- 1State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, Zhejiang, China
- 2Zhejiang Provincial Key Laboratory of Forest Aromatic Plants-based Healthcare Functions, Zhejiang A&F University, Hangzhou, Zhejiang, China
- 3College of Science and Technology, Ningbo University, Ningbo, Zhejiang, China
- 4College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu, China
Woody plants play a vital role in global ecosystems and serve as valuable resources for various industries and human needs. While many woody plant genomes have been fully sequenced, gene function research and biotechnological breeding advances have lagged behind. As a result, only a limited number of genes have been elucidated, making it difficult to use newer tools such as CRISPR-Cas9 for biotechnological breeding purposes. The use of Agrobacterium rhizogenes as a transformative tool in plant biotechnology has received considerable attention in recent years, particularly in the research field on woody plants. Over the past three decades, numerous woody plants have been effectively transformed using A. rhizogenes-mediated techniques. Some of these transformed plants have successfully regenerated. Recent research on A. rhizogenes-mediated transformation of woody plants has demonstrated its potential for various applications, including gene function analysis, gene expression profiling, gene interaction studies, and gene regulation analysis. The introduction of the Ri plasmid has resulted in the emergence of several Ri phenotypes, such as compact plant types, which can be exploited for Ri breeding purposes. This review paper presents recent advances in A. rhizogenes-mediated basic research and Ri breeding in woody plants. This study highlights various aspects of A. rhizogenes-mediated transformation, its multiple applications in gene function analysis, and the potential of Ri lines as valuable breeding materials
1 Introduction
The woodiness proportion among the global vascular plant population ranges from 45% to 48% (FitzJohn et al., 2014). Woody plants play a vital role in meeting human needs by serving as a source of energy, construction materials, and sustenance, while also offering essential ecosystem services such as carbon sequestration, biodiversity support, and climate regulation (Trumbore et al., 2015). Woody plants, encompassing both trees and shrubs, exhibit comparatively lengthier generation times than their herbaceous counterparts, which typically display shorter generation times. Understanding the fundamental biology of woody plants is of utmost importance to enhance their environmental resilience, productivity, and other desirable traits through technological advancements, thus facilitating the cultivation of novel varieties. Among the various methods available, Agrobacterium-mediated transformation is the most commonly employed approach. Both Agrobacterium tumefaciens and Agrobacterium rhizogenes possess the ability to infect plant cells and facilitate the transfer of a DNA segment, referred to as T-DNA, which carries oncogenes from the pathogen to the plant cells. Subsequently, this T-DNA integrates into the genome of the host plant. In the case of A. tumefaciens-mediated transformation, there are two obstacles for woody plants: in vitro (tissue culture-dependent) manipulation and regeneration from explants. Conversely, A. rhizogenes-mediated transformation allows for ex vitro (tissue culture-independent) manipulation, resulting in the production of composite plants within a shorter time frame (Meng et al., 2019; Cao et al., 2023). A. rhizogenes is a Gram-negative bacterium, which contains the root-inducing (Ri) plasmid that induces plants to produce hairy roots from wounds (Moore et al., 1979; White and Nester, 1980a; Chilton et al., 1982; Lothar et al., 1982). Currently, a wide range of plant species, including angiosperms (both dicotyledonous and monocotyledonous plants), gymnosperms, and even moss, have been found to be susceptible to successful infection by A. rhizogenes (De Cleene and De Ley, 1981; Porter and Flores, 1991; Spiess et al, 1977; Masako et al., 2004). The T-DNA fragment on the Ri plasmid integrates into the host plant genome, causing hairy root formation. Plants regenerate from hairy roots, while Ri T-DNA transmits through meiosis (Tepfer, 1984). Therefore, A. rhizogenes has become a useful tool for plant biotechnology. The principal steps and factors involved in A. rhizogenes-mediated plant transformation are similar to those of A. tumefaciens and have been comprehensively examined in previous studies (Gelvin, 2009; Gelvin, 2010; Pitzschke, 2013). It can be suggested that due to similarity in the linear organization of genetic loci that perform equivalent functions during T-DNA transfer, Ti and Ri plasmids appear to be very similar in structure and function in regard to mobilization and transfer of T-DNA (White and Nester, 1980b; Risuleo et al., 1982; Hooykaas et al., 1984; Huffman et al., 1984; Jouanin, 1984). This is confirmed by the genomic sequence of A. rhizogenes strain LBA9402 (Hooykaas and Hooykaas, 2021). Four rol (for root locus) genes, rolA, rolB, rolC, and rolD, have been the object of intense study and are mainly responsible for inducing hairy root formation (Mauro et al., 2017). Analyses have been largely performed in dicotyledonous plants such as tobacco, carrot, tomato, and kiwi, transformed with single genes or in combination (Capone et al., 1989; Christophe et al., 1991; Rugini et al., 1991; Van Altvorst et al., 1992). Auxin synthesis genes, such as aux1 and aux2, collaborate with rol genes to facilitate the induction of hairy root formation by supplying auxin. However, the absence of the auxin biosynthetic genes tms1 and tms2 in mannopine-type Ri plasmids does not affect root induction, suggesting that Ri plasmids may also alter the development of transformed explants through signaling other than auxin synthesis (Hansen et al., 1991). Opine synthesis genes including ags and mas can synthesize different opines. The classification of A. rhizogenes strains is primarily determined by the specific type of opine they produce. These strains can be divided into four groups: agropine (Petit et al., 1983), mannopine (Petit et al., 1983), mikimopine (Akira et al., 1990), and cucumopine (Elisabeth et al., 1988). According to the opine genes present in Ri plasmids, more than 20 strains of A. rhizogenes used for genetic transformation have been assigned to these aforementioned categories (Bahramnejad et al., 2019).
Almost three decades after the initial successful transformation of A. rhizogenes in grapevine (Guellec et al., 1990), numerous successful transformations have been performed in woody plants, typically in the form of hairy roots or composite plants. In addition, the regeneration of transgenic hairy roots in various plants has further demonstrated the applicability of the A. rhizogenes transformation system in molecular plant breeding. Hairy roots cultivated in vitro serve as a primary means for the synthesis of secondary metabolites, particularly within medicinal plants (Sharma et al., 2013). Furthermore, the utilization of hairy roots and composite plants in scientific investigations to explore gene functionality is extensive, encompassing various aspects such as root development, wood formation (Plasencia et al., 2016), interactions between roots and soil microbes (Plett et al., 2014), and plant allelopathy (Stanisic et al., 2019). This review aims to delve into the process of transformation of woody plants through the application of A. rhizogenes, while also discussing the potential implications of such transformations for basic research and biotechnological advancement.
2 Agrobacterium rhizogenes as a root agent
The propagation of plants through stem cuttings is commonly used in the commercial production of ornamental plants, medicinal plants, and timber trees. However, some commercially important tree species exhibit a low root formation rate. As far back as 1930, the induction of hairy roots was observed in nursery apple trees (Riker et al., 1930), and it was subsequently determined that the root induction was caused by A. rhizogenes (Hildebrand, 1934). Since then, A. rhizogenes has been used to enhance root formation in plants that are difficult to propagate through stem cuttings (Zavattieri et al., 2016).
The use of A. rhizogenes as a rooting inducer for difficult-to-root plants has been found to be remarkably effective and suitable for a wide range of plants. The determination of the root-promoting ability of A. rhizogenes is contingent upon the combination of bacterial strains and plant genotypes. Within woody plant species, there exists variation in rooting efficiency. Additionally, the specific strain of A. rhizogenes employed is a critical factor. For instance, in the case of the challenging-to-root woody species, “Golden Delicious” apple, the induction of rooting rates ranges from 0% to 20%, depending on the strain of A. rhizogenes utilized. Notably, strains A4 and 232 successfully induce adventitious root formation, whereas strains 178 X A4T and R1000 are unable to do so (Pateña et al., 1988). Cuttings from mature jujube trees (Ziziphus jujuba Mill.) exhibit a significant challenge in rooting. However, the application of A. rhizogenes, specifically strain TR105, resulted in the highest root formation percentage (65%), which was twice as high as that of the uninoculated cuttings (32.5%). Conversely, strain A4 did not show any significant difference in root formation (Mochammad et al., 1996). Interestingly, in the case of Corylus avellana, the rooting rate reached 100% when inoculated with a combination of A. rhizogenes (A7 + 22) (Bassil et al., 1991). In addition to bacterial strains, the success of inoculation and production of hairy roots is significantly influenced by plant genotypes and states, as evidenced by multiple studies. For instance, in hazelnut, the stimulation of rooting of cuttings was found to be influenced by both the cultivar and the date of cutting collection (Bassil et al., 1991). Similar results have also been observed in other research groups (Magnussen et al., 1994; Mihaljevic et al., 1996; Sarmast et al., 2019). The infectivity and adventitious root production of A. rhizogenes in host plant tissue are contingent upon the compatibility between A. rhizogenes and host plants, the responsiveness of the plant tissues to the T-DNA, the production of phytohormones, and the juvenile state of the host tissues.
Auxin is a well-known root inducer, and exogenously applied auxins have been shown to accelerate the rooting process in cuttings of a wide variety of plant species. When auxin and A. rhizogenes are combined, they exhibit varying effects on branch rooting, including synergy, antagonism, or no effect. Previous studies have observed a synergistic action between IBA and A. rhizogenes in inducing rooting in radiata pine (Li and Leung, 2003) and walnut (Caboni et al., 1996). Conversely, an antagonistic action between IAA and A. rhizogenes has been observed in inducing rooting in Pinus monticola (McAfee et al., 1993). The combined effect of A. rhizogenes to stimulate rooting is contingent upon the specific species and genotypes of micro propagated fruit trees. In all tested genotypes, root formation was observed following infection with A. rhizogenes. Three distinct responses were observed: genotypes that rooted without the presence of auxins showed a decrease in rooting percentage when auxin and infection were combined; genotypes that rooted only with auxin exhibited either no effect or a synergistic effect between auxins and infection; genotypes that rooted solely with A. rhizogenes displayed either no effect or an antagonistic effect between auxins and infection (Carmine et al., 1998). According to Zarei et al. (2020), the induction of rooting in Picea abies, a species known for its reluctance in rooting, cannot be achieved solely through the use of A. rhizogenes. However, when A. rhizogenes is combined with auxin, successful rooting can be achieved (Zarei et al., 2020).
