et al. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell. 18 (12), 3535–3547. doi: 10.1105/tpc.106.046151
Li, Q., Zhou, L., Li, Y., Zhang, D., Gao, Y. (2021). Plant NIGT1/HRS1/HHO transcription factors: key regulators with multiple roles in plant growth, development, and stress responses. Int. J. Mol. Sci. 22, (16). doi: 10.3390/ijms22168685
Liu, H., Gao, J., Sun, J., Li, S., Zhang, B., Wang, Z., et al. (2022a). Dimerization of PtrMYB074 and PtrWRKY19 mediates transcriptional activation of PtrbHLH186 for secondary xylem development in populus trichocarpa. New Phytol. 234 (3), 918–933. doi: 10.1111/nph.18028
Liu, Q., Luo, L., Zheng, L. (2018). Lignins: biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19, (2). doi: 10.3390/ijms19020335
Liu, M., Peng, Y., Li, H., Deng, L., Wang, X., Kang, Z. (2016). TaSYP71, a qc-SNARE, contributes to wheat resistance against puccinia striiformis f. sp. tritici. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00544
Liu, L., Wang, X., Chang, C. (2022b). Toward a smart skin: harnessing cuticle biosynthesis for crop adaptation to drought, salinity, temperature, and ultraviolet stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.961829
Liu, L., Zhao, L., Chen, P., Cai, H., Hou, Z., Jin, X., et al. (2020). ATP binding cassette transporters ABCG1 and ABCG16 affect reproductive development via auxin signalling in Arabidopsis. Plant J. 102 (6), 1172–1186. doi: 10.1111/tpj.14690
Luo, S., Tang, Z., Yu, J., Liao, W., Xie, J., Lv, J., et al. (2020). Hydrogen sulfide negatively regulates cd-induced cell death in cucumber (Cucumis sativus l) root tip cells. BMC Plant Biol. 20 (1), 480. doi: 10.1186/s12870-020-02687-8
Marjamaa, K., Kukkola, E. M., Fagerstedt, K. V. (2009). The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60 (2), 367–376. doi: 10.1093/jxb/ern278
Marmagne, A., Rouet, M. A., Ferro, M., Rolland, N., Alcon, C., Joyard, J., et al. (2004). Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics MCP 3 (7), 675–691. doi: 10.1074/mcp.M400001-MCP200
MartiniEre, A., Moreau, P. (2020). Complex roles of rabs and SNAREs in the secretory pathway and plant development: a never-ending story. J. Microsc. 280 (2), 140–157. doi: 10.1111/jmi.12952
McFarlane, H. E., Shin, J. J., Bird, D. A., Samuels, A. L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell. 22 (9), 3066–3075. doi: 10.1105/tpc.110.077974
Meents, M. J., Watanabe, Y., Samuels, A. L. (2018). The cell biology of secondary cell wall biosynthesis. Ann. Bot. 121 (6), 1107–1125. doi: 10.1093/aob/mcy005
Meng, G., Fan, W., Rasmussen, S. K. (2021). Characterisation of the class III peroxidase gene family in carrot taproots and its role in anthocyanin and lignin accumulation. Plant Physiol. Biochem. 167, 245–256. doi: 10.1016/j.plaphy.2021.08.004
Merali, Z., Mayer, M. J., Parker, M. L., Michael, A. J., Smith, A. C., Waldron, K. W. (2007). Metabolic diversion of the phenylpropanoid pathway causes cell wall and morphological changes in transgenic tobacco stems. Planta 225 (5), 1165–1178. doi: 10.1007/s00425-006-0427-5
Meyer, D., Pajonk, S., Micali, C., O'Connell, R., Schulze-Lefert, P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57 (6), 986–999. doi: 10.1111/j.1365-313X.2008.03743.x
Miao, Y. C., Liu, C. J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U S A 107 (52), 22728–22733. doi: 10.1073/pnas.1007747108
Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J. P., Hartmann, M. A., et al. (2004). Lipid rafts in higher plant cells: purification and characterization of triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279 (35), 36277–36286. doi: 10.1074/jbc.M403440200
Morel, J., Claverol, S., Mongrand, S., Furt, F., Fromentin, J., Bessoule, J. J., et al. (2006). Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics MCP 5 (8), 1396–1411. doi: 10.1074/mcp.M600044-MCP200
