- 1Dipartimento di Scienze Agrarie, Alimentari e Ambientali (DSA3), Università degli Studi di Perugia, Perugia, Italy
- 2Environmental Genomics and Systems Biology Research Group, Institute of Natural Resource Sciences (IUNR), Zurich University of Applied Sciences ZHAW, Wädenswil, Switzerland
- 3Bioinformatics and Systems Biology, Justus-Liebig University Giessen, Giessen, Germany
Pseudomonas syringae pv. tomato is the causal agent of bacterial speck of tomato, an important disease that results in severe crop production losses worldwide. Currently, two races within phylogroup 01a (PG01a) are described for this pathogen. Race 0 strains have avirulence genes for the expression of type III system-associated effectors AvrPto1 and AvrPtoB, that are recognized and targeted by the effector-triggered immunity in tomato cultivars having the pto race-specific resistance gene. Race 1 strains instead lack the avrPto1 and avrPtoB genes and are therefore capable to aggressively attack all tomato cultivars. Here, we have performed the complete genome sequencing and the analysis of P. syringae pv. tomato strain DAPP-PG 215, which was described as a race 0 strain in 1996. Our analysis revealed that its genome comprises a 6.2 Mb circular chromosome and two plasmids (107 kb and 81 kb). The results indicate that the strain is phylogenetically closely related to strains Max13, K40, T1 and NYS-T1, all known race 1 strains. The chromosome of DAPP-PG 215 encodes race 1-associated genes like avrA and hopW1 and lacks race 0-associated genes like hopN1, giving it a race 1 genetic background. However, the genome harbors a complete ortholog of avrPto1, which allows the strain to display a race 0 phenotype. Comparative genomics with several PG01a genomes revealed that mobile DNA elements are rather involved in the evolution of the two different races.
1 Introduction
Bacterial speck is one of the most widespread and economically important disease of tomato (Schneider & Grogan, 1977). Its symptoms affect leaves, flowers, fruits and stems of the plant and consist in small, irregular dark brown to black necrotic spots (Goode & Sasser, 1980). Usually surrounded by a yellow halo (green in the fruit), these spots can converge in the leaves as they appear, increasing the necrotic area of the foliage, and thus leading to plant death. Yield losses for outbreaks of this disease could be around 20-25% for tomato seedlings and up to 75% for tomato fruits (Yunis et al., 1980; Basim et al., 2004). The causative agent of the disease is Pseudomonas syringae pv. tomato, a Gram-negative, rod-shaped bacteria ubiquitously widespread in all tomato crop growing areas. It is a member of the P. syringae species complex, one of the most relevant groups of phytopathogenic bacteria (Mansfield et al., 2012) with its 15 species assigned to 13 different phylogroups (PGs) (Berge et al., 2014). P. syringae pv. tomato falls into one of the two clades of the phylogroup 01 (PG01a), while in the other clade (PG01b), Pseudomonas avellanae and P. syringae pv. actinidiae are included.
In order to begin its cycle of infection and cause disease, P. syringae pv. tomato can occur as a consequence of natural phenomena like rain and wind, water irrigation, and also due to the use of contaminated seeds. As an epiphyte, P. syringae pv. tomato can survive and grow on the surface of the leaves and the seeds. Then, when the environmental conditions are favorable, it can rapidly increase its population and enter the host plant trough natural openings or wounds exploiting its polar flagella.
Inside the plant, the apoplast is the niche used by P. syringae pv. tomato for its colonization and subsequentially infection (Melotto et al., 2008). Pathogenicity and virulence of P. syringae pv. tomato largely rely on the use of the Hrp (hypersensitive response and pathogenicity) Type III Secretion System (T3SS) and its Type III Effectors (T3Es) like Hop (Hrp outer protein) or Avr (avirulence) proteins, as well as the production of the phytotoxin coronatine. In P. syringae pv. tomato DC3000, the genes hopM1 and avrE1 are both required for the establishment of an aqueous environment in the apoplast that promotes growth and thus colonization of the pathogen, while avrPto1 and avrPtoB suppress the pattern-triggered immunity (PTI) of the plant, usually elicited by a structural component of the bacteria flagellum, flagellin, encoded by fliC (He et al., 2016).
The discovery and subsequent cloning in cultivated tomato strains of the pto gene, encoding a serine-threonine kinase capable of recognizing both the AvrPto1 and AvrPtoB protein effectors and activating the effector-triggered immunity (ETI) of the host (Xiao et al., 2007; Kunkeaw et al., 2010), allowed some control of the disease for a few decades. However, the emergence of P. syringae pv. tomato strains lacking functioning AvrPto1 and AvrPtoB effectively rendered the protection offered by Pto ineffective and allowed the establishment of a new race (race 1) of P. syringae pv. tomato capable of causing disease even in plants that carries the pto gene. Even today, the capability to cause disease in resistant tomato plants is the basis of the race assignment process for P. syringae pv. tomato, although there are useful genotypic markers to differentiate between the two different races (Jones et al., 2015). Moreover, strains of P. syringae pv. tomato expressing an intermediate phenotype between race 0 and race 1 strains have been characterized as well (Kraus et al., 2017).
P. syringae pv. tomato strain DAPP-PG 215 was obtained by isolation from an individual lesion on a diseased tomato plant, randomly selected from a field with tomato cultivar ‘Erminia F1’ (Petoseed) in Pontenure (Piacenza, Italy) in 1995 by Buonaurio et al. (1996) and described as race 0 following inoculation on resistant Ontario 7710 plants. In this study, the sequencing and genome analysis of this strain was performed and, according to the in silico results, we propose a different base for P. syringae pv. tomato race determination.
2 Materials and methods
2.1 Bacterial strain and growth conditions
Pseudomonas syringae pv. tomato strain DAPP-PG 215 belongs to the bacterial collection of the Plant Protection Unit, Department of Agricultural, Food and Environmental Sciences, University of Perugia, Italy, where it is stored in a -80°C ultrafreezer as glycerol stocks in 50% (vol/vol) King’s broth (King et al., 1954). For genome sequencing, strain DAPP-PG 215 was transferred on Luria-Bertani (LB) agar plates (Miller, 1972) and incubated at 28°C overnight; then, an isolated colony was streaked onto the same medium and grown in the same manner. Bacterial strains used for screening were instead grown on nutrient agar (NA) plates and then incubated at 27°C for 12 h.
2.2 Whole genome sequencing, assembly, and annotation
For genomic DNA (gDNA) extraction, P. syringae pv. tomato strain DAPP-PG 215 was inoculated in LB broth (Miller, 1972) from single colony of a pure bacterial culture on a LB plate and incubated overnight at 28°C. The following day, whole gDNA extraction was performed using the NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol with the following specifications: elution buffer was preheated at 70°C before use, and the gDNA was eluted in 60 μL. The gDNA was quantified using a high sensitivity double-stranded DNA (dsDNA) assay (DeNovix, Wilmigton, DE) with a Fluo-100B fluorometer (Allsheng, Hangzhou, China).
Library preparation for short-read sequencing was done using the Nextera XT DNA library prep kit (Illumina, San Diego, CA) following the manufacturer’s instructions. Sequencing was performed on a MiSeq Illumina sequencer with 2 × 300-bp paired-end reads using a MiSeq reagent kit version 3 (Illumina, San Diego, CA) according to the manufacturer’s instructions.
For long-read sequencing, the Gentra PureGene Yeast/Bact kit protocol (Qiagen, Hilden, Germany) was used on an overnight bacterial culture for the gDNA extraction. The gDNA was quantified as described above. Library preparation and sequencing were performed with the ligation sequencing kit (catalog no. SQK-LSK109; Oxford Nanopore Technologies, Oxford, United Kingdom) and run on an R9.4.1 Flongle Flow Cell with a MinION sequencer. The native barcoding expansion kit (catalog no. XP-NBD114; Oxford Nanopore Technologies, Oxford, United Kingdom) was used for multiplexing. Base calling was performed using Guppy version 5.0.11.