A histological investigation revealed that the development of auxin-induced roots differs from that of A. rhizogenes. In the presence of NAA, adventitious roots are generated endogenously, originating within the vascular tissues of the stem. Conversely, adventitious roots formed in response to A. rhizogenes infection exhibit both endogenous and exogenous growth patterns. In the process of endogenous root formation, calli are generated within the cortex, leading to the subsequent formation of tracheid nests, which results in a bulge in the stem. Additionally, exogenous callus, formed at the base of shoot, also gives rise to tracheid nests. Consequently, roots form from both of these callus structures (Ellen and Juvenal, 1993). The rooting process in walnut was found to be influenced by the combined action of IBA and A. rhizogenes, as well as the antagonistic effect of IAA and A. rhizogenes. Notably, a significant reduction in the roots containing bacteria was observed when A. rhizogenes was combined with either IAA or IBA (Falasca et al., 2000).
It is imperative to conduct strain screening to optimize the rooting rate for each specific plant species. The utilization of multiple strains of A. rhizogenes offers a captivating approach to enhance rooting. The modulation of plant sensitivity to auxin by A. rhizogenes is believed to contribute to the promotion of rooting (Shen et al., 1988; Spanò et al., 1988; Petersen et al., 1989; Christophe et al., 1991). This modulation is thought to occur through variations in endogenous auxin levels and auxin sensitivities across different plant species (Carmine et al., 1998).
3 Agrobacterium rhizogenes as genetic engineers
3.1 Transformation
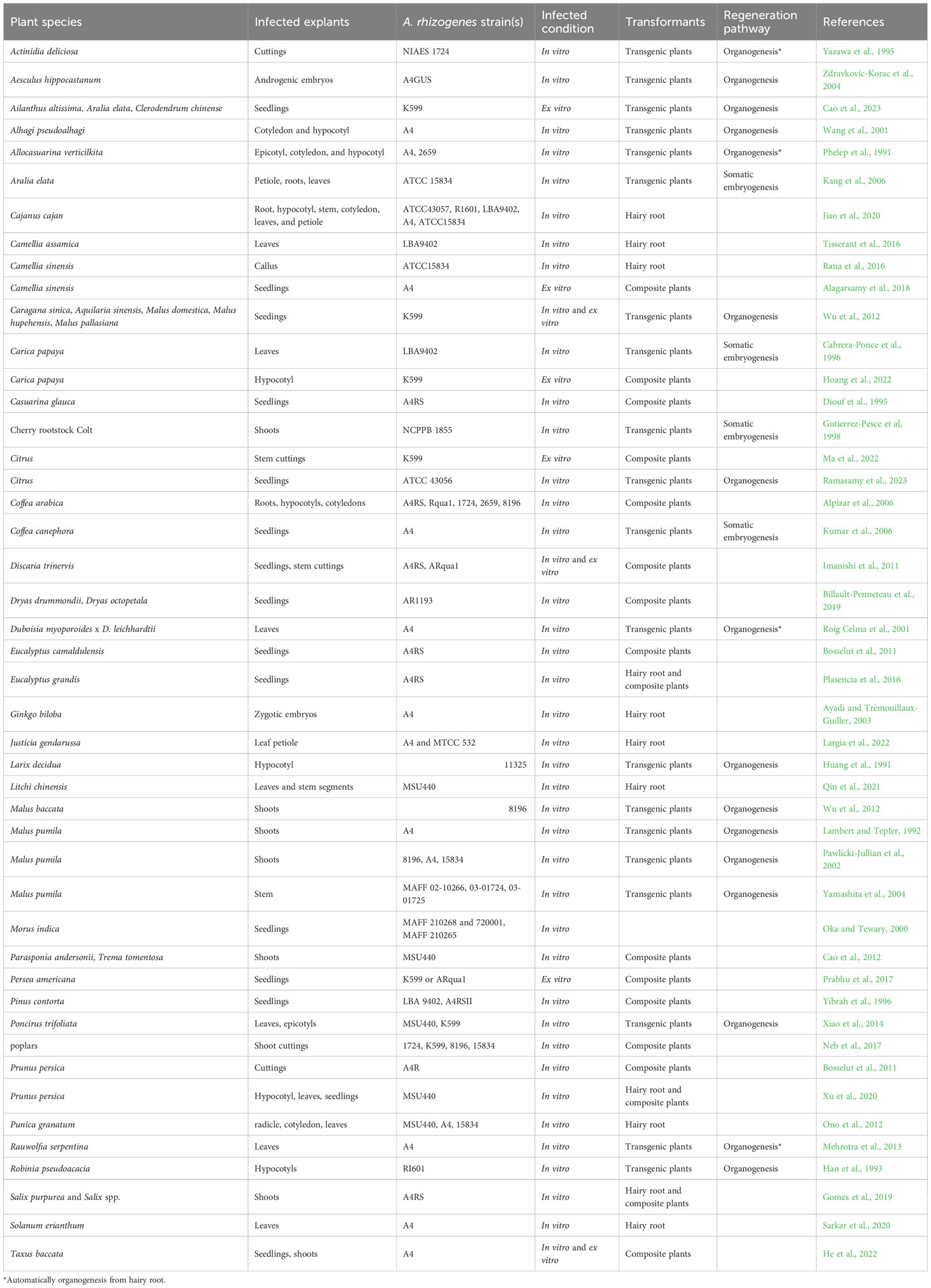
A. rhizogenes not only facilitates the rooting of difficult-to-propagate plants, but also allows for the integration of foreign genes into the plant genome through binary vectors. Researchers have successfully transferred foreign genes into various woody plant species, including Larix decidua (Huang et al., 1991), Alhagi pseudoalhagi, Eucalyptus camaldulensis (Balasubramanian et al., 2011), Prunus (Bosselut et al., 2011; Xu et al., 2020), Parasponia (Cao et al., 2012), Trema (Cao et al., 2012), Poncirus trifoliata (Xiao et al., 2014), Solanum erianthum (Sarkar et al., 2020), Populus (Neb et al., 2017), Salix purpurea (Gomes et al., 2019), Malus prunifolia (Yamashita et al., 2004), and Litchi chinensis (Qin et al., 2021), by infecting plant organs such as cotyledons, hypocotyls, stem segments, root segments, leaves, petioles, callus, and in vitro shoots with A. rhizogenes (Table 1). The use of in vitro shoots as explants has enabled the development of composite plants, which have numerous biological applications, such as nutrient absorption, biotic and abiotic stress tolerance, and signal exchange between aboveground and underground plant parts. However, the tissue culture of woody plants presents several technical challenges, such as browning and the need for aseptic conditions, which require skilled operators and the identification of suitable bacteriostatic agents for different strains and explants. To address these challenges, Collier et al (2005) developed a novel method to induce A. rhizogenes infection in plants without the use of tissue culture, thereby generating composite plants (Collier et al., 2005). This approach not only is cost-effective and efficient, but also enables the production of composite plants in a short time frame without the need for tissue culture. Furthermore, it is simple to execute, boasts a short cycle, and does not require complex sterilization procedures. Collier and colleagues successfully applied this technique to 14 dicotyledonous herbs from nine genera spanning four families. Following infection with the same strain, 14 composite dicotyledonous plants were generated, with transformation efficiencies ranging from 56% to 100% (Collier et al., 2005). This method has also been successfully implemented in woody plants. Using seedlings as explants, composite plants have been generated from a wide range of woody plants, including Camellia sinensis (Alagarsamy et al., 2018), Discaria trinervis (Imanishi et al., 2011), Persea americana (Prabhu et al., 2017), Taxus baccata (He et al., 2022), Carica papaya (Hoang et al., 2022), Citrus (Ma et al., 2022), Ailanthus altissima (Cao et al., 2023), Aralia elata (Cao et al., 2023), Clerodendrum chinense (Cao et al., 2023), Caragana sinica (Diouf et al., 1995), and Malus pumila (Pawlicki-Jullian et al., 2002; Yamashita et al., 2004) (Table 1). Although seedlings can be used as explants for woody plants, the genetic heterozygosity of these plants requires the use of a sufficient number of transformed lines to ensure the accuracy of experimental results, particularly in studies of stress resistance and other biological phenomena. In addition, stem cutting represents a crucial means of reproducing woody plants. In this regard, Ma et al (2022) successfully generated composite plants using citrus stem cuttings as explants, resulting in composite plants that possess the same genetic background, and are therefore more suitable for biological research applications (Ma et al., 2022).
3.2 Impact factors
The core factors that impact the transformation efficiency of A. rhizogenes are the same as those that influence root growth promotion when A. rhizogenes is employed as a rooting agent. These factors include the type of A. rhizogenes strains and the genetic or genotype or states of the host plants. For instance, in the context of poplar stem segment transformation, the four strains (1724, K599, 8196, and 15834, representing mikimopine, cucumopine, mannopine, and agropine strains, respectively) exhibit varying differences in hairy root formation time, number of formations, and transformation efficiency. Hence, it is essential to identify suitable strains for specific plants (Neb et al., 2017). Moreover, variations can also arise between species, varieties, and clones. For instance, a study involving five woody plants found that Aquilaria sinensis failed to produce hairy roots, while the remaining five plants generated composite plants with efficiencies ranging from 30% to 85% (Meng et al., 2019). In 22 citrus species, transformation efficiencies fluctuated from 0 to 95% (Ma et al., 2022).
Other factors, such as the tissue and physiological conditions of explants, bacterial concentration, acetosyringone concentration, hormone recipes, and additional treatments such as vacuuming will affect transformation efficiency. Leaves, petioles, stem segments, and adventitious buds can be used as explants. Suitable explants vary from plant to plant. For example, in Cajanus cajan, leaves have the highest transformation efficiency, reaching 70.92% (Jiao et al., 2020). In L. chinensis, there is no significant difference between the transformation efficiency of stem segments and leaves (Qin et al., 2021). In Coffea arabica, hypocotyls of cultivars of Caturra and IAPAR-59 had the highest infection efficiencies, reaching 82% and 51%, respectively (Alpizar et al., 2006). The age of the plants also affects the transformation efficiency; the transformation efficiency of 8-week-old seedlings was significantly higher than that of 20-week-old P. americana (Prabhu et al., 2017). After A. rhizogenes infection, hairy roots are often produced through cortical cells (Falasca et al., 2000). The older the physiological age, the higher the degree of lignification, the lower the proportion of cortical cells, and the longer the time used for hairy root induction. In addition, the concentration of the bacterial solution also affects transformation efficiency. In general, the concentration of A. rhizogenes infecting plants is between 0.4 and 1.0 (Ma et al., 2022; Cao et al., 2023). At the same time, full induction of the vir gene in the strain requires acetosyringone, a chemoattractant for Agrobacterium, at a commonly used concentration of 100–200 µM (Ma et al., 2022; Cao et al., 2023). To improve the transformation efficiency, additional treatments such as vacuum treatment have been applied to facilitate the infiltration A. rhizogenes into plant cells (Ma et al., 2022).