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21, (15). doi: 10.3390/ijms21155208
Opazo, M. C., Lizana, R., Stappung, Y., Davis, T. M., Herrera, R., Moya-León, M. A. (2017). XTHs from fragaria vesca: genomic structure and transcriptomic analysis in ripening fruit and other tissues. BMC Genomics 18 (1), 852. doi: 10.1186/s12864-017-4255-8
Pacifici, E., Di Mambro, R., Dello Ioio, R., Costantino, P., Sabatini, S. (2018). Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J. 37, (16). doi: 10.15252/embj.201899134
Pandey, S. K., Nookaraju, A., Fujino, T., Pattathil, S., Joshi, C. P. (2016). Virus-induced gene silencing (VIGS)-mediated functional characterization of two genes involved in lignocellulosic secondary cell wall formation. Plant Cell Rep. 35 (11), 2353–2367. doi: 10.1007/s00299-016-2039-2
Pilon, M., Ravet, K., Tapken, W. (2011). The biogenesis and physiological function of chloroplast superoxide dismutases. Biochim. Biophys. Acta 1807(8), 989–998. doi: 10.1016/j.bbabio.2010.11.002
Qin, G., Ma, J., Chen, X., Chu, Z., She, Y. M. (2017). Methylated-antibody affinity purification to improve proteomic identification of plant RNA polymerase pol V complex and the interacting proteins. Sci. Rep. 7, 42943. doi: 10.1038/srep42943
Roepke, J., Bozzo, G. G. (2015). Arabidopsis thaliana beta-glucosidase BGLU15 attacks flavonol 3-O-beta-glucoside-7-O-alpha-rhamnosides. Phytochemistry 109, 14–24. doi: 10.1016/j.phytochem.2014.10.028
Rolland, F., Sheen, J. (2005). Sugar sensing and signalling networks in plants. Biochem. Soc. Trans. 33 (Pt 1), 269–271. doi: 10.1042/bst0330269
Sanderfoot, A. A., Assaad, F. F., Raikhel, N. V. (2000). The Arabidopsis genome. an abundance of soluble n-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124 (4), 1558–1569. doi: 10.1104/pp.124.4.1558
Schuetz, M., Benske, A., Smith, R. A., Watanabe, Y., Tobimatsu, Y., Ralph, J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166 (2), 798–807. doi: 10.1104/pp.114.245597
Segal, A. W. (2016). NADPH oxidases as electrochemical generators to produce ion fluxes and turgor in fungi, plants and humans. Open Biol. 6, (5). doi: 10.1098/rsob.160028
Shanmugarajah, K., Linka, N., Grafe, K., Smits, S. H. J., Weber, A. P. M., Zeier, J., et al. (2019). ABCG1 contributes to suberin formation in Arabidopsis thaliana roots. Sci. Rep. 9 (1), 11381. doi: 10.1038/s41598-019-47916-9
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24, (13). doi: 10.3390/molecules24132452
Shigeto, J., Itoh, Y., Hirao, S., Ohira, K., Fujita, K., Tsutsumi, Y. (2015). Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J. Integr. Plant Biol. 57 (4), 349–356. doi: 10.1111/jipb.12334
Singh, M. K., Jurgens, G. (2018). Specificity of plant membrane trafficking - ARFs, regulators and coat proteins. Semin. Cell Dev. Biol. 80, 85–93. doi: 10.1016/j.semcdb.2017.10.005
Su, T., Xu, J., Li, Y., Lei, L., Zhao, L., Yang, H., et al. (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell. 23 (1), 364–380. doi: 10.1105/tpc.110.079145
Suwastika, I. N., Uemura, T., Shiina, T., Sato, M. H., Takeyasu, K. (2008). SYP71, a plant-specific qc-SNARE protein, reveals dual localization to the plasma membrane and the endoplasmic reticulum in Arabidopsis. Cell structure Funct. 33 (2), 185–192. doi: 10.1247/csf.08024
Tang, R. J., Zhao, F. G., Yang, Y., Wang, C., Li, K., Kleist, T. J., et al. (2020). A calcium signalling network activates vacuolar k(+) remobilization to enable plant adaptation to low-K environments. Nat. plants 6 (4), 384–393. doi: 10.1038/s41477-020-0621-7
Tobimatsu, Y., Wagner, A., Donaldson, L., Mitra, P., Niculaes, C., Dima, O., et al. (2013). Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 76 (3), 357–366. doi: 10.1111/tpj.12299
Tyrrell, M., Campanoni, P., Sutter, J. U., Pratelli, R., Paneque, M., Sokolovski, S., et al. (2007). Selective targeting of plasma membrane and tonoplast traffic by inhibitory (dominant-negative) SNARE fragments. Plant J. 51 (6), 1099–1115. doi: 10.1111/j.1365-313X.2007.03206.x
Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., Sato, M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-golgi network in plant cells. Cell structure Funct. 29 (2), 49–65. doi: 10.1247/csf.29.49
Wang, Y., Fan, C., Hu, H., Li, Y., Sun, D., Wang, Y., et al. (2016). Genetic modification of plant cell walls to enhance biomass yield and biofuel production in bioenergy crops. Biotechnol. advances 34 (5), 997–1017. doi: 10.1016/j.biotechadv.2016.06.001
Wang, J., Feng, J., Jia, W., Chang, S., Li, S., Li, Y. (2015). Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol. biofuels 8, 145. doi: 10.1186/s13068-015-0331-y
Wang, Z., Mao, Y., Guo, Y., Gao, J., Liu, X., Li, S., et al. (2020). MYB transcription Factor161 mediates feedback regulation of secondary wall-associated NAC-Domain1 family genes for wood formation. Plant Physiol. 184 (3), 1389–1406. doi: 10.1104/pp.20.01033
Wei, T., Zhang, C., Hou, X., Sanfaçon, H., Wang, A. (2013). The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PloS pathogens 9 (5), e1003378. doi: 10.1371/journal.ppat.1003378
Xiao, C., Zhang, T., Zheng, Y., Cosgrove, D. J., Anderson, C. T. (2016). Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in arabidopsis, altering cell growth and morphogenesis. Plant Physiol. 170 (1), 234–249. doi: 10.1104/pp.15.01395
Xing, D. H., Lai, Z. B., Zheng, Z. Y., Vinod, K. M., Fan, B. F., Chen, Z. X. (2008). Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 1 (3), 459–470. doi: 10.1093/mp/ssn020
Xu, Z., Escamilla-Treviño, L., Zeng, L., Lalgondar, M., Bevan, D., Winkel, B., et al. (2004). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55 (3), 343–367. doi: 10.1007/s11103-004-0790-1
Yoon, T. Y., Munson, M. (2018). SNARE complex assembly and disassembly. Curr. Biol. 28 (8), R397–R401. doi: 10.1016/j.cub.2018.01.005
You, J., Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01092
Zhang, A., Ren, H. M., Tan, Y. Q., Qi, G. N., Yao, F. Y., Wu, G. L., et al. (2016). S-type anion channels SLAC1 and SLAH3 function as essential negative regulators of inward k+ channels and stomatal opening in Arabidopsis. Plant Cell. 28 (4), 949–955. doi: 10.1105/tpc.16.01050
Zhao, J. (2015). Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20 (9), 576–585. doi: 10.1016/j.tplants.2015.06.007
Zhao, J., Dixon, R. A. (2010). The 'ins' and 'outs' of flavonoid transport. Trends Plant Sci. 15 (2), 72–80. doi: 10.1016/j.tplants.2009.11.006
Zhao, X., Guo, X., Tang, X., Zhang, H., Wang, M., Kong, Y., et al. (2018). Misregulation of ER-golgi vesicle transport induces ER stress and affects seed vigor and stress response. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00658
Zhao, Q., Nakashima, J., Chen, F., Yin, Y., Fu, C., Yun, J., et al. (2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 25 (10), 3976–3987. doi: 10.1105/tpc.113.117770
Keywords: AtSYP71, pH homeositasis, ROS homeostasis, cell wall biosynthesis and dynamics, root development, vesicle trafficking
Citation: Zhang H, Zhou J, Kou X, Liu Y, Zhao X, Qin G, Wang M, Qian G, Li W, Huang Y, Wang X, Zhao Z, Li S, Wu X, Jiang L, Feng X, Zhu J-K and Li L (2023) Syntaxin of plants71 plays essential roles in plant development and stress response via regulating pH homeostasis. Front. Plant Sci. 14:1198353. doi: 10.3389/fpls.2023.1198353
Received: 01 April 2023; Accepted: 02 May 2023;
Published: 05 June 2023.
Edited by:
Shaojun Dai, Shanghai Normal University, ChinaReviewed by:
Mohamed Fokar, Texas Tech University, United StatesCopyright © 2023 Zhang, Zhou, Kou, Liu, Zhao, Qin, Wang, Qian, Li, Huang, Wang, Zhao, Li, Wu, Jiang, Feng, Zhu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixin Li, bGl4aW5saTA1MTVAbmVmdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.