A hybrid assembly using the MiSeq and MinION reads was conducted with Unicycler version 0.4.9 (Wick et al., 2017). The genome was then annotated using Bakta version 1.2.4 (Schwengers et al., 2021) and the database version 3.0. All tools were run with default parameters unless otherwise specified. The BioCircos tool (Cui et al., 2016) was used to have a circular visualization of genomic data.
2.3 Identification of mobile regions
The genome of P. syringae pv. tomato strain DAPP-PG 215 was checked for the presence of potential prophage regions using the search tool PHASTER (Zhou et al., 2011; Arndt et al., 2016). It was thus possible to identify the prophage regions contained in the genome and to have them classified according to their level of completeness into incomplete, questionable, and intact. Genomic island (GI) prediction was carried out using IslandViewer 4 (Bertelli et al., 2017). The homology search of the proteins localized in these regions was performed by BLAST+ v. 2.13.0 (Camacho et al., 2009) using the National Center for Biotechnology Information (NCBI) database.
2.4 Selection of genome sequences and comparative analysis
Comparative genome analyses were performed on a total of 72 genomes taken from the P. syringae species complex and representatives of seven phylogroups according to Berge et al. (2014), plus P. syringae PDD-32b-74 and P. syringae pv. tomato DAPP-PG 215 (Table S1). Genome sequences were taken from the NCBI RefSeq database (http://www.ncbi.nlm.nih.gov) and added to an EDGAR 3.0 database (Dieckmann et al., 2021). Through the EDGAR platform, the core genome phylogenetic tree was constructed. Briefly, for the core genome of the 74 selected genomes, the EDGAR pipeline made an alignment for each of the 1,969 gene sets of the core genome using the MUSCLE software (Edgar, 2004). The resulting alignments for 145,706 genes in total were then concatenated into one multiple alignment of 753,574 AA-residues per genome, 55,764,476 AA-residues in total. This large alignment was submitted as input to FastTree 2 software (Price et al., 2010) within EDGAR 3.0, which processed a maximum likelihood phylogenetic tree and verified the tree topology using the Shimodaira-Hasegawa test.
Based on the phylogenetic tree obtained by comparing the core genomes of all strains in Table S1, the genomes belonging to PG01a were divided into five different subgroups (Figure 1). Using the subroutine in EDGAR 3.0, five meta-core genomes representing the core genome of all strains included in each subgroup were defined. Subsequently, the EDGAR pipeline was used to perform a comparison of the different meta-core genomes, in order to reveal the presence of orthologous genes for each subgroup. Protein sequences resulting from all the comparisons were then displayed in a Venn diagram.
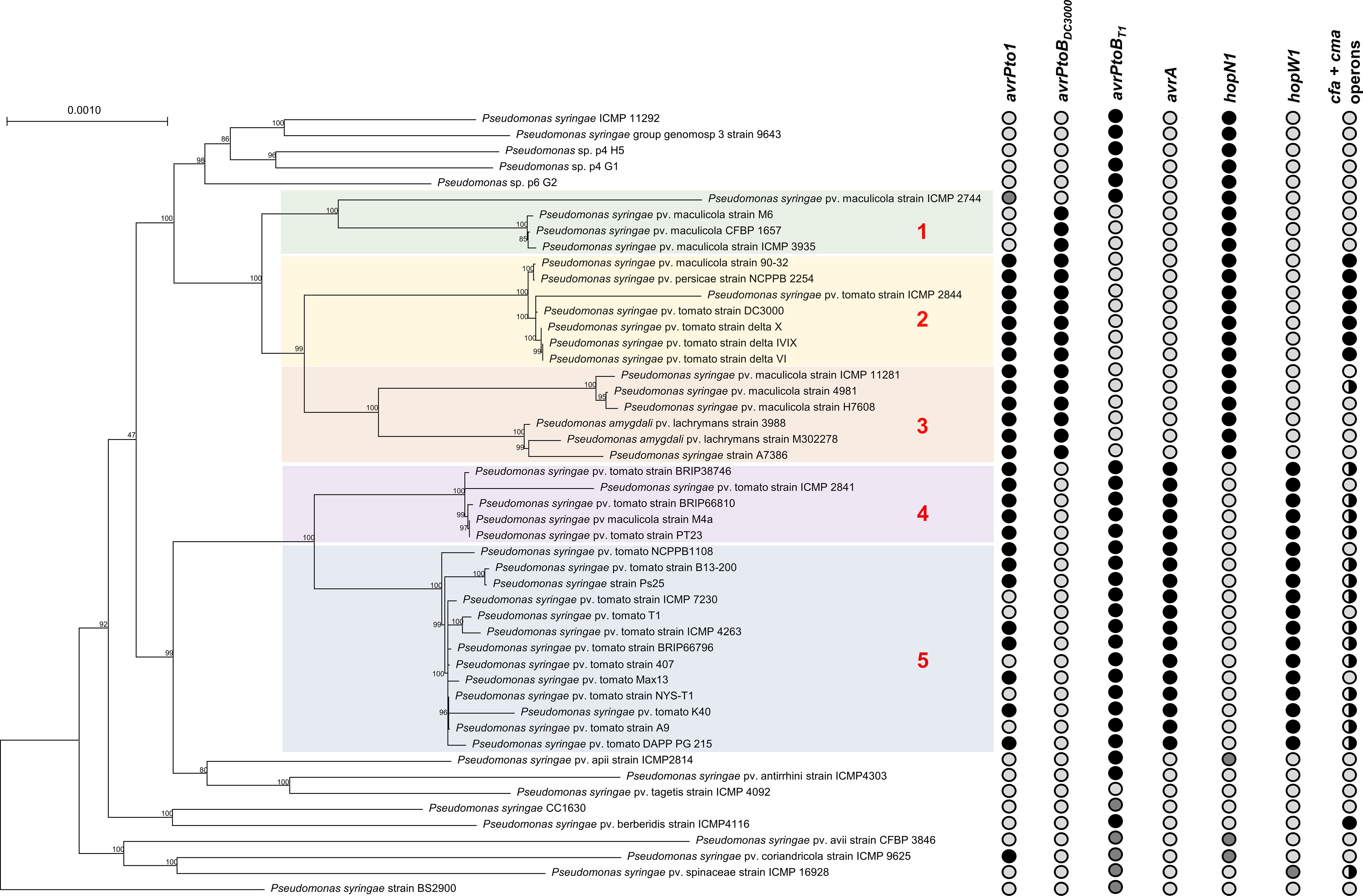
Figure 1 Subgroups defined according to core phylogeny for later comparative analysis. The green branch represents the first subgroup; the yellow branch represents the second subgroup; the orange branch represents the third subgroup; the pink branch represents the fourth subgroup; the blue branch represents the fifth subgroup. Dots in the right part of the figure are meant to show the absence or the presence of avirulence (avr) and Hrp-dependent outer protein (hop) genes avrPto1, avrPtoBDC3000, avrPtoBT1, avrA, hopN1 and hopW1, and of coronatine synthesis genes belonging to the cfa and cma clusters. Dot filling indicates the presence with high sequence identity (black), the presence with low sequence identity (dark grey) and absence of the genes (white); half-filled dots for the coronatine genes indicates presence of these genes with a plasmid origin.