3.3 Regeneration
Hairy roots induced by A. rhizogenes have the potential to facilitate the generation of transgenic plants through various ways, including spontaneous organ regeneration, organogenesis, or somatic embryogenesis (Figure 1). Nevertheless, many woody species were induced by regeneration plants via organogenesis or somatic embryogenesis. Some woody plants, such as Actinidia deliciosa (Yazawa et al., 1995), Allocasuarina verticillate (Phelep et al., 1991), Duboisia myoporoides x D. leichhardtii (Trovato et al., 2001), and Rauwolfia serpentina (Mehrotra et al., 2013), are capable of regeneration via spontaneous organ regeneration from their hairy roots, which can be completed in hormone-free medium. On the other hand, other woody plants such as Aesculus hippocastanum (Zdravkovic-Korac et al., 2004), A. pseudoalhagi (Wang et al., 2001), Larix decidua (Huang et al., 1991), M. pumila (Pawlicki-Jullian et al., 2002), P. trifoliata (Xiao et al., 2014), Robinia pseudoacacia (Han et al., 1993), and citrus (Ramasamy et al., 2023) require hormone ratios to induce the production of calli, from which regenerated plants can be obtained. It is worth noting that in the study of Malus baccata, regenerated plants were only obtained when the hairy roots remained attached to the mother plant (non-transformed aerial part) (Wu et al., 2012). In addition to organogenesis, some woody plants can also produce somatic embryos through the induction of hairy roots, which germinate into regenerated plants. Examples of such plants include A. elata (Kang et al., 2006), C. papaya (Cabrera-Ponce et al., 1996), cherry rootstock Colt (Gutierrez-Pesce et al., 1998), and Coffea canephora (Kang et al., 2006). However, despite multiple attempts, some studies have failed to obtain regenerated plants through hairy roots. Recently, a cut-dip-budding method has been successfully applied to three woody plants, namely, A. altissima, A. elata, and C. chinense, which allowed the generation of A. rhizogenes-mediated transgenic plants (Cao et al., 2023) (Figure 1). The regeneration of woody plants from roots is a common phenomenon and can be enhanced by pruning aboveground parts (Wan et al., 2006). These approaches would facilitate woody plant transformation mediated by A. rhizogenes.
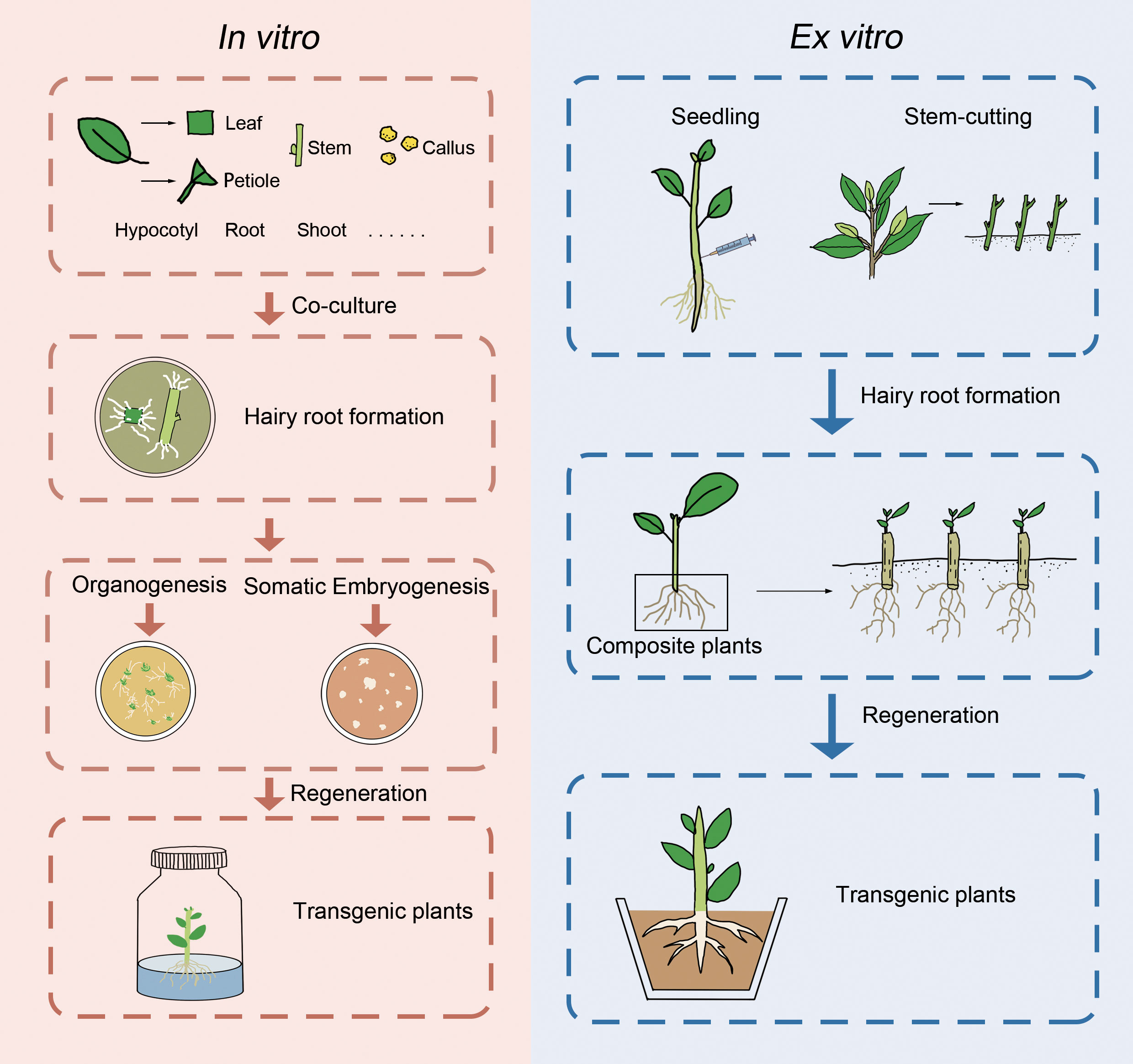
Figure 1 Presentation of in vitro and ex vitro genetic transformation of woody plants mediated by A. rhizogenes.
4 Applications in woody plants
4.1 Basic biological research
Currently, A. tumefaciens-mediated genetic transformation systems have been successfully implemented in several woody species, such as poplar (Wang et al., 2011), apple (Schropfer et al., 2022), kiwifruit (Uematsu et al., 1991), and walnut (McGranahan et al., 1988). However, the number of woody plants that have been sequenced far exceeds the number of plants that have established transformation systems. With advancements in sequencing technology and the enhanced level of genome assembly, there is a growing potential for the sequencing of numerous genetically intricate woody plant genomes. An example of this progress is the successful completion of the Chinese pine (Pinus tabuliformis) genome, which has been assembled at the chromosome level and spans a size of 25.4 gigabases (Gb) (Niu et al., 2022). Consequently, effective use of the substantial amount of sequence information will become an imperative endeavor. Transgenic technology for resolving gene function is emerging as a key tool to address this challenge. In recent years, the utilization of A. rhizogenes for the production of composite plants has gained significant traction in the realm of herbaceous plant investigation, particularly in the case of soybean. This development has provided a promising avenue for exploring woody plant research.
The application of A. rhizogenes-mediated transformation in soybean has proven to be highly successful in various biological contexts. A particularly efficient and rapid method for this transformation has been established (Kereszt et al., 2007). In soybean research, three main types of applications have been extensively utilized. These include the construction of composite plants through overexpression, RNAi, and CRISPR-Cas9 binary vectors for the purpose of gene function analysis (Traubenik et al., 2020; Li et al., 2021). Additionally, basic molecular analyses, such as promoter analysis (Yang et al., 2021), subcellular localization studies (Brear et al., 2020), ChIP-PCR (Pi et al., 2019), GST pull-down assays (Wang et al., 2015), Co-IP assays (Vadivel et al., 2021), protein ubiquitination and degradation studies (Zhang et al., 2021), and in vivo kinase assays (Gao et al., 2022), have been conducted. Of particular interest is mutant complementation (Feng et al., 2021; Jiang et al., 2021), which serves as a genetic validation of gene function.
A variety of studies have used A. rhizogenes-mediated transformation technology for basic biological investigations in woody plants, including binary vector generation of composite plants, basic molecular analysis, and genetic analysis. A. rhizogenes possesses the ability to carry binary vectors, which facilitates gene overexpression, RNAi, and gene editing. These transformants have been utilized to examine nutrient uptake, abiotic stresses, nodule development, mycorrhizal interactions, allelopathy, biosynthesis, and wood formation. For instance, in apple, the function of the MdPRP6 gene was elucidated under low nitrogen conditions through A. rhizogenes-mediated knockout or overexpression (Zhang et al., 2022). In Eucalyptus, the STOP-like gene was disrupted to assess its aluminum resistance function (Sawaki et al., 2014). In poplar, A. rhizogenes-mediated transformation yielded PtJAZ6 knockout and overexpression materials, which were subsequently employed to analyze the mutualistic interaction between PtJAZ6 and effectors in ectomycorrhizal Laccaria bicolor (Plett et al., 2014). This technology has also been applied to R. pseudoacacia to identify Rpf41 as a critical regulator of symbiotic nodulation in legumes (Chou et al., 2016). In apple, the function of BvSTI in allelopathy was examined using the hairy root system (Stanisic et al., 2019). Although genetic transformation of tea plants poses challenges, the function of CsTSI in theanine biosynthesis was successfully investigated using the A. rhizogenes system (She et al., 2022). Furthermore, it is worth noting that woody plant roots, similar to stems, undergo secondary growth, making them a valuable resource for investigating the process of wood formation (Plasencia et al., 2016).
Gene function studies involve analyzing tissue expression, protein interactions, and downstream target genes. The use of hairy roots enables researchers to conduct related research within homologous species, thereby providing a more accurate reflection of gene function. For instance, in citrus, the CsSUC2 promoter was analyzed in hairy roots (Roig Celma et al., 2001), while in Eucalyptus, the EgCCR1 and EgCAD2 promoters were studied in hairy roots (Rugini et al., 1991). Bimolecular fluorescence complementation (BiFC) was used to verify the interaction between CcCIPK14 and CcCBL1 in hairy roots of cowpea (Mehrotra et al., 2013). Furthermore, hairy roots can be utilized to screen downstream target genes of transcription factors. For instance, downstream target genes of MYB15 were identified in grape (White et al., 1985). In poplar, the A. rhizogenes system can be used to verify the downstream target genes of transcription factors through ChIP-PCR (Schmülling et al., 1988; Filippini et al., 1996). It is worth noting that genetic manipulation of woody plants, especially the genetic analysis of upstream and downstream genes, is challenging. The use of the A. rhizogenes system has overcome this challenge in some species. For example, Ma (2018) used A. rhizogenes to knock out MdSUT2.2 based on MdCIPK22 transgenic apple plants, which confirmed that MdCIPK22 depends on MdSUT2.2 for drought tolerance (Zhu et al., 2003).