2.5 Distribution of avirulence gene avrPto1 and avrPtoB
To test whether there was a different distribution of the avrPto1 and avrPtoB genes in the selected genomes, a pan-genome analysis of all strains was performed in EDGAR 3.0. Annotation gaps were checked using BLASTX (Altschul et al., 1990) and compared to a database of bacterial reference proteins (Refseq_proteins) restricted to “Pseudomonas syringae group (taxid:136849)”. Standard settings were used for all other parameters. Nucleotide sequences corresponding to each identified gene were used to analyze the phylogenetic relationship existing between the selected genomes and those containing avrPto1 and avrPtoB genes. The resulting phylogenetic trees were carried out with Molecular Evolutionary Genetics Analysis v11 (MEGA11) software (Tamura et al., 2021), according to the neighbor-joining method (Saitou & Nei, 1987). To model rates amongst sites, a gamma distribution (α = 5) was used and a partial deletion consisting in a 95% cut-off of the site coverage was applied to gaps and missing data. Furthermore, alignments from the MUSCLE algorithm were also checked for protein identity and coverage with NCBI Multiple Sequence Alignment Viewer 1.22.2 (MSA) using P. syringae pv. tomato DAPP-PG 215 sequences as anchor.
The presence of potential prophages was also assessed for all the strains carrying the gene avrPto1 using PHASTER (Zhou et al., 2011; Arndt et al., 2016) and visualized with MAUVE v2.4.0 (Darling et al., 2004).
2.6 Data availability
The genome sequence of P. syringae pv. tomato DAPP-PG 215 were submitted to EMBL and received the Assembly accession number GCA_949769235. Additional genome sequences analyzed within this study are available in the NCBI GenBank/DDJ/EMBL database under the accession numbers detailed in Table S1. The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJEB59188, OX458335-7.
3 Results
3.1 Whole genome general features
The complete genome of P. syringae pv. tomato DAPP-PG 215 consists of a 6,218,123 bp circular chromosome and two circular plasmids of 106,705 bp (p107) and 80,902 bp (p81), with a GC content of 59%, 58% and 57%, respectively (Figure 2). The 6.2 Mb chromosome comprises of 5,595 protein coding sequences (CDS), while plasmids p107 and p81 have 103 and 92 CDS, respectively. In the whole genome, 642 CDS were identified as hypothetical proteins, and 72 predicted tRNAs and 5 rRNA operons were found. Among the most prominent genomic features for the characterization of strain DAPP-PG 215 is the presence of the avirulence genes avrPto1 and avrPtoB, but also of the effector genes hopW1 and avrA, which are considered as diagnostic markers for race 1 (Jones et al., 2015), while the hopN1 gene (a diagnostic marker for race 0 strains), was missing (Figure 1). Gene clusters for the biosynthesis of coronafacic acid and coronamic acid, precursors of the phytotoxin coronatine, hopK1 and a type VI Hcp1 effector gene were detected within plasmid p107, while in plasmid p81, effector genes avrD1, hopD1 and hopQ1-1 were found.
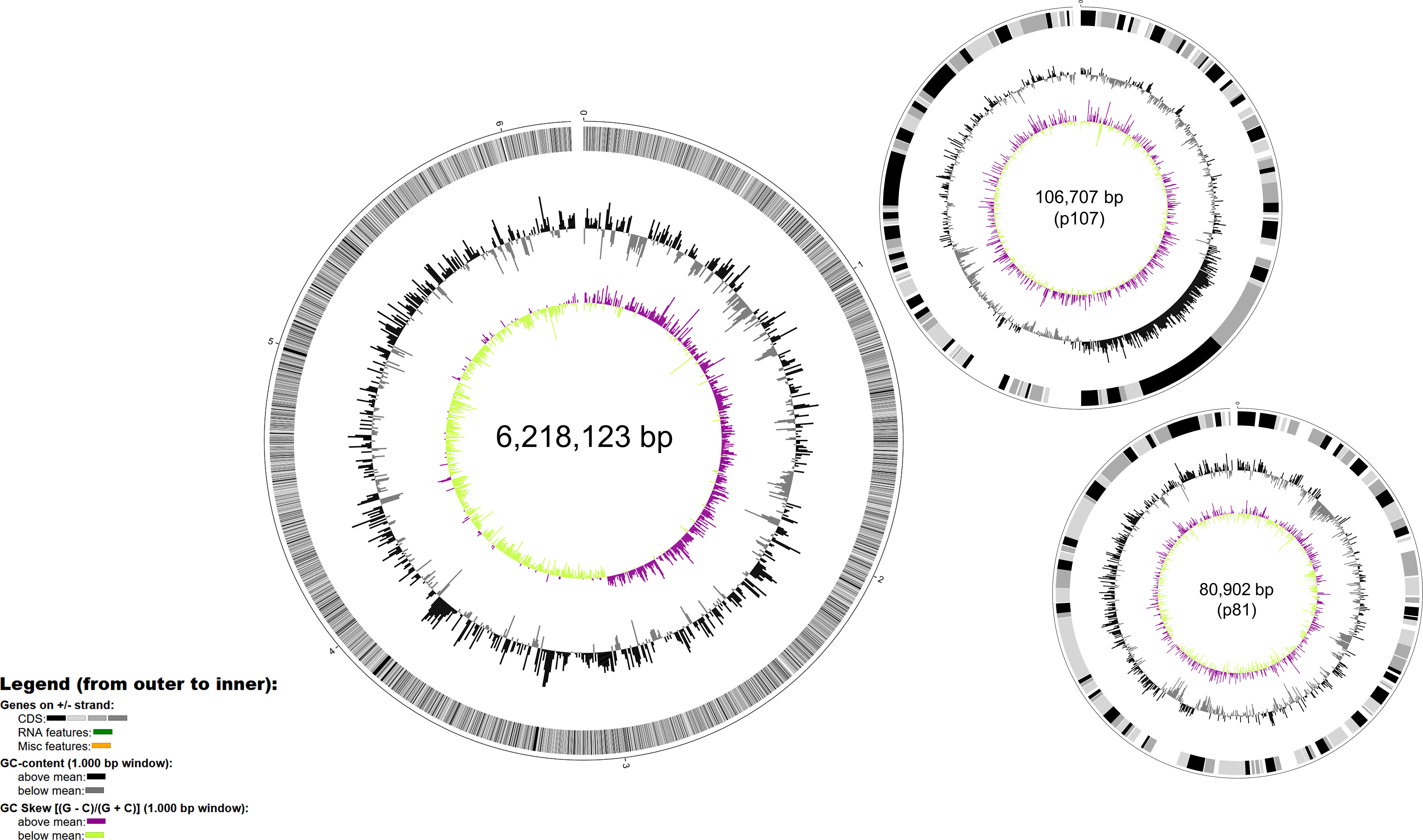
Figure 2 Map of the genome of Pseudomonas syringae pv. tomato DAPP-PG 215 generated with BioCircos. In the outermost ring, identified CDS are in grey, RNA features in green and miscellaneous features are in orange. In the intermediate ring GC content in grey and the innermost ring represents the GC skew above the mean in purple and GC skew below the mean in green.
Analysis of the DNA mobile regions using PHASTER indicated a total of nine prophages in the chromosome and two in plasmid p107. Information related to their size and their completeness level can be found in Table S2. Most of the prophages detected were taxonomically similar to phage families belonging to the class Caudoviricetes such as Myoviridae, Podoviridae and Siphoviridae (Iwasaki et al., 2018). Within prophage region 9 (PR-9, from position 5,843,821 to 5,884,716), the avrPto1 gene is located. Genomic islands (GIs), clusters of genes obtained by horizontal gene transfer, were also found in both the chromosome and the plasmids of strain DAPP-PG 215. In the chromosome, a total of 44 GIs were detected by IslandViewer 4 (with the SIGI-HMM prediction method); while two and a single GIs were predicted for plasmids p107 and p81, respectively (Table S2). Most of the GIs contained transposases, integrases, phage-related genes and hypothetical protein-coding genes. However, genes for protein effectors associated with the type III secretion system were annotated within the first GI on plasmid p81 and in six GIs on the chromosome. More specifically, effector genes hopD1 and hopQ1-1 within both GI-10 and GI-46, hop-T1 in GI-13, hopF2 in GI-16, hopC1 in GI-23, hopAF1 in GI-32, hopT1-2 and hopS in GI-35 were predicted. Genes associated with arsenic resistance such as arsC and arsN1 were found within GI-6, while in GI-4 the algorithm detected the presence of two fluoride exporters (eriC and crcB).
3.2 Phylogenomic analysis
Phylogenomic analysis conducted using the core genomes of the 74 strains listed in Table S1 has confirmed the correctness of the phylogroup clustering made by Berge et al. (2014) and revealed that strain DAPP-PG 215 is part of the PG01a, as well as the other P. syringae pv. tomato strains used for the comparison (Figure S1). Furthermore, looking more closely at the clustering of PG01a, it is possible to see that the P. syringae pv. tomato strains are divided into two different subgroups: in the first group, race 0 strain DC3000 was found along with strain ICMP 2844, P. syringae pv. persicae NCPPB 2254, P. syringae A7386 and a few strains of P. syringae pv. maculicola and P. amygdali pv. lachrymans while in the second group, several known strains of P. syringae pv. tomato race 1 such as strains T1, NYS-T1, Max13, K40 and NCPPB 1108, but also P. syringae pv. maculicola M4a, were identified. Strain DAPP-PG 215 was included within the second subgroup. This finding confirms again the greater genotypic proximity of this strain to race 1 P. syringae pv. tomato strains.
3.3 Comparative genomics
By comparing the five representative meta-core genomes of the subgroups defined in Figure 1, it was possible to identify which and how many orthologous genes were shared among the different subgroups and which genes were instead distinctive for each of them (Figure 3). A total number of 5,496 CDS were retrieved from the analysis, of which 3,177 (57.8%), 1,642 (29.9%) and 677 (12.3%) were assigned as core genome, disposable and singleton components, respectively. Among the latter, the distinctive proteins for each subset ranged from 74 (subgroup 1) to 269 (subgroup 4). Mostly, the singletons for each subgroup were hypothetical proteins and transposable elements. However, within the core genome of specific subgroups, distinctive genes were also found coding for: (a) the virulence effectors HopAD1 and HopK1, a TonB-dependent siderophore receptor, the coronafacic acid biosynthetic enzymes, and the proteins for biosynthesis of a putative bacteriocin in subset 2; (b) the virulence effector HopO1-3 in subgroup 3; (c) various ABC transporters, arsenic-resistance proteins (ArsB, ArsC, ArsH), the virulence effector AvrPphD, and colicin immunity protein in subset 4; and (d) copper resistance proteins (CopB, CopC and CopD) in subset 5. It is important to mention as well that the avrPtoB gene was observed to be one of the 3,177 core genes shared among all the subgroups.
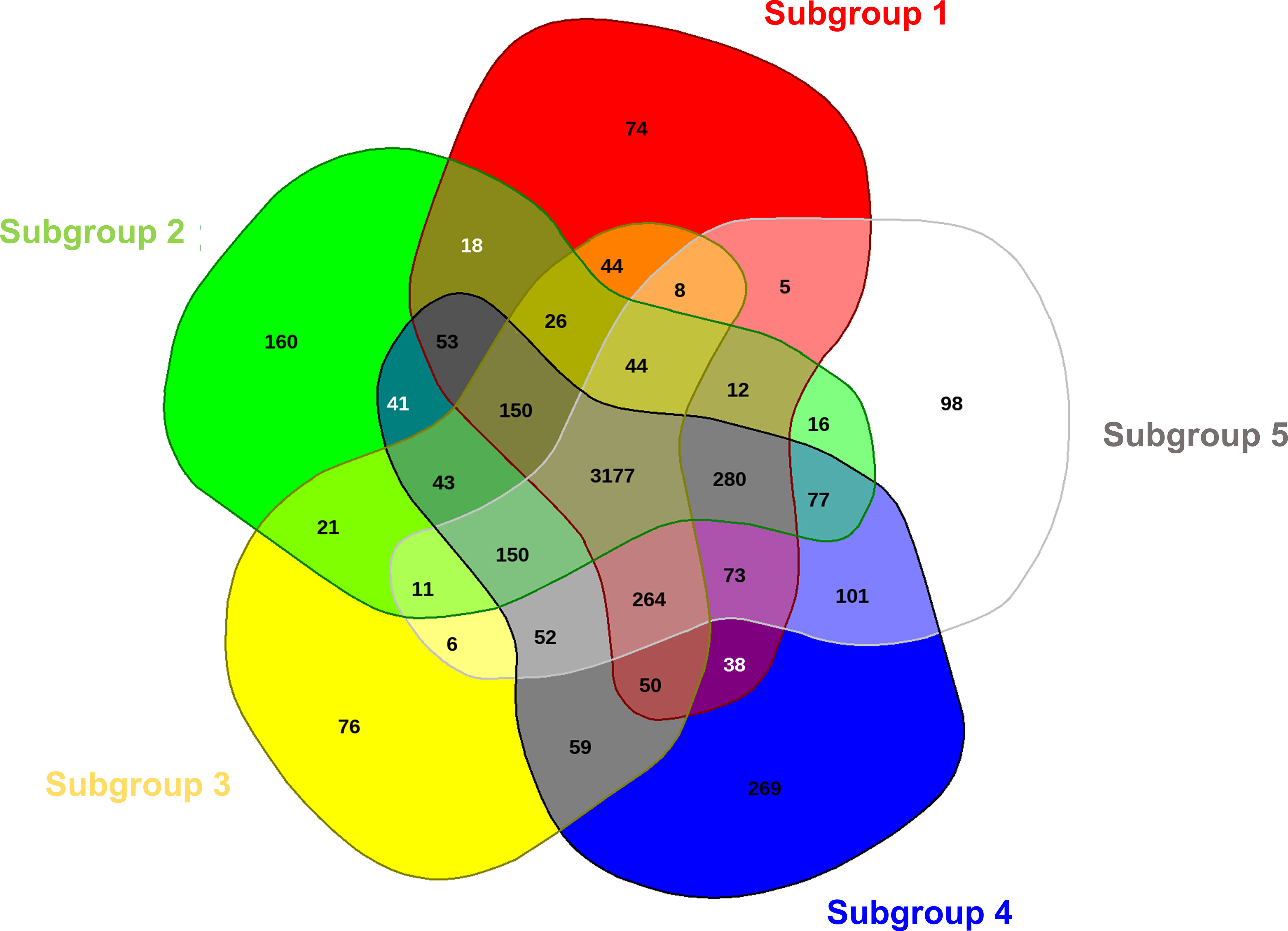
Figure 3 Comparative analysis of the meta-core-genomes of five different defined groups in PG01a. The Venn diagram, constructed with EDGAR 3.0, shows unique and shared genome components for all the five groups.
3.4 Avirulence gene avrPto1 is located in a prophage
Since the avrPto1 gene in P. syringae pv. tomato DAPP-PG 215 was found within an intact prophage region (PR-12, Pseudomonas phage phi3), the same analysis was also performed on all genomes that were part of the subgroups defined above (Figure 1). For all strains having the avrPto1 gene, PHASTER detected the presence of Pseudomonas phage phi3 prophage in the genome (Table 1). Despite the different degree of completeness of the region, for all strains, it was in a position overlapping with that of the avrPto1 gene. Strains lacking avrPto1 such as P. syringae pv. tomato race 1 strains A9, T1, NYS-T1, BRIP 66796 and 407, on the other hand, also did not have Pseudomonas phage phi3. It also appears that this avrPto1-containing prophage is located at different locations in the genome. Whereas in P. syringae pv. tomato DAPP-PG 215, it is inserted in the chromosome, between another prophage (Vibrio_vB_VpaM_MAR_NC_019722) and two contiguous transposons of the ISPsy4 family transposase, flanked by attL and attR sites of 11 bp (CCACGTAACAAG), in P. syringae DC3000, it is located in the chromosome as well, but about 1.35 Mb upstream of the attachment site attL of P. syringae pv. tomato DAPP-PG 215, flanked by different 11 bp attL and attR sites (CCTGACGATGAA).