4.2 Ri breeding
Plants regenerated from hairy roots exhibit phenotypes, including vigorous large root growth, lateral root development, root geotropism loss, loss of shoot apical dominance, internode shortening, and plant dwarfing (Han et al., 1993; Yazawa et al., 1995; Yamashita et al., 2004; Zdravkovic-Korac et al., 2004; Wu et al., 2012; Rugini et al., 2015). These characteristics have been observed in the regenerated plants of numerous species, such as R. pseudoacacia (Han et al., 1993), A. deliciosa (Rugini et al., 1991), papaya (Cabrera-Ponce et al., 1996), apple rootstock Jork 9 (Pawlicki-Jullian et al., 2002; Yamashita et al., 2004; Wu et al., 2012), and sweet cherry (Rugini et al., 2015). While the majority of research has concentrated on the initial characteristics of regenerated plants, Mehrotra et al (2013) conducted an investigation into the flowering traits of R. serpentina, a shrub characterized by a brief growth cycle (Mehrotra et al., 2013). Their findings revealed that transgenic plants, when compared to nontransformed plants, exhibited normal flowering patterns albeit with a reduced quantity of inflorescences and flowers. Another study conducted in cherries by Rugini et al (2015) tracked A. rhizogenes-transformed cherries for 10 years and observed a slight decrease in the number of flowers, while flower morphology, ovule differentiation, and flowering time remained unaltered. The fruit traits of transgenic materials did not change significantly, with the exception of reduced fruit yield due to the decreased number of flowers. The transgenic plants were grafted as rootstocks, reducing the size of the two plants to varying degrees, but fruit quality remained unchanged (Mehrotra et al., 2013). It should be noted that considerable differences in traits exist among the obtained transgenic plants due to strains of A. rhizogenes used in transformation and different copy numbers or insertion positions of T-DNA following transformation. Thus, a sufficient number of transgenic plants is required for stable, long-term observation.
The hairy root phenotype of transgenic plants is primarily regulated by rol genes. In 1985, White et al. identified and analyzed four rol genes, specifically rolA, rolB, rolC, and rolD, which play pivotal roles in neoplastic disease induction and hairy root formation (White et al., 1985). Transgenic plants obtained through transformation by A. rhizogenes exhibited phenotypes analogous to plants transformed by single or multiple rol genes. The rolB gene possesses tyrosine phosphatase activity and plays an essential role in root initiation and elongation, contributing significantly to the hairy root phenotype (Filippini et al., 1996). Plants transformed with rolB demonstrated typical A. rhizogenes-induced phenotypes including increased adventitious roots, loss of apical dominance, and shortened internodes. Examples of such transformed plants include Pyrus communis, Kalanchoe diagremontiana, and grape rootstocks “Richter 110” (Schmülling et al., 1988; Zhu et al., 2003; Geier et al., 2008). The rolB gene specifically upregulates ARF7 and ARF19 to promote root initiation in Nicotiana tabacum (Bose et al., 2022). The rolC is a glucosidase, and its expression correlates with enhanced cytokinin activity in tobacco (Estruch et al., 1991). Plants transformed with rolC exhibited dwarfing, dark green leaves, and increased branching, which were associated with reduced gibberellin levels. Furthermore, the introduction of rolC into plants such as P. trifoliata and Diospyros kaki displayed stronger rooting ability (Kaneyoshi and Kobayashi, 1999; Koshita et al., 2002). Similar to rolB and rolC genes, the rolA gene has the ability to induce leaf rooting in K. diagremontiana without the need for exogenous hormones (Schmülling et al., 1988). Plants transformed with rolA exhibited plant dwarfing and leaf shrinkage, which were associated with decreased gibberellin levels in tobacco (Schmülling et al., 1993). The rolA gene encodes a DNA-binding protein akin to the HPV-1 E2 DNA-binding protein (Rigden and Carneiro, 1999). The rolD gene promotes plant flowering, increases axillary inflorescence formation and elongation, and facilitates adventitious root formation in both tobacco and Arabidopsis (Mauro et al., 1996; Falasca et al., 2010). The rolD gene encodes ornithine cyclodeaminase, an enzyme that catalyzes the conversion of ornithine to proline (Trovato et al., 2001).
One notable characteristic of Ri plants is their compact plant structure, distinguished by shortened internodes, reduced plant height, increased branching, and enhanced axillary bud growth. These traits hold significant value in breeding of flowers and fruit trees. The Ri phenotype is heritable, with the inserted T-DNA being transmitted through meiosis (Tepfer, 1984) and inherited in a Mendelian dominant manner (Zhan et al., 1988; Christensen et al., 2019). Ri lines of K. blossfeldiana, characterized by increased branches, weakened apical dominance, and shortened internodes following backcross separation, have been successfully applied in commercial plant breeding (Christensen et al., 2019). It is widely recognized that rootstocks profoundly influence scion size. In fruit tree breeding, employing Ri plants as rootstocks is a good way to obtain dwarfed plants without compromising fruit quality (Rugini et al., 2015).
Another notable feature of Ri plants is the alteration of root morphology. Upon infection of host plants with A. rhizogenes, T-DNA insertion induces cell dedifferentiation, leading to the formation of hairy roots. Hairy roots exhibit characteristics such as rapid growth, a high degree of branching, and oblique development. Ri plants have demonstrated enhanced rooting ability under both greenhouse and field conditions in several plant species (Lambert and Tepfer, 1992; Pawlicki-Jullian et al., 2002; Casanova et al., 2005). This increased rooting ability offers several advantages. Firstly, it can lead to efficient asexual reproduction and improved adaptation to in vitro conditions. Secondly, Ri plants with enhanced root development hold significant promise in sustainable plant agriculture, as this can improve water and nutrient management, which can augment plant drought tolerance (Tepfer, 2017). Additionally, increased root biomass particularly benefits plants such that their roots were used for extracting specific metabolites (Mehrotra et al., 2013).
5 Conclusion and future perspectives
Over the past three decades, significant progress has been made in the transformation of woody plants by A. rhizogenes. Numerous woody plants have been transformed with A. rhizogenes, and regenerated plants can be obtained in vitro through hairy roots. The direct use of lignified hairy roots to acquire regenerated plants ex vitro offers broad prospects for the application of A. rhizogenes in woody plants. In addition, the incorporation of genome sequencing will significantly advance theoretical investigations in the field of woody plants. The utilization of diverse binary vectors with A. rhizogenes serves as a genetic tool for conducting in vivo investigations on gene functionality in woody plants (Bahramnejad et al., 2019).
The typical hairy root symptoms induced by A. rhizogenes, including compact plant types and enhanced rooting ability, have been used in flower breeding and rootstock modifications. Genome sequence analysis has revealed that A. rhizogenes undergoes horizontal gene transfer to plants, such as Nicotiana, Linaria, and Ipomoea species (White et al., 1983; Matveeva et al., 2012; Kyndt et al., 2015; Quispe-Huamanquispe et al., 2017). This implies that A. rhizogenes, during the process of pathogenesis, introduces multiple plasmid-encoded genes into its host through horizontal gene transfer, which occurs as a natural outcome. Currently, plants transformed with wild-type Agrobacterium strains are not classified as transgenic species (Lutken et al., 2012), which is advantageous for the application of Ri plants. The predominant natural A. rhizogenes strains used are listed in Table 2.
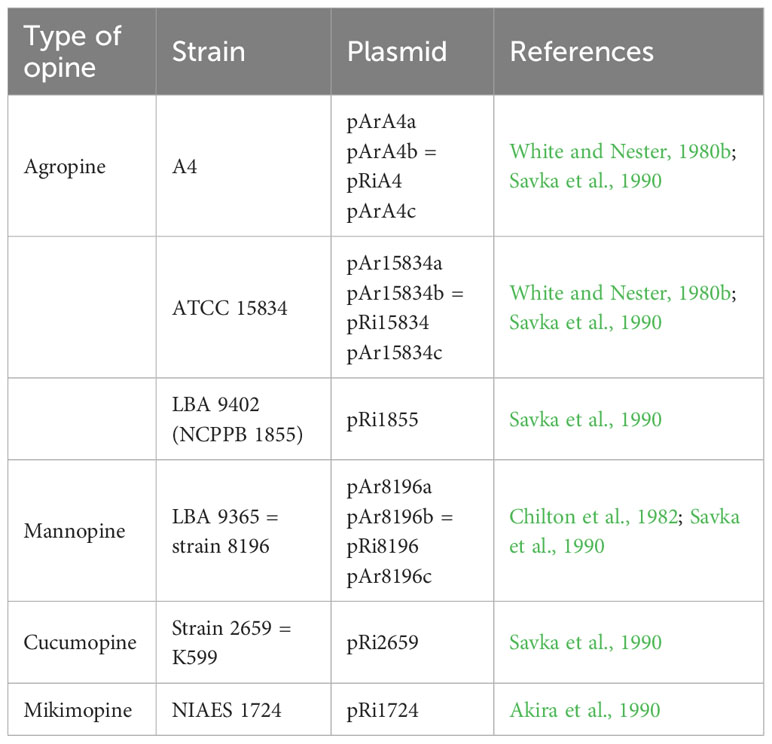
Table 2 The representative naturally occurring (wild type) strains of A. rhizogenes and their plasmids.
However, it is imperative to recognize the limitations associated with these applications. To enhance and exploit natural engineering capabilities, one promising avenue lies in the use of CRISPR-mediated base editing, which serves as a catalyst for “engineering the engineer” (Rodrigues et al., 2021). This approach holds promise for improving A. rhizogenes, thereby enabling more effective plant transformation and genome editing. According to the National Center for Biotechnology Information (NCBI) database, 96 strains of A. rhizogenes exist (https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=359) as of the search conducted on 26 September 2023. However, only four strains (LBA9402, A4, K599, and CA75/95) have complete genome sequences. The transformation capabilities of these strains vary, necessitating the sequencing and engineering of strain genomes to improve infectivity.