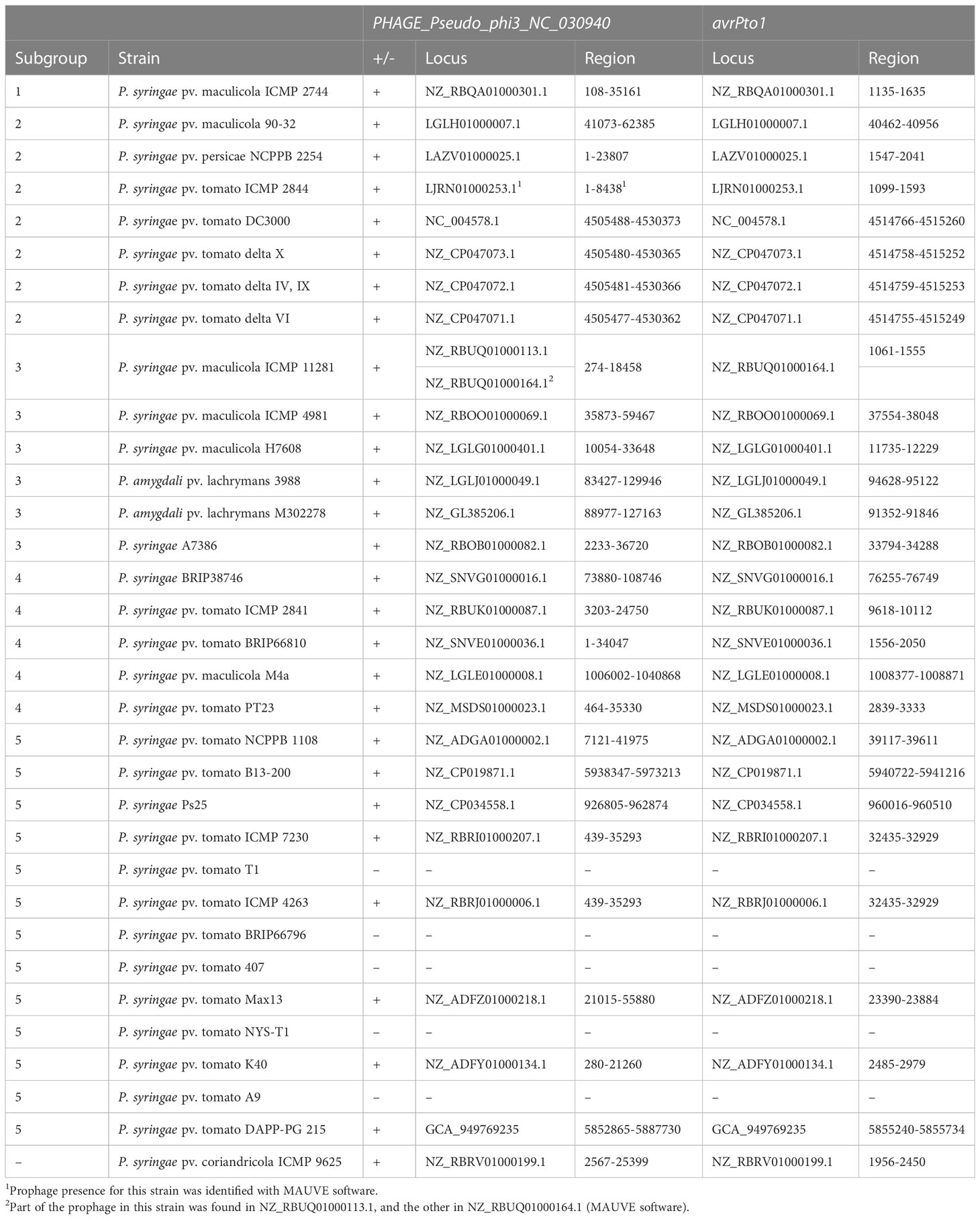
Table 1 PHASTER predictions of prophage Pseudo_phi3_NC_030940 locations in all Pseudomonas syringae pv. tomato strains used in this study and all the PG01a strains carrying avrPto1 in comparison with avrPto1 locations for each of these strains.
3.5 Distribution of the avirulence gene avrPto1 in genome sequences
A BLASTX search was performed using as queries the avrPto1 and avrPtoB sequences of P. syringae pv. tomato DAPP-PG 215 obtained with the aim of determining the evolutionary relationship existing between this specific strain and the other strains included in PG01a concerning these two avirulence genes. The BLASTX analysis revealed the presence of avrPto1 in 28 out of 49 strains within PG01a, while avrPtoB was found in 46 out of 49 genomes.
Comparing the dendrogram based on the presence or absence of avrPto1 with the core genome phylogeny, it was observed that although some of the major subgroups defined in PG01a by the phylogenetic analysis were confirmed, their positioning in the structure of these trees was not consistent (Figure S2). For AvrPto1, the sequence of P. syringae pv. tomato DAPP-PG 215 was found to cluster with those of strains of subgroup IV and V, in a branch of the tree that was only slightly distinct from subgroup II (98.82% protein identity, due to two substitutions: V49L and Y156A) and a part of subgroup III (P. syringae A7386 and P. amygdali pv. lachrymans strains 3988 and M302278), while the P. syringae pv. maculicola strains from subgroup III (ICMP 11281, 4981 and H7608, respectively) clustered together more distantly from the two groups (68.67% protein identity). The only strain of subgroup I carrying avrPto1 was P. syringae pv. maculicola ICMP 2744, the protein sequence of which was very distant to the AvrPto1 sequences of the other strains mentioned above (33.94% protein identity with P. syringae pv. tomato DAPP-PG 215). An AvrPto1 ortholog was also found in P. syringae pv. coriandricola ICMP 9625 (96.45% protein identity with P. syringae pv. tomato DAPP-PG 215). Among the five amino acid changes between the AvrPto1 protein sequences of P. syringae pv. coriandricola ICMP 9625 and P. syringae pv. tomato DAPP-PG 215 (I4M, D29G, A44S, Y90F and G97R), the fifth is occurring inside the GINP loop, a region known to be responsible for the interaction with plant Pto (Shan et al., 2000; Chang et al., 2001; Wulf et al., 2004). Lastly, it was observed that, unlike subgroups II and III, the avrPto1 gene was not observed in subgroups I and V: P. syringae pv. tomato race 1 strains T1, ICMP 7230, 407, NYS-T1 and A9 and P. syringae pv. maculicola strains M6, CFBP 1657 and ICMP 3935 were all lacking it.
3.6 Distribution of the avirulence gene avrPtoB in genome sequences
For orthologs of avrPtoB, there are two distinct major clades in the dendrogram (Figure S3). In the first clade, gene sequences from all strains included in the subgroups I, II and III were clustering together. However, subgroup III was divided with the P. syringae pv. maculicola strains and the P. syringae A7386 and P. amygdali pv. lachrymans3988 and M302278 positioned in two different parts of the subtree. In the other cluster, gene sequences from strains of subgroups IV and V were clustering together alongside P. syringae ICMP 11292, P. syringae group genomosp. 3 strain 9643, Pseudomonas sp. p4.H5, Pseudomonas sp. p4.G1, Pseudomonas sp. p6.G2, P. syringae pv. antirrhini ICMP 4303, P. syringae pv. apii ICMP 2814, P. syringae BS2900 and P. syringae CC1630. The tree also showed that strain P. syringae pv. avii CFBP 3846 also has an ortholog of avrPtoB, but it was found to be quite distinct from all other avrPtoB gene sequences. The existence of two major groups was consistent with previous discovery of P. syringae pv. tomato DC3000 avrPtoB orthologs with lower sequence identity in P. syringae pv. tomato PT23 (avrPtoBPT23) and T1 (avrPtoBT1) strains (Lin et al., 2006). The clear separation of race 0 and race 1 strains could again not be confirmed, as the avrPtoB sequence of P. syringae pv. tomato DAPP-PG 215 was observed to cluster with P. syringae pv. tomato T1 and PT23, rather than with race 0 strain P. syringae pv. tomato DC3000. From the amino acid alignment, avrPtoB of P. syringae pv. tomato DAPP-PG 215 resulted to have an almost identical amino acid sequence as the proteins of Pseudomonas sp. p6.G2 (99.64%), P. syringae pv. berberidis ICMP4116 and subgroup IV strains (99.29%); while its identity with the other major cluster with subgroups I, II and III strains was around 72.83%. Lower amino acid sequence identities were found with the AvrPtoB orthologs from P. syringae CC1630 (58.36% identity, 75.09% coverage), P. syringae BS2900 (36.30% identity, 75.98% coverage) and P. syringae pv. avii CFBP 3846 (44.84% identity, 89.32% coverage).
3.7 Localization of coronatine genes is divergent within PG01a
Coronatine (COR) is a phytotoxin produced by several pathovars of P. syringae, and consists in the conjunction of coronafacic acid (CFA) and coronamic acid (CMA), synthetized by the homonymous clusters. Because the presence of the cfa and cma clusters in plasmids has been observed in different pathovars of P. syringae (Bender et al., 1991), we made a comparison among the PG01a strains gene sequences of the cfa cluster (cfa1-9, and the cfl gene coding for coronofactate ligase), the cma cluster (the cmaABCDET cluster and cmaU) and the COR regulatory region (corRSP). Phylogenetic analysis of the sequences for these twenty genes, obtained using EDGARs ortholog retrieval function, allowed us to observe two different PG01a strains having the necessary clusters for coronatine production groups (Figure S4). Resuming the division made previously above (Figure 1), a first group includes all the strains from subgroup 4 except P. syringae pv. tomato ICMP 2841 and all the strains from subgroup 5 except P. syringae pv. tomato NCPPB 1108, T1 and Max13, but also P. syringae pv. maculicola ICMP 4891 (subgroup 3) and P. syringae pv. spinaceae ICMP 16928. A second group, on the other hand, contains the entire subgroup 2, and P. syringae pv. berberidis ICMP 4116. More specifically, the nucleotide sequences of P. syringae pv. tomato DAPP-PG 215 have approximately 96% identity with the group containing P. syringae pv. tomato DC3000 (90% with P. syringae pv. maculicola 90-32), 90% with P. syringae pv. berberidis ICMP 4116, 98% with P. syringae pv. maculicola ICMP 4891, and 99% with P. syringae pv. spinaceae ICMP 16928. Significant deletions (>50 bp) were observed in some genes carried by P. syringae pv. tomato K40 (cfa6, cfl, cmaT and corR), P. syringae pv. tomato NYS-T1 (cfa6, cmaT and corR), P. syringae pv. maculicola 90-32 (cfa4 and cmaT) and P. syringae pv. maculicola M4a (cfa4 and cmaT). In addition, it was possible to observe that both clusters plus the COR regulatory region were located on plasmid regions regarding strains P. syringae Ps25 (pPs252, RefSeq: NZ_CP034559.1) and P. syringae pv. tomato B13-200 (pB13-200A, RefSeq: NZ_CP019872.1). Moreover, checking the sequence upstream of cfl, the presence of a CorR-binding site identical to that found in P. syringae pv. glycinea PG4180 (Liyanage et al., 1995; Peñaloza-Vázquez & Bender, 1998) and considered to be essential for transcription of genes involved in coronatine synthesis was observed in P. syringae pv. tomato DAPP-PG 215.
4 Discussion
P. syringae pv. tomato DAPP-PG 215 was described as a race 0 strain (Buonaurio et al., 1996) based on its phenotype expressed upon inoculation on susceptible tomato plants cv. ‘Bonny Best’ (+/+) and resistant cv. ‘Ontario 7710’ (pto/pto). From the analysis of its genome reported here, it rather possesses the marker genes avrA and hopW1 used to distinguish race 1 from race 0 strains, and instead lacks the hopN1 gene, which is characteristic for race 0 strains (Jones et al., 2015). The presence of a race 1 genotypic background for P. syringae pv. tomato strain DAPP-PG 215 is also supported by the phylogenomic analysis of its core genome in comparison with other PG01a strains, in which it clustered with strains described as race 1, such as P. syringae pv. tomato T1 rather than with strains described as race 0, such as P. syringae pv. tomato DC3000, and by the sequence analysis of the avrPtoB gene, found to be identical to that possessed by strain T1.
Resistance to bacterial speck disease caused by P. syringae pv. tomato is due to the occurrence of the pto gene in the tomato plant. This genes encodes the protein Pto that is involved in recognition of the type III effectors AvrPto1 and AvrPtoB (HopAB2) expressed by the pathogen (Salmeron et al., 1996; Lin & Martin, 2007). Although it was long assumed that the presence or absence of the two genes was sufficient to distinguish a strain of P. syringae pv. tomato race 0 from a race 1, it was recently observed that specific mutations in the avrPto1 gene prevent the AvrPto1 protein effector from Pto-mediated recognition (Xing et al., 2007). On the other hand, some strains expressing a race 1 phenotype, such as P. syringae pv. tomato T1, are capable to evade recognition because they have orthologous avrPtoB genes that are weakly transcribed (Lin et al., 2006; Kunkeaw et al., 2010). However, an intact avrPto1 avirulence gene was also found in P. syringae pv. tomato DAPP-PG 215, which could be responsible for the expression of the target of Pto and therefore the reason for the race 0 phenotype expressed by this strain. This hypothesis is in agreement with what was reported by Kraus et al. (2017) who observed the presence of avrPto1 in some strains of P. syringae pv. tomato race 1 and noticed in these strains a growth profile in planta intermediate between a race 0 and a race 1. According to what was reported so far, therefore, P. syringae pv. tomato DAPP-PG 215 would result in a race 1 genotype and a race 0 strain for its expressed phenotype.
In addition, the detection of the avrPto1 gene within a prophage in P. syringae pv. tomato DAPP-PG 215 and in all other strains belonging to PG01a used in this work possessing this avirulence gene was only reported in this study. Prophages (viral DNA integrated within bacterial genome) are mobile genetic elements that can be responsible for horizontal gene transfer in bacteria. The presence within prophages of genes encoding for virulence effectors of the bacterium is well documented (Fortier & Sekulovic, 2013; Varani et al., 2013), however, since our results suggest that the avrPto1 gene is associated with this type of mobile regions, the genotype-based criterion for distinguishing race in P. syringae pv. tomato appears unstable and unreliable.
Since strains of P. syringae pv. tomato having avrPto1 express a race-typical 0 phenotype even when they carry a low expressed avrPtoB homologue such as avrPtoBT1 (Kraus et al., 2017), it is possible to advance a hypothesis that the phenotype expressed by P. syringae pv. tomato will result in a race 1 if the strain lacks a functioning avrPto1 and instead has the avrPtoB ortholog avrPtoBT1, capable of escaping the Pto recognition, and a race 0 if the P. syringae pv. tomato strain meets either of the two following conditions: (a) it lacks a functioning avrPto1 but has an avrPtoBDC3000 (Lin et al., 2006), which results in a normal expression of AvrPtoB; (b) it has a functioning avrPto1, which, regardless of which avrPtoB gene is present in its genome, will be recognized by Pto.