Author contributions
Conceptualization, XW and BZ. Data curation, WY and GW. Writing—original draft preparation, WY, GW, XW, and BZ. Writing—review and editing, WX, HL, YH, HY, DY, FC, and JH. Supervision, XW and BZ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02066-12), the Opening Project of Zhejiang Provincial Key Laboratory of Forest Aromatic Plants-based Healthcare Functions (2022E10008), the State Key Laboratory of Subtropical Silviculture (KF202007), the General Foundation of Education of Zhejiang (Y202250159 and Y202147311), and the Ningbo Natural Science Foundation (202003N4016).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akira, I., Naoyuki, F., Miki, H., Hiroshi, K., Hiroshi, H., Akinori, S. (1990). Mikimopine, an opine in hairy roots of tobacco induced by Agrobacterium rhizogenes. Phytochemistry 29 (10), 3131–3134. doi: 10.1016/0031-9422(90)80171-c
Alagarsamy, K., Shamala, L. F., Wei, S. (2018). Protocol: high-efficiency in-planta Agrobacterium-mediated transgenic hairy root induction of Camellia sinensis var. sinensis. Plant Methods 14, 17. doi: 10.1186/s13007-018-0285-8
Alpizar, E., Dechamp, E., Espeout, S., Royer, M., Lecouls, A. C., Nicole, M., et al. (2006). Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell Rep. 25 (9), 959–967. doi: 10.1007/s00299-006-0159-9
Ayadi, R., Trémouillaux-Guiller, J. (2003). Root formation from transgenic calli of Ginkgo biloba. Tree Physiol. 23, 713–718. doi: 10.1093/treephys/23.10.713
Bahramnejad, B., Naji, M., Bose, R., Jha, S. (2019). A critical review on use of Agrobacterium rhizogenes and their associated binary vectors for plant transformation. Biotechnol. Adv. 37 (7), 107405. doi: 10.1016/j.bioteChadv.2019.06.004
Balasubramanian, A., Venkatachalam, R., Selvakesavan, K. R., Mary, A. S., Gherbi, H., Svistoonoff, S., et al. (2011). Optimisation of methods for Agrobacterium rhizogenes mediated generation of composite plants in Eucalyptus camaldulensis. BMC Proc. 5 (7), 1–3. doi: 10.1186/1753-6561-5-s7-o45
Bassil, N. V., Proebsting, W. M., Moore, L. W., Lightfoot, D. A. (1991). Propagation of hazelnut stem cuttings using Agrobacterium rhizogenes. HortScience 26 (8), 1058–1060. doi: 10.21273/hortsci.26.8.1058
Billault-Penneteau, B., Sandré, A., Folgmann, J., Parniske, M., Pawlowski, K. (2019). Dryas as a model for studying the root symbioses of the rosaceae. Front. Plant Sci. 10, 661. doi: 10.3389/fpls.2019.00661
Bose, R., Sengupta, M., Basu, D., Jha, S. (2022). The rolB-transgenic Nicotiana tabacum plants exhibit upregulated ARF7 and ARF19 gene expression. Plant Direct 6 (6), e414. doi: 10.1002/pld3.414
Bosselut, N., Van Ghelder, C., Claverie, M., Voisin, R., Onesto, J. P., Rosso, M. N., et al. (2011). Agrobacterium rhizogenes-mediated transformation of Prunus as an alternative for gene functional analysis in hairy-roots and composite plants. Plant Cell Rep. 30 (7), 1313–1326. doi: 10.1007/s00299-011-1043-9
Brear, E. M., Bedon, F., Gavrin, A., Kryvoruchko, I. S., Torres-Jerez, I., Udvardi, M. K., et al. (2020). GmVTL1a is an iron transporter on the symbiosome membrane of soybean with an important role in nitrogen fixation. New Phytol. 228, 667–681. doi: 10.1111/nph.16734
Caboni, E., Lauri, P., Tonelli, M., Falasca, G., Damiano, C. (1996). ). Root induction by Agrobacterium rhizogenes in walnut. Plant Sci. 118, 203–208. doi: 10.1016/0168-9452(96)04449-4
Cabrera-Ponce, J. L., Vegas-Garcia, A., Herrera-Estrella, L. (1996). Regeneration of transgenic papaya plants via somatic embryogenesis induced by Agrobacterium rhizogenes. In Vitro Cell. Dev. Biol.-Plant 32, 86–90. doi: 10.1007/bf02823136
Cao, Q., Op Den Camp, R., Seifi Kalhor, M., Bisseling, T., Geurts, R. (2012). Efficiency of Agrobacterium rhizogenes-mediated root transformation of Parasponia and Trema is temperature dependent. Plant Growth Regul. 68 (3), 459–465. doi: 10.1007/s10725-012-9734-y
Cao, X., Xie, H., Song, M., Lu, J., Ma, P., Huang, B., et al. (2023). Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture. Innovation (Camb) 4 (1), 100345. doi: 10.1016/j.xinn.2022.100345
Capone, I., Spanò, L., Cardarelli, M., Bellincampi, D., Petit, A., Costantino, P. (1989). Induction and growth properties of carrot roots with different complements of Agrobacterium rhizogenes T-DNA. Plant Mol. Biol. 13, 43–52. doi: 10.1007/bf00027334
Carmine, D., Simona, M., Monticelli, S. (1998). In vitro fruit trees rooting by Agrobacterium rhizogenes wild type infection. Electron. J. Biotechn. 1 (3), 12–13. doi: 10.4067/s0717-34581998000300008
Casanova, E., Trillas, M. I., Moysset, L., Vainstein, A. (2005). Influence of rol genes in floriculture. Biotechnol. Adv. 23 (1), 3–39. doi: 10.1016/j.bioteChadv.2004.06.002
Chilton, M.-D., Tepfer, D. A., Petit, A., David, C., Casse-Delbart, F., Tempé, J. (1982). Agrobacterium rhizogenes inserts T-DNA into the genomes of the host plant root cells. Nature 295, 432–434. doi: 10.1038/295432a0
Chou, M., Xia, C., Feng, Z., Sun, Y., Zhang, D., Zhang, M., et al. (2016). A translationally controlled tumor protein gene Rpf41 is required for the nodulation of Robinia pseudoacacia. Plant Mol. Biol. 90 (4), 389–402. doi: 10.1007/s11103-015-0424-9
Christensen, E. M., Müller, R. P., Lütken, H. V., Hegelund, J. N., Jensen, L., Christensen, B., et al. (2019). Agrobacterium rhizogenes transformation and expression of rol genes in Kalanchoë. U. S. Patent No.9,253,952 (Washington, DC: U.S. Patent and Trademark Ofiice).
Christophe, M., Hélène, B.-B., Angelo, S., Jacques, T., Jean, G. (1991). Single rol genes from the Agrobacterium rhizogenes TL-DNA alter some of the cellular responses to auxin in Nicotiana tabacum. Plant Physiol. 97 (1), 212–216. doi: 10.1104/pp.97.1.212
Collier, R., Fuchs, B., Walter, N., Kevin Lutke, W., Taylor, C. (2005). Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J. 43 (3), 449–457. doi: 10.1111/j.1365-313X.2005.02454.x
De Cleene, M., De Ley, J. (1981). The host range of infectious hairy-root. Bot. Rev. 47, 147–194. doi: 10.1007/bf02868853
Diouf, D., Gherbi, H., Prin, Y., Franche, C., Duhoux, E., Bogusz, D., et al. (1995). Hairy root nodulation of Casuarina glauca: a system for the study of symbiotic gene expression in an actinorhizal tree. Plant Microbe Interact. 8 (4), 532–537. doi: 10.1094/mpmi-8-0532
Elisabeth, D., Annik, P., Max, E. T., Max, E. T., Maarten, H. R., Maarten, H. R., et al. (1988). Cucumopine—a new T-DNA-encoded opine in hairy root and crown gall. Phytochemistry 27, 2429–2433. doi: 10.1016/0031-9422(88)87007-9
Ellen, G. S., Juvenal, G. L. (1993). Development anatomy of roots induced by Agrobacterium rhizogenes in Malus pumila 'M.26' shoots grown in vitro. Int. J. Plant Sci. 154, 59–67. doi: 10.1086/297090
Estruch, J. J., Chriqui, D., Grossmann, K., Schell, J., Spena, A. (1991). The plant oncogene rolC is responsible for the release of cytokinins from glucoside conjugates. EMBO J. 10 (10), 2889–2895. doi: 10.1002/j.1460-2075.1991.tb07838.x
Falasca, G., Altamura, M. M., D'angeli, S., Zaghi, D., Costantino, P., Mauro, M. L. (2010). The rolD oncogene promotes axillary bud and adventitious root meristems in Arabidopsis. Plant Physiol. Biochem. 48 (9), 797–804. doi: 10.1016/j.plaphy.2010.06.002
Falasca, G., Reverberi, M., Lauri, P., Caboni, E., De Stradis, A., Altamura, M. M., et al. (2000). How Agrobacterium rhizogenes triggers de novo root formation in a recalcitrant woody plant: an integrated histological, ultrastructural and molecular analysis. New Phytol. 145 (1), 77–93. doi: 10.1046/j.1469-8137.2000.00558.x
Feng, J., Lee, T., Schiessl, K., Oldroyd, G. E. D. (2021). Processing of NODULE INCEPTION controls the transition to nitrogen fixation in root nodules. Science 374 (6567), 629–632. doi: 10.1126/science.abg2804
Filippini, F., Rossi, V., Marin, O., Trovato, M., Costantino, P., Downey, P. M., et al. (1996). A plant oncogene as a phosphatase. Nature 379 (6565), 499–500. doi: 10.1038/379499a0
FitzJohn, R. G., Pennell, M. W., Zanne, A. E., Stevens, P. F., Tank, D. C., Cornwell, W. K. (2014). How much of the world is woody? J. Ecol. 102 (5), 1266–1272. doi: 10.1111/1365-2745.12260
Gao, H., Jiang, L., Du, B., Ning, B., Ding, X., Zhang, C., et al. (2022). GmMKK4-activated GmMPK6 stimulates GmERF113 to trigger resistance to Phytophthora sojae in soybean. Plant J. 111, 473–495. doi: 10.1111/tpj.15809
Geier, T., Eimert, K., Scherer, R., Nickel, C. (2008). Production and rooting behaviour of rolB-transgenic plants of grape rootstock ‘Richter 110’ (Vitis berlandieri×V. rupestris). Plant Cell Tiss. Org. 94, 269–280. doi: 10.1007/s11240-008-9352-6
Gelvin, S. B. (2009). Agrobacterium in the genomics age. Plant Physiol. 150, 1665–1676. doi: 10.1104/pp.109.139873
Gelvin, S. B. (2010). Finding a way to the nucleus. Curr. Opin. Microbiol. 13, 53–58. doi: 10.1016/j.mib.2009.11.003
Gomes, C., Dupas, A., Pagano, A., Grima-Pettenati, J., Paiva, J. (2019). Hairy root transformation: a useful tool to explore gene function and expression in Salix spp. recalcitrant to transformation. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01427
Guellec, V., David, C., Branchard, M., Tempé, J. (1990). Agrobacterium rhizogenes mediated transformation of grapevine (Vitis vinifera L.). Plant Cell Tiss. Org. 20, 211–215. doi: 10.1007/BF00041883
Gutierrez-Pesce, P., Taylor, K., Muleo, R., Rugini, E. (1998). Somatic embryogenesis and shoot regeneration from transgenic roots of the cherry rootstock Colt (Prunus avium×P. pseudocerasus) mediated by pRi 1855 T-DNA of Agrobacterium rhizogenes. Plant Cell Rep. 17 (6-7), 574–580. doi: 10.1007/s002990050445
Han, K.-H., Keathley, D. E., Davis, J. M., Gordon, M. P. (1993). Regeneration of a transgenic woody legume (Robinia pseudoacacia L., black locust) and morphological alterations induced by Agrobacterium rhizogenes-mediated transformation. Plant Sci. 88 (2), 149–157. doi: 10.1016/0168-9452(93)90086-f
Hansen, G., Larribe, M., Vaubert, D., Tempé, J., Biermann, B. J., Montoya, A. L., et al. (1991). Agrobacterium rhizogenes pRi8196 T-DNA: mapping and DNA sequence of functions involved in mannopine synthesis and hairy root differentiation. Proc. Natl. Acad. Sci. U. S. A. 88 (17), 7763–7767. doi: 10.1073/pnas.88.17.7763
He, J., Plácido, J. P. A., Pateraki, I., Kampranis, S., Favero, B. T., Lütken, H. (2022). Hairy root induction of Taxus baccata L. by natural transformation with Rhizobium rhizogenes. Horticulturae 9 (1), 4. doi: 10.3390/horticulturae9010004
Hildebrand, E. (1934). Life history of the hairy-root organism in relation to its pathogenesis on nursery apple trees. J.Agr.Res. 48 (10), 857–885.