Another difference between strains having a race 0 genomic background and those having a race 1 is the type of coronatine biosynthesis genes. Coronatine is a phytotoxin that interferes with stomata closure in response to pathogen-associated molecular patterns (PAMPs) and salicylic acid-mediated plant defense, also contributing to the development of chlorotic spots on plant leaves (Geng et al., 2014). Produced by strains of various pathovars of P. syringae, such as the pathovars atropurpurea, glycinea, maculicola, morsprunorum, and tomato (Bereswill et al., 1994), the genes involved in its synthesis are either located on plasmids or integrated in the chromosome (Preston, 2000). More specifically, the cfa and cma clusters, required for the synthesis of coronafacic acid and coronamic acid, respectively, and the regulatory region separating them containing the three genes corP, corS, and corR, are found in the chromosome in several strains of P. syringae pv. maculicola (including ICMP 2744) and in P. syringae pv. tomato DC3000, while in other strains such as P. syringae pv. tomato PT23 and P. syringae pv. maculicola 4981, they are found on a plasmid (Bender et al., 1989; Cuppels & Ainsworth, 1995).
From our analyses, genes for coronatine were also found on a plasmid in strains P. syringae Ps25, P. syringae pv tomato B13-200, and P. syringae pv tomato DAPP-PG 215. In addition, between the latter named strains and P. syringae pv. tomato DC3000, the cfa and cma clusters have about 96% identity at the nucleotide level, a finding in agreement with what was observed by in P. syringae pv. glycinea PG4180, which also had the genes for coronatine located in a plasmid (Wang et al., 2002). The subdivision in the phylogenetic analysis showed that all strains having the coronatine genes in the plasmid mentioned earlier clustered in one group, while P. syringae pv. tomato DC3000, which has the genes on the chromosome, was part of the other group. Although it has not been possible to verify for P. syringae pv. maculicola ICMP 2744 belonging to the same group as P. syringae pv. tomato DC3000, it is possible to assume that all strains belonging to the group with P. syringae pv. tomato DAPP-PG 215, B13-200, PT23, P. syringae Ps25 and P. syringae pv. maculicola 4981 have the genes for coronatine located on a plasmid, while P. syringae pv. tomato ICMP 2844, P. syringae pv. persicae NCPPB 2254 and P. syringae pv. maculicola 90-32 have them on the chromosome. Although it is currently unclear what the implications are for having the gene clusters for coronatine located on the plasmid rather than on the chromosome, a different regulatory mechanism for the synthesis of this phytotoxin was proposed for P. syringae pv. tomato DC3000 (Fouts et al., 2002), which lacks the CorR-binding site upstream cfl (Wang et al., 2002).
The results of this study confirm what has been previously observed regarding the distribution of genes for coronatine in some strains of P. syringae pv. tomato and P. syringae pv. maculicola and proposes new hypotheses in the distinction between strains carrying the coronatine biosynthetic genes on plasmids or on chromosomes, although molecular investigations are needed to better understand the significance of this difference. The existence of strains of P. syringae pv. tomato having an intermediate phenotype between a race 0 and a race 1 is also confirmed. Subsequent studies that can estimate the ability of the respective strains to grow in planta and the severity of the disease on tomato plants in comparison with other non-intermediate strains are required to understand the role of avrPto1 in the various genomic assets. Lastly, the presence of the avrPto1 gene in a prophage may result in a different awareness of the concept of race in P. syringae pv. tomato, which is therefore, on a phenotypic basis, to be intended as potentially less permanent.
Whereas the race concept may thus become obsolete, it still needs to be commented that strains of both races were and will remain pathogens of tomato, irrespective of their genetic background.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
BO and TS have conceptualized and designed the study. JP has carried out the bacterial cultivation, DNA extraction and NGS sequencing. JP, LF, JB and TS have performed the bioinformatic and software analysis. CM conducted the molecular investigations. RB and TS provided the resources needed for the analysis. BO and TS have prepared the original draft preparation. RB, CM and TS have contributed to the funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
JP and TS were supported by the Department of Life Sciences and Facility Management of the Zurich University of Applied Sciences (ZHAW) in Wädenswil. The EDGAR platform is funded by the BMBF grant FKZ031A533 within the de.NBI network. CM and RB were supported by the research fund “Ricerca di Base” of Department of Agricultural, Food and Environmental Sciences (DSA3) of the University of Perugia, Italy.
Acknowledgments
We thank Dr. M. Orfei and Mr. L. Bonciarelli (DSA3, University of Perugia) for technical assistance. The authors would also like to thank the HPC team of the School for Life Sciences and Facility Management at ZHAW for providing computing resources and support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1197706/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Basim, H., Basim, E., Yilmaz, S., Dickstein, E. R., Jones, J. B. (2004). An outbreak of bacterial speck caused by Pseudomonas syringae pv. tomato on tomato transplants grown in commercial seedling companies located in the western mediterranean region of Turkey. Plant Dis. 88, 1050. doi: 10.1094/PDIS.2004.88.9.1050A
Bender, C. L., Malvick, D. K., Mitchell, R. E. (1989). Plasmid-mediated production of the phytotoxin coronatine in Pseudomonas syringae pv. tomato. J. Bacteriol. 171, 807–812. doi: 10.1128/jb.171.2.807-812.1989
Bender, C. L., Young, S. A., Mitchell, R. E. (1991). Conservation of plasmid DNA sequences in xoronatine-producing pathovars of Pseudomonas syringae. Appl. Environ. Microbiol. 57, 993–999. doi: 10.1128/aem.57.4.993-999.1991
Bereswill, S., Bugert, P., Völksch, B., Ullrich, M., Bender, C. L., Geider, K. (1994). Identification and relatedness of coronatine-producing Pseudomonas syringae pathovars by PCR analysis and sequence determination of the amplification products. Appl. Environ. Microbiol. 60, 2924–2930. doi: 10.1128/aem.60.8.2924-2930.1994
Berge, O., Monteil, C. L., Bartoli, C., Chandeysson, C., Guilbaud, C., Sands, D. C., et al. (2014). A user’s guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS One 9, e105547. doi: 10.1371/journal.pone.0105547
Bertelli, C., Laird, M. R., Williams, K. P., Fraser, S., Lau, B. Y., Hoad, G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Buonaurio, R., Stravato, V. M., Cappelli, C. (1996). Occurrence of Pseudomonas syringae pv. tomato race 1 in Italy on Pto gene-bearing tomato plants. J. Phytopathol. 144, 437–440. doi: 10.1111/j.1439-0434.1996.tb00320.x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Chang, J. H., Tobias, C. M., Staskawicz, B. J., Michelmore, R. W. (2001). Functional studies of the bacterial avirulence protein AvrPto by mutational analysis. Mol. Plant-Microbe Interact. 14, 451–459. doi: 10.1094/MPMI.2001.14.4.451
Cui, Y., Chen, X., Luo, H., Fan, Z., Luo, J., He, S., et al. (2016). BioCircos.js: an interactive circos JavaScript library for biological data visualization on web applications. Bioinformatics 32, 1740–1742. doi: 10.1093/bioinformatics/btw041
Cuppels, D. A., Ainsworth, T. (1995). Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl. Environ. Microbiol. 61, 3530–3536. doi: 10.1128/aem.61.10.3530-3536.1995
Darling, A. C. E., Mau, B., Blattner, F. R., Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Dieckmann, M. A., Beyvers, S., Nkouamedjo-Fankep, R. C., Hanel, P. H. G., Jelonek, L., Blom, J., et al. (2021). EDGAR3.0: comparative genomics and phylogenomics on a scalable infrastructure. Nucleic Acids Res. 49, W185–W192. doi: 10.1093/nar/gkab341
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fortier, L.-C., Sekulovic, O. (2013). Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4, 354–365. doi: 10.4161/viru.24498
Fouts, D. E., Abramovitch, R. B., Alfano, J. R., Baldo, A. M., Buell, C. R., Cartinhour, S., et al. (2002). Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. U. S. A. 99, 2275–2280. doi: 10.1073/pnas.032514099
Geng, X., Jin, L., Shimada, M., Kim, M. G., Mackey, D. (2014). The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 240, 1149–1165. doi: 10.1007/s00425-014-2151-x
Goode, M. J., Sasser, M. (1980). Prevention-the key to controlling bacterial spot and bacterial speck of tomato. Plant Dis. 64, 831. doi: 10.1094/PD-64-831
He, D., Zhan, J., Xie, L. (2016). Problems, challenges and future of plant disease management: from an ecological point of view. J. Integr. Agric. 15, 705–715. doi: 10.1016/S2095-3119(15)61300-4
Iwasaki, T., Yamashita, E., Nakagawa, A., Enomoto, A., Tomihara, M., Takeda, S. (2018). Three-dimensional structures of bacteriophage neck subunits are shared in Podoviridae, Siphoviridae and Myoviridae. Genes Cells Devoted Mol. Cell. Mech. 23, 528–536. doi: 10.1111/gtc.12594
Jones, L. A., Saha, S., Collmer, A., Smart, C. D., Lindeberg, M. (2015). Genome-assisted development of a diagnostic protocol for distinguishing high virulence Pseudomonas syringae pv. tomato strains. Plant Dis. 99, 527–534. doi: 10.1094/PDIS-08-14-0833-RE
King, E. O., Ward, M. K., Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307.
Kraus, C. M., Mazo-Molina, C., Smart, C. D., Martin, G. B. (2017). Pseudomonas syringae pv. tomato strains from new York exhibit virulence attributes intermediate between typical race 0 and race 1 strains. Plant Dis. 101, 1442–1448. doi: 10.1094/PDIS-03-17-0330-RE
Kunkeaw, S., Tan, S., Coaker, G. (2010). Molecular and evolutionary analyses of Pseudomonas syringae pv. tomato race 1. Mol. Plant-Microbe Interact. 23, 415–424. doi: 10.1094/MPMI-23-4-0415
Lin, N.-C., Abramovitch, R. B., Kim, Y. J., Martin, G. B. (2006). Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72, 702–712. doi: 10.1128/AEM.72.1.702-712.2006
Lin, N.-C., Martin, G. B. (2007). Pto- and prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse Pseudomonas syringae pathovars to infect tomato. Mol. Plant-Microbe Interact. 20, 806–815. doi: 10.1094/MPMI-20-7-0806
Liyanage, H., Palmer, D. A., Ullrich, M., Bender, C. L. (1995). Characterization and transcriptional analysis of the gene cluster for coronafacic acid, the polyketide component of the phytotoxin coronatine. Appl. Environ. Microbiol. 61, 3843–3848. doi: 10.1128/aem.61.11.3843-3848.1995
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Melotto, M., Underwood, W., He, S. Y. (2008). Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46, 101–122. doi: 10.1146/annurev.phyto.121107.104959
Miller, J. H. (1972). Experiments in molecular genetics (Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory).
Peñaloza-Vázquez, A., Bender, C. L. (1998). Characterization of CorR, a transcriptional activator which is required for biosynthesis of the phytotoxin coronatine. J. Bacteriol. 180, 6252–6259. doi: 10.1128/JB.180.23.6252-6259.1998
Preston, G. M. (2000). Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol. Plant Pathol. 1, 263–275. doi: 10.1046/j.1364-3703.2000.00036.x
Price, M. N., Dehal, P. S., Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. doi: 10.1371/journal.pone.0009490
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Salmeron, J. M., Oldroyd, G. E. D., Rommens, C. M. T., Scofield, S. R., Kim, H.-S., Lavelle, D. T., et al. (1996). Tomato prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the pto kinase gene cluster. Cell 86, 123–133. doi: 10.1016/S0092-8674(00)80083-5
Schneider, R. W., Grogan, R. G. (1977). Bacterial speck of tomato: sources of inoculum and establishment of a resident population. Phytopathology 67, 388–394. doi: 10.1094/Phyto-67-388
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genomics 7, 000685. doi: 10.1099/mgen.0.000685
Shan, L., Thara, V. K., Martin, G. B., Zhou, J.-M., Tang, X. (2000). The Pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12, 2323–2337. doi: 10.1105/tpc.12.12.2323
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Varani, A. M., Monteiro-Vitorello, C. B., Nakaya, H. I., Van Sluys, M.-A. (2013). The role of prophage in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 51, 429–451. doi: 10.1146/annurev-phyto-081211-173010
Wang, X., AlarcÓn-Chaidez, F., PeÑaloza-VÁzquez, A., Bender, C. L. (2002). Differential regulation of coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000 and P. syringae pv. glycinea PG4180. Physiol. Mol. Plant Pathol. 60, 111–120. doi: 10.1006/pmpp.2002.0382
Watson, M. (2018) Indels are not ideal: quick test for interrupted ORFs in bacterial/microbial. Available at: https://github.com/mw55309/ideel.
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wulf, J., Pascuzzi, P. E., Fahmy, A., Martin, G. B., Nicholson, L. K. (2004). The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure 12, 1257–1268. doi: 10.1016/j.str.2004.04.017
Xiao, F., He, P., Abramovitch, R. B., Dawson, J. E., Nicholson, L. K., Sheen, et al. (2007). The n-terminal region of Pseudomonas type III effector AvrPtoB elicits pto-dependent immunity and has two distinct virulence determinants. Plant J. 52, 595–614. doi: 10.1111/j.1365-313X.2007.03259.x
Xing, W., Zou, Y., Liu, Q., Liu, J., Luo, X., Huang, Q., et al. (2007). The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449, 243–247. doi: 10.1038/nature06109
Yunis, H., Bashan, Y., Okon, Y., Henis, Y. (1980). Weather dependence, yield losses, and control of bacterial speck of tomato caused by Pseudomonas tomato. Plant Dis. 64, 937–939. doi: 10.1094/PD-64-937
Keywords: Pseudomonas avellanae, Pseudomonas syringae pv. tomato, comparative genomics, AvrPto1, AvrPtoB, race shift
Citation: Orfei B, Pothier JF, Fenske L, Blom J, Moretti C, Buonaurio R and Smits THM (2023) Race-specific genotypes of Pseudomonas syringae pv. tomato are defined by the presence of mobile DNA elements within the genome. Front. Plant Sci. 14:1197706. doi: 10.3389/fpls.2023.1197706
Received: 31 March 2023; Accepted: 19 June 2023;
Published: 05 July 2023.
Edited by:
Prem Lal Kashyap, Indian Institute of Wheat and Barley Research (ICAR), IndiaReviewed by:
Rikky Rai, University of Allahabad, IndiaSatyendra Pratap Singh, National Botanical Research Institute (CSIR), India
Yasmina Jaufeerally Fakim, University of Mauritius, Mauritius
Copyright © 2023 Orfei, Pothier, Fenske, Blom, Moretti, Buonaurio and Smits. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiaraluce Moretti, Y2hpYXJhbHVjZS5tb3JldHRpQHVuaXBnLml0; Theo H. M. Smits, dGhlby5zbWl0c0B6aGF3LmNo
 Benedetta Orfei
Benedetta Orfei Joël F. Pothier
Joël F. Pothier Linda Fenske3
Linda Fenske3 Jochen Blom
Jochen Blom Chiaraluce Moretti
Chiaraluce Moretti Roberto Buonaurio
Roberto Buonaurio Theo H. M. Smits
Theo H. M. Smits