Hoang, T. H. T., Nguyen, N. H., Nguyen, L. T., Bui, T. P., Le, N. T., Dao, N. T., et al. (2022). Developing a robust in vivo hairy root system for assessing transgene expression and genome editing efficiency in papaya. Plant Cell Tiss. Org. 152, 661–667. doi: 10.1007/s11240-022-02421-2
Hooykaas, P. J., Hofker, M., Den Dulk-Ras, H., Schilperoort, R. A. (1984). A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid 11, 195–205. doi: 10.1016/0147-619x(84)90026-x
Hooykaas, M. J. G., Hooykaas, P. J. J. (2021). The genome sequence of hairy root Rhizobium rhizogenes strain LBA9402: Bioinformatics analysis suggests the presence of a new opine system in the agropine Ri plasmid. Microbiologyopen 10 (2), e1180. doi: 10.1002/mbo3.1180
Huang, Y., Diner, A. M., Karnosky, D. F. (1991). Agrobacterium rhizogenes-mediated genetic transformation and regeneration of a conifer:Larix decidua. In Vitro Cell. Dev. Biol.-Plant 27, 201–207. doi: 10.1007/bf02632217
Huffman, G. A., White, F. F., Gordon, M. P., Nester, E. W. (1984). Hairy-root-inducing plasmid: physical map and homology to tumor-inducing plasmids. J. Bacteriol. 157, 269–276. doi: 10.1128/jb.157.1.269-276.1984
Imanishi, L., Vayssieres, A., Franche, C., Bogusz, D., Wall, L., Svistoonoff, S. (2011). Transformed hairy roots of Discaria trinervis: a valuable tool for studying actinorhizal symbiosis in the context of intercellular infection. Mol. Plant-Microbe Interact. 24 (11), 1317–1324. doi: 10.1094/MPMI-03-11-0078
Jiang, S., Jardinaud, M.-F., Gao, J., Pecrix, Y., Wen, J., Mysore, K., et al. (2021). NIN-like protein transcription factors regulate leghemoglobin genes in legume nodules. Science 374 (6567), 625–628. doi: 10.1126/science.abg5945
Jiao, J., Gai, Q. Y., Wang, X., Liu, J., Lu, Y., Wang, Z. Y., et al. (2020). Effective production of phenolic compounds with health benefits in pigeon pea [Cajanus cajan (L.) Millsp.] hairy root cultures. J. Agric. Food Chem. 68 (1), 8350–8361. doi: 10.1021/acs.jafc.0c02600
Jouanin, L. (1984). Restriction map of an agropine-type Ri plasmid and its homologies with Ti plasmids. Plasmid 12, 91–102. doi: 10.1016/0147-619x(84)90055-6
Kaneyoshi, J., Kobayashi, S. (1999). Characteristics of transgenic trifoliate orange (Poncirus trifoliata Raf.) possessing the rolC gene of Agrobacterium rhizogenes Ri plasmid. J. Jpn. Soc Hortic. Sci. 68 (4), 734–738. doi: 10.2503/jjshs.68.734
Kang, H. J., Anbazhagan, V. R., You, X. L., Moon, H. K., Yi, J. S., Choi, Y. E. (2006). Production of transgenic Aralia elata regenerated from Agrobacterium rhizogenes-mediated transformed roots. Plant Cell Tiss. Org. 85 (2), 187–196. doi: 10.1007/s11240-005-9070-2
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Koshita, Y., Nakamura, Y., Kobayashi, S., Morinaga, K. (2002). Introduction of the rolC gene into the genome of the Japanese persimmon causes dwarfism. J. Jpn. Soc Hortic. Sci. 71 (4), 529–531. doi: 10.2503/jjshs.71.529
Kumar, V., Satyanarayana, K., Sarala Itty, S., Indu, E., Giridhar, P., Chandrashekar, A., et al. (2006). Stable transformation and direct regeneration in coffea canephora p ex. fr. by agrobacterium rhizogenes mediated transformation without hairy-root phenotype. Plant Cell Rep. 25 (3), 214–222. doi: 10.1007/s00299-005-0045-x
Kyndt, T., Quispe, D., Zhai, H., Jarret, R., Ghislain, M., Liu, Q., et al. (2015). The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc. Natl. Acad. Sci. U. S. A. 112 (18), 5844–5849. doi: 10.1073/pnas.1419685112
Lambert, C., Tepfer, D. (1992). Use of Agrobacterium rhizogenes to create transgenic apple trees having an altered organogenic response to hormones. Theor. Appl. Genet. 85 (1), 105–109. doi: 10.1007/BF00223851
Largia, M. J. V., Pandian, S., Shilpha, J., Chitradevi, M., Kavikkuil, M., Sohn, S.-I., et al. (2022). Improved in vitro regeneration, genetic fidelity analysis, antioxidant potential, and hairy root induction of Justicia gendarussa burm. f. Plant Biotechn. Rep. 16 (6), 621–632. doi: 10.1007/s11816-022-00775-9
Li, M., Chen, R., Jiang, Q., Sun, X., Zhang, H., Hu, Z. (2021). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105, 333–345. doi: 10.1007/s11103-020-01091-y
Li, M., Leung, D. (2003). Root induction in radiata pine using Agrobacterium rhizogenes. Electron J. Biotechn. 6 (3), 254–261. doi: 10.4067/S0717-34582003000300010
Lothar, W., Jose, J. S.-S., Ellen, B., Jeff, S. (1982). DNA from Agrobacterium-Rhizogenes is transferred to and expressed in axenic hairy root plant-tissues. Mol. Genet. Genomics 186, 16–22. doi: 10.1007/bf00422906
Lutken, H., Clarke, J. L., Muller, R. (2012). Genetic engineering and sustainable production of ornamentals: current status and future directions. Plant Cell Rep. 31 (7), 1141–1157. doi: 10.1007/s00299-012-1265-5
Ma, H., Meng, X., Xu, K., Li, M., Gmitter, F., Liu, N., et al. (2022). Highly efficient hairy root genetic transformation and applications in citrus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1039094
Magnussen, D., Clapham, D. E., Grönroos, R., Arnold, S. V. (1994). Induction of hairy and normal roots on Picea abies, Pinus sylvestris and Pinus contorta by Agrobacterium rhizogenes. Scand. J. For. Res. 9, 46–51. doi: 10.1080/02827589409382811
Masako, A., Masako, A., Takuma, I., Takuma, I., Hiroji, S. (2004). Transformation of the monocot Alstroemeria by Agrobacterium rhizogenes. Mol. Breeding. 13, 69–78. doi: 10.1023/b:molb.0000012860.29731.9c
Matveeva, T. V., Bogomaz, D. I., Pavlova, O. A., Nester, E. W., Lutova, L. A. (2012). Horizontal gene transfer from genus agrobacterium to the plant linaria in nature. Mol. Plant-Microbe Interact. 25, 1542–1551. doi: 10.1094/MPMI-07-12-0169-R
Mauro, M. L., Costantino, P., Bettini, P. P. (2017). The never ending story of rol genes: a century after. Plant Cell Tiss.Org. 131, 201–212. doi: 10.1007/s11240-017-1277-5
Mauro, M. L., Trovato, M., Paolis, A. D., Gallelli, A., Costantino, P., Altamura, M. M. (1996). The plant oncogene rolD stimulates flowering in transgenic tobacco plants. Dev. Biol. 180 (2), 693–700. doi: 10.1006/dbio.1996.0338
McAfee, B. J., White, E. E., Pelcher, L. E., Lapp, M. (1993). Root induction in pine (Pinus) and larch (Larix) spp. using Agrobacterium rhizogenes. Plant Cell Tiss. Org. 34, 53–62. doi: 10.1007/BF00048463
McGranahan, G. H., Leslie, C. A., Uratsu, S. L., Martin, L. A., Dandekar, A. M. (1988). Agrobacterium-Mediated transformation of walnut somatic embryos and regeneration of transgenic plants. Nat. Biotechnol. 6 (7), 800–804. doi: 10.1038/nbt0788-800
Mehrotra, S., Goel, M. K., Rahman, L. U., Kukreja, A. K. (2013). ). Molecular and chemical characterization of plants regenerated from Ri-mediated hairy root cultures of Rauwolfia serpentina. Plant Cell Tiss. Org. 114 (1), 31–38. doi: 10.1007/s11240-013-0302-6
Meng, D., Yang, Q., Dong, B., Song, Z., Niu, L., Wang, L., et al. (2019). Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol. J. 17, 1804–1813. doi: 10.1111/pbi.13101
Mihaljevic, S., Stipkovic, S., Jelaska, S. (1996). Increase of root induction in Pinus nigra explants using agrobacteria. Plant Cell Rep. 15 (8), 610–614. doi: 10.1007/BF00232463
Mochammad, H., Caula, A. B., Garton, S., Diner, A. M. (1996). Induction of roots on jujube softwood cuttings using Agrobacterium rhizogenes. J. Hortic. Sci. 71 (6), 881–886. doi: 14620316.1996.11515470
Moore, L., Warren, G., Strobel, G. (1979). Involvement of a plasmid in the hairy root disease of plants caused by Agrobacterium rhizogenes. Plasmid 2, 617–626. doi: 10.1016/0147-619x(79)90059-3
Neb, D., Das, A., Hintelmann, A., Nehls, U. (2017). Composite poplars: a novel tool for ectomycorrhizal research. Plant Cell Rep. 36 (12), 1959–1970. doi: 10.1007/s00299-017-2212-2
Niu, S., Li, J., Bo, W., Yang, W., Zuccolo, A., Giacomello, S., et al. (2022). The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 185, 204–217.e214. doi: 10.1016/j.cell.2021.12.006
Oka, S., Tewary, P. K. (2000). Induction of hairy roots from hypocotyls of mulberry (Morus indica) by japanese wild strains of Agrobacterium rhizogenes. J. Seric. Sci. Jpn. 69, 13–19. doi: 10.11416/kontyushigen1930.69.13
Ono, N. N., Bandaranayake, P. C., Tian, L. (2012). Establishment of pomegranate (Punica granatum) hairy root cultures for genetic interrogation of the hydrolyzable tannin biosynthetic pathway. Planta 236 (3), 931–941. doi: 10.1007/s00425-012-1706-y
Pateña, L., Sutter, E. G., Dandekar, A. M. (1988). Root induction by Agrobacterium rhizogenes in a difficult-to-root woody species. Acta Hortic. 227, 324–329. doi: 10.17660/ActaHortic.1988.227.59
Pawlicki-Jullian, N., Sedira, M., Welander, M. (2002). The use of Agrobacterium rhizogenes transformed roots to obtain transgenic shoots of the apple rootstock Jork 9. Plant Cell Tiss. Org. 70 (2), 163–171. doi: 10.1023/a:1016387004712
Petersen, S. G., Stummann, B. M., Olesen, P., Henningsen, K. W. (1989). Structure and function of root-inducing (Ri) plasmids and their relation to tumor-inducing (Ti) plasmids. Physiol. Plant 77 (3), 427–435. doi: 10.1111/j.1399-3054.1989.tb05664.x
Petit, A., David, C., Dahl, G. A., Ellis, J. G., Guyon, P., Casse-Delbart, F., et al. (1983). Further extension of the opine concept: plasmids in Agrobacterium rhizogenes cooperate for opine degradation. Mol. Gen. Genet. 190, 204–214. doi: 10.1007/bf00330641
Phelep, M., Petit, A., Martin, L., Duhoux, E., Tempé, J. (1991). Transformation and regeneration of a nitrogen-fixing tree, Allocasuarina Verticillata Lam. Nat. Biotechnol. 9 (5), 461–466. doi: 10.1038/nbt0591-461
Pi, E., Xu, J., Li, H., Fan, W., Zhu, C., Zhang, T., et al. (2019). Enhanced salt tolerance of Rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis. Mol. Cell Proteomics 18, 2225–2243. doi: 10.1074/mcp.RA119.001704
Pitzschke, A. (2013). Agrobacterium infection and plant defense-transformation success hangs by a thread. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00519
Plasencia, A., Soler, M., Dupas, A., Ladouce, N., Silva-Martins, G., Martinez, Y., et al. (2016). Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation. Plant Biotechnol. J. 14 (6), 1381–1393. doi: 10.1111/pbi.12502
Plett, J. M., Daguerre, Y., Wittulsky, S., Vayssieres, A., Deveau, A., Melton, S. J., et al. (2014). Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. U. S. A. 111 (22), 8299–8304. doi: 10.1073/pnas.1322671111
Porter, J. R., Flores, H. (1991). Host range and implications of plant infection by Agrobacterium rhizogenes. Crit. Rev. Plant Sci. 10 (4), 387–421. doi: 10.1080/07352689109382318
Prabhu, S. A., Ndlovu, B., Engelbrecht, J., Van Den Berg, N. (2017). Generation of composite Persea americana (Mill.) (avocado) plants: A proof-of-concept-study. PloS One 12 (10), e0185896. doi: 10.1371/journal.pone.0185896
Qin, Y., Wang, D., Fu, J., Zhang, Z., Qin, Y., Hu, G., et al. (2021). Agrobacterium rhizogenes-mediated hairy root transformation as an efficient system for gene function analysis in Litchi chinensis. Plant Methods 17 (1), 103. doi: 10.1186/s13007-021-00802-w
Quispe-Huamanquispe, D. G., Gheysen, G., Kreuze, J. F. (2017). Horizontal gene transfer contributes to plant evolution: the case of Agrobacterium T-DNAs. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02015
Ramasamy, M., Dominguez, M. M., Irigoyen, S., Padilla, C. S., Mandadi, K. K. (2023). Rhizobium rhizogenes-mediated hairy root induction and plant regeneration for bioengineering citrus. Plant Biotechnol. J. 21, 1728–1730. doi: 10.1111/pbi.14096
Rana, M. M., Han, Z., Song, D., Liu, G., Li, D., Wan, X., et al. (2016). Effect of medium supplements on Agrobacterium rhizogenes mediated hairy root induction from the callus tissues of Camellia sinensis var. sinensis. Int. J. Mol. Sci. 17 (7), 1132. doi: 10.3390/ijms17071132
Rigden, D. J., Carneiro, M. (1999). ). A structural model for the rolA protein and its interaction with DNA. Proteins: Struct. Funct. Genet. 37 (4), 697–708. doi: 10.1002/(sici)1097-0134(19991201)37:4<697::Aid-prot18>3.0.Co;2-y
Riker, A. J., Banfield, W. M., Wright, W. H., Keitt, G. W., Sagen, H. E. (1930). Studies on infectious hairy root of nursery apple trees. J. Agric. Res. 41 (7), 507–540.
Risuleo, G., Battistoni, P., Costantino, P. (1982). ). Regions of homology between tumorigenic plasmids from Agrobacterium rhizogenes and Agrobacterium tumefaciens. Plasmid 7, 45–51. doi: 10.1016/0147-619X(82)90025-7
Rodrigues, S. D., Karimi, M., Impens, L., Van Lerberge, E., Coussens, G., Aesaert, S., et al. (2021). Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc. Natl. Acad. Sci. U. S. A. 118 (2), e2013338118. doi: 10.1073/pnas.2013338118
Roig Celma, C., Palazon, J., Cusido, R. M., Pinol, M. T., Keil, M. (2001). Decreased scopolamine yield in field-grown Duboisia plants regenerated from hairy roots. Planta Med. 67 (3), 249–253. doi: 10.1055/s-2001-12006
Rugini, E., Pellegrineschi, A., Mencuccini, M., Mariotti, D. (1991). Increase of rooting ability in the woody species kiwi (Actinidia deliciosa A. Chev.) by transformation with Agrobacterium rhizogenes rol genes. Plant Cell Rep. 10 (6), 291–295. doi: 10.1007/BF00193144
Rugini, E., Silvestri, C., Cristofori, V., Brunori, E., Biasi, R. (2015). Ten years field trial observations of ri-TDNA cherry Colt rootstocks and their effect on grafted sweet cherry cv Lapins. Plant Cell Tiss. Org. 123 (3), 557–568. doi: 10.1007/s11240-015-0860-x
Sarkar, J., Misra, A., Banerjee, N. (2020). Genetic transfection, hairy root induction and solasodine accumulation in elicited hairy root clone of Solanum erianthum D. Don. J. Biotechnol. 323, 238–245. doi: 10.1016/j.jbiotec.2020.09.002
Sarmast, M. K., Kordkatoli, R., Rezaei, Z., Ghasemnezhad, A. (2019). Effect of seasons, gender and Agrobacterium rhizogenes strains on adventitious root induction of male and female Juniperus communis L. J. Ornamental Plants 9, 23–31.
Savka, M. A., Ravillion, B., Noel, G. R., Farrand, S. K. (1990). ). Induction of hairy roots on cultivated soybean genotypes and their use to propagate the soybean cyst nematode. Phytopathology 80 (5), 503–508.
Sawaki, Y., Kobayashi, Y., Kihara-Doi, T., Nishikubo, N., Kawazu, T., Kobayashi, M., et al. (2014). Identification of a STOP1-like protein in Eucalyptus that regulates transcription of Al tolerance genes. Plant Sci. 223, 8–15. doi: 10.1016/j.plantsci.2014.02.011
Schmülling, T., Fladung, M., Grossmann, K., Schell, J. (1993). Hormonal content and sensitivity of transgenic tobacco and potato plants expressing single rol genes of Agrobacterium rhizogenes T-DNA. Plant J. 3, 371–382. doi: 10.1046/j.1365-313X.1993.t01-20-00999.x
Schmülling, T., Schell, J., Spena, A. (1988). Single genes from Agrobacterium rhizogenes influence plant development. EMBO J. 7 (9), 2621–2629. doi: 10.1002/j.1460-2075.1988.tb03114.x
Schropfer, S., Lempe, J., Emeriewen, O. F., Flachowsky, H. (2022). Recent Developments and Strategies for the Application of Agrobacterium-Mediated Transformation of Apple Malus x domestica Borkh. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.928292
Sharma, P., Padh, H., Shrivastava, N. (2013). Hairy root cultures: A suitable biological system for studying secondary metabolic pathways in plants. Eng. Life Sci. 13, 62–75. doi: 10.1002/elsc.201200030
She, G., Yu, S., Li, Z., Peng, A., Li, P., Li, Y., et al. (2022). Characterization of CsTSI in the biosynthesis of theanine in tea plants (Camellia sinensis). J. Agric. Food Chem. 70 (3), 826–836. doi: 10.1021/acs.jafc.1c04816
Shen, W. H., Petit, A., Guern, J., Tempe, J. (1988). ). Hairy roots are more sensitive to auxin than normal roots. Proc. Natl. Acad. Sci. U. S. A. 85, 3417–3421. doi: 10.1073/pnas.85.10.3417
Spanò, L., Mariotti, D., Cardarelli, M., Branca, C., Costantino, P. (1988). Morphogenesis and auxin sensitivity of transgenic tobacco with different complements of Ri T-DNA. Plant Physiol. 87, 479–483. doi: 10.1104/pp.87.2.479
Spiess, L. D., Turner, J. C., Mahlberg, P. G., Lippincott, B. B., Lippincott, J. A. (1977). Adherence of Agrobacteria to moss protonema and gametophores viewed by scanning electron microscopy. Am. J. Bot. 64 (10), 1200–1208. doi: 10.2307/2442482
Stanisic, M., Cosic, T., Savic, J., Krstic-Milosevic, D., Misic, D., Smigocki, A., et al. (2019). Hairy root culture as a valuable tool for allelopathic studies in apple. Tree Physiol. 39 (5), 888–905. doi: 10.1093/treephys/tpz006
Tepfer, D. (1984). Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell 37, 959–967. doi: 10.1016/0092-8674(84)90430-6
Tepfer, D. (2017). “DNA Transfer to plants by Agrobacterium rhizogenes: a model for genetic communication between species and biospheres,” in Transgenesis and secondary metabolism. Ed. Jha, S. (Cham: Springer). doi: 10.1007/978-3-319-28669-3_19
Tisserant, L. P., Aziz, A., Jullian, N., Jeandet, P., Clement, C., Courot, E., et al. (2016). Enhanced stilbene production and excretion in Vitis vinifera cv pinot noir hairy root cultures. Molecules 21 (12), 1703. doi: 10.3390/molecules21121703
Traubenik, S., Reynoso, M. A., Hobecker, K., Lancia, M., Hummel, M., Rosen, B., et al. (2020). Reprogramming of root cells during nitrogen-fixing symbiosis involves dynamic polysome association of coding and noncoding RNAs. Plant Cell 32, 352–373. doi: 10.1105/tpc.19.00647
Trovato, M., Maras, B., Linhares, F., Costantino, P. (2001). The plant oncogene rolD encodes a functional ornithine cyclodeaminase. Proc. Natl. Acad. Sci. U. S. A. 98, 13449–13453. doi: 10.1073/pnas.231320398
Trumbore, S., Brando, P., Hartmann, H. (2015). Forest health and global change. Science 349, 814–818. doi: 10.1126/science.aac6759
Uematsu, C., Murase, M., Ichikawa, H., Imamura, J. (1991). Agrobacterium-mediated transformation and regeneration of kiwi fruit. Plant Cell Rep. 10 (6-7), 286–290. doi: 10.1007/BF00193143
Vadivel, A. K. A., Mcdowell, T., Renaud, J. B., Dhaubhadel, S. (2021). A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max). Commun. Biol. 4 (1), 356. doi: 10.1038/s42003-021-01889-6
Van Altvorst, A. C., Bino, R. J., Van Dijk, A. J., Lamers, A. M. J., Lindhout, W. H., van der Mark, F., et al. (1992). Effects of the introduction of Agrobacterium rhizogenes rol genes on tomato plant and flower development. Plant Sci. 83, 77–85. doi: 10.1016/0168-9452(92)90064-s
Wan, X., Landhausser, S. M., Lieffers, V. J., Zwiazek, J. J. (2006). Signals controlling root suckering and adventitious shoot formation in aspen (Populus tremuloides). Tree Physiol. 26 (5), 681–687. doi: 10.1093/treephys/26.5.681
Wang, F., Chen, H., Li, Q., Wei, W., Li, W., Zhang, W., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, H., Wang, C., Liu, H., Tang, R., Zhang, H. (2011). An efficient Agrobacterium-mediated transformation and regeneration system for leaf explants of two elite aspen hybrid clones Populus alba× P. berolinensis and Populus davidiana× P. bolleana. Plant Cell Rep. 30, 2037–2044. doi: 10.1007/s00299-011-1111-1
Wang, Y., Wang, J., Luo, D., Jia, J. (2001). Regeneration of plants from callus tissues of hairy roots induced by Agrobacterium rhizogenes on Alhagi pseudoalhagi. Cell Res. 11 (4), 279–284. doi: 10.1038/sj.cr.7290097
White, F. F., Garfinkel, D. J., Huffman, G. A., Gordon, M. P., Nester, E. W. (1983). Sequences homologous to Agrobacterium rhizogenes T-DNA in the genomes of uninfected plants. Nature 301, 348–350. doi: 10.1038/301348a0
White, F. F., Nester, E. W. (1980a). Hairy root: plasmid encodes virulence traits in Agrobacterium rhizogenes. J. Bacteriol. 141 (3), 1134–1141. doi: 10.1128/jb.141.3.1134-1141.1980
White, F. F., Nester, E. W. (1980b). Relationship of plasmids responsible for hairy root and crown gall tumorigenicity. J. Bacteriol. 144 (2), 710–720. doi: 10.1128/jb.144.2.710-720.1980
White, F. F., Taylor, B. H., Huffman, G. A., Gordon, M. P., Nester, E. W. (1985). Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 164 (1), 33–44. doi: 10.1128/jb.164.1.33-44.1985
Wu, J., Wang, Y., Zhang, L., Zhang, X., Kong, J., Lu, J., et al. (2012). High-efficiency regeneration of Agrobacterium rhizogenes-induced hairy root in apple rootstock Malus baccata (L.) Borkh. Plant Cell Tiss. Org. 111, 183–189. doi: 10.1007/s11240-012-0182-1
Xiao, X., Ma, F., Chen, C., Guo, W. (2014). High efficient transformation of auxin reporter gene into trifoliate orange via Agrobacterium rhizogenes-mediated co-transformation. Plant Cell Tiss. Org. 118, 137–146. doi: 10.1007/s11240-014-0469-5
Xu, S., Lai, E., Zhao, L., Cai, Y., Ogutu, C., Cherono, S., et al. (2020). Development of a fast and efficient root transgenic system for functional genomics and genetic engineering in peach. Sci. Rep. 10 (1), 2836. doi: 10.1038/s41598-020-59626-8
Yamashita, H., Daimon, H., Akasaka-Kennedy, Y., Masuda, T. (2004). Plant regeneration from hairy roots of apple rootstock, Malus prunifolia Borkh. var. ringo Asami, strain Nagano No.1, transformed by Agrobacterium rhizogenes. J. Jpn. Soc Hortic. Sci. 73 (6), 505–510. doi: 10.2503/jjshs.73.505
Yang, Z., Gao, Z., Zhou, H., He, Y., Liu, Y., Lai, Y., et al. (2021). GmPTF1 modifies root architecture responses to phosphate starvation primarily through regulating GmEXPB2 expression in soybean. Plant J. 107, 525–543. doi: 10.1270/jsbbs1951.45.241
Yazawa, M., Suginuma, C., Ichikawa, K., Kamada, H., Akihama, T. (1995). Regeneration of transgenic plants from hairy root of kiwi fruit (Actinidia deliciosa) induced by Agrobacterium rhizogenes. Jpn. J. Breed. 45 (2), 241–244. doi: 10.1270/jsbbs1951.45.241
Yibrah, H. S., Grönroos, R., Lindroth, A., Franzén, H., Clapham, D. E., Arnold, S. (1996). Agrobacterium rhizogenes-mediated induction of adventitious rooting from Pinus contorta hypocotyls and the effect of 5-azacytidine on transgene activity. Transgenic Res. 5, 75–85. doi: 10.1007/BF01969425
Zarei, M., Salehi, H., Jowkar, A. (2020). Controlling the barriers of cloning mature Picea abies (L.) H. Karst. via tissue culture and co-cultivation with Agrobacterium rhizogenes. Trees 34, 637–647. doi: 10.1007/s00468-019-01945-z
Zavattieri, M. A., Ragonezi, C., Klimaszewska, K. (2016). Adventitious rooting of conifers: influence of biological factors. Trees 30, 1021–1032. doi: 10.1007/s00468-016-1412-7
Zdravkovic-Korac, S., Muhovski, Y., Druart, P., Calic, D., Radojevic, L. (2004). ). Agrobacterium rhizogenes-mediated DNA transfer to Aesculus hippocastanum L. and the regeneration of transformed plants. Plant Cell Rep. 22, 698–704. doi: 10.1007/s00299-004-0756-4
Zhan, X., Jones, D. A., Kerr, A. (1988). Regeneration of flax plants transformed by Agrobacterium rhizogenes. Plant Mol. Biol. 11, 551–559. doi: 10.1007/BF00017455
Zhang, C., Cheng, Q., Wang, H., Gao, H., Fang, X., Chen, X., et al. (2021). GmBTB/POZ promotes the ubiquitination and degradation of LHP1 to regulate the response of soybean to Phytophthora sojae. Commun. Biol. 4 (1), 372. doi: 10.1038/s42003-021-01907-7
Zhang, X., Gong, X., Cheng, S., Yu, H., Li, D., Su, X., et al. (2022). Proline-rich protein MdPRP6 alters low nitrogen stress tolerance by regulating lateral root formation and anthocyanin accumulation in transgenic apple (Malus domestica). Environ. Exp. Bot. 197, 104841. doi: 10.1016/j.envexpbot.2022.104841
Keywords: gene function analysis, hairy root, regeneration, Ri plasmid, transformation
Citation: Ying W, Wen G, Xu W, Liu H, Ding W, Zheng L, He Y, Yuan H, Yan D, Cui F, Huang J, Zheng B and Wang X (2023) Agrobacterium rhizogenes: paving the road to research and breeding for woody plants. Front. Plant Sci. 14:1196561. doi: 10.3389/fpls.2023.1196561
Received: 30 March 2023; Accepted: 20 October 2023;
Published: 14 November 2023.
Edited by:
Quanzi Li, Chinese Academy of Forestry, ChinaReviewed by:
Jose Luis Cabrera Ponce, Unidad Irapuato (CINVESTAV), MexicoJens Stougaard, Aarhus University, Denmark
Copyright © 2023 Ying, Wen, Xu, Liu, Ding, Zheng, He, Yuan, Yan, Cui, Huang, Zheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingsong Zheng, YnN6aGVuZ0B6YWZ1LmVkdS5jbg==; Xiaofei Wang, eGZ3YW5nQHphZnUuZWR1LmNu
†These authors have contributed equally to this work
 Wei Ying
Wei Ying Guangchao Wen1,2†
Guangchao Wen1,2† Wona Ding
Wona Ding Luqing Zheng
Luqing Zheng Huwei Yuan
Huwei Yuan Fuqiang Cui
Fuqiang Cui Jianqin Huang
Jianqin Huang Bingsong Zheng
Bingsong Zheng Xiaofei Wang
Xiaofei Wang